
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 18 December 2023
Sec. Veterinary Infectious Diseases
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1260433
Introduction: The extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, such as Escherichia coli, are emerging as a serious threat to global health due to their rapid spread and their multidrug-resistant (MDR) phenotypes. However, limited information is available regarding the prevalence and antimicrobial resistance (AMR) profile of ESBL-E. coli in the United States dairy farms. This study aimed to determine the prevalence and AMR pattern of ESBL-E. coli in East Tennessee dairy cattle farms.
Methods: Rectal fecal samples from dairy cattle (n = 508) and manure (n = 30), water (n = 19), and feed samples (n = 15) were collected from 14 farms. The presumptive E. coli was isolated on CHROMagar™ ESBL and confirmed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS). Antimicrobial susceptibility testing was performed on the ESBL-E. coli isolates.
Results and discussion: From 572 fecal and farm environmental samples, a total of 233 (41%, n = 572) ESBL-E. coli were identified. The prevalence of fecal ESBL-E. coli was 47.5% (95% CI: 46.2–49.2). The within-farm prevalence of ESBL-E. coli ranged from 8 to 100%. Recent treatment history with third-generation cephalosporins (3GC), cow parity ≥3, and calves were the independent risk factors associated (P < 0.05) with fecal carriage of ESBL-E. coli. Overall, 99.6% (n = 231) ESBL-E. coli tested were phenotypically resistant to at least one of the 14 antimicrobial agents tested. The most common AMR phenotypes were against beta-lactam antibiotics, ampicillin (99.1%; n = 231 isolates), and ceftriaxone (98.7%, n = 231). Most ESBL-E. coli isolates (94.4%) were MDR (resistance to ≥3 antimicrobial classes), of which 42.6% showed co-resistance to at least six classes of antimicrobials. ESBL-E. coli isolates with concurrent resistance to ceftriaxone, ampicillin, streptomycin, tetracycline, sulfisoxazole, and chloramphenicol are widespread and detected in all the farms. The detection of MDR ESBL-E. coli suggests that dairy cattle can be a reservoir for these bacteria, highlighting the associated public health risk.
Antimicrobial resistance (AMR) is recognized as one of the top five global public health threats of this century (1). Of great concern are extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae, including Escherichia coli, due to their rising prevalence in food-producing animals (2, 3). Some studies identified the use of β-lactam antibiotics, especially the third generation cephalosporins (3GC) in dairy cattle farms, as a major risk factor for the rise of ESBL-producing Enterobacteriaceae (4, 5).
In the United States, β-lactam antibiotics such as 3GC (e.g., ceftiofur), first-generation cephalosporins (e.g., cephapirin), and penicillin are the top three most frequently used antibiotics in dairy cattle (6, 7). These antibiotics are mainly used to treat or prevent mastitis, metritis, endometritis, lameness, pneumonia or bovine respiratory disease complex, and neonatal calf diarrhea (8–10). According to FDA’s 2019 report, from 29,830 kg of cephalosporins sold and approved for use in food-producing animals, the vast majority (81%) was distributed to cattle production (11). Similarly, Nora et al. (12) also reported that dairy cattle use the largest amount of cephalosporins and penicillin, both of which are β-lactam antibiotics.
According to the WHO’s risk-based classification systems, third-generation cephalosporins are categorized as a ‘critically important class of antibiotics (CIAs) (13). They are used as antimicrobial drugs of choice for the treatment of severe infections caused by E. coli and Salmonella in humans (14, 15). The use of the same generation of cephalosporins in dairy cattle farms and human health settings may lead to cross-resistance to similar cephalosporins used for the treatment of human infections or vice versa if resistant bacteria transfer from carrier animals to humans through direct contact or indirectly through the food chain or environmental sources (16, 17).
In Enterobacteriaceae, resistance to 3GC is mainly mediated by the production of ESBL, a group of enzymes that hydrolyze the β-lactam ring of the 3GCs (18). ESBL-producing Enterobacteriaceae can be multidrug-resistant (MDR) and display resistance to other classes of antibiotics such as tetracycline, aminoglycosides, fluoroquinolones, sulfonamides, macrolide, and phenicols (19–24). E. coli, the most frequent colonizer of the gastrointestinal tracts of cattle (25), are frequently exposed to β-lactam antibiotics that exert selection pressure, and certain strains of E. coli can cause severe infections in humans (17, 25–29). Recent studies in the United States (16, 30–34) increasingly reported the emergence of ESBL-producing E. coli, in dairy cattle since its first report in Ohio in 2010 (32), posing a significant threat to both animal and human health.
Similarly, reports from the United States Centers for Disease Control and Prevention (CDC) indicated a continuous rise in human infections caused by community-associated ESBL-producing Enterobacteriaceae (35). The CDC report revealed a yearly rise of approximately 9% in hospitalizations and an increase in deaths attributed to ESBL-producing bacteria. There is speculation that dairy farms may serve as reservoirs of ESBL-producing human pathogens because of the frequent use of 3GC (29, 36–38). The CDC has not directly implicated dairy farms as a source of ESBL-producing Enterobacteriaceae infection. However, the frequent use of 3GCs and other β-lactam antibiotics in dairy farms may play a role in the overall increase of ESBL-producing Enterobacteriaceae.
Previous studies on the United States dairy cattle farms showed an increasing trend in the occurrence of ESBL-producing E. coli (4, 30, 32, 33, 37, 39). A recent review of available literature on United States dairy cattle farms and recent global reviews on the status of ESBL-producing Enterobacteriaceae in cattle indicated that there is limited information on the status of ESBL-producing Enterobacteriaceae, including ESBL-producing E. coli in the United States dairy farms (16, 40). The prevalence of ESBL-producing E. coli and factors affecting its occurrence in dairy farms are poorly understood. Understanding the status (prevalence, risk factors and resistance profile) of ESBL-producing E. coli is crucial to informing the associated public health risks and devising effective control measures. The overall objective of this study was to determine the prevalence and antimicrobial susceptibility patterns of ESBL-producing E. coli, in East Tennessee dairy cattle and farm environmental samples.
This study was approved by the University of Tennessee’s Institutional Animal Care and Use Committee (IACUC) Registration Number: 2782–0720. The study farms were conventional (non-organic) dairy farms in East Tennessee. A cross-sectional study was conducted in 14 dairy farms across eight counties in East Tennessee.
East Tennessee was selected due to its proximity to the University of Tennessee, and most of the dairy farms (59%, n = 183) in Tennessee are found in this part of the State. Dairy farms were randomly selected (using a random number generator) from a sampling frame of 108 dairy farms found at the beginning of this study in East Tennessee. Since some farmers may not be interested in the study, the first 60 dairy farms were randomly selected. Dairy farmers on the list were then contacted one at a time by phone and asked whether they were willing to participate in the study. When the first selected farm refused to participate or could not be contacted, the next farms on the list were contacted and asked for their willingness to participate in the study. In the end, of the contacted dairy farms (n = 50), only 14 agreed to participate in the study.
To estimate the sample size, 50% expected prevalence of ESBL-E. coli at an animal level, 0.05 desired precision at a 95% confidence interval was used. Accordingly, the minimum required sample size was 384. To account for the possible clustering effect (design effect), the minimum sample size (n = 384) was multiplied by 1.34 (design effect) (4, 41). Accordingly, 507 dairy cattle were sampled (9–74 animals per farm). The herd size of the farms ranges from 14 to 1700 dairy cattle. Convenience sampling was used to select an individual animal in the herd. On the day of the farm visits, a brief questionnaire survey was conducted with a dairy farmer (producer) before sample collection. The questions included farm-related information such as herd size, predominant breed of cows, major disease problems in the farm, common antibiotics used (for therapeutic and prophylactic purposes), use of a blanket dry cow therapy (BDCT) versus selective dry cow therapy (SDCT), management of waste milk (milk from antibiotic-treated cows), use of medicated milk replacer, management of manure, and type of farm (open or closed), if open the recent introduction of animals from other herds. Individual sampled animal-related information such as age, breed, parity, physiological status (dry/lactating), recent treatment history, and use of ceftiofur within the last 6 months before sample collection.
