
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Allergy, 28 February 2025
Sec. Mechanisms in Allergy
Volume 6 - 2025 | https://doi.org/10.3389/falgy.2025.1548338
This article is part of the Research TopicUpdate on Eosinophil-associated diseasesView all 3 articles
Eosinophils are immune cells that are crucial for the pathogenesis of allergic diseases, such as asthma. These cells play multifunctional roles in various situations, including infection. They are activated during viral infections and exert antiviral activity. Pattern recognition receptors, toll-like receptor 7 and retinoic acid inducible gene-I, are important for the recognition and capture of RNA viruses. In addition, intracellular granule proteins (eosinophil cationic protein and eosinophil-derived neurotoxin) and intracellular nitric oxide production inactivate and/or degrade RNA viruses. Interestingly, eosinophil-synthesizing specialized pro-resolving mediators possess antiviral properties that inhibit viral replication. Thus, eosinophils may play a protective role during respiratory virus infections. Notably, antiviral activities are impaired in patients with asthma, and eosinophil activities are perturbed in proportion with the severity of asthma. The exact roles of eosinophils in RNA virus (rhinovirus, respiratory syncytial virus, and influenza virus)-induced type 2 inflammation-based asthma exacerbation remain unclear. Our research demonstrates that interferons (IFN-α and IFN-γ) stimulate human eosinophils to upregulate antiviral molecules, including guanylate-binding proteins and tripartite motifs. Furthermore, IFN-γ specifically increases the expression of IL5RA, ICAM-1, and FCGR1A, potentially enhancing cellular responsiveness to IL-5, ICAM-1-mediated adhesion to rhinoviruses, and IgG-induced inflammatory responses, respectively. In this review, we have summarized the relationship between viral infections and asthma and the mechanisms underlying the development of antiviral functions of human and mouse eosinophils in vivo and in vitro.
Eosinophils are a type of granulocytes that contain abundant intracellular granule proteins that can be stained using acidic aniline dyes (1). This cell type performs various cellular functions, including degranulation, reactive oxygen species (ROS) production, cytokine release, inflammatory lipid mediator synthesis, and EETs (Eosinophil extra-cellular traps). Eosinophils play a central role in the pathogenesis of predominantly allergic disorders, as well as collagen vascular diseases, neoplastic conditions, and infectious diseases, acting as effector cells that mediate inflammatory responses in local tissues (2–4). In contrast to pro-inflammatory eosinophils that promote allergic inflammation, a subset known as tissue-resident eosinophils has been identified, which contributes to maintaining physiological homeostasis (5). This cell population is involved in glucose metabolism; prevention of obesity; muscle regeneration; immunoglobulin A production; induction of regulatory T cell differentiation; and synthesis of specialized pro-resolving mediators to suppress inflammatory responses and promote the resolution of inflammation (6–9). Tissue-resident eosinophils with these homeostatic cellular functions reside mainly in the gastrointestinal tracts and substantially in respiratory and lower urinary tracts (8, 9). Interestingly, eosinophils also participate in the clearance of microorganisms in vivo and in vitro. They respond to parasite invasion by degranulating (2, 10). In addition, they exhibit phagocytic activity via their granule proteins against microorganisms, including Staphylococcus aureus and Escherichia coli (11–13). Recent studies have demonstrated that eosinophil ETosis (EETosis) in which an eosinophil lyses and releases their DNA components and intracellular proteins is involved in bacteriostatic activity against S. aureus and Aspergillus fumigatus (14, 15). Additionally, previous reports have demonstrated the antiviral role of eosinophils in humans and mice. Eosinophil cationic protein (ECP) and eosinophil-derived neurotoxin (EDN), major granule proteins of eosinophils, function as ribonucleases that degrade RNA viruses (16). Human eosinophils can capture influenza virus and respiratory syncytial virus (RSV) and inactivate them (17). Interestingly, eosinophils isolated from asthma patients have lower antiviral abilities than those isolated from healthy subjects (17). In addition, asthma severity correlates with reduced capacity of capturing viruses. Experimental viral infection models using mice have shown eosinophil-mediated protection against influenza virus, parainfluenza virus, and RSV (18–20). In contrast, blood eosinophils from patients with severe asthma have been reported to suppress IFN-α production from airway epithelial cells and the functions of plasmacytoid dendritic cells (pDC) cells, contributing to asthma exacerbation in viral infections (21). Another study demonstrated that coculture of eosinophils with BEAS-2B cells induced transforming growth factor β secretion, which may suppress human rhinovirus (HRV)-induced IFN expression (22). These findings highlight the biological significance of the pro-viral or anti-viral roles of eosinophils.
In this review, we have focused on the antiviral activity of eosinophils. We have discussed the immunological mechanisms underlying their antiviral activities that are mediated by specific molecules and their receptors. The relevance and involvement of eosinophils in viral infections in asthma have also been discussed.
Asthma is a chronic airway inflammation that causes paroxysmal cough, dyspnea, wheezing, and chest discomfort (23). Among patients with asthma, 50-70% patients present with type 2 inflammation characterized by eosinophil accumulation in the airway (24). Type 2 cytokines include interleukin-4 (IL-4), IL-5, and IL-13, which are optimal therapeutic targets to treat severe eosinophilic asthma (25). From the perspective of viral infections, elevated fractional exhaled nitric oxide levels and sputum eosinophil counts are associated with an increased risk of future virus-induced exacerbations (26). T helper type 2 (Th2) cells and group 2 innate lymphoid cells (ILC2) mainly produce type 2 cytokines (27). IL-5 prolongs eosinophil survival and activates eosinophil functions, including degranulation, superoxide generation, and cytokine release, leading to airway epithelium injury with increased airway hyper-responsiveness (AHR) (28). Also, EETosis causes mucus plug formation and deposition of Charcot-Leyden crystals (3, 29–31). The numbers of EET+ eosinophils and ILC2s were significantly elevated in severe asthma, with a positive correlation between these cell populations. Furthermore, mice injected with EETs exhibited a significant increase in eosinophil and ILC2 counts (32). Eosinophils isolated from patients with asthma have altered expression patterns of surface antigens, including CD69, indicative of their activated status (33). CD69 is a marker of tissue-resident T cells involved in the production of type 2 cytokines (34), suggesting that CD69-high eosinophils may contribute to the maintenance of tissue autonomy. Approximately 5%–10% patients with asthma have severe disease, with resistance to standard treatments and are on other treatments such as inhalation of corticosteroids and add-on agents. Blood eosinophils display EETosis more frequently in patients with severe asthma than in those with mild to moderate asthma (35). Airway eosinophils obtained from patients with atopic asthma also induce EETosis (36). In addition, eosinophils isolated from patients with severe asthma or eosinophilic chronic rhinosinusitis, its comorbidity, showed dysregulated fatty acid metabolism (37, 38). These findings are indicative of cellular changes in eosinophils due to systemic and/or local inflammatory milieu in severe asthma. Recently, biologics targeting type 2 cytokines have become available for the treatment of severe asthma. IL-5, a strong activating cytokine for eosinophils, is an optimal target for reducing eosinophilic inflammation. Mepolizumab, an IL-5-neutralizing antibody, and benralizumab, an IL-5 receptor α (IL-5Rα)-targeting antibody, dramatically reduced the number of eosinophils in the blood and airways (39, 40). Anti-IL-5 treatment restores rhinovirus-induced IFN-α secretion by pDCs in patients with asthma (21). Omalizumab, an IgE-neutralizing antibody, also enhances IFN-α responses and reduces FcεRIα protein expression in pDC, providing evidence that these effects are related (41). These biologics have been shown to reduce asthma exacerbation, frequently caused by viral infection, and spare the intake of oral corticosteroids with long-term safety (42–49). These therapeutic effects indicate that eosinophils play an important role in asthma as an inflammatory immune cells.
Viral infections account for 40%–80% cases of asthma exacerbations (50, 51). Among these, rhinovirus, RSV, and influenza virus account for most cases of asthma exacerbations. Parainfluenza viruses, human metapneumoviruses, and enteroviruses also induce asthma exacerbation at low frequencies (50, 51). Rhinoviruses and respiratory syncytial virus (RSV) suppress IFN production by airway epithelial cells and basal cells, contributing to asthma exacerbations (52, 53). In murine models of asthma, infections with these viruses and influenza viruses also induce eosinophilic airway inflammation. Elevated levels of type 2 cytokines, eosinophil-derived granule proteins, and leukotrienes have been detected in the airways of asthmatic patients infected with these viruses (54–58). In the following sections, we will discuss previous reports on the pathogenic relationship between viral infections and asthma.
