- 1Michael Smith Laboratories, University of British Columbia, Vancouver, BC, Canada
- 2Department of Microbiology and Immunology, University of British Columbia, Vancouver, BC, Canada
- 3Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, BC, Canada
In addition to numerous clinical studies, research using experimental models have contributed extensive evidence to the link between antibiotic exposure and atopic disease. A number of mouse models of allergy have been developed and used to uncover the specific effects of various microbiota members and perturbations on allergy development. Studies in mice that lack microbes entirely have also demonstrated the various components of the immune system that require microbial exposure. The importance of the early-life period and the mechanisms by which atopy “protective” species identified in human cohorts promote immune development have been elucidated in mice. Finally, non-animal models involving human-derived cells shed light on specific effects of bacteria on human epithelial and immune responses. When considered alongside clinical cohort studies, experimental model systems have provided crucial evidence for the link between the neonatal gut microbiota and allergic disease, immensely supporting the stewardship of antibiotic administration in infants. The following review aims to describe the range of experimental models used for studying factors that affect the relationship between the gut microbiota and allergic disease and summarize key findings that have come from research in animal and in vitro models.
Introduction
Experimental models enable the characterization of complex host responses ranging from cellular to systemic, contributing vitally to major advancements in immunology. Mice have long been the model of choice when studying host immunity and infection responses. The murine genome overlaps substantially with that of humans, and many key immune pathways are conserved. However, human and mouse immune systems are not identical, and findings in mouse models do not always translate directly to human application (1, 2). Non-animal models using cultured intestinal epithelial and immune cells, or miniature organ-like structures, have also been developed to determine specific effects of bacteria on various cell types (3, 4). Although this enables the study of cells derived specifically from humans, the response of cells grown in culture does not always reflect what occurs in the complex environment of a mammalian host. However, when used in conjunction with clinical human studies, both animal and in vitro models are critical to defining the mechanisms of host-microbiota interactions that affect immune development and atopic disease.
Mouse models for studying the gut microbiota
Mouse models enable the manipulation of a microbial community growing within the complex environment of a mammalian host. Standard laboratory mice are considered “specific-pathogen free” or SPF, because they have tested negative for a set of disease-causing pathogens. These mice are raised in clean laboratory conditions and have a microbiota that is lower in diversity than that of humans, but complex enough to simulate a stable gut community with some colonization resistance capacity (5). Microbiota composition differs depending on the mouse vendor and housing conditions, which can lead to differences in baseline immunity and response to treatments (6). Additionally, it can be difficult to tease apart direct effects of microbes on the host from indirect effects that act through the existing microbiota when studying SPF mice. Pretreatment with antibiotics is often required to promote stable colonization with a newly introduced microbe, which can confound results. Despite these caveats, SPF mice are used extensively to demonstrate the effects of antibiotics and microbiota alterations on immune development and disease (7–11).
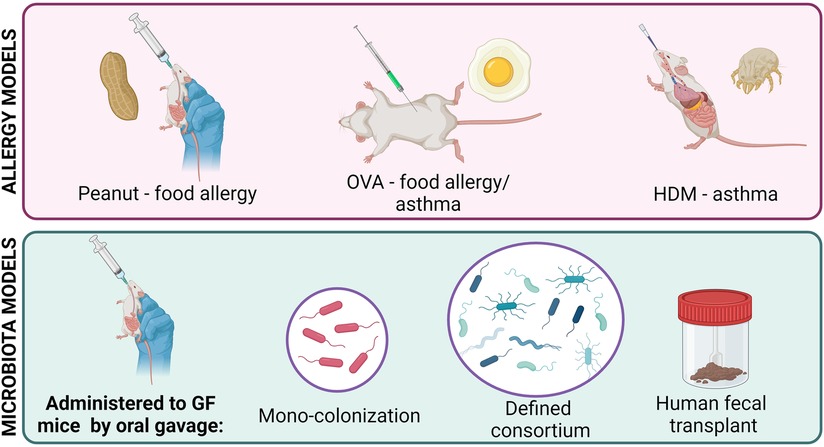
As the microbiome field has grown, a variety of animal models have been developed to combat the issues faced with SPF mice (Figure 1). Germ-free (GF) mice, which are completely sterile, do not have a microbiota and can be more easily colonized than SPF mice (12). Mono-colonization of germ-free mice is commonly used to study the effects of one bacterial species at a time, independent of any confounding effects of other microbes in the environment. GF mice can also be colonized with a defined consortia of microbes to study simplified, specific communities and microbe-microbe interactions within the gut environment (5, 13, 14). Humanized mice, which are created by transferring human feces to GF mice, display a microbiota that is more diverse and complex than that of SPF mice and more similar to that of humans, facilitating the study of clinically relevant human-colonizing microbes in vivo (15, 16). However, some microbes found in humans require a host-specific niche and fail to colonize mice, limiting the utility of humanized mice (17). Lastly, transient colonization of mice with microbes that decline and disappear demonstrate the lasting effects of microbial exposure at specific developmental stages (18).
