- 1Department of Biology, College of Life Sciences, Shanghai Normal University, Shanghai, China
- 2State Key Laboratory of South China Sea Marine Resource Utilization, Hainan University, Haikou, China
- 3State Key Laboratory of Bioreactor Engineering, East China University of Science and Technology, Shanghai, China
This article summarizes the current knowledge regarding omics approaches, which include genomics, transcriptomics, proteomics and metabolomics, in the context of bio-hydrogen production in Chlamydomonas reinhardtii. In this paper, critical genes (HydA1, Hyd A2, Sulp, Tla1, Sta7, PFL1) involved in H2 metabolism were identified and analyzed for their function in H2 accumulation. Furthermore, the advantages of gene microarrays and RNA-seq were compared, as well as their applications in transcriptomic analysis of H2 production. Moreover, as a useful tool, proteomic analysis could identify different proteins that participate in H2 metabolism. This review provides fundamental theory and an experimental basis for H2 production, and further research effort is needed in this field.
Introduction
With the increase of the exploitation and utilization of traditional energy, the supply of fossil fuels gradually decreases and the cost of exploiting new energy increases, eventually leading to the gradual rise of energy prices (Hwang et al., 2014; Fakhimi et al., 2019). Traditional energy sources tend to cause environmental problems and air pollution, access to renewable and environmentally sustainable fuels and energy sources may be the greatest challenge of this century. Hydrogen is considered one of the ideal clean-energy alternatives because its only combustion product is H2O as well as it has a high heating value (Ramadass et al., 2019). Currently, hydrogen production is expensive, and the energy output hardly exceeds the input. Since Gaffron and his coworkers discovered the presence of hydrogen metabolism in the green algae Scenedesmus obliquus, hydrogen biosynthesis from algae attracted many attentions of researchers (Gaffron, 1939; Gaffron and Rubin, 1942). There are many advantages to the use of algae as carrier of hydrogen production, such as the lack of requirement for occupational farmland and the high efficiency of solar energy conversion (Georgianna and Mayfield, 2012).
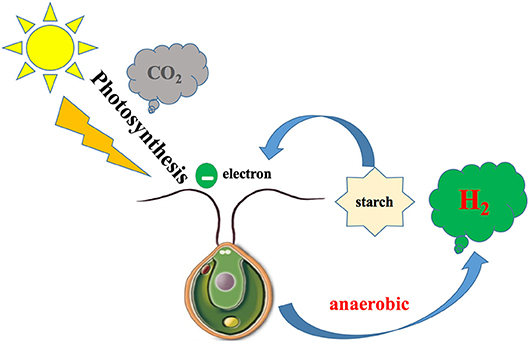
Chlamydomonas reinhardtii, as a species of unicellular green algae, has been chosen as a model species for studying biohydrogen production because of its sequenced genome; ease of cultivation and low cost; especially, high hydrogenase activity, which catalyzes the reaction of hydrogen formation (Das and Veziroglu, 2001, 2008). The hydrogenase of C. reinhardtii could be activated only under anaerobic condition and receive electrons from “photosynthetic electron transport chain” or decomposing intracellular organics, which constitute one-fourth of the photosynthetic electron chain, then reduced to H2 and released out of the cell (Melis et al., 2000; Melis, 2007) (Figure 1). The hydrogenase of C. reinhardtii is sensitive to oxygen, while oxygen is an inevitable product of photosynthesis, thus the hydrogen production of C. reinhardtii in natural state is very low, which is a bottleneck for commercialization of hydrogen production (Roessler and Lein, 1984; Happe and Naber, 1993). To overcome these limitations, some approaches have been used, such as controlling the conditions of cultivation, investigating the structure of hydrogenase to enhance activity, transferring exogenous genes into cells, and screening mutants with high hydrogen yields (Antal et al., 2003; Kruse et al., 2005; Kosourov et al., 2007; Beer et al., 2009; Eroglu and Melis, 2011; Srirangan et al., 2011; Alena et al., 2013; Dubini and Ghirardi, 2015). Low output of hydrogen yield in C. reinhardtii under purely anaerobic conditions results in and restricts the commercialization of hydrogen production (Melis et al., 2000; Melis, 2007). Before 2000, Melis proposed that depletion of sulfur from cultures led to sustainable hydrogen production (Melis et al., 2000), which is a landmark discovery for bio-hydrogen production by C. reinhardtii. The activity of PSII is inhibited by sulfur deficiency, resulting in a decrease in the rate of oxygen evolution of photosynthesis to rates lower than the rate of oxygen uptake by respiration, and the algal medium changes from an aerobic to anaerobic condition, which activates the hydrogenase (Happe and Naber, 1993; Melis et al., 2000; Antal et al., 2003; Melis, 2007). Hydrogenase-received electrons originate from the photosynthetic electron chain or from decomposing intracellular organics, which constitute one-fourth of the photosynthetic electron chain (Figure 2).
