- 1School of Biotechnology and Biomolecular Sciences, University of New South Wales, Sydney, NSW, Australia
- 2Recombinant Products Facility, University of New South Wales, Sydney, NSW, Australia
- 3Department of Chemistry, King’s College London, London, United Kingdom
- 4Nuclear Magnetic Resonance (NMR) Facility, Mark Wainwright Analytical Centre, University of New South Wales, Sydney, NSW, Australia
- 5School of Molecular Sciences, Arizona State University, Tempe, AZ, United States
- 6School of Biological Earth and Environmental Sciences, University of New South Wales, Sydney, NSW, Australia
Here, we review the processes involved in producing and assessing the quality of recombinant spider silk proteins (spidroins) and the challenges associated with their synthesis and spinning into robust fibres. We provide an overview of the techniques used to produce the proteins, from gene synthesis to expression in various host organisms. Evidence suggests that the N- and C-terminal regions of spidroins are of utmost importance for fibre assembly and the repetitive domains are responsible for the unique mechanical properties in both native and recombinant versions of spider silks. We describe the role of liquid–liquid phase separation (LLPS) in spidroin assembly and its importance in subsequent fibre formation. Recent developments in recombinant spidroin production and co-expression strategies for improving yield and scalability are highlighted. Techniques such as mass photometry and size exclusion chromatography (SEC) for analysing protein purity and assembly behaviour are thereupon detailed. Finally, we address the role that predictive computational methods play in the future of designing novel and high-performing materials inspired by spidroins.
Introduction
Spider silk has unique physical properties that make it attractive to utilise in applications that require lightweight materials with high tensile strength. Its properties include not just exceptional strength and toughness, but also high flexibility, unique anisotropy, and in situ biological inertness (Heim et al., 2009; Vollrath et al., 2013; Blamires et al., 2020; Blamires, 2022; Linklater et al., 2023). The successful replication of spider silk’s properties within fibres spun in the laboratory would undoubtedly open exciting opportunities for the production of extremely high-performing materials for applications in medicine, textiles, wearables, and materials engineering (Kluge et al., 2008; Leal-Egaña and Scheibel, 2010; Humenik et al., 2011; Heidebrecht et al., 2015; Blamires, 2022, Blamires, 2024).
Spiders are solitary and cannibalistic, and they need to be anaesthetised for silk to be spooled from their spinnerets for several hours at a time. Moreover, individual spiders yield proportionally smaller masses of silk than can be conventionally attained from an individual silkworm cocoon. Accordingly, it is not feasible to harvest spider silk by the same methods as silkworm silk is harvested (Blamires, 2022). The only way to possibly generate large amounts of spider silk, or spider silk-like material, is by the recombinant production of spider silk proteins (so called spidroins) followed by purification and the utilisation of advanced wet or dry fibre spinning methodologies to facilitate spinning the proteins into robust fibres (Lewis et al., 1996; Scheller et al., 2001; Wong Po Foo and Kaplan, 2002).
Recombinant spider silk proteins (or more strictly speaking proteins inspired by spider silk sequences) are produced using techniques that involve, firstly, the synthesis of a gene encoding the spidroin-like construct, insertion of this construct into a synthetic expression vector (such as plasmid DNA) and then transforming an expression host (such as Escherichia coli, yeast (Saccharomyces cerevisiae), plants (e.g. potato, tobacco) or animals (e.g. silkworms, hamsters, goats) (Scheller et al., 2001; Menassa et al., 2004; Miao et al., 2006; Chung et al., 2012; Teulé et al., 2012; Mi et al., 2023). Spidroins are produced by the host’s protein expression system, which can then be isolated and concentrated into a solution called dope. Generating recombinantly engineered synthetic fibres with physical qualities that resemble native spider silk fibres, however, remains one of material science’s grandest challenges (Humenik et al., 2011; Jastrzebska et al., 2016; Blamires, 2024). Some specific issues associated with producing recombinant spider silk proteins, and the spinning of fibres from those proteins, include inducing the hosts to express recombinant spidroins, and obtaining a sufficient quantity of proteins at concentrations high enough to spin the resulting dope into exceptionally robust fibres (Copeland et al., 2015; Harris et al., 2016)
Inducing host organisms to express full-length spidroins necessitates the use of sophisticated techniques, some of which we overview herein. Recent advances in DNA sequencing, DNA synthesis and improved protein structural prediction have enabled an acceleration in the construction of genes encoding recombinant proteins inspired by spider silk proteins. However, many difficulties remain in terms of expressing and purifying these constructs in a scalable manner (Scheller et al., 2001; Schacht and Scheibel, 2014; Rising and Johansson, 2015; Koeppel and Holland, 2017; Edlund et al., 2018).
Difficulties associated with expressing the large repetitive amino acid sequences of the spidroins include plasmid stability, difficulties in host-cell translation, the potential of protein misfolding that accompanies the expression of large and repetitive proteins and the potential toxicity of certain sequences to the host cells (Heim et al., 2010; Rising and Johansson, 2015). Purification of adequate quantities of the proteins yielded is another challenge. Indeed, efficient purification of high-quality samples of recombinant proteins is critical for the implementation of all of the downstream applications (Oliveira and Domingues, 2018; Tripathi and Shrivastava, 2019). This is all the more critical for the synthesis of spider dragline, or major ampullate, silks (i.e.,those silk whose toughness has been compared to that of Kevlar and carbon fibres; Blamires, 2022) because the quality of the fibres spun from these proteins is a product of the concentrations of the spidroins and factors associated with spinning protocols (Humenik et al., 2011; Liu et al., 2014; Doblhofer et al., 2015; Peng et al., 2016). Thus, we detail herein how the gene construction, protein expression and purification, structural characterisation protocols, and downstream fibre fabrication methodologies are performed in vitro to generate native-like spidroin-like fibres.
Recombinant expression of spidroins
Much of the reported work on recombinant spider silk protein expression has focused on the spider silk fibroins (known as spidroins). The first reported construction of a recombinant spider silk protein was in 1995, when constructs of varying length inspired by both Major Ampullate spidroin 1 and 2 (shortened conventionally to MaSp1 and Masp2) proteins from the golden orb web spider Triconephila clavipes were created using the gram-negative bacteria E. coli as the host (Prince et al., 1995). Since then, there have been many reports describing strategies to produce recombinant silks for research and commercial development (Tokareva et al., 2014). The primary advantage of spider silk protein expression by recombinant techniques is that it has a greater capacity for larger scale production than does collecting the silk directly from spiders.
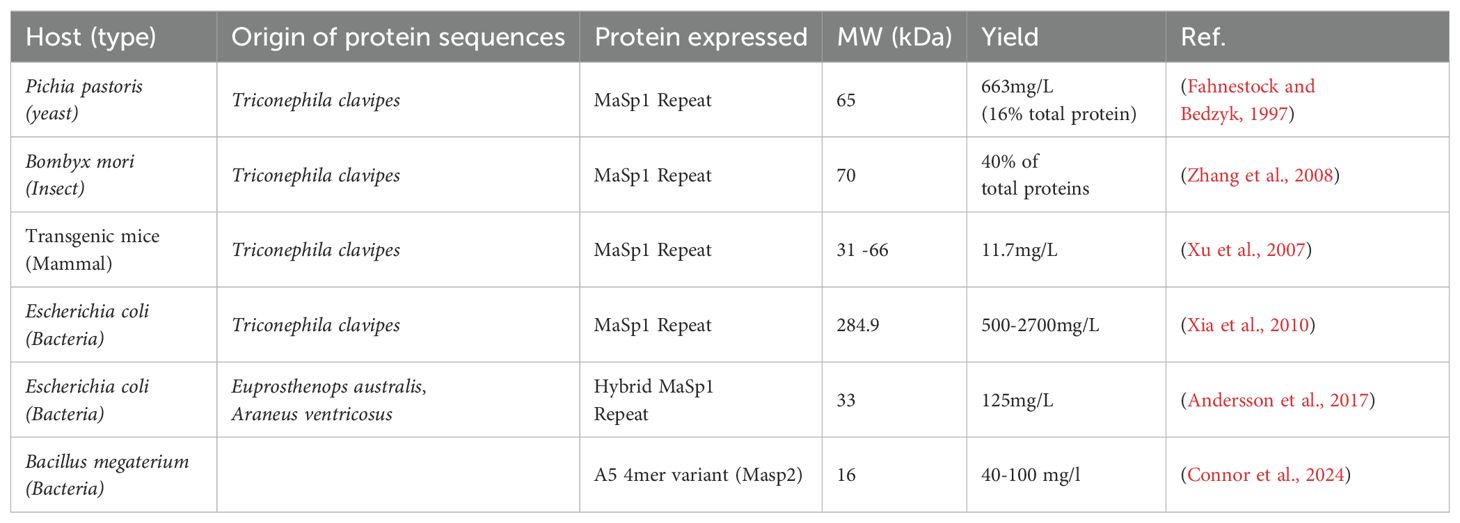
Much of the work thus far has focused on Major Ampullate Spidroin Proteins (MaSp), MaSp1 and MaSp2 due to their contribution to the strength and elasticity of dragline silk fibres. The remarkable properties of MaSp1 and MaSp2 can be attributed to their composition, i.e. a non-repetitive and partially conserved N- and C-terminus that flanks a repetitive core domain (Ayoub et al., 2007a; Römer and Scheibel, 2008; Gaines et al., 2010; Andersson et al., 2017; Xu et al., 2017). Recombinantly-derived sequences have hence been generated using conserved N- and C-terminal domains from single or different spider species, creating so called “chimeric” structures. These constructs also commonly have varying repeating numbers of conserved glycine- and alanine-rich domains, which create polypeptides of shorter length than the native proteins (Figure 1A).