Rhinoviruses most frequently cause respiratory tract infections and subsequent asthma exacerbations. Rhinovirus is correlated with asthma exacerbations in school-aged children in age-stratified time-series analysis (59). The ability to induce apoptosis for inhibiting viral replication is reduced in airway epithelial cells of asthma patients infected with rhinoviruses (52) in general, rhinovirus infection causes the production of type 1 interferons (IFN), IFN-α and IFN-β, and type 3 interferon, IFN-λ, in airway epithelial cells. However, the levels of IFNs in airway epithelial cells of patients with asthma were decreased compared with healthy subjects and those levels correlate negatively with blood eosinophil count and serum IL-4 concentration (52, 54). In contrast, human airway epithelial cells infected with rhinoviruses secrete Regulated on Activation Normal T Cell Expressed and Secreted (RANTES), resulting in accumulation of eosinophils in the airway (60). Rhinovirus infection induces pulmonary type 2 inflammation in mice. Compared to that observed after rhinovirus type A infections, rhinovirus type C infection increases the production of IL-5, IL-13, IL-25, IL-33, and thymic stromal lymphopoietin (TSLP), with abundant eosinophil infiltration in the airways (48). Airway eosinophilic inflammation did not occur in Roraflox/flox Il7r-Cre mice lacking ILC2, indicative of the importance of ILC2 in this model (61). Compared to that observed in non-sensitized mice, infection with rhinovirus type 1B resulted in eosinophilic inflammation with increased expression of IL-33 and IL-13 in a house dust-sensitized asthma model (62). These results suggested that rhinovirus infection induces type 2-high airway inflammation, which is mainly dependent on ILC2 in mice. In bronchoalveolar lavage fluid (BALF) from 11 rhinovirus-infected patients with moderate asthma, the concentrations of IL-33 and IL-25 correlated positively with those of IL-4, IL-5, and IL-13, suggesting the involvement of innate immunity-related type 2 inflammation (63, 64). ILC2 levels in BALF are predominantly higher in rhinovirus-infected asthma patients than in healthy subjects (65). In addition, the ILC2:ILC1 ratio increased in patients with asthma and correlated positively with the clinical score of asthma exacerbations and the concentration of type 2 cytokines in nasal mucosal lining fluid (65). In total, these findings indicated that rhinovirus infection in asthma diminishes type I IFN production and involves ILC2-mediated type 2 inflammation in human.
RSV infection, the major cause of bronchiolitis in children, is closely associated with the development of childhood-onset asthma. Among children who were infected with RSV within the first year of their lives, 30% of the children were diagnosed with asthma and/or wheezing within 7 years after infection (66). Non-structural protein 1 (NS1) of RSV inhibited IFN production with impaired IFN-related signaling and reduced antiviral immunity in A549 cells, a human epithelial cell line (53). In murine experiments, NS1 increased serum levels of TSLP and OX40l, inhibited the induction of regulatory T cells, and disrupted immune tolerance mechanisms (55). In an ovalbumin (OVA)-induced asthma model, RSV infection enhanced AHR, a persistent mucus production, and subepithelial fibrosis (67). In the RSV-infected asthma model, high levels of eosinophilic extracellular DNA were detected in the BALF, which is indicative of EETosis induction in the airway (68). Overall, these results suggested that RS virus infection can exacerbate asthma in mice. When stimulated with major basic protein (MBP), eosinophil peroxidase (EPO), and eosinophil granule proteins, RSV-infected human type II alveolar epithelial cells showed necrotic changes due to their cytotoxicity (57). The concentrations of ECP and EDN, other types of eosinophil granule proteins, and eosinophil chemoattractants (MIP-1α and RANTES) in nasal secretions of children with RSV-induced bronchiolitis were higher than those in nasal secretions of normal subjects (69, 70). Also, high levels of cysteinyl leukotrienes (cys-LTs) were detected in the sputum of asthma patients with RSV infection (56). Thus, eosinophils in patients with RSV infection can enhance airway inflammation and contribute tissue damage in human.
Influenza viruses are RNA viruses belonging to the Orthomyxoviridae family that cause seasonal infections in humans (71). A previous report has shown that influenza A virus was the most common cause for asthma exacerbation in 79 hospitalized patients with asthma, suggesting a close association of this virus with asthma (72). Animal experiments using asthma models have demonstrated an association between influenza virus and asthma. The OVA-sensitized asthma model of mice with influenza virus infection showed higher levels of OVA-specific IgE and AHR with airway epithelial injury than uninfected mice (73). House dust-sensitized mice infected with influenza virus showed increased AHR due to airway epithelial cell-derived IL-33-mediated responses (74). House dust-sensitized mice infected with influenza virus (X31 H3N2) showed increase in the number of ILC2 in the airway, along with high levels of IL-5 and IL-13 in BALF, especially upon virus clearance (58). Interestingly, infection with influenza type A virus induced the production of large amounts of IFN-γ in the lungs during the early phase of infection and enhanced type 2 inflammatory responses in the late phase (75). These findings emphasize the importance of influenza virus infection as a cause of asthma exacerbation with type 2 inflammation in mice. In human, among 34 children aged 2–11 years infected with H1N1 influenza A virus, 21 patients with pneumonia had higher serum levels of IFN-γ and IL-5 than patients without pneumonia. Also, patients with severe pneumonia had significantly higher serum levels of IL-4, IL-5, and IL-13 than those with mild pneumonia (76), indicative of the role of influenza virus infection in airway type 2 inflammation. In contrast, lower rates of pneumonia, mechanical ventilation, and mortality were observed in asthma patients infected with H1N1 influenza virus than in non-asthmatics (77, 78), suggesting that type 2 inflammation in the airways might provide protection from fatal H1N1 infection in human.
Parainfluenza virus is a single-stranded RNA virus belonging to the paramyxovirus family. The concentrations of cysLTs in the sputum of patients infected with parainfluenza during mild asthma exacerbations were higher than those in the sputum of patients with non-viral asthma exacerbations (79). ILC2 remains in the lungs of mice infected with parainfluenza virus after the virus has cleared and may be associated with the development of asthma (80). Studies evaluating the relationship between viral infection, asthma, and type 2 inflammation are summarized in Table 1.
Coronavirus disease 2019 (COVID-19) is an emerging infection caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that has become a global threat since its outbreak in Wuhan in December 2019. Coronaviruses, including SARS-CoV-2, are RNA viruses that cause mild to severe respiratory failure. Although some studies have reported their involvement in asthma exacerbations, the frequency of asthma patients with SARS-CoV-2 infection is lower than that of asthma patients with rhinovirus infection (81). Chronic obstructive pulmonary disease (82) and patients with asthma prescribed high-dose ICS were associated with an increased risk of death (83). Also, patients with nonallergic asthma had worse clinical outcomes that patients with allergic asthma (84). Type 2 cytokines decreased the mRNA level of angiotensin converting enzyme 2 (ACE2), a receptor for SARS-CoV-2, in epithelial cells, which may reduce the risk of infection (85). In addition, ICS decreased the expression of ACE2 in alveolar epithelial cells (86). Furthermore, the use of ICS was associated with low expression of ACE2 (87). However, patients with asthma who were prescribed high-dose ICS were at an increased risk of death. This finding indicates that high-dose ICS is possibly associated with high risk of COVID-19-related death (83), although severity of asthma may influence the outcome of COVID-19 infection. A recent study demonstrated that biological therapeutics for severe asthma can be used safely, with low risk of developing severe COVID-19 (88, 89). Another report showed that patients with severe asthma treated with anti-IL-5 receptor antibody may be at low risk of developing severe COVID-19 (90, 91). Thus, therapeutics should be used to obtain better control of the disease symptoms in patients with severe asthma during the COVID-19 pandemic.
Some clinical studies have indicated that blood eosinophil count (BEC) is a potential prognostic biomarker for COVID-19. In 140 patients hospitalized with COVID-19, more than half presented with eosinopenia during the early stage of infection (90, 92). A comparison of BEC between survivors and non-survivors during the disease course of COVID-19 showed that survivors had higher BEC than non-survivors during the recovery period (93). Among patients with asthma, eosinophils were not detected in the blood of 85% patients with COVID-19 on admission, and a BEC lower than 150/µl predicted a higher mortality rate during the disease course of COVID-19 (94). Autopsy of cases with severe COVID-19 demonstrated the absence of eosinophilic infiltration in the inflamed lung (95). Some studies have shown the activated status of blood eosinophils in COVID-19 patients. The number of blood eosinophils with high expression of CD62l increased on days 2‒6 of hospitalization. IFN-γ is suggested to be an activator of eosinophils during SARs-CoV-2 infection (96). Another report also demonstrated that CD62l-high eosinophils in severe COVID-19 cases expressed high levels of CXC chemokine receptor 4 (CXCR4) and programmed cell death 1-ligand 1 (PD-L1) (97). Interestingly, ECP and EDN concentrations in the sputum and BALF of patients with severe COVID-19 were higher in the first 10 days of severe infection than those in patients with a mild case (98). A case of suspected vasculitis, in which eosinophils accumulated around blood vessels in lung tissue with increased BEC, has been reported in COVID-19 (99). These findings suggest that eosinophils may play a protective role in COVID-19. Further studies are needed to confirm the role of eosinophils in the pathogenesis and progression of COVID-19.
