
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 31 January 2025
Sec. Molecular Bacterial Pathogenesis
Volume 15 - 2025 | https://doi.org/10.3389/fcimb.2025.1509037
This article is part of the Research Topic Mechanisms of microbial persistence and strategies to counter them View all 3 articles
The PhoP response regulator and the cognate sensor kinase PhoQ form one of the two-component signal transduction systems that is highly conserved in bacteria. The PhoP/PhoQ system is a crucial mediator of signal transduction. It regulates the expression of bacterial environmental tolerance genes, virulence factors, adhesion, and invasion-related genes by sensing various environmental signals in the host, including Mg2+, low pH, antimicrobial peptides, and osmotic pressure. In this review, we describe the PhoP/PhoQ system-induced signal composition and its feedback mechanism, and the abundance of PhoP phosphorylation in the activated state directly or indirectly controls the transcription and expression of related genes, regulating bacterial stability. Then, we discuss the relationship between the PhoP/PhoQ system and other components of the TCS system. Under the same induction conditions, their interaction relationship determines whether bacteria can quickly restore their homeostasis and exert virulence effects. Finally, we investigate the coordinated role of the PhoP/PhoQ system in acquiring pathogenic virulence.
Bacteria may encounter various environmental pressures, affecting their survival and virulence (Yao et al., 2024; He et al., 2025). In response to environmental pressure, many strategies have been evolved to fight against external pressure. The two-component signal transduction systems (TCSs) play an essential role in signal transduction during the change of bacterial environment (Xie et al., 2022). It enables bacterial pathogens to sense various environmental conditions such as light, temperature, pH, osmotic pressure, nutrients, small molecule metabolites, antibiotics, antimicrobial peptides, and other host-derived signals. This ability allows pathogens to determine when they have reached the microenvironment of a host or host interior. Subsequently, specific genes are activated or repressed to adapt, evade, or attack (Xie et al., 2022). The two-component signal transduction systems consist of conserved signal receivers: histidine kinases (HKs) and their cognate response regulators (RRs) (Xie et al., 2020). Studies have shown that TCS usually uses positive and negative feedback mechanisms to regulate gene expression in HK, RR, and downstream genes (Figure 1). In this way, PhoP/PhoQ system responds positively to environmental stress (Lippa et al., 2009). Depending on the structural domain is divided into six families, respectively: the OmpR family, the family of NarL, the NtrC family, the LuxR family, the CitB family, and Che (Pina et al., 2021), TCSs control the various components of the phosphate transferring principle almost similar; they form between complex signal transportation network (Véscovi et al., 1996; Pina et al., 2021; Shao et al., 2021). The composition of bacterial TCS is rich, forming a complex information transportation network between them. However, the TCS system has not been found in animal hosts, and as a signal transduction system, TCS could be a new target for developing new antibacterial therapeutic agents (Chen and Groisman, 2013).
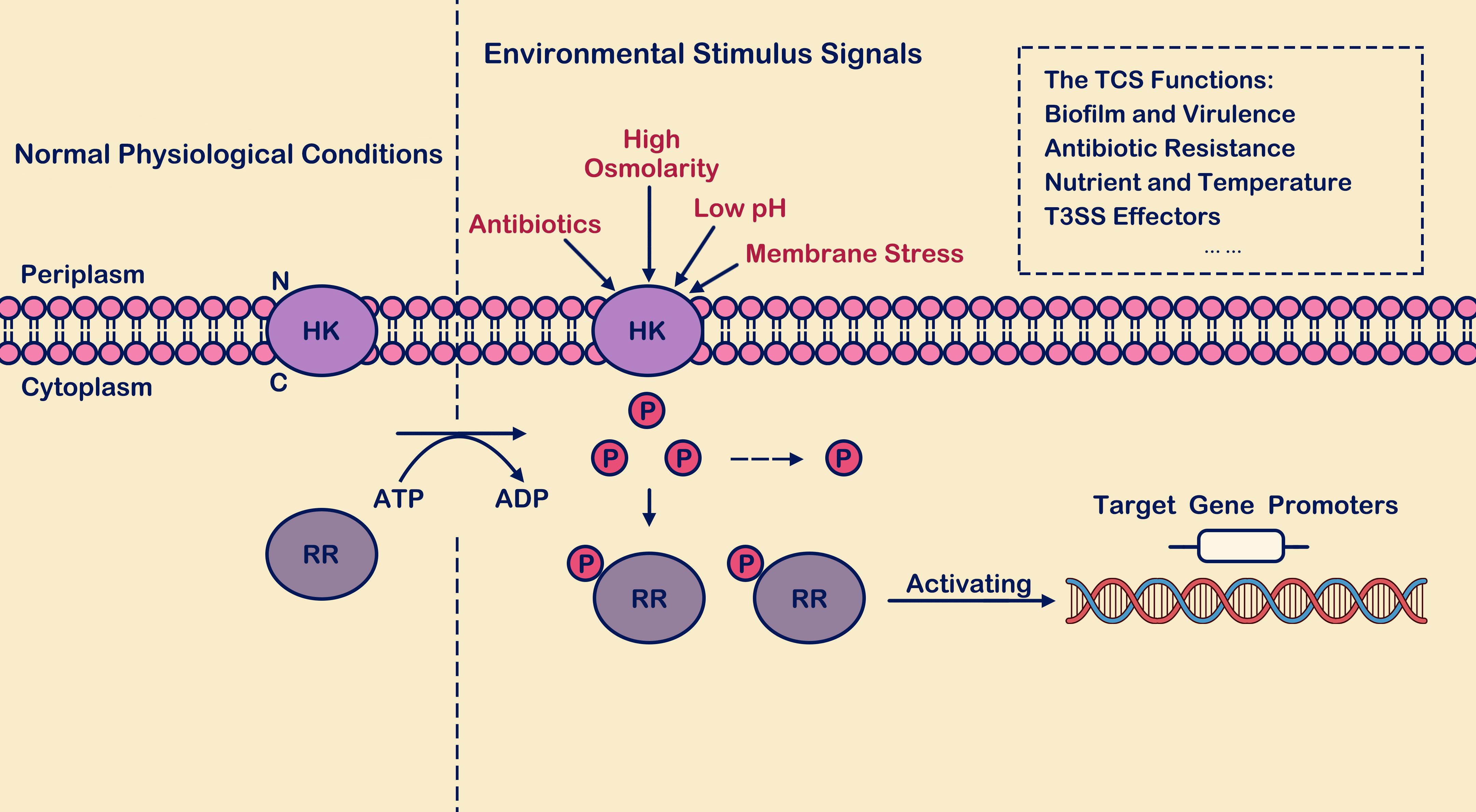
Figure 1. The process of two-component signal transduction. Under environmental stimulus conditions, histidine kinase (HK) interacts with its signaling ligand, leading to phosphorylation of histidine residue. The phosphate group is recognized and captured by response regulator (RR) in the cytoplasm, activating the output domains of the response regulator and inducing conformational changes. Subsequently, the regulator binds to the promoter regions of downstream target genes, thereby activating transcription levels of both themselves and downstream target genes. Here, “P” denotes the phosphate group, “HK” stands for histidine kinase, and “RR” represents response regulator.
The PhoP/PhoQ system, a member of the OmpR family, has been thoroughly studied in Salmonella enterica (S. enterica), Escherichia coli (E. coli), Shigella flexneri and other bacterial strains (Lin et al., 2017; Yuan et al., 2017; Pina et al., 2021; Guo et al., 2022), and plays an important role in the entire regulatory network (Pina et al., 2021). In S. enterica, PhoP/PhoQ is involved in regulating the transcription and expression of a variety of virulence genes, including invasion of non-phagocytic cells (such as epithelial cells), anti-phagosome killing, resistance to antimicrobial peptides (AMPs), and release of virulence proteins (Pina et al., 2021). The PhoP/PhoQ system consists of two parts: PhoQ belongs to transmembrane proteins, and its structural composition mainly includes the N-terminal conserved periplasmic sensor domain, two transmembrane (TM) domains, the histidine adenosine monophosphate associated protein (HAMP) domain located in the cytosol for signal transmission, the dimerization and histidine phosphotransfer (DHp) domain required for dimerization, and the catalytic adenosine (CA) domain that binds to catalytic adenosine triphosphate (ATP) (Mensa et al., 2021). PhoP, a homologous regulatory factor (RRs) located downstream of PhoQ, consists of two domains: the N-terminal regulatory domain, which has the necessary aspartate residue site, and one is the C-terminal effect domain, which is involved in binding to the specific DNA sequence in the target promoter (Ali and Abdel Aziz, 2024). The catalytic and regulatory structures of these two proteins are relatively conserved. PhoQ is commonly used as a sensor to recognize environmental stimuli, promote self-phosphorylation of histidine residues under the catalysis of ATP, and deliver phosphate groups to its cognate regulator PhoP (Yamamoto et al., 2002; Pathak et al., 2010). After the N-terminal aspartic acid residue of PhoP is captured and recognized, the phosphorylation reaction (PhoP-P) occurs, and the PhoP conformation changes (Yamamoto et al., 2002; Pathak et al., 2010). PhoQ controls PhoP phosphorylation and influences the transcription of PhoP-regulating genes (Gall et al., 2016; Mattos-Graner and Duncan, 2017). Following phosphorylation of PhoP, on the one hand, it can promote its own transcription and activate the expression of downstream gene targets (such as mgtA, slyB, pmrD, pagP). More so, it can competitively bind with other transcription factors, resulting in down-regulation of specific gene targets (Goldberg et al., 2010). The activated PhoP/PhoQ system mediates various phenotypic modifications, regulates bacterial homeostasis, and reduces the adverse effects of external environmental pressure (Goldberg et al., 2010).
The PhoP/PhoQ system, as a classic two-component system, involves the dual-function protein PhoQ, which senses environmental changes such as divalent cations (Véscovi et al., 1996), antibacterial (Yu and Guo, 2011), low pH (Bader et al., 2010), circumcellular redox (Choi and Groisman, 2016), and osmotic pressure (Yuan et al., 2017). These factors regulate the phosphorylation-mediated phenotypic modification of the response regulator PhoP. The PhoP/PhoQ system plays a crucial regulatory role in virulence of in the virulence of several pathogenic bacteria. Therefore, elucidating the response mechanisms of the PhoP/PhoQ system to various stimuli and its transcriptional regulation of downstream target genes provides fundamental insights into the PhoP/PhoQ system.
Divalent cations play a crucial role in organisms, serving as essential cofactors for numerous enzymes. They are vital for maintaining the integrity of biological membranes and facilitating various physiological functions (Lippa and Goulian, 2012). Mg2+ was initially identified as the environmental stimulus factor for the PhoP/PhoQ system, which plays a crucial role in maintaining Mg2+ homeostasis (Véscovi et al., 1996). When the cytoplasmic Mg2+ concentration falls below a certain threshold (e.g., when Salmonella typhimurium concentration below 0.5 mM Mg2+) (Véscovi et al., 1996), bacteria generally reduce the assembly of functional ribosomes and undergo auto-phosphorylation of the periplasmic PhoQ. PhoP is phosphorylated to PhoP-P, and PhoP-P specifically binds to the promoter region of Mg2+ transport-related genes (such as mgtA, mgtB, and mgtC), and thus activating gene transcription (Cromie and Groisman, 2010; Yeom et al., 2020; Yeom and Groisman, 2021). In the case of E. coli, when Mg2+ levels decrease to levels impairing protein production (below 10 μM Mg2+), PhoP-P promotes the expression of the iraP gene. This increases the intracellular content of RpoS, reducing the rate of protein synthesis to maintain essential cellular functions (Yin et al., 2019). Meanwhile, the expression level of the Mg2+ transporter protein MgtA is significantly upregulated, facilitating the transport of Mg2+ from the periplasm to the cytoplasm, thereby maintaining stable cellular Mg2+ concentrations (Park et al., 2018). When PhoQ is activated by cationic antimicrobial peptides or acidic environmental conditions, MgtA remains unaffected (Subramani et al., 2016).
