- 1Institute of Preventive Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Key Laboratory of Animal Diseases and Human Health of Sichuan Province, Chengdu, China
- 3Avian Diseases Research Center, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 4Department of Microbiology and Immunology, North Sichuan Medical College, Nanchong, China
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins provide acquired genetic immunity against the entry of mobile genetic elements (MGEs). The immune defense provided by various subtypes of the CRISPR-Cas system has been confirmed and is closely associated with the formation of immunological memory in CRISPR arrays, called CRISPR adaptation or spacer acquisition. However, whether type II-C CRISPR-Cas systems are also involved in spacer acquisition remains largely unknown. This study explores and provides some definitive evidence regarding spacer acquisition of the type II-C CRISPR-Cas system from Riemerella anatipestifer (RA) CH-2 (RA-CH-2). Firstly, introducing an exogenous plasmid into RA-CH-2 triggered spacer acquisition of RA CRISPR-Cas system, and the acquisition of new spacers led to plasmid instability in RA-CH-2. Furthermore, deletion of cas1 or cas2 of RA-CH-2 abrogated spacer acquisition and subsequently stabilized the exogenous plasmid, suggesting that both Cas1 and Cas2 are required for spacer acquisition of RA-CH-2 CRISPR-Cas system, consistent with the reported role of Cas1 and Cas2 in type I-E and II-A systems. Finally, assays for studying Cas1 nuclease activity and the interaction of Cas1 with Cas2 contributed to a better understanding of the adaptation mechanism of RA CRISPR-Cas system. This is the first experimental identification of the naïve adaptation of type II-C CRISPR-Cas system.
Introduction
Riemerella anatipestifer (RA) is a gram-negative bacterium that causes infection with high morbidity and mortality (Segers et al., 1993). RA infection occurs worldwide, primarily in domestic ducks, turkeys, and other birds, especially in 1–8-week-old ducklings (Ruiz and Sandhu, 2013). To date, more than 21 serotypes of RA have been isolated (Pathanasophon et al., 2002; Cheng et al., 2003). The complete genomic sequences of some RA strains have been submitted to the GenBank database in recent years (Wang X. et al., 2012; Wang et al., 2014; Zhu et al., 2016). Among these strains, R. anatipestifer CH-2 (RA-CH-2) is highly virulent and is one of the common serotype 2 strains in China (Wang et al., 2014). Research reports on RA are increasing and include studies on genomics (Wang et al., 2014; Liu J. et al., 2016; Song et al., 2016; Zhu et al., 2016), antibiotic resistance (Zhong et al., 2009, 2013; Luo et al., 2015, 2017; Huang et al., 2017; Zhang et al., 2017), energy metabolism (Liao et al., 2015, 2016; Liu et al., 2017a), physiology (Li et al., 2012; Yi et al., 2017), and virulence (Wang M. et al., 2017) of this organism and studies for method establishment (Wang X. P. et al., 2012; Gao et al., 2016; Liu M. et al., 2016; Liu et al., 2017b). However, studies on the physiology and pathogenic mechanism of RA are rare.
In many bacteria (~50%) and most archaea (~87%), a novel immune system comprised of clustered regularly interspaced short palindromic repeats (CRISPR) and the CRISPR-associated (Cas) protein has been identified, and this system can provide acquired genetic immunity against the entry of mobile genetic elements (MGEs), such as viruses (phage) and plasmids (Makarova et al., 2015). The immune system relies on the Cas protein (the effector module) to cleave specific sequences of MGEs, guided by CRISPR RNA (crRNA) (Barrangou, 2013). According to the current classification based on multiple criteria, CRISPR-Cas systems are divided into two classes that comprise six types and 19 subtypes (Koonin et al., 2017). Among these systems, class II CRISPR-Cas system is the simplest and contains a single effector protein. Among the class II systems, type II CRISPR-Cas systems are the most common and best understood, especially type II effectors (Shmakov et al., 2017). The subtype II-C CRISPR-Cas system comprises ~41% of type II systems, contains a mini cas operon (Shmakov et al., 2017), uses Cas9 protein as an effector and lacks Csn2 protein (Koonin et al., 2017). Systems with only three cas genes (cas1, cas2, and cas9) in the operon were originally defined as type II-C CRISPR-Cas systems (Chylinski et al., 2014). Burstein et al. reported a type II-C system variant with cas4 and considered it to be a fusion of type II-C and II-B systems (Burstein et al., 2017). The numerous and varied type II-C CRISPR-Cas systems remain to be explored.
Despite the extreme diversity in the organization of CRISPR-Cas loci, the CRISPR/Cas-mediated immune defense includes three phases: adaptation (spacer acquisition), expression (crRNA biogenesis), and interference (cleavage of target DNA or RNA) (Marraffini, 2015). It is generally known that the nearly universal Cas1 and Cas2 function in the early stage of the diverse CRISPR-Cas systems as adaptation proteins (Yosef et al., 2012; Arslan et al., 2014; Heler et al., 2015; Nuñez et al., 2015; Wei et al., 2015b; Fagerlund et al., 2017; Xiao et al., 2017). In addition to the functions of Cas1 and Cas2, various requirements for spacer acquisition must be met in the diverse CRISPR-Cas systems. For example, Cas adaptation proteins recognize only protospacers (the sequences from which spacers are derived) that are closest to the protospacer adjacent motif (PAM) to select and insert into the CRISPR array (Mojica et al., 2009; Wang et al., 2015; Leenay and Beisel, 2017). In addition, the leader sequence (generally A/T-rich) (Yosef et al., 2012; Wei et al., 2015a; McGinn and Marraffini, 2016), CRISPR repeat (Yosef et al., 2012; Wei et al., 2015a), host factor (Nuñez et al., 2016; Yoganand et al., 2017), and/or other Cas proteins (Heler et al., 2015; Wei et al., 2015b) are also required for spacer acquisition in various CRISPR-Cas systems.
Our previous study revealed that RA encoded the type II-C CRISPR-Cas system harboring a Cas9 effector (Zhu et al., 2016). Two CRISPR arrays were found in RA-CH-2. In addition, CRISPR1 was shown to be flanked on one side by cas genes, while CRISPR2 was designated an orphan. The organization of CRISPR1-Cas locus of RA-CH-2 (Figure 1) demonstrates that RA-CH-2 CRISPR-Cas system is a type II-C system with the Cas9 effector protein and lacking both Cas4 and Csn2. Only 2 spacers (2/32) were homologous with MGEs (phage sequences) and were located in RA-CH-2 CRISPR1 array. Putative protospacers were present in the genomes of Riemerella phage RAP44 (NC_019490.1, 100% similarity) and Enterobacteria phage phi92 (NC_023693.1, 87% similarity). However, no experimental evidence has been provided regarding RA CRISPR-Cas system. In this study, assays of spacer acquisition and plasmid loss were carried out in RA-CH-2 strain. In addition, the nuclease activity of RA Cas1 and the interaction of Cas1 with Cas2 were investigated. The above assays contribute to a better understanding of the adaptation of RA CRISPR-Cas system.
