- 1College of Animal Science, Jilin University, Changchun, China
- 2College of Public Health, Jilin University, Changchun, China
MicroRNAs let-7c and let-7f, two members of the let-7 family, were involved in regulating osteoblast differentiation and have an important role in bone formation. Let-7e-5p, which also belonged to the let-7 family, presented in the differentiation of adipose-derived stem cells and mouse embryonic stem cells. However, the role of let-7e-5p in osteoblast differentiation was unclear. Thus, this study aimed to elucidate the function of let-7e-5p in osteoblast differentiation and its mechanism. Firstly, we found that the let-7e-5p mimic promoted osteoblast differentiation but not the proliferation of MC3T3-E1 cells by positively regulating the expression levels of osteogenic-associated genes (RUNX2, OCN, OPN, and OSX), the activity of ALP, and formation of mineralized nodules. Moreover, we ascertained that the let-7e-5p mimic downregulated the post-transcriptional expression of SOCS1 by specifically binding to the 3′ untranslated region of SOCS1 mRNA. Also, let-7e-5p-induced SOCS1 downregulation increased the protein levels of p-STAT5 and IGF-1, which were both modulated by SOCS1 molecules. Furthermore, let-7e-5p abrogated the inhibition of osteogenic differentiation mediated by SOCS1 overexpression. Therefore, these results suggested that let-7e-5p regulated the differentiation of MC3T3-E1 cells through the JAK2/STAT5 pathway to upregulate IGF-1 gene expression by inhibiting SOCS1. These findings may provide a new insight into the regulatory role of let-7e-5p in osteogenic differentiation and imply the existence of a novel mechanism underlying let-7e-5p-mediated osteogenic differentiation.
Introduction
Bone repair in trauma, congenital malformations, infections, surgery, and radiotherapy largely depends on the capability of bone formation (Majidinia et al., 2018; Wang T. et al., 2020). Bone formation and resorption are affected by the balance between osteoblast and osteoclast activities, and osteoblast differentiation is a key stage in bone formation that is closely regulated by factors, such as microRNAs (miRNAs) (Dong et al., 2012; Chen et al., 2015; Castano et al., 2020). MiRNAs, which are small non-coding RNA molecules that participate in gene regulation (Ambros, 2004; Bartel, 2004), represent a potential class of bone repair therapeutics because they can enhance the capacity of osteoblast differentiation (Behera and Tyagi, 2018; Chen et al., 2019; Yang et al., 2019; Castano et al., 2020; Won Lee et al., 2020). Numerous miRNAs, including miR-196a, miR-125b, miR-141, miR-29b, miR-335-5p, and let-7, have an essential role in osteoblast differentiation (Mizuno et al., 2008; Itoh et al., 2009; Kim et al., 2009; Li et al., 2009; Zhang et al., 2011). Osteoblasts are the main cells responsible for bone formation through proliferation and differentiation (Salhotra et al., 2020), during which process, the MAPK/ERK (Sun et al., 2015; Lavoie et al., 2020) and PI3K/AKT (Liu et al., 2020; Meng et al., 2021) pathways participate in the regulation of cell proliferation. Type I collagen, osteopontin (OPN), osteocalcin (OCN), and alkaline phosphatase (ALP) are secreted by cells and have important roles in osteoblast differentiation (Long, 2011). In addition, several signaling pathways, including Wnt/β-catenin (Oh et al., 2020; Park et al., 2020), MAPK (Ge et al., 2007), and JAK/STAT (Levy et al., 1996; Joung et al., 2012, 2013; Darvin et al., 2013; Yu et al., 2018), promote osteogenesis in vivo and in vitro. Furthermore, the persistent activation of the JAK2/STAT5 signaling pathway increases osteoblastic differentiation (Joung et al., 2012, 2013; Darvin et al., 2013; Yu et al., 2018). Meanwhile, insulin-like growth factor 1 (IGF-1) is regulated by JAK2/STAT5, the downstream signaling pathway of GHR (Darvin et al., 2013). In this process, SOCS1 proteins bind to phosphoserine residues on JAK2 or GHR and then suppress GH signaling by inhibiting JAK2 activity or inducing GHR complex degradation (Pass et al., 2009). IGF-1 promotes bone repair by enhancing bone formation (Blumenfeld et al., 2002; Srouji et al., 2004; He et al., 2006). In vitro, IGF-1 is essential for the regulation of osteoblast differentiation (Darvin et al., 2013).
The let-7 family has 10 mature members that are produced from 13 precursor sequences, which are highly conserved across species in terms of sequence and function (Roush and Slack, 2008). An initial small-scale screening attempt using miRNA mimics showed that all let-7 family members can enhance the osteogenic differentiation in human adipose-derived mesenchymal stem cells, suggesting that these miRNAs have similar functions (Wei et al., 2014). Let-7c and let-7f all promote osteogenic differentiation and bone formation (Wei et al., 2014; Shen et al., 2019). However, the function of the other let-7 family members in bone formation has yet to be clarified. Let-7e-5p, which also belonged to the let-7 family, participated in the differentiation of adipose-derived stem cells (Ventayol et al., 2014) and mouse embryonic stem cells (Vinas et al., 2013). Thus, the potential functions of let-7e-5p in bone formation should be elucidated.
Therefore, this study aimed to explore the effect of let-7e-5p on the differentiation of osteoblasts and further clarify the molecular mechanism of this effect by detecting the signaling pathways that involve let-7e-5p. This work is expected to provide a new insight into the regulatory role of let-7e-5p in osteogenic differentiation.
Materials and Methods
Cell Culture
The mouse progenitor osteoblast cell line MC3T3-E1 was provided by the Hospital of Stomatology, Jilin University. The MC3T3-E1 cells were cultured in Dulbecco’s minimal essential medium (DMEM, Sigma, United States) supplemented with 10% fetal bovine serum (FBS, Hyclone, United States) and 1% streptomycin/penicillin (Gibco, United States) at 37°C in a humidified 5% CO2 incubator. The differentiation medium contained 10 mmol/L β-glycerophosphate (Sigma, United States), 50 mg/mL ascorbic acid, and 10 nM dexamethasone (Sigma, United States) (Wang C. et al., 2020; Wei et al., 2020). HEK293T cells were cultured in DMEM (Hyclone, China) supplemented with 10% FBS and 1% penicillin/streptomycin.
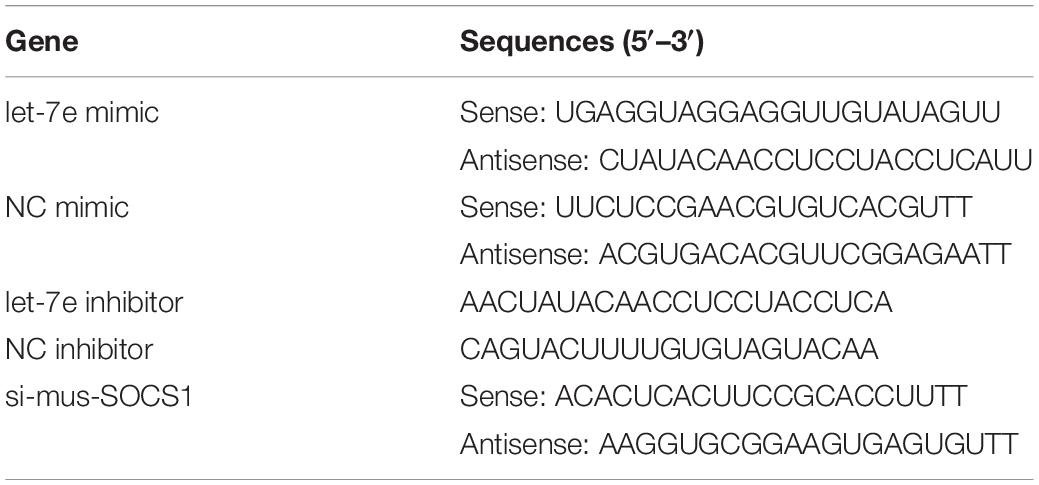
miRNA and siRNA Transfection
The let-7e-5p mimic; let-7e-5p inhibitor; mimic NC; inhibitor NC; and si-mus-SOCS1, the small interfering RNA used for the knockdown of SOCS1, were synthesized by Genepharma Co., Ltd (Suzhou, China). The sequences of these constructs are shown in Table 1. The nucleotides were transfected with LipoPlusTM Reagent (Sage creation, China) on the basis of the protocol of the manufacturer. MC3T3-E1 cells were seeded on a 6-cm-dish for 24 h before transfection and then transfected with the let-7e-5p mimic, let-7e-5p inhibitor, mimic NC, and inhibitor NC. The cells were then incubated in a complete culture medium or differentiation medium 6 h after transfection.
Cell Proliferation Assay
Cell Counting Kit-8 (Apexbio, United States) was used for the cell proliferation assay. Briefly, MC3T3-E1 cells were seeded at a density of 5.0 × 103 cells/well into 96-well plates containing 100 μL of culture medium. Then, the cells were cultured for 50 min in a complete culture medium with 10 μL of CCK-8 reagent at 37°C after transfection with the let-7e-5p mimic, let-7e-5p inhibitor, mimic NC, and inhibitor NC at 0, 24, 48, 72, and 96 h. Absorbance at a wavelength of 450 nm was measured by using a microplate reader (TECAN, Switzerland).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from the cells by using the RNAiso Plus reagent (Takara, United States) in accordance with the instructions of the manufacturer. Then, cDNA was generated with a reverse transcription kit. An ABI PRISM 7900HT thermocycler (Applied Biosystems, United States) was used for quantitative real-time PCR (qRT-PCR) with SYBR Select Master Mix (Roche). The primer sequences are displayed in Table 2. The relative expression levels of let-7e-5p and other genes were normalized against those of small nuclear RNA U6 and β-actin, respectively, by using the 2–ΔΔCt method.
Western Blot Analysis
A BCA protein assay kit (KeyGEN BioTECH, China) was applied to detect protein concentrations. Western blot assays were performed as previously reported (Wang C. et al., 2020). In this study, antibodies against AKT (1:2000, CST, United States), phosphor-AKT (1:2000, CST, United States), ERK1/2 (1:2000, CST, United States), and phosphor-ERK1/2 (1:2000, CST, United States) were used after 48 h of transfection to investigate the effects of let-7e-5p on MC3T3-E1 cell proliferation. For the evaluation of the effect of let-7e-5p on target gene expression, the antibody against SOCS1 (1:1000, Abcam, England) was used after 48 h of transfection. Antibodies against phosphor-STAT5 (1:1,000, Abcam, England), STAT5 (1:2,000, Abcam, England), OPN (1:500, WanLeiBio, China), collagen I (1:500, WanLeiBio, China), phosphor-JAK2 (1:500, WanLeiBio, China), and JAK2 (1:500, WanLeiBio, China) were used after 72 h of differentiation culture to evaluate the function of let-7e-5p in the osteogenic differentiation of MC3T3-E1 cells. The protein quantities detected by each antibody were analyzed by using the GenoSens gel analysis software and normalized on the basis of β-actin (1:2,000, BBI, China).