Mouse models of allergic disease
Several mouse models have been developed to study the atopic immune response, with allergic asthma and food allergy being most common (Figure 1). Since all atopic diseases involve the same key pathways (Th2 cell activation, IgE production, mast cell degranulation, basophil hyperplasia), the basic process of allergy induction is similar between models: mice are first sensitized and then challenged with an allergen to initiate and then stimulate a Th2-mediated response (19–21).
There are two popular models of allergic asthma. The OVA model involves intraperitoneal injection, followed by intratracheal or intranasal administration, of ovalbumin derived from chicken egg (22). The house dust mite (HDM) model involves a series of intranasal exposures to protein from a common HDM species such as Dermatophagoides pteronyssinus (23). Both models stimulate a Th2-mediated response and lung histopathology (22, 24).
Food allergy models are difficult to develop because oral exposure to even concentrated allergenic food proteins results in oral tolerance and fails to induce a response (25). For oral induction, food allergens must be administered with adjuvants to induce a Th2-mediated response (26). For example, concentrated peanut extract can be administered orally with cholera toxin, resulting in elevated IgE production and anaphylaxis (27). Peanut allergens can also be administered epicutaneously to mice to induce Th2-mediated responses which can be further stimulated orally in the absence of an adjuvant, which may be explained by the link between skin barrier function and food allergy (28–30). Although peanut proteins show the highest allergenicity in mice, egg and milk allergies have been induced using adjuvants as well (26).
There are also models of atopic dermatitis and allergic rhinitis, the common forms of which also involve sensitization and challenge with ovalbumin, but are less common (31, 32). In humans, atopic dermatitis often precedes the development of food allergy and asthma in a sequential pattern termed the “atopic march” (33). The atopic march has been demonstrated in the NC/Nga mouse, commonly used to study atopic dermatitis (34). Epicutaneous OVA or peanut protein sensitization can also induce allergic manifestations at other sites (35). Our understanding of the microbiota's role in the atopic march is lacking, and murine models that display multiple allergies initiated via the skin should be utilized to further investigate factors that affect the progression of disease.
Studies of the microbiota and antibiotics using allergy models will be highlighted below.
Atopic disease—insights from germ-free mice
GF mice, which lack a microbiota entirely, display a dramatically altered immune system (12). Intestinal and systemic immune compartments are largely underdeveloped or missing in GF mice. While some of these phenotypes can be rescued by colonization of germ-free mice at any age, others require colonization within the critical early-life window.
As in newborns, the immune systems of GF mice are skewed toward Th2 responses. Th1 and Th2 cells reciprocally regulate each other, and in the absence of microbial stimulation of Th1 pathways, Th2 responses go uncontrolled (36). This phenomenon supports the hygiene hypothesis, which posits that increased Th2 responses can be attributed to reduced microbial exposure and Th1 stimulation in modern, more hygienic environments (37, 38). Although mono-colonization of germ-free mice with certain commensals can reduce Th2 skewing, full restoration of normal T cell proportions requires colonization with a diverse microbiota (39).
Germ-free mice also display increased invariant natural killer T (iNKT) cells in the lung and colon. iNKT cells are found in higher proportions in individuals with severe and uncontrolled asthma (40). Concordantly, germ-free mice display increased iNKT-mediated inflammation in response to asthma models (41). Exposure to a specific antigen produced by the common commensal Bacteroides fragilis within the first two weeks of life is sufficient to rescue iNKT cell levels (42). iNKT cell levels cannot be restored in adult mice, implicating the preweaning period as an important moment in immune imprinting by the colonizing microbiota (12).
Serum IgE titers are also elevated in germ-free mice (43). This contributes to more severe anaphylaxis in food allergy models, and can only be rescued by colonization in the first few weeks of life (12, 43). Regulatory T cell development also requires antigenic exposure specifically during the early life period (44, 45). Many studies have demonstrated that these immunological features make GF mice more susceptible to asthma models (46–48). However, others have shown that certain aspects of the GF mouse allergy response are dampened (49), and that colonization of certain species can worsen allergy symptoms of germ-free mice (50). Together, studies in germ-free mice have shed light on the numerous pathways by which microbes in the gut can protect against or contribute to asthma and allergy development.
