- Center for Cancer Research, Medical University of Vienna, Vienna, Austria
The early microbial colonization of human mucosal surfaces is essential for the development of the host immune system. Already during pregnancy, the unborn child is prepared for the postnatal influx of commensals and pathogens via maternal antibodies, and after birth this protection is continued with antibodies in breast milk. During this critical window of time, which extends from pregnancy to the first year of life, each encounter with a microorganism can influence children's immune response and can have a lifelong impact on their life. For example, there are numerous links between the development of allergies and an altered gut microbiome. However, the exact mechanisms behind microbial influences, also extending to how viruses influence host-microbe interactions, are incompletely understood. In this review, we address the impact of infants’ first microbial encounters, how the immune system develops to interact with gut microbiota, and summarize how an altered immune response could be implied in allergies.
1 Introduction
The gut microbiome is composed of a diverse community comprising bacteria, archaea, eukaryotes, and viruses. These microorganisms maintain intricate relationships, not only among themselves but also with the human host, spanning from symbiotic to parasitic interactions, thereby exerting a profound influence on the host's immune system (1). This crucial interplay between the human immune system and these microbial communities begins early in life, extending influences into adulthood (2).
The preparation of the immature immune system for exposure to this vast number of microorganisms already begins in utero. Here, IgG antibodies, which are the only antibody class able to be transferred via the neonatal Fc receptor (FcRn), cross the placenta to the umbilical cord, reach systemic circulation in the infant (3). After birth, primarily IgA antibodies are transferred from the mother to the infant via breastmilk. These antibodies support the infant in keeping the balance between protection from pathogens by providing passive immunity and tolerance to non-threating, beneficial commensals (4). The human gut microbiota in early life is shaped by various factors, including the mode of delivery, feeding mode (breast milk vs. formula), antibiotics treatments, maternal health status prior to conception, maternal body mass index and diet, as well as the geographical region (Figure 1) (5–10). During the first years of life, the composition of the microbiome exhibits the highest levels of variability and host-microorganism interactions in prenatal and early postnatal life can have permanent effects on the immune development. The microbiome composition ultimately stabilizes into an adult-like configuration around the age of three (9, 11).
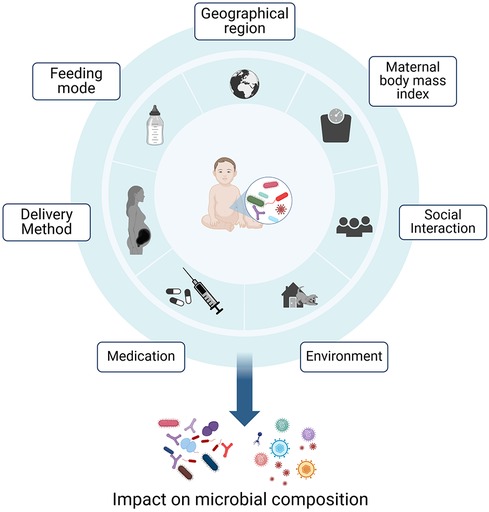
Figure 1 Early life factors influencing the composition of the gut microbiome. The microbial composition is influenced by feeding mode, medications, maternal-, geographical- and social factors. Host-microorganism interactions in prenatal and early postnatal life can have permanent effects on the immune development (5–7, 9, 10). Created with BioRender.com.
The most critical time period therein, is the switch from being breastfed to the introduction to solid food at approximately six months of age (12). This developmental period, known as the “window of opportunity”, in which the microbial diversity is settling, could be seen as a double-edged sword (13, 14). While it creates a fertile ground for microbial colonization, it also leaves infants more vulnerable to external factors that can disrupt the delicate balance of their microbiota. These external factors disturbing the microbiota include antibiotics and malnutrition, which can have long-lasting adverse effects on the function of the immune system (15, 16). Any dysbiosis could therefore result in the development of diseases including necrotizing enterocolitis, inflammatory bowel disease, obesity and allergy (17–21). Interestingly, also allergic diseases have been associated with microbial dysbiosis and over the last years, several studies have suggested an involvement of gut microbiota in the development of allergies and asthma (22–24). These effects have been associated with environmental factors, such as mode of delivery, breastfeeding and early exposure of antibiotics (25–29). Although our understanding of the exact mechanisms of how certain microbes provide protection is still limited, recent research suggests that antibodies play a role in shaping the microbiome and thus indirectly contribute to the development of allergies (22, 30, 31). In this review, we aim to summarize recent work on the development of the microbiota-immune axis in early life and its potential involvement in the onset of allergies in early life.
2 The microbiome in early life
2.1 Development of bacterial communities
The bacterial microbiome composition in early life has a high microbial diversity and strain heterogeneity, as well as a high turnover, as only about 11% of bacterial colonizers persist past the first year of life (32). Research suggests that strains of microorganisms contributing to the infant microbiome originate from a variety of maternal sources and include vaginal, dermal, oral and intestinal communities (9, 33, 34). Those microbial species are thereby mirroring the mode of delivery, which results in a noteworthy variance in microbial gut composition post-birth and has a distinct colonization pattern characterized by a limited number of species (6, 9, 35, 36). Nevertheless, the influence of the maternal microbiome decreases within a few days after birth (37). The microbial composition is declining dramatically within the first week after birth, followed by a recovery and gradually increases again over the following months (37). In vaginal deliveries, Lactobacillus is the predominant species in the infant's oral, cutaneous and intestinal environment, accompanied by Senathia and Prevotella, exhibiting a similar microbiota profile to the vaginal environment (38, 39). Conversely, the gut microbiome in neonates delivered by cesarean section (C-section) is associated with reduced microbial diversity, and mainly colonization by Staphylococcus, followed by Propionibacterium and Corynebacterium, resembling the microbial composition of maternal skin (6, 40). In one study, infants born vaginally shared 72% (135/187) of gut microbes with their mother, whereas those born by C-section only shared 41% (55/135) (9). Beyond maternal transfer of microbes, the newborn undergoes a gradual colonization by environmental microbes while also being introduced to potential pathogens (41). While the mode of delivery has a significant impact on the initial seeding of the gut microbiome, soon the feeding mode rapidly becomes more influential in shaping its composition. Breastfeeding promotes colonizing of beneficial microorganisms, but does not seem to fully compensate the deficiency of Bifidobacteria in infants delivered through C-section (42, 43). However, generalizing infants’ microbial compositions remains challenging due to the high variability across different cohorts, considering the differences in geographic and cultural settings (Table 1).
2.1.1 Longitudinal development of the gut microbiota and links to diseases
A longitudinal study by Stewart et al. (36) analyzing the bacterial gut microbiome of over 900 children from four high income countries (Germany, Finland, Sweden and the United States) from age three months to 46 months has shown, that the developing gut microbiome undergoes three different waves of development. The first wave is called developmental phase (month 3–14), followed by a transitional phase (month 15–30) and a stable phase (months 31–46). The birth mode has a significant association with the gut microbiota composition in the developmental phase, clearly indicated by the higher levels of Bacteroides spp. in infants born vaginally (36). Overall, the first gut colonizers are aerobic and facultative anaerobic bacteria (Enterobacter, Enterococcus and Escherichia) as well as Firmicutes (Streptococcus and Staphylococcus). As mentioned before, breastfeeding is the most significant factor influencing the gut microbiota composition and is associated with high levels of obligate anaerobe Actinobacteria such as Bifidobacterium species. In the developmental phase of the gut microbiome, the infants’ gut is colonized with primary Bifidobacteria which are responsible for human milk oligosaccharides (HMOs) catabolism and have genes involved in plant polysaccharide metabolism (44). These high levels of Bifidobacteria in this phase of development protect against allergy (45). Overall, an increased alpha diversity (describing the diversity of species within an individual), and a reduced beta diversity (defined as the diversity between different individuals), can be observed in the growing infant, with the microbiota becoming more complex over time (9, 11, 46, 47). Cessation of breastfeeding can be linked to a faster maturation of the gut microbiome, marked by the phyla of Firmicutes (Lachnospiraceae and Ruminococcaceae) and Bacteroidetes (Bacteroidaceae) (11, 36, 44). The introduction to solid food is marked by an increase in alpha diversity, mainly Actinobacteria, Bacteroides, Firmicutes, Proteobacteria and Verrucomicrobiota (36). The transitional phase (months 15–30) is characterized by a decrease of Proteobacteria and a strong increase in Bacteroidetes that proceeds to the stable phase (months 31–46), which, as the name would suggest, is characterized by unchanged alpha diversity and composition (36). These observations are in line with other metagenomic studies (9, 32, 46). Overall, the infant microbiome is subject to constant fluctuations and gradually approaches the adult microbiome at around 2–3 years of age (Figure 2) (48).
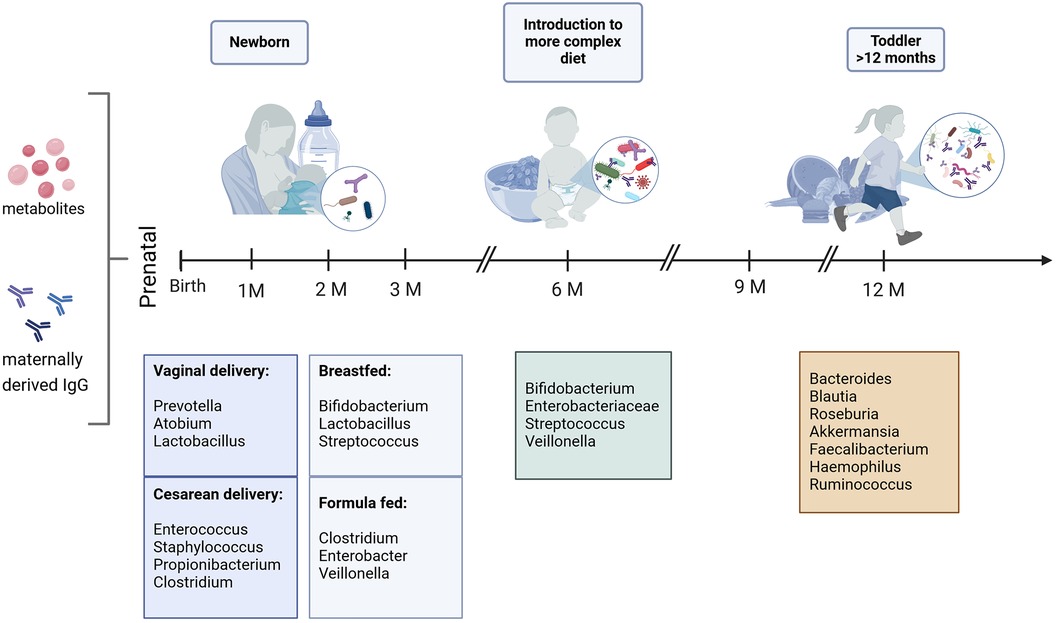
Figure 2 Predominant bacterial genera in the first year of life. Bacterial composition is subject to constant fluctuations, mostly influenced by delivery- and feeding mode in the first months of life. It gradually approaches the adult state starting with the introduction to solid food (9, 36, 46). Created with BioRender.com.
The composition of the bacterial gut microbiome in early life is linked to various diseases and, presumably, the starting point of immunological imprinting of allergies, highlighted by various studies with Bifidobacterium spp (22, 30). Isolation of Bifidobacterium breve and Bifidobacterium longum subsp. infantis from human feces stimulated T-regulatory (Treg) cell accumulation and thereby protected against allergy in mice. Moreover, B. longum subsp. infantis supplementation resulted in elevated levels of indole-3-lactic acid (ILA), which has the ability to dampen T-helper 17 (Th17) and T-helper 2 (Th2) cell responses (49). Furthermore, B. longum subsp. infantis not only has the ability to increase the number of Tregs, but has also been shown to have protective effects against asthma by mitigating the effect antibiotics in early life (45, 49). Furthermore, within the first year of life, Bifidobacterium breve colonization is linked to a reduced risk of atopic dermatitis, whereas Bifidobacterium catenulatum is associated with an increase (50). Interestingly, several studies observed that having a furry pet living in the household was associated with a lower relative abundance of the genus Bifidobacterium (36, 46, 51). Moreover, an immature microbial composition in the gut has been linked to an increased risk of asthma at age five, with lower abundances of the genera Firmicutes families like Lachnospiraceae and Ruminococcaceae (25, 52).
2.2 The virome in early life
In addition to bacteria, the human gut contains an enormous number of viruses, including eukaryotic viruses as well as bacteriophages (viruses infecting bacteria) (18, 19, 33). The complex interplay between the vast array of intestinal viruses (termed the “virome”) and the immunological maturation of children during their first years of life is an increasing area of research (18, 33, 53–57). Despite the overwhelming prevalence of viral entities, especially when compared to the bacteriome, our understanding of their role is limited. The maternal virome is stable in its composition, at least in the time from late pregnancy and after birth (33, 53). As more and more research focuses on influences of the intestinal virome in early life development, it has been observed that virus composition underlies strong fluctuations depending on various environmental factors such as age at gestation, older siblings, type of birth, feeding pattern, and geographical location or being born during summer (33, 58–61).
