
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bacteriol., 03 March 2025
Sec. Molecular Bacteriology and Microbiome
Volume 4 - 2025 | https://doi.org/10.3389/fbrio.2025.1541085
Cardiometabolic diseases (CMDs), particularly cardiovascular disease (CVD), type 2 diabetes mellitus (T2DM), and chronic kidney disease (CKD), emerged as primary contributors to global morbidity and mortality. In addition to traditional factors, recent studies demonstrated that blood microbiomes may also promote the development or progression of these CMDs. Traditionally, blood was considered sterile; however, the notion of blood as a sterile environment has been challenged by findings demonstrating the presence of a microbiome in both healthy and disease states. Although there has been a tremendous expansion in human microbiome research, with hundreds of projects underway globally the blood microbiome has not received the same level of attention as its gut and oral counterparts. The circulating microbiome is an emerging trend that has drawn a high level of interest in the biomedical field, given its potential to generate predictive biomarkers and the means to screen for potential pathogens. This comprehensive review explores the latest advancements in blood microbiome research, emphasizing biomarker identification, diagnostic tools, treatment modalities, and prevention in CMDs. We also delve into existing challenges and present a future-oriented treatment strategy using advanced methods. Deciphering the blood microbiome’s role in disease could lead to the classification of patient subgroups, enabling precision microbiota-based therapies.
Cardiometabolic disorders (CMDs), including type 2 diabetes mellitus (T2DM), chronic kidney disease (CKD), and CVD represent a major global health challenge characterized by their rising prevalence and profound social and economic burdens (Ralston and Nugent, 2019). The global burden of cardiometabolic disorders is substantial, with an estimated 523 million individuals affected by CVD (Fuster), 422 million by T2DM (Zhou et al., 2016), and 847 million by CKD (Jager et al., 2019). By 2030, the global economic burden of cardiometabolic diseases is projected to reach approximately US$6.3 trillion, effectively doubling current costs (Arena et al., 2015). These diseases are driven by a complex interplay of genetic, behavioral, and environmental factors, including genetic predispositions, sedentary lifestyles, poor dietary habits, and environmental exposures such as air pollution (Ralston and Nugent, 2019). In recent years, the gut microbiota has emerged as a key player in the pathogenesis of numerous diseases, with extensive research elucidating its role in modulating metabolic, immune, and inflammatory pathways (Velmurugan et al., 2020). These staggering figures highlight the widespread prevalence of these conditions and their significant impact on global health, emphasizing the critical need for effective management and intervention strategies. In this study, we aim to comprehensively characterize the composition and diversity of the blood microbiome in patients with CMD. Additionally, we provide a focused discussion on the potential mechanisms through which the blood microbiome may influence the pathogenesis and progression of CMD, offering insights into its role in disease development.
Nearly 70% of medical decision-making relies on laboratory findings, underscoring the critical role of diagnostic testing in clinical practice. Key laboratory results, such as biochemistry and hematology profiles, are often available on the day of hospital admission, providing essential insights into a patient’s health status and guiding immediate therapeutic interventions (Bissonnette and Bergeron, 2010). These tests serve as foundational tools for diagnosing conditions, monitoring disease progression, and tailoring treatment strategies, highlighting their indispensable value in modern healthcare. Triggering microbiome analysis is critical for enabling timely clinical decisions and interventions, particularly within the first 6 hours of sepsis presentation, to effectively reduce morbidity and mortality. Rapid identification of microbial profiles and their associated pathogenic mechanisms can guide targeted antimicrobial therapy and personalized treatment strategies, improving patient outcomes. Integrating microbiome analysis into early sepsis management protocols represents a promising approach to enhancing the precision and efficacy of care in this life-threatening condition (Kumar et al., 2006). Although blood has traditionally been considered a sterile environment, recent advances in high-throughput sequencing technologies have challenged this notion, revealing the presence of a low-abundance but diverse blood microbiome even in healthy individuals (Tan et al., 2023). In 2001, Nikkari and colleagues were among the first to detect bacterial DNA in the blood, challenging the long-held notion of blood sterility (Nikkari et al., 2001). This finding was further supported by Moriyama et al., who confirmed the presence of a blood microbiome in healthy individuals (Moriyama et al., 2008). Additionally, Castillo et al. investigated the interactions between the blood microbiome and other human microbiomes, shedding light on its potential systemic role (Castillo et al., 2019). In recent years, research has increasingly focused on elucidating the composition and functional significance of the blood microbiome and its association with human health and disease. Emerging evidence suggests that the blood microbiome may contribute to the development and progression of various conditions, including T2DM, colorectal cancer, inflammatory bowel diseases (IBD), hypertension, myocardial infarction (MI), coronary heart disease, kidney complications, and HIV infection (Potgieter et al., 2015; Amar et al., 2019; Shah et al., 2019; Søby et al., 2020; Gedgaudas et al., 2022; Khan et al., 2022a; Guo et al., 2023; Jagare et al., 2023; Sampaio et al., 2023). These findings underscore the importance of further exploring the blood microbiome as a potential biomarker and therapeutic target in a wide range of diseases.
Sequencing technologies have unveiled novel biomarkers like the blood microbiome, advancing disease diagnosis and prevention. Moving beyond traditional culture-based methods, modern molecular techniques such as 16S rRNA sequencing, qPCR, and shotgun metagenomics have confirmed the presence of a blood microbiome in healthy individuals. These tools enable precise characterization of microbial communities, offering new insights into their role in health and disease and their potential as diagnostic and therapeutic targets (Tan et al., 2023). It is crucial to acknowledge the limitations in study design, including small sample sizes, limited taxonomic resolution, methodological variability, and the difficulty in differentiating cell-free microbial DNA from viable microbial cells. Additionally, environmental contamination remains a common challenge that can confound results. Addressing these limitations is essential for ensuring the accuracy and reliability of blood microbiome research and its translation into clinical applications (Castillo et al., 2019; Berg et al., 2020). To ensure an accurate definition of the blood microbiome, it is critical to minimize microbial DNA contamination by adhering to strict aseptic techniques during sample collection and implementing thorough disinfection of the skin puncture site (Doern et al., 2019). DNA sequencing methods are susceptible to contamination from microbial DNA present in laboratory reagents and kits (Salter et al., 2014), a challenge further amplified by the low microbial biomass and high host DNA background in blood samples, which increases the noise-to-signal ratio (Glassing et al., 2016). However, evidence indicates that contamination alone cannot fully account for the observed microbial signals; instead, the blood microbiome is thought to originate, at least in part, from microbial translocation from sites such as the gut (Castillo et al., 2019). Despite significant advancements, the biology of the blood microbiome remains poorly understood, particularly its functional interactions with human health and disease. Further research is needed to elucidate its role and potential as a diagnostic or therapeutic target.
In this review, we examine the latest advancements in understanding the composition and role of the blood microbiome in human health and disease, supported by specific disease-related insights. We delve into the identification of blood-based biomarkers, address methodological and interpretative challenges, and explore innovative therapeutic strategies enabled by cutting-edge technologies. Furthermore, we propose future research directions aimed at elucidating the functional mechanisms of the blood microbiome, emphasizing an integrative approach to unravel its complex interactions and potential clinical applications.
