
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cardiovasc. Med. , 07 May 2021
Sec. Cardiovascular Genetics and Systems Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fcvm.2021.659628
Despite treatments being improved and many risk factors being identified, cardiovascular disease (CVD) is still a leading cause of mortality and disability worldwide. N6-methyladenosine (m6A) is the most common, abundant, and conserved internal modification in RNAs and plays an important role in the development of CVD. Many studies have shown that aabnormal m6A modifications of coding RNAs are involved in the development of CVD. In addition, non-coding RNAs (ncRNAs) exert post-transcriptional regulation in many diseases including CVD. Although ncRNAs have also been found to be modified by m6A, the studies on m6A modifications of ncRNAs in CVD are currently lacking. In this review, we summarized the recent progress in understanding m6A modifications in the context of coding RNAs and ncRNAs, as well as their regulatory roles in CVD.
Cardiovascular disease (CVD) is a leading cause of mortality and disability worldwide despite recent improvements in health care, with many risk factors identified (1). Therefore, the mechanisms underlying CVD development remain to be elucidated. Recently, abnormal modifications in RNA have been identified in CVD and have attracted attention to our understanding of the mechanism underlying CVD development (2).
Currently, over 100 chemical modifications of RNA have been identified. Among them, N6-methyladenosine (m6A) is the most common conserved internal modification in RNA and is activated by the m6A methyltransferases (m6A writers), reversed by m6A demethylases (m6A erasers), and recognized by m6A-binding proteins (m6A readers) (3). m6A is enriched in the 3′ untranslated regions (3′-UTRs), stop codons, internal long exons, and consensus sequence RRACH (where R: A or G and H: A, C, or U), thus affecting mRNA splicing, export, translation, and decay (4). m6A modification is also present in the 5′ cap, which is required for RNA stability or degradation (5). m6A modification accounts for ~50% of the mRNA modifications in mammals (6). In addition to mRNAs, the m6A modification is also found in non-coding RNAs (nc RNAs) including micro RNAs (miRNAs), long non-coding RNAs (lnc RNAs), and circular RNAs (circ RNAs), which have been found to regulate transcription in many diseases including CVD (6–9).
Abnormal m6A RNA modifications have been found in the CVD risk conditions and to regulate the CVD development. However, research regarding the underlying mechanism is still lacking. In this review, we summarize the recent progress on the m6A modifications of mRNAs and ncRNAs, as well as their regulatory roles in CVD.
The m6A RNA modification is dynamically regulated by diverse functional proteins, including m6A methyltransferases, demethylases, and others (m6A writers, erasers, and readers, respectively). m6A writers include methyltransferase-like 3/14/16 (METTL3, METTL14, METTL16), Wilm's tumor-associated protein (WTAP), RNA-binding motif protein 15 (RBM15)—and its paralog RBM15B—and KIAA1429, NOP2/Sun RNA methyltransferase 2 (NSun2), and zinc finger CCHC domain-containing protein 4 (ZCCHC4) (10–13). METTL3 and METTL14 are the core components of the m6A writers; WTAP, RBM15, and KIAA1429 are also important components of the m6A methylase complex to enhance methyltransferase activity (10, 11). METTL16, NOP2/Sun RNA methyltransferase 2 (NSun2), and zinc finger CCHC domain-containing protein 4 (ZCCHC4) are other components of the m6A methylase complex, which are indispensable for m6A deposition (12, 13). m6A erasers consist of fat mass and obesity-associated protein (FTO) and AlkB family member 5 (ALKBH5) and can mediate m6A demethylation (14). m6A readers include YT521-B homology (YTH) domain family proteins (YTHDF1-3), insulin-like growth factor 2 mRNA-binding protein (IGF2BP), YTH domain-containing proteins (YTHDC), heterogeneous nuclear ribonucleoprotein (HNRNP), and eukaryotic translation initiation factor 3 (eIF3), which are also involved in m6A modification (15–19). YTHDC1 can also regulate target gene transcription (16, 17). YTHDF1 can bind to the 3′UTRs and the stop codon of m6A-containing RNAs, and interact with eIF3 to initiate translation (18). IGF2BPs, HNRNPC, and HNRNPG affect the stability, storage, the structure of RNA (18, 19). Therefore, m6A writers, erasers, and readers affect the translation, export, degradation, and structure of RNAs to regulate the development of many diseases (Figure 1).
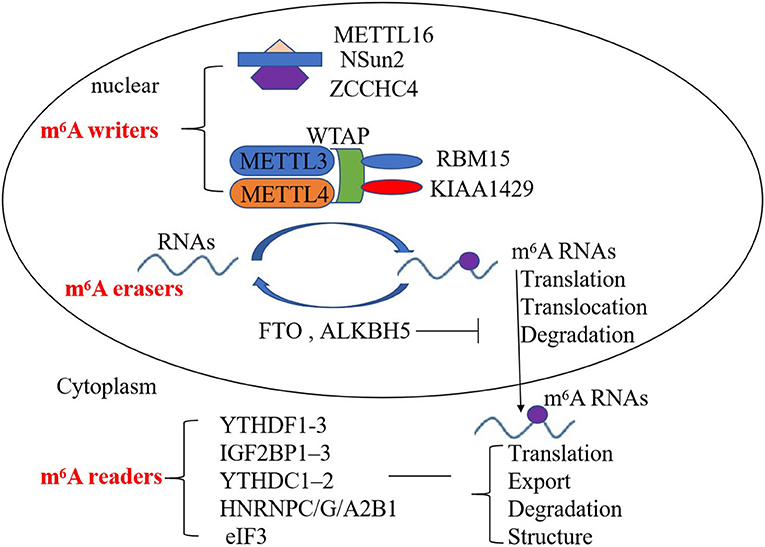
Figure 1. Mechanism of the deposition of m6A modification on mRNA and non-coding RNAs. m6A writers, erasers, and readers regulate the deposition of m6A modification and affect the translocation, translation, stability, degradation, and structure of RNA.
m6A methylation is the most prevalent internal post-transcriptional modification of mammalian mRNA and can affect mRNA splicing, translocation, translation, stability, and structure (20). m6A modification frequently occurs in the introns of pre-mRNAs and promotes the nuclear export of mRNAs and facilitates mRNA transcription in the cytoplasm (21). m6A writer, METTL3 accelerates mRNA translocation from the nucleus to the cytoplasm and enhances translation of target mRNAs (22, 23). Moreover, METTL3 and METTL14 also reduce mRNA stability and promote mRNA degradation efficiency (5). In contrast, m6A eraser, ALKBH5 inhibits mRNA export and stability (14, 24). m6A readers are also found to regulate mRNA in many ways. For example, YTHDF1, YTHDF3, and IGF2BP1/2/3 can drive mRNA translation and promote translation efficiency (25, 26). Furthermore, IGF2BP also can enhance the stability of mRNA by binding to mRNA-stabilizing proteins such as human antigen R (HuR) (27). HuR is an RNA binding protein and can increase RNA stability (28). However, m6A modification can interact with HuR and inhibit its ability of enhancing RNA stability (28). YTHDF2 can promote the degradation of m6A-containing mRNA by recruiting RNA-degrading enzymes or adaptor proteins CCR4/NOT or HRSP12-RNase (29). Similarly, HNRNPC and HNRNPG can recognize specific sites on mRNA, thereby altering the structure of mRNAs (19). m6A writers, erasers, and readers affect mRNA expression in many aspects.
