- 1College of Medicine, Yichun University, Yichun, China
- 2Jiangxi Provincial Key Laboratory of Active Component of Natural Drugs, Poster-Doctoral Research Center, Yichun, China
Streptococcus suis serotype 2 (S. suis type 2, SS2) is one of the zoonotic pathogens known to induce meningitis, septicemia, and arthritis in both pigs and humans, resulting in public health concerns. CbpD, also termed CrfP, is one of the choline-binding proteins (CBPs) that was found as a murein hydrolase in SS2 and plays crucial roles in natural genetic transformation under the control of ComRS-ComX regulatory system by a previous study. Nonetheless, the possible functions of CbpD in virulence and pathogenesis in SS2 remain unclear. In this study, a cbpD gene mutant (ΔcbpD) with its complemental strain (cΔcbpD) was constructed and further used to examine the pathogenic roles of CbpD in SS2 infection. The results showed that the CbpD deficiency leads to increased bacterial chain elongation and aggregation with little impact on the growth capability of SS2. The ΔcbpD strain represented more vulnerable to a thermo, acid, or oxidative stress. Elevated adhesion to human epithelial HEp-2 cells, decreased invasion into bEND3.0 cells, and more easily phagocytosed by murine RAW264.7 macrophages of ΔcbpD were found. The virulence of cbpD mutant was attenuated in a mouse infection model. Enhanced susceptibility within mice blood and impaired ability to colonize organs with alleviated histopathological lesions were also demonstrated as compared with wild-type SS2. It is noteworthy that the discrepant expression of multiple virulence-associated factors including serine/threonine phosphorylase Stp, anti-phagocytosis factor of transglutaminase TGase and adhesin of chaperon DnaJ, were examined resulting from the deletion of cbpD. Overall, these findings provided evidence that the CbpD factor contributes to SS2 infection and is involved in bacterial adhesion, invasion, and anti-phagocytosis processes by modulating crucial virulence-associated factors expression.
1 Introduction
Streptococcus suis (S. suis), a globally distributed and underestimated emerging Gram-positive pathogen, is zoonotic and can infect both swine and humans, and lead to symptoms such as septicemia, arthritis, meningitis, and streptococcal toxic shock-like syndrome (1–4). According to the polymorphism among the capsular polysaccharide antigen, S. suis has been classified into at least 56 serotypes, i.e., authentic serotypes 1–19, 21, 23–25, 27–31, 1/2, Chz, and 26 novel capsular polysaccharide loci (NCL) (5, 6). S. suis serotype 2 (SS2) is the most common and frequently reported isolate, capable of causing significant economic losses in the swine industry and has induced three large-scale outbreaks among humans in China (7–9). Currently, over 100 putative virulence factors of S. suis have been reported, including the typical suilysin (Sly), muramidase release protein (MRP), superoxide dismutase SodA, enolase, transglutaminase (TGase), heat shock protein of DnaJ, and serine/threonine kinase (Stk). However, the complex roles of various virulence factors with their potential regulatory networks and molecular interactions with the host are not yet fully understood (2, 10).
Choline-binding proteins (CBPs) are clusters of the important surface-anchored and conservatively distributed proteins in Streptococcus (11, 12). In Streptococcus pneumoniae (S. pneumoniae), the cell wall-associated CBPs typically consist of at least two domains: a highly conserved choline-binding domain (CBD) with characteristic repeat motifs and a functional domain (FD) (13, 14). The CBD functions to bind phosphorylcholine molecules on the cell wall through non-covalent interactions and is essential for microbe interaction with host cells for bacteria colonization in the upper respiratory tract. Fundamental divergence of the three-dimensional structure leading to the physiological discrepancy of FD contributes to CBP varieties (11, 15). To date, CBPs have been reported to be implicated in multiple roles in pneumococcal infection and pathogenesis (16, 17). The conventional CBPs of LytA, LytB, LytC, PspA, PspC, PcpA, Pce, and CbpD within S. pneumoniae are found to be an extracellular pneumococcal virulence determinant which can mediate not only for bacterial cell wall construction, colonization to host epithelial cells but also for antibiotics susceptibility and biofilm formation (18–25). Horizontal gene transfer contributes to pathogens’ adaptive evolution and genomic diversity that enables the acquisition of beneficial genes involved in pathogenesis and immune evasion (24, 26, 27). CBPs are also murein hydrolases that are involved in cellular autolysis during the stationary phase and fratricide during the competence process (28–30).
CbpD, one of the well-studied CBPs in S. pneumoniae, is characterized by an N-terminal cysteine and a histidine-dependent amidohydrolase/peptidase (CHAP) domain, followed by one or two Src homology 3b (SH3b) domains, and a C-terminal CBD with four choline-binding repeats (26). The CHAP domain is responsible for cleaving peptide bonds within the bridges of streptococcal peptidoglycan, while the SH3b domain plays an essential role in the critical biological function of CbpD, possibly by binding to the peptidoglycan portion of the cell wall. Moreover, CbpD can be involved in inducing virulence factor release from non-competent S. pneumoniae upon competent condition (31, 32). Recently, CrfP, a CbpD-like murein hydrolase, first found in S. suis type 2, has been identified as a key component in SS2’s competence, enhancing transcription by inducing peptides (33). This study aims to investigate the roles of the underlying pathogenesis of CbpD in SS2 infection.
2 Materials and methods
2.1 Bacterial agents, plasmids, and culture conditions
Bacterial strains and plasmids utilized for cloning and mutant construction are detailed in Table 1. The SS2 virulent wild-type (WT) strain of HA9801, isolated from diseased pigs which mainly manifested septicemia among SS2 outbreak in Jiangsu, China, was kindly offered by Professor Weihuan Fang at Zhejiang University. The S. suis strains were cultured in Brain heart infusion (BHI; Oxoid, UK) at 37°C. Escherichia coli DH5α cells were cultured in Luria–Bertani medium at 37°C. The various antibiotics (Solarbio, Beijing, China) used for cloning and the selection of mutants were incorporated into the culture media at specific concentrations: 50 μg/mL of ampicillin for E. coli, 100 μg/mL and 50 μg/mL of spectinomycin for SS2 and E. coli, respectively.
2.2 Construction of the cbpD mutation and complementation strains
The cbpD deletion mutant (ΔcbpD) based on wild-type strain HA9801 was constructed as previously described (34). In brief, the upstream and downstream flanking fragments of the cbpD gene were amplified by PCR based on the template of the HA9801 genome and the primers listed in Table 2. The flanking fragments were then fused by overlapping PCR. The product was subjected to endonuclease enzymatic digestion, ligated with the thermo-sensitive vector of pSET4s, and then transformed into the DH5α competent cells to generate the knockout plasmid of pSET4s::cbpD after PCR identification and DNA sequencing. The recombinant pSET4s::cbpD shuttle vector was introduced into HA9801 competent cells via electroporation. The resulting ΔcbpD mutant was obtained through a series of screening steps including repeated SS2 passage, targeted gene double exchange, and positive mutant colony isolation, and confirmed by PCR verification and DNA re-sequencing. To construct the allelic cbpD complementation strain of cΔcbpD, the previously reported impdh promoter was fused with the cbpD-ORF and subsequently integrated into pSET2 to yield the complemental plasmid pSET2::cbpD. The recombinant plasmid was electroporated into the ΔcbpD competent cells and the resulting cΔcbpD isolate was screened by spectinomycin resistance and PCR confirmation.
