
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Big Data , 14 February 2025
Sec. Data-driven Climate Sciences
Volume 8 - 2025 | https://doi.org/10.3389/fdata.2025.1507036
Accurate identification of pollen grains from Abies (fir), Picea (spruce), and Pinus (pine) is an important method for reconstructing historical environments, past landscapes and understanding human-environment interactions. However, distinguishing between pollen grains of conifer genera poses challenges in palynology due to their morphological similarities. To address this identification challenge, this study leverages advanced deep learning techniques, specifically transfer learning models, which are effective in identifying similarities among detailed features. We evaluated nine different transfer learning architectures: DenseNet201, EfficientNetV2S, InceptionV3, MobileNetV2, ResNet101, ResNet50, VGG16, VGG19, and Xception. Each model was trained and validated on a dataset of images of pollen grains collected from museum specimens, mounted and imaged for training purposes. The models were assessed on various performance metrics, including accuracy, precision, recall, and F1-score across training, validation, and testing phases. Our results indicate that ResNet101 relatively outperformed other models, achieving a test accuracy of 99%, with equally high precision, recall, and F1-score. This study underscores the efficacy of transfer learning to produce models that can aid in identifications of difficult species. These models may aid conifer species classification and enhance pollen grain analysis, critical for ecological research and monitoring environmental changes.
The scientific study of pollen is a key step in studies of historical and contemporary environmental analysis. Researchers use pollen data to reconstruct past vegetation patterns and understand changes in landscapes in paleoecology and climate change studies (Willard and Bernhardt, 2011; Shennan, 2015; Balmaki et al., 2019, 2024). By examining pollen grains preserved in sediments or peat bogs, paleoecologists can identify the types of vegetation that existed in a particular area at different times in the past. This information is used to help reconstruct historical climate conditions, as the distribution of plants is closely linked to specific climate parameters such as temperature and rainfall. Through this method, scientists can trace the ecological impacts of climate fluctuations over centuries, providing insights into how ecosystems responded to changes in the environment and aiding predictions of future ecological responses to climate change (Balmaki et al., 2019). Moreover, pollen grains can help to identify the interactions between human activities and environmental factors that significantly shape these landscape patterns (Sadori et al., 2010; Kobe et al., 2020).
Coniferous genera, in particular, are representative of specific ecological and climatic adaptations making them important to help map past landscapes and climate conditions. For example, fir trees (Abies) are sensitive to moisture changes, spruce trees (Picea) are adapted to cold environments, and pine trees (Pinus) are known to be resilient to various environmental stresses including fire. Collectively, data on these taxa provide a comprehensive understanding of historical humidity, precipitation patterns, climatic fluctuations, and fire regimes (Latałowa and van der Knaap, 2006; Balmaki and Wigand, 2019; Larson et al., 2020). Beyond its significance in reconstructive studies, pollen analysis, drawing from these conifers, is also critical to health and allergy research, helping to identify allergenic species and predict pollen-related health issues, thus playing a key role in public health management and allergen treatment (Gastaminza et al., 2009; Frisk et al., 2024). Traditional palynology, the study of pollen grains and spores, depends on morphological characters of pollen grains to identify taxa. Typical traits include shape, polarity, symmetry, apertures, size, and ornamentation. However, the subtle morphological differences between closely related pollen grains make it challenging to distinguish species accurately and quickly. Identifying pollen grains under the microscope is time-consuming, expensive, and dependent on subjective criteria, resulting in error rates as high as 33% (Langford et al., 1990; Gonçalves et al., 2016; Sevillano et al., 2020). Although digital imaging techniques and graphical software have been used to enhance analysis, these tasks largely rely on human visual inspection, which is prone to classification errors, particularly for novice palynologists. Such limitations highlight the need for more efficient, objective, and accurate methods of pollen identification.
Identification of pollen grains is a difficult task, requiring both expert knowledge and high-resolution micrographs as well as a large number of reference slides for accurate comparison and identification. In particular, pollen grains of conifer species such as fir, spruce and pine, are difficult to identify because they all have two air sacs with a central body, causing the grains of these groups to look very similar with little morphological distinctness (Figure 1). This issue has been extensively documented, notably in a detailed study by Bagnell (1975), where distinctions among several species of Abies, Picea, and Pinus were meticulously examined using scanning electron microscopy. This study highlighted the subtle morphological differences critical for species identification, reinforcing observations from other research that document the morphological overlap among these species and the consequent challenges in their microscopic identification. Due to these morphological similarities, accurately identifying these species under a microscope is challenging. Deep learning is a great technique for enhancing pollen analysis by identifying species from a model trained on thousands of images (e.g., Daood et al., 2016). Having a trained model may help to improve our identification of these species and reduce the need for extensive morphological training in palynology. Deep learning approaches not only improve the accuracy of pollen classification but also dramatically reduce the time required for identification compared to traditional methods. While manual identification of a single pollen sample may take several minutes to hours depending on expertise and sample complexity, ML/DL models can process thousands of images in seconds once trained, providing an exponential improvement in speed (Balmaki et al., 2022b; Rostami et al., 2023). This makes deep learning particularly valuable for large-scale ecological and environmental studies.
