
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 17 February 2025
Sec. Parasite and Host
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1524197
This article is part of the Research Topic Impact of landscape and feeding on the bees’ gut microbiome shaping and pathogens development View all 4 articles
In natural ecosystems, parasites often infect multiple host species, particularly when hosts share habitats, facilitating host-to-host transmission and altering traditional host-parasite coevolution dynamics. This study examines the microsporidian parasite Nosema ceranae in Eastern honey bees (Apis cerana) and Western honey bees (Apis mellifera), assessing its virulence and proliferation dynamics. Using inoculation experiments, we measured bee mortality and parasite spore loads to infer virulence and proliferation. Additionally, time-series transcriptome analysis of both bees and parasites provide insights into host-pathogen interactions. The results reveal that N. ceranae produces more spores with lower mortality in A. mellifera but causes higher mortality with lower spore production in A. cerana. The parasite also suppresses host gene expression, with stronger suppression observed in A. cerana. These findings suggest that N. ceranae is adapted for low virulence and high proliferation in A. mellifera but exhibits high virulence and limited proliferation in A. cerana. This study highlights the evolution of distinct trade-offs between virulence and proliferation in a multi-host system, offering valuable insights into parasite-host dynamics and their ecological implications.
Host mortality typically assesses parasite virulence, which is not static but evolves depending on host ecology (Gowler et al., 2023). Most evolutionary virulence theories connect the trade-off between the parasite and host fitness. For example, high parasite proliferation increases the transmission to other individuals. However, high proliferation may kill the host rapidly and reduce parasite transmission (de Roode et al., 2008). Thus, balanced transmission and virulence were expected in co-evolved host-parasite (Acevedo et al., 2019).
In natural ecosystems, parasites migrate and explore alternative host organisms. The phylogenetic relationship and proximity of the host species influence the success of host shifting, where closely related species most likely share parasites (Engelstädter and Fortuna, 2019). For example, Nosema ceranae infects both the Asian honey bee Apis cerana and the European honey bee Apis mellifera (Fries et al., 1996; Higes et al., 2007). The infection starts from ingesting spore-contaminated nectar. The spores germinate in the midgut lumen and inject the sporoplasm into the epithelial cells through the polar tube (Gisder et al., 2011). The infected honey bees show suppressed apoptosis (Higes et al., 2013), immature aging (Paris et al., 2018), shortened life span (Eiri et al., 2015), and impaired flight (Gage et al., 2018).
In Asia, the two honey bee species compete for habitats and shelter resources, and the prevalence of the parasite N. ceranae has been over 70% in both bee species (Li et al., 2012; Yang et al., 2013; Jack et al., 2016). Thus, the chance is high that the parasite switches between the two honey bee species back and forth (Graystock et al., 2015). N. ceranae infection changes the global gene expression in both European and Asia honey bees (Holt et al., 2013; Fan et al., 2022). The parasite infection causes colony failure in European honey bees (Higes et al., 2008; Botías et al., 2013). Comparatively, its virulence in Asia honey bees is unclear. Previously, we found that the host habitat sharing increases the parasite gene flow (Ke et al., 2022). In this follow-up study, we use two honey bee species and a microsporidian parasite to investigate how host habitat sharing shapes the parasite virulence. We find a distinct trade-off between virulence and proliferation in the two closely related honey bees in a shared habitat.
The honey bees Apis mellifera and Apis cerana are neither protected nor endangered species. Ethical approval is not required for this study.
We designed a two-by-two factorial experiment to study parasite virulence and proliferation in two honey bee species. We used two parasite sources PAcer (Parasite spores purified from the honey bee Apis cerana) and PAmel (Parasite spores purified from the honey bee Apis mellifera), as well as two host species HAcer (Host honey bee Apis cerana) and HAmel (Host honey bees Apis mellifera) (Supplementary Table S1). We combined bees from different hives for the inoculation, and uninfected bees (HAcer and HAmel) as controls for this multi-host-parasite experiment. The honey bee colonies are maintained in the experimental apiary at Jiangxi Agricultural University.
Three hundred honey bees of each A. mellifera and A. cerana were captured near the hive entrance using an insect net. The midgut was dissected and homogenized to isolate N. ceranae spores that were further purified using the Percoll gradient (Chen et al., 2013). The spores were counted under the light microscope using a hemacytometer. The sealed brood frames from three hives of HAmel and HAcer were kept in an incubator to collect newly emerged bees (35°C, 75% humidity).