2.3 Growth curves
Growth capability was examined by the protocol instructed previously (35). A single isolated colony of the WT, ΔcbpD, and cΔcbpD strains was inoculated into BHI broth for a 12 h incubation at 37°C with 150 rpm shaking. The cultures were centrifuged to form a precipitate and then adjusted to an OD600nm of 0.4 in BHI. Subsequently, 250 μL of the bacterial suspension each was transferred into 25 mL of BHI medium. The cultures were used for spectrophotometric measurement at OD600nm every 2 h after 37°C incubation at 150 rpm. Triplicate assays with three wells each were conducted and determined for this experiment.
2.4 Morphological change and stress tolerance analysis
The SS2-WT, ΔcbpD, and cΔcbpD strains were Gram-stained on the glass slides following the staining guide and observed by microscopy under oil immersion. The numbers of cocci among these strains in 50 chains each under light microscopy were counted and average bacterial cells per chain were examined. Stress tolerance phenotypes of SS2 strains upon thermo-induced treatment at 42°C, acetic acid treatment at pH5.5 for 37°C or oxidative treatment with 20 mM H2O2 at 37°C for 1 h, were conducted and examined as previously described, respectively (36).
2.5 In vitro cell adhesion and invasion assays
The adhesion to and invasion of host cells by SS2 was assessed in accordance with a previous study by using the human laryngeal epithelial HEp-2 cells and the murine brain microvascular endothelial bEND3.0 cells, respectively (36). Briefly, bacterial cultures were added to the cells in a 24-well culture plate at a multiplicity of infection (MOI) of 50, with three wells for each strain. The plates were incubated for 1 h at 37°C, 5% CO2. To remove non-adherent bacteria, the wells were washed four times with 1 mL of 10 mM PBS. For the adherence assay, the sterile ddH2O was added to lyse infected cells and then plated to BHI agar by 10-fold dilution in PBS. After 24 h incubation at 37°C, the bacteria colonies were enumerated. The adhesion rate was defined as (viable CFUAdh / inoculated CFUTotal) × 100%. The adhesive inhibition assay was conducted the same as the aforementioned procedure (37), with the exception of the 10% inactivated rabbit polyclonal targeted or pre-immune negative sera treatment of the WT and ΔcbpD for 1 h prior to infection.
For the invasion assay, DMEM consisting of gentamicin (300 μg/mL) was introduced into the bacterial infection cells following adhesion assay to eradicate extracellular dissociated SS2 for 1 h at 37°C. The cells underwent subsequently triple washes with PBS and were further lysed by using sterile distilled water. The intracellular evasive bacteria were determined by BHI plate counting. The invasion rate was defined as (viable intracellular CFUInv / inoculated CFUTotal) × 100%.
2.6 Phagocytosis test
Murine macrophage cell line RAW264.7 was used for the phagocytosis assay according to a previously reported protocol (34, 38). Cells were subcultured and inoculated into 24-well plates at 37°C, 5% CO2. The cells were cultivated for 12 h until the cell confluence reached above 90%, washed with PBS, and incubated in DMEM containing 10% inactivated fetal bovine serum. The suspensions of both the WT and ΔcbpD strains were pre-adjusted to an OD600nm of 0.4 and then added to the cells at an MOI of 50. After 1 h incubation, the cells were subjected to PBS washing and then treated with gentamicin (300 μg/mL) for another 1 h. Subsequently, the mixture was washed three times by PBS, lysed with sterile distilled water, and plated on BHI agar to count the viable SS2 colonies. The phagocytosis rate was determined by calculating the percentage of surviving CFU in the BHI medium compared to the CFU in the original inoculum. This assay was performed three times for each strain, with triplicate wells tested on each occasion.
2.7 Susceptibility in the murine ex vivo blood culture
Survival assay of SS2 within mice whole blood was executed following the established protocol (35). Cultures in the mid-exponential growth phase were standardized to an optical density of 0.4 at 600 nm. A 50 μL aliquot of either WT or ΔcbpD, was added to 450 μL of fresh whole blood collected from clinically healthy mice and then incubated at 37°C for 1 h. The viable bacterial counts were determined via BHI agar plate enumeration. Each assay was conducted in triplicate and replicated three times.
2.8 Virulence determination in mice model
In each designated group, 12 female BALB/c mice, aged 4 weeks, were randomly chosen and subjected to intraperitoneal injection with 1 mL of WT, ΔcbpD, and cΔcbpD at approximately 5 × 108 CFU/mL, respectively. An equal volume of PBS inoculation served as a negative control. The clinical manifestations and mortality of mice were monitored every 12 h and continuously recorded for 1 week. Furthermore, the heart, liver, spleen, lung, kidney, and brain organs from infected mice showing clinical symptoms were aseptically collected, homogenized, and subsequently subjected to BHI plate counting for bacterial load determination in another SS2 infection experiment. Meanwhile, 4% paraformaldehyde was used to fix tissue samples and then paraffin-embedded sections of the lesion tissues were prepared, stained by Hematoxylin and Eosin, and the histopathological changes of the tissues were examined under a microscope as described previously (37). Animal infection experiments conformed to the animal ethics guidelines from the Medical Ethics Committee of Yichun University in Yichun City, Jiangxi Province, China (2022018).
2.9 Quantitative real-time PCR (qPCR) analysis
Bacterial cultures, upon reaching the mid-logarithmic growth phase, were harvested. The cells were then pelleted through centrifugation at 5,000 rpm for 10 min. Total RNA extraction from the bacterial pellets was carried out following the manufacturer’s protocol by the RNA Extraction Kit (TransGen, China) (39). Subsequently, synthesis and amplification of cDNA were performed via a reverse transcription kit (Vazyme, China). To quantify the transcriptional levels of the target genes listed in Table 2, relative qPCR was conducted using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) with Bio-Rad CFX96 thermocycler. The 16S rRNA gene was determined as an internal reference to normalize the gene expression data.
2.10 Western blotting
The effect of CbpD deficiency on the virulence-related factors expression levels was analyzed by Western blotting. Overnight cultures of WT and ΔcbpD strains were centrifuged at 5,000 rpm for 10 min to collect the bacteria suspension and supernatants. Total protein samples from bacteria suspension, except for two secreted proteins Sly and TGase, which were directly determined from the harvested bacteria supernatant, were utilized for the analysis of protein expression changes. The total SS2 cellular proteins were extracted by cell wall cleavage buffer including 3 mg/mL of lysozyme in 37°C for 1.5 h and homogenizing the mixture following the previously established protocol, and the protein concentration was quantified by the Bradford method (40). SDS-PAGE was performed, followed by protein transfer onto PVDF membranes, blocking, and incubation with rabbit polyclonal antibodies (prepared in-house) at 4°C for 12 h. Then, the PVDF membranes underwent incubation of HRP-conjugated goat anti-rabbit monoclonal IgG, washed five times with TBST containing 0.5% skimmed milk powder, and visualized by chemiluminescent imaging system after a chemiluminescence reaction kit (GLPBIO, United States). Protein changes in grayscale were accomplished via ImageJ software (ImageJ 1.5, NIH, United States).