Nc RNAs exert post-transcriptional regulation in many diseases and mainly include miRNAs, lnc RNAs, and circ RNAs (30). miRNAs are small nc RNA molecules ~22 nucleotides in length that bind with the 3′-UTR of mRNA to post-transcriptionally regulate genes (31). Lnc RNAs are ncRNAs that are longer than 200 nucleotides in length and circ RNAs are a specific class of ncRNA that form a covalently closed loop, and they interfere with gene expression and signaling pathways at various stages, such as the sponging of miRNAs (32, 33). Recently, many studies showed that miRNAs, lnc RNAs, and circ RNAs are modified by m6A (28, 34–45).
The m6A writers, METTL3 and METTL14 affect miRNA maturation by interacting with DiGeorge critical region 8 (DGCR8), which can bind to pri-miRNAs and promote miRNA maturation (34, 35). HuR is also found to increase miRNA stability by interfering with the binding of miRNAs to the Ago complex (28). The m6A eraser, FTO can enhance the stability of hsa-miR-6505-5p, hsa-miR-651-5p, and hsa-miR-493-5p, and reduce the stability of hsa-miR-7-5p, hsa-miR-92a-1-5p, and hsa-miR-6769a-3p, but the underlying molecular mechanism is not clear (36). While m6A modifies miRNA, miRNAs can also target m6A independently. For example, the miRNA let-7g binds to the 3′-UTR of METTL3 mRNA to inhibit its expression (37). Similarly, miR-145 targets the mRNA encoding YTHDF2 and inhibits YTHDF2 expression, which can stabilize m6A-modified mRNAs (38). Therefore, there is crosstalk of m6A modification with miRNAs.
Many m6A-methylated lnc RNA transcripts have been identified in mouse transcriptome (46). For example, METTL3 can increase the nuclear accumulation of lnc RNA RP11 to enhance its expression in colon cancer (39). METTL16 can methylate 68 lnc RNAs in human embryonic kidney 293 cells (40). By contrast, the m6A eraser, ALKBH5, can demethylate lnc RNA KCNK15-AS1 and nuclear paraspeckle assembly transcript 1 (NEAT1) (41). ALKBH5 was also found to reduce the m6A level and increase the stability of lnc RNA growth arrest-specific 5 (GAS5) (42). The m6A readers, YTHDF2 and YTHDF3 were found to promote the degradation of GAS5 (42, 43). The m6A reader IGF2BP2 interacts with the lnc RNA DANCR and stabilizes DANCR RNA (44). In addition, YTHDC1 and YTHDF2 are found to regulate the export and stability of circ RNAs (29, 45). Thus, the m6A modification exerts the regulatory effect through regulating the expressions of lnc RNAs and circ RNAs.
Many risk factors of CVD such as hyperlipidaemia, diabetes, and inflammation have been identified, but their molecular mechanisms in regulating CVD are still investigated (47–49). Recently, it has been found that m6A RNA methylations are dysregulated in risk conditions, and involved in the pathology of CVD (50, 51). These findings may provide insight into the molecular mechanisms underlying CVD development.
Hyperlipidaemia and obesity are risk factors for CVD development, and m6A functional enzymes are dysregulated and involved in lipid metabolism (47, 50). Oscillations in mRNA m6A methylation in the murine liver depend on a functional circadian clock, which is essential for lipid metabolic homeostasis (52). m6A methylation of peroxisome proliferator-activated receptor α (PPaRα) mRNA that codes for a nuclear receptor can accelerate lipid metabolism (22). m6A modification of PPaRα mRNA was decreased by METTL3 knockdown, causing the reduction of cellular lipid accumulation (53). In addition, m6A erasers and readers are also involved in lipid metabolism. FTO facilitates the adipogenesis of 3T3-L1 cells by interacting with YTHDF2 to maintain FTO-induced m6A demethylation (54). Consistently, FTO inhibition suppresses adipogenesis through an m6A-YTHDF2-dependent mechanism (54, 55). YTHDF2 is found to promote lipid accumulation by directly binding to the m6A modification site to promote the translation of 6-phosphogluconate dehydrogenase, which can increase the level of cholesterol FAM134B (56). Similarly, YTHDF1 is also found to promote adipogenesis in intramuscular preadipocytes by enhancing the translation of mitochondrial carrier homolog 2, which limited energy utilization and promoted diet-induced obesity (57). These evidences show that m6A writers, erasers, and readers can regulate lipid metabolism genes, which are involved in the development of CVD.
Diabetes is another risk factor positively correlated with the incidence of CVD (48). In patients with type 2 diabetes (T2D), m6A levels were reduced, while the mRNA levels of FTO, METTL3, METTL14, WTAP were significantly elevated and involved in the pathogenesis of diabetes (51). However, high glucose was found to enhance FTO levels in HepG2 cells (58). FTO can participate in glucose and insulin metabolism by inducing the expression of forkhead box O1 (FOXO1), glucose-6-phosphatase catalytic subunit, and diacylglycerol O-acyltransferase 2 mRNA (51). METTL3 and METTL14 were found to regulate insulin secretion in human β-cells. METTL14 inhibition can inhibit β-cell proliferation and promote insulin dysregulation (58). These findings indicate that FTO, METTL3, and METTL14 play important roles in the development of diabetes or CVD by regulating glucose metabolism and insulin secretion.
Inflammation was found in all phenomena associated with CVD including vascular and cardiac dysfunction (49). For example, M1-type macrophage-mediated inflammation plays an important role in the development of atherosclerosis (59). METTL3 expression is increased in M1-type macrophages and can directly methylate the mRNA of signal transducer and activator of transcription 1 to increase its expression (60). METTL3 can also promote the activation of dendritic cells by activating toll-like receptor 4 (TLR4)/nuclear factor-κB (NF-κB) signaling and increasing the expression of CD40, CD80, and IL-12 (61). METTL3 also can stimulate T cells and promote their differentiation (61, 62). METTL14 was found to promote an inflammatory response in endothelial cell (EC) and atherosclerotic plaque formation by interacting with FOXO1 and mediating its m6A modification (63). FOXO1 is an important transcription factor that acts directly on the promoter regions of VCAM-1 and ICAM-1 to promote their transcription (63). This evidence indicates that METTL3 and METTL14 can promote inflammation to regulate the development of CVD.
