
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol. , 16 August 2024
Sec. Synthetic Biology
Volume 12 - 2024 | https://doi.org/10.3389/fbioe.2024.1423935
This article is part of the Research Topic Microorganisms and Microbial Technologies for Industry and Environmental Protection View all 5 articles
Since their first industrial application in the acetone-butanol-ethanol (ABE) fermentation in the early 1900s, Clostridia have found large application in biomass biorefining. Overall, their fermentation products include organic acids (e.g., acetate, butyrate, lactate), short chain alcohols (e.g., ethanol, n-butanol, isobutanol), diols (e.g., 1,2-propanediol, 1,3-propanediol) and H2 which have several applications such as fuels, building block chemicals, solvents, food and cosmetic additives. Advantageously, several clostridial strains are able to use cheap feedstocks such as lignocellulosic biomass, food waste, glycerol or C1-gases (CO2, CO) which confer them additional potential as key players for the development of processes less dependent from fossil fuels and with reduced greenhouse gas emissions. The present review aims to provide a survey of research progress aimed at developing Clostridium-mediated biomass fermentation processes, especially as regards strain improvement by metabolic engineering.
Clostridia include a large group of anaerobic gram-positive bacteria which have found large application in biomass biorefining (Cruz-Morales et al., 2019; Yang et al., 2022). Actually, the ABE (that stands for acetone, n-butanol and ethanol, in 3:6:1 ratio) fermentation of starch or sugar by Clostridium acetobutylicum was one of the largest fermentation industries until the 1960s when it was essentially replaced by cheaper oil-based technologies (Jones and Woods, 1986; Green, 2011; Jiang et al., 2015). Interest in biotechnological generation of n-butanol (hereinafter referred to as butanol) and other valuable chemicals has been revived in the last decades as a means to reduce dependence on fossil fuels, reduce CO2 emissions and ultimately improve the environmental sustainability of these productions (Azambuja and Goldbeck, 2020; Bao et al., 2020; Wen et al., 2020c; Ferreira et al., 2020; Nawab et al., 2020; Li et al., 2021). Within this framework, Clostridia are among the candidates with the greatest potential. A number of Clostridia can grow using inexpensive substrates such as lignocellulosic biomass (Mazzoli and Olson, 2020) or one carbon (C1) gases (CO, CO2) (Zhang et al., 2020b). Clostridium fermentation products include several compounds with important industrial application such as organic acids (e.g., lactate), short chain alcohols (e.g., ethanol, butanol, isobutanol, isopropanol), diols (e.g., 1,2-propanediol, 1,3-propanediol), acetone and H2 (Figures 1–4) (Altaras et al., 2001; Mazzoli, 2012; Wilkens et al., 2012; Mazzoli and Olson, 2020).

Figure 1. Fermentative pathways in Clostridia, production of C2-compounds and H2. Pyruvate decarboxylase (Pdc) has mainly been engineered in Clostridia (Tian et al., 2017a). However, a pdc gene has been identified on the pSOL1 megaplasmid of C. acetobutylicum (Lehmann and Lütke-Eversloh, 2011). Abbreviations: Acetyl-P, acetyl phosphate; Ack, acetate kinase; Adh, alcohol dehydrogenase; Aldh, aldehyde dehydrogenase; Aor, acetaldehyde ferredoxin oxidoreductase; cH2ase, confurcating hydrogenase; DHAP, dihydroxyacetone phosphate; EMP pathway, Embden Meyerhof Parnas pathway; Fd, ferredoxin; H2ase, hydrogenase; Pdc, pyruvate decarboxylase; Pfl, pyruvate formate lyase; Pfor, pyruvate ferredoxin oxidoreductase; Pta, phosphotransacetylase; WL pathway, Wood Ljungdahl pathway.
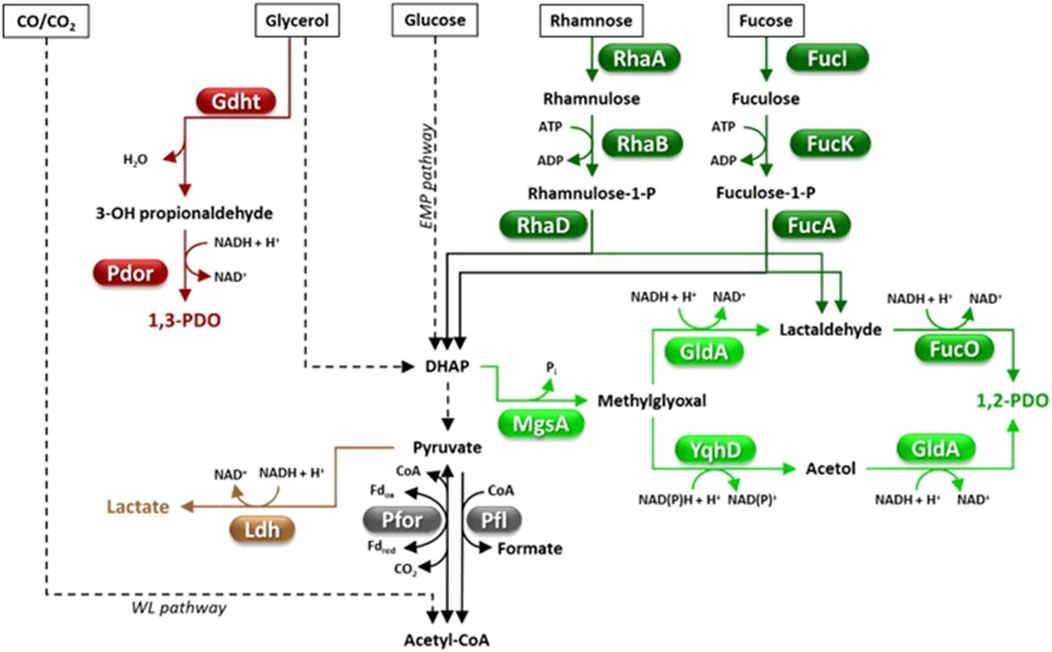
Figure 2. Fermentative pathways in Clostridia, production of C3-compounds. 1,2-PDO biosynthesis can occur through the methylglyoxal (light green) or the deoxyhexose pathway (dark green). Abbreviations: 1,2-PDO, 1,2-propanediol; 1,3-PDO, 1,3-propanediol; DHAP, dihydroxyacetone phosphate; EMP pathway, Embden Meyerhof Parnas pathway; Fd, ferredoxin; FucA, fuculose-1-phosphate aldolase; FucI, fucose isomerase; FucK, fuculokinase; FucO, 1,2-PDO oxidoreductase; Gdht, glycerol dehydratase; GldA, glycerol dehydrogenase; Ldh, lactate dehydrogenase; MgsA, methylglyoxal synthase; Pdor, 1,3-propanediol oxidoreductase; Pfl, pyruvate-formate lyase; Pfor, pyruvate ferredoxin oxidoreductase; RhaA, rhamnose isomerase; RhaB, rhamnulokinase; RhaD, rhamnulose-1-phosphate aldolase; WL pathway, Wood Ljungdahl pathway; YqhD, aldehyde reductase.
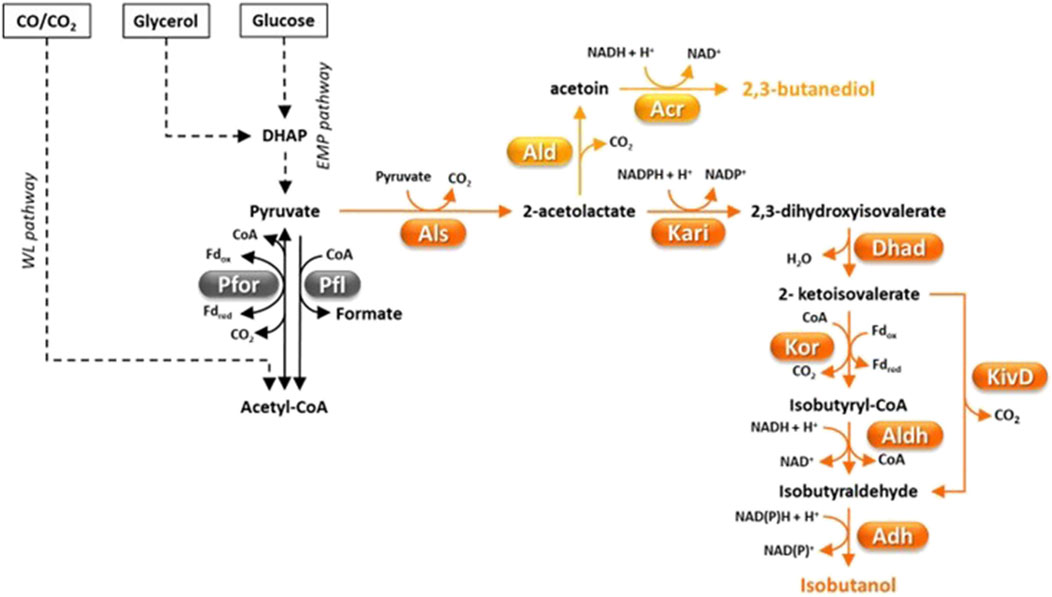
Figure 3. Fermentative pathways in Clostridia, production of C4-compounds. 2-ketoisovalerate decarboxylase (KivD) was not found in Clostridia but a Lactococcus lactis gene coding for it was engineered in C. thermocellum (Lin et al., 2015). Abbreviations: Acr, acetoin reductase; Adh, alcohol dehydrogenase; Ald, α-acetolactate decarboxylase; Aldh, aldehyde dehydrogenase; Als, α-acetolactate synthase; Dhad, dihydroxy acid dehydratase; DHAP, dyhydroxyacetone phosphate; Fd, ferredoxin; Kari, keto acid reductoisomerase; KivD, L. lactis 2-ketoisovalerate decarboxylase; Kor, ketoisovalerate ferrodoxin-dependent reductase; Pfl, pyruvate-formate lyase; Pfor, pyruvate:ferredoxin oxidoreductase.
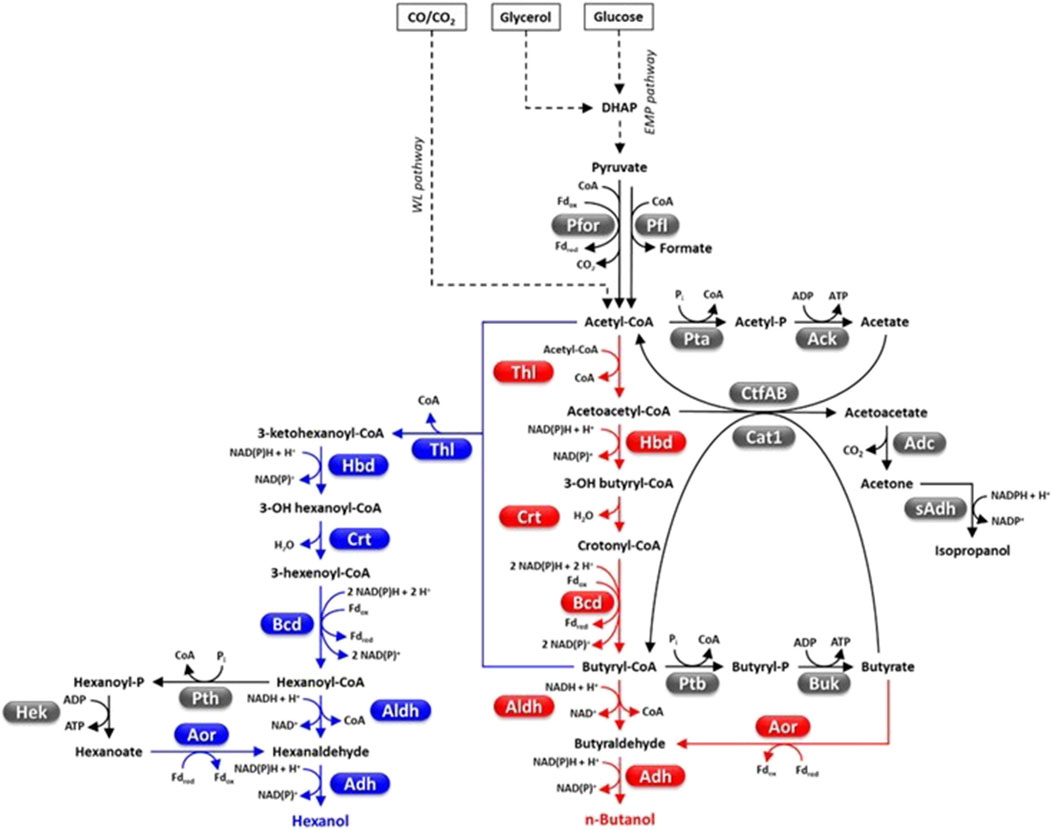
Figure 4. Fermentative pathways in Clostridia, production of C4-C6 alcohols. Abbreviations: Acetyl-P, acetyl phosphate; Ack, acetate kinase; Adc, acetoacetate decarboxylase; Adh, alcohol dehydrogenase; Aldh, aldehyde dehydrogenase; Aor, aldehyde ferredoxin oxidoreductase; Bcd, butyryl-CoA dehydrogenase; Buk, butyrate kinase; Butyryl-P, butyryl phosphate; Cat1, butyryl-CoA−acetate CoA transferase; Crt, crotonase; CtfAB, CoA transferase; DHAP, dyhydroxyacetone phosphate; Fd, ferredoxin; Hbd, 3-hydroxybutyryl-CoA dehydrogenase; Hek, hexanoate kinase; Pfl, pyruvate-formate lyase; Pfor, pyruvate ferredoxin oxidoreductase; Pta, phosphotransacetylase; Ptb, phosphotransbutyrylase; Pth, phosphotranshexanoylase; sAdh, secondary alcohol dehydrogenase; Thl, thiolase.
Development of universal systems for genetic manipulation of Clostridia (Minton et al., 2016; Yang et al., 2016; Wen et al., 2020d) has enabled significant enhancement of their natural potential as microbial cell factories (Charubin et al., 2018). This constantly expanding tool box also includes editing clostridial genome based on Clustered Regularly-Interspaced Short Palindromic Repeat (CRISPR)/cas (CRISPR associated) technology (Walker et al., 2020; Wilding-Steele et al., 2021; Husaini et al., 2023) or fine tuning gene expression by riboswitches (Marcano-Velazquez et al., 2019) or CRISPR interference (Ganguly et al., 2020). Consistently, significant improvement has been obtained as regards Clostridium production of a number of valuable chemicals and fuels such as ethanol (Tian et al., 2016; Hon et al., 2017), butanol (Re and Mazzoli, 2023), isobutanol (Higashide et al., 2011; Lin et al., 2015) and medium chain esters (Guo et al., 2023; Seo et al., 2023).
The very next sections will provide a survey of the main raw materials for Clostridium fermentation (Section 2) and the metabolic pathways involved (Section 3). The following sections will summarize research progress in production of some of the most valuable fuels and chemicals by Clostridia (Table 1), with special focus on strain improvement by metabolic engineering.
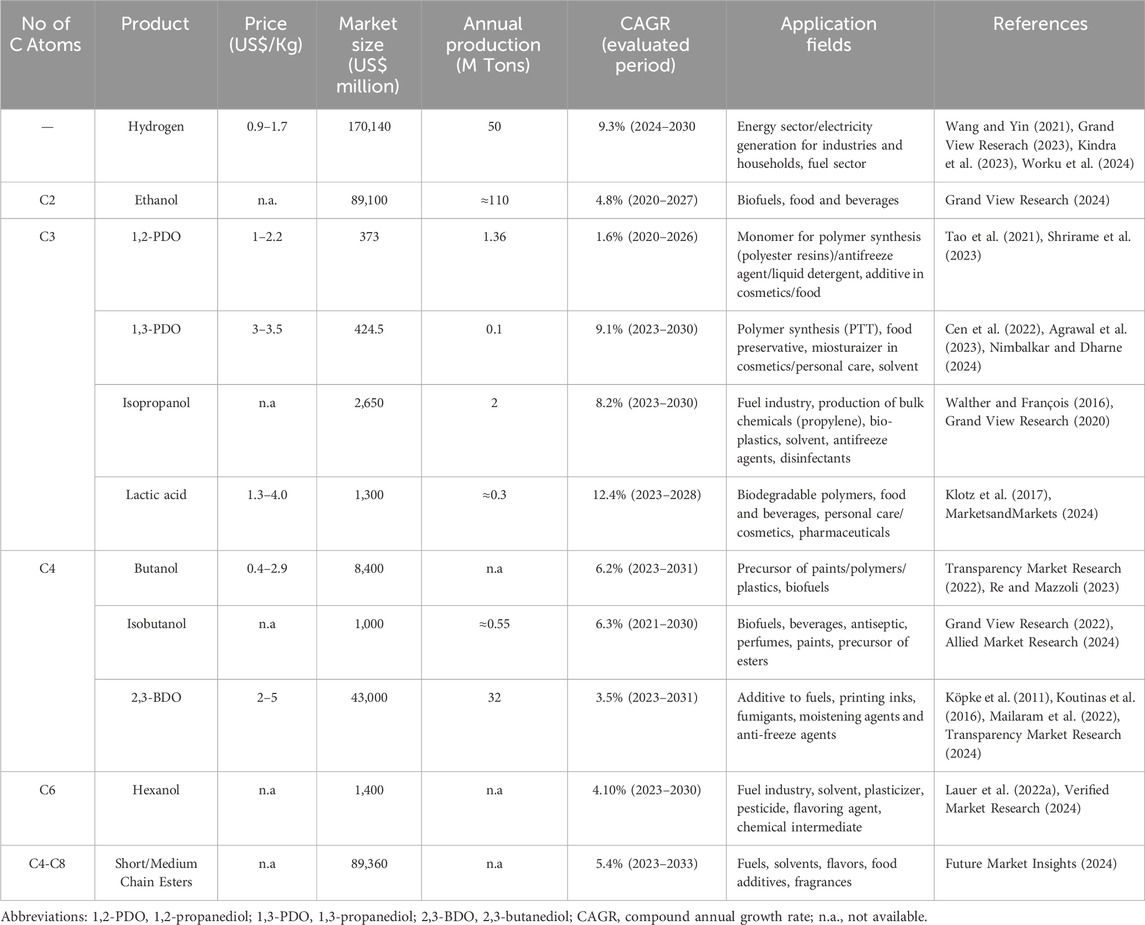
Table 1. Most recent estimation of economic parameters and applications of some top value chemicals produced by Clostridia.
Overall, Clostridia can ferment a wide range of substrates comprising soluble sugars (e.g., glucose, xylose, fructose, lactose, cellobiose), polysaccharides (e.g., starch, cellulose), glycerol and gaseous carbon compounds (CO, CO2) (Wang and Yin, 2021; Fernández-Blanco et al., 2023). Since feedstock expenditure may account for more than 70% of the total fermentation cost, the use of waste biomass is preferable to more expensive substrates such as pure sugars or edible crops, especially for producing bulk compounds such as fuels and platform chemicals (Gu et al., 2011; Abo et al., 2019; Rawoof et al., 2021). As a comparison, sugar costs about 460 US$/ton, pulp grade wood (a lignocellulosic biomass) can be calculated at 43–54 US$/ton of fermentable sugars and no cost can be referred to food waste (Nuss and Gardner, 2013; Gharehkhani et al., 2015; International Sugar Organization, 2019; Qureshi et al., 2020).
Carbohydrate-rich feedstocks include: i) lignocellulosic biomass; ii) (micro) algae biomass; iii) food waste; iv) municipal waste; v) agro-industrial effluents. Large availability and low cost of lignocellulosic feedstocks, such as wastes from agriculture (e.g., straws, stalks, wood fibers) and food processing (e.g., bagasse, mushroom compost) make it an ideal raw material for biorefining processes (Wang and Yin, 2021). However, this is offset by its complex composition (mainly consisting of cellulose, hemicellulose and lignin) and innate recalcitrance to biodegradation (Lynd, 2017). (Micro) algae biomass has gained considerable attention owing to its fast growth, no requirement of farmland, low demand for growth conditions and no or reduced lignin content which makes its hydrolysis and fermentation easier than plant biomass (Liu et al., 2012a; Ortigueira et al., 2015a; Ortigueira et al., 2015b; Fonseca et al., 2020). Globally, food waste (a very abundant biomass with high starch content) represents almost one-third of food produced for human consumption, and its disposal significantly contributes to greenhouse gas emission (Parthiba Karthikeyan et al., 2018; Qin et al., 2018; Zhang et al., 2020a; Su et al., 2022). Carbohydrate-rich raw materials also include some municipal wastes such as sewage sludge (Yin and Wang, 2016; Yin and Wang, 2019), paper waste (Liu et al., 2006) and garden wastes (leaves, branches) (Yang et al., 2019a). However, most of them have a complex composition (e.g., organics are mostly encapsulated in microbial cells in sewage sludge, paper and garden wastes are rich in lignocellulose) and generally need pretreatment prior to fermentation (Wang and Yin, 2018). Carbohydrate-rich agro-industrial effluents comprise sugarcane juice, molasses, cassava wastewater and cheese whey (Wang and Yin, 2021).
Glycerol is the main by-product (10% w/w) obtained by transesterification or saponification reactions aimed at producing oleochemicals such as biodiesel (Yang et al., 2012). The substantial increase in the biodiesel industry in the recent years has led to massive production of crude glycerol and drop in prices, hence, turned glycerol into a waste stream rather than a by-product (Ciriminna et al., 2014; Russmayer et al., 2019).
C1 gases (CO, CO2) are part of the greenhouse gases (mainly CO2) contributing to global warming and climate change (Canatoy et al., 2022). The use of these compounds as fermentation feedstocks can decrease their emission into the atmosphere by human activities. Among the industries that use fossil fuels for generating power and heat, steelmaking process emits about 50% of the carbon used as CO (Bengelsdorf et al., 2018). CO, CO2 (and H2) are also the major components of syn (thesis) gas which can be generated from natural gas, by gasification of coal, oil, biomass (e.g., agricultural and municipal waste) and by recycling used plastics (Köpke et al., 2010; Arslan et al., 2019; Zhang et al., 2020b). Syngas has extensively been used as a feedstock in the chemical industry, but this requires precise CO/H2 ratio and expensive gas purification from interfering contaminants (Köpke et al., 2010). Chemoautotrophic Clostridia are far more tolerant to such contaminants and already industrially used for ethanol production from these feedstocks (by companies such as Coskata, INEOS Bio, LanzaTech) (Köpke et al., 2010). However, low gas-liquid mass transfer rate (due to poor solubility of these gases in water) results in low cell densities and fermentation efficiency and is the main limit of this technology (Fernández-Blanco et al., 2023).
Clostridia include bacteria with heterotrophic and autotrophic metabolism. In saccharolytic strains, glucose is generally converted to pyruvate through the Embden Meyerhof Parnas (EMP) pathway, since most Clostridia lack the oxidative part of the pentose phosphate pathway (Crown et al., 2011; Koendjbiharie et al., 2020; Foulquier et al., 2022). It is worth noting that clostridial EMP pathway may contain a number of atypical reactions with respect to the traditional glycolysis which affect electron distribution among the different redox cofactors [pyridine cofactors, ferredoxin (Fd)], cellular pools of energy carriers (e.g., adenine/guanine nucleotides, pyrophosphate) and pathway thermodynamics (Iddar et al., 2002; Zhou et al., 2013; Scott et al., 2019; Jacobson et al., 2020). Some heterotrophic Clostridia can also ferment more reduced substrates than carbohydrates such as glycerol which is converted to dihydroxyacetone phosphate (DHAP) and enters the EMP pathway (Yoo et al., 2016; Agu et al., 2019; Sarma et al., 2019).