Our understanding of the role of the microbiota in human health and disease is built on epidemiological, observational, molecular, and animal-modeling experiments. The microbiota plays an important role in human health and disease. Healthy individuals’ blood microbiome composition and diversity are largely overlooked (Païssé et al., 2016; Castillo et al., 2019; Tsafarova et al., 2023; Di Gloria et al., 2024). The major blood phyla with their respective roles are summarized in Table 1. Emerging evidence highlights the association between dysbiosis in the human blood microbiome and the pathogenesis of various diseases, including chronic kidney disease, diabetes mellitus, and cardiovascular disease (Figure 1).
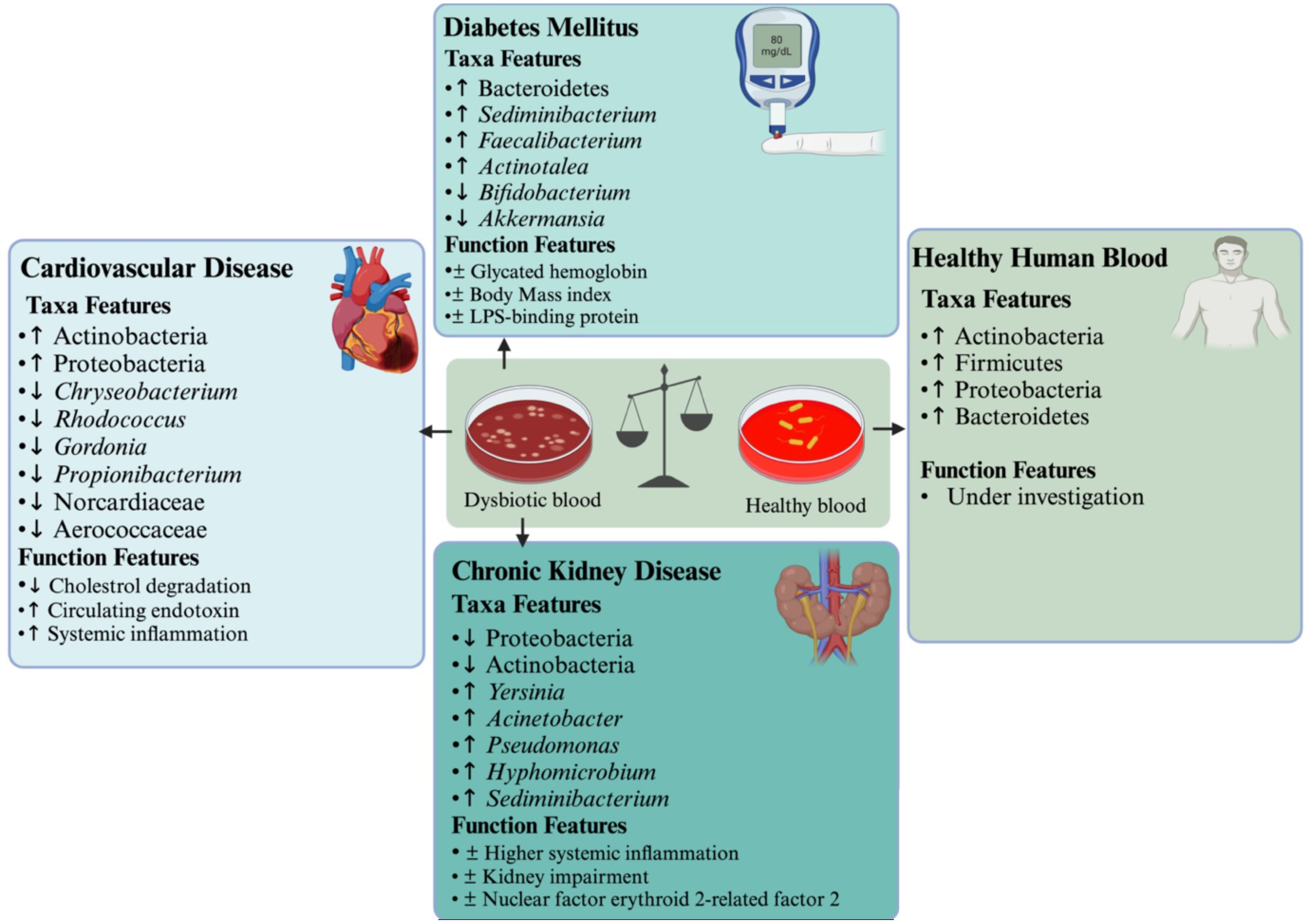
Figure 1. Perturbations in the blood microbiome and their association with cardiovascular disease, diabetes, and chronic kidney disease contribute to systemic inflammation and impaired endotoxin clearance. The (↑) symbol indicates higher species abundance, (↓) represents lower species abundance, an asterisk (*) denotes statistical significance, and (±) marks correlation.
Chronic kidney disease is a growing healthcare burden affecting about 13.4% of the population worldwide (Guo et al., 2025). In the last few decades, CKD patients have steadily increased. In adults, hypertension and diabetes are the leading causes of CKD, while congenital anomalies of the kidney and urogenital tract account for the majority of CKD etiologies in children. In children, CKD affects neurocognitive abilities, school performance, growth, quality of life, and the cost of medical care (Wehedy et al., 2022). CKD is associated with the development of severe health conditions like cardiovascular diseases, neurological complications, adverse pregnancy outcomes, and hyperkalemia. Factors contributing to CKD progression include activation of the renin-angiotensin-aldosterone system, proteinuria, chronic inflammation, and repetitive acute kidney injury (Wehedy et al., 2022). In addition, existing evidence showed an association between gut dysbiosis, inflammation, and CKD (Evenepoel et al., 2017). Gut dysbiosis disrupting intestinal barrier function in CKD permits the translocation of gut-derived toxins, bacterial products, and intact bacteria into the circulation, resulting in inflammation (Vaziri et al., 2016). This circulating microbiome in CKD is indirectly evidenced by higher levels of endotoxins, LPS levels, and gut uremic toxins measured in the blood (Andersen et al., 2017).
A previous study has shown higher levels of Legionella, Serratia, Yersinia, Acinetobacter, Pseudomonas, Lysobacter, Hyphomicrobium, Bacillus, Sediminibacterium, and Pseudarcicella, and lower levels of Stenotrophomonas, Paracoccus, Sphingomonas, and Tyzzer were found in CKD patients (Shah et al., 2019). Demmer et al. reported the association between fewer Proteobacteria levels with subgingival plaque and elevated systemic inflammation, suggesting that the oral microbiome could impact the circulating microbiome in hemodialysis patients (Demmer et al., 2017). Shah et al. examined blood fractions with leukocytes, revealed different microbes than cell-free blood, and showed a strong negative correlation between glomerular filtration and higher circulating Proteobacteria in CKD patients (Shah et al., 2021). Another study found higher levels of Staphylococcus and Streptococcus and predominant Legionella and Enhydrobacter in the blood of kidney patients (Shah et al., 2021). Intriguingly, most of the blood bacteria in CKD patients are not typical urinary tract commensals, suggesting that disturbances in urinary mucosa may not contribute to the dysbiotic blood microbiome (Perez-Carrasco et al., 2021). Sumida et al. used 16S rRNA sequencing and ITS analysis to compare the circulating microbiome in hemodialysis patients and healthy individuals. They found elevated Actinobacteria and reduced Proteobacteria levels in patients who died from cardiovascular events, with these microbial changes significantly correlated to Nrf2, a key regulator of antioxidative responses. This correlation persisted across demographic and clinical factors and was linked to a slight increase in cardiovascular mortality risk. By focusing on the “cell-free” circulating microbiome in serum or plasma, the study highlights its direct impact on immune cells and cardiac myocytes, offering a minimally invasive approach for assessing cardiovascular risk in end-stage renal disease (Sumida et al., 2021a). In another study, Sumida et al. reported depleted Proteobacteria levels in CVD patients, alongside elevated Staphylococcus genus, suggesting a link between altered microbiome composition and cardiovascular comorbidities in hemodialysis patients (Sumida et al., 2021b).
