- 1Laboratorio de Biología Celular y Tisular, Departamento de Morfología, Centro de Ciencias Básicas, Universidad Autónoma de Aguascalientes, Aguascalientes, Mexico
- 2Laboratorio de Estudios Ambientales, Departamento de Fisiología y Farmacología, Centro de Ciencias Básicas, Universidad Autónoma de Aguascalientes, Aguascalientes, Mexico
- 3Departamento de Ciencias Veterinaria, Centro de Ciencias Agropecuarias, Universidad Autónoma de Aguascalientes, Aguascalientes, Mexico
Actinobacillus pleuropneumoniae (App) is a globally distributed Gram-negative bacterium that produces porcine pleuropneumonia. This highly contagious disease produces high morbidity and mortality in the swine industry. However, no effective vaccine exists to prevent it. The infection caused by App provokes characteristic lesions, such as edema, inflammation, hemorrhage, and necrosis, that involve different virulence factors. The colonization and invasion of host surfaces involved structures and proteins such as outer membrane vesicles (OMVs), pili, flagella, adhesins, outer membrane proteins (OMPs), also participates proteases, autotransporters, and lipoproteins. The recent findings on surface structures and proteins described in this review highlight them as potential immunogens for vaccine development.
Introduction
Porcine pleuropneumonia, caused by Actinobacillus pleuropneumoniae (App), is one of the respiratory diseases with the greatest economic impact worldwide (1). This disease is the source of great economic losses because it produces high morbidity, mortality, and costs for treating infected animals. Despite the clinical and sanitary management carried out within pig farms, the transmission of the disease is very easy to spread due to the portability of the bacteria from animals that survived the infection or that were asymptomatic (2–4).
To date, 19 serotypes have been recognized. The classification of biovarieties by their growth in vitro is of two types. Biovar 1, requires nicotinamide adenine dinucleotide (NAD) exogenously for growth, and Biovar 2, which is independent of NAD. Generally, serotypes 1 to 12 and 15 to 19 are biovar 1, and serotypes 13 and 14 are biovar 2 (1, 5, 6).
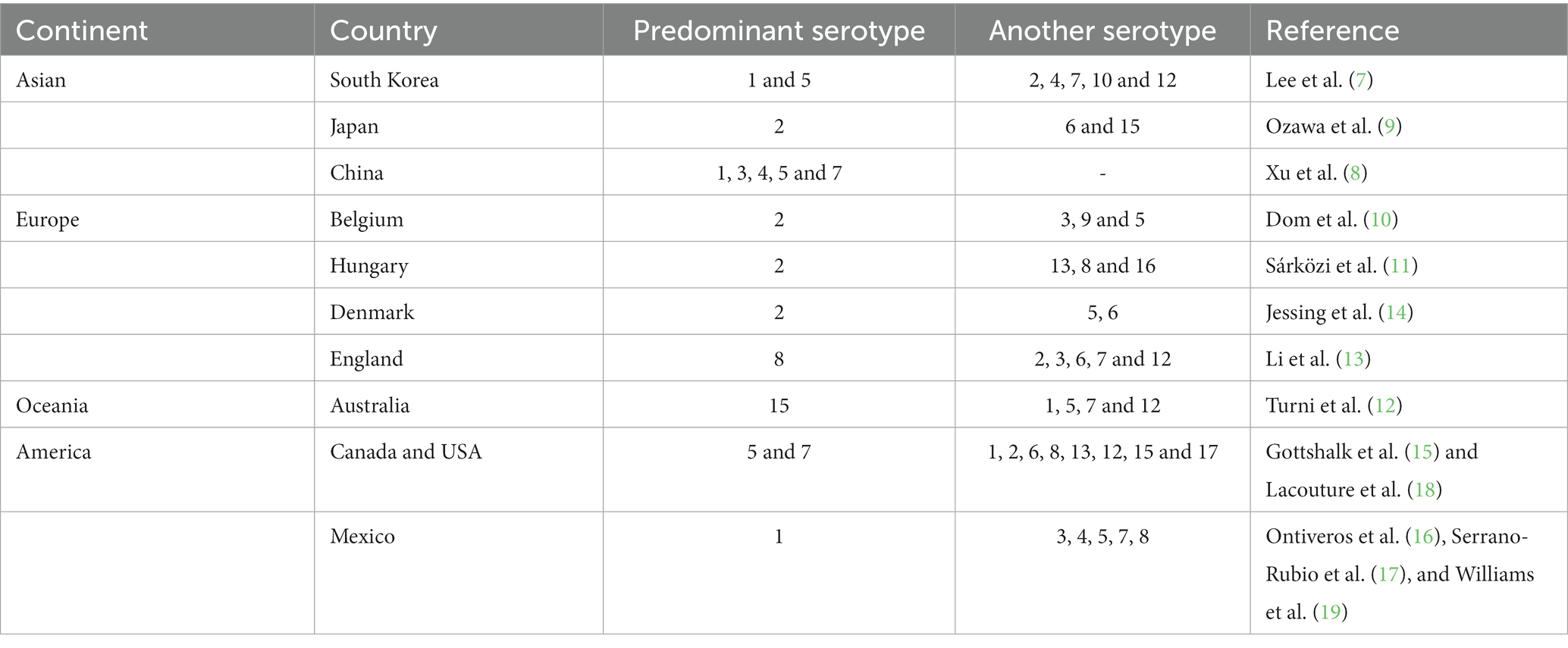
The prevalence of serotypes worldwide is very varied (7–19), making the infection very difficult to control since vaccines developed for some regions are not effective for others (9) (Table 1).
Transmission of App was thought to be solely due to direct contact of mucus and aerosol from one pig to another, but it has now been shown that App may be viable in water samples from pig farms, suggesting that it could function as a reservoir, Inoculum or vehicle to transmit the bacteria (20). Its viability in nasal secretions outside the pig is 3–4 days; it can survive at −20°C for 17 weeks and is maintained at −70°C in prolonged storage (21).
The App bacterium is usually considered a strict pathogen of the respiratory tract of pigs. However, there are scattered cases of it being detected as a causative agent in cases of arthritis, osteomyelitis, hepatitis, meningitis, and nephritis (22). The disease can range from hyperacute to chronic, depending on the number of bacteria that reach the lung. The main signs are fever, increased respiratory rate, coughing, sneezing, dyspnea, anorexia, ataxia, vomiting, diarrhea, and respiratory distress. The primary lesions occur at the level of the lungs, finding edema, inflammation, hemorrhage, and necrosis. In some cases, we can find hemorrhagic foam in the nose or mouth of animals that die. We can also find bloody fluid and fibrin in the thoracic cavity (23).
For App to install itself, persist in the host, and cause the characteristic lesions of the infection, it uses virulence factors. At present, bacterial virulence factors can be divided according to their mechanism of action: factors that involve adhesion to the host, factors in the acquisition of essential nutrients, factors involved in the induction of lesions, and the persistence of the infection (3). The bacterial membrane is composed of 50% of surface proteins, which participate diversely in virulence factors (24).
Due to the worldwide distribution of the serotypes, the high mortality and morbidity, the persistence in infected animals, the easy transmission of the infection due to the survival of the bacteria in the environment, its antigenic variability, including virulence factors, it has been difficult to find a vaccine that manages to control and eradicate the presentation of the infection caused by App within pig farms. This is why alternatives for potential immunogens must be sought to develop new vaccines. In this review article, a global analysis of the structures and surface proteins, general characteristics, transport, and secretion is carried out. It lists surface proteins described to date and how they intervene in the virulence factors and a review of potential vaccines based on these proteins. Finally, techniques for the isolation and identification of a variety of proteins are classified, allowing the establishment of protocols that support the advancement of knowledge of the pathogenesis of the infection caused by App.
Bacterial envelope and proteins transport
App bacterium is a Gram-negative, with typical coccobacillus morphology, hemolytic, urease positive, positive for mannitol, xylose, ribose, and lactose fermentation, and negative for glycerol, indole, and esculin (4, 25), of an approximate size of 0.4 × 1.0 μm, for its growth in vitro, requires aerobic or anaerobic conditions at a temperature of 37° C for a period of 24 to 72 h, as morphological characteristics the colonies are partially transparent, circular smooth, shiny and convex (26).
The membrane of Gram-negative bacteria is composed of an inner and an outer membrane; between the two membranes, there is a peptidoglycan mesh that gives it shape and rigidity. The outer membrane is a phospholipid bilayer composed of many proteins, surface structures, and lipopolysaccharides (LPS); the inner membrane is in contact with newly synthesized proteins, where some are transported to the extracellular medium (27–29) (Figure 1).
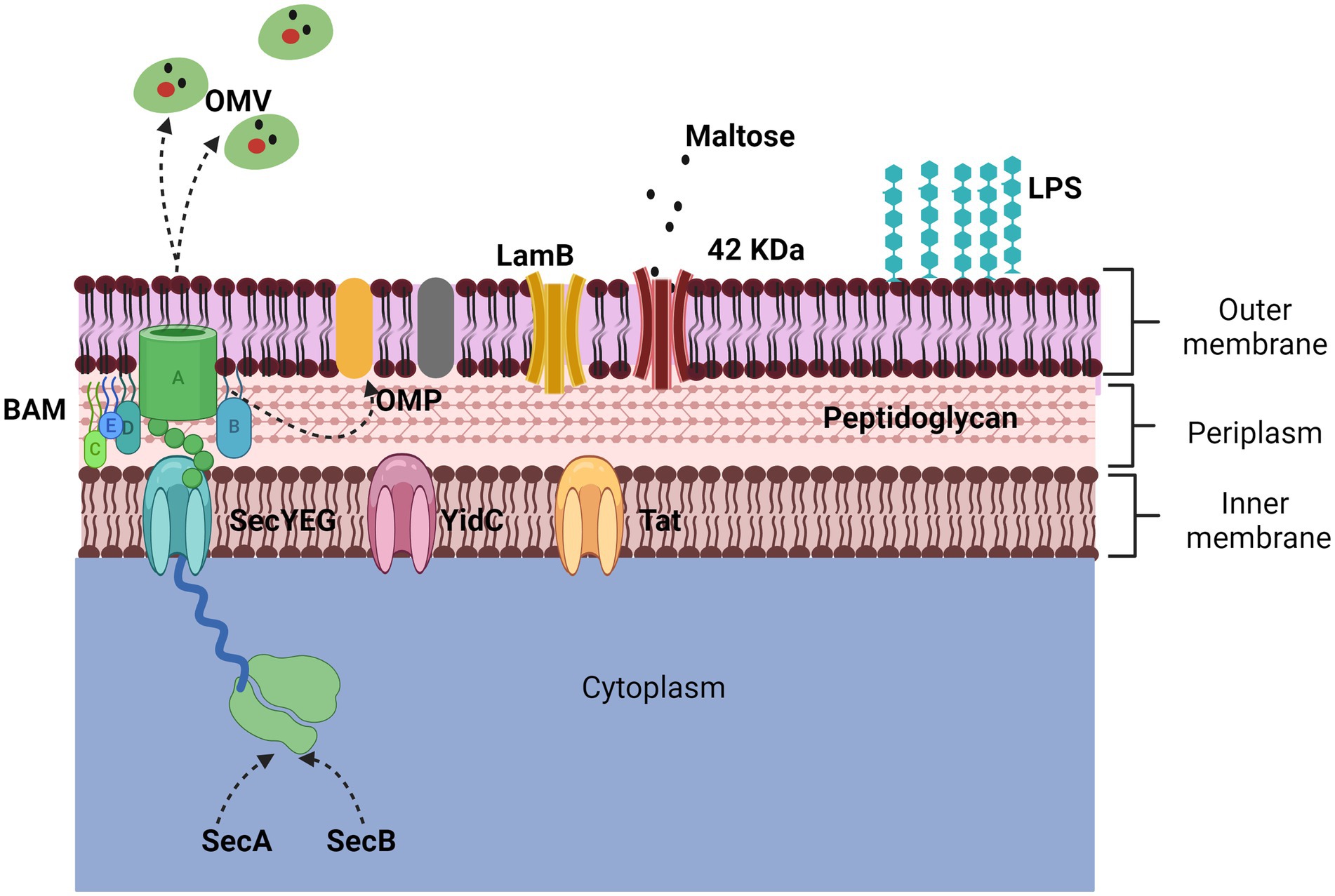
Figure 1. Representation of the outer membrane. The bacterial membrane is composed of a bilayer, the inner membrane, and the outer membrane. The two main proteins that make it up are porins and outer membrane proteins (OMPs). The 42 KDa protein identified in App is porin-like and is expressed in the presence of maltose while the LamB porin aids in antimicrobial resistance. The secretion of OMPs is regulated by three transporters (Sec, YidC, and Tad) through the BAM complex. The secretion mechanism of OMVs is not defined but they harbor numerous proteins and toxins.
One of the main proteins on the cell surface is the outer membrane proteins (OMPs), which can only be found in Gram-negative bacteria, mitochondria, and chloroplasts. The OMPs are composed of a β barrel fold, with an approximate size of 8 to 26 strands, and each protein is synthesized in the cytoplasm for a location and environment of the outer membrane (24, 30). Trafficking to the outer membrane begins with the secretion of their proteins, which are synthesized in the cytoplasmic ribosome, crossing the inner membrane. The inner membrane contains three types of transporters, Sec translocon, the YidC insertase, and the Tat system (31). The Sec translocon (SecYEG), through a small pore in the center, remains unfolded using chaperones (SecA and SecB), which emit a signal peptide to be delivered to the BAM complex for refolding and insertion into the outer membrane. However, it is still unknown in detail how this process happens; it is known that in some cases, the synthesis of some proteins is strongly induced when they are needed (Figure 1) (24, 28).
