- 1Department of Dermatology and Allergology, RWTH University Hospital Aachen, Aachen, Germany
- 2Interfaculty Institute of Microbiology and Infection Medicine, University of Tübingen, Tübingen, Germany
- 3Cluster of Excellence EXC 2124 “Controlling Microbes to Fight Infections”, University of Tübingen, Tübingen, Germany
Staphylococci are commensals of human skin and mucous membranes, but some species can also cause serious infections. Host niches during both colonization and infection differ greatly and are characterized by specific environmental conditions (pH, temperature, oxygen, nutrient availability, and microbiota) that can affect gene expression and virulence of microbes. To successfully occupy extremely different habitats at different anatomical sites, Staphylococci are equipped with a variety of regulatory elements that allow specific adaptation to the changing environments. Not surprisingly, gene expression in vivo can be significantly different from the expression pattern observed in vitro. Niche specific stimuli that influence the bacterial ability to either cause infection or maintain colonization are only partially understood. Here, we describe habitat specific conditions and discuss the available literature analyzing staphylococcal gene expression, focusing on Staphylococcus aureus and S. epidermidis during colonization of the nose and skin.
Introduction
Staphylococci are commensals of human skin and mucous membranes but can also cause serious infections (Gordon and Lowy, 2008). The versatility to colonize and infect various human body sites is facilitated by a complex transcriptional regulatory network. Regulation is achieved by > 100 regulatory elements, including two component systems (TCSs), alternative sigma factors, transcription factors and small regulatory RNAs (sRNAs) (Bleul et al., 2021). Recent advances in transcriptomics and molecular analyses revealed a close link between metabolic adaptation and virulence gene expression (Prince and Wong Fok Lung, 2020; Rudra and Boyd, 2020). For Staphylococcus aureus co-regulated genes were grouped into 29 independently modulated sets of genes (i-modulons) (Poudel et al., 2020), and for many regulators, prototypic target genes are well defined based on known binding motifs (Novichkov et al., 2013). However, which signals are perceived and how they are transmitted is often less clear (Bleul et al., 2021).
To obtain a better understanding of the adaptive processes in vivo, several approaches were chosen. First, the in vivo conditions can be defined e.g. through metabolomics and the information used to establish adapted growth media to mimic in vivo conditions (Krismer et al., 2014). Second, organoids or the use of explants are useful tools to unravel host-bacterial interactions (Burian et al., 2021; Cruz et al., 2021). Third, analyses of gene expression in ex vivo samples can decipher which regulatory circuits are active and allow conclusions about the growth conditions encountered in vivo (Burian et al., 2010b; Chaves-Moreno et al., 2016). The major limitation of such analyses in the authentic human environment is the difficulty in obtaining enough RNA at high purity and data normalization. Here, we summarize the current knowledge of conditions prevailing in two important staphylococcal habitats - the nose and the skin - and describe how in vivo gene expression may be determined by these conditions.
Nasal colonization
The nose environment
Vestibulum nasi forms the main ecological niche for S. aureus (Wertheim et al., 2005). According to the traditional view, the nasal epithelium consists of basal, secretory and ciliated cells. However, single-cell RNA sequencing revealed more than 10 different cell types, some of which were highly specialized. Club and goblet cells form nasal secretions (Hewitt and Lloyd, 2021), which are mainly composed of water (95%), mucin glycoproteins (2%), salt (1%), lipids (1%) and various proteins (1%) (Kaliner et al., 1984). Nasal secretions, along with the nasal microbiome, contribute to the first layer of host defense. Mucin glycoproteins provide binding sites for interactions with microbial structures and thus contribute to the sequestration of pathogens (Fahy and Dickey, 2010). In nasal secretions, in addition to the numerous antimicrobial proteins, of which lysozyme, lactoferrin, and the secretory leukoprotease inhibitor are the most abundant, immunoglobulins (IgA, IgE and IgG), and α- and β-defensins are also present (Cole et al., 1999; Tomazic et al., 2020). However, the detectability of defensins is highly donor dependent (Cole et al., 1999; Preiano et al., 2018).
Krismer and colleagues determined the metabolites in nasal secretions (Krismer et al., 2014) and found that nutrients were present in rather low amounts compared to the amounts in plasma (Nasset et al., 1979) and sputum from cystic fibrosis patients (Palmer et al., 2007). Of the carbohydrates in nasal secretions, glucose is the major monosaccharide (35 µM - 1 mM), and urea is the most abundant organic substance (2.5 - 7.5 mM). Interestingly, while most amino acids in nasal secretions are present at an average concentration of 50 - 150 µM, methionine, glutamine, tyrosine, isoleucine, asparagine and aspartate were nondetectable. In addition, no lactate and only trace amounts of fatty acids were detected (Krismer et al., 2014). The levels of essential metals, such as iron, zinc, and manganese, are also low (Krismer et al., 2014). Sodium chloride was present in nasal secretions at physiological concentrations (~150 mM) (Vanthanouvong and Roomans, 2004), and the mean nasal pH was 6.5 (± 0.5) (Kim et al., 2013). Based on these data, a synthetic nasal medium (SNM) was composed and gene expression in SNM versus BM complex medium compared (GSE43712) (Krismer et al., 2014). Key genes were expressed in SNM in a similar way as in the human nose, indicating that SNM represents a suitable surrogate environment for in vitro simulation studies.
