- 1Department of Respiratory and Critical Care Medicine, The Second Hospital of Jilin University, Changchun, Jilin, China
- 2Department of Nuclear Medicine, The Second Hospital of Jilin University, Changchun, Jilin, China
Corynebacterium striatum (C. striatum) is a Gram-positive bacterium commonly colonizing the skin and mucosa in healthy individuals and hospitalized patients. Traditionally regarded as a contaminant, C. striatum is now increasingly recognized as a potential cause of clinical infections, especially after the coronavirus disease pandemic. It has emerged as a pathogen implicated in severe infections, including pneumonia, bacteremia, meningitis, artificial joint infections, abdominal infections, and endocarditis. C. striatum has been reported in lower respiratory tract infections, mostly as a conditioned pathogen in immunocompromised individuals, particularly in those with chronic structural lung diseases, such as chronic obstructive pulmonary disease, leading to severe pneumonia or exacerbation of the existing disease and high mortality. Additionally, C striatum has been implicated in the community-acquired pneumonia among immunocompetent individuals and nosocomial lung infections, with evidence of person-to-person transmission through caregivers. C. striatum may exhibit multidrug resistance. Vancomycin, alone or in combination, is currently considered the most effective treatment for C. striatum. This review highlights the epidemiological characteristics, drug resistance mechanisms, diagnostics approaches, and treatment options for C. striatum lower respiratory tract infections to enhance clinician awareness and improve patient management strategies.
1 Introduction
Corynebacterium species are characterized by the presence of arabinose, galactose, and meso-diaminovaleric acid in their cell walls. Some strains may also contain corynomycolic acids and metachromatic granules, which serve as reserves of the high-energy phosphate. Corynebacterium striatum (C. striatum) is a non-diphtheriae corynebacterium that is Gram-positive, non-lipophilic, glucose-fermentative or non-fermentative, aerobic or facultatively anaerobic, non-sporulating, and non-motile bacilli. Under a light microscope, it can appear in various forms, including ball, rod-like, and filamentous shapes, occurring singly, in pairs, and/or palisade (Funke et al., 1997; Silva-Santana et al., 2021). C. striatum is typically a normal flora of the skin and oropharynx and is rarely pathogenic. However, it can opportunistically cause infections in hospitalized patients with underlying conditions (Martinez-Martinez et al., 1997). However, it can be isolated from wound swabs, respiratory specimens, tissue and bone samples, and blood cultures (Funke et al., 1997). In recent decades, there has been increasing reports of invasive infections caused by C. striatum, including lower respiratory tract infections, intracranial infections, joint infections, and abdominal infections. While they primarily affect immunocompromised individuals, cases have also been reported in patients with normal immune function, albeit rarely (Martinez-Martinez et al., 1997; Lee et al., 2005; Otsuka et al., 2006). C. striatum mainly causes severe infections in patients with immunocompromised state, including those with end-stage renal disease, structural lung diseases, and advanced cancers. In addition, surgical or invasive procedures, long hospital stays, advanced age, neoplastic disease, organ transplantation, acquired immune deficiency syndrome (AIDS), diabetes, long-term antibiotic use, and procedures requiring continuous or long-term medical devices, such as catheterization, heart valve implantation, and prosthesis placement, for chronic diseases, long intensive care unit stays and reduced hemoglobin levels have also been identified as risk factors associated with C. striatum infection (Martinez-Martinez et al., 1997; Superti et al., 2009; Chen et al., 2012; Verroken et al., 2014; Carvalho et al., 2018; Silva-Santana et al., 2021). There have been documented cases of C. striatum transmission between patients, leading to serious nosocomial outbreaks. Typically, the isolation of C. striatum from a culture is regarded as contamination unless repeated cultures yield consistent results (Kang et al., 2018). Among reported cases of invasive C. striatum infections, the most commonly isolated specimens were blood, followed by bone and joint tissues, sputum, and others. The most commonly employed detection method was biochemical analysis alone. However, 16S rRNA gene sequencing and mass spectrometry (mostly via the MALDI-TOF system) were also used as stand alone methods in approximately 17% and 20% of cases, respectively. Biochemical methods combined with MALDI-TOF, or MALDI-TOF combined with 16S rRNA sequencing were also used as detection methods in some cases. In one case, invasive infection caused by C. striatum was confirmed using all three methods. Different detection methods can not only identify C. striatum as the dominant bacterium causing invasive infection but also determine its drug resistance and virulence genes, providing valuable insights for treatment strategies. One study reported C. striatum as the predominant bacterium, with a detection rate of 11.76% in patients with pulmonary infections in the intensive care unit (Zhang et al., 2023). Osawa R and colleagues reported that among patients with bacteremia caused by C. striatum, the 90-day mortality rate reached 34% (Yamamuro et al., 2021). Another study indicated that the overall 30-day mortality rate was 34.7% (Abe et al., 2021). Currently, there is no established standard for the treatment of C. striatum infections. Given the invasive nature of the bacterium and its high rates of drug resistance—including cases of multidrug resistance—antibiotic selection should be guided by drug resistance gene detection. Vancomycin can be used as a treatment for multidrug-resistant C. striatum infections. However, no consensus exists regarding the duration of treatment, which should be tailored based on the severity of the infection and the affected site (Milosavljevic et al., 2021).
In this review, we conducted a comprehensive search of the MEDLINE (PubMed), EBSCO (Discovery Service), SCOPUS, SCIndex (Serbian Citation Index), and Cochrane Central (Wiley Online Library) databases. The search terms included: “corynebacterium striatum”[Supplementary Concept] OR “corynebacterium striatum”[All Fields] OR “corynebacterium striatum”[All Fields]) AND (((“invasibility”[All Fields] OR “invasible”[All Fields] OR “invasion”[All Fields] OR “invasions”[All Fields] OR “invasive”[All Fields] OR “invasively”[All Fields] OR “invasiveness”[All Fields] OR “invasives”[All Fields] OR “invasivity”[All Fields]) AND (“lower airway infect”[All Fields] OR “lower airway infection”[All Fields] OR “lower airway infective”[All Fields] OR”lung infect”[All Fields] OR “lung infection”[All Fields] OR “lung infective”[All Fields] OR “pneumonia”[All Fields]).We summarize the literature on the clinical characteristics, diagnosis, and treatment of C. striatum-associated lower respiratory tract infections.
2 Clinical characteristics of respiratory infections caused by Corynebacterium striatum
In recent years, the incidence of C. striatum lower respiratory tract infections has increased, likely owing to advancements in detection technology. Studies have revealed that, following the coronavirus disease (COVID-19) pandemic, the infection rate of C. striatum in the lower respiratory tract has markedly increased (Orosz et al., 2022). Furthermore, C. striatum, somtimes presented as a superdominant pathobiontic bacterial genus (defined as a genus comprising more than 50% of nasopharyngeal swab sequences), within the nasopharyngeal microbiota, may contribute to severe secondary infections in affected patients (Qin et al., 2020). Common pathogens detected in cases of aspiration pneumonia are C. striatum, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Candida albicans (DiBardino and Wunderink, 2015; Neill and Dean, 2019; Xu et al., 2024). Nosocomial infections and outbreaks caused by C. striatum primarily occur in the respiratory tract. Research from various countries has confirmed that mechanical ventilation is a risk factor for nosocomial infections (Campanile et al., 2009; Wong et al., 2010). A recent review (1976–2020) analyzing the clinical epidemiology and microbiology of 218 studies confirmed the emergence of multidrug-resistant and multidrug-sensitive C. striatum strains in 254 reported cases worldwide, which can cause hospital - and community-acquired infections (Silva-Santana et al., 2021). Prolonged hospital stays, advanced chronic obstructive pulmonary disease, recent antibiotic use, and invasive diagnostic procedures are the most common risk factors for patients with C. striatum pneumonia. A positive correlation has been observed between C. striatum infections and decreased lung function in chronic obstructive pulmonary disease (forced expiratory volume [FEV1]%) (Renom et al., 2014; Verroken et al., 2014). C. striatum resists infection control measures, as it can adhere to non-living surfaces and form biofilms on various medical devices, such as feeding tubes, endotracheal tubes, and ventilators (Souza et al., 2015; Ramos et al., 2019; Lee et al., 2022). The incidence rates of C. striatum lower respiratory tract infections are as follows: hospital-acquired pneumonia accounts for 96.3%, including healthcare-associated pneumonia (14.8%) and ventilator-associated pneumonia (11.1%), while community-acquired pneumonia represents 3.7%. C. striatum can cause severe pneumonia, which has a mortality rate of up to 60% and is likely to develop into solid cancers, diabetes, and structural lung diseases such as chronic obstructive onary disease (Lee et al., 2022).
