- 1Center of Excellence for Companion Animal Cancer, Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand
- 2Veterinary Diagnostic Laboratory, Michigan State University, Lansing, MI, United States
Background: The histopathological classification of T-cell lymphoma (TCL) in humans has distinctive mutational genotyping that suggests different lymphomagenesis. A similar concept is assumed to be observed in dogs with different TCL phenotypes.
Objective: This study aimed to identify the previously reported single-nucleotide polymorphisms (SNPs) in both human beings and dogs in canine TCLs and null-cell lymphomas (NCLs) and to design compatible oligonucleotides from each variant based on the multiplex polymerase chain reaction.
Methods: Genomic DNA was extracted from 68 tumor specimens (62 TCLs and 6 NCLs) and 5 buffy coat samples from dogs with TCL. Four TCL subtypes and NCL were analyzed in 44 SNPs from 21 genes using the MassARRAY.
Results: The greatest incidences of SNPs observed in all TCL subtypes and NCL ware SATB1 c.1259A > C, KIT c.1275A > G, SEL1L c.2040 + 200C > G, and TP53 c.1024C > T, respectively. Some SNP locations were statistically significant associated with NCL, including MYC p.S75F (p = 0.0003), TP53 p.I149N (p = 0.030), PDCD1 p.F37LX (p = 0.012), and POT1 p.R583* (p = 0.012).
Conclusion: Each TCL histological subtype and NCL are likely to contain distinctive mutational genetic profiles, which might play a role in lymphoma gene-risk factors and might be useful for selecting therapeutic target drugs for each canine patient.
Introduction
Canine lymphoma can be categorized into B-cell, T-cell, and null-cell (non-B, non-T-cell) lymphomas (1). Regardless of the prevalence of TCL and rare NCLs, a frequent histopathological subtype of TCL is peripheral TCL, not otherwise specified (PTCL-NOS) (2). Other less common subtypes include T-zone lymphoma (TZL), enteropathy-associated T-cell lymphoma (EATCL), and cutaneous T-cell lymphoma (CTCL) (3). CTCL can be either epitheliotropic (ECTCL) or non-epitheliotropic (NECTCL). Canine TCL often has a worse prognosis than B-cell lymphoma (4); however, each TCL subtype often shows a variable prognosis and disease outcome. TZL has a better prognosis because of its indolent disease course (5). Based on the anatomic locations of TCL, patients with hepatosplenic and gastrointestinal lymphoma often have shorter survival times than those with certain forms of multicentric or cutaneous lymphoma (6–10). Moreover, the overall survival time of dogs with ECTCL is shorter than that of dogs with NECTCL (11). Due to their differences in prognosis, a unique therapy for each TCL subtype may be required. Many studies have suggested multidrug chemotherapy for TCL, such as a combination of mechlorethamine, vincristine, prednisone, and procarbazine (12); lomustine, vincristine, procarbazine, and prednisolone (13), or vincristine, L-asparaginase, doxorubicin, cyclophosphamide, actinomycin-D, procarbazine, prednisolone, and lomustine (14), to improve progression-free survival and overall survival time in lymphoma dogs. However, CHOP-based induction (vincristine, cyclophosphamide, doxorubicin, and prednisolone) is still preferred by veterinary oncologists, and lomustine-based protocols are used as rescue treatments (15).
Specific molecular signatures of PTCL-NOS in humans can be classified into TBX21 and GATA3 subgroups, which are T helpers (TH)1 and TH2-cell differentiation regulators, respectively (16). PTCL-NOS-GATA3 frequently exhibited mutations of PTEN and TP53, with co-occurring amplifications of MYC and STAT3. For PTCL-NOS-TBX21, mutations of DNA methylation regulator genes were noted in TET1, TET3, and DNMT3A (17). For human CTCL, specific genetic mutations were involved in T-cell activation (CD28 and RHOA), cell apoptosis (FAS), NF-κB pathways (NFKB2 and STAT5B), chromatin remodeling (DNMT3A and ARD1A), and DNA damage response (TP53 and CDKN2A) (18). Moreover, the loss-of-function mutation of PDCD1—the gene encoding an inhibitory receptor program death protein 1 (PD-1)—was driven by aggressive behavior in CTCL (19). For EATCL, the most frequently mutated genes were associated with the JAK–STAT pathway in SETD2, STAT5B, JAK1, JAK3, and STAT3 (20–22). Hence, these findings are useful for selective potential therapeutic opportunities in each TCL subtype as a single agent or in combination with anti-neoplastic drugs. For example, the NF-κB inhibitor bortezomib and the PI3K-δ,λ inhibitor duvelisib were applied in clinical trials treating PTCL and CTCL, and the overall response rate ranged from 31 to 67% (23, 24).
In canine TCLs, Labadie et al. reported SNPs in TZL using genome-wide association mapping (25). Several SNPs of hyaluronidase genes were associated with disease risk for TZL, including SPAM1K482R, HYALP1M463T, and HYAL4G454S, S434F, L378I. Another study investigated somatic mutations in a variable group of TCL with limited subtyping in Golden retrievers and Boxers using whole-exome sequencing (26). The common mutated genes in both breeds were noted on SATB1 c.1259 A > G p.Q420R, while PTEN c.975 C > T p.L325 = was found in Boxer-TCLs (26, 27). Similar variants using RNA sequencing were found in two dogs with PTCL (27). Three somatic missense mutations were described in canine TCL, including MYC c.185C > T p.Ser62Phe, TP53 c.715G > A p.Arg239Trp, and MET c.3804C > G p.Asp1268Glu (28). Nonetheless, no study has examined novel SNPs associated with human TCL in dogs. Thus, our study aimed to investigate SNP genotyping patterns in canine PTCL, ECTCL, NECTCL, EATCL, and NCL based on the important gene-related lymphoma risks in human and canine TCLs (19, 29–31) and to obligate primer design for multiplex PCR, using the MassARRAY platform (Agena Bioscience, CA, United States).
Materials and methods
Study samples and immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) block archives were retrieved from the Department of Pathology, Faculty of Veterinary Science, Chulalongkorn University, between 2008 and 2021 and from three private veterinary labs (SQ Reference Lab, China; Vet Central Lab, Thailand; and Vet Clinical Center, Thailand) between 2020 and 2021. Only patients who had not previously received chemotherapeutic drugs were enrolled in this study. According to physical examination, abdominal ultrasonography, and buffy-coated smears, all lymphoma dogs were in at least WHO clinical stage III (15). The selected cases included multicentric, cutaneous, and alimentary lymphomas that were recut into 3-μm thickness, stained with hematoxylin and eosin, and immunostained against CD3 (Peter F. Moore, United States), CD20 (ab27093, Abcam, United States), and CD79a (HM57, Abcam) or Pax5 (1EW, Leica, United Kingdom). Immunohistochemistry was performed as described elsewhere (30). Only lymphoma samples that had no immunoreactivity for B- and T-cell markers were further investigated with CD18 (histiocyte marker, Peter F. Moore), CD117 (mast cell marker, MIB1, Dako), and MUM1 (plasma cell marker, ab133590, Abcam) to rule out the possibility of other hematopoietic lineages. Finally, cases that showed negative to specific leucocyte markers but positively immunostained with CD45 (common leucocyte antigen) were assigned as NCL.
To evaluate the effects of missense SATB1Q420P mutation on programmed death ligand 1 (PD-L1) and missense TP53I149N mutation on p53, five samples from each group (four SATB1 mutants and one SATB1 wild type vs. two TP53 mutants and three TP53 wild types) were immunolabeled against PD-L1 and p53, respectively. In brief, the tissue section was preheated by citrate buffer pH 6 at 95°C for 20 min and blocked endogenous peroxidase and non-specific binding protein by 0.3% (v/v) H2O2 for 30 min and 5% (w/v) bovine serum albumin for 20 min, respectively. Anti-CD45 antibody (Peter F. Moore) was incubated in a 1:10 dilution at room temperature (RT) for 1.5 h, while PD-L1 (1:100, clone 5D2) (32) and p53 (1: 500, PA5-27822, Invitrogen, United States) were incubated at RT for 2 h. Envision mouse/rabbit-HRP (Dako, Hilden, Germany) was used as a secondary antibody, and 3,3′-diaminobenzidine was used as a substrate. Histopathological diagnosis was made by a board-certified veterinary pathologist (TK) according to WHO classifications (3). Signalment and clinical history of all selected cases were recorded; however, most cases were lost due to a lack of treatment follow-up.
