- 1Center for Yunnan Plateau Biological Resources Protection and Utilization, Qujing Normal University, Qujing, Yunnan, China
- 2Faculty of Graduate Studies, Sabaragamuwa University of Sri Lanka, Belihuloya, Sri Lanka
- 3Department of Bioprocess Technology, Faculty of Technology, Rajarata University of Sri Lanka, Mihintale, Sri Lanka
- 4Tropical Microbiology Research Foundation, Colombo, Sri Lanka
- 5Department of Natural Resources, Faculty of Applied Sciences, Sabaragamuwa University of Sri Lanka, Belihuloya, Sri Lanka
- 6Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 7Vishnugupta Vishwavidyapeetam, Gokarna, India
Endophytic fungi associated with selected aquatic plants, Eichhornia crassipes, Nymphaea nouchali, Salvinia minima and S. molesta were evaluated. Ectophoma salviniae sp. nov. and Neottiosporina mihintaleensis sp. nov. are introduced as novel taxa from Salvinia spp. from Sri Lanka. Chaetomella raphigera is reported as a new geographical record, Colletotrichum siamense and C. truncatum are reported as novel host records in aquatic plants, while Phyllosticta capitalensis has been identified on the same host (Nymphaea nouchali) in the North-Central Province of Sri Lanka. Identification of the fungi was based on morphological characteristics and multi-locus phylogenetic analyses using ITS, LSU, SSU, ACT, CHS-1, GAPDH, tub2, rpb2, and tef1-α molecular markers. The identified fungi were analysed for extracellular enzymatic properties. According to the qualitative analysis, Ectophoma salviniae sp. nov. exhibited the highest amylase production, Chaetomella raphigera exhibited the highest cellulase enzyme production, and Neottiosporina mihintaleensis sp. nov. exhibited the highest laccase production. The results demonstrate the aquatic fungal diversity in this region and their extracellular enzymatic potentials, providing valuable insights for future biotechnological approaches.
1 Introduction
The magnitude of the fungal kingdom has been a debatable topic for decades and several studies have revised the prevailing estimations based on the advancements in estimation methods and technologies (e.g., Hawksworth, 1991, 2001; May, 2000; Tedersoo et al., 2015, 2022; Hyde et al., 2023). However, Wu et al. (2019) estimated the global fungal species diversity to be around 12 million, based on culture-independent approaches, while culture-dependent methods yielded a more conservative estimate of 2.2 to 3.8 million species (Hawksworth and Lücking, 2017). Niskanen et al. (2023) revisited the species number estimated by Hawksworth and Lücking (2017) and concluded that it would be 2–3 million species, however, the best estimation is at 2.5 million. Nevertheless, only 160,000 fungal species have been accepted in Species Fungorum (2024; accession date: 06 June 2024, https://www.speciesfungorum.org/names/names.asp), thus, a large number of taxa are yet to be described. Additionally, it has reported understudied geographical regions and well-studied hosts but biodiversity regions (temperate and tropical) would harbour more novel taxa (Wijayawardene et al., 2021). It is considered a challenge to reveal the unknown fungal diversity with traditional methods, such as morphological or cultural characteristics. The recent advances in molecular techniques (such as high-throughput sequencing of environmental samples) are accelerating the explorations and further trying to reveal the understudied fungal habitats, life modes and geographical regions (Wijayawardene et al., 2023).
Endophytes are widespread and have been reported in plants from diverse ecosystems such as deserts, temperate zones, arctic tundra, tropical forests, grasslands, and croplands (Arnold, 2007; Arnold and Lutzoni, 2007; Zheng et al., 2015; Rana et al., 2019; Harrison and Griffin, 2020; Dar et al., 2022; Hashem et al., 2023). Based on the ratio of the host (vascular plants) to species current estimates suggest that there are approximately one million species of fungal endophytes (Sun and Guo, 2012; Lugtenberg et al., 2016; Rashmi et al., 2019; Wu et al., 2019; Bhunjun et al., 2024). Endophytic fungi belong to both mitosporic and meiosporic ascomycetes, which reside within plants without causing symptoms and colonize healthy tissue beneath the epidermal cell layer through quiet infections (Lu et al., 2012; Ali et al., 2018; Abdel-Wareth, 2022). Further, it has been reported that comparing endophytic basidiomycetes and basal fungi with endophytic ascomycetes shows that almost 90% of the identified endophytes are Ascomycota (Rungjindamai and Gareth Jones, 2024).
Endophytes’ ecology, evolution, and applications are interesting topics; however, knowledge about their diversity, geographic and ecological distributions in most plant communities remains limited and unexplored (Gao et al., 2019; Zheng et al., 2021). Previous research has primarily focused on endophytes in terrestrial plants, while endophytic fungal studies related to aquatic plants received little attention (Li et al., 2010; Sandberg et al., 2014; Dissanayake et al., 2016; Myovela et al., 2024). However, the richness of endophytic fungal diversity has been reported in marine ecosystems (Kamat et al., 2020; El-Bondkly et al., 2021), mangrove ecosystems (Deshmukh et al., 2020; Jia et al., 2020; da Silveira Bastos et al., 2024; Myovela et al., 2024), and freshwater ecosystems (You et al., 2015; Chen et al., 2023; Pramanic et al., 2023). Aquatic plants (including emergent plants, floating-leaved plants, free-floating plants, submerged plants, and wet plants (Ismail et al., 2021; Zhou et al., 2023) serve as hosts for a diverse array of endophytic fungi (Zheng et al., 2021; Wimalasena et al., 2024). Many researchers have studied the diversity and ecological roles of aquatic plants (O’Hare et al., 2018; Kamat et al., 2020; Zheng et al., 2022; Ji et al., 2024). However, most studies have overlooked how endophytes affect these plants and their wider ecological functions. Accordingly, there is a significant gap in the understanding of endophytic fungal communities in aquatic plants (Sandberg et al., 2014; Zheng et al., 2021; Wimalasena et al., 2024).
Sri Lanka is a tropical biodiversity hotspot (Gunawardene et al., 2007; Surasinghe et al., 2019; Sarathchandra et al., 2021; De Zoysa, 2022) and harbours a diverse range of aquatic ecosystems, including both coastal and inland areas (Gunatilleke et al., 2008). Inland freshwater habitats (rivers, streams, marshes, swamp forests, villus, and man-made reservoirs) collectively cover approximately 202,435 hectares (Gunatilleke et al., 2008; Yakandawala, 2012). Sri Lanka is home to over 370 species of aquatic and wetland plants, with 12% being endemic to the country (Yakandawala, 2012). These plants serve as habitats for various fungi, including endophytes (Ratnaweera, 2019; Wimalasena et al., 2024). As highlighted by Wimalasena et al. (2024), Sri Lanka offers significant potential for the identification and study of endophytic fungi. Previous research on endophytic fungi in freshwater plants has been relatively limited in Sri Lanka. For instance, Dissanayake et al. (2016) isolated 20 distinct endophytic fungi from Nymphaea nouchali. More recently, Wimalasena et al. (2024) reported the ongoing study on the isolation of endophytic fungi from freshwater plants in Sri Lanka.
In this study, an effort was made to document the endophytic fungi associated with three freshwater plant taxa, viz., Eichhornia crassipes, Nymphaea nouchali, Salvinia minima and S. molesta, found in the lentic freshwater habitats of the Mihintale area (in Anuradhapura district, North-Central Province), Sri Lanka. We isolated six fungal species that belong to Colletotrichum, Chaetomella, Ectophoma, Neottiosporina and Phyllosticta. Among these taxa, two new species, Ectophoma salviniae sp. nov. and Neottiosporina mihintaleensis sp. nov. are introduced. Chaetomella raphigera has been reported as a new geographical record for Sri Lanka. Colletotrichum siamense and C. truncatum have been identified as new host records on Eichhornia crassipes. Isolation of Phyllosticta capitalensis on Nymphaea nouchali is in confirmative with the findings of Dissanayake et al. (2016) based on multilocus phylogenetic analyses. Furthermore, the study assessed the potential of these endophytic fungi to produce various extracellular enzymes by qualitative assays for amylolytic, cellulolytic, and laccase activities.
2 Materials and methods
2.1 Sampling, isolation and characterization of endophytic fungi
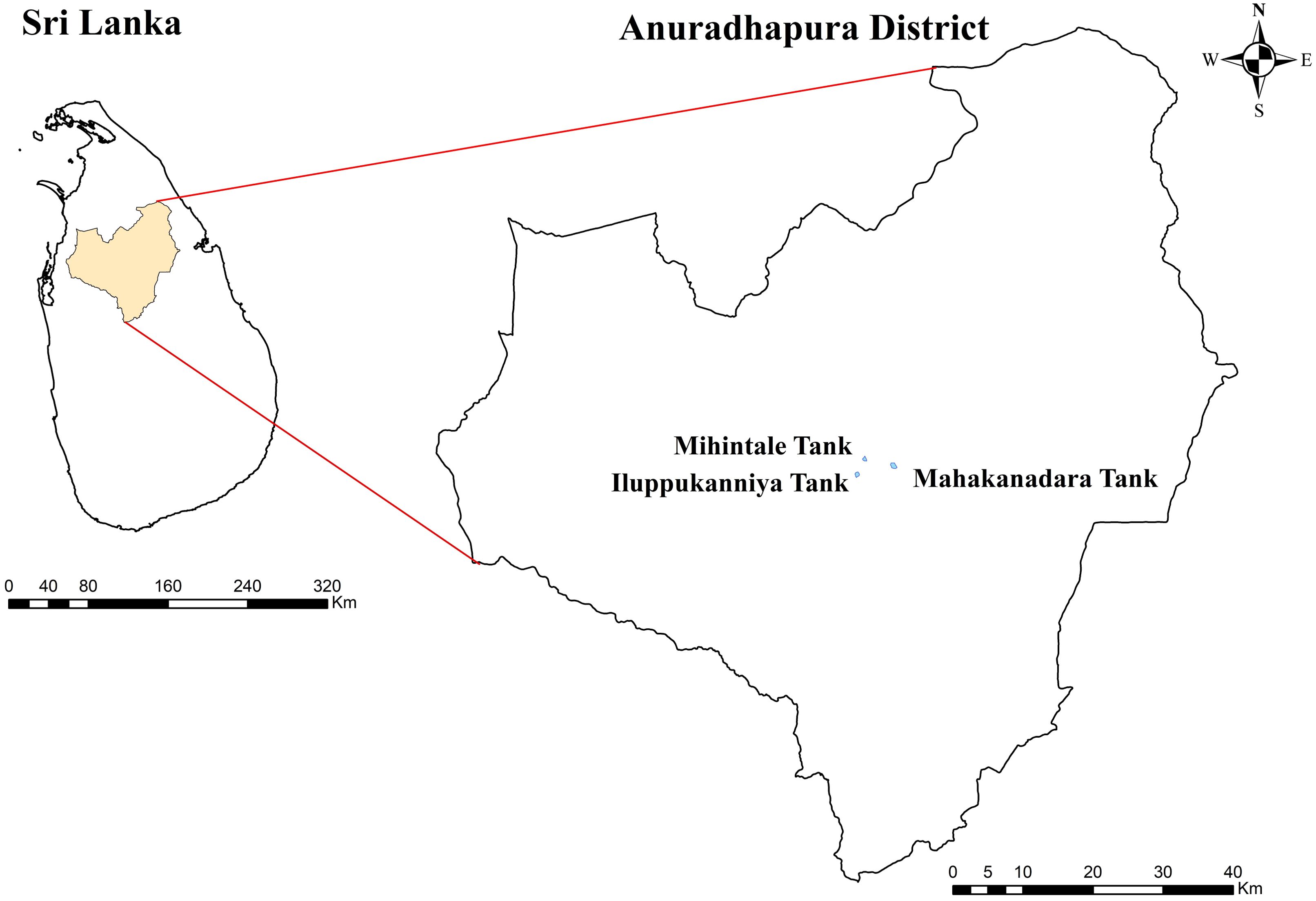
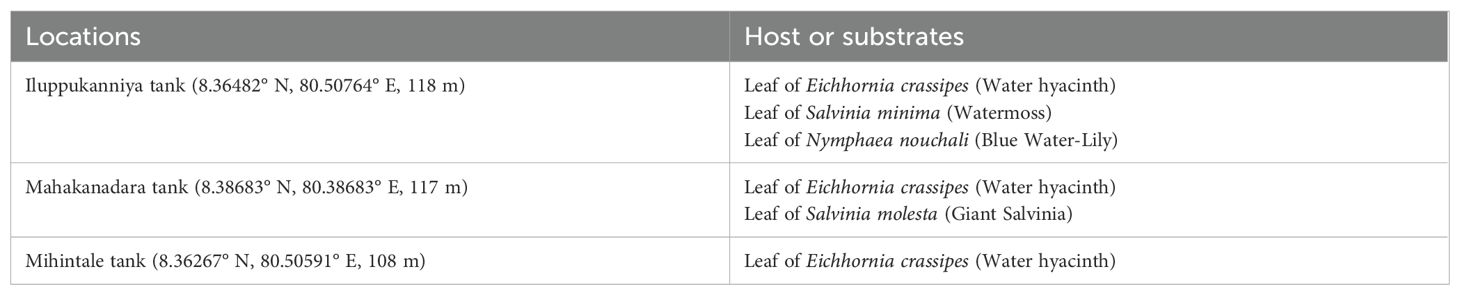
From November to December 2023, healthy aquatic plants were sampled from three lentic freshwater habitats in Mihintale, located in the Anuradhapura district of Sri Lanka including the Iluppukanniya tank (8.36482° N, 80.50764° E, 118 m), Mahakanadara tank (8.38683° N, 80.38683° E, 117 m), and Mihintale tank (8.36267° N, 80.50591° E, 108 m) (Figure 1; Table 1). Mature plants with undamaged leaves of Eichhornia crassipes, Nymphaea nouchali, Nymphaea pubescens, Salvinia minima, and Salvinia molesta (Figure 2), were carefully uprooted and brought to the laboratory within one hour in ziplock plastic bags containing fresh water. The samples were maintained separately in freshwater until the isolation process began immediately.
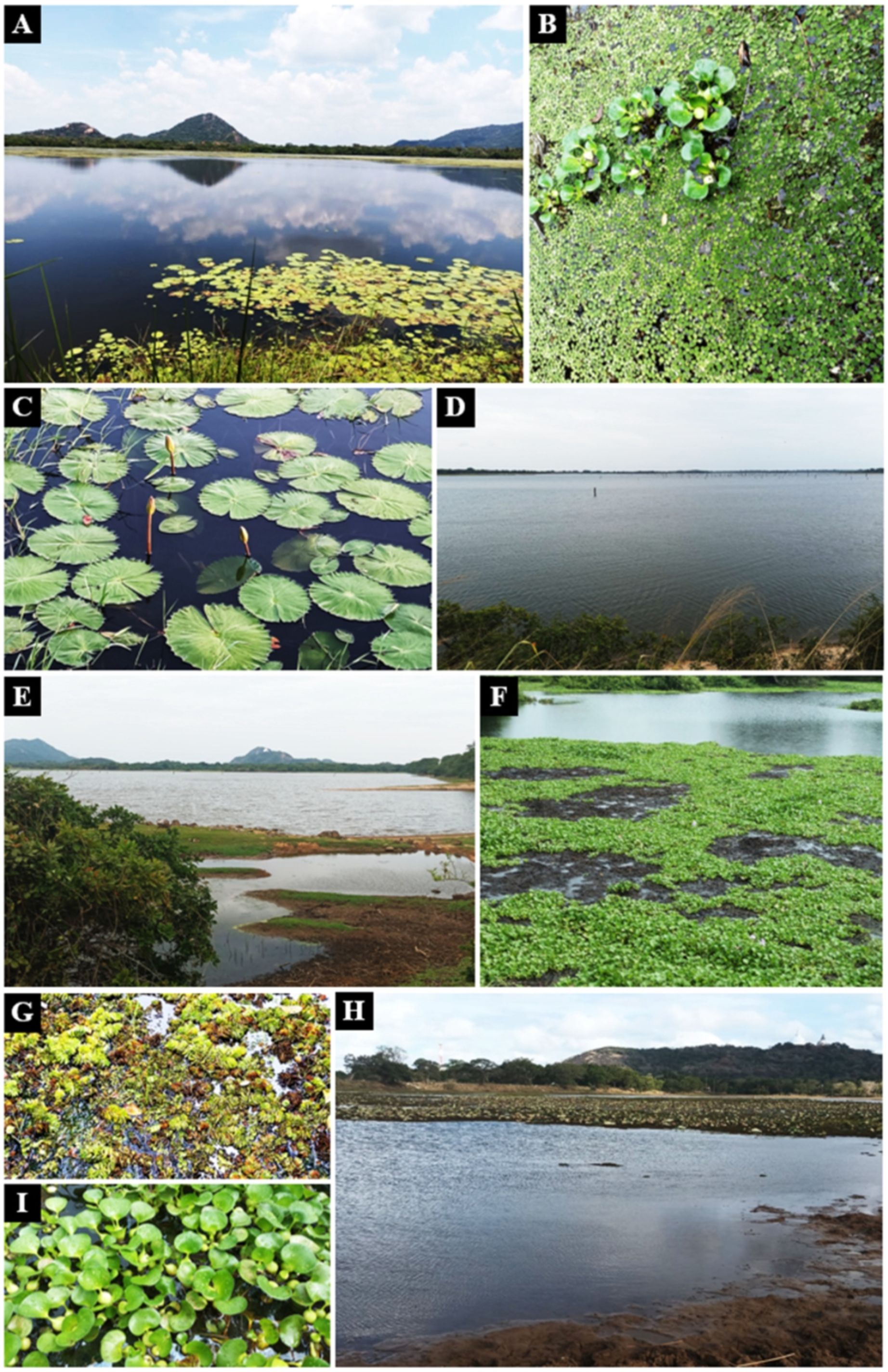
Figure 2. Aquatic plants in lentic freshwater habitats in Mihintale area were used to isolate endophytic fungal species. (A) Iluppukanniya tank. (B) Eichhornia crassipes (Water hyacinth) and Salvinia minima (Watermoss) in the Iluppukanniya tank. (C) Nymphaea sp. in the Iluppukanniya tank. (D, E) Mahakanadara tank. (F) Eichhornia crassipes in the Mahakanadara tank. (G) Salvinia molesta (Giant Salvinia) in the Mahakanadara tank. (H) Mihintale tank. (I) Eichhornia crassipes in Mihintale tank.
Each plant sample was thoroughly rinsed for 30 seconds under running tap water to remove the debris and adhered mud contaminants. The plants were cut into roots, stems, and leaves and processed in the following sequential surface sterilization steps: an initial immersion in 0.5% sodium hypochlorite for 2 minutes, followed by a rinse in sterile distilled water for 1 minute, immersion in 75% ethanol for 2 minutes, and a final rinse in sterile distilled water for 1 minute. After the final wash, the samples were dried using sterile paper towels under a laminar airflow cabinet (Li et al., 2010; Zheng et al., 2021). However, during surface sterilization of the plant material, the duration of treatment for each plant species depended on its tissue sensitivity. Specifically, being very spongy, the leaves of Salvinia molesta were processed with an initial immersion of 1 minute in 0.5% sodium hypochlorite, followed by a rinse in sterile distilled water for one minute, immersion in 70% ethanol one minute, and a final rinse in sterile distilled water for one minute.
Surface-sterilized plant tissues were cut into 5 × 5 mm segments and placed in Potato Dextrose Agar (PDA; 20% potato, 2% dextrose, 2% agar) media supplemented with tetracycline (50 mg/L). The plates were incubated under aseptic conditions at room temperature (28–30°C) and observed for fungal growth every second day for seven days. Once fungal mycelium emerged from the edges of the plant segments, a portion of the growing colony was aseptically cut and transferred to a freshly prepared PDA plate (Li et al., 2010).
The growth of the subcultures was monitored daily, and colony characteristics, including colour, form, elevation, margin, texture, and dimensions, were recorded for two weeks. The colony colour was identified by a colour guide (Colour guide; ArtyClick Colors, 2024). The stock cultures were stored in sterile water and agar slants and preserved at 4°C at Rajarata University Fungal Culture Collection (RUFCC) in Sri Lanka.
The fungal cultures were induced to sporulate using different stress stimulation techniques. These included reducing the normal growth temperature (cold treatment), reducing the quantity of PDA volume for each plate to create starvation media (Mattoo and Nonzom, 2022), exposing the plates to UV light for 15 minutes, and placing a double-sterilized pine needle or toothpick on the fungal colony to induce conidiomatal formation (Su et al., 2012). Post-stressors, the plates were incubated for a week under normal light conditions.
The sporulating cultures were studied using a Nikon SMZ18 stereomicroscope, a Nikon TS2R-FL inverted trinocular microscope and a Nikon ECLIPSE Ci upright microscope. Morphological features were examined and documented (Senanayake et al., 2018).
2.2 Molecular identification of endophytic fungi
2.2.1 Genomic DNA extraction
The genomic DNA was extracted from the freshly cultured fungi using trimethylammonium bromide (CTAB) method, following the protocol described by Gontia-Mishra et al. (2014) and Hatamzadeh et al. (2024).
2.2.2 Polymerase chain reaction
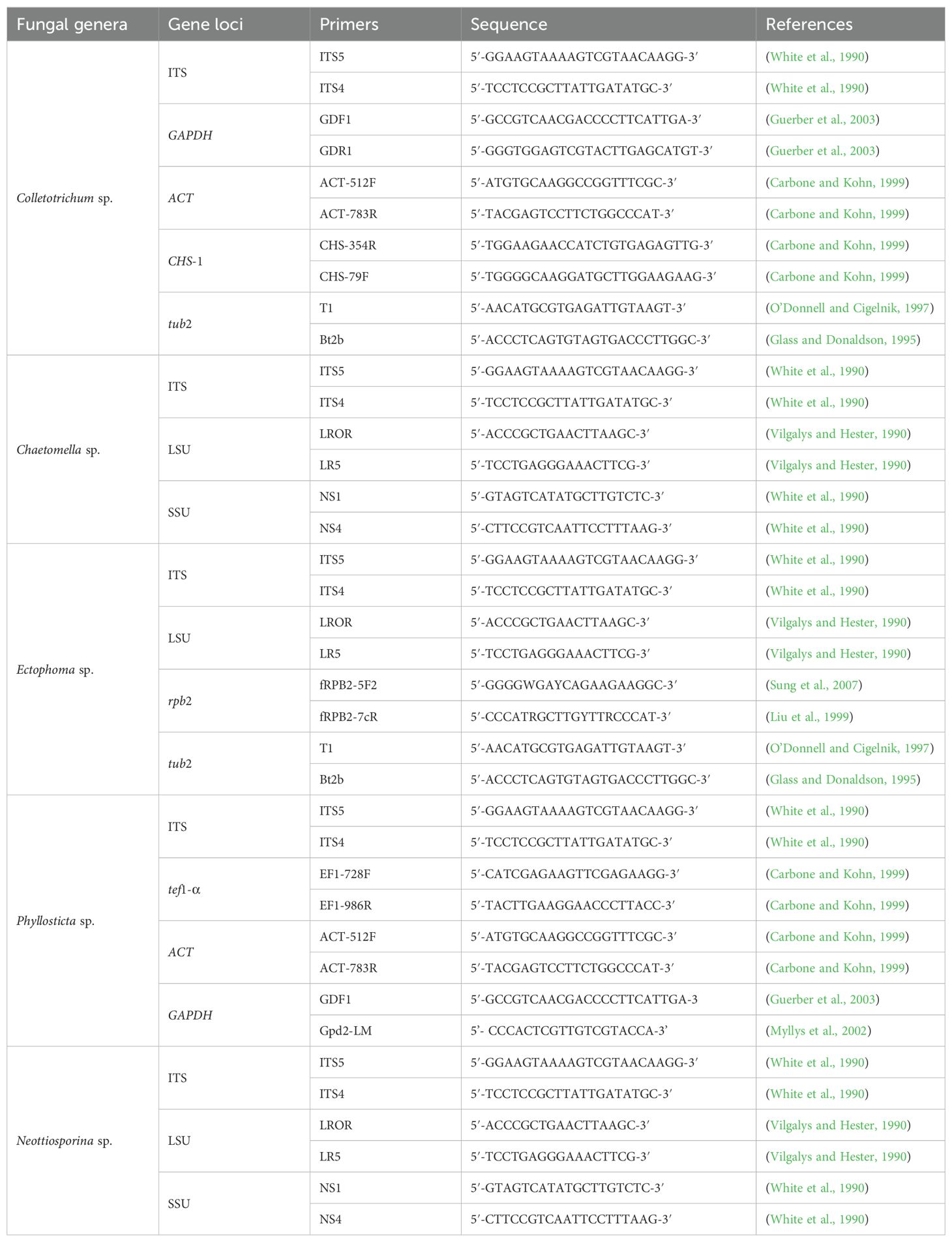
The targeted primers used in the polymerase chain reaction (PCR) included; Actin (ACT; ACT-512F/ACT-783R), Chitin (CHS-1; CHS-354R/CHS-79F), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; GDF1/GDR1/Gpd2-LM), Internal transcribed spacers (ITS; ITS5/ITS4), Large subunit nuclear ribosomal DNA (LSU; LROR/LR5R), RNA polymerase II subunit 2 (rpb2; fRPB2-5F2/fRPB2-7cR), Small-subunit ribosomal RNA (SSU; NS1/NS4), Translation elongation factor 1 (tef1-α; EF1-728F/EF1-986R), and Beta-tubulin (tub2; T1/Bt2b), (Table 2), each with specific annealing temperatures. The final volume of the PCR reaction was 25 μl, containing 5 μl of DNA template, 2.5 μl of each forward and reward primer, 12.5 μl of HIMEDIA MBT061-100R 2× PCR TaqMixture (mixture of Taq DNA Polymerase, dNTPs, and optimized buffer) and 2.5 μl of ddH2O.