Due to environmental stress, the PhoP/PhoQ cascade activates the transcription of downstream genes, which requires significant ATP consumption. The availability of ATP directly correlates with changes in the abundance of ClpXP (Groisman, 2016). Under normal conditions, upon binding with adaptor proteins, RpoS is transported to ClpXP for degradation, rapidly reducing RpoS levels (Groisman, 2016). The transcription factor RpoS regulates the expression of numerous bacterial genes, with its synthesis and degradation tightly controlled, varying in response to cellular growth stresses (Battesti and Gottesman, 2013; Schellhorn, 2020). In Salmonella enterica serovar Typhimurium (S. Typhimurium) under low Mg2+ conditions (≤20 μM Mg2+), the stability of the sigma factor RpoS plays a crucial role in the PhoP/PhoQ system cascade (Bougdour et al., 2008). PhoP-P promotes the upregulation of RssB anti-adaptors (IraM/IraP/IraD) expression (Bougdour et al., 2008). Acting as an intermediary in regulating RpoS stability, it interferes with RssB-mediated degradation of RpoS by interacting with RssB. Moreover, the PhoP/PhoQ cascade promotes the regulation of RpoS stability by iraP, and high levels of RpoS mediate transcriptional expression of its dependent genes (such as katE and esrB genes) (Bougdour et al., 2008).
SlyB, located in the outer membrane, is regulated by PhoP-P under decreased Mg2+ concentration (such as in Yersinia pestis when Mg2+ is below 50 μM) or increased osmotic pressure (such as in E. coli when stimulated with 300 mM NaCl) (Tu et al., 2006; Perez et al., 2009; Yuan et al., 2017). In addition, SlyB plays a negative regulatory role in some bacteria on PhoP/PhoQ (Tu et al., 2006; Perez et al., 2009). For instance, in Salmonella typhimurium, deletion of the slyB gene leads to decreased transcription levels of genes activated by PhoP-P. In contrast, such a negative regulatory mechanism is not observed in E. coli (Lippa et al., 2009). Additionally, SlyB can respond to outer membrane (OM) biogenesis defects by sensing the accumulation of lipopolysaccharide (LPS) and periplasmic unfolded outer membrane proteins (OMPs). The modification of LPS plays a crucial role in the PhoP/PhoQ cascade (Janssens et al., 2024). LPS modifications help bacteria reduce the electrostatic repulsion of phosphorylated residues and releases a certain amount of Mg2+ for MgtA-related proteins to transfer Mg2+ from the periplasmic space into the cytoplasm (Janssens et al., 2024). Studies on S. Typhimurium demonstrate that under low Mg2+ conditions (less than 50 μM Mg2+), the mgtA gene is activated in a PhoP-P-dependent manner, independent of other environmental stimuli (Yeom et al., 2020). When PhoQ detects signals like low pH or antimicrobial peptides, the expression level of the Mg2+ transporter gene mgtA remains unaffected (Yeom et al., 2020; Groisman et al., 2021). Additionally, Ca2+ and Mn2+ can serve as ligands for PhoQ with similar mechanisms of action, neutralizing electrostatic repulsion between negatively charged residues at the divalent cation binding sites (Regelmann et al., 2002; Barchiesi et al., 2008). Conversely, as the concentration of divalent cations increases, the expression levels of regulatory proteins produced by the PhoP/PhoQ cascade (such as PgtE, PhoN, MgtA, MgtB, and IraP) gradually decrease (Cho et al., 2006; Bougdour et al., 2008). When the Mg2+ concentration exceeds 50 μM, a stable bridge forms between the negatively charged outer and inner membranes, thereby inhibiting the PhoP/PhoQ cascade reaction (Regelmann et al., 2002).
Under conditions of low Mg2+ concentration (such as S. Typhimurium in a minimal medium containing 10 μM Mg2+), the PhoP/PhoQ system interacts with PmrA/PmrB (Hu et al., 2016). PmrA serves as the sensor responding to external stimulus signals, while PmrB acts as the downstream responder to PmrA (Kato et al., 2003; Paredes et al., 2023). PhoP-P stimulates the transcription of PmrD, which mediates the phosphorylation of another response regulator, PmrA. Sufficient PmrA-P is produced to promote the expression levels of genes such as pmrC, pmrE, pmrHFIJKLM, collectively modifying the outer membrane LPS (Chen and Groisman, 2013; Shprung et al., 2021; Paredes et al., 2023; Janssens et al., 2024). In pmrD deletion strains, it was found that the expression level of PmrA was significantly reduced compared to wild-type strains (Cho et al., 2006). Additionally, under high Fe3+ conditions, (such as S. enterica in a minimal medium containing 100 μM Fe3+) activate the PmrA/PmrB system (Bolard et al., 2019). PmrD also plays a role in promoting the activation of PmrA (Cho et al., 2006), serving as a crucial bridge between the PhoP/PhoQ and PmrA/PmrB systems, directly influencing the regulatory mechanism and abundance of PmrA (Cho et al., 2006). The above content is briefly described in Figure 2.
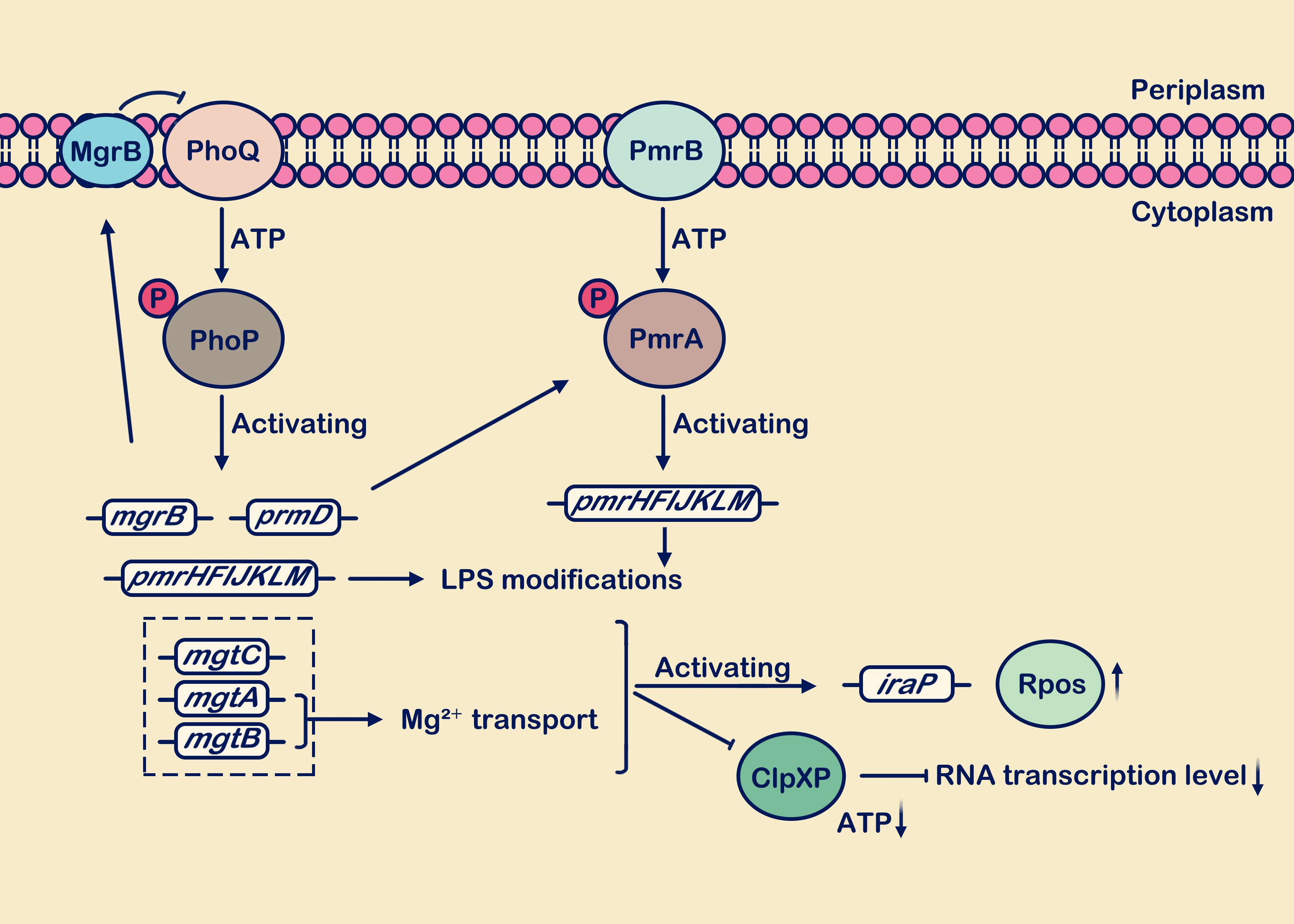
Figure 2. Low Mg2+ levels stimulate the activation of the PhoP/PhoQ and PmrA/PmrB systems. During growth under low Mg2+ conditions, the PhoP/PhoQ system induces transcription of target genes including mgrB, pmrD, pmrHFIJKLM, mgtC, mgtA, and mgtB. The mgrB gene is transcriptionally upregulated, and the synthesized MgrB membrane protein exerts negative feedback on PhoQ. Activation of the pmrD gene positively regulates the PmrA/PmrB system. The mgtA and mgtB genes facilitate the transport of extracellular Mg2+ into the cell. Presence of the mgtC, mgtA, and mgtB genes reduces ATP consumption and decreases protein synthesis rates. Activation of pmrHFIJKLM is involved in LPS modification. In the figure legend, a circle with “P” denotes a phosphate group, an upward vertical arrow indicates upregulation, a downward vertical arrow indicates downregulation, and the arrow from MgrB to PhoQ signifies “inhibition”.
Antimicrobial peptides are widely sourced from diverse origins, including animals, plants, microorganisms, and synthetic production, serving as integral components of the innate immune systems in most multicellular organisms (Kato et al., 2012). AMPs are predominantly concentrated within phagosomes, where they exert antimicrobial effects in macrophages (Li et al., 2022). AMPs are rich in positive charges, enabling them to bind with negatively charged molecules on bacterial surfaces (Yan et al., 2021; Zhu et al., 2022). They swiftly penetrate lipid membranes, forming pores in bacterial cell membranes and disrupting membrane permeability, ultimately causing bacterial cell lysis (Yan et al., 2021; Zhu et al., 2022). For pathogens, resistance to antimicrobial peptides is crucial for exerting their toxicity. It has been established that antimicrobial peptides serve as direct signals for activating the PhoQ histidine kinase (Ramezanifard et al., 2023). Cationic antimicrobial peptides competitively bind to the periplasmic domain of PhoQ with divalent cations, inducing a conformational change in the cytoplasmic dimer (Ramezanifard et al., 2023). This promotes the phosphorylation of PhoP and alters the total charge of the lipid A portion of bacterial lipopolysaccharide (LPS), modifying LPS to increase bacterial resistance (Bader et al., 2005; Yu and Guo, 2011; Ramezanifard et al., 2023). The inner membrane protein Mig-14 in extraintestinal pathogenic E. coli (ExPEC) and S. typhimurium) play a crucial role within macrophages, significantly enhancing bacterial resistance against AMPs (Zhuge et al., 2018; Martynowycz et al., 2019).
