
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 22 December 2021
Sec. Synthetic Biology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.770248
This article is part of the Research TopicSynthetic Live TherapyView all 7 articles
Bifidobacterium is a non-spore-forming, Gram-positive, anaerobic probiotic actinobacterium and commonly found in the gut of infants and the uterine region of pregnant mothers. Like all probiotics, Bifidobacteria confer health benefits on the host when administered in adequate amounts, showing multifaceted probiotic effects. Examples include B. bifidum, B. breve, and B. longum, common Bifidobacterium strains employed to prevent and treat gastrointestinal disorders, including intestinal infections and cancers. Herein, we review the latest development in probiotic Bifidobacteria research, including studies on the therapeutic impact of Bifidobacterial species on human health and recent efforts in engineering Bifidobacterium. This review article would provide readers with a wholesome understanding of Bifidobacteria and its potentials to improve human health.
Probiotic microorganisms are defined as living microorganisms that confer health benefiting properties to the host when administered adequately. Probiotics exert beneficial functions mainly through producing antimicrobial peptides, assimilating dietary fibers, regulating fat storage, modulating mucosal immunity, or regulating gut microbiota (Ku et al., 2016). For centuries, probiotics have been widely used in various functional foods, e.g., yoghurt, milk, cheese, infant formula, and dietary supplements. The most common probiotics include Lactobacilli and Bifidobacteria, which predominantly inhabit the animal or human intestinal tract (Hudault et al., 1994). Bifidobacteria are V- or Y-type branched, rod-shaped, immobile, non-spore-forming, Gram-positive, anaerobic, catalase-negative bacteria that belong to the family Bifidobacteriaceae and the phylum Actinobacteria. The Bifidobacterium genus currently includes over 90 species, excluding the unclassified species (Supplementary Table S1). Bifidobacterium was first isolated from breast-fed infant feces, but so far have been discovered from various ecological niches including sewage, fermented milk and anaerobic digestion facilities; nevertheless, the most frequent isolates are associated with the gastrointestinal tracts of humans and animals. The growth conditions (e.g., temperature, pH, oxygen level) of Bifidobacteria do not vary significantly among strains (Ruiz et al., 2011). For instance, the optimal growth temperature ranges between 36 and 38°C and 41–43°C for human- and animal-isolated strains, respectively. Additionally, the optimal growth pH is around pH 6.5–7.0, where B. animalis and B. thermacidophilum were found to be also metabolically active at pH 3.5–4.0. Most Bifidobacterial species are strict anaerobes, with a few exceptions, such as B. boum, B. thermophilum, B. dentium and B. psychraerophilum that tolerate microaerophilic environment. Hitherto, many probiotic Bifidobacteria have shown beneficial effects on humans or animals, e.g., antiinfection, anti-depression, regulating the host immune system, and facilitating host nutrition adsorption (Figure 1).
Furthermore, some probiotic Bifidobacteria are engineered to include other beneficial cellular functionalities and/or remove the disadvantageous properties. Herein, we provide a review of the various findings of Bifidobacterium probiotics’ therapeutic functions, metabolic pathways, and methods to engineer them. This review would provide readers with a better understanding of the beneficial functions of Bifidobacterium probiotics, including the genetically engineered strains.
One prerequisite for becoming a probiotic strain is the capability of colonizing at a specific location, e.g., in the gastrointestinal tract, such that the probiotic strain can interact effectively with the host and the host microbiome. The colonization of probiotics outcompetes some pathogens and thus confers the host some protection against pathogenic infections. Various studies demonstrated that different species of Bifidobacteria exert antiinfection properties (Table 1). B. longum ATCC 15708 showed antimicrobial activity against many pathogens, including Escherichia coli O157:H7 ATTC 35150, Salmonella typhimurium ATTC 13311 and Listeria monocytogenes ATTC 19115 (Igbafe et al., 2020). B. longum BB536 protects against gut-derived sepsis caused by Pseudomonas aeruginosa, likely through interfering with the adherence of pathogens to intestinal epithelial cells (Matsumoto et al., 2008); BB536 ameliorates the upper respiratory infections in healthy pre-school children probably through modulating gut microbiota, i.e., increasing the abundance of the genus Faecalibacterium (Lau et al., 2018); also, another randomized, double-blind, placebo-controlled trial reveals that administration of BB536 in combination with the standard triple therapy (esomeprazole, amoxicillin, clarithromycin) improves the eradication rate of Helicobacter pylori infection in 63 patients (Chitapanarux et al., 2015). The administration of B. lactis BB-12 in early childhood reduces respiratory tract infections (Taipale et al., 2016). An administration of 5 billion colony-forming units of BB-12 twice a day for 1.5 years observed improved resistance to respiratory tract infections and low rates of developing fever throughout the infant’s growth period. B. animalis AHC7 was found to protect mice against S. typhimurium infection and prevent acute diarrhoea in dogs (Kelley et al., 2009; O’Mahony et al., 2009). The underlying mechanisms of the acute diarrhoea prevention of B. animalis AHC7 are due to the attenuation of proinflammatory transcription factor activation in response to infection (O'Mahony et al., 2010). Bifidobacteria are also used in displacing latent and chronic infectious strains. For instance, B. longum ATCC 15707 can prevent Clostridium difficile-infection (Yun et al., 2017), while B. longum 51A confers protection against Klebsiella pneumoniae-induced lung infection (Vieira et al., 2016). This protection conferred by B. longum 51A is due to the activation of the Toll-like receptor-signaling pathway, resulting in reactive oxygen species production. Similarly, B. longum 51A was found to reduce the Giardia-parasitic load in Mongolian gerbils (Meriones unguiculatus), making this strain a suitable prophylactic and therapeutic probiotic for promoting human and animal health (Fonseca et al., 2019).
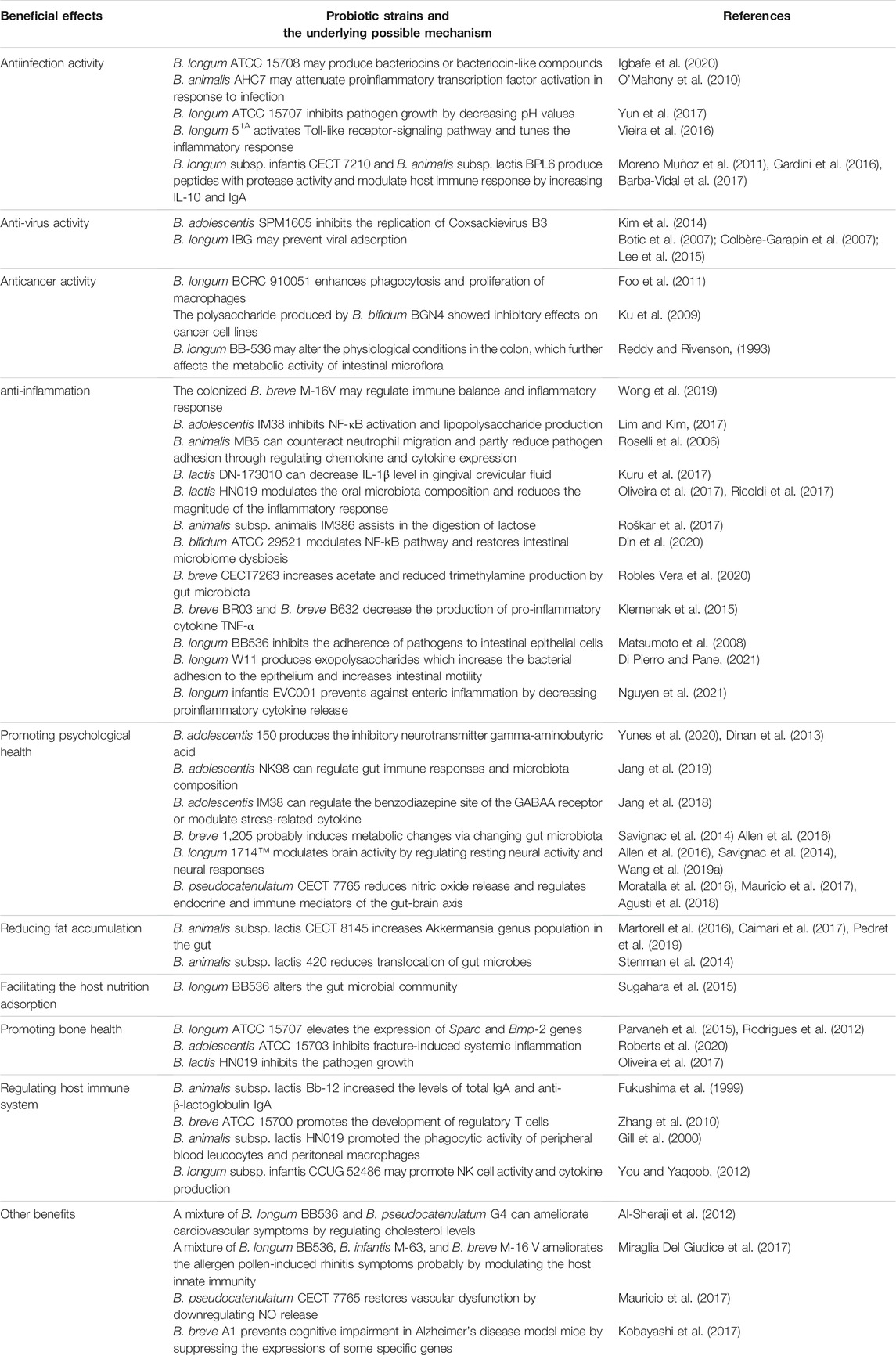
TABLE 1. Beneficial effects displayed by common probiotic Bifidobacteria and the mechanisms involved.
Additionally, multi-strain or multi-species probiotic formulations have greater efficacy in fighting infections compared to single strain administration due to the complementary or even synergistic effects of the multi-strain/species formulation (Timmerman et al., 2004; Collado et al., 2007; Chapman et al., 2011). A combination of B. longum subsp. infantis CECT 7210 and B. animalis subsp. lactis BPL6 enhances gut health and ameliorates S. typhimurium-infection in the porcine model (Barba-Vidal et al., 2017). A mixture of B. longum BB536 and L. rhamnosus HN001 significantly reduced potentially harmful bacteria and enriched beneficial ones (Toscano et al., 2017) in the gut microbiota. A cocktail of probiotic Lactobacilli and Bifidobacteria showed antimicrobial and anti-biofilm activities against multidrug-resistant E. coli (Abdelhamid et al., 2018), and pretreatment with yogurt containing Lactobacillus acidophilus La5 or B. lactis BB-12 suppresses H. pylori infections effectively in humans (Wang et al., 2004; Sheu et al., 2006). A fermented formula containing B. breve c50 and Streptococcus thermophilus 065 reduces the severity of acute diarrhea among healthy young infants (Thibault et al., 2004).
Aside from the common protection against pathogenic bacteria, Bifidobacteria also exhibit antiviral activities. For example, B. adolescentis SPM1605 inhibits human enterovirus Coxsackievirus B3, thus preventing the virus infection-related acute heart failure and aseptic meningitis (Kim et al., 2014); B. longum IBG inhibits infection by rotavirus in vitro and decreases the duration of diarrhoea in pediatric patients (Lee et al., 2015).
Probiotics have been employed to prevent and treat cancers for decades (Rowland et al., 1998; Lee et al., 2004; Paolillo et al., 2009; Ohara et al., 2010), where Bifidobacteria can effectively inhibit cancers in animal models. As an example, dietary supplementation of B. longum BB-536 significantly inhibits the 2-amino-3-methylimidazo [4,5-f]quinoline (IQ)-induced incidence of the colon (100% inhibition) and liver (80% inhibition) tumours in male rats and suppresses the IQ-induced mammary carcinogenesis (50% inhibition) and liver carcinogenesis (27% inhibition) of female rats (Reddy and Rivenson, 1993). B. longum BCRC 910051 prevents the development of 1,2-dimethylhydrazine-induced colonic tumorigenesis (Foo et al., 2011). An in vitro study shows that B. bifidum BGN4 inhibits the growth of several human colon cancer cell lines such as HT-29 and HCT-116 (Ku et al., 2009).
Inflammation is a physiological response generally triggered by damage to the living tissues. The inflammatory response is a defense mechanism that protects the host from infection and injury. Bifidobacteria colonize primarily in the oral cavity and intestinal tracts. They thus have been applied to suppress and prevent some oral and enteric inflammations including irritable bowel syndrome (Guglielmetti et al., 2011; Ringel-Kulka et al., 2011; O'Mahony et al., 2005), intestinal barrier functions (Krumbeck et al., 2018), and infant colic impairment (Kobayashi et al., 2019; Xiao et al., 2020). However, some strains also inhibit cutaneous inflammations, e.g., a mixture of B. breve M-16V and B. longum BB536 reduces the development of eczema and atopic dermatitis in infants (Enomoto et al., 2014).
