- 1Department of Biochemistry, University of Toronto, Toronto, ON, Canada
- 2Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada
Proteasomes, essential protease complexes in protein homeostasis, adapt to metabolic changes through intracellular movements. As the executive arm of the ubiquitin-proteasome system, they selectively degrade poly-ubiquitinated proteins in an ATP-dependent process. The primary proteasome configuration involved in this degradation is the 26S proteasome, which is composed of a proteolytically active core particle flanked by two regulatory particles. In metabolically active cells, such as proliferating yeast and mammalian cancer cells, 26S proteasomes are predominantly nuclear and actively engaged in protein degradation. However, during nutrient deprivation or stress-induced quiescence, proteasome localization changes. In quiescent yeast, proteasomes initially accumulate at the nuclear envelope. During prolonged quiescence with decreased ATP levels, proteasomes exit the nucleus and are sequestered into cytoplasmic membraneless organelles, so-called proteasome storage granules (PSGs). In mammalian cells, starvation and stress trigger formation of membraneless organelles containing proteasomes and poly-ubiquitinated substrates. The proteasome condensates are motile, reversible, and contribute to stress resistance and improved fitness during aging. Proteasome condensation may involve liquid-liquid phase separation, a mechanism underlying the assembly of membraneless organelles.
Proteasomal protein breakdown is ubiquitin- and ATP-dependent
Protein homeostasis describes the equilibrium between protein synthesis and degradation, and involves dynamic assembly and disassembly of proteins, and their trafficking between cellular compartments (Wolf and Menssen, 2018). Newly synthesized proteins can be misfolded, be supernumerary, or missing their native interaction partner due to heterologous expression. If these proteins expose hydrophobic regions prone to random aggregation, circuits of protein quality control make triage decisions. The question arises: Should these proteins be refolded by chaperones, eliminated by degradation, or deposited into organelles? Stress complicates triage decisions. To cope with stress, chaperones are activated to preserve proteins from being degraded. Up to the early 1980s, it was not plausible that peptide bonds, which require large amounts of energy to be built, could be reverted. Only the discovery of ubiquitin-mediated protein degradation triggered a paradigm shift that peptide bonds are broken under ATP consumption, which was awarded with the Nobel Prize in 2004 (Giles, 2004).
In the lab environment, yeast and mammalian cancer cells are easily cultured and have plenty of energy. ATP-dependent proteolysis was recognized as an advantage to eliminate unwanted short-lived proteins. By this, biological activities of proteins, i.e., regulating cell cycle progression and gene expression, are irreversibly switched off (Goldberg, 2003). Their shutdown is achieved by protein degradation through proteasomes, the key proteases of the ubiquitin-proteasome system (UPS) (Hershko and Ciechanover, 1998). Ubiquitin serves as a death signal and is conjugated in multiple copies to protein substrates for recognition by the proteasome. The ubiquitin moieties are linked to the substrate through reiterating cycles of ATP-consuming ubiquitin activation and ligation (Ciechanover, 2015). Thus, poly-ubiquitination of protein substrates is highly ATP demanding. The unfolding and translocation of protein substrates into the proteolytic cavity of proteasomes further consume hundreds of ATP molecules (Benaroudj et al., 2003; Peth et al., 2013). Ubiquitination is also involved in the elimination of proteins by the vacuole/lysosome, which engulfs cytoplasmic constituents and cell surface receptors via autophagic and endosomal vesicles (Ciechanover, 2005). Lysosomal degradation targets long-lived proteins, membrane-associated proteins, protein aggregates, and macromolecular machineries such as proteasomes (Ballabio and Bonifacino, 2020; Hoeller and Dikic, 2016; Marshall and Vierstra, 2019).
Proteasome structure
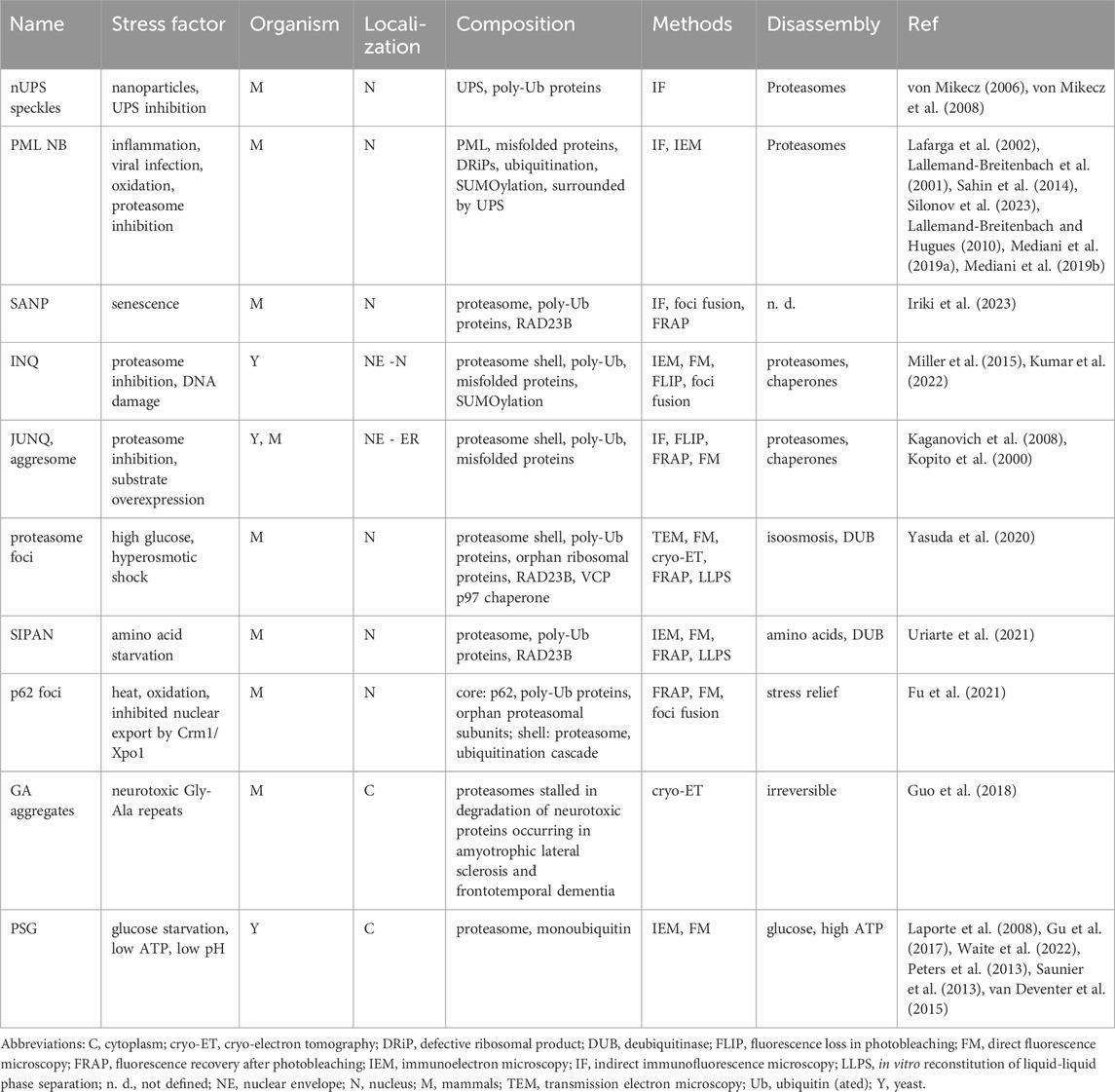
Proteasome biogenesis requires huge amounts of energy, as the proteasome is the second most abundant protein complex composed of ∼33 different subunits. Thus, proteasome biogenesis only takes place in proliferating cells with high metabolic activity (Marguerat et al., 2012). Subunit incorporation into the proteasome complex requires transient interactions (Gu and Enenkel, 2014). Their concerted action yields the proteolytic core particle (CP) with two adjacent regulatory particles (RP), known as RP-CP-RP configured 26S proteasome (Figure 1) (Hershko and Ciechanover, 1998; Tanaka, 2009). Asymmetric RP-CP configurations also exist under the name of 26S proteasomes but are not further dealt with in this review.