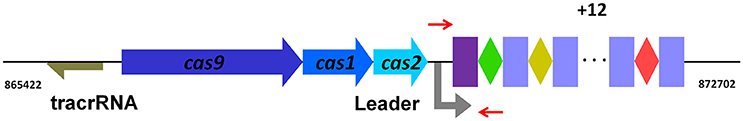
Figure 1. Organization of the type II-C CRISPR-Cas system from RA-CH-2. The CRISPR1 locus from RA-CH-2 comprises three cas genes (cas9, cas1, and cas2), 16 repeats (purple or blue rectangles, 47 bp long) and 15 spacers (colored diamonds, 30 bp long). The tracrRNA (trans-activating crRNA, brown arrow) containing the region with complementarity to the repeats is located upstream of cas9. Between cas2 and the first repeat (purple rectangle), the A/T-rich region is the presumed leader sequence. The primers (short red arrows) were used for PCR amplification to detect CRISPR1 expansion.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
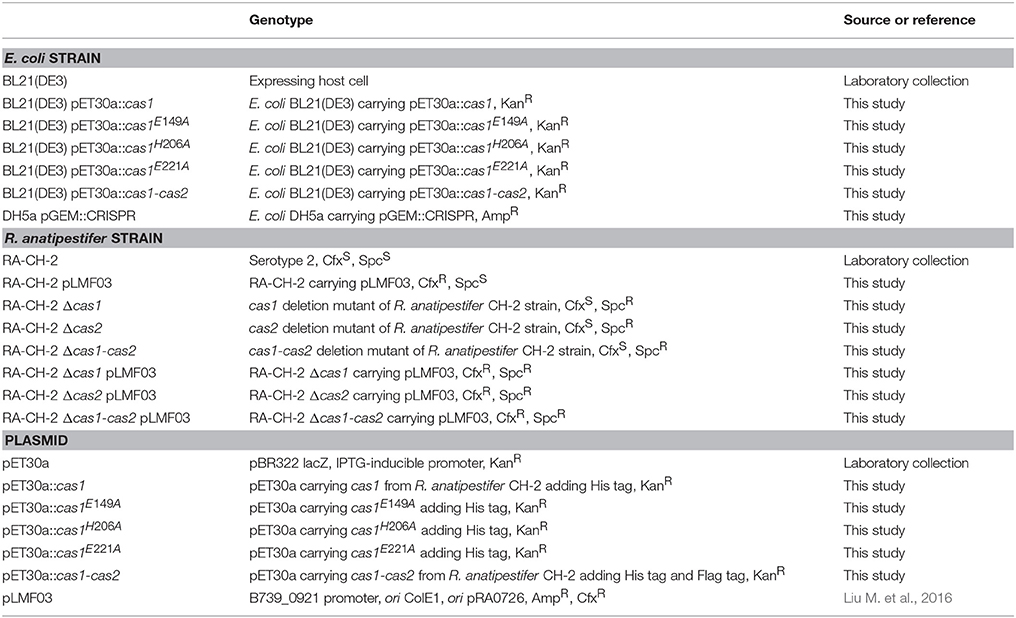
All strains and plasmids used in this study are listed in Table 1, and primers are listed in Table 2. RA cells were grown at 37°C in tryptic soy broth (TSB) or on tryptic soy agar (TSA) (Difco Laboratories, USA) supplemented with 5% calf serum (Biologos, USA) in a 5% CO2 atmosphere. Escherichia coli cells were cultured at 37°C in Luria-Bertani (LB) broth or on LB agar (LBA) plates. Isopropyl β-D-1-thiogalactopyranoside (0.5 mM; IPTG; Sigma, USA) was used to induce promoter expression. When necessary, antibiotics were added at the following concentrations: ampicillin (Amp) at 100 μg/ml and kanamycin (Kan) at 50 μg/ml for E. coli and spectinomycin (Spc) at 50 μg/ml and cefoxitin (Cfx) at 1 μg/ml for RA.
Construction of Deletion Mutant Strains
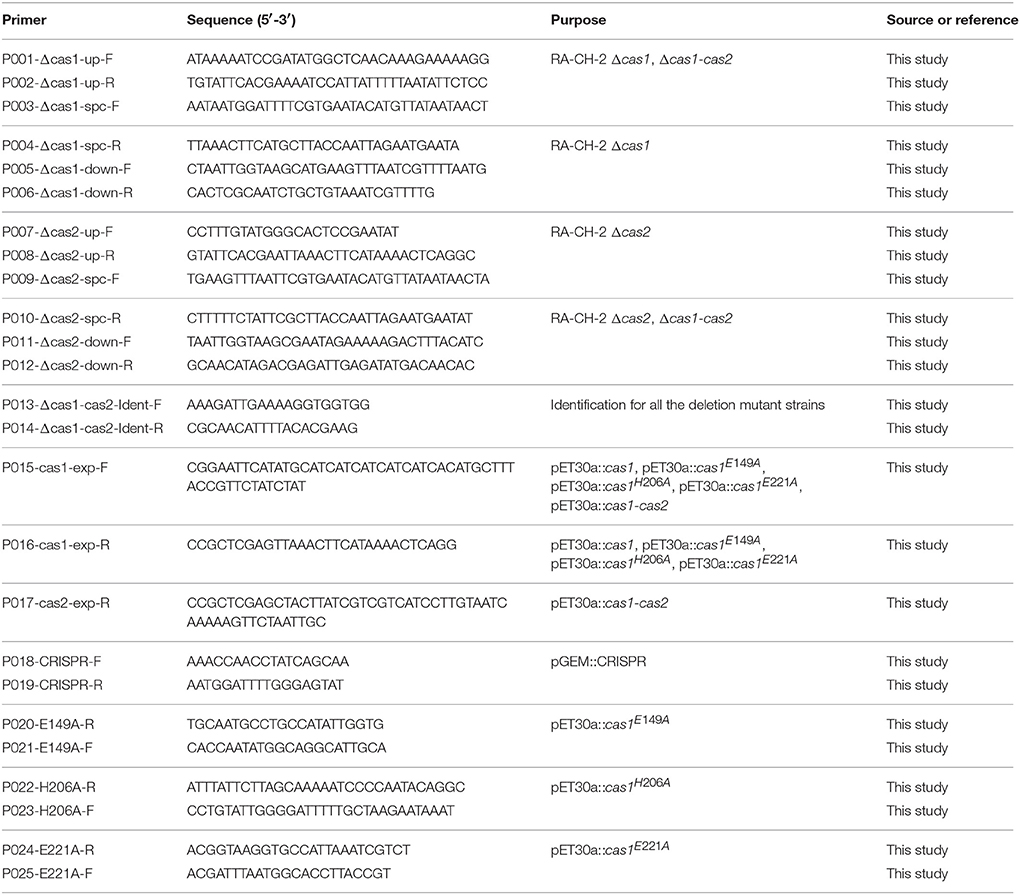
To generate deletion mutant strains, ~600-bp-long flanking sequences upstream and downstream of RA-CH-2 cas1 or cas2 were amplified by PCR using the primers described in Table 2. The sequence containing the SpcR cassette was amplified from the plasmid pAM238 (Fournier et al., 2011) and used as a selectable marker. The three PCR fragments (upstream, downstream, and SpcR cassette) were amplified using the overlap PCR method (Xiong et al., 2006). The mutant strains Δcas1, Δcas2, and Δcas1-cas2 were constructed by homologous recombination using the natural transformation method described by Liu et al. (2017b). Briefly, the PCR fragment (up-Spc-down) was mixed with 300 μl of RA-CH-2 (OD600 = 1). After incubation for 1 h at 37°C, the mixture was spread onto plates supplemented with Cfx and incubated overnight at 37°C until the appearance of colonies for PCR screening. Using the same method, the shuttle plasmid pLMF03 (Liu M. et al., 2016) was introduced into RA strains by natural transformation.
Construction of Expression Plasmids
To prepare expression plasmids, the cas1 gene of RA-CH-2 was amplified by PCR, digested by NdeI/XhoI and cloned into a similarly digested pET30a plasmid. Site-directed mutagenesis was performed by overlap PCR (Xiong et al., 2006) to substitute the residue E149, H206, or E221 of Cas1 with an alanine (Cas1E149A, Cas1H206A, or Cas1E221A). The cas1-mutant DNA fragment was obtained by two rounds of PCR and similarly cloned into pET30a to produce pET30a::cas1E149A, pET30a::cas1H206A, and pET30a::cas1E221A. The cas1-cas2 sequence was amplified by PCR from RA-CH-2 genomic DNA and cloned into the pET30a plasmid. E. coli BL21 (DE3) cells were transformed with the above plasmids using the calcium chloride method as described by Sambrook and Russell (2001).