An in vitro study shows that B. animalis MB5 protects intestinal Caco-2 cells from the inflammation-associated response by counteracting neutrophil migration and partly decreasing pathogen adhesion (Roselli et al., 2006). Many animal studies also validate the anti-inflammatory potentials. For example, a combination of B. bifidum and B. longum was found to effectively prevent devastating necrotizing enterocolitis (NEC) in an animal model (Wu et al., 2013); B. adolescentis IM38 ameliorates high fat diet-induced colitis by inhibiting NF-κB activation and lipopolysaccharide production by gut microbiota (Lim and Kim, 2017); B. bifidum ATCC 29521 restores the colon mucus layer of mice with ulcerative colitis by modulating NF-κB signalling pathway and rebuilding the gut intestinal microbiome equilibrium (Din et al., 2020). B. breve CECT7263 attenuates endothelial dysfunction by regulating the levels of acetate and trimethylamine produced by gut microbiota (Robles-Vera et al., 2020).
Human studies show that B. breve M-16V potentially protects infants from developing NEC (Wong et al., 2019). Mechanistic studies indicated that M-16V can promote early gut microbial colonization, thus regulating the host immunity and preventing the inflammatory response. The combination of 2 B. breve strains (BR03 and B632) coupled to a gluten-free diet has shown a positive effect on decreasing the production of pro-inflammatory cytokine TNF-α in children with celiac disease (Klemenak et al., 2015). A clinical study shows that the probiotics mixture containing B. bifidum BGN4, B. lactis AD011, L. acidophilus AD031 and L. casei IBS041 can effectively relieve irritable bowel syndrome (Hong et al., 2009). A synbiotic formula composing probiotics (B. breve and L. casei) and the prebiotics galactooligosaccharides improved the intestinal absorptive function and motility of patients with short bowel syndrome (Kanamori et al., 2004; Bongers et al., 2010). The combination of B. breve Yakult and L. casei Shirota can prevent infant enterocolitis (Kanamori et al., 2010), reduce NEC incidence and improve intestinal motility in infants (Braga et al., 2011). An open-label pilot study revealed that the administration of B. longum BB536 effectively induced remission of patients with ulcerative colitis (Takeda et al., 2009). Three B. breve strains and a B. longum strain show potential in treating enteric disorders in newborns such as infantile colics (Aloisio et al., 2012).
Lactose intolerance occurs in patients that produce insufficient lactase in the small intestine to digest dietary lactose, usually derived from dairy food. Undigested lactose flows into the colon, where the lactose is catabolized by the gut microbes, triggering the lactose intolerance symptoms, including diarrhoea, flatulence, nausea, stomach cramps, and vomiting. B. animalis subsp. animalis IM386 ameliorates diarrhoea and flatulence in lactose-intolerant individuals (Roškar et al., 2017) because it facilitates lactose degradation in the small intestines. Mixed probiotics containing B. animalis subsp. animalis IM386 and L. plantarum MP2026 also alleviated some gastrointestinal symptoms in lactose-intolerant subjects (Roškar et al., 2017).
Constipation usually results from changes in diet or inadequate intake of fibre, where treatment using a multi-component probiotics formula consisting of B. bifidum, B. infantis, B. longum, L. casei, L. plantarum, and L. rhamnosus has positive effects on alleviating symptoms of constipation (Bekkali et al., 2007). B. longum W11 was also found to relieve the constipation symptoms of patients with irritable bowel syndrome, synergize with rifaximin as an adjuvant antibiotic treatment, and treat minimal hepatic encephalopathy (Di Pierro and Pane, 2021).
Aside from preventing enteric inflammations, Bifidobacteria show some inhibitory effects on oral inflammations. B. lactis DN-173010-fermented yogurt fed to patients elicit a positive effect on gingival inflammatory parameters (Kuru et al., 2017) because it can decrease both concentration and the total amount of IL-1β in gingival crevicular fluid. Oral administration of B. lactis HN019 as an adjunct potentiates the effects of scaling and root planing (SRP) in treating experimental periodontitis in rats (Ricoldi et al., 2017) and patients (Invernici et al., 2018).
Psychobiotics are a category of probiotics that confers mental health benefits, and many Bifidobacteria are functionally considered psychobiotics. Studies show that the gut microbiota of stress-resilient mice has lower Bifidobacteria than control and susceptible mice, and supplementation of Bifidobacteria to the susceptible mice significantly increased the resilience compared with vehicle-treated mice (Yang et al., 2017a). These findings suggest that Bifidobacteria may confer resistance to stress. B. breve CCFM1025 showed antidepressant-like effect in chronically stressed mice due probably to the capacity of utilizing various carbohydrates and producing neuroactive metabolites, such as tryptophan, hypoxanthine, and nicotinate (Tian et al., 2021). B. breve CCFM1025 can also reverse chronic stress-induced depressive symptoms (Tian et al., 2020). B. adolescentis 150 shows anti-depression properties when fed to mice (Yunes et al., 2020) due to the ability to produce gamma-aminobutyric acid, a neurotransmitter inhibitor of the central nervous system (Dinan et al., 2013). B. adolescentis NK98 alleviates anxiety/depression symptoms through regulating gut immune responses and microbiota composition (Jang et al., 2019). B. adolescentis IM38 can attenuate anxiety by regulating the benzodiazepine site of the GABAA receptor and modulating stress-related cytokine expression (Jang et al., 2018). B. longum 1714 reduces stress, anxiety and depression-related behaviours in anxious mice (Allen et al., 2016). The potential probiotic B. pseudocatenulatum CECT 7765 ameliorates depression comorbid with obesity via regulating endocrine and immune mediators of the gut-brain axis (Agusti et al., 2018). B. breve 1,205 reduces general anxiety behaviours in mice (Savignac et al., 2014). The combinatorial use of Lactobacillus helveticus R0052 and B. longum R0175 has anxiolytic-like activity (Messaoudi et al., 2011a) and reduces post-myocardial infarction depression symptoms (Arseneault-Bréard et al., 2012) in rats.
Various clinical trials have further supported these claims on the psychological health promotion effects. For instance, the oral administration of B. longum NCC3001 to depression patients reduces depression scores and alters patients’ brain activity (Pinto-Sanchez et al., 2017). B. longum 1714™ was found to modulate brain activity by regulating resting neural activity and neural responses (Wang et al., 2019a). B. longum 1714 modulates electrophysiology and neurocognition in healthy humans (Allen et al., 2016). The mixed probiotics of B. longum and L. helveticus significantly reduced the depressive symptoms of patients with Major Depression Disorder (Kazemi et al., 2019), presented beneficial psychological effects in healthy human volunteers (Messaoudi et al., 2011a), and decreased stress-induced gastrointestinal discomfort (Messaoudi et al., 2011b).
Obesity is a complex disease and increases the risk of other diseases and health problems, such as heart disease, diabetes, high blood pressure and certain cancers. Some Bifidobacteria probiotics can reduce the host fat accumulation. B. lactis CECT 8145 reduces fat content and modulates lipid metabolism and antioxidant response in Caenorhabditis elegans (Martorell et al., 2016). Another study using heat-treated B. lactis CECT8145 found an increased lean mass and ameliorated metabolic syndrome in cafeteria-fed obese rats (Caimari et al., 2017). Similar studies using the same probiotic also show that administering either living or heat-treated B. lactis CECT8145 can reduce anthropometric adiposity biomarkers linked to changes in host immune system regulation and enrichment of Akkermansia genus in the gut of abdominally obese individuals (Pedret et al., 2019). B. lactis 420 is another strain found to reduce fat mass and glucose intolerance in both obese and diabetic mice (Stenman et al., 2014) by reducing the translocation of gut microbes.
Consumption of probiotics has been reported to facilitate the absorption of nutrients, e.g., vitamins and calcium ions (Ballini et al., 2019). B. longum BB536 can increase the abundance of nutrients including pimelate, biotin and butyrate, by facilitating the fermentation processes resulting from the microbial crosstalk between B. longum BB536 and human gut-derived microbiota (Sugahara et al., 2015). Additionally, B. pseudocatenulatum is commonly found in human faecal samples throughout their lifetime, where some of these strains have shown beneficial properties, such as the production of enterolignan, urolithin, and conjugated linoleic acid (Yang et al., 2017b; Vickers, 2017; Gaya et al., 2018).
Osteoporosis, a common bone metabolic disorder caused by low bone mass and deterioration of the bone tissue, results in the individual being prone to fractures. Using probiotic B. longum ATCC 15707 (Parvaneh et al., 2015) or in combination with yacon flour (Rodrigues et al., 2012) can increase bone mass density by elevating the expression of Sparc and Bmp-2 genes. Additionally, B. adolescentis ATCC 15703 can modulate bone repair by dampening fracture-induced systemic inflammation (Roberts et al., 2020), and the topical use of B. lactis HN019 promotes a protective effect against alveolar bone (Oliveira et al., 2017).
These probiotics can regulate the host immune system for the amelioration or prevention of diseases. B. animalis subsp. lactis BB-12 was found to protect murine pups and dams from exposure to food antigens by increasing total IgA and anti-β-lactoglobulin IgA levels in fecal extracts (Fukushima et al., 1999). B. breve ATCC 15700 suppresses the skewed T helper 2 pattern responses by promoting Treg development (Zhang et al., 2010). B. animalis subsp. lactis HN019 can enhance natural immunity in healthy elderly subjects by increasing the anti-inflammatory cytokine IFN-α and phagocytic activity (Arunachalam et al., 2000). HN019 also enhances several natural and acquired immunity indices in healthy mice, including NK-cell activity, IFN-γ production, antibody responses to antigens, and the phagocytic activity of peripheral blood leukocytes and macrophages (Gill et al., 2000). L. helveticus Bar13 and B. longum Bar33 can synergistically improve the physiologic status and immunity of older adults by increasing regulatory T (Treg and Tr1) cells and decreasing γδ T cells (Finamore et al., 2019). In vitro studies show that B. bifidum BGN4 can activate differentiation of host macrophages and stimulate the production of IL-10 and IL-6 (Lee et al., 2002; Kim and Ji, 2006). B. longum subsp. infantis CCUG 52486 shows strong immunomodulatory potential comparable with well-known commercial strains (e.g., B. longum SP 07/3, L. rhamnosus GG and L. casei Shirota) based on the IL-10/IL-12 ratios (You and Yaqoob, 2012).
A combination of B. longum BB536 and B. pseudocatenulatum G4 ameliorates cardiovascular symptoms by decreasing total cholesterol, LDL-cholesterol, triglyceride levels, malondialdehyde, and increasing HDL-cholesterol levels concentrations (Al-Sheraji et al., 2012). A Bifidobacteria mixture of B. longum BB536, B. infantis M-63, and B. breve M-16 V prevents allergen pollen-induced rhinitis symptoms (Miraglia Del Giudice et al., 2017). B. lactis BB-12 alters the colonization of cariogenic bacteria, prevents dental caries (Çaglar et al., 2008) and decreases plaque and gingival indexes (Toiviainen et al., 2015). B. pseudocatenulatum CECT 7765 can restore the obesity-induced vascular dysfunction by reducing nitric oxide release (Mauricio et al., 2017) and prevent gut-derived complications in experimental chronic liver disease via maintaining gut homeostasis (Moratalla et al., 2016). B. breve A1 exhibits therapeutic potential for preventing cognitive impairment in Alzheimer’s disease model mice by suppressing the hippocampal expressions of amyloid-β-induced specific genes (Kobayashi et al., 2017). While the studies on Bifidobacterium as probiotics may be considered extensive, it is inevitable that with the latest multi-omics approaches, more strains of Bifidobacteria and its numerous therapeutic activities are soon to be discovered in future with the advent of highly efficient isolation and determination methods.
The Gram-positive Bifidobacterium is an anaerobic branched rod-shaped actinobacterium often associated with symbiotic bacterial-host relationships with mammals, particularly humans. Given the anaerobic nature of the microbe, the natural microbial growth rate is relatively slower than their other counterparts in the microbiota. Thus, Bifidobacterium is usually administered at a high cell count to overwhelm the microbiota and displace the pathogens in the host to elicit its therapeutic properties.
Microbes from the Actinobacteria phyla, including Bifidobacterium, are generally recognized as natural product producers with the basic biochemical makeup for producing valuable metabolites used in pharmaceuticals, agricultural, environmental, and industrial applications (Hazarika et al., 2020). The use of Bifidobacterium as a chassis for genetic manipulation requires a better understanding of microbial biochemistry. Understanding the pathways regulating microbial behaviour in synthetic biology and metabolic engineering allows streamlining cellular processes to elicit the appropriate responses. These biochemical attributes include producing polyketides, short-chain fatty acids, conjugated linoleic acid, and metabolizing fructose, lactose, and cholesterol.
Bifidobacteria can be considered a powerful workhorse for producing polyketide products. This ability can be attributed to the Bifid shunt that can effectively produce more CoA derivatives than other conventional production methods of CoA derivatives (Wang et al., 2019b). The acetyl-CoA, malonyl-CoA, and other CoA derivatives are the building blocks involved in the biosynthesis of polyketide, fatty acids, butanol, isoprenoids, and amino acids (Figure 2) (O’Callaghan and Van Sinderen, 2016). Additionally, some Bifidobacteria can metabolize the byproducts and waste of the host body to produce a higher pool of CoA derivatives, providing these microbes with an additional edge in generating a larger pool of CoA derivatives for the biosynthesis of value-added chemicals. For instance, B. animalis subspecies lactis was found to degrade oxalate (a cellular byproduct secreted by the host cells in the amino acid metabolism), providing the microbe with a higher pool of oxalyl CoA while reducing the risk of oxalate toxicity to the host (Turroni et al., 2010). There have been efforts to use these probiotic strains to treat patients with weak kidney functions and at high risk of developing kidney stones. Another study investigating the role of Bifidobacteria in producing short-chain fatty acid (SCFA) further revealed that these microbes could metabolize complex sugar such as glucomannan to produce higher pools of SCFA, including lactic, acetic, propionic, and butyric acid (Usta-Gorgun and Yilmaz-Ersan, 2020).
