
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 February 2021
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.610781
 Hailong Dong1
Hailong Dong1 Bingxian Liu2
Bingxian Liu2 Aoyun Li3*
Aoyun Li3* Mudassar Iqbal4
Mudassar Iqbal4 Khalid Mehmood4
Khalid Mehmood4 Tariq Jamil5
Tariq Jamil5 Yung-Fu Chang6
Yung-Fu Chang6 Hui Zhang2*
Hui Zhang2* Qingxia Wu1*
Qingxia Wu1*Domestic yaks (Bos grunniens) are indigenous to the Tibetan Plateau and display a high diarrhea rate due to poor habitat and husbandry conditions. Lactobacillus has been shown to exert beneficial effects as antimicrobial, growth promotion, and gut microbiota in humans and/or murine models, but the relevant data regarding Lactobacillus isolated from yaks was unavailable. Therefore, this study aimed to investigate the effects of Lactobacillus from yaks on the intestinal microbial community in a mouse model and determine whether Lactobacillus supplementation contributed in alleviating diarrhea by modulating gut microbiota. A total of 12 ileac samples from four groups were collected for 16S rRNA gene amplicon sequencing of V3-V4 region. Results revealed that although Lactobacillus supplementation did not change the diversity of gut microbiota in mice, the proportion of some intestinal microbiota significantly changed. Specifically, the proportion of Lactobacillus and Sphingomonas in the Lactobacillus treated-group (L-group) were increased as compared to control group (C-group), whereas Pantoea, Cutibacterium, Glutamicibacter, Turicibacter, Globicatella, Microbacterium, Facklamia, unidentified_Corynebacteriaceae, Brachybacterium, and Staphylococcus were significantly decreased in the L-group. In contrast, Escherichia coli (E. coli) infection significantly decreased the proportion of beneficial bacteria such as Globicatella, Acinetobacter, Aerococcus, and Comamonas, while loads of pathogenic bacteria significantly increased including Roseburia and Megasphaera. Interestingly, Lactobacillus administration could ameliorate the microbial community structure of E. coli-induced diarrheal mice by reducing the relative abundance of pathogenic bacteria such as Paenibacillus, Aerococcus, Comamonas, Acinetobacter, Corynebacterium, Facklamia, and Globicatella. Results in this study revealed that Lactobacillus supplementation not only improved the gut microbiota but also alleviated diarrhea in mice, which may be mediated by modulating the composition and function of gut microbiota. Moreover, this study is expected to provide a new theoretical basis for the establishment of a preventive and treatment system for diarrhea in yaks.
Animal gut microbiota is one of the largest and most complicated existing micro-ecosystems that provides an important barrier to bacterial infections (Lynch and Pedersen, 2016; Li et al., 2020; Liu et al., 2020b). Additionally, it helps in providing mucosal immunity, material metabolism, and nutrient absorption and regulation (Wu and Wu, 2012; Yue et al., 2020). Generally, ongoing competition and interaction of microorganisms may gradually change in microbial community structure from simple to a complicated and eventually a dynamic and balanced ecosystem (Jami et al., 2013; Zhao et al., 2015). This community’s consistency is a precondition for maintaining normal physiological functions (Li et al., 2018a; Ritz et al., 2020). Previous research has shown that constipation, colitis, diabetes, and obesity may be related to alternation in intestinal flora (Cani et al., 2008; de La Serre et al., 2010; Kootte et al., 2012). Recent studies on gut microbiota have provided evidence that dysbacteriosis may be one of the reasons of diarrhea (Han et al., 2017; Yue et al., 2019).
Domestic yak (Bos grunniens) is an indigenous breed of the high-elevation hypoxic environment in the Tibetan plateau of China (3000 m above sea level or higher), spanning Mongolia and Siberia (Li K. et al., 2014; Li et al., 2018a). This biome is characterized by plants and animals adapted to cold and dry adverse environments (Qiu et al., 2012). Statistics indicate that approximately 90% of the word’s yaks reside in western China, where this animal is an important source of milk, meat, and draft power (Li et al., 2017; Li et al., 2019a). Any infectious disease in these yaks may result in substantial economic losses to the herdsmen relying on them. However, due to a lack of corresponding supervision and weak awareness of environmental issues, yaks (particularly juveniles) in Tibet typically display higher incidence of bacterial diarrhea (Gao et al., 2013; Han et al., 2017).
Diarrhea poses a significant threat to animal productivity in many countries (Pepin et al., 2004; Heuer et al., 2007). Statistical analyses indicate that yaks, especially juveniles, display a higher prevalence of diarrhea due to a lack of corresponding protocols and practices designed to sustain animal health (Pepin et al., 2004; Heuer et al., 2007). The aggressive use of antimicrobials as therapeutic agents has resulted in increased drug resistance and intestinal microbial imbalance (van den Bogaard and Stobberingh, 2000; Zhang et al., 2017). The importance of probiotics including Lactobacillus johnsonii, Bacillus subtilis, and Pediococcus acidilactici have been proverbially acknowledged because of their effects as potential antibacterial, immunity, and production enhancing performances (Li et al., 2018; Wang et al., 2018a; Khan, 2019; Li et al., 2019b). Moreover, Lactobacillus can alleviate bacterial diarrhea by competing with pathogenic bacteria for adhesion sites and nutrients, producing antimicrobial peptides, and improving gut microbiota (Sadowska et al., 2010; Huang et al., 2020; Wang et al., 2020). The probiotic effect of Lactobacillus has been understood and applied in chicken and other domestic animals. In yaks, probiotic applications are not well studied, so this species is a relatively new subject for probiotic research (Dudik et al., 2020: Sobanbua et al., 2020). Given the importance of yak husbandry in the Tibetan plateau, such research might be a worthy endeavor. Taking advantage of this gap, we investigated the effects of Lactobacillus (isolated from yaks) systematically on mice’s gut microbiota. Additionally, we also established diarrheal model using E. coli to identify Lactobacillus administrative effect on the intestinal microbiota in diarrheal mice.
Twenty-five-day-old healthy Kunming mice (n=40, initial weight 30 ± 3 g) were obtained from an experimental animal center, South China Agricultural University (Guangzhou, China). The study was permitted by the ethics committee of Tibet Agriculture & Animal Husbandry University. The mice used in this study were self-propagated and showed a higher degree of genetic uniformity. The selected mice were randomly divided into four groups, each comprising 10 mice (n=10) viz. control group (C-group), Lactobacillus-treated group (L-group), E. coli-induced group (E-group), and prevention group (EL-group). The mice were raised in plastic cages for 14 days under a recommended standard illumination time (12 h/12 h light/dark cycle), breeding temperature (33°C~35°C), and humidity (53%–57%). Furthermore, sufficient water and feed were provided ad libitum for all groups throughout the entire experiment. The E group was provided with same diet as control group but with the addition of E. coli at 1 × 109 CFU/day on day 14 post-hatch to induce diarrhea. The L and EL group were treated with Lactobacillus at 1 × 109 CFU/day from day 1 to day 14, but mice in the EL group were compulsively supplemented with E. coli at 1 × 109 CFU/day on day 14. Mice in the C group were provided with the same volumes of saline as the L group to minimize stress response. Moreover, the overall performance of mice was recorded three times a day and death, diarrhea, dullness, and tiredness were considered abnormal. Three mice from each group were euthanized by injecting pentobarbital (25 mg/kg). Subsequently, the intestines were removed from the abdominal cavity, and the mesentery was stripped using a sterilized surgical knife. The intestines including duodenal, ileum, jejunal, and cecum were knotted using cotton ropes to minimize the potential cross-contamination among different intestine samples. The contents in the ileum were immediately collected, snap-frozen using liquid nitrogen, and finally stored at -80°C until further study.
Bacterial DNA from all samples was extracted by using QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) as per manufacturer’s guidelines. The quality of DNA was evaluated by 0.8% (w/v) agarose gel electrophoresis. A Nanodrop™ Spectrophotometer (Thermo Scientific, Massachusetts, USA) was used to quantify the DNA.
Universal V3/V4 16S rRNA gene primers (338F: ACTCCTACGGGAGGCAGCA and 806R: GGACTACHVGGGTWTCTAAT) were used along with the barcode sequences for amplification of the conserved regions of the bacteria followed by a 2% agarose gel electrophoresis procedure to evaluate the quality of the polymerase chain reaction (PCR) products. The PCR product was then purified and recycled by using AxyPrep DNA Gel Extraction Kit (Axygen, Corning, New York, USA). The fluorescent quantitation of PCR-recycled products was conducted on FLx800™ Multi-Detection Microplate Reader (BioTek Instruments, Inc., Winooski, Vermont, USA) by using Quant-iT™ PicoGreen™ dsDNA Assay Kit (Invitrogen, Carlsbad, California, USA). The sequencing library was prepared by using TruSeq Nano DNA Low Throughput Library Prep Kit (Illumina, Inc., San Diego, California, USA) following the manufacturer’s protocol. The amplified products were repaired by End Repair Mix. Simultaneously, a magnetic bead screening procedure was used for removing the self-connecting fragments of the linker, and the sequencing library was purified. The PCR amplification was performed, and the library enrichment was performed by AMPure XP Beads (Beckman Coulter Inc., Brea, California, USA). The final fragment-selection and purification of the library were conducted on 2% agarose gel electrophoresis.