Most of the proteins transported by the Sec and Tat systems remain in the cell, either in the periplasm or on the inner membrane; some proteins can be transported out of the cell with the help of another secretion system, such as the In the case of exoproteins that use the Sec and Tat-dependent system and ultimately use systems that are based on the β-barrel channel that forms a ring in the outer membrane (Type I, II, III, and IV systems) (32).
On the other hand, other structures are also secreted by bacteria, which contain many proteins. Outer membrane vesicles (OMVs) are structures that derive from the envelopment of the bacterium (33), and were first described in Gram-negative bacteria, specifically in Escherichia coli, and subsequently in all Gram-negative bacteria related so far. OMVs are spherical particles that vary from 20 to 300 nm in size; the production mechanism of these vesicles is not yet fully elucidated (34).
Adhesion proteins
Once the bacterium enters the host, the latter activates different defense mechanisms, such as mucus secretion, mucociliary action, and the use of immune system components to prevent adherence. On the contrary, the bacterium uses mechanisms that allow it to evade the host’s response and will enable it to remain inside it. Adherence of bacteria to the host mucosal epithelium is the first step in colonization and establishing pathogenesis. The adhesion of App to the tracheal cells of the porcine respiratory tract has been mainly associated with the lipopolysaccharides (LPS) present in the cell membrane of the different serotypes of this bacterium (35) mechanisms allow them to adhere firmly to the host mucosa are pili, flagella, fimbriae and specific adhesins (36, 37).
The pili are filamentous structures typical of Gram-negative bacteria and are composed of large biopolymers in the form of helices attached to the outer membrane (38, 39). Pili types and functions will vary depending on the type of bacteria; for example, type IV pili have a sliding motility function, while type 1 pili and P-pili from uropathogenic Escherichia coli and CS pili from enterotoxigenic E. coli have a function adhesion (40). The pili in App is a pili IV type (Tfp) or IVb type (Flp), which is formed in defined chemical media; in addition, this pili is essential for adherence and biofilm formation (41, 42). In interactions with epithelial cells, it has been shown that the Tfp protein is upregulated after adhesion has occurred (43).
In the case of adhesins that enhance surface binding for App is the ApfA adhesin, which has been suggested as an important virulence factor for the ability to colonize the lung (44). Adherence of the bacteria to alveolar epithelial cells has been demonstrated in vitro associated with the expression of fimbrial proteins and OMPs of 55 kDa, a protein not yet described (45) has also been proposed that a 60 kDa protein could be related to adhesion to extracellular matrix components such as collagen and fibrinogen (46).
Trimeric autotransporter adhesins (TAAs) are a family of proteins that are secreted by the outer membrane of Gram-negative bacteria; their structure comprises an N-terminal “passenger” domain followed by a C-terminal translocation unit which functions as an anchor of the membrane. These proteins have been identified in various pathogenic bacteria, attributing to them diverse functions such as adhesion, hemagglutination, and serum resistance (47). A few TAAs have been identified in App; the adhesin Adh the YadA-like head region of Apa1 has been identified as the optimal adhesion in vitro adhesion assays, in addition to regulating auto-agglutination, biofilm formation, and maintenance of bacterial cell morphology (48), Apa2H1 is the primary domain of the trimeric autotransporter adhesin Apa2 which has been shown to have an effect of activating an immune response and protecting against infection by App (49).
To prevent the adhesion of the bacteria to the host and stop the onset of infections, alternative vaccines have been chosen using pili, fimbriae, and adhesins. Although in App, the results of vaccines with fimbrial antigens and adhesins have only induced a strong and non-protective immune response, other studies against various bacterial pathogens such as Moraxella bovis, Dichelobacter nodosus, Proteus mirabilis y E. coli enterotoxigenic have been successful (50). Conducting trials with vaccines made with these proteins and others could provide a more significant immune response against App. A clear example of success was a vaccine with a chimeric protein called Ap97, which comprises a deletion derivative of the N-terminal region of the ApxIII toxin from App and the R1 and R2 repeats of the P97 adhesin from Mycoplasma hyopneumoniae P97, which conferred protection to challenged pigs against these pathogens (51). Vaccines made with adhesins allow the stimulation of the protection of protective antibodies, allowing the neutralization of the bacteria, inhibiting the attachment to the host tissue, and, through opsonization, causing microbial death by phagocytosis or complement activity (52).
Nutrients acquisition
Bacteria need nutritional requirements such as sugars, lipids, proteins, peptides, oligonucleotides, xenobiotics, vitamins, minerals, and inorganic elements to survive, and the main routes for obtaining them are through anabolism and catabolism depending on the resources found around them.
One of the main cellular structures that function as transporters is the so-called porins, which, as their name suggests, form a pore in the bacterium’s outer membrane (53). Porins are now known to play an important role in energy production, photosynthesis, and nutrient transport (54). The OmpC and OmpF porins in E. coli were the first to be identified in Gram-negative bacteria; later, they were identified in other bacteria, including Gram-positive bacteria. Its structure can be trimeric or monomeric, which will depend on its physicochemical properties, its conformation, and the environment; for example, some Gram-negative bacteria, in case of nutrient limitation, adapt their permeability through the expression of OmpF and LamB porins, which are important for the absorption of sugar (55). Porins were described by Nakab in 1975 in S. typhimurium (56).
In App serotype 1, it has been possible to identify a 42 kDa protein expressed in growth conditions with maltose. This protein was described as a porin-type protein because it is associated with peptidoglycan and can resist trypsin proteolysis. Other maltose-inducible proteins have been described in S. typhimurium and Vibrio cholerae (57).
A study conducted by Ma et al. (58) found 12 porins in a strain of App (SC1810) and, through a transcriptome analysis, found that genes coding for the LamB and OmpP2B porins were downregulated, deletion of LamB led to a decrease in sensitivity to carbapenem drugs.
There is currently a commercial vaccine with a 42 kDa protein in combination with Apx toxins (Porcilis APP from MSD Animal Health and Pleurostar APP from Novartis Animal Health Canada Inc.); this vaccine has obtained favorable results as it reduces the prevalence of symptoms and respiratory lesions, in addition to reducing mortality and the use of antimicrobials (59). This reduction in the use of drugs could be precisely because porins have been related to this resistance to antimicrobials. One of the most impactful benefits of continuing to carry out this type of vaccine is that it helps prevent the establishment of infections with resistant strains in immunized individuals and reduces the resistance in other microorganisms by reducing the use of antibiotics (60).
Iron is another essential factor for growth, metabolism, and bacterial colonization in Gram-negative bacteria. It is also an indispensable factor in eukaryotic cells, leading to competition for this nutritional factor (61). When the bacterium is in contact with the host, it activates an immune response called nutritional immunity, which limits the production of iron and other metals to prevent absorption by pathogenic organisms (62). Several proteins in App contribute to the host’s defense of this immunity mechanism, which acts as virulence factors regarding iron acquisition.
The FhuA lipoprotein is a 75 kDa protein that acts as a ferrichrome receptor (63) is expressed in vivo (64) and is in all App serotypes (65). This protein encodes the receptor for ferrichrome TbpA/TbpB, of 110 and 60 kDa, respectively, lipoproteins related to transferrin binding (63) and is also associated with three other proteins FhuD (35.6 kDa), FhuC (28.5 kDa) and FhuB (69.4 kDa) proteins related to the translocation and transport of ferric hydroxamate (66). The FhuA protein has also been described in other bacteria (67), such as E. coli. It is attributed an important role not only in the transport of ferrichrome, but it can also become a high conductivity channel by binding to phage T5, T1, phi 80, colicin M and the antibiotic albomycin (68) so that it functions as a mediator for the transport of phage DNA (69) while in Salmonella enterica and Salmonella bongori it participates as a receptor for several bacteriophages and toxins (70).
The TbpA and TbpB proteins form a bipartite transferrin receptor; Tpbs are host-specific proteins and, therefore, will only bind transferrin from their host (71). The bacterium App expresses these proteins under iron-restricted conditions, which allows it to use porcine transferrin as the only source of iron and play an important role as a virulence factor during infection (72). Both proteins have also been found exposed on the surface of Neisseria meningitidis, forming a single complex (73) Haemophilus (Glässerella) parasuis. An increase in the genes of these two proteins has also been detected when they are under iron restriction stress (74).
The HgbA protein weighs approximately 105 kDa, is required for App growth on hemoglobin as the sole source of iron (64), and, like FhuA, is conserved in all App serotypes (65). This protein is essential in obtaining iron from other pathogenic microorganisms, such as Haemophilus ducreyi. The HgbA protein presents homology with HutA of V. cholerae, Tbp1, and Lbp1 of Neisseria spp., which also function as receptors for heme, transferrin, and lactoferrin groups (75).
The protective efficacy that the proteins involved with the iron acquisition factor in App can produce has been verified. A vaccine made with bacterial lysates containing the proteins TbpA, TbpB, HgbA, other membrane proteins, and exoproteins resulted in a humoral and protective response against serotypes 2 and 9 (76). There are no recent studies that evaluate the effectiveness of these proteins in App, but there are in other bacteria such as H. parasuis (74), N. meningitidis (77), and Salmonella spp. (78). The lack of up-to-date studies using these proteins is essential since reverse vaccinology studies suggest they could be potential immunogens for developing effective vaccines (67, 79). It has also been observed that the combination of some of these proteins involved in acquiring iron with others usually has satisfactory results in protecting against pathogenic infections (78, 80, 81).
Induction of injuries and evasion of defense mechanisms
Pathogenic bacteria also use virulence factors to cause injury and thus ensure their permanence within the host. Bacterial species combine virulence factors to induce disease, and depending on the combinations, they can cause characteristic lesions; for example, enteropathogenic E. coli (EPEC) destroys intestinal microvilli, which induce infection and the production of diarrhea (38).
In App, there are characteristic pulmonary lesions that are caused by pore-forming toxins (PFT) called Apx toxins, which allow dissemination and colonization within the host; their mechanism will enable them to embed themselves in the membrane to perforate it later, causing an ionic imbalance and nutritional (82) having proteolytic, cytotoxic and edemogenic effect (83, 84).
To date, the 19 known serovar (5) produce four toxins in different ways (ApxI, ApxII, ApxIII, and ApxIV) (84). The ApxI, ApxII, and ApxIII toxins have a cytotoxic and hemolytic effect and, when released, can cause lysis of epithelial cells, alveolar cells, endothelial cells, red blood cells, neutrophils, and macrophages even in the presence of neutralizing antibodies, where the serotype relates the production of these toxins and not by the App strains (3, 85), the ApxIV toxin is expressed in infected pigs and not in vitro and is a determinant to confirm the infection caused by App (86).
The ApxI, Apx II, and ApxIII toxins have been widely cataloged with a rather important role as a virulence factor of App; it has been shown that they are responsible for the presence of respiratory symptoms in addition to the typical lesions of porcine pleuropneumonia both in endobronchial inoculations as in the analysis of tissues with necrosis of clinically diseased animals (87, 88). The ApxI toxin secreted by serotypes 1, 5, 9, and 11 is the most virulent due to the cytolysis and hemolysis it causes in experimental models (84, 89).
In the case of respiratory tract invasion by infectious agents, as the primary defense, the immune system responds with the activation of alveolar macrophages and neutrophils. It has been verified that even in the presence of antibodies against Apx toxins, App can destroy alveolar macrophages, unlike neutrophils protected by these antibodies (90). Specifically, ApxI can induce necrosis, apoptosis in porcine alveolar macrophages (PAM) (91), and pyroptosis by caspase-1-dependent cell death (92). The use of transposon mutagenesis in different animals has shown that Apx toxins are the main virulence factors of the infection caused by App (93).
Within the group of toxins of the RTX family, other exotoxins have also been identified in bacteria such as Aggregatibacter actinomycetemcomitans (LtxA), Mannheimia haemolytica (LktA), Bordetella pertussis (CyaA), uropathogenic E. coli (HlyA) where their central receptors are β2 integrins found in leukocytes (94).
The App bacterium has a great diversity of serotypes, and, as we have already mentioned, they secrete a variety of Apx toxins diversely. This antigenic variability has not allowed the development of a single vaccine for the various existing App serotypes. The investigation of the different virulence factors of App has focused on the development of vaccines that allow the creation of optimal immunity for the other serotypes of this bacterium, the most studied being the development of vaccines with Apx toxins. However, they have also developed polysaccharide and liposaccharide vaccine conjugates involving OMPs combined with hemolysin and OMPs (95).
There are other proteins involved with the induction of injury. VacJ was first described as an outer membrane lipoprotein of the bacterium Shigella flexneri, with a molecular weight of 28 kDa, as a protein capable of allowing intercellular dissemination and causing a protrusion in infected cells (96). Roier et al. (97) demonstrated that, as a virulence factor, the VacJ lipoprotein of the bacteria Haemophilus influenzae and V. cholerae contributes to the increase in OMVs, obtaining the same result Antenucci et al. (98), who was confirming its presence in App serotypes 3, 5b and 7, in addition to demonstrating that VacJ is highly conserved and expressed in App during natural infection. It has been verified that the OMVs also contain toxins, proteases (99), and antigenic proteins, which are attributed to the damping of the response of alveolar macrophages at the beginning of the infection by App (91).
Proteases allow bacteria to degrade misfolded proteins, act as regulators of their environment, and actively participate as virulence factors (100). Within this group of proteins, the HtrA protein, also initially described in E. coli, has been described in App as a serine protease that allows quality control of other proteins and is active in response to secretion stress in animal models. It has been proven to induce an immune response through the secretion of IgG, IFN-γ, and IL-2 and activates the proliferation of lymphocytes in the spleen (101).
In the past, it was believed that infectious diseases were controlled by discovering chemotherapeutic agents. However, this was not the case for toxin-producing infections where vaccination has not met expectations (102), as with infection by App. There are currently commercial vaccines made from toxins such as Porcilis APP (MSD Animal Health), Ingelvac® APPX (Boehringer Ingelheim), and Coglapix® (Ceva), but none have managed to control infection by App (95). Therefore, it is necessary to continue looking for alternative immunogens to develop new vaccines.
