
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Bioeng. Biotechnol., 08 April 2020
Sec. Synthetic Biology
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00293
This article is part of the Research TopicDevelopment and Application of Novel Genome Engineering Tools in Microbial BiotechnologyView all 22 articles
Filamentous fungi are considered as unique cell factories for protein production due to the high efficiency of protein secretion and superior capability of post-translational modifications. In this review, we firstly introduce the secretory pathway in filamentous fungi. We next summarize the current state-of-the-art works regarding how various genetic engineering strategies are applied for enhancing protein expression and secretion in filamentous fungi. Finally, in a future perspective, we discuss the great potential of genome engineering for further improving protein expression and secretion in filamentous fungi.
Protein production has a broad application in life sciences, biotechnology, medicine and material sciences. Filamentous fungi are powerful and efficient cell factories for protein production at the industrial scale, and over half of the commercially available proteins were produced by filamentous fungi1. Many species of filamentous fungi are generally regarded as safe (GRAS), and exhibit superior protein secretory capability. For example, 25–30 g/L of glucoamylase was obtained from fermentation medium of Aspergillus niger, while Trichoderma reesei was able to secret 100 g/L of cellulose (Ward, 2012). Compared to prokaryotes, filamentous fungi own the mature systems for post-translational processing (e.g., glycosylation, protease cleavage, and disulfide bond formation) (Karnaukhova et al., 2007), which are indispensable for protein function and activity. Although yeasts are able to perform post-translational modification, they tend to produce proteins in the form of high mannose-type glycosylation. In contrast, filamentous fungi have less extensive hyper-mannosylation of glycoproteins, which could be directly converted to mammalian type of glycoproteins with pharmaceutical potential (Punt et al., 2002; Deshpande et al., 2008). In addition, due to the metabolic diversity, filamentous fungi can efficiently utilize many types of monosaccharides including xylose, arabinose, and galactose, while yeasts can only metabolize glucose and mannose (Cavka and Jönsson, 2014).
To further improve the production of various proteins by filamentous fungi, traditional strategies including optimization of fermentation process and obtaining beneficial mutants via random mutagenesis, were widely adopted in the past. Here, rather than providing a comprehensive view of achieving efficient protein production, we focus on summarizing the strategies based on genetic engineering of this particular cell factory to enhance protein expression and secretion. We also provide new ideas in terms of cell factory engineering.
Protein secretion pathway in filamentous fungi involves three major steps including: polypeptide transfer from ribosome to endoplasmic reticulum (ER), protein folding and modification in ER, transportation of the folded protein vesicles to the Golgi apparatus and extracellular environment (Figure 1). In the first step, the co- or post-translational transport pathway is responsible for the polypeptide transfer from the ribosome to ER. In the co-translational transport pathway, the signal peptide recognition particle (SRP) first binds to the signal peptide sequence to block translation (Halic et al., 2006). Then, SRP directs the ribosome-mRNA-nascent peptide complex to target the ER membrane and binds to the SRP receptor. Subsequently, SRP is released from the complex, translation resumes, and the nascent polypeptide enters ER lumen through the Sec61p transport complex (Conesa et al., 2001). In the post-translational transport pathway, the nascent polypeptide is translated in the cytosol, and kept unfolded by interacting with Hsp70 chaperone and co-chaperones (Conesa et al., 2001). This complex is able to target ER through interaction with the membrane receptor Sec62p-Sec72p-Sec73p subcomplex (Conesa et al., 2001). The ER luminal chaperone binding immunoglobulin protein (BiP) and the membrane protein Sec63p assist the aforementioned complex to enter ER (Haßdenteufel et al., 2018).
The second step is protein folding and modification in ER, which requires the assistance of a series of molecular chaperones and folding enzymes, including calnexin (ClxA), BiP, and protein disulfide isomerase (PDI) (Saloheimo and Pakula, 2012). For nascent peptides with correct folding, they are subjected to modifications such as glycosylation. As one of the most common and important post-translational modifications, glycosylation can significantly affect protein stability, localization, and secretion (Mitra et al., 2006). After proper folding and glycosylation, secreted proteins are transported extracellularly. On the other hand, the unfolded protein response (UPR) and ER-associated protein degradation (ERAD) are responsible for dealing with nascent peptides with incorrect folding (Bernasconi and Molinari, 2011; Wang et al., 2014). The UPR detects the presence of unfolded proteins in ER and induces the biosynthesis of chaperones and folding enzymes, while the ERAD degrades the misfolded proteins.
The third step is to transport the folded protein vesicles to the Golgi apparatus by fusion with target membrane, and secrete it to the extracellular environment (Spang, 2008). In filamentous fungi, Golgi-derived secretory vesicles are transmitted to the apical plasma membrane through apical vesicle clusters in Spitzenkörper (Virag and Harris, 2006). The formation, transportation and fusion of vesicles are mediated by a large number of proteins, including GTP-binding proteins (e.g., Sar, ARF) for vesicle budding, and Rab GTPases for fusion with Golgi (Hutagalung and Novick, 2011), etc. Specific fusion of vesicles with the target membrane is the critical process, which is mediated by soluble N-ethylmaleimide-sensitive factor-associated protein receptor (SNARE). Based on the localization, SNARE is divided into two categories: the vesicle SNARE (v-SNARE) and the target membrane SNARE (t-SNARE) (Söllner et al., 1993). In filamentous fungi, v-SNARE protein SNC1, and t-SNARE proteins SSO1 and SSO2, are involved in bubble fusion (Valkonen et al., 2007).