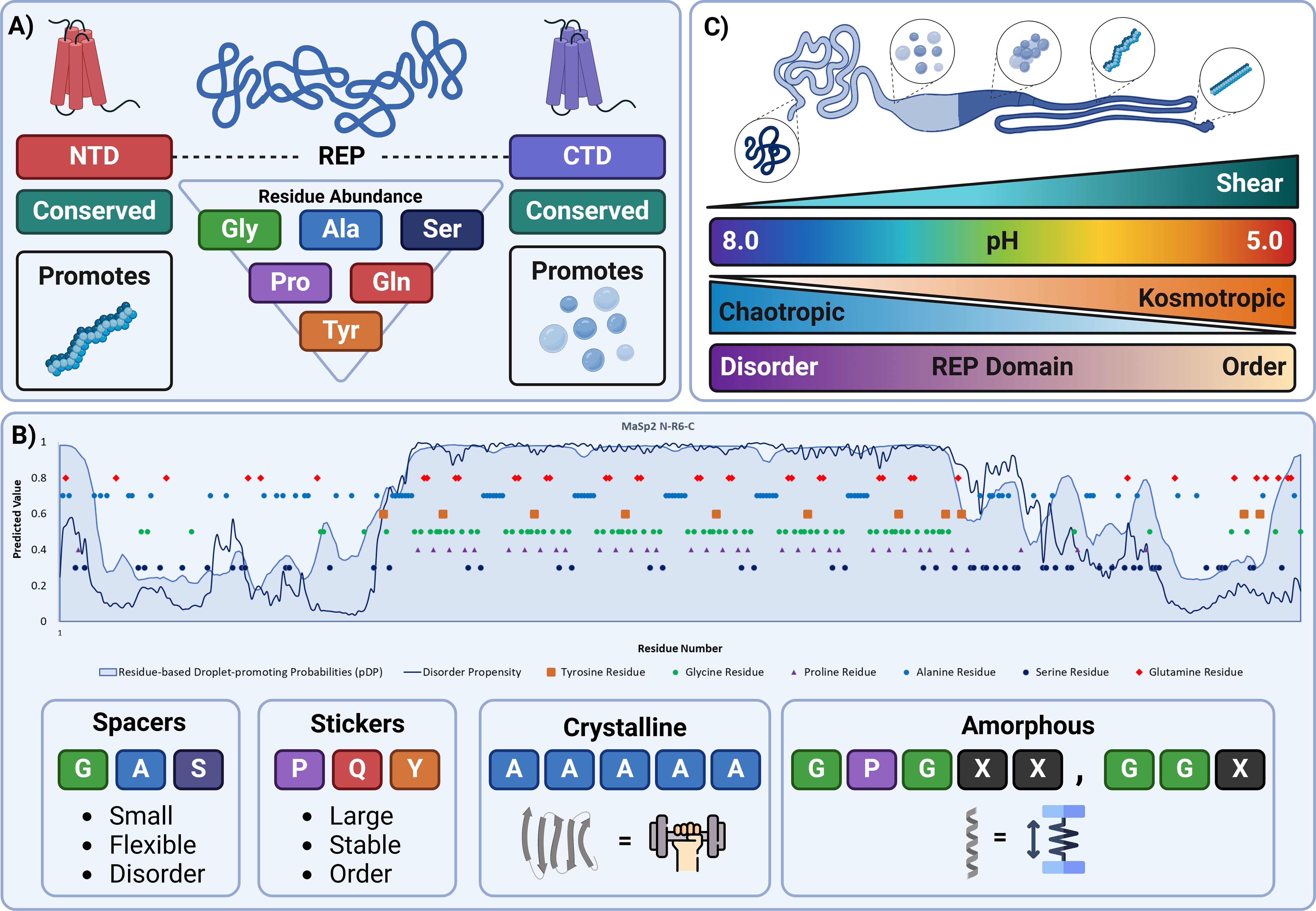
Figure 1. Sequence-structure-environment relationship of recombinant spidroin proteins. (A) Spidroin proteins are comprised of structurally resolved and conserved N- and C-terminal domains, responsible for promoting fibrillation and liquid-liquid phase separation respectively. These domains flank an intrinsically disordered repetitive domain. The abundance of residues commonly found within the repetitive domain are organised from most abundant (top) to least abundant(bottom). (B) (Top) The sequence map of a published recombinant MaSp2 protein highlighting the relationship between residue occurrence (orange-tyrosine, green-glycine, purple-proline, alanine-blue, serine-indigo, glutamine-red), structural disorder (Hu et al., 2021) (black line) and phase separation propensity (Hardenberg et al., 2020; Vendruscolo and Fuxreiter, 2022) (blue shaded region). (Bottom) The relationship of residues found within a spidroin sequence are shown categorised to their respective sticker (proline, glutamine and tyrosine) and spacer (glycine, alanine and serine) model role, as well as the motifs commonly found within the repetitive region and the corresponding structural and mechanical property they confer. (C) The anatomy of the spidroin duct and corresponding environmental shifts. Protein is synthesised in the tail (left of light blue) in a high pH, low shear, chaotropic environment. Free monomer then travels down to the sac (right of light blue) where liquid-liquid phase separation occurs due to an influx of kosmotropic ions. As shear and kosmotropic ion concentration increases and a subsequent decrease in pH occurs, condensates begin to coacervate and form nanofibril structures, progressively becoming more aligned until they are extruded out of the duct (dark blue). Figures created using BioRender.
Many biotech companies are now developing recombinant spider silk fibres using genetically modified organisms such as bacteria, yeast, and silkworms with some success. However, there are challenge associated with the limitations of such hosts to express recombinant proteins at high levels and/or consistently produce quality fibres.
Impact of N- and C-termini composition
The solubility and spinning characteristics are heavily influenced by both the N-terminus (NT) and the C-terminus (CT) of the silk proteins. The NT and CT regions are not only conserved amongst species but also spidroin types (Dicko et al., 2004; Ayoub et al., 2007a; Kronqvist et al., 2014). It was determined that across 12 different species, and multiple fibre types, more than 50% of the amino acids were congruent (Ayoub et al., 2007a). Such similarity amongst N- and C-termini between species and spidroins suggest they are integral in the assembly of spider silk proteins which must be considered when designing constructs for recombinant expression (Dicko et al., 2004; De Oliveira et al., 2024). This has indeed been confirmed by studies inducing recombinant expression of the N- and C-terminal domains of major ampullate spidroins, whereby their role in fibre assembly solubilising effect and structural solving have been accomplished (Figure 1A) (Askarieh et al., 2010; Malay et al., 2020; De Oliveira et al., 2024).
Structural motifs and importance of the repetitive domain
While the terminal domains are responsible for polymerisation and drive a portion of the assembly process, the repetitive domain is predominantly responsible for the unique mechanical properties and intrinsically disordered behaviour of spidroin fibres in solution. As a result, repetitive domain sequences have been the source of extensive investigation due to their direct relationship to fibre mechanical properties.
Spidroin paralog and phylogenetic analyses have confirmed the convergence of key residues across sequences (Ayoub et al., 2007a; Craig et al., 2020; Arakawa et al., 2022). The abundance of comparatively smaller glycine, alanine, and serine residues within the low complexity repetitive region contribute most heavily to fibre structural flexibility as a consequence of disorder of the domains within the highly soluble regions of the silk gland, i.e., in and around the tail and sac (Figures 1A, B) (Oktaviani et al., 2018; Yang et al., 2023).
Consistent with many other proteins that have a high degree of intrinsic disorder, spidroins have demonstrated the ability to phase separate, a phenomenon where proteins separate into two phases: a proteinaceous dense phase and a buffer light phase (Malay et al., 2020; Mohammadi et al., 2020). The sticker-spacer model is often used to describe residue-residue interactions that drive liquid-liquid phase separation (LLPS). Stickers are residues which act as molecular glues and hold proteins together, whereas spacers are flexible residues that have low interaction strengths with other residues. In the context of spidroins abundant residues like glycine and alanine act as spacers, providing flexibility to the protein chain. Additionally, spacer residues also spatially control the interactions between less abundant but more reactive sticker residues like tyrosine, glutamine and proline which can enter π- π, π-cation and charge interactions ultimately driving phase separation. Understanding the relationship between sticker-spacers and their patterning provides a platform to develop de novo biomaterials for a diverse range of applications not limited to tissue engineering, synthetic organelles and encapsulants (Martin and Mittag, 2018; Martin et al., 2020; Maraldo et al., 2024).
Considering the repetitive nature and low complexity of the repetitive domain, several motifs can be observed and used to classify the subspecies of MaSp to which they belong (Ayoub et al., 2007a; Arakawa et al., 2022). Most MaSp constructs are primarily formed with two major subunits; a glycine rich GGX or GPGXX where “X” denotes any amino acid and often a sticker residue (Tokareva et al., 2013). These motifs are then typically followed by a poly-alanine sequence spanning 4-7 residues on average (Figure 1B) (Gaines et al., 2010). The spatial arrangement of these residues ensures that sticker-sticker interactions are finely balanced preventing premature aggregation, and the formation of highly concentrated dope.
As fibre formation proceeds, the role of these residues transforms. These repetitive motifs in response to stimuli changes like pH, shear and kosmotropic buffer exchange in the duct begin to form defined secondary structures that are responsible for the unique mechanical properties spidroins are well-known for (Dong et al., 1991; Gosline et al., 1999; Hayashi et al., 1999; Gu et al., 2020). The comparatively smaller size of the most abundant residues enables the formation of highly compact β-sheet content due to minimised interchain distance (Van Beek et al., 2002; Keten et al., 2010). Evidence suggests that poly-alanine sequences form hydrophobic β-sheet crystalline domains provide silk with high tensile strength, whereas the glycine-rich regions form hydrophilic extensible 310-helical structures and account for the elasticity of silk– (Figure 1B) (Dong et al., 1991; Kümmerlen et al., 1996; Ayoub et al., 2007a). Recent work comparing over 1000 spider silk species transcriptomic data has provided unparalleled insight into the relationship between the sequence-property relationship and serves as a platform for designing and modifying the aforementioned motifs to create fibres with tuneable properties (Arakawa et al., 2022).
Impact of the repetitive domain on recombinant expression
Native spidroins are polypeptides of approximately 200–350 kDa (Kümmerlen et al., 1996; Hayashi et al., 1999; Van Beek et al., 2002; Chaw et al., 2017). Due to their length and the repetitive nature of MaSps, several challenges arise when utilising heterologous hosts for expression. When considering the two most common subclasses, MaSp1 repetitive sequences have been shown to be more homogenous than those of MaSp2 (Ayoub et al., 2007a; Craig et al., 2020). As a result, recombinant forms of MaSp1 have historically been more difficult to develop compared to MaSp2, in part because of limitations on producing highly repetitive synthetic gene sequence constructs.
Thankfully, several molecular biology techniques have been developed to enable the assembly of full length spidroin proteins from a single motif repeat, namely recursive directional ligation (McDaniel et al., 2010). Alternatively, recent circular RNA production serves as another promising avenue for generating native-length templates without the requirement for duplicating a single repeat, instead translational units loop over a single construct and insert fragments to generate multimeric repeats (Lee et al., 2021; Liu et al., 2022).
However, despite the ability to reliably produce vectors for recombinant expression, the main challenge lies in the lack of specialised tissue and translational machinery in the commonly used heterologous hosts (Candelas et al., 1990; Xia et al., 2010; Sonavane et al., 2024). As residue diversity is low within MaSp sequences, spiders have evolved to overexpress tRNAs for highly abundant residues like glycine and alanine to prevent translational bottlenecking (Candelas et al., 1990; Ayoub et al., 2007a; Xia et al., 2010; Tokareva et al, 2013). Comparatively, when expressing “native like” spidroins in heterologous hosts, a decrease or complete lack of expression compared to truncated forms is observed due to the exponential increase in the translational stress it places on the host (Xia et al., 2010).
Solutions for heterologous expression systems particularly within E. coli systems, have taken inspiration from spider physiology. Co-expression of recombinant forms of MaSp with a plasmid that supplies additional glycine and alanine t-RNAs showed significant improvement in expression (Xia et al., 2010; Cao et al., 2017). This co-expression strategy along with incubating the bacteria in media with elevated glycine and alanine levels allows expression of large “native like” spidroins with comparable yields to those of smaller-sized repeat proteins (Cao et al., 2017). Proteomic analyses have confirmed such expression patterns (Ramezaniaghdam et al., 2022). In conjunction with further codon optimisation, challenges pertaining to sequence repetition and size, on recombinant expression, can be mitigated when attempting to produce recombinant MaSps.
Liquid–liquid phase separation as an emerging pre-assembly paradigm
Liquid–liquid phase separation (LLPS) of proteins is a mechanism for the organisation and regulation of the intracellular environment with underlying driving forces, which typically include hydrophobic, aromatic, and/or electrostatic interactions regulated by temperature, (osmotic) pressure, or ion concentration (i.e., osmolyte, pH, salt concentration) (Dai et al., 2022). Protein phase separation is also known to be sensitive to protein concentration, sequence, and length. The aqueous solvent based LLPS of biopolymers such as proteins, leads to a dense phase in thermodynamic equilibrium with a dilute phase. The dispersed dense phase domains (biopolymer rich phase) are often referred to as coacervates or biomolecular condensates.