Additionally, Merino-Ribas et al. observed significant alterations in the genera levels, including Cutibacterium, Pajaroellobacter, Devosia, Hyphomicrobium, and Pelomonas in the CKD peritoneal dialysis patients with and without vascular calcification. The study has also shown the association between vascular calcification and an increased risk of all-cause mortality in CKD peritoneal dialysis patients, highlighting the potential role of microbial composition as a factor linked to vascular calcification and mortality risk in patients (Merino-Ribas et al., 2022). Patients at a higher mortality risk showed changes in Eubacterium eligens in the gut and the presence of the Devosia genus in the blood. Despite no disparities in uremic toxins, intestinal translocation markers, and inflammatory parameters between CKD-peritoneal dialysis patients with and without vascular calcification, soluble CD14 (sCD14), a nonspecific marker of monocyte activation, positively correlated with the severity of vascular calcification (Merino-Ribas et al., 2022). Hence, the gut Eubacterium eligens group, blood Devosia, and circulating sCD14 merit further exploration as potential biomarkers for vascular calcification, CVD, and CKD. Simões-Silva and colleagues explored the intricate world of peritoneal bacteria, pitting it against counterparts in various body regions, with blood emerging as the closest contender. Despite this close resemblance, their compelling results underscored substantial distinctions between peritoneal and blood bacteria (Simoes-Silva et al., 2020). In another study, Simões-Silva proposed that bloodstream bacteria primarily stem from the dysbiotic gut microbiome in end-stage renal disease, and hemodialysis, to some extent, exacerbates microinflammation by promoting gut microbiota translocation due to impaired gut barrier function (Simoes-Silva et al., 2018). Recently, Sampaio et al. investigated kidney transplant recipients (KTRs) and found evidence supporting the existence of a circulating microbiome that differs from the profiles of the gut and oral microbiomes. This circulating microbiome is characterized by a limited number of operational taxonomic units, representing a shared microbiome. Notably, the blood of KTRs contains a distinctive, relatively low-abundance microbiome, with a dominant of Proteobacteria and Firmicutes (Sampaio et al., 2023). The blood microbiome is now a hot topic. Hence, its composition and leveraging technological advancements are crucial for enhancing our understanding of the blood microbiome and its role in the development of kidney and other human-related diseases.
Type 2 diabetes mellitus is a metabolic disorder characterized by abnormally high blood glucose levels resulting essentially from the resistance of body cells to the effect of insulin (Sedighi et al., 2017). Diabetes is described by a state of chronic low-grade inflammation with abnormal expression and production of multiple inflammatory cytokines such as interleukins and tumor necrosis factor (Dandona et al., 2004). The prevalence of T2DM has reached epidemic proportions worldwide (Sedighi et al., 2017). Recent WHO statistics state that the number of individuals with T2DM around the world was approximately 382 million in 2013, and it is projected to be more than 590 million by 2035 year (Upadhyaya and Banerjee, 2015). In addition, 85% of premature deaths from metabolic syndromes happen in developing countries, of which about 80% are associated with diabetes (Upadhyaya and Banerjee, 2015). Based on WHO reports, T2DM is anticipated to be the seventh leading cause of death by 2030. T2DM is the consequence of a complex interaction between different degrees of genetic susceptibility and environmental factors such as stress, diet, and infections (Sedighi et al., 2017). Circulating microbiota refers to a complex community of microorganisms present in the blood of humans and other animals, primarily associated with immune system modulation and inflammation, as well as potential contributions to certain pathological conditions.
A previous D.E.S.I.R. cohort study has shown a higher 16S rDNA concentration in those who eventually developed T2DM and abdominal adiposity (Amar et al., 2011). A prospective cohort study with primarily Chinese volunteers uncovered the predictive value of the blood genus Bacteroides for T2DM (Qiu et al., 2019). Bacteroides is a probiotic that is available in the human intestine, yet when it drifts to other parts of the body, it can become pathogenic, causing infections in the mouth, nervous system, and bloodstream (Amar et al., 2011). The enrichment of Bacteroides observed in both T2DM and control groups, with a higher prevalence in T2DM, highlights its potential role in T2DM pathology. However, the mechanisms by which Bacteroides contribute to the development or progression of T2DM remain poorly understood. Given its dual role as a gut commensal and a potential systemic pathogen, further research is needed to elucidate the mechanisms through which Bacteroides influence systemic inflammation and contribute to T2DM pathology.
Subsequent analysis revealed a 28% elevation in gut microbiome levels among diabetes patients compared to a 4% level in healthy individuals, with T2DM patients exhibiting increased gram-positive bacteria, including the Atopobium cluster and Clostridium coccoides, similar to type 1 diabetes (T1D) (Sato et al., 2014). Another study has shown a higher abundance of Proteobacteria in the blood of patients with T1D (Yuan et al., 2024). A European study has revealed the association between elevated blood sugar and insulin levels with higher levels of blood bacterial DNA (D’Aquila et al., 2021). Donath et al. revealed significant correlations among glycated hemoglobin, body mass index, inflammatory markers, and LPS-binding protein, highlighting a clear association between diabetes and the blood microbiome, with notably elevated levels of the LPS-binding protein in patients with T2DM (Donath et al., 2019). Ghaemi et al. found that patients with diabetes had higher levels of Ralstonia spp. compared to non-diabetics and observed higher Proteobacteria in T2DM than in those without diabetes. Genus-level variations were observed in the gut microbiome, blood plasma, and cellular levels. In diabetic patients, Akkermansia, Bifidobacterium, and Faecalibacterium decreased in leukocytes, while the pre-diabetic group exhibited higher levels of Lactobacillus, E. coli, and Bacteroides fragilis genera compared to healthy individuals; all these bacteria were elevated in the T2DM group (Ghaemi et al., 2021). Moreover, a recent study found no significant differences at the phyla levels, however, the genera, including Aquabacterium, Xanthomonas, and Pseudonocardia, were depleted, while genera, e.g., Sediminibacterium, Pseudoclavibacter, Alishewanella, and Actinotalea were enriched in the patient with T2DM compared to the healthy group (Qiu et al., 2019). The involvement of the phylum Proteobacteria and the genus Sediminibacterium in the pathophysiology of T2DM remains insufficiently characterized. Future research should emphasize longitudinal investigations to establish causality, multi-omics approaches to integrate functional genomics with metabolomics, and validation studies in larger, diverse cohorts to enhance the generalizability of findings.
