- 1Department of Bioinformatics, Biocenter, University of Würzburg, Würzburg, Germany
- 2Division of Microbiology, Biology/Chemistry, University of Osnabrück, Osnabrück, Germany
The human-pathogenic bacterium Salmonella enterica adjusts and adapts to different environments while attempting colonization. In the course of infection nutrient availabilities change drastically. New techniques, “-omics” data and subsequent integration by systems biology improve our understanding of these changes. We review changes in metabolism focusing on amino acid and carbohydrate metabolism. Furthermore, the adaptation process is associated with the activation of genes of the Salmonella pathogenicity islands (SPIs). Anti-infective strategies have to take these insights into account and include metabolic and other strategies. Salmonella infections will remain a challenge for infection biology.
Introduction
Salmonella enterica is a Gram-negative enterobacterium closely related to Escherichia coli (Neidhardt, 1996). Salmonellae reside in humans, a range of animals as well as in the environment and hence are facultative pathogens, often taken up by contaminated food and causing self-limited gastrointestinal disease. In weakened conditions the non-typhoidal serovars may lead to severe bloodstream infections, with high fatality rates in developing countries (Feasey et al., 2012) while typhoidal forms (S. enterica serovars Typhi, Paratyphi) strike with endotoxins, typhoid fever, and severe systemic illness. The millions of infections and thousands of fatal cases every year are an important reason for a better understanding and control of Salmonella infection (Feasey et al., 2012). To capture the diversity of the Salmonella lifestyle in infection is a challenging task. In this review, we will focus on metabolic aspects as well as on insights from “-omics” data, systems biology, and new technologies studying Salmonella infection. Salmonella, like several other Gamma-proteobacteria, are found in various environments including soils, water systems, and sewage, as well as in the gut flora of various animals. To survive and multiply in this large variety of environments, their metabolism has to adapt well (Rosenkrantz et al., 2013). The large genome of Salmonella contains more than 4000 genes encoding a large range of metabolic pathways, for instance an S. Typhi chromosome comprises 4,809,037 bp corresponding to 4599 ORFs (including 204 pseudogenes; Parkhill et al., 2001). The pseudogene complement of S. Typhi is involved in the tight host restriction of this important human pathogen. There is no zoonotic reservoire. The S. Typhi genome reveals an unexpectedly large diversity compared to its relatives E. coli and non-typhoidal Salmonella.
The lifestyle of Salmonella, featuring intestinal colonization, environmental survival, and transmission is reflected in unique gene clusters for adaptation to environmental niches and pathogenicity such as inside the host cell the Salmonella-containing vacuole (SCV). Adaptations include multiple abilities for oxygen and nitrate respiration (Rowley et al., 2012). Many further substrates can be used in multiple pathways, depending on environmental conditions. As a food-borne pathogen, various sugars such as D-glucosaminate can be used, supported by suitable permeases (Miller et al., 2013). A vivid picture emerges from data gained by recently established methodologies. Still, not enough is known about regulatory networks around the Salmonella Pathogenicity Island, the impact of effector proteins and transport processes and their role in shaping the conditions in the SCV.
In the following we will present established and new approaches of studying Salmonella infections, after which we address new perspectives on systems biology including postgenomic modeling techniques and functional genomics. We next discuss stress conditions and specific nutrient supplies and their impact on Salmonella metabolism, in particular amino acids and carbohydrate metabolism. Furthermore, connections between metabolism and virulence are discussed. These include SPI1 and SPI2 inducing conditions and their interplay with metabolism. New anti-infective Salmonella strategies take these aspects into account. In particular, one has to refine metabolic targeting and drug strategies accordingly. Salmonella infection is a particular challenging aspect of its versatile, highly adaptive life style.
Techniques for Studying the Intracellular Lifestyle of Salmonella
Systems biology provides a new technological perspective on Salmonella metabolism and virulence: this includes scarless mutation techniques, metabolic flux measurements by isotopologs and sophisticated -omics techniques allowing to study all aspects of the intracellular lifestyle of Salmonella in unprecedented detail.
Genetics
The very first step in the analysis of the importance of different metabolic enzymes is the generation of mutant strains. For Salmonella, the preferred method to rapidly delete chromosomal genes is the phage λ Red deletion technique (Datsenko and Wanner, 2000). Defined single or multiple gene deletion collections for S. Typhimurium have recently been published, covering deletions of 3517 genes (Porwollik et al., 2014). Double or multiple mutations, often needed to delete all isoenzymes of a given metabolic pathway, are commonly generated by repeated rounds of Red deletion, combined with phage P22 transduction (Zinder and Lederberg, 1952) and curing of antibiotic resistance. Since the sequential mutagenesis may lead to accumulation of recombination scars and generation of genomic chimera, newer approaches are based on scarless Red recombinase-mediated deletion (Blank et al., 2011).
Phenotyping
Before testing the influence of a deactivated metabolic enzyme on Salmonella virulence, a primary phenotypic characterization is often performed via determination of growth kinetics. By using minimal medium with different C-sources, Paterson et al. could reveal the ability of a Tpi (triosephosphate-isomerase) deficient strain to utilize gluconate, but not other sources such as glucose (Paterson et al., 2009). Additionally, one can perform growth kinetics with media which mimic different in vivo conditions. For instance Wallrodt et al. studied the role of the sulfurtransferases GlpE and PspE for resistance against NO radicals via growth kinetics in minimal medium with S-nitrosoglutathion supplementation (Wallrodt et al., 2013). To investigate the adaptation of Salmonella to life within the SCV, conditions inducing SPI2 genes are frequently used, such as minimal medium with low phosphate concentrations (Deiwick et al., 1999).
After these first phenotypic characterizations, the impact of defined gene deletions on Salmonella virulence is tested most commonly in cell culture experiments, such as gentamicin protection assays, which provide first clues about the role of metabolic enzymes, transporters, etc., on virulence. In this kind of assays the inability of gentamicin to penetrate into eukaryotic cells is used to kill extracellular bacteria, whereas internalized bacteria do not come into contact with the antibiotic substance (Lobo, 1973). With this method not only Salmonella's ability to enter host cells by invasion or phagocytosis but also the intracellular replication ability can be examined (Hölzer and Hensel, 2012).
Animal models
Comprehensive Salmonella infection models are animals and specific mouse strains are often used. In mice, Salmonella enterica serovars pathogenic for humans have been reported (Mathur et al., 2012) not to cause any disease due to an additional Toll-like receptor in mice (TLR11) but further studies have to further confirm this. However, S. enterica serovar Typhimurium, which can cause human diarrhea, causes a systemic infection in mice with pathology and disease progression similar to human typhoid fever in mice defective in Slc11a1 (or NRAMP) encoding a Fe2+/Mn2+/Zn2+transporter. Thus, to study the mechanisms of systemic disease caused by Salmonella, infection models using Salmonella-susceptible inbred mouse strains such as BALB/c or C57BL/6 with defective Slc11a1 allele are frequently used (Steeb et al., 2013). To understand gastroenteritis caused by Salmonella, a major breakthrough was the advent of the Streptomycin-pretreated mouse model. Application of Streptomycin reduces the intestinal microbiota and renders mice susceptible to Salmonella-induced intestinal inflammation. For this, C57BL/6 or similar mouse laboratory strains can be used and the Salmonella have to be Streptomycin resistant, e.g., S. enterica serovar Typhimurium SL1344 (reviewed in Kaiser et al., 2012).