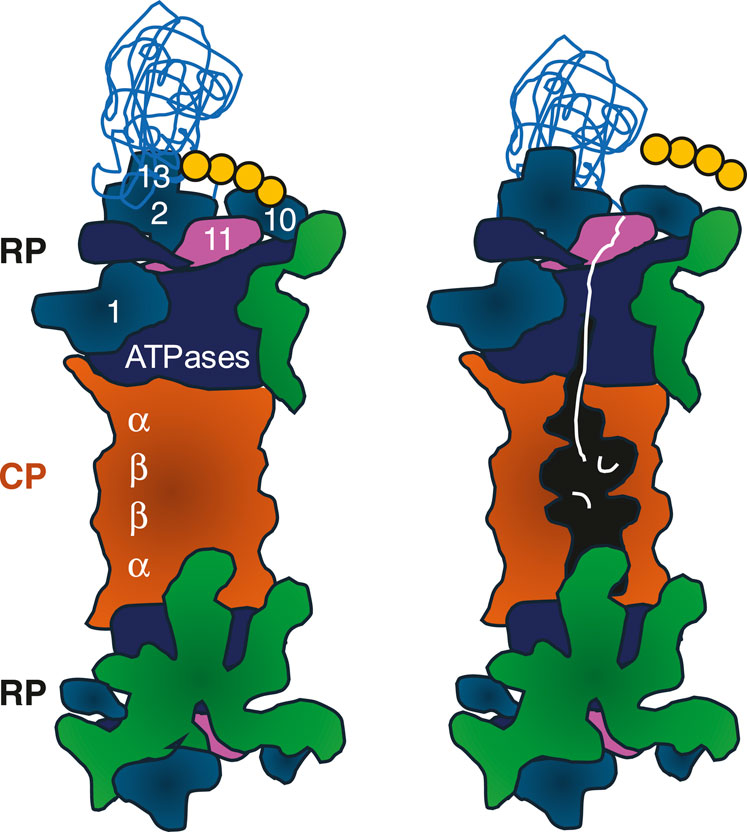
Figure 1. Cartoon of 26S proteasomes with RP-CP-RP configuration. Left: RP base ATPases (marine blue) and RP base subunits Rpn1, Rpn2, Rpn10, and Rpn13 recognizing the poly-ubiquitinated protein substrate (sky blue) are depicted. Four ubiquitin molecules represent the minimal poly-ubiquitin chain (yellow). RP lid subunits are depicted in green. Right: Once the poly-ubiquitin chain is cleaved off by Rpn11 (pink), the ATPases are committed to translocate the unfolded protein (white) through the outer α-rings of the CP. The unfolded polypeptide is degraded into peptides in the catalytic cavity located between the inner β-rings of the CP (orange). CP, core particle; RP, regulatory particle. The model is adopted from Matyskiela et al. (2013).
The RP is divided into a base and lid subcomplex (Glickman et al., 1998). The RP base contains, among other subunits, Rpn1, Rpn10 and Rpn13, both bridged by Rpn2, which recognizes the poly-ubiquitin chain of the substrate. In addition, several ubiquitin receptors exist, such as RAD23 in mammals and Rad23 in yeast. They transiently interact with the RP to hand over poly-ubiquitinated substrates for degradation (Shi et al., 2016). Proteasomes do not care about the nature of the substrates, just the presence of the ubiquitin death signal.
The RP base also contains a six-membered ATPase ring, which is responsible for opening/gating of the CP, substrate unfolding, and translocation. Since branched poly-ubiquitin chains are bulky, they are removed from the protein substrate prior to degradation. The isopeptide bond between the ubiquitin chain and the substrate is cleaved by the deubiquitinase Rpn8-Rpn11 module located in the RP lid (Wehmer et al., 2017; Bard et al., 2019). The release of the poly-ubiquitin chain induces a conformational switch by which the RP base ATPase ring snaps into place on the adjacent CP gate (Bard et al., 2019).
Over the last years, single-particle cryo-electron microscopy enabled the deconvolution of coexisting 26S proteasome conformations and their delineation in the degradation of poly-ubiquitinated substrates (Wehmer et al., 2017; Unverdorben et al., 2014; Schweitzer et al., 2016). The recognition of the poly-ubiquitinated substrate occurs in the inactive s1 ground state of the 26S proteasome. The 26S proteasome then adopts several commitment states until substrate degradation becomes irreversible (Wehmer et al., 2017; Unverdorben et al., 2014; Dong et al., 2019; de la Pena et al., 2018; Eisele et al., 2018). These proteasome rearrangements resulting in conformational heterogeneity prevented crystallographic analyses of the 26S proteasome and RP. It is worth mentioning that 26S proteasomes are further able to cleave proteins with intrinsically disordered regions in an ubiquitin-independent manner (Erales and Coffino, 2014), sometimes leading to protein processing through limited proteolysis (Rape and Jentsch, 2002). Sophisticated in vitro experiments revealed that a folded protein is spared from degradation although being modified by a poly-ubiquitin chain. Instead, an intrinsically disordered protein lacking poly-ubiquitination but interacting with the folded poly-ubiquitinated protein was degraded (Inobe and Matouschek, 2014). On one hand, this suggests that poly-ubiquitination is not necessarily leading to proteasomal degradation (Collins and Goldberg, 2017). On the other hand, proteins with intrinsically disordered regions are sensitive to proteasomal degradation and can have shorter half-life (Tsvetkov et al., 2009; van der Lee et al., 2014).
Furthermore, 26S proteasome assembly is sensitive to oxidative stress. Oxidative stress, e.g., induced by perhydrol, results in the dissociation of 26S proteasomes into the CP and RP, and an accumulation of poly-ubiquitinated substrates. Under these conditions, an increased association of the proteasome-interacting protein Ecm29 with purified RP was detected (Wang et al., 2010), consistent with the finding that Ecm29 fulfills quality control functions in proteasome assembly (Lehmann et al., 2010; Park et al., 2011). The resulting free CP has closed gates and thus latent enzyme activity (Eytan et al., 1989). However, the CP gates are accessible for intrinsically disordered and oxidatively damaged proteins that are in vitro degraded by the CP (Tsvetkov et al., 2009; Liu et al., 2003; Ben-Nissan and Sharon, 2014).
In contrast to the 26S holoenzyme, the CP with its more static global structure is resolved at atomic resolution by x-ray crystallography. The CP is composed of a stack of two inner β-subunit rings and two outer α-subunit rings. The inner β-rings harbor the active sites for endoproteolytic peptide bond cleavage. Outer α-rings serve as gates into the CP cavity, which are opened by the adjacent ATPase rings of the RP base. Thus, ATPase activity is required for α-ring opening, unfolding, and translocating of substrates into the CP (Groll et al., 1997).
Nuclear proteasome localization in dividing cells
On top of conformational plasticity, 26S proteasomes are highly dynamic regarding their intracellular localization (Enenkel, 2014a; Tomita et al., 2019; de Almeida et al., 2021). Our understanding of proteasome localization in cells was debated for decades before a consensus was reached.
In mammalian cells, intracellular proteasome localizations by indirect immunofluorescence microscopy had been controversially discussed as they varied depending on antibodies, cell lines, and culture conditions used (Brooks et al., 2000). At high confluency, when nutrients became limiting in the cell culture medium, proteasomes appeared to be cytoplasmic, while proteasomes appeared to be more nuclear in cancer cells grown at low confluency (Wojcik and DeMartino, 2003). Early indirect immunofluorescence microscopy using antibodies with cross-reactivity for proteasomes from different organisms revealed intracellular distributions of proteasomes. Proteasomes were localized to the nucleus in Xenopus laevis oocytes and HeLa cells (Peters et al., 1994). Particularly in the prophase of rat granulosa cells, proteasomes accumulated with chromatin, where also cyclins localize before being degraded (Amsterdam et al., 1993). Cyclins are short-lived proteins regulating cell cycle progression and one of the first identified proteasomal substrates (Glotzer et al., 1991). At that time, the detection of nuclear proteasomes was consistent with Varshavsky’s and co-workers’ discovery that ubiquitin-dependent protein degradation plays a critical role in cell cycle control and gene expression (Finley et al., 1984). Four decades later, proteasome abundance in the nucleus is still attracting attention, with quantification by nuclear fractionations and proteomics analyses confirming cell cycle-dependent recruitment of proteasomes to chromatin (Kito et al., 2020). Meanwhile, monoclonal antibodies that enable the co-immunoprecipitation of 26S proteasomes are commercially available and suitable for proteasome localization by indirect immunofluorescence microscopy (Hendil et al., 1995). Complementary to this classical approach, the labeling of proteins with green fluorescent protein (GFP) and related variants became an invaluable technique to correlate cell cycle-dependent dynamics of protein concentrations and their localizations using live-cell imaging (Litsios et al., 2024). In yeast, the chromosomal replacement of proteasomal subunits by GFP-labeled versions is standardized and yields reliable fluorescent reporter subunits that are fully incorporated into proteasomes. Almost every proteasomal subunit is functionally replaceable by a GFP-labeled version consistently showing the same intracellular distribution in yeast (Enenkel, 2014a). In mammalian cells, an increasing number of GFP reporter subunits for live-cell imaging of proteasomes is emerging. Dantuma and co-workers were one of the first who aimed for the stable expression of GFP-labeled CP subunit α4 in cancer cell lines (Gierisch et al., 2020). The efficiency of the reporter subunit incorporation into proteasomes was verified by glycerol gradient ultracentrifugation, which separates 26S proteasomes in fast-migrating fractions from not fully incorporated subunits in slow-migrating fractions (Salomons et al., 2010). Direct fluorescence microscopy of GFP-labeled α4 in Mel JuSo cells revealed significant nuclear localization (Enenkel, 2014a). We adopted this expression system to U2OS cells and confirmed major nuclear proteasome localization for GFP-labeled α4, consistent with indirect immunofluorescence microscopy using commercial MCP444 antibodies (unpublished results). Similar observations were reported by Murata and co-workers, who established RP lid subunit Rpn11-Flag-EGFP tag-exchangeable knock-in mice. Their approach allows one to distinguish between young, in other words, newly synthesized, proteasomes in the nucleus and old proteasomes in the cytoplasm of embryonic fibroblasts. Thus, this cell system is suited to monitor age-related proteasome dynamics in mammalian cells (Tomita et al., 2019). More recently, Zuber and co-workers developed an elegant approach by ectopic expression of fluorescent mCherry-labeled proteasomal subunits in CRISPR-Cas9 induced RKO knockdown cells. Their approach yielded the full replacement of the endogenous CP β4 subunit by a fluorescent-labeled version. Again, the fluorescent reporter subunit of the proteasome revealed nuclear localization in dividing RKO cells (de Almeida et al., 2021).