The newly emerged honey bee workers (HAcer and HAmel) were individually inoculated with 2 µL of sugar solution with 105 spores. Additional freshly emerged honey bees (< 24 h after emerging) were each fed 2 µL of sugar solution without spores as the uninfected control group. One hundred fifty honey bees were inoculated in each treatment group, and the cohorts were divided into 3 rearing cups (50 bees per cup) in an incubator (35°C, 75% humidity) (3 replicates, Supplementary Table S1) to investigate the general bee response and parasite proliferation. During the experiment, sucrose (50% w/w) was provided ad libitum as the only food. In each rearing cup, three bees were collected at 24 h intervals from 1 to 5 dpi (day post-inoculation) for RNA-seq. The remaining bees were dissected to count spores at 14 dpi.
As the parasite infects the epithelial cell in the midgut, we dissected midgut tissue for RNA-seq. Three bees per cup per day were dissected and pooled for RNA extraction using Trizol. The library was prepared using the NEBNext Ultra RNA Kit. In total, 90 RNA libraries (5 days * 6 treatments * 3 replicates) were sequenced on the Illumina NovaSeq 6000 platform.
The quality of RNA reads (150bp, paired-end) was first viewed using Fastqc (Andrews, 2010) and trimmed using the Seqtk package with default parameters (Li, 2022). The processed reads were aligned to N. ceranae (Version Ncer 3.0), A. cerana (version CC1.0), and A. mellifera (version Hav3.1) genome, respectively, with the Hisat2 package with default parameters (Kim et al., 2015; Diao et al., 2018; Wallberg et al., 2019; Huang et al., 2021). The read count per gene was retrieved using bedtools (Quinlan and Hall, 2010). The within-group dispersion was calculated from the three replicates to determine the significantly regulated genes with edgeR package and adjusted for FDR (false discovery rate) (Robinson et al., 2010). The genes with FDR < 0.05 were defined as significantly regulated ones. Gene Ontology (GO) terms were retrieved using EggNOG-mapper, and the enrichment analysis was performed using the TopGo package with an adjusted weighted ks test (Robinson et al., 2010; Alexa and Rahnenfuhrer, 2021). Bee survival was analyzed using the Kaplan-Meier estimate in the survival package, adjusted for FDR (R Core Team, 2013; Therneau, 2022). The variance of the spore load among the treatment groups was analyzed with the Wilcoxon rank test, and the p values were adjusted with FDR to reduce false positives. The impact of the spore source and day on the gene expression was analyzed using ANONA, where the day and parasite source were fixed factors, and the replicates were random factors. The number of up and down-regulated genes was analyzed using Pearson’s Chi-squared test.
The uninfected HAmel shows the highest survival (99.4% survival), followed by HAmel infected with PAcer (HAmel_PAcer, 98.5% survival) and the HAmel_PAmel group (96.9% survival). The bees in the HAcer_PAcer group show the lowest survival (86.6% survival). The parasite source (PAmel and PAcer) shows a minor impact on the survival of HAmel (HAmel_PAcer vs HAmel_PAmel, Kaplan-Meier test, P > 0.05) (Figure 1A, Supplementary Table S2). Comparatively, the parasite PAcer causes significantly higher mortality in HAcer than PAmel (Coxph test, P < 0.001). The uninfected HAmel survives substantially better than the uninfected HAcer (Coxph test, P < 0.01). Overall, the parasites cause a higher mortality in HAcer than in HAmel. Additionally, we normalize the mortality variance between the two honey bee species using uninfected_HAmel and uninfected_HAcer. Again, the parasites cause higher mortality in HAcer than HAmel (P < 0.001, Supplementary Figure S1).
