- 1Department of Microbiology and Plant Pathology, University of California, Riverside, CA, United States
- 2Department of Plant Pathology, University of California, Davis, Davis, CA, United States
- 3United States Department of Agriculture-APHIS-Biotechnology Risk Analysis Program, Riverdale, MD, United States
- 4Texas A&M, Department of Entomology Agrilife Research, Amarillo, TX, United States
- 5California Department of Food and Agriculture Nursery Services Program, Sacramento, CA, United States
Citrus germplasm programs can benefit from high-throughput polymerase chain reaction (PCR)-based methods for the detection of graft-transmissible pathogens in propagative materials. These methods increase diagnostic capacity, and thus contribute to the prevention of disease spread from nurseries to citrus orchards. High quality nucleic acids, as determined by purity, concentration, and integrity, are a prerequisite for reliable PCR detection of citrus pathogens. Citrus tissues contain high levels of polyphenols and polysaccharides, which can affect nucleic acid quality and inhibit PCR reactions. Various commercially available RNA isolation methods are used for citrus and include: phenol-chloroform (TRIzol®, Thermo Fisher Scientific); silica columns (RNeasy® Plant Mini Kit, Qiagen); and magnetic beads-based methods (MagMAX™-96 Viral RNA Isolation Kit, Thermo Fisher Scientific). To determine the quality of RNA and its impact on the detection of graft-transmissible citrus pathogens in reverse transcription (RT) PCR-based assays, we compared these three RNA isolation methods. We assessed RNA purity, concentration, and integrity from citrus inoculated with different viruses and viroids. All three RNA isolation methods produced high quality RNA, and its use in different RT-PCR assays resulted in the detection of all targeted citrus viruses and viroids with no false positive or negative results. TRIzol® yielded RNA with the highest concentration and integrity values but some samples required serial dilutions to remove PCR inhibitors and detect the targeted pathogens. The RNeasy® kit produced the second highest concentration and purity of RNA, and similar integrity to TRIzol®. MagMAX™ isolation also provided high quality RNA but most importantly produced RNA with consistent results clustered around a median value for concentration, purity, and integrity. Subsequently, MagMAX™-96 was combined with the semi-automated MagMAX™ Express-96 Deep Well Magnetic Particle Processor, for high-throughput sample processing. MagMAX™-96 enabled the diagnostic laboratory of the Citrus Clonal Protection Program-National Clean Plant Network at the University of California, Riverside to process over 16,500 samples from citrus budwood source trees between 2010 and 2019. This high-throughput approach dramatically reduced the incidence of viroids in citrus nurseries and was key to the successful implementation of the mandatory Citrus Nursery Stock Pest Cleanliness Program in California.
1 Introduction
Several citrus virus and viroid detection methods have been developed over the years, including biological indexing (Roistacher, 1991; Garnsey et al., 2005), enzyme-linked immunosorbent assay (ELISA) (Bar-Joseph et al., 1979), imprint hybridization (de Noronha Fonseca et al., 1996; Palacio-Bielsa et al., 1999), sequential polyacrylamide gel electrophoresis (Rivera-Bustamante et al., 1986), and direct blot immunoassay (Garnsey et al., 1993). Currently, nucleic acid-based molecular methods such as reverse transcription polymerase chain reaction (RT-PCR) and RT-quantitative PCR (RT-qPCR) are extensively used as standard detection methods for citrus viruses and viroids, because of their high sensitivity, specificity, and reproducibility (Bertolini et al., 2008; Rizza et al., 2009; Ruiz-Ruiz et al., 2009; Loconsole et al., 2010; Yokomi et al., 2010; Papayiannis, 2014; Osman et al., 2015; Osman et al., 2017). These methods are of utmost importance for citrus disease management programs, including citrus germplasm programs. Indeed, they are routinely used to monitor the sanitary status of budwood source trees and propagative materials, nursery stock, and commercial plantings (Bostock et al., 2014; Gergerich et al., 2015; Osman et al., 2015; Albrecht et al., 2020; Fuchs et al., 2021).
Various RNA isolation methods have been successfully used with different plant tissues for downstream detection of plant pathogens (MacKenzie et al., 1997; Portillo et al., 2006; Osman et al., 2012; Sun et al., 2014; Martinelli et al., 2015; Ali et al., 2017; Inglis et al., 2018; Liu et al., 2018; Vennapusa et al., 2020). More specifically, for citrus tissues, low-throughput phenol-chloroform based methods with cetrimonium bromide (CTAB) or TRIzol® (Thermo Fisher Scientific, Waltham, MA), and silica column-based kits such as the RNeasy® Plant Mini Kit (Qiagen, Valencia, CA) or Spectrum™ Plant Total RNA (Sigma-Aldrich, St. Louis, MO) have been effectively utilized for RNA isolation and detection of citrus viruses and viroids using RT-PCR and RT-qPCR assays (Li et al., 2008; Saponari et al., 2008; Damaj et al., 2009; Wang et al., 2013a; Wang et al., 2013b; Tan et al., 2019; Bester et al., 2021; Benítez-Galeano et al., 2021). More recently, high-throughput semi-automated total nucleic acid isolation systems such as the MagMAX™ Express-96 (Thermo Fisher Scientific, Waltham, MA) and BioSprint® 96 (Qiagen, Valencia, CA), capable of isolating RNA from up to 96 samples at once, have been used successfully in citrus pathogen detection protocols including but not limited to viruses and viroids (Osman et al., 2015; Osman et al., 2017; Braswell et al., 2020; Dang et al., 2022).
Regardless of the RNA isolation approach used, the common goal of all methods is to produce good quality nucleic acids for downstream molecular analysis, including RT-PCR based assays (Die and Román, 2012; Thatcher, 2015). For example, RNA quality as determined by purity, concentration, and integrity, is one of the most critical factors of RT-qPCR performance (Fleige and Pfaffl, 2006; Becker et al., 2010; Taylor et al., 2010; Die and Román, 2012). Therefore, for a properly designed, validated, and executed RT-PCR based detection assay (Bustin et al., 2009; Broeders et al., 2014; Tan et al., 2019), RNA quality becomes the decisive factor for the successful identification of a sample testing positive or negative for a target citrus virus or viroid.
RNA isolated from pulverized plant tissues is purified by removing PCR inhibitors, such as polyphenols and polysaccharides, which are commonly found in various woody and perennial plants including citrus (Newbury and Possingham, 1977; Porebski et al., 1997; Gasic et al., 2004; Gambino et al., 2008). Phenolic compounds can bind nucleic acids (Salzman et al., 1999) while polysaccharides can co-precipitate with RNA, thus hindering absorbance readings from spectrophotometers, and can inhibit enzymatic reactions (Wilkins and Smart, 1996). This may eventually lead to the inhibition of polymerase activity during PCR amplification and compromise the accuracy of pathogen detection (Sipahioglu et al., 2006; Schrader et al., 2012).
Optimal RNA concentrations, commonly measured via UV absorbance (Manchester, 1996), are important, because: low concentrations may not be sufficient for pathogen detection, and high concentrations could inhibit the PCR reaction, producing false negative results (Altshuler, 2006; Lorenz, 2012). Since the maximum absorbance of nucleic acids is at a wavelength of 260 nm (Manchester, 1996), while that of proteins is at 280 nm (Teare et al., 1997), and organic solvents and chaotropic salts common to RNA extraction protocols have an absorption maximum at 220–230 nm (Imbeaud et al., 2005; von Ahlfen and Schlumpberger, 2010; Koetsier and Cantor, 2019), the absorbance ratios of 260/280 and 260/230 provide insight into the purity of extracted RNA. For RNA, 260/280 ratios of 1.9 to 2.0 indicate highly purified preparations; ratios above 1.8 typically indicate an acceptable level of RNA purity, whereas ratios lower than 1.8 indicate the presence of contaminants such as proteins. The 260/230 absorption ratio can be used as a secondary measurement of RNA purity for the detection of contaminants such as buffer salts, solvents and other impurities. Even though the ratio of 1.8 has been used as a minimum RNA purity metric by some researchers (Imbeaud et al., 2005), a generally accepted range of optimum or minimum 260/230 ratio values has not been defined (Cicinnati et al., 2008; von Ahlfen and Schlumpberger, 2010; Gallagher, 2017; Zepeda and Verdonk, 2022).
Integrity of RNA is commonly measured by the 2100 Bioanalyzer Instrument (Agilent Technologies, Inc., Santa Clara, CA), which computes an RNA integrity number (RIN). The RIN software algorithm assigns a numerical score to the analyzed RNA sample, ranging from 1 to 10, where 1 indicates completely degraded and 10 the most intact RNA. The method is reliable, independent of RNA concentration- and instrument-based variabilities, but expensive for large sample sets (Mueller et al., 2004; Schroeder et al., 2006). A more affordable option to assess RNA integrity involves estimating the expression levels of endogenous housekeeping genes by RT-qPCR (Imbeaud et al., 2005).
In this study, we compared three commonly used RNA isolation methods for citrus tissues: the MagMAX™-96 Viral RNA Isolation Kit, the RNeasy® Plant Mini Kit, and TRIzol® to determine the quality of the RNA isolated by each kit as measured by its purity, concentration, and integrity, and its suitability for downstream detection of citrus viruses and viroids using various RT-PCR and RT-qPCR assays. In addition, we show the application of the selected RNA isolation protocol in a high-throughput sample processing system at the Citrus Clonal Protection Program-National Clean Plant Network (CCPP-NCPN), at the University of California (UC), Riverside and the California Department of Food and Agriculture (CDFA), which allowed testing of over 16,500 samples from citrus budwood source trees between 2010 and 2019, and the successful implementation of the mandatory “Citrus Nursery Stock Pest Cleanliness Program” (California Senate Bill SB-140, 2009-10; California Code of Regulations, Title 3, Section §3701).
2 Materials and methods
2.1 Plant materials and sample collection and handling
Thirty-three citrus trees infected with 11 different viruses and viroids, in single and mixed infections, and ten non-infected trees of nine different citrus varieties were used for the comparative analysis of the tested RNA isolation methods (Tables 1, 2). Trees were maintained in the greenhouses and screenhouses of the Citrus Clonal Protection Program (CCPP) at the Rubidoux Quarantine Facility, UC Riverside and at the CCPP Lindcove Foundation Facility, UC Agriculture and Natural Resources (UC ANR), Lindcove Research and Extension Center (LREC).
