
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Allergy, 16 August 2023
Sec. Mechanisms in Allergy
Volume 4 - 2023 | https://doi.org/10.3389/falgy.2023.1240727
This article is part of the Research TopicSpotlight on Allergy Research in AsiaView all 5 articles
Understanding the house dust mites (HDMs) microbiome is crucial due to its potential effects on the development of allergic diseases. In 1998, our laboratory collected Dermatophagoides farinae and D. pteronyssinus from beds in a Korean household and began cultivating these HDMs. Our laboratory has been actively investigating several topics about HDMs in recent years, including the bacterial and fungal microbiome and their interactions, as well as the impact of the HDM microbiome on airway inflammation. To study the D. farinae microbiome, we employed high-throughput sequencing of the 16S rDNA amplicons. The results revealed that the two most abundant bacteria were Enterococcus faecalis and Bartonella spp. In contrast, we found almost no bacteria in D. pteronyssinus. By inoculating bacteria to HDMs, we found that D. farinae is more susceptible to bacteria than D. pteronyssinus. This susceptibility was associated with the presence of certain fungal species in D. pteronyssinus. Additionally, we have recently made efforts to produce HDMs with reduced levels of symbiotic bacteria. We believe that standardizing and controlling the microbiome in HDMs are crucial steps for the future development and improvement of allergic immunotherapies.
House dust mites (HDMs) are widely recognized as the primary sources of indoor allergens, playing a crucial role in triggering allergic diseases such as allergic rhinitis, asthma, and atopic dermatitis (1–3). Approximately 50% of individuals with asthma have an allergic reaction to HDM (4). More than 30 specific HDM allergens have been identified as key triggers in allergic responses (5). Professor Ree in our department identified 23 species of mites from house dusts in South Korea in 1997 of which Dermatophagoides farinae and Dermatophagoides pteronyssinus were reported to be the most predominant HDM species (6). The predominant species of mites found in Korean households were determined to be D. farinae, present in 65%–77% of homes, and D. pteronyssinus, found in 8%–20% of homes (6, 7). HDMs are present in over 90% of households in Korea, and the degree of exposure to HDM is medically noteworthy (8). Approximately 40%–60% of Korean individuals with respiratory allergies are affected by HDM (9–12), and more than 40% of patients with atopic dermatitis exhibit sensitivity to HDM (13–19).
Afterward, in 1998, our research lab collected D. farinae and D. pteronyssinus from beds in a Korean household and cultivated these HDMs. We have been continuously cultivating them and conducting research on HDM allergens and allergic diseases up to the present. Furthermore, our laboratory also serves as a resource bank, where we cultivate various mite and cockroach species and supply these to other institutions for their research needs. In our laboratory, we have been actively investigating the following topics using HDM in recent years: bacterial and fungal microbiome and their interaction, and the impact of the HDM microbiome on airway inflammation. Therefore, in this manuscript, we would like to provide an overview of our lab's recent research.
Immune responses are modulated by the properties of the allergen itself and by the adjuvant-like substances that are concomitantly administered with the antigens. Characterizing the HDMs microbiome is crucial due to its potential immunomodulatory effects on allergic disease pathogenesis, through the generation of microbe-associated molecules such as lipopolysaccharides (LPS), lipoteichoic acid, and bacterial DNA (20, 21).
In 2018 we studied the D. farinae microbiome using the 16S rRNA amplicon deep sequencing (22). In this study, 113 bacterial species were found, and the most abundant bacteria was Enterococcus faecalis (53.6%) and the second most abundant bacteria was Bartonella HQ806746 (39.0%). We confirmed these results using the E. coil TA cloning vector system targeting the full sequence of 16S rDNA in mite's DNA. In addition, we found that E. faecalis were distributed throughout the intestine and the stool using Gram staining, and Bartonella spp. was detected in the hemocoel using silver staining (Figure 1).
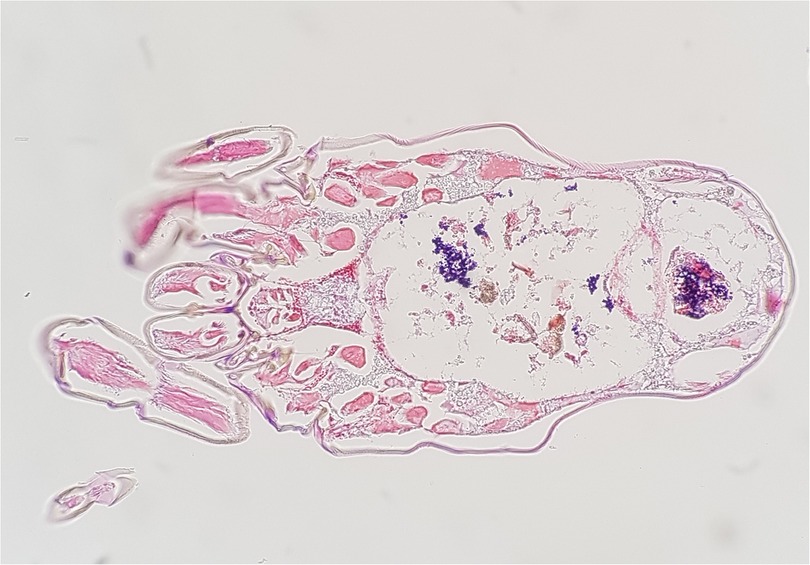
Figure 1. Enterococcus faecalis (gram-positive cocci, stained blue) observed in the intestine and stool of dermatophaoides farinae, Gram staining (magnification ×200).