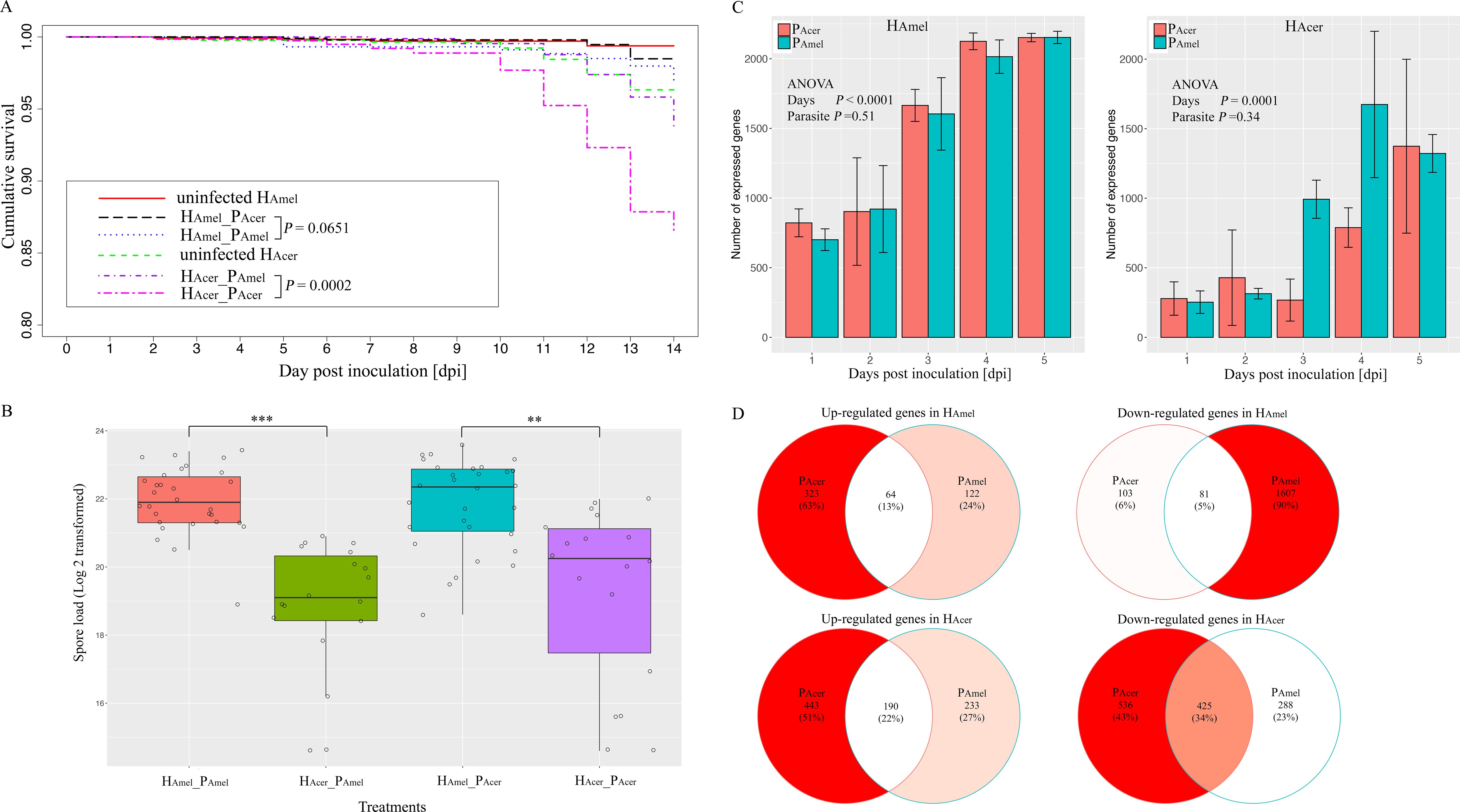
Figure 1. Transmission and virulence of the parasites and the bee responses towards the parasite in a two-parasite and two-host system. (A) Cumulative survival of the honey bees. The impact of the parasite source on the survival of bees was minor. The PAcer caused substantially higher mortality than PAmel in HAcer. (B) parasite proliferation variance in two honey bee species. The parasite produced more spores in HAmel than HAcer. Additionally, the impacts of the parasite sources on the spore load were minor in either host. (C) The number of expressed parasite genes in two hosts. Fewer parasite genes were expressed in A. cerana than A. mellifera, irrespective of the parasite sources. (D) Venn diagram of the shared and unique regulated host genes responding to the two parasite isolates. Overall, the bee genes were down-regulated by the infection, and a subset of genes responded to both parasite sources. ** indicates the significance level at P < 0.01; *** indicates the significance level at P < 0.001. The error bar indicates the standard deviation.
The spores are not found in the uninfected bees. The spore load is not evenly distributed among the four infected honey bee groups (Kruskal-Wallis test, χ2 = 43.1, df=3, P < 0.0001, Figure 1B). The parasite produces more spores in HAmel than HAcer, when infected by either PAmel or PAcer (Wilcoxon rank sum test, df=1, P < 0.001) (Supplementary Table S3). Thus, host species substantially impact the parasite proliferation (F=32.3, df=2, ANOVA, P < 0.0001).
To investigate the parasite gene expression profile, we quantify the parasite transcriptome in the two host species (Figure 1C). HAcer shows a stronger tendency to suppress the parasite gene expression than HAmel (F=77.1, df=1, ANOVA, P < 0.0001). A significantly lower number of up-regulated genes are observed in PAmel than PAcer when infecting the HAmel (Pearson’s Chi-squared test, χ2 = 20.8, df=2, P < 0.0001). The highest variance was at four dpi, and the number of up-regulated genes is threefold higher in PAcer (36 genes) than in PAmel (12 genes) in HAmel (Supplementary Table S4). Thus, the parasites express a higher number of genes and transcript levels in HAmel than in HAcer.