DNA extraction
A total of 70–100 μm FFPE tissue scrolls were collected into a sterile 1.5 mL microtube. The genomic DNA from each sample was then extracted using a DNeasy Blood and Tissue Kit (Qiagen, Germany). After deparaffinization with xylene and absolute ethanol, the sample was processed according to the manufacturer’s instructions. The DNA concentration of each sample was then measured with a NanoDrop Lite Spectrophotometer (Thermo Scientific, United States). The requirement of high-quality DNA for targeted SNP genotyping using the Agena Bioscience MassARRAY system is a 260/280 ratio of >1.8 and a concentration of >10 ng/μL.
iPLEX genotyping process
The dog reference genomes were obtained from Dog10K_Boxer_Tasha and UMICH_Zoey_3.1.1 MassARRAY was used for genotyping of the 44 targeted SNPs in 21 different genes, as shown in Table 1. The SNP locations were selected based on the previous studies (19, 26, 27, 29–31, 36). A primer pair for each location was designed by AgenaCx software (Agena Bioscience). First, multiplex PCR primers were amplified in the target regions by using a 5-uL reaction of 0.5 μL of 10X PCR buffer (Agena Bioscience), 2 mM MgCl2, 500 μM dNTPs, 0.5 μM of each amplification primer, and 0.2 U DNA polymerase. Second, 1.7 U shrimp alkaline phosphatase (SAP, Agena Bioscience) and 0.17 μL of 10X SAP buffer were added to the first-step PCR product to dephosphorylate residual nucleotides. Third, the iPLEX extension reaction was composed of 0.2 μL of 10X iPLEX buffer (Agena Bioscience), 0.2 μL of 10X dNTP/ddNTP combination, 0.142 U iPLEX enzyme, and 0.5–1.57 μM extend primer mix. These three steps were carried out by T100 thermal cycler (Bio-Rad, United States), and the cycling conditions were followed by Sirivisoot et al. (30). The final extension products were desalted and transferred onto a SpectroChip with an automated nano-dispenser. The different sizes of SNP variations were identified using a matrix-assisted laser desorption/ionization time-of-flight mass spectrometer. Genotyping of each target site was presented as mass spectrum peaks using a MassARRAY analyzer. The percentage of determinate 44 variants (%call rate) for each sample must be achieved by ≥90%.
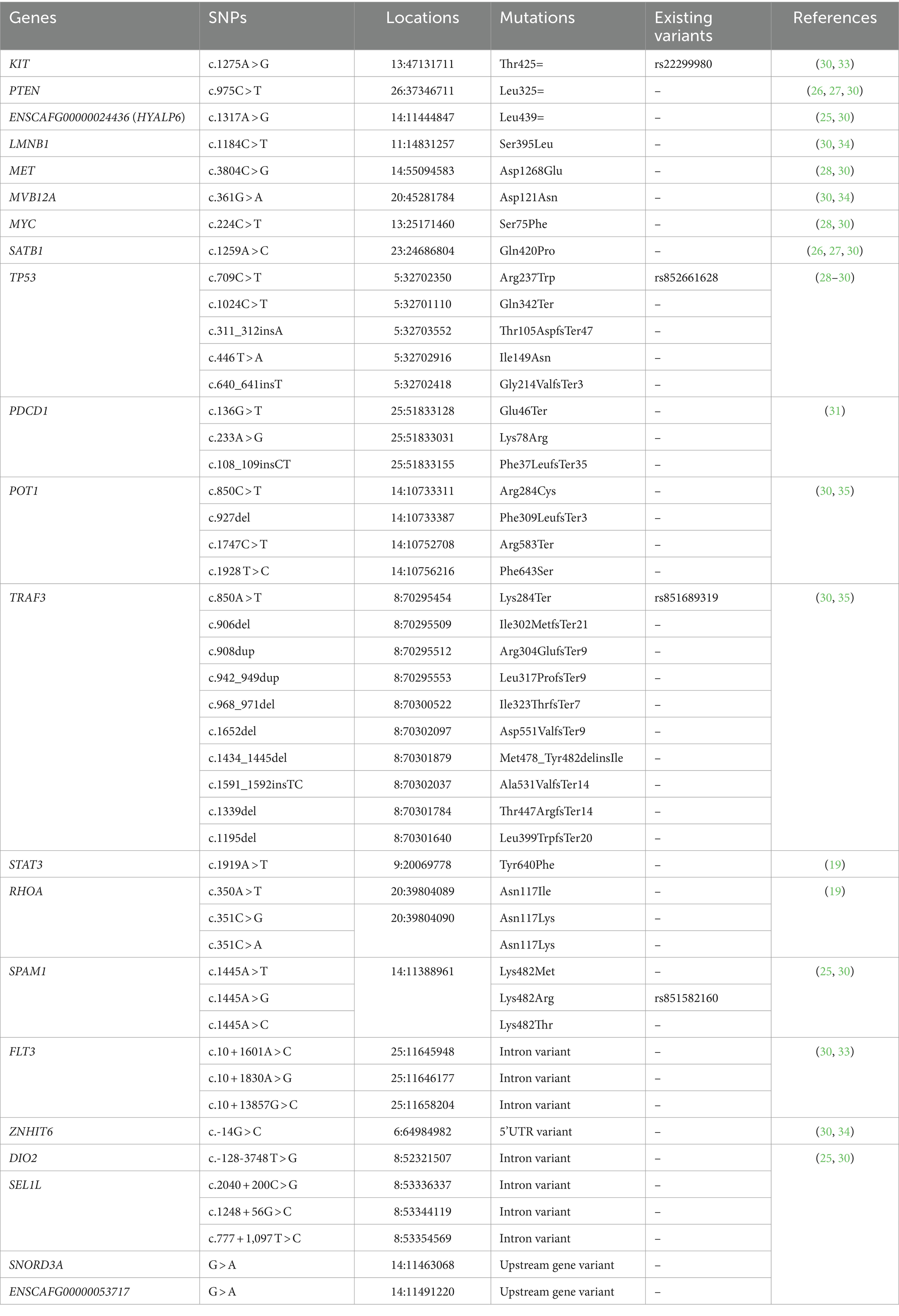
Table 1. A total of 44 specified single nucleotide polymorphisms (SNPs) were selected and investigated in canine T-cell and null-cell lymphomas.
Statistical analysis
A total of 44 SNP locations were calculated for the correlation with anatomical forms and histological subtypes of TCL/NCL using the chi-square test by GraphPad Prism version 9.0 for macOS (GraphPad Software, United States).
Results
Patient information
The demographics of the 68 dogs are shown in Table 2. The median age of dogs with TCL/NCL was 8 years (ranging from 1 to 15 years). Among the 35 purebred dogs, the most prevalent breeds were Golden retriever (28.57%, 10/35), Shih Tzu (14.29%, 5/35), Poodle (8.57%, 3/35), and Labrador retriever (8.57%, 3/35).