The weaning reaction in mice
While colonization of GF mice at any age can rescue some of their immune alterations, exposure to microbes during the early-life window is required for complete restoration of proper immune function. Recently, this window has been well-defined in mice to be the first 3 weeks of life. At weaning, mice undergo a dramatic immune reaction to the influx of microbes and loss of passive immune factors in milk, which imprints their immune systems and susceptibility to disease for life (51). Although it is not clear whether the “weaning reaction” occurs in humans, the associations of breastfeeding and microbiota maturation in infancy with immune health later in life suggest that the human weaning period is also critical for microbiota and immune development (52, 53).
At weaning, the mouse microbiota diversifies and expands, displaying a bloom in Clostridia and Bacteroides which replace gamma-Proteobacteria and Lactobacillus species. This is similar to the microbiota shifts observed in humans from birth until 6 months of age (21, 54, 55). Milk contains several anti-inflammatory molecules, which gradually decline in concentration over time post-gestation. Epidermal growth factor (EGF) in milk delays the opening of Goblet-cell-associated antigen passages (GAPs) in the gut. As EGF levels decrease over the course of the first few weeks of life, GAPs open and allow for microbes to interact with and stimulate the immune system. In response to the developing microbiota and mucosa, GAPs close shortly after weaning. Antigenic exposure through GAPs drives regulatory T cell development, promoting tolerance of the gut microbiota and imprinting the immune system. This process is unique to the early-life period and cannot be induced in adult mice. Importantly, the weaning reaction protects against allergic inflammation later in life, and antibiotic exposure before but not after weaning significantly disrupts immune programming and contributes to worsened health outcomes (51).
The weaning reaction has not been demonstrated in humans, and while the exact mechanism may not be the same, the weaning reaction theory provides a potential explanation for the relationship between neonatal microbiota disruption and adverse health outcomes that is well-observed in humans. Accessing human neonatal tissue and blood to define a human weaning reaction would be immensely difficult. However, the deep characterization of this response in mice along with ample evidence of long-term effects of early-life antibiotics in infants provide clues to the process of microbe-mediated immune imprinting that occurs in humans.
Antibiotics, the microbiota, and mouse models of asthma
Prompted by associations observed in human cohorts, the relationship between the gut microbiota and asthma has been studied and characterized extensively through colonization of GF mice or supplementation of SPF mice with different combinations of bacteria.
Numerous animal studies have identified specific immunomodulatory bacteria that protect against allergies. As mentioned above, colonization of germ-free mice with allergic infants is sufficient to increase anaphylaxis susceptibility (56). Through sequencing and host gene expression analysis, this effect was explained by protective effects of the Lachnospiraceae species, Anaerostipes caccae. Mono-colonization experiments confirmed that this species alone, which is elevated in healthy infants, contributes to oral tolerance and protects against anaphylactic responses. In another study, four genera of bacteria inversely associated with allergies in human infants were administered to mice and shown to ameliorate OVA-induced asthma (57). This was linked to the effects of specific bacterial metabolites on immune development.
Species of Bifidobacteria and Lactobacillus, which are known to be promoted by breastmilk and are inversely associated with allergies, have also been tested in animal models (58–60). Supplementation with Bifidobacterium longum and Bifidobacterium breve have been shown numerous times to limit allergic responses to various models by inducing regulatory T cells and dampening Th2 responses (48, 61–63). Lactobacillus species have also been shown to improve intestinal barrier integrity and promote Th1 responses, which limit allergic phenotypes (64, 65).
In all of these studies, bacteria known to be associated with health in humans were investigated and causally linked to allergy protection. Animal models have been key to moving from correlation to causation in our understanding of the multiple roles of the microbiota in immune development and have helped identify the specific bacteria and pathways that should not be disrupted during the neonatal period. In addition to providing insight into protective and beneficial bacteria, animal studies have demonstrated the effects of early-life antibiotic exposure on asthma.
Vancomycin treatment during neonatal but not adult life increases susceptibility to OVA-induced asthma in mice (8). This is linked to alterations in gut microbiota composition and diversity, and reduced colonic Treg cell levels. Both of these phenotypes were more drastic in neonatal than adult vancomycin-treated mice, and the period between birth and weaning was identified as the window during which antibiotic induced dysbiosis was found to significantly affect adult asthma outcomes (66). Similarly, Azithromycin treatment in early life increased IgE and Th2 responses to HDM-induced asthma (67). Interestingly, in this study, transfer of the azithromycin perturbed microbiota to adult germ-free mice did not transfer the phenotype. However, the offspring of these mice displayed worsened asthma outcomes, indicating that the effects of microbiota alterations on immune development must occur in early life. Dysbiosis induced by a combination of antibiotics has also been shown to impair oral tolerance by disrupting dendritic cell development in the gut (68).
