- 1Department of Otolaryngology Head and Neck Surgery, Beijing Tongren Hospital, Capital Medical University, Beijing, China
- 2Beijing Laboratory of Allergic Diseases, Beijing Municipal Education Commission and Beijing Key Laboratory of Nasal Diseases, Beijing Institute of Otolaryngology, Beijing, China
- 3Department of Allergy, Beijing Tongren Hospital, Capital Medical University, Beijing, China
- 4Research Unit of Diagnosis and Treatment of Chronic Nasal Diseases, Chinese Academy of Medical Sciences, Beijing, China
Background: Long noncoding RNAs (lncRNAs) have been implicated in a diverse array of human immune diseases; however, a comprehensive understanding of the expression and function of lncRNAs in the peripheral blood leukocytes of individuals suffering from house dust mite (HDM)-induced allergic rhinitis (AR) remains elusive.
Objective: To explore the potential roles and functions of long noncoding RNAs (lncRNAs) in the pathogenesis of AR.
Methods: Sequencing analysis was performed on peripheral blood leukocytes collected from patients with HDM-induced AR and healthy controls (HCs) to elucidate the expression patterns of lncRNAs. Differentially expressed (DE) lncRNAs were identified and validated, and further correlation analyses were conducted to explore their associations with visual analog scale (VAS) scores and cytokine levels in the serum and nasal secretions. Additionally, bioinformatics analyses were performed to predict the potential pathways influenced by DE lncRNAs. Finally, the diagnostic potential of these lncRNAs in AR was assessed via receiver operating characteristic (ROC) curve analysis.
Results: Significant differences in the expression profiles of lncRNAs and mRNAs were detected between AR patients and HCs. Four lncRNAs were markedly upregulated in AR patients. AC011524.2 was positively correlated with nasal pruritus (r = 0.4492, P = 0.0411). AL133371.3 was positively correlated with runny nose (r = 0.4889, P = 0.0245). AC011524.2 was positively correlated with CXCL8 (r = 0.4504, P = 0.0035). AL133371.3 was significantly positively correlated with only IL-17 (r = 0.4028, P = 0.0100). IL-4 in the serum was positively related to IL-17 in the serum (r = 0.4163, P = 0.0002). CXCL5 in the serum was positively correlated with IFN-γ (r = 0.3336, P = 0.0354) in nasal secretions. The area under the curve (AUC) of the ROC curve resulting from the integration of the 4 lncRNAs exhibited a remarkable value of 0.940 for AR diagnosis.
Conclusions: Our results identified several lncRNAs associated with AR symptoms and inflammatory cytokines. Specifically, AC011524.2 and AL133371.3 exhibited strong correlations with diverse AR manifestations and serum cytokines, suggesting their pivotal role in the pathogenesis of AR, likely via neutrophil- and Th17-related pathways. However, the precise underlying mechanisms are still elusive, necessitating further exploration.
Introduction
Allergic rhinitis (AR), a prevalent condition, frequently coexists with chronic rhinosinusitis, asthma, conjunctivitis and other diseases (1). In China, the self-reported incidence of AR has increased from 11.1% in 2005 to 17.6% in 2011, significantly affecting people's quality of life (2). The etiology of AR, akin to other allergic conditions, encompasses intricate and multifactorial elements (3). AR can be categorized as persistent or seasonal, with house dust mites (HDMs) being the most common allergen (4). In particular, more than 90% of AR cases in central and southern China are attributed to HDMs (5). These allergens are ubiquitous and present throughout the year. Therefore, patients with HDM-induced AR have persistent allergic inflammation (6). Consequently, further research on HDM-induced AR is imperative to enhance our comprehension of its pathogenesis and improve strategies.
Long noncoding RNAs (lncRNAs) exceeding 200 nucleotides in length, occupy an important position in cellular regulation (7). They are integral to modulating gene expression, chromatin remodeling, cell cycle progression, and numerous other essential biological functions (8). Recent research has underscored the significant involvement of lncRNAs in various pathological processes, including malignancies, cardiovascular diseases, and neurodegenerative disorders (9). Their crucial regulatory role extends to the immune response, inflammatory processes and allergic reactions. For example, the lncRNA ANRIL is positively correlated with nasal symptom severity and the expression of inflammatory cytokines such as TNF-α, IL-4, IL-6, IL-13, and IL-17, while inversely correlated with the levels of anti-inflammatory cytokines like IL-10 and IFN-γ in the nasal mucosal cells of AR patients and healthy controls (HCs) (10). Yue et al. found that LNC-000632 expression was downregulated in nasal mucosal samples from AR patients and in IL-13-stimulated nasal epithelial cells (11). The lncRNA GATA3-AS1 and the GATA3 gene are regulated by the same transcriptional regulatory element, which may play important roles in the Th2 immune response (12). These findings towards the significance of differentially expressed (DE) lncRNAs in the pathogenesis of AR. Further exploration of lncRNA expression and function in AR, as well as elucidation of the underlying mechanism, could contribute to novel therapeutic strategies. The investigation of the interplay between lncRNAs and HDM-induced AR is still in its infancy (13).
Obtaining nasal mucosa samples poses clinical challenges, and for AR, specific IgE testing remains the primary diagnostic tool. Although peripheral blood from AR patients is readily accessible, the relationship between lncRNAs in AR peripheral blood leukocytes and various serum inflammatory cytokines remains unexplored. Therefore, investigating AR peripheral blood lncRNAs, their regulation of cytokines, and their clinical correlations is imperative. This study aimed to elucidate the role of lncRNAs in AR by contrasting their expression in peripheral blood leukocytes from AR patients with that in HCs. Furthermore, we determined the correlations among DE lncRNAs, clinical manifestations, and serum inflammatory factors. This research holds significant value, as identifying lncRNAs associated with AR symptoms could deepen our understanding of the molecular basis of this disease, ultimately contributing to enhanced diagnostic and therapeutic strategies.
Methods
Patient population
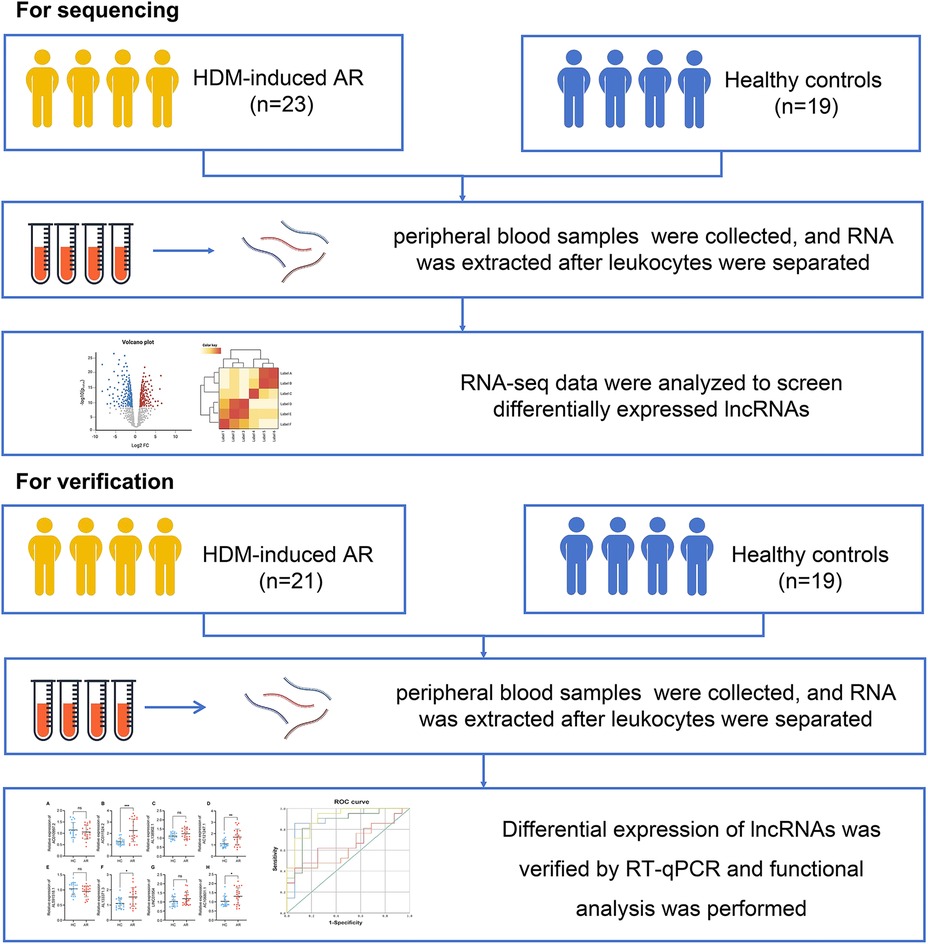
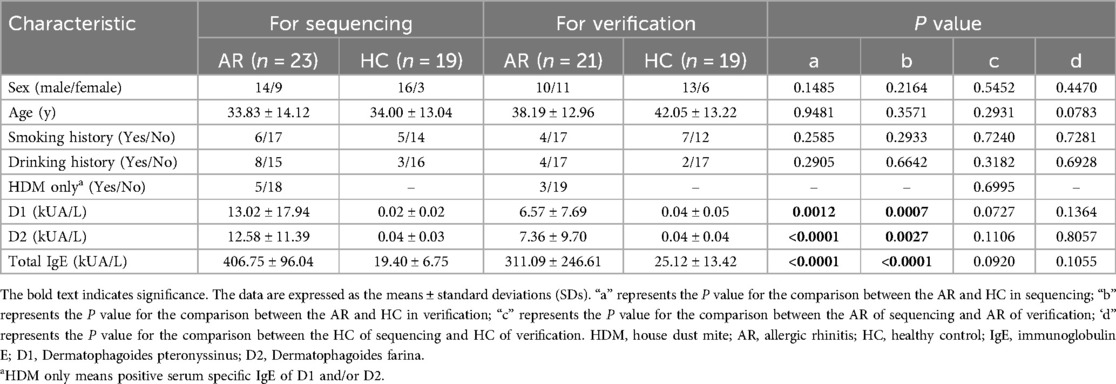
A total of 23 HDM-induced AR patients and 19 HCs were included in this study. All patients were diagnosed with AR in the Department of Otolaryngology Head and Neck Surgery, Beijing Tongren Hospital, Capital Medical University, from May 2018 to May 2019. For verification, information on 21 patients with HDM-induced AR and 19 HCs were collected under the same conditions from September 2020 to October 2021 (Figure 1).