The relationship between the blood microbiome and CVD has emerged as a prominent area of research, highlighting the critical role of the microbiome in systemic health. However, significant gaps remain in our understanding of the precise mechanisms and the multifactorial nature of these interactions. A previous study has shown that depletion in blood bacterial DNA content and enrichment in the Proteobacteria levels in the blood microbiology predicts long-term cardiovascular prognosis (Amar et al., 2013). Building on this, another study conducted a year later further reported a higher abundance of the phyla Actinobacteria and Proteobacteria in patients with CVD compared to the control group (Dinakaran et al., 2014), reinforcing the potential role of these microbial shifts in cardiovascular health. Amar et al. (2013) reported significant reductions in cholesterol-degrading bacterial families and genera (e.g., Norcardiaceae, Aerococcaceae, Gordonia, Propionibacterium, Chryseobacterium, and Rhodococcus) in patients with myocardial infarction compared to healthy individuals (Amar et al., 2019). Liberale et al. (2020) found elevated levels of the gut commensal bacterium Escherichia coli, specifically Escherichia coli lipopolysaccharides, in the blood of coronary thrombosis patients (Liberale et al., 2020). The Escherichia coli lipopolysaccharides strain was linked to disease pathology, including low-grade endotoxemia, proinflammatory cytokine release, elevated soluble P-selectin (indicating platelet activation), and zonulin, a marker of gut permeability (Carnevale et al., 2020). The link between Escherichia coli lipopolysaccharides and zonulin indicates the strain’s potential role in enabling bacterial translocation from the gut to the bloodstream. Consistent with the findings of Amar et al. (2013) and Dinakaran et al. (2014), we observed a higher abundance of the phyla Proteobacteria and Actinobacteria in MI patients compared to healthy individuals (Khan et al., 2022a). In another study, we analyzed the blood microbiome composition of healthy controls, acute MI patients, and chronic MI patients, finding a higher abundance of Proteobacteria in acute MI patients compared to chronic MI patients and controls (Khan et al., 2022b). Lawrence et al. reported that Kokuria, Pseudomonadota, and Enhydrobacter from the Actinobacteria were significantly associated with cardiovascular mortality risk, including an inverse correlation between Enhydrobacter and CVD mortality risk (Lawrence et al., 2022). Furthermore, a recent study reported an enrichment of Gammaproteobacteria, Proteobacteria, Ralstonia pickettii, Ralstonia, Burkholderiaceae, and Burkholderiales in the unstable angina group. In contrast, the AMI group exhibited a higher abundance of Bacteroidales, Bacteroidia, Bacteroidota, Clostridia, and Firmicutes (Chen et al., 2024). These findings suggest that elevated levels of Proteobacteria are associated with CVD, as Proteobacteria produce lipopolysaccharides (LPS), which play a critical role in the pathogenesis of CVD (Lin et al., 2020). LPS can induce inflammation indirectly by generating molecular patterns associated with non-microbial risk factors and directly through microbial-associated molecular patterns (MAMPs) (Bonhomme et al., 2024). LPS might also contribute to endotoxemia, which drives the development of inflammatory conditions such as obesity, liver dysfunction, and CVD (Sciarra et al., 2023).
In a recent study integrating blood microbiome and metabolomics, we found reduced AMP, L-leucine, and L-arginine levels in the mTOR pathway in MI patients compared to healthy controls (Khan et al., 2025). AMP regulates cellular energy balance by activating AMPK to inhibit mTOR signaling. Its downregulation may impair energy sensing, leading to unchecked mTOR activity despite energy deficits (Khan et al., 2025). This aberrant signaling can result in excessive energy consumption, ultimately driving cellular energy depletion and apoptosis. L-leucine activates mTORC1, which is crucial for protein synthesis and cardiac repair. Its reduction weakens mTORC1 signaling, leading to muscle atrophy and impaired recovery. Similarly, L-arginine, essential for nitric oxide synthesis and mTOR signaling, supports blood flow and cellular repair. Its depletion increases oxidative stress, and endothelial dysfunction, and hinders tissue recovery, exacerbating cellular damage (Khan et al., 2025). These findings enhance cardiovascular research by uncovering microbiome-metabolome interactions with implications for immunology, metabolism, and microbiology. By linking basic and translational science, they support personalized diagnostics and therapies. Future studies should focus on longitudinal designs, multi-omics integration, and diverse cohort validation to establish causality and improve generalizability.
Herein, we briefly explored the mechanisms underlying CKD, T2DM, and CVD, as shown in Figure 2. Microbiome dysbiosis has been implicated in the pathogenesis of CKD through mechanisms involving microbial metabolites and systemic inflammation (Noce et al., 2022). Dysbiosis often leads to an overabundance of bacteria producing trimethylamine N-oxide (TMAO), a metabolite derived from dietary choline and carnitine (Valenbreder et al., 2023). Elevated TMAO levels promote oxidative stress and inflammation in renal tissues, contributing to endothelial dysfunction and fibrosis (Florea et al., 2024). Additionally, microbial translocation of lipopolysaccharides (LPS) from gram-negative bacteria into the bloodstream can trigger immune activation, further exacerbating kidney damage (Wang et al., 2023). These processes impair renal filtration and function, leading to the accumulation of uremic toxins and a progressive decline in kidney health (Lim et al., 2021). The interplay between dysbiosis, TMAO, and inflammation highlights the blood microbiome’s role in CKD progression.
In addition, the blood microbiome influences diabetes mellitus through mechanisms that disrupt metabolic homeostasis and promote insulin resistance. Dysbiosis often results in increased levels of LPS, a component of gram-negative bacterial cell walls, which enters the bloodstream and triggers chronic low-grade inflammation (Di Vincenzo et al., 2024). This inflammation impairs insulin signaling pathways in peripheral tissues, leading to insulin resistance and hyperglycemia (Chen et al., 2015). Furthermore, dysbiosis can alter the production of short-chain fatty acids (SCFAs), which play a role in regulating glucose metabolism and insulin secretion. Reduced SCFA levels exacerbate pancreatic beta-cell dysfunction, further compromising glucose regulation (Rekha et al., 2024). The combined effects of inflammation, insulin resistance, and beta-cell dysfunction underscore the contribution of blood microbiome dysbiosis to the development and progression of DM.
Subsequently, blood microbiome dysbiosis contributes to CVD through pathways involving microbial metabolites, endothelial dysfunction, and systemic inflammation. Elevated levels of TMAO, produced by gut microbiota from dietary precursors, are strongly associated with atherosclerosis and plaque formation (Zhu et al., 2020). TMAO promotes endothelial inflammation, foam cell formation, and platelet hyperreactivity, all of which accelerate atherosclerotic progression (Oktaviono et al., 2023). Additionally, dysbiosis can lead to increased LPS translocation into the bloodstream, triggering systemic inflammation and oxidative stress, further damaging vascular tissues (Violi et al., 2023). These processes result in endothelial dysfunction, arterial stiffness, and increased risk of myocardial infarction or stroke (Tuttolomondo et al., 2020). The role of microbiomes in modulating TMAO and inflammation highlights its significance in CVD pathogenesis.
