- Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom
Campylobacter jejuni is the leading cause of bacterial foodborne gastroenteritis world wide and represents a major public health concern. Over the past two decades, significant progress in functional genomics, proteomics, enzymatic-based virulence profiling (EBVP), and the cellular biology of C. jejuni have improved our basic understanding of this important pathogen. We review key advances in our understanding of the multitude of emerging virulence factors that influence the outcome of C. jejuni–mediated infections. We highlight, the spatial and temporal dynamics of factors that promote C. jejuni to sense, adapt and survive in multiple hosts. Finally, we propose cohesive research directions to obtain a comprehensive understanding of C. jejuni virulence mechanisms.
Introduction
Campylobacters are the leading cause of bacterial foodborne gastroenteritis in the world. There are 31 different species1 and 10 sub-species within the genus Campylobacter (Garcia-Sanchez et al., 2018; Wilkinson et al., 2018). The Campylobacter genus encompasses several clinically relevant species, such as Campylobacter jejuni subsp. jejuni, Campylobacter coli, Campylobacter fetus, Campylobacter lari, and Campylobacter upsaliensis (Kaakoush et al., 2015; Garcia-Sanchez et al., 2018). This review focuses on C. jejuni subsp. jejuni which is the most relevant clinically (Skirrow, 1977; Skirrow, 2006). C. jejuni is responsible for 80%–90% of the diagnosed cases of Campylobacter infections (Facciola et al., 2017). C. jejuni colonizes the gastrointestinal (GI) tract of a wide variety of food-producing animals such as poultry, cattle, sheep and swine (Figure 1). However, poultry, particularly chickens are the major source of human infection (Humphrey et al., 2014; Ijaz et al., 2018; McKenna et al., 2020). Outbreaks of C. jejuni infections are also associated with exposure to contaminated soil, unpasteurized milk and untreated water sources (Korlath et al., 1985; Hudson et al., 1999; Bronowski et al., 2014; Artursson et al., 2018). Clinical symptoms of C. jejuni infection can be watery or bloody diarrhea accompanied by abdominal cramps, nausea, fever and sometimes vomiting (Blaser, 1997; Hansson et al., 2018; Igwaran and Okoh, 2019). Although C. jejuni infection is acute and self-limiting, in a small number of patients (1:1000) post infection sequalae can lead to severe neurological disorders such as Guillain-Barré syndrome (Yuki et al., 1993; Nachamkin et al., 1998; Sheikh et al., 1998a; Sheikh et al., 1998b; Houliston et al., 2011). According to a recent report by the World Health Organization (WHO), C. jejuni is responsible for 96 million cases of enteric infection globally each year (Havelaar et al., 2015; Bailey et al., 2018). In the United Kingdom, C. jejuni is responsible for more than 700,000 cases, of which 22,000 hospitalisations and more than 100 deaths occur each year (Bronowski et al., 2014; John et al., 2017). The economic burden associated with C. jejuni infection in the United Kingdom is estimated to be £1 billion per year (Bronowski et al., 2014). Moreover, in the European Union (EU), C. jejuni is responsible for estimated cases of 9 million with an economic burden of around €2.4 billion each year (https://www.efsa.europa.eu/en/topics/topic/campylobacter). According to the United States Centers for Disease Control, C. jejuni is responsible for an estimated 1.5 million human infections each year2 with a staggering economic burden of between $1.3 to 6.8 billion dollars per year.
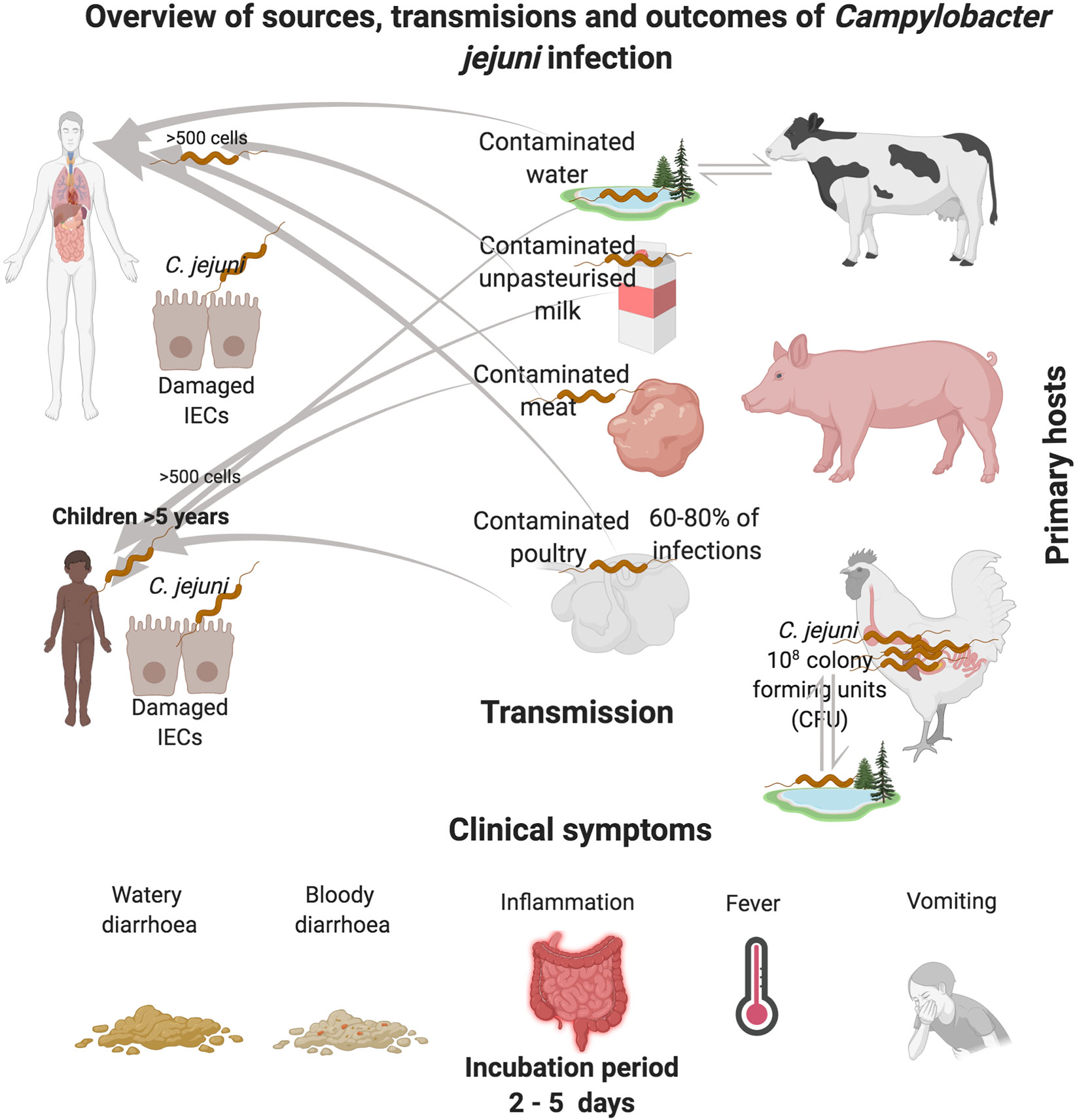
Figure 1 Overview of C. jejuni reservoirs and transmission routes of infection. C. jejuni reside in the GI tract of chickens, where the bacteria can be spread through consumption of contaminated poultry products. C. jejuni transmission can also occur via the consumption of contaminated raw cows drinking milk (RDM) which can occur during the milking process, most commonly via fecal contamination of udders. Pigs are also recognized as reservoirs of C. jejuni. Contamination of the environment can also occur via host fecal contamination. C. jejuni can persist for long periods in feces, milk and water, especially at temperatures close to 4℃. In adverse conditions, C jejuni converts to a viable nonculturable form that can be reactivated when ingested.
C. jejuni does not possess classical virulence factors observed in bacterial enteropathogens such as enterotoxigenic Escherichia coli and Salmonella spp. (Gaytan et al., 2016; Park et al., 2018). However, C. jejuni has a complex array of fitness and virulence factors (Crόinín and Backert, 2012; Backert and Hofreuter, 2013; Backert et al., 2013) which aid the bacterium to respond to the defense mounted by the host; C. jejuni can adhere, invade and temporarily survive inside human intestinal epithelial cells (IECs) in vitro. We review recent progress made in understanding C. jejuni pathogenesis. We highlight findings from several approaches that pioneered the integration of selective mutagenesis, phenotypic assays, high-resolution proteomics and ‘omics. Finally, we describe challenges ahead for successful research in understanding how C. jejuni causes disease in humans.
C. jejuni Virulence Factors, a Breakthrough in Understanding the Missing Link
In early 2000, the availability of the full genome sequence of C. jejuni NCTC 11168, isolated from the feces of a diarrheic patient in 1977 by Martin Skirrow, marked a new era in the study of the pathogenesis of this major enteric pathogen (Skirrow, 1977; Parkhill et al., 2000). The annotation of the full genome sequence revealed the absence of genes encoding for a non-flagellar type 3 protein secretion system (NF-T3SS). This finding has raised an intriguing question: Does C. jejuni sense, inject and secrete putative virulence factors into host cells? In contrast to the absence of NF-T3SS, the genome sequence shed light on the presence of a genomic locus encoding a novel bacterial protein N-glycosylation (pgl) system, absent in other enteropathogens (Szymanski et al., 1999; Linton et al., 2005). This 11 gene locus encodes for all the necessary enzymes for N-linked pgl system to produce a conserved heptasaccharide consisting of GalNAc–α1,4-GalNAc–α1,4(Glcβ1,3)-GalNAc–α1,4-GalNAc–α1,4-GalNAc–α1,3-Bac (Bac is bacillosamine or 2,4,-diacetamido-2,4,6-trideoxyglucose (Young et al., 2002; Jervis et al., 2012). C. jejuni conserved heptasaccharide has been found to modify up to 100 periplasmic and membrane-bound proteins while it also appears to be responsible for multiple cell functions (Cain et al., 2019; Abouelhadid et al., 2019; Abouelhadid et al., 2020). A feature of the availability of C. jejuni genome sequence was the identification and characterization of different glycostructures. In addition to the N-linked pgl system, other studies have facilitated systematic analysis of genes encoding for flagellar biosynthesis and modification (Jagannathan et al., 2001; Hendrixson and DiRita, 2003; Konkel et al., 2004), lipooligosaccharide (LOS) (Parker et al., 2005; Parker et al., 2008; Kanipes et al., 2008; Hameed et al., 2020) and capsule polysaccharide (CPS) (Karlyshev et al., 2001; Karlyshev et al., 2005). In parallel, the genome sequence of C. jejuni identified a large repertoire of phase-variable genes (Guerry et al., 2002; Aidley et al., 2018). The genome sequence of C. jejuni further accelerated characterization of repertoire of virulence and fitness factors such as putative adhesins (Konkel et al., 2005), proteases (Brondsted et al., 2005), autotransporters (Ashgar et al., 2007), chemotaxis regulatory genes (Marchant et al., 2002) and the cytolethal distending toxin (CDT) (Purdy et al., 2000). Sequencing the genomes of various C. jejuni isolates have also elucidated strain-specific genetic diversity, noticeably the finding of the putative pVir plasmid in C. jejuni strain 81–176 (Bacon et al., 2000). Because of the high genome plasticity of C. jejuni, genome sequencing also facilitated genome-wide association studies (GWAS) which provided insight into the prevalence of C. jejuni virulence genes, antimicrobial resistance markers as well as relatedness of human clinical isolates (Sheppard et al., 2013; Buchanan et al., 2017). Understanding the genetic variability of C. jejuni isolates is important for defining key factors that contribute to its ability to host adaptation and evolution. Some C. jejuni strains are restricted to specific host while there are C. jejuni strains with multi-host lineages. Defining how C. jejuni adapts to hosts is an enduring challenge. However, study has demonstrated that one factor that is driving rapid C. jejuni host adaptation is gain and loss of panBCD genes encoding for vitamin B5 biosynthesis pathway (Sheppard et al., 2013). Recently, the advent of large scale genome sequencing has also identified C. jejuni isolates possessing Type VI Secretion System (T6SS) (Corcionivoschi et al., 2015; Ugarte-Ruiz et al., 2015), offering the potential to better understand the role of T6SS in C. jejuni pathogenesis (Liaw et al., 2019).