Inclusion and exclusion criteria
The following inclusion criteria were used for HDM-induced AR: (1) met the diagnostic criteria for persistent AR (14); (2) patients with AR were primarily allergic to Dermatophagoides pteronyssinus (D1), Dermatophagoides farina (D2) or both, and the symptoms were caused by HDM; the serum specific immunoglobulin E (IgE) to D1 or D2 was equal to or greater than grade 2, as measured with ImmunoCAP Phadiatop (Thermo Fisher Scientific) (14, 15); (3) aged between 18 and 60 years, regardless of sex; and (4) did not use anti-allergic or steroid medications within the past 3 months. The following exclusion criteria were used: (1) patients who had undergone specific immunotherapy in the past; (2) patients with chronic rhinosinusitis, asthma, etc.; (3) patients with other immune-related diseases, tumors, chronic infectious diseases, hematologic disorders, etc.; and (4) pregnant or lactating women.
The following inclusion criteria were used for HCs: (1) absence of AR symptoms or history; (2) aged between 18 and 60 years, regardless of sex; and (3) not having used antiallergic or steroid medications within the past 3 months. The following exclusion criteria were used for HCs: (1) had chronic rhinosinusitis, asthma, etc.; (2) had other immune-related diseases, tumors, chronic infectious diseases, hematologic disorders, etc.; and (3) were pregnant or lactating.
Visual analog scale (VAS)
The VAS was used to assess the overall severity of symptoms during allergic rhinitis episodes in the past week and included 12 assessment items: nasal congestion, nasal itchiness, sneezing, runny nose, itchy eyes, tearing, red eyes, eye pain, cough, breath holding, wheezing, and pressure sensation. The VAS score ranged from 0 (absence of symptoms) to 10 cm (very severe symptoms) for all combined nasal symptoms. All the subjects completed the VAS score at enrollment. The more severe the symptoms, the higher the score, with a maximum score of 120.
Methods of nasal secretion collection
Nasal secretions were collected from AR patients and HCs via a polyvinyl alcohol medical sponge (Medtronic, 400402) into each patient's left and right nasal total nasal passages and left in place for 5–10 min to be removed when the sponge adsorbed nasal secretions until fully expanded. The sponges were subsequently placed into a centrifuge tube, which was weighed before and after collection to calculate the secretion content. One milliliter of saline was added, and the mixture was refrigerated at 4°C for 2 hours and centrifuged at 1500 × g for 15 min. The supernatant was retained and stored at −80°C.
Serum-specific IgE detection
Blood samples were collected in 5-ml vacuum tubes and centrifuged to separate the serum. An ImmunoCAP Phadiatop (Thermo Fisher Scientific) was used to detect allergen-specific IgE antibodies in human serum, including for HDM (D1 and D2), cat dander, dog dander, Quercus alba, Ulmus americana, Platanus acerifolia, Salix caprea, Populus deltoides, mountain juniper, common silver birch, mugwort, Japanese Hop, ragweed, Penicillium chrysogenum, Cladosporium herbarum, Aspergillus fumigatus and Alternaria alternata. The instrument used was Phadia information and data management software, and all calculations were performed automatically. The qualitative results were positive or negative, and the clinical threshold was 0.35 kU/L with 6 grades (grade 0: 0.00–0.34 kUA/L; grade 1: 0.35–0.69 kUA/L; grade 2: 0.70–3.49 kUA/L; grade 3: 3.50–17.49 kUA/L; grade 4: 17.5–49.9 kUA/L; grade 5: 50.0–100.0 kUA/L; and grade 6: >100.0 kUA/L) (14,15).
Generation of sequencing libraries from total RNA
Blood samples were collected in 5-ml EDTA vacuum tubes, and the leukocytes were separated via red blood cell lysis solution (Solarbio, R1010). Total RNA was extracted from leukocytes with Tri®-Reagent (Sigma, T9424) within 2 h. All procedures were performed according to the manufacturer's instructions. An RNA-seq library was constructed and sequenced by Novogene Company Limited. The NEBNext® UltraTM RNA Library Prep Kit (New England Biolabs) was used for library construction. After the library was constructed, an Agilent 2100 Bioanalyzer (Agilent Technologies) was used to determine the size of the library insert. Reverse transcription‒quantitative polymerase chain reaction (RT‒qPCR) was used to quantify the effective concentration of the library accurately to ensure quality. After qualified library inspection, Illumina sequencing was performed for the different pooled libraries according to the requirements of the effective concentration and target onboard data amount, and a 150 bp matching end reading was produced. The basic principle of sequencing is sequencing by synthesis.
Analysis of DE genes
Differential expression analysis was performed between two comparisons via the DESeq2 R package (1.16.1). DESeq2 provides statistical procedures to determine differential expression in digital gene expression data using a model based on a negative binomial distribution. The method of Benjamini and Hochberg was used to adjust the P value to control the error detection rate. Genes with adjusted P < 0.05 were considered DE according to DESeq2. For each sequenced library, the read count was adjusted via a scale normalization factor in the edgeR package prior to differential expression analysis. Differential expression analysis of the two conditions was performed via the edgeR software package. The Benjamini and Hochberg methods were used to adjust the P values. After correction, the P value and fold change were used as thresholds for significant differential expression.
Functional enrichment analyses
The Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases were used for functional enrichment analysis of the DE lncRNAs. The significance of the biological pathways is indicated by the P value. The P value was corrected via the Benjamin and Hochberg multiple tests, and the Q value was obtained.
RT-qPCR validation
The quality of the total RNA was assessed via a Nanodrop 2000 (Thermo Fisher Scientific), and complementary DNA was synthesized from 1 μg of total RNA via PrimeScript RT Master Mix (Takara, RR036A). RT‒qPCR was performed via SYBR Green mix (ABclonal Biotechnology, RK21203) to determine gene expression levels. The reaction was performed using a StepOnePlusTM Real-Time PCR System (Applied Biosystems). The average transcript levels of the genes were normalized to those of GAPDH. The primers used are listed in Supplementary Table S1.
Measurement of cytokine expression in serum and nasal secretions
We used the Human Cytokine A Premixed Magnetic Luminex® Performance Assay (R&D Systems, FCSTM03) to quantitatively detect a variety of cytokines and chemokines in serum and nasal secretions. Before Luminex testing, the sample was slowly thawed and pretreated according to the manufacturer's instructions. Each sample and standard were double-replicated in a 96-well plate. The concentration of each target protein in the serum samples was calculated via a standard curve. The results are expressed in pg/ml and were used for subsequent statistical analysis. The factors detected included C-X-C motif chemokine ligand 5 (CXCL5)/epithelial neutrophil-activating protein 78 (ENA-78), basic fibroblast growth factor (FGF basic), granulocyte colony stimulating factor (G-CSF), granulocyte‒macrophage colony stimulating factor (GM-CSF), interferon-γ (IFN-γ), interleukin (IL)-1β/IL-1F2, IL-2, IL-4, IL-5, IL-6, CXCL8/IL-8, IL-10, IL-17, CC chemokine ligand 2 (CCL2)/monocyte chemoattractant protein-1 (MCP-1), CCL3/macrophage inflammatory protein (MIP-1ɑ), CCL4/MIP-1β, CCL5/RANTES, tumor necrosis factor-ɑ (TNF-ɑ), thrombopoietin (Tpo), and vascular endothelial cell growth factor (VEGF).