The PCR amplification was performed with an initial denaturing step at 95°C for 5 min., followed by 40 amplification cycles consisting of a denaturation step at 95°C for 1 min., an annealing step for 1 min., and a final extension step at 72°C for 10 min. The annealing temperatures were set for the gene loci, with the optimum for each: ACT: 58°C, CHS-1: 58°C, GAPDH: 60°C, tub2: 55°C, ITS: 54°C, LSU: 55°C, rpb2: 56°C, SSU: 55°C, and tef1-α: 54°C. All PCR products were visualized by 1% agarose gel (stained with Diamond TM Nucleic Acid Dye) electrophoresis at 80 V/cm for 30 minutes. The gel was visualized under a UV transilluminator to estimate the fragment size.
2.2.3 DNA sequencing
Amplicons were sequenced using both PCR primers and DNA sequencing results were acquired through Sanger bidirectional sequencing (GeneLabs Medicals Pvt. Ltd., Sri Lanka). The obtained nucleotide sequences were checked for their quality by reviewing the chromatograms using BioEdit version 7.2. After confirming the quality, the sequences were compared with entries in the GenBank database using the Basic Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov; accessed on 03 April 2024) to identify significant alignments with similarity percentages.
2.3 Phylogenetic analysis
Closely related sequences were downloaded from GenBank based on blast similarity and recent publications. Multiple gene phylogenetic analyses were conducted for endophytic fungi for Colletotrichum sp., ITS, GAPDH, ACT, CHS-1, HIS 3, and tub2 (Damm et al., 2009). Chaetomella raphigera was analyzed using ITS, LSU, and SSU (Suwannarach et al., 2018). Ectophoma salviniae sp. nov. underwent analysis with ITS, LSU, rpb2 and tub2 (Hou et al., 2020a). Phyllosticta capitalensis was analyzed by using ITS, ACT, tef1-α, and GAPDH (Glienke et al., 2011; Wang et al., 2012). Neottiosporina mihintaleensis sp. nov. was analyzed using ITS, LSU, and SSU (de Gruyter et al., 2009) (see Table 2 for the primer details). The phylogenetic trees were constructed via Maximum likelihood (ML) and Bayesian analyses. Maximum likelihood (ML) analysis was constructed by the online portal CIPRES Science Gateway v. 3.3 (Miller et al., 2010), using RAxML-HPC v.8 on XSEDE (8.2.12) tool, with the default settings but adapted: with the GAMMA nucleotide substitution model and 1000 rapid bootstrap replicates. Bayesian analysis was generated from MrBayes v. 3.0b4 (Ronquist and Huelsenbeck, 2003), and the model of evolution was estimated with MrModeltest v. 2.2 (Nylander, 2004). The posterior probabilities (PP) (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002) were determined by the following Markov chain Monte Carlo sampling (MCMC) in MrBayes v.3.0b4 (Huelsenbeck and Ronquist, 2001). Six simultaneous Markov chains were run for 1,000,000 generations, with trees sampled every 100th generation. The preburn was set to 0.25 and the run was automatically stopped when the mean standard deviation of the split frequency reached below 0.01 (Maharachchikumbura et al., 2015). The bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.95, are given at the respective branches of each phylogenetic trees (See the Supplementary Tables 1–5). GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis.
2.4 Taxonomic classification
The higher-level taxonomic classification of each freshwater endophytic fungi was based on Wijayawardene et al. (2022a). Index Fungorum identifiers were obtained from Index Fungorum (2024) for the newly introduced taxa following the requirements mentioned in Art. F5.1 of International Code of Nomenclature for Algae, Fungi, and Plant.
2.5 Extracellular enzymatic assay of endophytic fungi
The qualitative analysis of amylolytic, cellulolytic, and laccase enzymatic activities of the endophyte isolates conducted using colourimetric changes in the PDA medium. Petri dishes containing PDA supplemented with tetracycline (1600 µg/mL), preventing bacterial contamination (Elshafie et al., 2019). The specific substrates were incorporated into the PDA media for each enzymatic identification, excluding laccase enzymatic activity. Each enzymatic assay included both negative and positive controls. The negative controls consisted of uninoculated fungal PDA plates supplemented with each substrate and treated with the specific chemicals used in each enzymatic assay. The positive controls, involved inoculating fungal PDA plates supplemented with each substrate and treated with the specific chemicals used in each enzymatic assay. All positive and negative controls were incubated at the required incubation temperatures and time periods (see methodology sections 2.5.1, 2.5.2, and 2.5.3).
2.5.1 Qualitative identification of amylase enzymatic activity
Petri dishes containing PDA supplemented with 1% starch were employed for the experiment. The fungal inoculum, comprising small fragments of mycelium (0.5 × 0.5 cm), was carefully placed in the centre of the PDA Petri dishes. Subsequently, the dishes were incubated at 28–30°C for a duration of seven days to facilitate fungal growth and development. Following the incubation period, 1–2 mL of iodine solution was added to each dish. The dishes were then incubated for an additional hour at 28–30°C. Following this incubation, the dishes were thoroughly washed with distilled water to remove any excess iodine solution. The success of the experiment was determined by observing colourimetric changes. A distinct halo appearing around the fungal colony was indicative of a positive result, highlighting the presence of cellulolytic activity (Robledo-Mahón et al., 2020).
2.5.2 Qualitative identification of cellulase enzymatic activity
The 0.5% (w/v) sodium carboxymethyl cellulose (CMC) (Central Drug House Pvt. Ltd., India) was added to the PDA media to evaluate cellulolytic activity. Small pieces of mycelium (0.5 × 0.5 cm) were then positioned on PDA petri dishes. The prepared fungal plates underwent incubation at 28–30°C for a period of three to five days. The qualitative cellulase activity of fungal isolates was assessed based on their ability to proliferate and create cleared zones around colonies on a solid medium. The surface of the media containing the developed fungal colonies was flooded with 0.1% (w/v) Congo red (Himedia Laboratories Pvt. Ltd., Mumbai, India) solution and incubated for 15 minutes at 28–30°C. Afterwards, the dye was removed with sterile distilled water, and the plates underwent an additional 10-minute incubation period at 28–30°C. Subsequently, the plates were further treated by flooding with 1M NaCl (Daejung Chemicals and Metals Co., Ltd., South Korea) for 5 minutes.
2.5.3 Qualitative identification of laccase enzymatic activity
The small pieces of mycelium (0.5 × 0.5 cm) were placed on a PDA medium and incubated for approximately five days at 28–30°C temperature. The colonized Petri dishes were utilized for adding solutions by droplets at the edge of each colony. Laccase activity was determined using a 0.1 M 1-Naphthol (Research Lab Fine Chem. Industries, Mumbai, India). Following the addition of droplets, Petri dishes were incubated for 24 hours at 28–30°C temperature and changes in the colour of the edge of the colony were considered positive results. The blue-purple appearance was displayed for laccase activity at the edge of each fungal colony (Gramss et al., 1998; Robledo-Mahón et al., 2020).
3 Results
3.1 Phylogenetic analyses
The taxa for each analysis were selected based on blast similarity and related publications and closely related sequences were downloaded from GenBank (See the Supplementary Tables 1–5).
3.1.1 Multi-gene analyses for Ectophoma
The concatenated dataset of LSU, ITS, rpb2, and tub2 regions contained 14 isolates, which comprised 2434 characters with gaps. Single gene analysis was carried out to compare the topology of the tree and clade stability. Didymella exigua (CBS 183.55) was used as the outgroup taxon. The best-scoring RAxML tree with a final likelihood value of -4415.606556 is presented in Figure 3. The matrix had 127 distinct alignment patterns, with 13.83% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.236991, C = 0.247923, G = 0.273055, T = 0.242031; substitution rates AC = 2.940086, AG = 5.756609, AT = 1.806664, CG = 1.455718, CT = 18.313095, GT = 1.000000; gamma distribution shape parameter alpha = 0.020000. In the phylogenetic analysis, our new strains (RUFCC2458 and RUFCC2462) form the sister clade to Ectophoma multirostrata (CBS 274.60 (ex-type) and CBS 380.67) and E. iranica (SCUATK1G1 (ex-type) and SCUAK1) with moderate statistical values (95% ML), 96 PP with BP values more than 95%.
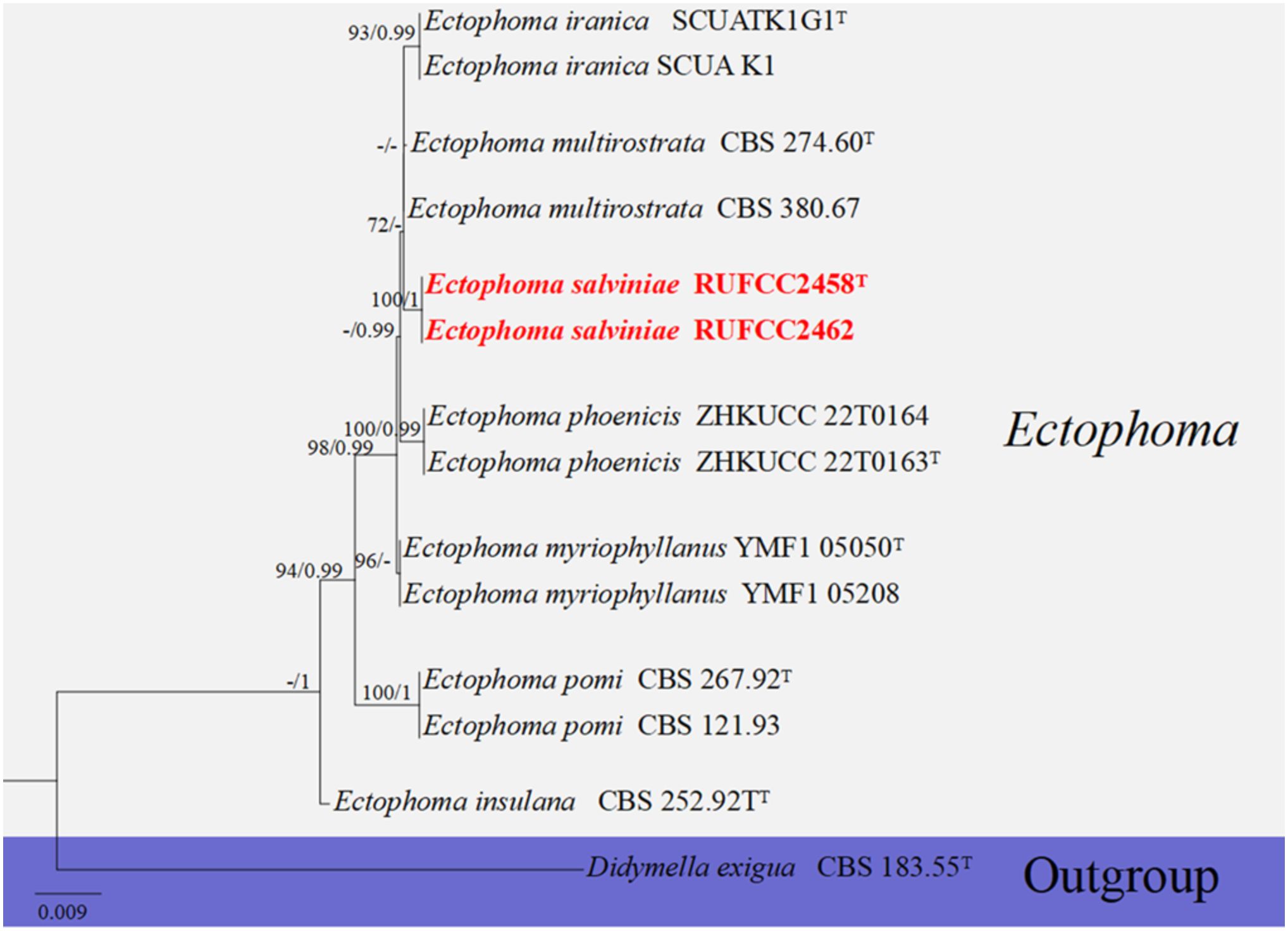
Figure 3. Phylogenetic tree from the best scoring of the RAxML analysis based on combined (ITS, LSU, rpb2 and tub2) is rooted to Didymella exigua (CBS 183.55). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.95, are given at the respective branches. Hyphen (-) means a value lower than 75% (BS) or 0.95 (PP). Ex-types are marked in “T”. New isolates are labeled in bold and red.
3.1.2 Multi-gene analyses for Phyllosticta
The concatenated ITS, tef1-α, ACT and GADPH region dataset contained 38 isolates, comprising 1698 characters with gaps. Single gene analysis was carried out to compare the topology of the tree and clade stability. Botryosphaeria obtusa (CMW 8232) and B. stevensii (CBS 112553) were used as the outgroup taxa. The best-scoring RAxML tree with a final likelihood value of -9726.371339 is presented in Figure 4. The matrix had 727 distinct alignment patterns, with 20.00% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.200267, C = 0.311861, G = 0.264840, T = 0.223032; substitution rates AC = 0.893796, AG = 2.729183, AT = 1.229527, CG = 1.044891, CT = 6.112001, GT = 1.000000; gamma distribution shape parameter alpha = 0.344610. The GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis. In the phylogenetic analysis, our new strain (RUFCC2452) clustered with Phyllosticta capitalensis (CBS 128856) with high statistical support (75% ML, 0.99 PP).

Figure 4. Phylogenetic tree from the best scoring of the RAxML analysis based on combined (ITS, tef1-α, ACT and GADPH) is rooted to Botryosphaeria obtusa (CMW 8232) and B. stevensii (CBS 112553). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 75% and 0.95, are given at the respective branches. Hyphen (-) means a value lower than 50% (BS) or 0.95 (PP). Ex-types are marked in “T”. New isolates are labelled in bold and red.
3.1.3 Multi-gene analyses for Colletotrichum
The concatenated dataset of ITS, tub2, ACT, CHS-1 and GADPH regions contained 39 isolates comprising 1818 characters with gaps. Single gene analysis was carried out to compare the topology of the tree and clade stability. Colletotrichum boninense (CBS 123755) and C. chamaedoreae (LC:13868) were used as the outgroup taxa. The best-scoring RAxML tree with a final likelihood value of -8361.798032 is presented in Figure 5. The matrix had 644 distinct alignment patterns, with 3.72% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.232029, C = 0.290209, G = 0.245763, T = 0.231999; substitution rates AC = 1.179354, AG = 3.249038, AT = 1.464001, CG = 0.806083, CT = 5.743800, GT = 1.000000; gamma distribution shape parameter alpha = 0.281325. The GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis. In the phylogenetic analysis, our new strains (RUFCC2457 and RUFCC2455) clustered in the clade that comprises Colletotrichum siamense (CBS 125378 (ex-type), C. australianum (BRIP 63698), and C. queenslandicum (ICMP 1778) with moderate statistical values (79% ML, 0.95 PP). We compared the conidial morphologies of the new collection against the three species and confirmed that our collections belong to Colletotrichum siamense (See the taxonomy section). While another new collection (RUFCC2451) clustered in the clade Colletotrichum truncatum with high statistical values (100% ML, 1 PP).

Figure 5. Phylogenetic tree from the best scoring of the RAxML analysis based on combined (ITS, tub2, ACT, CHS-1 and GADP) is rooted to Colletotrichum boninense (CBS 123755) and C. chamaedoreae (LC:13868). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.95, are given at the respective branches. Hyphen (-) means a value lower than 75% (BS) or 0.95 (PP). Ex-types are marked in “T”. New isolates are labelled in bold and red.
3.1.4 Multi-gene analyses for Chaetomella
The concatenated dataset of LSU, ITS and SSU regions contained 24 isolates, which comprised 2977 characters with gaps. Single gene analysis was carried out to compare the topology of the tree and clade stability. Hymenoscyphus scutula (CBS 101.66) and H. fructigenus (CBS 186.47) were used as the outgroup taxa. The best-scoring RAxML tree with a final likelihood value of -7269.420511 is presented in Figure 6. The matrix had 353 distinct alignment patterns, with 37.79% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.263303, C = 0.209012, G = 0.275987, T = 0.251698; substitution rates AC = 1.805316, AG = 2.136673, AT = 0.949986, CG = 0.780819, CT = 4.656160, GT = 1.000000; gamma distribution shape parameter alpha = 0.020000. The GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis. In the phylogenetic analysis, our new strain (RUFCC2453) clustered in the clade Chaetomella raphigera with high statistical values (100% ML, 1 PP).
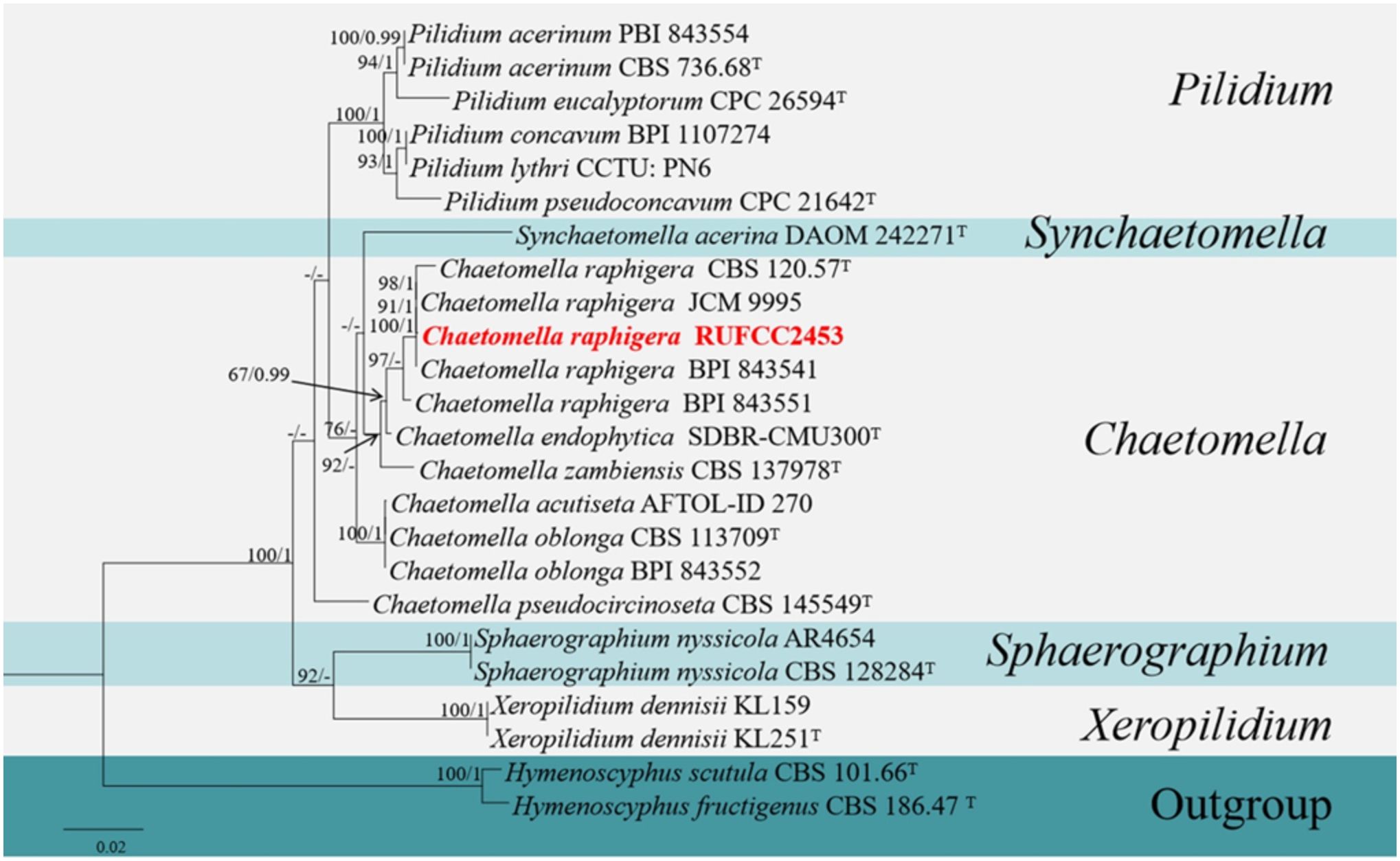
Figure 6. Phylogenetic tree from the best scoring of the RAxML analysis based on combined (LSU, ITS and SSU) is rooted to Hymenoscyphus scutula (CBS 101.66) and H. fructigenus (CBS 186.47). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.95, are given at the respective branches. Hyphen (-) means a value lower than 50% (BS) or 0.95 (PP). Ex-types are marked in “T”. New isolates are labelled in bold and red.
3.1.5 Multi-gene analyses for Neottiosporina
The concatenated dataset of ITS regions contained eleven isolates, which comprised 539 characters with gaps. Single gene analysis was carried out to compare the topology of the tree and clade stability. Suttonomyces rosae (MFLUCC 15-0051) was used as the outgroup taxon. The best-scoring RAxML tree with a final likelihood value of -1790.869339 is presented in Figure 7. The matrix had 138 distinct alignment patterns, with 3.39% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.220321, C = 0.266236, G = 0.234288, T = 0.279155; substitution rates AC = 4.232102, AG = 5.947883, AT = 5.637501, CG = 0.698220, CT = 13.019587, GT = 1.000000; gamma distribution shape parameter alpha = 0.164930. The GTR+I+G model was selected as the best model based on MrModeltest and was used for the Bayesian analysis. In the phylogenetic analysis, our new strain (RUFCC2454 (ex-type), and RUFCC2461) form a sister clustered with Neottiosporina cylindrica (BRIP 14187 (ex-type) and BRIP (16231) with high statistical values (84% ML, 0.95 PP). Based on the phylogenetic analyses and morphological characters, we confirm our strains differ from Neottiosporina cylindrica. Herein, we report our strains as a novel species viz. Neottiosporina mihintaleensis sp. nov.
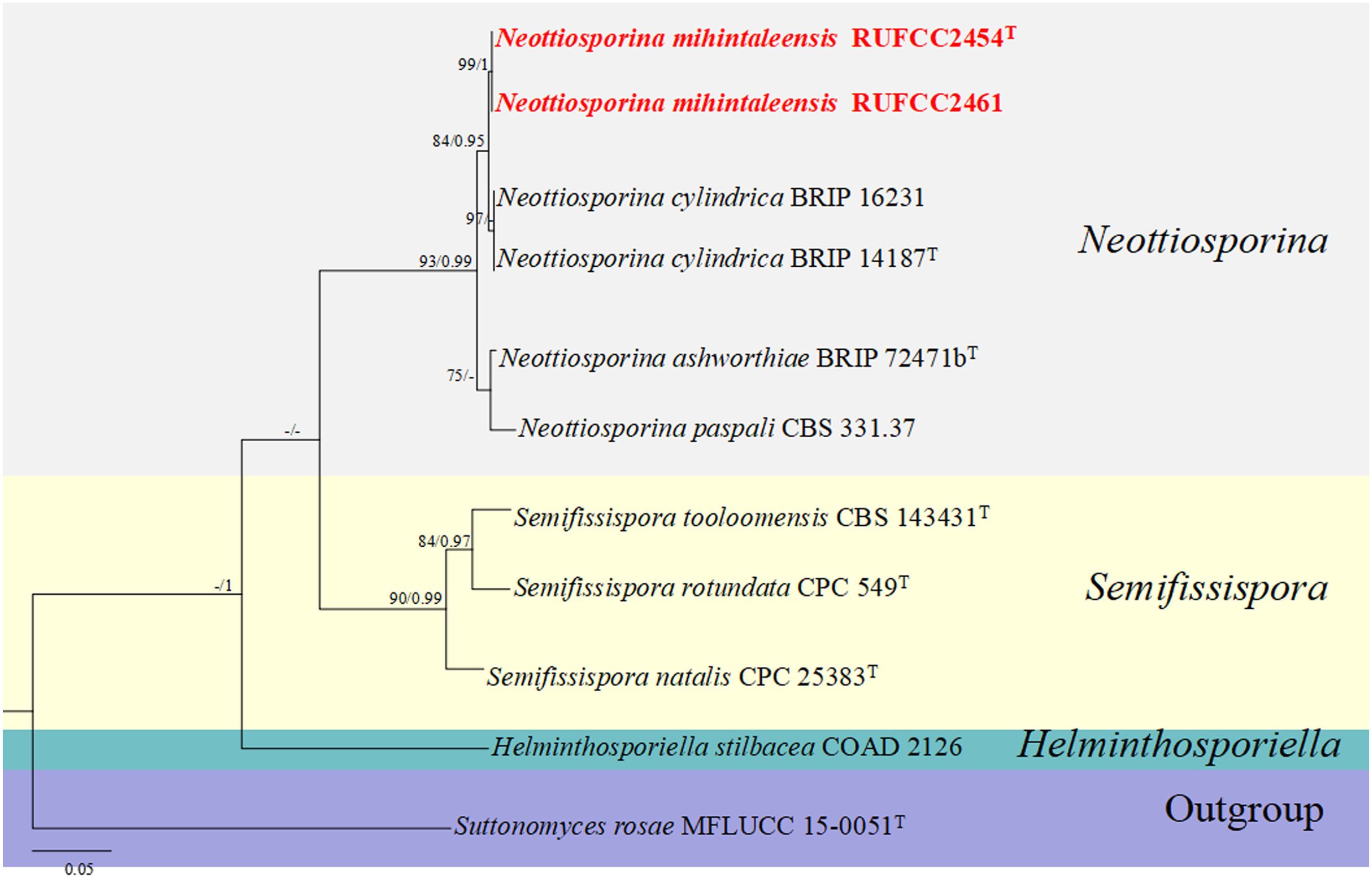
Figure 7. Phylogenetic tree from the best scoring of the RAxML analysis based on combined (ITS) is rooted to Suttonomyces rosae (MFLUCC 15-0051). Bootstrap values for maximum likelihood (MLBP) and Bayesian posterior probabilities (BYPP) equal to or greater than 50% and 0.95, are given at the respective branches. Hyphen (-) means a value lower than 50% (BS) or 0.95 (PP). Cultures from holotype and specimens are marked in “T”. New isolates are labelled in bold and red.
