
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Allergy, 27 February 2025
Sec. Therapies and Therapeutic Targets
Volume 6 - 2025 | https://doi.org/10.3389/falgy.2025.1497914
Background: A reduction in biodiversity and alterations in the microbiota composition are relevant to allergic diseases. However, combined analyses of the skin, nasal and gut microbiotas are lacking in the literature. In addition, in previous studies, microbiota were detected mainly by V3–V4 sequencing, but other sequences might be missed with this technique.
Methods: In this case–control study, we enrolled 3–12-year-old children with allergic rhinitis combined with atopic dermatitis and food allergy (AR-AD-FA group), children with allergic rhinitis only (AR-only) and healthy controls (HC group). We employed full-length 16S rRNA gene amplification and sequencing for the detection of gut, nasal and skin microbiota.
Results: Samples with an average sequence length of 1,459 bp were obtained in this study. Significant differences in beta diversity in the three compartments were found between the disease groups and the HC group. Differentially expressed genera were present mainly in the gut compartment. Peptoniphilus, Prevotella and Anaerococcus were abundant in the gut in the disease groups. Specifically, Streptomyces, Thermus and Pseudomonas showed differential expression in both the nasal and skin compartments of children in the disease groups.
Conclusion: Some meaningful differences in the abundance of some microbiota from the three compartments were observed between the disease groups and the HC group. These findings could provide new insights into the prevention and treatment of allergic diseases through the regulation of specific microbiota in the future.
The prevalence of allergic diseases has continued to increase across developing countries in recent years (1). The biodiversity hypothesis suggests that a reduction in biodiversity and alterations in microbiota composition are relevant to allergic diseases and that exposure to nonpathogenic commensal bacteria could regulate immunity to some extent (1). Therefore, identifying the specific microbiota features associated with allergic diseases might provide novel mechanisms and targets, thus improving the prevention and treatment of allergic diseases.
The gut, skin, and airway harbour the most abundant microbiota in the human body (2, 3). Recent studies have shown that the microbiota located in the gut, skin and airway play important roles in both human health and disease. The composition and structure of the gut flora reach a stable state at 2–3 years of age (4), and the skin flora is similar to that of adults at 12–18 months of age (5). Major changes in the airway flora always occur in the year following birth (6).
The gut‒skin axis theory suggests that disruptions in gut barrier integrity and imbalances in the gut microbial community could significantly impact skin homeostasis (7). Changes in the composition of the gut microbiota due to diet, disease, and medications are associated with alterations in airway homeostasis, which supports the concept of the “gut‒lung axis” (8). However, most of the previously published research has studied the gut, skin, and airway microbiota separately instead of studying all three compartments as an ecological community.
The progression from atopic dermatitis (AD) and food allergy in infancy to asthma and allergic rhinitis (AR) in childhood is known as the “allergic march”. Several studies have found that alterations in the microbiota can influence the allergic process as well. It has been proposed that early skin dysbiosis is more likely to lead to the development of AD, which can be corrected and improved with colonization by commensal staphylococci (9). Furthermore, patients with decreased gut flora diversity in infancy have been shown to be at increased risk of developing atopic diseases (10–12). To our knowledge, few studies have compared microbiota differences in children with AR with or without food allergy (FA) and AD.
In recent years, 16S rRNA gene sequencing has been widely used for the identification of microbiota because of its widespread distribution, high abundance and low detection cost. Currently, the V1–V3 variable region sequences of 16S rRNA are used to identify skin microbiota (13, 14), and the V3–V4 variable region sequences are used to identify gut microbiota (13, 15). In previous studies evaluating both skin and gut compartments, microbiota were detected via V3–V4 sequencing (13). Therefore, it is possible that specific skin microbiota expressing other sequences might be missed. To avoid missing important microbiota, full-length 16S rRNA gene sequencing of microbiota was employed in our study.
In this study, we explored the differential expression of microbiota in children with different allergies and delineated the clinical relevance of the gut, skin, and nasal microbiota to the allergy phenotype in children with allergic rhinitis combined with atopic dermatitis and food allergy (AR-AD-FA group), children with allergic rhinitis only (AR-only group) and healthy controls (HC group).
Children aged 3–12 years treated for FA, AD or AR from December 2019 to January 2021 at the paediatric outpatient clinic of Ruijin Hospital were included in the disease groups. Fifteen age- and sex-matched healthy children were recruited as controls. The inclusion criteria for the subjects were as follows: (1) aged between 3 and 12 years; (2) diagnosed with AD and FA; and (3) diagnosed with AR. The exclusion criteria were as follows: (1) had asthma; (2) had diarrhoea or constipation in the previous months; (3) had used antibiotics or probiotics in the previous months; (4) had received immunotherapy; (5) had respiratory infections, gastrointestinal infections or skin infections; and (6) had other immune disorders or gastrointestinal disorders. HC had no history of atopic disease. The exclusion criteria for the controls were the same as those for the subjects. Ultimately, 9 patients with AR combined with AD and FA, 12 patients with AR only, and 15 HCs were included in the study. The study was approved by the Ethics Committee of Ruijin Hospital, and informed consent forms were signed by the guardians of all participants.
This study employed the same questionnaire that was applied in the China, Children, Homes, Health (CCHH) programme (16). The questionnaire included questions on individual characteristics, such as sex, age, birth history, feeding history, term birth status, birth weight, mode of delivery, and family characteristics, such as parental education level and history of allergy. Questions on factors that might increase the risk of FA and AR were also considered in the questionnaire.
AD was diagnosed according to the Williams criteria (17). Patients who met at least 3 of the major criteria were diagnosed with AD.
The diagnosis of FA was made according to the criteria outlined in the European Academy of Allergy & Clinical Immunology (EAACI) Food Allergy Guidelines (18).
The diagnosis of AR was made according to the criteria outlined in the Clinical Practice Guidelines for Diagnosis and Treatment of Allergic Rhinitis in Pediatrics (19).
All children were diagnosed by paediatric allergists who had more than 5 years of experience in the diagnosis and treatment of allergic diseases.
Fecal samples were collected from selected children. Skin flora samples were collected by rubbing nonlesional skin behind the ear with a sterile swab for 10 s. Nasal flora samples were collected by rotating a swab in the nose 10 times. All the collected samples were stored at −80°C.