Statistical analysis
SPSS 16.0 for Windows was used for receiver operating characteristic (ROC) analyses, and other statistical analyses were performed via GraphPad Prism 7.0 software. R studio software was used to construct heatmaps and volcano plots. For normally distributed data, the mean ± standard deviation (SD) was used. Correlational analyses were performed via Spearman rank correlation analysis. Comparisons between the 2 groups were made via the independent samples t test or Mann‒Whitney U test. In all the cases, a P value < 0.05 was considered statistically significant.
Results
Characteristics of the study population
A total of 19 HCs and 23 AR patients were included in the first stage of this study. All AR patients were diagnosed in the Department of Otolaryngology, Beijing Tongren Hospital, Capital Medical University, from May 2018 to May 2019. The patients were diagnosed with HDM-induced AR via the UniCAP system. The total IgE, D1 and D2 values of the AR group were all positive (Table 1). There were no significant differences in age or sex distribution between the AR patients and HCs (P > 0.05). The serum D1, D2 and total IgE values in the AR group were significantly different from those in the HC group (P < 0.05).
The expression profiles of lncRNAs and mRNAs in the peripheral blood leukocytes of AR patients and HCs differed
To investigate differences in the peripheral blood leukocyte lncRNA expression profiles between the HDM-induced AR group and the HC group, each RNA sample extracted from peripheral blood leukocytes was sequenced via the Illumina HiSeq platform, and the data were calculated as feature counts. The expression levels of lncRNAs and mRNAs in each sample were calculated, and DE lncRNAs were identified via multiple differential comparisons. There were obvious differences between the 350 lncRNAs (|log2 fold change| >1, P < 0.05), including 170 downregulated and 180 upregulated lncRNAs (Supplementary Table S2) (Figures 2A,B). There were obvious differences between 298 mRNAs (|log2 fold change| >1, P < 0.05), including 125 downregulated and 173 upregulated mRNAs (|log2 fold change| >1, P < 0.05) (Supplementary Table S3) (Figures 2C,D).
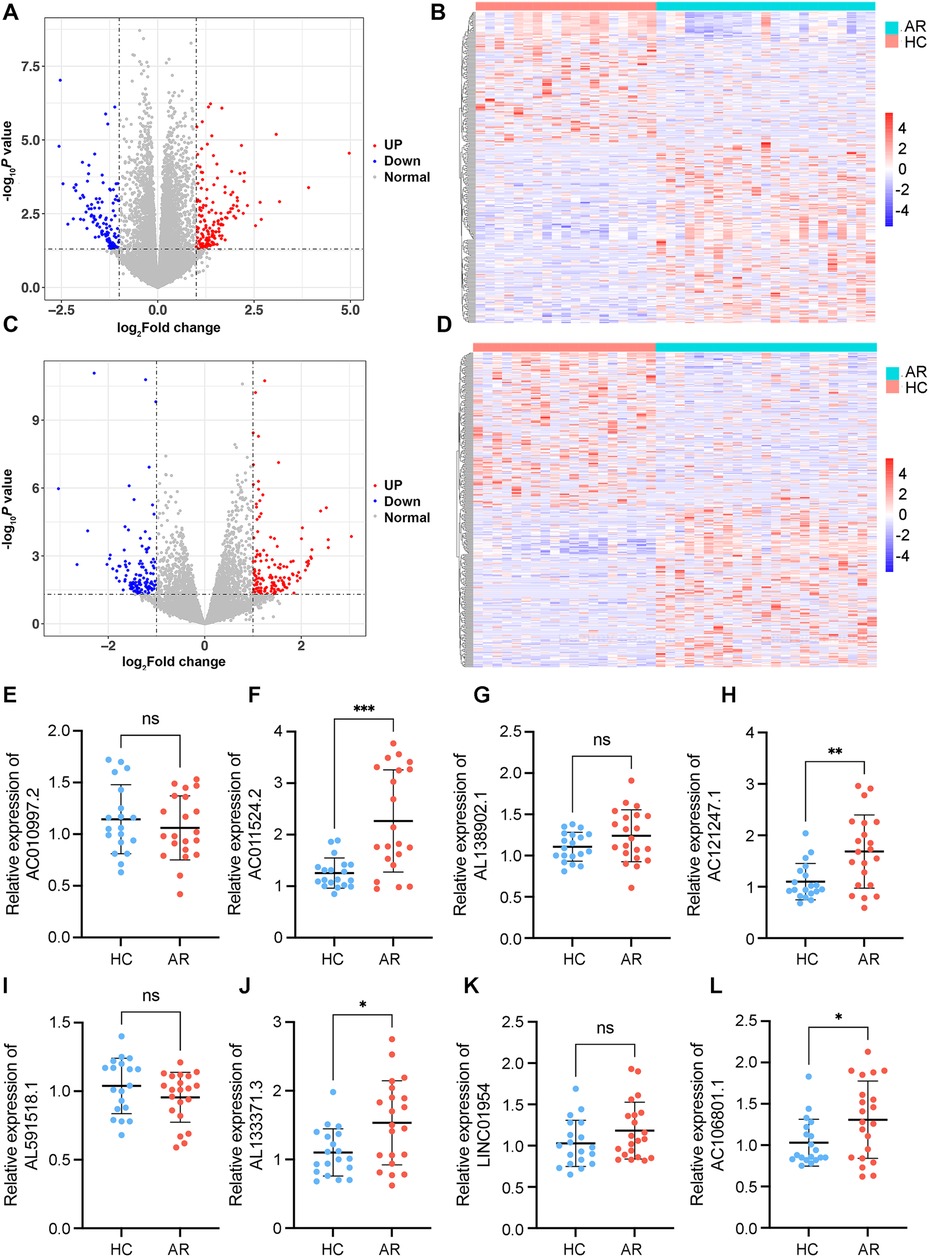
Figure 2. Identification of DE mRNAs and lncRNAs. (A) Volcano plot of DE mRNAs. (B) Heatmaps of DE mRNAs between the AR and healthy control groups (|log2 fold change | >1 and P < 0.05). (C) Volcano plot of DE lncRNAs. (D) Heatmaps of DE lncRNAs between the AR and healthy control groups (|log2 fold change| >1 and P < 0.05). In each heatmap, AR indicates the HDM-induced AR group, and HC indicates the healthy control group. In the volcano map, the red dots represent upregulated genes, and the blue dots represent downregulated genes. (E–L) Relative expression levels of the top 8 DE lncRNAs determined via RT–qPCR. AR indicates the HDM-induced AR group, and HC indicates the healthy control group. *P < 0.05, **P < 0.01, ***P < 0.001.
Validation of DE lncRNAs
On the basis of the results of differential gene analysis, the 8 most significantly upregulated lncRNAs were selected as validation targets for further experimental confirmation. A total of 21 AR patients and 19 HCs were further included according to the same inclusion and exclusion criteria (Table 1). The 8 most significantly upregulated lncRNAs were detected via RT‒qPCR: AC010997.2, AC011524.2, AL138902.1, AC121247.1, AL591518.1, AL133371.3, LINC01954, and AC106801.1. AC011524.2, AL138902.1, AC121247.1, AL133371.3, LINC01954, and AC106801.1 were upregulated in the peripheral blood leukocytes of AR patients compared with those of HCs (Figures 2E–L). The results were consistent with the RNA-seq results, with AC011524.2, AC121247.1, AL133371.3, and AC106801.1 showing significant differences (P < 0.05) (Figures 2F,H,J,L).