2.11 Statistical analysis
Each experimental assay was performed in triplicate and repeated on at least three distinct occasions, and the resulting data were presented as mean ± standard deviations (SDs). Statistical significance was determined using a paired, two-tailed Student’s t-test, facilitated by the GraphPad Prism 5.0 software. For animal infection experiments, the log-rank test was used to assess the differences in survival rate. The p-value threshold of 0.05 was established and considered as the criterion for statistical significance.
3 Results
3.1 The cbpD gene-deficient with its complementary strain construction in SS2
The construction of the cbpD mutant and complementation strains was performed using homologous recombination with the shuttle vectors pSET4s for the mutant and pSET2 for the complementation strain. The specific gene knockout plasmid of recombinant pSET4s::cbpD was generated as shown in Figure 1A. After electroporation, repeated subculture, and screening, the suspected ΔcbpD mutant and allelic cΔcbpD colonies were identified by PCR using the outer-, inner-, and S. suis-specific 16S rRNA gene primers, and subjected to verification by DNA sequencing. The cbpD mutant with its allelic complementary strain of cΔcbpD was successfully obtained, as evidenced by PCR identification (Figure 1B).
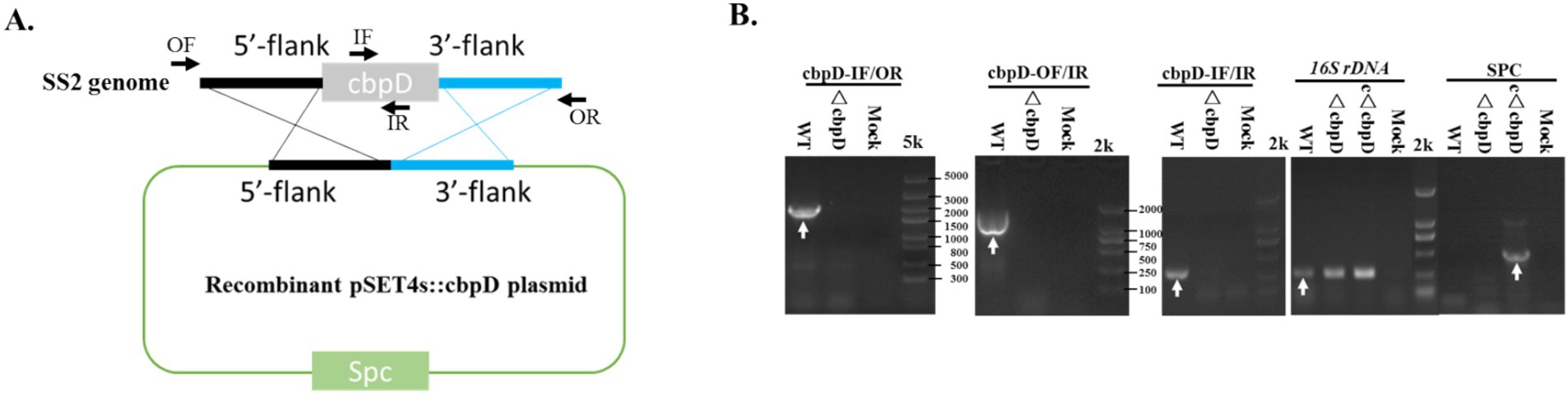
Figure 1. CbpD-deficient and complementary strain generation in S. suis type 2. (A) Schematic diagram for the recombinant pSET4s::cbpD construction. (B) PCR identification of the cbpD gene deletion and the complementation strains. OF/OR primers and IF/IR primers represented the outer-flank and inner-gene amplification regions for cbpD and were used for gene deletion identification. Antimicrobial resistance gene primers of SPC-F/R were used for cbpD-deficient and complementary identification. The 16S rDNA was used for SS2 identification.
3.2 Biological phenotypic changes for the cbpD gene deficiency
Figure 2A demonstrated that the growth patterns between the WT and ΔcbpD strains were predominantly congruent, and the ablation of cbpD does not impinge upon the growth competence of SS2. Morphological observations disclosed that in contrast to the WT strain, the ΔcbpD possessed enhanced chain-forming capability, with an average of 3–5 bacterial cells per chain, and manifested a propensity to aggregate (Figures 2B,C). The stress tolerance analysis discovered that the deletion of the cbpD gene would significantly decrease the survival rate of ΔcbpD and render this strain more susceptible to cultures at 42°C, pH5.5, or 20 mM H2O2 stress conditions (Figure 2D). The abovementioned deviations could be restored in phenotype after cbpD replenishment in cΔcbpD strain.
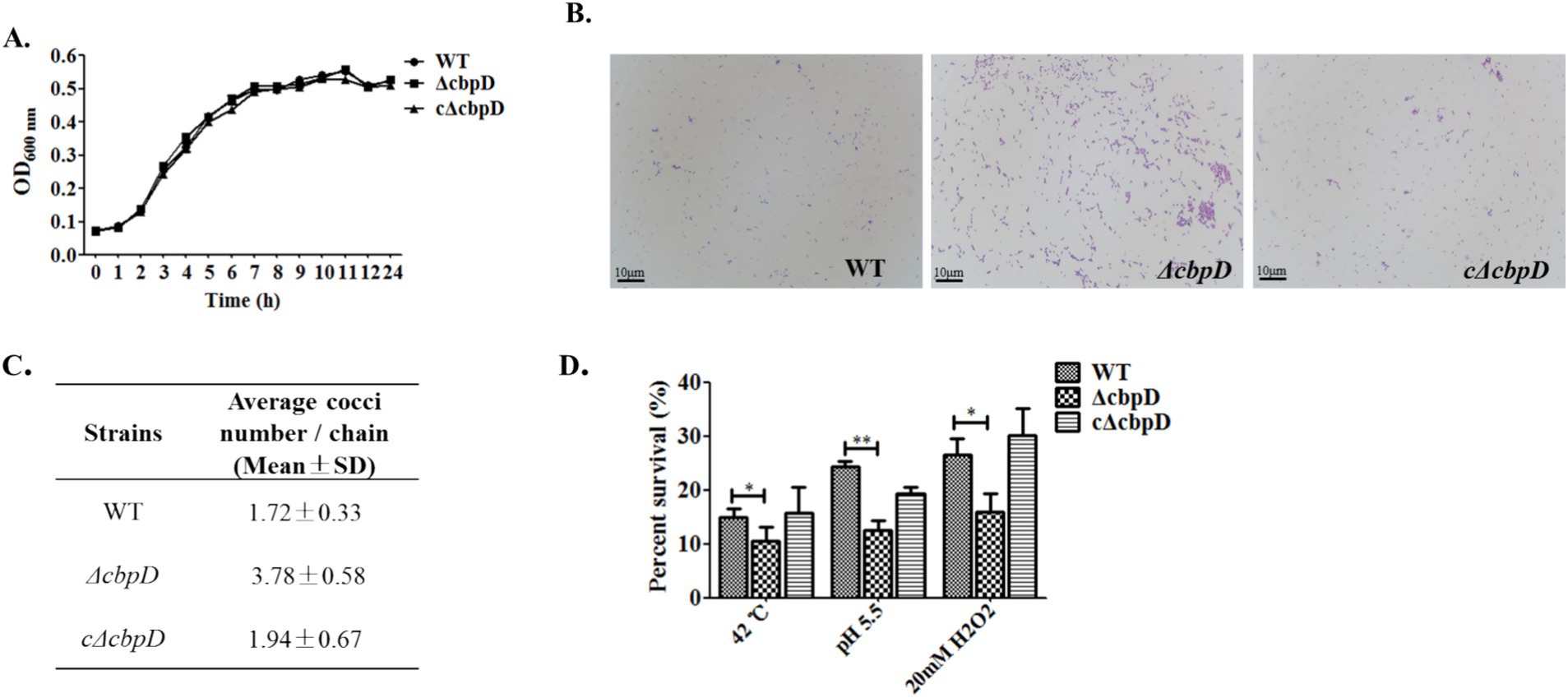
Figure 2. Phenotypic changes for the cbpD gene deficiency. (A) Growth characteristic among WT, ΔcbpD, and cΔcbpD strains. (B) Morphological observation under microscopy with a magnification of ×1,000. (C) Average bacteria cells in each chain between the WT and ΔcbpD. (D) SS2 survival assays at different stress conditions.