Exposure to a single course of macrolide antibiotics at a clinically relevant dose during neonatal life is sufficient to reduce microbial diversity and shift community composition, which persists into adulthood. This was accompanied by permanent dampening effects on local and systemic immunity (69). In contrast, adult mice treated with the same antibiotic course displayed rapid microbiota recovery and did not show immune aberrations. Microbiota transfer from antibiotic-treated mice to germ-free mice conferred the altered immune phenotype in this study, demonstrating that the perturbed microbiota is sufficient to drive immune alterations associated with antibiotic exposure. Lynn et al. treated mice with ampicillin and neomycin until weaning and then aged them to 700 days (70). They found that early-life antibiotic exposure affects immune status, longevity, and metabolism even long after antibiotic exposure.
The studies highlighted above demonstrate the multitude of detrimental effects that antibiotic exposure in early life have on long-term health. Mouse studies have been vital in our understanding of both the specific effects of “protective” microbes and antibiotics on different facets of immune development, and the uniqueness of the infancy period in these processes. They provide strong support for limiting antibiotic administration in infants whenever possible.
Non-animal models for studying antibiotics and allergic asthma
In addition to animal models, cultured human cells are frequently used to investigate the microbiota and allergic responses. Epithelial responses to microbial products can by studied by monoculture of human intestinal or lung epithelial cells. For example, Bifidobacteria species grown on human milk oligosaccharides induce anti-inflammatory responses in human-derived intestinal cells (71). Some commensal species and their metabolites have also affect barrier integrity of intestinal cell monolayers (72–75), either promoting or impairing barrier function, which has implications for allergy and asthma susceptibility (76, 77). However, allergic disease involves communication between the epithelium, the mucosal immune system, and systemic circulation, and it is difficult to define the mechanisms and effects of microbe-immune crosstalk using only one cell type. Co-culture systems involving epithelial cells grown with dendritic cells, macrophages, and lymphocytes have been developed to combat this issue.
In one study, peripheral blood mononuclear cells (PBMCs) were isolated from atopic or non-atopic individuals and co-cultured with epithelial cells exposed to microbial antigenic stimulation to demonstrate that commensal microbe exposure limits the allergic response (78). Co-culture of PBMCs with intestinal epithelial cells has also been used to mechanistically link Bifidobacteria and other commensal species to regulatory T cell responses (79, 80). In another study, human-derived naïve T cells were exposed to fecal water from infants that received Bifidobacterium infantis supplementation or fecal water from control infants. This enabled the identification of a specific microbial metabolite produced by B. infantis in the infant gut that skews T cell phenotypes away from allergic (Th2 and Th17) and towards Th1 phenotypes (81). More recently, co-culture systems involving many cell types have been developed. Zuurveld et al. designed a model involving epithelial cells, dendritic cells, T and B cells, and mast cells and were able to simulate the entire allergic pathway from epithelial cell allergen exposure to mast cell degranulation and IgE production in vitro (4).
Finally, in addition to mono- and co-culture systems, 3D structures that better simulate organ-level biology can model microbe-immune communication. Organoids and gut-on-a-chip devices are derived from human intestinal stem cells and morphologically mimic 3D intestinal tissue, displaying crypts, villi, Paneth cells, mucous production, and distinct apical and basolateral compartments (3, 82). These models are only beginning to be used to study microbe-host interactions that influence systemic health but offer a promising physiologically relevant alternative to animal models (83, 84).
Conclusions
Experimental models have characterized the complex and dynamic processes of microbe-mediated immune development that occurs in early-life and validated key taxa that drive protective and beneficial immune processes. They have provided ample evidence that antibiotics have detrimental effects on gut microbiota composition, long-term health, and allergy in a controlled setting. They have also defined the features of immunologic development that occur during the critical early-life window, and illustrated the imprinting effects of the microbiota during this period. While lab rodents and cultured cells do not replicate the human gut and immune system identically, the findings highlighted above strengthen and complement conclusions from human association studies, drawing clear mechanistic links between antibiotic exposure, reduced microbiota diversity, and allergic outcomes.