In recent years, polymyxins have garnered significant attention from researchers due to the rapid increase in bacterial antibiotic resistance (Brodsky et al., 2005; Wang et al., 2020). Polymyxins are important cyclic peptide antibiotics isolated from Bacillus species (Gahlot et al., 2024). They disrupt membrane integrity and induce bacterial outer membrane damage by interacting with negatively charged surface structures such as LPSs in Gram-negative bacteria and lipoteichoic acids in Gram-positive bacteria, thereby exhibiting bactericidal activity (Storm et al., 1977; Brodsky et al., 2005). The cascade of PhoP/PhoQ system modifying bacterial outer membrane LPS can lead to increased resistance of bacteria to polymyxins (Guo et al., 2022). Meanwhile, PhoP-P regulates the expression of PmrD, which effectively inhibits the dephosphorylation of PmrA-P, thereby mediating the involvement of the PmrA/PmrB system in the modification process of LPS (Brodsky et al., 2005; Zhang et al., 2022). PhoQ promotes the binding of PhoP-P to its downstream pmrHFIJKLM promoter (also known as arnBCADTEF or pbgPE operator) through the recognition of polymyxin (Chandler et al., 2020). Concurrently, the upregulation of PmrD expression indirectly enhances the cascade reaction of the PmrA/PmrB system. PmrA also binds to downstream pmrC, pmrE, and pmrHFIJKLM promoters (Shprung et al., 2012; Poirel et al., 2017; Paredes et al., 2023). The overexpression products of pmrC, pmrE, and pmrHFIJKLM are utilized for the synthesis of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoethanolamine (PEA) (Figure 3). These two components modify LPS by increasing the negative charge on the outer membrane (Hiroshi, 2003), reducing membrane permeability, thereby limiting the entry of antimicrobial peptides and playing a crucial role in promoting polymyxin resistance (Phan et al., 2017; Liu et al., 2021; Shahzad et al., 2023).
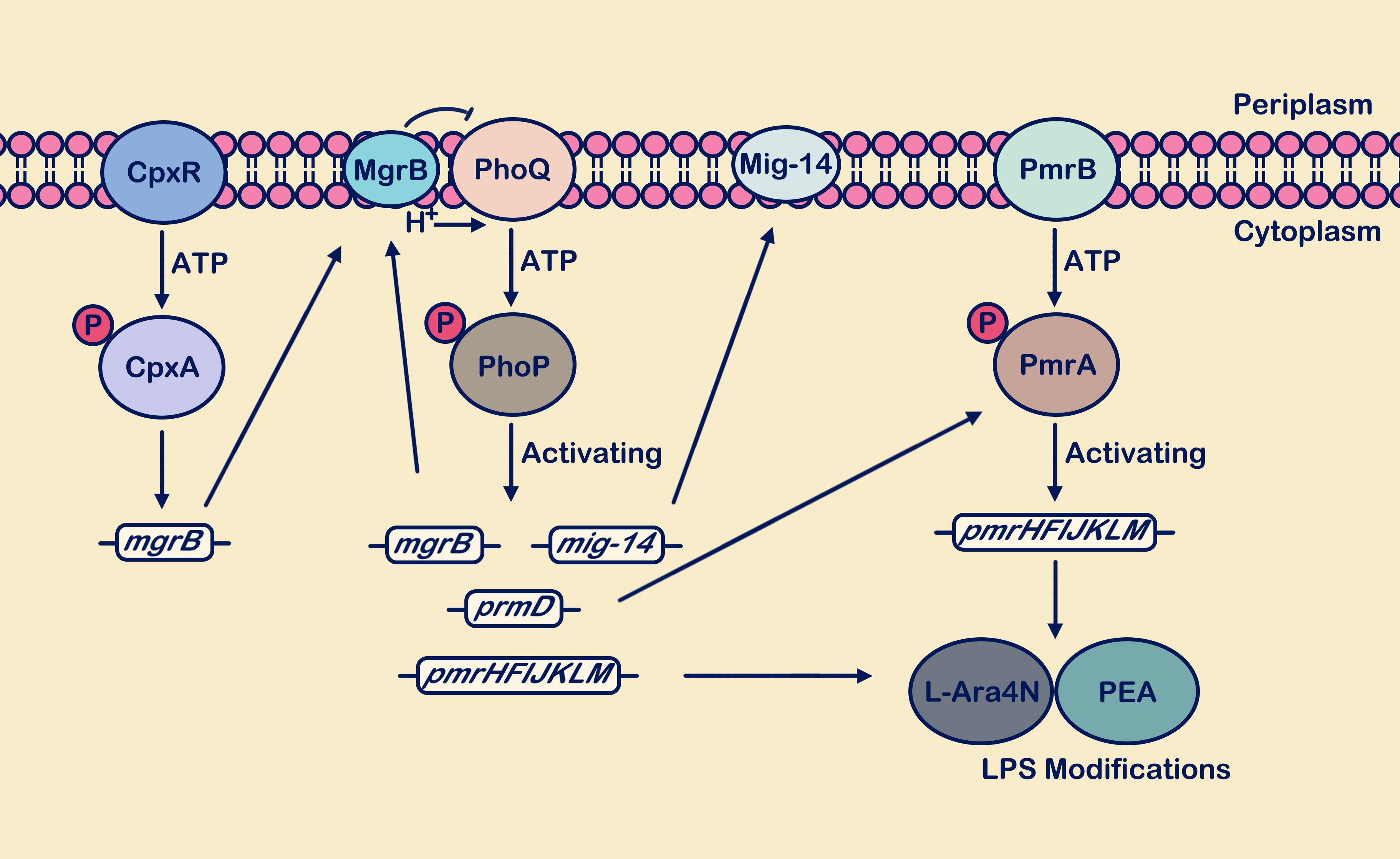
Figure 3. The PhoP/PhoQ and PmrA/PmrB systems synergistically respond to antimicrobial peptide attacks. Activation of the PhoP/PhoQ system promotes transcription of mgrB, mig-14, pmrD, and pmrHFIJKLM. The mgrB gene is upregulated, and the synthesized MgrB membrane protein exerts negative feedback on PhoQ. Upregulation of the mig-14 gene supports the synthesis of the inner membrane protein Mig-14. Activation of the pmrD gene positively regulates the PmrA/PmrB system. Activation of pmrHFIJKLM facilitates the synthesis of L-Ara4N and PEA, which are used for LPS site modification, enhancing bacterial resistance. In the figure legend, a circle with “P” denotes a phosphate group, and the arrow from MgrB to PhoQ signifies “inhibition”.
Typically, the regulation of its own and downstream target genes by the PhoP/PhoQ system to resist external stimuli is called positive feedback regulation. Negative feedback regulatory mechanisms collectively contribute in maintaining cellular homeostasis, and adverse feedback effects also play a crucial role in reducing intra-population cellular variability (Lippa et al., 2009). The membrane protein MgrB activation occurs through PhoP phosphorylation (Lippa et al., 2009). Subsequently, it binds to the periplasmic domain of PhoQ in order to attenuate its interaction with other stimulus signals, thereby inhibiting the phosphorylation of PhoP and establishing a negative feedback mechanism (Lippa et al., 2009; Poirel et al., 2017). The combination of positive and negative feedback in the PhoP/PhoQ system enhances bacterial sensitivity to signals and plays a crucial role in maintaining intracellular homeostasis (Lippa et al., 2009). When MgrB undergoes functional changes or is lost, the negative feedback regulation of PhoPQ is disrupted (Zafer et al., 2019; Kong et al., 2021). Although the absence of MgrB indirectly affects the activation of PmrD, MgrB appears to specifically target the PhoQ domain (Zafer et al., 2019). In PhoQ-deficient strains, MgrB does not exert its inhibitory effect and does not influence PmrD-mediated resistance to polymyxin B (Zafer et al., 2019). Recent studies have suggested that the CpxR/CpxA system may indirectly influence the antibiotic sensitivity of the PhoP/PhoQ and PmrA/PmrB systems by regulating the activity levels of MgrB (Wang et al., 2020).
pH regulates crucial biological processes such as genes expression, energy generation, and various enzyme functions. Many bacteria, including E. coli, S. enterica, P. aeruginosa, and Edwardsiella, have evolved distinct acid resistance mechanisms (Du et al., 2021; Mallick and Das, 2023). In addition to combating AMPs pressure within phagosomes, bacteria also face the challenge of phagosomal acidification that needs to be overcome (Di et al., 2017). The regulatory response to acid stress is achieved through the coordinated action of various regulators and regulatory systems (Krin et al., 2010). Two-component systems (TCS), such as PhoP/PhoQ, PmrA/PmrB, EvgS/EvgA, SsrA/SsrB, RstA/RstB, and CpxA/CpxR system consist of multiple components that enable bacteria to sense acidic environments and respond to acid stress (Perez and Groisman, 2007; Lin et al., 2017; Sen et al., 2017; Xu et al., 2020; Li and Yao, 2022; Wan et al., 2024). Deletion of phoPQ in E. coli leads to reduced expression levels of various acid-regulated proteins, highlighting the importance of PhoPQ under mildly acidic conditions. Multiple studies indicate that PhoPQ is an effective bacterial defense mechanism against phagosomal killing. PhoPQ directly regulates the lipid A deacylase PagL and the putative dehydrogenase/reductase (SDR) HlyF (Elhenawy et al., 2016; Martynowycz et al., 2019). The upregulation of their transcription levels mediates LPS modification and reshaping of lipid structures (formation of OMV) (Martynowycz et al., 2019). The periplasmic sensor PhoQ detects acidic stimuli and initiates a positive phosphorylation response, thereby activating the transcription of PhoP and its downstream acid resistance-related genes (Tu et al., 2006). Moreover, under acidic environmental conditions, PhoQ does not affect its response to other environmental stimuli (Prost et al., 2007; Choi and Groisman, 2017; Roggiani et al., 2017). For example, within macrophage phagosomes, the PhoP/PhoQ system in bacteria can simultaneously sense stimuli from cationic AMPs and mildly acidic environmental conditions. This capability reduces the damage caused by these stimuli to the outer membrane and maintains normal physiological functions of the bacteria (Han et al., 2023). PhoQ simultaneously sensing both signals rather than individually responding to one of them results in a significant increase in the abundance of PhoP and its downstream target genes (Prost et al., 2007). Under conditions of high or low concentrations of Mg2+, low pH can still be sensed by PhoQ (Prost et al., 2007). Be more specific about the concentration of Mg2+ and low pH. Research has reported that in S. enterica, low pH conditions lead to an increase in PhoP-P levels, which indirectly promotes transcription of pmrD (Perez and Groisman, 2007). This mediation enhances LPS modification effectiveness under acidic conditions, thereby strengthening bacterial resistance (Perez and Groisman, 2007). It can be seen that low pH can cooperate with other stimulating conditions to activate PhoQ, but currently, there is limited research on this aspect.
In S. enterica under weakly acidic conditions (pH 4.9), the UgtL protein is essential for activation of the PhoP/PhoQ system (Choi and Groisman, 2017). UgtL interacts with PhoQ, enhancing its autophosphorylation and increasing the intracellular abundance of phosphorylated PhoP (Choi and Groisman, 2017). This leads to the activation of downstream gene transcription by PhoP. However, under other stimulating conditions, the effect of UgtL on PhoQ is not significant (Choi and Groisman, 2017). Recent studies have shown that UgtS, a novel inner membrane protein homologous to UgtL, is upregulated at the transcriptional level by PhoP phosphorylation (Salvail et al., 2022). It acts as an antagonist to UgtL within macrophages of S. Typhimurium (Salvail et al., 2022). Following activation of the SsrB/SsrA system in response to weak acid conditions, further enhancement of ugtL gene expression can increase PhoP phosphorylation (Choi and Groisman, 2020a). Conversely, PhoP phosphorylation can also increase transcription of the ssrB gene (Choi and Groisman, 2020a). The PhoP/PhoQ and SsrB/SsrA systems play crucial regulatory roles in controlling genes within the (S. Typhimurium) pathogenicity island.