The libraries’ quality was accessed by using Agilent High Sensitivity DNA Kit on Agilent Bioanalyzer (Agilent Technologies, Inc., Santa Clara, California, USA) prior to the sequencing process. Moreover, the libraries having only one single peak without a linker was selected. The libraries were quantified by using Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, California, USA) on the QuantiFluor® RNA System (Promega Corporation, Madison, Wisconsin, USA), with a concentration >2 nM. The qualified libraries were gradient diluted and mixed in a proportion to the required amount of sequencing. Finally, the MiSeq Reagent Kit V3 (600 cycles) was used to perform the 2×300 bp paired-end sequencing on the MiSeq sequencing system (Illumina, Inc., San Diego, California, USA).
QIIME software (Qiime1.9.1) was used to screen and analyze the 16S rRNA preliminary data quality. The interrogative and short sequences (<200 bp) were removed by using QIIME software. The obtained sequences were clustered and operational taxonomic unit (OTU) were partitioned at ≥97% sequence similarity by program VSEARCH (1.9.6). The Ribosomal Database Program (RDP) classifier was used to classify the representative sequences of each OTU at confidence threshold of 0.8. The MUSCLE software was used for phylogenetic analysis and multiple sequence alignments of each OTU. The multiple alpha diversity indices including Shannon, Simpson, Chao1, and Good’s coverage were calculated to evaluate the alpha diversity. Moreover, the sparse curves were used for assessing the sequencing depth of each sample prior to the evaluation of alpha and beta diversity. The beta diversity was also calculated to assess the similarity of community structure in the samples. GraphPad Prism (version 6.0c) and R (v3.0.3) software were used to perform the statistical analysis. In addition, the criterion of significance was conducted at p-values <0.05. The values were expressed as means ± standard deviation (SD).
Clinical observation results showed that the mice in the C and L group had a normal feed intake and displayed active behavior. However, mice in the E group showed dullness, messy hair, watery feces, and pasting. Conversely, mice in the EL group and the control group were in good mental state and without diarrhea symptoms.
In the microbiome analysis, a total of 274,247, 265,341, 263,510, and 253,825 original sequences were acquired from C, L, E, and EL-group, respectively (Table 1). After eliminating the unqualified data, a total number of 961,904 high-quality reads were achieved from all the samples, with an average of 80,158 (ranging from 75,598 to 90,528) reads per sample. Following taxonomic assignment, a total of 2,668 OTUs (C=709, L=691, E=633, EL=635) were identified on the basis of 97% nucleotide-sequence similarity and 139 OTUs have shared all the samples, accounting for approximately 5.21% of the total OTUs (Figures 1A–E). Moreover, the number of unique OTUs in the C, L, E, and EL group was 87, 122, 67, and 68, respectively and 16 core OTUs were recognized in all the samples (Figure 1F). Both rarefaction and species accumulation curves for all samples tend to be stable. The number of qualified sequences reached over 10,000 and 50,000, respectively, suggesting that sequencing’s depth and quantity met the demands for sequencing and analysis (Figures 1G, H). Furthermore, the rank abundance curve is wide and falling relaxedly, showing excellent abundance and evenness (Figure 1I).
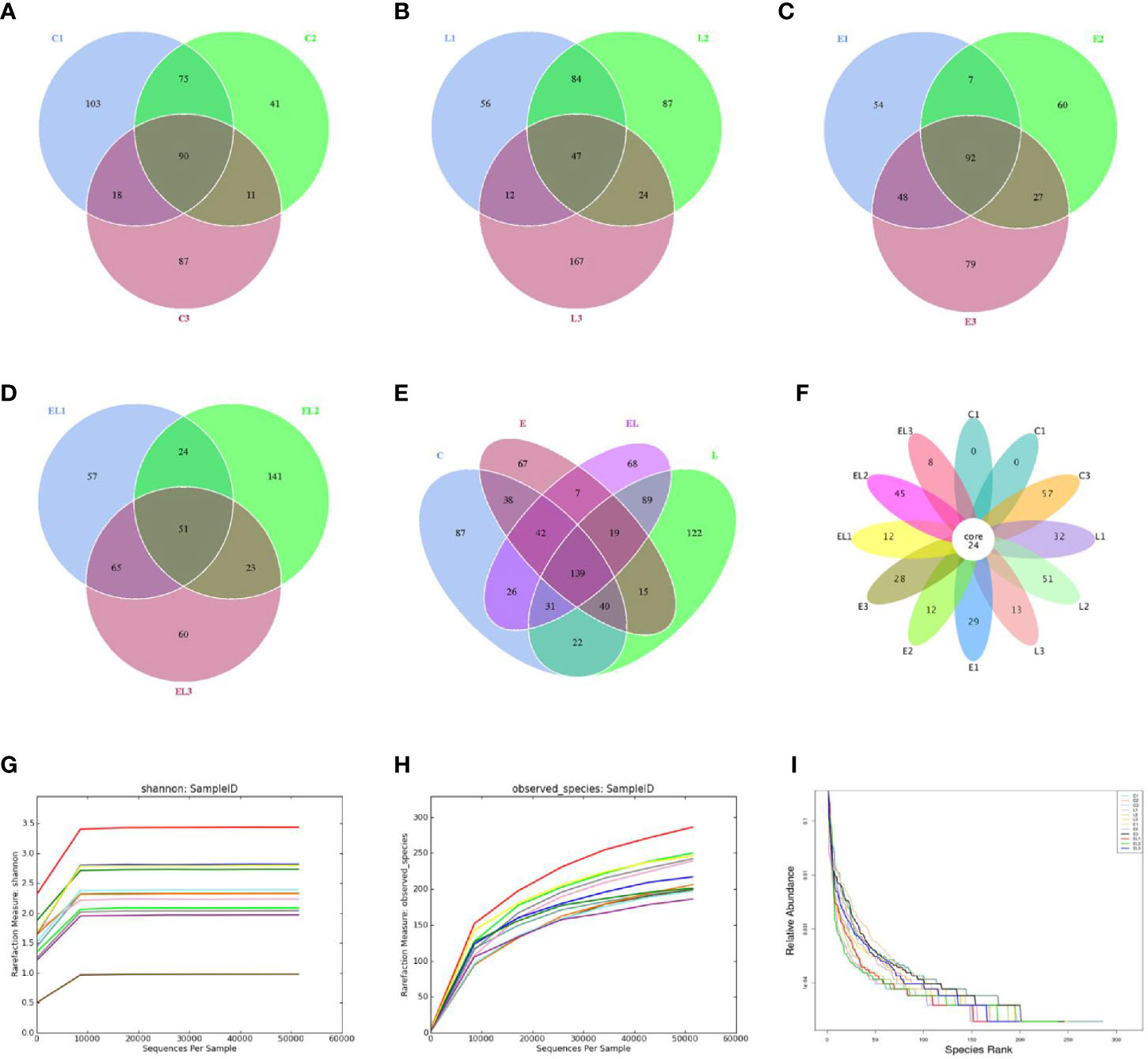
Figure 1 Venn diagrams and sample feasibility analysis. Venn diagrams for bacterial OTUs compositions in C (A), L (B), E (C) and EL (D) groups. (E) Venn diagram for unique and shared bacterial OTUs in four groups. (F) Venn diagrams for core OTUs compositions. The rarefaction (G) and species accumulation curve (H) and rank abundance curve (I) were used to assess the adequacy, evenness and richness of sequencing of each sample. Each curve with a different color shown in the figures indicates a sample.
To assess the differences in intestinal microbial community diversity among the four groups, the qualified sequences obtained in the sequencing were aligned to estimate alpha and beta indices. The alpha diversity of gut microbiota can be reflected by community abundance (Chao1 and ACE), diversity index (Simpson), and sequencing depth (Good’s coverage). Good’s coverage estimates in all the samples were approximately 100%, indicating excellent coverage (Figure 2A). The control mice showed the highest Chao1 and ACE indexes as compared to other groups, whereas the Chao1 and ACE indexes in mice infected with E. coli were the lowest (Figures 2B, C). The average of Chao1 and ACE indices in the L-group ranged from 218.71 to 277.02 and 226.022 to 289.15, respectively, while Simpson index ranged from 0.22 to 0.62. The analysis of alpha diversity indicated no statistically significant differences in the Chao1, ACE, and Simpson between the C and L groups, which indicated that Lactobacillus administration had no effect on the diversity and richness of the gut microbiota of mice (Figure 2D). However, intergroup analysis of alpha diversity intuitively indicated that gut microbiota’s richness and diversity in EL-group mice were higher than those in E-group, indicating that supplementing with Lactobacillus alleviated the gut microbiota imbalance of mice induced by E. coli. The beta diversity analysis showed that samples in the C, L, E, and EL groups were clustered closely, indicating that gut microbiota in the four groups was not different (Figure 3).
