
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 09 February 2021
Sec. Industrial Biotechnology
Volume 9 - 2021 | https://doi.org/10.3389/fbioe.2021.633028
This article is part of the Research Topic Biosynthesis of Amino Acids and their Derived Chemicals from Renewable Feedstock View all 12 articles
Bioproduction of 5-aminovalerate (5AVA) from renewable feedstock can support a sustainable biorefinery process to produce bioplastics, such as nylon 5 and nylon 56. In order to achieve the biobased production of 5AVA, a 2-keto-6-aminocaproate-mediated synthetic pathway was established. Combination of L-Lysine α-oxidase from Scomber japonicus, α-ketoacid decarboxylase from Lactococcus lactis and aldehyde dehydrogenase from Escherichia coli could achieve the biosynthesis of 5AVA from biobased L-Lysine in E. coli. The H2O2 produced by L-Lysine α-oxidase was decomposed by the expression of catalase KatE. Finally, 52.24 g/L of 5AVA were obtained through fed-batch biotransformation. Moreover, homology modeling, molecular docking and molecular dynamic simulation analyses were used to identify mutation sites and propose a possible trait-improvement strategy: the expanded catalytic channel of mutant and more hydrogen bonds formed might be beneficial for the substrates stretch. In summary, we have developed a promising artificial pathway for efficient 5AVA synthesis.
Increasing concerns over global water pollution, climate change, public health, and petroleum shortages have attracted considerable attention to sustainable development as promising green alternatives to traditional petrochemical-derived chemicals renewable feedstock (Tsuge et al., 2016). Recently a variety of valuable chemicals such as 6-aminocaproate (Cheng et al., 2019), fructose (Yang et al., 2016), mandelic acid (Youn et al., 2020), vitamin B12 (Fang et al., 2018), naringenin (Gao et al., 2020b), p-coumaric acid (Gao et al., 2020a), breviscapine (Liu et al., 2018), 4-hydroxybenzoic acid (Klenk et al., 2020), curcuminoids (Rodrigues et al., 2020) and hydroxytyrosol (Zeng et al., 2020) have been produced in microorganisms. As a kind of green alternative to petrochemical products, microbial bioplastics are composed of monomers containing appropriate functional groups, which have become the focus of metabolic engineering research. These compounds include amino acids such as methionine (Kromer et al., 2006) and leucine (Zhang et al., 2008), organic acids such as adipic acid (Zhao et al., 2018a) and glutarate (Zhao et al., 2018b), diamines such as 1,3-diaminopropane (Chae et al., 2015) and diaminopentane (Kind et al., 2010; Rui et al., 2020), as well as diols like 1,3-propanediol (Nakamura and Whited, 2003) and 1,2-propanediol (Niimi et al., 2011). It is worth mentioning that two straight-chain amino acids—5-aminovalerate (5AVA) and 4-aminobutyrate—are promising platform compounds for the synthesis of polyimides, serving as raw materials for disposable goods, clothes and automobile parts like nylon 5 (Adkins et al., 2013) and nylon 4 (Park et al., 2013a) because of its high temperature and organic solvent resistance.
Due to the high demand in the animal feed industry, the production of L-Lysine (L-lys) is saturated today and may even be in oversupply (Vassilev et al., 2018). As one of the most important bulk chemicals, 5AVA has become the precursor for the synthesis of δ-valerolactam (Zhang et al., 2017), glutarate (Rohles et al., 2016; Hong et al., 2018), nylon 5 (Adkins et al., 2013), 5-hydroxyvalerate (Liu et al., 2014) and 1,5-pentanediol (Park et al., 2014). 5AVA is currently produced from petroleum feedstocks with aerobic oxidation of piperidine catalyzed by ceria-supported nanogold (Dairo et al., 2016). However, this chemical synthesis method not only requires higher temperature, but results in greater pollution (Dairo et al., 2016), so it is necessary to discover alternative approaches to produce 5AVA. Recently, with the rapid development of biotechnology, the synthesis of 5AVA by means of metabolic engineering and synthetic biology has attracted more and more attention (Hong et al., 2018).
In nature, 5AVA synthesis is closely related to L-lys catabolism in Pseudomonas putida (Ying et al., 2017). As seen in Figure 1A, 5AVA was produced through the overexpression of L-lys 2-monooxygenase (DavB) and 5AVA amidohydrolase (DavA) (Joo et al., 2017). According to Park’s report (Park et al., 2013b), 3.6 g/L of 5AVA was succsessfully produced in WL3110/DavA-DavB, but the titer was relatively low. 33.1 g/L of 5AVA was produced under a novel artificial H36 promoter in Corynebacterium glutamicum (Shin et al., 2016). Interestingly, L-lys specific permease (LysP) has been shown to increase 5AVA titer to 63.2 g/L (Table 1; Li et al., 2016). As seen from Figure 1B, 5AVA has been successfully produced from L-lys via cadaverine-mediated and 5-aminopentanal-mediated pathway (Jorge et al., 2017). With the expression of L-lys α-oxidase (RaiP) from Scomber japonicus (S. japonicus), 29.12 g/L of 5AVA could be successfully formed from L-lys hydrochloride (L-lys HCl) via 2-keto-6-aminocaproate (2K6AC) as intermediate as seen in Figure 1C (Cheng et al., 2018b). However, the addition of ethanol and H2O2 were unsafe and uneconomical (Cheng et al., 2018b). 13.4 g/L 5AVA could be successfully obtained with RaiP immobilized on a solid support (Pukin et al., 2010). In addition, 5AVA could be effectively separated by macroporous adsorption resin AK-1 from bioconversion liquid with the purity of 99.3% (Xu et al., 2019).
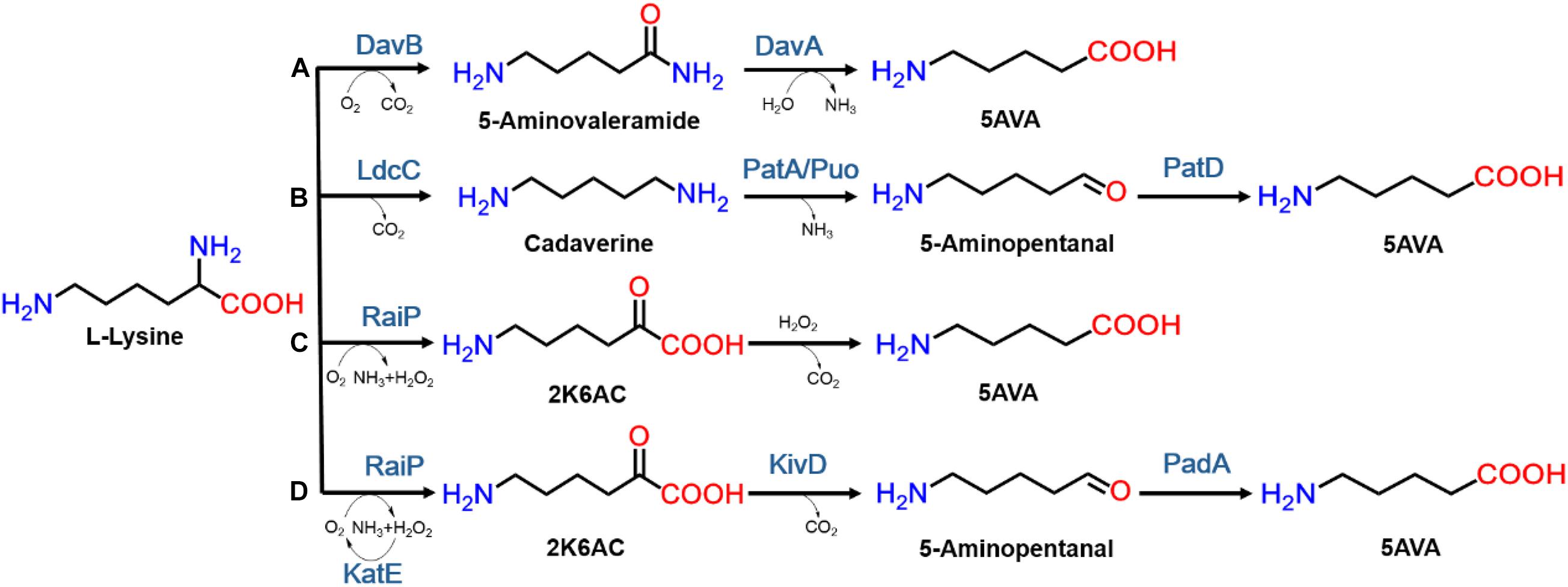
Figure 1. The biosynthesis routes of 5AVA from L-Lysine in microorganisms. The enzymes included in those routes are: (A) lysine 2-monooxygenase (DavB), δ-aminovaleramidase (DavA); (B) L-Lysine decarboxylase (LdcC), putrescine transaminase (PatA), monooxygenase putrescine oxidase (Puo), and γ-aminobutyraldehyde dehydrogenase (PatD); (C) L-Lysine α-oxidase (RaiP); (D) L-Lysine α-oxidase (RaiP), α-ketoacid decarboxylase (KivD), catalyze (KatE), and aldehyde dehydrogenase (PadA).