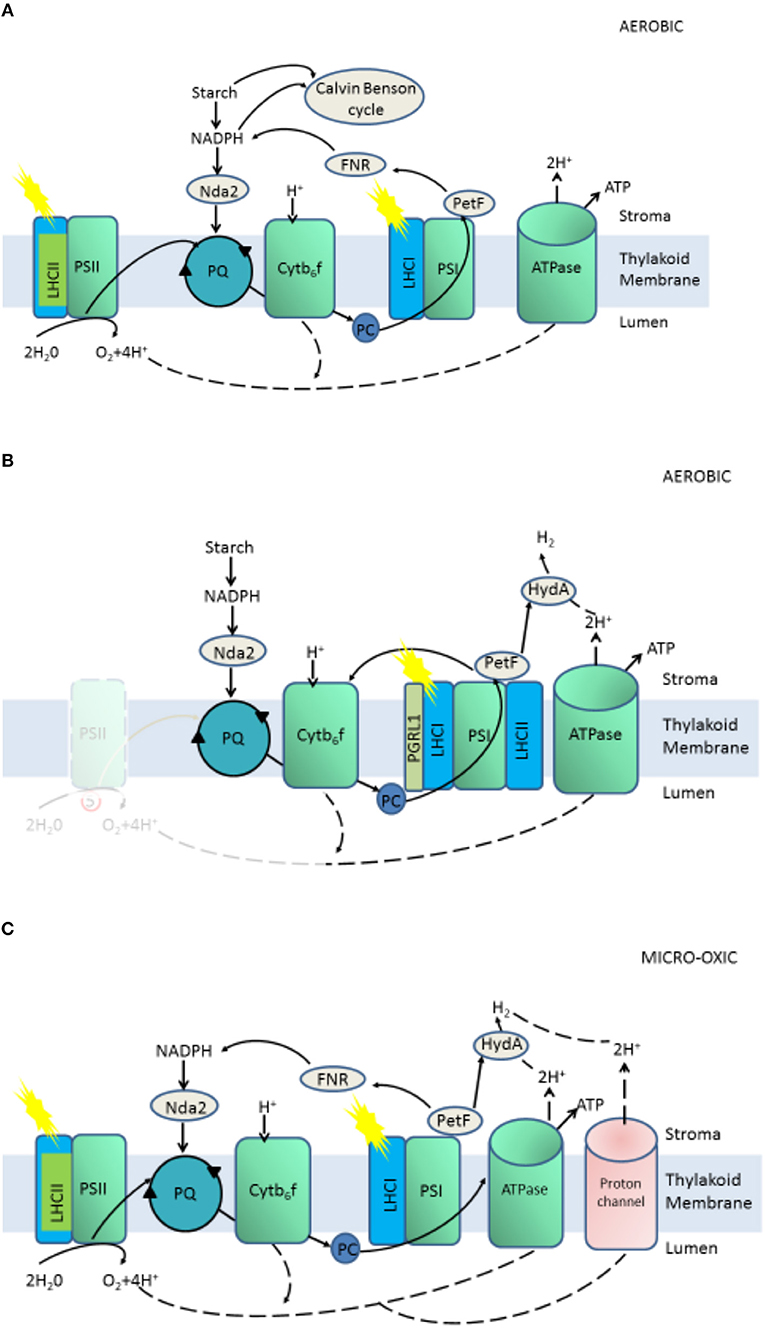
Figure 2. Aerobic (A) and anaerobic (B) stages of two phases H2 production and Micro-oxic continuous H2 production (C). Proton(H+) flow is marked with dashed lines, electron flow with continuous lines.
A “two-stage” protocol of hydrogen production proposed by Melis, resulted to oxygen and the production of hydrogen temporally separate by sulfur deficiency (Melis et al., 2000; Melis, 2007). Subsequently, many reports on the hydrogen production of C. reinhardtii are based on sulfur deficiency.
Since the human genome has been sequenced, “-omics” techniques used for visualizing entire cell processing has clarified biosynthesis and regulatory networks and developed rapidly. This new technology has created an exciting new frontier for high-throughput, predictable engineering of biofuels. Therefore, considerable attention has been paid to omics analyses of model organism C. reinhardtii in different pretreated conditions, which can reveal the molecular and metabolic mechanism comprehensively by three-dimensional analysis (Merchant et al., 2007; Winck et al., 2013; Schmollinger et al., 2014; Smitha et al., 2014; Kleessen et al., 2015; Strenkert et al., 2019). A great number of research regrading omics studies in bio-hydrogen production by C. reinhardtii in sulfur-deprived culture have been done. Investigation into the promising production carrier of C. reinhardtii and its mutants with these powerful techniques has improved predictability and understanding of the unique, complex interactions within model organisms.
In this paper, we summarize these omics approaches, which include genomic, transcriptomic, proteomic and metabolomics approaches, applied to the biohydrogen production of C. reinhardtii in sulfur-deprived culture with the aim of investigating the molecular and mechanism and pathways underlying hydrogen metabolism and regulation at the gene, transcription, protein, and metabolic levels.
Genomic Analysis of Hydrogen Evolution in C. reinhardtii
Hydrogenases Catalyze Hydrogen Evolution in C. reinhardtii
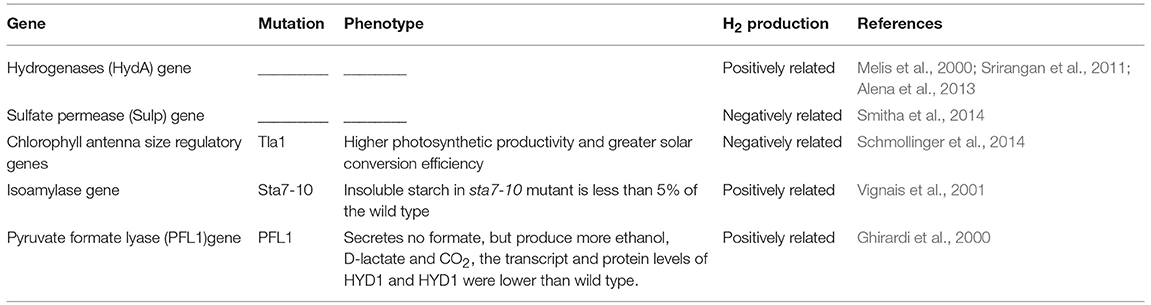
Hydrogen metabolism in C. reinhardtii is a complex biochemical reaction catalyzed by hydrogenases, and these enzymes that not only catalyze but also uptake or production of hydrogen, were confirmed to sensitive to O2 (Bamberger et al., 1982; Happe and Naber, 1993; Happe et al., 1994; Vignais et al., 2001; Forestier et al., 2003; Boichenko et al., 2004). Hydrogenases are classified into three groups: iron [Fe] only, nickel-iron [Ni-Fe], and no metal ion (Vignais et al., 2001; Boichenko et al., 2004). Hydrogenases of C. reinhardtii belong to the [Fe]-hydrogenases group. Isolation and identification of the [Fe]-hydrogenases and their associated genes could improve hydrogen production of C. reinhardtii by the manipulation of these genes and proteins. There are two hydrogenases that belong to the [Fe]-hydrogenase group in C. reinhardtii. The first of these hydrogenases is termed HydA1 and is encoded by the HydA1 gene (Forestier et al., 2003). Similarly, the second hydrogenase of C. reinhardtii is termed HydA2 and is encoded by the HydA2 gene. The protein encoded by HydA1 was 74% similar and 68% identical to HydA2. Both HydA1 and HydA2 contain all motifs and conserved residues which are present in the catalytic cores of the [Fe]-hydrogenase family members. The cDNA of HydA1 and HydA2 have a 158- and 139-nucleotide 5′-UTR, and a 747- and an 873-nucleotide 3′-UTR, respectively (Forestier et al., 2003) (Table 1). The ORFs of HydA1 gene and HydA2 gene encode proteins with 497 and 505 amino acid residues, respectively. HydA1 contains 8 exons, while hydA2 contains 10 exons (Happe and Kaminski, 2002) (Table 1). The analysis of the structure and characteristics of hydrogenase in C. reinhardtii provides a theoretical basis for improving the oxygen tolerance by modifying the genetic information, and it will become a promising research direction of hydrogen production.