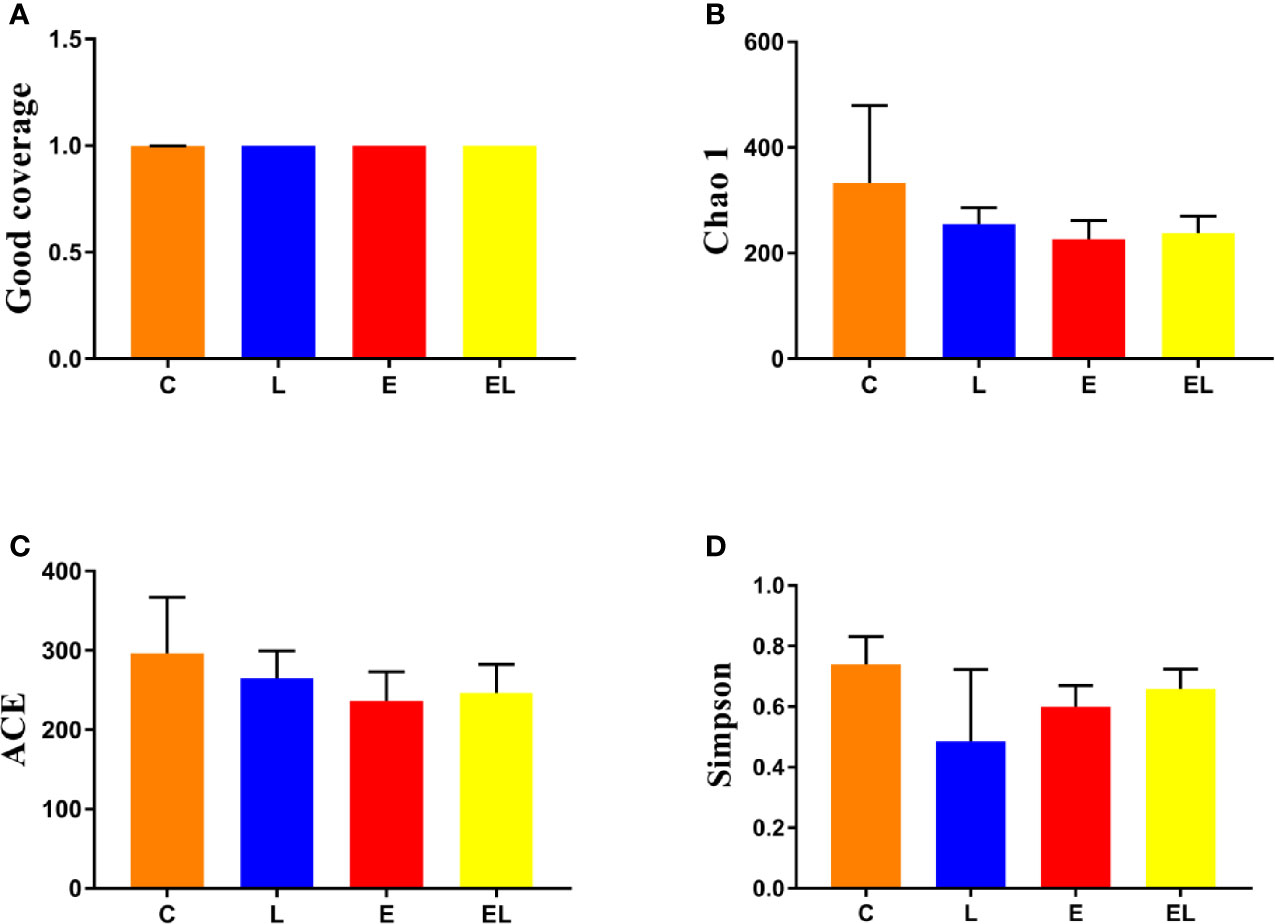
Figure 2 Comparison of alpha diversity of mice gut microbiota in different groups. Four indices such as Good's coverage (A), Chao1 (B), ACE (C), and Simpson (D) were used to assess the alpha diversity of gut microbiota. The data used in this study were expressed as the mean ± SD.
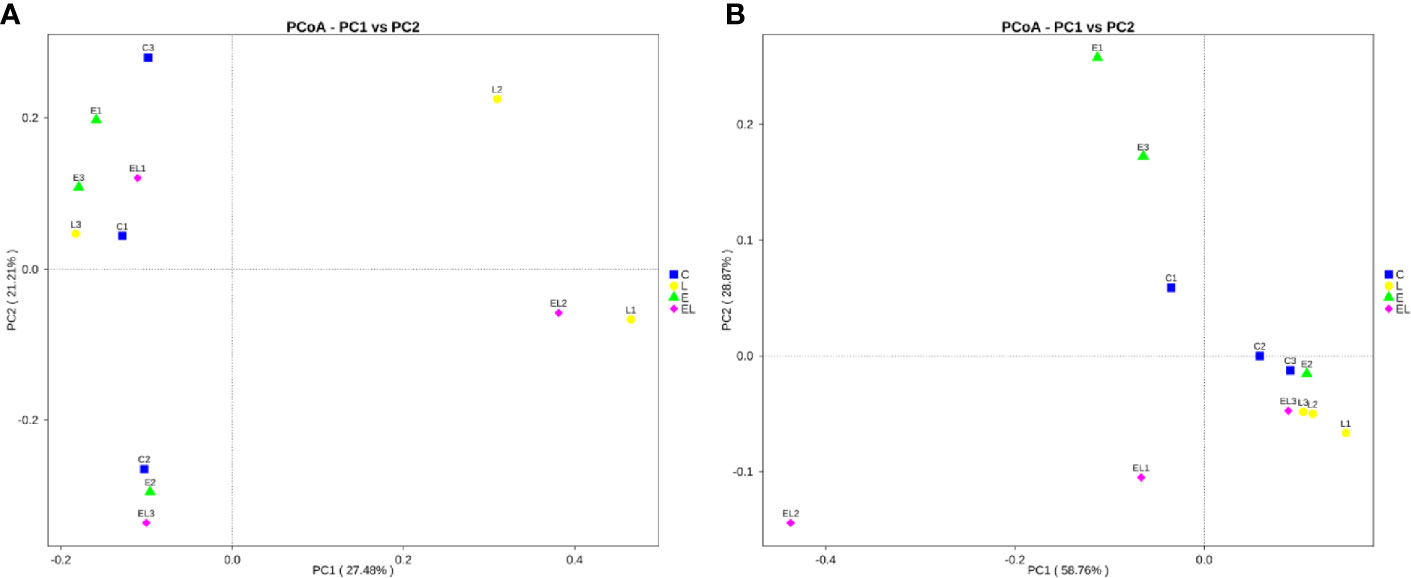
Figure 3 Principal coordinate (PCoA) analysis of gut microbiota in different groups. (A, B) represent PCoA map based on unweighted and weighted uniFrac distance, respectively. Each colored point indicates one sample and the difference in the different groups can be evaluate by the distance between the points.
The proportion of dominant phyla and genera were assessed by microbial taxa assignment in C, L, E, and EL groups (Figure 4). According to the phylum assignment result, phyla Firmicutes (91.08%, 97.82%, and 91.58%) and Actinobacteria (7.64%, 1.04%, and 5.51%) were the most preponderant bacteria in the mice of C, L, and E group, which accounted for approximately 97% of the taxonomic groups identified (Figures 4A, B). Remarkably, the predominant phylum in the EL-group was Firmicutes (72.01%), whereas phylum Proteobacteria was subsidiary (26.69%), slightly different from the other groups. Other phyla such as Fusobacteria, Acidobacteria, Chloroflexi, unidentified_Bacteria, and Tenericutes were represented with a lower abundance. At the genus level, Lactobacillus (68.30% and 91.98%) was the most predominant bacterium in the mice of C-group and L-group followed by the Staphylococcus (17.92% and 4.07%), which together made up 85% and 95% of the overall bacterial composition, respectively (Figures 4C, D). Moreover, Nosocomiicoccus (43.94%) and Lactobacillus (34.97%) were the most prevalent bacteria in the E-group, whereas Lactobacillus (63.23%) and Ralstonia (24.47%) were observed to be predominant in the EL-group. The relative richness of these bacteria was also displayed by a heat map produced by clustering analysis (Figure 5).
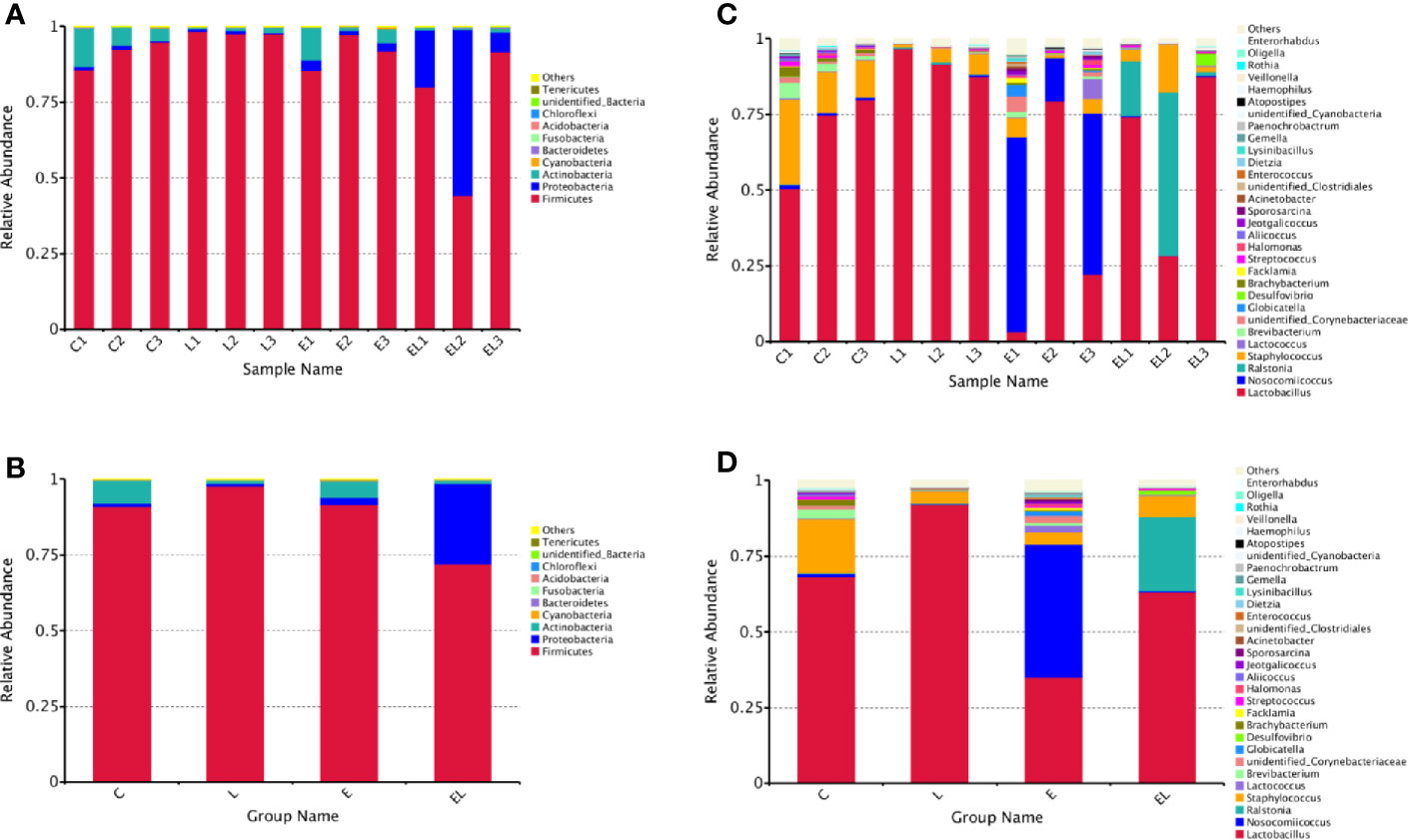
Figure 4 Relative abundance of the most preponderant (top 10 and 30) gut microbial taxa at phylum (top 10) and genus (top 30) levels for bacteria among four groups. (A, C) Relative abundance of gut microbiota in each sample at the phylum and genus levels. (B, D) Relative abundance of gut microbiota on the basis of the average number of each subfamily at the phylum and genus levels.
