
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 24 January 2025
Sec. Extra-intestinal Microbiome
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1499203
This article is part of the Research Topic Impact of oral and gut microbiome on health and diseases View all 14 articles
Breast cancer is the most common malignancy in women worldwide. Changes in the microbiota and their metabolites affect the occurrence and development of breast cancer; however, the specific mechanisms are not clear. Gut microbes and their metabolites influence the development of breast cancer by regulating the tumor immune response, estrogen metabolism, chemotherapy, and immunotherapy effects. It was previously thought that there were no microorganisms in breast tissue, but it is now thought that there are microorganisms in breast cancer that can affect the outcome of the disease. This review builds on existing research to comprehensively analyze the role of gut and intratumoral microbiota and their metabolites in the development and metastasis of breast cancer. We also explore the potential function of the microbiota as biomarkers for prognosis and therapeutic response, highlighting the need for further research to clarify the causal relationship between the microbiota and breast cancer. We hope to provide new ideas and directions for the development of new methods for breast cancer treatment.
Global cancer statistics for 2022 indicate that breast cancer will be the most commonly diagnosed cancer among women, with an estimated 2.3 million new cases every year, representing 11.6% of all cancer cases (Bray et al., 2024). With the continuous increase in research on breast cancer and the tumor microenvironment, treatment methods for the disease have become increasingly targeted, typically involving a combination of traditional therapy and novel immune therapy guided by the cancer molecular subtype. Although treatment regimens for breast cancer have been continuously optimized, the therapeutic effect for highly malignant breast cancer is still not ideal, with challenges such as drug resistance, recurrence, and distant metastasis. Therefore, identifying new treatment directions is helpful for the clinical selection of more effective treatment plans.
The large microbiota, composed of bacteria, viruses, and eukaryotes that inhabit the human body, play an important role in maintaining health and disease development. In a healthy human body, the microbiota coexists peacefully with the organism and assists in maintaining health. However, when the composition of the microbiota is unbalanced, diseases, including tumors, may occur (Lynch and Pedersen, 2016; Fan and Pedersen, 2021). These microbial organisms indirectly influence cancer through mechanisms, such as metabolite production and immune system modulation, which affect both distant and proximal tumor tissues (Yang et al., 2023a). Currently, research on tumor-related microorganisms mainly focuses on intestinal microorganisms. These microorganisms and their metabolites are crucial for maintaining the integrity of the intestinal mucosa, nutrient metabolism, immune regulation, and other functions (Li S. et al., 2024). This is particularly evident in colorectal cancer (CRC), where the gut microbiota directly affects the tumor microenvironment (TME) by regulating the immune system, thereby influencing CRC prognosis (Garrett, 2015; Xavier et al., 2020).
Relatively few studies have addressed the involvement of microbiota in breast cancer progression, particularly the impact of intratumoral microorganisms. Significant alterations in the breast microbiota have been detected in patients with malignancy, with notable differences between cancerous tissues and healthy controls, and between benign and malignant breast tissues (Bobin-Dubigeon et al., 2021; Ma et al., 2022). Microbial dysbiosis in other organs may also contribute to breast cancer development. For example, oral dysbiosis-mediated periodontal disease is involved in the development of breast cancer (Jia et al., 2020; Zheng et al., 2022). This indicates that both intratumoral microorganisms and microorganisms in other parts of the body can affect the progression of breast cancer.
This study systematically summarizes the roles of intestinal and intratumoral microorganisms in the development of breast cancer and seeks new ideas for the prevention and treatment of breast cancer.
Advancements in high-throughput sequencing technology have continuously improved the sequencing accuracy of microorganisms while reducing costs. 16S rRNA and metagenomic sequencing are the most widely employed approaches for detecting the distribution and characteristics of microorganisms in patients (Clarridge, 2004; Goodrich et al., 2014; Knight et al., 2018; Liu et al., 2021b). The 16S rRNA gene, which is common to both bacteria and archaea, consists of nine variable regions (V1-V9) and 10 conserved regions arranged alternately. Conserved regions facilitate primer design for gene amplification, whereas variable regions reflect evolutionary differences between species, making the 16S rRNA gene a widely used molecular marker for prokaryotic identification, classification, phylogenetic analysis, and diversity studies.
Compared to 16S rRNA gene sequences, metagenomics provides a broader spectrum of microbial information. An effective approach in metagenomics is the recovery of metagenome-assembled genomes (MAGs), which allow the reconstruction of microbial genomes from metagenomic data (Zhou et al., 2022). Additionally, novel microbiome analysis methods, such as machine learning-based multiomics analysis, have shown promise in predicting the characteristics of the human microbiome related to complex host diseases (Asnicar et al., 2024).
Other sensitive microbial detection techniques, including immunohistochemistry (IHC), fluorescence in situ hybridization (FISH), D-alanine labeling, and tissue isolation culture, can also be used to detect intratumoral bacterial biomass (Xue et al., 2023). IHC detects bacterial samples using antibodies against bacteria-derived lipopolysaccharides (LPS) or lipoteichoic acid (LTA). FISH can identify bacterial DNA in tissues using fluorescent dye-labeled probes that target the 16S rRNA gene (Lin et al., 2021). Furthermore, most bacteria exhibit alanine racemase activity, which is essential for D-alanine biosynthesis and peptidoglycan formation in bacterial cell walls, making D-alanine labeling a useful method for detecting live bacteria in situ.
Alterations in the intestinal microbial diversity can lead to intestinal microecological dysbiosis, thereby promoting the onset of various diseases (Sekirov et al., 2010). Intestinal microecological dysbiosis can affect the prognosis of diseases, including tumors, through multiple mechanisms, such as activating innate immune responses, pro-inflammatory responses, changes in metabolites, regulation of estrogen levels, and alteration of drug metabolism (Goedert et al., 2018; Arnone and Cook, 2022; Niekamp and Kim, 2023; Wang et al., 2023; Bernardo et al., 2023a; Giampazolias et al., 2024). Compared with healthy people, the diversity of intestinal microorganisms in patients with breast cancer is reduced, characterized by a depletion of Bacteroidetes, Odoribacter, Butyricimonas, and Coprococcus and an enrichment of Firmicutes, Acidaminococcus, Tyzzerella, Hungatella, Porphyromonas, and Peptoniphilus (Bobin-Dubigeon et al., 2021; Ma et al., 2022; Song et al., 2022; Altinok Dindar et al., 2023; Amaro-da-Cruz et al., 2024). These studies indicated that intestinal microecological dysbiosis may affect the occurrence, development, and therapeutic effects of breast cancer through multiple pathways.
Alterations in anti-tumor immune responses and chronic inflammation are important factors influencing the occurrence of various tumors. Microbial dysbiosis (gut microbiota that express the enzyme β-glucuronidase), characterized by chronic inflammation and immune evasion, can promote tumorigenesis in breast cancer (Buchta Rosean et al., 2019; Arnone and Cook, 2022). Gut microorganisms (e.g. Proteobacteria and Firmicutes) can modulate the immune system through regulating lymphocyte proliferation via bacterial metabolites and influencing chronic inflammation and estrogen metabolism (Van der Merwe et al., 2021; Zhang et al., 2021). Furthermore, commensal bacteria (e.g. Helicobacter hepaticus) also promotes the occurrence and distant metastasis of breast cancer by influencing the anti-tumor immune functions of IL-6 and neutrophils in the tumor microenvironment (Lakritz et al., 2015; Rutkowski et al., 2015). These studies indicate that immune system disorders induced by intestinal microecological dysbiosis play an important role in the development of breast cancer.
Toll-like receptors (TLRs) are essential components of innate immune responses. They serve as pattern recognition receptors (PRRs) that detect various pathogens, including commensal microbiota. TLR4/MyD88 stimulation by Fusobacterium nucleatum (F. nucleatum) promotes tumor development via NF-κB activation in CRC (Yang et al., 2017; Yu et al., 2017; Zhang et al., 2019). Similar TLR4 and NF-κB activation has been reported in breast cancer cells stimulated by bacterial LPS, leading to the expression of inflammatory factors and apoptotic proteins (Rajput et al., 2013). Moreover, microorganisms can drive malignant progression at extra-mucosal sites via TLR5-dependent signals to increase the expression of systemic IL-6 and immunosuppressive γδ T cells to regulate tumor-promoting inflammation (Rutkowski et al., 2015). These studies indicate that intestinal microorganisms and their metabolites promote the growth and metastasis of breast cancer cells by regulating TLR-mediated innate immune responses.
Estrogen exposure is an important factor that affects the occurrence and development of breast cancer. The “estrobolome” refers to the collection of intestinal bacterial genes whose metabolites can metabolize estrogen (Plottel and Blaser, 2011). Estrogen is mainly produced by the ovaries, adrenal glands, and adipose tissues, circulates in the blood in the form of free or conjugated estrogen, and combines with its metabolites in the liver to form conjugated estrogen. Conjugated estrogen is metabolized into water-soluble molecules and excreted in urine or bile. The conjugated estrogen in bile can be decomposed by bacteria with β-glucuronidase activity in the intestine, and after reabsorption and re-entry into the circulation, it increases the bioavailability of estrogen. Circulating estrogen stimulates growth and proliferation of breast cells (Kwa et al., 2016). Therefore, the enterohepatic circulation of estrogen can affect the levels of estrogen and its metabolites in the circulatory system and may ultimately contribute to the risk of hormone-driven breast cancer.
The process by which the intestinal “estrobolome” regulates the enterohepatic circulation and reabsorption of estrogen is also influenced by host factors, such as age, diet, and antibiotics. The effects of antibiotics and diet are discussed below.