Detection of Spacer Acquisition Using PCR
The strains carrying the shuttle plasmid were grown for 18 h at 37°C in TSB containing 1 μg/ml Cfx. The cells were then collected and frozen at −80°C in 20% (v/v) glycerol to be used as inocula in subsequent experiments. Each strain was inoculated into 5 ml of TSB without antibiotics and grown for 16 h with shaking (defined as one growth cycle). This process was performed for 20 growth cycles. At the same time, a volume of the culture (from cycles 1, 5, 10, 15, and 20) equivalent to 0.3 ml of the culture with an OD600 of 1.0 was pelleted by centrifugation, resuspended in 40 μl of water, boiled at 98°C for 10 min and centrifuged at 12,000 rpm for 1 min. The supernatant was used as the template for PCR amplification of the CRISPR1 locus from RA-CH-2. PCR products were analyzed on 2% TAE agarose gels stained with GoldView (SBS Genetech, China) to detect CRISPR1 array expansion. In addition, each single colony on antibiotic-free TSA plates was diluted in 20 μl of water and used as a PCR template. The PCR products of colonies with expanded arrays were submitted for sequencing.
In vivo Plasmid Loss Assays
Serial dilutions of the indicated culture at growth cycles 1, 5, 10, 15, or 20 were spread on TSA with or without 1 μg/ml Cfx, and the plates were incubated for 24 h at 37°C until colonies were visible. The plasmid loss rate was calculated by the following equation: 1-(the number of CFUs on the TSA-Cfx plate)/(the number of CFUs on the TSA plate). Experiments were performed in triplicate.
Analysis of New Spacers and the Protospacer Adjacent Motif (PAM)
Sequences of the expanded CRISPR1 arrays were analyzed using the CRISPRFinder program (http://crispr.i2bc.paris-saclay.fr/) (Grissa et al., 2007), and new spacer sequences were compiled and aligned to the target plasmid or genome sequence using SnapGene software (version 2.3.2, from GSL Biotech; available at snapgene.com) to determine the protospacer location and target strand. The protospacers with 10-bp-long flanking sequences were aligned to identify PAMs by using WebLogo (http://weblogo.berkeley.edu/logo.cgi/) (Crooks et al., 2004).
Expression and Purification of the R. anatipestifer Cas1 and Cas1 Mutant Proteins
Strains E. coli BL21 (DE3) pET30a::cas1, E. coli BL21 (DE3) pET30a::cas1E149A, E. coli BL21 (DE3) pET30a::cas1H206A, and E. coli BL21 (DE3) pET30a::cas1E221A were grown overnight in LB medium containing 50 μg/ml Kan. LB broth containing Kan was inoculated to an OD600 of 0.05 with the overnight culture, and the cells were grown at 37°C with shaking until the culture density reached an OD600 of 0.5. Expression was induced with 0.4 mM (final concentration) IPTG, and the cells were grown for an additional 6 h at 37°C. The induced cultures were harvested, and the cells were resuspended in 50 ml of buffer (pH 8.0, 300 mM NaCl, 50 mM NaH2PO4, 5 mM imidazole) and sonicated. The supernatant of cell lysates was collected by centrifugation and filtered through a 0.45-μm Millex syringe filter unit (Millipore, USA). The filtrate was subjected to nickel-chelate affinity chromatography (Ni-NTA agarose, Bio-Rad, USA) by following the manufacturer's instructions. Elution of the target protein was performed by competition with increasing concentrations of imidazole. Products from the course of purification were collected and measured by SDS-PAGE, and the protein concentration was determined using the Pierce BSA Protein Assay Kit (Pierce Biotechnology, USA). The purified target protein was dialyzed in 50 mM NaH2PO4 buffer (pH 8.0).
Nuclease Activity Assays
The CRISPR1 sequence from RA-CH-2, including the leader region, 16 repeats and 15 spacers, was cloned into the pGEM-T vector (Promega, USA), serving as the circular double-stranded DNA (dsDNA) substrate. Meanwhile, the pGEM::CRISPR vector was linearized with SacI (4.4 kb), serving as the linear dsDNA substrate. Circular or linear dsDNA substrates (0.15 μg) were incubated with the purified recombinant RA Cas1 protein or Cas1 mutants (1 μg) at 25°C for 90 min in the presence of 0.1 M KCl and 0.02 M HEPES (pH 7.5) and with either no metal ion or 2.5 mM manganese (Mn2+); deoxyribonuclease I (DNase I, 1 mg/ml) was used as a positive control for the nuclease reaction. The reaction products were extracted with phenol and analyzed by 1.5% agarose gel electrophoresis as described for the Pseudomonas aeruginosa Cas1 nuclease assay (Wiedenheft et al., 2009).
Preparation of Polyclonal Antibody
Five hundred microliters of an emulsion containing purified RA Cas1 (0.5 mg) and Freund's adjuvant (250 μl) was inoculated three times (at 1-week intervals) into 2-month-old rabbits (1.5 kg/rabbit). One week after the third immunization, blood samples were collected and centrifuged twice (5,000 rpm for 10 min) to obtain serum, which was then stored at −20°C. Before the serum was used, non-specific antibodies were removed by incubating the immune serum with E. coli cell extract for 1 h at 4°C, followed by centrifugation for 10 min at 8,000 rpm. The supernatant was then used as a polyclonal antibody.
Western Blotting Assays
Protein bands on SDS-PAGE gels were transferred to a polyvinylidene fluoride membrane (PVDF membrane, Millipore). The membrane was blocked with 5% skim milk and incubated with the rabbit polyclonal antibody against RA Cas1 protein or the FLAG-tag and then with goat antirabbit IgG-HRP (ZSGB-BIO, China). Protein bands on the membrane were visualized by the HRP-DAB Chromogenic Substrate Kit (Tiangen Biotech, China).
Ni-NTA Pull-Down Assays
The Cas1-His and Cas2-FLAG proteins were expressed from pET30a::cas1-cas2 in E. coli BL21 (DE3). Cell lysates of induced cultures (~108 CFU) were incubated with 50 μl of Ni-NTA agarose (Bio-Rad) that was pre-equilibrated with binding buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) for 16 h at 4°C. After incubation, the resin was washed four times with binding buffer containing 10 mM imidazole to remove non-specifically bound proteins. The resin was then resuspended in 30 μl of SDS loading buffer, heated to 100°C for 5 min and used for Western blot analysis using the anti-FLAG (Beyotime, China) as a primary antibody.
Immunoprecipitation Assays
Cell lysates with pET30a::cas1-cas2 (~108 CFU) were prepared as described above and incubated with rabbit anti-Cas1 polyclonal antibody overnight at 4°C with gentle shaking, followed by the addition of protein A+G agarose (Beyotime) for 2 h at 4°C. The samples above were washed five times with PBS, separated by SDS-PAGE and detected by Western blotting using anti-FLAG (Beyotime) or anti-Cas1 as primary antibodies.
Ethics Statement
Animal experiments were performed in strict compliance with the guidelines of the National Institutes of Health, and all procedures were approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University in Sichuan, China (Protocol Permit Number: XF2014-18).
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6 software. Statistical significance of the data was ascertained using Student's t-test. P-values < 0.05 were considered statistically significant.
Results
R. anatipestifer CH-2 Was Able to Acquire a New Spacer After the Introduction of an Exogenous Plasmid
The organization of the type II-C CRISPR1-Cas system from RA-CH-2 is shown in Figure 1. The CRISPR adaptation process of the type I-E system (Yosef et al., 2012; Díez-Villaseñor et al., 2013; Arslan et al., 2014; Nuñez et al., 2015) and the type II-A system (Heler et al., 2015; Wei et al., 2015b; Xiao et al., 2017) has been well studied; however, whether the type II-C CRISPR-Cas system is also capable of acquiring new spacers by itself remains unknown. To detect spacer acquisition of RA CRISPR-Cas system, the shuttle plasmid pLMF03 (Liu M. et al., 2016) was introduced into RA-CH-2 using natural transformation (Liu et al., 2017b). In addition, the 5′ end of RA-CH-2 CRISPR1 array was monitored by PCR using the primers shown in Figure 1 and Table 2 for the cultures sampled after the 1st−5th, 10, 15, and 20th growth cycles as described in the Materials and Methods section. As evidenced by the bands shown in Figure 2, two PCR products were amplified from the cultures sampled after two or more growth cycles but not from the culture sampled after one growth cycle. The higher band (~280 bp) was predicted to be the result of one spacer-repeat unit (~80 bp) being added to the CRISPR1 array, while the lower band represented the unexpanded array (Figure 2). Moreover, the higher band gained intensity as the growth cycles increased; in contrast, the lower band became increasingly weaker. This finding suggested that RA-CH-2 was able to acquire new spacers after the introduction of an exogenous plasmid.