The promiscuous α-ketoacid decarboxylase (KivD) has been demonstrated in the decarboxylation of α-ketoacids (Atsumi et al., 2008; Chen et al., 2017). In its native pathway, KivD catalyzes a wide variety of α-ketoacids into aldehydes (Xiong et al., 2012; Jambunathan and Zhang, 2014; Wang et al., 2017). Compared with the substrates of wild-type KivD, are mainly smaller substrates, such as 2-ketoisovalerate and α-ketoadipate (Zhang et al., 2008; Wang et al., 2017), KivD mutants are relatively longer, such as 2-keto-4-methylhexanoate and 2-keto-3-methylvalerate (Zhang et al., 2008). Overexpression of KivD from Lactococcus lactis (L. lactis) and alcohol dehydrogenase 2 (ADH2) in Escherichia coli, 1-propanol could be successfully produced from 2-ketobutyrate with a final titer of 2 g/L (Shen and Liao, 2008).
In this study, 5AVA was synthesized using 2-keto-6-aminocaproate as intermediate, which is related to the involvement of three key enzymes—RaiP, KivD, and aldehyde dehydrogenase (PadA)—as seen in Figure 1D. Compared with the wild type, the two mutants of KivD in residues F381 and M461 showed higher substrate recognition and catalytic efficiency. Moreover, the overexpression of KatE and LysP, contributes to the removal of H2O2 and the transport of L-lys, thereby increasing the production of 5AVA, respectively. As can be expected, this artificial pathway has a potential prospect in industrial application, which enhances the value of L-lys and produces 5AVA efficiently in engineered E. coli.
The strains and plasmids involved in this work are listed in Table 2. The nucleotide sequences of genes raiP from S. japonicus, kivD from L. lactis and padA from E. coli are available in the GenBank database with the accession numbers of MG423617 (Cheng et al., 2018a), AIS03677.1 (McCulloch et al., 2014) and NP_415903.4 (Riley et al., 2006), respectively. In order to establish the synthetic pathway, the raiP, padA, and kivD genes were inserted into pET21a, and then the plasmid pET21a-raiP-padA-kivD was generated, which was also named as pETaRPK. Primers for saturation mutation of KivD are listed in Supplementary Table 1. kivD was replaced by kivD# (kivD with F381A/V461A mutations) to form the engineered pET21a-raiP-padA-kivD#, also named as pETaRPK#. The lysine permease gene lysP from E. coli (GenBank accession No. WP_000253273.1) was amplified from plasmid pLMAIP-04 (Cheng et al., 2018a), and the catalase gene katE (GenBank accession No. AAT48137.1) from E. coli MG1655. In order to remove H2O2, accelerate transportation of L-lys and reduce energy consumption, the katE, and lysP genes were firstly constructed in another single operon with the transcriptional order of katE-lysP, and then the engineered pZA22-katE-lysP was produced, also named as pZAKL. In addition, E. coli BL21 (DE3) with knocked out cadA was transformed with the plasmid pCJ01, pETaRPK, pETaRPK#, pETakatE, or pETaKL to obtain the strains CJ02, CJ06, CJ07, CJ08, or CJ09, respectively.
The E. coli strains harboring the corresponding plasmids were streaked onto Luria-Bertani (LB) agar plates with appropriate antibiotics at 37°C for overnight. Engineering strains used for shake flask fermentation were cultured in the medium containing 5 g/L yeast extract, 10 g/L tryptone, 15 g/L glucose, 0.1 g/L FeCl3, 2.1 g/L citric acid⋅H2O, 2.5 g/L (NH4)2SO4, 0.5 g/L K2PO4⋅3H2O, 1.0 mM MgSO4, 3 g/L KH2PO4, and 0.5 mM thiamine diphosphate (ThDP) with appropriate antibiotics. After the OD600 of the strains reached 0.5, 0.5 mM of isopropyl β-D-thiogalactoside (IPTG) and 6.5 g/L of L-lys HCl were added.
Fed-batch biotransformation of engineering strains were conducted in a 5.0 L fermenter. The composition of the medium was described in our previous report as follows: glucose, 55 g/L; MgSO4⋅7H2O, 1.6 g/L; FeSO4⋅7H2O, 0.00756 g/L; (NH4)2SO4, 1.6 g/L; citric acid, 2 g/L; K2HPO4⋅3H2O, 7.5 g/L; Na2SO4, 0.02 g/L; ZnSO4, 0.0064 g/L; Cu2SO4⋅5H2O, 0.0006 g/L; CoCl2⋅6H2O, 0.004 g/L (Cheng et al., 2018a). The pH was controlled at 6.7–6.9 by the automatic addition of NH3⋅H2O, and the temperature was set at 30°C. Antifoam 289 was gradually added to prevent the formation of foam during biotransformation. The initial concentration of L-lys HCl was 40 g/L. The concentration of glucose and L-lys were maintained around 15 and 20 g/L during the whole fermentation process, respectively.
The media for protein expression was supplemented by 0.5 mM ThDP in LB at 37°C. At an OD600 of 0.5, 0.5 mM of IPTG was added and then cultured at 20°C for 16 h, cells were washed with potassium phosphate buffer (KPB, 50 mM, pH 8.0) and disrupted by sonication in an ice bath of 50 mM KPB. The enzymes were purified with AKTA Purifier 10 using a Ni-NTA column (Cheng et al., 2019). The concentration of protein was measured by SpectraMax M2e at 280 nm. The detections of 5AVA and L-lys were reported in our previous work (Cheng et al., 2018b).
The oxidation activity of RaiP was measured according to the concentration of hydrogen peroxide (Cheng et al., 2018b). The decarboxylation activity of KivD and KivD mutations (KivD∗) were determined at 30°C, using a coupled enzymatic assay (Wang et al., 2017). The reaction mixture contained 1.0 mM NAD+, 1.1 μM PadA, 1.1 μM RaiP, 0.85 μM KivD, or KivD∗ and different concentrations of L-lys in assay buffer (50 mM KPB, pH 8.0, 1 mM MgSO4, 1.0 mM TCEP, 0.5 mM ThDP). The reactions began with the addition of the substrate L-lys, and the formation of NADH was monitored at 340 nm with the extinction coefficient of 6.22 mM–1 cm–1.
The theoretical structure of native KivD and mutant KivD# (KivD with F381A/V461A mutations) (PDB: 2VBF), both were generated by SWISS-MODEL online Server1. The 3D structural comparison between KivD and KivD# was revealed using PyMOL 2.2. The ligand 2K6AC was docked into the pocket of KivD or KivD# using AutoDock 4.2.6 package, where the lowest energy conformation in the largest cluster was considered to be the approximately natural complex model (Xie et al., 2019; Tahara et al., 2020). Molecular dynamic (MD) simulation was used to simulate the relationship between structure and function of biomacromolecules in solution in this study (Wu et al., 2020). Two comparative MD simulations at 300 K were executed for KivD and KivD-2K6AC systems with AMBER 18 package (Zuo et al., 2017; Wu et al., 2020).