The formation of spider silk fibrils from soluble spidroin precursors during spinning is a complex process involving spidroin proteins encountering multiple chemical and physical gradients (e.g., numerous ion fluxes, water dehydration, CO2, shear force). There is mounting evidence that LLPS plays a critical role in mediating the self-assembly and aggregation processes involved in producing the unique molecular and hierarchical structures involved in spider silk fibre spinning (Yarger et al., 2018; Mohammadi et al., 2020; Perera et al., 2023). Much of our current understanding of spider silk assembly via LLPS comes from experimental studies of recombinant spidroins (Lemetti et al., 2019, Lemetti et al., 2022). For example, probing of a recombinant MaSp2 showed phosphate ion-induced LLPS at neutral pH which resulted in the formation of coacervates (Malay et al., 2020). Furthermore, if the pH is lowered during spinning, as happens within the silk gland of spiders, the coacervates become fibrous (Figure 1C). It has also been shown through the microfluidic spinning of recombinant MaSp2 that polyalanine blocks have limited influence on the occurrence of LLPS and hierarchical structure (Chen et al., 2024). The role of the N- and C-terminal domains and repetitive domains in LLPS and resulting fibril formation have also been explored using recombinant proteins, providing strong evidence that the C-terminal domain, repetitive domain and several key residues play a central role in LLPS, and the N-terminal domain is essential for subsequent fibril assembly (Malay et al., 2020; Watanabe and Arakawa, 2023).
LLPS is also being explored as a potentially useful mechanism to control protein solubility during expression and accumulation in the cytoplasm of E. coli as well as a property fit for developing non-canonical materials, enabling the further development of recombinant spidroin proteins for biomimetic spider silk materials and biomedical applications (Gabryelczyk et al., 2022; Dai et al., 2022; Maraldo et al., 2024). Evidence of such has already been shown through the generation of a synthetic organelle capable of performing enzymatically driven reactions within puncta of the cell using a recombinantly expressed MaSp2 (Wei et al., 2020). The importance of sequence design in both the terminal and repetitive domains is therefore integral in dictating the assembly behaviour and subsequent mechanical properties of formed fibres.
Formation of fibres in vitro
The methodologies involved in spinning spider silk fibres from recombinantly expressed spidroins are complex and multifaceted, particularly when aiming to replicate the natural spinning process of spiders. As stated, the success of any methodology at reproducing the properties of native silk is dependent on producing a spinning dope at appropriate concentrations. The spinning process itself aims to emulate the conditions under which spiders naturally spin their silk. This includes an aqueous medium, ambient temperature, low hydrostatic pressure, slow extrusion rates, and low post-spinning draw-down ratios (Figure 1C). Researchers have used various methods to spin silk fibres from recombinant spider silk proteins, including microfluidic spinning and post-stretching techniques. These methods aim to align the spidroins and induce the formation of β-sheet structures (Chung et al., 2012; Leclerc et al., 2013; Peng et al., 2016).
Studies have shown that factors such as temperature, pH, and ionic balance within the spinning column, and the speed at which the fibres are spooled, play significant roles in affecting the mechanical properties of the silk (Figure 1C). Higher reeling speeds tend to increase the orientation of spidroin molecules, leading to stiffer and stronger, but less extensible, fibres (Shao and Vollrath, 2002; Shao et al., 2003; Gronau et al., 2013; Oktaviani et al., 2023). Additionally, the acidification of the spinning dope during extrusion aids in the transition of spidroins from random coil and α-helix conformations to β-sheet structures (Gaines et al., 2010; Peng et al., 2016; Oktaviani et al., 2023). The incorporation of post-treatment steps, such as post-drawing in air and ethanol solutions, has been shown to improve the crystalline and molecular structure of the fibres, enhancing their mechanical properties (Hudspeth et al., 2012; Doblhofer et al., 2015; Peng et al., 2016).
Host selection for recombinant spidroin expression
Many different hosts have been used for expressing recombinant spidroins (Table 1, Figure 2A). The majority of reports describe the use of E. coli (Prince et al., 1995; Fahnestock and Bedzyk, 1997; Xu et al., 2007; Teulé et al., 2012). Reasons for the selection of E. coli include: (i) its ease of manipulation utilising plasmid transformation, (ii) extremely short regeneration times, and (iii) low cost and proven scale-up potential (Figure 2B). The methylotrophic yeast Pichia pastoris (now Komagatella phaffi) has an advantage over E. coli of being able to secrete the proteins and facilitate more complex post-translational modifications (Cregg et al., 2000; dos Santos-Pinto et al., 2014).
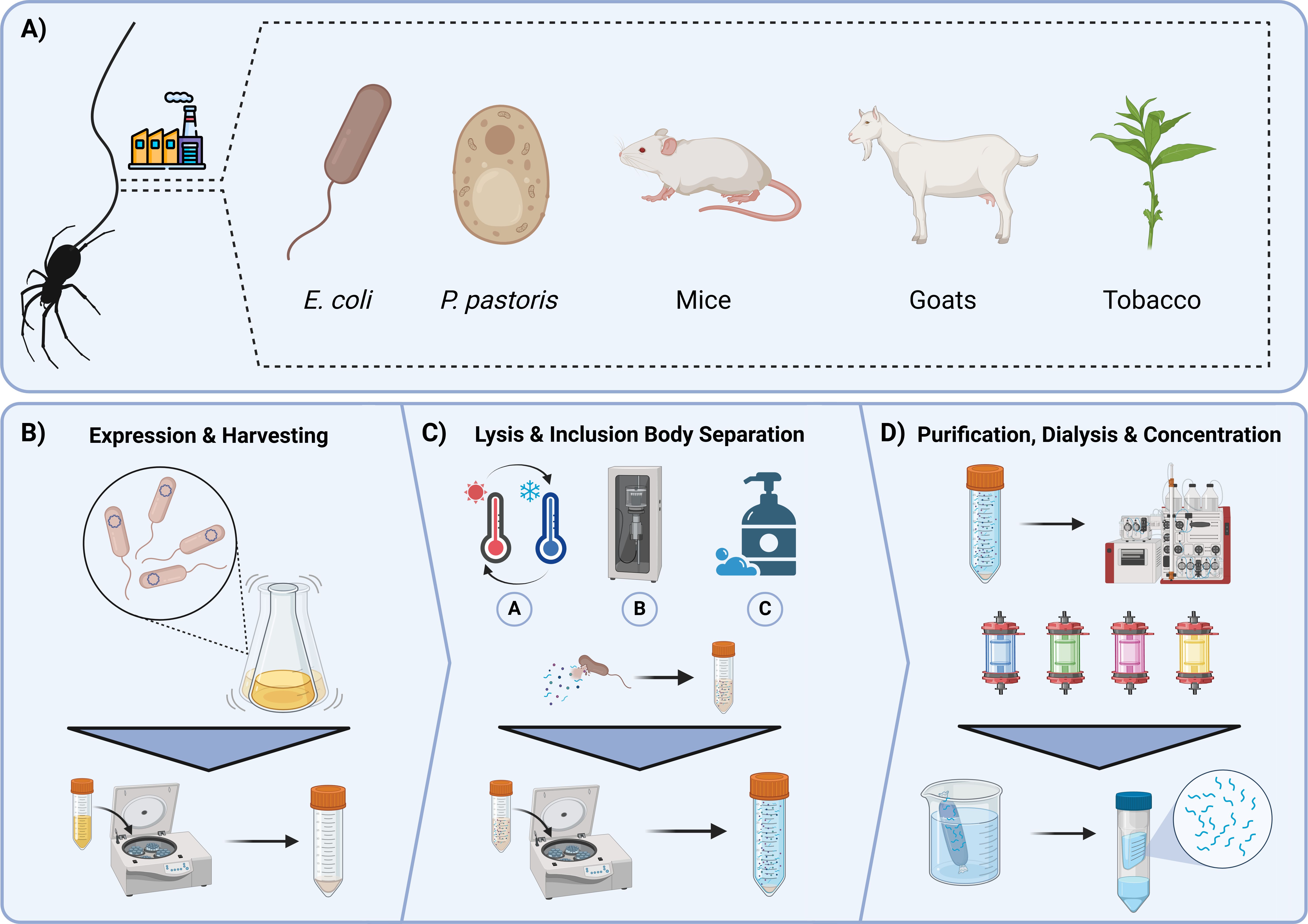
Figure 2. A common bioprocessing pathway to expressing and isolating spidroins. (A) Several well documented heterologous hosts capable of expressing recombinant spidroin proteins, E. coli, P. pastoris, mice, goats, tobacco (left to right). (B) Expression and harvesting of recombinantly expressing E. coli. Transformed E. coli cells are cultured in shaking flasks and later induced. Cells are harvested and pelleted via centrifugation. (C) Harvested cells can be lysed using several different methods, freeze-thaw cycling (A), sonication (B), and chemical lysing agents (C). Inclusion bodies and soluble protein are separated through centrifugation. (D) Soluble proteins are passed through an affinity column of choice. The purified protein is then dialysed into a desired storage buffer, which can then be concentrated for analytical or fibre-producing purposes. Figures created using BioRender.
Many studies have reported a decrease in replication fidelity when expressing proteins with sizes larger than ~43 kDa (Xia et al., 2010; Teulé et al., 2012; Cao et al., 2017). However, for synthesising spider silks, expression of proteins of this size or greater is required to create fibres that approximate the mechanical properties of the natively spun fibres. While solutions we highlighted previously may address this problem, resolving expression remains a major challenge for recombinant spider silk production (Blamires, 2024).
Recombinant protein production and purification
For spinning spider silk fibres, a high level of purity is desirable for optimum downstream assembly. Contaminants from E. coli or other expression systems could either cause damage to the protein of interest, e.g., by degradation or obstruction of the assembly of fibres.
Before purification, the cells of the expression system, most commonly E. coli, must be lysed to release their contents. There are a variety of ways to go about this and the approach taken depends on the downstream use of the proteins (Figure 2C) (Islam et al., 2017). Commonly used methods include ultrasonication, which harnesses the cavitation of air bubbles, pressure homogenisation, multiple freeze/thaw cycles, or the use of chemical lysing agents (e.g. BugBuster) to “melt” the cells. Some methods may result in damage to the sample protein (e.g. when using ultrasonication due to sample warming) whereas others could be prohibitive when dealing with larger samples (e.g. bigger volumes may require more freeze/thaw cycles). It is therefore important to tailor the method used to the specific case and its requirements. The resultant cell lysate from the lysis step then undergoes a series of downstream purification steps to reach a desired level of purity. The overexpressed proteins may be soluble and isolated from the cell cytosol following cell lysis. Alternatively, during expression the protein may form insoluble “inclusion bodies” and require subsequent solubilisation and/or refolding (Tsumoto et al., 2003; Singh et al., 2015). The propensity to form these insoluble inclusion bodies is very protein and/or system dependent and thus a significant degree of optimisation and trial-and-error may be required during recombinant protein expression and purification experiments.