Bacterial adaptation to the nose environment
S. aureus and coagulase-negative staphylococci (CoNS), such as S. epidermidis, are core members of the nasal microbiome (Liu et al., 2015) and thus have evolved to cope with that specific environment. In contrast to S. epidermidis, only approximately 20% of the healthy human population is persistently colonized with S. aureus in the nose (Van Belkum et al., 2009). Whereas S. aureus usually has only one strain colonizing the host (Vandenbergh et al., 1999; Van Belkum et al., 2009), recent metagenomics studies for S. epidermidis show a large heterogeneity at the strain level within a host niche (Both et al., 2021; Severn and Horswill, 2022). Nevertheless, virulence regulators such as the agr quorum sensing system are conserved among staphylococci (Thoendel et al., 2011). Individual virulence factors such as the sphingomyelinase gene of S. epidermidis are also highly conserved, as demonstrated in a large cohort of skin isolates from healthy volunteers (Zheng et al., 2022).
Ex vivo gene expression analyses are promising approaches to gain insight into niche adaptation. There are a few studies describing gene expression during nasal colonization of S. aureus (Burian et al., 2010b; Burian et al., 2012; Song et al., 2012; Krismer et al., 2014; Chaves-Moreno et al., 2016) or S. epidermidis (Teichmann et al., 2022). In most studies, transcript analyses were performed directly on nasal swabs from persistently colonized individuals. Gene expression was measured by qRT-PCR and compared to the expression pattern of the isogenic strain(s) grown in vitro. Similar in vivo transcriptional profiles were observed for most of the genes analyzed when specimens from different volunteers or follow-up specimens from the same volunteer were compared (Burian et al., 2010b). A similar in vivo expression pattern was also detected in a cotton rat model (Burian et al., 2010a) and in a human airway epithelial coculture model (Kiedrowski et al., 2016).
In one study, meta-transcriptomics was applied to obtain a more comprehensive overview of the in vivo gene expression of S. aureus (GSE73485) (Chaves-Moreno et al., 2016). Reads were compared to data obtained for two non-isogenic reference strains (USA300 LAC or IPL32) grown in vitro. Cluster analysis revealed that all in vivo transcriptomes differed substantially from those of the in vitro-grown S. aureus strains. However, large differences were obvious between the five in vivo transcriptomes, with them sharing only >55% similarity (Chaves-Moreno et al., 2016). Based on the known large strain differences between S. aureus isolates (Lindsay, 2010; Lindsay, 2014), it is not surprising that the in vivo transcription differs from transcription of laboratory strains. Thus, comparison with the isogenic strains grown in vitro (Burian et al., 2010b) is more informative about habitat specific changes in gene expression.
Nevertheless, comparison of the analyses performed thus far revealed some common themes (Figure 1). Nasal colonization of S. aureus is clearly linked to increased expression of adhesin genes (clfB, fnbA, sdrCDE, isdA, sasF, ebpS, atlA, and eap) and wall teichoic acid (WTA) biosynthesis genes (measured by tagO (Burian et al., 2010b) and by tagA (Chaves-Moreno et al., 2016)). For S. epidermidis, genes encoding the fibrinogen binding protein SdrG and WTA (measured by tagB) were upregulated (Teichmann et al., 2022). Some of the genes that encode host defense subversion, such as staphylokinase (sak), chemotaxis inhibitory protein (chp) and protein A (spa), were also expressed in vivo (Burian et al., 2010b; Chaves-Moreno et al., 2016). Expression of the S. aureus cap operon (encoding enzymes for capsular polysaccharide synthesis) was variable between and within specimens (Burian et al., 2010b; George et al., 2015). The capBCAD operon of S. epidermidis is responsible for the production of poly-γ-glutamic acid (γ-PGA) and highly expressed during colonization (Teichmann et al., 2022). Since γ-PGA is also present in other CoNS (Kocianova et al., 2005; Watanabe et al., 2018), this, together with the observed high transcription in S. epidermidis, suggests a species-wide protective mechansims for CoNS.