Figure 1D showed a heterogeneous artificial route for the bioconversion of L-lys to 5AVA. The designed artificial biosynthetic pathway of 5AVA consists of three steps: (1) deamination of L-lys to form intermediate 2K6AC via RaiP; (2) decarboxylation of 2K6AC to produce 5-aminopentanal via KivD; (3) oxidation of 5-aminopentanal to 5AVA via PadA. Firstly, a plasmid pETaRPK was constructed and introduced into E. coli ML03 to obtain the strain CJ05, with the co-expression of RaiP, KivD, and PadA under a T7 promoter. To reduce the degradation of L-lys to cadaverine, the lysine decarboxylase gene cadA was knocked out to obtain the strain CJ06. The maximum-likelihood tree was displayed in Figure 2A. Notably, 5AVA could be produced in strains CJ01, CJ02, CJ05, and CJ06. As shown in Figure 2B, the control strain CJ00 only produced 0.06 g/L 5AVA from 6.5 g/L L-lys HCl with the consumption of 0.01 g/g L-lys. For engineered strain CJ01, a titer of 0.23 g/L 5AVA was acquired. Moreover, the strain CJ05 produced 1.66 g/L of 5AVA by this artificial pathway (see Figure 1D), with a yield increase of 774% compared to the single gene pathway (see Figure 1C). These results demonstrate the feasibility of this proposed artificial 5AVA pathway.
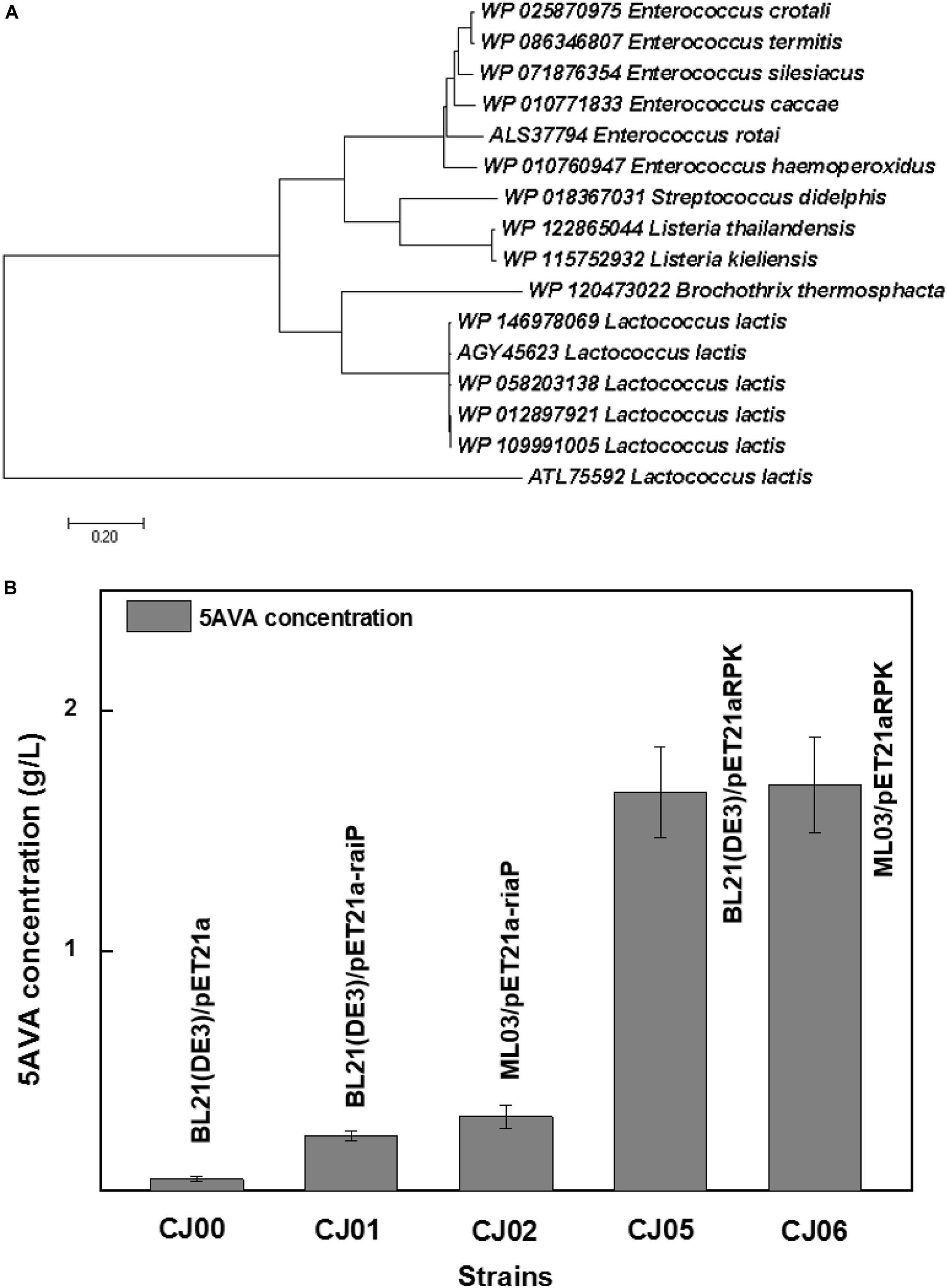
Figure 2. Function identification of α-ketoacid decarboxylase subfamily. (A) The phylogenetic relationship of α-ketoacid decarboxylase subfamily. WP 012897921 Lactococcus lactis was the α-ketoacid decarboxylase used in this study. All α-ketoacid decarboxylase genes were downloaded from NCBI by blastp against the nr database. The genes were from the species, L. lactis, Enterococcus crotali, Enterococcus termitis, Enterococcus Silesiacus, Enterococcus Caccae, Enterococcus rotai, Enterococcus haemoperoxidus, Streptococcus didelphis, Listeria thailandensis, Listeria kieliensis, and Brochothrix thermosphacta. The maximum-likelihood tree was constructed by MEGA (Li et al., 2019a, b). (B) An artificial pathway confirmed for the biosynthesis of 5AVA. 6.5 g/L of L-Lysine HCl was as substrate. All experiments were performed a minimum of three independent sets. All error bars represent standard deviations with n ≥ 3 independent reactions.
In order to explore the mechanism of the 5AVA increase in mutants, molecular docking and MD simulation were discussed (Xiang et al., 2019). The structures of KivD and KivD# both are mainly composed of 23 α-helices and 17 β-strands, containing a large activity pocket. Compared with that of KivD, the structure of KivD# remains almost unchanged. Nevertheless based on homology modeling analysis, the catalytic channel of mutant KivD# was enlarged. According to bioinformatics and crystal structure information (PDB: 2VBF) (Berthold et al., 2007), residues F381 and V461 are the two key residues for KivD catalysis (see Figure 3). Modeling and molecular docking of KivD with ligand 2K6AC further highlight the residues involved in substrate recognition. As shown in Figure 3, the substrate docking results indicated that the distances of ligand 2K6AC with F381A, V461A active sites both became farther. The docking results of KivD and 2K6AC showed that 2K6AC formed eight hydrogen bonds with the side chain Q377, T379, N456, T460, and V461. 2K6AC formed nine hydrogen bonds with the side chain D429, N456, G458, T460, A461, and E462 of KivD# (Figure 4). At the same time, the surface hydrophobicity of the catalytic pocket in mutated protein KivD# has also changed (Figure 4). We speculated that the increase in catalytic activity of KivD# may be due to the expansion of catalytic channel and the formation of more hydrogen bonds, the expansion that is likely to result in a change in the conformation of the small molecule 2K6AC which was beneficial to stretch. Through the MD simulations, the results of the root mean squared deviation (RMSD) showed that the RMSD of the KivD system and the complex system KivD-2K6AC were basically maintained at 1.639 ± 0.240 Å and 1.738 ± 0.152 Å (see Figure 5B), which indicated that the MD simulation process was reliable (Zuo et al., 2017). As seen in Figure 5A, there are four fragments of the KivD with lower root mean squared fluctuation (RMSF) values, that is G58-L69, T212-N223, T379-F388, and D457-H466. These four fragments are located near ThDP, which may be related to the activity of KivD (Zuo et al., 2017; Liu et al., 2019).