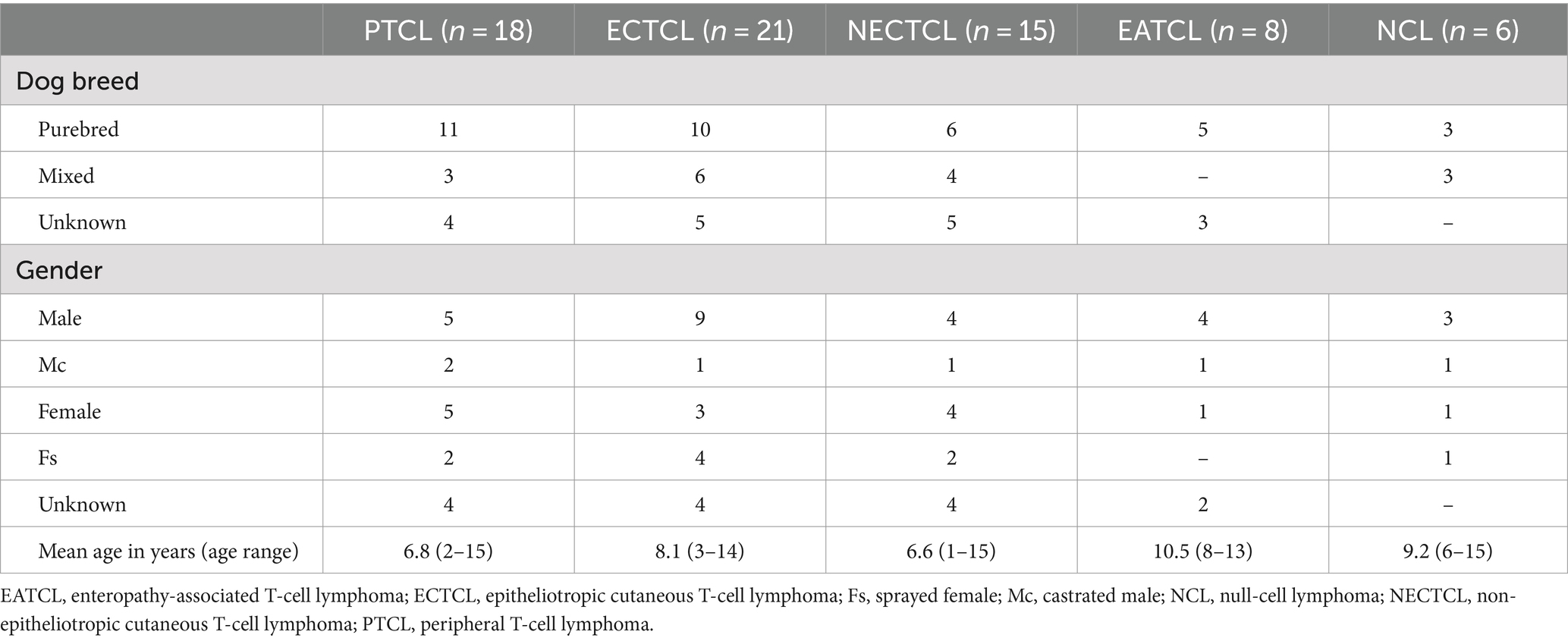
Table 2. Demographic information of 68 dogs with each T-cell lymphoma subtype and null-cell lymphoma.
Based on the anatomical locations, 21 cases were multicentric, 39 cases were cutaneous, and 8 cases were alimentary. According to histopathological and immunophenotyping results of 68 cases, there were 18 PTCLs, 21 ECTCLs, 15 NECTCLs, 8 EATCLs type I, and 6 cases of NCL (CD3-, CD20-, CD79a-, CD117-, CD18-, MUM1-, and C45+) (Figure 1). Among four aberrant TCL cases (CD3+, CD20+, and Pax5-), one dog had nodal PTCL, and three dogs had ECTCL. For two biphenotypic TCLs (CD3+, CD20+, and Pax5+), one case had nodal PTCL and the other case had EATCL.
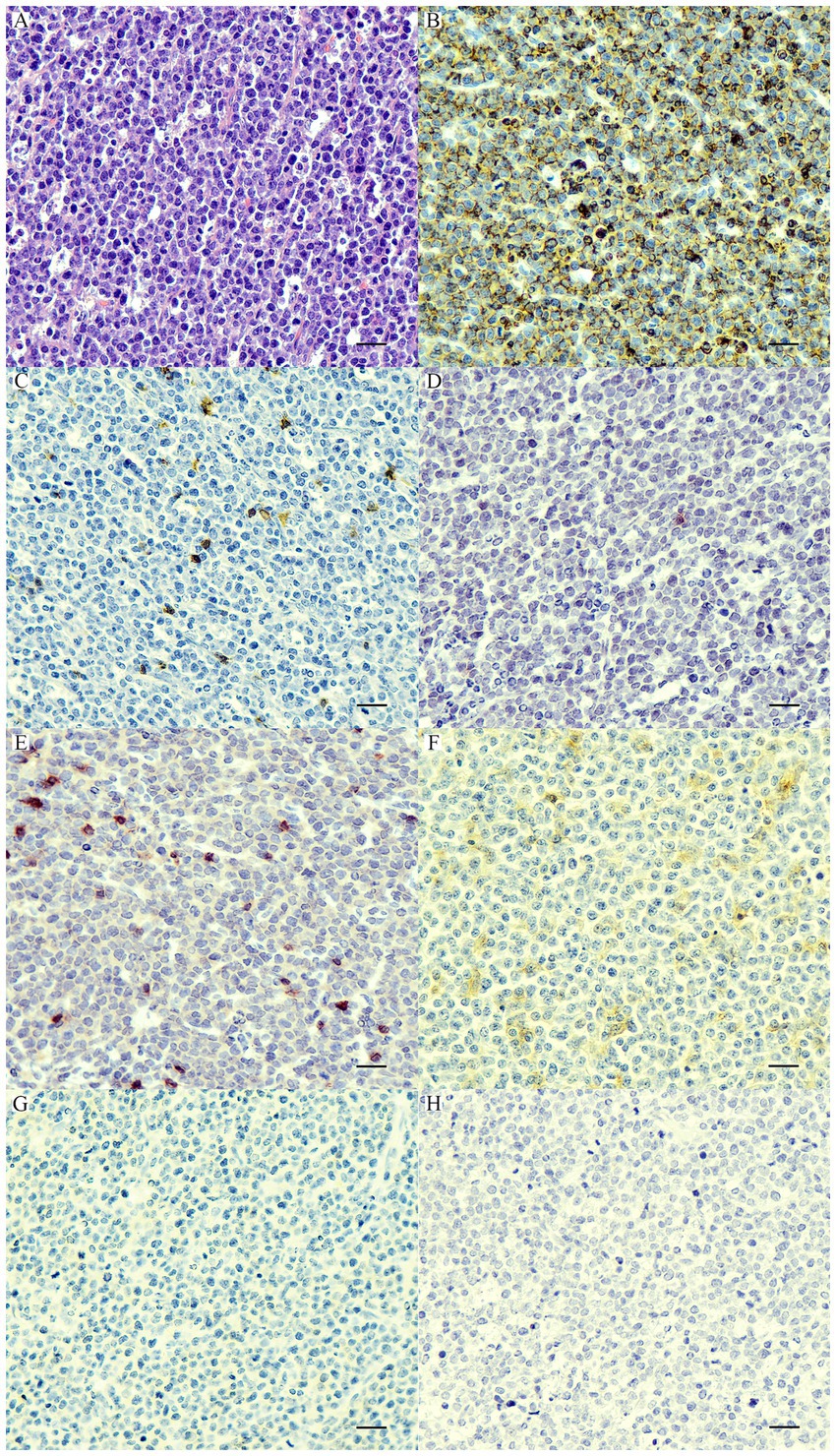
Figure 1. Nodal null-cell lymphoma. (A) The nuclei of neoplastic cells are large >2× of a red blood cell, finely stipple to vesiculate chromatin, and contain 1–2 prominent nucleoli with a scant amount of cytoplasm. H&E. Bar = 10 μm. The neoplastic cells show cytoplasmic immunolabeling with CD45 (B) and are negative for CD20 (C), CD79a (D), CD3 (E), CD18 (F), CD117 (G), and MUM1 (H). IHC. Bar = 10 μm.