As soon as the infant is born, the varying impact of different factors becomes apparent. At birth the delivery mode has the highest impact on the predominance of the gut virome (infants born vaginally showed higher diversity compared to infants born by C-section). After one and three months after birth, the largest effect was the gestational age at birth (preterm vs. term) (57). Overall, this first phase of viral colonization is characterized by the induction of prophages from pioneering bacteria followed by the colonization of viruses infecting human cells, which is regulated by breastfeeding (55). Breastfed infants showed lower numbers of viral operational taxonomic units (vOTUs) based on bulk metagenomes, compared to partially or exclusively formula-fed infants. Formula-fed infants also show a higher alpha diversity in their virome (33, 57). The feeding mode had a comparable effect size to the delivery method at month three but overtakes in the following months in breastfed infants. The geographical region becomes relevant from month six onward, with influences manifesting through the introduction of solid food to the infants’ diet (57). While the transmission of eukaryotic viruses such as cytomegalovirus, herpes simplex and rubella virus is known, transmission of bacteriophages from mothers to their offspring has still rarely been systematically addressed (62).
Bacteriophages constitute the main source of viruses in the gut (63). They demonstrate remarkable diversity within the gut milieu, mirroring the prevailing composition of gut bacteria and being key players of the modulation of the bacterial gut microbiome (56). Following birth and extending through the first two years of life, bacteriophage composition undergoes significant changes and a rapid expansion linked to the increase of bacterial communities in the infants gut (54, 57). However, this expansion is followed by a notable reduction shortly thereafter, resembling an adult like state by two to three years of age, accompanied by a decrease in diversity being inversely correlated with the bacterial diversity (33, 64, 65). While the maternal virome predominantly comprises bacteriophages and exogenous viruses from the environment and diet, the neonatal counterpart is composed of a unique set of bacteriophages (53). Intriguingly, depending on the study cohort, only 15%–32.3% of this virome diversity is likely to be maternally derived, for instance during vaginal delivery and breastfeeding (7, 33, 58, 66). Some distinct bacteriophages, for example, those that infect Bifidobacteria, can be transmitted from mother to child (67). Nevertheless, environmental transmission seems to be the major route in transmission (53). They most likely originate from maternal skin, infectious exposures as well as contaminated surfaces (53, 58, 68). Overall, each infant's virome exhibits a distinct signature, although monozygotic twins manifest a higher interspecies similarity compared to unrelated infants (58, 65, 69).
Recent research showed, that the infant gut virome is dominated by a rich temperate phage community, which is able to integrate their genomes into the chromosomes of their bacterial host. Deficiencies in certain temperate phage families could increase the risk of asthma development by one year of age, independent of their bacterial hosts (70, 71). The constant changes in the composition of the gut virome during early life significantly shape the gut bacterial microbiome, a factor known to influence the development of allergies. Despite this potential link, our understanding of the specific role of the gut virome in the development of allergies is still limited. However, there are correlative hints. Leal Rodríguez et al. have reported, that an exposure to cats, not dogs, in early life was associated with an asthma virome fingerprint, along with a negative correlation with being born in summer and having older siblings (61).
3 Pre- and postnatal factors in early life immune development
3.1 Prenatal factors of immune development
During pregnancy, rejection of the semi-allogeneic fetus is avoided by an immune privileged status of the placental trophoblast, a vascular separation from the mother and various maternal tolerance mechanisms (72, 73). Thus, the success of human pregnancy depends on maintaining a subtle balance between two conflicting aspects of the immune system. The fetus must acquire the ability to accept both its own and maternal antigens, while building protective immunity in anticipation of birth (74, 75).
Therefore, the placenta plays an essential role in the development of the fetus. The existence of a placental microbiome still remains a debated topic within the scientific community and it is currently rather believed that the infant encounters the first microorganisms during delivery (76–78). In the womb, the fetus is assumed to live in a largely sterile environment and is protected from infection by the maternal immune system (79). Chorionic villi, forming at the end of the first trimester of pregnancy, create a maternal-fetal interface. These villous structures, resulting in a hemochorial placenta, allow maternal blood to directly come into contact with the fetus via its fetal derived placenta. These structures not only provide the developing child with nutrients and oxygen, but they also facilitate the transfer of antibodies from maternal blood flow across the syncytiotrophoblast layer of the chorion into the inner layer of cytotrophoblast precursor cells (80, 81). From there, antibodies can travel via the FcRn into the fetal capillaries providing a layer of defense against pathogens (82).
In addition, microbial antigens and metabolites are transferred across the placenta, and thereby able to prime the fetal immune system (83, 84). Transferred metabolites are currently getting attention for their role in atopy protection (85). They have been demonstrated to affect transcription of the target gene Foxp3 in the lung, which has been associated with asthma development (86). The fetal innate immune system is being prepared for the subsequent influx of microbes that will later colonize the infants’ intestine and thereby influence the leukocyte development (87–89).
3.2 Adaptive immune response in early life—how the mother prepares the unborn child for a life with microbes
During the final trimester of pregnancy, a significant increase in maternally transferred IgG levels can be observed (90). Maternal antibodies, spanning all subclasses of IgG, are then transported across the placenta to the developing fetus. This transfer is facilitated by the FcRn ensuring the efficient passage of antibodies from maternal circulation to the fetus (91–93). Although the FcRn binds to the CH3 domain of the Fc fragment of IgG antibodies, previous studies reported a hierarchical transfer of different IgG subclasses independent of their specificity in with IgG1 is preferentially transferred across the placenta (IgG1 > IgG4 > IgG3 > IgG2) (94, 95).
More recent studies suggest a preferential transfer depending on Fc-glycosylations to the fetus, and thereby the transfer of more functionally enhanced antibodies (96–99). Thus, glycosylations could mediate the binding of IgG to certain placental Fc receptors, including FcγRI, FcγRII, and FcγRIII, allowing a more efficient transfer (99, 100). One indication of this can be seen in premature infants, which, despite having an overall limited number of total maternal IgG, possess similar anti-viral and neutralizing antibodies mirroring a robust transfer of broadly reactive and functional relevant antibodies (101). This finding suggests that the transfer of the most functional antibodies occurs very early in pregnancy, although the exact mechanisms of selection are not yet known. However, it appears, that the placenta preferentially transfers antibodies eliciting innate immune effector functions activating natural killer cells in the earliest days of life after birth (98).
Recently, Dolatshahi et al. (82) conducted a longitudinal study investigating the humoral immune response against vaccine or pathogen derived antigens in 12 full-term (FT, gestational age 37–40 weeks) and 11 preterm infants (PT, gestational age 24–29 weeks). Among mothers of both groups, they observed significant heterogeneity in IgG, IgM, and IgA levels, as well as Fc receptor binding antibodies. IgG subclasses were detectable in cord blood but declined in the following weeks, consistent with expectations. Comparing mother-baby dyads, differences in transfer of antigen-specific antibodies from the mothers to the children across different gestational ages could be observed. Cord blood of FT babies showed an enrichment of certain antibody populations, with enhanced levels of total IgG against norovirus, the tetanus toxin, Streptococcus pneumoniae, poliovirus, hepatitis A, the mumps toxin and the allergens Ara h 2 (peanut allergen) and Bet v 1 (birch pollen allergen). The predominant allergen-specific antibodies in preterm babies, were specific against Bos d 8 (bovine milk allergen). Notably, one week after birth, the only significantly enriched antibodies were specific for cow milk in PT compared to FT, setting a potential link for antibodies as a surrogate of food allergy (82). Furthermore, these findings suggest the presence of antigen-specific differences in transfer rates across the umbilical cord, possibly selected not only by the Fc but in collaboration with the Fab domain (82, 102, 103). Although the antibody profile across infants began to normalize after three months, the antibody profiles of FT and PT infants still differed. Few but potentially important antibody specificities were observed in FT vs. PT infants. These were the persistence of S. pneumoniae-and peanut-specific antibodies, as well as antibodies against adenovirus, cytomegalovirus and polio. This observation may suggest an explanation for the enhanced susceptibility to infections in PT infants. Maternally derived antibodies decreased to very low levels by three months of age, with a more pronounced decline in PT infants. However, the decline in transferred functional antibodies was similar in FT and PT infants. The FcRn-binding antibodies against the bovine milk allergen increased slightly in both infant groups by that time, along with an increase in IgM and IgA1 concentrations (82). It can thereby be concluded that elevated risks of viral infections in premature infants cannot be attributed to the lack of maternal antibodies, but rather to the weaker mucosal barriers and contact to disease-associated environmental exposures, e.g., due to longer stays in the hospitals, especially in intensive care units (101). Despite an adjustment of the adaptive immune response comparing PT and FT, very PT (<31 weeks) seem to have a more elevated risk in asthma (approximately 3.6 times higher than FT) (104). Overall, any observed differences in the composition of the microbiome, disappear after six months when the infant is getting introduced to a more complex diet (9).
3.3 Lactation
3.3.1 Breastmilk
Breastmilk is a vital factor in shaping the composition of the gut microbiota and thereby influencing the development of the immune system in infants (36, 105). Milk produced during the first days after infant birth is called colostrum and is characterized by its notably higher protein content and reduced levels of carbohydrates and fat compared to more mature milk, indicating a primary immunological role rather than a nutritional one. After four to five days, the content ratios start to change, by lower concentrations of immunoglobulins and proteins, however, the overall composition remains similar. Human milk is characterized by the presence of essential components altering cellular differentiation and gene expression such as lactoferrin, and short-chain fatty acids, thereby directly influencing immune cells to a more tolerogenic-like response via transforming growth factor β (TGF-ß) and interleukin 10 (IL-10) (106, 107). Moreover, breastmilk contains leukocytes and vital immunoglobulins, including IgA and IgM, as well as IgG. Soluble IgA (sIgA) stands as the predominant immunoglobulin in human milk, constituting over 90% of the total amount. It is followed by IgM and IgG, whose concentrations increase in more mature milk (108–111). As reviewed by Rio-Aige et al. (111), concentrations of sIgA are in a wide range from around 7.5 g/L in colostrum and 1.6–2 g/L in more mature milk. It is generated by plasma cells that migrate from the mesenteric lymph nodes to the mammary glands during the later stages of pregnancy and throughout the lactation period. Common in all mucosal secretions, it has the ability to neutralize pathogens before they come in contact with epithelial cells, thus preventing inflammation and damage to tissues (112). IgA antibodies have been shown to anchor beneficial bacteria in the mucus layer of the intestine, thereby promoting the colonization of the child's gut with a diverse set of microbiota (113).
Nonetheless, human breastmilk is highly personalized as its composition varies highly between mothers. The microbial composition of breastmilk is reflected by differences dependent on the mode of breast milk feeding, nursing directly from the breast vs. using pumps or by bottle-feeding (114). The precise origins of milk microbiota are currently subject to debate, although evidence suggests that the entero-mammary pathway or retrograde translocation may serve as significant routes for microbial colonization (115).
The HMOs found in breast milk not only serve as an optimal source for bacteria, but also have other important functions. Some HMOs provide support in maintaining structure and function of mucosal gut tissues, as well as, together with sIgA, preventing necrotizing enterocolitis (NEC), an acute inflammatory bowel necrosis, affecting the colon in neonates, especially in PT children (116, 117). Bifidobacteria, the most extensively studied among HMO-fermenters, are closely linked to breastfeeding. These microorganisms have the unique capability to convert aromatic amino acids like tryptophan, tyrosine, and phenylalanine into their respective lactic acid derivatives using aromatic lactate dehydrogenase (ALDH). Indoleacetic acid derived from tryptophan has been demonstrated to activate the aryl hydrocarbon receptor (AhR), which plays a pivotal role in regulating intestinal homeostasis and immune responses. In addition, it drives the upregulation of immunoregulatory molecules in CD4+ T-cells, which reduces the differentiation of Th2 and Th17 cells (45, 118). Moreover, depletion of Bifidobacteria was shown to be a marker of systemic and intestinal inflammation and increased the risk of developing autoimmune diseases and atopic wheeze (45, 119, 120).
3.3.2 The introduction of solid food and the weaning reaction
The first major immune response to colonizing microbiota after birth begins at the time of weaning and the introduction of solid food (121). It is resulting in a notable augmentation in both the quantity and diversity of bacterial taxa in the gastrointestinal tract. Notably, murine studies have demonstrated that throughout the weaning period, when levels of the epidermal growth factor (EGF) in breastmilk start to decline, goblet cells exhibit an enhanced capacity for the translocation of antigens from the intestinal lumen into the lamina propria (122). In response to this influx of luminal antigens, the neonatal immune system orchestrates a robust production of cytokines and T-cells lean towards an immunoregulatory phenotype, thereby helping to establish long-term tolerance to commensal microorganisms (121, 122). Increased food diversity is negatively correlated with the development of asthma and food allergy up to year six. Furthermore, increased isotype switching to IgE and a reduced expression of Foxp3, which is associated with Treg expression and moreover associated with a low food diversity score (123). In addition, certain dietary vitamins appear to be of great importance in the prevention of pathological imprinting in early life. Dietary vitamin A-derived retinoic acid holds a protective effect by inducing RORγt+ Tregs during the weaning response, as well as riboflavin metabolites during the neonatal period by generating mucosal-associated invariant T (MAIT) cells (121, 124). This immunological response can be modulated through temporary antibiotic intervention or excessive caloric intake, as has been shown in mice. These disturbances lead to a higher susceptibility to inflammatory pathology characterized by high release of cytokines (121, 125).
