- 1College of Animal Science and Technology, Inner Mongolia Minzu University, Tongliao, China
- 2Inner Mongolia Engineering Technology Research Center for Prevention and Control of Beef Cattle Diseases, Tongliao, China
- 3Beef Cattle Industry School of Inner Mongolia Autonomous Region, Tongliao, China
- 4Hinggan League Animal Disease Control Center, Hinggan League, China
- 5Zhalantun Vocational College, Hulunbeier, China
- 6Tongliao Vocational College, Tongliao, China
- 7College of Animal Science and Technology, Jilin Agricultural University, Changchun, China
Introduction: Escherichia coli is a major pathogen responsible for calf diarrhea, which has been exacerbated by the irrational and unscientific use of antimicrobial drugs, leading to significant drug resistance.
Methods: This study focused on the isolation and identification of E. coli from calf diarrhea samples in the Tongliao area of China. Isolation was conducted using selective media, Gram staining, and 16S rRNA sequencing. The minimum inhibitory concentration (MIC) of E. coli was determined through the microbroth dilution method. Additionally, the presence of antibiotic-resistant genes was detected, and multidrug-resistant strains were selected for whole-genome sequencing (WGS).
Results: The results revealed that all 40 isolated strains of E. coli exhibited resistance to sulfadiazine sodium, enrofloxacin, and ciprofloxacin, with 90% of the strains being susceptible to polymyxin B. Notably, strains 11, 23, and 24 demonstrated severe resistance. The detection rates of the antibiotic resistance genes TEM-1, TEM-206, strA, strB, qacH, and blaCTX were 100%, indicating a high prevalence of these genes. Moreover, the majority of strains carried antibiotic resistance genes consistent with their resistance phenotypes. WGS of strains 11, 23, and 24 revealed genome sizes of 4,897,185 bp, 4,920,234 bp, and 4,912,320 bp, respectively. These strains carried two, one, and two plasmids, respectively. The prediction of antibiotic resistance genes showed a substantial number of these genes within the genomes, with strain 24 harboring the highest number, totaling 77 subspecies containing 88 antibiotic resistance genes.
Discussion: In conclusion, all 40 isolated strains of E. coli from calf diarrhea in this study were multidrug-resistant, exhibiting a broad distribution of antibiotic resistance genes and mobile components. This poses a significant risk of horizontal gene transfer, highlighting the critical situation of antibiotic resistance in this region.
1 Introduction
Neonatal calf diarrhea (NCD) presents a formidable challenge to the global livestock industry, significantly hindering sector growth. This disease incurs substantial economic losses due to its high rates of morbidity and mortality, growth retardation in affected animals, and the associated treatment costs, along with severe long-term consequences (1–3). NCD is a leading cause of mortality in calves within their first month of life, with approximately 57% of pre-weaning calves succumbing to diarrhea (4–6). The etiology of NCD is multifactorial, with infections by various pathogens playing a critical role, compounded by factors such as genetics, age, environmental conditions, and nutritional status. The predominant pathogens causing NCD include Bovine Viral Diarrhea Virus(BVDV), Bovine Rotavirus (BRV), Bovine Coronavirus (BCoV), E. coli, Salmonella, Clostridium perfringens, Cryptosporidium, and Eimeriidae. E. coli, in particular, is highly contagious and can cause severe diarrhea and septicemia in calves, with higher morbidity and mortality rates observed in younger animals (7, 8).
Pathogenic E. coli is typically categorized into intestinal and extraintestinal pathogenic strains. The intestinal pathogenic strains are further divided into six types: Enterotoxigenic E. coli (ETEC), Enterohaemorrhagic E. coli (EHEC)/Shiga toxin-producing E. coli (STEC), Enteropathogenic E. coli (EPEC), Enteroinvasive E. coli (EIEC), Enteroaggregative E. coli (EAEC), and Diffusely adherent E. coli (DAEC). Among these, ETEC is the primary strain responsible for diarrhea in calves (9–12). ETEC’s pathogenic mechanism involves bacterial adhesion and colonization of the small intestinal epithelial cells via fimbriae, followed by the transfer of small peptides or secretion of enterotoxins that trigger a cascade of intracellular signals. This leads to a rapid loss of electrolytes from the intracellular environment to the intestinal lumen, resulting in watery diarrhea in calves (13–15).
In veterinary practice, while antibiotics are often used to treat bacterial NCD, the application of antibiotic therapy for ETEC infections remains controversial. In this respect, most authors only justify its use in cases that are evolving toward systemic disease, both for the prevention of bacteriemia and to reduce the number of ETEC in the gut. However, antimicrobials are commonly used by veterinary surgeons for teating NCD, even in the absence of demonstrated ETEC involvement. This increased use of antibiotics has accelerated the emergence and spread of bacterial resistance, giving rise to multidrug-resistant (MDR) strains that pose a significant threat to global public health (16, 17). The emergence of bacterial resistance has promoted the development of colostrums and probiotics. Several studies have suggested the benefits of using bovine colostrum and probiotics as prophylactics to prevent diarrhea in calves by the administration of colostrum and probiotics after the first day of life. However, there is a lack of evidence supporting the use of maternally derived bovine colostrum and colostrum replacements as a therapy for diarrhea in calves (18). E. coli, serving as a potential source, intermediate carrier, and crucial reservoir of antibiotic resistance genes (ARGs), plays a pivotal role in the dissemination of bacterial resistance (19). Horizontal gene transfer (HGT) via plasmid-mediated conjugation is one of the primary mechanisms for the spread of ARGs (20, 21). HGT is facilitated by several well-known mechanisms including transduction, transformation, and conjugation, with conjugation being the most significant.
HGT is mediated by mobile genetic elements (MGEs), which are DNA molecules capable of moving between replicons (intracellular mobility) or between bacterial cells (intercellular mobility). MGEs include conjugative DNA elements (plasmids and integrative and conjugative elements [ICEs]), transposable DNA elements (transposons and integrons), and bacteriophages. These elements carry various genes, including those for antimicrobial and metal resistance, virulence, and catabolism. Recently, antimicrobial-resistant (AMR) bacteria have emerged as a critical public health threat. The spread of ARGs through HGT-associated MGEs, coupled with the role of MGEs in promoting the diversification of AMR bacteria, exacerbates this issue (22–24). This study aimed to understand the drug resistance of E. coli causing diarrhea in calves in the Tongliao area of China. A preliminary assessment was conducted through the isolation and identification of bacteria, drug resistance analysis, detection of drug resistance genes, and analysis of mobile components. The findings provide guidance for the clinical treatment of calf diarrhea caused by E. coli in the Tongliao area. They also establish a research basis for further study and control of the spread of bacterial resistance.