Sample microbial DNA was extracted with an OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, USA), and the extracted DNA was stored at −20°C. The quantity and quality of the extracted DNA were assessed with a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and electrophoresis on a 1% agarose gel.
The forward primer 27F (5′-AGAGTTTGA TCMTGGCTCAG-3′) and reverse primer 1492R (5′-ACCTTGTTACGACTT-3′) were used for PCR amplification of the full-length bacterial 16S rRNA gene. The extracted DNA was amplified via two-step PCR, and the sample-specific 16 bp barcode was integrated into the forwards and reverse primers for multiple sequencing in the second PCR step. All the PCR amplicons were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified with a PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, equal amounts of amplicons were pooled. Single-molecule real-time (SMRT) sequencing was performed with the PacBio Sequel platform from Shanghai Parsonage Biotech (Shanghai, China).
Sequencing data analyses were performed via a platform named “personalbio”, which provides statistic analysis via QIIME2 and the R software (v4.1.1). The relative abundance at the genus level is presented in stacked histograms. Generally, alpha diversity reflects the variety of microbial species within a single sample, indicating how diverse the microbial community is in that particular environment. In contrast, beta diversity compares the microbial composition between different samples, highlighting how the microbial communities differ from one sample to another. Alpha diversity indices, including the Chao1, observed species, and Shannon and Simpson indices, were calculated with QIIME2 software and visualized in box plots. The beta diversity comparison was evaluated via principal coordinate analysis (PCoA) on the basis of the Bray‒Curtis distances. Differential microbiota between groups were identified via linear discriminant analysis (LDA) and LDA effect size (LEfSe) with an LDA threshold of 2 to identify biomarkers (20).
Clinical data were analysed via SPSS 26.0, with an alpha level of 0.05. P values were false discovery rate (FDR)-corrected for multiple comparisons. The quantitative data were analyzed via t tests or analysis of variance (ANOVA), whereas the qualitative data were analyzed via the chi-square test or Fisher's exact test.
Ultimately, 9 patients with AR combined with AD and FA (AR-AD-FA group), 12 patients with AR only (AR-only group) and 15 HCs (HC group) were included in this study. There were no statistically significant differences in the participants' individual or family baseline characteristics (Table 1).
We obtained the nucleotide sequences of 102 samples, with an average sequence length of 1,459 bp.
The Chao1 index and observed species index of the gut microbiota of children in the AR-AD-FA and AR-only groups were significantly lower than those in the HC group. These findings suggested that the alpha diversity decreased in the disease groups, including the AR-AD-FA and AR-only groups (Figure 1A).
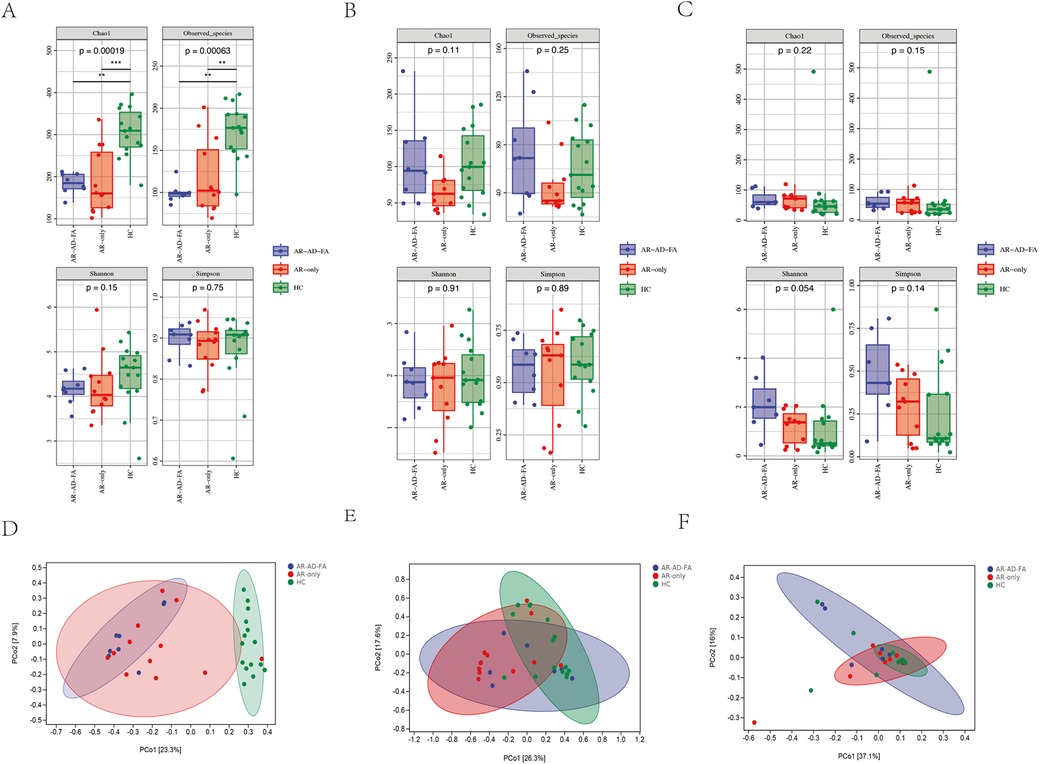
Figure 1. Alpha diversity of the gut (A), nasal (B) and skin (C) microbiota in children in the AR-AD-FA, AR only and HC groups. The p-values annotated in the figure represent the p-values of the three group comparisons. Beta diversity of the gut (D), nasal (E) and skin (F) microbiota in children in the AR-AD-FA, AR only and HC groups. F—faeces from the gut, N-nasal, E—the skin behind the ear, AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
There were significant differences in the community structure of the gut microbiota among the AR-AD-FA, AR-only and HC groups (p = 0.001). In addition, differences in the community structure of the gut microbiota were observed between the AR-AD-FA and HC groups, as well as between the AR-only and HC groups (p = 0.015, corrected). Significant differences in beta diversity were observed between the disease groups and the HC group (Figure 1D).
At the genus level, the gut community was more abundant in Prevotella, Peptoniphilus and Anaerococcus in the disease groups than in the HC group. However, Faecalibacterium, Bacteroides, Roseburia, Blautia and Mediterraneibacter were more abundant in the HC group.