C. jejuni in the Host-Pathogen Crosstalk: Virulence and Fitness Factors
In its natural environment C. jejuni adapts, survives and proliferates in the nutrient-rich mucous layer of the avian GI tract. C. jejuni growth in chicken ceca exceeds 108 colony-forming units per g of cecal contents (CFU)/g (Dhillon et al., 2006; Hermans et al., 2011; Gormley et al., 2014). The transition of C. jejuni from nutrient-rich chicken ceca to the environment exposes C. jejuni to perturbations. These perturbations unveil C. jejuni to atmospheric oxygen (ca. 21% O2) and temperature fluctuations which thus alter C. jejuni nutrient acquisition and metabolism. In the context of human infection, C. jejuni faces additional stresses such as peristalsis and expulsion in the GI tract. C. jejuni also faces endogenous stresses ranging from oxidative, nitrosative, pH fluctuations and cationic stresses. The ability to persist in spite of various stresses indicate C. jejuni harbors complex virulence and fitness factors (Hermans et al., 2012). These virulence and fitness factors do not only confer protection but also play a role in the ability of C. jejuni to sense, adapt and compete the constantly changing host microenvironments, working for example as sensors and/signal molecules, adhesins for host receptors, and/or effectors for invasion and intracellular survival.
C. jejuni interaction and invasion of human IECs induce numerous downstream host signaling pathways. C. jejuni activates mitogen-activated protein kinases (MAPKs), extracellular signal-regulated kinase (ERK) and p38, leading to the induction of a potent pro-inflammatory cytokine interleukin-8 (IL-8) (MacCallum et al., 2005). IL-8 is an important pro-inflammatory cytokine of IECs and acts as a chemotactic factor of immune cells. However, it is hypothesized that induction IL-8 from human IECs which is found to correlate with an increase in circulating neutrophils to the site of infection can inadvertently exacerbate the classical acute inflammatory symptoms. C. jejuni induction of Erk and p38 signaling pathways is dependent on bacterial de novo protein synthesis and a functional flagellum (Jin et al., 2003; Watson and Galan, 2005).
C. jejuni Flagella: Function and Virulence
C. jejuni produces two polar flagella at each pole of the cell, termed as amphitrichous flagellation. C. jejuni flagella is a multifunctional organelle which enables the bacterium to avoid hostile environments including forceful peristalsis and expulsion from the GI tract. C. jejuni flagella also enable the bacterium to penetrate the viscous mucosa lining of the human IECs, and to reach the distal ileum, jejunum and colon. Thus, C. jejuni flagella promotes bacteria motility, chemotaxis and avian colonization. Besides mediating these virulence attributes, C. jejuni flagellar also promotes adhesion and invasion into human IECs in vitro (Black et al., 1988; Grant et al., 1993; Szymanski et al., 1995; Konkel et al., 1999), biofilm formation (Svensson et al., 2014) and non-flagella protein export. The latter enables C. jejuni to secrete ∼18 putative virulence-associated proteins termed Campylobacter invasion antigens (Cia) (Konkel et al., 2004; Christensen et al., 2009). Some of C. jejuni Cia proteins are required for invading human IECs in vitro, for instance CiaC plays a role in invasion whereas CiaI is required for intracellular survival in human IECs (Buelow et al., 2011; Neal-McKinney and Konkel, 2012). Interestingly study showed CiaD involves in maximal activation of the MAP kinase signaling pathways Erk 1/2 and p38 resulting in the secretion of IL-8 (Samuelson et al., 2013).
C. jejuni flagella synthesis and glycan modification involves over 50 flagellum-related genes. The flagellum is composed of three major parts, the basal body, which crosses the bacterial cell membrane, as well as a flagellar-associated cytoplasmic ring, the hook complex and the flagellar filament. Debates had focused on finding relationships between C. jejuni flagellum, motility, colonization and secretion. C. jejuni flagellar filament contributes to bacterial motility (Wassenaar et al., 1991; Guerry et al., 1991), adherence and colonization. The flagellar filament is composed of subunits of FlaA and FlaB proteins. C. jejuni flagellin proteins are O-linked glycosylated and the O-linked glycosylation is specific to the serine and threonine residues on a flagellin subunit which is modified by pseudaminic acid (Pse) and derivatives containing acetyl and acetamindino groups (PseAcOAc or PseAm, respectively (Thibault et al., 2001; Schirm et al., 2005). Sometimes C. jejuni flagellin subunits are modified with legionaminic acid (Leg), moieties (Logan et al., 2009; Schoenhofen et al., 2009; Howard et al., 2009). C. jejuni flagellar subunit FlaA rather than FlaB is essential for C. jejuni motility. This is supported by evidence that showed a mutation of the flaA gene led to the generation of non-flagellated and non-motile cells (Nuijten et al., 1990; Wassenaar et al., 1991). By contrast, the mutation of flaB, has no impact on C. jejuni flagella synthesis and motility. These findings suggest that FlaA protein, rather than motility, is essential for C. jejuni optimal colonization in chickens (Wassenaar et al., 1993). However, subsequent studies have identified C. jejuni mutant with normal but paralyzed flagella that is also non-motile and had a reduced ability to colonize chickens (Yao et al., 1994). The role of C. jejuni flagella in chicken colonization is further confirmed through mutation of the flagellar motor genes MotA and MotB which are essential for the rotation of the flagella. A motAB mutant produced non-motile cells with a full-length flagellum that is unable to rotate, thus unable to colonize chickens (Hendrixson and DiRita, 2004). Other C. jejuni flagella genes that have been studied include the flagellar sigma factor σ28 (fliA) and the alternative sigma factor σ54 (rpoN). These two sigma factors regulate a large number of genes that are responsible for the expression and function of C. jejuni flagella. For example, sigma σ28 is known to regulate the major flagellin gene flaA and some other late flagellar genes which control synthesis of proteins forming motor and chemotaxis proteins. On the other hand, C. jejuni σ54 involves the transcription of genes encoding for the hook, basal body, and minor flagellin flaB. In the context of host colonization and infection, mutation of σ54 (rpoN) gene results a non-motile cells that are unable to colonize chickens (Fernando et al., 2007), adhere to and invade into human IECs in vitro (Wassenaar et al., 1991). Also, C. jejuni flagellar functions as an organelle to secrete flagellar co-expressed determinants (Feds) which are required for efficient invasion of human IECs in vitro (Song et al., 2004; Barrero-Tobon and Hendrixson, 2012). A unique feature of C. jejuni flagellar filament is its mechanism to escape immune interaction with Toll-like receptor 5 (TLR5). TLR5s are found at the basolateral side of the human IECs and recognize a highly conserved epitope in bacterial flagellin. However, C. jejuni flagellar filament evades TLR5 activation because it fails to make complementary contacts with the TLR5 LRR9 loop (Song et al., 2017). This is attributed to sequence divergence of C. jejuni flagellin particularly the highly conserved epitope found in most γ-proteobacteria and Firmicutes bacterial flagellin. Recently, specific amino acids found in C. jejuni flagellar filament have been shown to mediate weakened binding to human TLR5 (Kreutzberger et al., 2020).
C. jejuni Capsular Polysaccharide (CPS)
The first evidence of a CPS at the surface of C. jejuni was reported in 2001 (Karlyshev et al., 2001). C. jejuni CPS is found on the outermost layer of the cell surface of the bacterium and it is composed of a rare structure of diverse repeating units of sugars (Karlyshev et al., 2005; McNally et al., 2005; Gilbert et al., 2007). C. jejuni CPS possess a heptoses sugar with an unusual configuration (e.g., ido, gulo, and altro) and nonstoichiometric modifications on the sugars, including ethanolamine, aminoglycerol, and O-methyl phosphoramidate (MeOPN). Unsurprisingly, C. jejuni CPS is the major sero-determinant of the Penner serotyping scheme of C. jejuni strains (Karlyshev et al., 2000). Currently, there are more than 47 different C. jejuni Penner serotypes of the bacterial CPS with some forming related serotype complexes (Poly et al., 2015). The structural variations of C. jejuni CPS reflects differences in the genetic content of the genomic locus that drives CPS biosynthesis (Karlyshev et al., 2005). C. jejuni CPS contains homopolymeric tracts which are prone to phase variation. As expected, homopolymeric tracts allow a rapid on/off switching of the C. jejuni CPS genes resulting in variations in CPS arrangements even in C. jejuni isolates that have identical gene contents. In addition to the phase variation observed in CPS sugar composition, C. jejuni CPS is also modified with ethanolamine, glycerol, and nonstoichiometric MeOPN modifications in approximately 75% of C. jejuni strains (Thota et al., 2018).
C. jejuni CPS plays a role in bacteria pathogenicity (Guerry et al., 2012; Bolton, 2015). C. jejuni CPS is required to resist complement-mediated killing (Bacon et al., 2001; Keo et al., 2011), invade into human IECs in vitro (Bachtiar et al., 2007; Corcionivoschi et al., 2009), colonization of chickens (Jones et al., 2004), and diarrheal disease in ferrets (Bacon et al., 2001). Consistently, the nonstoichiometric modification of CPS with MeOPN has also been demonstrated to be essential for complement resistance. The role of CPS in C. jejuni resistance to complement-mediated killing is supported by evidence showing C. jejuni expressing full CPS structure but lacking MeOPN, displayed the same pattern of serum killing as a nonencapsulated kpsM mutant, which lacked CPS. Also, study, using Galeria mellonella larvae infection model demonstrated C. jejuni expressing full CPS but lacking specific MeOPN modification to be significantly attenuated in virulence (Champion et al., 2010). This same study suggested the structure of the MeOPN moiety has a remarkable similarities to the active structures of organophosphorous pesticides (McNally et al., 2007), therefore, the virulence attenuation of C. jejuni expressing full CPS but lacking specific MeOPN may be due to a consequence of toxicity provided by the MeOPN. However, from virulence perspective, the role of C. jejuni CPS in serum resistance is still unclear as C. jejuni induces human β-defensins 2 and 3 (hBD2 and hBD3) from human IECs in vitro (Zilbauer et al., 2005).
C. jejuni Putative Adhesins
Adhesins play an important role in the pathogenesis of bacteria to adhere, colonize, and invade into hosts. C. jejuni adherence to human IECs in vitro involves putative adhesins decorated on its outer membrane (OM) surface. C. jejuni adhesins seem to have alternate primary functions, yet some can target the same host receptor such as fibronectin. Once C. jejuni adheres to fibronectin on the basolateral side of human IECs, it is preceded by secondary steps that orchestrate cellular invasion (Konkel et al., 2020). The most highly investigated adhesins in C. jejuni that exist almost in mutually exclusive fashion are Campylobacter adhesion to fibronectin (CadF) and fibronectin-like protein A (FlpA). C. jejuni adhesins (CadF and FlpA) are highly conserved among C. jejuni strains. CadF and FlpA proteins are important for C. jejuni adherence to human IECs and colonization of chickens (Konkel et al., 2020). A C. jejuni cadF mutant displays reduced ability to adhere to human IECs and chicken hepatoma cell line, LMH cells. C. jejuni cadF mutant is also unable to adhere to immobilized fibronectin (Talukdar et al., 2020). C. jejuni FlpA also promotes C. jejuni adherence to human IECs in vitro and plays a role in C. jejuni colonization of chickens (Flanagan et al., 2009; Konkel et al., 2010; Larson et al., 2013). There are additional C. jejuni surface-exposed adhesins, such as Campylobacter adhesion protein A (CapA), PEB1 (Kervella et al., 1993; Pei et al., 1998) and PEB4 (Asakura et al., 2007). These adhesins which also play a role in C. jejuni adherence to human and chicken IECs in vitro represent the multifactorial ability of C. jejuni virulence mechanisms. However, study suggested that PEB1 is not required for adhering to chicken LMH cells but rather as a transporter of amino acids aspartate and glutamate (Leon-Kempis Mdel et al., 2006). Unfortunately, an important gap in our current knowledge is the lack of mechanistic insight as to how C. jejuni orchestrates adherence steps to IECs. This is due in part to the fact that some of the adhesins identified to date display an overlap in binding mechanisms, a factor that confounds straightforward analysis of C. jejuni adhesion mechanisms. It is hypothesized that these C. jejuni different adhesins are required in the multiple steps of infection. First, to adhere to the mucosal layer at the luminal side of human IECs and then to adhere to the fibronectin receptor at the basolateral side of IECs.