To infer how strong is the impact of the infection on the host, the honey bee transcriptomes responding to each of the two parasite sources are quantified as well. We find the expression of bee (HAmel and HAcer) genes is suppressed, where more down-regulated genes than up-regulated ones when infected by either PAmel or PAcer (Table 1, Pearson’s Chi-squared test, χ2 = 51.0, df=4, P < 0.001). Comparatively, more genes were regulated by the infection in HAcer than HAmel. The bee hosts share a common subset of genes that respond to infection (Figure 1D). The immune response toward the infection is small in HAmel, reflected by only two down-regulated immune genes in Toll pathway involved in pathogen recognition (LOC412536) and melanization (LOC406115). We found both up and down regulated immune genes in HAcer. Particularly, the Toll pathway is generally suppressed, including the genes involved in parasite recognition PGRP (APICC_00292), signal transport SPZ (APICC_01852), and antimicrobial peptides defensin and lysozyme (APICC_08301, APICC_03572, APICC_08272). These suppressed genes have previously been confirmed using qPCR (Ke et al., 2022). A few genes are continuously down-regulated in HAmel, reflecting genes inhibited by the infection (Supplementary Figure S2). GO enrichment analysis indicates that apoptosis regulation (GO:0043066, FDR< 0.0001), cell cycle phase transition (GO:0044843, FDR < 0.0001), and a few other biological functions are altered in the infected honey bee genes (Figure 2).
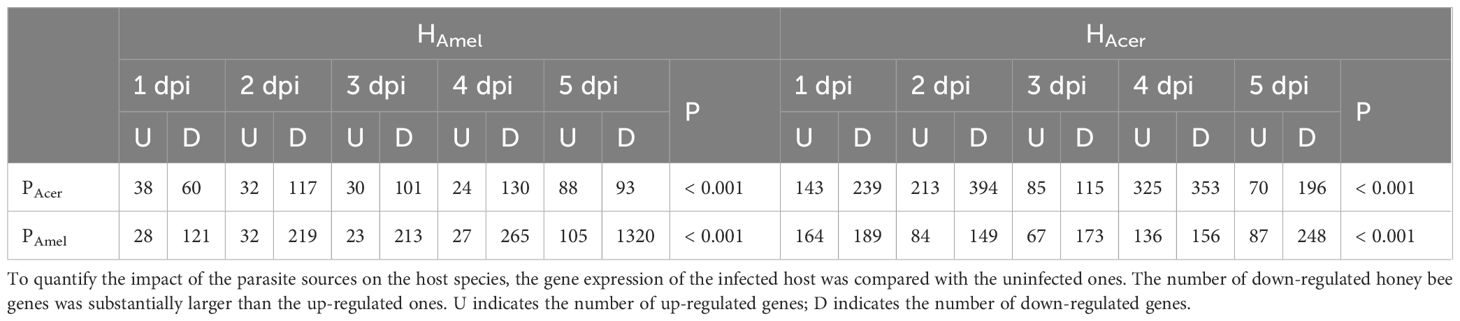
Table 1. The number of significantly regulated honey bee genes between infected and uninfected bees.
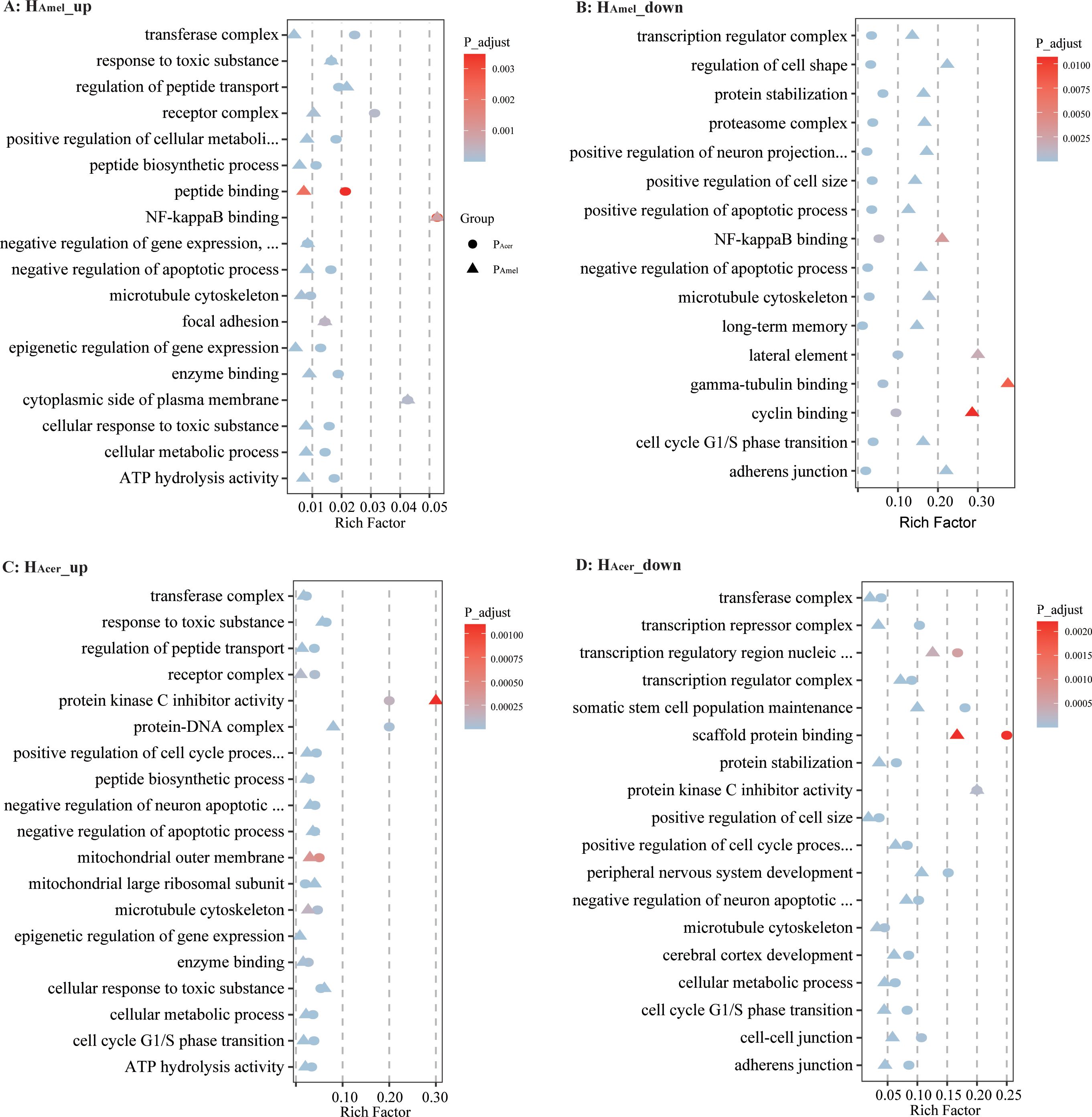
Figure 2. Bubble chart for GO enrichment of differentially expressed genes. The intersection of up-regulated genes at five time points in HAmel infected with PAmel and PAcer (A). The intersection of down-regulated genes at five time points in HAmel (B). Up-regulated genes at five time points in HAcer (C) and down-regulated genes at five time points in HAcer (D). Upon parasitic infection, the honey bee enzyme binding and metabolism-related genes were up-regulated, while genes related to the cell cycle and transcriptional regulation were suppressed.