Peptoniphilus, Anaerococcus and Prevotella were abundant in the gut microbiota of both the AR-AD-FA group and the AR-only group (Figures 2A,B). In addition, Finegoldia and Porphyromonas were specifically abundant in the gut microbiota of the AR-AD-FA group (Figure 2A), whereas Dialister and Fenollaria were more abundant in the AR-only group (Figure 2B). Faecalibacterium, Phocaeicola, Bacteroides, Akkermansia and Roseburia were the top five most abundant genera in the HC group (Figure 2C).
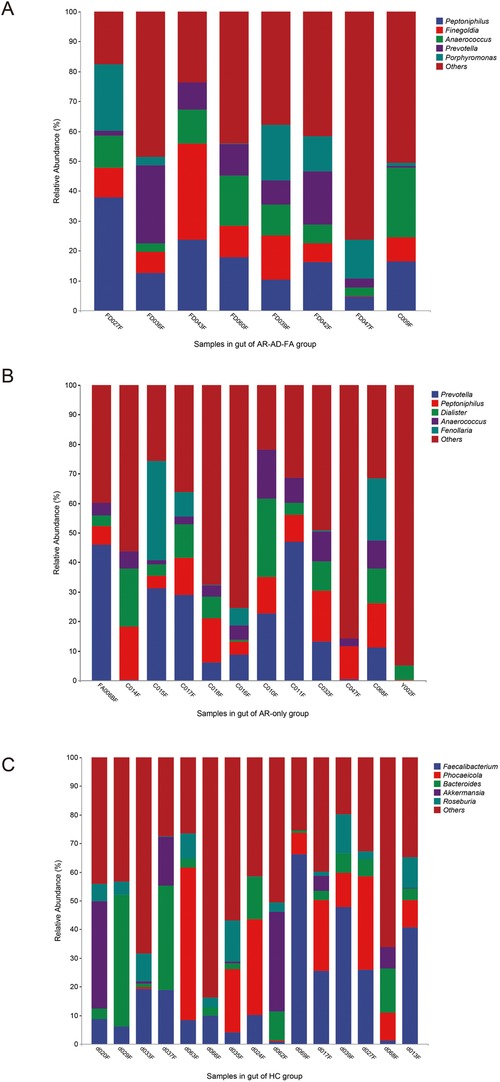
Figure 2. (A–C) The top five most abundant genera in the gut of the three groups (AR-AD-FA, AR only and HC groups). AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
In the LEfSe analysis (LDA threshold of 2), the differentially expressed genera of the gut flora of the AR-AD-FA and AR-only groups were compared with those of the HC group (Figures 3A,B). After the genera that differed consistently between the two disease groups were removed (Figure 3C), Dermabacter, Fenollaria, Howardella and Anaerostipes were highly expressed specifically in the AR-AD-FA group, whereas Burkholderia, Arcanobacterium, Schaalia, Winkia, Atopobium, Slackia, Gemella, Facklamia, Peptostreptococcus, Ezakiella, Parvimonas, Afipia, Methylobacterium and Pelomonas were highly expressed specifically in the AR-only group. Furthermore, in the LEfSe analysis of th AR-AD-FA and AR-only groups, the differentially expressed genera were Varibaculum, Erysipelatoclostridium, Burkholderia, Gordonibacter, Butyricicoccus, Clostridium, Evtepia, Lachnoclostridium, Roseburia, Oscillibacter, Ruthenibacterium, Afipia, Ralstonia, Enterocloster, Lacrimispora and Bilophila.
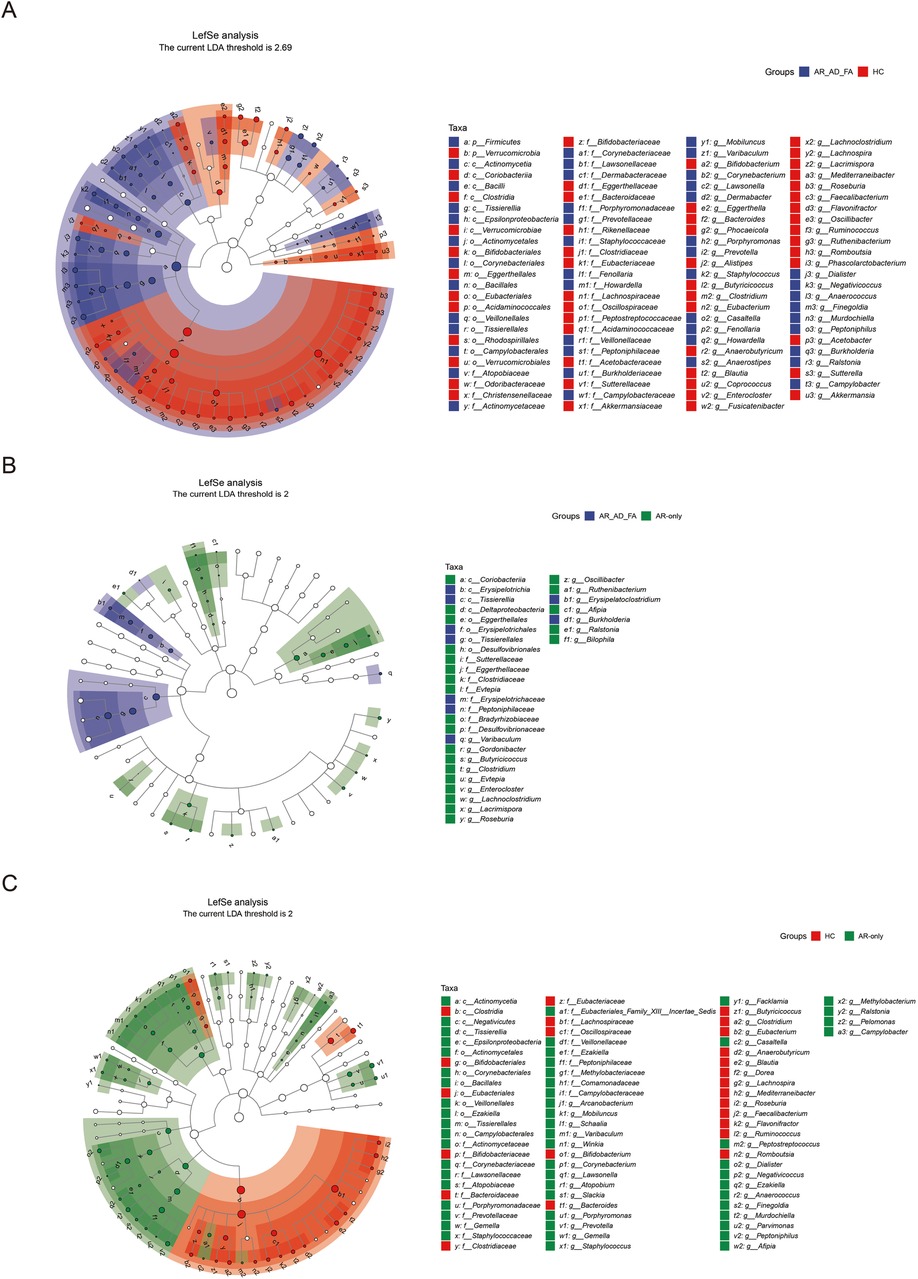
Figure 3. (A–C) The differentially expressed genera and pairwise comparisons among the three groups (AR-AD-FA, AR only and HC groups) from LefSe analysis of the gut are presented. F—faeces from the gut; AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
There were no significant differences in alpha diversity between the disease groups and the HC group (Figure 1B). A significant difference in the beta diversity of nasal microbiota was observed between the AR-only group and the HC group (p = 0.006, corrected), which suggested there was a significant difference in the community structure of the AR-only group and the HC group, whereas no significant differences were detected between the two disease groups (Figure 1E).