In the next study, we investigated the bacterial microbiomes of D. farinae and D. pteronyssinus, both of which have been continuously cultured and maintained in our laboratory for over 20 years (23). It was found that these mite species have distinctly different microbiomes. As per our previous study, in D. farinae two dominant bacteria: Bartonella and Enterococcus faecalis accounted for 99.51%. In contrast, the bacterial sequence read was almost not detected in D. pteronyssinus except for several Klebsiella pneumonia reads. In quantitative PCR, D. pteronyssinus was found to have 600 times less total bacterial DNA than D. farinae. Interestingly, we found that the D. farinae collected from a house in Japan, which had been maintained in our laboratory for two decades, had the same Bartonella species as the Korean D. farinae. Thus, when bred in the same environment, mites of the same species showed almost identical microbiome patterns regardless of their regional origin.
There have been reports that D. farinae extracts produced by different countries and companies may have varying levels of included LPS, with differences of up to 9-fold (24, 25). This can be attributed to the variations in the methods employed by each company in breeding HDMs, which in turn can lead to differences in the microbiome of the mites. In fact, the microbiomes within HDMs vary across countries and institutions. The studies on HDMs microbiomes in the world were summarized in Table 1. A study of the draft genome of D. farinae in Hong Kong, China reported Enterobacter spp. was the most predominant (63.5%) and Bartonella spp. accounted for only 1.7% (27). Staphylococcus, Escherichia, Rhizobium, Klebsiella, Endophytic, Bacillus, and Salmonella were found in this study too in D. farinae (27). While another study using 16S rRNA cloning in the US reported that Bartonella was the most abundant bacterial taxon (20). In another study using similar methods in Czech, many bacteria were found including Sphingobacteriales, Rhizobiales, Neisseriales, Enterobacteriales, Pseudomonadales, Bacillales, Lactobacillales, Clostridiales, and Actinomycetales, but Bartonella was not detected (26, 33). Among these Enterococcus spp. was abundant bacterial species in D. farinae (26). In 2020 in China, a study using the 16S rRNA gene metabarcoding, the same methods we used, reported that Klebsiella, Acinetobacter, Raoultella, Aeromonas, Streptococcus, Lactococcus, and Escherichia were found in D. farinae (28). According to a recent 2020 study conducted in the Czech Republic, there is an association between Cardinium and D. farinae, indicating that Cardinium might have biological functions within the D. farinae (29). Other bacteria found in this study were Cloacibacterium, Streptococcus termophilus, Streptococcus dentisani, Dolosigranulum pigrum, Lactobacillus fermentum, Staphylococcus saprophyticus, Bacillus anthracis, Propionibacterium acnes, Corynebacterium tuberculostearicum, Lawsonella clevelandensis, Ottowia beijingensis, Curvibacter fontanus, Acinetobacter vivianii, Moraxella osloensis, and Alphaproteobacteria. Another study in Czech in 2020, detected Staphylococcus arlettae, Lactobacillus fermentum, Cardinium, Pantoea, Acinetobacter radioresistens, and Lactobacillus amylovorus. In the study conducted in France in 2021, Bartonellaceae, Sphingomonadaceae, Enterobacteriaceae, Amoebophilaceae, Caulobacteraceae, Patubacteraceae, and Myxococcales were found in the D. farinae.
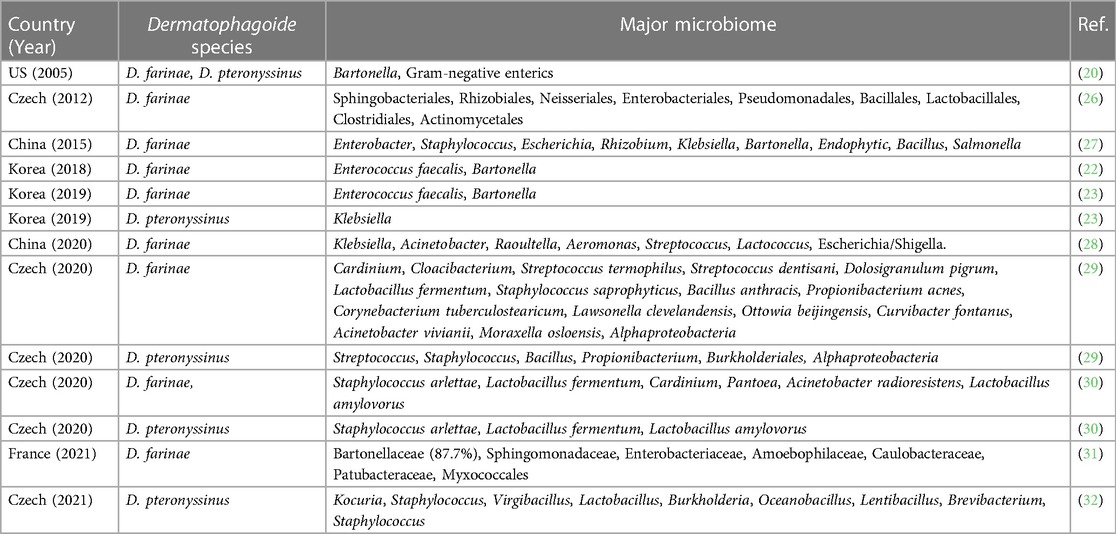
Table 1. Summary of studies on the microbiome of house dust mites (Dermatophagoide farinae and D. pteronyssinus).