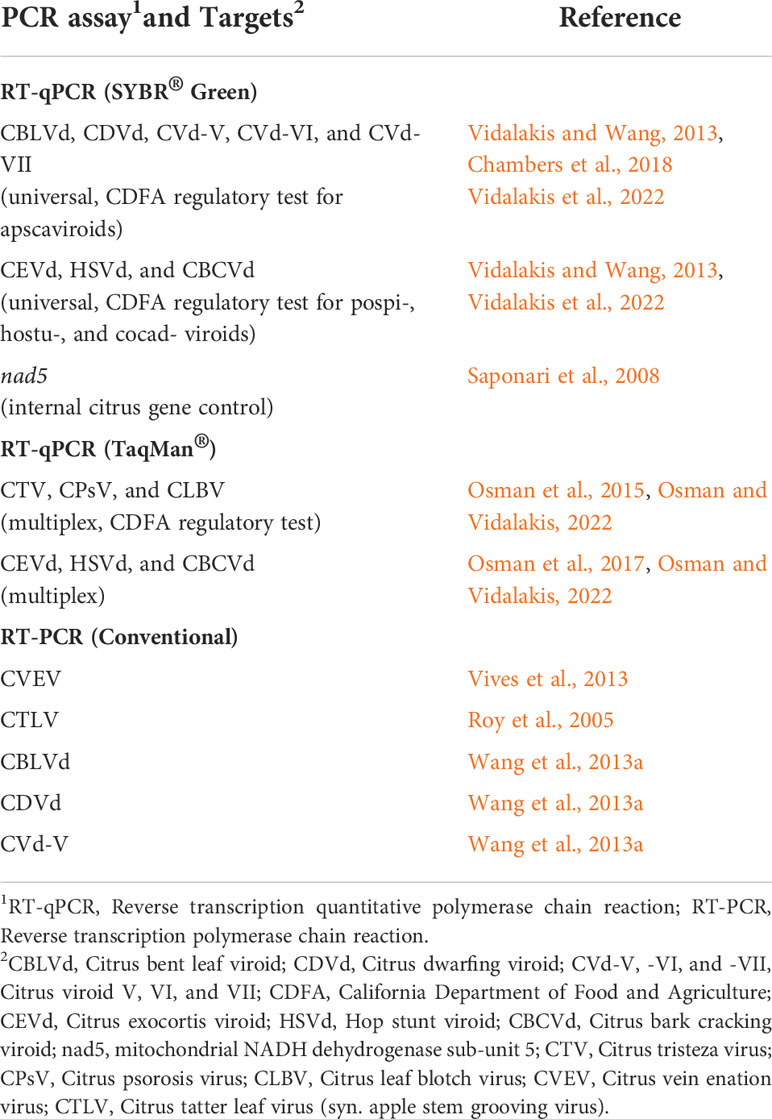
Table 1 List of polymerase chain reaction based assays, their targets and respective references, used in this study.
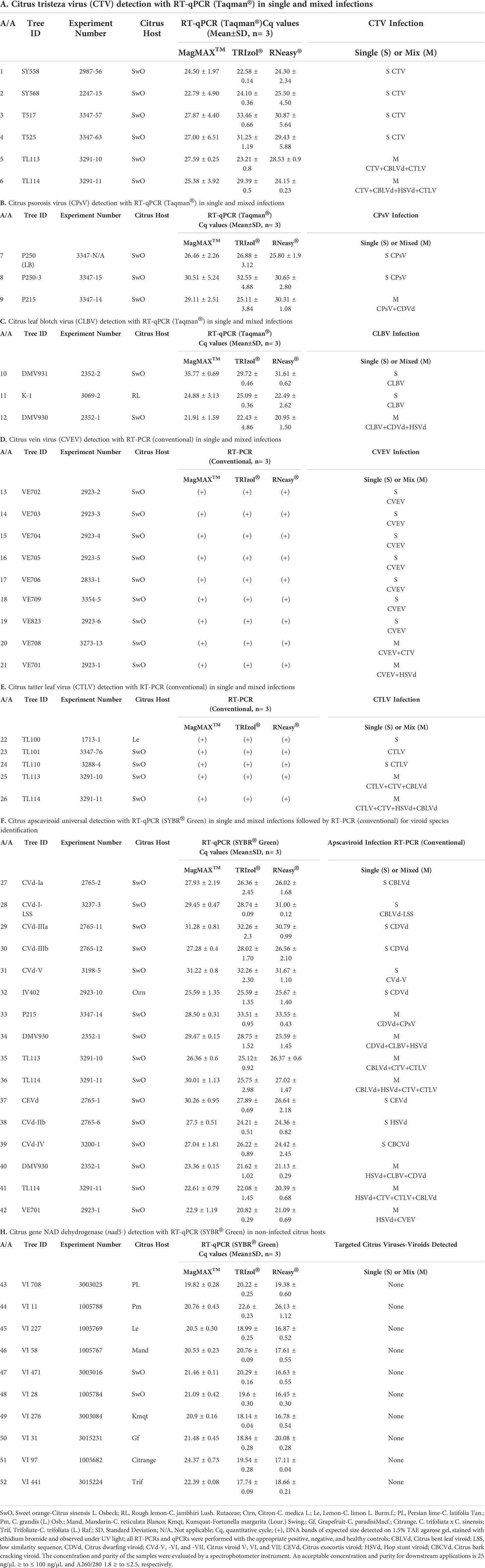
Table 2 Comparison of three RNA isolation methods for the detection of citrus viruses and viroids using reverse transcription (RT) polymerase chain reaction (PCR) and quantitative PCR (qPCR).
Citrus budwood samples (i.e., stems without leaves and thorns) were collected from the last mature vegetative flush (approximately 12 to 18 months old), and at multiple locations around the tree canopy to account for any unequal distribution of the viruses or viroids in the plant. The pruners were sanitized with a 10% household bleach solution (0.5% sodium hypochlorite) and dried with a paper towel to avoid cross contamination between the sampling of each tree. Budwood samples were packaged into a resealable plastic bag, placed on ice, transported to the CCPP, and immediately stored at 4°C until further processing within 10-14 days from collection.
Citrus budwood samples for regulatory virus and viroid testing under California’s Citrus Nursery Stock Pest Cleanliness Program were collected by the CDFA from over 7,000 budwood tree sources in 39 commercially licensed production citrus nurseries throughout the state. Budwood was sampled, shipped, and handled at the CCPP as described above. The RT-qPCR regulatory testing for citrus viruses began in 2014 (CDFA Permit, QC 1388). The citrus viroid regulatory testing included RT-qPCR and bioindexing combined with imprint hybridization for 2010 and 2011. After 2012 the regulatory citrus viroid test was performed by RT-qPCR (CDFA Permit, QC 1354). For viroid bioindexing, bark patches (i.e., blind buds) from the budwood samples were graft-inoculated by T-cut on ‘Etrog’ citron (Citrus medica L.) ‘Arizona-861-S-1’grown on rough lemon (Citrus jambhiri Lush. Rutaceae) rootstock, the bioindicator and bio-amplification host for citrus viroids. Two bark patches from each of two different budwood samples were inoculated onto one ‘Etrog’ citron, for a total of four grafts, to accommodate for the large number of samples (i.e., 3,600) required for bioindexing, limited greenhouse space, and the number of bioindicators. Graft-inoculations, care of bioindicators, and monitoring for symptoms under warm conditions (32–40°C day/24–27°C night) were performed as previously described (Roistacher, 1991; Krueger and Vidalakis, 2022). Approximately 6-8 weeks post inoculation, the ‘Etrog’ citrons were cut back to 5-6 buds, and the second flush was analyzed via imprint hybridization for the detection of the citrus variants of the hop stunt viroid (HSVd, i.e., citrus viroid II) that often causes very mild (i.e., faint leaf tip browning) or no symptoms on ‘Etrog’ citron (Roistacher, 1991; Palacio-Bielsa et al., 1999; Krueger and Vidalakis, 2022).
2.2 Sample processing and pulverization for RNA isolation
For all budwood samples, the phloem-rich bark tissue was peeled using a disposable, single edge razor blade. The peeled bark tissue was finely chopped into small pieces (4-5 mm) on small disposable chipboards, and 250 mg was then placed into a 2 mL safe-lock tube (Eppendorf, Hamburg, Germany). The chipboards were discarded after each sample, and the bench working area was decontaminated with 10% household bleach followed by application of 70% ethanol to remove any residual sodium hypochlorite. For each CCPP sample, three tubes were prepared, one for each of the three different RNA isolation methods tested (see below). For each CDFA sample, one tube was prepared for RNA isolation and regulatory pathogen testing.
All sample tubes were barcoded, kept on ice during processing, sanitized externally by dipping in a series of 10% household bleach and water baths, and placed in a -80°C freezer for at least two hours prior to lyophilization. Samples were lyophilized for 24-26 hours in a FreeZone® Triad™ 74000 freeze-dryer (Labconco®, Kansas City, MO). After lyophilization, a single sterile 4 mm stainless steel grinding ball was added into each sample tube and stored at -80°C until the tissue pulverization and RNA isolation steps.
Sample tubes were placed in stainless steel Cryo-Blocks (SPEX SamplePrep, Metuchen, NJ) and chilled with liquid nitrogen using a Cryo-Station (SPEX SamplePrep) for 20 minutes. Samples were ground into a fine powder using a Geno/Grinder® 2010 (SPEX SamplePrep) at 1,680 rpm for 20 seconds, in two cycles.
The pulverized citrus tissue samples were processed with three different RNA isolation methods as described below.
2.2.1 Modified MagMax™ 96 viral RNA isolation kit - magnetic bead-based method
The pulverized citrus tissues were treated with the MagMAX™ 96 Viral RNA Isolation Kit, utilized with the MagMAX™ Express-96 Deep Well Magnetic Particle Processor (ThermoFisher Scientific, Waltham, MA) following the manufacturer’s recommended protocol adjusted and optimized for citrus tissue. The protocol was as follows; 750 µL of 4 M guanidine lysis buffer (4 M guanidine thiocyanate, 0.2 M sodium acetate pH 5.0, 2 mM EDTA, 2.5% (w/v) PVP-40 at pH 5.0) was added to each sample. Samples were homogenized using the Geno/Grinder® 2010 at 1,680 rpm for 20 seconds, twice. The crude homogenized extracts were incubated at 4°C for 15 minutes and centrifuged at 4°C for 45 minutes at 17,200 x g. RNA was isolated using the default MagMAX™ program “AM1836_DW_50_V2” of the magnetic particle processor, as recommended by the manufacturer. Two mL deep well plates were used for the MagMAX™ Express-96 and were prepared as follows; lysis plate (position 1) which consisted of 139 µL of Lysis/Binding Solution Concentrate (premixed with 40 mL of isopropanol), 22 µL of Bead Mix (10 µL of RNA Binding Beads, 10 µL of Lysis/Binding Enhancer, and 2 µL of Carrier RNA), 139 µL of isopropanol, and 150 µL of the processed supernatant; a first set of wash plates (positions 2-3) which consisted of 500 µL of MagMAX™ Wash Solution 1; a second set of wash plates (positions 4-5); which consisted of 500 µL of MagMAX™ Wash Solution 2; the elution plate (position 6) which consisted of 100 µL of elution buffer; and the tip comb plate (position 7) loaded with the MagMAX™ Express-96 Deep Well Tip Comb. Upon completion of the magnetic particle processor run, the elution plate was placed on a magnetic rack for 5 minutes to collect any residual beads. The isolated RNA was transferred to individual 1.5 µL microcentrifuge tubes and stored in a -80°C freezer.