To enhance the protein expression and secretion in filamentous fungi, enhancing the intracellular protein production by optimization of the transcription and/or the codon of the target protein, is an effective strategy, as summarized in a few of reviews (Saunders et al., 1989; Jeenes et al., 1991; Nevalainen et al., 2005; Su et al., 2012). In order to bring new insights, we will discuss other genetic engineering strategies, including replacing original signal peptide with a more efficient one, fusion of heterologous protein to a naturally secreted one, regulation of UPR and ERAD, optimization of the intracellular transport process, construction of a protease-deficient strain, regulation of mycelium morphology, and optimization of the sterol regulatory element binding protein (SREBP) in this section (Table 1).
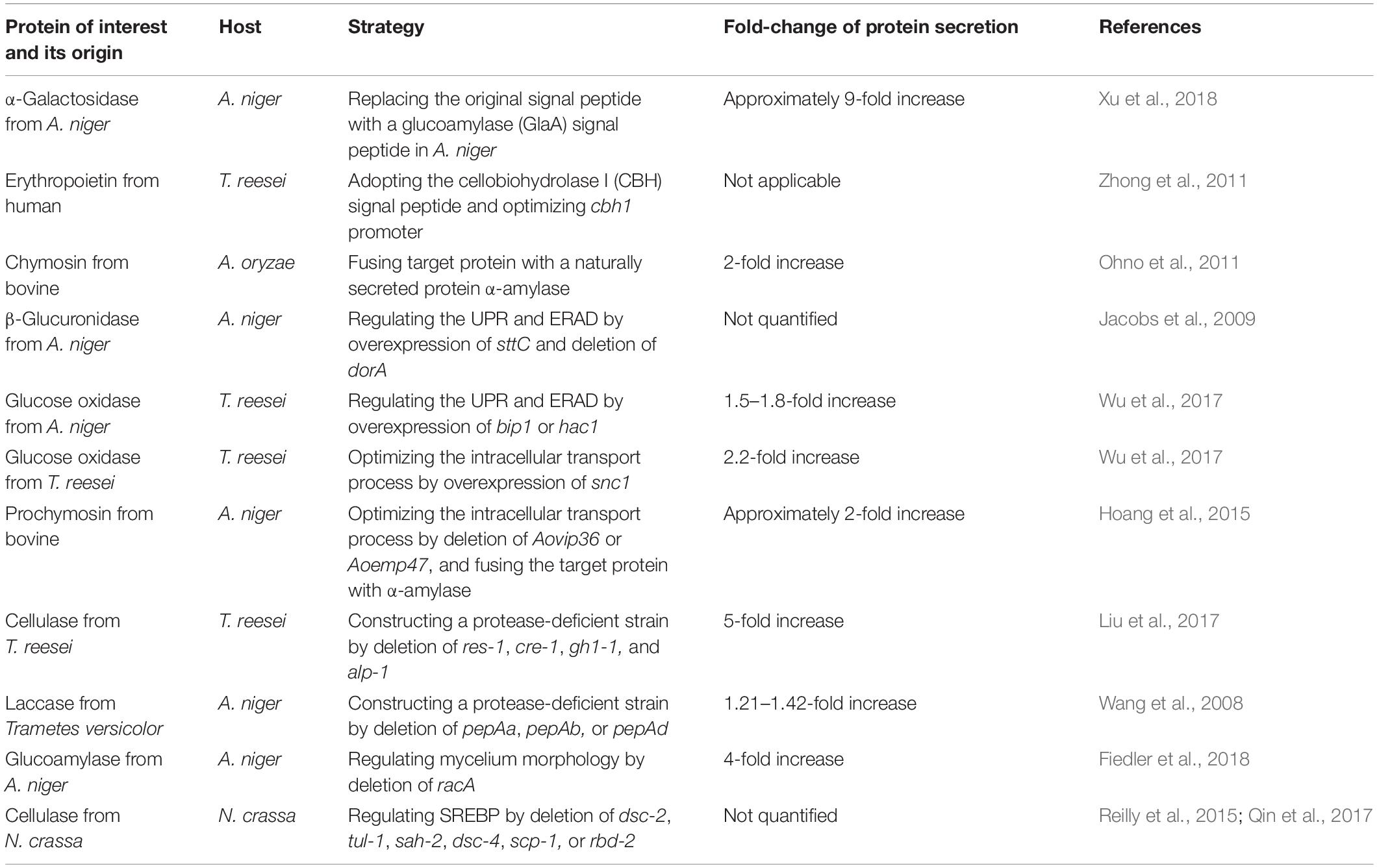
Table 1. Typical examples for genetic engineering of filamentous fungi for enhanced protein secretion.
The signal peptide sequence plays vital role in protein secretion. Replacing with a more efficient peptide in target protein tends to increase its secretion efficiency. Xu et al. replaced the original signal peptide AglB of α-galactosidase with a glucoamylase (GlaA) signal peptide in A. niger, and the activity of extracellular α-galactosidase increased nearly ninefold (Xu et al., 2018). Wang et al. used green fluorescent protein as a reporter gene in P. oxalicum to test the secretion efficiency of three signal peptides, PoxGA15A, PoxAmy13A, and PoxCbhCel7A-2. Then they selected the optimal signal peptide PoxGA15A to drive the secretion of endogenous raw starch-degrading enzymes, which was 3.4 times higher than the parental strain (Wang et al., 2018).