Staphylococcus, Moraxella, Dolosigranulum and Burkholderia were the top five most abundant genera in the nasal cavity and were observed in all the groups (Figures 4A–C). Streptococcus was specifically abundant in the nasal cavity of the AR-AD-FA group (Figure 4A).
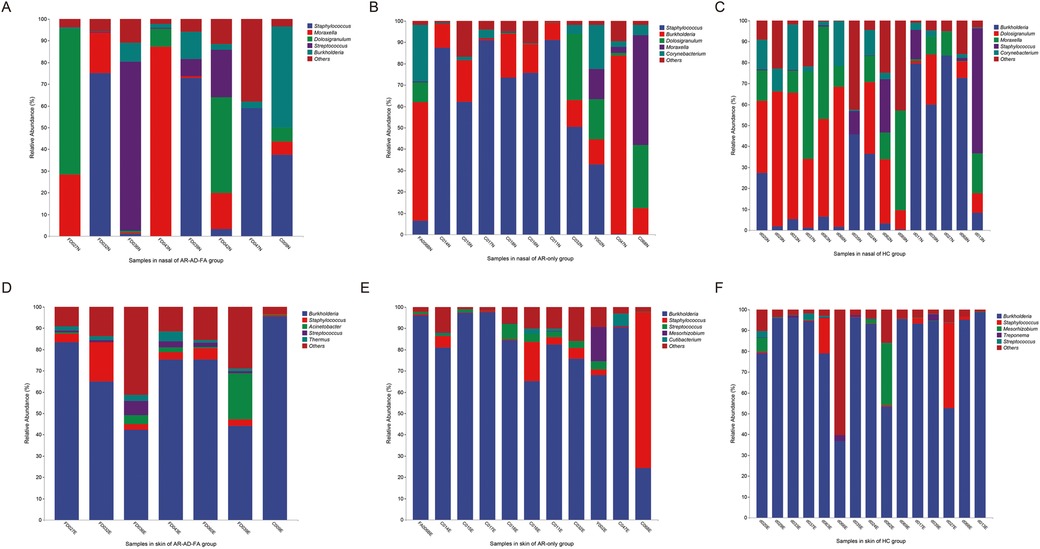
Figure 4. (A–F) The top five most abundant genera in two compartments (nasal and skin) of the three groups (AR-AD-FA, AR only and HC groups). AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
The differentially expressed genera identified via LEfSe analysis of the nasal flora of the AR-AD-FA and AR-only groups were compared with those identified in the HC group (Figures 5A,B). After the genera that differed consistently between the two disease groups were removed (Figure 5C), Streptomyces, Thermus, Kingella, Pseudomonas and Staphylococcus were highly expressed specifically in the AR-AD-FA group. LEfSe analysis confirmed that Streptomyces, Thermus, Kingella, Pseudomonas, Burkholderia and Mesorhizobium were the differentially expressed genera between the two disease groups.
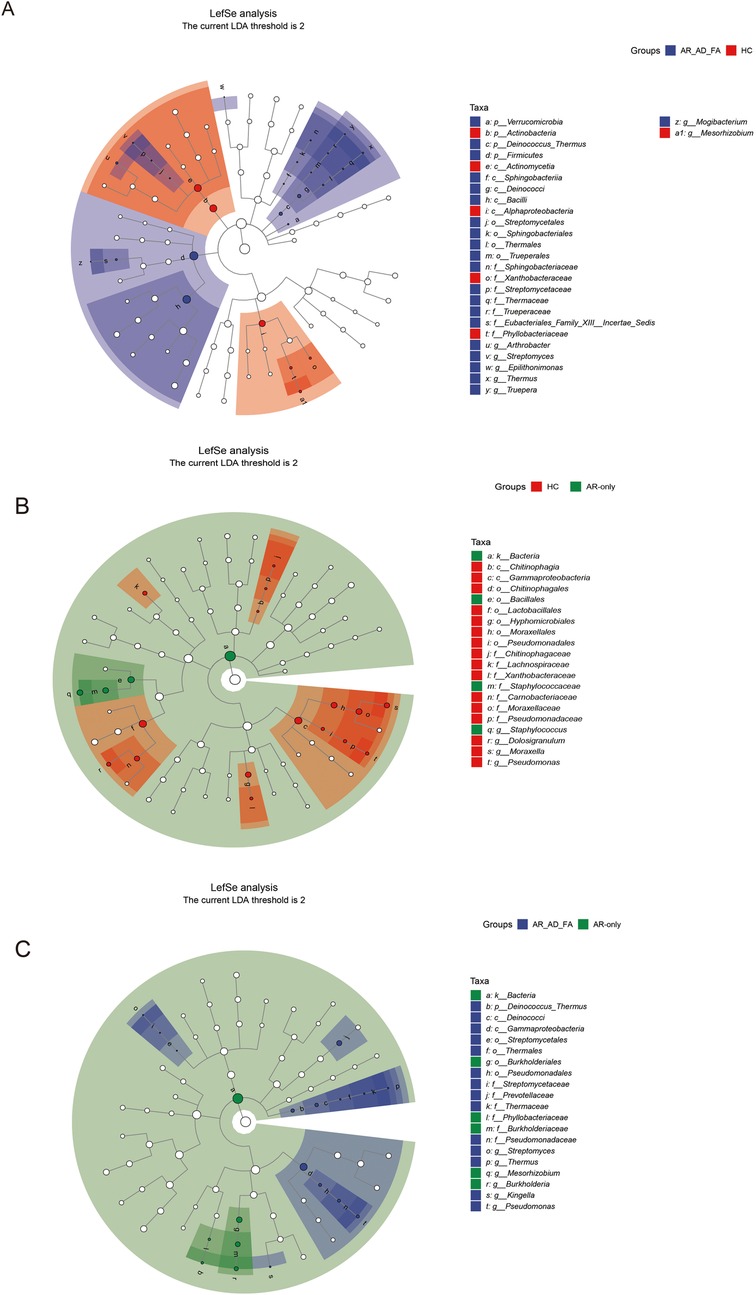
Figure 5. (A–C) The differentially expressed genera and pairwise comparisons among the three groups (AR-AD-FA, AR only and HC groups) from LefSe analysis of the nasal are presented. N-nasal; AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
There was no significant difference in alpha diversity between the disease groups and the HC groups (Figure 1C). However, a significant difference in the community structure of the skin samples was observed among the three groups (p = 0.046). Significant differences in beta diversity were observed among the three groups (Figure 1F). Furthermore, the comparisons between the two disease groups and the HC group revealed differences only between the AR-AD-FA and HC groups (p = 0.026).