2 Materials and methods
2.1 Sample collection and strain isolation
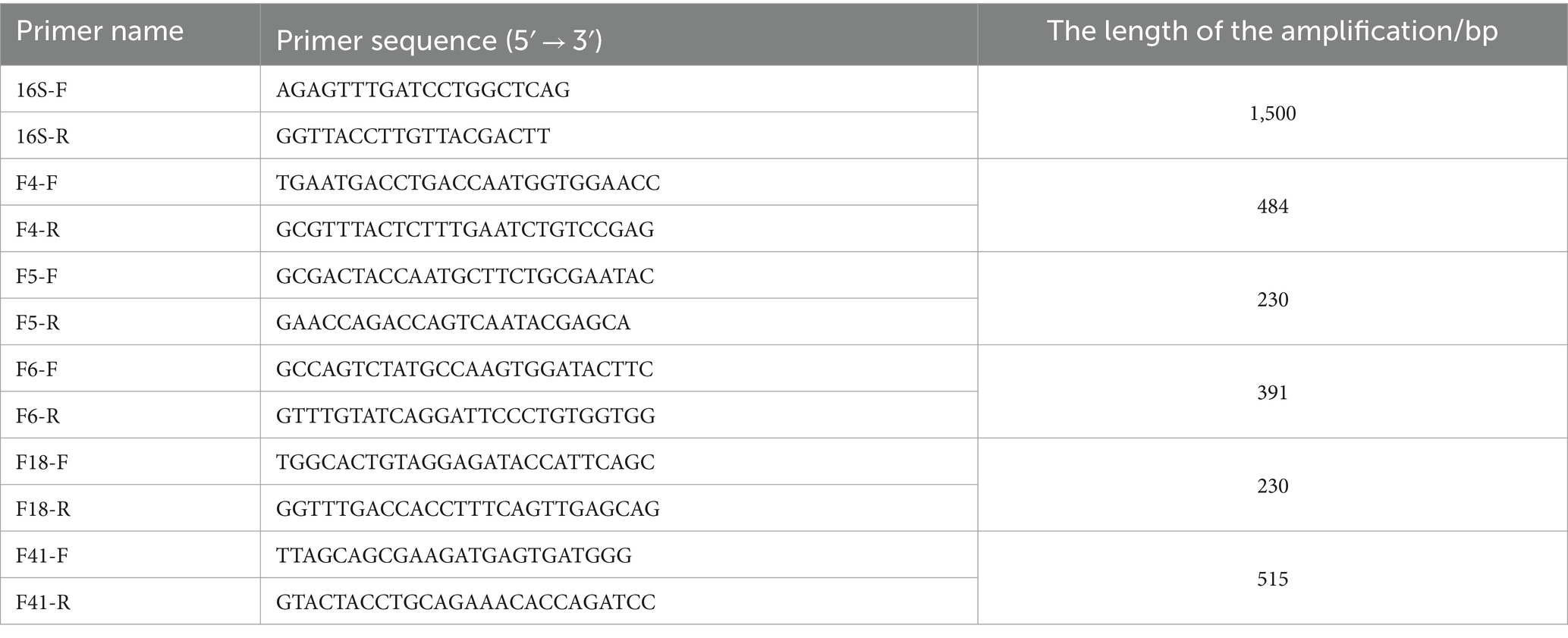
Between May 2021 and May 2022, fecal samples from 50 calves with diarrhea symptoms were collected from cattle farms in Tongliao City, Inner Mongolia. The sample group consisted of 28 female and 22 male Simmental calves within 1 month of age. None of the sampled calves had been treated with antibiotics. The samples were immediately streaked onto E. coli Chromogenic Medium and incubated at 37°C for 18 h. This process was repeated two to three times to ensure purity. Single colonies were obtained by isolating and purifying differently colored colonies. The isolated strains were identified by Gram staining and 16S rRNA sequencing. Genomic DNA was extracted from the bacteria using a previously described protocol, and the 16S rRNA gene was amplified using universal polymerase chain reaction (PCR) primers (Table 1). The PCR products were sequenced at Comate Bioscience Co., Ltd. (Jilin, China) and analyzed using BLASTN on the National Center for Biotechnology Information (NCBI) website (25). The E. coli ATCC 25922 quality control strain was kindly provided by the Laboratory of Pharmacology and Toxicology, School of Animal Medicine, Jilin Agricultural University.
2.2 Detection of Escherichia coli fimbria adhesin
PCR was used to detect the fimbria adhesin gene of E. coli isolates. The primer sequences and reaction conditions are shown in Table 1. The PCR products were detected by 1.3% agarose gel electrophoresis.
2.3 Drug sensitivity testing
Antimicrobial susceptibility was tested using the broth microdilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (26). Minimum inhibitory concentrations (MIC) of each antibiotic were classified as resistant (R), intermediate (I), or susceptible (S) based on CLSI breakpoints, or the National Antibiotic Resistance Monitoring System (NARMS) breakpoints for intestinal bacteria when CLSI breakpoints were unavailable. E. coli isolates resistant to three or more antibacterial agents were considered multidrug-resistant (MDR) (27). Tests were conducted in triplicate for each strain, with E. coli ATCC 25922 as the quality control strain.
2.4 Detection of resistance genes
Primer 5.0 software was used to design primers for the resistance genes. The primer sequences for E. coli antibiotic resistance genes are listed in Supplementary Table S1. Extraction of E. coli genomic DNA and detection of drug resistance genes were carried out as previously described (28).
2.5 Whole genome sequencing of multidrug-resistant strains
The selection of severely drug-resistant strains for whole-genome sequencing was based on drug susceptibility testing. Whole-genome sequencing was conducted on the Nanopore sequencing platform by Biomarker Technologies Corporation (Qingdao, China). High-quality genomic DNA was extracted, and its concentration, purity, and integrity were assessed using a NanoDrop spectrophotometer and Qubit fluorometer (Thermo Fisher, Waltham, MA, United States). Large DNA fragments were filtered using the BluePippin system. A library was prepared using the Oxford Nanopore Technologies (ONT) Template Prep Kit (SQK-LSK109; Oxford, United Kingdom) and the NEB Next FFPE DNA Repair Mix Kit (Ipswich, MA, United States). The high-quality library was sequenced on the ONT PromethION platform, yielding raw sequencing data (29). Filtered subreads were assembled using Canu v1.5 software (30). The assembly results were corrected by Racon v3.4.3 software using three generations of subreads, with further error correction performed using Pilon v1.22 software with second-generation data to obtain a more accurate genome for subsequent analysis.
2.6 Data analysis
Gene prediction was conducted using Prodigal v2.6.3 software (31). Gene sequences were cross-referenced with the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional databases for annotation (32, 33). Drug resistance genes were identified by comparing sequences with the antibiotic resistance database using ResFinder software (34–36). Plasmid typing was predicted using PlasmidFinder 2.1 software (36, 37). MobileElementFinder software was employed to predict mobile elements within the genomes (38).
3 Results
3.1 Identification of Escherichia coli
A total of 40 E. coli strains were isolated from 50 fecal samples. The isolates showed blue-green colonies on E. coli Chromogenic Medium and specific bands close to the size of the target fragment 16Sr RNA (1,500 bp) were amplified in all test samples. The sequencing results, compared via BLAST on the NCBI database, confirmed the isolates were E. coli.
3.2 Detection of Escherichia coli fimbria adhesin
The fimbria adhesin gene in the 40 E. coli genomes was detected using PCR. The detection rates for the F5 gene and the F41 gene were 55% (22/40) and 45%, respectively, while the F4, F6, and F18 genes were not detected. The PCR results for the fimbria adhesin gene are shown in Figure 1.