To achieve the required level of purity, a multistep purification protocol is generally recommended (Figure 2D). Most commonly this would begin with an affinity step, utilising a tag that has been incorporated into the expression construct. Many options are available (Pan et al., 2019), including the traditional 6-His tag or its newer equivalents HAT and NCTR25 (based on a human protein to avoid immunogenicity in downstream applications; Young et al., 2012; Pan et al., 2019), which selectively binds to immobilised divalent metal cations like Ni2+ and Co2+ and can be released using an imidazole gradient. Others include the streptavidin-based tags eluted with biotin, maltose binding protein (MBP), which binds to amylose resin and is eluted using maltose, and glutathione-S-transferase, which binds to glutathione resin and is eluted with reduced glutathione. Recently a new generation of tags has been released including the Protein Select™ system by Cytiva™ (Clifford et al., 2024), in which the desired protein self-cleaves from its tag during purification leaving no residual amino acids. Alternative strategies to affinity chromatography include ion-exchange or hydrophobic interaction chromatography, which exploit the natural polarity of the protein construct in order to facilitate the separation of the target from the host cell contaminants. The initial steps, which involve either affinity, ion-exchange or hydrophobic interaction chromatography, all serve to achieve the bulk cleaning of the protein sample from contaminants prior to a later size-exclusion step (McCue, 2009; Cummins et al., 2017)
In-solution analyses
Analytical and preparative mass determination
When producing recombinant protein subunits as building blocks for silk fibre assembly, it is useful to identify conditions in which the proteins are reliably monodispersed. Hence, the purification will almost always require size exclusion chromatography to investigate the oligomeric entities within the proteins and isolate the monomeric fraction.
Size exclusion chromatography (SEC), also known as gel filtration, relies on a separation matrix whose pores proportionally restrict the flow of proteins small enough to penetrate them to varying extents, while leaving larger proteins to flow straight through in the ‘‘void volume’’ with further proteins eluting by decreasing size (Figure 3). In this way, larger proteins may be separated out from smaller proteins. Appropriate SEC columns should be selected based on the size range of the protein mixture being separated. Some SEC matrices allow for the separation of a larger range of sizes, but with correspondingly lower resolution, and vice-versa. Elution is monitored spectroscopically at a range of wavelengths; fractions are collected, and oligomeric state can be estimated by comparison to standards. In this way SEC serves as a useful check, and it allows for the protein target to be placed into the appropriate buffer for downstream uses (Burgess, 2018). It may also be coupled to other instrumentation to allow for further analysis (e.g. SEC-MALS (Thomsen, 2020) or SEC-SAXS). This could be useful, for example, in instances of silk fibre formation whereby the production of higher-order oligomers and protofilaments may be followed (Sahin and Roberts, 2012).
Figure 3. Analytical processes for quantifying protein quality, assembly behaviour and structural conformation. Conditions observed in the duct are shown at the top. Ions present in the duct transition from chaotropic to kosmotropic, pH decreases from high to low, whereas shear increases from low to high resulting in a structural transformation from a disordered to ordered state. Tail – Size exclusion chromatography is used as an analytical and preparatory method to quantify and separate proteins of various sizes. Similarly, Mass photometry can be used to rapidly quantify size distributions of protein samples using the scattering light of protein molecules falling onto a glass slide. Sac – Absorbance measures can be used to generate maps that illustrate protein-environment conditions. Points of interest on these maps can be visualised using microscopy to determine whether phase separation is occurring. Droplet dynamics can then be quantified using Fluorescent Recovery after Photobleaching (FRAP) to understand protein diffusion within phase separation entities. Duct – More advanced spectroscopic methods can be used to determine structural transformations at various stages in the assembly process like Circular Dichroism (CD). Kinetics of this assembly can be monitored through UV-Vis spectroscopy where monitoring of ThT Fluorescence can indicate β-sheet formation. Once fibres are generated morphology can be monitored and assessed using electron microscopy methods.
SEC is an established method for fractionating a polydisperse spidroin solution, nonetheless its efficiency and ability to capture real time assembly is limited. A newly developed light-based method called mass photometry is able to accurately and efficiently determine the molecular weights of a single molecule, and protein assemblies with extremely low volumes and concentrations (Cole et al., 2017; Young et al., 2018). As a result, mass photometry has been used to quantify assembly properties, polydispersity and protein behaviour in various buffers at reading intervals relevant to native assembly processing (Cole et al., 2017; Qi et al., 2023). The potential to utilise mass photometry as a powerful analytical tool has been highlighted in amyloid fibril research – with a particular focus on neurodegenerative-associated peptides like α-synuclein. Methodological developments have enabled high-resolution fibril growth kinetics to be quantified, thus equating the impact of various stimuli on assembly growth at the molecular level (Paul et al., 2022; Ray et al., 2023). Adopting such methods to analyse spidroin assembly will undoubtedly be a time- and resource-effective process for facilitating a better understanding of fibril assembly.
Spectrophotometric analysis of assembly
Whilst separation methods are commonplace for determining sample quality and isolating monomeric forms of spidroins, their use beyond these applications is limited. The nature of spidroins to rapidly aggregate and the need to determine the underlying sequence–environment–assembly mechanics behind this phenomenon have resulted in the use of high-frequency spectroscopic methods.
Absorbance readings and turbidimetry, are commonly used to study macroscale assembly, particularly in the context of liquid-liquid phase separation. High-throughput multifactorial analysis may be done to reveal the relationship between environmental stimuli and protein assembly (Figure 3). As such, the impact of ion type and concentration on spidroin phase separation has been detailed, where increases in absorbance correlates with environments that exhibit phase separation, highlighting the crucial role of kosmotropic ions in driving fibril assembly (Malay et al., 2020; Mohammadi et al., 2020). As such, implementation of this method enables the rapid comparison of engineered recombinant spidroins under duct-like conditions to determine the best candidates worth pursuing, with respect to fibre formation, whilst simultaneously benchmarking against native spidroin proteins.
Similarly, spectrofluorimetric analysis is often used to accurately determine β-sheet aggregation kinetics. Originally adopted from neurodegenerative research into amyloid fibre forming peptides, Thioflavin T (ThT) is frequently used in recombinant spidroin biochemical assays to determine β-sheet formation behaviour (Figure 3) (Xue et al., 2016) Functionally, ThT reports amyloid aggregation through fluorescence, which only occurs once the small molecular dye has intercalated into the β-sheet structure. The sigmoidal increase in fluorescence intensity overtime can be used to determine molecular scale assembly kinetics, as well as the ability to compare and make inferences on the β-sheet content of recombinant constructs to form aggregates with richer β-sheet content from function maxima when seeding protein concentration is equal (Kaldmäe et al., 2020; Arndt et al., 2022; Qi et al., 2023; De Oliveira et al., 2024). As such the use of ThT fluorescence has and will continue to serve as an effective low-cost and reproducible method for assessing β-sheet formation in selectable environments.
Circular dichroism (CD), Raman, vibrational, and nuclear magnetic resonance (NMR) spectroscopy are commonly used techniques to gain insights into protein assembly, kinetics, and molecular interactions of proteins within silk fibres. Accordingly, these techniques have been reviewed (in the context of spidroins) extensively elsewhere (Blamires et al., 2023).
Microscopic analyses
Spectroscopic analyses coupled with microscopy-based analytical methods offer a means for researchers to visualise protein structural organisation in silk fibres across hierarchical scales and spatial resolutions, thus, facilitating a greater understanding of how environmental factors can influence silk fibre properties.
Bright-field and confocal microscopy, in particular, have been deployed to effectively characterise spidroin phase separation (Zhang et al., 2023). Firstly, serving as a binary method to determine whether a two-phase state is observed, image analysis techniques such as circle detection offer valuable insights into droplet formation, growth, and coacervation kinetics, shedding light on the crucial LLPS pre-assembly step in fibril development (Figure 3).
An extension of these methods is Fluorescence Recovery after Photobleaching (FRAP). Unlike image analysis methods, which assess overall droplet characteristics such as size, morphology, and fusion rates, FRAP is specifically used to determine the diffusion properties of proteins within the dense proteinaceous droplets they form, providing insights into the relationship between these proteins and their environment (Figure 3). In this technique, a high-intensity laser bleaches a specific region within the imaging field, removing the fluorescence of a conjugated fluorophore or fluorescently expressed tag. The time it takes for fluorescence to recover indicates the diffusion rate and, ultimately, the mobility of proteins within the droplet (Meyvis et al., 1999; Sprague et al., 2004). Recent work has utilised FRAP analysis of a recombinantly expressed MaSp2 protein to determine physical property shifts when buffered into a series of environments with progressively decreasing pH (Lemetti et al., 2019; Malay et al., 2020; Feng et al., 2023). Samples with lower pH environments showed a significant decrease in fluorescence recovery compared to higher pH environments, characterising the transition from pre-assembly droplets into more rigid nanofibril structures (Malay et al., 2020). Methods like FRAP will become increasingly utilised to effectively measure changes in the physical properties of spidroins at a molecular level, allowing inferences on residue-residue interactions that drive structural development to be made.
In contrast to the dynamic, in situ observations provided by confocal microscopy scanning electron microscopy (SEM) and transmission electron microscopy (TEM) offer high-resolution, static imaging that enables precise quantification of the structural arrangement and fine details of nano to microscale entities allowing for a more detailed analysis of their architecture and organisation at the nanoscale (Henini, 2000; Putthanarat et al., 2000). TEM, which utilises electrons passing through a sample, has enabled the characterisation of spidroin protein arrangement in the pre-fibril phase, providing a better understanding of the structural organisation of various domains and the interactions between monomers within the network (Parent et al., 2018). Conversely, SEM provides high resolution images of the surface of a sample by scanning samples with an electron beam. Imaging fibre topography is extremely useful as it can rapidly determine stress, fractures and void formation in fibres (Stark et al., 2007; dos Santos-Pinto et al., 2018; Arakawa et al., 2022; Craig et al., 2022). SEM can be implemented as a quick and effective quality control step to ensure fibre uniformity during production.
Machine learning and AI approaches to predicting and evaluating structure
Protein structure prediction programs are frequently employed to aid the design of protein constructs for expression and purification. As these predictions provide critical information about domain boundaries and allow researchers to determine the effect of mutations, it is imperative that these programs can predict protein structures with a high degree of accuracy.
The release of AlphaFold2 in 2021 has revolutionised the field of protein structure prediction with an increase in accuracy of the predicted structures even in instances where there are no known structures of homologous proteins (Jumper et al., 2021). Figures 4A, B provides an example of its use for predicting the structure of spider silk proteins. The accurate and reliable protein structure predictions afforded by AlphaFold2 have proven to be invaluable as a replacement or complement to X-ray crystallography and cryo-EM (Gupta et al., 2021; Pereira et al., 2021), thus enabling researchers to make predictions about protein structure by combining experimental data with integrative structural modelling.
