- 1Department of Gynecology, Peking University First Hospital Ningxia Women and Children’s Hospital, Yinchuan, Ningxia, China
- 2School of Health Science and Engineering, University of Shanghai for Science and Technology, Shanghai, China
- 3Department of Pharmacy, Peking University First Hospital Ningxia Women and Children’s Hospital, Yinchuan, Ningxia, China
- 4Department of Gynecology and Obstetrics, Yinchuan Second People’s Hospital, Yinchuan, Ningxia, China
For over a century, scientists have been harnessing the therapeutic potential of bacteria in treating diseases. The advent of synthetic biology in recent years has propelled the development of genetically engineered bacteria with enhanced intelligence. These bacteria can autonomously detect environmental cues and relay them to pivotal promoters, leading to the expression of functional proteins. By utilizing modular components, they are capable of executing a range of functions, including sensing, transmitting, and outputting signals. Based on these principles, a series of intelligent diagnostic and therapeutic engineered bacteria have emerged. These bacteria are capable of targeting diseased sites, sensing disease-specific signals, and producing reporter and therapeutic drugs. Furthermore, the integration of intelligent diagnostic and therapeutic engineered bacteria with advanced technologies such as artificial intelligence, nanomaterials, and optics has paved the way for diverse clinical applications. Three critical stages are explored in this article, which include the selection of strains, the design of biosensing systems, and the planning of release strategies. The application of intelligent diagnosis and treatment engineering bacteria in metabolic diseases, inflammatory diseases, tumors and infectious diseases is reviewed.
1 Introduction
The use of bacteria for treating diseases by humans dates back to the 19th century, when Dr. William Coley discovered that injecting patients with Streptococcus pyogenes and Serratia marcescens could lead to a reduction in tumor size. At the end of the 20th century, with the development of molecular biology and genetic engineering technology, scientists began to try to transform bacteria through genetic engineering to diagnose and treat diseases. Living engineered bacterial therapy emerged as the times require (Srivastava and Lesser, 2024). This therapy has the advantages of low cost, sensitivity and robustness, and can detect and treat diseases noninvasively and in situ, which is in line with the sustainable development goals of the World Health Organization (Tanniche and Behkam, 2023). In recent years, with the advancement of synthetic biology, genetically engineered bacteria have become increasingly intelligent, capable of sensing environmental signals and transmitting them to key promoters, ultimately leading to the expression of functional proteins (Zhang X. E. et al., 2023). The cell factors involved in the aforementioned processes are categorized into three modules based on their functions: the reception module (proteins that can sense specific chemical or physical signals inside or outside the cell and convert the signals to downstream modules); the transmission control module (one or a series of transcription factors and their target promoters); and the output module (proteins that can serve as markers or change the bacteria themselves and the surrounding environment) (Chen and Elowitz, 2021). These three modules constitute the core component of the intelligent engineered bacteria’s biosensing system (Brophy and Voigt, 2014; Gordley et al., 2016). Intelligent engineered bacteria have been applied in agriculture, energy, manufacturing, biology, and basic medical research (Smanski et al., 2016; Dobrin et al., 2016; Clarke and Kitney, 2020; Gao et al., 2019). This article reviews intelligent engineered bacteria used for disease diagnosis and treatment, introducing their general construction and application in disease research.
2 Construction of intelligent diagnosis and treatment engineering bacteria
Engineered bacteria for disease diagnosis and treatment are introduced into the host’s body through various routes such as oral administration (Steidler et al., 2003), intravenous injection (Ryan et al., 2009), enema (Ricci et al., 2003), vaginal administration (Liu et al., 2006), and intratumoral injection (Din et al., 2016). The environmental compatibility of the bacterial strains with the lesion site allows them to effectively reach the target location, moderately colonize at the lesion site, and sense specific physical and chemical signals in the environment. These signals are then transmitted into the engineered bacteria, where they are conducted through transcription factors and their corresponding promoters to controllably express diagnostic reporting factors or therapeutic factors. These factors can be effectively released from the engineered bacteria into the lesion site, thereby playing roles in real-time diagnosis, metabolic regulation, inflammation suppression, tumor cell killing, and anti-infection, and can be cleared from the host’s body after fulfilling their functions (Riglar and Silver, 2018). From the entire process, the selection of bacterial strains, the construction of the biosensing system, and the design of the release mechanism are three key aspects in constructing intelligent engineered bacteria for disease diagnosis and treatment.
2.1 Selection of bacterial strains
When selecting chassis strains for disease diagnosis and treatment, its biosafety and non pathogenicity should be considered first. In addition, genetic manipulation difficulties of chassis strains, that is, the adaptability to DNA transformation and the availability of genome manipulation tools, must also be considered. Finally, the survival and colonization ability of strains in the disease environment are also the key factors to achieve the purpose of diagnosis and treatment.
2.1.1 Nonpathogenicity and biosafety
Nonpathogenicity are prerequisites for the selection of engineering bacteria chassis for diagnosis and treatment. Therefore, safe non pathogenic probiotics should be selected as chassis strains as far as possible. For example, Escherichia coli Nissle 1917 (EcN) is a gram negative probiotic that is sensitive to serum and does not produce enterotoxins or cytotoxins associated with pathogenic Escherichia coli (Escherichia coli) strains (Yu et al., 2023). However, the virulence of some chassis strains is inevitable, and the virulence of bacteria can be reduced by modifying the relevant genes of bacteria, so as to enhance its safety. For example, attenuated Salmonella strain ΔppGpp was engineered by regulating endotoxin gene expression. This strain is a double mutant (rela-, spot-), which has defects in ppGpp synthesis, resulting in downregulation of endotoxin gene expression (Na et al., 2006). Biosafety is a key factor in the approval of clinical trials and the use of engineered bacterial therapies. Preventing engineered bacteria from surviving after leaving the experimental and clinical environment is an important method to improve biosafety. Scientists have designed a killing switch in response to temperature changes. When the engineered bacteria leave the host, the cold induced promoter will express the toxin, thus reducing the survival rate of the strain (Stirling et al., 2017).
2.1.2 Genetic manipulation difficulties
Gene modification technology plays a pivotal role in the in-depth exploration of biotechnology. The precise gene modification is essential for accurately regulating the functions of organisms and serves as an indispensable cornerstone in the construction of “intelligent” diagnostic engineering bacteria. Successful gene modification relies on reliable DNA transformation tools, effective control of gene expression, and post-translational control of protein processes, such as secretion (Arnold et al., 2023). In addition to these fundamental elements, the design of engineering bacteria also necessitates the integration of various advanced functional components, including sensors, circuit components, and actuators. E. coli, due to its clear genetic background, rapid propagation, low cost, and robust ability to express foreign proteins, has been employed as the primary strain for engineering development (Rosano et al., 2019). Lactobacillus, being easy to cultivate and cost-effective, possesses a variety of mature gene tools and vector systems, and can effectively target single membrane proteins of eukaryotic cells, making it a promising candidate chassis (Frelet-Barrand, 2022). Furthermore, thanks to the successful development of supportive genetic tools, including promoters, ribosome binding sites (RBS), vector backbones, and genome integration systems, the intestinal commensal bacterium Bacteroides polymorphus has also been gradually utilized as a new probiotic chassis (Lai et al., 2022).
2.1.3 Survival and colonization ability
The survival and colonization capabilities of chassis strains at the lesion site are crucial for disease diagnosis and treatment. Taking ulcerative colitis, a challenging condition in gastroenterology, as an example, if the chassis strain can effectively colonize the ileocolonic region, it may provide significant assistance in the treatment of such diseases. Given that Bacteroides and other intestinal bacteria have a higher distribution in the colon, they can be considered as potential strains for the treatment of ulcerative colitis (Aitken et al., 2024). In contrast, since the pathological changes in Crohn’s disease can extend from the mouth to the anus, implying that the corresponding treatment strategies need to cover a broader intestinal area, the use of Lactobacillus, which can colonize both the small intestine and the colon, is likely to enhance the therapeutic effect on Crohn’s disease (Li et al., 2023). Furthermore, the nanoscale outer membrane vesicles (OMVs) produced by symbiotic gut bacteria Bacteroides thetaiotaomicron can penetrate the intestinal mucosa and directly interact with the innate layer immune cells, making it highly suitable as an engineered bacterial chassis for the diagnosis and treatment of intestinal diseases (Durant et al., 2020; Taketani et al., 2020).
2.2 Design of biosensing system
With the advancement of synthetic biology, biosensing systems are often integrated into the functional components of engineered bacteria (Gui et al., 2017; Ali et al., 2020). Therapeutic engineered bacteria have evolved from simply expressing therapeutic factors to becoming more intelligent, capable of first receiving disease signals (physical signals: temperature, pH; chemical signals: oxygen, ions, small molecule metabolites, peptides, sugars, etc.) and then transferring these signals to the transmission control module for the controllable output of therapeutic factors (Tabor et al., 2009). The entire biosensing system can be functionally divided into three modules: reception, transmission control, and output (Figure 1).
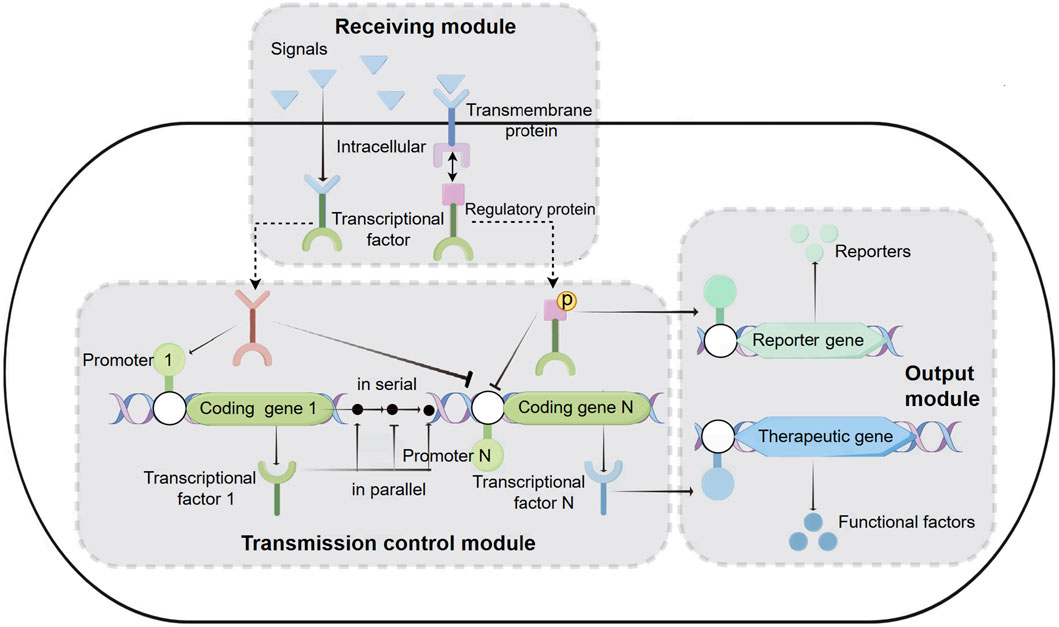
Figure 1. Biosensing System of Intelligent Diagnosis and Treatment Engineering Bacteria. Receiving module: intracellular reception: Physical and chemical signals from the disease environment enter the cell, interact with transcription factors, and cause conformational changes in transcription factors. Subsequently, transcription factors activate or inhibit inducible promoters on the transport control module or output module. Transmembrane reception: including a transmembrane protein and a transcriptional regulatory protein. Transmembrane proteins receive extracellular signals and phosphorylate transcription regulatory proteins. Then, transcriptional regulatory proteins bind to the promoters of transport or output modules, activating or inhibiting the transcription of coding genes. Transmission control module: located between the receiving module and the output module, it is composed of one or more transcription units connected in series or parallel. Output module: It can express reporter genes or therapeutic genes to achieve diagnostic and therapeutic purposes.