2.2.2 TRIzol®reagent- phenol chloroform-based method
The pulverized citrus tissues were treated with TRIzol® following the manufacturer’s recommended protocol adjusted and optimized for citrus tissue. The protocol was as follows; 2.5 mL of TRIzol® reagent was added to each sample. Samples were homogenized with a vortex for 20 seconds, centrifuged at 4°C for 5 minutes at 12,000 x g, and the supernatant was then transferred to a new 5 mL tube (Eppendorf, Hamburg, Germany). Five hundred µL (500 µL) of chloroform was added to each sample and incubated at room temperature for 3 minutes. Samples were centrifuged at 4°C for 15 minutes at 12,000 x g. The aqueous phase was transferred to a new 1.5 mL microcentrifuge tube and 1.25 mL of isopropanol was added to each sample. Samples were then incubated at room temperature for 10 minutes and subsequently centrifuged at 4°C for 10 minutes at 12,000 x g. The supernatant was discarded, and 2.5 mL of 75% ethanol was added to each sample to wash the RNA pellet. The samples were vortexed briefly and centrifuged at 4°C for 5 minutes at 7,500 x g. Ethanol was discarded and the pellet was left to air dry for 30 minutes to 1 hour. The RNA pellet was resuspended in 100 µL of UltraPure™ DNase/RNase-free distilled water (Thermo Fisher Scientific, Waltham, MA) and stored at -80°C.
2.2.3 Qiagen RNeasy® plant mini kit - silica column-based method
The pulverized citrus tissues were treated with the Qiagen RNeasy® Plant Mini Kit following the manufacturer’s protocol adjusted and optimized for citrus tissue. The protocol was as follows; 1,125 µL of RLT buffer and 11.25 µL (0.01%) of β-mercaptoethanol were added to each sample and subsequently vortexed. The lysate was transferred to a QIA shredder spin column and centrifuged at room temperature for 2 minutes at 17,200 x g. The flow-through was transferred to a clean 2 mL collection tube and 562.5 µL of 200 proof ethanol was added and mixed by pipetting. Six hundred and fifty µL (650 µL) of sample were transferred to the RNeasy Mini spin column and centrifuged for 15 seconds at 8,000 x g; this step was repeated until the remaining sample was used up. The RNeasy spin column was washed with 700 µL RW1 Buffer and centrifuged for 15 seconds at 8,000 x g at room temperature. A second wash with 500 µL of RPE was added to the column and centrifuged for 15 seconds 8,000 x g, twice. The column was centrifuged at room temperature for 1 minute at 12,000 x g to remove the excess wash buffer. The column was transferred to a clean 1.5 mL standard microcentrifuge tube and 100 µL of UltraPure™ DNase/RNase-free distilled water were added to each sample and incubated at room temperature for 2 minutes. The samples were centrifuged at room temperature for 1 minute at 8,000 x g and the eluted RNA was stored at -80°C.
2.3 RNA quality assessment and RT-qPCR pathogen detection
For all samples, the RNA concentration and purity (ratio of absorbance at a wavelength of 260 nm and 280 nm, 260/280, and 260 nm and 230 nm, 260/230) was assessed using the Infinite M1000 Pro plate reader (Tecan, Männedorf, Switzerland). Integrity of RNA was assessed by an RT-qPCR (SYBR® Green) assay targeting the single-copy endogenous citrus mitochondrial NADH dehydrogenase sub-unit 5 (nad5) gene and a subset of 15 samples (five randomly chosen samples per RNA isolation methodology) were processed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA) using the RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA) to obtain RNA integrity number (RIN) and visualize the RNA qualitative profile.
For the three RNA isolation methods comparison, 10 assays of conventional RT-PCR and RT-qPCR (SYBR®Green and TaqMan®), developed previously by various researchers, were performed to detect five citrus-infecting viruses and seven citrus-infecting viroids following the PCR protocols as described in their respective publications (for specific PCR references see Table 1).
For the regulatory testing of citrus nursery budwood tree sources, all samples were processed by the MagMAX™ protocol. Universal RT-qPCR (SYBR® Green) and multiplex RT-qPCR (TaqMan®) assays were performed for the detection of citrus-infecting viroids and viruses, respectively, according to California’s Citrus Nursery Stock Pest Cleanliness Program (for specific PCR references see Table 1).
Universal RT-qPCR reactions (Saponari et al., 2008; Vidalakis and Wang, 2013; Chambers et al., 2018; Vidalakis et al., 2022) were performed using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA) and multiplex RT-qPCR reactions (Osman et al., 2015; Osman et al., 2017; Osman and Vidalakis, 2022) were carried out using the QuantStudio 12K Flex System (Thermo Fisher Scientific, Waltham, MA).RT-qPCR data was collected and analyzed with the Bio-Rad CFX Manager version 3.1 and the QuantStudio Flex software version 1.3, respectively.
Conventional RT-PCR reactions (Roy et al., 2005; Vives et al., 2013; Wang et al., 2013a) were performed with the ProFlex PCR System (Thermo Fisher Scientific, Waltham, MA). PCR products were analyzed using electrophoresis on a 1.5% Tris-acetate-EDTA (TAE) agarose gel, stained with ethidium bromide and visualized with the ChemiDoc™ Imaging System (Bio-Rad, Hercules, CA).
All RT-PCR and RT-qPCR assays were repeated at least twice for each sample and performed with inclusion of the appropriate positive, negative, and non-template (water) PCR controls.
2.4 Statistical analysis
Mean differences in the concentration (ng/μL), purity (260/280 and 260/230 absorbance ratios), and integrity (Cq values) of RNA obtained from the different RNA isolation methods were analyzed by ANOVA. Prior to performing statistical analysis, the RNA concentration data was log transformed to reduce skewing the original data. Furthermore, Dunn’s test of multiple comparisons was performed to determine significant pair wise differences between different treatment groups (i.e., RNA isolation methods). Interquartile range (IQR), was defined as the distance between the upper (Q3) and lower quartiles (Q1) of the box plots, and calculated as IQR= Q3-Q1, where Q1 (or 25th percentile) is the median of the lower half of the dataset and Q3 (or 75th percentile) is the median of the upper half of the dataset.
3 Results
3.1 Quality assessment of RNA isolated from citrus tissues using three different methods
The concentration, purity, and integrity of the RNA obtained from the three RNA isolation methods tested, were assessed to determine RNA quality and its suitability for downstream use in RT-PCR and RT-qPCR assays for citrus pathogen detection.
Dunn’s multiple comparison test showed significant differences between the RNA concentrations among the three isolation methods tested (adjusted P values < 0.0002). On average, MagMAX™ yielded lower but more consistent RNA concentrations around a median value of 54.4 ng/μL (mean= 57.74 ± 15.93 ng/μL, IQR= 22.65, n= 43) in comparison to TRIzol® and RNeasy® (Figure 1A). The MagMAX™ RNA concentration values clustered within the narrow range of 84.4 ng/μL, with a minimum and maximum of 28.0 and 112.40 ng/μL (Figure 1A). On average, TRIzol® (mean= 473.59 ± 132.44 ng/μL, IQR= 167.4, n= 43) and RNeasy® (mean= 147.83 ± 72.69 ng/μL, IQR= 100.96, n= 43) yielded RNA at higher (ANOVA P < 0.0001) but more variable concentrations than MagMAX™ (Figure 1A). TRIzol® RNA concentrations clustered around a median value of 466.16 ng/μL and ranged within 733.76 ng/μL between a minimum and a maximum of 160.88 and 894.64 ng/μL, respectively (Figure 1A). RNeasy® RNA concentration values were clustered around a median value of 142.00 ng/μL and ranged within 278.88 ng/μL between a minimum and a maximum of 19.12 and 298.00 ng/μL (Figure 1A).
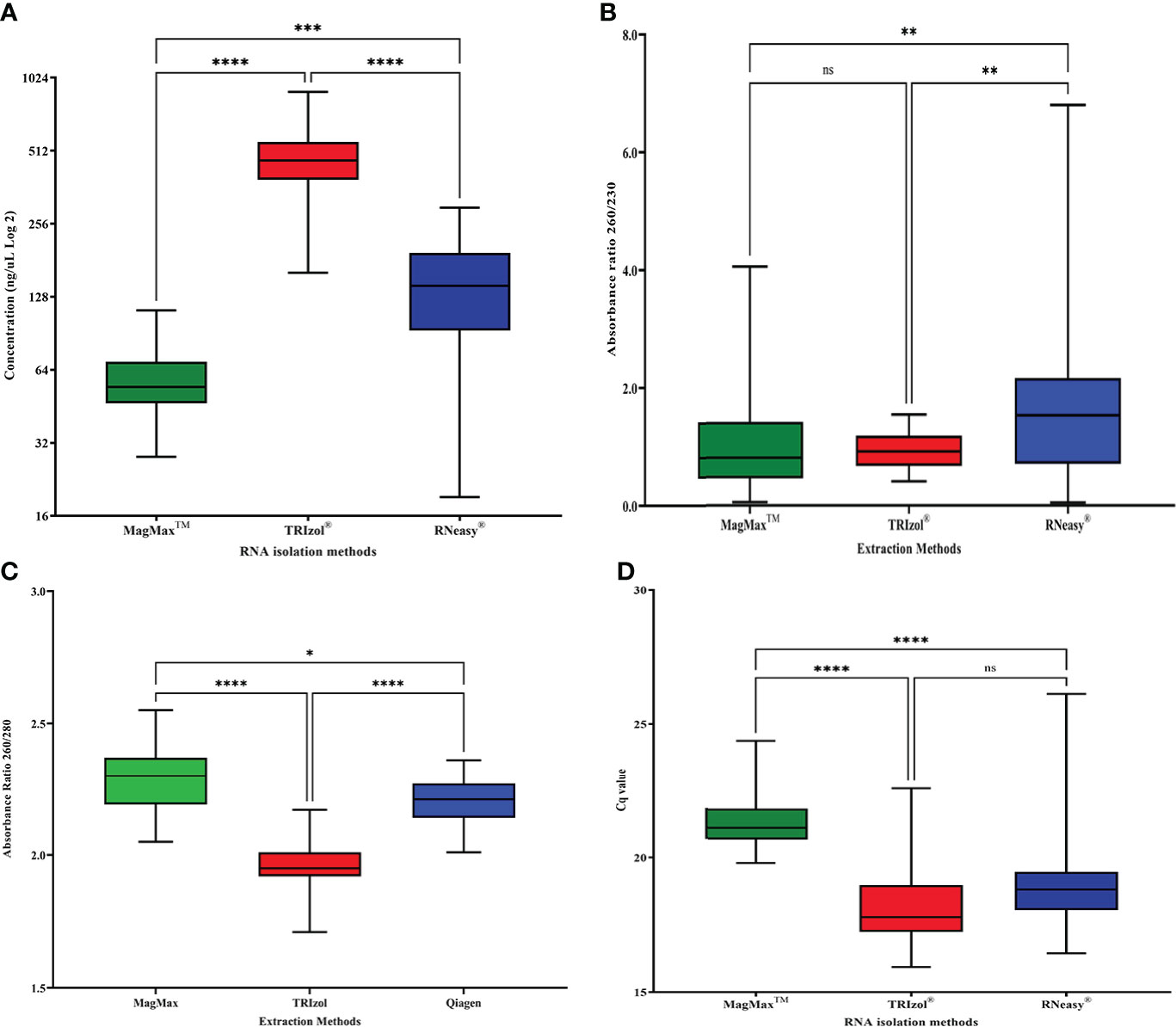
Figure 1 Comparison of (A) RNA concentration (ng/μL) (data transformed to Log 2) (B) RNA purity (absorbance ratio 260/280), (C) RNA purity (absorbance ratio 260/230), and (D) RNA integrity (Cq values for RT-qPCR targeting the NAD dehydrogenase, nad5, citrus housekeeping gene) for three different RNA isolation methods: MagMAX™, TRIzol®, and RNeasy® (n=43). Error bars represent the minimum and maximum values. For the box plot, the lower, middle, and upper lines represent the 25th quartile, median value, and the 75th quartile, respectively. Asterisk (*) indicate statistically significant differences based on Dunn’s test of multiple comparisons (p<0.05), while “ns” indicate not statistically significant.