Correlation analysis between serum IgE levels, DE lncRNAs, and clinical symptoms
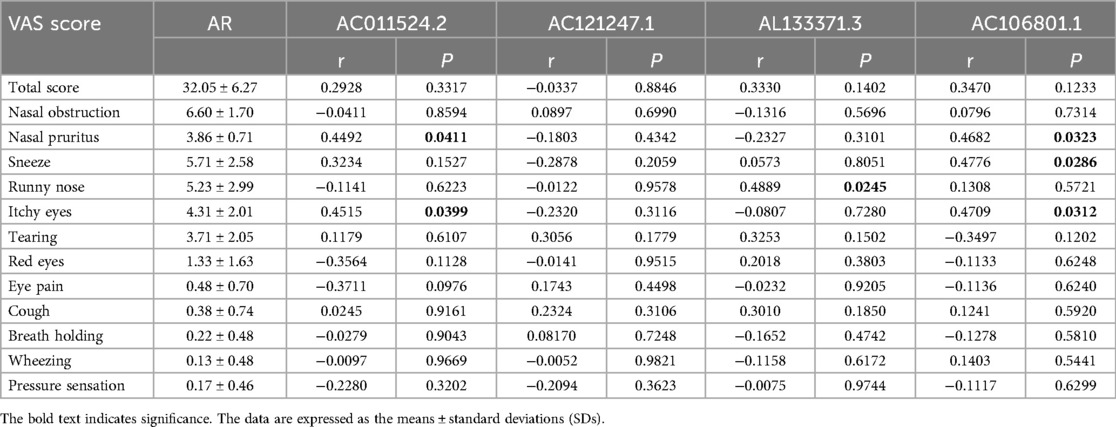
To understand the associations between DE lncRNAs, serum IgE levels, and clinical symptoms and to evaluate whether these lncRNAs are involved in AR and may influence patient symptom presentation, we conducted a correlation analysis. The correlation analysis results revealed that AC011524.2 was positively correlated with nasal pruritus (r = 0.4492, P = 0.0411) and itchy eyes (r = 0.4515, P = 0.0399). There was no significant correlation between AC121247.1 and symptom scores (P > 0.05). AL133371.3 was positively correlated with runny nose (r = 0.4889, P = 0.0245) but not significantly correlated with other symptoms (P > 0.05). AC106801.1 was positively correlated with nasal pruritus (r = 0.4682, P = 0.0323), sneezing (r = 0.4776, P = 0.0286), and itching (r = 0.4709, P = 0.0312) but was not significantly correlated with other symptoms (Table 2). However, no significant correlation was found between the 4 DE lncRNAs and total serum IgE, D1 or D2 (Supplementary Table S4). Similarly, correlation analysis between the VAS score and total IgE, D1, and D2 also revealed no significant correlations (Supplementary Table S5).
Correlation analysis of DE lncRNAs and inflammatory cytokines in the serum
To investigate the potential involvement of lncRNAs in AR and understand their associations with serum inflammatory cytokines, we conducted an analysis. The analysis of serum inflammatory cytokine levels revealed that the levels of basic FGF, IL-4, CXCL8, IL-17 and CCL3 were significantly greater in the AR group than in the HC group, whereas the other cytokines did not show significant differences (Figure 3A; Supplementary Table S6). The expression of AC011524.2 was positively correlated with that of IL-1β (r = 0.3362, P = 0.0349), CXCL8 (r = 0.4504, P = 0.0035), CCL3 (r = 0.3344, P = 0.0349), and CCL4 (r = 0.4010, P = 0.0103). AC121247.1 was significantly positively correlated with FGF basic (r = 0.4697, P = 0.0022), G-CSF (r = 0.5130, P = 0.0007), IL-4 (r = 0.3903, P = 0.0128), IL-6 (r = 0.4415, P = 0.0044), and CXCL8 (r = 0.3210, P = 0.0434). AL133371.3 was significantly positively correlated with only IL-17 (r = 0.4028, P = 0.0100). AC106801.1 was significantly positively correlated with IL-1β (r = 0.3274, P = 0.0392) and CCL3 (r = 0.3138, P = 0.0487) (Figure 3B; Table 3).
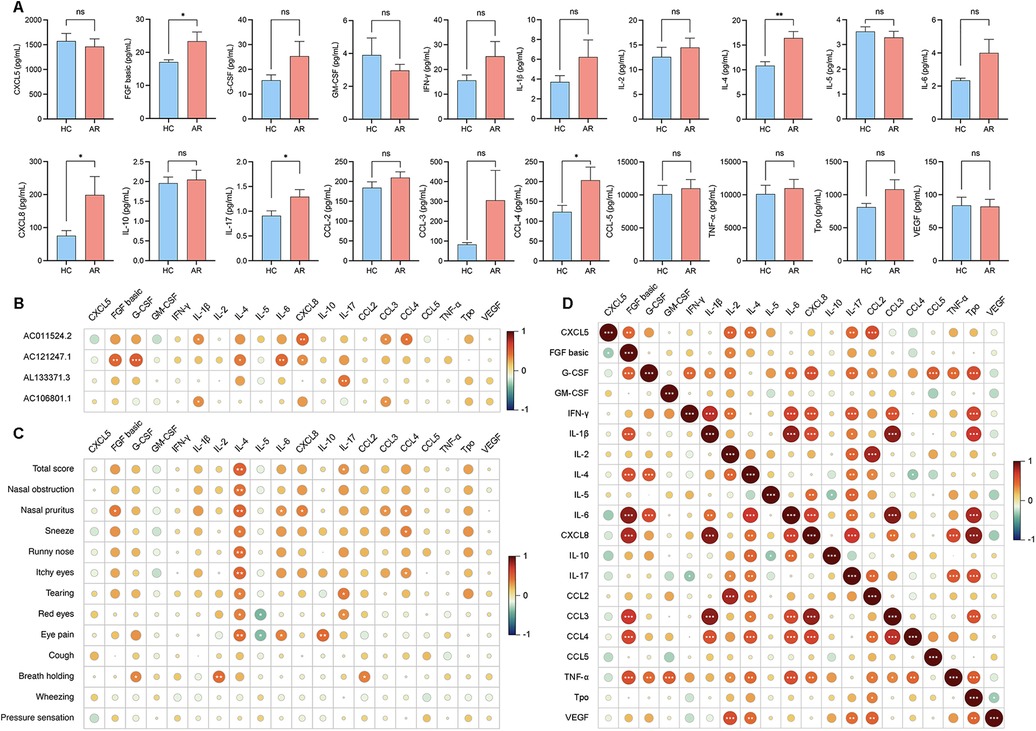
Figure 3. (A) Levels of cytokine expression in serum. (B) Correlation analysis of the 4 DE lncRNAs with serum inflammatory cytokines. (C) Correlation analysis between the VAS score and the levels of inflammatory cytokines in the serum. (D) Correlation analysis between each cytokine in the serum.
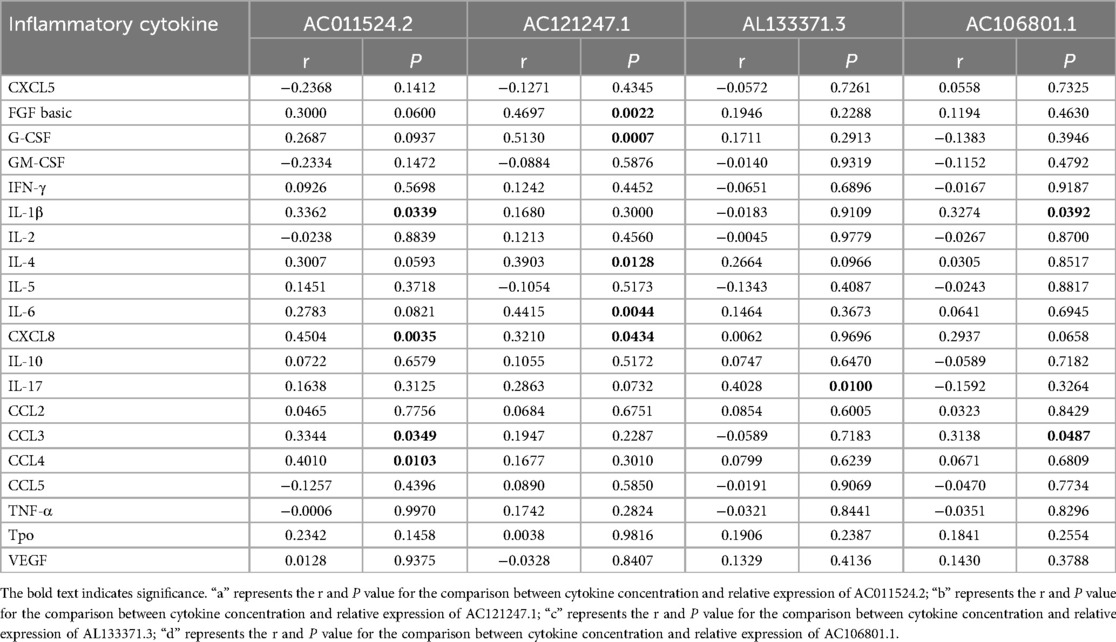
Table 3. Correlation analysis of serum inflammatory cytokines and differential expression of lncRNA.