Additionally, the TCS EvgS/EvgA system is also a major player in acid resistance, activating the expression of numerous acid-resistant genes (Dan et al., 2021; Zeng et al., 2021). It primarily branches into two pathways: EvgSA-YdeO and EvgSA-SafA (Zeng et al., 2021). YdeO is a critical component of glutamate-dependent acid resistance AR2, whose transcriptional upregulation activates the expression of the gadE gene, mediating the upregulation of AR2 effector genes (gadABC) (Roggiani et al., 2017). The membrane protein SafA acts as a connector between the EvgS/EvgA system and the PhoP/PhoQ system (Schellhorn, 2020). SafA and UgtL are both short membrane proteins (65 and 132 residues, respectively) that interact with PhoQ to facilitate network regulation of PhoP/PhoQ (Choi and Groisman, 2017; Yoshitani et al., 2019). However, they lack sequence similarity between each other and independently exert their functions (Choi and Groisman, 2017; Yoshitani et al., 2019). Through binding with anti-adaptor proteins, RssB reduces its interaction with RpoS, thereby mediating the upregulation of RpoS expression levels and ultimately promoting the transcriptional upregulation of gadE (Xu et al., 2019). As mentioned above, the MgrB protein acts as a feedback inhibitor in the PhoP/PhoQ system. The expression of the mgrB gene may be associated with acid resistance, as its deletion can increase the transcription levels of iraM, thereby promoting the activation of the acid resistance gene gadE (Yuan et al., 2017; Xu et al., 2019). Figure 4 depicts a brief description of the PhoP/PhoQ system responding to acidic pH stimulation.
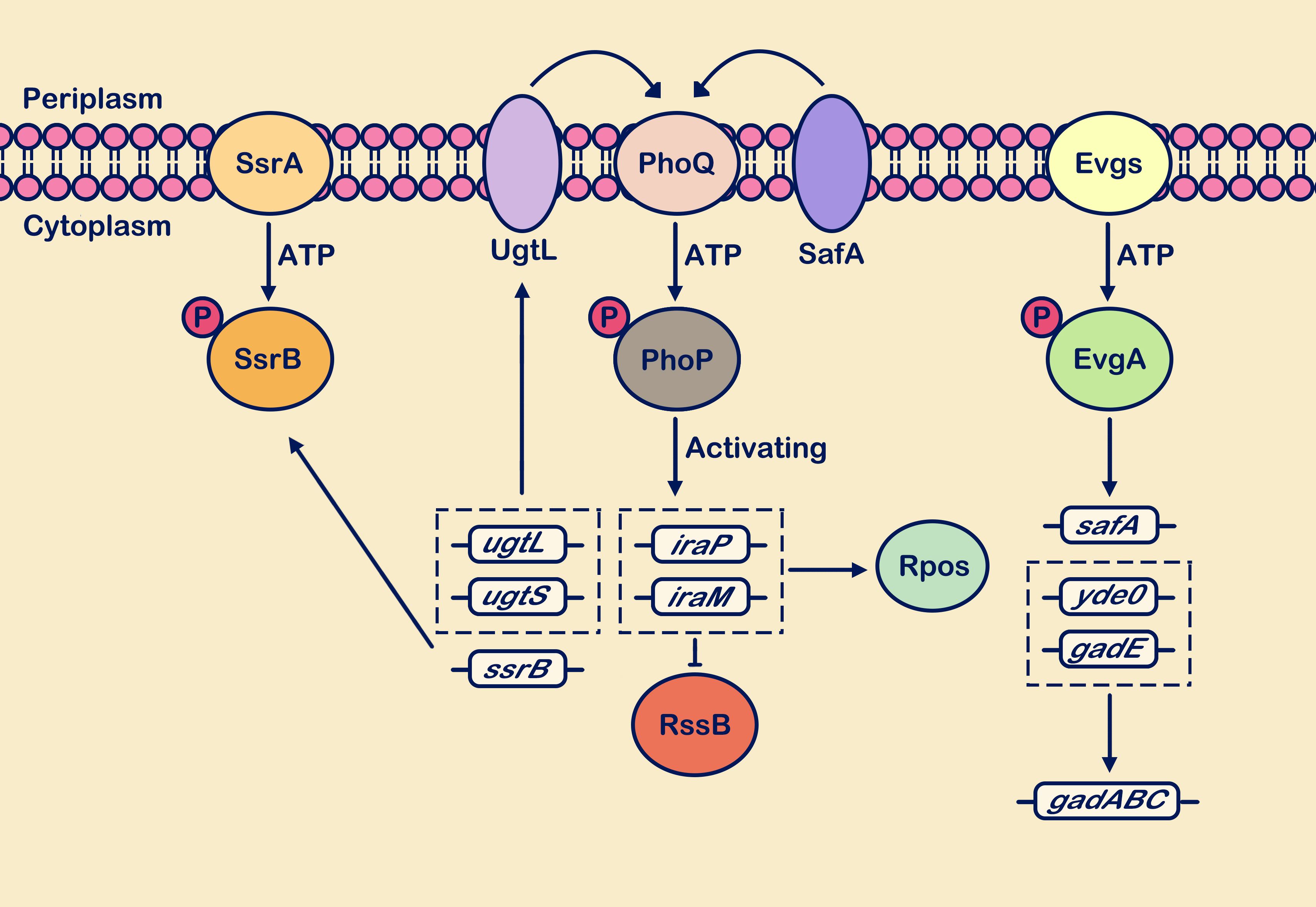
Figure 4. Weakly acidic pH activates multiple TCSs. Sensing the weakly acidic stimulus, the PhoP/PhoQ, EvgS/EvgA, and SsrA/SsrB systems respond positively. The PhoP/PhoQ system cascades to enhance transcription levels of genes including ugtL, ugtS, iraP, iraM, and ssrB. Upregulation of ssrB gene expression enhances the SsrA/SsrB system’s response to weakly acidic stimuli. Activation of ugtL and ugtS genes strengthens the interaction of the membrane protein UgtL with PhoQ. Upregulation of iraP and iraM gene transcription interferes with RssB protein degradation of RpoS. Within the EvgS/EvgA system cascade, transcriptional expression of acid resistance genes gadABC is promoted, along with upregulation of the safA gene. This gene interacts with PhoQ and positively regulates PhoQ. The circle with “P” represents a phosphate group.
Bacteria encounter oxidative stress responses induced by reactive oxygen species (ROS) during both natural environments and host infection (Hillion and Antelmann, 2015). Bacteria have evolved complex oxidative stress regulatory networks (Hillion and Antelmann, 2015). Before oxidative repair can occur, there is a need for oxidative redox sensors to transmit oxidative information directly or indirectly between them, thereby further regulating the expression of relevant proteins (Hillion and Antelmann, 2015). DsbA, a member of the Dsb family of oxidoreductases involved in disulfide bond formation, is commonly found in the bacterial inner membrane and periplasmic space (Eckels et al., 2021). Its transcriptional regulation depends on the PhoP/PhoQ system cascade, and it plays a crucial role in disulfide bond synthesis (Cardenal-Muñoz and Ramos-Morales, 2013; Choi and Groisman, 2016; Eckels et al., 2021). DsbA acts as a potent oxidase involved in disulfide bond formation, while DsbB re-oxidizes DsbA to form new disulfide bonds (Kadokura and Beckwith, 2014; Santos-Martin et al., 2021). The DsbA-DsbB pathway functions as a redox cycle, continuously driving proper folding and function of substrate proteins in the bacterial envelope and periplasmic space (Kadokura and Beckwith, 2014; Santos-Martin et al., 2021). Studies have shown that the transcription of dsbA and mgrB is regulated by PhoP-P (Cardenal-Muñoz and Ramos-Morales, 2013). In E. coli, the absence of dsbA directly leads to the activation of the PhoP/PhoQ system (Cardenal-Muñoz and Ramos-Morales, 2013). Meanwhile, MgrB, a membrane protein, has been shown to interact with PhoQ, exerting a dephosphorylating effect on PhoP and thereby inhibiting the activation of the PhoP/PhoQ system (Cardenal-Muñoz and Ramos-Morales, 2013). Furthermore, deletion of mgrB in dsbA-deficient strains has been found to reduce the activation of DsbA by the PhoP/PhoQ system (Cardenal-Muñoz and Ramos-Morales, 2013). However, there is currently limited research on the regulatory role of the PhoP/PhoQ system on the Dsb family of proteins.
Osmotic pressure is also among the environmental factors encountered during microbial growth (Brauer et al., 2023). High osmotic pressure caused by excessive or insufficient extracellular solutes that may have detrimental effects on bacteria (Sun et al., 2021; Brauer et al., 2023). The TCSs are important regulatory mechanisms in prokaryotic microbes for coping with osmotic stress. Components involved in osmotic regulation include OmpR/EnvZ, CpxA/CpxR, and the PhoP/PhoQ system (Yuan et al., 2017). The OmpR/EnvZ system can perceive stimuli of both low and high osmotic pressure across the outer membrane (Yuan et al., 2017). Meanwhile, the CpxAR system, responds to signals of outer membrane stress, whereas the PhoP/PhoQ system is specifically associated with high osmotic pressure (Yuan et al., 2017). During hyperosmotic stress (300 mM NaCl), cells experience water loss, growth stagnation, and an increase in the thickness of the lipid bilayer (Poolman et al., 2002). When PhoQ senses high osmolarity, there is a reorganization of lipid bilayers and transmembrane domain conformations, promoting the accumulation of osmoregulatory proteins through PhoP-P (Yuan et al., 2017). In E. coli, mutants lacking PhoPQ show decreased sensitivity to high osmolarity (Xu et al., 2019). PhoP-P mediates the activation of the iraM gene, whose increased expression prevents the binding of RssB to RpoS, thereby further enhancing PhoP/PhoQ activation (Xu et al., 2019). This process regulates the balance of intracellular osmotic pressure in bacteria (Xu et al., 2019). In E. coli, through individual knockout studies of phoQ, phoP, envZ, and ompR, it was found that the PhoP/PhoQ and OmpR/EnvZ systems independently perceive and respond to osmotic pressure stimuli (Xu et al., 2019).
Exogenous long-chain unsaturated fatty acids (LCUFAs) are transported across the bacterial outer membrane and converted into acyl-CoA derivatives, which serve as substrates for β-oxidation or membrane phospholipid synthesis (Viarengo et al., 2013; Xia et al., 2024). LCUFAs inhibit the activity of the PhoP/PhoQ system by interacting with the PhoQ periplasmic sensor, disrupting its autophosphorylation activity, and subsequently downregulating the expression of PhoP-P and its downstream target genes (Viarengo et al., 2013). However, previous studies have shown that LCUFAs do not compete for binding sites with other stimuli (Viarengo et al., 2013; Carabajal et al., 2020). In S. Typhimurium, the PhoP/PhoQ system is inhibited in response to LCUFAs stimulation. LCUFAs may bind to Ca2+, aiding in the distinction between intracellular and extracellular environmental conditions (Viarengo et al., 2013). Furthermore, as signaling molecules, LCUFAs play a regulatory role in coordinating bacterial virulence expression (Xia et al., 2024). For instance, in S. enterica, their presence can interact with the transcription regulators HilC/HilD, leading to the expression of the type III secretion system (Xia et al., 2024).