A static environment favors decreased genetic diversity. In contrast, a fluctuating environment favors a high genetic diversity (Abdul-Rahman et al., 2021). The two bee species in Asia compete for the habitat, and the parasite can shift between them. To adapt to both bees, the parasite may favor a large gene pool to survive. Indeed, a higher diversity in the sympatric population than in the allopatric population was observed (Ke et al., 2022). Parasites show elevated genetic diversity by infecting diverse host populations, suggesting host diversity shapes parasite diversity (Ekroth et al., 2021).
Previous studies suggest that N. ceranae causes 40% ~ 90% of bee mortality, and the spore load is at 106 levels in HAmel two weeks post-inoculation (Higes et al., 2007; Paxton et al., 2007; Martín-Hernández et al., 2011; Suwannapong et al., 2011; Eiri et al., 2015). In our data, low mortality (4.1%) is observed in HAmel, and slightly higher mortality (13.4%) is observed in HAcer. The parasite shows low virulence in the primary host when the alternative host species are less common (Manzoli et al., 2018). Historically, the parasite is first described in A. cerana and does not necessarily indicate that A. cerana is the primary host (Fries et al., 1996). Subsequently, the parasite is identified in A. mellifera (Higes et al., 2007). The anthropogenic-driven contact enhanced the gene flow of the parasites (Pelin et al., 2015). If A. cerana is the primary host, a balanced transmission and virulence are expected between HAcer and PAcer. In our data, the parasite shows high virulence and low spore load in HAcer, which do not follow the conventional host-parasite evolution. Thus, additional studies are needed to investigate whether HAmel or HAcer is the primary host of N. ceranae.
