
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell Dev. Biol. , 22 January 2025
Sec. Embryonic Development
Volume 12 - 2024 | https://doi.org/10.3389/fcell.2024.1491394
The spindle assembly checkpoint (SAC) is a surveillance mechanism that prevents uneven segregation of sister chromatids between daughter cells during anaphase. This essential regulatory checkpoint prevents aneuploidy which can lead to various congenital defects observed in newborns. Many studies have been carried out to elucidate the role of proteins involved in the SAC as well as the function of the checkpoint during gametogenesis and embryogenesis. In this review, we discuss the role of SAC proteins in regulating both meiotic and mitotic cell division along with several factors that influence the SAC strength in various species. Finally, we outline the role of SAC proteins and the consequences of their absence or insufficiency on proper gametogenesis and embryogenesis in vivo.
Cell division is a critical feature of embryonic development (Kumar et al., 2015). Fertilization of a secondary oocyte, a haploid cell arrested in metaphase II of meiosis, by a spermatozoon results in a diploid zygote, which then divides into 2 cells through the process of mitosis (Yeste et al., 2017). This is followed by extensive cell division, which combined with various cellular signaling and morphogenic events promotes embryogenesis including blastulation, gastrulation, neurulation, and organ development (Zhai et al., 2022). Cell division during embryonic development must be carefully monitored since error prone division produces aneuploidy that has severe consequences for fecundity (Jia et al., 2015). The eukaryotic cell cycle consists of interphase (comprising the G1, S, and G2 phases) and mitosis, with each phase orchestrated by regulatory checkpoints mediated by cyclins and cyclin-dependent kinases (CDKs) (Wang, 2021). During the G1 phase, the restriction checkpoint ensures the cell is prepared to enter the cell cycle by ensuring any DNA damage is properly repaired and the cell is receiving the appropriate growth signals before committing to division (Bertoli et al., 2013). Cyclin D-CDK4/6 complexes drive progression through G1 by phosphorylating the retinoblastoma (Rb) protein, releasing E2F transcription factors to activate genes necessary for the transition into S phase and carrying out DNA synthesis (Bertoli et al., 2013; Topacio et al., 2019). During the G1/S phase transition, Cyclin E-CDK2 initiates DNA replication by activating origins of replication. During S phase, Cyclin A replaces Cyclin E, terminating DNA synthesis and facilitating the transition into the G2 phase (Ding et al., 2020). The G2 phase is regulated by the G2/M checkpoint, which ensures the integrity of replicated DNA before mitotic entry (Krempler et al., 2007; Kousholt et al., 2012). Cyclin A-CDK1 activity orchestrates the preparatory events for mitosis by promoting the nuclear translocation of Cyclin B1-CDK1 (Gong et al., 2007; Gong and Ferrell, 2010). Once in the nucleus, Cyclin A and Cyclin B1-CDK1 collaborate to drive key mitotic processes, including chromosome condensation and nuclear envelope breakdown (Petrone et al., 2016; Enserink and Kolodner, 2010; Gong and Ferrell, 2010). To ensure fidelity in chromosome segregation and prevent aneuploidy, the spindle assembly checkpoint (SAC) operates during M phase, monitoring the attachment of chromosomes to the spindle apparatus to guarantee proper segregation between daughter cells (Lara-Gonzalez et al., 2012; Lara-Gonzalez et al., 2021b). A variety of studies have been published over the last decade that have produced intriguing insights into the role of the SAC, and the factors that regulate SAC function, in gametogenesis and embryogenesis. In this review, we will explore the role of SAC proteins during these crucial biological processes.
Segregation of genetic material between daughter cells begins during the anaphase stage of mitosis (Musacchio and Desai, 2017). However, the mechanisms that ensure each daughter cell receives an equal and full complement of DNA, begin earlier in the cell division process. During S phase, cells fully duplicate each chromosome (Musacchio and Desai, 2017). Additionally, centrosome duplication also occurs during S phase (Tsou and Stearns, 2006; Holland et al., 2010) and errors in centrosome duplication can cause chromosome missegregation leading to aneuploid daughter cells (Silkworth et al., 2009; Ganem et al., 2009). During prometaphase of mitosis, a complex of proteins called the kinetochore assembles at the centromere of each chromosome, serving as the attachment site for microtubules from the mitotic spindle (Musacchio and Desai, 2017). Kinetochore associated proteins regulate the movement of each sister chromatid in opposing directions along spindle fiber microtubules during anaphase (Musacchio and Desai, 2017). Therefore, it is imperative for microtubules from the bipolar spindle to attach to kinetochores of both chromatids (amphitelic attachment) for the divergent movement and even distribution of chromosomes to each daughter cell (Lampson and Grishchuk, 2017). However, various challenges can compromise the fidelity of chromosome segregation, such as syntelic attachment where both sister chromatids connect to the same spindle pole, merotelic attachment in which a single chromatid or kinetochore attaches to both spindle poles, or monotelic attachment in which only one sister chromatid is attached to spindle microtubules (Lampson and Grishchuk, 2017; Khodjakov and Pines, 2010; Taylor et al., 2004). If these issues are not resolved, they can lead to chromosome missegregation (Lampson and Grishchuk, 2017). While the SAC monitors unattached kinetochores, it does not reliably detect all attachment errors, such as merotelic and syntelic attachments (Cimini et al., 2001; Jin et al., 2012). Loss or gain of chromosomes, termed aneuploidy, results in severe consequences such as developmental defects and cancer (Shahbazi et al., 2020; Boveri, 2008). Trisomy 21 is one such instance in which an extra copy of chromosome 21 is acquired during abnormal cell division leading to Down’s syndrome (Roizen and Patterson, 2003). Therefore, each sister chromatid must bi-orient and attach to microtubules during metaphase before anaphase initiates. The SAC, also known as the mitotic checkpoint, fulfills this need as a surveillance mechanism to confirm all chromosomes are properly aligned and bound by bipolar spindles to ensure anaphase fidelity (Lara-Gonzalez et al., 2012).
The SAC prevents the progression to anaphase until all chromosomes are properly attached to the mitotic spindle via their kinetochores (Musacchio, 2015b; London and Biggins, 2014; Cheeseman, 2014). The SAC is activated by unattached or improperly attached kinetochores, which generate a ‘wait’ signal that halts anaphase progression. This signal prevents the activation of the anaphase-promoting complex/cyclosome (APC/C), ensuring that cells do not prematurely segregate their chromosomes (McAinsh and Kops, 2023). A multitude of studies have been carried out to decipher what exactly the SAC senses leading to its activation (McVey et al., 2021; Khodjakov and Pines, 2010). Traditionally, it is believed that the SAC acts as a quality control mechanism, preventing the cell from segregating its chromosomes (progressing from metaphase to anaphase) if there are errors in chromosome attachment to the bipolar spindle (Rieder et al., 1995). According to this view, the SAC is primarily concerned with whether the chromosomes are correctly attached to the spindle microtubules. This suggests that the SAC is unable to monitor for proper orientation of the chromatids. For instance, chromatids can be mono-oriented or bi-oriented regardless of proper kinetochore attachment (amphitelic) (Khodjakov and Pines, 2010). This view is supported by studies using laser beam-mediated ablation of unattached kinetochore on a monotelic chromosome that resulted in a normal progression of mitosis suggesting that the SAC only detects the presence or absence of microtubule attachment on the kinetochores (Rieder et al., 1995). Another mechanistic explanation behind SAC activation that has been extensively studied is based on the lack of tension across unattached kinetochores (Mukherjee et al., 2019; Chen et al., 2021). For instance, applying tension to an unattached kinetochore through a micromanipulation needle releases cells from metaphase and they progress into anaphase suggesting that tension between kinetochores of sister chromatids regulates SAC activation (Li and Nicklas, 1995; Nicklas and Koch, 1969). Moreover, there is a correlation between the level of tension and metaphase delay time suggesting that the cell is highly sensitive to tension levels, which may serve as a critical signal to prevent chromosome segregation errors during mitosis (Mukherjee et al., 2019). Hence, the SAC is thought to sense either the presence of unattached kinetochores or the lack of tension across the kinetochores of sister chromatids. Although strong evidence exist for both explanations, the ability of the kinase Aurora B to contribute to both mechanisms may connect these mechanisms together (McVey et al., 2021). Aurora B has been proposed to sense the lack of tension in the kinetochores and translate this into a biochemical signal by phosphorylating members of the kinetochore such as Knl1 and Mis12 that belong to the KMN network (Akiyoshi et al., 2010; Welburn et al., 2010; Cheeseman et al., 2002). Another study suggests that Aurora B activity is not directly regulated by tension (Liu et al., 2009). Instead, phosphorylation depends on the spatial proximity of substrates to Aurora B at the inner centromere. This proximity or distance, is affected by tension which indirectly affects the phosphorylation of the substrates by Aurora B (Liu et al., 2009). Such a spatial model underscores a nuanced mechanism by which Aurora B may modulate kinetochore signaling.
The KMN network is a large assembly of proteins composed of three main subcomplexes, namely, the Mis12 complex, the Ndc80 complex, and the Knl1 complex (Varma and Salmon, 2012). The Mis12 complex, consisting of four proteins (Dsn1, Mis12, Nnf1, and Nsl1), is responsible for the assembly of the Ndc80 and Knl1 complexes along the outer kinetochore and has been described as a protein interaction hub for outer kinetochore assembly (Cheeseman and Desai, 2008; Maskell et al., 2010; Petrovic et al., 2010). The Knl1 and Ndc80 complexes are tethered by Nsl1 aiding in their recruitment to the kinetochore (Petrovic et al., 2010). The Ndc80 complex has four subunits (Hec1, Nuf2, Spc24, and Spc25) in a coiled coil fashion and is responsible for bridging the KMN network to microtubules (Cheeseman and Desai, 2008; Ciferri et al., 2008; Wei et al., 2007). The Ndc80 complex provides a docking site for the N-terminal region of the SAC protein Mps1 (Kemmler et al., 2009; Nijenhuis et al., 2013). Studies have shown that the Mps1 and microtubules compete against each other to bind to Ndc80 (Pleuger et al., 2024). According to this competition model, Mps1 directly interacts with the HEC1 domain of the NDC80 complex and initiates the SAC (Pleuger et al., 2024; Ji et al., 2015). However, the presence of end-on microtubule attachment to the NDC80 complex disrupts this interaction (Pleuger et al., 2024). This competitive binding mechanism serves as a direct sensor for kinetochore-microtubule attachment status, regulating SAC signaling (Ji et al., 2015; Pleuger et al., 2024). However, recent findings challenge this direct competition model showing that Mps1 autophosphorylation is responsible for its release from kinetochores at least in the context of native kinetochores isolated from yeast (Koch et al., 2019). These findings suggest that while direct competition between Mps1 and microtubules for Ndc80 binding may contribute to SAC regulation, Mps1 autophosphorylation likely plays a key role in modulating its kinetochore association.
Once docked on Ndc80, Mps1 phosphorylates Knl1 which is one of the two subunits of the Knl1 complex, the other being Zwint1. Phosphorylation of Knl1 occurs on threonine residues present on its MELT repeats, triggering Knl1 to act as a docking site for SAC proteins BubR1, Bub1, and Bub3 (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012; Krenn et al., 2014; Overlack et al., 2015; Vleugel et al., 2013; Zhang et al., 2014). The dephosphorylation of Knl1 is prevented by Aurora B which phosphorylates the SILK and RVSF motifs at the N-terminus of Knl1 (Agarwal and Varma, 2015). These phosphorylation events prevent protein phosphatase 1 (PP1) from binding to Knl1. It has also been speculated that Aurora B promotes the recruitment of Mps1 to the KMN network (Figure 1) (Agarwal and Varma, 2015). The coordinated assembly and regulation of the KMN network, particularly through the actions of key protein complexes such as Mis12, Ndc80, and Knl1, are crucial for the proper activation of the SAC (Agarwal and Varma, 2015). These interactions ensure accurate chromosome segregation by tightly controlling kinetochore-microtubule attachments and signaling pathways, highlighting the intricate molecular mechanisms that maintain genomic stability during mitosis (Figure 1A).
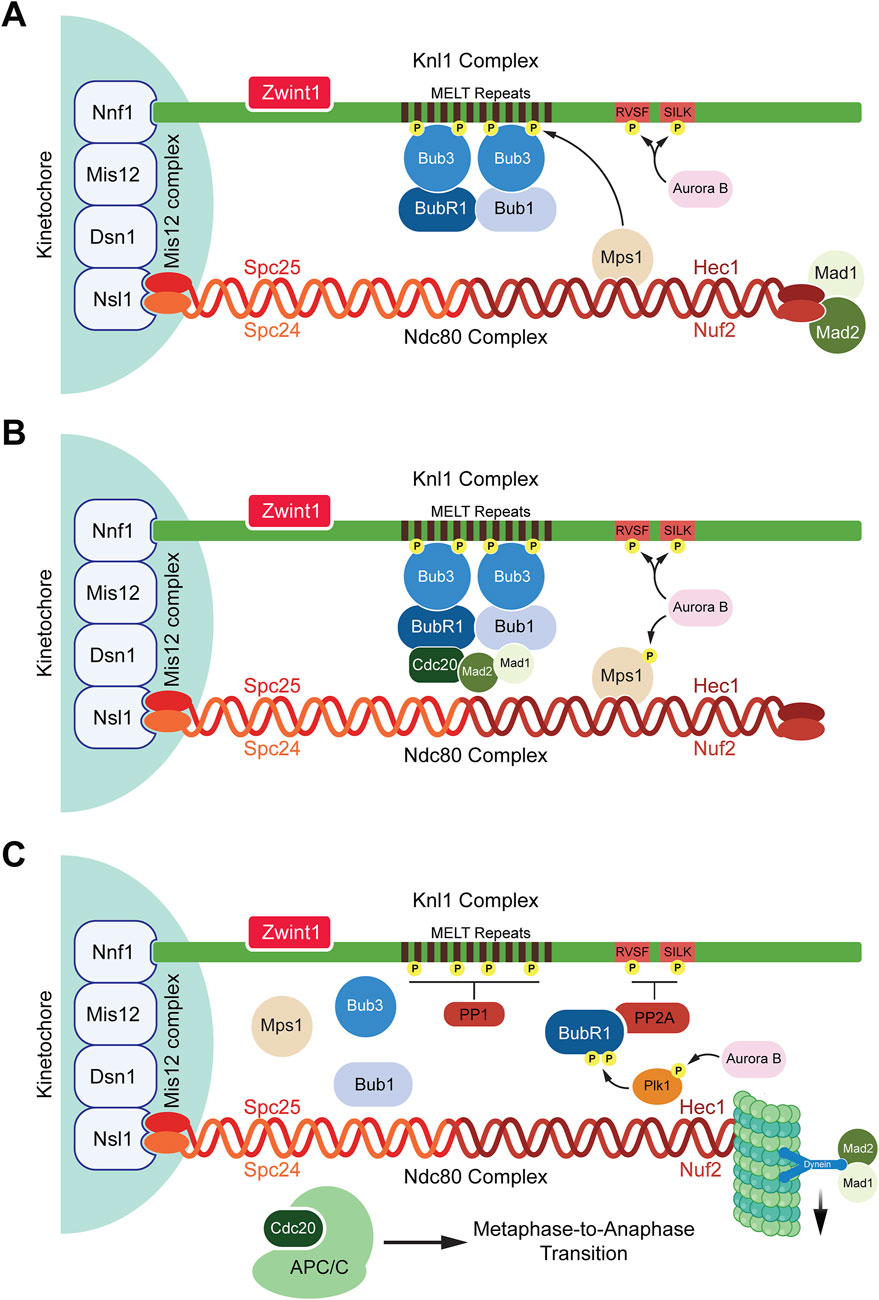
Figure 1. Spindle assembly checkpoint activation and deactivation. (A) SAC activation: When kinetochores are unattached to microtubules, the SAC is activated. Aurora B recruits Mps1 that binds to the Ndc80 complex and phosphorylates the MELT repeats present on the Knl1 complex. Once phosphorylated, the MELT repeats recruit the SAC protein Bub3 which further recruits BubR1 and Bub1. Aurora B also phosphorylates the RVSF and SILK domains preventing the phosphatase PP1 from binding to Knl1 complex. (B) SAC maintenance: Bub1 interacts with Cdc20 and the Mad1:Mad2 complex bringing Mad2 and Cdc20 in close proximity to form a subcomplex inhibiting the interactions between APC/C and Cdc20. The assembled MCC now consists of the Bub3:BubR1 and Mad2:Cdc20 subcomplexes. (C) SAC deactivation: Once microtubules attach to the Ndc80 complex, Mps1 is displaced from the Ndc80 complex, Aurora B activates Plk1 which stimulates the binding of PP2A to BubR1 by phosphorylating BubR1. PP2A dephosphorylates the RVSF and SILK domains allowing for PP1 to bind to Knl1 where it dephosphorylates the MELT repeats. This causes the release of MCC components which are stripped away from the Knl1 complex by dynein proteins. Hence, the SAC is deactivated and the binding of APC/C to Cdc20 occurs to allow for the metaphase-to-anaphase transition.