Lysine acetylation is a typical post-translational modification in bacteria that can regulate various cellular functions (Weinert et al., 2013). Acetylation utilizes acetyl coenzyme A as a cofactor, transferring acetyl groups via acetyltransferases (Weinert et al., 2013). During aerobic microbial growth, acetate is secreted as part of metabolic processes. Acetate can be converted into acetyl coenzyme A, mediating the occurrence of PhoP acetylation During aerobic microbial growth, acetate is secreted as part of metabolic processes. Acetate can be converted into acetyl coenzyme A, mediating the occurrence of PhoP acetylation (Ren et al., 2019). Research indicates that acetylation plays a crucial role in modulating PhoP activity, regulating changes in bacterial virulence (Ren et al., 2016). In S. Typhimurium, PhoP undergoes acetylation at three lysine residues (K201, K88, and K102), which inhibits the binding of PhoP-P to downstream gene promoters (Ren et al., 2016). PhoP K201 undergoes acetylation and deacetylation mediated by Pat and CobB, while PhoP K88 and PhoP K102 are acetylated by non-enzymatic acetyl phosphate (AcP) modification (Ren et al., 2019; Li et al., 2021). Acetylation of PhoP inhibits its phosphorylation (Ren et al., 2019), resulting in a 2- to 5-fold reduction in transcriptional activation of PhoP-regulated genes (Ren et al., 2019).
The two-component systems regulate the activity of their sensors, response regulators, and subsequent proteins through feedback mechanisms to maintain the stability of the bacterial internal environment (Chen et al., 2021; Pina et al., 2021). The PhoP/PhoQ system, in response to various environmental signals, is also influenced by components of other two-componentsystems (Chen et al., 2021; Pina et al., 2021). It interacts with the PmrA/PmrB, EvgS/EvgA, RstA/RstB, SsrB/SsrA, and CpxR/CpxAsystems (Wang et al., 2020; Pina et al., 2021). They are interconnected through intermediate connectors (such as PmrD, SafA, MgrB), forming a complex regulatory network (Yoshitani et al., 2019; Yadavalli et al., 2020; Chen et al., 2021). PmrD, known as a connector protein, is a small regulatory RNA that acts as a connector and is activated by the PhoP phosphorylation mechanism (Zafer et al., 2019; Chen et al., 2021). It mediates the activation pathway of PhoP-PmrD-PmrA (Zafer et al., 2019; Chen et al., 2021). In many members of the Enterobacteriaceae family, the regulation of polymyxin resistance is primarily governed by two two-component systems: PmrA/PmrB and PhoP/PhoQ (Chen et al., 2021). These systems modulate the modification of bacterial outer membrane LPS through intricate signal transduction networks, thereby influencing bacterial resistance to polymyxins (Chen et al., 2021). Simultaneously, under conditions of magnesium deficiency, low pH environment, or strong stimulation of PhoQ, the PmrD protein also functions as a connector (Kox et al., 2000; Luo et al., 2010). Therefore, the PmrD protein plays a crucial role in the two-component signal transduction process by facilitating important information transfer. Moreover, the PhoP/PhoQ system can also act as an inhibitor of iron uptake proteins, synergizing with the PmrA/PmrB system to mount an immune response against high Fe3+ (Cho et al., 2006).
As stated above, SafA serves as a connector between the EvgS/EvgA and PhoP/PhoQ systems (Yoshitani et al., 2019). When E. coli is in a weakly acidic environment, it regulates acid resistance gene networks through the EvgS/EvgA and PhoP/PhoQ systems (Yoshitani et al., 2019). The sensor kinase EvgS detects low pH signals and activates the response regulator EvgA, subsequently initiating a cascade of gene transcription (Yamanaka et al., 2013). This pathway primarily bifurcates into two branches: one involving EvgA-YdeO-GadE, where YdeO activates GadE, leading to the regulation of various decarboxylases and providing resistance to acid stress (Yamanaka et al., 2013); the other branch includes SafA-PhoPQ-IraM-RpoS, with the membrane protein SafA acting as a connector, interacting with PhoQ to initiate a phosphorylation cascade. PhoP activates IraM to promote an increase in RpoS levels (Yamanaka et al., 2013). RpoS serves as a central regulator in response to external stresses, and its regulation of the gadE gene is a key strategy for combating weakly acidic environments (Chattopadhyay et al., 2015). Research has shown that E. coli significantly upregulates the expression levels of gadA, gadB, and gadE genes when exposed to low pH (pH 6) values (Hao et al., 2004).
Similar to SafA, UgtL is a membrane protein essential for PhoQ-mediated weakly acidic environmental signals, acting between the PhoP/PhoQ and SsrB/SsrA systems (Choi and Groisman, 2017; Choi and Groisman, 2020a). Under low pH conditions, UgtL interacts with the periplasmic domain of PhoQ, promoting the transcriptional levels of phosphorylated PhoP (Janssens et al., 2024). Research indicates that PhoP is a key regulator of the S. Typhimurium. Pathogenicity Island 2 (SPI-2) gene cluster, facilitating the cascade response of the SsrB/SsrA system (Shetty and Kenney, 2023; Janssens et al., 2024). Simultaneously, SsrB can also enhance the transcriptional expression of phoP and ugtL, thereby augmenting the network regulatory function of the PhoP/PhoQ system (Choi and Groisman, 2020a; Janssens et al., 2024). However, under low Mg2+ conditions (10 μM Mg2+), the expression of UgtL does not significantly change despite activation signals for PhoQ (Choi and Groisman, 2017; Janssens et al., 2024).
Meanwhile, there is cross-regulation of environmental stress between RstA/RstB and PhoP/PhoQ (Tran et al., 2016). Upon activation of PhoQ by low Mg2+ concentration (10 μM Mg2+) and low pH (the pH range is 5.0 to 6.5) signals, PhoP-P binds to the rstA promoter region, activating rstA gene transcription and influencing the cascade response of the RstA/RstB system (Tran et al., 2016). The RstA/RstB system specifically regulates purine metabolism, iron acquisition, biofilm formation, and tolerance to acidic environments (Tran et al., 2016). PhoP/PhoQ controls the function of RstA and mediates the transcriptional level regulation of acid-resistant genes (i.e. asr gene), curli-regulatory gene (i.e. csgD gene), and iron transport genes (i.e. feoB gene) (Ogasawara et al., 2007; Jeon et al., 2008; Tran et al., 2016). Environmental conditions influence the degree of cross-regulation between PhoQ/PhoP and other regulatory systems. Overall, the PhoP/PhoQ system does not solely respond to specific stimuli but is intricately interconnected with other TCS systems and regulatory networks.
When activated, the PhoP/PhoQ system enables various bacteria to tolerate stresses such as low Mg2+ (10-50 μM Mg2 +), antimicrobial peptides, Mildly acidic pH (the pH range is 5.0 to 6.5), and high osmolarity. Multiple studies in the research process have shown that the PhoP/PhoQ cascade plays a crucial role in regulating virulence in various pathogenic bacteria, including Salmonella, E. coli, Shigella, Yersinia, and P. aeruginosa (Lin et al., 2017; Martynowycz et al., 2019; Fukuto et al., 2020; Xu et al., 2020; Cabezudo et al., 2022; Zhang et al., 2022). Deletion of the phoP or phoQ genes significantly reduces the virulence of these pathogens (Lin et al., 2017; Martynowycz et al., 2019; Fukuto et al., 2020; Xu et al., 2020; Cabezudo et al., 2022; Zhang et al., 2022). In Shigella strains with PhoPQ deletion, a reduced ability to withstand environmental stresses was observed, with the key virulence factor icsA being regulated by the PhoP/PhoQ system (Lin et al., 2017). SPI-1 and SPI-2 encode two type III secretion systems (T3SS), which are crucial for the pathogenicity of S. enterica (Jennings et al., 2017). PhoP/PhoQ mediates virulence by activating downstream target genes that modulate the expression of SPI-1 and SPI-2 (Lou et al., 2019). HilA acts as a positive regulator controlling the expression of SPI-1 genes, coordinated by the combined action of three AraC-like transcriptional activators: HilC, HilD, and RtsA (Lou et al., 2019). Studies have shown that S. Typhimurium lacking the hilA gene exhibit a phenotype equivalent to SPI-1 functionality deficiency (Lou et al., 2019). HilE is the most critical negative regulator of the hilA expression (Lou et al., 2019). Under conditions of low Mg2+ concentration (low magnesium was at 8 μM), PhoP binds to the hilE promoter, increasing hilE gene expression, which mediates inhibition of hilA gene expression and indirectly affects transcription of hilD and rtsA genes (Lou et al., 2019). These transcriptional changes in these genes highlight the significant role of PhoP in SPI-1 (Bijlsma and Groisman, 2005; Pérez-Morales et al., 2017). As mentioned earlier, the PhoP/PhoQ system activates and enhances the kinase activity of SsrB, concurrently boosting the transcriptional levels of its downstream gene cluster SpiCBA (Bijlsma and Groisman, 2005). Additionally, PhoPQ induces two small RNAs: MgrR and PinT (Westermann et al., 2016; Kim et al., 2019; Yeom and Groisman, 2021). The former responds to low Mg2+ levels by upregulating expression to influence Mg2+ homeostasis (Yeom and Groisman, 2021). The latter, under mildly acidic conditions, mediates the expression of SPI-1 and SPI-2 genes by regulating the transcription levels of hilA and rtsA (Kim et al., 2019). The mgtC gene plays a crucial role in pathogen virulence, and its transcription levels are upregulated during activation of the PhoP/PhoQ system, surpassing the expression levels of the virulence factor CigR (Yeom et al., 2018). MgtC inhibits ATP synthesis by suppressing the F1Fo ATP synthase, thereby reducing transcription of ribosomal RNA and simultaneously protecting PhoP from degradation (Yeom et al., 2017; Yeom et al., 2018). Previous studies have shown that the S. Typhimurium genes cigR and mgtC are located within SPI-3 and are part of the same transcriptional unit under MgtC-inducing conditions (Yeom et al., 2018). Research has shown that in S. Typhimurium, when exposed to low Mg2+ (10 μM Mg2+), the PhoP/PhoQ system indirectly regulates the expression of the pagM gene by affecting the transcription levels of mgtA and mgtC (Park et al., 2015). The PagM secreted protein, in turn, mediates a flagella-independent mode of motility (Park et al., 2015). This process helps the bacteria adapt to low Mg2+ environmental conditions by altering their mode of movement.
The H-NS nucleoid protein is a common negative regulatory protein that readily binds to AT-rich sequences, leading to silencing of associated genes (Choi and Groisman, 2020b). In S. Typhimurium, the SPI gene clusters exhibit higher AT content in their promoter sequences compared to ancestral genes, enhancing the pronounced negative regulatory role of H-NS, which plays a crucial role in virulence expression (Hu et al., 2019). Upon activation of the PhoP/PhoQ system, the transcription levels of downstream target genes, ssrB and slyB are upregulated. PhoP interacts with SsrB and SlyB to counteract H-NS-mediated silencing (Choi and Groisman, 2020a). Under weakly acidic conditions, the abundance of H-NS is significantly lower compared to neutral pH states (Krin et al., 2010; Choi and Groisman, 2020b). This suggests that activation of the PhoP/PhoQ system plays a crucial regulatory role in relieving H-NS-mediated gene silencing mechanisms. Previous studies have found that the PhoP/PhoQ system negatively regulates bacterial flagella (Adams et al., 2001; Janssens et al., 2024). When acid-adapted Salmonella (pH 5.0) is exposed to pH 3.0 conditions, the transcription level of the fliC gene is significantly downregulated, inhibiting flagella expression (Adams et al., 2001). This may help Salmonella avoid excessive activation of the host immune system (Adams et al., 2001). Overall, the PhoP/PhoQ system influences bacterial virulence systems directly or indirectly, adjusting the expression of relevant genes under different stimulus signals to maintain bacterial internal environmental stability.