Various bacteria were found in D. pteronyssinus including Kocuria, Staphylococcus, Virgibacillus, Lactobacillus, Burkholderia, Oceanobacillus, Lentibacillus, Brevibacterium, Streptococcus, Bacillus, and Propionibacterium (29, 32), but it is considered that the number of bacteria in D. pteronyssinus is far less than those in D. farinae (23). The previous studies support the notion that D. farinae extracts contain higher concentrations of LPS compared to D. pteronyssinus extracts (20, 29, 34–39).
In our previous study, we discovered that D. farinae contained significantly higher levels of bacterial microbiome compared to D. pteronyssinus, predominantly consisting of E. faecalis and Bartonella spp. (23). To better understand this phenomenon, E. faecalis carrying a gene for green fluorescence protein was introduced into both D. farinae and D. pteronyssinus (40). After a 10-day inoculation period, the colony-forming units (CFU) per mite were measured. The results showed that D. farinae had 7,950 CFU/mite, while D. pteronyssinus had 6.65 CFU/mite, which demonstrates that D. farinae can maintain over 1,000 times more E. faecalis compared to D. pteronyssinus. In the time series analysis, the amount of E. faecalis kept decreasing exponentially in D. pteronyssinus after inoculation, but the amount of E. faecalis was maintained in D. farinae from day 6. Therefore, these results indicate that D. farinae has a higher susceptibility to bacteria compared to D. pteronyssinus, suggesting that D. pteronyssinus may possess factors that make it less suitable for bacterial symbiosis or parasitism.
Transcriptomic analysis of airway epithelial cells (BEAS-2B) treated with D. farinae and D. pteronyssinus extracts revealed enrichment of the antibiotic metabolic process pathway in D. pteronyssinus-treated cells, not D. farinae-treated cells (40). We hypothesized that the reason for the enrichment of this antibiotic metabolic process could be due to the abundance of specific fungus in D. pteronyssinus and measured fungal gene levels in two HDM species. Fungal genes were found to be 1,300 times more abundant in D. pteronyssinus than in D. farinae, with Aspergillus penicillioides being the predominant species identified through mycobiome analysis using fungus-specific ITS2 amplicons. In our study, A. penicillioides and Pencillium cinerascens were exclusively detected in D. pteronyssinus, while they were not found in D. farinae.
Meanwhile, bacterial genes were 12,000 times more found in D. farinae than in D. pteronyssinus in the study. Then, we hypothesized that these fungi in D. pteronyssinus could inhibit bacterial growth in the mites. To prove this, D. pteronyssinus were inoculated with E. faecalis and treated with amphotericin B, the antifungal agent. After a 10-day period, it was observed that the CFU of E. faecalis per mite was 40 times higher in D. pteronyssinus treated with amphotericin B compared to untreated D. pteronyssinus. Furthermore, the extract of A. penicillioides, which was isolated from D. pteronyssinus, demonstrated inhibitory effects on the growth of E. faecalis in vitro.
A. penicillioides is a xerophilic fungus commonly found in low-moisture environments such as indoor air and house dust (41, 42). Its presence has been suggested to contribute to the reproduction of D. pteronyssinus (43). Notably, both A. penicillioides and Penicillium cinerascens, found in D. pteronyssinus, possess antibacterial properties (34–46). Similar to our study, Saccharomyces cerevisiae, Aspergillus, and Candida were reported in D. pteronyssinus in other studies (30, 32, 47, 48). In addition, there have been several studies suggesting that the presence of fungi in insects can impact bacterial growth, which is consistent with our findings (49–51).
To determine whether changes in the HDM microbiome affect allergy development, D. farinae were cultured under ampicillin and the number of bacteria in D. farinae was reduced by 25-fold (23). Moreover, the use of antibiotics led to a significant 100-fold reduction in the concentration of LPS. However, there was no difference observed in the concentration of the Der f 1 allergen in the HDM extract following antibiotic treatment. Notably, treating the BEAS-2B cells with the antibiotic-treated D. farinae extract resulted in a significant decrease in the secretion levels of proinflammatory cytokines IL-6 and IL-8. These findings indicate that alterations in the microbiome can impact the HDM's ability to trigger allergic diseases.
In addition, in the transcriptomic analysis of the response of airway epithelial cells to D. farinae and D. pteronyssinus extracts, we found that the “response to bacterium” and “pattern recognition receptor signaling pathway” were enriched in the cells treated with D. farinae compared with D. pteronyssinus (40). We were able to interpret that these results were due to the higher abundance of bacteria in D. farinae compared to D. pteronyssinus. D. farinae displayed a 14,000-fold higher bacterial DNA level, along with a 60-fold higher LPS concentration, compared to D. pteronyssinus. Treatment with an LPS inhibitor resulted in a decrease in the secretion of IL-6 and IL-8 in cells treated with D. farinae extracts but not in cells treated with D. pteronyssinus extracts. These results indicate that bacterial-derived substances such as LPS in HDMs have a synergistic impact on cytokine secretion in airway epithelial cells.