Measurement of ALP Activity
Confluent MC3T3-E1 cells were transfected with the let-7e-5p mimic, let-7e-5p inhibitor, and NC in 24-well plates. Six hours after transfection, the cells were incubated in differentiation media and finally collected on 3, 5, 7, 14, and 21 days post-transfection. An ALP kit (Nanjing Jiancheng, China) was utilized to measure ALP activities in accordance with the instructions of the manufacturer. A BCA protein assay kit (Beyotime, China) was used to detect the protein content on the basis of the protocols of the manufacturer. A microplate reader (TECAN, China) was applied to detect absorbance at 520 nm for the determination of enzyme activity. ALP activity was normalized by using the total protein content.
Alizarin Red Staining
Mineralization is the last stage of osteogenic differentiation, and the formation of mineralized nodules is a crucial marker of osteoblast mineralization (Wang C. et al., 2020). The chelation of Alizarin Red (AR) with the calcium ions that form mineralized nodules is used to determine the osteogenic mineralization capability in tissues or cells (Puchtler et al., 1969; Xi et al., 2016; Zheng et al., 2018). Confluent MC3T3-E1 cells were transfected with the let-7e-5p mimic, let-7e-5p inhibitor, and negative controls in a six-well plate. Six hours after transfection, the cells were cultured in differentiation media for 21 days. Then, AR staining (ARS) was performed as previously reported (Wang C. et al., 2020). The results were replicated in at least three independent experiments.
Bioinformatics Analysis
TargetScan1 and miRanda2 were applied to predict the target of let-7e-5p. In this research, SOCS1, one of the targets, was selected as the candidate. The secondary structure formed by the binding of let-7e-5p to the 3′ untranslated region (UTR) of the SOCS1 gene, and the minimum free energy (MFE) value were predicted by using mfold.3
Plasmid Construction and Transfection
In accordance with the seed sequence of let-7e-5p, the complementary oligonucleotides of the wild-type (WT) or seed-mutated (MUT) 3′UTR of the SOCS1 gene were synthesized by Genewiz (Tianjin, China) and inserted into a psiCHECKTM2 vector (Promega, United States) to construct the recombinant vectors. The constructs were named SOCS1-WT and SOCS1-MUT.
The full coding sequence of SOCS1 was amplified via PCR by using primers (forward 5′CGCGGATCCCTC GAGTAGGATGGTAGCACG3′; reverse 5′ CCGGATATCTCAA ATGAAGCCAGAGACCCTCC3′) and cloned into the pcDNA3.1 expression vector with BamHI and EcoRV. The SOCS1 overexpression vector was named pcDNA3.1-SOCS1. MC3T3-E1 cells were seeded onto a 6-cm dish or 24-well plates for 24 h before transfection to study the effect of SOCS1 in osteoblast differentiation. Then, pcDNA3.1, pcDNA3.1-SOCS1, the mixtures of pcDNA3.1 and mimic NC, let-7e-5p mimic and pcDNA3.1, or let-7e-5p mimic and pcDNA3.1-SOCS1 were transfected into MC3T3-E1 cells with the LipoPlusTM Reagent (Sage Creation, China). Six hours after transfection, the cells were incubated in a differentiation medium.
Dual-Luciferase Reporter Assay
HEK-293T cells were plated on 96-well culture plates at a density of 1 × 104 cells/well and cotransfected with SOCS1-WT and SOCS1-MUT and miRNA NC or mimic duplexes (10 pM) for 48 h with the LipoPlusTM Reagent (Sage creation, China). The cells were harvested, and luciferase activities were detected by the means of a Dual-Luciferase Reporter Assay system (TECAN, Switzerland) in accordance with the instructions of the manufacturer.
Statistics Analysis
All experimental results were shown as mean ± S.E.M with at least three independent replications. One-way and two-way ANOVA were applied to test the statistical differences among different groups. All statistical analyses were performed by using the GraphPad Prism 6.0. Here, p < 0.05 was regarded as statistically significant (∗p < 0.05, ∗∗p < 0.01).
Results
Let-7e-5p Did Not Affect the Proliferation of MC3T3-E1 Cells
Given that previous studies have shown that let-7e-5p has an important effect on the proliferation of many types of cancer cells (Lin et al., 2017; Li et al., 2018; Wang et al., 2019), the let-7e-5p mimic and inhibitor were first transfected into MC3T3-E1 cells, and cell proliferation was then detected. As expected, in the transfected MC3T3-E1 cells, the let-7e-5p mimic markedly increased the let-7e-5p levels compared with the mimic NC (p < 0.01), whereas the let-7e-5p inhibitor reduced the let-7e-5p levels compared with the inhibitor NC (p < 0.05) (Figure 1A). Furthermore, the increase and decrease in the let-7e-5p expression continued for 21 days (Figure 1A). Cell proliferation did not significantly differ after let-7e-5p mimic transfection (Figures 1B,C) (p > 0.05). Possible signaling pathways (MAPK/ERK and PI3K/AKT signaling pathways) were detected via Western blot analysis to further explore the effect of let-7e-5p on cell proliferation. The results demonstrated that the phosphorylation of AKT and ERK1/2 in the let-7e-5p mimic group did not significantly differ from that in the mimic NC group (Figure 2) (p > 0.05), suggesting that let-7e-5p did not enhance the growth potential of MC3T3-E1 cells.
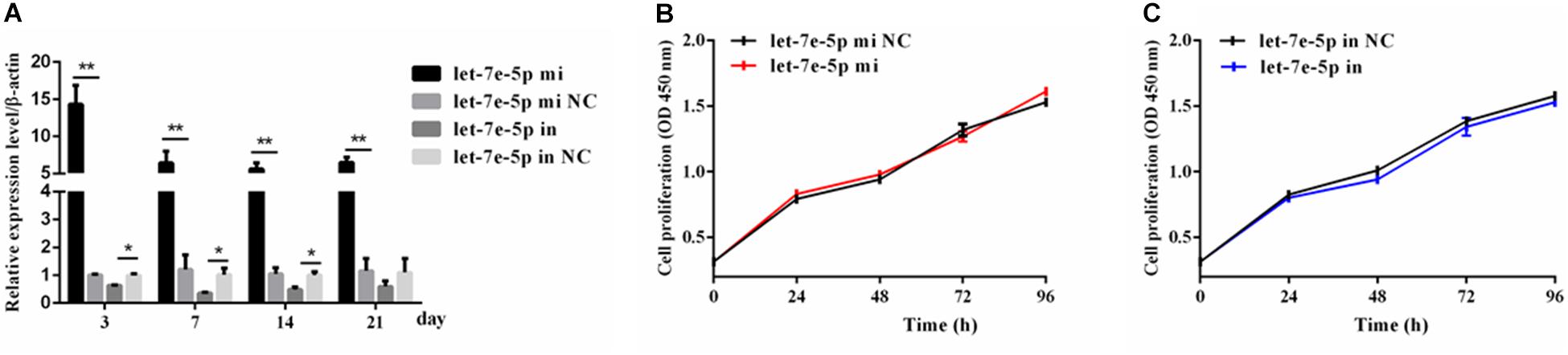
Figure 1. Let-7e-5p had no significant effect on MC3T3-E1 cell proliferation. (A) qRT-PCR detection of the transfection efficiency of the let-7e-5p mimic or inhibitor in MC3T3-E1 cells on days 3, 7, 14, and 21. (B,C) Proliferation of MC3T3-E1 cells transfected with the let-7e-5p mimic (B) or inhibitors (C) (*p < 0.05; **p < 0.01).
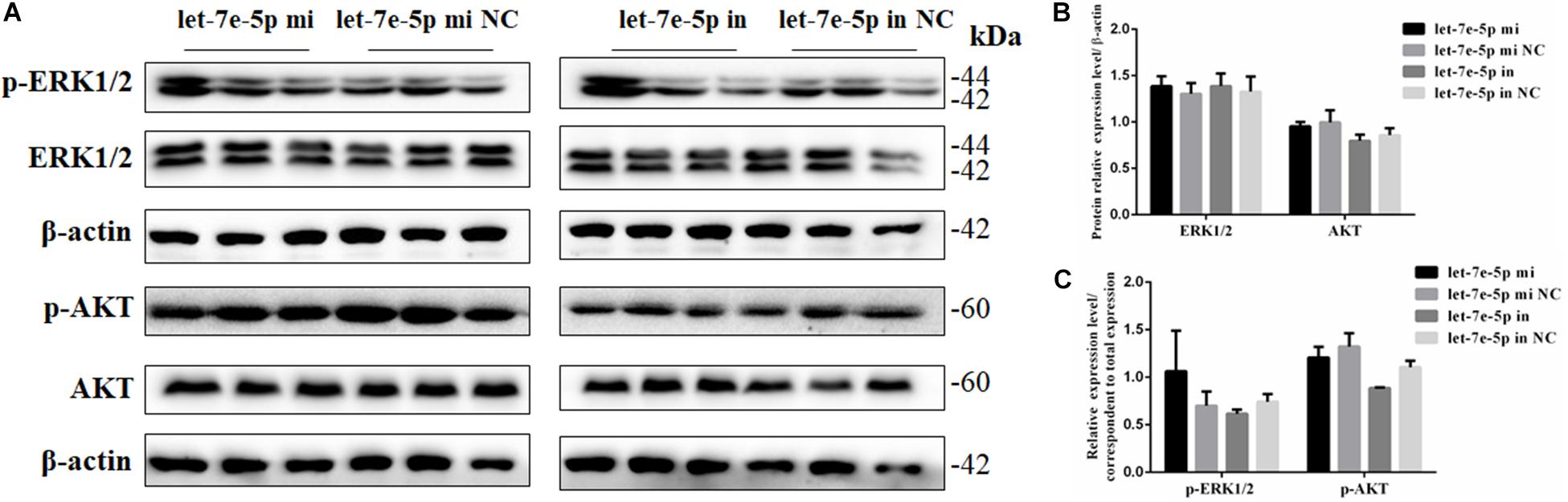
Figure 2. Let-7e-5p had no significant effect on proliferation-related signal pathways. (A) Western blot analysis results for p-ERK1/2, ERK1/2, p-AKT, and AKT protein expression following transfection with the let-7e-5p mimic or inhibitor. (B,C) Quantification of Western blot analysis results. Data are presented as the mean ± S.E.M of three lanes of the panel (∗p < 0.05).