The use of antibiotics can affect the diversity and quantity of the flora in the body. Improper use can lead to dysbiosis of the intestinal flora and various diseases, including breast cancer (Velicer et al., 2004; Garcia Rodriguez and Gonzalez-Perez, 2005; Sorensen et al., 2005; Friedman et al., 2006). Ampicillin and oxytetracycline can increase the content of conjugated estrogen in the feces of women and men, respectively, while reducing estrogen in the urine (Adlercreutz et al., 1975; Martin et al., 1975; Hamalainen et al., 1987). Increased antibiotic exposure may also increase the risk of breast cancer (Velicer et al., 2004; Friedman et al., 2006). These studies indicate that certain antibiotics ultimately affect the risk of breast cancer by regulating estrogen excretion and influencing the deconjugative activity of intestinal bacteria. However, the mechanism by which antibiotics affect the development of breast cancer through the intestinal flora remains unclear and requires further research.
Although factors such as lifestyle, exercise, and supplements can affect estrogen levels, diet remains a major factor influencing the overall estrogen concentration, potentially through the modulation of gut microbiome composition and function (Muegge et al., 2011). As early as 1982, Goldin et al. found that vegetarians excreted higher levels of conjugated estrogens in feces than non-vegetarians, resulting in lower plasma estrogen levels (Goldin et al., 1982).
Adiposity has been linked to higher serum estrogen levels in postmenopausal women, which are correlated with an increased risk of multiple malignancies (Keum et al., 2015). High-fat diet (HFD) disrupts gastrointestinal metabolism and immune homeostasis and contributes to disease states. Soto-Pantoja et al. found that HFD mice and mice that received fecal transplantation from HFD-fed mice exhibited an increased Firmicutes/Bacteroidetes (F/B) ratio (Soto-Pantoja et al., 2021). Microbiota changes observed in genetically obese mice are consistent with those observed in obese humans (Ley et al., 2005, 2006). This altered ratio increases the abundance of harmful bacteria, leading to the release of enterotoxins and chronic low-grade inflammation (Mikó et al., 2019; Parida and Sharma, 2019). The HFD promotes cancer progression by inducing gut microbiota-mediated leucine production and polymorphonuclear myeloid-derived suppressor cell differentiation (Chen et al., 2024b). The above studies indicate that the gut-breast signaling axis is involved in regulating the influence of diet on breast cancer risk, which provides a reference for guiding the daily dietary intake of breast cancer patients and women at a high risk of breast cancer.
The gut microbiome plays a significant regulatory role in modulating responses to both traditional and immune therapies (Battaglia et al., 2024). They can regulate local inflammation and gut barrier function by targeting drug metabolism, modulating immune responses, and secreting different metabolites, ultimately affecting chemotherapy outcomes (Alexander et al., 2017; Sampsell et al., 2020). Chemotherapy drugs may also affect chemotherapy-induced weight gain and neurological side effects by regulating microbial diversity (Terrisse et al., 2021).
HER2 inhibitors such as trastuzumab have a good therapeutic effect on HER2-positive breast cancer. However, antibiotic treatment changes the composition of the intestinal flora, reduces dendritic cell activation and IL12p70 secretion, leads to changes in anti-tumor immunity in the tumor microenvironment, and ultimately reduces the therapeutic activity of trastuzumab (Di Modica et al., 2021). This study indicates that intestinal microbial dysregulation can regulate the host immune system and ultimately affect the therapeutic effects of chemotherapeutic drugs.
The immune checkpoint blockade (ICB) is a new-generation immunotherapeutic strategy for various cancers. Commensal gut bacteria can suppress inflammation, reshape primary and acquired immune responses, and reprogram the TME in murine models and patients, thereby influencing ICB efficacy (Xue et al., 2024). Jia et al. reported that the gut microbial metabolite indolepropionic acid (IPA) enhances immunotherapy efficacy by modulating T cell stemness in cancers (Jia et al., 2024). Additionally, using a murine model of gut microbiota dysbiosis, Shi et al. found that Lactobacillus and its metabolite lactic acid promote breast cancer progression, particularly triple-negative breast cancer (TNBC), by affecting the anti-tumor activities of immune cells in the TME (Shi et al., 2023). These results indicate that intestinal microbiota is expected to become a potential target for breast cancer treatment.
A large part of the physiological regulatory function of microorganisms is exerted through their metabolites such as short-chain fatty acids (SCFAs), bile acids, and inosine. These metabolites enter blood circulation, serve as significant modulators of the TME, and influence immune cell differentiation signals and the release of substances from both immune cells and tumors (Jaye et al., 2022a; Yang et al., 2023b).
SCFAs, primarily butyrate, propionate, and acetate, are produced by microbiotal dietary fiber digestion. They are tumor suppressors in various cancer types, particularly colon cancer. SCFAs are the most common gut microbial metabolites and are mainly produced by intestine-colonizing species, such as Eubacterium rectale, Clostridium leptum, and Faecalibacterium prausnitzii (Williams et al., 2017; Jaye et al., 2022b). The total concentration of SCFAs exceeding 100 mM in the intestine include propionate, acetate, and butyrate (Mirzaei et al., 2021). Butyrate, one of the most abundant SCFAs, has a dual effect on cancer cell proliferation, which is largely dependent on its concentration; low concentrations may promote carcinogenesis, whereas higher concentrations may inhibit tumorigenesis (Jaye et al., 2022b). Butyrate has demonstrated strong inhibitory effects on various breast cancer cell lines (Rodrigues et al., 2015; Semaan et al., 2020; Jaye et al., 2023). For example, sodium butyrate suppresses breast cancer cells by inducing cell cycle arrest at the G2/M phase, increasing caspase-10 levels, promoting apoptosis, and initiating intracellular calcium influx (Jaye et al., 2022a). Microbiota-derived butyrate can influence tumor progression by reshaping the TME. Butyrate’s interaction with its receptor Gpr109a exerts anti-inflammatory effects on colonic macrophages and dendritic cells, inducing regulatory T cell (Treg) differentiation and T cell production of IL-10, thereby suppressing colonic inflammation and cancer progression (Singh et al., 2014). Additionally, butyrate enhances ICB efficacy by modulating T cell receptor signaling in CD8+ T cells, indicating its potential as a therapeutic biomarker (Zhu et al., 2023). Despite butyrate’s considerable anti-tumor effects, low bioavailability and dose-dependent side effects have limited its clinical application. Nanoparticle-based delivery systems for butyrate may overcome these challenges (Yu et al., 2023).
Most primary bile acids are reabsorbed in the small intestine, returned to the liver via the portal vein, and secreted back into the bile through enterohepatic circulation. Primary bile acids can also be metabolized by gut microbiota into secondary bile acids through deconjugation and dehydrogenation. Because breast cells do not produce bile acids, the presence of secondary bile acids in breast cancer tissues likely results from minimal leakage from the enterohepatic circulation or local production by the microbiome within the breast tissue. Wu et al. analyzed the transcriptomic and clinical case information of three large open primary breast cancer cohorts as well as the microbiome data of 16S rRNA gene sequences in TCGA breast cancer tissues. They found that breast tumors with low bile acid metabolism were more aggressive, and that there were a large number of microorganisms related to aggressive tumor biology in the TME. In breast tumors with high bile acid metabolism, oxidative stress-induced apoptosis leads to a significant increase in survival rate (Wu et al., 2022). This study indicates that bile acid metabolism usually inhibits tumor growth in breast cancer.
Sodium deoxycholate (DC), synthesized by intestinal bacteria, typically maintains a serum concentration of 5-10 μmol/L. However, in breast cyst fluid, DC levels can increase to more than 50 μmol/L. Although DC has been implicated in the promotion of colon carcinogenesis (Debruyne et al., 2002), its effects on breast cancer are complex.
DC plays a dual role in breast cancer epithelial cells, with lower concentrations promoting cell proliferation, likely through AKT phosphorylation and cyclin D1 expression, and higher concentrations inducing apoptosis and sustained activation of p38 and AKT (Gándola et al., 2020). This study suggests that the effects of bile salts on breast cancer cells are concentration-dependent. In a metastatic murine breast cancer model, DC act as natural tumor promoters by increasing Flk-1 and reducing ceramide-mediated progenitor cell apoptosis (Krishnamurthy et al., 2008).
Moreover, DC has been implicated in paclitaxel treatment-related peripheral neuropathy in patients with breast cancer. Paclitaxel treatment leads to the ingrowth of Clostridium species and increased DC levels. DC appear to elevate serum levels of CCL5/CCR5 in the dorsal root ganglion through the bile acid receptor TGR5, contributing to neuronal hyperexcitability and neuropathic pain (Zhong et al., 2023).
Breast tissue was once considered a sterile environment. However, with an increasing number of studies focusing on the relationship between microorganisms and breast diseases, it has been found that breast tissue contains a variety of microbiota. The breast is primarily composed of adipose tissue with a rich vascular and lymphatic network, creating a conducive environment for bacterial growth, particularly of the phyla Proteobacteria and Firmicutes. Streptococcus, Enterococcus, and Staphylococcus can be isolated from both breast cancer and normal breast tissues, but the relative abundance at the genus level is very different, the expression of these microbiota in breast cancer tissue is significantly increased (Urbaniak et al., 2014, 2016; Thompson et al., 2017; Nejman et al., 2020; Fu et al., 2022). At the phylum level, Proteobacteria is predominant, followed by Firmicutes, Bacteroidetes, and Actinobacteria. The breast microbiome maintains healthy breast tissues by stimulating the resident immune cells (Xuan et al., 2014). Although the biomass of the intratumoral microbiota is very low, it plays an important role in promoting breast cancer progression.
The origin and colonization of intratumoral microbiota remain uncertain. Microbiota may colonize tumor tissues through three primary mechanisms: mucosal destruction, hematogenic invasion, and adjacent tissue migration (Cao et al., 2024; Guo et al., 2024). Additionally, the immunosuppressive, hypoxic, and nutrient-rich environments in tumors may facilitate microbial colonization and reproduction (Figure 1).