Burkholderia, Staphylococcus and Streptococcus were the top five most abundant genera in the skin samples among the three groups (Figures 4D–F). Treponema was more abundant at the genus level within the skin community of the HC group. Thermus, Pseudomonas and Halomonas were more abundant in the AR-AD-FA group. There were significant differences in Mesorhizobium and Rhodopseudomonas abundance between the two disease groups.
In the LEfSe analysis, the differentially expressed genera of the skin flora of the AR-AD-FA and AR-only groups were compared with those of the HC group (Figures 6A,B). After the genera that differed consistently between the two disease groups were removed (Figure 6C), Arachnia, Streptomyces, Thermus, Bacillus, Erysipelothrix, Delftia, Acinetobacter, Pseudomonas, Vibrio, Actinomyces, Corynebacterium, Anaerococcus, Rhodopseudomonas, Sphingomonas and Haemophilus were highly expressed specifically in the AR-AD-FA group. Streptomyces, Thermus, Dolosigranulum, Erysipelothrix, Halomonas, Pseudomonas, Anaerococcus, Rhodopseudomonas and Mesorhizobium were considered differentially expressed genera between the two disease groups according to the LEfSe analysis.
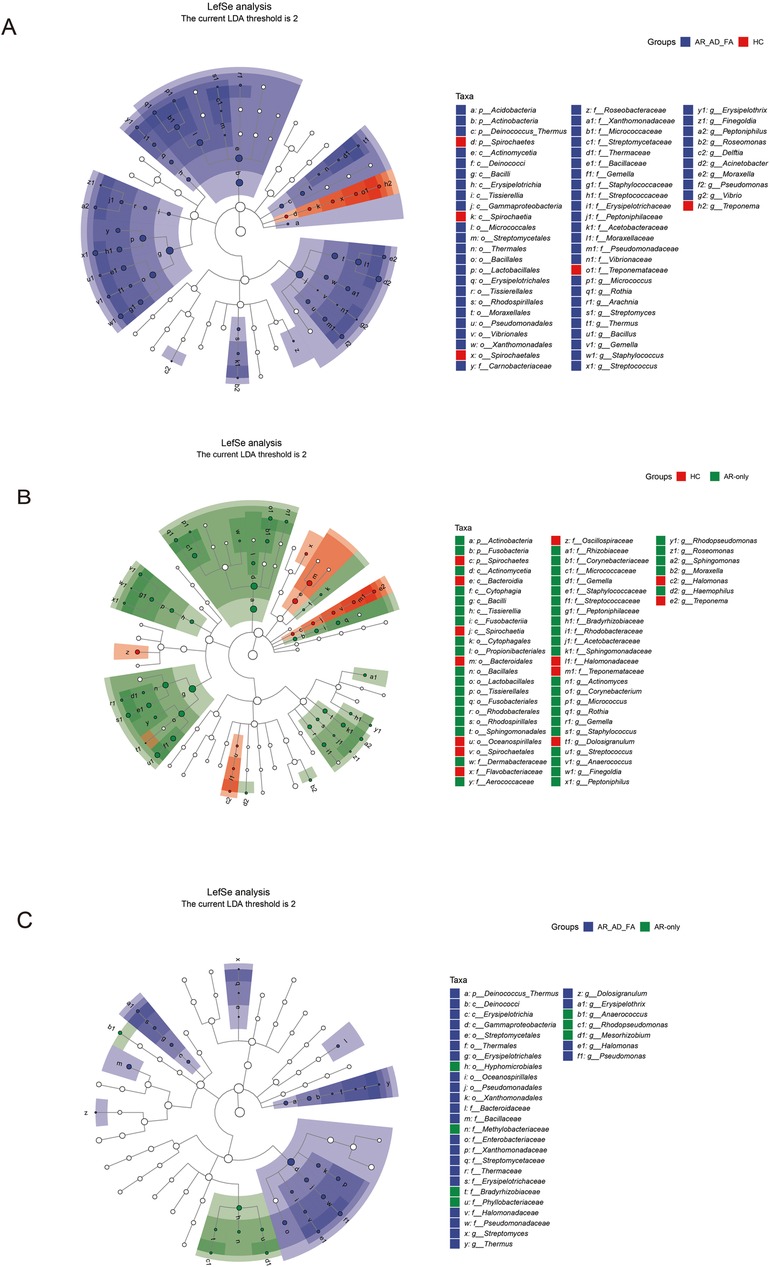
Figure 6. (A–C) The differentially expressed genera and pairwise comparisons among the three groups (AR-AD-FA, AR only and HC groups) from LefSe analysis of the skin are presented. E—the skin behind the ear; AR-AD-FA—allergic rhinitis combined with atopic dermatitis and food allergy, AR only—allergic rhinitis, HC—healthy controls.