Pyruvate can be fermented to a variety of compounds such as organic acids (e.g., acetate, butyrate, formate, lactate), short chain alcohols (e.g., ethanol, n-butanol, isobutanol), acetone, CO2 and H2 (Figures 1–4) (Mazzoli, 2012; Mazzoli and Olson, 2020). Metabolic flux distribution among fermentative pathways significantly differs from one strain to another and is affected by the growth conditions (e.g., the kind and amount of carbon sources, agitation, H2 partial pressure, pH, bioreactor operation mode) (Łukajtis et al., 2018; Yang et al., 2020; Wang and Yin, 2021; Fernández-Blanco et al., 2023; Julkipli et al., 2023). A major regulator of carbon flux distribution is the redox-responsive protein Rex (Wietzke and Bahl, 2012; Schwarz et al., 2017). Rex can affect gene transcription in response to changes of intracellular NADH/NAD+ ratio (Ravcheev et al., 2012) and is involved in modulating central carbon metabolism, solvent and organic acid production, H2 generation, tolerance to oxidative stress, biofilm formation, and sulfate and nitrate reduction (Hu et al., 2016; Sander et al., 2019). Furthermore, a network of enzymes known as ferredoxin:NAD oxidoreductases (Fnor) catalyze re-distribution of electrons deriving from substrate oxidation among redox cofactors (ferredoxin, NAD, NADP) and, finally, to the fermentation end-products (Mazzoli and Olson, 2020).
Some Clostridia can grow chemoautotrophically using CO and/or CO2 as the carbon source(s) (Liew et al., 2017; Zhang et al., 2020b) which are reduced to acetyl-CoA through the Wood–Ljungdahl (WL) pathway (Müller, 2003; Schuchmann and Müller, 2014). If CO2 is used as the sole carbon substrate, H2 is required as the reductant. The WL pathway requires eight reducing equivalents and one ATP. Energy is provided by specialized version of Fnor which is proton translocating reduced ferredoxin:NAD+ oxidoreductase (Rnf) and ATP synthase (which uses the proton gradient generated by Rnf for ATP synthesis). In most Clostridia growing autotrophically, acetyl-CoA is mainly converted to acetic acid but some strains such as C. autoethanogenum, C. ragsdalei, C. ljungdahlii and C. carboxidivorans can also produce other chemicals such as ethanol, butyrate, butanol, 2,3-butanediol (2,3-BDO), hexanoate, hexanol and lactate (Köpke et al., 2011; Arslan et al., 2019; Arslan et al., 2022) (Figures 1–4). In the latter strains, the production of solvents (e.g., ethanol) through gas fermentation generally occurs in two steps. First, CO/CO2 are converted to acids (usually acetic acid) in a step called acetogenesis, then the accumulated acids are reduced to alcohols (solventogenesis) (Arslan et al., 2019; Arslan et al., 2022). Biosynthesis of lactate and 2,3-BDO occurs through pyruvate formation (Figures 2, 3). In fact, in autotrophic Clostridia, pyruvate ferredoxin oxidoreductase (Pfor) can also catalyze acetyl-CoA reductive carboxylation to pyruvate (acetyl-CoA + CO2 + Fdred → pyruvate + CoA + Fdox) (Figures 2, 3), a reaction that is coupled to CO oxidation by CO dehydrogenase/acetyl-CoA synthase complex (Codh/Acs) (Furdui and Ragsdale, 2000).
Hydrogen gas (H2) is an optimal energy carrier featuring high energy content (122 kJ/g) and clean combustion product (Table 1) (Akhlaghi and Najafpour-Darzi, 2020). Biological production of H2 has gained attention over traditional technologies (e.g., steam reforming of CH4, coal gasification) (Valle et al., 2019; Akhlaghi and Najafpour-Darzi, 2020) because it does not rely upon usage of fossil fuels and has reduced CO2 emissions (Lepage et al., 2021). However, current technologies for biological production of H2 need to improve their yield and cost competitiveness (Lepage et al., 2021; Nirmala et al., 2023).
Bio-H2 production by the so called dark processes, that is anaerobic fermentation of organic compounds by a number of heterotrophic microbes (mainly bacteria), is generally considered more effective than light-driven processes (i.e., direct and indirect biophotolysis, photofermentation by means of photosynthetic microorganisms) (Das and Veziroglu, 2008; Mudhoo et al., 2011; Arizzi et al., 2021; Cao et al., 2022). Among microorganisms catalyzing dark fermentation, obligate anaerobes such as Clostridium spp. feature 2-fold higher maximum theoretical H2 yield (i.e., 4 mol/mol hexose) compared to facultative anaerobes (e.g., Escherichia coli, Enterobacter sp.) (Wang et al., 2011a). Clostridial genomes generally encode multiple hydrogenases (H2ases) likely involved in different functions (e.g., redox balancing, derivation of energy from H2 oxidation, proton respiration and/or proton-gradient build-up) (Calusinska et al., 2010; Arizzi et al., 2021). These include monomeric FeFe H2ases that catalyze proton reduction to H2 by oxidation of reduced ferredoxin or flavodoxin (Demuez et al., 2007; Flamholz et al., 2012; Therien et al., 2017; Morra, 2022) and multimeric electron-confurcating H2ases that catalyze reduction of protons to H2 via the oxidation of reduced ferredoxin and NAD(P)H (Figure 1) (Wang et al., 2013; Latifi et al., 2019). In Clostridia, reducing equivalents for proton reduction by H2ases mainly derive from glyceraldehyde-3-phosphate (GAP) oxidation by GAP dehydrogenase (Gapdh) and pyruvate oxidation by Pfor (Figure 1).
Metabolic engineering strategies aimed to increase H2 production in Clostridia have focused on different targets that include: i) overexpression of (native and/or heterologous) H2-producing enzymes and/or downregulation of uptake H2ases (Nakayama et al., 2008; Cha et al., 2016; Sarma et al., 2019; Son et al., 2021); ii) impairment of metabolic pathways competing for reducing equivalents (e.g., production of ethanol, butyrate, formate, lactate) (Wang et al., 2011a; Jiang et al., 2011; Lo et al., 2015; Rydzak et al., 2015); iii) improvement of substrate catabolism (Sarma et al., 2019; Son et al., 2021; Kim et al., 2023); iv) optimization of electron/redox metabolism (Lo et al., 2017; Nguyen et al., 2018; Foulquier et al., 2022). It is worth remembering that maximum H2 yield can be obtained when sugars are fermented to acetate (glucose + 4 ADP + 4 Pi → 2 acetate + 2 CO2 + 4 ATP + 2 H2O + 4 H2), while it is lower when more reduced products (e.g., propionate, butyrate, lactate, ethanol) are accumulated (Wang et al., 2011a; Wang et al., 2021b; Ortigueira et al., 2015a; Islam et al., 2015). Studies aiming at improving monosaccharide catabolism include diversion of glucose towards the pentose phosphate (PP) pathway (Son et al., 2021). In fact, glucose fermentation through the PP pathway could increase the maximum theoretical H2 yield by 33% (1 glucose + 3.33 ADP + 3.33 Pi → 1.67 acetate + 2.67 CO2 + 3.33 ATP + 5.33 H2) (Singh et al., 2019).
So far, most of these investigations achieved limited H2 yield enhancement (generally comprised between 15% and 80%) with H2 yield still lower than 2 mol/mol hexose in most engineered strains (Mazzoli et al., 2024). One likely reason is that only one gene has generally been down- or upregulated in each engineered strain. For instance, in Clostridia multiple hydrogenase-encoding genes are present, with different roles and expression levels, and the balance of their activity can be a key to improved performances (Land et al., 2020; Arizzi et al., 2021; Morra, 2022; Fasano et al., 2024). Combination of multiple advantageous metabolic modifications (e.g., overexpression of evolving H2ases, elimination of alternative pathways, optimization of sugar metabolism) in one strain seems among the most obvious implementation. More extensive application of adaptive laboratory evolution could be an additional tool that avoid or implement complex rational genetic engineering to tackle all these issues and enhance H2 production of microorganisms (Tunca et al., 2023). Along with strain optimization, the use of improved fermentation conditions (e.g., pH, temperature, substrate concentration, H2 partial pressure), mode (e.g., continuous fermentation) and bioreactor configuration is pivotal (Cai et al., 2013; Wang et al., 2014; Łukajtis et al., 2018; Son et al., 2021; Julkipli et al., 2023). From this standpoint, the improvement of systems for reducing H2 partial pressure in the bioreactor (e.g., stirring the growth medium, sparging the growth medium with inert gas, removing gas by a vacuum pump, selectively removing H2 by active membranes) is essential for overcoming thermodynamic barriers of biological H2 production. The ΔrG’m (i.e., the change in Gibbs free energy associated with a metabolic reaction/pathway when all the reactants have a concentration of 1 mM) for the production of H2 by oxidation of glucose to acetate (through the EMP pathway) can vary from −205.1 to 0.2 kJ/mol for dissolved H2 concentration ranging from 10–9 M–1 M (Flamholz et al., 2012). Promising results have also been reported through the development of processes based on syntrophic microbial chains, such as two-step fermentation (e.g., dark fermentation + photofermentation) (Ramprakash et al., 2022) or co-cultures (e.g., H2 producing bacteria + bacteria able to perform anaerobic respiration) (Zhang et al., 2023b) providing complete oxidation of Clostridium fermentation by-products. Improvement of H2 yield obtained by combining dark- and photo-fermentation was significantly higher (≈100–200%) (Ramprakash et al., 2022) than those reported for co-cultures (≈30–45%) (Zhang et al., 2023b). The efficiency of syntrophic co-cultures could be enhanced within the framework of microbial electrolysis cells in which additional electric voltage is used to increase H2 yield from organic compound oxidation (Bora et al., 2022).
In summary, the high potential of Clostridia for H2 production has been so far improved to a limited extent by a number of studies employing metabolic engineering. More intense efforts in this direction are desirable. Process optimization (e.g., reduction of H2 partial pressure, synthrophic microbial chains for complete substrate oxidation) is also necessary for achieving suitable efficiency for industrial application.
Ethanol is the most broadly produced biofuel today, with over 16 billion gallons produced in the United States alone in 2019 (Table 1) (Ethanolrfa, 2019). It is typically blended with gasoline for use in spark-ignited engines (10% in the United States, 27% in Brazil), but can also be catalytically upgraded to longer-chain fuel molecules, such as jet fuel and gasoline (Hannon et al., 2020; Mazzoli and Olson, 2020). 3.1 EJ/year of ethanol is currently obtained from sugar cane (30%) and cereals (50%) (Lynd et al., 2017). However, projected future global demand of bioethanol and the need for more massive reduction of greenhouse gas emission will require a significant contribution by bioethanol derived from other feedstocks (e.g., lignocellulose, CO2) (Wang et al., 2015; Lynd et al., 2017). Today, bioethanol is primarily produced by fermentation of mono- or disaccharides using Saccharomyces cerevisiae or Zymomonas mobilis. Nonetheless, these organisms cannot directly grow on cheap feedstocks such as lignocellulose or syngas (Himmel et al., 2007). Substantial research has been dedicated to develop recombinant cellulolytic yeast or Z. mobilis strains, yet the maximum cellulosic ethanol titer obtained through direct biomass fermentation by these strains (≤10 g/L) is far lower than what is generally considered as necessary for commercial application (titer = 40 g/L, yield = 1.8 mol/mol hexose) (Dien et al., 2003; Kojima et al., 2013; Todhanakasem et al., 2019; Anandharaj et al., 2020). Clostridium thermocellum and Thermoanaerobacterium saccharolyticum have been important alternative microbial paradigms for one-step production of ethanol from cellulose or hemicellulose, respectively (Figure 1) (Mazzoli and Olson, 2020). On the other hand, acetogenic Clostridia such as C. ljungdahlii (Phillips et al., 1993) and C. ragsdalei (Sun et al., 2018) have been investigated as promising ethanol producers through C1-gas fermentation.
C. thermocellum naturally produces low ethanol yield (typically 12%–34% of the theoretical maximum, i.e., 2 mol/mol hexose) (Olson et al., 2015), but extensive metabolic engineering efforts have significantly increased this efficiency (Mazzoli and Olson, 2020). These studies have included: i) elimination of pathways that compete for carbon and electron flux (namely, production of acetate, lactate, formate, and H2) (Biswas et al., 2015; Papanek et al., 2015; Rydzak et al., 2015; Holwerda et al., 2020); ii) improvement of electron metabolism (Hon et al., 2017; Hon et al., 2018; Lo et al., 2017); iii) improvement of glycolytic flux (Deng et al., 2013; Zhou et al., 2013; Tian et al., 2017b; Hon et al., 2022); iv) overexpression of autologous and heterologous genes for ethanol production (e.g., alcohol dehydrogenase, aldehyde dehydrogenase, pyruvate decarboxylase) (Tian et al., 2017a; Zheng et al., 2017; Hon et al., 2018). Eventually, these efforts led to strains which can produce ethanol at titers of 25–30 g/L and high yields (75%–80% of the theoretical maximum) (Olson et al., 2023). Still, this is too low for commercial application. In particular, ethanol titer in these strains seems to be limited by ethanol tolerance. The latter has been improved in the wild type C. thermocellum to 50–80 g/L (Williams et al., 2007; Brown et al., 2011; Shao et al., 2011). However, a recent investigation has highlighted a tradeoff between ethanol tolerance and production, namely, C. thermocellum strains with enhanced ethanol tolerance generally show decreased ethanol production (Olson et al., 2023). The main reason for C. thermocellum inhibition by ethanol accumulation seems to be related to redox, i.e., NADH/NAD+ ratio, imbalance (which affect alcohol dehydrogenase and GAP dehydrogenase activities) but other yet elusive mechanisms could be involved.
T. saccharolyticum is a thermophilic anaerobic bacterium which can ferment xylan (the main polymer in hemicellulose) and all the most abundant monosaccharides of plant biomass (e.g., glucose, mannose, xylose, galactose, and arabinose) although it cannot use cellulose (Herring et al., 2016). The wild-type T. saccharolyticum accumulates ethanol as its main fermentation product (yield = 1 mol/mol xylose) but also significant amounts of acetate and lactate (Shaw et al., 2008). Disruption of genes involved in production of acetate (phosphotransacetylase, pta, and acetate kinase, ack) and adaptation to high xylose concentration resulted in homoethanologenic phenotype with a maximum ethanol titer = 37 g/L (Shaw et al., 2008). In a following study, higher performing T. saccharolyticum strains were obtained by introducing additional genetic modifications such as the expression of a urease and the disruption of an operon involved in exopolysaccharide biosynthesis (Herring et al., 2016). These strains were able to produce up to 70 g/L of ethanol from a mixture of cellobiose and maltodextrin. However, much lower ethanol titer (26 g/L) was obtained by fermentation of a hemicellulose extract (Herring et al., 2016). Interestingly, co-culture of C. thermocellum and T. saccharolyticum (or other hemicellulolytic microbes such as (Thermoanaerobacterium thermosaccharolyticum and Herbinix spp.) has been performed which enabled one-pot fermentation of both the cellulose and hemicellulose components of plant biomass (He et al., 2011; Jiang et al., 2013; Froese et al., 2019; Beri et al., 2020).
As mentioned above, acetogenic Clostridia have been studied for ethanol production via C1-gas fermentation. This generally occurs in two steps, that is acetogenesis precedes ethanol formation (solventogenesis) (Fernández-Blanco et al., 2023). Gas fermenting Clostridia can produce ethanol through two pathways that is: i) reduction of acetyl-CoA by aldehyde/alcohol dehydrogenase or; ii) reduction of acetate to acetaldehyde by acetaldehyde ferredoxin oxidoreductase (Aor) and then reduction of acetaldehyde by alcohol dehydrogenase (Figure 1) (Zhang et al., 2020b). Ethanol production is triggered by stress conditions limiting cell growth such as acidic pH and/or lack of nutrients (Daniell et al., 2012; Fernández-Naveira et al., 2016; Al-Shorgani et al., 2018). The highest ethanol titer reported through C1-gas fermentation (48 g/L) was achieved in 1993 by Philips and co-workers by using a C. ljungdahlii strain growing on syngas (Phillips et al., 1993). This result was obtained after 560 h fermentation in a stirred tank bioreactor with cell recirculation, using an optimized growth medium and high gas-liquid mass transfer. This study indicated that a medium with pH range of 4.0–4.5 promoted ethanol accumulation. However, far lower ethanol titers were obtained in more recent investigations (Fernández-Blanco et al., 2023). As far as we know, the highest ethanol titer (16.25 g/L) reported by the latest studies was obtained through syngas fermentation by C. ragsdalei in a medium supplemented with poultry litter biochar (Sun et al., 2018). As regards strain improvement by metabolic engineering, significant increase of ethanol production (50%–180%) by C. autoethanogenum or C. carboxidivorans was obtained by either inactivation (Liew et al., 2017) or overexpression (Lu et al., 2019) of adhE genes (encoding bifunctional alcohol/aldehyde dehydrogenase). However, the maximum ethanol titers obtained through autotrophic growth of these strains were ≤ 3 g/L (Liew et al., 2017; Cheng et al., 2019).
In conclusion, Clostridia provide promising paradigms for production of ethanol from low-cost feedstocks, such as lignocellulose and C1-gas. Research progress is at a more advanced stage as regards lignocellulose fermentation (also in reason of the higher number of studies), while investigation of C1-gas fermentation still needs substantial efforts.
1,2-propanediol (1,2-PDO) is a bulk chemical with applications in antifreeze agents, cosmetics, nutrition, medicine and polyester resins (Table 1) (Siebert and Wendisch, 2015). Currently, 1,2-PDO is mostly produced through chemical hydration of fossil-derived propylene which leads to a racemic mixture of R- and S-1,2-PDO (Tao et al., 2021). Fermentative production of 1,2-PDO benefits from the use of renewable feedstocks and can generate pure 1,2-PDO stereoisomers (Tao et al., 2021). However, native bacterial producers of 1,2-PDO (Prevotella, Salmonella, Klebsiella, Corynebacterium, Clostridium) show low yield and productivity, hindering their application in industrial processes (Turner and Robertson, 1979; Badia et al., 1985; Cameron and Cooney, 1986; Siebert and Wendisch, 2015).
Natural 1,2-PDO producing Clostridia include Clostridium sp. AK1 (Ingvadottir et al., 2018), C. beijerinckii DSM 6423 (Diallo et al., 2019), C. phytofermentans (Petit et al., 2013), C. sphenoides and C. thermosaccharolyticum (Tran-Din and Gottschalk, 1985; Cameron and Cooney, 1986; Altaras et al., 2001). Clostridia can produce 1,2-PDO through two alternative metabolic pathways, the deoxyhexose (DXH) or the methylglyoxal (MGL) pathway (Figure 2) (Tao et al., 2021). The main limit of the DHX pathway is that it requires expensive feedstocks such as L-fucose and L-rhamnose (Figure 2) (Tran-Din and Gottschalk, 1985; Cameron and Cooney, 1986). The MGL route allows the conversion of a much larger panel of cheaper sugars (e.g., glucose, fructose, mannose, galactose, xylose, arabinose, lactose or cellobiose), however, suffers from possible accumulation of toxic intermediates (e.g., methylglyoxal) which limit bacterial growth and 1,2-PDO production (Tao et al., 2021). Efforts have been made to find cheaper natural sources of L-rhamnose, such as the macroalgae Ulva lactuca which contains L-rhamnose and D-glucose as the major sugars (Diallo et al., 2019). C. beijerinckii DSM 6423 was able to accumulate up to 5.96 g/L 1,2-PDO by fermenting (DHX pathway) a U. lactuca hydrolysate with a yield of 0.41 g/g of rhamnose (Diallo et al., 2019). C. thermosaccharolyticum HG-8 is equipped with the MGL pathway and is currently the highest performing 1,2-PDO producer among Clostridia (Sánchez-Riera et al., 1987). This strain was reported to produce 1,2-PDO from a variety of feedstocks (e.g., glucose, xylose, mannose, cellobiose or whey permeate) up to a titer = 9.05 g/L (yield = 0.20 g/g hexose) (Table 2) (Sánchez-Riera et al., 1987). As far as we know, no study has attempted to increase 1,2-PDO production in Clostridia by metabolic engineering strategies which, to date, have mainly targeted E. coli (Tao et al., 2021).
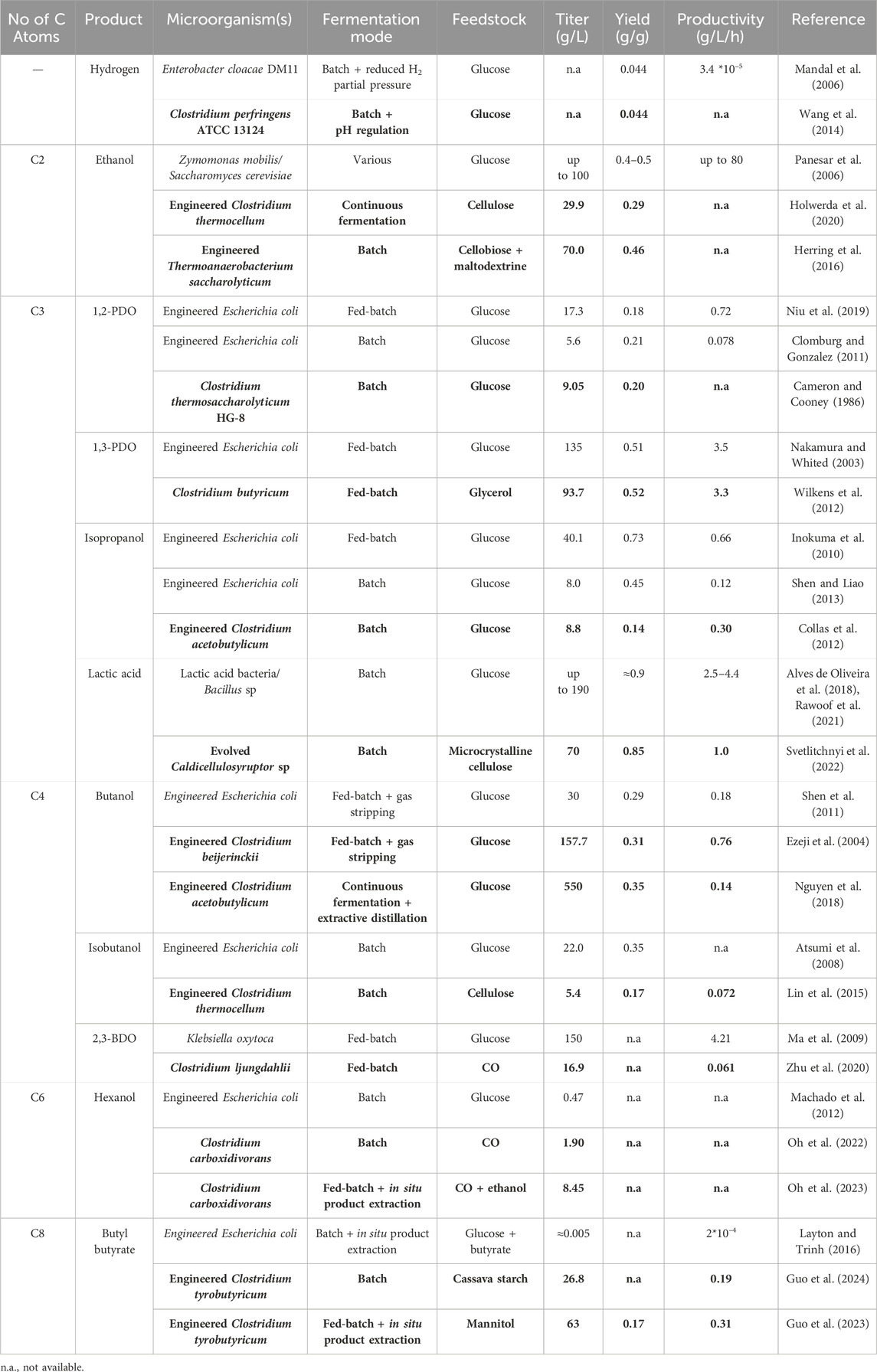
Table 2. Comparison between production of top value chemicals by Clostridia (bold) and other high performing microorganisms.