Fusion of heterologous protein to a naturally secreted one can enhance protein stability, promote translocation, and prevent protein from degradation. The in-frame fusion of human protein granulocyte colony stimulating factor (G-CSF) with an endogenous highly secreted glucoamylase allowed secretion of 5–10 mg/L of G-CSF by A. niger (Kraševec et al., 2014). When bovine chymosin (CHY) was fused with alpha-amylase (AmyB), the engineered A. oryzae was able to produce two times higher amount of CHY than that with none fused CHY, while multiple genes involved in ER folding and protein secretion pathway increased significantly in the fused CHY producing strain (Ohno et al., 2011). It should be noted that the fusion carrier protein could greatly affect the secretion. In order to secrete Escherichia coli β-glucuronidase (GUS) protein in Penicillium funiculosum, researchers attempted to use xylanase as a carrier. The modular structure, a catalytic domain separated from the cellulose-binding domain by a linker with serine and threonine rich sequence, enables some xylanases as a group of unique protein carrier (Alcocer et al., 2003). It was reported that xylanase A (XYNA) is an effective carrier protein, while XYNB and XYNC are ineffective (Alcocer et al., 2003).
Correct protein folding is one of the many prerequisites to protein secretion. Abnormal folding proteins could form toxic aggregates exerting pressure on the ER, and trigger the feedback regulation called repression under secretion stress (RESS) to affect protein secretion (Pakula et al., 2003). UPR and ERAD are considered as two important ways to regulate protein folding, and enhanced protein secretion could be achieved via regulation of UPR and ERAD. For example, overexpression of the transcription factor hac1 in Aspergillus awamori led to 7- and 2.8-fold increases in laccase and bovine prechymotrypsin production, respectively (Valkonen et al., 2003). Overexpression of bip1 and hac1 in T. reesei exhibited 1.5- and 1.8-fold improvement on secretion of an A. niger glucose oxidase (Wu et al., 2017).
To avoid degradation of some heterologous proteins or semi-folded proteins, deleting key genes involved in ERAD is a solution. Deletion of the ERAD factor doaA and overexpression of the oligosaccharyltransferase sttC responsible for glycosylation of secretory proteins (Yan and Lennarz, 2002) in A. niger caused an increase in β-glucuronidase yield (Jacobs et al., 2009). In addition, autophagy is considered as another way to degrade the misfolded proteins (Kario et al., 2011). Disruption of autophagy-related gene aoatg15 in A. oryzae caused a threefold increase in secretion of bovine chymosin (Yoon et al., 2013).
Of particular note, manipulation of certain gene may cause quite different effects in different strains. For example, overexpression of bip1 promoted protein secretion in T. reesei (Wu et al., 2017) and A. awamori (Lombraña et al., 2004), while reduced protein secretion was observed in A. niger by adopting the same strategy (Conesa et al., 2002). These effects could be attributed to the multifunction of BiP. BiP is able to promote protein translocation and folding, as well as to promote ER-associated protein degradation. Similarly, overexpression of hac1 could promote protein secretion, which may also affect cell growth in certain strains (Valkonen et al., 2003; Carvalho et al., 2012). In addition, deletion of derA in A. niger can promote protein production (Carvalho et al., 2011), while deletion of the same gene affected the cell growth of Aspergillus fumigatus (Richie et al., 2011). It’s not difficult to see that the effect of protein secretion by regulating UPR and ERAD is host-dependent. Thus, a deep understanding of the complexity and specificity of the interactions between the components of the secretory pathway in a particular host is required prior to the manipulation.
Before being secreted outside, proteins are transported between ER and Golgi tendencies via vesicles. In this process, the ER-Golgi cargo receptor recruits the secreted proteins into the vesicles, thereby facilitating their transport (Dancourt and Barlowe, 2010). Optimization of the intracellular protein transport process allows enhanced protein secretion. In A. oryzae, the cargo receptor AoVip36 is localized in the ER and AoEmp47 is localized in the Golgi compartment. Deletion of AoVip36, responsible for anterograde transport, caused a 30% reduction of the endogenous α-amylase activity, and overexpression of this gene led to the increased secretion of EGFP (Hoang et al., 2015). In addition, deletion of Aovip36 or Aoemp47 increased the secretion of bovine prochymosin by approximately twofold (Hoang et al., 2015). In Aspergillus nidulans, gene podB is predicted to encode the subunit of the Golgi-conserved oligomeric complex (Gremillion et al., 2014), which is involved in Golgi retrograde vesicle transport, and affects cell polar growth, germination, and protein glycosylation (Wuestehube et al., 1996; Harris et al., 1999; Suvorova et al., 2002; Gremillion et al., 2014). A G-to-T mutation at nucleotide #751 in podB1 led to significant increase in cellulase and xylanase activities (Boppidi et al., 2018). Wu et al. overexpressed snc1 gene, which is involved in fusion of vesicles and plasma membrane, and observed a 2.2-fold increase in secretion of an A. niger glucose oxidase in T. reesei (Wu et al., 2017).