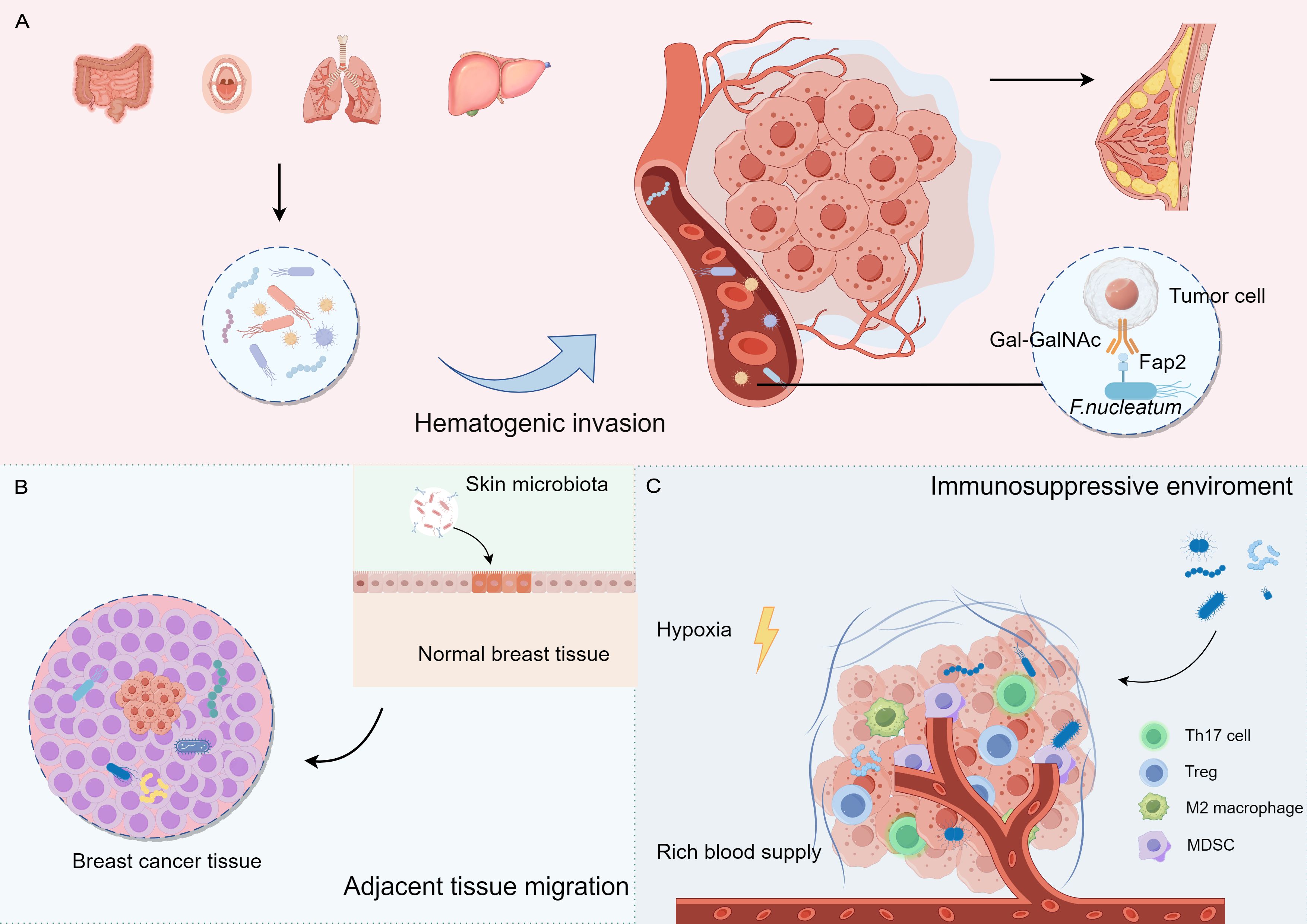
Figure 1. The potential origins and colonization of intratumoural microbiota in breast cancer. (A) Hematogenic invasion: Microorganisms from distant organs may reach breast tissue through the blood. (B) Adjacent tissue migration: Microorganisms from adjacent tissues may migrate to breast tissue (e.g., skin surface microorganisms). (C) Tumor microenvironment: tumor microenvironment (e.g., hypoxia, immunosuppression) may contribute to the colonization of microorganisms in the tumor tissue.
F. nucleatum primarily presented in the oral cavity and gastrointestinal tract, has been implicated in various cancers, including breast cancer. This bacterium may promote tumorigenesis by affecting the infiltration and function of immune cells. Transient bacteremia during periodontal disease can facilitate oral F. nucleatum invasion into the bloodstream, leading to its translocation to the mammary glands. F. nucleatum can dock to tumor tissues via its surface-exposed lectin, Fap2, which recognizes the host Gal-GalNAc and enhances breast tumor growth and metastatic progression (Abed et al., 2016; Parhi et al., 2020). In addition, Bacteroides have been detected in canine breast tumors, as well as in the oral and gut microbiomes, suggesting a potential route of dissemination from the mouth to the gastrointestinal tract and ultimately to distant mammary tissue (Zheng et al., 2022). These studies indicate that oral microorganisms are a source of colonizing microorganisms in the breasts.
Adjacent normal tissues may also serve as potential sources of the intratumoral microbiota. The skin-related bacteria Staphylococcus epidermidis and Micrococcus luteus have been identified in mammary tumors, suggesting that these microbes may access the mammary duct through the nipple and disseminate to the mammary gland via the lobules and ducts (Urbaniak et al., 2014; Bernardo et al., 2023a). The similarities between the microbiome communities in tumors and adjacent normal tissues support the possibility of adjacent tissue invasion.
Methylobacterium radiotolerans, Bacillus, Enterobacteriaceae, Staphylococcus, Ralstonia, Bacillaceae, and Burkholderiaceae are more abundant in breast cancer and their adjacent tissues, whereas Sphingomonas yanoikuyae, Acetobacter aceti, Lactobacillus vini, and Lactobacillus paracasei are more abundant in healthy breast tissues (Xuan et al., 2014; Urbaniak et al., 2016; Hoskinson et al., 2022; German et al., 2023). Analysis of microbiome-immune networks in the breast has revealed that Anaerococcus, Caulobacter, and Streptococcus are crucial hubs in benign tissues but are absent in tumor tissues (Tzeng et al., 2021). Taken together, these studies indicate that both healthy breast tissues and breast cancer tissues have unique microbial environments.
The intratumoral microbiota of patients with breast cancer also varies according to race and sex. Metagenomic analyses have identified race-associated microbial biomarkers, such as Pseudomonas and Methylobacter in tumors from Asian women, and Amycolatopsis in tumors from black women (Siddharth et al., 2021; Parida et al., 2023b). Notably, Ralstonia was most enriched in non-Hispanic Black patients, whereas Xanthomonadales were more prevalent in non-Hispanic white patients (Smith et al., 2019). Differences in the breast microbiomes of men and women have also been reported. Tenericutes, particularly Mesoplasma and Mycobacterium, are implicated in breast carcinogenesis in both sexes (Niccolai et al., 2023). Dysbiosis extends throughout the breast tissue in females and is more localized to the tumor site in males.
Microbial signatures differ among breast cancer subtypes. Actinomyces signatures have been detected across all breast cancer types, with higher signal intensities observed in patients with HER2+ breast cancer (Banerjee et al., 2018). A larger cohort analysis revealed that each subtype had unique microbial signatures, with ER+ breast cancer showing the most diverse tumor microbiome and TNBC exhibiting the least diversity. Notably, higher abundances of Bacillus, Mucor, Nodaviridae, Toxocara, and Trichophyton in TNBC samples were significantly correlated with a better prognosis (Banerjee et al., 2021).
Although the direct effects of the intratumoral microbiota on breast cancer have been less frequently reported, evidence suggests that the intratumoral microbiota may regulate the development of breast cancer through inducing genomic instability and DNA mutations, activating carcinogenic pathways, promoting inflammatory responses, and modulating the local immune microenvironment to facilitate invasion (Xie et al., 2022; Yang et al., 2023a; Cao et al., 2024) (Figure 2).