The SAC ensures accurate chromosome segregation by preventing anaphase onset until all kinetochores are properly attached to spindle microtubules and tension is generated across sister chromatids. Upon detecting unattached kinetochores, the SAC activates and assembles a complex of proteins collectively known as the mitotic checkpoint complex (MCC) (Lara-Gonzalez et al., 2012; Jia et al., 2013; Musacchio, 2015b; Musacchio and Desai, 2017). The MCC consists of four proteins, Mad2, BubR1, Bub3, and Cdc20 (Chao et al., 2012; Hardwick et al., 2000; Sudakin et al., 2001; Tang et al., 2001). The primary function of the MCC is to inhibit the activation of the APC/C, an E3 ubiquitin ligase responsible for initiating anaphase (Lara-Gonzalez et al., 2021b; Barford, 2020). Several proteins aid in the assembly of the MCC complex such as Bub1, Mad1, and Mps1 (London and Biggins, 2014; Fischer et al., 2021; Kulukian et al., 2009; Hewitt et al., 2010; Abrieu et al., 2001). The process begins with Mps1 kinase phosphorylating the MELT repeats of the kinetochore protein Knl1 followed by recruitment of Bub1 and Bub3 to the kinetochores (London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012). Bub1-Bub3 binding increases the affinity of Bub3 towards the phosphorylated MELT repeats (Primorac et al., 2013). Bub3 also interacts with BubR1 which leads to the formation of a heterotetrametric complex containing Bub1-Bub3 and BubR1-Bub3 dimers present on the kinetochores (Overlack et al., 2015; Zhang et al., 2015). While BubR1 also interacts with Bub1, studies have shown that the recruitment of BubR1 to the kinetochores by Bub1 is dispensable for SAC activation (Vleugel et al., 2015). The recruitment of the other two MCC proteins, Mad2 and Cdc20, occurs through the interaction of multiple proteins (Ge et al., 2009). Bub1 recruits and stabilizes the Mad1-Mad2 complex at unattached kinetochores. This occurs via Bub1s CM1 motif, which binds the RLK motif of Mad1 (London and Biggins, 2014; Zhang et al., 2017; Lara-Gonzalez et al., 2021b; Klebig et al., 2009). Simultaneously, ABBA motif of Bub1 interacts with the WD40 domain of Cdc20 (Piano et al., 2021; Lara-Gonzalez et al., 2021b). These events bring Mad2 and Cdc20 in close proximity (Piano et al., 2021; Lara-Gonzalez et al., 2021a). Mad1, when phosphorylated by Mps1 on its RWD domain, also interacts with Cdc20 which exposes the Mad2 binding motif present within the Cdc20 N-terminus (Faesen et al., 2017; Ji et al., 2017; Lara-Gonzalez et al., 2021a; Piano et al., 2021). BubR1 also interacts with Cdc20 through its KEN box (K1), ABBA motif, and D box (D2) domains (Burton and Solomon, 2007; King et al., 2007; Lara-Gonzalez et al., 2011). Together, these events bring Mad2, BubR1, Bub3, and Cdc20 into close proximity, culminating in the formation of a complete MCC complex (Figure 1B).
A primary function of the MCC is to inhibit APC/C activation by sequestering its coactivator, Cdc20. Without APC/C activity, key substrates such as Securin and Cyclin B1 remain stabilized (Barford, 2020). The APC/C ubiquitinates Securin and Cyclin B1, marking them for proteasome-dependent degradation (Barford, 2020). Securin inhibits Separase, an enzyme required for cleaving the cohesin subunit RAD21, which holds sister chromatids together (Anderson et al., 2002; Haering et al., 2002). Cyclin B1, on the other hand, is the regulatory subunit of the mitotic kinase CDK1 which is critical for the G2 to M phase transition (Zur and Brandeis, 2002). During anaphase, APC/C-mediated ubiquitination of Cyclin B1 marks it for degradation, leading to the inactivation of CDK1 (Zur and Brandeis, 2002). This inactivation is crucial for reversing key mitotic processes including nuclear envelope breakdown (NEB) and chromosome condensation (Malumbres, 2014; Qian et al., 2015). Thus, inhibition of the APC/C by the MCC prevents the degradation of its substrates, Securin and Cyclin B1, until the SAC is satisfied, ensuring proper chromosome alignment and attachment to bipolar spindles prior to anaphase onset.
It has long been accepted that the primary function of the MCC is to prevent the formation of the APC/CCdc20 complex by binding to Cdc20 (Sudakin et al., 2001). This implies that the MCC competes with the APC/C for available Cdc20. Recent observations have forced revisions to this model and have provided an elegant explanation for how the MCC inhibits the activity of APC/C (Herzog et al., 2009; Alfieri et al., 2016; Yamaguchi et al., 2016; Kelly et al., 2014). First, the MCC consists of BubR1:Bub3:Cdc20:Mad2 in a 1:1:1:1 stoichiometric ratio (Sudakin et al., 2001; Alfieri et al., 2016; Yamaguchi et al., 2016; Izawa and Pines, 2015). One Cdc20 is associated with the MCC (Cdc20M). It is bound to the MCC through its interaction with BubR1 by binding to the KEN box, TPR motifs, ABBA motifs, and the D box that Cdc20M recognizes within BubR1 (Di Fiore et al., 2015; Diaz-Martinez et al., 2015; Lischetti et al., 2014). Cdc20M also binds to the MCC through interactions with Mad2 and Mad3 (Izawa and Pines, 2015; Yamaguchi et al., 2016). Second, BubR1 consists of a second set of KEN box (K2), ABBA motifs (A1), and the D box (D1) that is recognized by another Cdc20 which is associated with the APC/C and is termed Cdc20A (Alfieri et al., 2016; Yamaguchi et al., 2016). Recent studies have demonstrated a direct interaction between Cdc20A and Cdc20M (Zhang et al., 2024). The CRY box, an unconventional degron motif within Cdc20, plays a critical role in this interaction. Electrostatic interactions are present between the CRY box residues (R162, K163) of Cdc20M and acidic residues (E180, D203) of Cdc20A (Zhang et al., 2024). Mutations in the CRY box, such as R162E/K163E or E180R/D203R, disrupt these interactions, abolishing SAC function and accelerating mitosis. Additionally, the CRY box in Cdc20M interacts with the KNOT domain of BubR1, consisting of a D-box pseudo-degron sequence (residues 224–232) and a hydrophobic loop, further stabilizing the MCC. Mutations that unfold the KNOT domain lead to SAC defects as well (Zhang et al., 2024). These findings suggest the functional implications of a Cdc20A and Cdc20M interaction for SAC function.
The interaction of Cdc20A with BubR1 shifts it away from the D-box receptor, which is formed by Cdc20A and APC10 within the APC/C (Izawa and Pines, 2015; Alfieri et al., 2016; Yamaguchi et al., 2016). Cdc20A and APC10 together recognize APC/C substrates and target them for ubiquitination (Buschhorn et al., 2011; da Fonseca et al., 2011). Therefore, preventing the interaction of Cdc20A and APC10 prevents substrate recognition. An additional mechanism by which BubR1 in the MCC prevents APC/C activity is through blocking recruitment of ubiquitination machinery. Since APC/C is an E3 ubiquitin ligase, it requires an E2 ubiquitin conjugating enzyme to add ubiquitin to substrates (Yang et al., 2021). The APC/C recruits two E2s, namely, UbcH10 (also known as UBE2C) and UBE2S, to its catalytic core composed of APC2 and APC11 (Brown et al., 2016; Brown et al., 2014). BubR1 utilizes its TPR binding motifs to interact with APC2 and sterically hinder UbcH10 from binding to the catalytic subunit on APC/C (Yamaguchi et al., 2016). Hence, the MCC functions to inhibit the APC/C via multiple mechanisms during the SAC.
Once bipolar microtubules attach to kinetochores of all chromosomes, the SAC must be silenced which involves multiple mechanisms. Three major processes have been characterized that silence the SAC.
1) Cdc20 ubiquitination: Since the MCC inhibits the APC/C, its disassembly is necessary to initiate anaphase. Specifically, Cdc20 that is a part of the MCC, Cdc20M, is ubiquitinated by the APC/C itself (Nilsson et al., 2008). Recent studies have clarified how Cdc20M gets ubiquitinated by the APC/C and is preferred over Cdc20A (Alfieri et al., 2016; Yamaguchi et al., 2016). The catalytic core of the APC/C consists of APC2, which contains a Cullin domain, and APC11, which has a RING domain involved in recruiting the two E2 ubiquitin-conjugating enzymes, UBE2C and UBE2S (Yamaguchi et al., 2016). UBE2C is essential for substrate ubiquitination, while UBE2S is required for ubiquitin chain elongation (Yamaguchi et al., 2016). The MCC inhibits the recruitment of these E2 enzymes by binding to APC2 and APC11. Specifically, BubR1 binds to the WHB domain of APC2 and the RING domain of APC11 (Yamaguchi et al., 2016). This represents the closed state of the APC/C, where its catalytic cavity is blocked. In its open conformation, the MCC complex shifts away from the catalytic cavity, allowing UBE2C to bind to the WHB domain of APC2 and the RING domain of APC11 (Alfieri et al., 2016; Yamaguchi et al., 2016). This positioning places Cdc20M near the APC2-APC11 Cullin-RING complex, activated by UBE2C, which then ubiquitinates Cdc20M (Alfieri et al., 2016; Yamaguchi et al., 2016). This is supported by the observation that deletion of APC15, another component of the APC/C, enhances the MCC:APC/C interaction and reduces Cdc20M ubiquitination suggesting the presence of complexes within the APC/C that target Cdc20M (Foster and Morgan, 2012). Overall, Cdc20M ubiquitination releases it from the MCC (Alfieri et al., 2016; Yamaguchi et al., 2016). However, whether it is degraded or not remains unclear. This also leads to the proper interaction between Cdc20A and APC10 which allows for APC/C to interact with its substrates, including Securin and Cyclin B1, for ubiquitination and subsequent degradation (Alfieri et al., 2016; Yamaguchi et al., 2016).
2) Mad2:Cdc20 disassembly: Like the MCC:APC/C:Cdc20 super complex, Mad2 exists in either an open or closed conformation (Habu et al., 2002). In its closed form, Mad2 interacts with Mad1 and Cdc20 to participate in the MCC (Habu et al., 2002). Therefore, it is vital to reestablish Mad2 in its open conformation to silence the SAC. TRIP13, an AAA-ATPase, uses an adaptor protein termed p31comet to promote the conversion of the closed Mad2 to an open conformation (Wang et al., 2014; Eytan et al., 2014; Ye et al., 2015; Musacchio, 2015a). Interestingly, the reestablished open form of Mad2 is also essential for SAC activation since TRIP13-deficient cells, which contain only the closed form of Mad2, cannot undergo SAC activation (Ma and Poon, 2018).
3) Phosphatases: Phosphorylation plays a key role in the initiation of the SAC. The phosphorylation of Knl1 at the MELT repeats by Mps1 initiates SAC signaling (Zhang et al., 2014). Knl1 is also phosphorylated at the SILK and RVSF motifs by Aurora B which abrogates the recruitment of phosphatases preventing SAC silencing (Nasa et al., 2018; Caldas and DeLuca, 2014). Once kinetochores are attached to microtubules on Ndc80, this dislodges Mps1 from kinetochores (Ji et al., 2015; Ji et al., 2017). While phosphorylation events initiate SAC assembly, dephosphorylation is crucial for SAC silencing. Two phosphatases, PP2A and PP1, are key players in this process (Foley et al., 2011; Liu et al., 2010; Posch et al., 2010). Plk1 triggers the binding of PP2A to BubR1 by further phosphorylating Serine-676 and Threonine-680 residues present on the kinetochore attachment regulatory domain (KARD) motif of BubR1 (Elowe et al., 2007; Suijkerbuijk et al., 2012; Kruse et al., 2013; Wang et al., 2016a; Wang et al., 2016b). The regulatory subunit of PP2A, B56, binds to BubR1 on its phosphorylated KARD domain (Suijkerbuijk et al., 2012; Ghongane et al., 2014). PP2A then dephosphorylates the SILK and RVSF motifs of Knl1 (Espert et al., 2014). This dephosphorylation counters the kinase activity of Aurora B allowing access for the phosphatase PP1 to dock on Knl1 and dephosphorylate the MELT repeats (Nijenhuis et al., 2014). Recent studies have further elucidated the role of PP2A/B56 in dephosphorylation processes critical for SAC signaling. PP2A/B56 directly targets the MELT repeats of Knl1, as demonstrated by the dephosphorylation of Knl1 at Threonine-875, a conserved Mps1 phosphorylation site (Espert et al., 2014). Loss of PP2A/B56 activity results in increased Threonine-875 phosphorylation (Espert et al., 2014). Additionally, PP2A/B56 bound to BubR1 can dephosphorylate Bub1 at Threonine-461 (Wang et al., 2023). This residue is phosphorylated by Mps1 creating a binding site for Mad1 on Bub1 which is crucial for SAC signaling (Zhang et al., 2017). Therefore, dephosphorylation events by these phosphatases initiate SAC silencing leading to the disassembly of the MCC from the kinetochore. In order to remove the MCC proteins, the microtubule motor dynein recognizes Mad1–Mad2 at the kinetochore as cargoes and transports them away from the kinetochores towards the spindle poles along microtubules (Gassmann et al., 2010; Howell et al., 2001; Ide et al., 2023; Buffin et al., 2005). Although Bub1, BubR1, and Bub3 have been previously confirmed as dynein cargoes, a recent study suggests that SAC proteins maybe evicted from kinetochores due to dephosphorylation of kinases such as Mps1 rather than through motor activity of dynein (Ide et al., 2023) (Figure 1C). Overall, SAC signaling is a complex process involving multiple kinases and phosphatases tightly regulated to ensure an even separation of sister chromatids during mitosis.
Gametogenesis is defined as the process by which mature haploid gametes develop from precursor cells known as primordial germ cells. This process involves a series of mitotic and meiotic cell divisions, followed by cell differentiation to form gametes (Larose et al., 2019). Gametogenesis occurs in the testes in males and is termed spermatogenesis during which spermatozoa are produced while females undergo oogenesis in the ovaries to produce oocytes (Larose et al., 2019). It has been observed that defective meiotic cell division during gametogenesis can lead to aneuploid embryos and embryonic lethality (Syrjanen et al., 2014). For instance, female mice lacking synaptonemal complex protein 3 (Sycp3) have aneuploid oocytes due to meiotic non-disjunction events (Syrjanen et al., 2014). The synaptonemal complex is active during prophase of meiosis-I during which homologous chromosomes are brought into proximity for synapsis and homologous recombination (Kouznetsova et al., 2005; Westergaard and von Wettstein, 1972). Deletion of Sycp3 in mice triggers a sexually dimorphic phenotype where males are infertile while the females are sub fertile and produce a high number of aneuploid secondary oocytes (Yuan et al., 2002). Despite a similar rate of fertilization, the Sycp3 null oocytes display a significantly higher level of embryonic lethality (Lightfoot et al., 2006). While these chromosomally abnormal embryos are able to implant into the uterine lining and undergo gastrulation, they fail to develop beyond E8.0. Cytological analysis show that about 57% of E3.5 blastocyst cells derived from Sycp3 null females are aneuploid with 91% of those displaying mosaic aneuploid karyotypes (Yuan et al., 2002). Histological analysis of the E7.0 aneuploid embryos have pycnotic bodies in the ectodermal layer. Pycnotic bodies show as darkly stained nuclei due to nuclear condensation during cell death (Magrassi and Graziadei, 1995). At E8.0, aneuploid embryos are severely disordered and have higher levels of pycnotic bodies (Yuan et al., 2002). TUNEL staining show that aneuploid embryos are eliminated through apoptosis. However, loss of p53 does not rescue embryonic lethality in Sycp3 null females suggesting that apoptosis occurs through a p53-independent mechanism (Lightfoot et al., 2006; Yuan et al., 2002). Furthermore, apoptosis is not due to unfaithful chromosomal segregation during mitosis as both wild-type and Sycp3 null embryos contain an equal number of cells at different stages of mitosis suggesting that mitosis is unaffected in the mosaic aneuploid cells (Yuan et al., 2002). Therefore, faithful chromosomal segregation is integral to meiosis.
The spindle assembly checkpoint is active during meiosis in mouse oocytes as demonstrated by microtubule disruption studies using nocodazole (Eichenlaub-Ritter and Boll, 1989). Treatment of metaphase I oocytes with nocodazole causes microtubule depolymerization, leading to spindle disassembly and loss of equatorial chromosome alignment. Upon recovery from nocodazole treatment, oocytes proceed successfully through meiosis I, indicating that SAC activation delays anaphase onset in response to spindle disruption (Eichenlaub-Ritter and Boll, 1989). Further support for SAC functionality comes from studies examining the SAC protein Bub1. In mouse oocytes, Bub1 localizes to kinetochores and undergoes phosphorylation during anaphase I and II (Brunet et al., 2003). This phosphorylation is crucial for SAC activity and is consistent with observations in Xenopus oocytes, where Bub1 is also phosphorylated and localized to kinetochores (Schwab et al., 2001). However, in contrast to Bub1 in Xenopus oocytes, Bub1 in mouse oocytes is not dependent on the MAPK-Rsk pathway (Schwab et al., 2001). The MAPK pathway has been previously implicated in SAC regulation, as its activity is required for nocodazole-induced metaphase arrest in Xenopus egg extracts (Minshull et al., 1994; Takenaka et al., 1997; Wang et al., 1997). In mammalian cells, active MAPK localizes to spindle poles and kinetochores during mitosis (Zecevic et al., 1998). In Xenopus oocytes, MAPK-Rsk directly phosphorylates Bub1, enabling its kinetochore localization and interaction with other SAC proteins, which is essential for checkpoint activation and metaphase arrest (Schwab et al., 2001). Furthermore, Rsk activation alone has been shown to directly phosphorylate Bub1 both in vitro and in vivo in Xenopus oocytes (Schwab et al., 2001). Another key SAC protein, Mad2, is recruited to kinetochores during early metaphase I in mammalian oocytes (Schwab et al., 2001). Functional studies with dominant-negative Mad2 mutants reveal that disrupting Mad2 activity prevents metaphase I arrest upon nocodazole treatment, underscoring its role in SAC-mediated checkpoint enforcement (Schwab et al., 2001). Taken together, these studies demonstrate that the SAC, including key proteins such as Bub1 and Mad2, is functional during gametogenesis.
Interestingly, the strength of SAC signaling appears to be stronger in male spermatocytes compared to female oocytes. In female oocytes, treatment with low concentrations of nocodazole (0.01–0.1 µM) induces spindle abnormalities and aneuploidy without significantly arresting meiosis I, suggesting weaker SAC activation (Everett and Searle, 1995). Intermediate concentrations of nocodazole (0.2–2 µM) induces a robust SAC response, resulting in meiosis I arrest in approximately 90% of oocytes (Everett and Searle, 1995). High concentrations of nocodazole (≥10 µM) completely depolymerizes microtubules, causing chromosome scattering and a total disruption of spindle integrity (Eichenlaub-Ritter and Boll, 1989). These results suggest that lower nocodazole concentrations may cause only partial destabilization or mild spindle abnormalities, without fully disassembling microtubules. Furthermore, when oocytes treated with high nocodazole treatments are allowed to recover, they reestablish the spindle apparatus and proceed through anaphase. However, this recovery process is highly error-prone and leads to a dramatic increase in aneuploidy (Eichenlaub-Ritter and Boll, 1989), suggesting a compromised or weaker SAC function in oocytes. Interestingly, there are insufficient studies carried out with respect to nocodazole effects in spermatocytes to make direct comparisons with the oocyte data described above, and future studies in this area could help to define differences between oocytes and spermatocytes with regard to SAC response.