3.3 The CbpD factor of SS2 is engaged in the host cell infection in vitro
To examine the potential role of CbpD in SS2 infection, in vitro cell infection models previously developed were used to investigate the differences in adhesion, invasion, resistance to phagocytosis, and survival capability among WT, ΔcbpD, and cΔcbpD strains. The findings revealed a prominent increase in the adhesion rate of the cbpD-deficient strain to HEp-2 epithelial cells (p < 0.05, Figure 3A), while the colonies of ΔcbpD invading endothelial bEND3.0 cells were notably decreased (p < 0.05, Figure 3B). These cbpD-deficient SS2 were prone to be phagocytosed by macrophage cells of RAW264.7 and showed an 80% reduction in survival when incubated in ex vivo mouse whole blood (p < 0.01, Figures 3C,D). The introduction of the cbpD gene could partially restore these phenotypic changes. These data implied that the CbpD factor was involved in SS2 infection by mediating the immune invasion and phagocytosis process.
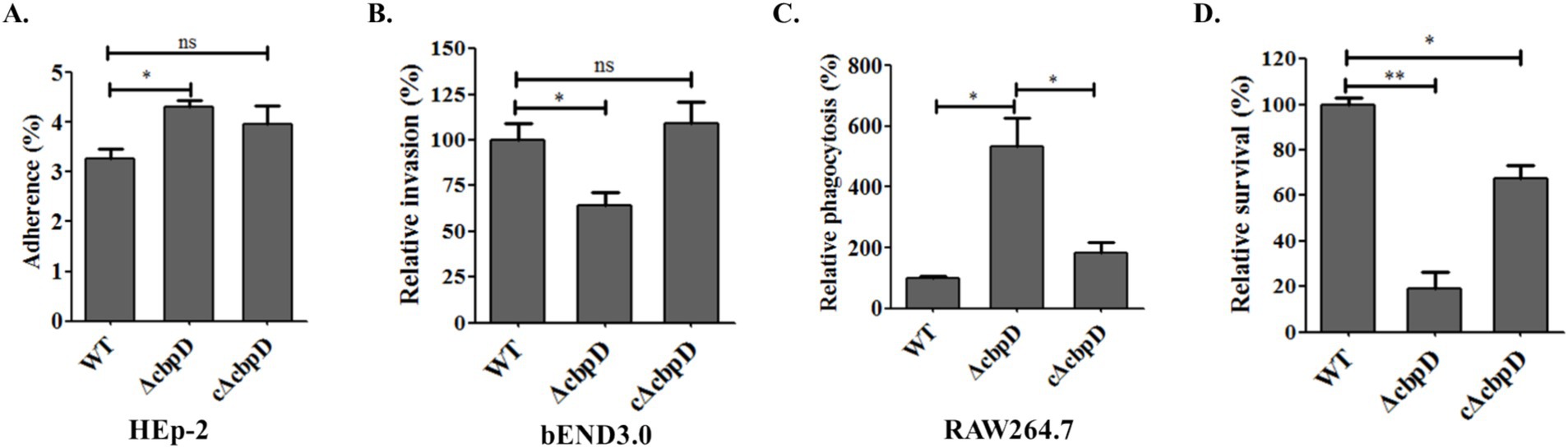
Figure 3. Roles of the CbpD factor among host cell infection. (A) Adhesion assay in HEp-2 cells. (B) Invasion to bEND.3 cells. (C) Phagocytosis rate in murine macrophage RAW264.7. (D) Survival capability in mice whole blood. Data were shown as mean ± SD of three independent experiments and the significant differences indicated as *, p < 0.05; **, p < 0.01.
3.4 The CbpD is a potential virulence factor involved in the pathogenicity of SS2
An in vivo mouse infection model was used to evaluate the impact of cbpD gene deletion on SS2 virulence at a lethal dose. The findings indicated that 50% of the mice in the WT infection group died within 24 h, and all died within 72 h. The clinical symptoms in the WT-infected mice were noticeable, with common manifestations such as distinguished disheveled hair, tearing and blindness, mental signs, and movement disorders. Nevertheless, a significant decrease in the virulence of the cbpD deletion strain was shown as none of the mice died 3 days after infection, resulting in a survival rate of 41.67% as well as strikingly alleviated clinical symptoms (Figure 4A). The results of bacterial load in mouse organs affirmed that the susceptible strain of the ΔcbpD was easier to clear since the prominently reduced colonization colonies of the ΔcbpD were shown in mice organs in Figure 4B. In accordance with the results of bacterial burden in organs, the milder and relieved pathological damages, such as myolysis and disruption of cardiac muscle fibers in the heart, hepatocellular necrosis, and inflammatory cell infiltration with hemorrhage in the liver, disintegration of the splenic nodules along with bleeding in the spleen, congestion, and rupture of the alveoli accompanied by hemorrhage in the lung, glomerular enlargement, and infiltration with inflammatory cells in the kidney, and purulent meningitis, neuronal cell swelling and degeneration in the brain in the WT infection group, were induced by the CbpD deficiency SS2 infection based on the microscopic histopathological observation (Figure 4C). The results illustrated that cbpD deletion attenuated the virulence of S. suis 2.