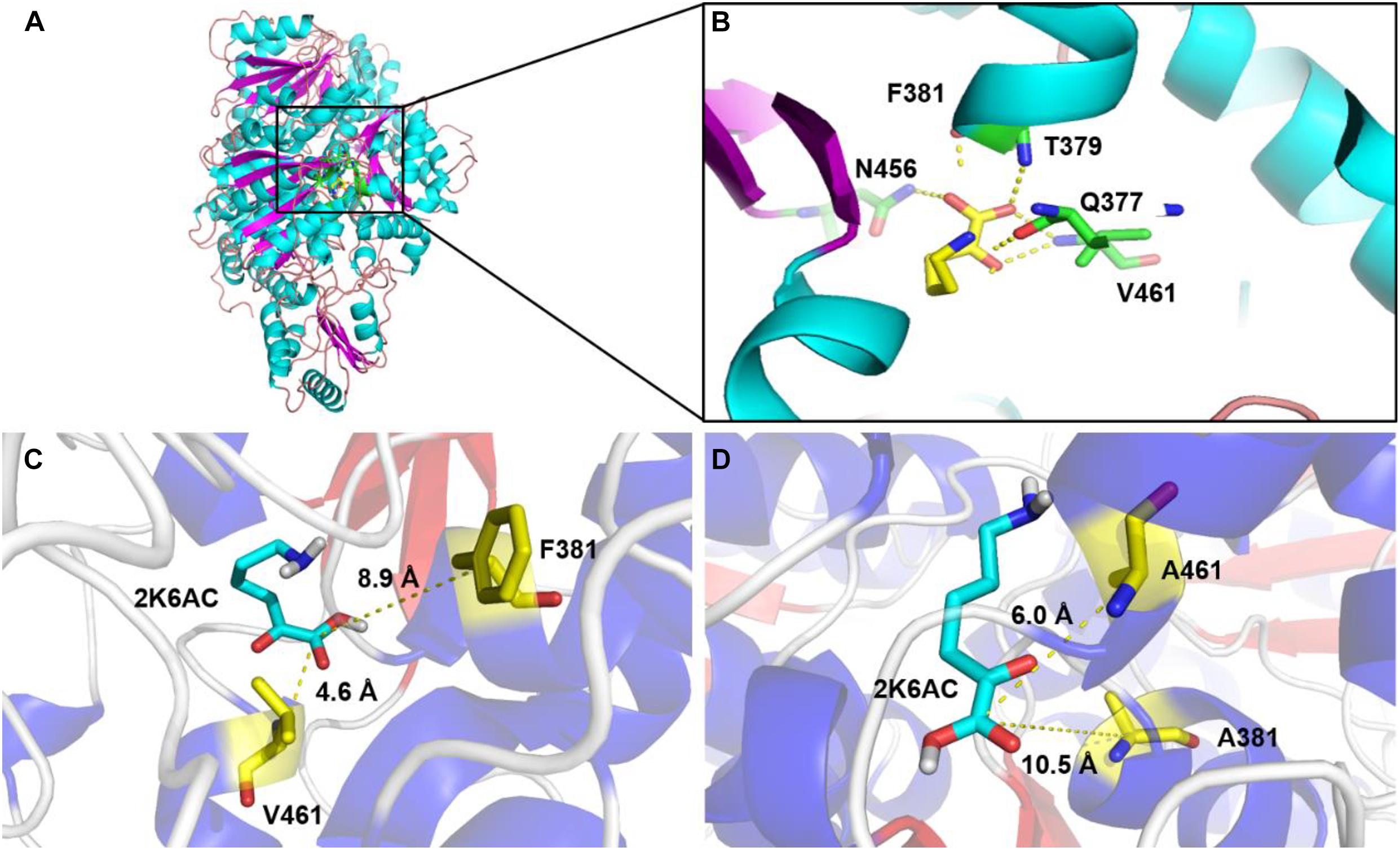
Figure 3. Homology modeling and structure comparison between KivD and KivD#(F381A/V461A). (A) Overall architecture of the KivD system; (B) Interactions of the ligand 2K6AC with their surroundings in KivD system; Binding pocket of L. lactis KivD (PDB: 2VBF) (C) and KivD#(F381A/V461A) (D) complexed with its substrate 2K6AC. The active pocket of KivD which is constituted by a number of hydrophobic residues, including F381, T379, and V461. KivD, α-ketoacid decarboxylase.
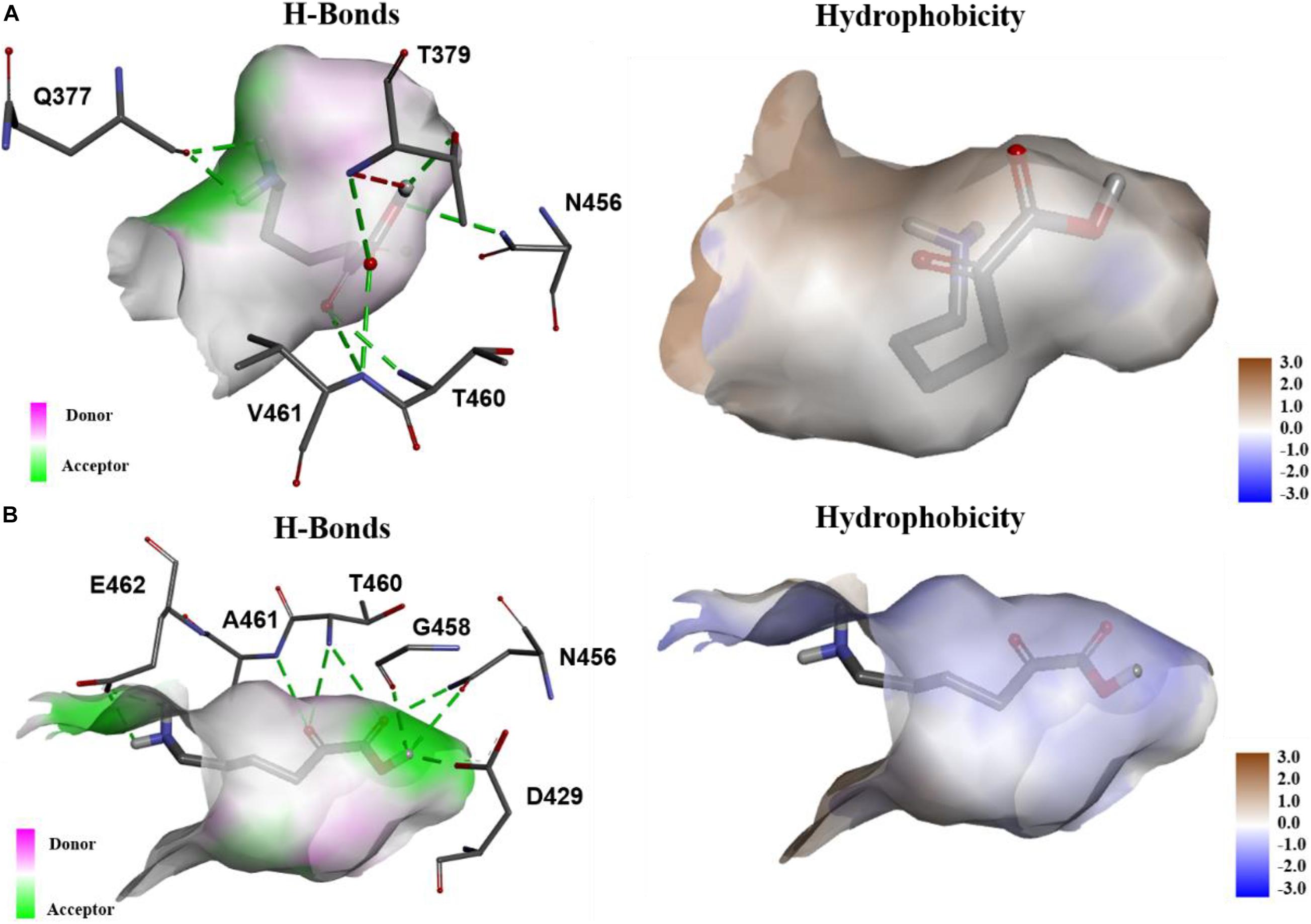
Figure 4. The analysis of hydrogen bonds and hydrophobicity. (A) The hydrogen bonds formed of KivD and 2K6AC, and the hydrophobicity of the active pocket in KivD; (B) The hydrogen bonds formed of KivD#(F381A/V461A) and 2K6AC, and the hydrophobicity of the active pocket in KivD#(F381A/V461A).