Other C. jejuni Outer Membrane Channels
C. jejuni produces numerous virulence and/or fitness proteins that function as major outer membrane proteins (MOMPs). Two of the most well characterized MOMPs in C. jejuni are MOMP and OMP50. C. jejuni MOMP is also referred to as PorA. In contrast to E. coli, C. jejuni possesses only one MOMP that is present in all isolates and is highly (but not absolutely) conserved in other Campylobacters (Ferrara et al., 2016). C. jejuni, MOMP, is a 44-kDa protein, with sequence signature typical of β-barrel porin seen in other enteropathogens (Amako et al., 1996; Ferrara et al., 2016). C. jejuni, MOMP is relatively well characterized compared to OMP50. As might be expected, considering its association with the outer surface of the bacterial cell, C. jejuni MOMP exhibits substrate selectivity and functions as a control channel for the entry/exit of nutrients and other specific molecules (Dhanasekar et al., 2017). Mutation of porA have been thought to be lethal due to critical structural and transport functions. However, inactivation of porA enhances sensitivity to certain hydrophilic antibiotics (Iovine, 2013). Unlike MOMP, which is present in most Campylobacters, Omp50 is only found in C. jejuni and C. lari strains, but not in C. coli (Dedieu et al., 2008). The synthesis of Omp50 is tightly regulated by the host microenvironment. For example, C. jejuni Omp50 is down-regulated in chicken cecum and up-regulated in rabbit ileal loop (Stintzi et al., 2005; Woodall et al., 2005). Mutation of Omp50 substantially reduced C. jejuni motility and invasion, while it also involves bacterium decreased Nox1-dependent ROS generation (Corcionivoschi et al., 2012).
C. jejuni Putative Proteases: New Perspective in Virulence Involvement
Recent characterization of C. jejuni putative proteases represent an important step forward in the efforts to dissect C. jejuni pathogenesis. As opposed to traditional candidate-mutant experimental approaches, a proteomics analysis coupled with enzymatic-based virulence profiling (EBVP) have shed light on the specific role of C. jejuni putative proteases in adhesion to and invasion into human IECs in vitro. C. jejuni secretes outer membrane vesicles (OMVs) that contain three active serine proteases (HtrA, Cj0511, and Cj1365c) (Elmi et al., 2012). The mechanism responsible for the abundance of these serine proteases in OMVs remains elusive. However, C. jejuni proteases have been demonstrated to contribute targeted damage to human IECs in vitro (Elmi et al., 2016). Treatment of human IECs with active protease result in cleavage of IECs tight and adherens junction proteins, namely occludin and E‐cadherin. The targeted proteolytic activity of C. jejuni OMVs also enhance C. jejuni adhesion to and invasion into IECs in vitro (Elmi et al., 2016). Moreover, follow-up study has shown that bile salt sodium taurocholate (ST) upregulates C. jejuni expression of htrA, Cj0511, Cj1365, and the cdtABC operon, highlighting the importance of bacterium adaptation to host metabolites (Elmi et al., 2018). Furthermore, recent study has demonstrated that physiological concentrations of ST regulates C. jejuni OMVs production through changes in expression of the maintenance of lipid asymmetry (MLA) pathway (Davies et al., 2019). Although most of the examples discussed above had focused on the role of serine proteases in virulence, it should be remembered that C. jejuni OMVs also contain a cocktail of virulence and fitness factors, including stress response enzymes, adhesins, CDT, lipoproteins and other metalloproteases, which also play an important role in bacterial virulence. Thus, suggestions have been raised that C. jejuni OMVs might also function as fitness and survival factors, allowing the bacterium to adapt new niches, adhere to surfaces, translocate rapidly across IECs, and resist antibiotics and other deleterious circumstances.
C. jejuni Fitness and Virulence Factors: Role in Stress Adaptation, Temperature, Nutrient Sensing, and Metabolic Rewiring
As C. jejuni transitions from nutritionally rich ceca in the GI tract of chickens to accidentally infect humans, the bacterium faces formidable stresses. Here, the term “stress” refers to environmental and human host stresses that reduce C. jejuni fitness or negatively impact on its virulence. Unlike other entero-pathogens, C. jejuni does not have homologs of the classical stress response regulators such as SoxRS and OxyR found in E. coli and Salmonella spp. respectively. SoxRS regulates response to redox-active compounds while OxyR responds to hydrogen peroxide (Nunoshiba et al., 1992; Zheng et al., 1998). In addition, C. jejuni lacks transcription regulators such as cold shock protein A (CspA) and leucine-responsive regulatory protein (Lrp) (Calvo and Matthews, 1994; Murphy et al., 2006; Keto-Timonen et al., 2016). Besides, C. jejuni does not possess the classical alternative sigma factors such as RpoS (σ38) although it has limited sigma factors including RpoD (σ70), RpoN (σ54), and RpoF/FliA (σ28). Interestingly, C. jejuni possesses unique and yet unresolved mechanisms to survive under various stress conditions. C. jejuni utilizes OmpR‐type response regulators such as Campylobacter oxidative stress regulator (CosR) (Hwang et al., 2011), peroxide-sensing regulator (PerR) (Palyada et al., 2009) and Multiple Antibiotic Resistance Regulator, MarR‐type regulators designated for response to peroxide stress (Gundogdu et al., 2016). C. jejuni CosR is a pleiotropic regulator that controls the expression of genes involved in various cellular processes, especially genes that involve in macromolecule biosynthesis, metabolism, and oxidative stress response (Kim et al., 2015b). The genes that CosR regulates mostly encode for stress response-related proteins such as the DNA binding protein from starved cells (Dps), rubredoxin oxidoreductase/rubrerythrin (Rrc), alkyl hydroperoxide reductase (AhpC), and superoxide dismutase (SodB). On the other hand PerR, non-OxyR-dependent regulator, controls transcription of peroxide as well as the superoxide defense genes particularly under oxidative stress conditions. For instance, perR mutation abrogates the transcriptional response of ahpC, katA, and sodB to oxidants (Kim et al., 2015a).
C. jejuni also possesses global transcriptional regulators such as carbon starvation regulator (CsrA), ortholog of the E. coli global posttranscriptional regulator CsrA. In addition, C. jejuni has two-component regulatory systems (TCRS) such as Campylobacter planktonic growth regulator (CprRS) (Svensson et al., 2015; El Abbar et al., 2019). Mutation of csrA results in C. jejuni cells with altered motility, biofilm formation, adherence to and invasion of human IECs cells and resistance to oxidative stress (Fields and Thompson, 2008). CprRS is two‐component systems regulator typically consisting of a sensor kinase and a response regulator. The CprR response regulator is essential and mutation to the cprR, is lethal to C. jejuni, but a cprS mutation, results in decreased expression of SodB, Rrc and LuxS. C. jejuni also possesses a ferric uptake regulator (Fur) to control the expression of a range of oxidative stress genes, to prevent the build-up of toxic levels of iron within the cell (Butcher et al., 2012). In addition to the stress-responsive regulators, C. jejuni KatA and SodB proteins play critical roles in detoxification, SodB detoxifies free radicals while KatA contributes for the detoxification of H2O2 (Atack and Kelly, 2009). SodB also contributes to C. jejuni chicken colonization and intracellular survival in human IECs in vitro (Palyada et al., 2009; Novik et al., 2010). C. jejuni cell surface structures such as flagella, CPS, LOS and OM also can act at the interface between the bacterium and the extracellular environment. These cellular surface structures assist C. jejuni to sense environmental and host stresses, in principle, inducing a collective response to protect the bacterium from damage caused by stresses.
Environmental Stress Survival and Adaptation
In light of its relatively small genome (1.6–1.7 Mb), it remains enigmatic how C. jejuni senses, adapts and persists in diverse environmental stresses. C. jejuni requires optimal oxygen concentrations of approximately 5%–10% for growth; however, the bacterium can survive in the environment, which is rich in oxygen (ca. 21% O2). This variation in oxygen concentration constraints C. jejuni to rewire its physiology to adapt flexible metabolic pathways. The requirement of 5%–10% O2 for growth is governed by single class I-type Ribonucleotide Reductase (RNR) (Burnham and Hendrixson, 2018). This is an oxygen-dependent enzyme that catalyses the de novo conversion of ribonucleotides diphosphates (NDPs) to deoxyribonucleotides diphosphates (dNDPs), and therefore plays a pivotal role in maintaining C. jejuni synthesis of deoxyribonucleotide (dNTP). Besides, C. jejuni also possesses a highly-branched respiratory chain feature that facilitates the use of oxygen as an electron acceptor for one of two respiratory oxidases, cytochrome c oxidase (CcoNOQP), a cbb3-type cytochrome c oxidoreductase and a bd-type (CioAB or CydAB) quinol oxidase (Guccione et al., 2017; van der Stel and Wosten, 2019). The sensitivity of C. jejuni pyruvate: acceptor oxidoreductase (POR) and the TCA cycle 2-oxoglutarate: acceptor oxidoreductase (OOR) to oxygen has been suggested as one of the explanations of the so-called ‘C. jejuni-oxygen paradox’ - that is, why C. jejuni is unable to proliferate in aerobic environment. Also, atmospheric levels of oxygen inactivate C. jejuni L-serine dehydratase (SdaA), which catalyses the deamination of serine and converts serine into pyruvate which is further converted to acetyl CoA, which is oxidized via the TCA cycle to carbon dioxide and free energy. SdaA is essential for colonization of the avian gut (Velayudhan et al., 2004). The ability of C. jejuni to tolerate oxygen in the environment can also vary between strains. For instance, study has found a higher prevalence of some strain genotypes in environmental samples attributing these variations in oxygen tolerance (Champion et al., 2005; Bronowski et al., 2014). Besides, another study has reported atypical C. jejuni Bf strain that is oxygen tolerant (Rodrigues et al., 2015; Bronnec et al., 2016a). This strain has been demonstrated to have protective mechanisms against oxidative stress which is thought to be mediated by regulation of genes involved in oxidative stress response and biofilm formation (Bronnec et al., 2016b). Interestingly, recent assessment of C. jejuni phospholipidome profile has indicated that C. jejuni phospholipidome have an unusually high percentage of lysophospholipid. Lysophospholipids are small bioactive lipid molecules characterized by a single carbon chain and a polar head group. It is hypothesized lysophospholipid allows C. jejuni to be motile under low O2 conditions (Cao et al., 2020a). This is a significant observation considering the requirement of C. jejuni to adapt to the low oxygen deep in the mucus layer of the human GI tract. This could give C. jejuni competitive advantage when competing with other microbiota that colonize the mucosal layer as it transitions into the IECs. In addition, the ability of C. jejuni to sense environmental oxygen have been thought to correlate altering its membrane lipid composition which could be crucial for biofilm formation.