In addition to being successfully secreted outside, some heterologous proteins may be transported to the vacuole for degradation (Masai et al., 2003). Disruption of the vacuolar sorting receptor encoding gene Aovps10 resulted in three and twofold increases in the production yields of bovine chymosin and human lysozyme in A. oryzae, respectively (Yoon et al., 2010).
The efficient production of certain endogenous protein in filamentous fungi disturbs the secretion of the protein of interest, and construction of a protease-deficient strain can strongly support the modification and secretion of the target protein. Disruption of alkaline serine protease SPW in T. reesei reduced the extracellular total protease activity by about 50%, and improved the production and stability of the heterologous alkaline endoglucanase EGV from Humicola insolens (Zhang et al., 2014). To construct a cellulase hyper-producing strain, β-glucosidase encoding gene gh1-1, alkaline protease encoding gene alp-1, and cellulase production related genes cre-1 and res-1 were simultaneously deleted in Myceliophthora thermophile. The secreted cellulase of the resulted strain was five times higher than that of the original strain (Liu et al., 2017).
Proteins are mainly secreted at vigorously growing mycelial tips in filamentous fungi (Wessels, 1993), and the mycelium morphology is especially important to protein secretion. The increased branching of the mycelium tip usually facilitates endogenous protein secretion. Lin et al. screened 90 morphological mutants of Neurospora crassa and found that disruption of gul-1 led to a marked decrease in viscosity of the culture medium, while overexpression of gul-1 led to a sharp increase in viscosity. In the gul-1 disrupted strain, 25% and 56% increases were observed in the total extracellular protein concentration and β-glucosidase activity, respectively (Lin et al., 2018), suggesting that cell wall integrity has a significant effect on protein secretion. In A. niger, the Rho GTPase RacA regulates the polymerization and depolymerization of actin at the tip of mycelium (Kwon et al., 2011). When racA was deleted, the mycelial tip increased by about 20%, the number of secreted vesicles increased, and the secretion of glucoamylase increased 4 times as compared to the wild type strain (Fiedler et al., 2018). Similarly, deletion of racA resulted in a hyperbranched phenotype and three folds increase of cellulase activity in T. reesei (Fitz et al., 2019).
In filamentous fungi, SREBP, responsible for regulating sterol homeostasis under challenging environments, is strongly associated with protein secretion, including linkages to the UPR (Qin et al., 2017) and formation of hyphae branches (Willger et al., 2008). After analysis the phenotype of a 567 single-gene deletion collection of N. crassa, researchers found that deletion of dsc-2 and tul-1 (dsc-1) significantly increased the secretion of proteins (Reilly et al., 2015). In Schizosaccharomyces pombe and A. fumigatus, homologs of Dsc-2 and Tul-1 are part of the Golgi E3 ligase complex (Dsc complex), which can activate SREBP orthologs Sre1 and SreA through proteolytic cleavage (Lloyd et al., 2013). In addition, deletion of the unit of Dsc complex Dsc-4 and the Sre1/SreA homolog SAH-2 also showed a high secretion phenotype of cellulases (Reilly et al., 2015). Homologs of SAH-2 and TUL-1 from N. crassa are discovered in T. reesei, and their deletions enhanced the capability of protein secretion (Reilly et al., 2015). In a follow-up study, deletion of gene scp-1 and rbd-2, encoding SREBP cleavage activating protein and rhomboid protease respectively, also led to the high producing phenotype of cellulose (Qin et al., 2017).
Owing to the powerful protein secretion pathway, filamentous fungi are attractive cell factories for protein expression and secretion. For all the discussed strategies, replacing original signal peptide with a more efficient one, regulation of UPR and ERAD, optimization of the intracellular transport process, and construction of a protease-deficient strain have been successfully applied to improve the production of endogenous and heterologous proteins by filamentous fungi, while fusion of heterologous protein to a naturally secreted one is extremely effective for production of heterologous protein (Table 1). As for regulation of mycelium morphology and optimization of SREBP, although they were mainly adopted for production of endogenous protein, we believe that they are also applicable for production of heterologous protein. However, most efforts in genetic engineering of filamentous fungi for enhanced protein expression and secretion were solely based on the protein of interest, the secretory pathway or the host. Although these engineering strategies significantly improved target protein production, they were mainly related to single gene or pathway.
With the aid of multiple gene editing technologies (e.g., DNA recombination, RNAi, CRISPR-Cas), genome engineering strategies introduce deletion, insertion and/or point mutations across the genome via a trackable manner to accelerate strain evolution (Si et al., 2015). Compared with traditional metabolic engineering strategies, genome engineering allows rapid tracking and discovery of novel determinants (Xiao and Zhao, 2014; Si et al., 2017), editing of key determinant with single-nucleotide precision (Garst et al., 2017; Bao et al., 2018), or simultaneous manipulating multiple pathways (Barbieri et al., 2017; Liang et al., 2017). Apart from the unicellular model organisms (e.g., Saccharomyces cerevisiae), many filamentous fungi, particularly the mushroom-forming fungi, contain two different nuclei with different genetic contents (Gehrmann et al., 2018). In addition to the heterogeneity, many important medicinal mushrooms also exhibit low efficiency on gene transformation and homologous recombination (HR), which pose a great challenge to establish gene editing tools for genome engineering (Wang et al., 2020). To circumvent these difficulties, developing effective technologies for single spore isolation, gene delivery and/or improving HR efficiency are highly required in these filamentous fungi. It is notable that the target performances of the engineered strains, which are greatly improved by the aforementioned genome engineering strategies, can usually be screened out via cell growth or color. Thus, high-throughput screening methods are highly required to ensure the success of genome engineering. In fact, the fluorescence-activated cell sorting (FACS) assisted the intracellular protein production has been extensively adopted in filamentous fungi, but such strategy is difficult to screen out the beneficial mutants with enhanced protein secretion capacity (Throndset et al., 2010). To solve this problem, displaying the fluorescence protein on the cell surface, coupled by FACS, allows screening of the cellulose hypersecretors from T. reesei (Gao et al., 2018). As a promising alternative, the droplet-based microfluidic high-throughput screening platform has been established in T. reesei and A. niger (Beneyton et al., 2016; He et al., 2019). In future, we believe that harnessing the great potential of genome engineering will further increase protein expression and secretion by filamentous fungi.