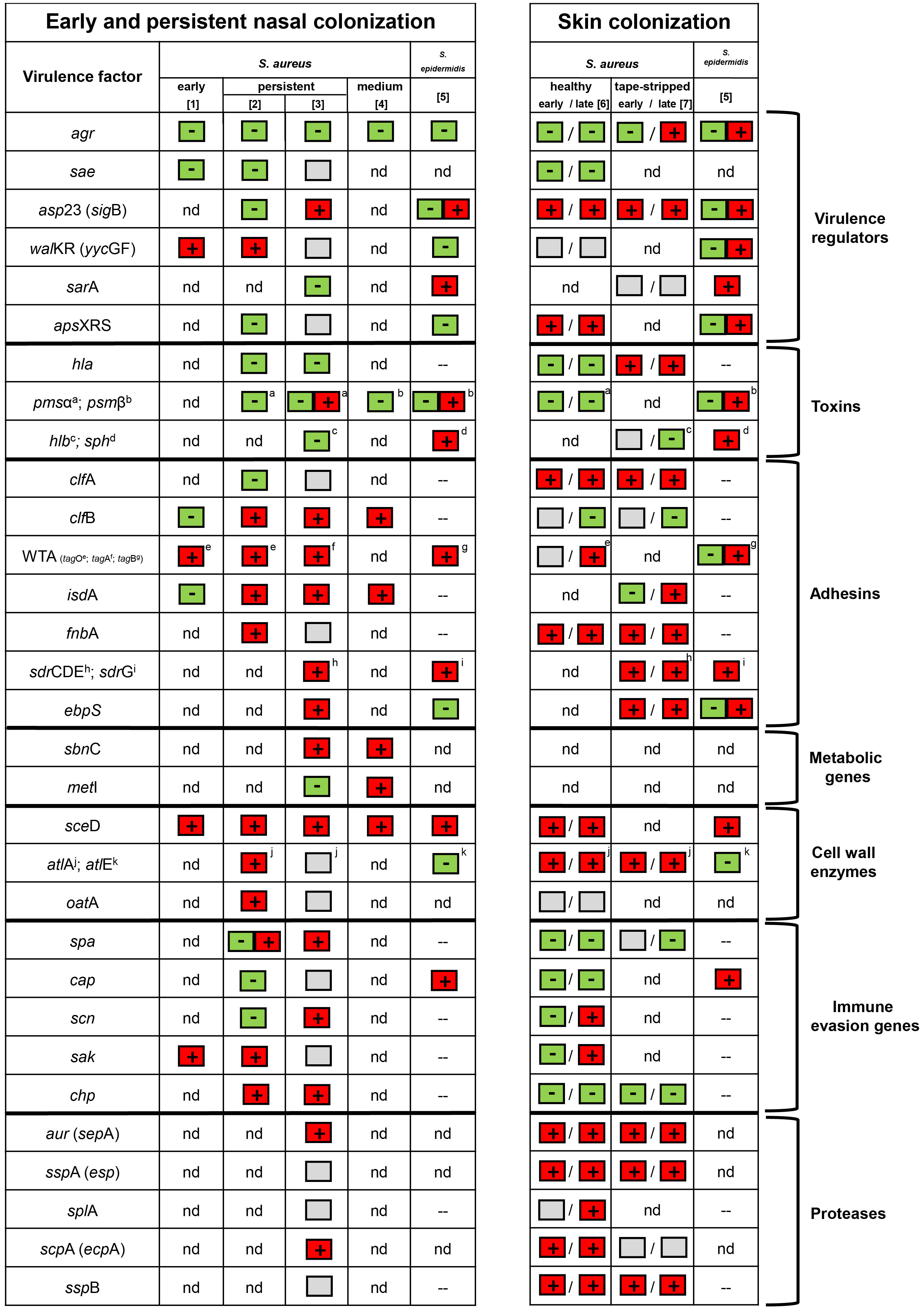
Figure 1 Transcriptional patterns in the nose and skin of S. aureus and S. epidermidis based on the following literature: 1 = (Burian et al., 2010a); 2 = (Burian et al., 2010b); 3 = (Chaves-Moreno et al., 2016); 4 = (Krismer et al., 2014); 5 = (Teichmann et al., 2022); 6 = (Burian et al., 2021); and 7 = (Cruz et al., 2021). Genes upregulated were marked with a + (box with red background), while genes downregulated were marked with a – (box with green background). The box with a gray background indicates no regulation. A box with + and – (red and green background) indicates heterogeneous transcription. nd = not determined. – = not present.
It can be assumed that bacteria encounter specific stress conditions in the nasal environment. There is a clear indication that S. aureus is iron-restricted in vivo. Iron-regulated genes, such as isdA, were found to be highly expressed in all studies (Burian et al., 2010b; Krismer et al., 2014; Chaves-Moreno et al., 2016) (Figure 1). Further indications of iron restriction are the high transcription of genes encoding enzymes for siderophore synthesis and their respective transport systems (sir and hts) (Chaves-Moreno et al., 2016).
The induction of genes protecting against reactive oxygen species (katA, ahpC) and genes forming the compatible solute glycine betaine (betA, betB) indicate that S. aureus is exposed to oxidative and osmotic stress, respectively (Chaves-Moreno et al., 2016).