Following the recruitment of the farms, one-time visits were made to each farm between August 2020 and July 2021 for survey data and sample collections. Fresh rectal fecal samples were collected from individual cows and calves using sterile disposable rectal long-arm gloves. Farm environmental samples such as manure (pooled from different pen surfaces and slurry), feed (pooled feed), and water (from troughs) were collected from the study farms. Approximately 100 g of individual cow and 20 g of individual calf rectal feces were collected into 50 mL sterile falcon tubes (Thermo Fischer Scientific, Waltham, MA, United States) and labeled with individual animal ID. Similarly, about 200 g of manure and feed samples and 50 mL of water from water troughs were collected into 50 mL sterile tubes. The samples were immediately placed on ice in an icebox, transported to the Lab, and processed in less than 24 h of collection.
The study recruited 14 dairy farms (designated A-N) across eight counties in East Tennes(see Table 1). A total of 424 dairy cows (average parity: 2.3 lactations, range: 1–8 lactations; average age: 3.9 years, range: 2–13 years) and 84 calves (average age: 2.7 months old, range: 12 days to 8 months). Pooled manure (n = 30), pooled water samples (n = 19) from water troughs, and polled feed samples (n = 15) were collected from the farms (Table 1). Two samples (from farm N), one collected from a lagoon and the other one collected from runoff from the farm to the environment, were included as manure. A feed sample was not collected from one of the farms (Farm M) as the farmer was less comfortable. One of the two collected feed samples in farm G was a liquid whey fed to cattle.
About 10 g of individual animal rectal feces, feed, and manure samples were combined with 90 mL of Tryptic soy broth (TSB-PO4, MG Scientific, Pleasant Prairie, WI, United States) in a Whirl-Pak bag (Whirl-Pak, Pleasant Prairie, WI, United States) and massaged by hand for 15–20 s to mix the samples with the TSB-PO4. For water samples, 20 mL of sample was mixed with 80 mL TSB-PO4. The homogenized sample was placed at room temperature for about 2 h. Then, 50 μL of the homogenized sample was spirally platted on CHROMagar ESBL plates (DRG International Inc., Springfield, NJ, United States) and incubated at 37°C for 24 h to isolate ESBL-E. coli. Two to three presumptive ESBL-E. coli (dark pink to reddish) colonies were subcultured onto a new CHROMagar ESBL plate and incubated for another 24 h at 37°C. Well-isolated pure colonies were transferred to 1 mL Luria-Bertani broth (LBB) (Thermo Fischer Scientific) into a sterile 2 mL 96 well-serum block and incubated at 37°C for 24 h. A 0.5 mL of the culture was transferred to a new 96-well serum block, mixed with an equal volume of sterile 80% glycerol, and stored at −80°C for further analysis.
Presumptive E. coli isolates stored at −80°C were thawed and plated on CHROMagar ESBL plates ESBL plates and incubated at 37°C for about 18 h. A single colony of E. coli was picked and subcultured on blood agar (Thermo Fischer Scientific) at 37°C for about 18 h. Subsequently, E. coli was identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) as described by the manufacturer (Bruker Daltonics, Billerica, MA, United States) at the University of Tennessee, College of Veterinary Medicine, Diagnostic Bacteriology and Mycology Lab. The samples were prepared using a formic acid (FA) extraction method (42) and as described in detail in our previous publication (43).
Antimicrobial susceptibility testing (AST) was performed on MALDI-TOF MS confirmed E. coli isolates against 14 antimicrobials representing ten classes of antimicrobials using the broth microdilution method. Commercially available 96-well microtiter plates containing the 14 antimicrobial panels (Sensitire™ CMV4AGNF) (Thermo Fisher Scientific) described in our previous publication were used (43). The minimum inhibitory concentrations (MIC) of the 14 antimicrobials were determined following the manufacturer’s recommended protocol and following CLSI M100 Clinical and Laboratory Standards Institute guidelines (CLSI M100: Enterobacteriaceae) (44) and summarized (Supplementary Table S1). E. coli ATCC 25922, Staphylococcus aureus ATCC 29213, Streptococcus pneumoniae ATCC 49619, E. coli ATCC 25922, and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains.
The results were reported as susceptible (S), Intermediate(I), resistant (R) along with the MIC values, the lowest concentration (μg/mL) of an antibiotic that completely inhibits visible growth of the tested isolates, using CLSI M100 guidelines for interpretation.
An MIC breakpoint was not available on CLSI M100 guidelines for streptomycin and, thus, National Antimicrobial Resistance Monitoring System (NARMS) interpretive criteria (breakpoints) for E. coli antimicrobial susceptibility testing1 was used. Accordingly, an MIC ≥32 μg/mL was defined as resistant to streptomycin. For sulfisoxazole, CLSI defines an MIC ≤256 μg/mL as susceptible and an MIC ≥512 mL as resistant, and there are no breakpoints for intermediate resistance. Thus, E. coli isolates that have an MIC ≥256 μg/mL were reported as resistant to sulfisoxazole in this study. In addition, for analyses, a few E. coli isolates that displayed intermediate resistance, mostly to ciprofloxacin, were recategorized as resistant. Multidrug resistance (MDR) was defined as acquired resistance to at least one antimicrobial agent in three or more antimicrobial classes (45).
Raw data was entered in Microsoft Excel for Windows (2010, Microsoft Corp., Redmond, WA). Data was imported to SPSS for Windows and analyzed using IBM SPSS Statistics for Windows Version 27.0. (IBM Corp, Armonk, NY, United States). For all statistical analyses, the unit of analysis was the sample obtained from a given source (cow, calf, manure, water, and feed), the farms, and the bacterial isolates. Descriptive statistics were done to summarize the animal and farm-level prevalence of ESBL-E. coli and the prevalence of the bacteria from different sources (fecal, water, feed, and manure samples) of the farms. The individual animal was classified as ESBL-positive when ESBL-producing E. coli isolates were identified phenotypically on the CHROMagar ESBL™ plate and confirmed by MALDI-TOF MS (46, 47) in the feces of the dairy cow or calf and the other samples.
A farm (herd) was classified as ESBL-positive when ESBL-producing E. coli was detected from at least one animal. Descriptive statistics were used to calculate the prevalence of ESBL-producing E. coli as the number of samples tested positive for ESBL-producing E. coli divided by the total number of samples tested for each sample category. Sampling weight was used to calculate the within-herd and individual animal level prevalence to account for the unequal probability of sampling employed during the sample collection. The weighted sample for a specific farm was determined by dividing the actual number of animals sampled on the farm by the herd size of that farm. The reciprocal of this quotient was then taken as the weighted sample for the farm. To calculate the overall weighted prevalence, the unweighted prevalence of ESBL-producing E. coli in each of the 14 herds (farms) was multiplied by the corresponding herd proportion. These products were then summed up, and the result was divided by 100. A univariate and multivariable mixed-effects logistic regression model, which accounts for the clustering effect of cattle or isolates within farms, was used in the analysis. A mixed effects logistic regression analysis (weighted) was performed to evaluate the association between selected predictors (farm or animal level data) and the odds of ESBL-producing E. coli fecal carriage (dependent variable) in cattle.
In addition, the multidrug resistance (MDR) status of each bacterial isolate obtained from the study was treated as a dichotomous outcome variable (MDR vs. non-MDR), and their possible variation between sample sources was evaluated. The MDR isolates were further dichotomized (whether they were resistant to at least six classes of antibiotics or not) and assessed if there was a variation between the sample sources. A Pearson χ2 or Fisher exact test (as appropriate) was used to test the association between different categorical variables (ignoring clustering) when the mixed effect model fails to converge. A value of p = 0.05 was used for all statistical analyses to determine the significance level.