Genomics
Methods useful in analyzing the global impact of gene deletions on Salmonella and “-omics” techniques (genomics, proteomics, transcriptomics, metabolomics) facilitate studies on virulence mechanisms and metabolic activities on a molecular level and allow a detailed picture of host-pathogen interactions. Comparative genomics was used for example to identify the presence of different metabolic pathways for non-typhoidal and typhoidal pathovars of Salmonella (Nuccio et al., 2014). Several recent studies use next generation sequencing (NGS) to understand non-typhoidal Salmonella genomes (reviewed by Wain et al., 2013). A broad collection of African isolates showed that they share a common ancestry with S. Typhimurium ST313. The study furthermore implies antibiotic resistances were acquired independently in two lineages of S. Typhimurium. These data are complemented by phage typing and pulse field gel electrophoresis (PFGE) for additional high resolution typing of Salmonella isolates by phage types and different PFGE patterns. This allows investigation in unprecedented detail of virulent strains as well as their correlation with metabolic resistance features such as pathways for degradation of antibiotics.
Transcriptomics
The second “-omics” level, namely transcriptomics including microarrays and high throughput sequencing approaches, gives insights into how Salmonella regulates its metabolic pathways in response to changing nutritional environments. A study performed by Blair et al. focused on changes in transcriptomic profiles when using LB or various minimal media for growth. Transcription profiles were established and the article instructively starts from microarray experiments (pan-Salmonella generation IV microarray) and verifies putative differences by quantitative real-time PCR (Blair et al., 2013). RNA sequencing was applied by Shah (2014) in a recent comparative study of global transcriptomes of high and low pathogenicity (LP) S. enterica serovar Enteritidis strains. This technique reveals important links between metabolism and virulence: in LP strains, reduced expression of virulence genes in SPI1 and SPI5 and defensive virulence factors were observed. Interestingly, this was combined with down regulation of metabolic defense pathways, in particular osmotic (glycine betaine/choline transport), oxidative (katE, sodC), and iron-limiting metabolic protection. In the four ferritins, bacterioferritin (Bfr) was found to be down-regulated in LP strains.
Proteomics and metabolomics
Mass spectroscopy (MS)-based proteomics is a method of choice when analyzing gene products: with this approach protein expression is directly measured. Typically only several matching peptides from a protein are identified applying the knowledge of the genome sequence and identified reading frames. This only partial peptide coverage for a given Salmonella protein is a challenge for MS analyses. Nevertheless, with more effort even quantification of proteins is possible applying different labeling techniques and standards. A good example for the application of the technique to Salmonella is the enzyme quantifications of ex vivo purified Salmonella performed by Steeb et al. (2013), also illustrating that many proteins can be fast analyzed in this way.
Metabolomics is an upcoming technique as it provides at the same time a global as well as direct view on Salmonella metabolism. In particular, isotopolog profiling (IP) allows analysis of current metabolic fluxes under defined conditions. For a detailed method explanation see the study by Härtel et al. (2012), demonstrating the technique on the central carbon metabolism and how individual fluxes are deduced by isotopolog patterns. Furthermore, Götz et al. used this technique to analyze the carbon metabolism of enterobacteria infecting CaCo cells and analyzed which carbon sources are used during intracellular growth (Götz et al., 2010). Metabolic measurements have also been improved by other new techniques such as engineering genetically encoded nanosensors from citrate binding proteins such as the histidine sensor kinase CitA to achieve in vivo measurements of changing citrate concentrations in E. coli by FRET. This system is readily applicable to Salmonella (Ewald et al., 2011).
In general, imaging techniques promote and complement the above approaches to studying the intracellular lifestyle of Salmonella. Non-invasive imaging techniques like radioisotope-labeled nucleosides, bioluminescence or the use of microscopy (e.g., advanced light microscopy such as with polarized light) coupled to different cell culture techniques (including establishing tissue infection models) offer here a wealth of information. A nice example including bioluminescent Salmonella, the Streptomycin mouse model and bioimaging is Pontier-Bres et al. (2014). Here metabolism and virulence are investigated on possibly the highest level: the protective effect of a pro-biotic food, Saccharomyces boulardii and its effect on Salmonella clearance in mice.
“-Omics” Data Integration and Systems Biology for Studying Salmonella During Infection
Data repositories
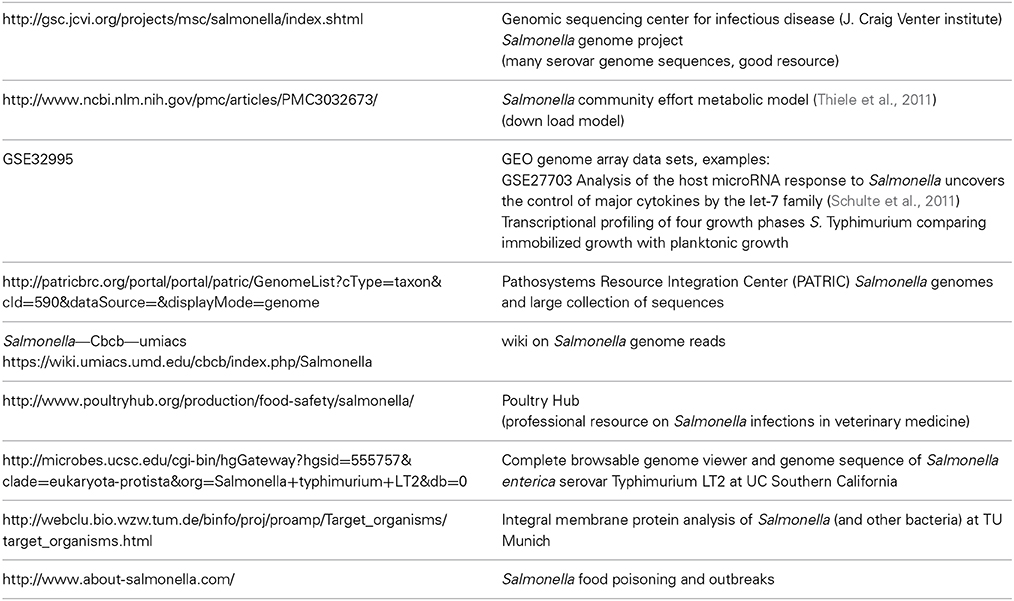
The combination of the various “-omics” approaches provides an integrated view on the adaptation of a pathogen to its host, ranging of from understanding of the genetic basis of virulence to the control of metabolic functions within a host organism or host cell. To describe infection processes on a holistic level, multi-omics strategies are required. Large “-omics” datasets on pathogens have become more readily available and have until now shaped the vivid picture of Salmonella infection. We present resources of “-omics” data which can be used to integrate and study different levels of systems biology of Salmonella infection (Table 1). This list compiles several useful resources but it is of course not exhaustive. Many “-omics” studies rely on large-scale sequence analysis using next-generation sequencing techniques on the genome or on RNA (RNAseq). This includes genome information from the Venter institute, different transcriptome data on gene expression and miRNAs from the Gene Expression Omnibus databank (GEO), proteomics data on membrane proteins from TU Munich, a Salmonella wiki on genome information as well as links for veterinary and medical resources on Salmonella infection.
Integrated analysis
Integration of high dimensional “-omics” datasets improves genome annotations, discovers novel virulence-related factors, and models Salmonella growth under infectious states (Ansong et al., 2012).