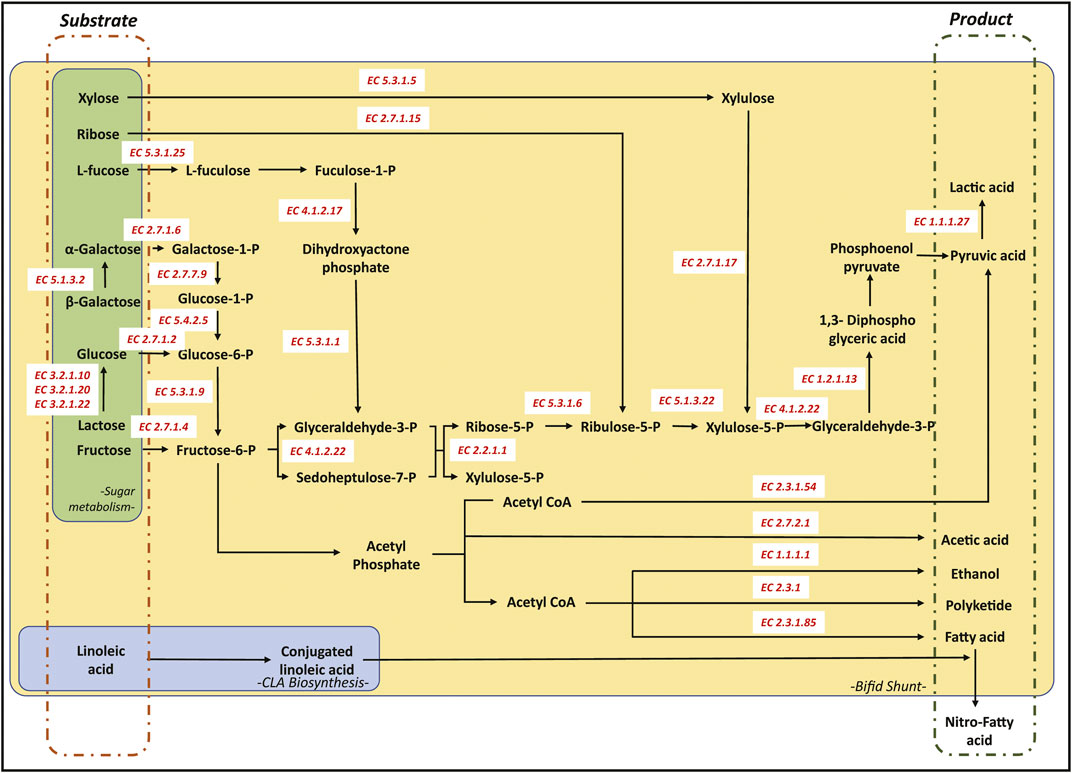
FIGURE 2. Schematic representation of the Bifid shunt pathway (yellow), CLA pathway (blue) and other carbohydrate degradation pathways (green). All corresponding enzymes in the pathway were annotated using the Enzyme Commission numbers as recommended by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (IUBMB).
The produced CoA derivatives are used to produce secondary metabolites by modular enzymes encoded in the Bifidobacterium gene (Figure 2). The gene sequences of various Bifidobacterium revealed several polyketide synthase (PKS) gene clusters, including those from the Type 1 and Type 3 PKSs. A study comparing the Bifidobacteria isolated from humans and marmosets showed that Type 1 PKS was conserved across Bifidobacteria, including B. myosotis, B. reuteri, B. breve, B. longum, B. tissieri, B. bifidum, and B. callitrichos (Brown et al., 2019). Closer investigation on the metabolomics of B. breve UCC2003 showed that under the presence of high bile salt concentrations, the Bifidobacteria reverts to the sessile biofilm state through the expression of polyketide synthase Bbr_0,204/0,205. The study hypothesized the role of PKS in the microbe’s adaptive behaviour in the presence of bile salts (Kelly et al., 2020).
Like polyketide production, the SCFAs, medium-chain fatty acid (MCFA), and fatty acid ethyl esters are produced through the fermentation of complex sugars in Bifidobacteria to generate the various CoA-derivatives (Figure 2). These SCFAs produced include butyrate, propionate, and so forth that play an essential role in host health, including boosting the metabolism and reducing the risk of developing diabetes (Usta-Gorgun and Yilmaz-Ersan, 2020). These SCFAs are generated as a byproduct of the Bifid shunt and can be used for the biosynthesis of MCFA using a homologous protein to polyketide synthases, known as fatty acid synthase (FAS) (Gu et al., 2016). FASs are modular enzymatic proteins that catalyze the CoA derivatives to produce the various fatty acids.
Thus, it is evident that the presence of the Bifid shunt and the various metabolic processes within Bifidobacterium strains provides the necessary metabolic precursors needed for engineering PKS, FAS and other CoA related pathways. This unique characteristic facilitates the biosynthesis of value-added compounds through the use of multi-domain modular enzymes. So far, there has been no reported use of Bifidobacterium in the bioproduction of polyketides or fatty acids due to the limited gene engineering tools. However, leveraging this attribute would help improve the productivity of the bioconversion (the substrate/product ratio).
Many Bifidobacteria are natural producers of conjugated linoleic acids (CLA), resulting from the biohydrogenation of linoleic acid (LA) and other unsaturated fatty acids (Figure 2). The first reported strain that produces CLA is B. breve with a 66% endpoint conversion, where the converted LA to CLA was secreted into the surrounding medium (Park et al., 2011). Other strains showing a similar ability to convert LA to CLA includes B. infantis, B. dentium, B. catenulatum and B. pseudocatenatalum (Raimondi et al., 2016). The biosynthesis of CLA from LA involves a three-step process, relying on LA isomerase enzymes, direct reduction of the unstable variant, and delta-9 desaturase enzyme (Coakley et al., 2003). While CLA is directly linked to many health benefits, the CLA is a preferred substrate for producing nitrated fatty acids (Bonacci et al., 2012). The various CLA health benefits include anti-inflammation, antiinfection, anticancer, and increasing host metabolism. On the other hand, nitro-fatty acids are often used as an agent to reduce blood pressure and prevent inflammation and other fibrotic diseases (Schopfer et al., 2018). The natural ability of Bifidobacterium to convert and produce CLA generates a perpetual pool of CLA for the biosynthesis of nitro-fatty acids and their B. animalis subsp. lactis various derivatives.
Fructose is often linked to various metabolic and heart diseases, where increased dietary levels of fructose are linked to an increased risk of intestinal inflammation (Tan et al., 2021). Thus, probiotic strains that can absorb and utilize fructose as a carbon source is considerably preferred. Conversely, various Bifidobacterium strains have been shown to possess fructokinase (fructose phosphotransferase) activity, crucial for fructose metabolism (Figure 2) (Mazé et al., 2007). The phosphorylated fructose (fructose-1-phosphate) is then assimilated into the Bifid shunt to increase CoA derivatives production. This fructokinase activity is encoded by the fruA gene in B. breve UCC2003 that is homologous to the EIIBCA gene of the phosphoenolpyruvate: sugar phosphotransferase system of B. longum NCC2705. Both the encoded FruA and EIIBCA are known to play an integral part in the breakdown and assimilation of fructose (Mazé et al., 2007).
Similarly, many people suffer from lactose intolerance, where symptoms can range from mild indigestion to severe water loss and malnutrition. To this extent, the ability of Bifidobacterium to metabolize lactose can be considered as a potential health benefiting property. Additionally, lactose utilization should not produce biochemicals that are toxic to the human host. Interestingly, studies have shown that Bifidobacterium has a higher preference for lactose than other simple sugars like glucose. For instance, in B. longum NCC2705, the gene glcP, a putative glucose transporter, showed that the presence of lactose reduces the influx of glucose into the Bifidobacterial cytosol conferring a preference to lactose instead (Parche et al., 2006). Additionally, studies using B. bifidum indicate that the microbe presents some lactase activity in a slightly acidic environment within the gastrointestinal tract, resulting in the breakdown of lactose to glucose and galactose that are then assimilated into the Bifid shunt (Passerat et al., 1995).
Similar to the observations discussed earlier, the intake of the various sugars fructose and lactose is highly dependent on the concentrations of other sugars in the surrounding environment. The synergistic interactions of the various sugars result in a change in the preference of certain sugar types that enters the Bifid shunt. In some instances, such as in B. adolescentis MB239, the Bifidobacterium prefers the uptake of fructose and lactose depending on other types of sugars where the studies suggest some form of synergistic interactions between the sugars (Amaretti et al., 2006; Tan et al., 2021).
However, not all strains of Bifidobacterium have the desired properties of being a probiotic strain. A probiotic strain needs several attributes, including the ability to survive the human host environment and provide host-benefiting properties while showing no pathogenicity to the host. The following subsection discusses the limitations of using Bifidobacterium as an engineering chassis and methods of circumventing these limitations.
Some studies found that certain Bifidobacterium could not survive in the presence of bile salt due to the lack of conjugated bile salt hydrolase activity. In particular, the presence of glycoconjugate bile salt such as glycodeoxycholic acid presents higher toxicity to Bifidobacterium under an acidic environment. A study concluded that the susceptibility of Bifidobacteria to the glycoconjugated bile salt is dependent on the conjugated bile salt hydrolase activity (CSBH) (Grill et al., 2000). The presence of glycodeoxycholic and taurodeoxycholic acids was found to interfere with the survivability of the B. animalis ATCC25527, B. breve ATCC15700, B. longum ATCC15707 and B. coryneforme under lower pH conditions. These bile salt-sensitive Bifidobacteria are either CBSH or lacking the deconjugating properties, where either one of the two properties allows the Bifidobacteria to protonate and deconjugate the bile salt prior to export to the surrounding (Grill et al., 2000). Currently, many variants of Bifidobacteria naturally resistant to bile salts are considered good engineering chassis. However, should there be a need to develop a novel strain of Bifidobacterium that does not have the bile salt tolerance, engineering these microbes to have CSBH activities can be considered an excellent strategy to engineer the microbial cell.
The mucin layer forms a protective barrier system crucial for preventing the adhesion and penetration of pathogens, toxins, and other damaging agents in the gut. The mucin layer comprises highly glycosylated O-linked glycoproteins secreted primarily by the exocrine glands and mucosa (Karav et al., 2018). The protection conferred by the mucin layer is attributed to the glycoprotein structure within the mucus layer that retains a large body of water (>95%), where the retain water forms a reservoir of electrolytes, antibodies, and nucleic acids. Mucin degradation is commonly facilitated by mucolytic taxa such as those from the Bacteroides that carries a large variety of enzymes, including proteases, sulfatases, fucosidases, neuramidases, β-galactosidases, α-N-acetylgalactosaminidases, α-N-acetylglucosaminidases, and exo/endo-β-N-acetyl-glucosamindases (Karav et al., 2018). Moreover, the depleted mucin layers might result in the translocation of the gut microflora and other toxins across the exposed gut lining tissues (Abe et al., 2010). Several intestinal Bifidobacterium isolates can degrade the mucin as a form of sustenance through endo-α-N-acetylgalatosaminidase and 1,2-α-fucosidase (Ruas-Madiedo et al., 2008). A study conducted on 22 different Bifidobacterium strains isolated from the human host found that most B. longum and B. bifidum isolates can break down the mucin layers in the host gut (Ruas-Madiedo et al., 2008). Conversely, the use of non-mucin degrading variants of Bifidobacterium such as B. longum subsp. infantis EVC001 showed that the probiotic strain prevents colonic mucin degradation in breastfed infants. This characteristic of the probiotic strain is due to the displacement of other mucin-degrading Bacteroides population (Karav et al., 2018). While there is limited proof to support the adverse effects of mucin-degrading Bifidobacterium, it is preferable that the engineered Bifidobacterium does not have the mucin-degrading capabilities as it would be deemed safer for consumption. The selection of Bifidobacterium as a suitable engineering host could leverage on naturally occurring Bifidobacteria or through knockout of the corresponding mucin-degrading genes.