2.2.1 Receiving module
Currently, the reception modules used in therapeutic engineered bacteria are divided into two types: intracellular reception modules and transmembrane reception modules (Figure 1). Intracellular reception modules typically refer to transcription factors that can sense signals within the cytoplasm of engineered bacteria. The physicochemical signals in the disease environment directly enter the cytoplasm, causing the transcription factors to undergo conformational changes. The conformationally changed transcription factors activate or inhibit the inducible promoters on the transmission control module or the output module, thereby controlling the expression of diagnostic reporting factors or therapeutic factors. For example, the zinc-responsive transcription factors Zur and ZntR can serve as reception modules that react with zinc ions entering the cell, and their integration into E. coli allows for the sensing of serum zinc levels and the production of visible pigments to identify zinc deficiency (McNerney et al., 2019). A protein, lasR, which reacts with acyl-homoserine lactone (AHL), has been used as an intracellular reception module in engineered E. coli to monitor the presence of pathogenic Pseudomonas aeruginosa (P.aeruginosa) in solid agar (Gupta et al., 2013), biofilms (Saeidi et al., 2011), and animal models (Hwang et al., 2017). Another transcriptional regulator, NorR, which is activated by nitric oxide (NO), has also become an intracellular reception module for engineered bacteria to detect inflammatory signals (Archer et al., 2012; Chen et al., 2021). Finally, various orally administered gut-adapted bacteria have been implanted with intracellular reception modules to sense lactose (Drouault et al., 2002), xylan (Hamady et al., 2010), and rhamnose (Mimee et al., 2015) in the diet, as well as chemical substances produced by the gut microbiota in mice (Pickard et al., 2014). Although intracellular reception modules play an important role in disease diagnosis, they also have limitations because they cannot sense extracellular disease signals.
Transmembrane reception modules are the primary means for engineered bacteria to sense extracellular stimuli (Gao and Stock, 2009; Galperin, 2010). The classic transmembrane reception module features a histidine kinase transmembrane protein that, upon stimulation by specific signals in the environment, phosphorylates histidine on the intracellular side of the membrane. It then phosphorylates a transcriptional regulatory protein, which can bind to the promoters of the transmission control module or the output module, activating or inhibiting the transcription of the encoded genes (Beier and Gross, 2006). Many transmembrane reception modules have been utilized for disease diagnosis and treatment (Lazar and Tabor, 2021). Two early-developed transmembrane reception modules from the marine bacterium Shewanella, thiosulfate sensor ThsS-ThsR and tetrathionate sensor TtrS-TtrR, respectively receive thiosulfate and tetrathionate signals across the membrane, and these modules have been designed to be installed in E. coli as a potential therapeutic means for intestinal inflammation (Daeffler et al., 2017; Riglar et al., 2017). Recently, researchers designed a NarX-NarL-based transmembrane reception module to detect nitrate levels, and in EcN, the combination of NarX-NarL with ThsS-ThsR modules across the membrane receives nitrate and thiosulfate, enhancing the specificity of the engineered bacteria in diagnosing intestinal inflammation (Woo et al., 2020). Another study integrated the transmembrane reception elements CqsS-LuxU-LuxO derived from Vibrio cholerae with a dCAS9-based green fluorescent protein (GFP) reporter gene system into E. coli, constructing a biosensor with high sensitivity to quorum sensing (QS) ligand cholera CAI-1, indicating the presence and proliferation of Vibrio cholerae (Holowko et al., 2016). The reception module can act directly on the output module or can be combined with the output module through the transmission control module to form a more intelligent biosensing system (Anderson et al., 2007).
2.2.2 Transmission control module
The transmission control module is located between the reception module and the output module, and it is composed of several serial or parallel transcriptional units (Figure 1). The efficiency of the transmission control module is equal to the sum of the rates of all transcriptional units. The efficiency of each transcriptional unit depends on its promoter, terminator, and ribosome binding site (RBS) components. The promoter marks the starting point of transcription and can ensure the transcription rate of mRNA through a series of well-designed constitutive promoter libraries or chemically inducible promoters (Kelly et al., 2009; Mutalik et al., 2013; Yao et al., 2013; Lutz and Bujard, 1997; Cox et al., 2007; Armetta et al., 2021). Terminating the transcription process is crucial to avoid unintended interactions between different transcriptional units in the engineered DNA sequence. Previous studies have established numerous high-efficiency transcription terminator resource libraries, which include a large number of high-performing terminators (Chen et al., 2013; Cambray et al., 2013). The RBS determines the efficiency and location of ribosome binding, and it can regulate the rate of translation initiation, thereby controlling the expression level of proteins. By using RBS calculation tools to design specific ribosome binding sequences (Salis et al., 2009; Espah Borujeni et al., 2014; Farasat et al., 2014), or employing a more flexible “dual-cistron design” method with RBS (Mutalik et al., 2013), the translation efficiency of target genes in engineered bacteria can be conveniently adjusted.
The transmission control module, based on Boolean logic, is divided into three basic modes: “AND gate transmission control” (functional factors are expressed only when both signals are present simultaneously) (Figure 2A) (Woo et al., 2020; Anderson et al., 2007), “OR gate transmission control” (functional factors are expressed as long as one of the two signals is present) (Figure 2B) (Wong et al., 2015), and “NOT gate transmission control” (functional factors are not expressed when a certain signal is present, otherwise they are expressed) (Figure 2C) (Bartoli et al., 2020). These three basic modes can also be combined to form advanced transmission control modules such as NAND (Wang et al., 2011), NOR (Tamsir et al., 2011), and XOR gates (Wong et al., 2015).
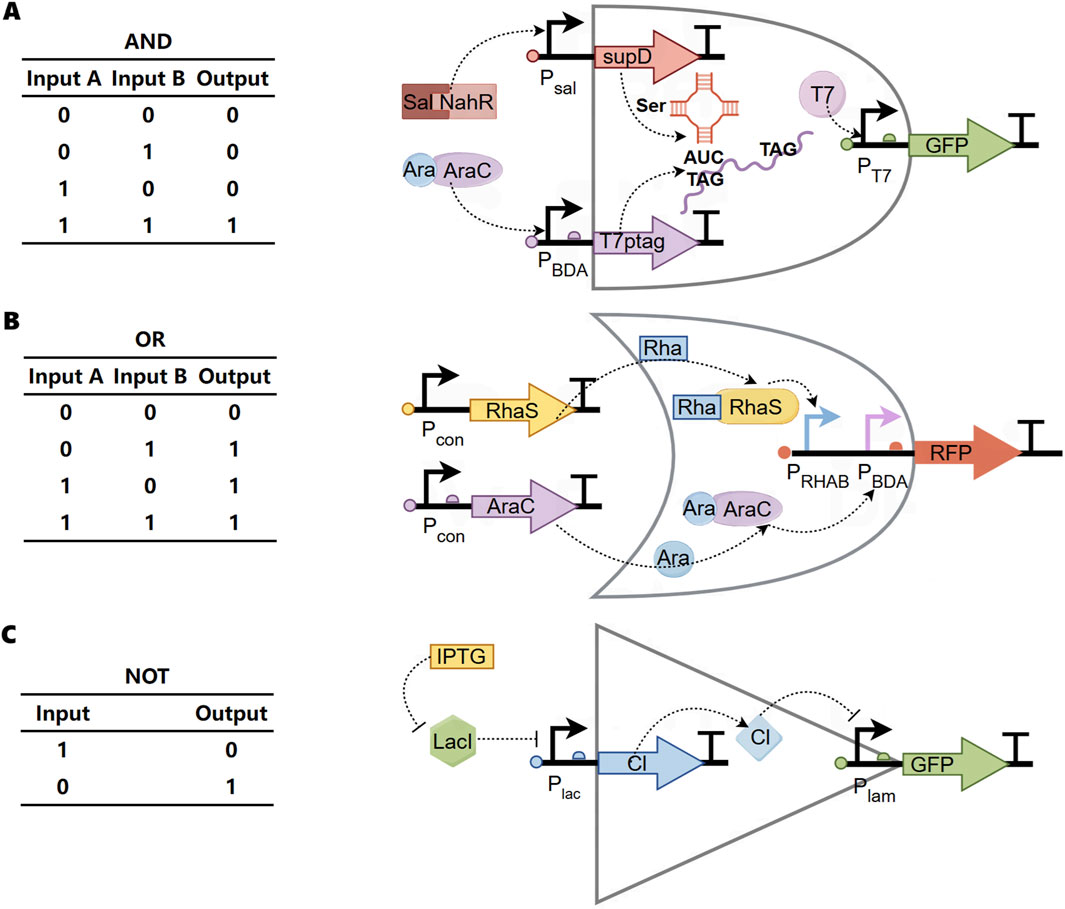
Figure 2. Boolean logic of transmission control module (A) Biological “AND gate”. Two input promoters were used that react with small molecule salicylates (Sal) and arabinose (Ara). The first promoter Psal is related to the transcription of the amber inhibitory factor tRNA supD. The second promoter PBAD drives transcription of T7 RNA polymerase. The polymerase gene has been modified to contain two amber stop codons (T7ptag). When supD is also transcribed, these stop codons are translated into serine. The polymerase T7 is only expressed in the presence of supd and T7ptag mRNA. Therefore, GFP can be output only when Sal and Ara are input at the same time. (B) Biological “OR gate”. PBAD and PRHAB promoters were designed upstream of an RFP reporter gene with strong RBS. When rhamnose (Rha) or arabinose (Ara) existed in the environment, they could combine with the corresponding receiving module, and then induce the expression of RFP. (C) Biological “NOT gate”. In the absence of isopropyl-β-d-thiogalactoside (IPTG), the repressor protein lacI will prevent the polymerase from binding to the promoter Plac and cannot express CI. At this time, GFP can be expressed. When IPTG is present, IPTG inhibits lacI, CI repressor protein can be expressed, and then interacts with the promoter Plam to inhibit the expression of GFP.
2.2.3 Output module
The output module of intelligent therapeutic engineered bacteria is the functional gene that ultimately exerts its effect, which can be divided into two types: diagnosis and treatment (Figure 1).
Diagnostic output modules often express pigments, luciferases, or fluorescent proteins because the intensity of this form of output is easily observable and measurable. The advantage of pigments as diagnostic outputs is that they are visible to the naked eye without the need for special equipment. The earliest example is the enzymatic activity of the lacZ gene on X-gal, which produces a blue pigment (Horwitz et al., 1964). Violacein, lycopene, and carotenoids are induced to express by Zur and ZntR zinc-responsive transcription factors (TFs) to sense serum zinc levels for identifying zinc deficiency (McNerney et al., 2019). Luciferase, due to its very low background luminescence in fecal samples, typically produces higher sensitivity than pigment reporter genes. Although the use of specific equipment and substrates is required, these limitations do not hinder their significant role in improving the reliability of detection (Corthier et al., 1998). Fluorescent proteins are also commonly used as output proteins for disease diagnosis. Researchers have combined GFP and its variants (such as RFP, YFP, CFP) with transmembrane reception modules for the diagnosis of inflammatory bowel disease (IBD) (Daeffler et al., 2017; Riglar et al., 2017; Woo et al., 2020). This allows researchers to more accurately observe the morphology, growth status, and interactions of engineered bacteria at the single-cell level.
Therapeutic output modules express functional factors such as interleukins, hormones, enzymes, and bacterial toxins. Lactobacillus expressing interleukin-27 (IL-27) can be used to treat inflammatory bowel disease (Hanson et al., 2014). Interleukin-10 (IL-10) and proinsulin are co-expressed in Lactobacillus for the treatment of type 1 diabetes (Takiishi et al., 2017). Engineering EcN can bind to heparin sulfate proteoglycans (HSPG) on the surface of cancer cells and secrete a substance called myrosinase, which converts glucosinolates in food into sulforaphanes with anticancer activity (Ho et al., 2018). E. coli loaded with the bacterial toxin antimicrobial protein (S5 pyocin) and biofilm-degrading enzyme (DspB) has shown a synergistic effect in eliminating P. aeruginosa, improving the efficiency of pathogen clearance (Hwang et al., 2017).
2.3 Design of release method
The release of functional factors from engineered bacteria into the lesion environment is essential for their pharmacological efficacy. These factors are typically released through mechanisms of bacterial lysis or via excretion systems (Figure 3). The cutting-edge lytic system derived from bacteriophage iEPS5 has been used to induce lysis of Salmonella typhimurium, promoting the release of mitochondrial targeted domain protein (MTD) (Jeong et al., 2014) and immunotoxin TGFα- PE38 (Lim et al., 2017) for cancer treatment (Figure 3A).