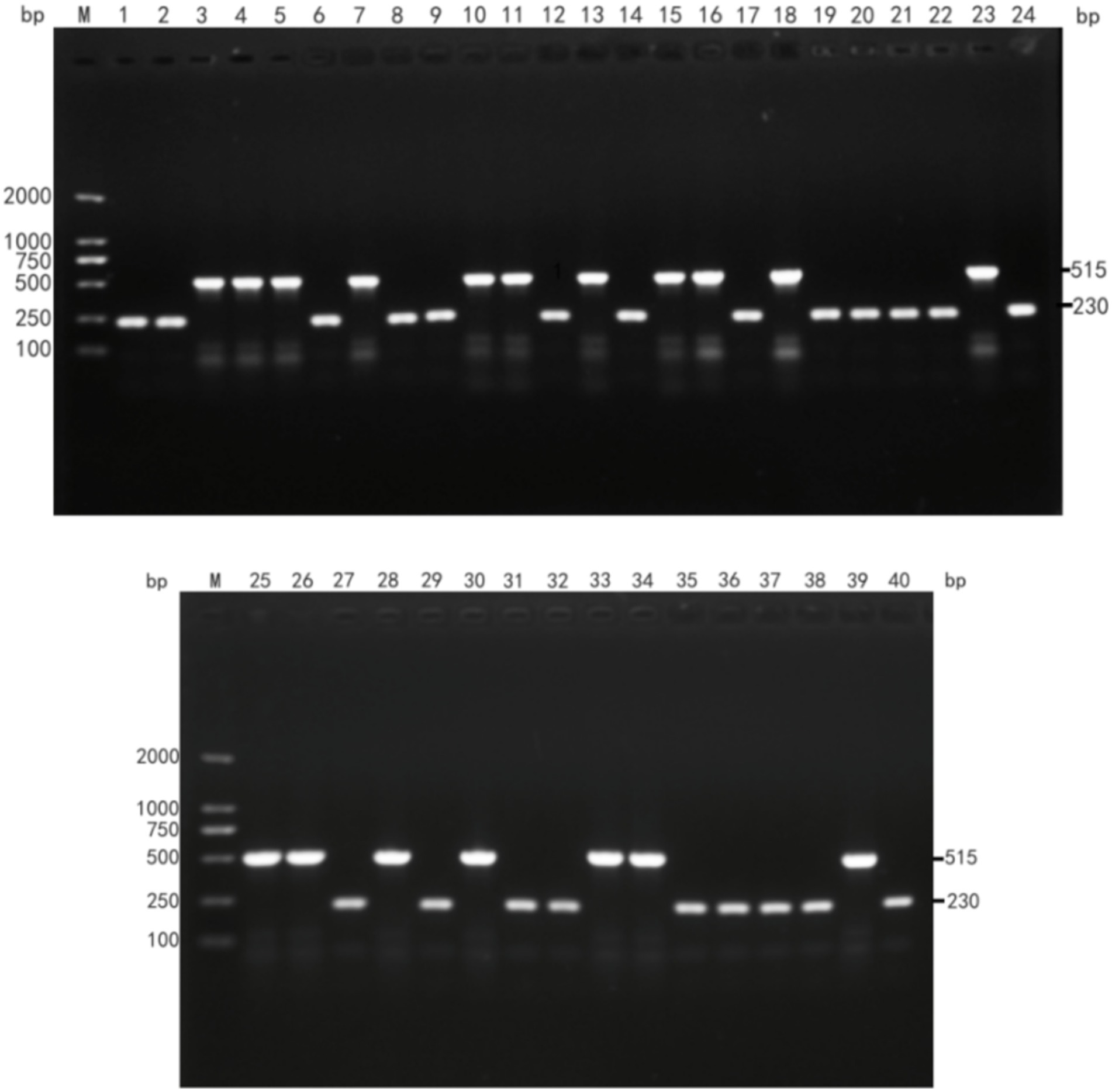
Figure 1. Detection results of E. coli fimbria adhesin. (M: DL2000DNA Maeker; 1–40 PCR amplification product of E. coli fimbria adhesin).
3.3 Drug sensitivity tests
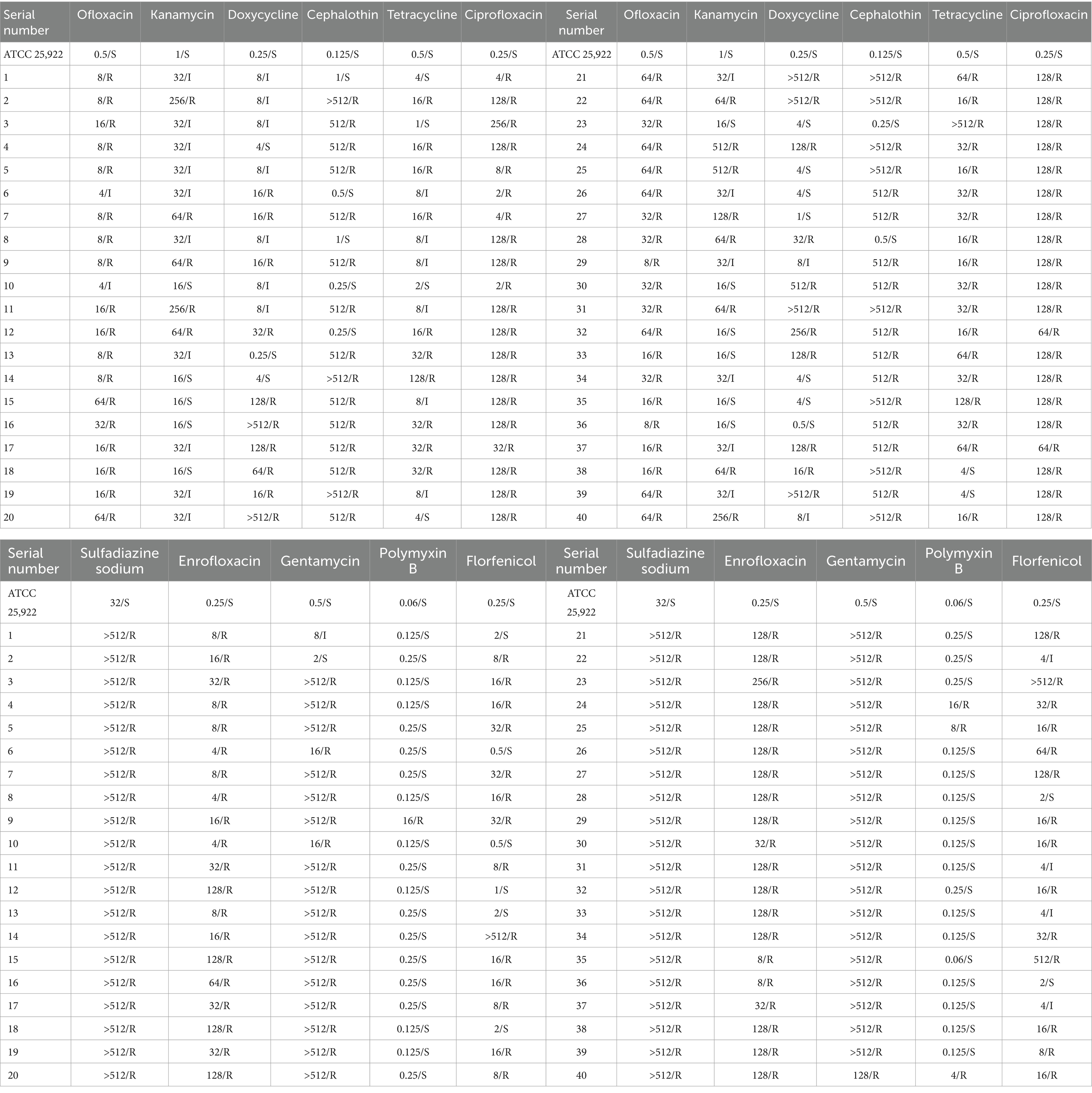
The results of drug sensitivity tests for the 40 E. coli isolates are presented in Table 2. The highest resistance rates were observed for sulfadiazine sodium, enrofloxacin, and ciprofloxacin at 100%. In contrast, polymyxin B showed the lowest resistance rate at 10%. E. coli strain 24 exhibited resistance to all 11 antibiotics tested, while strains 11 and 23 also demonstrated high resistance. Therefore, these three strains were selected for whole-genome sequencing.