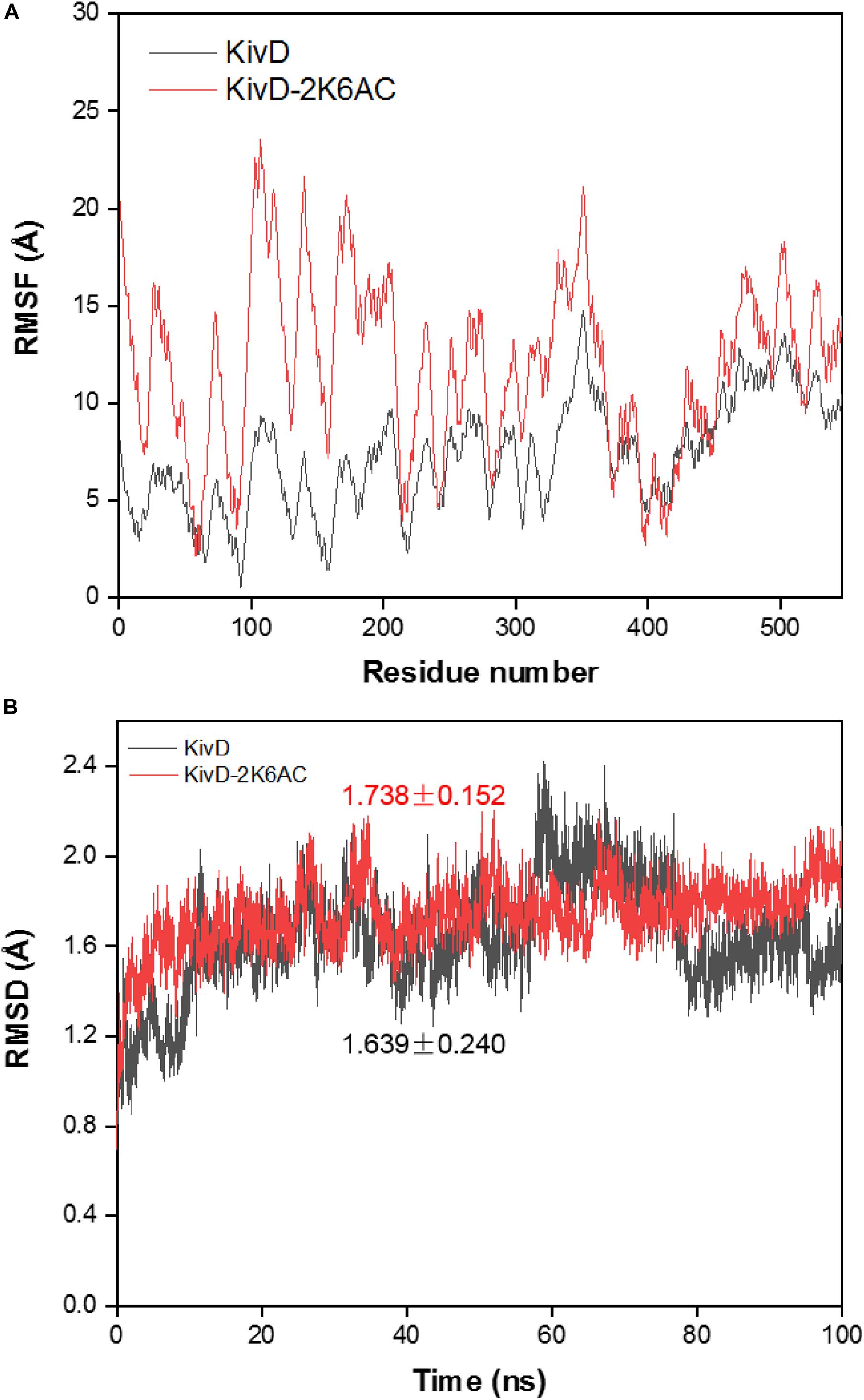
Figure 5. Molecular dynamic simulation of KivD and KivD-2K6AC. (A) RMSD of the Cα atoms in the KivD and KivD-2K6AC versus simulation time. (B) RMSF distribution of the Cα atom in the KivD and KivD-2K6AC. RMSD, Root mean squared deviation; RMSF, Root mean squared fluctuation.
KivD mutations (F381A/V461, F381L/V461, F381/V461A, F381/V461L, and F381A/V461A) displayed enhanced activities in Table 3. The KivD F381A/V461A (KivD#) showed the greatest activity shown in Table 3. KivD# displays a Km value of 2.52 mM, a Kcat value of 562.16 s–1 and a Kcat/Km value of 223.08 mM–1s–1 with 2K6AC used as the substrate shown in Table 3.
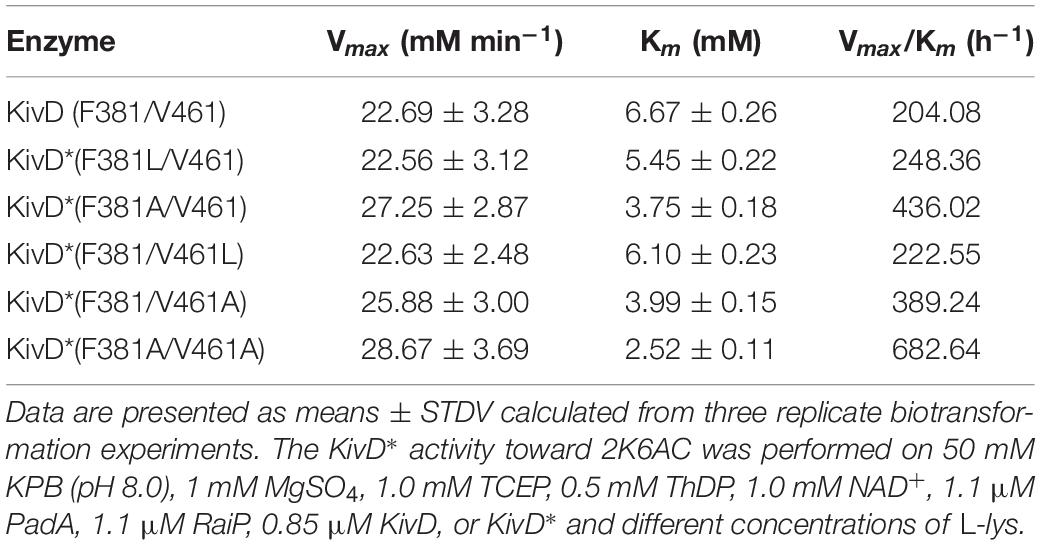
Table 3. Kinetic parameters of α-ketoacid decarboxylase KivD mutants (KivD*) on 2-keto-6-aminocaproate (2K6AC).
There are four strategies used in this study to increase the production of 5AVA. Firstly, lysine decarboxylase gene cadA was knocked out and L-lys HCl was selected as the industrial substrate for enhancing the utilization of L-lys (Cheng et al., 2018a, b, 2020). Thirdly, H2O2 could inhibit cell growth, thus affecting the production of goal production (Niu et al., 2014). In Liu’s experiments, through the expression of catalase, the content of H2O2 was significantly reduced, and the output of α-ketoglutarate was greatly increased (Liu et al., 2017). In this study, the co-expression of katE, raiP, kivD#, and padA in strain CJ08 yielded 1.88 g/L of 5AVA without addition of catalase, there was no significant difference compared to strain CJ07 (Table 4). In fact, the H2O2 generated by RaiP in this work was instantly eliminated by KatE. The data in rows 5 and 7 of Table 4 showed that the overexpression of katE did not significantly increase the OD600 and the production of 5AVA during shake flask fermentation. On the contrary, it decreased the OD600, possibly because the increase in gene expression caused an increase in cell burden (Camara et al., 2017). However, in the fermentation tank, H2O2 could significantly inhibit cell growth, resulting in limited production of 5AVA (Cheng et al., 2018b, 2020). In addition, a lysine transporter gene lysP was overexpressed and inserted into the plasmid pZAkatE to form a new plasmid pZAKL. As shown in Table 4, strain CJ09 produced 1.93 g/L of 5AVA.