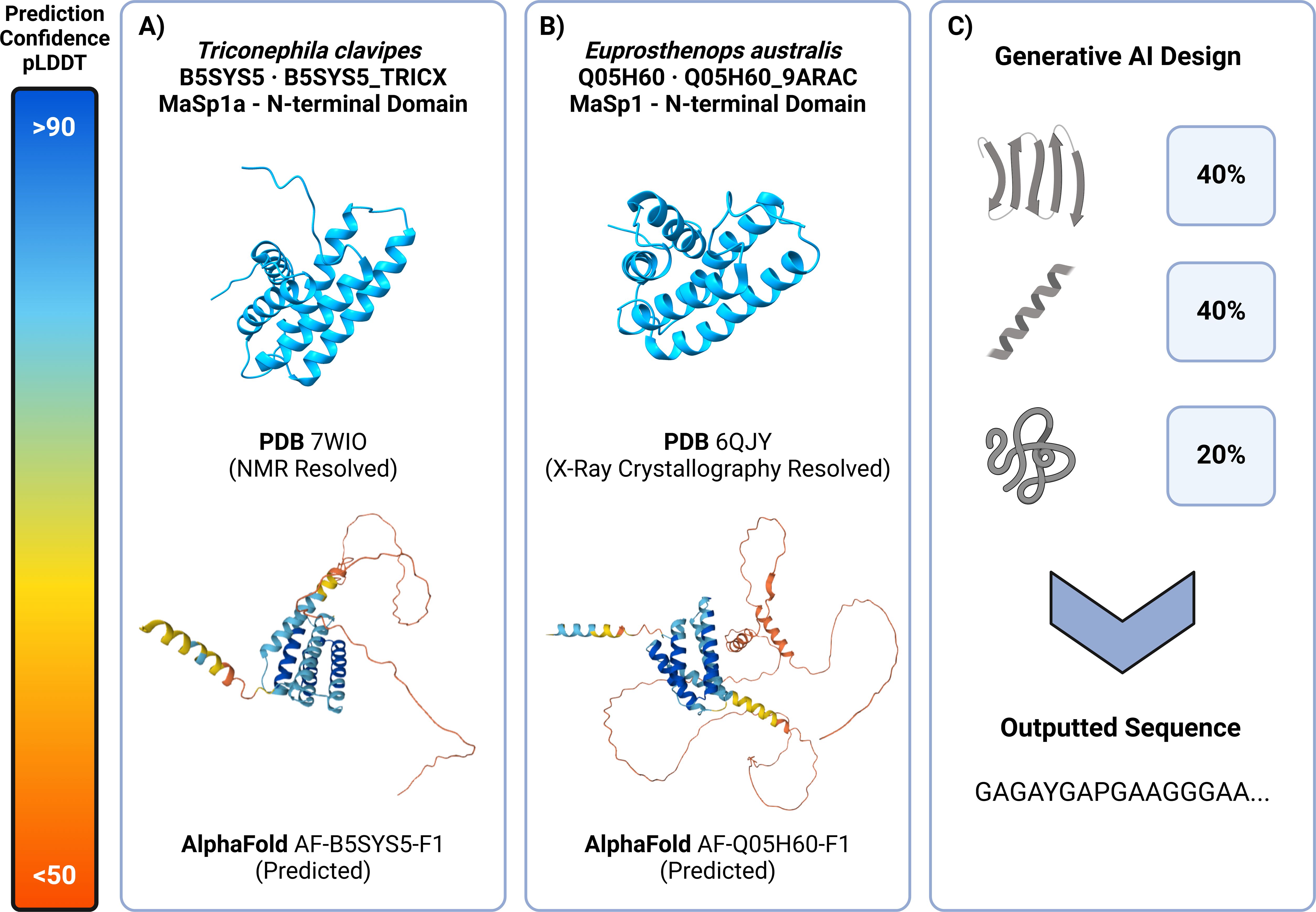
Figure 4. Predictive and generative AI tools can be used to inform and design spidroin materials. (A) (Top) Triconephila clavipes MaSp1 N-terminal domain (PDB – 7WIO) experimentally solved using NMR depicts a conserved and well formed 5 helical domain (Oktaviani et al., 2023). (Bottom) AlphaFold prediction (AF-B5S7S5-F1) of the same terminal domain shows a highly confident 5 helical structure in blue with similarities to that of the NMR solved structure with the exception of an intrinsically disordered low confidence “noodle-like” structure in red representing the start of the repetitive domain. (B) (Top) Euprosthenops australis N-terminal MaSp1 N-terminal domain (PDB – 6QJY) experimentally solved using X-Ray Crystallography with a similar helical domain conformation that is conserved amongst all spidroin proteins (RCSB PDB – 6QJY: Solution NMR structure of a mutant major ampullate spidroin 1 N-terminal domain, https://www.rcsb.org/structure/6qjy). (Bottom) AlphaFold prediction (AF-Q05H60-F1) shows a similar well conserved helical domain in blue and start of the repetitive domain in red.
ColabFold (Mirdita et al., 2022) further enhanced the accessibility of fast, accurate protein structure prediction by opening up AlphaFold2 and RosettaFold to the wider scientific community (Mirdita et al., 2022). ColabFold is five times faster than for single predictions than AlphaFold2 and AlphaFold-Colab. These improvements, coupled with AlphaFold multimer that opens up the possibility of investigating macromolecular complexes, meaning that complex and previously intractable protein systems – including spider silk proteins and fibres – are more explorable than ever (Evans et al., 2022). AlphaFold3, which was released during the preparation of this manuscript, offers even more exciting improvements, with increased speed and ease of use (Abramson et al., 2024).
As a result of these innovations, structural coverage of large proteomes has been estimated to have increased from 48% to 76% (Porta-Pardo et al., 2022). AlphaFold has also democratised the use of protein structures with millions of structure predictions for a range of species readily available via the AlphaFold database to researchers who had not routinely considered protein structure when presenting their findings (Varadi et al., 2022). Current limitations of AlphaFold include an inclination to overpredict folded regions. With the algorithm being largely trained on X-ray derived crystalline structures, there is an inability to include ligands in the predictions (although AlphaFold3 now offers the inclusion of certain ligands, ions, and post-translational modifications), and size limitations exist in the complexes that can be predicted. AlphaFold3 has improved this situation significantly but is still limited to 5000 ‘‘tokens’’ per run which equates to a maximum of about 14 MaSp1 units.
Many computational researchers have built on the foundations of AlphaFold introducing new algorithms that combine it with larger templating and other molecular dynamics-based methods to explore larger complexes and potential conformers and movements within them (Bryant et al., 2022; Mirabello et al., 2023). These experiments are producing vast, and difficult to interpret, arrays of data and offer opportunities for iterative and symbiotic marriages between experimental and computational work. The recombinant spider silk field seems ripe for such collaborations given the modular nature and unique properties of these materials.
Fold prediction tools are capable of estimating terminal domain structures and binding interactions of spidroins with high confidence. Their ability to confidently estimate the structure of the repetitive domain of spidroin proteins is, nevertheless, only capable of outputting low confidence and minimal structural conformations (Figures 4A, B). Recent developments, allow for comparative queries of sequences with residue-per-residue data output on the structure promoting and disorder promoting residues with a protein (Linding et al., 2003; Hu et al., 2021). Further to this, applying deep learning on proteome datasets has enabled accurate and efficient predictions of radius of gyration, end-to-end distance, polymer-scaling exponents, asphericity and phase separation propensity of proteins (Vendruscolo and Fuxreiter, 2022; Lotthammer et al., 2024) The ability to computationally determine these values provides an opportunity to compare experimentally compare native spidroins to recombinantly produced variants and allow more informed sequence design. While the surge in popularity of deep learning implementation at a generalised proteomic level has occurred, specialised spidroin tools have also been developed. These range from coarse-grained modelling of silk synthesis and behaviour throughout the gland to generative design of novel spidroin sequences with advanced mechanical properties trained on a dataset of over 1,000 silk sequences and motifs, annotated with physical properties such as elasticity, tensile strength, and water content (Figure 4C) (Lin et al., 2015; Ni et al., 2023; Lu et al., 2024). The wide range of in silico tools now available provides unprecedented potential to high-throughput screening and development of high-performing recombinant spidroins.
Conclusion
The production of recombinant spider silk proteins represents a promising yet complex endeavour that leverages advances in molecular biology, protein engineering, and materials science. While significant strides have been made in expressing and purifying the requisite spidroins using heterologous hosts, challenges remain in replicating the natural silk spinning process, particularly with regard to maintaining protein stability, scaling up production, and emulating the hierarchical structural features of native spider silk. Continued research into the roles of the N- and C-termini, as well as the repetitive domains of spidroins, will be vital in overcoming these obstacles. Further investigation into liquid–liquid phase separation and other pre-assembly processes offers exciting avenues for enhancing the quality and consistency of the fibres produced. Coupled with advances in analytical techniques such as mass photometry and spectrofluorimetric analysis, the future of recombinant spider silk production lies in optimising both expression systems and spinning methodologies to develop economically viable methods to create fibres with properties approaching those of their natural counterparts.
Addressing the urgency and direction for future research should prioritise understanding the unique silk gland environments across spider species, as this may hold key insights into the optimisation of synthetic spinning techniques. How factors such as the roles of pH shifts, shear forces, ion concentrations and duct physiology influence the silks mechanical properties requires closer examination. Comparative studies across species could reveal crucial differences in gland morphology and function, offering clues to refining recombinant processes. Additionally, advancements in bioengineering approaches, such as gene editing and expression systems, alongside improvements in synthetic spinning methodologies, are likely to drive advancements in fibre quality and production scalability.
Author contributions
AM: Visualization, Conceptualization, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. JT: Methodology, Validation, Writing – review & editing. SE: Methodology, Validation, Writing – original draft. AR: Funding acquisition, Methodology, Project administration, Writing – original draft, Writing – review & editing. JY: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. RI: Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Writing – original draft, Writing – review & editing. CM: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. SB: Funding acquisition, Project administration, Resources, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by a Plus Alliance seed grant to SB, AR, CM, RI, and JY, and support from the USA National Science Foundation (NSF) grant DMR-BMAT-2105312. AM is supported by the Australian Research Training Program. JT and SE were supported by the UKRI Biotechnology and Biological Sciences Research Council (BB/S006877/1 and BB/X001415/1 to RI).