Many studies have reported a higher concentration of LPS in D. farinae compared to D. pteronyssinus (20, 29, 34–39). LPS plays a crucial role in triggering various immunological and allergic responses (52–55). LPS has been implicated in the development and severity of asthma (53, 56, 57). In a mouse model, depletion of LPS in HDM extract led to a reduction in features of allergic asthma (58). LPS can stimulate the development of type 2 immune responses to inhaled allergens through Toll-like receptor 4 (TLR4) signaling (59, 60). The HDM's LPS acts as a TLR4 activator in airway epithelial cells, leading to the induction of allergic inflammation through the activation of mucosal dendritic cells (55). Previous research demonstrated molecular-level differences in the lung transcriptome of mice challenged with D. pteronyssinus extracts containing high or low levels of LPS (61). HDMs trigger a type 2 immune response characterized by increased expression of IL-6 and IL-8 in airway epithelial cells (62, 63). Specifically, group 1 allergens and LPS from HDMs stimulate the expression of IL-6 and IL-8 by activating protease-activated receptor 2 (PAR-2) and TLR4, respectively (64). Furthermore, in a recent mouse asthma model, it was discovered that Gram-negative bacteria associated with HDM exacerbated the severity of the disease through nucleotide-binding oligomerization domain-containing protein (NOD) (31). Moreover, the presence of certain muropeptides from Gram-negative bacteria in the HDM extract potentially contributes to airway inflammation. Additionally, lipoteichoic acid has been found to influence the development of allergies through TLR2 (65). It has also been reported that LPS from Bartonella acts as an antagonist of TLR4 (66). Therefore, it is important to consider the composition of the microbiome within the mite to accurately evaluate the immunotherapeutic effectiveness of the extract.
As observed earlier, the microbiome of HDMs and their associated substances can have an impact on immune responses in humans. Furthermore, symbiotic bacteria within HDMs can exist as different species depending on the country or rearing environment. Hence, accurately predicting all the immunological side effects associated with each bacterium poses a significant challenge, particularly in the clinical use of immunotherapeutic agents.
We recently conducted a study where we treated D. farinae with ampicillin during their rearing process. We published our findings, demonstrating that despite multiple sub-culturing events, the bacterial levels in D. farinae remained low for 18 weeks following a single ampicillin treatment (67). In this study, we administered ampicillin at an appropriate concentration to ensure it did not affect the growth of HDMs. As a result, we successfully achieved a 150-fold reduction in symbiotic bacteria levels and a 33-fold reduction in LPS levels over an 18-week period. In the ampicillin-treated and untreated D. farinae, the LPS concentrations were measured as 155.6 and 5,135.5 EU/mg, respectively. Previous research found LPS concentrations of 8,740 EU/mg in a Korean D. farinae extract and 3,890 EU/mg in a D. farinae extract from the United States (8). The protein bands pattern in SDS PAGE analysis of D. farinae extract remained unchanged and the concentration of Der f 1 and Der f 2 were not changed by ampicillin treatment.
Then, the D. farinae extracts were used to induce asthma in a mouse model (67). The extracts were injected intraperitoneally twice with a 2-week interval, along with alum adjuvant, and challenged intranasally. In our study, we found that the level of lung function, airway inflammation, and serum-specific immunoglobulin did not differ between the mouse asthma models developed using ampicillin-treated D. farinae and those developed using ampicillin-untreated D. farinae. These findings suggest that D. farinae with low bacterial content was sufficient to induce allergic sensitization and immune response. However, another study we conducted demonstrated that administering the extract intranasally at two-day intervals for a total of nine treatments, without the use of an adjuvant, resulted in less pronounced airway inflammation in mice treated with mites that had reduced bacterial levels (68). In this study, the mouse sensitized with the antibiotics-treated mite compared to ampicillin-untreated mite showed less eosinophils in the bronchoalveolar lavage and less immune cell infiltration in lung histopathology. Serum IgE and IgG1 were not different among the groups.
Since bacteria within HDMs can act as adjuvants and exacerbate inflammatory responses, it is crucial to exercise caution when using HDM extract in both clinical and experimental settings. A study has indicated that HDMs can act as carriers of bacteria, leading to the induction of IgE sensitization to bacterial antigens (69). Bacterial IgE sensitization has been associated with different allergic disease conditions, such as the severity of atopic dermatitis and the co-occurrence of rhinitis (70–75). Therefore, reducing bacteria in HDMs would be beneficial, provided that the effectiveness of allergen immunotherapy remains unchanged. Furthermore, standardizing mite production, extract preparation, and controlling bacterial levels are crucial steps to facilitate the development and improvement of diagnostic and therapeutic agents for allergic diseases (76).
Further research is needed to better understand the immunological role of symbiotic bacteria in the HDMs. In particular, future studies should be conducted to investigate whether the bacterial components derived from HDM extract exhibit enhanced or diminished efficacy when utilized as immunotherapeutic agents.
Furthermore, additional research is warranted to explore the impact of symbiotic bacteria on the growth of HDMs. Based on our own observations, we have discovered that excessive antibiotic treatment can significantly reduce the growth rate of mites. Further research is required to determine whether this effect is a result of the direct toxicity of antibiotics on HDMs or the consequence of reduced bacterial populations in HDMs.