Author contributions
KD: Writing – original draft, Writing – review & editing. BF: Writing – original draft, Funding acquisition, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. KD is supported by the Four Year Fellowship Tuition Award, President’s Academic Excellence Initiative PhD Award and International Tuition Award at the University of British Columbia.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J Immunol. (2004) 172:2731–8. doi: 10.4049/jimmunol.172.5.2731
2. Masopust D, Sivula CP, Jameson SC. Of mice, dirty mice, and men: using mice to understand human immunology. J Immunol. (2017) 199:383–8. doi: 10.4049/jimmunol.1700453
3. Valiei A, Aminian-dehkordi J, Mofrad MRK. Gut-on-a-chip models for dissecting the gut microbiology and physiology. APL Bioeng. (2023) 7(1):011502. doi: 10.1063/5.0126541
4. Zuurveld M, Díaz CB, Redegeld F, Folkerts G, Garssen J, van’t Land B, et al. An advanced in vitro human mucosal immune model to predict food sensitizing allergenicity risk: a proof of concept using ovalbumin as model allergen. Front Immunol. (2023) 13:1–12. doi: 10.3389/fimmu.2022.1073034
5. Thomson CA, Morgan SC, Ohland C, McCoy KD. From germ-free to wild: modulating microbiome complexity to understand mucosal immunology. Mucosal Immunol. (2022) 15:1085–94. doi: 10.1038/s41385-022-00562-3
6. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous Bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
7. Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. (2011) 9:233–43. doi: 10.1038/nrmicro2536
8. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. (2012) 13:440–7. doi: 10.1038/embor.2012.32
9. Jin Y, Wu Y, Zeng Z, Jin C, Wu S, Wang Y, et al. Exposure to oral antibiotics induces gut microbiota dysbiosis associated with lipid metabolism dysfunction and low-grade inflammation in mice. Toxicol Sci. (2016) 154:140–52. doi: 10.1093/toxsci/kfw150
10. Leclercq S, Mian FM, Stanisz AM, Bindels LB, Cambier E, Ben-Amram H, et al. Low-dose penicillin in early life induces long-term changes in murine gut microbiota, brain cytokines and behavior. Nat Commun. (2017) 8:15062. doi: 10.1038/ncomms15062
11. Adami AJ, Bracken SJ, Guernsey LA, Rafti E, Maas KR, Graf J, et al. Early-life antibiotics attenuate regulatory T cell generation and increase the severity of murine house dust mite-induced asthma. Pediatr Res. (2018) 84:426–34. doi: 10.1038/s41390-018-0031-y
12. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352:539–44. doi: 10.1126/science.aad9378
13. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
14. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. (2013) 500:232–5. doi: 10.1038/nature12331
15. Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. (2013) 7:1933–43. doi: 10.1038/ismej.2013.89
16. Arrieta M, Sadarangani M, Brown EM, Russell SL, Nimmo M, Dean J, et al. A humanized microbiota mouse model of ovalbumin-induced lung inflammation. Gut Microbes. (2016) 7:342–52. doi: 10.1080/19490976.2016.1182293
17. Chung H, Pamp J, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut immune maturation Depends on colonization with a host-specific Microbiota. Cell. (2012) 149:1578–93. doi: 10.1016/j.cell.2012.04.037
18. De Agüero MG, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351:1296–302. doi: 10.1126/science.aad2571
19. Barcik W, Boutin RCT, Sokolowska M, Finlay BB. The role of lung and gut microbiota in the pathology of asthma. Immunity. (2020) 52:241–55. doi: 10.1016/j.immuni.2020.01.007
20. Luu M, Monning H, Visekruna A. Exploring the molecular mechanisms underlying the protective effects of microbial SCFAs on intestinal tolerance and food allergy. Front Immunol. (2020) 11:16–21. doi: 10.3389/fimmu.2020.01225
21. Donald K, Finlay BB. Early-life interactions between the microbiota and immune system: impact on immune system development and atopic disease. Nat Rev Immunol. (2023) 23:735–48. doi: 10.1038/s41577-023-00874-w
22. Kim DI, Song M, Lee K. Comparison of asthma phenotypes in OVA- induced mice challenged via inhaled and intranasal routes. BMC Pulm Med. (2019) 19:1–11. doi: 10.1186/s12890-019-1001-9
23. Foray AP, Dietrich C, Pecquet C, Machavoine F, Chatenoud L, Leite-de-Moraes M. IL-4 and IL-17 are required for house dust mite-driven airway hyperresponsiveness in autoimmune diabetes-prone non-obese diabetic mice. Front Immunol. (2021) 11:1–8. doi: 10.3389/fimmu.2020.595003