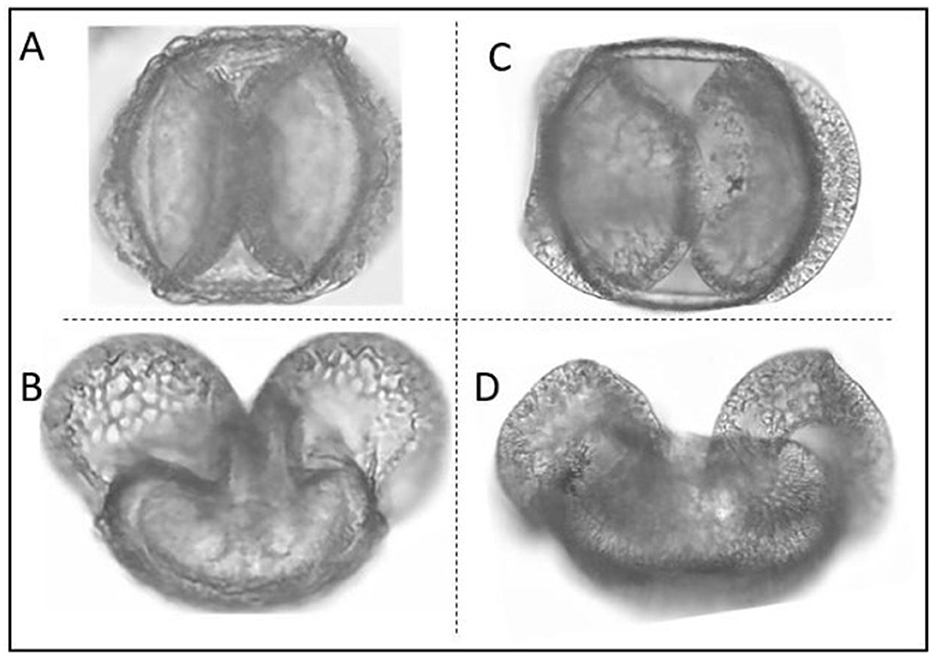
Figure 1. Morphological Comparison of Pinus monophylla (A, B) and Abies concolor (C, D). In this image each grain is visualized from two different angles. Conifer species tend to have morphologically similar pollen grains making their identification difficult.
In this study, we examine the ability of deep learning to identify pollen grains from conifer species. Deep learning techniques enhance accuracy, efficiency, and reduce manual effort and errors across image classification, object detection, and task recognition, as evidenced by multiple studies (Wäldchen and Mäder, 2018; Buddha et al., 2019; Afonso et al., 2020; Norouzzadeh et al., 2021; Jabbar et al., 2024). Specifically, deep learning has been highly effective in pollen taxonomic classification, utilizing transfer learning to achieve notable advancements (Daood et al., 2016; Khanzhina et al., 2018; Sevillano and Aznarte, 2018; Sevillano et al., 2020; Jaccard et al., 2020; Polling et al., 2021; Olsson et al., 2021; Zeng et al., 2021; Balmaki et al., 2022b; Rostami et al., 2023). Transfer learning, a technique where a model developed for one task is adapted for another, is particularly valuable as it leverages pre-trained models on large, diverse datasets to enhance learning efficiency and prediction accuracy, even with limited data specific to conifer species. This approach is cost-effective and less time-consuming, addressing the challenges of data scarcity in this domain.
For this research, we selected six common pollen species from the Pinaceae family: Abies concolor, Picea pungens, Picea wilsonii, Pinus flexilis, Pinus monophylla, and Pinus sabiniana, all sourced from the University of Nevada, Reno Museum of Natural History (UNRMNH). We manually collected pollen grains from the herbarium collections of a historical museum using entomological pins under a binocular microscope. We checked the pollen grains to ensure that no contaminant grains were present (e.g., grains from other plant species, potentially brought in by wind or insects). Pollen grains were prepared on glass slides by applying two drops of 2,000 cs silicone oil. This suspension allowed for the pollen grains to be rotated under the microscope, making it easier to examine their dimensions and shapes from various angles. All pollen grains are arranged on the slide to prevent them from sticking together, making it easier to take pictures for creating models. Each slide contained at least 400 individual pollen grains. Slides were secured with cover slips and sealed with nail polish as in Balmaki et al. (2019, 2022a, 2022b). A ZEISS Axiolab 5 light microscope paired with an Axiocam 208 color microscope camera was used to photograph the pollen grains. Images were taken using 20 × objective lenses and 10 × ocular lenses. We collected a dataset of nearly 1,400 images of pollen grains. The dataset includes images from the six pollen species, with each class containing between 96 and 487 images. All images were standardized to a resolution of 224 × 224 pixels and saved in JPEG format. For further details, see Table 1.
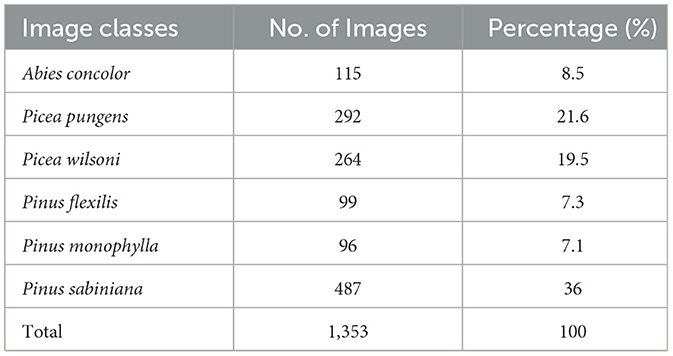
Table 1. Distribution of pollen grain images across different species and percentage of images each species had in the model.