Bacteria perceive different ecological niches within the host to evade attacks from the host immune system by regulating the expression levels of relevant genes. The PhoP/PhoQ system is the most extensively studied TCS to date, and it is highly conserved across both pathogenic and non-pathogenic bacteria. The PhoP/PhoQ system senses external environmental stimuli through the dual-function membrane protein PhoQ, which, upon phosphorylation, transfers phosphate groups to the response regulator, PhoP. PhoP then regulates the abundance of downstream target genes in response to external environmental signals until the components return to stable levels upon restoration of bacterial physiological balance (Xu et al., 2019). The gene products obtained at different levels during the cascade reaction of the PhoP/PhoQ system integrate into the regulatory circuit, influencing changes in closely associated regulatory proteins and phenotype modifications (Choi and Groisman, 2020a). As mentioned earlier, the cascade reaction of PhoP/PhoQ reduces the modification of LPS, decreasing the overall negative charge of the bacterial membrane. This enhances bacterial tolerance to extreme environments, including increased resistance to antibiotics, stabilizing cytoplasmic pH, and releasing Mg2+ ions, among other effects. The interaction of the PhoP/PhoQ system with other TCS systems forms a complex regulatory network, collectively controlling bacterial cellular activities and virulence. Theoretically, this strategy establishes resilience and infection capabilities that can harm host cells without negative effects on the bacteria.
In summary, the PhoP/PhoQ system regulates the physiological, biochemical, antibiotic resistance, and virulence characteristics of bacteria across various environments. Moreover, it exhibits intricate synergistic interactions with other components of the TCS regulatory network. Although the PhoP/PhoQ system has received considerable attention in the past, research on its signal transduction mechanisms has primarily focused on enteric pathogens, with studies in other bacteria being relatively scarce. Studying the specific mechanisms of action of the PhoP/PhoQ system in other pathogenic or non-pathogenic bacteria, as well as its interactions with other regulatory networks, contributes to the development of effective antimicrobial therapies and mitigates the negative impacts of antibiotic use.
MM: Conceptualization, Writing – original draft. LH: Conceptualization, Writing – original draft. QY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China under contract No. 32373181, Science and Technology Plan Project of Fujian Province under contract No. 2022L3059.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adams, P., Fowler, R., Kinsella, N., Howell, G., Farris, M., Coote, P., et al. (2001). Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1, 597–607. doi: 10.1002/1615-9861(200104)1:4<597::AID-PROT597>3.0.CO;2-P
Ali, L., Abdel Aziz, M. H. (2024). Crosstalk involving two-component systems in Staphylococcus aureus signaling networks. J. Bacteriol. 206, e00418–e00423. doi: 10.1128/jb.00418-23
Bader, M. W., Navarre, W. W., Shiau, W., et al. (2010). Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50, 219–230. doi: 10.1046/j.1365-2958.2003.03675.x
Bader, M. W., Sanowar, S., Daley, M. E., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. doi: 10.1016/j.cell.2005.05.030
Barchiesi, J., Castelli, M. E., Soncini, F. C., et al. (2008). mgtA Expression is induced by rob overexpression and mediates a Salmonella enterica resistance phenotype. J. Bacteriol. 190, 4951–4958. doi: 10.1128/JB.00195-08
Battesti, A., Gottesman, S. (2013). Roles of adaptor proteins in regulation of bacterial proteolysis. Curr. Opin. Microbiol. 16, 140–147. doi: 10.1016/j.mib.2013.01.002
Bijlsma, J. J. E., Groisman, E. A. (2005). The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57, 85–96. doi: 10.1111/j.1365-2958.2005.04668.x
Bolard, A., Schniederjans, M., Haüssler, S., Triponney, P., Valot, B., Plésiat, P., et al. (2019). Production of Norspermidine Contributes to Aminoglycoside Resistance in pmrAB Mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, 10. doi: 10.1128/aac.01044-19
Bougdour, A., Cunning, C., Baptiste, P. J., Elliott, T., Gottesman, S. S. (2008). Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68, 298–313. doi: 10.1111/j.1365-2958.2008.06146.x
Brauer, A. M., Shi, H., Levin, P. A., Huang, K. C. (2023). Physiological and regulatory convergence between osmotic and nutrient stress responses in microbes. Curr. Opin. Cell Biol. 81, 102170. doi: 10.1016/j.ceb.2023.102170
Brodsky, I. E., Ghori, N., Falkow, S., Monack, D. (2005). Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol. Microbiol. 55, 954–972. doi: 10.1111/j.1365-2958.2004.04444.x
Cabezudo, I., Lobertti, C. A., Véscovi, E. G., Furlan, R. L. E. (2022). Effect-directed synthesis of PhoP/PhoQ inhibitors to regulate Salmonella virulence. J. Agric. Food Chem. 70, 6755–6763. doi: 10.1021/acs.jafc.2c01087
Carabajal, M. A., Viarengo, G., Yim, L., Martínez-Sanguiné, A., Mariscotti, J. F., Chabalgoity, J. A., et al. (2020). PhoQ is an unsaturated fatty acid receptor that fine-tunes Salmonella pathogenic traits. Sci. Signaling 13, eaaz3334. doi: 10.1126/scisignal.aaz3334
Cardenal-Muñoz, E., Ramos-Morales, F. (2013). DsbA and MgrB regulate steA expression through the two-component system PhoQ/PhoP in Salmonella enterica. J. Bacteriol. 195, 2368–2378. doi: 10.1128/jb.00110-13
Chandler, C. E., Harberts, E. M., Pelletier, M. R., Thaipisuttikul, I., Jones, J. W., Hajjar, A. M., et al. (2020). Early evolutionary loss of the lipid A modifying enzyme PagP resulting in innate immune evasion in Yersinia pestis. Proc. Natl. Acad. Sci. 117, 22984–22991. doi: 10.1073/pnas.1917504117
Chattopadhyay, M. K., Keembiyehetty, C. N., Chen, W., Tabor, H. (2015). Polyamines stimulate the level of the σ38 subunit (RpoS) of Escherichia coli RNA polymerase, resulting in the induction of the glutamate decarboxylase-dependent acid response system via the gadE regulon. J. Biol. Chem. 290, 17809–17821. doi: 10.1074/jbc.M115.655688
Chen, A. I., Albicoro, F. J., Zhu, J., Goulian, M. (2021). Effects of Regulatory Network Organization and Environment on PmrD Connector Activity and Polymyxin Resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob. Agents Chemother. 3, 65. doi: 10.1128/AAC.00889-20
Chen, H. D., Groisman, E. A. (2013). The biology of the pmrA/pmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu. Rev. Microbiol. 67, 83–112. doi: 10.1146/annurev-micro-092412-155751
Cho, U. S., Bader, M. W., Amaya, M. F., Daley, M. E., Klevit, R. E., Miller, S. I., et al. (2006). Metal bridges between the phoQ sensor domain and the membrane regulate transmembrane signaling. J. Mol. Biol. 356, 1193–1206. doi: 10.1016/j.jmb.2005.12.032
Choi, J., Groisman, E. A. (2016). Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Mol. Microbiol. 101, 1024–1038. doi: 10.1111/mmi.13439
Choi, J., Groisman, E. A. (2017). Activation of master virulence regulator PhoP in acidic pH requires the Salmonella-specific protein UgtL. Sci. Signaling 10, eaan6284. doi: 10.1126/scisignal.aan6284
Choi, J., Groisman, E. A. (2020a). Horizontally acquired regulatory gene activates ancestral regulatory system to promote Salmonella virulence. Nucleic Acids Res. 48, 10832–10847. doi: 10.1093/nar/gkaa813
Choi, J., Groisman, E. A. (2020b). Salmonella expresses foreign genes during infection by degrading their silencer. Proc. Natl. Acad. Sci. 117, 8074–8082. doi: 10.1073/pnas.1912808117
Cromie, M. J., Groisman, E. A. (2010). Promoter and riboswitch control of the mg2+ Transporter mgtA from salmonella enterica. J. Bacteriol. 192, 604–607. doi: 10.1128/JB.01239-09
Dan, G., Han, X., Xiaohui, Y., Yu, J. Y., Xu, X. M., Huang, Y., et al. (2021). Genome-wide identification of genes involved in acid stress resistance of salmonella derby. Genes 12, 476–476. doi: 10.3390/genes12040476
Di, A., Kiya, T., Gong, H., Gao, X. P., Malik, A. B. (2017). Role of the phagosomal redox-sensitive TRP channel TRPM2 in regulating bactericidal activity of macrophages. J. Cell Sci. 130, 735–744. doi: 10.1242/jcs.196014
Du, C., Huo, X., Gu, H., Wu, D. M., Hu, Y. H. (2021). Acid resistance system CadBA is implicated in acid tolerance and biofilm formation and is identified as a new virulence factor of Edwardsiella tarda. Vet. Res. 52, 117. doi: 10.1186/s13567-021-00987-x
Eckels, E. C., Chaudhuri, D., Chakraborty, S., Echelman, D. J., Haldar, S. (2021). DsbA is a redox-switchable mechanical chaperone. Chem. Sci. 12, 11109–11120. doi: 10.1039/D1SC03048E
Elhenawy, W., Bording-Jorgensen, M., Valguarnera, E., Haurat, M. F., Wine, E., Feldman, M. F., et al. (2016). LPS remodeling triggers formation of outer membrane vesicles in Salmonella. MBio 7, 10. doi: 10.1128/mbio.00940-16
Fukuto, H. S., Viboud, G. I., Vadyvaloo, V. (2020). The diverse roles of the global transcriptional regulator PhoP in the lifecycle of Yersinia pestis. Pathogens 9, 1039. doi: 10.3390/pathogens9121039
Gahlot, K. D., Patkowski, B. J., Santaella, D. F. J., Allsopp, L. P., Pan, Z. Q., Filloux, A., et al. (2024). Cpx-signalling in Yersinia pseudotuberculosis modulates Lipid-A remodelling and resistance to last-resort antimicrobials. NPJ Antimicrob. Resist. 2, 39. doi: 10.1038/s44259-024-00059-y
Gall, A. R., Datsenko, K. A., Figueroa-Bossi, N., Bossi, L., Masuda, I., Hou, Y. M., et al. (2016). Mg2+ regulates transcription of mgtA in Salmonella Typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. 113, 15096–15101. doi: 10.1073/pnas.1612268113
Goldberg, S. D., Clinthorne, G. D., Goulian, M., Goulian, M., DeGrado, W. F. (2010). Transmembrane polar interactions are required for signaling in the Escherichia coli sensor kinase PhoQ. Proc. Natl. Acad. Sci. 107, 8141–8146. doi: 10.1073/pnas.1003166107
Groisman, E. A. (2016). Feedback control of two-component regulatory systems. Annu. Rev. Microbiol. 70, 103–124. doi: 10.1146/annurev-micro-102215-095331
Groisman, E. A., Duprey, A., Choi, J. (2021). How the phoP/phoQ system controls virulence and mg2+ Homeostasis: lessons in signal transduction, pathogenesis, physiology, and evolution. Microbiol. Mol. Biol. Rev. 85, 10. doi: 10.1128/MMBR.00176-20
Guo, H., Zhao, T., Huang, C., Chen, J. Y. (2022). The role of the two-component system phoP/phoQ in intrinsic resistance of yersinia enterocolitica to polymyxin. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.758571
Han, J., Gao, X., Luo, X., Zhu, L. X., Zhang, Y. M., Dong, P. C. (2023). The role of PhoP/PhoQ system in regulating stress adaptation response in Escherichia coli O157: H7. Food Microbiol. 112, 104244. doi: 10.1016/j.fm.2023.104244
Hao, Y. S., Wei, W., Qi, J. H., Juan, T. S., Qi, L., Guo, X. J., et al. (2004). Preparation and characterization of the monoclonal antibodies against enterohemahagic E.coli O157:H7. Chin. J. Zoon.