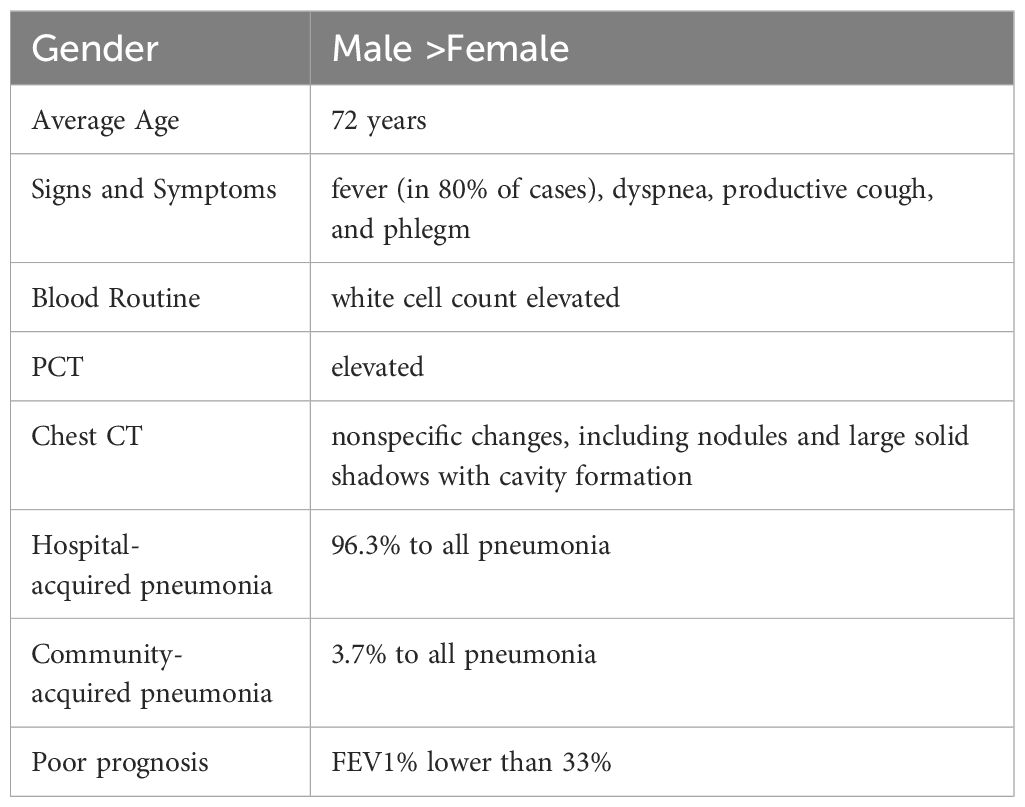
Patients with positive cultures from sputum, airway extracts, or bronchoalveolar lavage fluid are typically referred to infectious disease physicians for consultation to determine whether they are pathogenic bacteria. This evaluation includes repeated culture testing, evaluation of infection symptoms and markers, and identification of pathogenic bacteria based on infection guidelines from the Centers for Disease Control and Prevention (Martinez-Martinez et al., 1997; Yamamuro et al., 2021). Invasive infections caused by C. striatumare reported more frequently in men than women, with a median patient age of 72 years in cases of severe pneumonia. Notably, 51.9% of the patients are immunocompromised, especially those with structural lung diseases (Martinez-Martinez et al., 1997; Lee et al., 2022). Common clinical symptoms (Table 1) include fever (in 80% of cases), dyspnea, productive cough, and phlegm. Serum leukocyte and procalcitonin levels in peripheral blood are often significantly elevated. Pulmonary computed tomography may reveal various nonspecific changes, including nodules and large solid shadows with cavity formation (Severo et al., 2014). Among reported cases of C. striatum invasive infections, 75.3% of patients underwent antimicrobial susceptibility testing, primarily due to critical and progressively worsening conditions (Milosavljevic et al., 2021). Many hospitals also conduct routine antimicrobial susceptibility testing on positive pathogenic cultures to guide appropriate clinical therapy. In addition, according to European Committee for Antimicrobial Susceptibility Testing (EUCAST) guidelines, laboratory antimicrobial susceptibility testing for C. striatum requires special Settings, as well as broth microdilution, MIC assessment and AGAR diffusion using equine defibrination blood and beta-NAD (Marino et al., 2022).Among patients with severe pneumonia, a high proportion (67%) experienced septic shock, and the 30-day mortality rate was as high as 40.7%. Further analysis of deceased patients revealed a mean FEV1% of 33%, suggesting that decreased lung function may be associated with poor prognosis (Lee et al., 2022).
3 Examination method
C. striatum is most frequently identified in cultures from clinical specimens, including blood, pus, urine, and pleural effusion (Bao et al., 2017). C. striatum as a cause of infection was most often identified exclusively by biochemical methods (Milosavljevic et al., 2021). However, several reports indicate that C. striatum may be misidentified or not identified at all using biochemical methods alone. In such cases, alternative techniques like 16S rRNA sequencing are used to confirm the diagnosis (Iaria et al., 2007; Milosavljevic et al., 2021). Application of the VITEK 2 system (bioMerieux,USA), a biochemical method commonly used in clinical practice to detect C. striatum, has shown instances of misidentification, including cases where C. striatum was mistaken for other bacteria like Clostridium striatum. However, the reliability of detection can be improved by including the pathogen in the VITEK system (VITEK®2 ANC ID card) database. It can be used to identify C. striatum, especially if no alternative test is available (Lee et al., 2011).With advancements in molecular technology, C. striatum can be identified using various methods, including 16SrRNA sequencing, complex infection detection chips, metagenomic next-generation sequencing, high-resolution melting analysis, and conventional microbiological tests. These techniques provide simple, rapid, sensitive, and specific options for detecting C. striatum, supporting early diagnosis, epidemiological surveillance, and rapid outbreak response (Xu et al., 2021). Mass spectrometry techniques, such as matrix-assisted laser desorption ionization, combined with time of flight and mass spectrometry (MALDI-TOF MS), are also used for bacterial species identification, providing faster and more practical diagnostic solutions (Gomila et al., 2012; Diez-Aguilar et al., 2013). In a hospital outbreak in Belgium, biochemical methods and 16sRNA sequencing techniques were used to identify C. striatum. Additionally, MALDI-TOF MS and combined 16S rRNA sequencing have been successfully used in other cases (Verroken et al., 2014). In one case, invasive C. striatum infection was identified using a combination of biochemical, MALDI-TOF, and 16S rRNA sequencing techniques. Several studies have shown that gene sequencing is the most reliable method for identifying C. striatum (Gomila et al., 2012; Alibi et al., 2015; Suh et al., 2019). Additionally, MALDI-TOF mass spectrometry offers a cost-effective, simple, and reliable alternative for identifying C. striatum (Gomila et al., 2012; Suh et al., 2019).