A multi-omics view on Salmonella in intestinal infection helps to better understand the interdependence of regulation and virulence vs. metabolic change, specific techniques and examples are given in Table 2. Thus, proteome, metabolome, glycome, and metagenome all change during the murine infection by S. enterica serovar Typhimurium. After multiplication in the mouse gut inflammation occurs and the whole microbiome changes: Bacteroidetes and Firmicutes are suppressed, Salmonella and Enterococcus grow (Deatherage Kaiser et al., 2013). In response to S. enterica serovar Typhimurium infection, potential novel innate immune factors can be discovered, there is transmigration and activation of neutrophils and up-regulation of cell surface molecules. Coordinate murine immune responses include complement activation and inflammatory antibacterial response. Salmonella metabolism reacts by induction of stress response proteins, synthesis of outer membrane proteins and lipoproteins.
The combination of integrated analysis of the different data sets shows that Salmonella reshapes its metabolism for its adaptation to different host environments. Virulence-associated remodeling adapts Salmonella to new niches and locations in the host, there nutrient-poor conditions are encountered and a strong protection against hostile environments of the host is mounted (Figure 1).
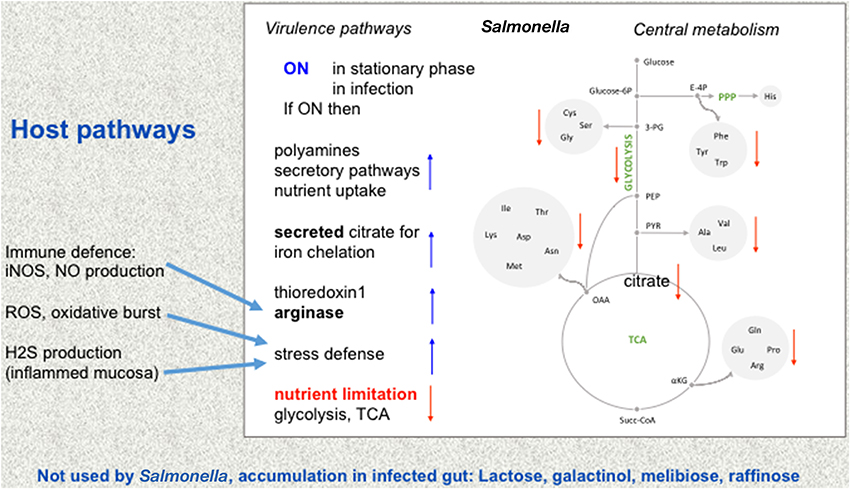
Figure 1. Metabolic adaptation of Salmonella. Changes from intestinal to intracellular lifestyle in the mucosa and the resulting adaptations are depicted. The central metabolism of Salmonella is shown in the mid panel, while virulence pathways are shown on the left. Environments may change from rather nutrient-rich conditions to nutrient-restricted conditions (red letters) such as in infection, for instance when Salmonella is ingested with contaminated food. Amino acids are abbreviated by their three letter code. Other abbreviations: PPP, pentose phosphate cycle; E-4P, erythrose 4-phosphate; Glucose-6P, glucose 6-phosphate; TCA, tricarbonic acid cycle (citric acid cycle). Some of the ensuing changes in Salmonella pathways are indicated (right, blue arrows up or red arrows down compared to rich nutrient environment, e.g., TCA goes down while some of the now less used citrate is used to chelate iron). Some metabolic changes from the host that influence Salmonella metabolism (Winter et al., 2010; blue arrows) are given on the left. Bottom: these sugars which are not used by Salmonella nor by the host accumulate in the infected gut.
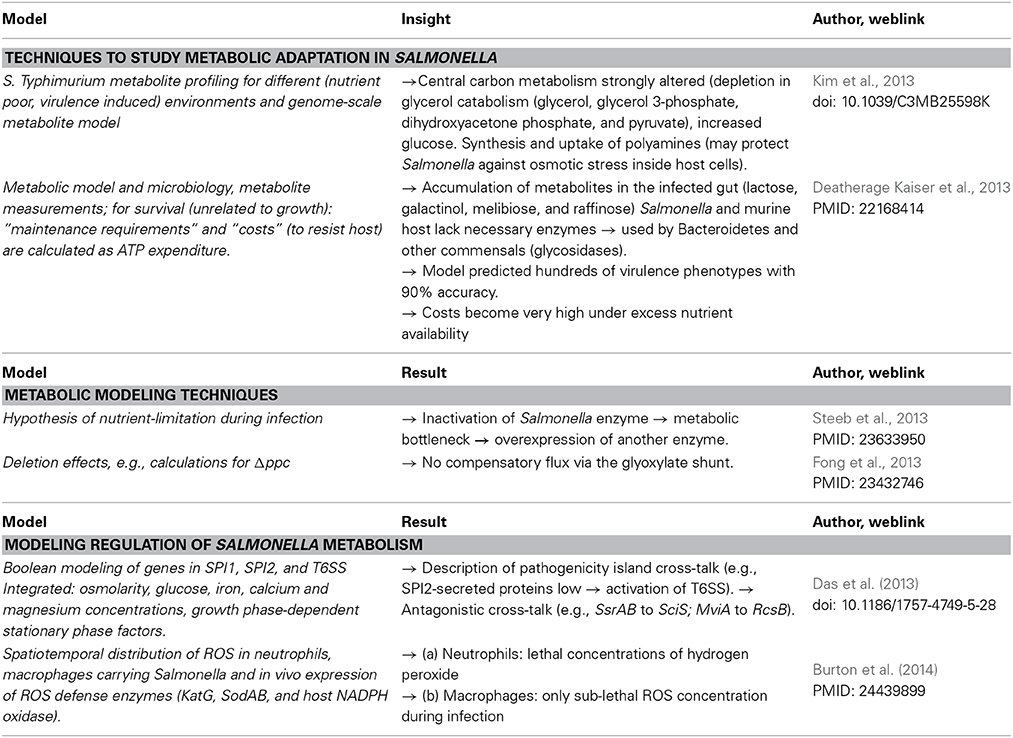
Metabolic modeling
Metabolic modeling of Salmonella in infection reveals an integrated picture of Salmonella adaptation processes. Furthermore, in the past few years, several groups established extensive, well-curated, models of Salmonella metabolism (Raghunathan et al., 2009; Thiele et al., 2011). Metabolic models are refined by considering additional energy required for stress defense mechanisms and adaptation during infection (Steeb et al., 2013) or considering metabolic bottlenecks (Table 2).
Modeling regulation of salmonella metabolism
Several studies analyzed Salmonella regulatory networks of genes in various SPI by means of mathematical models (Temme et al., 2008; Bailly-Bechet et al., 2011). Current results allow to model close to observation the sequential activation of virulence gene clusters in adaptation to distinct host environments (Table 2).
In the analysis of Salmonella-human interactions, large-scale cellular networks can already be described by looking at their structure, without attempting a dynamical simulation. Such graph-based methods mainly focusing on the topology to predict the chain of events in signaling or estimate metabolic capabilities. Here, cellular modules for different functions are identified as sub-graphs (sub-networks) with proteins mediating only this function in the complete network. Furthermore, hubs, central nodes in the network receiving many connections and indicating strongly connected genes or proteins, are of interest. For instance, interactome networks describing protein-protein interactions are built up and serve as scaffolds for further analysis (Schleker et al., 2012).
Rosenkrantz et al. (2013) compared two types of networks for S. Typhimurium strain LT2 regarding stress response and metabolic adaptation: a transcriptional data network using transcriptional data for 425 selected genes under different growth and stress conditions identifying the significantly and strongly regulated genes (transcriptional network) for each condition. This was compared to a genome-scale network connecting genes with metabolic pathways and cellular functions. Looking at the top five connecting hub proteins from the transcriptional network (wraB, ygaU, uspA, cbpA, and osmC) as well as the hubs in the genome scale metabolic pathway and cellular function network (ychN, siiF, yajD, ybeB, and dcoC), all these hubs were found to be dispensable for virulence in mutation studies. However, double mutants of these two sets of regulatory proteins showed clear effects on virulence in mouse infection experiments (Rosenkrantz et al., 2013). This is a particular strong example confirming the robust and well-buffered Salmonella regulation of metabolism and cellular function with virulence factors having partly redundant, overlapping functions.