C. jejuni Biofilm: Environmental Adaptation and Persister Phenomena
C. jejuni adaptation to an oxygen-rich environment such as contaminated freshwater, poultry meat or raw milk can be attributed to the ability of the bacterium to form biofilms on different substrates. C. jejuni can attach and persist on a variety of abiotic and biotic surfaces, and several studies have reported on the viable but non-culturable (VBNC) state (Teh et al., 2014; Magajna and Schraft, 2015). C. jejuni cells switch to VBNC state to survive better under adverse environmental conditions. In the environments, C. jejuni is exposed to high oxygen tension, limited nutrient availability, heat, acidic pH, temperatures fluctuations and antimicrobials. These environmental constraints are known to stimulate increased C. jejuni biofilm formation to a relatively high level, supporting the proposal that C. jejuni forms biofilm as a survival strategy outside of the avian host. C. jejuni forms increased biofilm in oxygen-rich conditions compared to microaerobic conditions (Reuter et al., 2010). It is commonly agreed that all C. jejuni strains form biofilm, however, the ability of C. jejuni to form biofilm appears to be strain-dependent (Melo et al., 2017). Interestingly, C. jejuni mutant strains deficient in genes encoding for key oxidative stress resistance enzymes such as alkyl hydroperoxide reductase (AhpC) or C. jejuni’s sole catalase enzyme (KatA) have been shown to have an increased ability to form biofilm (Oh and Jeon, 2014). This is attributed to the accumulation of reactive oxygen species (ROS) which may serve as a trigger to increase the level of biofilm formed in response to increased oxidative stress. Overexpression of ahpC is correlated with decreased biofilm formation, and treatment of the ahpC mutant with antioxidants reduces biofilm formation (Oh and Jeon, 2014). C. jejuni lacks the classical two-component regulatory systems involved in biofilm formation found in other bacteria, such as GacSA in Pseudomonas aeruginosa, however, C. jejuni biofilm formation is thought to be under the control of a complex array of regulatory factors that respond to a variety of environmental signals. These complex regulatory factors include global regulator CsrA, Campylobacter oxidative stress regulator (CosR), stringent response regulator (SpotT) and CprRS, which have been shown to play an important role in biofilm formation in C. jejuni under aerobic conditions (Gaynor et al., 2005; Fields and Thompson, 2008; Svensson et al., 2015; El Abbar et al., 2019). Mutations of cosR, cprRS, and, spotT increase biofilm formation under aerobic conditions, while mutation of the gene encoding for global regulator (CsrA) decreases the ability of C. jejuni to form biofilms when grown in static culture as well as increased sensitivity to oxidative stresses (Fields and Thompson, 2008). Interestingly, in other enteric bacteria spoT mutation decreases biofilm formation (He et al., 2012). In C. jejuni, the mutation of spoT alters the expression of genes related to redox balance, metabolism, energy production, and conversion pathways while CosR, a key orphan regulator in the maturation of biofilm, has also been shown to affect the expression of the antimicrobial efflux pump CmeABC (Turonova et al., 2015). CprRS is two‐component systems regulator typically consisting of a sensor kinase and a response regulator. The CprR response regulator is essential and deletion of the cprS sensor kinase enhances biofilms. Current evidence suggests that CprRS likely regulates genes related to aspects of the C. jejuni surface structures (Svensson et al., 2015). The molecular mechanism of C. jejuni biofilm formation also appears to indirectly correlate with factors required for fitness and virulence. For instance, mutation of the flagella genes flaA, flaB and the cell surface modification genes pgp1 and waaF have been shown to increase biofilm formation (Reeser et al., 2007). This indicates that C. jejuni increases biofilm formation as a survival strategy during stress. Interestingly, a recent study suggests C. jejuni does not form biofilms under conditions encountered in the environment but attaches to surfaces or biofilms of other species (Teh et al., 2014; Teh et al., 2019). This is an attractive proposal supporting the notion that C. jejuni is a poor biofilm initiator, and is likely to form enhanced biofilms in a “mixed-species biofilm” with other bacteria such as P. aeruginosa, Enterococcus faecalis and Staphylococcus simulans.
C. jejuni Temperature Stress Adaptation
Temperature is a prominent signal used by many enteric pathogens. The strategies enteric pathogens use to sense temperature variation across space, hosts and time broadly acts as a mechanism to adjust bacterial survival and virulence. For C. jejuni, the transition from its primary chicken host (42°C) to the environment, the bacterium experiences temperature variation. This temperature variation confines proliferation and shifts C. jejuni physiology forcing the bacterium to coordinate fitness and virulence regulatory systems. It is puzzling that C. jejuni lacks classical RpoS homolog (Parkhill et al., 2000) and cold shock proteins (Oh et al., 2019), yet C. jejuni has the ability to survive in low and/or high nonpermissive temperature growth conditions before reaching human host. C. jejuni doesn’t also grow temperatures below ~ 30°C, however the bacterium survives temperature growth range between 4°C to 33°C (Hazeleger et al., 1998). C. jejuni survives better at 4°C in various biological milieu than at 25°C (Murphy et al., 2006). C. jejuni also survives in water, at low temperatures, for up to 4 months (Oberheim et al., 2020). The ability of C. jejuni to survive in cold temperatures is different among strains, with C. jejuni strains isolated from human infection being significantly more capable of prolonged survival at 4°C than poultry‐derived strains (Chan et al., 2001). Intriguingly C. jejuni also survives extreme freezing temperatures (−20°C) for several weeks (Bhaduri and Cottrell, 2004).
C. jejuni genes associated with oxygen tolerance, starvation and osmotic stress are essential for the bacterium to survive in the low temperature. This perplexing physiology of C. jejuni seems to be the bottleneck to the efforts aimed to eradicate the risk of C. jejuni to human health. The ability of C. jejuni to rapidly sense and adapt to cold temperature is largely driven at the transcriptional level (Bronowski et al., 2017). Studies focusing on human infections, use in vitro human IECs grown at 37°C to mimic the temperature that the bacteria encounters inside human host. C. jejuni ability to sense 37°C is crucial to optimize its fitness and adjust expression of its virulence genes. C. jejuni is more invasive into human IECs cultured at 37°C than IECs cultures at 42°C (Aroori et al., 2013). Although the exact mechanism of C. jejuni response to temperature stress is not yet explicitly known, changes in temperature are known to affect expression of bacterial heat shock proteins (HSP). C. jejuni possesses two-component regulatory systems (TCSs) such as reduced ability to colonize response regulator (RacRS). RacRS function to assist the bacteria to overcome stresses associated with heat shock response. In addition, C. jejuni RacR is required for avian colonization and growth while mutation of racR alters the expression of selected proteins in both temperature-dependent and independent manners (Hazeleger et al., 1998; Wouters et al., 2000).
C. jejuni Acid Stress Adaptation
C. jejuni grows at optimal pH range of 6.5–7.5, while it is also able to survive pH range as low as 5.5 and as high as 8.5. However, C. jejuni encounters acidic conditions either in the environment or within the gut of the various hosts that it colonizes. In the context of human infection, C. jejuni survives passage through the stomach, where the concentration of acid is high and the pH ranges 1.5–3.5. The molecular strategies that C. jejuni uses to sense, adapt and survive the luminal acid concentration in the stomach upon ingestion and within the phagosomes and phagolysosomes of human IECs is not currently known. However, C. jejuni tolerance to human GI tract luminal acid is important for disease development. So far, it is hypothesized C. jejuni lacks proteins required for acid tolerance such as urease protein found in Helicobacter pylori. However, it is intriguing that with low infectious dose of (500–800 bacteria), C. jejuni cells survive the gastric acid of the human stomach and continue down to reach the small intestine. Study has demonstrated some C. jejuni strains can survive acid exposure at pH 3.5 and above for up to 30 min (Le et al., 2012). Another study has suggested adaptation of C. jejuni to the luminal acid concentration in humans requires genes mediating various cellular processes, including those involved in motility, metabolism, stress response, DNA repair and surface polysaccharide biosynthesis (Reid et al., 2008). For instance, C. jejuni RpoN, a classical flagellar transcriptional regulator, which is historically known to play an important role in motility has been demonstrated to be important for the resistance of C. jejuni to various stresses including acid stress. This suggested flagella mediated motility is critical for both initial navigation through the acid environment in the GI tract lumen and mucus layer to IECs attachment. C. jejuni adaptation to low pH stress also involved the differential expression of genes involve in respiratory pathways, the upregulation of genes for phosphate transport, and the repression of energy generation and intermediary metabolism genes (Reid et al., 2008). Recent study that investigated acid-stressed adaptation of C. jejuni under iron-enriched conditions has shown the capacity of C. jejuni to survive acid stress is greatly enhanced in presence of iron (Askoura et al., 2020). However, limited information is available about the role which human host microbiota plays in the pathophysiology of C. jejuni adaptation in acidity along the gut, although it is evident that many species of the microbiota are able to generate metabolites that have bearing on the composition of GI tract luminal acidity. For example, lactate which is an organic acid that is found in the upper GI tract of human and avian species can act as a chemoattractant signal of C. jejuni (Bernalier-Donadille, 2010; Hofreuter, 2014).
C. jejuni Metabolic Sensing and Adaptation
While, as discussed above, C. jejuni has complex stress response mechanisms, its ability to resist stresses overlaps its ability to adapt to different metabolic requirements. C. jejuni sequenced strain NCTC11168 lacks the glycolytic enzymes glucokinase (Glk) and phosphofructokinase (PfkA) of the classical Embden-Meyerhof-Parnas (EMP) pathway (Parkhill et al., 2000; Guccione et al., 2008; Hofreuter, 2014). C. jejuni was once considered to be non-saccharolytic since C. jejuni sequenced strain NCTC11168 lacks genes encoding for the complete pentose phosphate (PPP) or Entner-Doudoroff (ED) pathway. Interestingly, few isolates of C. jejuni subsp. doylei encode a complete ED pathway which suggests the potential to catabolize glucose (Vegge et al., 2016; Garber et al., 2020). The inability to utilize glucose has necessitated C. jejuni to utilize amino acids such as serine, aspartate, glutamate, glutamine, proline and asparagine as carbon and energy sources (Stahl et al., 2012; Hofreuter, 2014; Szymanski, 2015). Most C. jejuni strains preferentially use serine, aspartate, glutamate, and proline, although certain C. jejuni strains can also utilize asparagine and glutamine (Thompson and Gaynor, 2008; van der Hooft et al., 2018). This unique ability to metabolize only a few amino acids allows the bacterium to utilize efficient strategies to include host nutrients into its anabolic processes, to fuel its metabolic pathways and to support its survival and adaptation in hosts with largely commensalism outcome in avian species or pathogenesis in humans. For instance, C. jejuni catabolism of serine and aspartate enhances the ability of the bacterium to colonize the avian gut (Hermans et al., 2011), while a C. jejuni mutant that is lacking an oxygen-labile serine dehydratase and unable to catabolize serine is demonstrated to be incapable of colonizing chickens (Velayudhan et al., 2004). Furthermore, C. jejuni rewires its metabolic requirements during avian colonization and human infection. C. jejuni has the ability to adopt an asaccharolytic lifestyle, likely as a strategy to evade microbiome competition. It is known that certain C. jejuni strains metabolize sugars such as L‐fucose (Stahl et al., 2011). These C. jejuni strains possess an operon for L‐fucose utilisation which until recently has been known to be limited to some C. coli and C. jejuni subsp. doylei strains. L-fucose acts as a chemoattractant for C. jejuni (Dwivedi et al., 2016). Interestingly, C. jejuni binds to α1, 2-fucosylated glycans, however the L-fucose catabolism is not essential for C. jejuni colonization of avian species (Muraoka and Zhang, 2011; Stahl et al., 2011). Furthermore, C. jejuni lacks fucosidase enzyme which is essential for the release of the L-fucose from glycosylated host proteins such as mucin. A study recently demonstrated that C. jejuni fucose positive strain utilisation of L-fucose is dependent on the fucosidase activity of the gastrointestinal bacterium Bacteriodes fragilis (Garber et al., 2020). This same study also revealed that C. jejuni becomes more invasive toward human Caco-2 cells in the presence of an exogenous fucosidases from B. fragilis.
Recently, examining the idea of a host nutritional role in C. jejuni adaptation and pathogenesis, studies showed that C. jejuni senses and utilizes catabolic end products of the intestinal microbiota such as short-chain fatty acids (SCFAs) butyrate and acetate, CO2-derived hydrogen carbonate, and free amino acids and di-/or oligopeptides, which are released by microbiota from dietary or endogenous proteins (Gao et al., 2017). The ability of C. jejuni to sense SCFAs positively regulates many C. jejuni amino acids uptake and catabolism systems that are essential for host colonization. SCFAs are found in abundance in the lower regions of the intestinal tracts of avian species and humans where they play a major role in host physiology through nutritional, regulatory, and immunomodulatory functions. However, in the context of C. jejuni avian and human colonization, the abundance of butyrate and acetate in the lower GI tract provides the bacterium with a competitive advantage to thrive in this niche (Burnham and Hendrixson, 2018). A prevailing belief is that C. jejuni has the ability to spatially differentiate between sections of the GI tract by sensing the presence of acetate and butyrate, and thereby modifying the transcription of its colonization factors (Goodman et al., 2020). This enables C. jejuni to obtain sufficient nutrients and resources to allow for optimal survival and persistence in both avian and human intestinal tracts. C. jejuni specifically senses butyrate via a noncanonical TCS termed BumSR (Goodman et al., 2020). BumS functions as a phosphatase, via a noncanonical mechanism for signal transduction in place of a sensor kinase, to control the activity of the cognate BumR response regulator. BumS phosphorylates BumR in response to the presence of butyrate. C. jejuni genes known to be induced after sensing butyrate and acetate include genes encoding for nutrient acquisition systems, energy generation pathways, and colonization factors (Goodman et al., 2020). In addition, acetate which is more abundant in the gut is preferred metabolite for C. jejuni once the rate-limiting step of carbohydrate metabolism is surpassed in stationary phase. C. jejuni also catabolizes organic acids such as lactate which is abundant in the upper gut of avian hosts (Luethy et al., 2017).