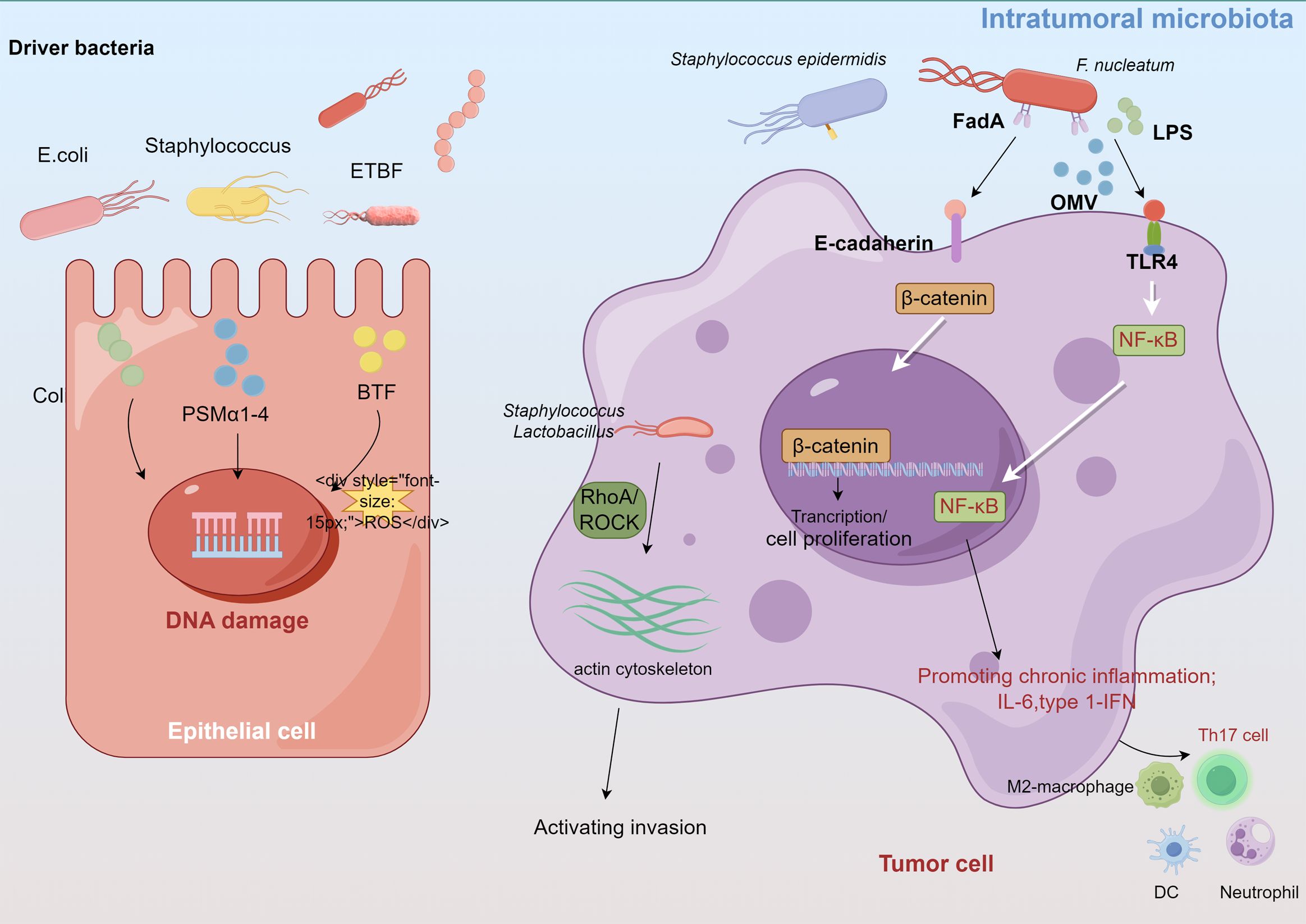
Figure 2. Intratumoral microbiota affect tumor development through several proposed mechanisms, such as DNA damage, promoting immune and inflammation, activating invasion and metastasis.
Certain carcinogenic bacteria, such as pks+ Escherichia coli and Staphylococcus aureus, encode and secrete cytolytic toxins (e.g., colibactin, PSMα1-4) that cause ROS-mediated DNA damage and accelerate tumor onset (Dejea et al., 2018; Krueger et al., 2022). These bacteria, which belong to the Enterobacteriaceae and Staphylococcus genera, are enriched in breast cancer tissues. Guo et al. found that F. nucleatum, abundant in the human breast cancer microbiome, secretes adhesin FadA to activate the E-cadherin/β-catenin pathway, upregulate Chk2 levels and induce DNA damage (Guo et al., 2020a). Moreover, the gut-colonizing bacterium enterotoxigenic Bacteroides fragilis (ETBF) in the mammary gland increases the expression of spermine oxidase in intestinal epithelial cells, leading to ROS production and γ-H2A activation that causes DNA damage (Goodwin et al., 2011; Parida et al., 2021).
Dysregulated innate immunity in cancer often causes persistent chronic inflammation, which promotes cancer progression and anti-tumor immune resistance. Intratumoral microbiomes can activate inflammatory signals by interacting with pattern recognition receptors (PRRs) such as TLRs in the TME. The intratumoral microbiota in breast cancer significantly influence TLR signaling, particularly through the LPS/TLR4 pathway cascade (Afroz et al., 2022; Wilkie et al., 2022). F. nucleatum and A. actinomycetemcomitans also activate diverse TLRs and NF-κB in bone marrow-derived macrophages, increasing IL-6 production (Park et al., 2014). Untreated murine mammary tumors display increased Staphylococcus abundance, of which isolated Staphylococcus epidermidis demonstrates significant inflammatory activity (Bernardo et al., 2023b). Moreover, bioinformatics analysis revealed that Propionibacterium and Staphylococcus, which are decreased in tumors, correlate negatively with oncogenic immune signatures, whereas Streptococcus and Propionibacterium correlate positively with T cell activation (Tzeng et al., 2021). Mammary metabolism-related microbiota are related to T cell exclusion and immunotherapy responses (Chen et al., 2023).
Intratumoral microorganisms influence both intercellular interactions and the external microenvironment, facilitating distant metastasis (Cao et al., 2024). Fu et al. demonstrated that in mouse models of spontaneous breast tumors (MMTV-PyMT), the intratumoral bacteria, carried by circulating tumor cells, can modulate RhoA/ROCK signaling pathways, alternate the actin cytoskeleton, enhance resistance of host cells to fluid shear stress, and promote lung metastasis in breast cancer, thereby promoting the survival and metastasis of host cells (Fu et al., 2022).
The colonic oncogenic microorganism ETBF, found in carcinogenic breast tissues, secretes a toxin that induces hyperplasia in breast epithelial cells (Parida et al., 2021). ETBF is not present in normal breast tissues. Compared with non-toxigenic Bacteroides fragilis, ETBF colonizing in the breast and intestinal ducts can affect epithelial-mesenchymal transition (EMT) by activating the β-catenin and Notch1 signaling pathways, significantly promoting tumor growth and metastasis. ETBF infection can also trigger systemic inflammation in breast cancer, increasing the levels of proinflammatory and tumorigenic cytokines such as IL-17A and IL-6 (Parida et al., 2023a). These inflammatory changes reshape the TME, create a pre-metastatic niche in the target organs, and promote metastasis to the lungs and liver. F. nucleatum-derived outer membrane vesicles also promote lung metastasis in tumor-bearing mice by altering EMT-related protein levels and activating intracellular autophagy pathways (Chen et al., 2024a). Small extracellular vesicles derived from F. nucleatum in breast cancer facilitate tumor growth and metastasis via TLR4 signaling (Li et al., 2023). Thus, extracellular vehicles play a role in modulating communication between cancer cells and the surrounding microenvironment and distant organs.
A study by Gao et al. demonstrated that F. nucleatum can induce PD-L1 expression through activating STING signals, leading to accumulation of IFN-γ+ CD8+ lymphocytes (Gao et al., 2021). This could potentially improve the efficacy of ICBs in the treatment of CRC. Similarly, the pathogenic bacterium Salmonella typhimurium exerts potent anti-tumor effects by activating the immune system (Zheng et al., 2017; Guo et al., 2020b). These studies indicate that symbiotic microorganisms play important roles in regulating host immune responses and therapeutic effects.
Tumor-resident intracellular microbiome (TRIM) can enhance tumor cell proliferation and metastatic colonization while decreasing chemotherapy efficacy (Sears et al., 2014; Geller et al., 2017; Fu et al., 2022). ETBF-secreted toxins enhance cancer cell stemness and chemoresistance by activating NUMB phosphorylation, which leads to lysosomal degradation and Notch1 activation (Ma et al., 2024). In CRC, F. nucleatum induces resistance to oxaliplatin and 5-fluorouracil (5-FU) by upregulating autophagy through TLR4/MYD88-dependent signals and preventing apoptosis via the upregulation of ANO1/BIRC3 (Yu et al., 2017; Zhang et al., 2019). In addition, F. nucleatum is involved in promoting chemotherapy resistance in esophageal squamous cell carcinoma (Liang et al., 2022) and oral squamous cell carcinoma (Liu et al., 2021a). In an animal model of breast cancer, F. nucleatum colonization was linked to reduced chemotherapy efficacy by activating autophagy-related pathways in cancer cells (Su et al., 2024).
Combining targeted antibacterial treatments with chemotherapy can suppress both TRIM and tumor cells, promoting M2-to-M1-like tumor-associated macrophage repolarization and achieving long-term survival in animal models with no recurrence (Wang et al., 2024). F. nucleatum-mimicking nanovehicles, which fuse cytoplasmic membranes with antibiotic-loaded liposomes, have been shown to selectively target and eradicate tumor-resident bacteria, thereby significantly restoring the effectiveness of chemotherapy (Chen et al., 2024c). The biomimetic nanocarriers improve the immunosuppressive TME induced by intratumoral F. nucleatum and enhance the therapeutic effect of PD-L1 (Geng et al., 2024). These studies indicate that targeting pathogenic microorganisms in tumors may help improve the efficacy of chemotherapy and prevent recurrence.
Additionally, nanoparticles coated with bacteria-derived outer membrane vesicles (OMVs) convert intratumor F. nucleatum into immunopotentiators, releasing pathogen-associated molecular patterns (PAMPs) and enhancing immunochemodynamic therapy efficacy in TNBC (Liu et al., 2023). Encapsulating bacteria-derived extracellular vesicles (BEVs) in nanocloaks can increase immunogenicity and facilitate DC maturation by activating cGAS-STING signaling. This approach, in combination with anti-PD-L1 antibodies, elicits a potent immune response and synergistically inhibits tumor progression and lung metastasis (Zhang et al., 2024). Therefore, bacteria-derived vesicles can effectively improve the immunosuppressive state of the TME, and represent a potential treatment method.
Intratumoral microbiome profiles may serve as valuable tools for predicting patient prognosis and assessing the clinical efficacy of specific drugs. A recent study by Li et al. identified four immune-related intratumor microbiomes (IRIM; e.g., Acidibacillus and Succinimonas) that have potential prognostic value (Li J. et al., 2024). These microbiomes were correlated with immune gene levels and sensitivity to chemotherapeutic agents, particularly tamoxifen and docetaxel.