CZ and HX designed this manuscript. QW and HX wrote this manuscript. QW, CZ, and HX revised this manuscript.
This work was supported by the National Natural Science Foundation of China (No. 31971344) and Municipal Natural Science Foundation of Shanghai (Nos. 17ZR1448900 and 18ZR1420300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alcocer, M. J. C., Furniss, C. S. M., Kroon, P. A., Campbell, M., and Archer, D. B. (2003). Comparison of modular and non-modular xylanases as carrier proteins for the efficient secretion of heterologous proteins from Penicillium funiculosum. Appl. Microbiol. Biotechnol. 60, 726–732. doi: 10.1007/s00253-002-1184-4
Bao, Z., HamediRad, M., Xue, P., Xiao, H., Tasan, I., Chao, R., et al. (2018). Genome-scale engineering of Saccharomyces cerevisiae with single-nucleotide precision. Nat. Biotechnol. 36, 505–508. doi: 10.1038/nbt.4132
Barbieri, E. M., Muir, P., Akhuetie-Oni, B. O., Yellman, C. M., and Isaacs, F. J. (2017). Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell 171, 1453–1467.e1413. doi: 10.1016/j.cell.2017.10.034
Beneyton, T., Wijaya, I. P. M., Postros, P., Najah, M., Leblond, P., Couvent, A., et al. (2016). High-throughput screening of filamentous fungi using nanoliter-range droplet-based microfluidics. Sci. Rep. 6:27223. doi: 10.1038/srep27223
Bernasconi, R., and Molinari, M. (2011). ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 23, 176–183. doi: 10.1016/j.ceb.2010.10.002
Boppidi, K. R., Ribeiro, L. F. C., Iambamrung, S., Nelson, S. M., Wang, Y., Momany, M., et al. (2018). Altered secretion patterns and cell wall organization caused by loss of PodB function in the filamentous fungus Aspergillus nidulans. Sci. Rep. 8:11433. doi: 10.1038/s41598-018-29615-z
Carvalho, N. D. S. P., Arentshorst, M., Kooistra, R., Stam, H., Sagt, C. M., van den Hondel, C. A., et al. (2011). Effects of a defective ERAD pathway on growth and heterologous protein production in Aspergillus niger. Appl. Microbiol. Biotechnol. 89, 357–373. doi: 10.1007/s00253-010-2916-5
Carvalho, N. D. S. P., Jørgensen, T. R., Arentshorst, M., Nitsche, B. M., van den Hondel, C. A., Archer, D. B., et al. (2012). Genome-wide expression analysis upon constitutive activation of the HacA bZIP transcription factor in Aspergillus niger reveals a coordinated cellular response to counteract ER stress. BMC Genomics 13:350. doi: 10.1186/1471-2164-13-350
Cavka, A., and Jönsson, L. J. (2014). Comparison of the growth of filamentous fungi and yeasts in lignocellulose-derived media. Biocatal. Agric. Biotechnol. 3, 197–204. doi: 10.1016/j.bcab.2014.04.003
Conesa, A., Jeenes, D., Archer, D. B., van den Hondel, C. A. M., and Punt, P. J. (2002). Calnexin overexpression increases manganese peroxidase production in Aspergillus niger. Appl. Environ. Microbiol. 68, 846–851. doi: 10.1128/AEM.68.2.846-851.2002
Conesa, A., Punt, P. J., van Luijk, N., and van den Hondel, C. A. (2001). The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 33, 155–171. doi: 10.1006/fgbi.2001.1276
Dancourt, J., and Barlowe, C. (2010). Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79, 777–802. doi: 10.1146/annurev-biochem-061608-091319
Deshpande, N., Wilkins, M. R., Packer, N., and Nevalainen, H. (2008). Protein glycosylation pathways in filamentous fungi. Glycobiology 18, 626–637. doi: 10.1093/glycob/cwn044
Fiedler, M. R. M., Barthel, L., Kubisch, C., Nai, C., and Meyer, V. (2018). Construction of an improved Aspergillus niger platform for enhanced glucoamylase secretion. Microb. Cell Fact. 17, 95–95. doi: 10.1186/s12934-018-0941-8