Conclusions and Future Directions
Recent developments in the understanding of C. jejuni pathogenesis have combined several experimental approaches that link the functional characterization of various putative genes. Although this is important, characterizing C. jejuni virulence and fitness factors requires an integrative approach. In the future, an ideal experiment should involve the use of single-gene inactivations and phenotypic assays, incorporated with integrative multi-omics approach including, transcriptomics, proteomics and metabolomics. This should reveal comprehensive findings that would contribute to the characterization of C. jejuni pathogenesis. This approach will also guide us to re-focus on re-characterization of many C. jejuni virulence-associated genes that have not yet been fully characterized. From our perspective, the incorporation of integrative multi-omics and phenotypic assays in C. jejuni research promises enormous potential. However, there are many challenges and thus, opportunities for further development of experiments involving multi-omics technology. Also, future studies of C. jejuni should include refining, optimisation and normalization of experimental design and protocols that represent ideal settings for C. jejuni and host cells, allowing researchers to reproduce data. Unsurprisingly, there are a plethora of C. jejuni studies that use experimental approaches that give an insight into the selected role of C. jejuni putative virulence associate genes. For instance, in stress survival, adhesion, invasion and intracellular survival, however, few studies provide information about the function of such putative genes. Also, integration of C. jejuni virulence characterizations with spatial analysis at the various time point and C. jejuni strains variability is needed to improve our understanding of C. jejuni pathogenesis.
Author’s Note
For the purpose of this review, we define a virulence factor as a protein (such as a toxin) or macromolecular structure (such as flagellum) that contribute to the ability of the bacteria to cause disease and a fitness factor as a protein or macromolecular structure that, while not required for virulence, offers a competitive advantage during infection.
Author Contributions
AE: Conceived and designed the structure of the manuscript; AE Created Figure 1; AE, OG, and FN: Wrote the manuscript; AE, BW, ND, OG, and FN: Read and edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
FN and BW gratefully acknowledge the support of the Biotechnology and Biological Sciences Research Council Institute Strategic Programme BB/R012504/1 constituent project BBS/E/F/000PR10349.
Footnotes
References
Abouelhadid S., North S. J., Hitchen P., Vohra P., Chintoan-Uta C., Stevens M., et al. (2019). Quantitative Analyses Reveal Novel Roles for N-Glycosylation in a Major Enteric Bacterial Pathogen. mBio 10. doi: 10.1128/mBio.00297-19
Abouelhadid S., Raynes J., Bui T., Cuccui J., Wren B. W. (2020). Characterization of Posttranslationally Modified Multidrug Efflux Pumps Reveals an Unexpected Link between Glycosylation and Antimicrobial Resistance. mBio 11. doi: 10.1128/mBio.02604-20
Aidley J., Wanford J. J., Green L. R., Sheppard S. K., Bayliss C. D. (2018). PhasomeIt: an ‘omics’ approach to cataloguing the potential breadth of phase variation in the genus Campylobacter. Microb. Genom. 4. doi: 10.1099/mgen.0.000228
Amako K., Wai S. N., Umeda A., Shigematsu M., Takade A. (1996). Electron microscopy of the major outer membrane protein of Campylobacter jejuni. Microbiol. Immunol. 40, 749–754. doi: 10.1111/j.1348-0421.1996.tb01136.x
Aroori S. V., Cogan T. A., Humphrey T. J. (2013). The effect of growth temperature on the pathogenicity of Campylobacter. Curr. Microbiol. 67, 333–340. doi: 10.1007/s00284-013-0370-1
Artursson K., Schelin J., Thisted Lambertz S., Hansson I., Olsson Engvall E. (2018). Foodborne pathogens in unpasteurized milk in Sweden. Int. J. Food Microbiol. 284, 120–127. doi: 10.1016/j.ijfoodmicro.2018.05.015
Asakura H., Yamasaki M., Yamamoto S., Igimi S. (2007). Deletion of peb4 gene impairs cell adhesion and biofilm formation in Campylobacter jejuni. FEMS Microbiol. Lett. 275, 278–285. doi: 10.1111/j.1574-6968.2007.00893.x
Ashgar S. S., Oldfield N. J., Wooldridge K. G., Jones M. A., Irving G. J., Turner D. P., et al. (2007). CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut. J. Bacteriol. 189, 1856–1865. doi: 10.1128/JB.01427-06
Askoura M., Youns M., Halim Hegazy W. A. (2020). Investigating the influence of iron on Campylobacter jejuni transcriptome in response to acid stress. Microb. Pathog. 138, 103777. doi: 10.1016/j.micpath.2019.103777
Atack J. M., Kelly D. J. (2009). Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol. 4, 677–690. doi: 10.2217/fmb.09.44
Bachtiar B. M., Coloe P. J., Fry B. N. (2007). Knockout mutagenesis of the kpsE gene of Campylobacter jejuni 81116 and its involvement in bacterium-host interactions. FEMS Immunol. Med. Microbiol. 49, 149–154. doi: 10.1111/j.1574-695X.2006.00182.x
Backert S., Hofreuter D. (2013). Molecular methods to investigate adhesion, transmigration, invasion and intracellular survival of the foodborne pathogen Campylobacter jejuni. J. Microbiol. Methods 95, 8–23. doi: 10.1016/j.mimet.2013.06.031
Backert S., Boehm M., Wessler S., Tegtmeyer N. (2013). Transmigration route of Campylobacter jejuni across polarized intestinal epithelial cells: paracellular, transcellular or both? Cell Commun. Signal 11, 72. doi: 10.1186/1478-811X-11-72
Bacon D. J., Alm R. A., Burr D. H., Hu L., Kopecko D. J., Ewing C. P., et al. (2000). Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68, 4384–4390. doi: 10.1128/IAI.68.8.4384-4390.2000
Bacon D. J., Szymanski C. M., Burr D. H., Silver R. P., Alm R. A., Guerry P. (2001). A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40, 769–777. doi: 10.1046/j.1365-2958.2001.02431.x
Bailey R. A., Kranis A., Psifidi A., Watson K. A., Rothwell L., Hocking P. M., et al. (2018). Colonization of a commercial broiler line by Campylobacter is under limited genetic control and does not significantly impair performance or intestinal health. Poult. Sci. 97, 4167–4176. doi: 10.3382/ps/pey295
Barrero-Tobon A. M., Hendrixson D. R. (2012). Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol. Microbiol. 84, 352–369. doi: 10.1111/j.1365-2958.2012.08027.x
Bernalier-Donadille A. (2010). [Fermentative metabolism by the human gut microbiota]. Gastroenterol. Clin. Biol. 34 (Suppl 1), S16–S22. doi: 10.1016/S0399-8320(10)70016-6
Bhaduri S., Cottrell B. (2004). Survival of cold-stressed Campylobacter jejuni on ground chicken and chicken skin during frozen storage. Appl. Environ. Microbiol. 70, 7103–7109. doi: 10.1128/AEM.70.12.7103-7109.2004
Black R. E., Levine M. M., Clements M. L., Hughes T. P., Blaser M. J. (1988). Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157, 472–479. doi: 10.1093/infdis/157.3.472
Blaser M. J. (1997). Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176 (Suppl 2), S103–S105. doi: 10.1086/513780
Bolton D. J. (2015). Campylobacter virulence and survival factors. Food Microbiol. 48, 99–108. doi: 10.1016/j.fm.2014.11.017
Brondsted L., Andersen M. T., Parker M., Jorgensen K., Ingmer H. (2005). The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl. Environ. Microbiol. 71, 3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005
Bronnec V., Haddad N., Cruveiller S., Hernould M., Tresse O., Zagorec M. (2016a). Draft Genome Sequence of Campylobacter jejuni Bf, an Atypical Strain Able To Grow under Aerobiosis. Genome Announc. 4. doi: 10.1128/genomeA.00120-16
Bronnec V., Turonova H., Bouju A., Cruveiller S., Rodrigues R., Demnerova K., et al. (2016b). Adhesion, Biofilm Formation, and Genomic Features of Campylobacter jejuni Bf, an Atypical Strain Able to Grow under Aerobic Conditions. Front. Microbiol. 7, 1002. doi: 10.3389/fmicb.2016.01002
Bronowski C., James C. E., Winstanley C. (2014). Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 356, 8–19. doi: 10.1111/1574-6968.12488
Bronowski C., Mustafa K., Goodhead I., James C. E., Nelson C., Lucaci A., et al. (2017). Campylobacter jejuni transcriptome changes during loss of culturability in water. PloS One 12, e0188936. doi: 10.1371/journal.pone.0188936
Buchanan C. J., Webb A. L., Mutschall S. K., Kruczkiewicz P., Barker D. O. R., Hetman B. M., et al. (2017). A Genome-Wide Association Study to Identify Diagnostic Markers for Human Pathogenic Campylobacter jejuni Strains. Front. Microbiol. 8, 1224. doi: 10.3389/fmicb.2017.01224
Buelow D. R., Christensen J. E., Neal-Mckinney J. M., Konkel M. E. (2011). Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol. Microbiol. 80, 1296–1312. doi: 10.1111/j.1365-2958.2011.07645.x
Burnham P. M., Hendrixson D. R. (2018). Campylobacter jejuni: collective components promoting a successful enteric lifestyle. Nat. Rev. Microbiol. 16, 551–565. doi: 10.1038/s41579-018-0037-9
Butcher J., Sarvan S., Brunzelle J. S., Couture J. F., Stintzi A. (2012). Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U. S. A. 109, 10047–10052. doi: 10.1073/pnas.1118321109
Cain J. A., Dale A. L., Niewold P., Klare W. P., Man L., White M. Y., et al. (2019). Proteomics Reveals Multiple Phenotypes Associated with N-linked Glycosylation in Campylobacter jejuni. Mol. Cell Proteomics 18, 715–734. doi: 10.1074/mcp.RA118.001199
Calvo J. M., Matthews R. G. (1994). The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58, 466–490. doi: 10.1128/MR.58.3.466-490.1994
Cao X., Brouwers J., Van Dijk L., Van De Lest C. H. A., Parker C. T., Huynh S., et al. (2020a). Dataset of the phospholipidome and transcriptome of Campylobacter jejuni under different growth conditions. Data Brief 33, 106349. doi: 10.1016/j.dib.2020.106349
Cao X., Brouwers J. F. H. M., Van Dijk L., van de Lest C. H. A., Parker C. T., Huynh S., et al. (2020b). Pathogen Campylobacter jejuni: Lysophosholipids Are Required for Motility atLow Oxygen Availability. J. Mol. Biol. 432 (19), 5244-5258. doi: 10.1016/j.jmb.2020.07.012
Champion O. L., Gaunt M. W., Gundogdu O., Elmi A., Witney A. A., Hinds J., et al. (2005). Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc. Natl. Acad. Sci. U. S. A. 102, 16043–16048. doi: 10.1073/pnas.0503252102
Champion O. L., Karlyshev A. V., Senior N. J., Woodward M., La Ragione R., Howard S. L., et al. (2010). Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201, 776–782. doi: 10.1086/650494
Chan K. F., Le Tran H., Kanenaka R. Y., Kathariou S. (2001). Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4 degrees C). Appl. Environ. Microbiol. 67, 4186–4191. doi: 10.1128/AEM.67.9.4186-4191.2001
Christensen J. E., Pacheco S. A., Konkel M. E. (2009). Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol. Microbiol. 73, 650–662. doi: 10.1111/j.1365-2958.2009.06797.x
Corcionivoschi N., Clyne M., Lyons A., Elmi A., Gundogdu O., Wren B. W., et al. (2009). Campylobacter jejuni cocultured with epithelial cells reduces surface capsular polysaccharide expression. Infect. Immun. 77, 1959–1967. doi: 10.1128/IAI.01239-08
Corcionivoschi N., Alvarez L. A., Sharp T. H., Strengert M., Alemka A., Mantell J., et al. (2012). Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe 12, 47–59. doi: 10.1016/j.chom.2012.05.018
Corcionivoschi N., Gundogdu O., Moran L., Kelly C., Scates P., Stef L., et al. (2015). Virulence characteristics of hcp (+) Campylobacter jejuni and Campylobacter coli isolates from retail chicken. Gut Pathog. 7, 20. doi: 10.1186/s13099-015-0067-z
Cróinín T. O., Backert S. (2012). Host epithelial cell invasion by Campylobacter jejuni: trigger or zipper mechanism? Front. Cell Infect. Microbiol. 2, 25. doi: 10.3389/fcimb.2012.00025
Davies C., Taylor A. J., Elmi A., Winter J., Liaw J., Grabowska A. D., et al. (2019). Sodium Taurocholate Stimulates Campylobacter jejuni Outer Membrane Vesicle Production via Down-Regulation of the Maintenance of Lipid Asymmetry Pathway. Front. Cell. Infect. Microbiol. 9, 177. doi: 10.3389/fcimb.2019.00177
Dedieu L., Pages J. M., Bolla J. M. (2008). The omp50 gene is transcriptionally controlled by a temperature-dependent mechanism conserved among thermophilic Campylobacter species. Res. Microbiol. 159, 270–278. doi: 10.1016/j.resmic.2008.03.002
Dhanasekar N. N., Aliouane S., Winterhalter M., Pages J. M., Bolla J. M. (2017). Peptide translocation across MOMP, the major outer membrane channel from Campylobacter jejuni. Biochem. Biophys. Rep. 11, 79–83. doi: 10.1016/j.bbrep.2017.06.007
Dhillon A. S., Shivaprasad H. L., Schaberg D., Wier F., Weber S., Bandli D. (2006). Campylobacter jejuni infection in broiler chickens. Avian Dis. 50, 55–58. doi: 10.1637/7411-071405R.1
Dwivedi R., Nothaft H., Garber J., Xin Kin L., Stahl M., Flint A., et al. (2016). L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 101, 575–589. doi: 10.1111/mmi.13409
El Abbar F. M., Li J., Owen H. C., Daugherty C. L., Fulmer C. A., Bogacz M., et al. (2019). RNA Binding by the Campylobacter jejuni Post-transcriptional Regulator CsrA. Front. Microbiol. 10, 1776. doi: 10.3389/fmicb.2019.01776
Elmi A., Watson E., Sandu P., Gundogdu O., Mills D. C., Inglis N. F., et al. (2012). Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 80, 4089–4098. doi: 10.1128/IAI.00161-12
Elmi A., Nasher F., Jagatia H., Gundogdu O., Bajaj-Elliott M., Wren B., et al. (2016). Campylobacter jejuni outer membrane vesicle-associated proteolytic activity promotes bacterial invasion by mediating cleavage of intestinal epithelial cell E-cadherin and occludin. Cell Microbiol. 18, 561–572. doi: 10.1111/cmi.12534
Elmi A., Dorey A., Watson E., Jagatia H., Inglis N. F., Gundogdu O., et al. (2018). The bile salt sodium taurocholate induces Campylobacter jejuni outer membrane vesicle production and increases OMV-associated proteolytic activity. Cell Microbiol. 20. doi: 10.1111/cmi.12814
Facciola A., Riso R., Avventuroso E., Visalli G., Delia S. A., Lagana P. (2017). Campylobacter: from microbiology to prevention. J. Prev. Med. Hyg. 58, E79–E92.