Reverse vaccinology analyses have also made it possible to establish the VacJ protein as a potential immunogen (98). Studies where vaccines made with OMVs have been used with this protein have resulted in a high titer of IgG antibodies but have not allowed effective protection against infection caused by App to be established (91, 103). Similar results were obtained with a vaccine against Glaesserella parasuis (104) In contrary cases, the combination of VacJ with other proteins has given 100% protection for controlling Pasteurella multocida (105).
In the case of the HtrA protein, it has only been possible to carry out clinical trials where it has been possible to establish that the absence of this protein in the bacteria decreases the colonization capacity, in addition to the fact that conditions of oxidative stress and high temperature potentiate infection by App (106). This result has been attributed to the fact that HtrA can cleave cell-to-cell binding factors, causing damage to the extracellular matrix, disrupting the epithelial barrier, and thus finally causing damage to the host cell. This protein is proposed as a possible antibacterial treatment or for vaccine development (100).
Cell integrity
As previously mentioned, the cell membrane plays one of the most important roles concerning the integrity of the bacteria when it encounters the external environment with a diversity of physical, chemical, and nutritional environments. Some proteins that contribute to maintaining the cell membrane have been identified in App.
The OmlA lipoprotein is an outer membrane protein first described by Gerlach et al. (107) with a molecular weight of 40 kDa; in addition to being also found within the outer membrane vesicles (OMVs), it is detected in serotypes 1, 2, 8, 9, 11 and 12. Subsequently, allelic variations between the OmlA genes between different App serotypes were identified (108, 109). It was not until years later that App serotypes 1-15 showed a phylogeny of up to 10 variants (110). This protein has also been studied in other bacterial species such as Pseudomonas aeruginosa, Pseudomonas fluorescens (111), and Xanthomonas campestris pv. phaseoli (112), having a role in maintaining cell integrity, in addition to allowing iron acquisition by the host (72).
The OmlA protein has been related to the AasP protein. AasP is a 104 kDa subsistilin-like autotransporter conserved in the outer membrane, similar to a serine protease. It is experimentally transcribed in porcine lung tissue between 7 and 21 days after infection and is associated and expressed in membrane fractions. It is regulated by a global anaerobic regulator (HlyX) (113). Ali et al. (114) used recombinant AasP protein, and immunoblot analysis showed that AasP does not undergo proteolytic cleavage. Using an AasP mutant derivative and a complemented AasP mutant led to identifying OmlA fragments, suggesting that AasP is involved in OmlA modification. On the other hand, AasP is also related to other proteins of the SPATE family of serine proteases. Plasencia-Muñoz et al. (115) identified in immunoblot a band with a molecular weight of 100 kDa using a polyclonal antibody against Vat toxins, weight like the AasP protein. In a cellular interaction, the internalization of the bacteria is observed as the formation of vacuoles in endothelial cells in an in vitro cell model. However, it is not established whether AasP is responsible for said vacuoles.
There are other proteins related to membrane integrity. PalA lipoprotein is analogous to the OMPs associated with the PAL peptidoglycan of E. coli. This protein was initially purified from App serotype 2, with a molecular weight of 14 kDa, and later identified in 12 App serotypes, in addition to having a high similarity with the E. coli TolB protein (116). TolB interacts with other OMPs such as TolA, TolQ, and TolR and plays a role in maintaining the integrity of the outer membrane of E. coli (117, 118). PalA has a similarity with the P6 protein of H. influenzae. With an OMPs of 16 kDa of P. multocida, it has been shown that an antiserum against the P6 protein acts as a bactericide against clinical isolates of highly pathogenic non-typeable strains (118).
Polyamines are a group of small polycationic compounds that participate in aspects of cell physiology, stabilizing the structure of the cell membrane, in addition to participating in stress tolerance, signaling, biofilm formation, and immune system evasion. In App, the PotD periplasmic protein binds to polyamines before transport, it has been identified in 15 serotypes, and deletion of the potD2 gene can lead to defects in extracellular polyamine transport in the early period of growth, leading to cell damage (119).
The outer membrane lipoprotein VacJ in the pathogenicity of App has an important role in maintaining cell integrity and serum resistance. It has been shown that bacteria mutated with the VacJ gene show greater sensitivity to EDTA and SDS detergents, allowing greater hydrophobicity of the bacterial membrane. These same results have been seen in S. flexneri and H. influenzae. In the case of serum resistance, it has been observed that bacteria mutated with the VacJ gene are much more sensitive to porcine serum in vitro, reflecting the loss of the ability to resist complement factor (120).
There are few trials of vaccines with proteins involved in the integrity of the bacterial membrane. OmlA has been proposed as a potential immunogenic agent, but currently, there are no vaccines with this protein against infection caused by App (95, 121).
The only vaccine prototype for the AasP autotransporter was the study by Oldfield et al. (122). They hypothesized that the vaccine would protect pigs from infections against App, but the results were unfavorable since it did not inhibit colonization and severe clinical disease. There is no evidence of the presence of AasP in other bacterial species. However, a wide variety of other autotransporters exist in E. coli, Citrobacter rodentium, Salmonella, Neisseria, Shigella, and Edwardsiella species (123, 124), including the serine proteases (SPATEs) of E. coli the most studied, whose immunoprotective effect results have allowed us to establish that it inhibits colonization (125–128). With the results of investigations on the virulence of the group of autocarrier proteins, it has been concluded that the widespread presence of conserved and highly immunogenic epitopes suggests that they may be the target of vaccine development for infections caused by Gram-negative bacteria (128, 129).
Although in the case of the PalA lipoprotein, there are studies of vaccination in combination with other proteins, the results have not been favorable since the protective effect of different proteins decreases (118). This same result was obtained in a murine model that corroborates that in the case of this protein, the product is immunogenic but not protective (130). In the case of the PotD protein, no vaccine was developed for App, but it should be noted that there is evidence that this protein has had an immunoprotective effect against H. parasuis (131).
Biofilm formation
Biofilms are structured communities of different bacterial cells within a polymeric matrix produced by themselves, which are embedded in a surface, allowing them to survive in the surrounding environment (132). It has been found that App is a bacterium capable of forming biofilms in pigs in natural infection processes (133) and that it can form multispecies biofilms with other porcine respiratory pathogens under laboratory conditions (134).
Studies of molecular factors involved in forming biofilms have focused on lipopolysaccharides (LPS) and exopolysaccharides. Still, there are also important studies concerning the role played by proteins in the adhesion and formation of biofilms. This close relationship between these last two virulence factors could be vital to understanding the pathogenicity of App (133, 134).
Tegetmeyer et al. (2) found that the AasP autotransporter does not have adhesion properties. Still, they propose that it participates in the biosynthesis and degradation of poly-N-acetyl-glucosamine (PGA), which are the main and indispensable components for the formation of the biofilm matrix of App. Xie et al. used a VacJ lipoprotein mutant, resulting in poor in vitro biofilm formation at 36 h compared to the wild-type strain (120).
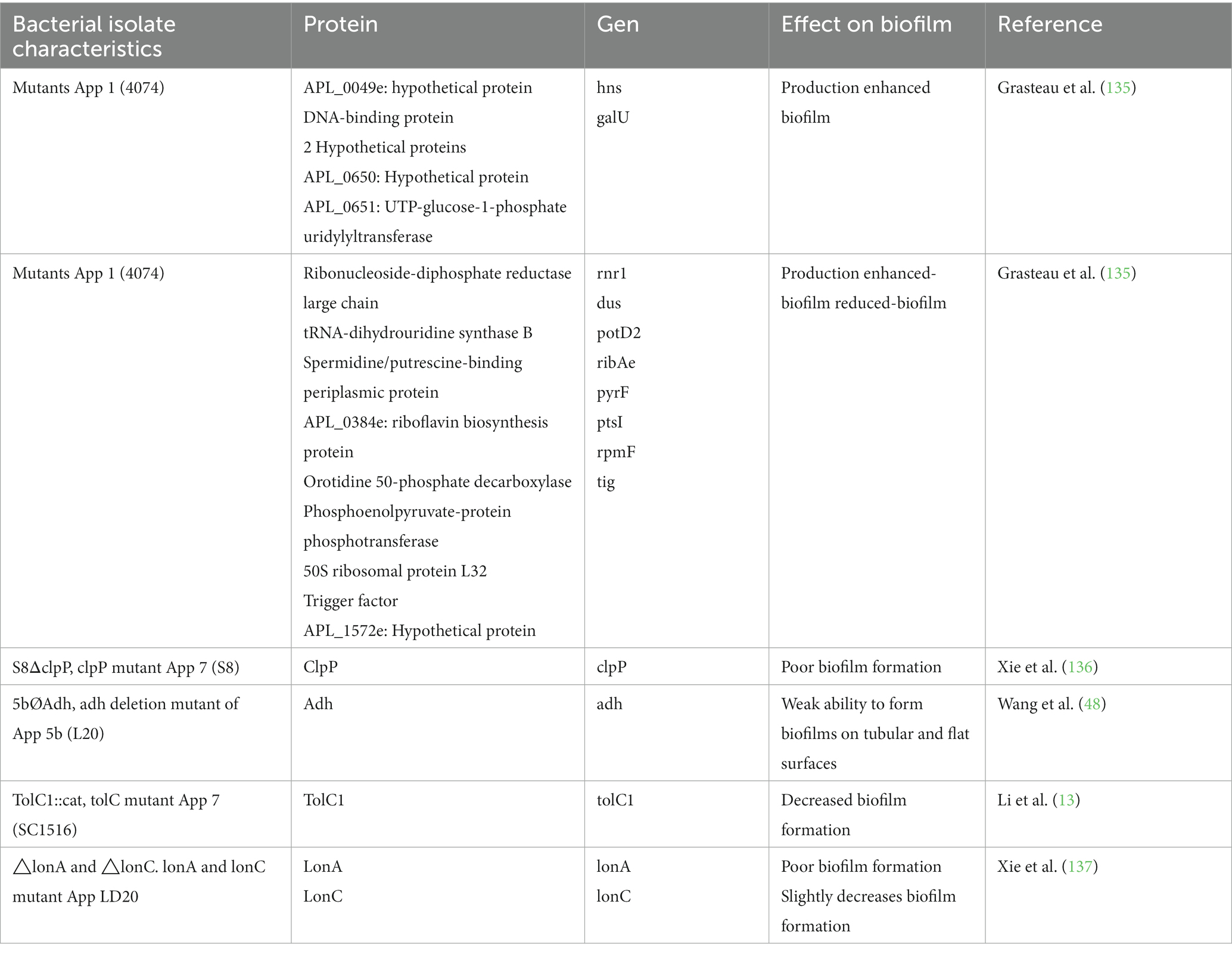
Assays with mutants and gene deletion have been used to evaluate the role of specific proteins in biofilm formation, the results of which have detected the participation of surface proteins (48, 135–138) (Table 2).
Although there is very little information about vaccines made with biofilm-related immunogens, some proposals suggest that they could effectively prevent and control App infections. Still, more studies are required to identify relevant molecules expressed in biofilms during infection (139).
Actinobacillus pleuropneumoniae proteome
To understand the molecular interactions a little better, it has been decided to carry out proteomic analyses of the cell surface. Proteomic studies, in addition to identifying many proteins, also reveal information about their internal and external structures, their composition, mechanisms of action including biofilm formation, antimicrobial resistance, and their way of adapting to the external environment (140).
Over the last decades, advances have been made that allow a better understanding of the pathobiology of the infection, and genetic tools have also allowed a better understanding of the molecular mechanisms of App. However, despite this, many of the molecular functions of this bacterium are still unknown (27).
Regarding the identification of OMPS of the bacterium App. Chung et al. (141) identified 47 OMPS in serotype 5b, representing 50% of the proteome and finding that the proteome is mostly hydrophilic. Liao et al. (30) identified 30 proteins for serotype 3, where 6 antigens were already known (MomP1, MomP2, ApxIIA, ApxIIIA, NqrA, and FhuA), and 24 more were identified for the first time. Subsequently, Zhang et al. (108) conducted a genomic sequencing analysis for serotype 1, where they identified 32 total proteins (two of them being outer membrane protein protective surface antigen D15 and outer membrane proteins P5). Chung et al. (109) also developed a work to identify new App antigens through the outer membrane proteome and under iron restriction conditions; in this work, they were able to identify the proteins TbpA, TbpB, PotD2, HbpA, CpxD, and OmlA.
Zhu et al. (91) carried out a study of the OMVs proteome and determined the content within them where they found 82 proteins whose origin is mainly from the outer membrane, periplasmatic proteins, and extracellular toxins (MomP2, OmlA, FkpA, OmpP1, LpoA, and ApxIII extracellular toxin), they also found other proteins that are involved with the absorption and restriction of iron (HbpA1, TbpB1, FrpB, HbpA2, YfeA, AfuA2, FhuA, TbpA) and a periplasmic protein of Fe type ABC3+. It has been shown that some proteins, such as PalA and VacJ, increase the secretion of OMVs (96–98).
Many proteins can also be secreted into the external environment of the bacterium. Stancheva et al. (142) identified 593 exoproteins, of which 104, through a prediction study, were proposed to be important virulence factors. Among the most found proteins were the Apx toxins.
The large number of proteins currently identified (Figure 2) opens a window of opportunity to increase knowledge about the role they play individually, how they could be related to each other at a molecular level, and how they participate in virulence factors, being able to propose them as possible immunogens for the elaboration of future vaccines.