While our studies indicated that the reduction of bacteria in HDMs did not impact the production of Der f 1 and Der f 2, it is necessary to assess any changes in the expression of other allergens. The composition of the microbiome in storage mites might have an influence on allergen expression (77). Notably, recent findings have suggested that Cardinium in D. farinae could potentially modulate gene expressions in mites related to immunity and metabolism (78).
Conceptualization: JK; Data curation: MK; Investigation: JK; Supervision: T-SY; Writing—original draft: M-hY; Writing—review & editing: JK. All authors contributed to the article and approved the submitted version.
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST; Number NRF-2020R1I1A2074562). In addition, this study was supported by a faculty research grant from the Yonsei University College of Medicine (6-2022-0125).
We express gratitude to all the past and present researchers of our departments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Fernández-Caldas E, Puerta L, Caraballo L. Mites and allergy. Chem Immunol Allergy. (2014) 100:234–42. doi: 10.1159/000358860
2. Thomas WR, Hales BJ, Smith WA. House dust mite allergens in asthma and allergy. Trends Mol Med. (2010) 16:321–8. doi: 10.1016/j.molmed.2010.04.008
3. Park KH, Sim DW, Lee SC, Moon S, Choe E, Shin H, et al. Effects of air purifiers on patients with allergic rhinitis: a multicenter, randomized, double-blind, and placebo-controlled study. Yonsei Med J. (2020) 61:689–97. doi: 10.3349/ymj.2020.61.8.689
4. Calderón MA, Linneberg A, Kleine-Tebbe J, De Blay F, Hernandez fernandez de rojas D, Virchow JC, et al. Respiratory allergy caused by house dust mites: what do we really know? J Allergy Clin Immunol. (2015) 136:38–48. doi: 10.1016/j.jaci.2014.10.012
5. Vrtala S. Allergens from house dust and storage mites. Allergo J Int. (2022) 31:267–71. doi: 10.1007/s40629-022-00226-5
6. Ree HI, Jeon SH, Lee IY, Hong CS, Lee DK. Fauna and geographical distribution of house dust mites in Korea. Korean J Parasitol. (1997) 35:9–17. doi: 10.3347/kjp.1997.35.1.9
7. Jeong KY, Lee IY, Lee J, Ree HI, Hong CS, Yong TS. Effectiveness of health education for the control of house dust mites and cockroaches in Seoul, Korea. Korean J Parasitol. (2006) 44:73–9. doi: 10.3347/kjp.2006.44.1.73
8. Jeong KY, Park JW, Hong CS. House dust mite allergy in Korea: the most important inhalant allergen in current and future. Allergy Asthma Immunol Res. (2012) 4:313–25. doi: 10.4168/aair.2012.4.6.313
9. Ahu SD, Kim HJ. The allergen skin test and the effect of specific desensitising vaccination therapy in allergic rhinitis and bronchial asthma. Allergy. (1983) 3:159–67.
10. Kang SY, Choi BW, Moon HB, Min KU, Kim YY. The prevalence of immediate skin reactions in patients with respiratory allergies. Allergy. (1984) 4:49–56.
11. Cho HS, Lee KH, Choi YK, Cho DK, Kim NS. Result of allergen skin tests in type I hypersensitivity. Allergy. (1985) 5:14–22.
12. Rhee YG, Oh YI. Results of skin test, peripheral eosinophil count, total and specific IgE in allergic patients in chonbuk area. Allergy. (1985) 5:147–55.
13. Min TH, Hong CK, Ro BI, Chang CY. Allergen prick test reactivity in the patients with urticaria and atopic dermatitis. Korean J Dermatol. (1987) 25:587–98.
14. Kang SJ, Kim KH, Kim SH. A study on type I allergy to house dust mite in patients with atopic dermatitis. Korean J Dermatol. (1991) 29:285–91.
15. Cho HJ, Choi HJ, Kim DK, Lee KH. Dermatophagoides farinae-specific IgE and IgG4 antibodies in atopic dermatitis patients. Korean J Dermatol. (1998) 36:16–22.
16. Yim YS, Park CW, Lee CH. A comparative study of atopy patch test using house dust mite antigens with skin prick test and specific serum IgE level in atopic dermatitis. Korean J Dermatol. (2001) 39:1072–9.
17. Kim SS, Bang HD, Kim KH, Kim KJ. Evaluation of pharmacia CAP system FEIA for house dust mites in patients with atopic dermatitis. Korean J Dermatol. (2003) 41:1455–62.