Let-7e-5p Promoted the Osteogenic Differentiation of MC3T3-E1 Cells
The effect of let-7e-5p on osteoblast differentiation was further detected. ALP activity was used to assess the effect of let-7e-5p on cell differentiation because it is regarded as a marker of early osteogenic differentiation (Bancroft et al., 2002; Hosseini et al., 2019). In brief, the let-7e-5p mimic and inhibitor were first transfected into the MC3T3-E1 cells. The cells were then cultured in a differentiation medium, and ALP activities were separately measured on days 3, 5, 7, 14, and 21. The results showed that let-7e-5p significantly enhanced ALP activities compared with the mimic NC on day 5 (p < 0.05), day 14 (p < 0.01), and day 21 (p < 0.05); this effect was counteracted on day 14 by the let-7e-5p inhibitor (Figure 3A) (p < 0.05). The expression of osteogenic-differentiation-related genes in each group on day 3 after differentiation culture was detected. Comparison with the let-7e-5p mimic NC showed that the let-7e-5p mimic did not significantly influence the mRNA expression of collagen type I, alpha 1 (COL-1-α1) (Figure 3B) (p > 0.05), and instead significantly increased the mRNA expression of runt-related transcription factor 2 (RUNX2), OPN, OCN, and osterix (OSX) (Figure 3B) (p < 0.01). By contrast, the knockdown of let-7e-5p reduced OPN and RUNX2 expression (Figure 3B) (p < 0.05). The protein levels of COL-1 and OPN were further detected via Western blot analysis considering their vital roles in osteoblast differentiation (Chai et al., 2019; Hosseini et al., 2019). The results revealed that COL-1 was expressed at slightly higher levels in the let-7 mimic group than in the let-7e-5p mimic NC group (p > 0.05) (Figures 3C,D) and that the expression levels of OPN were significantly higher in the let-7 mimic group than in the let-7e-5p mimic NC group (Figures 3C,D) (p < 0.05). However, the knockdown of let-7e-5p reduced OPN expression (Figures 3C,E) (p < 0.05). In accordance with these results, ARS demonstrated that the staining intensity of mineralization nodes was markedly increased in the let-7e-5p mimic group (p < 0.01) and reduced in the let-7e-5p inhibitor group (Figures 3F,G) (p < 0.05). The consistency of these results with the results presented in the previous section indicated that let-7e-5p contributed a positive regulation potential to osteoblast differentiation.
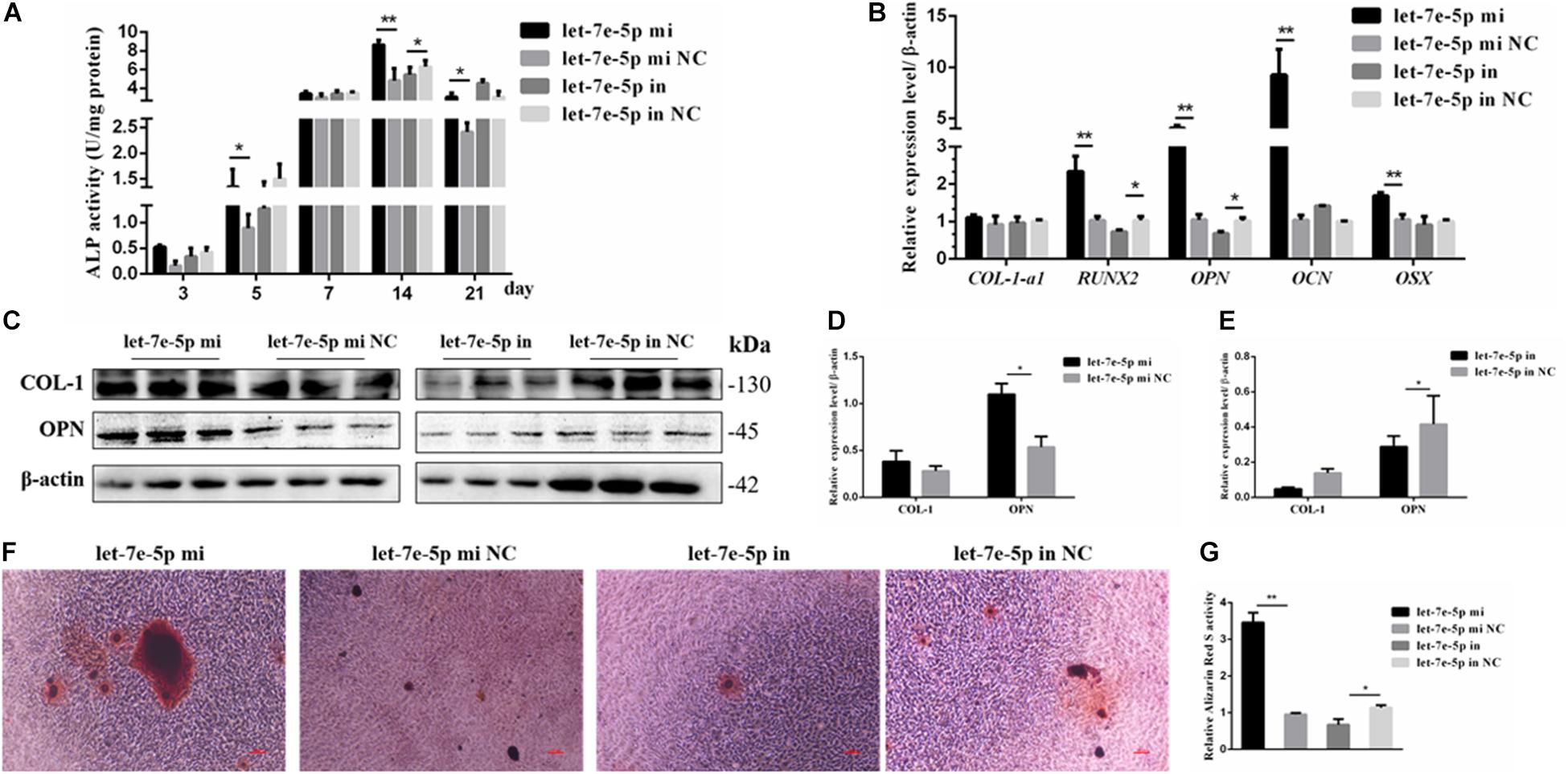
Figure 3. Let-7e-5p promoted the differentiation of MC3T3-E1 cells. (A) Effect of let-7e-5p on ALP activity in MC3T3-E1 cells on days 3, 5, 7, 14, and 21. (B) Effect of let-7e-5p on the mRNA levels of osteogenic-differentiation-related genes in MC3T3T-E1 cells. (C) Western blot analysis of COL-1 and OPN protein levels in MC3T3-E1 cells. (D,E) Columnar charts of (C) after grayscale analyses. Data are presented as the mean ± S.E.M of the quantified values of three lanes (*p < 0.05). (F) Effect of let-7e-5p on the mineralization of MC3T3-E1 cells on day 21. Images were captured with an inverted phase microscope (scale bar, 100 μm). (G) Mineralization results quantified with 10% cetylpyridinium chloride (**p < 0.01).
Let-7e-5p Directly Targeted the SOCS1 3′UTR and Inhibited SOCS1 Expression
TargetScan and RNAhybrid were used to predict and screen the let-7e-5p target genes to determine the potential target through which let-7e-5p exerts its effect on MC3T3-E1 cells. SOCS1, which is involved in the JAK2/STAT5 pathway, was selected as the candidate target in this research. The predictive software showed that the SOCS1 3′UTR contained putative binding sites for the let-7e-5p seed sequence (Figure 4A). The MFE formed by let-7e-5p and the SOCS1 gene was 19.0 kcal/mol. The secondary structure formed by the binding of let-7e-5p to the 3′UTR of the SOCS1 gene is shown in Figure 4B. The SOCS1-WT or SOCS1-MUT vectors and the let-7e-5p mimic or let-7e-5p mimic NC were cotransfected into HEK293T cells. The results showed that the luciferase activity of the cells transfected with the let-7e-5p mimic and SOCS1 3′UTR was markedly reduced compared with that of the cells cotransfected with the let-7e-5p mimic NC and SOCS1 3′UTR (p < 0.05) (Figure 4C). These results were consistent with previous findings showing that SOCS1 is a direct target gene of let-7e-5p in RAW264.7 cells (Li et al., 2021). Then, we explored the effect of the let-7e-5p mimic on the expression of the SOCS1 protein. Western blot analysis revealed that in contrast to the let-7e-5p mimic NC, the let-7e-5p mimic reduced SOCS1 protein levels in MC3T3-E1 cells; this effect was counteracted by the inhibitor (Figures 5A,B) (p < 0.05). This result was consistent with the relative luciferase activity measurements obtained in this work and in a previous study (Li et al., 2021).