Figure 6 showed the results of the fed-batch biotransformation in E. coli strain CJ09. Recombinant E. coli strain CJ09 grew quickly throughout the biotransformation, reaching the highest cell concentration of an OD600 of 142 in 18 h. After the addition of L-lys HCl, 5AVA was accumulated to 48.3 g/L between 18 and 36 h. With the fermentation time increasing to 48 h, 52.24 g/L of 5AVA was successfully acquired. The productivity and yield of 5AVA were 1.09 g/L/h and 0.65 g/g L-lys, respectively. The control strain CJ02 just produced 9.16 g/L 5AVA with a yield of 0.11 g/g L-lys. Interestingly, the expression of KatE in strain CJ08 had no effect on the production of 5AVA in shake flask (Table 4), but it could significantly improve the production of 5AVA to 45.92 g/L in fermentation tank compared to strain CJ07 with a titer of 16.48 g/L. This is because H2O2 can significantly inhibit the growth of strain CJ07, resulting in OD600 of only 40. The above results advocated that the synthetic route developed in this work can effectively produce 5AVA.
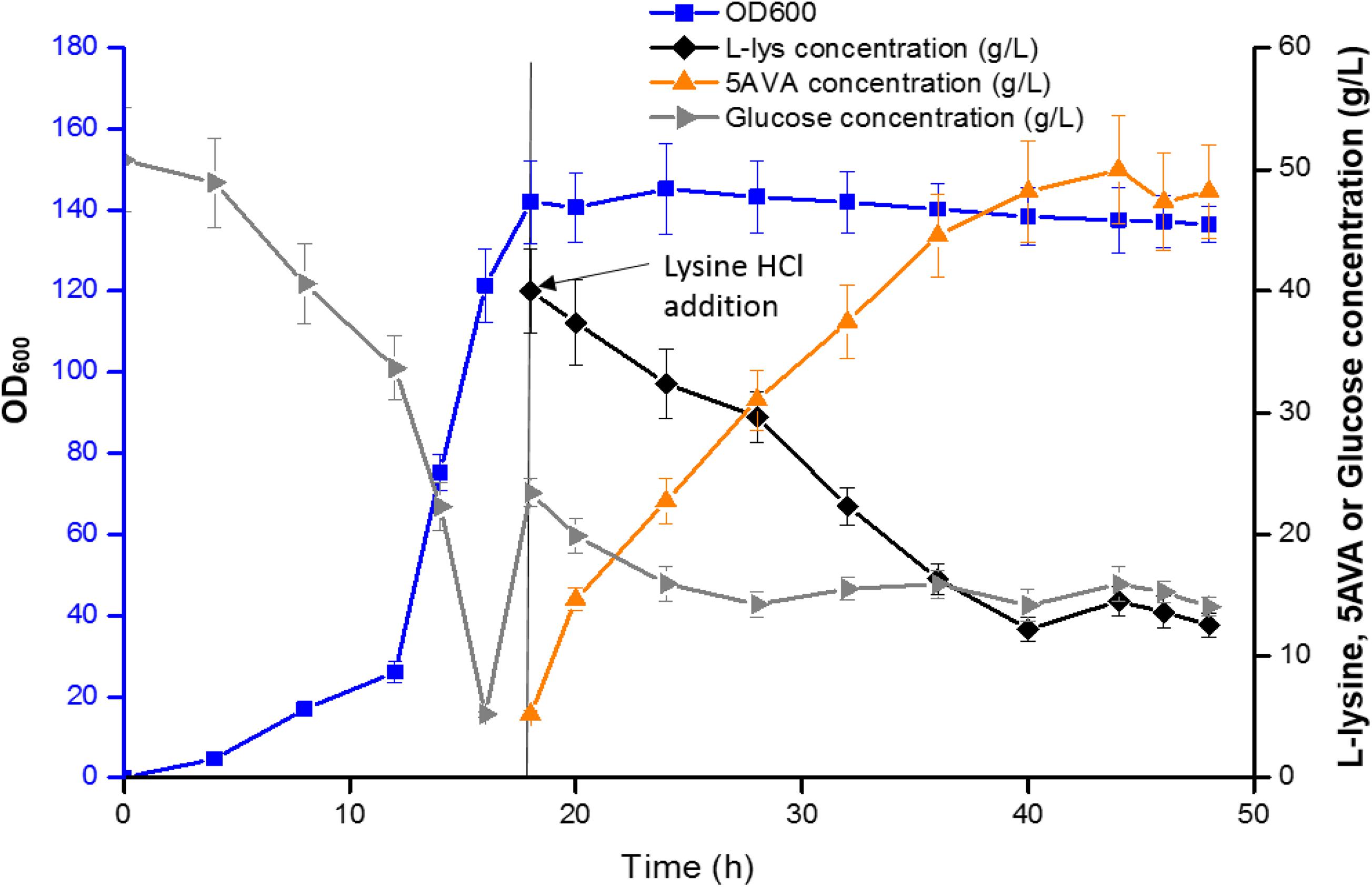
Figure 6. 5AVA synthesis by engineered strain CJ09 in a 5 L fermenter. Values and error bars represent the mean and the standard deviation of triplicate cultivations.
Compared with a previous whole cell transformation, the titer of 5AVA based on this synthesis pathway increased by about 79.4% from 29.12 to 52.24 g/L as seen in Table 1; the inhibition of cell growth and enzyme activity by H2O2 both resulted in the lower yields of 5AVA (Cheng et al., 2018b). Compared with another new synthesis pathway for the fermentative production of 5AVA, in which the titer was only 5.1 g/L (seen in Table 1; Jorge et al., 2017), and the titer was greatly increased in this study. Compared with another whole-cell catalysis work, this synthetic pathway increased the titer of 5AVA by about 3.20% from 50.62 to 52.24 g/L (Cheng et al., 2020). Importantly, the industrial production of 5AVA without the addition of ethanol and H2O2 was more safe and economical in this study. In terms of reaction mechanism, the new 5AVA synthesis strategy proposed in this work mainly includes three steps: (1) the accumulation of intermediate 6A2KCA by RaiP; (2) the decarboxylation of 6A2KCA to 5-aminopentanal by KivD; (3) the oxidization of 5-aminopentanal to 5AVA by PadA.
From renewable feedstocks, an artificial pathway in E. coli was proposed and optimized to produce 5AVA in this study. Since the inhibition of enzyme activity and cell growth by H2O2 is the main limiting factor in the production of 5AVA, catalase KatE was overexpressed to decompose H2O2 to achieve high yield of 5AVA. Finally, an engineered strain CJ09 with RaiP, KivD, PadA, KatE, and LysP overexpression successfully produced 5AVA from biobased L-lys HCl at a final titer of 52.24 g/L. The renewable substrate and simple culture conditions were adopted in this work, while possessing higher yield and less environmental pollution. The improvement of substrate utilization and H2O2 decomposition efficiency contributes to the increase in the yield of 5AVA, which has the potential to become a common strategy for the sustainable production of other chemicals.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
JC and WT performed the experiments, analyzed the data, and drafted the manuscript. ZL, QL, and XG analyzed the data. JC and DW conceived and coordinated the study. JC, DW, and JZ finalized the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Open Funding Project of the Key Laboratory of Meat Processing of Sichuan Province (20-R-06), the Open Funding Project of Key Laboratory of Coarse Cereal Processing, Ministry of Agriculture and Rural Affairs (2020CC010), the National Key Research and Development Program of China (2018YFC1602101), the National Natural Science Foundation of China (21978027), and the Project of Chongqing Key Laboratory of Environmental Materials and Restoration Technology (CEK1803).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2021.633028/full#supplementary-material
Adkins, J., Jordan, J., and Nielsen, D. R. (2013). Engineering Escherichia coli for renewable production of the 5-carbon polyamide building-blocks 5-aminovalerate and glutarate. Biotechnol. Bioeng. 110, 1726–1734. doi: 10.1002/bit.24828
Atsumi, S., Hanai, T., and Liao, J. C. (2008). Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89. doi: 10.1038/nature06450
Berthold, C. L., Gocke, D., Wood, M. D., Leeper, F. J., Pohl, M., and Schneider, G. (2007). Structure of the branched-chain keto acid decarboxylase (KdcA) from Lactococcus lactis provides insights into the structural basis for the chemoselective and enantioselective carboligation reaction. Acta Crystallogr. D. Biol. Crystallogr. 63(Pt 12), 1217–1224. doi: 10.1107/s0907444907050433
Camara, E., Landes, N., Albiol, J., Gasser, B., Mattanovich, D., and Ferrer, P. (2017). Increased dosage of AOX1 promoter-regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris. Sci. Rep. 7:44302.