Metabolic Adaptation of Salmonella During Stress Conditions
Stress factors linking virulence and metabolism
When Salmonella enters into an intestinal epithelial cell, environmental factors such as high osmolarity and neutral pH lead to an activation of HilD, which in turn induces HilA and invF gene expression (Altier, 2005). HilA as transcriptional regulator in turn activates all SPI1 genes necessary for assembly of the T3SS (Ellermeier and Slauch, 2007) and translocation of various SPI1 effector and host interaction proteins (Sop proteins, SipA) as well as DksA to coordinate NAD(P)H/NAD(P)(+) redox balance under nutrient limitation (Henard et al., 2010). For instance, SopB protein changes host cell exocytosis (Perret and Zhou, 2013). SPI1 gene expression is dependent on the growth phase (e.g., there is highest SPI1 induction after 3.5 h of growth in rich medium, Cossart and Sansonetti, 2004). Effector protein activity leads to reorganization of the host cell actin cytoskeleton, followed by membrane ruffling and internalization of Salmonella (Haraga et al., 2008). Next key factors influencing SPI2 expression (Haraga et al., 2008) such as detection of low osmolarity, low calcium concentrations and acidic pH by the two-component systems Envz/OmpR and SsrAB lead to activation of SPI2 gene expression (Garmendia et al., 2003) with factors such as SifA, SseJ, PipB2, and SseG (Núñez-Hernández et al., 2014) and result in a SCV containing multiplying Salmonella and inducing filaments. The combined action of these regulatory mechanisms ensures that sufficient nutrients are available for Salmonella during the infection (Figure 2).
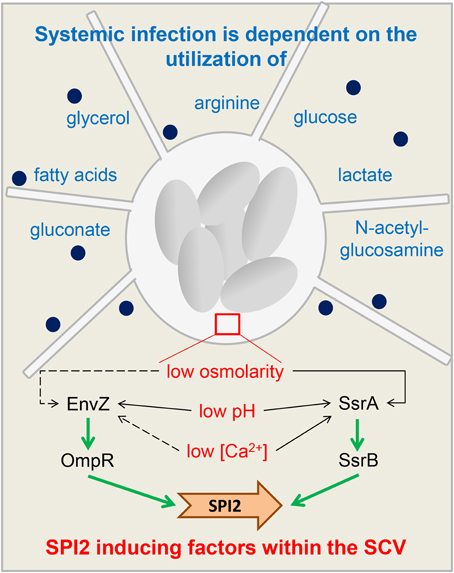
Figure 2. Regulatory adaptations and nutritional requirements of Salmonella. A schematic overview of functions of SPI2 during host cell infection and nutritional requirements to manifest a systemic infection. The Salmonella-containing vacuole (SCV) with replicating Salmonella (gray ovals) is connected to Salmonella-induced filaments. Blue writing: substrates Salmonella depends on to manifest systemic infections (Steeb et al., 2013). Dark blue spots: SPI2 effectors. The insert (brown rectangle) shows key factors influencing SPI2 expression (Haraga et al., 2008). Detection of low osmolarity, low calcium concentrations and acidic pH by the two-component systems Envz/OmpR and SsrAB inside the SCV leads to activation of SPI2 gene expression (Garmendia et al., 2003). Dotted arrow: minor effect on EnvZ/OmpR activity and SsrAB activity. Non-dotted arrow: strong effect on EnvZ/OmpR activity and SsrAB activity.
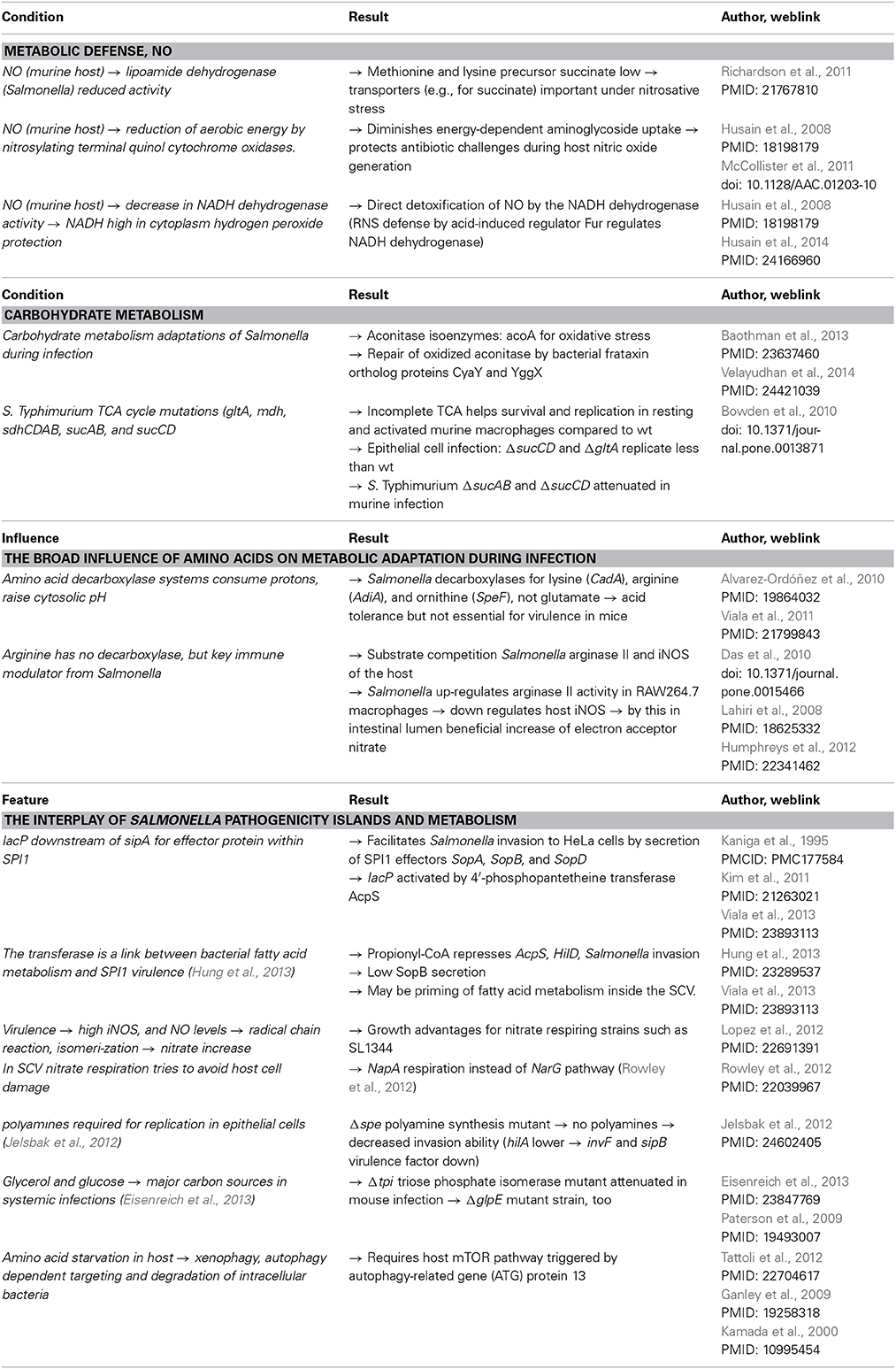
Metabolic defense pathways
Salmonella has to adapt its metabolism to different environmental stresses and niches when entering the human host, starting with the challenging acidic environment of the stomach (Table 3). Furthermore, immune defense reactions from the host involve free radicals, complement reaction, enzymatic degradation and autophagy reactions. Individual examples for these biochemical assaults on Salmonella have been studied in detail. Nitric oxide (NO) produced by the NO synthase of several immune cells of the host has a severe impact on central carbon metabolism of Salmonella. NO targets the pyruvate and α-ketoglutarate dehydrogenase complexes (Richardson et al., 2011).