Acknowledgments
Figures were generated using BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The corresponding author (SB) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abramson J., Adler J., Dunger J., Evans R., Green T., Pritzel A., et al. (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 8016 630, 493–500. doi: 10.1038/s41586-024-07487-w
Andersson M., Jia Q., Abella A., Lee X. Y., Landreh M., Purhonen P., et al. (2017). Biomimetic spinning of artificial spider silk from a chimeric minispidroin. Nat. Chem. Biol. 13, 3 13, 262–264. doi: 10.1038/nchembio.2269
Arakawa K., Kono N., Malay A. D., Tateishi A., Ifuku N., Masunaga H., et al. (2022). 1000 spider silkomes: Linking sequences to silk physical properties. Sci. Adv. 8, 6043. doi: 10.1126/SCIADV.ABO6043/SUPPL_FILE/SCIADV.ABO6043_DATA_S1_TO_S4.ZIP
Arndt T., Jaudzems K., Shilkova O., Francis J., Johansson M., Laity P. R., et al. (2022). Spidroin N-terminal domain forms amyloid-like fibril based hydrogels and provides a protein immobilization platform. Nat. Commun. 13, 46595. doi: 10.1038/S41467-022-32093-7
Askarieh G., Hedhammar M., Nordling K., Saenz A., Casals C., Rising A., et al. (2010). Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 465, 7295 465, 236–238doi: 10.1038/nature08962
Ayoub N. A., Garb J. E., Tinghitella R. M., Collin M. A., Hayashi C. Y. (2007a). Blueprint for a high-performance biomaterial: full-length spider dragline silk genes. PloS One 2, e514. doi: 10.1371/JOURNAL.PONE.0000514
Blamires S. J. (2024). Grand challenges in arachnid genetics and biomaterials. Front. Arachnid Sci. 3. doi: 10.3389/FRCHS.2024.1356170
Blamires S. J., Little D. J., White T. E., Kane D. M. (2020). Photoreflectance/scattering measurements of spider silks informed by standard optics. R Soc. Open Sci. 7, 192174. doi: 10.1098/RSOS.192174
Blamires S. J., Rawal A., Edwards A. D., Yarger J. L., Oberst S., Allardyce B. J., et al. (2023). Methods for silk property analyses across structural hierarchies and scales. Molecules 28, 2120. doi: 10.3390/MOLECULES28052120
Bryant P., Pozzati G., Zhu W., Shenoy A., Kundrotas P., Elofsson A. (2022). Predicting the structure of large protein complexes using AlphaFold and Monte Carlo tree search. . bioRxiv 03, 12.484089. doi: 10.1101/2022.03.12.484089
Burgess R. R. (2018). A brief practical review of size exclusion chromatography: Rules of thumb, limitations, and troubleshooting. Protein Expr Purif 150, 81–85. doi: 10.1016/J.PEP.2018.05.007
Candelas G. C., Arroyo G., Carrasco C., Dompenciel R. (1990). Spider silkglands contain a tissue-specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev. Biol. 140, 215–220. doi: 10.1016/0012-1606(90)90069-U
Cao H., Parveen S., Ding D., Xu H., Tan T., Liu L. (2017). Metabolic engineering for recombinant major ampullate spidroin 2 (MaSp2) synthesis in Escherichia coli. Sci. Rep. 7, 1 7, 1–1 7, 8. doi: 10.1038/s41598-017-11845-2
Chaw R. C., Saski C. A., Hayashi C. Y. (2017). Complete gene sequence of spider attachment silk protein (PySp1) reveals novel linker regions and extreme repeat homogenization. Insect Biochem. Mol. Biol. 81, 80–90. doi: 10.1016/J.IBMB.2017.01.002
Chen J., Tsuchida A., Malay A. D., Tsuchiya K., Masunaga H., Tsuji Y., et al. (2024). Replicating shear-mediated self-assembly of spider silk through microfluidics. Nat. Commun. 15, 1 15, 1–11. doi: 10.1038/s41467-024-44733-1
Chung H., Kim T. Y., Lee S. Y. (2012). Recent advances in production of recombinant spider silk proteins. Curr. Opin. Biotechnol. 23, 957–964. doi: 10.1016/J.COPBIO.2012.03.013
Clifford R., Lindman S., Zhu J., Luo E., Delmar J., Tao Y., et al. (2024). Production of native recombinant proteins using a novel split intein affinity technology. J. Chromatogr A 1724, 464908. doi: 10.1016/J.CHROMA.2024.464908
Cole D., Young G., Weigel A., Sebesta A., Kukura P. (2017). Label-free single-molecule imaging with numerical-aperture-shaped interferometric scattering microscopy. ACS Photonics 4, 211–216. doi: 10.1021/ACSPHOTONICS.6B00912/SUPPL_FILE/PH6B00912_SI_004.PDF
Connor A., Zha R. H., Koffas M. (2024). Production and secretion of recombinant spider silk in Bacillus megaterium. Microb. Cell Fact. 23 (1), 35. doi: 10.1186/s12934-024-02304-5
Copeland C. G., Bell B. E., Christensen C. D., Lewis R. V. (2015). Development of a process for the spinning of synthetic spider silk. ACS Biomater Sci. Eng. 1, 577–584. doi: 10.1021/ACSBIOMATERIALS.5B00092/SUPPL_FILE/AB5B00092_SI_001.PDF
Craig H. C., Piorkowski D., Nakagawa S., Kasumovic M. M., Blamires S. J. (2020). Meta-analysis reveals materiomic relationships in major ampullate silk across the spider phylogeny. J. R. Soc. Interface 17, 20200471. doi: 10.1098/RSIF.2020.0471
Craig H. C., Yao Y., Ariotti N., Setty M., Remadevi R., Kasumovic M. M., et al. (2022). Nanovoid formation induces property variation within and across individual silkworm silk threads. J. Mater Chem. B 10, 5561–5570. doi: 10.1039/D2TB00357K
Cregg J. M., Cereghino J. L., Shi J., Higgins D. R. (2000). Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 16, 23–52. doi: 10.1385/MB:16:1:23
Cummins P. M., Rochfort K. D., O’Connor B. F. (2017). Ion-exchange chromatography: basic principles and application. Methods Mol. Biol. 1485, 209–223. doi: 10.1007/978-1-4939-6412-3_11
Dai Y., You L., Chilkoti A. (2022). Engineering synthetic biomolecular condensates. Nature. Rev. Bioengineering 1, 7 1, 466–480. doi: 10.1038/s44222-023-00052-6
De Oliveira D. H., Gowda V., Sparrman T., Gustafsson L., Sanches Pires R., Riekel C., et al. (2024). Structural conversion of the spidroin C-terminal domain during assembly of spider silk fibers. Nat. Commun. 15, 1 15, 1–14. doi: 10.1038/s41467-024-49111-5
Dicko C., Knight D., Kenney J. M., Vollrath F. (2004). Secondary structures and conformational changes in flagelliform, cylindrical, major, and minor ampullate silk proteins. Temperature and concentration effects. Biomacromolecules 5, 2105–2115. doi: 10.1021/BM034486Y/ASSET/IMAGES/LARGE/BM034486YF00007.JPEG
Doblhofer E., Heidebrecht A., Scheibel T. (2015). To spin or not to spin: spider silk fibers and more. Appl. Microbiol. Biotechnol. 99, 9361–9380. doi: 10.1007/S00253-015-6948-8
Dong Z., Lewis R. V., Middaugh C. R. (1991). Molecular mechanism of spider silk elasticity. Arch. Biochem. Biophys. 284, 53–57. doi: 10.1016/0003-9861(91)90262-H
dos Santos-Pinto J. R. A., Arcuri H. A., Esteves F. G., Palma M. S., Lubec G. (2018). Spider silk proteome provides insight into the structural characterization of Triconephila clavipes flagelliform spidroin. Sci. Rep. 8, 1 8, 1–1 8,12. doi: 10.1038/s41598-018-33068-9
dos Santos-Pinto J. R. A., Lamprecht G., Chen W. Q., Heo S., Hardy J. G., Priewalder H., et al. (2014). Structure and post-translational modifications of the web silk protein spidroin-1 from Nephila spiders. J. Proteomics 105, 174–185. doi: 10.1016/J.JPROT.2014.01.002
Edlund A. M., Jones J., Lewis R., Quinn J. C. (2018). Economic feasibility and environmental impact of synthetic spider silk production from escherichia coli. N Biotechnol. 42, 12–18. doi: 10.1016/J.NBT.2017.12.006
Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., et al. (2022). Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021, 10.04.463034. doi: 10.1101/2021.10.04.463034
Fahnestock S. R., Bedzyk L. A. (1997). Production of synthetic spider dragline silk protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 47, 33–39. doi: 10.1007/S002530050884
Feng J., Gabryelczyk B., Tunn I., Osmekhina E., Linder M. B. (2023). A minispidroin guides the molecular design for cellular condensation mechanisms in S. cerevisiae. ACS Synth Biol. 12, 3050–3063. doi: 10.1021/ACSSYNBIO.3C00374/SUPPL_FILE/SB3C00374_SI_001.PDF
Gabryelczyk B., Sammalisto F. E., Gandier J. A., Feng J., Beaune G., Timonen J. V. I., et al. (2022). Recombinant protein condensation inside E. coli enables the development of building blocks for bioinspired materials engineering – Biomimetic spider silk protein as a case study. Mater Today Bio 17, 100492. doi: 10.1016/J.MTBIO.2022.100492
Gaines W. A., Sehorn M. G., Marcotte W. R. (2010). Spidroin N-terminal Domain Promotes a pH-dependent Association of Silk Proteins during Self-assembly. J. Biol. Chem. 285, 40745. doi: 10.1074/JBC.M110.163121
Gosline J. M., Guerette P. A., Ortlepp C. S., Savage K. N. (1999). The mechanical design of spider silks: from fibroin sequence to mechanical function. J. Exp. Biol. 202, 3295–3303. doi: 10.1242/JEB.202.23.3295
Gronau G., Qin Z., Buehler M. J. (2013). Effect of sodium chloride on the structure and stability of spider silk’s N-terminal protein domain. Biomater Sci. 1, 276. doi: 10.1039/C2BM00140C
Gu Y., Yu L., Mou J., Wu D., Zhou P., Xu M. (2020). Mechanical properties and application analysis of spider silk bionic material. E-Polymers 20, 443–457. doi: 10.1515/EPOLY-2020-0049/ASSET/GRAPHIC/J_EPOLY-2020-0049_FIG_009.JPG
Gupta M., Azumaya C. M., Moritz M., Pourmal S., Diallo A., Merz G. E., et al. (2021). CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes. bioRxiv 05, 10. doi: 10.1101/2021.05.10.443524
Hardenberg M., Horvath A., Ambrus V., Fuxreiter M., Vendruscolo M. (2020). Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. U.S.A. 117, 33254–33262. doi: 10.1073/PNAS.2007670117/SUPPL_FILE/PNAS.2007670117.SD08.XLSX
Harris T. I., Gaztambide D. A., Day B. A., Brock C. L., Ruben A. L., Jones J. A., et al. (2016). Sticky situation: an investigation of robust aqueous-based recombinant spider silk protein coatings and adhesives. Biomacromolecules 17, 3761–3772. doi: 10.1021/ACS.BIOMAC.6B01267
Hayashi C. Y., Shipley N. H., Lewis R. V. (1999). Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int. J. Biol. Macromol 24, 271–275. doi: 10.1016/S0141-8130(98)00089-0
Heidebrecht A., Eisoldt L., Diehl J., Schmidt A., Geffers M., Lang G., et al. (2015). Biomimetic fibers made of recombinant spidroins with the same toughness as natural spider silk. Advanced Materials 27, 2189–2194. doi: 10.1002/ADMA.201404234