1,3-propanediol (1,3-PDO) has various industrial uses in cosmetics (e.g., solvent, moisturizer), food (e.g., preservative) and polymer synthesis (e.g., polytrimethylene terephthalate) (Table 1). Since 2003, 1,3-PDO has mainly been produced via fermentation of corn-derived glucose by an engineered E. coli strain through a collaboration of DuPont, Genencor and Tate & Lyle group (Agrawal et al., 2023). Advantageously, a number of microorganisms including some Clostridia can naturally produce 1,3-PDO through glycerol fermentation (Figure 2) (Nimbalkar and Dharne, 2024). When glycerol is the only carbon source, a part of it is dehydrated and reduced to 1,3-PDO, however, another part is oxidized to DHAP which enters the glycolytic flux, thus providing the reducing power for generating 1,3-PDO (Figure 2). Glycerol oxidative metabolism leads to accumulation of by-products such as organic acids (mainly acetic, butyric, and lactic), alcohols (e.g., ethanol, butanol, 1,2-propanediol, 2,3-butanediol), H2 and CO2, depending on the species and culture conditions (Nimbalkar and Dharne, 2024). The most studied natural producers of 1,3-PDO are Klebsiella pneumoniae and Clostridium butyricum owing to their high glycerol consumption rate and 1,3-PDO production (titer up to 100 g/L, yield ≈ 0.5 g/g, productivity ≈ 2 g/L/h) (Wilkens et al., 2012; Zhu et al., 2022). Natural 1,3-PDO producing-Clostridia offer many advantages with respect to K. pneumoniae since they are non-pathogenic, produce lower amounts of by-products and require cheaper fermentation systems (Yazdani and Gonzalez, 2007). For instance, C. butyricum and other Clostridium strains biosynthesize a vitamin B12-independent glycerol dehydratase (GDHt, glycerol → 3-hydroxypropionaldehyde + H2O) (Figure 2), hence, do not require exogenous supplementation of this expensive vitamin in the growth medium (Raynaud et al., 2003). Yet, optimization of 1,3-PDO production by C. butyricum is hindered by low tolerance to product accumulation, large amounts of by-products (e.g., butyric acid, acetic acid) and lack of efficient gene modification tools (Yang et al., 2019b).
This has prompted research on other native or engineered 1,3-PDO producing Clostridia such as C. acetobutylicum (González-Pajuelo et al., 2005), C. beijerinckii (Wischral et al., 2016; Schoch et al., 2023), C. diolis (Otte et al., 2009; Li et al., 2020a) or C. perfringens (Guo et al., 2017) for which maximum 1,3-PDO titers range between 40-84 g/L. Particularly noteworthy are the results obtained with an engineered C. acetobutylicum (González-Pajuelo et al., 2005). In fact, the C. acetobutylicum ATCC824 DG1 mutant (unable to produce solvents and sporulate) overexpressing the genes encoding C. butyricum GDHt and 1,3-propanediol oxidoreductase (Pdor, 3-hydroxypropionaldehyde + NADH + H+ → 1,3-PDO + NAD+) (Figure 2) was able to ferment glycerol and produce 1,3-PDO as the major product in fed-batch fermentation (titer = 84 g/L 1,3-PDO, yield = 0.53 g/g, maximum productivity = 1.70 g/L/h). Interestingly, a two-step fermentation involving C. acetobutylicum was developed which was able to convert glucose or molasses to 1,3-PDO (Mendes et al., 2011). In the first fermentation, the recombinant S. cerevisiae HC42 (adapted to grow on high glucose concentration) converted sugars into glycerol, which was subsequently fermented to 1,3-PDO by a 1,3-PDO hyperproducing C. acetobutylicum DG1 (final titer = 25.5 g/L, yield = 0.56 g/g of glycerol and 0.24 g/g of glucose, maximum productivity = 0.16 g/L/h). This study is paradigmatic of alternative strategies for extending the panel of feedstocks for fermentative production of 1,3-PDO by Clostridia beyond glycerol.
Lactic acid (LA) is among the chemicals with the largest worldwide industrial demand by sectors that include the food, cosmetic, pharmaceutical industry and the synthesis of biodegradable solvents and plastics (e.g., polylactide) (Table 1) (Abdel-Rahman et al., 2011; Abdel-Rahman et al., 2013; Alves de Oliveira et al., 2018). About 90% of global LA production is obtained by microbial fermentation producing pure L- or D-LA enantiomer (chemical synthesis generates a racemic mixture) (Abdel-Rahman et al., 2013; Alves de Oliveira et al., 2018) which is required for plastic polymer synthesis (Abdel-Rahman and Sonomoto, 2016), and food and pharmaceutical applications (Jem et al., 2010). Currently, industrial production of LA mainly rely on lactic acid bacteria (LAB) (Sauer et al., 2008) based on their high LA yield, productivity and GRAS (generally regarded as safe) status (Abdel-Rahman et al., 2013; Rawoof et al., 2021). However, LAB also have drawbacks such as limited acid tolerance, requirement for complex nutrients (amino acids, nucleotides, vitamins) and inability to directly ferment cheap feedstocks (e.g., lignocellulose). This has stimulated research on alternative microbial platforms for LA production (Poudel et al., 2016; Tarraran and Mazzoli, 2018). Within this perspective, a major advantage of using Clostridia for LA production is the ability of (hemi) cellulolytic strains to directly ferment lignocellulosic biomass. One-step fermentation of lignocellulose to LA could dramatically decrease the cost of LA production and its current dependence from food crops (Nuss and Gardner, 2013; Gharehkhani et al., 2015; International Sugar Organization, 2019; Qureshi et al., 2020).
So far, the number of studies aimed at enhancing LA production in natural (hemi) cellulolytic microorganisms is limited. Research on bacteria such as C. thermocellum (Lo et al., 2015; Mazzoli et al., 2020), C. bescii (Williams-Rhaesa et al., 2018), Thermoanaerobacter mathranii (Yao and Mikkelsen, 2010), T. saccharolyticum (Zhou et al., 2015), Thermoanaerobacterium aotearoense (Yang et al., 2013) and Thermoanaerobacterium thermosaccharolyticum (Bhandiwad et al., 2013) has suggested valuable metabolic engineering strategies to increase LA accumulation in these strains. These include impairment of alternative fermentative pathways (e.g., production of H2, acetate, ethanol, formate) (Mazzoli, 2020), increasing the expression of lactate dehydrogenase (Ldh) (Figure 2) (Williams-Rhaesa et al., 2018; Mazzoli et al., 2020), or engineering the redox state of the cell (Ravcheev et al., 2012; Lo et al., 2017; Sander et al., 2019). Studies on C. thermocellum (Mazzoli et al., 2020), Caldicellulosiruptor saccharolyticus (Willquist and van Niel, 2010) and Thermoanaerobacter ethanolicus (Bryant, 1991) have indicated that the catalytic activity of their Ldh is modulated by a number of compounds such fructose-1,6-bisphosphate, nicotinamide cofactors (e.g., NADH, NAD+) and/or, energy carriers (e.g., ATP, PPi), which need to be taken into account in metabolic engineering strategies.
One of the main hurdles in engineering LA hyper-production in anaerobic (hemi) cellulolytic microbes is their limited acid tolerance (Yang et al., 2013; Mazzoli et al., 2022; Svetlitchnyi et al., 2022). Growth at regulated pH through base addition was essential for major increase of the LA production of a Thermoanaerobacterium aotearoense strain deficient in acetate production (Yang et al., 2013). This growth condition resulted in nearly homolactic fermentation (LA yield = 0.93 g/g glucose) with a final LA titer up to 47 g/L (Yang et al., 2013). As base addition is complicated and expensive at the industrial scale, use of strains with high acid tolerance is recommended (Singhvi et al., 2018). Ideally, a microbial host should tolerate pH conditions around the pKa(s) of the produced acid (typically in the range 3–5 for organic acids) (Skoog et al., 2018). So far, C. thermocellum and Caldicellulosyruptor sp. strains have been obtained by adaptive laboratory evolution which have increased tolerance to LA (Mazzoli et al., 2022; Svetlitchnyi et al., 2022). An evolved Caldicellulosyruptor sp. strain showed more than 10-fold increase in LA titer and was able to produce up to 70 g/L LA through batch fermentation of microcrystalline cellulose (Svetlitchnyi et al., 2022). This titer is comparable to that obtained through fermentation of cellulosic biomass hydrolysate by lactic acid bacteria (Rawoof et al., 2021). However, it should be noted that pH regulation of Caldicellulosyruptor sp. cultures was still necessary (Svetlitchnyi et al., 2022). In fact, acid stress depends on both the decrease of pH and the nature of the acid, i.e., more hydrophobic carboxylic acids generally are more toxic (Jarboe et al., 2013; Wilbanks and Trinh, 2017). Irrespective of the strategy used (random mutagenesis, strain evolution, rational engineering), no research could lower the acidic pH limit allowing a microorganism to grow beyond 0.5 pH unit such as in the case of the anaerobic cellulolytic bacteria Clostridium cellulovorans and Fibrobacter succinogenes (Wu et al., 2017; Wen et al., 2020a; Mazzoli, 2021). More intense effort in this direction is needed as regards LA hyper-producing cellulolytic Clostridia.
Among the C3 compounds considered here, production of 1,3-PDO by C. butyricum is the most established (Table 2), although the lack of efficient genetic tools hampers strain improvement (e.g., increase product tolerance, reduce by-products formation) and increase in process efficiency which has stimulated research on other clostridial strains (e.g., C. acetobutylicum). A few engineered clostridial strains have shown promising potential for direct fermentation of lignocellulose to lactic acid although their limited acid tolerance constitutes a major issue. As reported also for other microorganisms (Table 2), 1,2-PDO production by Clostridia is intrinsically hindered by the high cost of substrates or by accumulation of toxic intermediates which need to be addressed (e.g., by metabolic engineering).
Butanol has high potential as a drop-in fuel (namely, it can be fed to spark ignited engines without any modification) which adds to its applications as paint, polymer and plastic precursor (Table 1) (Gu et al., 2011; Campos-Fernández et al., 2012; Jiang et al., 2015). ABE fermentation is still the most economically viable route for biobutanol production, yet, it is affected by several drawbacks especially under industrial conditions: i) high cost of feedstock and substrate inhibition; ii) low butanol titer (≤20 g/L), yield (≈0.33 g/g), and productivity (<0.5 g/L/h); iii) important formation of by-products which increase butanol purification costs; iv) poor understanding of Clostridium physiology (Green, 2011; Gu et al., 2011; Abo et al., 2019; Li et al., 2020c). Current research aimed to improve butanol production by native solventogenic Clostridia includes: i) enhancing butanol yield, titer and tolerance; ii) expanding fermentation substrates to low-cost feedstocks (e.g., food waste, lignocellulosic biomass and C1-gases) (Zhang et al., 2020a; Fernández-Blanco et al., 2023; Re and Mazzoli, 2023).
Four Clostridium species, C. acetobutylicum, C. beijerinckii, C. saccaroperbutylacetonicum and C. saccharoacetobutylicum, can biosynthesize significant butanol amounts (Figure 4) (Keis et al., 2001; Huang et al., 2010). The growth of these strains develops through three phases that is acidogenesis, solventogenesis and sporogenesis which is subjected to complex metabolic regulation whose molecular details are still poorly understood (Li et al., 2020c). The exponential phase is characterized by accumulation of acids (mainly acetic and butyric acid), ATP and reduced pyridine cofactors. Growth medium acidification and accumulation of ATP, NAD(P)H and possibly other metabolites (e.g., butyryl-phosphate, formic acid) contribute to shift the metabolism towards solvent (acetone, butanol, ethanol) production and is accompanied by acid re-assimilation (Zhao et al., 2005; Wang et al., 2011b; Li et al., 2020c). Biomass production, non-assimilation of acids, and accumulation of other carbohydrates contribute to lower the actual butanol yield with respect to the theoretical maximum (1 mol/mol of glucose, that is 0.41 g/g) (Li et al., 2020c).
Significant enhancement of butanol production by natural solventogenic Clostridia has been obtained by: i) strain improvement (by mutagenesis, metabolic engineering, adaptive laboratory evolution); ii) optimization of the growth medium; iii) optimization of the fermentation process (e.g., high cell density fermentation, use of in situ product recovery techniques) [extensively reviewed by Li et al. (2020c)]. Further improvement of bacterial strains through mutagenesis is hampered by the complex and unknown metabolic/phenotypic changes generated by random gene mutations (Palsson and Zengler, 2010). Rational metabolic engineering has targeted genes involved in: i) butanol biosynthetic pathway; ii) pathways competing for carbon and electrons; iii) butanol tolerance (illustrated in Section 4.4.1.4); iv) redox homeostasis; v) energy homeostasis; vi) regulation of acidogenesis-solventogenesis shift and solvent production (Wietzke and Bahl, 2012; Nguyen et al., 2018; Li et al., 2020c; Dai et al., 2021; Re and Mazzoli, 2023).
The acetyl-CoA-butanol production pathway is intrinsically hampered by its high NADH consumption (5 mol/mol butanol) (Figure 4) and is not very exergonic (∆rG’m = −50.1 ± 13.7 KJ/mol) (Flamholz et al., 2012). Metabolic engineering strategies have been used to enhance the cellular NADH levels by up-regulating NADH formation pathways [e.g., by overexpressing heterologous ferredoxin-NAD(P) oxidoreductase] (Qi et al., 2018) or reducing NADH consumption by the butanol pathway (Lee et al., 2012; Nguyen et al., 2018; Qi et al., 2018; Li et al., 2019; Li et al., 2020b). As regards the latter strategy, increased butanol flux in C. acetobutylicum was obtained by replacing NADH-dependent enzymes (e.g., 3-hydroxybutyryl-CoA dehydrogenase, Hbd; alcohol-aldehyde dehydrogenase, AdhE) with NADPH-dependent counterparts (Figure 4) (Lee et al., 2012; Nguyen et al., 2018). C. acetobutylicum thiolase (CaThl, 2 acetyl-CoA → acetoacetyl-CoA + CoA, ∆rG’m = 25.0 ± 1.7 kJ/mol) is among the most critical nodes of the butanol pathway. CaThl is subject to redox-switch, namely, oxidized cell conditions lead to enzyme inactivation and repression of butanol (and butyric acid) biosynthesis (Flamholz et al., 2012; Kim et al., 2015). Furthermore, CaThl is inhibited by low concentration of CoA (Nguyen et al., 2018). Engineering CaThl protein to alleviate feedback inhibition (Mann and Lütke-Eversloh, 2013) or avoid enzyme inactivation by oxidized conditions (Kim et al., 2015), or replacing CaThl with E. coli thiolase (AtoB) (that shows higher catalytic efficiency, lower sensitivity to CoA and is not redox-switch modulated) (Nguyen et al., 2018) increased Thl activity and/or butanol flux to varying degrees (18%–64%). Recently, early activation (that is during acidogenesis) of butanol production leading to increased titer has accidentally been obtained by trying to engineer a complete WL pathway in C. acetobutylicum (Jang et al., 2023).
The impairment of biosynthetic pathways for alternative fermentation products (e.g., acetate, acetone, butyrate, ethanol, lactate) (Figures 1–4) has generally decreased by-product accumulation and enhanced butanol yield (Lee et al., 2012; Nguyen et al., 2018) but sometimes generated undesired phenotypes (e.g., defective acid assimilation or growth rate) (Li et al., 2020c). Elimination of acetone production (a non-fuel solvent with high corrosivity) (dos Santos Vieira et al., 2019) has been reported to increase acetate (and ethanol) generation and decrease butanol accumulation (Jiang et al., 2009; Lehmann et al., 2012). Instead, overexpression of secondary-alcohol dehydrogenases (sAdh) (coupled with upregulation of acetone pathway and disruption of butyrate production) in C. acetobutylicum resulted in efficient conversion of acetone to isopropanol (up to 8 g/L) (Dusséaux et al., 2013; dos Santos Vieira et al., 2019; Zhang et al., 2023a). This strategy generated isopropanol, butanol, ethanol (IBE) mixture that, differently from the traditional ABE fermentation, can be directly applied to spark-ignition engines without the need to remove by-products (Peralta-Yahya and Keasling, 2010; Zhang et al., 2023a). Engineering of C. acetobytylicum aldehyde/alcohol dehydrogenases has been used to increase substrate specificity for butyryl-CoA and increase butanol over ethanol production (Cho et al., 2019).
So far, the largest increases in butanol titer and/or productivity have been obtained by engineering the fermentation process, that is by using fed-batch or continuous configuration, and/or using immobilized cells and/or using in situ product recovery (e.g., pervaporation, adsorption, liquid–liquid extraction, gas stripping, vacuum fermentation) (Nguyen et al., 2018; Li et al., 2020c), leading to alleviation of substrate and/or product inhibition. So far, the highest butanol titer (550 g/L) was obtained by an engineered C. acetobutylicum strain in a continuous high cell density bioreactor with in situ alcohol extractive distillation (Table 2) (Nguyen et al., 2018). However, these improved fermentation systems are generally associated with higher technical complexity and cost (Nguyen et al., 2018; Li et al., 2020c).
Few natural Clostridia can produce low butanol titer (≤3.6 g/L) through direct fermentation of cellulose or hemicellulose (Mendez et al., 1991; Virunanon et al., 2008; Li et al., 2018). The development of a recombinant cellulolytic C. acetobutylicum has been hampered by severe issues in heterologous cellulase expression (Mingardon et al., 2011; Kovács et al., 2013; Willson et al., 2016). However, the replacement of the inactive catalytic module of C. acetobutylicum cellulase Cel48A with a homologous domain from C. cellulolyticum Cel48F enabled direct fermentation of phosphoric acid swollen cellulose (although no butanol titer, yield or productivity was reported) (Soucaille et al., 2010). (Hemi)cellulolytic and solvent-producing Clostridia have been used to develop artificial consortia or generate fusant strains (Re and Mazzoli, 2023) leading to production of 11–14 g/L butanol through direct fermentation of lignocellulosic feedstocks (Wen et al., 2014; Begum and Dahman, 2015; Jiang et al., 2020). These titers are close to those generated through fermentation of lignocellulose hydrolysates by solventogenic Clostridia (Gu et al., 2011) but still significantly lower than those obtained by conversion of starch or soluble sugars (≈20 g/L) (Gu et al., 2011; Wen et al., 2014; Abo et al., 2019). Among C1-gas fermenting Clostridia, only C. carboxidivorans can naturally produce low amounts (≤2 g/L) of butanol in addition to hexanol and ethanol (Fernández-Naveira et al., 2017; Fernández-Blanco et al., 2023). Recently, metabolic engineering has resulted in minor increase of butanol titer (18%) in this bacterium (Cheng et al., 2019). Higher butanol titer (2.6–6.8 g/L) was obtained by mixotrophic growth (i.e., using both sugars and C1-gas as growth substrates), or by natural or artificial microbial co-cultures (Fernández-Blanco et al., 2023).
Engineering butanol production in non-native hosts has at least two possible advantages: i) no production of other solvents which allows simplified downstream processing for product purification; ii) selection of hosts that can naturally ferment low-cost substrates (e.g., lignocellulose, CO2). Much interest has been attracted by the hyper-butyrate producer C. tyrobutyricum (Bao et al., 2020). In fact, this microorganism has high metabolic flux toward butyryl-CoA and a high butanol tolerance (>15 g/L) (Yu et al., 2011). In addition, C. tyrobutycum has rarely been reported to be subjected to bacteriophage infection, which is a common issue of industrial ABE fermentation (Bao et al., 2020). A butanol hyper-producing C. tyrobutyricum was developed by overexpressing the C. acetobutylicum bifunctional acetaldehyde-alcohol dehydrogenase AdhE2 and disrupting the gene encoding butyrate:acetate CoA transferase (Cat1, which catalyzes butyrate production) which can generate 26.2 g/L butanol through glucose fermentation (Figure 4) (Lee et al., 2016; Zhang et al., 2018). This butanol titer is actually higher than those reported for native butanol producers. Hence, C. tyrobutyricum appears to be a microbial platform with high potential for butanol production, at a level similar or higher than other non-native butanol producers such as E. coli (Shen et al., 2011) or the yeast Arxula adeninivorans (Kunze and Haehnel, 2011).
Butanol production has mainly been engineered in three cellulolytic Clostridia, C. cellulolyticum (Gaida et al., 2016) C. cellulovorans (Yang et al., 2015) and C. thermocellum (Tian et al., 2019b). Reduced or imbalanced biosynthesis of butanol pathway enzymes, enzyme instability, insufficient availability of co-factors and unfavorable reaction thermodynamics have probably contributed to a variable extent to modest butanol titer (<0.5 g/L) in C. cellulolyticum and C. thermocellum (Gaida et al., 2016; Tian et al., 2019b). Fewer genetic modifications enabled butanol production in C. cellulovorans (it is naturally equipped with a butyryl-CoA biosynthetic pathway) (Yang et al., 2015; Wen et al., 2019) which likely contributed to its higher butanol titer (4.96 g/L) from lignocellulosic biomass (Wen et al., 2020b). Similar metabolic engineering strategies used for ABE fermenting strains [e.g., impairing alternative fermentative pathways, enhancing NAD(P)H availability, dysregulating redox homeostasis] are likely to succeed also in further improving butanol formation in C. cellulovorans (and other cellulolytic Clostridia) (Bao et al., 2021; Re and Mazzoli, 2023). However, the efficiency of genetic tools for manipulating C. cellulovorans is still limited (Wen et al., 2017). Apart from the native ability to produce butanol of C. carboxidivorans, the butanol pathway has been engineered in another C1-gas fermenting Clostridium, namely, C. ljungdahlii (Köpke et al., 2010; Lauer et al., 2022b). However, butanol titer < 0.2 g/L was obtained by this engineered strain through autotrophic growth. An alternative approach to bioconvert C1-gases into alcohols with longer chain than C2 consists in co-culturing gas fermenting strains with chain-elongating strains (Fernández-Blanco et al., 2023). Different gas fermenting Clostridia (e.g., C. aceticum, C. autoethanogenum, C. carboxidivorans, C. ljungdahlii) have been co-cultured with the chain-elongating model strain Clostridium kluyvery resulting in production of C4-C8 alcohol mixtures (maximum alcohol titer generally ≤ 1 g/L). (Diender et al., 2016; Richter et al., 2016; Bäumler et al., 2022; Fernández-Blanco et al., 2022). In fact, C. kluyvery is able to elongate the chain of acetate and ethanol (produced by acetogenic Clostridia) to butyrate, hexanoate and octanoate by using reverse β-oxidation. On the other side, acetogenic strains can reduce these fatty acids to their corresponding alcohols (Figure 4). One of the main limitations of this approach is that acetogenic bacteria usually have an optimum pH for growth close to 6, while that of C. kluyveri is close to neutrality (Fernández-Blanco et al., 2023).