Numerous studies have used Bifidobacterium as an engineering chassis, mainly to respond to various environmental triggers. These engineered Bifidobacterium have been designed to facilitate various roles, such as biosensing (Cronin et al., 2012), treatment of diseases (Cronin et al., 2012) or improved bioprocessing. Other approaches have been used mainly to elucidate the Bifidobacterium cellular function and improve microbial tolerance and survivability in the environment (Watson et al., 2008; He et al., 2012a). An example of conventional Bifidobacterium engineering chassis is B. breve UCC 2003, which was used for improving tolerance to bile salt and survival under the gastrointestinal tract physiological environment. This improved tolerance was achieved by expressing the Listeria monocytogenes bile resistance gene, BilE (Watson et al., 2008). A similar Bifidobacterium was engineered to sense tumours and set the stage for later anticancer treatment using the microbial cell line (Cronin et al., 2012). Another commonly used strain of Bifidobacterium is the B. longum 105-A that was previously used in investigating the inducible and constitutive promoters. The isolated promoter sequences were reintroduced into B. longum 105-A with the α-galatosidase reporter gene (Sakanaka et al., 2014). This particular strain is easier to manipulate compared to other variants of Bifidobacterium due to the higher transformation efficiency used primarily in knocking out/down genes for closer study of the biochemical pathway (Kanesaki et al., 2014). Similar to B. breve UCC 2003, B. longum 105-A was also used in improving its tolerance to oxidative stress through the expression of the katE catalase gene isolated from Bacillus subtilis (He et al., 2012a). Another study used a similar approach to confer oxidative stress tolerance in B. thermophilum RBL67 through transferring the B. longum gene bl_1,404 (Stevens et al., 2017). The various Bifidobacterium used as an engineering chassis, their culture collection bank, and their source are described in Table 2.
Genetic tools used to regulate the Bifidobacterial gene expression could be generally divided into the promoters, ribosomal binding sites (RBS), and terminator sequence. The various elements regulating gene expression will be discussed in the following sections. These various elements can be used individually or concertedly to optimize the regulated microbial function.
Promoters are gene sequences that recruit the RNA polymerase to trigger mRNA transcription for downstream cellular processes. These sequences are located upstream of the coding region, including the ribosomal binding site and the gene sequences. The naturally occurring promoter sequences contains promoter core motifs located at the -35 to -10 region from the start of the coding sequence. These sequences conventionally contain the TTGNNN and the TANNNT conserved sequences respectively (Kozakai et al., 2021).
Promoter sequences are generally divided into two categories, namely the constitutive promoters and the inducible promoters. Constitutive promoters piggyback on the cellular metabolic function of the Bifidobacterium, facilitating a gene expression depending on the microbial metabolism. On the other hand, inducible promoters often carry a repressor/activator protein that binds on the operator site at either side of the -35 sequence. The repressor protein will bind on the operator site, inhibiting the recruitment of the RNA polymerase complex. In the presence of the corresponding ligand, binding of the ligand to the repressor causes conformational changes triggering the release from the operator site. On the other hand, activators will bind on the operator site after binding to the target ligand, facilitating the recruitment of the RNA polymerase complex.
Various constitutive promoters for Bifidobacterium expression were developed by sifting through the gene sequences within the Bifidobacterium genome (Sakanaka et al., 2014; Kozakai et al., 2021). The promoter core motif sequences were identified in silico using the hidden Markov model on the upstream sequences of the transcriptional start site of various coding genes in Bifidobacterium. This approach was used to identify the various putative constitutive promoter sequences in B. longum NCC2705 (Kozakai et al., 2020; Kozakai et al., 2021) and B. longum 105-A (Sakanaka et al., 2014). Additionally, the space between the -35 and -10 sequences can range from 11 to 18 nucleotides in length, where a shorter space length was found to improve the transcription levels (Kozakai et al., 2020).
Inducible expression within Bifidobacterium is mainly linked to the sugar-related inducers that are linked to the Bifid shunt. Some of these promoters were found to function in other microbes, whereas others were specific to Bifidobacterium. Methods of identifying these inducible promoters are through transcriptomic analysis of genes that are upregulated in the presence of the inducer molecules, where through annotating the upregulated genes, the corresponding promoter region is investigated. This approach identified sucrose and raffinose inducible promoters from B. lactis (Trindade et al., 2003) and B. longum NCC2705 (Kullin et al., 2006). Another example would be using the pNZ8048 nisin inducible promoter initially used for expression within Lactobacillus (Landete et al., 2014). In a similar study, the elongation factor Tu from B. longum subsp. infantis ATCC15697 was used for stable anaerobic expression of green fluorescent protein in both B. longum Reuter 1963 CECT4551 and B. breve INIA P734 (Landete et al., 2014).
In order to facilitate a better understanding of the various working promoters and the −35 and −10 sequences, Table 3 provides the information of the various types of promoters, the source plasmids, sequence origin and the promoter motifs.
In the efforts to optimize the RBS for efficient protein, two primary considerations are taken into account. First, the flanking sequences of the RBS site influence the recruitment of the ribosomal subunits to the mRNA sequences (Fukiya et al., 2018). Second, the optimal distance between the RBS site and the start codon influences the rate of protein expression (Fukiya et al., 2018). Interestingly, certain studies have shown that the predominant conserved Shine-Dalgarno sequence in Bifidobacterium differs from conventional microbes where the most common of 6-mer consensus RBS in B. longum is AAGGAG as compared to the common AGGAGG (He et al., 2012b; Kozakai et al., 2020). Additionally, the space between the RBS and the start codon was found to be optimal at 5 nucleotides apart, where fewer than 5 nucleotides apart result in translation repression. While 5 nucleotides space showed the best expression outcome, changes in expression level were minimal when the RBS is 6–9 nucleotides from the start codon (He et al., 2012b).
In investigating the various terminators used in Bifidobacterium, the gene sequences were identified using WebGeSTer DB terminator database (Kozakai et al., 2021). These include the canonical (L-shaped hairpin structure) and non-canonical (I-, U-, V-, and X-shaped hairpin structures) terminators used across different phyla of microorganisms (Mitra et al., 2011). Other approaches rely on pre-existing terminator sequences found in the Bifidobacterial genome.
Although wild-type Bifidobacteria have exhibited many therapeutic applications in treating diseases (Section 1), they are endowed with more functions via synthetic biology tools such as integrating new pathways or modifying the original metabolic pathways. These engineered Bifidobacteria are wildly applied to treat inflammatory diseases. For example, B. longum HB15 expressing α-melanocyte-stimulating hormone (α-MSH) was used to combat ulcerative colitis (Wei et al., 2016a). α-MSH is a tridecapeptide that exhibits anti-inflammatory properties by regulating the production of inflammatory mediators. B. longum NCC2705 expressing interleukin-12 was used to treat Coxsackie virus B3-induced myocarditis in mice (Yu et al., 2012). The oral administration of B. longum NCC 2705 expressing oxyntomodulin can reduce food intake, body weight and plasma lipid level in overweight mice (Long et al., 2010). Oxyntomodulin is a gut hormone that reduces food intake and body weight. B. longum HB25 expressing the antibacterial peptide LL-37 was used for treating bacterial diarrhea (Guo et al., 2017).
Additionally, since Bifidobacteria can germinate and proliferate in the hypoxic regions of solid tumors (Yazawa et al., 2000; Yazawa et al., 2001; Cronin et al., 2010), Bifidobacteria are currently widely applied as an in situ delivery and production of various anticancer agents for treating tumors (Taniguchi, 2021). For example, B. longum 105-A was used to deliver cytosine deaminase that catalytically converts the non-toxic prodrug 5-fluorocytosine to the anticancer drug 5-fluorouracil, to the rat mammary tumors (Taniguchi et al., 2016) or mice metastatic breast tumors (Fujimori, 2006). A similar approach using B. breve I-53-8w was used to deliver cytosine deaminase to mice lung cancer tumors (Zu and Wang, 2014). Other approaches uses B. longum NCC2705 to express tumstatin protein (a powerful angiostatin that inhibits proliferation and induces apoptosis of tumorous vascular endothelial cells), used as an antitumor therapy in tumor-bearing mice (Wei et al., 2016b).
Bifidobacterium is considered a vital composition of the probiotic mix that essentially has various health benefiting properties when administered at the appropriate dosage. The commercial use of the various types of these probiotics have been found to exert various therapeutic properties including antiinfection, antiinflammation, anticancer, promoting host psychological and physical health, and regulating host immune system.
On the other hand, given the nature of Bifidobacteria and their natural biochemical properties, the microbe presents a suitable host for cellular engineering. The engineering of Bifidobacterium can facilitate the increased bioproduction of value-added chemicals while consuming lesser resources compared to other microbial workhorses. One of the vital biochemical processes is the Bifid shunt that produces higher CoA precursors for the bioproduction of polyketide products and fatty acid biosynthesis (Wang et al., 2019b). On top of this, the probiotic microbe readily consumes other forms of sugars that further expands the ability of the microbial cell to function as a microbial cell factory (Mazé et al., 2007; Parche et al., 2006). Additionally, with the natural ability of the microbe to produce conjugated linoleic acids (Park et al., 2011), the microbe can be used in general to produce nitro-fatty acids that have recently been gaining interest as a potential treatment for metabolic diseases (Bonacci et al., 2012). Furthermore, coupled to the natural therapeutic properties of the microbial cell, the use of Bifidobacteria as an engineering chassis presents an interesting alternative for metabolic engineers, synthetic biologists, and evolutionary biologists to develop in-situ treatment of various ailments in the human host.
However, as earlier discussed, not all Bifidobacterium is considered a probiotic strain due to the lack of microbial resistance to the harsh environment within the human host. Further, some of the microbes are considered unsuited for use as a probiotic owing to the microbe’s ability to break down the host mucin layers (Ruas-Madiedo et al., 2008). The mucin layer essentially functions as a protective layer within the human host, preventing the infiltration of other pathogens and the absorption of toxic compounds by the human host. Thus, to increase the list of engineerable Bifidobacterium, supplementing or removing certain genes would help develop better-suited probiotics for the human host.
Current known Bifidobacterium chassis and gene tools are limited, hampering the progress of developing engineering Bifidobacterium. Currently, most engineering efforts of Bifidobacterium are centred on B. longum and B. breve, with a few exceptions of other suitable microbial chassis. Additionally, various constitutive and inducible promoters were identified that were directly isolated from the Bifidobacterial genome. Interestingly, these gene promoters were functional in other microbial chassis such as Bacillus and Escherichia coli. The current inducible promoters used in Bifidobacterial engineering use sugar complexes as inducers to trigger genes regulated in the Bifid shunt pathway (Wang et al., 2019b).
While the current studies show promise of further developing Bifidobacterium as an engineering host, there is an increasing need for identifying more genetic tools that are better suited for detecting and responding to the various triggers in the host body. These efforts include identifying various regulatory elements from the pre-existing Bifidobacterial genome sequences and other closely related microbial species from the Actinobacterial taxa. Furthermore, the role of these engineered Bifidobacteria in the host-microbiome is scarcely studied and would require further investigation to better understand the impact and safety of using these microbes to treat diseases in the future.
Conceptualization- JC, and CLH; Formal analysis- JC, and CLH; Investigation- JC, and CLH; Resources- CLH; Writing-original draft preparation, JC, and CLH; Writing-review and editing- JC, XC, and CLH; Visualization- JC, and CLH; Supervision- XC, and CLH; Project administration- JC, and CLH; Fund acquisition- CLH; All authors discussed, commented and agreed to publish this version of the manuscript.