In our case‒control study, we compared the expression of genes associated with the gut, nasal and skin microbiota in children with AR combined with AD and FA, AR only and healthy children. Significant differences in beta diversity were found between the disease groups and the HC group in the three compartments. Differentially expressed genera were present mainly in the gut compartment. Peptoniphilus, Prevotella and Anaerococcus were abundant in the gut in the disease groups. Streptomyces, Thermus, and Pseudomonas were highly expressed in the nasal and skin compartments of children in the disease groups. To our knowledge, this study is the first to evaluate the gut, nasal and skin compartments to investigate the interactions among the microbiota in these compartments in individuals with allergic diseases. Furthermore, full-length 16S rRNA gene sequencing of microbiota was employed to avoid missing other sequences.
The alpha diversity of the gut microbiota was significantly lower in both disease groups than in the HC group, which was also consistent with previous findings (13). Previous studies have shown that gut microbiota maintain the balance between type 1 (Th1) and type 2 (Th2) T-helper cell immune responses by regulating various lymphocyte subpopulations, especially regulatory T (Treg) cells (4, 21–23). In the absence of gut microbiota, the levels of both IgE and circulating basophils tend to increase (24), thus leading to a Th1/Th2 cell immune response imbalance that could cause allergic symptoms.
However, the skin microbiota of the three groups did not differ significantly in terms of microbial alpha diversity, which was different from previous studies showing that patients with AD had reduced skin microbial diversity (25, 26). Possible reasons for this discrepancy between our study findings and those of previous studies were the differences in disease phenotypes, sampling sites and detection methods. Children with FA were enrolled in this study, whose AD was associated with FA. In other words, the severity of AD was significantly reduced after avoiding the allergic food. The relatively reduced Th1 cell immune response might result in the skin being chronically infected with bacteria. A previous study showed that patients with AD have greater biodiversity in nonlesioned areas than in lesioned areas, and the microbial diversity in lesioned areas gradually increases with therapeutic intervention (27). In the present study, the microbiota of nonlesional postauricular skin from patients with AD was sampled, amplified and sequenced with the full-length 16S rRNA gene, unlike in previous analyses of skin flora fragments.
Notably, we found that Streptomyces, Thermus and Pseudomonas were highly expressed in the nasal cavity and skin of patients with AR combined with AD and FA. To date, few studies on Streptomyces in patients with allergies have been reported. Lu et al. reported a significant increase in Streptomyces abundance in the skin lesions of patients with active vitiligo (28). Streptomyces can produce numerous secondary metabolites similar to the calcineurin inhibitor tacrolimus, which is widely used for the treatment of AD (29). The significantly increased abundance of Streptomyces in the skin and nasal cavities of patients with AR combined with AD and FA might constitute a self-regulatory mechanism of the skin or mucosa. However, the confirmed correlation between Thermus abundance and FA and AD remains unclear. A study focusing on the characterization of the ocular and nasopharyngeal microbiomes in individuals with allergic rhinoconjunctivitis revealed that Thermus was more abundant on the ocular surface than on the nasal surface, especially in healthy children (30). This difference might be attributed to the different phenotypes of the allergic diseases. Pseudomonas is an accepted pathogenic bacterium associated with allergic diseases. A high abundance of Pseudomonas during pregnancy is a risk factor for the development of AD (31). A higher abundance of Pseudomonas had been found in the gut of children with FA than in that of healthy children (32). Furthermore, a study showed that infants who had developed allergies at the age of 7 years had a significant increase of Pseudomonas abundance in breastmilk during the first month of life (33). Peptoniphilus abundance is high in the sinonasal cavity, and Peptoniphilus is always found in patients with chronic rhinosinusitis (34, 35); however, Peptoniphilus is seldom found in the gut. In contrast to our study, previous studies have observed Prevotella in rural healthy children and in children with a lower occurrence of FA (36, 37). Dietary prebiotics were found to promote intestinal Prevotella in a murine oxazolone-induced model of AD (38). Furthermore, fructo-oligofructose treatment was found to increase the relative abundance of Prevotella in the gut microbiota of mice with AD (39). In this study, participants with AD were in the stable phase, and the recovery process may have led to the increase in Prevotella abundance.
The direct associations between specific microbiota, including Burkholderia, Afipia, Kingella or Rhodopseudomonas, and allergic diseases, including FA, AD or the allergic march, have not been reported previously. Burkholderia belongs to the phylum Proteobacteria, with an outer membrane composed of lipopolysaccharides (LPS) (40). Elevated LPS could increase the risk of inflammation and immune abnormalities (41–43). The home environmental microbiota in children with eczema is characterized by a greater abundance of Anaerococcus than that in healthy children (44). In our study, Anaerococcus was significantly more abundant in the skin compartment of the AR-only group than in those of the AR-AD-FA and HC groups.
In the classic allergic march, allergic diseases progress from AD and FA in infancy to allergic respiratory diseases in childhood, such as AR or asthma, while the mechanisms underlying the allergic march remain to be fully investigated. Recent studies have revealed the role of the microbiome in the allergic march. On the basis of the current literature, our study is the first in which a systematic comparative analysis of the combined gut, nasal and skin microbiomes in allergic children in mainland China was performed.
The V3–V4 hypervariable region of the 16S rRNA gene was analysed via high-throughput DNA sequencing in previous studies, reducing the detection cost, but some important microbiota in other sequences could be missed. To overcome this problem, we chose to amplify and sequence the full-length 16S rRNA gene of microbiota. The final average length of the analysed sequences was 1,459 bp, which was essentially comparable to the theoretical length of the full-length sequences and met the requirements for amplification and sequencing. We considered the characteristics of microbiota expressed in different compartments that were close to the accurate results and real conditions, which is another strength of this study.
However, there are also several limitations of this study. Our sample size was small, and the sample size should be increased in future studies to verify these findings. Future studies should consider disease state and severity when grouping patients. The lack of gut, nasal, and skin samples from early life limited our ability to verify the changes in the microbiota caused by allergic diseases. Further studies could be designed as cohort studies, and more than 2 years of follow-up are necessary.