RNA purity was assessed by the 260/280 absorbance ratio focusing on the optimum value of 2.0 associated with highly purified RNA preparations (Gallagher, 2017), and the lowest acceptable value of 1.8 for RNA preparations with low protein contamination (Imbeaud et al., 2005). Dunn’s multiple comparison test showed significant differences in the 260/280 ratio for the three RNA isolation methods tested (adjusted P value < 0.0179) (Figure 1B). MagMAX™ (mean= 2.30 ± 0.12, n= 43) and RNeasy® (mean= 2.20 ± 0.08, n= 43) isolated RNA with higher 260/280 absorbance ratios than TRIzol® (mean= 1.97 ± 0.09, n= 43) (ANOVA P value: < 0.0001). MagMAX™ yielded RNA with 260/280 ratios around a median value of 2.3 (IQR= 0.18) consistently higher than the optimum ratio of 2.0. The MagMAX™ RNA purity values did not drop below the acceptable 260/280 ratio of 1.8 (minimum=2.05) (Figure 1B). TRIzol® RNA 260/280 ratios clustered around a median value of 1.95 (IQR= 0.09) and the minimum absorbance ratio recorded was 1.71 (Figure 1B). RNeasy® yielded RNA with 260/280 ratios around a median value of 2.21 (IQR= 0.13) consistently higher than the optimum 2.0 value. The RNeasy® RNA purity values did not decrease the acceptable 260/280 ratio of 1.8 (minimum=2.01) (Figure 1B).
A secondary assessment of RNA purity was performed by measuring the 260/230 absorbance ratio, using 1.8 as the acceptable value for the RNA preparations with a small carryover from the extraction reagents (Imbeaud et al., 2005). MagMAX™ (mean= 1.01 ± 0.84, n= 43) and RNeasy® (mean= 1.61 ± 1.19, n= 43) isolated RNA with higher 260/230 absorbance than TRIzol® (mean= 0.92 ± 0.30, n= 43). MagMAX™ yielded RNA with 260/230 ratios around a median value of 0.77 (IQR= 0.96), RNeasy® yielded ratios around a median value of 1.64 (IQR= 1.45), and TRIzol® yielded ratios around a median value of 0.92 (IQR= 0.52). Regardless of extraction method, the average 260/230 ratios for the three extraction protocols fell below one of the recommended values of 1.8 for RNA preparations (Figure 1C).
To determine the integrity of the isolated RNA, RT-qPCR of the citrus housekeeping gene mitochondrial NADH dehydrogenase sub-unit 5 (nad5) was performed. Dunn’s multiple comparison test showed significant differences between nad5 Cq values for RNA isolated by MagMAX™ vs. TRIzol® and MagMAX™ vs. RNeasy® (adjusted P value: <0.0001). There was no significant difference between TRIzol® vs. RNeasy® (adjusted P value: 0.172). MagMAX™ yielded higher but more consistent Cq values for the citrus housekeeping gene nad5 around a median value of 21.12 (mean= 21.30 ± 0.89, IQR= 1.15) in comparison to TRIzol® and RNeasy® (Figure 1D). The MagMAX™ RNA integrity Cq values clustered within the narrow range of 4.55 with a minimum and maximum of 19.82 and 24.37, respectively (Figure 1D). TRIzol® (mean= 18.18 ± 1.46, IQR= 1.74) and RNeasy® (mean= 18.97 ± 1.69, IQR= 1.43) yielded RNA with lower (ANOVA P < 0.0001) but more variable Cq values for nad5 than MagMAX™ (Figure 1D). TRIzol® RNA Cq values for nad5 were lower and more consistent than those of RNeasy® clustered around a median Cq value of 17.8 and ranged within 6.66 Cq values with a minimum and maximum value of 15.94 and 22.60, respectively (Figure 1D). RNeasy® RNA Cq values for nad5 were clustered around a median of 18.83 and had the widest range of 9.67 Cq values with a minimum and maximum of 16.45 and 26.12, respectively (Figure 1D).
RNA integrity of a subset of samples was analyzed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA). Fifteen samples of RNA were randomly selected across the three RNA isolation methodologies (five samples per RNA isolation method) and subjected to analysis to obtain their RIN values and a visual profile of RNA quality. In the case of the five samples extracted by TRIzol®, due to their high concentrations, the RNA was diluted prior to loading into the RNA 6000 Nano chip. Distinct bands were observed at the 18S and 28S in the majority of the samples isolated with TRIzol® (4/5) and MagMAX™ (3/5). Samples isolated with RNeasy® were degraded and the average RIN score was the lowest (mean= 4.32 ± 2.42, n= 5) compared to TRIzol® (mean= 4.36 ± 3.01, n= 5) and MagMAX™ (mean= 5.40 ± 2.69, n= 5) methods. The bioanalyzer results are summarized in Supplementary Figure 1.
3.2 RT-PCR and RT-qPCR citrus pathogen and housekeeping gene detection using RNA isolated by three different methods
RNA obtained from MagMAX™, TRIzol®, and RNeasy® was used in 52 tests with different types of conventional RT-PCR and RT-qPCR assays (i.e., universal, multiplex, singleplex, TaqMan®, and SYBR® Green), targeting 11 citrus-infecting viruses and viroids and one citrus housekeeping gene in various citrus hosts (Table 2).
RNA isolated via the MagMAX™ and RNeasy® protocols was used in PCR reactions without any further manipulation. Some of the TRIzol®isolated RNA had very high concentrations (Figure 1A), and as a result, PCR reactions were inhibited (data not shown). Ten of these samples required a dilution of 1:100 for successful PCR reactions.
Regardless of the RNA isolation method, all types of PCR assays detected consistently their targeted citrus-infecting viruses (Tables 2A-E) and viroids (Tables 2F, G) in single or mixed infections in four different citrus host types. For citrus-infecting viruses, both quantitative (Tables 2A-C) and conventional (Tables 2D, E) PCR assays detected all targeted pathogens using RNA from all three isolation methods tested. For citrus-infecting viroids, the RNA isolated from all three methods was successfully used for both the universal detection and the species-specific identification of the targeted viroids (Tables 2F, G).
The citrus housekeeping gene nad5 was reliably detected in nine different non-infected citrus hosts using RNA from all three isolation methods tested in this study. No pathogens were detected in the non-infected citrus by any of the PCR assays using the RNA isolated by the three tested methods (Table 2H). The positive, negative, and non-template controls used in over 500 PCR tests presented in Table 2 produced the expected results with no evidence of cross contamination or false negative and false positive results (data not shown).
3.3 Application of MagMAX™ RNA isolation method for high-throughput RT-qPCR pathogen detection in California’s citrus nursery stock pest cleanliness program
MagMAX™ was selected for application and further large-scale evaluation in the California’s Citrus Nursery Stock Pest Cleanliness Program for two reasons. Firstly, MagMAX™ produced RNA with the most consistent values of concentration, purity, and integrity without large fluctuations between minimum and maximum values (Figure 1). In addition, MagMAX™ could be used in conjunction with the semi-automated MagMAX™ Express-96 Deep Well Magnetic Particle Processor for high-throughput RNA isolation from citrus tissues.
Between 2010 and 2019, 16,656 samples from citrus nursery budwood source trees, were processed with MagMAX™ and the RNA quality of multiple samples was assessed. Purity of the isolated RNA was high (260/280 ratio 2.22 ± 0.28, n= 6,461) with 85.6% of the 260/280 ratios ranging from 1.8 to 2.5, which is within the desirable values denoting low protein contamination (Figure 2A). Secondary purity assessment for extraction buffer salts and reagents contamination using the 260/230 absorption ratio indicated high amounts of reagent carryover (260/230 ratio mean= 0.93 ± 0.77, n= 6,461). Even though there are no generally accepted values for optimum 260/230 ratios for RNA extraction protocols (Cicinnati et al., 2008; Ahlfen and Schlumpberger, 2010; Gallagher, 2017; Zepeda and Verdonk, 2022), taking into consideration one of the recommended ratio values of 1.8 Imbeaud et al., 2005, 87.5% of the ratios were below that value (Figure 2B). The concentration of the isolated RNA ranged from 8.16 to 256.96 ng/μL, with a mean of 67.97 (± 33.13 ng/μL, n= 6,461), while most of the samples (78.1%) had concentrations from 25 to 100 ng/μL (Figure 2C). RNA integrity was also high and the nad5 citrus gene-targeting RT-qPCR Cq values ranged from 15.4 to 25.43 with a mean of 19.39 (± 1.54, n= 255), regardless of the RNA concentration (Figure 2D).
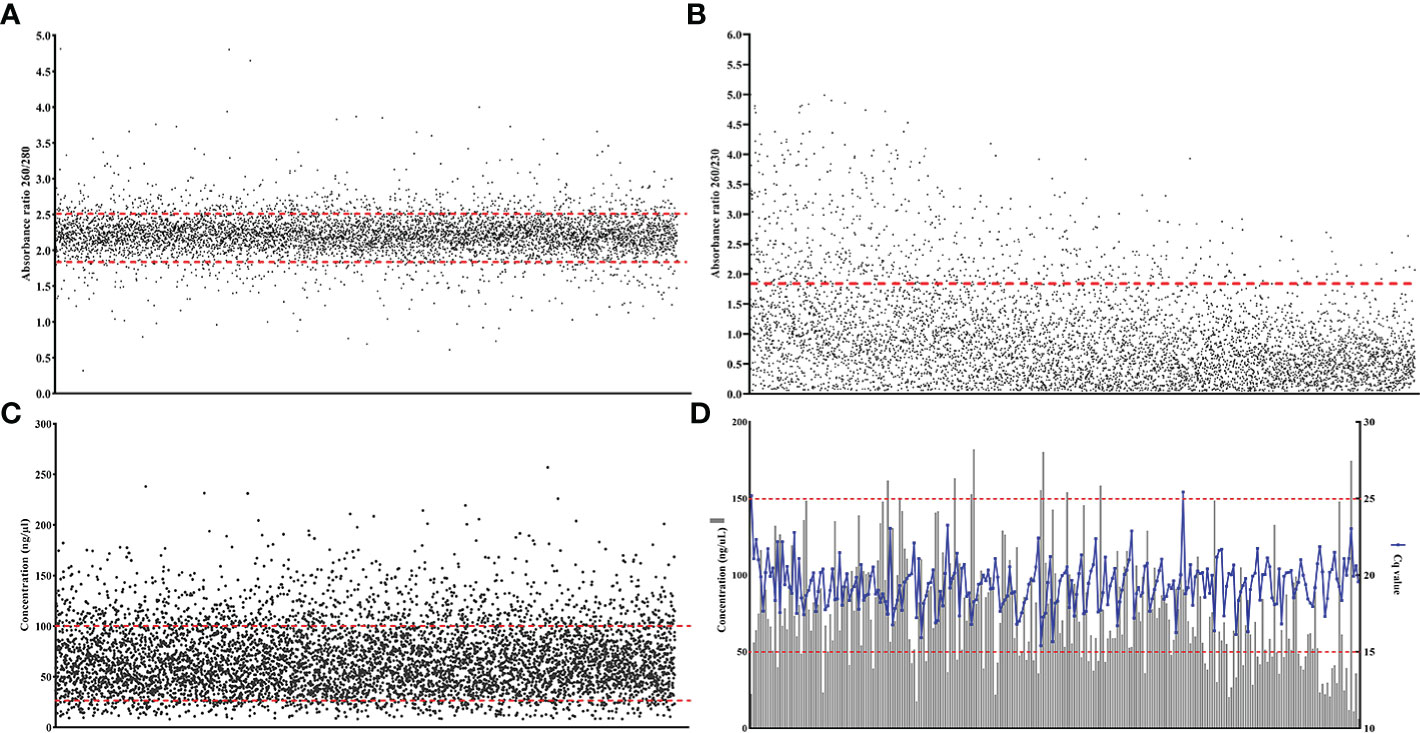
Figure 2 Large scale evaluation of MagMAX™ with citrus nursery samples (A) RNA purity (absorbance ratio 260/280)(n= 6,461) the red lines indicates the optimum range for RNA purity (1.8–2.2, minimal protein contamination), (B) RNA purity (absorbance ratio 260/230)(n= 6,461) the red line indicates one acceptable metric for RNA purity (1.8, buffer salt contamination) (Imbeaud et al., 2005) (C) RNA concentration (ng/μL) (n= 6,461), and (D) RNA integrity (Cq values for RT-qPCR targeting the NAD dehydrogenase, nad5, citrus housekeeping gene)(n= 255).