iPLEX genotyping results
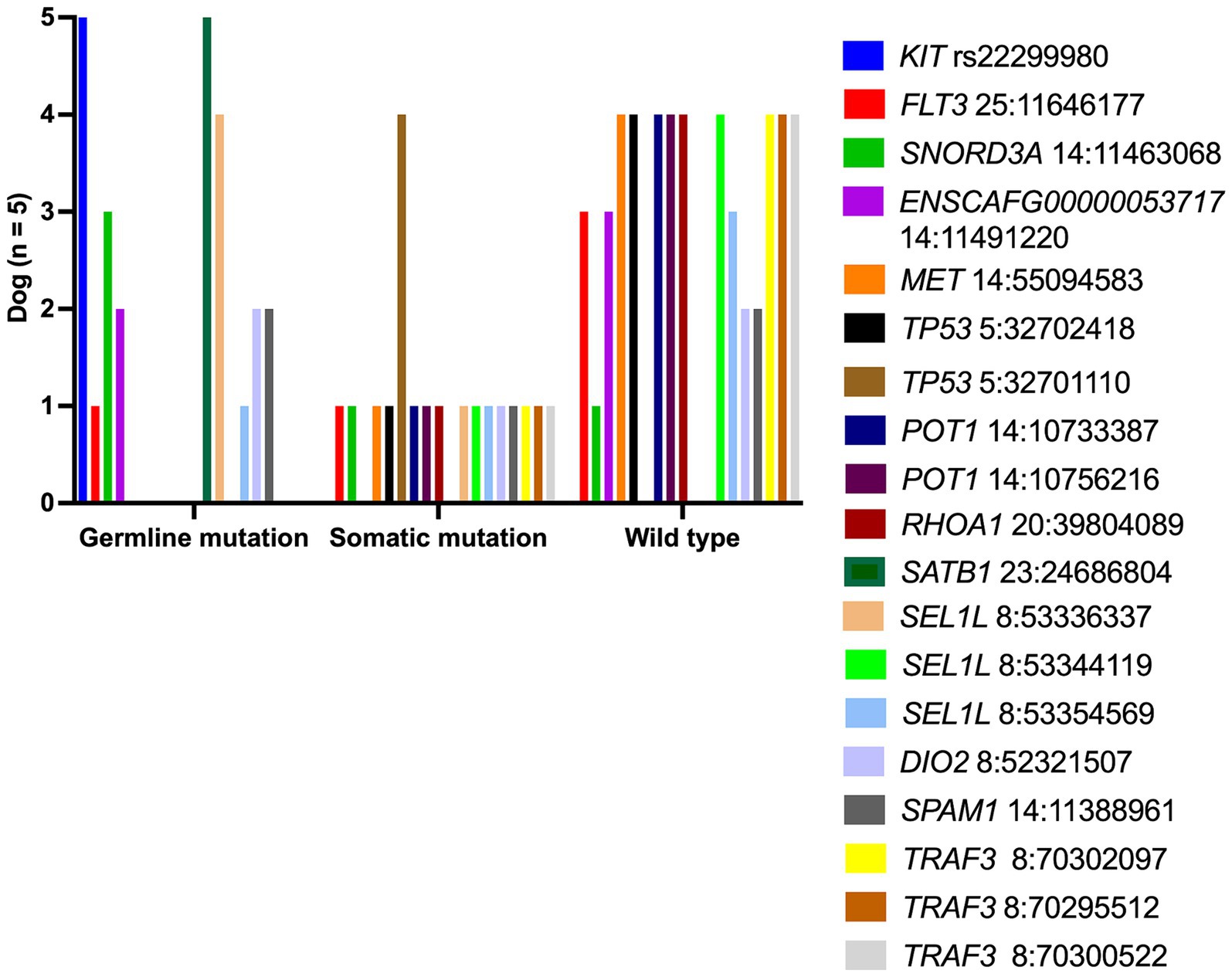
Each lymphoma dog had a different mutational genotyping profile, even though they had a similar TCL subtype (Figure 2). The highest genetic variations frequently observed in all TCL/NCL subtypes were SATB1 c.1259A > C p.Gln420Pro (89.71%, 61/68), KIT rs22299980 p.Thr425 = (89.39%, 59/66), SEL1L c.2040 + 200C > G (82.35%, 56/68), and TP53 c.1024C>T p.Gln342Ter (80.39%, 41/51). When comparing each SNP across anatomic locations, some SNP variants were frequently observed in specific forms, regardless of statistical significance. For instance, the SNPs mainly found in cutaneous TCL were TP53 c.640_641insT (53.85%, p = 0.08). SEL1L c.1248 + 56G > C was noted in 50% of intestinal TCL (p = 0.16). SNORD3A 14:11463068 G > A was regularly mutated in nodal TCL (40%, p = 0.12).
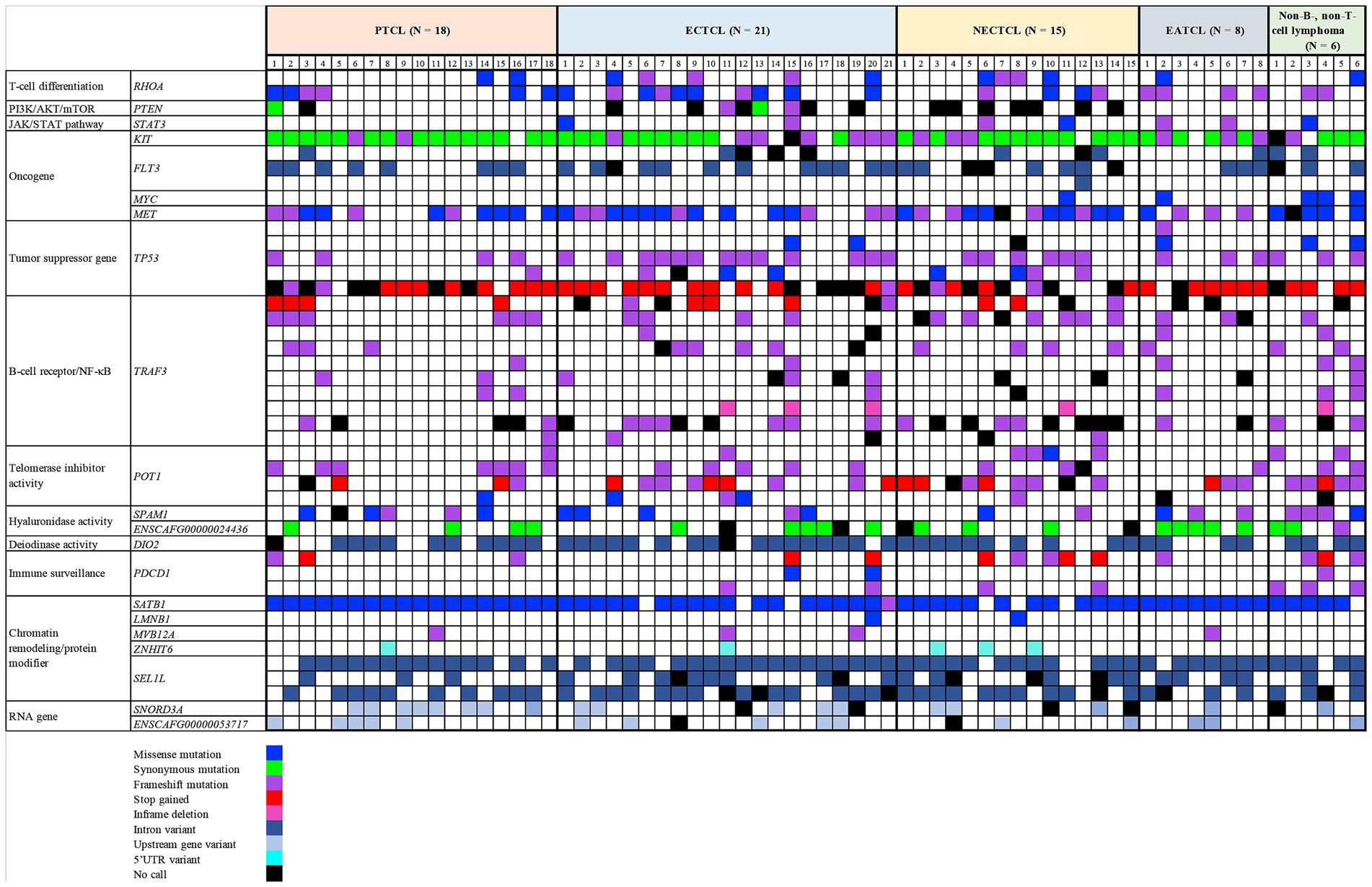
Figure 2. Landscape of single-nucleotide polymorphisms in canine T-cell and null-cell lymphomas. Genomic analysis of 21 mutated genes was harbored in 68 dogs with 4 T-cell lymphoma variants: peripheral T-cell lymphoma (PTCL), epitheliotropic cutaneous T-cell lymphoma (ECTCL), non-epitheliotropic cutaneous T-cell lymphoma (NECTCL), enteropathy-associated T-cell lymphoma (EATCL), and null-cell lymphoma (NCL). *No call indicates a failure to detect either a wild-type or mutant peak.