Taken together, direct and indirect fluorescence microscopy in proliferating yeast and mammalian cells with high metabolic activity reveal nuclear localization of proteasomes. It is not surprising that the localization of an essential and abundant protease complex is evolutionarily conserved (Botstein and Fink, 2011). However, even in yeast as model organism of eukaryotic cells, it was puzzling that proteasomes were primarily nuclear (Russell et al., 1999; Enenkel et al., 1998; Wilkinson et al., 1998). At this point, we would like to point out again that proteasomes in the cytoplasm are proteolytically active. Cryo-electron tomography (cryo-ET), a non-invasive imaging technology that preserves protein structures in their native cellular environment (Baumeister, 2022), of cytoplasmic volumes of neuronal cells revealed that ∼20% of the cytoplasmic proteasomes were engaged in substrate degradation. The remainder of 26S proteasomes was in the substrate-accepting ground state (Asano et al., 2015). Without stress, the reservoir of cytoplasmic 26S proteasomes appears to be far from exhausted.
Nuclear import of proteasomes
The answer to the question of how proteasomes are imported into the nucleus is that several pathways are used. Our previous reviews have recapitulated in detail the discoveries on nuclear import of proteasomes over the last decades (Enenkel, 2014b; Wendler and Enenkel, 2019; Enenkel et al., 2022). We briefly summarize the basic concepts of nuclear import of proteasomes. In proliferating yeast, inactive CP precursor complexes and RP subcomplexes are imported by the conventional import receptor importin/karyopherin αβ, suggesting that holoenzymes are assembled in the nucleus (Lehmann et al., 2002; Isono et al., 2007). Alternatively, proteasomes are imported as matured enzymes by importins/karyopherins with transient accessory proteins such as Sts1 binding to the RP lid in yeast (Chen et al., 2011; Budenholzer et al., 2020), and AKIRIN2 binding to the CP α-ring in mammalian cells (de Almeida et al., 2021). Intriguingly, Sts1 and AKIRIN2 are short-lived. Their proteasomal degradation is triggered upon arrival in the nucleus by a mechanism not fully understood.
When yeast cells rest in quiescence, a temporary halt of proliferation, CP precursor complexes are unavailable due to stalled proteasome biogenesis. Upon exit from quiescence, nuclear proteasome assembly from newly synthesized precursor complexes takes time (Laporte et al., 2008). Thus, matured proteasomes are immediately transported from the cytoplasm into the nucleus. Sudden changes in metabolic activities, i.e., from low state in quiescence to high state in proliferation, require quick adaptations. With the resumption of cell proliferation, Blm10 facilitates nuclear import of the CP in yeast (Weberruss et al., 2013). PA200, the mammalian counterpart of Blm10, is similarly involved in nuclear proteasome activation (Ustrell et al., 2002). Which nuclear import pathway prevails over another depends on the availability of proteasomal transport cargoes, importins/karyopherins, and adaptor proteins, showcasing the plasticity of proteasome configurations under different growth conditions. To put it simply, all nuclear import pathways have in common that 26S proteasomes do not pass the nuclear pore as active enzymes. The fact that 26S proteasomes and free CP are able to degrade intrinsically disordered proteins would make the passage of active enzymes detrimental to nuclear pore proteins, because nuclear pore proteins with repetitive hydrophobic Gly-Leu-Phe-Gly motifs are intrinsically disordered (Denning et al., 2003; Denning and Rexach, 2007). To avoid collateral damage to nuclear pore proteins, proteasomes are translocated as inactive enzymes. On the way through the nuclear pore, proteasome activity is inhibited either as a precursor complex or by binding to accessory proteins, such as Blm10, which seals the CP gate.
Proteasome condensates in response to stress and metabolic challenges
In the 2000s, the UPS field predominantly focused on cytoplasmic protein degradation by proteasomes because the scientific community was interested in endoplasmic reticulum (ER)-associated protein degradation, antigen processing, and the removal of newly synthesized proteins (Sommer and Wolf, 1997; Kloetzel and Ossendorp, 2004; Yewdell, 2005). Experiments were designed to study newly synthesized proteins that were often more expressed than their binding partners. However, Hartl and colleagues found that endogenous nascent polypeptides remain largely protected from proteolysis due to the abundance of cytoplasmic chaperones (Vabulas and Hartl, 2005). Moreover, misfolded proteins were found to be delivered into the nucleus for proteasomal degradation (Park et al., 2013), while tumor suppressor protein p53 was proposed to be exported into the cytoplasm for proteasomal degradation (Hirayama et al., 2018). The fate of p53 is intriguing because it is controlled by mono- or poly-ubiquitination, the latter has been shown to promote degradation in the nucleus (Li et al., 2003). 26S proteasomes are also engaged in cytoplasmic protein breakdown. Since 26S proteasomes are enzymes, quality counts over quantity. Sites of proteasome localizations may not correlate with major sites of proteolysis. Furthermore, the activities of nucleo- and cytoplasmic 26S proteasomes are differently regulated by post-translational modifications (Sha et al., 2011; VerPlank and Goldberg, 2017). Therefore, based on our current knowledge, it is difficult to decide in which compartment proteasomes are most active in protein degradation.
Proteasome condensates in the nucleus of mammalian cells
Based on indirect immunofluorescence localization studies, von Mikecz and co-workers shifted the research focus back to the UPS within nuclear speckles. Intrigued by the observation that ubiquitin, the ubiquitin-activating enzyme, ubiquitin ligases, and proteasomes accumulate in nuclear speckles, also known as foci, bodies, and granules, the question arose: What is the function of these UPS conglomerations (von Mikecz, 2006)? When all UPS players are in place, it is conceivable that short-lived proteins that regulate cell cycle progression and gene expression, such as cyclins and transcription factors, are instantaneously poly-ubiquitinated and degraded on site. If so, these conglomerations represent enhanced UPS activities and differ from pathological aggregates that accumulate undegradable proteins linked to neurodegenerative disorders and can be caused by nanoparticles (von Mikecz et al., 2008).
Stress-adaptable Promyelocytic leukemia protein (PML)-associated nuclear bodies (PML NB) or clastosomes (Lafarga et al., 2002; Lallemand-Breitenbach et al., 2001) have multifaceted roles by recruiting a variety of unrelated proteins in response to stress, i.e., oxidation. PML NB are archetypes of membraneless organelles with a diameter of ∼1 µm (Sahin et al., 2014). Membraneless organelles are fascinating subcompartments, as they are thought to float as dense phase in the dilute phase of an aqueous environment. Their components often condense in response to cellular stress and dilute upon stress relief (van Leeuwen and Rabouille, 2019).