4 Postnatal immunity
In the waves of fetal hematopoiesis, from the yolk sac (126, 127) (in humans at four weeks, in mice at embryonic day seven to nine) to the fetal liver (127, 128) (in humans at six weeks, in mice at embryonic day twelve) and to the bone marrow (128) (in humans at 10 weeks, in mice at embryonic day 7–15), specific immune cells develop that are important for early life tolerance against microbiota. Neonatal B and T-cells show more innate-like functions, as they have the ability to respond to antigens more quickly than the adult version. Unconventional B- and T- cell subsets have been shown to be particularly responsive by early life microbial exposure and metabolites, whereas the exposure to pathogens in early life can have a strong influences on the development and the functionality of the immune system (129).
4.1 Immune interactions by gut microbiota, infectious diseases and metabolic effects
Once the infant is getting colonized with microbiota, the adaptive and innate immune system must adjust to tolerate this intricate interaction. In order to recognize bacteria, the innate immune system uses various receptors, including pattern recognition receptors (PRR) like Toll like receptors (TLRs) and nucleotide-binding oligomerization domain/caspase recruitment domain (NOD/CARD) isoforms. These receptors are expressed by surface enterocytes and dendritic cells and are crucial for bacteria- host communication (130). Thus, PRRs can detect conserved microbe-associated molecular patterns (MAMPs), such as components of the bacterial cell wall, like lipopolysaccharides (LPS), peptidoglycan and flagellin. By binding TLRs, the microbiota can suppress inflammatory responses and promote immunological tolerance (131, 132). This sensor function of TLR's as well as downstream signaling molecules are fully developed in newborns (133). Following initial exposure to LPS shortly after birth, intestinal epithelial cells exhibit a reduced response to subsequent TLR stimulation, thereby favoring microbial colonization and maintaining host-microbe homeostasis (134). Following stimulation by TLRs, a spectrum of cytokines is synthesized that regulate both the adaptive and innate immune systems during ontogeny (135). In PT infants, the predominant cytokine profile is biased toward the production of anti-inflammatory mediators, particularly IL-10. In contrast, the predominant cytokine in term infants tends to promote T-helper 17 (Th17) cells, characterized by elevated levels of interleukin-6 (IL-6) and interleukin-23 (IL-23) (136–138). IL-12p70 is one of the final cytokines to reach adult levels following TLR stimulation as it is promoting the development of Th1 cell immune responses (138).
Regarding adaptive immunity, a reduced diversity in the gut microbiota has been linked to the development of allergy (139, 140). This reduction can be provoked by treatments with antibiotics, but the exact mechanisms are not yet known (15, 141). One possibility could be that microbes induce the T-helper 1 (Th1) pathway as well as Tregs, thereby counteracting the Th2 cell responses associated with allergy (142, 143). For example, endotoxin, produced by gut bacteria in early life is linked with Th1 maturation and prevents from Th2-mediated responses in a mouse model of asthma (31).
Exposure to pathogens in early life can have a great influence on the development and the functionality of the immune system. Certain infections can lead to life-long pathologies affecting all organs and influence the onset of various diseases (144). For instance, early life infection with Listeria monocytogenes can cause long term-organ specific alterations in both the innate and adaptive immune system (145). Similarly, some infections by enteroviruses contribute to the onset of autoimmune diseases, such as diabetes (146–148), while others, like Respiratory syncytial virus (RSV) (149, 150), Streptococcus pneumoniae (151) and Rhinovirus (152, 153) show a strong association with increased allergic airway diseases in adulthood.
In addition to the immune system, also metabolites play an important role in early life microbiota-host interaction. Alongside to the aforementioned HMOs (see previous section “Breastmilk”), SCFAs (short chain fatty acids) produced by gut microbiota (including acetate, propionate and butyrate), are key metabolites linked to gut colonization and immune maturation. SCFAs are produced by anaerobic bacteria that ferment complex carbohydrates originating from diet and colonic mucus (154). Reduced levels of SCFA-producing bacteria (such as Ruminococcus bromii and Faecalibacterium prausnitzii) have been shown to be associated with an increased risk of allergic diseases in infants, as they promote anti-inflammatory and tolerogenic immune responses (20, 155). Additionally, they are involved in Treg differentiation, as demonstrated in both murine (156–158) and human cells (159).
4.2 Environmental influences and the hygiene hypothesis
Several epidemiological studies have demonstrated a link between growing up in specific agricultural environments and being protected from allergies in childhood. This protective effect has been attributed to respiratory exposure to certain environmental microbes (160–163). The hygiene hypothesis, first postulated by Strachan (164) more than 30 years ago, gives an explanation for this connection. According to this hypothesis, a decreased frequency of infections directly contributes to the increase in allergic and autoimmune diseases (165). Repeated low grade acute immune responses triggered by infectious and even harmless microbes in early life are associated with lower prevalence of chronic inflammatory disorders in adulthood (160, 166). Prenatal exposure to household pets, especially dogs, has been shown to lower the risk for asthma and atopic dermatitis until 2 years of age (167). As it was shown by Panzer et al. (168), prenatal, as well as early life dog exposure is associated with an altered gut microbiome during infancy, supporting a potential link between dog-keeping and a decreased allergy risk.
Several farm derived bacteria have been reported to contribute to the protection against asthma, including Lactococcus lactis (169), Staphylococcus sciuri (170), Bacillus licheniformis (171) or Acinetobacter lwoffii (172–174). Acinetobacter lwoffii has been suggested to induce pro-inflammatory responses in airways by increasing IL-6 (172). Epigenetic modifications in CD4+ T cells then result in IL-10 induction. The combined activity of IL-6 and IL-10 influences the gastrointestinal microbiome, with specific taxa being significantly associated with either disease activity or protection (172).
Furthermore, certain biochemical modifications of the genetic information carrying chromatin, called epigenetic modifications, have been associated with the increase of allergic diseases. They are not changing the nucleotide sequence of the genome, but are best known for changing the accessibility of genes, thereby regulate the gene expression (14, 175). Those modifications can be induced by several extrinsic factors interacting with the genetic background (14). In particular, dietary components in breast milk and bovine milk, exposure to microbial components, house dust, as well as the production of SCFA by gut microbiota are associated with several epigenetic modifications in the gene expression (153, 176). Moreover, maternal exposures during pregnancy have been shown to influence the immune development in utero by epigenetic mechanisms and thereby affecting the onset of allergy (177–179). The differentiation of Th cell populations is strictly controlled by epigenetic mechanisms, which control the differentiation into the with allergy associated Th cell populations (14, 180). Allergy is specifically associated with changes in DNA methylation patterns in the Th2, Th1, Th17 and Treg subsets (180).
In the next chapters, we highlight lymphoid immune cells which contribute to an establishment of mucosal immune-microbiota homeostasis such as B-1 cells, RORγt+ Tregs, MAIT cells and invariant natural killer T (iNKT) cells (181).
4.3 B-cells in early life
Mature fetal B-cells develop in the fetal liver from post conception week nine and later in the bone marrow (127, 182). These B-cells achieve their repertoire in early stages, which is defined by somatic recombination of immunoglobulin genes and Ig heavy chain class-switching, although the formation of germinal centers and the accompanied somatic hypermutation starts by antigen exposure after birth (183–185). In early gestation, innate-like B-1 cells prevail, being the most abundant B-cell population in the peritoneal and pleural cavities. B-1 cells are thought to recognize surface epitopes of common pathogens and self-antigens and most importantly, are thought to represent the only B-cells in adult repertoires shaped by early life antigen exposures (186–189). Furthermore, B-1 cells have an pre-activated phenotype that is also conserved under germ free conditions, making them prepared for antibody secretion (190). B-1 cells can spontaneously differentiate into plasma cells and are believed to be the major source of secreted IgM in unchallenged mice (189). New et al. (186) showed, that neonatal immunization with group A Streptococcus recruits unique B-1 memory cells, which cannot be found when mice were immunized in adulthood. This result was confirmed by Vergani et al. (190), who showed that mice orally infected with a murine rotavirus as five-day-old-neonates- in contrast to those infected as adults- exclusively generated IgA plasma cells originating from early life origin (ELO)-B-cells nine weeks after infection. These plasma cells arose from the same hematopoietic progenitor cells as B-1a cells, suggesting that neonatal exposure to antigens uniquely primes immune responses later in life and ELO-B-cells harbor the memory of neonatal antigen exposure in the gut (190).
4.3.1 Mucosal IgA serves as the first line of defense against invaders
IgA serves as the first immune protection against invading pathogens and represents the predominant antibody isotype in mammals at mucosal surfaces, while IgG dominates systemically in blood. It is secreted across mucosal surfaces and the intestinal epithelium, especially in the small intestines. Polymeric IgA is transcytosed through epithelial cells from the basolateral surface by the polymeric immunoglobulin receptor (pIgR) (191). As early as 1996, it has been shown that a significant portion of bacteria found in feces is bound by IgA, showing a continuous presence of IgA antibodies in response to the persistent resident microbial population (192). This type of antibody coats and agglutinates microbiota and antigens coming from components of the lumen as well as toxins of the intestine to prevent direct interaction with the host (193, 194). Moreover, it has the ability to preferentially coat colitogenic bacteria, and thereby prevents inflammation and maintain intestinal health. Impairment of IgA secretion is associated with increased susceptibility to various diseases of the gut, e.g., enterocolitis (195). IgA assumes a crucial role in the regulation of bacterial gene transcription, ultimately influencing the composition, invasiveness, and immunometabolic functions of bacterial communities within the gut (196–198). Conversely, commensal bacteria have the capacity to stimulate the production of IgA antibodies (199). These IgA antibodies play a crucial role in boosting the humoral mucosal immune defense system. Notably, they are significantly decreased in germ-free (GF) animal models, which can be reversed by initiation of microbial colonization (200).
IgA can be produced via two distinct pathways. The first, the T-cell-dependent pathway, yields high-affinity antibodies that are predominantly directed against specific protein antigens, particularly from pathogens. These responses occur in germinal centers in gut-associated lymphoid tissues, e.g., Peyer's patches and mesenteric lymph nodes. The second, termed as the T-cell-independent pathway, operates primarily through specialized B-cells in the small intestine. These B-cells in both, organized lymphoid tissues and non-lymphoid tissues, are utilizing innate immune receptors, for example TLRs (201–203). The T-cell-independent pathway generates lower-affinity antibodies that recognize a wider variety of microbial antigens. IgAs yielded from both pathways are directed against commensals, but to different strains and species (204).
Infants initiate the production the of IgA between two and four weeks of age (205). However, it appears that the T-cell independent pathway holds greater significance in children until they develop an adult-like microbiome and establish germinal center-dependent IgA production (206). Moreover, experiments in mice have demonstrated that the production of neonatal IgA in pre-weaning immunocompetent pups is notably increased when they are fed with milk of immunodeficient mothers. Furthermore, enrichment of special maternal derived microbiota will induce early enhanced IgA production in the intestines, as it has been specifically observed with Limosilactobacillus reuteri (207). L.reuteri is known for its antimicrobial activity as it produces a variety of substances against gram-positive and gram-negative bacteria, fungi and parasites. This finding demonstrates that certain bacteria have the ability to influence immune responses against potentially hazardous microbiota in infants by both, direct and indirect ways (208). Beyond that, IgA has the ability to interact and thereby neutralize harmless food antigens by preventing their penetration of the gut epithelium (209). As such, it plays an important role in creating tolerance, thereby preventing allergic sensitization (210).
4.4 T-cells
Within the intestinal tract, T-cells play a key role in balancing the immune responses to commensal microbes by inhibiting inflammatory responses targeted against them while at the same time also preventing them to break mucosal barriers. It has been shown in mouse experiments that intestinal microorganisms are transported from the intestines to the thymus by CX3CR1+ dendritic cells, which present microbially-derived antigens to T-cells and thereby initiate their expansion (211). As fetal T-cells are hyperresponsive to foreign antigens, it is believed that those components are priming fetal memory T-cell differentiation (212, 213). Compared to adult T-cells, neonatal ones seem to be more sensitive upon antigen exposure, having distinct gene expression profiles and, being capable of shifting quickly from a pro-tolerant state to self- and non-self-antigens, while being able to mount rapid effector function in case of injury or infection (214, 215).