Isobutanol is an attractive vehicle fuel with energy density similar to butanol and higher octane number, which is advantageous for blending into gasoline (Chen and Liao, 2016). Moreover, isobutanol can be dehydrated to isobutene, which can then be converted to C8-C12 alkenes to be used as jet fuel (Lin et al., 2015). Additional uses of isobutanol and its derivatives are as solvents, additives in paints, ink ingredients, and extractants for organic compounds (Table 1) (Nawab et al., 2024). A few microorganisms such as Lactococcus lactis, S. cerevisiae, Pichia pastoris and Candida sp. can naturally produce very little isobutanol amounts (≤0.44 g/L) (Nawab et al., 2024). In S. cerevisiae and lactic acid bacteria, isobutanol is biosynthesized through the diversion of 2-ketoisovalerate (an intermediate of valine and isoleucine biosynthesis) which is decarboxylated to isobutyraldehyde by 2-ketoisovalerate decarboxylase and finally reduced (Figure 3) (Hazelwood et al., 2008). Higher levels of isobutanol have been produced by engineered microbial platforms (Nawab et al., 2024) with studies targeting Corynebacterium glutamicum (Yamamoto et al., 2013; Hasegawa et al., 2020) and E. coli (Atsumi et al., 2008) reporting the highest titers (≥20 g/L) through glucose fermentation. However, much lower isobutanol titer (1.88 g/L) was obtained through fermentation of cheaper feedstocks such as pretreated corn stover (Minty et al., 2013).
Advantageously, native production of higher isobutanol levels (1.6 g/L) has been observed in the cellulolytic bacterium C. thermocellum (Holwerda et al., 2014). Isobutanol biosynthetic pathway in C. thermocellum differs from that found in lactic acid bacteria or S. cerevisiae in that 2-ketoisovalerate is converted to isobutyraldehyde in two steps (Figure 3) (Lin et al., 2015). First, ferredoxin-dependent ketoisovalerate reductase (KOR) catalyzes oxidative decarboxylation of 2-ketoisovalerate to isobutyryl-CoA by which is then reduced to isobutyraldehyde (Figure 3). Isobutanol production of C. thermocellum was enhanced by overexpressing autologous acetohydroxy acid synthase (Als), keto acid reductoisomerase (Kari) and dihydroxy acid dehydratase (Dhad) and introducing L. lactis 2-ketoisovalerate decarboxylase (KivD) (Figure 3) (Lin et al., 2015). In optimized growth conditions, the most efficient engineered C. thermocellum strain produced 5.4 g/L of isobutanol from cellulose, which corresponds to 41% of the theoretical yield (Lin et al., 2015). Since this concentration is close to the isobutanol tolerance of the wild type C. thermocellum (Tian et al., 2019a), it is likely that isobutanol production in this microorganism is limited by tolerance. Less successful engineering of isobutanol production (titer ≤ 0.66 g/L) was reported in other cellulolytic Clostridia, i.e., C. cellulolyticum (Higashide et al., 2011) and C. cellulovorans (Wen et al., 2022). Recently, an original approach was reported which consisted in producing a mixture of butanol and isobutanol through direct fermentation of alkali extracted deshelled corn cobs (Wen et al., 2022). To this aim, artificial consortia were developed that included engineered strains of C. cellulovorans and C. beijerinckii which were able to generate up to 1.05 g/L isobutanol and 6.22 g/L butanol in the same fermentation (Wen et al., 2022). Efforts to engineer the isobutanol pathway into autotrophic Clostridia have also been reported (Weitz et al., 2021). However, fermentation of a syngas mixture (50% CO, 45% H2, 5% CO2) by an engineered C. ljungdahlii strain could only produce ≈ 70 mg/L isobutanol (Weitz et al., 2021).
Butanol (and isobutanol) toxicity is among the main issues of biological production of this chemical(s). These compounds are inherently more noxious than other established biofuels, such as ethanol, owing to their higher hydrophobicity (Heipieper et al., 2007; Wilbanks and Trinh, 2017). The toxicity of butanol is mainly due to impairment of structure and functions of biological membranes, dissipation of proton motive force and ATP pools and protein denaturation (Bowles and Ellefson, 1985; Tomas et al., 2004; Alsaker et al., 2010; Venkataramanan et al., 2015). Even native butanol-producing strains, e.g., C. acetobutylicum, can typically tolerate up to 1%–2% v/v butanol (Huang et al., 2010; Nicolaou et al., 2010).
Increasing microbial tolerance to butanol/isobutanol is a key aspect for enhancing their biological production especially as regards final titer. To this aim, responses to butanol stress have been studied in several microbial species that showed the implication of a very complex network of mechanisms only partially understood (Re and Mazzoli, 2023). Hence, increasing butanol tolerance by targeted gene manipulation (e.g., overexpression of protein chaperones) has so far attained only limited results. Random approaches (e.g., random mutagenesis, genome shuffling, adaptive evolutionary engineering) proved to be more suitable strategies to select for multiple-gene trait combinations conferring higher butanol resistance such as for mutant C. acetobutylicum strains able to tolerate up to 3%–4% (v/v) butanol (Liu et al., 2012b; Liu et al., 2013). Interestingly, the adaptive evolution strategy used to develop a butanol-hypertolerant C. thermocellum also enhanced tolerance to isobutanol to the same extent (i.e., 15 g/L), suggesting that similar physiological mechanisms allow cells to cope with both compounds (Tian et al., 2019a). Interestingly, most of the strains characterized by increased tolerance (and equipped with butanol pathway) also showed higher butanol production which highlights the importance of this line of research in improving biobutanol production (Re and Mazzoli, 2023).
Metabolic engineering strategies targeting global gene regulators involved in stress response could advantageously contribute to future development of butanol/isobutanol hypertolerant Clostridia (Jones et al., 2016; Mazzoli, 2021; Xu et al., 2021). Recent studies have indicated that small non-coding RNAs (sRNAs) and RNA chaperones (e.g., Hfq) have important roles in the ability of microorganisms to tolerate a variety of stresses such as butanol exposure (Venkataramanan et al., 2013; Jones et al., 2016; Sun et al., 2017; Costa et al., 2021).
2,3-butanediol (2,3-BDO) plays a critical role in numerous industrial sectors (Table 1) (Köpke et al., 2011). 2,3-BDO is an important starting material in the production of solvents like methyl-ethyl-ketone (MEK) and 1,3-butadiene, which are essential for creating fuel additives, resins, rubbers, printing inks, and lubricating oils. Additionally, 2,3-BDO is used in cosmetics and food industry and as antifreezing agent (Soltys et al., 2001). Nowadays, 2,3-BDO is mostly produced through cheap petrochemical processes, although a pilot plant for its biological production using Klebsiella oxytoca and Paenibacillus polymyxa was operating during the World War II (Blackwood et al., 1949; Celińska and Grajek, 2009). The most efficient natural producers of bio-2,3-BDO include Paenibacillus polymyxa, Klebsiella pneumoniae, K. oxytoca, P. polymyxa, Serratia marcescens, Enterobacter aerogenes and S. cerevisiae (Köpke et al., 2011). In particular, Klebsiella species show fast growth, can use a wide variety of simple sugars and produce up to 150 g/L 2,3-BDO through fed-batch fermentation of glucose (Köpke et al., 2011; Hakizimana et al., 2020). 2,3-BDO biosynthesis is primed by condensation of two pyruvate units to form acetolactate which is subsequently decarboxylated to acetoin (by α-acetolactate decarboxylase, Ald) and reduced to 2,3-BDO by acetoin reductase (Acr) (Figure 3) (Xiao and Xu, 2007; Caspi et al., 2008).
Although C. acetobutylicum is naturally equipped with acetoin biosynthetic pathway, overexpression of C. beijerinckii Acr led to modest 2,3-BDO production (≈2 g/L) (Siemerink et al., 2011). Instead, some C1-gas fermenting Clostridia such as C. autoethanogenum, C. ljungdahlii, C. ragsdalei can naturally produce 2,3-BDO (Köpke et al., 2011; Ricci et al., 2021). Nonetheless, autotrophic growth on CO-rich steel mill waste gas (44% CO, 32% N2, 22% CO2, and 2% H2) led to accumulation of small amounts of 2,3-BDO (<0.2 g/L) especially as compared to the predominant metabolic end products (1.7–1.9 g/L acetate, 0.9–1 g/L ethanol) (Köpke et al., 2011). A recent in silico study has suggested some metabolic engineering strategies for increasing 2,3-BDO biosynthesis by C. autoethanogenum (Ghadermazi et al., 2022). However, so far, the most significant progress in 2,3-BDO production by gas-fermenting Clostridia has been obtained through optimization of the growth medium, substrate composition and fermentation conditions (Zhu et al., 2020; Ricci et al., 2021). Improved gas-liquid mass transfer, gas mixture composition (CO:CO2 4:1) and growth medium (Zinc and Iron supplementation) was able to significantly shift carbon flux of C. ljungdahlii from acetate/ethanol to 2,3-BDO production up to a titer ≈ 2 g/L (batch fermentation) (Ricci et al., 2021). Use of fed-batch fermentation technology with pH and gas (CO) pressure control dramatically increased 2,3-BDO titers up to ≈ 17 g/L (Zhu et al., 2020). It is worth noting that pH regulation is essential for energy conservation in C. ljungdahlii since it affects the Rnf-ATPase responsible for ATP generation (Zhu et al., 2020). It is likely that further optimization of the bioreactor configuration (e.g., continuous fermentation, optimized gas flow rate, cell recycling) can enable further enhancement of 2,3-BDO production by gas fermentation (Simpson et al., 2009; Simpson et al., 2014; Ricci et al., 2021).
Extensive research has been dedicated to improving butanol production by ABE fermenting strains and non-native butanol producers. Although native solventogenic Clostridia are still the most established microorganisms for industrial production of butanol, other engineered strains are promising alternative candidates on traditional substrates (e.g., C. tyrobutyricum) or lignocellulosic feedstocks (e.g., C. cellulovorans). More limited progress has been reported as regards the generation of isobutanol through direct fermentation of lignocellulose or C1-gases. Yet, a major challenge in the biological production of butanol/isobutanol is the development of hyper-tolerant strains which still requires substantial efforts. Despite the high commercial interest in 2,3-BDO, the levels so far produced by Clostridia are modest both from gaseous substrates and sugars.
Hexanol has different industrial applications (e.g., solvent, pesticide, flavoring agent, platform chemical) (Table 1). Currently, it is mainly obtained by petrochemical production systems, although it can also be generated through fermentation of sugars or gaseous substrates by a number of microorganisms, such as C. carboxydivorans (Lauer et al., 2022b). C. carboxydivorans can condense acetyl-CoA units to generate hexanoyl-CoA which is then reduced to hexanol (Figure 4) (Shen et al., 2017; Lauer et al., 2022b). C. carboxidivorans can produce hexanol also through the reduction of hexanoate by Aor (Wirth and Dürre, 2021). The highest hexanol titers obtained so far through CO fermentation by the wild-type C. carboxidivorans are 1.4–1.9 g/L (Shen et al., 2017; Oh et al., 2022). Other Clostridia such as C. kluyvery instead convert hexanoyl-CoA mainly into hexanoic acid (caproic acid) and produce only traces of hexanol (Figure 4) (Shen et al., 2017; Lauer et al., 2022b). However, as mentioned in the Section 4.4.1.2., C. kluyvery has been used as chain-elongating partner in co-cultures with gas-fermenting Clostridia (e.g., C. carboxidivorans, C. ljungdahlii) for fermenting C1 substrates into butanol-hexanol-octanol mixtures (Richter et al., 2016; Fernández-Blanco et al., 2023).
Recently, hexanol production was engineered in another gas fermenting Clostridium (C. ljungdhalii) (Lauer et al., 2022b). Two sets of genes encoding the acetyl-CoA-to-hexanoyl-CoA pathway of C. kluyvery, and the gene coding for the aldehyde-alcohol dehydrogenase AdhE2 of C. acetobutylicum (for the reduction of hexanoyl-CoA to hexanol) were integrated into the C. ljungdahlii chromosome (Figure 4). The engineered C. ljungdahlii showed improvement of both butanol and hexanol production (Lauer et al., 2022b). Fermentation in 2 L bioreactor, with continuous CO2-H2 supplementation and pH regulation (pH = 6) resulted in the production of 0.122 g/L hexanol. Limited hexanol titer was attributed to inefficient biosynthesis of some enzymes of the acetyl-CoA-to-hexanoyl-CoA pathway. Therefore, additional genes from C. carboxidivorans (encoding thiolase, crotonase, 3-hydroxybutyryl-CoA dehydrogenase and butyryl-CoA dehydrogenase complex) (Figure 4) were introduced in the genome of the engineered C. ljungdahlii. The final strain produced 0.251 g/L hexanol through fermentation of 20% CO2, 80% H2 gas mixture (Oh et al., 2022).
As for other products obtained through gas fermentation, significant research towards higher hexanol production has been focused on optimizing the fermentation process such as growth conditions, media composition, gas composition or supply (Oh et al., 2022). Substantial improvement was obtained by using in situ extraction of hexanol, which reduces hexanol accumulation in the fermentation medium (Kottenhahn et al., 2021; Oh et al., 2023). It is worth remembering that hexanol is even more toxic than butanol because of its longer carbon chain (Heipieper et al., 2007). Supplementation of 1 g/L hexanol is enough for reducing C. ljungdahlii growth and 5 g/L caused total growth cessation (Oh et al., 2023). Supplementation of small quantities of a biocompatible extractant (i.e., oleyl alcohol) and ethanol (as a precursor) to C. carboxidivorans P7 cultures growing on CO enabled production of 8.45 g/L hexanol, which is the highest titer reported so far (Oh et al., 2023).
Short- and medium-chain (C2-C12) esters such as ethyl acetate, butyl acetate, isobutyl acetate and butyl butyrate, have a broad range of application as flavors, fragrances, pharmaceuticals, green solvents and advanced biofuels (Table 1) (Lee and Trinh, 2020; Wang et al., 2021a). The traditional methods for synthesizing short- and medium-chain fatty acid esters are mainly based on concentrated sulfuric acid-mediated esterification of acids and alcohols and are affected by serious health and environmental issues (Cull et al., 2000; Jermy and Pandurangan, 2005). Alternative chemical strategies based on ionic liquid catalysis can reduce these problems to some extent, but they are expensive and not stable (Tankov et al., 2017). In nature, microbes and plants can biosynthesize several esters which encounters the increasing consumer preference for natural and sustainable products (Lee and Trinh, 2020; Seo et al., 2021). Some yeasts and lactic acid bacteria can naturally form esters but with limited efficiency (Abeijón Mukdsi et al., 2009; Kruis et al., 2018). This has stimulated research aimed to engineer more efficient producers (Rodriguez et al., 2014; Kruis et al., 2017).
Clostridia can synthesize a variety of volatile organic acids (e.g., acetic and butyric acid) and alcohols (e.g., ethanol, butanol and isobutanol) (Tracy et al., 2012; Cho et al., 2015; Lin et al., 2015). Therefore, significant amounts of acyl-CoA, organic acids, and alcohols are available in cells that can serve as precursors for ester generation. Biological synthesis of esters is mainly catalyzed by esterases/lipases (alcohol + acid → ester + H2O) or by alcohol acyltransferases (AAT, alcohol + acyl-CoA→ester + CoA) (Kruis et al., 2019; Noh et al., 2019). In a number of studies, exogenous lipases have been supplemented to clostridial fermentation broth to convert acids and alcohols to esters (Cui et al., 2020; Wang et al., 2021a). The main issue of these approaches is the cost of exogenously added lipases (Wang et al., 2021a). Alternatively, overexpression of heterologous lipases in Clostridia has proven to be challenging (Wen et al., 2020c). The AAT-dependent pathway is more thermodynamically favorable (Flamholz et al., 2012; Noh et al., 2019). Overexpression of heterologous AATs has enabled different mesophilic Clostridia (e.g., C. acetobutylicum, C. diolis, C. beijerinckii, C. saccharoperbutylacetonicum) to produce esters such as butyl acetate and butyl butyrate from glucose or xylose (Horton and Bennett, 2006; Noh et al., 2018; Li et al., 2020a; Fang et al., 2020; Feng et al., 2021). Efforts have been made also for producing esters from cheaper feedstocks. By introducing an engineered heterologous chloramphenicol acetyltransferase and inactivating endogenous esterases, C. thermocellum was able to produce C4-C8 esters (e.g., ethyl acetate, ethyl isobutyrate, isobutyl acetate, and isobutyl isobutyrate) through direct fermentation of cellulose (Seo et al., 2023). As far as we know, only one study has reported engineering medium chain ester production in a gas fermenting Clostridium (namely, C. autoethagenum) resulting in accumulation of traces of ethyl-acetate and butyl-acetate (Dykstra et al., 2022).
One of the key aspects of engineering ester production in Clostridia relies on overproduction of stable AATs with desired substrate specificity (Seo et al., 2021). Most characterized AATs derive from plants or yeasts which results in poor biosynthesis, solubility and/or thermostability in prokaryotes (Horton and Bennett, 2006; Noh et al., 2018; Li et al., 2020a; Fang et al., 2020). Studies have been dedicated to optimize AATs structure and/or biosynthesis (Seo et al., 2021). In addition, limited knowledge is available on substrate specificities of AATs (Noh et al., 2018; Kruis et al., 2019). Both these aspects were tackled to develop an ester overproducing C. saccharoperbutylaceticum (Feng et al., 2021). In addition, systems metabolic engineering was used to increase availability of NADH and ester precursor(s) (namely, acetyl-CoA). The most efficient strain was able to produce up to 20.3 g/L butyl acetate (and 0.9 g/L butyl butyrate) through glucose fermentation and up to 17.8 g/L butyl acetate by fermentation of corn stover hydrolysate (Feng et al., 2021). Few metabolic modifications were able to convert C. tyrobutyricum (an efficient butyrate producer) into high performing butyl butyrate producing strain, namely, enhancing butyryl-CoA pathway (by upregulating CtfAB encoding CoA transferase, Figure 4), engineering butyryl-CoA reduction to butanol (by expressing C. acetobutylicum AdhE2), and introducing the wild strawberry alcohol acyltransferase (VAAT) (Guo et al., 2023). Additional reducing power for ester production was supplied by using a reduced growth substrate, that is mannitol, instead of glucose. Fed batch fermentation of mannitol at regulated pH and supplementation of hexadecane for in situ extraction of esters led to ≈ 63 g/L butyl butyrate (yield = 0.21 mol/mol, theoretical maximum is 0.5 mol/mol) (Table 2) (Guo et al., 2023). This is by far the highest medium chain ester titer obtained by microbial fermentation. Additional introduction of heterologous α-amylase and α-glucosidase enabled the engineered C. tyrobutyricum to produce high amounts of butyl butyrate (26.8 g/L) through batch fermentation of starch from a non-food crop (i.e., cassava) (Guo et al., 2024). As mentioned above for other hydrophobic products (e.g., butanol, hexanol), in situ extraction of esters significantly contributed to increase fermentation efficiencies of the engineered strains by limiting toxicity linked to product accumulation in the growth medium (Fang et al., 2020; Feng et al., 2021; Guo et al., 2023; Guo et al., 2024).
In conclusion, the variety of acyl-CoA, acids and alcohols produced by Clostridia confers them the ability to produce a panel of commercially interesting medium-chain esters in high amounts. Yet, challenges of this research area reside in efficient expression of AATs with desired substrate specificity and improving the availability of carbon precursors and reducing power.
The studies summarized in the present review illustrate the key role of Clostridia in the development of sustainable biomass biorefining processes. Clostridia are already the reference cell factories for fuels such as butanol and among the most promising microbial platforms for production of valuable chemicals (e.g., ethanol, lactate, H2, hexanol, medium chain esters) through direct fermentation of low cost biomasses (e.g., lignocellulose, C1-gases) (Table 2). Significant progress has also been reported as regards clostridial production of 1,2-PDO and 1,3-PDO, whose titers are not far from those obtained by the highest performing microorganisms (e.g., engineered E. coli) (Table 2). However, for the full deployment of Clostridium potential in commercial applications further advances at multiple levels are necessary.
Extensive investigations on the physiology and metabolism of Clostridia over the last decades have revealed peculiar characteristics (e.g., atypical pathway reactions, sophisticated regulation of enzyme activity) and subtle differences between the different strains which affect carbon and electron flux and pathway thermodynamics. We believe that further advances in detailed understanding the metabolic network of Clostridia will be major sources of inspiration for improving current metabolic engineering strategies. To this aim, further development of genetic tools for manipulating Clostridia is also necessary, especially for strains which are recalcitrant to the currently available methods (e.g., Clostridium diolis) or which shows low genetic tractability (e.g., C. cellulovorans).
Apart from strain improvement by metabolic engineering (enhancement of product yield, titer, productivity; increase of tolerance to toxic products), research aimed to optimize the fermentation process (e.g., growth media and conditions, fermentation mode, bioreactor configuration, use of syntrophic co-cultures) including extraction and purification of products will be essential to achieve the efficiency required by industrial application. Coping with the different feedstocks (e.g., insoluble solids such as lignocellulose versus gaseous substrates) and products (e.g., acids versus hydrophobic solvents versus gaseous products) described in the present review will certainly require far different process optimization. The efficiency of gas fermentation in inherently limited by the low solubility of CO/CO2 in water. Developing suitable systems for improving gas-liquid mass transfer is among the priority of this research area. Accumulation of acids (e.g., lactic acid) or solvents (e.g., butanol, hexanol, medium chain esters) reduces Clostridium fermentation efficiency due to limited tolerance of cells these chemicals. Both strain improvement by metabolic engineering and process engineering (e.g., pH regulation, in situ recovery of solvents) should synergistically provide a cost sustainable solution. As regards biological production of H2, the improvement of systems for reducing H2 partial pressure in the bioreactor (e.g., stirring the growth medium, sparging the growth medium with inert gas, removing gas by a vacuum pump, selectively removing H2 by active membranes) is essential for overcoming thermodynamic barriers of biological H2 production. We are confident that thanks to the contribution of these different approaches a rapid improvement of Clostridium-mediated biorefinery processes suitable for their industrial application will be possible.
PP: Writing–original draft, Writing–review and editing. ES: Writing–original draft, Writing–review and editing. RM: Conceptualization, Supervision, Visualization, Writing–original draft, Writing–review and editing. FV: Supervision, Writing–review and editing. GG: Supervision, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdel-Rahman, M. A., and Sonomoto, K. (2016). Opportunities to overcome the current limitations and challenges for efficient microbial production of optically pure lactic acid. J. Biotechnol. 236, 176–192. doi:10.1016/j.jbiotec.2016.08.008
Abdel-Rahman, M. A., Tashiro, Y., and Sonomoto, K. (2011). Lactic acid production from lignocellulose-derived sugars using lactic acid bacteria: overview and limits. J. Biotechnol. 156, 286–301. doi:10.1016/j.jbiotec.2011.06.017
Abdel-Rahman, M. A., Tashiro, Y., and Sonomoto, K. (2013). Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 31, 877–902. doi:10.1016/j.biotechadv.2013.04.002
Abeijón Mukdsi, M. C., Medina, R. B., Alvarez, M. de F., and González, S. N. (2009). Ester synthesis by lactic acid bacteria isolated from goat’s and Ewe’s milk and cheeses. Food Chem. 117, 241–247. doi:10.1016/j.foodchem.2009.03.105
Abo, B. O., Gao, M., Wang, Y., Wu, C., Wang, Q., and Ma, H. (2019). Production of butanol from biomass: recent advances and future prospects. Environ. Sci. Pollut. Res. 26, 20164–20182. doi:10.1007/s11356-019-05437-y
Agrawal, D., Budakoti, M., and Kumar, V. (2023). Strategies and tools for the biotechnological valorization of glycerol to 1, 3-propanediol: challenges, recent advancements and future outlook. Biotechnol. Adv. 66, 108177. doi:10.1016/j.biotechadv.2023.108177
Agu, C. V., Ujor, V., and Ezeji, T. C. (2019). Metabolic engineering of Clostridium beijerinckii to improve glycerol metabolism and furfural tolerance. Biotechnol. Biofuels 12, 50–19. doi:10.1186/S13068-019-1388-9
Akhlaghi, N., and Najafpour-Darzi, G. (2020). A comprehensive review on biological hydrogen production. Int. J. Hydrogen Energy 45, 22492–22512. doi:10.1016/J.IJHYDENE.2020.06.182
Allied Market Research (2024). Isobutanol market size, share | industry growth & forecast, 2030. Available at: https://www.alliedmarketresearch.com/isobutanol-market-A13034 (Accessed April 16, 2024).