N. ceranae infection causes global gene expression changes in HAmel and HAcer (Holt et al., 2013; Fan et al., 2022). A few studies suggest the Toll pathway is the primary immune response to the N. ceranae infection (Huang and Evans, 2016; Li et al., 2017; Ke et al., 2022). In our data, we find the infection caused minor immune stress in HAmel. Comparatively, the infection suppresses the Toll pathway from pathway recognition to the antimicrobial peptides in HAcer. Additionally, the infection strongly regulates the transcripts, and more genes are altered by the infection, suggesting intense stress in HAcer. Remarkably, the infection causes higher mortality in Asia honey bees, which has been overlooked for decades. Lipid metabolism is important for microsporidians to establish infection (El Alaoui et al., 2001; Jeon, 2021). In our data, the up-regulated parasite genes are enriched in lipid metabolism, which might be necessary for N. ceranae to establish infection. The apoptosis pathway is also enriched in regulated honey bee genes, confirming apoptosis is an essential defense mechanism in bees against N. ceranae infection (Higes et al., 2013; Kurze et al., 2015). In bumblebees, the microsporidian parasite Nosema bombi shows distinctive virulence toward hosts in a sympatric population (Rutrecht and Brown, 2009). In Daphnia, the microsporidian shows reduced infection intensity with increased geographic distance (Ebert, 1994). In our case, bee genetics and co-evolutionary status may shape the distinct trade-offs between virulence and proliferation in N. ceranae. Future studies to identify the genome diversification of N. ceranae may help to determine its primary host. Additional gene functional studies help to understand the host-parasite co-evolution in this multi-host system, also as target genes for this parasite control.
The microsporidian parasite N. ceranae evolves a balanced virulence and transmission with the honey bee A. mellifera in Asia. Comparatively, the parasite shows high virulence and low transmission in the honey bee A. cerana, supported by the time series transcripts. Thus, additional study is needed to investigate whether A. cerana or A. mellifera is the primary host of this microsporidian parasite.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The sequencing reads are available in NCBI Bio-project PRJNA822678 and PRJNA784016.
XW: Writing – original draft. QH: Funding acquisition, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the National Natural Science Foundation of China #32260862.
The authors declare the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1524197/full#supplementary-material
Abdul-Rahman, F., TranChina, D., Gresham, D. (2021). Fluctuating environments maintain genetic diversity through neutral fitness effects and balancing selection. Mol. Biol. Evol. 38, 4362–4375. doi: 10.1093/molbev/msab173
Acevedo, M. A., Dillemuth, F. P., Flick, A. J., Faldyn, M. J., Elderd, B. D. (2019). Virulence-driven trade-offs in disease transmission: A meta-analysis. Evolution 73, 636–647. doi: 10.1111/evo.2019.73.issue-4
Alexa, A., Rahnenfuhrer, J. (2021). TopGO: Enrichment Analysis for Gene Ontology. R Packag. version 2.46.0. Available online at: https://rdrr.io/bioc/topGO/ (Accessed November 2024).
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed November 2024).
Botías, C., Martín-Hernández, R., Barrios, L., Meana, A., Higes, M. (2013). Nosema spp. infection and its negative effects on honey bees (Apis mellifera iberiensis) at the colony level. Vet. Res. 44, 25. doi: 10.1186/1297-9716-44-25
Chen, Y.p., Pettis, J. S., Zhao, Y., Liu, X., Tallon, L. J., Sadzewicz, L. D., et al. (2013). Genome sequencing and comparative genomics of honey bee microsporidia, Nosema apis reveal novel insights into host-parasite interactions. BMC Genomics 14, 451. doi: 10.1186/1471-2164-14-451
de Roode, J. C., Yates, A. J., Altizer, S. (2008). Virulence-transmission trade-offs and population divergence in virulence in a naturally occurring butterfly parasite. Proc. Natl. Acad. Sci. 105, 7489–7494. doi: 10.1073/pnas.0710909105
Diao, Q., Sun, L., Zheng, H., Zeng, Z., Wang, S., Xu, S., et al. (2018). Genomic and transcriptomic analysis of the Asian honeybee Apis cerana provides novel insights into honeybee biology. Sci. Rep. 8, 822. doi: 10.1038/s41598-017-17338-6