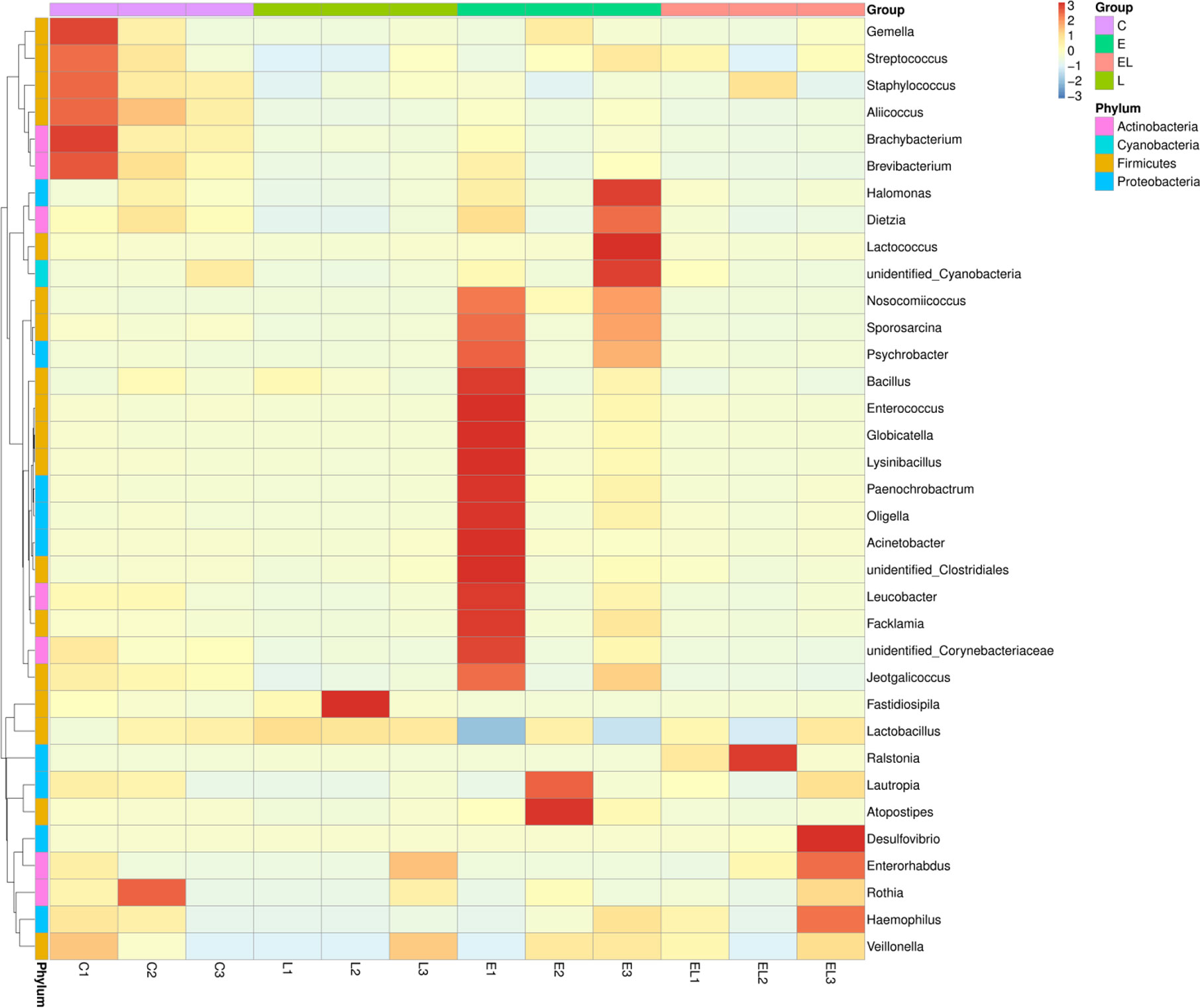
Figure 5 Heatmap of the relative abundance of bacterial communities at the genus level in each sample. Each color-block in the heatmap indicates the relative abundance of a bacterial genus in a sample.
To further compare the differences in intestinal microflora among the four groups, Linear discriminant analysis effect size (LEfSe) analysis coupled with Linear discriminant analysis (LDA) was performed for different classification levels (Figures 6 and 7). At the phylum level, Firmicutes was obviously more abundant in L-group than in the C-group, whereas the abundance of Cyanobacteria and Actinobacteria was lower. Additionally, the abundance of the Proteobacteria was significantly increased in E-group in comparison with C-group. At the genus level, Lactobacillus and Sphingomonas levels tended to be higher in the L-group than C-group, whereas the Pantoea, Cutibacterium, Glutamicibacter, Turicibacter, Globicatella, Microbacterium, Facklamia, unidentified_Corynebacteriaceae, Brachybacterium, and Staphylococcus showed the opposite trend (Figures 6A and 7A). Moreover, a comparison of the E and C groups displayed a significant increase in the abundance of Globicatella, Acinetobacter, Aerococcus, and Comamonas as well as a distinct decrease in the abundance of Roseburia and Megasphaera (Figures 6B and 7B). Meanwhile, the E-group was significantly enriched for Paenibacillus, Aerococcus, Comamonas, Acinetobacter, Corynebacterium, Facklamia, and Globicatella in comparison with EL-group (Figures 6C and 7C).
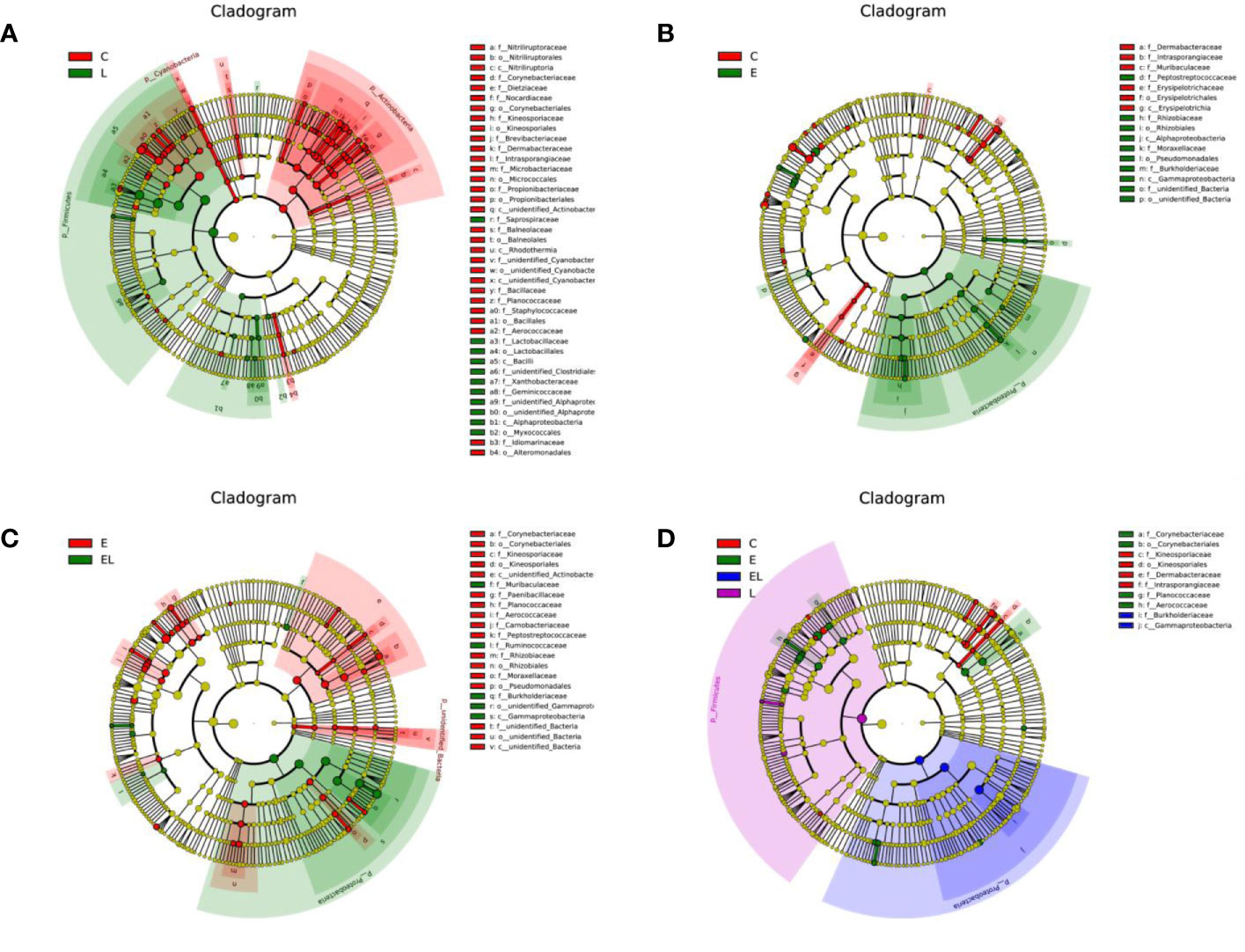
Figure 6 Differentially abundant phylotypes in different groups on the basis of LEfSe analysis. Cladogram obtained from LEfSe analysis displayed the different taxa in microbiota of different groups of mice. (A) Cladogram indicating the phylogenetic distribution of microbiota correlated with the C and L groups. (B) Cladogram indicating the phylogenetic distribution of microbiota correlated with the C and E groups. (C) Cladogram indicating the phylogenetic distribution of microbiota correlated with the E and EL groups. (D) Cladogram indicating the phylogenetic distribution of microbiota correlated with the C, E, EL and L groups. The colored circles from the inside to the outside represent different taxonomic levels (phylum, class, order, family, and genus levels). The yellow circles in the cladogram indicate the taxa with no evident differences.