The development of gene sequencing technology has increased detection sensitivity enabling identification of virulence and resistance genes as well as the dominant cloning of bacteria. The core genome multilocus sequence typing (cgMLST) method, based on whole-genome sequencing, can be used to track transmission routes of C. striatum infections in hospital settings, which holds significant clinical and epidemiological value (Trost et al., 2011; Torres Lde et al., 2013; Kang et al., 2023). Whole-genome sequencing has also been applied to characterize multidrug-resistant and non-multidrug-resistant clinical isolates of C. striatum, providing insights into molecular epidemiology, global transmission, and virulence mechanisms of pathogens (Nudel et al., 2018; Ramos et al., 2018; Wang et al., 2021).
4 Virulence and resistance
4.1 Virulence
C. striatum is highly invasive and exhibits strong genetic plasticity, with a robust iron acquisition genetic library, independent determinants, and antimicrobial resistance genes (Jesus et al., 2022). C. striatum strains from different sources demonstrate high diversity, with those isolated from skin tissues being relatively stable and more conserved. The pan-genome analysis of an emerging multi-drug-resistant C. striatum isolate showed an open pan-genome, consisting of 5,692 gene families, 1,845 core gene families, 2,362 auxiliary gene families, and 1,485 unique gene families (Figure 1). This analysis further identified 53 resistance genes and 42 virulence factors. Notably, 77.7% of these strains carried two or more resistance genes exhibiting resistance to aminoglycosides, tetracycline, lincomycin, macrolides, and streptomycin. Virulence factors are primarily associated with the survival of pathogenic bacteria in the host, iron uptake, and early biofilm formation (Qiu et al., 2023). The SpaD and SpaE genes are involved in the formation of pili, which allow the strain to adhere specifically to human pharyngeal epithelial cells (Kang et al., 2014). Additionally, SrtC and SrtB genes were identified in 48.7% of the strains. SrtC is a sorting enzyme associated with pili production through a sorting enzyme mechanism. The SrtB gene encodes a collagen-binding protein that binds to the human complement C1q, potentially involved in host immune escape mechanisms and playing an important role in early biofilm formation (Donahue et al., 2014; Chambers et al., 2015). Biofilms have been identified on various human tissues and abiotic surfaces, including medical devices such as catheters, central venous catheters, and endoscopes. The formation of biofilms enhances the survivability of microorganisms under adverse conditions (Ramirez de Arellano et al., 1995; Al Akhrass et al., 2012; Ocalan et al., 2023). Bacteria within biofilms are highly resistant to components of the human immune system and a variety of antibiotics. Moreover, the ability of bacterial cells to transfer genes horizontally is enhanced in biofilm communities, which can promote the spread of antibiotic resistance. The formation of biofilms allows bacteria to adhere to different substrates, enabling their survival in hospital environments. Studies have confirmed that increased biofilm production in C. striatum infection group is a common virulence factor, and it has been observed that C. striatum isolates adhere to various abiotic surfaces to form biofilms in vitro, which is the cause of hospital infection outbreak (Souza et al., 2015; Alibi et al., 2021; Ocalan et al., 2023). Research indicates that at 37°C, C. striatum is capable of producing biofilms on abiotic surfaces, including polystyrene, glass, and tracheostomy tubes made of polyvinyl chloride, silicone, and stainless steel. These biofilms adhere to both human epithelial cells and abiotic surfaces, enhancing the viability of C. striatum in host tissues and the hospital environment. The ability to form biofilms increases the likelihood of hospital-acquired infections while simultaneously elevating bacterial resistance to antimicrobial agents and immune responses (Alibi et al., 2021).Furthermore, several virus-associated proteins, including acyl-CoA carboxylase β-subunits (DtsR1, DtsR2, and AccD3), cell wall-associated hydrolase (CwlH), nonribosomal peptide synthetase (NrpS2), nitric oxide reductase (Nor), resuscitation promoting factors RpfA and RpfB, subtilisin-like serine protease (MycP), SGNh-hydrolase (SgnH), and venom serine protease (Vsp2) identified in C. striatum strains are also associated with their pathogenicity (Sangal et al., 2024).
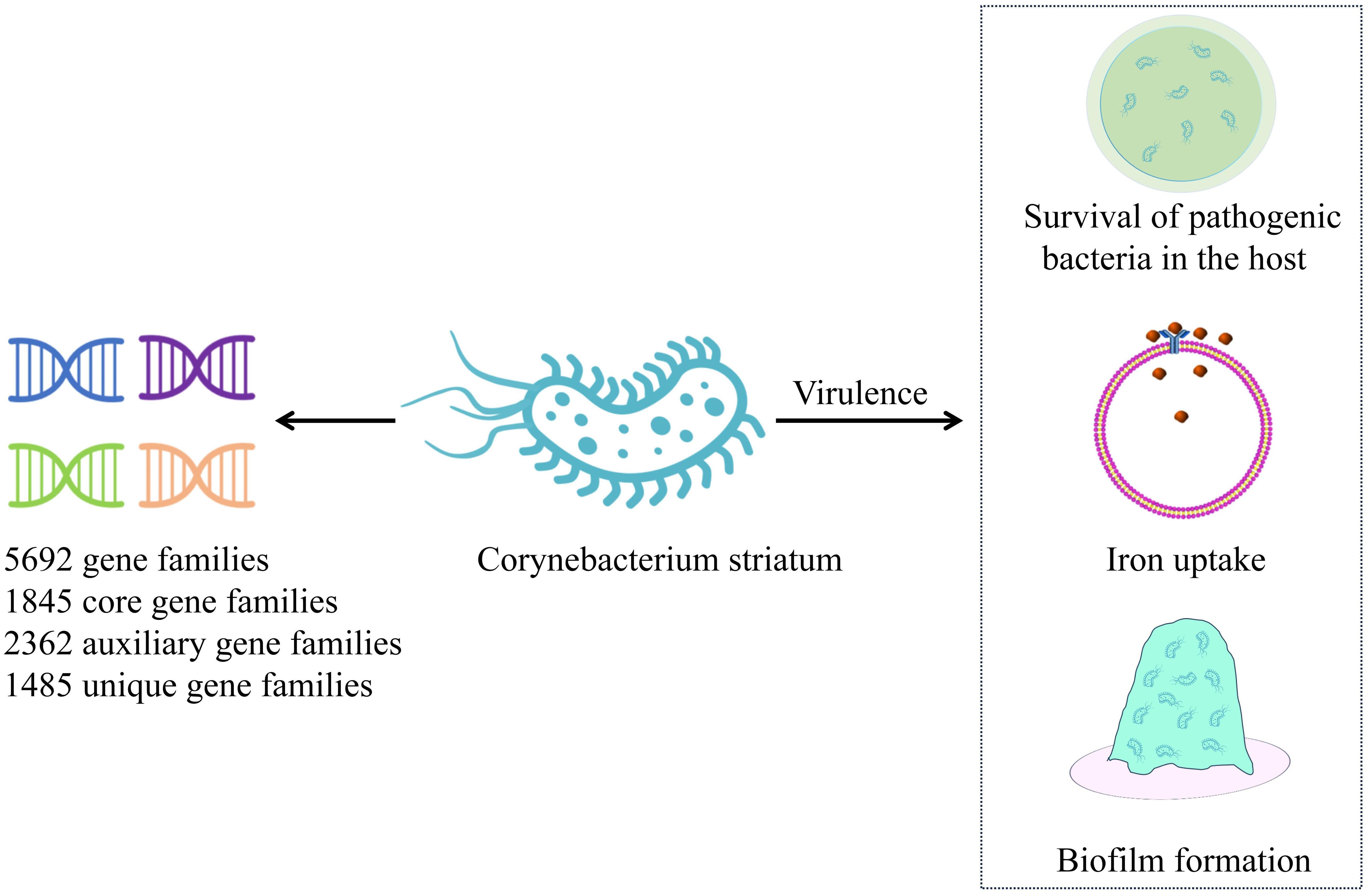
Figure 1. Pan-genome of Corynebacterium striatum. This figure illustrates the pan-genome of recently isolated C. striatum, highlighting key mechanisms of virulence. C. striatum – Corynebacterium striatum.