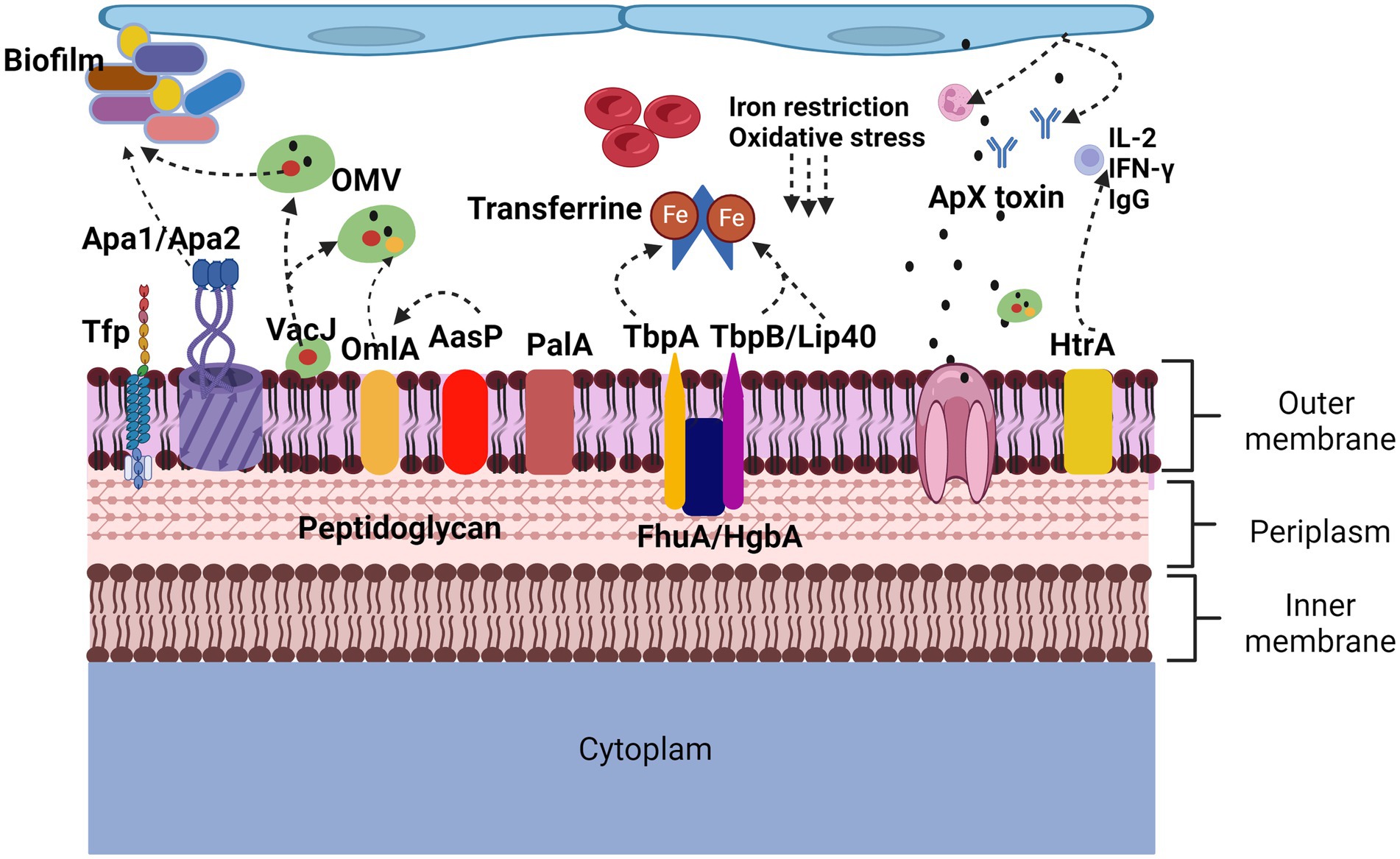
Figure 2. Proteins involved in virulence factors of Actinobacillus pleuropneumoniae. App adherence is regulated by lipopolysaccharides but also by proteins such as ApfA adhesins that allow adherence to collagen and fibrinogen, Apa1 and Apa2 that, like type IV Tfp pili, allow biofilm formation. In the induction of lesions and evasion of defense mechanisms, the Apx toxins cause neutralization of antibodies, lysis of neutrophils and red blood cells, the HtrA protein induces the secretion of IgG, IL-2 and IFN-γ. The OmlA protein plays an important role in the cell integrity of bacteria, in addition to being found within the vesicles of the outer membrane, VacJ, in addition to being found within the vesicles of the outer membrane, increases their secretion, it also participates in the formation of biofilms and plays an important role. Important role in serum resistance. PalA binds peptidoglycan to the outer membrane. AasP modifies the secretion of OmlA. The TbpA, TbpB, Lip40 and FhuA proteins are related to the iron acquisition virulence factor, are upregulated under anaerobic conditions and oxidative stress.
Potential vaccines
A wide variety of vaccines have contributed to reducing mortality and morbidity caused by App in pig farms. In the review work carried out by Loera et al. (95) we found that there is an excellent diversity of live attenuated and inactivated vaccines made with Apx toxins, membrane proteins, and combinations of these, DNA-based vaccines, which provide a protective effect in pigs and mice, but the search for new alternatives continues.
Highlighting the importance that proteins have acquired as potential immunogens for the preparation of future vaccines, the existence of moonlighting proteins has also been found. These proteins are expressed by the same gene and perform two or more functions in an organism in addition to having molecular links in different biological processes, whose role in bacteria is to participate as chaperones in response to stress and their ability to participate in pathogenesis as a factor of virulence once it is secreted into the external environment (143, 144).
The participation of these proteins in virulence factors focuses on adhesion to epithelial cells, especially to mucus and components of the ECM. One of the moonlighting proteins that have been found most frequently is the GAPDH protein; in Streptococcus pyogenes, it participates in the binding to cellular components such as plasmin, actin, myosin, and fibronectin; in Streptococcus pneumoniae, it also engages in the imperative to plasmin, in Streptococcus agalactiae functions as a stimulator of B lymphocyte activity, in Bacillus anthracis it functions as a plasminogen-binding protein. It may have NAD-ribosylating activity in enteropathogenic E. coli and enterohemorrhagic E. coli. Other moonlight proteins found in bacteria that also have similar activities are Enolase, 17 glycolytic enzymes, FbaA, Pgk, Eno, EF-Tu, PdhB, malate synthase, SodA, DnaK, GroEL, Tig, TP1 and GS, where their secretion and localization are regulated by specific stimuli (144).
In a study by Stancheva et al. (142) in a proteomic analysis in App serotype 2, moonlighting proteins were found, highlighting EF-Tu as a potential immunogen for new therapies and vaccines. However, there are still no studies that evaluate its effectiveness. These proteins have been considered as candidates for vaccines against fungi, such as the Fba1, Pgk, and Pk proteins of Candida albicans and Candida glabrata, conferring protection in a murine model, although in vivo studies are still lacking (145). A prototype vaccine with the moonlighting protein Fba (a protein with a central position in the glycolytic pathway) has resulted in a higher survival status in fish and a decrease in bacterial concentration in the spleen of the bacterium Photobacterium damselae subsp. piscicide, considered one of the most dangerous pathogens in aquaculture (146).
In general, many membrane proteins, pili, OMPs, and OMVs, among others, have been proposed as potential candidates for the preparation of future vaccines. Their participation in virulence factors has been studied. However, there is still a need to design and apply vaccines in vivo models to determine their effectiveness against infections against App.
Isolation and identification of proteins
An essential step in carrying out studies on the pathogenesis of infectious agents is to develop protocols that allow the isolation and identification of immunogens, where molecular factors, specifically surface proteins, play an important role.
As previously mentioned, the bacterial membrane has many components, so it is sometimes easy for samples to be contaminated with cellular by-products during protein extraction or for proteins to be easily denatured during the experiment (147). Therefore, it is important to be clear about the target proteins to be isolated to handle the samples properly.
Cell lysing methods used for subsequent protein identification include physical processes, such as heat treatment, sonication, osmotic shock, and chemical methods, including treatment with a detergent solution (EDTA), polymyxin, and extraction with chloroform. These techniques allow the recovery of up to 90% of specific proteins (147, 148).
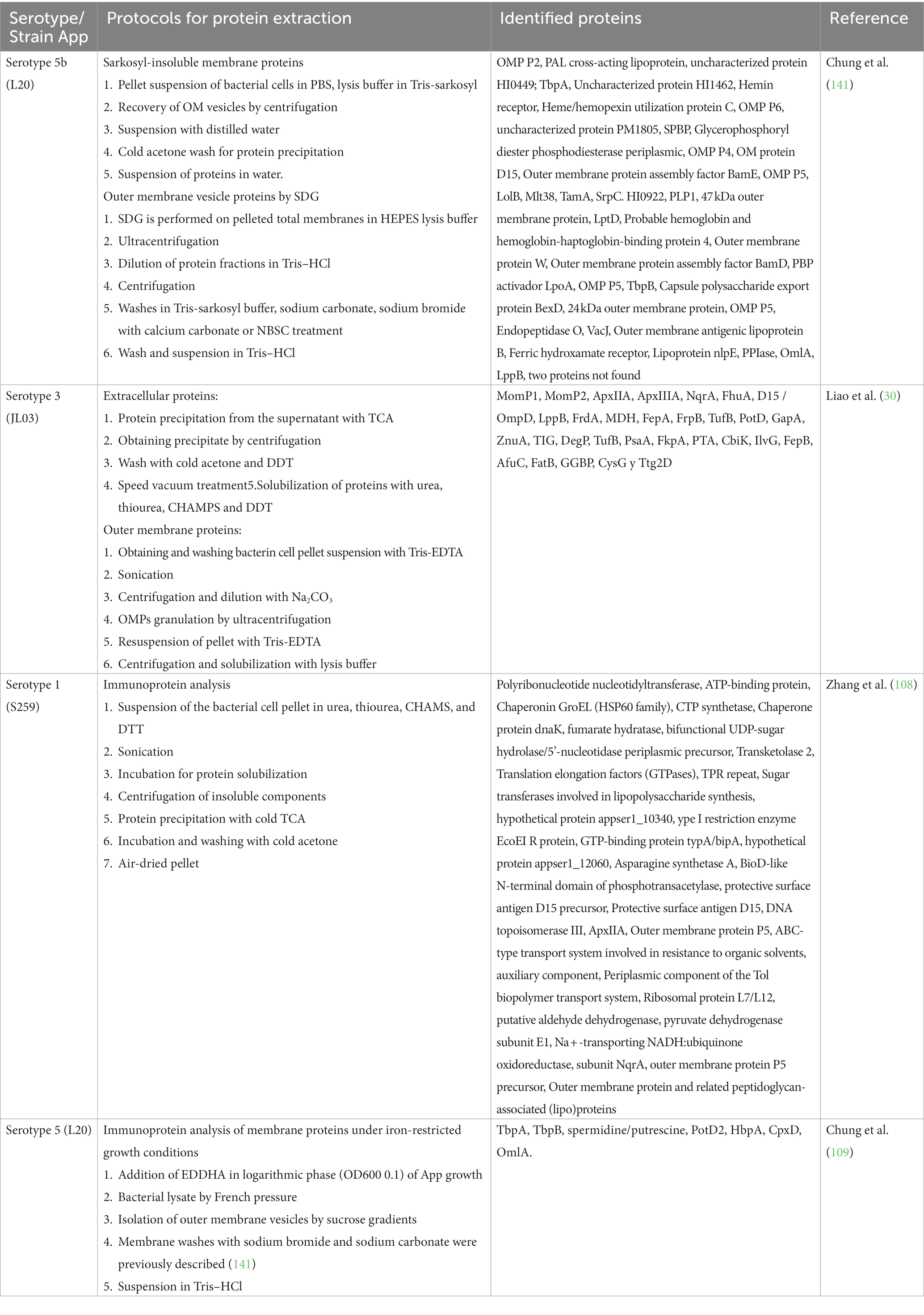
Most of the protein extractions in the App use sodium lauroyl sarcosinate (Sarkosyl). This detergent has been shown to have a milder effect than sodium dodecyl sulfate (SDS) and can identify proteins with resistance to SDS (149). Trichloroacetic acid (TCA) can be used to search for target proteins through isoelectric precipitation and solvents such as methanol and acetone to promote the precipitation of said proteins (147). TCA is mainly used in proteomic studies to search for isoelectric points in 2D gels where proteins must be free of contaminating residues (150). These last two techniques used in the reviewed studies identify many proteins found on the surface of the App bacterium (21, 28–30) (Table 3).
Since proteins cannot be chemically synthesized, it was decided to elaborate them by biological processes with other bacteria (151), by obtaining recombinant proteins, or by mutants where the deletion or insertion of protein genes to be studied is carried out. These techniques have also been widely used, allowing the screening of specific proteins and the antigenic enrichment of the outer membrane of App (141, 152) as well as the isolation of OMVs (98) (Table 4).