The skin, nasal, and gut flora should be analysed in combination to investigate the differences in microbiota between children with and without allergies. Some important microbiota could be missed when sequencing only partial regions of the 16S rRNA gene. Significant differences in beta diversity were found between the disease groups and the HC group in the three compartments. Peptoniphilus, Prevotella and Anaerococcus were abundant in the gut in the disease groups. Specifically, Streptomyces, Thermus and Pseudomonas showed high differential expression in both the nasal and skin compartments of children in the disease groups. These findings could provide new insights into the prevention and treatment of allergic diseases through the regulation of specific microbiota in the future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving humans were approved by Ruijin Hospital Ethics Committee Shanghai JiaoTong University School of Medicine. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants' legal guardians/next of kin.
JH: Conceptualization, Project administration, Software, Validation, Visualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. ZT: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – review & editing. DZ: Data curation, Investigation, Methodology, Project administration, Validation, Writing – review & editing. CM: Investigation, Methodology, Validation, Writing – review & editing, Visualization. WS: Methodology, Visualization, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources, Software, Supervision. JS: Conceptualization, Project administration, Resources, Software, Supervision, Visualization, Writing – review & editing, Validation.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We thank the team of Prof. Zhuohui Zhao from Fudan University for help. We thank Ruijin Hospital Affiliated with Shanghai Jiaotong University School of Medicine for help, as well as all the children enrolled in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Akdis CA. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol. (2021) 21:739–51. doi: 10.1038/s41577-021-00538-7
2. Zhao J, Cheng W, Lu H, Shan A, Zhang Q, Sun X, et al. High fiber diet attenuate the inflammation and adverse remodeling of myocardial infarction via modulation of gut microbiota and metabolites. Front Microbiol. (2022) 13:1046912. doi: 10.3389/fmicb.2022.1046912
3. Pattaroni C, Macowan M, Chatzis R, Daunt C, Custovic A, Shields MD, et al. Early life inter-kingdom interactions shape the immunological environment of the airways. Microbiome. (2022) 10:34. doi: 10.1186/s40168-021-01201-y
4. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
5. Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. (2011) 131:2026–32. doi: 10.1038/jid.2011.168
6. Zubeldia-Varela E, Barker-Tejeda TC, Obeso D, Villasenor A, Barber D, Perez-Gordo M. Microbiome and allergy: new insights and perspectives. J Investig Allergol Clin Immunol. (2022) 32:327–44. doi: 10.18176/jiaci.0852
7. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
8. Dang AT, Marsland BJ. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. (2019) 12:843–50. doi: 10.1038/s41385-019-0160-6
9. Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WHI, Murray D, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. (2017) 139:166–72. doi: 10.1016/j.jaci.2016.07.029
10. Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. (2015) 45:632–43. doi: 10.1111/cea.12487
11. Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. (2014) 44:842–50. doi: 10.1111/cea.12253
12. Yang L, Fu J, Zhou Y. Research progress in atopic march. Front Immunol. (2020) 11:1907. doi: 10.3389/fimmu.2020.01907
13. Los-Rycharska E, Golebiewski M, Sikora M, Grzybowski T, Gorzkiewicz M, Popielarz M, et al. A combined analysis of gut and skin microbiota in infants with food allergy and atopic dermatitis: a pilot study. Nutrients. (2021) 13(5):1682. doi: 10.3390/nu13051682
14. Meisel JS, Hannigan GD, Tyldsley AS, SanMiguel AJ, Hodkinson BP, Zheng Q, et al. Skin microbiome surveys are strongly influenced by experimental design. J Invest Dermatol. (2016) 136:947–56. doi: 10.1016/j.jid.2016.01.016
15. Zheng H, Liang H, Wang Y, Miao M, Shi T, Yang F, et al. Altered gut microbiota composition associated with eczema in infants. PLoS One. (2016) 11:e0166026. doi: 10.1371/journal.pone.0166026
16. Chen F, Lin Z, Chen R, Norback D, Liu C, Kan H, et al. The effects of PM(2.5) on asthmatic and allergic diseases or symptoms in preschool children of six Chinese cities, based on China, children, homes and health (CCHH) project. Environ Pollut. (2018) 232:329–37. doi: 10.1016/j.envpol.2017.08.072
17. Williams HC, Burney PG, Hay RJ, Archer CB, Shipley MJ, Hunter JJ, et al. The U.K. working party’s diagnostic criteria for atopic dermatitis. I. Derivation of a minimum set of discriminators for atopic dermatitis. Br J Dermatol. (1994) 131:383–96. doi: 10.1111/j.1365-2133.1994.tb08530.x
18. Muraro A, Werfel T, Hoffmann-Sommergruber K, Roberts G, Beyer K, Bindslev-Jensen C, et al. EAACI food allergy and anaphylaxis guidelines: diagnosis and management of food allergy. Allergy. (2014) 69:1008–25. doi: 10.1111/all.12429
19. SGo R, So P, Association C. Clinical practice guidelines for diagnosis and treatment of allergic rhinitis in pediatric. Chinese Journal of Practical Pediatics. (2019) 34:169–75. doi: 10.19538/j.ek2019030601
20. Fazlollahi M, Lee TD, Andrade J, Oguntuyo K, Chun Y, Grishina G, et al. The nasal microbiome in asthma. J Allergy Clin Immunol. (2018) 142:834–43 e2. doi: 10.1016/j.jaci.2018.02.020
21. Ai C, Ma N, Zhang Q, Wang G, Liu X, Tian F, et al. Immunomodulatory effects of different lactic acid Bacteria on allergic response and its relationship with in vitro properties. PLoS One. (2016) 11:e0164697. doi: 10.1371/journal.pone.0164697