This work was supported by the Shenzhen Special Fund for Innovation and Entrepreneurship of Overseas High-level Talents Peacock Team (KQTD20170810111314625), Shenzhen Institutes of Advanced Technology External Funds (DWKF20190,001), National Natural Science Foundation of China’s Research Fund for International Young Scientists (22,050,410,270), and Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y191).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank the advice provided by Professor Dai Lei and Assistant Professor Tan Yang from the Shenzhen Institute of Advanced Technology (SIAT), and Associate Professor Tang Bin from the Southern University of Science and Technology.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.770248/full#supplementary-material
Abdelhamid, A. G., Esaam, A., and Hazaa, M. M. (2018). Cell Free Preparations of Probiotics Exerted Antibacterial and Antibiofilm Activities against Multidrug Resistant E. coli. Saudi Pharm. J. 26, 603–607. doi:10.1016/j.jsps.2018.03.004
Abe, F., Muto, M., Yaeshima, T., Iwatsuki, K., Aihara, H., Ohashi, Y., et al. (2010). Safety Evaluation of Probiotic Bifidobacteria by Analysis of Mucin Degradation Activity and Translocation Ability. Anaerobe 16, 131–136. doi:10.1016/j.anaerobe.2009.07.006
Agusti, A., Moya-Pérez, A., Campillo, I., Montserrat-De La Paz, S., Cerrudo, V., Perez-Villalba, A., et al. (2018). Bifidobacterium Pseudocatenulatum CECT 7765 Ameliorates Neuroendocrine Alterations Associated with an Exaggerated Stress Response and Anhedonia in Obese Mice. Mol. Neurobiol. 55, 5337–5352. doi:10.1007/s12035-017-0768-z
Al-Sheraji, S. H., Ismail, A., Manap, M. Y., Mustafa, S., Yusof, R. M., and Hassan, F. A. (2012). Hypocholesterolaemic Effect of Yoghurt Containing Bifidobacterium Pseudocatenulatum G4 or Bifidobacterium Longum BB536. Food Chem. 135, 356–361. doi:10.1016/j.foodchem.2012.04.120
Allen, A. P., Hutch, W., Borre, Y. E., Kennedy, P. J., Temko, A., Boylan, G., et al. (2016). Bifidobacterium Longum 1714 as a Translational Psychobiotic: Modulation of Stress, Electrophysiology and Neurocognition in Healthy Volunteers. Transl Psychiatry 6, e939. doi:10.1038/tp.2016.191
Aloisio, I., Santini, C., Biavati, B., Dinelli, G., Cencič, A., Chingwaru, W., et al. (2012). Characterization of Bifidobacterium Spp. Strains for the Treatment of Enteric Disorders in Newborns. Appl. Microbiol. Biotechnol. 96, 1561–1576. doi:10.1007/s00253-012-4138-5
Amaretti, A., Tamburini, E., Bernardi, T., Pompei, A., Zanoni, S., Vaccari, G., et al. (2006). Substrate Preference of Bifidobacterium Adolescentis MB 239: Compared Growth on Single and Mixed Carbohydrates. Appl. Microbiol. Biotechnol. 73, 654–662. doi:10.1007/s00253-006-0500-9
Arseneault-Bréard, J., Rondeau, I., Gilbert, K., Girard, S.-A., Tompkins, T. A., Godbout, R., et al. (2012). Combination of Lactobacillus Helveticus R0052 and Bifidobacterium Longum R0175 Reduces post-myocardial Infarction Depression Symptoms and Restores Intestinal Permeability in a Rat Model. Br. J. Nutr. 107, 1793–1799. doi:10.1017/s0007114511005137
Arunachalam, K., Gill, H., and Chandra, R. (2000). Enhancement of Natural Immune Function by Dietary Consumption of Bifidobacterium Lactis (HN019). Eur. J. Clin. Nutr. 54, 263–267. doi:10.1038/sj.ejcn.1600938
Ballini, A., Gnoni, A., De Vito, D., Dipalma, G., Cantore, S., Gargiulo Isacco, C., et al. (2019). Effect of Probiotics on the Occurrence of Nutrition Absorption Capacities in Healthy Children: A Randomized Double-Blinded Placebo-Controlled Pilot Study. Eur. Rev. Med. Pharmacol. Sci. 23, 8645–8657. doi:10.26355/eurrev_201910_19182
Barba-Vidal, E., Castillejos, L., Roll, V. F. B., Cifuentes-Orjuela, G., Moreno Muñoz, J. A., and Martín-Orúe, S. M. (2017). The Probiotic Combination of Bifidobacterium Longum Subsp. Infantis CECT 7210 and Bifidobacterium Animalis Subsp. Lactis BPL6 Reduces Pathogen Loads and Improves Gut Health of Weaned Piglets Orally Challenged with Salmonella Typhimurium. Front. Microbiol. 8, 1570. doi:10.3389/fmicb.2017.01570
Bekkali, N.-L. -H., Bongers, M. E., Van den Berg, M. M., Liem, O., and Benninga, M. A. (2007). The Role of a Probiotics Mixture in the Treatment of Childhood Constipation: a Pilot Study. Nutr. J. 6, 17. doi:10.1186/1475-2891-6-17
Bonacci, G., Baker, P. R. S., Salvatore, S. R., Shores, D., Khoo, N. K. H., Koenitzer, J. R., et al. (2012). Conjugated Linoleic Acid Is a Preferential Substrate for Fatty Acid Nitration. J. Biol. Chem. 287, 44071–44082. doi:10.1074/jbc.m112.401356
Bongers, M. E. J., van Wijk, M. P., Reitsma, J. B., and Benninga, M. A. (2010). Long-term Prognosis for Childhood Constipation: Clinical Outcomes in Adulthood. Pediatrics 126, e156–e162. doi:10.1542/peds.2009-1009
Botic, T., Klingberg, T., Weingartl, H., and Cencic, A. (2007). A Novel Eukaryotic Cell Culture Model to Study Antiviral Activity of Potential Probiotic Bacteria. Int. J. Food Microbiol. 115, 227–234. doi:10.1016/j.ijfoodmicro.2006.10.044
Braga, T. D., da Silva, G. A. P., de Lira, P. I. C., and de Carvalho Lima, M. (2011). Efficacy of Bifidobacterium Breve and Lactobacillus Casei Oral Supplementation on Necrotizing Enterocolitis in Very-Low-Birth-Weight Preterm Infants: a Double-Blind, Randomized, Controlled Trial. Am. J. Clin. Nutr. 93, 81–86. doi:10.3945/ajcn.2010.29799
Brown, C. J., Mtui, D., Oswald, B. P., Van Leuven, J. T., Vallender, E. J., Schultz-Darken, N., et al. (2019). Comparative Genomics of Bifidobacterium Species Isolated from Marmosets and Humans. Am. J. Primatol 81, e983. doi:10.1002/ajp.22983
Çaglar, E., Onder Kuscu, O., Selvi Kuvvetli, S., Kavaloglu Cildir, S., Sandalli, N., and Twetman, S. (2008). Short-term Effect of Ice-Cream containingBifidobacterium lactisBb-12 on the Number of Salivary Mutans Streptococci and Lactobacilli. Acta Odontologica Scand. 66, 154–158. doi:10.1080/00016350802089467
Caimari, A., del Bas, J. M., Boqué, N., Crescenti, A., Puiggròs, F., Chenoll, E., et al. (2017). Heat-killed Bifidobacterium Animalis Subsp. Lactis CECT 8145 Increases Lean Mass and Ameliorates Metabolic Syndrome in Cafeteria-Fed Obese Rats. J. Funct. Foods 38, 251–263. doi:10.1016/j.jff.2017.09.029
Chapman, C. M. C., Gibson, G. R., and Rowland, I. (2011). Health Benefits of Probiotics: Are Mixtures More Effective Than Single Strains? Eur. J. Nutr. 50, 1–17. doi:10.1007/s00394-010-0166-z
Chitapanarux, T., Thongsawat, S., Pisespongsa, P., Leerapun, A., and Kijdamrongthum, P. (2015). Effect of Bifidobacterium Longum on PPI-Based Triple Therapy for Eradication of Helicobacter pylori: a Randomized, Double-Blind Placebo-Controlled Study. J. Funct. Foods 13, 289–294. doi:10.1016/j.jff.2015.01.003
Coakley, M., Ross, R. P., Nordgren, M., Fitzgerald, G., Devery, R., and Stanton, C. (2003). Conjugated Linoleic Acid Biosynthesis by Human-Derived Bifidobacterium Species. J. Appl. Microbiol. 94, 138–145. doi:10.1046/j.1365-2672.2003.01814.x
Colbère-Garapin, F., Martin-Latil, S., Blondel, B., Mousson, L., Pelletier, I., Autret, A., et al. (2007). Prevention and Treatment of Enteric Viral Infections: Possible Benefits of Probiotic Bacteria. Microbes Infect. 9, 1623–1631. doi:10.1016/j.micinf.2007.09.016
Collado, M. C., Meriluoto, J., and Salminen, S. (2007). Development of New Probiotics by Strain Combinations: Is it Possible to Improve the Adhesion to Intestinal Mucus? J. Dairy Sci. 90, 2710–2716. doi:10.3168/jds.2006-456
Cronin, M., Akin, A. R., Collins, S. A., Meganck, J., Kim, J.-B., Baban, C. K., et al. (2012). High Resolution In Vivo Bioluminescent Imaging for the Study of Bacterial Tumour Targeting. PLoS One 7, e30940. doi:10.1371/journal.pone.0030940
Cronin, M., Morrissey, D., Rajendran, S., El Mashad, S. M., Van Sinderen, D., O'sullivan, G. C., et al. (2010). Orally Administered Bifidobacteria as Vehicles for Delivery of Agents to Systemic Tumors. Mol. Ther. 18, 1397–1407. doi:10.1038/mt.2010.59
Di Pierro, F., and Pane, M. (2021). Bifidobacterium Longum W11: Uniqueness and Individual or Combined Clinical Use in Association with Rifaximin. Clin. Nutr. ESPEN 42, 15–21. doi:10.1016/j.clnesp.2020.12.025
Din, A. U., Hassan, A., Zhu, Y., Zhang, K., Wang, Y., Li, T., et al. (2020). Inhibitory Effect of Bifidobacterium Bifidum ATCC 29521 on Colitis and its Mechanism. J. Nutr. Biochem. 79, 108353. doi:10.1016/j.jnutbio.2020.108353
Dinan, T. G., Stanton, C., and Cryan, J. F. (2013). Psychobiotics: A Novel Class of Psychotropic. Biol. Psychiatry 74, 720–726. doi:10.1016/j.biopsych.2013.05.001
Enomoto, T., Sowa, M., Nishimori, K., Shimazu, S., Yoshida, A., Yamada, K., et al. (2014). Effects of Bifidobacterial Supplementation to Pregnant Women and Infants in the Prevention of Allergy Development in Infants and on Fecal Microbiota. Allergol. Int. 63, 575–585. doi:10.2332/allergolint.13-oa-0683
Finamore, A., Roselli, M., Donini, L., Brasili, D. E., Rami, R., Carnevali, P., et al. (2019). Supplementation with Bifidobacterium Longum Bar33 and Lactobacillus Helveticus Bar13 Mixture Improves Immunity in Elderly Humans (Over 75 years) and Aged Mice. Nutrition 63-64, 184–192. doi:10.1016/j.nut.2019.02.005
Fonseca, J. F., Alvim, L. B., Nunes, Á. C., Oliveira, F. M. S., Amaral, R. S., Caliari, M. V., et al. (2019). Probiotic Effect of Bifidobacterium Longum 5 1A and Weissella Paramesenteroides WpK4 on Gerbils Infected with Giardia Lamblia. J. Appl. Microbiol. 127, 1184–1191. doi:10.1111/jam.14338
Foo, N.-P., Ou Yang, H., Chiu, H.-H., Chan, H.-Y., Liao, C.-C., Yu, C.-K., et al. (2011). Probiotics Prevent the Development of 1,2-Dimethylhydrazine (DMH)-Induced Colonic Tumorigenesis through Suppressed Colonic Mucosa Cellular Proliferation and Increased Stimulation of Macrophages. J. Agric. Food Chem. 59, 13337–13345. doi:10.1021/jf203444d
Fujimori, M. (2006). Genetically Engineeredbifidobacterium as a Drug Delivery System for Systemic Therapy of Metastatic Breast Cancer Patients. Breast cancer 13, 27–31. doi:10.2325/jbcs.13.27
Fukiya, S., Sakanaka, M., and Yokota, A. (2018). “Genetic Manipulation and Gene Modification Technologies in Bifidobacteria,” in The Bifidobacteria and Related Organisms. Editor P. Mattarelliet al. (Academic Press), 243–259. doi:10.1016/b978-0-12-805060-6.00015-6
Fukushima, Y., Kawata, Y., Mizumachi, K., Kurisaki, J.-i., and Mitsuoka, T. (1999). Effect of Bifidobacteria Feeding on Fecal flora and Production of Immunoglobulins in Lactating Mouse. Int. J. Food Microbiol. 46, 193–197. doi:10.1016/s0168-1605(98)00183-4
Gardini, F., Özogul, Y., Suzzi, G., Tabanelli, G., and Özogul, F. (2016). Technological Factors Affecting Biogenic Amine Content in Foods: a Review. Front. Microbiol. 7, 1218. doi:10.3389/fmicb.2016.01218
Gaya, P., Peirotén, Á., Medina, M., Álvarez, I., and Landete, J. M. (2018). Bifidobacterium Pseudocatenulatum INIA P815: the First Bacterium Able to Produce Urolithins A and B from Ellagic Acid. J. Funct. Foods 45, 95–99. doi:10.1016/j.jff.2018.03.040
Gill, H. S., Rutherfurd, K. J., Prasad, J., and Gopal, P. K. (2000). Enhancement of Natural and Acquired Immunity byLactobacillus rhamnosus(HN001),Lactobacillus acidophilus(HN017) and Bifidobacterium lactis(HN019). Br. J. Nutr. 83, 167–176. doi:10.1017/s0007114500000210
Grill, J. P., Perrin, S., and Schneider, F. (2000). Bile Salt Toxicity to Some Bifidobacteria Strains: Role of Conjugated Bile Salt Hydrolase and pH. Can. J. Microbiol. 46, 878–884. doi:10.1139/w00-066
Gu, H., Jinkerson, R. E., Davies, F. K., Sisson, L. A., Schneider, P. E., and Posewitz, M. C. (2016). Modulation of Medium-Chain Fatty Acid Synthesis in Synechococcus Sp. PCC 7002 by Replacing FabH with a Chaetoceros Ketoacyl-ACP Synthase. Front. Plant Sci. 7, 690. doi:10.3389/fpls.2016.00690
Guglielmetti, S., Mora, D., Gschwender, M., and Popp, K. (2011). Randomised Clinical Trial: Bifidobacterium Bifidum MIMBb75 Significantly Alleviates Irritable Bowel Syndrome and Improves Quality of Life -- a Double-Blind, Placebo-Controlled Study. Aliment. Pharmacol. Ther. 33, 1123–1132. doi:10.1111/j.1365-2036.2011.04633.x
Guo, Q., Li, S., Xie, Y., Xu, Z., Liu, M., Zhang, Q., et al. (2017). Bifidobacterium Longum as an Orally Administered Carrier of LL-37 to Treat Bacterial Diarrhea. J. Pharm. Biomed. Sci. 7.