3.2 Taxonomy
In this section, we listed all the collected taxa according to the higher-level classification referenced by Wijayawardene et al. (2022a).
Ascomycota Caval.-Sm.
Dothideomycetes O.E. Erikss. & Winka
Pleosporales Luttr. ex M.E. Barr
Didymellaceae Gruyter, Aveskamp & Verkley
Ectophoma Valenz.-Lopez, Cano, Crous, Guarro and Stchigel, Stud. Mycol. 90: 34 (2017)
Index Fungorum Registration Identifier: 819952
Notes: The genus Ectophoma was introduced by Valenzuela-Lopez et al. (2018) with E. multirostrata (basionym: Sphaeronaema multirostratum P.N. Mathur et al.) as the type species. Ectophoma comprises six species in diverse habitats (Valenzuela-Lopez et al., 2018) and a well-defined genus in Didymellaceae (Hou et al., 2020a, b). In this study, we introduce Ectophoma salviniae sp. nov. from a healthy leaf of Salvinia minima (Watermoss).
Ectophoma salviniae Wimalasena, Wijayaw. & Bamunuarachchige sp. nov.
Index Fungorum Registration Identifier: IF902503 (Figure 8).

Figure 8. Ectophoma salviniae sp. nov. (A, B). Salvinia minima in the lake. (A) and in the lab (B) (Watermoss). (C) Top view of the PDA culture plate (diameter; 9.5 cm). (D) Downside of the PDA culture plate (diameter; 9.5 cm) after five days. (E) Sporulated culture after seven days. (F) Stereo microscopic view of sporulation in culture. (G, H) Squashed pycnidium. (I) Inner wall of pycnidium. (J) Conidial production. (K) Conidiogenous cells and conidia. (L, M) Conidia. Scale bars: (F) = 1000 µm, (G–M) = 100 µm.
Etymology: The name is derived from the host genus Salvinia, from which the fungus was isolated.
Holotype: RUSLH/240
Description: Endophytic of healthy leaf of Salvinia minima. Sexual morph: Undetermined. Asexual morph on the culture: Colonies on PDA slow growing, effuse, with a regular margin, flat, white to grey olivaceous reaching a diam of 1.5–2 cm after 7 days at 28°C. Mycelium regular, composed of filamentous, septate, branched, smooth, pale olivaceous hyphae 0.5 µm wide. Conidiomata pycnidial, 140–300 µm long × 50–80 µm wide, brown to dark brown, solitary or confluent, abundant, ostiolate, with one or more short necks. Pycnidial wall, glabrous, with globose to subglobose or irregular cells of textura angularis. Conidiogenous cells holoblastic to phialidic, minute. Conidia 6.1–9.6 × 3.6–6.7 ( = 8.0 × 5.0 µm; n = 30) µm, aseptate, hyaline, smooth-walled, oblong to ellipsoidal, end of conidia is acute, guttulate: two guttules are inside the conidial cytoplasm.
Culture characteristics: Colonies on PDA reached a diameter of 1.5–2 cm after 7 days at 28°C, with a regular margin and flat, colourless to weak olivaceous, poorly developed, white to grey olivaceous aerial mycelium. The centre of the colony is olivaceous, followed by a black circle formed by abundant pycnidia. The reverse of the colony was dark black and featured some radially furrowed zones and concentric circles of greyish-black colours.
Material examined: SRI LANKA, North Central Province, Mihintale, Iluppukanniya Tank (8.36482° N, 80.50764° E, 118 m), on healthy leaf of Salvinia minima (Watermoss), 28 November 2023, Madhara K. Wimalasena, RUSLH/240 (dried culture as the holotype), RUFCC2462 (ex-type).
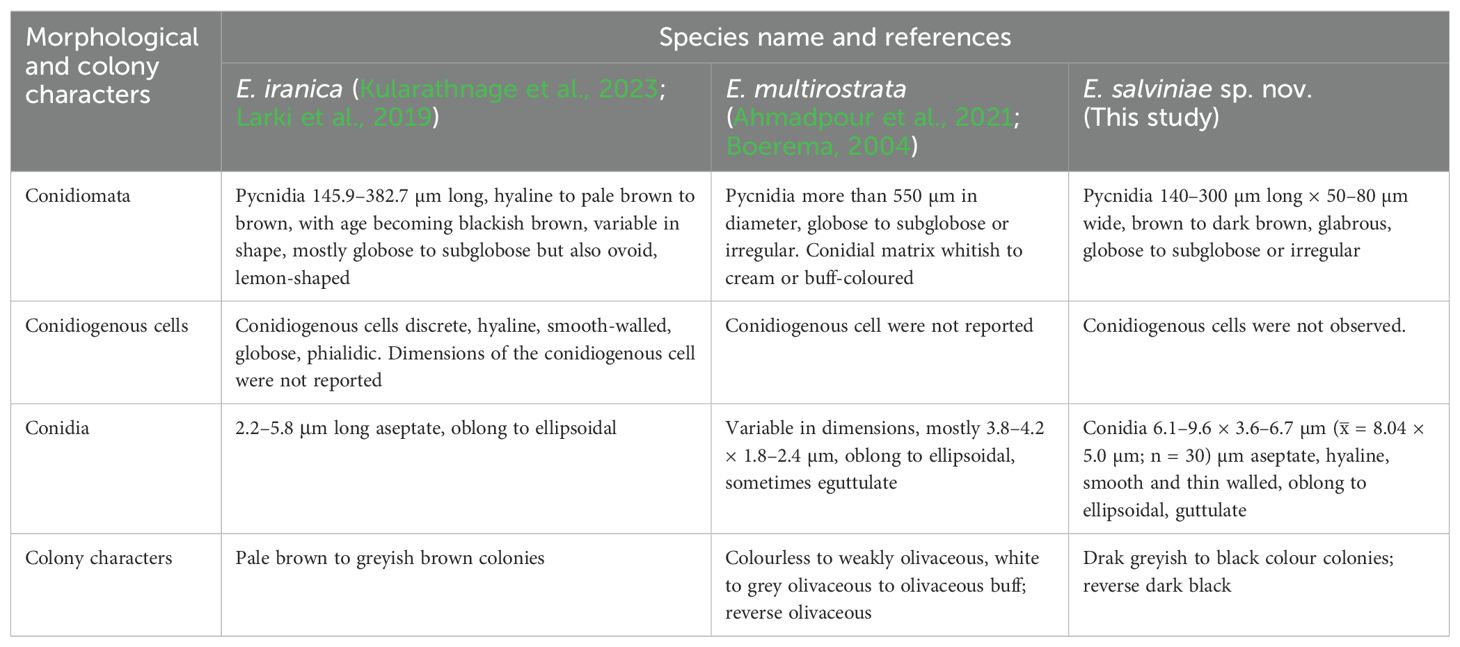
Notes: The multi-locus analyses of combined data set of ITS, LSU, rpb2, and tub2 sequence data revealed that our isolates (RUFCC2458, RUFCC2462) are clustered within the Ectophoma s. str., forming a sister clade to E. iranica (SCUATK1G1 (ex-type) and SCUAK1) and E. multirostrata (CBS 274.60 (ex-type) and CBS 380.67). The conidiomata of E. salviniae show a slight similarity in shape and the dimensions to those of E. iranica and E. multirostrata. However, E. salviniae has larger conidia than in both E. iranica and E. multirostrata (Table 3). Furthermore, its cultural characteristics, including dark greyish to black colonies, are also distinct from E. iranica and E. multirostrata (Table 3). Moreover, phylogenetic analysis further confirms that E. salviniae is distinct from E. iranica and E. multirostrata (Figure 3). Herein, the taxon, represented by RUFCC2458 (ex-type) and RUFCC2462 is introduced as a novel species viz., Ectophoma salviniae sp. nov.
Botryosphaeriales C.L. Schoch, Crous & Shoemaker
Phyllostictaceae Fr.
Phyllosticta Pers., Traité champ. Comest. (Paris): 55, 147 (1818)
Index Fungorum Registration Identifier: 9384
= Guignardia Viala & Ravaz, Bull. Soc. mycol. Fr. 8(2): 63 (1892)
Notes: Phyllosticta is a well-established genus in Phyllostictaceae, Botryosphaeriales with over 3000 species epithets in the Index Fungorum 2024 (accession date: 14 of May 2024). This genus currently includes 1,499 recognized species (Sui et al., 2023). Recently, Gong et al. (2024) introduced two novel species (P. savannaensis and P. ovalina), and Jiang et al. (2024) added three new species (P. fujianensis, P. saprophytica, and P. turpiniae) to this genus. The members of Phyllosticta have been mostly reported as pathogens, saprobes, and endophytes of different hosts worldwide (Wikee et al., 2013; Rodrigues et al., 2019; Sui et al., 2023; Jiang et al., 2024). In this study, we isolated Phyllosticta capitalensis as an endophytic taxon from Nymphaea nouchali.
Phyllosticta capitalensis Henn., Hedwigia 48: 13 (1908) [1909]
Index Fungorum Registration Identifier: 168326 (Figure 9).
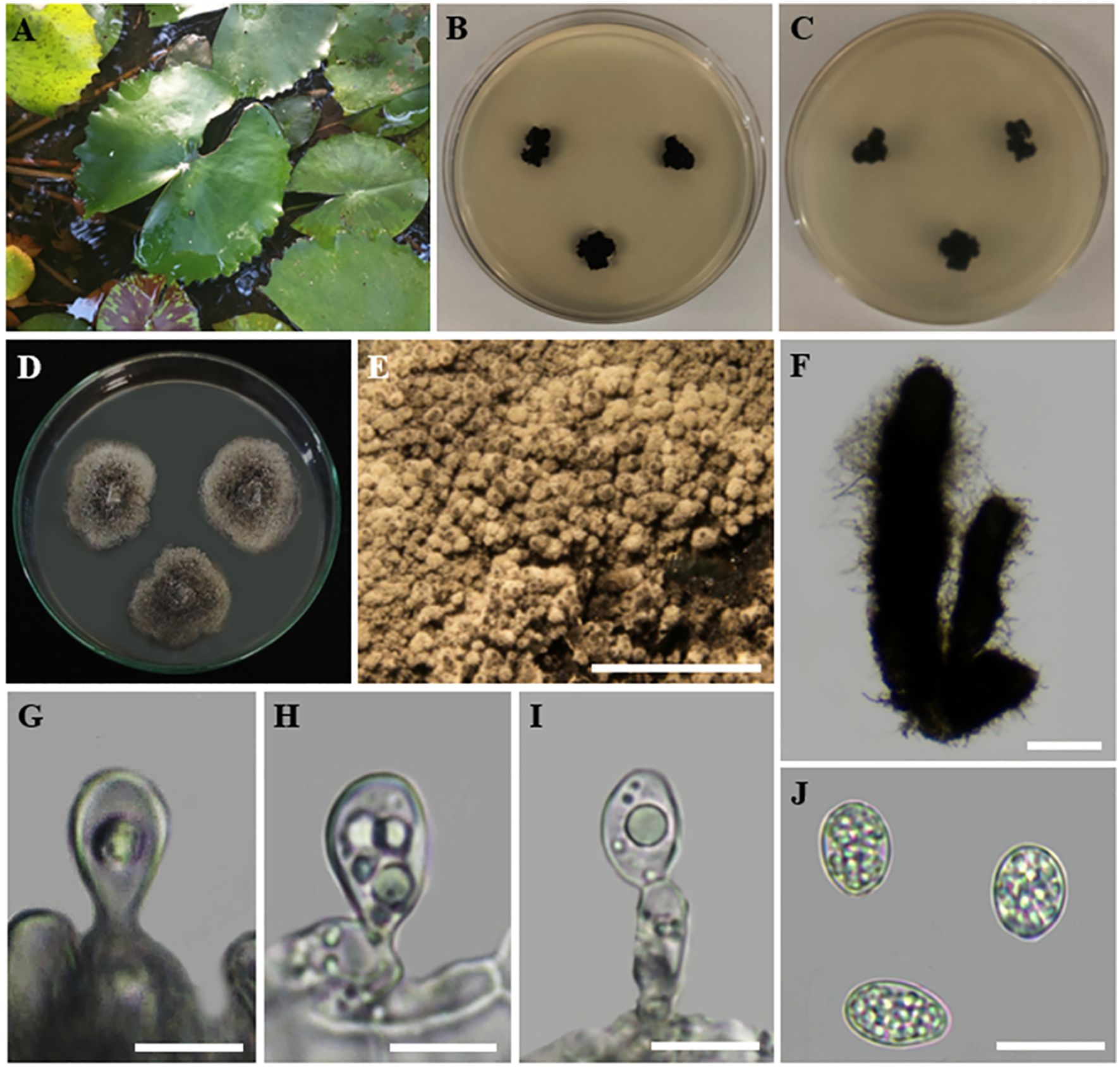
Figure 9. Phyllosticta capitalensis (A) Host freshwater plant leaf of Nymphaea nouchali (Blue Water-Lily). (B) Upside of the PDA culture plate (diameter; 9.5 cm). (C) Downside of the PDA culture plate (diameter; 9.5 cm) after five days. (D) Sporulated culture after seven days. (E) Stereo microscopic view of sporulation in culture. (F) Conidiomata. (G-I) Conidiogenous cells. (J) Conidia. Scale bars: (E) = 1000 µm, (F) = 50 µm (G-J) = 100 µm.
Description: Endophytic of healthy leaf of Nymphaea nouchali. Sexual morph: Undetermined; Asexual morph: Pycnidia up to 300 µm diam, 250 µm tall, black, aggregated, erumpent, globose to ampulliform, ostiolate, exuding a colourless, glossy, slimy conidial mass. Pycnidial wall consisting of 6–8 layers, up to 40 μm thick, with cells of textura angularis. Ostiole single, central, 5–15 μm diam. Conidiophores subcylindrical to ampulliform, frequently reduced to conidiogenous cells, or branching from a basal supporting cell, coated in mucoid layer, 7–20 × 3–7 μm. Conidiogenous cells subcylindrical to ampulliform to doliiform, holoblastic, polyblastic, hyaline, smooth, 7–10 × 3–5 μm; percurrently proliferating 1–2 times near apex. Conidia 10–11 × 6–7 μm ( = 9.5 × 6.5 µm; n = 30), solitary, hyaline, aseptate, thin- and smooth-walled, coarsely guttulate, ellipsoid to obovoid, tapering toward a narrowly truncate base, enclosed in a mucilaginous 2–4 μm thick sheath, bearing a hyaline, mucoid, straight to curved, unbranched, 6–8 × 1–1.5 μm, apical appendage on a bluntly rounded apex.
Culture characteristics: Colonies incubated for 14 days at 28–30°C in darkness on PDA initially appear woolly and white with abundant mycelium. Over the next 2–3 days, they transform from greenish to dark green, with white hyphae visible along the undulating margins, eventually becoming black. Over two weeks of incubation in darkness at 28°C, the mycelium extends to the edge of the Petri dish.
Material examined: SRI LANKA, North Central Province, Mihintale, Iluppukanniya tank (8.36482° N, 80.50764° E, 118 m), on healthy leaf of Nymphaea nouchali (Blue Water-Lily), 02 December 2023, Madhara K. Wimalasena, RUFCC2452 (living culture), RUSLH/242 (dried culture as the herbarium specimen).
Notes: Phyllosticta capitalensis is often found as an endophyte on a wide range of hosts and exhibits a broad geographic distribution (Silva and Pereira, 2007; Silva et al., 2008; Glienke et al., 2011; Wang et al., 2023). It was reported from 70 plant families and is considered a weak plant pathogen (Wikee et al., 2013). Phyllosticta capitalensis has been previously reported in Sri Lanka as a rubber foliar pathogen (Herath et al., 2019) and as an endophytic fungus in the leaves of Camellia sinensis (Thambugala et al., 2018). Dissanayake et al. (2016) reported Phyllosticta capitalensis on healthy specimens of Nymphaea nouchali collected from an unpolluted natural freshwater pond in the Western Province of Sri Lanka, based on a single gene locus (ITS) study. In this study, we reconfirmed the occurrence of Phyllosticta capitalensis on healthy leaves of Nymphaea nouchali in lentic freshwater habitats in the North-Central Province of Sri Lanka, based on four gene loci (ITS, tef1-α, ACT, and GADPH) study, a polyphasic approach. Previous reports of Phyllosticta species in freshwater plants worldwide include Phyllosticta aquatica (on Lemna minor fide (Spegazzini, 1881), P. fatiscens (on Nymphaea odorata fide (Anonymous, 1960), and P. nymphaeacea (on Nymphaea sp. fide (Ellis and Everhart, 1900). According to (Farr et al., 2012), P. capitalensis has not been reported from Nymphaea nouchali so far, and thus, this is the first confirmative report of P. capitalensis on Nymphaea nouchali. When comparing the recently isolated P. capitalensis (SDBR-CMU497 and SDBR-CMU498) (Chaiwong et al., 2024) isolates with P. capitalensis RUFCC2452, their morphological features, such as pycnidia, conidiophores, and conidiogenous cells, are similar. However, the asexual conidia of P. capitalensis RUFCC2452 are slightly larger than those of the SDBR-CMU497 and SDBR-CMU498 (5.2 to 9.4 × 3.6 to 7.5 µm (n = 50) isolates.
Leotiomycetes O.E. Erikss. & Winka
Chaetomellales Crous & Denman
Chaetomellaceae Baral, P.R. Johnst. & Rossman
Chaetomella Fuckel, Jb. nassau. Ver. Naturk. 23–24: 401 (1870) [1869–70]
Index Fungorum Registration Identifier: 7575
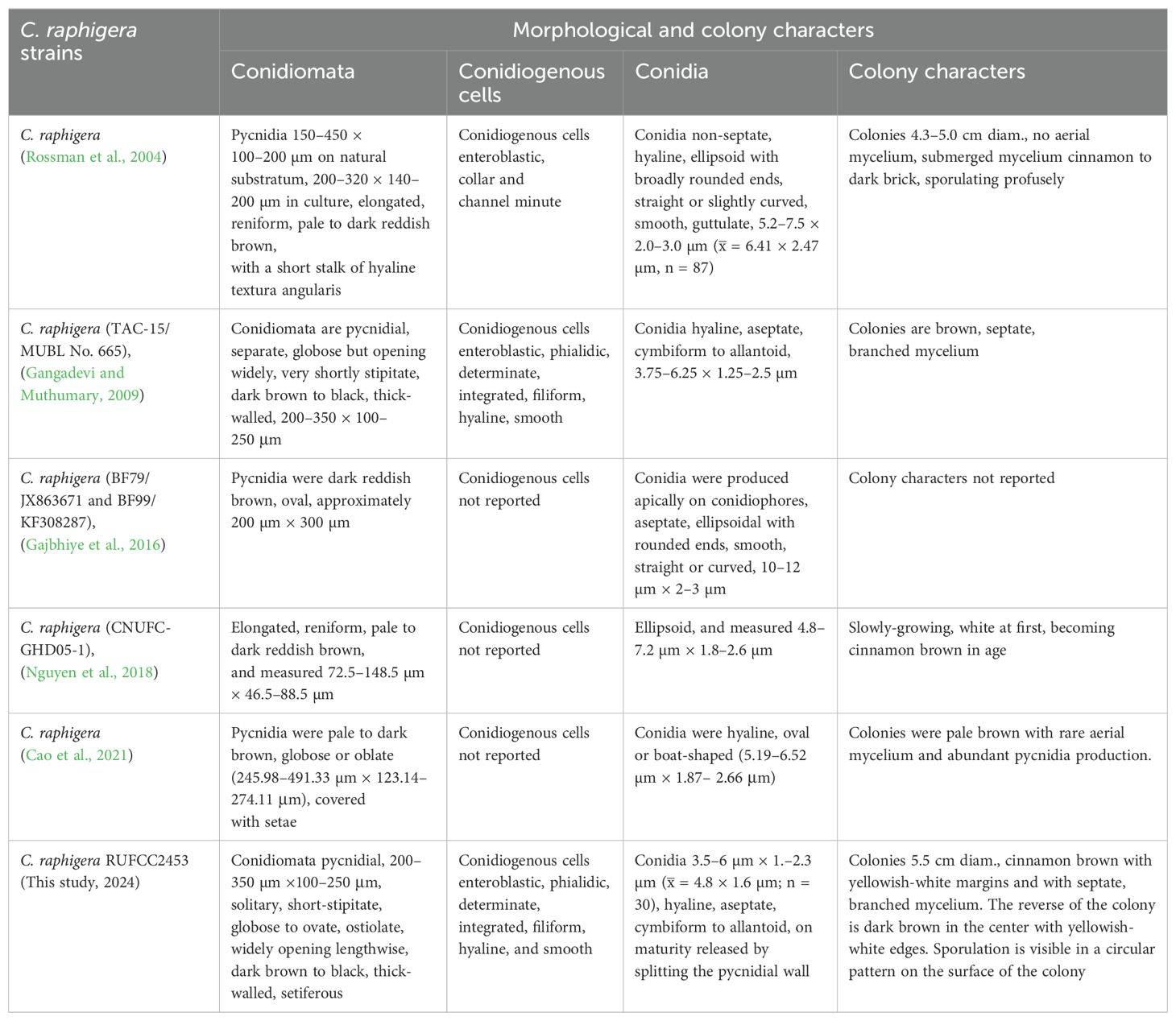
Notes: Fuckel (1869), established the genus Chaetomella, including two species; C. oblonga, characterized by hyaline spores, and C. atra, characterized by olivaceous spores. Among these, the type species was C. oblonga (Rossman et al., 2004). The members of the genus were reported as plant pathogens (Gajbhiye et al., 2016; Pärtel et al., 2017; Nguyen et al., 2018; Suwannarach et al., 2018; Cao et al., 2021), saprophytes and as endophyte (Suwannarach et al., 2018). As of June 2024, the Index Fungorum lists 61 records for the genus Chaetomella (https://www.indexfungorum.org/names/Names.asp). In this study, we report C. raphigera from the healthy leaf of Eichhornia crassipes as an endophytic species.
Chaetomella raphigera Swift, Mycologia 22(4): 165 (1930)
= Volutellospora raphigera (Swift) Thirum. & P.N. Mathur, Sydowia 18 (1–6):38 (1965)
= Chaetomella terricola P.Rama Rao, Mycopathologia et Mycologia Applicata 19 (3):255 (1963)
Index Fungorum Registration Identifier: 163400 (Figure 10).

Figure 10. Chaetomella raphigera (A) Host freshwater plant leaf of Eichhornia crassipes (Water hyacinth). (B) Upside of the PDA culture plate (diameter; 9.5 cm). (C) Downside of the PDA culture plate (diameter; 9.5 cm) after five days. (D) Sporulated culture after seven days. (E, F) Stereo microscopic view of sporulation in culture. (G) Pycnidia. (H, I) Types of setae on pycnidium. (J, K) Pycnidia release the conidia. (L) Conidiogenous cells. (M) Conidia. Scale bars: (E, F) = 1000 µm (G–M) = 100 µm.
Description: Endophytic of healthy leaf of Eichhornia crassipes. Sexual morph: Undetermined. Asexual morph: Conidiomata pycnidial, 200–350×100–250 μm, solitary, short-stipitate, globose to ovate, ostiolate, widely opening lengthwise, dark brown to black, thick-walled, setiferous. Basal stipe short, composed of hyaline, pseudoparenchymatous cells. Setae 50–100×2.5–5 μm, brown, smooth, thick-walled, septate, unbranched, with hooked apices. Conidiophores hyaline, short, branched, filiform, septate, and smooth. Conidiogenous cells enteroblastic, phialidic, determinate, integrated, filiform, hyaline, and smooth. Conidia 3.5–6 × 1.–2.3 µm ( = 4.8 × 1.6 µm; n = 30), hyaline, aseptate, cymbiform to allantoid, on maturity released by splitting the pycnidial wall along the thin-walled cells of the raphe, in mass becoming amber-coloured on aging.
Culture characteristics: Colonies on incubation for 14 days at 28–30°C in darkness on PDA media, attaining 5.5 cm diam., cinnamon brown with yellowish-white margins and with septate, branched mycelium. The reverse of the colony is dark brown in the center with yellowish-white edges. Sporulation is visible in a circular pattern on the surface of the colony.