Ebert, D. (1994). Virulence and local adaptation of a horizontally transmitted parasite. Science 265, 1084–1086. doi: 10.1126/science.265.5175.1084
Eiri, D. M., Suwannapong, G., Endler, M., Nieh, J. C. (2015). Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS One 10, e0126330. doi: 10.1371/journal.pone.0126330
Ekroth, A. K. E., Gerth, M., Stevens, E. J., Ford, S. A., King, K. C. (2021). Host genotype and genetic diversity shape the evolution of a novel bacterial infection. ISME J. 15, 2146–2157. doi: 10.1038/s41396-021-00911-3
El Alaoui, H., Bata, J., Bauchart, D., Doré, J. C., Vivarès, C. P. (2001). Lipids of three microsporidian species and multivariate analysis of the host-parasite relationship. J. Parasitol. 87, 554–559. doi: 10.2307/3285092
Engelstädter, J., Fortuna, N. Z. (2019). The dynamics of preferential host switching: Host phylogeny as a key predictor of parasite distribution. Evolution 73, 1330–1340. doi: 10.1111/evo.2019.73.issue-7
Fan, Y., Wang, J., Yu, K., Zhang, W., Cai, Z., Sun, M., et al. (2022). Comparative transcriptome investigation of Nosema ceranae infecting Eastern honey bee workers. Insects. 13, 241. doi: 10.3390/insects13030241
Fan, X., Zhang, W., Zhang, K., Zhang, J., Long, Q., Wu, Y., et al. (2022). In-depth investigation of microRNA-mediated cross-kingdom regulation between Asian honey bee and microsporidian. Front. Microbiol. 13, 1003294. doi: 10.3389/fmicb.2022.1003294
Fries, I., Feng, F., da Silva, A., Slemenda, S. B., Pieniazek, N. J. (1996). Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae). Eur. J. Protistol. 32, 356–365. doi: 10.1016/S0932-4739(96)80059-9
Gage, S. L., Kramer, C., Calle, S., Carroll, M., Heien, M., DeGrandi-Hoffman, G. (2018). Nosema ceranae parasitism impacts olfactory learning and memory and neurochemistry in honey bees (Apis mellifera). J. Exp. Biol. 221, jeb161489. doi: 10.1242/jeb.161489
Gisder, S., Mockel, N., Linde, A., Genersch, E. (2011). A cell culture model for Nosema ceranae and Nosema apis allows new insights into the life cycle of these important honey bee-pathogenic microsporidia. Environ. Microbiol. 13, 404–413. doi: 10.1111/j.1462-2920.2010.02346.x
Gowler, C. D., Essington, H., O’Brien, B., Shaw, C. L., Bilich, R. W., Clay, P. A., et al. (2023). Virulence evolution during a naturally occurring parasite outbreak. Evol. Ecol. 37, 113–129. doi: 10.1007/s10682-022-10169-6
Graystock, P., Goulson, D., Hughes, W. O. H. (2015). Parasites in bloom: flowers aid dispersal and transmission of pollinator parasites within and between bee species. Proc. Biol. Sci. 282, 20151371. doi: 10.1098/rspb.2015.1371
Higes, M., Garcia-Palencia, P., Martin-Hernandez, R., Meana, A. (2007). Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94, 211–217. doi: 10.1016/j.jip.2006.11.001
Higes, M., Juarranz, A., Dias-Almeida, J., Lucena, S., Botias, C., Meana, A., et al. (2013). Apoptosis in the pathogenesis of Nosema ceranae (Microsporidia: Nosematidae) in honey bees (Apis mellifera). Environ. Microbiol. Rep. 5, 530–536. doi: 10.1111/emi4.2013.5.issue-4
Higes, M., Martín-Hernández, R., Botías, C., Bailón, E. G., González-Porto, A. V., Barrios, L., et al. (2008). How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10, 2659–2669. doi: 10.1111/j.1462-2920.2008.01687.x
Holt, H. L., Aronstein, K. A., Grozinger, C. M. (2013). Chronic parasitization by Nosema microsporidia causes global expression changes in core nutritional, metabolic and behavioral pathways in honey bee workers (Apis mellifera). BMC Genomics 14, 799. doi: 10.1186/1471-2164-14-799
Huang, Q., Evans, J. D. (2016). Identification of microRNA-like small RNAs from fungal parasite Nosema ceranae. J. Invertebr. Pathol. 133, 107–109. doi: 10.1016/j.jip.2015.12.005
Huang, Q., Wu, Z. H., Li, W. F., Guo, R., Xu, J. S., Dang, X. Q., et al. (2021). Genome and evolutionary analysis of Nosema ceranae: A microsporidian parasite of honey bees. Front. Microbiol. 12, 645353. doi: 10.3389/fmicb.2021.645353
Jack, C. J., Lucas, H. M., Webster, T. C., Sagili, R. R. (2016). Colony level prevalence and intensity of Nosema ceranae in honey bees (Apis mellifera L.). PloS One 11, e0163522. doi: 10.1371/journal.pone.0163522
Jeon, J. (2021). Characterizing the Role of Host Lipid Metabolism on Microsporidia Infection in Caenorhabditis elegans. (master's thesis). University of Toronto. Available at: https://tspace.library.utoronto.ca/handle/1807/105004 (Accessed November 2024).