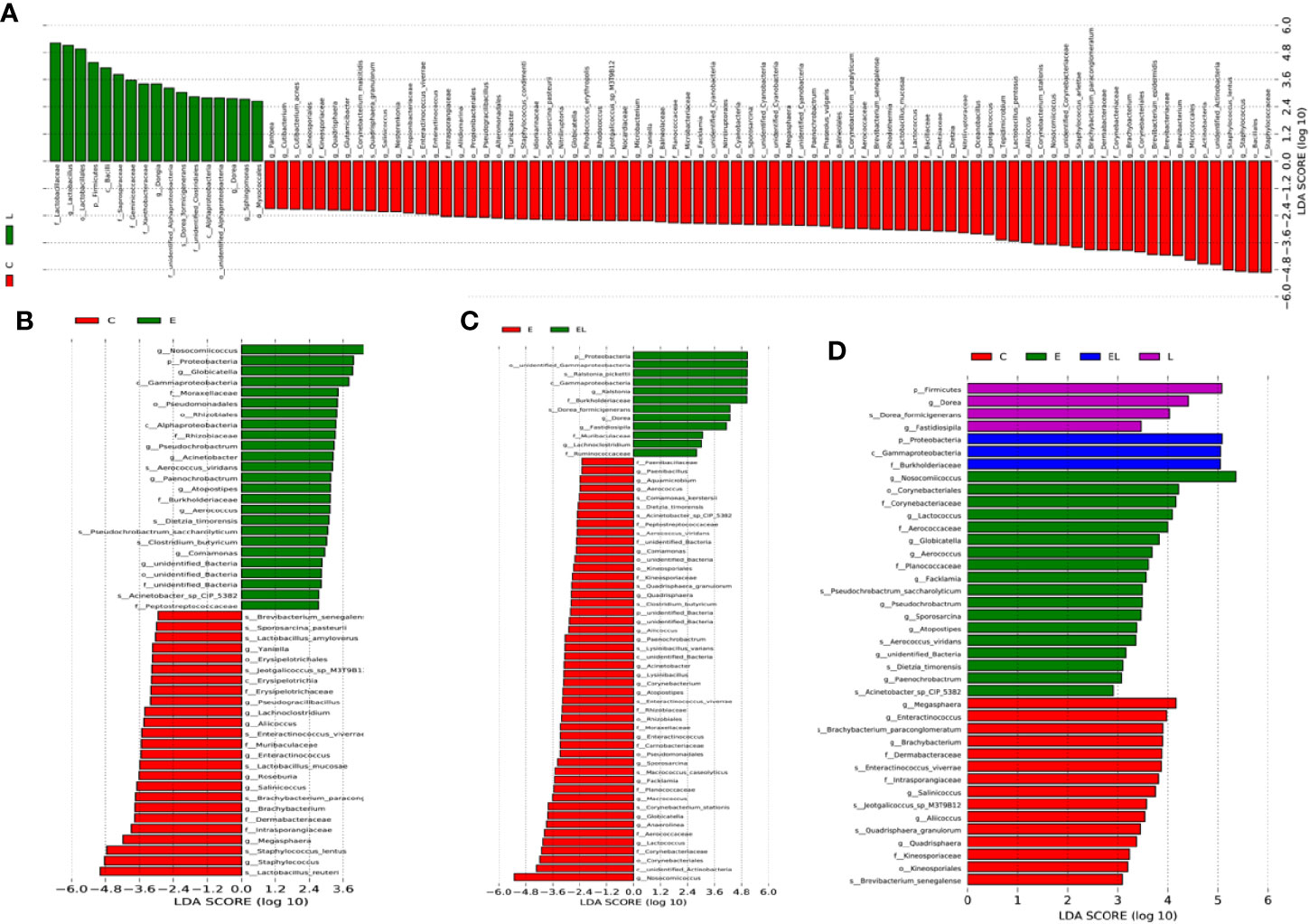
Figure 7 The differences in abundance between the different groups were evaluated using LDA scores. (A) The differences in abundance between the C and L groups. (B) The differences in abundance between the C and E groups. (C) The differences in abundance between the E and EL groups. (D) The differences in abundance between the C, E, EL and L groups. LDA scores > 2 was considered statistically significant.
In livestock industry, diarrhea is widely prevalent in juvenile animals, which is deemed as a crucial factor resulting in the reduction of global animal productivity (Pepin et al., 2004; Diao et al., 2020). Multiple measures have been performed to prevent diarrhea, but it still occurs from time to time. Recently, role of gut microbiota is revealed in the development of diarrhea (Huang et al., 2020). Therefore, the improvement of the intestinal microbial community structure may contribute to alleviate diarrhea (Yue et al., 2019). The significance of the Lactobacillus has been widely acknowledged as a result of its role in gut microbiota, metabolism, immunity, and health maintenance, but few reports have been published on the Lactobacillus from yaks inhabiting the Tibet Plateau (Li et al., 2018b; Wang et al., 2020). In this study, we analyzed the influence of Lactobacillus isolated from yaks on the gut microbiota and investigate whether it could improve the microbial community structure of mice with E. coli-induced diarrhea. Our results indicated that the Lactobacillus administration alleviated the intestinal microbial community of diarrheal mice colonized by E. coli.
Previous studies revealed that mammalian gut microbiota was dynamically varied during development and reached stability at maturity (Poroyko et al., 2011; David et al., 2014, Yang et al., 2020). Diarrheal diseases were widespread in the childhood of animals, which may be closely related to their immature gut microbiota (Wang et al., 2018b). Therefore, probiotics supplementation in the juvenile period of animals may reduce diarrhea by improving the structure in the intestinal microbial community (Wang et al., 2018a). Our study found the phylum Firmicutes as the most dominant bacteria in all samples regardless of the treatment. Consistent with previous observations, this phylum was also found to be widely distributed in camels, sheep, goats, and roe deer, indicating its important role in intestinal ecology and function (Li Z. et al., 2014; Zeng et al., 2017; Lei et al., 2018). The Firmicutes is responsible for digestion of cellulose and its richness in the gut contributes to meet the energy and nutrition requirements of animals in the growth and development process (Sun et al., 2016). Additionally, Firmicutes is mainly composed of gram-positive bacteria including Lactococcus, Bacillus, and Lactobacillus and most of them are perceived as beneficial bacteria, which are conducive to inhibit the proliferation of pathogenic bacteria and improve the intestinal environment (Garneau et al., 2008).