IFNs are produced as antiviral molecules during viral infection in the body. Among the IFNs, IFN-γ, a type 2 interferon, is a potent activator of eosinophils. Viruses are recognized by toll-like receptors (TLRs) 3/7/8/9 and specific types of pattern recognition receptors (PRRs) expressed on immune cells that recognize microorganisms. Retinoic acid inducible gene-I (RIG-I) and melanoma differentiation-associated protein 5 are RIG-I-like receptors (RLRs) that are also virus-associated molecules. Among these receptors of human eosinophils, TLR7 and RIG-I play major roles in the cellular response to viruses (100–102). Previous studies have shown that human and murine eosinophils are capable of capturing and inactivating viruses (17). Activated eosinophils release ECP and EDN, which possess ribonuclease activity. The ribonuclease activities of these granule proteins contribute to their antiviral effects on RNA viruses. Furthermore, eosinophils can synthesize fatty acid metabolites with antiviral activity. However, the significance of eosinophils in vivo as antiviral effector cells has not been completely elucidated. In the following sections, we discuss the eosinophil-related factors associated with antiviral activity.
IFNs are proteins with virus-interfering effects that are categorized into type 1 (IFN-α and IFN-β), type 2 (IFN-γ), and type 3 IFNs (IFN-λ); type 1 IFNs show the strongest antiviral activity. In contrast, IFN-γ robustly enhances inflammatory responses and is mainly produced by Th1 cells, CD8+ T cells, macrophages, and natural killer (NK) cells. IFN-γ receptors, IFN-γR1 and IFN-γR2, mediate downstream signaling via the Janus kinase-signal transducer and activator of transcription 1 (JAK-STAT1) pathway (103). IFN-γ induces interferon-stimulated gene expression. Protein kinase R and adenosine deaminases acting on RNA (ADARs) function as antiviral proteins (104). Guanylate-binding proteins (GBPs) and tripartite motifs (TRIMs) with antiviral properties against RNA viruses, influenza virus and RSV, are also induced by IFNs (105–109). Eosinophils express functional IFN receptors, especially for IFN-γ. IFN-γ prolongs eosinophil survival more strongly than type 1 IFNs do (110–112). Blood eosinophils express CD69, an activation marker, via JAK2 upon IFN-γ stimulation (110). IFN-γ stimulation activates eosinophils that function as effector cells (113). IFN-γ stimulation induces ROS production and degranulation of eosinophils via mitogen-activated protein kinase (MAPK) (114, 115). Notably, eosinophil-derived IFN-γ enhances AHR in a murine model of asthma and stimulates the release of ECP from eosinophils (116). IFN-γ-induced antiviral activity of eosinophils has not yet been completely elucidated. IFN-γ-stimulated eosinophils exert antiviral activity via intracellular nitric oxide (NO) production (18). This promotes virus elimination in mouse models of parainfluenza infection. In addition, eosinophils bind to RV via intercellular adhesion molecule-1 (ICAM-1) and present viral antigens to RV-specific T cells to induce their IFN-γ production (117). Adhesion of eosinophils to ICAM-1 can further activate the functions of eosinophils including ROS production (118). Our research groups identified that eosinophils stimulated with IFN-α and IFN-γ upregulate the antiviral molecules GBPs and TRIMs. IFN-γ-stimulated eosinophils specifically upregulate expression of ICAM-1 and Fc gamma receptor-1A (FCGR1A) which may result in enhanced ICAM-1-mediated adhesion to rhinoviruses and enhanced inflammatory functions in response to virus with IgG cross linking (119). C-X-C motif chemokine ligand 10 (CXCL10), a ligand for CXCR3, is associated with IFN-γ. CXCL10 production induced by IFN-γ has been identified as a biomarker for rhinovirus-induced asthma exacerbations. CXCL10 stimulation increases ICAM-1 expression and reactive oxygen species (ROS) production in eosinophils, underscoring the pro-inflammatory role of IFN-γ in eosinophils (120, 121). However, further studies are required to elucidate the detailed mechanism.
TLRs are a type of PRR that recognize pathogen-associated molecular patterns. TLRs are type I transmembrane proteins with external, transmembrane, and intracytoplasmic regions. TLRs of the intracytoplasmic region activates downstream signaling pathways, including nuclear factor-kappa B (NF-κB), MAPK, and interferon regulatory factors (IRF)-3, to induce the expression of cytokines and chemokines, including IFNs (122). TLR3, TLR7, and TLR8 are localized intracellularly and recognize RNA viruses. TLR3 recognizes dsRNA, while TLR7 and TLR8 recognize single-stranded RNA (ssRNA), respectively (123). Among TLRs, eosinophils highly express TLR7. Stimulation with TLR7 ligand changes the adhesion molecule expression, ROS generation, cytokine production, and prosurvival pathways (100, 124). TLR7 expression in eosinophils was upregulated upon stimulation with IFN-γ but not with IL-4 and IL-5 (100). TLR7 signaling is mediated by p38-MAPK, phosphoinositide 3-kinase, extracellular signal-regulated kinase, and NF-κB (124). In mouse models infected with RSV and parainfluenza virus, activated eosinophils thorough TLR7 eliminate these viruses via IRF-7-mediated induction of intracellular NO and eosinophil-associated ribonucleases (EAR)-1 and EAR-2, which possess ribonuclease activity (18, 19). Interestingly, TLR7 expression in immune cells is reduced in patients with asthma (125). In murine asthma models, administration of the TLR7 ligand suppressed allergic airway inflammation (126, 127). TLR7 governs IFN-related responses to rhinovirus and its expression is suppressed by IL-5-induced lung eosinophilia (128). These results suggested that eosinophil-expressing TLR7 exerts antiviral and/or anti-allergic effects that might be impaired in asthma.
RIG-I is a cytoplasmic RNA helicase that is a retinoic acid-induced RLR. RIG-I recognizes double-stranded viral RNA. RIG-I signals are transmitted by NF-κB and IRF-3 to produce type 1 IFNs (129). RIG-I is expressed in virus-infected cells, including airway epithelial cells and macrophages. Human airway epithelial cells infected with rhinovirus express RIG-I via TLR3 (130). Eosinophils also express RIG-I and exert an RIG-I-dependent antiviral effect. In human eosinophils, α2–6 and α2–3-linked sialic acids reduce titers of the H1N1 influenza type A virus and express RIG-I mRNA while these precise mechanisms are not fully uncovered (20). In a murine model of fungus-sensitized asthma, exposure to a novel influenza virus (A/California/04/2009) increases intracellular RIG-I expression in bone marrow-derived eosinophils, leading to CD8+ T cell expansion (102). However, the mechanism underlying the RIG-I-mediated antiviral effect in eosinophils is unclear.
Eosinophils harbor abundant intracellular granule proteins, including MBP, EDN, ECP, and EPO (131). EDN and ECP exert antiviral effects on RNA viruses due to their ribonuclease activity (16). Sequences orthologous to human EDN and ECP have been identified in higher primate genomes. EDN/RNase2 and its divergent ortholog, mouse eosinophil-associated RNases (mEars), are prominent secretory proteins of eosinophils within the RNase A-type ribonuclease family (132). A previous study has revealed that the antiviral activity of EDN was higher than that of ECP (133). As mentioned above, murine eosinophils recognize the RSV via the TLR7-Myd88 system, express the genes encoding granule proteins (EAR1 and EAR2), and release ECP (19). Also, recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against RSV (134) However, the importance of granule proteins in RNA virus clearance in human remains unclear.
Eosinophils can synthesize fatty acid-derived bioactive mediators (135). During allergic inflammation, eosinophils produce large amounts of cys-LTs that are converted from arachidonic acid released from nuclear membranes. Our research revealed that human eosinophils stimulated with IL-5 produce LTD4, a ligand with high affinity to CysLT1 and CysLT2, and the combined stimulation with IL-5 and IL-4 further augments its production (136). Cys-LTs induce airway constriction, increase vascular permeability, and enhance mucus production and accumulation of inflammatory cells (137). On the other hands, eosinophils can synthesize SPMs via the fatty acid metabolizing enzyme, 15-lipoxygenase (15-LOX), which inhibits allergic airway inflammation and promotes the resolution of inflammation (138, 139). In murine tissue, tissue-resident eosinophils specifically express this enzyme, which possibly contributes to the maintenance of homeostasis (140, 141). Previous studies have shown that SPMs also play important roles in infection (142). The production of a docosahexaenoic acid (DHA)-derived SPM, protectin D1 (PD1), decreased following lethal influenza virus infection (143, 144). In a murine model of influenza virus infection, PD1 prevented fatal infection by inhibiting viral replication in airway epithelial cells (144). Results of an in vitro lipid screening assay demonstrated that 15-LOX-derived metabolites, including PD1, exerted similar inhibitory effects on viral replication (144). A recent study also showed that PD1 and another SPM, protectin conjugates in tissue regeneration 1 (PCTR1), induced the production of IFN-λ from airway epithelial cells and inhibited RSV replication (145). These results indicated that SPMs may function as antiviral molecules in vivo. Our study showed that human eosinophils are capable of synthesizing sufficient amounts of PD1 (37, 146). Interestingly, blood eosinophils isolated from patients with severe asthma have reduced capacity of producing PD1 and other 15-LOX-derived metabolites (37). Reduced PD-1 production may result from impaired DHA utilization in eosinophils and defective 15-LOX metabolic synthesis, although no change in 15-LOX was observed in eosinophils stimulated with IL-4 or IL-4 plus IL-5 compared to non-stimulated cells (136). These findings suggest that antiviral activity via 15-LOX metabolism in eosinophils may be reduced in refractory eosinophilic diseases, and further investigation is required to elucidate the balance between the pro-inflammatory and anti-inflammatory roles of eosinophils. The antiviral activities of eosinophils have been summarized in Figure 1.