4.2 Resistance
Drug-susceptibility testing through bacterial culture remains a widely used classical method for selecting antimicrobial agents against C. striatum. Gene sequencing technology can identify drug resistance genes of bacteria, facilitating the understanding of resistance mechanisms in C. striatum isolates and the selection of appropriate antimicrobial agents. In the following paragraphs, we discuss the drug resistance mechanisms identified in C. striatum isolates reported in this study and the antimicrobial agents to which resistance has been observed.
(A) Efflux pump: The ATP-binding cassette transporter encoded by the tetA/B gene transports antibiotics out of the cell membrane, leading to tetracycline and beta-lactam resistance (Kono et al., 1983) (Figure 2). Specific major facilitator superfamily transporters and cmx genes encoded efflux pumps are associated with chloramphenicol resistance (Leyton et al., 2021).
(B) Biofilm formation: C. striatum produces mature biofilms in vitro, similar to other pathogenic organisms (Souza et al., 2015). These isolates also exhibited enhanced biofilm formation in the presence of human fibrinogen. Multidrug-resistant strains are strong biofilm producers. Interestingly, isolates from intubated patients showed the highest biofilm production. Other clinical studies on C. striatum have also confirmed the production of biofilms (Galimand et al., 2015; Wang et al., 2016; Shariff et al., 2018). The SrtB gene encodes a collagen-binding protein in Clostridium difficile, which binds to the human complement C1q and may be involved in the host immune escape mechanisms while playing an important role in early biofilm formation (Chambers et al., 2015; Kang et al., 2020; Qiu et al., 2023).
(C) Changing the target of antibiotics: Changes in drug targets due to gyrA mutations, including mutations in the quinolone resistance-determining region of the gyrA gene (Nudel et al., 2018; Ramos et al., 2018; Asgin and Otlu, 2020), mutations at positions 87 and 91 in the gyrA peptide sequence (including those in Ser87Phe, Asp91Ala, and Asp91Gly) (Ramos et al., 2018; Dragomirescu et al., 2020; Ramos et al., 2020). New mutation sites er95Thr, Asp94Ala, Glu88Ala, and Asp87Gly (Qin et al., 2020) convert polar amino acids to nonpolar (except at the 95th position), thereby reducing affinity for fluoroquinolones and preventing their binding to topoisomerase (Nudel et al., 2018). Resistance to macrolides is due to modification of the target by the erm gene, which methylates the 23S subunit of the ribosome. Similarly, the tet gene, which encodes the ribosome-protective protein tet, confers resistance to tetracycline drugs.
(D) Antibiotic inactivation: The aminoglycoside N-acetyltransferase-coding genes AAC(6) and AAC(6 ‘)-Ib-cr, which inactivate aminoglycoside antibiotics by acetylating their 6-amino groups, were identified in 57% of the strains (Smith et al., 2017). The APH(6’) and APH(3’) genes encoding aminoglycoside O-phosphotransferase were detected in 32% of strains, which can inactivate antibiotics, especially streptomycin. The ANT(3”) gene family, which encodes a class of aminoglycoside O-nucleotidyl transferases, that are regionally specific according to the 3”-hydroxyl modification of antibiotics, was detected in 38.9% of strains (Qiu et al., 2023). These enzymes inactivate aminoglycoside antibiotics by transferring the AMP group from the ATP substrate to the 3”-hydroxyl group of the compound (Ramirez and Tolmasky, 2010). Beta-lactam resistance is attributed to beta-lactamases encoded by the bla and ampC genes, which degrade beta-lactam antibiotics (Leyton et al., 2021).
(E) Prevention of antibiotic binding to bacteria: In Bacillus subtilis, the loss of phosphoglyceride (PG) in membranes can be fatal; thus, PG is essential for membrane integrity (Hachmann et al., 2011). The pgsA2 gene of Fusarium striatum encodes phosphatidylglycerol synthetase A, responsible for synthesizing diglyceride diphosphate in the PG synthesis pathway (Hines et al., 2017; Goldner et al., 2018; Hagiya et al., 2019). A mutation in the pgsA2 gene leads to a deficiency of glycerol phosphate in the membrane, preventing daptomycin from binding to the cell membrane. However, Goldner et al. (2018). demonstrated that the striatal C. HLDR phenotype was sufficient in the absence of functional loss of the pgsA2 gene. This suggests that C. striatum is a more persistent and adaptable bacterium than B. subtilis. Other resistance mechanisms include various ARGs associated with mobile genetic elements such as plasmids, integrons, insertion sequences, and transposons (Nesvera et al., 1998; Tauch et al., 2003; Hennart et al., 2020). The resistance of C. striatum is primarily determined by transposons, insertion sequences, and plasmids. The Macrolide-lincosamide-streptogramin B (MLS) phenotype is a common resistance mechanism in coagulase-negative staphylococci and also in C. striatum related to the erm(X) gene transported by the transposon Tn5432 (Szemraj et al., 2018; Szemraj et al., 2019).
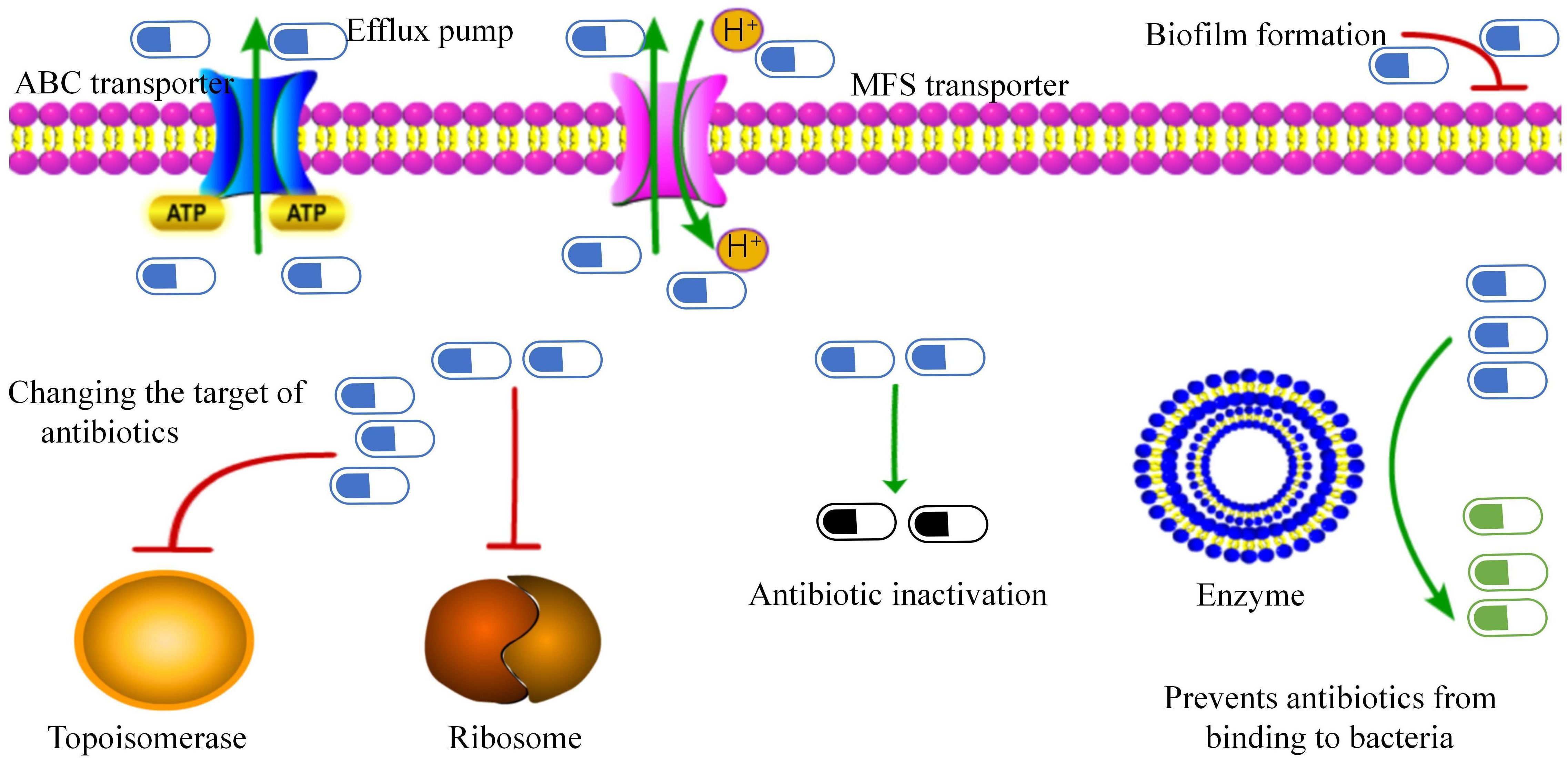
Figure 2. Antibiotic resistance mechanisms in Corynebacterium striatum. This figure presents the identified mechanisms contributing to antibiotic resistance.