The current model to describe the phenomenon of condensed mixtures of macromolecules is liquid-liquid phase separation (LLPS). LLPS is driven by concentration gradients, the promiscuity and multivalency of macromolecules, i.e., variable weak interactions through proteins with repetitive sequences of hydrophobic amino acids, low complexity or intrinsically disordered regions, and proteins in folding transitions (Alberti and Hyman, 2021; Vernon and Forman-Kay, 2019).
PML has an intrinsically disordered C-terminal domain and interacts with multiple proteins. According to immunogold electron microscopy and confocal microscopy, PML surrounds PML NB (Silonov et al., 2023; Lallemand-Breitenbach and Hugues, 2010). Like a sponge, PML NB serves as overflow compartment for nuclear quality control and hosts misfolded proteins under conditions of proteotoxic stress, such as proteasome inhibition. Defective ribosomal products (DRiPs), in other words, aberrant newly synthesized proteins of low molecular mass, constantly escape the cytoplasmic quality control system. They diffuse through nuclear pores into the nucleus, are ubiquitinated, and transiently stored in PML NB (Silonov et al., 2023; Lallemand-Breitenbach and Hugues, 2010). The condensed PML NB core could be envisioned as a unique solvent continuously extracting and exchanging proteins from the environment. Heat shock proteins, chaperones, and proteasomes around PML NB reduce the influx of DRiPs by refolding and degradation, respectively. Thus, PML NB have a highly dynamic DRiP composition, and prevent unintended interactions of DRiPs with nuclear proteins. Failures of DRiP clearance under conditions of prolonged stress, such as critical energy shortage and irreversible proteasome inhibition, result in PML NB solidification. Immobilization of UPS components in solidifying PML NB leads to depletion of ubiquitin and proteasomes, which jeopardizes cell vitality (Mediani et al., 2019a; Mediani et al., 2019b). The age-related and thus irreversible decline of proteasome activities (Chondrogianni et al., 2003; Torres et al., 2006) causes challenges in senescent cells that cope with the burden of poly-ubiquitinated proteins by uptake into nuclear proteasome bodies (senescence-associated nuclear proteasome foci; SANPs) using the ubiquitin receptor RAD23B (Iriki et al., 2023).
Meanwhile, evidence has increased that membraneless organelles containing proteasomes and a diversity of poly-ubiquitinated substrates originate from various stress conditions. The simplest explanation for this phenomenon is that different kinds of stress cause an energy crisis and force the energy-consuming UPS into quality control compartments. As mentioned above, stress-adaptable PML NB and SANP might represent overflow compartments for UPS clearance (Mediani et al., 2019a; Mediani et al., 2019b; Iriki et al., 2023). Additional stressful situations leading to UPS condensation are listed in Table 1. For example hyperosmotic shock induces the formation of nuclear organelles containing proteasomes, poly-ubiquitinated proteins, chaperone VCP p97, and ubiquitin receptor RAD23B. Orphan ribosomal subunits that failed to be incorporated into nascent ribosomes represent an abundant source of proteasomal substrates and are part of these organelles. These organelles were one of the first to be resolved by cryo-ET analysis. 26S Proteasomes were found to be randomly distributed within the organelle. In vitro, LLPS was mediated by multivalent interactions between two ubiquitin-associated domains of RAD23B and tetraubiquitin chains, two components of this UPS organelle (Yasuda et al., 2020).
Heat, oxidation, and inhibition of nuclear export through the canonical export receptor Crm1/Xpo1 are alternative stressors. They trigger nuclear LLPS of poly-ubiquitinated proteins, including orphan proteasomal subunits that escaped the incorporation into precursor complexes in the cytoplasm. Notably, the receptor p62 for transport of ubiquitinated cargo into autophagosomes is sequestered into these nuclear foci. Confocal immunofluorescence localization studies revealed that proteasomes and enzymes of the ubiquitination cascade are at the periphery of these foci, suggesting a local enhancement of UPS activities (Fu et al., 2021).
Acute amino acid deprivation is another stressor. It triggers the reversible formation of starvation-induced proteasome assemblies in the nucleus (SIPAN) with poly-ubiquitinated proteins shuffled by RAD23B. RAD23B is highly intrinsically disordered and undergoes LLPS in the presence of crowding agents, i.e., Ficoll, dextran or polyethylene glycol (Uriarte et al., 2021). In vivo, SIPAN is dissolved by amino acid replenishment and contributes to stress resilience and fitness under pathological conditions (Uriarte et al., 2021). Following the starvation of amino acids, specifically of the mTOR-agonistic aromatic amino acids Phe, Tyr, and Trp, the proteasome moves from its large nuclear pool to the cytoplasm (Livneh et al., 2023). This phenomenon mirrors early observations of proteasome movements from the nucleus to the cytoplasm in response to increased confluency of mammalian cells in culture (Wojcik and DeMartino, 2003).
Proteasome condensates in the nuclear periphery of mammalian cells
At both the nucleo- and cytoplasmic side of the nuclear envelope (NE), the latter connected with the ER, proteasomes and substrates were found to be condensed in intranuclear quality (INQ) and juxta-nuclear quality (JUNQ/CytoQ) speckles, respectively. These cellular ‘junkyards’ were initially observed when proteasomal degradation was overwhelmed through heterologous expression of model substrates, e.g., the von Hippel Lindau protein in yeast (Kaganovich et al., 2008; Kopito, 2000; Miller et al., 2015). JUNQ formation is fostered by proteasome inhibition either chemically in mammalian cells or by UPS-specific mutations in yeast. When stress relieves, e.g., by proteasome and chaperone activation, the ‘junkyards’ dissolve (Kaganovich et al., 2008). Interestingly, INQ and PML NB are discussed to represent counterparts in yeast and humans, respectively, and depend on SUMO-ylation, an ubiquitin-like modifier, which distinguishes INQ and PML NB from JUNQ (Kumar et al., 2022). Failures in the clearance of these quality control compartments are thought to be related to neurodegenerative disorders and premature aging (Kopito, 2000; Miller et al., 2015).
To understand the occurrence of UPS-containing organelles on either nucleo- or cytoplasmic side of the NE, the kinetics of UPS transport through nuclear pores might be considered. Cryo-ET of Chlamydomonas cells revealed 26S proteasomes tethered to the basket of the nuclear pore, at the inner NE (Albert et al., 2017) and in clusters at the outer NE/ER for ER-associated protein degradation (Albert et al., 2020). As the diameter of nuclear pores shrinks upon stress and energy depletion and dilates with stress relief and energy replenishment (Zimmerli et al., 2021), UPS components and poly-ubiquitinated substrates tend upon stress to be concentrated on either side of the bottleneck formed by nuclear pores. Thus, phase separation of proteasomes and poly-ubiquitinated substrates could be the result of molecular crowding due to impaired nuclear transport. In the event of extreme stress, UPS organelles may even fragment nuclear pore components, but this hypothesis remains to be tested (Lee et al., 2021).
To gain an overview of the repeating patterns of UPS organelles that harbor proteasomes or are surrounded by a proteasome shell (Figure 2), the common theme is stress, which ultimately requires energy management. Macromolecular machineries such as proteasomes may simply throttle their motor once energy is scarce. Further intriguing questions are: How many different UPS organelles coexist? To answer this question, we will focus future research on the analysis of proteasome configurations rather than individual substrates within different organelles. And how many more energy-dependent macromolecular machineries simultaneously condense into membraneless organelles to overcome stressful conditions (Gu et al., 2017)?
Figure 2. Models for membraneless organelles containing the ubiquitin-proteasome system (UPS) that form in response to nutrient limitations and stress. For simplicity, only proteasomes, the key proteases of the UPS, are depicted. Left: A shell of proteasomes surrounds a core of poly-ubiquitinated proteins. Middle: UPS components are randomly distributed between poly-ubiquitinated proteins. Both types of organelles are proposed to serve as proteolysis centers and to be driven by liquid-liquid phase separation (LLPS). Right: In quiescent yeast, proteasome storage granules (PSGs) behave differently as they contain no poly-ubiquitinated proteins. PSG formation requires the presence of mono-ubiquitin above a threshold, for details see Table 1. Proteasomes are depicted as in Figure 1. The variety of poly-ubiquitinated proteins is symbolized by green, violet and blue colors. Ubiquitin is yellow.
Proteasome condensates in the cytoplasm of mammalian cells
In the cytoplasm of mammalian cells, the presence of proteasome condensates is less explored. Instead, stress granules are known to be cytoplasmic organelles formed by LLPS. Stress granules contain non-translating RNA, a plethora of RNA-binding proteins and stalled preinitiation 40S ribosomes (Protter and Parker, 2016). Again, stress relief triggers stress granule clearance and the resumption of protein biogenesis. Stress granules do not contain proteasomes (Jain et al., 2016) but relieve the burden on the nuclear UPS by hosting misfolded proteins in the cytoplasm (Xu et al., 2023).