Heim M., Ackerschott C. B., Scheibel T. (2010). Characterization of recombinantly produced spider flagelliform silk domains. J. Struct. Biol. 170, 420–425. doi: 10.1016/J.JSB.2009.12.025
Heim M., Keerl D., Scheibel T. (2009). Spider silk: from soluble protein to extraordinary fiber. Angew Chem. Int. Ed Engl. 48, 3584–3596. doi: 10.1002/ANIE.200803341
Henini M. (2000). Scanning electron microscopy: an introduction. III-Vs Rev. 13, 40–44. doi: 10.1016/S0961-1290(00)80006-X
Hu G., Katuwawala A., Wang K., Wu Z., Ghadermarzi S., Gao J., et al. (2021). flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 12, 1 12, 1–8. doi: 10.1038/s41467-021-24773-7
Hudspeth M., Nie X., Chen W., Lewis R. (2012). Effect of loading rate on mechanical properties and fracture morphology of spider silk. Biomacromolecules 13, 2240–2246. doi: 10.1021/BM3003732/ASSET/IMAGES/LARGE/BM-2012-003732_0010.JPEG
Humenik M., Smith A. M., Scheibel T. (2011). Recombinant spider silks—Biopolymers with potential for future applications. Polymers 3, 640–661. doi: 10.3390/POLYM3010640
Islam M. S., Aryasomayajula A., Selvaganapathy P. R. (2017). A review on macroscale and microscale cell lysis methods. Micromachines 8, 83. doi: 10.3390/mi8030083
Jastrzebska K., Felcyn E., Kozak M., Szybowicz M., Buchwald T., Pietralik Z., et al. (2016). The method of purifying bioengineered spider silk determines the silk sphere properties. Sci. Rep. 6, 1 6, 1–1 6,15doi: 10.1038/srep28106
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kaldmäe M., Leppert A., Chen G., Sarr M., Sahin C., Nordling K., et al. (2020). High intracellular stability of the spidroin N-terminal domain in spite of abundant amyloidogenic segments revealed by in-cell hydrogen/deuterium exchange mass spectrometry. FEBS J. 287, 2823. doi: 10.1111/FEBS.15169
Keten S., Xu Z., Ihle B., Buehler M. J. (2010). Nanoconfinement controls stiffness, strength and mechanical toughness of beta-sheet crystals in silk. Nat. Mater 9, 359–367. doi: 10.1038/NMAT2704
Kluge J. A., Rabotyagova O., Leisk G. G., Kaplan D. L. (2008). Spider silks and their applications. Trends. Biotechnol 26, 244–251. doi: 10.1016/J.TIBTECH.2008.02.006
Koeppel A., Holland C. (2017). Progress and trends in artificial silk spinning: A systematic review. ACS Biomater Sci. Eng. 3, 226–237. doi: 10.1021/ACSBIOMATERIALS.6B00669/ASSET/IMAGES/LARGE/AB-2016-006697_0005.JPEG
Kronqvist N., Otikovs M., Chmyrov V., Chen G., Andersson M., Nordling K., et al. (2014). Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 5, 1 5, 1–1 5,11. doi: 10.1038/ncomms4254
Kümmerlen J., Van Beek J. D., Vollrath F., Meier B. H. (1996). Local structure in spider dragline silk investigated by two-dimensional spin-diffusion nuclear magnetic resonance. Macromolecules 29, 2920–2928. doi: 10.1021/MA951098I/ASSET/IMAGES/LARGE/MA951098IF00009.JPEG
Leal-Egaña A., Scheibel T. (2010). Silk-based materials for biomedical applications. Biotechnol. Appl. Biochem. 55, 155–167. doi: 10.1042/BA20090229
Leclerc J., Lefèvre T., Gauthier M., Gagné S. M., Auger M. (2013). Hydrodynamical properties of recombinant spider silk proteins: Effects of pH, salts and shear, and implications for the spinning process. . Biopolymers 99, 582–593. doi: 10.1002/BIP.22218
Lee S. O., Xie Q., Fried S. D. (2021). Optimized loopable translation as a platform for the synthesis of repetitive proteins. ACS Cent Sci. 7, 1736–1750. doi: 10.1021/ACSCENTSCI.1C00574/ASSET/IMAGES/LARGE/OC1C00574_0007.JPEG
Lemetti L., Hirvonen S. P., Fedorov D., Batys P., Sammalkorpi M., Tenhu H., et al. (2019). Molecular crowding facilitates assembly of spidroin-like proteins through phase separation. Eur. Polym J. 112, 539–546. doi: 10.1016/J.EURPOLYMJ.2018.10.010
Lemetti L., Scacchi A., Yin Y., Shen M., Linder M. B., Sammalkorpi M., et al. (2022). Liquid-liquid phase separation and assembly of silk-like proteins is dependent on the polymer length. Biomacromolecules 23, 3142–3153. doi: 10.1021/ACS.BIOMAC.2C00179/ASSET/IMAGES/LARGE/BM2C00179_0011.JPEG
Lewis R. V., Hinman M., Kothakota S., Fournier M. J. (1996). Expression and purification of a spider silk protein: a new strategy for producing repetitive proteins. Protein Expr Purif 7, 400–406. doi: 10.1006/PREP.1996.0060
Lin S., Ryu S., Tokareva O., Gronau G., Jacobsen M. M., Huang W., et al. (2015). Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres. Nat. Commun. 6, 161–162doi: 10.1038/ncomms7892
Linding R., Jensen L. J., Diella F., Bork P., Gibson T. J., Russell R. B. (2003). Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459. doi: 10.1016/J.STR.2003.10.002
Linklater D., Vailionis A., Ryu M., Kamegaki S., Morikawa J., Mu H., et al. (2023). Structure and optical anisotropy of spider scales and silk: the use of chromaticity and azimuth colors to optically characterize complex biological structures. Nanomaterials 13, 1894. doi: 10.3390/NANO13121894/S1
Liu L., Wang P., Zhao D., Zhu L., Tang J., Leng W., et al. (2022). Engineering circularized mRNAs for the production of spider silk proteins. Appl. Environ. Microbiol. 88, e00028–22. doi: 10.1128/AEM.00028-22
Liu X., Zhang K.-Q., Liu X., Zhang K.-Q. (2014). Silk fiber — Molecular formation mechanism, structure- property relationship and advanced applications. Oligom. Chem. Biol. Comps. 3, 69–102. doi: 10.5772/57611
Lotthammer J. M., Ginell G. M., Griffith D., Emenecker R. J., Holehouse A. S. (2024). Direct prediction of intrinsically disordered protein conformational properties from sequence. Nat. Methods 21, 465–476doi: 10.1038/s41592-023-02159-5
Lu W., Kaplan D. L., Buehler M. J. (2024). Generative modeling, design, and analysis of spider silk protein sequences for enhanced mechanical properties. Adv. Funct. Mater 34, 2311324. doi: 10.1002/ADFM.202311324
Malay A. D., Suzuki T., Katashima T., Kono N., Arakawa K., Numata K. (2020). Spider silk self-assembly via modular liquid-liquid phase separation and nanofibrillation. Sci. Adv. 6, 45. doi: 10.1126/SCIADV.ABB6030/SUPPL_FILE/ABB6030_SM.PDF
Maraldo A., Rnjak-Kovacina J., Marquis C. (2024). Tyrosine - a structural glue for hierarchical protein assembly. Trends Biochem. Sci. 49, 633–648. doi: 10.1016/J.TIBS.2024.03.014
Martin E. W., Holehouse A. S., Peran I., Farag M., Incicco J. J., Bremer A., et al. (2020). Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367, 694–699. doi: 10.1126/SCIENCE.AAW8653/SUPPL_FILE/AAW8653_S6.MP4
Martin E. W., Mittag T. (2018). Relationship of sequence and phase separation in protein low-complexity regions. Biochemistry 57, 2478–2487. doi: 10.1021/ACS.BIOCHEM.8B00008/ASSET/IMAGES/LARGE/BI-2018-00008N_0005.JPEG
McCue J. T. (2009). Theory and use of hydrophobic interaction chromatography in protein purification applications. Methods Enzymol. 463, 405–414. doi: 10.1016/S0076-6879(09)63025-1
McDaniel J. R., MacKay J. A., Quiroz F. G., Chilkoti A. (2010). Recursive directional ligation by plasmid reconstruction allows rapid and seamless cloning of oligomeric genes. Biomacromolecules 11, 944–952. doi: 10.1021/BM901387T/ASSET/IMAGES/LARGE/BM-2009-01387T_0005.JPEG
Menassa R., Zhu H., Karatzas C. N., Lazaris A., Richman A., Brandle J. (2004). Spider dragline silk proteins in transgenic tobacco leaves: accumulation and field production. Plant Biotechnol. J. 2, 431–438. doi: 10.1111/J.1467-7652.2004.00087.X
Meyvis T. K. L., De Smedt S. C., Van Oostveldt P., Demeester J. (1999). Fluorescence recovery after photobleaching: A versatile tool for mobility and interaction measurements in pharmaceutical research. Pharm. Res. 16, 1153–1162. doi: 10.1023/A:1011924909138/METRICS
Mi J., Zhou Y., Ma S., Zhou X., Xu S., Yang Y., et al. (2023). High-strength and ultra-tough whole spider silk fibers spun from transgenic silkworms. Matter 6, 3661–3683. doi: 10.1016/J.MATT.2023.08.013/ATTACHMENT/5F32AAA5-A883-4F88-BBF5-D7D2EA03B12C/MMC1.PDF
Miao Y., Zhang Y., Nakagaki K., Zhao T., Zhao A., Meng Y., et al. (2006). Expression of spider flagelliform silk protein in Bombyx mori cell line by a novel Bac-to-Bac/BmNPV baculovirus expression system. Appl. Microbiol. Biotechnol. 71, 192–199. doi: 10.1007/S00253-005-0127-2/FIGURES/6
Mirabello C., Wallner B., Nystedt B., Azinas S., Carroni M. (2023). Unmasking AlphaFold: integration of experiments and predictions with a smarter template mechanism. bioRxiv 09, 20. doi: 10.1101/2023.09.20.558579
Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., Steinegger M. (2022). ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682. doi: 10.1038/s41592-022-01488-1
Mohammadi P., Christopher J., Beaune G., Engelhardt P., Kamada A., Timonen J. V. I., et al. (2020). Controllable coacervation of recombinantly produced spider silk protein using kosmotropic salts. J. Colloid Interface Sci. 560, 149–160. doi: 10.1016/J.JCIS.2019.10.058
Ni B., Kaplan D. L., Buehler M. J. (2023). Generative design of de novo proteins based on secondary-structure constraints using an attention-based diffusion model. Chem 9, 1828–1849. doi: 10.1016/J.CHEMPR.2023.03.020/ATTACHMENT/3A17589E-1482-472F-BAB4-7EA232BAA4B4/MMC4.PDF
Oktaviani N. A., Malay A. D., Matsugami A., Hayashi F., Numata K. (2023). Unusual pKa values mediate the self-assembly of spider dragline silk proteins. Biomacromolecules 24, 1604–1616. doi: 10.1021/ACS.BIOMAC.2C01344/SUPPL_FILE/BM2C01344_SI_001.PDF
Oktaviani N. A., Matsugami A., Malay A. D., Hayashi F., Kaplan D. L., Numata K. (2018). Conformation and dynamics of soluble repetitive domain elucidates the initial β-sheet formation of spider silk. Nat. Commun. 9, 2121. doi: 10.1038/s41467-018-04570-5
Oliveira C., Domingues L. (2018). Guidelines to reach high-quality purified recombinant proteins. Appl. Microbiol. Biotechnol. 102, 81–92. doi: 10.1007/S00253-017-8623-8
Pan W., Wang Y., Wang N. (2019). A new metal affinity NCTR25 tag as a better alternative to the His-tag for the expression of recombinant fused proteins. Protein Expr. Purif. 164, 105477. doi: 10.1016/J.PEP.2019.105477