The resistance in C. striatum is dynamic and expansive, which is also related to the transposon Tn5432, the MLS phenotype, and the insertion sequence, among others (Nesvera et al., 1998; Hennart et al., 2020). This leads to cross-infection, spread, and evolution of pathogenic bacteria in hospitals, necessitating that hospital staff be aware of nosocomial infections caused by C. striatum (Souza et al., 2020; Wang et al., 2022).
5 Treatment
Several studies have documented resistance in isolated strains of C. striatum. As mentioned earlier, bacterial culture and drug susceptibility testing can be effective methods for guiding antibiotic selection. Additionally, gene sequencing technology can accurately identify drug-resistance genes in isolated strains and is increasingly being utilized to inform clinical decisions regarding antibiotic selection.
In China, isolates were collected from three hospitals across three regions, with 260 isolates from patients with respiratory infections. Nearly all isolates (96.2%, 250/260) showed multidrug resistance, although they remained sensitive to vancomycin or linezolid, which aligns with findings from other countries (Zhang et al., 2023). Of these strains, 77.7% harbored two or more resistance genes and showed primary resistance to aminoglycosides, tetracycline, lincomycin, macrolides, and streptomycin (Qiu et al., 2023). Another study reported that 54 C. striatum isolates exhibited multidrug resistance to three or more antibiotics, with a resistance rate of 85.2% to lincomines and 93.5% to quinolones and tetracyclines. Sensitivity to vancomycin and linezolid was 100% (Wang et al., 2022).
Given its efficacy, vancomycin should be considered the antibiotic of choice for treating C. striatum infections. Vancomycin monotherapy, or combined with other antibiotics, such as piperacillin-tazobactam, may be the most prudent approach for multidrug-resistant C. striatum lung infections. Based on resistance predictions, vancomycin may remain one of the few effective agents currently in use by 2030 (Tarr et al., 2003; Babay and Kambal, 2004; Orosz et al., 2022). Alternatively, linezolid, teicoplanin, or daptomycin may be considered for treating severe lung infections caused by C. striatum, while amoxicillin-clavulanate may be used in mild infection cases (Milosavljevic et al., 2021).
However, a study has reported no difference in hospital mortality among patients with severe pneumonia who received antimicrobials targeting C. striatum, including vancomycin and linezolid. This outcome was similar to that in patients who did not receive anti-infective treatment. This may be attributed to the severity of illness in the C. striatum infection group, which included more critically ill patients with APACHE II scores >15. A subgroup analysis indicated that vancomycin or linezolid use reduced all-cause mortality (Zhang et al., 2023).
Daptomycin is also effective in resistant Gram-positive bacterial infections; however, reports of daptomycin-resistant C. striatum strains have emerged, potentially leading to treatment failure (Chauvelot et al., 2020).
Dalbavancin may serve as a successful and safe alternative for C. striatum infections, particularly in soft tissue infections. Approved for bacterial skin and soft tissue infections, dalbavancin’s optimal dosing and interval remain to be determined, and therapeutic drug monitoring may help in guiding treatment (Soderquist et al., 2023; Camara-Rodriguez et al., 2024).
Recently, new treatments for multidrug-resistant C. striatum infections have also shown promise. CSP1, a novel temperate bacteriophage and the first phage identified to target C. striatum strains could offer new possibilities in bacteriophage therapy research (Wang et al., 2024). Niclosamide, which reduces biofilm viability in a dose-dependent manner, has been approved for degrading biofilm biomass, and drastically reducing cell viability. Therefore, niclosamide is emerging as a promising therapeutic agent against multidrug-resistant C. striatum infections (Folliero et al., 2022).
6 Conclusion
Reports indicate that C. striatum lower respiratory tract infections are increasing, particularly during the COVID-19 pandemic. C. striatum can cause both community-acquired and hospital-acquired pneumonia in healthy and immunocompromised individuals. It can lead to severe pneumonia with high mortality, particularly in patients with structural lung diseases, such as chronic obstructive pulmonary disease. The prognosis of C. striatum pneumonia correlates with a decline in FEV1%. Despite potential detection errors, biochemical methods remain reliable for clinical identification of C. striatum. It has been reported that gene sequencing technology (16sRNA) is the most reliable method for detection of invasive infections caused by this pathogen, and can be considered as the gold standard for diagnosis. Whole-genome sequencing offers additional insights by identifying bacterial virulence factors and drug resistance genes. Furthermore, MALDI-TOF mass spectrometry is increasingly being used in clinical settings due to its rapid, cost-effective, and reliable detection capabilities. Isolated strains of C. striatum exhibit high rates of drug resistance, complex resistance mechanisms, including efflux pumps, biofilm formation, target modification of antibiotics, antibiotic inactivation, and prevention of antibiotic binding to bacteria. These factors also contribute to its potential for nosocomial transmission. At present, there is no standardized antibiotic regimen for treating lower respiratory tract infections caused by C. striatum. Antibiotic selection should be guided by the severity of the patient’s condition, as well as the virulence and drug resistance profile of the isolated strain. Vancomycin remains the most effective treatment, either alone or in combination with other agents. For mild cases, drugs such as piperacillin and sulbactam may be considered. New therapies, including temperate bacteriophages such as CSP1, hold promise for managing drug-resistant C. striatum in the future.
Author contributions
WL: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing, Software. MG: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – original draft, Project administration, Visualization, Formal analysis. JY: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant no. 81800080), Natural Science Foundation of Jilin Province (grant no. YDZJ202201ZYTS120).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AIDS, Acquired Immune Deficiency Syndrome; C. striatum, Corynebacterium striatum; COPD, Chronic Obstructive Pulmonary Disease; COVID-19, Coronavirus Disease; FEV1%, Forced Expiratory Volume; cgMLST, Core Genome Multilocus Sequence Typing; MALDI-TOF MS, Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry; MLS, Macrolide-lincosamide-streptogramin B.
References
Abe, M., Kimura, M., Maruyama, H., Watari, T., Ogura, S., Takagi, S., et al. (2021). Clinical characteristics and drug susceptibility patterns of Corynebacterium species in bacteremic patients with hematological disorders. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2095–2104. doi: 10.1007/s10096-021-04257-8
Al Akhrass, F., Al Wohoush, I., Chaftari, A. M., Reitzel, R., Jiang, Y., Ghannoum, M., et al. (2012). Rhodococcus bacteremia in cancer patients is mostly catheter related and associated with biofilm formation. PloS One 7, e32945. doi: 10.1371/journal.pone.0032945
Alibi, S., Ferjani, A., Gaillot, O., Marzouk, M., Courcol, R., Boukadida, J. (2015). Identification of clinically relevant Corynebacterium strains by Api Coryne, MALDI-TOF-mass spectrometry and molecular approaches. Pathol. Biol. (Paris) 63, 153–157. doi: 10.1016/j.patbio.2015.07.007
Alibi, S., Ramos-Vivas, J., Ben Selma, W., Ben Mansour, H., Boukadida, J., Navas, J. (2021). Virulence of clinically relevant multidrug resistant Corynebacterium striatum strains and their ability to adhere to human epithelial cells and inert surfaces. Microb. Pathog. 155, 104887. doi: 10.1016/j.micpath.2021.104887
Asgin, N., Otlu, B. (2020). Antimicrobial resistance and molecular epidemiology of corynebacterium striatum isolated in a tertiary hospital in Turkey. Pathogens 9. doi: 10.3390/pathogens9020136
Babay, H. A., Kambal, A. M. (2004). Isolation of coryneform bacteria from blood cultures of patients at a University Hospital in Saudi Arabia. Saudi Med. J. 25, 1073–1079.