Between 2004 and 2010, the yearly diagnostic capacity of the citrus nursery testing program in California was on average 455 samples (Figure 3). This was primarily because of the voluntary nature of the program and the limited sample throughput of the approved regulatory test of bioindexing (Mather and McEachern, 1974; Calavan et al., 1978). This low diagnostic capacity in combination with the ease of viroid transmission by grafting or even by a single slash with a knife blade (Barbosa et al., 2005), resulted in viroid infections that persisted in nurseries budwood source trees at an average of 5.67% (Figure 3).
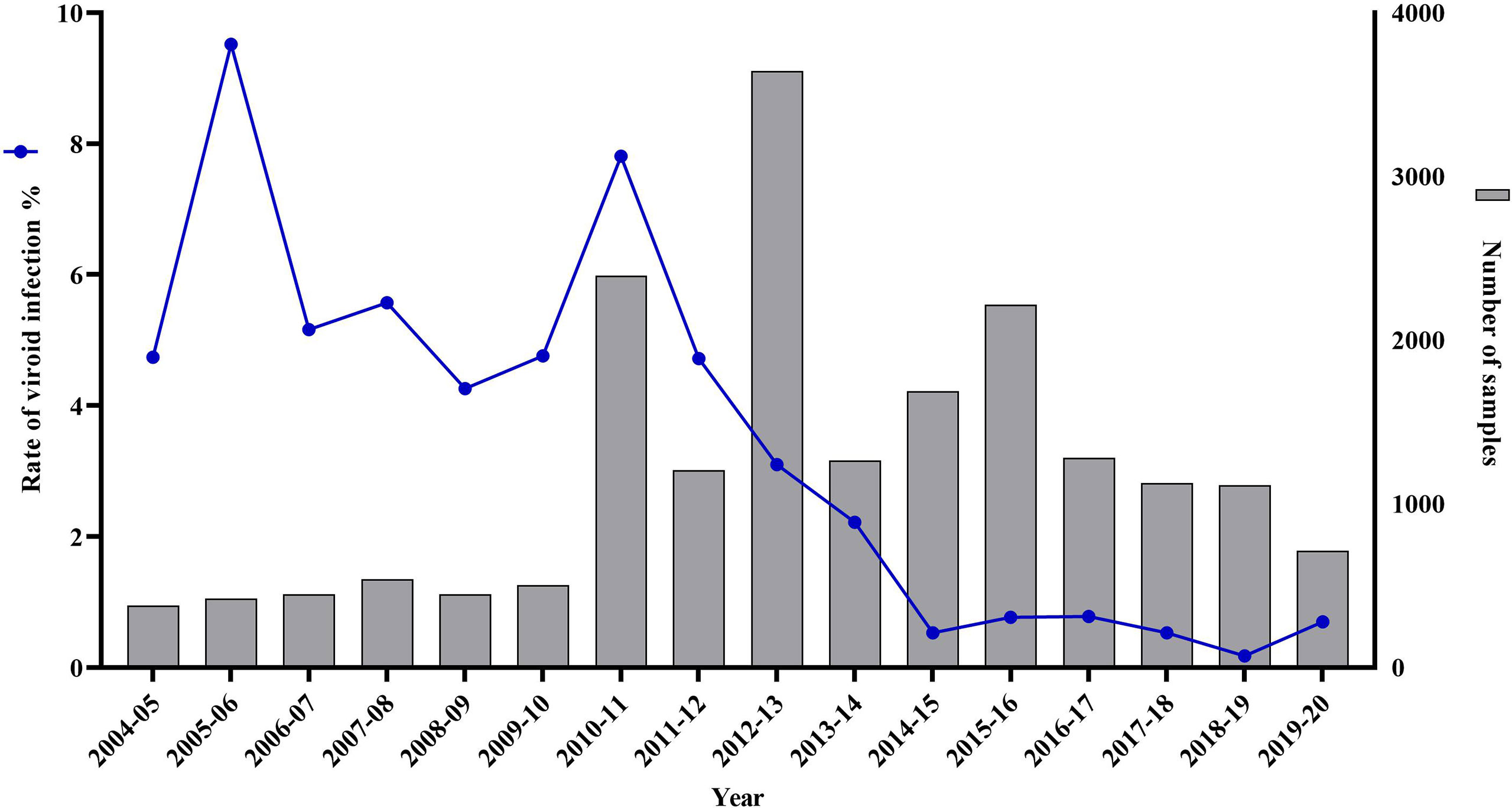
Figure 3 Citrus viroid infection rate and number of citrus nursery samples tested by the Citrus Clonal Protection Program for a 16-year period. From 2004 to 2009 (n= 2,735), viroid testing was limited to biological indexing, combined with imprint hybridization, using ‘Etrog’ citron, Arizona 861-S-1 (Citrus medica L.) indicator plants. From 2010 to 2019 (n= 16,656), viroid testing was performed with the high-throughput MagMAX™-RNA isolation method followed by the RT-qPCR SYBR®Green assays for the universal detection of citrus viroids. For 2010 and 2011 (n= 3,600) both bioindexing and RT-qPCR citrus viroid detection assays were performed.
Between 2010 and 2011, the first two years of the implementation of the mandatory Citrus Nursery Stock Pest Cleanliness Program, the CCPP received 3,600 citrus nursery samples for viroid testing. Bioindexing and imprint hybridization identified 237 samples as viroid-positive with ‘Etrog’ citron expressing the typical symptoms of stem, petiole, and midvein necrosis resulting in various degrees of stunting and leaf epinasty (data not shown). MagMAX™ and the two RT-qPCR SYBR® Green assays for the universal detection of citrus-infecting viroids (Table 1) identified as positive, both in single and mixed infections, the same 237 samples as those identified as positive by bioindexing (Cq apsca viroids: 27.29 ± 4.41, n= 384 and Cq pospi-, hostu-, and cocad- viroids: 27.53 ± 3.34, n= 276).
Following the successful evaluation of the high-throughput MagMAX™ - RT-qPCR system, against bioindexing, the CCPP processed with MagMAX™ and tested with RT-qPCR 13,056 additional samples for citrus-infecting viroids between 2012 and 2019 (Figure 3). The first year of the application of the high-throughput testing (i.e., 2010), viroid infection rate was at 7.81%, then progressively decreased each year. By 2013, the viroid infection rate dropped to 2.22% and from 2014 onwards it decreased further and remained below 0.78% (Figure 3).
Since 2014, when the RT-qPCR regulatory test for citrus viruses was approved, only three samples tested positive for citrus psorosis virus (CPsV, Cq: 24.39 ± 8.74, n= 10) using the MagMAX™ - RT-qPCR system. No citrus tristeza virus (CTV) or citrus leaf blotch virus (CLBV) has been detected in the budwood tree sources of California citrus nurseries. In 2020, after a series of workshops and ring tests, the CCPP completed the technology transfer to CDFA, and the MagMAX™ - RT-qPCR system for the Citrus Nursery Stock Pest Cleanliness Program is now operated by CDFA’s Nursery, Seed, and Cotton Program.
4 Discussion
The California citrus industry has tripled in size in the last 20 years, and it was recently valued at $3.63 billion with an estimated economic impact of $7.6 billion (Babcock, 2022). The spread of the deadly Huanglongbing (HLB) disease in conjunction with a long list of other graft-transmissible diseases (e.g., tristeza stem pitting-CTV, exocortis-CEVd and cachexia-HSVd) all threatening the economy of the industry, make reliable citrus pathogen detection tools an absolute requirement to ensure the production and maintenance of pathogen-tested citrus propagative materials for use by the citrus nurseries (Gottwald, 2010; Gergerich et al., 2015; da Graça et al., 2016; Zhou et al., 2020; Graham et al., 2020).
With the advent of new technologies in recent years, tools for the detection of graft-transmissible pathogens of citrus have significantly evolved. More specifically, the adoption of PCR-based assays in citrus pathogen detection plays a critical role in disease management programs monitoring the sanitary status of germplasm and propagative materials, nursery stock, and commercial plantings of citrus and other agriculturally significant crops (Bostock et al., 2014; Gergerich et al., 2015; Osman et al., 2015; Albrecht et al., 2020; Fuchs et al., 2021). While the protocols for the development and validation of PCR assays are focused primarily on primer and probe sequence design, conditions, composition and efficiency of the reaction as well as the specificity, sensitivity, transferability and robustness of the PCR assay (Bustin et al., 2009; Broeders et al., 2014; Tan et al., 2019), the importance of the quality of nucleic acids isolated from a specific type of plant tissue that is used in the PCR assays cannot be overlooked (Fleige and Pfaffl, 2006; Becker et al., 2010; Taylor et al., 2010; Die and Román, 2012; Huma et al., 2020).