The questionnaire survey showed that half of the farms practice closed production systems where replacement heifers originated from the same farm, whereas the other half buy replacement heifers/cows from outside sources in addition to raising their own. Thirteen of the fourteen participating dairy farms use cow manure as fertilizer, mainly on pasture, and one farm (Farm G) uses manure for biogas production. Most of the farms (71%) discard waste milk from cows on antibiotic treatment (dump down the drain or dump on the pasture field), whereas the remaining four farms (Farm C, F, G, and I) feed it to calves. A blanket dry cow treatment program was routinely practiced in all the farms except three (B, F, and N), which used selective dry cow therapy and teat sealant. The most frequently used antibiotics for dry cow therapy were Spectromast® DC (ceftiofur hydrochloride), toMORROW (cephapirin benzathine), and Quartermaster (procaine penicillin G and dihydrostreptomycin). Eleven farms predominantly have Holstein-breed cows, whereas Jersey cows dominate the remaining three. The herd size of the 14 farms ranged from 14 to 1700 cattle, with a median herd size of 750 animals.
All farmers mentioned mastitis as the most frequent herd health problem, followed by hoof problems. Beta-lactam antibiotics (penicillin and cephalosporins) were the most common antibiotics used in all farms. All study farms frequently use different generations and formulations of cephalosporins. Major disease problems and frequently used antibiotics in each farm are listed in Table 2.
According to CHROMagar™ ESBL result, a total of 306 presumptive ESBL-E. coli were isolated from rectal feces, manure, water, and feed samples. Out of 306 presumptive ESBL-E. coli isolates, 233 (76%) were confirmed to be E. coli by MALDI-TOF MS. The remaining bacteria include Citrobacter sedlakii (n = 69 isolates), Citrobacter freundii (n = 2), and Acinetobacter baumanii and Enterococcus faecalis. One each. Except for AST, which was conducted on 231 isolates, all statistical analyses and inferences were based on the 233 E. coli isolates. Most of the E. coli isolates (90%, n = 233) were isolated and identified from rectal fecal samples, whereas the remaining (10%) were from farm environmental samples (manure, water, and feed samples).
The weighted prevalence of fecal ESBL-producing E. coli in dairy cattle was 47.5% (95% CI: 46.2–49.2). ESBL-producing E. coli was detected from all farms. The prevalence varies widely between farms, with the highest within-herd weighted prevalence of 100% from farm A, whereas the lowest prevalence of 8% was from two farms (farms E and N). ESBL-producing E. coli was isolated from fecal samples of all fourteen farms, resulting in a herd-level prevalence of 100%. The herd size of the farms, the number of animals tested from each farm, the number of animals that tested positive and within-herd weighted prevalence, and the 95% confidence interval (95% CI) of the estimate for each farm are presented in Table 3.
The Chi-square test of independence was performed (without accounting for within farm clustering effect of the isolates) to test if there was a statistically significant difference in the weighted prevalence of ESBL-producing E. coli between farms. The test showed an overall statistically significant variation (χ2 = 21; p < 0.001) of within-herd prevalence of ESBL-producing E. coli. Similarly, a mixed-effect logistic regression (controlling the random effect of the farm) showed an overall significant variation in the fecal ESBL-producing E. coli prevalence between farms (p < 0.001). A comparison of the within-herd prevalence of fecal ESBL-producing E. coli between individual farms is shown in Figure 1.
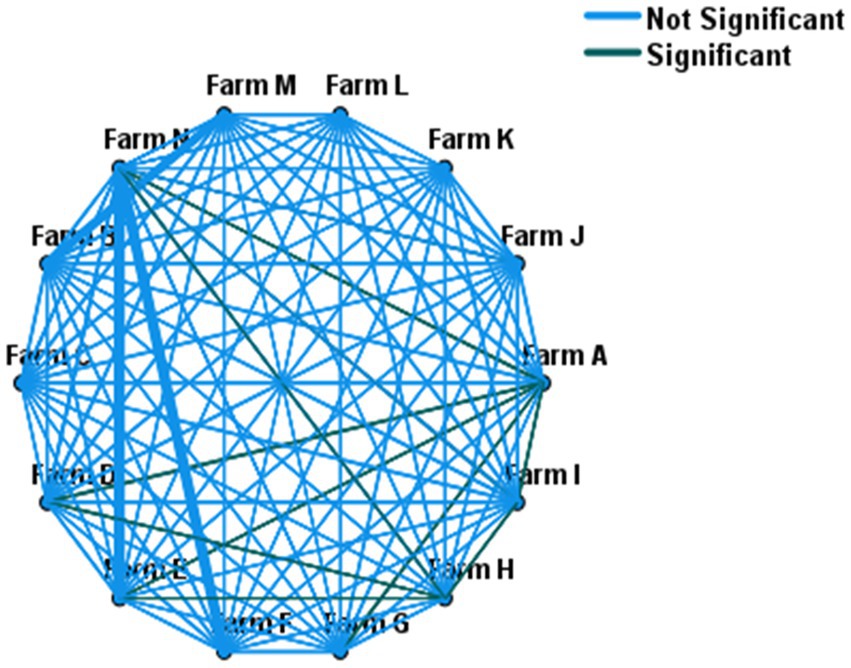
Figure 1. Pairwise comparison of within-herd fecal ESBL-E. coli prevalence across 14 farms. Farms connected with green lines (—) showed statistically significant variation in their within-herd prevalence, whereas no difference for those farms connected with light blue lines (—).
The weighted prevalence of fecal ESBL-producing E. coli was 44 and 61.7% in cows and calves, respectively. Calves were twice [OR = 2.12 (95% CI: 1.78–2.53), p < 0.001] as likely to carry ESBL-E. coli in their feces compared to cows using mixed effect logistic regression analysis with farms as a random variable and herd structure (calf vs. cow) as the sole fixed variable (Table 4).
ESBL-producing E. coli was recovered from over a quarter of all sample types (26%). The highest proportion of ESBL-producing E. coli was obtained from pooled manure samples (46.7%), whereas the least (26.3%) was from water samples (Supplementary Table S2).
After matching the farm-level survey data with the findings of the MALDI-TOF MS results from individual animals, a mixed-effect logistic regression analysis was carried out to determine the association between the variables collected during the survey and the probability of isolating ESBL-producing E. coli in samples. The farm-level collected data include herd size, farm type (closed vs. open), predominant breed in the farm, use of blanket dry cow therapy (BDCT) vs. selective dry cow therapy (SDCT), types of antibiotics used for DCT, presence of treatment ward in the farm, management of waste milk from treated cows (discard vs. fed to calves). None of these farm-level variables were associated with the detection of ESBL-producing E. coli in fecal samples, in univariate and multivariable analysis (p > 0.05), and, thus, were not included in this report.
The relationship between individual animal-related surveys such as the recent (up to 6 months prior to sampling collection) use of beta-lactam antibiotics such as ceftiofur, parity, lactation status of the cows (lactating vs. dry or non-lactating), age (of cows), and the breed was assessed for possible association with fecal ESBL-producing E. coli carriage. First, a univariate analysis was conducted to test the association between independent variables and the likelihood of isolating ESBL-producing E. coli in fecal samples. Then, only those variables with a p ≤ 0.2 in univariate analyses were used in a multivariable analysis controlling the random effect of the farm. Overall, recent treatment with ceftiofur, cow parity, and the lactation status of the cows was independently associated (p < 0.001) with the prevalence of fecal ESBL-producing E. coli. The odds of ESBL-producing E. coli fecal carriage in cows recently treated with ceftiofur was higher than [ORadj = 1.35 (95% CI: 1.1–1.65), p = 0.004] those which did not receive the treatment. Lactating cows were more than three times [ORadj = 3.42 (95% CI: 2.79–4.10), p < 0.001] more likely to carry ESBL-E. coli in their feces compared to dry cows.