Bao, R., Gao, X., Hu, B., Zhou, Z. (2017). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: a powerful tool for identification of Corynebacterium species. J. Thorac. Dis. 9, 3239–3245. doi: 10.21037/jtd.2017.09.69
Camara-Rodriguez, M., Jover-Diaz, F., Delgado-Sanchez, E., Infante-Urrios, A., Peris-Garcia, J. (2024). Dalbavancin as long-term treatment in Corynebacterium striatum Infections: a literature review. Rev. Esp Quimioter 37, 276–278. doi: 10.37201/req
Campanile, F., Carretto, E., Barbarini, D., Grigis, A., Falcone, M., Goglio, A., et al. (2009). Clonal multidrug-resistant Corynebacterium striatum strains, Italy. Emerg. Infect. Dis. 15, 75–78. doi: 10.3201/eid1501.080804
Carvalho, R. V., Lima, F., Santos, C. S. D., Souza, M. C., Silva, R. S. D., Mattos-Guaraldi, A. L. (2018). Central venous catheter-related infections caused by Corynebacterium amycolatum and other multiresistant non-diphtherial corynebacteria in paediatric oncology patients. Braz. J. Infect. Dis. 22, 347–351. doi: 10.1016/j.bjid.2018.07.004
Chambers, C. J., Roberts, A. K., Shone, C. C., Acharya, K. R. (2015). Structure and function of a Clostridium difficile sortase enzyme. Sci. Rep. 5, 9449. doi: 10.1038/srep09449
Chauvelot, P., Ferry, T., Tafani, V., Diot, A., Tasse, J., Conrad, A., et al. (2020). Bone and joint infection involving corynebacterium spp.: from clinical features to pathophysiological pathways. Front. Med. (Lausanne) 7, 539501.
Chen, F. L., Hsueh, P. R., Teng, S. O., Ou, T. Y., Lee, W. S. (2012). Corynebacterium striatum bacteremia associated with central venous catheter infection. J. Microbiol. Immunol. Infect. 45, 255–258. doi: 10.1016/j.jmii.2011.09.016
DiBardino, D. M., Wunderink, R. G. (2015). Aspiration pneumonia: a review of modern trends. J. Crit. Care 30, 40–48. doi: 10.1016/j.jcrc.2014.07.011
Diez-Aguilar, M., Ruiz-Garbajosa, P., Fernandez-Olmos, A., Guisado, P., Del Campo, R., Quereda, C., et al. (2013). Non-diphtheriae Corynebacterium species: an emerging respiratory pathogen. Eur. J. Clin. Microbiol. Infect. Dis. 32, 769–772. doi: 10.1007/s10096-012-1805-5
Donahue, E. H., Dawson, L. F., Valiente, E., Firth-Clark, S., Major, M. R., Littler, E., et al. (2014). Clostridium difficile has a single sortase, SrtB, that can be inhibited by small-molecule inhibitors. BMC Microbiol. 14, 219. doi: 10.1186/s12866-014-0219-1
Dragomirescu, C. C., Lixandru, B. E., Coldea, I. L., Corneli, O. N., Pana, M., Palade, A. M., et al. (2020). Antimicrobial susceptibility testing for corynebacterium species isolated from clinical samples in Romania. Antibio. (Basel) 9. doi: 10.3390/antibiotics9010031
Folliero, V., Dell’Annunziata, F., Roscetto, E., Cammarota, M., De Filippis, A., Schiraldi, C., et al. (2022). Niclosamide as a Repurposing Drug against Corynebacterium striatum Multidrug-Resistant Infections. Antibio. (Basel). doi: 10.3390/antibiotics11050651
Funke, G., von Graevenitz, A., Clarridge, J. E., 3rd, Bernard, K. A. (1997). Clinical microbiology of coryneform bacteria. Clin. Microbiol. Rev. 10, 125–159. doi: 10.1128/CMR.10.1.125
Galimand, M., Fishovitz, J., Lambert, T., Barbe, V., Zajicek, J., Mobashery, S., et al. (2015). AAC(3)-XI, a new aminoglycoside 3-N-acetyltransferase from Corynebacterium striatum. Antimicrob. Agents Chemother. 59, 5647–5653. doi: 10.1128/AAC.01203-15
Goldner, N. K., Bulow, C., Cho, K., Wallace, M., Hsu, F. F., Patti, G. J., et al. (2018). Mechanism of high-level daptomycin resistance in corynebacterium striatum. mSphere 3. doi: 10.1128/mSphereDirect.00371-18
Gomila, M., Renom, F., Gallegos Mdel, C., Garau, M., Guerrero, D., Soriano, J. B., et al. (2012). Identification and diversity of multiresistant Corynebacterium striatum clinical isolates by MALDI-TOF mass spectrometry and by a multigene sequencing approach. BMC Microbiol. 12, 52. doi: 10.1186/1471-2180-12-52
Hachmann, A. B., Sevim, E., Gaballa, A., Popham, D. L., Antelmann, H., Helmann, J. D. (2011). Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55, 4326–4337. doi: 10.1128/AAC.01819-10
Hagiya, H., Kimura, K., Okuno, H., Hamaguchi, S., Morii, D., Yoshida, H., et al. (2019). Bacteremia due to high-level daptomycin-resistant Corynebacterium striatum: A case report with genetic investigation. J. Infect. Chemother. 25, 906–908. doi: 10.1016/j.jiac.2019.04.009
Hennart, M., Panunzi, L. G., Rodrigues, C., Gaday, Q., Baines, S. L., Barros-Pinkelnig, M., et al. (2020). Population genomics and antimicrobial resistance in Corynebacterium diphtheriae. Genome Med. 12, 107. doi: 10.1186/s13073-020-00805-7
Hines, K. M., Waalkes, A., Penewit, K., Holmes, E. A., Salipante, S. J., Werth, B. J., et al. (2017). Characterization of the mechanisms of daptomycin resistance among gram-positive bacterial pathogens by multidimensional lipidomics. mSphere 2. doi: 10.1128/mSphere.00492-17
Iaria, C., Stassi, G., Costa, G. B., Biondo, C., Gerace, E., Noto, A., et al. (2007). Outbreak of multi-resistant Corynebacterium striatum infection in an Italian general intensive care unit. J. Hosp Infect. 67, 102–104. doi: 10.1016/j.jhin.2007.07.002
Jesus, H. N. R., Ramos, J. N., Rocha, D., Alves, D. A., Silva, C. S., Cruz, J. V. O., et al. (2022). The pan-genome of the emerging multidrug-resistant pathogen Corynebacterium striatum. Funct. Integr. Genomics 23, 5. doi: 10.21203/rs.3.rs-1666801/v1