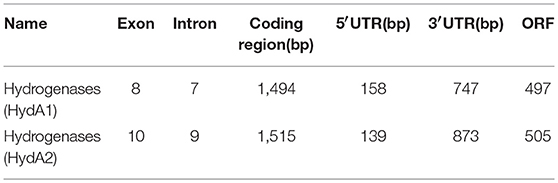
Table 1. Structural characteristics of algal Fe-hydrogenase from C. reinhardtii (Forestier et al., 2003).
Genes Associated With Hydrogen Production in C. reinhardtii
DNA insertion and nuclear transformation to construct mutants is an effective method for screening novel genes associated with hydrogen production, and the gene families regulated of hydrogen production have been described by many researchers (Table 2).
Sulfate Permease Gene
The production of hydrogen in C. reinhardtii could be increased by deficiency of sulfur from cultures and inhibition of the activity of PSII (Melis et al., 2000; Melis, 2007). Therefore, the metabolism of sulfur is closely associated with hydrogen production and it is essential to reveal the sulfur metabolism mechanism and identify genes associated with sulfur metabolism. A novel gene encoded a sulfate permease (Sulp) in C. reinhardtii was first reported by Chen et al. (2003). The Sulp gene of C. reinhardtii is nucleus encoded and consists of five exons and four introns, which is different from other Sulp genes that are encoded by chloroplasts and lack introns. SulP takes an important part in the uptake of sulfur in the chloroplasts of C. reinhardtii. RNAi-generated mutants with low levels of SulP or lacking SulP are expected to be good tools with low rates of H2O oxidation but high H2 production (Melis et al., 2000; Melis, 2007).
Chlorophyll Antenna Size-Regulating Genes
Hydrogenase could accept electrons from the “photosynthetic electron transport chain” and its activity inhibited by the release of oxygen from photosynthesis; therefore, hydrogen metabolism is associated with photosynthesis. Screening photosynthesis regulatory genes is significant and essential for understanding and regulation of hydrogen metabolism in C. reinhardtii. The size of the chlorophyll antenna is important for the function of the antenna in photosynthesis. The Tla1 gene was the first to be identified to regulate the chlorophyll antenna size in photosynthesis and encodes a protein which contains 213 amino acids. The Tla1 gene-deficient mutant, with a truncated light-harvesting antenna and functional chlorophyll antenna sizes of PSI and PSII of ~65 and 50% that of the control (wild type), respectively, being chlorophyll deficient (Polle et al., 2003). Moreover, the Tla1 algal strain showed higher photosynthetic productivity and greater solar conversion efficiencies than the control under most culture conditions (Polle et al., 2003). The Tla1 gene is a promising target to solve the problem of low light utilization efficiency in photosynthesis under biohydrogen production in C. reinhardtii. The Tla1 gene and functionally similar genes will be identified and used in mass cultivation of C. reinhardtii for H2 production and bioenergy accumulation.
Isoamylase Gene
The starch content in C. reinhardtii cells is associated with H2 production, as starch breakdown could produce the endogenous substrate could donate electrons to the photosynthetic electron chain and mitochondrial electron transport chain, while the electrons of the photosynthetic electron transport chain can supply to hydrogen production (Ghirardi et al., 2000; Melisa and Happe, 2001; Zhang et al., 2002; Kosourov et al., 2003). The Sta7 gene was identified in a C. reinhardtii mutant from a library of 6,000 colonies, and the protein encoded by this gene is similar to the isoamylase enzyme found in other species of plant (Flynn et al., 2002). This enzyme takes an important part in starch metabolism (Dauvillée et al., 2001), and the insoluble starch content in the sta7-10 mutant is <5% of the wild-type C. reinhardtii strain.
The Sta7 gene may be an important target in research on H2 metabolism. It has been shown that hydrogen can be produced continuously as long as the cell contains starch under conditions of sulfur deficiency (Zhang et al., 2002). Consequently, it is hypothesized that the more starch content in the cell, the higher the hydrogen yield of C. reinhardtii could be increased accumulation of starch in cells might be achieved by overexpression of the Sta7 gene of C. reinhardtii; thus, increased starch accumulation in C. reinhardtii cells could be achieved by normal photosynthesis.
Pyruvate Formate Lyase Gene
The procedure of hydrogen metabolism in C. reinhardtii is a multiphase process that is not only associated with photosynthesis but also closely associated with dark fermentation (Bamberger et al., 1982). The first step of the dark fermentation reaction is activated by pyruvate lyase (PFL1), which catalyzes pyruvate to form formate and acetyl CoA in this process (Gfeller and Gibbs, 1984; Kreuzberg, 1984; Mus et al., 2007; Hemschemeier et al., 2008). A special C. reinhardtii mutant strain 48F5 was screened from library of 5000 colonies. The mutant strain 48F5, in which the affected gene was PFL1, does not secrete formate but produces more CO2, ethanol and D-lactate than the control, meanwhile the transcription and protein expression levels of HYD1 and HYD1 in mutant strain were lower than those of the control (Philipps et al., 2011). Consequently, the PFL1 gene may take an important part in H2 production related metabolism in C. reinhardtii. Additionally, it is hypothesized that increased hydrogen yields might be achieved by overexpression of the PFL1 gene of C. reinhardtii.