CVD risk factors, such as hyperlipidaemia, hyperglycaemia, and inflammation can lead to vascular dysfunction, which ultimately results in cardiomyocyte ischemic injury and myocardial infarction (MI) (64, 65). The fibroblasts are activated and extracellular matrix (ECM) components are over-produced after MI; these compensate for cardiomyocyte loss and maintain the structural integrity of the ECM (66). Excessive cardiac remodeling and fibrosis following the cardiac injury can cause cardiomyocyte hypertrophy, which ultimately leads to heart failure (67). Dysregulated m6A RNA methylation has also been found to be responsible for vascular or cardiac dysfunction (Figure 2).
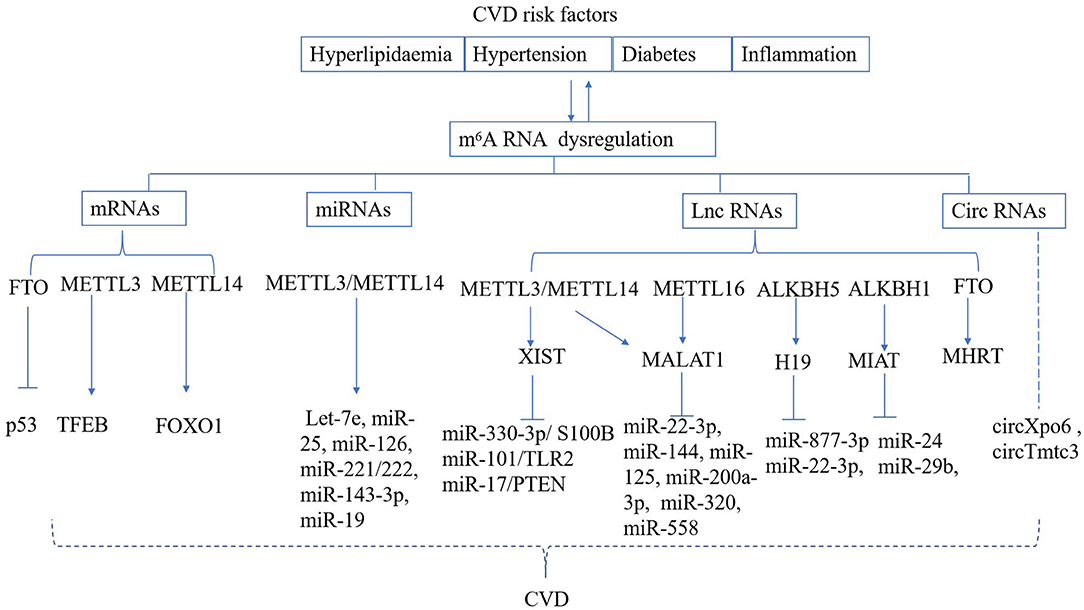
Figure 2. Regulation of m6A RNA modification in CVD. CVD risk factors can result in deregulated m6A deposition on mRNA, miRNA, lnc RNA, circ RNA, which are known to play a role in the development of cardiac and vascular disease.
METTL3 and FTO have been found to regulate vascular or cardiac dysfunction under stress conditions (68). METTL3 induced by hypoxic stress can promote the differentiation of adipose-derived stem cells into vascular smooth muscle cells (VSMCs) by increasing the expression of paracrine factors, including VEGF, and TGF-β (69). Similarly, METTL3 was also found to promote the differentiation of mouse embryonic stem cells into cardiomyocytes (70). In addition, METTL3 promoted the apoptosis of hypoxia and reperfusion (H/R)-treated cardiomyocytes by regulating the expression of transcription factor EB, which is a master regulator of lysosomal biogenesis and autophagy genes (71). Moreover, METTL3 promotes cardiac remodeling and hypertrophy by catalyzing the m6A methylation of certain subsets of mRNAs (70). In contrast, METTL3 knockout hearts develop maladaptive eccentric remodeling and cardiac functional defects with aging and rapid progressive dysfunction following acute pressure-overload stress (72). Cardiac FTO expression is decreased in cardiomyocytes under conditions of hypoxia, ischemia, and heart failure (73). It has been observed that FTO overexpression attenuates hypoxia-induced cardiomyocyte dysfunction and restores calcium handling and sarcomere dynamics (73). FTO has been shown to attenuate ischemia-induced cardiac remodeling and improve cardiac contractility by demethylating the m6A modifications of p53, thereby inhibiting the expression of p53 (74, 75). Thus, m6A writers, erasers, and readers can regulate the developments of vascular and cardiac diseases via the methylation of target mRNAs.
miRNAs are a determinant of cardiovascular pathology and could be modified by m6A (31). For example, m6A modification and METTL14 are significantly up-regulated in atherosclerotic vascular endothelial cells and promote their proliferation (76). The underlying mechanism is that the METTL14 inhibits the expression of pri-miR-19a but increases the expression of mature miR-19a by binding to DGCR8 (76). Similarly, METTL3 homolog, mRNA adenosine methylase (MTA) can accumulate primary pri-miRNAs but inhibits the expression of mature miRNAs In Arabidopsis (77). In addition, many miRNAs are found to be mediated the deposition of m6A modification by METTL3 or METTL14, and some of them play important roles in CVD development (78). For example, METTL3 affect the stability of Let-7e, miR-25, miR-126, miR-221/222, and miR-143-3p (78). METTL14 modulates the primary processing of miR-126 and miR-375 by interacting with DGCR8 in hepatocellular carcinoma or colon cancer, respectively (34, 79). Let-7, miR-126, miR-221/222, and miR-143-3p are key vascular biology players that are involved in the development of atherosclerosis and angiogenesis via their effects on ECs and VSMCs (80–85). Let-7, miR-25, and miR-375 play an important role in the development of cardiac diseases, including arrhythmia, dilated cardiomyopathy, MI, cardiac hypertrophy, fibrosis, and heart failure by regulating apoptosis, autophagy, oxidative stress, inflammation, and calcium handling (80, 86, 87). Those pieces of evidence indicate that m6A modifications are involved in the development of CVD by affecting the expressions of miRNAs.
Similar to miRNAs, lnc RNAs and circ RNAs have been thoroughly investigated in the context of CVD and have recently been found to be m6A-methylated (32, 88). For example, the m6A modification is enriched on lnc RNA 1281 and the m6A modification of lnc RNA 1281 affects the differentiation of embryonic stem cells (ESC) via sponging Let-7, which has been reported to play an important role in the cardiovascular differentiation of ESCs and the development of CVD (80, 89).
The lnc RNA H19 is highly expressed in human atherosclerotic lesions and promotes the development of atherosclerosis by regulating the mitogen-activated protein kinase and NF-kB signaling pathways (90). Additionally, H19 ameliorates ischemia-reperfusion (I/R)-induced myocardial apoptosis or MI-induced myocardial injury by sponging miR-877-3p or miR-22-3p, respectively (91, 92). In H9c2 cells with H2O2-induced senescence, H/R enhanced the level of m6A methylation and increased the expression of lnc RNA H19 by upregulating ALKBH5 (93). Therefore, the m6A modification of H19 is involved in the development of CVD.