Correlation analysis of inflammatory cytokines in the serum with clinical symptoms
To investigate the associations between inflammatory cytokines in serum and nasal secretions and clinical symptoms and to understand how these cytokines influence symptom manifestation in AR patients, we conducted a correlation analysis. IL-4 in the serum was positively correlated with the total score (r = 0.4912, P = 0.0013) and nasal pruritus (r = 0.4731, P = 0.0020), IL-17 in the serum was positively correlated with the total score (r = 0.3127, P = 0.0045), CCL3 in the serum was positively correlated with nasal pruritus (r = 0.3141, P = 0.0484), and CCL4 in the serum was positively correlated with nasal pruritus (r = 0.3596, P = 0.0227) (Figure 3C; Supplementary Table S7).
Correlation analysis between inflammatory factors in the serum
To explore the interactions between inflammatory factors in the serum and to understand how they collectively influence immune and inflammatory responses in AR patients, we conducted a correlation analysis. FGF basic was positively correlated with CXCL8 (r = 0.7850, P < 0.0001), IL-4 was positively correlated with IL-17 (r = 0.4163, P = 0.0002), CXCL8 was positively correlated with IL-1β (r = 0.8885, P < 0.0001), and CCL3 was positively correlated with CXCL8 (r = 0.8580, P < 0.0001) (Figure 3D; Supplementary Table S8).
Correlation analysis of inflammatory cytokines in nasal secretions and serum
To explore the associations between inflammatory factors in nasal secretions and serum and to understand the interactions and underlying mechanisms of these factors in AR patients, we performed a correlation analysis. The analysis results revealed that G-CSF, IFN-γ, IL-4, IL-6 and TNF-ɑ were significantly elevated in patients with AR nasal secretions (Figure 4A; Supplementary Table S9). IFN-γ in nasal secretions was positively correlated with nasal obstruction (r = 0.5025, P = 0.0010) and nasal pruritus (r = 0.3264, P = 0.0398). IL-4 in nasal secretions was positively correlated with nasal obstruction (r = 0.4492, P = 0.0036). IL-6 in nasal secretions was positively correlated with nasal obstruction (r = 0.3516, P = 0.0261). TNF-α was positively correlated with nasal obstruction (r = 0.4389, P = 0.0046) (Figure 4B; Supplementary Table S10). CXCL5 in the serum was positively correlated with G-CSF (r = 0.3274, P = 0.0392), IFN-γ (r = 0.3336, P = 0.0354), IL-4 (r = 0.3209, P = 0.0435), and IL-6 (r = 0.3182, P = 0.0454) in nasal secretions. Serum IL-4 was positively correlated with Tpo (r = 0.3242, P = 0.0416) in nasal secretions. Serum IL-5 was negatively correlated with CXCL5 (r = −0.3372, P = 0.0164) in nasal secretions. Serum CCL4 was positively correlated with IL-6 (r = 0.3342, P = 0.0351) in nasal secretions. Serum TNF-ɑ was negatively correlated with CCL4 (r = −0.3350, P = 0.0346) in nasal secretions. Serum Tpo was positively correlated with CXCL5 (r = 0.3327, P = 0.0360) in nasal secretions (Figure 4C; Supplementary Table S11).
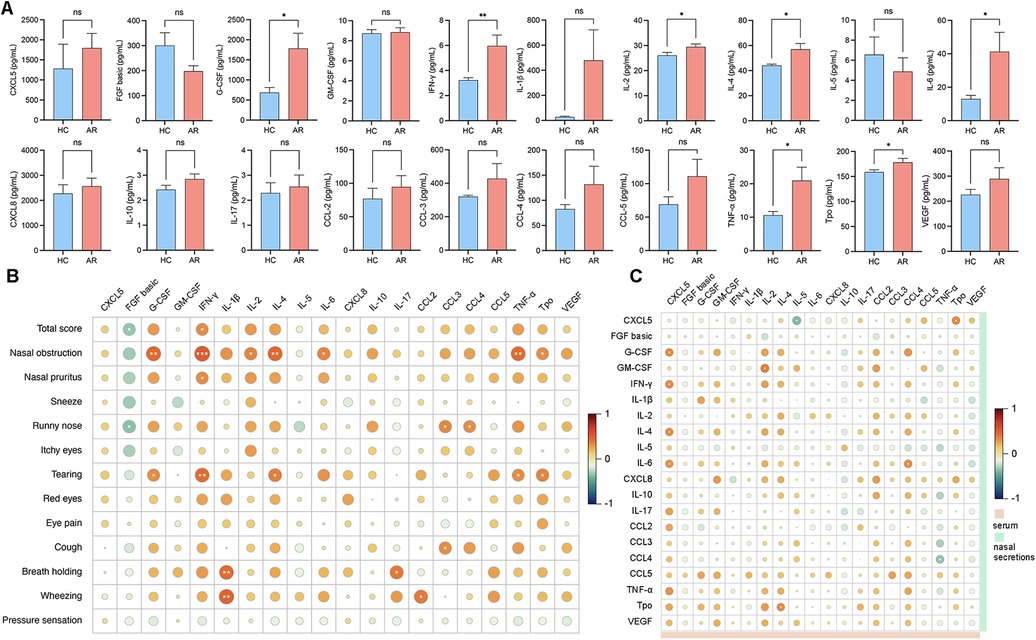
Figure 4. (A) Levels of cytokine expression in nasal secretions. (B) Correlation analysis between the VAS score and the levels of inflammatory cytokines in nasal secretions. (C) Correlation analysis between serum cytokines and nasal secretion cytokines.
Biological pathway analysis of significantly DE lncRNAs
To investigate the potential metabolic pathways and biological processes in which these lncRNAs may be involved, the potential functions of the lncRNAs were predicted via the GO and KEGG pathway annotations of their coexpressed mRNAs via the DAVID online tool, and possible biological pathways were identified. GO and KEGG analyses revealed that the DE genes were significantly enriched in multiple pathways involved in the immune response, such as the Toll-like receptor signaling pathway, the IL-17 signaling pathway and neutrophil activation. Overall, AC121247.1, AC106801.1, AL133371.3, and AC011524.2 may participate in the immune response (Figure 5).
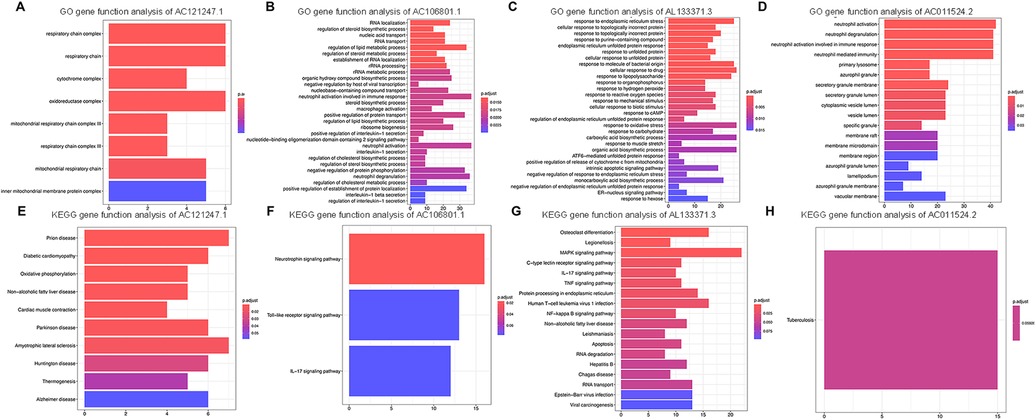
Figure 5. Go and KEGG analyses of DE lncRNAs. (A) GO gene function analysis of AC121247.1. (B) GO gene function analysis of AC106801.1. (C) GO gene function analysis of AL133371.3. (D) GO gene function analysis of AC011524.2. (E) KEGG gene function analysis of AC121247.1. (F) KEGG gene function analysis of AC106801.1. (G) KEGG gene function analysis of AL133371.3. (H) KEGG gene function analysis of AC011524.2.