18. Kim YJ, Lee MY, Yang AR, Sol IS, Kwak JH, Jung HL, et al. Trends of sensitization to inhalant allergens in Korean children over the last 10 years. Yonsei Med J. (2020) 61:797–804. doi: 10.3349/ymj.2020.61.9.797
19. Park HJ, Lim HS, Park KH, Lee JH, Park JW, Hong CS. Changes in allergen sensitization over the last 30 years in Korea respiratory allergic patients: a single-center. Allergy Asthma Immunol Res. (2014) 6:434–43. doi: 10.4168/aair.2014.6.5.434
20. Valerio CR, Murray P, Arlian LG, Slater JE. Bacterial 16S ribosomal DNA in house dust mite cultures. J Allergy Clin Immunol. (2005) 116:1296–300. doi: 10.1016/j.jaci.2005.09.046
21. Stewart GA. Studies of house dust mites can now fully embrace the “-omics” era. J Allergy Clin Immunol. (2015) 135:549–50. doi: 10.1016/j.jaci.2014.12.934
22. Kim JY, Yi MH, Hwang Y, Lee JY, Lee IY, Yong D, et al. 16S rRNA profiling of the Dermatophagoides farinae core microbiome: enterococcus and Bartonella. Clin Exp Allergy. (2018) 48:607–10. doi: 10.1111/cea.13104
23. Lee J, Kim JY, Yi MH, Hwang Y, Lee IY, Nam SH, et al. Comparative microbiome analysis of Dermatophagoides farinae, Dermatophagoides pteronyssinus, and Tyrophagus putrescentiae. J Allergy Clin Immunol. (2019) 143:1620–3. doi: 10.1016/j.jaci.2018.10.062
24. Jeong KY, Choi SY, Lee JH, Lee IY, Yong TS, Lee JS, et al. Standardization of house dust mite extracts in Korea. Allergy Asthma Immunol Res. (2012) 4:346–50. doi: 10.4168/aair.2012.4.6.346
25. Kim JT, Kim H, Kim SH, Kim DJ, Shin Y, Kim JD, et al. Comparison of allergenic properties among commercially available house dust mite allergen extracts in Korea. Yonsei Med J. (2021) 62:86–90. doi: 10.3349/ymj.2021.62.1.86
26. Hubert J, Kopecký J, Perotti MA, Nesvorná M, Braig HR, Ságová-Marečková M, et al. Detection and identification of species-specific bacteria associated with synanthropic mites. Microb Ecol. (2012) 63:919–28. doi: 10.1007/s00248-011-9969-6
27. Chan TF, Ji KM, Yim AK, Liu XY, Zhou JW, Li RQ, et al. The draft genome, transcriptome, and microbiome of Dermatophagoides farinae reveal a broad spectrum of dust mite allergens. J Allergy Clin Immunol. (2015) 135:539–48. doi: 10.1016/j.jaci.2014.09.031
28. Guo Y, Wang R, Zhao Y, Niu D, Gong X, Hu L. Study on the relationship between microbial composition and living environment in important medical mites based on illumina MiSeq sequencing technology. J Med Entomol. (2020) 57:1049–56. doi: 10.1093/jme/tjaa034
29. Erban T, Klimov P, Molva V, Hubert J. Whole genomic sequencing and sex-dependent abundance estimation of Cardinium sp., a common and hyperabundant bacterial endosymbiont of the American house dust mite Dermatophagoides farinae. Exp Appl Acarol. (2020) 80:363–80. doi: 10.1007/s10493-020-00475-5
30. Molva V, Bostlova M, Nesvorna M, Hubert J. Do the microorganisms from laboratory culture spent growth medium affect house dust mite fitness and microbiome composition? Insect Sci. (2020) 27:266–75. doi: 10.1111/1744-7917.12636
31. Ait Yahia S, Audousset C, Alvarez-Simon D, Vorng H, Togbe D, Marquillies P, et al. NOD1 sensing of house dust mite-derived microbiota promotes allergic experimental asthma. J Allergy Clin Immunol. (2021) 148:394–406. doi: 10.1016/j.jaci.2020.12.649
32. Nesvorna M, Pekar S, Shcherbachenko E, Molva V, Erban T, Green SJ, et al. Microbiome variation during culture growth of the European house dust mite, Dermatophagoides pteronyssinus. FEMS Microbiol Ecol. (2021) 97:fiab039. doi: 10.1093/femsec/fiab039
33. Kopecký J, Nesvorná M, Hubert J. Bartonella-like bacteria carried by domestic mite species. Exp Appl Acarol. (2014) 64:21–32. doi: 10.1007/s10493-014-9811-1
34. Trivedi B, Valerio C, Slater JE. Slater endotoxin content of standardized allergen vaccines. J Allergy Clin Immunol. (2003) 111:777–83. doi: 10.1067/mai.2003.1338
35. Valerio C, Murray P, Arlian LG, Slater JE. Endotoxin in dust mite allergen extracts. J Allergy Clin Immunol. (2004) 113:S136–7. doi: 10.1016/j.jaci.2003.12.489
36. Arlian LG, Elder BL, Morgan MS. House dust mite extracts activate cultured human dermal endothelial cells to express adhesion molecules and secrete cytokines. J Med Entomol. (2009) 46:595–604. doi: 10.1603/033.046.0326
37. Valerio C, Arlian LG, Slater JE. Bacterial DNA sequences isolated from standardized dust mite extracts and wild mites. J Allergy Clin Immunol. (2009) 123:S216. doi: 10.1016/j.jaci.2008.12
38. Arlian LG, Morgan MS. Immunomodulation of skin cytokine secretion by house dust mite extracts. Int Arch Allergy Immunol. (2011) 156:171–8. doi: 10.1159/000323351