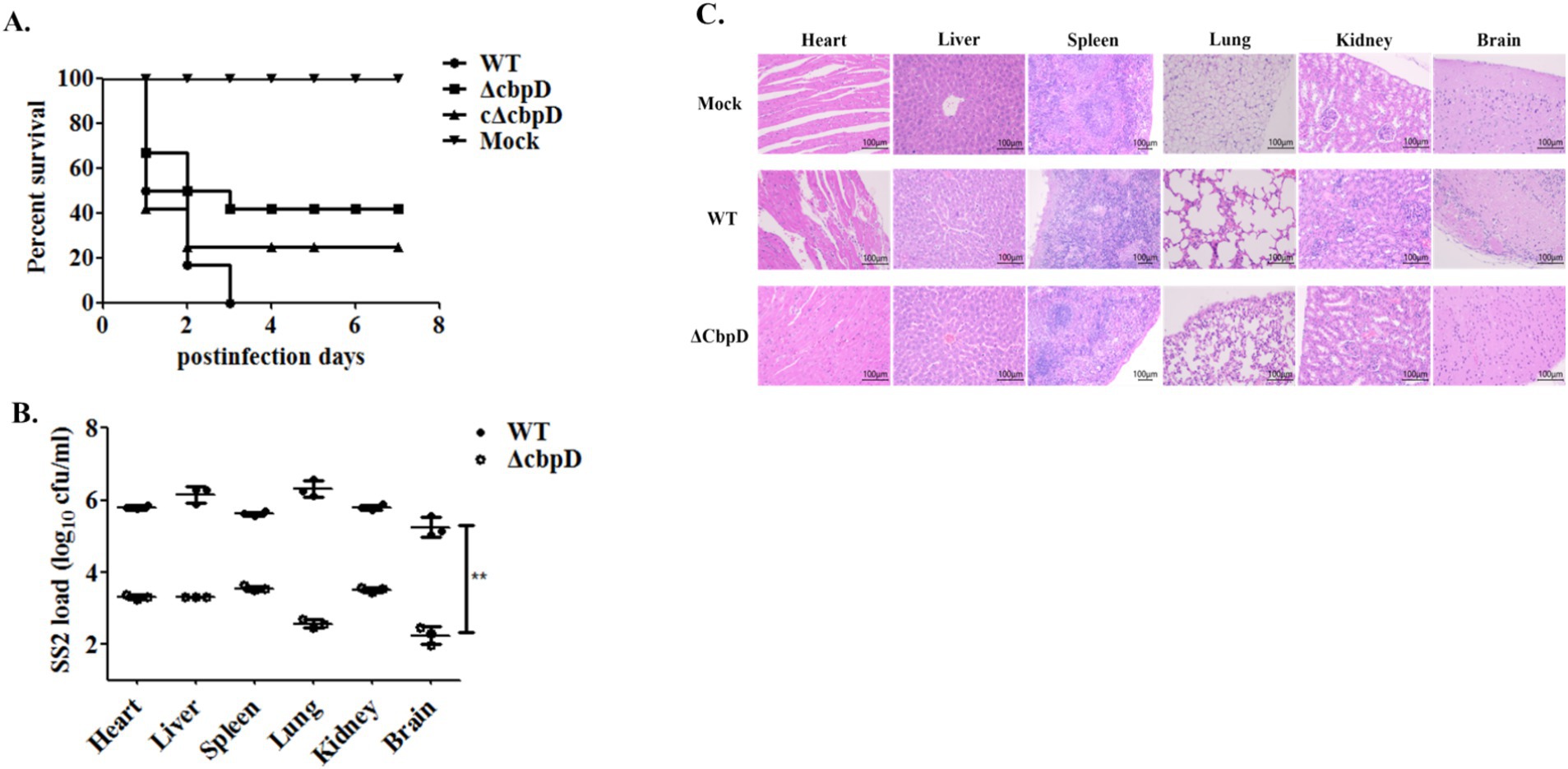
Figure 4. Animal infection model for virulence evaluation. (A) Survival curve of the mice challenged with WT, cbpD gene mutant, and its complemental strains. (B) Bacterial burden in mice organs. Significant difference of ** is indicated as p-value of < 0.01. (C) Histopathological observation of mice organs infected with the WT, cbpD gene mutant and its complemental strain.
3.5 The CbpD deficiency alters the expression of virulence-related factors in SS2
To dissect the impact of the cbpD mutation on bacterial virulence factors expression, seven previously reported virulence-associated factors including the Sly, TGase, SodA, adhesion factor of chaperone DnaJ, Stp, membrane-associated protein of enolase, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were examined by transcriptional qPCR and Western blotting analysis. The qPCR results indicated that the deletion of the cbpD gene induces the upregulated expression of dnaJ and the conspicuously downregulated expression of three genes of tgase, enolase, and stp (Figure 5A). Further protein level detection by Western blotting proved the corresponding differential alterations of these targeted factors in transcriptional levels (Figures 5B,C).
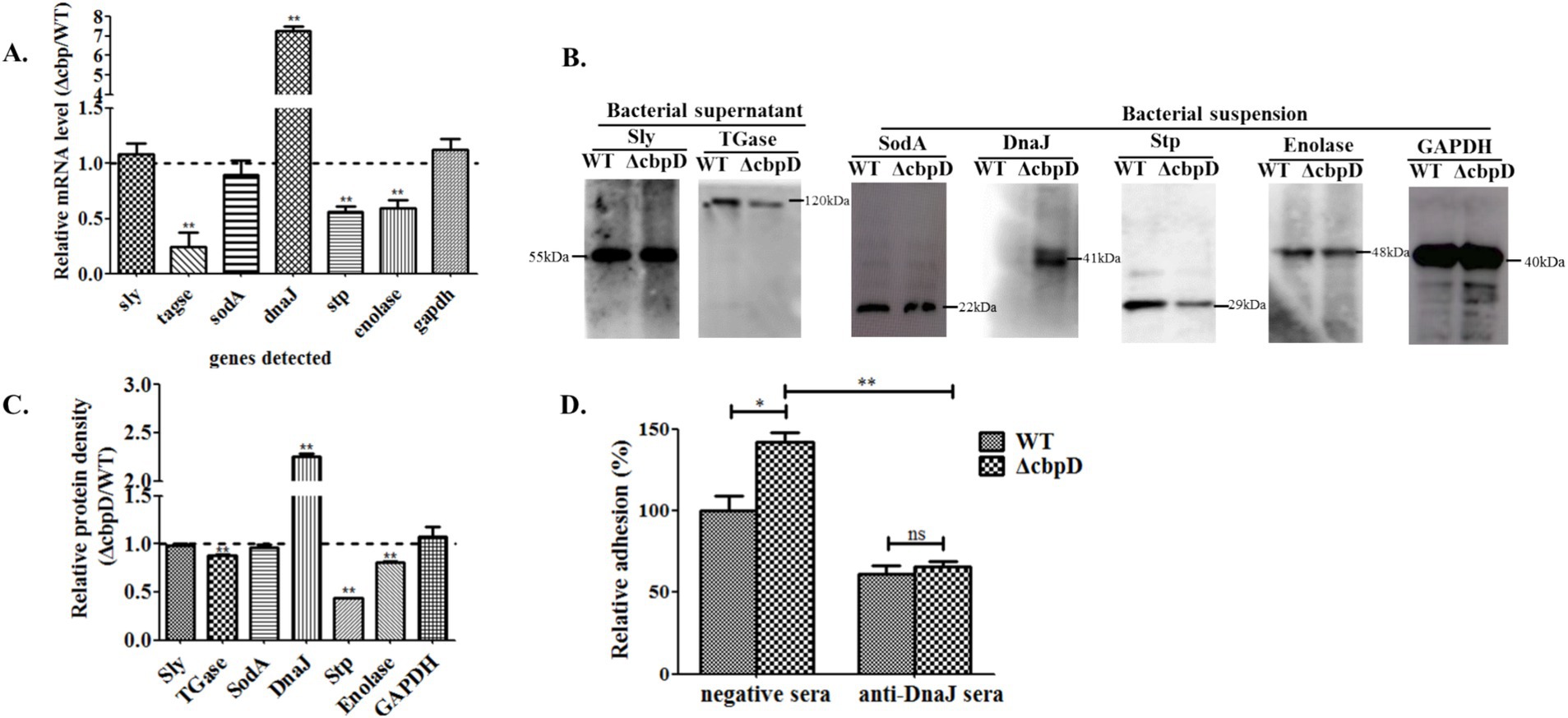
Figure 5. Distinctive virulence-associated factors expression of SS2 in the CbpD factor mutant strain. (A) Relative mRNA fold changes for virulence-associated genes of ΔcbpD mutant in relation to the WT strain. Significant difference of ** is indicated as p < 0.01. (B) Western blotting analysis of the correspondingly detected genes in (A). Two secreted proteins of Sly and TGase, and total proteins of SodA, DnaJ, Stp, enolase, and GAPDH were examined from bacteria supernatant and suspension, respectively. (C) The densitometric difference analysis for Western blotting result. (D) Adhesion inhibition assay for the WT and ΔcbpD mutant strains pretreated with antisera against DnaJ. Significant differences of * indicated as p < 0.05 and ** as p < 0.01.