Each SNP location was analyzed to determine the association among five histological subtypes by chi-square test. MYC c.224C > T (p.Ser75Phe, p = 0.0003), TP53 c.446 T > A (p.Ile149Asn, p = 0.030), PDCD1 c.108_109insCT (p.Phe37LeufsTer35, p = 0.012), and POT1 c.1747C > T (p.Arg583Ter, p = 0.012) were significantly mutated in NCLs (Figure 3).
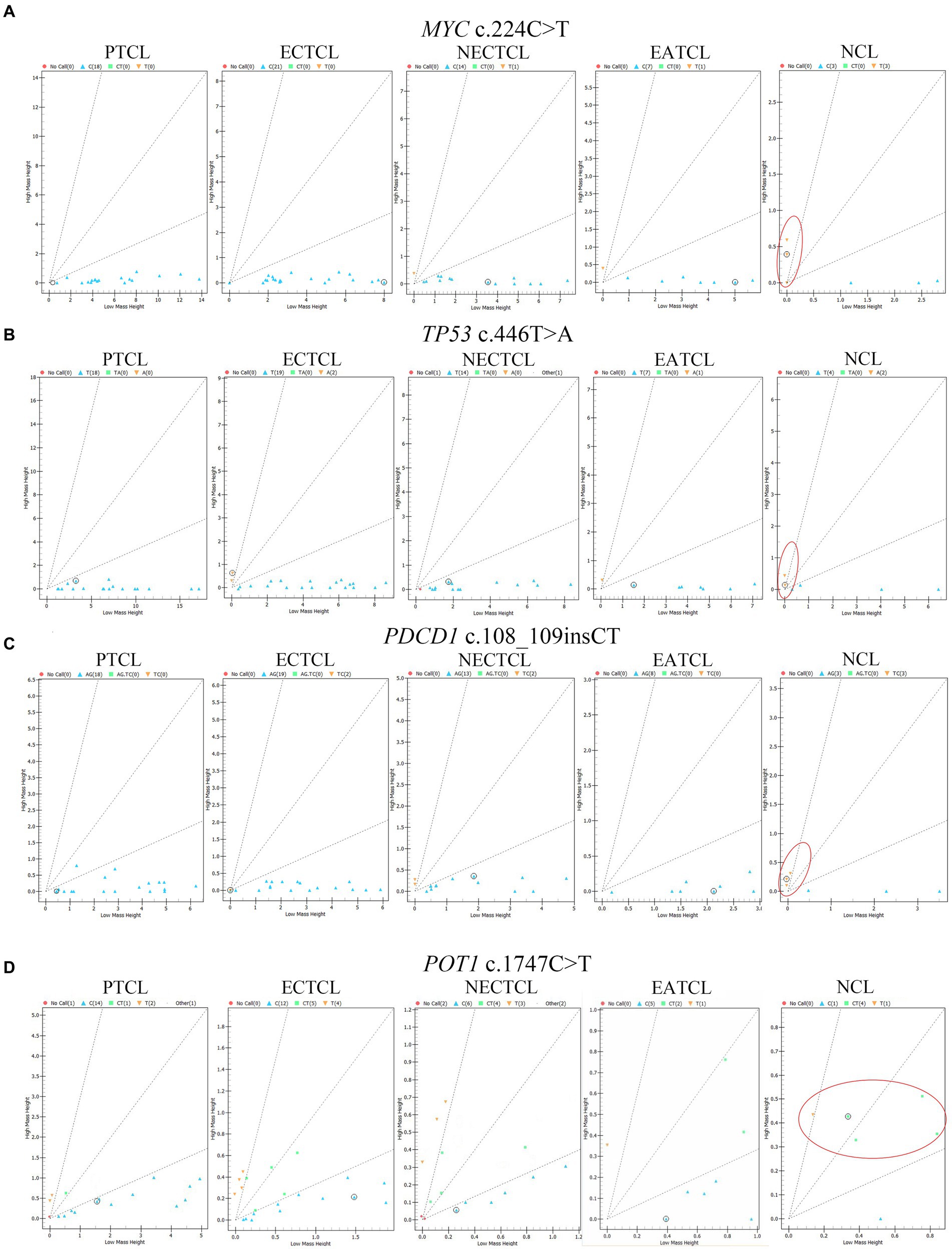
Figure 3. Cluster plots present mass spectra from each lymphoma dog in four single nucleotide polymorphisms significantly associated with the null-cell lymphoma subtype. (A) MYC c.224C > T (p = 0.0003), (B) TP53 c.446 T > A (p = 0.03), (C) PDCD1 c.108_109insCT (p = 0.012), and (D) POT1 c.1747C > T (p = 0.012) were generally seen in NCL comparing to other subtypes (red oval). Blue triangle = wild type; green square; and yellow inverted triangle = mutant; red circle = no call; EATCL, enteropathy associated T-cell lymphoma; ECTCL, epitheliotropic cutaneous T-cell lymphoma; NCL, null-cell lymphoma; NECTCL, non-epitheliotropic cutaneous T-cell lymphoma; PTCL, peripheral T-cell lymphoma.
All primer pairs of each SNP location had amplification efficiency in multiplex PCR of more than 90%, except for TP53 c.1024C > T, TRAF3 c.942_949dup (p.Leu317ProfsTer9), PTEN c.975C > T (p.Leu325=), and TRAF3 rs851689319 (p.Lys284Ter), which had 75% (51/68), 79% (54/68), 81% (55/68), and 90% (61/68), respectively.
To distinguish a germline from a somatic mutational profile, the authors selected five archival-matched buffy coat samples from PTCL dogs (dog no.7–10 and dog no.14) to assess this SNPs panel compared to TCL specimens. Somatic SNPs were uniquely varied in each dog, as shown in Figure 4. The frequent somatic mutation was seen in TP53Q342*(4 out of 4 dogs), while other somatic variants, for instance, MET, TP53, and RHOA were found in 20% (1 out of 5 dogs).
This study investigated the effect of SATB1Q420P on PD-L1 expression in canine neoplastic T-cells. Four cases of mutant and one case of wild-type SATB1 were immunolocalized for PD-L1. In four mutant dogs, neoplastic T lymphocytes expressed strong cytoplasmic intensity against PD-L1 compared to the wild type (Supplementary Figure S1). Therefore, T-cell anergy and exhaustion might be one of the lymphomageneses in dogs that are related to SATB1 mutation.
Two NCLs having TP53I149N were evaluated for their p53 expression status. Interestingly, both dogs with TP53I149N had a loss of p53 expression (<1% of positive nuclei) when compared to three dogs with wild-typed TP53, which showed weak and heterogenous intensity of positive cells <10% (Supplementary Figure S2).
