- 1Department of Solar Materials, Helmholtz Centre for Environmental Research GmbH – UFZ, Leipzig, Germany
- 2Engineering and Technology, Bayer AG, Leverkusen, Germany
The implementation of biocatalytic steroid hydroxylation processes at an industrial scale still suffers from low conversion rates. In this study, we selected variants of the self-sufficient cytochrome P450 monooxygenase BM3 from Bacillus megaterium (BM3) for the hydroxylation of testosterone either at the 2β- or 15β-position. Recombinant Escherichia coli cells were used as biocatalysts to provide a protective environment for recombinant enzymes and to ensure continuous cofactor recycling via glucose catabolism. However, only low initial whole-cell testosterone conversion rates were observed for resting cells. Results obtained with different biocatalyst formats (permeabilized cells, cell-free extracts, whole cells) indicated a limitation in substrate uptake, most likely due to the hydrophilic character of the outer membrane of E. coli. Thus, we co-expressed nine genes encoding hydrophobic outer membrane proteins potentially facilitating steroid uptake. Indeed, the application of four candidates led to increased initial testosterone hydroxylation rates. Respective whole-cell biocatalysts even exceeded activities obtained with permeabilized cells or cell-free extracts. The highest activity of 34 U gCDW−1 was obtained for a strain containing the hydrophobic outer membrane protein AlkL from Pseudomonas putida GPo1 and the BM3 variant KSA14m. Overall, we show that the straightforward application of hydrophobic outer membrane pores can boost whole-cell steroid conversion rates and thus be game-changing with regard to industrial steroid production efficiency.
Introduction
Steroid-based drugs account for one of the largest and still expanding sectors in the pharmaceutical industry (Tong and Dong, 2009). Their broad spectrum of pharmaceutical activities is reflected in the prominent role they play, e.g., in contraceptives, anti-inflammatory agents, or cancer treatments (Bureik and Bernhardt, 2007; Donova and Egorova, 2012; Donova, 2017). Hydroxylated steroid derivatives frequently show higher biological activities than their more hydrophobic non-hydroxylated analogs, rendering steroid hydroxylations particularly relevant (Donova and Egorova, 2012). As number and position of hydroxyl groups are decisive for the biological activity of steroids, highly selective hydroxylation is crucial (Bureik and Bernhardt, 2007; Donova and Egorova, 2012). Desired reactions often are not accessible by chemical means, making biocatalysis a promising highly selective and often eco-efficient alternative for steroid functionalization (Bureik and Bernhardt, 2007; Julsing et al., 2008; Donova, 2017).
Several microorganisms show native steroid-modifying activities and are regularly applied at industrial scale. However, respective processes typically suffer from low productivities, defined as amount of product produced per time and volume, and undesired side-reactions (Bureik and Bernhardt, 2007; Fernández-Cabezón et al., 2018). Thus, enzyme discovery as well as enzyme and strain engineering have been of immense interest in recent decades (Donova, 2017; Fernández-Cabezón et al., 2018), giving rise to a huge variety of steroid hydroxylating cytochrome P450 monooxygenases (CYP450s) covering virtually all possible specificities (Bernhardt, 2006; Bureik and Bernhardt, 2007; Donova, 2017; Szaleniec et al., 2018).
Employing CYP450s of mammalian origin appears attractive due to their natural role in steroid synthesis and conversion (Donova and Egorova, 2012; Donova, 2017). However, they often suffer from low enzyme activities severely hampering application (Bernhardt, 2006; Urlacher and Schmid, 2006; Julsing et al., 2008). Bacterial CYP450s typically provide higher activities and high levels of soluble expression in recombinant microorganisms. The vast majority of CYP450s features separate components involved in the shuttling of electrons from NAD(P)H to the monooxygenase subunit (Bernhardt, 2006). Subunits need to be expressed in a stable and active form and in the optimal ratio (Murdock et al., 1993; Staijen et al., 2000) which is likely to confine the biotransformation efficiency (Duetz et al., 2001). Biocatalyst efficiency, namely the enzyme turnover rate kcat (s−1), may therefore benefit from a fusion of subunits as it is the case for native CYP450 BM3 (BM3, in CYP450 nomenclature: CYP102A1) from Bacillus megaterium ATC14581 featuring the highest known CYP450 activity (Narhi and Fulco, 1986, 1987; Noble et al., 1999). For this fatty acid monooxygenase, which does not accept steroids as substrates (van Vugt-Lussenburg et al., 2006), Kille et al. developed a set of variants with promising activities as well as regio- and diastereoselectivities for testosterone hydroxylation (Kille et al., 2011), constituting an interesting foundation for efficient steroid hydroxylation.
CYP450s are cofactor-dependent and instable in isolated form and thus are preferably applied in metabolically active cells (Duetz et al., 2001; Schrewe et al., 2013), with self- and enzyme regeneration and reactive oxygen species (ROS) degradation as important stabilizing factors (Woodley, 2006). However, microbial cell envelopes typically feature a hydrophilic outer face, established by a lipopolysaccharide layer and the hydrophilic nature of outer membrane porins, e.g., in Gram-negatives (Leive, 1974; Nikaido, 2003; Chen, 2007; Schrewe et al., 2013). This restrains the uptake of hydrophobic substrates (Carter et al., 2003; Schrewe et al., 2011; Grant et al., 2014), including steroids (Zehentgruber et al., 2010a; Zehentgruber et al., 2010b; Putkaradze et al., 2019). The heterologous co-expression of genes encoding hydrophobic outer membrane pores can facilitate hydrophobic substrate uptake into metabolically active cells, as it has been shown for the bioconversion of alkanes (Julsing et al., 2012; Grant et al., 2014; Call et al., 2016; Hsieh et al., 2018), long-chain fatty acids (Schneider et al., 1998; Jeon et al., 2018), fatty acid methyl esters (Julsing et al., 2012; Ladkau et al., 2016; van Nuland et al., 2016), terpenes (Cornelissen et al., 2013; Ruff et al., 2016), and aromatics (Ruff et al., 2016).
In this study, we evaluate the steroid hydroxylation performance of highly active BM3 variants in living Escherichia coli cells and follow up the hypotheses that the outer membrane of Gram-negative bacteria forms an effective barrier for steroids and that this barrier can be relieved by introducing hydrophobic pores. Thereby, 2β- or 15β-hydroxylations of testosterone served as model reactions and the performance of permeabilized and intact cells was compared (Figure 1). Further, different outer membrane proteins were evaluated for their potential to improve specific steroid hydroxylation activities of living cells (Supplementary Table S1).
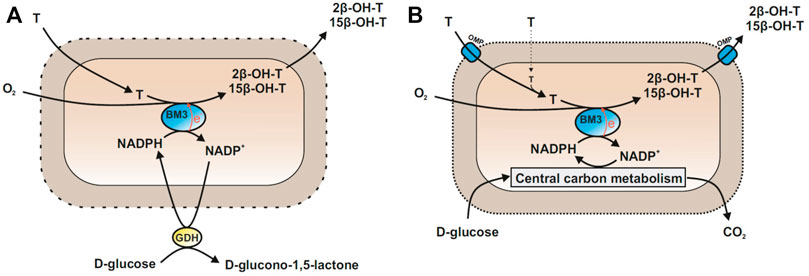
FIGURE 1. Biocatalyst formats for the hydroxylation of testosterone (T) to 2β- and 15β-hydroxytestosterone (2/15β-OH-T) using self-sufficient CYP450 BM3 variants (BM3). (A) Permeabilized cells supplied with glucose and glucose dehydrogenase (GDH) for NADPH regeneration. Efficient steroid diffusion into cells can be assumed. (B) Intact living cells recycling NADPH via active glucose catabolism. They are hypothesized to require hydrophobic outer membrane proteins (OMP) for efficient steroid uptake and secretion.
Materials and Methods
Gene Synthesis, Oligonucleotides, and Chemicals
Custom synthesized genes and oligonucleotides were purchased from Eurofins (Ebersberg, Germany). 2β- and 15β-hydroxytestosterone were purchased from Steraloids Inc. (Newport, RI, United States). All other chemicals were obtained from AppliChem (Darmstadt, Germany), Carl Roth (Karlsruhe, Germany), Chemsolute (Renningen, Germany), or Sigma-Aldrich (Steinheim, Germany) in the highest purity available.
Generation of Recombinant Bacterial Strains
Enzymes (Phusion High-Fidelity Polymerase, restriction enzymes, T5 exonuclease, Taq DNA ligase), dNTPS, and the corresponding buffers were purchased from Thermo Scientific Molecular Biology (St. Leon-Rot, Germany) or New England Biolabs (Frankfurt/Main, Germany). Plasmid isolation and purification of DNA from agarose gels was performed according to the manufacturer’s instructions of the respective kits from Macherey-Nagel (Düren, Germany). Electro-competent E. coli cells were prepared according to standard procedures (Sambrook and Russell, 2001) and transformed via electroporation (2500 V, Eppendorf Eporator®, Hamburg, Germany).
Microbial strains and plasmids used in this work are listed in Supplementary Table S2. E. coli DH5α was used for cloning purposes, whereas E. coli BL21-Gold(DE3) was used for expression and biotransformation studies. The plasmids pETM11_KSA1, pETM11_KSA2, pETM11_KSA3, and pETM11_KSA14 were constructed according to the publication of Kille et al. (2011). Plasmids harboring genes encoding the respective outer membrane proteins were constructed as shown in Supplementary Table S3. The respective genes were either ordered as gene synthesis constructs or obtained from genomic DNA. Successful cloning was confirmed by sequencing (Genewiz Germany GmbH, Leipzig, Germany).
Cultivation of E. coli Strains
For bacterial growth and heterologous protein synthesis, cells were grown in lysogeny broth (LB) medium (Sambrook and Russell, 2001), modified terrific broth (TB) (Kille et al., 2011), or M9 medium (Sambrook and Russell, 2001) supplemented with 0.1% (v/v) US* trace element solution (Panke et al., 1999), 2 mM MgSO4, and 0.5% (w/v) d-glucose as sole carbon and energy source. Cultivations were conducted in a Multitron shaker (Infors, Bottmingen, Switzerland).