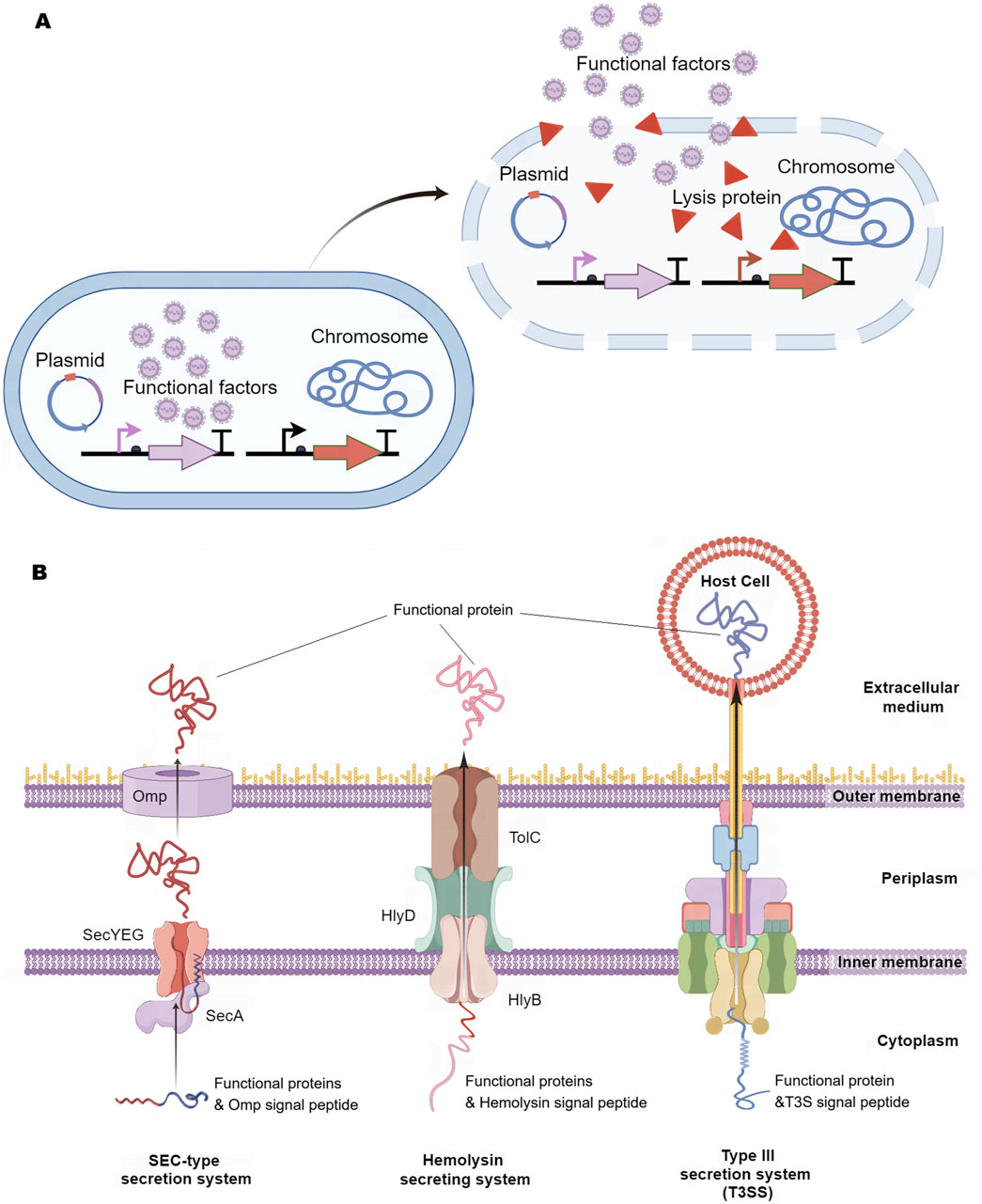
Figure 3. Release Method of Intelligent Diagnosis and Treatment Engineering Bacteria. (A) Bacterial lysis: Engineering bacteria first express functional factors, and when they reach the lesion, special signals in the disease environment induce engineering bacteria to express lytic proteins, followed by the release of bacterial lytic functional factors. (B) Extracellular secretion system: SEC-type secretion system, which first secretes functional proteins with signal peptides into the cytoplasm, and then secretes them out of the cell through outer membrane proteins. Hemolysin secretion system is comprised of three components (HlyB, HlyD, and TolC), which can secrete functional proteins carrying the HlyA signal peptide outside the cell. Type III secretion system (T3SS) can directly inject functional factors into host cells. Omp, outer membrane proteins; Hly, Hemolysin.
Among the various excretion systems, the SEC-type secretion system plays a pivotal role by linking a signal peptide to the functional factor, which aids in the translocation of the factor across the inner membrane of the engineered bacteria from the cytoplasm to the periplasm, ultimately secreting it into the surrounding environment (Zheng et al., 2017; Lim et al., 2017; Pöhlmann et al., 2012) (Figure 3B). The hemolysin secretion system is a continuous tunnel that spans the inner and outer membranes (composed of HlyB/HlyD/TolC), which can induce functional proteins with signal peptides to be excreted from the intracellular space (Gentschev et al., 2002; Zou et al., 2023) (Figure 3B). Researchers have implanted the Type III secretion system (T3SS) from pathogenic bacteria into non-pathogenic E. coli, which can directly inject functional factors into host cells (Reeves et al., 2015; Ruano-Gallego et al., 2015; González-Prieto and Lesser, 2018) (Figure 3B). Although these release systems have been proven to be effective in animal models, their safety for human disease applications still requires further validation.
3 Application of intelligent diagnosis and treatment engineering bacteria
Compared to other therapies, engineered bacteria can perform sensitive, noninvasive, and in situ detection and treatment of diseases at low cost (Tanniche and Behkam, 2023), and the rapid advancement of synthetic biology has further enhanced their stability and intelligence (Landry and Tabor, 2017). Consequently, the application of intelligent diagnostic and therapeutic engineered bacteria has been extended from gastrointestinal diseases to metabolic disorders, oncology, infectious diseases, and other areas (Lim and Song, 2019; Gurbatri et al., 2022).
3.1 Metabolic disorders
Metabolic disorders can lead to diseases such as diabetes and obesity. Most of these disease belongs to chronic diseases, and current treatment methods include long-term scheduled medication or injections. These therapies have significant side effects, high costs, and require patients to strictly follow the treatment plan. Consequently, the deployment of intelligent diagnostic and therapeutic engineered bacteria presents itself as a potential treatment strategy for metabolic diseases.
3.1.1 Diabetes
Type 1 diabetes is an autoimmune disease that leads to insulin deficiency and ultimately the loss of function of pancreatic β-cells, necessitating lifelong treatment, particularly with long-term, scheduled insulin injections (Subramanian et al., 2024). Researchers have utilized Lactobacillus gasseri to express glucagon-like peptide 1 (GLP-17–37) and engineered it to be released via a type III secretion system. Data from diabetic rats treated orally provide evidence for the use of engineered commensal bacteria in the treatment of diabetes (Duan et al., 2015). Oral administration of engineered Lactococcus lactis expressing both human proinsulin (PINS) and human interleukin-10 (hIL-10), in combination with low-dose systemic anti-CD3 monoclonal antibodies, can stably restore blood glucose levels in non-obese diabetic (NOD) mice (Takiishi et al., 2017; Takiishi et al., 2012). Similarly, engineered Lactococcus lactis co-expressing human glutamic acid decarboxylase (GAD65) and human interleukin-10 (hIL-10) has shown the same therapeutic efficacy (Robert et al., 2014).
Type 2 diabetes is widely concerned because of its incidence rate and complications (Tao et al., 2020). GLP-1 is an incretin hormone produced by intestinal cells that stimulates insulin secretion from the pancreas in a glucose-dependent manner. Exogenous GLP-1 analogs are used for the treatment of type 2 diabetes. Lactobacillus lactis (LL-pUBGLP-1) containing a plasmid encoding GLP-1 was used as a delivery system for GLP-1, and its effectiveness was demonstrated by the reduction in plasma glucose levels and increment insulin concentrations in obese type 2 diabetic rats (Zucker Diabetic Fatty rats, ZDF) following oral administration (Agarwal et al., 2014). Researchers have designed a GLP-1 mutant pentamer (5 × GLP-1), which is expressed and extracellularly anchored on the cell wall of lactic acid bacteria, and retains its in vitro bioactivity after being digested by intestinal trypsin into GLP-1 monomers (Lin et al., 2016). To induce a significant insulinotropic effect in diabetic animal models, further enhancement of the expression level of GLP-1 by lactic acid bacteria is required. Consequently, researchers have further mutated GLP-1 and linked it to form a GLP-1 hexamer (6 × mGLP-1), and the hexamer expressed by E. coli has been shown to exert a long-lasting hypoglycemic effect in type 2 diabetic mice (Xu et al., 2017). Additionally, exendin-4, a GLP-1 receptor agonist with a more prolonged bioactivity compared to GLP-1, has been effectively expressed and secreted by Lactobacillus paracasei transformed with a plasmid encoding the exendin-4 gene. The secreted exendin-4 significantly enhanced insulin secretion from INS-1 β-cells, providing a novel strategy for the treatment of diabetes with engineered bacteria (Zeng et al., 2016). Recently, researchers have orally administered optogenetic engineered Lactococcus lactis in the gut, which secretes GLP-1 to regulate metabolism under the control of a wearable optical device, and this strategy has effectively controlled blood glucose, body weight, and other characteristics in rat and mouse models (Zhang X. et al., 2023).
3.1.2 Phenylketonuria
Phenylketonuria (PKU; also known as phenylalanine hydroxylase deficiency) is an autosomal recessive genetic disorder of phenylalanine (Phe) metabolism, where excessive Phe levels can lead to brain dysfunction (van Spronsen et al., 2021). To reduce Phe levels in the body, Synlogic Corporation has designed a strain, EcN (SYNB1618), capable of expressing genes for phenylalanine transport protein (PheP), phenylalanine ammonia lyase (PAL), and L-amino acid deaminase (LAAD), which can convert Phe to trans-cinnamic acid (TCA) and phenylacetate (PP) in the gastrointestinal tract, effectively reducing blood Phe levels in mice and crab-eating macaques (Isabella et al., 2018). Subsequently, researchers conducted human trials with EcN (SYNB1618), which demonstrated safety and good tolerability (Puurunen et al., 2021). Building upon the success of EcN (SYNB1618), the entire cell PAL activity was optimized, resulting in a more effective strain, EcN (SYNB 1934), which showed approximately twice the PAL activity in vivo compared to its predecessor (Adolfsen et al., 2021).
3.1.3 Obesity
Obesity is a major global issue that significantly impacts public health (Perdomo et al., 2023). In recent years, researchers have also made attempts to treat obesity using smart engineered bacteria. N-acylphosphatidylethanolamines (NAPEs) synthesized in the small intestine can enhance satiety and reduce food intake. Engineered EcN, through genetic modification, can synthesize NAPEs in the gut, thereby treating obesity. This approach is more effective and enduring than traditional dietary and lifestyle interventions (Chen et al., 2014). Butyrate can reduce appetite and alleviate fatty liver through gut-brain neural circuits. Transgenic Bacillus subtilis SCK6 can increase butyrate production, which has beneficial effects on obesity, fasting blood glucose, insulin resistance, hepatic steatosis, and fat accumulations (Bai et al., 2020). Engineered Lactobacillus expressing GLP-1 has an ameliorative effect on obesity in mice induced by a high-fat diet (HFD) (Wang et al., 2021).