Hazarika, S. N., and Thakur, D. (2020). “Actinobacteria,” in Beneficial Microbes in Agro-Ecology. Editor N. Amaresanet al. (Academic Press), 443–476. doi:10.1016/b978-0-12-823414-3.00021-6
He, J., Sakaguchi, K., and Suzuki, T. (2012). Acquired Tolerance to Oxidative Stress in Bifidobacterium Longum 105-A via Expression of a Catalase Gene. Appl. Environ. Microbiol. 78, 2988–2990. doi:10.1128/aem.07093-11
He, J., Sakaguchi, K., and Suzuki, T. (2012). Determination of the Ribosome-Binding Sequence and Spacer Length between Binding Site and Initiation Codon for Efficient Protein Expression in Bifidobacterium Longum 105-A. J. Biosci. Bioeng. 113, 442–444. doi:10.1016/j.jbiosc.2011.11.019
Hong, K. S., Kang, H. W., Im, J. P., Ji, G. E., Kim, S. G., Jung, H. C., et al. (2009). Effect of Probiotics on Symptoms in Korean Adults with Irritable Bowel Syndrome. Gut Liver 3, 101–107. doi:10.5009/gnl.2009.3.2.101
Hudault, S., Bridonneau, C., Raibaud, P., Chabanet, C., and Vial, M. F. (1994). Relationship between Intestinal Colonization of Bifidobacterium Bifidum in Infants and the Presence of Exogenous and Endogenous Growth-Promoting Factors in Their Stools. Pediatr. Res. 35, 696–700. doi:10.1203/00006450-199406000-00015
Igbafe, J., Kilonzo-Nthenge, A., Nahashon, S. N., Mafiz, A. I., and Nzomo, M. (2020). Probiotics and Antimicrobial Effect of Lactiplantibacillus Plantarum, Saccharomyces cerevisiae, and Bifidobacterium Longum against Common Foodborne Pathogens in Poultry. Agriculture 10, 368. doi:10.3390/agriculture10090368
Invernici, M. M., Salvador, S. L., Silva, P. H. F., Soares, M. S. M., Casarin, R., Palioto, D. B., et al. (2018). Effects of Bifidobacterium Probiotic on the Treatment of Chronic Periodontitis: a Randomized Clinical Trial. J. Clin. Periodontol. 45, 1198–1210. doi:10.1111/jcpe.12995
Jang, H.-M., Lee, K.-E., and Kim, D.-H. (2019). The Preventive and Curative Effects of Lactobacillus Reuteri NK33 and Bifidobacterium Adolescentis NK98 on Immobilization Stress-Induced Anxiety/depression and Colitis in Mice. Nutrients 11, 819. doi:10.3390/nu11040819
Jang, H. M., Jang, S.-E., Han, M. J., and Kim, D.-H. (2018). Anxiolytic-like Effect of Bifidobacterium Adolescentis IM38 in Mice with or without Immobilisation Stress. Beneficial microbes 9, 123–132. doi:10.3920/bm2016.0226
Kanamori, Y., Iwanaka, T., Sugiyama, M., Komura, M., Takahashi, T., Yuki, N., et al. (2010). Early Use of Probiotics Is Important Therapy in Infants with Severe Congenital Anomaly. Pediatr. Int. 52, 362–367. doi:10.1111/j.1442-200X.2009.02963.x
Kanamori, Y., Sugiyama, M., Hashizume, K., Yuki, N., Morotomi, M., and Tanaka, R. (2004). Experience of Long-Term Synbiotic Therapy in Seven Short Bowel Patients with Refractory Enterocolitis. J. Pediatr. Surg. 39, 1686–1692. doi:10.1016/j.jpedsurg.2004.07.013
Kanesaki, Y., Masutani, H., Sakanaka, M., Shiwa, Y., Fujisawa, T., Nakamura, Y., et al. (2014). Complete Genome Sequence of Bifidobacterium Longum 105-A, a Strain with High Transformation Efficiency. Genome Announc 2. doi:10.1128/genomea.01311-14
Karav, S., Casaburi, G., and Frese, S. A. (2018). Reduced Colonic Mucin Degradation in Breastfed Infants Colonized by Bifidobacterium Longum Subsp . Infantis EVC001. FEBS open bio 8, 1649–1657. doi:10.1002/2211-5463.12516
Kazemi, A., Noorbala, A. A., Azam, K., Eskandari, M. H., and Djafarian, K. (2019). Effect of Probiotic and Prebiotic vs Placebo on Psychological Outcomes in Patients with Major Depressive Disorder: A Randomized Clinical Trial. Clin. Nutr. 38, 522–528. doi:10.1016/j.clnu.2018.04.010
Kelley, R. L., Minikhiem, D., Kiely, B., O'Mahony, L., O'Sullivan, D., Boileau, T., et al. (2009). Clinical Benefits of Probiotic Canine-Derived Bifidobacterium Animalis Strain AHC7 in Dogs with Acute Idiopathic Diarrhea. Vet. Ther. 10, 121–130.
Kelly, S. M., Lanigan, N., O’Neill, I. J., Bottacini, F., Lugli, G. A., Viappiani, A., et al. (2020). Bifidobacterial Biofilm Formation Is a Multifactorial Adaptive Phenomenon in Response to Bile Exposure. Sci. Rep. 10, 11598. doi:10.1038/s41598-020-68179-9
Kim, M. J., Lee, D. K., Park, J. E., Park, I. H., Seo, J. G., and Ha, N. J. (2014). Antiviral Activity ofBifidobacterium adolescentisSPM1605 against Coxsackievirus B3. Biotechnol. Biotechnological Equipment 28, 681–688. doi:10.1080/13102818.2014.945237
Kim, N.-J., and Ji, G.-E. (2006). Modulatory Activity of Bifidobacterium Sp. BGN4 Cell Fractions on Immune Cells. J. Microbiol. Biotechnol. 16, 584–589.
Klemenak, M., Dolinšek, J., Langerholc, T., Di Gioia, D., and Mičetić-Turk, D. (2015). Administration of Bifidobacterium Breve Decreases the Production of TNF-α in Children with Celiac Disease. Dig. Dis. Sci. 60, 3386–3392. doi:10.1007/s10620-015-3769-7
Kobayashi, Y., Kinoshita, T., Matsumoto, A., Yoshino, K., Saito, I., and Xiao, J. Z. (2019). Bifidobacterium Breve A1 Supplementation Improved Cognitive Decline in Older Adults with Mild Cognitive Impairment: an Open-Label, Single-Arm Study. J. Prev. Alzheimers Dis. 6, 70–75. doi:10.14283/jpad.2018.32
Kobayashi, Y., Sugahara, H., Shimada, K., Mitsuyama, E., Kuhara, T., Yasuoka, A., et al. (2017). Therapeutic Potential of Bifidobacterium Breve Strain A1 for Preventing Cognitive Impairment in Alzheimer's Disease. Sci. Rep. 7, 13510–10. doi:10.1038/s41598-017-13368-2
Kozakai, T., Izumi, A., Horigome, A., Odamaki, T., Xiao, J. Z., Nomura, I., et al. (2020). Structure of a Core Promoter in Bifidobacterium Longum NCC2705. J. Bacteriol. 202, 202. doi:10.1128/JB.00540-19
Kozakai, T., Shimofusa, Y., Nomura, I., and Suzuki, T. (2021). Construction of a Reporter System for Bifidobacteria Using Chloramphenicol Acetyltransferase and its Application for Evaluation of Promoters and Terminators. Biosci. microbiota, Food Health 40, 115–122. doi:10.12938/bmfh.2020-070
Krumbeck, J. A., Rasmussen, H. E., Hutkins, R. W., Clarke, J., Shawron, K., Keshavarzian, A., et al. (2018). Probiotic Bifidobacterium Strains and Galactooligosaccharides Improve Intestinal Barrier Function in Obese Adults but Show No Synergism when Used Together as Synbiotics. Microbiome 6, 1–16. doi:10.1186/s40168-018-0494-4
Ku, S., Park, M., Ji, G., and You, H. (2016). Review on Bifidobacterium Bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. Ijms 17, 1544. doi:10.3390/ijms17091544
Ku, S., You, H., and Ji, G. (2009). Enhancement of Anti-tumorigenic Polysaccharide Production, Adhesion, and Branch Formation of Bifidobacterium Bifidum BGN4 by Phytic Acid. Food Sci. Biotechnol. 18, 749–754.
Kullin, B., Abratt, V. R., and Reid, S. J. (2006). A Functional Analysis of the Bifidobacterium Longum cscA and scrP Genes in Sucrose Utilization. Appl. Microbiol. Biotechnol. 72, 975–981. doi:10.1007/s00253-006-0358-x
Kuru, B. E., Laleman, I., Yalnızoğlu, T., Kuru, L., and Teughels, W. (2017). The Influence of a Bifidobacterium Animalis Probiotic on Gingival Health: a Randomized Controlled Clinical Trial. J. Periodontol. 88, 1115–1123. doi:10.1902/jop.2017.170213
Landete, J. M., Peirotén, Á., Rodríguez, E., Margolles, A., Medina, M., and Arqués, J. L. (2014). Anaerobic green Fluorescent Protein as a Marker of Bifidobacterium Strains. Int. J. Food Microbiol. 175, 6–13. doi:10.1016/j.ijfoodmicro.2014.01.008
Lau, A. S.-Y., Yanagisawa, N., Hor, Y.-Y., Lew, L.-C., Ong, J.-S., Chuah, L.-O., et al. (2018). Bifidobacterium Longum BB536 Alleviated Upper Respiratory Illnesses and Modulated Gut Microbiota Profiles in Malaysian Pre-school Children. Beneficial microbes 9, 61–70. doi:10.3920/bm2017.0063
Lee, D. K., Park, J. E., Kim, M. J., Seo, J. G., Lee, J. H., and Ha, N. J. (2015). Probiotic Bacteria, B. Longum and L. Acidophilus Inhibit Infection by Rotavirus In Vitro and Decrease the Duration of Diarrhea in Pediatric Patients. Clin. Res. Hepatol. Gastroenterol. 39, 237–244. doi:10.1016/j.clinre.2014.09.006
Lee, J. W., Shin, J. G., Kim, E. H., Kang, H. E., Yim, I. B., Kim, J. Y., et al. (2004). Immunomodulatory and Antitumor Effects In Vivo by the Cytoplasmic Fraction of Lactobacillus Casei and Bifidobacterium Longum. J. Vet. Sci. 5, 41–48. doi:10.4142/jvs.2004.5.1.41
Lee, M.-J., Zang, Z.-L., Choi, E.-Y., Shin, H.-K., and Ji, G.-E. (2002). Cytoskeleton Reorganization and Cytokine Production of Macrophages by Bifidobacterial Cells and Cell-free Extracts. J. Microbiol. Biotechnol. 12, 398–405.