Microorganisms from a frozen glycerol stock were cultivated in LB medium (supplemented with 50 μg mL−1 kanamycin for plasmid-containing cells) at 37°C and 200 rpm for 6–8 h. From the LB culture, 10 mL of either M9 or TB medium were inoculated and incubated at 30°C for 14–16 h. These precultures were used to inoculate the same media to an optical density of 0.2 at 450 nm (OD450). Heterologous gene expression was induced in the early exponential phase (OD450 ∼0.6) by addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). In case of cultivation in M9 medium, 0.5 mM of the heme precursor 5-aminolevulinic acid was simultaneously added for enhanced heme synthesis. Cells were harvested 5 h after induction by centrifugation (5,000 g, 5 min, 4°C) for the preparation of permeabilized cells, resting cells, or cell-free extracts. For the isolation of total membrane fractions, cell pellets were stored at −20°C until further processing.
Biotransformation With Permeabilized Cells
Specific activities of permeabilized cells were determined according to a modified version of the protocol described by Kille et al. (2011). Cells were harvested from TB cultures 5 h after induction, washed once with P450 washing buffer (100 mM potassium phosphate, pH 7.4, 5% (v/v) glycerol), and resuspended to a cell concentration of 1 gCDW L−1 in P450 reaction buffer (100 mM potassium phosphate, pH 7.4, 5% (v/v) glycerol, 5% (w/v) glucose, 5 mM EDTA, 0.25 mM NADP+, 1 U mL−1 glucose dehydrogenase). The cell suspension was immediately frozen in liquid nitrogen and then thawed at room temperature. One mL of sample was transferred to screw-capped glass tubes (12 mL) and equilibrated in a water bath at 30°C and 250 rpm for 15 min. The biotransformation was then started by adding 10 µL of a 100 mM testosterone stock solution in DMSO, resulting in final concentrations of 1 mM testosterone and 1% (v/v) DMSO. Reactions were terminated after a specific incubation time (given in figures and corresponding descriptions) by adding 12.5 µL HCl (1 M) per 100 µL resting cell suspension. Acetonitrile was added to the samples (50% v/v) to dissolve precipitated steroids, followed by mixing (2000 rpm, 5 min, 4°C) and centrifugation (17,000 g, 5 min, 4°C) for biomass removal. The resulting supernatant was stored at −20°C until further analysis. Specific activities are given in U gCDW−1, with 1 U defined as the activity forming 1 µmol of product per min, and were retrieved via quantification of accumulated product divided by the biomass applied.
In Vivo Biotransformation With Resting Cells
Fresh cells harvested 5 h after induction from an exponentially growing M9 culture were washed once and resuspended to a cell concentration of 1 gCDW L−1 in 100 mM potassium phosphate buffer (pH 7.4) supplemented with 1% (w/v) glucose as the source for energy and reduction equivalents. One mL of resting cell suspension was filled into screw-capped glass tubes (12 mL) and equilibrated at 30°C and 250 rpm for 15 min. Reaction start and termination as well as sampling were performed as described above for biotransformations with permeabilized cells.
In Vitro Biotransformation With Cell-Free Extracts
Cells were harvested from induced exponentially growing M9 cultures, and cell pellets were resuspended in 100 mM potassium phosphate buffer (pH 7.4) to a concentration of 3 gCDW L−1. Subsequently, cells were disrupted by three passages through a French Press (6.9 MPa, Glenn Mills Inc., Clifton, NJ, United States). Cell debris was removed by centrifugation at 12,000 g for 15 min at 4°C. One mL of supernatant was transferred to screw-capped glass tubes (12 mL), equilibrated at 30°C and 250 rpm for 15 min, and supplied with 10 µL of a 50 mM NADPH stock solution in the same buffer. The reaction was started by adding 10 µL of a 100 mM testosterone stock solution in DMSO, resulting in final concentrations of 0.5 mM NADPH, 1 mM testosterone, and 1% (v/v) DMSO. Reactions were terminated and samples were treated as described above.
Isolation of Total Membrane Fractions
Cell pellets were thawed in ice, resuspended in a 100 mM potassium phosphate buffer (pH 7.4) to a concentration of 3–5 gCDW L−1, and disrupted by two passages through a French Press (6.9 MPa, Glenn Mills Inc.). The resulting crude cell extract was centrifuged at 12,000 g for 15 min at 4°C followed by centrifugation of the supernatant at 200,000 g and 4°C for 2 h. The resulting pellet was resuspended in the same buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, see below).
Analytical Methods
Biomass concentrations were determined photometrically as the optical density at a wavelength of 450 nm (Libra S11, Biochrom Ltd., Cambridge, United Kingdom) with one OD450 unit corresponding to 0.166 gCDW L−1 (Blank et al., 2008).
Protein concentrations were analyzed via Bradford Dye Reagent (Alfa Aesar, Kandel, Germany) according to the manufacturer’s instructions using bovine serum albumin as protein standard. Monitoring of protein synthesis was performed by harvesting 80 µg of cell dry weight (CDW) from cultures followed by SDS-PAGE according to Laemmli (1970). Proteins extracted from 15 µgCDW were loaded per lane and stained with Coomassie Brilliant Blue R-250. For cell-free extract and membrane fraction analysis, 7.5 µg of total protein were loaded per lane. PageRuler™ Prestained Protein Ladder (Thermo Fisher Scientific, Waltham, MA, United States) was used as reference.
Steroid concentrations were determined by HPLC using a Dionex Ultimate 3,000 system (Thermo Fisher Scientific) equipped with an Accucore C18 column (150 × 3 mm, 2.6 µm particle size, Thermo Fisher Scientific). An eluent consisting of 40% acetonitrile in ultrapure water was used at a flow rate of 0.6 mL min−1. The column oven temperature was set to 40°C and a sample volume of 5 µL was injected. Steroids were detected at 245 nm using an UV detector and quantified based on calibration curves.
Results
Testosterone Hydroxylation by E. coli BL21-Gold(DE3) Harboring Engineered BM3 Variants
In a previous study, Kille et al. developed a library of engineered BM3 variants capable of regio- and stereoselective hydroxylation of testosterone either at the 2β- or the 15β-position (Kille et al., 2011). Genes encoding the respective BM3 variants were heterologously expressed in E. coli BL21-Gold(DE3) via the lac-based T7 expression system. Promising variants showed good conversions and regioselectivities, from which volumetric activities of up to 0.58 U L−1 can be estimated (Supplementary Table S4). An important remark is that cell preparation included the addition of EDTA and a freeze-thawing step which is prone to involve cell membrane permeabilization (Felix, 1982; Chen, 2007). Thus, cells presumably were incapable of intracellular cofactor regeneration (Julsing et al., 2008; Julsing et al., 2012; Schrewe et al., 2013), which is corroborated by the use of an extracellular cofactor regeneration system (Kille et al., 2011). The study presents long-term data (24 h of biotransformation). In order to assess biocatalyst efficiency in detail, we set out to determine initial and specific catalytic rates for permeabilized as well as living cells equipped with the promising BM3 variants KSA1, KSA2, KSA3, or KSA14.
Constructed strains were based on pETM11 and E. coli BL21-Gold(DE3) according to Kille et al. (2011) (Supplementary Table S2, Supplementary Figure S1A). For cell cultivation and heterologous protein synthesis in TB medium, the protocol for microplate cultivation (Kille et al., 2011) was adapted to shake flask scale. As expected, permeabilized cells containing KSA1, KSA2, and KSA3 primarily produced 2β-hydroxytestosterone, whereas 15β-hydroxytestosterone was detected as main product of KSA14 (Supplementary Table S5). Volumetric activities for 24 h of biotransformation were in a similar range as previously reported (Supplementary Figure S2). Initial activities within the first 10 min were found to exceed long-term activities by factors of 7.8–19.8, indicating that testosterone hydroxylation rates do not remain constant in the experimental setup chosen. Interestingly, one particular KSA14 clone exhibited a 2-fold higher initial activity and similar 15β-regioselectivity (Supplementary Table S5). Sequencing of the plasmid from this clone revealed four additional nucleotide exchanges compared to the original ksa14 gene, leading to two missense mutations (K224E, V314I) and two silent mutations (L78L, L871L). Both amino acid exchanges are located in the BM3 monooxygenase subunit and thus are likely to affect the catalytic properties of the enzyme. For example, K224 forms salt bridges with the residues involved in the opening and closing of the substrate channel (Whitehouse et al., 2012). This variant is hereafter designated as KSA14m.
Compared to the wildtype strain (specific growth rate µ = 0.85 ± 0.04 h−1 at 30°C in TB medium), the recombinant strains after induction showed a shorter phase of exponential growth, followed by a growth rate reduction which may be due to a metabolic burden of BM3 expression (Figure 2A). However, growth of the empty vector control was even slower than that of BM3-containing strains and involved the synthesis of two unexpected 32 and 38 kDa proteins (Figure 2B). Analysis of open reading frames on the empty vector revealed that the so-called max dimerization protein 1 (NCBI accession number NP_002348) may be synthesized. However, its molecular weight is estimated to be only 25 kDa, which does not fit to the observed protein weights.
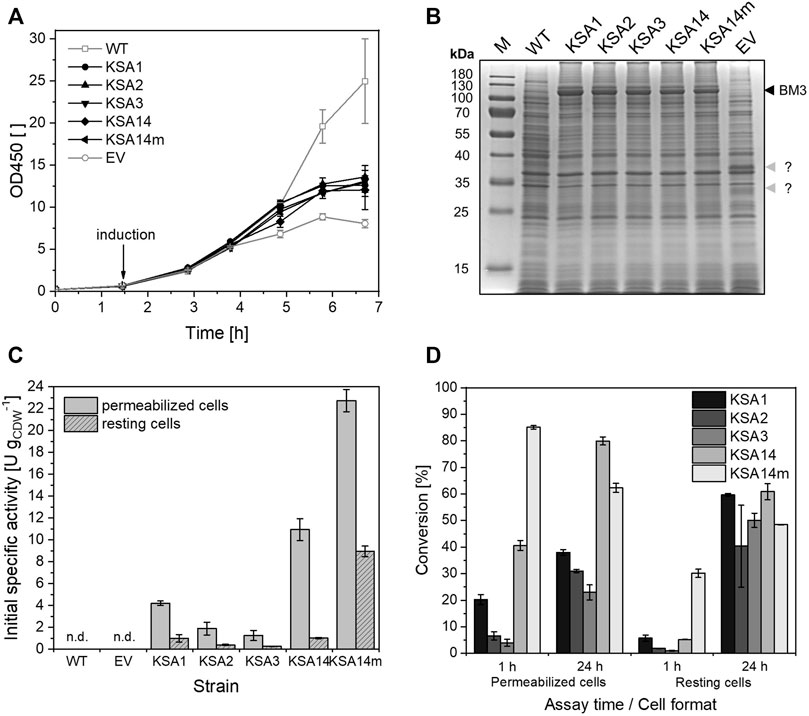
FIGURE 2. Performance of E. coli BL21-Gold(DE3) strains carrying pETM11 with genes encoding the respective BM3 variants compared to the wildtype (WT) and an empty vector control (EV) strain during and after cultivation in TB medium. (A) Bacterial growth in TB medium upon induction with 0.1 mM IPTG. (B) SDS-PAGE analysis of the different strains 5 h after induction, with the band at 119 kDa representing the respective BM3 variant (black arrow). For the empty vector strain, two unidentified proteins (32 and 38 kDa, grey arrows) were observed. (C) Initial (10 min) testosterone hydroxylation activities of permeabilized and resting cells. Activity assays were conducted as described in the Materials and Methods section. (D) Conversion of testosterone (based on product formation) by the different cell formats 1 and 24 h after substrate addition. The bars represent average values and standard deviations of two biological replicates (n.d., no product formation detected).