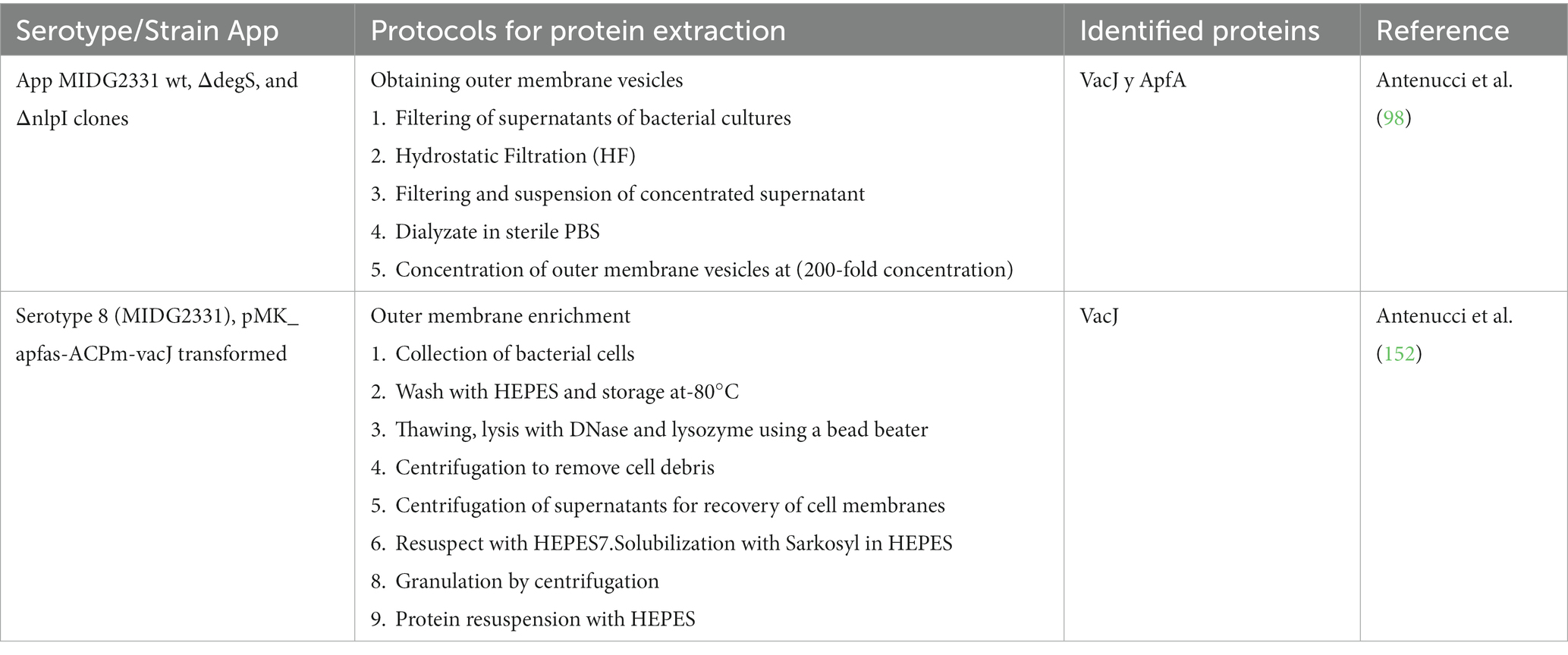
Table 4. Outer membrane enrichment and obtaining outer membrane vesicles for the identification of Actinobacillus pleuropneumoniae proteins.
To carry out said protein isolations, knowing under what growth conditions they are expressed is essential. It has been shown that some proteins are expressed in a greater or lesser proportion depending on the bacterial growth conditions in which they are immersed. In App, a difference in protein expression was found under normal growth conditions (153) compared to proteins obtained under nutrient addition conditions such as maltose (57), anaerobiosis, and different temperatures (154), as well as the restriction of iron (64) (Table 5). Protein expression under these growth conditions has also been tested in other bacteria, such as Salmonella typhimurium (155), E. coli (156), and Pasteurella haemolytica (157). Knowing how this bacterium behaves molecularly under stress conditions can help to find possible routes to improve management and treatments in App.
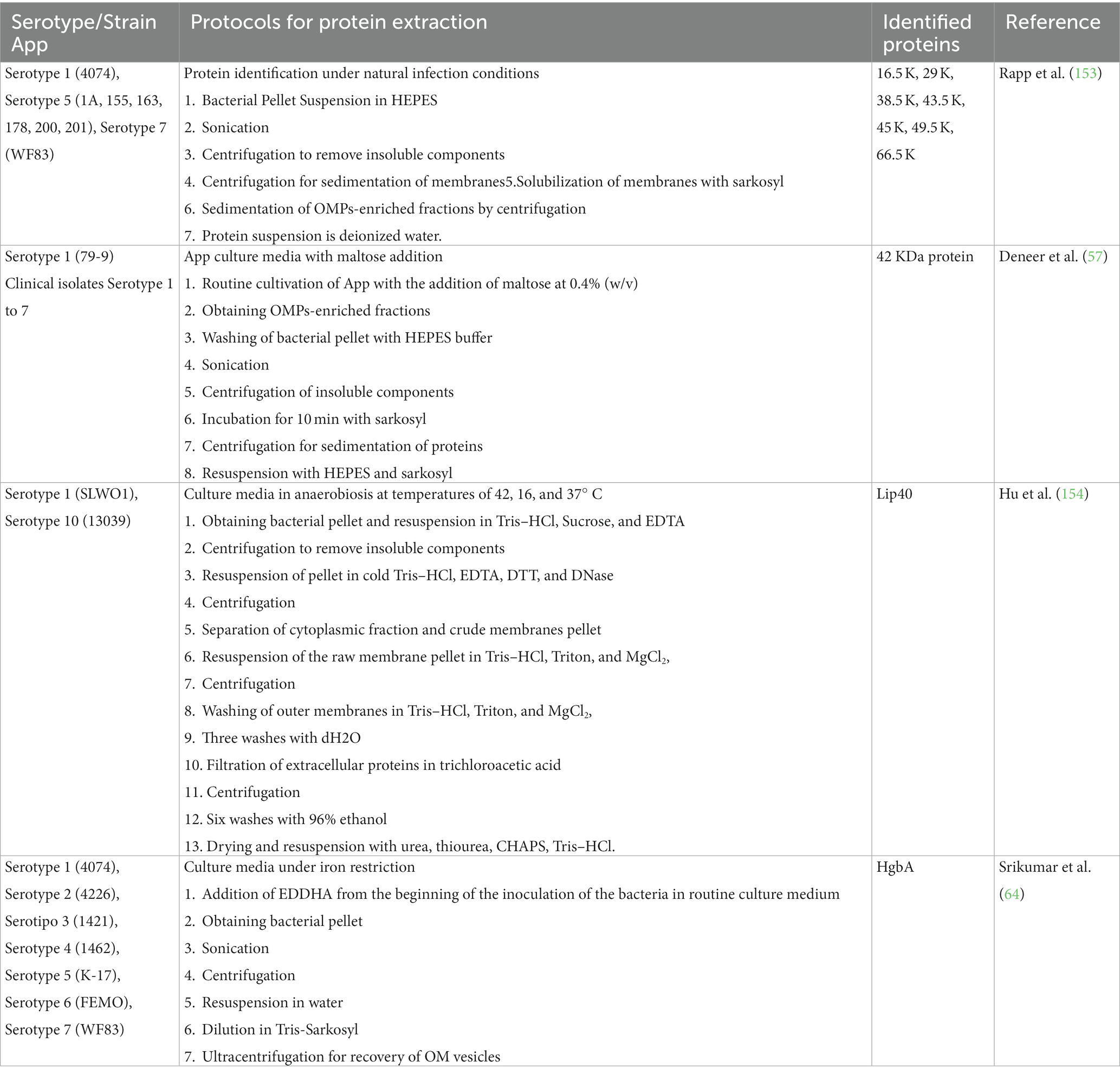
Table 5. Obtaining Actinobacillus pleuropneumoniae proteins under different bacterial growth conditions.
Conclusion
There is a great diversity of proteins that are not only secreted by the App, but also form part of the bacterial surface and actively participate during infection. The Tfp, ApfA, Adh, and Apa2 proteins participate in adhesion. A 42 kDa protein is actively expressed in the presence of maltose. FhuA, FhuB, FhuC, FhuD, and HgbA are proteins related to the iron acquisition factor. LamB is the porin-associated protein involved in drug resistance. The ApxI, ApxII, ApxIII, and ApxIV toxins are the proteins most related to the production of lesions and stimulation of the immune system. However, other proteins, such as VacJ and HtrA, have also been found to cause lesions. Proteins such as OmlA, PalA, and PotD participate in the maintenance of cell integrity. The AasP protein, in addition to participating in the modification of OmlA, is also related to the biosynthesis of other adhesion factors in the formation of biofilms; in the same way, VacJ engages in the maintenance of cell integrity, these last two proteins are related in more of a virulence factor. Many proteins have only been identified, but their role in the infection caused by App has not been established. It is essential to continue the search for new immunogens that allow the establishment of an effective vaccine against all App serotypes. Using computer tools that contain App proteomics information has permitted the development of reverse or in silico vaccinology techniques, finding new immunogens focused on surface proteins. In this study, we found that combining different surface proteins could be the key to developing an effective vaccine against App, although in vitro and in vivo studies are necessary to establish the efficacy of these vaccines.
Author contributions
MMSP: Investigation, Methodology, Writing – original draft. ALGB: Conceptualization, Formal analysis, Funding acquisition, Resources, Writing – review & editing, Project administration. FJAG: Writing – review & editing. TQT: Writing – review & editing. OMM: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financed through the support of the CONACyT scholarship (615420), through the support of PIB19-3 Host-pathogen relationship between bacteria and eukaryotic cells, and Resources of the Doctorate in Biological Sciences of the Autonomous University of Aguascalientes, Mexico.
Acknowledgments
We thank for the technical assistance provided by Flor Yazmín Ramirez-Castillo, Juan Manuel Díaz, Adriana Moreno-Flores, and Fabiola Galindo-Guerrero, as well as the Universidad Autonoma de Aguascalientes for the support to this paperwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sassu, EL, Bossé, JT, Tobias, TJ, Gottschalk, M, Langford, PR, and Hennig-Pauka, I. Update on Actinobacillus pleuropneumoniae-knowledge, gaps and challenges. Transbound Emerg Dis. (2018) 65:72–90. doi: 10.1111/tbed.12739
2. Tegetmeyer, HE, Fricke, K, and Baltes, N. An isogenic Actinobacillus pleuropneumoniae AasP mutant exhibits altered biofilm formation but retains virulence. Vet Microbiol. (2009) 137:392–6. doi: 10.1016/j.vetmic.2009.01.026
3. Chiers, K, De Waele, T, Pasmans, F, Ducatelle, R, and Haesebrouck, F. Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet Res. (2010) 41:65. doi: 10.1051/vetres/2010037
4. Stygar, AH, Niemi, JK, Oliviero, C, Laurila, T, and Heinonen, M. Economic value of mitigating Actinobacillus pleuropneumoniae infections in pig fattening herds. Agric Syst. (2016) 144:113–21. doi: 10.1016/j.agsy.2016.02.005
5. Stringer, OW, Bossé, JT, Lacouture, S, Gottschalk, M, Fodor, L, Angen, Ø, et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet Microbiol. (2021) 255:109021. doi: 10.1016/j.vetmic.2021.109021
6. Niven, DF, and Lévesque, M. V-factor-dependent growth of Actinobacillus pleuropneumoniae biotype 2 (Bertschinger 2008/76). Int J Syst Evol Microbiol. (1988) 38:319–20. doi: 10.1099/00207713-38-3-319
7. Lee, KE, Choi, HW, Kim, HH, Song, JY, and Yang, DK. Prevalence and characterization of Actinobacillus pleuropneumoniae isolated from Korean pigs. jbv. (2015) 45:19–25. doi: 10.4167/jbv.2015.45.1.19
8. Xu, Z, Zhou, Y, Li, L, Zhou, R, Xiao, S, Wan, Y, et al. Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serotype 3 prevalent in China. PLoS One. (2008) 3:e1450. doi: 10.1371/journal.pone.0001450
9. Ozawa, M, Kawano, M, Abo, H, Issiki, Y, Kumakawa, M, Kawanishi, M, et al. Characterization of Actinobacillus pleuropneumoniae isolated from pigs in Japan using whole genome sequencing. Comp Immunol Microbiol Infect Dis. (2023) 102:102062. doi: 10.1016/j.cimid.2023.102062
10. Dom, P, Hommez, J, Castryck, F, Devriese, LA, and Haesebrouck, F. Serotyping and quantitative determination of in vitro antibiotic susceptibility of Actinobacillus pleuropneumoniae strains isolated in Belgium (July 1991–August 1992). Vet Quart. (1994) 16:10–3. doi: 10.1080/01652176.1994.9694407
11. Sárközi, R, Makrai, L, and Fodor, L. Actinobacillus pleuropneumoniae serotypes in Hungary. Acta Vet Hung. (2018) 66:343–9. doi: 10.1556/004.2018.031
12. Turni, C, Singh, R, Schembri, MA, and Blackall, PJ. Evaluation of a multiplex PCR to identify and serotype Actinobacillus pleuropneumoniae serovars 1, 5, 7, 12 and 15. Lett Appl Microbiol. (2014) 59:362–9. doi: 10.1111/lam.12287
13. Li, Y, Bossé, JT, Williamson, SM, Maskell, DJ, Tucker, AW, Wren, BW, et al. Actinobacillus pleuropneumoniae serovar 8 predominates in England and Wales. Vet Rec. (2016) 179:276. doi: 10.1136/vr.103820
14. Jessing, SG, Angen, Ø, and Inzana, TJ. Evaluation of a multiplex PCR test for simultaneous identification and serotyping of Actinobacillus pleuropneumoniae serotypes 2, 5, and 6. J Clin Microbiol. (2003) 41:4095–100. doi: 10.1128/JCM.41.9.4095-4100.2003
15. Gottschalk, M, and Lacouture, S. Canada: distribution of Streptococcus suis (from 2012 to 2014) and Actinobacillus pleuropneumoniae (from 2011 to 2014) serotypes isolated from diseased pigs. Can Vet J. (2015) 56:1093–4.
16. Ontiveros-Corpus, L, Camacho-Machin, J, and Alvarez de la Cuadra-J, J. Correlación entre la serotipifiación de Actinobacillus pleuropneumoniae y su aislamiento en cerdos de rastro. Tee Peeu Mex. (1995) 33:1–7.
17. Serrano-Rubio, LE, Tenorio-Gutiérrez, V, Suárez-Güemes, F, Reyes-Cortés, R, Rodríguez-Mendiola, M, Arias-Castro, C, et al. Identification of Actinobacillus pleuropneumoniae biovars 1 and 2 in pigs using a PCR assay. Mol Cell Probes. (2008) 22:305–12. doi: 10.1016/j.mcp.2008.09.001
18. Lacouture, S, and Gottschalk, M. Distribution of Actinobacillus pleuropneumoniae (from 2015 to June 2020) and Glaesserella parasuis (from 2017 to June 2020) serotypes isolated from diseased pigs in Quebec. Can Vet J. (2020) 61:1261–3.