The methodology pipeline is illustrated in Figure 2, encompassing the stages of dataset collection, data preprocessing, model training with image augmentation, and evaluation through hyperparameter optimization and performance metrics. For model training, each image was cropped to ensure consistency and to focus on relevant details. We used the Python-based OpenCV (Version 4.9) package for segmenting images containing multiple pollen grains into individual images and then converting to grayscale. Thresholding and morphological operations were then applied to highlight and clean up the particles within the images. Contours of these particles were identified and filtered based on a specified diameter range to avoid segmenting particles that were either too small, which could be dust or bubbles, or too large, which might result from overlapping grains creating an abnormally large particle. For each particle that met the criteria, the region of interest was cropped with an added margin, and the resulting cropped images were saved (Figure 3). Challenges such as overlapping grains and misidentified dust particles required manual exclusion from the final dataset.
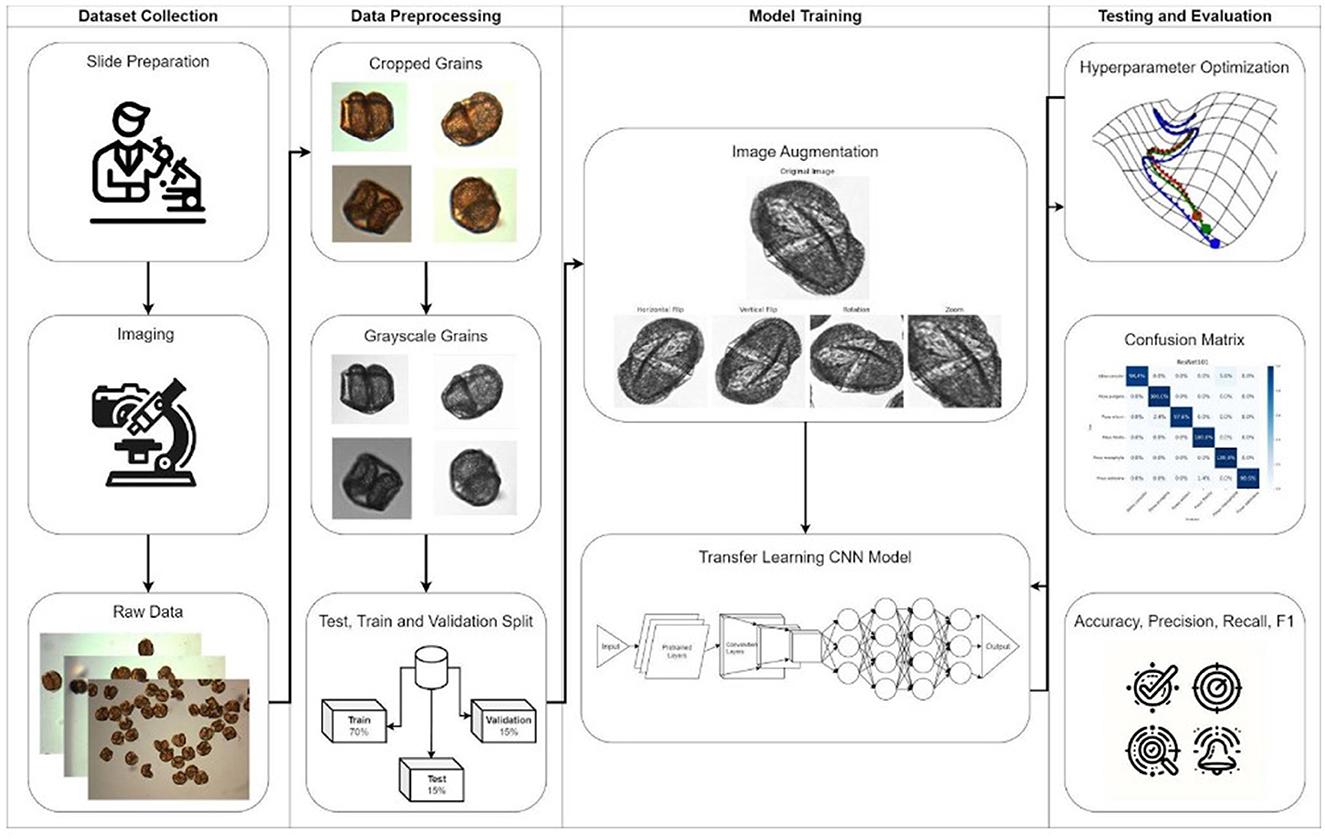
Figure 2. Methodological Pipeline for Pollen Analysis: This figure illustrates the comprehensive process used for pollen data analysis, starting from dataset collection through slide preparation and imaging. It details data preprocessing steps including cropping and converting pollen grains to grayscale, followed by a split into training, validation, and test sets. The model training phase incorporates image augmentation techniques to enhance the robustness of the transfer learning CNN model. The final evaluation phase is depicted through different metrics, including a confusion matrix to illustrate the model's performance.

Figure 3. Cropped Pollen Grains from Microscopic Imaging: This figure displays individual pollen grains (A–E) extracted from a single slide. The cropping process isolates each grain for detailed analysis and further study in pollen classification.
Before training the models, the dataset underwent comprehensive preprocessing. This included normalizing pixel values within a specified range and resizing all images to align with model input requirements. To increase model robustness and reduce the likelihood of overfitting, data augmentation techniques such as random rotations, flipping, and zooming were utilized. The complete dataset consisted of 1,344 images and was systematically divided into three subsets: training, testing, and validation. The training subset contained 70% of the total images (944 images), while the testing and validation subsets each comprised 15%, with 200 images per subset. This structure was chosen to optimize the effectiveness of model training.