Alsaker, K. V., Paredes, C., and Papoutsakis, E. T. (2010). Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol. Bioeng. 105, 1131–1147. doi:10.1002/bit.22628
Al-Shorgani, N. K. N., Kalil, M. S., Yusoff, W. M. W., and Hamid, A. A. (2018). Impact of pH and butyric acid on butanol production during batch fermentation using a new local isolate of Clostridium acetobutylicum YM1. Saudi J. Biol. Sci. 25, 339–348. doi:10.1016/j.sjbs.2017.03.020
Altaras, N. E., Etzel, M. R., and Cameron, D. C. (2001). Conversion of sugars to 1,2-propanediol by Thermoanaerobacterium thermosaccharolyticum hg-8. Biotechnol. Prog. 17, 52–56. doi:10.1021/bp000130b
Alves de Oliveira, R., Komesu, A., Vaz Rossell, C. E., and Maciel Filho, R. (2018). Challenges and opportunities in lactic acid bioprocess design—from economic to production aspects. Biochem. Eng. J. 133, 219–239. doi:10.1016/j.bej.2018.03.003
Anandharaj, M., Lin, Y. J., Rani, R. P., Nadendla, E. K., Ho, M. C., Huang, C. C., et al. (2020). Constructing a yeast to express the largest cellulosome complex on the cell surface. Proc. Natl. Acad. Sci. U. S. A. 117, 2385–2394. doi:10.1073/pnas.1916529117
Arizzi, M., Morra, S., Gilardi, G., Pugliese, M., Gullino, M. L., and Valetti, F. (2021). Improving sustainable hydrogen production from green waste: [FeFe]-hydrogenases quantitative gene expression RT-qPCR analysis in presence of autochthonous consortia. Biotechnol. Biofuels 14, 182. doi:10.1186/s13068-021-02028-3
Arslan, K., Bayar, B., Nalakath Abubackar, H., Veiga, M. C., and Kennes, C. (2019). Solventogenesis in Clostridium aceticum producing high concentrations of ethanol from syngas. Bioresour. Technol. 292, 121941. doi:10.1016/j.biortech.2019.121941
Arslan, K., Schoch, T., Höfele, F., Herrschaft, S., Oberlies, C., Bengelsdorf, F., et al. (2022). Engineering Acetobacterium woodii for the production of isopropanol and acetone from carbon dioxide and hydrogen. Biotechnol. J. 17, 2100515. doi:10.1002/biot.202100515
Atsumi, S., Hanai, T., and Liao, J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi:10.1038/nature06450
Azambuja, S. P. H., and Goldbeck, R. (2020). Butanol production by Saccharomyces cerevisiae: perspectives, strategies and challenges. World J. Microbiol. Biotechnol. 36, 48. doi:10.1007/s11274-020-02828-z
Badia, J., Ros, J., and Aguilar, J. (1985). Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J. Bacteriol. 161, 435–437. doi:10.1128/jb.161.1.435-437.1985
Bao, T., Feng, J., Jiang, W., Fu, H., Wang, J., and Yang, S. T. (2020). Recent advances in n-butanol and butyrate production using engineered Clostridium tyrobutyricum. World J. Microbiol. Biotechnol. 36, 138. doi:10.1007/s11274-020-02914-2
Bao, T., Hou, W., Wu, X., Lu, L., Zhang, X., and Yang, S. T. (2021). Engineering Clostridium cellulovorans for highly selective n-butanol production from cellulose in consolidated bioprocessing. Biotechnol. Bioeng. 118, 2703–2718. doi:10.1002/bit.27789
Bäumler, M., Schneider, M., Ehrenreich, A., Liebl, W., and Weuster-Botz, D. (2022). Synthetic co-culture of autotrophic Clostridium carboxidivorans and chain elongating Clostridium kluyveri monitored by flow cytometry. Microb. Biotechnol. 15, 1471–1485. doi:10.1111/1751-7915.13941
Begum, S., and Dahman, Y. (2015). Enhanced biobutanol production using novel clostridial fusants in simultaneous saccharification and fermentation of green renewable agriculture residues. Biofuels, Bioprod. Biorefining 9, 529–544. doi:10.1002/BBB.1564
Bengelsdorf, F. R., Beck, M. H., Erz, C., Hoffmeister, S., Karl, M. M., Riegler, P., et al. (2018). “Bacterial anaerobic synthesis gas (syngas) and CO2 + H2 fermentation,” in Advances in applied microbiology (Academic Press), 143–221. doi:10.1016/bs.aambs.2018.01.002
Beri, D., York, W. S., Lynd, L. R., Peña, M. J., and Herring, C. D. (2020). Development of a thermophilic coculture for corn fiber conversion to ethanol. Nat. Commun. 11, 1937. doi:10.1038/s41467-020-15704-z
Bhandiwad, A., Guseva, A., and Lynd, L. (2013). Metabolic engineering of Thermoanaerobacterium thermosaccharolyticum for increased n-butanol production. Adv. Microbiol. 3, 46–51. doi:10.4236/aim.2013.31007
Biswas, R., Zheng, T., Olson, D. G., Lynd, L. R., and Guss, A. M. (2015). Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol. Biofuels 8, 20. doi:10.1186/s13068-015-0204-4
Blackwood, A. C., Simpson, F. J., Wheat, J. A., Leslie, J. D., and Ledingham, G. A. (1949). Production and properties of 2,3-butanediol: XXXI. Pilot plant studies on the fermentation of wheat by Aerobacillus polymyxa. Can. J. Res. 27f, 199–210. doi:10.1139/cjr49f-020
Bora, A., Mohanrasu, K., Angelin Swetha, T., Ananthi, V., Sindhu, R., Chi, N. T. L., et al. (2022). Microbial electrolysis cell (MEC): reactor configurations, recent advances and strategies in biohydrogen production. Fuel 328, 125269. doi:10.1016/j.fuel.2022.125269
Bowles, L. K., and Ellefson, W. L. (1985). Effects of butanol on Clostridium acetobutylicum. Appl. Environ. Microbiol. 50, 1165–1170. doi:10.1128/aem.50.5.1165-1170.1985
Brown, S. D., Guss, A. M., Karpinets, T. V., Parks, J. M., Smolin, N., Yang, S., et al. (2011). Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 108, 13752–13757. doi:10.1073/pnas.1102444108
Bryant, F. O. (1991). Characterization of the fructose 1,6-bisphosphate-activated, L(+) lactate dehydrogenase from thermoanaerobacter ethanolicus. J. Enzyme Inhib. Med. Chem. 5, 235–248. doi:10.3109/14756369109080062
Cai, G., Jin, B., Monis, P., and Saint, C. (2013). A genetic and metabolic approach to redirection of biochemical pathways of Clostridium butyricum for enhancing hydrogen production. Biotechnol. Bioeng. 110, 338–342. doi:10.1002/bit.24596
Calusinska, M., Happe, T., Joris, B., and Wilmotte, A. (2010). The surprising diversity of clostridial hydrogenases: a comparative genomic perspective. Microbiology 156, 1575–1588. doi:10.1099/mic.0.032771-0
Cameron, D. C., and Cooney, C. L. (1986). A novel fermentation: the production of R(–)–1,2–Propanediol and acetol by Clostridium thermosaccharolyticum. Bio/Technology 4, 651–654. doi:10.1038/nbt0786-651
Campos-Fernández, J., Arnal, J. M., Gómez, J., and Dorado, M. P. (2012). A comparison of performance of higher alcohols/diesel fuel blends in a diesel engine. Appl. Energy 95, 267–275. doi:10.1016/j.apenergy.2012.02.051
Canatoy, R. C., Jeong, S. T., Galgo, S. J. C., Kim, P. J., and Cho, S. R. (2022). Biochar as soil amendment: syngas recycling system is essential to create positive carbon credit. Sci. Total Environ. 809, 151140. doi:10.1016/j.scitotenv.2021.151140
Cao, Y., Liu, H., Liu, W., Guo, J., and Xian, M. (2022). Debottlenecking the biological hydrogen production pathway of dark fermentation: insight into the impact of strain improvement. Microb. Cell Fact. 21, 166. doi:10.1186/s12934-022-01893-3
Caspi, R., Foerster, H., Fulcher, C. A., Kaipa, P., Krummenacker, M., Latendresse, M., et al. (2008). The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 36, D623–D631. doi:10.1093/nar/gkm900
Celińska, E., and Grajek, W. (2009). Biotechnological production of 2,3-butanediol-Current state and prospects. Biotechnol. Adv. 27, 715–725. doi:10.1016/j.biotechadv.2009.05.002
Cen, X., Dong, Y., Liu, D., and Chen, Z. (2022). New pathways and metabolic engineering strategies for microbial synthesis of diols. Curr. Opin. Biotechnol. 78, 102845. doi:10.1016/j.copbio.2022.102845
Cha, M., Chung, D., and Westpheling, J. (2016). Deletion of a gene cluster for [Ni-Fe] hydrogenase maturation in the anaerobic hyperthermophilic bacterium Caldicellulosiruptor bescii identifies its role in hydrogen metabolism. Appl. Microbiol. Biotechnol. 100, 1823–1831. doi:10.1007/s00253-015-7025-z
Charubin, K., Bennett, R. K., Fast, A. G., and Papoutsakis, E. T. (2018). Engineering Clostridium organisms as microbial cell-factories: challenges & opportunities. Metab. Eng. 50, 173–191. doi:10.1016/j.ymben.2018.07.012
Chen, C. T., and Liao, J. C. (2016). Frontiers in microbial 1-butanol and isobutanol production. FEMS Microbiol. Lett. 363, fnw020. doi:10.1093/femsle/fnw020
Cheng, C., Li, W., Lin, M., and Yang, S. T. (2019). Metabolic engineering of Clostridium carboxidivorans for enhanced ethanol and butanol production from syngas and glucose. Bioresour. Technol. 284, 415–423. doi:10.1016/J.BIORTECH.2019.03.145
Cho, C., Hong, S., Moon, H. G., Jang, Y. S., Kim, D., and Lee, S. Y. (2019). Engineering clostridial aldehyde/alcohol dehydrogenase for selective butanol production. MBio 10, e02683-18. doi:10.1128/mBio.02683-18
Cho, C., Jang, Y. S., Moon, H. G., Lee, J., and Lee, S. Y. (2015). Metabolic engineering of clostridia for the production of chemicals. Biofuels, Bioprod. Biorefining 9, 211–225. doi:10.1002/bbb.1531
Ciriminna, R., Pina, C. D., Rossi, M., and Pagliaro, M. (2014). Understanding the glycerol market. Eur. J. Lipid Sci. Technol. 116, 1432–1439. doi:10.1002/ejlt.201400229
Clomburg, J. M., and Gonzalez, R. (2011). Metabolic engineering of Escherichia coli for the production of 1,2-propanediol from glycerol. Biotechnol. Bioeng. 108, 867–879. doi:10.1002/bit.22993
Collas, F., Kuit, W., Clément, B., Marchal, R., López-Contreras, A. M., and Monot, F. (2012). Simultaneous production of isopropanol, butanol, ethanol and 2,3-butanediol by Clostridium acetobutylicum ATCC 824 engineered strains. Amb. Express 2, 45–10. doi:10.1186/2191-0855-2-45
Costa, P., Usai, G., Re, A., Manfredi, M., Mannino, G., Bertea, C. M., et al. (2021). Clostridium cellulovorans proteomic responses to butanol stress. Front. Microbiol. 12, 674639. doi:10.3389/fmicb.2021.674639
Crown, S. B., Indurthi, D. C., Ahn, W. S., Choi, J., Papoutsakis, E. T., and Antoniewicz, M. R. (2011). Resolving the TCA cycle and pentose-phosphate pathway of Clostridium acetobutylicum ATCC 824: isotopomer analysis, in vitro activities and expression analysis. Biotechnol. J. 6, 300–305. doi:10.1002/biot.201000282
Cruz-Morales, P., Orellana, C. A., Moutafis, G., Moonen, G., Rincon, G., Nielsen, L. K., et al. (2019). Revisiting the evolution and taxonomy of clostridia, a phylogenomic update. Genome Biol. Evol. 11, 2035–2044. doi:10.1093/gbe/evz096
Cui, Y., He, J., Yang, K. L., and Zhou, K. (2020). Production of isopropyl and butyl esters by Clostridium mono-culture and co-culture. J. Ind. Microbiol. Biotechnol. 47, 543–550. doi:10.1007/s10295-020-02279-3
Cull, S. G., Holbrey, J. D., Vargas-Mora, V., Seddon, K. R., and Lye, G. J. (2000). Room-temperature ionic liquids as replacements for organic solvents in multiphase bioprocess operations. Biotechnol. Bioeng. 69, 227–233. doi:10.1002/(SICI)1097-0290(20000720)69:2<227::AID-BIT12>3.0.CO;2-0
Dai, Z., Zhu, Y., Dong, H., Zhao, C., Zhang, Y., and Li, Y. (2021). Enforcing ATP hydrolysis enhanced anaerobic glycolysis and promoted solvent production in Clostridium acetobutylicum. Microb. Cell Fact. 20, 149. doi:10.1186/s12934-021-01639-7
Daniell, J., Köpke, M., and Simpson, S. D. (2012). Commercial biomass syngas fermentation. Energies 5, 5372–5417. doi:10.3390/en5125372
Das, D., and Veziroglu, T. N. (2008). Advances in biological hydrogen production processes. Int. J. Hydrogen Energy 33, 6046–6057. doi:10.1016/j.ijhydene.2008.07.098
Demuez, M., Cournac, L., Guerrini, O., Soucaille, P., and Girbal, L. (2007). Complete activity profile of Clostridium acetobutylicum [FeFe]-hydrogenase and kinetic parameters for endogenous redox partners. FEMS Microbiol. Lett. 275, 113–121. doi:10.1111/J.1574-6968.2007.00868.X
Deng, Y., Olson, D. G., Zhou, J., Herring, C. D., Joe Shaw, A., and Lynd, L. R. (2013). Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab. Eng. 15, 151–158. doi:10.1016/j.ymben.2012.11.006
Diallo, M., Simons, A. D., van der Wal, H., Collas, F., Houweling-Tan, B., Kengen, S. W. M., et al. (2019). L-Rhamnose metabolism in Clostridium beijerinckii strain DSM 6423. Appl. Environ. Microbiol. 85, e02656-18. doi:10.1128/AEM.02656-18
Dien, B. S., Cotta, M. A., and Jeffries, T. W. (2003). Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63, 258–266. doi:10.1007/s00253-003-1444-y
Diender, M., Stams, A. J. M., and Sousa, D. Z. (2016). Production of medium-chain fatty acids and higher alcohols by a synthetic co-culture grown on carbon monoxide or syngas. Biotechnol. Biofuels 9, 82. doi:10.1186/s13068-016-0495-0
dos Santos Vieira, C. F., Maugeri Filho, F., Maciel Filho, R., and Pinto Mariano, A. (2019). Acetone-free biobutanol production: past and recent advances in the Isopropanol-Butanol-Ethanol (IBE)fermentation. Bioresour. Technol. 287, 121425. doi:10.1016/j.biortech.2019.121425
Dusséaux, S., Croux, C., Soucaille, P., and Meynial-Salles, I. (2013). Metabolic engineering of Clostridium acetobutylicum ATCC 824 for the high-yield production of a biofuel composed of an isopropanol/butanol/ethanol mixture. Metab. Eng. 18, 1–8. doi:10.1016/j.ymben.2013.03.003
Dykstra, J. C., van Oort, J., Yazdi, A. T., Vossen, E., Patinios, C., van der Oost, J., et al. (2022). Metabolic engineering of Clostridium autoethanogenum for ethyl acetate production from CO. Microb. Cell Fact. 21, 243. doi:10.1186/S12934-022-01964-5
Ethanolrfa (2019). Ethanol industry outlook. Powered with renewed energy. Available at: https://ethanolrfa.org/wp-content/uploads/2019/02/RFA2019Outlook.pdf.
Ezeji, T. C., Qureshi, N., and Blaschek, H. P. (2004). Acetone butanol ethanol (ABE) production from concentrated substrate: reduction in substrate inhibition by fed-batch technique and product inhibition by gas stripping. Appl. Microbiol. Biotechnol. 63, 653–658. doi:10.1007/s00253-003-1400-x
Fang, D., Wen, Z., Lu, M., Li, A., Ma, Y., Tao, Y., et al. (2020). Metabolic and process engineering of Clostridium beijerinckii for butyl acetate production in one step. J. Agric. Food Chem. 68, 9475–9487. doi:10.1021/acs.jafc.0c00050
Fasano, A., Fourmond, V., and Eger, C. L. ´ (2024). Outer-sphere effects on the O2 sensitivity, catalytic bias and catalytic reversibility of hydrogenases. Chem. Sci. 15, 5418–5433. doi:10.1039/D4SC00691G
Feng, J., Zhang, J., Ma, Y., Feng, Y., Wang, S. S., Guo, N., et al. (2021). Renewable fatty acid ester production in Clostridium. Nat. Commun. 12, 4368. doi:10.1038/S41467-021-24038-3
Fernández-Blanco, C., Robles-Iglesias, R., Naveira-Pazos, C., Veiga, M. C., and Kennes, C. (2023). Production of biofuels from C<sub>1</sub>-gases with Clostridium and related bacteria—recent advances. Microb. Biotechnol. 16, 726–741. doi:10.1111/1751-7915.14220
Fernández-Blanco, C., Veiga, M. C., and Kennes, C. (2022). Efficient production of n-caproate from syngas by a co-culture of Clostridium aceticum and Clostridium kluyveri. J. Environ. Manage. 302, 113992. doi:10.1016/j.jenvman.2021.113992
Fernández-Naveira, Á., Abubackar, H. N., Veiga, M. C., and Kennes, C. (2016). Efficient butanol-ethanol (B-E) production from carbon monoxide fermentation by Clostridium carboxidivorans. Appl. Microbiol. Biotechnol. 100, 3361–3370. doi:10.1007/s00253-015-7238-1
Fernández-Naveira, Á., Veiga, M. C., and Kennes, C. (2017). Glucose bioconversion profile in the syngas-metabolizing species Clostridium carboxidivorans. Bioresour. Technol. 244, 552–559. doi:10.1016/j.biortech.2017.07.174
Ferreira, S., Pereira, R., Wahl, S. A., and Rocha, I. (2020). Metabolic engineering strategies for butanol production in Escherichia coli. Biotechnol. Bioeng. 117, 2571–2587. doi:10.1002/bit.27377
Flamholz, A., Noor, E., Bar-Even, A., and Milo, R. (2012). EQuilibrator - the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775. doi:10.1093/nar/gkr874
Fonseca, B. C., Dalbelo, G., Gelli, V. C., Carli, S., Meleiro, L. P., Zimbardi, A. L. R. L., et al. (2020). Use of algae biomass obtained by single-step mild acid hydrolysis in hydrogen production by the β-glucosidase-producing Clostridium beijerinckii Br21. Waste Biomass Valorization 11, 1393–1402. doi:10.1007/s12649-018-0430-7
Foulquier, C., Rivière, A., Heulot, M., Dos Reis, S., Perdu, C., Girbal, L., et al. (2022). Molecular characterization of the missing electron pathways for butanol synthesis in Clostridium acetobutylicum. Nat. Commun. 13, 4691. doi:10.1038/s41467-022-32269-1
Froese, A., Schellenberg, J., and Sparling, R. (2019). Enhanced depolymerization and utilization of raw lignocellulosic material by co-cultures of ruminiclostridium thermocellum with hemicellulose-utilizing partners. Can. J. Microbiol. 65, 296–307. doi:10.1139/cjm-2018-0535
Furdui, C., and Ragsdale, S. W. (2000). The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J. Biol. Chem. 275, 28494–28499. doi:10.1074/JBC.M003291200
Future Market Insights (2024). Esters market | global sales analysis report - 2033. Available at: https://www.futuremarketinsights.com/reports/esters-market (Accessed April 16, 2024).
Gaida, S. M., Liedtke, A., Jentges, A. H. W., Engels, B., and Jennewein, S. (2016). Metabolic engineering of Clostridium cellulolyticum for the production of n-butanol from crystalline cellulose. Microb. Cell Fact. 15, 6. doi:10.1186/s12934-015-0406-2
Ganguly, J., Martin-Pascual, M., and van Kranenburg, R. (2020). CRISPR interference (CRISPRi) as transcriptional repression tool for Hungateiclostridium thermocellum DSM 1313. Microb. Biotechnol. 13, 339–349. doi:10.1111/1751-7915.13516
Ghadermazi, P., Re, A., Ricci, L., and Chan, S. H. J. (2022). Metabolic engineering interventions for sustainable 2,3-butanediol production in gas-fermenting Clostridium autoethanogenum. mSystems 7, e0111121. doi:10.1128/msystems.01111-21
Gharehkhani, S., Sadeghinezhad, E., Kazi, S. N., Yarmand, H., Badarudin, A., Safaei, M. R., et al. (2015). Basic effects of pulp refining on fiber properties - a review. Carbohydr. Polym. 115, 785–803. doi:10.1016/j.carbpol.2014.08.047
González-Pajuelo, M., Meynial-Salles, I., Mendes, F., Andrade, J. C., Vasconcelos, I., and Soucaille, P. (2005). Metabolic engineering of Clostridium acetobutylicum for the industrial production of 1,3-propanediol from glycerol. Metab. Eng. 7, 329–336. doi:10.1016/j.ymben.2005.06.001
Grand View Research (2020). Isopropyl alcohol market size, trends | industry report, 2020-2027. Mark. Anal. 110. Available at: https://www.grandviewresearch.com/industry-analysis/isopropyl-alcohol-market (Accessed April 16, 2024).
Grand View Research (2022). Isobutanol market size, share | global industry report. Available at: https://www.grandviewresearch.com/industry-analysis/isobutanol-market (Accessed April 16, 2024).
Grand View Research (2024). Ethanol market size, share & trends report, 2020-2027. Available at: https://www.grandviewresearch.com/industry-analysis/ethanol-market (Accessed April 16, 2024).
Grand View Reserach (2023). Hydrogen generation market size and share report, 2030. Available at: https://www.grandviewresearch.com/industry-analysis/hydrogen-generation-market (Accessed April 16, 2024).
