
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Virol., 02 March 2023
Sec. Fundamental Virology
Volume 3 - 2023 | https://doi.org/10.3389/fviro.2023.1111335
Visualization of viruses and their hosts has been paramount to their study and understanding. The direct observation of the morphological dynamics of infection is a highly desired capability and the focus of instrument development across a variety of microscopy technologies. This study demonstrates progress that has been made in exploiting the capabilities offered by HS-AFM to characterise the interactions between coccolithoviruses and their globally important coccolithophore hosts. We observe whole Emiliania huxleyi Virus capsids, transient binding to Emiliania huxleyi derived supported lipid bilayers, and host-virus binding in real-time in an environmentally relevant, aqueous environment.
Emiliania huxleyi (E. huxleyi) is the most numerically dominant and widespread coccolithophore in Earth’s oceans, making it a key player in global biogeochemical cycles (1–6). E. huxleyi algal blooms grow rapidly before abruptly disappearing. Implicated in this process are giant, specific, lytic double-stranded DNA viruses, belonging to the Phycodnaviridae group (Emiliania huxleyi Virus, EhV) (7–11).
EhV is a nucleocytoplasmic large dsDNA virus (NCLDV) thought to be roughly spherical shaped with icosahedral arrangement of their capsid protein subunits and 170 – 190 nm in diameter (9). The model species, EhV-86 has a genome size of 407,339 bp and is an enveloped virus (Figure 1).
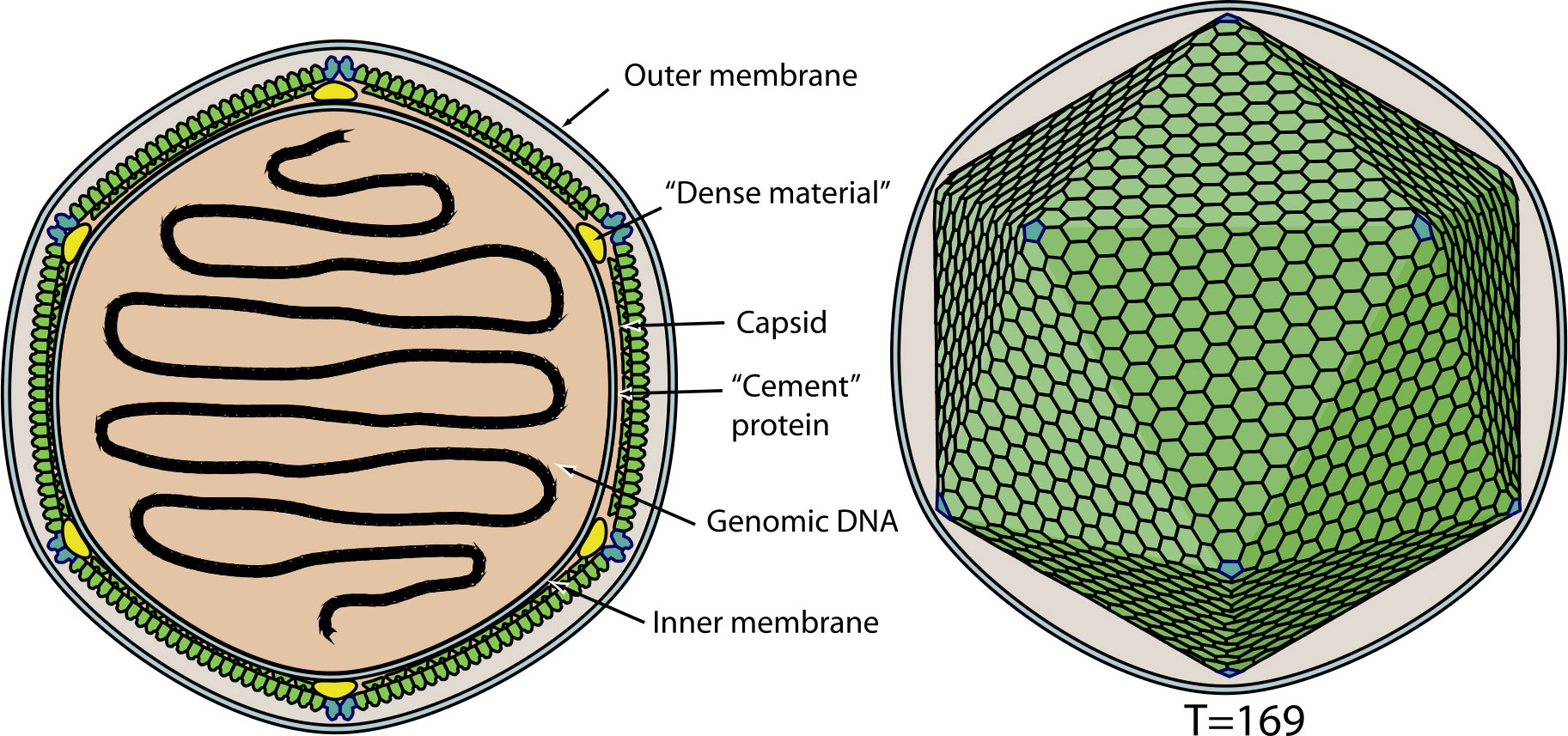
Figure 1 Schematic representation of Emiliania huxleyi Virus. source: Coccolithovirus, ViralZone (www.expasy.ch/viralzone, Swiss Institute of Bioinformatics).
The literature suggests that lipid rafts in cell membranes can be targeted by pathogens (12). For E. huxleyi, it has been hypothesised that these rafts could function as specific points of viral attachment, entry assembly and budding (13, 14). Indeed, EhV induce and employ virus encoded glycosphingolipids (GSLs) during E. huxleyi infection (14–16).
Atomic Force Microscopy (AFM) is a physical probe microscopy technique that measures deflection in a cantilever possessing a sharp tip as it passes over, and interacts with a surface. These measurements can occur in vacuum, air or liquid environments and produces topographical and material property data for the sample. A negative of AFM can be the slow data collection speeds and small field of view resulting from the serial nature of acquisition as the tip moves from pixel to pixel. Improving data acquisition speeds led to High-Speed Atomic Force Microscopy (HS-AFM), essentially producing ‘real-time’ or ‘video’ AFM. HS-AFM provides the ability to visualise individually unique particles, surfaces, and structures of this microalgae-virus system under environmentally and physiologically relevant conditions. Data is collected at high spatial and temporal resolution and without the requirement for labels or fixatives.
In this study we used contact mode HS-AFM to first characterize the viral particle along with isolated Supported Lipid Bilayers (SLBs) made from detergent resistant membrane components, and thereafter whole cell, live membranes. We then introduced purified viral particles to these membranes during live imaging under physiologically relevant conditions to visualize real-time physical interactions between the microalgae, E. huxleyi, and its virus, EhV.
Non-calcifying Emiliania huxleyi cells (CCMP 2090 or 374, National Culture Collection of Marine Phytoplankton for the USA, Bigelow Laboratory for Ocean Sciences, Boothbay, ME, USA) were maintained at 17°C under a 16:8 h light:dark cycle in f/2 media (17) (enriched Atlantic seawater, salinity ~33 ppt determined by refractometer) in a Versatile Environmental Test Chamber (Sanyo MLR-350, Osaka, Japan).