Transcriptomic Analysis of Hydrogen Production in C. reinhardtii
Gene Microarray and RNA-seq
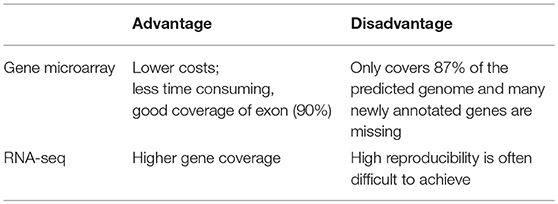
Both gene microarrays and RNA-seq are powerful tools for studying transcription in eucaryons (Table 3). Compared to RNA-seq, gene microarrays are associated with low costs, good coverage of exon-based transcript levels (90%) and low time consumption (Bradford et al., 2010). However, the current microarray platform covers only 87% of the existing genome, and many newly annotated genes have not been identified (Eberhard et al., 2006; Voss et al., 2011). There are expected to be up to 17,000 transcript models in C. reinhardtii based on the known genome information (Jain et al., 2007; Prochnik et al., 2010). Therefore, to achieve the same high transcript coverage as RNA-seq, deep sequencing is required. RNA-seq has greater sensitivity for differential gene expression and higher gene coverage than gene microarray (Fu et al., 2009; Wilhelm and Landry, 2009; Feng et al., 2010; Nagalakshmi et al., 2010; Tang et al., 2010; van Vliet, 2010). However, the results and repeatability of RNA-seq are difficult to detect, and good reproducibility is ususlly difficult to obtain. A typical result is that the use of RNA-seq often overestimates the abundance and length-dependent amplification of highly expressed genes (Marioni et al., 2008; Bradford et al., 2010; Liu et al., 2011). These internal challenges associated with data normalization and data analysis clearly should be addressed.
Some researchers have used gene microarray and RNA-seq approach to analyze the expression levels of genes in C. reinhardtii with sulfur-deprived H2 production conditions. Transcriptional analysis of C. reinhardtii by the microarray approach was first reported by Nguyen et al. Finally, 166 markedly differentially expressed genes were identified, and these genes were divided into different functional groups by genome comparison: 22 genes related to in photosynthesis, 8 genes related to sulfur metabolism, 4 genes related to carbon metabolism, 5 genes related to proteolysis, 4 genes related to amino acid synthesis, 10 genes were related to transcription and translation, 3 genes related to redox cycling, 27 genes were involved in other processes and pathways, functions of 83 genes were unknown (Nguyen et al., 2008). This study provides a novel insight for the expression and regulation of genes involved in sulfur metabolism, photosynthesis and carbon metabolism in the process of sulfur-deficient hydrogen production.
Toepel et al. analyzed the transcript levels of C. reinhardtii in nitrogen- and sulfate-limited cultures by the new microarray method, and 813 downregulated genes and 100 upregulated genes were identified under sulfur-deprived conditions (Toepel et al., 2011). The new microarray data were highly similar to those for many genes in terms of changes in differential gene expression patterns between the first-generation conditions and sulfur-deprived conditions (Toepel et al., 2013). The author of this paper stated that microarrays are inexpensive and reliable tools for detecting the expression levels of changes in transcription, although RNA-seq analysis provide detailed information regarding the transcriptome, have low costs, and exhibit good reproducibility; moreover, this work provides a new microarray platform for transcriptome analysis of C. reinhardtii and the established analysis systems can be used with microarrays for routine applications.
RNA-seq technology was used to detect the response of hydrogen production of C. reinhardtii cells and mutant cells by Authur's group under sulfur-deficient conditions. Sulfur- deficient C. reinhardtii cells gathering transcripts involved in the synthesis of sulfur-containing metabolites, sulfur acquisition, sulfur assimilation, sulfur recycling, and Cys degradation (González-Ballester et al., 2010). Furthermore, changes in cellular structures could occur during the process of sulfur deprivation, including the structure of photosynthetic complexes and the cell wall (González-Ballester et al., 2010). Additionally, this research shows that the protein accumulated by the cells under sulfur deficiency conditions has less sulfur content (González-Ballester et al., 2010).
Since most of the sulfur-deficient reactions are controlled by the SNRK2.1 protein kinase, the mutant exhibits a number of reactions different from those of wild-type cells under conditions of sulfur-deficient and hydrogen production. The mutant strain could not adapt to the increased oxygen content and oxidative stress caused by sulfur deficiency, which led to cell death (González-Ballester et al., 2010). The results of transcriptome analysis showed that the mutants and wild algae have significant changes in the physiology and metabolism under the condition of sulfur deficiency and hydrogen production. This change is important for maintaining the survival of cells under sulfur deficiency stress.
MicroRNA (miRNA)-seq
MicroRNA (miRNAs) regulates many important metabolic processes in eukaryotes Hu's group has identified expression level of changes in miRNA through a deep sequencing platform and extensively profile in the process of H2 production and sulfur-deficient culture (Shu and Hu, 2012). The results indicated that sulfur-deprived conditions may have an appreciable impact on miRNA expression patterns. In this study, the expression levels of 47 miRNAs were significantly different under sulfur-deprived conditions; meanwhile, 310 miRNAs, including 225 novel miRNAs and 85 known miRNAs, were predicted and analyzed. In particular, 13 miRNAs were closely associated with the response to sulfur-deprived conditions; moreover, based on the published information of the transcriptome, target gene associated with metabolic response to the deficiency of sulfur stress were identified in C. reinhardtii (Shu and Hu, 2012). The differential interactions and expression of miRNAs and their potential targets could reveal the molecular mechanism of hydrogen metabolism response to sulfur deprivation in C. reinhardtii.
Genome-wide Long Non-coding RNA (IncRNA) Screening and Characterization
A considerable portion of the genome of eukaryotes can be transcribed to RNAs, but will not be translated to proteins. These non-coding RNAs (ncRNAs) consist of housekeeping, regulatory and functional unknown ncRNAs. Regulatory ncRNAs are usually classified as small non-coding RNAs and long non-coding RNAs (lncRNAs) according to their lengths (Shu and Hu, 2010; Pauli et al., 2011). lncRNAs play important roles in cell differentiation and development (Guttman et al., 2011; Pauli et al., 2011; Fatica and Bozzoni, 2014) silencing gene expression in X-chromosome (Dimond and Fraser, 2013), neurological diseases occurrence (Ponting et al., 2009), cancer progression (Wapinski and Chang, 2011; Yang et al., 2014) and immune response genes mediation (Carpenter et al., 2013; Wang et al., 2016). In plants, lncRNAs express differentially in various organs and under different treatment conditions, which indicates lncRNAs can modulate gene activity during development and in response to external stimuli (Kim and Sung, 2012). Based on the regulatory functions found in other higher plants, lncRNA investigation and manipulation may provide new insights and solutions for green algae. Transcriptome and proteome analyses indicated that sulfur deprivation affects massive pathways including sulfur metabolism, cell wall structure, photosystems, protein biosynthetic apparatus, molecular chaperones and 20 S proteasomal components (Nguyen et al., 2008; Chen et al., 2010; González-Ballester et al., 2010). It was previously demonstrated that hydrogen production can be regulated by an artificial non-coding RNA miRNA (amiRNA) targeting OEE2 encoded gene (a photosystem II related protein, oxygen evolving enhancer) (Li et al., 2015). RNA sequencing in C. reinhardtii under hydrogen production and sulfur deficient condition has been done and obtained totally 3,574 putative lncRNAs. 1440 were considered as high-confidence lncRNAs, including 936 large intergenic, 310 intronic and 194 anti-sense lncRNAs. The average transcript length, ORF length and numbers of exons for lncRNAs are much less than for genes in this green algae (Li et al., 2016). In addition, 367 lncRNAs responsive to sulfur deprivation was identified, including 36 photosynthesis-related lncRNAs (Li et al., 2016). lncRNAs used to reveal the molecular and metabolism mechanisms in C. reinhardtii are very rare, nevertheless the lncRNA data could provide new insights into C. reinhardtii hydrogen production under sulfur deprivation.