Green, E. M. (2011). Fermentative production of butanol-the industrial perspective. Curr. Opin. Biotechnol. 22, 337–343. doi:10.1016/j.copbio.2011.02.004
Gu, Y., Jiang, Y., Wu, H., Liu, X., Li, Z., Li, J., et al. (2011). Economical challenges to microbial producers of butanol: feedstock, butanol ratio and titer. Biotechnol. J. 6, 1348–1357. doi:10.1002/biot.201100046
Guo, X., Li, X., Feng, J., Yue, Z., Fu, H., and Wang, J. (2024). Engineering of Clostridium tyrobutyricum for butyric acid and butyl butyrate production from cassava starch. Bioresour. Technol. 391, 129914. doi:10.1016/J.BIORTECH.2023.129914
Guo, X., Zhang, H., Feng, J., Yang, L., Luo, K., Fu, H., et al. (2023). De novo biosynthesis of butyl butyrate in engineered Clostridium tyrobutyricum. Metab. Eng. 77, 64–75. doi:10.1016/J.YMBEN.2023.03.009
Guo, Y., Dai, L., Xin, B., Tao, F., Tang, H., Shen, Y., et al. (2017). 1,3-Propanediol production by a newly isolated strain, Clostridium perfringens GYL. Bioresour. Technol. 233, 406–412. doi:10.1016/j.biortech.2017.02.116
Hakizimana, O., Matabaro, E., and Lee, B. H. (2020). The current strategies and parameters for the enhanced microbial production of 2,3-butanediol. Biotechnol. Rep. 25, e00397. doi:10.1016/j.btre.2019.e00397
Hannon, J. R., Lynd, L. R., Andrade, O., Benavides, P. T., Beckham, G. T., Biddy, M. J., et al. (2020). Technoeconomic and life-cycle analysis of single-step catalytic conversion of wet ethanol into fungible fuel blendstocks. Proc. Natl. Acad. Sci. U. S. A. 117, 12576–12583. doi:10.1073/pnas.1821684116
Hasegawa, S., Jojima, T., Suda, M., and Inui, M. (2020). Isobutanol production in Corynebacterium glutamicum: suppressed succinate by-production by pckA inactivation and enhanced productivity via the Entner–Doudoroff pathway. Metab. Eng. 59, 24–35. doi:10.1016/j.ymben.2020.01.004
Hazelwood, L. A., Daran, J. M., Van Maris, A. J. A., Pronk, J. T., and Dickinson, J. R. (2008). The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266. doi:10.1128/AEM.02625-07
He, Q., Hemme, C. L., Jiang, H., He, Z., and Zhou, J. (2011). Mechanisms of enhanced cellulosic bioethanol fermentation by co-cultivation of Clostridium and Thermoanaerobacter spp. Bioresour. Technol. 102, 9586–9592. doi:10.1016/j.biortech.2011.07.098
Heipieper, H. J., Neumann, G., Cornelissen, S., and Meinhardt, F. (2007). Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74, 961–973. doi:10.1007/s00253-006-0833-4
Herring, C. D., Kenealy, W. R., Joe Shaw, A., Covalla, S. F., Olson, D. G., Zhang, J., et al. (2016). Strain and bioprocess improvement of a thermophilic anaerobe for the production of ethanol from wood. Biotechnol. Biofuels 9, 125. doi:10.1186/s13068-016-0536-8
Higashide, W., Li, Y., Yang, Y., and Liao, J. C. (2011). Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl. Environ. Microbiol. 77, 2727–2733. doi:10.1128/AEM.02454-10
Himmel, M. E., Ding, S. Y., Johnson, D. K., Adney, W. S., Nimlos, M. R., Brady, J. W., et al. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807. doi:10.1126/science.1137016
Holwerda, E. K., Olson, D. G., Ruppertsberger, N. M., Stevenson, D. M., Murphy, S. J. L., Maloney, M. I., et al. (2020). Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production. Biotechnol. Biofuels 13, 40. doi:10.1186/s13068-020-01680-5
Holwerda, E. K., Thorne, P. G., Olson, D. G., Amador-Noguez, D., Engle, N. L., Tschaplinski, T. J., et al. (2014). The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol. Biofuels 7, 155. doi:10.1186/s13068-014-0155-1
Hon, S., Holwerda, E. K., Worthen, R. S., Maloney, M. I., Tian, L., Cui, J., et al. (2018). Expressing the Thermoanaerobacterium saccharolyticum pforA in engineered Clostridium thermocellum improves ethanol production. Biotechnol. Biofuels 11, 242. doi:10.1186/s13068-018-1245-2
Hon, S., Jacobson, T., Stevenson, D. M., Maloney, M. I., Giannone, R. J., Hettich, R. L., et al. (2022). Increasing the thermodynamic driving force of the phosphofructokinase reaction in Clostridium thermocellum. Appl. Environ. Microbiol. 88, e0125822. doi:10.1128/aem.01258-22
Hon, S., Olson, D. G., Holwerda, E. K., Lanahan, A. A., Murphy, S. J. L., Maloney, M. I., et al. (2017). The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab. Eng. 42, 175–184. doi:10.1016/j.ymben.2017.06.011
Horton, C. E., and Bennett, G. N. (2006). Ester production in E. coli and C. acetobutylicum. Enzyme Microb. Technol. 38, 937–943. doi:10.1016/J.ENZMICTEC.2005.08.025
Hu, L., Huang, H., Yuan, H., Tao, F., Xie, H., and Wang, S. (2016). Rex in Clostridium kluyveri is a global redox-sensing transcriptional regulator. J. Biotechnol. 233, 17–25. doi:10.1016/j.jbiotec.2016.06.024
Huang, H., Liu, H., and Gan, Y. R. (2010). Genetic modification of critical enzymes and involved genes in butanol biosynthesis from biomass. Biotechnol. Adv. 28, 651–657. doi:10.1016/j.biotechadv.2010.05.015
Husaini, C. U. N., Roslan, R., Ramzi, A. B., Luthfi, A. A. I., Tan, J. P., Lim, S. S., et al. (2023). The CRISPR technology: a promising strategy for improving dark fermentative biohydrogen production using Clostridium spp. Int. J. Hydrogen Energy 48, 23498–23515. doi:10.1016/J.IJHYDENE.2023.03.162
Iddar, A., Valverde, F., Serrano, A., and Soukri, A. (2002). Expression, purification, and characterization of recombinant nonphosphorylating NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Clostridium acetobutylicum. Protein Expr. Purif. 25, 519–526. doi:10.1016/S1046-5928(02)00032-3
Ingvadottir, E. M., Scully, S. M., and Orlygsson, J. (2018). Production of (S)-1,2-propanediol from L-rhamnose using the moderately thermophilic Clostridium strain AK1. Anaerobe 54, 26–30. doi:10.1016/j.anaerobe.2018.07.003
Inokuma, K., Liao, J. C., Okamoto, M., and Hanai, T. (2010). Improvement of isopropanol production by metabolically engineered Escherichia coli using gas stripping. J. Biosci. Bioeng. 110, 696–701. doi:10.1016/j.jbiosc.2010.07.010
International Sugar Organization (2019). Daily sugar prices | international sugar organization. Available at: https://www.isosugar.org/prices.php (Accessed July 27, 2021).
Islam, R., Sparling, R., Cicek, N., and Levin, D. B. (2015). Optimization of influential nutrients during direct cellulose fermentation into hydrogen by Clostridium thermocellum. Int. J. Mol. Sci. 16, 3116–3132. doi:10.3390/IJMS16023116
Jacobson, T. B., Korosh, T. K., Stevenson, D. M., Foster, C., Maranas, C., Olson, D. G., et al. (2020). In vivo thermodynamic analysis of glycolysis in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum using 13 C and 2 H tracers. mSystems 5, e00736-19. doi:10.1128/msystems.00736-19
Jang, Y. S., Kim, W. J., Im, J. A., Palaniswamy, S., Yao, Z., Lee, H. L., et al. (2023). Efforts to install a heterologous Wood-Ljungdahl pathway in Clostridium acetobutylicum enable the identification of the native tetrahydrofolate (THF) cycle and result in early induction of solvents. Metab. Eng. 77, 188–198. doi:10.1016/j.ymben.2023.04.005
Jarboe, L. R., Royce, L. A., and Liu, P. (2013). Understanding biocatalyst inhibition by carboxylic acids. Front. Microbiol. 0, 272. doi:10.3389/FMICB.2013.00272
Jem, K. J., van der Pol, J. F., and de Vos, S. (2010). “Microbial lactic acid, its polymer poly(lactic acid), and their industrial applications,” in Plastics from bacteria. Editors Q. Chen, and G. Guo (Berlin, Heidelberg: Springer), 323–346. doi:10.1007/978-3-642-03287-5_13
Jermy, B. R., and Pandurangan, A. (2005). A highly efficient catalyst for the esterification of acetic acid using n-butyl alcohol. J. Mol. Catal. A Chem. 237, 146–154. doi:10.1016/j.molcata.2005.04.034
Jiang, H. L., He, Q., He, Z., Hemme, C. L., Wu, L., and Zhou, J. (2013). Continuous cellulosic bioethanol fermentation by cyclic fed-batch cocultivation. Appl. Environ. Microbiol. 79, 1580–1589. doi:10.1128/AEM.02617-12
Jiang, L., Wang, J., Liang, S., Cai, J., Xu, Z., Cen, P., et al. (2011). Enhanced butyric acid tolerance and bioproduction by Clostridium tyrobutyricum immobilized in a fibrous bed bioreactor. Biotechnol. Bioeng. 108, 31–40. doi:10.1002/bit.22927
Jiang, Y., Liu, J., Jiang, W., Yang, Y., and Yang, S. (2015). Current status and prospects of industrial bio-production of n-butanol in China. Biotechnol. Adv. 33, 1493–1501. doi:10.1016/j.biotechadv.2014.10.007
Jiang, Y., Lv, Y., Wu, R., Lu, J., Dong, W., Zhou, J., et al. (2020). Consolidated bioprocessing performance of a two-species microbial consortium for butanol production from lignocellulosic biomass. Biotechnol. Bioeng. 117, 2985–2995. doi:10.1002/bit.27464
Jiang, Y., Xu, C., Dong, F., Yang, Y., Jiang, W., and Yang, S. (2009). Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 11, 284–291. doi:10.1016/j.ymben.2009.06.002
Jones, A. J., Venkataramanan, K. P., and Papoutsakis, T. (2016). Overexpression of two stress-responsive, small, non-coding RNAs, 6S and tmRNA, imparts butanol tolerance in Clostridium acetobutylicum. FEMS Microbiol. Lett. 363, fnw063. doi:10.1093/femsle/fnw063
Jones, D. T., and Woods, D. R. (1986). Acetone-butanol fermentation revisited. Microbiol. Rev. 50, 484–524. doi:10.1128/mr.50.4.484-524.1986
Julkipli, J., Babel, S., Bilyaminu, A. M., and Rene, E. R. (2023). Hydrogen and biodiesel production from food waste: a review. Environ. Chem. Lett. 1, 585–607. doi:10.1007/S10311-023-01674-3
Keis, S., Shaheen, R., and Jones, D. T. (2001). Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2095–2103. doi:10.1099/00207713-51-6-2095
Kim, S., Jang, Y. S., Ha, S. C., Ahn, J. W., Kim, E. J., Hong Lim, J., et al. (2015). Redox-switch regulatory mechanism of thiolase from Clostridium acetobutylicum. Nat. Commun. 6, 8410–8411. doi:10.1038/ncomms9410
Kim, S. H., Hwang, J. H., Kim, H. J., Oh, S. J., Kim, H. J., Shin, N., et al. (2023). Enhancement of biohydrogen production in Clostridium acetobutylicum ATCC 824 by overexpression of glyceraldehyde-3-phosphate dehydrogenase gene. Enzyme Microb. Technol. 168, 110244. doi:10.1016/j.enzmictec.2023.110244
Kindra, V., Maksimov, I., Oparin, M., Zlyvko, O., and Rogalev, A. (2023). Hydrogen technologies: a critical review and feasibility study. Energies 16, 5482. doi:10.3390/en16145482
Klotz, S., Kuenz, A., and Prüße, U. (2017). Nutritional requirements and the impact of yeast extract on the d-lactic acid production by Sporolactobacillus inulinus. Green Chem. 19, 4633–4641. doi:10.1039/c7gc01796k
Koendjbiharie, J. G., Hon, S., Pabst, M., Hooftman, R., Stevenson, D. M., Cui, J., et al. (2020). The pentose phosphate pathway of cellulolytic clostridia relies on 6-phosphofructokinase instead of transaldolase. J. Biol. Chem. 295, 1867–1878. doi:10.1074/JBC.RA119.011239
Kojima, M., Okamoto, K., and Yanase, H. (2013). Direct ethanol production from cellulosic materials by Zymobacter palmae carrying Cellulomonas endoglucanase and Ruminococcus β-glucosidase genes. Appl. Microbiol. Biotechnol. 97, 5137–5147. doi:10.1007/s00253-013-4874-1
Köpke, M., Held, C., Hujer, S., Liesegang, H., Wiezer, A., Wollherr, A., et al. (2010). Clostridium ljungdahlii represents a microbial production platform based on syngas. Proc. Natl. Acad. Sci. U. S. A. 107, 13087–13092. doi:10.1073/PNAS.1004716107
Köpke, M., Mihalcea, C., Liew, F. M., Tizard, J. H., Ali, M. S., Conolly, J. J., et al. (2011). 2,3-Butanediol production by acetogenic bacteria, an alternative route to chemical synthesis, using industrial waste gas. Appl. Environ. Microbiol. 77, 5467–5475. doi:10.1128/AEM.00355-11
Kottenhahn, P., Philipps, G., and Jennewein, S. (2021). Hexanol biosynthesis from syngas by Clostridium carboxidivorans P7 – product toxicity, temperature dependence and in situ extraction. Heliyon 7, e07732. doi:10.1016/j.heliyon.2021.e07732
Koutinas, A. A., Yepez, B., Kopsahelis, N., Freire, D. M. G., de Castro, A. M., Papanikolaou, S., et al. (2016). Techno-economic evaluation of a complete bioprocess for 2,3-butanediol production from renewable resources. Bioresour. Technol. 204, 55–64. doi:10.1016/j.biortech.2015.12.005
Kovács, K., Willson, B. J., Schwarz, K., Heap, J. T., Jackson, A., Bolam, D. N., et al. (2013). Secretion and assembly of functional mini-cellulosomes from synthetic chromosomal operons in Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 6, 117. doi:10.1186/1754-6834-6-117
Kruis, A. J., Bohnenkamp, A. C., Patinios, C., van Nuland, Y. M., Levisson, M., Mars, A. E., et al. (2019). Microbial production of short and medium chain esters: enzymes, pathways, and applications. Biotechnol. Adv. 37, 107407. doi:10.1016/j.biotechadv.2019.06.006
Kruis, A. J., Gallone, B., Jonker, T., Mars, A. E., van Rijswijck, I. M. H., Wolkers–Rooijackers, J. C. M., et al. (2018). Contribution of Eat1 and other alcohol acyltransferases to ester production in Saccharomyces cerevisiae. Front. Microbiol. 9, 3202. doi:10.3389/fmicb.2018.03202
Kruis, A. J., Levisson, M., Mars, A. E., van der Ploeg, M., Garcés Daza, F., Ellena, V., et al. (2017). Ethyl acetate production by the elusive alcohol acetyltransferase from yeast. Metab. Eng. 41, 92–101. doi:10.1016/j.ymben.2017.03.004
Kunze, G., and Haehnel, U. (2011). Production of butanol by fermentation in Arxula sp. EP2508597A1. European Patent Office.
Land, H., Senger, M., Berggren, G., and Stripp, S. T. (2020). Current state of [FeFe]-Hydrogenase research: biodiversity and spectroscopic investigations. ACS Catal. 10, 7069–7086. doi:10.1021/ACSCATAL.0C01614
Latifi, A., Avilan, L., and Brugna, M. (2019). Clostridial whole cell and enzyme systems for hydrogen production: current state and perspectives. Appl. Microbiol. Biotechnol. 103, 567–575. doi:10.1007/S00253-018-9514-3
Lauer, I., Philipps, G., and Jennewein, S. (2022a). Metabolic engineering of Clostridium ljungdahlii for the production of hexanol and butanol from CO(2) and H(2). Microb. Cell Fact. 21, 85. doi:10.1186/s12934-022-01802-8
Lauer, I., Philipps, G., and Jennewein, S. (2022b). Metabolic engineering of Clostridium ljungdahlii for the production of hexanol and butanol from CO2 and H2. Microb. Cell Fact. 21, 85–18. doi:10.1186/S12934-022-01802-8
Layton, D. S., and Trinh, C. T. (2016). Expanding the modular ester fermentative pathways for combinatorial biosynthesis of esters from volatile organic acids. Biotechnol. Bioeng. 113, 1764–1776. doi:10.1002/bit.25947
Lee, J., Jang, Y. S., Han, M. J., Kim, J. Y., and Lee, S. Y. (2016). Deciphering Clostridium tyrobutyricum metabolism based on the whole-genome sequence and proteome analyses. MBio 7, e00743-16. doi:10.1128/mBio.00743-16
Lee, J. W., and Trinh, C. T. (2020). Towards renewable flavors, fragrances, and beyond. Curr. Opin. Biotechnol. 61, 168–180. doi:10.1016/j.copbio.2019.12.017
Lee, S. Y., Jang, Y. S., Lee, J. Y., Lee, J., Park, J. H., Im, J. A., et al. (2012). Enhanced butanol production obtained by reinforcing the direct butanol-forming route in Clostridium acetobutylicum. MBio 3, e00314-12. doi:10.1128/mBio.00314-12
Lehmann, D., Hönicke, D., Ehrenreich, A., Schmidt, M., Weuster-Botz, D., Bahl, H., et al. (2012). Modifying the product pattern of Clostridium acetobutylicum: physiological effects of disrupting the acetate and acetone formation pathways. Appl. Microbiol. Biotechnol. 94, 743–754. doi:10.1007/s00253-011-3852-8
Lehmann, D., and Lütke-Eversloh, T. (2011). Switching Clostridium acetobutylicum to an ethanol producer by disruption of the butyrate/butanol fermentative pathway. Metab. Eng. 13, 464–473. doi:10.1016/j.ymben.2011.04.006
Lepage, T., Kammoun, M., Schmetz, Q., and Richel, A. (2021). Biomass-to-hydrogen: a review of main routes production, processes evaluation and techno-economical assessment. Biomass Bioenergy 144, 105920. doi:10.1016/J.BIOMBIOE.2020.105920
Li, A., Wen, Z., Fang, D., Lu, M., Ma, Y., Xie, Q., et al. (2020a). Developing Clostridium diolis as a biorefinery chassis by genetic manipulation. Bioresour. Technol. 305, 123066. doi:10.1016/j.biortech.2020.123066
Li, J., Du, Y., Bao, T., Dong, J., Lin, M., Shim, H., et al. (2019). n-Butanol production from lignocellulosic biomass hydrolysates without detoxification by Clostridium tyrobutyricum Δack-adhE2 in a fibrous-bed bioreactor. Bioresour. Technol. 289, 121749. doi:10.1016/j.biortech.2019.121749
Li, Q., Wu, M., Wen, Z., Jiang, Y., Wang, X., Zhao, Y., et al. (2020b). Optimization of n-butanol synthesis in Lactobacillus brevis via the functional expression of thl, hbd, crt and ter. J. Ind. Microbiol. Biotechnol. 47, 1099–1108. doi:10.1007/s10295-020-02331-2
Li, Q., Zhang, J., Yang, J., Jiang, Y., and Yang, S. (2021). Recent progress on n-butanol production by lactic acid bacteria. World J. Microbiol. Biotechnol. 37, 205. doi:10.1007/s11274-021-03173-5
Li, S., Huang, L., Ke, C., Pang, Z., and Liu, L. (2020c). Pathway dissection, regulation, engineering and application: lessons learned from biobutanol production by solventogenic clostridia. Biotechnol. Biofuels 13, 39–25. doi:10.1186/s13068-020-01674-3
Li, T., Zhang, C., Yang, K. L., and He, J. (2018). Unique genetic cassettes in a Thermoanaerobacterium contribute to simultaneous conversion of cellulose and monosugars into butanol. Sci. Adv. 4, e1701475. doi:10.1126/sciadv.1701475
Liew, F., Henstra, A. M., Kӧpke, M., Winzer, K., Simpson, S. D., and Minton, N. P. (2017). Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab. Eng. 40, 104–114. doi:10.1016/j.ymben.2017.01.007
Lin, P. P., Mi, L., Morioka, A. H., Yoshino, K. M., Konishi, S., Xu, S. C., et al. (2015). Consolidated bioprocessing of cellulose to isobutanol using Clostridium thermocellum. Metab. Eng. 31, 44–52. doi:10.1016/j.ymben.2015.07.001
Liu, C. H., Chang, C. Y., Cheng, C. L., Lee, D. J., and Chang, J. S. (2012a). Fermentative hydrogen production by Clostridium butyricum CGS5 using carbohydrate-rich microalgal biomass as feedstock. Int. J. Hydrogen Energy 37, 15458–15464. doi:10.1016/j.ijhydene.2012.04.076
Liu, D., Liu, D., Zeng, R. J., and Angelidaki, I. (2006). Hydrogen and methane production from household solid waste in the two-stage fermentation process. Water Res. 40, 2230–2236. doi:10.1016/j.watres.2006.03.029
Liu, X. B., Gu, Q. Y., and Yu, X. B. (2013). Repetitive domestication to enhance butanol tolerance and production in Clostridium acetobutylicum through artificial simulation of bio-evolution. Bioresour. Technol. 130, 638–643. doi:10.1016/j.biortech.2012.12.121
Liu, X. B., Gu, Q. Y., Yu, X. B., and Luo, W. (2012b). Enhancement of butanol tolerance and butanol yield in Clostridium acetobutylicum mutant NT642 obtained by nitrogen ion beam implantation. J. Microbiol. 50, 1024–1028. doi:10.1007/s12275-012-2289-9
Lo, J., Olson, D. G., Murphy, S. J. L., Tian, L., Hon, S., Lanahan, A., et al. (2017). Engineering electron metabolism to increase ethanol production in Clostridium thermocellum. Metab. Eng. 39, 71–79. doi:10.1016/j.ymben.2016.10.018
Lo, J., Zheng, T., Hon, S., Olson, D. G., and Lynd, L. R. (2015). The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J. Bacteriol. 197, 1386–1393. doi:10.1128/JB.02450-14
Lu, H., Wang, L., Li, S., Pan, C., Cheng, K., Luo, Y., et al. (2019). Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO. Nucleic Acids Res. 47, 9925–9933. doi:10.1093/nar/gkz720
Łukajtis, R., Hołowacz, I., Kucharska, K., Glinka, M., Rybarczyk, P., Przyjazny, A., et al. (2018). Hydrogen production from biomass using dark fermentation. Renew. Sustain. Energy Rev. 91, 665–694. doi:10.1016/j.rser.2018.04.043
Lynd, L. R. (2017). The grand challenge of cellulosic biofuels. Nat. Biotechnol. 35, 912–915. doi:10.1038/nbt.3976
Lynd, L. R., Liang, X., Biddy, M. J., Allee, A., Cai, H., Foust, T., et al. (2017). Cellulosic ethanol: status and innovation. Curr. Opin. Biotechnol. 45, 202–211. doi:10.1016/j.copbio.2017.03.008
Ma, C., Wang, A., Qin, J., Li, L., Ai, X., Jiang, T., et al. (2009). Enhanced 2,3-butanediol production by Klebsiella pneumoniae SDM. Appl. Microbiol. Biotechnol. 82, 49–57. doi:10.1007/s00253-008-1732-7
Machado, H. B., Dekishima, Y., Luo, H., Lan, E. I., and Liao, J. C. (2012). A selection platform for carbon chain elongation using the CoA-dependent pathway to produce linear higher alcohols. Metab. Eng. 14, 504–511. doi:10.1016/j.ymben.2012.07.002
Mailaram, S., Narisetty, V., Ranade, V. V., Kumar, V., and Maity, S. K. (2022). Techno-economic analysis for the production of 2,3-butanediol from brewers’ spent grain using pinch technology. Ind. Eng. Chem. Res. 61, 2195–2205. doi:10.1021/acs.iecr.1c04410
Mandal, B., Nath, K., and Das, D. (2006). Improvement of biohydrogen production under decreased partial pressure of H2 by Enterobacter cloacae. Biotechnol. Lett. 28, 831–835. doi:10.1007/s10529-006-9008-8
Mann, M. S., and Lütke-Eversloh, T. (2013). Thiolase engineering for enhanced butanol production in Clostridium acetobutylicum. Biotechnol. Bioeng. 110, 887–897. doi:10.1002/bit.24758
Marcano-Velazquez, J. G., Lo, J., Nag, A., Maness, P. C., and Chou, K. J. (2019). Developing riboswitch-mediated gene regulatory controls in thermophilic bacteria. ACS Synth. Biol. 8, 633–640. doi:10.1021/acssynbio.8b00487
MarketsandMarkets (2024). Global lactic acid market size, trends, advantages, and forecast report - 2028. Available at: https://www.marketsandmarkets.com/Market-Reports/polylacticacid-387.html?gclid=Cj0KCQjw6s2IBhCnARIsAP8RfAgISU79DUg8my_rRKY3ly98GZIdnJuh6gogfiiwScOqI9j0iRsPWmIaAvs-EALw_wcB (Accessed April 16, 2024).