Variability in the blood microbiome may be attributed to a combination of host-related, environmental, and methodological factors, as shown in Figure 3. Host-specific characteristics, such as genetic background, age, immune status, and underlying health conditions, including disease severity and treatment histories, play a significant role in shaping microbial profiles (Alanazi et al., 2024). Additionally, environmental and demographic factors, such as geographic location and lifestyle differences, further contribute to this variability by influencing microbial exposure and colonization dynamics (Parizadeh and Arrieta, 2023). Methodological inconsistencies across studies, including variations in sample collection protocols, DNA extraction techniques, sequencing platforms, and bioinformatics pipelines, can also introduce technical biases, affecting the detection, quantification, and characterization of microbial communities. These methodological disparities, coupled with the heterogeneity of studied populations, may impact the reproducibility and comparability of findings, underscoring the need for standardized approaches in blood microbiome research (Mancin et al., 2024). Temporal fluctuations, driven by changes in health status, treatment regimens, or external exposures (Karwowska et al., 2024), further complicate the interpretation of microbial profiles, highlighting the multifaceted nature of factors influencing the blood microbiome.
To enhance the reliability and reproducibility of blood microbiome research, we advocate for standardized methodologies across studies. This includes harmonizing protocols for sample collection, storage, DNA extraction, sequencing, and bioinformatics analysis to minimize technical variability. Rigorous quality control measures, such as the use of negative and positive controls, are essential to mitigate contamination and ensure data accuracy. Collaborative efforts, including large-scale consortia and shared databases, should facilitate data integration from diverse studies, enabling robust meta-analyses and cross-validation. Advanced computational approaches—such as machine learning, multi-omics integration, single-cell sequencing, metatranscriptomics, and metabolomics can further enhance microbial characterization, improve disease prediction, and refine diagnostic and therapeutic strategies. By promoting standardization, fostering collaboration, and leveraging cutting-edge analytical tools, we aim to establish a more comprehensive understanding of the blood microbiome’s role in health and disease, ultimately unlocking its potential as a diagnostic and therapeutic target in personalized medicine.
Blood acts as a liquid medium carrying the elements necessary for host life. In recent years, the concept of human blood microbiota has been developed, and this has caused serious debates as it may shelve the idea that “blood is a sterile environment” that has been in existence for years. Although there are different hypotheses that the blood microbiota originates from the gut microbiota, its origin is not clearly understood because it contains different phyla (Castillo et al., 2019). The concept of human blood microbiota and its potential therapeutic implications represents a groundbreaking advancement in medicine. If the blood microbiome is confirmed as a key factor in health and disease, it could unlock a range of innovative treatment strategies with transformative clinical implications (Almeida et al., 2022). For instance, (i). In today’s medicine, where fecal microbiota transplantation is being considered for treatment, blood transplantation is much more easily feasible. (ii). Potential therapies include targeted probiotic formulations designed to modulate blood microbiota, microbial metabolite-based treatments utilizing postbiotics, and immunomodulation strategies to regulate cytokine production and enhance immune responses. (iii). Dietary and lifestyle interventions, including nutraceuticals and lifestyle modifications, could support a healthy blood microbiota. Advanced diagnostic tools for microbiota profiling and real-time monitoring could enable early disease detection and personalized interventions. (iv). Antimicrobial therapies, such as selective antimicrobials and phage therapy, could eliminate harmful microbes while preserving beneficial ones. (v). Personalized blood microbiota transplants, both autologous and allogeneic, could restore microbial balance, while CRISPR-based microbial engineering may enable gene editing to enhance beneficial properties or reduce pathogenicity. (vi). Nanotechnology-based delivery systems, such as targeted drug delivery and microbial carriers, could ensure precise therapeutic applications, making blood microbiota manipulation a promising avenue for future medicine. (vii). Combination therapies offer a multi-modal approach by integrating blood microbiota modulation with treatments like immunotherapy, chemotherapy, or antibiotics to enhance efficacy. A holistic strategy also considers the interplay between blood and gut microbiota for comprehensive health benefits. (viii) Microbial vaccines could target harmful blood microbes, preventing their colonization and priming the immune system for improved responses to infections and cancers. (ix). Additionally, microbiome banking could enable the preservation of healthy blood microbiota samples for future therapeutic use, with donor-matching systems optimizing transplantation outcomes (Figure 4).
Microbiome research highlights the integral role of coexisting microbes in human biology, challenging the notion of human blood sterility. Rare microbial entry into the bloodstream can trigger severe complications, including sepsis and septic shock (Hotchkiss et al., 2016). Controversy persists among researchers on whether the blood hosts a core microbiome or whether these microbes disseminate from other body niches like the gut, mouth, and skin. For instance, Tan and colleagues have lately been unable to identify the microbiome in healthy human blood. They contradicted the hypothesis of a structured blood microbiome by detecting 117 random microbial species in healthy human blood, most of which originated from different body regions and some of which demonstrated evidence of replication outside the bloodstream (Tan et al., 2023). Although studies have begun to uncover the blood microbiome composition and its mechanisms by which blood microbiome promotes the development and progression of diverse human diseases, however, there are many details remain unknown. Some of the most important unanswered questions that remained include: Why do some healthy individuals have microbes in their blood while others do not, and what is the underlying mechanism for this phenomenon? How do microbes in the blood “deceive” the immune system and live in harmony with the immune system? What is the specific role of bacteria in the onset of a particular disease? What are the advantages and disadvantages of the blood microbes? The blood microbiome in the blood of the diseased population is much higher than that of the healthy population. Is it related to immunity?
Multiple human microbiome studies actively correlate disease states with the structure of the blood microbial community. Interpreting the significance of these cross-sectional studies is challenging due to numerous confounding variables in patient populations. Factors such as structural variations, standardized procedures, incomplete microbial databases, contamination concerns, limited reference databases, unquantified metabolome read-outs, the necessity for functional characterization, and the need for longitudinal studies complicate the analysis. Fortunately, many of these issues can be addressed by conducting intervention studies, standardized protocols, decontamination, improved sequencing technologies, larger sample sizes, rigorous study designs, and strain identification. Further research is necessary to understand the correlation between specific blood bacteria, certain diseases, and their underlying mechanisms.
Different viewpoints exist on the presence of microbiome in the blood of healthy individuals and its association with various human diseases. Some authors support the concept of a healthy blood microbiome, while others suggest that the microbiome disseminates into the blood from other body sites, especially the gut and oral niches. However, it is undeniable that over the past decade, multiple studies have detailed blood microbiome composition and diversity, revealing their association with human health and diseases. Current understanding recognizes the presence of microbes in the blood, emphasizing the complexity of detection and caution needed when associating their genetic material with live bacteria. Observing shifts in the blood microbiome in diabetes, CKD, and CVD suggests their potential role in disease development. Crucial for establishing the blood microbiome as viable biomarkers and innovative therapeutic targets is future research on microbial translocation and physiological mechanisms, with significant implications for healthcare and disease management.