Importantly, our study also found a higher variation in some bacterial phyla and genera of different treatment groups, and this variation may play a crucial role in the intestinal ecosystem and function. Cyanobacteria comprises a great quantity of cyanotoxin-producing bacteria, posing a great threat to animal and human health (Carmichael, 1992). Wang et al. (2018b) observed that the proportion of Actinobacteria in the gut of diarrheal goat was significantly increased. Moreover, the synergy of Actinobacteria with one partner or host can easily be transformed into a pathogenic interaction with another (Miao and Davies, 2010). Mice in the L-group displayed increased Firmicutes and decreased Cyanobacteria and Actinobacteria abundance when compared to C-group, indicating a possible reduction in disease risk through Lactobacillus supplementation. Moreover, Lactobacillus and Sphingomonas were enriched in mice treated with Lactobacillus, whereas Pantoea, Cutibacterium, Glutamicibacter, Turicibacter, Globicatella, Microbacterium, Facklamia, unidentified_Corynebacteriaceae, Brachybacterium, and Staphylococcus were reduced in the control group. Previously, Lactobacillus had improved the intestinal mucosal immunity and interact with intestinal epithelial cells against entero-invasive E. coli (Johnson-Henry et al., 2007; Wang et al., 2019; Dong et al., 2019). Studies have also reported that supplementing diet with Lactobacillus daily can prevent non-alcoholic fatty liver disease by ameliorating the intestinal environment and attenuating inflammation in obese mice (Zhang et al., 2020a; Zhang et al., 2020b). Aside from improving immunity and regulating gut microbiota, Lactobacillus supplementation enhanced digestive enzyme activity and intestinal antioxidant ability benefiting the host (Chen F. et al., 2020; Zhang et al., 2020). Sphingomonas can degrade multiple organic matter, displaying the great application potential in environmental protection and industrial production (Leys et al., 2004). Pantoea, a gram-negative pathogenic bacterium, is associated with disease in plants, humans, and rarely in domestic animals (Silva-Rojas et al., 2012). Henker et al. (2020) indicated that Pantoea could induce fibrinonecrotic placentitis and abortions in mare. Zaccone et al. (2020) reported that Pantoea was closely related to bacteremia in humans. Cutibacterium was considered as skin flora contaminant, which can result in pericarditis with serious complications (Fakhri et al., 2020). Moreover, Cutibacterium was also closely related to multiple postoperative complications, including persistent postoperative pain, chronic inflammation, and endoprosthesis involving bacterial biofilms because it ubiquitously colonizes the skin and resides in various other locations in the human body (Patel et al., 2009; Achermann et al., 2013; Hudek et al., 2021). Turicibacter is a pro-inflammatory bacterium whose level rises during enteritis (Bretin et al., 2018). Globicatella was previously reported to associate with meningitis and bacteremia (Lau et al., 2006; Seegmuller et al., 2007). Furthermore, Globicatella was also observed in the purulent joint and lung infections in calves and sheep (Vandamme et al., 2001). Microbacterium, a novel bacterial pathogen, was also closely related to bacteremia (Hodgkin et al., 2000; Lau et al., 2002). Facklamia may be relevant to invasive disease such as meningitis and septicemia (Hughes, 2014; Parvataneni et al., 2015). Corynebacteriaceae can lead to endocarditis (Prada et al., 1994). Brachybacterium can cause bloodstream infection (Tamai et al., 2018). Most pathogenic Staphylococcus can produce coagulase, staphylolysin, enterotoxin, and toxic shock syndrome toxin1, resulting in fever, emesis, diarrhea, acute gastroenteritis, and even shock (Gemeinder et al., 2020; White et al., 2020). Moreover, Staphylococcus can invade the host through multiple ways and cause local and systemic infections as well as various invasive diseases such as pneumonia, meningitis, blood poisoning, and septic pyemia (Chen H. A. et al., 2020; Ranzani et al., 2020). Remarkably, E. coli infection significantly increased Acinetobacter, Aerococcus, and Comamonas levels and decreased Roseburia and Megasphaera content as compared to control group. Acinetobacter, a common opportunistic pathogen, is widely colonized in the digestive tract, skin, respiratory tract, and genitourinary tract, which can cause bacteremia, pneumonia, endocarditis, as well as urinary and skin infections (Go et al., 1994; Livermore and Woodford, 2006; Lima et al., 2020). Aerococcus can result in endocarditis and urinary tract infections (Yabes et al., 2018; Sous et al., 2019). Comamonas may be closely related to bacteremia (Liu et al., 2020a; Palacio et al., 2020). Roseburia, a butyrate-producing bacterium, can utilize sucrose, cellobiose, galactose, and glycogen (Duncan et al., 2002). Megasphaera was previously reported to produce short-chain fatty acids (SCFAs), displaying a positive regulatory effect to physiological functioning of gut and intestinal permeability (Kim et al., 2002). Conversely, Lactobacillus supplementation significantly reduced Paenibacillus, Aerococcus, Comamonas, and Corynebacterium levels in the ileum of mice induced by E. coli. Although reports of Paenibacillus infections are exceedingly rare, the infection may result in meningitis in some cases (Hunt et al., 2020). Corynebacterium, an acclimatized pathogen, can cause respiratory disease (Samies et al., 1986). This study conveyed a message that Lactobacillus supplementation resulted in an increase in beneficial bacteria and decreased pathogenic bacteria, whereas E. coli infection increased the ratio of harmful and beneficial bacteria. Additionally, Lactobacillus administration effectively ameliorate the microbial community structure of mice induced by E. coli and decreased the proportion of pathogenic bacteria.
In summary, this study revealed that the gut microbiota in diarrheal mice induced by E. coli undergoes striking changes, characterized by an increased proportion of harmful bacteria. Conversely, Lactobacillus administration not only improves the microbial community structure of normal mice but also alleviates E. coli-induced diarrhea by mediating gut microbiota. These results expand our understanding of the potential benefits of Lactobacillus from yaks and convey an important message that Lactobacillus may be one of effective methods to attenuate diarrhea in yaks. Importantly, these findings also enriched the knowledge of disease prevention and control system in yaks. However, several limitations in this study need to be noticed, such as individual variation, experimental environment, and small sample size.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA665922.
The animal study was reviewed and approved by the ethics committee of Tibet Agriculture & Animal Husbandry University.
QW, HZ, HD and AL conceived and designed the experiments. AL, YC, QW, HZ, HD and BL contributed reagents, materials, and analysis tools. HD wrote the manuscript. HZ, QW, MI, KM and TJ revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Key research, development, and transformation program of Tibet autonomous region (XZ201902NB05), the Key laboratory of clinical veterinary medicine in Tibet, and the Research and demonstration of technologies for prevention and control of major infectious diseases in characteristic livestock (XZ201901NA02).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Achermann Y., Sahin F., Schwyzer H. K., Kolling C., Wust J., Vogt M. (2013). Characteristics and outcome of 16 periprosthetic shoulder joint infections. Infection 41, 613–620. doi: 10.1007/s15010-012-0360-4
Bretin A., Lucas C., Larabi A., Dalmasso G., Billard E., Barnich N., et al. (2018). AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci. Rep. 8, 12301. doi: 10.1038/s41598-018-30055-y
Cani P. D., Bibiloni R., Knauf C., Waget A., Neyrinck A. M., Delzenne N. M., et al. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 57, 1470–1481. doi: 10.2337/db07-1403
Carmichael W. W. (1992). Cyanobacteria secondary metabolites–the cyanotoxins. J. Appl. Bacteriol. 72, 445–459. doi: 10.1111/j.1365-2672.1992.tb01858.x
Chen F., Wang H., Chen J., Liu Y., Wen W., Li Y., et al. (2020). Lactobacillus delbrueckii Ameliorates Intestinal Integrity and Antioxidant Ability in Weaned Piglets after a Lipopolysaccharide Challenge. Oxid. Med. Cell Longev. 2020, 6028606. doi: 10.1155/2020/6028606
Chen H. A., Yang C. J., Tsai M. S., Liao C. H., Lee C. H. (2020). Linezolid as salvage therapy for central nervous system infections due to methicillin-resistant Staphylococcus aureus at two medical centers in Taiwan. J. Microbiol. Immunol. Infect. 53, 909–915. doi: 10.1016/j.jmii.2020.08.004
David L. A., Materna A. C., Friedman J., Campos-Baptista M. I., Blackburn M. C., Perotta A., et al. (2014). Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89. doi: 10.1186/gb-2014-15-7-r89
de La Serre C. B., Ellis C. L., Lee J., Hartman A. L., Rutledge J. C., Raybould H. E., et al. (2010). Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G440–G448. doi: 10.1152/ajpgi.00098.2010
Diao N. C., Gong Q. L., Li J. M., Zhao D., Li D., Zhao B., et al. (2020). Prevalence of bovine viral diarrhea virus (BVDV) in yaks between 1987 and 2019 in mainland China: A systematic review and meta-analysis. Microb. Pathog. 144, 104185. doi: 10.1016/j.micpath.2020.104185
Dong H., Ijaz M., Mehmood K., Ali M., Tian F., Li J., et al. (2019). Protective Effects of Salidroside and Dexamethasone against E.coli-Induced Inflammatory Response on Endometrial Epithelium Cells in Yaks. Pak Vet. J. 39, 101–105. doi: 10.29261/pakvetj/2018.116
Dudik B., Kinova S. H., Bilka F., Paskova L., Bilkova A. (2020). Mucin pre-cultivated Lactobacillus reuteri E shows enhanced adhesion and increases mucin expression in HT-29 cells. Antonie Van Leeuwenhoek 113, 1191–1200. doi: 10.1007/s10482-020-01426-1
Duncan S. H., Hold G. L., Barcenilla A., Stewart C. S., Flint H. J. (2002). Roseburia intestinalis sp. Nov., A novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52, 1615–1620. doi: 10.1099/00207713-52-5-1615
Fakhri G., Tayeh C., Dbaibo G., El S. O., Abdul H. N., Bitar F., et al. (2020). Cardiac tamponade caused by cutibacterium acnes: An updated and comprehensive review of the literature. Can. J. Infect. Dis. Med. Microbiol. 2020, 9598210. doi: 10.1155/2020/9598210
Gao J., Liu M., Meng X., Han Z., Zhang D., Hou B., et al. (2013). Seroprevalence of bovine viral diarrhea infection in Yaks (Bos grunniens) on the Qinghai-Tibetan Plateau of China. Trop. Anim. Health Prod. 45, 791–793. doi: 10.1007/s11250-012-0290-2