Microbiota dysbiosis can trigger various immune-mediated diseases by regulating the derived metabolites and host environmental factors. miRNAs have emerged as critical mediators of host-microbiome interactions, with bidirectional effects observed between the microbiome and miRNAs in carcinogenesis (Yang et al., 2017; Yuan et al., 2019; Wang et al., 2021).Yang et al. Found that patients with both high amount of tissue Fusobacterium nucleatum DNA and miR21 had a higher risk of poor prognosis (Yang et al., 2017). Laborda-Illanes et al. revealed an increase in the expression of miR-149-5p, miR-20b-5p, and miR-342-5p in metastatic breast cancer (Met-BC) patients, compare with non-metastatic breast cancer (nonMet-BC) patients (Laborda-Illanes et al., 2024). The Met-BC group exhibited an increase in several pathogenic and pro-inflammatory species, including Streptococcus epidermidis, Haemophilus influenzae, Corynebacterium aurimucosum, and Corynebacterium kroppenstedtii, while the NonMet-BC group displayed higher levels of probiotic bacteria, such as Parabacteroides distasonis, Lactobacillus iners, Blautia obeum, and Faecalibacterium prausnitzii (Laborda-Illanes et al., 2024). These studies suggest that consideration of both intratumoral miRNA expression and microbiota changes could aid in precision treatment for breast cancer.
In summary, the influence of the gut and local microbiota on breast cancer development requires further investigation to elucidate the underlying mechanisms and identify key microbial players (Bernardo et al., 2023a). Challenges such as low microbial biomass, environmental contamination from non-microbial genomes, and antibiotic perturbations persist. Additionally, animal models used in bacterial research may not fully represent the human microbiome owing to differences in diet, genetics, and age. Co-culturing specific bacterial taxa (e.g., Streptococcus, Lactobacillus, Salmonella, Bifidobacterium) with pluripotent stem cells or organoids may be a promising approach for studying host-microbe interactions (Puschhof et al., 2021; Kim et al., 2022; Morelli et al., 2023). Regarding microbiota-related metabolites, patient-derived tumor spheroids could provide insights into the potential therapeutic use of these metabolites; however, effective therapeutic doses need to be determined. Beneficial bacterial or microbiota-based therapies may enhance hormonal, metabolic, and immune regulation in hormone receptor-positive cancers. Strategies such as the administration of prebiotics, probiotics, and fecal FMT could be beneficial (Zitvogel et al., 2008; Terrisse et al., 2023). Microbes that promote tumors can be eradicated using antibiotics or phage therapies.
Future research involving human subjects is crucial to unravel the complex interactions among the microbiome, disease, and the host. The goal is to explore new therapeutic avenues that modulate the gut and/or local microbiota to create a more favorable TME (Fessler et al., 2019; Laborda-Illanes et al., 2020). Personalized assessment of a patient’s gut and tumor microbiome composition could serve as a diagnostic and prognostic tool, potentially improving treatment outcomes and patient prognosis.
HG: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The author declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Natural Science Foundation of Zhejiang Province (grant no. LQ20H160009), the Health Research Foundation of Shaanxi Province (grant no. 2022E030) and the Science and Technology Development Incubation Foundation of Shaanxi Provincial People’s Hospital (grant no. 2021HL-2).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CRC, colorectal cancer; TME, tumor microenvironment; MAGs, metagenome-assembled genomes; TLRs, Toll-like receptors; F. nucleatum, Fusobacterium nucleatum; HFD, High-fat diet; ICB, immune checkpoint blockade; DC, deoxycholate; TNBC, triple-negative breast cancer; SCFAs, short-chain fatty acids; ETBF, enterotoxigenic Bacteroides fragilis; PRRs, pattern recognition receptors; EMT, epithelial-mesenchymal transition.
Abed, J., Emgård, J. E., Zamir, G., Faroja, M., Almogy, G., Grenov, A., et al. (2016). Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galNAc. Cell Host Microbe 20, 215–225. doi: 10.1016/j.chom.2016.07.006
Adlercreutz, H., Martin, F., Tikkanen, M. J., Pulkkinen, M. (1975). Effect of ampicillin administration on the excretion of twelve estrogens in pregnancy urine. Acta Endocrinol. (Copenh) 80, 551–557. doi: 10.1530/acta.0.0800551
Afroz, R., Tanvir, E. M., Tania, M., Fu, J., Kamal, M. A., Khan, M. A. (2022). LPS/TLR4 pathways in breast cancer: insights into cell signaling. Curr. Med. Chem. 29, 2274–2289. doi: 10.2174/0929867328666210811145043
Alexander, J. L., Wilson, I. D., Teare, J., Marchesi, J. R., Nicholson, J. K., Kinross, J. M. (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365. doi: 10.1038/nrgastro.2017.20
Altinok Dindar, D., Chun, B., Palma, A., Cheney, J., Krieger, M., Kasschau, K., et al. (2023). Association between gut microbiota and breast cancer: diet as a potential modulating factor. Nutrients 15, 4628. doi: 10.3390/nu15214628
Amaro-da-Cruz, A., Rubio-Tomás, T., Álvarez-Mercado, A. I. (2024). Specific microbiome patterns and their association with breast cancer: the intestinal microbiota as a potential biomarker and therapeutic strategy. Clin. Transl. Oncol. 18. doi: 10.1007/s12094-024-03554-w
Arnone, A. A., Cook, K. L. (2022). Gut and breast microbiota as endocrine regulators of hormone receptor-positive breast cancer risk and therapy response. Endocrinology 164(1), bqac177. doi: 10.1210/endocr/bqac177
Asnicar, F., Thomas, A. M., Passerini, A., Waldron, L., Segata, N. (2024). Machine learning for microbiologists. Nat. Rev. Microbiol. 22, 191–205. doi: 10.1038/s41579-023-00984-1
Banerjee, S., Tian, T., Wei, Z., Shih, N., Feldman, M. D., Peck, K. N., et al. (2018). Distinct microbial signatures associated with different breast cancer types. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00951
Banerjee, S., Wei, Z., Tian, T., Bose, D., Shih, N. N. C., Feldman, M. D., et al. (2021). Prognostic correlations with the microbiome of breast cancer subtypes. Cell Death Dis. 12, 831. doi: 10.1038/s41419-021-04092-x
Battaglia, T. W., Mimpen, I. L., Traets, J. J. H., van Hoeck, A., Zeverijn, L. J., Geurts, B. S., et al. (2024). A pan-cancer analysis of the microbiome in metastatic cancer. Cell 187, 2324–2335.e19. doi: 10.1016/j.cell.2024.03.021
Bernardo, G., Le Noci, V., Di Modica, M., Montanari, E., Triulzi, T., Pupa, S. M., et al. (2023a). The emerging role of the microbiota in breast cancer progression. Cells 12, 1945. doi: 10.3390/cells12151945
Bernardo, G., Le Noci, V., Ottaviano, E., De Cecco, L., Camisaschi, C., Guglielmetti, S., et al. (2023b). Reduction of Staphylococcus epidermidis in the mammary tumor microbiota induces antitumor immunity and decreases breast cancer aggressiveness. Cancer Lett. 555, 216041. doi: 10.1016/j.canlet.2022.216041
Bobin-Dubigeon, C., Luu, H. T., Leuillet, S., Lavergne, S. N., Carton, T., Le Vacon, F., et al. (2021). Fecal microbiota composition varies between patients with breast cancer and healthy women: A comparative case-control study. Nutrients 13, 2705. doi: 10.3390/nu13082705
Bray, F., Laversanne, M., Sung, H., Ferlay, J., Siegel, R. L., Soerjomataram, I., et al. (2024). Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263. doi: 10.3322/caac.21834
Buchta Rosean, C., Bostic, R. R., Ferey, J. C., Feng, T.-Y., Azar, F. N., Tung, K. S., et al. (2019). Preexisting commensal dysbiosis is a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor–positive breast cancer. Cancer Res. 79, 3662–3675. doi: 10.1158/0008-5472.CAN-18-3464
Cao, Y., Xia, H., Tan, X., Shi, C., Ma, Y., Meng, D., et al. (2024). Intratumoral microbiota: a new frontier in cancer development and therapy. Signal Transduct. Target Ther. 9, 15. doi: 10.1038/s41392-023-01693-0
Chen, G., Gao, C., Jiang, S., Cai, Q., Li, R., Sun, Q., et al. (2024a). Fusobacterium nucleatum outer membrane vesicles activate autophagy to promote oral cancer metastasis. J. Adv. Res. 56, 167–179. doi: 10.1016/j.jare.2023.04.002
Chen, F., Yang, J., Guo, Y., Su, D., Sheng, Y., Wu, Y. (2023). Integrating bulk and single-cell RNA sequencing data reveals the relationship between intratumor microbiome signature and host metabolic heterogeneity in breast cancer. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1140995