The high expression levels of the respective BM3 variants (Figure 2B) enabled initial specific activities ranging from 1.25 ± 0.47 to 10.93 ± 1.00 U gCDW−1 for permeabilized cells of the different strains (Figure 2C), whereas KSA14m hydroxylated testosterone at a significantly higher rate of 22.73 ± 1.02 U gCDW−1. To investigate if intact cells enable similar specific testosterone hydroxylation activities as permeabilized cells, non-growing living (i.e., resting) cells were prepared after cultivation and heterologous gene expression in TB medium and employed in activity assays. Direct comparison revealed 4.2 to 10.7-fold lower initial activities in the range of 0.26 ± 0.01 to 1.02 ± 0.06 U gCDW−1 for KSA1, KSA2, KSA3, and KSA14 (Figure 2C). KSA14m containing cells again showed the highest testosterone hydroxylation rate (8.94 ± 0.49 U gCDW−1, 2.5-fold lower than in permeabilized cell format). Regioselectivity was not or only slightly reduced compared to permeabilized cells (Supplementary Table S5). Furthermore, permeabilized cells generally demonstrated higher conversions than resting cells 1 hour after substrate addition (Figure 2D). This however changed during the following 23 h, after which resting cells of some strains showed higher conversions than permeabilized cells. It has to be mentioned that solubility limits of testosterone and the 15β-hydroxylated product in the aqueous assay setup (RCA buffer supplemented with glucose and 1% (v/v) DMSO) amounted to 66 and 565 μM, respectively. Depicted conversions after 1 and 24 h of biotransformation were calculated based on product formation and need to be interpreted carefully when exceeding the aqueous solubility limit (conversions >56%).
Heterologous BM3 Synthesis in Defined Media
High CYP450 levels led to noticeably impaired growth of the recombinant bacterial strains in TB medium, pointing to metabolic burden effects. High oxygenase levels have been reported to affect host cell physiology (van Beilen et al., 2003; Bühler et al., 2008) and thus do not necessarily lead to higher whole-cell activities (Schäfer et al., 2020). In contrast to complex media like TB, defined media usually lead to slower bacterial growth and lower metabolic activity, including heterologous gene expression and protein synthesis, but are more feasible for process scale up. We thus also tested M9 medium for cultivation.
In contrast to modified TB medium, all recombinant strains showed exponential growth in M9 medium, which was not impaired compared to the wildtype, also not upon induction (Figure 3A). Even though BM3 levels after 5 h of induction were lower in M9 medium compared to TB medium (Figure 3B), resting cells showed similar initial specific activities, ranging from 0.26 ± 0.01 U gCDW−1 to 1.06 ± 0.02 U gCDW−1. The KSA14m strain again displayed a noticeably higher activity of 9.75 ± 1.60 U gCDW−1 (Figure 3C). Interestingly, regioselectivity was slightly lower (Supplementary Table S5).
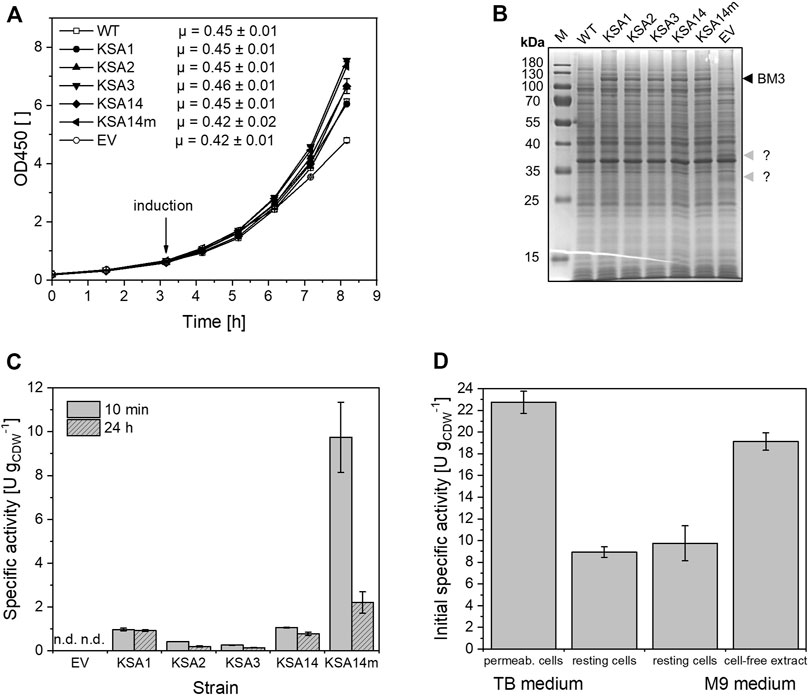
FIGURE 3. Performance of E. coli BL21-Gold(DE3) strains carrying pETM11 with genes encoding the respective BM3 variants compared to the wildtype and an empty vector control strain during and after cultivation in M9 minimal medium supplemented with 0.5% (w/v) glucose. (A) Bacterial growth in M9 medium upon induction with 0.1 mM IPTG and addition of 0.5 mM 5-aminolevulinic acid. (B) SDS-PAGE analysis of the different strains 5 h after induction, with the band at 119 kDa representing the respective BM3 variant (black arrow). For the empty vector strain, two unidentified proteins (32 and 38 kDa, grey arrows) were observed. (C) Initial (10 min) and long-term (24 h) testosterone hydroxylation activities of resting cells. (D) Activities of the KSA14m strain in different biocatalyst formats are compared. Activity assays were performed as described in the Materials and Methods section. The bars represent average values and standard deviations of two biological replicates (n.d., no product formation detected).
In summary, the choice of cultivation medium did not affect the activity of intact cells. However, selection of a suitable biocatalyst format proved to be pivotal for higher specific testosterone hydroxylation rates. The resting-cell format appears less suitable for BM3-catalyzed testosterone hydroxylation than the permeabilized-cell format, suffering from impaired substrate uptake. It however is clearly superior regarding cofactor regeneration and biocatalyst stability, making cell engineering for improved substrate uptake a strategy of choice.
Testosterone Hydroxylation by KSA14m in Cell-Free Extracts Versus Whole Cells
As the strain containing KSA14m displayed the highest specific testosterone hydroxylation activity, it was used to investigate the possible substrate uptake limitation. Cells cultivated in M9 medium were either used for the preparation of intact resting cells or disrupted to generate cell-free extracts, for which steroid hydroxylation is not subject to mass transfer limitation over cellular membranes. A specific activity of 34.8 ± 1.5 U per g of total protein was obtained with cell-free extracts, which, assuming that standard E. coli cells consist of 55% total protein (Schmidt et al., 2016), is 2-fold higher than that of intact cells (Figure 3D). Obviously, the activity obtained for intact cells does not reflect the maximum BM3 activity available in the cells which corroborates the findings with permeabilized cells and clearly indicates steroid uptake limitation.
Hydrophobic Outer Membrane Pores Boost Testosterone Hydroxylation by Living Cells
The significant difference between the activities of cell-free extracts, permeabilized, and intact cells indicates that the uptake of testosterone as a large hydrophobic substrate limits its hydroxylation by intact Gram-negative cells. We tested nine different outer membrane proteins for an improvement of testosterone uptake by E. coli BL21-Gold(DE3) containing the BM3 variant KSA14m. Candidates were selected from operons that encode pathways enabling growth on or degradation of hydrophobic compounds (Wang et al., 1995; Ayoubi and Harker, 1998; Kasai et al., 2001; Hearn et al., 2008; Leu et al., 2011; Kothari et al., 2016; Mounier et al., 2018; Olivera and Luengo, 2019) or based on previous studies employing hydrophobic outer membrane proteins to facilitate hydrophobic substrate uptake (Julsing et al., 2012; Cornelissen et al., 2013; Grant et al., 2014; Call et al., 2016; Ruff et al., 2016; Jeon et al., 2018). Corresponding genes were placed downstream of the ksa14m gene in a bicistronic operon (Supplementary Figure S1B).
The co-synthesis of four outer membrane proteins significantly improved the specific whole-cell testosterone hydroxylation activity of living cells. XylN, FhuA Δ1-160, and TodX resulted in 3.3-, 4.6-, and 4.9-fold specific activity increases, respectively (Figure 4A). AlkL emerged as the most effective candidate enabling a 5.6-fold increase in whole-cell testosterone hydroxylation activity, corresponding to an even higher protein-specific rate than obtained with KSA14m containing cell-free extract. Induction of heterologous gene expression slightly affected bacterial growth of only the strains carrying aupA/aupB, fhuAΔ1-160, todX, and xylN compared to strains without an additional outer membrane protein (Supplementary Figure S3). SDS-PAGE analysis of isolated total membrane fractions further confirmed AlkL, FhuA Δ1-160, TodX, and XylN synthesis, whereas bands representing the remaining outer membrane proteins were not clearly visible (Figure 4B) which may explain why an effect on whole-cell activity was not observed in these cases. Interestingly, a protein band representing the fatty acid pore FadL was clearly visible, but the respective strain did not show an improved activity. To conclude, whole living cells equipped with hydrophobic outer membrane proteins facilitating testosterone uptake allowed even higher specific turnover rates than obtained with permeabilized cells and cell-free extracts.