Fernando U., Biswas D., Allan B., Willson P., Potter A. A. (2007). Influence of Campylobacter jejuni fliA, rpoN and flgK genes on colonization of the chicken gut. Int. J. Food Microbiol. 118, 194–200. doi: 10.1016/j.ijfoodmicro.2007.07.038
Ferrara L. G. M., Wallat G. D., Moynie L., Dhanasekar N. N., Aliouane S., Acosta-Gutierrez S., et al. (2016). MOMP from Campylobacter jejuni Is a Trimer of 18-Stranded beta-Barrel Monomers with a Ca(2+) Ion Bound at the Constriction Zone. J. Mol. Biol. 428, 4528–4543. doi: 10.1016/j.jmb.2016.09.021
Fields J. A., Thompson S. A. (2008). Campylobacter jejuni CsrA mediates oxidative stress responses, biofilm formation, and host cell invasion. J. Bacteriol. 190, 3411–3416. doi: 10.1128/JB.01928-07
Flanagan R. C., Neal-Mckinney J. M., Dhillon A. S., Miller W. G., Konkel M. E. (2009). Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 77, 2399–2407. doi: 10.1128/IAI.01266-08
Gao B., Vorwerk H., Huber C., Lara-Tejero M., Mohr J., Goodman A. L., et al. (2017). Metabolic and fitness determinants for in vitro growth and intestinal colonization of the bacterial pathogen Campylobacter jejuni. PloS Biol. 15, e2001390. doi: 10.1371/journal.pbio.2001390
Garber J. M., Nothaft H., Pluvinage B., Stahl M., Bian X., Porfirio S., et al. (2020). The gastrointestinal pathogen Campylobacter jejuni metabolizes sugars with potential help from commensal Bacteroides vulgatus. Commun. Biol. 3, 2. doi: 10.1038/s42003-019-0727-5
Garcia-Sanchez L., Melero B., Rovira J. (2018). Campylobacter in the Food Chain. Adv. Food Nutr. Res. 86, 215–252. doi: 10.1016/bs.afnr.2018.04.005
Gaynor E. C., Wells D. H., Mackichan J. K., Falkow S. (2005). The Campylobacter jejuni stringent response controls specific stress survival and virulence-associated phenotypes. Mol. Microbiol. 56, 8–27. doi: 10.1111/j.1365-2958.2005.04525.x
Gaytan M. O., Martinez-Santos V. II, Soto E., Gonzalez-Pedrajo B. (2016). Type Three Secretion System in Attaching and Effacing Pathogens. Front. Cell Infect. Microbiol. 6, 129. doi: 10.3389/fcimb.2016.00129
Gilbert M., Mandrell R. E., Parker C. T., Li J., Vinogradov E. (2007). Structural analysis of the capsular polysaccharide from Campylobacter jejuni RM1221. Chembiochem 8, 625–631. doi: 10.1002/cbic.200600508
Goodman K. N., Powers M. J., Crofts A. A., Trent M. S., Hendrixson D. R. (2020). Campylobacter jejuni BumSR directs a response to butyrate via sensor phosphatase activity to impact transcription and colonization. Proc. Natl. Acad. Sci. U. S. A. 117, 11715–11726. doi: 10.1073/pnas.1922719117
Gormley F. J., Bailey R. A., Watson K. A., Mcadam J., Avendano S., Stanley W. A., et al. (2014). Campylobacter colonization and proliferation in the broiler chicken upon natural field challenge is not affected by the bird growth rate or breed. Appl. Environ. Microbiol. 80, 6733–6738. doi: 10.1128/AEM.02162-14
Grant C. C., Konkel M. E., Cieplak W. Jr., Tompkins L. S. (1993). Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61, 1764–1771. doi: 10.1128/IAI.61.5.1764-1771.1993
Guccione E., Leon-Kempis Mdel R., Pearson B. M., Hitchin E., Mulholland F., Van Diemen P. M., et al. (2008). Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69, 77–93. doi: 10.1111/j.1365-2958.2008.06263.x
Guccione E. J., Kendall J. J., Hitchcock A., Garg N., White M. A., Mulholland F., et al. (2017). Transcriptome and proteome dynamics in chemostat culture reveal how Campylobacter jejuni modulates metabolism, stress responses and virulence factors upon changes in oxygen availability. Environ. Microbiol. 19, 4326–4348. doi: 10.1111/1462-2920.13930
Guerry P., Alm R. A., Power M. E., Logan S. M., Trust T. J. (1991). Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173, 4757–4764. doi: 10.1128/JB.173.15.4757-4764.1991
Guerry P., Szymanski C. M., Prendergast M. M., Hickey T. E., Ewing C. P., Pattarini D. L., et al. (2002). Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70, 787–793. doi: 10.1128/IAI.70.2.787-793.2002
Guerry P., Poly F., Riddle M., Maue A. C., Chen Y. H., Monteiro M. A. (2012). Campylobacter polysaccharide capsules: virulence and vaccines. Front. Cell. Infect. Microbiol. 2, 7. doi: 10.3389/fcimb.2012.00007
Gundogdu O., Da Silva D. T., Mohammad B., Elmi A., Wren B. W., Van Vliet A. H., et al. (2016). The Campylobacter jejuni Oxidative Stress Regulator RrpB Is Associated with a Genomic Hypervariable Region and Altered Oxidative Stress Resistance. Front. Microbiol. 7, 2117. doi: 10.3389/fmicb.2016.02117
Hameed A., Woodacre A., Machado L. R., Marsden G. L. (2020). An Updated Classification System and Review of the Lipooligosaccharide Biosynthesis Gene Locus in Campylobacter jejuni. Front. Microbiol. 11, 677. doi: 10.3389/fmicb.2020.00677
Hansson I., Sandberg M., Habib I., Lowman R., Engvall E. O. (2018). Knowledge gaps in control of Campylobacter for prevention of campylobacteriosis. Transbound. Emerg. Dis. 65 (Suppl 1), 30–48. doi: 10.1111/tbed.12870
Havelaar A. H., Kirk M. D., Torgerson P. R., Gibb H. J., Hald T., Lake R. J., et al. (2015). World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PloS Med. 12, e1001923. doi: 10.1371/journal.pmed.1001923
Hazeleger W. C., Wouters J. A., Rombouts F. M., Abee T. (1998). Physiological activity of Campylobacter jejuni far below the minimal growth temperature. Appl. Environ. Microbiol. 64, 3917–3922. doi: 10.1128/AEM.64.10.3917-3922.1998
He H., Cooper J. N., Mishra A., Raskin D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. doi: 10.1128/JB.00014-12
Hendrixson D. R., Dirita V. J. (2003). Transcription of sigma54-dependent but not sigma28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50, 687–702. doi: 10.1046/j.1365-2958.2003.03731.x
Hendrixson D. R., Dirita V. J. (2004). Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52, 471–484. doi: 10.1111/j.1365-2958.2004.03988.x
Hermans D., Van Deun K., Martel A., Van Immerseel F., Messens W., Heyndrickx M., et al. (2011). Colonization factors of Campylobacter jejuni in the chicken gut. Vet. Res. 42, 82. doi: 10.1186/1297-9716-42-82
Hermans D., Pasmans F., Heyndrickx M., Van Immerseel F., Martel A., Van Deun K., et al. (2012). A tolerogenic mucosal immune response leads to persistent Campylobacter jejuni colonization in the chicken gut. Crit. Rev. Microbiol. 38, 17–29. doi: 10.3109/1040841X.2011.615298
Hofreuter D. (2014). Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell. Infect. Microbiol. 4, 137. doi: 10.3389/fcimb.2014.00137
Houliston R. S., Vinogradov E., Dzieciatkowska M., Li J., St Michael F., Karwaski M. F., et al. (2011). Lipooligosaccharide of Campylobacter jejuni: similarity with multiple types of mammalian glycans beyond gangliosides. J. Biol. Chem. 286, 12361–12370. doi: 10.1074/jbc.M110.181750
Howard S. L., Jagannathan A., Soo E. C., Hui J. P., Aubry A. J., Ahmed I., et al. (2009). Campylobacter Jejuni Glycosylation Island Important In Cell Charge, Legionaminic Acid Biosynthesis, And Colonization Of Chickens. Infect. Immun. 77, 2544–2556. doi: 10.1128/IAI.01425-08
Hudson J. A., Nicol C., Wright J., Whyte R., Hasell S. K. (1999). Seasonal Variation Of Campylobacter Types From Human Cases, Veterinary Cases, Raw Chicken, Milk And Water. J. Appl. Microbiol. 87, 115–124. doi: 10.1046/j.1365-2672.1999.00806.x
Humphrey S., Chaloner G., Kemmett K., Davidson N., Williams N., Kipar A., et al. (2014). Campylobacter Jejuni Is Not Merely A Commensal In Commercial Broiler Chickens And Affects Bird Welfare. Mbio 5, E01364–E01314. doi: 10.1128/mBio.01364-14
Hwang S., Kim M., Ryu S., Jeon B. (2011). Regulation of oxidative stress response by CosR, an essential response regulator in Campylobacter jejuni. PloS One 6, e22300. doi: 10.1371/journal.pone.0022300
Igwaran A., Okoh A. II (2019). Human campylobacteriosis: A public health concern of global importance. Heliyon 5, e02814. doi: 10.1016/j.heliyon.2019.e02814
Ijaz U. Z., Sivaloganathan L., McKenna A., Richmond A., Kelly C., Linton M., et al. (2018). Comprehensive Longitudinal Microbiome Analysis of the Chicken Cecum Reveals a Shift from Competitive to Environmental Drivers and a Window of Opportunity for Campylobacter. Front. Microbiol. 9, 2452. doi: 10.3389/fmicb.2018.02452
Iovine N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. doi: 10.4161/viru.23753
Jagannathan A., Constantinidou C., Penn C. W. (2001). Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183, 2937–2942. doi: 10.1128/JB.183.9.2937-2942.2001
Jervis A. J., Butler J. A., Lawson A. J., Langdon R., Wren B. W., Linton D. (2012). Characterization of the structurally diverse N-linked glycans of Campylobacter species. J. Bacteriol. 194, 2355–2362. doi: 10.1128/JB.00042-12
Jin S., Song Y. C., Emili A., Sherman P. M., Chan V. L. (2003). JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90alpha and triggers signalling pathways leading to the activation of NF-kappaB and p38 MAP kinase in epithelial cells. Cell Microbiol. 5, 165–174. doi: 10.1046/j.1462-5822.2003.00265.x
John D. A., Williams L. K., Kanamarlapudi V., Humphrey T. J., Wilkinson T. S. (2017). The Bacterial Species Campylobacter jejuni Induce Diverse Innate Immune Responses in Human and Avian Intestinal Epithelial Cells. Front. Microbiol. 8, 1840. doi: 10.3389/fmicb.2017.01840
Jones M. A., Marston K. L., Woodall C. A., Maskell D. J., Linton D., Karlyshev A. V., et al. (2004). Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72, 3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004
Kaakoush N. O., Castano-Rodriguez N., Mitchell H. M., Man S. M. (2015). Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 28, 687–720. doi: 10.1128/CMR.00006-15
Kanipes M. II, Tan X., Akelaitis A., Li J., Rockabrand D., Guerry P., et al. (2008). Genetic analysis of lipooligosaccharide core biosynthesis in Campylobacter jejuni 81-176. J. Bacteriol. 190, 1568–1574. doi: 10.1128/JB.01696-07