Material examined: SRI LANKA, North Central Province, Mihintale, Iluppukanniya tank (8.36482° N, 80.50764° E, 118 m), on healthy leaf of Eichhornia crassipes (Water hyacinth), 5 December 2023, Madhara K. Wimalasena, RUFCC2453 (living culture), RUSLH/243 (dried culture as the herbarium specimen).
Notes: Chaetomella raphigera has been reported as a plant pathogen from India (Gajbhiye et al., 2016). However, this is the first report of C. raphigera as a new geographical record in Sri Lanka, found in the freshwater plant Eichhornia crassipes. Besides, Chaetomella species have not been reported in Sri Lanka thus this is the first genus report from the country. Morphological similarities of C. raphigera (RUFCC2453) with previous studies are mentioned in Table 4.
Sordariomycetes O.E. Erikss. & Winka
Glomerellales Chadef. ex Réblová, W. Gams & Seifert
Glomerellaceae Locq. ex Seifert & W. Gams
Colletotrichum Corda
Index Fungorum Registration Identifier: 7737
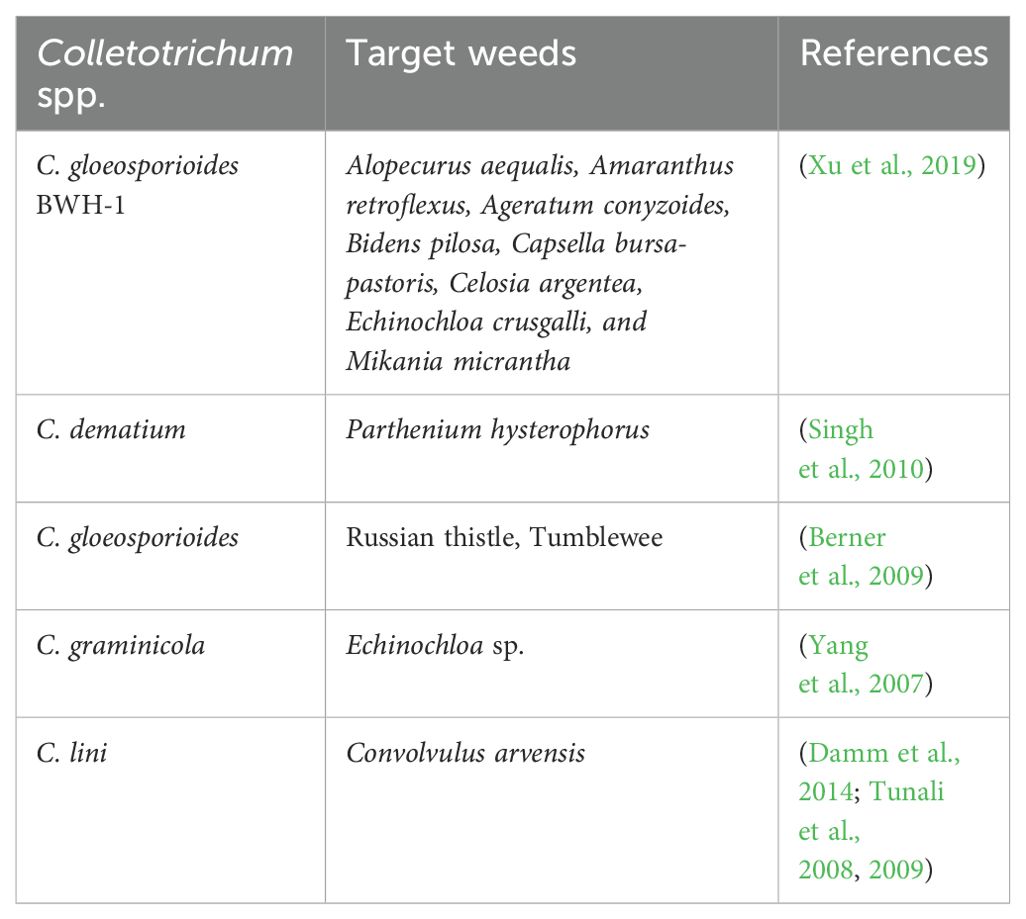
Note: Colletotrichum represents a diverse and complex genus, with currently 344 recognized species grouped into 20 species complexes (Talhinhas and Baroncelli, 2021). These species often lead to considerable economic losses, mostly infecting economically important crops (Peng et al., 2023; Peralta-Ruiz et al., 2023; Zhang et al., 2023). The members of Colletotrichum exhibit different lifestyles that are found in varied environments and host species (Jayawardena et al., 2016a, b; Samarakoon et al., 2018; Talhinhas and Baroncelli, 2023). These include necrotrophic (Vargas et al., 2012; De Silva et al., 2017; Talhinhas and Baroncelli, 2021; Páez Redondo et al., 2022), biotrophic and hemibiotrophic (De Silva et al., 2017; Páez Redondo et al., 2022; Jia et al., 2023), quiescent (De Silva et al., 2017; Fu et al., 2022), and endophytic (De Silva et al., 2017; Páez Redondo et al., 2022; Lin et al., 2023; Liu et al., 2023; Zhang et al., 2023; Barreto Ramos et al., 2024) lifestyles. Among these lifestyles, endophytic Colletotrichum spp. have been documented in marine environments such as mangroves (Grano-Maldonado et al., 2021; Norphanphoun and Hyde, 2023; Aumentado et al., 2024) and freshwater habitats (Zheng et al., 2022). In this study, we isolated C. siamense and C. truncatum as two endophytic taxa of Eichhornia crassipes in freshwater environments.
Colletotrichum siamense Prihast., L. Cai & K.D. Hyde, Fungal Diversity 39: 98 (2009)
Index Fungorum Registration Identifier: 515410 (Figure 11).

Figure 11. Colletotrichum siamense (A) Host freshwater plant leaf of Eichhornia crassipes (Water hyacinth). (B) Upside of the PDA culture plate (diameter; 9.5 cm). (C) Downside of the PDA culture plate (diameter; 9.5 cm) after seven days. (D) Sporulated colony. (E) Acervuli with orange conidial ooze. (F, G) Conidiomata and conidiogenous cells. (H) Conidiogenous cells. (I–P), Maturity levels of conidiogenous cells. (P) Conidia. Scale bars: (E) = 500 µm, (F–P) = 100 µm.
Description: Endophytic of healthy leaf of Eichhornia crassipes. Sexual morph: Undetermined. Asexual morph: Conidiomata acervular, pulvinate, with erect conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores maconematous, hyaline, septate, branched. Conidiogenous cells hyaline, cylindrical to ampulliform, phialidic, smooth, guttulate, 7–15.5 μm long, 1–2.5 μm wide at apex. Conidia 9.89–15.95 × 3.73–5.67 µm ( = 13.75 × 4.75 µm; n = 30), hyaline, aseptate, smooth-walled, cylindrical, bluntly rounded at both ends, guttulate.
Culture characteristics: Colonies on incubating for ten days at 28–30°C in dark on PDA media, reaching a diameter of 1–1.5 cm Eichhornia crassipes. The aerial mycelium is white, cottony, and sparse. The colony surface features numerous, small acervuli with orange conidial ooze, and the reverse side remain pale yellowish.
Material examined: SRI LANKA, North Central Province, Mihintale tank (8.36267° N, 80.50591° E, 108 m), Mihintale, on healthy leaf of Eichhornia crassipes (Water hyacinth), 30 November 2023, Madhara K. Wimalasena, RUFCC2455 and RUFCC2457 (living cultures), RUSLH/244 (dried culture as the herbarium specimen).
Notes: Colletotrichum siamense and C. truncatum are important plant pathogens causing a wide range of diseases worldwide (Talhinhas and Baroncelli, 2023). Several studies reported C. siamense and C. truncatum from different hosts and habitats in Sri Lanka and these include Allium cepa (Herath et al., 2021), Hevea brasiliensis (Herath et al., 2019), Musa sp. (Kurera et al., 2023), and Persea americana (Dissanayake et al., 2021) as hosts for Colletotrichum siamense, while Begonia sp. (Wickramasinghe et al., 2019), Capsicum annuum (Welideniya et al., 2019), and Hevea brasiliensis (Herath et al., 2019) have been identified as hosts for Colletotrichum truncatum. De Silva et al. (2019) reported isolates of Colletotrichum siamense from different countries showed noticeable differences in growth rates and culture morphology. However, conidial measurements from isolates in distinct subclades of the phylogenetic tree were consistent, and the morphological traits within each subclade were highly uniform within each country (De Silva et al., 2019) (see Table 5 for the morphological comparison).
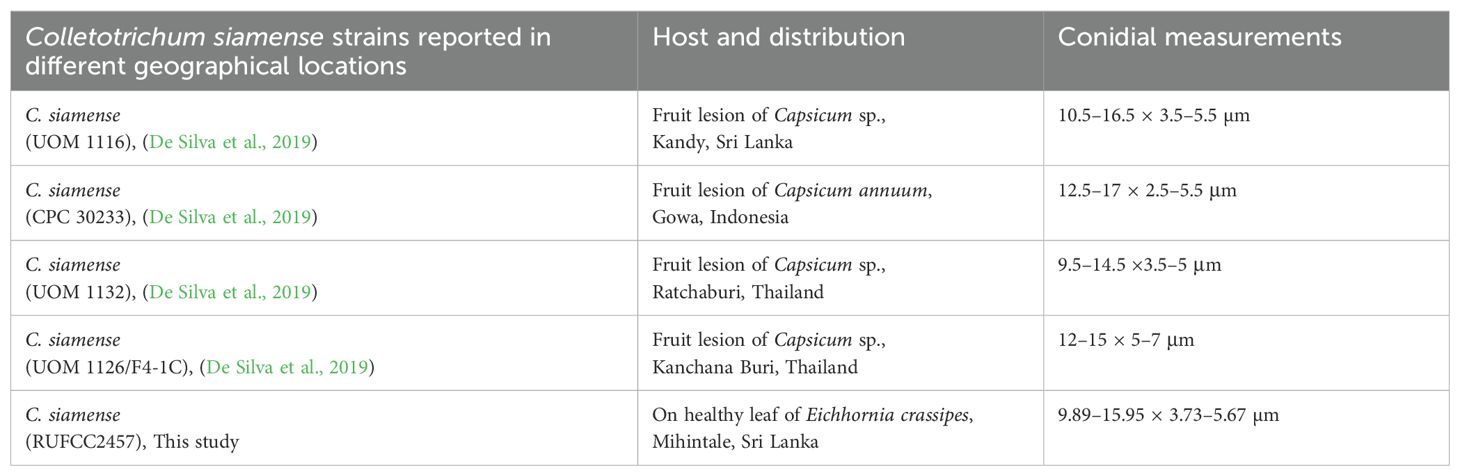
Table 5. Colletotrichum siamense strains reported in different geographical locations and their conidial measurements (CPC-Culture collection of P.W. Crous, housed at Westerdijk Fungal Biodiversity Institute, RUFCC-Rajarata University Fungal Culture Collection, UOM-University of Melbourne culture collection, Victoria, Australia).
Huang et al. (2021) reported C. fructicola (which causes irregular necrotic lesions on leaves, stems, and crown and petiole rot symptoms) from Eichhornia crassipes in China. However, as far as we know, there are no hitherto reports of C. siamense or C. truncatum on Eichhornia crassipes in Sri Lanka or elsewhere (2024; accession date: 06 June 2024, https://fungi.ars.usda.gov/). Hence, this is the first host report of C. siamense and C. truncatum from Eichhornia crassipes from Sri Lanka.
Colletotrichum truncatum (Schwein.) Andrus & W.D. Moore, Phytopathology 25: 121 (1935)
Index Fungorum Registration Identifier: 280780 (Figure 12).

Figure 12. Colletotrichum truncatum (A) Host freshwater plant leaf of Eichhornia crassipes (Water hyacinth). (B) Upside of the PDA culture plate (diameter; 9.5 cm). (C) Downside of the PDA culture plate (diameter; 9.5 cm) after five days. (D) Sporulated culture after ten days. (E) Stereo microscopic view of sporulation in culture. (F) Stereo microscopic observation of acervuli. (G, H) Acervuli. (J) Aseta. (I, K) Conidiogenesis. (L–P) Maturity levels of conidiogenus cells. (Q) Conidia. Scale bars: (E, F) = 1000 µm, (G–Q) = 100 µm.
Description: Endophytic of healthy leaf of Eichhornia crassipes. Sexual morph: Undetermined. Asexual morph: Conidiomata acervular, with conidiophores and setae formed directly on hyphae. Setae subhyaline to moderately brown, smooth to verruculose, 2 to 5-septate, cylindrical to conical at base, tapering towards the slightly acute to roundish tip, 4–6 µm diam. Conidiophores up to 90 µm long, hyaline to pale brown, septate, densely branched, clustered, Conidiogenous cells enteroblastic, phialidic, hyaline to pale brown, cylindrical, 6–20 × 2.5–4 µm, with invisible collarette, periclinal thickening not observed. Conidia 20.35–28.39 × 2.23–4 µm ( = 25 × 3 µm; n = 30), hyaline, cylindric-fusiform, elongated, smooth-walled, aseptate, curved at tapering apex, truncate at base, guttulate with granular content.
Culture characteristics: Colonies on incubating for seven days at 28–30°C in dark on PDA, exhibit a diameter of 1.5–2 cm. The colonies are flat with an entire margin, devoid of aerial mycelium, bluff at surface and covered by olivaceous-grey to iron-grey acervuli. The reverse of the colony is buff to pale olivaceous-grey. Conidia in mass are whitish, buff to pale saffron.
Material examined: SRI LANKA, North Central Province, Mihintale, Mahakanadara tank (8.38683° N, 80.38683° E, 117 m), on healthy leaf of Eichhornia crassipes (Water hyacinth), 8 December 2023, Madhara K. Wimalasena, RUFCC2451 (living culture), RUSLH/245 (dried culture as the herbarium specimen).
Notes: De Silva et al. (2019) found that Colletotrichum isolates with curved conidia and ITS sequences matching the ex-type of C. truncatum were the most common, making up 44% of all isolates. These isolates came from Indonesia, Malaysia, Sri Lanka, and Thailand, while species with straight conidia were identified separately. The remaining 56% were species with straight conidia, mostly from other complexes within the Colletotrichum genus. Liu et al. (2022) reported that the C. truncatum species complex produces curved conidia. Interestingly, species with curved conidia appear throughout the phylogenetic tree, suggesting this trait evolved multiple times. While ITS is useful for identifying Colletotrichum species complexes (Cannon et al., 2012), other loci like GAPDH, ACT, CHS-1, HIS 3, and tub2 are increasingly used to better define species boundaries, including in the C. truncatum complex (Damm et al., 2009, 2014; Liu et al., 2022). See the notes under Colletotrichum siamense.
Dothideomycetes genera incertae sedis
Neottiosporina Subram., Proc. Natl. Inst. Sci. India, B 27: 238 (1961)
Index Fungorum Registration Identifier: 9117
Notes: Sutton and Alcorn (1974) revisited the genus Neottiosporina, typified by N. apoda (Speg.) Subram. (1961), which is characterized by pycnidia that are solitary, dark brown, globose to subglobose, thin-walled, and ostiolate; conidiogenous cells are holoblastic, solitary, hyaline, determinate, and originate from the inner wall of the pycnidium. The conidia are acrogenous, solitary, hyaline, multiseptate, smooth-walled, cylindrical to cymbiform, obtuse at apex, and truncate at base. The genus comprises ten species viz., N. apoda (Speg.) Subram (Sutton and Alcorn, 1974), N. ashworthiae (From Scleria: Queensland fide Tan and Shivas, 2022), N. asymmetrica (on Themeda australis fide Sutton and Alcorn, 1974), N. australiensis (on Phragmites australis fide Sutton and Alcorn, 1974), N. clavata (on Phragmites australis fide Sutton, 1981), N. masonii (on Pinus caribaea fide Sutton and Sarbhoy, 1976), N. cylindrica (on Cyperus brevifolius fide Sutton and Alcorn, 1985), N. paspali (Sutton and Alcorn, 1974), N. phragmiticola (in Ethiopia, Sudan, and Uganda fide (Nag Raj, 1993), and N. sorghicola in China (Sutton and Wu, 1995). In our study of endophytic fungi inhabiting aquatic plant species, we isolated a novel taxon of Neottiosporina.
Neottiosporina mihintaleensis Wimalasena, Wijayaw. & Bamunuarachchige sp. nov.
Index Fungorum Registration Identifier: IF902502 (Figure 13).
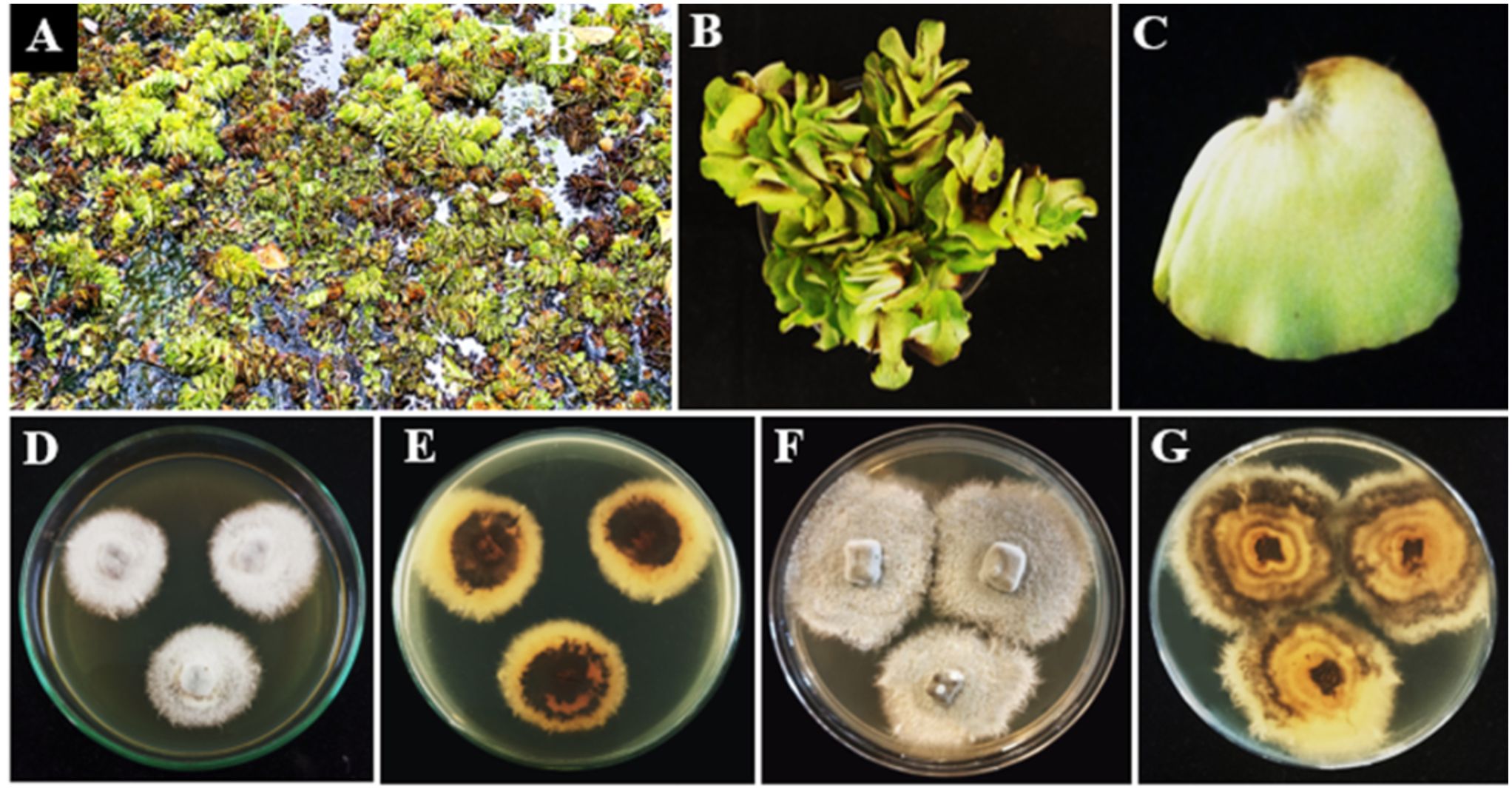
Figure 13. Neottiosporina mihintaleensis sp. nov. (A–C) Host freshwater plant leaf of Salvinia molesta (Giant Salvinia). (D) Upside of the PDA culture plate (diameter; 9.5 cm). (E) Downside of the PDA culture plate (diameter; 9.5 cm) after five days. (F) Upside of the PDA culture plate (diameter; 9.5 cm). (G) Downside of the PDA culture plate (diameter; 9.5 cm) after ten days.
Etymology: The name is derived from the locality from where the fungus was collected.
Holotype: RUSLH/241
Description: Endophytic of healthy leaf of Salvinia molesta. Sexual and asexual morphs undetermined. Despite efforts to induce the asexual morph using sporulation techniques described in the materials and methods section (2.1), the fungal cultures did not sporulate. Thus, we conclude it is sterile mycelia.
Culture characteristics: Colonies grown on PDA at 28–30°C in a 2 cm Petri dish over a two-week period show that the superficial mycelium is abundant and dark grey at the centre, with white patches towards the periphery. The immersed mycelium appears pale to medium brown with an irregular margin. On the reverse side, the colony displays a centre ranging from yellowish-brown to dark brown, transitioning to a yellowish-white colour at the margin.
Material examined: SRI LANKA, North Central Province, Mihintale, Mahakanadara tank (8.38683° N, 80.38683° E, 117 m), on healthy leaf of Salvinia molesta (Giant Salvinia), 10 December 2023, Madhara K. Wimalasena, RUSLH/241 (holotype as the dry culture), RUFCC2454 (ex-type); ibid RUFCC2461 (living culture).
Notes: Neottiosporina mihintaleensis sp. nov. is a newly identified species within the genus Neottiosporina, discovered in freshwater environments in Sri Lanka. Phylogenetic analysis indicates that it is closely related to N. cylindrica and N. ashworthiae (Figure 7). Neottiosporina cylindrica produces cylindrical to slightly clavate conidia (Sutton and Alcorn, 1985; Li et al., 2020) whereas N. ashworthiae has not reported its micromorphological characters. However, Neottiosporina mihintaleensis did not produce asexual morph in culture, despite the use of sporulation techniques (see sporulation techniques in materials and methods 2.1), making it impossible to compare its morphological features along with phylogenetically related species. Hence, we introduce Neottiosporina mihintaleensis as sterile mycelia.
3.3 Qualitative enzymatic assay for extracellular enzymes production by endophytic fungi
This study shows that fungal isolates can produce amylase, cellulase, and laccase enzymes. This enhances our understanding of their ecological roles and opens avenues for future biotechnological applications in diverse industries, thereby addressing the ongoing demand for enzymatic solutions in global markets.
3.3.1 Production of amylase enzyme by fungal isolates
The amylase activity shown by these endophytes can help break down starch when plants start to age (Mahfooz et al., 2017). As biotechnology advances, the significance of amylases in the production of various commodities, such as food and starch-based products, continues to grow. Given the widespread utilization of these enzymes across numerous industries, there exists a persistently high demand for amylases (Khokhar et al., 2011; Bilal and Iqbal, 2019; Patil et al., 2021). As a result, there is an ongoing search for new microbial strains that can produce these enzymes (Khokhar et al., 2011; Patil et al., 2021). The emergence of these newly identified fungal isolates suggests promising prospects for large-scale amylase production.
To assess amylase production, the positive control contained fungal endophytes cultured on PDA media supplemented with 1% soluble starch. Following a seven-day incubation period at 28–30°C (range of the room temperature), 1–2 mL of iodine solution was applied to flood the culture plates, resulting in a blue-black coloration. Observations were recorded at 15 minute and 30 minute intervals. A change from blue-black to a colorless medium indicated the presence of amylase activity, as the enzyme catalyzed the hydrolysis of starch. Among the isolates, Ectophoma salviniae sp. nov. exhibited the highest amylase production, forming a clear zone with a diameter of 2.5 cm around the fungal colony and decolorizing the medium completely within 10 minutes, while Phyllosticta capitalensis produced a clear zone of 1.5 cm in diameter around its colony after 15 minutes. Other fungal isolates also displayed amylase production, with discernible effects after 15 minutes. All endophyte isolates, except Neottiosporina mihintaleensis sp. nov. and C. truncatum, have exhibited the ability to produce extracellular amylase, underscoring their notable enzymatic capabilities (Table 6; Figure 14). Prior research has documented amylase activity in species like Phyllosticta spp. (Wikee et al., 2017; Reyes et al., 2021). and Colletotrichum spp. (Prajapati et al., 2013; Armesto et al., 2020; da Silva et al., 2021). The comparison with the negative control, which consisted of PDA media supplemented with 1% soluble starch and without the inoculation of endophytic fungi, involved a seven-day incubation period at 28–30°C. Following this incubation, 1–2 mL of iodine solution was applied to flood the culture plates, resulting in a blue-black coloration. Observations were recorded at 15 minute and 30minute intervals; however, no color change occurred, and the blue-black coloration remained (Figure 14).