Age and parity of the cows were highly correlated (p < 0.001; R = 0.73) since parity increases with age. Thus, the age of the cows was not included in the analysis, and parity was retained in the model. Odds of retrieving ESBL-producing E. coli was 33% higher [ORadj = 1.33 (95% CI: 1.13–1.63), p = 0.01] in cows with parity greater than two compared to those cows with parity less than or equal to two (Table 5).
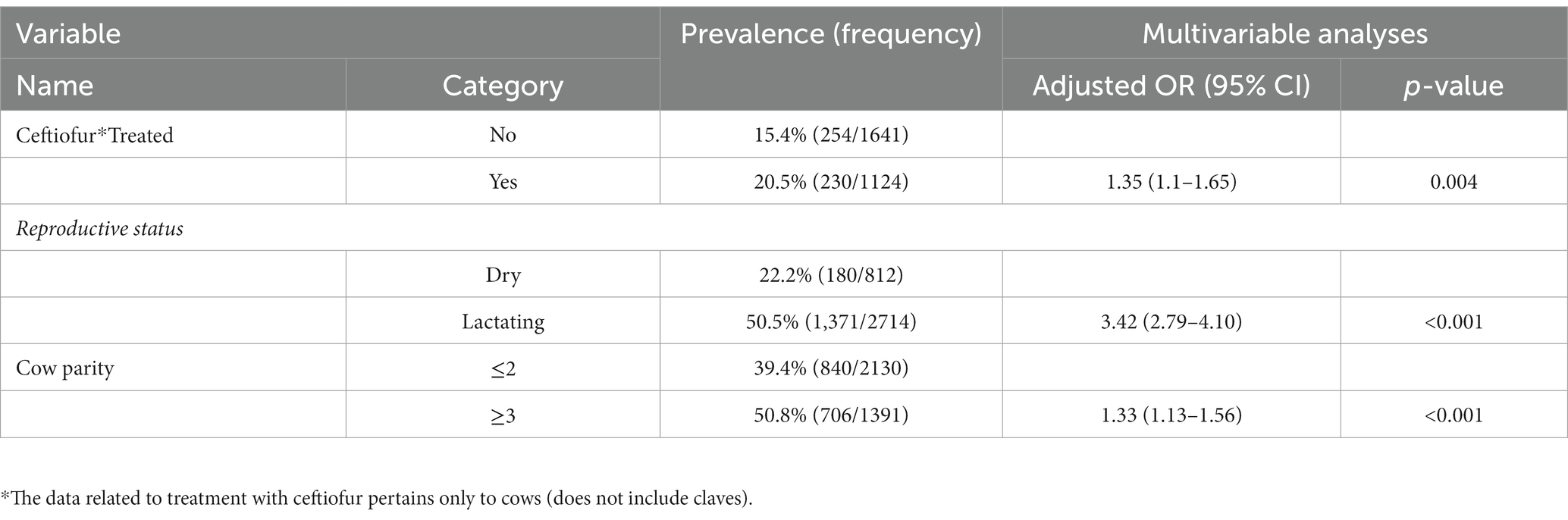
Table 5. Association between cow-level characteristics and weighted fecal ESBL Escherichia coli prevalence.
Overall, 230 (99.6%) of 231 E. coli tested for phenotypic resistance were resistant to at least one of the 14 antimicrobial agents tested (Table 6). One isolate was susceptible to all tested antimicrobials. The most common resistance phenotypes were against beta-lactam antibiotics, ampicillin (99.1%; n = 231), and ceftriaxone (98.7%, n = 231). Only three isolates showed susceptibility to ceftriaxone; two of them were recovered from cows. After ceftriaxone and ampicillin, the isolates showed the highest resistance to tetracycline (80.1%), sulfisoxazole (60.2%), streptomycin (55%), and chloramphenicol (46.8%).
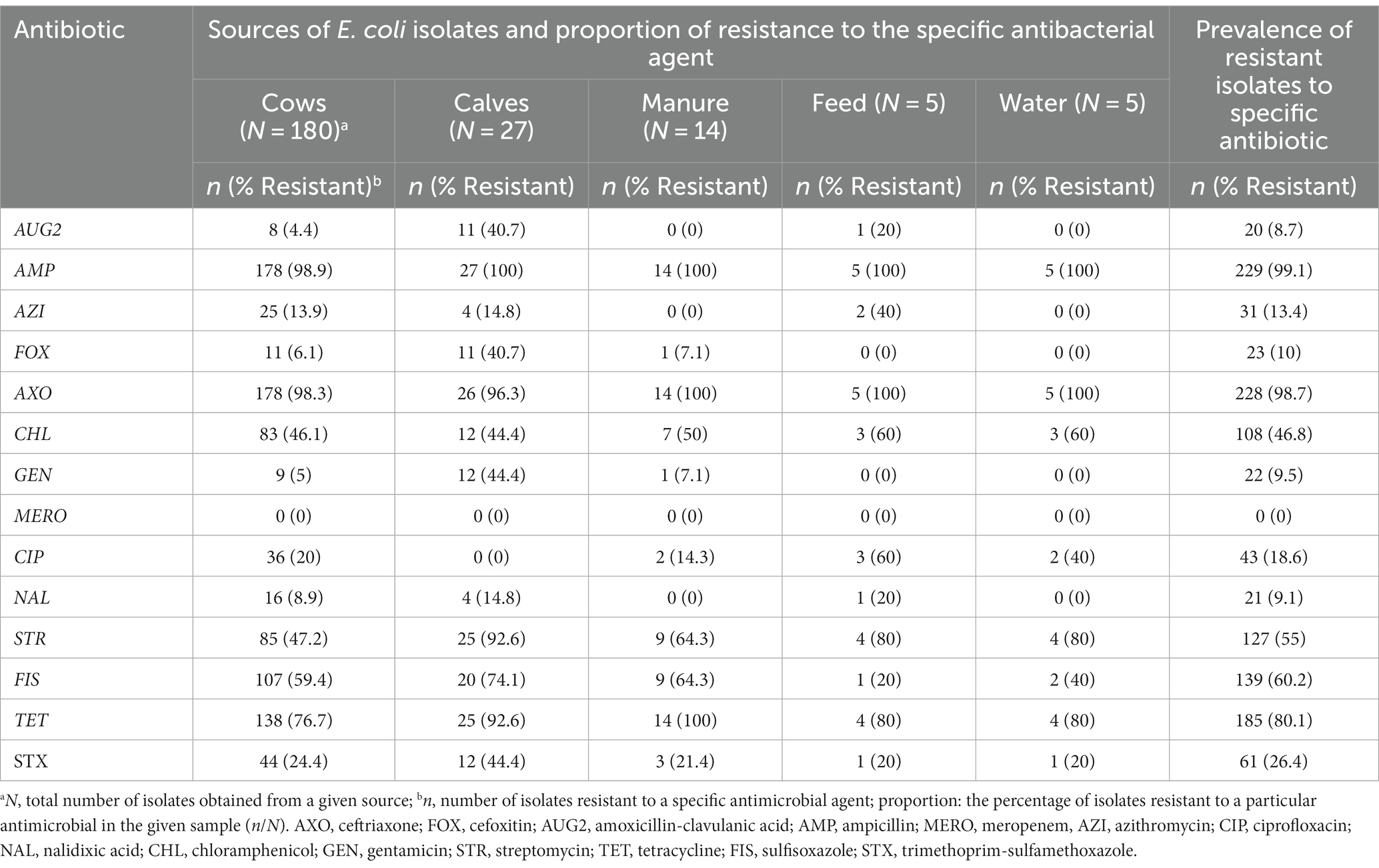
Table 6. Number and percentage of resistance of ESBL Escherichia coli isolates (N = 231) obtained from various sources.