Oocytes derived from an XO mouse model of monosomy, which lack second sex chromosome, proceed through meiosis I even in the presence of an unpaired X chromosome (LeMaire-Adkins et al., 1997). In contrast, XO spermatocytes elicit a strong meiotic arrest during metaphase I leading to apoptosis (de Boer et al., 1991; Sutcliffe et al., 1991). Similar sex differences in SAC function are observed in Mlh1 knockout models. Mlh1 is a mismatch repair gene essential for proper DNA replication prior to cell division in meiosis I, and mutant Mlh1 leads to univalent chromosomes during meiosis (LeMaire-Adkins et al., 1997). Male spermatocytes that are Mlh1-null halt meiosis. In contrast, Mlh-1 null female oocytes proceed through meiosis even in the presence of univalent chromosomes and initiate anaphase (Gorbsky, 2015). This suggests that mammalian oocytes require stable bipolar attachment of some, but not all, chromosomes which increases aneuploidy and errors in female oogenesis (Nagaoka et al., 2011).
However, conclusions from these observations regarding the differential strength of SAC signaling between male spermatocytes and female oocytes is challenged by another study which utilized spermatocytes from Robertsonian centric fusion heterozygous mice (Eaker et al., 2001). Robertsonian translocations involve Robertsonian (ROB) chromosomes that are acrocentric with centromeres located towards the chromosomal ends. Individuals who are heterozygous for Robertsonian chromosomes have a higher chance of producing aneuploid gametes during meiosis I due to unbalanced segregation (Yip, 2014). Spermatocytes from Robertsonian heterozygous chromosomes display an increased level of apoptosis in meiosis I suggesting that abnormal spermatocytes are eliminated (Eaker et al., 2001). Interestingly, staining for centromeric proteins CENP-E and CENP-F in Robertsonian heterozygous spermatocytes revealed an increase in fluorescence intensity at the kinetochores of lagging or malattached chromosomes (Eaker et al., 2001). CENP-E and CENP-F facilitate the attachment of spindle microtubules to the kinetochores and interact with BubR1 and Bub1 (Ciossani et al., 2018). However, this SAC signaling is not entirely infallible, as it fails to eliminate all aneuploid gametes efficiently, leading to the production of abnormal gametes (Ciossani et al., 2018). Nevertheless, all evidence points to the fact that the SAC is active during gamete formation to ensure proper meiosis although differences in SAC strength are present in oogenesis and spermatogenesis (Vogt et al., 2008). SAC proteins and their localization dynamics differ between spermatocytes and oocytes. In oocytes undergoing meiosis I, Mad2 localizes to unattached kinetochores and dissipates once proper kinetochore-microtubule attachments are formed, similar to its behavior in mitotic cells (Kallio et al., 2000). In contrast, in spermatocytes, immunofluorescence studies on rats and mice reveal that Mad2 remains localized to kinetochores throughout meiosis I (Kallio et al., 2000). Despite these differences in Mad2 dynamics, its depletion has significant consequences in both cell types. In oocytes, reduced Mad2 levels accelerate meiosis I and lead to chromosomal missegregation, whereas Mad2 insufficiency in spermatocytes results in aneuploidy (Niault et al., 2007; Lane and Jones, 2017). The benefits behind altered SAC protein dynamics in spermatocytes is yet to be made clear.
The SAC plays a pivotal role in ensuring accurate chromosome segregation during mitosis. However, its role in meiosis is comparatively less well understood. What is evident is that SAC proteins are both present and functionally relevant in meiosis. Mammalian oocytes, for example, express key SAC components, including MAD1, MAD2, BubR1, Bub1, Bub3, Mps1, and Aurora B (Zhang et al., 2005; Homer et al., 2005; Tsurumi et al., 2004; McGuinness et al., 2009; Li et al., 2009b; Hached et al., 2011; Lane et al., 2010; Yang et al., 2010). These SAC proteins execute similar roles in meiosis as they do in mitosis. For instance, Mad2 depletion in mouse oocytes accelerates the degradation of Securin and Cyclin B1, increases aneuploidy rates, and shortens the duration of meiosis I (Homer et al., 2005). This outcome parallels observations in mitotic cells, where Mad2 depletion in HeLa cells leads to a similar reduction in mitotic duration (Meraldi et al., 2004; Michel et al., 2001). Likewise, depletion of Bub1 in oocytes speeds up meiosis I and results in chromosome segregation errors, a phenotype mirrored in HeLa cells with Bub1 depletion, which experience significant sister chromatid segregation errors (McGuinness et al., 2009; Tang et al., 2004b). Despite these functional similarities, notable differences exist between SAC function in mitosis and meiosis. One key distinction is that the SAC response is weaker or less sensitive during meiosis. This is evident from multiple studies showing that manipulations causing chromosome misalignment in oocytes often fail to halt anaphase onset (Gui and Homer, 2012; Kolano et al., 2012; Lane and Jones, 2017; Sebestova et al., 2012; Kyogoku and Kitajima, 2017). Therefore, mammalian oocytes are prone to chromosomal segregation errors (Bennabi et al., 2016). In meiosis I, homologous chromosomes pair and recombine to form bivalents, which are held together by chiasmata. Non-aligned bivalents, which fail to properly position at the metaphase plate, risk missegregation (Lane et al., 2012). However, the presence of such non-aligned bivalents often fails to maintain SAC activation in oocytes, allowing the APC/C to remain active leading to nondisjunction (Lane et al., 2012).
The reduced SAC sensitivity in meiosis I is further exemplified by studies on NuMA (nuclear mitotic apparatus protein), which anchors microtubules to spindle poles in acentrosomal oocytes. NuMA deletion disrupts spindle assembly and chromosome alignment, yet the SAC remains silent, permitting oocytes to proceed through anaphase I despite severe defects, leading to aneuploid oocytes and infertility (Kolano et al., 2012). Similarly, in meiosis II, misaligned chromosomes fail to trigger SAC activation, allowing oocytes to progress through anaphase II resulting in further aneuploidy (Mihajlovic et al., 2023). Another significant difference is the timing and duration of SAC activity. Mitosis is a relatively fast process, lasting minutes to a few hours in human somatic cells (Rieder and Maiato, 2004; Herbert et al., 2015). Consequently, SAC activation and silencing occur rapidly, ensuring efficient progression through mitosis. In contrast, meiosis is a prolonged process that can span hours to days leading to delayed SAC activation and silencing (Musacchio and Salmon, 2007; Homer et al., 2009; Gui and Homer, 2012; McGuinness et al., 2009). This delayed SAC response in meiosis may allow oocytes more time to correct alignment errors and promote the production of high-quality gametes (Lane et al., 2012). In summary, while SAC proteins share conserved roles in mitosis and meiosis, key differences in SAC sensitivity, timing, and response exist.
Embryogenesis is a dynamic intricate process that encapsulates a multitude of temporal and spatial changes across the entire embryo for proper differentiation and maturation (Zhai et al., 2022). It occurs in all species and has been studied extensively in model organisms such as roundworms, fruit flies, frogs, fish, mice, and humans, although the process varies depending on the species (D'Costa and Shepherd, 2009; Technau, 1987). Cell division plays a critical role in embryogenesis as the zygote begins as a unicellular organism which undergoes multiple rounds of mitosis to promote multicellularity (Cinalli et al., 2008). Faithful segregation of chromosomes is equally essential during mitosis as it is during meiosis where aneuploid oocytes can lead to an increase in embryonic lethality as described above (Pan and Li, 2019; Jia et al., 2015). Interestingly, some organisms such as the fruit fly Drosophila melanogaster can tolerate aneuploidy. The loss of a copy of chromosome 4 is not lethal to Drosophila embryos, but results in smaller body size compared to wild-type flies (Bridges, 1921). Similarly, unicellular organisms like the fungus Batrachochytrium dendrobatidis exhibit extensive aneuploidy, suggesting that tolerance to aneuploidy varies across different organisms (Rosenblum et al., 2013). In contrast, mammals generally display lower tolerance towards aneuploidy, such as mosaic aneuploidy, where some cells in the body have an abnormal number of chromosomes (Garcia-Castillo et al., 2008). In mouse embryos, mosaic aneuploidy promotes embryo lethality by E8.0 through a p53-independent apoptotic mechanism (Lightfoot et al., 2006). Therefore, proteins that ensure proper chromosomal segregation during mitosis are essential for successful embryogenesis.
Once the male and female germ cells have fertilized, this produces a unicellular zygote which undergoes several rounds of cleavage to reach an 8-cell stage followed by an apical-basal polarization that allows for the segregation of distinct cell lineages (Zhu et al., 2021). Asymmetric cell divisions and the process of cavitation give rise to the blastocyst which then hatches from the zona pellucida and implants itself in the uterine lining (Zhu et al., 2021; Dey et al., 2004). In humans, this preimplantation development prior to hatching occurs within 5 days post fertilization (Dey et al., 2004). The SAC is active during preimplantation development (Wei et al., 2011). For instance, simultaneous overexpression of three SAC proteins (Bub3, BubR1, and Mad2) in one-cell embryos leads to inhibition of the metaphase to anaphase transition (Wei et al., 2011). Co-localization studies further revealed that all three SAC proteins are localized at the kinetochores. Conversely, RNAi-mediated depletion of Bub3, BubR1, or Mad2 disrupts the SAC and leads to anaphase onset with no apparent metaphase stage (Wei et al., 2011). The bypass of the SAC is further evidenced by the ability of RNAi-depleted embryos to progress to the two-cell stage despite nocodazole treatment, which normally arrests control embryos at the one-cell stage. These data indicate that SAC proteins are necessary for mitotic arrest in embryos (Wei et al., 2011). Karyotyping of embryos depleted of these SAC factors show a heightened level of aneuploidy and an accumulation of micronuclei. When these embryos are implanted in the uterine horn of mice, their viability are similar to control embryos (Wei et al., 2011). However, a substantially higher percentage of SAC depleted embryos display delayed development as compared to control embryos, demonstrating that the SAC is critical for mitotic progression and preimplantation development during embryogenesis (Wei et al., 2011).
SAC strength and response has been shown to vary between species which became apparent when assessing responses of 2-cell stage embryos from nine different species, including both chordates and non-chordates, to nocodazole treatment (Chenevert et al., 2020). The chordates (Phallusia mammillata) continue to carry out mitosis even in the presence of nocodazole as suggested by oscillations in phosphorylated-Histone H3 levels (Chenevert et al., 2020). Whereas nocodazole induces a significant delay in mitotic progression in all other species that were either echinoderms or mollusks which was shown to be mediated by the SAC kinase Mps1 (Chenevert et al., 2020). Further examination of chordate embryos using live microscopy showed no difference in the duration of mitosis between control and nocodazole treated embryos (Chenevert et al., 2020). Since the chordate group continue to progress through mitosis even in the presence of spindle disruption, it indicates that the SAC is not efficient or active during the early embryonic developmental phase in chordates. Similar results were also observed in other species, including the invertebrates Ciona intestinalis and Branchiostoma lanceolatum (Chenevert et al., 2020). It is plausible to hypothesize that SAC kinases are not expressed during the early embryonic development which leads to a lack of SAC activity or mitotic delay in response to nocodazole treatment. However, the SAC proteins Mad1, Mad2, Bub1, Bub3, and Mps1 are all detected at the mRNA level during and after fertilization in Phallusia mammillata (Chenevert et al., 2020). Interestingly, fluorescence imaging revealed that Mps1, Mad1, and Mad2 are not recruited to unattached kinetochores in P. mammillata 2-cell embryos treated with nocodazole, suggesting that SAC assembly at the kinetochore is prevented during early embryonic development in chordates (Chenevert et al., 2020). It is important to note that BubR1 was not assessed in this study, leaving its role in this process unclear.
There are various hypotheses that have been proposed to address the differential SAC activity between various species. The first hypothesis postulates that the SAC response is dependent on cell size (Galli and Morgan, 2016). Evidence for this comes from a study conducted in Caenorhabditis elegans where the duration of mitotic arrests following nocodazole treatment was longer after each embryonic cell division from the 2-cell stage to 8-cell stage which correlates with a decline in the cell volume (Galli and Morgan, 2016). Interestingly, there was also a strong correlation between time from nuclear envelope breakdown to nuclear envelope reformation and cell size (Galli and Morgan, 2016). As the cell volume reduced with each consecutive division, SAC mediated mitotic delay became longer. Depletion of ani-2, an anillin homolog, produces C. elegans embryos of various sizes (Galli and Morgan, 2016). When ani-2 was depleted alongside zyg-1, the Plk4 homolog that causes monopolar spindles, mitotic delays were longer in smaller size embryos further supporting the hypothesis that cell size may influence SAC activity. Along these same lines, since the ratio of unattached kinetochores to cytoplasmic volume increases as cell size decreases, it is possible that this kinetochore-to-cytoplasm ratio determines the strength of the SAC response (Galli and Morgan, 2016). In C. elegans heterozygous for rec-8(ok978), which produces triploid embryos with 50% more kinetochores than wild-type diploid embryos, the SAC signal was found to be stronger than in control diploid embryos of the same size. These results suggest that the kinetochore-to-cytoplasm ratio may influence the SAC response strength. However, this hypothesis is not supported by observations in other species (Vazquez-Diez et al., 2019). Live imaging studies of mouse embryos in the presence of nocodazole showed that the SAC was more efficient in the much larger 2-cell stage blastocyst than in smaller 4- or 8-cell stage morulae. Furthermore, when 40% of cytoplasm was removed to alter cell volume, nocodazole prolonged mitotic arrest in the 2-cell stage but did not extend the arrest in 4-cell stage (Vazquez-Diez et al., 2019). Similarly, this lack of correlation is evident in studies where SAC response and cell size were compared between species that produce embryos of various sizes. For instance, the blastomere of P. mammilata is 130 µm while a blastomere of Drosophila is 500 µm. Upon nocodazole treatment, the latter delays mitotic progression suggesting an efficient SAC response while the former fails to do so even with its smaller size (Chenevert et al., 2020). To rationalize these contradicting observations, it was proposed that the spindle architectural features should be considered along with cell size to explain mitotic timing and SAC efficiency (Bloomfield et al., 2020). Cytokinesis failure was induced in human diploid colon cancer DLD-1 cells to produce tetraploid cells of both small and large sizes. Smaller size 4N clones had longer mitotic arrest although the arrest time varied between the smaller size clones indicating that cell size itself may not be the primary factor (Bloomfield et al., 2020). Furthermore, the spindle pole size, spindle height, and spindle microtubule density were considered alongside cell size with results showing that all these factors influence SAC timing to various extents and thus may together control SAC efficiency (Bloomfield et al., 2020).
Another hypothesis that has been recently postulated asserts that the difference between SAC acquisition and SAC modulation is what influences SAC efficiency (Roca et al., 2023). Embryos from P. mammillata, which show weak SAC activity in the presence of nocodazole, exhibit similar mitotic duration up until the 7th cell cycle between control and nocodazole treated embryos (Roca et al., 2023). However, nocodazole treatment induced a prolonged mitotic delay from the 8th cycle onward suggesting that the SAC initiates at the 8th cycle (Roca et al., 2023). A dominant negative Mad2 hampered this SAC initiation at the 8th cycle while it had no effect prior to the 8th cycle. While Mad1 expression is similar throughout development, immunofluorescence staining revealed that it does not accumulate on the mitotic chromosomes during the 4- to 32-cell embryos (Roca et al., 2023). However, Mad1 starts accumulating on the mitotic chromosomes during the 128- and 256-cell stage (Roca et al., 2023). This suggests that proper localization of SAC factors may define SAC proficiency in these embryos indicating that there may be an initial SAC-deficient phase followed by a later SAC-proficient phase during early embryonic development (Roca et al., 2023). The evolutionary benefit, if any, of the lack of SAC activity during early cell divisions remains to be determined.
Since embryogenesis leads to the development of various tissues, it is plausible that SAC efficiency could be influenced by different cell fates (Gerhold et al., 2018). This is supported by the observation that the anterior and posterior ventral ectodermal cells of P. mammillata embryos vary in their SAC strength (Roca et al., 2023). Moreover, cells within these two different regions are of the same size suggesting that the differences in SAC strength could be due to the different cell fates rather than cell size (Roca et al., 2023). The anterior cells can be induced to acquire posterior cell like identity through the inhibition of glycogen synthase kinase-3 (GSK3) (Feinberg et al., 2019). Interestingly, posterization of the anterior cells by GSK3 inhibition leads to a significant reduction in SAC efficiency in the presence of nocodazole (Roca et al., 2023). Similarly, posterior cells can be induced to undergo anterior fate by injecting the transcription factor Ci-FoxA-a (Lamy et al., 2006). Upon nocodazole treatment, the mitotic delay in Ci-FoxA-a injected posterior cells is 2.4-fold longer, further supporting the notion that cell fate may play a crucial part in regulating SAC strength (Roca et al., 2023). The relationship between cell fate and SAC efficiency has also been observed in C. elegans where germline fated cells display longer mitotic delays compared to somatic fated cells when spindle formation is perturbed in blastomeres with nocodazole (Gerhold et al., 2018). When germline and somatic cells of comparable sizes are treated with nocodazole, the germline cells still display a significantly longer mitotic delay suggesting that germline fated cells have a stronger SAC response (Gerhold et al., 2018). While the contribution of cell size cannot be completely eliminated in this model organism since the SAC strength increases as the blastomere undergoes cleavage and the cell size decreases, cell fate may have a greater role in governing SAC strength (Gerhold et al., 2018).