Karlyshev A. V., Linton D., Gregson N. A., Lastovica A. J., Wren B. W. (2000). Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35, 529–541. doi: 10.1046/j.1365-2958.2000.01717.x
Karlyshev A. V., Mccrossan M. V., Wren B. W. (2001). Demonstration of polysaccharide capsule in Campylobacter jejuni using electron microscopy. Infect. Immun. 69, 5921–5924. doi: 10.1128/IAI.69.9.5921-5924.2001
Karlyshev A. V., Champion O. L., Churcher C., Brisson J. R., Jarrell H. C., Gilbert M., et al. (2005). Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55, 90–103. doi: 10.1111/j.1365-2958.2004.04374.x
Keo T., Collins J., Kunwar P., Blaser M. J., Iovine N. M. (2011). Campylobacter capsule and lipooligosaccharide confer resistance to serum and cationic antimicrobials. Virulence 2, 30–40. doi: 10.4161/viru.2.1.14752
Kervella M., Pages J. M., Pei Z., Grollier G., Blaser M. J., Fauchere J. L. (1993). Isolation and characterization of two Campylobacter glycine-extracted proteins that bind to HeLa cell membranes. Infect. Immun. 61, 3440–3448. doi: 10.1128/IAI.61.8.3440-3448.1993
Keto-Timonen R., Hietala N., Palonen E., Hakakorpi A., Lindstrom M., Korkeala H. (2016). Cold Shock Proteins: A Minireview with Special Emphasis on Csp-family of Enteropathogenic Yersinia. Front. Microbiol. 7, 1151. doi: 10.3389/fmicb.2016.01151
Kim J. C., Oh E., Hwang S., Ryu S., Jeon B. (2015a). Non-selective regulation of peroxide and superoxide resistance genes by PerR in Campylobacter jejuni. Front. Microbiol. 6, 126. doi: 10.3389/fmicb.2015.00126
Kim J. C., Oh E., Kim J., Jeon B. (2015b). Regulation of oxidative stress resistance in Campylobacter jejuni, a microaerophilic foodborne pathogen. Front. Microbiol. 6, 751. doi: 10.3389/fmicb.2015.00751
Konkel M. E., Kim B. J., Rivera-Amill V., Garvis S. G. (1999). Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32, 691–701. doi: 10.1046/j.1365-2958.1999.01376.x
Konkel M. E., Klena J. D., Rivera-Amill V., Monteville M. R., Biswas D., Raphael B., et al. (2004). Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186, 3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004
Konkel M. E., Christensen J. E., Keech A. M., Monteville M. R., Klena J. D., Garvis S. G. (2005). Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol. Microbiol. 57, 1022–1035. doi: 10.1111/j.1365-2958.2005.04744.x
Konkel M. E., Larson C. L., Flanagan R. C. (2010). Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 192, 68–76. doi: 10.1128/JB.00969-09
Konkel M. E., Talukdar P. K., Negretti N. M., Klappenbach C. M. (2020). Taking Control: Campylobacter jejuni Binding to Fibronectin Sets the Stage for Cellular Adherence and Invasion. Front. Microbiol. 11, 564. doi: 10.3389/fmicb.2020.00564
Korlath J. A., Osterholm M. T., Judy L. A., Forfang J. C., Robinson R. A. (1985). A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152, 592–596. doi: 10.1093/infdis/152.3.592
Kreutzberger M. A. B., Ewing C., Poly F., Wang F., Egelman E. H. (2020). Atomic structure of the Campylobacter jejuni flagellar filament reveals how epsilon Proteobacteria escaped Toll-like receptor 5 surveillance. Proc. Natl. Acad. Sci. U.S.A. 117, 16985–16991. doi: 10.1073/pnas.2010996117
Larson C. L., Samuelson D. R., Eucker T. P., O’loughlin J. L., Konkel M. E. (2013). The fibronectin-binding motif within FlpA facilitates Campylobacter jejuni adherence to host cell and activation of host cell signaling. Emerg. Microbes Infect. 2, e65. doi: 10.1038/emi.2013.65
Le M. T., Porcelli I., Weight C. M., Gaskin D. J., Carding S. R., Van Vliet A. H. (2012). Acid-shock of Campylobacter jejuni induces flagellar gene expression and host cell invasion. Eur. J. Microbiol. Immunol. (Bp) 2, 12–19. doi: 10.1556/EuJMI.2.2012.1.3
Leon-Kempis Mdel R., Guccione E., Mulholland F., Williamson M. P., Kelly D. J. (2006). The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60, 1262–1275. doi: 10.1111/j.1365-2958.2006.05168.x
Liaw J., Hong G., Davies C., Elmi A., Sima F., Stratakos A., et al. (2019). The Campylobacter jejuni Type VI Secretion System Enhances the Oxidative Stress Response and Host Colonization. Front. Microbiol. 10, 2864. doi: 10.3389/fmicb.2019.02864
Linton D., Dorrell N., Hitchen P. G., Amber S., Karlyshev A. V., Morris H. R., et al. (2005). Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55, 1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x
Logan S. M., Hui J. P., Vinogradov E., Aubry A. J., Melanson J. E., Kelly J. F., et al. (2009). Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276, 1014–1023. doi: 10.1111/j.1742-4658.2008.06840.x
Luethy P. M., Huynh S., Ribardo D. A., Winter S. E., Parker C. T., Hendrixson D. R. (2017). Microbiota-Derived Short-Chain Fatty Acids Modulate Expression of Campylobacter jejuni Determinants Required for Commensalism and Virulence. mBio 8. doi: 10.1128/mBio.00407-17
Maccallum A., Haddock G., Everest P. H. (2005). Campylobacter jejuni activates mitogen-activated protein kinases in Caco-2 cell monolayers and in vitro infected primary human colonic tissue. Microbiol. (Reading) 151, 2765–2772. doi: 10.1099/mic.0.27979-0
Magajna B., Schraft H. (2015). Evaluation of Propidium Monoazide and Quantitative PCR To Quantify Viable Campylobacter jejuni Biofilm and Planktonic Cells in Log Phase and in a Viable but Nonculturable State. J. Food Prot. 78, 1303–1311. doi: 10.4315/0362-028X.JFP-14-583
Marchant J., Wren B., Ketley J. (2002). Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol. 10, 155–159. doi: 10.1016/S0966-842X(02)02323-5
Mcnally D. J., Jarrell H. C., Li J., Khieu N. H., Vinogradov E., Szymanski C. M., et al. (2005). The HS:1 serostrain of Campylobacter jejuni has a complex teichoic acid-like capsular polysaccharide with nonstoichiometric fructofuranose branches and O-methyl phosphoramidate groups. FEBS J. 272, 4407–4422. doi: 10.1111/j.1742-4658.2005.04856.x
McKenna A., Ijaz U. Z., Kelly C., Linton M., Sloan W. T., Green B. D., et al. (2020). Impact of industrial production system parameters on chicken microbiomes: mechanisms to improve performance and reduce Campylobacter. Microbiome 8, 128. doi: 10.1186/s40168-020-00908-8
Mcnally D. J., Lamoureux M. P., Karlyshev A. V., Fiori L. M., Li J., Thacker G., et al. (2007). Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282, 28566–28576. doi: 10.1074/jbc.M704413200
Melo R. T., Mendonca E. P., Monteiro G. P., Siqueira M. C., Pereira C. B., Peres P., et al. (2017). Intrinsic and Extrinsic Aspects on Campylobacter jejuni Biofilms. Front. Microbiol. 8, 1332. doi: 10.3389/fmicb.2017.01332
Muraoka W. T., Zhang Q. (2011). Phenotypic and genotypic evidence for L-fucose utilization by Campylobacter jejuni. J. Bacteriol. 193, 1065–1075. doi: 10.1128/JB.01252-10
Murphy C., Carroll C., Jordan K. N. (2006). Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 100, 623–632. doi: 10.1111/j.1365-2672.2006.02903.x
Nachamkin I., Allos B. M., Ho T. (1998). Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11, 555–567. doi: 10.1128/CMR.11.3.555
Neal-Mckinney J. M., Konkel M. E. (2012). The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front. Cell Infect. Microbiol. 2, 31. doi: 10.3389/fcimb.2012.00031
Novik V., Hofreuter D., Galan J. E. (2010). Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 78, 3540–3553. doi: 10.1128/IAI.00109-10
Nuijten P. J., Van Asten F. J., Gaastra W., Van Der Zeijst B. A. (1990). Structural and functional analysis of two Campylobacter jejuni flagellin genes. J. Biol. Chem. 265, 17798–17804. doi: 10.1016/S0021-9258(18)38234-6
Nunoshiba T., Hidalgo E., Amabile Cuevas C. F., Demple B. (1992). Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174, 6054–6060. doi: 10.1128/JB.174.19.6054-6060.1992
Oberheim J., Hoser C., Luchters G., Kistemann T. (2020). Small-scaled association between ambient temperature and campylobacteriosis incidence in Germany. Sci. Rep. 10, 17191. doi: 10.1038/s41598-020-73865-9
Oh E., Jeon B. (2014). Role of alkyl hydroperoxide reductase (AhpC) in the biofilm formation of Campylobacter jejuni. PloS One 9, e87312. doi: 10.1371/journal.pone.0087312
Oh E., Andrews K. J., Mcmullen L. M., Jeon B. (2019). Tolerance to stress conditions associated with food safety in Campylobacter jejuni strains isolated from retail raw chicken. Sci. Rep. 9, 11915. doi: 10.1038/s41598-019-48373-0
Palyada K., Sun Y. Q., Flint A., Butcher J., Naikare H., Stintzi A. (2009). Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10, 481. doi: 10.1186/1471-2164-10-481
Park D., Lara-Tejero M., Waxham M. N., Li W., Hu B., Galan J. E., et al. (2018). Visualization of the type III secretion mediated Salmonella-host cell interface using cryo-electron tomography. Elife 7. doi: 10.7554/eLife.39514.018
Parker C. T., Horn S. T., Gilbert M., Miller W. G., Woodward D. L., Mandrell R. E. (2005). Comparison of Campylobacter jejuni lipooligosaccharide biosynthesis loci from a variety of sources. J. Clin. Microbiol. 43, 2771–2781. doi: 10.1128/JCM.43.6.2771-2781.2005
Parker C. T., Gilbert M., Yuki N., Endtz H. P., Mandrell R. E. (2008). Characterization of lipooligosaccharide-biosynthetic loci of Campylobacter jejuni reveals new lipooligosaccharide classes: evidence of mosaic organizations. J. Bacteriol. 190, 5681–5689. doi: 10.1128/JB.00254-08
Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Pei Z., Burucoa C., Grignon B., Baqar S., Huang X. Z., Kopecko D. J., et al. (1998). Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66, 938–943. doi: 10.1128/IAI.66.3.938-943.1998
Poly F., Serichantalergs O., Kuroiwa J., Pootong P., Mason C., Guerry P., et al. (2015). Updated Campylobacter jejuni Capsule PCR Multiplex Typing System and Its Application to Clinical Isolates from South and Southeast Asia. PloS One 10, e0144349. doi: 10.1371/journal.pone.0144349
Purdy D., Buswell C. M., Hodgson A. E., Mc A. K., Henderson I., Leach S. A. (2000). Characterisation of cytolethal distending toxin (CDT) mutants of Campylobacter jejuni. J. Med. Microbiol. 49, 473–479. doi: 10.1099/0022-1317-49-5-473