Among the critically important classes of antibiotics, the higher level of resistance after ceftriaxone was to streptomycin (55%), followed by ciprofloxacin (18.6%) and azithromycin (13.4%). The prevalence of resistance to gentamycin and nalidixic acid, the other critically important antimicrobials tested in this study, were 10 and 9%, respectively.
The resistance level to other β-lactam antibiotics was 10% for cefoxitin and 9.2% for amoxicillin-clavulanic acid. All ESBL-producing E. coli isolates in this study were susceptible to meropenem.
Most of the ESBL-producing E. coli isolates (94.4%; 218/231) displayed multidrug resistance (MDR) phenotypes (acquired resistance to at least one agent in three or more antimicrobial classes) (45). All isolates recovered from manure (n = 5) and water samples (n = 5) showed MDR phenotypes. Most of the isolated from calves (96.3%; n = 27), cows (93.9%; n = 180), and feed (80%; n = 4) were MDR. Among the 14 farms, all isolates obtained from 10 of the farms were MRD. From 218 MDR isolates, 42.6% showed concurrent resistance to at least one antibiotic in six classes of antibiotics. ESBL-producing E. coli isolates that showed concurrent resistance to ≥6 antimicrobial classes were obtained from all sample sources (Supplementary Table S3).
Overall, resistance to multiple classes of antibiotics is widespread across the study farms (Table 7). The most common (52.8%, 122/231) and widespread multidrug resistance pattern was concurrent resistance to ceftriaxone, ampicillin, streptomycin, and tetracycline, detected in all farms and sample types. Similarly, simultaneous resistance to ceftriaxone, ampicillin, sulfisoxazole, and tetracycline was frequent (48.5%, 112/231) and detected in all farms and sample types. Concurrent resistance to at least one antimicrobial agent in all critically important classes of antibiotics (ceftriaxone, streptomycin, azithromycin, and ciprofloxacin) was relatively less frequent (6.5%, 15/231) and limited in the scope of spread among farms (detected in only three farms). Fourteen of fifteen E. coli isolates with this resistance pattern were retrieved from cow fecal samples. Seventeen (7.4%) of E. coli isolates were co-resistant to all beta-lactam antibiotics tested in this study (ceftriaxone, ampicillin, cefoxitin, and amoxicillin-clavulanic acid). E. coli isolates with this co-resistance phenotype were detected in six farms, and all of them were isolated from fecal samples except one recovered from a feed sample.
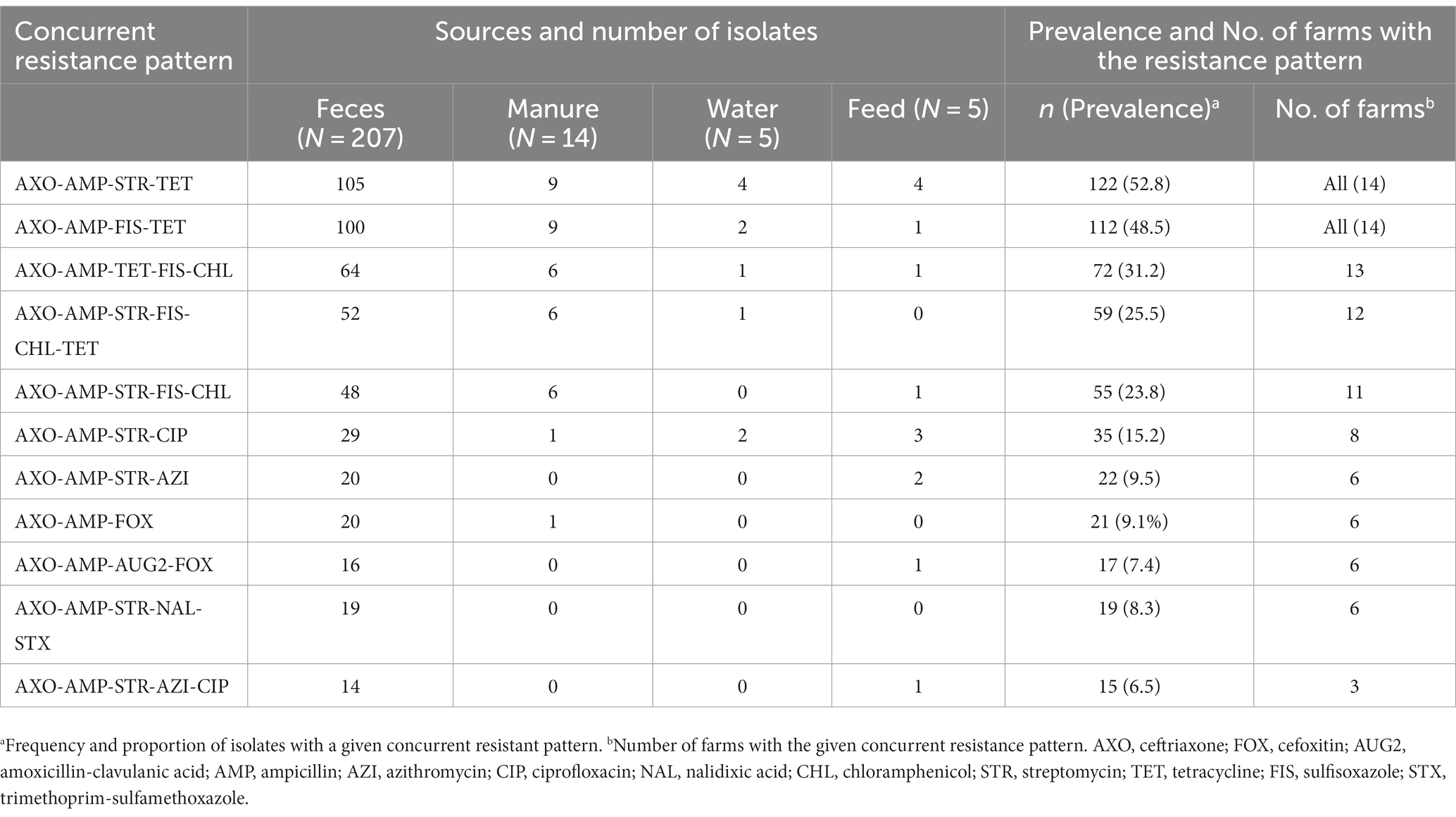
Table 7. Distribution of concurrent resistance of ESBL Escherichia coli to multiple classes of antibiotics across farms and sample sources.
In this study, all farms identified mastitis as the most frequent health problem of dairy cows and a primary driving factor for antibiotic use. This finding is consistent with other studies in the United States that also reported mastitis as the most common disease and the main reason for antibiotics use in dairy farms (8, 11, 46, 48, 49). Similar to previous studies in the United States (50, 51), most (86%, n = 14) of the farms in the present study used blanket dry cow therapy to manage mastitis. This practice may expose many animals and their gut commensal bacteria to antibiotics as portions of the administered dose or metabolites may enter the bloodstream and reach the gut (52).
Beta-lactam antibiotics such as cephalosporins (ceftiofur, cephapirin benzathine, and cephapirin sodium) and penicillin were the most frequently used antibiotics in the study farms. This finding concurs with a recent study by Nora et al. (12), who reported the consumption of large quantities of cephalosporins and penicillin in dairy cattle farms compared to other antibiotics. Similarly, the latest USDA survey reports (50, 53) and two other studies in the United States (8, 54) also showed that cephalosporins and penicillin are the most commonly used antibiotics to treat or prevent mastitis and other common diseases of dairy cattle. The previous studies demonstrated that frequent use of a given antibiotic leads to the emergence of resistance to antibiotics (47, 55–59). Regularly using cephalosporins, particularly ceftiofur, in the study farms is concerning as it could lead to the emergence of ESBL-producing bacteria such as E. coli (60).