We selected these models based on their varied architectures and proven effectiveness in image recognition tasks, adapting each to our dataset's unique requirements through a systematic process of feature adjustment and fine-tuning. In this study, we tested nine different transfer learning models to tackle the challenge of distinguishing similar pollen grains from Abies, Picea, and Pinus species. Each of these models—DenseNet201, EfficientNetV2S, InceptionV3, MobileNetV2, ResNet101, ResNet50, VGG16, VGG19, and Xception has been pre-trained on large-scale image datasets, making them well-suited for feature extraction in complex image recognition tasks. The use of multiple models allows us to compare their effectiveness and robustness across similar images, ensuring that we can identify the most effective architecture for our specific application.
Each model employed a combination of feature transfer, parameter transfer, and layer fine-tuning. The convolutional bases of each model were utilized as fixed feature extractors where only the top layers were retrained to adapt to the nuances of our pollen dataset. Parameters from select layers were finely adjusted to better suit the detailed features of pollen grains. This adaptation was crucial for enhancing the model's sensitivity to subtle inter-species variations. After the initial adaptation phase, several layers were progressively unfrozen and fine-tuned with a reduced learning rate to allow precise adjustments, optimizing the models for high accuracy in pollen classification.
This architecture utilizes dense connections, where each layer receives inputs from all preceding layers and passes on its feature-maps to subsequent layers. This effectively reduces the vanishing gradient problem, enhances feature propagation, and facilitates feature reuse. This design simplifies training and increases parameter efficiency by ensuring that each layer can access feature maps from every other layer, enhancing the learning and feature-utilization efficiency (Huang et al., 2017).
This architecture employs a compound scaling method that uniformly scales the depth, width, and resolution of the network. This approach optimizes both accuracy and efficiency, reducing training time and improving scalability across various devices. Fine-tuning was particularly focused on the scaling parameters to match the complexity of pollen images (Tan and Le, 2021).
This architecture builds on its predecessor, InceptionV2, by incorporating factorization into its modules to reduce computational load. It uses asymmetric convolutions that split larger convolutions into smaller ones, effectively reducing operations and complexity. This optimization enhances performance in large-scale image recognition tasks and was fine-tuned to improve the model's capacity to process the unique textural features of pollen (Szegedy et al., 2016).
This architecture introduces inverted residuals and linear bottlenecks with shortcut connections that maintain a compact and efficient model structure, ideal for mobile environments with limited computational resources. This model was adapted for rapid processing, enabling efficient deployment in field studies (Sandler et al., 2018).
The ResNet series enhances deep learning architectures using deep residual frameworks. These models use skip connections to facilitate training deeper networks by preventing vanishing gradients and degradation. ResNet50 offers a balance between performance and computational cost, whereas ResNet101 provides greater depth for enhanced feature learning, crucial for distinguishing closely similar pollen types (He et al., 2016).
The VGG series standardizes deep learning architectures using repetitive 3 × 3 convolutions and pooling layers. VGG16 focuses on building rich feature hierarchies' layer by layer, while VGG19, an extension with more convolutional layers, captures more complex features, benefiting tasks requiring detailed feature differentiation and was particularly useful in identifying subtle morphological differences between pollen species (Simonyan and Zisserman, 2014).
This architecture refines Inception by introducing depthwise separable convolutions, decoupling the learning of spatial hierarchies from channel-wise correlations. This improvement boosts both performance and efficiency, making it ideal for handling large-scale, high-dimensional data and was adapted to enhance feature extraction capabilities specific to the textural and shape-related characteristics of different pollen grains (Chollet, 2017).
All models were trained and tested on an NVIDIA GeForce RTX 3060 with 12GB of memory using Python 3.10.6 and TensorFlow. Each model was configured with a learning rate of 0.001, a batch size of 128, and the number of epochs adjusted between 30 and 100 to match the complexity and convergence behavior of each model. The total parameters and disk storage varied significantly across models, for instance, DenseNet201 had ~96 million parameters with a disk size of 369.53 MB, while EfficientNetV2S utilized about 131 million parameters with a disk size of 500.05 MB. This design and approach were critical for reliable and efficient training, essential for precise classification of closely similar pollen species. Table 2 provides an overview of the hyperparameter configurations, detailing the specific settings for each model. This includes the use of pre-trained ImageNet weights, the ADAM optimizer, and early stopping mechanisms (implemented if no improvement is observed), with the loss function set to sparse categorical cross entropy. The variation in the number of epochs and the strategic implementation of early stopping are tailored to optimize each model's learning process effectively.
In our evaluation of various transfer learning models for the classification of similar pollen grains from Abies, Picea, and Pinus species, the ResNet models, particularly ResNet50 and ResNet101, outperformed the other models (Table 3). The confusion matrix for ResNet101 had the highest accuracy in pollen classification, with minimal misclassifications (Figure 4). This performance can be attributed primarily to the architectural advantages of the ResNet models. ResNet architectures, such as ResNet50 and ResNet101, are particularly effective for tasks like conifer species classification due to their deep residual learning framework (He et al., 2016). This framework mitigates the vanishing gradient problem through the use of skip connections that facilitate direct gradient flow across multiple layers. This enhancement not only accelerates training but also improves learning capabilities as network depth increases, which is essential for distinguishing subtle features in highly similar classes. The effectiveness of these models is demonstrated by the high-performance metrics, as detailed in Table 1, with ResNet101 achieving nearly perfect scores in accuracy, precision, recall, and F1-score. A key strength of ResNet models is their scalability, which allows them to effectively manage hundreds of layers without performance degradation. This attribute is key for addressing the challenges posed by high intra-class variation and subtle inter-class differences observed in pollen grain images (He et al., 2016).