Carbohydrate metabolism
Citrate is a TCA cycle intermediate (Figure 1) and is an important regulatory molecule in the control of glycolysis and lipid metabolism (Neidhardt, 1996). Furthermore, acetylation and deacetylation regulate the amount of glycolysis vs. gluconeogenesis as well as branching between citrate cycle and glyoxylate (Wang et al., 2010; Table 3). Moreover, citrate is a crucial iron-chelator which is involved in the homeostasis of iron in the pathogen, as well as the host. Iron is an essential component for several enzymes, but in high concentrations, it may cause damage. Citrate is consumed during NO exposure and other stress conditions because the export pump IctE (iron citrate efflux transporter, former called MdtD) transports iron chelated with citrate out of the cell. Export of citrate leads to growth arrest (Frawley et al., 2013), a status that allows it to survive antibiotic challenges as observed in persister bacteria. This function decreases harmful cellular iron content and reduces growth of Salmonella making it more stress resistant (Figure 1).
The broad influence of amino acids on metabolic adaptation during infection. The work on acetylation regulation in Salmonella by Wang et al. (2010) also underlines also that the central carbon as well as connected amino acid metabolism, including the TCA cycle, can directly be linked to stress response (Figure 1).
In particular, the bacterial arginine permease ArgT is an essential virulence determinant which decreases the host's cellular arginine content and reduces by this way the NO production of the host (Das et al., 2010; Table 3). In contrast, arginine degradation by Salmonella appears to be without influence on NO production. Although arginine degradation pathways are up-regulated in Salmonella during infection of macrophage and essential for virulence, this is due to other mechanisms but not related to substrate degradation of iNOS (Choi et al., 2012).
Cysteine is a key amino acid during oxidative stress response in Salmonella. In a study on cysteine biosynthesis during oxidative stress, cysteine biosynthesis regulation was blocked in ΔcysB and ΔcysE mutants and oxidative defense pathways encoded by katG and soxS were up-regulated compared to the wild-type strain (Turnbull and Surette, 2010). Consequently, the cysteine biosynthesis and cysteine-derived molecules such as thioredoxin play an important role for intracellular Salmonella survival and replication (Bjur et al., 2006). In this regard, the oxidoreductase thioredoxin 1 (TrxA) was found to be co-induced and essential for SPI2-T3SS activity under conditions that mimic life in the SCV (Negrea et al., 2009).
The Richness of Salmonella Metabolism and its Influence on Virulence
The SsrAB virulon controlling SPI2 gene expression is induced under nutrient-poor conditions (e.g., presence in the phagosome, Kuhle and Hensel, 2004).
The interplay of salmonella pathogenicity islands and metabolism
Various metabolic pathways which have an impact on the SPI1 activity of Salmonella enterica (Table 3). One example is the interaction between the invasion acyl carrier protein (IacP; Viala et al., 2013) and secretion of SPI1 effector proteins into the host cell to achieve rearrangement of the host cytoskeleton and engulfment of the bacterium (reviewed in Cossart and Sansonetti, 2004).
There are also indications for the influence of SPI1 functions on the host's metabolism in order to facilitate survival in the intestine and subsequently intracellular to promote the infection process. Thus, the SPI1-T3SS effector protein SopE is known to increase Salmonella invasiveness and to induce strong inflammatory host responses (Humphreys et al., 2012).
Although SPI1 and SPI2 are induced under very distinct nutritional environments (SPI1 in a nutrient rich environment, SPI2 by nutrient starvation, Kuhle and Hensel, 2004), there are some bacterial metabolites which effect SPI1 as well as SPI2 activity and have a general impact on Salmonella virulence (Table 3). One example are polyamines, short cationic amines, of which spermidine and putrescine are mostly common in bacteria (Jelsbak et al., 2012).
Intracellular adaptation and metabolism of Salmonella. While conditions in the intestinal lumen are nutrient rich, the situation changes after Salmonella invades into the epithelial cells and is phagocytosed at the basolateral cell side by macrophages or dendritic cells. Staying inside the SCV, the pathogen has to deal with nutrient limitations. To investigate which metabolites could interact with expression of genes in SPI1 or mainly SPI2, one issue is to define the nutritional situation of Salmonella gain inside the SCV and to figure out which metabolites Salmonella has access to. Mouse infection experiments showed on the one hand that intracellular Salmonella get access to a wide range of nutrients, including nearly all amino acids except proline. On the other hand, it was shown that the ability to manifest a full systemic infection is dependent on the utilization of “glycerol, fatty acids, N-acetylglucosamine, gluconate, glucose, lactate, and arginine” (Steeb et al., 2013). However, Salmonella is able to counteract various defense mechanisms in order to facilitate growth or reduce immune responses (Table 3). Invasion of pathogens into epithelial cells is followed by cytosolic amino acid starvation in host cells, which seems to be explained by membrane damage during the invasion process (Tattoli et al., 2012).
However, in contrast to Shigella-infected cells, amino acid levels of epithelial cells invaded by Salmonella normalized 3 h after infection, which leads to relocalization of mTor invasion sustaining pathway to the SCV, phosphorylation of ATG protein 13, leading to a low ATG protein 1 activity and thus reduced autophagy (Ganley et al., 2009). By this, Salmonella is able to avoid autophagy in epithelial cells. Further investigations are required to clarify if normalization of amino acid levels is directly induced by Salmonella. At least the invasion-induced membrane disturbance is only severe in the first hour of infection and somehow repaired faster than in cases of invasion by other intracellular pathogens (Tattoli et al., 2012).