Our study demonstrated that the three tested RNA isolation methods could be reliably used in citrus surveys or germplasm programs as they produced high quality RNA from citrus phloem-rich tissues (Figure 1). MagMAX™ yielded lower but more consistent RNA concentrations while TRIzol® and RNeasy® yielded RNA at a higher, but far more variable concentration range that often required serial dilution for downstream application. All three isolation methods produced RNA with desirable 260/280 absorbance ratios, averaging very close to the target value of 2.0, indicating low protein contamination (Figure 1B). The low values of the 260/230 absorption ratios also indicated that all three isolation methods performed comparably with extraction reagents carryover into the isolated RNA (Figure 1C). Since all three tested RNA isolation methods are guanidine-based, the observed low 260/230 absorption ratio values were most likely due to guanidine carryover, which absorbs very strongly at 220–230 nm (Ahlfen and Schlumpberger, 2010). Despite these low 260/230 ratio values, all three isolation methods produced RNA that was successfully used for the detection of all targeted graft-transmissible pathogens of citrus using various RT-PCR-based assays without any reaction inhibition (Table 2). Our comparative and regulatory testing results using over 6,000 samples are in agreement with multiple studies that have demonstrated that there is no correlation between 260/230 absorbance ratios of RNA extracts and performance of downstream RT-PCR or RT-qPCR analysis (Figure 2B) (Cicinnati et al., 2008; Ahlfen and Schlumpberger, 2010; Gallagher, 2017; Zepeda and Verdonk, 2022). Specifically for guanidine, it has been calculated that up to 100 mM in an RNA sample does not compromise RT-PCR reactions (Ahlfen and Schlumpberger, 2010). Therefore in the case of RNA isolation from citrus tissues using guanidine, the 260/230 absorption ratios should be used as a complimentary RNA purity measurement, and should never be used as a major determinant of a sample’s suitability for pathogen testing using RT-PCR and RT-qPCR based assays (Zepeda and Verdonk, 2022).
Although all three RNA isolation methods were proven valuable and could have different citrus applications, the consistency of the RNA quality isolated from citrus tissues with the semi-automated MagMAX™ method, as previously reported for grapevine and lily (Osman et al., 2012; Sun et al., 2014) allowed its application in the high-throughput Citrus Nursery Stock Pest Cleanliness Program in California. Beyond the development and validation of the MagMAX™ - RT-qPCR system, its successful implementation was supported by the integration of robotic pipetting systems and a series of standard operating procedures for swab testing and decontamination protocols allowing the technical personnel, regardless of experience level, to use the method with consistent results with little or no troubleshooting needs (Osman and Vidalakis, 2022; Vidalakis et al., 2022; Dang et al., 2022). This systems approach allowed the use of MagMAX™ isolated nucleic acids not only for pathogen detection but also for high-throughput sequencing-based microbiome field and greenhouse studies for different citrus tissue types (i.e., stems, leaves, and roots) (Ginnan et al., 2018; Ginnan et al., 2020; Pagliaccia et al., 2020).
It is worth noting here that this project was just a small but important piece of a complex collaborative effort among industry, university, and regulators. Following the first Asian citrus psyllid (ACP) (Diaphorinacitri Kuwayama) detection in southern California in 2008 (Kumagai et al., 2013), the CDFA in collaboration with the citrus industry of the state, organized a series of stakeholder’s meetings to discuss actions to protect nurseries and citrus propagative materials. In 2009, the draft regulations for the “Citrus Nursery Stock Pest Cleanliness Program”, as prepared by the California Citrus Nursery Board, Registration and Certification Protocol Committee, which included pathogen testing and insect protective structures, were finalized at the “Meeting the Challenge of the Asian Citrus Psyllid in California Nurseries” workshop (June 11-12, 2009, https://acpnurseryworkshop.ucr.edu/), a meeting organized by the California Citrus Nursery Society, UC Riverside and the United States Department of Agriculture (USDA). Subsequently, the 2009 California Senate Bill 140 was approved on November 2, 2009. On May 17, 2010, the regulations were filed as an emergency action based on the authority conveyed to CDFA by the California Food and Agricultural Code Sections 6940-6946. The importance of a high-throughput nucleic acid isolation protocol from citrus tissues for the success of the Citrus Nursery Stock Pest Cleanliness Program was not fully appreciated at the time.
The timely testing for viruses and viroids of the 7,245 samples from nursery owned budwood source trees of commercially used citrus varieties in the first three years of the program (i.e., 2010-12, Figure 3) became the key for the successful implementation of the new citrus nursery testing program, which, in addition to testing for HLB, included mandatory testing for CTV, CPsV and all known citrus viroid species. From 1933, when the viral nature of the citrus psorosis disease was discovered by Dr. H. S. Fawcett at the Citrus Experiment Station at Riverside, California, until 2010, when the MagMAX™ RNA isolation protocol for citrus tissues was developed by the CCPP, California’s citrus nursery testing program relied on symptom observation and biological indexing for the detection of viruses and viroids (Hiltebrand, 1957; Mather, 1968; Mather McEachern, 1974; Calavan et al., 1978; Dang et al., 2022). Therefore, the number of samples tested was limited by the availability of appropriate greenhouse space for plant indicator growth, graft-inoculation, and observations for symptom development for six or more months (Roistacher, 1991; Krueger and Vidalakis, 2022). During this study, in 2010-11, the diagnostic capacity of the bioindexing program increased from approximately 500 to 1,800 samples per year with the use of double nursery sample inoculation on ‘Etrog’ citron. However, even with such an increase, it would have taken five years or more to test the 7,000 nursery owned budwood source trees without the high-throughput MagMAX™ - RT-qPCR system, significantly delaying the rollout of the new critical citrus nursery program. By 2015-16, the risk for the citrus propagative material in California would have increased dramatically since the ACP had already spread throughout southern California by 2014, and the first HLB-positive tree was detected in Los Angeles in 2012 (Kumagai et al., 2013; Bayles et al., 2017).
The collaborative efforts of the industry in adopting the idea and funding the research, the university that performed the research and provided extension and outreach services, and the regulatory agencies that approved the new MagMAX™ - RT-qPCR protocols and participated in the technology transfer that the research and regulatory efforts described here were successful. Every collaborative step happened in a timely manner and helped the California citrus industry to transition successfully into the HLB era with a minimal risk of graft-transmissible diseases contaminating its germplasm sources and nursery production. The success of the Citrus Nursery Stock Pest Cleanliness Program was an integral part and complemented the successful implementation of the ACP and HLB management programs by the CDFA and the California Citrus Pest and Disease Prevention Committee (California Assembly Bill AB-281, 2009) (Albrecht et al., 2020; Graham et al., 2020; Garcia-Figuera et al., 2021; McRoberts et al., 2019).
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
GV conceived the study, designed and supervised the experiments. JW, FO, SB and TD designed the experiments. TD, JW, TR, SB, FO, DP, S-HT, AH and GV performed experiments. SC, PQ-L, BR, GU, AS, SH, IM, RC, SA, MV, JB, ED, BN, XC, NS, YH, SA-H processed nursery and other citrus samples and performed experiments as directed by SB, TD, S-HT, IL-C, JW and GV. JK and KW organized, administered and executed the citrus nursery testing program with the CFDA personnel. TD, JW, TR, S-HT, SB, AM, JK, KW and GV analyzed the data and developed the tables and figures. TD, GV, IL-C, KG, PQ-L, AM and SC wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by four California Citrus Nursery Board projects from 2005-2020: “Cooperative Registration Testing of Nursery Owned Citrus Scion and Seed Source Trees”, “The Future of the Cooperative Registration Program of Nursery Owned Citrus Source Trees”, “Citrus Nursery Stock Pest Cleanliness Program-Annual Research, Troubleshoot and Program Review Period” and “Implementation and Streamlining of the Newly Developed High-Throughput Diagnostic System for Citrus Nurseries Registration” awarded to GV. Additionally, funded by two CDFA projects from 2011-2020: “From the Cooperative Registration Testing of Nursery Owned Citrus Scion and Seed Source Trees to the Citrus Nursery Stock Pest Cleanliness Program” and “The Citrus Nursery Stock Pest Cleanliness Program” awarded to GV. Additional support was provided in part by the Citrus Research Board (project 6100), the USDA National Institute of Food and Agriculture (Hatch project 1020106), and the National Clean Plant Network-USDA Animal and Plant Health Inspection Service (11-8100-1544-CA, 12-8100-1544-CA, 14-8130-0419-CA,15-8130-0419-CA, 16-8130-0419-CA, AP17PPQS&T00C118, AP18PPQS&T00C107, AP19PPQS&T00C148) awarded to GV.
Acknowledgments
The authors are acknowledging the Cahuilla people as the Traditional Custodians of the Land on which the experimental work was completed. We are grateful to all past and current, CCPP personnel for their dedicated work and especially for creating and maintaining the in planta CCPP disease bank and for their close collaboration with the CDFA personnel for the execution of the citrus nursery testing program in California since 1962. We acknowledge the vision, forward thinking, support and collaborative spirit of the California citrus nurseries and growers with the University of California and regulatory agencies that made this type of research possible and successfully transferred new technologies from the lab into the field. We would like to acknowledge all the graduate and undergraduate students at the Vidalakis Laboratory, UC Riverside, German Torres-Contreras, Issac Menchaca, Bailey R. Van Zanten, Erika Varady, Jocelyn Sun, for their hard and diligent work in processing thousands of samples and preparing nucleic acids for the PCR reactions. The findings and conclusions in this publication are those of the author(s) and should not be construed to represent any official USDA, CDFA, US, or State government determination or policy. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the UC, CDFA or USDA. UC is an equal opportunity provider and employer.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fagro.2022.911627/full#supplementary-material
Supplementary Figure 1 | Electronic gel image of RNA isolated by the MagMAX™, TRIzol®, and RNeasy® extraction protocols. RNA integrity was analyzed using the RNA 6000 Nano kit in the Agilent 2100 Bioanalyzer system. First lane on the left is the RNA molecular size ladder, followed by the RNA samples. Five samples per extraction method (four virus/viroid-infected and one healthy control) were analyzed. TRIzol®-extracted RNA samples were diluted prior to analysis. RNA quality assessment for each sample summarized below the gel image. The RNA integrity can be evaluated based on the 18S (2kb) and 28S (5kb) rRNA. Well defined bands and a high RNA integrity number (RIN) indicate intact RNA, while faint and shorter fragment size would indicate degradation. Samples extracted with TRIzol® maintained intact RNA, while the RNeasy® extraction showed the most degradation.
References
Ahlfen S. V., Schlumpberger M. (2010). “Effects of low A260/A230 ratios in RNA preparations on downstream applications,” in Qiagen gene expression newsletter.
Albrecht C., Hicks A., Hornbaker V., Khalid S., Kumagai L., Morgan D., et al. (2020). Action plan for Asian citrus psyllid and huanglongbing (Citrus greening) in California. J. Citrus Pathol. 7 (1), 1–56. doi: 10.5070/C471049054
Ali N., Rampazzo R., de C. P., Costa A. D. T., Krieger M. A. (2017). Current nucleic acid extraction methods and their implications to point-of-Care diagnostics. BioMed. Res. Int. 2017, 9306564. doi: 10.1155/2017/9306564
Altshuler M. L. (2006). PCR troubleshooting: The essential guide (Wymondham: Caister Academic Press).