Chae, T. U., Kim, W. J., Choi, S., Park, S. J., and Lee, S. Y. (2015). Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine. Sci. Rep. 5:13040.
Chen, G. S., Siao, S. W., and Shen, C. R. (2017). Saturated mutagenesis of ketoisovalerate decarboxylase V461 enabled specific synthesis of 1-pentanol via the ketoacid elongation cycle. Sci. Rep. 7:11284.
Cheng, J., Hu, G., Xu, Y., Torrens-Spence, M. P., Zhou, X., Wang, D., et al. (2019). Production of nonnatural straight-chain amino acid 6-aminocaproate via an artificial iterative carbon-chain-extension cycle. Metab. Eng. 55, 23–32. doi: 10.1016/j.ymben.2019.06.009
Cheng, J., Huang, Y., Mi, L., Chen, W., Wang, D., and Wang, Q. (2018a). An economically and environmentally acceptable synthesis of chiral drug intermediate L-pipecolic acid from biomass-derived lysine via artificially engineered microbes. J. Ind. Microbiol. Biot. 45, 405–415. doi: 10.1007/s10295-018-2044-2
Cheng, J., Luo, Q., Duan, H., Peng, H., Zhang, Y., Hu, J., et al. (2020). Efficient whole-cell catalysis for 5-aminovalerate production from L-lysine by using engineered Escherichia coli with ethanol pretreatment. Sci. Rep. 10:990.
Cheng, J., Zhang, Y., Huang, M., Chen, P., Zhou, X., Wang, D., et al. (2018b). Enhanced 5-aminovalerate production in Escherichia coli from L-lysine with ethanol and hydrogen peroxide addition. J. Chem. Technol. Biot. 93, 3492–3501. doi: 10.1002/jctb.5708
Dairo, T. O., Nelson, N. C., Slowing, I. I., Angelici, R. J., and Woo, L. K. (2016). Aerobic oxidation of cyclic amines to lactams catalyzed by ceria-supported nanogold. Catal Lett. 146, 2278–2291. doi: 10.1007/s10562-016-1834-2
Fang, H., Li, D., Kang, J., Jiang, P., Sun, J., and Zhang, D. (2018). Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12. Nat. Commun. 9:4917.
Gao, S., Lyu, Y., Zeng, W., Du, G., Zhou, J., and Chen, J. (2020a). Efficient biosynthesis of (2S)-Naringenin from p-Coumaric Acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 68, 1015–1021. doi: 10.1021/acs.jafc.9b05218
Gao, S., Zhou, H., Zhou, J., and Chen, J. (2020b). Promoter-library-based pathway optimization for efficient (2S)-naringenin production from p-Coumaric acid in Saccharomyces cerevisiae. J. Agric. Food Chem. 68, 6884–6891. doi: 10.1021/acs.jafc.0c01130
Haupka, C., Delépine, B., Irla, M., Heux, S., and Wendisch, V. F. (2020). Flux enforcement for fermentative production of 5-aminovalerate and glutarate by Corynebacterium glutamicum. Catalysts 10:1065. doi: 10.3390/catal10091065
Hong, Y. G., Moon, Y. M., Hong, J. W., No, S. Y., Choi, T. R., Jung, H. R., et al. (2018). Production of glutaric acid from 5-aminovaleric acid using Escherichia coli whole cell bio-catalyst overexpressing GabTD from Bacillus subtilis. Enzyme Microb. Technol. 118, 57–65. doi: 10.1016/j.enzmictec.2018.07.002
Jambunathan, P., and Zhang, K. (2014). Novel pathways and products from 2-keto acids. Curr. Opin. Biotechnol. 29, 1–7. doi: 10.1016/j.copbio.2014.01.008
Joo, J. C., Oh, Y. H., Yu, J. H., Hyun, S. M., Khang, T. U., Kang, K. H., et al. (2017). Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process. Bioresour. Technol. 245(Pt B), 1692–1700. doi: 10.1016/j.biortech.2017.05.131
Jorge, J. M. P., Perez-Garcia, F., and Wendisch, V. F. (2017). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245(Pt B), 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kind, S., Jeong, W. K., Schroder, H., and Wittmann, C. (2010). Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 12, 341–351. doi: 10.1016/j.ymben.2010.03.005
Klenk, J. M., Ertl, J., Rapp, L., Fischer, M.-P., and Hauer, B. (2020). Expression and characterization of the benzoic acid hydroxylase CYP199A25 from Arthrobacter sp. Mol. Catal. 484:110739. doi: 10.1016/j.mcat.2019.110739
Kromer, J. O., Wittmann, C., Schroder, H., and Heinzle, E. (2006). Metabolic pathway analysis for rational design of L-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab. Eng. 8, 353–369. doi: 10.1016/j.ymben.2006.02.001
Li, Q., Ren, Y., Shi, X., Peng, L., Zhao, J., Song, Y., et al. (2019a). Comparative mitochondrial genome analysis of two ectomycorrhizal fungi (Rhizopogon) reveals dynamic changes of intron and phylogenetic relationships of the subphylum agaricomycotina. Int. J. Mol. Sci. 20:1567.
Li, Q., Wang, Q., Jin, X., Chen, Z., Xiong, C., Li, P., et al. (2019b). Characterization and comparison of the mitochondrial genomes from two Lyophyllum fungal species and insights into phylogeny of Agaricomycetes. Int. J. Biol. Macromol. 121, 364–372. doi: 10.1016/j.ijbiomac.2018.10.037
Li, Z., Xu, J., Jiang, T., Ge, Y., Liu, P., Zhang, M., et al. (2016). Overexpression of transport proteins improves the production of 5-aminovalerate from L-lysine in Escherichia coli. Sci. Rep. 6:30884.
Liu, B. B., Qu, G., Li, J. K., Fan, W. C., Ma, J. A., Xu, Y., et al. (2019). Conformational dynamics-guided loop engineering of an alcohol dehydrogenase: capture, turnover and enantioselective transformation of difficult-to-reduce ketones. Adv. Synth. Catal. 361, 3182–3190. doi: 10.1002/adsc.201900249
Liu, P., Zhang, H., Lv, M., Hu, M., Li, Z., Gao, C., et al. (2014). Enzymatic production of 5-aminovalerate from L-lysine using L-lysine monooxygenase and 5-aminovaleramide amidohydrolase. Sci. Rep. 4:5657.
Liu, Q., Ma, X., Cheng, H., Xu, N., Liu, J., and Ma, Y. (2017). Co-expression of L-glutamate oxidase and catalase in Escherichia coli to produce alpha-ketoglutaric acid by whole-cell biocatalyst. Biotechnol. Lett. 39, 913–919. doi: 10.1007/s10529-017-2314-5
Liu, X., Cheng, J., Zhang, G., Ding, W., Duan, L., Yang, J., et al. (2018). Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 9:448.
McCulloch, J. A., de Oliveira, V. M., de Almeida Pina, A. V., Pérez-Chaparro, P. J., de Almeida, L. M., de Vasconcelos, J. M., et al. (2014). Complete genome sequence of Lactococcus lactis strain AI06, an endophyte of the amazonian açaí palm. Genome Announc. 2, e1225–e1214.
Nakamura, C. E., and Whited, G. M. (2003). Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459. doi: 10.1016/j.copbio.2003.08.005
Niimi, S., Suzuki, N., Inui, M., and Yukawa, H. (2011). Metabolic engineering of 1,2-propanediol pathways in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 90, 1721–1729. doi: 10.1007/s00253-011-3190-x
Niu, P., Dong, X., Wang, Y., and Liu, L. (2014). Enzymatic production of alpha-ketoglutaric acid from l-glutamic acid via l-glutamate oxidase. J. Biotechnol. 179, 56–62. doi: 10.1016/j.jbiotec.2014.03.021
Park, S. J., Kim, E. Y., Noh, W., Oh, Y. H., Kim, H. Y., Song, B. K., et al. (2013a). Synthesis of nylon 4 from gamma-aminobutyrate (GABA) produced by recombinant Escherichia coli. Bioprocess Biosyst. Eng. 36, 885–892. doi: 10.1007/s00449-012-0821-2
Park, S. J., Kim, E. Y., Noh, W., Park, H. M., Oh, Y. H., Lee, S. H., et al. (2013b). Metabolic engineering of Escherichia coli for the production of 5-aminovalerate and glutarate as C5 platform chemicals. Metab. Eng. 16, 42–47. doi: 10.1016/j.ymben.2012.11.011
Park, S. J., Oh, Y. H., Noh, W., Kim, H. Y., Shin, J. H., Lee, E. G., et al. (2014). High-level conversion of L-lysine into 5-aminovalerate that can be used for nylon 6,5 synthesis. Biotechnol. J. 9, 1322–1328. doi: 10.1002/biot.201400156
Pukin, A. V., Boeriu, C. G., Scott, E. L., Sanders, J. P. M., and Franssen, M. C. R. (2010). An efficient enzymatic synthesis of 5-aminovaleric acid. J. Mol. Catal. B Enzym 65, 58–62. doi: 10.1016/j.molcatb.2009.12.006
Riley, M., Abe, T., Arnaud, M. B., Berlyn, M. K., Blattner, F. R., Chaudhuri, R. R., et al. (2006). Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 34, 1–9. doi: 10.1093/nar/gkj405
Rodrigues, J. L., Gomes, D., and Rodrigues, L. R. (2020). A combinatorial approach to optimize the production of curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 8:59.