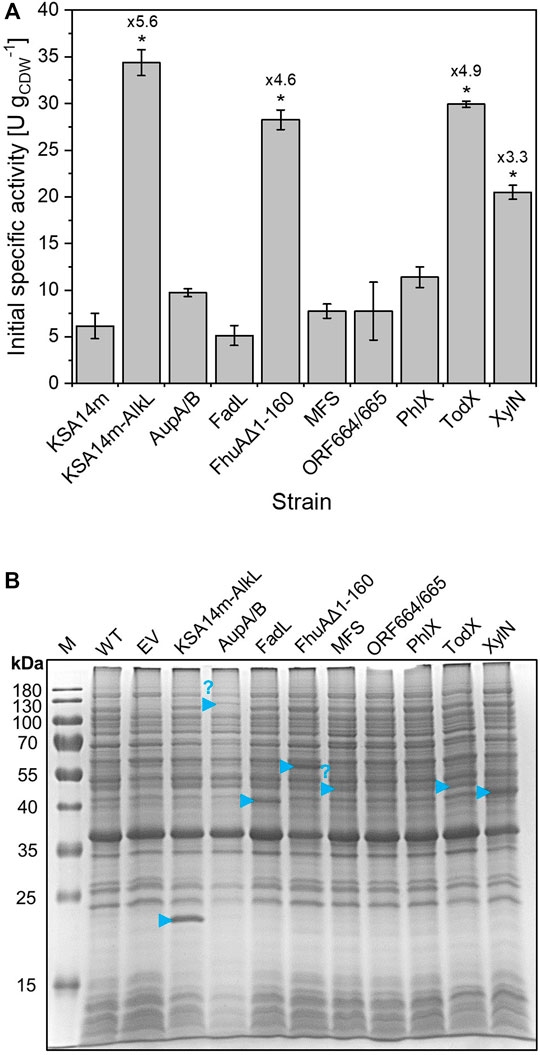
FIGURE 4. Initial specific testosterone hydroxylation activities (A) of resting E. coli BL21-Gold(DE3) cells carrying pETM11 with ksa14m and the genes encoding different membrane proteins after cultivation in M9 medium supplemented with 0.5% (w/v) glucose. Resting cell preparation and activity assays were conducted as described in the Materials and Methods section. Average values and standard deviations of two biological replicates are given. Statistical analysis was performed using Student’s t test, and the values were taken as significantly different when p ≤ 0.05. (B) SDS-PAGE analysis of total membrane fractions isolated from the respective bacterial strains 5 h after induction with 0.1 mM IPTG and addition of 0.5 mM 5-aminolevulinic acid. Blue arrows point to the protein bands at the sizes expected for the respective membrane proteins.
Potential of Engineered BM3 Variants for Testosterone Hydroxylation
Incorporation of AlkL into the whole-cell biocatalyst turned out as the most promising strategy to increase whole-cell testosterone hydroxylation activities by a facilitated steroid uptake. Thus, the testosterone hydroxylation capabilities of different BM3 variants were investigated with cells co-expressing alkL. Following this strategy, increased whole-cell testosterone hydroxylation rates also were obtained with the variants KSA1, KSA2, KSA3, and KSA14 (Figure 5A) emphasizing the high efficiency, with which these variants can catalyze steroid hydroxylation in living cells. Thereby, AlkL increased specific activities 6- to 28-fold depending on the BM3 variant. Again, bacterial growth was barely affected by additional alkL expression (Figure 5B). Cells carrying KSA14m and AlkL emerged as the most active whole-cell biocatalysts and enabled almost complete conversion of 1 mM testosterone in the course of 1 h as it is shown in Supplementary Figure S4. Substrate depletion appeared to cause a conversion rate decrease towards the end of the biotransformation experiment. Catalyst instability issues possibly caused by inherent enzyme instability and/or substrate/product inhibitions may however also contribute to this effect and will be subject of future research. In conclusion, incorporation of hydrophobic outer membrane proteins, such as AlkL, led to a significant increase of testosterone hydroxylation activities of recombinant E. coli cells and thus proved to be a valuable strategy to overcome steroid uptake limitation.
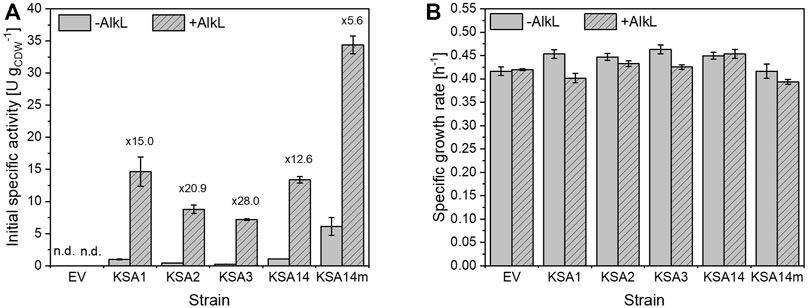
FIGURE 5. Initial specific testosterone hydroxylation activities (A) of resting E. coli BL21-Gold(DE3) cells carrying pETM11 with genes encoding different BM3 variants together with or without alkL after 5 h of induction in M9 medium supplemented with 0.5% (w/v) glucose. Resting cell preparation and activity assays were performed as described in the Materials and Methods section. (B) Specific growth rates of the respective strains in M9 medium upon induction with 0.1 mM IPTG and addition of 0.5 mM 5-aminolevulinic acid. Average values and standard deviations of two biological replicates are given.
Discussion
Biotechnological steroid hydroxylation at industrial scale with the typically employed wildtype microorganisms still suffers from low transformation rates and poor selectivities (Fernández-Cabezón et al., 2018). Steroid-hydroxylating CYP450s are generally characterized by low catalytic turnover rates, the need for electron transfer partners, as well as limitations in the electron transfer from the redox cofactor to the active site (Bernhardt, 2006; Donova and Egorova, 2012; Donova, 2017). These bottlenecks considerably impede the generation of recombinant production strains for commercial applications (Duetz et al., 2001; van Beilen et al., 2003; Urlacher and Eiben, 2006; Julsing et al., 2008; Bernhardt and Urlacher, 2014). By engineering the self-sufficient and highly active CYP450 BM3 from Bacillus megaterium ATC14581 towards regio- and stereoselective hydroxylation of testosterone, Kille and colleagues laid the foundation for efficient steroid-hydroxylation biocatalysts (Kille et al., 2011). In this study, we set out to evaluate and improve the applicability of selected BM3 variants in intact living cells as the preferred biocatalyst format for oxygenase-based biocatalysis (Duetz et al., 2001; Schrewe et al., 2013) with a focus on increasing the catalytic turnover rates.
Introduction of Hydrophobic Outer Membrane Proteins as Promising Strategy to Improve Whole-Cell Steroid Turnover Rates
Former studies demonstrated that hydrophobic substrate uptake across cellular membranes constitutes a major bottleneck in whole-cell biotransformations (Carter et al., 2003; Fontanille and Larroche, 2003; Zehentgruber et al., 2010b; Schrewe et al., 2011; Grant et al., 2014). Contrary to small hydrophobic molecules crossing the outer membrane of Gram-negatives through hydrophilic porins and readily diffusing through the cytoplasmic membrane, the uptake of larger hydrophobic molecules is restrained by polar and charged lipopolysaccharides on the cell surface and the hydrophilic nature of porins in the outer membrane (Hancock, 1997; Nikaido, 2003). Although strategies targeting membrane destruction or permeabilization may relieve this limitation, they are destructive to the cell and its functionalities and thus not suitable for biocatalytic reactions depending on an active metabolism for cofactor and enzyme regeneration as well as ROS degradation (Julsing et al., 2008; Julsing et al., 2012; Schrewe et al., 2013).
Alternatively, the supply of cells with membrane pores has been established as an approach enabling the conversion of a variety of highly hydrophobic substrates by intact metabolically active cells. Table 1 shows a comparison of the best results obtained in this study for steroid hydroxylation with those obtained in earlier studies for the conversion of other hydrophobic substrates facilitated by outer membrane proteins. Applied pores range from members of aliphatic and aromatic hydrocarbon degradation pathways to artificially created pores, all with a typically uncharacterized substrate spectrum. In this study, the feasibility of this approach was tested for and extended to steroids. It is clear that such uptake limitation only becomes relevant, if high intracellular steroid conversion activities are established. Our results obtained with the recently published testosterone hydroxylating BM3 variants show that their high activity becomes limited by steroid uptake into intact living cells. The introduction of 4 out of 9 tested outer membrane proteins significantly increased the activity of E. coli containing the highly active BM3 variant KSA14m.
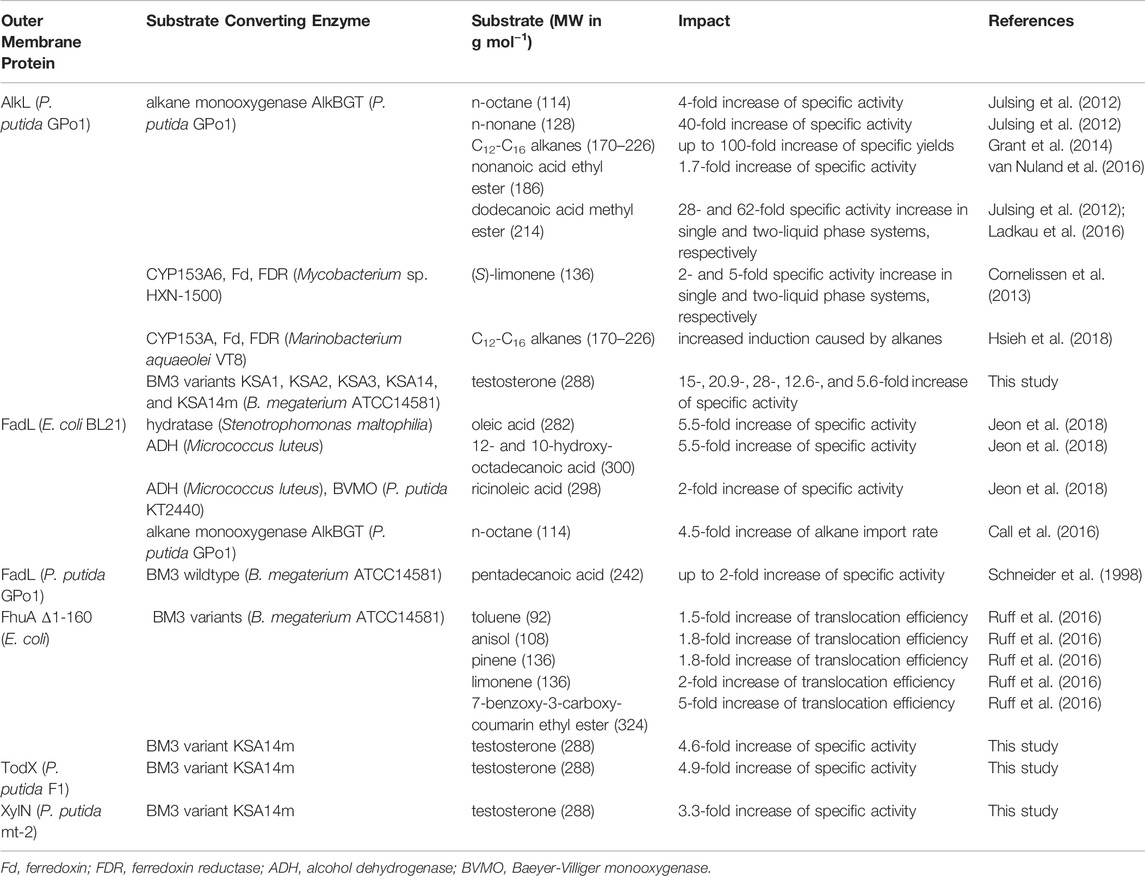
TABLE 1. Studies reporting enhanced uptake or biotransformation of hydrophobic substrates via heterologous outer membrane proteins.