Chen, J., Liu, X., Zou, Y., Gong, J., Ge, Z., Lin, X., et al. (2024b). A high-fat diet promotes cancer progression by inducing gut microbiota-mediated leucine production and PMN-MDSC differentiation. Proc. Natl. Acad. Sci. U.S.A. 121, e2306776121. doi: 10.1073/pnas.2306776121
Chen, L., Shen, J., Kang, Z., Zhang, Z., Zheng, Z., Zhang, L., et al. (2024c). Fusobacterium nucleatum-mimicking nanovehicles to overcome chemoresistance for breast cancer treatment by eliminating tumor-colonizing bacteria. Chem 10, 1783–1803. doi: 10.1016/j.chempr.2024.01.030
Clarridge, J. E., 3rd (2004). Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17, 840–862. doi: 10.1128/cmr.17.4.840-862.2004
Debruyne, P. R., Bruyneel, E. A., Karaguni, I. M., Li, X., Flatau, G., Müller, O., et al. (2002). Bile acids stimulate invasion and haptotaxis in human colorectal cancer cells through activation of multiple oncogenic signaling pathways. Oncogene 21, 6740–6750. doi: 10.1038/sj.onc.1205729
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Di Modica, M., Gargari, G., Regondi, V., Bonizzi, A., Arioli, S., Belmonte, B., et al. (2021). Gut microbiota condition the therapeutic efficacy of trastuzumab in HER2-positive breast cancer. Cancer Res. 81, 2195–2206. doi: 10.1158/0008-5472.CAN-20-1659
Fan, Y., Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55–71. doi: 10.1038/s41579-020-0433-9
Fessler, J., Matson, V., Gajewski, T. F. (2019). Exploring the emerging role of the microbiome in cancer immunotherapy. J. Immunother. Cancer 7, 108. doi: 10.1186/s40425-019-0574-4
Friedman, G. D., Oestreicher, N., Chan, J., Quesenberry, C. P., Jr., Udaltsova, N., Habel, L. A. (2006). Antibiotics and risk of breast cancer: up to 9 years of follow-up of 2.1 million women. Cancer Epidemiol. Biomarkers Prev. 15, 2102–2106. doi: 10.1158/1055-9965.EPI-06-0401
Fu, A., Yao, B., Dong, T., Chen, Y., Yao, J., Liu, Y., et al. (2022). Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185, 1356–1372.e1326. doi: 10.1016/j.cell.2022.02.027
Gándola, Y. B., Fontana, C., Bojorge, M. A., Luschnat, T. T., Moretton, M. A., Chiapetta, D. A., et al. (2020). Concentration-dependent effects of sodium cholate and deoxycholate bile salts on breast cancer cells proliferation and survival. Mol. Biol. Rep. 47, 3521–3539. doi: 10.1007/s11033-020-05442-2
Gao, Y., Bi, D., Xie, R., Li, M., Guo, J., Liu, H., et al. (2021). Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target Ther. 6, 398. doi: 10.1038/s41392-021-00795-x
Garcia Rodriguez, L. A., Gonzalez-Perez, A. (2005). Use of antibiotics and risk of breast cancer. Am. J. Epidemiol. 161, 616–619. doi: 10.1093/aje/kwi087
Geller, L. T., Barzily-Rokni, M., Danino, T., Jonas, O. H., Shental, N., Nejman, D., et al. (2017). Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160. doi: 10.1126/science.aah5043
Geng, S., Guo, P., Li, X., Shi, Y., Wang, J., Cao, M., et al. (2024). Biomimetic Nanovehicle-Enabled Targeted Depletion of Intratumoral Fusobacterium nucleatum Synergizes with PD-L1 Blockade against Breast Cancer. ACS Nano 18, 8971–8987. doi: 10.1021/acsnano.3c12687
German, R., Marino, N., Hemmerich, C., Podicheti, R., Rusch, D. B., Stiemsma, L. T., et al. (2023). Exploring breast tissue microbial composition and the association with breast cancer risk factors. Breast Cancer Res. 25, 82. doi: 10.1186/s13058-023-01677-6
Giampazolias, E., Costa, M. P., Lam, K. C., Lim, K. H. J., Cardoso, A., Piot, C., et al. (2024). Vitamin D regulates microbiome-dependent cancer immunity. Science 384, 428–437. doi: 10.1126/science.adh7954
Goedert, J. J., Hua, X., Bielecka, A., Okayasu, I., Milne, G. L., Jones, G. S., et al. (2018). Postmenopausal breast cancer and estrogen associations with the IgA-coated and IgA-noncoated fecal microbiota. Br. J. Cancer 118, 471–479. doi: 10.1038/bjc.2017.435
Goldin, B. R., Adlercreutz, H., Gorbach, S. L., Warram, J. H., Dwyer, J. T., Swenson, L., et al. (1982). Estrogen excretion patterns and plasma levels in vegetarian and omnivorous women. N Engl. J. Med. 307, 1542–1547. doi: 10.1056/nejm198212163072502
Goodrich, J. K., Di Rienzi, S. C., Poole, A. C., Koren, O., Walters, W. A., Caporaso, J. G., et al. (2014). Conducting a microbiome study. Cell 158, 250–262. doi: 10.1016/j.cell.2014.06.037
Goodwin, A. C., Destefano Shields, C. E., Wu, S., Huso, D. L., Wu, X., Murray-Stewart, T. R., et al. (2011). Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 15354–15359. doi: 10.1073/pnas.1010203108
Guo, Y., Chen, Y., Liu, X., Min, J. J., Tan, W., Zheng, J. H. (2020b). Targeted cancer immunotherapy with genetically engineered oncolytic Salmonella typhimurium. Cancer Lett. 469, 102–110. doi: 10.1016/j.canlet.2019.10.033
Guo, P., Tian, Z., Kong, X., Yang, L., Shan, X., Dong, B., et al. (2020a). FadA promotes DNA damage and progression of Fusobacterium nucleatum-induced colorectal cancer through up-regulation of chk2. J. Exp. Clin. Cancer Res. 39, 202. doi: 10.1186/s13046-020-01677-w
Guo, X., Yu, K., Huang, R. (2024). The ways Fusobacterium nucleatum translocate to breast tissue and contribute to breast cancer development. Mol. Oral. Microbiol. 39, 1–11. doi: 10.1111/omi.12446
Hamalainen, E., Korpela, J. T., Adlercreutz, H. (1987). Effect of oxytetracycline administration on intestinal metabolism of estrogens and on plasma sex hormones in healthy men. Gut 28, 439–445. doi: 10.1136/gut.28.4.439
Hoskinson, C., Zheng, K., Gabel, J., Kump, A., German, R., Podicheti, R., et al. (2022). Composition and functional potential of the human mammary microbiota prior to and following breast tumor diagnosis. mSystems 7, e0148921. doi: 10.1128/msystems.01489-21
Jaye, K., Alsherbiny, M. A., Chang, D., Li, C. G., Bhuyan, D. J. (2023). Mechanistic Insights into the Anti-Proliferative Action of Gut Microbial Metabolites against Breast Adenocarcinoma Cells. Int. J. Mol. Sci. 24, 15053. doi: 10.3390/ijms242015053
Jaye, K., Chang, D., Li, C. G., Bhuyan, D. J. (2022a). Gut metabolites and breast cancer: the continuum of dysbiosis, breast cancer risk, and potential breast cancer therapy. Int. J. Mol. Sci. 23, 9490. doi: 10.3390/ijms23169490
Jaye, K., Li, C. G., Chang, D., Bhuyan, D. J. (2022b). The role of key gut microbial metabolites in the development and treatment of cancer. Gut Microbes 14, 2038865. doi: 10.1080/19490976.2022.2038865
Jia, D., Wang, Q., Qi, Y., Jiang, Y., He, J., Lin, Y., et al. (2024). Microbial metabolite enhances immunotherapy efficacy by modulating T cell stemness in pan-cancer. Cell 187, 1651–1665.e1621. doi: 10.1016/j.cell.2024.02.022
Jia, M., Wu, Z., Vogtmann, E., O’Brien, K. M., Weinberg, C. R., Sandler, D. P., et al. (2020). The association between periodontal disease and breast cancer in a prospective cohort study. Cancer Prev. Res. (Phila) 13, 1007–1016. doi: 10.1158/1940-6207.CAPR-20-0018
Keum, N., Greenwood, D. C., Lee, D. H., Kim, R., Aune, D., Ju, W., et al. (2015). Adult weight gain and adiposity-related cancers: a dose-response meta-analysis of prospective observational studies. J. Natl. Cancer Inst. 107, djv088. doi: 10.1093/jnci/djv088
Kim, M. B., Hwangbo, S., Jang, S., Jo, Y. K. (2022). Bioengineered Co-culture of organoids to recapitulate host-microbe interactions. Mater. Today Bio 16, 100345. doi: 10.1016/j.mtbio.2022.100345
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analyzing microbiomes. Nat. Rev. Microbiol. 16, 410–422. doi: 10.1038/s41579-018-0029-9
Krishnamurthy, K., Wang, G., Rokhfeld, D., Bieberich, E. (2008). Deoxycholate promotes survival of breast cancer cells by reducing the level of pro-apoptotic ceramide. Breast Cancer Res. 10, R106. doi: 10.1186/bcr2211
Krueger, A., Mohamed, A., Kolka, C. M., Stoll, T., Zaugg, J., Linedale, R., et al. (2022). Skin cancer-associated S. aureus strains can induce DNA damage in human keratinocytes by downregulating DNA repair and promoting oxidative stress. Cancers (Basel) 14, 2143. doi: 10.3390/cancers14092143
Kwa, M., Plottel, C. S., Blaser, M. J., Adams, S. (2016). The intestinal microbiome and estrogen receptor-positive female breast cancer. J. Natl. Cancer Inst. 108, djw029. doi: 10.1093/jnci/djw029
Laborda-Illanes, A., Aranega-Martín, L., Sánchez-Alcoholado, L., Boutriq, S., Plaza-Andrades, I., Peralta-Linero, J., et al. (2024). Exploring the relationship between microRNAs, intratumoral microbiota, and breast cancer progression in patients with and without metastasis. Int. J. Mol. Sci. 25, 7091. doi: 10.3390/ijms25137091
Laborda-Illanes, A., Sanchez-Alcoholado, L., Dominguez-Recio, M. E., Jimenez-Rodriguez, B., Lavado, R., Comino-Méndez, I., et al. (2020). Breast and gut microbiota action mechanisms in breast cancer pathogenesis and treatment. Cancers (Basel) 12, 2465. doi: 10.3390/cancers12092465