Lim, S.-M., and Kim, D.-H. (2017). Bifidobacterium Adolescentis IM38 Ameliorates High-Fat Diet-Induced Colitis in Mice by Inhibiting NF-Κb Activation and Lipopolysaccharide Production by Gut Microbiota. Nutr. Res. 41, 86–96. doi:10.1016/j.nutres.2017.04.003
Long, R. T., Zeng, W. S., Chen, L. Y., Guo, J., Lin, Y. Z., Huang, Q. S., et al. (2010). Bifidobacterium as an Oral Delivery Carrier of Oxyntomodulin for Obesity Therapy: Inhibitory Effects on Food Intake and Body Weight in Overweight Mice. Int. J. Obes. 34, 712–719. doi:10.1038/ijo.2009.277
Martorell, P., Llopis, S., González, N., Chenoll, E., López-Carreras, N., Aleixandre, A., et al. (2016). Probiotic Strain Bifidobacterium Animalis Subsp. Lactis CECT 8145 Reduces Fat Content and Modulates Lipid Metabolism and Antioxidant Response in Caenorhabditis elegans. J. Agric. Food Chem. 64, 3462–3472. doi:10.1021/acs.jafc.5b05934
Matsumoto, T., Ishikawa, H., Tateda, K., Yaeshima, T., Ishibashi, N., and Yamaguchi, K. (2008). Oral Administration of Bifidobacterium Longum Prevents Gut-Derived Pseudomonas aeruginosa Sepsis in Mice. J. Appl. Microbiol. 104, 672–680. doi:10.1111/j.1365-2672.2007.03593.x
Mauricio, M. D., Serna, E., Fernández-Murga, M. L., Portero, J., Aldasoro, M., Valles, S. L., et al. (2017). Bifidobacterium Pseudocatenulatum CECT 7765 Supplementation Restores Altered Vascular Function in an Experimental Model of Obese Mice. Int. J. Med. Sci. 14, 444–451. doi:10.7150/ijms.18354
Mazé, A., O'Connell-Motherway, M., Fitzgerald, G. F., Deutscher, J., and van Sinderen, D. (2007). Identification and Characterization of a Fructose Phosphotransferase System in Bifidobacterium Breve UCC2003. Appl. Environ. Microbiol. 73, 545–553. doi:10.1128/aem.01496-06
Messaoudi, M., Lalonde, R., Violle, N., Javelot, H., Desor, D., Nejdi, A., et al. (2011). Assessment of Psychotropic-like Properties of a Probiotic Formulation (Lactobacillus helveticusR0052 andBifidobacterium longumR0175) in Rats and Human Subjects. Br. J. Nutr. 105, 755–764. doi:10.1017/s0007114510004319
Messaoudi, M., Violle, N., Bisson, J.-F., Desor, D., Javelot, H., and Rougeot, C. (2011). Beneficial Psychological Effects of a Probiotic Formulation (Lactobacillus helveticusR0052 andBifidobacterium longumR0175) in Healthy Human Volunteers. Gut microbes 2, 256–261. doi:10.4161/gmic.2.4.16108
Miraglia Del Giudice, M., Indolfi, C., Capasso, M., Maiello, N., Decimo, F., and Ciprandi, G. (2017). Bifidobacterium Mixture (B Longum BB536, B Infantis M-63, B Breve M-16V) Treatment in Children with Seasonal Allergic Rhinitis and Intermittent Asthma. Ital. J. Pediatr. 43, 25. doi:10.1186/s13052-017-0340-5
Mitra, A., Kesarwani, A. K., Pal, D., and Nagaraja, V. (2011). WebGeSTer DB-A Transcription Terminator Database. Nucleic Acids Res. 39, D129–D135. doi:10.1093/nar/gkq971
Moratalla, A., Gómez-Hurtado, I., Moya-Pérez, Á., Zapater, P., Peiró, G., González-Navajas, J. M., et al. (2016). Bifidobacterium Pseudocatenulatum CECT7765 Promotes a TLR2-dependent Anti-inflammatory Response in Intestinal Lymphocytes from Mice with Cirrhosis. Eur. J. Nutr. 55, 197–206. doi:10.1007/s00394-015-0837-x
Moreno Muñoz, J. A., Chenoll, E., Casinos, B., Bataller, E., Ramón, D., Genovés, S., et al. (2011). Novel Probiotic Bifidobacterium Longum Subsp. Infantis CECT 7210 Strain Active against Rotavirus Infections. Appl. Environ. Microbiol. 77, 8775–8783. doi:10.1128/aem.05548-11
Nguyen, M., Holdbrooks, H., Mishra, P., Abrantes, M. A., Eskew, S., Garma, M., et al. (2021). Impact of Probiotic B. Infantis EVC001 Feeding in Premature Infants on the Gut Microbiome, Nosocomially Acquired Antibiotic Resistance, and Enteric Inflammation. Front. Pediatr. 9, 24. doi:10.3389/fped.2021.618009
Nunoura, N., Ohdan, K., Yamamoto, K., and Kumagai, H. (1997). Expression of the β-d-glucosidase I Gene in Bifidobacterium Breve 203 during Acclimation to Cellobiose. J. Ferment. Bioeng. 83, 309–314. doi:10.1016/s0922-338x(97)80134-1
O'Callaghan, A., and Van Sinderen, D. (2016). Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 7, 925. doi:10.3389/fmicb.2016.00925
O'Mahony, D., Murphy, K. B., MacSharry, J., Boileau, T., Sunvold, G., Reinhart, G., et al. (2009). Portrait of a Canine Probiotic Bifidobacterium-Ffrom Gut to Gut. Vet. Microbiol. 139, 106–112. doi:10.1016/j.vetmic.2009.05.002
O'Mahony, D., Murphy, S., Boileau, T., Park, J., O'Brien, F., Groeger, D., et al. (2010). Bifidobacterium Animalis AHC7 Protects against Pathogen-Induced NF-Κb Activation In Vivo. BMC Immunol. 11, 63–69. doi:10.1186/1471-2172-11-63
O'Mahony, L., McCarthy, J., Kelly, P., Hurley, G., Luo, F., Chen, K., et al. (2005). Lactobacillus and Bifidobacterium in Irritable Bowel Syndrome: Symptom Responses and Relationship to Cytokine Profiles. Gastroenterology 128, 541–551. doi:10.1053/j.gastro.2004.11.050
Oberg, T. S., Ward, R. E., Steele, J. L., and Broadbent, J. R. (2015). Transcriptome Analysis of Bifidobacterium Longum Strains that Show a Differential Response to Hydrogen Peroxide Stress. J. Biotechnol. 212, 58–64. doi:10.1016/j.jbiotec.2015.06.405
Ohara, T., Yoshino, K., and Kitajima, M. (2010). Possibility of Preventing Colorectal Carcinogenesis with Probiotics. Hepatogastroenterology 57, 1411–1415.
Oliveira, L. F. F., Salvador, S. L., Silva, P. H. F., Furlaneto, F. A. C., Figueiredo, L., Casarin, R., et al. (2017). Benefits ofBifidobacterium animalissubsp.lactisProbiotic in Experimental Periodontitis. J. Periodontol. 88, 197–208. doi:10.1902/jop.2016.160217
Paolillo, R., Romano Carratelli, C., Sorrentino, S., Mazzola, N., and Rizzo, A. (2009). Immunomodulatory Effects of Lactobacillus Plantarum on Human colon Cancer Cells. Int. immunopharmacology 9, 1265–1271. doi:10.1016/j.intimp.2009.07.008
Parche, S., Beleut, M., Rezzonico, E., Jacobs, D., Arigoni, F., Titgemeyer, F., et al. (2006). Lactose-over-Glucose Preference in Bifidobacterium Longum NCC2705: glcP , Encoding a Glucose Transporter, Is Subject to Lactose Repression. J. Bacteriol. 188, 1260–1265. doi:10.1128/jb.188.4.1260-1265.2006
Park, H. G., Heo, W., Kim, S. B., Kim, H. S., Bae, G. S., Chung, S. H., et al. (2011). Production of Conjugated Linoleic Acid (CLA) by Bifidobacterium Breve LMC520 and its Compatibility with CLA-Producing Rumen Bacteria. J. Agric. Food Chem. 59, 984–988. doi:10.1021/jf103420q
Parvaneh, K., Ebrahimi, M., Sabran, M. R., Karimi, G., Hwei, A. N., Abdul-Majeed, S., et al. (2015). Probiotics (Bifidobacterium Longum) Increase Bone Mass Density and Upregulate Sparc and Bmp-2 Genes in Rats with Bone Loss Resulting from Ovariectomy. Biomed. Res. Int. 2015, 897639. doi:10.1155/2015/897639
Passerat, B., Desmaison, A.-M., Pharmaceutiques, D. e. S., and de Conférences, M. (1995). Lactase Activity of Bifidobacterium Bifidum. Nutr. Res. 15, 1287–1295. doi:10.1016/0271-5317(95)02004-f
Pedret, A., Valls, R. M., Calderón-Pérez, L., Llauradó, E., Companys, J., Pla-Pagà, L., et al. (2019). Effects of Daily Consumption of the Probiotic Bifidobacterium Animalis Subsp. Lactis CECT 8145 on Anthropometric Adiposity Biomarkers in Abdominally Obese Subjects: a Randomized Controlled Trial. Int. J. Obes. 43, 1863–1868. doi:10.1038/s41366-018-0220-0
Pinto-Sanchez, M. I., Hall, G. B., Ghajar, K., Nardelli, A., Bolino, C., Lau, J. T., et al. (2017). Probiotic Bifidobacterium Longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients with Irritable Bowel Syndrome. Gastroenterology 153, 448–459. e8. doi:10.1053/j.gastro.2017.05.003
Raimondi, S., Amaretti, A., Leonardi, A., Quartieri, A., Gozzoli, C., and Rossi, M. (2016). Conjugated Linoleic Acid Production by Bifidobacteria: Screening, Kinetic, and Composition. Biomed. Res. Int. 2016, 8654317. doi:10.1155/2016/8654317
Reddy, B. S., and Rivenson, A. (1993). Inhibitory Effect of Bifidobacterium Longum on colon, Mammary, and Liver Carcinogenesis Induced by 2-Amino-3-Methylimidazo[4,5-F]quinoline, a Food Mutagen. Cancer Res. 53, 3914–3918.
Ricoldi, M. S. T., Furlaneto, F. A. C., Oliveira, L. F. F., Teixeira, G. C., Pischiotini, J. P., Moreira, A. L. G., et al. (2017). Effects of the Probiotic Bifidobacterium Animalis Subsp. Lactis on the Non-surgical Treatment of Periodontitis. A Histomorphometric, Microtomographic and Immunohistochemical Study in Rats. PloS one 12, e0179946. doi:10.1371/journal.pone.0179946
Ringel-Kulka, T., Palsson, O. S., Maier, D., Carroll, I., Galanko, J. A., Leyer, G., et al. (2011). Probiotic Bacteria Lactobacillus Acidophilus NCFM and Bifidobacterium Lactis Bi-07 versus Placebo for the Symptoms of Bloating in Patients with Functional Bowel Disorders. J. Clin. Gastroenterol. 45, 518–525. doi:10.1097/mcg.0b013e31820ca4d6
Roberts, J. L., Liu, G., Darby, T. M., Fernandes, L. M., Diaz-Hernandez, M. E., Jones, R. M., et al. (2020). Bifidobacterium Adolescentis Supplementation Attenuates Fracture-Induced Systemic Sequelae. Biomed. Pharmacother. 132, 110831. doi:10.1016/j.biopha.2020.110831
Robles‐Vera, I., de la Visitación, N., Toral, M., Sánchez, M., Romero, M., Gómez‐Guzmán, M., et al. (2020). Probiotic Bifidobacterium Breve Prevents DOCA‐salt Hypertension. FASEB J. 34, 13626–13640.
Rodrigues, F. C., Castro, A. S. B., Rodrigues, V. C., Fernandes, S. A., Fontes, E. A. F., de Oliveira, T. T., et al. (2012). Yacon Flour andBifidobacterium longumModulate Bone Health in Rats. J. Med. Food 15, 664–670. doi:10.1089/jmf.2011.0296
Roselli, M., Finamore, A., Britti, M. S., and Mengheri, E. (2006). Probiotic bacteriaBifidobacterium animalisMB5 andLactobacillus rhamnosusGG Protect Intestinal Caco-2 Cells from the Inflammation-Associated Response Induced by enterotoxigenicEscherichia coliK88. Br. J. Nutr. 95, 1177–1184. doi:10.1079/bjn20051681
Roškar, I., Švigelj, K., Štempelj, M., Volfand, J., Štabuc, B., Malovrh, Š., et al. (2017). Effects of a Probiotic Product Containing Bifidobacterium Animalis Subsp. Animalis IM386 and Lactobacillus Plantarum MP2026 in Lactose Intolerant Individuals: Randomized, Placebo-Controlled Clinical Trial. J. Funct. Foods 35, 1–8.