Stock EhV-86/E. huxleyi CCMP 2090 or EhV-99B1/E. huxleyi CCMP 374 lysate stored in low light conditions at 4°C was dead-end filtered through 0.8 µm, followed by 0.45 µm and 0.2 µm filters. Using Amicon Ultra-15 100 KDa molecular weight cut off centrifuge filter tubes (Millipore) viruses were concentrated around 200-fold by centrifugation (4000 x g at 4°C for 60 mins). The membranes were washed with salinity adjusted seawater for maximum viral recovery. Virus number was enumerated using analytical flow cytometry (18, 19). Briefly, samples are fixed in glutaraldehyde (0.5% final concentration) and flash frozen in liquid N2. Samples were then diluted in standard TE buffer (pH 8.0, 10 mM Tris-HCl, 1 mM EDTA) and stained with SYBR Green I (Invitrogen) in the dark at 80°C and examined with a 488 nm laser equipped flow cytometer.
EhV-86 or EhV-99B1 concentrate was added to E. huxleyi CCMP 2090 or CCMP 374 at a 5:1 multiplicity of infection, infections were consistent in their dynamics with full collapse of cell culture after 72 hours, with EhV-99B1/E. huxleyi CCMP 2090 infection systems resulting in a more efficient, lab infection cycle (EhV-99B1/E. huxleyi CCMP 2090 example infection seen in Supplementary Figure 1).
25 µL of 5 mg L-1 poly-l-lysine was incubated on freshly cleaved mica for 30 mins. Next, 25 µL of filtered viral (EhV-86 or EhV-99B1) lysate was incubated overnight (~ 16 hours) at room temperature. The sample was washed with 3 × 1 mL of 0.1 M Phosphate buffer and followed by addition of 25 µL of 0.1% glutaraldehyde for 30 min. Finally, the sample was washed with 5 × 100 µL of MilliQ and allowed to dry at room temperature. This preparation was applied to viruses with and without detergent (Triton X-100) treatments to remove outer lipid envelopes. For imaging of this preparation in liquid, it was ensured that the virus remained hydrated by shorted viral incubation time (30 min) on mica, and included no drying between preparation steps, washing and imaging in 0.1 M phosphate buffer.
E. huxleyi CCMP 374 culture was centrifuged at 5,000 × g to generate a cell pellet and resuspended in 1 mL of lysis buffer (0.5% Brij-96, 25 mM Tris-HCl, 150 mM NaCl, 1 mM PMSF) for 30 min on ice. The lysate was clarified by centrifugation at 4,000 × g at 4°C. The supernatant generated during lysate clarification was loaded into a 5 - 35% discontinuous OptiPrep density gradient. OptiPrep gradient was ultracentrifuged for 16 hrs at 39,000 × g at 4°C. 12 × 1 mL fractions were collected post ultracentrifugation (Method adapted from (20)). These 12 fractions were further purified by dialysis with a 3.5 KDa MWCO membrane for 16 hrs at 4°C into adsorption buffer to remove residual OptiPrep and/or possible residual detergent. The fractions were sonicated for 30 mins and extruded 15 times through a 50 nm nanopore nucleopore membrane. These were then prepared for HS-AFM analysis, with 25 µL of sample incubated on freshly cleaved mica for 90 mins at ambient room temperature. The sample was then washed with 5 × 100 µL of imaging buffer. For imaging in air, this washed sample was then allowed to dry before imaging. For imaging in liquid, a sample covering droplet of imaging buffer was maintained. Fractionated SLB samples and controls were imaged with contact mode HS-AFM in both an air and liquid environment.
For viral recognition either E. huxleyi origin detergent resistant SLBs were prepared, or E. huxleyi CCMP 374 cells were prepared in bespoke 3D printed cell arrays as previously described (21). For the live cell arrays, suitable cells were located using optical and fluorescence microscopy and transferred to HS-AFM for imaging in liquid. 0.2 µm filtered and concentrated (Amicon Ultra 100 KDa MWCO centrifuge Spin Tubes, MERCK) EhV-99B1 lysate was loaded into a 1.1 g cm-1 – 1.4 g cm-3 continuous Caesium Chloride gradient and centrifuged (Ultra-Clear 14 x 89 mm, Beckman) for 2 hours at 25000 RPM at 20 °C with a slow deceleration speed. The generated viral band was isolated by direct syringe extraction through the wall of the centrifuge tube. This collected virus was dialysed against imaging buffer with a 3.5 KDa MWCO dialysis membrane. Virus was flowed into the liquid imaging environment during HS-AFM data collection.
The instrument used was a Bristol Nano Dynamics Ltd. High-Speed Atomic Force Microscope (BND, Bristol, UK) (21) with removable liquid cell. Data was collected in contact mode, with passive mechanical feedback loop, using low spring constant, triangle cantilevers (MSNL-10, 0.01 N m-1 nominal spring constant, Bruker).
Data was collected as video files using BND’s acquisition software. Individual frames were exported for further processing and analysis with Gwyddion (http://gwyddion.net). Data has been processed including basic functions, fixing zero, adjustment to the colour scale, levelling data by mean plane subtraction, 3-point plane, or intersections, correction of horizontal scars, alignment of rows, removal of polynomial background, and integral transforms of the data.
A number of viruses have been studied by AFM including recently Sars-CoV-2 (22, 23), with PBCV-1 being the most well studied algal virus (24). Fewer whole viruses have been studied by HS-AFM, with more commonly viral components such as protein complexes (25) and their interactions with larger objects such as exosomes (26) being investigated. No viruses have been previously imaged by our contact mode HS-AFM prior to this study. Before studying the live virus and algae targets in dynamic systems, we began by confirming virus could be successfully imaged in isolation in both air and liquid conditions.
Seen in Figure 2 is an example of collected viral EhV-86 HS-AFM data in an air environment. In these images the expected icosahedral shape expected from these double stranded NCLDV viruses (27) can be seen. EhV has an external lipid membrane (13) and Figure 2 shows the virus with this external membrane present, due to its smooth topography and lack of distinct texture in the outer surface.
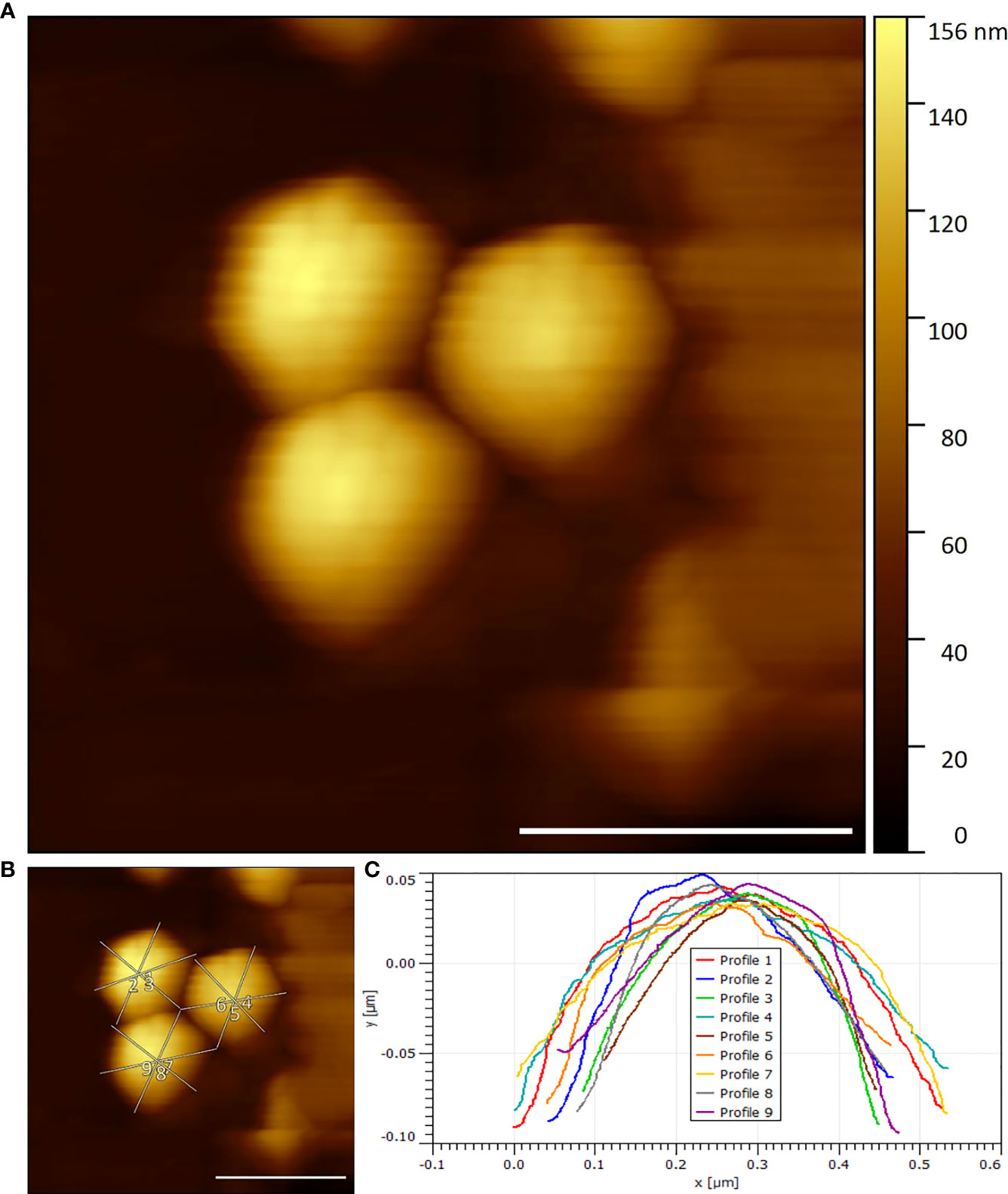
Figure 2 (A) Three EhV-86 viruses visualised by HS-AFM in air (Scale bar 400 nm, Data captured at 2 fps) (B) Example width and height measurement transects (Scale bar 400nm) (C) Extracted transects marked in (B).