3.1.4 Hyperammonemia
Ammonia (NH3) is a toxic metabolite, which is produced in the small intestine and colon. The main route of ammonia (NH3) is to synthesize urea in the liver and excrete it in the urine from the kidney. Hyperammonemia occurs when there is a defect in ammonia metabolism. A large amount of ammonia that is not metabolized into urea by the liver enters the brain through the blood circulation, which can lead to brain injury and death (Ribas et al., 2022). Kurtz et al. Modified the metabolism of probiotic ECN, deleted the gene encoding arginine repressor ArgR, and then integrated the gene Arga215 encoding ArgAY19C into the intergenic region of malE and malK genes under the control of endogenous anaerobic inducible promoter fnrS (Pfnrs) of Escherichia coli. ArgAY19C is the feedback resistant version of N-acetylglutamate synthase ArgA (argAfbr). This engineered strain, SYNB1020, can overproduce arginine, thereby sequestering some ammonia produced by intestinal bacteria into amino acid molecules. The experiment proved that synb1020 could reduce blood ammonia, improve the survival rate of mouse hyperammonemia model, and showed repeated dose tolerance in non-human primates. A phase 1 dose escalation study in healthy human volunteers did not result in serious adverse events and showed that the bacterium was metabolically active in vivo, indicating that SYNB1020 needs further clinical development (Kurtz et al., 2019). Subsequently, the researchers conducted experiments using two engineered EcN strains, S-ARG (SYNB1020) and S-ARG + BUT (SYNB1536), in a rat model of chronic liver injury (CLD) and hepatic encephalopathy induced by bile duct ligation (BDL). The S-ARG (SYNB1020) strain showed positive benefits in reducing hyperammonemia, thereby alleviating memory impairment in BDL rats. The S-ARG + BUT (SYNB1536) strain exhibits similar therapeutic effects in hyperammonemia and shows additional positive effects in systemic inflammation and endotoxemia, leading to adequate protection against memory deterioration (Ochoa-Sanchez et al., 2021).
3.1.5 Hyperuricemia and gout
Uric acid is the final product of purine degradation in the human body, which can be excreted from the body through the kidneys and intestines. Excess uric acid will lead to hyperuricemia. The chronic form of arthritis caused by precipitation of sodium urate crystals is called gout. Gout afflicts hundreds of millions of people worldwide, with a high incidence rate in developed countries (Borghi et al., 2020). Yang et al. discovered a new method to eliminate purines from the diet by engineering gut bacteria, integrating a set of enzymes and transporters capable of degrading purines under anaerobic conditions into the EcN chromosome, which can bypass the formation of uric acid. By orally supplementing these engineered probiotics or purified recombinant enzymes, it is possible to effectively treat hyperuricemia fruit fly models, providing a new approach for developing novel probiotic therapies for the treatment of hyperuricemia and gout (Tong et al., 2023). Zou et al. also developed a probiotic EcN called YES301, which significantly improved the ability of bacteria to uptake xanthine and hypoxanthine by rationally designing overexpression of the xanthine transporter XanQ. As a result, serum uric acid levels were reduced and kidney damage was improved in a hyperuricemia mouse model. YES301 has shown comparable efficacy to traditional uric acid lowering drugs such as allopurinol, but with fewer side effects and higher biosafety, highlighting the potential of engineered probiotics in reducing intestinal purine levels to manage hyperuricemia (Zou et al., 2024).
3.2 Inflammatory bowel disease
Inflammatory bowel disease (IBD), which includes Crohn’s disease and ulcerative colitis (UC), is a type of incurable inflammatory condition (McDowell et al., 2024; Lu et al., 2022; Taku et al., 2020). The oral administration of smart diagnostic and therapeutic engineered bacteria, equipped with an internal biosensing system that can directly deliver therapeutic molecules to the gut, offers a highly promising approach for the diagnosis and treatment of inflammatory bowel disease (IBD).
In the early diagnosis of IBD, Archer et al. (2012) used the transcriptional regulator NorR as a receiving module that reacts with nitric oxide (NO) released during intestinal inflammation, thereby relieving the repression of the promoter and activating the expression of the DNA recombinase FilmE. FilmE can reverse the expression type of the fluorescent protein, from yellow fluorescent protein (YFP) to cyan fluorescent protein (CFP), and the change in fluorescence color can serve as a basis for the diagnosis of intestinal inflammation. Another study designed an E. coli strain (NGF1) that uses the transmembrane receiving module TtrS-TtrR to sense tetrathionate in the inflammatory intestinal environment, then remembers the presence of tetrathionate through the phage CI/Cro control module, and finally expresses β-galactosidase (β-gal) as an output. The β-gal in the feces reacts with a reagent to produce a color change, which can serve as a diagnostic basis for intestinal inflammation. This smart diagnostic engineered bacterium is effective for a long period within mice (Riglar et al., 2017).
Engineered bacterial strains that directly or indirectly express anti-inflammatory factors to inhibit pro-inflammatory factors show great promise in the treatment of IBD. Cui et al. constructed a photosensitive E. coli strain that uses YF1/FixJ as a transmembrane receiving module, capable of sensing blue light to secrete IL-10, and this system demonstrated real-time monitoring and long-term alleviation effects in acute and chronic UC mouse models (Cui et al., 2021). Recently, Zou et al. designed an EcN named i-ROBOT for real-time diagnosis and treatment of IBD. The transmembrane receiving module thsS/thsR of i-ROBOT senses the inflammatory signal thiosulfate, triggering the expression of the diagnostic factor superfolder green fluorescent protein (sfGFP) and the therapeutic factor immunomodulator AvCystatin. The therapeutic factor AvCystatin, connected with the Hly signal peptide, is released into the lesion through the implanted hemolysin secretion system to exert its pharmacological effect. At the same time, the inserted codon ACG in the chromosome is edited by BE2 and sgRNA to become the start codon ATG, initiating the expression of LacZ, which serves as a signal recording function. DSS-induced colitis in mice treated with i-ROBOT by gavage effectively alleviated the disease (Zou et al., 2023). Chua et al. designed an EcN strain using the endogenous regulatory factor NorR as a receiving module to respond to the colorectal inflammatory marker nitric oxide (NO), then activating the inducible promoter pNorV to express interferon lambda 1 (IFNL1). IFNL1 is excreted from the bacteria with the help of the signal peptide YebF, acting on immune cells to regulate inflammatory pathways. This engineered bacterium played an anti-inflammatory role in two in vitro IBD models (Chua et al., 2023). Researchers administered Lactobacillus expressing the antimicrobial peptide (LL37) to mice by continuous gavage for 10 days, with a single rectal injection of the inflammatory inducer DNBS on the 7th day, and the results proved that this engineered bacterium could prevent the occurrence of colitis and alleviate colitis symptoms (Noguès et al., 2022). In addition, Lactobacillus has been engineered to express IL-10 (Steidler et al., 2000), IL-27 (Hanson et al., 2014), IL-35 (Wang et al., 2019), thymic stromal lymphopoietin (TSL) (Aubry et al., 2015), and heme oxygenase (PHO-1) (Shigemori et al., 2015), among other immune factors, for the treatment of IBD.
Restoring intestinal mucosal healing and epithelial integrity is also a strategy for the treatment of IBD. Liang et al. (2022) directly inserted the nattokinase gene, connected with a signal peptide, into the natural plasmid pMUT1 of EcN. As a therapeutic factor, nattokinase alleviates DSS-induced intestinal epithelial injury and restores mucosal integrity by upregulating the levels of tight junction proteins (including ZO-1 and occludin). Praveschotinunt et al. (2019) linked the trefoil factor family (TFF) with a membrane protein (CsgA) to form a mucosal component that can protect the intestinal epithelium. This component, expressed by EcN and secreted onto the intestinal mucosa through the curli secretion system, can promote the repair of DSS-induced intestinal epithelial cells in mice.
The production of reactive oxygen species (ROS) leads to oxidative stress, which can damage intestinal cells. Increasing the activity of enzymes that can scavenge ROS becomes another strategy for the treatment of IBD (Montero-Blay et al., 2023). Zhou et al. (2022) engineered EcN to express superoxide dismutase (SOD) and catalase (CAT), which are capable of scavenging ROS. In a mouse model of IBD, this engineered bacterium, coated with chitosan/alginate, effectively alleviated colonic inflammation and unexpectedly increased the abundance of important microorganisms that maintain intestinal homeostasis.
3.3 Cancer
The use of engineered bacteria for cancer therapy is an emerging biotherapeutic approach that leverages genetically modified bacteria to combat cancer cells. This method primarily operates through the following three mechanisms: releasing natural toxins to induce apoptosis in cancer cells, activating the immune system to recognize and eliminate cancer cells, and delivering anticancer drugs directly to the tumor site (Weerakkody and Witharana, 2019; Li et al., 2021; Yang et al., 2021).
3.3.1 Releasing natural toxins promotes cancer cell apoptosis
Firstly, many bacteria contain natural toxins with potential anti-cancer properties. To better utilize these bacterial toxins for cancer treatment, scientists have recombined the natural toxin genes to express and purify them through engineered bacteria. This not only enhances the targeting of cancer cells but also reduces damage to normal tissues. For instance, researchers have mutated the P. aeruginosa exotoxin A (PE) and fused it with anti-CD25 to form a new recombinant toxin (CD25-PE38KDELKQK). The recombinant toxin, expressed and purified by E. coli, significantly reduced the natural toxin’s toxicity to the pulmonary vessels in the lungs of mice with lung cancer (Wang et al., 2008). Researchers have also linked the B subunit of the natural bacterial toxin Shiga-like toxin (STXB) with the DT389 fragment of the natural diphtheria toxin (DT538) to form a fusion toxic protein (DT389-STXB). STXB is responsible for recognizing the cancer cell surface receptor GB3, while the DT389 toxin is responsible for eliminating cancer cells, showing high affinity for breast cancer cells (Mohseni et al., 2021).
3.3.2 Activate the immune system to recognize and eliminate cancer cells
Engineering bacteria can activate immune cells or guide them to recognize and kill cancer cells by expressing specific immune-stimulating molecules, such as cytokines or chemokines. This approach enhances the immune memory effect, preventing the recurrence of cancer.
Interleukin-2 (IL-2) can activate immune cells infiltrating tumor tissues, including T cells, NK cells, and ILC cells (Raeber et al., 2022). By electroporation of plasmids carrying the human IL-2 gene into the attenuated Salmonella typhimurium strain x455O, a drug delivery system x455O (pIL2) is constructed. Gavage of this engineered bacteria in MC-38 adenocarcinoma mice not only reduced liver metastasis of MC-38 adenocarcinoma but also did not observe toxicity of IL-2 or Salmonella (Saltzman et al., 1996). Subsequently, researchers successively attempted to use Clostridium butyricum secreting rat interleukin-2 (rIL2) (Barbé et al., 2005), and Bacillus cereus secreting mouse interleukin-2 (mIL2) as engineered bacteria for tumor treatment (Kubiak et al., 2021). Recently, scientists have progressively optimized the bioactivity of IL-2 using EcN, and mice with colon cancer models treated with intravenous injection of this engineered bacterial strain showed a moderate reduction in tumor growth rate and a significant increase in IL-2 levels in the tumor (Tumas et al., 2023).
Components of the interleukin-18 (IL-18) signaling pathway are upregulated on tumor-infiltrating lymphocytes, indicating that IL-18 can enhance the immune system’s ability to combat tumors (Zhou et al., 2020). Researchers synthesized IL-18 using an engineered attenuated Salmonella typhimurium strain (VNP20009), and after intravenous injection of this engineered bacteria into a mouse model, the growth of colon cancer and breast cancer cells was significantly inhibited, as well as lung metastasis in immunocompetent mice (Loeffler et al., 2008).
L-arginine in tumor tissue can enhance the antitumor capability of immune checkpoint inhibitors. To increase the production of L-arginine, scientists deleted the arginine repressor gene (ArgR) of EcN and mutated the N-acetylglutamate synthase gene (ArgAfbr). The engineered bacteria, when co-injected with anti-PD-L1, significantly reduced tumor growth rate (Canale et al., 2021).
Researchers used EcN with the lysis gene (ΦX174E) to express nanobodies targeting immune checkpoints PD-L1nb and CTLA-4nb. After intratumoral injection of this engineered bacteria, the tumor partially or completely regressed in the mouse model, and no visible liver metastasis was observed (Gurbatri et al., 2020).
Another study utilized engineered Salmonella (△ppGpp) to secrete the Del-1 protein (a potent inhibitor of neutrophil recruitment, antagonizing the antigen-1 associated with lymphocyte function on vascular endothelial cells). After tail vein injection of this engineered bacteria into a mouse model, the number of neutrophils in the tumor decreased, the number of M1-type macrophages increased, and tumor elimination was observed in some samples (Tian et al., 2022).