19. Williams, JJ, Torres-León, MA, Echeverria-Coello, P, and Matos-Medina, MC. Aislamiento e identificación de Actinobacillus pleuropneumoniae en pulmones de cerdos con pleuroneumonía crónica sacrificados en el rastro municipal de Mérida, Yucatán. México Rev Biomed. (2002) 11:175–81. doi: 10.32776/revbiomed.v11i3.234
20. Loera-Muro, VM, Loera-Muro, A, Morfín-Mata, M, Jacques, M, Avelar-González, FJ, Ramírez-Castillo, F, et al. Porcine respiratory pathogens in swine farms environment in Mexico. Open J Anim Sci. (2014) 4:196–205. doi: 10.4236/ojas.2014.44025
21. Assavacheep, P, and Rycroft, AN. Survival of Actinobacillus pleuropneumoniae outside the pig. Res Vet Sci. (2013) 94:22–6. doi: 10.1016/j.rvsc.2012.07.024
22. Hoeltig, D, Rohde, J, Frase, R, Nietfeld, F, Waldmann, KH, Valentin-Weigand, P, et al. Multi-organ spreading of Actinobacillus pleuropneumoniae serovar 7 in weaned pigs during the first week after experimental infection. Vet Res. (2018) 49:97. doi: 10.1186/s13567-018-0592-0
23. Bossé, JT, Janson, H, Sheehan, BJ, Beddek, AJ, Rycroft, AN, Simon Kroll, J, et al. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. (2002) 4:225–35. doi: 10.1016/S1286-4579(01)01534-9
24. Rollauer, SE, Sooreshjani, MA, Noinaj, N, and Buchanan, SK. Outer membrane protein biogenesis in gram-negative bacteria. Philos Trans Royal Soc B Biol Sci. (2015) 370:20150023. doi: 10.1098/rstb.2015.0023
25. Sheehan, BJ, Bossé, JT, Beddek, AJ, Rycroft, AN, Kroll, JS, and Langford, PR. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect Immun. (2003) 71:3960–70. doi: 10.1128/IAI.71.7.3960-3970.2003
26. Žutić, M, Ašanin, R, Milić, N, Ivetić, V, Vidić, B, Žutić, J, et al. Isolation and identification of Actinobacillus pleuropneumoniae in pig’s lungs at farms and their sensitivity. Acta Vet Brno. (2008) 58:499–507. doi: 10.2298/avb0806499z
27. Giordano, NP, Cian, MB, and Dalebroux, ZD. Outer membrane lipid secretion and the innate immune response to gram-negative Bacteria. Infect Immun. (2020) 88:e00920–19. doi: 10.1128/IAI.00920-19
28. Koebnik, R, Locher, KP, and Van Gelder, P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol Microbiol. (2000) 37:239–53. doi: 10.1046/j.1365-2958.2000.01983.x
29. Dalbey, RE, and Kuhn, A. Protein traffic in gram-negative bacteria – how exported and secreted proteins find their way. FEMS Microbiol Rev. (2012) 36:1023–45. doi: 10.1111/j.1574-6976.2012.00327.x
30. Liao, Y, Deng, J, Zhang, A, Zhou, M, Hu, Y, Chen, H, et al. Immunoproteomic analysis of outer membrane proteins and extracellular proteins of Actinobacillus pleuropneumoniae JL03 serotype 3. BMC Microbiol. (2009) 9:172. doi: 10.1186/1471-2180-9-172
31. Kudva, R, Denks, K, Kuhn, P, Vogt, A, Müller, M, and Koch, HG. Protein translocation across the inner membrane of gram-negative bacteria: the sec and tat dependent protein transport pathways. Res Microbiol. (2013) 164:505–34. doi: 10.1016/j.resmic.2013.03.016
32. Green, ER, and Joan, M. Bacterial secretion systems: an overview. Microbiol Spectr. (2016) 4:12. doi: 10.1128/microbiolspec.vmbf-0012-2015
33. Schwechheimer, C, and Kuehn, MJ. Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. (2015) 13:605–19. doi: 10.1038/nrmicro3525
34. Kim, JY, Suh, JW, Kang, JS, Kim, SB, Yoon, YK, and Sohn, JW. Gram-negative Bacteria’s Outer Membrane Vesicles. Infect Chemother. (2023) 55:145. doi: 10.3947/ic.2022.0145
35. Bélanger, M, Dubreuil, D, Harel, J, Girard, C, and Jacques, M. Role of lipopolysaccharides in adherence of Actinobacillus pleuropneumoniae to porcine tracheal rings. Infect Immun. (1990) 58:3523–30. doi: 10.1128/iai.58.11.3523-3530.1990
36. Bavington, C, and Page, C. Stopping bacterial adhesion: a novel approach to treating infections. Theatr Res Int. (2005) 72:335–44. doi: 10.1159/000086243
37. Berne, C, Ducret, A, Hardy, GG, and Brun, YV. Adhesins involved in attachment to abiotic surfaces by gram-negative Bacteria. In: Microbial Biofilms. (2015) 3:163–99. doi: 10.1128/9781555817466.ch9
38. Pizarro-Cerdá, J, and Cossart, P. Bacterial adhesion and entry into host cells. Cell. (2006) 124:715–27. doi: 10.1016/j.cell.2006.02.012
39. Lukaszczyk, M, Pradhan, B, and Remaut, H. The biosynthesis and structures of bacterial pili In: A Kuhn, editor. Bacterial cell walls and membranes. Cham: Springer International Publishing (2019). 369–413.
40. Epler Barbercheck, CR, Bullitt, E, and Andersson, M. Bacterial adhesion pili In: JR Harris and EJ Boekema, editors. Membrane protein complexes: Structure and function. Singapore: Springer Singapore (2018). 1–18.
41. Liu, F, Peng, W, Liu, T, Zhao, H, Yan, K, Yuan, F, et al. Biological role of Actinobacillus pleuropneumoniae type IV pilus proteins encoded by the apf and pil operons. Vet Microbiol. (2018) 224:17–22. doi: 10.1016/j.vetmic.2018.08.006
42. Li, T, Zhang, Q, Wang, R, Zhang, S, Pei, J, Li, Y, et al. The roles of flp1 and tadD in Actinobacillus pleuropneumoniae pilus biosynthesis and pathogenicity. Microb Pathog. (2019) 126:310–7. doi: 10.1016/j.micpath.2018.11.010
43. Boekema, BKHL, Van Putten, JPM, Norbert, SZ, and Smith, HE. Host cell contact-induced transcription of the type IV Fimbria gene cluster of Actinobacillus pleuropneumoniae. Infect Immun. (2004) 72:691–700. doi: 10.1128/IAI.72.2.691-700.2004
44. Yang, Z, Lu, L, Zhaohui, C, Hong, Y, Huanchun, C, and Rui, Z. Adhesion protein ApfA of Actinobacillus pleuropneumoniae is required for pathogenesis and is a potential target for vaccine development. Clin Vaccine Immunol. (2013) 20:287–94. doi: 10.1128/CVI.00616-12
45. van Overbeke, I, Chiers, K, Charlier, G, Vandenberghe, I, van Beeumen, J, Ducatelle, R, et al. Characterization of the in vitro adhesion of Actinobacillus pleuropneumoniae to swine alveolar epithelial cells. Vet Microbiol. (2002) 88:59–74. doi: 10.1016/S0378-1135(02)00080-9
46. Enríquez-Verdugo, I, Guerrero, AL, Serrano, JJ, Godínez, D, Rosales, JL, Tenorio, V, et al. Adherence of Actinobacillus pleuropneumoniae to swine-lung collagen. Microbiology (Reading). (2004) 150:2391–400. doi: 10.1099/mic.0.27053-0
47. Łyskowski, A, Leo, JC, and Goldman, A. Structure and biology of trimeric autotransporter Adhesins In: D Linke and A Goldman, editors. Bacterial adhesion: Chemistry, biology and physics. Dordrecht: Springer Netherlands (2011). 143–58.
48. Wang, L, Qin, W, Yang, S, Zhai, R, Zhou, L, Sun, C, et al. The Adh adhesin domain is required for trimeric autotransporter Apa1-mediated Actinobacillus pleuropneumoniae adhesion, autoaggregation, biofilm formation and pathogenicity. Vet Microbiol. (2015) 177:175–83. doi: 10.1016/j.vetmic.2015.02.026
49. Qin, W, Wang, L, Zhai, R, Ma, Q, Liu, J, Bao, C, et al. Apa2H1, the first head domain of Apa2 trimeric autotransporter adhesin, activates mouse bone marrow-derived dendritic cells and immunization with Apa2H1 protects against Actinobacillus pleuropneumoniae infection. Mol Immunol. (2017) 81:108–17. doi: 10.1016/j.molimm.2016.12.004
50. Lu, YC, Li, MC, Chen, YM, Chu, CY, Lin, SF, and Yang, WJ. DNA vaccine encoding type IV pilin of Actinobacillus pleuropneumoniae induces strong immune response but confers limited protective efficacy against serotype 2 challenge. Vaccine. (2011) 29:7740–6. doi: 10.1016/j.vaccine.2011.07.127
51. Lee, SH, Lee, S, Chae, C, and Ryu, DY. A recombinant chimera comprising the R1 and R2 repeat regions of M. hyopneumoniae P97 and the N-terminal region of A. pleuropneumoniae ApxIII elicits immune responses. BMC Vet Res. (2014) 10:43. doi: 10.1186/1746-6148-10-43
52. Raynes, JM, Young, PG, Proft, T, Williamson, DA, Baker, EN, and Moreland, NJ. Protein adhesins as vaccine antigens for group a Streptococcus. Pathog Dis. (2018) 76:fty016. doi: 10.1093/femspd/fty016
53. Maresso, AW. The acquisition and consumption of host nutrients In: AW Maresso, editor. Bacterial virulence: A conceptual primer. Cham: Springer International Publishing (2019). 131–44.
54. Weatherill, EE, Fahie, MA, Marshall, DP, Andvig, RA, Cheetham, MR, Chen, M, et al. Fast slow folding of an outer membrane porin. Proc Natl Acad Sci. (2022) 119:e2121487119. doi: 10.1073/pnas.2121487119
55. Achouak, W, Heulin, T, and Pagès, JM. Multiple facets of bacterial porins. FEMS Microbiol Lett. (2001) 199:1–7. doi: 10.1111/j.1574-6968.2001.tb10642.x
56. Welte, W, Nestel, U, Wacker, T, and Diederichs, K. Structure and function of the porin channel. Kidney Int. (1995) 48:930–40. doi: 10.1038/ki.1995.374
57. Deneer, HG, and Potter, AA. Identification of a maltose-inducible major outer membrane protein in Actinobacillus (Haemophilus) pleuropneumoniae. Microb Pathog. (1989) 6:425–32. doi: 10.1016/0882-4010(89)90084-3
58. Ma, X, Zheng, B, Wang, J, Li, G, Cao, S, Wen, Y, et al. Quinolone resistance of Actinobacillus pleuropneumoniae revealed through genome and transcriptome analyses. Int J Mol Sci. (2021) 22:3535. doi: 10.3390/ijms222413535
59. Del Pozo, SR, Michiels, A, Martens, M, Haesebrouck, F, and Maes, D. Efficacy of vaccination against Actinobacillus pleuropneumoniae in two Belgian farrow-to-finish pig herds with a history of chronic pleurisy. Vet Rec. (2014) 174:302. doi: 10.1136/vr.101961
60. López-Siles, M, Corral-Lugo, A, and McConnell, MJ. Vaccines for multidrug resistant gram negative bacteria: lessons from the past for guiding future success. FEMS Microbiol Rev. (2021) 45:fuaa054. doi: 10.1093/femsre/fuaa054
61. Klebba, PE, Newton, SMC, Six, DA, Kumar, A, Yang, T, Nairn, BL, et al. Iron Acquisition Systems of Gram-negative Bacterial Pathogens Define ton B-dependent pathways to novel antibiotics. Chem Rev. (2021) 121:5193–239. doi: 10.1021/acs.chemrev.0c01005
62. Cornelissen, CN. Subversion of nutritional immunity by the pathogenic Neisseriae. Pathog Dis. (2018) 76:ftx112. doi: 10.1093/femspd/ftx112
63. Mikael, LG, Srikumar, R, Coulton, JW, and Jacques, M. fhuA of Actinobacillus pleuropneumoniae encodes a Ferrichrome receptor but is not regulated by Iron. Infect Immun. (2003) 71:2911–5. doi: 10.1128/IAI.71.5.2911-2915.2003
64. Srikumar, R, Mikael, LG, Pawelek, PD, Khamessan, A, Gibbs, BF, Jacques, M, et al. Molecular cloning of haemoglobin-binding protein HgbA in the outer membrane of Actinobacillus pleuropneumoniae. Microbiology (NY). (2004) 150:1723–34. doi: 10.1099/mic.0.27046-0
65. Shakarji, L, Mikael, LG, Srikumar, R, Kobisch, M, Coulton, JW, and Jacques, M. Fhua and HgbA, outer membrane proteins of Actinobacillus pleuropneumoniae: their role as virulence determinants. Can J Microbiol. (2006) 52:391–6. doi: 10.1139/w05-135
66. Mikael, LG, Pawelek, PD, Labrie, J, Sirois, M, Coulton, JW, and Jacques, M. Molecular cloning and characterization of the ferric hydroxamate uptake (fhu) operon in Actinobacillus pleuropneumoniae the GenBank accession number for the sequence of the fhuCDBA operon of Actinobacillus pleuropneumoniae serotype 1 reference strain 4074 described in this study is AF351135. Microbiology (NY). (2002) 148:2869–82. doi: 10.1099/00221287-148-9-2869
67. Alshammari, A, Alharbi, M, Alghamdi, A, Alharbi, SA, Ashfaq, UA, Ul Qamar, M, et al. Computer-aided multi-epitope vaccine design against Enterobacter xiangfangensis. Int J Environ Res Public Health. (2022) 19:7723. doi: 10.3390/ijerph19137723
68. Braun, V, Killmann, H, and Benz, R. Energy-coupled transport through the outer membrane of Escherichia coli small deletions in the gating loop convert the Fhu a transport protein into a diffusion channel. FEBS Lett. (1994) 346:59–64. doi: 10.1016/0014-5793(94)00431-5
69. Bonhivers, M, Plançon, L, Ghazi, A, Boulanger, A, le Maire, M, Lambert, O, et al. FhuA, an Escherichia coli outer membrane protein with a dual function of transporter and channel which mediates the transport of phage DNA. Biochimie. (1998) 80:363–9.