39. Rockwood J, Morgan MS, Arlian LG. Proteins and endotoxin in house dust mite extracts modulate cytokine secretion and gene expression by dermal fibroblasts. Exp Appl Acarol. (2013) 61:311–25. doi: 10.1007/s10493-013-9703-9
40. Kim JY, Yi MH, Lee S, Lee IY, Yong D, Yoon SS, et al. Microbiome and mycobiome interaction in house dust mites and impact on airway cells. Clin Exp Allergy. (2021) 51:1592–602. doi: 10.1111/cea.13962
41. Gock MA, Hocking AD, Pitt JI, Poulos PG. Influence of temperature, water activity and pH on growth of some xerophilic fungi. Int J Food Microbiol. (2003) 81:11–9. doi: 10.1016/s0168-1605(02)00166-6
42. Williams JP, Hallsworth JE. Limits of life in hostile environments: no barriers to biosphere function? Environ Microbiol. (2009) 11:3292–308. doi: 10.1111/j.1462-2920.2009.02079.x
43. Hay DB, Hart BJ, Douglas AE. Effects of the fungus aspergillus penicillioides on the house dust mite Dermatophagoides pteronyssinus: an experimental re-evaluation. Med Vet Entomol. (1993) 7:271–4. doi: 10.1111/j.1365-2915.1993.tb00687.x
44. Tamokou Jde D, Kuiate JR, Tene M, Kenla Nwemeguela TJ, Tane P. The antimicrobial activities of extract and compounds isolated from Brillantaisia lamium. Iran J Med Sci. (2011) 36:24–31.23365474
45. Tamokou Jde D, Simo Mpetga DJ, Keilah Lunga P, Tene M, Tane P, Kuiate JR. Antioxidant and antimicrobial activities of ethyl acetate extract, fractions and compounds from stem bark of Albizia adianthifolia (mimosoideae). BMC Complement Altern Med. (2012) 18:99. doi: 10.1186/1472-6882-12-99
46. Bracken A, Raistrick H. Studies in the biochemistry of micro-organisms; dehydrocarolic acid, a metabolic product of Penicillium cinerascens biourge. Biochem J. (1947) 41:569–75. doi: 10.1042/bj0410569
47. Lustgraaf BV. Ecological relationships between xerophilic fungi and house-dust mites (acarida: pyroglyphidae). Oecologia. (1978) 33:351–9. doi: 10.1007/BF00348118
48. Douglas AE, Hart BJ. The significance of the fungus Aspergillus penicillioides to the house dust mite Dermatophagoides pteronyssinus. Symbiosis. (1989) 7:105–16. doi: 10.1890/07-0815.1
49. Little AE, Currie CR. Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants. Ecology. (2008) 89:1216–22. doi: 10.1890/07-0815
50. McFrederick QS, Mueller UG, James RR. Interactions between fungi and bacteria influence microbial community structure in the Megachile rotundata larval gut. Proc Biol Sci. (2014) 281:20132653. doi: 10.1098/rspb.2013.2653
51. Fan Y, Liu X, Keyhani NO, Tang G, Pei Y, Zhang W, et al. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc Natl Acad Sci U S A. (2017) 114:E1578–86. doi: 10.1073/pnas.1616543114
52. Platts-Mills TA, Woodfolk JA. Allergens and their role in the allergic immune response. Immunol Rev. (2011) 242:51–68. doi: 10.1111/j.1600-065X.2011.01021.x
53. Arora M, Poe SL, Ray A, Ray P. LPS-induced CD11b+ Gr1(int)F4/80+ regulatory myeloid cells suppress allergen-induced airway inflammation. Int Immunopharmacol. (2011) 11:827–32. doi: 10.1016/j.intimp.2011.01.034
54. Kim YM, Kim YS, Jeon SG, Kim YK. Immunopathogenesis of allergic asthma: more than the Th2 hypothesis. Allergy Asthma Immunol Res. (2013) 5:189–96. doi: 10.4168/aair.2013.5.4.189
55. Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via toll-like receptor 4 triggering of airway structural cells. Nat Med. (2009) 15:410–6. doi: 10.1038/nm.1946
56. Braun-Fahrländer C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. (2002) 347:869–77. doi: 10.1056/NEJMoa020057
57. Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. (2002) 196:1645–51. doi: 10.1084/jem.20021340
58. Ryu JH, Yoo JY, Kim MJ, Hwang SG, Ahn KC, Ryu JC, et al. Distinct TLR-mediated pathways regulate house dust mite-induced allergic disease in the upper and lower airways. J Allergy Clin Immunol. (2013) 131:549–61. doi: 10.1016/j.jaci.2012.07.050
59. Trompette A, Divanovic S, Visintin A, Blanchard C, Hegde RS, Madan R, et al. Allergenicity resulting from functional mimicry of a toll-like receptor complex protein. Nature. (2009) 457:585–8. doi: 10.1038/nature07548
60. Lambrecht BN, Hammad H. The role of dendritic and epithelial cells as master regulators of allergic airway inflammation. Lancet. (2010) 376:835–43. doi: 10.1016/S0140-6736(10)61226-3
61. Pascoe CD, Jha A, Basu S, Mahood T, Lee A, Hinshaw S, et al. The importance of reporting house dust mite endotoxin abundance: impact on the lung transcriptome. Am J Physiol Lung Cell Mol Physiol. (2020) 318:L1229–36. doi: 10.1152/ajplung.00103.2020