22. Dabrowska K, Witkiewicz W. Correlations of host genetics and gut microbiome composition. Front Microbiol. (2016) 7:1357. doi: 10.3389/fmicb.2016.01357
23. Hua X, Goedert JJ, Pu A, Yu G, Shi J. Allergy associations with the adult fecal microbiota: analysis of the American gut project. EBioMedicine. (2016) 3:172–9. doi: 10.1016/j.ebiom.2015.11.038
24. Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. (2012) 18:538–46. doi: 10.1038/nm.2657
25. Paller AS, Kong HH, Seed P, Naik S, Scharschmidt TC, Gallo RL, et al. The microbiome in patients with atopic dermatitis. J Allergy Clin Immunol. (2019) 143:26–35. doi: 10.1016/j.jaci.2018.11.015
26. Koh LF, Ong RY, Common JE. Skin microbiome of atopic dermatitis. Allergol Int. (2022) 71:31–9. doi: 10.1016/j.alit.2021.11.001
27. Kwon S, Choi JY, Shin JW, Huh CH, Park KC, Du MH, et al. Changes in lesional and non-lesional skin microbiome during treatment of atopic dermatitis. Acta Derm Venereol. (2019) 99:284–90. doi: 10.2340/00015555-3089
28. Lu H, Xu J, Hu Y, Luo H, Chen Y, Xie B, et al. Differences in the skin microbial community between patients with active and stable vitiligo based on 16S rRNA gene sequencing. Australas J Dermatol. (2021) 62:e516–23. doi: 10.1111/ajd.13721
29. Yan L, Zhang Z, Zhang Y, Yang H, Qiu G, Wang D, et al. Improvement of tacrolimus production in Streptomyces tsukubaensis by mutagenesis and optimization of fermentation medium using Plackett-Burman design combined with response surface methodology. Biotechnol Lett. (2021) 43:1765–78. doi: 10.1007/s10529-021-03144-8
30. Yau JW, Hou J, Tsui SKW, Leung TF, Cheng NS, Yam JC, et al. Characterization of ocular and nasopharyngeal microbiome in allergic rhinoconjunctivitis. Pediatr Allergy Immunol. (2019) 30:624–31. doi: 10.1111/pai.13088
31. Fan X, Zang T, Dai J, Wu N, Hope C, Bai J, et al. The associations of maternal and children’s gut microbiota with the development of atopic dermatitis for children aged 2 years. Front Immunol. (2022) 13:1038876. doi: 10.3389/fimmu.2022.1038876
32. Wang S, Wei Y, Liu L, Li Z. Association between breastmilk microbiota and food allergy in infants. Front Cell Infect Microbiol. (2022) 11:770913. doi: 10.3389/fcimb.2021.770913
33. Dzidic M, Mira A, Artacho A, Abrahamsson TR, Jenmalm MC, Collado MC. Allergy development is associated with consumption of breastmilk with a reduced microbial richness in the first month of life. Pediatr Allergy Immunol. (2020) 31:250–7. doi: 10.1111/pai.13176
34. Hernaiz-Leonardo JC, Ryu C, Pascual A, Fan J, Caray M, Pezato R, et al. Comparison between upper and lower airway microbiome profiles in chronic rhinosinusitis patients. Int Forum Allergy Rhinol. (2024) 14:1294–301. doi: 10.1002/alr.23335
35. Kim RJ, Biswas K, Hoggard M, Taylor MW, Douglas RG. Paired analysis of the microbiota of surface mucus and whole-tissue specimens in patients with chronic rhinosinusitis. Int Forum Allergy Rhinol. (2015) 5:877–83. doi: 10.1002/alr.21600
36. Cho H, Kim J, Kim S, Jeong HI, Kwon M, Kim HM, et al. Postpartum maternal anxiety affects the development of food allergy through dietary and gut microbial diversity during early infancy. Allergy Asthma Immunol Res. (2024) 16:154–67. doi: 10.4168/aair.2024.16.2.154
37. Yang Z, Chen Z, Lin X, Yao S, Xian M, Ning X, et al. Rural environment reduces allergic inflammation by modulating the gut microbiota. Gut Microbes. (2022) 14:2125733. doi: 10.1080/19490976.2022.2125733
38. Laigaard A, Krych L, Zachariassen LF, Ellegaard-Jensen L, Nielsen DS, Hansen AK, et al. Dietary prebiotics promote intestinal Prevotella in association with a low-responding phenotype in a murine oxazolone-induced model of atopic dermatitis. Sci Rep. (2020) 10:21204. doi: 10.1038/s41598-020-78404-0
39. Chen S, Tang L, Nie T, Fang M, Cao X. Fructo-oligofructose ameliorates 2,4-dinitrofluorobenzene-induced atopic dermatitis-like skin lesions and psychiatric comorbidities in mice. J Sci Food Agric. (2023) 103:5004–18. doi: 10.1002/jsfa.12582
40. Cheng R, Guo J, Pu F, Wan C, Shi L, Li H, et al. Loading ceftriaxone, vancomycin, and bifidobacteria bifidum TMC3115 to neonatal mice could differently and consequently affect intestinal microbiota and immunity in adulthood. Sci Rep. (2019) 9:3254. doi: 10.1038/s41598-018-35737-1
41. Bufe A, Holst O. LPS-binding protein as a target molecule in allergy and asthma. J Allergy Clin Immunol. (2004) 114:583–5. doi: 10.1016/j.jaci.2004.06.034
42. Liu X, Tao J, Li J, Cao X, Li Y, Gao X, et al. Dysbiosis of fecal microbiota in allergic rhinitis patients. Am J Rhinol Allergy. (2020) 34:650–60. doi: 10.1177/1945892420920477
43. Wang Z, Lai Z, Zhang X, Huang P, Xie J, Jiang Q, et al. Altered gut microbiome compositions are associated with the severity of asthma. J Thorac Dis. (2021) 13:4322–38. doi: 10.21037/jtd-20-2189
Keywords: food allergy, atopic dermatitis, allergic rhinitis, gut microbiota, nasal microbiota, skin microbiota
Citation: Hong J, Tang Z, Zhang D, Mo C, Su W and Shao J (2025) Profiling of the gut, skin and nasal microbiotas revealed clinically relevant microbial taxa from children with allergies: a pilot study. Front. Allergy 6:1497914. doi: 10.3389/falgy.2025.1497914
Received: 2 October 2024; Accepted: 13 February 2025;
Published: 27 February 2025.
Edited by:
Claudio Rhyner, Christine-Kühne-Center for Allergy Research and Education (CK-Care), SwitzerlandReviewed by:
Cristina Benito-Villalvilla, Complutense University of Madrid, SpainCopyright: © 2025 Hong, Tang, Zhang, Mo, Su and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Su, c3cxMTgzMUByamguY29tLmNu; Jie Shao, ZHIuc2hhb2ppZUAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.