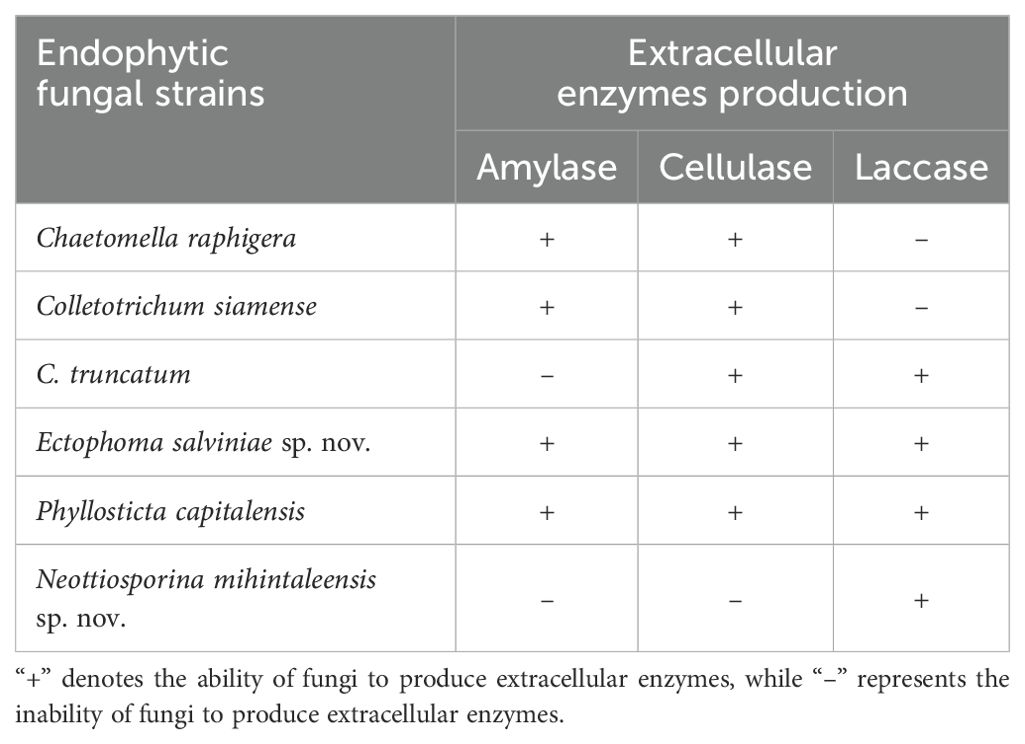
Table 6. The ability for extracellular enzymes production by endophytic fungi isolated from freshwater plants.
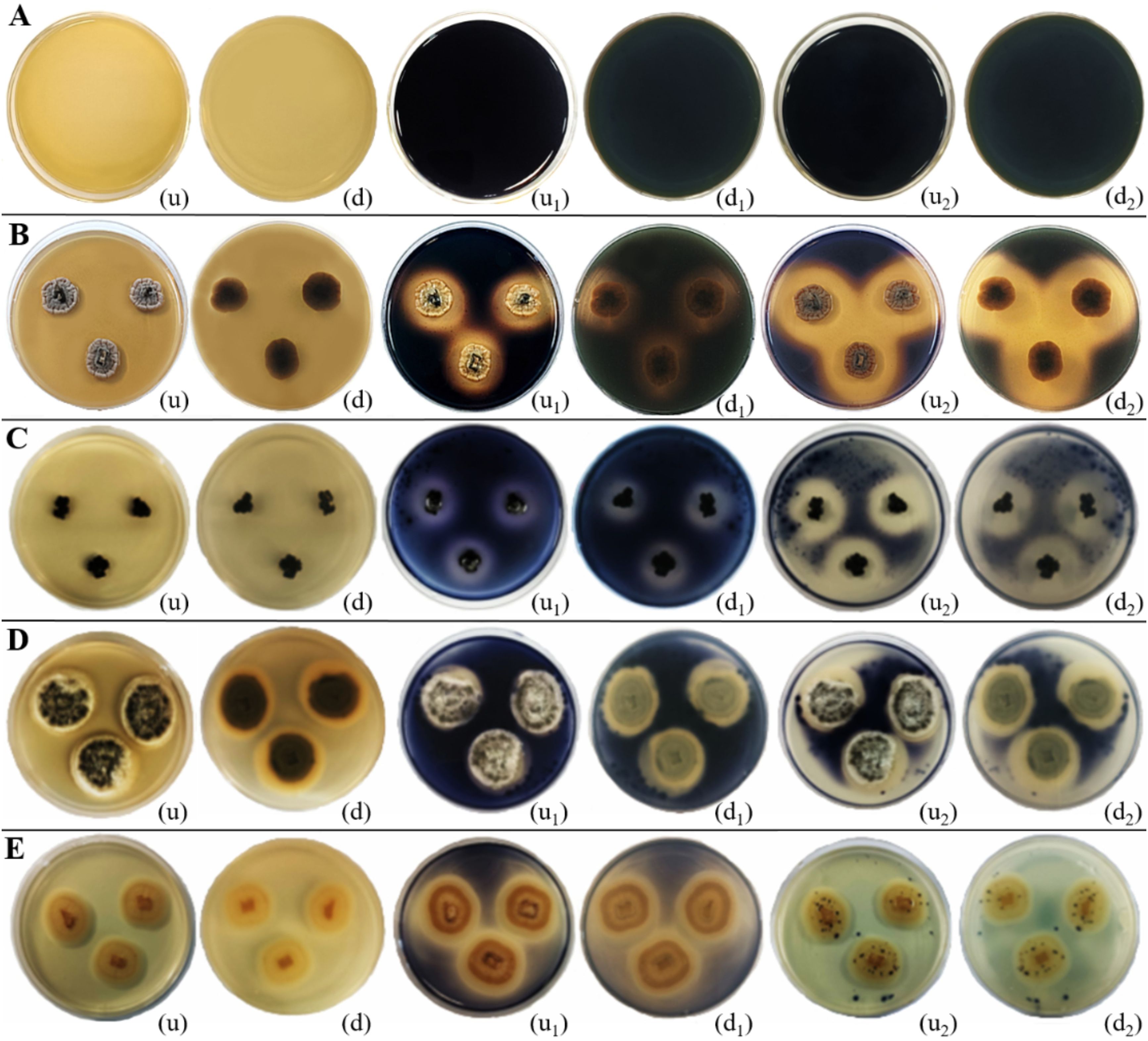
Figure 14. Amylase enzymatic activity of endophytic fungi isolated from freshwater plants. (A) Negative control. (B) Ectophoma salviniae sp. nov. (C) Phyllosticta capitalensis. (D) Colletotrichum siamense. (E) Chaetomella raphigera. (u, d) Upside and downside of the plate before adding 1–2 mL of iodine solution. (u1, d1) Upside and downside of the plate after adding 1–2 mL of iodine solution for 15 minutes (u2, d2) Upside and downside of the plate after adding 1–2 mL of iodine solution for 30 minutes, respectively. Amylase enzymatic activity was indicated by the clear zone appearance of the fungal colony on PDA media supplemented with 1% starch. (B–E) Positive control.
3.3.2 Production of cellulase enzyme by fungal isolates
Cellulase has significant applications across various industries, making it a highly researched enzyme in academic and industrial settings. It is particularly valuable in the pulp and paper, textile industry, bio-ethanol production, wine and brewery sectors, food industry, extraction of pigments and bioactive compounds, pharmaceutical industries, and waste management (Srivastava et al., 2018; Dhevagi et al., 2021; Ejaz et al., 2021; Maravi and Kumar, 2021; Singh et al., 2021; Łubek-Nguyen et al., 2022). Due to its broad utility, cellulase is in high demand, accounting for approximately 20% of the global enzyme market (Srivastava et al., 2015; Singh et al., 2021). Fungal cellulase enzymes are particularly effective in breaking down the cellulose component of lignocellulosic materials into hexose sugars, making fungi good producers of cellulase enzymes among microorganisms (Singh et al., 2021).
In this study, the positive control consisted of endophytic fungi inoculated into PDA media supplemented with 0.5% (w/v) sodium carboxymethyl cellulose and incubated for five days at 28–30°C. Following incubation, 0.1% (w/v) Congo red was applied, followed by 1M NaCl for 5 minutes to visualize the enzymatic activity (clear halo around the colonies). Chaetomella raphigera exhibited the highest cellulase enzyme production, as evidenced by the red media turning colorless with a clear halo. Phyllosticta capitalensis formed clear halos around each colony, indicating significant cellulase enzyme production, second only to Chaetomella raphigera. The other isolates (Colletotrichum truncatum, C. siamense, and Ectophoma salviniae sp. nov) exhibited only minimal cellulase enzyme production on solid media, whereas Neottiosporina mihintaleensis sp. nov. displayed no cellulase enzyme activity (Table 6; Figure 15). In the comparison between the positive and negative controls, the negative control consisted of PDA media supplemented with 0.5% (w/v) sodium carboxymethyl cellulose, without the inoculation of endophytic fungi, and incubated for five days at 28–30°C. After incubation, 0.1% (w/v) Congo red was applied, followed by a 5-minute treatment with 1M NaCl. No enzymatic activity (clear halo) was observed, leaving only the Congo red stain visible on the plates (Figure 15).
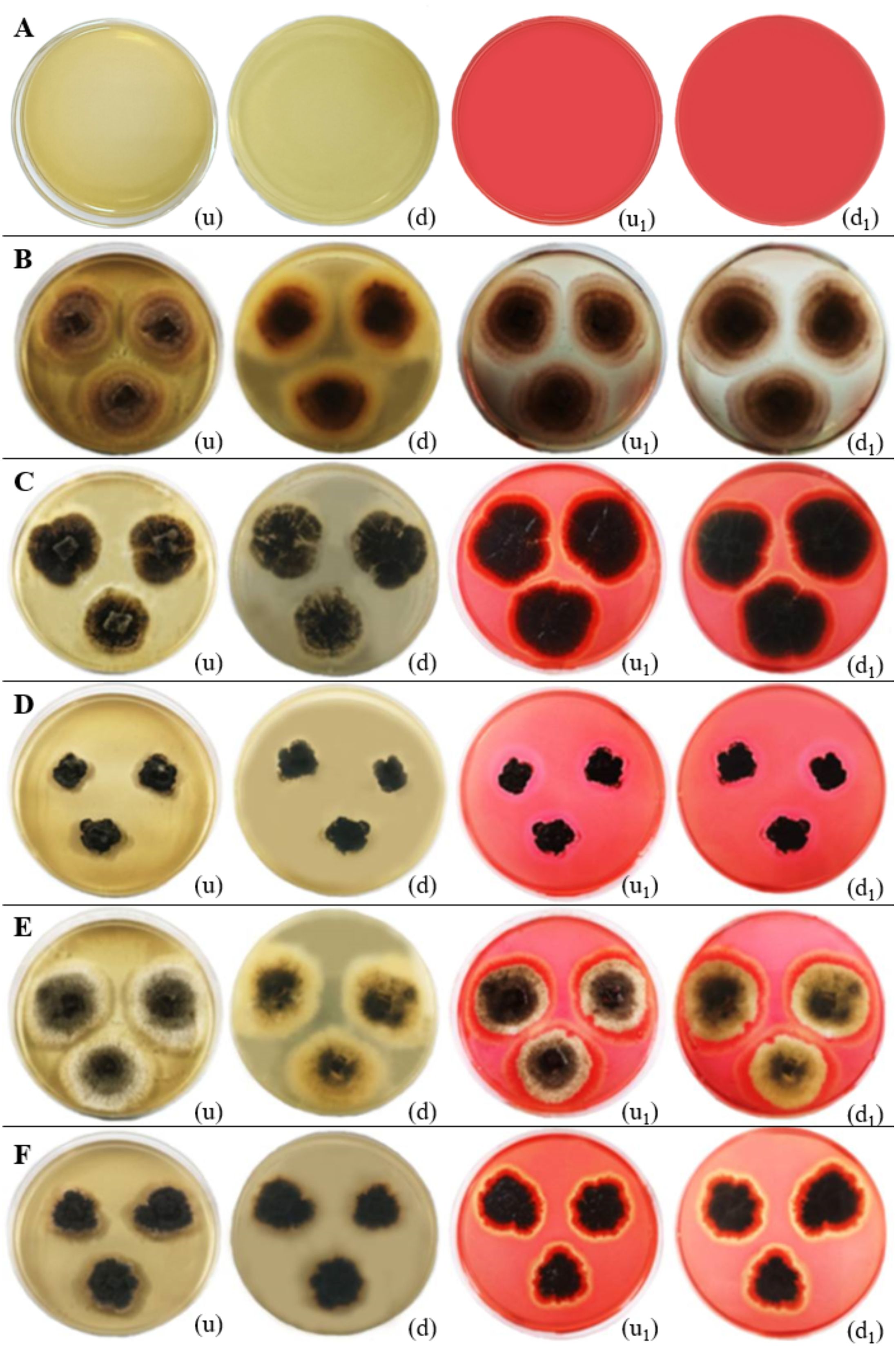
Figure 15. Cellulase enzymatic activity of endophytic fungi isolated from freshwater plants. (A) Negative control. (B) Chaetomella raphigera. (C) Colletotrichum truncatum. (D) Phyllosticta capitalensis. (E) Colletotrichum siamense. (F) Ectophoma salviniae sp. nov. (u, d) Upside and downside of the plate before adding 0.1% (w/v) Congo red solution respectively. (u1, d1) Upside and downside of the plate after adding 0.1% (w/v) Congo red solution respectively. Cellulase enzymatic activity was indicated by the clear zone appearance of the fungal colony on PDA media. (B–F) Positive control.
Previously, Kao et al. (2019) and Singh et al. (2023) have found that Chaetomella sp. exhibits a high capacity for producing glucose-tolerant cellulase enzymes. Similarly, Amirita et al. (2012); Yopi et al. (2017), and Wikee et al. (2017) have highlighted the significant potential of Phyllosticta sp. for cellulase enzyme production.
3.3.3 Production of laccase enzyme by fungal isolates
Laccases, classified as blue multicopper oxidases, catalyze the one-electron oxidation of a wide range of substrates and play a crucial role in lignin degradation (Abdel-Hamid et al., 2013; Viswanath et al., 2014; Singh and Gupta, 2020; Kyomuhimbo and Brink, 2023; Sharma et al., 2024). These enzymes are extensively used in various industries, including industrial effluent decolorization and detoxification, wastewater treatment, paper and pulp production, textiles, xenobiotic degradation, bioremediation, and as biosensors, owing to their key role in the breakdown of lignin and phenolic compounds (Shraddha et al., 2011; Viswanath et al., 2014; Singh and Gupta, 2020; Khatami et al., 2022). Laccase have been identified in approximately 60 fungal strains from the genera Ascomycetes, Deuteromycetes, and Basidiomycetes (Leonowicz et al., 2001; Albu et al., 2019; Abo Nahas et al., 2021; Mahuri et al., 2023). Fungal laccases are categorized into two types: true laccase and false laccase (De Jesus et al., 2009; Mahuri et al., 2023). True laccases can oxidize phenols and aminophenols but cannot oxidize the amino acid residue tyrosine. On the other hand, false laccases can oxidize tyrosine (De Jesus et al., 2009; Chauhan et al., 2017; Jayaram et al., 2023; Mahuri et al., 2023). Jayaram et al. (2023) highlighted that laccase production by fungal endophytes is a promising area of research due to its potential industrial applications, such as bioremediation and detoxification of pollutants.
As per the findings of this study, upon comparing the negative and positive controls in the laccase assay, no color change (blue-purple coloration) was observed in the negative control after the addition of 1-Naphthol solution droplets and 24 hours of incubation at 28–30°C (Figure 16). In the positive control, the addition of 1-Naphthol solution droplets to each colony of endophyte isolates, followed by subsequent incubation for 24 hours at 28–30°C, resulted in a blue-purple coloration at the edges of the colonies, indicating the presence of laccase enzymes. This reaction was observed in Colletotrichum truncatum, Ectophoma salviniae, Neottiosporina mihintaleensis, and Phyllosticta capitalensis. Among these, Neottiosporina mihintaleensis exhibited the highest laccase production, with a 3.5 cm diameter blue-purple circle around the colony. Phyllosticta capitalensis showed the second-highest laccase production, with a 1 cm diameter blue-purple circle around the colony, compared to its appearance before applying 1-Naphthol droplets. In contrast, Chaetomella raphigera, and C. siamense did not exhibit laccase enzyme production in this qualitative assay (Table 6; Figure 16).
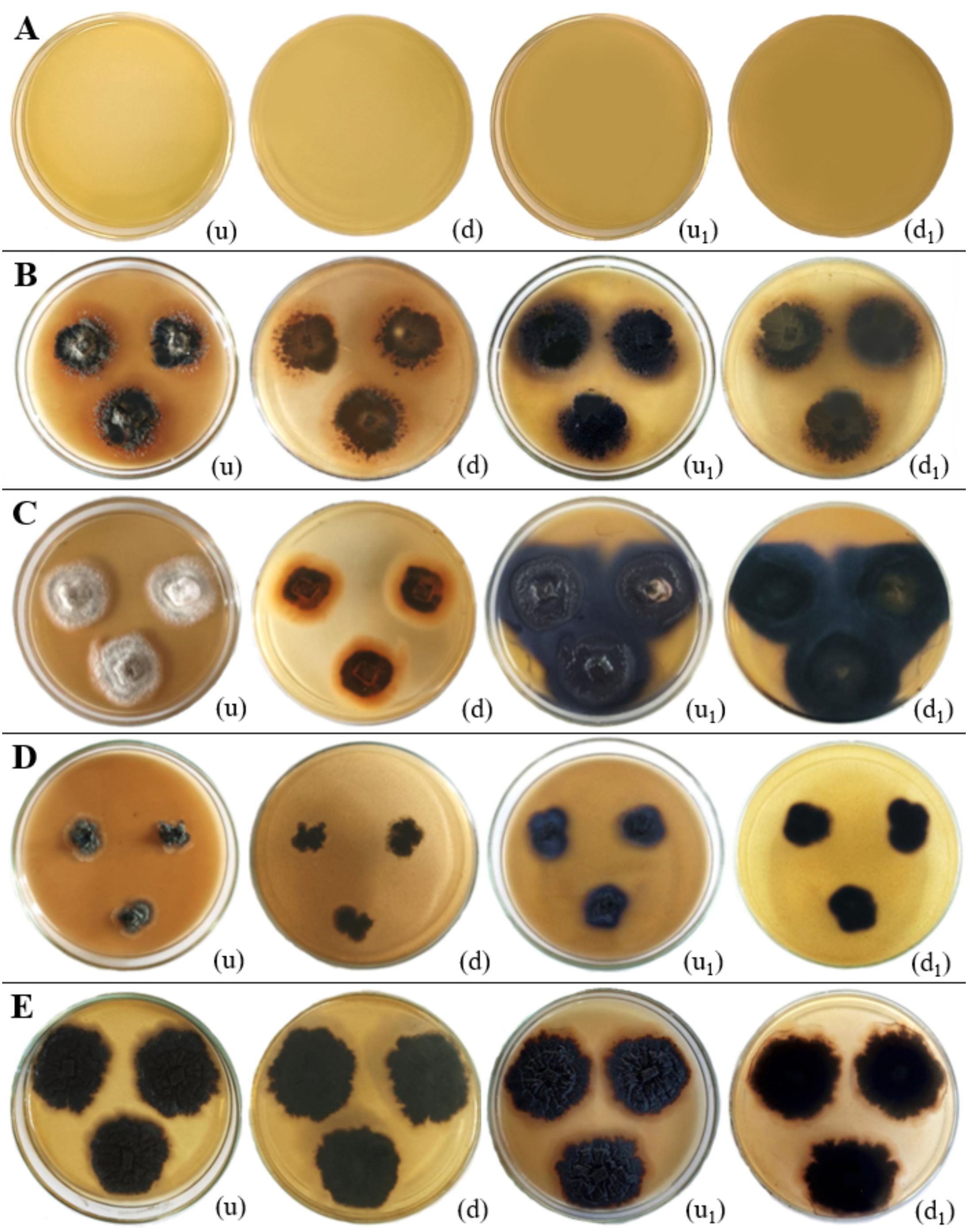
Figure 16. Laccase enzymatic activity of endophytic fungi isolated from freshwater plants. (A) Negative control. (B) Colletotrichum truncatum. (C) Neottiosporina mihintaleensis sp. nov. (D) Phyllosticta capitalensis. (E) Ectophoma salviniae sp. nov. (u, d) Upside and downside of the plate before adding 0.1M 1-Naphthol respectively. (u1, d1) Upside and downside of the plate after adding after adding 0.1M 1-Naphthol respectively. Laccase enzymatic activity was indicated by the blue purple colour appearance of the fungal colony on PDA media. (B–E) Positive control.
This research has shown the novel discovery of laccase production in N. mihintaleensis and E. salviniae. These findings contribute to an expanding understanding of fungal laccases. Notably, prior studies have extensively documented laccase production capabilities in Phyllosticta spp. (Wikee et al., 2017; Shankar Naik et al., 2019), C. truncatum (Levin et al., 2007; Núñez et al., 2023).
4 Discussion
4.1 Exploration of endophytic fungi in Sri Lanka’s freshwater environments
4.1.1 Current status and research gaps on endophytic fungal study in Sri Lanka
Currently, only around 3,000 fungal species are known in Sri Lanka, with an estimated 31,000 plant-associated species still to be described (Adikaram and Yakandawala, 2020; Wijayawardene et al., 2022b). Several of the known fungi in the island nation remain unpublished and have not been sufficiently studied or documented (Adikaram and Yakandawala, 2020; Karunarathna et al., 2022; Wijayawardene et al., 2022b, 2023; Wimalasena et al., 2024). In the Sri Lankan context, particularly regarding endophytic fungi, many studies have focused on terrestrial plants (e.g (Alwis et al., 2021; Pathmanathan et al., 2022; Koshila et al., 2023; Undugoda et al., 2023). Studies on endophytic fungi associated with aquatic plants in Sri Lanka are relatively scarce (Rajagopal et al., 2018; Ravimannan and Sepali, 2020; Ekanayake et al., 2021). Hitherto, the identification of endophytic fungi in freshwater plants was based largely on morphological characters. For instance, Hettiarachchi et al. (1983) reported 15 fungi (Alternaria sp., Cephalosporium sp., Cercospora piaropi, Curvularia tuberculata, Fusarium sp., Idriella lunata, Mucor sp., Myrothecium roridum, Neurospora sp., Penicillium oxalicum, Phaeotrichoconis crotalariae, and Septofusidium elegantulum) isolated from Eichhornia crassipes, with identification based only on morphologal charcteristics. In some of the Sri Lankan studies, endophytic fungal identification was based on a single gene locus, which is inadequate for accurate identification. For example, Dissanayake et al. (2014, 2016) identified Chaetomium globosum from healthy Nymphaea nouchali using only the ITS locus.
4.1.2 Identification of endophytic fungi in Sri Lankan freshwater habitats: potential for novel species discovery
Sri Lanka harbors over 370 aquatic and wetland plant species, with 12% being unique to the country (Yakandawala, 2012; Bambaranda et al., 2024). These endemic plants serve as essential habitats for fungi, including freshwater fungi and endophytic species that have adapted to unique environmental conditions (Ratnaweera, 2019; Wimalasena et al., 2024). Wimalasena et al. (2024), highlighted that these habitats offer substantial potential for the collection, identification, and utilization of endophytic fungi found in freshwater plants.
This study focused on isolation of freshwater endophytic fungi in three lentic freshwater habitats (Iluppukanniya tank, Mahakanadara tank, and Mihintale tank) located in Mihintale area within the Anuradhapura district. Using polyphasic approaches, six endophytic fungi were isolated including two novel taxa, Ectophoma salviniae sp. nov. and Neottiosporina mihintaleensis sp. nov. These fungal species were identified in their endophytic life modes, occurring within healthy freshwater plant tissues, particularly in healthy leaves, isolated by a culture-dependent method. In this study, the invasive plant species Eichhornia crassipes (Ayanda et al., 2020; Maulidyna et al., 2021; Bayu et al., 2024) provided a wider range of host substrates for fungi compared to other freshwater plants such as Salvinia and Nymphaea, highlighting its value for biodiversity. Hence, expanding such studies to cover more freshwater habitats could lead to the identification of additional novel species, contributing significantly to the field of mycology and biotechnology worldwide.
4.2 Reference cultures of pathologically important taxa, Colletotrichum siamense, C. truncatum and Ectophoma sp. in Sri Lanka
Precise identification of fungi is an important step in taxonomy. DNA sequence analyses and morphological characters play an important role in modern taxonomy which aids in identifying species and providing their classification (Wijayawardene et al., 2023). A large number of species originally described from Sri Lanka lack sequence data and were identified based on only morphological characteristics (Wijayawardene et al., 2022b). Nevertheless, delineating species boundaries of species complexes of specious genera would depend only on DNA sequence data analyses.
Adikaram and Yakandawala (2020) listed pathologically important Colletotrichum species in Sri Lanka, including C. siamense and C. truncatum. However, either C. siamense or C. truncatum have not been reported as a pathogenic species from aquatic plants. Table 7 lists the studies that provided phylogenetic identifications of C. siamense and C. truncatum.