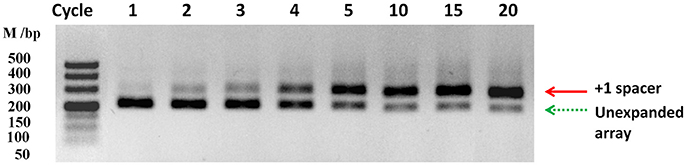
Figure 2. Spacer acquisition assays. The 5′ end of the CRISPR1 locus in RA-CH-2 population (harboring the shuttle plasmid pLMF03) was monitored by PCR amplification using the primers indicated in Figure 1 for the culture at the indicated growth cycle. Each growth cycle represents a 3:1,000 dilution of a previous 16-h culture grown for an additional 16 h in TSB without antibiotics. The red solid arrow represents a new spacer integrated into the CRISPR1 array (~280 bp), while the green dotted arrow represents the unexpanded array (~200 bp). The image is representative of multiple experiments.
The Active Process of Spacer Acquisition Contributed to the Decreased Stability of the Shuttle Plasmid in R. anatipestifer CH-2
Previous studies have shown that CRISPR-Cas system provides resistance against the entry of MGEs by acquiring spacers that match the MGEs (Barrangou et al., 2007; Deveau et al., 2008; Garneau et al., 2010; Sapranauskas et al., 2011). The shuttle plasmid pLMF03 triggered spacer acquisition of RA-CH-2 CRISPR-Cas system (Figure 2), raising the question of whether new spacers contribute to CRISPR/Cas-mediated interference with the plasmid. Thus, plasmid loss assay of the abovementioned cultures (cycles 1, 5, 10, 15, and 20, shown in Figure 2) was carried out by counting the colonies on TSA plates with or without antibiotics (Cfx), as described in the Materials and Methods section. The number of colonies on the TSA plate did not change significantly as the number of growth cycles increased. However, the number of colonies on the TSA plate containing Cfx decreased significantly (Figure 3A). After 20 growth cycles, the number of colonies on the Cfx-containing plate decreased to ~105 CFUs from ~109 CFUs (Figure 3A). Simultaneously, the rate of plasmid loss of the cultures was calculated. As shown in Figure 3B, after five or more growth cycles, more than 70% of the bacteria lost the shuttle plasmid pLMF03 (Figure 3B).
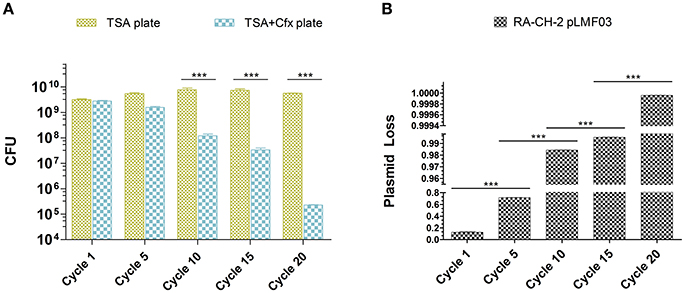
Figure 3. Plasmid loss assays. (A) Colony count of the cultures at growth cycles 1, 5, 10, 15, and 20 on TSA plates with or without antibiotics. (B) Plasmid loss rates of the cultures. The plasmid loss rate was calculated by the following equation: 1-(the number of CFUs on the TSA-Cfx plate)/(the number of CFUs on the TSA plate). Data were obtained from three independent experiments and analyzed using Student's t-test. The asterisks above the line indicate significance between the two indicated groups. ***p < 0.001.
Cas1 and Cas2 of R. anatipestifer CH-2 Were Required for Spacer Acquisition
Previous studies have indicated that Cas1 and Cas2 participate in the adaptation phase of CRISPR-Cas systems (Yosef et al., 2012; Arslan et al., 2014; Heler et al., 2015; Nuñez et al., 2015; Wei et al., 2015b; Fagerlund et al., 2017; Xiao et al., 2017). To determine the role of RA-CH-2 Cas1 and Cas2, the mutant strains RA-CH-2 Δcas1, RA-CH-2 Δcas2, and RA-CH-2 Δcas1-cas2 were constructed as described in the Materials and Methods section. The shuttle plasmid pLMF03 was introduced into the wild-type and mutant strains. These strains were cultured in TSB without antibiotics, and spacer acquisition assay of the cultures through passages for 10 growth cycles was performed using PCR. The expanded CRISPR1 arrays (with increased sizes of PCR products, ~280 bp) were observed in the wild-type strain (Figure 4, Lane 1). However, the PCR products amplified from the mutant strains had sizes corresponding to the unexpanded CRISPR1 array (~200 bp) (Figure 4, Lanes 2–4), indicating the absence of CRISPR expansion in the mutant strains. Spacer acquisition of the cas1 or cas2 deletion mutant was rescued by the introduction of the shuttle plasmid expressing both Cas1 and Cas2 (data not shown). These results suggested that Cas1 and Cas2 were required for spacer acquisition of RA-CH-2 CRISPR-Cas system.
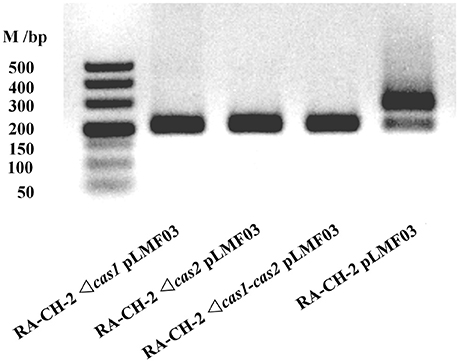
Figure 4. Cas1 and Cas2 were essential for RA-CH-2 spacer acquisition. Cultures of RA-CH-2 strains (wild-type and cas gene deletion mutant strains) harboring the shuttle plasmid pLMF03 were grown in TSB without antibiotics for 10 growth cycles. A PCR assay of the cultures was performed with the primers indicated in Figure 1 to detect spacer acquisition. The image is representative of multiple experiments.
Deletion of the Cas1 or Cas2 Gene of R. anatipestifer CH-2 Increased the Stability of the Shuttle Plasmid
To identify whether inactivating spacer acquisition affects the stability of the shuttle plasmid, plasmid loss assay of the abovementioned cultures (wild-type and mutant strains with pLMF03, as shown in Figure 4) was performed as described previously. The plasmid loss rate of the mutant strains was significantly lower than that of the wild-type strain (Figure 5). This result revealed that deletion of RA-CH-2 cas1 or cas2 gene largely eliminated spacer acquisition of the CRISPR-Cas system and subsequently stabilized the shuttle plasmid.
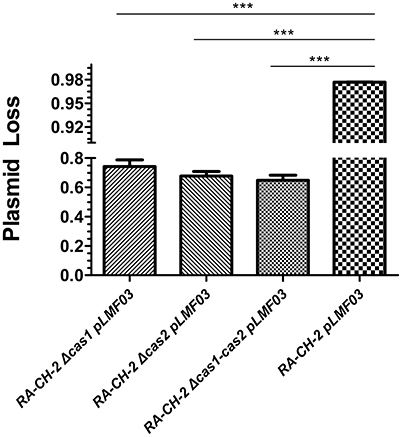
Figure 5. Deletion of RA-CH-2 cas1 or cas2 stabilized the shuttle plasmid. Plasmid loss from RA-CH-2 strains (wild-type and cas gene deletion mutant strains harboring pLMF03) were analyzed by counting the colonies on plates with or without antibiotics (Cfx). The rate of plasmid loss was calculated, and data obtained from three independent experiments were analyzed using Student's t-test. ***p < 0.001.