4.4.1 Regulatory T- cells
The timing of antigen exposure is important. In general, fetal and neonate T-cells tend towards a more tolerogenic phenotype with more innate-like cytokine production than proinflammatory responses (212). Fetus-derived CD4+ T-cells preferentially produce Th2 cytokines when stimulated with low amounts of antigen (216, 217). Antigenic encounters in the phase before weaning (the window of opportunity) have the ability to induce the differentiation of neonatal CD8+ T-cells to RORγt+ Tregs (218, 219). Upon re-exposure to the same antigen later in life, the infant is more likely to elicit a more tolerant immune reactivity (220). T-cell responses to gut commensal bacteria may be dominated by a relatively small number of microorganisms and the induction of colonic Tregs depends on various commensal bacteria with different properties, including Bacteroides fragilis and the Clostridium clusters IV and XIVa. B. fragilis has the ability to produce polysaccharide A, thereby inducing Treg cell development via the TLR 2 (132, 221, 222). Moreover, early life mouse models have shown that these particular clostridial species are able to induce Treg accumulation in the colon following oral inoculation by inducing the release of TGF-ß and other Treg inducing factors from intestinal epithelial cells (221, 222). Furthermore, these spore forming bacteria are able to protect from colitis and elevated systemic IgE levels in adult mice (221). In humans similar effects can be observed, as children who lack Tregs in early life develop severe inflammations of the skin and the intestines following microbial colonization (223). By the time of weaning, certain dietary components, such as the vitamin A-derived retinoic acid and SCFA are linked to gut colonization and subsequent immune maturation, as they skew Tregs towards expressing RORγt+ and are associated with the development of asthma in later life (Figure 3) (121).
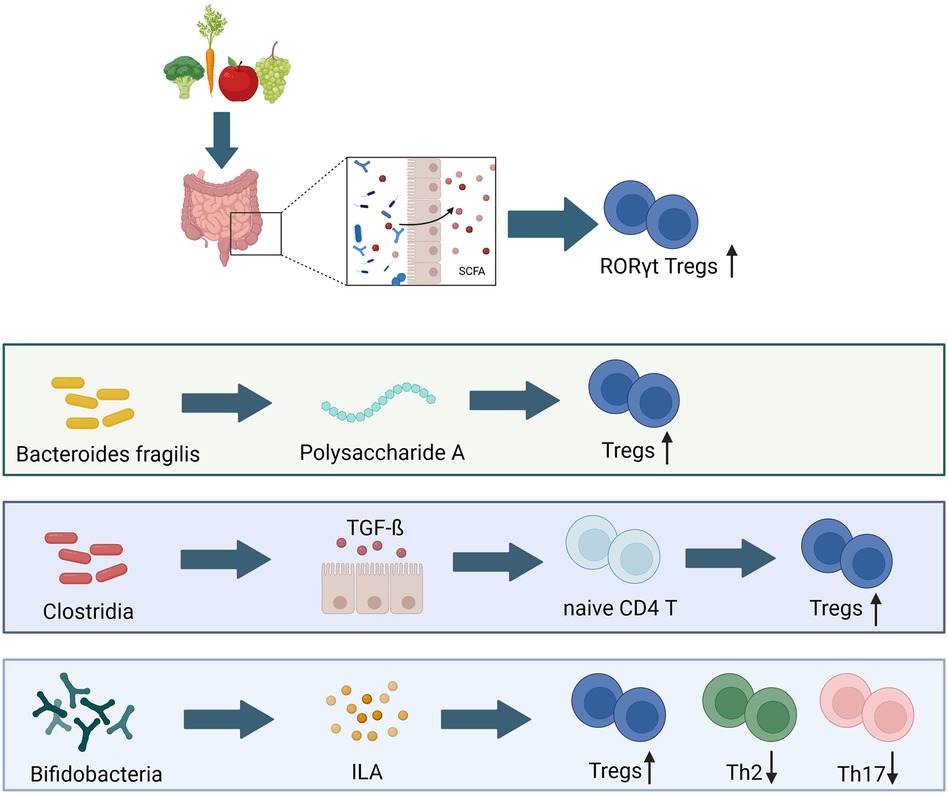
Figure 3 Bacterial composition has long lasting effects on early life immune development. Tregs, T regulatory cells; Th2, T-helper 2 cells; Th 17, T-helper 17 cells; SCFA, shorth chain fatty acids; TGF-ß, transforming growth factor β; ILA, indole-3-lactic acid (49, 122, 132, 221, 222). Created with BioRender.com.
4.4.2 Mucosal-associated invariant T (MAIT) cells and invariant natural killer T (iNKT) cells
The mucosal homeostasis is additionally maintained by the innate like MAIT cells and iNKT cells, both responding to glycolipids derived from early life microbial colonization. These distinct cells bridge the randomly generated T- and B-cell receptors within the adaptive immune system with the innate germline-encoded immune receptors (224). Moreover, they acquire tissue tropism, such as lung tropism, and the ability to release cytokines as part of their developmental process in the thymus before exiting the latter and accumulating in the tissue ahead of the arrival of conventional effector cells (225, 226).
iNKT cells are a rare subset of T-cells, having the ability to recognize self and microbial lipid antigens presented by CD1d molecules (227). They are important in influencing the outcomes of infectious and autoimmune diseases, as well as in neoplastic disorders and have been implicated in several mouse models of allergic asthma (228–231). In the first weeks of life, they migrate from the thymus to the colon and lung. Their development and residency is controlled by embryonic macrophages during a specific early life window (232, 233). The colonization by microbiota can prevent this iNKT cell migration (230). iNKT cells were shown to be the dominant CD4+ T-cell subset in airways of both, non-allergic and allergic patients with severe asthma, while not being present in the healthy population (233). An increased number of iNKT cells have been shown to being associated with asthma, possibly due to similar functions as Th2 cells (234).
MAIT cells can recognize small microbial molecules as riboflavin (vitamin B2 precursor derivates (5-(2-oxopropylideneamino)-6-d-ribitylaminouracil (5-OP-RU)) presented by major histocompatibility complex class 1b (MHC-Ib) molecule MR1 (235). As the human body cannot synthesize vitamin B2, it serves as a marker for “non-self” and has been implicated in various diseases together with bacterial dysbiosis (236–238). MAIT cells respond to various strains of bacteria and yeasts, but not to viruses (239, 240). MHC-Ib molecules have the capacity to present antigens characterized by specific amino acid sequences or chemical motifs originating from a wide range of microbiota. This highlights the potential of MAIT cells as ideal candidates for regulating communication between the microbiota and the immune system in early life (241). MAIT cells are enriched in the mucosal tissues in the intestinal tract and in the lungs, as well as in the blood and in the periphery (242–245). Although a number of immature MAIT cells can be detected in GF mice, those were not able to fully mature and expand in the periphery. Microbial colonization (and peripheral B-cells) seem to be required for the maturation and expansion, but not for the initial selection of MAIT cells (225, 246). Increased levels of MAIT cells in one-year-old children were associated with a reduced risk of asthma by year seven and a potent Th1 immune response (247).
5 Concluding remarks on the interplay between the microbiota and allergy
The rising prevalence of allergies has been striking over the last few decades. Several theories have been proposed to give an explanation, one of which points to higher rates of caesarean birth, formula feeding, misuse of antibiotics and general dietary changes as contributing factors. All these factors can be linked to a dysbiosis of the gut microbiome (15, 248–250).
The first potential links between allergy and microbiota appear already during pregnancy, as maternal carriage of Prevotella copri is associated with a decreased risk of food allergy in infants. The association is related to maternal diet, which is high in fat and fiber, the absence of antibiotics and an increased house hold size (251). Additionally, caesarean birth increases the risk of developing asthma at the age of six if the microbiota still has a C-section signature by the age of one year. However, children whose microbiota showed a non-C-section signature by one year of age, appeared to be comparable to those who were born vaginally (252). Furthermore, in the first one to three months after birth, an absence of certain bacterial taxa can be linked to an increased risk of atopy and is associated with the absence of polyunsaturated fatty acids (253, 254).
Antibiotics are one of the most commonly prescribed drugs given to children in the Western world (255). Even short administrations can cause microbial dysbiosis in the gut, which, when administered in early life, may lead to long term immunological consequences (256, 257). For instance, Vancomycin administration in early life murine models induces increased IgE levels, reduced Tregs and an overall increased risk of developing allergic asthma (258).
Legumes, such as soy, peanuts and sesame, show a significant effect on the microbiota at 12 months of age, suggesting that potential allergenic sources may contribute to allergy protective effects (259, 260). Furthermore, children with IgE mediated food hypersensitivity have a significant reduced gut microbiota diversity and richness compared to healthy children. This dysbiosis is characterized by high abundances of the phylum Firmicutes and low abundance of Bacteroidetes within a cohort of children between the age of 18–36 months. Moreover, certain enrichments of the bacterial families Clostridiaceae, Ruminococcaceae, Lachnospiraceae, or Erysipelotrichaceae were associated with milk, egg white and peanut hypersensitivities in those children (261). Taken together, there is substantial evidence that gut microbiota in early life influence the development of the immune system and dysbiosis may be involved in the onset of allergies. Future studies, both in human cohorts as well as mouse models, may shed light on the underlying mechanisms and help to establish causation.