3.3.3 Transporting anti-cancer drugs directly to the tumor microenvironment
Engineering bacteria can serve as drug delivery systems, directly delivering anticancer drugs to tumor cells. This approach can increase the local concentration of drugs and reduce systemic side effects. Researchers have used E. coli DH5α that can specifically target tumor tissues to express beta-glucuronidase (βG), which converts inactive prodrugs into anticancer active drugs at the tumor site, significantly inhibiting tumor growth (Cheng et al., 2008; Afkhami-Poostchi et al., 2020).
Bacterial spores can also serve as carriers for anticancer drugs because obligate anaerobic bacterial spores can only germinate in the hypoxic necrotic areas present in solid tumors, while remaining dormant in other parts of the body (Barbé et al., 2006; Kubiak and Minton, 2015). However, the original bacterial spores cannot eliminate cancer cells and require genetic modification or combination with other anticancer strategies. Scientists have covalently attached the chemotherapeutic drug gemcitabine (MGEM) to Clostridium butyricum spores (SPORE), and after oral administration, the drug (SPORE-MGEM) migrates from the upper gastrointestinal tract to pancreatic tumors, with a threefold increase in drug accumulation within the tumor compared to MGEM alone. In a mouse model of pancreatic cancer, SPORE-MGEM inhibited tumor growth without significant side effects (Han et al., 2022).
3.4 Infectious diseases
3.4.1 AIDS
The primary mode of transmission for HIV/AIDS is sexual contact, with the cervicovaginal mucosa being the main route of HIV infection in women. Scientists have engineered Lactobacillus jensenii, isolated from vaginal mucosa, to express a bispecific CD4 (2D CD4) molecule on its surface that can bind to HIV proteins, which can impede heterosexual transmission of HIV (Chang et al., 2003; Liu et al., 2008). Subsequently, a study used engineered Lactobacillus acidophilus (ATCC 4356) to express human CD4, with the expressed CD4 protein anchored on the outer wall of the engineered bacteria to adsorb HIV-1 virus. In a humanized mouse model, the engineered bacteria, when stimulated rectally, prevented HIV-1 infection, but the vaginal effect was suboptimal (Wei et al., 2019). Similarly, the probiotic EcN was designed to secrete HIV-gp41-lysin peptides, thereby preventing HIV fusion and subsequent invasion. This engineered bacterial strain can colonize persistently and stably in the colon and cecum, releasing anti-HIV peptides in mice for several months (Rao et al., 2005).
3.4.2 Cholera
Cholera is a life-threatening gastrointestinal infectious disease caused by the infection of toxigenic Vibrio cholerae (Kanungo et al., 2022). Engineered bacterial interventions have been used for the diagnosis and treatment of cholera. Scientists have constructed an intelligent engineered bacterium using Lactococcus lactis, whose recombinant transmembrane receptor module HR4M can specifically detect the quorum-sensing signal CAI-1 of Vibrio cholerae in the intestine and trigger the expression of the reporter gene β-lactamase. The expressed product is easily detectable in fecal samples, thereby assisting in the diagnosis of cholera (Mao et al., 2018). Holowko et al. (2016) developed an intelligent diagnostic engineered bacterium based on the quorum-sensing mechanism of Vibrio cholerae and CRISPRi technology for detecting Vibrio cholerae. It uses E. coli as a vector, and when Vibrio cholerae is at a high density, the transmembrane receptor module CqsS-LuxU-LuxO reacts with the signal molecule CAI-1, and the CRISPRi-based transmission control system triggers the expression of the diagnostic factor GFP, which has been successfully used for detecting Vibrio cholerae. Similarly, Jayaraman et al. (2017) replaced the GFP from the previous study with the lytic protein YebF-Art-085, and introduced the expression of the killing protein Art-085. The lysed engineered bacteria release the killing protein, thereby eliminating Vibrio cholerae. In vitro experiments have shown that this intelligent engineered bacterium can effectively inhibit the growth of Vibrio cholerae cells.
3.4.3 Pseudomonas aeruginosa infection
P.aeruginosa primarily colonizes the respiratory and gastrointestinal tracts (Ungor and Apidianakis, 2024). It is one of the main causes of hospital-acquired infections due to its resistance to many antibiotics and antimicrobial agents (Jurado-Martín et al., 2021). Utilizing engineered bacteria to effectively sense and kill P. aeruginosa provides a novel antibacterial strategy for controlling infections caused by this bacterium. Researchers have designed a smart E. coli that can detect acyl-homoserine lactone 3OC12HSL produced by P. aeruginosa using the intracellular receptor protein lasR. Upon detection, the bacteriocin Pyocin S5 is produced, which can be released by the E7 lysis protein after lysing E. coli, and then kills P. aeruginosa in the infected environment (Saeidi et al., 2011). Building on the aforementioned research, another study changed the release mode of the smart engineered bacteria from lysis to secretion. When lasR binds to the P. aeruginosa infection signal 3OC12HSL, the pathogen-specific toxin CoPy, which carries the signal peptide Flgm, is expressed and can be released through the flagellar secretion system. This design can obviously kill P. aeruginosa more effectively and for a longer duration (Gupta et al., 2013). Subsequently, a similar study used the probiotic strain EcN as a host and added the expression of the biofilm-degrading enzyme DspB to the bactericidal protein Pyocin S5. This engineered bacterium showed both preventive and therapeutic activity in Caenorhabditis elegans and mouse models of P. aeruginosa intestinal infection (Hwang et al., 2017). Recently, Gao et al. (2023) constructed a chimeric pyocin (ChPy) to specifically kill P. aeruginosa and designed a near-infrared (NIR) light-responsive strain to produce and deliver this drug. This smart engineered bacterial strain can continuously produce ChPy without light and release it to kill P. aeruginosa upon NIR light-induced bacterial lysis.
4 Discussion
Although intelligent engineered bacteria have achieved preliminary research results in disease diagnosis and treatment, there are still a series of challenges and issues to be faced before they can be applied clinically. First, most of the research on therapeutic engineered bacteria is still focused on in vitro testing and animal experiments. While these experiments are important, they do not fully simulate the human body’s environment. Therefore, more rigorous clinical trials are needed to prove the actual efficacy and safety of therapeutic engineered bacteria. Second, the biosensing systems of the diagnostic and therapeutic engineered bacteria that have been developed are relatively simple, with some having only a single input and output, and some only containing basic logic control, which is not intelligent enough. Faced with complex disease environments, it is necessary to design biosensing systems with signal storage, automatic calibration, and self-learning capabilities to improve the specificity and targeting of intelligent diagnostic and therapeutic engineered bacteria. Finally, the ethics, legal issues, and social acceptance of intelligent diagnostic and therapeutic engineered bacteria are also issues that cannot be ignored. How to balance innovation and risk, ensure that technological development complies with ethical standards, and gain public understanding and support are necessary conditions for promoting the development of intelligent diagnostic and therapeutic engineered bacteria.
Despite the many challenges, with the comprehensive progress and in-depth research in various fields such as synthetic biology, artificial intelligence, and nanomaterials, interdisciplinary cooperation has injected new vitality and hope into the development of intelligent diagnostic and therapeutic engineered bacteria. The rapidly developing field of synthetic biology continuously provides various components for intelligent diagnostic and therapeutic engineered bacteria. These include easily manipulated engineered bacterial strains, biosensing systems with computational capabilities, storage capacity, and calibration functions. These standardized module libraries make the development of intelligent diagnostic and therapeutic engineered bacteria more convenient (Zhang C. et al., 2023). With the integration of artificial intelligence and big data technology in the field of bioinformatics, researchers have built numerous computational platforms for predicting protein structures and simulating drug-target interactions (Gupta et al., 2021). These platforms have greatly accelerated the research and development and evolution of intelligent diagnostic and therapeutic engineered bacteria. Innovations in fields such as optics (Gao et al., 2023), ultrasound technology (Bourdeau et al., 2018; Lakshmanan et al., 2020), and nanomaterials (Cao et al., 2023; Ma et al., 2023) have opened up multiple paths for the widespread clinical application of intelligent diagnostic and therapeutic engineered bacteria.
In summary, intelligent diagnostic and therapeutic engineered bacteria will play an important role in the future medical field. It will open up a new method of treatment and inject new vitality into the cause of human health.
Author contributions
NZ: Conceptualization, Funding acquisition, Project administration, Supervision, Writing–original draft, Writing–review and editing. JC: Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. JS: Data curation, Formal Analysis, Investigation, Writing–original draft. YG: Investigation, Writing–original draft. LL: Investigation, Writing–original draft. LD: Investigation, Writing–original draft.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by Yinchuan Science and Technology Innovation Project, Grant/Award Number:2023SF14.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adolfsen, K. J., Callihan, I., Monahan, C. E., Greisen, P. J., Spoonamore, J., Momin, M., et al. (2021). Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215. doi:10.1038/s41467-021-26524-0
Afkhami-Poostchi, A., Mashreghi, M., Iranshahi, M., and Matin, M. M. (2020). Use of a genetically engineered E. coli overexpressing β-glucuronidase accompanied by glycyrrhizic acid, a natural and anti-inflammatory agent, for directed treatment of colon carcinoma in a mouse model. Int. J. Pharm. 579, 119159. doi:10.1016/j.ijpharm.2020.119159
Agarwal, P., Khatri, P., Billack, B., Low, W. K., and Shao, J. (2014). Oral delivery of glucagon like peptide-1 by a recombinant Lactococcus lactis. Pharm. Res. 31, 3404–3414. doi:10.1007/s11095-014-1430-3
Aitken, J. M., Aitken, J. E., and Agrawal, G. (2024). Mycobacterium avium ssp. paratuberculosis and Crohn's disease-diagnostic microbiological investigations can inform new therapeutic approaches. Antibiot. (Basel) 13, 158. doi:10.3390/antibiotics13020158
Ali, A. A., Altemimi, A. B., Alhelfi, N., and Ibrahim, S. A. (2020). Application of biosensors for detection of pathogenic food bacteria: a review. Biosens. (Basel) 10, 58. doi:10.3390/bios10060058
Anderson, J. C., Voigt, C. A., and Arkin, A. P. (2007). Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 3, 133. doi:10.1038/msb4100173
Archer, E. J., Robinson, A. B., and Süel, G. M. (2012). Engineered E. coli that detect and respond to gut inflammation through nitric oxide sensing. ACS Synth. Biol. 1, 451–457. doi:10.1021/sb3000595
Armetta, J., Schantz-Klausen, M., Shepelin, D., Vazquez-Uribe, R., Bahl, M. I., Laursen, M. F., et al. (2021). Escherichia coli promoters with consistent expression throughout the murine gut. ACS Synth. Biol. 10, 3359–3368. doi:10.1021/acssynbio.1c00325