Ke, L., Yan, W. Y., Zhang, L. Z., Zeng, Z. J., Evans, J. D., Huang, Q. (2022). Honey bee habitat sharing enhances gene flow of the parasite Nosema ceranae. Microb. Ecol. 83, 1105–1111. doi: 10.1007/s00248-021-01827-3
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357. doi: 10.1038/nmeth.3317
Kurze, C., Le Conte, Y., Dussaubat, C., Erler, S., Kryger, P., Lewkowski, O., et al. (2015). Nosema tolerant honeybees (Apis mellifera) escape parasitic manipulation of apoptosis. PloS One 10, e0140174. doi: 10.1371/journal.pone.0140174
Li, H. (2022).seqtk: a fast and lightweight tool for processing sequences in the FASTA or FASTQ format. Available online at: https://github.com/lh3/seqtk (Accessed November 2024).
Li, W., Evans, J., Li, J., Su, S., Hamilton, M., Chen, Y. (2017). Spore load and immune response of honey bees naturally infected by Nosema ceranae. Parasitol. Res. 116, 1–10. doi: 10.1007/s00436-017-5630-8
Li, J., Qin, H., Wu, J., Sadd, B. M., Wang, X., Evans, J. D., et al. (2012). The prevalence of parasites and pathogens in Asian honeybees Apis cerana in China. PloS One 7, e47955. doi: 10.1371/journal.pone.0047955
Manzoli, D. E., Saravia-Pietropaolo, M. J., Antoniazzi, L. R., Barengo, E., Arce, S. I., Quiroga, M. A., et al. (2018). Contrasting consequences of different defence strategies in a natural multihost–parasite system. Int. J. Parasitol. 48, 445–455. doi: 10.1016/j.ijpara.2017.11.001
Martín-Hernández, R., Botías, C., Barrios, L., Martínez-Salvador, A., Meana, A., Mayack, C., et al. (2011). Comparison of the energetic stress associated with experimental Nosema ceranae and Nosema apis infection of honeybees (Apis mellifera). Parasitol. Res. 109, 605–612. doi: 10.1007/s00436-011-2292-9
Paris, L., El Alaoui, H., Delbac, F., Diogon, M. (2018). Effects of the gut parasite Nosema ceranae on honey bee physiology and behavior. Curr. Opin. Insect Sci. 26, 149–154. doi: 10.1016/j.cois.2018.02.017
Paxton, R. J., Klee, J., Korpela, S., Fries, I. (2007). Nosema ceranae has infected Apis mellifera in Europe since at least 1998 and may be more virulent than Nosema apis. Apidologie 38, 558–565. doi: 10.1051/apido:2007037
Pelin, A., Selman, M., Aris-Brosou, S., Farinelli, L., Corradi, N. (2015). Genome analyses suggest the presence of polyploidy and recent human-driven expansions in eight global populations of the honeybee pathogen Nosema ceranae. Environ. Microbiol. 17, 4443–4458. doi: 10.1111/emi.2015.17.issue-11
Quinlan, A. R., Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
R Core Team (2013). R: A language and environment for statistical computing (Vienna, Austria: R Found. Stat. Comput). Available at: http://www.R-project.org/ (Accessed November 2024).
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rutrecht, S. T., Brown, M. J. F. (2009). Differential virulence in a multiple-host parasite of bumble bees: resolving the paradox of parasite survival? Oikos 118, 941–949. doi: 10.1111/j.1600-0706.2009.17392.x
Suwannapong, G., Yemor, T., Boonpakdee, C., Benbow, M. E. (2011). Nosema ceranae, a new parasite in Thai honeybees. J. Invertebr. Pathol. 106, 236–241. doi: 10.1016/j.jip.2010.10.003
Therneau, T. (2022). A package for survival analysis in R. R Packag. version 3.4-0. Available online at: https://CRAN.R-project.org/package=survival (Accessed November 2024).
Wallberg, A., Bunikis, I., Pettersson, O. V., Mosbech, M.-B., Childers, A. K., Evans, J. D., et al. (2019). A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genomics 20, 275. doi: 10.1186/s12864-019-5642-0
Keywords: Nosema ceranae, host switch, habitat competition, selection, mortality
Citation: Wei X and Huang Q (2025) Distinct virulence of the microsporidian parasite in honey bees competing habitat. Front. Cell. Infect. Microbiol. 15:1524197. doi: 10.3389/fcimb.2025.1524197
Received: 07 November 2024; Accepted: 31 January 2025;
Published: 17 February 2025.
Edited by:
Sudhir Kumar, Iowa State University, United StatesReviewed by:
Chinmay V. Tikhe, Johns Hopkins University, United StatesCopyright © 2025 Wei and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Huang, cWlhbmctaHVhbmdAbGl2ZS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.