The effect of dehydration and air imaging on the viral dimensions or compression of the virus under imaging conditions combined with the tip convolution effects likely results in an increased measured viral width and reduced viral height compared to theoretical values.
Figure 3 gives examples of detergent treated EhV-86 virus samples in an air environment. In this images, contact mode HS-AFM has clearly resolved five high points (the data is true topography rather than pixel intensity). These represent a pentamer of the trimeric capsid proteins at the apex vertex of the viral icosahedral structure.
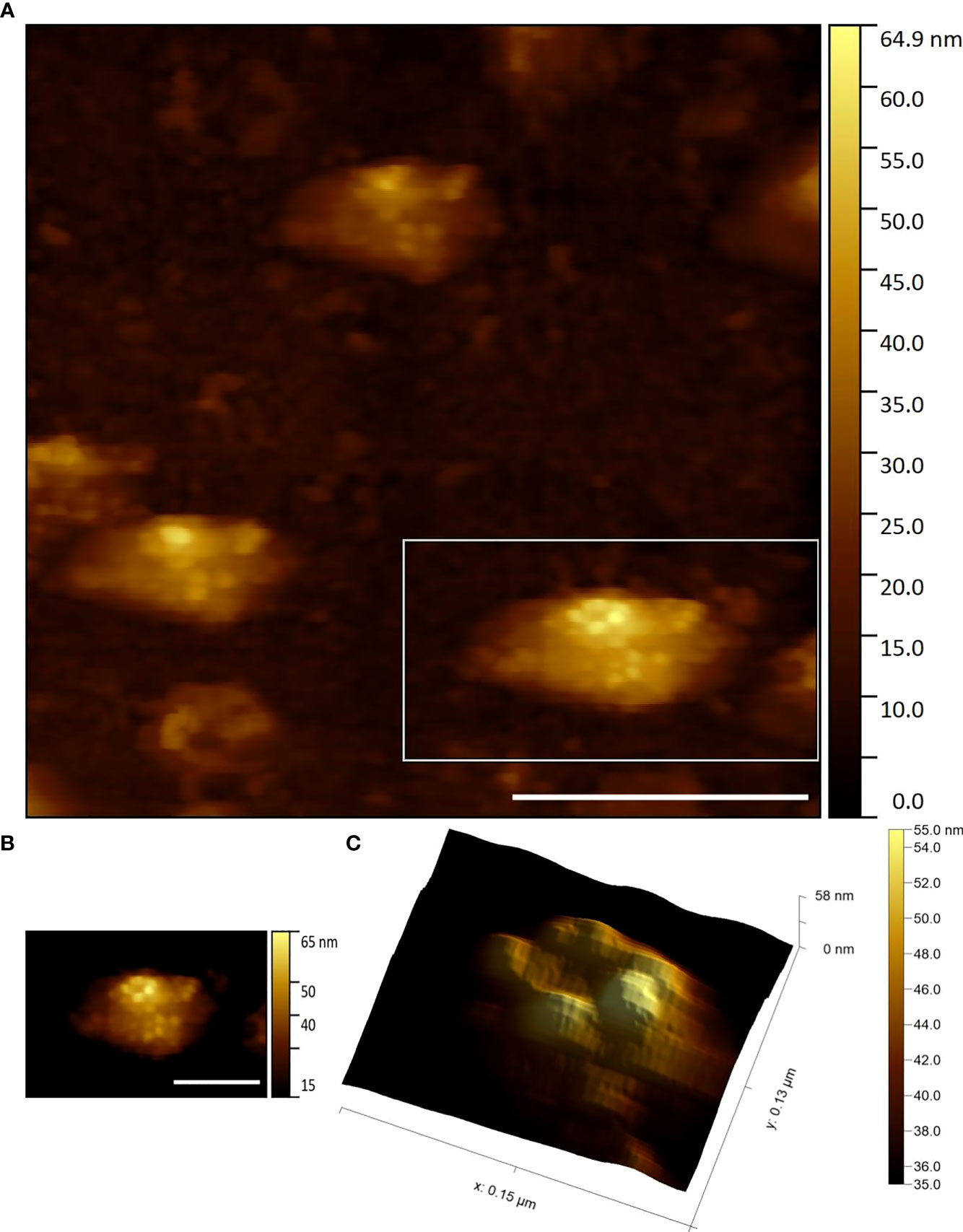
Figure 3 (A) Three EhV-86 viruses showing capsid protein structure visualised by HS-AFM in air (Scale bar 400 nm, Data captured at 2 fps) (B) Zoom of highlighted bottom right virus seen in (A) (Scale bar 200 nm) (C) Zoom of (B) data displaying potential apical pentasymmetron structure (3D render with lighting).
Shown in Figure 4 is the dimensional measurements of EhV-86 and EhV-99B1 under air and liquid conditions. We measured an average EhV viral diameter (N = 103) of 333.6 nm (Min = 147.5, Max = 842.8, SD = 123.8 nm) and average viral height of 50.6 nm (Min = 10.0, Max = 144.6, SD = 31.6 nm). On average, EhV’s diameter was measured higher than theoretically expected and average height was lower than theoretically expected.
The data was manually classified. ‘Large’ viral examples (potentially flattened or completely different particles), potential empty ‘shells’ of viruses and ‘broken’ viruses were removed to generate more precise and accurate viral proportional data and used in the below statistical analysis (Supplementary Table 1).
There is no statistical significance between EhV-86 and EhV-99B1 heights and widths (Two tailed T test assuming unequal variances p = 0.301, p = 0.340 respectively). There is statistical significance between measured virus heights in air vs liquid environments (Two tailed T test assuming unequal variances p = <0.001). There is no statistical significance between air versus liquid widths (Two tailed T test assuming unequal variances p = 0.169). This data gave us the foundations for viral particle identification in later studies.