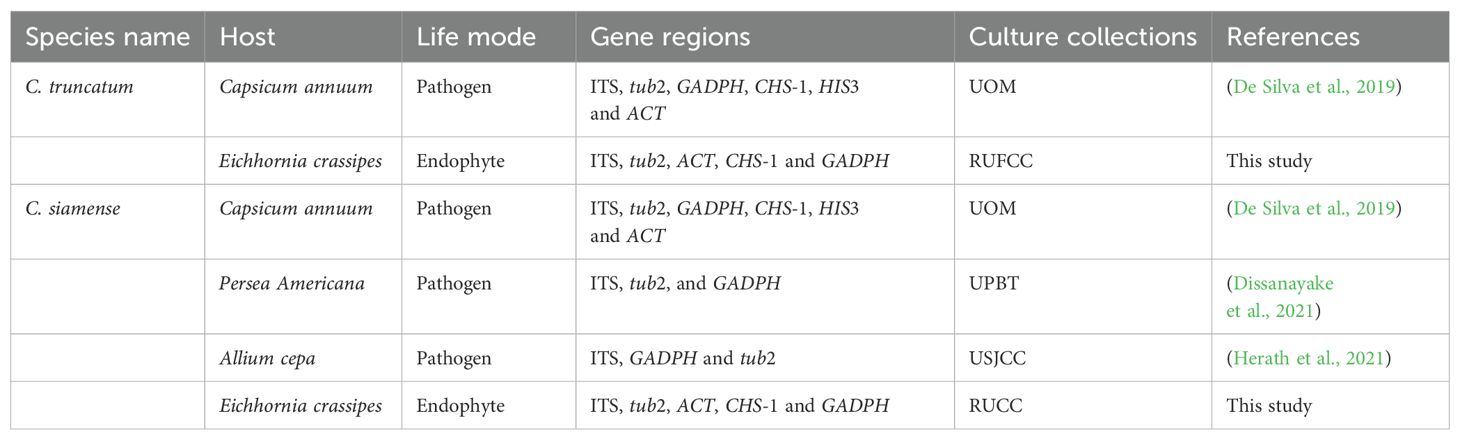
Table 7. Studies provided phylogenetic analyses for Colletotrichum siamense and C. truncatum species in Sri Lanka (RUFCC-Rajarata University Fungal Culture Collection, UOM-University of Melbourne culture collection, Victoria, Australia, UPBT-University of Peradeniya, Department of Biotechnology, USJCC-University of Sri Jayewardenepura Culture Collection, Department of Botany, University of Sri Jayewardenepura, Nugegoda, Sri Lanka).
In a previous study, Dissanayake et al. (2016) reported C. siamense from Nymphaea nouchali but they used only the ITS region to identify the taxon. Use of one locus is not recommended for Colletotrichum thus, we used ITS, tub2, ACT, CHS-1 and GADPH regions in our phylogenetic analyses following Armand et al. (2023) and Armand and Jayawardena (2024). We have not observed any disease symptoms in the leaves of Eichhornia crassipes. Hence, it is concluded that both C. siamense and C. truncatum are endophytic species of Eichhornia crassipes. This is the first study that provided multi-locus phylogenetic evidence to identify two pathologically important (but endophytic in this study) Colletotrichum species (e.g., C. siamense and C. truncatum) in aquatic habitats. It is important to maintain the reference living cultures of both species; thus, it has been deposited at the Rajarata University Culture Collection.
Ectophoma species have been reported as important plant pathogens. Ectophoma multirostrata, the type species of Ectophoma (Valenzuela-Lopez et al., 2018), has been originally reported as a soil-inhabiting fungi in India (as Sphaeronaema multirostratum fide (Mathur and Thirumalachar, 1959). Later, this species was reported as a pathogen of different plants worldwide (e.g (Aveskamp et al., 2010; Valenzuela-Lopez et al., 2018; Chobe et al., 2020; Ahmadpour et al., 2021; Kularathnage et al., 2023). Lee et al. (2022) reported Ectophoma multirostrata as a pathogenic agent infecting the aquatic plant water spinach (Ipomoea aquatica) in Korea. Ectophoma myriophyllana Huang Y. and Yu Z. F. was recently introduced as an epiphyte of leaves of Myriophyllum spicatum (Chen et al., 2023). Our novel species, Ectophoma salviniae did not cause any diseased symptoms on the leaves of Salvinia minima and thus, we conclude it is an endophytic taxon inhabiting the host. As far as we know, this is the first report of Ectophoma species from Salvinia species in Sri Lanka (Farr and Rossman, 2024). We have not observed Ipomoea aquatica (which was affected by Ectophoma multirostrata) in the same aquatic environment, and the distribution of Ectophoma salviniae sp. nov. is unknown. Future studies would be essential to recognise the potential host jumping and life mode switching of Ectophoma salviniae sp. nov. and its impact on Ipomoea aquatica since it is a widely-used leafy vegetable in Sri Lanka.
4.3 The possibility of endophytic fungi being used as mycoherbicides against invasive weed management in wetland environments
4.3.1 Threat of invasive aquatic plants
Invasive plant species pose a major threat to natural ecosystems by reducing biological diversity (Rodríguez-Merino, 2023; Xiong et al., 2023). Over the past few decades, the spread of aquatic alien plant species in the lentic water bodies of Sri Lanka has created significant ecological, environmental, and economic problems (Dissanayake, 2020; Kariyawasam et al., 2021). Thus, fungi can be used as mycoherbicides for a more effective solution to eradicate invasive aquatic plant species due to their pathogenic activity and host-specific targeting.
4.3.2 Application of fungi as mycoherbicides
Bioherbicides are biological products used to control weed species and are typically formulated using microbiological agents, especially fungi, and are often referred to as mycoherbicides (Golijan et al., 2023; Ravlić and Baličević, 2014). The concept of mycoherbicides emerged during the 1980s and 1990s, as documented by TeBeest and Templeton (1985); Templeton (1987); Templeton (1992) and Wall et al. (1992). Mycoherbicides are considered environmentally friendly alternatives to chemical herbicides because they are harmless to the environment, eco-friendly, and specifically target certain types of plants (Chakraborty and Ray, 2021; Hasan et al., 2021; Keshamma, 2022). The efficiency of fungi in weed management is exemplified by the reported potential of Colletotrichum species (Table 8). Colletotrichum siamense and C. truncatum have previously been identified as pathogenic fungi across various plant species. According to this study, these Colletotrichum spp. may demonstrate potential for managing the invasive weed Eichhornia crassipes in Sri Lanka.
5 Conclusion
This study identified the culturable mycobiota in three lentic freshwater habitats located in Mihintale, within the Anuradhapura District of Sri Lanka, revealing a rich fungal diversity. Through identification, six endophytic fungal species were found, including two novel endophytic fungal species: Ectophoma salviniae sp. nov. and Neottiosporina mihintaleensis sp. nov., recorded on the freshwater plant Salvinia. The identification was confirmed using a polyphasic approach. The next step involved qualitatively assessing the extracellular enzymatic potentials of these endophytic isolates. Ectophoma salviniae sp. nov. exhibited the highest amylase production, Chaetomella raphigera showed the highest cellulase enzyme production, and Neottiosporina mihintaleensis sp. nov. demonstrated the highest laccase production, offering novel insights for future biotechnological applications. Besides, this study discussed the potential of fungi as mycoherbicides for managing invasive freshwater weeds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/genbank/, ITS: PP989214, PP989215, PP989216, PP989217, PP989218, PP989219, PP989220, PP989221, PP989222; LSU: PP989223, PP989224, PP989225, PP989226, PP989227; SSU: PP989228, PP989229, PP989230; GAPDH: PQ014240, PQ014241, PQ014242, PQ014243; CHS-1: PQ014237, PQ014238, PQ014239; ACT: PQ014233, PQ014234, PQ014235, PQ014236; tub2: PQ014246, PQ014247, PQ014248; rpb2: PQ014244, PQ014245; tef1-α: PQ014249.
Author contributions
MW: Writing – original draft, Writing – review & editing. NW: Writing – original draft, Writing – review & editing. TB: Writing – original draft, Writing – review & editing. G-QZ: Writing – original draft, Writing – review & editing. RJ: Writing – original draft, Writing – review & editing. DB: Writing – original draft, Writing – review & editing. TD: Writing – original draft, Writing – review & editing. HZ: Writing – original draft, Writing – review & editing. D-QD: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors are grateful to the High-Level Talent Recruitment Plan of Yunnan Province (“Young Talents” Program and “High-End Foreign Experts” Program), the Key Laboratory of Yunnan Provincial Department of Education of the Deep-Time Evolution on Biodiversity from the Origin of the Pearl River, Meemann Chang Academician Workstation in Yunnan Province (202225AF150002), the Yunnan Province Young and Middle-aged Academic and Technical Leaders Reserve Talents Program (Grant No. 202305AC350252), and the General Programs of the Provincial Department of Science and Technology (Grant No. 202101BA070001-076). This work was supported by the Science and Technology Human Resource Development Project, Ministry of Higher Education, Sri Lanka, funded by the Asian Development Bank (Grant number R3-RJ2) and Tropical Microbiology Research Foundation (TMRF). Jayarama D. Bhat and Turki M. Dawoud gratefully acknowledge the financial support under the Distinguished Scientist Fellowship Programme (DSFP), King Saud University, Riyadh, Saudi Arabia.
Acknowledgments
We extend our gratitude to R.M.K. Rathnayaka for creating the map of sampling locations for lentic freshwater habitats in the Mihintale area. We acknowledge ChatGPT-4, one of OpenAI’s language models (https://chat.openai.com), available online, for language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1475114/full#supplementary-material
References
Abdel-Hamid, A. M., Solbiati, J. O., Cann, I. K. O. (2013). “Insights into Lignin Degradation and its Potential Industrial Applications,” in Advances in applied microbiology, vol. 82 . Eds. Geoffrey Michael, G., Sariaslani, S. (Elsevier), 1–28). doi: 10.1016/B978-0-12-407679-2.00001-6
Abdel-Wareth, M. T. A. (2022). “Phyllosphere of submerged plants: A reservoir of mycobiota,” in Freshwater mycology. Eds. Bandh, S. A., Shafi, S. (Elsevier), 73–89. doi: 10.1016/B978-0-323-91232-7.00005-2
Abo Nahas, H. H., Mansour, S. A., Abo Nouh, F. A., Landa-Acuña, D., Abo Nahas, Y. H., Nieto-Taype, M. A., et al. (2021). “Fungal laccases to where and where?,” in Industrially important fungi for sustainable development. Fungal biology. Eds. Abdel-Azeem, A. M., Yadav, A. N., Yadav, N., Sharma, M. (Switzerland: Springer Nature), 205–262. doi: 10.1007/978-3-030-85603-8_6
Adikaram, N. K. B., Yakandawala, D. M. D. (2020). A checklist of plant pathogenic fungi and Oomycota in Sri Lanka. Ceylon J. Sci. 49, 123. doi: 10.4038/CJS.V49I1.7709
Ahmadpour, S. A., Mehrabi-Koushki, M., Farokhinejad, R., Asgari, B., Javadi Estahbanati, A., Mirabolfathy, M., et al. (2021). New records of fungal species of the family Didymellaceae from Iran. Mycologia Iranica 8, 119–133. doi: 10.22043/MI.2022.359850.1229
Albu, C., Encea, R., Diguță, C., Matei, F., Cornea, C. (2019). Laccase: macro and microbial sources, production, purification and biotechnological applications. Sci. Bulletin. Ser. F. Biotechnologies 23, 128–136.
Ali, A. H., Radwan, U., El-Zayat, S., El-Sayed, M. A. (2018). Desert Plant-Fungal Endophytic Association: The Beneficial Aspects to their Hosts. Biol. Forum – Int. J. 10, 138–145. Available at: www.researchtrend.net.
Alwis, Y. V., Wethalawe, A. N., Udukala, D. N., Paranagama, P. A. (2021). “Endophytic microflora of Sri Lankan plants: an overview of the therapeutic and agricultural applications of the secondary metabolites,” in Endophytes: potential source of compounds of commercial and therapeutic applications. Eds. Patil, R. H., Maheshwari, V. L. (Springer, Singapore), 153–175. doi: 10.1007/978-981-15-9371-0_8
Amirita, A., Sindhu, P., Swetha, J., Vasanthi, N. S., Kannan, K. P. (2012). Enumeration of endophytic fungi from medicinal plants and screening of extracellular enzymes. World J. Sci. Technol. 2, 13–19. Available at: https://www.academia.edu/download/32012212/12432-34875-1-PB.pdf.
Anonymous (1960). “Index of plant diseases in the United States,” in Agriculture handbook, vol. 165 . Ed. Anonymous (U.S.D.A). Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=Anonymous.+%281960%29.+Index+of+Plant+Diseases+in+the+United+States.+U.S.D.A.+Agriculture+Handbook%2C+165%2C+1-531.andbtnG=d=gs_citandt=1719662667729andu=%2Fscholar%3Fq%3Dinfo%3ApeNYHP4Pq1IJ%3Ascholar.google.com%2F%26output%3Dcite%26scirp%3D1%26hl%3Den.
Armand, A., Jayawardena, R. S. (2024). Morphomolecular identification and pathogenicity of Colletotrichum species associated with avocado anthracnose in northern Thailand. Plant Pathology 73 (1), 186–197.
Armand, A., Hyde, K. D., Jayawardena, R. S. (2023). First report of Colletotrichum fructicola causing fruit rot and leaf-tip dieback on pineapple in Northern Thailand. Plants 12 (4), 971.
Armesto, C., Maia, F. G. M., Monteiro, F. P., de Abreu, M. S. (2020). Exoenzymes as a pathogenicity factor for Colletotrichum gloeosporioides associated with coffee plants. Summa Phytopathologica 45, 368–373. doi: 10.1590/0100-5405/191071
Arnold, A. E. (2007). Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol. Rev. 21, 51–66. doi: 10.1016/J.FBR.2007.05.003
Arnold, A. E., Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: Are tropical leaves biodiversity hotspots? Ecology 88, 541–549. doi: 10.1890/05-1459
Aumentado, H. D. R., Armand, A., Phukhamsakda, C., Hyde, K. D., Jones, E. B. G., Jayawardena, R. S. (2024). Polyphasic identification of two novel Colletotrichum species causing leaf spots on mangroves in Thailand. Eur. J. Plant Pathol. 169, 1–27. doi: 10.1007/S10658-024-02819-Y/METRICS
(2024). Colour guide; ArtyClick Colors. Available online at: https://colors.artyclick.com/color-name-finder/ (accessed April 02, 2024).
(2024). Index fungorum. Available online at: https://www.indexfungorum.org/names/Names.asp (accessed January to August, 2024).
Aveskamp, M. M., de Gruyter, J., Woudenberg, J. H. C., Verkley, G. J. M., Crous, P. W. (2010). Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud. Mycology 65, 1–60. doi: 10.3114/SIM.2010.65.01
Ayanda, O. I., Ajayi, T., Asuwaju, F. P. (2020). Eichhornia crassipes (Mart.) solms: uses, challenges, threats, and prospects. Sci. World J. 2020, 3452172. doi: 10.1155/2020/3452172
Bambaranda, M., Jayamanne, S. C., Yakandawala, D. M., Tsusaka, T. W. (2024). “Aquatic plants in different reservoir types of uva province,” in Diversity and impacts on reservoir fishery (SSRN, Sri Lanka), 1–29. Available at: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4741490.
Barreto Ramos, D. G., Gurgel Amaral, A. G., Duarte, I. G., Carlos da Silva, A., Anderson dos Santos Vieira, W., Castlebury, L. A., et al. (2024). Endophytic species of Colletotrichum associated with cashew tree in northeastern Brazil. Fungal Biol. 128, 1780–1789. doi: 10.1016/J.FUNBIO.2024.04.002
Bayu, T., Soeprobowati, T. R., Adissu, S., Warsito, B. (2024). Effect of climate change on “Eichhornia crassipes” infestation in Lake Tana Sub-Basin, North Western Ethiopia. Sustain. Environ. 10, 1–13. doi: 10.1080/27658511.2024.2314399
Berner, D. K., Bruckart, W. L., Cavin, C. A., Michael, J. L., Carter, M. L., Luster, D. G. (2009). Best linear unbiased prediction of host-range of the facultative parasite Colletotrichum gloeosporioides f. sp. salsolae, a potential biological control agent of Russian thistle. Biol. Control 51, 158–168. doi: 10.1016/J.BIOCONTROL.2009.06.003
Bhunjun, C. S., Phukhamsakda, C., Hyde, K. D., McKenzie, E. H. C., Saxena, R. K., Li, Q. (2024). Do all fungi have ancestors with endophytic lifestyles? Fungal Diversity 125, 73–98. doi: 10.1007/S13225-023-00516-5
Bilal, M., Iqbal, H. M. (2019). Sustainable bioconversion of food waste into high-value products by immobilized enzymes to meet bio-economy challenges and opportunities–A review. Food Res. Int. 123, 226–240. Available at: https://www.sciencedirect.com/science/article/pii/S0963996919302984.
Boerema, G. (2004). Phoma identification manual: differentiation of specific and infra-specific taxa in culture. Available online at: https://books.google.com/books?hl=enandlr=andid=Pm-BCwAAQBAJandoi=fndandpg=PR3anddq=Phoma+Identification+Manual:+Differentiation+of+Specific+and+Infra-Specific+Taxa+in+Culture.+Wallingford,+UK:+CAB+International.andots=pkvMo9H0jWandsig=LkK3Ngb2UF1-XU93Tu1jJhq0TBE (accessed June 2, 2024).
Cannon, P. F., Damm, U., Johnston, P. R., Weir, B. S. (2012). Colletotrichum: Current status and future directions. Stud. Mycology 73, 181–213. doi: 10.3114/sim0014
Cao, X. X., Wei, J. G., Hou, J. G., Fan, C. L., Luo, J., Wu, Y. J., et al. (2021). First report of Chaetomella raphigera causing brown spot on Eucalyptus urophylla × E. grandis in China. J. Plant Pathol. 103, 403. doi: 10.1007/S42161-020-00737-6/METRICS
Carbone, I., Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Chaiwong, K., Khuna, S., Senwanna, C., Kumla, J., Suwannarach, N. (2024). First report of Phyllosticta capitalensis causing postharvest brown spot disease on guava fruits in Thailand. Plant Dis. 108, 2930. doi: 10.1094/PDIS-06-24-1314-PDN
Chakraborty, A., Ray, P. (2021). Mycoherbicides for the noxious meddlesome: can colletotrichum be a budding candidate? Front. Microbiol. 12. doi: 10.3389/FMICB.2021.754048/BIBTEX
Chauhan, P. S., Goradia, B., Saxena, A. (2017). Bacterial laccase: recent update on production, properties and industrial applications. 3 Biotech. 7, 1–20. doi: 10.1007/S13205-017-0955-7/METRICS
Chen, T., Wang, S., Jiang, X., Huang, Y., Mo, M., Yu, Z. (2023). New species of didymellaceae within aquatic plants from southwestern China. J. Fungi 9, 1–34. doi: 10.3390/jof9070761
Chobe, D. R., Tarafdar, A., Sharath Chandra, U. S., Sudharani, Sharma, M. (2020). First report of ectophoma multirostrata causing root rot in chickpea. Plant Dis. 104, 1866–1866. doi: 10.1094/PDIS-01-19-0121-PDN
Damm, U., O’Connell, R. J., Groenewald, J. Z., Crous, P. W. (2014). The Colletotrichum destructivum species complex - hemibiotrophic pathogens of forage and field crops. Stud. Mycology 79, 49–84. doi: 10.1016/J.SIMYCO.2014.09.003
Damm, U., Woudenberg, J., Cannon, P. F., Crous, P. W. (2009). Colletotrichum species with curved conidia from herbaceous hosts. Fungal Diversity 39, 1–87. Available at: http://www.ascofrance.com/uploads/forum_file/damm-etal-2009-0001.pdf.
Dar, Z. A., Rifat, B., Bhat, J. I. A., Bhatti, A. A., Haq, S., Amin, A., et al. (2022). Potential role of endophytes for sustainable environment. Research anthology on emerging techniques in environmental remediation. IGI Global., 177–194. doi: 10.4018/978-1-6684-3714-8.CH009
da Silva, M. A., Santos, C., Pérez-Nevado, F., Lima, N., da Silva Bentes, J. L., Silva, M. A., et al. (2021). Enzymatic activity and process of initial infection of guarana plant (Paullinia cupana) by pathogenic and endophytic strains of Colletotrichum guaranicola. Rev. Fitotecnia Mexicana 44, 67–75. doi: 10.35196/RFM.2021.1.67
da Silveira Bastos, I. M. A., Cadamuro, R. D., de Freitas, A. C. O., da Silva, I. T., Stoco, P. H., Sandjo, L. P., et al. (2024). Diversity of fungal endophytes from mangrove plants of Santa Catarina Island, Brazil. Braz. J. Microbiol. 55, 1477–1487. doi: 10.1007/S42770-023-01234-5/METRICS
de Gruyter, J., Aveskamp, M. M., Woudenberg, J. H. C., Verkley, G. J. M., Groenewald, J. Z., Crous, P. W. (2009). Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycological Res. 113, 508–519. doi: 10.1016/J.MYCRES.2009.01.002
De Jesus, M., Nicola, A. M., Rodrigues, M. L., Janbon, G., Casadevall, A. (2009). Capsular localization of the Cryptococcus neoformans polysaccharide component galactoxylomannan. Eukaryotic Cell 8, 96–103. doi: 10.1128/EC.00331-08/SUPPL_FILE/EC331_08SUPP.DOC
De Silva, D. D., Crous, P. W., Ades, P. K., Hyde, K. D., Taylor, P. W. (2017). Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biology Reviews 31 (3), 155–168.
De Silva, D. D., Groenewald, J. Z., Crous, P. W., Ades, P. K., Nasruddin, A., Mongkolporn, O., et al (2019). Identification, prevalence and pathogenicity of Colletotrichum species causing anthracnose of Capsicum annuum in Asia. IMA Fungus 10, 1–32.
Deshmukh, S. K., Agrawal, S., Prakash, V., Gupta, M. K., Reddy, M. S. (2020). Anti-infectives from mangrove endophytic fungi. South Afr. J. Bot. 134, 237–263. doi: 10.1016/J.SAJB.2020.01.006
De Zoysa, M. (2022). Ecotourism development and biodiversity conservation in Sri Lanka: objectives, conflicts and resolutions. Open J. Ecol. 12, 638–666. doi: 10.4236/OJE.2022.1210037
Dhevagi, P., Ramya, A., Priyatharshini, S., Geetha Thanuja, K., Ambreetha, S., Nivetha, A. (2021). “Industrially important fungal enzymes: productions and applications,” in Recent trends in mycological research. Fungal biology. Ed. Yadav, A. N. (Springer, Cham), 263–309. doi: 10.1007/978-3-030-68260-6_11
Dissanayake, N. P. (2020). Weed; an invasive plant species in Sri Lanka and its control measures. J. Univ. Ruhuna 8, 93–103. doi: 10.4038/jur.v8i2.7937
Dissanayake, D. M. S., Adikaram, N. K. B., Yakandawala, D. M. D., Jayasinghe, L. (2021). Molecular phylogeny-based identification of Colletotrichum endophytica and C. siamense as causal agents of avocado anthracnose in Sri Lanka. Ceylon J. Sci. 50, 449–458. doi: 10.4038/cjs.v50i4.7943
Dissanayake, D. M. R. K., Ratnaweera, P. B., De Silva, E. D., Williams, D. E., Wijayarathne, C. D., Wijesundera, R. L. C., et al. (2014). “Isolated from Nymphaea nouchali endemic to Sri Lanka and the isolation of chaetoglobosin C from the endophytic Chaetomium sp,” in Proceedings of the Young Scientist Forum Symposium. 57–60. Available at: https://www.academia.edu/download/52431117/Investigation_of_antibacterial_propertie20170402-28544-yqsfpl.pdf.
Dissanayake, R. K., Ratnaweera, P. B., Williams, D. E., Wijayarathne, C. D., Wijesundera, R. L. C., Andersen, R. J., et al. (2016). Antimicrobial activities of endophytic fungi of the Sri Lankan aquatic plant Nymphaea nouchali and chaetoglobosin A and C, produced by the endophytic fungus Chaetomium globosum. Mycology 7, 1–8. doi: 10.1080/21501203.2015.1136708
Ejaz, U., Sohail, M., Ghanemi, A. (2021). Cellulases: from bioactivity to a variety of industrial applications. Biomimetics 6, 44. doi: 10.3390/BIOMIMETICS6030044
Ekanayake, M. S., Udayanga, D., Wijesekara, I., Manage, P. (2021). Phytoremediation of synthetic textile dyes: biosorption and enzymatic degradation involved in efficient dye decolorization by Eichhornia crassipes (Mart.) Solms and Pistia stratiotes L. Environ. Sci. pollut. Res. 28, 20476–20486. doi: 10.1007/S11356-020-11699-8/METRICS
El-Bondkly, E. A. M., El-Bondkly, A. A. M., El-Bondkly, A. A. M. (2021). Marine endophytic fungal metabolites: A whole new world of pharmaceutical therapy exploration. Heliyon 7, 1–15. doi: 10.1016/J.HELIYON.2021.E06362
Ellis, J., Everhart, B. (1900). The North American Phyllostictas with descriptions of the species published up to August. Available online at: https://agris.fao.org/search/en/providers/122376/records/647472d52d5d435c424edc8a (accessed May 10, 2024).
Elshafie, H. S., Grul’ová, D., Baranová, B., Caputo, L., De Martino, L., Sedlák, V., et al. (2019). Antimicrobial activity and chemical composition of essential oil extracted from solidago canadensis L. Growing wild in Slovakia. Molecules 24, 1206. doi: 10.3390/MOLECULES24071206
Farr, D. F., Rossman, A. Y. (2024). Fungal databases, systematic mycology and microbiology laboratory (ARS, USDA). Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=Farr%2C+D.+F.%2C+%26+Rossman%2C+A.+Y.+%282024%29.+Fungal+Databases.+Systematic+Mycology+and+Microbiology+Laboratory%2C+ARS%2C+USDA.+Retrieved+January+2024%2C+from+andbtnG.