24. Woo LN, Guo WY, Wang X, Young A, Salehi S, Hin A, et al. A 4-week model of house dust Mite (HDM) induced allergic airways inflammation with airway remodeling. Sci Rep. (2018) 8:1–11. doi: 10.1038/s41598-018-24574-x
25. Kanagaratham C, Sallis BF, Fiebiger E. Experimental models for studying food allergy. Cell Mol Gastroenterol Hepatol. (2018) 6:356–69.e1. doi: 10.1016/j.jcmgh.2018.05.010
26. Smeekens JM, Kulis MD. Mouse models of food allergy in the pursuit of novel treatment modalities. Front Allergy. (2021) 2:1–7. doi: 10.3389/falgy.2021.810067
27. Orgel K, Kulis M. A mouse model of peanut allergy induced by sensitization through the gastrointestinal tract. Methods Mol Biol. (2018) 1799:39–47. doi: 10.1007/978-1-4939-7896-0_4
28. Strid J, Hourihane J, Kimber I, Callard R, Strobel S. Epicutaneous exposure to peanut protein prevents oral tolerance and enhances allergic sensitization. Clin Exp Allergy. (2005) 35:757–66. doi: 10.1111/j.1365-2222.2005.02260.x
29. Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. (2015) 133:291–307.e5. doi: 10.1016/j.jaci.2013.11.020
30. Walker M, Green J, Ferrie R, Queener A, Kaplan MH, Cook-mills JM. Mechanism for initiation of food allergy: dependence on skin barrier mutations and environmental allergen co-stimulation. J Allergy Clin Immunol. (2018) 141:1711–25. doi: 10.1016/j.jaci.2018.02.003
31. Jin H, He R, Oyoshi M, Geha R. Animal models of atopic dermatitis. J Invest Deratol. (2009) 129:31–40. doi: 10.1038/jid.2008.106.Animal
32. Zhu Y, Yu J, Zhu X, Yuan J, Dai M, Bao Y, et al. Experimental observation of the effect of immunotherapy on CD4+T cells and Th1/Th2 cytokines in mice with allergic rhinitis. Sci Rep. (2023) 13:4–12. doi: 10.1038/s41598-023-32507-6
33. Tran MM, Lefebvre DL, Dharma C, Dai D, Lou WYW, Subbarao P, et al. Atopic dermatitis and inflammatory skin disease predicting the atopic march: results from the Canadian healthy infant longitudinal development study study population. J Allergy Clin Immunol. (2018) 141(2):601–7.e8. doi: 10.1016/j.jaci.2017.08.024
34. Lee HJ, Lee NR, Jung M, Kim DH, Choi EH. Atopic march from atopic dermatitis to asthma-like lesions in NC/nga mice is accelerated or aggravated by neutralization of stratum corneum but partially inhibited by acidification. J Invest Dermatol. (2015) 135:3025–33. doi: 10.1038/jid.2015.333
35. Han H, Roan F, Ziegler SF. The atopic march: current insights into skin barrier dysfunction and epithelial cell-derived cytokines. Immunol Rev. (2017) 278:116–30. doi: 10.1111/imr.12546
36. Mazmanian SK, Cui HL, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. (2005) 122:107–18. doi: 10.1016/j.cell.2005.05.007
37. Okada H, Kuhn C, Feillet H, Bach J-F. The “hygiene hypothesis” for autoimmune and allergic diseases: an update. Clin Exp Immunol. (2010) 160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x
38. Garn H, Potaczek DP, Pfefferle PI. The hygiene hypothesis and new perspectives—current challenges meeting an old postulate. Front Immunol. (2021) 12:1–7. doi: 10.3389/fimmu.2021.637087
39. Gaboriau-Routhiau V, Rakotobe S, Emelyne L, Mulder I, Bridonneau C, Rochet V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. (2009) 31:677–89. doi: 10.1016/j.immuni.2009.08.020
40. Matangkasombut P, Pichavant M, DeKruyff RH, Umetsu DT. Natural killer T cells and the regulation of asthma. Mucosal Immunol. (2009) 2:383–92. doi: 10.1038/mi.2009.96
41. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 23:1–7. doi: 10.1126/science.1219328
42. An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. (2014) 156:123–33. doi: 10.1016/j.cell.2013.11.042
43. Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. (2013) 14:559–70. doi: 10.1016/j.chom.2013.10.004
44. Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. (2014) 20:642–7. doi: 10.1038/nm.3568
45. Reynolds HM, Bettini ML. Early-life microbiota-immune homeostasis. Front Immunol. (2023) 14:1–8. doi: 10.3389/fimmu.2023.1266876
46. Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. (2011) 184:198–205. doi: 10.1164/rccm.201010-1574OC
47. Balzola F, Cullen G, Ho GT, Russell RK, Wehkamp J. Microbial exposure during early life has persistent effects on natural killer T cell function. Inflamm Bowel Dis Monit. (2012) 13:29–30. doi: 10.1126/science.1219328