62. King C, Brennan S, Thompson PJ, Stewart GA. Dust mite proteolytic allergens induce cytokine release from cultured airway epithelium. J Immunol. (1998) 161:3645–51. doi: 10.4049/jimmunol.161.7.3645
63. Ullah MA, Revez JA, Loh Z, Simpson J, Zhang V, Bain L, et al. Allergen-induced IL-6 trans-signaling activates γδ T cells to promote type 2 and type 17 airway inflammation. J Allergy Clin Immunol. (2015) 136:1065–73. doi: 10.1016/j.jaci.2015.02.032
64. Asokananthan N, Graham PT, Stewart DJ, Bakker AJ, Eidne KA, Thompson PJ, et al. House dust mite allergens induce proinflammatory cytokines from respiratory epithelial cells: the cysteine protease allergen, der p 1, activates protease-activated receptor (PAR)-2 and inactivates PAR-1. J Immunol. (2002) 169:4572–8. doi: 10.4049/jimmunol.169.8.4572
65. Kaesler S, Volz T, Skabytska Y, Köberle M, Hein U, Chen KM, et al. Toll-like receptor 2 ligands promote chronic atopic dermatitis through IL-4-mediated suppression of IL-10. J Allergy Clin Immunol. (2014) 134:92–9. doi: 10.1016/j.jaci.2014.02.017
66. Malgorzata-Miller G, Heinbockel L, Brandenburg K, van der Meer JW, Netea MG, Joosten LA. Bartonella quintana lipopolysaccharide (LPS): structure and characteristics of a potent TLR4 antagonist for in-vitro and in-vivo applications. Sci Rep. (2016) 6:34221. doi: 10.1038/srep34221
67. Kim JY, Yi MH, Kim M, Choi JH, Lee S, Yong TS. Production of Dermatophagoides farinae having low bacterial content using ampicillin. J Immunol Res. (2023) 2023:9024595. doi: 10.1155/2023/9024595
68. Kim JY. Microbiome and mycobiome profiling of house dust mites and their significance in allergic inflammation [Dissertation/doctoral thesis]. Seoul (KR): Yonsei University (2019).
69. Dzoro S, Mittermann I, Resch-Marat Y, Vrtala S, Nehr M, Hirschl AM, et al. House dust mites as potential carriers for IgE sensitization to bacterial antigens. Allergy. (2018) 73:115–24. doi: 10.1111/all.13260
70. Hales BJ, Pearce LJ, Kusel MM, Holt PG, Sly PD, Thomas WR. Differences in the antibody response to a mucosal bacterial antigen between allergic and non-allergic subjects. Thorax. (2008) 63:221–7. doi: 10.1136/thx.2006.069492
71. Hales BJ, Martin AC, Pearce LJ, Rueter K, Zhang G, Khoo SK, et al. Anti-bacterial IgE in the antibody responses of house dust mite allergic children convalescent from asthma exacerbation. Clin Exp Allergy. (2009) 39:1170–8. doi: 10.1111/j.1365-2222.2009.03252.x
72. Hollams EM, Hales BJ, Bachert C, Huvenne W, Parsons F, de Klerk NH, et al. Th2-associated immunity to bacteria in teenagers and susceptibility to asthma. Eur Respir J. (2010) 36:509–16. doi: 10.1183/09031936.00184109
73. Reginald K, Westritschnig K, Linhart B, Focke-Tejkl M, Jahn-Schmid B, Eckl-Dorna J, et al. Staphylococcus aureus fibronectin-binding protein specifically binds IgE from patients with atopic dermatitis and requires antigen presentation for cellular immune responses. J Allergy Clin Immunol. (2011) 128:82–91.e8. doi: 10.1016/j.jaci.2011.02.034
74. Hales BJ, Chai LY, Elliot CE, Pearce LJ, Zhang G, Heinrich TK, et al. Antibacterial antibody responses associated with the development of asthma in house dust mite-sensitised and non-sensitised children. Thorax. (2012) 67:321–7. doi: 10.1136/thoraxjnl-2011-200650
75. Bachert C, van Steen K, Zhang N, Holtappels G, Cattaert T, Maus B, et al. Specific IgE against Staphylococcus aureus enterotoxins: an independent risk factor for asthma. J Allergy Clin Immunol. (2012) 130:376–81.e8. doi: 10.1016/j.jaci.2012.05.012
76. Jeong KY, Hong CS, Lee JS, Park JW. Optimization of allergen standardization. Yonsei Med J. (2011) 52:393–400. doi: 10.3349/ymj.2011.52.3.393
77. Hubert J, Nesvorna M, Klimov P, Dowd SE, Sopko B, Erban T. Differential allergen expression in three Tyrophagus putrescentiae strains inhabited by distinct microbiome. Allergy. (2019) 74:2502–7. doi: 10.1111/all.13921
Keywords: house dust mites, microbiome, Dermatophagoides farinae, 16S rDNA, Bartonella, Enterococcus
Citation: Yi M-h, Kim M, Yong T-S and Kim JY (2023) Investigating the microbiome of house dust mites in South Korea. Front. Allergy 4:1240727. doi: 10.3389/falgy.2023.1240727
Received: 15 June 2023; Accepted: 8 August 2023;
Published: 16 August 2023.
Edited by:
Andreas L. Lopata, James Cook University, AustraliaReviewed by:
Wayne Robert Thomas, University of Western Australia, Australia© 2023 Yi, Kim, Yong and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ju Yeong Kim anlraW0wODAyQHl1aHMuYWM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.