Mazzoli, R. (2012). Development of microorganisms for cellulose-biofuel consolidated bioprocessings: metabolic engineers’ tricks. Comput. Struct. Biotechnol. J. 3, e201210007. doi:10.5936/csbj.201210007
Mazzoli, R. (2020). Metabolic engineering strategies for consolidated production of lactic acid from lignocellulosic biomass. Biotechnol. Appl. Biochem. 67, 61–72. doi:10.1002/bab.1869
Mazzoli, R. (2021). Current progress in production of building-block organic acids by consolidated bioprocessing of lignocellulose. Fermentation 7, 248. doi:10.3390/fermentation7040248
Mazzoli, R., and Olson, D. G. (2020). Clostridium thermocellum: a microbial platform for high-value chemical production from lignocellulose. Adv. Appl. Microbiol. 113, 111–161. doi:10.1016/bs.aambs.2020.07.004
Mazzoli, R., Olson, D. G., Concu, A. M., Holwerda, E. K., and Lynd, L. R. (2022). In vivo evolution of lactic acid hyper-tolerant Clostridium thermocellum. N. Biotechnol. 67, 12–22. doi:10.1016/j.nbt.2021.12.003
Mazzoli, R., Olson, D. G., and Lynd, L. R. (2020). Construction of lactic acid overproducing Clostridium thermocellum through enhancement of lactate dehydrogenase expression. Enzyme Microb. Technol. 141, 109645. doi:10.1016/j.enzmictec.2020.109645
Mazzoli, R., Pescarolo, S., Gilli, G., Gilardi, G., and Valetti, F. (2024). Hydrogen production pathways in Clostridia and their improvement by metabolic engineering. Biotechnol. Adv. 73, 108379. doi:10.1016/j.biotechadv.2024.108379
Mendes, F. S., González-Pajuelo, M., Cordier, H., François, J. M., and Vasconcelos, I. (2011). 1,3-Propanediol production in a two-step process fermentation from renewable feedstock. Appl. Microbiol. Biotechnol. 92, 519–527. doi:10.1007/s00253-011-3369-1
Mendez, B. S., Pettinari, M. J., Ivanier, S. E., Ramos, C. A., and Sineriz, F. (1991). Clostridium thermopapyrolyticum sp. nov., a cellulolytic thermophile. Int. J. Syst. Bacteriol. 41, 281–283. doi:10.1099/00207713-41-2-281
Mingardon, F., Chanal, A., Tardif, C., and Fierobe, H. P. (2011). The issue of secretion in heterologous expression of Clostridium cellulolyticum cellulase-encoding genes in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 77, 2831–2838. doi:10.1128/AEM.03012-10
Minton, N. P., Ehsaan, M., Humphreys, C. M., Little, G. T., Baker, J., Henstra, A. M., et al. (2016). A roadmap for gene system development in Clostridium. Anaerobe 41, 104–112. doi:10.1016/j.anaerobe.2016.05.011
Minty, J. J., Singer, M. E., Scholz, S. A., Bae, C. H., Ahn, J. H., Foster, C. E., et al. (2013). Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc. Natl. Acad. Sci. U. S. A. 110, 14592–14597. doi:10.1073/pnas.1218447110
Morra, S. (2022). Fantastic [FeFe]-Hydrogenases and where to find them. Front. Microbiol. 13, 853626. doi:10.3389/fmicb.2022.853626
Mudhoo, A., Forster-Carneiro, T., and Sánchez, A. (2011). Biohydrogen production and bioprocess enhancement: a review. Crit. Rev. Biotechnol. 31, 250–263. doi:10.3109/07388551.2010.525497
Müller, V. (2003). Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 69, 6345–6353. doi:10.1128/AEM.69.11.6345-6353.2003
Nakamura, C. E., and Whited, G. M. (2003). Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459. doi:10.1016/j.copbio.2003.08.005
Nakayama, S. I., Kosaka, T., Hirakawa, H., Matsuura, K., Yoshino, S., and Furukawa, K. (2008). Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1-4. Appl. Microbiol. Biotechnol. 78, 483–493. doi:10.1007/s00253-007-1323-z
Nawab, S., Wang, N., Ma, X., and Huo, Y. X. (2020). Genetic engineering of non-native hosts for 1-butanol production and its challenges: a review. Microb. Cell Fact. 19, 79–16. doi:10.1186/s12934-020-01337-w
Nawab, S., Zhang, Y. F., Ullah, M. W., Lodhi, A. F., Shah, S. B., Rahman, M. U., et al. (2024). Microbial host engineering for sustainable isobutanol production from renewable resources. Appl. Microbiol. Biotechnol. 108, 33–18. doi:10.1007/s00253-023-12821-9
Nguyen, N. P. T., Raynaud, C., Meynial-Salles, I., and Soucaille, P. (2018). Reviving the Weizmann process for commercial n-butanol production. Nat. Commun. 9, 3682. doi:10.1038/s41467-018-05661-z
Nicolaou, S. A., Gaida, S. M., and Papoutsakis, E. T. (2010). A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 12, 307–331. doi:10.1016/j.ymben.2010.03.004
Nimbalkar, P. R., and Dharne, M. S. (2024). A review on microbial 1, 3-propanediol production: emerging strategies, key hurdles and attainable solutions to re-establish its commercial interest. Ind. Crops Prod. 209, 117961. doi:10.1016/j.indcrop.2023.117961
Nirmala, N., Praveen, G., AmitKumar, S., SundarRajan, P., Baskaran, A., Priyadharsini, P., et al. (2023). A review on biological biohydrogen production: outlook on genetic strain enhancements, reactor model and techno-economics analysis. Sci. Total Environ. 896, 165143. doi:10.1016/j.scitotenv.2023.165143
Niu, W., Kramer, L., Mueller, J., Liu, K., and Guo, J. (2019). Metabolic engineering of Escherichia coli for the de novo stereospecific biosynthesis of 1,2-propanediol through lactic acid. Metab. Eng. Commun. 8, e00082–e00089. doi:10.1016/j.mec.2018.e00082
Noh, H. J., Lee, S. Y., and Jang, Y. S. (2019). Microbial production of butyl butyrate, a flavor and fragrance compound. Appl. Microbiol. Biotechnol. 103, 2079–2086. doi:10.1007/s00253-018-09603-z
Noh, H. J., Woo, J. E., Lee, S. Y., and Jang, Y. S. (2018). Metabolic engineering of Clostridium acetobutylicum for the production of butyl butyrate. Appl. Microbiol. Biotechnol. 102, 8319–8327. doi:10.1007/s00253-018-9267-z
Nuss, P., and Gardner, K. H. (2013). Attributional life cycle assessment (ALCA) of polyitaconic acid production from northeast US softwood biomass. Int. J. Life Cycle Assess. 18, 603–612. doi:10.1007/s11367-012-0511-y
Oh, H. J., Gong, G., Ahn, J. H., Ko, J. K., Lee, S. M., and Um, Y. (2023). Effective hexanol production from carbon monoxide using extractive fermentation with Clostridium carboxidivorans P7. Bioresour. Technol. 367, 128201. doi:10.1016/j.biortech.2022.128201
Oh, H. J., Ko, J. K., Gong, G., Lee, S. M., and Um, Y. (2022). Production of hexanol as the main product through syngas fermentation by Clostridium carboxidivorans P7. Front. Bioeng. Biotechnol. 10, 850370. doi:10.3389/fbioe.2022.850370
Olson, D. G., Maloney, M. I., Lanahan, A. A., Cervenka, N. D., Xia, Y., Pech-Canul, A., et al. (2023). Ethanol tolerance in engineered strains of Clostridium thermocellum. Biotechnol. Biofuels Bioprod. 16, 137. doi:10.1186/s13068-023-02379-z
Olson, D. G., Sparling, R., and Lynd, L. R. (2015). Ethanol production by engineered thermophiles. Curr. Opin. Biotechnol. 33, 130–141. doi:10.1016/j.copbio.2015.02.006
Ortigueira, J., Alves, L., Gouveia, L., and Moura, P. (2015a). Third generation biohydrogen production by Clostridium butyricum and adapted mixed cultures from Scenedesmus obliquus microalga biomass. Fuel 153, 128–134. doi:10.1016/J.FUEL.2015.02.093
Ortigueira, J., Pinto, T., Gouveia, L., and Moura, P. (2015b). Production and storage of biohydrogen during sequential batch fermentation of Spirogyra hydrolyzate by Clostridium butyricum. Energy 88, 528–536. doi:10.1016/j.energy.2015.05.070
Otte, B., Grunwaldt, E., Mahmoud, O., and Jennewein, S. (2009). Genome shuffling in Clostridium diolis DSM 15410 for improved 1,3-propanediol production. Appl. Environ. Microbiol. 75, 7610–7616. doi:10.1128/AEM.01774-09
Palsson, B., and Zengler, K. (2010). The challenges of integrating multi-omic data sets. Nat. Chem. Biol. 6, 787–789. doi:10.1038/nchembio.462
Panesar, P. S., Marwaha, S. S., and Kennedy, J. F. (2006). Zymomonas mobilis: an alternative ethanol producer. J. Chem. Technol. Biotechnol. 81, 623–635. doi:10.1002/jctb.1448
Papanek, B., Biswas, R., Rydzak, T., and Guss, A. M. (2015). Elimination of metabolic pathways to all traditional fermentation products increases ethanol yields in Clostridium thermocellum. Metab. Eng. 32, 49–54. doi:10.1016/j.ymben.2015.09.002
Parthiba Karthikeyan, O., Trably, E., Mehariya, S., Bernet, N., Wong, J. W. C., and Carrere, H. (2018). Pretreatment of food waste for methane and hydrogen recovery: a review. Bioresour. Technol. 249, 1025–1039. doi:10.1016/j.biortech.2017.09.105
Peralta-Yahya, P. P., and Keasling, J. D. (2010). Advanced biofuel production in microbes. Biotechnol. J. 5, 147–162. doi:10.1002/biot.200900220
Petit, E., LaTouf, W. G., Coppi, M. V., Warnick, T. A., Currie, D., Romashko, I., et al. (2013). Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 8, e54337. doi:10.1371/journal.pone.0054337
Phillips, J. R., Klasson, K. T., Clausen, E. C., and Gaddy, J. L. (1993). Biological production of ethanol from coal synthesis gas - medium development studies. Appl. Biochem. Biotechnol. 39–40, 559–571. doi:10.1007/bf02919018
Poudel, P., Tashiro, Y., and Sakai, K. (2016). New application of Bacillus strains for optically pure L-lactic acid production: general overview and future prospects. Biosci. Biotechnol. Biochem. 80, 642–654. doi:10.1080/09168451.2015.1095069
Qi, F., Thakker, C., Zhu, F., Pena, M., San, K. Y., and Bennett, G. N. (2018). Improvement of butanol production in Clostridium acetobutylicum through enhancement of NAD(P)H availability. J. Ind. Microbiol. Biotechnol. 45, 993–1002. doi:10.1007/S10295-018-2068-7
Qin, Z., Duns, G. J., Pan, T., and Xin, F. (2018). Consolidated processing of biobutanol production from food wastes by solventogenic Clostridium sp. strain HN4. Bioresour. Technol. 264, 148–153. doi:10.1016/j.biortech.2018.05.076
Qureshi, N., Lin, X., Liu, S., Saha, B. C., Mariano, A. P., Polaina, J., et al. (2020). Global view of biofuel butanol and economics of its production by fermentation from sweet sorghum bagasse, food waste, and yellow top presscake: application of novel technologies. Fermentation 6, 58. doi:10.3390/FERMENTATION6020058
Ramprakash, B., Lindblad, P., Eaton-Rye, J. J., and Incharoensakdi, A. (2022). Current strategies and future perspectives in biological hydrogen production: a review. Renew. Sustain. Energy Rev. 168, 112773. doi:10.1016/J.RSER.2022.112773
Ravcheev, D. A., Li, X., Latif, H., Zengler, K., Leyn, S. A., Korostelev, Y. D., et al. (2012). Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor rex. J. Bacteriol. 194, 1145–1157. doi:10.1128/JB.06412-11
Rawoof, S. A. A., Kumar, P. S., Vo, D. V. N., Devaraj, K., Mani, Y., Devaraj, T., et al. (2021). Production of optically pure lactic acid by microbial fermentation: a review. Environ. Chem. Lett. 19, 539–556. doi:10.1007/S10311-020-01083-W
Raynaud, C., Sarçabal, P., Meynial-Salles, I., Croux, C., and Soucaille, P. (2003). Molecular characterization of the 1,3-propanediol (1,3-PD) operon of Clostridium butyricum. Proc. Natl. Acad. Sci. U. S. A. 100, 5010–5015. doi:10.1073/pnas.0734105100
Re, A., and Mazzoli, R. (2023). Current progress on engineering microbial strains and consortia for production of cellulosic butanol through consolidated bioprocessing. Microb. Biotechnol. 16, 238–261. doi:10.1111/1751-7915.14148
Ricci, L., Agostino, V., Fino, D., and Re, A. (2021). Screening of gas substrate and medium effects on 2,3-butanediol production with C. Ljungdahlii and C. autoethanogenum aided by improved autotrophic cultivation technique. Fermentation 7, 264. doi:10.3390/fermentation7040264
Richter, H., Molitor, B., Diender, M., Sousa, D. Z., and Angenent, L. T. (2016). A narrow pH range supports butanol, hexanol, and octanol production from syngas in a continuous co-culture of Clostridium ljungdahlii and Clostridium kluyveri with in-line product extraction. Front. Microbiol. 7, 1773. doi:10.3389/fmicb.2016.01773
Rodriguez, G. M., Tashiro, Y., and Atsumi, S. (2014). Expanding ester biosynthesis in Escherichia coli. Nat. Chem. Biol. 10, 259–265. doi:10.1038/nchembio.1476
Russmayer, H., Egermeier, M., Kalemasi, D., and Sauer, M. (2019). Spotlight on biodiversity of microbial cell factories for glycerol conversion. Biotechnol. Adv. 37, 107395. doi:10.1016/j.biotechadv.2019.05.001
Rydzak, T., Lynd, L. R., and Guss, A. M. (2015). Elimination of formate production in Clostridium thermocellum. J. Ind. Microbiol. Biotechnol. 42, 1263–1272. doi:10.1007/s10295-015-1644-3
Sánchez-Riera, F., Cameron, D. C., and Cooney, C. L. (1987). Influence of environmental factors in the production of R(-)-1, 2-propanediol by clostridium thermosaccharolyticum. Biotechnol. Lett. 9, 449–454. doi:10.1007/BF01027450
Sander, K., Chung, D., Hyatt, D., Westpheling, J., Klingeman, D. M., Rodriguez, M., et al. (2019). Rex in Caldicellulosiruptor bescii: novel regulon members and its effect on the production of ethanol and overflow metabolites. Microbiologyopen 8, e00639. doi:10.1002/mbo3.639
Sarma, S., Ortega, D., Minton, N. P., Dubey, V. K., and Moholkar, V. S. (2019). Homologous overexpression of hydrogenase and glycerol dehydrogenase in Clostridium pasteurianum to enhance hydrogen production from crude glycerol. Bioresour. Technol. 284, 168–177. doi:10.1016/j.biortech.2019.03.074
Sauer, M., Porro, D., Mattanovich, D., and Branduardi, P. (2008). Microbial production of organic acids: expanding the markets. Trends Biotechnol. 26, 100–108. doi:10.1016/j.tibtech.2007.11.006
Schoch, T., Baur, T., Kunz, J., Stöferle, S., and Dürre, P. (2023). Heterologous 1,3-propanediol production using different recombinant Clostridium beijerinckii DSM 6423 strains. Microorganisms 11, 784. doi:10.3390/microorganisms11030784
Schuchmann, K., and Müller, V. (2014). Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nat. Rev. Microbiol. 12, 809–821. doi:10.1038/nrmicro3365
Schwarz, K. M., Grosse-Honebrink, A., Derecka, K., Rotta, C., Zhang, Y., and Minton, N. P. (2017). Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab. Eng. 40, 124–137. doi:10.1016/j.ymben.2017.01.009
Scott, I. M., Rubinstein, G. M., Poole, F. L., Lipscomb, G. L., Schut, G. J., Williams-Rhaesa, A. M., et al. (2019). The thermophilic biomass-degrading bacterium Caldicellulosiruptor bescii utilizes two enzymes to oxidize glyceraldehyde 3-phosphate during glycolysis. J. Biol. Chem. 294, 9995–10005. doi:10.1074/JBC.RA118.007120
Seo, H., Lee, J. W., Giannone, R. J., Dunlap, N. J., and Trinh, C. T. (2021). Engineering promiscuity of chloramphenicol acetyltransferase for microbial designer ester biosynthesis. Metab. Eng. 66, 179–190. doi:10.1016/J.YMBEN.2021.04.005
Seo, H., Singh, P., Wyman, C. E., Cai, C. M., and Trinh, C. T. (2023). Rewiring metabolism of Clostridium thermocellum for consolidated bioprocessing of lignocellulosic biomass poplar to produce short-chain esters. Bioresour. Technol. 384, 129263. doi:10.1016/j.biortech.2023.129263
Shao, X. J., Raman, B., Zhu, M. J., Mielenz, J. R., Brown, S. D., Guss, A. M., et al. (2011). Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 92, 641–652. doi:10.1007/s00253-011-3492-z
Shaw, A. J., Podkaminer, K. K., Desai, S. G., Bardsley, J. S., Rogers, S. R., Thorne, P. G., et al. (2008). Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc. Natl. Acad. Sci. U. S. A. 105, 13769–13774. doi:10.1073/PNAS.0801266105
Shen, C. R., Lan, E. I., Dekishima, Y., Baez, A., Cho, K. M., and Liao, J. C. (2011). Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 77, 2905–2915. doi:10.1128/AEM.03034-10
Shen, C. R., and Liao, J. C. (2013). Synergy as design principle for metabolic engineering of 1-propanol production in Escherichia coli. Metab. Eng. 17, 12–22. doi:10.1016/j.ymben.2013.01.008
Shen, S., Gu, Y., Chai, C., Jiang, W., Zhuang, Y., and Wang, Y. (2017). Enhanced alcohol titre and ratio in carbon monoxide-rich off-gas fermentation of Clostridium carboxidivorans through combination of trace metals optimization with variable-temperature cultivation. Bioresour. Technol. 239, 236–243. doi:10.1016/j.biortech.2017.04.099
Shrirame, B. S., Varma, A. R., Sahoo, S. S., Gayen, K., and Maity, S. K. (2023). Techno-commercial viability of glycerol valorization to 1,2- and 1,3-propanediol using pinch technology. Biomass Bioenergy 177, 106943. doi:10.1016/j.biombioe.2023.106943
Siebert, D., and Wendisch, V. F. (2015). Metabolic pathway engineering for production of 1,2-propanediol and 1-propanol by Corynebacterium glutamicum. Biotechnol. Biofuels 8, 91–13. doi:10.1186/s13068-015-0269-0
Siemerink, M. A. J., Kuit, W., Contreras, A. M. L., Eggink, G., van der Oost, J., and Kengen, S. W. M. (2011). D-2,3-butanediol production due to heterologous expression of an acetoin reductase in Clostridium acetobutylicum. Appl. Environ. Microbiol. 77, 2582–2588. doi:10.1128/AEM.01616-10
Simpson, S. D., Collet, C., Mihalcea, C. D., Conolly, J. J., and Waters, G. W. (2014). Fermentation process for controlling butanediol production. Available at: https://patents.google.com/patent/US8673603B2/en (Accessed April 10, 2024).
Simpson, S. D., Tran, P. L., Mihalcea, C. D., Fung, J. M. Y., and Liew, F. M. (2009). Production of butanediol by anaerobic microbial fermentation. Available at: https://patents.google.com/patent/WO2009151342A1/enIt.pdf (Accessed April 10, 2024).
Singh, R., Tevatia, R., White, D., Demirel, Y., and Blum, P. (2019). Comparative kinetic modeling of growth and molecular hydrogen overproduction by engineered strains of Thermotoga maritima. Int. J. Hydrogen Energy 44, 7125–7136. doi:10.1016/j.ijhydene.2019.01.124
Singhvi, M., Zendo, T., and Sonomoto, K. (2018). Free lactic acid production under acidic conditions by lactic acid bacteria strains: challenges and future prospects. Appl. Microbiol. Biotechnol. 102, 5911–5924. doi:10.1007/s00253-018-9092-4
Skoog, E., Shin, J. H., Saez-Jimenez, V., Mapelli, V., and Olsson, L. (2018). Biobased adipic acid – the challenge of developing the production host. Biotechnol. Adv. 36, 2248–2263. doi:10.1016/j.biotechadv.2018.10.012
Soltys, K. A., Batta, A. K., and Koneru, B. (2001). Successful nonfreezing, subzero preservation of rat liver with 2,3-butanediol and type I antifreeze protein. J. Surg. Res. 96, 30–34. doi:10.1006/jsre.2000.6053
Son, Y. S., Jeon, J. M., Kim, D. H., Yang, Y. H., Jin, Y. S., Cho, B. K., et al. (2021). Improved bio-hydrogen production by overexpression of glucose-6-phosphate dehydrogenase and FeFe hydrogenase in Clostridium acetobutylicum. Int. J. Hydrogen Energy 46, 36687–36695. doi:10.1016/J.IJHYDENE.2021.08.222
Soucaille, P., Croux, C., and Meynial, I. (2010). Cellulolytic Clostridium acetobutylicum. EP2436698A1 European Patent Office.