Anti-Infective Strategies in the Face of Robust Salmonella Metabolism
As Salmonella adapts rapidly and successfully to changing conditions including intracellular survival in macrophages, in epithelia and in the gut, we will now examine which antibiotic strategies are nevertheless available for Salmonella infections. A seminal work by Becker and co-workers showed that the robust metabolism of Salmonella limits possibilities for new antibiotics (Becker et al., 2006) and Bumann stressed this point asking “has nature already identified all useful antibacterial targets?” (Bumann, 2008). It is of course important to mention the billions of years sampling time to test and select bacteria and bacterial metabolism during evolution. Furthermore, the parallel exploitation of diverse host nutrients often enhances often Salmonella virulence (Steeb et al., 2013) and persistent Salmonella are highly resilient (Barat et al., 2012). On the other hand, as many medical areas such as cancer research or aging research also make clear, any medical intervention happened only very recently in evolutionary times. Hence, additional medical interventions are not limited by evolutionary constraints such as positive epistatic selection or direct metabolic energy costs. There are many potential targets still in stock, both by targeting metabolic pathways in pathogenic bacteria and Salmonella in particular, as well as by exploring novel ways of anti-infectives. One inspiring example is metabolic engineering of Salmonella vaccine bacteria in the mevalonate pathway to boost human Vγ2Vδ2 T cell immunity (Workalemahu et al., 2014). As reviewed earlier (Dandekar and Dandekar, 2010), anti-infective action starts furthermore from typical hygienic measures such as isolation of patients with multi-resistant strains including silent clinical carriers, but also includes targeted disturbance of metabolic pathways for example by sulfonamides. In particular, both targeted therapy (direct delivery of an antibiotic to only the location it should act, e.g., in the intestine) as well as targeted modification of standard drugs (so that they are more detrimental to the pathogen even if the host shares similar proteins) are options which have high potential and are not much explored. Our own research highlights the interconnectivity of metabolism. This renders Salmonella also vulnerable also in conserved and well investigated pathways, such as TCA cycle and its anaplerotic reactions. Thus, Salmonella Typhimurium is controlled by host NO production as shown in mice experiments in vivo. Methionine or lysine auxotrophy results from reduced succinyl-CoA availability as the lipoamide dehydrogenase activity is targeted by NO while compensatory Salmonella pathways to achieve more succinyl-CoA are again blocked by NO (Richardson et al., 2011). Here it also becomes obvious why the therapeutic strategies are not easy exhausted, for instance by direct delivery of NO-increasing drugs to the severely infected gut. Anesthetic drugs are membrane modifiers yielding even multi-resistant pathogens again vulnerable to additional antibiotics, just to cite another possibility (Dandekar and Dandekar, 2010). Furthermore, novel vaccination strategies may proof successful. Hence, the task is more to implement some of the many open alleys for novel antibiotic therapies in clinic. This includes targeting of the metabolism. Furthermore, clinical studies are required for each novel antibiotic strategy, these are currently too expensive for high patient numbers and the prize margin for antibiotics is low so antibiotic development pipelines dry out. However, the prices for such clinical studies could be drastically lowered by modern patient hospital information systems, and furthermore, public awareness and willingness to have better protection against infections is currently increasing.
Conclusions: Salmonella General Metabolic Lifestyle During Infection
We saw that multiple “-omics” and especially metabolomic data are currently used to determine the needs for Salmonella to facilitate intracellular survival within the SCV in host cells and its nutrient supply.
Salmonella's generalist metabolic lifestyle meets all types of environmental challenges, be it ROS or nutrient limitation by its broad metabolic capabilities. The broad metabolism suggests nevertheless potential for novel anti-infective strategies. However, under severe conditions Salmonella regulation and metabolism are spiked up by input from SPI1, SPI2, T3SS and T6SS, modified invasion abilities, redox protection and central metabolism to turn the neutral environmental lifestyle of Salmonella into a pathogenic lifestyle for its host. On top of this such genetic modules catalyze rapid genetic exchange between Salmonella strains showing that only an integrated picture will help to sustain antibiotic efficiency against Salmonella infections.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the German Research Foundation (DFG), grants Da 208/13-1 and He 1964/14-2. We thank Jennifer Heilig for critical revision of the manuscript.
References
Altier, C. (2005). Genetic and environmental control of Salmonella invasion. J. microbiol. 43 Spec No, 85–92.
Alvarez-Ordóñez, A., Fernández, A., Bernardo, A., and López, M. (2010). Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int. J. Food Microbiol. 136, 278–282. doi: 10.1016/j.ijfoodmicro.2009.09.024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ansong, C., Deatherage, B. L., Hyduke, D., Schmidt, B., McDermott, J. E., Jones, M. B., et al. (2012). Studying Salmonellae and yersiniae host-pathogen interactions using integrated'-omics' and modeling. Curr. Top. Microbiol. Immunol. 363, 21–41. doi: 10.1007/82_2012_247
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailly-Bechet, M., Benecke, A., Hardt, W. D., Lanza, V., Sturm, A., and Zecchina, R. (2011). An externally modulated, noise-driven switch for the regulation of SPI1 in Salmonella enterica serovar Typhimurium. J. Math. Biol. 63, 637–662. doi: 10.1007/s00285-010-0385-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Baothman, O. A., Rolfe, M. D., and Green, J. (2013). Characterization of Salmonella enterica serovar Typhimurium aconitase A. Microbiology 159, 1209–1216. doi: 10.1099/mic.0.067934-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Barat, S., Steeb, B., Mazé, A., and Bumann, D. (2012). Extensive in vivo resilience of persistent Salmonella. PLoS ONE 7:e42007. doi: 10.1371/journal.pone.0042007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Becker, D., Selbach, M., Rollenhagen, C., Ballmaier, M., Meyer, T. F., Mann, M., et al. (2006). Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440, 303–307. doi: 10.1038/nature04616
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bjur, E., Eriksson-Ygberg, S., Aslund, F., and Rhen, M. (2006). Thioredoxin 1 promotes intracellular replication and virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 74, 5140–5151. doi: 10.1128/IAI.00449-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blair, J. M. A., Richmond, G. E., Bailey, A. M., Ivens, A., and Piddock, L. J. V. (2013). Choice of bacterial growth medium alters the transcriptome and phenotype of Salmonella enterica serovar Typhimurium. PLoS ONE 8:e63912. doi: 10.1371/journal.pone.0063912
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Blank, K., Hensel, M., and Gerlach, R. G. (2011). Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS ONE 6:e15763. doi: 10.1371/journal.pone.0015763
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowden, S. D., Ramachandran, V. K., Knudsen, G. M., Hinton, J. C., and Thompson, A. (2010). An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS ONE 5:e13871. doi: 10.1371/journal.pone.0013871
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bumann, D. (2008). Has nature already identified all useful antibacterial targets? Curr. Opin. Microbiol. 11, 387–392. doi: 10.1016/j.mib.2008.08.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Burton, N. A., Schürmann, N., Casse, O., Steeb, A. K., Claudi, B., Zankl, J., et al. (2014). Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15, 72–83. doi: 10.1016/j.chom.2013.12.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Choi, Y., Choi, J., Groisman, E. A., Kang, D.-H., Shin, D., and Ryu, S. (2012). Expression of STM4467-encoded arginine deiminase controlled by the STM4463 regulator contributes to Salmonella enterica serovar Typhimurium virulence. Infect. Immun. 80, 4291–4297. doi: 10.1128/IAI.00880-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cossart, P., and Sansonetti, P. J. (2004). Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304, 242–248. doi: 10.1126/science.1090124
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dandekar, T., and Dandekar, G. (2010). Pharmacogenomic strategies against microbial resistance: from bright to bleak to innovative. Pharmacogenomics 11, 1193–1196. doi: 10.2217/pgs.10.18