Lnc RNA myocardial infarction associated transcript (MIAT) is also found to inhibit EC proliferation, migration, and tube formation in diabetes via the sponging of miR-29b (94). MIAT levels were also increased in MI and deregulated some fibrosis-related regulators by sponging miR-24 and increasing the expression of furin and TGF-β1 (95). Similarly, the MIAT levels increase in response to hypoxia, and MIAT is involved in cardiac interstitial fibrosis (96). Oxidized low-density lipoprotein (ox-LDL)-induced m6A demethylation was found to facilitate the binding of HIF1α to the ALKBH1-demethylated MIAT promoter and the transactivation of MIAT, indicating that MIAT is a target gene of ALKBH1-related m6A methylation (97).
Lnc RNA X-inactive specific transcript (XIST) was reported to play an important role in CVD development and is highly m6A-methylated (3). XIST was highly expressed in human thoracic aortic dissection and promoted the apoptosis of VSMCs by sponging miR-17 (98). Consistently miR-17 was reported to promote mitochondria-dependent apoptosis by targeting at phosphatase and tensin homolog deleted on chromosome 10 (PTEN) (98). In addition, XIST inhibited myocardial cell proliferation by sponging miR-130a-3p, which targets phosphodiesterase 4D (99). XIST was also found to promote phenylephrine-induced cardiac hypertrophy via the miR-330-3p/S100B and miR-101/TLR2 axis (100, 101). The METTL3/METTL14 complex deposited the 78 m6A-methylation on XIST RNA by interacting with the MACOM complex, comprising WTAP, VIRMA, and RBM15 proteins, and inhibited the expression of XIST (102). YTHDC1 and YTHDF2 bind to XIST and mediate its degradation (102). This evidence indicated that the m6A modification of XIST might regulate the development of CVD.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) regulates the development of CVD and contains several m6A motifs (103, 104). MALAT1 protects against endothelial injury induced by ox-LDL, hyperglycaemia, and oxidative stress via the sponging of miR-22-3p or activation of nuclear factor erythroid 2 (105, 106). MALAT1 levels were increased in the serum and myocardial tissue of AMI and promoted cardiomyocyte apoptosis or myocardial tissue injury induced by hypoxia, H/R, or I/R by targeting miR-144, miR-125, miR-200a-3p, or miR-320, respectively (107–110). However, it was also found that MALAT1 inhibits isoproterenol-induced cardiomyocyte apoptosis by sponging miR-558 (111). Additionally, MALAT1 promoted angiotensin II-induced cardiac fibrosis by sponging miR-145, thereby enhancing target growth factor-β1 activity (112). Recently, the m6A-deposition sites of MALAT1 have been identified. For example, m6A modification at the A2577 or A2515 site of MALAT1 can destabilize the RNA hairpin, release the poly(U) tract, and increase binding with HNRNPC or HNRNPG, respectively (103, 104). METTL16 specifically binds to the 3′-end of a triple-helix and enhances the stability of MALAT1 transcripts (113). This evidence indicates that MALAT1 could be m6A modified to regulate the development of CVD.
The levels of GAS5 were increased in atherosclerotic rats and aggravated ox-LDL-induced inflammation by inhibiting the expression of miR-221 or miR-135a (114, 115). GAS5 was also found to accelerate myocardial I/R injury by sponging miR-532-5p (116). In contrast, other studies showed that GAS5 could attenuate homocysteine-induced cardiac microvascular ECs by inhibiting miR-33a-5p and reverse cardiac apoptosis and fibrosis via the inhibition of semaphorin-3A or miR-21 expression, respectively (117–119). The m6A modifications of GAS5 have also been identified. ALKBH5 reduced the m6A level and increased the stability of GAS5. m6A induced GAS5 RNA degradation in a YTHDF2-dependent manner (42). Knockdown of YTHDF3 was also found to prolong the degradation of GAS5. This evidence indicates that m6A-deposited GAS5 might be involved in the development of CVD (42, 43).
Lnc RNA, Myheart (MHRT), plays an important role in cardiac disease. MHRT protects against the H2O2 or H/R -induced apoptosis of cardiomyocytes (120). In addition, MHRT is found to regulate cardiac hypertrophy and is associated with the outcome of heart failure (121, 122). Over-expression of FTO protects against H/R-induced apoptosis of myocardial cells by regulating m6A modification of MHRT, indicating that m6A modification of MHRT participates in the development of cardiac disease (123).
Certain circ RNAs, such as circXpo6 and circTmtc3, have also demonstratedm6A-methylation in the lungs of rats with hypoxia-induced pulmonary hypertension, as well as in pulmonary artery smooth muscle cells, and, finally, in pulmonary arterial ECs exposed to hypoxia. This suggests that m6A-methylated circXpo6 and circTmtc3 might be involved in the development of CVD (124). However, the role of m6A-methylated circ RNAs in the development of CVD requires further study.
Modulation of m6A could be a strategy for CVD treatment. For example, silencing of METTL reduced I/R-induced cardiac injury and H/R-induced apoptosis of cardiomyocytes by inducing autophagy (71). Moreover, METTL3 inhibition reduced cardiomyocyte remodeling under the hypertrophic stimuli (125). Similarly, inhibition of METTL14 was found to decrease the calcification and enhance the vascular repair function (126). It was favored that inhibition of METTL14 inhibited the proliferation of atherosclerotic vascular endothelial cells by affecting the expression of miR-19 (76). Over-expression of FTO by adeno-associated virus serotype 9 (AAV9) significantly prevented the formation of atherosclerotic plaques by reducing total cholesterol (127). Furthermore, FTO over-expression significantly improved cardiac function by reducing fibrosis and increasing angiogenesis at the chronic stage of post-myocardial infarction (73). Moreover, the protective effect of FTO in cardiac disease is associated with the regulation of m6A modification of MHRT (123).
Non-coding RNAs also can regulate m6A Micro RNAs such as miR-33a and miR-4429 were found to inhibit METTL3 in the field of tumor studies, indicating that those miRNAs might be as therapeutic agents for CVD (128, 129). In addition, lnc RNA H19 has been reported to protect against H2O2-induced H9c2 cell apoptosis by up-regulating ALKBH5 (93). Thus, ncRNAs might be used for the regulation of m6A and CVD treatment.
CVD is a leading cause of death worldwide, but the underlying mechanism remains unknown. m6A is the most common, abundant, and conserved internal modification in RNAs, including mRNA and ncRNAs. In this review, we summarized the current research on m6A RNA modification on CVD risk conditions and development, which may help elucidate the molecular mechanism underlying CVD development. In addition, inhibition of MELL3/14 or over-expression of FTO could be used for the treatment of CVD. Notably, some ncRNAs also can regulate m6A modifications and could be therapeutic molecules for CVD, However, m6A modifications of ncRNAs in CVD require further study.