Predictive properties of lncRNA levels in the diagnosis of AR
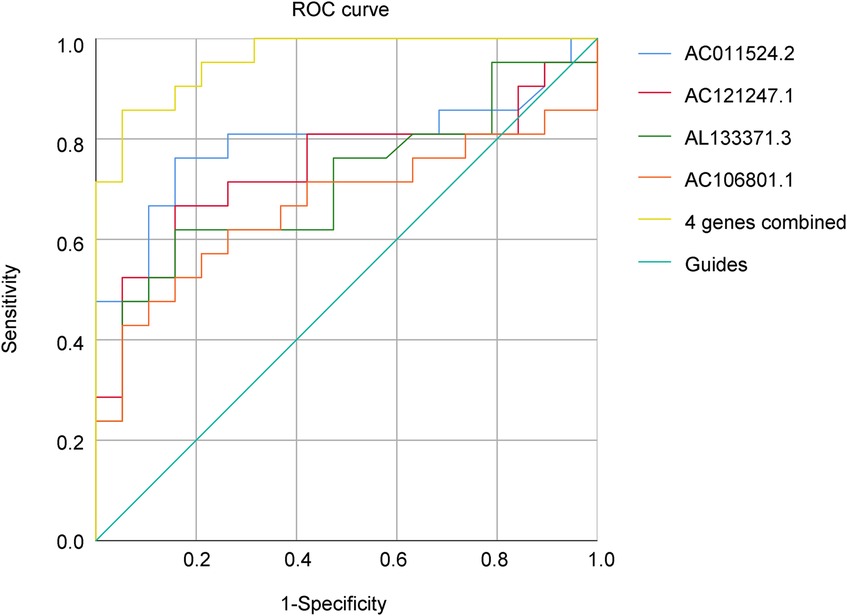
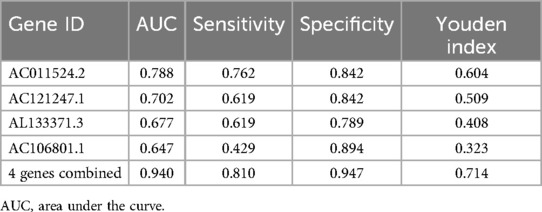
The area under the curve (AUC) of a single lncRNA was between 0.667 and 0.788; the highest AUC was 0.788 for AC011524.2, and the sensitivity and specificity were 0.762 and 0.842, respectively. When the 4 genes were combined, the AUC was significantly increased to 0.940, indicating greater sensitivity and specificity, and the Youden index reached 0.714, indicating that this combination method had greater accuracy in identifying AR (Figure 6; Table 4).
Discussion
The positive findings of specific IgE in serum offer a clear indication of the primary allergens, while their levels serve as an objective gauge of the body's sensitization status. However, in clinical practice, the level of a specific IgE does not always align with the severity of the condition, and positive results do not necessarily align with the gravity of clinical symptoms (14–17). Our study concurs with this, finding no significant correlation. However, our research has uncovered numerous DE lncRNAs that exhibit a connection with AR symptoms.
Our study revealed that AC011524.2 was positively correlated with nasal pruritus and smell dysfunction. Additionally, AC011524.2 was positively correlated with CXCL8 and CCL3. Meanwhile, CXCL8 was positively correlated with nasal pruritus. Moreover, AC011524.2 was enriched in pathways related to neutrophil activation, neutrophil degranulation, and other immune response pathways. These findings indicate that AC011524.2 might be a pivotal factor in the pathogenesis of AR by influencing neutrophil-related processes. Historically, neutrophils have been considered relatively understated white blood cells within the intricate immune system (18). However, an increasing number of studies have identified their significant involvement in allergic respiratory diseases (19–24), especially in AR (24–26). Following continuous, low-dose exposure to HDM allergens, neutrophils in the nasal mucosa undergo notable elevation, culminating in a peak count during the heightened state of HDM allergic inflammation, which suggests a correlation between the level of neutrophils and the severity of AR (25, 26). CXCL8 and CCL3 are both neutrophil-associated factors (27), and CXCL8 significantly increases in various bodily fluids after sensitization, such as sputum (28, 29), nasal secretions (30), and serum (31), which may be related to neutrophil chemotaxis during allergic reactions. AC011524.2 may affect the activation and degranulation process of neutrophils in peripheral blood by regulating the expression of these cytokines, thus contributing to the occurrence and development of AR. While we observed a correlation between AC011524.2 and various clinical symptoms and inflammatory factors, the specific regulatory mechanism involved remains elusive. Our previous studies revealed that neutrophils can express matrix metalloproteinases (MMPs), such as MMP-8, in nasal polyps (32), which indicates a similar remodeling process by neutrophils in AR combined with the above findings in the present study. Therefore, future research should focus on the intricate molecular mechanisms governing how AC011524.2 modulates neutrophils and their related pathways, ultimately fostering the emergence and progression of AR.
Our findings indicated that AL133371.3 was positively correlated with runny nose. Additionally, AL133371.3 was significantly positively correlated with only IL-17. Furthermore, AL133371.3 was enriched in the IL-17 signaling pathway in the present study, suggesting that it may be involved in the expression of IL-17 and the development of AR. The discovery of Th17 and regulatory T cells has provided a valuable addition to the established Th1/Th2 imbalance theory, especially as Th17 cells are believed to be linked to chronic allergic reactions (33–35). IL-17 actively contributes to inflammatory responses, specifically by promoting the recruitment and activation of neutrophils and triggering the generation of significant amounts of IL-6, CXCL8 and granulocyte colony-stimulating factor (34, 36). Many studies have shown that IL-17 is associated with the severity of AR symptoms (37–39). It has also been shown that the Th17 cytokines IL-17 and TGF-β1 promote the expression of Th2-related factors (40). Therefore, AL133371.3 may be involved in the development of AR by affecting the expression of IL-17.
Concurrently, we observed significant increases in the serum IL-4, CXCL8/IL-8, and IL-17 levels in AR patients. IL-4, a pivotal cytokine in type 2 inflammatory responses, plays a role in the formation of polyclonal IgE, facilitating the class switch of B cells toward IgE production (41). CXCL8/IL-8 and IL-17 are key cytokines involved in the type 3 inflammatory response, which can promote neutrophil recruitment and participate in angiogenesis. These key cytokines can promote chemotactic eosinophils and T lymphocytes, thereby promoting the occurrence and development of AR (31, 33–35). In addition, the level of the chemokine CCL3, a key regulator of the immune microenvironment, is significantly elevated in the serum of AR patients (42), which has also been confirmed in animal experiments (43). CCL3 synthesis is important for maintaining the recruitment of inflammatory cells during episodes of inflammation, as well as for activating eosinophils and T cells and regulating Ig production. CCL3 constitutes a second signal for the degranulation of mast cells, acting as a direct costimulatory signal via CCR1, making this chemokine crucial for inducing acute-phase AR (42–44).
By correlation analysis of serum inflammatory factors, IL-4 and IL-17 were found to be positively correlated in the serum of AR patients. The interaction between the Th2 and Th17 pathways in patients with AR is very complex. Sun et al. demonstrated in a mouse model that Th17 cytokines positively affect the differentiation of CD4+ cells and promote the production of IL-4 (45), which aggravates the symptoms associated with AR, a result that is consistent with the results of the present study. However, Choy et al. reported an inhibitory effect of IL-4 and IL-13 on the Th17 response in a mouse model of asthma (46), which is consistent with our previous findings in patients with CRSwNP (40), indicating that the Th2 and Th17 pathways are mutually exclusive and regulate each other.
Meanwhile, we found that G-CSF, IFN-γ, IL-4, IL-6 and TNF-ɑ were significantly increased in AR nasal secretions, suggesting the occurrence of local inflammatory response in the nasal cavity (47). In addition, all of these significantly elevated factors were positively correlated with the symptoms of nasal obstruction in AR, which may be due to the stimulation of HDM to the monocyte-macrophage in the nasal mucosa to produce G-CSF, IFN-γ and TNF-ɑ, and increased secretion of IL-4, IL-6 by upregulating B lymphocytes, mast cells and basophils, and induced VCAM-1 on vascular endothelium, thus directing inflammatory cells to migrate to the site of inflammation and causing exacerbation of nasal symptoms (48, 49).
Research on lncRNAs in AR has been widely reported. Ma et al. investigated the changes in the expression of lncRNAs in nasal mucosal samples from 4 HCs and 4 patients with AR. The study revealed a total of 2,259 lncRNAs detected in these samples, and GO and pathway analyses revealed enrichment in several biological processes and cellular signaling pathways related to AR development, such as positive regulation of IL-13 secretion, the Fc epsilon RI signaling pathway and the NF-kappa B signaling pathway (50). Peripheral blood mononuclear cells (PBMCs) from 3 AR patients and 3 HCs were collected and analyzed via microarray detection and verified in 16 AR patients and 18 HCs. They demonstrated that 31 lncRNAs were DE in the PBMCs of patients with AR and that 4 DE lncRNAs may be related to inflammation and the immune response (51). While prior investigations delved into the significance of lncRNAs in AR, they focused primarily on PBMCs, potentially neglecting the crucial role played by granulocytes, notably neutrophils. Our decision to focus on peripheral blood leukocytes offers a broader perspective on the immune and inflammatory profiles of AR patients. This study marks the first instance of uncovering the potential of lncRNAs in blood to modulate the severity of AR through neutrophils, thereby augmenting the knowledge base of previous research.