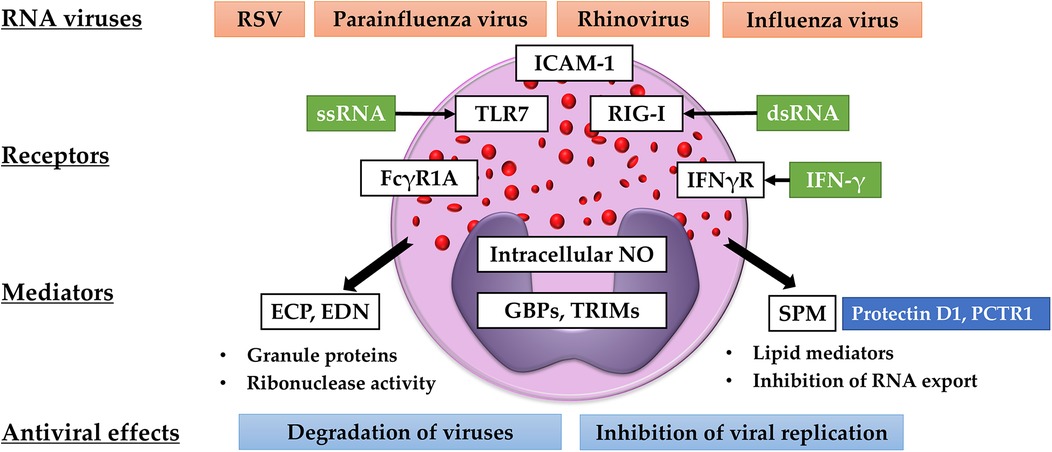
Figure 1. Antiviral activities of eosinophils. Eosinophils recognize RNA viruses via TLR-7 and RIG-I. The expression of ICAM-1, FCGR1A, and TLR-7 is upregulated by IFN-γ. Eosinophils produce ribonucleases (ECP and EDN), intracellular NO, SPMs, GBPs, and TRIMs. These mediators are believed to degrade viruses and inhibit viral replication and transport.
Eosinophils accumulate in the airways of patients with asthma due to type 2 cytokine-dependent inflammation and enhance type 2 inflammation with tissue injury. Eosinophils can also be activated by IFN-γ and can recognize RNA viruses via TLR7 and RIG-I (18, 20). They can clear viruses via ECP and EDN granule proteins and intracellular NO (16, 18). Interestingly, non-eosinophilic exacerbations triggered by viral or bacterial infections were observed in patients treated with mepolizumab (147), an anti-IL-5 antibody, suggesting that removal of eosinophils may cause expansion of virus and/or bacteria. However, patients with asthma have reduced capacity of producing IFNs and are vulnerable to viral infections (52, 54). In addition, the antiviral activity of eosinophils is impaired in asthma patients, especially in patients with severe asthma (17). Our study also reported that the expression of antiviral molecules up-regulated by IFN-γ in human eosinophils was attenuated by co-stimulation with IFN-γ and IL-5 (119). Further investigations are required to better understand the role of eosinophils in viral infection. Additionally, novel therapeutic strategies for severe asthma are required to suppress allergic inflammation and enhance antiviral defense.
HS: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. JM: Conceptualization, Methodology, Writing – original draft. AK: Supervision, Writing – review & editing. KF: Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
ECP, eosinophil cationic protein; EDN, eosinophil derived neurotoxin; dsRNA, double-stranded RNA; FCGR1A, Fc gamma receptor-1A; GBPs, guanylate binding proteins; PCTR1, protectin conjugates in tissue regeneration 1; RIG-I, retinoic acid-inducible gene-I; RSV, respiratory syncytial virus; SPM, specialized pro-resolving mediators; ssRNA, single-stranded RNA; TLR7, toll like receptor 7; TRIMs, tripartite motifs.
1. O’Sullivan JA, Bochner BS. Eosinophils and eosinophil-associated diseases: an update. J Allergy Clin Immunol. (2018) 141(2):505–17. doi: 10.1016/j.jaci.2017.09.022
2. Klion AD, Ackerman SJ, Bochner BS. Contributions of eosinophils to human health and disease. Annu Rev Pathol. (2020) 15:179–209. doi: 10.1146/annurev-pathmechdis-012419-032756
3. Ueki S, Melo RCN, Ghiran I, Spencer LA, Dvorak AM, Weller PF, et al. Eosinophil extracellular DNA trap cell death mediates lytic release of free secretion-competent eosinophil granules in humans. Blood. (2013) 121(11):2074–83. doi: 10.1182/blood-2012-05-432088
4. Gigon L, Fettrelet T, Yousefi S, Simon D, Simon HU. Eosinophils from A to Z. Allergy. (2023) 78(7):1810–46. doi: 10.1111/all.15751
5. Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. (2016) 126(9):3279–95. doi: 10.1172/JCI85664
6. Wu D, Molofsky AB, Liang HE, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. (2011) 332(6026):243–7. doi: 10.1126/science.1201475
7. Heredia JE, Mukundan L, Chen FM, Mueller AA, Deo RC, Locksley RM, et al. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. (2013) 153(2):376–88. doi: 10.1016/j.cell.2013.02.053
8. Arnold IC, Artola-Borán M, de Lara P T, Kyburz A, Taube C, Ottemann K, et al. Eosinophils suppress Th1 responses and restrict bacterially induced gastrointestinal inflammation. J Exp Med. (2018) 215(8):2055–72. doi: 10.1084/jem.20172049
9. Jung Y, Wen T, Mingler MK, Caldwell JM, Wang YH, Chaplin DD, et al. IL-1β in eosinophil-mediated small intestinal homeostasis and IgA production. Mucosal Immunol. (2015) 8(4):930–42. doi: 10.1038/mi.2014.123
10. Klion AD, Nutman TB. The role of eosinophils in host defense against helminth parasites. J Allergy Clin Immunol. (2004) 113(1):30–7. doi: 10.1016/j.jaci.2003.10.050
11. Hosoki K, Nakamura A, Nagao M, Hiraguchi Y, Tanida H, Tokuda R, et al. Staphylococcus aureus directly activates eosinophils via platelet-activating factor receptor. J Leukoc Biol. (2012) 92(2):333–41. doi: 10.1189/jlb.0112009
12. Kato M, Kephart GM, Talley NJ, Wagner JM, Sarr MG, Bonno M, et al. Eosinophil infiltration and degranulation in normal human tissue. Anat Rec. (1998) 252(3):418–25.<418::AID-AR10>3.0.CO;2-19811220
13. Persson T, Andersson P, Bodelsson M, Laurell M, Malm J, Egesten A, et al. Bactericidal activity of human eosinophilic granulocytes against Escherichia coli. Infect Immun. (2001) 69(6):3591–6. doi: 10.1128/IAI.69.6.3591-3596.2001
14. Fujieda S, Imoto Y, Kato Y, Ninomiya T, Tokunaga T, Tsutsumiuchi T, et al. Eosinophilic chronic rhinosinusitis. Allergol Int. (2019) 68(4):403–12. doi: 10.1016/j.alit.2019.07.002
15. Barroso MV, Gropillo I, Detoni MAA, Thompson-Souza GA, Muniz VS, Vasconcelos CRI, et al. Structural and signaling events driving Aspergillus fumigatus-induced human eosinophil extracellular trap release. Front Microbiol. (2021) 12:633696. doi: 10.3389/fmicb.2021.633696
16. Rosenberg HF, Domachowske JB. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. J Leukoc Biol. (2001) 70(5):691–8. doi: 10.1189/jlb.70.5.691
17. Piñeros YS S, Bal SM, Dijkhuis A, Majoor CJ, Dierdorp BS, Dekker T, et al. Eosinophils capture viruses, a capacity that is defective in asthma. Allergy. (2019) 74(10):1898–909. doi: 10.1111/all.13802
18. Drake MG, Bivins-Smith ER, Proskocil BJ, Nie Z, Scott GD, Lee JJ, et al. Human and mouse eosinophils have antiviral activity against parainfluenza virus. Am J Respir Cell Mol Biol. (2016) 55(3):387–94. doi: 10.1165/rcmb.2015-0405OC
19. Phipps S, Lam CE, Mahalingam S, Newhouse M, Ramirez R, Rosenberg HF, et al. Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood. (2007) 110(5):1578–86. doi: 10.1182/blood-2007-01-071340
20. Flores-Torres AS, Rendon A, Salinas-Carmona MC, Salinas E, Rosas-Taraco AG. Human eosinophils reduce viral titer, secrete IL-8, and increase RIG-I expression in response to influenza A H1N1 pdm09. Viral Immunol. (2021) 34(8):573–8. doi: 10.1089/vim.2021.0004
21. Dill-McFarland KA, Schwartz JT, Zhao H, Shao B, Fulkerson PC, Altman MC, et al. Eosinophil-mediated suppression and anti-IL-5 enhancement of plasmacytoid dendritic cell interferon responses in asthma. J Allergy Clin Immunol. (2022) 150(3):666–75. doi: 10.1016/j.jaci.2022.03.025
22. Mathur SK, Fichtinger PS, Kelly JT, Lee WM, Gern JE, Jarjour NN, et al. Interaction between allergy and innate immunity: model for eosinophil regulation of epithelial cell interferon expression. Ann Allergy Asthma Immunol. (2013) 111(1):25–31. doi: 10.1016/j.anai.2013.05.010
23. Miller RL, Grayson MH, Strothman K. Advances in asthma: new understandings of asthma’s natural history, risk factors, underlying mechanisms, and clinical management. J Allergy Clin Immunol. (2021) 148(6):1430–41. doi: 10.1016/j.jaci.2021.10.001
24. Ray A, Raundhal M, Oriss TB, Ray P, Wenzel SE. Current concepts of severe asthma. J Clin Invest. (2016) 126(7):2394–403. doi: 10.1172/JCI84144