3.4 Detection of resistance genes in Escherichia coli
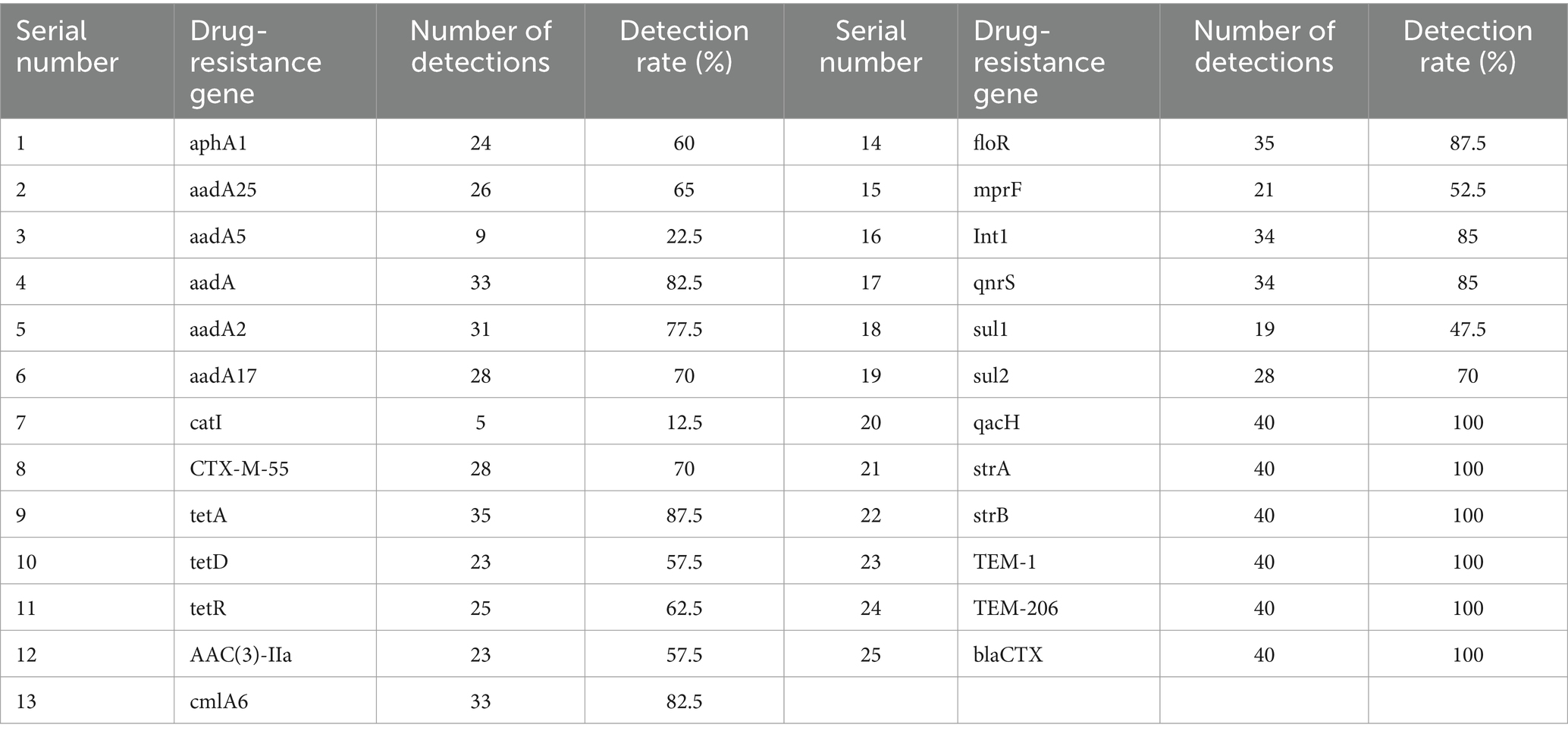
The detection results of AMR genes are shown in Table 3. Resistance genes for 25 antibiotics were identified. The highest detection rates were for β-lactam antibiotic resistance genes TEM-1, TEM-206, and blaCTX; aminoglycoside resistance genes strA and strB; and the quaternary amine compound efflux pump gene qacH, all at 100%. The lowest detection rate was for the phenylpropanol resistance gene catI at 12.5%. The aminoglycoside resistance gene aadA5 and the sulfonamide resistance gene sul1 had detection rates above 50%. This indicates that the predominant resistance genes in E. coli from calf diarrhea in Tongliao are qacH, strA, strB, TEM-1, TEM-206, and blaCTX. Among the 40 E. coli strains, 24 carried the highest number of resistance genes, accounting for 84.62% of the total detected resistance genes. Most resistance genes correlated with their resistance phenotype.
3.5 Whole genome sequencing analysis of multi-drug resistant strains
3.5.1 Genome assembly statistics
Whole genome sequencing of the three multidrug-resistant strains revealed the following genome lengths: strain 24 had 4,912,320 bp with two plasmids and a G + C content of 50.31%; strain 11 had 4,897,185 bp with two plasmids and a G + C content of 50.68%; and strain 23 had 4,920,234 bp with one plasmid and a G + C content of 50.62%. All sequences have been deposited in the NCBI Sequence Read Archive and are publicly accessible under the accession numbers CP157955-CP157957, CP103295-CP103297 and CP157958-CP157989, respectively. The circular whole genome maps of the three E. coli strains are shown in Figure 2. Comparative analysis indicated that strains 11 and 24 showed significant advantages in amino acid transport and metabolism, energy production and conversion, and DNA replication, recombination, and repair. Strain 24 had superior capabilities in energy production and conversion, amino acid transport and metabolism, and carbohydrate transport and metabolism compared to strains 11 and 23, but was less efficient in lipid transport and metabolism, biosynthesis of secondary metabolites, transport and catabolism, and signal transduction mechanisms.
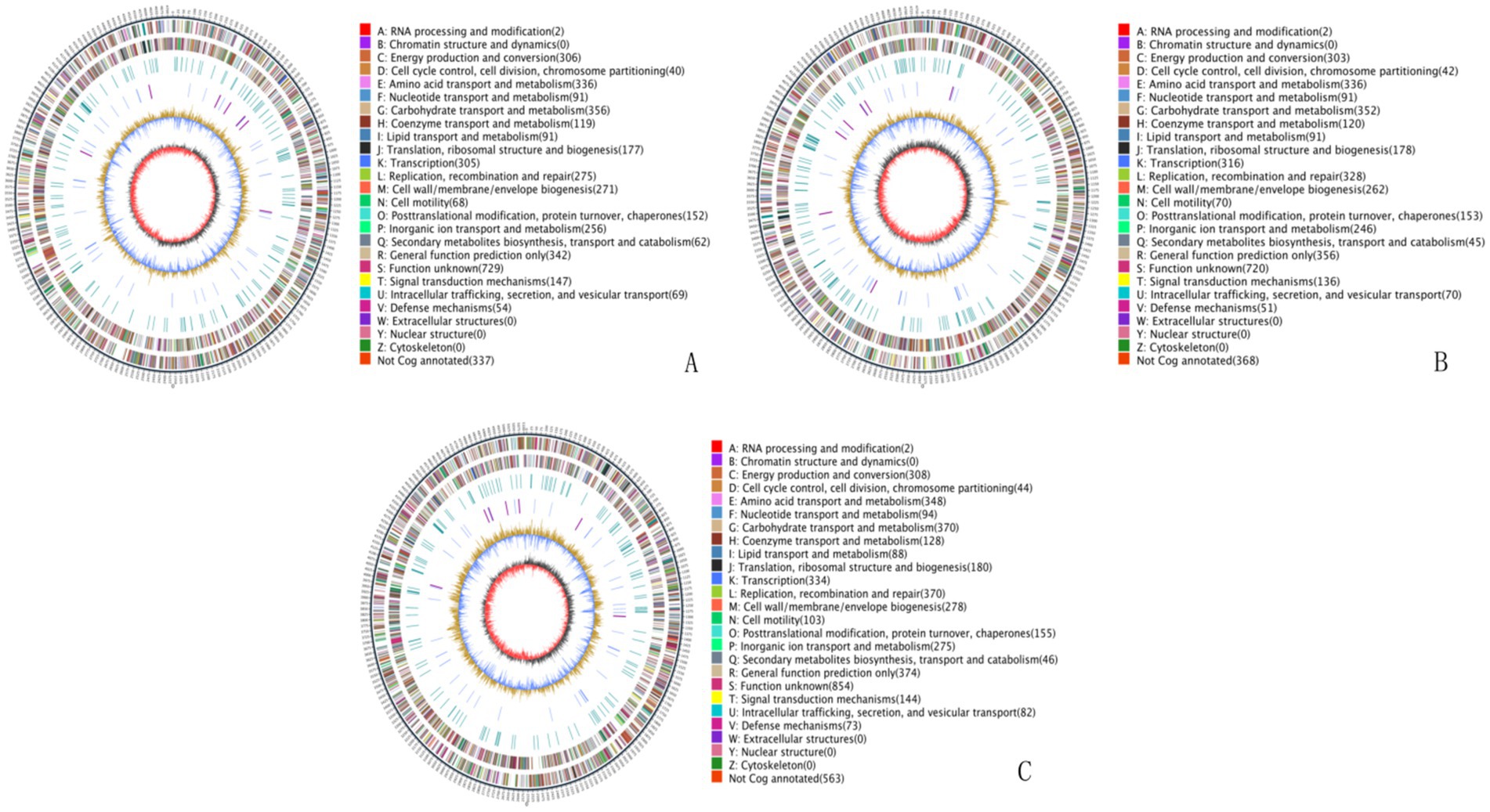
Figure 2. Whole genome maps. (A) Whole genome map of strain 11. (B) Whole genome map of strain 23. (C) Whole genome map of strain 24. The outermost circle is the genome size indication, each scale is 5 kb; the second and third circles are the genes on the positive and negative strands of the genome, respectively, with different colors representing different COG functional classifications; the fourth circle is the repetitive sequences; the fifth circle is the tRNAs and rRNAs, with tRNAs in blue and rRNAs in purple; the sixth circle is the GC-content, and the light yellow portion indicates that the GC-content of the region is higher than the average GC content of the genome, the higher the peak the greater the difference with the average GC content, and the blue part indicates that the GC content of the region is lower than the average GC content of the genome; the innermost circle is the GC-skew, the dark gray represents the region where the G content is greater than the C, and the red represents the region where the C content is greater than the G.