However, the introduction of outer membrane proteins also can be harmful to host cell physiology, either due to metabolic burden of the expression of additional heterologous genes and/or energy demanding, inefficient, and/or improper folding and translocation of membrane proteins (van Beilen et al., 2003; Kadisch et al., 2017b). The expression level of pores can be another critical factor as consequentially increased intracellular substrate concentrations may become toxic (Julsing et al., 2012; Call et al., 2016; Kadisch et al., 2017a). For example, biocatalyst destabilization related to high alkL expression has been reported during biotransformation of dodecanoic acid methyl ester (Schrewe et al., 2014) and octane (Grant et al., 2014). Similar findings have been reported for high level fadL expression employed for the conversion of octane (Call et al., 2016) and (hydroxy) fatty acids (Jeon et al., 2018). Strains expressing aupAB, fhuAΔ1-160, todX, and xylN displayed slightly lower specific growth rates compared to a strain harboring only the BM3 variant. Effects on biocatalyst stability remain to be investigated. A fine-tuning of membrane protein expression levels may become necessary to control substrate influx, for example by the use of tightly controlled expression systems (Grant et al., 2014; Call et al., 2016), variation of promoter strength (Kadisch et al., 2017a), or appropriate gene copy number (Jeon et al., 2018). Furthermore, several technical solutions can be applied to avoid toxic effects of substrate and product molecules, such as the introduction of an organic phase as substrate reservoir and product sink (van Beilen et al., 2003; Schrewe et al., 2014).
Substrate Spectrum of Outer Membrane Proteins
Introduction or overexpression of outer membrane proteins has become an increasingly relevant tool for biotransformations of hydrophobic substrates with whole microbial cells (Table 1). Several investigated candidates feature a broad substrate range reaching beyond the natural substrates of the pathways they are associated to. In this study, we report an extension of the so far-known substrate spectrum of several hydrophobic pores towards steroids. Several studies showed that AlkL promotes alkane uptake by E. coli (Julsing et al., 2012; Grant et al., 2014). This outer membrane protein has been proposed to be structured as an eight-stranded β-barrel traversing the lipopolysaccharide layer similar to OmpW of E. coli, with which it shares 27% amino acid sequence homology (van Beilen et al., 1992; Hong et al., 2006). The extracellular domain has a high affinity for hydrophobic molecules so that substrates are easily channeled across the lipopolysaccharide layer into the hydrophobic interior of AlkL in the outer membrane (Grant et al., 2014). As for OmpW, the passage is hindered by a narrowing in the channel, which directs molecules to exit through a putative lateral opening into the outer membrane (Hong et al., 2006). Apparently, this mechanism also works for fatty acid (m)ethyl esters (Julsing et al., 2012; Ladkau et al., 2016; van Nuland et al., 2016) and monoterpenes (Cornelissen et al., 2013). Interestingly, the radius of the lateral opening (1.3 Å) has been described as incompatible with the size of these known AlkL substrates but has recently been shown to be dynamic due to a continuous restructuring of the barrel dimension and release into the membrane through ephemeral openings (Schubeis et al., 2020). In this study, AlkL relieved the constrained testosterone uptake into E. coli and boosted specific testosterone hydroxylation activities, thus expanding the AlkL substrate spectrum to steroids, which are even larger than the substrates known so far (Table 1).
The pivotal role of FadL as a long-chain fatty acid transporter in E. coli has already been reported decades ago (Nunn and Simons, 1978; Black, 1988). In whole-cell biotransformations of long-chain fatty acids, increased FadL levels significantly improved specific rates (Schneider et al., 1998; Jeon et al., 2018). Similar results were obtained for atypical FadL substrates, i.e., long-chain hydroxyl fatty acids (Jeon et al., 2018). Interestingly, overexpression of fadL did not lead to improved steroid hydroxylation rates in this study. Even though the FadL basic structure has been described as similar to AlkL (van den Berg, 2010), the proposed uptake mechanism differs significantly. FadL is described as a long barrel composed of 14 antiparallel β-strands that is plugged by a central hatch domain and possesses an opening based on an inward-pointing kink in one of the β-strands (van den Berg et al., 2004). Between two extracellular loops, a hydrophobic groove comprises a low-affinity binding site for substrates, which then diffuse into a high-affinity binding pocket inside the barrel close the N-terminus. Conformational changes in the N-terminus and the hatch result in an affinity decrease and generate a lateral channel for the substrate to move into the outer membrane (Hearn et al., 2009; van den Berg, 2010). Thereby, substrate binding has been suggested as prerequisite (Black and Zhang, 1995; Lepore et al., 2011), with the affinity strongly varying with fatty acid chain length (Black, 1990) and long-chain fatty acids (≥C12) being preferred over mid-chain fatty acids (C8-C11) (Maloy et al., 1981; Black, 1990). Generally, electrostatic forces between negatively charged carboxyl groups and positively charged amino acid residues within the FadL binding pocket have been considered necessary for substrate binding (van den Berg, 2005), which would exclude non-charged compounds as FadL substrates. The variation of FadL expression levels however was recently shown to modulate the import of medium-chain alkanes into E. coli (Call et al., 2016), suggesting that substrate binding is not necessarily charge-dependent. The missing facilitation of steroid import by FadL observed in this study is in line with a possible charge dependency and/or a rather confined substrate specificity.
In contrast, truncated FhuA was found to facilitate testosterone uptake. The native, complete protein is responsible for the active import of ferric hydroxamate into E. coli. Removal of the first 160 amino acids (N-terminal cork domain) converted the active iron transporter into a large passive diffusion channel (i.e., FhuA Δ1-160) differing from AlkL in terms of uptake mechanism (Ruff et al., 2016). FhuA Δ1-160 has been shown to improve the uptake of terpenes and aromatic compounds into E. coli up to 5-fold (Ruff et al., 2016). It also was found to facilitate testosterone uptake resulting in a 4.6-fold specific whole-cell activity increase, and thus can be considered a valuable outer membrane protein with a broad substrate spectrum.
Among the putative hydrophobic substrate uptake pores from hydrocarbon-degrading bacteria, TodX from P. putida F1 and XylN from P. putida mt-2 were found to significantly enhance whole-cell testosterone hydroxylation. Both have been reported to be involved in the uptake of alkylaromatic compounds such as toluene or xylene (Wang et al., 1995; Kasai et al., 2001). Interestingly, they share sequence homology with each other and further putative outer membrane proteins from hydrocarbon-degrading Gram-negative bacteria, e.g., TbuX, CymD, PorA, and CumH (Kahng et al., 2000; Kasai et al., 2001; Hearn et al., 2008), which thus also constitute interesting candidates for the enhancement of hydrophobic substrate uptake.
Few bacteria have been reported to be capable of metabolizing steroids, which generally are considered as highly biodegradation-resistant. Known uptake systems mostly do not involve only one, but a set of proteins (Olivera and Luengo, 2019), e.g., ten ATP-dependent proteins encoded in the actinobacterial mce4 locus (Mohn et al., 2008). A putative, but not yet characterized Major Facilitator Superfamily Transporter (MFS) originating from the testosterone-degrading strain Comamonas thiooxydans (NCBI Reference Sequence WP_041743963) did not increase testosterone hydroxylation rates. While AupA and AupB have been described as outer and inner membrane proteins, respectively, involved in alkane uptake in Marinobacter hydrocarbonoclasticus SP17 (Mounier et al., 2018), the exact function of proteins encoded by ORF664 and ORF665 in Acinetobacter venetianus RAG-1 is currently uncertain, with a possible role in dodecane uptake (Kothari et al., 2016). Besides an unsuitable substrate specificity, expression issues such as improper folding may be the reason for the lacking positive effect on testosterone hydroxylation in these cases.
The increase in testosterone hydroxylation activities not only depended on the type of outer membrane protein applied, but, interestingly, also deferred for different BM3 variants applying the same pore. This high variation may be due to differing Km values of respective BM3 variants. For example, the Km of KSA2 for testosterone has been reported to be 86-times higher than the Km of KSA14 (Kille et al., 2011). Such a high Km may increase the substrate limitation effect without AlkL. The impact of facilitated substrate uptake via an outer membrane protein thus would be much higher as it was the case for KSA2 as compared to KSA14 in combination with AlkL (Figure 5A).
Benchmarking BM3-Catalyzed Testosterone Hydroxylation in the Whole-Cell Biocatalyst Format
In this study, we investigated the steroid hydroxylation performance of highly active BM3 variants in living E. coli cells equipped with different hydrophobic outer membrane proteins to relieve substrate uptake limitation. In vivo testosterone hydroxylation rates obtained with these whole-cell biocatalysts even exceeded activities obtained with permeabilized cells or cell-free extracts (for KSA14m), which can be ascribed to the stabilizing cellular environment and efficient cell-, enzyme-, and cofactor regeneration qualifying such whole-cell biocatalysts for successful future bioprocess development (Duetz et al., 2001; van Beilen et al., 2003; Woodley, 2006; Schrewe et al., 2013). During the past decades, many CYP450s with diverse specificities have been described and engineered for steroid hydroxylations (Bureik and Bernhardt, 2007; Donova and Egorova, 2012; Szaleniec et al., 2018; Zhang et al., 2020), with the discovery of novel products and selectivity improvement as main arguments for high industrial potential (Fernandes et al., 2003; Zhang et al., 2020). However, only few studies went beyond typical screening experiments towards the evaluation of biocatalytic performance in terms of final product titer, volumetric productivity, and maximum specific activity. An overview of the latter is given in Table 2 based on data provided in references and estimations derived therefrom. The summarized studies differ in employed CYP450 and desired hydroxylation as well as applied redox partner proteins, host organism, and biocatalyst format.