Babcock B. A. (2022). Economic impact of california’s citrus industry in 2020. J. Citrus Pathol. 9, 1–19. doi: 10.5070/C49156433
Barbosa C. J., Pina J. A., Pérez-Panadés J., Bernad L., Serra P., Navarro L., et al. (2005). Mechanical transmission of citrus viroids. Plant Dis. 89, 749–754. doi: 10.1094/PD-89-0749
Bar-Joseph M., Garnsey S. M., Gonsalves D., Moscovitz M., Purcifull D. E., Clark M. F., et al. (1979). The use of enzyme-linked immunosorbent assay for detection of citrus tristeza virus. Phytopathology 69, 190–194. doi: 10.5070/C586v8r487
Bayles B. R., Thomas S. M., Simmons G. S., Grafton-Cardwell E. E., Daugherty M. P. (2017). Spatiotemporal dynamics of the Southern California Asian citrus psyllid (Diaphorinacitri) invasion. PLoS One 12, e0173226. doi: 10.1371/journal.pone.0173226
Becker C., Hammerle-Fickinger A., Riedmaier I., Pfaffl M. W. (2010). mRNA and microRNA quality control for RT-qPCR analysis. Methods 50, 237–243. doi: 10.1016/j.ymeth.2010.01.010
Benítez-Galeano M. J., Hernández-Rodríguez L., Dalmao F., Bertoni E., Bertalmío A., Rubio L., et al. (2021). First comprehensive sanitary report of citrus-infecting viruses and viroids in Uruguay. J. Citrus Pathol. 8 (1), 1–11. doi: 10.5070/C481049181
Bertolini E., Moreno A., Capote N., Olmos A., de Luis A., Vidal E., et al. (2008). Quantitative detection of citrus tristeza virus in plant tissues and single aphids by real-time RT-PCR. Eur. J. Plant Pathol. 120, 177–188. doi: 10.1007/s10658-007-9206-9
Bester R., Karaan M., Cook G., Maree H. J. (2021). First report of citrus virus a in citrus in south Africa. J. Citrus Pathol. 8 (1), 1–11. doi: 10.5070/C481049000
Bostock R. M., Thomas C. S., Hoenisch R. W., Golino D. A., Vidalakis G. (2014). Plant health: How diagnostic networks and interagency partnerships protect plant systems from pests and pathogens. Calif. Agric. 68, 117–124. doi: 10.3733/ca.v068n04p117
Braswell W. E., Park J.-W., Stansly P. A., Kostyk B. C., Louzada E. S., da Graça J. V., et al. (2020). Root samples provide early and improved detection of CandidatusLiberibacter asiaticus in citrus. Sci. Rep. 10, 16982. doi: 10.1038/s41598-020-74093-x
Broeders S., Huber I., Grohmann L., Berben G., Taverniers I., Mazzara M., et al. (2014). Guidelines for validation of qualitative real-time PCR methods. Trends Food Sci. Technol. 37, 115–126. doi: 10.1016/j.tifs.2014.03.008
Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Calavan C. E., Mather S. M., Mceachern E. H. (1978). “Registration, certification, and indexing of citrus trees,” in The citrus industry vol. IV. crop protection. Eds. Reuther W., Calavan E. C., Carman G. E. (Berkeley, CA: University of California, Division of Agricultural Sciences), 185–222.
Chambers G. A., Donovan N. J., Bodaghi S., Jelinek S. M., Vidalakis G. (2018). A novel citrus viroid found in Australia, tentatively named citrus viroid VII. Arch. Virol. 163, 215–218. doi: 10.1007/s00705-017-3591-y
Cicinnati V. R., Shen Q., Sotiropoulos G. C., Radtke A., Gerken G., Beckebaum S. (2008). Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer 8. doi: 10.1186/1471-2407-8-350
da Graça J. V., Douhan G. W., Halbert S. E., Keremane M. L., Lee R. F., Vidalakis G., et al. (2016). Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 58, 373–387. doi: 10.1111/jipb.12437
Damaj M. B., Beremand P. D., Buenrostro-Nava M. T., Riedel B., Molina J. J., Kumpatla S. P., et al. (2009). Reproducible RNA preparation from sugarcane and citrus for functional genomic applications. Int. J. Plant Genomics 2009, 765367. doi: 10.1155/2009/765367
Dang T., Osman F., Wang J., Rucker T., Bodaghi S., Tan S.-H., et al. (2022). “High-throughput RNA extraction from citrus tissues for the detection of viroids,” in Viroids: Methods and protocols. Eds. Rao A. L. N., Lavagi-Craddock I., Vidalakis G. (New York, NY: Springer US), 57–64.
de Noronha Fonseca M. E., Marcellino L. H., Gander E. (1996). A rapid and sensitive dot-blot hybridization assay for the detection of citrus exocortis viroid in citrus medica with digoxigenin-labelled RNA probes. J. Virol. Methods 57, 203–207. doi: 10.1016/0166-0934(95)01976-6
Die J. V., Román B. (2012). RNA Quality assessment: A view from plant qPCR studies. J. Exp. Bot. 63, 6069–6077. doi: 10.1093/jxb/ers276
Fleige S., Pfaffl M. W. (2006). RNA Integrity and the effect on the real-time qRT-PCR performance. Mol. Aspects Med. 27, 126–139. doi: 10.1016/j.mam.2005.12.003
Fuchs M., Almeyda C. V., Al Rwahnih M., Atallah S. S., Cieniewicz E. J., Farrar K., et al. (2021). Economic studies reinforce efforts to safeguard specialty crops in the United States. Plant Dis. 105, 14–26. doi: 10.1094/PDIS-05-20-1061-FE
Gallagher S. R. (2017). Quantitation of DNA and RNA with absorption and fluorescence spectroscopy. Curr. Protoc. Immunol. 116, A.3L.1–A.3L.14.
Gambino G., Perrone I., Gribaudo I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525. doi: 10.1002/pca.1078
Garcia-Figuera S., Deniston-Sheets H., Grafton-Cardwell E., Babcock B., Lubell M., McRoberts N. (2021). Perceived vulnerability and propensity to adopt best management practices for huanglongbing disease of citrus in California. Phytopathology 111, 1758–1773. doi: 10.1094/PHYTO-12-20-0544-R
Garnsey S. M., Civerolo E. L., Gumpf D. J., Paul C., Hilf M. E., Lee R. F., et al. (2005). “Biological characterization of an international collection of citrus tristeza virus (CTV) isolates,” in International Organization of Citrus Virologists Conference Proceedings (Riverside, CA: IOCV 1957-2010), 16, 75–93. doi: 10.5070/C53nj1r1gt
Garnsey S. M., Permar T. A., Cambra M., Henderson C. T. (1993). “Direct tissue blot immunoassay (DTBIA) for detection of citrus tristeza virus (CTV),” in International Organization of Citrus Virologists Conference Proceedings, (Riverside, CA: IOCV. 1957-2010) 12, 39–50. doi: 10.5070/C55t45z8fb. Available at: https://escholarship.org/uc/item/5t45z8fb
Gasic K., Hernandez A., Korban S. S. (2004). RNA Extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol. Biol. Rep. 22, 437–438. doi: 10.1007/BF02772687
Gergerich R. C., Welliver R. A., Osterbauer N. K., Kamenidou S., Martin R. R., Golino D. A., et al. (2015). Safeguarding fruit crops in the age of agricultural globalization. Plant Dis. 99, 176–187. doi: 10.1094/PDIS-07-14-0762-FE
Ginnan N. A., Dang T., Bodaghi S., Ruegger P. M., McCollum G., England G., et al. (2020). Disease-induced microbial shifts in citrus indicate microbiome-derived responses to huanglongbing across the disease severity spectrum. Phytobiomes J. 4, 375–387. doi: 10.1094/PBIOMES-04-20-0027-R
Ginnan N. A., Dang T., Bodaghi S., Ruegger P. M., Peacock B. B., McCollum G, et al. (2018). Bacterial and fungal next generation sequencing datasets and metadata from citrus infected with “CandidatusLiberibacter asiaticus”. Phytobiomes J. 2, 64–70. doi: 10.1094/PBIOMES-08-17-0032-A
Gottwald T. R. (2010). Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 48, 119–139. doi: 10.1146/annurev-phyto-073009–114418
Graham J., Gottwald T., Setamou M. (2020). Status of huanglongbing (HLB) outbreaks in Florida, California and Texas. Trop. Plant Pathol. 45, 265–278. doi: 10.1007/s40858-020-00335-y
Hiltebrand W. F. (1957). “Certification program for maintenance of virus-free propagation sources of citrus in California” in International Organization of Citrus Virologists Conference Proceedings, (Riverside, CA: IOCV. 1957-2010) 1, 229–231. doi: 10.5070/C56qm794kp
Huma N., Sahar S., Iftikhar Y., Azhar A., Galani S. (2020). RNA Isolation efficacy of commercial and modified conventional methods for citrus tristeza virus and mRNA internal control amplification. Biologia 75, 1195–1202.
Imbeaud S., Graudens E., Boulanger V., Barlet X., Zaborski P., Eveno E., et al. (2005). Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 33, e56. doi: 10.1093/nar/gni054
Inglis P. W., Pappas M., de C. R., Resende L. V., Grattapaglia D. (2018). Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS One 13, e0206085. doi: 10.1371/journal.pone.0206085
Koetsier G., Cantor E. (2019). A practicle guide to analysing nucleic acid concentration and purity with microvolume spectrophotometer (Ipswich, MA, USA: New England Biolabs Inc).
Krueger R. R., Vidalakis G. (2022). Study and detection of citrus viroids in woody hosts. Methods Mol. Biol. 2316, 3–21. doi: 10.1007/978-1-0716-1464-8_1
Kumagai L. B., LeVesque C. S., Blomquist C. L., Madishetty K., Guo Y., Woods P. W., et al. (2013). First report of CandidatusLiberibacter asiaticus associated with citrus huanglongbing in California. Plant Dis. 97, 283–283. doi: 10.1094/PDIS-09-12-0845-PDN
Li R., Mock R., Huang Q., Abad J., Hartung J., Kinard G. (2008). A reliable and inexpensive method of nucleic acid extraction for the PCR-based detection of diverse plant pathogens. J. Virol. Methods 154, 48–55. doi: 10.1016/j.jviromet.2008.09.008
Liu L., Han R., Yu N., Zhang W., Xing L., Xie D., et al. (2018). A method for extracting high-quality total RNA from plant rich in polysaccharides and polyphenols using dendrobium huoshanense. PLoS One 13, e0196592. doi: 10.1371/journal.pone.0196592
Loconsole G., Saponari M., Savino V. (2010). Development of real-time PCR based assays for simultaneous and improved detection of citrus viruses. Eur. J. Plant Pathol. 128, 251–259. doi: 10.1007/s10658-010-9653-6
Lorenz T. C. (2012). Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. doi: 10.3791/3998
MacKenzie D. J., McLean M. A., Mukerji S., Green M. (1997). Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis. 81, 222–226. doi: 10.1094/pdis.1997.81.2.222
Manchester K. L. (1996). Use of UV methods for measurement of protein and nucleic acid concentrations. Biotechniques 20, 968–970. doi: 10.2144/96206bm05
Martinelli F., Scalenghe R., Davino S., Panno S., Scuderi G., Ruisi P., et al. (2015). Advanced methods of plant disease detection. a review. Agron. Sustain. Dev. 35, 1–25. doi: 10.1007/s13593-014-0246-1
Mather S. M. (1968). Regulations affecting control of citrus virus diseases in California, in International Organization of Citrus Virologists Conference Proceedings, (Riverside, CA: IOCV. 1957- 2010) 4, 369–373. doi: 10.5070/C52sx375f4
Mather S. M., McEachern E. H. (1974). Recent improvements in citrus registration and certification programs in California, in International Organization of Citrus Virologists Conference Proceedings, (Riverside, CA: IOCV. 1957- 2010) 6, 227–230. doi: 10.5070/C589z369g4
McRoberts N., Figuera S. G., Olkowski S., McGuire B., Luo W., Posny D., et al. (2019). Using models to provide rapid programme support for california’s efforts to suppress huanglongbing disease of citrus. Philos. Trans. R. Soc Lond. B Biol. Sci. 374, 20180281. doi: 10.1098/rstb.2018.0281
Mueller O., Lightfoot S., Schroeder A. (2004). “RNA Integrity number (RIN) – standardization of RNA quality control,” in Aligent technologies.