Author contributions
A-LP: Writing – original draft, Writing – review & editing. TV: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
Funded by the European Union (ERC STG, EarlyMicroAbs, project number 101075733 to TV). Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, et al. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. (2009) 7(1):61–71. doi: 10.1038/nrmicro2047
2. Martino C, Dilmore AH, Burcham ZM, Metcalf JL, Jeste D, Knight R. Microbiota succession throughout life from the cradle to the grave. Nat Rev Microbiol. (2022) 20(12):707–20. doi: 10.1038/s41579-022-00768-z
3. Roopenian DC, Akilesh S. FcRn: the neonatal fc receptor comes of age. Nat Rev Immunol. (2007) 7(9):715–25. doi: 10.1038/nri2155
4. Brodin P. Immune-microbe interactions early in life: a determinant of health and disease long term. Science. (2022) 376(6596):945–50. doi: 10.1126/science.abk2189
5. Arnoldini M, Cremer J, Hwa T. Bacterial growth, flow, and mixing shape human gut microbiota density and composition. Gut Microbes. (2018) 9(6):559–66. doi: 10.1080/19490976.2018.1448741
6. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. (2010) 107(26):11971–5. doi: 10.1073/pnas.1002601107
7. Lim ES, Wang D, Holtz LR. The bacterial microbiome and virome milestones of infant development. Trends Microbiol. (2016) 24(10):801–10. doi: 10.1016/j.tim.2016.06.001
8. Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun. (2014) 5(1):3889. doi: 10.1038/ncomms4889
9. Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. (2015) 17(5):690–703. doi: 10.1016/j.chom.2015.04.004
10. Stephenson J, Heslehurst N, Hall J, Schoenaker DAJM, Hutchinson J, Cade JE, et al. Before the beginning: nutrition and lifestyle in the preconception period and its importance for future health. Lancet. (2018) 391(10132):1830–41. doi: 10.1016/S0140-6736(18)30311-8
11. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. (2012) 486(7402):222–7. doi: 10.1038/nature11053
12. Sordillo JE, Zhou Y, McGeachie MJ, Ziniti J, Lange N, Laranjo N, et al. Factors influencing the infant gut microbiome at age 3–6 months: findings from the ethnically diverse vitamin D antenatal asthma reduction trial (VDAART). J Allergy Clin Immunol. (2017) 139(2):482. doi: 10.1016/j.jaci.2016.08.045
13. Hornef MW, Torow N. ‘Layered immunity’ and the ‘neonatal window of opportunity’—timed succession of non-redundant phases to establish mucosal host–microbial homeostasis after birth. Immunology. (2020) 159(1):15–25. doi: 10.1111/imm.13149
14. Acevedo N, Alashkar Alhamwe B, Caraballo L, Ding M, Ferrante A, Garn H, et al. Perinatal and early-life nutrition, epigenetics, and allergy. Nutrients. (2021) 13(3):724. doi: 10.3390/nu13030724
15. Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. (2012) 13(5):440–7. doi: 10.1038/embor.2012.32
16. Fragkou PC, Karaviti D, Zemlin M, Skevaki C. Impact of early life nutrition on children’s immune system and noncommunicable diseases through its effects on the bacterial microbiome, virome and mycobiome. Front Immunol. (2021) 12:644269. doi: 10.3389/fimmu.2021.644269
17. Gensollen T, Iyer SS, Kasper DL, Blumberg RS. How colonization by microbiota in early life shapes the immune system. Science. (2016) 352(6285):539–44. doi: 10.1126/science.aad9378
18. Kaelin EA, Rodriguez C, Hall-Moore C, Hoffmann JA, Linneman LA, Ndao IM, et al. Longitudinal gut virome analysis identifies specific viral signatures that precede necrotizing enterocolitis onset in preterm infants. Nat Microbiol. (2022) 7(5):653–62. doi: 10.1038/s41564-022-01096-x
19. Liang G, Gao H, Bushman FD. The pediatric virome in health and disease. Cell Host Microbe. (2022) 30(5):639–49. doi: 10.1016/j.chom.2022.04.006
20. Boutin RCT, Sbihi H, Dsouza M, Malhotra R, Petersen C, Dai D, et al. Mining the infant gut microbiota for therapeutic targets against atopic disease. Allergy. (2020) 75(8):2065–8. doi: 10.1111/all.14244
21. Qiu P, Ishimoto T, Fu L, Zhang J, Zhang Z, Liu Y. The gut microbiota in inflammatory bowel disease. Front Cell Infect Microbiol. (2022) 12:733992. doi: 10.3389/fcimb.2022.733992
22. Feehley T, Plunkett CH, Bao R, Hong SMC, Culleen E, Belda-Ferre P, et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat Med. (2019) 25(3):448–53. doi: 10.1038/s41591-018-0324-z
23. Stephen-Victor E, Crestani E, Chatila TA. Dietary and microbial determinants in food allergy. Immunity. (2020) 53(2):277–89. doi: 10.1016/j.immuni.2020.07.025
24. Depner M, Taft DH, Kirjavainen PV, Kalanetra KM, Karvonen AM, Peschel S, et al. Maturation of the gut microbiome during the first year of life contributes to the protective farm effect on childhood asthma. Nat Med. (2020) 26(11):1766–75. doi: 10.1038/s41591-020-1095-x
25. Stokholm J, Blaser MJ, Thorsen J, Rasmussen MA, Waage J, Vinding RK, et al. Maturation of the gut microbiome and risk of asthma in childhood. Nat Commun. (2018) 9(1):141. doi: 10.1038/s41467-017-02573-2
26. Zheng W, Zhao W, Wu M, Song X, Caro F, Sun X, et al. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature. (2020) 577(7791):543–8. doi: 10.1038/s41586-019-1898-4
27. Uzan-Yulzari A, Turta O, Belogolovski A, Ziv O, Kunz C, Perschbacher S, et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun. (2021) 12(1):443. doi: 10.1038/s41467-020-20495-4
28. Li X, Stokholm J, Brejnrod A, Vestergaard GA, Russel J, Trivedi U, et al. The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host Microbe. (2021) 29(6):975–987.e4. doi: 10.1016/j.chom.2021.03.017
29. Atyeo C, Alter G. The multifaceted roles of breast milk antibodies. Cell. (2021) 184(6):1486–99. doi: 10.1016/j.cell.2021.02.031
30. Renz H, Skevaki C. Early life microbial exposures and allergy risks: opportunities for prevention. Nat Rev Immunol. (2021) 21(3):177–91. doi: 10.1038/s41577-020-00420-y
31. Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. (2015) 349(6252):1106–10. doi: 10.1126/science.aac6623
32. Lou YC, Olm MR, Diamond S, Crits-Christoph A, Firek BA, Baker R, et al. Infant gut strain persistence is associated with maternal origin, phylogeny, and traits including surface adhesion and iron acquisition. Cell Rep Med. (2021) 2(9):100393. doi: 10.1016/j.xcrm.2021.100393
33. Garmaeva S, Sinha T, Gulyaeva A, Kuzub N, Spreckels JE, Andreu-Sánchez S, et al. Transmission and dynamics of mother-infant gut viruses during pregnancy and early life. Nat Commun. (2024) 15:1945. doi: 10.1038/s41467-024-45257-4
34. Zhang C, Li L, Jin B, Xu X, Zuo X, Li Y, et al. The effects of delivery mode on the gut microbiota and health: state of art. Front Microbiol. (2021) 12:724449. doi: 10.3389/fmicb.2021.724449
35. Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. (2014) 80(9):2889–900. doi: 10.1128/AEM.00342-14
36. Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. (2018) 562(7728):583–8. doi: 10.1038/s41586-018-0617-x
37. Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-Infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. (2018) 24(1):133–145.e5. doi: 10.1016/j.chom.2018.06.005
38. Tamburini S, Shen N, Wu HC, Clemente JC. The microbiome in early life: implications for health outcomes. Nat Med. (2016) 22(7):713–22. doi: 10.1038/nm.4142
39. Yao Y, Cai X, Ye Y, Wang F, Chen F, Zheng C. The role of microbiota in infant health: from early life to adulthood. Front Immunol. (2021) 12:708472. doi: 10.3389/fimmu.2021.708472
40. Montoya-Williams D, Lemas DJ, Spiryda L, Patel K, Carney OO, Neu J, et al. The neonatal microbiome and its partial role in mediating the association between birth by cesarean section and adverse pediatric outcomes. Neonatology. (2018) 114(2):103–11. doi: 10.1159/000487102
41. Fulde M, Hornef MW. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol Rev. (2014) 260(1):21–34. doi: 10.1111/imr.12190
42. Akagawa S, Tsuji S, Onuma C, Akagawa Y, Yamaguchi T, Yamagishi M, et al. Effect of delivery mode and nutrition on gut Microbiota in neonates. Ann Nutr Metab. (2019) 74(2):132–9. doi: 10.1159/000496427
43. Reyman M, van Houten MA, van Baarle D, Bosch AATM, Man WH, Chu MLJN, et al. Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat Commun. (2019) 10:4997. doi: 10.1038/s41467-019-13014-7
44. Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. (2011) 108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107
45. Henrick BM, Rodriguez L, Lakshmikanth T, Pou C, Henckel E, Arzoomand A, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. (2021) 184(15):3884–3898.e11. doi: 10.1016/j.cell.2021.05.030
46. Wernroth ML, Peura S, Hedman AM, Hetty S, Vicenzi S, Kennedy B, et al. Development of gut microbiota during the first 2 years of life. Sci Rep. (2022) 12(1):9080. doi: 10.1038/s41598-022-13009-3
47. Kers JG, Saccenti E. The power of microbiome studies: some considerations on which alpha and Beta metrics to use and how to report results. Front Microbiol. (2022) 12:796025. doi: 10.3389/fmicb.2021.796025
48. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut Microbiota on human health: an integrative view. Cell. (2012) 148(6):1258–70. doi: 10.1016/j.cell.2012.01.035
49. Lyons A, O’Mahony D, O’Brien F, MacSharry J, Sheil B, Ceddia M, et al. Bacterial strain-specific induction of Foxp3+ T regulatory cells is protective in murine allergy models. Clin Exp Allergy. (2010) 40(5):811–9. doi: 10.1111/j.1365-2222.2009.03437.x
50. Ismail IH, Boyle RJ, Licciardi PV, Oppedisano F, Lahtinen S, Robins-Browne RM, et al. Early gut colonization by Bifidobacterium breve and B. catenulatum differentially modulates eczema risk in children at high risk of developing allergic disease. Pediatr Allergy Immunol. (2016) 27(8):838–46. doi: 10.1111/pai.12646
51. Tun HM, Konya T, Takaro TK, Brook JR, Chari R, Field CJ, et al. Exposure to household furry pets influences the gut microbiota of infant at 3–4 Months following various birth scenarios. Microbiome. (2017) 5:40. doi: 10.1186/s40168-016-0226-6
52. Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. (2009) 11(10):2574–84. doi: 10.1111/j.1462-2920.2009.01982.x
53. Walters WA, Granados AC, Ley C, Federman S, Stryke D, Santos Y, et al. Longitudinal comparison of the developing gut virome in infants and their mothers. Cell Host Microbe. (2023) 31(2):187–198.e3. doi: 10.1016/j.chom.2023.01.003
54. Taboada B, Morán P, Serrano-Vázquez A, Iša P, Rojas-Velázquez L, Pérez-Juárez H, et al. The gut virome of healthy children during the first year of life is diverse and dynamic. PLoS ONE. (2021) 16(4):e0240958. doi: 10.1371/journal.pone.0240958
55. Liang G, Zhao C, Zhang H, Mattei L, Sherrill-Mix S, Bittinger K, et al. Step-wise assembly of the neonatal virome modulated by breastfeeding. Nature. (2020) 581(7809):470–4. doi: 10.1038/s41586-020-2192-1
56. Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, Bry L, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe. (2019) 25(6):803–814.e5. doi: 10.1016/j.chom.2019.05.001
57. Zeng S, Almeida A, Li S, Ying J, Wang H, Qu Y, et al. A metagenomic catalog of the early-life human gut virome. Nat Commun. (2024) 15(1):1864. doi: 10.1038/s41467-024-45793-z
58. Maqsood R, Rodgers R, Rodriguez C, Handley SA, Ndao IM, Tarr PI, et al. Discordant transmission of bacteria and viruses from mothers to babies at birth. Microbiome. (2019) 7:156. doi: 10.1186/s40168-019-0766-7
59. Jansen D, Matthijnssens J. The emerging role of the gut virome in health and inflammatory bowel disease: challenges, covariates and a viral imbalance. Viruses. (2023) 15(1):173. doi: 10.3390/v15010173
60. Kennedy EA, Holtz LR. Gut virome in early life: origins and implications. Curr Opin Virol. (2022) 55:101233. doi: 10.1016/j.coviro.2022.101233
61. Rodríguez CL, Shah SA, Rasmussen MA, Thorsen J, Boulund U, Pedersen CET, et al. The infant gut virome is associated with preschool asthma risk independently of bacteria. Nat Med. (2024) 30(1):138–48. doi: 10.1038/s41591-023-02685-x
62. Megli CJ, Coyne CB. Infections at the maternal–fetal interface: an overview of pathogenesis and defence. Nat Rev Microbiol. (2022) 20(2):67–82. doi: 10.1038/s41579-021-00610-y
63. Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. (2021) 19(8):514–27. doi: 10.1038/s41579-021-00536-5
64. Gregory AC, Zablocki O, Zayed AA, Howell A, Bolduc B, Sullivan MB. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell Host Microbe. (2020) 28(5):724–740.e8. doi: 10.1016/j.chom.2020.08.003
65. Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. (2015) 21(10):1228–34. doi: 10.1038/nm.3950