Su, G., Chan, C., and He, J. (2022). Enhanced biobutanol production from starch waste via orange peel doping. Renew. Energy 193, 576–583. doi:10.1016/j.renene.2022.04.096
Sun, T., Chen, L., and Zhang, W. (2017). Quantitative proteomics reveals potential crosstalk between a small RNA CoaR and a two-component regulator Slr1037 in Synechocystis sp. PCC6803. J. Proteome Res. 16, 2954–2963. doi:10.1021/acs.jproteome.7b00243
Sun, X., Atiyeh, H. K., Kumar, A., and Zhang, H. (2018). Enhanced ethanol production by Clostridium ragsdalei from syngas by incorporating biochar in the fermentation medium. Bioresour. Technol. 247, 291–301. doi:10.1016/j.biortech.2017.09.060
Svetlitchnyi, V. A., Svetlichnaya, T. P., Falkenhan, D. A., Swinnen, S., Knopp, D., and Läufer, A. (2022). Direct conversion of cellulose to l-lactic acid by a novel thermophilic Caldicellulosiruptor strain. Biotechnol. Biofuels Bioprod. 15, 44. doi:10.1186/s13068-022-02137-7
Tankov, I., Mitkova, M., Nikolova, R., Veli, A., and Stratiev, D. (2017). n-Butyl acetate synthesis in the presence of pyridinium-based acidic ionic liquids: influence of the anion nature. Catal. Lett. 147, 2279–2289. doi:10.1007/s10562-017-2135-0
Tao, Y. M., Bu, C. Y., Zou, L. H., Hu, Y. L., Zheng, Z. J., and Ouyang, J. (2021). A comprehensive review on microbial production of 1,2-propanediol: micro-organisms, metabolic pathways, and metabolic engineering. Biotechnol. Biofuels 14, 216–16. doi:10.1186/s13068-021-02067-w
Tarraran, L., and Mazzoli, R. (2018). Alternative strategies for lignocellulose fermentation through lactic acid bacteria: the state of the art and perspectives. FEMS Microbiol. Lett. 365. doi:10.1093/femsle/fny126
Therien, J. B., Artz, J. H., Poudel, S., Hamilton, T. L., Liu, Z., Noone, S. M., et al. (2017). The physiological functions and structural determinants of catalytic bias in the [FeFe]-hydrogenases CpI and CpII of Clostridium pasteurianum strain W5. Front. Microbiol. 8, 1305. doi:10.3389/fmicb.2017.01305
Tian, L., Cervenka, N. D., Low, A. M., Olson, D. G., and Lynd, L. R. (2019a). A mutation in the AdhE alcohol dehydrogenase of Clostridium thermocellum increases tolerance to several primary alcohols, including isobutanol, n-butanol and ethanol. Sci. Rep. 9, 1736. doi:10.1038/s41598-018-37979-5
Tian, L., Conway, P. M., Cervenka, N. D., Cui, J., Maloney, M., Olson, D. G., et al. (2019b). Metabolic engineering of Clostridium thermocellum for n-butanol production from cellulose. Biotechnol. Biofuels 12, 186. doi:10.1186/s13068-019-1524-6
Tian, L., Papanek, B., Olson, D. G., Rydzak, T., Holwerda, E. K., Zheng, T., et al. (2016). Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol. Biofuels 9, 116. doi:10.1186/s13068-016-0528-8
Tian, L., Perot, S. J., Hon, S., Zhou, J., Liang, X., Bouvier, J. T., et al. (2017a). Enhanced ethanol formation by Clostridium thermocellum via pyruvate decarboxylase. Microb. Cell Fact. 16, 171. doi:10.1186/s12934-017-0783-9
Tian, L., Perot, S. J., Stevenson, D., Jacobson, T., Lanahan, A. A., Amador-Noguez, D., et al. (2017b). Metabolome analysis reveals a role for glyceraldehyde 3-phosphate dehydrogenase in the inhibition of C. thermocellum by ethanol. Biotechnol. Biofuels 10, 276. doi:10.1186/s13068-017-0961-3
Todhanakasem, T., Sowatad, A., Kanokratana, P., Havanapan, P. O., and Champreda, V. (2019). Expression and extracellular secretion of endo-glucanase and xylanase by Zymomonas mobilis. Appl. Biochem. Biotechnol. 187, 239–252. doi:10.1007/s12010-018-2821-4
Tomas, C. A., Beamish, J., and Papoutsakis, E. T. (2004). Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J. Bacteriol. 186, 2006–2018. doi:10.1128/JB.186.7.2006-2018.2004
Tracy, B. P., Jones, S. W., Fast, A. G., Indurthi, D. C., and Papoutsakis, E. T. (2012). Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr. Opin. Biotechnol. 23, 364–381. doi:10.1016/j.copbio.2011.10.008
Tran-Din, K., and Gottschalk, G. (1985). Formation of d(-)-1,2-propanediol and d(-)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Arch. Microbiol. 142, 87–92. doi:10.1007/BF00409243
Transparency Market Research (2022). n-Butanol market size, share, growth by 2031. Available at: https://www.transparencymarketresearch.com/n-butanol-market.html (Accessed April 16, 2024).
Transparency Market Research (2024). 2,3-Butanediol market size, share, sale & industry demand-2031. Available at: https://www.transparencymarketresearch.com/2-3-butanediol-market.html (Accessed April 16, 2024).
Tunca, B., Kutlar, F. E., Kas, A., and Yilmazel, Y. D. (2023). Enhanced biohydrogen production from high loads of unpretreated cattle manure by cellulolytic bacterium Caldicellulosiruptor bescii at 75 °C. Waste Manag. 171, 401–410. doi:10.1016/j.wasman.2023.09.028
Turner, K. W., and Robertson, A. M. (1979). Xylose, arabinose, and rhamnose fermentation by Bacteroides ruminicola. Appl. Environ. Microbiol. 38, 7–12. doi:10.1128/aem.38.1.7-12.1979
Valle, A., Cantero, D., and Bolívar, J. (2019). Metabolic engineering for the optimization of hydrogen production in Escherichia coli: a review. Biotechnol. Adv. 37, 616–633. doi:10.1016/j.biotechadv.2019.03.006
Venkataramanan, K. P., Jones, S. W., McCormick, K. P., Kunjeti, S. G., Ralston, M. T., Meyers, B. C., et al. (2013). The Clostridium small RNome that responds to stress: the paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genomics 14, 849. doi:10.1186/1471-2164-14-849
Venkataramanan, K. P., Min, L., Hou, S., Jones, S. W., Ralston, M. T., Lee, K. H., et al. (2015). Complex and extensive post-transcriptional regulation revealed by integrative proteomic and transcriptomic analysis of metabolite stress response in Clostridium acetobutylicum. Biotechnol. Biofuels 8, 81. doi:10.1186/s13068-015-0260-9
Verified Market Research (2024). 1-Hexanol market size, share, trends, growth, scope, & forecast. Available at: https://www.verifiedmarketresearch.com/product/1-hexanol-market/(Accessed April 16, 2024).
Virunanon, C., Chantaroopamai, S., Denduangbaripant, J., and Chulalaksananukul, W. (2008). Solventogenic-cellulolytic clostridia from 4-step-screening process in agricultural waste and cow intestinal tract. Anaerobe 14, 109–117. doi:10.1016/J.ANAEROBE.2007.11.001
Walker, J. E., Lanahan, A. A., Zheng, T., Toruno, C., Lynd, L. R., Cameron, J. C., et al. (2020). Development of both type I–B and type II CRISPR/Cas genome editing systems in the cellulolytic bacterium Clostridium thermocellum. Metab. Eng. Commun. 10, e00116. doi:10.1016/j.mec.2019.e00116
Walther, T., and François, J. M. (2016). Microbial production of propanol. Biotechnol. Adv. 34, 984–996. doi:10.1016/j.biotechadv.2016.05.011
Wang, H., Ma, S., and Bu, H. (2014). Fermentative hydrogen production by newly isolated Clostridium perfringens ATCC 13124. J. Renew. Sustain. Energy 6, 013130. doi:10.1063/1.4863085
Wang, J., and Yin, Y. (2018). Fermentative hydrogen production using various biomass-based materials as feedstock. Renew. Sustain. Energy Rev. 92, 284–306. doi:10.1016/J.RSER.2018.04.033
Wang, J., and Yin, Y. (2021). Clostridium species for fermentative hydrogen production: an overview. Int. J. Hydrogen Energy 46, 34599–34625. doi:10.1016/j.ijhydene.2021.08.052
Wang, M., Han, J., Dunn, J. B., Cai, H., and Elgowainy, A. (2015). “Well-to-wheels energy use and greenhouse gas emissions of ethanol from corn, sugarcane and cellulosic biomass for US use,” in Efficiency and sustainability in biofuel production: environmental and land-use research. Editor B. Gikonyo (Palm Bay, Florida, United States: Apple Academic Press, Inc.), 249–280. doi:10.1088/1748-9326/7/4/045905
Wang, Q., Al Makishah, N. H., Li, Q., Li, Y., Liu, W., Sun, X., et al. (2021a). Developing clostridia as cell factories for short- and medium-chain ester production. Front. Bioeng. Biotechnol. 9, 661694. doi:10.3389/fbioe.2021.661694
Wang, R., Zong, W., Qian, C., Wei, Y., Yu, R., and Zhou, Z. (2011a). Isolation of Clostridium perfringens strain W11 and optimization of its biohydrogen production by genetic modification. Int. J. Hydrogen Energy 36, 12159–12167. doi:10.1016/j.ijhydene.2011.06.105
Wang, S., Huang, H., Kahnt, H. H., Mueller, A. P., Köpke, M., and Thauer, R. K. (2013). NADP-Specific electron-bifurcating [FeFe]-hydrogenase in a functional complex with formate dehydrogenase in clostridium autoethanogenum grown on CO. J. Bacteriol. 195, 4373–4386. doi:10.1128/JB.00678-13
Wang, S., Zhang, Y., Dong, H., Mao, S., Zhu, Y., Wang, R., et al. (2011b). Formic acid triggers the “acid crash” of acetone-butanol-ethanol fermentation by Clostridium acetobutylicum. Appl. Environ. Microbiol. 77, 1674–1680. doi:10.1128/AEM.01835-10
Wang, Y., Yang, G., Sage, V., Xu, J., Sun, G., He, J., et al. (2021b). Optimization of dark fermentation for biohydrogen production using a hybrid artificial neural network (ANN) and response surface methodology (RSM) approach. Environ. Prog. Sustain. Energy 40, e13485. doi:10.1002/EP.13485
Weitz, S., Hermann, M., Linder, S., Bengelsdorf, F. R., Takors, R., and Dürre, P. (2021). Isobutanol production by autotrophic acetogenic bacteria. Front. Bioeng. Biotechnol. 9, 657253. doi:10.3389/fbioe.2021.657253
Wen, Z., Ledesma-Amaro, R., Lin, J., Jiang, Y., and Yangd, S. (2019). Improved n-butanol production from Clostridium cellulovorans by integrated metabolic and evolutionary engineering. Appl. Environ. Microbiol. 85, e02560-18. doi:10.1128/AEM.02560-18
Wen, Z., Ledesma-Amaro, R., Lu, M., Jiang, Y., Gao, S., Jin, M., et al. (2020a). Combined evolutionary engineering and genetic manipulation improve low pH tolerance and butanol production in a synthetic microbial Clostridium community. Biotechnol. Bioeng. 117, 2008–2022. doi:10.1002/bit.27333
Wen, Z., Ledesma-Amaro, R., Lu, M., Jin, M., and Yang, S. (2020b). Metabolic engineering of Clostridium cellulovorans to improve butanol production by consolidated bioprocessing. ACS Synth. Biol. 9, 304–315. doi:10.1021/acssynbio.9b00331
Wen, Z., Li, Q., Liu, J., Jin, M., and Yang, S. (2020c). Consolidated bioprocessing for butanol production of cellulolytic Clostridia: development and optimization. Microb. Biotechnol. 13, 410–422. doi:10.1111/1751-7915.13478
Wen, Z., Lu, M., Ledesma-Amaro, R., Li, Q., Jin, M., and Yang, S. (2020d). TargeTron technology applicable in solventogenic clostridia: revisiting 12 Years’ advances. Biotechnol. J. 15, e1900284. doi:10.1002/biot.201900284
Wen, Z., Minton, N. P., Zhang, Y., Li, Q., Liu, J., Jiang, Y., et al. (2017). Enhanced solvent production by metabolic engineering of a twin-clostridial consortium. Metab. Eng. 39, 38–48. doi:10.1016/j.ymben.2016.10.013
Wen, Z., Tu, Z., Al Makishah, N., Fang, D., Ledesma-Amaro, R., Zhu, J., et al. (2022). Genetic manipulation of a twin clostridia consortium for Co-production of n-butanol and isobutanol by consolidated bioprocessing. ACS Sustain. Chem. Eng. 10, 14671–14684. doi:10.1021/acssuschemeng.1c08486
Wen, Z., Wu, M., Lin, Y., Yang, L., Lin, J., and Cen, P. (2014). A novel strategy for sequential co-culture of Clostridium thermocellum and Clostridium beijerinckii to produce solvents from alkali extracted corn cobs. Process Biochem. 49, 1941–1949. doi:10.1016/j.procbio.2014.07.009
Wietzke, M., and Bahl, H. (2012). The redox-sensing protein Rex, a transcriptional regulator of solventogenesis in Clostridium acetobutylicum. Appl. Microbiol. Biotechnol. 96, 749–761. doi:10.1007/s00253-012-4112-2
Wilbanks, B., and Trinh, C. T. (2017). Comprehensive characterization of toxicity of fermentative metabolites on microbial growth Mike Himmel. Biotechnol. Biofuels 10, 1–11. doi:10.1186/s13068-017-0952-4
Wilding-Steele, T., Ramette, Q., Jacottin, P., and Soucaille, P. (2021). Improved crispr/cas9 tools for the rapid metabolic engineering of clostridium acetobutylicum. Int. J. Mol. Sci. 22, 3704. doi:10.3390/ijms22073704
Wilkens, E., Ringel, A. K., Hortig, D., Willke, T., and Vorlop, K. D. (2012). High-level production of 1,3-propanediol from crude glycerol by Clostridium butyricum AKR102a. Appl. Microbiol. Biotechnol. 93, 1057–1063. doi:10.1007/s00253-011-3595-6
Williams, T. I., Combs, J. C., Lynn, B. C., and Strobel, H. J. (2007). Proteomic profile changes in membranes of ethanol-tolerant Clostridium thermocellum. Appl. Microbiol. Biotechnol. 74, 422–432. doi:10.1007/s00253-006-0689-7
Williams-Rhaesa, A. M., Awuku, N. K., Lipscomb, G. L., Poole, F. L., Rubinstein, G. M., Conway, J. M., et al. (2018). Native xylose-inducible promoter expands the genetic tools for the biomass-degrading, extremely thermophilic bacterium Caldicellulosiruptor bescii. Extremophiles 22, 629–638. doi:10.1007/s00792-018-1023-x
Willquist, K., and van Niel, E. W. J. (2010). Lactate formation in Caldicellulosiruptor saccharolyticus is regulated by the energy carriers pyrophosphate and ATP. Metab. Eng. 12, 282–290. doi:10.1016/j.ymben.2010.01.001
Willson, B. J., Kovács, K., Wilding-Steele, T., Markus, R., Winzer, K., and Minton, N. P. (2016). Production of a functional cell wall-anchored minicellulosome by recombinant Clostridium acetobutylicum ATCC 824. Biotechnol. Biofuels 9, 109. doi:10.1186/s13068-016-0526-x
Wirth, S., and Dürre, P. (2021). Investigation of putative genes for the production of medium-chained acids and alcohols in autotrophic acetogenic bacteria. Metab. Eng. 66, 296–307. doi:10.1016/j.ymben.2021.04.010
Wischral, D., Zhang, J., Cheng, C., Lin, M., De Souza, L. M. G., Pessoa, F. L. P., et al. (2016). Production of 1,3-propanediol by Clostridium beijerinckii DSM 791 from crude glycerol and corn steep liquor: process optimization and metabolic engineering. Bioresour. Technol. 212, 100–110. doi:10.1016/j.biortech.2016.04.020
Worku, A. K., Ayele, D. W., Deepak, D. B., Gebreyohannes, A. Y., Agegnehu, S. D., and Kolhe, M. L. (2024). Recent advances and challenges of hydrogen production technologies via renewable energy sources. Adv. Energy Sustain. Res. 5, 2300273. doi:10.1002/aesr.202300273
Wu, C. W., Spike, T., Klingeman, D. M., Rodriguez, M., Bremer, V. R., and Brown, S. D. (2017). Generation and characterization of acid tolerant Fibrobacter succinogenes S85. Sci. Rep. 7, 2277. doi:10.1038/s41598-017-02628-w
Xiao, Z., and Xu, P. (2007). Acetoin metabolism in bacteria. Crit. Rev. Microbiol. 33, 127–140. doi:10.1080/10408410701364604
Xu, G., Xiao, L., Wu, A., Han, R., and Ni, Y. (2021). Enhancing n-butanol tolerance of Escherichia coli by overexpressing of stress-responsive molecular chaperones. Appl. Biochem. Biotechnol. 193, 257–270. doi:10.1007/s12010-020-03417-4
Yamamoto, S., Suda, M., Niimi, S., Inui, M., and Yukawa, H. (2013). Strain optimization for efficient isobutanol production using corynebacterium glutamicum under oxygen deprivation. Biotechnol. Bioeng. 110, 2938–2948. doi:10.1002/bit.24961
Yang, F., Hanna, M. A., and Sun, R. (2012). Value-added uses for crude glycerol--a byproduct of biodiesel production. Biotechnol. Biofuels 5, 13. doi:10.1186/1754-6834-5-13
Yang, G., Hu, Y., and Wang, J. (2019a). Biohydrogen production from co-fermentation of fallen leaves and sewage sludge. Bioresour. Technol. 285, 121342. doi:10.1016/j.biortech.2019.121342
Yang, J., Wei, Y., Li, G., Zhou, S., and Deng, Y. (2020). Computer-aided engineering of adipyl-CoA synthetase for enhancing adipic acid synthesis. Biotechnol. Lett. 42, 2693–2701. doi:10.1007/s10529-020-02978-y
Yang, M., An, Y., Zabed, H. M., Guo, Q., Yun, J., Zhang, G., et al. (2019b). Random mutagenesis of Clostridium butyricum strain and optimization of biosynthesis process for enhanced production of 1,3-propanediol. Bioresour. Technol. 284, 188–196. doi:10.1016/j.biortech.2019.03.098
Yang, X., Lai, Z., Lai, C., Zhu, M., Li, S., Wang, J., et al. (2013). Efficient production of L-lactic acid by an engineered Thermoanaerobacterium aotearoense with broad substrate specificity. Biotechnol. Biofuels 6, 124. doi:10.1186/1754-6834-6-124
Yang, X., Xu, M., and Yang, S. T. (2015). Metabolic and process engineering of Clostridium cellulovorans for biofuel production from cellulose. Metab. Eng. 32, 39–48. doi:10.1016/j.ymben.2015.09.001
Yang, X., Xu, M., and Yang, S. T. (2016). Restriction modification system analysis and development of in vivo methylation for the transformation of Clostridium cellulovorans. Appl. Microbiol. Biotechnol. 100, 2289–2299. doi:10.1007/s00253-015-7141-9
Yang, Z., Leero, D. D., Yin, C., Yang, L., Zhu, L., Zhu, Z., et al. (2022). Clostridium as microbial cell factory to enable the sustainable utilization of three generations of feedstocks. Bioresour. Technol. 361, 127656. doi:10.1016/j.biortech.2022.127656
Yao, S., and Mikkelsen, M. J. (2010). Identification and overexpression of a bifunctional aldehyde/alcohol dehydrogenase responsible for ethanol production in thermoanaerobacter mathranii. J. Mol. Microbiol. Biotechnol. 19, 123–133. doi:10.1159/000321498
Yazdani, S. S., and Gonzalez, R. (2007). Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr. Opin. Biotechnol. 18, 213–219. doi:10.1016/j.copbio.2007.05.002
Yin, Y., and Wang, J. (2016). Fermentative hydrogen production from waste sludge solubilized by low-pressure wet oxidation treatment. Energy Fuels 30, 5878–5884. doi:10.1021/acs.energyfuels.6b01034
Yin, Y., and Wang, J. (2019). Enhanced sewage sludge disintegration and hydrogen production by ionizing radiation pretreatment in the presence of Fe2+. ACS Sustain. Chem. Eng. 7, 15548–15557. doi:10.1021/acssuschemeng.9b03362
Yoo, M., Croux, C., Meynial-Salles, I., and Soucaille, P. (2016). Elucidation of the roles of adhE1 and adhE2 in the primary metabolism of Clostridium acetobutylicum by combining in-frame gene deletion and a quantitative system-scale approach. Biotechnol. Biofuels 9, 92. doi:10.1186/s13068-016-0507-0
Yu, M., Zhang, Y., Tang, I. C., and Yang, S. T. (2011). Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metab. Eng. 13, 373–382. doi:10.1016/j.ymben.2011.04.002
Zhang, C., Li, T., Su, G., and He, J. (2020a). Enhanced direct fermentation from food waste to butanol and hydrogen by an amylolytic Clostridium. Renew. Energy 153, 522–529. doi:10.1016/j.renene.2020.01.151
Zhang, F., Zhang, K., Zhang, Z., Chen, H. Q., Chen, X. W., Xian, X. Y., et al. (2023a). Efficient isopropanol-butanol-ethanol (IBE) fermentation by a gene-modified solventogenic Clostridium species under the co-utilization of Fe(III) and butyrate. Bioresour. Technol. 373, 128751. doi:10.1016/j.biortech.2023.128751
Zhang, J., Zong, W., Hong, W., Zhang, Z. T., and Wang, Y. (2018). Exploiting endogenous CRISPR-Cas system for multiplex genome editing in Clostridium tyrobutyricum and engineer the strain for high-level butanol production. Metab. Eng. 47, 49–59. doi:10.1016/j.ymben.2018.03.007
Zhang, L., Zhao, R., Jia, D., Jiang, W., and Gu, Y. (2020b). Engineering Clostridium ljungdahlii as the gas-fermenting cell factory for the production of biofuels and biochemicals. Curr. Opin. Chem. Biol. 59, 54–61. doi:10.1016/j.cbpa.2020.04.010
Zhang, Y., Zheng, S., Hao, Q., Wang, O., and Liu, F. (2023b). Respiratory electrogen Geobacter boosts hydrogen production efficiency of fermentative electrotroph Clostridium pasteurianum. Chem. Eng. J. 456, 141069. doi:10.1016/j.cej.2022.141069
Zhao, Y., Tomas, C. A., Rudolph, F. B., Papoutsakis, E. T., and Bennett, G. N. (2005). Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl. Environ. Microbiol. 71, 530–537. doi:10.1128/AEM.71.1.530-537.2005
Zheng, T., Cui, J., Bae, H. R., Lynd, L. R., and Olson, D. G. (2017). Expression of adhA from different organisms in Clostridium thermocellum. Biotechnol. Biofuels 10, 251. doi:10.1186/s13068-017-0940-8
Zhou, J., Olson, D. G., Argyros, D. A., Deng, Y., van Gulik, W. M., van Dijken, J. P., et al. (2013). Atypical glycolysis in Clostridium thermocellum. Appl. Environ. Microbiol. 79, 3000–3008. doi:10.1128/AEM.04037-12
Zhou, J., Olson, D. G., Lanahan, A. A., Tian, L., Murphy, S. J. L., Lo, J., et al. (2015). Physiological roles of pyruvate ferredoxin oxidoreductase and pyruvate formate-lyase in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Biotechnol. Biofuels 8, 138. doi:10.1186/s13068-015-0304-1
Zhu, F., Liu, D., and Chen, Z. (2022). Recent advances in biological production of 1,3-propanediol: new routes and engineering strategies. Green Chem. 24, 1390–1403. doi:10.1039/d1gc04288b
Keywords: hydrogen, ethanol, lactate, propanediol, butanol, isobutanol, medium chain esters, hexanol
Citation: Ponsetto P, Sasal EM, Mazzoli R, Valetti F and Gilardi G (2024) The potential of native and engineered Clostridia for biomass biorefining. Front. Bioeng. Biotechnol. 12:1423935. doi: 10.3389/fbioe.2024.1423935
Received: 26 April 2024; Accepted: 06 August 2024;
Published: 16 August 2024.
Edited by:
Hongbo Hu, Shanghai Jiao Tong University, ChinaReviewed by:
Yu-Sin Jang, Gyeongsang National University, Republic of KoreaCopyright © 2024 Ponsetto, Sasal, Mazzoli, Valetti and Gilardi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roberto Mazzoli, cm9iZXJ0by5tYXp6b2xpQHVuaXRvLml0
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.