The adhesive inhibition assay was executed to investigate whether the elevated expression of the adhesion factor of DnaJ in ΔcbpD strain prompted the augmented bacterial adherence to HEp-2 cells. The surprising results in Figure 5D revealed that pre-blocked treatment of the ΔcbpD mutant with monoclonal DnaJ antisera significantly attenuated its adhesion to HEp-2 cells when compared to the negative pre-immune antisera treatment group (p < 0.01).
4 Discussion
CbpD is one of the choline-binding proteins, which are major surface-anchored components that are involved in the microbe’s physiological processes and engagement with host cells. CbpD plays a role in the growth, autolysis, and biofilm formation, they are important for the pathogenicity of S. pneumoniae (11, 12, 20). Both in S. thermophilus and S. mutants, CbpD exerts a pivotal function in fratricide as a murein hydrolase and deletion of the CbpD factor would reduce the transformability (12, 29). A recent investigation has confirmed the presence of CbpD in SS2 and its involvement in enhancing transcription by stimulating peptides that expedite the process of transformation (33). Nevertheless, the specific contribution of CbpD to SS2 pathogenicity remains unknown. In this study, a cbpD gene knockout mutant and its allelic complementation strain based on the lethal zoonotic strain of HA9801 were obtained by typical homologous recombination method and utilized to explore the prospective phenotypes changes and pathogeneses induced by the CbpD factor in SS2 (Figure 1). As a result, the data unveiled that the cbpD deletion mutant exhibited a tendency toward aggregation and a noticeable increase in chain length, and decreased resistance to thermal, acid, and oxidative stress, though no visible growth capability variation was found (Figure 2). Similar morphological abnormality such as cell elongation and septum misplacement was examined and verified in S. pneumoniae (41). This implied the probable impact of CbpD on bacterial cell division and stress tolerance and required further investigation.
Stable adherence to host cells constitutes a crucial step in pathogenic microbe colonization and invasion (42, 43). Zoonotic S. suis strains primarily colonize through the bodies’ respiratory epithelial tissue and the virulent strain possesses the capacity to release a variety of virulence factors, such as suilysin, to compromise the integrity of the epithelial cells and invade the bloodstream (3, 44). They propagate and multiply through immune evasion and other sophisticated modalities (10). Moreover, these virulent strains have the potential to transgress the blood–brain barrier and thereby instigate the occurrence of meningitis (45). In the current study, the inactivation of CbpD was found to enhance the SS2 adherence to the HEp-2 cells and reduce their capability of invasion into bEND.3 cells (Figure 3). High adhesion indicated a stronger colonization for SS2, which may be related to the differential expression of adhesins. Similar results have been found in prsA gene mutant and vaccine candidate strain of SS2 (34, 37), while the invasiveness of pathogens which can destroy the host immune barrier is closely related to virulence factors penetration. Additionally, it was noted that the deletion of cbpD rendered SS2 unable to induce a systemic infection as shown by lower lethality compared to the WT strain in BALB/c mice (Figure 4). The CbpD deficiency resulted in an increased phagocytosed SS2 by macrophage RAW264.7 and a significantly reduced survival capability in mice whole blood (Figures 3, 4). A higher phagocytosis rate owing to the CbpL deficiency in S. pneumoniae was investigated (46). Notably, mice infected with ΔcbpD mutant exhibited a significant reduction in bacterial loads within organs compared to those infected with the WT strain and an effectively mitigated pathological lesion (Figure 4). These findings indicated that CbpD plays a crucial role for SS2 in counteracting host-killing mechanisms by potentially regulating the adhesion process and contributing to anti-phagocytosis as part of its immune evasion response to the host.
Microorganisms encounter diverse microenvironments during the process of host infection, and they can respond to stimuli by modulating the production of proteins essential for a specific process (47). It is well known that the pathogenicity of SS2 demands the complicated and coordinated action of a multitude of virulence-associated factors (42, 45, 48). GAPDH and Enolase have been found to act as crucial adhesins for S. suis, which bind to the extracellular matrix of the host and facilitate adhesion (42, 43). Stp, one of the Ser/thr protein phosphatase, is verified to participate in SS2 infection by regulating capsule thickness and virulence factors translocation (49). TGase is an important virulence factor which can mediate the crucial pathogenic process of antiphagocytosis (50). Sly is also instrumental in anti-phagocytic processes, facilitating bacterial evasion of phagocytosis, countering phagocytosis, and ensuring intracellular viability (44). DnaJ, a pivotal element situated within the DnaK chaperone molecular operon, was identified to act as an adhesin factor contributing to cell adhesion and thermotolerance in SS2 (40). The main function of SodA is to catalyze the superoxide ions released by the respiratory bursts in phagocytes, thus affecting pathogenicity (51). The qRT-PCR analysis and Western blotting confirmation results revealed that CbpD factor deficiency could lead to the discrepant alteration of virulence-associated gene expression. The proteins of enolase, Stp, and the anti-phagocytosis factor of TGase, were discovered to be downregulated, whereas the adhesin of the DnaJ factor was conspicuously induced to high expression, in the cbpD gene mutant strain (Figure 5). Furthermore, the adhesion inhibition test further verified the result of a prominent adhesion rate in ΔcbpD strain owing to its enhanced expression of DnaJ, and these data prompt us to suggest that the differential expression of the corresponding virulence-associated factors possibly contribute to the cbpD gene-deficient strain being more susceptible to phagocytic clearance and a significant attenuation in pathogenicity.