IkK: Conceptualization, Data curation, Investigation, Writing – original draft. ImK: Data curation, Writing – review & editing. AB: Investigation, Writing – review & editing. YX: Data curation, Investigation, Validation, Writing – review & editing. PX: Data curation, Investigation, Writing – review & editing. XX: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. LZ: Conceptualization, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alanazi A., Younas S., Ejaz H., Zainab Mazhari B. B., Abosalif K., Abdalla A. E., et al. (2024). Exploration of the human microbiome’s role in health and disease through the lens of genetics. J. Pure Appl. Microbiol. 18. doi: 10.22207/JPAM.18.3.61
Almeida C., Oliveira R., Baylina P., Fernandes R., Teixeira F. G., Barata P. (2022). Current trends and challenges of fecal microbiota transplantation—an easy method that works for all? Biomedicines 10, 2742. doi: 10.3390/biomedicines10112742
Amar J., Lange C., Payros G., Garret C., Chabo C., Lantieri O., et al. (2013). Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the DESIR study. PloS One 8, e54461. doi: 10.1371/journal.pone.0054461
Amar J., Lelouvier B., Servant F., Lluch J., Burcelin R., Bongard V., et al. (2019). Blood microbiota modification after myocardial infarction depends upon low-density lipoprotein cholesterol levels. J. Am. Heart Assoc. 8, e011797. doi: 10.1161/JAHA.118.011797
Amar J., Serino M., Lange C., Chabo C., Iacovoni J., Mondot S., et al. (2011). Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia 54, 3055–3061. doi: 10.1007/s00125-011-2329-8
Andersen K., Kesper M. S., Marschner J. A., Konrad L., Ryu M., Vr S. K., et al. (2017). Intestinal dysbiosis, barrier dysfunction, and bacterial translocation account for CKD–related systemic inflammation. J. Am. Soc. nephrol. 28, 76–83. doi: 10.1681/ASN.2015111285
Arena R., Guazzi M., Lianov L., Whitsel L., Berra K., Lavie C. J., et al. (2015). Healthy lifestyle interventions to combat noncommunicable disease—a novel nonhierarchical connectivity model for key stakeholders: a policy statement from the American Heart Association, European Society of Cardiology, European Association for Cardiovascular Prevention and Rehabilitation, and American College of Preventive Medicine. Eur. Heart J. 36, 2097–2109. doi: 10.1093/eurheartj/ehv207
Berg G., Rybakova D., Fischer D., Cernava T., Vergès M. C., Charles T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 1–22. 10.1186/s40168-020-00875-0
Bissonnette L., Bergeron M. G. (2010). Diagnosing infections—current and anticipated technologies for point-of-care diagnostics and home-based testing. Clin. Microbiol. Infect. 16, 1044–1053. doi: 10.1111/j.1469-0691.2010.03282.x
Bonhomme D., Cavaillon J. M., Werts C. (2024). The dangerous liaisons in innate immunity involving recombinant proteins and endotoxins: Examples from the literature and the Leptospira field. J. Biol. Chem. 300 (1), 105506. doi: 10.1016/j.jbc.2023.105506
Carnevale R., Sciarretta S., Valenti V., di Nonno F., Calvieri C., Nocella C., et al. (2020). Low-grade endotoxaemia enhances artery thrombus growth via Toll-like receptor 4: implication for myocardial infarction. Eur. Heart J. 41, 3156–3165. doi: 10.1093/eurheartj/ehz893
Castillo D. J., Rifkin R. F., Cowan D. A., Potgieter M. (2019). The healthy human blood microbiome: fact or fiction? Front. Cell. infect. Microbiol. 9, 449041. doi: 10.3389/fcimb.2019.00148
Chen L., Chen R., Wang H., Liang F. (2015). Mechanisms linking inflammation to insulin resistance. Int. J. endocrinol. 2015, 508409. doi: 10.1155/2015/508409
Chen R., Ye Y., Ding Y., Wan Z., Ye X., Liu J. (2024). Potential biomarkers of acute myocardial infarction based on the composition of the blood microbiome. Clinica Chimica Acta 556, 117843. doi: 10.1016/j.cca.2024.117843
D’Aquila P., Giacconi R., Malavolta M., Piacenza F., Bürkle A., Villanueva M. M., et al. (2021). Microbiome in blood samples from the general population recruited in the MARK-AGE project: a pilot study. Front. Microbiol. 12, 707515. doi: 10.3389/fmicb.2021.707515
Dandona P., Aljada A., Bandyopadhyay A. (2004). Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 25, 4–7. doi: 10.1016/j.it.2003.10.013
Demmer R. T., Breskin A., Rosenbaum M., Zuk A., LeDuc C., Leibel R., et al. (2017). The subgingival microbiome, systemic inflammation and insulin resistance: the oral infections, glucose intolerance and insulin resistance study. J. Clin. periodontol. 44, 255–265. doi: 10.1111/jcpe.2017.44.issue-3
Di Gloria L., Baldi S., Curini L., Bertorello S., Nannini G., Cei F., et al. (2024). Experimental tests challenge the evidence of a healthy human blood microbiome. FEBS J. doi: 10.1111/febs.17362
Dinakaran V., Rathinavel A., Pushpanathan M., Sivakumar R., Gunasekaran P., Rajendhran J. (2014). Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PloS One 9, e105221. doi: 10.1371/journal.pone.0105221
Di Vincenzo F., Del Gaudio A., Petito V., Lopetuso L. R., Scaldaferri F. (2024). Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Internal Emergency Med. 19, 275–293. doi: 10.1007/s11739-023-03374-w
Doern G. V., Carroll K. C., Diekema D. J., Garey K. W., Rupp M. E., Weinstein M. P., et al. (2019). Practical guidance for clinical microbiology laboratories: a comprehensive update on the problem of blood culture contamination and a discussion of methods for addressing the problem. Clin. Microbiol. Rev. 33, 10–128. doi: 10.1128/CMR.00009-19
Donath M. Y., Meier D. T., Böni-Schnetzler M. (2019). Inflammation in the pathophysiology and therapy of cardiometabolic disease. Endocr. Rev. 40, 1080–1091. doi: 10.1210/er.2019-00002
Evenepoel P., Poesen R., Meijers B. (2017). The gut–kidney axis. Pediatr. Nephrol. 32, 2005–2014. doi: 10.1007/s00467-016-3527-x