This study has notable limitations. First, it was conducted with a restricted sample size confined to a single center, excluding patients from diverse geographical locations. Consequently, the small sample size hindered the ability to further categorize patients into subgroups. Additionally, the study duration was prolonged due to the COVID-19 pandemic, which hindered its continuation for an extended period. Furthermore, the study lacked an in-depth exploration of the role of lncRNAs, and the intricate mechanism between DE lncRNAs and AR remains to be elucidated.
Our study revealed novel upregulated lncRNAs in the peripheral blood leukocytes of AR patients, highlighting their potential significance in AR, particularly via neutrophil-related processes and the IL-17 signaling pathway. Notably, AC011524.2 and AL133371.3 displayed remarkable correlations with AR symptoms and inflammatory cytokines, indicating their involvement in AR severity. In addition, many neutrophil-related cytokines are significantly elevated in the serum, and these cytokines are associated with various clinical symptoms of AR. These findings suggest that cytokines in serum may be influenced by the expression of specific lncRNAs. Despite the inevitable limitations of our study, these findings significantly contribute to the current understanding of lncRNAs in AR and lay the groundwork for future endeavors to decipher the intricate mechanisms and therapeutic implications of these lncRNAs in AR.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by Ethics Review Board of Beijing TongRen Hospital and the Beijing Institute of Otorhinolaryngology. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
JZ: Data curation, Validation, Writing – original draft. XP: Data curation, Validation, Writing – original draft. XW: Funding acquisition, Investigation, Writing – review & editing. LZ: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the High Level Public Health Technical Talent Training Plan (Lingjunrencai-01-08, Lingjunrencai-02-09), the National Key R&D Program of China (2022YFC2504100), the Program for the Changjiang Scholars and Innovative Research Team (IRT13082), the National Natural Science Foundation of China (82171110), the Beijing Natural Science Fund (7222024), the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-022), the Beijing Municipal Science and Technology Project (Z181100001618002), the Beijing New-Star Plan of Science and Technology (20220484226 and 20230484476), and the Public Welfare Development and Reform Pilot Project (2019-10).
Acknowledgments
We thank all participants for their work which made this study possible.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision. The reviewer HW declared a shared affiliation with the authors at the time of the review.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/falgy.2024.1466480/full#supplementary-material
References
1. Brożek JL, Bousquet J, Agache I, Agarwal A, Bachert C, Bosnic-Anticevich S, et al. Allergic rhinitis and its impact on asthma (ARIA) guidelines—2016 revision. J Allergy Clin Immunol. (2017) 140(4):950–8. doi: 10.1016/j.jaci.2017.03.050
2. Wang XD, Zheng M, Lou HF, Wang CS, Zhang Y, Bo MY, et al. An increased prevalence of self-reported allergic rhinitis in major Chinese cities from 2005 to 2011. Allergy. (2016) 71(8):1170–80. doi: 10.1111/all.12874
3. Bjermer L, Westman M, Holmström M, Wickman MC. The complex pathophysiology of allergic rhinitis: scientific rationale for the development of an alternative treatment option. Allergy Asthma Clin Immunol. (2019) 15:1–15. doi: 10.1186/s13223-018-0314-1
4. Nurmatov U, van Schayck CP, Hurwitz B, Sheikh A. House dust mite avoidance measures for perennial allergic rhinitis: an updated cochrane systematic review. Allergy. (2012) 67(2):158–65. doi: 10.1111/j.1398-9995.2011.02752.x
5. Wang J, Wu Y, Li J, Huang X, Zhu R. Eight aeroallergen skin extracts may be the optimal panel for allergic rhinitis patients in central China. Int Arch Allergy Immunol. (2017) 173(4):193–8. doi: 10.1159/000479429
6. Ciprandi G, Tosca M A. House dust mites–driven allergic rhinitis: could its natural history be modified? Expert Rev Clin Immunol. (2021) 17(2):109–14. doi: 10.1080/1744666X.2021.1879642
7. Ip JY, Nakagawa S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ. (2012) 54(1):44–54. doi: 10.1111/j.1440-169X.2011.01303.x
8. Sigdel KR, Cheng A, Wang Y, Duan L, Zhang Y. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J Immunol Res. (2015) 2015:1. doi: 10.1155/2015/848790
9. Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. (2010) 220(2):126–39. doi: 10.1002/path.2638
10. Ye S, Zhu S, Feng L. LncRNA ANRIL/miR-125a axis exhibits potential as a biomarker for disease exacerbation, severity, and inflammation in bronchial asthma. J Clin Lab Anal. (2020) 34(3):e23092. doi: 10.1002/jcla.23092
11. Yue L, Yin X, Hao F, Dong J, Ren X, Xu O, et al. Long noncoding RNA Linc00632 inhibits interleukin-13-induced inflammatory cytokine and mucus production in nasal epithelial cells. J Innate Immun. (2020) 12(1):116–28. doi: 10.1159/000500420
12. Zhang H, Nestor CE, Zhao S, Lentini A, Bohle B, Benson M, et al. Profiling of human CD4+ T-cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J Allergy Clin Immunol. (2013) 132(4):1005–8. doi: 10.1016/j.jaci.2013.05.033
13. Wei X, Xu M, Wang C, Fang S, Zhang Y, Wang W. Genetic association between CDKN2B/CDKN2B-AS1 gene polymorphisms with primary glaucoma in a North Indian cohort: an original study and an updated meta-analysis. BMC Med Genomics. (2021) 14:1–12. doi: 10.1186/s12920-020-00855-1
14. Cheng L, Dong Z, Kong WJ, Li HB, Liu Z, Shi Li, et al. Chinese guideline for diagnosis and treatment of allergic rhinitis (2015, Tianjin). Chin J Otorhinolaryngol Head Neck Surg. (2016) 51(1):6–24. doi: 10.3760/cma.j.issn.1673-0860.2016.01.004
15. Chen JJ, Cheng L, Kong WJ, Li HB, Li L, Liu Z, et al. Chinese Medical Association. Chinese guideline for diagnosis and treatment of allergic rhinitis (2022, revision). Chin J Otorhinolaryngol Head Neck Surg. (2022) 57(2):106–29. doi: 10.3760/cma.j.cn115330-20211228-00828
16. Pepper AN, Ledford DK. Allergic rhinitis: diagnosis and treatment. In: Mahmoudi M, editor. Allergy and Asthma: Practical Diagnosis and Management. Cham: Springer (2016) p. 63–85. doi: 10.1007/978-3-319-30835-7_6
17. Bousquet J, Van Cauwenberge P, Khaltaev N, Aria Workshop Group, World Health Organization. Allergic rhinitis and its impact on asthma. J Allergy Clin Immunol. (2001) 108(5):S147–334. doi: 10.1067/mai.2001.118891
18. Rigby KM, DeLeo FR. Neutrophils in innate host defense against Staphylococcus aureus infections. Semin Immunopathol. (2012):237–59. doi: 10.1007/s00281-011-0295-3
19. Jatakanon A, Uasuf C, Maziak W, Lim S, Chung KF, Barnes PJ. Neutrophilic inflammation in severe persistent asthma. Am J Resp Crit Care Med. (1999) 160(5):1532–9. doi: 10.1164/ajrccm.160.5.9806170
20. Carlson M, Håkansson L, Kämpe M, Stålenheim G, Peterson C, Venge P. Degranulation of eosinophils from pollen-atopic patients with asthma is increased during pollen season†. J Allergy Clin Immunol. (1992) 89(1):131–9. doi: 10.1016/S0091-6749(05)80050-8
21. Carlson M, Håkansson L, Peterson C, Stålenheim G. Secretion of granule proteins from eosinophils and neutrophils is increased in asthma. J Allergy Clin Immunol. (1991) 87(1):27–33. doi: 10.1016/0091-6749(91)90209-7