Garneau J. E., Tremblay D. M., Moineau S. (2008). Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373, 298–309. doi: 10.1016/j.virol.2007.12.002
Gemeinder J. L. P., de Barros N. R., Pegorin G. S., Singulani J. L., Borges F. A., Arco M. C. G. D., et al. (2020). Gentamicin encapsulated within a biopolymer for the treatment of Staphylococcus aureus and Escherichia coli infected skin ulcers. J. Biomater. Sci. Polym. Ed. 32, 93–111. doi: 10.1080/09205063.2020.1817667
Go E. S., Urban C., Burns J., Kreiswirth B., Eisner W., Mariano N., et al. (1994). Clinical and molecular epidemiology of acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet 344, 1329–1332. doi: 10.1016/s0140-6736(94)90694-7
Han Z., Li K., Shahzad M., Zhang H., Luo H., Qiu G., et al. (2017). Analysis of the intestinal microbial community in healthy and diarrheal perinatal yaks by high-throughput sequencing. Microb. Pathog. 111, 60–70. doi: 10.1016/j.micpath.2017.08.025
Henker L. C., Lorenzett M. P., Keller A., Siqueira F. M., Driemeier D., Pavarini S. P. (2020). Fibrinonecrotic placentitis and abortion associated with pantoea agglomerans infection in a mare. J. Equine Vet. Sci. 92, 103156. doi: 10.1016/j.jevs.2020.103156
Heuer C., Healy A., Zerbini C. (2007). Economic effects of exposure to bovine viral diarrhea virus on dairy herds in New Zealand. J. Dairy Sci. 90, 5428–5438. doi: 10.3168/jds.2007-0258
Hodgkin J., Kuwabara P. E., Corneliussen B. (2000). A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. Elegans. Curr. Biol. 10, 1615–1618. doi: 10.1016/s0960-9822(00)00867-8
Huang W. C., Pan C. H., Wei C. C., Huang H. Y. (2020). Lactobacillus plantarum PS128 improves physiological adaptation and performance in triathletes through gut microbiota modulation. Nutrients 12, 2315. doi: 10.3390/nu12082315
Hudek R., Brobeil A., Bruggemann H., Sommer F., Gattenlohner S., Gohlke F. (2021). Cutibacterium acnes is an intracellular and intra-articular commensal of the human shoulder joint. J. Shoulder Elbow Surg. 30, 16–26. doi: 10.1016/j.jse.2020.04.020
Hughes T. (2014). Anti-NMDA receptor encephalitis and Facklamia languida septicaemia: Initially unpronounceable but eventually treatable. Age Ageing 43, 588–589. doi: 10.1093/ageing/afu102
Hunt B., Rogers C., Blais R. M., Adachi K., Sathyavagiswaran L. (2020). Paenibacillus sepsis and meningitis in a premature infant: A case report. Am. J. Foren. Med. Pathol. doi: 10.1097/PAF.0000000000000610
Jami E., Israel A., Kotser A., Mizrahi I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. Isme J. 7, 1069–1079. doi: 10.1038/ismej.2013.2
Johnson-Henry K. C., Hagen K. E., Gordonpour M., Tompkins T. A., Sherman P. M. (2007). Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell Microbiol. 9, 356–367. doi: 10.1111/j.1462-5822.2006.00791.x
Khan I. (2019). Isolation and in vitro Characterization of Anti-Salmonella Enteritidis Probiotic Potential of Indigenous Lactobacilli from Poultry. Pak Vet. J. 39, 563–567. doi: 10.29261/pakvetj/2019.022
Kim Y. J., Liu R. H., Rychlik J. L., Russell J. B. (2002). The enrichment of a ruminal bacterium (Megasphaera elsdenii YJ-4) that produces the trans-10, cis-12 isomer of conjugated linoleic acid. J. Appl. Microbiol. 92, 976–982. doi: 10.1046/j.1365-2672.2002.01610.x
Kootte R. S., Vrieze A., Holleman F., Dallinga-Thie G. M., Zoetendal E. G., de Vos W. M., et al. (2012). The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 14, 112–120. doi: 10.1111/j.1463-1326.2011.01483.x
Lau S. K., Woo P. C., Woo G. K., Yuen K. Y. (2002). Catheter-related Microbacterium bacteremia identified by 16S rRNA gene sequencing. J. Clin. Microbiol. 40, 2681–2685. doi: 10.1128/jcm.40.7.2681-2685.2002
Lau S. K., Woo P. C., Li N. K., Teng J. L., Leung K. W., Ng K. H. L., et al. (2006). Globicatella bacteraemia identified by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 59, 303–307. doi: 10.1136/jcp.2005.028878
Lei Y., Zhang K., Guo M., Li G., Li C., Li B., et al. (2018). Exploring the Spatial-Temporal microbiota of compound stomachs in a pre-weaned goat model. Front. Microbiol. 9, 1846. doi: 10.3389/fmicb.2018.01846
Leys N. M., Ryngaert A., Bastiaens L., Verstraete W., Top E. M., Springael D. (2004). Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70, 1944–1955. doi: 10.1128/aem.70.4.1944-1955.2004
Li K., Zhang L., Zhang H., Lei Z., Luo H., Mehmood K., et al. (2017). Epidemiological investigation and risk factors of Echinococcus granulosus in yaks (Bos grunniens), Tibetan pigs and Tibetans on Qinghai Tibetan plateau. Acta Trop. 173, 147–152. doi: 10.1016/j.actatropica.2017.06.019
Li N., Wang Q., Wang Y., Sun A., Lin Y., Jin Y., et al. (2018). Oral probiotics ameliorate the behavioral deficits induced by chronic mild stress in mice via the gut Microbiota-Inflammation axis. Front. Behav. Neurosci. 12, 266. doi: 10.3389/fnbeh.2018.00266
Li K., Mehmood K., Zhang H., Jiang X., Shahzad M., Dong X., et al. (2018a). Characterization of fungus microbial diversity in healthy and diarrheal yaks in Gannan region of Tibet Autonomous Prefecture. Acta Trop. 182, 14–26. doi: 10.1016/j.actatropica.2018.02.017
Li K., Shahzad M., Zhang H., Jiang X., Mehmood K., Zhao X., et al. (2018b). Socio-economic burden of parasitic infections in yaks from 1984 to 2017 on Qinghai Tibetan Plateau of China-A review. Acta Trop. 183, 103–109. doi: 10.1016/j.actatropica.2018.04.011
Li A., Wang Y., Li Z., Qamar H., Mehmood K., Zhang L., et al. (2019a). Probiotics isolated from yaks improves the growth performance, antioxidant activity, and cytokines related to immunity and inflammation in mice. Microb. Cell Fact. 18, 112. doi: 10.1186/s12934-019-1161-6
Li A., Wang Y., Pei L., Mehmood K., Li K., Qamar H., et al. (2019b). Influence of dietary supplementation with Bacillus velezensis on intestinal microbial diversity of mice. Microb. Pathog. 136, 103671. doi: 10.1016/j.micpath.2019.103671
Li S., He Y., Zhang H., Zheng R., Xu R., Liu Q., et al. (2020). Formulation of traditional Chinese medicine and its application on intestinal flora of constipated rats. Microb. Cell Fact. 19, 212. doi: 10.1186/s12934-020-01473-3
Li K., Gao J., Shahzad M., Han Z., Nabi F., Liu M., et al. (2014). Seroprevalence of Toxoplasma gondii infection in yaks (Bos grunniens) on the Qinghai-Tibetan Plateau of China. Vet. Parasitol. 205, 354–356. doi: 10.1016/j.vetpar.2014.07.014
Li Z., Zhang Z., Xu C., Zhao J., Liu H., Fan Z., et al. (2014). Bacteria and methanogens differ along the gastrointestinal tract of Chinese roe deer (Capreolus pygargus). PloS One 9, e114513. doi: 10.1371/journal.pone.0114513
Lima W. G., Brito J., Da C. N. W. (2020). Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: Two problems, one solution? Med. Hypotheses 144:110139. doi: 10.1016/j.mehy.2020.110139
Liu X. J., Qiao X. W., Huang T. M., Li L., Jiang S. P. (2020a). Comamonas kerstersii bacteremia. Med. Mal. Infect. 50, 288–290. doi: 10.1016/j.medmal.2019.12.005
Liu Z., Li A., Wang Y., Iqbal M., Zheng A., Zhao M., et al. (2020b). Comparative analysis of microbial community structure between healthy and Aeromonas veronii-infected Yangtze finless porpoise. Microb. Cell Fact. 19, 123. doi: 10.1186/s12934-020-01383-4
Livermore D. M., Woodford N. (2006). The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 14, 413–420. doi: 10.1016/j.tim.2006.07.008
Lynch S. V., Pedersen O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Miao V., Davies J. (2010). Actinobacteria: The good, the bad, and the ugly. Antonie Van Leeuwenhoek 98, 143–150. doi: 10.1007/s10482-010-9440-6
Palacio R., Cabezas L., Cornejo C., Seija V. (2020). [Comamonas kerstersii bacteremia in a young man with acute appendicitis]. Rev. Chil. Infectol. 37, 182–185. doi: 10.4067/s0716-10182020000200182
Parvataneni K. C., Iyer S., Khatib R., Saravolatz L. D. (2015). Facklamia species and streptococcus pneumoniae meningitis: A case report and review of the literature. Open Forum Infect. Dis. 2, v29. doi: 10.1093/ofid/ofv029
Patel A., Calfee R. P., Plante M., Fischer S. A., Green A. (2009). Propionibacterium acnes colonization of the human shoulder. J. Shoulder Elbow Surg. 18, 897–902. doi: 10.1016/j.jse.2009.01.023
Pepin J., Valiquette L., Alary M. E., Villemure P., Pelletier A., Forget K., et al. (2004). Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: A changing pattern of disease severity. Cmaj 171, 466–472. doi: 10.1503/cmaj.1041104
Poroyko V., Morowitz M., Bell T., Ulanov A., Wang M., Donovan S., et al. (2011). Diet creates metabolic niches in the “immature gut” that shape microbial communities. Nutr. Hosp. 26, 1283–1295. doi: 10.1590/S0212-16112011000600015
Prada J. L., Qillanueva J. L., Torre-Cisneros J., Rodriguez F., Espinosa M., Anguita M., et al. (1994). Endocarditis due to corynebacteriaceae in a kidney transplant patient. Nephrol. Dial. Transplant. 9, 1185–1187. doi: 10.1093/ndt/9.8.1185
Qiu Q., Zhang G., Ma T., Qian W., Wang J., Ye Z., et al. (2012). The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949. doi: 10.1038/ng.2343
Ranzani O. T., Motos A., Chiurazzi C., Ceccato A., Rinaudo M., Bassi G. L., et al. (2020). Diagnostic accuracy of Gram staining when predicting staphylococcal hospital-acquired pneumonia and ventilator-associated pneumonia: A systematic review and meta-analysis. Clin. Microbiol. Infect. 26, 1456–1463. doi: 10.1016/j.cmi.2020.08.015
Ritz S., Hahn D., Wami H., Tegelkamp K., Dobrindt U., Schnekenburger J. (2020). Gut microbiome as a response marker for pancreatic enzyme replacement therapy in a porcine model of exocrine pancreas insufficiency. Microb. Cell Fact. 19, 221. doi: 10.1186/s12934-020-01482-2
Sadowska B., Walencka E., Wieckowska-Szakiel M., Rozalska B. (2010). Bacteria competing with the adhesion and biofilm formation by Staphylococcus aureus. Folia Microbiol. (Praha.) 55, 497–501. doi: 10.1007/s12223-010-0082-x
Samies J. H., Hathaway B. N., Echols R. M., Veazey J. J., Pilon V. A. (1986). Lung abscess due to Corynebacterium equi. Report of the first case in a patient with acquired immune deficiency syndrome. Am. J. Med. 80, 685–688. doi: 10.1016/0002-9343(86)90825-9
Seegmuller I., van der Linden M., Heeg C., Reinert R. R. (2007). Globicatella sanguinis is an etiological agent of ventriculoperitoneal shunt-associated meningitis. J. Clin. Microbiol. 45, 666–667. doi: 10.1128/JCM.01774-06
Silva-Rojas H. V., Esker P. D., Mahuku G., Peralta-Nava J. A., Cortes-Cueto A. L., Rodriguez-Herrera J. A., et al. (2012). Pantoea ananatis, a maize plant pathogenic bacterium associated with chlorotic streaks and rolling upper leaves. Phytopathology 1026, 6.