ESBL-producing E. coli was detected from fecal samples of at least one animal in all 14 study farms resulting in 100% herd-level prevalence. This is higher than previous studies from the United States that reported 20% (5/25 farms) from Ohio (37), 85% (18/21) from Washington (30), and 4% (3/80) from Pennsylvania (61) dairy herds. However, this comparison should be interpreted with caution as these studies may differ in the criteria that they used to define a herd as positive (use of pooled manure samples vs. rectal fecal sample), sample processing steps (use or not using enrichment steps, type of media used for bacterial isolation) involved to determine the herd level status of ESBL-producing E. coli, which has different detection sensitivity and specificity. The widespread occurrence of ESBL-producing E. coli across the present study farms could be related to the frequent use of beta-lactam antibiotics, such as ceftiofur, that may subject them to selection pressure and other farm management related factors that favor the spread of the ESBL-producing E. coli or their ESBL genes.
The weighted within-herd prevalence of fecal ESBL-producing E. coli ranged from 8.0 to 100%, and the difference between within-herd prevalence was statistically significant across the farms. Detection of statistically significant variation in prevalence between different farms could be related to the differences in the type and level of beta-lactam antibiotics use and the presence of other influencing farm management related factors that were not within this study’s scope.
The within-herd prevalence of 8–100% in the present study is higher than a few previous studies conducted in the United States, including 3.3–100% (37) in Washington state and 0–33% in Ohio (33). Studies from other countries also reported a within-herd prevalence ranging from 5.2–86.7% (62, 63). However, it should be noted that these studies differ from the present study and each other in the microbiological techniques they used to isolate and identify the bacteria and the study farms’ selection criteria, which makes the comparison less plausible.
The prevalence of fecal ESBL-producing E. coli in the present study was 47.5% at an animal level, indicating a high level of colonization. The current prevalence report is higher than the previous report from Ohio dairy cattle (9.4%,70/747) (37) but closely similar to a report from European dairy cattle, 41% (37/90) (63). The fecal prevalence of ESBL-producing E. coli in this study is higher than the recently reported prevalence of 4.6% in United States cow-calf operations (64). This variation could be related to the differences in the type and frequency of beta-lactam antibiotics use and other managemental factors that may favor the occurrence and spread of ESBL-producing E. coli in the farms.
The fecal ESBL-producing E. coli prevalence in dairy calves was significantly (p < 0.001) higher than in cows (61.7% vs. 44%). This finding agrees with recent studies from North America (56, 61, 64–68) and Europe (63, 69–71) that reported a higher proportion of ESBL-producing or -third-generation cephalosporins (3GC) resistant-E. coli in calves compared to older cattle. Conclusive evidence as to why the higher prevalence of resistant bacteria in calves is not available and needs further study. It is hypothesized that feeding medicated milk replacers, colostrum, or milk from cows treated with beta-lactam antibiotics might have exerted selective pressure that led to the increased fecal carriage of ESBL-producing E. coli in calves (56, 63, 66, 69, 72). However, a recent study in the United States and Canada dairy calves showed a high prevalence of antimicrobial resistant E. coli in young calves that were not exposed to antimicrobials-containing milk replacers or did not feed milk from cows treated with antibiotics (66). Another alternative hypothesis for a higher prevalence of ESBL-producing E. coli in calves is related to the fitness of the resistant bacteria due to less bacterial diversity in the calves’ gut compared to adult dairy cattle with more diverse gut bacteria where competition is strong for ESBL-producing E. coli to thrive (71, 73, 74). A controlled study is needed to identify factors that promote a higher fecal ESBL-producing E. coli carriage in dairy calves than in adult cattle. Whatever the driving factor may be, this study suggests that calves may be an important source or reservoir of ESBL-producing E. coli and can be considered as sentinel animals for surveillance.
The detection of a higher proportion of ESBL-producing E. coli in manure is not surprising as manure contains a mixture of fecal samples, bedding, and wastewater pooled from different animal sources and has a high chance of containing the bacteria. Previous studies in the United States also reported a comparable level (53.5%; 62/116) of ESBL-producing E. coli from pooled manure samples (33). Most dairy farms in this study use manure as a fertilizer on pasture fields, which can contaminate the environment and water. Thus, the presence of ESBL-producing E. coli in the manure sample has significant public and environmental health implications. In our previous study, we detected a higher proportion of 3GC-resistant E. coli in dairy manure-amended soils compared to prairie soils not impacted by dairy cattle suggesting the need for proper management of manure to reduce the spread of resistant bacteria (16). The detection of ESBL-producing E. coli in feed (33.3%) and water (26%) samples in this study suggest that the bacteria may remain circulating in dairy farms through oral-fecal transmission via contaminated feed and water (75). Frequent cleaning of water and feed troughs may help to reduce the chance of maintenance of ESBL-producing E. coli in the dairy farm (5, 67).
This study also assessed animal-level factors that may influence the fecal prevalence of ESBL-producing E. coli. Recent use of ceftiofur (for treatment or prophylaxis), parity, and lactation status of the cow were significant predictors for the probability of ESBL-producing E. coli fecal carriage (p < 0.05). Cows recently receiving ceftiofur treatment (up to 6 months before sample collection) were 35% more likely (OR = 1.35) to carry ESBL-producing E. coli in their feces compared to cows not exposed to this antibiotic during the same period. Previous studies (4, 72, 76–79) also showed ceftiofur use leads to an increased probability of ESBL-producing E. coli detection or decreased susceptibility to 3GC, such as ceftriaxone in dairy cattle. Controlled studies in the United States (78) and Europe (80) showed a significant increase in ESBL mediating genes and mobile genetic elements (plasmids and prophages) in ceftiofur treated compared to non-treated dairy cows. In addition, a European study showed a restrictive use of ceftiofur and cefquinome (4th generation cephalosporin) in cattle significantly decreased the prevalence of fecal ESBL-producing E. coli in cattle (71).
Nevertheless, existing literature does not fully agree on the association between ceftiofur use and the probability of detection of E. coli with reduced susceptibility to 3GC. Some authors reported a lack of association between the two (37, 68), and others reported a decreased chance of recovering resistant E. coli following treatment with ceftiofur (72). However, most controlled and observational studies (37, 68) (including the present study) indicated a stronger association between ceftiofur use and the probability of detecting ESBL-producing E. coli. Thus, considering ways to reduce ceftiofur use (e.g., using only when sensitivity test result justifies) may help to reduce the ESBL-producing E. coli emergence and spread.
The likelihood of lactating cows having ESBL-producing E. coli in their feces were more than three times greater (OR = 3.42) than that of dry cows. The difference in the prevalence of fecal ESBL-producing E. coli may be related to the increased risk of mastitis and other diseases in lactating cows, prompting antibiotic use and favoring the emergence of ESBL-producing E. coli strains. This finding is significant because lactating cows are more likely to come into contact with humans than dry cows, and they also produce milk that can be contaminated. This increases the risk of transmission of ESBL-producing E. coli from cows to humans during the lactation period.
Cows with higher parity (≥3) were associated with an increased probability (OR = 1.33) of fecal ESBL-producing E. coli recovery compared to those cows with lower parity (≤2). This could be related to frequent exposure to beta-lactam antibiotics for managing mastitis and other diseases whose prevalence likely increases with age. Previous studies (81–83) showed a strong association between the prevalence or incidence of mastitis and parity (higher risk in older cows), which is also correlated with increased use of antibiotics that promote selection pressure on commensal E. coli.