The metabolic state of in vivo-grown bacteria is still not well understood. For S. epidermidis, sphingomyelinase (sph) activity provides nutrients to the bacterium by cleaving sphingomyelin into phosphocholine and ceramide (Zheng et al., 2022). This was supported by the unusual high sph expression in S. epidermidis nose and skin specimens (Teichmann et al., 2022). Interestingly, the expression of genes encoding tricarboxylic acid cycle enzymes of S. epidermidis was low in vivo. Since these enzymes are usually suppressed under nutrient-rich conditions (Somerville and Proctor, 2009), the results indicate good nutrient supply for S. epidermidis in its natural habitat (Teichmann et al., 2022). This seems to contrast with the observation that in SNM medium, the growth of S. epidermidis is inferior to that of S. aureus (Krismer et al., 2014). One can speculate that the high activity of sphingomyelinase contributes to the growth advantage of S. aureus in vivo since the substrate, sphingomyelin, is missing in SNM.
The host nasal environment activates specific metabolic pathways required for long-term colonization. For example de novo synthesis of methionine and significant upregulation of several amino acid biosynthesis genes was observed during S. aureus nose colonization (Krismer et al., 2014). A shift toward lipid and amino acid metabolism was also detected in an airway epithelial coculture model (Kiedrowski et al., 2016).
The activity of pleiotropic regulators should be informative to obtain further insights into the environmental conditions encountered in vivo. Major regulatory systems driving the expression of virulence genes, such as the agr quorum-sensing system or the virulence gene regulatory system saePQRS, were found to be inactive during nasal colonization (Burian et al., 2010a; Burian et al., 2010b; Pynnonen et al., 2011; Song et al., 2012; Teichmann et al., 2022). The inactivity of the SaePQRS system might be due to the low abundance of α-defensins, which were shown to be important ligands for the activation of the histidine kinase SaeS (Geiger et al., 2008). The Agr system might be inactive due to the low bacterial density, the inhibition by interfering staphylococcal species (Jenul and Horswill, 2019) or the presence of hemoglobin (Pynnonen et al., 2011). The inactivity of both virulence regulators indicates that S. aureus is kept in a nontoxic state during colonization.
The essential two-component system WalKR seems to be active during colonization (Burian et al., 2010a; Burian et al., 2010b). To date, the signal for WalKR activation is still not well defined but probably involves some disturbance of cell-wall metabolism (Bleul et al., 2021). The defined WalKR target gene sceD coding for a lytic transglycosylase (Dubrac et al., 2007) is the most prominent and reproducible in vivo activated gene in S. aureus (Burian et al., 2010a; Burian et al., 2010b; Krismer et al., 2014; Chaves-Moreno et al., 2016; Kiedrowski et al., 2016) and S. epidermidis (Teichmann et al., 2022). Given the clear involvement of WalKR and especially its target gene sceD (Figure 1), this could be a useful target to prevent colonization/infection. Therefore, further research is needed to decipher the exact role of sceD and its regulatory system WalKR. For S. epidermidis the accessory staphylococcal regulator A (sarA) also seems to play an important role during colonization (Teichmann et al., 2022).
Small RNAs are involved in the posttranscriptional regulation of metabolic pathways and in responses to stress and virulence (Menard et al., 2021). The expression levels of five sRNAs of S. aureus were quantified during human colonization and infection. The expression level of the Agr effector molecule RNAIII was again much lower in vivo, supporting that the system is largely kept inactive (Song et al., 2012).
One important question is whether the bacteria divide actively and at what rate of growth. Evidence, such as the high expression of cell envelope components (tagO, tarK, atlA, sceD and oatA)indicates that S. aureus is not in a dormant state. Moreover, genes expressed during the exponential growth phase in vitro are highly expressed in the human nose (Burian et al., 2010b). The expression levels of the sRNAs in vivo also resembled those obtained at the exponential phase or late exponential phase of growth in vitro (Song et al., 2012). The assumption that S. aureus is rapidly dividing during colonization is also supported by the distribution of sequencing coverage along the staphylococcal chromosome and the rate of mutational accumulation (Szafranska et al., 2019). This indicates that colonization of the human upper respiratory tract is characterized by a highly dynamic equilibrium between bacterial growth and removal.
Skin colonization
The skin environment
Human skin represents a highly variable organ that varies in temperature, pH, moisture and sebum content, creating different niches for microorganisms (Grice and Segre, 2011). The outermost layer, the stratum corneum, consists of the upper layers of corneocytes and is rich in ceramides, cholesterol, and free fatty acids. The hydrophobic and viscous sebum produced by sebaceous glands located in the dermis consists of a mixture of nonpolar lipids, such as triglycerides, wax esters, squalene, fatty acids and smaller amounts of cholesterol and diglycerides (Pappas, 2009).
The microenvironment on the skin is also influenced by sweat produced by eccrine and apocrine glands. Eccrine glands excrete ions and various proteins and peptides, some of which are also involved in innate host defense mechanisms, such as DNase I, lysozyme, and dermcidin (for review see (Wilke et al., 2007)). The molecular composition together with the secreted products of the microbiota results in a pH range of the stratum corneum between 4.1 and 5.8 (Proksch, 2018). Acidification of the skin’s surface is critical to maintaining a healthy skin environment, as antimicrobial peptides, such as dermcidin, require an acidic pH for their action (Malik et al., 2016). In inflammatory diseases, such as atopic dermatitis (AD), the skin exhibits an elevated pH value, which contributes to the inability to form a healthy skin environment (Panther and Jacob, 2015).