A model cell membrane system intended to recapitulate live E. huxleyi cell membranes was designed and characterised prior to live cell imaging. SLBs generated from E. huxleyi detergent resistant membrane components were laid over mica surface. This overlays the soft biological feature for analysis over a supporting solid surface in a controlled manner. These bilayers have been used previously in various biophysical research examples as model systems (28).
Vesicle deposition (29) for SLB models relies on the generation of small unilamellar vesicles (SUVs). Upon initial extraction the lipids are expected to form multilamellar vesicles. Further processing of this suspension through sonication and extrusion generates SUVs which can be fused, ruptured, and adsorbed to a solid support to form SLBs.
SLBs were first prepared from model purified lipids. These were 12:0 PC (DLPC, 1,2-dilauroyl-sn-glycero-3-phosphocholine, Avanti Polar Lipids) and 16:0-18:1 PC (POPC, 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine, Avanti Polar Lipids) prepared in a 1:1 ratio. This was a trial of our contact mode HS-AFM suitability for SLB analysis in air and liquid environments. Confident we could clearly distinguish the height differential between the two lipids in the bilayer (Supplementary Figures 2–4) we followed with generation of SLBs by vesicle deposition from E. huxleyi derived cell membrane components as described in the methods (Figure 5).
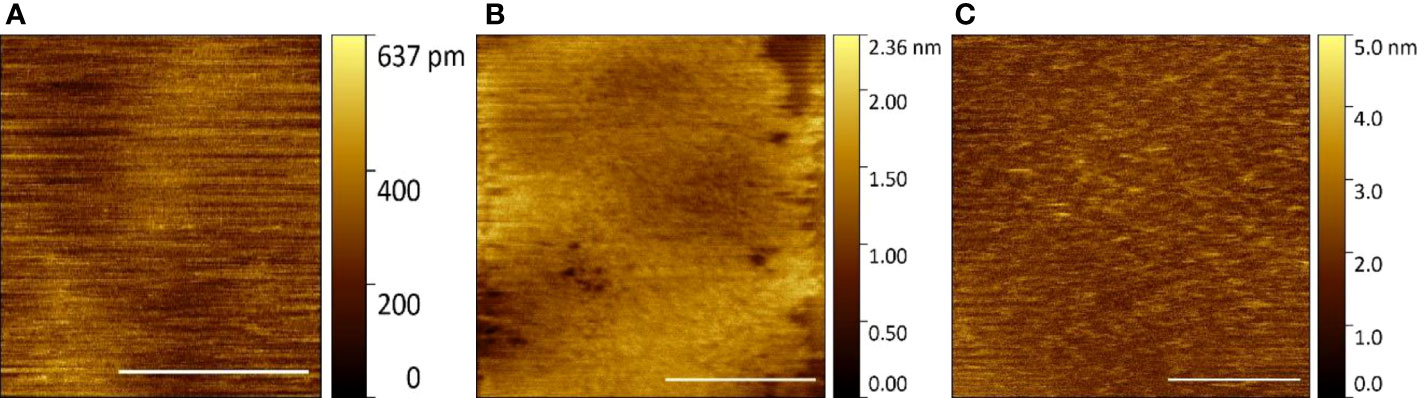
Figure 5 E huxleyi component SLBs visualised by HS-AFM (Scale bar 1 μm, Data captured at 2 fps) (A) Control surface (B) Fraction 2 in air (C) Fraction 2 in liquid.
In air, using fraction 2 of the OptiPrep gradient and its further processing, a bilayer is produced (Figure 5B). However, due to dehydration, this forms a pitted peeling surface that does not completely cover the data collection field of view. In liquid imaging, this fraction shows a uniform complete SLB with nanoscale features that should represent any associated membrane proteins co-purified during preparation (Figure 5C). Fraction 2 was confirmed to generate a SLB on a solid substrate made from natural cell components and is relatively stable with carefully applied tip force during contact mode HS-AFM imaging in a liquid environment.
Upon establishment and characterization of a suitable SLB model of E. huxleyi’s cell membrane, EhV was introduced to the imaging environment via the liquid cell during data collection.
Figure 6 shows a summary graph of the collected data. In total, there were 27 examples of suspected viral binding. These data were isolated, processed and assigned a likelihood score from 0 – 5 (0 being non-virus, and 5 being confirmation of viral presence). The likelihood score is a qualitative measure of the confidence with which the data sequence in question is of EhV-99B1/SLB binding interaction. These interactions ranged from 1 to 25 frames of potential viral presence.
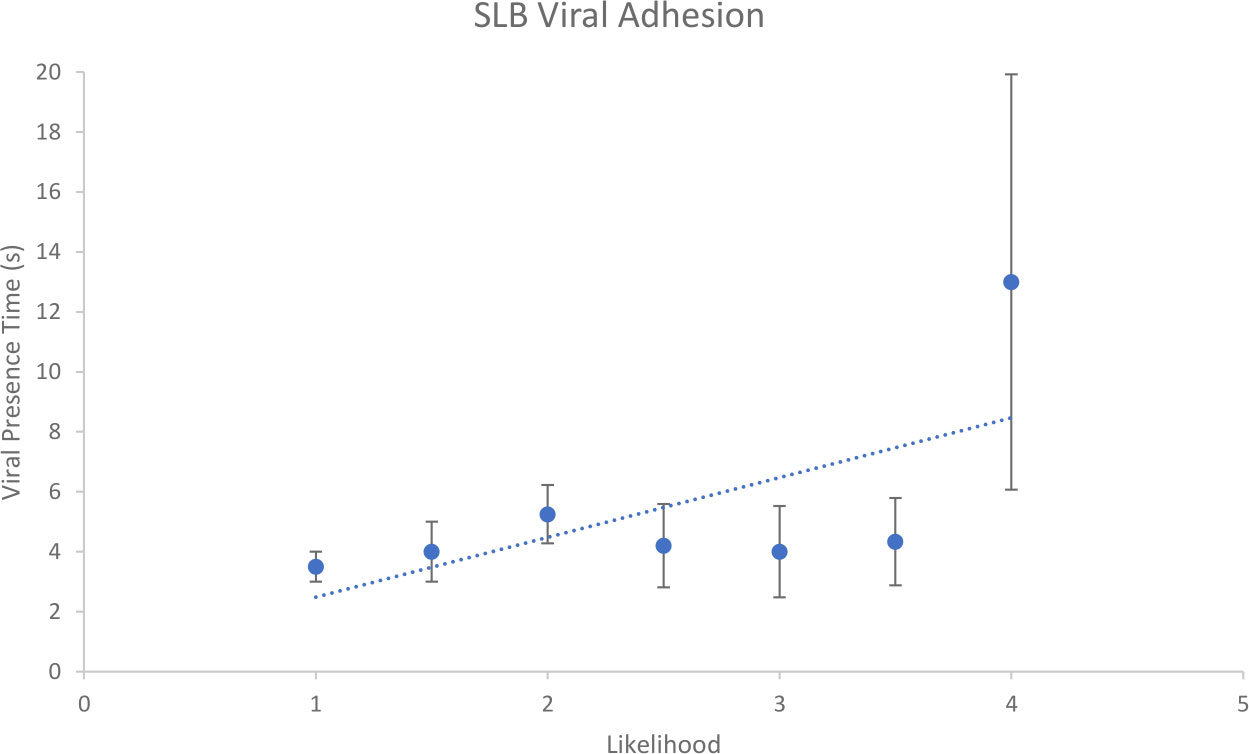
Figure 6 Relationship between likelihood assignment and viral presence time for suspected EhV-99B1/E. huxleyi derived SLB binding interactions.