Proteomic Analysis of H2 Photoproduction in C. reinhardtii
Since the completion of C. reinhardtii genome sequencing and annotation (Merchant et al., 2007), the research of C. reinhardtii has opened up a new era. DNA-microarray analysis and transcriptome analysis indicated that more than 100 genes were up- or down-regulated throughout the process of sulfur-depleted H2 photoproduction (Nguyen et al., 2008). These findings had revealed the mechanism of H2 metabolism in C. reinhardtii at the genetic level. Meanwhile proteomic analysis has been regarded as a powerful tool to study global translational profiles for biological processes. At the protein level, however, experimental data about H2 metabolism in C. reinhardtii are still limited. Comparative proteomics to assess the expression level of proteomic changes in C. reinhardtii under sulfur-depleted H2 released conditions has been achieved (Chen et al., 2010). Total 159 different protein spots were identified, 105 were found enhanced or reduced significantly, corresponding to 82 unique genes, throughout H2 production under sulfur deprivation. Meanwhile the photosynthetic machinery, molecular chaperones and protein biosynthetic apparatus were changed significantly in the process. Additionally, many proteins associated with anti-oxidative reactions and sulfate, acetate, and nitrogen metabolism also changed markedly. Furthermore, other proteins involved in cell wall and flagellum metabolism showed changes under sulfur-depleted H2 released conditions. These data provide not only detailed information regarding the complex interactions between photosynthesis and hydrogen metabolism in switching the organism from O2-Generating to H2 production, but also more candidate genes for targeted genetic engineering of C. reinhardtii that would lead to further elucidation of the mechanisms of H2 production and its large-scale utilization.
Metabolomics Analysis of H2 Photoproduction in C. reinhardtii
The understanding of the metabolic pathways essential to the hydrogen production metabolism is fundamental to identify metabolites and proteins which may relate to the cell acclimation to environmental changes. Based on the genome sequence it is possible to develop models of the metabolic network for some organisms and to predict their behavior under defined conditions. Metabolic network models may serve as a basis for in hydrogen metabolic engineering, and their complementation with experimental data will certainly improve the prediction capacity of the available models (Pauli et al., 2011). In order to elucidate the role of the metabolic pathways and metabolites into the biological systems, theoretical and experimental approaches have been performed. Single metabolite analysis has not been reported, metabolomics analysis of H2 photoproduction in C. reinhardtii were combined with transcriptomic and proteomics analysis (May et al., 2008; Subramanian et al., 2014).
Application of Multiomics Technique in H2 Photoproduction by C. reinhardtii
The unique complexity and adaptability of C. reinhardtii to maintain its cellular activity under H2 production and sulfur deficiency conditions is well-known. H2 production metabolism, fermentative pathways and photosynthesis become active during the process (May et al., 2008; Subramanian et al., 2014). A combination of multiple omics techniques, transcriptomics, proteomics and metabolomics analyses, is a useful tool to study H2 metabolism in C. reinhardtii under sulfur-depleted H2 released conditions. Up-regulated proteins and down-regulated proteins were detected by transcriptomics and proteomics analysis and those proteins belong to flagellum proteins, light-harvesting complex proteins, glyoxylate cycle proteins, nitrogen reorganization proteins, putative redox proteins and ATP synthase proteins by functional annotation (Subramanian et al., 2014). Additionally, combined with the analysis of metabolites by GC-MS, strong evidence showed that the glyoxylate pathway could reverse TCA reactions, which helps in conserving carbon within the cell, while simultaneously reoxidizing NADH. Meanwhile, the presence of the serine-isocitrate lyase pathway has been reported to be active in this process. Finally, C. reinhardtii appears to cope with the reduced cellular energy levels under sulfur-depleted H2 released conditions, by relying on glycolysis and fermentation in order to generate more ATP and regenerate NAD+, respectively, for continuity of catabolic processes. Future work would require measurement of energy allocation to different metabolite pathways within the cell over longer periods of sulfur-depleted H2 production, and understanding the alteration of the metabolism in sulfur-depleted H2 production conditions.
Conclusions and Prospects
H2 production by C. reinhardtii may provide an effective method to solve the energy crisis and is a complex physiological process involved in photosynthesis, respiration, and fermentation. Many researchers have studied this physiological process. Currently, omics technologies (genomics, transcriptomics, proteomics, and metabolomics) have applications in the bioproduction of H2 in C. reinhardtii and could be used to comprehensively investigate the underlying mechanisms, leading to the utilization of this process on an industrial scale.
Author Contributions
LX and QW conceived and designed the paper. LX and JF wrote and revised the paper.