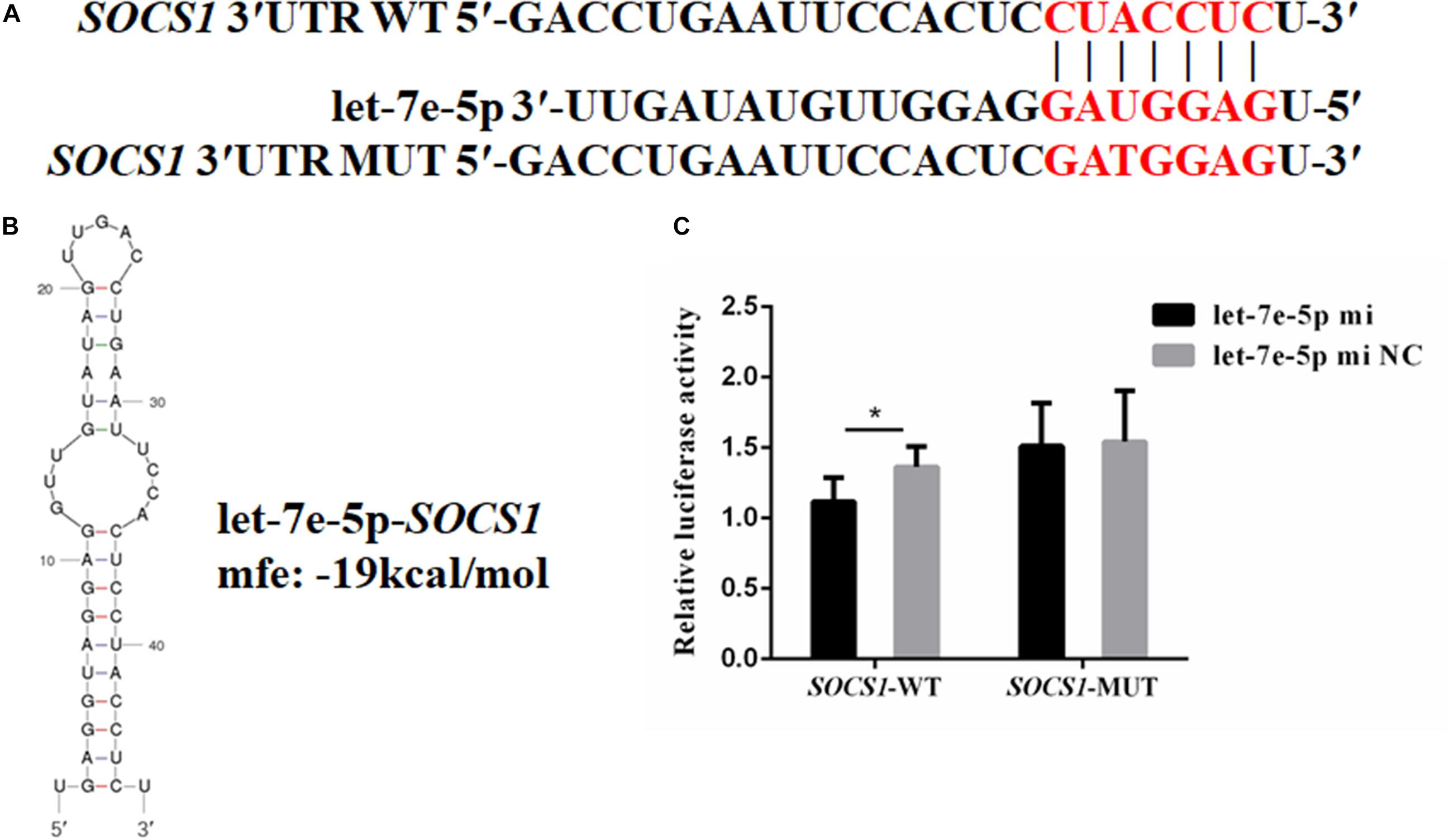
Figure 4. SOCS1 was the direct target gene of let-7e-5p. (A) Seed regions of let-7e-5p and the seed-recognizing sites in the 3′UTR (WT and MUT) of the SOCS1 gene. The seed-recognizing sites were marked in red font. (B) RNA secondary structure and MFE formed by the binding of let-7e-5p to the 3′UTR of the SOCS1 gene predicted by mfold and RNAhybrid. (C) Relative luciferase activities in HEK-293T cells cotransfected with the constructed SOCS1-WT and SOCS1-MUT vectors and the let-7e-5p mimic or let-7e-5p mimic NC. (*p < 0.05).
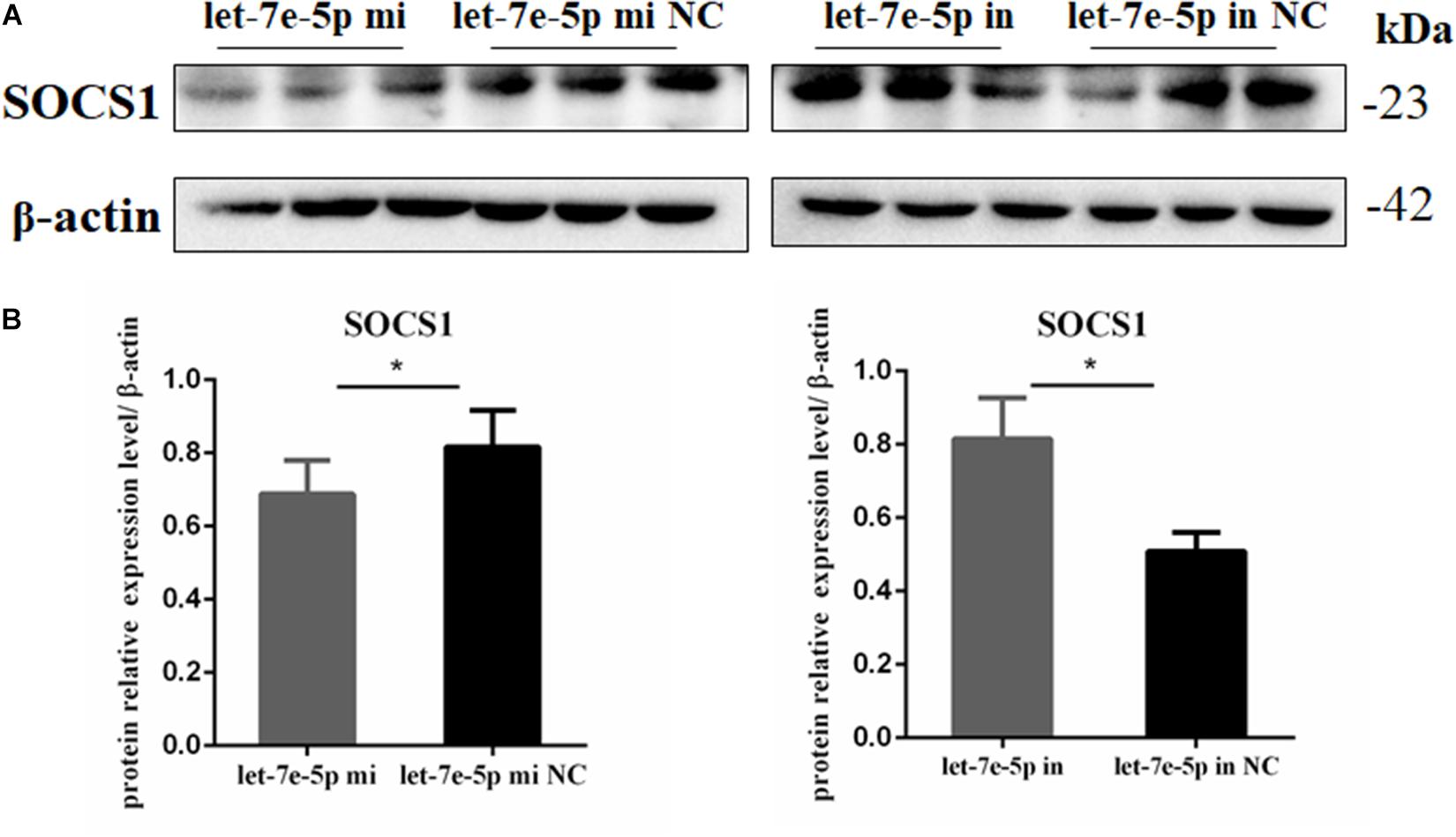
Figure 5. Let-7e-5p reduced the expression of SOCS1. (A) Western blot analysis results for SOCS1 expression in MC3T3-E1 cells transfected with the let-7e-5p mimic or inhibitor (*p < 0.05). (B) Quantification of Western blot analysis results. Data are presented as the mean ± S.E.M of the quantified values of three lanes (*p < 0.05).
SOCS1 Inhibited the Osteogenic Differentiation of MC3T3-E1 Cells
The efficiencies of the overexpression plasmid (pcDNA3.1-SOSC1) and si-SOCS1 were detected via qRT-PCR and Western blot analyses. The results confirmed that SOCS1 was successfully overexpressed and knocked-down in MC3T3-E1 cells (Figures 6A,G). The expression of osteogenic-differentiation-related genes in each group on day 3 after differentiation culture was detected. The results showed that compared with the si-NC, si-SOCS1 did not significantly influence the mRNA expression levels of RUNX2, OSX, and COL-1-a1 (Figures 6B,C,F) (p > 0.05) but significantly increased the mRNA expression levels of OCN and OPN (Figures 6D,E) (p < 0.01). Furthermore, compared with pc.DNA3.1, pc.DNA3.1-SOCS1 significantly reduced the mRNA expression levels of RUNX2, OCN, and OPN. The protein levels of COL-1 and OPN were further detected through Western blot analysis due to their vital roles in osteoblast differentiation (Chai et al., 2019; Hosseini et al., 2019). The results revealed that COL-1 expression in the si-SOCS1 and pc.DNA3.1-SOCS1 groups had slightly changed compared with that in the si-NC and pc.DNA3.1 groups (p > 0.05). OPN protein expression was inhibited by SOCS1 overexpression and upregulated after SOCS1 knockdown (Figures 6G–I) (p < 0.05). Also, the activity of ALP in MC3T3-E1 cells on day 14 was inhibited by SOCS1 overexpression and promoted after SOCS1 knockdown (Figure 6J) (p < 0.05). These results suggested that SOCS1 could serve as an inhibitor of osteoblast differentiation.