The SAC response might also be influenced by whether cells are undergoing constant division, as seen in rapidly dividing cells during early embryogenesis, or whether they are in a differentiated state with limited or no division. In early embryogenesis, rapidly dividing cells exhibit a more error-prone and less robust SAC response (Horakova et al., 2024). Potential reasons for this could be the rapid pace of division and incomplete checkpoint maturation. Early embryonic cells, particularly those undergoing cleavage divisions, lack cell cycle phases associated with interphase such as G1 and G2 that provide checkpoints to monitor and rectify DNA damage (Kumar et al., 2015). These cells undergo rapid cycling, often without sufficient time for quality control, leading to a higher incidence of aneuploidy (Horakova et al., 2024). For instance, aneuploidy rates are extremely high in oocytes and embryos compared to somatic cells (Vazquez-Diez and FitzHarris, 2018; Horakova et al., 2024). The cell cycle phases also vary significantly in Xenopus, Drosophila, and C. elegans embryos (Masui and Wang, 1998; Foe and Alberts, 1983; Edgar and McGhee, 1988). The durations of these phases lengthen as development progresses due to the acquisition of the G1 and G2 phases. Since the SAC operates during the mitotic phase, determining the duration of mitosis during development is of particular interest. A study on Helobdella triserialis embryos suggests that the duration of the mitotic phase remains constant during development, indicating that the SAC has sufficient time to function despite changes in other cell cycle phases (Bissen and Weisblat, 1989). In contrast, differentiated cells such as neurons do not undergo cell division, which might suggest that the SAC is inactive in these cells (Aranda-Anzaldo and Dent, 2017). However, SAC proteins such as Bub1 and Mad2 are present in neurons and influence microtubule dynamics suggesting that SAC proteins may functions beyond mitosis (Hertzler et al., 2020; Zhao et al., 2019; Cheerambathur et al., 2019). More research is needed to elucidate the roles of SAC proteins in embryonic versus differentiated cells and their potential non-mitotic functions in differentiated tissues.
Another intriguing perspective on SAC response is through the lens of evolution. Across eukaryotes, many species have retained SAC proteins and their conserved functions, such as Saccharomyces cerevisiae and Homo sapiens (Rudner and Murray, 1996). However, there are notable exceptions, including Trypanosoma brucei, which lacks a BubR1/Mad3 homolog, and Giardia intestinalis, which has lost the APC/C at some point in its evolutionary history (Akiyoshi and Gull, 2013; Gourguechon et al., 2013; van Hooff et al., 2017). In T. brucei, spindle assembly perturbation does not trigger a SAC response, as the cells proceed to cytokinesis despite the disruption, suggesting that the absence of key SAC components compromises SAC proficiency (Ploubidou et al., 1999; Akiyoshi and Gull, 2013). Interestingly, while such observations suggest a correlation between the presence of SAC proteins and SAC strength, there are exceptions. For example, the flatworm Schmidtea mediterranea, which lacks Mad2, BubR1/Mad3, and Bub1, still exhibits a delay in G2/M progression in response to nocodazole, implying the existence of alternative mechanisms to ensure chromosomal fidelity (Grohme et al., 2018). In addition, species such as G. intestinalis rely on Cyclin B degradation for cell cycle progression; however, unlike most organisms, the degradation of Cyclin B in G. intestinalis is not mediated by the APC/C. Instead, degradation is regulated through phosphorylation by unidentified upstream mechanisms, as G. intestinalis lacks the APC/C machinery (Gourguechon et al., 2013). Interestingly, ubiquitin ligases beyond APC/C have been identified as regulators of Cyclin B1 degradation, including CRL2ZYG-11 (Balachandran et al., 2016). This ligase, also found in C. elegans and human cells, operates redundantly with APC/C in targeting Cyclin B1 (Balachandran et al., 2016). Under normal conditions, when the APC/C is active, CRL2ZYG-11 inactivation has minimal effects. However, in the absence of APC/C activity, CRL2ZYG-11 compensates by degrading Cyclin B1, facilitating normal progression through metaphase (Balachandran et al., 2016). Cyclin B1 is essential for SAC function, as it serves as a scaffold for Mad1, anchoring it to the kinetochore corona. This localization enables Mps1 to phosphorylate Mad1 on its C-terminus (Allan et al., 2020). Once proper kinetochore-microtubule attachments are formed, Cyclin B1 is degraded by the APC/C to allow anaphase progression (Allan et al., 2020). However, in scenarios of sustained mitotic arrest, such as during nocodazole treatment, cells may bypass the SAC and exit mitosis via a process called mitotic slippage (Brito and Rieder, 2006). The mechanisms behind mitotic slippage have been debated, with evidence pointing to a gradual weakening of SAC signaling due to the loss of Mad2 at kinetochores (Lok et al., 2020). The discovery of CRL2ZYG-11 as a redundant Cyclin B1 regulator provides further insight into this phenomenon. Notably, depletion of CRL2ZYG-11 significantly reduces mitotic slippage in human cells, even under nocodazole treatment (Balachandran et al., 2016). Future studies exploring whether other species possess mechanisms beyond APC/C to degrade Cyclin B could shed light on the evolutionary diversity of SAC regulation and its implications for SAC proficiency.
The evolutionary origins of SAC components are particularly intriguing, as their presence across diverse eukaryotic species suggests that they likely originated in a common ancestor or the last eukaryotic common ancestor (LECA) (Kops et al., 2020). A recent review provides a comprehensive overview of the evolutionary branches emanating from the LECA and highlights how SAC components have diverged across eukaryotic lineages (Kops et al., 2020). Understanding the diversity of SAC components and their structural domains across species could shed light on the broad spectrum of SAC proficiency observed today. Such studies could offer valuable insights into the evolutionary adaptations that shape mitotic regulation across species.
Overall, the efficiency of the SAC during development is dictated by various factors whose level of contribution varies among species. It has also been speculated that SAC silencing is a by-product of changes that occur in reproductive regulation (Chenevert et al., 2020). It remains to be determined what exactly causes SAC function in the initial stages of embryonic development and what are the evolutionary benefits of the varied SAC activity in different species during early embryogenesis (Table 1).
The proteins involved in the SAC are broadly categorized into two groups: sensor and signal transducer proteins. Sensor proteins consist of Bub1, Mad1, and Mps1 while signal transducer proteins consist of the MCC proteins BubR1, Bub3, Mad2, and Cdc20 (Peters, 2006). The remainder of this review will discuss each of these SAC proteins and their role in gametogenesis and embryogenesis.
Bub1 (budding uninhibited by benzimidazoles 1) is a serine/threonine kinase that is recruited to unattached kinetochores where it plays a crucial role in the assembly of the signal transducers (Kim and Gartner, 2021). Bub1 is recruited to the kinetochore by Bub3, along with BubR1. Both Bub1 and BubR1 also bind to the KI1 and KI2 motifs of Knl1, respectively (Kiyomitsu et al., 2007; Kiyomitsu et al., 2011; Krenn et al., 2012). Bub1 is one of the first proteins that arrives at unattached kinetochores where it activates the SAC by recruiting the Mad1-Mad2 complex (Kim and Gartner, 2021; Tang et al., 2004a). Therefore, Bub1 is vital for proper SAC function. The importance of Bub1 in embryogenesis and tissue homeostasis has been highlighted through the use of Bub1 knockout mouse models (Perera et al., 2007). Homozygous Bub1 null mice are embryonic lethal suggesting that Bub1 is essential for embryogenesis (Perera et al., 2007). Bub1 null embryos die between E3.5 and E8.5 (Perera et al., 2007). Mouse embryonic fibroblasts with inactivated Bub1 display reduced proliferation, improper chromosome alignment, and aberrant mitosis, suggesting that Bub1 is essential for maintaining proper mitosis in developing embryos (Perera et al., 2007). E7.5 Bub1 deficient embryos also have reduced phosphorylated-histone H3 positive cells and mature spermatids in their seminiferous tubules (Perera et al., 2007). Deletion of Bub1 in post implantation embryos at E10.5 also leads to an arrested development within 48 h which indicates that Bub1 is essential to organogenesis (Perera et al., 2007; Tilston et al., 2009). Histological analysis of these embryos revealed an extensive hemorrhaging within the brain and the body cavity (Tilston et al., 2009). At a cellular level, Bub1 deleted embryos have a heightened level of apoptosis and a reduction in the number of miotic cells as demonstrated by reduced phosphorylated-histone H3 positive cells (Tilston et al., 2009). Overall, Bub1 is indispensable for normal embryogenesis.
In humans, studies of spontaneous miscarriages carried out in patients from Chongqing Medical University Affiliated Hospitals showed that almost half of embryos had reduced levels of Bub1 and aneuploidy (Shi et al., 2011). RNAi-mediated depletion of Bub1 in primary embryonic villus cells suppresses cell proliferation and increases aneuploidy (Shi et al., 2011). Biallelic germline mutations in Bub1 that leads to a reduction in the total protein level and kinase activity have been identified in two patients (Carvalhal et al., 2022). These patients suffer from microcephaly, growth retardation, and cardiovascular defects, and impaired mitotic fidelity (Carvalhal et al., 2022). Overall, these studies suggest that Bub1 is essential for both embryogenesis and organogenesis.
Mad1 is an adaptor protein that plays crucial role in the assembly of the MCC by recruiting Mad2 (Devigne and Bhalla, 2021). Mad2 is in an inactive (or open conformation) as it is recruited to the kinetochore (Musacchio, 2015a; Luo et al., 2018). Once it binds to Cdc20, Mad2 changes its conformation to its closed, active, conformation. This complex then functions with other SAC proteins to inhibit the APC/C (Luo et al., 2018). Mad1 knockout embryos do not survive embryogenesis while overexpression of Mad1 correlates with an increased frequency of early-lethality related to aneuploidy (Iwanaga et al., 2007; Zhao et al., 2021). This suggests that Mad1 expression needs to be finely tuned for effective SAC function during embryogenesis. Mad1 heterozygous knockout animals show a higher incidence of tumors such as melanoma, rhabdomyosarcoma, and osteosarcoma along with aneuploidy (Iwanaga et al., 2007). Factors that regulate Mad1 levels include miR-125b that binds to the 3′UTR region of Mad1 and negatively regulates Mad1 expression levels (Zhao et al., 2021). Aneuploid embryos that undergo miscarriages have higher expression of Mad1 and a downregulation of miR-125b, suggesting that upstream regulators of Mad1 are essential for effective SAC response and prevention of aneuploidy (Zhao et al., 2021). Interestingly, the same study noted a decrease in expression of Bub3 protein which is another essential player of the SAC (Zhao et al., 2021).
Mad2 knockout mice were utilized to study its role during embryogenesis (Dobles et al., 2000). When Mad2 heterozygous mice were mated, no Mad2 homozygous null pups were born suggesting that Mad2 is embryonic lethal and essential for embryogenesis (Dobles et al., 2000). Histological analysis of E7.5 Mad2 null embryos revealed that they were smaller in size and had abnormal gross morphology compared to wild-type embryos (Dobles et al., 2000). TUNEL staining indicated that these embryos show enhanced apoptosis between E6.5-E7.5. They also displayed aberrant mitosis as demonstrated by reduced mitotic cells and a higher number of cells with abnormal anaphases (Dobles et al., 2000). Mad2 null E5.5 embryos cultured in vitro also failed to arrest in mitosis in response to nocodazole treatment (Dobles et al., 2000). Overall, Mad2 is essential for embryonic viability and is required for proper chromosomal segregation in mice (Dobles et al., 2000). In humans, similar to Bub1, reduced levels of Mad2 are observed in cases of spontaneous miscarriages (Shi et al., 2011). Depleting Mad2 in embryonic villus cells leads to a higher percentage of cells with abnormal chromosomes and reduces cell proliferation (Shi et al., 2011).
Mad2 overexpression has also been studied extensively in mice as Mad2 is overexpressed in many different tumor types such as Burkitt lymphomas and Plasmacytomas (Sotillo et al., 2007). Mouse embryonic fibroblasts (MEFs) that overexpress Mad2 show reduced proliferation and display tetraploidy with a higher incidence of apoptosis compared to control cells (Sotillo et al., 2007). Therefore, chromosomal instability is observed both when Mad2 is lost or overexpressed suggesting that its level must be precisely regulated for accurate cell division.
Since loss of Mad2 leads to embryonic lethality, conditional Mad2 knockout mice were used to assess Mad2 deletion in adult mice (Schukken et al., 2021). Systemic knockout of Mad2 in 9–10-week-old mice leads to atrophy in the intestines specifically in the jejunum and ileum, coinciding with an increase in apoptosis and mitotic abnormalities such as lagging chromosomes and anaphase bridges (Schukken et al., 2021). Interestingly, other organs including the lung, liver, and kidneys do not display any phenotype in response to systemic loss of Mad2 (Schukken et al., 2021). This suggests that various cell and tissue types respond differently to loss of SAC proteins, potentially due to differences in the frequency or rate of cell division in different organs. Tissues with higher rates of cell turnover, such as the intestine, may be more susceptible to mitotic defects caused by the loss of SAC proteins such as Mad2, while other organs with relatively slower or less frequent cell division may tolerate SAC protein loss. Therefore, Mad2 expression is crucial post embryogenesis especially in tissues that divide rapidly.
The interaction of Mad1 and Mad2 is also essential in gametogenesis specifically during synapsis which is the crossover between homologous chromosomes (Devigne and Bhalla, 2021). For instance, mutations that prevent Mad1:Mad2 interaction delays synapsis formation and abolishes the synapsis checkpoint in C. elegans (Devigne and Bhalla, 2021). Similarly, vertebrate oocytes require Mad1 and Mad2 for metaphase arrest during meiosis II (Tunquist et al., 2003). Overall, Mad1 and Mad2 are essential proteins whose precise regulation is critical for maintaining chromosomal stability during gametogenesis and embryogenesis. Their roles in the spindle assembly checkpoint are vital, with dysregulation leading to deleterious consequences, including aneuploidy and embryonic lethality.
The kinase Mps1 is responsible for the initiation of the SAC and promoting MCC assembly (Lara-Gonzalez et al., 2021a; Fischer et al., 2021; Ji et al., 2017). Through its phosphorylation of Knl1, it provides a docking site for the SAC proteins at unattached kinetochores to successfully inhibit the activation of the APC/C (Ji et al., 2017). Mps1 activity is essential in both oogenesis and spermatogenesis. Mps1 deletion in Drosophila causes chromosomal segregation errors (Gilliland et al., 2007). Similarly, oocyte specific deletion of Mps1 impairs the segregation of chromosome in oocytes during meiosis I and negatively impacts the fertility of mice. In male mice, germ cell specific deletion of Mps1 depletes spermatocytes and causes infertility suggesting that Mps1 is essential for spermatogenesis (Fang et al., 2021). Mps1 is also crucial in embryogenesis. For instance, inhibition of Mps1 during the preimplantation stage of mouse embryos significantly disturbs the development of embryos from the 2-cell stage to the blastocyst stage (Ju et al., 2021). These embryos display misaligned chromosomes and an absence of kinetochore-microtubule attachment along with DNA damage and an increase in oxidative stress followed by heightened apoptosis (Ju et al., 2021). Overall, Mps1 is crucial for gametogenesis and embryogenesis which underscores the importance of Mps1 in ensuring accurate cell division and chromosomal stability during these processes.
Budding uninhibited by benzimidazole-related 1 (BubR1), encoded by the Bub1b gene, is a component of the MCC along with Bub3, Mad2, and Cdc20 (Bolanos-Garcia and Blundell, 2011). The MCC blocks the activation of the APC/C complex thereby preventing ubiquitination and degradation of key anaphase onset substrates such as Securin and Cyclin B1 (Sudakin et al., 2001; Reddy et al., 2007). Homozygous BubR1 knockout embryos fail to survive beyond E6.5 in utero (Wang et al., 2004). Blastocysts from wild-type and heterozygous knockout mice are able to hatch from the zona pellucida, attach to culture plates, and proliferate by E6.5 (Wang et al., 2004). BubR1 knockout embryos also display slower proliferation and atrophy by E6.5 with heightened levels of cells undergoing apoptosis as revealed by TUNEL staining (Wang et al., 2004) Adult BubR1+/− mice also show splenomegaly and extramedullary megakaryopoiesis (Wang et al., 2004). Aneuploidy was not examined in the knockout embryos although it can be speculated that the higher level of cell death may be due to chromosomal instability in the BubR1 knockout embryos (Wang et al., 2004). BubR1 also plays a crucial role in regulating kinetochore-microtubule attachments. Cells depleted of BubR1 fail to form stable microtubule attachments to kinetochores (Lampson and Kapoor, 2005). Similarly, depletion of Bub1 also disrupts these attachments, suggesting that the deletion of either Bub1 or BubR1 contributes to aneuploidy not only by impairing SAC function but also by causing unstable kinetochore-microtubule interactions (Lampson and Kapoor, 2005).
Mammalian oocytes undergo two phases of meiosis to produce secondary oocytes that can fertilize with a mature spermatozoon (MacLennan et al., 2015). Oocytes depleted of BubR1 cannot carry out prophase I arrest during meiosis I and undergo germinal vesicle breakdown (MacLennan et al., 2015). BubR1 depletion in oocytes leads to a significant reduction in CDH1, a co-activator of APC/C, which is required for APC/C led arrest during prophase I (Homer et al., 2009). Interestingly, these BubR1 depleted oocytes become arrested before the completion of meiosis I and fail to produce polar bodies (Homer et al., 2009). In contrast, another study demonstrated that BubR1 depleted oocytes do not undergo cell cycle arrest even in the presence of nocodazole and successfully produce polar bodies, which might be attributed to various degrees of BubR1 depletion achieved in these contradictory studies (Wei et al., 2010). Using mouse oocytes completely devoid of BubR1 revealed that BubR1 is not required for prophase I arrest during meiosis (Touati et al., 2015). As a result, BubR1 knockout oocytes undergo germinal vesicle breakdown and readily carry out meiosis I in an accelerated manner suggesting that SAC function is impaired in the absence of BubR1 (Touati et al., 2015). Kinetochore-microtubule interactions that are resistant to depolymerization when exposed to cold temperatures are absent in BubR1 deleted oocytes indicating that BubR1 loss reduces stability of spindle fibers in oocytes (Touati et al., 2015). Furthermore, BubR1 depleted oocytes that complete meiosis I and II are found to be aneuploid (Touati et al., 2015). Overall, these studies indicate that BubR1 is essential for regulating the timing of oogenesis and the production of healthy oocytes.