Funding
This work was supported by the National Natural Science Foundation of China 31600284, Shanghai Engineering Research Center of Plant Germplasm Resources 17DZ2252700, Shanghai Pujiang Program (18PJD008), Open Funding Project of the State Key Laboratory of Bioreactor Engineering 2018021, and Open Funding Project of the State Key Laboratory of Marine Resource Utilization in South China Sea (Hainan University) 2019002.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alena, V., Stenbjörn, S., and Fikret, M. (2013). Increased photosystem II stability promotes H2 production in sulfur-deprived Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 110, 7223–7228. doi: 10.1073/pnas.1220645110
Antal, T. K., Krendeleva, T. E., Laurinavichene, T. V., Makarova, V. V., Ghirardi, M. L., Rubin, A. B., et al. (2003). The dependence of algal H2 production on photosystem II and O2-consumption activity in sulfur-deprived Chlamydomonas reinhardtii cells. Biochim. Biophys. Acta. 1607, 153–160. doi: 10.1016/j.bbabio.2003.09.008
Bamberger, E. S., King, D., Erbes, D. L., and Gibbs, M. (1982). H2 and CO2 evolution by anaerobically adapted Chlamydomonas reinhardtii F-60. Plant Physiol. 69, 1268–1273. doi: 10.1104/pp.69.6.1268
Beer, L. L., Boyd, E. S., Peters, J. W., and Posewitz, M. C. (2009). Engineering algae for biohydrogen and biofuel production. Curr. Opin. Biotechnol. 20, 264–271. doi: 10.1016/j.copbio.2009.06.002
Boichenko, V. A., Greenbaum, E., and Seibert, M. (2004). Hydrogen production by photosynthetic microorganisms. Mol. Glob. Photosynt. 2, 397–451. doi: 10.1142/9781860945496_0008
Bradford, J. R., Hey, Y., Yates, T., Li, Y., Pepper, S. D., and Miller, C. J. (2010). A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics 11:282. doi: 10.1186/1471-2164-11-282
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792. doi: 10.1126/science.1240925
Chen, H. C., Yokthongwattana, K., Newton, A. J., and Melis, A. (2003). SulP, a nuclear gene encoding a putative chloroplast-targeted sulfate permease in Chlamydomonas reinhardtii. Planta 218, 98–106. doi: 10.1007/s00425-003-1076-6
Chen, M., Zhao, L., Sun, Y. L., Cui, S. X., Zhang, L. F., Yang, B., et al. (2010). Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J. Proteome Res. 9, 3854–3866. doi: 10.1021/pr100076c
Das, D., and Veziroglu, T. N. (2001). Hydrogen production by biological processes: a survey of literature. Int. J. Hydrogen Energy. 26, 13–28. doi: 10.1016/S0360-3199(00)00058-6
Das, D., and Veziroglu, T. N. (2008). Advances in biological hydrogen production processes. Int. J. Hydrogen Energy. 33, 6046–6057. doi: 10.1016/j.ijhydene.2008.07.098
Dauvillée, D., Colleoni, C., Mouille, G., Buléon, A., Gallant, D. J., Bouchet, B., et al. (2001). Two loci control phytoglycogen produiction in the monocellular green alga Chlamydomonas reinhardtii. Plant Physiol. 125, 1710–1722. doi: 10.1104/pp.125.4.1710
Dimond, A., and Fraser, P. (2013). Long noncoding RNAs X- in three dimensions. Science 341, 720–721. doi: 10.1126/science.1243257
Dubini, A., and Ghirardi, M. L. (2015). Engineering photosynthetic organisms for the production of biohydrogen. Photosyn. Res. 123, 241–253. doi: 10.1007/s11120-014-9991-x
Eberhard, S., Jain, M., Im, C. S., Pollock, S., Shrager, J., Lin, Y., et al. (2006). Generation of an oligonucleotide array for analysis of gene expression in Chlamydomonas reinhardtii. Curr Genet. 49, 106–124. doi: 10.1007/s00294-005-0041-2
Eroglu, E., and Melis, A. (2011). Photobiological hydrogen production: recent advances and state of the art. Bioresour. Technol. 102, 8403–8413. doi: 10.1016/j.biortech.2011.03.026
Fakhimi, N., Dubini, A., Tavakoli, O., and González-Ballester, D. (2019). Acetic acid is key for synergetic hydrogen production in Chlamydomonas-bacteria co-cultures. Bioresour. Technol. 289:121648. doi: 10.1016/j.biortech.2019.121648
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Feng, L., Liu, H., Liu, Y., Lu, Z., Guo, G., Guo, S., et al. (2010). Power of deep sequencing and agilent microarray for gene expression profiling study. Mol. Biotechnol. 45, 101–110. doi: 10.1007/s12033-010-9249-6
Flynn, T., Ghirardi, M., and Seibert, M. (2002). Accumulation of O2-tolerant phenotypes in H2-producing strains of Chlamydomonas reinhardtii by sequential applications of chemical mutagenesis and selection. Int. J. Hydrogen Energy 27, 1421–1430. doi: 10.1016/S0360-3199(02)00117-9
Forestier, M., King, P., Zhang, L. P., Posewitz, M., Schwarzer, S., Happe, T., et al. (2003). Expression of two [Fe]-hydrogenase in Chlamydomonas reinhardtii under anaerobic conditions. Eur. J. Biochem. 270, 2750–2758. doi: 10.1046/j.1432-1033.2003.03656
Fu, X., Fu, N., Guo, S., Yan, Z., Xu, Y., Hu, H., et al. (2009). Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genom. 10:161. doi: 10.1186/1471-2164-10-161
Gaffron, H. (1939). Reduction of CO2 with H2 in green plants. Nature 143, 204–205. doi: 10.1038/143204a0
Gaffron, H., and Rubin, J. (1942). Fermentative and photochemical production of hydrogen in algae. J. Gen. Physiol. 26, 219–240. doi: 10.1085/jgp.26.2.219
Georgianna, D. R., and Mayfield, S. P. (2012). Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 488, 329–335. doi: 10.1038/nature11479
Gfeller, R. P., and Gibbs, M. (1984). Fermentative metabolism of Chlamydomonas reinhardtii. I. Analysis of fermentative products from starch in dark and light. Plant Physiol. 75, 212–218. doi: 10.1104/pp.75.1.212
Ghirardi, M. L., Zhang, L., Lee, J. W., Flynn, T., Seibert, M., Greenbaum, E., et al. (2000). Microalgae: a green source of renewable H2. Trends Biotechnol. 18, 506–511. doi: 10.1016/S0167-7799(00)01511-0