Discussion
In total, 44 targeted SNP panels were designed and evaluated using the MassARRAY in 68 FFPE specimens of canine TCLs and NCLs. The overall pathways of the targeted genes in this study are shown in Figure 5. Most variants were observed in TCL/NCL subtypes except for MYCS75F, LMNB1S395L, or STAT3Y640F which were absent in PTCL. The highest frequency of genetic mutations in canine TCL was found at SATB1Q420P and KITT425=, respectively, and similar locations were reported in canine nodal B-cell lymphomas (30). However, the significance of the KIT mutation was synonymous, causing a low impact on carcinogenesis. The variants in hyaluronidase (SPAM1 and ENSCAFG00000024436/HYALP6) and thyroid hormone regulation (DIO2) genes reported in canine TZL (25) were also detected in other histologic TCLs, including PTCL, ECTCL, NECTCL, EATCL, and NCL. Low molecular weight hyaluronan (a byproduct of ligand activation) has pro-inflammatory and pro-oncogenic effects that might be associated with cell proliferation, angiogenesis, and metastasis (37, 38). Moreover, three intron variants of SEL1L were present in 34–82% of the TCL/NCL subtypes. Intron mutations impact as a genetic modifier. SEL1L is an unfolded protein response gene that is stimulated during the accumulation of unfolded and misfolded proteins in endoplasmic reticulum stress and plays a role in the protein degradation pathway through the ubiquitin-proteosome system (39). In one study, downregulation of SEL1L significantly decreased the expression of TIMP and PTEN involving tumor invasion in human pancreatic cancer (40). Hence, the effect pathway of SEL1L, the mutation function of hyaluronidase genes, and hyaluronan expression levels need further investigation to confirm their contribution to lymphoma pathogenesis in dogs.
Mutations of PTEN and SATB1 were frequently found in 12–25% of canine TCL (26, 27). In a recent study, a missense mutation of SATB1Q420P was noted in TCL and NCL, whereas a synonymous mutation of PTENL325= was exhibited in one PTCL (5%), three ECTCLs (18%), and one EATCL (12%). SATB1, a global chromatin organizer, is dysregulated in human cutaneous TCL by promoting the expression of TH-2 cytokines (IL-5 and IL-9), which were appropriate for the tumor microenvironment (41). SATB1 also demonstrated an inhibitory effect on PD-L1 expression in T lymphocytes (42). A low SATB1 mRNA expression level was related to an unfavorable prognosis in human mycosis fungoides and revealed decreased eosinophil infiltration, increased large-cell transformation, a high Ki-67 index, and elevated PD-1 expression (43). The SNPs of MYCS75F, MET (c.3804C > G, p.Asp1268Glu), and TP53 (c.709C > T, p.Arg237Trp; c.1024C > T, p.Gln342Ter; c.311_312insA, p.Thr105AspfsTer47; c.446 T > A, p.Ile149Asn; and c.640_641insT, p.Gly214ValfsTer3) from previous publications (19, 28, 29) were also observed in the current study. TP53Q342* (80.39%) and METD1268E (69.7%) were highly mutated in canine TCLs/NCLs. The oncogenic MET gene was upregulated in human PTCL and EATCL, and its gain-of-function contributed to T-cell lymphomagenesis (44, 45); therefore, it may play an important role in dogs as well.
Even though POT1 and TRAF3 polymorphisms were not described in four cases of canine TCL (35), we included them in our genotyping panel due to their detection in preliminary samples. A total of 4 variants of POT1 and 10 variants of TRAF3 were identified in 7–41% and 4–29% of all TCL/NCL specimens, respectively. A tumor necrosis factor receptor-associated factor 3 (TRAF3) serves as a tumor suppressor in B-cell lymphoma; nevertheless, it is required for the cell proliferation of anaplastic large T-cell lymphoma by activating the PI3K/AKT and JAK/STAT pathways (46). Another study of mutations affecting the protection of telomere 1 (POT1) explained that POT1 inhibition in CTCL induced telomere fragility, replication fork stalling, and telomere elongation, which led to defective telomere replication during tumorigenesis (47). Thus, a significant mechanism of lymphoma development in dogs caused by TRAF3 and POT1 mutations pivotally requires further study.
Some SNP locations in RHOA, STAT3, and PDCD1 were frequently described in human TCLs; however, no study has investigated them in dogs with TCL. RHOA encodes a small GTPase that switches signal transduction cascades and promotes cytoskeleton organization, cell migration, and the cell cycle (48). The missense mutations of RHOA p.Asn117Lys and p.Asn117Ile were specifically altered in human CTCLs (36); therefore, we selected these variants and investigated them in four canine TCL/NCL subtypes. Surprisingly, RHOAN117I, N117K was found in all TCL subtypes with the highest frequency in EATCL (4/8, 50%), NCL (3/6, 50%), and ECTCL (10/21, 47%). Another SNP, STAT3 p.Tyr640Phe, was recently discovered to be mutated in human NK-cell lymphoma and CTCL (19, 49). STAT3 is a transcription factor that plays a key role in many cellular processes, including cell growth and apoptosis. Unlike in humans, STAT3Y640F was rarely observed in canine TCLs/NCLs; it was found in four cases of CTCL (11%), one case of EATCL (12%), and one NCL (16%). Three SNP locations of missense (p.Lys78Arg), stop gained (p.Glu46Ter), and frameshift (p.Phe37LeufsTer35) variants of PDCD1 were found to be loss-of-function mutations in human PTCLs (31). In addition, PD-1 deletions were related to a worse prognosis because they could revoke T-cell exhaustion and drive aggressive behavior in CTCL (19). In dogs, PDCD1E46*, K78R, F37LX was detected in NCL (4/6, 66%) and NECTCL (5/15, 33%). Besides, the lack of complete clinical information in the current study concealed the association between the PDCD1 mutation and prognosis. Thus, a prospective study of these SNPs affecting clinical stage, survival time, and disease progression is required to confirm their significance in prognosis.
Our study demonstrated that each canine patient with a specific subtype of TCL showed a different targeted SNP genotype. Hence, specific target therapy based on the SNP information may be advantageous in dogs with a certain type of lymphoma to increase treatment efficiency concurrent with chemotherapy. A study of CTCL in mice illustrated that methyltransferase inhibitors could restore SATB1 function in the Sézary cell line (50), and the usage of this target drug possibly abrogates malignant expansion in CTCL dogs. A STAT3 mutation could cause constitutive activation and might be a possible STAT3 inhibitor target. Napabucasin, a novel STAT3 inhibitor, has shown promising effects in inducing intrinsic and extrinsic apoptosis and downregulating the expression of STAT3 target genes against neoplastic B-cell lineages (51). Furthermore, napabucasin displayed a synergistic effect when administered with doxorubicin in vitro and in vivo experiments and may provide therapeutic implications in human B-cell lymphoma. Immunotherapy of PD-1 is an attractive cancer target. In human clinical trials, PD-1 blockage demonstrated substantial therapeutic activity to treat relapsed/refractory Hodgkin’s lymphoma as a single agent and advanced NK/TCL concurrent with chemotherapy (52, 53). The effectiveness of novel targeted drugs, particularly somatic SNPs, for treating canine lymphoma is necessitated in preclinical studies.
A small sample size of EATCL and NCL in the present study affected the estimated prevalence of each SNP between the two groups. Moreover, the lack of medical records for each patient hindered the valuable significance of SNPs and prognostic indicators. Another recently critical concern was FFPE-induced mutational artifacts, predominantly excessive T > C mutations, that may be affected in this study.
In conclusion, the MassARRAY platform revealed diverse mutational profiles, exhibiting significant variations across different TCL/NCL subtypes and their anatomical locations. Mutations in SATB1, KIT, SEL1L, and TP53 are frequently observed. Somatic mutations, particularly in TP53, are detected, implying a potential disparity between germline and somatic mutational patterns, although this analysis requires further validation in a larger number of cases and their contribution if they are cancer-associated mutations in dogs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal studies were approved by the Institutional Animal Care and Use Committee (IACUC), Chulalongkorn University Laboratory Animal Center. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
SS: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. TK: Conceptualization, Investigation, Writing – review & editing. ST: Conceptualization, Supervision, Writing – review & editing. AR: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Thailand Science Research and Innovation Fund Chulalongkorn University (FOOD66310015), 2023, the Chulalongkorn University–Veterinary Science Research Fund, 2022, and the Center of Excellence for Companion Animal Cancer, Faculty of Veterinary Science, Chulalongkorn University. SS was supported by the Postdoctoral Fellowship, Ratchadapisek Somphot Fund, and Chulalongkorn University.