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Das, C., Dutta, A., Rajasingh, H., and Mande, S. S. (2013). Understanding the sequential activation of type III and type VI secretion systems in Salmonella typhimurium using boolean modeling. Gut Pathog. 5, 28. doi: 10.1186/1757-4749-5-28
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Das, P., Lahiri, A., Lahiri, A., Sen, M., Iyer, N., Kapoor, N., et al. (2010). Cationic amino acid transporters and Salmonella Typhimurium ArgT collectively regulate arginine availability towards intracellular Salmonella growth. PLoS ONE 5:e15466. doi: 10.1371/journal.pone.0015466
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deatherage Kaiser, B. L., Li, J., Sanford, J. A., Kim, Y. M., Kronewitter, S. R., Jones, M. B., et al. (2013). A multi-omic view of host-pathogen-commensal interplay in Salmonella-mediated intestinal infection. PLoS ONE 8:e67155. doi: 10.1371/journal.pone.0067155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Deiwick, J., Nikolaus, T., Erdogan, S., and Hensel, M. (1999). Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31, 1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenreich, W., Heesemann, J., Rudel, T., and Goebel, W. (2013). Metabolic host responses to infection by intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 3:24. doi: 10.3389/fcimb.2013.00024
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ellermeier, J. R., and Slauch, J. M. (2007). Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10, 24–29. doi: 10.1016/j.mib.2006.12.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ewald, J. C., Reich, S., Baumann, S., Frommer, W. B., and Zamboni, N. (2011). Engineering genetically encoded nanosensors for real-time in vivo measurements of citrate concentrations. PLoS ONE 6:e28245. doi: 10.1371/journal.pone.0028245
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Feasey, N. A., Dougan, G., Kingsley, R. A., Heyderman, R. S., and Gordon, M. A. (2012). Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499. doi: 10.1016/S0140-6736(11)61752-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fong, N. L., Lerman, J. A., Lam, I., Palsson, B. O., and Charusanti, P. (2013). Reconciling a Salmonella enterica metabolic model with experimental data confirms that overexpression of the glyoxylate shunt can rescue a lethal ppc deletion mutant. FEMS Microbiol. Lett. 342, 62–69. doi: 10.1111/1574-6968.12109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frawley, E. R., Crouch, M. L., Bingham-Ramos, L. K., Robbins, H. F., Wang, W., Wright, G. D., et al. (2013). Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc. Natl. Acad. Sci. U.S.A. 110, 12054–12059. doi: 10.1073/pnas.1218274110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ganley, I. G., Lam, D. H., Wang, J., Ding, X., Chen, S., and Jiang, X. (2009). ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305. doi: 10.1074/jbc.M900573200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Garmendia, J., Beuzon, C. R., Ruiz-Albert, J., and Holden, D. W. (2003). The roles of SsrA-SsrB and OmpR-EnvZ in the regulation of genes encoding the Salmonella Typhimurium SPI-2 type III secretion system. Microbiology 149, 2385–2396. doi: 10.1099/mic.0.26397-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Götz, A., Eylert, E., Eisenreich, W., and Goebel, W. (2010). Carbon metabolism of enterobacterial human pathogens growing in epithelial colorectal adenocarcinoma (Caco-2) cells. PLoS ONE 5:e10586. doi: 10.1371/journal.pone.0010586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Haraga, A., Ohlson, M. B., and Miller, S. I. (2008). Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66. doi: 10.1038/nrmicro1788
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Härtel, T., Eylert, E., Schulz, C., Petruschka, L., Gierok, P., Grubmüller, S., et al. (2012). Characterization of central carbon metabolism of Streptococcus pneumoniae by isotopologue profiling. J. Biol. Chem. 287, 4260–4274. doi: 10.1074/jbc.M111.304311
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Henard, C. A., Bourret, T. J., Song, M., and Vázquez-Torres, A. (2010). Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J. Biol. Chem. 285, 36785–36793. doi: 10.1074/jbc.M110.160960
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hölzer, S. U., and Hensel, M. (2012). Divergent roles of Salmonella pathogenicity island 2 and metabolic traits during interaction of S. enterica serovar typhimurium with host cells. PLoS ONE 7:e33220. doi: 10.1371/journal.pone.0033220
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Humphreys, D., Davidson, A., Hume, P. J., and Koronakis, V. (2012). Salmonella virulence effector SopE and host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 11, 129–139. doi: 10.1016/j.chom.2012.01.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hung, C.-C., Garner, C. D., Slauch, J. M., Dwyer, Z. W., Lawhon, S. D., Frye, J. G., et al. (2013). The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol. 87, 1045–1060. doi: 10.1111/mmi.12149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Husain, M., Bourret, T. J., McCollister, B. D., Jones-Carson, J., Laughlin, J., and Vázquez-Torres, A. (2008). Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J. Biol. Chem. 283, 7682–7689. doi: 10.1074/jbc.M708845200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Husain, M., Jones-Carson, J., Liu, L., Song, M., Saah, J. R., Troxell, B., et al. (2014). Ferric uptake regulator-dependent antinitrosative defenses in Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 82, 333–340. doi: 10.1128/IAI.01201-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jelsbak, L., Thomsen, L. E., Wallrodt, I., Jensen, P. R., and Olsen, J. E. (2012). Polyamines are required for virulence in Salmonella enterica serovar Typhimurium. PLoS ONE 7:e36149. doi: 10.1371/journal.pone.0036149
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaiser, P., Diard, M., Stecher, B., and Hardt, W. D. (2012). The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen's virulence factors, and the host's mucosal immune response. Immunol. Rev. 245, 56–83. doi: 10.1111/j.1600-065X.2011.01070.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513. doi: 10.1083/jcb.150.6.1507
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaniga, K., Trollinger, D., and Galan, J. E. (1995). Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella Typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177, 7078–7085.
Kim, J. S., Eom, J. S., Jang, J. I., Kim, H. G., Seo, D. W., Bang, I.-S., et al. (2011). Role of Salmonella Pathogenicity Island 1 protein IacP in Salmonella enterica serovar Typhimurium pathogenesis. Infect. Immun. 79, 1440–1450. doi: 10.1128/IAI.01231-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kim, Y.-M., Schmidt, B. J., Kidwai, A. S., Jones, M. B., Deatherage Kaiser, B. L., Brewer, H. M., et al. (2013). Salmonella modulates metabolism during growth under conditions that induce expression of virulence genes. Mol. Biosyst. 9, 1522–1534. doi: 10.1039/c3mb25598k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuhle, V., and Hensel, M. (2004). Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell. Mol. Life Sci. 61, 2812–2826. doi: 10.1007/s00018-004-4248-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lahiri, A., Das, P., and Chakravortty, D. (2008). Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 10, 1166–1174. doi: 10.1016/j.micinf.2008.06.008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lobo, M. C. (1973). The effect of antibiotics on E. coli ingested by macrophages. Proc. Soc. Exp. Biol. Med. 142, 1048–1050. doi: 10.3181/00379727-142-37173
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopez, C. A., Winter, S. E., Rivera-Chavez, F., Xavier, M. N., Poon, V., Nuccio, S.-P., et al. (2012). Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. MBio 3:e00143–12. doi: 10.1128/mBio.00143-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mathur, R., Oh, H., Zhang, D., Park, S. G., Seo, J., Koblansky, A., et al. (2012). A mouse model of Salmonella typhi infection. Cell 51, 590–602. doi: 10.1016/j.cell.2012.08.042
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCollister, B. D., Hoffman, M., Husain, M., and Vázquez-Torres, A. (2011). Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob. Agents Chemother. 55, 2189–2196. doi: 10.1128/AAC.01203-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miller, K. A., Phillips, R. S., Mrázek, J., and Hoover, T. R. (2013). Salmonella utilizes D-glucosaminate via a mannose family phosphotransferase system permease and associated enzymes. J. Bacteriol. 195, 4057–4066. doi: 10.1128/JB.00290-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Negrea, A., Bjur, E., Puiac, S., Ygberg, S. E., Aslund, F., and Rhen, M. (2009). Thioredoxin 1 participates in the activity of the Salmonella enterica serovar Typhimurium pathogenicity island 2 type III secretion system. J. Bacteriol. 191, 6918–6927. doi: 10.1128/JB.00532-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Neidhardt, F. C. (1996). Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Edn. Washington, DC: ASM Press.