Arnold, J., Glazier, J., and Mimee, M. (2023). Genetic engineering of resident bacteria in the gut microbiome. J. Bacteriol. 205, e0012723. doi:10.1128/jb.00127-23
Aubry, C., Michon, C., Chain, F., Chvatchenko, Y., Goffin, L., Zimmerli, S. C., et al. (2015). Protective effect of TSLP delivered at the gut mucosa level by recombinant lactic acid bacteria in DSS-induced colitis mouse model. Microb. Cell Fact. 14, 176. doi:10.1186/s12934-015-0367-5
Bai, L., Gao, M., Cheng, X., Kang, G., Cao, X., and Huang, H. (2020). Engineered butyrate-producing bacteria prevents high fat diet-induced obesity in mice. Microb. Cell Fact. 19, 94. doi:10.1186/s12934-020-01350-z
Barbé, S., Van Mellaert, L., and Anné, J. (2006). The use of clostridial spores for cancer treatment. J. Appl. Microbiol. 101, 571–578. doi:10.1111/j.1365-2672.2006.02886.x
Barbé, S., Van Mellaert, L., Theys, J., Geukens, N., Lammertyn, E., Lambin, P., et al. (2005). Secretory production of biologically active rat interleukin-2 by Clostridium acetobutylicum DSM792 as a tool for anti-tumor treatment. FEMS Microbiol. Lett. 246, 67–73. doi:10.1016/j.femsle.2005.03.037
Bartoli, V., Meaker, G. A., Di Bernardo, M., and Gorochowski, T. E. (2020). Tunable genetic devices through simultaneous control of transcription and translation. Nat. Commun. 11, 2095. doi:10.1038/s41467-020-15653-7
Beier, D., and Gross, R. (2006). Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9, 143–152. doi:10.1016/j.mib.2006.01.005
Borghi, C., Agabiti-Rosei, E., Johnson, R. J., Kielstein, J. T., Lurbe, E., Mancia, G., et al. (2020). Hyperuricaemia and gout in cardiovascular, metabolic and kidney disease. Eur. J. Intern Med. 80, 1–11. doi:10.1016/j.ejim.2020.07.006
Bourdeau, R. W., Lee-Gosselin, A., Lakshmanan, A., Farhadi, A., Kumar, S. R., Nety, S. P., et al. (2018). Acoustic reporter genes for noninvasive imaging of microorganisms in mammalian hosts. Nature 553, 86–90. doi:10.1038/nature25021
Brophy, J. A., and Voigt, C. A. (2014). Principles of genetic circuit design. Nat. Methods 11, 508–520. doi:10.1038/nmeth.2926
Cambray, G., Guimaraes, J. C., Mutalik, V. K., Lam, C., Mai, Q. A., Thimmaiah, T., et al. (2013). Measurement and modeling of intrinsic transcription terminators. Nucleic Acids Res. 41, 5139–5148. doi:10.1093/nar/gkt163
Canale, F. P., Basso, C., Antonini, G., Perotti, M., Li, N., Sokolovska, A., et al. (2021). Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 598, 662–666. doi:10.1038/s41586-021-04003-2
Cao, F., Jin, L., Gao, Y., Ding, Y., Wen, H., Qian, Z., et al. (2023). Artificial-enzymes-armed Bifidobacterium longum probiotics for alleviating intestinal inflammation and microbiota dysbiosis. Nat. Nanotechnol. 18, 617–627. doi:10.1038/s41565-023-01346-x
Chang, T. L., Chang, C. H., Simpson, D. A., Xu, Q., Martin, P. K., Lagenaur, L. A., et al. (2003). Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl. Acad. Sci. U. S. A. 100, 11672–11677. doi:10.1073/pnas.1934747100
Chen, X. J., Wang, B., Thompson, I. P., and Huang, W. E. (2021). Rational design and characterization of nitric oxide biosensors in E. coli Nissle 1917 and mini SimCells. ACS Synth. Biol. 10, 2566–2578. doi:10.1021/acssynbio.1c00223
Chen, Y. J., Liu, P., Nielsen, A. A., Brophy, J. A., Clancy, K., Peterson, T., et al. (2013). Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659–664. doi:10.1038/nmeth.2515
Chen, Z., and Elowitz, M. B. (2021). Programmable protein circuit design. Cell 184, 2284–2301. doi:10.1016/j.cell.2021.03.007
Chen, Z., Guo, L., Zhang, Y., Walzem, R. L., Pendergast, J. S., Printz, R. L., et al. (2014). Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J. Clin. Invest. 124, 3391–3406. doi:10.1172/jci72517
Cheng, C. M., Lu, Y. L., Chuang, K. H., Hung, W. C., Shiea, J., Su, Y. C., et al. (2008). Tumor-targeting prodrug-activating bacteria for cancer therapy. Cancer Gene Ther. 15, 393–401. doi:10.1038/cgt.2008.10
Chua, K. J., Ling, H., Hwang, I. Y., Lee, H. L., March, J. C., Lee, Y. S., et al. (2023). An engineered probiotic produces a type III interferon IFNL1 and reduces inflammations in in vitro inflammatory bowel disease models. ACS Biomater. Sci. Eng. 9, 5123–5135. doi:10.1021/acsbiomaterials.2c00202
Clarke, L., and Kitney, R. (2020). Developing synthetic biology for industrial biotechnology applications. Biochem. Soc. Trans. 48, 113–122. doi:10.1042/bst20190349
Corthier, G., Delorme, C., Ehrlich, S. D., and Renault, P. (1998). Use of luciferase genes as biosensors to study bacterial physiology in the digestive tract. Appl. Environ. Microbiol. 64, 2721–2722. doi:10.1128/aem.64.7.2721-2722.1998
Cox, R. S., Surette, M. G., and Elowitz, M. B. (2007). Programming gene expression with combinatorial promoters. Mol. Syst. Biol. 3, 145. doi:10.1038/msb4100187
Cui, M., Pang, G., Zhang, T., Sun, T., Zhang, L., Kang, R., et al. (2021). Optotheranostic nanosystem with phone visual diagnosis and optogenetic microbial therapy for ulcerative colitis at-home care. ACS Nano 15, 7040–7052. doi:10.1021/acsnano.1c00135
Daeffler, K. N., Galley, J. D., Sheth, R. U., Ortiz-Velez, L. C., Bibb, C. O., Shroyer, N. F., et al. (2017). Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol. 13, 923. doi:10.15252/msb.20167416
Din, M. O., Danino, T., Prindle, A., Skalak, M., Selimkhanov, J., Allen, K., et al. (2016). Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85. doi:10.1038/nature18930
Dobrin, A., Saxena, P., and Fussenegger, M. (2016). Synthetic biology: applying biological circuits beyond novel therapies. Integr. Biol. (Camb) 8, 409–430. doi:10.1039/c5ib00263j
Drouault, S., Anba, J., and Corthier, G. (2002). Streptococcus thermophilus is able to produce a β-galactosidase active during its transit in the digestive tract of germ-free mice. Appl. Environ. Microbiol. 68, 938–941. doi:10.1128/aem.68.2.938-941.2002
Duan, F. F., Liu, J. H., and March, J. C. (2015). Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes 64, 1794–1803. doi:10.2337/db14-0635
Durant, L., Stentz, R., Noble, A., Brooks, J., Gicheva, N., Reddi, D., et al. (2020). Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 8, 88. doi:10.1186/s40168-020-00868-z
Espah Borujeni, A., Channarasappa, A. S., and Salis, H. M. (2014). Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res. 42, 2646–2659. doi:10.1093/nar/gkt1139
Farasat, I., Kushwaha, M., Collens, J., Easterbrook, M., Guido, M., and Salis, H. M. (2014). Efficient search, mapping, and optimization of multi-protein genetic systems in diverse bacteria. Mol. Syst. Biol. 10, 731. doi:10.15252/msb.20134955
Frelet-Barrand, A. (2022). Lactococcus lactis, an attractive cell factory for the expression of functional membrane proteins. Biomolecules 12, 180. doi:10.3390/biom12020180
Galperin, M. Y. (2010). Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13, 150–159. doi:10.1016/j.mib.2010.01.005
Gao, C., Xu, P., Ye, C., Chen, X., and Liu, L. (2019). Genetic circuit-assisted smart microbial engineering. Trends Microbiol. 27, 1011–1024. doi:10.1016/j.tim.2019.07.005
Gao, R., and Stock, A. M. (2009). Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63, 133–154. doi:10.1146/annurev.micro.091208.073214
Gao, Y., Wei, J., Pu, L., Fu, S., Xing, X., Zhang, R., et al. (2023). Remotely controllable engineered bacteria for targeted therapy of Pseudomonas aeruginosa infection. ACS Synth. Biol. 12, 1961–1971. doi:10.1021/acssynbio.2c00655
Gentschev, I., Dietrich, G., and Goebel, W. (2002). The E. coli α-hemolysin secretion system and its use in vaccine development. Trends Microbiol. 10, 39–45. doi:10.1016/s0966-842x(01)02259-4
González-Prieto, C., and Lesser, C. F. (2018). Rationale redesign of type III secretion systems: toward the development of non-pathogenic E. coli for in vivo delivery of therapeutic payloads. Curr. Opin. Microbiol. 41, 1–7. doi:10.1016/j.mib.2017.10.011
Gordley, R. M., Bugaj, L. J., and Lim, W. A. (2016). Modular engineering of cellular signaling proteins and networks. Curr. Opin. Struct. Biol. 39, 106–114. doi:10.1016/j.sbi.2016.06.012
Gui, Q., Lawson, T., Shan, S., Yan, L., and Liu, Y. (2017). The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors (Basel) 17, 1623. doi:10.3390/s17071623
Gupta, R., Srivastava, D., Sahu, M., Tiwari, S., Ambasta, R. K., and Kumar, P. (2021). Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol. Divers 25, 1315–1360. doi:10.1007/s11030-021-10217-3
Gupta, S., Bram, E. E., and Weiss, R. (2013). Genetically programmable pathogen sense and destroy. ACS Synth. Biol. 2, 715–723. doi:10.1021/sb4000417
Gurbatri, C. R., Arpaia, N., and Danino, T. (2022). Engineering bacteria as interactive cancer therapies. Science 378, 858–864. doi:10.1126/science.add9667
Gurbatri, C. R., Lia, I., Vincent, R., Coker, C., Castro, S., Treuting, P. M., et al. (2020). Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 12, eaax0876. doi:10.1126/scitranslmed.aax0876
Hamady, Z. Z., Scott, N., Farrar, M. D., Lodge, J. P., Holland, K. T., Whitehead, T., et al. (2010). Xylan-regulated delivery of human keratinocyte growth factor-2 to the inflamed colon by the human anaerobic commensal bacterium Bacteroides ovatus. Gut 59, 461–469. doi:10.1136/gut.2008.176131
Han, Z. Y., Chen, Q. W., Fu, Z. J., Cheng, S. X., and Zhang, X. Z. (2022). Probiotic spore-based oral drug delivery system for enhancing pancreatic cancer chemotherapy by gut-pancreas-axis-guided delivery. Nano Lett. 22, 8608–8617. doi:10.1021/acs.nanolett.2c03131
Hanson, M. L., Hixon, J. A., Li, W., Felber, B. K., Anver, M. R., Stewart, C. A., et al. (2014). Oral delivery of IL-27 recombinant bacteria attenuates immune colitis in mice. Gastroenterology 146, 210–221.e13. doi:10.1053/j.gastro.2013.09.060
Ho, C. L., Tan, H. Q., Chua, K. J., Kang, A., Lim, K. H., Ling, K. L., et al. (2018). Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat. Biomed. Eng. 2, 27–37. doi:10.1038/s41551-017-0181-y
Holowko, M. B., Wang, H., Jayaraman, P., and Poh, C. L. (2016). Biosensing Vibrio cholerae with genetically engineered Escherichia coli. ACS Synth. Biol. 5, 1275–1283. doi:10.1021/acssynbio.6b00079
Horwitz, J. P., Chua, J., Curby, R. J., Tomson, A. J., Da Rooge, M. A., Fisher, B. E., et al. (1964). Substrates for cytochemical demonstration of enzyme activity. I. Some substituted 3-Indolyl-β-D-glycopyranosides1a. J. Med. Chem. 7, 574–575. doi:10.1021/jm00334a044
Hwang, I. Y., Koh, E., Wong, A., March, J. C., Bentley, W. E., Lee, Y. S., et al. (2017). Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8, 15028. doi:10.1038/ncomms15028
Isabella, V. M., Ha, B. N., Castillo, M. J., Lubkowicz, D. J., Rowe, S. E., Millet, Y. A., et al. (2018). Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864. doi:10.1038/nbt.4222
Jayaraman, P., Holowko, M. B., Yeoh, J. W., Lim, S., and Poh, C. L. (2017). Repurposing a two-component system-based biosensor for the killing of Vibrio cholerae. ACS Synth. Biol. 6, 1403–1415. doi:10.1021/acssynbio.7b00058
Jeong, J. H., Kim, K., Lim, D., Jeong, K., Hong, Y., Nguyen, V. H., et al. (2014). Anti-tumoral effect of the mitochondrial target domain of Noxa delivered by an engineered Salmonella typhimurium. PLoS One 9, e80050. doi:10.1371/journal.pone.0080050
Jurado-Martín, I., Sainz-Mejías, M., and Mcclean, S. (2021). Pseudomonas aeruginosa: an audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 22, 3128. doi:10.3390/ijms22063128
Kanungo, S., Azman, A. S., Ramamurthy, T., Deen, J., and Dutta, S. (2022). Cholera. Lancet 399, 1429–1440. doi:10.1016/s0140-6736(22)00330-0
Kelly, J. R., Rubin, A. J., Davis, J. H., Ajo-Franklin, C. M., Cumbers, J., Czar, M. J., et al. (2009). Measuring the activity of BioBrick promoters using an in vivo reference standard. J. Biol. Eng. 3, 4. doi:10.1186/1754-1611-3-4
Kubiak, A. M., Bailey, T. S., Dubois, L. J., Theys, J., and Lambin, P. (2021). Efficient secretion of murine IL-2 from an attenuated strain of Clostridium sporogenes, a novel delivery vehicle for cancer immunotherapy. Front. Microbiol. 12, 669488. doi:10.3389/fmicb.2021.669488