3.5.2 Gene ontology and KEGG function annotation
The genomic sequences of the three E. coli strains were annotated for KEGG metabolic pathways. The results showed significant gene involvement in metabolic pathways, particularly in amino acid biosynthesis and carbon metabolism. GO functional annotation indicated that most genes were related to biological processes, especially metabolic processes, cellular processes, and single-organism processes, which are crucial to the strains’ life activities. Strain 24 had more genes related to catalytic activity, metabolic processes, cell membranes, and cell membrane components compared to strains 11 and 23 (see Supplementary Figure S1).
3.5.3 Analysis of resistance genes
Whole genome sequencing of E. coli strains 11, 23, and 24 revealed 68, 63, and 77 isoform types of resistance genes, respectively, with total counts of 71, 78, and 88 resistance genes. Chromosomal genomes of strains 11, 23, and 24 carried 51, 57, and 58 types of resistance genes, with counts of 53, 72, and 59, respectively. Plasmid genomes carried 17, 6, and 21 types of resistance genes, with counts of 18, 6, and 29, respectively (see Supplementary Tables S2–S4). Resistance genes in the chromosomal genomes of all three strains mediated resistance to quinolones, aminoglycosides, tetracyclines, macrolides, β-lactams, and lincosamide antibiotics. The primary mechanisms included antibiotic efflux, modification-induced inactivation, target site substitution, and alteration of antibiotic targets.
3.5.4 Analysis of mobile components
Mobile elements in the genomes were analyzed using Mobile Element Finder. The analysis predicted that mobile elements in the genomes of the three E. coli strains mainly existed as transposons and insertion sequences, with numerous reverse repetitive sequences (Figure 3). The mobile elements carrying drug resistance genes were primarily composite transposons, mostly from the IS6 family. Only the insertion sequence in plasmid 1 of strain 24 carried a drug resistance gene. Strain 11 contained a compound transposon (TN4352) with no predicted family, while strain 23 had a single transposon (Tn2) with no predicted family (see Supplementary Tables S5–S7).
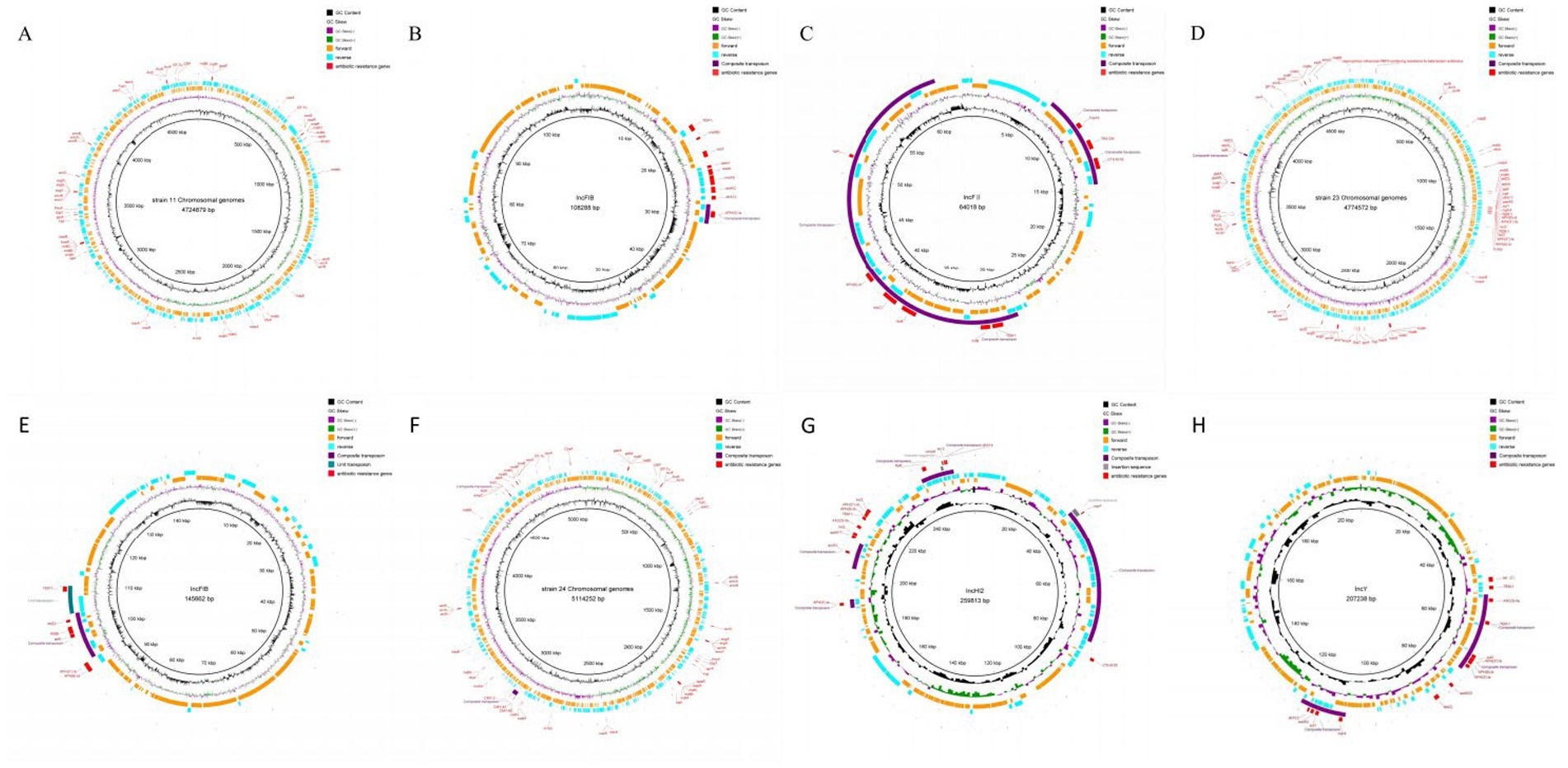
Figure 3. Analysis result of mobile components. (A) Result of mobile components analysis of strain 11 Chromosomal genomes. (B) Result of mobile components analysis of strain 11 plasmid 1. (C) Result of mobile components analysis of strain 11 plasmid 2. (D) Result of mobile components analysis of strain 23 Chromosomal genomes. (E) Result of mobile components analysis of strain 23 plasmid 1. (F) Result of mobile components analysis of strain 24 Chromosomal genomes. (G) Result of mobile components analysis of strain 24 plasmid 1, (H) Result of mobile components analysis of strain 24 plasmid 2.
4 Discussion
Calf diarrhea is prevalent in many farms worldwide, characterized by rapid disease progression and high mortality, leading to significant economic losses for the global farming industry (39). Reports indicate that 39% of calves in the United States suffer from diarrhea (40), with similarly high rates observed in Korea, where 53.4% of calf deaths are attributed to this condition (41). ETEC represents a major cause of diarrhea in calves, as well as an important zoonotic pathogen (42). Diarrhea remains a leading cause of morbidity and mortality throughout the world, especially in developing countries, with ETEC being responsible for hundreds of millions of cases of childhood diarrhea each year, accounting for between 1.6 and 2.5 million deaths (43). Since diarrheic calves are a major source of ETEC transmission to humans, effective prevention and treatment of calf diarrhea remain a significant global challenge (44, 45). Antibiotics have traditionally been effective in managing this disease; however, the extensive and often inappropriate use of antibiotics has led to the emergence of antibiotic-resistant E. coli strains, posing a severe public health problem globally.