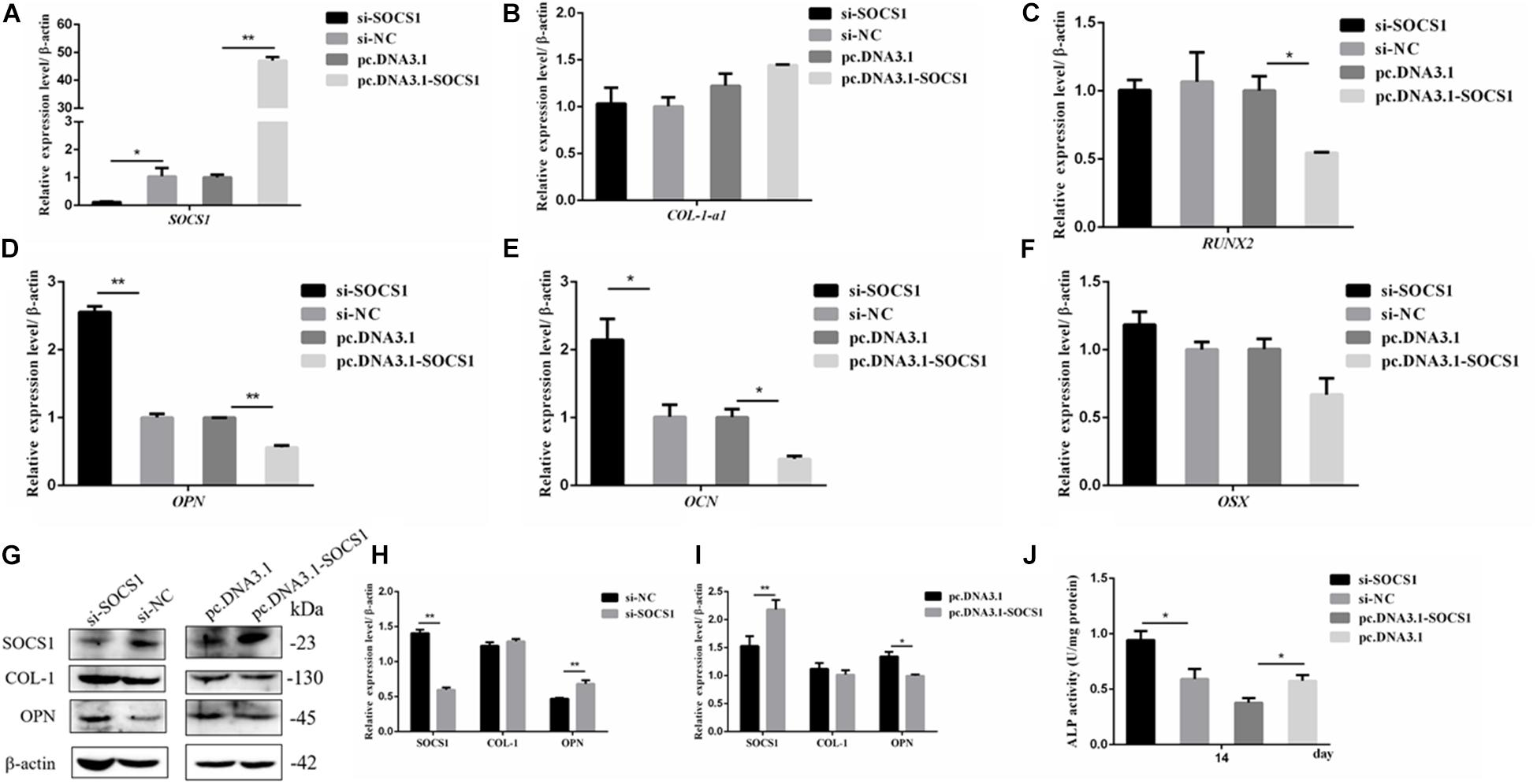
Figure 6. SOCS1 suppressed the differentiation of MC3T3-E1 cells. (A) SOCS1 mRNA level after SOCS1 overexpression and knockdown. (B–F) Effect of SOCS1 on ALP activity and mRNA amounts of differentiation-related genes. (G) Western blot analysis of COL-1 and OPN protein levels in MC3T3-E1 cells. (H,I) Columnar charts of (G) after grayscale analyses. Data are presented as the mean ± S.E.M of the values of three lanes (*p < 0.05). (J) Effect of SOCS1 on ALP activity in MC3T3-E1 cells on day 14. (*p < 0.05; **p < 0.01).
Let-7e-5p Regulated the JAK2/STAT5 Signal Pathway and IGF-1 Expression
Considering the effect of let-7e-5p on osteogenic differentiation, the possible signaling pathways (JAK2/STAT5 signaling pathways) involved in this effect were examined to investigate how let-7e-5p regulated the differentiation of MC3T3-E1 cells. The results demonstrated that compared with the let-7e-5p mimic NC, the let-7e-5p mimic significantly increased the phosphorylation of STAT5 (p < 0.05) but did not obviously increase the phosphorylation of JAK2 (Figure 7) (p > 0.05). These results suggested that let-7e-5p regulated osteogenic differentiation, at least partially, through the JAK2/STAT5 signaling pathway.
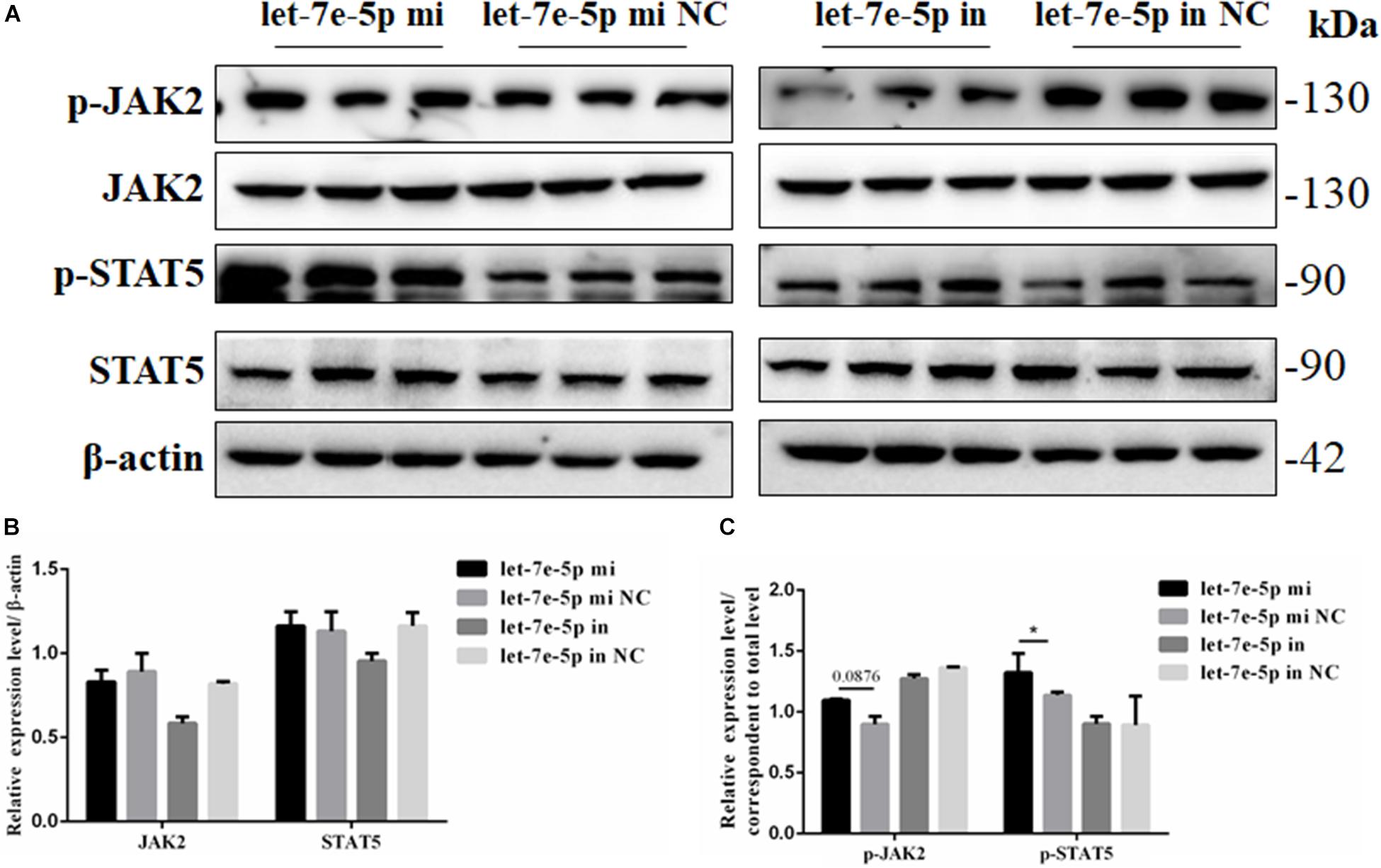
Figure 7. Let-7e-5p activated the differentiation-related signal pathway. (A) Western blot analysis results for p-JAK2, JAK2, p-STAT5, and STAT5 protein expression following transfection with the let-7e-5p mimic or inhibitor. (B,C) Quantification of the Western blot analysis results (*p < 0.05).
IGF-1 is regulated as a typical downstream target of STAT5 (Darvin et al., 2013). The mRNA and protein expression levels of IGF-1 were further detected through the transfection of the let-7e-5p mimic and inhibitor into MC3T3-E1 cells. The results showed that compared with the let-7e-5p mimic NC, the let-7e-5p mimic significantly increased the expression of IGF-1 mRNA (Figure 8A) (p < 0.05). Although the Western blot analysis revealed that IGF-1 protein expression was not obvious, the same trend was identified (Figures 8B,C) (p = 0.0768). These effects were counteracted by the inhibitors (Figures 8B,C) (p < 0.05).
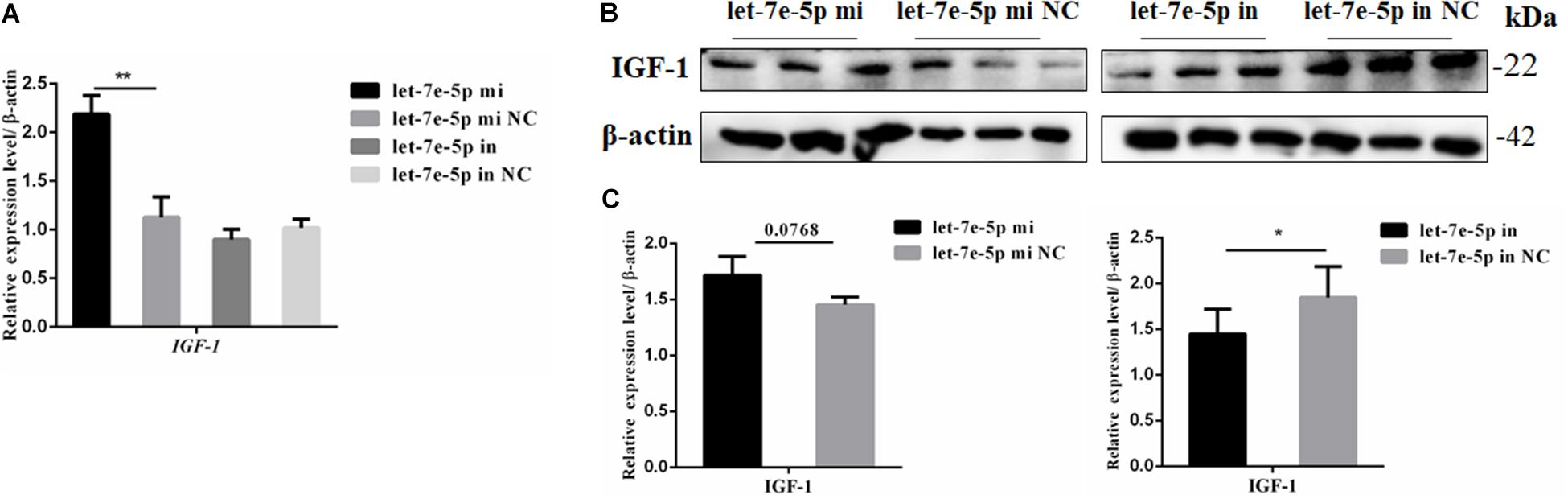
Figure 8. Let-7e-5p decreased the expression of IGF-1. (A) mRNA level of IGF-1 after transfection with the let-7e-5p mimic or inhibitor. (B) IGF-1 expression in MC3T3-E1 cells was determined via Western blot analysis after transfection with the let-7e-5p mimic and inhibitor. (C) Quantification of the Western blot analysis results (*p < 0.05; **p < 0.01).