Parent L. R., Onofrei D., Xu D., Stengel D., Roehling J. D., Bennett Addison J., et al. (2018). Hierarchical spidroin micellar nanoparticles as the fundamental precursors of spider silks. Proc. Natl. Acad. Sci. U.S.A. 115, 11507–11512. doi: 10.1073/PNAS.1810203115/SUPPL_FILE/PNAS.1810203115.SM08.MOV
Paul S. S., Lyons A., Kirchner R., Woodside M. T. (2022). Quantifying oligomer populations in real time during protein aggregation using single-molecule mass photometry. ACS Nano 16, 16462–16470. doi: 10.1021/ACSNANO.2C05739/ASSET/IMAGES/MEDIUM/NN2C05739_M001.GIF
Peng Q., Zhang Y., Lu L., Shao H., Qin K., Hu X., et al. (2016). Recombinant spider silk from aqueous solutions via a bio-inspired microfluidic chip. Sci. Rep. 6, 36473. doi: 10.1038/srep36473
Pereira J., Simpkin A. J., Hartmann M. D., Rigden D. J., Keegan R. M., Lupas A. N. (2021). High-accuracy protein structure prediction in CASP14. Proteins: Structure. Function Bioinf. 89, 1687–1699. doi: 10.1002/prot.26171
Perera D., Li L., Walsh C., Silliman J., Xiong Y., Wang Q., et al. (2023). Natural spider silk nanofibrils produced by assembling molecules or disassembling fibers. Acta Biomater 168, 323–332. doi: 10.1016/J.ACTBIO.2023.06.044
Porta-Pardo E., Ruiz-Serra V., Valentini S., Valencia A. (2022). The structural coverage of the human proteome before and after AlphaFold. PloS Comput. Biol. 18, e1009818. doi: 10.1371/JOURNAL.PCBI.1009818
Prince J. T., McGrath K. P., DiGirolamo C. M., Kaplan D. L. (1995). Construction, cloning, and expression of synthetic genes encoding spider dragline silk. Biochemistry 34, 10879–10885. doi: 10.1021/BI00034A022
Putthanarat S., Stribeck N., Fossey S. A., Eby R. K., Adams W. W. (2000). Investigation of the nanofibrils of silk fibers. Polymer (Guildf) 41, 7735–7747. doi: 10.1016/S0032-3861(00)00036-7
Qi X., Wang Y., Yu H., Liu R., Leppert A., Zheng Z., et al. (2023). Spider silk protein forms amyloid-like nanofibrils through a non-nucleation-dependent polymerization mechanism. Small 19, 2304031. doi: 10.1002/SMLL.202304031
Ramezaniaghdam M., Nahdi N. D., Reski R. (2022). Recombinant spider silk: promises and bottlenecks. Front. Bioeng Biotechnol. 10. doi: 10.3389/FBIOE.2022.835637/BIBTEX
Ray S., Mason T. O., Boyens-Thiele L., Farzadfard A., Larsen J. A., Norrild R. K., et al. (2023). Mass photometric detection and quantification of nanoscale α-synuclein phase separation. Nat. Chem. 15, 9 15, 1306–1316. doi: 10.1038/s41557-023-01244-8
Rising A., Johansson J. (2015). Toward spinning artificial spider silk. Nat. Chem. Biol. 11, 309–315. doi: 10.1038/NCHEMBIO.1789
Römer L., Scheibel T. (2008). The elaborate structure of spider silk: Structure and function of a natural high performance fiber. Prion 2, 154. doi: 10.4161/PRI.2.4.7490
Sahin E., Roberts C. J. (2012). Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms. Methods Mol. Biol. 899, 403–423. doi: 10.1007/978-1-61779-921-1_25
Schacht K., Scheibel T. (2014). Processing of recombinant spider silk proteins into tailor-made materials for biomaterials applications. Curr. Opin. Biotechnol. 29, 62–69. doi: 10.1016/J.COPBIO.2014.02.015
Scheller J., Gührs K. H., Grosse F., Conrad U. (2001). Production of spider silk proteins in tobacco and potato. Nat. Biotechnol. 19, 573–577. doi: 10.1038/89335
Shao Z., Vollrath F. (2002). Surprising strength of silkworm silk. Nature 418, 741. doi: 10.1038/418741A
Shao Z., Vollrath F., Yang Y., Thøgersen H. C. (2003). Structure and behaviour of regenerated spider silk. Macromolecules 36, 1157–1161. doi: 10.1021/ma0214660
Singh A., Upadhyay V., Upadhyay A. K., Singh S. M., Panda A. K. (2015). Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact 14, 1–10. doi: 10.1186/S12934-015-0222-8/TABLS/3
Sonavane S., Hassan S., Chatterjee U., Soler L., Holm L., Mollbrink A., et al. (2024). Origin, structure, and composition of the spider major ampullate silk fiber revealed by genomics, proteomics, and single-cell and spatial transcriptomics. Sci. Adv. 10, eadn0597. doi: 10.1126/sciadv.adn0597
Sprague B. L., Pego R. L., Stavreva D. A., McNally J. G. (2004). Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86, 3473–3495. doi: 10.1529/biophysj.103.026765
Stark M., Grip S., Rising A., Hedhammar M., Engström W., Hjälm G., et al. (2007). Macroscopic fibers self-assembled from recombinant miniature spider silk proteins. Biomacromolecules 8, 1695–1701. doi: 10.1021/BM070049Y/SUPPL_FILE/BM070049YSI20070220_064756.PDF
Teulé F., Miao Y. G., Sohn B. H., Kim Y. S., Hull J. J., Fraser M. J., et al. (2012). Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc. Natl. Acad. Sci. U.S.A. 109, 923–928. doi: 10.1073/PNAS.1109420109/SUPPL_FILE/PNAS.201109420SI.PDF
Thomsen M. (2020). Determination of the molecular mass of membrane proteins using size-exclusion chromatography with multiangle laser light scattering (SEC-MALLS). Methods Mol. Biol. 2168, 263–269. doi: 10.1007/978-1-0716-0724-4_12/FIGURES/2
Tokareva O. S., Lin S., Jacobsen M. M., Huang W., Rizzo D., Li D., et al. (2014). Effect of sequence features on assembly of spider silk block copolymers. J. Struct. Biol. 186, 412–419. doi: 10.1016/j.jsb.2014.03.004
Tokareva O., Michalczechen-Lacerda V. A., Rech E. L., Kaplan D. L. (2013). Recombinant DNA production of spider silk proteins. Microb. Biotechnol. 6, 651–663. doi: 10.1111/1751-7915.12081
Tripathi N. K., Shrivastava A. (2019). Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front. Bioeng Biotechnol. 7. doi: 10.3389/FBIOE.2019.00420/BIBTEX
Tsumoto K., Ejima D., Kumagai I., Arakawa T. (2003). Practical considerations in refolding proteins from inclusion bodies. Protein Expr Purif 28, 1–8. doi: 10.1016/S1046-5928(02)00641-1
Van Beek J. D., Hesst S., Vollrath F., Meier B. H. (2002). The molecular structure of spider dragline silk: Folding and orientation of the protein backbone. Proc. Natl. Acad. Sci. U.S.A. 99, 10266–10271. doi: 10.1073/PNAS.152162299/SUPPL_FILE/1622FIG6.PDF
Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/NAR/GKAB1061
Vendruscolo M., Fuxreiter M. (2022). ). Sequence determinants of the aggregation of proteins within condensates generated by liquid-liquid phase separation. J. Mol. Biol. 434, 167201. doi: 10.1016/J.JMB.2021.167201
Vollrath F., Porter D., Holland C. (2013). The science of silks. MRS Bull. 38, 73–80. doi: 10.1557/MRS.2012.314
Watanabe Y., Arakawa K. (2023). Molecular mechanisms of the high performance of spider silks revealed through multi-omics analysis. Biophys. Physicobiol. 20, e200014. doi: 10.2142/BIOPHYSICO.BPPB-V20.0014
Wei S. P., Qian Z. G., Hu C. F., Pan F., Chen M. T., Lee S. Y., et al. (2020). Formation and functionalization of membraneless compartments in Escherichia coli. Nat. Chem. Biol. 16, 10 16, 1143–1148. doi: 10.1038/s41589-020-0579-9
Wong Po Foo C., Kaplan D. L. (2002). Genetic engineering of fibrous proteins: spider dragline silk and collagen. Adv. Drug Delivery Rev. 54, 1131–1143. doi: 10.1016/S0169-409X(02)00061-3
Xia X. X., Qian Z. G., Ki C. S., Park Y. H., Kaplan D. L., Lee S. Y. (2010). Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. U.S.A. 107, 14059–14063. doi: 10.1073/PNAS.1003366107/-/DCSUPPLEMENTAL
Xu H. T., Fan B. L., Yu S. Y., Huang Y. H., Zhao Z. H., Lian Z. X., et al. (2007). Construct synthetic gene encoding artificial spider dragline silk protein and its expression in milk of transgenic mice. Anim. Biotechnol. 18, 1–12. doi: 10.1080/10495390601091024
Xu L., Lefèvre T., Orrell K. E., Meng Q., Auger M., Liu X. Q., et al. (2017). Structural and mechanical roles for the C-terminal non-repetitive domain become apparent in recombinant spider aciniform silk. Biomacromolecules 18, 3678. doi: 10.1021/ACS.BIOMAC.7B01057
Xue C., Lin T. Y., Chang D., Guo Z. (2016). Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc. Open Sci. 4, 160696. doi: 10.1098/RSOS.160696
Yang Y., Gao Z., Yang D. (2023). pH-dependent self-assembly mechanism of a single repetitive domain from a spider silk protein. Int. J. Biol. Macromol 242, 124775. doi: 10.1016/J.IJBIOMAC.2023.124775
Yarger J. L., Cherry B. R., van der Vaart A. (2018). Uncovering the structure–function relationship in spider silk. Nat. Rev. Materials 3, 3 3, 1–3 3,11. doi: 10.1038/natrevmats.2018.8
Young C. L., Britton Z. T., Robinson A. S. (2012). Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol. J. 7, 620–634. doi: 10.1002/BIOT.201100155
Young G., Hundt N., Cole D., Fineberg A., Andrecka J., Tyler A., et al. (2018). Quantitative mass imaging of single biological macromolecules. Science 360, 423–427. doi: 10.1126/SCIENCE.AAR5839
Zhang Y., Hu J., Miao Y., Zhao A., Zhao T., Wu D., et al. (2008). Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline proteins yield. Mol. Biol. Rep. 35, 329–335. doi: 10.1007/s11033-007-9090-6
Keywords: recombinant spider silk, spidroin synthesis, fibre assembly, liquid–liquid phase separation, predictive computational model
Citation: Maraldo A, Torpey JH, Evans SL, Rawal A, Yarger JL, Isaacson RL, Marquis CP and Blamires SJ (2025) Biochemical methods for producing and characterising recombinant spider silks. Front. Arachn. Sci. 3:1488680. doi: 10.3389/frchs.2024.1488680
Received: 30 August 2024; Accepted: 16 December 2024;
Published: 21 January 2025.
Edited by:
Matjaz Kuntner, National Institute of Biology (NIB), SloveniaReviewed by:
Sandra M. Correa-Garhwal, Suffolk University, United StatesMatjaž Gregorič, Research Centre of the Slovenian Academy of Sciences and Arts, Slovenia
Copyright © 2025 Maraldo, Torpey, Evans, Rawal, Yarger, Isaacson, Marquis and Blamires. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean J. Blamires, c2Vhbi5ibGFtaXJlc0B1bnN3LmVkdS5hdQ==
 Anton Maraldo
Anton Maraldo James H. Torpey3
James H. Torpey3 Jeffery L. Yarger
Jeffery L. Yarger Rivka L. Isaacson
Rivka L. Isaacson Christopher P. Marquis
Christopher P. Marquis Sean J. Blamires
Sean J. Blamires