Rohles, C. M., Giesselmann, G., Kohlstedt, M., Wittmann, C., and Becker, J. (2016). Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb. Cell Fact. 15:154.
Rui, J., You, S., Zheng, Y., Wang, C., Gao, Y., Zhang, W., et al. (2020). High-efficiency and low-cost production of cadaverine from a permeabilized-cell bioconversion by a Lysine-induced engineered Escherichia coli. Bioresour. Technol. 302:122844. doi: 10.1016/j.biortech.2020.122844
Shen, C. R., and Liao, J. C. (2008). Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 10, 312–320. doi: 10.1016/j.ymben.2008.08.001
Shin, J. H., Park, S. H., Oh, Y. H., Choi, J. W., Lee, M. H., Cho, J. S., et al. (2016). Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb. Cell Fact. 15:174.
Tahara, T., Watanabe, A., Yutani, M., Yamano, Y., Sagara, M., Nagai, S., et al. (2020). STAT3 inhibitory activity of naphthoquinones isolated from Tabebuia avellanedae. Bioorg. Med. Chem. 28:115347. doi: 10.1016/j.bmc.2020.115347
Tsuge, Y., Kawaguchi, H., Sasaki, K., and Kondo, A. (2016). Engineering cell factories for producing building block chemicals for bio-polymer synthesis. Microb. Cell Fact. 15:19.
Vassilev, I., Giesselmann, G., Schwechheimer, S. K., Wittmann, C., Virdis, B., and Kromer, J. O. (2018). Anodic electro-fermentation: anaerobic production of L-Lysine by recombinant Corynebacterium glutamicum. Biotechnol. Bioeng. 115, 1499–1508. doi: 10.1002/bit.26562
Wang, J., Wu, Y. F., Sun, X. X., Yuan, Q. P., and Yan, Y. J. (2017). De novo biosynthesis of glutarate via alpha-keto acid carbon chain extension and decarboxylation pathway in Escherichia coli. ACS Synth. Biol. 6, 1922–1930. doi: 10.1021/acssynbio.7b00136
Wang, X., Cai, P., Chen, K., and Ouyang, P. (2016). Efficient production of 5-aminovalerate from L-lysine by engineered Escherichia coli whole-cell biocatalysts. J. Mol. Catal. B Enzym 134, 115–121. doi: 10.1016/j.molcatb.2016.10.008
Wu, Z., Peng, L., Hu, Y., Xie, T., Yan, H., Wan, H., et al. (2020). BP[dG]-induced distortions to DNA polymerase and DNA duplex: a detailed mechanism of BP adducts blocking replication. Food Chem. Toxicol. 140:111325. doi: 10.1016/j.fct.2020.111325
Xiang, L., Lu, Y., Wang, H., Wang, M., and Zhang, G. (2019). Improving the specific activity and pH stability of xylanase XynHBN188A by directed evolution. Bioresour. Bioprocess. 6:25.
Xie, T., Wu, Z., Gu, J., Guo, R., Yan, X., Duan, H., et al. (2019). The global motion affecting electron transfer in Plasmodium falciparum type II NADH dehydrogenases: a novel non-competitive mechanism for quinoline ketone derivative inhibitors. Phys. Chem. Chem. Phys. 21, 18105–18118. doi: 10.1039/c9cp02645b
Xiong, M., Deng, J., Woodruff, A. P., Zhu, M., Zhou, J., Park, S. W., et al. (2012). A bio-catalytic approach to aliphatic ketones. Sci. Rep. 2:311.
Xu, S., Lu, X., Li, M., Wang, J., Li, H., He, X., et al. (2019). Separation of 5-aminovalerate from its bioconversion liquid by macroporous adsorption resin: mechanism and dynamic separation. J. Chem. Technol. Biot. 95, 686–693. doi: 10.1002/jctb.6249
Yang, J., Zhu, Y., Men, Y., Sun, S., Zeng, Y., Zhang, Y., et al. (2016). Pathway construction in Corynebacterium glutamicum and strain engineering to produce rare sugars from glycerol. J. Agric. Food Chem. 64, 9497–9505. doi: 10.1021/acs.jafc.6b03423
Ying, H., Tao, S., Wang, J., Ma, W., Chen, K., Wang, X., et al. (2017). Expanding metabolic pathway for de novo biosynthesis of the chiral pharmaceutical intermediate L-pipecolic acid in Escherichia coli. Microb. Cell Fact. 16:52.
Youn, J.-W., Albermann, C., and Sprenger, G. A. (2020). In vivo cascade catalysis of aromatic amino acids to the respective mandelic acids using recombinant E. coli cells expressing hydroxymandelate synthase (HMS) from Amycolatopsis mediterranei. Mol. Catal. 483:110713. doi: 10.1016/j.mcat.2019.110713
Zeng, B., Lai, Y., Liu, L., Cheng, J., Zhang, Y., and Yuan, J. (2020). Engineering Escherichia coli for high-yielding hydroxytyrosol synthesis from biobased L-tyrosine. J. Agric. Food Chem. 68, 7691–7696. doi: 10.1021/acs.jafc.0c03065
Zhang, J., Barajas, J. F., Burdu, M., Wang, G., Baidoo, E. E., and Keasling, J. D. (2017). Application of an Acyl-CoA ligase from Streptomyces aizunensis for Lactam biosynthesis. ACS Synth. Biol. 6, 884–890. doi: 10.1021/acssynbio.6b00372
Zhang, K., Sawaya, M. R., Eisenberg, D. S., and Liao, J. C. (2008). Expanding metabolism for biosynthesis of nonnatural alcohols. Proc. Natl. Acad. Sci. U.S.A. 105, 20653–20658. doi: 10.1073/pnas.0807157106
Zhao, M., Huang, D., Zhang, X., Koffas, M. A. G., Zhou, J., and Deng, Y. (2018a). Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metab. Eng. 47, 254–262. doi: 10.1016/j.ymben.2018.04.002
Zhao, M., Li, G., and Deng, Y. (2018b). Engineering Escherichia coli for glutarate production as the C5 platform backbone. Appl. Environ. Microbiol. 84, e814–e818.
Keywords: 5-aminovalerate, L-Lysine HCl, artificial pathway, molecular dynamic simulation, molecular docking
Citation: Cheng J, Tu W, Luo Z, Gou X, Li Q, Wang D and Zhou J (2021) A High-Efficiency Artificial Synthetic Pathway for 5-Aminovalerate Production From Biobased L-Lysine in Escherichia coli. Front. Bioeng. Biotechnol. 9:633028. doi: 10.3389/fbioe.2021.633028
Received: 24 November 2020; Accepted: 20 January 2021;
Published: 09 February 2021.
Edited by:
K. Madhavan Nampoothiri, National Institute for Interdisciplinary Science and Technology (CSIR), IndiaReviewed by:
Volker F. Wendisch, Bielefeld University, GermanyCopyright © 2021 Cheng, Tu, Luo, Gou, Li, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Cheng, amNoZW5nQGNxdS5lZHUuY24=; Dan Wang, ZHdhbmdAY3F1LmVkdS5jbg==; Jingwen Zhou, emhvdWp3MTk4MkBqaWFuZ25hbi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.