To study the role of BubR1 in adult mice, BubR1 hypomorphic mice expressing approximately 10% of BubR1 protein found in wild type mice were engineered (Baker et al., 2004). BubR1 hypomorphic mice display severe phenotypes including aneuploidy and premature aging phenotypes such as growth retardation, cataracts, sarcopenia, and lordokyphosis (Baker et al., 2004). BubR1 hypomorphic mice also exhibit cardiac anomalies, including prolonged QT syndrome, decreased tolerance to cardiac stress following injection with the β-adrenergic agonist isoproterenol, and a tendency to die in a manner reminiscent of sudden cardiac death (Baker et al., 2004; Baker et al., 2013; North et al., 2014). Mutation of BubR1 in humans leads to Mosaic Variegated Aneuploidy (MVA) syndrome, which is characterized by increased incidences of tumorigenesis, and progeroid traits such as shortened lifespan, cataracts, congenital heart defects, and facial dysmorphism (Suijkerbuijk et al., 2010). Overall, these studies indicate that BubR1 is essential for oogenesis, embryogenesis, and maintenance of tissue/organs in adulthood.
Budding uninhibited by benomyl (Bub3) is a component of the MCC complex along with BubR1, Cdc20, and Mad2. Bub3 is a crucial component of the SAC since it binds to phosphorylated MELT repeats on the outer kinetochore subunit Knl1 that leads to the recruitment of Bub1 and BubR1 to kinetochores (Kiyomitsu et al., 2007; Krenn et al., 2012; Yamagishi et al., 2012; Primorac et al., 2013). Bub3 binds to Bub1 and BubR1 through their GLEBS motifs (Taylor et al., 1998). Mice that harbor Bub3 null alleles were generated, and similar to other MCC components, they are embryonic lethal and die between E6.5-E7.5 (Kalitsis et al., 2000). Blastocysts obtained at day E3.5 were cultured in vitro and examined for morphological abnormalities (Kalitsis et al., 2000). While control embryos rapidly divided and increased their inner cell mass, Bub3 null embryos remained smaller in size and degenerated, suggesting they are alive at E3.5 but eventually die (Kalitsis et al., 2000). These embryos were also found to be resorbed prior to E8.5 in utero (Kalitsis et al., 2000). Furthermore, mitotic abnormalities are present in the Bub3 null embryos starting at E3.5 and as the embryos progress, they accumulate more mitotic abnormalities including the formation of micronuclei, nuclear bridging, and abnormal nuclei (Kalitsis et al., 2000). These data suggest that Bub3 is required for proper early embryogenesis.
Bub3 shares extensive sequence homology with Rae1 (Babu et al., 2003). Rae1, although not a well-characterized SAC component, has been implicated in processes that influence mitotic progression and checkpoint functions (Martinez-Exposito et al., 1999; Taylor et al., 1998; Babu et al., 2003). Rae1 was originally found to be involved in mRNA export during interphase where Rae1 binds to the GLEBS motif of the nucleoporin Nup98 (Brown et al., 1995; Wang et al., 2001). Similar to Bub3, Rae1 also binds to the GLEBS domain in both Bub1 and BubR1 to regulate the SAC complex (Wang et al., 2001). Rae1 null embryos die by E8.5 (Babu et al., 2003). At E3.5, Rae1 null embryos are morphologically indistinguishable from wild type and heterozygous littermates (Babu et al., 2003). Blastocysts harvested at E3.5 continue to hatch from the zona pellucida, but Rae1 null embryos fail to divide their inner cell mass and degenerate by E8.5 (Babu et al., 2003). Interestingly, Rae1 null embryos are able to carry out nuclear mRNA export suggesting that failure to export nuclear mRNA due to loss of Rae1 is not responsible for the observed embryonic lethality (Babu et al., 2003). When MEFs isolated from heterozygous Rae1 null embryos were challenged with nocodazole to examine the effect of Rae1 loss on the response to spindle damage, Rae1 haploinsufficient cells were unable to arrest in mitosis and showed significantly higher levels of chromosomal missegregation (Babu et al., 2003). Similarly, Bub3 haploinsufficiency also leads to failure in mitotic checkpoint arrest and errors in chromosome segregation when treated with nocodazole (Babu et al., 2003). When Rae1 and Bub3 haploinsufficiency are combined, an increase in aneuploidy both in vitro and in vivo was observed (Babu et al., 2003). Interestingly, Rae1 overexpression reduces the mitotic checkpoint defects observed in Bub3 haploinsufficient MEFs suggesting that Rae1 can compensate for the reduction of Bub3 with respect to SAC function (Babu et al., 2003). Furthermore, combined haploinsufficiency of Rae1 and Bub3 leads to increased tumorigenesis in mice due to an increase in chromosomal instability (Babu et al., 2003). Taken together, these findings suggest that both Bub3 and Rae1 are essential for SAC function during embryogenesis.
Cdc20 is another member of the MCC complex and also serves as a substrate recognition subunit of the E3 ubiquitin ligase APC/C promoting ubiquitination of its substrates (Qiao et al., 2016). The phosphorylation of the APC/C by CDK1 promotes the binding of Cdc20 to the APC/C (Golan et al., 2002; Kramer et al., 2000). The formation of APC/C-Cdc20 complex allows for the recognition of APC/C substrates important for the metaphase-to-anaphase transition. The WD-40 domain in Cdc20 recognizes degron motifs in substrate proteins allowing for their recruitment to the APC/C and subsequent ubiquitination (Kraft et al., 2005; Glotzer et al., 1991). Cdc20 is essential for successful embryogenesis, as its deletion in mouse embryos leads to embryonic lethality. Analysis of Cdc20 deleted embryos showed that these embryos arrest at E3.5 at the two-cell stage (Li et al., 2007). When E1.5 embryos were harvested and cultured in vitro, the Cdc20 null embryos failed to progress from the 2-cell stage to the 4-cell stage (Li et al., 2007). Cdc20 bound to the APC/C mediates the ubiquitination of Securin and Cyclin B1 followed by the activation of Separase and the onset of anaphase (Li et al., 2007). Cdc20 null embryos at E1.5 demonstrate a high level of Cyclin B1 and Securin suggesting that Cdc20 null embryos fail to transition to anaphase causing metaphase arrest (Li et al., 2007). Meanwhile, Cdc20 and Securin double mutants were unable to maintain the metaphase arrest suggesting that Securin is important for the metaphase arrest caused by Cdc20 deletion (Li et al., 2007). Homozygous mutant embryos that have Cdc20 lacking the Mad2 binding domain are also not viable and die before E14.5 (Li et al., 2009a). Morphologically, the mutant embryos are smaller and suffer from massive apoptosis (Li et al., 2009a). Upon nocodazole treatment, mutant cells fail to undergo mitotic arrest (Li et al., 2009a). Moreover, reduction in Cdc20 activity, using heterozygous Cdc20 mice, leads to chromosomal instability and an increase in tumorigenesis (Li et al., 2009a).
In humans, mutations in Cdc20 were identified in a cohort of infertile individuals who displayed oocyte maturation arrest, fertilization failure, and early embryonic arrest (Zhao et al., 2020). These mutations included a missense mutation c.965G > A (p. Arg322Gln) and nonsense or frameshift mutations (c.544C > T [p.Arg182*], c.813_814ins AGTG [p.Gly272Serfs*24], and c.1176_1179del TCTG [p.Cys392*]) (Zhao et al., 2020). The missense mutation (p.Arg322Gln) showed normal kinetochore localization of Cdc20 in oocytes, similar to the wild type protein (Zhao et al., 2020). However, the frameshift and nonsense mutations (p.Arg182*, p.Gly272Serfs*24, and p.Cys392*) caused defective kinetochore localization, leading to a loss-of-function phenotype (Zhao et al., 2020). In transfected chinese hamster ovary (CHO) cells, these mutations resulted in reduced Cdc20 protein levels or truncated Cdc20 proteins. Additionally, these mutations caused a decrease in Cyclin B1 degradation, which is a critical function of Cdc20 during the metaphase-to-anaphase transition (Zhao et al., 2020). The mutations also resulted in reduced mRNA expression of Cdc20 further indicating that these mutations lead to unstable Cdc20 protein and degraded mRNA. Injection of Cdc20 cRNA rescued these phenotypes in the mutated oocytes indicating that Cdc20 is essential for proper oocyte development and fertility in humans (Zhao et al., 2020). Furthermore, treatment with the Cdc20 inhibitor Apcin interfered with embryo implantation in mice (Guo et al., 2020). Cdc20 expression reaches its peak during the estrus phase of menstrual cycle in mouse endometrial tissue and during the mid-secretory phase in human endometrial tissue (Guo et al., 2020). Apcin treatment reduced the proliferation and adhesion rate of human uterine epithelial cells lines HEC-1A and RL95-2, and significantly reduced the number of implanted embryos as compared to control groups (Guo et al., 2020). Taken together, Cdc20 appears to be essential for embryogenesis, embryo implantation, and fertility in mammals.
In summary, SAC proteins play a vital role in gametogenesis and embryogenesis. Knocking out most SAC proteins leads to embryonic lethality while inducible systemic knockout in adult animals negatively impacts the functions of various organs (Table 2). SAC function and efficiency seem to vary between species and are affected by different factors including cell fate, cell size, and kinetochore-to-cytoplasm ratio. The molecular basis and functional consequences of this species variation in SAC function should be studied in more depth to decipher what factors regulate SAC efficiency during embryogenesis. Moreover, cellular pathways that regulate the activity level of SAC is a substantial gap in knowledge that is yet to be explored in detail which is imperative since SAC response appears to undergo a period of silencing during the initial period of embryogenesis in some organisms. Such analysis may also shed light on the potential evolutionary benefit of regulating SAC activity in some species that holds potential for clinical therapy such as in aneuploidy and cancer that arise as a result of chromosomal instability.
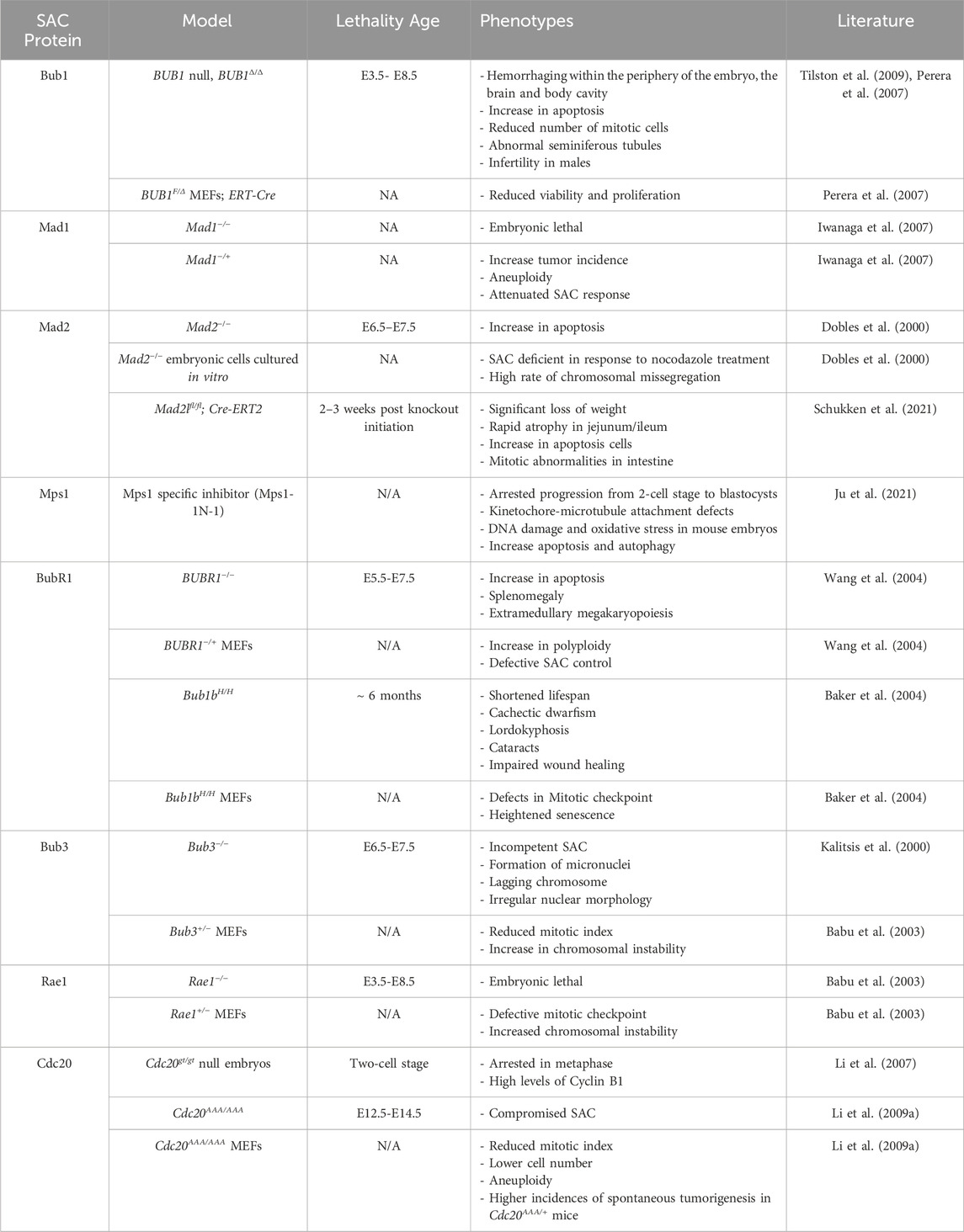
Table 2. Lethality age and phenotypes displayed by various in vivo and in vitro models of SAC proteins.
RP: Writing–original draft, Writing–review and editing. BN: Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Health Science Strategic Investment Fund Faculty Development Grant from Creighton University and the National Institute on Aging/National Institutes of Health (R01AG077574) (BN).