66. Pannaraj PS, Ly M, Cerini C, Saavedra M, Aldrovandi GM, Saboory AA, et al. Shared and distinct features of human milk and infant stool viromes. Front Microbiol. (2018) 9:1162. doi: 10.3389/fmicb.2018.01162
67. Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. (2017) 5(1):66. doi: 10.1186/s40168-017-0282-6
68. Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. (2008) 159(5):367–73. doi: 10.1016/j.resmic.2008.04.006
69. Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A. (2015) 112(38):11941–6. doi: 10.1073/pnas.1514285112
70. Gummalla VS, Zhang Y, Liao YT, Wu VCH. The role of temperate phages in bacterial pathogenicity. Microorganisms. (2023) 11(3):541. doi: 10.3390/microorganisms11030541
71. Korpela K, Hurley S, Ford SA, Franklin R, Byrne S, Lunjani N, et al. Beyond bacteria: early-life gut virome link with childhood asthma development. Nat Med. (2024) 30(1):43–4. doi: 10.1038/s41591-023-02747-0
72. Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. (2012) 150(1):29–38. doi: 10.1016/j.cell.2012.05.031
73. Nancy P, Tagliani E, Tay CS, Asp P, Levy DE, Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. (2012) 336(6086):1317–21. doi: 10.1126/science.1220030
74. Rackaityte E, Halkias J. Mechanisms of fetal T cell tolerance and immune regulation. Front Immunol. (2020) 11:588. doi: 10.3389/fimmu.2020.00588
75. Frascoli M, Coniglio L, Witt R, Jeanty C, Fleck-Derderian S, Myers DE, et al. Alloreactive fetal T cells promote uterine contractility in preterm labor via IFN-γ and TNF-α. Sci Transl Med. (2018) 10(438):eaan2263. doi: 10.1126/scitranslmed.aan2263
76. Kennedy KM, de Goffau MC, Perez-Muñoz ME, Arrieta MC, Bäckhed F, Bork P, et al. Questioning the fetal microbiome illustrates pitfalls of low-biomass microbial studies. Nature. (2023) 613(7945):639–49. doi: 10.1038/s41586-022-05546-8
77. Rackaityte E, Halkias J, Fukui EM, Mendoza VF, Hayzelden C, Crawford ED, et al. Viable bacterial colonization is highly limited in the human intestine in utero. Nat Med. (2020) 26(4):599–607. doi: 10.1038/s41591-020-0761-3
78. de Goffau MC, Lager S, Sovio U, Gaccioli F, Cook E, Peacock SJ, et al. Human placenta has no microbiome but can contain potential pathogens. Nature. (2019) 572(7769):329–34. doi: 10.1038/s41586-019-1451-5
79. Koren O, Konnikova L, Brodin P, Mysorekar IU, Collado MC. The maternal gut microbiome in pregnancy: implications for the developing immune system. Nat Rev Gastroenterol Hepatol. (2024) 21(1):35–45. doi: 10.1038/s41575-023-00864-2
80. Knöfler M, Haider S, Saleh L, Pollheimer J, Gamage TKJB, James J. Human placenta and trophoblast development: key molecular mechanisms and model systems. Cell Mol Life Sci. (2019) 76(18):3479–96. doi: 10.1007/s00018-019-03104-6
81. Burton GJ, Charnock-Jones DS, Jauniaux E. Regulation of vascular growth and function in the human placenta. Reproduction. (2009) 138(6):895–902. doi: 10.1530/REP-09-0092
82. Dolatshahi S, Butler AL, Pou C, Henckel E, Bernhardsson AK, Gustafsson A, et al. Selective transfer of maternal antibodies in preterm and fullterm children. Sci Rep. (2022) 12(1):14937. doi: 10.1038/s41598-022-18973-4
83. Husso A, Pessa-Morikawa T, Koistinen VM, Kärkkäinen O, Kwon HN, Lahti L, et al. Impacts of maternal microbiota and microbial metabolites on fetal intestine, brain, and placenta. BMC Biol. (2023) 21(1):207. doi: 10.1186/s12915-023-01709-9
84. van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. (2014) 508(7494):123–7. doi: 10.1038/nature13158
85. Kimura I, Miyamoto J, Ohue-Kitano R, Watanabe K, Yamada T, Onuki M, et al. Maternal gut microbiota in pregnancy influences offspring metabolic phenotype in mice. Science. (2020) 367(6481):eaaw8429. doi: 10.1126/science.aaw8429
86. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. (2015) 6:7320. doi: 10.1038/ncomms8320
87. Macpherson AJ, de Agüero MG, Ganal-Vonarburg SC. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol. (2017) 17(8):508–17. doi: 10.1038/nri.2017.58
88. Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. (1999) 69(5):1035S–45. doi: 10.1093/ajcn/69.5.1035s
89. de Agüero MG, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al. The maternal microbiota drives early postnatal innate immune development. Science. (2016) 351(6279):1296–302. doi: 10.1126/science.aad2571
90. Malek A, Sager R, Schneider H. Maternal—fetal transport of immunoglobulin G and its subclasses during the third trimester of human pregnancy. Am J Reprod Immunol. (1994) 32(1):8–14. doi: 10.1111/j.1600-0897.1994.tb00873.x
91. Simister NE. Placental transport of immunoglobulin G. Vaccine. (2003) 21(24):3365–9. doi: 10.1016/S0264-410X(03)00334-7
92. Simister NE, Rees AR. Isolation and characterization of an fc receptor from neonatal rat small intestine. Eur J Immunol. (1985) 15(7):733–8. doi: 10.1002/eji.1830150718
93. Wilcox CR, Holder B, Jones CE. Factors affecting the FcRn-mediated transplacental transfer of antibodies and implications for vaccination in pregnancy. Front Immunol. (2017) 8:1294. doi: 10.3389/fimmu.2017.01294
94. Pereira RA, de Almeida VO, Vidori L, Colvero MO, Amantéa SL. Immunoglobulin G and subclasses placental transfer in fetuses and preterm newborns: a systematic review. J Perinatol. (2023) 43(1):3–9. doi: 10.1038/s41372-022-01528-w
95. Vidarsson G, Dekkers G, Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front Immunol. (2014) 5:520. doi: 10.3389/fimmu.2014.00520
96. Fu C, Lu L, Wu H, Shaman J, Cao Y, Fang F, et al. Placental antibody transfer efficiency and maternal levels: specific for measles, coxsackievirus A16, enterovirus 71, poliomyelitis I–III and HIV-1 antibodies. Sci Rep. (2016) 6:38874. doi: 10.1038/srep38874
97. Palmeira P, Quinello C, Silveira-Lessa AL, Zago CA, Carneiro-Sampaio M. IgG placental transfer in healthy and pathological pregnancies. J Immunol Res. (2011) 2012:e985646. doi: 10.1155/2012/985646
98. Jennewein MF, Goldfarb I, Dolatshahi S, Cosgrove C, Noelette FJ, Krykbaeva M, et al. Fc glycan-mediated regulation of placental antibody transfer. Cell. (2019) 178(1):202–215.e14. doi: 10.1016/j.cell.2019.05.044
99. Martinez DR, Fong Y, Li SH, Yang F, Jennewein MF, Weiner JA, et al. Fc characteristics mediate selective placental transfer of IgG in HIV-infected women. Cell. (2019) 178(1):190–201.e11. doi: 10.1016/j.cell.2019.05.046
100. Mahan AE, Jennewein MF, Suscovich T, Dionne K, Tedesco J, Chung AW, et al. Antigen-specific antibody glycosylation is regulated via vaccination. PLOS Pathog. (2016) 12(3):e1005456. doi: 10.1371/journal.ppat.1005456
101. Pou C, Nkulikiyimfura D, Henckel E, Olin A, Lakshmikanth T, Mikes J, et al. The repertoire of maternal anti-viral antibodies in human newborns. Nat Med. (2019) 25(4):591–6. doi: 10.1038/s41591-019-0392-8
102. Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. (2018) 18(1):46–61. doi: 10.1038/nri.2017.106
103. Dugast AS, Stamatatos L, Tonelli A, Suscovich TJ, Licht AF, Mikell I, et al. Independent evolution of fc- and fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur J Immunol. (2014) 44(10):2925–37. doi: 10.1002/eji.201344305
104. Trønnes H, Wilcox AJ, Lie RT, Markestad T, Moster D. The association of preterm birth with severe asthma and atopic dermatitis: a national cohort study. Pediatr Allergy Immunol. (2013) 24(8):782–7. doi: 10.1111/pai.12170
105. Hanson LA. Breastfeeding provides passive and likely long-lasting active immunity. Ann Allergy Asthma Immunol. (1998) 81(6):523–33. quiz 533–4, 537. doi: 10.1016/S1081-1206(10)62704-4
106. Gridneva Z, Lai CT, Rea A, Tie WJ, Ward LC, Murray K, et al. Human milk immunomodulatory proteins are related to development of infant body composition during the first year of lactation. Pediatr Res. (2021) 89(4):911–21. doi: 10.1038/s41390-020-0961-z
107. Dawod B, Marshall JS. Cytokines and soluble receptors in breast milk as enhancers of oral tolerance development. Front Immunol. (2019) 10:16. doi: 10.3389/fimmu.2019.00016
108. Ballard O, Morrow AL. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. (2013) 60(1):49–74. doi: 10.1016/j.pcl.2012.10.002
109. Sriraman NK. The nuts and bolts of breastfeeding: anatomy and physiology of lactation. Curr Probl Pediatr Adolesc Health Care. (2017) 47(12):305–10. doi: 10.1016/j.cppeds.2017.10.001
110. Goldsmith SJ, Dickson JS, Barnhart HM, Toledo RT, Eiten-Miller RR. IgA, IgG, IgM and lactoferrin contents of human milk during early lactation and the effect of processing and storage. J Food Prot. (1983) 46(1):4–7. doi: 10.4315/0362-028X-46.1.4
111. Rio-Aige K, Azagra-Boronat I, Castell M, Selma-Royo M, Collado MC, Rodríguez-Lagunas MJ, et al. The breast milk immunoglobulinome. Nutrients. (2021) 13(6):1810. doi: 10.3390/nu13061810
112. Brandtzaeg P. Secretory IgA: designed for anti-microbial defense. Front Immunol. (2013) 4:222. doi: 10.3389/fimmu.2013.00222
113. Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. (2018) 360(6390):795–800. doi: 10.1126/science.aaq0926
114. Moossavi S, Azad MB. Origins of human milk microbiota: new evidence and arising questions. Gut Microbes. (2020) 12(1):1667722. doi: 10.1080/19490976.2019.1667722
115. Fernández L, Langa S, Martín V, Maldonado A, Jiménez E, Martín R, et al. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res. (2013) 69(1):1–10. doi: 10.1016/j.phrs.2012.09.001
116. Rich BS, Dolgin SE. Necrotizing enterocolitis. Pediatr Rev. (2017) 38(12):552–9. doi: 10.1542/pir.2017-0002
117. Jantscher-Krenn E, Zherebtsov M, Nissan C, Goth K, Guner YS, Naidu N, et al. The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut. (2012) 61(10):1417–25. doi: 10.1136/gutjnl-2011-301404
118. Laursen MF, Sakanaka M, von Burg N, Mörbe U, Andersen D, Moll JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. (2021) 6(11):1367–82. doi: 10.1038/s41564-021-00970-4
119. Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. (2016) 165(4):842–53. doi: 10.1016/j.cell.2016.04.007
120. Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. (2018) 142(2):424–434.e10. doi: 10.1016/j.jaci.2017.08.041
121. Al Nabhani Z, Dulauroy S, Marques R, Cousu C, Al Bounny S, Déjardin F, et al. A weaning reaction to Microbiota is required for resistance to immunopathologies in the adult. Immunity. (2019) 50(5):1276–1288.e5. doi: 10.1016/j.immuni.2019.02.014
122. Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol. (2015) 8(1):198–210. doi: 10.1038/mi.2014.58
123. Roduit C, Frei R, Depner M, Schaub B, Loss G, Genuneit J, et al. Increased food diversity in the first year of life is inversely associated with allergic diseases. J Allergy Clin Immunol. (2014) 133(4):1056–1064.e7. doi: 10.1016/j.jaci.2013.12.1044
124. Constantinides MG, Link VM, Tamoutounour S, Wong AC, Perez-Chaparro PJ, Han SJ, et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. (2019) 366(6464):eaax6624. doi: 10.1126/science.aax6624
125. Al Nabhani Z, Dulauroy S, Lécuyer E, Polomack B, Campagne P, Berard M, et al. Excess calorie intake early in life increases susceptibility to colitis in adulthood. Nat Metab. (2019) 1(11):1101–9. doi: 10.1038/s42255-019-0129-5
126. Stremmel C, Schuchert R, Wagner F, Thaler R, Weinberger T, Pick R, et al. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat Commun. (2018) 9(1):75. doi: 10.1038/s41467-017-02492-2
127. Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, et al. Decoding human fetal liver haematopoiesis. Nature. (2019) 574(7778):365–71. doi: 10.1038/s41586-019-1652-y
128. Park JE, Jardine L, Gottgens B, Teichmann SA, Haniffa M. Prenatal development of human immunity. Science. (2020) 368(6491):600–3. doi: 10.1126/science.aaz9330
129. Constantinides MG, Belkaid Y. Early-life imprinting of unconventional T cells and tissue homeostasis. Science. (2021) 374(6573):eabf0095. doi: 10.1126/science.abf0095
130. Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. J Immunol Baltim Md 1950. (2005) 174(8):4453–60. doi: 10.4049/jimmunol.174.8.4453
131. Cario E. Bacterial interactions with cells of the intestinal mucosa: toll-like receptors and NOD2. Gut. (2005) 54(8):1182–93. doi: 10.1136/gut.2004.062794
132. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. (2011) 332(6032):974–7. doi: 10.1126/science.1206095
133. Strunk T, Currie A, Richmond P, Simmer K, Burgner D. Innate immunity in human newborn infants: prematurity means more than immaturity. J Matern Fetal Neonatal Med. (2011) 24(1):25–31. doi: 10.3109/14767058.2010.482605