He, L., Mao, M. Q., Zhao, L. M., Li, Q., Ge, H., Zhang, J. N., et al. (2025). sRNA113 regulates the motility of Pseudomonas plecoglossicida to affect the immune response against infection in pearl gentian grouper. Zool. Res. 46, 1–10. doi: 10.24272/j.issn.2095-8137.2024.333
Hillion, M., Antelmann, H. (2015). Thiol-based redox switches in prokaryotes. Biol. Chem. 396, 415–444. doi: 10.1515/hsz-2015-0102
Hiroshi, N. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. MMBR 67, 593–656. doi: 10.1128/mmbr.67.4.593-656.2003
Hu, L., Kong, W., Yang, D., Han, Q. Q., Guo, L., Shi, Y. X. (2019). Threonine phosphorylation fine-tunes the regulatory activity of histone-like nucleoid structuring protein in Salmonella transcription. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01515
Hu, L. Z., Zhang, W. P., Zhou, M. T., Han, Q. Q., Gao, X. L., Zeng, H. L., et al. (2016). Analysis of Salmonella PhoP/PhoQ regulation by dimethyl-SRM-based quantitative proteomics. Biochim. Biophys. Acta (BBA)-Proteins Proteomics 1864, 20–28. doi: 10.1016/j.bbapap.2015.10.003
Janssens, A., Nguyen, V. S., Cecil, A. J., Verren, S. E. V. D., Timmerman, E., Deghelt, M., et al. (2024). SlyB encapsulates outer membrane proteins in stress-induced lipid nanodomains. Nature 7999, 626. doi: 10.1038/s41586-023-06925-5
Jennings, E., Thurston, T. L. M., Holden, D. W. (2017). Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231. doi: 10.1016/j.chom.2017.07.009
Jeon, J., Kim, H., Yun, J., Ryu, S., Groisman, E. A., Shin, D. (2008). RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J. Bacteriol. 190, 7326–7334. doi: 10.1128/jb.00903-08
Kadokura, H., Beckwith, J. (2014). Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 21, 2354–2363. doi: 10.1093/emboj/21.10.2354
Kato, A., Chen, H. D., Latifi, T., Groisman, E. A. (2012). Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol. Cell 47, 897–908. doi: 10.1016/j.molcel.2012.07.017
Kato, A., Latifi, T., Groisman, E. A. (2003). Closing the loop: The PmrA/PmrB two-component system negatively controls expression of its posttranscriptional activator PmrD. Proc. Natl. Acad. Sci. United States America 100, 4706–4711. doi: 10.1073/pnas.0836837100
Kim, K., Palmer, A. D., Vanderpool, K., Slauch, J. M. (2019). The small RNA PinT contributes to PhoP-mediated regulation of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 201, 201. doi: 10.1128/jb.00312-19
Kong, Y., Li, C., Chen, H., Zheng, W., Sun, Q. Y., Xie, X. Y., et al. (2021). In vivo Emergence of Colistin Resistance in Carbapenem-Resistant Klebsiella pneumoniae Mediated by Premature Termination of the mgrB Gene Regulator. Front. Microbiol. 12). doi: 10.3389/fmicb.2021.656610
Kox, L. F. F., Wösten, M. M. S. M., Groisman, E. A. (2000). A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. doi: 10.1093/emboj/19.8.1861
Krin, E., Danchin, A., Soutourina, O. (2010). Decrypting the H-NS-dependent regulatory cascade of acid stress resistance in Escherichia coli. BMC Microbiol. 10, 1–9. doi: 10.1186/1471-2180-10-273
Li, J., Liu, S., Su, Y., Ren, J., Sang, Y., Ni, J. J., et al. (2021). Acetylation of PhoP K88 is involved in regulating Salmonella virulence. Infect. Immun. 89, 10. doi: 10.1128/iai.00588-20
Li, G., Yao, Y. (2022). TorR/TorS two-component system resists extreme acid environment by regulating the key response factor RpoS in Escherichia coli. Gene 821, 146295. doi: 10.1016/j.gene.2022.146295
Li, X., Zuo, S., Wang, B., Wang, Y. (2022). Antimicrobial mechanisms and clinical application prospects of antimicrobial peptides. Molecules 27, 2675. doi: 10.3390/molecules27092675
Lin, Z. W., Cai, X., Chen, M. L., Ye, L., Wu, Y., Wang, X. F., et al. (2017). Virulence and stress responses of shigella flexneri regulated by phoP/phoQ. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02689
Lippa, A. M., Goulian, M. (2012). Perturbation of the oxidizing environment of the periplasm stimulates the phoQ/phoP system in escherichia coli. J. Bacteriol. 194, 1457–1463. doi: 10.1128/JB.06055-11
Lippa, A. M., Goulian, M., Burkholder, W. F. (2009). Feedback inhibition in the phoQ/phoP signaling system by a membrane peptide. PloS Genet. 5, e1000788. doi: 10.1371/journal.pgen.1000788
Liu, J., Wu, P., Wang, F., Niu, W. C., Ahmed, Z., Chen, M. Q., et al. (2021). Differential regulation and the underlying mechanisms of clay minerals to Escherichia coli under the stress of polymyxin B: Comparing halloysite with kaolinite. Chemosphere 265, 129095. doi: 10.1016/j.chemosphere.2020.129095
Lou, L., Zhang, P., Piao, R., Wang, Y. (2019). Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00270
Luo, S. C., Lou, Y. C., Cheng, H. Y., Pan, Y. R., Peng, H. L., Chen, C. P. (2010). Solution structure and phospho-PmrA recognition mode of PmrD from Klebsiella pneumoniae. J. Struct. Biol. 172, 319–330. doi: 10.1016/j.jsb.2010.06.007
Mallick, S., Das, S. (2023). Acid-tolerant bacteria and prospects in industrial and environmental applications. Appl. Microbiol. Biotechnol. 107, 3355–3374. doi: 10.1007/s00253-023-12529-w
Martynowycz, M. W., Rice, A., Andreev, K., Nobre, T. M., Kuzmenko, I., Wereszczynski, J., et al. (2019). Salmonella membrane structural remodeling increases resistance to antimicrobial peptide LL-37. ACS Infect. Dis. 5, 1214–1222. doi: 10.1021/acsinfecdis.9b00066
Mattos-Graner, R. O., Duncan, M. J. (2017). Two-component signal transduction systems in oral bacteria. J. Oral. Microbiol. 9, 1400858. doi: 10.1080/20002297.2017.1400858
Mensa, B., Polizzi, N. F., Molnar, K. S., Natale, A. M., Lemmin, T., DeGrado, W. F. (2021). Allosteric mechanism of signal transduction in the two-component system histidine kinase PhoQ. ELife. doi: 10.1101/2021.09.03.458835
Ogasawara, H., Hasegawa, A., Kanda, E., Miki, T., Yamamoto, K., Ishihama, A. (2007). Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189, 4791–4799. doi: 10.1128/jb.00319-07
Paredes, A., Iheacho, C., Smith, A. T. (2023). Metal messengers: communication in the bacterial world through transition-metal-sensing two-component systems. Biochemistry 62, 2339–2357. doi: 10.1021/acs.biochem.3c00296
Park, M., Nam, D., Kweon, D. H., Shin, D. W. (2018). ATP reduction by MgtC and Mg2+ homeostasis by MgtA and MgtB enables Salmonella to accumulate RpoS upon low cytoplasmic Mg2+ stress. Mol. Microbiol. 110, 283–295. doi: 10.1111/mmi.14105
Park, S. Y., Pontes, M. H., Groisman, E. A. (2015). Flagella-independent surface motility in Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. United States America 112. doi: 10.1073/pnas.1422938112
Pathak, A., Goyal, R., Sinha, A., Sarkar, D. (2010). Domain structure of virulence-associated response regulator PhoP of Mycobacterium tuberculosis: role of the linker region in regulator-promoter interaction(s). J. Biol. Chem. 285, 34309–34318. doi: 10.1074/jbc.M110.135822
Perez, J. C., Groisman, E. A. (2007). Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63, 283–293. doi: 10.1111/j.1365-2958.2006.05512.x
Perez, J. C., Shin, D., Zwir, I., Latifi, T., Hadley, T. J., Groisman, E. A. (2009). Evolution of a bacterial regulon controlling virulence and mg2+ Homeostasis. PloS Genet. 5, e1000428. doi: 10.1371/journal.pgen.1000428
Pérez-Morales, D., Banda, M. M., Chau, N. Y. E., Salgado, H., Martínez-Flores, I., Ibarra, J. A., et al. (2017). The transcriptional regulator SsrB is involved in a molecular switch controlling virulence lifestyles of Salmonella. PloS Pathog. 13, e1006497. doi: 10.1371/journal.ppat.1006497
Phan, M. D., Nhu, N. T. K., Achard, M. E. S., Forde, B. M., Hong, K. W., Chong, T. M., et al. (2017). Modifications in the pmrB gene are the primary mechanism for the development of chromosomally encoded resistance to polymyxins in uropathogenic Escherichia coli. J. Antimicrob. Chemother. 72, 2729–2736. doi: 10.1093/jac/dkx204
Pina, L. C., Silva, F. S. H., Galvao, T. C., Pauer, H., Ferreira, R. B. R., Antunes, L. C. M., et al. (2021). The role of two-component regulatory systems in environmental sensing and virulence in Salmonella. Crit. Rev. Microbiol. 47, 397–434. doi: 10.1080/1040841X.2021.1895067
Poirel, L., Jayol, A., Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/cmr.00064-16
Poolman, B., Blount, P., Folgering, J. H. A., Friesen, R. H. E., Moe, P. C., Heide, T. V. D. (2002). How do membrane proteins sense water stress? Mol. Microbiol. 44. doi: 10.1046/j.1365-2958.2002.02894.x
Prost, L. R., Daley, M. E., Le Sage, V., Bader, M. W., Moual, H. L., Klevit, R. E., et al. (2007). Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26, 165–174. doi: 10.1016/j.molcel.2007.03.008
Ramezanifard, R., Golubeva, Y. A., Palmer, A. D., Slauch, J. M. (2023). TamAB is regulated by PhoPQ and functions in outer membrane homeostasis during Salmonella pathogenesis. J. Bacteriol. 205, e00183–e00123. doi: 10.1128/jb.00183-23
Regelmann, A. G., Lesley, J. A., Mott, C., Stokes, L., Waldburger, C. D. (2002). Mutational Analysis of the Escherichia coli PhoQ Sensor Kinase: Differences with the Salmonella enterica Serovar Typhimurium PhoQ Protein and in the Mechanism of Mg2+ and Ca2+ Sensing. J. Bacteriol. 184, 5468–5478. doi: 10.1128/JB.184.19.5468-5478.2002
Ren, J., Sang, Y., Qin, R., Su, Y., Cui, Z. L., Mang, Z. G., et al. (2019). Metabolic intermediate acetyl phosphate modulates bacterial virulence via acetylation. Emerg. Microbes Infect. 8, 55–69. doi: 10.1080/22221751.2018.1558963
Ren, J., Sang, Y., Tan, Y., Tao, J., Ni, J. J., Liu, S. T., et al. (2016). Acetylation of lysine 201 inhibits the DNA-binding ability of PhoP to regulate Salmonella virulence. PloS Pathog. 12, e1005458. doi: 10.1371/journal.ppat.1005458
Roggiani, M., Yadavalli, S. S., Goulian, M. (2017). Natural variation of a sensor kinase controlling a conserved stress response pathway in Escherichia coli. PloS Genet. 13, e1007101. doi: 10.1371/journal.pgen.1007101
Salvail, H., Choi, J., Groisman, E. A. (2022). Differential synthesis of novel small protein times Salmonella virulence program. PloS Genet. 18, e1010074. doi: 10.1371/journal.pgen.1010074