We are grateful for the North lab members and the manuscript reviewers for their feedback and helpful suggestions that improved the quality of this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abrieu, A., Magnaghi-Jaulin, L., Kahana, J. A., Peter, M., Castro, A., Vigneron, S., et al. (2001). Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106, 83–93. doi:10.1016/s0092-8674(01)00410-x
Agarwal, S., and Varma, D. (2015). How the SAC gets the axe: integrating kinetochore microtubule attachments with spindle assembly checkpoint signaling. Bioarchitecture 5, 1–12. doi:10.1080/19490992.2015.1090669
Akiyoshi, B., and Gull, K. (2013). Evolutionary cell biology of chromosome segregation: insights from trypanosomes. Open Biol. 3, 130023. doi:10.1098/rsob.130023
Akiyoshi, B., Sarangapani, K. K., Powers, A. F., Nelson, C. R., Reichow, S. L., Arellano-Santoyo, H., et al. (2010). Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 468, 576–579. doi:10.1038/nature09594
Alfieri, C., Chang, L., Zhang, Z., Yang, J., Maslen, S., Skehel, M., et al. (2016). Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature 536, 431–436. doi:10.1038/nature19083
Allan, L. A., Camacho Reis, M., Ciossani, G., Huis In 'T Veld, P. J., Wohlgemuth, S., Kops, G. J., et al. (2020). Cyclin B1 scaffolds MAD1 at the kinetochore corona to activate the mitotic checkpoint. EMBO J. 39, e103180. doi:10.15252/embj.2019103180
Anderson, D. E., Losada, A., Erickson, H. P., and Hirano, T. (2002). Condensin and cohesin display different arm conformations with characteristic hinge angles. J. Cell Biol. 156, 419–424. doi:10.1083/jcb.200111002
Aranda-Anzaldo, A., and Dent, M. A. (2017). Why cortical neurons cannot divide, and why do they usually die in the attempt? J. Neurosci. Res. 95, 921–929. doi:10.1002/jnr.23765
Babu, J. R., Jeganathan, K. B., Baker, D. J., Wu, X., Kang-Decker, N., and VAN Deursen, J. M. (2003). Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160, 341–353. doi:10.1083/jcb.200211048
Baker, D. J., Dawlaty, M. M., Wijshake, T., Jeganathan, K. B., Malureanu, L., VAN Ree, J. H., et al. (2013). Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat. Cell Biol. 15, 96–102. doi:10.1038/ncb2643
Baker, D. J., Jeganathan, K. B., Cameron, J. D., Thompson, M., Juneja, S., Kopecka, A., et al. (2004). BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36, 744–749. doi:10.1038/ng1382
Balachandran, R. S., Heighington, C. S., Starostina, N. G., Anderson, J. W., Owen, D. L., Vasudevan, S., et al. (2016). The ubiquitin ligase CRL2ZYG11 targets cyclin B1 for degradation in a conserved pathway that facilitates mitotic slippage. J. Cell Biol. 215, 151–166. doi:10.1083/jcb.201601083
Barford, D. (2020). Structural interconversions of the anaphase-promoting complex/cyclosome (APC/C) regulate cell cycle transitions. Curr. Opin. Struct. Biol. 61, 86–97. doi:10.1016/j.sbi.2019.11.010
Bennabi, I., Terret, M. E., and Verlhac, M. H. (2016). Meiotic spindle assembly and chromosome segregation in oocytes. J. Cell Biol. 215, 611–619. doi:10.1083/jcb.201607062
Bertoli, C., Skotheim, J. M., and De Bruin, R. A. (2013). Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14, 518–528. doi:10.1038/nrm3629
Bissen, S. T., and Weisblat, D. A. (1989). The durations and compositions of cell cycles in embryos of the leech, Helobdella triserialis. Development 106, 105–118. doi:10.1242/dev.106.1.105
Bloomfield, M., Chen, J., and Cimini, D. (2020). Spindle architectural features must Be considered along with cell size to explain the timing of mitotic checkpoint silencing. Front. Physiol. 11, 596263. doi:10.3389/fphys.2020.596263
Bolanos-Garcia, V. M., and Blundell, T. L. (2011). BUB1 and BUBR1: multifaceted kinases of the cell cycle. Trends Biochem. Sci. 36, 141–150. doi:10.1016/j.tibs.2010.08.004
Boveri, T. (2008). Concerning the origin of malignant tumours by theodor Boveri. Translated and annotated by henry harris. J. Cell Sci. 121 (Suppl. 1), 1–84. doi:10.1242/jcs.025742
Bridges, C. B. (1921). Genetical and cytological proof of non-disjunction of the fourth chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 7, 186–192. doi:10.1073/pnas.7.7.186
Brito, D. A., and Rieder, C. L. (2006). Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr. Biol. 16, 1194–1200. doi:10.1016/j.cub.2006.04.043
Brown, J. A., Bharathi, A., Ghosh, A., Whalen, W., Fitzgerald, E., and Dhar, R. (1995). A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 270, 7411–7419. doi:10.1074/jbc.270.13.7411
Brown, N. G., Vanderlinden, R., Watson, E. R., Weissmann, F., Ordureau, A., Wu, K. P., et al. (2016). Dual RING E3 architectures regulate multiubiquitination and ubiquitin chain elongation by APC/C. Cell 165, 1440–1453. doi:10.1016/j.cell.2016.05.037
Brown, N. G., Watson, E. R., Weissmann, F., Jarvis, M. A., Vanderlinden, R., Grace, C. R. R., et al. (2014). Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol. Cell 56, 246–260. doi:10.1016/j.molcel.2014.09.009
Brunet, S., Pahlavan, G., Taylor, S., and Maro, B. (2003). Functionality of the spindle checkpoint during the first meiotic division of mammalian oocytes. Reproduction 126, 443–450. doi:10.1530/rep.0.1260443
Buffin, E., Lefebvre, C., Huang, J., Gagou, M. E., and Karess, R. E. (2005). Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15, 856–861. doi:10.1016/j.cub.2005.03.052
Burton, J. L., and Solomon, M. J. (2007). Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 21, 655–667. doi:10.1101/gad.1511107
Buschhorn, B. A., Petzold, G., Galova, M., Dube, P., Kraft, C., Herzog, F., et al. (2011). Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1. Nat. Struct. Mol. Biol. 18, 6–13. doi:10.1038/nsmb.1979
Caldas, G. V., and Deluca, J. G. (2014). KNL1: bringing order to the kinetochore. Chromosoma 123, 169–181. doi:10.1007/s00412-013-0446-5
Carvalhal, S., Bader, I., Rooimans, M. A., Oostra, A. B., Balk, J. A., Feichtinger, R. G., et al. (2022). Biallelic BUB1 mutations cause microcephaly, developmental delay, and variable effects on cohesion and chromosome segregation. Sci. Adv. 8, eabk0114. doi:10.1126/sciadv.abk0114
Chao, W. C., Kulkarni, K., Zhang, Z., Kong, E. H., and Barford, D. (2012). Structure of the mitotic checkpoint complex. Nature 484, 208–213. doi:10.1038/nature10896
Cheerambathur, D. K., Prevo, B., Chow, T. L., Hattersley, N., Wang, S., Zhao, Z., et al. (2019). The kinetochore-microtubule coupling machinery is repurposed in sensory nervous system morphogenesis. Dev. Cell 48, 864–872. doi:10.1016/j.devcel.2019.02.002
Cheeseman, I. M. (2014). The kinetochore. Cold Spring Harb. Perspect. Biol. 6, a015826. doi:10.1101/cshperspect.a015826
Cheeseman, I. M., Anderson, S., Jwa, M., Green, E. M., Kang, J., Yates, J. R., et al. (2002). Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172. doi:10.1016/s0092-8674(02)00973-x
Cheeseman, I. M., and Desai, A. (2008). Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46. doi:10.1038/nrm2310
Chen, G. Y., Renda, F., Zhang, H., Gokden, A., Wu, D. Z., Chenoweth, D. M., et al. (2021). Tension promotes kinetochore-microtubule release by Aurora B kinase. J. Cell Biol. 220, e202007030. doi:10.1083/jcb.202007030
Chenevert, J., Roca, M., Besnardeau, L., Ruggiero, A., Nabi, D., Mcdougall, A., et al. (2020). The spindle assembly checkpoint functions during early development in non-chordate embryos. Cells 9, 1087. doi:10.3390/cells9051087
Ciferri, C., Pasqualato, S., Screpanti, E., Varetti, G., Santaguida, S., Dos Reis, G., et al. (2008). Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133, 427–439. doi:10.1016/j.cell.2008.03.020
Cimini, D., Howell, B., Maddox, P., Khodjakov, A., Degrassi, F., and Salmon, E. D. (2001). Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153, 517–527. doi:10.1083/jcb.153.3.517
Cinalli, R. M., Rangan, P., and Lehmann, R. (2008). Germ cells are forever. Cell 132, 559–562. doi:10.1016/j.cell.2008.02.003
Ciossani, G., Overlack, K., Petrovic, A., Huis In 'T Veld, P. J., Koerner, C., Wohlgemuth, S., et al. (2018). The kinetochore proteins CENP-E and CENP-F directly and specifically interact with distinct BUB mitotic checkpoint Ser/Thr kinases. J. Biol. Chem. 293, 10084–10101. doi:10.1074/jbc.RA118.003154
Da Fonseca, P. C., Kong, E. H., Zhang, Z., Schreiber, A., Williams, M. A., Morris, E. P., et al. (2011). Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature 470, 274–278. doi:10.1038/nature09625
D’Costa, A., and Shepherd, I. T. (2009). Zebrafish development and genetics: introducing undergraduates to developmental biology and genetics in a large introductory laboratory class. Zebrafish 6, 169–177. doi:10.1089/zeb.2008.0562
De Boer, P., De Jong, J. H., and VAN Der Hoeven, F. A. (1991). Meiosis in a sterile male mouse with an isoYq marker chromosome. Cytogenet Cell Genet. 56, 36–39. doi:10.1159/000133042
Devigne, A., and Bhalla, N. (2021). Mad1's ability to interact with Mad2 is essential to regulate and monitor meiotic synapsis in C. elegans. PLoS Genet. 17, e1009598. doi:10.1371/journal.pgen.1009598
Dey, S. K., Lim, H., Das, S. K., Reese, J., Paria, B. C., Daikoku, T., et al. (2004). Molecular cues to implantation. Endocr. Rev. 25, 341–373. doi:10.1210/er.2003-0020
Diaz-Martinez, L. A., Tian, W., Li, B., Warrington, R., Jia, L., Brautigam, C. A., et al. (2015). The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 290, 2431–2443. doi:10.1074/jbc.M114.616490
Di Fiore, B., Davey, N. E., Hagting, A., Izawa, D., Mansfeld, J., Gibson, T. J., et al. (2015). The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell 32, 358–372. doi:10.1016/j.devcel.2015.01.003
Ding, L., Cao, J., Lin, W., Chen, H., Xiong, X., Ao, H., et al. (2020). The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 21, 1960. doi:10.3390/ijms21061960
Dobles, M., Liberal, V., Scott, M. L., Benezra, R., and Sorger, P. K. (2000). Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101, 635–645. doi:10.1016/s0092-8674(00)80875-2
Eaker, S., Pyle, A., Cobb, J., and Handel, M. A. (2001). Evidence for meiotic spindle checkpoint from analysis of spermatocytes from Robertsonian-chromosome heterozygous mice. J. Cell Sci. 114, 2953–2965. doi:10.1242/jcs.114.16.2953
Edgar, L. G., and McGhee, J. D. (1988). DNA synthesis and the control of embryonic gene expression in C. elegans. Cell 53, 589–599. doi:10.1016/0092-8674(88)90575-2
Eichenlaub-Ritter, U., and Boll, I. (1989). Nocodazole sensitivity, age-related aneuploidy, and alterations in the cell cycle during maturation of mouse oocytes. Cytogenet Cell Genet. 52, 170–176. doi:10.1159/000132871
Elowe, S., Hummer, S., Uldschmid, A., Li, X., and Nigg, E. A. (2007). Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219. doi:10.1101/gad.436007
Encalada, S. E., Willis, J., Lyczak, R., and Bowerman, B. (2005). A spindle checkpoint functions during mitosis in the early Caenorhabditis elegans embryo. Mol. Biol. Cell 16, 1056–1070. doi:10.1091/mbc.e04-08-0712
Enserink, J. M., and Kolodner, R. D. (2010). An overview of Cdk1-controlled targets and processes. Cell Div. 5, 11. doi:10.1186/1747-1028-5-11
Espert, A., Uluocak, P., Bastos, R. N., Mangat, D., Graab, P., and Gruneberg, U. (2014). PP2A-B56 opposes Mps1 phosphorylation of Knl1 and thereby promotes spindle assembly checkpoint silencing. J. Cell Biol. 206, 833–842. doi:10.1083/jcb.201406109
Evans, T., Rosenthal, E. T., Youngblom, J., Distel, D., and Hunt, T. (1983). Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division. Cell 33, 389–396. doi:10.1016/0092-8674(83)90420-8
Everett, C. A., and Searle, J. B. (1995). Pattern and frequency of nocodazole induced meiotic nondisjunction in oocytes of mice carrying the 'tobacco mouse' metacentric Rb(16.17)7Bnr. Genet. Res. 66, 35–43. doi:10.1017/s0016672300034376
Eytan, E., Wang, K., Miniowitz-Shemtov, S., Sitry-Shevah, D., Kaisari, S., Yen, T. J., et al. (2014). Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet). Proc. Natl. Acad. Sci. U S A. 111, 12019–12024. doi:10.1073/pnas.1412901111
Faesen, A. C., Thanasoula, M., Maffini, S., Breit, C., Muller, F., VAN Gerwen, S., et al. (2017). Basis of catalytic assembly of the mitotic checkpoint complex. Nature 542, 498–502. doi:10.1038/nature21384
Fang, Q., Chen, X. L., Zhang, L., Li, Y. B., Sun, T. Z., Yang, C. X., et al. (2021). The essential roles of Mps1 in spermatogenesis and fertility in mice. Cell Death Dis. 12, 531. doi:10.1038/s41419-021-03815-4
Feinberg, S., Roure, A., Piron, J., and Darras, S. (2019). Antero-posterior ectoderm patterning by canonical Wnt signaling during ascidian development. PLoS Genet. 15, e1008054. doi:10.1371/journal.pgen.1008054
Fischer, E. S., Yu, C. W. H., Bellini, D., Mclaughlin, S. H., Orr, C. M., Wagner, A., et al. (2021). Molecular mechanism of Mad1 kinetochore targeting by phosphorylated Bub1. EMBO Rep. 22, e52242. doi:10.15252/embr.202052242
Foe, V. E., and Alberts, B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31–70. doi:10.1242/jcs.61.1.31
Foley, E. A., Maldonado, M., and Kapoor, T. M. (2011). Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13, 1265–1271. doi:10.1038/ncb2327
Foster, S. A., and Morgan, D. O. (2012). The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Mol. Cell 47, 921–932. doi:10.1016/j.molcel.2012.07.031
Galli, M., and Morgan, D. O. (2016). Cell size determines the strength of the spindle assembly checkpoint during embryonic development. Dev. Cell 36, 344–352. doi:10.1016/j.devcel.2016.01.003
Ganem, N. J., Godinho, S. A., and Pellman, D. (2009). A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282. doi:10.1038/nature08136
Garcia-Castillo, H., Vasquez-Velasquez, A. I., Rivera, H., and Barros-Nunez, P. (2008). Clinical and genetic heterogeneity in patients with mosaic variegated aneuploidy: delineation of clinical subtypes. Am. J. Med. Genet. A 146A, 1687–1695. doi:10.1002/ajmg.a.32315
Gassmann, R., Holland, A. J., Varma, D., Wan, X., Civril, F., Cleveland, D. W., et al. (2010). Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24, 957–971. doi:10.1101/gad.1886810
Gerhart, J., Wu, M., and Kirschner, M. (1984). Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. J. Cell Biol. 98, 1247–1255. doi:10.1083/jcb.98.4.1247
Gerhold, A. R., Poupart, V., Labbe, J. C., and Maddox, P. S. (2018). Spindle assembly checkpoint strength is linked to cell fate in the Caenorhabditis elegans embryo. Mol. Biol. Cell 29, 1435–1448. doi:10.1091/mbc.E18-04-0215
Ge, S., Skaar, J. R., and Pagano, M. (2009). APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell Cycle 8, 167–171. doi:10.4161/cc.8.1.7606
Ghongane, P., Kapanidou, M., Asghar, A., Elowe, S., and Bolanos-Garcia, V. M. (2014). The dynamic protein Knl1 - a kinetochore rendezvous. J. Cell Sci. 127, 3415–3423. doi:10.1242/jcs.149922
Gilliland, W. D., Hughes, S. E., Cotitta, J. L., Takeo, S., Xiang, Y., and Hawley, R. S. (2007). The multiple roles of mps1 in Drosophila female meiosis. PLoS Genet. 3, e113. doi:10.1371/journal.pgen.0030113
Glotzer, M., Murray, A. W., and Kirschner, M. W. (1991). Cyclin is degraded by the ubiquitin pathway. Nature 349, 132–138. doi:10.1038/349132a0
Golan, A., Yudkovsky, Y., and Hershko, A. (2002). The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 277, 15552–15557. doi:10.1074/jbc.M111476200
Gong, D., and Ferrell, J. E. (2010). The roles of cyclin A2, B1, and B2 in early and late mitotic events. Mol. Biol. Cell 21, 3149–3161. doi:10.1091/mbc.E10-05-0393
Gong, D., Pomerening, J. R., Myers, J. W., Gustavsson, C., Jones, J. T., Hahn, A. T., et al. (2007). Cyclin A2 regulates nuclear-envelope breakdown and the nuclear accumulation of cyclin B1. Curr. Biol. 17, 85–91. doi:10.1016/j.cub.2006.11.066
Gorbsky, G. J. (2015). The spindle checkpoint and chromosome segregation in meiosis. FEBS J. 282, 2471–2487. doi:10.1111/febs.13166
Gourguechon, S., Holt, L. J., and Cande, W. Z. (2013). The Giardia cell cycle progresses independently of the anaphase-promoting complex. J. Cell Sci. 126, 2246–2255. doi:10.1242/jcs.121632
Grohme, M. A., Schloissnig, S., Rozanski, A., Pippel, M., Young, G. R., Winkler, S., et al. (2018). The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms. Nature 554, 56–61. doi:10.1038/nature25473
Gui, L., and Homer, H. (2012). Spindle assembly checkpoint signalling is uncoupled from chromosomal position in mouse oocytes. Development 139, 1941–1946. doi:10.1242/dev.078352
Guo, C., Kong, F., Lv, Y., Gao, N., Xiu, X., and Sun, X. (2020). CDC20 inhibitor Apcin inhibits embryo implantation in vivo and in vitro. Cell Biochem. Funct. 38, 810–816. doi:10.1002/cbf.3550
Habu, T., Kim, S. H., Weinstein, J., and Matsumoto, T. (2002). Identification of a MAD2-binding protein, CMT2, and its role in mitosis. EMBO J. 21, 6419–6428. doi:10.1093/emboj/cdf659
Hached, K., Xie, S. Z., Buffin, E., Cladiere, D., Rachez, C., Sacras, M., et al. (2011). Mps1 at kinetochores is essential for female mouse meiosis I. Development 138, 2261–2271. doi:10.1242/dev.061317
Haering, C. H., Lowe, J., Hochwagen, A., and Nasmyth, K. (2002). Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell 9, 773–788. doi:10.1016/s1097-2765(02)00515-4
Hardwick, K. G., Johnston, R. C., Smith, D. L., and Murray, A. W. (2000). MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J. Cell Biol. 148, 871–882. doi:10.1083/jcb.148.5.871