Acknowledgments
The authors would like to thank Ken Chen and Tzu-yin Lin from the SQ Reference Lab and Sayamon Srisuwatanasagul from the Veterinary Clinical Center for recruiting some samples used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1439706/full#supplementary-material
Footnotes
References
1. Valli, VE, Kass, PH, San Myint, M, and Scott, F. Canine lymphomas: association of classification type, disease stage, tumor subtype, mitotic rate, and treatment with survival. Vet Pathol. (2013) 50:738–48. doi: 10.1177/0300985813478210
2. Valli, VE, San Myint, M, Barthel, A, Bienzle, D, Caswell, J, Colbatzky, F, et al. Classification of canine malignant lymphomas according to the World Health Organization criteria. Vet Pathol. (2011) 48:198–211. doi: 10.1177/0300985810379428
3. Valli, VE, Bienzle, D, and Meuten, DJ. Tumors of the hemolymphatic system In: DJ Meuten, editor. Tumors in domestic animals. Ames, IA, USA: John Wiley & Sons, Inc. (2016). 203–321.
4. Ponce, F, Magnol, JP, Ledieu, D, Marchal, T, Turinelli, V, Chalvet-Monfray, K, et al. Prognostic significance of morphological subtypes in canine malignant lymphomas during chemotherapy. Vet J. (2004) 167:158–66. doi: 10.1016/j.tvjl.2003.10.009
5. Seelig, DM, Avery, P, Webb, T, Yoshimoto, J, Bromberek, J, Ehrhart, EJ, et al. Canine T-zone lymphoma: unique immunophenotypic features, outcome, and population characteristics. J Vet Intern Med. (2014) 28:878–86. doi: 10.1111/jvim.12343
6. Deravi, N, Berke, O, Woods, JP, and Bienzle, D. Specific immunotypes of canine T cell lymphoma are associated with different outcomes. Vet Immunol Immunopathol. (2017) 191:5–13. doi: 10.1016/j.vetimm.2017.07.008
7. Couto, KM, Moore, PF, Zwingenberger, AL, Willcox, JL, and Skorupski, KA. Clinical characteristics and outcome in dogs with small cell T-cell intestinal lymphoma. Vet Comp Oncol. (2018) 16:337–43. doi: 10.1111/vco.12384
8. Dettwiler, M, Mauldin, EA, Jastrebski, S, Gillette, D, Stefanovski, D, and Durham, AC. Prognostic clinical and histopathological features of canine cutaneous epitheliotropic T-cell lymphoma. Vet Pathol. (2023) 60:162–71. doi: 10.1177/03009858221140818
9. Keller, SM, Vernau, W, Hodges, J, Kass, PH, Vilches-Moure, JG, McElliot, V, et al. Hepatosplenic and hepatocytotropic T-cell lymphoma: two distinct types of T-cell lymphoma in dogs. Vet Pathol. (2013) 50:281–90. doi: 10.1177/0300985812451625
10.Sogame, N, Risbon, R, and Burgess, KE. Intestinal lymphoma in dogs: 84 cases (1997-2012). J Am Vet Med Assoc (2018). 252:440–447. doi: 10.2460/javma.252.4.440
11. Azuma, K, Ohmi, A, Goto-Koshino, Y, Tomiyasu, H, Ohno, K, Chambers, JK, et al. Outcomes and prognostic factors in canine epitheliotropic and nonepitheliotropic cutaneous T-cell lymphomas. Vet Comp Oncol. (2022) 20:118–26. doi: 10.1111/vco.12752
12. Brodsky, EM, Maudlin, GN, Lachowicz, JL, and Post, GS. Asparaginase and MOPP treatment of dogs with lymphoma. J Vet Intern Med. (2009) 23:578–84. doi: 10.1111/j.1939-1676.2009.0289.x
13. Brown, PM, Tzannes, S, Nguyen, S, White, J, and Langova, V. LOPP chemotherapy as a first-line treatment for dogs with T-cell lymphoma. Vet Comp Oncol. (2018) 16:108–13. doi: 10.1111/vco.12318
14. Goodman, IH, Moore, AS, and Frimberger, AE. Treatment of canine non-indolent T cell lymphoma using the VELCAP-TSC protocol: a retrospective evaluation of 70 dogs (2003-2013). Vet J. (2016) 211:39–44. doi: 10.1016/j.tvjl.2016.03.003
15. Vail, DM, Thamm, DH, and Liptak, JM. Hematopoietic tumors In: Withrow and MacEwen's small animal clinical oncology. St. Louis, MO, USA: Elsevier, Inc. (2019). 688–772.
16. Iqbal, J, Wright, G, Wang, C, Rosenwald, A, Gascoyne, RD, Weisenburger, DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. (2014) 123:2915–23. doi: 10.1182/blood-2013-11-536359
17. Heavican, TB, Bouska, A, Yu, J, Lone, W, Amador, C, Gong, Q, et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood. (2019) 133:1664–76. doi: 10.1182/blood-2018-09-872549
18. Choi, J, Goh, G, Walradt, T, Hong, BS, Bunick, CG, Chen, K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. (2015) 47:1011–9. doi: 10.1038/ng.3356
19. Park, J, Daniels, J, Wartewig, T, Ringbloom, KG, Martinez-Escala, ME, Choi, S, et al. Integrated genomic analyses of cutaneous T-cell lymphomas reveal the molecular bases for disease heterogeneity. Blood. (2021) 138:1225–36. doi: 10.1182/blood.2020009655
20. Moffitt, AB, Ondrejka, SL, McKinney, M, Rempel, RE, Goodlad, JR, Teh, CH, et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J Exp Med. (2017) 214:1371–86. doi: 10.1084/jem.20160894
21. Nairismagi, ML, Tan, J, Lim, JQ, Nagarajan, S, Ng, CC, Rajasegaran, V, et al. JAK-STAT and G-protein-coupled receptor signaling pathways are frequently altered in epitheliotropic intestinal T-cell lymphoma. Leukemia. (2016) 30:1311–9. doi: 10.1038/leu.2016.13
22. Roberti, A, Dobay, MP, Bisig, B, Vallois, D, Boechat, C, Lanitis, E, et al. Type II enteropathy-associated T-cell lymphoma features a unique genomic profile with highly recurrent SETD2 alterations. Nat Commun. (2016) 7:12602. doi: 10.1038/ncomms12602
23. Horwitz, SM, Koch, R, Porcu, P, Oki, Y, Moskowitz, A, Perez, M, et al. Activity of the PI3K-delta,gamma inhibitor duvelisib in a phase 1 trial and preclinical models of T-cell lymphoma. Blood. (2018) 131:888–98. doi: 10.1182/blood-2017-08-802470
24. Zinzani, PL, Musuraca, G, Tani, M, Stefoni, V, Marchi, E, Fina, M, et al. Phase II trial of proteasome inhibitor bortezomib in patients with relapsed or refractory cutaneous T-cell lymphoma. J Clin Oncol. (2007) 25:4293–7. doi: 10.1200/JCO.2007.11.4207
25. Labadie, JD, Elvers, I, Feigelson, HS, Magzamen, S, Yoshimoto, J, Dossey, J, et al. Genome-wide association analysis of canine T zone lymphoma identifies link to hypothyroidism and a shared association with mast-cell tumors. BMC Genomics. (2020) 21:464. doi: 10.1186/s12864-020-06872-9
26. Elvers, I, Turner-Maier, J, Swofford, R, Koltookian, M, Johnson, J, Stewart, C, et al. Exome sequencing of lymphomas from three dog breeds reveals somatic mutation patterns reflecting genetic background. Genome Res. (2015) 25:1634–45. doi: 10.1101/gr.194449.115
27. Harris, LJ, Hughes, KL, Ehrhart, EJ, Labadie, JD, Yoshimoto, J, and Avery, AC. Canine CD4+ T-cell lymphoma identified by flow cytometry exhibits a consistent histomorphology and gene expression profile. Vet Comp Oncol. (2019) 17:253–64. doi: 10.1111/vco.12460