The Newly Inserted Spacers Were Derived From the Shuttle Plasmid
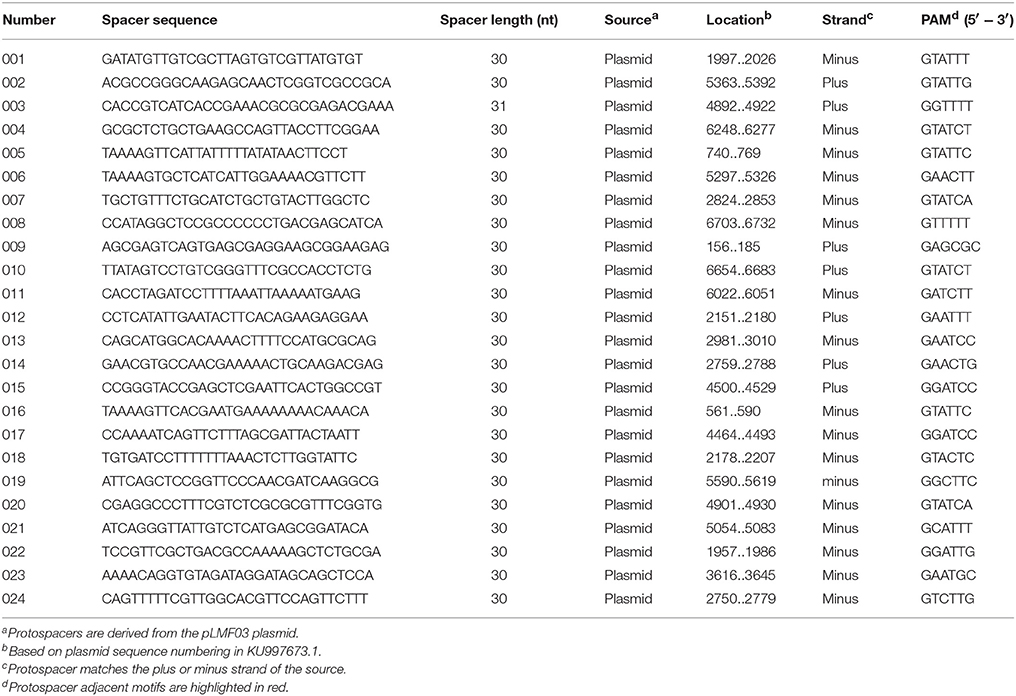
To further characterize the newly acquired spacers, expanded CRISPR1 arrays of 34 colonies were sequenced and analyzed. Sequencing data showed that the new spacer-repeat units were inserted into RA-CH-2 CRISPR1 array at the leader-proximal terminus (between cas2 and the first repeat, shown in Figure 1). Twenty-four unique spacers were obtained and are listed in Table 3. The length of most of the spacers was 30 bp, which is consistent with the observed spacer length in RA-CH-2 CRISPR1 array, with the exception of one spacer that was 31 bp in length. All spacers were derived from pLMF03 via sequence analysis. The positions and orientations of the protospacers are indicated on the pLMF03 plasmid map (Figure 6). Previous studies have shown that PAM plays an important role in the selection of the protospacer (Mojica et al., 2009; Wang et al., 2015; Leenay and Beisel, 2017). To identify the PAM sequence, 10-bp-long flanking sequences upstream or downstream of the protospacers were aligned by WebLogo. The observed PAMs were located immediately upstream of each protospacer, and the conserved motif was predicted as 5′-GWATTN-3′ (W represents A or T) (Figure 7 and Table 3). The first nucleotide of the PAMs, G, was significantly more conserved than the other bases.
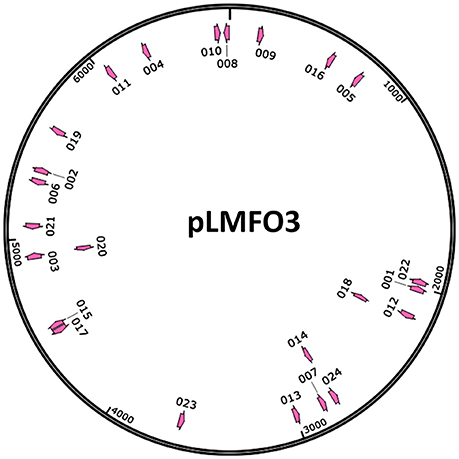
Figure 6. Distribution of the protospacers. The protospacers were numbered based on the sequenced spacers (as listed in Table 3) and are indicated on the pLMF03 plasmid map according to their positions and orientations (pink arrows). Arrows pointing in the clockwise direction represent protospacers matching the plus strand of the pLMF03 vector, and those pointing in the counterclockwise direction represent protospacers matching the minus strand.

Figure 7. Prediction of the PAM using WebLogo. The flanking sequences upstream of the protospacers (10 nt) were aligned using WebLogo. The first nucleotide (at position −1) was adjacent to the 5′ end of the protospacer. The sizes of the letters indicate the relative frequency of the corresponding base at that position, and the conserved PAM was predicted to be 5′-GWATTN-3′ (W represents A or T).
The Interaction of Cas1 and Cas2 of R. anatipestifer CH-2 in Vitro
The above results show that deletion of the cas1 or cas2 gene largely eliminated spacer acquisition. Correspondingly, the plasmid loss rate of the cas1 or cas2 deletion mutant strain was lower than that of the wild-type strain. It has been shown that in CRISPR-Cas systems from E. coli (Nuñez et al., 2014, 2015; Wang et al., 2015; Künne et al., 2016; Yoganand et al., 2017), Pectobacterium atrosepticum (Fagerlund et al., 2017), Streptococcus pyogenes (Heler et al., 2015), and Enterococcus faecalis (Xiao et al., 2017), interaction of Cas1 and Cas2 is required for these proteins to function in spacer acquisition. Thus, we asked whether Cas1 interacts with Cas2 to function in RA-CH-2 CRISPR-Cas system in the same pathway. To test the interaction of Cas1 and Cas2 in vitro, expression of Cas1-His and Cas2-FLAG fusion proteins was induced in BL21 (DE3) cells, and cell lysates were used to conduct Ni-NTA pull-down assays and immunoprecipitation assays. The pull-down sample from the Ni-NTA agarose assay exhibited two protein bands of different sizes by Western blotting using the anti-FLAG antibody (Figure 8, Lane 2). The protein with a molecular weight of ~50 kDa was predicted to be the complex of Cas1-His (35.5 kDa) and Cas2-FLAG (14.4 kDa), and the other protein, with a molecular weight of ~15 kDa, was the Cas2-FLAG fusion protein and was the same size as that observed in the untreated control sample (cell lysates before assay, Figure 8, Lane 3). The sample immunoprecipitated by the anti-Cas1 polyclonal antibody also presented the band corresponding to the His-Cas1-Cas2-FLAG complex (with a molecular weight of ~50 kDa) by Western blotting using either anti-FLAG or anti-Cas1 antibody (Figure 8, Lane 1 and 4), which was consistent with the pull-down assay. These results revealed that RA Cas1 and Cas2 interacted directly.
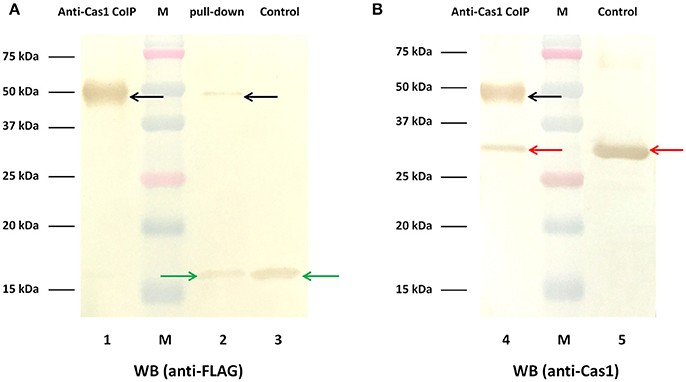
Figure 8. Interaction of RA Cas1 and Cas2 in vitro. (A,B) Western blotting using anti-FLAG and anti-Cas1 antibodies, respectively. Lanes 1 and 4, the samples immunoprecipitated by anti-Cas1. Lane 2, the pull-down sample obtained by using Ni-NTA agarose. Lanes 3 and 5, the untreated control samples (cell lysates before assay).