Figure 7 shows an area of E. huxleyi SLB model membrane three frames apart post-viral flow. The first example frame (Figure 7A) highlights a ~ 20 nm height ~ 300 nm diameter, roughly surfaced, hemisphere shaped particle, that is not seen in control bilayer samples. This feature is strongly believed to be introduced EhV-99B1 virus. To maintain SLB integrity of these natural component bilayers, operator applied minimal tip interaction force is instrumental to produce reliable imaging data. Lower imaging forces can result in poorer tracking of the surface contours, leading to textures appearing blurred and less distinct and is therefore harder to measure true heights. This can produce poor background quality and a potential for misrepresented relative height feature to background measurements; but we believe offers the highest percentage chance to capture dynamic, random, delicate viral adhesion events safely.
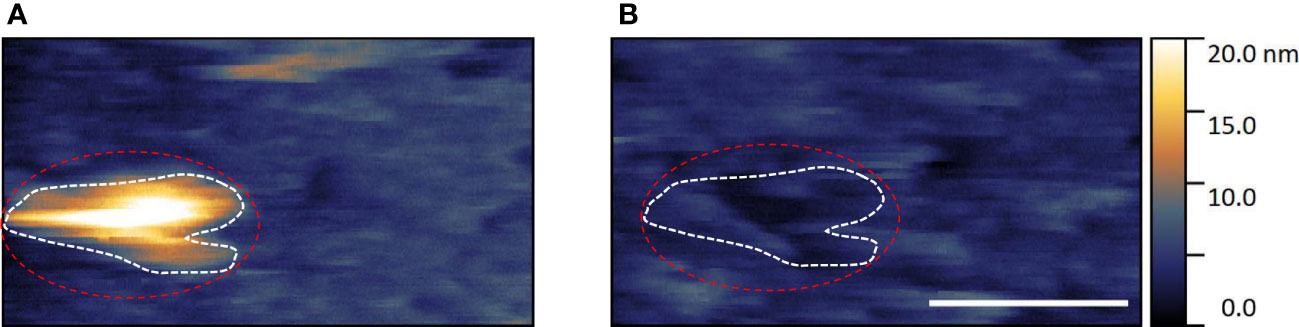
Figure 7 Example of suspected EhV-99B1/E. huxleyi derived SLB binding interaction visualised by HS-AFM in liquid (Scale bar 500 nm, data captured at 2 fps, dashed outlines highlights viral location).
In the second frame (Figure 7B) there is a clear ~ 5 nm depth ‘hole’ in the SLB that, relative to surrounding SLB features and by shape, matches the two dimensional ‘silhouette’ of the virus seen in the first frame (Red and white dashed outlines). It seems the adhered virus has been dislodged, bringing the bound area of lipid bilayer with it, potentially due to viral adhesion forces being greater than intrinsic SLB forces. We believe these high temporal resolution datasets indicate contact mode HS-AFM capturing transient binding of EhV virus to E. huxleyi derived SLBs in aqueous environment.
Characterisation of live cell membranes has been reported previously (21). This cell array method was used to mechanically isolate and identify suitable E. huxleyi cells for analysis. EhV was introduced during imaging to try and capture binding events to complete, live microalgal cell membranes.
Figure 8 Shows a summary graph of the collected data. In total, there were 105 examples of suspected viral binding. These data were isolated, processed and assigned a likelihood score as above. These interactions ranged from 1 to 29 frames of potential viral presence. In both this case, and the model membrane case, HS-AFM has facilitated acquisition and investigation of a high number of possible adhesion events.
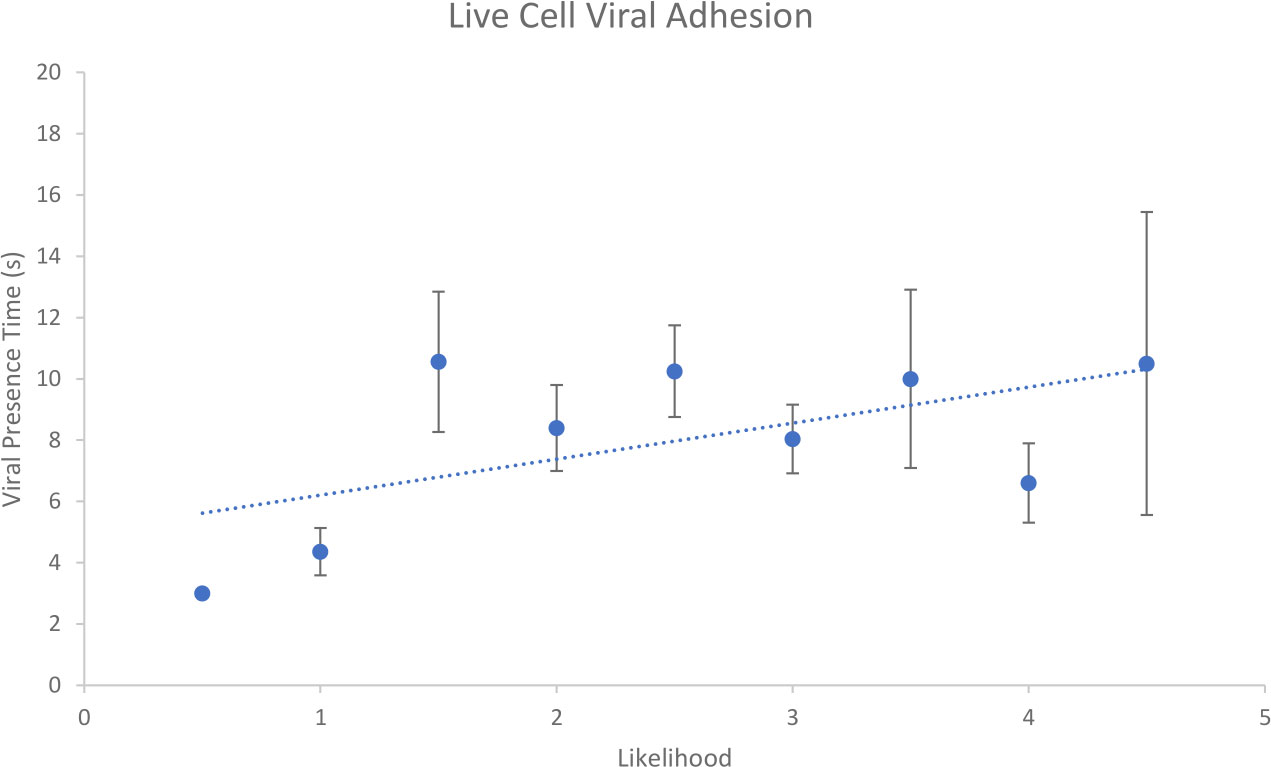
Figure 8 Relationship between likelihood assignment and viral presence time for suspected EhV-99B1/E. huxleyi binding interactions.
When live cell binding events were compared with SLB binding events there was a statistical difference between average viral presence time (Two tailed T test assuming unequal variances p = 0.020) with live cell being higher (8.1 s) than SLB (5.4 s) average. From this it could be concluded that live cell interactions are more robust, being imaged for longer times on average than those interactions with SLB. This suggests live cell membranes are better substrates for viral adhesion studies than model SLB membranes.
Figure 9 shows an example of a ‘likelihood 4.5’ assigned data sequence consisting of 22 data frames with the suspected virus adhering to the cell membrane towards the centre of the imaging frame.