70. Yejun, W, Xiongbin, C, Yueming, H, Guoqiang, Z, White, AP, and Wolfgang, K. Evolution and sequence diversity of FhuA in Salmonella and Escherichia. Infect Immun. (2018) 86:e00573–18. doi: 10.1128/IAI.00573-18
71. Pogoutse, AK, and Moraes, TF. Transferrin binding protein B and transferrin binding protein A2 expand the transferrin recognition range of Histophilus somni. J Bacteriol. (2020) 202:e00177–20. doi: 10.1128/JB.00177-20
72. Baltes, N, Hennig-Pauka, I, and Gerlach, GF. Both transferrin binding proteins are virulence factors in Actinobacillus pleuropneumoniae serotype 7 infection. FEMS Microbiol Lett. (2002) 209:283–7. doi: 10.1111/j.1574-6968.2002.tb11145.x
73. Pintor, M, Gomez, JA, Ferron, L, Ferreiros, CM, and Criado, MT. Analysis of TbpA and TbpB functionality in defective mutants of Neisseria meningitidis. J Med Microbiol. (1998) 47:757–60. doi: 10.1099/00222615-47-9-757
74. Álvarez-Estrada, Á, Gutiérrez-Martín, CB, Rodríguez-Ferri, EF, and Martínez-Martínez, S. Transcriptomics of Haemophilus (Glässerella) parasuis serovar 5 subjected to culture conditions partially mimetic to natural infection for the search of new vaccine antigens. BMC Vet Res. (2018) 14:326. doi: 10.1186/s12917-018-1647-1
75. Elkins, C, Chen, CJ, and Thomas, CE. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect Immun. (1995) 63:2194–200. doi: 10.1128/iai.63.6.2194-2200.1995
76. Maas, A, Meens, J, Baltes, N, Hennig-Pauka, I, and Gerlach, GF. Development of a DIVA subunit vaccine against Actinobacillus pleuropneumoniae infection. Vaccine. (2006) 24:7226–37. doi: 10.1016/j.vaccine.2006.06.047
77. Fegan, JE, Calmettes, C, Islam, EA, Ahn, SK, Chaudhuri, S, Yu, R, et al. Utility of hybrid transferrin binding protein antigens for protection against pathogenic Neisseria species. Front Immunol. (2019) 10. doi: 10.3389/fimmu.2019.00247
78. Li, J, Qiu, J, Huang, Z, Liu, T, Pan, J, Zhang, Q, et al. Reverse vaccinology approach for the identifications of potential vaccine candidates against Salmonella. Int J Med Microbiol. (2021) 311:151508. doi: 10.1016/j.ijmm.2021.151508
79. Fernández, AG, Martín, CBG, Rilo, MP, Fernández, EP, Pérez, RM, Frandoloso, R, et al. Phylogenetic study and comparison of different TbpB obtained from Glaesserella parasuis present in Spanish clinical isolates. Res Vet Sci. (2023) 157:35–9. doi: 10.1016/j.rvsc.2023.02.003
80. Qamsari, MM, Rasooli, I, Chaudhuri, S, Astaneh, SDA, and Schryvers, AB. Hybrid antigens expressing surface loops of Znu D from Acinetobacter baumannii is capable of inducing protection against infection. Front Immunol. (2020):11. doi: 10.3389/fimmu.2020.00158
81. Akbari, Z, Rasooli, I, Ghaini, MH, Chaudhuri, S, Farshchi Andisi, V, Jahangiri, A, et al. Bau a and Omp34 surface loops trigger protective antibodies against Acinetobacter baumannii in a murine sepsis model. Int Immunopharmacol. (2022) 108:108731. doi: 10.1016/j.intimp.2022.108731
82. Ulhuq, FR, and Mariano, G. Bacterial pore-forming toxins. Microbiology (NY). (2022) 168:001154. doi: 10.1099/mic.0.001154
83. Bertram, TA. Actinobacillus pleuropneumoniae: molecular aspects of virulence and pulmonary injury. Can J Vet Res. (1990) 54:S53–6.
84. Frey, J, Bosse, JT, Chang, YF, Cullen, JM, Fenwick, B, Gerlach, GF, et al. Actinobacillus pleuropneumoniae RTX-toxins: uniform designation of haemolysins, cytolysins, pleurotoxin and their genes. J Gen Microbiol. (1993) 139:1723–8. doi: 10.1099/00221287-139-8-1723
85. Kamp, EM, and van Leengoed, LA. Serotype-related differences in production and type of heat-labile hemolysin and heat-labile cytotoxin of Actinobacillus (Haemophilus) pleuropneumoniae. J Clin Microbiol. (1989) 27:1187–91. doi: 10.1128/jcm.27.6.1187-1191.1989
86. Schaller, A, Djordjevic, SP, Eamens, GJ, Forbes, WA, Kuhn, R, Kuhnert, P, et al. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet Microbiol. (2001) 79:47–62. doi: 10.1016/S0378-1135(00)00345-X
87. Choi, C, Kwon, D, Min, K, and Chae, C. Detection and localization of ApxI, -II, and-III genes of Actinobacillus pleuropneumoniae in natural porcine pleuropneumonia by in situ hybridization. Vet Pathol. (2001) 38:390–5. doi: 10.1354/vp.38-4-390
88. Kamp, EM, Stockhofe-Zurwieden, N, van Leengoed, LA, Smits, MA, van Leengoed, LA, and Smits, MA. Endobronchial inoculation with Apx toxins of Actinobacillus pleuropneumoniae leads to pleuropneumonia in pigs. Infect Immun. (1997) 65:4350–4. doi: 10.1128/iai.65.10.4350-4354.1997
89. Kamp, EM, Popma, JK, Anakotta, J, and Smits, MA. Identification of hemolytic and cytotoxic proteins of Actinobacillus pleuropneumoniae by use of monoclonal antibodies. Infect Immun. (1991) 59:3079–85. doi: 10.1128/iai.59.9.3079-3085.1991
90. Haesebrouck, F, Chiers, K, Van Overbeke, I, and Ducatelle, R. Actinobacillus pleuropneumoniae infections in pigs: the role of virulence factors in pathogenesis and protection. Vet Microbiol. (1997) 58:239–49.
91. Zhu, Z, Antenucci, F, Winther-Larsen, HC, Skovgaard, K, and Bojesen, AM. Outer membrane vesicles of Actinobacillus pleuropneumoniae exert immunomodulatory effects on porcine alveolar macrophages. Microbiol Spectr. (2022) 10:e01819–22. doi: 10.1128/spectrum.01819-22
92. Hernandez-Cuellar, E, Guerrero-Barrera, AL, Avelar-Gonzalez, FJ, Díaz, JM, Chávez-Reyes, J, and Salazar de Santiago, A. An in vitro study of ApxI from Actinobacillus pleuropneumoniae serotype 10 and induction of NLRP3 inflammasome-dependent cell death. Vet Rec Open. (2021) 8:e20. doi: 10.1002/vro2.20
93. Tascón, RI, Vázquez-Boland, JA, Gutiérrez-Martín, CB, Rodríguez-Barbosa, I, and Rodríguez-Ferri, EF. The RTX haemolysins ApxI and ApxII are major virulence factors of the swine pathogen Actinobacillus pleuropneumoniae: evidence from mutational analysis. Mol Microbiol. (1994) 14:207–16. doi: 10.1111/j.1365-2958.1994.tb01282.x
94. Ristow, LC, and Welch, RA. RTX toxins ambush Immunity’s first cellular responders. Toxins (Basel). (2019) 11:720. doi: 10.3390/toxins11120720
95. Loera-Muro, A, and Angulo, C. New trends in innovative vaccine development against Actinobacillus pleuropneumoniae. Vet Microbiol. (2018) 217:66–75. doi: 10.1016/j.vetmic.2018.02.028
96. Suzuki, T, Murai, T, Fukuda, I, Tobe, T, Yoshikawa, M, and Sasakawa, C. Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol Microbiol. (1994) 11:31–41. doi: 10.1111/j.1365-2958.1994.tb00287.x
97. Roier, S, Zingl, FG, Cakar, F, Durakovic, S, Kohl, P, Eichmann, TO, et al. A novel mechanism for the biogenesis of outer membrane vesicles in gram-negative bacteria. Nat Commun. (2016) 7:10515. doi: 10.1038/ncomms10515
98. Antenucci, F, Fougeroux, C, Bossé, JT, Magnowska, Z, Roesch, C, Langford, P, et al. Identification and characterization of serovar-independent immunogens in Actinobacillus pleuropneumoniae. Vet Res. (2017) 48:74. doi: 10.1186/s13567-017-0479-5
99. Negrete-Abascal, E, García, RM, Reyes, ME, Godínez, D, and de la Garza, M. Membrane vesicles released by Actinobacillus pleuropneumoniae contain proteases and Apx toxins. FEMS Microbiol Lett. (2000) 191:109–13. doi: 10.1111/j.1574-6968.2000.tb09326.x
100. Backert, S, Bernegger, S, Skórko-Glonek, J, and Wessler, S. Extracellular HtrA serine proteases: an emerging new strategy in bacterial pathogenesis. Cell Microbiol. (2018) 20:e12845. doi: 10.1111/cmi.12845
101. Xu, K, Zhao, Q, Jiang, HZ, Mou, XR, Chang, YF, Cao, YQ, et al. Molecular and functional characterization of HtrA protein in Actinobacillus pleuropneumoniae. Vet Microbiol. (2021) 257:109058. doi: 10.1016/j.vetmic.2021.109080
102. Dorner, F, and McDonel, JL. Bacterial toxin vaccines. Vaccine. (1985) 3:94–102. doi: 10.1016/0264-410X(85)90056-8
103. Antenucci, F, Fougeroux, C, Deeney, A, Ørskov, C, Rycroft, A, Holst, PJ, et al. In vivo testing of novel vaccine prototypes against Actinobacillus pleuropneumoniae. Vet Res. (2018) 49:4. doi: 10.1186/s13567-017-0502-x
104. Hau, SJ, Luan, SL, Loving, CL, Nicholson, TL, Wang, J, Peters, SE, et al. Evaluation of the recombinant proteins RlpB and VacJ as a vaccine for protection against Glaesserella parasuis in pigs. BMC Vet Res. (2020) 16:167. doi: 10.1186/s12917-020-02377-5
105. Li, Y, Xiao, J, Chang, YF, Zhang, H, Teng, Y, Lin, W, et al. Immunogenicity and protective efficacy of the recombinant Pasteurella multocida lipoproteins VacJ and PlpE, and outer membrane protein H from P. multocida a: 1 in ducks. Front Immunol. (2022) 13:2573. doi: 10.3389/fimmu.2022.1072573
106. Zhang, L, Zhao, F, Xu, H, Chen, Y, Qi, C, and Liu, J. HtrA of Actinobacillus pleuropneumoniae is a virulence factor that confers resistance to heat shock and oxidative stress. Gene. (2022) 841:146771. doi: 10.1016/j.gene.2022.146771
107. Gerlach, GF, Anderson, C, Klashinsky, S, Rossi-Campos, A, Potter, AA, and Willson, PJ. Molecular characterization of a protective outer membrane lipoprotein (OmlA) from Actinobacillus pleuropneumoniae serotype 1. Infect Immun. (1993) 61:565–72. doi: 10.1128/iai.61.2.565-572.1993
108. Zhang, W, Shao, J, Liu, G, Tang, F, Lu, Y, Zhai, Z, et al. Immunoproteomic analysis of bacterial proteins of Actinobacillus pleuropneumoniae serotype 1. Proteome Sci. (2011) 9:32. doi: 10.1186/1477-5956-9-32
109. Chung, JW, Küster-Schöck, E, Gibbs, BF, Jacques, M, and Coulton, JW. Immunoproteomic analyses of outer membrane antigens of Actinobacillus pleuropneumoniae grown under iron-restricted conditions. Vet Microbiol. (2012) 159:187–94. doi: 10.1016/j.vetmic.2012.03.038
110. Rossi, CC, de Araújo, EF, de Queiroz, MV, and Bazzolli, DMS. Characterization of the omlA gene from different serotypes of Actinobacillus pleuropneumoniae: a new insight into an old approach. Genet Mol Biol. (2013) 36:243–51. doi: 10.1590/S1415-47572013005000012
111. Ochsner, UA, Vasil, AI, Johnson, Z, and Vasil, ML. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J Bacteriol. (1999) 181:1099–109. doi: 10.1128/JB.181.4.1099-1109.1999
112. Fuangthong, M, Sallabhan, R, Atichartpongkul, S, Rangkadilok, N, Sriprang, R, Satayavivad, J, et al. The omlA gene is involved in multidrug resistance and its expression is inhibited by coumarins in Xanthomonas campestris pv. Phaseoli. Arch Microbiol. (2008) 189:211–8. doi: 10.1007/s00203-007-0310-1
113. Baltes, N, Buettner, FFR, and Gerlach, GF. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Vet Microbiol. (2007) 123:110–21. doi: 10.1016/j.vetmic.2007.03.026
114. Ali, T, Oldfield, NJ, Wooldridge, KG, Turner, DP, and Ala’Aldeen, DAA. Functional characterization of AasP, a maturation protease autotransporter protein of Actinobacillus pleuropneumoniae. Infect Immun. (2008) 76:5608–14. doi: 10.1128/IAI.00085-08
115. Plasencia-Muñoz, B, Avelar-González, FJ, De la Garza, M, Jacques, M, Moreno-Flores, A, and Guerrero-Barrera, AL. Actinobacillus pleuropneumoniae interaction with swine endothelial cells. Front Vet Sci. (2020) 7:569370. doi: 10.3389/fvets.2020.569370
116. Frey, J, Kuhnert, P, Villiger, L, and Nicolet, J. Cloning and characterization of an Actinobacillus pleuropneumoniae outer membrane protein belonging to the family of PAL lipoproteins. Res Microbiol. (1996) 147:351–61.