As the national core of beef cattle producing area of China, Tongliao has a large number of beef and the diarrhea caused by E. coli is frequently occurring. However, there are few studies on the drug resistance of E. coli in this area. In this study, drug sensitivity testing revealed that the 40 E. coli isolates from calves with diarrhea exhibited varying degrees of resistance to 11 antibiotics. Sodium sulfadiazine, enrofloxacin, and ciprofloxacin showed the highest resistance rates at 100%, followed by gentamicin, ofloxacin, and thiramycin with resistance rates exceeding 80%. Multidrug resistance was also prevalent. Previous studies, such as those by Yassin et al. and Jia et al., have reported high resistance rates to tetracycline, ciprofloxacin, enrofloxacin, and gentamicin in E. coli isolates from different regions in China (46, 47). Similarly, He et al. reported high resistance rates in the Xinjiang region (48). Comparatively, Algammal et al. found lower resistance rates in Egyptian isolates, while Khawaskar et al. reported varied resistance in Indian isolates (11, 15). These variations highlight the regional differences in antibiotic resistance, likely influenced by local therapeutic practices. The high resistance rates observed in this study underscore the severe antibiotic resistance in bovine E. coli in Tongliao, China. Although the sample size of strains in our study is relatively small, but in the follow-up study, we will continuous attention to the resistance of E. coli strains isolate from calf diarrhea in Tongliao.
Polymyxin B has become a last-resort antibiotic for treating Gram-negative multidrug-resistant bacterial infections (49). In this study, the E. coli isolates were generally sensitive to polymyxin B, although 10% showed resistance, emphasizing the need for judicious antibiotic use to prevent further resistance development. The detection of resistance genes revealed that β-lactam resistance genes (blaCTX, TEM-1, TEM-206), aminoglycoside resistance genes (strA, strB), and the quaternary amine compound efflux pump gene (qacH) were prevalent, with a 100% detection rate. This high prevalence of resistance genes, coupled with the presence of multiple resistance genes in individual strains, correlates with their resistance phenotypes and indicates severe resistance issues in the Tongliao area.
Whole genome sequencing (WGS) has been increasingly used to analyze the genetic information of drug-resistant strains and the transfer mechanisms of resistance genes (50–52). WGS of three multidrug-resistant E. coli strains in this study revealed significant genetic insights. Strains 11, 23, and 24 carried two, one, and two plasmids, respectively. GO functional annotation showed an abundance of genes related to metabolic processes, cellular processes, and single-organism processes. Strain 24, in particular, had more genes associated with catalytic activity, metabolic processes, and cell membrane components, suggesting a role in antibiotic efflux and reduced absorption. KEGG pathway analysis indicated significant enrichment of genes involved in amino acid biosynthesis and carbon metabolism, aligning with the GO annotation results. Previous studies have shown that E. coli can develop antibiotic resistance through various mechanisms, with antibiotic resistance mediated by efflux pumps, biofilm formation, and enzymatic modification of antibiotics, among other mechanisms (23, 47, 53). The three E. coli strains in this study exhibited multiple resistance mechanisms, with antibiotic efflux being predominant, especially in strain 24, which carried more resistance genes on plasmids compared to strains 11 and 23.
Previous studies have shown that E. coli can develop antibiotic resistance through various mechanisms, including efflux pumps, biofilm formation, and enzymatic modification of antibiotics (23, 47, 53). As predicted by the CARD database, the three E. coli strains in this study exhibited multiple resistance mechanisms, with antibiotic efflux being the predominant mechanism, especially in strain 24, which carried more resistance genes on plasmids compared to strains 11 and 23. Insertion sequences, the simplest mobile elements in bacterial genomes, play a crucial role in the transfer and dissemination of antibiotic resistance genes (54). Studies have shown that composite transposons, formed by identical or related insertion sequence elements, can facilitate the movement of resistance genes, promoting their spread (55). This study integrated the results of whole-genome sequencing with prediction of mobile genetic elements (MGEs) to analyze the genomes of three E. coli strains. It was found that these genomes carried a large number of transposons and insertion sequences containing antibiotic-resistance genes, and Type I integrase genes were discovered in some composite transposons carrying antibiotic-resistance genes. The Tn4352 complex transposon in plasmid 1 of strain 11 carried both an integrase gene and the aminoglycoside-resistance gene APH(3′)-Ia. One complex transposon containing a Type I integrase gene was found in both plasmid 1 and plasmid 2 of strain 24, and the complex transposon in plasmid 1 contained the chloramphenicol efflux pump protein gene cmlA6, the ADP-ribosyltransferase gene arr-2, and the dihydrofolate reductase gene dfrA14. Furthermore, this composite transposon overlapped with the cmlA6 gene in a Tn3 composite transposon carrying the florfeniol-resistance gene floR. In the composite transposon of plasmid 2, in addition to the Type I integrase and aadA2 and dfrA12 resistance genes, a gene island was observed containing the resistance genes mphA and sul1, although the Type I integrase gene was not located within the gene island. Unlike strains 11 and 24, strain 23 carried the Type I integrase gene in the gene island of the chromosome, which contained the dfrA17, aadA5, and sul1 resistance genes, and its gene structure was more similar to that of the gene island in plasmid 2 of strain 24. Based on the above results and integrase structural analysis, it was inferred that the composite integrase on the plasmids and chromosomal gene islands contained Type I integrase, with some of these integrases carrying the dfrA-aadA resistance gene cassette. Due to the specific structure of the integrons’ multi-gene cassettes, the gene cassettes can be inserted at specific sites under the action of integrase, and the unique structure allows the integrons to capture a large number of exogenous mobile drug resistance genes and then to express these captured exogenous genes. At the same time, the integrons can be carried by transposons, gene islands, and other mobile elements, thus contributing to the horizontal transfer of drug-resistant genes, resulting in the intensification of bacterial drug resistance.
5 Conclusion
In this study, the isolation and identification of E. coli isolates from calf diarrhea from the Tongliao region, together with analysis of drug resistance, revealed that E. coli in this region is severely resistant to drugs, with 100% resistance to sulfadiazine sodium, enrofloxacin and ciprofloxacin. The detection rates of the antibiotic-resistance genes TEM-1, TEM-206, strA, strB, qacH, and blaCTX were 100%. Whole-genome sequencing of the three multi-drug resistant E. coli strains demonstrated that all three strains carried plasmids containing resistance genes. These resistance genes and MGE prediction showed a large number of transposons and insertion sequences in these genes in the strains. Furthermore, the presence of integrase genes was found in the plasmids and chromosomal genomes of several strains, contributing significantly to the horizontal transfer of drug-resistance genes, and thus leading to significantly increased resistance against antimicrobial drugs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, CP157955-CP157957, CP157958-CP157959, CP103295-CP103297.
Ethics statement
The animal study was approved by Laboratory Animal Welfare and Ethics Committee, College of Animal Science and Technology, Inner Mongolia MINZU University, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZW: Writing – review & editing. MS: Writing – original draft. SG: Writing – review & editing. YW: Data curation, Writing – review & editing. LM: Data curation, Writing – review & editing. JS: Data curation, Writing – review & editing. CG: Data curation, Writing – review & editing. DH: Data curation, Writing – review & editing. XF: Data curation, Writing – review & editing. JX: Data curation, Writing – review & editing. HM: Funding acquisition, Project administration, Supervision, Writing – review & editing. KL: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (U23A20242, China), the Inner Mongolia Autonomous Region Colleges and Universities Basic Scientific Research Operating Expenses Program (GXKY22097 and GXKY22082, Inner Mongolia, China), the Inner Mongolia Autonomous Region Natural Science Foundation (2021BS03015, Inner Mongolia, China), the Inner Mongolia Autonomous Region Central Guiding Local Development Fund Project (2022ZY0149, Inner Mongolia, China), the Inner Mongolia Xilin Gol League Science and Technology Project (202213, Inner Mongolia, China).