Fitz, E., Gamauf, C., Seiboth, B., and Wanka, F. (2019). Deletion of the small GTPase rac1 in Trichoderma reesei provokes hyperbranching and impacts growth and cellulase production. Fungal Biol. Biotechnol. 6:16. doi: 10.1186/s40694-019-0078-5
Gao, F., Hao, Z., Sun, X., Qin, L., Zhao, T., Liu, W., et al. (2018). A versatile system for fast screening and isolation of Trichoderma reesei cellulase hyperproducers based on DsRed and fluorescence-assisted cell sorting. Biotechnol. Biofuels 11:261. doi: 10.1186/s13068-018-1264-z
Garst, A. D., Bassalo, M. C., Pines, G., Lynch, S. A., Halweg-Edwards, A. L., Liu, R., et al. (2017). Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55. doi: 10.1038/nbt.3718
Gehrmann, T., Pelkmans, J. F., Ohm, R. A., Vos, A. M., Sonnenberg, A. S. M., Baars, J. J. P., et al. (2018). Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs. Proc. Natl. Acad. Sci. U.S.A. 115, 4429–4434. doi: 10.1073/pnas.1721381115
Gremillion, S. K., Harris, S. D., Jackson-Hayes, L., Kaminskyj, S. G. W., Loprete, D. M., Gauthier, A. C., et al. (2014). Mutations in proteins of the conserved oligomeric Golgi complex affect polarity, cell wall structure, and glycosylation in the filamentous fungus Aspergillus nidulans. Fungal Genet. Biol. 73, 69–82. doi: 10.1016/j.fgb.2014.10.005
Halic, M., Blau, M., Becker, T., Mielke, T., Pool, M. R., Wild, K., et al. (2006). Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444, 507–511. doi: 10.1038/nature05326
Harris, S. D., Hofmann, A. F., Tedford, H. W., and Lee, M. P. (1999). Identification and characterization of genes required for hyphal morphogenesis in the filamentous fungus Aspergillus nidulans. Genetics 151, 1015–1025.
Haßdenteufel, S., Johnson, N., Paton, A. W., Paton, J. C., High, S., and Zimmermann, R. (2018). Chaperone-mediated Sec61 channel gating during ER import of small precursor proteins overcomes Sec61 inhibitor-reinforced energy barrier. Cell Rep. 23, 1373–1386. doi: 10.1016/j.celrep.2018.03.122
He, R., Ding, R., Heyman, J. A., Zhang, D., and Tu, R. (2019). Ultra-high-throughput picoliter-droplet microfluidics screening of the industrial cellulase-producing filamentous fungus Trichoderma reesei. J. Ind. Microbiol. Biotechnol. 46, 1603–1610. doi: 10.1007/s10295-019-02221-2
Hoang, H. D., Maruyama, J. I., and Kitamoto, K. (2015). Modulating endoplasmic reticulum-Golgi cargo receptors for improving secretion of carrier-fused heterologous proteins in the filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 81, 533–543. doi: 10.1128/AEM.02133-14
Hutagalung, A. H., and Novick, P. J. (2011). Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91, 119–149. doi: 10.1152/physrev.00059.2009
Jacobs, D. I., Olsthoorn, M. M. A., Maillet, I., Akeroyd, M., Breestraat, S., Donkers, S., et al. (2009). Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet. Biol. 46(1, Suppl.), S141–S152. doi: 10.1016/j.fgb.2008.08.012
Jeenes, D. J., Mackenzie, D. A., Roberts, I. N., and Archer, D. B. (1991). Heterologous protein production by filamentous fungi. Biotechnol. Genet. Eng. Rev. 9, 327–367. doi: 10.1080/02648725.1991.10750006
Kario, E., Amar, N., Elazar, Z., and Navon, A. (2011). A new autophagy-related checkpoint in the degradation of an ERAD-M target. J. Biol. Chem. 286, 11479–11491. doi: 10.1074/jbc.M110.177618
Karnaukhova, E., Ophir, Y., Trinh, L., Dalal, N., Punt, P. J., Golding, B., et al. (2007). Expression of human α1-proteinase inhibitor in Aspergillus niger. Microb. Cell Fact. 6:34. doi: 10.1186/1475-2859-6-34
Kraševec, N., Milunović, T., Lasnik, M. A., Lukančič, I., Komel, R., and Porekar, V. G. (2014). Human granulocyte colony stimulating factor (G-CSF) produced in the filamentous fungus Aspergillus niger. Acta Chim. Slov. 61, 709–717.