Florea C. M., Rosu R., Moldovan R., Vlase L., Toma V., Decea N., et al. (2024). The impact of chronic Trimethylamine N-oxide administration on liver oxidative stress, inflammation, and fibrosis. Food Chem. Toxicol. 184, 114429. doi: 10.1016/j.fct.2023.114429
Fuster V. (2019). The global burden of cardiovascular diseases and risk factors: 2020 and beyond. doi: 10.1111/trf.13477
Gedgaudas R., Bajaj J. S., Skieceviciene J., Varkalaite G., Jurkeviciute G., Gelman S., et al. (2022). Circulating microbiome in patients with portal hypertension. Gut Microbes 14, 2029674. doi: 10.1080/19490976.2022.2029674
Ghaemi F., Fateh A., Sepahy A. A., Zangeneh M., Ghanei M., Siadat S. D. (2021). Blood microbiota composition in Iranian pre-diabetic and type 2 diabetic patients. Hum. antibod. 29, 243–248. doi: 10.3233/HAB-210450
Glassing A., Dowd S. E., Galandiuk S., Davis B., Chiodini R. J. (2016). Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut pathog. 8, 1–2. doi: 10.1186/s13099-016-0103-7
Guo J., Liu Z., Wang P., Wu H., Fan K., Jin J., et al. (2025). Global, regional, and national burden inequality of chronic kidney disease, 1990–2021: a systematic analysis for the global burden of disease study 2021. Front. Med. 11, 1501175. doi: 10.3389/fmed.2024.1501175
Guo X., Wang Z., Qu M., Guo Y., Yu M., Hong W., et al. (2023). Abnormal blood microbiota profiles are associated with inflammation and immune restoration in HIV/AIDS individuals. Msystems 8, e00467–e00423. doi: 10.1128/msystems.00467-23
Hotchkiss R. S., Moldawer L. L., Opal S. M., Reinhart K., Turnbull I. R., Vincent J. L. (2016). Sepsis and septic shock. Nat. Rev. Dis. prime. 2, 1–21. doi: 10.1038/nrdp.2016.45
Jagare L., Rozenberga M., Silamikelis I., Ansone L., Elbere I., Briviba M., et al. (2023). Metatranscriptome analysis of blood in healthy individuals and irritable bowel syndrome patients. J. Med. Microbiol. 72, 001719. doi: 10.1099/jmm.0.001719
Jager K. J., Kovesdy C., Langham R., Rosenberg M., Jha V., Zoccali C. (2019). A single number for advocacy and communication—worldwide more than 850 million individuals have kidney diseases. Nephrol. Dialysis Transplant. 34, 1803–1805. doi: 10.1093/ndt/gfz174
Karwowska Z., Szczerbiak P., Kosciolek T. (2024). Microbiome time series data reveal predictable patterns of change. Microbiol. Spectr. 12, e04109–e04123. doi: 10.1128/spectrum.04109-23
Khan I., Khan I., Kakakhel M. A., Xiaowei Z., Ting M., Ali I., et al. (2022a). Comparison of microbial populations in the blood of patients with myocardial infarction and healthy individuals. Front. Microbiol. 13, 845038. doi: 10.3389/fmicb.2022.845038
Khan I., Khan I., Usman M., Jianye Z., Wei Z. X., Ping X., et al. (2022b). Analysis of the blood bacterial composition of patients with acute coronary syndrome and chronic coronary syndrome. Front. Cell. Infect. Microbiol. 12, 943808. doi: 10.3389/fcimb.2022.943808
Khan I., Stefan P., Kotb A. A., Arif A. M., Uzair M., Imran K., et al. (2025). Integrated analysis of blood microbiome and metabolites reveals key biomarkers and functional pathways in myocardial infarction. J. Transl. Med., 1–34. doi: 10.1186/s1267-025-06165-3
Kumar A., Roberts D., Wood K. E., Light B., Parrillo J. E., Sharma S., et al. (2006). Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34, 1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9
Lawrence G., Midtervoll I., Samuelsen S. O., Kristoffersen A. K., Enersen M., Håheim L. L. (2022). The blood microbiome and its association to cardiovascular disease mortality: case-cohort study. BMC Cardiovasc. Disord. 22, 344. doi: 10.1186/s12872-022-02791-7
Liberale L., Gaul D. S., Akhmedov A., Bonetti N. R., Nageswaran V., Costantino S., et al. (2020). Endothelial SIRT6 blunts stroke size and neurological deficit by preserving blood–brain barrier integrity: a translational study. Eur. Heart J. 41, 1575–1587. doi: 10.1093/eurheartj/ehz712
Lim Y. J., Sidor N. A., Tonial N. C., Che A., Urquhart B. L. (2021). Uremic toxins in the progression of chronic kidney disease and cardiovascular disease: mechanisms and therapeutic targets. Toxins 13, 142. doi: 10.3390/toxins13020142
Lin T. L., Shu C. C., Chen Y. M., Lu J. J., Wu T. S., Lai W. F., et al. (2020). Like cures like: pharmacological activity of anti-inflammatory lipopolysaccharides from gut microbiome. Front. Pharmacol. 11, 554. doi: 10.3389/fphar.2020.00554
Mancin L., Paoli A., Berry S., Gonzalez J. T., Collins A. J., Lizarraga M. A., et al. (2024). Standardization of gut microbiome analysis in sports. Cell Rep. Med. 5. doi: 10.1016/j.xcrm.2024.101759
Merino-Ribas A., Araujo R., Pereira L., Campos J., Barreiros L., Segundo M. A., et al. (2022). Vascular calcification and the gut and blood microbiome in chronic kidney disease patients on peritoneal dialysis: a pilot study. Biomolecules 12, 867. doi: 10.3390/biom12070867
Moriyama K., Ando C., Tashiro K., Kuhara S., Okamura S., Nakano S., et al. (2008). Polymerase chain reaction detection of bacterial 16S rRNA gene in human blood. Microbiol. Immunol. 52, 375–382. doi: 10.1111/j.1348-0421.2008.00048.x
Nikkari S., McLaughlin I. J., Bi W., Dodge D. E., Relman D. A. (2001). Does blood of healthy subjects contain bacterial ribosomal DNA? J. Clin. Microbiol. 39, 1956–1959. doi: 10.1128/JCM.39.5.1956-1959.2001
Noce A., Marchetti M., Marrone G., Di Renzo L., Di Lauro M., Di Daniele F., et al. (2022). Link between gut microbiota dysbiosis and chronic kidney disease. Eur. Rev. Med. Pharmacol. Sci. 26.
Oktaviono Y. H., Lamara A. D., Saputra P. B., Arnindita J. N., Pasahari D., Saputra M. E., et al. (2023). The roles of trimethylamine-N-oxide in atherosclerosis and its potential therapeutic aspect: A literature review. Biomol. Biomed. 23, 936. doi: 10.17305/bb.2023.8893
Païssé S., Valle C., Servant F., Courtney M., Burcelin R., Amar J., et al. (2016). Comprehensive description of blood microbiome from healthy donors assessed by 16 S targeted metagenomic sequencing. Transfusion 56, 1138–1147.