SOCS1 Inhibited the Let-7e-5p-Induced Osteogenic Differentiation of MC3T3-E1 Cells
SOCS1 overexpression by using an expression vector was confirmed via qRT-PCR (Figure 9A) to further confirm that SOCS1 was involved in a let-7e-5p-induced osteoblast differentiation. Subsequently, MC3T3-E1 cells were cotransfected with the let-7e-5p mimic-NC and pc.DNA.3.1, let-7e-5p mimic and pc.DNA.3.1, or let-7e-5p mimic and pc.DNA.3.1-SOCS1 expression vector. The results showed that compared with the let-7e-5p mimic and pc.DNA.3.1, the let-7e-5p mimic and pc.DNA.3.1-SOCS1 did not significantly influence the expression of OSX and COL-1-a1 mRNA (Figures 9B,F) (p > 0.05) but instead significantly decreased the expression of RUNX2, OCN, and OPN mRNA and OPN protein (Figures 9C–E) (p < 0.01). As a result, the overexpression of SOCS1 inhibited the let-7e-5p-induced osteogenic differentiation of MC3T3-E1 cells. All these results further indicated that let-7e-5p mediated the induction of osteogenic differentiation by targeting SOCS1.
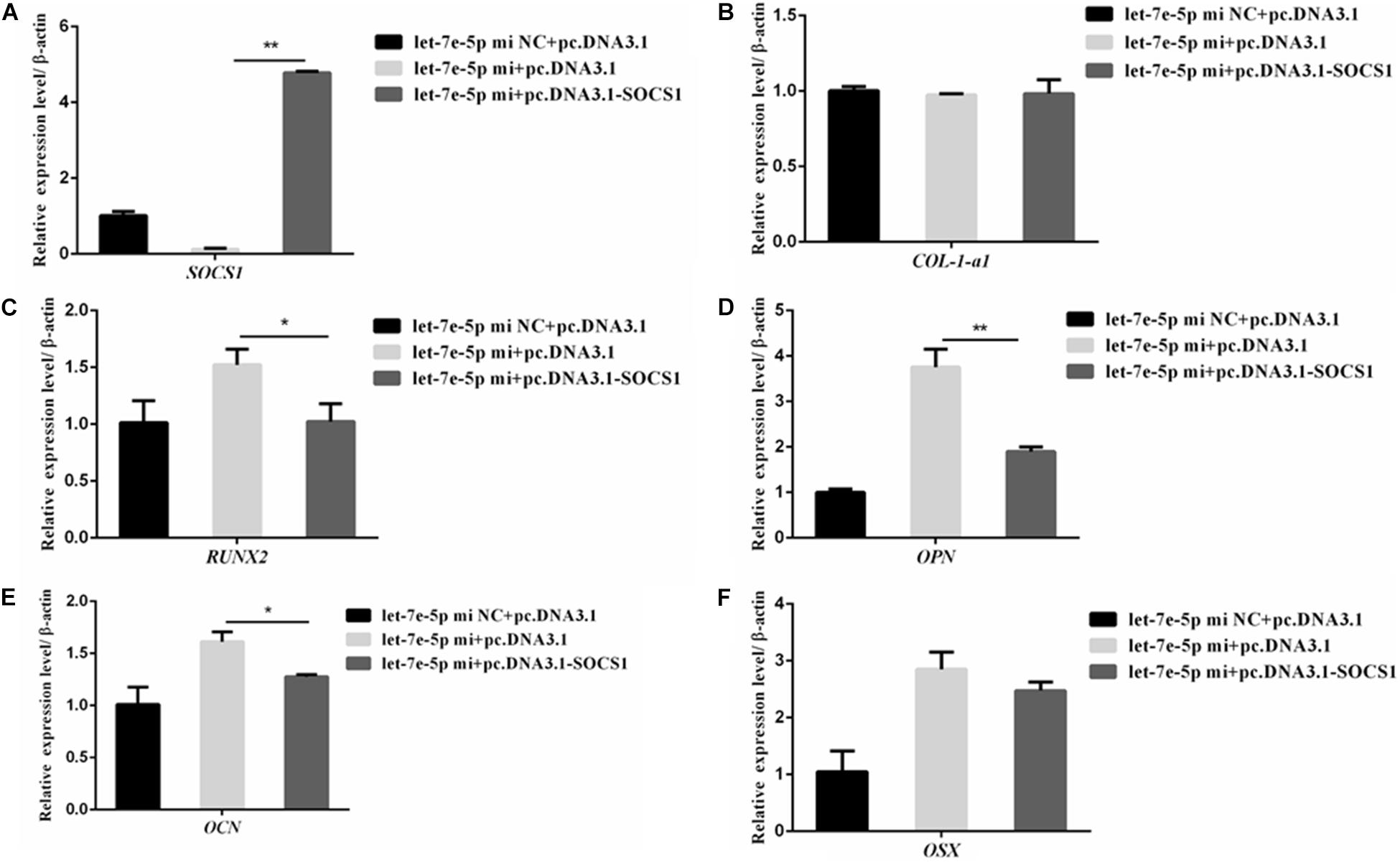
Figure 9. SOCS1 inhibited let-7e-5p-induced osteogenic differentiation in MC3T3-E1 cells. (A) mRNA level of SOCS1 after transfection with the let-7e-5p mimic NC and pc.DNA3.1, let-7e-5p mimic and pc.DNA3.1, or let-7e-5p mimic and pc.DNA3.1-SOCS1. (B–F) mRNA levels of osteogenic-differentiation-related genes after transfection with the let-7e-5p mimic NC and pc.DNA3.1, let-7e-5p mimic and pc.DNA3.1, or let-7e-5p mimic and pc.DNA3.1-SOCS1 (*p < 0.05; **p < 0.01).
Discussion
In recent years, miRNAs have been regarded as the pivotal regulators that participate in bone repair and regeneration caused by trauma, congenital malformations, infections, surgery, and radiotherapy (Zhang et al., 2017; Chen et al., 2019; Hosseinpour et al., 2019; Zhai et al., 2020). During these processes, many miRNAs exert a considerable effect on osteoblast differentiation and bone formation by increasing the activity of ALP and regulating the expression of genes related to osteogenic differentiation (COL-1-α1, OPN, OCN, RUNX2, and OSX) (Itoh et al., 2009; Kim et al., 2009; Li et al., 2009). The let-7 family, which consists of several members, has an important influence on various cellular activities, including the committed differentiation of multiple cell types (Wulczyn et al., 2007; Boyerinas et al., 2010; Eguchi et al., 2013). An initial small-scale screening study involving miRNA mimics indicated that all the members of the let-7 family can enhance the osteogenic differentiation of hADSCs, thus demonstrating that these miRNAs have similar functions; for example, let-7c promotes the osteogenic differentiation of hADSCs by increasing RUNX2, OSX, and ALP, and let-7f can rescue the Dex-inhibited osteogenic differentiation of murine BMSCs (Wei et al., 2014; Shen et al., 2019). Let-7e-5p is involved in the differentiation of adipose-derived stem cells (Ventayol et al., 2014) and mouse embryonic stem cells (Vinas et al., 2013), However, the effect and mechanism of let-7e-5p in osteogenic differentiation have yet to be clarified. We suspected that let-7e-5p likely plays a vital role in regulating the differentiation of osteoblasts and then participates in bone formation. The activity of ALP and the expression levels of genes related to osteogenic differentiation (RUNX2, OPN, OCN, and OSX) regulate osteoblast differentiation and bone formation in growing bones; the expression levels of the above genes gradually increase during osteoblast differentiation and are important factors for evaluating cell differentiation (Wang C. et al., 2020; Wei et al., 2020). Similar to the results of previous works, our results for the transfection of the let-7e-5p mimic confirmed that let-7e-5p enhanced the osteoblast differentiation potential by increasing the ALP activity (Xi et al., 2016) and regulating RUNX2, OCN, OPN, and OSX expression in MC3T3-E1 cells. The above results further verified that these miRNAs have similar functions (Roush and Slack, 2008). Further studies involving animal models are warranted to prove this conclusion.
The differentiation potential of osteoblasts is subject to various internal and external factors that are regulated by signaling pathways. These signaling pathways include the Wnt/β-catenin (Oh et al., 2020; Park et al., 2020), MAPK (Ge et al., 2007), and JAK/STAT (Levy et al., 1996; Joung et al., 2012, 2013; Darvin et al., 2013; Yu et al., 2018) pathways, which have all been reported to promote osteogenesis in vivo and in vitro. Furthermore, let-7 participates in the JAK2/STAT signaling pathway in cancer cells by targeting SOCS4 and activates the JAK1/STAT3 signaling pathway (Patel et al., 2014; Chen et al., 2017). Meanwhile, SOCS1 proteins participate in the JAK2/STAT signaling pathway by inhibiting JAK2 activity or inducing GHR complex degradation (Pass et al., 2009). The potential target gene of let-7e-5p was predicted via bioinformatics analysis to further explore the regulatory mechanism of let-7e-5p in osteogenic differentiation. The SOCS1 gene, which is involved in the JAK2/STAT5 pathway, was predicted to be targeted by let-7e-5p. This result was further confirmed through an analysis with the dual luciferase reporter system and was consistent with the findings of Wu et al., who found that SOCS-1 is the direct target gene of let-7e-5p (Li et al., 2021). A growing number of studies have revealed that SOCS1 negatively affects the downstream JAK2/STAT5 signaling pathway (Oh et al., 2009; Zhao et al., 2016; Liau et al., 2018; Zhang X. H. et al., 2018), and the reduction in SOCS1 expression enhances the expression of p-JAK2 and p-STAT5 (Zhao et al., 2016; Zhang X. H. et al., 2018). Furthermore, the JAK2/STAT5B pathway enhances the osteoblast differentiation of osteoblast-like cells and MSCs (Joung et al., 2012, 2013). In this study, we found that SOCS1 could suppress the differentiation of MC3T3-E1 cells by inhibiting RUNX2, OCN, and OPN expression and ALP activity. Meanwhile, transfection with the let-7e-5p mimic activated the JAK2/STAT5 pathway, suggesting that the activation of this pathway was related to let-7e-5p-regulated osteoblast differentiation. This result was consistent with the capability of let-7e-5p to reduce the expression of SOCS1. Moreover, the JAK2/STAT5 signaling pathway has considerable effects on the expression of various differentiation markers (Darvin et al., 2013; Yu et al., 2018), and the activation of the JAK2/STAT5 signaling pathway promotes the expression of IGF-1 (Moon et al., 2015; Zhang M. et al., 2018). The administration of IGF-1, a typical downstream target of STAT5, with nanoparticles to promote osteogenesis and angiogenesis, which play a pivotal role in the regulation of bone formation, has become increasingly practiced (Ishida et al., 2010; Zhang et al., 2020). IGF-1 treatment almost completely rescues all of the effects of GHR(–/–) on bone growth and remodeling, suggesting that IGF-1 directly affects the osteoblasts (Sims et al., 2000). As expected, our present findings demonstrated that let-7e-5p regulated osteogenic differentiation by inhibiting SOCS1. This effect resulted in the activation of the JAK2/STAT5 signaling pathway and then in the upregulation of IGF-1 expression.