Kwon, M. J., Arentshorst, M., Roos, E. D., van den Hondel, C. A., Meyer, V., and Ram, A. F. J. (2011). Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol. Microbiol. 79, 1151–1167. doi: 10.1111/j.1365-2958.2010.07524.x
Liang, L., Liu, R., Garst, A. D., Lee, T., Nogué, V. S. I., Beckham, G. T., et al. (2017). CRISPR EnAbled trackable genome engineering for isopropanol production in Escherichia coli. Metab. Eng. 41, 1–10. doi: 10.1016/j.ymben.2017.02.009
Lin, L., Sun, Z., Li, J., Chen, Y., Liu, Q., Sun, W., et al. (2018). Disruption of gul-1 decreased the culture viscosity and improved protein secretion in the filamentous fungus Neurospora crassa. Microb. Cell Fact. 17:96. doi: 10.1186/s12934-018-0944-5
Liu, Q., Gao, R., Li, J., Lin, L., Zhao, J., Sun, W., et al. (2017). Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels 10:1. doi: 10.1186/s13068-016-0693-9
Lloyd, S. J.-A., Raychaudhuri, S., and Espenshade, P. J. (2013). Subunit architecture of the Golgi Dsc E3 ligase required for sterol regulatory element-binding protein (SREBP) cleavage in fission Yeast. J. Biol. Chem. 288, 21043–21054. doi: 10.1074/jbc.M113.468215
Lombraña, M., Moralejo, F. J., Pinto, R., and Martín, J. F. (2004). Modulation of Aspergillus awamori thaumatin secretion by modification of bipA gene expression. Appl. Environ. Microbiol. 70, 5145–5152. doi: 10.1128/AEM.70.9.5145-5152.2004
Masai, K., Maruyama, J.-I., Nakajima, H., and Kitamoto, K. (2003). In vivo visualization of the distribution of a secretory protein in Aspergillus oryzae hyphae using the RntA-EGFP fusion protein. Biosci. Biotechnol. Biochem. 67, 455–459. doi: 10.1271/bbb.67.455
Mitra, N., Sinha, S., Ramya, T. N. C., and Surolia, A. (2006). N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 31, 156–163. doi: 10.1016/j.tibs.2006.01.003
Nevalainen, K. M. H., Te’o, V. S. J., and Bergquist, P. L. (2005). Heterologous protein expression in filamentous fungi. Trends Biotechnol. 23, 468–474. doi: 10.1016/j.tibtech.2005.06.002
Ohno, A., Maruyama, J.-I., Nemoto, T., Arioka, M., and Kitamoto, K. (2011). A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 92, 1197–1206. doi: 10.1007/s00253-011-3487-9
Pakula, T. M., Laxell, M., Huuskonen, A., Uusitalo, J., Saloheimo, M., and Penttilä, M. (2003). The effects of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei: evidence for down-regulation of genes that encode secreted proteins in the stressed cells. J. Biol. Chem. 278, 45011–45020. doi: 10.1074/jbc.m302372200
Punt, P. J., van Biezen, N., Conesa, A., Albers, A., Mangnus, J., and van den Hondel, C. (2002). Filamentous fungi as cell factories for heterologous protein production. Trends Biotechnol. 20, 200–206. doi: 10.1016/S0167-7799(02)01933-9
Qin, L., Wu, V. W., and Glass, N. L. (2017). Deciphering the regulatory network between the SREBP pathway and protein secretion in Neurospora crassa. mBio 8:e00233-17. doi: 10.1128/mBio.00233-17
Reilly, M. C., Qin, L., Craig, J. P., Starr, T. L., and Glass, N. L. (2015). Deletion of homologs of the SREBP pathway results in hyper-production of cellulases in Neurospora crassa and Trichoderma reesei. Biotechnol. Biofuels 8:121. doi: 10.1186/s13068-015-0297-9
Richie, D. L., Feng, X., Hartl, L., Aimanianda, V., Krishnan, K., Powers-Fletcher, M. V., et al. (2011). The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR). Virulence 2, 12–21. doi: 10.4161/viru.2.1.13345
Saloheimo, M., and Pakula, T. M. (2012). The cargo and the transport system: secreted proteins and protein secretion in Trichoderma reesei (Hypocrea jecorina). Microbiology 158(Pt 1), 46–57. doi: 10.1099/mic.0.053132-0
Saunders, G., Picknett, T. M., Tuite, M. F., and Ward, M. (1989). Heterologous gene expression in filamentous fungi. Trends Biotechnol. 7, 283–287. doi: 10.1016/0167-7799(89)90048-6
Si, T., Chao, R., Min, Y., Wu, Y., Ren, W., and Zhao, H. (2017). Automated multiplex genome-scale engineering in yeast. Nat. Commun. 8:15187. doi: 10.1038/ncomms15187
Si, T., Xiao, H., and Zhao, H. (2015). Rapid prototyping of microbial cell factories via genome-scale engineering. Biotechnol. Adv. 33, 1420–1432. doi: 10.1016/j.biotechadv.2014.11.007
Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., et al. (1993). SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324. doi: 10.1038/362318a0
Spang, A. (2008). Membrane traffic in the secretory pathway. Cell. Mol. Life Sci. 65, 2781–2789. doi: 10.1007/s00018-008-8349-y
Su, X., Schmitz, G., Zhang, M., Mackie, R. I., and Cann, I. K. O. (2012). “Chapter One - heterologous gene expression in filamentous fungi,” in Advances in Applied Microbiology, eds G. M. Gadd and S. Sariaslani (Cambridge, MA: Academic Press), 1–61.