Bacterial adaptation to the skin environment
Healthy human skin is rarely colonized with S. aureus (Shi et al., 2016), in contrast to the skin of AD patients (Schlievert et al., 2010). Therefore, to date, there are no gene expression analyses of S. aureus colonizing healthy human skin. However, some insights were gained using skin explant models (Burian et al., 2021; Cruz et al., 2021). For S. epidermidis expression data from healthy skin are available (Teichmann et al., 2022).
Using human skin explants cultivated at the air-liquid interface for up to 8 days, we could mimic skin colonization of S. aureus. Similar to the expression profile in the human nose, we provided evidence for significant downregulation of the global virulence regulator agr and its target genes hla and psm during co-culture (Burian et al., 2021) (Figure 1). In contrast, the alternative sigma factor B (sigB) and its target genes (clfA and fnbA) as well as the antimicrobial peptide-sensing system (graRS) were strongly upregulated upon skin contact. At later time points, transcription of molecules involved in immune evasion (scn and sak) and WTA synthesis (tagO) was induced. Similar to the expression profile in the nose, enzymes involved in cell wall metabolism (sceD and atlA) were highly transcribed during co-culture. Interestingly, proteases from all three catalytic classes were strongly induced during the entire colonization process (Burian et al., 2021).
Gene expression was also analyzed using a similar human skin explant model in which the stratum corneum was “tape-stripped” to mimic barrier dysfunction. This procedure allowed invasion of S. aureus from the epidermis to the dermis (Cruz et al., 2021). Similar to analysis of un-disturbed skin explants (Burian et al., 2021) asp23, clfA, atlA and the protease genes (aur, sspA, sspB) were upregulated upon skin contact (Figure 1). Additionally, genes encoding part of the ESAT-6 secretion system (esxA, esxB, esxC, esaA, and essB), immunodominant antigens (isaA and isaB), conserved staphylococcal antigens (csa1A and csa2) and adhesion proteins (ebpS and sasF) were found activated after skin inoculation. The highly similar gene expression pattern observed in both skin explant studies indicate that deeper invasion of the strains does not per se induce major changes in gene expression. Only, hla promoter activity was shown to be enhanced inside the sweat glands and ducts but not on the skin surface (Cruz et al., 2021).
Recently, S. epidermidis gene expression from skin and nose specimens from the same patients were compared (Teichmann et al., 2022). Gene expression was mostly congruent between both sides and characterized by strong induction of adhesion and immune evasion genes (sdrG, capC, dltA and sceD), as well as sph and a putative chitinase (SE0760) (Figure 1). However, agr activity was low in the nose but readily present on the skin. A similar expression profile was also identified for SE0760, whereas sceD and the wall teichoic acid (WTA) biosynthesis gene tagB were more pronounced in the nose specimens.
Conclusion and outlook
The still limited data on gene expression during colonization of the nose or healthy skin indicate that the bacteria are actively growing, adapted to adhere to the underlying tissue and are kept in a non-toxic state by down-regulation of major virulence regulators and their target genes. However, one may assume that gene expression drastically changes once the bacteria enter deeper tissues or the blood stream. Pulia and colleagues demonstrated that gene expression was significantly different when comparing pus samples and wound swabs (Pulia et al., 2022). For example, a relative increase in the expression of toxin genes and virulence regulators (agr and sae) was observed in purulent material (Pulia et al., 2022). Higher strain toxicity is also indicated by analyses of human cutaneous abscesses (Loughman et al., 2009; Date et al., 2014). Quorum and/or defensin sensing may be major triggers for the switch towards higher toxicity. One can also assume that colonization of non-healthy skin impact gene expression. Recently, Poh and colleagues analyzed the expression of S. aureus virulence factors on lesional and non-lesional skin of AD patients. Of the genes investigated, scn (encoding staphylococcal complement inhibitor) and the protein A-encoding gene spa were the two most highly expressed genes in atopic skin (Poh et al., 2022).
Each habitat is characterized by specific conditions (pH, temperature, nutrient availability, and microbiota) and the habitat can even be subdivided into microenvironments. Thus, gene expression is controlled by a variety of different stimuli. Various tools (metabolomics, improved cell culture techniques, global transcriptome analyses) have been developed to tackle this issue. However, we are still far from knowing the major triggers acting in vivo and which regulatory circuits and i-modulons determine specific niche adaptation. Controlled switches likely determine the severity and/or chronicity of infections. More comprehensive gene analyses, sophisticated imaging and metabolomics from different infection sites are required to understand the transition from commensal to pathogenic lifestyles. Such insight can guide new anti-infective strategies to suppress bacterial growth and virulence.