Some reported final product titers exceed the minimum process requirements of 0.001 and 0.1 g L−1 h−1 for pharmaceuticals and fine chemicals, respectively (Straathof et al., 2002). Mammalian CYP450s provided the least promising results (Zehentgruber et al., 2010a; Braun et al., 2012; Brixius-Anderko et al., 2015; Schiffer et al., 2015; König et al., 2020) as expected based on their discussed drawbacks for biotechnological use (Bernhardt, 2006; Urlacher and Schmid, 2006). With bacterial CYP450s generally acknowledged as more favorable, recent research has focused especially on the families CYP106, CYP154, CYP109, and CYP260 (Szaleniec et al., 2018). Comparable with mammalian CYP450s, good steroid hydroxylation efficiencies were not reported for wildtype CYP109 and CYP260 or their variants in the chosen setups (Litzenburger and Bernhardt, 2017; Putkaradze et al., 2019; Zhang et al., 2021). In contrast, CYP106A2 and CYP154C5 enabled ca. 10-fold higher final product titers (Hannemann et al., 2006; Zehentgruber et al., 2010b; Bracco et al., 2013; Schmitz et al., 2014). Final product titers in the same range have also been reported for BM3 variants engineered for steroid hydroxylation with differing regiospecificities (Venkataraman et al., 2015; Li et al., 2020; Peng et al., 2022). In the present study, such product titers were reached in a shorter period of time and with lower whole-cell biocatalyst concentrations. This was enabled by specific activities exceeding the so far reported values by factors of 8 up to 40 compared with the turnover rates of progesterone hydroxylation by Bracco and coworkers (Bracco et al., 2013), emphasizing the great potential of these BM3 variants in whole cells equipped with hydrophobic outer membrane pores such as AlkL for steroid hydroxylation. For high volumetric productivities and product titers, extended reaction times and high biomass concentrations are prerequisites and promise highly efficient steroid hydroxylation with the hydrophobic pore-containing whole-cell biocatalysts developed in this study.
Conclusion
In this study, we highlight whole-cell biocatalysts as preferable format for testosterone hydroxylation. The application of a highly active steroid-hydroxylating CYP450 such as the BM3 variant KSA14m and a suitable hydrophobic outer membrane protein such as AlkL brings whole-cell steroid hydroxylation activities to a new level (an up to 40-fold increase) with the potential to boost productivities and product titers of microbial steroid conversion processes. Furthermore, this study highlights the broad substrate spectrum of several hydrophobic outer membrane proteins and thus also contributes to the improvement of whole-cell bioconversions of hydrophobic substrates in general. Following studies will assess stability and kinetics of the constructed whole-cell biocatalysts aiming at their highly productive application on bioreactor scale.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
CB, MM, RK, AS, and BB were involved in the conception and design of the study. CB and MM performed experimental work and collected data. CB, MM, and BB did data curation and interpretation. CB wrote the original draft of the manuscript. BB contributed in terms of article structuring and editing. All authors were involved in final editing and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Bayer AG for financial support. We further acknowledge the use of the facilities of the Centre for Biocatalysis (MiKat) at the Helmholtz Centre for Environmental Research, which is supported by the European Regional Development Funds (EFRE, Europe founds Saxony) and the Helmholtz Association.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fctls.2022.887458/full#supplementary-material
References
Ayoubi, P. J., and Harker, A. R. (1998). Whole-cell Kinetics of Trichloroethylene Degradation by Phenol Hydroxylase in a Ralstonia eutropha JMP134 Derivative. Appl. Environ. Microbiol. 64 (11), 4353–4356. doi:10.1128/AEM.64.11.4353-4356.1998
Bernhardt, R., and Urlacher, V. B. (2014). Cytochromes P450 as Promising Catalysts for Biotechnological Application: Chances and Limitations. Appl. Microbiol. Biotechnol. 98 (14), 6185–6203. doi:10.1007/s00253-014-5767-7
Bernhardt, R. (2006). Cytochromes P450 as Versatile Biocatalysts. J. Biotechnol. 124 (1), 128–145. doi:10.1016/j.jbiotec.2006.01.026
Black, P. N., and Zhang, Q. (1995). Evidence that His110 of the Protein FadL in the Outer Membrane of Escherichia coli Is Involved in the Binding and Uptake of Long-Chain Fatty Acids: Possible Role of This Residue in Carboxylate Binding. Biochem. J. 310, 389–394. doi:10.1042/bj3100389
Black, P. N. (1988). The fadL Gene Product of Escherichia coli Is an Outer Membrane Protein Required for Uptake of Long-Chain Fatty Acids and Involved in Sensitivity to Bacteriophage T2. J. Bacteriol. 170 (6), 2850–2854. doi:10.1128/jb.170.6.2850-2854.1988
Black, P. N. (1990). Characterization of FadL-specific Fatty Acid Binding in Escherichia coli. Biochim. Biophys. Acta (Bba) - Lipids Lipid Metab. 1046 (1), 97–105. doi:10.1016/0005-2760(90)90099-j
Blank, L. M., Ebert, B. E., Bühler, B., and Schmid, A. (2008). Metabolic Capacity Estimation of Escherichia coli as a Platform for Redox Biocatalysis: Constraint-Based Modeling and Experimental Verification. Biotechnol. Bioeng 100 (6), 1050–1065. doi:10.1002/bit.21837
Bracco, P., Janssen, D. B., and Schallmey, A. (2013). Selective Steroid Oxyfunctionalisation by CYP154C5, a Bacterial Cytochrome P450. Microb. Cel. Fact. 12, 95. doi:10.1186/1475-2859-12-95
Braun, A., Geier, M., Bühler, B., Schmid, A., Mauersberger, S., and Glieder, A. (2012). Steroid Biotransformations in Biphasic Systems with Yarrowia lipolytica Expressing Human Liver Cytochrome P450 Genes. Microb. Cel. Fact. 11, 106. doi:10.1186/1475-2859-11-106
Brixius-Anderko, S., Schiffer, L., Hannemann, F., Janocha, B., and Bernhardt, R. (2015). A CYP21A2 Based Whole-Cell System in Escherichia coli for the Biotechnological Production of Premedrol. Microb. Cel. Fact. 14, 135. doi:10.1186/s12934-015-0333-2
Bühler, B., Park, J.-B., Blank, L. M., and Schmid, A. (2008). NADH Availability Limits Asymmetric Biocatalytic Epoxidation in a Growing Recombinant Escherichia coli Strain. Appl. Environ. Microbiol. 74 (5), 1436–1446. doi:10.1128/AEM.02234-07
Bureik, M., and Bernhardt, R. (2007). “Steroid Hydroxylation: Microbial Steroid Biotransformations Using Cytochrome P450 Enzymes,” in Modern Biooxidation. Enzymes, Reactions and Applications. Editors R. D. Schmid, and V. B. Urlacher (Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA), 155–176.
Call, T. P., Akhtar, M. K., Baganz, F., and Grant, C. (2016). Modulating the Import of Medium-Chain Alkanes in E. coli through Tuned Expression of FadL. J. Biol. Eng. 10, 5. doi:10.1186/s13036-016-0026-3
Carter, O. A., Peters, R. J., and Croteau, R. (2003). Monoterpene Biosynthesis Pathway Construction in Escherichia coli. Phytochemistry 64 (2), 425–433. doi:10.1016/s0031-9422(03)00204-8
Chen, R. R. (2007). Permeability Issues in Whole-Cell Bioprocesses and Cellular Membrane Engineering. Appl. Microbiol. Biotechnol. 74 (4), 730–738. doi:10.1007/s00253-006-0811-x
Cornelissen, S., Julsing, M. K., Volmer, J., Riechert, O., Schmid, A., and Bühler, B. (2013). Whole-cell-based CYP153A6-Catalyzed (S)-limonene Hydroxylation Efficiency Depends on Host Background and Profits from Monoterpene Uptake via AlkL. Biotechnol. Bioeng. 110 (5), 1282–1292. doi:10.1002/bit.24801
Donova, M. V., and Egorova, O. V. (2012). Microbial Steroid Transformations: Current State and Prospects. Appl. Microbiol. Biotechnol. 94 (6), 1423–1447. doi:10.1007/s00253-012-4078-0
Donova, M. V. (2017). Steroid Bioconversions. Methods Mol. Biol. 1645, 1–13. doi:10.1007/978-1-4939-7183-1_1
Duetz, W. A., van Beilen, J. B., and Witholt, B. (2001). Using Proteins in Their Natural Environment: Potential and Limitations of Microbial Whole-Cell Hydroxylations in Applied Biocatalysis. Curr. Opin. Biotechnol. 12 (4), 419–425. doi:10.1016/s0958-1669(00)00237-8
Falcioni, F., Blank, L. M., Frick, O., Karau, A., Bühler, B., and Schmid, A. (2013). Proline Availability Regulates Proline-4-Hydroxylase Synthesis and Substrate Uptake in Proline-Hydroxylating Recombinant Escherichia coli. Appl. Environ. Microbiol. 79 (9), 3091–3100. doi:10.1128/AEM.03640-12
Felix, H. (1982). Permeabilized Cells. Anal. Biochem. 120 (2), 211–234. doi:10.1016/0003-2697(82)90340-2
Fernández-Cabezón, L., Galán, B., and García, J. L. (2018). New Insights on Steroid Biotechnology. Front. Microbiol. 9, 958. doi:10.3389/fmicb.2018.00958