Kang, C. Y., Huang, I. H., Chou, C. C., Wu, T. Y., Chang, J. C., Hsiao, Y. Y., et al. (2020). Functional analysis of Clostridium difficile sortase B reveals key residues for catalytic activity and substrate specificity. J. Biol. Chem. 295, 3734–3745. doi: 10.1074/jbc.RA119.011322
Kang, H. J., Paterson, N. G., Kim, C. U., Middleditch, M., Chang, C., Ton-That, H., et al. (2014). A slow-forming isopeptide bond in the structure of the major pilin SpaD from Corynebacterium diphtheriae has implications for pilus assembly. Acta Crystallogr. D Biol. Crystallogr. 70, 1190–1201. doi: 10.1107/S1399004714001400
Kang, S. J., Choi, S. M., Choi, J. A., Choi, J. U., Oh, T. H., Kim, S. E., et al. (2018). Factors affecting the clinical relevance of Corynebacterium striatum isolated from blood cultures. PloS One 13, e0199454. doi: 10.1371/journal.pone.0199454
Kang, Y., Chen, S., Zheng, B., Du, X., Li, Z., Tan, Z., et al. (2023). Epidemiological investigation of hospital transmission of corynebacterium striatum infection by core genome multilocus sequence typing approach. Microbiol. Spectr. 11, e0149022. doi: 10.1128/spectrum.01490-22
Kono, M., Sasatsu, M., Aoki, T. (1983). R plasmids in Corynebacterium xerosis strains. Antimicrob. Agents Chemother. 23, 506–508. doi: 10.1128/AAC.23.3.506
Lee, E. H., Degener, J. E., Welling, G. W., Veloo, A. C. (2011). Evaluation of the Vitek 2 ANC card for identification of clinical isolates of anaerobic bacteria. J. Clin. Microbiol. 49, 1745–1749. doi: 10.1128/JCM.02166-10
Lee, P. P., Ferguson, D. A., Jr., Sarubbi, F. A. (2005). Corynebacterium striatum: an underappreciated community and nosocomial pathogen. J. Infect. 50, 338–343. doi: 10.1016/j.jinf.2004.05.005
Lee, Y. W., Huh, J. W., Hong, S. B., Jung, J., Kim, M. J., Chong, Y. P., et al. (2022). Severe pneumonia caused by corynebacterium striatum in adults, seoul, South Korea, 2014-2019. Emerg. Infect. Dis. 28, 2147–2154. doi: 10.3201/eid2811.220273
Leyton, B., Ramos, J. N., Baio, P. V. P., Veras, J. F. C., Souza, C., Burkovski, A., et al. (2021). Treat me well or will resist: uptake of mobile genetic elements determine the resistome of corynebacterium striatum. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22147499
Marino, A., Campanella, E., Stracquadanio, S., Ceccarelli, M., Zagami, A., Nunnari, G., et al. (2022). Corynebacterium striatum Bacteremia during SARS-CoV2 Infection: Case Report, Literature Review, and Clinical Considerations. Infect. Dis. Rep. 14, 383–390. doi: 10.3390/idr14030042
Martinez-Martinez, L., Suarez, A. I., Rodriguez-Bano, J., Bernard, K., Muniain, M. A. (1997). Clinical significance of Corynebacterium striatum isolated from human samples. Clin. Microbiol. Infect. 3, 634–639. doi: 10.1111/j.1469-0691.1997.tb00470.x
Milosavljevic, M. N., Milosavljevic, J. Z., Kocovic, A. G., Stefanovic, S. M., Jankovic, S. M., Djesevic, M., et al. (2021). Antimicrobial treatment of Corynebacterium striatum invasive infections: a systematic review. Rev. Inst Med. Trop. Sao Paulo 63, e49. doi: 10.1590/s1678-9946202163049
Neill, S., Dean, N. (2019). Aspiration pneumonia and pneumonitis: a spectrum of infectious/noninfectious diseases affecting the lung. Curr. Opin. Infect. Dis. 32, 152–157. doi: 10.1097/QCO.0000000000000524
Nesvera, J., Hochmannova, J., Patek, M. (1998). An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169, 391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x
Nudel, K., Zhao, X., Basu, S., Dong, X., Hoffmann, M., Feldgarden, M., et al. (2018). Genomics of Corynebacterium striatum, an emerging multidrug-resistant pathogen of immunocompromised patients. Clin. Microbiol. Infect. 24, 1016 e1017–1016 e1013. doi: 10.1016/j.cmi.2017.12.024
Ocalan, F., Mumcuoglu, I., Unaldi, O., Bakkaloglu, Z., Dinc, B. (2023). Evaluation of antibiotic susceptibility, biofilm production and clonal analysis of corynebacterium striatum isolates. Mikrobiyol Bul 57, 1–13.
Orosz, L., Soki, J., Kokai, D., Burian, K. (2022). Corynebacterium striatum-got worse by a pandemic? Pathogens 11. doi: 10.3390/pathogens11060685
Otsuka, Y., Ohkusu, K., Kawamura, Y., Baba, S., Ezaki, T., Kimura, S. (2006). Emergence of multidrug-resistant Corynebacterium striatum as a nosocomial pathogen in long-term hospitalized patients with underlying diseases. Diagn. Microbiol. Infect. Dis. 54, 109–114. doi: 10.1016/j.diagmicrobio.2005.08.005
Qin, T., Geng, T., Zhou, H., Han, Y., Ren, H., Qiu, Z., et al. (2020). Super-dominant pathobiontic bacteria in the nasopharyngeal microbiota as causative agents of secondary bacterial infection in influenza patients. Emerg. Microbes Infect. 9, 605–615. doi: 10.1080/22221751.2020.1737578
Qiu, J., Shi, Y., Zhao, F., Xu, Y., Xu, H., Dai, Y., et al. (2023). The Pan-Genomic Analysis of Corynebacterium striatum Revealed its Genetic Characteristics as an Emerging Multidrug-Resistant Pathogen. Evol. Bioinform. Online 19, 11769343231191481. doi: 10.1177/11769343231191481
Ramirez, M. S., Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Update 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ramirez de Arellano, E., Pascual, A., Martinez-Martinez, L., Perea, E. J. (1995). Antimicrobial activity in Staphylococcus epidermidis biofilms on polyvinyl chloride catheters. Enferm Infecc Microbiol. Clin. 13, 581–586.