Author contributions
MB, CW and AY contributed to the manuscript writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the START-Program of the Faculty of Medicine of the RWTH Aachen University and by the Deutsche Forschungsgemeinschaft (TRR 156, 246807620 and FOR2497, 289113135 to AY and SPP2225, 446507619 to CW and infrastructural funding of the Cluster of Excellence EXC 2124 “Controlling Microbes to Fight Infections).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bleul, L., Francois, P., Wolz, C. (2021). Two-component systems of S. aureus: Signaling and sensing mechanisms. Genes (Basel) 13, 34. doi: 10.3390/genes13010034
Both, A., Huang, J., Qi, M., Lausmann, C., Weisselberg, S., Buttner, H., et al. (2021). Distinct clonal lineages and within-host diversification shape invasive Staphylococcus epidermidis populations. PloS Pathog. 17, e1009304. doi: 10.1371/journal.ppat.1009304
Burian, M., Grumann, D., Holtfreter, S., Wolz, C., Goerke, C., Broker, B. M. (2012). Expression of staphylococcal superantigens during nasal colonization is not sufficient to induce a systemic neutralizing antibody response in humans. Eur. J. Clin. Microbiol. Infect. Dis. 31, 251–256. doi: 10.1007/s10096-011-1302-2
Burian, M., Plange, J., Schmitt, L., Kaschke, A., Marquardt, Y., Huth, L., et al. (2021). Adaptation of Staphylococcus aureus to the human skin environment identified using an ex vivo tissue model. Front. Microbiol. 12, 728989. doi: 10.3389/fmicb.2021.728989
Burian, M., Rautenberg, M., Kohler, T., Fritz, M., Krismer, B., Unger, C., et al. (2010a). Temporal expression of adhesion factors and activity of global regulators during establishment of Staphylococcus aureus nasal colonization. J. Infect. Dis. 201, 1414–1421. doi: 10.1086/651619
Burian, M., Wolz, C., Goerke, C. (2010b). Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PloS One 5, e10040. doi: 10.1371/journal.pone.0010040
Chaves-Moreno, D., Wos-Oxley, M. L., Jauregui, R., Medina, E., Oxley, A. P., Pieper, D. H. (2016). Exploring the transcriptome of Staphylococcus aureus in its natural niche. Sci. Rep. 6, 33174. doi: 10.1038/srep33174
Cole, A. M., Dewan, P., Ganz, T. (1999). Innate antimicrobial activity of nasal secretions. Infect. Immun. 67, 3267–3275. doi: 10.1128/IAI.67.7.3267-3275.1999
Cruz, A. R., Van Strijp, J. A. G., Bagnoli, F., Manetti, A. G. O. (2021). Virulence gene expression of Staphylococcus aureus in human skin. Front. Microbiol. 12, 692023. doi: 10.3389/fmicb.2021.692023
Date, S. V., Modrusan, Z., Lawrence, M., Morisaki, J. H., Toy, K., Shah, I. M., et al. (2014). Global gene expression of methicillin-resistant Staphylococcus aureus USA300 during human and mouse infection. J. Infect. Dis. 209, 1542–1550. doi: 10.1093/infdis/jit668
Dubrac, S., Boneca, I. G., Poupel, O., Msadek, T. (2007). New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269. doi: 10.1128/JB.00645-07
Fahy, J. V., Dickey, B. F. (2010). Airway mucus function and dysfunction. N Engl. J. Med. 363, 2233–2247. doi: 10.1056/NEJMra0910061
Geiger, T., Goerke, C., Mainiero, M., Kraus, D., Wolz, C. (2008). The virulence regulator sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190, 3419–3428. doi: 10.1128/JB.01927-07
George, S. E., Nguyen, T., Geiger, T., Weidenmaier, C., Lee, J. C., Liese, J., et al. (2015). Phenotypic heterogeneity and temporal expression of the capsular polysaccharide in Staphylococcus aureus. Mol. Microbiol. 98, 1073–1088. doi: 10.1111/mmi.13174
Gordon, R. J., Lowy, F. D. (2008). Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin. Infect. Dis. 46 Suppl 5, S350–S359. doi: 10.1086/533591
Grice, E. A., Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. doi: 10.1038/nrmicro2537
Hewitt, R. J., Lloyd, C. M. (2021). Regulation of immune responses by the airway epithelial cell landscape. Nat. Rev. Immunol. 21, 347–362. doi: 10.1038/s41577-020-00477-9
Jenul, C., Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Microbiol. Spectr. 7, 2. doi: 10.1128/microbiolspec.GPP3-0031-2018
Kaliner, M., Marom, Z., Patow, C., Shelhamer, J. (1984). Human respiratory mucus. J. Allergy Clin. Immunol. 73, 318–323. doi: 10.1016/0091-6749(84)90403-2