Fernandes, P., Cruz, A., Angelova, B., Pinheiro, H. M., and Cabral, J. M. S. (2003). Microbial Conversion of Steroid Compounds: Recent Developments. Enzyme Microb. Technol. 32 (6), 688–705. doi:10.1016/S0141-0229(03)00029-2
Fontanille, P., and Larroche, C. (2003). Optimization of Isonovalal Production from α-pinene Oxide Using Permeabilized Cells of Pseudomonas rhodesiae CIP 107491. Appl. Microbiol. Biotechnol. 60 (5), 534–540. doi:10.1007/s00253-002-1164-8
Grant, C., Deszcz, D., Wei, Y.-C., Martínez-Torres, R. J., Morris, P., Folliard, T., et al. (2014). Identification and Use of an Alkane Transporter Plug-In for Applications in Biocatalysis and Whole-Cell Biosensing of Alkanes. Sci. Rep. 4, 5844. doi:10.1038/srep05844
Hancock, R. E. W. (1997). The Bacterial Outer Membrane as a Drug Barrier. Trends Microbiol. 5 (1), 37–42. doi:10.1016/S0966-842X(97)81773-8
Hannemann, F., Virus, C., and Bernhardt, R. (2006). Design of an Escherichia coli System for Whole Cell Mediated Steroid Synthesis and Molecular Evolution of Steroid Hydroxylases. J. Biotechnol. 124 (1), 172–181. doi:10.1016/j.jbiotec.2006.01.009
Hearn, E. M., Patel, D. R., and van den Berg, B. (2008). Outer-membrane Transport of Aromatic Hydrocarbons as a First Step in Biodegradation. Proc. Natl. Acad. Sci. U S A. 105 (25), 8601–8606. doi:10.1073/pnas.0801264105
Hearn, E. M., Patel, D. R., Lepore, B. W., Indic, M., and van den Berg, B. (2009). Transmembrane Passage of Hydrophobic Compounds through a Protein Channel wall. Nature 458 (7236), 367–370. doi:10.1038/nature07678
Hong, H., Patel, D. R., Tamm, L. K., and van den Berg, B. (2006). The Outer Membrane Protein OmpW Forms an Eight-Stranded β-barrel with a Hydrophobic Channel. J. Biol. Chem. 281 (11), 7568–7577. doi:10.1074/jbc.M512365200
Hsieh, S.-C., Wang, J.-H., Lai, Y.-C., Su, C.-Y., and Lee, K.-T. (2018). Production of 1-dodecanol, 1-tetradecanol, and 1,12-dodecanediol through Whole-Cell Biotransformation in Escherichia coli. Appl. Environ. Microbiol. 84 (4), e01806–e01817. doi:10.1128/AEM.01806-17
Jeon, E.-Y., Song, J.-W., Cha, H.-J., Lee, S.-M., Lee, J., and Park, J.-B. (2018). Intracellular Transformation Rates of Fatty Acids Are Influenced by Expression of the Fatty Acid Transporter FadL in Escherichia coli Cell Membrane. J. Biotechnol. 281, 161–167. doi:10.1016/j.jbiotec.2018.07.019
Julsing, M. K., Cornelissen, S., Bühler, B., and Schmid, A. (2008). Heme-iron Oxygenases: Powerful Industrial Biocatalysts? Curr. Opin. Chem. Biol. 12 (2), 177–186. doi:10.1016/j.cbpa.2008.01.029
Julsing, M. K., Schrewe, M., Cornelissen, S., Hermann, I., Schmid, A., and Bühler, B. (2012). Outer Membrane Protein AlkL Boosts Biocatalytic Oxyfunctionalization of Hydrophobic Substrates in Escherichia coli. Appl. Environ. Microbiol. 78 (16), 5724–5733. doi:10.1128/AEM.00949-12
Kadisch, M., Julsing, M. K., Schrewe, M., Jehmlich, N., Scheer, B., von Bergen, M., et al. (2017a). Maximization of Cell Viability rather Than Biocatalyst Activity Improves Whole-Cell ω-Oxyfunctionalization Performance. Biotechnol. Bioeng. 114 (4), 874–884. doi:10.1002/bit.26213
Kadisch, M., Willrodt, C., Hillen, M., Bühler, B., and Schmid, A. (2017b). Maximizing the Stability of Metabolic Engineering-Derived Whole-Cell Biocatalysts. Biotechnol. J. 12 (8), 1600170. doi:10.1002/biot.201600170
Kahng, H.-Y., Byrne, A. M., Olsen, R. H., and Kukor, J. J. (2000). Characterization and Role of tbuX in Utilization of Toluene by Ralstonia pickettii PKO1. J. Bacteriol. 182 (5), 1232–1242. doi:10.1128/JB.182.5.1232-1242.2000
Kasai, Y., Inoue, J., and Harayama, S. (2001). The TOL Plasmid pWW0 xylN Gene Product from Pseudomonas putida Is Involved in M-Xylene Uptake. J. Bacteriol. 183 (22), 6662–6666. doi:10.1128/JB.183.22.6662-6666.2001
Kille, S., Zilly, F. E., Acevedo, J. P., and Reetz, M. T. (2011). Regio- and Stereoselectivity of P450-Catalysed Hydroxylation of Steroids Controlled by Laboratory Evolution. Nat. Chem. 3 (9), 738–743. doi:10.1038/nchem.1113
König, L., Brixius-Anderko, S., Milhim, M., Tavouli-Abbas, D., Hutter, M. C., Hannemann, F., et al. (2020). Identification and Circumvention of Bottlenecks in CYP21A2-Mediated Premedrol Production Using Recombinant Escherichia coli. Biotechnol. Bioeng. 117 (4), 901–911. doi:10.1002/bit.27246
Kothari, A., Charrier, M., Wu, Y.-W., Malfatti, S., Zhou, C. E., Singer, S. W., et al. (2016). Transcriptomic Analysis of the Highly Efficient Oil-Degrading Bacterium Acinetobacter Venetianus RAG-1 Reveals Genes Important in Dodecane Uptake and Utilization. FEMS Microbiol. Lett. 363 (20), fnw224. doi:10.1093/femsle/fnw224
Laemmli, U. K. (1970). Cleavage of Structural Proteins During the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. doi:10.1038/227680a0
Ladkau, N., Assmann, M., Schrewe, M., Julsing, M. K., Schmid, A., and Bühler, B. (2016). Efficient Production of the Nylon 12 Monomer ω-aminododecanoic Acid Methyl Ester from Renewable Dodecanoic Acid Methyl Ester with Engineered Escherichia coli. Metab. Eng. 36, 1–9. doi:10.1016/j.ymben.2016.02.011
Leive, L. (1974). The Barrier Function of the Gram-Negative Envelope. Ann. N. Y Acad. Sci. 235 (1), 109–129. doi:10.1111/j.1749-6632.1974.tb43261.x
Lepore, B. W., Indic, M., Pham, H., Hearn, E. M., Patel, D. R., and van den Berg, B. (2011). Ligand-gated Diffusion across the Bacterial Outer Membrane. Proc. Natl. Acad. Sci. U S A. 108 (25), 10121–10126. doi:10.1073/pnas.1018532108
Leu, Y.-L., Wang, P.-H., Shiao, M.-S., Ismail, W., and Chiang, Y.-R. (2011). A Novel Testosterone Catabolic Pathway in Bacteria. J. Bacteriol. 193 (17), 4447–4455. doi:10.1128/JB.00331-11
Li, A., Acevedo-Rocha, C. G., D'Amore, L., Chen, J., Peng, Y., Garcia-Borrás, M., et al. (2020). Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants. Angew. Chem. Int. Ed. Engl. 59 (30), 12499–12505. doi:10.1002/anie.202003139
Litzenburger, M., and Bernhardt, R. (2017). CYP260B1 Acts as 9α-Hydroxylase for 11-deoxycorticosterone. Steroids 127, 40–45. doi:10.1016/j.steroids.2017.08.006
Maloy, S. R., Ginsburgh, C. L., Simons, R. W., and Nunn, W. D. (1981). Transport of Long and Medium Chain Fatty Acids by Escherichia coli K12. J. Biol. Chem. 256 (8), 3735–3742. doi:10.1016/S0021-9258(19)69516-5
Mohn, W. W., van der Geize, R., Stewart, G. R., Okamoto, S., Liu, J., Dijkhuizen, L., et al. (2008). The Actinobacterial Mce4 Locus Encodes a Steroid Transporter. J. Biol. Chem. 283 (51), 35368–35374. doi:10.1074/jbc.M805496200
Mounier, J., Hakil, F., Branchu, P., Naïtali, M., Goulas, P., Sivadon, P., et al. (2018). AupA and AupB Are Outer and Inner Membrane Proteins Involved in Alkane Uptake in Marinobacter Hydrocarbonoclasticus SP17. mBio 9 (3), e00520–00518. doi:10.1128/mBio.00520-18
Murdock, D., Ensley, B. D., Serdar, C., and Thalen, M. (1993). Construction of Metabolic Operons Catalyzing the de novo Biosynthesis of Indigo in Escherichia coli. Biotechnology (N Y) 11 (3), 381–386. doi:10.1038/nbt0393-381
Narhi, L. O., and Fulco, A. J. (1986). Characterization of a Catalytically Self-Sufficient 119,000-dalton Cytochrome P-450 Monooxygenase Induced by Barbiturates in Bacillus megaterium. J. Biol. Chem. 261 (16), 7160–7169. doi:10.1016/S0021-9258(17)38369-2
Narhi, L. O., and Fulco, A. J. (1987). Identification and Characterization of Two Functional Domains in Cytochrome P-450BM-3, a Catalytically Self-Sufficient Monooxygenase Induced by Barbiturates in Bacillus megaterium. J. Biol. Chem. 262 (14), 6683–6690. doi:10.1016/S0021-9258(18)48296-8
Nikaido, H. (2003). Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 67 (4), 593–656. doi:10.1128/mmbr.67.4.593-656.2003
Noble, M. A., Miles, C. S., Chapman, S. K., Lysek, D. A., MacKay, A. C., Reid, G. A., et al. (1999). Roles of Key Active-Site Residues in Flavocytochrome P450 BM3. Biochem. J. 339, 371–379. doi:10.1042/bj3390371
Nunn, W. D., and Simons, R. W. (1978). Transport of Long-Chain Fatty Acids by Escherichia coli: Mapping and Characterization of Mutants in the fadL Gene. Proc. Natl. Acad. Sci. U S A. 75 (7), 3377–3381. doi:10.1073/pnas.75.7.3377
Olivera, E. R., and Luengo, J. M. (2019). Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways. Genes (Basel) 10 (7), 512. doi:10.3390/genes10070512
Panke, S., Meyer, A., Huber, C. M., Witholt, B., and Wubbolts, M. G. (1999). An Alkane-Responsive Expression System for the Production of fine Chemicals. Appl. Environ. Microbiol. 65 (6), 2324–2332. doi:10.1128/AEM.65.6.2324-2332.1999