Reeser R. J., Medler R. T., Billington S. J., Jost B. H., Joens L. A. (2007). Characterization of Campylobacter jejuni biofilms under defined growth conditions. Appl. Environ. Microbiol. 73, 1908–1913. doi: 10.1128/AEM.00740-06
Reid A. N., Pandey R., Palyada K., Whitworth L., Doukhanine E., Stintzi A. (2008). Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 74, 1598–1612. doi: 10.1128/AEM.01508-07
Reuter M., Mallett A., Pearson B. M., Van Vliet A. H. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76, 2122–2128. doi: 10.1128/AEM.01878-09
Rodrigues R. C., Pocheron A. L., Hernould M., Haddad N., Tresse O., Cappelier J. M. (2015). Description of Campylobacter jejuni Bf, an atypical aero-tolerant strain. Gut Pathog. 7, 30. doi: 10.1186/s13099-015-0077-x
Samuelson D. R., Eucker T. P., Bell J. A., Dybas L., Mansfield L. S., Konkel M. E. (2013). The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease. Cell Commun. Signal 11, 79. doi: 10.1186/1478-811X-11-79
Schirm M., Schoenhofen I. C., Logan S. M., Waldron K. C., Thibault P. (2005). Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem. 77, 7774–7782. doi: 10.1021/ac051316y
Schoenhofen I. C., Vinogradov E., Whitfield D. M., Brisson J. R., Logan S. M. (2009). The CMP-legionaminic acid pathway in Campylobacter: biosynthesis involving novel GDP-linked precursors. Glycobiology 19, 715–725. doi: 10.1093/glycob/cwp039
Sheikh K. A., Ho T. W., Nachamkin I., Li C. Y., Cornblath D. R., Asbury A. K., et al. (1998a). Molecular mimicry in Guillain-Barre syndrome. Ann. N. Y. Acad. Sci. 845, 307–321. doi: 10.1111/j.1749-6632.1998.tb09683.x
Sheikh K. A., Nachamkin I., Ho T. W., Willison H. J., Veitch J., Ung H., et al. (1998b). Campylobacter jejuni lipopolysaccharides in Guillain-Barre syndrome: molecular mimicry and host susceptibility. Neurology 51, 371–378. doi: 10.1212/WNL.51.2.371
Sheppard S. K., Didelot X., Meric G., Torralbo A., Jolley K. A., Kelly D. J., et al. (2013). Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc. Natl. Acad. Sci. U. S. A. 110, 11923–11927. doi: 10.1073/pnas.1305559110
Skirrow M. B. (1977). Campylobacter enteritis: a “new” disease. Br. Med. J. 2, 9–11. doi: 10.1136/bmj.2.6078.9
Skirrow M. B. (2006). John McFadyean and the centenary of the first isolation of Campylobacter species. Clin. Infect. Dis. 43, 1213–1217. doi: 10.1086/508201
Song Y. C., Jin S., Louie H., Ng D., Lau R., Zhang Y., et al. (2004). FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol. Microbiol. 53, 541–553. doi: 10.1111/j.1365-2958.2004.04175.x
Song W. S., Jeon Y. J., Namgung B., Hong M., Yoon S. II (2017). A conserved TLR5 binding and activation hot spot on flagellin. Sci. Rep. 7, 40878. doi: 10.1038/srep40878
Stahl M., Friis L. M., Nothaft H., Liu X., Li J., Szymanski C. M., et al. (2011). L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U. S. A. 108, 7194–7199. doi: 10.1073/pnas.1014125108
Stahl M., Butcher J., Stintzi A. (2012). Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2, 5. doi: 10.3389/fcimb.2012.00005
Stintzi A., Marlow D., Palyada K., Naikare H., Panciera R., Whitworth L., et al. (2005). Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 73, 1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005
Svensson S. L., Pryjma M., Gaynor E. C. (2014). Flagella-mediated adhesion and extracellular DNA release contribute to biofilm formation and stress tolerance of Campylobacter jejuni. PloS One 9, e106063. doi: 10.1371/journal.pone.0106063
Svensson S. L., Huynh S., Parker C. T., Gaynor E. C. (2015). The Campylobacter jejuni CprRS two-component regulatory system regulates aspects of the cell envelope. Mol. Microbiol. 96, 189–209. doi: 10.1111/mmi.12927
Szymanski C. M., King M., Haardt M., Armstrong G. D. (1995). Campylobacter jejuni motility and invasion of Caco-2 cells. Infect. Immun. 63, 4295–4300. doi: 10.1128/IAI.63.11.4295-4300.1995
Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. (1999). Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x
Szymanski C. M. (2015). Are campylobacters now capable of carbo-loading? Mol. Microbiol. 98, 805–808. doi: 10.1111/mmi.13162
Talukdar P. K., Negretti N. M., Turner K. L., Konkel M. E. (2020). Molecular Dissection of the Campylobacter jejuni CadF and FlpA Virulence Proteins in Binding to Host Cell Fibronectin. Microorganisms 8. doi: 10.3390/microorganisms8030389
Teh A. H., Lee S. M., Dykes G. A. (2014). Does Campylobacter jejuni form biofilms in food-related environments? Appl. Environ. Microbiol. 80, 5154–5160. doi: 10.1128/AEM.01493-14
Teh A. H. T., Lee S. M., Dykes G. A. (2019). Association of some Campylobacter jejuni with Pseudomonas aeruginosa biofilms increases attachment under conditions mimicking those in the environment. PloS One 14, e0215275. doi: 10.1371/journal.pone.0215275
Thibault P., Logan S. M., Kelly J. F., Brisson J. R., Ewing C. P., Trust T. J., et al. (2001). Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276, 34862–34870. doi: 10.1074/jbc.M104529200
Thompson S. A., Gaynor E. C. (2008). Campylobacter jejuni host tissue tropism: a consequence of its low-carb lifestyle? Cell Host Microbe 4, 409–410. doi: 10.1016/j.chom.2008.10.010
Thota V. N., Ferguson M. J., Sweeney R. P., Lowary T. L. (2018). Synthesis of the Campylobacter jejuni 81-176 Strain Capsular Polysaccharide Repeating Unit Reveals the Absolute Configuration of its O-Methyl Phosphoramidate Motif. Angew. Chem. Int. Ed. Engl. 57, 15592–15596. doi: 10.1002/anie.201810222
Turonova H., Briandet R., Rodrigues R., Hernould M., Hayek N., Stintzi A., et al. (2015). Biofilm spatial organization by the emerging pathogen Campylobacter jejuni: comparison between NCTC 11168 and 81-176 strains under microaerobic and oxygen-enriched conditions. Front. Microbiol. 6, 709. doi: 10.3389/fmicb.2015.00709
Ugarte-Ruiz M., Stabler R. A., Dominguez L., Porrero M. C., Wren B. W., Dorrell N., et al. (2015). Prevalence of Type VI Secretion System in Spanish Campylobacter jejuni Isolates. Zoonoses Public Health 62, 497–500. doi: 10.1111/zph.12176
Van Der Hooft J. J. J., Alghefari W., Watson E., Everest P., Morton F. R., Burgess K. E. V., et al. (2018). Unexpected differential metabolic responses of Campylobacter jejuni to the abundant presence of glutamate and fucose. Metabolomics 14, 144. doi: 10.1007/s11306-018-1438-5
Van Der Stel A. X., Wosten M. (2019). Regulation of Respiratory Pathways in Campylobacterota: A Review. Front. Microbiol. 10, 1719. doi: 10.3389/fmicb.2019.01719
Vegge C. S., Jansen Van Rensburg M. J., Rasmussen J. J., Maiden M. C., Johnsen L. G., Danielsen M., et al. (2016). Glucose Metabolism via the Entner-Doudoroff Pathway in Campylobacter: A Rare Trait that Enhances Survival and Promotes Biofilm Formation in Some Isolates. Front. Microbiol. 7, 1877. doi: 10.3389/fmicb.2016.01877
Velayudhan J., Jones M. A., Barrow P. A., Kelly D. J. (2004). L-serine catabolism via an oxygen-labile L-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72, 260–268. doi: 10.1128/IAI.72.1.260-268.2004
Wassenaar T. M., Bleumink-Pluym N. M., Van Der Zeijst B. A. (1991). Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10, 2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x
Wassenaar T. M., Van Der Zeijst B. A., Ayling R., Newell D. G. (1993). Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139 (Pt 6), 1171–1175. doi: 10.1099/00221287-139-6-1171
Watson R. O., Galan J. E. (2005). Signal transduction in Campylobacter jejuni-induced cytokine production. Cell Microbiol. 7, 655–665. doi: 10.1111/j.1462-5822.2004.00498.x
Wilkinson D. A., O’donnell A. J., Akhter R. N., Fayaz A., Mack H. J., Rogers L. E., et al. (2018). Updating the genomic taxonomy and epidemiology of Campylobacter hyointestinalis. Sci. Rep. 8, 2393. doi: 10.1038/s41598-018-20889-x
Woodall C. A., Jones M. A., Barrow P. A., Hinds J., Marsden G. L., Kelly D. J., et al. (2005). Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect. Immun. 73, 5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005
Wouters J. A., Rombouts F. M., Kuipers O. P., De Vos W. M., Abee T. (2000). The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23, 165–173. doi: 10.1016/S0723-2020(00)80001-6
Yao R., Burr D. H., Doig P., Trust T. J., Niu H., Guerry P. (1994). Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14, 883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x
Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., et al. (2002). Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539. doi: 10.1074/jbc.M206114200
Yuki N., Taki T., Inagaki F., Kasama T., Takahashi M., Saito K., et al. (1993). A bacterium lipopolysaccharide that elicits Guillain-Barre syndrome has a GM1 ganglioside-like structure. J. Exp. Med. 178, 1771–1775. doi: 10.1084/jem.178.5.1771
Zheng M., Aslund F., Storz G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721. doi: 10.1126/science.279.5357.1718
Keywords: Campylobacter jejuni, virulence, host-pathogen, sensing, adaptation, stress and survival
Citation: Elmi A, Nasher F, Dorrell N, Wren B and Gundogdu O (2021) Revisiting Campylobacter jejuni Virulence and Fitness Factors: Role in Sensing, Adapting, and Competing. Front. Cell. Infect. Microbiol. 10:607704. doi: 10.3389/fcimb.2020.607704
Received: 17 September 2020; Accepted: 11 December 2020;
Published: 03 February 2021.
Edited by:
Mohamed Elhadidy, University of Science and Technology at Zewail City, EgyptReviewed by:
Marc Heyndrickx, Institute for Agricultural, Fisheries and Food Research (ILVO), BelgiumCraig T. Parker, United States Department of Agriculture, United States
Copyright © 2021 Elmi, Nasher, Dorrell, Wren and Gundogdu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdi Elmi, YWJkaS5lbG1pQGxzaHRtLmFjLnVr
 Abdi Elmi
Abdi Elmi Fauzy Nasher
Fauzy Nasher Nick Dorrell
Nick Dorrell Brendan Wren
Brendan Wren Ozan Gundogdu
Ozan Gundogdu