Nuccio, S., Analysis, C., Identifies, S. G., Network, M., Growth, E., Gut, I., et al. (2014). Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5:e00929-14. doi: 10.1128/mBio.00929-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Núñez-Hernández, C., Alonso, A., Pucciarelli, M. G., Casadesús, J., and García-del Portillo, F. (2014). Dormant intracellular Salmonella enterica serovar Typhimurium discriminates among Salmonella pathogenicity island 2 effectors to persist inside fibroblasts. Infect. Immun. 82, 221–232. doi: 10.1128/IAI.01304-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Parkhill, J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., et al. (2001). Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852. doi: 10.1038/35101607
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Paterson, G. K., Cone, D. B., Northen, H., Peters, S. E., and Maskell, D. J. (2009). Deletion of the gene encoding the glycolytic enzyme triosephosphate isomerase (tpi) alters morphology of Salmonella enterica serovar Typhimurium and decreases fitness in mice. FEMS Microbiol. Lett. 294, 45–51. doi: 10.1111/j.1574-6968.2009.01553.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Perret, C. A., and Zhou, D. (2013). Salmonella type III effector SopB modulates host cell exocytosis. Emerg. Microbes Infect. 2, e32. doi: 10.1038/emi.2013.37
Pontier-Bres, R., Munro, P., Boyer, L., Anty, R., Imbert, V., Terciolo, C., et al. (2014). Saccharomyces boulardii Modifies Salmonella Typhimurium traffic and host immune responses along the intestinal tract. PLoS ONE 9:e103069. doi: 10.1371/journal.pone.0103069
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Porwollik, S., Santiviago, C. A., Cheng, P., Long, F., Desai, P., Fredlund, J., et al. (2014). Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS ONE 9:e99820. doi: 10.1371/journal.pone.0099820
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Raghunathan, A., Reed, J., Shin, S., Palsson, B., and Daefler, S. (2009). Constraint-based analysis of metabolic capacity of Salmonella Typhimurium during host-pathogen interaction. BMC Syst. Biol. 3:38. doi: 10.1186/1752-0509-3-38
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richardson, A. R., Payne, E. C., Younger, N., Karlinsey, J. E., Thomas, V. C., Becker, L. A., et al. (2011). Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10, 33–43. doi: 10.1016/j.chom.2011.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rosenkrantz, J. T., Aarts, H., Abee, T., Rolfe, M. D., Knudsen, G. M., Nielsen, M.-B., et al. (2013). Non-essential genes form the hubs of genome scale protein function and environmental gene expression networks in Salmonella enterica serovar Typhimurium. BMC Microbiol. 13:294. doi: 10.1186/1471-2180-13-294
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rowley, G., Hensen, D., Felgate, H., Arkenberg, A., Appia-Ayme, C., Prior, K., et al. (2012). Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem. J. 441, 755–762. doi: 10.1042/BJ20110971
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schleker, S., Sun, J., Raghavan, B., Srnec, M., Müller, N., Koepfinger, M., et al. (2012). The current Salmonella-host interactome. Proteomics Clin. Appl. 6, 117–133. doi: 10.1002/prca.201100083
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schulte, L. N., Eulalio, A., Mollenkopf, H. J., Reinhardt, R., and Vogel, J. (2011). Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 30, 1977–1989. doi: 10.1038/emboj.2011.94
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shah, D. H. (2014). RNA sequencing reveals differences between the global transcriptomes of Salmonella enterica serovar enteritidis strains with high and low pathogenicities. Appl. Environ. Microbiol. 80, 896–906. doi: 10.1128/AEM.02740-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Steeb, B., Claudi, B., Burton, N. A., Tienz, P., Schmidt, A., Farhan, H., et al. (2013). Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9:e1003301. doi: 10.1371/journal.ppat.1003301
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tattoli, I., Sorbara, M. T., Vuckovic, D., Ling, A., Soares, F., Carneiro, L., et al. (2012). Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575. doi: 10.1016/j.chom.2012.04.012
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Temme, K., Salis, H., Tullman-Ercek, D., Levskaya, A., Hong, S.-H., and Voigt, C. A. (2008). Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J. Mol. Biol. 377, 47–61. doi: 10.1016/j.jmb.2007.12.044
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thiele, I., Hyduke, D. R., Steeb, B., Fankam, G., Allen, D. K., Bazzani, S., et al. (2011). A community effort towards a knowledge-base and mathematical model of the human pathogen Salmonella Typhimurium LT2. BMC Syst. Biol. 5:8. doi: 10.1186/1752-0509-5-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Turnbull, A. L., and Surette, M. G. (2010). Cysteine biosynthesis, oxidative stress and antibiotic resistance in Salmonella Typhimurium. Res. Microbiol. 161, 643–650. doi: 10.1016/j.resmic.2010.06.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Velayudhan, J., Karlinsey, J. E., Frawley, E. R., Becker, L. A., Nartea, M., and Fang, F. C. (2014). Distinct roles of the Salmonella enterica serovar Typhimurium CyaY and YggX proteins in the biosynthesis and repair of iron-sulfur clusters. Infect. Immun. 82, 1390–1401. doi: 10.1128/IAI.01022-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Viala, J. P. M., Méresse, S., Pocachard, B., Guilhon, A.-A., Aussel, L., and Barras, F. (2011). Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS ONE 6:e22397. doi: 10.1371/journal.pone.0022397
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Viala, J. P. M., Puppo, R., My, L., and Bouveret, E. (2013). Posttranslational maturation of the invasion acyl carrier protein of Salmonella enterica serovar Typhimurium requires an essential phosphopantetheinyl transferase of the fatty acid biosynthesis pathway. J. Bacteriol. 195, 4399–4405. doi: 10.1128/JB.00472-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wain, J., Keddy, K. H., Hendriksen, R. S., and Rubino, S. (2013). Using next generation sequencing to tackle non-typhoidal Salmonella infections. J. Infect. Dev. Ctries. 7, 1–5. doi: 10.3855/jidc.3080
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wallrodt, I., Jelsbak, L., Thorndahl, L., Thomsen, L. E., Lemire, S., and Olsen, J. E. (2013). The putative thiosulfate sulfurtransferases PspE and GlpE contribute to virulence of Salmonella Typhimurium in the mouse model of systemic disease. PLoS ONE 8:e70829. doi: 10.1371/journal.pone.0070829
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007. doi: 10.1126/science.1179687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Workalemahu, G., Wang, H., Puan, K. J., Nada, M. H., Kuzuyama, T., Jones, B. D., et al. (2014). Metabolic engineering of Salmonella vaccine bacteria to boost human Vγ2Vδ T cell immunity. J. Immunol. 193, 708–721. doi: 10.4049/jimmunol.1302746
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: metabolism, Salmonella-containing vacuole (SCV), regulation, virulence, “-omics”
Citation: Dandekar T, Fieselmann A, Fischer E, Popp J, Hensel M and Noster J (2015) Salmonella—how a metabolic generalist adopts an intracellular lifestyle during infection. Front. Cell. Infect. Microbiol. 4:191. doi: 10.3389/fcimb.2014.00191
Received: 18 July 2014; Accepted: 21 December 2014;
Published online: 29 January 2015.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Giovanna Suzzi, Università degli Studi di Teramo, ItalyManuela Raffatellu, University of California, Irvine, USA
Copyright © 2015 Dandekar, Fieselmann, Fischer, Popp, Hensel and Noster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Dandekar, Biocenter, Am Hubland, 97074 Würzburg, Germany e-mail:ZGFuZGVrYXJAYmlvemVudHJ1bS51bmktd3VlcnpidXJnLmRl
 Thomas Dandekar
Thomas Dandekar Astrid Fieselmann
Astrid Fieselmann Eva Fischer
Eva Fischer Jasmin Popp
Jasmin Popp Michael Hensel
Michael Hensel Janina Noster
Janina Noster