JWa designed the article. DS, JH, JWu, and JWa wrote and critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Jilin Provincial Health and Family Planning Commission (2020C036-3) and the basic and clinical research of coronary heart disease and artificial intelligence diagnosis team in Jilin Province (20200301003RQ) and Talent Project of Jilin Provincial Finance (2020SCZT003 and 2019SCZT081).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CVD, Cardiovascular disease; m6A,N6, methyladenosine; 3′-UTRs, 3′ untranslated regions; miRNAs, micro RNAs; lnc RNAs, long non-coding RNAs; circ RNAs, circular RNAs; METTL3, methyltransferase-like 3; METTL14, methyltransferase-like 14; WTAP, Wilm's tumor-associated protein; RBM15, RNA-binding motif protein 15; IGF2BP, insulin-like growth factor 2 mRNA-binding protein; HNRNP, heterogeneous nuclear ribonucleoprotein; NSun2, NOP2/Sun RNA methyltransferase 2; ZCCHC4, zinc finger CCHC domain-containing protein 4; FTO, fat mass and obesity-associated protein; ALKBH5, AlkB family member 5; YTH, YT521-B homology; YTHDF, YTH domain family proteins; YTHDC, YTH domain-containing proteins; eIF3, eukaryotic translation initiation factor 3; DGCR8, DiGeorge Critical Region 8; T2D, type 2 diabetes; FOXO1, forkhead box O1; H/R, hypoxia and reperfusion; MI, myocardial infarction; ox-LDL, oxidized low-density lipoprotein; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; EC, endothelial cell; VSMCs, vascular smooth muscle cells; XIST, X-inactive specific transcript; ESC, embryonic stem cells; I/R, ischemia-reperfusion; GAS5, growth arrest-specific 5; MIAT, myocardial infarction associated transcript; TLR4, toll-like receptor 4; NF-κB, nuclear factor-κB; ECM, extracellular matrix.
1. Mc Namara K, Alzubaidi H, Jackson JK. Cardiovascular disease as a leading cause of death: how are pharmacists getting involved? Integr Pharm Res Pract. (2019) 8:1–11. doi: 10.2147/IPRP.S133088
2. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. (2017) 169:1187–200.doi: 10.1016/j.cell.2017.05.045
3. He Y, Xing J, Wang S, Xin S, Han Y, Zhang J. Increased m6A methylation level is associated with the progression of human abdominal aortic aneurysm. Ann Transl Med. (2019) 7:797. doi: 10.21037/atm.2019.12.65
4. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m (6) a RNA methylation. Nat Rev Genet. (2014) 15:293–306. doi: 10.1038/nrg3724
5. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m6Am in the 5' cap controls mRNA stability. Nature. (2017) 541:371–5. doi: 10.1038/nature21022
6. Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. (2019) 12:121. doi: 10.1186/s13045-019-0805-7
7. Molinie B, Wang J, Lim KS, Hillebrand R, Lu ZX, Van Wittenberghe N, et al. M (6) A-LAIC-seq reveals the census and complexity of the m (6) a epitranscriptome. Nat Methods. (2016) 13:692–8. doi: 10.1038/nmeth.3898
8. Wang J, Ishfaq M, Xu L, Xia C, Chen C, Li J. METTL3/m6A/miRNA-873-5p attenuated oxidative stress and apoptosis in colistin-induced kidney injury by modulating Keap1/Nrf2 pathway. Front Pharmacol. (2019) 10:517. doi: 10.3389/fphar.2019.00517
9. Gurha P. Noncoding RNAs in cardiovascular diseases. Curr Opin Cardiol. (2019) 34:241–5. doi: 10.1097/HCO.0000000000000615
10. Wu R, Jiang D, Wang Y, Wang X. N (6)-Methyladenosine (m(6)A) methylation in mRNA with a dynamic and reversible epigenetic modification. Mol Biotechnol. (2016) 58:450–9. doi: 10.1007/s12033-016-9947-9
11. Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Corrigendum: structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. (2017) 542:260. doi: 10.1038/nature21073
12. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. (2017) 18:2004–14. doi: 10.15252/embr.201744940
13. Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q, et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. (2014) 34:3630–41. doi: 10.1128/MCB.00243-14
14. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
15. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
16. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLoS Genet. (2018) 14:e1007412. doi: 10.1371/journal.pgen.1007412
17. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. (2015) 162:1299–308. doi: 10.1016/j.cell.2015.08.011
18. Liang Y, Zhan G, Chang KJ, Yang YP, Wang L, Lin J, et al. The roles of m6A RNA modifiers in human cancer. J Chin Med Assoc. (2020) 83:221–6. doi: 10.1097/JCMA.0000000000000251
19. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. (2017) 45:6051–63. doi: 10.1093/nar/gkx141
20. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. (2019) 20:608–24. doi: 10.1038/s41580-019-0168-5
21. Lesbirel S, Wilson SA. The m6A-methylase complex and mRNA export. Biochim Biophys Acta Gene Regul Mech. (2019) 1862:319–28. doi: 10.1016/j.bbagrm.2018.09.008
22. Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. (2013) 155:793–806. doi: 10.1016/j.cell.2013.10.026
23. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. (2018) 561:556–60. doi: 10.1038/s41586-018-0538-8
24. Li XC, Jin F, Wang BY, Yin XJ, Hong W, Tian FJ. The m6A demethylase ALKBH5 controls trophoblast invasion at the maternal-fetal interface by regulating the stability of CYR61 mRNA. Theranostics. (2019) 9:3853–65. doi: 10.7150/thno.31868
25. Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. (2020) 34:1354–63. doi: 10.1038/s41375-019-0656-9
26. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. (2017) 27:315–28. doi: 10.1038/cr.2017.15
27. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. (2018) 20:285–95. doi: 10.1038/s41556-018-0045-z
28. Ahuja D, Goyal A, Ray PS. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. (2016) 13:1152–65. doi: 10.1080/15476286.2016.1229734
29. Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. (2020) 36:177–88. doi: 10.1016/j.tig.2019.12.007
30. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. (2011) 12:861–74. doi: 10.1038/nrg3074
31. Colpaert RMW, Calore M. MicroRNAs in cardiac diseases. Cells. (2019) 8:737. doi: 10.3390/cells8070737
32. Hermans-Beijnsberger S, van Bilsen M, Schroen B. Long non-coding RNAs in the failing heart and vasculature. Noncoding RNA Res. (2018) 3:118–30. doi: 10.1016/j.ncrna.2018.04.002
33. Wang L, Meng X, Li G, Zhou Q, Xiao J. Circular RNAs in cardiovascular diseases. Adv Exp Med Biol. (2018) 1087:191–204. doi: 10.1007/978-981-13-1426-1 15
34. Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6 -methyladenosine-dependent primary microRNA processing. Hepatology. (2017) 65:529–43. doi: 10.1002/hep.28885
35. Faller M, Toso D, Matsunaga M, Atanasov I, Senturia R, Chen Y, et al. DGCR8 recognizes primary transcripts of microRNAs through highly cooperative binding and formation of higher-order structures. RNA. (2010) 16:1570–83. doi: 10.1261/rna.2111310
36. Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B. N6-adenosine methylation in miRNAs. PLoS ONE. (2015) 10:e0118438. doi: 10.1371/journal.pone.0118438
37. Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. (2018) 415:11–9. doi: 10.1016/j.canlet.2017.11.018
38. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3'-Untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. (2017) 292:3614–23. doi: 10.1074/jbc.M116.749689
39. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. (2019) 18:87. doi: 10.1186/s12943-019-1014-2
40. Wei W, Ji X, Guo X, Ji S. Regulatory role of N6 -methyladenosine (m6 A) methylation in RNA processing and human diseases. J Cell Biochem. (2017) 118:2534–43. doi: 10.1002/jcb.25967
41. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. (2018) 48:838–46. doi: 10.1159/000491915
42. Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. (2019) 11:4909–21.
43. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer. (2019) 18:143. doi: 10.1186/s12943-019-1079-y
44. Hu X, Peng WX, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. (2020) 27:1782–94. doi: 10.1038/s41418-019-0461-z
45. Zhou C, Molinie B, Daneshvar K, Pondick JV, Wang J, Van Wittenberghe N, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. (2017) 20:2262–76. doi: 10.1016/j.celrep.2017.08.027
46. Nie Y, Tian GG, Zhang L, Lee T, Zhang Z, Li J, et al. Identifying cortical specific long noncoding RNAs modified by m6A RNA methylation in mouse brains. Epigenetics. (2020) 23:1–17. doi: 10.1080/15592294.2020.1861170
47. Joseph P, Leong D, McKee M, Anand SS, Schwalm JD, Teo K, et al. Reducing the global burden of cardiovascular disease, part 1: the epidemiology and risk factors. Circ Res. (2017) 121:677–94. doi: 10.1161/CIRCRESAHA.117.308903
48. Archundia Herrera MC, Subhan FB, Chan CB. Dietary patterns and cardiovascular disease risk in people with type 2 diabetes. Curr Obes Rep. (2017) 6:405–13. doi: 10.1007/s13679-017-0284-5
49. Willerson JT, Ridker PM. Inflammation as a cardiovascular risk factor. Circulation. (2004) 109(21 Suppl. 1):II2–10. doi: 10.1161/01.CIR.0000129535.04194.38
50. Heng J, Tian M, Zhang W, Chen F, Guan W, Zhang S. Maternal heat stress regulates the early fat deposition partly through modification of m6A RNA methylation in neonatal piglets. Cell Stress Chaperones. (2019) 24:635–45. doi: 10.1007/s12192-019-01002-1
51. Yang Y, Shen F, Huang W, Qin S, Huang JT, Sergi C, et al. Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J Clin Endocrinol Metab. (2019) 104:665–73. doi: 10.1210/jc.2018-00619
52. Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. (2018) 25:1816–28.e4. doi: 10.1016/j.celrep.2018.10.068
53. Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, et al. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. (2019) 33:7529–44. doi: 10.1096/fj.201802644R
54. Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q, et al. FTO regulates adipogenesis by controlling cell cycle progression via m6A-YTHDF2 dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids. (2018) 1863:1323–30. doi: 10.1016/j.bbalip.2018.08.008
55. Wu R, Guo G, Bi Z, Liu Y, Zhao Y, Chen N, et al. m6A methylation modulates adipogenesis through JAK2-STAT3-C/EBPβ signaling. Biochim Biophys Acta Gene Regul Mech. (2019) 1862:796–806. doi: 10.1016/j.bbagrm.2019.06.008
56. Cai M, Liu Q, Jiang Q, Wu R, Wang X, Wang Y. Loss of m6 A on FAM134B promotes adipogenesis in porcine adipocytes through m6 A-YTHDF2-dependent way. IUBMB Life. (2019) 71:580–6. doi: 10.1002/iub.1974
57. Jiang Q, Sun B, Liu Q, Cai M, Wu R, Wang F, et al. MTCH2 promotes adipogenesis in intramuscular preadipocytes via an m6A-YTHDF1-dependent mechanism. FASEB J. (2019) 33:2971–81. doi: 10.1096/fj.201801393RRR
58. De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, et al. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab. (2019) 1:765–74. doi: 10.1038/s42255-019-0089-9
59. Xu H, Jiang J, Chen W, Li W, Chen Z. Vascular macrophages in atherosclerosis. J Immunol Res. (2019) 2019:4354786. doi: 10.1155/2019/4354786
60. Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie N, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol. (2019) 317:C762–75. doi: 10.1152/ajpcell.00212.2019
61. Wang H, Hu X, Huang M, Liu J, Gu Y, Ma L, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat Commun. (2019) 10:1898. doi: 10.1038/s41467-019-09903-6
62. Furlan M, Galeota E, de Pretis S, Caselle M, Pelizzola M. m6A-dependent RNA dynamics in T cell differentiation. Genes. (2019) 10:28. doi: 10.3390/genes10010028
63. Jian D, Wang Y, Jian L, Tang H, Rao L, Chen K, et al. METTL14 aggravates endothelial inflammation and atherosclerosis by increasing FOXO1 N6-methyladeosine modifications. Theranostics. (2020) 10:8939–56. doi: 10.7150/thno.45178
64. Sun HJ, Wu ZY, Nie XW, Bian JS. Role of endothelial dysfunction in cardiovascular diseases: the link between inflammation and hydrogen sulfide. Front Pharmacol. (2020) 10:1568. doi: 10.3389/fphar.2019.01568
65. Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. (2012) 298:229–317. doi: 10.1016/B978-0-12-394309-5.00006-7
66. Piek A, de Boer RA, Silljé HH. The fibrosis-cell death axis in heart failure. Heart Fail Rev. (2016) 21:199–211. doi: 10.1007/s10741-016-9536-9
67. Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. (2010) 122:2727–35. doi: 10.1161/CIRCULATIONAHA.110.942268
68. Engel M, Eggert C, Kaplick PM, Eder M, Röh S, Tietze L, et al. The role of m6A/m-RNA methylation in stress response regulation. Neuron. (2018) 99:389–403.e9. doi: 10.1016/j.neuron.2018.07.009
69. Lin J, Zhu Q, Huang J, Cai R, Kuang Y. Hypoxia promotes vascular smooth muscle cell (VSMC) differentiation of adipose-derived stem cell (ADSC) by regulating Mettl3 and paracrine factors. Stem Cells Int. (2020) 2020:2830565. doi: 10.1155/2020/2830565
70. Dorn LE, Lasman L, Chen J, Xu X, Hund TJ, Medvedovic M, et al. The N6-Methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. (2019) 139:533–45. doi: 10.1161/CIRCULATIONAHA.118.036146
71. Song H, Feng X, Zhang H, Luo Y, Huang J, Lin M, et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. (2019) 15:1419–37. doi: 10.1080/15548627.2019.1586246
72. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. (2014) 10:93–5. doi: 10.1038/nchembio.1432
73. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N, et al. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation. (2019) 139:518–32. doi: 10.1161/CIRCULATIONAHA.118.033794