Kiedrowski, M. R., Paharik, A. E., Ackermann, L. W., Shelton, A. U., Singh, S. B., Starner, T. D., et al. (2016). Development of an in vitro colonization model to investigate Staphylococcus aureus interactions with airway epithelia. Cell Microbiol. 18, 720–732. doi: 10.1111/cmi.12543
Kim, B. G., Kim, J. H., Kim, S. W., Kim, S. W., Jin, K. S., Cho, J. H., et al. (2013). Nasal pH in patients with chronic rhinosinusitis before and after endoscopic sinus surgery. Am. J. Otolaryngol. 34, 505–507. doi: 10.1016/j.amjoto.2013.04.015
Kocianova, S., Vuong, C., Yao, Y., Voyich, J. M., Fischer, E. R., Deleo, F. R., et al. (2005). Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest. 115, 688–694. doi: 10.1172/JCI200523523
Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PloS Pathog. 10, e1003862. doi: 10.1371/journal.ppat.1003862
Lindsay, J. A. (2010). Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300, 98–103. doi: 10.1016/j.ijmm.2009.08.013
Lindsay, J. A. (2014). Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 304, 103–109. doi: 10.1016/j.ijmm.2013.11.010
Liu, C. M., Price, L. B., Hungate, B. A., Abraham, A. G., Larsen, L. A., Christensen, K., et al. (2015). Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 1, e1400216. doi: 10.1126/sciadv.1400216
Loughman, J. A., Fritz, S. A., Storch, G. A., Hunstad, D. A. (2009). Virulence gene expression in human community-acquired Staphylococcus aureus infection. J. Infect. Dis. 199, 294–301. doi: 10.1086/595982
Malik, E., Dennison, S. R., Harris, F., Phoenix, D. A. (2016). pH dependent antimicrobial peptides and proteins, their mechanisms of action and potential as therapeutic agents. Pharm. (Basel) 9 (4), 67. doi: 10.3390/ph9040067
Menard, G., Rouillon, A., Ghukasyan, G., Emily, M., Felden, B., Donnio, P. Y. (2021). Galleria mellonella larvae as an infection model to investigate sRNA-mediated pathogenesis in Staphylococcus aureus. Front. Cell Infect. Microbiol. 11, 631710. doi: 10.3389/fcimb.2021.631710
Nasset, E. S., Heald, F. P., Calloway, D. H., Margen, S., Schneeman, P. (1979). Amino acids in human blood plasma after single meals of meat, oil, sucrose and whiskey. J. Nutr. 109, 621–630. doi: 10.1093/jn/109.4.621
Novichkov, P. S., Kazakov, A. E., Ravcheev, D. A., Leyn, S. A., Kovaleva, G. Y., Sutormin, R. A., et al. (2013). RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14, 745. doi: 10.1186/1471-2164-14-745
Palmer, K. L., Aye, L. M., Whiteley, M. (2007). Nutritional cues control pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189, 8079–8087. doi: 10.1128/JB.01138-07
Panther, D. J., Jacob, S. E. (2015). The importance of acidification in atopic eczema: An underexplored avenue for treatment. J. Clin. Med. 4, 970–978. doi: 10.3390/jcm4050970
Poh, S. E., Koh, W. L. C., Lim, S. Y. D., Wang, E. C. E., Yew, Y. W., Common, J. E. A., et al. (2022). Expression of Staphylococcus aureus virulence factors in atopic dermatitis. JID Innov. 2, 100130. doi: 10.1016/j.xjidi.2022.100130
Poudel, S., Tsunemoto, H., Seif, Y., Sastry, A. V., Szubin, R., Xu, S., et al. (2020). Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc. Natl. Acad. Sci. U.S.A. 117, 17228–17239. doi: 10.1073/pnas.2008413117
Preiano, M., Maggisano, G., Murfuni, M. S., Villella, C., Colica, C., Fregola, A., et al. (2018). Rapid detection and identification of antimicrobial peptide fingerprints of nasal fluid by mesoporous silica particles and MALDI-TOF/TOF mass spectrometry: From the analytical approach to the diagnostic applicability in precision medicine. Int. J. Mol. Sci. 19 (12), 4005. doi: 10.3390/ijms19124005
Prince, A., Wong Fok Lung, T. (2020). Consequences of metabolic interactions during Staphylococcus aureus infection. Toxins (Basel) 12 (9), 581. doi: 10.3390/toxins12090581
Proksch, E. (2018). pH in nature, humans and skin. J. Dermatol. 45, 1044–1052. doi: 10.1111/1346-8138.14489
Pulia, M. S., Anderson, J., Ye, Z., Elsayed, N. S., Le, T., Patitucci, J., et al. (2022). Expression of staphylococcal virulence genes In situ in human skin and soft tissue infections. Antibiotics (Basel) 11 (14), 527. doi: 10.3390/antibiotics11040527
Pynnonen, M., Stephenson, R. E., Schwartz, K., Hernandez, M., Boles, B. R. (2011). Hemoglobin promotes Staphylococcus aureus nasal colonization. PloS Pathog. 7, e1002104. doi: 10.1371/journal.ppat.1002104
Rudra, P., Boyd, J. M. (2020). Metabolic control of virulence factor production in Staphylococcus aureus. Curr. Opin. Microbiol. 55, 81–87. doi: 10.1016/j.mib.2020.03.004