Sobanbua S., Dolkittikul S., Nakphaichit M., Keawsompong S., Nitisinprasert S. (2020). Antimicrobial peptide presenting potential strain-specific real time polymerase chain reaction assay for detecting the probiotic Lactobacillus reuteri KUB-AC5 in chicken intestine. Poult Sci. 99, 526–535. doi: 10.3382/ps/pez549
Sous N., Piwoz J. A., Baer A. Z., Bhavsar S. M. (2019). Subacute aerococcus urinae infective endocarditis with mycotic aneurysms in a pediatric patient: Case report and literature review. J. Pediatr. Infect. Dis. Soc. 8, 492–494. doi: 10.1093/jpids/piz016
Sun B., Wang X., Bernstein S., Huffman M. A., Xia D. P., Gu Z., et al. (2016). Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci. Rep. 6, 26035. doi: 10.1038/srep26035
Tamai K., Akashi Y., Yoshimoto Y., Yaguchi Y., Takeuchi Y., Shiigai M., et al. (2018). First case of a bloodstream infection caused by the genus Brachybacterium. J. Infect. Chemother. 24, 998–1003. doi: 10.1016/j.jiac.2018.06.005
van den Bogaard A. E., Stobberingh E. E. (2000). Epidemiology of resistance to antibiotics. Links between animals and humans. Int. J. Antimicrob. Agents 14, 327–335. doi: 10.1016/s0924-8579(00)00145-x
Vandamme P., Hommez J., Snauwaert C., Hoste B., Cleenwerck I., Lefebvre K., et al. (2001). Globicatella sulfidifaciens sp. Nov., Isolated from purulent infections in domestic animals. Int. J. Syst. Evol. Microbiol. 51, 1745–1749. doi: 10.1099/00207713-51-5-1745
Wang Y., Li A., Jiang X., Zhang H., Mehmood K., Zhang L., et al. (2018a). Probiotic potential of leuconostoc pseudomesenteroides and lactobacillus strains isolated from yaks. Front. Microbiol. 9, 2987. doi: 10.3389/fmicb.2018.02987
Wang Y., Zhang H., Zhu L., Xu Y., Liu N., Sun X., et al. (2018b). Dynamic distribution of gut microbiota in goats at different ages and health states. Front. Microbiol. 9, 2509. doi: 10.3389/fmicb.2018.02509
Wang Y., Li A., Zhang L., Waqas M., Mehmood K., Iqbal M., et al. (2019). Probiotic potential of Lactobacillus on the intestinal microflora against Escherichia coli induced mice model through high-throughput sequencing. Microb. Pathog. 137, 103760. doi: 10.1016/j.micpath.2019.103760
Wang G., Huang S., Cai S., Yu H., Wang Y., Zeng X., et al. (2020). Lactobacillus reuteri ameliorates intestinal inflammation and modulates gut microbiota and metabolic disorders in dextran sulfate Sodium-Induced colitis in mice. Nutrients 12, 2298. doi: 10.3390/nu12082298
White J. K., Nielsen J. L., Larsen C. M., Madsen A. M. (2020). Impact of dust on airborne Staphylococcus aureus’ viability, culturability, inflammogenicity, and biofilm forming capacity. Int. J. Hyg. Environ. Health 230:113608. doi: 10.1016/j.ijheh.2020.113608
Wu H. J., Wu E. (2012). The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 3, 4–14. doi: 10.4161/gmic.19320
Yabes J. M., Perdikis S., Graham D. B., Markelz A. (2018). A rare case of Aerococcus urinae infective endocarditis in an atypically young male: Case report and review of the literature. BMC Infect. Dis. 18, 522. doi: 10.1186/s12879-018-3414-0
Yang F., DeLuca J., Menon R., Garcia-Vilarato E., Callaway E., Landrock K., et al. (2020). Effect of diet and intestinal AhR expression on fecal microbiome and metabolomic profiles. Microb. Cell Fact. 19, 219. doi: 10.1186/s12934-020-01463-5
Yue S. J., Liu J., Wang W. X., Wang A. T., Yang X. Y., Guan H., et al. (2019). Berberine treatment-emergent mild diarrhea associated with gut microbiota dysbiosis. BioMed. Pharmacother. 116, 109002. doi: 10.1016/j.biopha.2019.109002
Yue Y., Yang B., Lu J., Zhang S., Liu L., Nassar K., et al. (2020). Metabolite secretions of Lactobacillus plantarum YYC-3 may inhibit colon cancer cell metastasis by suppressing the VEGF-MMP2/9 signaling pathway. Microb. Cell Fact. 19, 213. doi: 10.1186/s12934-020-01466-2
Zaccone V., Lockwood M., Ticona J., Jouharian P., Zamora M., Hampton C., et al. (2020). An unusual cause of neutropenic fever: Spontaneous pantoea agglomerans bacteremia in an adult. Am. J. Med. Case Rep. 8, 289–292.
Zeng Y., Zeng D., Ni X., Zhu H., Jian P., Zhou Y., et al. (2017). Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using Illumina MiSeq sequencing revealed high microbial diversity. AMB Express 7, 75. doi: 10.1186/s13568-017-0378-1
Zhang H., Rehman M. U., Li K., Luo H., Lan Y., Nabi F., et al. (2017). Antimicrobial Resistance of Escherichia coli Isolated from Tibetan Piglets Suffering from White Score Diarrhea. Pakistan Vet. J. 37, 43–46.
Zhang H., Wang Q., Liu H., Kong B., Chen Q. (2020). In vitro growth performance, antioxidant activity and cell surface physiological characteristics of Pediococcus pentosaceus R1 and Lactobacillus fermentum R6 stressed at different NaCl concentrations. Food Funct. 11, 6376–6386. doi: 10.1039/c9fo02309g
Zhang Z., Zhou H., Guan M., Zhou X., Liang X., Lv Y., et al. (2020a). Lactobacillus casei YRL577 combined with plant extracts reduce markers of non-alcoholic fatty liver disease in mice. Br. J. Nutr. 1–31. doi: 10.1017/S0007114520003013
Zhang Z., Zhou H., Zhou X., Sun J., Liang X., Lv Y., et al. (2020b). Lactobacillus casei YRL577 ameliorates markers of non-alcoholic fatty liver and alters expression of genes within the intestinal bile acid pathway. Br. J. Nutr. 1–9. doi: 10.1017/S0007114520003001
Keywords: Tibet Plateau, yak, gut microbiota, Lactobacillus, Escherichia coli
Citation: Dong H, Liu B, Li A, Iqbal M, Mehmood K, Jamil T, Chang Y-F, Zhang H and Wu Q (2021) Microbiome Analysis Reveals the Attenuation Effect of Lactobacillus From Yaks on Diarrhea via Modulation of Gut Microbiota. Front. Cell. Infect. Microbiol. 10:610781. doi: 10.3389/fcimb.2020.610781
Received: 29 September 2020; Accepted: 21 December 2020;
Published: 16 February 2021.
Edited by:
Jie Yin, Hunan Agricultural University, ChinaReviewed by:
Yunxia Li, Hunan Agricultural University, ChinaCopyright © 2021 Dong, Liu, Li, Iqbal, Mehmood, Jamil, Chang, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Zhang, aHoyMzZAc2NhdS5lZHUuY24=; Qingxia Wu, d3VxeDIwMTRAc2luYS5jb20=; Aoyun Li, ODA3MjU4NTczQHFxLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.