74. Zhou P, Wu M, Ye C, Xu Q, Wang L. Meclofenamic acid promotes cisplatin-induced acute kidney injury by inhibiting fat mass and obesity-associated protein-mediated m6A abrogation in RNA. J Biol Chem. (2019) 294:16908–17. doi: 10.1074/jbc.RA119.011009
75. Si R, Zhang Q, Tsuji-Hosokawa A, Watanabe M, Willson C, Lai N, et al. Overexpression of p53 due to excess protein O-GlcNAcylation is associated with coronary microvascular disease in type 2 diabetes. Cardiovasc Res. (2020) 116:1186–98. doi: 10.1093/cvr/cvz216
76. Zhang BY, Han L, Tang YF, Zhang GX, Fan XL, Zhang JJ, et al. METTL14 regulates m6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci. (2020) 24:7015–23. doi: 10.26355/eurrev_202006_21694
77. Bhat SS, Bielewicz D, Gulanicz T, Bodi Z, Yu X, Anderson SJ, et al. mRNA adenosine methylase (MTA) deposits m6A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc Natl Acad Sci USA. (2020) 117:21785–95. doi: 10.1073/pnas.2003733117
78. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. (2015) 519:482–5. doi: 10.1038/nature14281
79. Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L, et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol Ther. (2020) 28:599–612. doi: 10.1016/j.ymthe.2019.11.016
80. Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y, Zhou HH. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci. (2013) 14:23086–102. doi: 10.3390/ijms141123086
81. Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. (2008) 15:272–84. doi: 10.1016/j.devcel.2008.07.008
82. Schober A, Nazari-Jahantigh M, Wei Y, Bidzhekov K, Gremse F, Grommes J, et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med. (2014) 20:368–76. doi: 10.1038/nm.3487
83. Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Human miR-221/222 in physiological and atherosclerotic vascular remodeling. Biomed Res Int. (2015) 2015:354517. doi: 10.1155/2015/354517
84. Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. (2009) 104:476–87. doi: 10.1161/CIRCRESAHA.108.185363
85. Zhao W, Zhao SP, Zhao YH. MicroRNA-143/-145 in Cardiovascular Diseases. Biomed Res Int. (2015) 2015:531740. doi: 10.1155/2015/531740
86. Sárközy M, Kahán Z, Csont T. A myriad of roles of miR-25 in health and disease. Oncotarget. (2018) 9:21580–612. doi: 10.18632/oncotarget.24662
87. Garikipati VNS, Verma SK, Jolardarashi D, Cheng Z, Ibetti J, Cimini M, et al. Therapeutic inhibition of miR-375 attenuates post-myocardial infarction inflammatory response and left ventricular dysfunction via PDK-1-AKT signalling axis. Cardiovasc Res. (2017) 113:938–49. doi: 10.1093/cvr/cvx052
88. Fazi F, Fatica A. Interplay between N6-methyladenosine (m6A) and non-coding RNAs in cell development and cancer. Front Cell Dev Biol. (2019) 7:116. doi: 10.3389/fcell.2019.00116
89. Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N 6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. (2018) 46:3906–20. doi: 10.1093/nar/gky130
90. Pan JX. LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur Rev Med Pharmacol Sci. (2017) 21:322−8.
91. Li X, Luo S, Zhang J, Yuan Y, Jiang W, Zhu H, et al. lncRNA H19 alleviated myocardial I/RI via suppressing miR-877-3p/Bcl-2-mediated mitochondrial apoptosis. Mol Ther Nucleic Acids. (2019) 17:297–309. doi: 10.1016/j.omtn.2019.05.031
92. Zhang BF, Chen J, Jiang H. LncRNA H19 ameliorates myocardial ischemia-reperfusion injury by targeting miR-22-3P. Int J Cardiol. (2019) 278:224. doi: 10.1016/j.ijcard.2018.11.017
93. Zhang X, Fu Q, Xu L, Yang Y, Zhao W, Zhang Y, et al. Dexmedetomidine postconditioning alleviates hypoxia/reoxygenation injury in senescent myocardial cells by regulating lncRNA H19 and m6A modification. Oxidative Med Cell Longevity. (2020) 2020:7059751. doi: 10.1155/2020/9250512
94. Zhang J, Chen M, Chen J, Lin S, Cai D, Chen C, et al. Long non-coding RNA MIAT acts as a biomarker in diabetic retinopathy by absorbing miR-29b and regulating cell apoptosis. Biosci Rep. (2017) 37:BSR20170036. doi: 10.1042/BSR20170036
95. Wu L, Pei Y, Zhu Y, Jiang M, Wang C, Cui W, et al. Association of N6-methyladenine DNA with plaque progression in atherosclerosis via myocardial infarction-associated transcripts. Cell Death Dis. (2019) 10:909. doi: 10.1038/s41419-019-2152-6
96. Qu X, Du Y, Shu Y, Gao M, Sun F, Luo S, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. (2017) 7:42657. doi: 10.1038/srep42657
97. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. (2015) 116:1143–56. doi: 10.1161/CIRCRESAHA.116.305510
98. Zhang X, Wu H, Mai C, Qi Y. Long non-coding RNA XIST/miR-17/PTEN axis modulates the proliferation and apoptosis of vascular smooth muscle cells to affect Stanford type A aortic dissection. J Cardiovasc Pharmacol. (2020) 76:53–62. doi: 10.1097/FJC.0000000000000835
99. Zhou T, Qin G, Yang L, Xiang D, Li S. LncRNA XIST regulates myocardial infarction by targeting miR-130a-3p. J Cell Physiol. (2019) 234:8659–67. doi: 10.1002/jcp.26327
100. Chen Y, Liu X, Chen L, Chen W, Zhang Y, Chen J, et al. The long noncoding RNA XIST protects cardiomyocyte hypertrophy by targeting miR-330-3p. Biochem Biophys Res Commun. (2018) 505:807–15. doi: 10.1016/j.bbrc.2018.09.135
101. Xiao L, Gu Y, Sun Y, Chen J, Wang X, Zhang Y, et al. The long noncoding RNA XIST regulates cardiac hypertrophy by targeting miR-101. J Cell Physiol. (2019) 234:13680–92. doi: 10.1002/jcp.28047
102. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m (6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
103. Liu XM, Zhou J, Mao Y, Ji Q, Qian SB. Programmable RNA N (6)- methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. (2019) 15:865–71. doi: 10.1038/s41589-019-0327-1
104. Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, et al. N (6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol. (2016) 428(5 Pt A):822–33. doi: 10.1016/j.jmb.2015.08.021