Schlievert, P. M., Strandberg, K. L., Lin, Y. C., Peterson, M. L., Leung, D. Y. (2010). Secreted virulence factor comparison between methicillin-resistant and methicillin-sensitive Staphylococcus aureus, and its relevance to atopic dermatitis. J. Allergy Clin. Immunol. 125, 39–49. doi: 10.1016/j.jaci.2009.10.039
Severn, M. M., Horswill, A. R. (2022). Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat. Rev. Microbiol. doi: 10.1038/s41579-022-00780-3
Shi, B., Bangayan, N. J., Curd, E., Taylor, P. A., Gallo, R. L., Leung, D. Y. M., et al. (2016). The skin microbiome is different in pediatric versus adult atopic dermatitis. J. Allergy Clin. Immunol. 138, 1233–1236. doi: 10.1016/j.jaci.2016.04.053
Somerville, G. A., Proctor, R. A. (2009). At The crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248. doi: 10.1128/MMBR.00005-09
Song, J., Lays, C., Vandenesch, F., Benito, Y., Bes, M., Chu, Y., et al. (2012). The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PloS One 7, e37294. doi: 10.1371/journal.pone.0037294
Szafranska, A. K., Junker, V., Steglich, M., Nubel, U. (2019). Rapid cell division of Staphylococcus aureus during colonization of the human nose. BMC Genomics 20, 229. doi: 10.1186/s12864-019-5604-6
Teichmann, P., Both, A., Wolz, C., Hornef, M. W., Rohde, H., Yazdi, A. S., et al. (2022). The Staphylococcus epidermidis transcriptional profile during carriage. Front. Microbiol. 13, 896311. doi: 10.3389/fmicb.2022.896311
Thoendel, M., Kavanaugh, J. S., Flack, C. E., Horswill, A. R. (2011). Peptide signaling in the staphylococci. Chem. Rev. 111, 117–151. doi: 10.1021/cr100370n
Tomazic, P. V., Darnhofer, B., Birner-Gruenberger, R. (2020). Nasal mucus proteome and its involvement in allergic rhinitis. Expert Rev. Proteomics 17, 191–199. doi: 10.1080/14789450.2020.1748502
Van Belkum, A., Verkaik, N. J., De Vogel, C. P., Boelens, H. A., Verveer, J., Nouwen, J. L., et al. (2009). Reclassification of Staphylococcus aureus nasal carriage types. J. Infect. Dis. 199, 1820–1826. doi: 10.1086/599119
Vandenbergh, M. F., Yzerman, E. P., Van Belkum, A., Boelens, H. A., Sijmons, M., Verbrugh, H. A. (1999). Follow-up of Staphylococcus aureus nasal carriage after 8 years: redefining the persistent carrier state. J. Clin. Microbiol. 37, 3133–3140. doi: 10.1128/JCM.37.10.3133-3140.1999
Vanthanouvong, V., Roomans, G. M. (2004). Methods for determining the composition of nasal fluid by X-ray microanalysis. Microsc. Res. Tech. 63, 122–128. doi: 10.1002/jemt.20020
Watanabe, S., Aiba, Y., Tan, X. E., Li, F. Y., Boonsiri, T., Thitiananpakorn, K., et al. (2018). Complete genome sequencing of three human clinical isolates of staphylococcus caprae reveals virulence factors similar to those of S. epidermidis and S. capitis. BMC Genomics 19, 810. doi: 10.1186/s12864-018-5185-9
Wertheim, H. F., Melles, D. C., Vos, M. C., Van Leeuwen, W., Van Belkum, A., Verbrugh, H. A., et al. (2005). The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762. doi: 10.1016/S1473-3099(05)70295-4
Wilke, K., Martin, A., Terstegen, L., Biel, S. S. (2007). A short history of sweat gland biology. Int. J. Cosmet. Sci. 29, 169–179. doi: 10.1111/j.1467-2494.2007.00387.x
Keywords: bacterial adaptation, gene expression, in vivo, global regulators, nasal colonization, skin colonization, virulence factors, host pathogen interaction
Citation: Burian M, Wolz C and Yazdi AS (2022) Transcriptional adaptation of staphylococci during colonization of the authentic human environment: An overview of transcriptomic changes and their relationship to physiological conditions. Front. Cell. Infect. Microbiol. 12:1062329. doi: 10.3389/fcimb.2022.1062329
Received: 05 October 2022; Accepted: 02 November 2022;
Published: 17 November 2022.
Edited by:
Nora Lía Padola, National University of Central Buenos Aires, ArgentinaReviewed by:
Kenneth L. Brockman, Medical College of Wisconsin, United StatesHolger Brüggemann, Aarhus University, Denmark
Copyright © 2022 Burian, Wolz and Yazdi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marc Burian, bWJ1cmlhbkB1a2FhY2hlbi5kZQ==
†These authors have contributed equally to this work and share senior authorship
 Marc Burian
Marc Burian Christiane Wolz
Christiane Wolz Amir S. Yazdi
Amir S. Yazdi