25. Walsh GM. Biologics targeting IL-5, IL-4 or IL-13 for the treatment of asthma - an update. Expert Rev Clin Immunol. (2017) 13(2):143–9. doi: 10.1080/1744666X.2016.1216316
26. Bjerregaard A, Laing A, Backer A, Sverrild A, Khoo S-K, Chidlow G, et al. High fractional exhaled nitric oxide and sputum eosinophils are associated with an increased risk of future virus-induced exacerbations: a prospective cohort study. Clin Exp Allergy. (2017) 47(8):1007–13. doi: 10.1111/cea.12935
27. Asano K, Ueki S, Tamari M, Imoto Y, Fujieda S, Taniguchi M, et al. Adult-onset eosinophilic airway diseases. Allergy. (2020) 75(12):3087–99. doi: 10.1111/all.14620
28. Lee JJ, McGarry MP, Farmer SC, Denzler KL, Larson KA, Carrigan PE, et al. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med. (1997) 185(12):2143–56. doi: 10.1084/jem.185.12.2143
29. Ueki S, Konno Y, Takeda M, Moritoki Y, Hirokawa M, Matsuwaki Y, et al. Eosinophil extracellular trap cell death-derived DNA traps: their presence in secretions and functional attributes. J Allergy Clin Immunol. (2016) 137(1):258–67. doi: 10.1016/j.jaci.2015.04.041
30. Muniz VS, Silva JC, Braga YAV, Melo RCN, Ueki S, Takeda M, et al. Eosinophils release extracellular DNA traps in response to Aspergillus fumigatus. J Allergy Clin Immunol. (2018) 141(2):571–585.e7. doi: 10.1016/j.jaci.2017.07.048
31. Ueki S, Tokunaga T, Melo RCN, Saito H, Honda K, Fukuchi M, et al. Charcot-Leyden crystal formation is closely associated with eosinophil extracellular trap cell death. Blood. (2018) 132(20):2183–7. doi: 10.1182/blood-2018-04-842260
32. Choi Y, Kim YM, Lee HR, Mun J, Sim S, Lee DH, et al. Eosinophil extracellular traps activate type 2 innate lymphoid cells through stimulating airway epithelium in severe asthma. Allergy. (2020) 75(1):95–103. doi: 10.1111/all.13997
33. Johansson MW. Activation states of blood eosinophils in asthma. Clin Exp Allergy. (2014) 44(4):482–98. doi: 10.1111/cea.12292
34. Kobayashi T, Iijima K, Matsumoto K, Lama JK, Kita H. Lung-resident CD69+ST2+ TH2 cells mediate long-term type 2 memory to inhaled antigen in mice. J Allergy Clin Immunol. (2023) 152(1):167–81. doi: 10.1016/j.jaci.2023.01.016
35. Choi Y, Pham DL, Lee DH, Lee SH, Kim SH, Park HS, et al. Biological function of eosinophil extracellular traps in patients with severe eosinophilic asthma. Exp Mol Med. (2018) 50(8):1–8. doi: 10.1038/s12276-018-0136-8
36. Dworski R, Simon HU, Hoskins A, Yousefi S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J Allergy Clin Immunol. (2011) 127(5):1260–6. doi: 10.1016/j.jaci.2010.12.1103
37. Miyata J, Fukunaga K, Iwamoto R, Isobe Y, Niimi K, Takamiya R, et al. Dysregulated synthesis of protectin D1 in eosinophils from patients with severe asthma. J Allergy Clin Immunol. (2013) 131(2):353–60.e1-2. doi: 10.1016/j.jaci.2012.07.048
38. Miyata J, Fukunaga K, Kawashima Y, Watanabe T, Saitoh A, Hirosaki T, et al. Dysregulated fatty acid metabolism in nasal polyp-derived eosinophils from patients with chronic rhinosinusitis. Allergy. (2019) 74(6):1113–24. doi: 10.1111/all.13726
39. Laviolette M, Gossage DL, Gauvreau G, Leigh R, Olivenstein R, Katial R, et al. Effects of benralizumab on airway eosinophils in asthmatic patients with sputum eosinophilia. J Allergy Clin Immunol. (2013) 132(5):1086–1096.e5. doi: 10.1016/j.jaci.2013.05.020
40. Kelly EA, Esnault S, Liu LY, Evans MD, Johansson MW, Mathur S, et al. Mepolizumab attenuates airway eosinophil numbers, but not their functional phenotype, in asthma. Am J Respir Crit Care Med. (2017) 196(11):1385–95. doi: 10.1164/rccm.201611-2234OC
41. Gill MA, Liu AH, Calatroni A, Krouse RZ, Shao B, Schiltz A, et al. Enhanced plasmacytoid dendritic cell antiviral responses after omalizumab. J Allergy Clin Immunol. (2018) 141(5):1735–1743.e9. doi: 10.1016/j.jaci.2017.07.035
42. Pavord ID, Korn S, Howarth P, Bleecker ER, Buhl R, Keene ON, et al. Mepolizumab for severe eosinophilic asthma (DREAM): a multicentre, double-blind, placebo-controlled trial. Lancet. (2012) 380(9842):651–9. doi: 10.1016/S0140-6736(12)60988-X
43. Bleecker ER, FitzGerald JM, Chanez P, Papi A, Weinstein SF, Barker P, et al. Efficacy and safety of benralizumab for patients with severe asthma uncontrolled with high-dosage inhaled corticosteroids and long-acting β2-agonists (SIROCCO): a randomised, multicentre, placebo-controlled phase 3 trial. Lancet. (2016) 388(10056):2115–27. doi: 10.1016/S0140-6736(16)31324-1
44. Ortega HG, Liu MC, Pavord ID, Brusselle GG, FitzGerald JM, Chetta A, et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med. (2014) 371(13):1198–207. doi: 10.1056/NEJMoa1403290
45. Chupp GL, Bradford ES, Albers FC, Bratton DJ, Wang-Jairaj J, Nelsen LM, et al. Efficacy of mepolizumab add-on therapy on health-related quality of life and markers of asthma control in severe eosinophilic asthma (MUSCA): a randomised, double-blind, placebo-controlled, parallel-group, multicentre, phase 3b trial. Lancet Respir Med. (2017) 5(5):390–400. doi: 10.1016/S2213-2600(17)30125-X
46. FitzGerald JM, Bleecker ER, Nair P, Korn S, Ohta K, Lommatzsch M, et al. Benralizumab, an anti-interleukin-5 receptor α monoclonal antibody, as add-on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet. (2016) 388(10056):2128–41. doi: 10.1016/S0140-6736(16)31322-8
47. Jackson DJ, Korn S, Mathur SK, Barker P, Meka VG, Martin UJ, et al. Safety of eosinophil-depleting therapy for severe, eosinophilic asthma: focus on benralizumab. Drug Saf. (2020) 43(5):409–25. doi: 10.1007/s40264-020-00926-3
48. Principe S, Porsbjerg C, Ditlev SB, Klein DK, Golebski K, Dyhre-Petersen N, et al. Treating severe asthma: targeting the IL-5 pathway. Clin Exp Allergy. (2021) 51(8):992–1005. doi: 10.1111/cea.13885
49. Ravanetti L, Dijkhuis A, Sabogal Pineros YS, Bal SM, Dierdorp BS, Dekker T, et al. An early innate response underlies severe influenza-induced exacerbations of asthma in a novel steroid-insensitive and anti-IL-5-responsive mouse model. Allergy. (2017) 72(5):737–53. doi: 10.1111/all.13057
50. Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, et al. Community study of role of viral infections in exacerbations of asthma in 9–11 year-old children. Br Med J. (1995) 310(6989):1225–9. doi: 10.1136/bmj.310.6989.1225
51. El-Sahly HM, Atmar RL, Glezen WP, Greenberg SB. Spectrum of clinical illness in hospitalized patients with “common cold” virus infections. Clin Infect Dis. (2000) 31(1):96–100. doi: 10.1086/313937
52. Wark PAB, Johnston SL, Bucchieri F, Powell R, Puddicombe S, Laza-Stanca V, et al. Asthmatic bronchial epithelial cells have a deficient innate immune response to infection with rhinovirus. J Exp Med. (2005) 201(6):937–47. doi: 10.1084/jem.20041901
53. Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G, et al. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat Med. (2005) 11(1):56–62. doi: 10.1038/nm1174
54. Rupani H, Busse WW, Howarth PH, Bardin PG, Adcock IM, Konno S, et al. Therapeutic relevance of eosinophilic inflammation and airway viral interactions in severe asthma. Allergy. (2024) 79(10):2589–604. doi: 10.1111/all.16242
55. Fan P, Liu Z, Zheng M, Chen M, Xu Y, Zhao D. Respiratory syncytial virus nonstructural protein 1 breaks immune tolerance in mice by downregulating tregs through TSLP-OX40/OX40l-mTOR axis. Mol Immunol. (2021) 138:20–30. doi: 10.1016/j.molimm.2021.07.019