Santos-Martin, C., Wang, G., Subedi, P., Hor, L., Totsika, M., Paxman, J. J., et al. (2021). Structural bioinformatic analysis of DsbA proteins and their pathogenicity associated substrates. Comput. Struct. Biotechnol. J. 19, 4725–4737. doi: 10.1016/j.csbj.2021.08.018
Schellhorn, H. E. (2020). Function, evolution, and composition of the rpoS regulon in escherichia coli. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.560099
Sen, H., Aggarwal, N., Ishionwu, C., Hussain, N., Parmar, C., Jamshad, M., et al. (2017). Structural and functional analysis of the Escherichia coli acid-sensing histidine kinase EvgS. J. Bacteriol. 199, 310–317. doi: 10.1128/jb.00310-17
Shahzad, S., Willcox, M. D. P., Rayamajhee, B. (2023). A review of resistance to Polymyxins and evolving Mobile Colistin resistance gene (mcr) among pathogens of clinical significance. Antibiotics 12, 1597. doi: 10.3390/antibiotics12111597
Shao, X. L., Tan, M. M., Xie, Y. P., Yao, C. Y., Wang, T. T., Huang, H., et al. (2021). Integrated regulatory network in Pseudomonas syringae reveals dynamics of virulence. Cell Rep. 34, 108920. doi: 10.1016/j.celrep.2021.108920
Shetty, D., Kenney, L. J. (2023). A pH-sensitive switch activates virulence in Salmonella. Elife 12, e85690. doi: 10.7554/eLife.85690
Shprung, T., Peleg, A., Rosenfeld, Y., Trieu-Cuot, P., Shai, Y. (2012). Effect of PhoP-PhoQ activation by broad repertoire of antimicrobial peptides on bacterial resistance. J. Biol. Chem. 287, 4544–4551. doi: 10.1074/jbc.M111.278523
Shprung, T., Wani, N. A., Wilmes, M., Mangoni, M. L., Bitler, A., Shimoni, E., et al. (2021). Opposing effects of PhoPQ and PmrAB on the properties of Salmonella enterica serovar Typhimurium: Implications on resistance to antimicrobial peptides. Biochemistry 60, 2943–2955. doi: 10.1021/acs.biochem.1c00287
Storm, D. R., Rosenthal, K. S., Swanson, P. E. (1977). Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46, 723–763. doi: 10.1146/annurev.bi.46.070177.003451
Subramani, S., Perdreau-Dahl, H., Morth, J. P. (2016). The magnesium transporter A is activated by cardiolipin and is highly sensitive to free magnesium in vitro. Elife 5. doi: 10.7554/eLife.11407
Sun, J., Shi, H., Huang, K. C. (2021). Hyperosmotic shock transiently accelerates constriction rate in Escherichia coli. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.718600
Tran, T. K., Han, Q. Q., Shi, Y., Guo, L. (2016). A comparative proteomic analysis of Salmonella typhimurium under the regulation of the RstA/RstB and PhoP/PhoQ systems. Biochim. Biophys. Acta (BBA)-Proteins Proteomics 1864, 1686–1695. doi: 10.1016/j.bbapap.2016.09.003
Tu, X., Latifi, T., Bougdour, A., Gottesman, S. S., Groisman, E. A. (2006). The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. United States America 103, 13503–13508. doi: 10.1073/pnas.0606026103
Véscovi, E. G., Soncini, F. C., Groisman, E. A. (1996). Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174. doi: 10.1016/S0092-8674(00)81003-X
Viarengo, G., Sciara, M. I., Salazar, M. O., Kieffer, P. M., Furlán, R. L. E., Véscovi, E. G. (2013). Unsaturated long chain free fatty acids are input signals of the Salmonella enterica PhoP/PhoQ regulatory system. J. Biol. Chem. 288, 22346–22358. doi: 10.1074/jbc.m113.472829
Wan, J., Gao, X., Liu, F. (2024). Regulatory role of the Cpx ESR in bacterial behaviours. Virulence 15, 2404951. doi: 10.1080/21505594.2024.2404951
Wang, M., Odom, T., Cai, J. (2020). Challenges in the development of next-generation antibiotics: opportunities of small molecules mimicking mode of action of host-defense peptides. Expert Opin. Ther. Patents 30, 303–305. doi: 10.1080/13543776.2020.1740683
Weinert, B. T., Iesmantavicius, V., Wagner, S. A., Christian, S., Bertil, G., Petra, B., et al. (2013). Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 51, 265–272. doi: 10.1016/j.molcel.2013.06.003
Westermann, A. J., Förstner, K. U., Amman, F., Barquist, L., Chao, Y. J., Schulte, L. N., et al. (2016). Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature 529, 496–501. doi: 10.1038/nature16547
Xia, F., Liu, Y., Wei, L., Shao, S., Zhang, Y. X., Ma, Y. (2024). Long-chain unsaturated fatty acids sensor controlling the type III/VI secretion system is essential for Edwardsiella piscicida infection. Microbiol. Res. 285, 127770. doi: 10.1016/j.micres.2024.127770
Xie, Y. P., Li, J. W., Ding, Y. Q., et al. (2022). An atlas of bacterial two-component systems reveals function and plasticity in signal transduction. Cell Rep. 41, 111502. doi: 10.1016/j.celrep.2022.111502
Xie, M. Q., Wu, M. Y., Han, A. D. (2020). Structural insights into the signal transduction mechanism of the K+-sensing two-component system KdpDE. Sci. Signaling 13, eaaz2970. doi: 10.1126/scisignal.aaz2970
Xu, J. T., Li, T., Gao, Y. R., Deng, J. Y., Gu, J. (2019). MgrB affects the acid stress response of Escherichia coli by modulating the expression of iraM. FEMS Microbiol. Lett. 366, fnz123. doi: 10.1093/femsle/fnz123
Xu, Q., Xu, T., Zhuang, Y., Liu, X. F., Li, Y., Chen, Y. J. (2020). In Vivo Development of Polymyxin B Resistance in Klebsiella pneumoniae owing to a 42 bp Deletion in the Sequence of phoQ. BioMed. Res. Int. 2020, 5868479. doi: 10.1155/2020/5868479
Xu, Y., Zhao, Z., Tong, W., Ding, Y. M., Shi, Y. X., Wang, J. C., et al. (2020). An acid-tolerance response system protecting exponentially growing Escherichia coli. Nat. Commun. 11, 1496. doi: 10.1038/s41467-020-15350-5
Yadavalli, S. S., Goh, T., Carey, J. N., Malengo, G., Vellappan, S., Nickels, B. E., et al. (2020). Functional determinants of a small protein controlling a broadly conserved bacterial sensor kinase. J. Bacteriol. 202. doi: 10.1128/JB.00305-20
Yamamoto, K., Ogasawara, H., Fujita, N., Utsumi, R., Ishihama, A. (2002). Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 45, 423–438. doi: 10.1046/j.1365-2958.2002.03017.x
Yamanaka, Y., Oshima, T., Ishihama, A., Yamamoto, K. (2013). Characterization of the YdeO regulon in Escherichia coli. PloS One 409, 151–157. doi: 10.1016/j.jcis.2013.07.049
Yan, Y. H., Li, Y. Z., Zhang, Z.W., Wang, X. H., Niu, Y. Z., Zhang, S. H., et al. (2021). Advances of peptides for antibacterial applications. Colloids Surf. B: Biointerf. 202, 111682. doi: 10.1016/j.colsurfb.2021.111682
Yao, Y. X., Wang, X., Lin, X. Y., Wu, J. S., Wang, P., Zhu, C. Z., et al. (2024). Isolation and characterization of probiotic Lysinibacillus species from the gastrointestinal tract of large yellow croaker (Larimichthys crocea). Front. Mar. Sci. 10. doi: 10.3389/fmars.2024.1408979
Yeom, J., Groisman, E. A. (2021). Low cytoplasmic magnesium increases the specificity of the lon and clpAP proteases. J. Bacteriol., 203(14). doi: 10.1128/jb.00143-21
Yeom, J., Pontes, M. H., Choi, J., Groisman, E. A. (2018). A protein that controls the onset of a Salmonella virulence program. EMBO J. 37, e96977. doi: 10.15252/embj.201796977
Yeom, J., Shao, Y., Groisman, E. A. (2020). Small proteins regulate Salmonella survival inside macrophages by controlling degradation of a magnesium transporter. Proc. Natl. Acad. Sci. 117, 20235–20243. doi: 10.1073/PNAS.2006116117
Yeom, J., Wayne, K. J., Groisman, E. A. (2017). Sequestration from protease adaptor confers differential stability to protease substrate. Mol. Cell 66, 234–246. doi: 10.1016/j.molcel.2017.03.009
Yin, X. F., Orr, M. M., Wang, H. B., Hobbs, E. C., Shabalina, S. A., Storz, G. (2019). The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol. Microbiol. 111, 131–144. doi: 10.1111/mmi.14143
Yoshitani, K., Ishii, E., Taniguchi, K., Sugimoto, H., Shiro, Y., Akiyama, Y., et al. (2019). Identification of an internal cavity in the PhoQ sensor domain for PhoQ activity and SafA-mediated control. Biosci. Biotechnol. Biochem. 83, 684–694. doi: 10.1080/09168451.2018.1562879
Yu, J. L., Guo, L. (2011). Quantitative Proteomic Analysis of Salmonella enterica Serovar Typhimurium under PhoP/PhoQ Activation Conditions. J. Proteome Res. 10, 2992–3002. doi: 10.1021/pr101177g
Yuan, J., Jin, F., Glatter, T., Sourjik, V. (2017). Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. United States America 114, 10792–10798. doi: 10.1073/pnas.1717272114
Zafer, M. M., El-Mahallawy, H. A., Abdulhak, A., Amin, M. A., Al-Agamy, M. H., Radwan, H. H. (2019). Emergence of colistin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli strains isolated from cancer patients. Ann. Clin. Microbiol. Antimicrob. 18, 1–8. doi: 10.1186/s12941-019-0339-4
Zeng, J., Wu, L., Liu, Z., Lv, Y. H., Feng, J. Z., Wang, W. J., et al. (2021). Gain-of-function mutations in acid stress response (evgS) protect Escherichia coli from killing by gallium nitrate, an antimicrobial candidate. Antimicrob. Agents Chemother. 65, 10. doi: 10.1128/aac.01595-20
Zhang, P., Ouyang, Q., Zhai, T., Sun, J., Wu, J., Qin, F., et al. (2022). An inflammation-targeted nanoparticle with bacteria forced release of polymyxin B for pneumonia therapy. Nanoscale 14, 15291–15304. doi: 10.1039/D2NR02026B
Zhu, Y., Hao, W., Wang, X., Ouyang, J. H., Deng, X. Y., Yu, H. N., et al. (2022). Antimicrobial peptides, conventional antibiotics, and their synergistic utility for the treatment of drug-resistant infections. Med. Res. Rev. 42, 1377–1422. doi: 10.1002/med.21879
Keywords: bacterial, TCS, PhoP/PhoQ, phosphorylation, virulence
Citation: Mao M, He L and Yan Q (2025) An updated overview on the bacterial PhoP/PhoQ two-component signal transduction system. Front. Cell. Infect. Microbiol. 15:1509037. doi: 10.3389/fcimb.2025.1509037
Received: 10 October 2024; Accepted: 08 January 2025;
Published: 31 January 2025.
Edited by:
Sara María Soto, Instituto Salud Global Barcelona (ISGlobal), SpainReviewed by:
Roberto Rosales-Reyes, National Autonomous University of Mexico, MexicoCopyright © 2025 Mao, He and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingpi Yan, eWFucXBAam11LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.