Farr, D. F., Rossman, A. Y., Palm, M. E., McCray, E. B. (2012). Fungal databases, systematic mycology and microbiology laboratory (ARS, USDA). Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=Farr%2C+D.+F.%2C+%26+Rossman%2C+A.+Y.+%282024%29.+Fungal+Databases.+Systematic+Mycology+and+Microbiology+Laboratory%2C+ARS%2C+USDA.+Retrieved+January+2024%2C+from+https%3A%2F%2Ffungi.ars.usda.gov%2FandbtnG=d=gs_citandt=1719683542249andu=%2Fscholar%3Fq%3Dinfo%3A80BRsnlOEQwJ%3Ascholar.google.com%2F%26output%3Dcite%26scirp%3D1%26hl%3Den.
Fu, M., Bai, Q., Zhang, H., Guo, Y., Peng, Y., Zhang, et al. (2022). Transcriptome analysis of the molecular patterns of pear plants infected by two colletotrichum fructicola pathogenic strains causing contrasting sets of leaf symptoms. Front. Plant Sci. 13. doi: 10.3389/FPLS.2022.761133/BIBTEX
Fuckel, K. (1869). Symbolae mycologicae: Beiträge zur Kenntniss der rheinischen Pilze. Available online at: https://books.google.com/books?hl=enandlr=andid=AN8UAAAAYAAJandoi=fndandpg=PA1anddq=Fuckel,+L.+(1870).+Symbolae+mycologicae.+Beitr%E2%82%ACage+zur+Kenntniss+der+Rheinischen+Pilze.+Jahrbucher+des+Nassauischen+Vereins+f%C3%BCr+Naturkunde,+23e24,+1e459.andots=LGH49WrBKsandsig=FNZoMA6uVULveK6Tb5d8MJxLurs (accessed May 10, 2024).
Gajbhiye, M., Sathe, S., Shinde, V., Kapadnis, B. (2016). Morphological and Molecular Characterization of Pomegranate Fruit Rot Pathogen, Chaetomella raphigera, and its Virulence Factors. Indian J. Microbiol. 56, 99–102. doi: 10.1007/S12088-015-0554-4/METRICS
Gangadevi, V., Muthumary, J. (2009). A novel endophytic taxol-producing fungus Chaetomella raphigera isolated from a medicinal plant, Terminalia arjuna. Appl. Biochem. Biotechnol. 158, pp.675–pp.684. doi: 10.1007/s12010-009-8532-0
Gao, Y.-H., Bai, W.-X., Sun, W.-H., Zhou, W.-N., Wu, G.-L., Zhu, Z.-Q., et al. (2019). Diversity of culturable endophytic fungi associated with bryophytes, pteridophytes and spermatophytes from Dawei mountain nature reserve, China. Chiang Mai J. Sci. 46, 626–638. Available at: https://www.thaiscience.info/Journals/Article/CMJS/10990618.pdf.
Glass, N. L., Donaldson, G. C. (1995). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/AEM.61.4.1323-1330.1995
Glienke, C., Pereira, O. L., Stringari, D., Fabris, J., Kava-Cordeiro, V., Galli-Terasawa, L., et al. (2011). Endophytic and pathogenic Phyllosticta species, with reference to those associated with Citrus Black spot. Persoonia: Mol. Phylogeny Evol. Fungi 26, 47–56. doi: 10.3767/003158511X569169
Golijan, P. J., Sečanski, M., Gordanić, S., Šarčević, T. L. (2023). Weed biological control with fungi-based bioherbicides. Acta Agriculturae Serbica 28, 23–37. doi: 10.5937/AASer2355023G
Gong, C. L., Wu, H. X., Li, J. C., Song, J.-Y. (2024). Morphological and multi-gene phylogenetic analyses of two new species of Phyllosticta (Phyllostictaceae, Botryosphaeriales) from Southwestern China. Phytotaxa 653, 20–36. doi: 10.11646/phytotaxa.653.1.2
Gontia-Mishra, I., Tripathi, N., Tiwari, S. (2014). A simple and rapid DNA extraction protocol for filamentous fungi efficient for molecular studies. Available online at: https://nopr.niscpr.res.in/handle/123456789/30483 (accessed May 5, 2024).
Gramss, G., Günther, T., Fritsche, W. (1998). Spot tests for oxidative enzymes in ectomycorrhizal, wood-, and litter decaying fungi. Mycological Res. 102, 67–72. doi: 10.1017/S095375629700436X
Grano-Maldonado, M. I., Ramos-Payan, R., Rivera-Chaparro, F., Aguilar-Medina, M., Romero-Quintana, J. G., Rodríguez-Santiago, A., et al. (2021). First Molecular Characterization of Colletotrichum sp. and Fusarium sp. Isolated from Mangrove in Mexico and the Antagonist Effect of Trichoderma harzianum as an Effective Biocontrol Agent. Plant Pathol. J. 37, 465. doi: 10.5423/PPJ.OA.03.2021.0048
Guerber, J. C., Liu, B., Correll, J. C., Johnston, P. R. (2003). Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95, 872–895. doi: 10.1080/15572536.2004.11833047
Gunatilleke, N., Pethiyagoda, R., Gunatilleke, S. (2008). Biodiversity of Sri Lanka. J. Natl. Sci. Foundation Sri Lanka 36, 25–61. doi: 10.4038/JNSFSR.V36I0.8047
Gunawardene, N. R., Dulip Daniels, A. E., Gunatilleke, I. A. U. N., Gunatilleke, C. V. S., Karunakaran, P. V., Nayak, K. G., et al. (2007). A brief overview of the Western Ghats–Sri Lanka biodiversity hotspot. Curr. Sci. 93, 1567–1572. Available at: https://wgbis.ces.iisc.ac.in/biodiversity/sahyadri_enews/newsletter/issue42/bibliography/A-brief-overview-of-the-Western-Ghats.PDF.
Harrison, J. G., Griffin, E. A. (2020). The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: how far have we come and where do we go from here? Environ. Microbiol. 22, 2107–2123. doi: 10.1111/1462-2920.14968
Hasan, M., Saiful Ahmad-Hamdani, M., Rosli, A. M., Hamdan, H. (2021). Bioherbicides: an eco-friendly tool for sustainable weed management. Plants 10, 1–21. doi: 10.3390/plants10061212
Hashem, A. H., Attia, M. S., Kandil, E. K., Fawzi, M. M., Abdelrahman, A. S., Khader, M. S., et al. (2023). Bioactive compounds and biomedical applications of endophytic fungi: a recent review. Microbial Cell Factories 22, 1–23. doi: 10.1186/S12934-023-02118-X
Hatamzadeh, S., Akbari Oghaz, N., Rahnama, K., Noori, F. (2024). Combination of a hierarchical and thermal shock process with a specific aqueous buffer: a safe, rapid and reliable DNA extraction method for plant endophytic fungi. Biologia 79, 597–604. doi: 10.1007/S11756-023-01579-0/METRICS
Hawksworth, D. L. (1991). The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycological Res. 95, 641–655. doi: 10.1016/S0953-7562(09)80810-1
Hawksworth, D. L. (2001). The magnitude of fungal diversity: the 1·5 million species estimate revisited. Mycological Res. 105, 1422–1432. doi: 10.1017/S0953756201004725
Hawksworth, D. L., Lücking, R. (2017). Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5, 10–1128. doi: 10.1128/MICROBIOLSPEC.FUNK-0052-2016/ASSET/A56131C0-D4D3-43FE-85ED-8C479B914E39/ASSETS/GRAPHIC/FUNK-0052-2016-FIG3.GIF
Herath, I. H. M. I. S., Manamgoda, D. S., Udayanga, D. (2019). “Morphological and molecular identification of fungal pathogens associated with cultivated rubber trees in Sri Lanka,” in Proceedings of the First National Symposium of Sri Lanka Association for Mycology and Plant Pathology. 12–12. Available at: http://dr.lib.sjp.ac.lk/bitstream/handle/123456789/11727/Morphological%20and%20molecular%20identification%20of%20fungal%20pathogens%20associated%20with%20cultivated%20rubber%20trees%20in%20sri%20Lanka.pdf?sequence=1.
Herath, I. S., Udayanga, D., Miriyagalla, S., Castlebury, L. A., Manamgoda, D. S. (2021). Colletotrichum siamense causing anthracnose-twister disease of onion (Allium cepa) in Sri Lanka. Australas. Plant Dis. Notes 16, 1–6. doi: 10.1007/S13314-021-00444-W/FIGURES/2
Hettiarachchi, S., Gunasekera, S. A., Balasooriya, I. (1983). Leaf spot diseases of water hyacinth in Sri Lanka. J. Aquat. Plant Manage. 21, 62–65. Available at: https://www.apms.org/wp-content/uploads/japm-21-02-062.pdf.
Hou, L. W., Groenewald, J. Z., Pfenning, L. H., Yarden, O., Crous, P. W., Cai, L. (2020a). The phoma-like dilemma. Stud. Mycology 96, 309–396. doi: 10.1016/J.SIMYCO.2020.05.001
Hou, L. W., Hernández-Restrepo, M., Groenewald, J. Z., Cai, L., Crous, P. W. (2020b). Citizen science project reveals high diversity in Didymellaceae (Pleosporales, Dothideomycetes). MycoKeys 65, 99. doi: 10.3897/MYCOKEYS.65.47704
Huang, W., Lin, Z., Zhang, Z., Chen, J. (2021). First report of colletotrichum fructicola causing anthracnose on water hyacinth in China. Plant Dis. 105. doi: 10.1094/PDIS-12-20-2621-PDN
Huelsenbeck, J. P., Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hyde, K. D., Abdel-Wahab, M. A., Abdollahzadeh, J., Abeywickrama, P. D., Absalan, S., Afshari, N., et al. (2023). Global consortium for the classification of fungi and fungus-like taxa. Mycosphere 14, 1960–2012. Available at: https://repozytorium.bg.ug.edu.pl/info/article/UOG15bdd94b159b4b96ab6bcf179a783d29/.
Ismail, S. N., Hamid, M. A., Mansor, M. (2021). An overview of macrophytes in the tropical wetland ecosystem. Indonesian J. Limnology 2, 25–34. doi: 10.51264/INAJL.V2I1.12
Jayaram, S., Biswas, S., Philip, I., Umesh, M., Sarojini, S. (2023). Differential Laccase Production among Diverse Fungal Endophytes in Aquatic Plants of Hulimavu Lake in Bangalore, India. J. Pure Appl. Microbiol., 1–12. Available at: https://drive.google.com/file/d/1Qz7SS96BzEwJCUm4bNsY68lHHQT_s7ur/view.
Jayawardena, R., Hyde, K., Damm, U., Cai, L., Liu, M., Li, X. H., et al. (2016a). Notes on currently accepted species of Colletotrichum. Mycosphere 7, 1192–1260. doi: 10.5943/mycosphere/si/2c/9
Jayawardena, R., Li, X., Liu, M., Zhang, W., Yan, J. Y. (2016b). Mycosphere essay 16: Colletotrichum: biological control, biocatalyst, secondary metabolites and toxins. Mycosphere 7, 1164–1176. doi: 10.5943/mycosphere/si/2c/7
Ji, M., Dong, R., Zhang, J., Shi, X., Wang, Y., Huang, Q., et al. (2024). Diversity and environmental determinants of aquatic plants across China. Hydrobiologia 851, 3453–3469. doi: 10.1007/S10750-024-05509-2
Jia, S. L., Chi, Z., Liu, G. L., Hu, Z., Chi, Z. M. (2020). Fungi in mangrove ecosystems and their potential applications. Crit. Rev. Biotechnol. 40, 852–864. doi: 10.1080/07388551.2020.1789063
Jia, M., Gong, X., Fan, M., Liu, H., Zhou, H., Gu, S., et al. (2023). Identification and analysis of the secretome of plant pathogenic fungi reveals lifestyle adaptation. Front. Microbiol. 14. doi: 10.3389/FMICB.2023.1171618/BIBTEX
Jiang, Y., Zhang, Z., Zhang, J., Wang, S., Zhang, X. (2024). Morphological and phylogenetic analyses reveal three new species of Phyllosticta (Botryosphaeriales, Phyllostictaceae) in China. J. Fungi 10, 7. doi: 10.3390/jof10010007
Kamat, S., Kumari, M., Taritla, S., Jayabaskaran, C. (2020). Endophytic fungi of marine alga from konkan coast, India—A rich source of bioactive material. Front. Mar. Sci. 7. doi: 10.3389/FMARS.2020.00031/BIBTEX
Kao, M. R., Kuo, H. W., Lee, C. C., Huang, K. Y., Huang, T. Y., Li, C. W., et al. (2019). Chaetomella raphigera β-glucosidase D2-BGL has intriguing structural features and a high substrate affinity that renders it an efficient cellulase supplement for lignocellulosic biomass hydrolysis. Biotechnol. Biofuels 12, 1–18. doi: 10.1186/S13068-019-1599-0/FIGURES/12
Kariyawasam, C. S., Kumar, L., Ratnayake, S. S. (2021). Potential distribution of aquatic invasive alien plants, Eichhornia crassipes and Salvinia molesta under climate change in Sri Lanka. Wetlands Ecol. Manage. 29, 531–545. doi: 10.1007/S11273-021-09799-4/METRICS
Karunarathna, A., Withee, P., Haituk, S., Mai Sci, C. J., Cheewangkoon, R. (2022). Worldwide checklist on Grass Fungi: What do we know so far in Ascomycota. Chiang Mai J. Sci. 49, 742–984. doi: 10.12982/CMJS.2022.058
Keshamma, E. (2022). Application of eco-friendly mycoherbicides: A need to develop a sustainable ecosystem. UGC Care Group 1 J. 52, 0378–4568. Available at: https://www.researchgate.net/profile/Priya-Lokare-2/publication/364905871_APPLICATION_OF_ECO-FRIENDLY_MYCOHERBICIDES_A_NEED_TO_DEVELOP_A_SUSTAINABLE_ECOSYSTEM/links/635e233512cbac6a3e0b449d/APPLICATION-OF-ECO-FRIENDLY-MYCOHERBICIDES-A-NEED-TO-DEVELOP-A-SUSTAINABLE-ECOSYSTEM.pdf.
Khatami, S. H., Vakili, O., Movahedpour, A., Ghesmati, Z., Ghasemi, H., Taheri-Anganeh, M. (2022). Laccase: Various types and applications. Biotechnol. Appl. Biochem. 69, 2658–2672. doi: 10.1002/BAB.2313
Khokhar, I., Mukhtar, I., Mushtaq, S. (2011). Isolation and screening of amylolytic filamentous fungi. J. Appl. Sci. Environ. Manage. 15, 203–206. doi: 10.4314/JASEM.V15I1.68442
Koshila, H. V. A. S., Wanigatunge, R. P., Dias, R. K. S., Edirisinghe, P. (2023). Morphological characterization and molecular identification of three fungal pathogens isolated from solanum melongena L. @ in Sri Lanka. J. Agric. Sciences-Sri Lanka 18, 251–260. doi: 10.4038/jas.v18i2.10257
Kularathnage, N. D., Senanayake, I. C., Wanasinghe, D. N., Doilom, M., Stephenson, S. L., Song, J., et al. (2023). Plant-associated novel didymellaceous taxa in the South China botanical garden (Guangzhou, China). J. Fungi 9, 182. doi: 10.3390/JOF9020182
Kurera, W. M. S., Adikaram, N. K. B., Yakandawala, D. M. D., Maharachchikumbura, S. S., Jayasinghe, L., Samarakoon, K. (2023). Plurivorum and Colletotrichum musae causing banana anthracnose disease in the Central Province of Sri Lanka. Ournal Natl. Sci. Foundation Sri Lanka 51, 311–326. Available at: http://www.nsf.ac.lk/images/nsf/jnsf/JNSF_Volume_51_No_2.pdfpage=133.
Kyomuhimbo, H. D., Brink, H. G. (2023). Applications and immobilization strategies of the copper-centred laccase enzyme; a review. Heliyon 9, 13156. doi: 10.1016/J.HELIYON.2023.E13156/ATTACHMENT/414D1A8F-E3AB-4AC6-9EDE-D045BB5D9B40/MMC1
Larki, R., Mehrabi-Koushki, M., Farokhinejad, R. (2019). Ectophoma Iranica sp. nov. and new hosts and records of Allophoma spp. in Iran. J. Phytopathol. 167, 538–545. doi: 10.1111/JPH.12840
Lee, G., Shim, H., Cho, W., Kim, W. (2022). First Report of Leaf Spot in Water Spinach Caused by Ectophoma multirostrata. J. Korean Soc. Mycology 50, 367–372. Available at: http://www.kjmycology.or.kr/view/N0320500414.pdf.
Leonowicz, A., Cho, N., Luterek, J., Wilkolazka, A., WojtasWasilewska, M., Matuszewska, A., et al. (2001). Fungal laccase: properties and activity on lignin. J. Basic Microbiology: Int. J. Biochemistry Physiology Genetics Morphology Ecol. Microorganisms 41, 185–227. doi: 10.1002/1521-4028(200107)41:3/4%3C185::AID-JOBM185%3E3.0.CO;2-T
Levin, L. N., Ramos, A. M., Parisi, M., Gally, M. E. (2007). Screening of Colletotrichum (Ascomycota) isolates, causal agents of Soybean Anthracnose, for Laccase production Vol. 42 (Argentine Botanical Society; Bulletin of the Argentine Botanical Society: Sociedad Argentina de Botánica; Boletín de la Sociedad Argentina de Botánica), 71–77. Available at: https://ri.conicet.gov.ar/handle/11336/41819.
Li, W. J., McKenzie, E. H. C., Liu, J. K., Bhat, D. J., Dai, D. Q., Camporesi, E., et al. (2020). Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Diversity 100, 279–801. doi: 10.1007/S13225-020-00440-Y
Li, H. Y., Zhao, C. A., Liu, C. J., Xu, X. F. (2010). Endophytic fungi diversity of aquatic/riparian plants and their antifungal activity in vitro. J. Microbiol. 48, 1–6. doi: 10.1007/S12275-009-0163-1/METRICS
Lin, W. L., Duan, C. H., Wang, C. L. (2023). Identification and virulence of Colletotrichum species causing anthracnose on mango. Plant Pathol. 72, 623–635. doi: 10.1111/PPA.13682
Liu, F., Ma, Z. Y., Hou, L. W., Diao, Y. Z., Wu, W. P., Damm, U., et al. (2022). Updating species diversity of Colletotrichum, with a phylogenomic overview. Stud. Mycology 101, pp.1–pp56. doi: 10.3114/sim.2022.101.01
Liu, J. W., Manawasinghe, I. S., Liao, X. N., Mao, J., Dong, Z. Y., Jayawardena, R. S., et al. (2023). Endophytic Colletotrichum (Sordariomycetes, Glomerellaceae) species associated with Citrus grandis cv. “Tomentosa” in China. MycoKeys 95, 163. doi: 10.3897/MYCOKEYS.95.87121
Liu, Y. J., Whelen, S., Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A026092
Lu, Y., Chen, C., Chen, H., Zhang, J., Chen, W. (2012). Isolation and identification of endophytic fungi from actinidia macrosperma and investigation of their bioactivities. Evidence-Based Complementary Altern. Med. 2012, 382742. doi: 10.1155/2012/382742
Łubek-Nguyen, A., Ziemichód, W., Olech, M. (2022). Application of enzyme-assisted extraction for the recovery of natural bioactive compounds for nutraceutical and pharmaceutical applications. Appl. Sci. 12, 3232. doi: 10.3390/APP12073232
Lugtenberg, B. J. J., Caradus, J. R., Johnson, L. J. (2016). Fungal endophytes for sustainable crop production. FEMS Microbiol. Ecol. 92, 1–17. doi: 10.1093/FEMSEC/FIW194
Maharachchikumbura, S. S. N., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Huang, S. K., Abdel-Wahab, M. A., et al. (2015). Towards a natural classification and backbone tree for Sordariomycetes. Fungal Diversity 72, 199–301. doi: 10.1007/s13225-015-0331-z
Mahfooz, M., Dwedi, S., Bhatt, A., Mahfooz, M., Bhatt, M., Kumar Agrawal, P. (2017). Evaluation of antifungal and enzymatic potential of endophytic Fungi isolated from Cupressus torulosa D. Don. Int. J. Curr. Microbiol. Appl. Sci. 6, 4084–4100. doi: 10.20546/ijcmas.2017.607.424
Mahuri, M., Paul, M., Thatoi, H. (2023). A review of microbial laccase production and activity toward different biotechnological applications. Syst. Microbiol. Biomanufacturing 3, 533–551. doi: 10.1007/S43393-023-00163-6/METRICS
Maravi, P., Kumar, A. (2021). Cellulase: distribution, production, characterization and industrial applications. Biotechnol. J. Int. 25, 36–71. doi: 10.9734/BJI/2021/V25I330143
Mathur, P. N., Thirumalachar, M. J. (1959). Studies on some Indian soil fungi 1. Some new or noteworthy Sphaeropsidales. Sydowia 13, 143–147. Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=Studies+on+some+Indian+soil+fungi+1.+Some+new+or+noteworthy+Sphaeropsidales.+Sydowia%2C+13%281-6%29%2C+143-146.andbtnG.
Mattoo, A. J., Nonzom, S. (2022). Investigating diverse methods for inducing sporulation in endophytic fungi. Stud. Fungi 7, 1–10. doi: 10.48130/SIF-2022-0016
Maulidyna, A., Alicia, F., Agustin, H. N., Dewi, I. R., Nurhidayah, I., Dewangga, A., et al. (2021). Review: Economic impacts of the invasive species water hyacinth (Eichhornia crassipes): Case study of Rawapening Lake, Central Java, Indonesia. Int. J. Bonorowo Wetlands 11, 18–31. doi: 10.13057/BONOROWO/W110103
May, R. M. (2000). The dimensions of life on Earth (Nature and Human Society), 30–45. Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=May%2C+R.+M.+%282000%29.+The+dimensions+of+life+on+Earth.+In+P.+H.+Raven+%26+T.+Williams+%28Eds.%29%2C+Nature+and+Human+Society%3A+The+Quest+for+a+Sustainable+World+%28pp.+30-45%29.+Washington%2C+DC%3A+National+Academy+Press.andbtnG.
Miller, M. A., Pfeiffer, W., Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in 2010 gateway computing environments workshop (GCE)(New Orleans, LA, USA), 1–8. doi: 10.1109/GCE.2010.5676129
Myllys, L., Stenroos, S., Thell, A. (2002). New genes for phylogenetic studies of lichenized fungi: glyceraldehyde-3-phosphate dehydrogenase and beta-tubulin genes. Lichenologist 34, 237–246. doi: 10.1006/LICH.2002.0390
Myovela, H., Hussein, J., Tibuhwa, D. (2024). The hidden diversity of mangrove endophytic fungi from Tanzania: insights from a preliminary study. Biologia 79, 669–683. doi: 10.1007/S11756-023-01587-0/METRICS
Nag Raj, T. R. (1993). Coelomycetous anamorphs with appendage-bearing conidia (Waterloo, Ontario: Mycologue Publications). Available at: https://cir.nii.ac.jp/crid/1130282272565879808.
Nguyen, T. T. T., Pangging, M., Lee, S. H., Lee, H. B. (2018). Four new records of ascomycete species from korea. Mycobiology 46, 328–340. doi: 10.1080/12298093.2018.1550169
Niskanen, T., Lücking, R., Dahlberg, A., Gaya, E., Suz, L. M., Mikryukov, et al. (2023). Pushing the frontiers of biodiversity research: unveiling the global diversity, distribution, and conservation of fungi. Annu. Rev. Environ. Resour. 48, 149–176. doi: 10.1146/ANNUREV-ENVIRON-112621-090937/1
Norphanphoun, C., Hyde, K. D. (2023). First Report of Colletotrichum fructicola, C. rhizophorae sp. nov. and C. Thailandica sp. nov. on Mangrove in Thailand. Pathogens 12, 1436. doi: 10.3390/PATHOGENS12121436
Núñez, M., del, P., Levin, L. N., Cinto, I. E. (2023). Variabilidad intraespecífica en el crecimiento y la producción in vitro de enzimas degradadoras de pared celular vegetal entre aislamientos argentinos de Colletotrichum graminicola, un patógeno de maíz Vol. 58 (Boletín de la Sociedad Argentina de Botánica), 187–194. doi: 10.31055/1851.2372.V58.N2.39049
O’Donnell, K., Cigelnik, E. (1997). Two Divergent Intragenomic rDNA ITS2 Types within a Monophyletic Lineage of the Fungus Fusarium Are Nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/MPEV.1996.0376
O’Hare, M. T., Aguiar, F. C., Asaeda, T., Bakker, E. S., Chambers, P. A., Clayton, J. S., et al. (2018). Plants in aquatic ecosystems: current trends and future directions. Hydrobiologia 812, 1–11. doi: 10.1007/S10750-017-3190-7/METRICS
Páez Redondo, A., Prusky, D., Hoyos Carvajal, L. M. (2022). Quiescence as a strategic stage for the infective process of Colletotrichum species. Rev. UDCA Actualidad Divulgación Científica 25, 17. Available at: https://dialnet.unirioja.es/servlet/articulo?codigo=9508805andinfo=resumenandidioma=ENG.