134. Lotz M, Gütle D, Walther S, Ménard S, Bogdan C, Hornef MW. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. J Exp Med. (2006) 203(4):973–84. doi: 10.1084/jem.20050625
135. Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. (2011) 470(7335):543–7. doi: 10.1038/nature09737
136. Kollmann TR, Crabtree J, Rein-Weston A, Blimkie D, Thommai F, Wang XY, et al. Neonatal innate TLR-mediated responses are distinct from those of adults. J Immunol Baltim Md 1950. (2009) 183(11):7150–60. doi: 10.4049/jimmunol.0901481
137. Nguyen M, Leuridan E, Zhang T, De Wit D, Willems F, Van Damme P, et al. Acquisition of adult-like TLR4 and TLR9 responses during the first year of life. PLoS One. (2010) 5(4):e10407. doi: 10.1371/journal.pone.0010407
138. Corbett NP, Blimkie D, Ho KC, Cai B, Sutherland DP, Kallos A, et al. Ontogeny of toll-like receptor mediated cytokine responses of human blood mononuclear cells. PLoS ONE. (2010) 5(11):e15041. doi: 10.1371/journal.pone.0015041
139. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. (2014) 44(6):842–50. doi: 10.1111/cea.12253
140. Reddel S, Del Chierico F, Quagliariello A, Giancristoforo S, Vernocchi P, Russo A, et al. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep. (2019) 9(1):4996. doi: 10.1038/s41598-019-41149-6
141. Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. (2015) 45(3):632–43. doi: 10.1111/cea.12487
142. Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. (2011) 69(6):465–72. doi: 10.1203/PDR.0b013e318217638a
143. Penders J, Stobberingh EE, van den Brandt PA, Thijs C. The role of the intestinal microbiota in the development of atopic disorders. Allergy. (2007) 62(11):1223–36. doi: 10.1111/j.1398-9995.2007.01462.x
144. Henneke P, Kierdorf K, Hall LJ, Sperandio M, Hornef M. Perinatal development of innate immune topology. eLife. (2021) 10:e67793. doi: 10.7554/eLife.67793
145. Zou M, Yang J, Wiechers C, Huehn J. Acute neonatal Listeria monocytogenes infection causes long-term, organ-specific changes in immune cell subset composition. Eur J Microbiol Immunol. (2020) 10(2):98–106. doi: 10.1556/1886.2020.00007
146. Nekoua MP, Mercier A, Alhazmi A, Sane F, Alidjinou EK, Hober D. Fighting enteroviral infections to prevent type 1 diabetes. Microorganisms. (2022) 10(4):768. doi: 10.3390/microorganisms10040768
147. Tracy S, Drescher KM, Jackson JD, Kim K, Kono K. Enteroviruses, type 1 diabetes and hygiene: a complex relationship. Rev Med Virol. (2010) 20(2):106–16. doi: 10.1002/rmv.639
148. Lloyd RE, Tamhankar M, Lernmark Å. Enteroviruses and type 1 diabetes: multiple mechanisms and factors? Annu Rev Med. (2022) 73:483–99. doi: 10.1146/annurev-med-042320-015952
149. Ruiz S, Calvo C, Pozo F, Casas I, García-García ML. Lung function, allergic sensitization and asthma in school-aged children after viral-coinfection bronchiolitis. Sci Rep. (2022) 12(1):7552. doi: 10.1038/s41598-022-11356-9
150. Malinczak CA, Fonseca W, Rasky AJ, Ptaschinski C, Morris S, Ziegler SF, et al. Sex-associated TSLP-induced immune alterations following early-life RSV infection leads to enhanced allergic disease. Mucosal Immunol. (2019) 12(4):969–79. doi: 10.1038/s41385-019-0171-3
151. Peng D, Shi Y, Pang J, Cui L, Xu Y, Meng H, et al. Early-life infection of the airways with Streptococcus pneumoniae exacerbates HDM-induced asthma in a murine model. Cell Immunol. (2022) 376:104536. doi: 10.1016/j.cellimm.2022.104536
152. Rajput C, Han M, Ishikawa T, Lei J, Jazaeri S, Bentley JK, et al. Early life heterologous rhinovirus infections induce an exaggerated asthma-like phenotype. J Allergy Clin Immunol. (2020) 146(3):571–582.e3. doi: 10.1016/j.jaci.2020.03.039
153. Yang Z, Mitländer H, Vuorinen T, Finotto S. Mechanism of rhinovirus immunity and asthma. Front Immunol. (2021) 12:731846. doi: 10.3389/fimmu.2021.731846
154. Pascale A, Marchesi N, Marelli C, Coppola A, Luzi L, Govoni S, et al. Microbiota and metabolic diseases. Endocrine. (2018) 61(3):357–71. doi: 10.1007/s12020-018-1605-5
155. Patrick DM, Sbihi H, Dai DLY, Al Mamun A, Rasali D, Rose C, et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: evidence from population-based and prospective cohort studies. Lancet Respir Med. (2020) 8(11):1094–105. doi: 10.1016/S2213-2600(20)30052-7
156. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504(7480):451–5. doi: 10.1038/nature12726
157. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. (2013) 341(6145):569–73. doi: 10.1126/science.1241165
158. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504(7480):446–50. doi: 10.1038/nature12721
159. Hu M, Alashkar Alhamwe B, Santner-Nanan B, Miethe S, Harb H, Renz H, et al. Short-chain fatty acids augment differentiation and function of human induced regulatory T cells. Int J Mol Sci. (2022) 23(10):5740. doi: 10.3390/ijms23105740
160. Garn H, Potaczek DP, Pfefferle PI. The hygiene hypothesis and new perspectives-current challenges meeting an old postulate. Front Immunol. (2021) 12:637087. doi: 10.3389/fimmu.2021.637087
161. von Mutius E, Smits HH. Primary prevention of asthma: from risk and protective factors to targeted strategies for prevention. Lancet Lond Engl. (2020) 396(10254):854–66. doi: 10.1016/S0140-6736(20)31861-4
162. von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. (2010) 10(12):861–8. doi: 10.1038/nri2871
163. von Mutius E. The “hygiene hypothesis” and the lessons learnt from farm studies. Front Immunol. (2021) 12:635522. doi: 10.3389/fimmu.2021.635522
164. Strachan DP. Hay fever, hygiene, and household size. Br Med J. (1989) 299(6710):1259–60. doi: 10.1136/bmj.299.6710.1259
165. Pfefferle PI, Postigo I, Garn H. Editorial: the immunological implications of the hygiene hypothesis. Front Immunol. (2021) 12:732127. doi: 10.3389/fimmu.2021.732127
166. McDade TW. Early environments and the ecology of inflammation. Proc Natl Acad Sci U S A. (2012) 109(Suppl 2):17281–8. doi: 10.1073/pnas.1202244109
167. Lodge CJ, Allen KJ, Lowe AJ, Hill DJ, Hosking CS, Abramson MJ, et al. Perinatal cat and dog exposure and the risk of asthma and allergy in the urban environment: a systematic review of longitudinal studies. Clin Dev Immunol. (2012) 2012:176484. doi: 10.1155/2012/176484
168. Panzer AR, Sitarik AR, Fadrosh D, Havstad SL, Jones K, Davidson B, et al. The impact of prenatal dog keeping on infant gut microbiota development. Clin Exp Allergy. (2023) 53(8):833–45. doi: 10.1111/cea.14303
169. Debarry J, Garn H, Hanuszkiewicz A, Dickgreber N, Blümer N, von Mutius E, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. (2007) 119(6):1514–21. doi: 10.1016/j.jaci.2007.03.023
170. Hagner S, Harb H, Zhao M, Stein K, Holst O, Ege MJ, et al. Farm-derived gram-positive bacterium Staphylococcus sciuri W620 prevents asthma phenotype in HDM- and OVA-exposed mice. Allergy. (2013) 68(3):322–9. doi: 10.1111/all.12094
171. Vogel K, Blümer N, Korthals M, Mittelstädt J, Garn H, Ege M, et al. Animal shed Bacillus licheniformis spores possess allergy-protective as well as inflammatory properties. J Allergy Clin Immunol. (2008) 122(2):307–12. 312.e1–8. doi: 10.1016/j.jaci.2008.05.016
172. Alhamwe BA, Gao Z, Alhamdan F, Harb H, Pichene M, Garnier A, et al. Intranasal administration of Acinetobacter lwoffii in a murine model of asthma induces IL-6-mediated protection associated with caecal microbiota changes. Allergy. (2023) 78(5):1245–57. doi: 10.1111/all.15606
173. Kang H, Bang JY, Mo Y, Shin JW, Bae B, Cho SH, et al. Effect of Acinetobacter lwoffii on the modulation of macrophage activation and asthmatic inflammation. Clin Exp Allergy. (2022) 52(4):518–29. doi: 10.1111/cea.14077
174. Brand S, Teich R, Dicke T, Harb H, Yildirim AÖ, Tost J, et al. Epigenetic regulation in murine offspring as a novel mechanism for transmaternal asthma protection induced by microbes. J Allergy Clin Immunol. (2011) 128(3):618–625.e7. doi: 10.1016/j.jaci.2011.04.035
175. Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. (2018) 14:39. doi: 10.1186/s13223-018-0259-4
176. van Esch BCAM, Porbahaie M, Abbring S, Garssen J, Potaczek DP, Savelkoul HFJ, et al. The impact of milk and its components on epigenetic programming of immune function in early life and beyond: implications for allergy and asthma. Front Immunol. (2020) 11:2141. doi: 10.3389/fimmu.2020.02141
177. Schaub B, Liu J, Höppler S, Schleich I, Huehn J, Olek S, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. (2009) 123(4):774–782.e5. doi: 10.1016/j.jaci.2009.01.056
178. Zuccarello D, Sorrentino U, Brasson V, Marin L, Piccolo C, Capalbo A, et al. Epigenetics of pregnancy: looking beyond the DNA code. J Assist Reprod Genet. (2022) 39(4):801–16. doi: 10.1007/s10815-022-02451-x
179. Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. (2009) 180(5):462–7. doi: 10.1164/rccm.200901-0135OC
180. Potaczek DP, Harb H, Michel S, Alhamwe BA, Renz H, Tost J. Epigenetics and allergy: from basic mechanisms to clinical applications. Epigenomics. (2017) 9(4):539–71. doi: 10.2217/epi-2016-0162
181. Jain N. The early life education of the immune system: moms, microbes and (missed) opportunities. Gut Microbes. (2020) 12(1):1824564. doi: 10.1080/19490976.2020.1824564
182. Nuñez C, Nishimoto N, Gartland GL, Billips LG, Burrows PD, Kubagawa H, et al. B cells are generated throughout life in humans. J Immunol. (1996) 156(2):866–72. doi: 10.4049/jimmunol.156.2.866
183. Rechavi E, Lev A, Lee YN, Simon AJ, Yinon Y, Lipitz S, et al. Timely and spatially regulated maturation of B and T cell repertoire during human fetal development. Sci Transl Med. (2015) 7(276):276ra25. doi: 10.1126/scitranslmed.aaa0072
184. Weller S, Mamani-Matsuda M, Picard C, Cordier C, Lecoeuche D, Gauthier F, et al. Somatic diversification in the absence of antigen-driven responses is the hallmark of the IgM+IgD+CD27+ B cell repertoire in infants. J Exp Med. (2008) 205(6):1331–42. doi: 10.1084/jem.20071555
185. Tonegawa S. Somatic generation of antibody diversity. Nature. (1983) 302(5909):575–81. doi: 10.1038/302575a0
186. New JS, Dizon BLP, Fucile CF, Rosenberg AF, Kearney JF, King RG. Neonatal exposure to commensal-bacteria-derived antigens directs polysaccharide-specific B-1 B cell repertoire development. Immunity. (2020) 53(1):172–186.e6. doi: 10.1016/j.immuni.2020.06.006
187. Graf R, Seagal J, Otipoby KL, Lam KP, Ayoub S, Zhang B, et al. BCR-dependent lineage plasticity in mature B cells. Science. (2019) 363(6428):748–53. doi: 10.1126/science.aau8475
188. Kroese FGM, Butcher EC, Stall AM, Lalor PA, Adams S, Herzenberg LA. Many of the IgA producing plasma cells in murine gut are derived from self-replenishing precursors in the peritoneal cavity a. Int Immunol. (1989) 1(1):75–84. doi: 10.1093/intimm/1.1.75
189. Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. (2011) 11(1):34–46. doi: 10.1038/nri2901
190. Vergani S, Muleta KG, Da Silva C, Doyle A, Kristiansen TA, Sodini S, et al. A self-sustaining layer of early-life-origin B cells drives steady-state IgA responses in the adult gut. Immunity. (2022) 55(10):1829–1842.e6. doi: 10.1016/j.immuni.2022.08.018
191. Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. (2005) 206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x
192. van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. (1996) 38(3):348–54. doi: 10.1136/gut.38.3.348
193. Roche AM, Richard AL, Rahkola JT, Janoff EN, Weiser JN. Antibody blocks acquisition of bacterial colonization through agglutination. Mucosal Immunol. (2015) 8(1):176–85. doi: 10.1038/mi.2014.55
194. Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol. (2012) 12(12):821–32. doi: 10.1038/nri3322
195. Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. (2014) 158(5):1000–10. doi: 10.1016/j.cell.2014.08.006
196. Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, et al. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. (2018) 10(439):eaan1217. doi: 10.1126/scitranslmed.aan1217
197. Reikvam DH, Derrien M, Islam R, Erofeev A, Grcic V, Sandvik A, et al. Epithelial-microbial crosstalk in polymeric ig receptor deficient mice. Eur J Immunol. (2012) 42(11):2959–70. doi: 10.1002/eji.201242543
198. Macpherson AJ, Yilmaz B, Limenitakis JP, Ganal-Vonarburg SC. IgA function in relation to the intestinal Microbiota. Annu Rev Immunol. (2018) 36:359–81. doi: 10.1146/annurev-immunol-042617-053238
199. Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. (2020) 13(1):12–21. doi: 10.1038/s41385-019-0227-4
200. Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. (2010) 328(5986):1705–9. doi: 10.1126/science.1188454
201. Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature. (2001) 413(6856):639–43. doi: 10.1038/35098100
202. Tsuji M, Suzuki K, Kitamura H, Maruya M, Kinoshita K, Ivanov II, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. (2008) 29(2):261–71. doi: 10.1016/j.immuni.2008.05.014
203. Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu Rev Immunol. (2010) 28:243–73. doi: 10.1146/annurev-immunol-030409-101314
204. Bunker JJ, Flynn TM, Koval JC, Shaw DG, Meisel M, McDonald BD, et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. (2015) 43(3):541–53. doi: 10.1016/j.immuni.2015.08.007
205. Relman DA. Maternal IgA: matchmaking in early childhood. Immunity. (2019) 51(2):211–3. doi: 10.1016/j.immuni.2019.07.010
206. Planer JD, Peng Y, Kau AL, Blanton LV, Ndao IM, Tarr PI, et al. Development of the gut microbiota and mucosal IgA responses in twins and gnotobiotic mice. Nature. (2016) 534(7606):263–6. doi: 10.1038/nature17940
207. Mu Q, Swartwout BK, Edwards M, Zhu J, Lee G, Eden K, et al. Regulation of neonatal IgA production by the maternal microbiota. Proc Natl Acad Sci. (2021) 118(9):e2015691118. doi: 10.1073/pnas.2015691118
208. Chung TC, Axelsson L, Lindgren SE, Dobrogosz WJ. In vitro studies on reuterin synthesis by Lactobacillus reuteri. Microb Ecol Health Dis. (1989) 2(2):137–44. doi: 10.3109/08910608909140211
209. Corthésy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol. (2013) 4:185. doi: 10.3389/fimmu.2013.00185
210. Brandtzaeg P. The secretory immunoglobulin system: regulation and biological significance. Focusing on human mammary glands. Adv Exp Med Biol. (2002) 503:1–16. doi: 10.1007/978-1-4615-0559-4_1
211. Zegarra-Ruiz DF, Kim DV, Norwood K, Kim M, Wu WJH, Saldana-Morales FB, et al. Thymic development of gut-microbiota-specific T cells. Nature. (2021) 594(7863):413–7. doi: 10.1038/s41586-021-03531-1
212. Mold JE, Michaëlsson J, Burt TD, Muench MO, Beckerman KP, Busch MP, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science. (2008) 322(5907):1562–5. doi: 10.1126/science.1164511
213. Michaëlsson J, Mold JE, McCune JM, Nixon DF. Regulation of T cell responses in the developing human fetus. J Immunol Baltim Md 1950. (2006) 176(10):5741–8. doi: 10.4049/jimmunol.176.10.5741
214. Wissink EM, Smith NL, Spektor R, Rudd BD, Grimson A. MicroRNAs and their targets are differentially regulated in adult and neonatal mouse CD8+ T cells. Genetics. (2015) 201(3):1017–30. doi: 10.1534/genetics.115.179176
215. Palin AC, Ramachandran V, Acharya S, Lewis DB. Human neonatal naive CD4+ T cells have enhanced activation-dependent signaling regulated by the microRNA miR-181a. J Immunol Baltim Md 1950. (2013) 190(6):2682–91. doi: 10.4049/jimmunol.1202534
216. Webster RB, Rodriguez Y, Klimecki WT, Vercelli D. The human IL-13 locus in neonatal CD4+ T cells is refractory to the acquisition of a repressive chromatin architecture. J Biol Chem. (2007) 282(1):700–9. doi: 10.1074/jbc.M609501200
217. Hebel K, Weinert S, Kuropka B, Knolle J, Kosak B, Jorch G, et al. CD4+ T cells from human neonates and infants are poised spontaneously to run a nonclassical IL-4 program. J Immunol Baltim Md 1950. (2014) 192(11):5160–70. doi: 10.4049/jimmunol.1302539
218. Reibke R, Garbi N, Ganss R, Hämmerling GJ, Arnold B, Oelert T. CD8 + regulatory T cells generated by neonatal recognition of peripheral self-antigen. Proc Natl Acad Sci U S A. (2006) 103(41):15142–7. doi: 10.1073/pnas.0602622103
219. Zhao X, Liang W, Wang Y, Yi R, Luo L, Wang W, et al. Ontogeny of RORγt+ cells in the intestine of newborns and its role in the development of experimental necrotizing enterocolitis. Cell Biosci. (2022) 12(1):3. doi: 10.1186/s13578-021-00739-6
220. Knoop KA, Gustafsson JK, McDonald KG, Kulkarni DH, Coughlin PE, McCrate S, et al. Microbial antigen encounter during a preweaning interval is critical for tolerance to gut bacteria. Sci Immunol. (2017) 2(18):eaao1314. doi: 10.1126/sciimmunol.aao1314
221. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium Species. Science. (2011) 331(6015):337–41. doi: 10.1126/science.1198469
222. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of clostridia strains from the human microbiota. Nature. (2013) 500(7461):232–6. doi: 10.1038/nature12331
223. Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. (2001) 27(1):18–20. doi: 10.1038/83707
224. Toubal A, Nel I, Lotersztajn S, Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. (2019) 19(10):643–57. doi: 10.1038/s41577-019-0191-y
225. Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, et al. Stepwise development of MAIT cells in mouse and human. PLoS Biol. (2009) 7(3):e1000054. doi: 10.1371/journal.pbio.1000054
226. Chandra S, Kronenberg M. Activation and function of iNKT and MAIT cells. Adv Immunol. (2015) 127:145–201. doi: 10.1016/bs.ai.2015.03.003
227. Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB. The burgeoning family of unconventional T cells. Nat Immunol. (2015) 16(11):1114–23. doi: 10.1038/ni.3298
228. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. (2002) 17(5):629–38. doi: 10.1016/S1074-7613(02)00453-3
229. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336(6080):489–93. doi: 10.1126/science.1219328
230. An D, Oh SF, Olszak T, Neves JF, Avci F, Erturk-Hasdemir D, et al. Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell. (2014) 156(0):123–33. doi: 10.1016/j.cell.2013.11.042
231. Wingender G, Rogers P, Batzer G, Lee MS, Bai D, Pei B, et al. Invariant NKT cells are required for airway inflammation induced by environmental antigens. J Exp Med. (2011) 208(6):1151–62. doi: 10.1084/jem.20102229
232. Gensollen T, Lin X, Zhang T, Pyzik M, See P, Glickman JN, et al. Embryonic macrophages function during early life to determine iNKT cell levels at barrier surfaces. Nat Immunol. (2021) 22(6):699–710. doi: 10.1038/s41590-021-00934-0
233. Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, et al. CD4+ invariant T-cell–receptor+ natural killer T cells in bronchial asthma. N Engl J Med. (2006) 354(11):1117–29. doi: 10.1056/NEJMoa053614
234. Matangkasombut P, Pichavant M, Dekruyff RH, Umetsu DT. Natural killer T cells and the regulation of asthma. Mucosal Immunol. (2009) 2(5):383–92. doi: 10.1038/mi.2009.96
235. Franciszkiewicz K, Salou M, Legoux F, Zhou Q, Cui Y, Bessoles S, et al. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunol Rev. (2016) 272(1):120–38. doi: 10.1111/imr.12423
236. Averianova LA, Balabanova LA, Son OM, Podvolotskaya AB, Tekutyeva LA. Production of vitamin B2 (riboflavin) by microorganisms: an overview. Front Bioeng Biotechnol. (2020) 8:570828. doi: 10.3389/fbioe.2020.570828
237. Serriari NE, Eoche M, Lamotte L, Lion J, Fumery M, Marcelo P, et al. Innate mucosal-associated invariant T (MAIT) cells are activated in inflammatory bowel diseases. Clin Exp Immunol. (2014) 176(2):266–74. doi: 10.1111/cei.12277
238. Rouxel O, Da silva J, Beaudoin L, Nel I, Tard C, Cagninacci L, et al. Cytotoxic and regulatory roles of mucosal-associated invariant T cells in type 1 diabetes. Nat Immunol. (2017) 18(12):1321–31. doi: 10.1038/ni.3854
239. Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat Immunol. (2010) 11(8):701–8. doi: 10.1038/ni.1890
240. Gold MC, Cerri S, Smyk-Pearson S, Cansler ME, Vogt TM, Delepine J, et al. Human mucosal associated invariant T cells detect bacterially infected cells. PLoS Biol. (2010) 8(6):e1000407. doi: 10.1371/journal.pbio.1000407
241. Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the Microbiota. Annu Rev Immunol. (2021) 39:449–79. doi: 10.1146/annurev-immunol-093019-112348
242. Gibbs A, Leeansyah E, Introini A, Paquin-Proulx D, Hasselrot K, Andersson E, et al. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. (2017) 10(1):35–45. doi: 10.1038/mi.2016.30
243. Hinks TSC, Wallington JC, Williams AP, Djukanović R, Staples KJ, Wilkinson TMA. Steroid-induced deficiency of mucosal-associated invariant T cells in the chronic obstructive pulmonary disease lung. Implications for nontypeable Haemophilus influenzae infection. Am J Respir Crit Care Med. (2016) 194(10):1208–18. doi: 10.1164/rccm.201601-0002OC
244. Jeffery HC, van Wilgenburg B, Kurioka A, Parekh K, Stirling K, Roberts S, et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J Hepatol. (2016) 64(5):1118–27. doi: 10.1016/j.jhep.2015.12.017
245. Sobkowiak MJ, Davanian H, Heymann R, Gibbs A, Emgård J, Dias J, et al. Tissue-resident MAIT cell populations in human oral mucosa exhibit an activated profile and produce IL-17. Eur J Immunol. (2019) 49(1):133–43. doi: 10.1002/eji.201847759
246. Koay HF, Gherardin NA, Enders A, Loh L, Mackay LK, Almeida CF, et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat Immunol. (2016) 17(11):1300–11. doi: 10.1038/ni.3565
247. Chandra S, Wingender G, Greenbaum JA, Khurana A, Gholami AM, Ganesan AP, et al. Development of asthma in inner-city children: possible roles of MAIT cells and variation in the home environment. J Immunol Baltim Md 1950. (2018) 200(6):1995–2003. doi: 10.4049/jimmunol.1701525
248. Wesemann DR, Nagler CR. Commensal bacteria, timing and barrier function in the context of allergic disease. Immunity. (2016) 44(4):728–38. doi: 10.1016/j.immuni.2016.02.002
249. Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, et al. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. (2014) 63(4):559–66. doi: 10.1136/gutjnl-2012-303249
250. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: a study of 91 term infants. Sci Rep. (2020) 10:15792. doi: 10.1038/s41598-020-72635-x
251. Vuillermin PJ, O’Hely M, Collier F, Allen KJ, Tang MLK, Harrison LC, et al. Maternal carriage of prevotella during pregnancy associates with protection against food allergy in the offspring. Nat Commun. (2020) 11(1):1452. doi: 10.1038/s41467-020-14552-1
252. Stokholm J, Thorsen J, Blaser MJ, Rasmussen MA, Hjelmsø M, Shah S, et al. Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci Transl Med. (2020) 12(569):eaax9929. doi: 10.1126/scitranslmed.aax9929
253. Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. (2015) 7(307):307ra152. doi: 10.1126/scitranslmed.aab2271
254. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. (2016) 22(10):1187–91. doi: 10.1038/nm.4176
255. Allwell-Brown G, Hussain-Alkhateeb L, Kitutu FE, Strömdahl S, Mårtensson A, Johansson EW. Trends in reported antibiotic use among children under 5 years of age with fever, diarrhoea, or cough with fast or difficult breathing across low-income and middle-income countries in 2005–17: a systematic analysis of 132 national surveys from 73 countries. Lancet Glob Health. (2020) 8(6):e799–807. doi: 10.1016/S2214-109X(20)30079-6
256. West CE, Rydén P, Lundin D, Engstrand L, Tulic MK, Prescott SL. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy. (2015) 45(9):1419–29. doi: 10.1111/cea.12566
257. Sjödin KS, Hammarström ML, Rydén P, Sjödin A, Hernell O, Engstrand L, et al. Temporal and long-term gut microbiota variation in allergic disease: a prospective study from infancy to school age. Allergy. (2019) 74(1):176–85. doi: 10.1111/all.13485
258. Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. (2013) 4(2):158–64. doi: 10.4161/gmic.23567
259. Logan K, Bahnson HT, Ylescupidez A, Beyer K, Bellach J, Campbell DE, et al. Early introduction of peanut reduces peanut allergy across risk groups in pooled and causal inference analyses. Allergy. (2023) 78(5):1307–18. doi: 10.1111/all.15597
260. Korpela K, Hurley S, Ford SA, Franklin R, Byrne S, Lunjani N, et al. Association between gut microbiota development and allergy in infants born during pandemic-related social distancing restrictions. Allergy. (2024) 534:263–6. doi: 10.1111/all.16069
Keywords: microbiome, early life, allergy, antibodies, pregnancy, breast milk
Citation: Pirker A-L and Vogl T (2024) Development of systemic and mucosal immune responses against gut microbiota in early life and implications for the onset of allergies. Front. Allergy 5: 1439303. doi: 10.3389/falgy.2024.1439303
Received: 27 May 2024; Accepted: 5 July 2024;
Published: 17 July 2024.
Edited by:
Amanda Atanasio, Regeneron Pharmaceuticals, Inc., United StatesReviewed by:
Ronald L. Rabin, United States Food and Drug Administration, United StatesDaniel P. Potaczek, University of Marburg, Germany
© 2024 Pirker and Vogl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Vogl, dGhvbWFzLnZvZ2xAbWVkdW5pd2llbi5hYy5hdA==
 Anna-Lena Pirker
Anna-Lena Pirker Thomas Vogl*
Thomas Vogl*