117. Clavel, T, Germon, P, Vianney, A, Portalier, R, and Lazzaroni, JC. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins pal, Lpp and OmpA. Mol Microbiol. (1998) 29:359–67. doi: 10.1046/j.1365-2958.1998.00945.x
118. van den Bosch, H, and Frey, J. Interference of outer membrane protein PalA with protective immunity against Actinobacillus pleuropneumoniae infections in vaccinated pigs. Vaccine. (2003) 21:3601–7. doi: 10.1016/s0264-410x(03)00410-9
119. Zhu, Z, Zhao, Q, Zhao, Y, Zhang, F, Wen, X, Huang, X, et al. Polyamine-binding protein PotD2 is required for stress tolerance and virulence in Actinobacillus pleuropneumoniae. Antonie Van Leeuwenhoek. (2017) 110:1647–57. doi: 10.1007/s10482-017-0914-7
120. Xie, F, Li, G, Zhang, W, Zhang, Y, Zhou, L, Liu, S, et al. Outer membrane lipoprotein VacJ is required for the membrane integrity, serum resistance and biofilm formation of Actinobacillus pleuropneumoniae. Vet Microbiol. (2016) 183:1–8. doi: 10.1016/j.vetmic.2015.11.021
121. Buettner, FFR, Konze, SA, Maas, A, and Gerlach, GF. Proteomic and immunoproteomic characterization of a DIVA subunit vaccine against Actinobacillus pleuropneumoniae. Proteome Sci. (2011) 9:23. doi: 10.1186/1477-5956-9-23
122. Oldfield, NJ, Worrall, KE, Rycroft, AN, Ali, T, Wooldridge, KG, and Ala’Aldeen, DAA. AasP autotransporter protein of Actinobacillus pleuropneumoniae does not protect pigs against homologous challenge. Vaccine. (2009) 27:5278–83. doi: 10.1016/j.vaccine.2009.06.047
123. Henderson, IR, and Nataro, JP. Autotransporter Proteins. EcoSal Plus. (2005) 1. doi: 10.1128/ecosalplus.8.7.3
124. Ruiz-Perez, F, and Nataro, JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Molecular Life Sci. (2014) 71:745–70. doi: 10.1007/s00018-013-1355-8
125. Harris, JA, Roy, K, Woo-Rasberry, V, Hamilton, DJ, Kansal, R, Qadri, F, et al. Directed evaluation of Enterotoxigenic Escherichia coli autotransporter proteins as putative vaccine candidates. PLoS Negl Trop Dis. (2011) 5:e1428. doi: 10.1371/journal.pntd.0001428
126. Xing, Y, Clark, JR, Chang, JD, Chirman, DM, Green, S, Zulk, JJ, et al. Broad protective vaccination against systemic Escherichia coli with autotransporter antigens. PLoS Pathog. (2023) 19:e1011082. doi: 10.1371/journal.ppat.1011082
127. Habouria, H, Pokharel, P, Maris, S, Garénaux, A, Bessaiah, H, Houle, S, et al. Three new serine-protease autotransporters of Enterobacteriaceae (SPATEs) from extra-intestinal pathogenic Escherichia coli and combined role of SPATEs for cytotoxicity and colonization of the mouse kidney. Virulence. (2019) 10:568–87. doi: 10.1080/21505594.2019.1624102
128. Pokharel, P, Habouria, H, Bessaiah, H, and Dozois, CM. Serine protease autotransporters of the Enterobacteriaceae (SPATEs): out and about and chopping it up. Microorganisms. (2019) 7:594. doi: 10.3390/microorganisms7120594
129. Wells, TJ, Tree, JJ, Ulett, GC, and Schembri, MA. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol Lett. (2007) 274:163–72. doi: 10.1111/j.1574-6968.2007.00833.x
130. Liu, C, Cao, Y, Yang, J, Zhao, H, Zahid, KR, Zhao, J, et al. Be aware of immunogenic but not protective antigens: the Actinobacillus pleuropneumoniae PalA as an example. Protein Pept Lett. (2018) 24:1558. doi: 10.2174/0929866524666170822121558
131. Dai, K, Ma, X, Yang, Z, Chang, YF, Cao, S, Zhao, Q, et al. Polyamine transport protein PotD protects mice against Haemophilus parasuis and elevates the secretion of pro-inflammatory cytokines of macrophage via JNK–MAPK and NF–κB signal pathways through TLR4. Vaccines (Basel). (2019) 7:216. doi: 10.3390/vaccines7040216
132. Jacques, M, Aragon, V, and Tremblay, YDN. Biofilm formation in bacterial pathogens of veterinary importance. Anim Health Res Rev. (2010) 11:97–121. doi: 10.1017/S1466252310000149
133. Tremblay, YDN, Labrie, J, Chénier, S, and Jacques, M. Actinobacillus pleuropneumoniae grows as aggregates in the lung of pigs: is it time to refine our in vitro biofilm assays? Microb Biotechnol. (2017) 10:756–60. doi: 10.1111/1751-7915.12432
134. Loera-Muro, A, Ramírez-Castillo, FY, Moreno-Flores, AC, Martin, EM, Avelar-González, FJ, and Guerrero-Barrera, AL. Actinobacillus pleuropneumoniae surviving on environmental multi-species biofilms in swine farms. Front Vet Sci. (2021) 8:1104. doi: 10.3389/fvets.2021.722683
135. Grasteau, A, Tremblay, YDN, Labrie, J, and Jacques, M. Novel genes associated with biofilm formation of Actinobacillus pleuropneumoniae. Vet Microbiol. (2011) 153:134–43. doi: 10.1016/j.vetmic.2011.03.029
136. Xie, F, Zhang, Y, Li, G, Zhou, L, Liu, S, and Wang, C. The ClpP protease is required for the stress tolerance and biofilm formation in Actinobacillus pleuropneumoniae. PLoS One. (2013) 8:e53600. doi: 10.1371/journal.pone.0053600
137. Xie, F, Li, G, Zhang, Y, Zhou, L, Liu, S, Liu, S, et al. The Lon protease homologue LonA, not LonC, contributes to the stress tolerance and biofilm formation of Actinobacillus pleuropneumoniae. Microb Pathog. (2016) 93:38–43. doi: 10.1016/j.micpath.2016.01.009
138. Li, Y, Cao, S, Zhang, L, Lau, GW, Wen, Y, Wu, R, et al. A TolC-like protein of Actinobacillus pleuropneumoniae is involved in antibiotic resistance and biofilm formation. Front Microbiol. (2016):7. doi: 10.3389/fmicb.2016.01618
139. Hathroubi, S, Loera-Muro, A, Guerrero-Barrera, AL, Tremblay, YDN, and Jacques, M. Actinobacillus pleuropneumoniae biofilms: role in pathogenicity and potential impact for vaccination development. Anim Health Res Rev. (2017) 19:17–30. doi: 10.1017/S146625231700010X
140. Ozma, MA, Khodadadi, E, Rezaee, MA, Asgharzadeh, M, Aghazadeh, M, Zeinalzadeh, E, et al. Bacterial proteomics and its application in pathogenesis studies. Curr Pharm Biotechnol. (2022) 23:1245–56. doi: 10.2174/1389201022666210908153234
141. Chung, JW, Ng-Thow-Hing, C, Budman, LI, Gibbs, BF, Nash, JHE, Jacques, M, et al. Outer membrane proteome of Actinobacillus pleuropneumoniae: LC-MS/MS analyses validate in silico predictions. Proteomics. (2007) 7:1854–65. doi: 10.1002/pmic.200600979
142. Stancheva, SG, Frömbling, J, Sassu, EL, Hennig-Pauka, I, Ladinig, A, Gerner, W, et al. Proteomic and immunoproteomic insights into the exoproteome of Actinobacillus pleuropneumoniae, the causative agent of porcine pleuropneumonia. Microb Pathog. (2022):105759. doi: 10.1016/j.micpath.2022.105759
143. Singh, N, and Bhalla, N. Moonlighting Proteins. Annu Rev Genet. (2020) 54:265–85. doi: 10.1146/annurev-genet-030620-102906
144. Wang, G, Xia, Y, Cui, J, Gu, Z, Song, Y, Chen, YQ, et al. The roles of moonlighting proteins in Bacteria. Curr Issues Mol Biol. (2014) 16:15–22. doi: 10.21775/cimb.016.015
145. Medrano-Díaz, CL, Vega-González, A, Ruiz-Baca, E, Moreno, A, and Cuéllar-Cruz, M. Moonlighting proteins induce protection in a mouse model against Candida species. Microb Pathog. (2018) 124:21–9. doi: 10.1016/j.micpath.2018.08.024
146. Pham, TH, Rao, S, Cheng, TC, Wang, PC, and Chen, SC. The moonlighting protein fructose 1,6-bisphosphate aldolase as a potential vaccine candidate against Photobacterium damselae subsp. piscicida in Asian sea bass (Lates calcarifer). Dev Comp Immunol. (2021) 124:104187. doi: 10.1016/j.dci.2021.104187
147. Lee, CH. A simple outline of methods for protein isolation and purification. Endocrinol Metab. (2017) 32:18–22. doi: 10.3803/EnM.2017.32.1.18
148. Quan, S, Hiniker, A, Collet, JF, and Bardwell, JCA. Isolation of Bacteria envelope proteins In: AH Delcour, editor. Bacterial cell surfaces: Methods and protocols. Totowa, NJ: Humana Press (2013). 359–66.
149. Thibeault, J, Patrick, J, Martin, A, Ortiz-Perez, B, Hill, S, Zhang, S, et al. Sarkosyl: a milder detergent than SDS for identifying proteins with moderately high hyperstability using gel electrophoresis. Anal Biochem. (2019) 571:21–4. doi: 10.1016/j.ab.2019.02.008
150. Herbert, B. Advances in protein solubilisation for two-dimensional electrophoresis. Electrophoresis. (1999) 20:660–663. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<660::AID-ELPS660>3.0.CO;2-Q
151. Overton, TW. Recombinant protein production in bacterial hosts. Drug Discov Today. (2014) 19:590–601. doi: 10.1016/j.drudis.2013.11.008
152. Antenucci, F, Ovsepian, A, Wrobel, A, Winther-Larsen, HC, and Bojesen, AM. Design and characterization of a novel tool for the antigenic enrichment of Actinobacillus pleuropneumoniae outer membrane. Pathogens. (2020) 9:1014. doi: 10.3390/pathogens9121014
153. Rapp, VJ, and Ross, RF. Antibody response of swine to outer membrane components of Haemophilus pleuropneumoniae during infection. Infect Immun. (1986) 54:751–60. doi: 10.1128/iai.54.3.751-760.1986
154. Hu, X, Yan, H, Liu, K, Hu, J, Qi, C, Yang, J, et al. Identification and characterization of a novel stress-responsive outer membrane protein Lip40 from Actinobacillus pleuropneumoniae. BMC Biotechnol. (2015) 15:106. doi: 10.1186/s12896-015-0199-8
155. Kenyon, WJ, Thomas, SM, Johnson, E, Pallen, MJ, and Spector, MP. Shifts from glucose to certain secondary carbon-sources result in activation of the extracytoplasmic function sigma factor σ E in Salmonella enterica serovar typhimurium. Microbiology (N Y). (2005) 151:2373–83. doi: 10.1099/mic.0.27649-0
156. Pakarian, P, and Pawelek, PD. A novel set of vectors for Fur-controlled protein expression under iron deprivation in Escherichia coli. BMC Biotechnol. (2016) 16:68. doi: 10.1186/s12896-016-0298-1
Keywords: Actinobacillus pleuropneumoniae, bacterial envelope, membrane proteins, proteome, vaccine, biofilm
Citation: Soto Perezchica MM, Guerrero Barrera AL, Avelar Gonzalez FJ, Quezada Tristan T and Macias Marin O (2023) Actinobacillus pleuropneumoniae, surface proteins and virulence: a review. Front. Vet. Sci. 10:1276712. doi: 10.3389/fvets.2023.1276712
Edited by:
Jerry William Simecka, University of North Texas Health Science Center, United StatesReviewed by:
Rosa Estela Quiroz Castañeda, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), MexicoHan Sang Yoo, Seoul National University, Republic of Korea
Copyright © 2023 Soto Perezchica, Guerrero Barrera, Avelar Gonzalez, Quezada Tristan and Macias Marin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alma L. Guerrero Barrera, YWxndWVycmVAY29ycmVvLnVhYS5teA==; bGlsaWFuLmd1ZXJyZXJvQGVkdS51YWEubXg=
†ORCID: Francisco J. Avelar Gonzalez, https://orcid.org/0000-0002-0777-9243
Teodulo Quezada Tristan, https://orcid.org/0000-0002-3793-4582
Osvaldo Macias Marin, https://orcid.org/0000-0001-9941-0971
 María M. Soto Perezchica1
María M. Soto Perezchica1 Alma L. Guerrero Barrera
Alma L. Guerrero Barrera