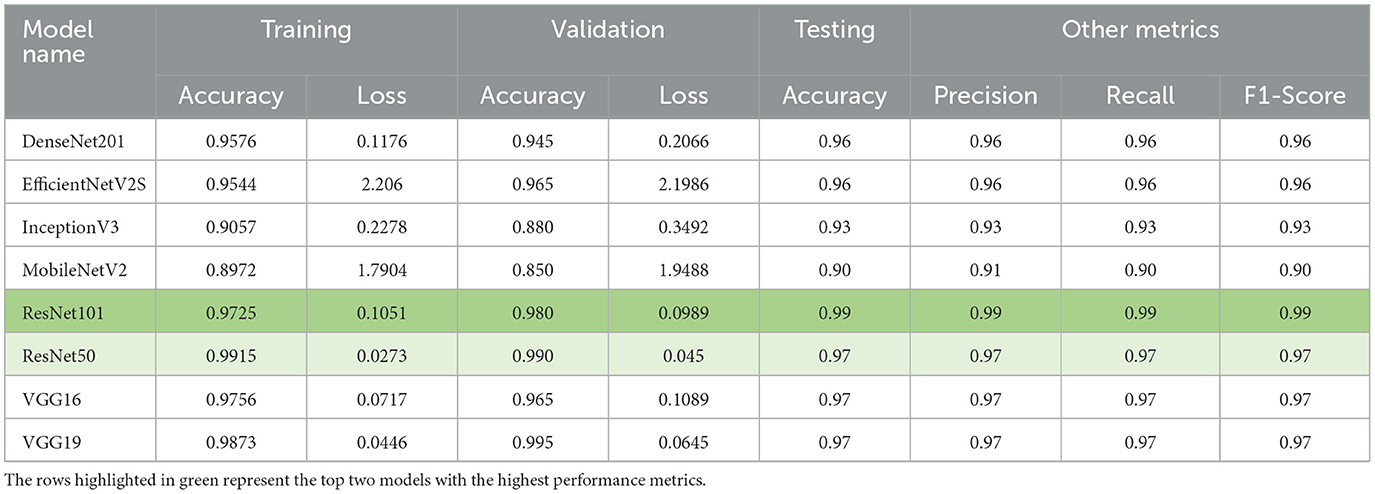
Table 3. Comparison of model performance metrics across various transfer learning models used in this study.
VGG-19 was also a top-performing model in this study, characterized by its deep convolutional layers that are able to capture intricate details (Simonyan and Zisserman, 2014). While it requires more computational resources, the depth of its architecture allows for the thorough extraction of features, which is instrumental in distinguishing between classes that share close similarities. VGG-19′s design focuses on increasing the depth with smaller convolution filters, which effectively increases the model's capacity to learn finer details in the Conifer pollen species without substantially widening the network (Simonyan and Zisserman, 2014; Shen et al., 2019). The training and testing performance of ResNet101 over 50 epochs, presented in Figure 5, demonstrates the model's convergence behavior. The accuracy plot indicates that the model achieves high accuracy early in the training process and maintains it throughout. This suggests that ResNet101 efficiently learns to generalize from the training data. The loss plot complements this by showing a rapid decrease in loss during the initial epochs, which stabilizes as training progresses, indicating that the model is effectively minimizing the error.
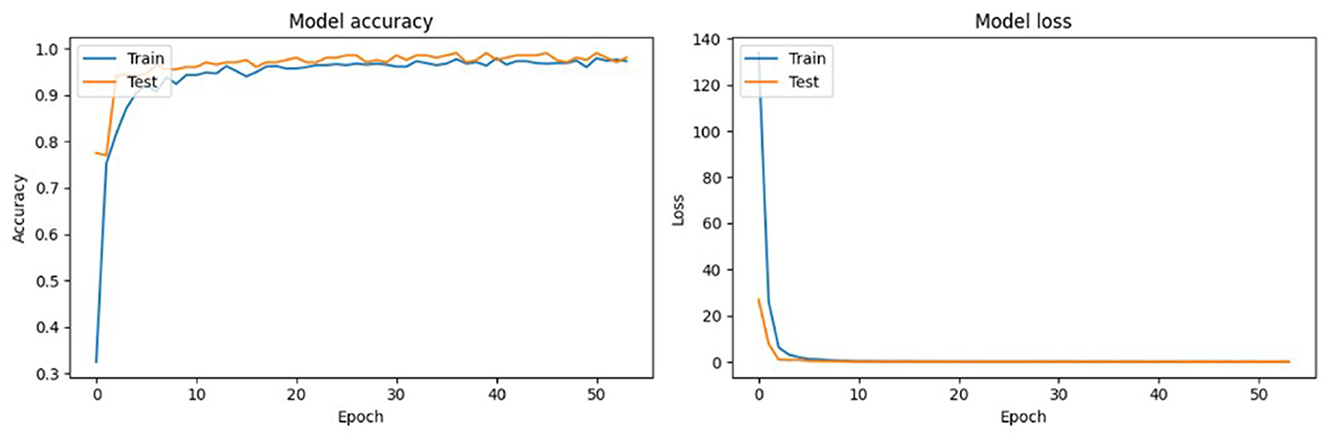
Figure 5. Training and testing performance of the ResNet-101 model over 50 epochs, presented in two plots: Model Accuracy and Model Loss.