28. McDonald, JT, Kritharis, A, Beheshti, A, Pilichowska, M, Burgess, K, Ricks-Santi, L, et al. Comparative oncology DNA sequencing of canine T cell lymphoma via human hotspot panel. Oncotarget. (2018) 9:22693–702. doi: 10.18632/oncotarget.25209
29. Koshino, A, Goto-Koshino, Y, Setoguchi, A, Ohno, K, and Tsujimoto, H. Mutation of p53 gene and its correlation with the clinical outcome in dogs with lymphoma. J Vet Intern Med. (2016) 30:223–9. doi: 10.1111/jvim.13807
30. Sirivisoot, S, Kasantikul, T, Techangamsuwan, S, Radtanakatikanon, A, Chen, K, Lin, TY, et al. Evaluation of 41 single nucleotide polymorphisms in canine diffuse large B-cell lymphomas using MassARRAY. Sci Rep. (2022) 12:5120. doi: 10.1038/s41598-022-09112-0
31. Watatani, Y, Sato, Y, Miyoshi, H, Sakamoto, K, Nishida, K, Gion, Y, et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. (2019) 33:2867–83. doi: 10.1038/s41375-019-0473-1
32. Sirivisoot, S, Boonkrai, C, Wongtangprasert, T, Phakham, T, Muanwein, P, Pisitkun, T, et al. Development and characterization of mouse anti-canine PD-L1 monoclonal antibodies and their expression in canine tumors by immunohistochemistry in vitro. Vet Q. (2023) 43:1–9. doi: 10.1080/01652176.2023.2240380
33. Bóna, G, and Šiviková, K. Detection of mutations in selected proto-oncogenes of canine lymphoma. Folia Vet. (2017) 61:34–9. doi: 10.1515/fv-2017-0036
34. Giannuzzi, D, Marconato, L, Cascione, L, Comazzi, S, Elgendy, R, Pegolo, S, et al. Mutational landscape of canine B-cell lymphoma profiled at single nucleotide resolution by RNA-seq. PLoS One. (2019) 14:e0215154. doi: 10.1371/journal.pone.0215154
35. Smith, PAD, Waugh, EM, Crichton, C, Jarrett, RF, and Morris, JS. The prevalence and characterisation of TRAF3 and POT1 mutations in canine B-cell lymphoma. Vet J. (2020) 266:105575. doi: 10.1016/j.tvjl.2020.105575
36. Park, J, Yang, J, Wenzel, AT, Ramachandran, A, Lee, WJ, Daniels, JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood. (2017) 130:1430–40. doi: 10.1182/blood-2017-02-768234
37. Girish, KS, and Kemparaju, K. The magic glue hyaluronan and its eraser hyaluronidase: a biological overview. Life Sci. (2007) 80:1921–43. doi: 10.1016/j.lfs.2007.02.037
38. Tian, X, Azpurua, J, Hine, C, Vaidya, A, Myakishev-Rempel, M, Ablaeva, J, et al. High-molecular-mass hyaluronan mediates the cancer resistance of the naked mole rat. Nature. (2013) 499:346–9. doi: 10.1038/nature12234
39. Kaneko, M, and Nomura, Y. ER signaling in unfolded protein response. Life Sci. (2003) 74:199–205. doi: 10.1016/j.lfs.2003.09.007
40. Cattaneo, M, Fontanella, E, Canton, C, Delia, D, and Biunno, I. SEL1L affects human pancreatic cancer cell cycle and invasiveness through modulation of PTEN and genes related to cell-matrix interactions. Neoplasia. (2005) 7:1030–8. doi: 10.1593/neo.05451
41. Fredholm, S, Willerslev-Olsen, A, Met, O, Kubat, L, Gluud, M, Mathiasen, SL, et al. SATB1 in malignant T cells. J Invest Dermatol. (2018) 138:1805–15. doi: 10.1016/j.jid.2018.03.1526
42. Stephen, TL, Payne, KK, Chaurio, RA, Allegrezza, MJ, Zhu, H, Perez-Sanz, J, et al. SATB1 expression governs epigenetic repression of PD-1 in tumor-reactive T cells. Immunity. (2017) 46:51–64. doi: 10.1016/j.immuni.2016.12.015
43. Gao, Y, Liu, F, Sun, J, Wen, Y, Tu, P, Kadin, ME, et al. Differential SATB1 expression reveals heterogeneity of cutaneous T-cell lymphoma. J Invest Dermatol. (2021) 141:607–618.e6. doi: 10.1016/j.jid.2020.05.120
44. Kikuma, K, Yamada, K, Nakamura, S, Ogami, A, Nimura, S, Hirahashi, M, et al. Detailed clinicopathological characteristics and possible lymphomagenesis of type II intestinal enteropathy-associated T-cell lymphoma in Japan. Hum Pathol. (2014) 45:1276–84. doi: 10.1016/j.humpath.2013.10.038
45. Mahadevan, D, Spier, C, Della Croce, K, Miller, S, George, B, Riley, C, et al. Transcript profiling in peripheral T-cell lymphoma, not otherwise specified, and diffuse large B-cell lymphoma identifies distinct tumor profile signatures. Mol Cancer Ther. (2005) 4:1867–79. doi: 10.1158/1535-7163.MCT-05-0146
46. Muro, I, Fang, G, Gardella, KA, Mahajan, IM, and Wright, CW. The TRAF3 adaptor protein drives proliferation of anaplastic large cell lymphoma cells by regulating multiple signaling pathways. Cell Cycle. (2014) 13:1918–27. doi: 10.4161/cc.28895
47. Pinzaru, AM, Hom, RA, Beal, A, Phillips, AF, Ni, E, Cardozo, T, et al. Telomere replication stress induced by POT1 inactivation accelerates tumorigenesis. Cell Rep. (2016) 15:2170–84. doi: 10.1016/j.celrep.2016.05.008
48. Schaefer, A, and Der, CJ. RHOA takes the RHOad less traveled to cancer. Trends Cancer. (2022) 8:655–69. doi: 10.1016/j.trecan.2022.04.005
49. Song, TL, Nairismagi, ML, Laurensia, Y, Lim, JQ, Tan, J, Li, ZM, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. (2018) 132:1146–58. doi: 10.1182/blood-2018-01-829424
50. Harro, CM, Perez-Sanz, J, Costich, TL, Payne, KK, Anadon, CM, Chaurio, RA, et al. Methyltransferase inhibitors restore SATB1 protective activity against cutaneous T cell lymphoma in mice. J Clin Invest. (2021) 131:e135711. doi: 10.1172/JCI135711
51. Li, X, Wei, Y, and Wei, X. Napabucasin, a novel inhibitor of STAT3, inhibits growth and synergises with doxorubicin in diffuse large B-cell lymphoma. Cancer Lett. (2020) 491:146–61. doi: 10.1016/j.canlet.2020.07.032
52. Ansell, SM, Lesokhin, AM, Borrello, I, Halwani, A, Scott, EC, Gutierrez, M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. (2015) 372:311–9. doi: 10.1056/NEJMoa1411087
Keywords: cutaneous T-cell lymphoma, dog, intestinal T-cell lymphoma, nodal T-cell lymphoma, null-cell lymphoma, SNPs
Citation: Sirivisoot S, Kasantikul T, Techangamsuwan S and Rungsipipat A (2024) Single nucleotide polymorphism profiles of canine T-cell and null-cell lymphomas. Front. Vet. Sci. 11:1439706. doi: 10.3389/fvets.2024.1439706
Edited by:
Maria Elena Turba, Genefast srl, ItalyReviewed by:
Fabio Gentilini, University of Bologna, ItalyMarkéta Dajbychová, Genomia s.r.o., Czechia
Copyright © 2024 Sirivisoot, Kasantikul, Techangamsuwan and Rungsipipat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anudep Rungsipipat, YW51ZGVwLnJAY2h1bGEuYWMudGg=
†ORCID: Tanit Kasantikul, https://orcid.org/0000-0002-1895-8992
 Sirintra Sirivisoot
Sirintra Sirivisoot Tanit Kasantikul2†
Tanit Kasantikul2† Somporn Techangamsuwan
Somporn Techangamsuwan Anudep Rungsipipat
Anudep Rungsipipat