Close to the cytoplasmic side of the NE, proteasomes are concentrated in aggresomes, which are structurally and functionally overlapping with JUNQs. The formation of aggresomes, as well as of JUNQs, is induced by proteasome inhibition and overexpression of neurotoxic proteins. Aggresomes were initially proposed to serve as proteolytic centers (Wojcik and DeMartino, 2003; Kaganovich et al., 2008; Kopito, 2000). How inhibited proteasomes accelerate the proteolysis of toxic and sometimes undegradable proteins remains a conundrum.
Cryo-ET was employed to understand the molecular architecture of neurotoxic protein aggregates within intact neurons. A genetic aberration in the C9orf72 gene leading to modifications with repetitive Gly-Ala motifs is responsible for the development of amyotrophic lateral sclerosis and frontotemporal dementia. The hydrophobic patches of poly-Gly-Ala peptides produced by this genetic aberration are prone to aggregation. Cryo-ET revealed that undegradable aggregates containing poly-Gly-Ala peptides trap 26S proteasomes in a substrate-processing conformation, causing them to be stuck in a dead-end road of protein degradation and severely compromising protein homeostasis (Guo et al., 2018).
Notably, fluorescence microscopy cannot distinguish between proteolytically active proteasomes and proteasomes stalled in degradation, and thus cannot differentiate between reversible UPS organelles and irreversible UPS aggregates. This may explain why previous histograms of centenarians’ brains showing aggregations and inclusions of proteasomes and ubiquitin were difficult to interpret. UPS organelles were not necessarily associated with a medical history of neurological diseases (Tanaka and Matsuda, 2014; Ciechanover and Kwon, 2017; Itoh et al., 1998). As long as UPS organelles remain reversible, they confer fitness during aging (Iriki et al., 2023).
Can we nowadays characterize reversible UPS organelles? Unfortunately, membraneless UPS organelles fall apart during cell disintegration, unless they are chemically fixed. They escape biochemical characterization by conventional means. Cryo-ET became the state-of-the-art technology to provide insight into the structure and function of UPS organelles without interfering with their native environment (Baumeister, 2022).
Proteasome storage granules in the cytoplasm of yeast cells
In yeast cells transitioning from logarithmic to stationary phase, cells start competing for nutrients. Due to glucose deprivation, cells become less metabolically active. Thus, the ATP concentration strikingly decreases (Laporte et al., 2011). Cell proliferation is temporarily halted, and cells enter quiescence (De Virgilio, 2012). During the transition to quiescence, yeast proteasomes uniformly move towards the NE. Proteasome clusters are detected close to nuclear pores, suggesting that proteasomes are piling up before being slowly translocated through nuclear pores (Laporte et al., 2008). During prolonged quiescence, which is marked by high cell density and low metabolic activity, yeast proteasomes eventually exit the nucleus. They then accumulate in proteasome storage granules (PSGs) in the cytoplasm. PSGs are also induced by mitochondrial malfunctions (Waite and Roelofs, 2022) and low pH due to deficient proton pumping (Peters et al., 2013), suggesting that ATP availability and further downstream metabolites of catabolic pathways influence PSG formation. Additionally, signaling cascades involving mitogen-activated protein kinase MAPK, and AMP kinase, named Snf1 in yeast, strengthen the importance of cellular energy homeostasis for PSG formation (Li et al., 2019). Acidification converts the aqueous protoplasm into a solid-like phase that restricts the mobility of macromolecules and fosters their condensation (Munder et al., 2016; Parry et al., 2014).
Upon metabolic reactivation, PSGs immediately dissolve. Within a few minutes, mature CP and RP stored in PSGs appear reassembled in the nucleus (Laporte et al., 2008; Weberruss et al., 2013). If quiescent yeast cells are disintegrated in buffer with ATP regeneration, intact 26S proteasomes are obtained. If quiescent cells are disintegrated without ATP supplement, 26S proteasomes are dissociated into RP and CP (Weberruss et al., 2013; Gu et al., 2017; Bajorek et al., 2003). Which buffer will mimic the physiological environment of PSGs? Only in the presence of a chemical cross-linker could PSGs be isolated as intact organelles (Gu et al., 2017). Mass spectrometry and biochemical analysis of cross-linked PSGs revealed a homogeneous composition of proteasomal subunits but no poly-ubiquitinated substrates, suggesting that PSGs are not active in the degradation of poly-ubiquitinated proteins. High-throughput screens using the collection of yeast null mutants suggested that proteasomal deubiquitination activities (Rpn11, Ubp6) and a threshold level of free ubiquitin promote PSG formation (Gu et al., 2017). This contrasts with the various UPS organelles containing poly-ubiquitinated substrates as mentioned above. Notably, rpn11-m1 and ubp6Δ mutants do not form PSGs (Gu et al., 2017; Saunier et al., 2013). Due to the RP’s deficient deubiquitinase activity in rpn11-m1, the Cullin-RING E3 ligase, accounting for one-fifth of the poly-ubiquitination of proteasomal substrates, remains activated by modification through the ubiquitin-like protein NEDD8/Rub1 (Bramasole et al., 2019). Thus, the burden of poly-ubiquitinated substrates upon entry into quiescence might be incompatible with PSG formation, since the negative feedback loop of reducing the pool of poly-ubiquitinated substrates by the RP is disturbed.
The reversibility of PSGs remains during prolonged quiescence (Saunier et al., 2013; van Deventer et al., 2015). Furthermore, PSGs seem to protect proteasome assemblies from autophagy (Marshall and Vierstra, 2019; Li and Hochstrasser, 2020). The question is whether LLPS is the underlying mechanism of PSG formation. Miscellaneous protein composition is a typical feature of LLPS organelles. However, mass spectrometry of cross-linked PSGs and stochastic optical reconstruction microscopy suggested dense packing of proteasomes within PSGs (Gu et al., 2017). Will few proteasomal subunits with intrinsically disordered regions support LLPS-driven PSG formation (Aufderheide et al., 2015)? Ubiquitin is a key component of the UPS, and mono-ubiquitin is required for PSG formation. Mono-ubiquitin disrupts multivalent interactions and modulates LLPS (Dao et al., 2018). In line with this, mono-ubiquitin is essential for the disassembly of stress granules in cells recovering from stress (Franzmann and Alberti, 2021). It will be exciting to learn about the structure of PSG-related organelles in comparison with proteasome condensates containing poly-ubiquitinated proteins.
Our review started with a paradigm shift in protein homeostasis based on cellular energy homeostasis: the hydrolysis of peptide bonds of short-lived proteins is achieved by ATP-consuming ubiquitination and proteasomal proteolysis in cells with high metabolic activity. Protein homeostasis is severely impacted by metabolic imbalances that are associated with aging and diseases such as diabetes, obesity, cancer, and neurodegeneration (Ciechanover and Kwon, 2017). The master regulator of metabolic stress and proteasome activity is the TOR complex1 (Rousseau and Bertolotti, 2018). Not only adenosine nucleotides but also nicotinamide adenine dinucleotide (NAD+) are essential as coenzymes in a myriad of bioenergetic pathways. Cells with balanced metabolism are well prepared for energy shortages and the systemic decline of coenzymes during aging and stress, to protect the UPS from autophagy and to regulate protein degradation within UPS condensates (Marshall and Vierstra, 2019; Li et al., 2019; Karmon and Ben Aroya, 2019).