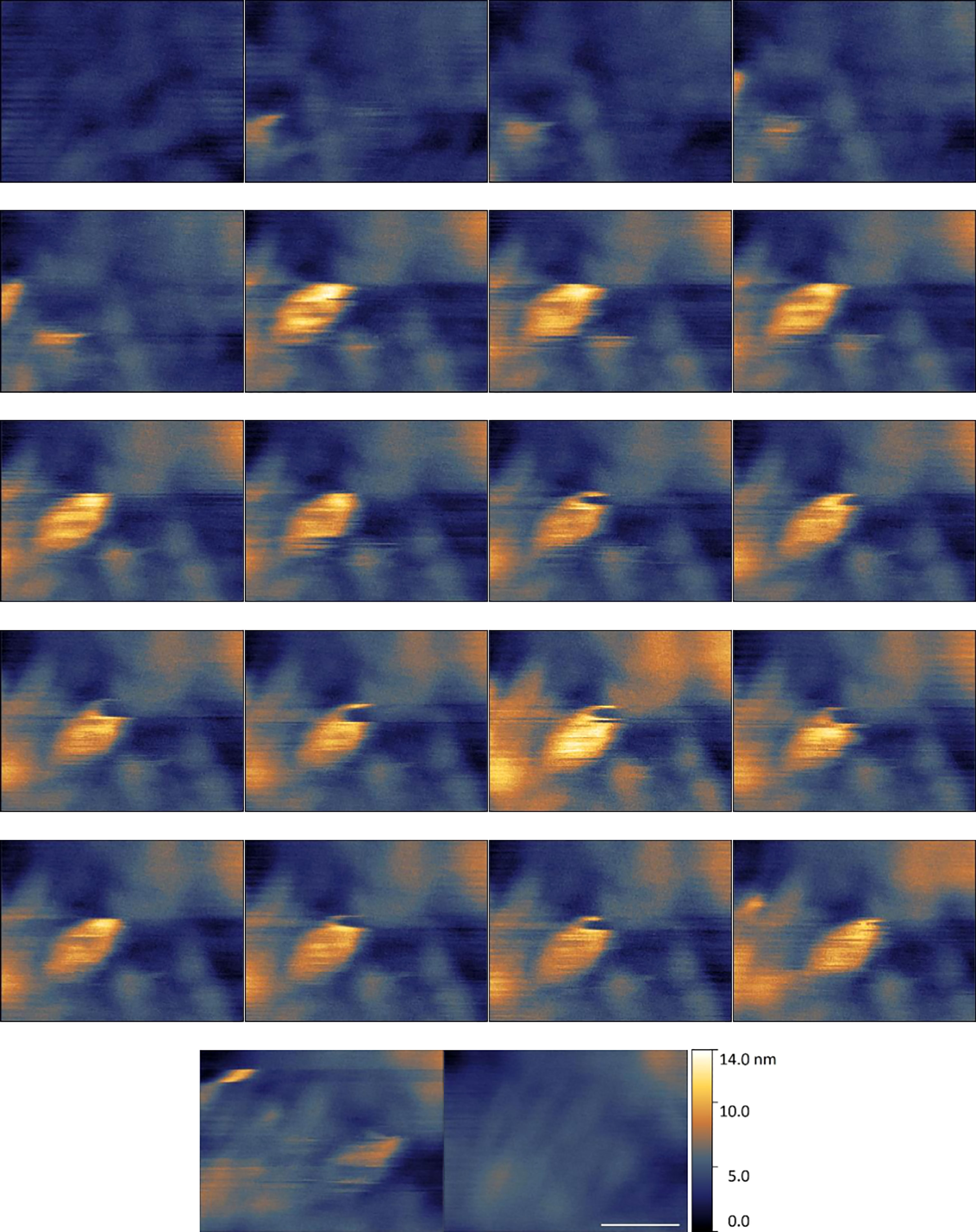
Figure 9 Example of suspected EhV-99B1/E. huxleyi binding interaction centre frame visualised by HS-AFM in liquid (Scale bar 300 nm, 1 s between frames, Data captured at 2 fps).
In this data set the virus particle appears as a maximal ~ 10 nm height, 200 – 400 nm width hemisphere (Figure 9). This height value is low, and width value at its maximum is large when compared to the hypothetical viral diameter. However, these measurements are in line with measurement range seen during particle characterisation. Deformability of the background surface and of the viral structure itself under imaging conditions is likely having a huge effect on these dimensional measurements. To reiterate, these strongly suspected viral structures are not present in control cell membrane surface examples. Their diameters and motility in comparison to cell membrane suggest these are highly likely to be virus.
There were a few examples of viral dimensions appearing closer to expected theoretical values in select data sequences. A short 5 frame sequence exhibiting this (Figure 10) was explored. Here, part of the virus is briefly imaged in the top right-hand corner of the data collection frame window for 3 frames. Two of these frames exhibit what would be expected from a partial data collection of an icosahedral shaped viral capsid with correct dimensions in height and extrapolated width. It is likely the virus is very quickly dislodged by the cantilever leaving doubts in the binding strength in this example. When compared to previous examples, this data series is one of 5 examples that exhibit high quality dimensional resolution, but particularly poor framing and time lengths.
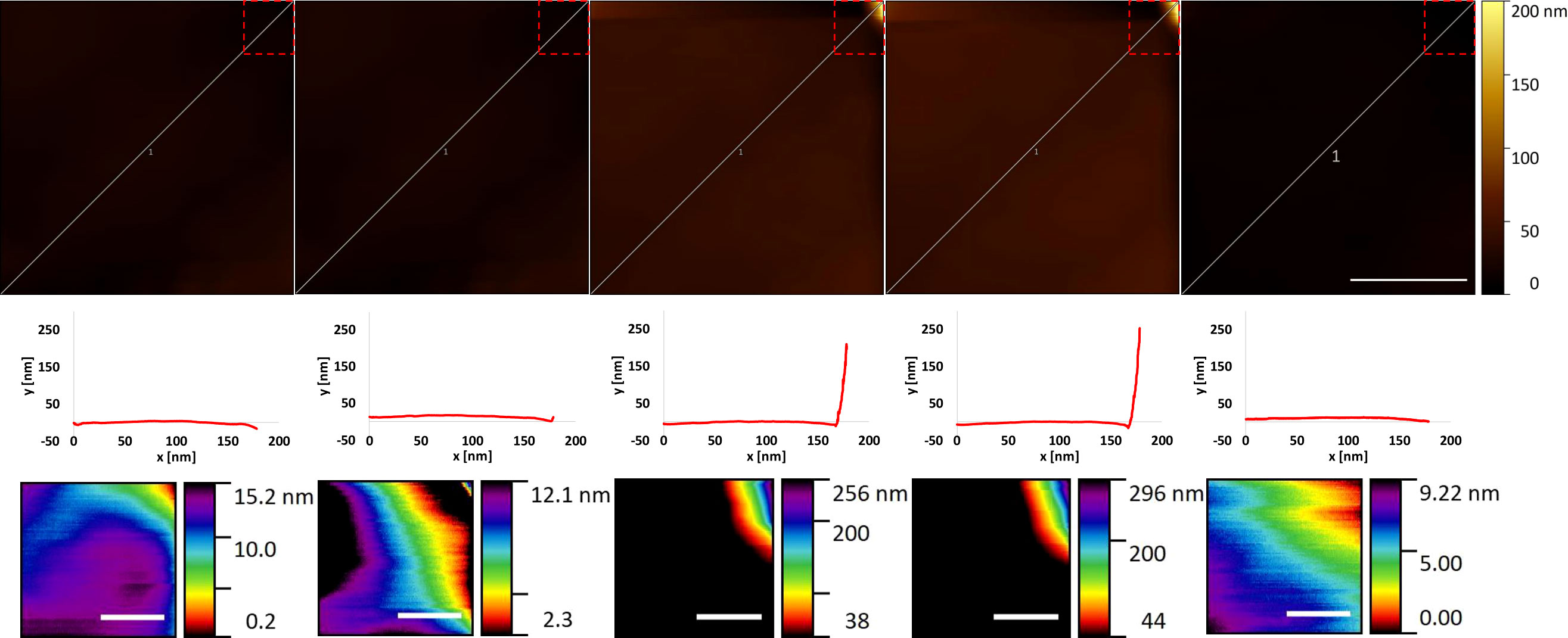
Figure 10 Example of suspected EhV-99B1/E. huxleyi binding interaction edge of frame visualised by HS-AFM in liquid (Data captured at 2 fps). For each frame top to bottom rows: Profile (scale bar 500 nm), Extracted profile marked in row 1 and Viral zoom (Scale bar 100 nm, row 1 dashed outline location).