Peng, Y., Gao, C., Zhang, Z., Wu, S., Zhao, J., and Li, A. (2022). A Chemoenzymatic Strategy for the Synthesis of Steroid Drugs Enabled by P450 Monooxygenase-Mediated Steroidal Core Modification. ACS Catal. 12 (5), 2907–2914. doi:10.1021/acscatal.1c05776
Putkaradze, N., Litzenburger, M., Hutter, M. C., and Bernhardt, R. (2019). CYP109E1 from Bacillus megaterium Acts as a 24- and 25-hydroxylase for Cholesterol. ChemBioChem 20 (5), 655–658. doi:10.1002/cbic.201800595
Ruff, A. J., Arlt, M., van Ohlen, M., Kardashliev, T., Konarzycka-Bessler, M., Bocola, M., et al. (2016). An Engineered Outer Membrane Pore Enables an Efficient Oxygenation of Aromatics and Terpenes. J. Mol. Catal. B Enzym 134, 285–294. doi:10.1016/j.molcatb.2016.11.007
Sambrook, J., and Russell, D. W. (2001). Molecular Cloning: A Laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Schäfer, L., Karande, R., and Bühler, B. (2020). Maximizing Biocatalytic Cyclohexane Hydroxylation by Modulating Cytochrome P450 Monooxygenase Expression in P. Taiwanensis VLB120. Front. Bioeng. Biotechnol. 8, 140. doi:10.3389/fbioe.2020.00140
Schiffer, L., Anderko, S., Hobler, A., Hannemann, F., Kagawa, N., and Bernhardt, R. (2015). A Recombinant CYP11B1 Dependent Escherichia coli Biocatalyst for Selective Cortisol Production and Optimization towards a Preparative Scale. Microb. Cel. Fact. 14, 25. doi:10.1186/s12934-015-0209-5
Schmidt, A., Kochanowski, K., Vedelaar, S., Ahrné, E., Volkmer, B., Callipo, L., et al. (2016). The Quantitative and Condition-dependent Escherichia coli Proteome. Nat. Biotechnol. 34 (1), 104–110. doi:10.1038/nbt.3418
Schmitz, D., Zapp, J., and Bernhardt, R. (2014). Steroid Conversion with CYP106A2 - Production of Pharmaceutically Interesting DHEA Metabolites. Microb. Cel. Fact. 13, 81. doi:10.1186/1475-2859-13-81
Schneider, S., Wubbolts, M. G., Sanglard, D., and Witholt, B. (1998). Biocatalyst Engineering by Assembly of Fatty Acid Transport and Oxidation Activities for In Vivo Application of Cytochrome P-450BM-3 Monooxygenase. Appl. Environ. Microbiol. 64 (10), 3784–3790. doi:10.1128/AEM.64.10.3784-3790.1998
Schrewe, M., Magnusson, A. O., Willrodt, C., Bühler, B., and Schmid, A. (2011). Kinetic Analysis of Terminal and Unactivated C-H Bond Oxyfunctionalization in Fatty Acid Methyl Esters by Monooxygenase-Based Whole-Cell Biocatalysis. Adv. Synth. Catal. 353 (18), 3485–3495. doi:10.1002/adsc.201100440
Schrewe, M., Julsing, M. K., Bühler, B., and Schmid, A. (2013). Whole-cell Biocatalysis for Selective and Productive C-O Functional Group Introduction and Modification. Chem. Soc. Rev. 42 (15), 6346–6377. doi:10.1039/c3cs60011d
Schrewe, M., Julsing, M. K., Lange, K., Czarnotta, E., Schmid, A., and Bühler, B. (2014). Reaction and Catalyst Engineering to Exploit Kinetically Controlled Whole-Cell Multistep Biocatalysis for Terminal FAME Oxyfunctionalization. Biotechnol. Bioeng. 111 (9), 1820–1830. doi:10.1002/bit.25248
Schubeis, T., Le Marchand, T., Daday, C., Kopec, W., Tekwani Movellan, K., Stanek, J., et al. (2020). A β-barrel for Oil Transport through Lipid Membranes: Dynamic NMR Structures of AlkL. Proc. Natl. Acad. Sci. U S A. 117 (35), 21014–21021. doi:10.1073/pnas.2002598117
Staijen, I. E., van Beilen, J. B., and Witholt, B. (2000). Expression, Stability and Performance of the Three-Component Alkane Mono-Oxygenase of Pseudomonas oleovorans in Escherichia coli. Eur. J. Biochem. 267 (7), 1957–1965. doi:10.1046/j.1432-1327.2000.01196.x
Straathof, A. J., Panke, S., and Schmid, A. (2002). The Production of fine Chemicals by Biotransformations. Curr. Opin. Biotechnol. 13 (6), 548–556. doi:10.1016/s0958-1669(02)00360-9
Szaleniec, M., Wojtkiewicz, A. M., Bernhardt, R., Borowski, T., and Donova, M. (2018). Bacterial Steroid Hydroxylases: Enzyme Classes, Their Functions and Comparison of Their Catalytic Mechanisms. Appl. Microbiol. Biotechnol. 102 (19), 8153–8171. doi:10.1007/s00253-018-9239-3
Tong, W.-Y., and Dong, X. (2009). Microbial Biotransformation: Recent Developments on Steroid Drugs. Recent Pat Biotechnol. 3 (2), 141–153. doi:10.2174/187220809788700157
Urlacher, V. B., and Eiben, S. (2006). Cytochrome P450 Monooxygenases: Perspectives for Synthetic Application. Trends Biotechnol. 24 (7), 324–330. doi:10.1016/j.tibtech.2006.05.002
Urlacher, V. B., and Schmid, R. D. (2006). Recent Advances in Oxygenase-Catalyzed Biotransformations. Curr. Opin. Chem. Biol. 10 (2), 156–161. doi:10.1016/j.cbpa.2006.02.001
van Beilen, J. B., Eggink, G., Enequist, H., Bos, R., and Witholt, B. (1992). DNA Sequence Determination and Functional Characterization of the OCT-Plasmid-Encoded alkJKL Genes of Pseudomonas oleovorans. Mol. Microbiol. 6 (21), 3121–3136. doi:10.1111/j.1365-2958.1992.tb01769.x
van Beilen, J. B., Duetz, W. A., Schmid, A., and Witholt, B. (2003). Practical Issues in the Application of Oxygenases. Trends Biotechnol. 21 (4), 170–177. doi:10.1016/S0167-7799(03)00032-5
van den Berg, B., Black, P. N., Clemons, W. M. J., and Rapoport, T. A. (2004). Crystal Structure of the Long-Chain Fatty Acid Transporter FadL. Science 304 (5676), 1506–1509. doi:10.1126/science.1097524
van den Berg, B. (2005). The FadL Family: Unusual Transporters for Unusual Substrates. Curr. Opin. Struct. Biol. 15 (4), 401–407. doi:10.1016/j.sbi.2005.06.003
van den Berg, B. (2010). Going Forward Laterally: Transmembrane Passage of Hydrophobic Molecules through Protein Channel walls. ChemBioChem 11 (10), 1339–1343. doi:10.1002/cbic.201000105
van Nuland, Y. M., Eggink, G., and Weusthuis, R. A. (2016). Application of AlkBGT and AlkL from Pseudomonas putida GPo1 for Selective Alkyl Ester ω-oxyfunctionalization in Escherichia coli. Appl. Environ. Microbiol. 82 (13), 3801–3807. doi:10.1128/AEM.00822-16
van Vugt-Lussenburg, B. M. A., Damsten, M. C., Maasdijk, D. M., Vermeulen, N. P., and Commandeur, J. N. M. (2006). Heterotropic and Homotropic Cooperativity by a Drug-Metabolising Mutant of Cytochrome P450 BM3. Biochem. Biophys. Res. Commun. 346 (3), 810–818. doi:10.1016/j.bbrc.2006.05.179
Venkataraman, H., te Poele, E. M., Rosłoniec, K. Z., Vermeulen, N., Commandeur, J. N. M., van der Geize, R., et al. (2015). Biosynthesis of a Steroid Metabolite by an Engineered Rhodococcus erythropolis Strain Expressing a Mutant Cytochrome P450 BM3 Enzyme. Appl. Microbiol. Biotechnol. 99 (11), 4713–4721. doi:10.1007/s00253-014-6281-7
Wang, Y., Rawlings, M., Gibson, D. T., Labbé, D., Bergeron, H., Brousseau, R., et al. (1995). Identification of a Membrane Protein and a Truncated LysR-type Regulator Associated with the Toluene Degradation Pathway in Pseudomonas putida F1. Mol. Gen. Genet. 246 (5), 570–579. doi:10.1007/BF00298963
Whitehouse, C. J. C., Bell, S. G., and Wong, L.-L. (2012). P450BM3 (CYP102A1): Connecting the Dots. Chem. Soc. Rev. 41 (3), 1218–1260. doi:10.1039/c1cs15192d
Woodley, J. M. (2006). Microbial Biocatalytic Processes and Their Development. Adv. Appl. Microbiol. 60, 1–15. doi:10.1016/S0065-2164(06)60001-4
Zehentgruber, D., Drǎgan, C. A., Bureik, M., and Lütz, S. (2010a). Challenges of Steroid Biotransformation with Human Cytochrome P450 Monooxygenase CYP21 Using Resting Cells of Recombinant Schizosaccharomyces pombe. J. Biotechnol. 146 (4), 179–185. doi:10.1016/j.jbiotec.2010.01.019
Zehentgruber, D., Hannemann, F., Bleif, S., Bernhardt, R., and Lütz, S. (2010b). Towards Preparative Scale Steroid Hydroxylation with Cytochrome P450 Monooxygenase CYP106A2. ChemBioChem 11 (5), 713–721. doi:10.1002/cbic.200900706
Zhang, X., Peng, Y., Zhao, J., Li, Q., Yu, X., Acevedo-Rocha, C. G., et al. (2020). Bacterial Cytochrome P450-Catalyzed Regio- and Stereoselective Steroid Hydroxylation Enabled by Directed Evolution and Rational Design. Bioresour. Bioproc. 7, 2. doi:10.1186/s40643-019-0290-4
Keywords: whole-cell biocatalysis, steroid hydroxylation, substrate uptake, cytochrome P450 monooxygenase, hydrophobic membrane pores, AlkL
Citation: Bertelmann C, Mock M, Koch R, Schmid A and Bühler B (2022) Hydrophobic Outer Membrane Pores Boost Testosterone Hydroxylation by Cytochrome P450 BM3 Containing Cells. Front. Catal. 2:887458. doi: 10.3389/fctls.2022.887458
Received: 01 March 2022; Accepted: 28 March 2022;
Published: 26 April 2022.
Edited by:
Dirk Tischler, Ruhr University Bochum, GermanyReviewed by:
Aitao Li, Hubei University, ChinaFlorian Rudroff, Vienna University of Technology, Austria
Copyright © 2022 Bertelmann, Mock, Koch, Schmid and Bühler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bruno Bühler, YnJ1bm8uYnVlaGxlckB1ZnouZGU=
 Carolin Bertelmann
Carolin Bertelmann Magdalena Mock1
Magdalena Mock1 Bruno Bühler
Bruno Bühler