48. Schwarzer M, Srutkova D, Schabussova I, Hudcovic T, Akgün J, Wiedermann U, et al. Neonatal colonization of germ-free mice with Bifidobacterium longum prevents allergic sensitization to major birch pollen allergen bet v 1. Vaccine. (2013) 31:5405–12. doi: 10.1016/j.vaccine.2013.09.014
49. Schwarzer M, Hermanova P, Srutkova D, Golias J, Hudcovic T, Zwicker C, et al. Germ-free mice exhibit mast cells with impaired functionality and gut homing and do not develop food allergy. Front Immunol. (2019) 10:1–12. doi: 10.3389/fimmu.2019.00205
50. Parrish A, Boudaud M, Grant ET, Willieme S, Neumann M, Wolter M, et al. Akkermansia muciniphila exacerbates food allergy in fibre-deprived mice. Nat Microbiol. (2023) 8:1863–79. doi: 10.1038/s41564-023-01464-1
51. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, et al. A weaning reaction to microbiota is required for resistance to immunopathologies in the adult. Immunity. (2019) 50:1276–88.e5. doi: 10.1016/j.immuni.2019.02.014
52. Nuzzi G, Elisa M, Cicco D, Peroni DG. Breastfeeding and allergic diseases: what’s new? J Funct Foods. (2021) 8(5):330. doi: 10.3390/children8050330
53. Hoskinson C, Dai DLY, Del Bel KL, Becker AB, Moraes TJ, Mandhane PJ, et al. Delayed gut microbiota maturation in the first year of life is a hallmark of pediatric allergic disease. Nat Commun. (2023):1–14. doi: 10.1038/s41467-023-40336-4
54. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17:690–703. doi: 10.1016/j.chom.2015.04.004
55. Jain N. The early life education of the immune system: moms, microbes and (missed) opportunities. Gut Microbes. (2020) 12:1–20. doi: 10.1080/19490976.2020.1824564
56. Feehley T, Plunkett CH, Bao R, Choi Hong SM, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. (2019) 25:448–53. doi: 10.1038/s41591-018-0324-z
57. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7(307):307ra152. doi: 10.1126/scitranslmed.aab2271
58. Galazzo G, van Best N, Bervoets L, Dapaah IO, Savelkoul PH, Hornef MW, et al. Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood. Gastroenterology. (2020) 158:1584–96. doi: 10.1053/j.gastro.2020.01.024
59. Vandenplas Y, Carnielli VP, Ksiazyk J, Luna MS, Migacheva N, Mosselmans JM, et al. Factors affecting early-life intestinal microbiota development. Nutrition. (2020) 78:110812. doi: 10.1016/j.nut.2020.110812
60. Boudry G, Charton E, Le Huerou-Luron I, Ferret-Bernard S, Le Gall S, Even S, et al. The relationship between breast milk components and the infant gut microbiota. Front Nutr. (2021) 8:1–21. doi: 10.3389/fnut.2021.629740
61. Inoue Y, Iwabuchi N, Xiao JZ, Yaeshima T, Iwatsuki K. Suppressive effects of Bifidobacterium breve strain M-16 V on T-helper type 2 immune responses in a murine model. Biol Pharm Bull. (2009) 32:760–3. doi: 10.1248/bpb.32.760
62. Hougee S, Vriesema AJM, Wijering SC, Knippels LMJ, Folkerts G, Nijkamp FP, et al. Oral treatment with probiotics reduces allergic symptoms in ovalbumin-sensitized mice: a bacterial strain comparative study. Int Arch Allergy Immunol. (2010) 151:107–17. doi: 10.1159/000236000
63. Lyons A, O’Mahony D, O’Brien F, MacSharry J, Sheil B, Ceddia M, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy. (2010) 40:811–9. doi: 10.1111/j.1365-2222.2009.03437.x
64. Schwarzer M, Repa A, Daniel C, Schabussova I, Hrncir T, Pot B, et al. Neonatal colonization of mice with Lactobacillus plantarum producing the aeroallergen bet v 1 biases towards Th1 and T-regulatory responses upon systemic sensitization. Allergy Eur J Allergy Clin. Immunol. (2011) 66:368–75. doi: 10.1111/j.1398-9995.2010.02488.x
65. Kozakova H, Schwarzer M, Tuckova L, Srutkova D, Czarnowska E, Rosiak I, et al. Colonization of germ-free mice with a mixture of three lactobacillus strains enhances the integrity of gut mucosa and ameliorates allergic sensitization. Cell Mol Immunol. (2016) 13:251–62. doi: 10.1038/cmi.2015.09
66. Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. (2013) 4:158–64. doi: 10.4161/gmic.23567
67. Borbet TC, Pawline MB, Zhang X, Wipperman MF, Reuter S, Maher T, et al. Influence of the early-life gut microbiota on the immune responses to an inhaled allergen. Mucosal Immunol. (2022) 15:1000–11. doi: 10.1038/s41385-022-00544-5