González-Ballester, D., Casero, D., Cokus, S., Pellegrini, M., Merchant, S. S., and Grossman, A. R. (2010). RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22, 2058–2084. doi: 10.1105/tpc.109.071167
Guttman, M., Donaghey, J., Carey, B. W., Garber, M., Grenier, J. K., Munson, G., et al. (2011). LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300. doi: 10.1038/nature10398
Happe, T., and Kaminski, A. (2002). Differential regulation of the [Fe]-hydrogenase during anaerobic adaptation in the gree alga Chlamydomonas reinhardtii. Eur. J. Biochem. 269, 1022–1032. doi: 10.1046/j.0014-2956.2001.02743.x
Happe, T., Mosler, B., and Naber, J. D. (1994). Induction, localization and metal content of hydrogenase in the green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 222, 769–774. doi: 10.1111/j.1432-1033.1994.tb18923.x
Happe, T., and Naber, J.D. (1993). Isolation, characterization and N-terminal amino acid sequence of hydrogenase from green alga Chlamydomonas reinhardtii. Eur. J. Biochem. 214, 475–481. doi: 10.1111/j.1432-1033.1993.tb17944.x
Hemschemeier, A., Jacobs, J., and Happe, T. (2008). Biochemical and physiological characterization of the pyruvate formate-lyase Pfl1 of Chlamydomonas reinhardtii, a typically bacterial enzyme in a eukaryotic alga. Eukaryot. Cell 7, 518–526. doi: 10.1128/EC.00368-07
Hwang, J. H., Kim, H. C., Choi, J. A., Abou-Shanab, R. A., Dempsey, B. A., Regan, J. M., et al. (2014). Photoautotrophic hydrogen production by eukaryotic microalgae under aerobic conditions. Nat. Commun. 5:3234. doi: 10.1038/ncomms4234
Jain, M., Shrager, J., Harris, E. H., Halbrook, R., Grossman, A. R., Hauser, C., et al. (2007). EST assembly supported by a draft genome sequence: an analysis of the Chlamydomonas reinhardtii transcriptome. Nucleic Acids Res. 35, 2074–2083. doi: 10.1093/nar/gkm081
Kim, E. D., and Sung, S. (2012). Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 17, 16–21. doi: 10.1016/j.tplants.2011.10.008
Kleessen, S., Irgang, S., Klie, S., Giavalisco, P., and Nikoloski, Z. (2015). Integration of transcriptomics and metabolomics data specifies the metabolic response of Chlamydomonas to rapamycin treatment. Plant J. 81, 822–835. doi: 10.1111/tpj.12763
Kosourov, S., Patrusheva, E., Ghirardi, M. L., Seibert, M., and Tsygankov, A. A. (2007). Comparison of hydrogen photoproduction by sulfur-deprived Chlamydomonas reinhardtii under different growth conditions. J. Biotech. 128, 776–787. doi: 10.1016/j.jbiotec.2006.12.025
Kosourov, S., Seibert, M., and Ghirardi, M. L. (2003). Effects of extracellular pH on the metabolic pathways in sulfurdeprived, H2-producing Chlamydomonas reinhardtii cultures. Plant Cell Physiol. 44, 146–155. doi: 10.1093/pcp/pcg020
Kreuzberg, K. (1984). Starch fermentation via a formate producing pathway in Chlamydomonas reinhardtii, Chlorogonium elongatum and Chlorella fusca. Physiol. Plant. 61, 87–94. doi: 10.1111/j.1399-3054.1984.tb06105.x
Kruse, O., Rupprecht, J., Bader, K. P., Thomas-Hall, S., Schenk, P. M., Finazzi, G., et al. (2005). Improved Photobiological H2 Production in engineered green algal cells. J. Biol. Chem. 280, 34170–34177. doi: 10.1074/jbc.M503840200
Li, H., Wang, Y., Chen, M., Xiao, P., Hu, C., Zeng, Z., et al. (2016). Genome-wide long non-coding RNA screening, identification and characterization in a model microorganism Chlamydomonas reinhardtii. Sci. Rep. 6:34109. doi: 10.1038/srep34109
Li, H., Zhang, L., Shu, L., Zhuang, X., Liu, L., Chen, J., et al. (2015). Sustainable photosynthetic H2-production mediated by artificial miRNA silencing of OEE2 gene in green alga Chlamydomonas reinhardtii. Int. J. Hydrogen Energy 40, 5609–5616. doi: 10.1016/j.ijhydene.2015.02.073
Liu, S., Lin, L., Jiang, P., Wang, D., and Xing, Y. (2011). A comparison of RNA-seq and high density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 39, 578–588. doi: 10.1093/nar/gkq817
Marioni, J. C., Mason, C. E., Mane, S. M., Stephens, M., and Gilad, Y. (2008). RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 18, 1509–1517. doi: 10.1101/gr.079558.108
May, P., Wienkoop, S., Kempa, S., Usadel, B., Christian, N., Rupprecht, J., et al. (2008). Metabolomics and proteomics assisted genome annotation and analysis of the draft metabolic network of Chlamydomonas reinhardtii. Genetics 179, 157–166. doi: 10.1534/genetics.108.088336
Melis, A. (2007). Photosynthetic H2 metabolism in Chlamydomonas reinahrdtii. Planta. 226, 1075–1086. doi: 10.1007/s00425-007-0609-9
Melis, A., Zhang, L., Forestier, M., Ghirardi, M. L., and Seibert, M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122, 127–136. doi: 10.1104/pp.122.1.127
Melisa, A., and Happe, T. (2001). Hydrogen production: green algae as a source of energy. Plant Physiol. 127, 740–748. doi: 10.1104/pp.010498
Merchant, S. S., Prochnik, S. E., Vallon, O., Harris, E. H., Karpowicz, S. J., and Witman, G. B. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–251. doi: 10.1126/science.1143609
Mus, F., Dubini, A., Seibert, M., Posewitz, M. C., and Grossman, A. R. (2007). Anaerobic acclimation in Chlamydomonas reinhardtii: anoxic gene expression, hydrogenase induction and metabolic pathways. J. Biol. Chem. 282, 25475–25486. doi: 10.1074/jbc.M701415200
Nagalakshmi, U., Waern, K., and Snyder, M. (2010). RNA-Seq: a method for comprehensive transcriptome analysis. Curr. Protoc. Mol. Biol. 11, 11–13. doi: 10.1002/0471142727.mb0411s89
Nguyen, A. V., Thomas-Hall, S. R., Malnoë, A., Timmins, M., Mussgnug, J. H., Rupprecht, J., et al. (2008). Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryotic Cell 7, 1965–1979. doi: 10.1128/EC.00418-07