AFM provides capability to visualise structures in appropriate buffers and the ability to examine individual particles without relying on a form of averaging or symmetry for structural information. Biological variation in these particles can be measured, especially with the substantial quantities of data generated by HS-AFM. This technique typically collects data at 2 million pixels per second being able to amass a volume of information in under a day that might take a ‘conventional’ AFM a year to collect. It was imperative that the dimensions and behaviour of EhV virus particles was understood when imaged by HS-AFM for preparation optimisation, easy identification, and further analysis in later experiments. This study is the first time that EhV has been imaged and investigated using AFM. Structural detail and information was measured whilst collecting diverse particle data extremely rapidly (0.5 s per individual frame).
Dimensions were recorded producing lower heights and larger widths than expected theoretical values. Statistical difference in heights was seen between air and liquid environment HS-AFM data collection. These effects could be due to a few factors.
In brief, with a typical pyramidal shaped tip and the contact, scanning, raster scan nature of HS-AFM data collection; high surface steps with imaged slopes steeper than the gradient of the probe tip appear far wider than their actual geometry. This could certainly apply to icosahedral viruses of this size. The heights could possibly be attributed to a few ideas. One could be the compression of the virus by the action of the imaging cantilever tip causing a flattening effect. Low spring constant cantilevers are used during contact mode HS-AFM to minimise differential vertical forces on features versus background substrate. Differences in prepared viral orientation (30) or build up of material around the base of the virus may also reduce apparent height. For air preparation and imaging versus fully hydrated samples, one issue could be dehydration. A more extreme example of dehydration effects during air imaging is seen in Supplementary Figure 5. This image shows an example of the pronounced central raised area as seen in previous conventional AFM enveloped viral studies (31). Fully hydrated viruses could be softer and more plastic allowing greater deformation along with changes in tip forces and dynamics in an aqueous environment.
A model SLB system was created in an attempt to effectively mimic E. huxleyi cell membrane surfaces for capturing associated dynamic processes. Transient viral binding to E. huxleyi derived SLBs was imaged using contact mode HS-AFM in a liquid environment. It is clear there are potential issues with this experiment in its currently presented form. It is unknown if SLB orientation is in the correct planar rotation for viral adhesion and if copurification of required receptors for viral recognition and adhesion of these reconstituted SLBs occurs. The targeted detergent resistant areas of the membrane during extraction are only thought to be involved with viral adhesion, entry and maybe exit in E. huxleyi. The SLB is ‘connected’ to the mica surface via a small hydration layer, and it is unknown if this hinders the processes we attempted to measure.
The adhesion force is critical and is not measured using our contact mode system with passive mechanical feedback. Viral binding is reversible (32) meaning imaged sequences are not guaranteed to reflect successful viral entry. Attempted improvement and optimisation of image quality with manual adjustments in the Z axis and therefore applied force could also result in virus being lost from cell surface. In providing physical space for viral adhesion to occur, intermittent contact with the cell surface was made during and post viral flow into the imaging environment. Although this should provide a higher chance of successfully capturing a viral adhesion event with HS-AFM imaging, this technique also increases the variability in imaging force without Z feedback resulting in background and viral resolution issues, relative height measurement issues and other imaging artefacts. This inconsistency in applied force could also increase inaccuracy of dimensional measurements during dynamic events.
Live cells are far more representative than the SLB membrane model, with one issue being a lack of solid support when compared with SLBs prepared on mica. Consequently, the background could flex under application of imaging forces effecting data collection. However, live cell data produced high likelihood viral binding sequences of statistically greater length than model SLBs.
The presented data show examples of viral adhesion. Residence time of virus measured by (33) was estimated at around 0.25 s, suggesting that the binding events seen by HS-AFM of EhV to E. huxleyi derived SLBs and live membranes are greater than or equal to those seen in the literature using a different technique.
In summary, HS-AFM has been instrumental in collecting sufficient data from the samples to generate quantifiable structural data. Without the speed of collection and ability to rapidly scan the surface area for sample examples, far fewer images would have been collected in the imaging timeframe. Geometry and, once the outer envelope is removed, general capsid protein position can be imaged successfully through contact mode HS-AFM.
Coccolithovirus EhV binding to live E. huxleyi cell membranes has been captured with contact mode HS-AFM in a physiologically relevant aqueous environment, however there is no evidence of transmembrane travel. It is most likely that the disappearance of virus is due to the structure being spatially lost from the data collection frame by small sample movements or dislodgment of the adhered virus under imaging forces.
Using current methods, having both high quality spatial resolution of the virus and the background membrane, with the virus centre frame, has extremely low capture percentage by contact mode HS-AFM. One or more of these factors had to be sacrificed in the examples collected during this study.
The data holds promise for future experiments using this technology platform. HS-AFM differs to other techniques such as forms of electron microscopy or fluorescence microscopy by being the only label-less method that can directly collect data with the required spatial and temporal resolution under physiologically or environmentally relevant conditions of these dynamic events. The speed and volume of data acquisition meant that 132 potential viral adhesion to SLB and live membranes sequences were extracted from 2fps video data, enabling many more measurements than classical AFM could capture over a similar time frame.
Atomic force microscopy is traditionally seen as a structural or mechanical property tool, but with increases in speed and availability shows promise as a method for dynamic observations. This work highlights advances in HS-AFM as an innovative tool for studying the mechanisms of characterization and interaction between marine viruses and their hosts during the infection process in a novel, dynamic way.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Conceptualization, CE, LP, OP and MA. Methodology, CE, LP, OP and MA. Software, LP, OP. Formal analysis, CE, LP, OP and MA. Investigation, CE. Data curation, CE. Writing—original draft preparation, CE. Writing—review and editing, CE, LP, OP and MA. Supervision, LP, OP and MA. Project administration, LP, OP and MA. Funding acquisition, LP, OP and MA. All authors contributed to the article and approved the submitted version.
CE was supported by an Engineering and Physical Sciences Research Council (EPSRC) and Bristol Nano Dynamics Ltd. as part of C.T.E.’s PhD studentship. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
The HS-AFM work was undertaken in the Plymouth Marine Laboratory and University of Exeter Environmental Single Cell Genomics Facility which was supported by grants from the UK’s Natural Environment Research Council (NERC) and The Wolfson Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1111335/full#supplementary-material
1. Paasche E. A review of the coccolithophorid emiliania huxleyi (prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia (2001) 40:503–29. doi: 10.2216/i0031-8884-40-6-503.1
2. Larsen SH. Solar variability, dimethyl sulphide, clouds, and climate. Global Biogeochem. Cycles (2005) 19:1–12. doi: 10.1029/2004GB002333
3. Winter A, Jordan RW, Roth P (1994) Biogeography of living coccolithophores in ocean waters. In: Coccolithophores Winter A, Siesser WG editors. Cambridge: Cambridge University Press, (1994) 161–177.