Herbert, M., Kalleas, D., Cooney, D., Lamb, M., and Lister, L. (2015). Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol. 7, a017970. doi:10.1101/cshperspect.a017970
Hertzler, J. I., Simonovitch, S. I., Albertson, R. M., Weiner, A. T., Nye, D. M. R., and Rolls, M. M. (2020). Kinetochore proteins suppress neuronal microtubule dynamics and promote dendrite regeneration. Mol. Biol. Cell 31, 2125–2138. doi:10.1091/mbc.E20-04-0237-T
Herzog, F., Primorac, I., Dube, P., Lenart, P., Sander, B., Mechtler, K., et al. (2009). Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323, 1477–1481. doi:10.1126/science.1163300
Hewitt, L., Tighe, A., Santaguida, S., White, A. M., Jones, C. D., Musacchio, A., et al. (2010). Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 190, 25–34. doi:10.1083/jcb.201002133
Holland, A. J., Lan, W., and Cleveland, D. W. (2010). Centriole duplication: a lesson in self-control. Cell Cycle 9, 2731–2736. doi:10.4161/cc.9.14.12184
Homer, H. A., Mcdougall, A., Levasseur, M., Yallop, K., Murdoch, A. P., and Herbert, M. (2005). Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 19, 202–207. doi:10.1101/gad.328105
Homer, H., Gui, L., and Carroll, J. (2009). A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science 326, 991–994. doi:10.1126/science.1175326
Horakova, A., Konecna, M., and Anger, M. (2024). Chromosome division in early embryos-is everything under control? And is the cell size important? Int. J. Mol. Sci. 25, 2101. doi:10.3390/ijms25042101
Howell, B. J., Mcewen, B. F., Canman, J. C., Hoffman, D. B., Farrar, E. M., Rieder, C. L., et al. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155, 1159–1172. doi:10.1083/jcb.200105093
Ide, A. H., Deluca, K. F., Wiggan, O., Markus, S. M., and Deluca, J. G. (2023). The role of kinetochore dynein in checkpoint silencing is restricted to disassembly of the corona. Mol. Biol. Cell 34, ar76. doi:10.1091/mbc.E23-04-0130
Ikegami, R., Zhang, J., Rivera-Bennetts, A. K., and Yager, T. D. (1997). Activation of the metaphase checkpoint and an apoptosis programme in the early zebrafish embryo, by treatment with the spindle-destabilising agent nocodazole. Zygote 5, 329–350. doi:10.1017/s0967199400003919
Iwanaga, Y., Chi, Y. H., Miyazato, A., Sheleg, S., Haller, K., Peloponese, J. M., et al. (2007). Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67, 160–166. doi:10.1158/0008-5472.CAN-06-3326
Izawa, D., and Pines, J. (2015). The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature 517, 631–634. doi:10.1038/nature13911
Jia, C. W., Wang, L., Lan, Y. L., Song, R., Zhou, L. Y., Yu, L., et al. (2015). Aneuploidy in early miscarriage and its related factors. Chin. Med. J. Engl. 128, 2772–2776. doi:10.4103/0366-6999.167352
Jia, L., Kim, S., and Yu, H. (2013). Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem. Sci. 38, 302–311. doi:10.1016/j.tibs.2013.03.004
Jin, F., Liu, H., Li, P., Yu, H. G., and Wang, Y. (2012). Loss of function of the Cik1/Kar3 motor complex results in chromosomes with syntelic attachment that are sensed by the tension checkpoint. PLoS Genet. 8, e1002492. doi:10.1371/journal.pgen.1002492
Ji, Z., Gao, H., Jia, L., Li, B., and Yu, H. (2017). A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. Elife 6, e22513. doi:10.7554/eLife.22513
Ji, Z., Gao, H., and Yu, H. (2015). Cell division cycle. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 348, 1260–1264. doi:10.1126/science.aaa4029
Ju, J. Q., Li, X. H., Pan, M. H., Xu, Y., Xu, Y., Sun, M. H., et al. (2021). Mps1 controls spindle assembly, SAC, and DNA repair in the first cleavage of mouse early embryos. J. Cell Biochem. 122, 290–300. doi:10.1002/jcb.29858
Kalitsis, P., Earle, E., Fowler, K. J., and Choo, K. H. (2000). Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14, 2277–2282. doi:10.1101/gad.827500
Kallio, M., Eriksson, J. E., and Gorbsky, G. J. (2000). Differences in spindle association of the mitotic checkpoint protein Mad2 in mammalian spermatogenesis and oogenesis. Dev. Biol. 225, 112–123. doi:10.1006/dbio.2000.9818
Kelly, A., Wickliffe, K. E., Song, L., Fedrigo, I., and Rape, M. (2014). Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol. Cell 56, 232–245. doi:10.1016/j.molcel.2014.09.010
Kemmler, S., Stach, M., Knapp, M., Ortiz, J., Pfannstiel, J., Ruppert, T., et al. (2009). Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 28, 1099–1110. doi:10.1038/emboj.2009.62
Khodjakov, A., and Pines, J. (2010). Centromere tension: a divisive issue. Nat. Cell Biol. 12, 919–923. doi:10.1038/ncb1010-919
Kim, T., and Gartner, A. (2021). Bub1 kinase in the regulation of mitosis. Anim. Cells Syst. Seoul. 25, 1–10. doi:10.1080/19768354.2021.1884599
King, E. M., Van Der Sar, S. J., and Hardwick, K. G. (2007). Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS One 2, e342. doi:10.1371/journal.pone.0000342
Kiyomitsu, T., Murakami, H., and Yanagida, M. (2011). Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell Biol. 31, 998–1011. doi:10.1128/MCB.00815-10
Kiyomitsu, T., Obuse, C., and Yanagida, M. (2007). Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13, 663–676. doi:10.1016/j.devcel.2007.09.005
Klebig, C., Korinth, D., and Meraldi, P. (2009). Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185, 841–858. doi:10.1083/jcb.200902128
Koch, L. B., Opoku, K. N., Deng, Y., Barber, A., Littleton, A. J., London, N., et al. (2019). Autophosphorylation is sufficient to release Mps1 kinase from native kinetochores. Proc. Natl. Acad. Sci. U S A. 116, 17355–17360. doi:10.1073/pnas.1901653116
Kolano, A., Brunet, S., Silk, A. D., Cleveland, D. W., and Verlhac, M. H. (2012). Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc. Natl. Acad. Sci. U S A. 109, E1858–E1867. doi:10.1073/pnas.1204686109
Kops, G., Snel, B., and Tromer, E. C. (2020). Evolutionary dynamics of the spindle assembly checkpoint in eukaryotes. Curr. Biol. 30, R589-R602–R602. doi:10.1016/j.cub.2020.02.021
Kousholt, A. N., Menzel, T., and Sorensen, C. S. (2012). Pathways for genome integrity in G2 phase of the cell cycle. Biomolecules 2, 579–607. doi:10.3390/biom2040579
Kouznetsova, A., Novak, I., Jessberger, R., and Hoog, C. (2005). SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J. Cell Sci. 118, 2271–2278. doi:10.1242/jcs.02362
Kraft, C., Vodermaier, H. C., Maurer-Stroh, S., Eisenhaber, F., and Peters, J. M. (2005). The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Mol. Cell 18, 543–553. doi:10.1016/j.molcel.2005.04.023
Kramer, E. R., Scheuringer, N., Podtelejnikov, A. V., Mann, M., and Peters, J. M. (2000). Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell 11, 1555–1569. doi:10.1091/mbc.11.5.1555
Krempler, A., Deckbar, D., Jeggo, P. A., and Lobrich, M. (2007). An imperfect G2M checkpoint contributes to chromosome instability following irradiation of S and G2 phase cells. Cell Cycle 6, 1682–1686. doi:10.4161/cc.6.14.4480
Krenn, V., Overlack, K., Primorac, I., VAN Gerwen, S., and Musacchio, A. (2014). KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats. Curr. Biol. 24, 29–39. doi:10.1016/j.cub.2013.11.046
Krenn, V., Wehenkel, A., Li, X., Santaguida, S., and Musacchio, A. (2012). Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196, 451–467. doi:10.1083/jcb.201110013
Kruse, T., Zhang, G., Larsen, M. S., Lischetti, T., Streicher, W., Kragh Nielsen, T., et al. (2013). Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126, 1086–1092. doi:10.1242/jcs.122481
Kulukian, A., Han, J. S., and Cleveland, D. W. (2009). Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell 16, 105–117. doi:10.1016/j.devcel.2008.11.005
Kumar, M., Pushpa, K., and Mylavarapu, S. V. (2015). Splitting the cell, building the organism: mechanisms of cell division in metazoan embryos. IUBMB Life 67, 575–587. doi:10.1002/iub.1404
Kyogoku, H., and Kitajima, T. S. (2017). Large cytoplasm is linked to the error-prone nature of oocytes. Dev. Cell 41, 287–298. doi:10.1016/j.devcel.2017.04.009
Lampson, M. A., and Grishchuk, E. L. (2017). Mechanisms to avoid and correct erroneous kinetochore-microtubule attachments. Biol. (Basel) 6, 1. doi:10.3390/biology6010001
Lampson, M. A., and Kapoor, T. M. (2005). The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7, 93–98. doi:10.1038/ncb1208
Lamy, C., Rothbacher, U., Caillol, D., and Lemaire, P. (2006). Ci-FoxA-a is the earliest zygotic determinant of the ascidian anterior ectoderm and directly activates Ci-sFRP1/5. Development 133, 2835–2844. doi:10.1242/dev.02448
Lane, S. I., Chang, H. Y., Jennings, P. C., and Jones, K. T. (2010). The Aurora kinase inhibitor ZM447439 accelerates first meiosis in mouse oocytes by overriding the spindle assembly checkpoint. Reproduction 140, 521–530. doi:10.1530/REP-10-0223
Lane, S. I., Yun, Y., and Jones, K. T. (2012). Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development 139, 1947–1955. doi:10.1242/dev.077040
Lane, S. I. R., and Jones, K. T. (2017). Chromosome biorientation and APC activity remain uncoupled in oocytes with reduced volume. J. Cell Biol. 216, 3949–3957. doi:10.1083/jcb.201606134
Lara-Gonzalez, P., Kim, T., Oegema, K., Corbett, K., and Desai, A. (2021a). A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science 371, 64–67. doi:10.1126/science.abc1424
Lara-Gonzalez, P., Pines, J., and Desai, A. (2021b). Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 117, 86–98. doi:10.1016/j.semcdb.2021.06.009
Lara-Gonzalez, P., Scott, M. I., Diez, M., Sen, O., and Taylor, S. S. (2011). BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J. Cell Sci. 124, 4332–4345. doi:10.1242/jcs.094763
Lara-Gonzalez, P., Westhorpe, F. G., and Taylor, S. S. (2012). The spindle assembly checkpoint. Curr. Biol. 22, R966–R980. doi:10.1016/j.cub.2012.10.006
Larose, H., Shami, A. N., Abbott, H., Manske, G., Lei, L., and Hammoud, S. S. (2019). Gametogenesis: a journey from inception to conception. Curr. Top. Dev. Biol. 132, 257–310. doi:10.1016/bs.ctdb.2018.12.006
Lemaire-Adkins, R., Radke, K., and Hunt, P. A. (1997). Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J. Cell Biol. 139, 1611–1619. doi:10.1083/jcb.139.7.1611
Lightfoot, D. A., Kouznetsova, A., Mahdy, E., Wilbertz, J., and Hoog, C. (2006). The fate of mosaic aneuploid embryos during mouse development. Dev. Biol. 289, 384–394. doi:10.1016/j.ydbio.2005.11.001
Li, M., Fang, X., Wei, Z., York, J. P., and Zhang, P. (2009a). Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J. Cell Biol. 185, 983–994. doi:10.1083/jcb.200904020
Li, M., Li, S., Yuan, J., Wang, Z. B., Sun, S. C., Schatten, H., et al. (2009b). Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. PLoS One 4, e7701. doi:10.1371/journal.pone.0007701
Li, M., York, J. P., and Zhang, P. (2007). Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell Biol. 27, 3481–3488. doi:10.1128/MCB.02088-06
Lischetti, T., Zhang, G., Sedgwick, G. G., Bolanos-Garcia, V. M., and Nilsson, J. (2014). The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat. Commun. 5, 5563. doi:10.1038/ncomms6563
Liu, D., Vader, G., Vromans, M. J., Lampson, M. A., and Lens, S. M. (2009). Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323, 1350–1353. doi:10.1126/science.1167000
Liu, D., Vleugel, M., Backer, C. B., Hori, T., Fukagawa, T., Cheeseman, I. M., et al. (2010). Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188, 809–820. doi:10.1083/jcb.201001006
Li, X., and Nicklas, R. B. (1995). Mitotic forces control a cell-cycle checkpoint. Nature 373, 630–632. doi:10.1038/373630a0
Lok, T. M., Wang, Y., Xu, W. K., Xie, S., Ma, H. T., and Poon, R. Y. C. (2020). Mitotic slippage is determined by p31(comet) and the weakening of the spindle-assembly checkpoint. Oncogene 39, 2819–2834. doi:10.1038/s41388-020-1187-6
London, N., and Biggins, S. (2014). Signalling dynamics in the spindle checkpoint response. Nat. Rev. Mol. Cell Biol. 15, 736–747. doi:10.1038/nrm3888
London, N., Ceto, S., Ranish, J. A., and Biggins, S. (2012). Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22, 900–906. doi:10.1016/j.cub.2012.03.052
Luo, Y., Ahmad, E., and Liu, S. T. (2018). MAD1: kinetochore receptors and catalytic mechanisms. Front. Cell Dev. Biol. 6, 51. doi:10.3389/fcell.2018.00051
Ma, H. T., and Poon, R. Y. C. (2018). TRIP13 functions in the establishment of the spindle assembly checkpoint by replenishing O-MAD2. Cell Rep. 22, 1439–1450. doi:10.1016/j.celrep.2018.01.027
Maclennan, M., Crichton, J. H., Playfoot, C. J., and Adams, I. R. (2015). Oocyte development, meiosis and aneuploidy. Semin. Cell Dev. Biol. 45, 68–76. doi:10.1016/j.semcdb.2015.10.005
Magrassi, L., and Graziadei, P. P. (1995). Cell death in the olfactory epithelium. Anat. Embryol. Berl. 192, 77–87. doi:10.1007/BF00186993
Martinez-Exposito, M. J., Kaplan, K. B., Copeland, J., and Sorger, P. K. (1999). Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc. Natl. Acad. Sci. U S A. 96, 8493–8498. doi:10.1073/pnas.96.15.8493
Maskell, D. P., Hu, X. W., and Singleton, M. R. (2010). Molecular architecture and assembly of the yeast kinetochore MIND complex. J. Cell Biol. 190, 823–834. doi:10.1083/jcb.201002059
Masui, Y., and Wang, P. (1998). Cell cycle transition in early embryonic development of Xenopus laevis. Biol. Cell 90, 537–548. doi:10.1016/s0248-4900(99)80011-2
Mcainsh, A. D., and Kops, G. (2023). Principles and dynamics of spindle assembly checkpoint signalling. Nat. Rev. Mol. Cell Biol. 24, 543–559. doi:10.1038/s41580-023-00593-z
Mcguinness, B. E., Anger, M., Kouznetsova, A., Gil-Bernabe, A. M., Helmhart, W., Kudo, N. R., et al. (2009). Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr. Biol. 19, 369–380. doi:10.1016/j.cub.2009.01.064
Mcvey, S. L., Cosby, J. K., and Nannas, N. J. (2021). Aurora B tension sensing mechanisms in the kinetochore ensure accurate chromosome segregation. Int. J. Mol. Sci. 22, 8818. doi:10.3390/ijms22168818
Meraldi, P., Draviam, V. M., and Sorger, P. K. (2004). Timing and checkpoints in the regulation of mitotic progression. Dev. Cell 7, 45–60. doi:10.1016/j.devcel.2004.06.006
Michel, L. S., Liberal, V., Chatterjee, A., Kirchwegger, R., Pasche, B., Gerald, W., et al. (2001). MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409, 355–359. doi:10.1038/35053094
Mihajlovic, A. I., Byers, C., Reinholdt, L., and Fitzharris, G. (2023). Spindle assembly checkpoint insensitivity allows meiosis-II despite chromosomal defects in aged eggs. EMBO Rep. 24, e57227. doi:10.15252/embr.202357227
Milani, H., Schwarting, R. K., Kumpf, S., Steiner, H., and Huston, J. P. (1990). Interaction between recovery from behavioral asymmetries induced by hemivibrissotomy in the rat and the effects of apomorphine and amphetamine. Behav. Neurosci. 104, 470–476. doi:10.1037//0735-7044.104.3.470
Minshull, J., Sun, H., Tonks, N. K., and Murray, A. W. (1994). A MAP kinase-dependent spindle assembly checkpoint in Xenopus egg extracts. Cell 79, 475–486. doi:10.1016/0092-8674(94)90256-9
Mukherjee, S., Sandri, B. J., Tank, D., Mcclellan, M., Harasymiw, L. A., Yang, Q., et al. (2019). A gradient in metaphase tension leads to a scaled cellular response in mitosis. Dev. Cell 49, 63–76. doi:10.1016/j.devcel.2019.01.018
Musacchio, A. (2015b). The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018. doi:10.1016/j.cub.2015.08.051
Musacchio, A., and Desai, A. (2017). A molecular view of kinetochore assembly and function. Biol. (Basel) 6, 5. doi:10.3390/biology6010005
Musacchio, A., and Salmon, E. D. (2007). The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8, 379–393. doi:10.1038/nrm2163
Nagaoka, S. I., Hodges, C. A., Albertini, D. F., and Hunt, P. A. (2011). Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr. Biol. 21, 651–657. doi:10.1016/j.cub.2011.03.003
Nasa, I., Rusin, S. F., Kettenbach, A. N., and Moorhead, G. B. (2018). Aurora B opposes PP1 function in mitosis by phosphorylating the conserved PP1-binding RVxF motif in PP1 regulatory proteins. Sci. Signal 11, eaai8669. doi:10.1126/scisignal.aai8669
Niault, T., Hached, K., Sotillo, R., Sorger, P. K., Maro, B., Benezra, R., et al. (2007). Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PLoS One 2, e1165. doi:10.1371/journal.pone.0001165
Nicklas, R. B., and Koch, C. A. (1969). Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 43, 40–50. doi:10.1083/jcb.43.1.40
Nijenhuis, W., Vallardi, G., Teixeira, A., Kops, G. J., and Saurin, A. T. (2014). Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 16, 1257–1264. doi:10.1038/ncb3065
Nijenhuis, W., Von Castelmur, E., Littler, D., De Marco, V., Tromer, E., Vleugel, M., et al. (2013). A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol. 201, 217–231. doi:10.1083/jcb.201210033
Nilsson, J., Yekezare, M., Minshull, J., and Pines, J. (2008). The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10, 1411–1420. doi:10.1038/ncb1799
North, B. J., Rosenberg, M. A., Jeganathan, K. B., Hafner, A. V., Michan, S., Dai, J., et al. (2014). SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 33, 1438–1453. doi:10.15252/embj.201386907
Overlack, K., Primorac, I., Vleugel, M., Krenn, V., Maffini, S., Hoffmann, I., et al. (2015). A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. Elife 4, e05269. doi:10.7554/eLife.05269
Pan, B., and Li, J. (2019). The art of oocyte meiotic arrest regulation. Reprod. Biol. Endocrinol. 17, 8. doi:10.1186/s12958-018-0445-8