In conclusion, this study demonstrated the crucial functions of CbpD with its underlying pathogenesis in SS2 infection. However, further validation is still required to ascertain whether this regulation is direct or indirect and to explore the underlying molecular mechanism of the CbpD involved in the pathogenesis of SS2. Moreover, the exploration of the anti-virulence strategies or treatments for SS2 infection, such as developing putative antagonists for the targeted CbpD or CbpD-associated virulence proteins, deserves to be studied in the future.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used. The animal study was approved by Animal infection experiments conformed to the animal ethics guidelines from the Medical Ethics Committee of Yichun University (2022018). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LZ: Data curation, Formal analysis, Methodology, Validation, Visualization, Writing – original draft. ML: Formal analysis, Investigation, Validation, Writing – review & editing. GY: Formal analysis, Investigation, Validation, Writing – review & editing. DZ: Data curation, Validation, Visualization, Writing – review & editing. WZ: Data curation, Validation, Visualization, Writing – review & editing. NF: Data curation, Validation, Visualization, Writing – review & editing. XJ: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study is supported by the Jiangxi Provincial Department of Science & Technology (20202BABL215025) and the Jiangxi Provincial Department of Education Project (No. GJJ2201748 and No. GJJ211634).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dutkiewicz, J, Zajac, V, Sroka, J, Wasinski, B, Cisak, E, Sawczyn, A, et al. Streptococcus suis: a re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II – pathogenesis. Ann Agric Environ Med. (2018) 25:186–203. doi: 10.26444/aaem/85651
2. Roodsant, TJ, Van Der Putten, BCL, Tamminga, SM, Schultsz, C, and Van Der Ark, KCH. Identification of Streptococcus suis putative zoonotic virulence factors: a systematic review and genomic meta-analysis. Virulence. (2021) 12:2787–97. doi: 10.1080/21505594.2021.1985760
3. Segura, M. Streptococcus suis research: Progress and challenges. Pathogens. (2020) 9:1–8. doi: 10.3390/pathogens9090707
4. Tang, J, Wang, C, Feng, Y, Yang, W, Song, H, Chen, Z, et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. (2006) 3:e151. doi: 10.1371/journal.pmed.0030151
5. Dong, X, Chao, Y, Zhou, Y, Zhou, R, Zhang, W, Fischetti, VA, et al. The global emergence of a novel Streptococcus suis clade associated with human infections. EMBO Mol Med. (2021) 13:e13810. doi: 10.15252/emmm.202013810
6. Segura, M, Aragon, V, Brockmeier, SL, Gebhart, C, Greeff, A, Kerdsin, A, et al. Update on Streptococcus suis research and prevention in the era of antimicrobial restriction: 4th international workshop on S. suis. Pathogens. (2020) 9:1–39. doi: 10.3390/pathogens9050374
7. Ji, L, Chen, Z, Li, F, Hu, Q, Xu, L, Duan, X, et al. Epidemiological and genomic analyses of human isolates of Streptococcus suis between 2005 and 2021 in Shenzhen, China. Front Microbiol. (2023) 14:1118056. doi: 10.3389/fmicb.2023.1118056
8. Neila-Ibáñez, C, Casal, J, Hennig-Pauka, I, Stockhofe-Zurwieden, N, Gottschalk, M, Migura-García, L, et al. Stochastic assessment of the economic impact of Streptococcus suis-associated disease in German, Dutch and Spanish swine farms. Front Vet Sci. (2021) 8:676002. doi: 10.3389/fvets.2021.676002
9. Weinert, LA, Chaudhuri, RR, Wang, J, Peters, SE, Corander, J, Jombart, T, et al. Genomic signatures of human and animal disease in the zoonotic pathogen Streptococcus suis. Nat Commun. (2015) 6:6740. doi: 10.1038/ncomms7740
10. Segura, M, Fittipaldi, N, Calzas, C, and Gottschalk, M. Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol. (2017) 25:585–99. doi: 10.1016/j.tim.2017.02.005
11. Galán-Bartual, S, Pérez-Dorado, I, García, P, and Hermoso, JA. Chapter 11 – Structure and function of choline-binding proteins In: J Brown, S Hammerschmidt, and C Orihuela, editors. Streptococcus pneumoniae. Amsterdam: Academic Press (2015). 207–30.
12. Maestro, B, and Sanz, J. Choline binding proteins from Streptococcus pneumoniae: a dual role as Enzybiotics and targets for the Design of new Antimicrobials. Antibiotics. (2016) 5:21. doi: 10.3390/antibiotics5020021
13. Galán-Bartual, S, Pérez-Dorado, I, García, P, and Hermoso, JA (2015). Structure and function of choline-binding proteins. 207–230. doi: 10.1016/B978-0-12-410530-0.00011-9
14. Lin, IH, Hsu, M-T, and Chang, C-H. Protein domain repetition is enriched in streptococcal cell-surface proteins. Genomics. (2012) 100:370–9. doi: 10.1016/j.ygeno.2012.08.001
15. Bergmann, S, and Hammerschmidt, S. Versatility of pneumococcal surface proteins. Microbiology. (2006) 152:295–303. doi: 10.1099/mic.0.28610-0
16. Hakenbeck, R, Madhour, A, Denapaite, D, and Brückner, R. Versatility of choline metabolism and choline-binding proteins in streptococcus pneumoniae and commensal streptococci. FEMS Microbiol Rev. (2009) 33:572–86. doi: 10.1111/j.1574-6976.2009.00172.x
17. Gosink, KK, Mann, ER, Guglielmo, C, Tuomanen, EI, and Masure, HR. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun. (2000) 68:5690–5. doi: 10.1128/IAI.68.10.5690-5695.2000
18. Desa, MN, Sekaran, SD, Vadivelu, J, and Parasakthi, N. Distribution of CBP genes in Streptococcus pneumoniae isolates in relation to vaccine types, penicillin susceptibility and clinical site. Epidemiol Infect. (2007) 136:940–2. doi: 10.1017/S0950268807009363
19. Domenech, M, Ruiz, S, Moscoso, M, and García, E. In vitro biofilm development of Streptococcus pneumoniae and formation of choline-binding protein–DNA complexes. Environ Microbiol Rep. (2015) 7:715–27. doi: 10.1111/1758-2229.12295
20. Ji, H, Zhou, Y, Zhang, L, Wang, Y, Zhang, F, Bai, J, et al. (2021). Function analysis of choline binding domains of LytA, LytC and CbpD in the biofilm formation process of Streptococcus pneumoniae. Microb Pathog. (2023) 174:105939. doi: 10.1016/j.micpath.2022.105939
21. Johnsborg, O, and HåVarstein, LS. Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J Bacteriol. (2009) 191:5859–64. doi: 10.1128/JB.00724-09
22. Luo, R, Mann, B, Lewis, WS, Rowe, A, Heath, R, Stewart, ML, et al. Solution structure of choline binding protein a, the major adhesin of Streptococcus pneumoniae. EMBO J. (2005) 24:34–43. doi: 10.1038/sj.emboj.7600490
23. Trappetti, C, Potter, AJ, Paton, AW, Oggioni, MR, Paton, JC, and Camilli, A. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun. (2011) 79:4550–8. doi: 10.1128/IAI.05644-11