22. Poto R, Shamji M, Marone G, Durham SR, Scadding GW, Varricchi G. Neutrophil extracellular traps in asthma: friends or foes? Cells. (2022) 11(21):3521. doi: 10.3390/cells11213521
23. Hosoki K, Itazawa T, Boldogh I, Sur S. Neutrophil recruitment by allergens contribute to allergic sensitization and allergic inflammation. Curr Opin Allergy Clin Immunol. (2016) 16(1):45–50. doi: 10.1097/ACI.0000000000000231
24. Göker AE, Ekincioglu E, Alagöz MH, Hummatov R, Arkan ME, Baskadem Yilmazer A, et al. The association of allergic rhinitis severity with neutrophil–lymphocyte and platelet–lymphocyte ratio in adults. Eur Arch Otorhinolaryngol. (2019) 276:3383–8. doi: 10.1007/s00405-019-05640-0
25. Gelardi M, Peroni DG, Incorvaia C, Quaranta N, De Luca C, Barberi S, et al. Seasonal changes in nasal cytology in mite-allergic patients. J Inflamm Res. (2014):39–44. doi: 10.2147/JIR.S54581
26. Ciprandi G, Buscaglia S, Pesce G, Pronzato C, Ricca V, Parmiani S, et al. Minimal persistent inflammation is present at mucosal level in patients with asymptomatic rhinitis and mite allergy. J Allergy Clin Immunol. (1995) 96(6):971–9. doi: 10.1016/S0091-6749(95)70235-0
27. Özcan A, Boyman O. Mechanisms regulating neutrophil responses in immunity, allergy, and autoimmunity. Allergy. (2022) 77(12):3567–83. doi: 10.1111/all.15505
28. Lavinskiene S, Bajoriuniene I, Malakauskas K, Jeroch J, Sakalauskas R. Sputum neutrophil count after bronchial allergen challenge is related to peripheral blood neutrophil chemotaxis in asthma patients. Inflamm Res. (2014) 63:951–9. doi: 10.1007/s00011-014-0770-0
29. Norzila MZ, Fakes K, Henry RL, Simpson J, Gibson PG. Interleukin-8 secretion and neutrophil recruitment accompanies induced sputum eosinophil activation in children with acute asthma. Am J Resp Crit Care. (2000) 161(3):769–74. doi: 10.1164/ajrccm.161.3.9809071
30. Pantin CT, Southworth T, Wetzel K, Singh D. Reproducibility of nasal allergen challenge responses in adults with allergic rhinitis. Clin Pharmacol. (2019):67–76. doi: 10.2147/CPAA.S184404
31. Kordulewska NK, Cieślińska A, Fiedorowicz E, Jarmołowska B, Piskorz-Ogórek K, Kostyra E. Cytokines concentrations in serum samples from allergic children—multiple analysis to define biomarkers for better diagnosis of allergic inflammatory process. Immunobiology. (2018) 223(11):648–57. doi: 10.1016/j.imbio.2018.07.010
32. Wang X, Sima Y, Zhao Y, Zhang N, Zheng M, Du K, et al. Endotypes of chronic rhinosinusitis based on inflammatory and remodeling factors. J Allergy Clin Immunol. (2023) 151(2):458–68. doi: 10.1016/j.jaci.2022.10.010
33. Oboki K, Ohno T, Saito H, Nakae S. Th17 and allergy. Allergol Int. (2008) 57(2):121–34. doi: 10.2332/allergolint.R-07-160
34. Roussel L, Houle F, Chan C, Yao Y, Bérubé J, Olivenstein R, et al. IL-17 promotes p38 MAPK-dependent endothelial activation enhancing neutrophil recruitment to sites of inflammation. J Immunol. (2010) 184(8):4531–7. doi: 10.4049/jimmunol.0903162
35. Ciprandi G, Fenoglio D, De Amici M, Quaglini S, Negrini S, Filaci G, et al. Serum IL-17 levels in patients with allergic rhinitis. J Allergy Clin Immunol. (2008) 122(3):650–651.e2. doi: 10.1016/j.jaci.2008.06.005
36. Mills KH. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol. (2023) 23(1):38–54. doi: 10.1038/s41577-022-00746-9
37. Amin K, Issa SM, Ali KM, Aziz MI, Hama Amieen HM, Bystrom J, et al. Evidence for eosinophil and IL-17 mediated inflammation in allergic rhinitis. Clin Mol Allergy. (2020) 18:1–7. doi: 10.1186/s12948-020-00117-6
38. Kudo M, Melton AC, Chen C, Engler MB, Huang KE, Ren X, et al. IL-17A produced by αβ T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat Med. (2012) 18(4):547–54. doi: 10.1038/nm.2684
39. Wang M, Zhang W, Shang J, Yang J, Zhang L, Bachert C. Immunomodulatory effects of IL-23 and IL-17 in a mouse model of allergic rhinitis. Clin Exp Allergy. (2013) 43(8):956–66. doi: 10.1111/cea.12123
40. Wang M, Zhang N, Zheng M, Li Y, Meng L, Ruan Y, et al. Cross-talk between TH2 and TH17 pathways in patients with chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. (2019) 144(5):1254–64. doi: 10.1016/j.jaci.2019.06.023
41. Wynn TA. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol. (2015) 15(5):271–82. doi: 10.1038/nri3831
42. Berghi O, Dumitru M, Caragheorgheopol R, Tucureanu C, Simioniuc-Petrescu A, Sfrent-Cornateanu R, et al. The relationship between chemokine ligand 3 and allergic rhinitis. Cureus. (2020) 12(4):e7783. doi: 10.7759/cureus.7783
43. Miyazaki D, Nakamura T, Toda M, Cheung-Chau KW, Richardson RM, Ono SJ. Macrophage inflammatory protein–1ɑ as a costimulatory signal for mast cell–mediated immediate hypersensitivity reactions. J Clin Invest. (2005) 115(2):434–42. doi: 10.1172/JCI18452
44. Albloushi S, Al-Ahmad M. Exploring the latest understanding on the role of immune mediators, genetic and environmental factors in pathogenesis of allergic rhinitis: a systematic review. Front in Allergy. (2023) 4:1223427. doi: 10.3389/falgy.2023.1223427
45. Sun H, Liu B, Ma W, Ji D, Guan B. Changing characteristics of treg/Th17 cells in the nasal mucosa of a mouse model of allergic rhinitis and the effect of intervention with anti-IL-17 antibody. Discov Med. (2024) 36(185):1260–7. doi: 10.24976/Discov.Med.202436185.116
46. Choy DF, Hart KM, Borthwick LA, Shikotra A, Nagarkar DR, Siddiqui S, et al. TH2 and TH17 inflammatory pathways are reciprocally regulated in asthma. Sci Transl Med. (2015) 7(301):301ra129. doi: 10.1126/scitranslmed.aab3142
47. Martynova E, Rizvanov A, Urbanowicz RA, Khaiboullina S. Inflammasome contribution to the activation of Th1, Th2, and Th17 immune responses. Front Microbiol. (2022) 13(13):851835. doi: 10.3389/fmicb.2022.851835
48. Ciprandi G, Tosca MA, Marseglia GL, Klersy C. Relationships between allergic inflammation and nasal airflow in children with seasonal allergic rhinitis. Ann Allergy Asthma Immunol. (2005) 94(2):258–61. doi: 10.1016/S1081-1206(10)61305-1
49. Scadding GK, Scadding GW. Innate and adaptive immunity: ILC2 and Th2 cells in upper and lower airway allergic diseases. J Allergy Clin Immunol Pract. (2021) 9(5):1851–7. doi: 10.1016/j.jaip.2021.02.013
50. Ma Z, Teng Y, Liu X, Li J, Mo J, Sha M, et al. Identification and functional profiling of differentially expressed long non-coding RNAs in nasal mucosa with allergic rhinitis. Tohoku J Exp Med. (2017) 242(2):143–50. doi: 10.1620/tjem.242.143
Keywords: allergic rhinitis, house dust mite, lncRNA, RNA-seq, peripheral leukocyte
Citation: Zhao J, Pu X, Wang X and Zhang L (2024) Altered expression of long noncoding RNAs regulating neutrophilic inflammation in peripheral blood was associated with symptom severity in patients with house dust mite-induced allergic rhinitis. Front. Allergy 5:1466480. doi: 10.3389/falgy.2024.1466480
Received: 18 July 2024; Accepted: 10 October 2024;
Published: 25 October 2024.
Edited by:
Nives Zimmermann, University of Cincinnati, United StatesReviewed by:
Hontian Wang, Capital Medical University, ChinaJuan Abonia, Cincinnati Children’s Hospital Medical Center, United States
Copyright: © 2024 Zhao, Pu, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangdong Wang, ZW50d3hkQHZpcC5zaW5hLmNvbQ==; Luo Zhang, ZHIubHVvemhhbmdAMTM5LmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Jinming Zhao
Jinming Zhao Xiaoyu Pu
Xiaoyu Pu Xiangdong Wang
Xiangdong Wang Luo Zhang
Luo Zhang