Perera, D., Tilston, V., Hopwood, J. A., Barchi, M., Boot-Handford, R. P., and Taylor, S. S. (2007). Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell 13, 566–579. doi:10.1016/j.devcel.2007.08.008
Perez-Mongiovi, D., Malmanche, N., Bousbaa, H., and Sunkel, C. (2005). Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster. Development 132, 4509–4520. doi:10.1242/dev.02028
Peters, J. M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656. doi:10.1038/nrm1988
Petrone, A., Adamo, M. E., Cheng, C., and Kettenbach, A. N. (2016). Identification of candidate cyclin-dependent kinase 1 (Cdk1) substrates in mitosis by quantitative phosphoproteomics. Mol. Cell Proteomics 15, 2448–2461. doi:10.1074/mcp.M116.059394
Petrovic, A., Pasqualato, S., Dube, P., Krenn, V., Santaguida, S., Cittaro, D., et al. (2010). The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J. Cell Biol. 190, 835–852. doi:10.1083/jcb.201002070
Piano, V., Alex, A., Stege, P., Maffini, S., Stoppiello, G. A., Huis In 'T Veld, P. J., et al. (2021). CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science 371, 67–71. doi:10.1126/science.abc1152
Pleuger, R., Cozma, C., Hohoff, S., Denkhaus, C., Dudziak, A., Kaschani, F., et al. (2024). Microtubule end-on attachment maturation regulates Mps1 association with its kinetochore receptor. Curr. Biol. 34, 2279–2293 e6. doi:10.1016/j.cub.2024.03.062
Ploubidou, A., Robinson, D. R., Docherty, R. C., Ogbadoyi, E. O., and Gull, K. (1999). Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci. 112 (Pt 24), 4641–4650. doi:10.1242/jcs.112.24.4641
Posch, M., Khoudoli, G. A., Swift, S., King, E. M., Deluca, J. G., and Swedlow, J. R. (2010). Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191, 61–74. doi:10.1083/jcb.200912046
Primorac, I., Weir, J. R., Chiroli, E., Gross, F., Hoffmann, I., VAN Gerwen, S., et al. (2013). Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. Elife 2, e01030. doi:10.7554/eLife.01030
Qian, J., Beullens, M., Huang, J., De Munter, S., Lesage, B., and Bollen, M. (2015). Cdk1 orders mitotic events through coordination of a chromosome-associated phosphatase switch. Nat. Commun. 6, 10215. doi:10.1038/ncomms10215
Qiao, R., Weissmann, F., Yamaguchi, M., Brown, N. G., Vanderlinden, R., Imre, R., et al. (2016). Mechanism of APC/CCDC20 activation by mitotic phosphorylation. Proc. Natl. Acad. Sci. U S A. 113, E2570–E2578. doi:10.1073/pnas.1604929113
Reddy, S. K., Rape, M., Margansky, W. A., and Kirschner, M. W. (2007). Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921–925. doi:10.1038/nature05734
Rieder, C. L., Cole, R. W., Khodjakov, A., and Sluder, G. (1995). The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130, 941–948. doi:10.1083/jcb.130.4.941
Rieder, C. L., and Maiato, H. (2004). Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7, 637–651. doi:10.1016/j.devcel.2004.09.002
Roca, M., Besnardeau, L., Christians, E., Mcdougall, A., Chenevert, J., and Castagnetti, S. (2023). Acquisition of the spindle assembly checkpoint and its modulation by cell fate and cell size in a chordate embryo. Development 150, dev201145. doi:10.1242/dev.201145
Roizen, N. J., and Patterson, D. (2003). Down's syndrome. Lancet 361, 1281–1289. doi:10.1016/S0140-6736(03)12987-X
Rosenblum, E. B., James, T. Y., Zamudio, K. R., Poorten, T. J., Ilut, D., Rodriguez, D., et al. (2013). Complex history of the amphibian-killing chytrid fungus revealed with genome resequencing data. Proc. Natl. Acad. Sci. U S A. 110, 9385–9390. doi:10.1073/pnas.1300130110
Rudner, A. D., and Murray, A. W. (1996). The spindle assembly checkpoint. Curr. Opin. Cell Biol. 8, 773–780. doi:10.1016/s0955-0674(96)80077-9
Schukken, K. M., Zhu, Y., Bakker, P. L., Koster, M. H., Harkema, L., Youssef, S. A., et al. (2021). Acute systemic loss of Mad2 leads to intestinal atrophy in adult mice. Sci. Rep. 11, 68. doi:10.1038/s41598-020-80169-5
Schwab, M. S., Roberts, B. T., Gross, S. D., Tunquist, B. J., Taieb, F. E., Lewellyn, A. L., et al. (2001). Bub1 is activated by the protein kinase p90(Rsk) during Xenopus oocyte maturation. Curr. Biol. 11, 141–150. doi:10.1016/s0960-9822(01)00045-8
Sebestova, J., Danylevska, A., Novakova, L., Kubelka, M., and Anger, M. (2012). Lack of response to unaligned chromosomes in mammalian female gametes. Cell Cycle 11, 3011–3018. doi:10.4161/cc.21398
Shahbazi, M. N., Wang, T., Tao, X., Weatherbee, B. A. T., Sun, L., Zhan, Y., et al. (2020). Developmental potential of aneuploid human embryos cultured beyond implantation. Nat. Commun. 11, 3987. doi:10.1038/s41467-020-17764-7
Shepperd, L. A., Meadows, J. C., Sochaj, A. M., Lancaster, T. C., Zou, J., Buttrick, G. J., et al. (2012). Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22, 891–899. doi:10.1016/j.cub.2012.03.051
Shi, Q., Hu, M., Luo, M., Liu, Q., Jiang, F., Zhang, Y., et al. (2011). Reduced expression of Mad2 and Bub1 proteins is associated with spontaneous miscarriages. Mol. Hum. Reprod. 17, 14–21. doi:10.1093/molehr/gaq065
Silkworth, W. T., Nardi, I. K., Scholl, L. M., and Cimini, D. (2009). Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One 4, e6564. doi:10.1371/journal.pone.0006564
Sotillo, R., Hernando, E., Diaz-Rodriguez, E., Teruya-Feldstein, J., Cordon-Cardo, C., Lowe, S. W., et al. (2007). Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11, 9–23. doi:10.1016/j.ccr.2006.10.019
Sudakin, V., Chan, G. K., and Yen, T. J. (2001). Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 154, 925–936. doi:10.1083/jcb.200102093
Suijkerbuijk, S. J., VAN Osch, M. H., Bos, F. L., Hanks, S., Rahman, N., and Kops, G. J. (2010). Molecular causes for BUBR1 dysfunction in the human cancer predisposition syndrome mosaic variegated aneuploidy. Cancer Res. 70, 4891–4900. doi:10.1158/0008-5472.CAN-09-4319
Suijkerbuijk, S. J., Vleugel, M., Teixeira, A., and Kops, G. J. (2012). Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell 23, 745–755. doi:10.1016/j.devcel.2012.09.005
Sutcliffe, M. J., Darling, S. M., and Burgoyne, P. S. (1991). Spermatogenesis in XY, XYSxra and XOSxra mice: a quantitative analysis of spermatogenesis throughout puberty. Mol. Reprod. Dev. 30, 81–89. doi:10.1002/mrd.1080300202
Syrjanen, J. L., Pellegrini, L., and Davies, O. R. (2014). A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife 3, e02963. doi:10.7554/eLife.02963
Takenaka, K., Gotoh, Y., and Nishida, E. (1997). MAP kinase is required for the spindle assembly checkpoint but is dispensable for the normal M phase entry and exit in Xenopus egg cell cycle extracts. J. Cell Biol. 136, 1091–1097. doi:10.1083/jcb.136.5.1091
Tang, Z., Bharadwaj, R., Li, B., and Yu, H. (2001). Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 1, 227–237. doi:10.1016/s1534-5807(01)00019-3
Tang, Z., Shu, H., Oncel, D., Chen, S., and Yu, H. (2004a). Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol. Cell 16, 387–397. doi:10.1016/j.molcel.2004.09.031
Tang, Z., Sun, Y., Harley, S. E., Zou, H., and Yu, H. (2004b). Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc. Natl. Acad. Sci. U S A. 101, 18012–18017. doi:10.1073/pnas.0408600102
Taylor, S. S., Ha, E., and Mckeon, F. (1998). The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142, 1–11. doi:10.1083/jcb.142.1.1
Taylor, S. S., Scott, M. I., and Holland, A. J. (2004). The spindle checkpoint: a quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 12, 599–616. doi:10.1023/B:CHRO.0000036610.78380.51
Technau, G. M. (1987). A single cell approach to problems of cell lineage and commitment during embryogenesis of Drosophila melanogaster. Development 100, 1–12. doi:10.1242/dev.100.1.1
Tilston, V., Taylor, S. S., and Perera, D. (2009). Inactivating the spindle checkpoint kinase Bub1 during embryonic development results in a global shutdown of proliferation. BMC Res. Notes 2, 190. doi:10.1186/1756-0500-2-190
Topacio, B. R., Zatulovskiy, E., Cristea, S., Xie, S., Tambo, C. S., Rubin, S. M., et al. (2019). Cyclin D-cdk4,6 drives cell-cycle progression via the retinoblastoma protein's C-terminal helix. Mol. Cell 74, 758–770. doi:10.1016/j.molcel.2019.03.020
Touati, S. A., Buffin, E., Cladiere, D., Hached, K., Rachez, C., VAN Deursen, J. M., et al. (2015). Mouse oocytes depend on BubR1 for proper chromosome segregation but not for prophase I arrest. Nat. Commun. 6, 6946. doi:10.1038/ncomms7946
Tsou, M. F., and Stearns, T. (2006). Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18, 74–78. doi:10.1016/j.ceb.2005.12.008
Tsurumi, C., Hoffmann, S., Geley, S., Graeser, R., and Polanski, Z. (2004). The spindle assembly checkpoint is not essential for CSF arrest of mouse oocytes. J. Cell Biol. 167, 1037–1050. doi:10.1083/jcb.200405165
Tunquist, B. J., Eyers, P. A., Chen, L. G., Lewellyn, A. L., and Maller, J. L. (2003). Spindle checkpoint proteins Mad1 and Mad2 are required for cytostatic factor-mediated metaphase arrest. J. Cell Biol. 163, 1231–1242. doi:10.1083/jcb.200306153
Van Hooff, J. J., Tromer, E., Van Wijk, L. M., Snel, B., and Kops, G. J. (2017). Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, 1559–1571. doi:10.15252/embr.201744102
Varma, D., and Salmon, E. D. (2012). The KMN protein network--chief conductors of the kinetochore orchestra. J. Cell Sci. 125, 5927–5936. doi:10.1242/jcs.093724
Vazquez-Diez, C., and Fitzharris, G. (2018). Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction 155, R63-R76–R76. doi:10.1530/REP-17-0569
Vazquez-Diez, C., Paim, L. M. G., and Fitzharris, G. (2019). Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Curr. Biol. 29, 865–873. doi:10.1016/j.cub.2018.12.042
Vleugel, M., Hoek, T. A., Tromer, E., Sliedrecht, T., Groenewold, V., Omerzu, M., et al. (2015). Dissecting the roles of human BUB1 in the spindle assembly checkpoint. J. Cell Sci. 128, 2975–2982. doi:10.1242/jcs.169821
Vleugel, M., Tromer, E., Omerzu, M., Groenewold, V., Nijenhuis, W., Snel, B., et al. (2013). Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J. Cell Biol. 203, 943–955. doi:10.1083/jcb.201307016
Vogt, E., Kirsch-Volders, M., Parry, J., and Eichenlaub-Ritter, U. (2008). Spindle formation, chromosome segregation and the spindle checkpoint in mammalian oocytes and susceptibility to meiotic error. Mutat. Res. 651, 14–29. doi:10.1016/j.mrgentox.2007.10.015
Wang, X. M., Zhai, Y., and Ferrell, J. E. (1997). A role for mitogen-activated protein kinase in the spindle assembly checkpoint in XTC cells. J. Cell Biol. 137, 433–443. doi:10.1083/jcb.137.2.433
Wang, J., Wang, Z., Yu, T., Yang, H., Virshup, D. M., Kops, G. J., et al. (2016a). Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization. Protein Cell 7, 516–526. doi:10.1007/s13238-016-0283-4
Wang, K., Sturt-Gillespie, B., Hittle, J. C., Macdonald, D., Chan, G. K., Yen, T. J., et al. (2014). Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J. Biol. Chem. 289, 23928–23937. doi:10.1074/jbc.M114.585315
Wang, L., Kruse, T., Lopez-Mendez, B., Zhang, Y., Song, C., Zhu, L., et al. (2023). Spatial separation of phosphatase and kinase activity within the Bub complex is required for proper mitosis. J. Mol. Cell Biol. 14, mjac062. doi:10.1093/jmcb/mjac062
Wang, Q., Liu, T., Fang, Y., Xie, S., Huang, X., Mahmood, R., et al. (2004). BUBR1 deficiency results in abnormal megakaryopoiesis. Blood 103, 1278–1285. doi:10.1182/blood-2003-06-2158
Wang, X., Babu, J. R., Harden, J. M., Jablonski, S. A., Gazi, M. H., Lingle, W. L., et al. (2001). The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBS)-containing proteins. J. Biol. Chem. 276, 26559–26567. doi:10.1074/jbc.M101083200
Wang, X., Bajaj, R., Bollen, M., Peti, W., and Page, R. (2016b). Expanding the PP2A interactome by defining a B56-specific SLiM. Structure 24, 2174–2181. doi:10.1016/j.str.2016.09.010
Wang, Z. (2021). Regulation of cell cycle progression by growth factor-induced cell signaling. Cells 10, 3327. doi:10.3390/cells10123327
Wei, R. R., Al-Bassam, J., and Harrison, S. C. (2007). The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat. Struct. Mol. Biol. 14, 54–59. doi:10.1038/nsmb1186
Wei, L., Liang, X. W., Zhang, Q. H., Li, M., Yuan, J., Li, S., et al. (2010). BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle 9, 1112–1121. doi:10.4161/cc.9.6.10957
Wei, Y., Multi, S., Yang, C. R., Ma, J., Zhang, Q. H., Wang, Z. B., et al. (2011). Spindle assembly checkpoint regulates mitotic cell cycle progression during preimplantation embryo development. PLoS One 6, e21557. doi:10.1371/journal.pone.0021557
Welburn, J. P., Vleugel, M., Liu, D., Yates, J. R., Lampson, M. A., Fukagawa, T., et al. (2010). Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38, 383–392. doi:10.1016/j.molcel.2010.02.034
Westergaard, M., and Von Wettstein, D. (1972). The synaptinemal complex. Annu. Rev. Genet. 6, 71–110. doi:10.1146/annurev.ge.06.120172.000443
Yamagishi, Y., Yang, C. H., Tanno, Y., and Watanabe, Y. (2012). MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14, 746–752. doi:10.1038/ncb2515
Yamaguchi, M., Vanderlinden, R., Weissmann, F., Qiao, R., Dube, P., Brown, N. G., et al. (2016). Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell 63, 593–607. doi:10.1016/j.molcel.2016.07.003
Yang, K. T., Li, S. K., Chang, C. C., Tang, C. J., Lin, Y. N., Lee, S. C., et al. (2010). Aurora-C kinase deficiency causes cytokinesis failure in meiosis I and production of large polyploid oocytes in mice. Mol. Biol. Cell 21, 2371–2383. doi:10.1091/mbc.e10-02-0170
Yang, Q., Zhao, J., Chen, D., and Wang, Y. (2021). E3 ubiquitin ligases: styles, structures and functions. Mol. Biomed. 2, 23. doi:10.1186/s43556-021-00043-2
Ye, Q., Rosenberg, S. C., Moeller, A., Speir, J. A., Su, T. Y., and Corbett, K. D. (2015). TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. Elife 4, e07367. doi:10.7554/eLife.07367
Yeste, M., Jones, C., Amdani, S. N., and Coward, K. (2017). Oocyte activation and fertilisation: crucial contributors from the sperm and oocyte. Results Probl. Cell Differ. 59, 213–239. doi:10.1007/978-3-319-44820-6_8
Yip, M. Y. (2014). Uniparental disomy in Robertsonian translocations: strategies for uniparental disomy testing. Transl. Pediatr. 3, 98–107. doi:10.3978/j.issn.2224-4336.2014.03.03
Yuan, L., Liu, J. G., Hoja, M. R., Wilbertz, J., Nordqvist, K., and Hoog, C. (2002). Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science 296, 1115–1118. doi:10.1126/science.1070594
Zecevic, M., Catling, A. D., Eblen, S. T., Renzi, L., Hittle, J. C., Yen, T. J., et al. (1998). Active MAP kinase in mitosis: localization at kinetochores and association with the motor protein CENP-E. J. Cell Biol. 142, 1547–1558. doi:10.1083/jcb.142.6.1547
Zhai, J., Xiao, Z., Wang, Y., and Wang, H. (2022). Human embryonic development: from peri-implantation to gastrulation. Trends Cell Biol. 32, 18–29. doi:10.1016/j.tcb.2021.07.008
Zhang, D., Li, M., Ma, W., Hou, Y., Li, Y. H., Li, S. W., et al. (2005). Localization of mitotic arrest deficient 1 (MAD1) in mouse oocytes during the first meiosis and its functions as a spindle checkpoint protein. Biol. Reprod. 72, 58–68. doi:10.1095/biolreprod.104.032987
Zhang, G., Kruse, T., Lopez-Mendez, B., Sylvestersen, K. B., Garvanska, D. H., Schopper, S., et al. (2017). Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat. Commun. 8, 15822. doi:10.1038/ncomms15822
Zhang, G., Lischetti, T., Hayward, D. G., and Nilsson, J. (2015). Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat. Commun. 6, 7162. doi:10.1038/ncomms8162
Zhang, G., Lischetti, T., and Nilsson, J. (2014). A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J. Cell Sci. 127, 871–884. doi:10.1242/jcs.139725
Zhang, Y., Young, R., Garvanska, D. H., Song, C., Zhai, Y., Wang, Y., et al. (2024). Functional analysis of Cdc20 reveals a critical role of CRY box in mitotic checkpoint signaling. Commun. Biol. 7, 164. doi:10.1038/s42003-024-05859-6
Zhao, G., Oztan, A., Ye, Y., and Schwarz, T. L. (2019). Kinetochore proteins have a post-mitotic function in neurodevelopment. Dev. Cell 48, 873–882. doi:10.1016/j.devcel.2019.02.003
Zhao, J., Li, H., Chen, G., Du, L., Xu, P., Zhang, X., et al. (2021). Aneuploid abortion correlates positively with MAD1 overexpression and miR-125b down-regulation. Mol. Cytogenet 14, 22. doi:10.1186/s13039-021-00538-1
Zhao, L., Xue, S., Yao, Z., Shi, J., Chen, B., Wu, L., et al. (2020). Biallelic mutations in CDC20 cause female infertility characterized by abnormalities in oocyte maturation and early embryonic development. Protein Cell 11, 921–927. doi:10.1007/s13238-020-00756-0
Zhu, M., Shahbazi, M., Martin, A., Zhang, C., Sozen, B., Borsos, M., et al. (2021). Human embryo polarization requires PLC signaling to mediate trophectoderm specification. Elife 10, e65068. doi:10.7554/eLife.65068
Keywords: spindle assembly checkpoint, embryogenesis, gametogenesis, mitotic checkpoint, mitosis
Citation: Pun R and North BJ (2025) Role of spindle assembly checkpoint proteins in gametogenesis and embryogenesis. Front. Cell Dev. Biol. 12:1491394. doi: 10.3389/fcell.2024.1491394
Received: 04 September 2024; Accepted: 17 December 2024;
Published: 22 January 2025.
Edited by:
Jiyue Zhu, Washington State University Health Sciences Spokane, United StatesReviewed by:
Gang Zhang, Qingdao University, ChinaCopyright © 2025 Pun and North. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian J. North, QnJpYW5Ob3J0aEBjcmVpZ2h0b24uZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.