Rowland, I., Rumney, C., Coutts, J., and Lievense, L. (1998). Effect of Bifidobacterium Longum and Inulin on Gut Bacterial Metabolism and Carcinogen-Induced Aberrant Crypt Foci in Rats. Carcinogenesis 19, 281–285. doi:10.1093/carcin/19.2.281
Ruas-Madiedo, P., Gueimonde, M., Fernández-García, M., de los Reyes-Gavilán, C. G., and Margolles, A. (2008). Mucin Degradation by Bifidobacterium Strains Isolated from the Human Intestinal Microbiota. Appl. Environ. Microbiol. 74, 1936–1940. doi:10.1128/aem.02509-07
Ruiz, L., Ruas-Madiedo, P., Gueimonde, M., de los Reyes-Gavilán, C. G., Margolles, A., and Sánchez, B. (2011). How Do Bifidobacteria Counteract Environmental Challenges? Mechanisms Involved and Physiological Consequences. Genes Nutr. 6, 307–318. doi:10.1007/s12263-010-0207-5
Sakanaka, M., Tamai, S., Hirayama, Y., Onodera, A., Koguchi, H., Kano, Y., et al. (2014). Functional Analysis of Bifidobacterial Promoters in Bifidobacterium Longum and Escherichia coli Using the α-galactosidase Gene as a Reporter. J. Biosci. Bioeng. 118, 489–495. doi:10.1016/j.jbiosc.2014.05.002
Savignac, H. M., Kiely, B., Dinan, T. G., and Cryan, J. F. (2014). Bifidobacteriaexert Strain-specific Effects on Stress-Related Behavior and Physiology in BALB/c Mice. Neurogastroenterol. Motil. 26, 1615–1627. doi:10.1111/nmo.12427
Schopfer, F. J., Vitturi, D. A., Jorkasky, D. K., and Freeman, B. A. (2018). Nitro-fatty Acids: New Drug Candidates for Chronic Inflammatory and Fibrotic Diseases. Nitric Oxide 79, 31–37. doi:10.1016/j.niox.2018.06.006
Sheu, B.-S., Cheng, H.-C., Kao, A.-W., Wang, S.-T., Yang, Y.-J., Yang, H.-B., et al. (2006). Pretreatment with Lactobacillus- and Bifidobacterium-Containing Yogurt Can Improve the Efficacy of Quadruple Therapy in Eradicating Residual Helicobacter pylori Infection after Failed Triple Therapy. Am. J. Clin. Nutr. 83, 864–869. doi:10.1093/ajcn/83.4.864
Stenman, L. K., Waget, A., Garret, C., Klopp, P., Burcelin, R., and Lahtinen, S. (2014). Potential Probiotic Bifidobacterium Animalis Ssp. Lactis 420 Prevents Weight Gain and Glucose Intolerance in Diet-Induced Obese Mice. Beneficial microbes 5, 437–445. doi:10.3920/bm2014.0014
Stevens, M. J. A., Venturini, A., Lacroix, C., and Meile, L. (2017). Enhancing Oxidative Stress Resistance in Bifidobacterium Thermophilum Using a Novel Overexpression Vector and Transformation Protocol. Plasmid 92, 43–48. doi:10.1016/j.plasmid.2017.06.002
Sugahara, H., Odamaki, T., Fukuda, S., Kato, T., Xiao, J.-z., Abe, F., et al. (2015). Probiotic Bifidobacterium Longum Alters Gut Luminal Metabolism through Modification of the Gut Microbial Community. Sci. Rep. 5, 13548. doi:10.1038/srep13548
Taipale, T. J., Pienihäkkinen, K., Isolauri, E., Jokela, J. T., and Söderling, E. M. (2016). Bifidobacterium Animalis Subsp. Lactis BB-12 in Reducing the Risk of Infections in Early Childhood. Pediatr. Res. 79, 65–69. doi:10.1038/pr.2015.174
Takeda, Y., Nakase, H., Namba, K., Inoue, S., Ueno, S., Uza, N., et al. (2009). Upregulation of T-Bet and Tight junction Molecules by Bifidobactrium Longum Improves Colonic Inflammation of Ulcerative Colitis. Inflamm. Bowel Dis. 15, 1617–1618. doi:10.1002/ibd.20861
Takeuchi, A., Matsumura, H., and Kano, Y. (2002). Cloning and Expression inEscherichia Coliof a Gene,hup, Encoding the Histone-like Protein HU ofBifidobacterium Longum. Biosci. Biotechnol. Biochem. 66, 598–603. doi:10.1271/bbb.66.598
Tan, R., Dong, H., Chen, Z., Jin, M., Yin, J., Li, H., et al. (2021). Intestinal Microbiota Mediates High-Fructose and High-Fat Diets to Induce Chronic Intestinal Inflammation. Front. Cell. Infect. Microbiol. 11, 654074. doi:10.3389/fcimb.2021.654074
Taniguchi, S. i. (2021). In Situ Delivery and Production System (iDPS) of Anti-cancer Molecules with Gene-Engineered Bifidobacterium. Jpm 11, 566. doi:10.3390/jpm11060566
Taniguchi, S. i., Shimatani, Y., and Fujimori, M. (2016). “Tumor-targeting Therapy Using Gene-Engineered Anaerobic-Nonpathogenic Bifidobacterium Longum,” in Bacterial Therapy of Cancer (Springer), 49–60. doi:10.1007/978-1-4939-3515-4_5
Thibault, H., Aubert-Jacquin, C., and Goulet, O. (2004). Effects of Long-Term Consumption of a Fermented Infant Formula (With Bifidobacterium Breve C50 and Streptococcus Thermophilus 065) on Acute Diarrhea in Healthy Infants. J. Pediatr. Gastroenterol. Nutr. 39, 147–152. doi:10.1097/00005176-200408000-00004
Tian, P., Bastiaanssen, T. F. S., Song, L., Jiang, B., Zhang, X., Zhao, J., et al. (2021). Unraveling the Microbial Mechanisms Underlying the Psychobiotic Potential of a Bifidobacterium Breve Strain. Mol. Nutr. Food Res. 65, 2000704. doi:10.1002/mnfr.202000704
Tian, P., O'Riordan, K. J., Lee, Y.-K., Wang, G., Zhao, J., Zhang, H., et al. (2020). Towards a Psychobiotic Therapy for Depression: Bifidobacterium Breve CCFM1025 Reverses Chronic Stress-Induced Depressive Symptoms and Gut Microbial Abnormalities in Mice. Neurobiol. Stress 12, 100216. doi:10.1016/j.ynstr.2020.100216
Timmerman, H. M., Koning, C. J. M., Mulder, L., Rombouts, F. M., and Beynen, A. C. (2004). Monostrain, Multistrain and Multispecies Probiotics-A Comparison of Functionality and Efficacy. Int. J. Food Microbiol. 96, 219–233. doi:10.1016/j.ijfoodmicro.2004.05.012
Toiviainen, A., Jalasvuori, H., Lahti, E., Gursoy, U., Salminen, S., Fontana, M., et al. (2015). Impact of Orally Administered Lozenges with Lactobacillus Rhamnosus GG and Bifidobacterium Animalis Subsp. Lactis BB-12 on the Number of Salivary Mutans Streptococci, Amount of Plaque, Gingival Inflammation and the Oral Microbiome in Healthy Adults. Clin. Oral Invest. 19, 77–83. doi:10.1007/s00784-014-1221-6
Toscano, M., De Grandi, R., Stronati, L., De Vecchi, E., and Drago, L. (2017). Effect ofLactobacillus rhamnosusHN001 andBifidobacterium longumBB536 on the Healthy Gut Microbiota Composition at Phyla and Species Level: A Preliminary Study. Wjg 23, 2696–2704. doi:10.3748/wjg.v23.i15.2696
Trindade, M. I., Abratt, V. R., and Reid, S. J. (2003). Induction of Sucrose Utilization Genes from Bifidobacterium Lactis by Sucrose and Raffinose. Appl. Environ. Microbiol. 69, 24–32. doi:10.1128/aem.69.1.24-32.2003
Turroni, S., Bendazzoli, C., Dipalo, S. C. F., Candela, M., Vitali, B., Gotti, R., et al. (2010). Oxalate-Degrading Activity in Bifidobacterium Animalis Subsp. Lactis : Impact of Acidic Conditions on the Transcriptional Levels of the Oxalyl Coenzyme A (CoA) Decarboxylase and Formyl-CoA Transferase Genes. Appl. Environ. Microbiol. 76, 5609–5620. doi:10.1128/aem.00844-10
Usta-Gorgun, B., and Yilmaz-Ersan, L. (2020). Short-chain Fatty Acids Production by Bifidobacterium Species in the Presence of Salep. Electron. J. Biotechnol. 47, 29–35. doi:10.1016/j.ejbt.2020.06.004
Vickers, N. J. (2017). Animal Communication: When I'm Calling You, Will You Answer Too? Curr. Biol. 27, R713–R715. doi:10.1016/j.cub.2017.05.064
Vieira, A. T., Rocha, V. M., Tavares, L., Garcia, C. C., Teixeira, M. M., Oliveira, S. C., et al. (2016). Control of Klebsiella pneumoniae Pulmonary Infection and Immunomodulation by Oral Treatment with the Commensal Probiotic Bifidobacterium Longum 51A. Microbes Infect. 18, 180–189. doi:10.1016/j.micinf.2015.10.008
Wang, H., Braun, C., Murphy, E. F., and Enck, P. (2019). Bifidobacterium Longum 1714 Strain Modulates Brain Activity of Healthy Volunteers during Social Stress. Am. J. Gastroenterol. 114, 1152–1162. doi:10.14309/ajg.0000000000000203
Wang, K. Y., Li, S. N., Liu, C. S., Perng, D. S., Su, Y. C., Wu, D. C., et al. (2004). Effects of Ingesting Lactobacillus- and Bifidobacterium-Containing Yogurt in Subjects with Colonized Helicobacter pylori. Am. J. Clin. Nutr. 80, 737–741. doi:10.1093/ajcn/80.3.737
Wang, Q., Xu, J., Sun, Z., Luan, Y., Li, Y., Wang, J., et al. (2019). Engineering an In Vivo EP-Bifido Pathway in Escherichia coli for High-Yield Acetyl-CoA Generation with Low CO2 Emission. Metab. Eng. 51, 79–87. doi:10.1016/j.ymben.2018.08.003
Watson, D., Sleator, R. D., Hill, C., and Gahan, C. G. (2008). Enhancing Bile Tolerance Improves Survival and Persistence of Bifidobacterium and Lactococcus in the Murine Gastrointestinal Tract. BMC Microbiol. 8, 176. doi:10.1186/1471-2180-8-176
Wei, C., Xun, A. Y., Wei, X. X., Yao, J., Wang, J. Y., Shi, R. Y., et al. (2016). Bifidobacteria Expressing Tumstatin Protein for Antitumor Therapy in Tumor-Bearing Mice. Technol. Cancer Res. Treat. 15, 498–508. doi:10.1177/1533034615581977
Wei, P., Yang, Y., Ding, Q., Li, X., Sun, H., Liu, Z., et al. (2016). Oral Delivery of Bifidobacterium Longum Expressing α-melanocyte-stimulating Hormone to Combat Ulcerative Colitis. J. Med. Microbiol. 65, 160–168. doi:10.1099/jmm.0.000197
Wong, C. B., Iwabuchi, N., and Xiao, J.-z. (2019). Exploring the Science behind Bifidobacterium Breve M-16V in Infant Health. Nutrients 11, 1724. doi:10.3390/nu11081724
Wu, S.-F., Chiu, H.-Y., Chen, A.-C., Lin, H.-Y., Lin, H.-C., and Caplan, M. (2013). Efficacy of Different Probiotic Combinations on Death and Necrotizing Enterocolitis in a Premature Rat Model. J. Pediatr. Gastroenterol. Nutr. 57, 23–28. doi:10.1097/mpg.0b013e3182929210
Xiao, J., Katsumata, N., Bernier, F., Ohno, K., Yamauchi, Y., Odamaki, T., et al. (2020). Probiotic Bifidobacterium Breve in Improving Cognitive Functions of Older Adults with Suspected Mild Cognitive Impairment: a Randomized, Double-Blind, Placebo-Controlled Trial. J. Alzheimers Dis. 77, 139–147. doi:10.3233/JAD-200488
Yang, B., Chen, H., Stanton, C., Chen, Y. Q., Zhang, H., and Chen, W. (2017). Mining Bifidobacteria from the Neonatal Gastrointestinal Tract for Conjugated Linolenic Acid Production. Bioengineered 8, 232–238. doi:10.1080/21655979.2016.1222996
Yang, C., Fujita, Y., Ren, Q., Ma, M., Dong, C., and Hashimoto, K. (2017). Bifidobacterium in the Gut Microbiota Confer Resilience to Chronic Social Defeat Stress in Mice. Sci. Rep. 7, 45942–45947. doi:10.1038/srep45942
Yazawa, K., Fujimori, M., Amano, J., Kano, Y., and Taniguchi, S. i. (2000). Bifidobacterium Longum as a Delivery System for Cancer Gene Therapy: Selective Localization and Growth in Hypoxic Tumors. Cancer Gene Ther. 7, 269–274. doi:10.1038/sj.cgt.7700122
Yazawa, K., Fujimori, M., Nakamura, T., Sasaki, T., Amano, J., Kano, Y., et al. (2001). Bifidobacterium Longum as a Delivery System for Gene Therapy of Chemically Induced Rat Mammary Tumors. Breast Cancer Res. Treat. 66, 165–170. doi:10.1023/a:1010644217648
You, J., and Yaqoob, P. (2012). Evidence of Immunomodulatory Effects of a Novel probiotic,Bifidobacterium longumbv.infantisCCUG 52486. FEMS Immunol. Med. Microbiol. 66, 353–362. doi:10.1111/j.1574-695x.2012.01014.x
Yu, Z., Huang, Z., Sao, C., Huang, Y., Zhang, F., Yang, J., et al. (2012). Bifidobacterium as an Oral Delivery Carrier of Interleukin-12 for the Treatment of Coxsackie Virus B3-Induced Myocarditis in the Balb/c Mice. Int. immunopharmacology 12, 125–130. doi:10.1016/j.intimp.2011.10.022
Yun, B., Song, M., Park, D.-J., and Oh, S. (2017). Beneficial Effect of Bifidobacterium Longum ATCC 15707 on Survival Rate of Clostridium difficile Infection in Mice. Korean J. Food Sci. Anim. Resour. 37, 368–375. doi:10.5851/kosfa.2017.37.3.368
Yunes, R. A., Poluektova, E. U., Vasileva, E. V., Odorskaya, M. V., Marsova, M. V., Kovalev, G. I., et al. (2020). A Multi-Strain Potential Probiotic Formulation of GABA-Producing Lactobacillus Plantarum 90sk and Bifidobacterium Adolescentis 150 with Antidepressant Effects. Probiotics Antimicro. Prot. 12, 973–979. doi:10.1007/s12602-019-09601-1
Zhang, L.-L., Chen, X., Zheng, P.-Y., Luo, Y., Lu, G.-F., Liu, Z.-Q., et al. (2010). OralBifidobacteriummodulates Intestinal Immune Inflammation in Mice with Food Allergy. J. Gastroenterol. Hepatol. 25, 928–934. doi:10.1111/j.1440-1746.2009.06193.x
Keywords: bifidobacteria, probiotic, therapeutic, genetic engineering, synthetic biology
Citation: Chen J, Chen X and Ho CL (2021) Recent Development of Probiotic Bifidobacteria for Treating Human Diseases. Front. Bioeng. Biotechnol. 9:770248. doi: 10.3389/fbioe.2021.770248
Received: 03 September 2021; Accepted: 08 December 2021;
Published: 22 December 2021.
Edited by:
Qing Sun, Texas A&M University, United StatesReviewed by:
Konstantinos Vavitsas, National and Kapodistrian University of Athens, GreeceCopyright © 2021 Chen, Chen and Ho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Loong Ho, aGVqbEBzdXN0ZWNoLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.