Almost all (99.6%, 230/231) ESBL-producing E. coli tested for AST exhibited resistance to at least one of the 14 antimicrobial agents tested. The isolates displayed the highest level of resistance to ampicillin (99.1%, n = 231) and ceftriaxone (98.7%) which is expected as the isolates produce ESBL-enzymes that hydrolyze penicillin and extended-spectrum cephalosporins (14, 84). Three ESBL-producing E. coli isolates were identified as ESBL-producers by the selective chromogenic media and were unexpectedly susceptible to ceftriaxone, a 3GC. One isolate was sensitive to all the tested antimicrobial agents. This finding was unexpected as the selective media used in this study contains a supplement that is supposed to select isolates resistant to beta-lactam antibiotics such as penicillin and 3GC. This could be related to the reduced specificity of the screening media used to isolate ESBL-producing E. coli or later loss of expression of the ESBL gene (85).
A high prevalence of co-resistance was detected between ceftriaxone and older antibiotics such as ampicillin (97.8%, n = 231), tetracycline (79.7%), sulfisoxazole (59.7%), and streptomycin (54.5%). This resistance trend and prevalence are consistent with previous studies from the United States dairy cattle (16, 30, 61). Recently, Masse et al. (65) also reported a similar trend but a lower prevalence of co-resistance to these antibiotics in Canada. However, some authors used different initial screening steps (selective media) to isolate E. coli, making the comparison challenging. Some of these antibiotics (streptomycin and sulfisoxazole) were not mentioned by farmers as predominantly used antibiotics in their farms, suggesting the high level of resistance detected may not be related to their utilization. Further investigation is important to explain if these resistance phenotypes were co-selected with ESBL phenotypes, common resistance phenotypes reported in this study.
ESBL-producing E. coli exhibited low resistance to amoxicillin-clavulanic acid (8.7%, n = 231) and cefoxitin (10%). Generally, the growth of ESBL-producing E. coli is expected to be inhibited by both amoxicillin-clavulanic acid and cefoxitin, a cephamycin (14, 84, 86). As reported in other studies, the detection of resistance to these two antibiotics suggests the possible expression of multiple resistance phenotypes in the same E. coli isolates (5, 30, 65).
ESBL-producing E. coli displayed a higher level of concurrent resistance between ceftriaxone and other members of CIAs, such as streptomycin (55%), ciprofloxacin (18.6%), and azithromycin (13.4%). The prevalence of concurrent resistance is similar to a previous study from the United States (30) but 5 to 20 times higher than the reported prevalence from Canadian dairy farms (65). Other previous studies reported ESBL-producing E. coli often exhibit co-resistance to multiple CIAs (3, 68).
The overall prevalence of MDR ESBL-producing E. coli isolates in this study were 94.4% (218/231), suggesting an association between MDR and ESBL-phenotypes in E. coli. This finding is consistent with previous studies that reported a high prevalence of MDR phenotype1 in ESBL-producing E. coli isolates from cattle rectal fecal and farm manure samples (30, 64, 68). About 42.6% of MDR E. coli isolates in this study showed concurrent resistance to at least six classes of antibiotics, suggesting the ESBL-producing E. coli is a host of resistance genes which may complicate treatment if these bacteria cause infection or transfer the resistance genes to pathogenic bacteria. Multidrug resistant ESBL-E. coli was detected from all farms and all sample types, suggesting widespread occurrence across farms. Unlike recent studies from Canadian dairy farms (65), no difference was detected in the prevalence of the MDR across the five sample sources and study farms. The lack of detection of variation indicates that ESBL phenotype is a phenotypic marker of the MDR pattern in E. coli, irrespective of sample sources (5, 86, 87).
The study found two specific patterns of multidrug resistance ESBL-producing E. coli, AXO-AMP-STR-TET (52.8%), and AXO-AMP-FIS-TET (48.5%) were prevalent and detected in all sample types and study farms. The widespread occurrence of these patterns is a cause for concern regarding the spread of antibiotic resistance. The study calls for further research to understand the mechanisms behind the spread of these patterns, and the potential impact on animal and human health to identify strategies for reducing their prevalence in dairy farms.
The study found that 6.5% of ESBL-producing E. coli had concurrent resistance to at least one antibiotic in all CIAs tested in this study. This is a serious cause for concern as these antibiotics are of the highest importance for human health, and their efficacy could be compromised. However, concurrent resistance was detected from only three study farms, highlighting the need for continuous monitoring with targeted and localized interventions. Another study in the United States reported that CIAs such as macrolides and fluoroquinolones are commonly used to treat diseases in dairy calves and other non-lactating dairy cattle under 2 years of age, which could possibly drive the concurrent presence of resistance to these antibiotics (88). Other studies in humans reported the frequent use of 3GC as a risk factor for the co-occurrence of resistance to CIAs such as fluoroquinolones (89–91) which could be the case as two of these farms were frequently using ceftiofur, a 3GC.
This study showed a high prevalence of ESBL-producing E. coli in dairy cattle farms, with almost half of the tested animals positive for the bacteria. The prevalence of ESBL-producing E. coli varied significantly within each farm. The study found that recent treatment with third generation cephalosporins, cows with higher parity, and being calves was linked to an increased risk of fecal carriage of ESBL-producing E. coli. These findings suggest that interventions should target these identified variables. Almost all the ESBL-producing E. coli isolates were MDR with the highest resistance to ampicillin and ceftriaxone. Over 40% of ESBL-E. coli showed concurrent resistance to at least six classes of antibiotics. Concurrent resistance to ceftriaxone, ampicillin, tetracycline, sulfisoxazole, streptomycin, and chloramphenicol was high and widespread across all farms. It is important to indicate that the study farms and animals sampled from each farm were based on farmers that agreed to participate in the study and individual animals were selected based on convenience sampling. In addition, we acknowledge that the use of CHROMagar ESBL plates, while a widely accepted method, may result in some misclassifications. Despite these limitations, the findings of this study particularly the high prevalence of MDR ESBL-producing E. coli in the study area is significant and provides baseline data for future intervention measures. Further investigation, such as the longitudinal monitoring of ESBL-E. coli colonization in dairy cattle and possible transmission between cattle and farm workers who are in direct and frequent contact with animals and their feces is required.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The animal study was approved by The University of Tennessee Institutional Animal Care and Use Committee (IACUC) registration number 2782-0722. The study was conducted in accordance with the local legislation and institutional requirements.
BG: Conceptualization, Writing – review & editing, Formal analysis, Investigation, Methodology, Writing – original draft. AG: Investigation, Writing – review & editing. OK: Writing – review & editing, Conceptualization, Project administration, Supervision.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors thank the dairy farmers in East Tennessee for willingly volunteering to participate in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1260433/full#supplementary-material
1. WHO . (2015). Available at:https://ahpsr.who.int/publications/i/item/global-action-plan-on-antimicrobial-resistance. (Accessed July 10, 2023) (Online).
2. Lee, S , Mir, RA , Park, SH , Kim, D , Kim, HY , Boughton, RK, et al. Prevalence of extended-spectrum beta-lactamases in the local farm environment and livestock: challenges to mitigate antimicrobial resistance. Crit Rev Microbiol. (2020) 46:1–14. doi: 10.1080/1040841X.2020.1715339
3. Gelalcha, BD , Ensermu, DB , Agga, GE , Vancuren, M , Gillespie, BE , D'Souza, DH, et al. Prevalence of antimicrobial resistant and extended-Spectrum Beta-lactamase-producing Escherichia coli in dairy cattle farms in East Tennessee. Foodborne Pathog Dis. (2022) 19:408–16. doi: 10.1089/fpd.2021.0101
4. Tragesser, LA , Wittum, TE , Funk, JA , Winokur, PL , and Rajala-Schultz, PJ . Association between ceftiofur use and isolation of Escherichia coli with reduced susceptibility to ceftriaxone from fecal samples of dairy cows. Am J Vet Res. (2006) 67:1696–700. doi: 10.2460/ajvr.67.10.1696
5. Gonggrijp, MA , Santman-Berends, IMGA , Heuvelink, AE , Buter, GJ , van Schaik, G , Hage, JJ, et al. Prevalence and risk factors for extended spectrum beta-lactamase and AmpC-producing Escherichia coli in dairy farms. J Dairy Sci. (2016) 99:9001–13. doi: 10.3168/jds.2016-11134
6. Gelalcha, BD , Agga, GE , and Kerro Dego, O . Antimicrobial Usage for the Management of Mastitis in the USA: Impacts on Antimicrobial Resistance and Potential Alternative Approaches. In the Mastitis in Dairy Cattle, Sheep and goats, ed. O. Kerro Dego (London, UK: IntechOpen), 117–137.