4. Poulton AJ, Adey TR, Balch WM, Holligan PM. Relating coccolithophore calcification rates to phytoplankton community dynamics: Regional differences and implications for carbon export. Deep. Res Part II Top Stud Oceanogr. (2007) 54:538–57. doi: 10.1016/j.dsr2.2006.12.003
5. Smith SV, Mackenzie FT. The role of CaCO3 reactions in the contemporary oceanic CO2 cycle. Aquat Geochemistry (2016) 22:153–75. doi: 10.1007/s10498-015-9282-y
6. Balch WM. The ecology, biogeochemistry, and optical properties of coccolithophores. Ann Rev Mar Sci (2018) 10:71–98. doi: 10.1146/annurev-marine-121916-063319
7. Bratbak G, Egge JK, Heldal M. Viral mortality of the marine alga emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser (1993) 93:39–48. doi: 10.3354/meps093039
8. Van Etten JL, Graves MV, Müller DG, Boland W, Delaroque N. Phycodnaviridae - Large DNA algal viruses. Arch Virol (2002) 147:1479–516. doi: 10.1007/s00705-002-0822-6
9. Wilson WH, Tarran GA, Schroeder D, Cox M, Oke J, Malin G. Isolation of viruses responsible for the demise of an emiliania huxleyi bloom in the English channel. J Mar Biol Assoc United Kingdom (2002) 82:369–77. doi: 10.1017/S002531540200560X
10. Wilson WH. Coccolithovirus-emiliania huxleyi dynamics: an introduction to the coccolithovirocell. Perspect Phycol (2015) 2:91–103. doi: 10.1127/pip/2015/0032
11. Laber CP, Hunter JE, Carvalho F, Collins JR, Hunter EJ, Schieler BM, et al. Coccolithovirus facilitation of carbon export in the north Atlantic. Nat Microbiol (2018) 3:537–47. doi: 10.1038/s41564-018-0128-4
12. Van Der Goot FG, Harder T. Raft membrane domains: From a liquid-ordered membrane phase to a site of pathogen attack. Semin Immunol (2001) 13:89–97. doi: 10.1006/smim.2000.0300
13. Mackinder LCM, Worthy CA, Biggi G, Hall M, Ryan KP, Varsani A, et al. A unicellular algal virus, emiliania huxleyi virus 86, exploits an animal-like infection strategy. J Gen Virol (2009) 90:2306–16. doi: 10.1099/vir.0.011635-0
14. Bidle KD, Vardi A. A chemical arms race at sea mediates algal host-virus interactions. Curr Opin Microbiol (2011) 14:449–57. doi: 10.1016/j.mib.2011.07.013
15. Vardi A, Van Mooy BAS, Fredricks HF, Popendorf KJ, Ossolinski JE, Haramaty L, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton. Sci (80-. ). (2009) 326:861–5. doi: 10.1126/science.1177322
16. Vardi A, Haramaty L, Van Mooy BAS, Fredricks HF, Kimmance SA, Larsen A, et al. Host-virus dynamics and subcellular controls of cell fate in a natural coccolithophore population. Proc Natl Acad Sci (2012) 109:19327–32. doi: 10.1073/pnas.1208895109
17. Guillard R.R.L. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL., Chanley MH editors Cult Mar Invertebr AnimBoston, MA: Springer. (1975) doi: 10.1007/978-1-4615-8714-9_3
18. Brussaard CPD, Marie D, Bratbak G. Flow cytometric detection of viruses. J Virol Methods (2000) 85:175–82. doi: 10.1016/S0166-0934(99)00167-6
19. Mojica KDA, Evans C, Brussaard CPD. Flow cytometric enumeration of marine viral populations at low abundances. Aquat Microb Ecol (2014) 71:203–9. doi: 10.3354/ame01672
20. Rose SL, Fulton JM, Brown CM, Natale F, Van Mooy BAS, Bidle K. D. Isolation and characterization of lipid rafts in emiliania huxleyi: A role for membrane microdomains in host-virus interactions. Environ Microbiol (2014) 16:1150–66. doi: 10.1111/1462-2920.12357
21. Evans CT, Baldock SJ, Hardy JG, Payton O, Picco L, Allen MJ. A non-destructive, tuneable method to isolate live cells for high-speed AFM analysis. Microorganisms (2021) 9(4):680. doi: 10.3390/microorganisms9040680
22. Kiss B, Kis Z, Pályi B, Kellermayer MSZ. Topography, spike dynamics, and nanomechanics of individual native SARS-CoV-2 virions. Nano Lett (2021) 21:2675–80. doi: 10.1021/acs.nanolett.0c04465
23. Lyonnais S, Hénaut M, Neyret A, Merida P, Cazevieille C, Gros N, et al. Atomic force microscopy analysis of native infectious and inactivated SARS-CoV-2 virions. Sci Rep (2021) 11(1):11885. doi: 10.1038/s41598-021-91371-4
24. Kuznetsov YG, Gurnon JR, Van Etten JL, McPherson A. Atomic force microscopy investigation of a chlorella virus, PBCV-1. J Struct Biol (2005) 149:256–63. doi: 10.1016/j.jsb.2004.10.007
25. Pan Y, Shlyakhtenko LS, Lyubchenko YL. High-speed atomic force microscopy directly visualizes conformational dynamics of the HIV vif protein in complex with three host proteins. J Biol Chem (2020) 295:11995–2001. doi: 10.1074/jbc.ra120.014442
26. Lim K, Kodera N, Wang H, Mohamed MS, Hazawa M, Kobayashi A, et al. High-speed AFM reveals molecular dynamics of human influenza a hemagglutinin and its interaction with exosomes. Nano Lett (2020) 20:6320–8. doi: 10.1021/acs.nanolett.0c01755
27. Xiao C, Rossmann MG. Structures of giant icosahedral eukaryotic dsDNA viruses. Curr Opin Virol (2011) 1:101–9. doi: 10.1016/j.coviro.2011.06.005
28. El Kirat K, Morandat S, Dufrêne YF. Nanoscale analysis of supported lipid bilayers using atomic force microscopy. Biochim Biophys Acta - Biomembr (2010) 1798:750–65. doi: 10.1016/j.bbamem.2009.07.026
29. Reviakine I, Brisson A. Formation of supported phospholipid bilayers from unilamellar vesicles investigated by atomic force microscopy. Langmuir (2000) 16:1806–15. doi: 10.1021/la9903043
30. Carrasco C, Carreira A, Schaap IAT, Serena PA, Gómez-Herrero J, Mateu MG, et al. DNA-Mediated anisotropic mechanical reinforcement of a virus. Proc Natl Acad Sci USA (2006) 103:13706–11. doi: 10.1073/pnas.0601881103
31. Malkin AJ, McPherson A, Gershon PD. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J Virol (2003) 77:6332–40. doi: 10.1128/JVI.77.11.6332-6340.2003
32. Agarkova I, Hertel B, Zhang X, Lane L, Tchourbanov A, Dunigan DD, et al. Dynamic attachment of chlorovirus PBCV-1 to chlorella variabilis. Virology (2014) 466–467:95–102. doi: 10.1016/j.virol.2014.07.002
Keywords: algae, virus, interactions, high-speed, atomic force microscopy
Citation: Evans CT, Payton O, Picco L and Allen MJ (2023) Visualisation of microalgal-viral interactions by high-speed atomic force microscopy. Front. Virol. 3:1111335. doi: 10.3389/fviro.2023.1111335
Received: 09 December 2022; Accepted: 20 February 2023;
Published: 02 March 2023.
Edited by:
Jean-Michel Claverie, Aix-Marseille Université, FranceReviewed by:
Rodrigo Araújo Lima Rodrigues, Federal University of Minas Gerais, BrazilCopyright © 2023 Evans, Payton, Picco and Allen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael J. Allen, TS5hbGxlbjVAZXhldGVyLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.