Parizadeh M., Arrieta M. C. (2023). The global human gut microbiome: genes, lifestyles, and diet. Trends Mol. Med. 29 (10), 789-801. doi: 10.1016/j.molmed.2023.07.002
Perez-Carrasco V., Soriano-Lerma A., Soriano M., Gutiérrez-Fernández J., Garcia-Salcedo J. A. (2021). Urinary microbiome: yin and yang of the urinary tract. Front. Cell. infect. Microbiol. 11, 617002. doi: 10.3389/fcimb.2021.617002
Potgieter M., Bester J., Kell D. B., Pretorius E. (2015). The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol. Rev. 39, 567–591. doi: 10.1093/femsre/fuv013
Qiu J., Zhou H., Jing Y., Dong C. (2019). Association between blood microbiome and type 2 diabetes mellitus: A nested case-control study. J. Clin. Lab. anal. 33, e22842. doi: 10.1002/jcla.2019.33.issue-4
Ralston J., Nugent R. (2019). Toward a broader response to cardiometabolic disease. Nat. Med. 25, 1644–1646. doi: 10.1038/s41591-019-0642-9
Rekha K., Venkidasamy B., Samynathan R., Nagella P., Rebezov M., Khayrullin M., et al. (2024). Short-chain fatty acid: An updated review on signaling, metabolism, and therapeutic effects. Crit. Rev. Food Sci. Nutr. 64, 2461–2489. doi: 10.1080/10408398.2022.2124231
Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., et al. (2014). Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 1–2. doi: 10.1186/s12915-014-0087-z
Sampaio S., Araujo R., Merino-Riba A., Lelouvier B., Servant F., Quelhas-Santos J., et al. (2023). Blood, gut, and oral microbiome in kidney transplant recipients. Indian J. Nephrol. 33, 366–370. doi: 10.4103/ijn.ijn_168_22
Sato J., Kanazawa A., Ikeda F., Yoshihara T., Goto H., Abe H., et al. (2014). Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 37, 2343–2350. doi: 10.2337/dc13-2817
Sciarra F., Franceschini E., Campolo F., Venneri M. A. (2023). The diagnostic potential of the human blood microbiome: are we dreaming or awake? Int. J. Mol. Sci. 24, 10422. doi: 10.3390/ijms241310422
Sedighi M., Razavi S., Navab-Moghadam F., Khamseh M. E., Alaei-Shahmiri F., Mehrtash A., et al. (2017). Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. pathogen. 111, 362–369. doi: 10.1016/j.micpath.2017.08.038
Shah N. B., Allegretti A. S., Nigwekar S. U., Kalim S., Zhao S., Lelouvier B., et al. (2019). Blood microbiome profile in CKD: a pilot study. Clin. J. Am. Soc. Nephrol. 14, 692–701. doi: 10.2215/CJN.12161018
Shah N. B., Nigwekar S. U., Kalim S., Lelouvier B., Servant F., Dalal M., et al. (2021). The gut and blood microbiome in IgA nephropathy and healthy controls. Kidney360 2, 1261–1274. doi: 10.34067/KID.0000132021
Shah J. M., Ramsbotham J., Seib C., Muir R., Bonner A. (2021). A scoping review of the role of health literacy in chronic kidney disease self-management. J. Renal Care 47, 221–233. doi: 10.1111/jorc.12364
Simoes-Silva L., Araujo R., Pestana M., Soares-Silva I., Sampaio-Maia B. (2018). The microbiome in chronic kidney disease patients undergoing hemodialysis and peritoneal dialysis. Pharmacol. Res. 130, 143–151. doi: 10.1016/j.phrs.2018.02.011
Simoes-Silva L., Araujo R., Pestana M., Soares-Silva I., Sampaio-Maia B. (2020). Peritoneal microbiome in end-stage renal disease patients and the impact of peritoneal dialysis therapy. Microorganisms 8, 173. doi: 10.3390/microorganisms8020173
Søby J. H., Watt S. K., Vogelsang R. P., Servant F., Lelouvier B., Raskov H., et al. (2020). Alterations in blood microbiota after colonic cancer surgery. BJS Open 4, 1227–1237. doi: 10.1002/bjs5.50357
Sumida K., Lau W. L., Kovesdy C. P., Kalantar-Zadeh K., Kalantar-Zadeh K. (2021a). Microbiome modulation as a novel therapeutic approach in chronic kidney disease. Curr. Opin. Nephrol. hypertens. 30, 75–84. doi: 10.1097/MNH.0000000000000661
Sumida K., Pierre J. F., Han Z., Mims T. S., Potukuchi P. K., Yuzefpolskaya M., et al. (2021b). Circulating microbial signatures and cardiovascular death in patients with ESRD. Kidney Int. Rep. 6, 2617–2628. doi: 10.1016/j.ekir.2021.07.023
Tan C. C., Ko K. K., Chen H., Liu J., Loh M., Chia M., et al. (2023). No evidence for a common blood microbiome based on a population study of 9,770 healthy humans. Nat. Microbiol. 8, 973–985. doi: 10.1038/s41564-023-01350-w
Tsafarova B., Hodzhev Y., Yordanov G., Tolchkov V., Kalfin R., Panaiotov S. (2023). Morphology of blood microbiota in healthy individuals assessed by light and electron microscopy. Front. Cell. Infect. Microbiol. 12, 1091341. doi: 10.3389/fcimb.2022.1091341
Tuttolomondo A., Daidone M., Pinto A. (2020). Endothelial dysfunction and inflammation in ischemic stroke pathogenesis. Curr. Pharm. Design. 26, 4209–4219. doi: 10.2174/1381612826666200417154126
Upadhyaya S., Banerjee G. (2015). Type 2 diabetes and gut microbiome: at the intersection of known and unknown. Gut Microbes 6, 85–92. doi: 10.1080/19490976.2015.1024918
Valenbreder I. M., Balăn S., Breuer M., Adriaens M. E. (2023). Mapping out the gut microbiota-dependent trimethylamine N-oxide super pathway for systems biology applications. Front. Syst. Biol. 3, 1074749. doi: 10.3389/fsysb.2023.1074749
Vaziri N. D., Zhao Y. Y., Pahl M. V. (2016). Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol. Dialysis Transplant. 31, 737–746. doi: 10.1093/ndt/gfv095
Velmurugan G., Dinakaran V., Rajendhran J., Swaminathan K. (2020). Blood microbiota and circulating microbial metabolites in diabetes and cardiovascular disease. Trends Endocrinol. Metab. 31, 835–847. doi: 10.1016/j.tem.2020.01.013
Violi F., Cammisotto V., Bartimoccia S., Pignatelli P., Carnevale R., Nocella C. (2023). Gut-derived low-grade endotoxemia, atherothrombosis, and cardiovascular disease. Nat. Rev. Cardiol. 20, 24–37. doi: 10.1038/s41569-022-00737-2
Wang M., Feng J., Zhou D., Wang J. (2023). Bacterial lipopolysaccharide-induced endothelial activation and dysfunction: a new predictive and therapeutic paradigm for sepsis. Eur. J. Med. Res. 28, 339. doi: 10.1186/s40001-023-01301-5
Wehedy E., Shatat I. F., Al Khodor S. (2022). The human microbiome in chronic kidney disease: a double-edged sword. Front. Med. 8, 790783. doi: 10.3389/fmed.2021.790783
Yuan X., Yang X., Xu Z., Li J., Sun C., Chen R., et al. (2024). The profile of blood microbiome in new-onset type 1 diabetes children. iScience. 27 (7).doi: 10.1016/j.isci.2024.110252
Zhou B., Lu Y., Hajifathalian K., Bentham J., Di Cesare M., Danaei G., et al. (2016). Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4· 4 million participants. Lancet 387, 1513–1530. doi: 10.1016/S0140-6736(16)00618-8
Keywords: CMDS, blood microbiome, dysbiosis, biomarkers, therapeutics
Citation: Khan I, Khan I, Bacha AS, Xiaohui Y, Xie P, Xie X-d and Li Z (2025) Blood microbiome and cardiometabolic disease: insights, therapeutic strategies, and future directions. Front. Bacteriol. 4:1541085. doi: 10.3389/fbrio.2025.1541085
Received: 09 December 2024; Accepted: 12 February 2025;
Published: 03 March 2025.
Edited by:
Darina Cejkova, Brno University of Technology, CzechiaReviewed by:
Santosh Dulal, Management Sciences for Health, United StatesCopyright © 2025 Khan, Khan, Bacha, Xiaohui, Xie, Xie and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-dong Xie, eGR4aWVAbHp1LmVkdS5jbg==; Zhiqiang Li, bGl6aGlxaWFuZzY3NjdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.