Newbury H. J., Possingham J. V. (1977). Factors affecting the extraction of intact ribonucleic acid from plant tissues containing interfering phenolic compounds. Plant Physiol. 60, 543–547. doi: 10.1104/pp.60.4.543
Osman F., Dang T., Bodaghi S., Vidalakis G. (2017). One-step multiplex RT-qPCR detects three citrus viroids from different genera in a wide range of hosts. J. Virol. Methods 245, 40–52. doi: 10.1016/j.jviromet.2017.03.007
Osman F., Hodzic E., Kwon S.-J., Wang J., Vidalakis G. (2015). Development and validation of a multiplex reverse transcription quantitative PCR (RT-qPCR) assay for the rapid detection of citrus tristeza virus, citrus psorosis virus, and citrus leaf blotch virus. J. Virol. Methods 220, 64–75. doi: 10.1016/j.jviromet.2015.04.013
Osman F., Olineka T., Hodzic E., Golino D., Rowhani A. (2012). Comparative procedures for sample processing and quantitative PCR detection of grapevine viruses. J. Virol. Methods 179, 303–310. doi: 10.1016/j.jviromet.2011.11.008
Osman F., Vidalakis G. (2022). Real-time detection of viroids using singleplex and multiplex quantitative polymerase chain reaction. Methods Mol. Biol. 2316, 181–194. doi: 10.1007/978-1-0716-1464-8_16
Pagliaccia D., Bodaghi S., Chen X., Stevenson D., Deyett E., De Francesco A., et al. (2020). Two food waste by-products selectively stimulate beneficial resident citrus host-associated microbes in a zero-runoff indoor plant production system. Front. Sustain. Food Syst. 4. doi: 10.3389/fsufs.2020.593568
Palacio-Bielsa A., Foissac X., Duran-Vila N. (1999). Indexing of citrus viroids by imprint hybridisation. Eur. J. Plant Pathol. 105, 897–903. doi: 10.1023/A:1008711402856
Papayiannis L. C. (2014). Diagnostic real-time RT-PCR for the simultaneous detection of citrus exocortis viroid and hop stunt viroid. J. Virol. Methods 196, 93–99. doi: 10.1016/j.jviromet.2013.11.001
Porebski S., Grant Bailey L., Baum B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.5402/2012/205049
Portillo M., Fenoll C., Escobar C. (2006). Evaluation of different RNA extraction methods for small quantities of plant tissue: Combined effects of reagent type and homogenization procedure on RNA quality-integrity and yield. physiol. Plant 128, 1–7. doi: 10.1111/j.1399-3054.2006.00716.x
Rivera-Bustamante R. F., Gin R., Semancik J. S. (1986). Enhanced resolution of circular and linear molecular forms of viroid and viroid-like RNA by electrophoresis in a discontinuous-pH system. Anal. Biochem. 156, 91–95. doi: 10.1016/0003-2697(86)90159-4
Rizza S., Nobile G., Tessitori M., Catara A. (2009). Real time RT-PCR assay for quantitative detection of citrus viroid III in plant tissues. Plant 58, 181–185. doi: 10.1111/j.1365-3059.2008.01941
Roistacher C. N. (1991). Graft-transmissible diseases of citrus: Handbook for detection and diagnosis (Food & Agriculture Org).
Roy A., Fayad A., Barthe G., Brlansky R. H. (2005). A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J. Virol. Methods 129, 47–55. doi: 10.1016/j.jviromet.2005.05.008
Ruiz-Ruiz S., Ambrós S., Vives M., del C., Navarro L., Moreno P., et al. (2009). Detection and quantitation of citrus leaf blotch virus by TaqMan real-time RT-PCR. J. Virol. Methods 160, 57–62. doi: 10.1016/j.jviromet.2009.04.012
Salzman R. A., Fujita T., Zhu-Salzman K., Hasegawa P. M., Bressan R. A. (1999). An improved RNA isolation method for plant tissues containing high levels of phenolic compounds or carbohydrates. Plant Mol. Biol. Rep. 17, 11–17. doi: 10.1023/A:1007520314478
Saponari M., Manjunath K., Yokomi R. K. (2008). Quantitative detection of citrus tristeza virus in citrus and aphids by real-time reverse transcription-PCR (TaqMan®). J. Virol. Methods 147, 43–53. doi: 10.1016/j.jviromet.2007.07.026
Schrader C., Schielke A., Ellerbroek L., Johne R. (2012). PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 113, 1014–1026. doi: 10.1111/j.1365-2672.2012.05384
Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., et al. (2006). The RIN: An RNA integrity number for assigning integrity values to RNA measurements. J. BMC Mol. Biol. 7, 1–14. doi: 10.1186/1471-2199-7-3
Sipahioglu H. M., Usta M., Ocak M. (2006). Use of dried high-phenolic laden host leaves for virus and viroid preservation and detection by PCR methods. J. Virol. Methods 137, 120–124. doi: 10.1016/j.jviromet.2006.06.009
Sun N., Deng C., Zhao X., Zhou Q., Ge G., Liu Y., et al. (2014). Extraction of total nucleic acid based on silica-coated magnetic particles for RT-qPCR detection of plant RNA virus/viroid. J. Virol. Methods 196, 204–211. doi: 10.1016/j.jviromet.2013.11.012
Tan S.-H., Osman F., Bodaghi S., Dang T., Greer G., Huang A., et al. (2019). Full genome characterization of 12 citrus tatter leaf virus isolates for the development of a detection assay. PLoS One 14, e0223958. doi: 10.1371/journal.pone.0223958
Taylor S., Wakem M., Dijkman G., Alsarraj M., Nguyen M. (2010). A practical approach to RT-qPCR–publishing data that conform to the MIQE guidelines. Methods 50, S1–S5. doi: 10.1016/j.ymeth.2010.01.005
Teare J. M., Islam R., Flanagan R., Gallagher S., Davies M. G., Grabau C. (1997). Measurement of nucleic acid concentrations using the DyNA quant and the GeneQuant. Biotechniques 22, 1170–1174. doi: 10.2144/97226pf02
Thatcher S. A. (2015). DNA/RNA preparation for molecular detection. Clin. Chem. 61, 89–99. doi: 10.1373/clinchem.2014.221374
Vennapusa A. R., Somayanda I. M., Doherty C. J., Jagadish S. V. K. (2020). A universal method for high-quality RNA extraction from plant tissues rich in starch, proteins and fiber. Sci. Rep. 10, 16887. doi: 10.1038/s41598-020-73958-5
Vidalakis G., Wang J. (2013). Molecular method for universal detection of citrus viroids (United States patent US).
Vidalakis G., Wang J., Dang T., Rucker T., Bodaghi S., Tan S.-H., et al. (2022). “SYBR® green RT-qPCR for the universal detection of citrus viroids,” in Viroids: Methods and protocols. Eds. Rao A. L. N., Lavagi-Craddock I., Vidalakis G. (New York, NY: Springer US), 211–217.
Vives M. C., Velázquez K., Pina J. A., Moreno P., Guerri J., Navarro L. (2013). Identification of a new enamovirus associated with citrus vein enation disease by deep sequencing of small RNAs. Phytopathology 103, 1077–1086. doi: 10.1094/PHYTO-03-13-0068-R
Wang J., Boubourakas I. N., Voloudakis A. E., Agorastou T., Magripis G., Rucker T. L., et al. (2013a). Identification and characterization of known and novel viroid variants in the Greek national citrus germplasm collection: threats to the industry. Eur. J. Plant Pathol. 137, 17–27. doi: 10.1007/s10658-013-0231-6
Wang J., Bozan O., Kwon S.-J., Dang T., Rucker T., Yokomi R. K., et al. (2013b). Past and future of a century old citrus tristeza virus collection: a California citrus germplasm tale. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00366
Yokomi R. K., Saponari M., Sieburth P. J. (2010). Rapid differentiation and identification of potential severe strains of citrus tristeza virus by real-time reverse transcription-polymerase chain reaction assays. Phytopathology 100, 319–327. doi: 10.1094/PHYTO-100-4-0319
Zepeda B., Verdonk J. C. (2022). RNA Extraction from plant tissue with homemade acid guanidinium thiocyanate phenol chloroform (AGPC). Curr. Protoc. 2, e351. doi: 10.1002/cpz1.351
Keywords: graft-transmissible pathogens of citrus, magnetic bead-based RNA isolation, RNA quality, Citrus Clonal Protection Program (CCPP), National Clean Plant Network (NCPN), California Department of Food and Agriculture (CDFA)
Citation: Dang T, Bodaghi S, Osman F, Wang J, Rucker T, Tan S-H, Huang A, Pagliaccia D, Comstock S, Lavagi-Craddock I, Gadhave KR, Quijia-Lamina P, Mitra A, Ramirez B, Uribe G, Syed A, Hammado S, Mimou I, Campos R, Abdulnour S, Voeltz M, Bae J, Dang E, Nguyen B, Chen X, Siddiqui N, Hsieh YT, Abu-Hajar S, Kress J, Weber K and Vidalakis G (2022) A comparative analysis of RNA isolation methods optimized for high-throughput detection of viral pathogens in California’s regulatory and disease management program for citrus propagative materials. Front. Agron. 4:911627. doi: 10.3389/fagro.2022.911627
Received: 02 April 2022; Accepted: 08 July 2022;
Published: 18 August 2022.
Edited by:
David Ezra, Agricultural Research Organization (ARO), IsraelReviewed by:
Giorgio Gambino, National Research Council (CNR), ItalyGarima Singh, Pachhunga University College, India
Copyright © 2022 Dang, Bodaghi, Osman, Wang, Rucker, Tan, Huang, Pagliaccia, Comstock, Lavagi-Craddock, Gadhave, Quijia-Lamina, Mitra, Ramirez, Uribe, Syed, Hammado, Mimou, Campos, Abdulnour, Voeltz, Bae, Dang, Nguyen, Chen, Siddiqui, Hsieh, Abu-Hajar, Kress, Weber and Vidalakis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgios Vidalakis, dmlkYWxnQHVjci5lZHU=
 Tyler Dang1
Tyler Dang1 Sohrab Bodaghi
Sohrab Bodaghi Fatima Osman
Fatima Osman Deborah Pagliaccia
Deborah Pagliaccia Stacey Comstock
Stacey Comstock Paulina Quijia-Lamina
Paulina Quijia-Lamina Jinhwan Bae
Jinhwan Bae Xingyu Chen
Xingyu Chen Noora Siddiqui
Noora Siddiqui Yi Tien Hsieh
Yi Tien Hsieh Georgios Vidalakis
Georgios Vidalakis