Lakritz, J. R., Poutahidis, T., Mirabal, S., Varian, B. J., Levkovich, T., Ibrahim, Y. M., et al. (2015). Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget 6, 9387–9396. doi: 10.18632/oncotarget.3328
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Turnbaugh, P. J., Klein, S., Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, S., Chen, M. F., Wang, Z. Y., Abudourexiti, W., Zhang, L., Ding, C., et al. (2024). Ant may well destroy a whole dam: glycans of colonic mucus barrier disintegrated by gut bacteria. Microbiol. Res. 281, 127599. doi: 10.1016/j.micres.2023.127599
Li, G., Sun, Y., Huang, Y., Lian, J., Wu, S., Luo, D., et al. (2023). Fusobacterium nucleatum-derived small extracellular vesicles facilitate tumor growth and metastasis via TLR4 in breast cancer. BMC Cancer 23, 473. doi: 10.1186/s12885-023-10844-z
Li, J., Zhang, Y., Cai, Y., Yao, P., Jia, Y., Wei, X., et al. (2024). Multi-omics analysis elucidates the relationship between intratumor microbiome and host immune heterogeneity in breast cancer. Microbiol. Spectr. 12, e0410423. doi: 10.1128/spectrum.04104-23
Liang, M., Liu, Y., Zhang, Z., Yang, H., Dai, N., Zhang, N., et al. (2022). Fusobacterium nucleatum induces MDSCs enrichment via activation the NLRP3 inflammosome in ESCC cells, leading to cisplatin resistance. Ann. Med. 54, 989–1003. doi: 10.1080/07853890.2022.2061045
Lin, L., Du, Y., Song, J., Wang, W., Yang, C. (2021). Imaging commensal microbiota and pathogenic bacteria in the gut. Acc. Chem. Res. 54, 2076–2087. doi: 10.1021/acs.accounts.1c00068
Liu, Y., Baba, Y., Ishimoto, T., Tsutsuki, H., Zhang, T., Nomoto, D., et al. (2021a). Fusobacterium nucleatum confers chemoresistance by modulating autophagy in esophageal squamous cell carcinoma. Br. J. Cancer 124, 963–974. doi: 10.1038/s41416-020-01198-5
Liu, Y. X., Qin, Y., Chen, T., Lu, M., Qian, X., Guo, X., et al. (2021b). A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 12, 315–330. doi: 10.1007/s13238-020-00724-8
Liu, X., Sun, M., Pu, F., Ren, J., Qu, X. (2023). Transforming intratumor bacteria into immunopotentiators to reverse cold tumors for enhanced immuno-chemodynamic therapy of triple-negative breast cancer. J. Am. Chem. Soc. 145, 26296–26307. doi: 10.1021/jacs.3c09472
Lynch, S. V., Pedersen, O. (2016). The human intestinal microbiome in health and disease. N Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Ma, Z., Qu, M., Wang, X. (2022). Analysis of gut microbiota in patients with breast cancer and benign breast lesions. Pol. J. Microbiol. 71, 217–226. doi: 10.33073/pjm-2022-019
Ma, W., Zhang, L., Chen, W., Chang, Z., Tu, J., Qin, Y., et al. (2024). Microbiota enterotoxigenic Bacteroides fragilis-secreted BFT-1 promotes breast cancer cell stemness and chemoresistance through its functional receptor NOD1. Protein Cell 15, 419–440. doi: 10.1093/procel/pwae005
Martin, F., Peltonen, J., Laatikainen, T., Pulkkinen, M., Adlercreutz, H. (1975). Excretion of progesterone metabolites and estriol in feces from pregnant women during ampicillin administration. J. Steroid Biochem. 6, 1339–1346. doi: 10.1016/0022-4731(75)90363-5
Mikó, E., Kovács, T., Sebő, É., Tóth, J., Csonka, T., Ujlaki, G., et al. (2019). Microbiome-microbial metabolome-cancer cell interactions in breast cancer-familiar, but unexplored. Cells 8, 293. doi: 10.3390/cells8040293
Mirzaei, R., Afaghi, A., Babakhani, S., Sohrabi, M. R., Hosseini-Fard, S. R., Babolhavaeji, K., et al. (2021). Role of microbiota-derived short-chain fatty acids in cancer development and prevention. BioMed. Pharmacother. 139, 111619. doi: 10.1016/j.biopha.2021.111619
Morelli, M., Kurek, D., Ng, C. P., Queiroz, K. (2023). Gut-on-a-chip models: current and future perspectives for host-microbial interactions research. Biomedicines 11, 619. doi: 10.3390/biomedicines11020619
Muegge, B. D., Kuczynski, J., Knights, D., Clemente, J. C., González, A., Fontana, L., et al. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332, 970–974. doi: 10.1126/science.1198719
Nejman, D., Livyatan, I., Fuka, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189
Niccolai, E., Baldi, S., Nannini, G., Gensini, F., Papi, L., Vezzosi, V., et al. (2023). Breast cancer: the first comparative evaluation of oncobiome composition between males and females. Biol. Sex Differ. 14, 37. doi: 10.1186/s13293-023-00523-w
Niekamp, P., Kim, C. H. (2023). Microbial metabolite dysbiosis and colorectal cancer. Gut Liver 17, 190–203. doi: 10.5009/gnl220260
Parhi, L., Alon-Maimon, T., Sol, A., Nejman, D., Shhadeh, A., Fainsod-Levi, T., et al. (2020). Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 11, 3259. doi: 10.1038/s41467-020-16967-2
Parida, S., Sharma, D. (2019). The power of small changes: Comprehensive analyses of microbial dysbiosis in breast cancer. Biochim. Biophys. Acta Rev. Cancer 1871, 392–405. doi: 10.1016/j.bbcan.2019.04.001
Parida, S., Siddharth, S., Gatla, H. R., Wu, S., Wang, G., Gabrielson, K., et al. (2023a). Gut colonization with an obesity-associated enteropathogenic microbe modulates the premetastatic niches to promote breast cancer lung and liver metastasis. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1194931
Parida, S., Siddharth, S., Xia, Y., Sharma, D. (2023b). Concomitant analyses of intratumoral microbiota and genomic features reveal distinct racial differences in breast cancer. NPJ Breast Cancer 9, 4. doi: 10.1038/s41523-023-00505-6
Parida, S., Wu, S., Siddharth, S., Wang, G., Muniraj, N., Nagalingam, A., et al. (2021). A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and β-catenin axes. Cancer Discovery 11, 1138–1157. doi: 10.1158/2159-8290.Cd-20-0537
Park, S. R., Kim, D. J., Han, S. H., Kang, M. J., Lee, J. Y., Jeong, Y. J., et al. (2014). Diverse Toll-like receptors mediate cytokine production by Fusobacterium nucleatum and Aggregatibacter actinomycetemcomitans in macrophages. Infect. Immun. 82, 1914–1920. doi: 10.1128/iai.01226-13
Plottel, C. S., Blaser, M. J. (2011). Microbiome and Malignancy. Cell Host Microbe 10, 324–335. doi: 10.1016/j.chom.2011.10.003
Puschhof, J., Pleguezuelos-Manzano, C., Clevers, H. (2021). Organoids and organs-on-chips: Insights into human gut-microbe interactions. Cell Host Microbe 29, 867–878. doi: 10.1016/j.chom.2021.04.002
Rajput, S., Volk-Draper, L. D., Ran, S. (2013). TLR4 is a novel determinant of the response to paclitaxel in breast cancer. Mol. Cancer Ther. 12, 1676–1687. doi: 10.1158/1535-7163.Mct-12-1019
Rodrigues, M. F., Carvalho, É., Pezzuto, P., Rumjanek, F. D., Amoêdo, N. D. (2015). Reciprocal modulation of histone deacetylase inhibitors sodium butyrate and trichostatin A on the energy metabolism of breast cancer cells. J. Cell Biochem. 116, 797–808. doi: 10.1002/jcb.25036
Rutkowski, M. R., Stephen, T. L., Svoronos, N., Allegrezza, M. J., Tesone, A. J., Perales-Puchalt, A., et al. (2015). Microbially driven TLR5-dependent signaling governs distal Malignant progression through tumor-promoting inflammation. Cancer Cell 27, 27–40. doi: 10.1016/j.ccell.2014.11.009
Sampsell, K., Hao, D., Reimer, R. A. (2020). The gut microbiota: A potential gateway to improved health outcomes in breast cancer treatment and survivorship. Int. J. Mol. Sci. 21, 9239. doi: 10.3390/ijms21239239
Sears, C. L., Geis, A. L., Housseau, F. (2014). Bacteroides fragilis subverts mucosal biology: from symbiont to colon carcinogenesis. J. Clin. Invest. 124, 4166–4172. doi: 10.1172/jci72334
Sekirov, I., Russell, S. L., Antune, L. C. M., Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Semaan, J., El-Hakim, S., Ibrahim, J. N., Safi, R., Elnar, A. A., El Boustany, C. (2020). Comparative effect of sodium butyrate and sodium propionate on proliferation, cell cycle and apoptosis in human breast cancer cells MCF-7. Breast Cancer 27, 696–705. doi: 10.1007/s12282-020-01063-6
Shi, Q., Wang, J., Zhou, M., Zheng, R., Zhang, X., Liu, B. (2023). Gut Lactobacillus contribute to the progression of breast cancer by affecting the anti-tumor activities of immune cells in the TME of tumor-bearing mice. Int. Immunopharmacol. 124, 111039. doi: 10.1016/j.intimp.2023.111039
Siddharth, S., Parida, S., Muniraj, N., Hercules, S., Lim, D., Nagalingam, A., et al. (2021). Concomitant activation of GLI1 and Notch1 contributes to racial disparity of human triple negative breast cancer progression. Elife 10, e70729. doi: 10.7554/eLife.70729
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Smith, A., Pierre, J. F., Makowski, L., Tolley, E., Lyn-Cook, B., Lu, L., et al. (2019). Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 9, 11940. doi: 10.1038/s41598-019-48348-1