In conclusion, let-7e-5p participated in osteogenic differentiation by mediating SOCS1 expression likely by promoting the JAK2/STAT5 signaling pathway activation and IGF-1 expression. Further studies involving animal models are warranted to prove this conclusion. The results of the present study may provide a new insight into the regulatory role of let-7e-5p in osteogenic differentiation.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
LH, CW, and SL conceived and designed the experiments. CW performed the experiments. CW and SL wrote the manuscript. All authors assessed the experiments, provided the data analysis, contributed to the manuscript, and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32072813) and the Natural Science Foundation of Jilin Province (20200201119JC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Bancroft, G. N., Sikavitsas, V. I., van den Dolder, J., Sheffield, T. L., Ambrose, C. G., Jansen, J. A., et al. (2002). Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 99, 12600–12605. doi: 10.1073/pnas.202296599
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/s0092-8674(04)00045-5
Behera, J., and Tyagi, N. (2018). Exosomes: mediators of bone diseases, protection, and therapeutics potential. Oncoscience 5, 181–195. doi: 10.18632/oncoscience.421
Blumenfeld, I., Srouji, S., Lanir, Y., Laufer, D., and Livne, E. (2002). Enhancement of bone defect healing in old rats by TGF-beta and IGF-1. Exp. Gerontol. 37, 553–565. doi: 10.1016/s0531-5565(01)00215-7
Boyerinas, B., Park, S. M., Hau, A., Murmann, A. E., and Peter, M. E. (2010). The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 17, F19–F36. doi: 10.1677/ERC-09-0184
Castano, I. M., Raftery, R. M., Chen, G., Cavanagh, B., Quinn, B., Duffy, G. P., et al. (2020). Rapid bone repair with the recruitment of CD206(+)M2-like macrophages using non-viral scaffold-mediated miR-133a inhibition of host cells. Acta Biomater. 109, 267–279. doi: 10.1016/j.actbio.2020.03.042
Chai, X., Zhang, W., Chang, B., Feng, X., Song, J., Li, L., et al. (2019). GPR39 agonist TC-G 1008 promotes osteoblast differentiation and mineralization in MC3T3-E1 cells. Artif. Cells Nanomed. Biotechnol. 47, 3569–3576. doi: 10.1080/21691401.2019.1649270
Chen, C. C., You, J. Y., Lung, J., Huang, C. E., Chen, Y. Y., Leu, Y. W., et al. (2017). Aberrant let7a/HMGA2 signaling activity with unique clinical phenotype in JAK2-mutated myeloproliferative neoplasms. Haematologica 102, 509–518. doi: 10.3324/haematol.2016.154385
Chen, J., Qiu, M., Dou, C., Cao, Z., and Dong, S. (2015). MicroRNAs in bone balance and osteoporosis. Drug. Dev. Res. 76, 235–245. doi: 10.1002/ddr.21260
Chen, S., Tang, Y., Liu, Y., Zhang, P., Lv, L., Zhang, X., et al. (2019). Exosomes derived from miR-375-overexpressing human adipose mesenchymal stem cells promote bone regeneration. Cell Prolif. 52:e12669. doi: 10.1111/cpr.12669
Darvin, P., Joung, Y. H., and Yang, Y. M. (2013). JAK2-STAT5B pathway and osteoblast differentiation. JAKSTAT 2:e24931. doi: 10.4161/jkst.24931
Dong, S., Yang, B., Guo, H., and Kang, F. (2012). MicroRNAs regulate osteogenesis and chondrogenesis. Biochem. Biophys. Res. Commun. 418, 587–591. doi: 10.1016/j.bbrc.2012.01.075
Eguchi, T., Watanabe, K., Hara, E. S., Ono, M., Kuboki, T., and Calderwood, S. K. (2013). OstemiR: a novel panel of microRNA biomarkers in osteoblastic and osteocytic differentiation from mesencymal stem cells. PLoS One 8:e58796. doi: 10.1371/journal.pone.0058796
Ge, C., Xiao, G., Jiang, D., and Franceschi, R. T. (2007). Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J. Cell. Biol. 176, 709–718. doi: 10.1083/jcb.200610046
He, J., Rosen, C. J., Adams, D. J., and Kream, B. E. (2006). Postnatal growth and bone mass in mice with IGF-I haploinsufficiency. Bone 38, 826–835. doi: 10.1016/j.bone.2005.11.021
Hosseini, S., Naderi-Manesh, H., Vali, H., Baghaban Eslaminejad, M., Azam Sayahpour, F., Sheibani, S., et al. (2019). Contribution of osteocalcin-mimetic peptide enhances osteogenic activity and extracellular matrix mineralization of human osteoblast-like cells. Colloids Surf. B Biointerfaces 173, 662–671. doi: 10.1016/j.colsurfb.2018.10.035
Hosseinpour, S., He, Y., Nanda, A., and Ye, Q. (2019). MicroRNAs involved in the regulation of angiogenesis in bone regeneration. Calcif. Tissue Int. 105, 223–238. doi: 10.1007/s00223-019-00571-8
Ishida, K., Matsumoto, T., Sasaki, K., Mifune, Y., Tei, K., Kubo, S., et al. (2010). Bone regeneration properties of granulocyte colony-stimulating factor via neovascularization and osteogenesis. Tissue Eng. Part A 16, 3271–3284. doi: 10.1089/ten.tea.2009.0268
Itoh, T., Nozawa, Y., and Akao, Y. (2009). MicroRNA-141 and -200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J. Biol. Chem. 284, 19272–19279. doi: 10.1074/jbc.M109.014001
Joung, Y. H., Lim, E. J., Darvin, P., Chung, S. C., Jang, J. W., Do Park, K., et al. (2012). MSM enhances GH signaling via the Jak2/STAT5b pathway in osteoblast-like cells and osteoblast differentiation through the activation of STAT5b in MSCs. PLoS One 7:e47477. doi: 10.1371/journal.pone.0047477
Joung, Y. H., Lim, E. J., Darvin, P., Jang, J. W., Park, K. D., Lee, H. K., et al. (2013). Hwanggeumchal sorghum extract enhances BMP7 and GH signaling through the activation of Jak2/STAT5B in MC3T3E1 osteoblastic cells. Mol. Med. Rep. 8, 891–896. doi: 10.3892/mmr.2013.1593
Kim, Y. J., Bae, S. W., Yu, S. S., Bae, Y. C., and Jung, J. S. (2009). miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner Res. 24, 816–825. doi: 10.1359/jbmr.081230
Lavoie, H., Gagnon, J., and Therrien, M. (2020). ERK signalling: a master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell. Biol. 21, 607–632. doi: 10.1038/s41580-020-0255-7
Levy, J. B., Schindler, C., Raz, R., Levy, D. E., Baron, R., and Horowitz, M. C. (1996). Activation of the JAK-STAT signal transduction pathway by oncostatin-M cultured human and mouse osteoblastic cells. Endocrinology 137, 1159–1165. doi: 10.1210/endo.137.4.8625884
Li, W., Zhang, W., Liu, J., Han, Y., Jiang, H., Ji, G., et al. (2021). Down-regulation of miR-let-7e attenuates LPS-induced acute lung injury in mice via inhibiting pulmonary inflammation by targeting SCOS1/NF-kappaB pathway. Biosci. Rep. 41:BSR20201089. doi: 10.1042/BSR20201089
Li, Z., Hassan, M. Q., Jafferji, M., Aqeilan, R. I., Garzon, R., Croce, C. M., et al. (2009). Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 284, 15676–15684. doi: 10.1074/jbc.M809787200
Li, Z., Pan, W., Shen, Y., Chen, Z., Zhang, L., Zhang, Y., et al. (2018). IGF1/IGF1R and microRNA let-7e down-regulate each other and modulate proliferation and migration of colorectal cancer cells. Cell Cycle 17, 1212–1219. doi: 10.1080/15384101.2018.1469873
Liau, N. P. D., Laktyushin, A., Lucet, I. S., Murphy, J. M., Yao, S., Whitlock, E., et al. (2018). The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 9:1558. doi: 10.1038/s41467-018-04013-1
Lin, Z., Ge, J., Wang, Z., Ren, J., Wang, X., Xiong, H., et al. (2017). Let-7e modulates the inflammatory response in vascular endothelial cells through ceRNA crosstalk. Sci. Rep. 7:42498. doi: 10.1038/srep42498
Liu, P., Chen, H., Yan, L., and Sun, Y. (2020). Laminin alpha5 modulates fibroblast proliferation in epidural fibrosis through the PI3K/AKT/mTOR signaling pathway. Mol. Med. Rep. 21, 1491–1500. doi: 10.3892/mmr.2020.10967
Long, F. (2011). Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell. Biol. 13, 27–38. doi: 10.1038/nrm3254
Majidinia, M., Sadeghpour, A., and Yousefi, B. (2018). The roles of signaling pathways in bone repair and regeneration. J. Cell. Physiol. 233, 2937–2948. doi: 10.1002/jcp.26042
Meng, X., Sun, R., Wang, W., Zhang, N., Cao, S., Liu, B., et al. (2021). ADFP promotes cell proliferation in lung adenocarcinoma via Akt phosphorylation. J. Cell. Mol. Med. 25, 827–839. doi: 10.1111/jcmm.16136
Mizuno, Y., Yagi, K., Tokuzawa, Y., Kanesaki-Yatsuka, Y., Suda, T., Katagiri, T., et al. (2008). miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 368, 267–272. doi: 10.1016/j.bbrc.2008.01.073