The confusion matrices for ResNet50, ResNet101, VGG-16, and VGG-19 further illustrate the models' performance by showing the classification results for particulate matter from six tree species (Figure 6). Each cell in the matrices represents the number of predictions made by the models. The diagonal cells indicate correct predictions, while off-diagonal cells represent misclassifications. The high values along the diagonal and low values off the diagonal underscore the models' accuracy in classifying the pollen grains. For instance, the matrices show perfect classification for several species, with only minor misclassifications, demonstrating the models' precision and recall. Our study achieved high accuracy in identifying closely related coniferous species, demonstrating the value of AI in palynology to advance our ability to rapidly identify and distinguish between similar pollen grains. Despite challenges such as extensive training needs and the time-intensive nature of creating training datasets, the integration of AI helps overcome the limitations of traditional methods, which often require meticulous manual effort and are prone to errors, particularly with similar pollen grains. Our models reduce these errors by effectively learning complex patterns and subtle distinctions, speeding up the research process and enabling large-scale analysis that would be impractical manually. This emphasizes the critical role of trained palynologists in ensuring precise image capturing to grow these models on more species in the future.
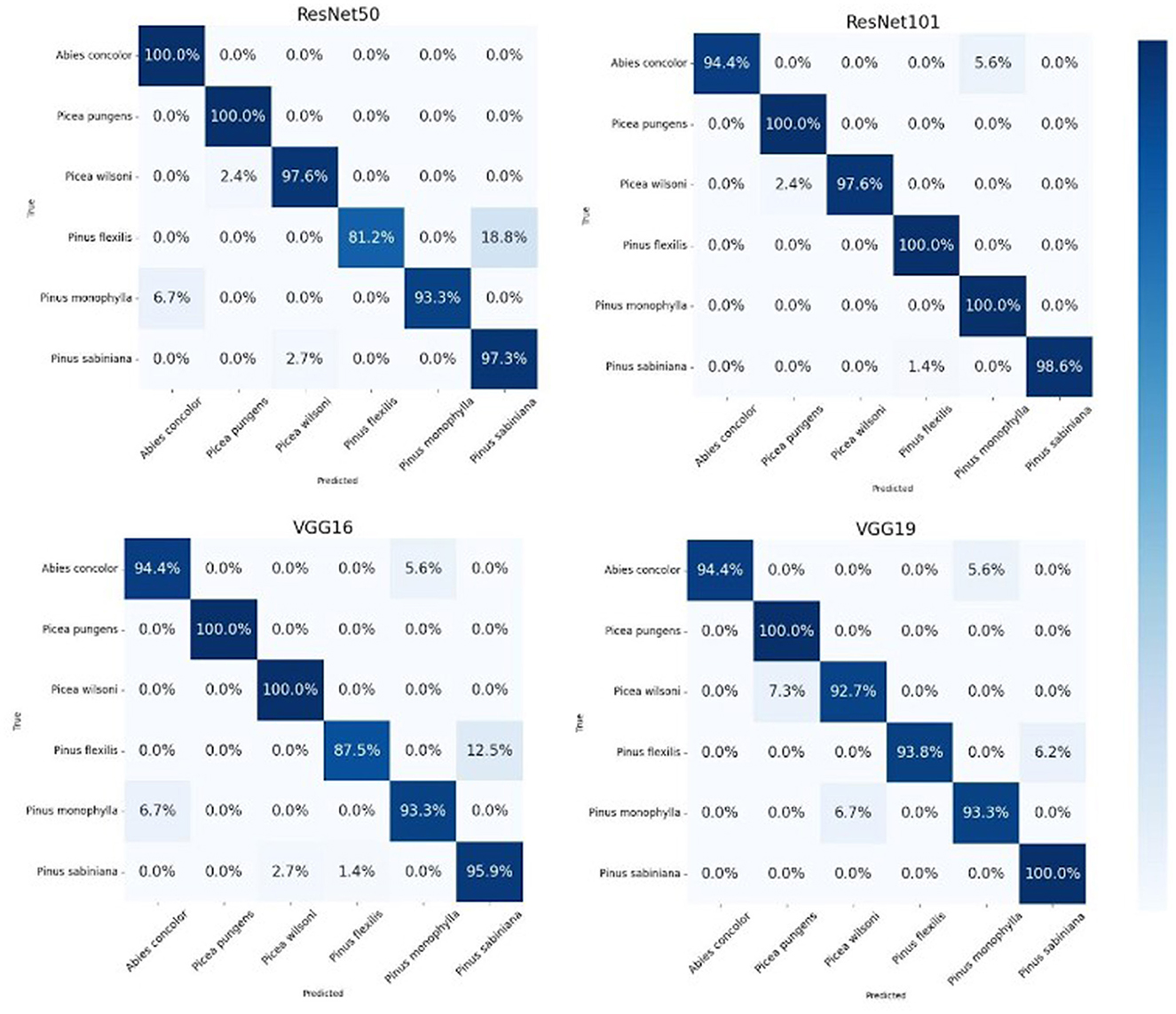
Figure 6. Confusion matrix for the ResNet-101 model illustrating classification results for particulate matter from six tree species.