Song, X. L., Wei, C. R., Li, X. Q. (2022). The relationship between microbial community and breast cancer. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.849022
Sorensen, H. T., Skriver, M. V., Friis, S., McLaughlin, J. K., Blot, W. J., Baron, J. A. (2005). Use of antibiotics and risk of breast cancer: a population-based case-control study. Br. J. Cancer 92, 594–596. doi: 10.1038/sj.bjc.6602313
Soto-Pantoja, D. R., Gaber, M., Arnone, A. A., Bronson, S. M., Cruz-Diaz, N., Wilson, A. S., et al. (2021). Diet alters entero-mammary signaling to regulate the breast microbiome and tumorigenesis. Cancer Res. 81, 3890–3904. doi: 10.1158/0008-5472.Can-20-2983
Su, J., Lin, X., Li, D., Yang, C., Lv, S., Chen, X., et al. (2024). Prevotella copri exhausts intrinsic indole-3-pyruvic acid in the host to promote breast cancer progression: inactivation of AMPK via UHRF1-mediated negative regulation. Gut Microbes 16, 2347757. doi: 10.1080/19490976.2024.2347757
Terrisse, S., Derosa, L., Iebba, V., Ghiringhelli, F., Vaz-Luis, I., Kroemer, G., et al. (2021). Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ. 28, 2778–2796. doi: 10.1038/s41418-021-00784-1
Terrisse, S., Zitvogel, L., Kroemer, G. (2023). Impact of microbiota on breast cancer hormone therapy. Cell Stress 7, 12–19. doi: 10.15698/cst2023.03.277
Thompson, K. J., Ingle, J. N., Tang, X., Chia, N., Jeraldo, P. R., Walther-Antomio, M. R., et al. (2017). A comprehensive analysis of breast cancer microbiota and host gene expression. PloS One 12, e0188873. doi: 10.1371/journal.pone.0188873
Tzeng, A., Sangwan, N., Jia, M., Liu, C. C., Keslar, K. S., Downs-Kelly, E., et al. (2021). Human breast microbiome correlates with prognostic features and immunological signatures in breast cancer. Genome Med. 13, 60. doi: 10.1186/s13073-021-00874-2
Urbaniak, C., Cummins, J., Brackstone, M., Macklaim, J. M., Gloor, G. B., Baban, C. K., et al. (2014). Microbiota of human breast tissue. Appl. Environ. Microbiol. 80, 3007–3014. doi: 10.1128/aem.00242-14
Urbaniak, C., Gloor, G. B., Brackstone, M., Scott, L., Tangney, M., Reid, G. (2016). The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 82, 5039–5048. doi: 10.1128/aem.01235-16
Van der Merwe, M., Van Niekerk, G., Botha, A., Engelbrecht, A. M. (2021). The onco-immunological implications of Fusobacterium nucleatum in breast cancer. Immunol. Lett. 232, 60–66. doi: 10.1016/j.imlet.2021.02.007
Velicer, C. M., Heckbert, S. R., Lampe, J. W., Potter, J. D., Robertson, C. A., Taplin, S. H. (2004). Antibiotic use in relation to the risk of breast cancer. JAMA 291, 827–835. doi: 10.1001/jama.291.7.827
Wang, Q., Ding, H., Dong, G., Xu, L., Jiang, F., Mao, Q. (2021). Bi-direction effects between microbiome and MiRNAs in carcinogenesis. J. Cancer Res. Clin. Oncol. 147, 1299–1305. doi: 10.1007/s00432-021-03567-w
Wang, Y., Han, Y., Yang, C., Bai, T., Zhang, C., Wang, Z., et al. (2024). Long-term relapse-free survival enabled by integrating targeted antibacteria in antitumor treatment. Nat. Commun. 15, 4194. doi: 10.1038/s41467-024-48662-x
Wang, Y. H., Wang, M., Chen, J. X., Li, Y., Kuang, Z., Dende, C., et al. (2023). The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 381, 851–857. doi: 10.1126/science.ade0522
Wilkie, T., Verma, A. K., Zhao, H., Charan, M., Ahirwar, D. K., Kant, S., et al. (2022). Lipopolysaccharide from the commensal microbiota of the breast enhances cancer growth: role of S100A7 and TLR4. Mol. Oncol. 16, 1508–1522. doi: 10.1002/1878-0261.12975
Williams, B. A., Grant, L. J., Gidley, M. J., Mikkelsen, D. (2017). Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int. J. Mol. Sci. 18, 2203. doi: 10.3390/ijms18102203
Wu, R., Yu, I., Tokumaru, Y., Asaoka, M., Oshi, M., Yan, L., et al. (2022). Elevated bile acid metabolism and microbiome are associated with suppressed cell proliferation and better survival in breast cancer. Am. J. Cancer Res. 12, 5271–5285.
Xavier, J. B., Young, V. B., Skufca, J., Ginty, F., Testerman, T., Pearson, A. T., et al. (2020). The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer 6, 192–204. doi: 10.1016/j.trecan.2020.01.004
Xie, Y., Xie, F., Zhou, X., Zhang, L., Yang, B., Huang, J., et al. (2022). Microbiota in tumors: from understanding to application. Adv. Sci. (Weinh) 9, e2200470. doi: 10.1002/advs.202200470
Xuan, C., Shamonki, J. M., Chung, A., Dinome, M. L., Chung, M., Sieling, P. A., et al. (2014). Microbial dysbiosis is associated with human breast cancer. PloS One 9, e83744. doi: 10.1371/journal.pone.0083744
Xue, C., Chu, Q., Zheng, Q., Yuan, X., Su, Y., Bao, Z., et al. (2023). Current understanding of the intratumoral microbiome in various tumors. Cell Rep. Med. 4, 100884. doi: 10.1016/j.xcrm.2022.100884
Xue, K., Li, J., Huang, R. (2024). The immunoregulatory role of gut microbiota in the incidence, progression, and therapy of breast cancer. Front. Cell Infect. Microbiol. 5)14. doi: 10.3389/fcimb.2024.1411249
Yang, L., Li, A., Wang, Y., Zhang, Y. (2023a). Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct. Target Ther. 8, 35. doi: 10.1038/s41392-022-01304-4
Yang, Q., Wang, B., Zheng, Q., Li, H., Meng, X., Zhou, F., et al. (2023b). A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv. Sci. (Weinh) 10, e2207366. doi: 10.1002/advs.202207366
Yang, Y., Weng, W., Peng, J., Hong, L., Yang, L., Toiyama, Y., et al. (2017). Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851–866.e824. doi: 10.1053/j.gastro.2016.11.018
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e516. doi: 10.1016/j.cell.2017.07.008
Yu, Y., Shen, X., Xiao, X., Li, L., Huang, Y. (2023). Butyrate modification promotes intestinal absorption and hepatic cancer cells targeting of ferroptosis inducer loaded nanoparticle for enhanced hepatocellular carcinoma therapy. Small 19, e2301149. doi: 10.1002/smll.202301149
Yuan, C., Steer, C. J., Subramanian, S. (2019). Host-MicroRNA-Microbiota interactions in colorectal cancer. Genes (Basel) 10, 270. doi: 10.3390/genes10040270
Zhang, J., Wan, S., Zhou, H., Du, J., Li, Y., Zhu, H., et al. (2024). Programmed nanocloak of commensal bacteria-derived nanovesicles amplify strong immunoreactivity against tumor growth and metastatic progression. ACS Nano 18, 9613–9626. doi: 10.1021/acsnano.3c13194
Zhang, J., Xia, Y., Sun, J. (2021). Breast and gut microbiome in health and cancer. Genes Dis. 8, 581–589. doi: 10.1016/j.gendis.2020.08.002
Zhang, S., Yang, Y., Weng, W., Guo, B., Cai, G., Ma, Y., et al. (2019). Fusobacterium nucleatum promotes chemoresistance to 5-fluorouracil by upregulation of BIRC3 expression in colorectal cancer. J. Exp. Clin. Cancer Res. 38, 14. doi: 10.1186/s13046-018-0985-y
Zheng, H. H., Du, C. T., Yu, C., Tang, X. Y., Huang, R. L., Zhang, Y. Z., et al. (2022). The relationship of tumor microbiome and oral bacteria and intestinal dysbiosis in canine mammary tumor. Int. J. Mol. Sci. 23, 10928. doi: 10.3390/ijms231810928
Zheng, J. H., Nguyen, V. H., Jiang, S. N., Park, S. H., Tan, W., Hong, S. H., et al. (2017). Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 9, eaak9537. doi: 10.1126/scitranslmed.aak9537
Zhong, S., Liu, F., Giniatullin, R., Jolkkonen, J., Li, Y., Zhou, Z., et al. (2023). Blockade of CCR5 suppresses paclitaxel-induced peripheral neuropathic pain caused by increased deoxycholic acid. Cell Rep. 42, 113386. doi: 10.1016/j.celrep.2023.113386
Zhou, Y., Liu, M., Yang, J. (2022). Recovering metagenome-assembled genomes from shotgun metagenomic sequencing data: Methods, applications, challenges, and opportunities. Microbiol. Res. 260, 127023. doi: 10.1016/j.micres.2022.127023
Zhu, X., Li, K., Liu, G., Wu, R., Zhang, Y., Wang, S., et al. (2023). Microbial metabolite butyrate promotes anti-PD-1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microbes 15, 2249143. doi: 10.1080/19490976.2023.2249143
Keywords: breast cancer, tumor microbiome, intestinal microbiota, intratumoral microbiota, microbial metabolites
Citation: Guo H (2025) Interactions between the tumor microbiota and breast cancer. Front. Cell. Infect. Microbiol. 14:1499203. doi: 10.3389/fcimb.2024.1499203
Received: 23 September 2024; Accepted: 11 December 2024;
Published: 24 January 2025.
Edited by:
Soumyadev Sarkar, Arizona State University, United StatesReviewed by:
Qingyao Shang, Chinese Academy of Medical Sciences, ChinaCopyright © 2025 Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Guo, MjI5NDYyMjQzQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.