Pärtel, K., Baral, H. O., Tamm, H., Põldmaa, K. (2017). Evidence for the polyphyly of Encoelia and Encoelioideae with reconsideration of respective families in Leotiomycetes. Fungal Diversity 82, 183–219. doi: 10.1007/S13225-016-0370-0/METRICS
Pathmanathan, N., Deshappriya, N., Manamgoda, D. S., Sandamali, T. G. I., Munasinghe, M. (2022). Comparison of colonization, diversity, and molecular phylogeny of endophytic fungi in selected traditional and newly improved rice (Oryza sativa L.) varieties in Sri Lanka Pathmanathan N1, Deshappriya N1, Manamgoda US1. Sandamali TGI2 and Munasinghe M1. Curr. Res. Environ. Appl. Mycology (Journal Fungal Biology) 12, 147–169. doi: 10.5943/cream/12/l/12
Patil, A. G., Khan, K., Aishwarya, S., Padyana, S., Huchegowda, R., Reddy, K. R., et al. (2021). “Fungal amylases and their industrial applications,” in Industrially important fungi for sustainable development. Fungal biology. Eds. Abdel-Azeem, A. M., Yadav, A. N., Yadav, N., Sharma, M. (Springer, Cham), 407–434. doi: 10.1007/978-3-030-85603-8_11
Peng, X. J., Wang, Q. C., Zhang, S. K., Gun, K., Zhou, X. D. (2023). Colletotrichum species associated with Camellia anthracnose in China. Mycosphere 14, 130–157. doi: 10.5943/mycosphere/14/si2/3
Peralta-Ruiz, Y., Rossi, C., Grande-Tovar, C. D., Chaves-López, C. (2023). Green management of postharvest anthracnose caused by colletotrichum gloeosporioides. J. Fungi 9, 623. doi: 10.3390/JOF9060623
Prajapati, V. S., Trivedi, U. B., Patel, K. C. (2013). Optimization of glucoamylase production by Colletotrichum sp. KCP1 using statistical methodology. Food Sci. Biotechnol. 22, 31–38. doi: 10.1007/S10068-013-0005-0/METRICS
Pramanic, A., Sharma, S., Dhanorkar, M., Prakash, O., Singh, P. (2023). Endophytic microbiota of floating aquatic plants: recent developments and environmental prospects. World J. Microbiol. Biotechnol. 39, 1–15. doi: 10.1007/S11274-023-03543-1/METRICS
Rajagopal, K., Binika, D., Joshila, D., Ps, T., Arulmathi, R., Kathiravan, G., et al. (2018). Mycodiversity and biotechnological potential of endophytic fungi isolated from hydrophytes. Curr. Res. Environ. Appl. Mycology (Journal Fungal Biology) 8, 172–182. doi: 10.5943/cream/8/2/2
Rana, K. L., Kour, D., Sheikh, I., Dhiman, A., Yadav, N., Yadav, A. N., et al. (2019). “Endophytic fungi: biodiversity, ecological significance, and potential industrial applications,” in Recent advancement in white biotechnology through fungi. Eds. Yadav, A., Mishra, S., Singh, S., Gupta, A. (Springer, Cham), 1–62. doi: 10.1007/978-3-030-10480-1_1
Rannala, B., Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Rashmi, M., Kushveer, J., Sarma, V. (2019). A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 10, 798–1079. doi: 10.5943/mycosphere/10/1/19
Ratnaweera, P. (2019). Novel and interesting antibiotic scaffolds from endophytic fungi of Sri Lankan origin. Chem. Sri Lanka 36, 1217. Available at: http://ichemcdr.com:8080/xmlui/handle/123456789/4.
Ravimannan, N., Sepali, E. (2020). Antibacterial activity of endophytic fungi isolated from mangroves of Jaffna Peninsula, Sri Lanka. Afr. J. Plant Sci. 14, 243–247. doi: 10.5897/AJPS2020.1987
Ravlić, M., Baličević, R. (2014). Biološka kontrola korova biljnim patogenima. Poljoprivreda 20, 34–40. Available at: https://urn.nsk.hr/urn:nbn:hr:151:066442.
Reyes, R., Parayao, A., Waing, K. (2021). Identification and evaluation of enzymatic ability of fungal endophytes from Citrofortunella microcarpa (Bunge) Wijnands. Stud. Fungi 6, 460–468. doi: 10.5943/SIF/6/1/35
Robledo-Mahón, T., Calvo, C., Aranda, E. (2020). Enzymatic potential of bacteria and fungi isolates from the sewage sludge composting process. Appl. Sci. 10, 7763. doi: 10.3390/APP10217763
Rodrigues, C. M., Takita, M. A., Silva, N. V., Ribeiro-Alves, M., MacHado, M. A. (2019). Comparative genome analysis of Phyllosticta citricarpa and Phyllosticta capitalensis, two fungi species that share the same host. BMC Genomics 20, 1–12. doi: 10.1186/S12864-019-5911-Y/TABLES/2
Rodríguez-Merino, A. (2023). Identifying and managing areas under threat in the iberian peninsula: an invasion risk atlas for non-native aquatic plant species as a potential tool. Plants, 1–18. doi: 10.3390/plants12173069
Ronquist, F., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rossman, A. Y., Aime, M. C., Farr, D. F., Castlebury, L. A., Peterson, K. R., Leahy, R. (2004). The coelomycetous genera Chaetomella and Pilidium represent a newly discovered lineage of inoperculate discomycetes. Mycological Prog. 3, 275–290. doi: 10.1007/s11557-006-0098-4
Rungjindamai, N., Gareth Jones, E. B. (2024). Why are there so few basidiomycota and basal fungi as endophytes? A review. J. Fungi 10, 1–30. doi: 10.3390/jof10010067
Samarakoon, M., Peršoh, D., Hyde, K., Bulgakov, T. S., Manawasinghe, I. S., Jayawardena, R. S., et al. (2018). Colletotrichum acidae sp. nov. from northern Thailand and a new record of C. dematium on Iris sp. Mycosphere 9, 583–597. Available at: https://www.academia.edu/download/67478435/Colletotrichum_acidae_sp._nov._from_northern_Thailand_and_a_new.pdf.
Sandberg, D. C., Battista, L. J., Arnold, A. E. (2014). Fungal endophytes of aquatic macrophytes: diverse host-generalists characterized by tissue preferences and geographic structure. Microbial Ecol. 67, 735–747. doi: 10.1007/S00248-013-0324-Y/METRICS
Sarathchandra, C., Abebe, Y. A., Wijerathne, I. L., Aluthwattha, S. T., Wickramasinghe, S., Ouyang, Z. (2021). An overview of ecosystem service studies in a tropical biodiversity hotspot, Sri Lanka: Key perspectives for future research. Forests 12, 540. doi: 10.3390/F12050540/S1
Senanayake, I. C., Jeewon, R., Chomnunti, P., Wanasinghe, D. N., Norphanphoun, C., Karunarathna, A., et al. (2018). Taxonomic circumscription of Diaporthales based on multigene phylogeny and morphology. Fungal Diversity 93, 241–443. doi: 10.1007/S13225-018-0410-Z/METRICS
Shankar Naik, B., Abrar, S., Krishnappa, M. (2019). “Industrially important enzymes from fungal endophytes,” in recent advancement in white biotechnology through fungi. Fungal biology. Eds. Yadav, A., Mishra, S., Singh, S., Gupta, A. (Springer, Cham), 263–280. doi: 10.1007/978-3-030-10480-1_7
Sharma, P., Tamang, S., Thakur, N., Das, S. (2024). “8 Application, current and future trends of bacterial and fungal polyphenol oxidases (laccases). Polyphenol Oxidases: Function, Wastewater Remediation, and Biosensors,” in Polyphenol oxidases: function, wastewater remediation, and biosensors. Eds. Sharma, P., Tamang, S., Thakur, N., Das, S.. doi: 10.1515/9783111033525-008
Shraddha, Shekher, R., Sehgal, S., Kamthania, M., Kumar, A. (2011). Laccase: microbial sources, production, purification, and potential biotechnological applications. Enzyme Res. 2011, 217861. doi: 10.4061/2011/217861
Silva, M., Pereira, O. (2007). First report of Guignardia endophyllicola leaf blight on Cymbidium (Orchidaceae) in Brazil. Australas. Plant Dis. Notes 2, 31–32. doi: 10.1071/DN07015
Silva, M., Pereira, O., Braga, I. F., Lelis, S. M. (2008). Leaf and pseudobulb diseases on Bifrenaria harrisoniae (Orchidaceae) caused by Phyllosticta capitalensis in Brazil. Australas. Plant Dis. Notes 3, 53–56. doi: 10.1007/BF03211238
Singh, A., Bajar, S., Devi, A., Pant, D. (2021). An overview on the recent developments in fungal cellulase production and their industrial applications. Bioresource Technol. Rep. 14, 100652. doi: 10.1016/J.BITEB.2021.100652
Singh, D., Gupta, N. (2020). Microbial Laccase: a robust enzyme and its industrial applications. Biologia 75, 1183–1193. doi: 10.2478/S11756-019-00414-9/METRICS
Singh, J., Quereshi, S., Banerjee, N., Pandey, A. K. (2010). Production and extraction of phytotoxins from Colletotrichum dematium FGCC20 effective against Parthenium hysterophorus L. Braz. Arch. Biol. Technol. 53, 669–678. doi: 10.1590/S1516-89132010000300023
Singh, N., Sithole, B., Govinden, R. (2023). Screening for cellulases and preliminary optimisation of glucose tolerant β-glucosidase production and characterisation. Mycology 14, 91–107. doi: 10.1080/21501203.2022.2155261
Spegazzini, C. (1881). Nova addenda ad mycologiam venetam. Michelia 5, 453–487. Available at: https://scholar.google.com/scholar?hl=enandas_sdt=0%2C5andq=Spegazzini%2C+C.+%281879%29.+Nova+addenda+ad+Mycologiam+Venetam.+Michelia%2C+1%285%29%2C+453-487.andbtnG.
Srivastava, N., Srivastava, M., Mishra, P. K., Gupta, V. K., Molina, G., Rodriguez-Couto, et al. (2018). Applications of fungal cellulases in biofuel production: Advances and limitations. Renewable Sustain. Energy Rev. 82, 2379–2386. doi: 10.1016/J.RSER.2017.08.074
Srivastava, N., Srivastava, M., Mishra, P. K., Singh, P., Ramteke, P. W. (2015). Application of cellulases in biofuels industries: an overview. J. Biofuels Bioenergy 1, 63. doi: 10.5958/2454-8618.2015.00007.3
Su, Y. Y., Qi, Y. L., Cai, L. (2012). Induction of sporulation in plant pathogenic fungi. Mycology 3, 195–200. doi: 10.1080/21501203.2012.719042
Sui, X. N., Guo, M. J., Zhou, H., Hou, C. L. (2023). Four new species of Phyllosticta from China based on morphological and phylogenetic characterization. Mycology 14, 190–203. doi: 10.1080/21501203.2023.2225552
Sun, X., Guo, L. D. (2012). Endophytic fungal diversity: review of traditional and molecular techniques. Mycology 3, 65–76. doi: 10.1080/21501203.2012.656724
Sung, G. H., Sung, J. M., Hywel-Jones, N. L., Spatafora, J. W. (2007). A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 44, 1204–1223. doi: 10.1016/J.YMPEV.2007.03.011
Surasinghe, T., Kariyawasam, R., Sudasinghe, H., Karunarathna, S. (2019). Challenges in biodiversity conservation in a highly modified tropical river basin in Sri Lanka. Water 12, 26. doi: 10.3390/W12010026
Sutton, B. (1981). “The coelomycetes,” in Fungi imperfecti with pycnidia, acervuli and stromata. doi: 10.5555/19801366283
Sutton, B. C., Alcorn, J. L. (1974). Neottiosporina. Aust. J. Bot. 22, 517–530. doi: 10.1071/BT9740517
Sutton, B. C., Alcorn, J. L. (1985). Undescribed species of Crinitospora gen. nov., Massariothea, Mycoleptodiscus and Neottiosporina from Australia. Trans. Br. Mycological Soc. 84, 437–445. doi: 10.1016/S0007-1536(85)80005-X
Sutton, B., Sarbhoy, A. K. (1976). Revision of Chaetomella, and comments upon Vermiculariopsis and Thyriochaetum. Trans. Br. Mycological Soc. 66, 297–303. Available at: https://www.sciencedirect.com/science/article/pii/S0007153676800563.
Sutton, B. C., Wu, W. P. (1995). Neottiosporina sorghicola sp. nov., associated with leaf lesions on Sorghum vulgare from China. Mycological Res. 99, 831–832. doi: 10.1016/S0953-7562(09)80736-3
Suwannarach, N., Kumla, J., Matsui, K., Lumyong, S. (2018). Morphological and molecular evidence support a new endophytic fungus, Chaetomella endophytica from Japan. Mycoscience 59, 473–478. doi: 10.1016/J.MYC.2018.05.001
Talhinhas, P., Baroncelli, R. (2021). Colletotrichum species and complexes: geographic distribution, host range and conservation status. Fungal Diversity 110, 109–198. doi: 10.1007/S13225-021-00491-9
Talhinhas, P., Baroncelli, R. (2023). Hosts of colletotrichum. Mycosphere 14, 158–261. doi: 10.5943/mycosphere/14/si2/4
TeBeest, D. O., Templeton, G. E. (1985). Mycoherbicides: progress in the biologic. Plant Dis. 69, 1–10. Available online at: https://www.apsnet.org/publications/plantdisease/backissues/Documents/1985Articles/PlantDisease69n01_6.pdf.
Tedersoo, L., Anslan, S., Bahram, M., Põlme, S., Riit, T., Liiv, I., et al. (2015). Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 1–43. doi: 10.3897/mycokeys.10.4852
Tedersoo, L., Bahram, M., Zinger, L., Nilsson, R. H., Kennedy, P. G., Yang, T., et al. (2022). Best practices in metabarcoding of fungi: From experimental design to results. Mol. Ecol. 31, 2769–2795. doi: 10.1111/MEC.16460
Templeton, G. (1987). Mycoherbicides-achievements, developments and prospects. doi: 10.5555/20083007589
Thambugala, K., Daranagama, D., Kannangara, S. (2018). Biocontrol potential of endophytic fungi in tea (Camellia sinensis (L.) Kuntze) leaves against selected fungal phytopathogens. Malaysian J. Microbiol. 18, 665–669. doi: 10.21161/mjm.220055
Tunali, B., Berner, D. K., Dubin, H. J. (2008). First Report of Leaf Spot Caused by Colletotrichum cf. linicola on Field Bindweed in Turkey. Plant Dis. 92, 316–316. doi: 10.1094/PDIS-92-2-0316A
Tunali, B., Kansu, B., Berner, D. K. (2009). Biological control studies on Convolvulus arvensis L. with fungal pathogens. J. Turkish Phytopathol. 38, 1–8. Available at: https://www.ars.usda.gov/research/publications/publication/?seqNo115=265287.
Undugoda, L., Thambugala, K., Kannangara, S., Munasinghe, J., Premarathna, N., Dharmasiri, N. (2023). Phylloremediation of pyrene and anthracene by endophytic fungi inhabiting tea leaves (Camellia sinensis (L.) Kuntze) in Sri Lanka. New Z. J. Bot., 1–14. doi: 10.1080/0028825X.2023.2258829
Valenzuela-Lopez, N., Cano-Lira, J. F., Guarro, J., Sutton, D. A., Wiederhold, N., Crous, P. W., et al. (2018). Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycology 90, 1–69. doi: 10.1016/J.SIMYCO.2017.11.003
Vargas, W. A., Sanz Martín, J. M., Rech, G. E., Rivera, L. P., Benito, E. P., Díaz-Mínguez, et al. (2012). Plant Defense Mechanisms Are Activated during Biotrophic and Necrotrophic Development of Colletotricum graminicola in Maize. Plant Physiol. 158, 1342–1358. doi: 10.1104/PP.111.190397
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriology 172, 4238–4246. doi: 10.1128/JB.172.8.4238-4246.1990
Viswanath, B., Rajesh, B., Janardhan, A., Kumar, A. P., Narasimha, G. (2014). Fungal laccases and their applications in bioremediation. Enzyme Res. 2014, 163242. doi: 10.1155/2014/163242
Wall, R. E., Prasad, R., Shamoun, S. F. (1992). The development and potential role of mycoherbicides for forestry. Forestry Chronicle 68, 736–741. doi: 10.5558/TFC68736-6
Wang, X., Chen, G., Huang, F., Zhang, J., Hyde, K. D., Li, H. (2012). Phyllosticta species associated with citrus diseases in China. Fungal Diversity 52, 209–224. doi: 10.1007/S13225-011-0140-Y/METRICS
Wang, Q. H., Ye, R., Zhou, Y., Pan, Y., Hu, T. Q., Zeng, Y. C., et al. (2023). First Report of Colletotrichum siamense Causing Leaf Spot on Manglietia decidua in the World. Plant Dis. 107, 562. doi: 10.1094/PDIS-04-22-0774-PDN
Welideniya, W. A., Rienzie, K. D. R. C., Wickramaarachchi, W. A. R. T., Aruggoda, A. G. B. (2019). Characterization of fungal pathogens causing anthracnose in capsicum pepper (Capsicum annuum L.) and their seed borne nature. Ceylon J. Sci. 48, 261–269. doi: 10.4038/cjs.v48i3.7650
White, T., Bruns, T., Lee, S. J. W. T., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications, 1st ed, vol. 18 . Eds. White, T., Bruns, T., Lee, S. J. W. T., Taylor, J., 315–322). Available at: https://books.google.com/books?hl=enandlr=andid=Z5jwZ2rbVe8Candoi=fndandpg=PA315anddq=White,+T.+J.,+Bruns,+T.,+Lee,+J.,+%26+Taylor,+J.+(1990).+Amplification+and+direct+sequencing+of+fungal+ribosomal+RNA+genes+for+phylogenetics.+In+M.+A.+Innis,+D.+H.+Gelfand,+J.+J.+Sninsky,+%26+T.+J.+White+(Eds.),+PCR+Protocols:+A+Guide+to+Methods+and+Applications+(pp.+315-322).+Academic+Press.andots=ICG-MiYM7zandsig=tZzDeKV4ajIVdZSoM6oz7l3YxyE.
Wickramasinghe, W. A. P. B., Yakandawala, D., Adikaram, N. K. B. (2019). “Morphological and molecular characterization of Colletotrichum causing anthracnose in Sri Lankan Begonia,” in Proceedings of the SLAMPP Symposium on Plant Health. doi: 10.4038/cjs.v49i1.7709
Wijayawardene, N. N., Boonyuen, N., Ranaweera, C. B., de Zoysa, H. K. S., Padmathilake, R. E., Nifla, F., et al. (2023). OMICS and other advanced technologies in mycological applications. J. Fungi 9, 1–27. doi: 10.3390/JOF9060688
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Magurno, F., et al. (2022a). Outline of Fungi and fungus-like taxa–2021. Mycosphere 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Wijayawardene, N. N., Phillips, A. J. L., Pereira, D. S., Dai, D. Q., Aptroot, A., Monteiro, J. S., et al. (2022b). Forecasting the number of species of asexually reproducing fungi (Ascomycota and Basidiomycota). Fungal Diversity 114, 463–490. doi: 10.1007/S13225-022-00500-5
Wijayawardene, N. N., Phillips, A. J. L., Tibpromma, S., Dai, D. Q., Selbmann, L., Monteiro, J. S., et al. (2021). Looking for the undiscovered asexual taxa: case studies from lesser studied life modes and habitats. Mycosphere 12, 1290–1333. doi: 10.5943/MYCOSPHERE/12/1/17
Wikee, S., Chomnunti, P., Kanghae, A., Chukeatirote, E., Lumyong, S., Faulds, C. (2017). Lignocellulolytic capability of endophytic Phyllostica sp. J. Bacteriology Mycology 4, 1–7. doi: 10.26420/JBACTERIOLMYCOL.2017.1047
Wikee, S., Lombard, L., Crous, P. W., Nakashima, C., Motohashi, K., Chukeatirote, E., et al. (2013). Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Diversity 60, 91–105. doi: 10.1007/S13225-013-0235-8/METRICS
Wimalasena, M. K., Wijayawardene, N. N., Bamunuarachchige, T. C., Jayalal, R. G. U., Bhat, D. J., Dawoud, T. M., et al. (2024). Ongoing study reveals high aquatic fungal diversity; an untouched component in Sri Lankan biodiversity. Chiang Mai J. Sci. 51, 1–31. doi: 10.12982/CMJS.2024.039
Wu, B., Hussain, M., Zhang, W., Stadler, M., Liu, X., Xiang, M. (2019). Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 10, 127–140. doi: 10.1080/21501203.2019.1614106
Xiong, W., Xie, D., Wang, Q., Wang, H., Wu, Z., Sun, H., et al. (2023). Non-native species in Poyang Lake Basin: status, threats and management. Aquat. Invasions 18, 119–134. doi: 10.3391/AI.2023.18.1.103610
Xu, Z., Shi, M., Tian, Y., Zhao, P., Niu, Y., Liao, M. (2019). Dirhamnolipid produced by the pathogenic fungus colletotrichum gloeosporioides BWH-1 and its herbicidal activity. Molecules 24, 1–12. doi: 10.3390/molecules24162969
Yakandawala, D. (2012). Present status of fresh water aquatic flora in Sri Lanka. In: Weerakoon, D. K., Wjesundera, S., editors. The national red list 2012 of Sri Lanka; conservation status of the fauna and flora. Colombo, Sri Lanka: Ministry of Environment, p. 186–196.
Yang, Y. K., Kim, S. O., Chung, H. S., Lee, Y. H. (2007). Use of colletotrichum graminicola KA001 to control barnyard grass. Plant Dis. 84, 55–59. doi: 10.1094/PDIS.2000.84.1.55
Yopi, Tasia, W., Melliawati, R. (2017). “Cellulase and xylanase production from three isolates of indigenous endophytic fungi,” in IOP Conference Series: Earth and Environmental Science, Vol. 101. 012035. doi: 10.1088/1755-1315/101/1/012035
You, Y. H., Park, J. M., Park, J. H., Kim, J. G. (2015). Diversity of endophytic fungi associated with the roots of four aquatic plants inhabiting two wetlands in korea. Mycobiology 43, 231–238. doi: 10.5941/MYCO.2015.43.3.231
Zhang, Y. X., Chen, J. W., Manawasinghe, I. S., Lin, Y. H., Jayawardena, R. S., Mckenzie, E. H. C., et al. (2023). Identification and characterization of Colletotrichum species associated with ornamental plants in Southern China. Mycosphere 14, 262–302. doi: 10.5943/mycosphere/14/si2/5
Zhaxybayeva, O., Gogarten, J. P. (2002). Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genomics 3, 4. doi: 10.1186/1471-2164-3-4
Zheng, Y. K., Qiao, X. G., Miao, C. P., Liu, K., Chen, Y. W., Xu, L. H., et al. (2015). Diversity, distribution and biotechnological potential of endophytic fungi. Ann. Microbiol. 66, 529–542. doi: 10.1007/S13213-015-1153-7
Zheng, H., Qiao, M., Xu, J., Yu, Z. (2021). Culture-based and culture-independent assessments of endophytic fungal diversity in aquatic plants in southwest China. Front. Fungal Biol. 2. doi: 10.3389/FFUNB.2021.692549/BIBTEX
Zheng, H., Yu, Z., Jiang, X., Fang, L., Qiao, M. (2022). Endophytic colletotrichum species from aquatic plants in southwest China. J. Fungi 8, 87. doi: 10.3390/JOF8010087/S1
Keywords: freshwater plants, fungal endophytes, hydrolytic and oxidative enzymes, new species, phylogeny, taxonomy
Citation: Wimalasena MK, Wijayawardene NN, Bamunuarachchige TC, Zhang G-Q, Udeni Jayalal RG, Bhat DJ, Dawoud TM, de Zoysa HKS and Dai D-Q (2025) Ectophoma salviniae sp. nov., Neottiosporina mihintaleensis sp. nov. and four other endophytes associated with aquatic plants from Sri Lanka and their extracellular enzymatic potential. Front. Cell. Infect. Microbiol. 14:1475114. doi: 10.3389/fcimb.2024.1475114
Received: 02 August 2024; Accepted: 11 November 2024;
Published: 08 January 2025.
Edited by:
Sinang Hongsanan, Shenzhen University, ChinaReviewed by:
K. W. Thilini Chethana, Mae Fah Luang University, ThailandArun Kumar Dutta, Gauhati University, India
Copyright © 2025 Wimalasena, Wijayawardene, Bamunuarachchige, Zhang, Udeni Jayalal, Bhat, Dawoud, de Zoysa and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thushara C. Bamunuarachchige, dGNiYW11bnVAdGVjLnJqdC5hYy5saw==; Dong-Qin Dai, Y2ljaWRhaWRvbmdxaW5AZ21haWwuY29t
†These authors have contributed equally to this work
 Madhara K. Wimalasena
Madhara K. Wimalasena Nalin N. Wijayawardene
Nalin N. Wijayawardene Thushara C. Bamunuarachchige
Thushara C. Bamunuarachchige Gui-Qing Zhang
Gui-Qing Zhang R. G. Udeni Jayalal5
R. G. Udeni Jayalal5 Heethaka K. S. de Zoysa
Heethaka K. S. de Zoysa Dong-Qin Dai
Dong-Qin Dai