Acknowledgments
We would like to thank the Laboratory of Pharmacology and Toxicology, School of Animal Medicine, Jilin Agricultural University for help with the experiments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1466690/full#supplementary-material
References
1. Wang, D, Gao, H, Zhao, L, Lv, C, Dou, W, Zhang, X, et al. Detection of the dominant pathogens in diarrheal calves of Ningxia, China in 2021-2022. Front Vet Sci. (2023) 10:1155061. doi: 10.3389/fvets.2023.1155061
2. Brunauer, M, Roch, FF, and Conrady, B. Prevalence of worldwide neonatal calf Diarrhoea caused by bovine rotavirus in combination with bovine coronavirus, Escherichia coli K99 and Cryptosporidium spp.: a meta-analysis. Animals (Basel). (2021) 11:1014. doi: 10.3390/ani11041014
3. Schwaiger, K, Storch, J, Bauer, C, and Bauer, J. Lactobacillus (Limosilactobacillus) reuteri: a probiotic candidate to reduce neonatal diarrhea in calves. Front Microbiol. (2023) 14:1266905. doi: 10.3389/fmicb.2023.1266905
4. Cho, YI, and Yoon, KJ. An overview of calf diarrhea - infectious etiology, diagnosis, and intervention. J Vet Sci. (2014) 15:1–17. doi: 10.4142/jvs.2014.15.1.1
5. Dall Agnol, AM, Lorenzetti, E, Leme, RA, Ladeia, WA, Mainardi, RM, Bernardi, A, et al. Severe outbreak of bovine neonatal diarrhea in a dairy calf rearing unit with multifactorial etiology. Braz J Microbiol. (2021) 52:2547–53. doi: 10.1007/s42770-021-00565-5
6. Wei, X, Wang, W, Dong, Z, Cheng, F, Zhou, X, Li, B, et al. Detection of infectious agents causing neonatal calf diarrhea on two large dairy farms in Yangxin County, Shandong Province, China. Front Vet Sci. (2021) 7:589126. doi: 10.3389/fvets.2020.589126
7. Carter, HSM, Renaud, DL, Steele, MA, Fischer-Tlustos, AJ, and Costa, JHC. A narrative review on the unexplored potential of colostrum as a preventative treatment and therapy for diarrhea in neonatal dairy calves. Animals (Basel). (2021) 11:2221. doi: 10.3390/ani11082221
8. Kolenda, R, Burdukiewicz, M, and Schierack, P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front Cell Infect Microbiol. (2015) 5:23. doi: 10.3389/fcimb.2015.00023
9. Ryu, JH, Kim, S, Park, J, and Choi, KS. Characterization of virulence genes in Escherichia coli strains isolated from pre-weaned calves in the Republic of Korea. Acta Vet Scand. (2020) 62:45. doi: 10.1186/s13028-020-00543-1
10. Berber, E, Çanakoğlu, N, Sözdutmaz, İ, Simsek, E, Sursal, N, Ekinci, G, et al. Seasonal and age-associated pathogen distribution in newborn calves with diarrhea admitted to ICU. Vet Sci. (2021) 8:128. doi: 10.3390/vetsci8070128
11. Khawaskar, DP, Sinha, DK, Lalrinzuala, MV, Athira, V, Kumar, M, Chhakchhuak, L, et al. Pathotyping and antimicrobial susceptibility testing of Escherichia coli isolates from neonatal calves. Vet Res Commun. (2022) 46:353–62. doi: 10.1007/s11259-021-09857-5
12. du, W, Wang, X, Hu, M, Hou, J, du, Y, Si, W, et al. Modulating gastrointestinal microbiota to alleviate diarrhea in calves. Front Microbiol. (2023) 14:1181545. doi: 10.3389/fmicb.2023.1181545
13. Osorio, JS. Gut health, stress, and immunity in neonatal dairy calves: the host side of host-pathogen interactions. J Anim Sci Biotechnol. (2020) 11:105. doi: 10.1186/s40104-020-00509-3
14. Karamzadeh-Dehaghani, A, Towhidi, A, Zhandi, M, Mojgani, N, and Fouladi-Nashta, A. Combined effect of probiotics and specific immunoglobulin Y directed against Escherichia coli on growth performance, diarrhea incidence, and immune system in calves. Animal. (2021) 15:100124. doi: 10.1016/j.animal.2020.100124
15. Algammal, AM, el-Kholy, A, Riad, EM, Mohamed, HE, Elhaig, MM, Yousef, SAA, et al. Genes encoding the virulence and the antimicrobial resistance in Enterotoxigenic and Shiga-Toxigenic E. coli isolated from diarrheic calves. Toxins (Basel). (2020) 12:383. doi: 10.3390/toxins12060383
16. Saeed, U, Insaf, RA, Piracha, ZZ, Tariq, MN, Sohail, A, Abbasi, UA, et al. Crisis averted: a world united against the menace of multiple drug-resistant superbugs -pioneering anti-AMR vaccines, RNA interference, nanomedicine, CRISPR-based antimicrobials, bacteriophage therapies, and clinical artificial intelligence strategies to safeguard global antimicrobial arsenal. Front Microbiol. (2023) 14:1270018. doi: 10.3389/fmicb.2023.1270018
17. Fernández, M, Casaux, ML, Fraga, M, Vignoli, R, Bado, I, Zunino, P, et al. Shiga toxin-producing Escherichia coli (STEC) associated with calf mortality in Uruguay. Microorganisms. (2023) 11:1704. doi: 10.3390/microorganisms11071704
18. Carter, HSM, Steele, MA, Costa, JHC, and Renaud, DL. Evaluating the effectiveness of colostrum as a therapy for diarrhea in preweaned calves. J Dairy Sci. (2022) 105:9982–94. doi: 10.3168/jds.2022-22187
19. Zhu, DM, Ding, Q, Li, PH, Wang, YL, Li, YZ, Yu Li, X, et al. Antimicrobial resistance in E. coli of animal origin and discovery of a novel ICE mobile element in Northeast China. BMC Vet Res. (2023) 19:828. doi: 10.1186/s12917-023-03828-5
20. Li, B, Qiu, Y, Song, Y, Lin, H, and Yin, H. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics. Environ Int. (2019) 131:105007. doi: 10.1016/j.envint.2019.105007
21. Meng, M, Li, Y, and Yao, H. Plasmid-mediated transfer of antibiotic resistance genes in soil. Antibiotics (Basel). (2022) 11:525. doi: 10.3390/antibiotics11040525
22. Dong, N, Yang, X, Chan, EW, Zhang, R, and Chen, S. Klebsiella species: taxonomy, hypervirulence and multidrug resistance. EBioMedicine. (2022) 79:103998. doi: 10.1016/j.ebiom.2022.103998
23. Partridge, SR, Kwong, SM, Firth, N, and Jensen, SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. (2018) 31:e00088–17. doi: 10.1128/CMR.00088-17
24. Tokuda, M, and Shintani, M. Microbial evolution through horizontal gene transfer by mobile genetic elements. Microb Biotechnol. (2024) 17:e14408. doi: 10.1111/1751-7915.14408
25. Wang, L, Lin, Z, Ali, M, Zhu, X, Zhang, Y, Li, S, et al. Effects of lactic acid bacteria isolated from Tibetan chickens on the growth performance and gut microbiota of broiler. Front Microbiol. (2023) 14:1171074. doi: 10.3389/fmicb.2023.1171074
26. CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals; clinical laboratory standard institution. Wayne, PA, USA: CLSI (2015).