Kubiak, A. M., and Minton, N. P. (2015). The potential of clostridial spores as therapeutic delivery vehicles in tumour therapy. Res. Microbiol. 166, 244–254. doi:10.1016/j.resmic.2014.12.006
Kurtz, C. B., Millet, Y. A., Puurunen, M. K., Perreault, M., Charbonneau, M. R., Isabella, V. M., et al. (2019). An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 11, eaau7975. doi:10.1126/scitranslmed.aau7975
Lai, Y., Hayashi, N., and Lu, T. K. (2022). Engineering the human gut commensal Bacteroides thetaiotaomicron with synthetic biology. Curr. Opin. Chem. Biol. 70, 102178. doi:10.1016/j.cbpa.2022.102178
Lakshmanan, A., Jin, Z., Nety, S. P., Sawyer, D. P., Lee-Gosselin, A., Malounda, D., et al. (2020). Acoustic biosensors for ultrasound imaging of enzyme activity. Nat. Chem. Biol. 16, 988–996. doi:10.1038/s41589-020-0591-0
Landry, B. P., and Tabor, J. J. (2017). Engineering diagnostic and therapeutic gut bacteria. Microbiol. Spectr. 5. doi:10.1128/microbiolspec.bad-0020-2017
Lazar, J. T., and Tabor, J. J. (2021). Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol. 28, 100398. doi:10.1016/j.coisb.2021.100398
Li, Q., Zheng, T., Ding, H., Chen, J., Li, B., Zhang, Q., et al. (2023). Exploring the benefits of probiotics in gut inflammation and diarrhea-from an antioxidant perspective. Antioxidants (Basel) 12, 1342. doi:10.3390/antiox12071342
Li, Z., Wang, Y., Liu, J., Rawding, P., Bu, J., Hong, S., et al. (2021). Chemically and biologically engineered bacteria-based delivery systems for emerging diagnosis and advanced therapy. Adv. Mater 33, e2102580. doi:10.1002/adma.202102580
Liang, M., Zhang, J., Yang, Y., Xia, Y., Liu, L., Liu, L., et al. (2022). Nattokinase enhances the preventive effects of Escherichia coli Nissle 1917 on dextran sulfate sodium-induced colitis in mice. World J. Microbiol. Biotechnol. 39, 8. doi:10.1007/s11274-022-03452-9
Lim, D., Kim, K. S., Kim, H., Ko, K. C., Song, J. J., Choi, J. H., et al. (2017). Anti-tumor activity of an immunotoxin (TGFα-PE38) delivered by attenuated Salmonella typhimurium. Oncotarget 8, 37550–37560. doi:10.18632/oncotarget.17197
Lim, D., and Song, M. (2019). Development of bacteria as diagnostics and therapeutics by genetic engineering. J. Microbiol. 57, 637–643. doi:10.1007/s12275-019-9105-8
Lin, Y., Krogh-Andersen, K., Pelletier, J., Marcotte, H., Östenson, C. G., and Hammarström, L. (2016). Oral delivery of pentameric glucagon-like peptide-1 by recombinant lactobacillus in diabetic rats. PLoS One 11, e0162733. doi:10.1371/journal.pone.0162733
Liu, X., Lagenaur, L. A., Lee, P. P., and Xu, Q. (2008). Engineering of a human vaginal Lactobacillus strain for surface expression of two-domain CD4 molecules. Appl. Environ. Microbiol. 74, 4626–4635. doi:10.1128/aem.00104-08
Liu, X., Lagenaur, L. A., Simpson, D. A., Essenmacher, K. P., Frazier-Parker, C. L., Liu, Y., et al. (2006). Engineered vaginal lactobacillus strain for mucosal delivery of the human immunodeficiency virus inhibitor cyanovirin-N. Antimicrob. Agents Chemother. 50, 3250–3259. doi:10.1128/aac.00493-06
Loeffler, M., Le'negrate, G., Krajewska, M., and Reed, J. C. (2008). IL-18-producing Salmonella inhibit tumor growth. Cancer Gene Ther. 15, 787–794. doi:10.1038/cgt.2008.48
Lu, Q., Yang, M. F., Liang, Y. J., Xu, J., Xu, H. M., Nie, Y. Q., et al. (2022). Immunology of inflammatory bowel disease: molecular mechanisms and therapeutics. J. Inflamm. Res. 15, 1825–1844. doi:10.2147/jir.s353038
Lutz, R., and Bujard, H. (1997). Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210. doi:10.1093/nar/25.6.1203
Ma, X., Liang, X., Li, Y., Feng, Q., Cheng, K., Ma, N., et al. (2023). Modular-designed engineered bacteria for precision tumor immunotherapy via spatiotemporal manipulation by magnetic field. Nat. Commun. 14, 1606. doi:10.1038/s41467-023-37225-1
Mao, N., Cubillos-Ruiz, A., Cameron, D. E., and Collins, J. J. (2018). Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 10, eaao2586. doi:10.1126/scitranslmed.aao2586
Mcdowell, C., Farooq, U., and Haseeb, M. (2024). “Inflammatory bowel disease,” in StatPearls (Treasure Island (FL): StatPearls Publishing).
Mcnerney, M. P., Michel, C. L., Kishore, K., Standeven, J., and Styczynski, M. P. (2019). Dynamic and tunable metabolite control for robust minimal-equipment assessment of serum zinc. Nat. Commun. 10, 5514. doi:10.1038/s41467-019-13454-1
Mimee, M., Tucker, A. C., Voigt, C. A., and Lu, T. K. (2015). Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71. doi:10.1016/j.cels.2015.06.001
Mohseni, Z., Sedighian, H., Halabian, R., Amani, J., Behzadi, E., and Imani Fooladi, A. A. (2021). Potent in vitro antitumor activity of B-subunit of Shiga toxin conjugated to the diphtheria toxin against breast cancer. Eur. J. Pharmacol. 899, 174057. doi:10.1016/j.ejphar.2021.174057
Montero-Blay, A., Blanco, J. D., Rodriguez-Arce, I., Lastrucci, C., Piñero-Lambea, C., Lluch-Senar, M., et al. (2023). Bacterial expression of a designed single-chain IL-10 prevents severe lung inflammation. Mol. Syst. Biol. 19, e11037. doi:10.15252/msb.202211037
Mutalik, V. K., Guimaraes, J. C., Cambray, G., Lam, C., Christoffersen, M. J., Mai, Q. A., et al. (2013). Precise and reliable gene expression via standard transcription and translation initiation elements. Nat. Methods 10, 354–360. doi:10.1038/nmeth.2404
Na, H. S., Kim, H. J., Lee, H. C., Hong, Y., Rhee, J. H., and Choy, H. E. (2006). Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine 24, 2027–2034. doi:10.1016/j.vaccine.2005.11.031
Noguès, E. B., Kropp, C., Bétemps, L., de Sousa, C., Chain, F., Auger, S., et al. (2022). Lactococcus lactis engineered to deliver hCAP18 cDNA alleviates DNBS-induced colitis in C57BL/6 mice by promoting IL17A and IL10 cytokine expression. Sci. Rep. 12, 15641. doi:10.1038/s41598-022-19455-3
Ochoa-Sanchez, R., Oliveira, M. M., Tremblay, M., Petrazzo, G., Pant, A., Bosoi, C. R., et al. (2021). Genetically engineered E. coli Nissle attenuates hyperammonemia and prevents memory impairment in bile-duct ligated rats. Liver Int. 41, 1020–1032. doi:10.1111/liv.14815
Perdomo, C. M., Cohen, R. V., Sumithran, P., Clément, K., and Frühbeck, G. (2023). Contemporary medical, device, and surgical therapies for obesity in adults. Lancet 401, 1116–1130. doi:10.1016/s0140-6736(22)02403-5
Pickard, J. M., Maurice, C. F., Kinnebrew, M. A., Abt, M. C., Schenten, D., Golovkina, T. V., et al. (2014). Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641. doi:10.1038/nature13823
Pöhlmann, C., Brandt, M., Mottok, D. S., Zschüttig, A., Campbell, J. W., Blattner, F. R., et al. (2012). Periplasmic delivery of biologically active human interleukin-10 in Escherichia coli via a sec-dependent signal peptide. J. Mol. Microbiol. Biotechnol. 22, 1–9. doi:10.1159/000336043
Praveschotinunt, P., Duraj-Thatte, A. M., Gelfat, I., Bahl, F., Chou, D. B., and Joshi, N. S. (2019). Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 10, 5580. doi:10.1038/s41467-019-13336-6
Puurunen, M. K., Vockley, J., Searle, S. L., Sacharow, S. J., Phillips, J. A., Denney, W. S., et al. (2021). Safety and pharmacodynamics of an engineered E. coli Nissle for the treatment of phenylketonuria: a first-in-human phase 1/2a study. Nat. Metab. 3, 1125–1132. doi:10.1038/s42255-021-00430-7
Raeber, M. E., Sahin, D., and Boyman, O. (2022). Interleukin-2–based therapies in cancer. Sci. Transl. Med. 14, eabo5409. doi:10.1126/scitranslmed.abo5409
Rao, S., Hu, S., Mchugh, L., Lueders, K., Henry, K., Zhao, Q., et al. (2005). Toward a live microbial microbicide for HIV: commensal bacteria secreting an HIV fusion inhibitor peptide. Proc. Natl. Acad. Sci. U. S. A. 102, 11993–11998. doi:10.1073/pnas.0504881102
Reeves, A. Z., Spears, W. E., Du, J., Tan, K. Y., Wagers, A. J., and Lesser, C. F. (2015). Engineering Escherichia coli into a protein delivery system for mammalian cells. ACS Synth. Biol. 4, 644–654. doi:10.1021/acssynbio.5b00002
Ribas, G. S., Lopes, F. F., Deon, M., and Vargas, C. R. (2022). Hyperammonemia in inherited metabolic diseases. Cell Mol. Neurobiol. 42, 2593–2610. doi:10.1007/s10571-021-01156-6
Ricci, S., Macchia, G., Ruggiero, P., Maggi, T., Bossù, P., Xu, L., et al. (2003). In vivo mucosal delivery of bioactive human interleukin 1 receptor antagonist produced by Streptococcus gordonii. BMC Biotechnol. 3, 15. doi:10.1186/1472-6750-3-15
Riglar, D. T., Giessen, T. W., Baym, M., Kerns, S. J., Niederhuber, M. J., Bronson, R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35, 653–658. doi:10.1038/nbt.3879
Riglar, D. T., and Silver, P. A. (2018). Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 16, 214–225. doi:10.1038/nrmicro.2017.172
Robert, S., Gysemans, C., Takiishi, T., Korf, H., Spagnuolo, I., Sebastiani, G., et al. (2014). Oral delivery of glutamic acid decarboxylase (GAD)-65 and IL10 by Lactococcus lactis reverses diabetes in recent-onset NOD mice. Diabetes 63, 2876–2887. doi:10.2337/db13-1236
Rosano, G. L., Morales, E. S., and Ceccarelli, E. A. (2019). New tools for recombinant protein production in Escherichia coli: a 5-year update. Protein Sci. 28, 1412–1422. doi:10.1002/pro.3668
Ruano-Gallego, D., Álvarez, B., and Fernández, L. (2015). Engineering the controlled assembly of filamentous injectisomes in E. coli K-12 for protein translocation into mammalian cells. ACS Synth. Biol. 4, 1030–1041. doi:10.1021/acssynbio.5b00080
Ryan, R. M., Green, J., Williams, P. J., Tazzyman, S., Hunt, S., Harmey, J. H., et al. (2009). Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther. 16, 329–339. doi:10.1038/gt.2008.188
Saeidi, N., Wong, C. K., Lo, T. M., Nguyen, H. X., Ling, H., Leong, S. S., et al. (2011). Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Mol. Syst. Biol. 7, 521. doi:10.1038/msb.2011.55
Salis, H. M., Mirsky, E. A., and Voigt, C. A. (2009). Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950. doi:10.1038/nbt.1568
Saltzman, D. A., Heise, C. P., Hasz, D. E., Zebede, M., Kelly, S. M., Curtiss, R., et al. (1996). Attenuated Salmonella typhimurium containing interleukin-2 decreases MC-38 hepatic metastases: a novel anti-tumor agent. Cancer Biother Radiopharm. 11, 145–153. doi:10.1089/cbr.1996.11.145
Shigemori, S., Watanabe, T., Kudoh, K., Ihara, M., Nigar, S., Yamamoto, Y., et al. (2015). Oral delivery of Lactococcus lactis that secretes bioactive heme oxygenase-1 alleviates development of acute colitis in mice. Microb. Cell Fact. 14, 189. doi:10.1186/s12934-015-0378-2
Smanski, M. J., Zhou, H., Claesen, J., Shen, B., Fischbach, M. A., and Voigt, C. A. (2016). Synthetic biology to access and expand nature's chemical diversity. Nat. Rev. Microbiol. 14, 135–149. doi:10.1038/nrmicro.2015.24
Srivastava, R., and Lesser, C. F. (2024). Living engineered bacterial therapeutics: emerging affordable precision interventions. Microb. Biotechnol. 17, e70057. doi:10.1111/1751-7915.70057
Steidler, L., Hans, W., Schotte, L., Neirynck, S., Obermeier, F., Falk, W., et al. (2000). Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289, 1352–1355. doi:10.1126/science.289.5483.1352
Steidler, L., Neirynck, S., Huyghebaert, N., Snoeck, V., Vermeire, A., Goddeeris, B., et al. (2003). Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 21, 785–789. doi:10.1038/nbt840
Stirling, F., Bitzan, L., O'keefe, S., Redfield, E., Oliver, J. W. K., Way, J., et al. (2017). Rational design of evolutionarily stable microbial kill switches. Mol. Cell 68, 686–697.e3. doi:10.1016/j.molcel.2017.10.033
Subramanian, S., Khan, F., and Hirsch, I. B. (2024). New advances in type 1 diabetes. Bmj 384, e075681. doi:10.1136/bmj-2023-075681
Tabor, J. J., Groban, E. S., and Voigt, C. A. (2009). “Performance characteristics for sensors and circuits used to program E. coli,” in Systems biology and biotechnology of Escherichia coli. Editor S. Y. Lee (Dordrecht: Springer), 401–439.