Pauli, A., Rinn, J. L., and Schier, A. F. (2011). Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149. doi: 10.1038/nrg2904
Philipps, G., Krawietz, D., Hemschemeier, A., and Happe, T. (2011). A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J. 66, 330–340. doi: 10.1111/j.1365-313X.2011.04494.x
Polle, J. E., Kanakagiri, S., and Melis, A. (2003). tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size. Planta 217, 49–59. doi: 10.1007/s00425-002-0968-1
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Prochnik, S. E., Umen, J., Nedelcu, A. M., Hallmann, A., Miller, S. M., Nishii, I., et al. (2010). Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223–226. doi: 10.1126/science.1188800
Ramadass, K., Sathish, C. I., Johns, A., Ruban, S. J., Singh, G., Lakhi, K. S., et al. (2019). Characterization and hydrogen storage performance of halloysite nanotubes. J. Nanosci. Nanotechnol. 19, 7892–7898. doi: 10.1166/jnn.2019.16751
Roessler, P. G., and Lein, S. (1984). Purification of hydrogenase from Chlamydomonas reinhardtii. Plant Physiol. 75, 705–709. doi: 10.1104/pp.75.3.705
Schmollinger, S., Mühlhaus, T., Boyle, N. R., Blaby, I. K., Casero, D., Mettler, T., et al. (2014). Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plan Cell 26, 1410–1435. doi: 10.1105/tpc.113.122523
Shu, L., and Hu, Z. (2012). Characterization and differential expression of microRNAs elicited by sulfur deprivation in Chlamydomonas reinhardtii. BMC Genom. 13:108. doi: 10.1186/1471-2164-13-108
Shu, L. F., and Hu, Z. L. (2010). Small silencing RNAs in Chlamydomonas reinhardtii. Minerva Biotec. 22, 29–37. Available online at: https://www.researchgate.net/publication/286305180
Smitha, P., Renata, B., Holger, N. S., Marc, J.-F., Laura, S., and Schirmer, K. (2014). Linking toxicity and adaptive responses across the transcriptome, proteome, and phenotype of Chlamydomonas reinhardtii exposed to silver. Proc. Natl. Acad. Sci. U.S.A. 111, 3490–3495. doi: 10.1073/pnas.1319388111
Srirangan, K., Pyne, M. E., and Perry, C. (2011). Biochemical and genetic engineering strategies to enhance hydrogen production in photosynthetic algae and cyanobacteria. Bioresour. Technol. 102, 8589–8604. doi: 10.1016/j.biortech.2011.03.087
Strenkert, D., Schmollinger, S., Gallaher, S. D., Salomé, P. A., Purvine, S. P., Nicora, C. D., et al. (2019). Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 116, 2374–2383. doi: 10.1073/pnas.1815238116
Subramanian, V., Dubini, A., Astling, D. P., Laurens, L. M., Old, W. M., Grossman, A. R., et al. (2014). Profiling Chlamydomonas metabolism under dark, anoxic H2-producing conditions using a combined proteomic, transcriptomic, and metabolomic approach. J. Proteome Res. 13, 5431–5451. doi: 10.1021/pr500342j
Tang, F., Barbacioru, C., Nordman, E., Li, B., Xu, N., Bashkirov, V. I., et al. (2010). RNA-seq analysis to capture the transcriptome landscape of a single cell. Nat. Protoc. 5, 516–535. doi: 10.1038/nprot.2009.236
Toepel, J., Albaum, S. P., Arvidsson, S., Goesmann, A., la Russa, M., Rogge, K., et al. (2011). Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics 12:579. doi: 10.1186/1471-2164-12-579
Toepel, J., Illmer-Kephalides, M., Jaenicke, S., Straube, J., May, P., Goesmann, A., et al. (2013). New insights into Chlamydomonas reinhardtii hydrogen production processes by combinedmicroarray/RNA-seq transcriptomics. Plant Biotechnol. J. 11, 1–17. doi: 10.1111/pbi.12062
van Vliet, A. H. (2010). Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol. Lett. 302, 1–7. doi: 10.1111/j.1574-6968.2009.01767.x
Vignais, P. M., Billoud, B., and Meyer, J. (2001). Classification and phylogeny of hydrogenases. FEMS Micro Rev. 25 455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x
Voss, B., Meinecke, L., Kurz, T., Al-Babili, S., and Beck, C. F. (2011). Hess WR, Hemin and magnesium-protoporphyrin IX induce global changes in gene expression in Chlamydomonas reinhardtii. Plant Physiol. 155, 892–905. doi: 10.1104/pp.110.158683
Wang, Y., Xue, S., Liu, X., Liu, H., Hu, T., Qiu, X., et al. (2016). Analyses of long non-coding RNA and mRNA profiling using RNA sequencing during the pre-implantation phases in pig endometrium. Sci. Rep. 6:20238. doi: 10.1038/srep20238
Wapinski, O., and Chang, H. Y. (2011). Long noncoding RNAs and human disease. Trends Cell Biol. 21, 354–361. doi: 10.1016/j.tcb.2011.04.001
Wilhelm, B. T., and Landry, J. R. (2009). RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48, 249–257. doi: 10.1016/j.ymeth.2009.03.016
Winck, F. V., Páez Melo, D. O., and González Barrios, A. F. (2013). Carbon acquisition and accumulation in microalgae Chlamydomonas: Insights from “omics” approaches. J. Proteomics. 94, 207–218. doi: 10.1016/j.jprot.2013.09.016
Yang, G., Lu, X., and Yuan, L. (2014). LncRNA: a link between RNA and cancer. Biochim Biophys Acta 1839, 1097–1109. doi: 10.1016/j.bbagrm.2014.08.012
Keywords: Chlamydomonas reinhardtii, hydrogen production, genomics, transcriptomics, proteomics, metabolomics
Citation: Xu L, Fan J and Wang Q (2019) Omics Application of Bio-Hydrogen Production Through Green Alga Chlamydomonas reinhardtii. Front. Bioeng. Biotechnol. 7:201. doi: 10.3389/fbioe.2019.00201
Received: 02 July 2019; Accepted: 06 August 2019;
Published: 21 August 2019.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2019 Xu, Fan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhua Fan, amhmYW5AZWN1c3QuZWR1LmNu; Quanxi Wang, d2FuZ3F4QHNobnUuZWR1LmNu
 Lili Xu
Lili Xu Jianhua Fan
Jianhua Fan Quanxi Wang1*
Quanxi Wang1*