27. Zhao, X, Lv, Y, Adam, FEA, Xie, Q, Wang, B, Bai, X, et al. Comparison of antimicrobial resistance, virulence genes, Phylogroups, and biofilm formation of Escherichia coli isolated from intensive farming and free-range sheep. Front Microbiol. (2021) 12:699927. doi: 10.3389/fmicb.2021.699927
28. Liu, H, Pan, S, Cheng, Y, Luo, L, Zhou, L, Fan, S, et al. Distribution and associations for antimicrobial resistance and antibiotic resistance genes of Escherichia coli from musk deer (Moschus berezovskii) in Sichuan, China. PLoS One. (2023) 18:e0289028. doi: 10.1371/journal.pone.0289028
29. Jain, M, Olsen, HE, Paten, B, and Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. (2016) 17:239. doi: 10.1186/s13059-016-1103-0
30. Koren, S, Walenz, BP, Berlin, K, Miller, JR, Bergman, NH, and Phillippy, AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. (2017) 27:722–36. doi: 10.1101/gr.215087.116
31. Hyatt, D, Chen, GL, LoCascio, PF, Land, ML, Larimer, FW, and Hauser, LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. (2010) 11:119. doi: 10.1186/1471-2105-11-119
32. Ashburner, M, Ball, CA, Blake, JA, Botstein, D, Butler, H, Cherry, JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
33. Kanehisa, M, Goto, S, Kawashima, S, Okuno, Y, and Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. (2004) 32:277D–2280D. doi: 10.1093/nar/gkh063
34. Jia, B, Raphenya, AR, Alcock, B, Waglechner, N, Guo, P, Tsang, KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2017) 45:D566–73. doi: 10.1093/nar/gkw1004
35. Bortolaia, V, Kaas, RS, Ruppe, E, Roberts, MC, Schwarz, S, Cattoir, V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. (2020) 75:3491–500. doi: 10.1093/jac/dkaa345
36. Clausen, PTLC, Aarestrup, FM, and Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics. (2018) 19:307. doi: 10.1186/s12859-018-2336-6
37. Carattoli, A, Zankari, E, García-Fernández, A, Voldby Larsen, M, Lund, O, Villa, L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
38. Johansson, MHK, Bortolaia, V, Tansirichaiya, S, Aarestrup, FM, Roberts, AP, and Petersen, TN. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J Antimicrob Chemother. (2021) 76:101–9. doi: 10.1093/jac/dkaa390
39. Das, SC, Khan, A, Panja, P, Datta, S, Sikdar, A, Yamasaki, S, et al. Dairy farm investigation on Shiga toxin-producing Escherichia coli (STEC) in Kolkata, India with emphasis on molecular characterization. Epidemiol Infect. (2005) 133:617–26. doi: 10.1017/s0950268805004000
40. Kim, HS, Whon, TW, Sung, H, Jeong, YS, Jung, ES, Shin, NR, et al. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat Commun. (2021) 12:161. doi: 10.1038/s41467-020-20389-5
41. Jang, JY, Kim, S, Kwon, MS, Lee, J, Yu, DH, Song, RH, et al. Rotavirus-mediated alteration of gut microbiota and its correlation with physiological characteristics in neonatal calves. J Microbiol. (2019) 57:113–21. doi: 10.1007/s12275-019-8549-1
42. Shi, Z, Lan, Y, Wang, Y, Yan, X, Ma, X, Hassan, F-u, et al. Multi-omics strategy reveals potential role of antimicrobial resistance and virulence factor genes responsible for Simmental diarrheic calves caused by Escherichia coli. mSystems. (2024) 9:e0134823. doi: 10.1128/msystems.01348-23
43. Shabana, II, Zaraket, H, and Suzuki, H. Molecular studies on diarrhea-associated Escherichia coli isolated from humans and animals in Egypt. Vet Microbiol. (2013) 167:532–9. doi: 10.1016/j.vetmic.2013.08.014
44. Klein-Jöbstl, D, Iwersen, M, and Drillich, M. Farm characteristics and calf management practices on dairy farms with and without diarrhea: a case-control study to investigate risk factors for calf diarrhea. J Dairy Sci. (2014) 97:5110–9. doi: 10.3168/jds.2013-7695
45. Eibl, C, Bexiga, R, Viora, L, Guyot, H, Félix, J, Wilms, J, et al. The antibiotic treatment of calf diarrhea in four European countries: a survey. Antibiotics (Basel). (2021) 10:910. doi: 10.3390/antibiotics10080910
46. Yassin, AK, Gong, J, Kelly, P, Lu, G, Guardabassi, L, Wei, L, et al. Antimicrobial resistance in clinical Escherichia coli isolates from poultry and livestock, China. PLoS One. (2017) 12:e0185326. doi: 10.1371/journal.pone.0185326
47. Jia, Y, Mao, W, Liu, B, Zhang, S, Cao, J, and Xu, X. Study on the drug resistance and pathogenicity of Escherichia coli isolated from calf diarrhea and the distribution of virulence genes and antimicrobial resistance genes. Front Microbiol. (2022) 13:992111. doi: 10.3389/fmicb.2022.992111
48. He, WY, Zhang, XX, Gao, GL, Gao, MY, Zhong, FG, Lv, LC, et al. Clonal spread of Escherichia coli O101: H9-ST10 and O101: H9-ST167 strains carrying fosA3 and blaCTX-M-14 among diarrheal calves in a Chinese farm, with Australian Chroicocephalus as the possible origin of O101: H9-ST10. Zool Res. (2021) 42:461–8. doi: 10.24272/j.issn.2095-8137.2021.153
49. Nang, SC, Azad, MAK, Velkov, T, Zhou, Q(T), and Li, J. Rescuing the last-line Polymyxins: achievements and challenges. Pharmacol Rev. (2021) 73:679–728. doi: 10.1124/pharmrev.120.000020
50. Grevskott, DH, Salvà-Serra, F, Moore, ERB, and Marathe, NP. Nanopore sequencing reveals genomic map of CTX-M-type extended-spectrum β-lactamases carried by Escherichia coli strains isolated from blue mussels (Mytilus edulis) in Norway. BMC Microbiol. (2020) 20:134. doi: 10.1186/s12866-020-01821-8
51. Peter, S, Bosio, M, Gross, C, Bezdan, D, Gutierrez, J, Oberhettinger, P, et al. Tracking of antibiotic resistance transfer and rapid plasmid evolution in a hospital setting by Nanopore sequencing. Clin Vaccine Immunol. (2020) 5:e00525–12. doi: 10.1128/mSphere.00525-20
52. Bouzouita, I, Cabibbe, AM, Trovato, A, Daroui, H, Ghariani, A, Midouni, B, et al. Whole-genome sequencing of drug-resistant Mycobacterium tuberculosis strains, Tunisia, 2012-2016. Emerg Infect Dis. (2019) 25:538–46. doi: 10.3201/eid2503.181370
53. Misawa, K, Tarumoto, N, Tamura, S, Osa, M, Hamamoto, T, Yuki, A, et al. Single nucleotide polymorphisms in genes encoding penicillin-binding proteins in β-lactamase-negative ampicillin-resistant Haemophilus influenzae in Japan. BMC Res Notes. (2018) 11:53. doi: 10.1186/s13104-018-3169-0
54. Iranzo, J, Gómez, MJ, López de Saro, FJ, and Manrubia, S. Large-scale genomic analysis suggests a neutral punctuated dynamics of transposable elements in bacterial genomes. PLoS Comput Biol. (2014) 10:e1003680. doi: 10.1371/journal.pcbi.1003680
Keywords: Escherichia coli , calf diarrhea, drug resistance, antimicrobial resistance genes, whole genome sequencing
Citation: Wang Z, Sun M, Guo S, Wang Y, Meng L, Shi J, Geng C, Han D, Fu X, Xue J, Ma H and Liu K (2024) Detection of drug resistance in Escherichia coli from calves with diarrhea in the Tongliao region: an analysis of multidrug-resistant strains. Front. Vet. Sci. 11:1466690. doi: 10.3389/fvets.2024.1466690
Edited by:
Getahun E. Agga, Food Animal Environmental Systems Research, Agricultural Research Service (USDA), United StatesReviewed by:
Aman Ullah Khan, University of Veterinary and Animal Sciences, PakistanDalia Hamza, Cairo University, Egypt
Copyright © 2024 Wang, Sun, Guo, Wang, Meng, Shi, Geng, Han, Fu, Xue, Ma and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Liu, bGl1a2FpNzIxMDI2QDE2My5jb20=; Hongxia Ma, bWFob25neGlhQGpsYXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Zi Wang
Zi Wang Miao Sun
Miao Sun Shuang Guo4
Shuang Guo4 Jiangdong Xue
Jiangdong Xue