Author contributions
CE: Writing–original draft, Writing–review and editing. OE: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Canadian Institutes of Health Research (CIHR) Operating Grants PJT-159464 and PJT-195648 to OPE and NSERC Operating Grant (RGPIN-2019-05974) to CE.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albert, S., Schaffer, M., Beck, F., Mosalaganti, S., Asano, S., Thomas, H. F., et al. (2017). Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 114, 13726–13731. doi:10.1073/pnas.1716305114
Albert, S., Wietrzynski, W., Lee, C. W., Schaffer, M., Beck, F., Schuller, J. M., et al. (2020). Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl. Acad. Sci. U. S. A. 117, 1069–1080. doi:10.1073/pnas.1905641117
Alberti, S., and Hyman, A. A. (2021). Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213. doi:10.1038/s41580-020-00326-6
Amsterdam, A., Pitzer, F., and Baumeister, W. (1993). Changes in intracellular localization of proteasomes in immortalized ovarian granulosa cells during mitosis associated with a role in cell cycle control. Proc. Natl. Acad. Sci. U. S. A. 90, 99–103. doi:10.1073/pnas.90.1.99
Asano, S., Fukuda, Y., Beck, F., Aufderheide, A., Förster, F., Danev, R., et al. (2015). Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442. doi:10.1126/science.1261197
Aufderheide, A., Unverdorben, P., Baumeister, W., and Forster, F. (2015). Structural disorder and its role in proteasomal degradation. FEBS Lett. 589, 2552–2560. doi:10.1016/j.febslet.2015.07.034
Bajorek, M., Finley, D., and Glickman, M. H. (2003). Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 13, 1140–1144. doi:10.1016/s0960-9822(03)00417-2
Ballabio, A., and Bonifacino, J. S. (2020). Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118. doi:10.1038/s41580-019-0185-4
Bard, J. A. M., Bashore, C., Dong, K. C., and Martin, A. (2019). The 26S proteasome utilizes a kinetic gateway to prioritize substrate degradation. Cell 177, 286–298. doi:10.1016/j.cell.2019.02.031
Baumeister, W. (2022). Cryo-electron tomography: a long journey to the inner space of cells. Cell 185, 2649–2652. doi:10.1016/j.cell.2022.06.034
Benaroudj, N., Zwickl, P., Seemuller, E., Baumeister, W., and Goldberg, A. L. (2003). ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 11, 69–78. doi:10.1016/s1097-2765(02)00775-x
Ben-Nissan, G., and Sharon, M. (2014). Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 4, 862–884. doi:10.3390/biom4030862
Botstein, D., and Fink, G. R. (2011). Yeast: an experimental organism for 21st Century biology. Genetics 189, 695–704. doi:10.1534/genetics.111.130765
Bramasole, L., Sinha, A., Harshuk, D., Cirigliano, A., Gurevich, S., Yu, Z., et al. (2019). The proteasome lid triggers COP9 signalosome activity during the transition of Saccharomyces cerevisiae cells into quiescence. Biomolecules 9, 449. doi:10.3390/biom9090449
Brooks, P., Fuertes, G., Murray, R. Z., Bose, S., Knecht, E., Rechsteiner, M. C., et al. (2000). Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 1, 155–161. doi:10.1042/bj3460155
Budenholzer, L., Breckel, C., Hickey, C. M., and Hochstrasser, M. (2020). The Sts1 nuclear import adapter uses a non-canonical bipartite nuclear localization signal and is directly degraded by the proteasome. J. Cell. Sci. 133 (jcs), 236158. doi:10.1242/jcs.236158
Chen, L., Romero, L., Chuang, S. M., Tournier, V., Joshi, K. K., Lee, J. A., et al. (2011). Sts1 plays a key role in targeting proteasomes to the nucleus. J. Biol. Chem. 286, 3104–3118. doi:10.1074/jbc.M110.135863
Chondrogianni, N., Stratford, F. L. L., Trougakos, I. P., Friguet, B., Rivett, A. J., and Gonos, E. S. (2003). Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J. Biol. Chem. 278, 28026–28037. doi:10.1074/jbc.M301048200
Ciechanover, A. (2005). Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87. doi:10.1038/nrm1552
Ciechanover, A. (2015). The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 16, 322–324. doi:10.1038/nrm3982
Ciechanover, A., and Kwon, Y. T. (2017). Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 11, 185. doi:10.3389/fnins.2017.00185
Collins, G. A., and Goldberg, A. L. (2017). The logic of the 26S proteasome. Cell 169, 792–806. doi:10.1016/j.cell.2017.04.023
Dao, T. P., Kolaitis, R. M., Kim, H. J., O'Donovan, K., Martyniak, B., Colicino, E., et al. (2018). Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978. doi:10.1016/j.molcel.2018.02.004
de Almeida, M., Hinterndorfer, M., Brunner, H., Grishkovskaya, I., Singh, K., Schleiffer, A., et al. (2021). AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature 599, 491–496. doi:10.1038/s41586-021-04035-8
de la Pena, A. H., Goodall, E. A., Gates, S. N., Lander, G. C., and Martin, A. (2018). Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362, eaav0725. doi:10.1126/science.aav0725
Denning, D. P., Patel, S. S., Uversky, V., Fink, A. L., and Rexach, M. (2003). Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. U. S. A. 100, 2450–2455. doi:10.1073/pnas.0437902100
Denning, D. P., and Rexach, M. F. (2007). Rapid evolution exposes the boundaries of domain structure and function in natively unfolded FG nucleoporins. Mol. Cell Proteomics 6, 272–282. doi:10.1074/mcp.M600309-MCP200
De Virgilio, C. (2012). The essence of yeast quiescence. FEMS Microbiol. Rev. 36, 306–339. doi:10.1111/j.1574-6976.2011.00287.x
Dong, Y., Zhang, S., Wu, Z., Li, X., Wang, W. L., Zhu, Y., et al. (2019). Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55. doi:10.1038/s41586-018-0736-4
Eisele, M. R., Reed, R. G., Rudack, T., Schweitzer, A., Beck, F., Nagy, I., et al. (2018). Expanded coverage of the 26S proteasome conformational landscape reveals mechanisms of peptidase gating. Cell Rep. 24, 1301–1315. doi:10.1016/j.celrep.2018.07.004
Enenkel, C. (2014a). Proteasome dynamics. Biochim. Biophys. Acta 1843, 39–46. doi:10.1016/j.bbamcr.2013.03.023
Enenkel, C. (2014b). Nuclear transport of yeast proteasomes. Biomolecules 4, 940–955. doi:10.3390/biom4040940
Enenkel, C., Kang, R. W., Wilfling, F., and Ernst, O. P. (2022). Intracellular localization of the proteasome in response to stress conditions. J. Biol. Chem. 298, 102083. doi:10.1016/j.jbc.2022.102083
Enenkel, C., Lehmann, A., and Kloetzel, P. M. (1998). Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 17, 6144–6154. doi:10.1093/emboj/17.21.6144
Erales, J., and Coffino, P. (2014). Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta 1843, 216–221. doi:10.1016/j.bbamcr.2013.05.008
Eytan, E., Ganoth, D., Armon, T., and Hershko, A. (1989). ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc. Natl. Acad. Sci. U. S. A. 86, 7751–7755. doi:10.1073/pnas.86.20.7751
Finley, D., Ciechanover, A., and Varshavsky, A. (1984). Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 37, 43–55. doi:10.1016/0092-8674(84)90299-x
Franzmann, T., and Alberti, S. (2021). Ubiquitin protein helps cells to recover from stress. Nature 597, 183–184. doi:10.1038/d41586-021-02197-z
Fu, A., Cohen-Kaplan, V., Avni, N., Livneh, I., and Ciechanover, A. (2021). p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc. Natl. Acad. Sci. U. S. A. 118, e2107321118. doi:10.1073/pnas.2107321118
Gierisch, M. E., Giovannucci, T. A., and Dantuma, N. P. (2020). Reporter-based screens for the ubiquitin/proteasome system. Front. Chem. 8, 64. doi:10.3389/fchem.2020.00064
Giles, J. (2004). Chemistry Nobel for trio who revealed molecular death-tag. Nature 431, 729. doi:10.1038/431729a
Glickman, M. H., Rubin, D. M., Coux, O., Wefes, I., Pfeifer, G., Cjeka, Z., et al. (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623. doi:10.1016/s0092-8674(00)81603-7
Glotzer, M., Murray, A. W., and Kirschner, M. W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. doi:10.1038/349132a0
Goldberg, A. L. (2003). Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899. doi:10.1038/nature02263