68. Fukaya T, Uto T, Mitoma S, Takagi H, Nishikawa Y, Tominaga M, et al. Gut dysbiosis promotes the breakdown of oral tolerance mediated through dysfunction of mucosal dendritic cells. Cell Rep. (2023) 42:112431. doi: 10.1016/j.celrep.2023.112431
69. Ruiz VE, Battaglia T, Kurtz ZD, Bijnens L, Ou A, Engstrand I, et al. A single early-in-life macrolide course has lasting effects on murine microbial network topology and immunity. Nat Commun. (2017) 8:518. doi: 10.1038/s41467-017-00531-6
70. Lynn MA, Eden G, Ryan FJ, Bensalem J, Wang X, Blake SJ, et al. The composition of the gut microbiota following early-life antibiotic exposure affects host health and longevity in later life. Cell Rep. (2021) 36:109564. doi: 10.1016/j.celrep.2021.109564
71. Chichlowski M, De Lartigue G, German BJ, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides. J Pediatr Gastroenerol Nutr. (2012) 55:321–7. doi: 10.1097/MPG.0b013e31824fb899.Bifidobacteria
72. Guo S, Al-Sadi R, Said HM, Ma TY. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am J Pathol. (2013) 182:375–87. doi: 10.1016/j.ajpath.2012.10.014
73. Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE, et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of claudin-2. J Immunol. (2017) 199:2976–84. doi: 10.4049/jimmunol.1700105
74. Nielsen DSG, Jensen BB, Theil PK, Nielsen TS, Knudsen KEB, Purup S. Effect of butyrate and fermentation products on epithelial integrity in a mucus-secreting human colon cell line. J Funct Foods. (2018) 40:9–17. doi: 10.1016/j.jff.2017.10.023
75. Mohebali N, Ekat K, Kreikemeyer B, Breitrück A. Barrier protection and recovery effects of gut commensal bacteria on differentiated intestinal epithelial cells in vitro. Nutrients. (2020) 12:1–23. doi: 10.3390/nu12082251
76. Price DB, Ackland ML, Burks W, Knight MI, Suphioglu C. Peanut allergens alter intestinal barrier permeability and tight junction localisation in caco-2 cell cultures. Cell Physiol Biochem. (2014) 33:1758–77. doi: 10.1159/000362956
77. Losol P, Sokolowska M, Hwang YK, Ogulur I, Mitamura Y, Yazici D, et al. Epithelial barrier theory: the role of exposome, microbiome, and barrier function in allergic diseases. Allergy Asthma Immunol Res. (2023) 15:705–24. doi: 10.4168/aair.2023.15.6.705
78. de Kivit S, Tobin MC, DeMeo MT, Fox S, Garssen J, Forsyth CB, et al. In vitro evaluation of intestinal epithelial TLR activation in preventing food allergic responses. Clin Immunol. (2014) 154:91–9. doi: 10.1016/j.clim.2014.07.002
79. López P, González-Rodríguez I, Sánchez B, Ruas-Madiedo P, Suárez A, Margolles A, et al. Interaction of Bifidobacterium bifidum LMG13195 with HT29 cells influences regulatory-T-cell-associated chemokine receptor expression. Appl Environ Microbiol. (2012) 78:2850–7. doi: 10.1128/AEM.07581-11
80. Kleniewska P, Kopa-Stojak PN, Hoffmann A, Pawliczak R. The potential immunomodulatory role of the gut microbiota in the pathogenesis of asthma: an in vitro study. Sci Rep. (2023) 13:1–8. doi: 10.1038/s41598-023-47003-0
81. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. (2021) 184:3884–98.e11. doi: 10.1016/j.cell.2021.05.030
82. Bozzetti V, Senger S. Organoids technologies for the study of intestinal microbiota-host interactions. Trends Mol Med. (2022) 28:290–303. doi: 10.1016/j.molmed.2022.02.001.Organoids
83. Gómez DP, Boudreau F. Organoids and their use in modeling gut epithelial cell lineage differentiation and barrier properties during intestinal diseases. Front Cell Dev Biol. (2021) 9:1–8. doi: 10.3389/fcell.2021.732137
Keywords: allergies, atopy & microbiome, gut microbiota, antibiotics – immune effect, animal models, cell culture models
Citation: Donald K and Finlay BB (2024) Experimental models of antibiotic exposure and atopic disease. Front. Allergy 5:1455438. doi: 10.3389/falgy.2024.1455438
Received: 26 June 2024; Accepted: 7 October 2024;
Published: 25 October 2024.
Edited by:
Petra Zimmermann, Université de Fribourg, SwitzerlandReviewed by:
Hontian Wang, Capital Medical University, ChinaMwenya Mubanga, Centre for Infectious Disease Research in Zambia, Zambia
Copyright: © 2024 Donald and Finlay. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. Brett Finlay, YmZpbmxheUBtc2wudWJjLmNh
 Katherine Donald
Katherine Donald B. Brett Finlay1,2,3*
B. Brett Finlay1,2,3*