This study highlights the benefits of integrating advanced AI technologies, specifically deep learning models like ResNet and VGG, into traditional pollen analysis using light microscopes. While AI applications are powerful, the expertise of trained palynologists remains essential for creating necessary datasets, emphasizing their indispensable role. Applying these models, we achieved high accuracy and efficiency in classifying pollen grains from closely related and hard-to-distinguish coniferous species. Using AI in pollen identification is not without challenges. It requires extensive training and expertise. Capturing high-quality images is important, as they need to be consistently scaled and properly focused, which is time-consuming and demands precision. Despite these difficulties, this research demonstrates the significant advantages of AI in environmental sciences. By merging traditional palynology and ecology research with AI technology, we can use these tools to better understand historical climate patterns, vegetation distributions, and the impacts of environmental changes on human and ecological health. For example, time series exist both in pollen from soil cores and in museum pollen from plant and insect specimens. These time series can be used to reconstruct plant communities (Balmaki et al., 2019; Balmaki and Wigand, 2019), and plant-pollinator interactions (Balmaki et al., 2022a,b, 2024) and then combined with historical climate or disturbance data to test hypotheses of the effects of climate and disturbance on plant communities and interactions. This balanced approach allows us to recognize both the potential and the challenges of using AI, paving the way for more effective and accurate environmental studies.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
MR: Writing – original draft, Writing – review & editing. LK: Writing – original draft. BB: Writing – original draft. LD: Writing – review & editing. JA: Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We appreciate University of Nevada Reno Museum of Natural History (UNRMNH) for supporting us and giving us this opportunity to conduct this research on these invaluable collections.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Afonso, M., Fonteijn, H., Fiorentin, F. S., Lensink, D., Mooij, M., Faber, N., et al. (2020). Tomato fruit detection and counting in greenhouses using deep learning. Front. Plant Sci. 11:571299. doi: 10.3389/fpls.2020.571299
Bagnell, C. R. (1975). Species distinction among pollen grains of Abies, Picea, and Pinus in the rocky mountain area (A scanning electron microscope study). Rev. Palaeobot. Palynol. 19, 203–220. doi: 10.1016/0034-6667(75)90041-X
Balmaki, B., Christensen, T., and Dyer, L. A. (2022a). Reconstructing butterfly-pollen interaction networks through periods of anthropogenic drought in the Great Basin (USA) over the past century. Anthropocene 37:100325. doi: 10.1016/j.ancene.2022.100325
Balmaki, B., Rostami, M. A., Allen, J. M., et al. (2024). Effects of climate change on Lepidoptera pollen loads and their pollination services in space and time. Oecologia 204, 751–759. doi: 10.1007/s00442-024-05533-y
Balmaki, B., Rostami, M. A., Christensen, T., Leger, E. A., Allen, J. M., Feldman, C. R., et al. (2022b). Modern approaches for leveraging biodiversity collections to understand change in plant-insect interactions. Front. Ecol. Evol. 10:924941. doi: 10.3389/fevo.2022.924941
Balmaki, B., Wigand, P., Frontalini, F., Shaw, A. T., Avnaim-Katav, S., Asgharian Rostami, M., et al. (2019). Late holocene paleoenvironmental changes in the seal beach wetland (California, USA): a micropaleontological perspective. Q. Int. 530–531, 14–24. doi: 10.1016/j.quaint.2019.10.012
Balmaki, B., and Wigand, P. E. (2019). Reconstruction of the Late Pleistocene to Late Holocene vegetation transition using packrat midden and pollen evidence from the Central Mojave Desert. Acta Bot. Brasilica 33, 539–547. doi: 10.1590/0102-33062019abb0155
Buddha, K., Nelson, H., Zermas, D., and Papanikolopoulos, N. (2019). “Weed detection and classification in high altitude aerial images for robot-based precision agriculture,” in Proceedings of the 27th Mediterranean Conference on Control and Automation (MED) (IEEE), 280–285.
Chollet, F. (2017). “Xception: Deep learning with depthwise separable convolutions,” in Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE), 1800–1807.
Daood, A., Ribeiro, E., and Bush, M. (2016). “Pollen grain recognition using deep learning,” in Advances in Visual Computing. Lecture Notes in Computer Science (Cham: Springer), 321–330.
Frisk, C. A., Brobakk, T. E., and Ramfjord, H. (2024). Allergenic pollen seasons and regional pollen calendars for Norway. Aerobiologia 40, 145–159. doi: 10.1007/s10453-023-09806-6
Gastaminza, G., Lombardero, M., Bernaola, G., Antepara, I., Munoz, D., Gamboa, P. M., et al. (2009). Allergenicity and cross-reactivity of pine pollen. Clin. Exp. Aller. 39, 1438–1446. doi: 10.1111/j.1365-2222.2009.03308.x
Gonçalves, A. B., Souza, J. S., Silva, G. G., Cereda, M. P., Pott, A., Naka, M. H., et al. (2016). Feature extraction and machine learning for the classification of Brazilian Savannah pollen grains. PLoS ONE 11:e0157044. doi: 10.1371/journal.pone.0157044
He, K., Zhang, X., Ren, S., and Sun, J. (2016). “Deep residual learning for image recognition,” in Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (IEEE).
Huang, G., Liu, Z., Van Der Maaten, L., and Weinberger, K. Q. (2017). “Densely connected convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR).
Jabbar, A., Naseem, S., Li, J., Mahmood, T., Jabbar, M. K., Rehman, A., et al. (2024). Deep transfer learning-based automated diabetic retinopathy detection using retinal fundus images in remote areas. Int. J. Comput. Intell. Syst. 17:135. doi: 10.1007/s44196-024-00557-x
Jaccard, P., Cosandey-Godin, A., Pernet, L., Rey, P., and Guisan, A. (2020). Improving the automation of pollen identification: A deep learning approach. Appl. Plant Sci. 8:e11372.