R. anatipestifer CH-2 Cas1 Exhibited DNA Nuclease Activity
To determine whether RA-CH-2 Cas1 exhibits DNA nuclease activity, DNase activity of the recombinant Cas1 protein was explored by adding this protein to circular or linear dsDNA substrates in the presence of 2.5 mM Mn2+ (or no metal ion), 0.1 M KCl, and 0.02 M HEPES (pH 7.5) at 25°C for 90 min. The electrophoretic profiles show that both the circular and linear dsDNA substrates were degraded by recombinant Cas1 to non-specific smear products in the presence of Mn2+ (Figure 9A, Lanes 3, 4, 8, and 9). However, the nuclease reactions without the Cas1 protein or Mn2+ abolished the cleavage of the dsDNA substrates (Figure 9A, Lanes 1, 2, 6, and 7), suggesting that Mn2+ assisted RA Cas1 recombinant protein in the cleavage of dsDNA.
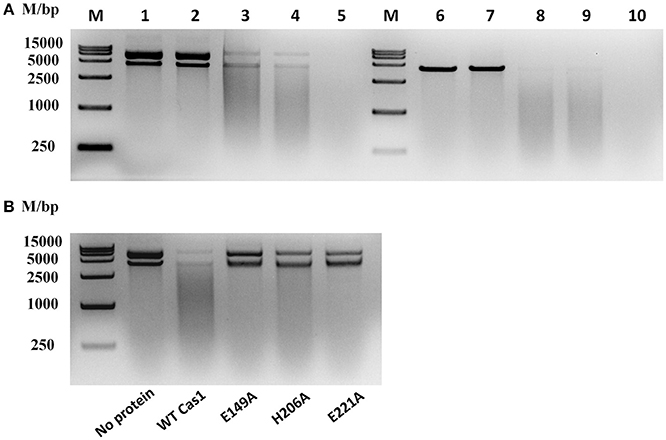
Figure 9. Nuclease activity assays for RA Cas1. (A) The recombinant RA Cas1 protein degraded circular and linear dsDNA. Lanes 1–5, circular dsDNA substrate. Lanes 6–10, linear dsDNA substrate. Lanes 1 and 6, no protein (no Cas1). Lanes 2 and 7, no metal ion (no Mn2+). Lanes 3, 4, 8, and 9, nuclease reactions of the recombinant Cas1 protein in the presence of Mn2+; Lanes 4 and 9, reaction products extracted with phenol. Lanes 5 and 10, deoxyribonuclease I (1 mg/ml DNase I) used for the nuclease reaction as a positive control. (B) Nuclease activity assays for Cas1 mutants; reaction products were extracted with phenol before electrophoresis. All nuclease reaction products were analyzed by 1.5% agarose gel electrophoresis.
By sequence analysis using the UniProt Knowledgebase (UniProtKB), three conserved residues of RA-CH-2 Cas1, namely, E149, H206, and E221, were predicted as functional sites for DNA binding and nuclease activity. To determine the roles of these residues in the nuclease activity of RA Cas1, Cas1 mutant proteins (Cas1E149A, Cas1H206A, and Cas1E221A, generated by site-directed mutagenesis) were expressed and purified for nuclease activity assays. In comparison with wild-type Cas1, the Cas1 mutants were unable to completely degrade dsDNA into smaller fragments under the conditions used in this study (Figure 9B). The above results suggested that RA Cas1 was a metal-dependent nuclease and was able to degrade circular and linear dsDNA. In addition, the results indicated that residues E149, H206, and E221 were the active sites of RA Cas1 nuclease.
Discussion
Subtype II-C CRISPR-Cas loci universally contain a trans-activating crRNA (tracrRNA), three cas genes (cas9, cas1, and cas2), a leader sequence (A/T-rich), and CRISPR (repeats and spacers) (Chylinski et al., 2014). In addition to containing these elements, subtype II-A and II-B loci contain Csn2 or Cas4, and these proteins have been found to be essential components for CRISPR adaptation (Li et al., 2014; Heler et al., 2015; Wei et al., 2015b). Subtype II-C CRISPR-Cas systems are prevalent in pathogens and commensals and are the most common subtypes among type II systems; however, these subtypes are the least well studied (Chylinski et al., 2014). Previously, the only report of type II-C adaptation was from Campylobacter jejuni, and the C. jejuni adaptation was unusual (Hooton and Connerton, 2014).
RA, the organism used in this study, is a pathogenic bacterium with high morbidity and mortality that primarily infects domestic ducks (Ruiz and Sandhu, 2013). Similar to C. jejuni, new spacer-repeat units from RA were inserted into the CRISPR array at the leader-proximal terminus (between cas2 and the first repeat, as shown in Figure 1), suggesting that spacer acquisition of type II-C system is generally directional (more data on the adaptation of type II-C systems from other organisms are needed to support this hypothesis). However, the adaptation of the C. jejuni CRISPR-Cas system requires a Cas4-like protein from the Campylobacter bacteriophage to activate spacer acquisition, and all the new spacers observed were derived from C. jejuni chromosomal DNA (Hooton and Connerton, 2014), which was quite different from the adaptation observed for RA CRISPR-Cas system.
The type II-C CRISPR-Cas system from RA-CH-2 was extremely active and was able to acquire new spacers after the introduction of an exogenous plasmid (containing no cas gene) on its own. All new spacers sequenced from RA-CH-2 were preferentially selected from the extrachromosomal DNA (the shuttle plasmid pLMF03), identical to E. coli and Streptococcus thermophilus. However, the adaptation of E. coli type I-E system (Yosef et al., 2012; Díez-Villaseñor et al., 2013) and S. thermophilus type II-A system (Wei et al., 2015b) was triggered by a plasmid expressing Cas1 and Cas2 (or Csn2) but not by an empty plasmid. One possibility is that the expression level of Cas1 and Cas2 was higher in RA-CH-2 but lower in E. coli and S. thermophilus. Another possibility is that Cas1 and Cas2 of RA-CH-2 were more active than those of E. coli and S. thermophilus. Interestingly, RNA-Seq data of RA-CH-2 showed that the transcription level of cas1 or cas2 was not particularly high compared to other genes of RA-CH-2 (our unpublished data). qRT-PCR analysis showed that expression levels of Cas1 and Cas2 in RA-CH-2 carrying the shuttle plasmid pLMF03 were both upregulated slightly, compared with wild type strain (Supplementary Figure 1).
As with the adaptation of E. coli and S. thermophilus CRISPR-Cas systems or other subtype systems, both Cas1 and Cas2 were required for the adaptation of RA CRISPR-Cas system, and deletion of either cas1 or cas2 from RA-CH-2 abrogated spacer acquisition, supporting the hypothesis that active adaptation contributes to CRISPR/Cas-mediated interference with the exogenous plasmid. In addition, RA-CH-2 Cas1 interacted with Cas2 directly. Moreover, RA Cas1 was able to degrade circular and linear dsDNA in the presence of Mn2+, and alanine substitution of residues E149, H206, and E221 abolished the nuclease activity of RA Cas1. It is hypothesized that the nuclease activity of RA Cas1 and the interaction of RA Cas1 with Cas2 are involved in the capture of prespacer substrates and integration of these substrates into the CRISPR array.
The conserved PAM of RA-CH-2 CRISPR-Cas system was predicted to be 5′-GWATTN-3′ and was located immediately upstream of the protospacer. The first nucleotide G of the observed PAM of RA-CH-2 system was highly conservative, consistent with the deduced PAM from the putative protospacer (100% similarity) of RA-CH-2 CRISPR1 spacer3. However, the consensus PAMs of other type II systems from S. pyogenes, S. thermophilus, Streptococcus aureus, and Neisseria meningitidis were located at the 3′ end of the protospacer (Leenay and Beisel, 2017). The location and sequence of the PAMs of those systems were quite different from those of the observed PAM of RA-CH-2 system. This is the first report of the PAM sequence of RA CRISPR-Cas system.