Ramos, J. N., Rodrigues, I. D. S., Baio, P. V. P., Veras, J. F. C., Ramos, R. T. J., Pacheco, L. G., et al. (2018). Genome sequence of a multidrug-resistant Corynebacterium striatum isolated from bloodstream infection from a nosocomial outbreak in Rio de Janeiro, Brazil. Mem Inst Oswaldo Cruz 113, e180051. doi: 10.1590/0074-02760180051
Ramos, J. N., Souza, C., Faria, Y. V., da Silva, E. C., Veras, J. F. C., Baio, P. V. P., et al. (2019). Bloodstream and catheter-related infections due to different clones of multidrug-resistant and biofilm producer Corynebacterium striatum. BMC Infect. Dis. 19, 672. doi: 10.1186/s12879-019-4294-7
Ramos, J. N., Valadao, T. B., Baio, P. V. P., Mattos-Guaraldi, A. L., Vieira, V. V. (2020). Novel mutations in the QRDR region gyrA gene in multidrug-resistance Corynebacterium spp. isolates from intravenous sites. Antonie Van Leeuwenhoek 113, 589–592. doi: 10.1007/s10482-019-01353-w
Renom, F., Gomila, M., Garau, M., Gallegos, M. D., Guerrero, D., Lalucat, J., et al. (2014). Respiratory infection by Corynebacterium striatum: epidemiological and clinical determinants. New Microbes New Infect. 2, 106–114. doi: 10.1002/nmi2.48
Sangal, V., Marrs, E. C. L., Nelson, A., Perry, J. D. (2024). Phylogenomic analyses of multidrug resistant Corynebacterium striatum strains isolated from patients in a tertiary care hospital in the UK. Eur. J. Clin. Microbiol. Infect. Dis. 43, 1495–1501. doi: 10.1007/s10096-024-04857-0
Severo, C. B., Guazzelli, L. S., Barra, M. B., Hochhegger, B., Severo, L. C. (2014). Multiple pulmonary nodules caused by Corynebacterium striatum in an immunocompetent patient. Rev. Inst Med. Trop. Sao Paulo 56, 89–91. doi: 10.1590/S0036-46652014000100015
Shariff, M., Aditi, A., Beri, K. (2018). Corynebacterium striatum: an emerging respiratory pathogen. J. Infect. Dev. Ctries 12, 581–586. doi: 10.3855/jidc.10406
Silva-Santana, G., Silva, C. M. F., Olivella, J. G. B., Silva, I. F., Fernandes, L. M. O., Sued-Karam, B. R., et al. (2021). Worldwide survey of Corynebacterium striatum increasingly associated with human invasive infections, nosocomial outbreak, and antimicrobial multidrug-resistance, 1976-2020. Arch. Microbiol. 203, 1863–1880. doi: 10.1007/s00203-021-02246-1
Smith, C. A., Bhattacharya, M., Toth, M., Stewart, N. K., Vakulenko, S. B. (2017). Aminoglycoside resistance profile and structural architecture of the aminoglycoside acetyltransferase AAC(6’)-Im. Microb. Cell 4, 402–410. doi: 10.15698/mic2017.12.602
Soderquist, B., Henningsson, T., Stegger, M. (2023). Corynebacterium striatum prosthetic joint infection successfully treated with long-term dalbavancin. Microorganisms 11. doi: 10.3390/microorganisms11030550
Souza, C., Faria, Y. V., Sant’Anna Lde, O., Viana, V. G., Seabra, S. H., Souza, M. C., et al. (2015). Biofilm production by multiresistant Corynebacterium striatum associated with nosocomial outbreak. Mem Inst Oswaldo Cruz 110, 242–248. doi: 10.1590/0074-02760140373
Souza, C., Mota, H. F., Faria, Y. V., Cabral, F. O., Oliveira, D. R., Sant’Anna, L. O., et al. (2020). Resistance to antiseptics and disinfectants of planktonic and biofilm-associated forms of corynebacterium striatum. Microb. Drug Resist. 26, 1546–1558. doi: 10.1089/mdr.2019.0124
Suh, J. W., Ju, Y., Lee, C. K., Sohn, J. W., Kim, M. J., Yoon, Y. K. (2019). Molecular epidemiology and clinical significance of Corynebacterium striatum isolated from clinical specimens. Infect. Drug Resist. 12, 161–171. doi: 10.2147/IDR.S184518
Superti, S. V., Martins Dde, S., Caierao, J., Soares, F., Prochnow, T., Cantarelli, V. V., et al. (2009). Corynebacterium striatum infecting a Malignant cutaneous lesion: the emergence of an opportunistic pathogen. Rev. Inst Med. Trop. Sao Paulo 51, 115–116. doi: 10.1590/S0036-46652009000200011
Szemraj, M., Czekaj, T., Kalisz, J., Szewczyk, E. M. (2019). Differences in distribution of MLS antibiotics resistance genes in clinical isolates of staphylococci belonging to species: S. epidermidis, S. hominis, S. haemolyticus, S. simulans and S. warneri. BMC Microbiol. 19, 124.
Szemraj, M., Kwaszewska, A., Szewczyk, E. M. (2018). New gene responsible for resistance of clinical corynebacteria to macrolide, lincosamide and streptogramin B. Pol. J. Microbiol. 67, 237–240. doi: 10.21307/pjm-2018-028
Tarr, P. E., Stock, F., Cooke, R. H., Fedorko, D. P., Lucey, D. R. (2003). Multidrug-resistant Corynebacterium striatum pneumonia in a heart transplant recipient. Transpl. Infect. Dis. 5, 53–58. doi: 10.1034/j.1399-3062.2003.00002.x
Tauch, A., Bischoff, N., Brune, I., Kalinowski, J. (2003). Insights into the genetic organization of the Corynebacterium diphtheriae erythromycin resistance plasmid pNG2 deduced from its complete nucleotide sequence. Plasmid 49, 63–74. doi: 10.1016/S0147-619X(02)00115-4
Torres Lde, F., Ribeiro, D., Hirata, R., Jr., Pacheco, L. G., Souza, M. C., dos Santos, L. S., et al. (2013). Multiplex polymerase chain reaction to identify and determine the toxigenicity of Corynebacterium spp with zoonotic potential and an overview of human and animal infections. Mem Inst Oswaldo Cruz 108, 272–279. doi: 10.1590/S0074-02762013000300003
Trost, E., Al-Dilaimi, A., Papavasiliou, P., Schneider, J., Viehoever, P., Burkovski, A., et al. (2011). Comparative analysis of two complete Corynebacterium ulcerans genomes and detection of candidate virulence factors. BMC Genomics 12, 383. doi: 10.1186/1471-2164-12-383
Verroken, A., Bauraing, C., Deplano, A., Bogaerts, P., Huang, D., Wauters, G., et al. (2014). Epidemiological investigation of a nosocomial outbreak of multidrug-resistant Corynebacterium striatum at one Belgian university hospital. Clin. Microbiol. Infect. 20, 44–50. doi: 10.1111/1469-0691.12197
Wang, J., Pei, J., Liu, M., Huang, R., Li, J., Liao, S., et al. (2022). Identification and evolutionary relationship of corynebacterium striatum clinical isolates. Pathogens 11. doi: 10.3390/pathogens11091012
Wang, J., Wang, Y., Du, X., Cui, J., Wang, K., Zhang, L., et al. (2016). Rapid transmission of multidrug-resistant Corynebacterium striatum among susceptible patients in a tertiary hospital in China. J. Infect. Dev. Ctries 10, 1299–1305. doi: 10.3855/jidc.7577
Wang, J., Zhang, M., Pei, J., Yi, W., Fan, L., Wang, C., et al. (2024). Isolation and identification of a novel phage targeting clinical multidrug-resistant Corynebacterium striatum isolates. Front. Cell Infect. Microbiol. 14, 1361045. doi: 10.3389/fcimb.2024.1361045
Wang, X., Zhou, H., Du, P., Lan, R., Chen, D., Dong, A., et al. (2021). Genomic epidemiology of Corynebacterium striatum from three regions of China: an emerging national nosocomial epidemic. J. Hosp Infect. 110, 67–75. doi: 10.1016/j.jhin.2020.10.005
Wong, K. Y., Chan, Y. C., Wong, C. Y. (2010). Corynebacterium striatum as an emerging pathogen. J. Hosp Infect. 76, 371–372. doi: 10.1016/j.jhin.2010.05.018
Xu, S., Qiu, X., Hou, X., Zhou, H., Chen, D., Wang, X., et al. (2021). Direct detection of Corynebacterium striatum, Corynebacterium propinquum, and Corynebacterium simulans in sputum samples by high-resolution melt curve analysis. BMC Infect. Dis. 21, 21. doi: 10.1186/s12879-020-05633-z
Xu, H., Zhang, R., Zhang, X., Cheng, Y., Lv, L., Lin, L. (2024). Microbiological profile of patients with aspiration pneumonia identified by combined detection methods. Infect. Drug Resist. 17, 2077–2088. doi: 10.2147/IDR.S461935
Yamamuro, R., Hosokawa, N., Otsuka, Y., Osawa, R. (2021). Clinical characteristics of corynebacterium bacteremia caused by different species, Japan, 2014-2020. Emerg. Infect. Dis. 27, 2981–2987. doi: 10.3201/eid2712.210473
Keywords: Corynebacterium striatum, drug resistance, lower respiratory tract infections, vancomycin therapy, multidrug resistance mechanisms, bacteriophage therapy
Citation: Li W, Gao M and Yu J (2025) Rising prevalence and drug resistance of Corynebacterium striatum in lower respiratory tract infections. Front. Cell. Infect. Microbiol. 14:1526312. doi: 10.3389/fcimb.2024.1526312
Received: 11 November 2024; Accepted: 16 December 2024;
Published: 07 January 2025.
Edited by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyReviewed by:
Diana Alame, Duke University, United StatesCassius Souza, Rio de Janeiro State University, Brazil
Copyright © 2025 Li, Gao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinyan Yu, eXVqaW55YW5Aamx1LmVkdS5jbg==
 Wei Li1
Wei Li1 Jinyan Yu
Jinyan Yu