56. Matsuse H, Tsuchida T, Fukahori S, Kawano T, Tomari S, Matsuo N, et al. Differential airway inflammatory responses in asthma exacerbations induced by respiratory syncytial virus and influenza virus A. Int Arch Allergy Immunol. (2013) 161(4):378–82. doi: 10.1159/000348381
57. Kato M, Ishioka T, Kita H, Kozawa K, Hayashi Y, Kimura H. Eosinophil granular proteins damage bronchial epithelial cells infected with respiratory syncytial virus. Int Arch Allergy Immunol. (2012) 158(Suppl 1):11–8. doi: 10.1159/000337752
58. Li BWS, de Bruijn MJW, Lukkes M, van Nimwegen M, Bergen IM, KleinJan A, et al. T cells and ILC2s are major effector cells in influenza-induced exacerbation of allergic airway inflammation in mice. Eur J Immunol. (2019) 49(1):144–56. doi: 10.1002/eji.201747421
59. Lee SW, Yon DK, James CC, Lee S, Koh HY, Sheen YH, et al. Short-term effects of multiple outdoor environmental factors on risk of asthma exacerbations: age-stratified time-series analysis. J Allergy Clin Immunol. (2019) 144(6):1542–1550.e1. doi: 10.1016/j.jaci.2019.08.037
60. Schroth MK, Grimm E, Frindt P, Galagan DM, Konno SI, Love R, et al. Rhinovirus replication causes RANTES production in primary bronchial epithelial cells. Am J Respir Cell Mol Biol. (1999) 20(6):1220–8. doi: 10.1165/ajrcmb.20.6.3261
61. Rajput C, Han M, Ishikawa T, Lei J, Goldsmith AM, Jazaeri S, et al. Rhinovirus C infection induces type 2 innate lymphoid cell expansion and eosinophilic airway inflammation. Front Immunol. (2021) 12:649520. doi: 10.3389/fimmu.2021.649520
62. Mehta AK, Croft M. Rhinovirus infection promotes eosinophilic airway inflammation after prior exposure to house dust mite allergen. Immunohorizons. (2020) 4(8):498–507. doi: 10.4049/immunohorizons.2000052
63. Southworth T, Pattwell C, Khan N, Mowbray SF, Strieter RM, Erpenbeck VJ, et al. Increased type 2 inflammation post rhinovirus infection in patients with moderate asthma. Cytokine. (2020) 125:154857. doi: 10.1016/j.cyto.2019.154857
64. Jackson DJ, Makrinioti H, Rana BMJ, Shamji BWH, Trujillo-Torralbo MB, Footitt J, et al. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med. (2014) 190(12):1373–82. doi: 10.1164/rccm.201406-1039OC
65. Dhariwal J, Cameron A, Wong E, Paulsen M, Trujillo-Torralbo MB, Del Rosario A, et al. Pulmonary innate lymphoid cell responses during rhinovirus-induced asthma exacerbations in vivo: a clinical trial. Am J Respir Crit Care Med. (2021) 204(11):1259–73. doi: 10.1164/rccm.202010-3754OC
66. Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B. Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am J Respir Crit Care Med. (2000) 161(5):1501–7. doi: 10.1164/ajrccm.161.5.9906076
67. Becnel D, You D, Erskin J, Dimina DM, Cormier SA. A role for airway remodeling during respiratory syncytial virus infection. Respir Res. (2005) 6(1):122. doi: 10.1186/1465-9921-6-122
68. Silveira JS, Antunes GL, Gassen RB, Breda RV, Stein RT, Pitrez PM, et al. Respiratory syncytial virus increases eosinophil extracellular traps in a murine model of asthma. Asia Pac Allergy. (2019) 9(4):e32. doi: 10.5415/apallergy.2019.9.e32
69. Harrison AM, Bonville CA, Rosenberg HF, Domachowske JB. Respiratory syncytial virus-induced chemokine expression in the lower airways: eosinophil recruitment and degranulation. Am J Respir Crit Care Med. (1999) 159(6):1918–24. doi: 10.1164/ajrccm.159.6.9805083
70. Chung HL, Kim SG. RANTES may be predictive of later recurrent wheezing after respiratory syncytial virus bronchiolitis in infants. Ann Allergy Asthma Immunol. (2002) 88(5):463–7. doi: 10.1016/S1081-1206(10)62383-6
71. Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. (2014) 14(5):315–28. doi: 10.1038/nri3665
72. Teichtahl H, Buckmaster N, Pertnikovs E. The incidence of respiratory tract infection in adults requiring hospitalization for asthma. Chest. (1997) 112(3):591–6. doi: 10.1378/chest.112.3.591
73. Looi K, Larcombe AN, Perks KL, Berry LJ, Zosky GR, Rigby P, et al. Previous influenza infection exacerbates allergen specific response and impairs airway barrier integrity in pre-sensitized mice. Int J Mol Sci. (2021) 22(16):8790. doi: 10.3390/ijms22168790
74. Ravanetti L, Dijkhuis A, Dekker T, Sabogal Pineros YS, Ravi A, Dierdorp BS, et al. IL-33 drives influenza-induced asthma exacerbations by halting innate and adaptive antiviral immunity. J Allergy Clin Immunol. (2019) 143(4):1355–70.e16. doi: 10.1016/j.jaci.2018.08.051
75. Dahl ME, Dabbagh K, Liggitt D, Kim S, Lewis DB. Viral-induced T helper type 1 responses enhance allergic disease by effects on lung dendritic cells. Nat Immunol. (2004) 5(3):337–43. doi: 10.1038/ni1041
76. Takano T, Tajiri H, Kashiwagi Y, Kimura S, Kawashima H. Cytokine and chemokine response in children with the 2009 pandemic influenza A (H1N1) virus infection. Eur J Clin Microbiol Infect Dis. (2011) 30(1):117–20. doi: 10.1007/s10096-010-1041-9
77. McKenna JJ, Bramley AM, Skarbinski J, Fry AM, Finelli L, Jain S, et al. Asthma in patients hospitalized with pandemic influenza A(H1N1) pdm09 virus infection-United States, 2009. BMC Infect Dis. (2013) 13:57. doi: 10.1186/1471-2334-13-57
78. Bramley AM, Dasgupta S, Skarbinski J, Kamimoto L, Fry AM, Finelli L, et al. Intensive care unit patients with 2009 pandemic influenza A (H1N1pdm09) virus infection - United States, 2009. Influenza Other Respir Viruses. (2012) 6(6):e134–42. doi: 10.1111/j.1750-2659.2012.00385.x
79. Matsuse H, Kondo Y, Saeki S, Nakata H, Fukushima C, Mizuta Y, et al. Naturally occurring parainfluenza virus 3 infection in adults induces mild exacerbation of asthma associated with increased sputum concentrations of cysteinyl leukotrienes. Int Arch Allergy Immunol. (2005) 138(3):267–72. doi: 10.1159/000088728
80. Castro IA, Yang Y, Gnazzo V, Kim D-H, Dyken SJV, López CB, et al. Murine parainfluenza virus persists in lung innate immune cells sustaining chronic lung pathology. Nat Microbiol. (2024) 9(11):2803–16. doi: 10.1038/s41564-024-01805-8
81. Padayachee Y, Faiez TS, Singanayagam A, Mallia P, Johnston SL. Asthma and viruses: a focus on rhinoviruses and SARS-CoV-2. J Allergy Clin Immunol. (2021) 147(5):1648–51. doi: 10.1016/j.jaci.2021.03.011
82. Zhao Q, Meng M, Kumar R, Wu Y, Huang J, Lian N, et al. The impact of COPD and smoking history on the severity of COVID-19: a systemic review and meta-analysis. J Med Virol. (2020) 92(10):1915–21. doi: 10.1002/jmv.25889
83. Schultze A, Walker AJ, MacKenna B, Morton CE, Bhaskaran K, Brown JP, et al. Risk of COVID-19-related death among patients with chronic obstructive pulmonary disease or asthma prescribed inhaled corticosteroids: an observational cohort study using the OpenSAFELY platform. Lancet Respir Med. (2020) 8(11):1106–20. doi: 10.1016/S2213-2600(20)30415-X
84. Yang JM, Koh HY, Moon SY, Yoo IK, Ha EK, You S, et al. Allergic disorders and susceptibility to and severity of COVID-19: a nationwide cohort study. J Allergy Clin Immunol. (2020) 146(4):790–8. doi: 10.1016/j.jaci.2020.08.008
85. Bradding P, Richardson M, Hinks TSC, Howarth PH, Choy DF, Arron JR, et al. ACE2, TMPRSS2, and furin gene expression in the airways of people with asthma-implications for COVID-19. J Allergy Clin Immunol. (2020) 146(1):208–11. doi: 10.1016/j.jaci.2020.05.013