Khanzhina, N., Putin, E., Filchenkov, A., and Zamyatina, E. (2018). “Pollen grain recognition using convolutional neural network,” in Proceedings of the ESANN 2018 proceedings, European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning, 25–27.
Kobe, F., Bezrukova, E. V., Leipe, C., Shchetnikov, A. A., Goslar, T., Wagner, M., et al. (2020). Holocene vegetation and climate history in Baikal Siberia reconstructed from pollen records and its implications for archaeology. Archaeol. Res. Asia 23, 100209. doi: 10.1016/j.ara.2020.100209
Langford, M., Taylor, G. E., and Flenley, J. R. (1990). Computerized identification of pollen grains by texture analysis. Rev. Palaeobot. Palynol., 64, 197–203. doi: 10.1016/0034-6667(90)90133-4
Larson, E. R., Kipfmueller, K. F., and Johnson, L. B. (2020). People, fire, and pine: linking human agency and landscape in the boundary waters canoe area wilderness and beyond. Ann. Am. Assoc. Geogr. 111, 1–25. doi: 10.1080/24694452.2020.1768042
Latałowa, M., and van der Knaap, W. O. (2006). Late Quaternary expansion of Norway spruce Picea abies (L.) Karst in Europe according to pollen data. Q. Sci. Rev. 25, 2780–2805. doi: 10.1016/j.quascirev.2006.06.007
Norouzzadeh, M. S., Morris, D., Beery, S., Joshi, N., Jojic, N., Clune, J., et al. (2021). A deep active learning system for species identification and counting in camera trap images. Meth. Ecol. Evol. 12, 150–161. doi: 10.1111/2041-210X.13504
Olsson, O., Karlsson, M., Persson, A. S., Smith, H. G., Varadarajan, V., Yourstone, J., et al. (2021). Efficient, automated and robust pollen analysis using deep learning. Meth. Ecol. Evol. 12, 850–862. doi: 10.1111/2041-210X.13575
Polling, M., Li, C., Cao, L., et al. (2021). Neural networks for increased accuracy of allergenic pollen monitoring. Sci. Rep. 11, 11357–11367. doi: 10.1038/s41598-021-90433-x
Rostami, M. A., Balmaki, B., Dyer, L. A., Allen, J. M., Sallam, M. F., Frontalini, F., et al. (2023). Efficient pollen grain classification using pre-trained Convolutional Neural Networks: a comprehensive study. J. Big Data 10:151. doi: 10.1186/s40537-023-00815-3
Sadori, L., Mercuri, A. M., and Mariotti Lippi, M. (2010). Reconstructing past cultural landscape and human impact using pollen and plant macroremains. Plant Biosyst. Int. J. Deal. Aspects Plant Biol. 144, 940–951. doi: 10.1080/11263504.2010.491982
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., and Chen, L. C. (2018). “MobileNetV2: Inverted residuals and linear bottlenecks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Computer Vision Foundation).
Sevillano, V., and Aznarte, J. L. (2018). Improving classification of pollen grain images of the polen23e dataset through three different applications of deep learning convolutional neural networks. PLoS ONE 13:e0201807. doi: 10.1371/journal.pone.0201807
Sevillano, V., Holt, K., and Aznarte, J. L. (2020). Precise automatic classification of 46 different pollen types with convolutional neural networks. PLoS ONE 15:e0229751. doi: 10.1371/journal.pone.0229751
Shen, L., Margolies, L. R., Rothstein, J. H., Fluder, E., McBride, R., Sieh, W., et al. (2019). Deep learning to improve breast cancer detection on screening mammography. Sci. Rep. 9:12495. doi: 10.1038/s41598-019-48995-4
Shennan, I. (2015). “Handbook of sea-level research: framing research questions,” in Handbook of Sea-Level Research, eds. I. Shennan, A. Long, and B. Horton (John Wiley and Sons).
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv [preprint] arXiv:1409, 1556. doi: 10.48550/arXiv.1409.1556
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., and Wojna, Z. (2016). “Rethinking the inception architecture for computer vision,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Computer Vision Foundation).
Tan, M., and Le, Q. (2021). EfficientNetV2: Smaller models and faster training. arXiv [preprint] arXiv:2104. doi: 10.48550/arXiv.2104.00298
Wäldchen, J., and Mäder, P. (2018). Plant species identification using computer vision techniques: a systematic literature review. Arch. Comput. Meth. Eng. 25, 507–543. doi: 10.1007/s11831-016-9206-z
Willard, D. A., and Bernhardt, C. E. (2011). Impacts of past climate and sea level change on Everglades wetlands: placing a century of anthropogenic change into a late-Holocene context. Clim. Change 107, 59–80. doi: 10.1007/s10584-011-0078-9
Keywords: deep learning, ecological research, environmental changes, palynology, transfer learning
Citation: Rostami MA, Kydd L, Balmaki B, Dyer LA and Allen JM (2025) Deep learning for accurate classification of conifer pollen grains: enhancing species identification in palynology. Front. Big Data 8:1507036. doi: 10.3389/fdata.2025.1507036
Received: 06 October 2024; Accepted: 28 January 2025;
Published: 14 February 2025.
Edited by:
Vyacheslav Lyubchich, University of Maryland Center for Environmental Science (UMCES), United StatesReviewed by:
Steffen M. Noe, Estonian University of Life Sciences, EstoniaCopyright © 2025 Rostami, Kydd, Balmaki, Dyer and Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Behnaz Balmaki, YmVobmF6LmJhbG1ha2lAdXRhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.