105. Tang Y, Jin X, Xiang Y, Chen Y, Shen CX, Zhang YC, et al. The lncRNA MALAT1 protects the endothelium against ox-LDL-induced dysfunction via upregulating the expression of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett. (2015) 589(20 Pt B):3189–96. doi: 10.1016/j.febslet.2015.08.046
106. Zeng R, Zhang R, Song X, Ni L, Lai Z, Liu C, et al. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem Biophys Res Commun. (2018) 495:2532–8. doi: 10.1016/j.bbrc.2017.12.105
107. Gong X, Zhu Y, Chang H, Li Y, Ma F. Long noncoding RNA MALAT1 promotes cardiomyocyte apoptosis after myocardial infarction via targeting miR-144-3p. Biosci Rep. (2019) 39:BSR20191103. doi: 10.1042/BSR20191103
108. Liu Z, Liu J, Wei Y, Xu J, Wang Z, Wang P, et al. LncRNA MALAT1 prevents the protective effects of miR-125b-5p against acute myocardial infarction through positive regulation of NLRC5. Exp Ther Med. (2020) 19:990–8. doi: 10.3892/etm.2019.8309
109. Sun R, Zhang L. Long non-coding RNA MALAT1 regulates cardiomyocytes apoptosis after hypoxia/reperfusion injury via modulating miR-200a-3p/PDCD4 axis. Biomed Pharmacother. (2019) 111:1036–45. doi: 10.1016/j.biopha.2018.12.122
110. Hu H, Wu J, Li D, Zhou J, Yu H, Ma L. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed Pharmacother. (2018) 106:738–46. doi: 10.1016/j.biopha.2018.06.122
111. Guo X, Wu X, Han Y, Tian E, Cheng J. LncRNA MALAT1 protects cardiomyocytes from isoproterenol-induced apoptosis through sponging miR-558 to enhance ULK1-mediated protective autophagy. J Cell Physiol. (2019) 234:10842–54. doi: 10.1002/jcp.27925
112. Huang S, Zhang L, Song J, Wang Z, Huang X, Guo Z, et al. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J Cell Physiol. (2019) 234:2997–3006. doi: 10.1002/jcp.27117
113. Brown JA, Kinzig CG, DeGregorio SJ, Steitz JA. Methyltransferase-like protein 16 binds the 3'-terminal triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA. (2016) 113:14013–8. doi: 10.1073/pnas.1614759113
114. Ye J, Wang C, Wang D, Yuan H. LncRBA GSA5, up-regulated by ox-LDL, aggravates inflammatory response and MMP expression in THP-1 macrophages by acting like a sponge for miR-221. Exp Cell Res. (2018) 369:348–55. doi: 10.1016/j.yexcr.2018.05.039
115. Zhang Y, Lu X, Yang M, Shangguan J, Yin Y. GAS5 knockdown suppresses inflammation and oxidative stress induced by oxidized low-density lipoprotein in macrophages by sponging miR-135a. Mol Cell Biochem. (2021). 476:949–57. doi: 10.1007/s11010-020-03962-w
116. Han Y, Wu N, Xia F, Liu S, Jia D. Long non-coding RNA GAS5 regulates myocardial ischemia-reperfusion injury through the PI3K/AKT apoptosis pathway by sponging miR-532-5p. Int J Mol Med. (2020) 45:858–72. doi: 10.3892/ijmm.2020.4471
117. Diao L, Bai L, Jiang X, Li J, Zhang Q. Long-chain noncoding RNA GAS5 mediates oxidative stress in cardiac microvascular endothelial cells injury. J Cell Physiol. (2019) 234:17649–62. doi: 10.1002/jcp.28388
118. Hao S, Liu X, Sui X, Pei Y, Liang Z, Zhou N. Long non-coding RNA GAS5 reduces cardiomyocyte apoptosis induced by MI through sema3a. Int J Biol Macromol. (2018) 120(Pt A):371–7. doi: 10.1016/j.ijbiomac.2018.08.039
119. Tao H, Zhang JG, Qin RH, Dai C, Shi P, Yang JJ, et al. LncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. (2017) 386:11–8. doi: 10.1016/j.tox.2017.05.007
120. Zhang J, Gao C, Meng M, Tang H. Long noncoding RNA MHRT protects cardiomyocytes against H2O2-induced apoptosis. Biomol Ther. (2016) 24:19–24. doi: 10.4062/biomolther.2015.066
121. Xu Y, Luo Y, Liang C, Zhang T. LncRNA-Mhrt regulates cardiac hypertrophy by modulating the miR-145a-5p/KLF4/myocardin axis[J]. J Mol Cell Cardiol. (2020) 139:47–61. doi: 10.1016/j.yjmcc.2019.12.013
122. Zhang L, Wu YJ, Zhang SL. Circulating lncRNA MHRT predicts survival of patients with chronic heart failure. J Geriatr Cardiol. (2019) 16:818−21. doi: 10.11909/j.issn.1671-5411.2019.11.006
123. Shen W, Li H, Su H, Chen K, Yan J. FTO overexpression inhibits apoptosis of hypoxia/reoxygenation-treated myocardial cells by regulating m6A modification of Mhrt. Mol Cell Biochem. (2021) 476:2171–9. doi: 10.1007/s11010-021-04069-6
124. Su H, Wang G, Wu L, Ma X, Ying K, Zhang R. Transcriptome-wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics. (2020) 21:39. doi: 10.1186/s12864-020-6462-y
125. Gao XQ, Zhang YH, Liu F, Ponnusamy M, Zhao XM, Zhou LY, et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N 6-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol. (2020) 22:1319–31. doi: 10.1038/s41556-020-0576-y
126. Chen J, Ning Y, Zhang H, Song N, Gu Y, Shi Y, et al. METTL14-dependent m6A regulates vascular calcification induced by indoxyl sulfate. Life Sci. (2019) 239:117034. doi: 10.1016/j.lfs.2019.117034
127. Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. (2014) 24:1403–19. doi: 10.1038/cr.2014.151
128. Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue Q, et al. MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem Biophys Res Commun. (2017) 482:582–9. doi: 10.1016/j.bbrc.2016.11.077
Keywords: M6A, mRNAs, non-coding RNAs, CVD, modification
Citation: Song D, Hou J, Wu J and Wang J (2021) Role of N6-Methyladenosine RNA Modification in Cardiovascular Disease. Front. Cardiovasc. Med. 8:659628. doi: 10.3389/fcvm.2021.659628
Received: 03 February 2021; Accepted: 15 April 2021;
Published: 07 May 2021.
Edited by:
Christoph Dieterich, Heidelberg University, GermanyReviewed by:
Cheng-Kai Huang, Hannover Medical School, GermanyCopyright © 2021 Song, Hou, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junnan Wang, amRleXdqbkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.