The CRISPR1 array of RA-CH-2 was active for inserting new spacers. However, no CRISPR amplification was observed in the CRISPR2 array (data not shown), probably because there are no cas genes flanking the CRISPR2 array of RA-CH-2, or the function of inserting new spacers into CRISPR2 array was lost. This study focused on the naïve adaptation of RA CRISPR-Cas system. It would be interesting to know whether primed adaptation is involved and the requirements in terms of Cas proteins for this process. Previous studies have revealed the role of type II CRISPR-Cas system in bacterial physiology and virulence (Sampson et al., 2013; Serbanescu et al., 2015). In this study, we observed that knockout of cas1 or/and cas2 did not have any effect on the growth of RA, and the effects on antibiotic resistance, energy metabolism, physiology, and virulence were not investigated in this study but should be investigated in the future.
The key feature of CRISPR/Cas-mediated immunity against MGEs is the ability to acquire new spacers to continuously update the CRISPR repertory (Jackson et al., 2017). Therefore, adaptation seems to be particularly important in CRISPR-Cas systems. Numerous and varied type II-C CRISPR-Cas systems remain to be explored, especially for their role in adaptation. Here, we provide the first experimental evidence of the naïve adaptation of type II-C CRISPR-Cas system, filling the knowledge gap in this field to a certain extent.
Author Contributions
AC, MW, and YH conceived and designed the project. LH, CL, and YH performed the research. XZ, HY, and DZ participated in the animal experiments. QY, YW, SZ, YL, YY, and LZ contributed to reagents, materials, analysis tools. XXZ, SC, RJ, and YH analyzed the data. MW and YH drafted the manuscript. AC and ML reviewed the manuscript. All authors have read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFD0500800), China Agricultural Research System (CARS-42-17), Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (CARS-SVDIP), and the Special Fund for the Key Laboratory of Animal Disease and Human Health of Sichuan Province (2016JPT0004).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00195/full#supplementary-material
References
Arslan, Z., Hermanns, V., Wurm, R., Wagner, R., and Pul, Ü. (2014). Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 42, 7884–7893. doi: 10.1093/nar/gku510
Barrangou, R. (2013). CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4, 267–278. doi: 10.1002/wrna.1159
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Burstein, D., Harrington, L. B., Strutt, S. C., Probst, A. J., Anantharaman, K., Thomas, B. C., et al. (2017). New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241. doi: 10.1038/nature21059
Cheng, A., Wang, M., Chen, X., Zhu, D., Huang, C., Liu, F., et al. (2003). Epidemiology and new serotypes of Riemerella anatipestifer isolated from ducks in China and studies on their pathogenic characteristics. Chinese J. Vet. Sci. 23, 320–323.
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Deveau, H., Barrangou, R., Garneau, J. E., Labonté, J., Fremaux, C., Boyaval, P., et al. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. doi: 10.1128/JB.01412-07
Díez-Villaseñor, C., Guzmán, N. M., Almendros, C., García-Martínez, J., and Mojica, F. J. (2013). CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 10, 792–802. doi: 10.4161/rna.24023
Fagerlund, R. D., Wilkinson, M. E., Klykov, O., Barendregt, A., Pearce, F. G., Kieper, S. N., et al. (2017). Spacer capture and integration by a type I-F Cas1-Cas2-3 CRISPR adaptation complex. Proc. Natl. Acad. Sci. U.S.A. 114, E5122–E5128. doi: 10.1073/pnas.1618421114
Fournier, C., Smith, A., and Delepelaire, P. (2011). Haem release from haemopexin by HxuA allows Haemophilus influenzae to escape host nutritional immunity. Mol. Microbiol. 80, 133–148. doi: 10.1111/j.1365-2958.2011.07562.x
Gao, Q., Tang, T., Cheng, A. C., Wang, M. S., Jia, R. Y., Zhu, D. K., et al. (2016). Development of an indirect ELISA using recombinant ompH protein for serological detection of Riemerella anatipestifer infection in ducks. Int. J. Clin. Exp. Med. 2, 1330–1337.
Garneau, J. E., Dupuis, M. E., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172
Heler, R., Samai, P., Modell, J. W., Weiner, C., Goldberg, G. W., Bikard, D., et al. (2015). Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202. doi: 10.1038/nature14245
Hooton, S. P., and Connerton, I. F. (2014). Campylobacter jejuni acquire new host-derived CRISPR spacers when in association with bacteriophages harboring a CRISPR-like Cas4 protein. Front. Microbiol. 5:744. doi: 10.3389/fmicb.2014.00744
Huang, L., Yuan, H., Liu, M. F., Zhao, X. X., Wang, M. S., Jia, R. Y., et al. (2017). Type B chloramphenicol acetyltransferases are responsible for chloramphenicol resistance in Riemerella anatipestifer, China. Front. Microbiol. 8:297. doi: 10.3389/fmicb.2017.00297
Jackson, S. A., McKenzie, R. E., Fagerlund, R. D., Kieper, S. N., Fineran, P. C., and Brouns, S. J. (2017). CRISPR-Cas: adapting to change. Science 356:eaal5056. doi: 10.1126/science.aal5056
Koonin, E. V., Makarova, K. S., and Zhang, F. (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 37, 67–78. doi: 10.1016/j.mib.2017.05.008
Künne, T., Kieper, S. N., Bannenberg, J. W., Vogel, A. I., Miellet, W. R., Klein, M., et al. (2016). Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol. Cell 63, 852–864. doi: 10.1016/j.molcel.2016.07.011
Leenay, R. T., and Beisel, C. L. (2017). Deciphering, communicating, and engineering the CRISPR PAM. J. Mol. Biol. 429, 177–191. doi: 10.1016/j.jmb.2016.11.024
Li, L., Zhu, D. K., Zhou, Y., Wang, M. S., Cheng, A. C., Jia, R. Y., et al. (2012). Adhesion and invasion to duck embryo fibroblast cells by Riemerella anatipestifer. Poult. Sci. 91, 3202–3208. doi: 10.3382/ps.2012-02552
Li, M., Wang, R., Zhao, D., and Xiang, H. (2014). Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492. doi: 10.1093/nar/gkt1154
Liao, H., Cheng, X., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2015). TonB energy transduction systems of Riemerella anatipestifer are required for iron and hemin utilization. PLoS ONE 10:e0127506. doi: 10.1371/journal.pone.0127506
Liao, H., Liu, M., Cheng, X., Zhu, D., Wang, M., Jia, R., et al. (2016). The detection of hemin-binding proteins in Riemerella anatipestifer CH-1. Curr. Microbiol. 72, 152–158. doi: 10.1007/s00284-015-0932-5
Liu, J., Zhu, D., Ma, G., Liu, M., Wang, M., Jia, R., et al. (2016). Genome-wide analysis of the synonymous codon usage patterns in Riemerella anatipestifer. Int. J. Mol. Sci. 17, 1304. doi: 10.3390/ijms17081304
Liu, M., Huang, M., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2017a). Identifying the genes responsible for iron-limited condition in Riemerella anatipestifer CH-1 through RNA-seq-based analysis. Biomed. Res. Int. 2017:8682057. doi: 10.1155/2017/8682057
Liu, M., Wang, M., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci. Rep. 6:37159. doi: 10.1038/srep37159
Liu, M., Zhang, L., Huang, L., Biville, F., Zhu, D., Wang, M., et al. (2017b). Use of natural transformation to establish an easy knockout method in Riemerella anatipestifer. Appl. Environ. Microbiol. 83, e00127–e00117. doi: 10.1128/AEM.00127-17
Luo, H. Y., Liu, M. F., Wang, M. S., Zhao, X. X., Jia, R. Y., Chen, S., et al. (2017). A novel resistance gene, lnu(H), conferring resistance to lincosamides in Riemerella anatipestifer CH-2. Int. J. Antimicrob. Agents 51, 136–139. doi: 10.1016/j.ijantimicag.2017.08.022
Luo, H., Liu, M., Wang, L., Zhou, W., Wang, M., Cheng, A., et al. (2015). Identification of ribosomal RNA methyltransferase gene ermF in Riemerella anatipestifer. Avian Pathol. 44, 162–168. doi: 10.1080/03079457.2015.1019828
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736. doi: 10.1038/nrmicro3569
Marraffini, L. A. (2015). CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. doi: 10.1038/nature15386
McGinn, J., and Marraffini, L. A. (2016). CRISPR-cas systems optimize their immune response by specifying the site of spacer integration. Mol. Cell 64, 616–623. doi: 10.1016/j.molcel.2016.08.038
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J., and Almendros, C. (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740. doi: 10.1099/mic.0.023960-0
Nuñez, J. K., Bai, L., Harrington, L. B., Hinder, T. L., and Doudna, J. A. (2016). CRISPR immunological memory requires a host factor for specificity. Mol. Cell 62, 824–833. doi: 10.1016/j.molcel.2016.04.027
Nuñez, J. K., Kranzusch, P. J., Noeske, J., Wright, A. V., Davies, C. W., and Doudna, J. A. (2014). Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534. doi: 10.1038/nsmb.2820
Nuñez, J. K., Lee, A. S., Engelman, A., and Doudna, J. A. (2015). Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198. doi: 10.1038/nature14237
Pathanasophon, P., Phuektes, P., Tanticharoenyos, T., Narongsak, W., and Sawada, T. (2002). A potential new serotype of Riemerella anatipestifer isolated from ducks in Thailand. Avian Pathol. 31, 267–270. doi: 10.1080/03079450220136576
Ruiz, J., and Sandhu, T. (2013). “Riemerella anatipestifer infection,” in Diseases of Poultry, ed Y. M. F. A. Saif (Iowa, LA: Blackwell Publishing), 823–828.