Suvorova, E. S., Duden, R., and Lupashin, V. V. (2002). The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J. Cell Biol. 157, 631–643. doi: 10.1083/jcb.200111081
Throndset, W., Kim, S., Bower, B., Lantz, S., Kelemen, B., Pepsin, M., et al. (2010). Flow cytometric sorting of the filamentous fungus Trichoderma reesei for improved strains. Enzyme Microb. Technol. 47, 335–341. doi: 10.1016/j.enzmictec.2010.09.003
Valkonen, M., Kalkman, E. R., Saloheimo, M., Penttilä, M., Read, N. D., and Duncan, R. R. (2007). Spatially segregated SNARE protein interactions in living fungal cells. J. Biol. Chem. 282, 22775–22785. doi: 10.1074/jbc.m700916200
Valkonen, M., Ward, M., Wang, H., Penttilä, M., and Saloheimo, M. (2003). Improvement of foreign-protein production in Aspergillus niger var. awamori by constitutive induction of the unfolded-protein response. Appl. Environ. Microbiol. 69, 6979–6986. doi: 10.1128/AEM.69.12.6979-6986.2003
Virag, A., and Harris, S. D. (2006). The Spitzenkörper: a molecular perspective. Mycol. Res. 110, 4–13. doi: 10.1016/j.mycres.2005.09.005
Wang, G., Zhang, D., and Chen, S. (2014). Effect of earlier unfolded protein response and efficient protein disposal system on cellulase production in Rut C30. World J. Microbiol. Biotechnol. 30, 2587–2595. doi: 10.1007/s11274-014-1682-4
Wang, L., Zhao, S., Chen, X., Deng, Q., Li, C., and Feng, J. (2018). Secretory overproduction of a raw starch-degrading glucoamylase in Penicillium oxalicum using strong promoter and signal peptide. Appl. Microbiol. Biotechnol. 102, 9291–9301. doi: 10.1007/s00253-018-9307-8
Wang, P., Xiao, H., and Zhong, J. (2020). CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 104, 1661–1671. doi: 10.1007/s00253-019-10298-z
Wang, Y., Xue, W., Sims, A. H., Zhao, C., Wang, A., Tang, G., et al. (2008). Isolation of four pepsin-like protease genes from Aspergillus niger and analysis of the effect of disruptions on heterologous laccase expression. Fungal Genet. Biol. 45, 17–27. doi: 10.1016/j.fgb.2007.09.012
Ward, O. P. (2012). Production of recombinant proteins by filamentous fungi. Biotechnol. Adv. 30, 1119–1139. doi: 10.1016/j.biotechadv.2011.09.012
Wessels, J. G. H. (1993). Tansley Review No. 45 Wall growth, protein excretion and morphogenesis in fungi. New Phytol. 123, 397–413. doi: 10.1111/j.1469-8137.1993.tb03751.x
Willger, S. D., Puttikamonkul, S., Kim, K. H., Burritt, J. B., Grahl, N., Metzler, L. J., et al. (2008). A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4:e1000200. doi: 10.1371/journal.ppat.1000200
Wu, Y., Sun, X., Xue, X., Luo, H., Yao, B., Xie, X., et al. (2017). Overexpressing key component genes of the secretion pathway for enhanced secretion of an Aspergillus niger glucose oxidase in Trichoderma reesei. Enzyme Microb. Technol. 106, 83–87. doi: 10.1016/j.enzmictec.2017.07.007
Wuestehube, L. J., Duden, R., Eun, A., Hamamoto, S., Korn, P., Ram, R., et al. (1996). New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics 142, 393–406.
Xiao, H., and Zhao, H. (2014). Genome-wide RNAi screen reveals the E3 SUMO-protein ligase gene SIZ1 as a novel determinant of furfural tolerance in Saccharomyces cerevisiae. Biotechnol. Biofuels 7:78. doi: 10.1186/1754-6834-7-78
Xu, Y., Wang, Y., Liu, T., Zhang, H., Zhang, H., and Li, J. (2018). The GlaA signal peptide substantially increases the expression and secretion of α-galactosidase in Aspergillus niger. Biotechnol. Lett. 40, 949–955. doi: 10.1007/s10529-018-2540-5
Yan, Q., and Lennarz, W. J. (2002). Studies on the function of oligosaccharyl transferase subunits: Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277, 47692–47700. doi: 10.1074/jbc.m208136200
Yoon, J., Aishan, T., Maruyama, J., and Kitamoto, K. (2010). Enhanced production and secretion of heterologous proteins by the filamentous fungus Aspergillus oryzae via disruption of vacuolar protein sorting receptor gene Aovps10. Appl. Environ. Microbiol. 76, 5718–5727. doi: 10.1128/AEM.03087-09
Yoon, J., Kikuma, T., Maruyama, J., and Kitamoto, K. (2013). Enhanced production of bovine chymosin by autophagy deficiency in the filamentous fungus Aspergillus oryzae. PLoS One 8:e62512. doi: 10.1371/journal.pone.0062512
Zhang, G., Zhu, Y., Wei, D., and Wang, W. (2014). Enhanced production of heterologous proteins by the filamentous fungus Trichoderma reesei via disruption of the alkaline serine protease SPW combined with a pH control strategy. Plasmid 71, 16–22. doi: 10.1016/j.plasmid.2014.01.001
Zhong, Y., Liu, X., Xiao, P., Wei, S., and Wang, T. (2011). Expression and secretion of the human erythropoietin using an optimized cbh1 promoter and the native CBH I signal sequence in the industrial fungus Trichoderma reesei. Appl. Biochem. Biotechnol. 165, 1169–1177. doi: 10.1007/s12010-011-9334-8
Keywords: filamentous fungi, genetic engineering, protein secretion, expression, genome engineering
Citation: Wang Q, Zhong C and Xiao H (2020) Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 8:293. doi: 10.3389/fbioe.2020.00293
Received: 15 January 2020; Accepted: 19 March 2020;
Published: 08 April 2020.
Edited by:
Jiazhang Lian, Zhejiang University, ChinaReviewed by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2020 Wang, Zhong and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Zhong, emhvbmdjaGFvQHNoYW5naGFpdGVjaC5lZHUuY24=; Han Xiao, c21hbGxoYW5Ac2p0dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.