Groll, M., Ditzel, L., Löwe, J., Stock, D., Bochtler, M., Bartunik, H. D., et al. (1997). Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386, 463–471. doi:10.1038/386463a0
Gu, Z. C., and Enenkel, C. (2014). Proteasome assembly. Cell. Mol. Life Sci. 71, 4729–4745. doi:10.1007/s00018-014-1699-8
Gu, Z. C., Wu, E., Sailer, C., Jando, J., Styles, E., Eisenkolb, I., et al. (2017). Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol. Biol. Cell 28, 2479–2491. doi:10.1091/mbc.E17-03-0162
Guo, Q., Lehmer, C., Martínez-Sánchez, A., Rudack, T., Beck, F., Hartmann, H., et al. (2018). In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705. doi:10.1016/j.cell.2017.12.030
Hendil, K. B., Kristensen, P., and Uerkvitz, W. (1995). Human proteasomes analysed with monoclonal antibodies. Biochem. J. 305 (Pt 1), 245–252. doi:10.1042/bj3050245
Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. doi:10.1146/annurev.biochem.67.1.425
Hirayama, S., Sugihara, M., Morito, D., Iemura, S. I., Natsume, T., Murata, S., et al. (2018). Nuclear export of ubiquitinated proteins via the UBIN-POST system. Proc. Natl. Acad. Sci. U. S. A. 115, E4199–E4208. doi:10.1073/pnas.1711017115
Hoeller, D., and Dikic, I. (2016). How the proteasome is degraded. Proc. Natl. Acad. Sci. U. S. A. 113, 13266–13268. doi:10.1073/pnas.1616535113
Inobe, T., and Matouschek, A. (2014). Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 24, 156–164. doi:10.1016/j.sbi.2014.02.002
Iriki, T., Iio, H., Yasuda, S., Masuta, S., Kato, M., Kosako, H., et al. (2023). Senescent cells form nuclear foci that contain the 26S proteasome. Cell Rep. 42, 112880. doi:10.1016/j.celrep.2023.112880
Isono, E., Nishihara, K., Saeki, Y., Yashiroda, H., Kamata, N., Ge, L., et al. (2007). The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell 18, 569–580. doi:10.1091/mbc.e06-07-0635
Itoh, Y., Yamada, M., Suematsu, N., Matsushita, M., and Otomo, E. (1998). An immunohistochemical study of centenarian brains: a comparison. J. Neurol. Sci. 157, 73–81. doi:10.1016/s0022-510x(98)00050-1
Jain, S., Wheeler, J. R., Walters, R. W., Agrawal, A., Barsic, A., and Parker, R. (2016). ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487–498. doi:10.1016/j.cell.2015.12.038
Kaganovich, D., Kopito, R., and Frydman, J. (2008). Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095. doi:10.1038/nature07195
Karmon, O., and Ben Aroya, S. (2019). Spatial organization of proteasome aggregates in the regulation of proteasome homeostasis. Front. Mol. Biosci. 6, 150. doi:10.3389/fmolb.2019.00150
Kito, Y., Matsumoto, M., Hatano, A., Takami, T., Oshikawa, K., Matsumoto, A., et al. (2020). Cell cycle-dependent localization of the proteasome to chromatin. Sci. Rep. 10, 5801. doi:10.1038/s41598-020-62697-2
Kloetzel, P. M., and Ossendorp, F. (2004). Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr. Opin. Immunol. 16, 76–81. doi:10.1016/j.coi.2003.11.004
Kopito, R. R. (2000). Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530. doi:10.1016/s0962-8924(00)01852-3
Kumar, A., Mathew, V., and Stirling, P. C. (2022). Nuclear protein quality control in yeast: the latest INQuiries. J. Biol. Chem. 298, 102199. doi:10.1016/j.jbc.2022.102199
Lafarga, M., Berciano, M. T., Pena, E., Mayo, I., Castaño, J. G., Bohmann, D., et al. (2002). Clastosome: a subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13, 2771–2782. doi:10.1091/mbc.e02-03-0122
Lallemand-Breitenbach, V., and Hugues, D. T. (2010). PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661. doi:10.1101/cshperspect.a000661
Lallemand-Breitenbach, V., Zhu, J., Puvion, F., Koken, M., Honoré, N., Doubeikovsky, A., et al. (2001). Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193, 1361–1371. doi:10.1084/jem.193.12.1361
Laporte, D., Lebaudy, A., Sahin, A., Pinson, B., Ceschin, J., Daignan-Fornier, B., et al. (2011). Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 192, 949–957. doi:10.1083/jcb.201009028
Laporte, D., Salin, B., Daignan-Fornier, B., and Sagot, I. (2008). Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 181, 737–745. doi:10.1083/jcb.200711154
Lee, J., Le, L., Kim, E., and Lee, M. J. (2021). Formation of non-nucleoplasmic proteasome foci during the late stage of hyperosmotic stress. Cells 10, 2493. doi:10.3390/cells10092493
Lehmann, A., Janek, K., Braun, B., Kloetzel, P. M., and Enenkel, C. (2002). 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J. Mol. Biol. 317, 401–413. doi:10.1006/jmbi.2002.5443
Lehmann, A., Niewienda, A., Jechow, K., Janek, K., and Enenkel, C. (2010). Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell 38, 879–888. doi:10.1016/j.molcel.2010.06.016
Li, J., Breker, M., Graham, M., Schuldiner, M., and Hochstrasser, M. (2019). AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet. 15, e1008387. doi:10.1371/journal.pgen.1008387
Li, J., and Hochstrasser, M. (2020). Microautophagy regulates proteasome homeostasis. Curr. Genet. 66, 683–687. doi:10.1007/s00294-020-01059-x
Li, M., Brooks, C. L., Wu-Baer, F., Chen, D., Baer, R., and Gu, W. (2003). Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302, 1972–1975. doi:10.1126/science.1091362
Litsios, A., Grys, B. T., Kraus, O. Z., Friesen, H., Ross, C., Masinas, M. P. D., et al. (2024). Proteome-scale movements and compartment connectivity during the eukaryotic cell cycle. Cell 187, 1490–1507.e21. doi:10.1016/j.cell.2024.02.014
Liu, C. W., Corboy, M. J., DeMartino, G. N., and Thomas, P. J. (2003). Endoproteolytic activity of the proteasome. Science 299, 408–411. doi:10.1126/science.1079293
Livneh, I., Cohen-Kaplan, V., Fabre, B., Abramovitch, I., Lulu, C., Nataraj, N. B., et al. (2023). Regulation of nucleo-cytosolic 26S proteasome translocation by aromatic amino acids via mTOR is essential for cell survival under stress. Mol. Cell 83, 3333–3346.e5. doi:10.1016/j.molcel.2023.08.016
Marguerat, S., Schmidt, A., Codlin, S., Chen, W., Aebersold, R., and Bähler, J. (2012). Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683. doi:10.1016/j.cell.2012.09.019
Marshall, R. S., and Vierstra, R. D. (2019). Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front. Mol. Biosci. 6, 40. doi:10.3389/fmolb.2019.00040
Matyskiela, M. E., Lander, G. C., and Martin, A. (2013). Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788. doi:10.1038/nsmb.2616
Mediani, L., Guillen-Boixet, J., Alberti, S., and Carra, S. (2019b). Nucleoli and Promyelocytic Leukemia Protein (PML) bodies are phase separated nuclear protein quality control compartments for misfolded proteins. Mol. Cell Oncol. 6, e1415624. doi:10.1080/23723556.2019.1652519
Mediani, L., Guillén-Boixet, J., Vinet, J., Franzmann, T. M., Bigi, I., Mateju, D., et al. (2019a). Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J. 38, e101341. doi:10.15252/embj.2018101341
Miller, S. B., Mogk, A., and Bukau, B. (2015). Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J. Mol. Biol. 427, 1564–1574. doi:10.1016/j.jmb.2015.02.006
Munder, M. C., Midtvedt, D., Franzmann, T., Nüske, E., Otto, O., Herbig, M., et al. (2016). A pH-driven transition of the cytoplasm from a fluid-to a solid-like state promotes entry into dormancy. eLife 5, e09347. doi:10.7554/eLife.09347
Park, S., Kim, W., Tian, G., Gygi, S. P., and Finley, D. (2011). Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 286, 36652–36666. doi:10.1074/jbc.M111.285924
Park, S. H., Kukushkin, Y., Gupta, R., Chen, T., Konagai, A., Hipp, M. S., et al. (2013). PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154, 134–145. doi:10.1016/j.cell.2013.06.003
Parry, B. R., Surovtsev, I. V., Cabeen, M. T., O'Hern, C. S., Dufresne, E. R., and Jacobs-Wagner, C. (2014). The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194. doi:10.1016/j.cell.2013.11.028
Peters, J. M., Franke, W. W., and Kleinschmidt, J. A. (1994). Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 269, 7709–7718. doi:10.1016/s0021-9258(17)37345-3
Peters, L. Z., Hazan, R., Breker, M., Schuldiner, M., and Ben-Aroya, S. (2013). Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J. Cell Biol. 201, 663–671. doi:10.1083/jcb.201211146