24. Vilhena, C, Du, S, Battista, M, Westermann, M, Kohler, T, Hammerschmidt, S, et al. The choline-binding proteins PspA, PspC, and LytA of Streptococcus pneumoniae and their interaction with human endothelial and red blood cells. Infect Immun. (2023) 91:e00154–23. doi: 10.1128/iai.00154-23
25. Wei, H, and Håvarstein, LS. Fratricide is essential for efficient gene transfer between pneumococci in biofilms. Appl Environ Microbiol. (2012) 78:5897–905. doi: 10.1128/AEM.01343-12
26. Eldholm, V, Johnsborg, O, Straume, D, Ohnstad, HS, Berg, KH, Hermoso, JA, et al. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol. (2010) 76:905–17. doi: 10.1111/j.1365-2958.2010.07143.x
27. Haleem Kashif, S, Ali Youssif, M, Yesilkaya, H, Kohler, T, Hammerschmidt, S, Andrew Peter, W, et al. The pneumococcal surface proteins PspA and PspC sequester host C4-binding protein to inactivate complement C4b on the bacterial surface. Infect Immun. (2018) 87:e00742-18. doi: 10.1128/iai.00742-00718
28. Berg, KH, Ohnstad, HS, and Håvarstein, LS. LytF, a novel competence-regulated Murein hydrolase in the genus streptococcus. J Bacteriol. (2012) 194:627–35. doi: 10.1128/JB.06273-11
29. Biørnstad, TJ, Ohnstad, HS, and Håvarstein, LS. Deletion of the murein hydrolase CbpD reduces transformation efficiency in Streptococcus thermophilus. Microbiology. (2012) 158:877–85. doi: 10.1099/mic.0.056150-0
30. Claverys, JP, Martin, B, and Håvarstein, LS. Competence-induced fratricide in streptococci. Mol Microbiol. (2007) 64:1423–33. doi: 10.1111/j.1365-2958.2007.05757.x
31. Eldholm, V, Johnsborg, O, Haugen, K, Ohnstad, HS, and Håvarstein, LS. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology. (2009) 155:2223–34. doi: 10.1099/mic.0.026328-0
32. Kausmally, L, Johnsborg, O, Lunde, M, Knutsen, E, and HåVarstein, LS. Choline-binding protein D (CbpD) in streptococcus pneumoniae is essential for competence-induced cell lysis. J Bacteriol. (2005) 187:4338–45. doi: 10.1128/JB.187.13.4338-4345.2005
33. Zhu, Y, Ma, J, Zhang, Y, Zhong, X, Bai, Q, Dong, W, et al. CrfP, a fratricide protein, contributes to natural transformation in Streptococcus suis. Vet Res. (2021) 52:50. doi: 10.1186/s13567-021-00917-x
34. Liu, H, Fu, H, Jiang, X, Liao, X, Yue, M, Li, X, et al. PrsA contributes to Streptococcus suis serotype 2 pathogenicity by modulating secretion of selected virulence factors. Vet Microbiol. (2019) 236:108375. doi: 10.1016/j.vetmic.2019.07.027
35. Jiang, X, Yang, Y, Zhou, J, Zhu, L, Gu, Y, Zhang, X, et al. Roles of the putative type IV-like secretion system key component VirD4 and PrsA in pathogenesis of Streptococcus suis type 2. Front Cell Infect Microbiol. (2016) 6:172. doi: 10.3389/fcimb.2016.00172
36. Jiang, X, Zhu, L, and Zhan, D. Deletion of lacD gene affected stress tolerance and virulence of Streptococcus suis serotype 2. J Microbiol. (2022) 60:948–59. doi: 10.1007/s12275-022-2146-4
37. Jiang, X, Yang, Y, Zhu, L, Gu, Y, Shen, H, Shan, Y, et al. Live Streptococcus suis type 5 strain XS045 provides cross-protection against infection by strains of types 2 and 9. Vaccine. (2016) 34:6529–38. doi: 10.1016/j.vaccine.2016.05.003
38. Jiang, X, Yang, Y, Zhou, J, Liu, H, Liao, X, Luo, J, et al. Peptidyl isomerase PrsA is surface-associated on Streptococcus suis and offers cross-protection against serotype 9 strain. FEMS Microbiol Lett. (2019) 366:fnz002. doi: 10.1093/femsle/fnz002
39. Jiang, X, Yu, G, Zhu, L, Siddique, A, Zhan, D, Zhou, L, et al. Flanking N- and C-terminal domains of PrsA in Streptococcus suis type 2 are crucial for inducing cell death independent of TLR2 recognition. Virulence. (2023) 14:1–13. doi: 10.1080/21505594.2023.2249779
40. Zhang, X, Jiang, X, Yang, L, Fang, L, Shen, H, Lu, X, et al. DnaJ of Streptococcus suis type 2 contributes to cell adhesion and Thermotolerance. J Microbiol Biotechnol. (2015) 25:771–81. doi: 10.4014/jmb.1408.08085
41. Straume, D, Stamsås, GA, Salehian, Z, and Håvarstein, LS. Overexpression of the fratricide immunity protein ComM leads to growth inhibition and morphological abnormalities in Streptococcus pneumoniae. Microbiology. (2017) 163:9–21. doi: 10.1099/mic.0.000402
42. Fittipaldi, N, Segura, M, Grenier, D, and Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. (2012) 7:259–79. doi: 10.2217/fmb.11.149
43. Xia, X, Qin, W, Zhu, H, Wang, X, Jiang, J, and Hu, J. How Streptococcus suis serotype 2 attempts to avoid attack by host immune defenses. J Microbiol Immunol Infect. (2019) 52:516–25. doi: 10.1016/j.jmii.2019.03.003
44. Tenenbaum, T, Asmat, TM, Seitz, M, Schroten, H, and Schwerk, C. Biological activities of suilysin: role in Streptococcus suis pathogenesis. Future Microbiol. (2016) 11:941–54. doi: 10.2217/fmb-2016-0028
45. Meurer, M, Öhlmann, S, Bonilla, MC, Valentin-Weigand, P, Beineke, A, Hennig-Pauka, I, et al. Role of bacterial and host DNases on host-pathogen interaction during Streptococcus suis meningitis. Int J Mol Sci. (2020) 21:5289. doi: 10.3390/ijms21155289
46. Gutiérrez-Fernández, J, Saleh, M, Alcorlo, M, Gómez-Mejía, A, Pantoja-Uceda, D, Treviño, MA, et al. Modular architecture and unique teichoic acid recognition features of choline-binding protein L (CbpL) contributing to pneumococcal pathogenesis. Sci Rep. (2016) 6:38094 doi: 10.1038/srep38094
47. Olive, AJ, and Sassetti, CM. Metabolic crosstalk between host and pathogen: sensing, adapting and competing. Nat Rev Microbiol. (2016) 14:221–34. doi: 10.1038/nrmicro.2016.12
48. Segura, M, Calzas, C, Grenier, D, and Gottschalk, M. Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett. (2016) 590:3772–99. doi: 10.1002/1873-3468.12364
49. Liu, H, Ye, C, Fu, H, Yue, M, Li, X, and Fang, W. Stk and Stp1 participate in Streptococcus suis serotype 2 pathogenesis by regulating capsule thickness and translocation of certain virulence factors. Microb Pathog. (2021) 152:104607. doi: 10.1016/j.micpath.2020.104607
50. Yu, J, Pian, Y, Ge, J, Guo, J, Zheng, Y, Jiang, H, et al. Functional and structural characterization of the Antiphagocytic properties of a novel transglutaminase from Streptococcus suis. J Biol Chem. (2015) 290:19081–92. doi: 10.1074/jbc.M115.643338
Keywords: Streptococcus suis type 2, choline-binding protein of CbpD, pathogenicity, virulence factors, modulation
Citation: Zhu L, Li M, Yu G, Zhan D, Zeng W, Fu N and Jiang X (2024) Investigation of choline-binding protein of CbpD in the pathogenesis of Streptococcus suis type 2. Front. Vet. Sci. 11:1486347. doi: 10.3389/fvets.2024.1486347
Edited by:
Camila Hamond, University of Connecticut, United StatesReviewed by:
Chun Fang, Yangtze University, ChinaCarlos Emilio Cabrera Matajira, University of São Paulo, Brazil
Copyright © 2024 Zhu, Li, Yu, Zhan, Zeng, Fu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowu Jiang, eHdqaWFuZ0B6anUuZWR1LmNu
 Lexin Zhu1
Lexin Zhu1 Xiaowu Jiang
Xiaowu Jiang