86. Finney LJ, Glanville N, Farne H, Aniscenko J, Fenwick P, Kemp SV, et al. Inhaled corticosteroids downregulate the SARS-CoV-2 receptor ACE2 in COPD through suppression of type I interferon. J Allergy Clin Immunol. (2021) 147(2):510–19.e5. doi: 10.1016/j.jaci.2020.09.034
87. Peters MC, Sajuthi S, Deford P, Christenson S, Rios CL, Montgomery MT, et al. COVID-19-related genes in sputum cells in asthma. Relationship to demographic features and corticosteroids. Am J Respir Crit Care Med. (2020) 202(1):83–90. doi: 10.1164/rccm.202003-0821OC
88. Morais-Almeida M, Aguiar R, Martin B, Ansotegui IJ, Ebisawa M, Arruda LK, et al. COVID-19, asthma, and biological therapies: what we need to know. World Allergy Organ J. (2020) 13(5):100126. doi: 10.1016/j.waojou.2020.100126
89. Izquierdo JL, Soriano JB. Biologics may have a beneficial effect in asthma patients with COVID-19. Eur Respir J. (2021) 58(2):2101076. doi: 10.1183/13993003.01076-2021
90. García-Moguel I, Díaz Campos R, Alonso Charterina S, Fernández Rodríguez C, Fernández Crespo J. COVID-19, severe asthma, and biologics. Ann Allergy Asthma Immunol. (2020) 125(3):357–359.e1. doi: 10.1016/j.anai.2020.06.012
91. Pala D, Pistis M. Anti-IL5 drugs in COVID-19 patients: role of eosinophils in SARS-CoV-2-induced immunopathology. Front Pharmacol. (2021) 12:622554. doi: 10.3389/fphar.2021.622554
92. Zhang JJ, Dong X, Cao YY, Yuan YD, Yang YB, Yan YQ, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy. (2020) 75(7):1730–41. doi: 10.1111/all.14238
93. Mateos González M, Sierra Gonzalo E, Casado Lopez I, Arnalich Fernández F, Beato Pérez JL, Monge Monge D, et al. The prognostic value of eosinophil recovery in COVID-19: a multicentre, retrospective cohort study on patients hospitalised in Spanish hospitals. J Clin Med. (2021) 10(2):305. doi: 10.3390/jcm10020305
94. Ferastraoaru D, Hudes G, Jerschow E, Jariwala S, Karagic M, de Vos G, et al. Eosinophilia in asthma patients is protective against severe COVID-19 illness. J Allergy Clin Immunol Pract. (2021) 9(3):1152–1162.e3. doi: 10.1016/j.jaip.2020.12.045
95. Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. COVID-19 autopsies, Oklahoma, USA. Am J Clin Pathol. (2020) 153(6):725–33. doi: 10.1093/ajcp/aqaa062
96. Rodriguez L, Pekkarinen PT, Lakshmikanth T, Tan Z, Rosat Consiglio C, Pou C, et al. Systems-level immunomonitoring from acute to recovery phase of severe COVID-19. Cell Rep Med. (2020) 1(5):100078. doi: 10.1016/j.xcrm.2020.100078
97. Lourda M, Dzidic M, Hertwig L, Bergsten H, Palma Medina LM, Sinha I, et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. Proc Natl Acad Sci U S A. (2021) 118(40):e2109123118. doi: 10.1073/pnas.2109123118
98. Kim DM, Kim Y, Seo JW, Lee J, Park U, Ha NY, et al. Enhanced eosinophil-mediated inflammation associated with antibody and complement-dependent pneumonic insults in critical COVID-19. Cell Rep. (2021) 37(1):109798. doi: 10.1016/j.celrep.2021.109798
99. Luecke E, Jeron A, Kroeger A, Bruder D, Stegemann-Koniszewski S, Jechorek D, et al. Eosinophilic pulmonary vasculitis as a manifestation of the hyperinflammatory phase of COVID-19. J Allergy Clin Immunol. (2021) 147(1):112–3. doi: 10.1016/j.jaci.2020.09.026
100. Nagase H, Okugawa S, Ota Y, Yamaguchi M, Tomizawa H, Matsushima K, et al. Expression and function of toll-like receptors in eosinophils: activation by toll-like receptor 7 ligand. J Immunol. (2003) 171(8):3977–82. doi: 10.4049/jimmunol.171.8.3977
101. Gebremeskel S, Schanin J, Coyle KM, Butuci M, Luu T, Brock EC, et al. Mast cell and eosinophil activation are associated with COVID-19 and TLR-mediated viral inflammation: implications for an anti-siglec-8 antibody. Front Immunol. (2021) 12:650331. doi: 10.3389/fimmu.2021.650331
102. LeMessurier KS, Rooney R, Ghoneim HE, Liu B, Li K, Smallwood HS, et al. Influenza A virus directly modulates mouse eosinophil responses. J Leukoc Biol. (2020) 108(1):151–68. doi: 10.1002/JLB.4MA0320-343R
103. Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. (2018) 18(9):545–58. doi: 10.1038/s41577-018-0029-z
104. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol. (2004) 75(2):163–89. doi: 10.1189/jlb.0603252
105. Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, et al. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One. (2013) 8(2):e55920. doi: 10.1371/journal.pone.0055920
106. Braun E, Hotter D, Koepke L, Zech F, Groß R, Sparrer KMJ, et al. Guanylate-Binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep. (2019) 27(7):2092–2104.e10. doi: 10.1016/j.celrep.2019.04.063
107. Li Z, Qu X, Liu X, Huan C, Wang H, Zhao Z, et al. GBP5 is an interferon-induced inhibitor of respiratory syncytial virus. J Virol. (2020) 94(21):e01407–20. doi: 10.1128/JVI.01407-20
108. Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, Mechti N, et al. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol. (2013) 87(8):4523–33. doi: 10.1128/JVI.02548-12
109. Wang Y, Chen Y, Lin Y, Quan Y, Xiao X, Zhang R. TRIM22 inhibits respiratory syncytial virus replication by targeting JAK-STAT1/2 signaling. J Med Virol. (2021) 93(6):3412–9. doi: 10.1002/jmv.26436
110. Ochiai K, Tanabe E, Ishihara C, Kagami M, Sugiyama T, Sueishi M, et al. Role of JAK2 signal transductional pathway in activation and survival of human peripheral eosinophils by interferon-gamma (IFN-gamma). Clin Exp Immunol. (1999) 118(3):340–3. doi: 10.1046/j.1365-2249.1999.01068.x
111. Mikami C, Ochiai K, Umemiya K, Matsumura R, Kagami M, Tomioka H, et al. Eosinophil activation and in situ interleukin-5 production by mononuclear cells in skin lesions of patients with drug hypersensitivity. J Dermatol. (1999) 26(10):633–9. doi: 10.1111/j.1346-8138.1999.tb02064.x
112. Létuvé S, Druilhe A, Grandsaigne M, Aubier M, Pretolani M. Involvement of caspases and of mitochondria in fas ligation-induced eosinophil apoptosis: modulation by interleukin-5 and interferon-gamma. J Leukoc Biol. (2001) 70(5):767–75. doi: 10.1189/jlb.70.5.767
113. Valerius T, Repp R, Kalden JR, Platzer E. Effects of IFN on human eosinophils in comparison with other cytokines. A novel class of eosinophil activators with delayed onset of action. J Immunol. (1990) 145(9):2950–8. doi: 10.4049/jimmunol.145.9.2950
114. Yamaguchi T, Kimura H, Kurabayashi M, Kozawa K, Kato M. Interferon-gamma enhances human eosinophil effector functions induced by granulocyte-macrophage colony-stimulating factor or interleukin-5. Immunol Lett. (2008) 118(1):88–95. doi: 10.1016/j.imlet.2008.03.005
115. Ishihara C, Ochiai K, Kagami M, Takashahi H, Matsuyama G, Yoshida S, et al. Human peripheral eosinophils express functional interferon-gamma receptors (IFN-gammaR). Clin Exp Immunol. (1997) 110(3):524–9. doi: 10.1046/j.1365-2249.1997.4511469.x
116. Kanda A, Driss V, Hornez N, Abdallah M, Roumier T, Abboud G, et al. Eosinophil-derived IFN-gamma induces airway hyperresponsiveness and lung inflammation in the absence of lymphocytes. J Allergy Clin Immunol. (2009) 124(3):573–82. doi: 10.1016/j.jaci.2009.04.031
117. Handzel ZT, Busse WW, Sedgwick JB, Vrtis R, Lee WM, Kelly EA, et al. Eosinophils bind rhinovirus and activate virus-specific T cells. J Immunol. (1998) 160(3):1279–84. doi: 10.4049/jimmunol.160.3.1279