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Sampson, T. R., Saroj, S. D., Llewellyn, A. C., Tzeng, Y. L., and Weiss, D. S. (2013). A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257. doi: 10.1038/nature12048
Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., and Siksnys, V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282. doi: 10.1093/nar/gkr606
Segers, P., Mannheim, W., Vancanneyt, M., De Brandt, K., Hinz, K. H., Kersters, K., et al. (1993). Riemerella anatipestifer gen. nov., comb. nov., the causative agent of septicemia anserum exsudativa, and its phylogenetic affiliation within the Flavobacterium-Cytophaga rRNA homology group. Int. J. Syst. Bacteriol. 43, 768–776. doi: 10.1099/00207713-43-4-768
Serbanescu, M. A., Cordova, M., Krastel, K., Flick, R., Beloglazova, N., Latos, A., et al. (2015). Role of the Streptococcus mutans CRISPR-Cas systems in immunity and cell physiology. J. Bacteriol. 197, 749–761. doi: 10.1128/JB.02333-14
Shmakov, S., Smargon, A., Scott, D., Cox, D., Pyzocha, N., Yan, W., et al. (2017). Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 15, 169–182. doi: 10.1038/nrmicro.2016.184
Song, X. H., Zhou, W. S., Wang, J. B., Liu, M. F., Wang, M. S., Cheng, A. C., et al. (2016). Genome sequence of Riemerella anatipestifer strain RCAD0122, a multidrug-resistant isolate from ducks. Genome Announc. 4, e00332–e00316. doi: 10.1128/genomeA.00332-16
Wang, J., Li, J., Zhao, H., Sheng, G., Wang, M., Yin, M., et al. (2015). Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853. doi: 10.1016/j.cell.2015.10.008
Wang, M., Zhang, P., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2017). Identification of the ferric iron utilization gene B739_1208 and its role in the virulence of R. anatipestifer CH-1. Vet. Microbiol. 201, 162–169. doi: 10.1016/j.vetmic.2017.01.027
Wang, X. P., Zhu, D. K., Wang, M. S., Cheng, A. C., Jia, R. Y., Chen, S., et al. (2012). Development and application of specific polymerase chain reaction assay targeting the gyrB gene for rapid detection of Riemerella anatipestifer. Poult. Sci. 91, 2450–2453. doi: 10.3382/ps.2012-02375
Wang, X., Liu, W., Zhu, D., Yang, L., Liu, M., Yin, S., et al. (2014). Comparative genomics of Riemerella anatipestifer reveals genetic diversity. BMC Genomics 15:479. doi: 10.1186/1471-2164-15-479
Wang, X., Zhu, D., Wang, M., Cheng, A., Jia, R., Zhou, Y., et al. (2012). Complete genome sequence of Riemerella anatipestifer reference strain. J. Bacteriol. 194, 3270–3271. doi: 10.1128/JB.00366-12
Wei, Y., Chesne, M. T., Terns, R. M., and Terns, M. P. (2015a). Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 43, 1749–1758. doi: 10.1093/nar/gku1407
Wei, Y., Terns, R. M., and Terns, M. P. (2015b). Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 29, 356–361. doi: 10.1101/gad.257550.114
Wiedenheft, B., Zhou, K., Jinek, M., Coyle, S. M., Ma, W., and Doudna, J. A. (2009). Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17, 904–912. doi: 10.1016/j.str.2009.03.019
Xiao, Y., Ng, S., Nam, K. H., and Ke, A. (2017). How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550, 137–141. doi: 10.1038/nature24020
Xiong, A. S., Yao, Q. H., Peng, R. H., Duan, H., Li, X., Fan, H. Q., et al. (2006). PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 1, 791–797. doi: 10.1038/nprot.2006.103
Yi, H., Yuan, B., Liu, J., Zhu, D., Wu, Y., Wang, M., et al. (2017). Identification of a wza-like gene involved in capsule biosynthesis, pathogenicity and biofilm formation in Riemerella anatipestifer. Microb. Pathog. 107, 442–450. doi: 10.1016/j.micpath.2017.04.023
Yoganand, K. N., Sivathanu, R., Nimkar, S., and Anand, B. (2017). Asymmetric positioning of Cas1-2 complex and Integration Host Factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res. 45, 367–381. doi: 10.1093/nar/gkw1151
Yosef, I., Goren, M. G., and Qimron, U. (2012). Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576. doi: 10.1093/nar/gks216
Zhang, X., Wang, M. S., Liu, M. F., Zhu, D. K., Biville, F., Jia, R. Y., et al. (2017). Contribution of RaeB, a putative RND-type transporter to aminoglycoside and detergent resistance in Riemerella anatipestifer. Front. Microbiol. 8:2435. doi: 10.3389/fmicb.2017.02435
Zhong, C. Y., Cheng, A. C., Wang, M. S., Zhu, D. K., Luo, Q. H., Chen, S., et al. (2013). Quantitative real-time PCR study of the expression and regulation of the tetracycline resistance gene in Riemerella anatipestifer. Poult. Sci. 92, 1552–1559. doi: 10.3382/ps.2012-02672
Zhong, C. Y., Cheng, A. C., Wang, M. S., Zhu, D. K., Luo, Q. H., Zhong, C. D., et al. (2009). Antibiotic susceptibility of Riemerella anatipestifer field isolates. Avian Dis. 53, 601–607. doi: 10.1637/8552-120408-ResNote.1
Keywords: Riemerella anatipestifer, CRISPR-Cas, Cas1, Cas2, spacer acquisition
Citation: He Y, Wang M, Liu M, Huang L, Liu C, Zhang X, Yi H, Cheng A, Zhu D, Yang Q, Wu Y, Zhao X, Chen S, Jia R, Zhang S, Liu Y, Yu Y and Zhang L (2018) Cas1 and Cas2 From the Type II-C CRISPR-Cas System of Riemerella anatipestifer Are Required for Spacer Acquisition. Front. Cell. Infect. Microbiol. 8:195. doi: 10.3389/fcimb.2018.00195
Received: 14 February 2018; Accepted: 24 May 2018;
Published: 12 June 2018.
Edited by:
Marcel Doerflinger, Walter and Eliza Hall Institute of Medical Research, AustraliaReviewed by:
Yuqing Li, Sichuan University, ChinaOlga Soutourina, UMR9198 Institut de Biologie Intégrative de la Cellule (I2BC), France
Copyright © 2018 He, Wang, Liu, Huang, Liu, Zhang, Yi, Cheng, Zhu, Yang, Wu, Zhao, Chen, Jia, Zhang, Liu, Yu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Yang He
Yang He Mingshu Wang1,2,3†
Mingshu Wang1,2,3† Mafeng Liu
Mafeng Liu Xin Zhang
Xin Zhang Anchun Cheng
Anchun Cheng Dekang Zhu
Dekang Zhu