Peth, A., Nathan, J. A., and Goldberg, A. L. (2013). The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 288, 29215–29222. doi:10.1074/jbc.M113.482570
Protter, D. S. W., and Parker, R. (2016). Principles and properties of stress granules. Trends Cell Biol. 26, 668–679. doi:10.1016/j.tcb.2016.05.004
Rape, M., and Jentsch, S. (2002). Taking a bite: proteasomal protein processing. Nat. Cell Biol. 4, E113–E116. doi:10.1038/ncb0502-e113
Rousseau, A., and Bertolotti, A. (2018). Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 19, 697–712. doi:10.1038/s41580-018-0040-z
Russell, S. J., Steger, K. A., and Johnston, S. A. (1999). Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274, 21943–21952. doi:10.1074/jbc.274.31.21943
Sahin, U., Lallemand-Breitenbach, V., and de The, H. (2014). PML nuclear bodies: regulation, function and therapeutic perspectives. J. Pathol. 234, 289–291. doi:10.1002/path.4426
Salomons, F. A., Acs, K., and Dantuma, N. P. (2010). Illuminating the ubiquitin/proteasome system. Exp. Cell Res. 316, 1289–1295. doi:10.1016/j.yexcr.2010.02.003
Saunier, R., Esposito, M., Dassa, E. P., and Delahodde, A. (2013). Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS One 8, e70357. doi:10.1371/journal.pone.0070357
Schweitzer, A., Aufderheide, A., Rudack, T., Beck, F., Pfeifer, G., Plitzko, J. M., et al. (2016). Structure of the human 26S proteasome at a resolution of 3.9 A. Proc. Natl. Acad. Sci. U. S. A. 113, 7816–7821. doi:10.1073/pnas.1608050113
Sha, Z., Peth, A., and Goldberg, A. L. (2011). Keeping proteasomes under control--a role for phosphorylation in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 108, 18573–18574. doi:10.1073/pnas.1115315108
Shi, Y., Chen, X., Elsasser, S., Stocks, B. B., Tian, G., Lee, B. H., et al. (2016). Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351, aad9421. doi:10.1126/science.aad9421
Silonov, S. A., Smirnov, E. Y., Kuznetsova, I. M., Turoverov, K. K., and Fonin, A. V. (2023). PML body biogenesis: a delicate balance of interactions. Int. J. Mol. Sci. 24, 16702. doi:10.3390/ijms242316702
Sommer, T., and Wolf, D. H. (1997). Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 11, 1227–1233. doi:10.1096/fasebj.11.14.9409541
Tanaka, K. (2009). The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36. doi:10.2183/pjab.85.12
Tanaka, K., and Matsuda, N. (2014). Proteostasis and neurodegeneration: the roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta 1843, 197–204. doi:10.1016/j.bbamcr.2013.03.012
Tomita, T., Hirayama, S., Sakurai, Y., Ohte, Y., Yoshihara, H., Saeki, Y., et al. (2019). Specific modification of aged proteasomes revealed by tag-exchangeable knock-in mice. Mol. Cell Biol. 39, e00426–e00418. doi:10.1128/MCB.00426-18
Torres, C., Lewis, L., and Cristofalo, V. J. (2006). Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J. Cell. physiology 207, 845–853. doi:10.1002/jcp.20630
Tsvetkov, P., Reuven, N., and Shaul, Y. (2009). The nanny model for IDPs. Nat. Chem. Biol. 5, 778–781. doi:10.1038/nchembio.233
Unverdorben, P., Beck, F., Śledź, P., Schweitzer, A., Pfeifer, G., Plitzko, J. M., et al. (2014). Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 111, 5544–5549. doi:10.1073/pnas.1403409111
Uriarte, M., Sen Nkwe, N., Tremblay, R., Ahmed, O., Messmer, C., Mashtalir, N., et al. (2021). Starvation-induced proteasome assemblies in the nucleus link amino acid supply to apoptosis. Nat. Commun. 12, 6984. doi:10.1038/s41467-021-27306-4
Ustrell, V., Hoffman, L., Pratt, G., and Rechsteiner, M. (2002). PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21, 3516–3525. doi:10.1093/emboj/cdf333
Vabulas, R. M., and Hartl, F. U. (2005). Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–1963. doi:10.1126/science.1121925
van der Lee, R., Lang, B., Kruse, K., Gsponer, J., Sánchez de Groot, N., Huynen, M. A., et al. (2014). Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 8, 1832–1844. doi:10.1016/j.celrep.2014.07.055
van Deventer, S., Menendez-Benito, V., van Leeuwen, F., and Neefjes, J. (2015). N-terminal acetylation and replicative age affect proteasome localization and cell fitness during aging. J. Cell. Sci. 128, 109–117. doi:10.1242/jcs.157354
van Leeuwen, W., and Rabouille, C. (2019). Cellular stress leads to the formation of membraneless stress assemblies in eukaryotic cells. Traffic 20, 623–638. doi:10.1111/tra.12669
Vernon, R. M., and Forman-Kay, J. D. (2019). First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 58, 88–96. doi:10.1016/j.sbi.2019.05.016
VerPlank, J. J. S., and Goldberg, A. L. (2017). Regulating protein breakdown through proteasome phosphorylation. Biochem. J. 474, 3355–3371. doi:10.1042/BCJ20160809
von Mikecz, A. (2006). The nuclear ubiquitin-proteasome system. J. Cell. Sci. 119, 1977–1984. doi:10.1242/jcs.03008
von Mikecz, A., Chen, M., Rockel, T., and Scharf, A. (2008). The nuclear ubiquitin-proteasome system: visualization of proteasomes, protein aggregates, and proteolysis in the cell nucleus. Methods Mol. Biol. 463, 191–202. doi:10.1007/978-1-59745-406-3_14
Waite, K. A., and Roelofs, J. (2022). Proteasome granule formation is regulated through mitochondrial respiration and kinase signaling. J. Cell. Sci. 135, jcs259778. doi:10.1242/jcs.259778
Wang, X., Yen, J., Kaiser, P., and Huang, L. (2010). Regulation of the 26S proteasome complex during oxidative stress. Sci. Signal. 3, ra88. doi:10.1126/scisignal.2001232
Weberruss, M. H., Savulescu, A. F., Jando, J., Bissinger, T., Harel, A., Glickman, M. H., et al. (2013). Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 32, 2697–2707. doi:10.1038/emboj.2013.192
Wehmer, M., Rudack, T., Beck, F., Aufderheide, A., Pfeifer, G., Plitzko, J. M., et al. (2017). Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 114, 1305–1310. doi:10.1073/pnas.1621129114
Wendler, P., and Enenkel, C. (2019). Nuclear transport of yeast proteasomes. Front. Mol. Biosci. 6, 34. doi:10.3389/fmolb.2019.00034
Wilkinson, C. R., Wallace, M., Morphew, M., Perry, P., Allshire, R., Javerzat, J. P., et al. (1998). Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 17, 6465–6476. doi:10.1093/emboj/17.22.6465
Wojcik, C., and DeMartino, G. N. (2003). Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 35, 579–589. doi:10.1016/s1357-2725(02)00380-1
Wolf, D. H., and Menssen, R. (2018). Mechanisms of cell regulation - proteolysis, the big surprise. FEBS Lett. 592, 2515–2524. doi:10.1002/1873-3468.13109
Xu, S., Gierisch, M. E., Schellhaus, A. K., Poser, I., Alberti, S., Salomons, F. A., et al. (2023). Cytosolic stress granules relieve the ubiquitin-proteasome system in the nuclear compartment. EMBO J. 42, e111802. doi:10.15252/embj.2022111802
Yasuda, S., Tsuchiya, H., Kaiho, A., Guo, Q., Ikeuchi, K., Endo, A., et al. (2020). Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300. doi:10.1038/s41586-020-1982-9
Yewdell, J. W. (2005). Serendipity strikes twice: the discovery and rediscovery of defective ribosomal products (DRiPS). Cell. Mol. Biol. 51, 635–641.
Keywords: metabolic regulation of proteasome localization, proteasome condensates in membraneless organelles, proteasome storage granules, protein homeostasis (proteostasis), ubiquitin 26S-proteasome system
Citation: Enenkel C and Ernst OP (2025) Proteasome dynamics in response to metabolic changes. Front. Cell Dev. Biol. 13:1523382. doi: 10.3389/fcell.2025.1523382
Received: 05 November 2024; Accepted: 03 February 2025;
Published: 03 March 2025.
Edited by:
Ralf Stohwasser, Brandenburg University of Technology Cottbus-Senftenberg, GermanyReviewed by:
Abhishek Sinha, Atria University, IndiaElena Panizza, Cornell University, United States
Copyright © 2025 Enenkel and Ernst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cordula Enenkel, Y29yZHVsYS5lbmVua2VsQHV0b3JvbnRvLmNh
 Cordula Enenkel
Cordula Enenkel Oliver P. Ernst1,2
Oliver P. Ernst1,2