118. Nagata M, Sedgwick JB, Bates ME, Kita H, Busse WW. Eosinophil adhesion to vascular cell adhesion molecule-1 activates superoxide anion generation. J Immunol. (1995) 155(4):2194–202. doi: 10.4049/jimmunol.155.4.2194
119. Sasaki H, Miyata J, Kawashima Y, Konno R, Ishikawa M, Hasegawa Y, et al. Distinct roles of types 1 and 2 interferons in human eosinophil regulation: a multi-omics analysis. Allergy. (2024) 79(11):3141–5. doi: 10.1111/all.16215
120. Osman HM, Basha NRE, Mansour AF, Hanna MOF. Serum IFNγ-induced protein 10 (IP10/CXCL10): association with asthma exacerbations and severity in children. J Asthma. (2022) 59(11):2135–42. doi: 10.1080/02770903.2021.1999465
121. Wark PAB, Bucchieri F, Johnston SL, Gibson PG, Hamilton L, Mimica J, et al. IFN-gamma-induced protein 10 is a novel biomarker of rhinovirus-induced asthma exacerbations. J Allergy Clin Immunol. (2007) 120(3):586–93. doi: 10.1016/j.jaci.2007.04.046
122. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. (2011) 34(5):637–50. doi: 10.1016/j.immuni.2011.05.006
123. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. (2010) 32(3):305–15. doi: 10.1016/j.immuni.2010.03.012
124. Wong CK, Cheung PFY, Ip WK, Lam CWK. Intracellular signaling mechanisms regulating toll-like receptor-mediated activation of eosinophils. Am J Respir Cell Mol Biol. (2007) 37(1):85–96. doi: 10.1165/rcmb.2006-0457OC
125. Roponen M, Yerkovich ST, Hollams E, Sly PD, Holt PG, Upham JW. Toll-like receptor 7 function is reduced in adolescents with asthma. Eur Respir J. (2010) 35(1):64–71. doi: 10.1183/09031936.00172008
126. Moisan J, Camateros P, Thuraisingam T, Marion D, Koohsari H, Martin P, et al. TLR7 ligand prevents allergen-induced airway hyperresponsiveness and eosinophilia in allergic asthma by a MYD88-dependent and MK2-independent pathway. Am J Physiol Lung Cell Mol Physiol. (2006) 290(5):987–95. doi: 10.1152/ajplung.00440.2005
127. Okuzumi S, Miyata J, Kabata H, Mochimaru T, Kagawa S, Masaki K, et al. TLR7 agonist suppresses group 2 innate lymphoid cell-mediated inflammation via IL-27-producing interstitial macrophages. Am J Respir Cell Mol Biol. (2021) 65(3):309–18. doi: 10.1165/rcmb.2021-0042OC
128. Hatchwell L, Collison A, Girkin J, Parsons K, Li J, Zhang J, et al. Toll-like receptor 7 governs interferon and inflammatory responses to rhinovirus and is suppressed by IL-5-induced lung eosinophilia. Thorax. (2015) 70(9):854–61. doi: 10.1136/thoraxjnl-2014-205465
129. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. (2004) 5(7):730–7. doi: 10.1038/ni1087
130. Slater L, Bartlett NW, Haas JJ, Zhu J, Message SD, Walton RP, et al. Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathog. (2010) 6(11):e1001178. doi: 10.1371/journal.ppat.1001178
131. Chopra A, Batra JK. Antimicrobial activity of human eosinophil granule proteins. Methods Mol Biol. (2014) 1178:267–81. doi: 10.1007/978-1-4939-1016-8_23
132. Rosenberg HF. Eosinophil-derived neurotoxin (EDN/RNase 2) and the mouse eosinophil-associated RNases (mEars): expanding roles in promoting host defense. Int J Mol Sci. (2015) 16(7):15442–55. doi: 10.3390/ijms160715442
133. Barker RL, Loegering DA, Ten RM, Hamann KJ, Pease LR, Gleich GJ. Eosinophil cationic protein cDNA. Comparison with other toxic cationic proteins and ribonucleases. J Immunol. (1989) 143(3):952–5. doi: 10.4049/jimmunol.143.3.952
134. Domachowske JB, Dyer KD, Bonville CA. Recombinant human eosinophil-derived neurotoxin/RNase 2 functions as an effective antiviral agent against respiratory syncytial virus. J Infect Dis. (1998) 177(6):1458–64. doi: 10.1086/515322
135. Miyata J, Fukunaga K, Kawashima Y, Ohara O, Arita M. Cysteinyl leukotriene metabolism of human eosinophils in allergic disease. Allergol Int. (2020) 69(1):28–34. doi: 10.1016/j.alit.2019.06.002
136. Sunata K, Miyata J, Kawashima Y, Konno R, Ishikawa M, Hasegawa Y, et al. Multiomics analysis identified IL-4-induced IL1RL1high eosinophils characterized by prominent cysteinyl leukotriene metabolism. J Allergy Clin Immunol. (2024) 154(5):1277–88. doi: 10.1016/j.jaci.2024.07.012
137. Yamamoto T, Miyata J, Arita M, Fukunaga K, Kawana A. Current state and future prospect of the therapeutic strategy targeting cysteinyl leukotriene metabolism in asthma. Respir Investig. (2019) 57(6):534–43. doi: 10.1016/j.resinv.2019.08.003
138. Miyata J, Arita M. Role of omega-3 fatty acids and their metabolites in asthma and allergic diseases. Allergol Int. (2015) 64(1):27–34. doi: 10.1016/j.alit.2014.08.003
139. Miyata J, Yokokura Y, Moro K, Arai H, Fukunaga K, Arita M, et al. 12/15-lipoxygenase regulates IL-33-induced eosinophilic airway inflammation in mice. Front Immunol. (2021) 12:687192. doi: 10.3389/fimmu.2021.687192
140. Zhu C, Weng QY, Zhou LR, Cao C, Li F, Wu YF, et al. Homeostatic and early-recruited CD101- eosinophils suppress endotoxin-induced acute lung injury. Eur Respir J. (2020) 56(5):1902354. doi: 10.1183/13993003.02354-2019
141. Andreev D, Liu M, Kachler K, Llerins Perez M, Kirchner P, Kölle J, et al. Regulatory eosinophils induce the resolution of experimental arthritis and appear in remission state of human rheumatoid arthritis. Ann Rheum Dis. (2021) 80(4):451–68. doi: 10.1136/annrheumdis-2020-218902
142. Chiang N, Fredman G, Bäckhed F, Oh SF, Vickery T, Schmidt BA, et al. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. (2012) 484(7395):524–8. doi: 10.1038/nature11042
143. Tam VC, Quehenberger O, Oshansky CM, Suen R, Armando AM, Treuting PM, et al. Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell. (2013) 154(1):213–27. doi: 10.1016/j.cell.2013.05.052
144. Morita M, Kuba K, Ichikawa A, Nakayama M, Katahira J, Iwamoto R, et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. (2013) 153(1):112–25. doi: 10.1016/j.cell.2013.02.027
145. Walker KH, Krishnamoorthy N, Brüggemann TR, Shay AE, Serhan CN, Levy BD. Protectins PCTR1 and PD1 reduce viral load and lung inflammation during respiratory syncytial virus infection in mice. Front Immunol. (2021) 12:704427. doi: 10.3389/fimmu.2021.704427
146. Miyata J, Fukunaga K, Kawashima Y, Ohara O, Kawana A, Asano K, et al. Dysregulated metabolism of polyunsaturated fatty acids in eosinophilic allergic diseases. Prostaglandins Other Lipid Mediat. (2020) 150:106477. doi: 10.1016/j.prostaglandins.2020.106477
147. McDowell PJ, Diver S, Yang F, Borg C, Busby J, Brown V, et al. The inflammatory profile of exacerbations in patients with severe refractory eosinophilic asthma receiving mepolizumab (the MEX study): a prospective observational study. Lancet Respir Med. (2021) 9(10):1174–84. doi: 10.1016/S2213-2600(21)00004-7
Keywords: asthma, asthma exacerbation, eosinophil, interferon, antiviral activities
Citation: Sasaki H, Miyata J, Kawana A and Fukunaga K (2025) Antiviral roles of eosinophils in asthma and respiratory viral infection. Front. Allergy 6:1548338. doi: 10.3389/falgy.2025.1548338
Received: 19 December 2024; Accepted: 7 February 2025;
Published: 28 February 2025.
Edited by:
Yoshiyuki Yamada, Tokai University, JapanReviewed by:
Tomoyuki Soma, Saitama Medical University, JapanCopyright: © 2025 Sasaki, Miyata, Kawana and Fukunaga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Miyata, anVubWl5YXRhLmEyQGtlaW8uanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.