Moon, P. D., Kim, M. H., Oh, H. A., Nam, S. Y., Han, N. R., Jeong, H. J., et al. (2015). Cysteine induces longitudinal bone growth in mice by upregulating IGF-I. Int. J. Mol. Med. 36, 571–576. doi: 10.3892/ijmm.2015.2257
Oh, J., Hur, M. W., and Lee, C. E. (2009). SOCS1 protects protein tyrosine phosphatases by thioredoxin upregulation and attenuates Jaks to suppress ROS-mediated apoptosis. Oncogene 28, 3145–3156. doi: 10.1038/onc.2009.169
Oh, Y., Ahn, C. B., and Je, J. Y. (2020). Blue mussel-derived peptides PIISVYWK and FSVVPSPK trigger Wnt/beta-catenin signaling-mediated osteogenesis in human bone marrow mesenchymal stem cells. Mar. Drugs. 18:510. doi: 10.3390/md18100510
Park, K. R., Kwon, Y. J., Park, J. E., and Yun, H. M. (2020). 7-HYB, a phenolic compound isolated from myristica fragrans houtt increases cell migration, osteoblast differentiation, and mineralization through BMP2 and beta-catenin signaling. Int. J. Mol. Sci. 21:8059. doi: 10.3390/ijms21218059
Pass, C., MacRae, V. E., Ahmed, S. F., and Farquharson, C. (2009). Inflammatory cytokines and the GH/IGF-I axis: novel actions on bone growth. Cell Biochem. Funct. 27, 119–127. doi: 10.1002/cbf.1551
Patel, K., Kollory, A., Takashima, A., Sarkar, S., Faller, D. V., and Ghosh, S. K. (2014). MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. 347, 54–64. doi: 10.1016/j.canlet.2014.01.020
Puchtler, H., Meloan, S. N., and Terry, M. S. (1969). On the history and mechanism of alizarin and alizarin red S stains for calcium. J. Histochem. Cytochem. 17, 110–124. doi: 10.1177/17.2.110
Roush, S., and Slack, F. J. (2008). The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516. doi: 10.1016/j.tcb.2008.07.007
Salhotra, A., Shah, H. N., Levi, B., and Longaker, M. T. (2020). Mechanisms of bone development and repair. Nat. Rev. Mol. Cell. Biol. 21, 696–711. doi: 10.1038/s41580-020-00279-w
Shen, G. Y., Ren, H., Shang, Q., Zhao, W. H., Zhang, Z. D., Yu, X., et al. (2019). Let-7f-5p regulates TGFBR1 in glucocorticoid-inhibited osteoblast differentiation and ameliorates glucocorticoid-induced bone loss. Int. J. Biol. Sci. 15, 2182–2197. doi: 10.7150/ijbs.33490
Sims, N. A., Clement-Lacroix, P., Da Ponte, F., Bouali, Y., Binart, N., Moriggl, R., et al. (2000). Bone homeostasis in growth hormone receptor-null mice is restored by IGF-I but independent of Stat5. J. Clin. Invest. 106, 1095–1103. doi: 10.1172/JCI10753
Srouji, S., Blumenfeld, I., Rachmiel, A., and Livne, E. (2004). Bone defect repair in rat tibia by TGF-beta1 and IGF-1 released from hydrogel scaffold. Cell Tissue Bank 5, 223–230. doi: 10.1007/s10561-004-0503-7
Sun, Y., Liu, W. Z., Liu, T., Feng, X., Yang, N., and Zhou, H. F. (2015). Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 35, 600–604. doi: 10.3109/10799893.2015.1030412
Ventayol, M., Vinas, J. L., Sola, A., Jung, M., Brune, B., Pi, F., et al. (2014). miRNA let-7e targeting MMP9 is involved in adipose-derived stem cell differentiation toward epithelia. Cell Death Dis. 5:e1048. doi: 10.1038/cddis.2014.2
Vinas, J. L., Ventayol, M., Brune, B., Jung, M., Sola, A., Pi, F., et al. (2013). miRNA let-7e modulates the Wnt pathway and early nephrogenic markers in mouse embryonic stem cell differentiation. PLoS One 8:e60937. doi: 10.1371/journal.pone.0060937
Wang, C., Wang, S., Liu, S., Cheng, Y., Geng, H., Yang, R., et al. (2020). Synonymous mutations of porcine igf1r extracellular domain affect differentiation and mineralization in MC3T3-E1 cells. Front. Cell Dev. Biol. 8:623. doi: 10.3389/fcell.2020.00623
Wang, S., Jin, S., Liu, M. D., Pang, P., Wu, H., Qi, Z. Z., et al. (2019). Hsa-let-7e-5p inhibits the proliferation and metastasis of head and neck squamous cell carcinoma cells by targeting chemokine receptor 7. J. Cancer 10, 1941–1948. doi: 10.7150/jca.29536
Wang, T., Zhang, C., Wu, C., Liu, J., Yu, H., Zhou, X., et al. (2020). miR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling pathway. Stem. Cell. Res. Ther. 11:62. doi: 10.1186/s13287-020-1579-0
Wei, J., Li, H., Wang, S., Li, T., Fan, J., Liang, X., et al. (2014). let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem. Cells Dev. 23, 1452–1463. doi: 10.1089/scd.2013.0600
Wei, W., Liu, S., Song, J., Feng, T., Yang, R., Cheng, Y., et al. (2020). MGF-19E peptide promoted proliferation, differentiation and mineralization of MC3T3-E1 cell and promoted bone defect healing. Gene 749:144703. doi: 10.1016/j.gene.2020.144703
Won Lee, G., Thangavelu, M., Joung Choi, M., Yeong Shin, E., Sol Kim, H., Seon Baek, J., et al. (2020). Exosome mediated transfer of miRNA-140 promotes enhanced chondrogenic differentiation of bone marrow stem cells for enhanced cartilage repair and regeneration. J. Cell. Biochem. 121, 3642–3652. doi: 10.1002/jcb.29657
Wulczyn, F. G., Smirnova, L., Rybak, A., Brandt, C., Kwidzinski, E., Ninnemann, O., et al. (2007). Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 21, 415–426. doi: 10.1096/fj.06-6130com
Xi, G., Rosen, C. J., and Clemmons, D. R. (2016). IGF-I and IGFBP-2 stimulate AMPK activation and autophagy, which are required for osteoblast differentiation. Endocrinology 157, 268–281. doi: 10.1210/en.2015-1690
Yang, C., Liu, X., Zhao, K., Zhu, Y., Hu, B., Zhou, Y., et al. (2019). miRNA-21 promotes osteogenesis via the PTEN/PI3K/Akt/HIF-1alpha pathway and enhances bone regeneration in critical size defects. Stem. Cell. Res. Ther. 10:65. doi: 10.1186/s13287-019-1168-2
Yu, X., Li, Z., Wan, Q., Cheng, X., Zhang, J., Pathak, J. L., et al. (2018). Inhibition of JAK2/STAT3 signaling suppresses bone marrow stromal cells proliferation and osteogenic differentiation, and impairs bone defect healing. Biol. Chem. 399, 1313–1323. doi: 10.1515/hsz-2018-0253
Zhai, M., Zhu, Y., Yang, M., and Mao, C. (2020). Human mesenchymal stem cell derived exosomes enhance cell-free bone regeneration by altering their miRNAs profiles. Adv. Sci. 7:2001334. doi: 10.1002/advs.202001334
Zhang, J., Tu, Q., Bonewald, L. F., He, X., Stein, G., Lian, J., et al. (2011). Effects of miR-335-5p in modulating osteogenic differentiation by specifically downregulating Wnt antagonist DKK1. J. Bone Miner Res. 26, 1953–1963. doi: 10.1002/jbmr.377
Zhang, L., Tang, Y., Zhu, X., Tu, T., Sui, L., Han, Q., et al. (2017). Overexpression of MiR-335-5p promotes bone formation and regeneration in mice. J. Bone Miner Res. 32, 2466–2475. doi: 10.1002/jbmr.3230
Zhang, M., Xu, J., Wang, T., Wan, X., Zhang, F., Wang, L., et al. (2018). The dipeptide pro-gly promotes IGF-1 expression and secretion in HepG2 and female mice via PepT1-JAK2/STAT5 pathway. Front. Endocrinol. 9:424. doi: 10.3389/fendo.2018.00424
Zhang, X., Xing, H., Qi, F., Liu, H., Gao, L., and Wang, X. (2020). Local delivery of insulin/IGF-1 for bone regeneration: carriers, strategies, and effects. Nanotheranostics 4, 242–255. doi: 10.7150/ntno.46408
Zhang, X. H., Yang, L., Liu, X. J., Zhan, Y., Pan, Y. X., Wang, X. Z., et al. (2018). Association between methylation of tumor suppressor gene SOCS1 and acute myeloid leukemia. Oncol. Rep. 40, 1008–1016. doi: 10.3892/or.2018.6508
Zhao, H. M., Xu, R., Huang, X. Y., Cheng, S. M., Huang, M. F., Yue, H. Y., et al. (2016). Curcumin suppressed activation of dendritic cells via JAK/STAT/SOCS signal in mice with experimental colitis. Front. Pharmacol. 7:455. doi: 10.3389/fphar.2016.00455
Keywords: bone defects, let-7e-5p, osteoblast differentiation, IGF-1, JAK2/STAT5
Citation: Wang C, Liu S, Li J, Cheng Y, Wang Z, Feng T, Lu G, Wang S, Song J, Xia P and Hao L (2021) Biological Functions of Let-7e-5p in Promoting the Differentiation of MC3T3-E1 Cells. Front. Cell Dev. Biol. 9:671170. doi: 10.3389/fcell.2021.671170
Received: 24 February 2021; Accepted: 03 August 2021;
Published: 08 September 2021.
Edited by:
Katiucia Batista Silva Paiva, University of São Paulo, BrazilReviewed by:
Quamarul Hassan, University of Alabama at Birmingham, United StatesAbbas Jafari, University of Copenhagen, Denmark
Copyright © 2021 Wang, Liu, Li, Cheng, Wang, Feng, Lu, Wang, Song, Xia and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linlin Hao, aGFvbGlubGluQGpsdS5lZHUuY24=
†These authors have contributed equally to this work
 Chunli Wang1†
Chunli Wang1† Songcai Liu
Songcai Liu Linlin Hao
Linlin Hao