Taketani, M., Zhang, J., Zhang, S., Triassi, A. J., Huang, Y. J., Griffith, L. G., et al. (2020). Genetic circuit design automation for the gut resident species Bacteroides thetaiotaomicron. Nat. Biotechnol. 38, 962–969. doi:10.1038/s41587-020-0468-5
Takiishi, T., Cook, D. P., Korf, H., Sebastiani, G., Mancarella, F., Cunha, J. P., et al. (2017). Reversal of diabetes in NOD mice by clinical-grade proinsulin and IL-10-secreting Lactococcus lactis in combination with low-dose anti-CD3 depends on the induction of foxp3-positive T cells. Diabetes 66, 448–459. doi:10.2337/db15-1625
Takiishi, T., Korf, H., Van Belle, T. L., Robert, S., Grieco, F. A., Caluwaerts, S., et al. (2012). Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J. Clin. Invest. 122, 1717–1725. doi:10.1172/jci60530
Taku, K., Britta, S., Chen, W. S., Ferrante, M., Shen, B., Bernstein, C. N., et al. (2020). Ulcerative colitis. Nat. Rev. Dis. Prim. 6, 74. doi:10.1038/s41572-020-0205-x
Tamsir, A., Tabor, J. J., and Voigt, C. A. (2011). Robust multicellular computing using genetically encoded NOR gates and chemical 'wires. Nature 469, 212–215. doi:10.1038/nature09565
Tanniche, I., and Behkam, B. (2023). Engineered live bacteria as disease detection and diagnosis tools. J. Biol. Eng. 17, 65. doi:10.1186/s13036-023-00379-z
Tao, Y. W., Gu, Y. L., Mao, X. Q., Zhang, L., and Pei, Y. F. (2020). Effects of probiotics on type II diabetes mellitus: a meta-analysis. J. Transl. Med. 18, 30. doi:10.1186/s12967-020-02213-2
Tian, S., Lin, G., Piao, L., and Liu, X. (2022). Del-1 enhances therapeutic efficacy of bacterial cancer immunotherapy by blocking recruitment of tumor-infiltrating neutrophils. Clin. Transl. Oncol. 24, 244–253. doi:10.1007/s12094-021-02679-6
Tong, Y., Wei, Y., Ju, Y., Li, P., Zhang, Y., Li, L., et al. (2023). Anaerobic purinolytic enzymes enable dietary purine clearance by engineered gut bacteria. Cell Chem. Biol. 30, 1104–1114.e7. doi:10.1016/j.chembiol.2023.04.008
Tumas, S., Meldgaard, T. S., Vaaben, T. H., Suarez Hernandez, S., Rasmussen, A. T., Vazquez-Uribe, R., et al. (2023). Engineered E. coli Nissle 1917 for delivery of bioactive IL-2 for cancer immunotherapy. Sci. Rep. 13, 12506. doi:10.1038/s41598-023-39365-2
Ungor, I., and Apidianakis, Y. (2024). Bacterial synergies and antagonisms affecting Pseudomonas aeruginosa virulence in the human lung, skin and intestine. Future Microbiol. 19, 141–155. doi:10.2217/fmb-2022-0155
Van Spronsen, F. J., Blau, N., Harding, C., Burlina, A., Longo, N., and Bosch, A. (2021). Phenylketonuria 7, 36. doi:10.1038/s41572-021-00267-0
Wang, B., Kitney, R. I., Joly, N., and Buck, M. (2011). Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nat. Commun. 2, 508. doi:10.1038/ncomms1516
Wang, H., Dai, J., Li, B., Fan, K., Peng, L., Zhang, D., et al. (2008). Expression, purification, and characterization of an immunotoxin containing a humanized anti-CD25 single-chain fragment variable antibody fused to a modified truncated Pseudomonas exotoxin A. Protein Expr. Purif. 58, 140–147. doi:10.1016/j.pep.2007.09.009
Wang, J., Tian, M., Li, W., and Hao, F. (2019). Preventative delivery of IL-35 by Lactococcus lactis ameliorates DSS-induced colitis in mice. Appl. Microbiol. Biotechnol. 103, 7931–7941. doi:10.1007/s00253-019-10094-9
Wang, L., Chen, T., Wang, H., Wu, X., Cao, Q., Wen, K., et al. (2021). Engineered bacteria of MG1363-pMG36e-GLP-1 attenuated obesity-induced by high fat diet in mice. Front. Cell Infect. Microbiol. 11, 595575. doi:10.3389/fcimb.2021.595575
Weerakkody, L. R., and Witharana, C. (2019). The role of bacterial toxins and spores in cancer therapy. Life Sci. 235, 116839. doi:10.1016/j.lfs.2019.116839
Wei, W., Wiggins, J., Hu, D., Vrbanac, V., Bowder, D., Mellon, M., et al. (2019). Blocking HIV-1 infection by chromosomal integrative expression of human CD4 on the surface of lactobacillus acidophilus ATCC 4356. J. Virol. 93, e01830. doi:10.1128/jvi.01830-18
Wong, A., Wang, H., Poh, C. L., and Kitney, R. I. (2015). Layering genetic circuits to build a single cell, bacterial half adder. BMC Biol. 13, 40. doi:10.1186/s12915-015-0146-0
Woo, S. G., Moon, S. J., Kim, S. K., Kim, T. H., Lim, H. S., Yeon, G. H., et al. (2020). A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron. 168, 112523. doi:10.1016/j.bios.2020.112523
Xu, F., Wang, K. Y., Wang, N., Li, G., and Liu, D. (2017). Modified human glucagon-like peptide-1 (GLP-1) produced in E. coli has a long-acting therapeutic effect in type 2 diabetic mice. PLoS One 12, e0181939. doi:10.1371/journal.pone.0181939
Yang, M., Yang, F., Chen, W., Liu, S., Qiu, L., and Chen, J. (2021). Bacteria-mediated cancer therapies: opportunities and challenges. Biomater. Sci. 9, 5732–5744. doi:10.1039/d1bm00634g
Yao, A. I., Fenton, T. A., Owsley, K., Seitzer, P., Larsen, D. J., Sit, H., et al. (2013). Promoter element arising from the fusion of standard BioBrick parts. ACS Synth. Biol. 2, 111–120. doi:10.1021/sb300114d
Yu, M., Hu, S., Tang, B., Yang, H., and Sun, D. (2023). Engineering Escherichia coli Nissle 1917 as a microbial chassis for therapeutic and industrial applications. Biotechnol. Adv. 67, 108202. doi:10.1016/j.biotechadv.2023.108202
Zeng, Z., Yu, R., Zuo, F., Zhang, B., Peng, D., Ma, H., et al. (2016). Heterologous expression and delivery of biologically active exendin-4 by lactobacillus paracasei L14. PLoS One 11, e0165130. doi:10.1371/journal.pone.0165130
Zhang, C., Liu, H., Li, X., Xu, F., and Li, Z. (2023a). Modularized synthetic biology enabled intelligent biosensors. Trends Biotechnol. 41, 1055–1065. doi:10.1016/j.tibtech.2023.03.005
Zhang, X., Ma, N., Ling, W., Pang, G., Sun, T., Liu, J., et al. (2023b). A micro-nano optogenetic system based on probiotics for in situ host metabolism regulation. Nano Res. 16, 2829–2839. doi:10.1007/s12274-022-4963-5
Zhang, X. E., Liu, C., Dai, J., Yuan, Y., Gao, C., Feng, Y., et al. (2023c). Enabling technology and core theory of synthetic biology. Sci. China Life Sci. 66, 1742–1785. doi:10.1007/s11427-022-2214-2
Zheng, J. H., Nguyen, V. H., Jiang, S. N., Park, S. H., Tan, W., Hong, S. H., et al. (2017). Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 9, eaak9537. doi:10.1126/scitranslmed.aak9537
Zhou, J., Li, M., Chen, Q., Li, X., Chen, L., Dong, Z., et al. (2022). Programmable probiotics modulate inflammation and gut microbiota for inflammatory bowel disease treatment after effective oral delivery. Nat. Commun. 13, 3432. doi:10.1038/s41467-022-31171-0
Zhou, T., Damsky, W., Weizman, O. E., Mcgeary, M. K., Hartmann, K. P., Rosen, C. E., et al. (2020). IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 583, 609–614. doi:10.1038/s41586-020-2422-6
Zou, Z. P., Du, Y., Fang, T. T., Zhou, Y., and Ye, B. C. (2023). Biomarker-responsive engineered probiotic diagnoses, records, and ameliorates inflammatory bowel disease in mice. Cell Host Microbe 31, 199–212.e5. doi:10.1016/j.chom.2022.12.004
Keywords: intelligent diagnosis and treatment, genetic engineering, engineering bacteria, biosensor system, synthetic biology
Citation: Zhao N, Chen J, Shi J, Gao Y, Li L and Dong L (2024) Research and application of intelligent diagnosis and treatment engineering bacteria. Front. Bioeng. Biotechnol. 12:1524376. doi: 10.3389/fbioe.2024.1524376
Received: 07 November 2024; Accepted: 09 December 2024;
Published: 18 December 2024.
Edited by:
Shashank Gupta, Norwegian University of Life Sciences, NorwayReviewed by:
Ivana Radojević, University of Kragujevac, SerbiaNidhin Thomas, Los Alamos National Laboratory (DOE), United States
Xu Yan, Tsinghua University, China
Copyright © 2024 Zhao, Chen, Shi, Gao, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Na Zhao, cmVtYXJuYV81MjVAMTYzLmNvbQ==
 Na Zhao1*
Na Zhao1* Junwei Chen
Junwei Chen