7. Ruegg, PL . What is success? A narrative review of research evaluating outcomes of antibiotics used for treatment of clinical mastitis. Front Vet Sci. (2021) 8:639641. doi: 10.3389/fvets.2021.639641
8. Redding, LE , Bender, J , and Baker, L . Quantification of antibiotic use on dairy farms in Pennsylvania. J Dairy Sci. (2019) 102:1494–507. doi: 10.3168/jds.2018-15224
9. USDA–APHIS–VS–CEAH–NAHMS . Dairy: Milk Quality, Milking Procedures, and Mastitis on U.S. Dairies, 2014. (2016). Fort Collins, CO, USA. Available at:https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy14/Dairy14_dr_Mastitis.pdf, (Accessed July 10, 2023) (Online).
10. USDA:APHIS:VS:CEAH . Dairy: Part II: Changes in the U.S. Dairy Cattle Industry, 1991–2007. (2007) Fort Collins, CO, USA. Available at:https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_is_PartII_Highlights_1.pdf, (Accessed July 10, 2023) (Online).
11. FDA . Antimicrobial Use and Resistance in Animal Agriculture in the United States 2016–2019. (2022) Summary Report of FDA Center for Veterinary Medicine, Laurel, MD, USA, Available at: https://www.fda.gov/media/159544/download, (Accessed July 10, 2023) (Online).
12. Nora, FD , SMG, S , Apley, MD , Singer, RS , and Brian, V . Lubbers antimicrobial use quantification in adult dairy cows – part 3 – use measured by standardized regimens and grams on 29 dairies in the United States. Wiley. (2020) 67:82–93. doi: 10.1111/zph.12773
13. WHO . Critically important Antimicrobials for Human Medicine. 6th Revision: Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance due to Non-Human Use. Geneva, Switzerland: WHO (2018).
14. Paterson, DL , and Bonomo, RA . Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. (2005) 18:657–86. doi: 10.1128/CMR.18.4.657-686.2005
15. Malloy, AM , and Campos, JM . Extended-spectrum beta-lactamases: a brief clinical update. Pediatr Infect Dis J. (2011) 30:1092–3. doi: 10.1097/INF.0b013e31823c0e9d
16. Gelalcha, B , and Kerro, DO . Extended-Spectrum Beta-lactamases producing Enterobacteriaceae in the USA dairy cattle farms and implications for public health. Antibiotics. (2022) 11:1313. doi: 10.3390/antibiotics11101313
17. Carattoli, A . Animal reservoirs for extended spectrum beta-lactamase producers. Clin Microbiol Infect. (2008) 14:117–23. doi: 10.1111/j.1469-0691.2007.01851.x
18. Bush, K . Proliferation and significance of clinically relevant beta-lactamases. Ann N Y Acad Sci. (2013) 1277:84–90. doi: 10.1111/nyas.12023
19. Founou, RC , Founou, LL , Allam, M , Ismail, A , and Essack, SY . Whole genome sequencing of extended Spectrum beta-lactamase (ESBL)-producing Klebsiella pneumoniae isolated from hospitalized patients in KwaZulu-Natal, South Africa. Sci Rep. (2019) 9:6266. doi: 10.1038/s41598-019-42672-2
20. Cormier, A , Zhang, PLC , Chalmers, G , Weese, JS , Deckert, A , Mulvey, M, et al. Diversity of CTX-M-positive Escherichia coli recovered from animals in Canada. Vet Microbiol. (2019) 231:71–5. doi: 10.1016/j.vetmic.2019.02.031
21. Tadesse, DA , Li, C , Mukherjee, S , Hsu, CH , Bodeis Jones, S , Gaines, SA, et al. Whole-genome sequence analysis of CTX-M containing Escherichia coli isolates from retail meats and cattle in the United States. Microb Drug Resist. (2018) 24:939–48. doi: 10.1089/mdr.2018.0206
22. Coque, TM , Baquero, F , and Canton, R . Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. (2008) 13:19044. doi: 10.2807/ese.13.47.19044-en
23. Gniadkowski, M . Evolution and epidemiology of extended-spectrum beta-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. (2001) 7:597–608. doi: 10.1046/j.1198-743x.2001.00330.x
24. Taylor, EA , Ossa-Trujillo, C , Vinasco, J , Jordan, ER , García Buitrago, JA , Hagevoort, R, et al. Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extended-spectrum beta-lactam use in dairy cattle. Lett Appl Microbiol. (2021) 72:220–4. doi: 10.1111/lam.13419
25. Gelalcha, BD , Brown, S , Crocker, H , Agga, GE , and Kerro Dego, O . Regulation mechanisms of virulence genes in Enterohemorrhagic Escherichia coli. Foodborne Pathog Dis. (2022) 19:598–612. doi: 10.1089/fpd.2021.0103
26. Schaufler, K , Semmler, T , Wieler, LH , Wohrmann, M , Baddam, R , Ahmed, N, et al. Clonal spread and interspecies transmission of clinically relevant ESBL-producing Escherichia coli of ST410--another successful pandemic clone? FEMS Microbiol Ecol. (2016) 92:fiv155. doi: 10.1093/femsec/fiv155
27. Mukerji, S , O'Dea, M , Barton, M , Kirkwood, R , Lee, T , and Abraham, S . Development and transmission of antimicrobial resistance among gram-negative bacteria in animals and their public health impact. Essays Biochem. (2017) 61:23–35. doi: 10.1042/EBC20160055
28. Lazarus, B , Paterson, DL , Mollinger, JL , and Rogers, BA . Do human Extraintestinal Escherichia coli infections resistant to expanded-Spectrum Cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis. (2015) 60:439–52. doi: 10.1093/cid/ciu785
29. Fey, PD , Safranek, TJ , Rupp, ME , Dunne, EF , Ribot, E , Iwen, PC, et al. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N Engl J Med. (2000) 342:1242–9. doi: 10.1056/NEJM200004273421703
30. Afema, JA , Ahmed, S , Besser, TE , Jones, LP , Sischo, WM , and Davis, MA . Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington state. Appl Environ Microbiol. (2018) 84:e02430-17. doi: 10.1128/AEM.02430-17
31. Agga, GE , Schmidt, JW , and Arthur, TM . Antimicrobial-resistant fecal bacteria from Ceftiofur-treated and non antimicrobial-treated comingled beef cows at a cow-calf operation. Microb Drug Resist. (2016) 22:598–608. doi: 10.1089/mdr.2015.0259
32. Wittum, TE , Mollenkopf, DF , Daniels, JB , Parkinson, AE , Mathews, JL , Fry, PR, et al. CTX-M-type extended-spectrum beta-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog Dis. (2010) 7:1575–9. doi: 10.1089/fpd.2010.0615
33. Davis, MA , Sischo, WM , Jones, LP , Moore, DA , Ahmed, S , Short, DM, et al. Recent emergence of Escherichia coli with cephalosporin resistance conferred by blaCTX-M on Washington state dairy farms. Appl Environ Microbiol. (2015) 81:4403–10. doi: 10.1128/AEM.00463-15
34. Berge, ACB , Adaska, JM , and Sischo, WM . Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multi-drug-resistant Salmonella enterica subsp enterica serovar Newport. Appl Environ Microbiol. (2004) 70:318–23. doi: 10.1128/AEM.70.1.318-323.2004
35. CDC . (2019). Antibiotic Resistance Threats in the United States. Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf, (Accessed August, 2023).
36. Thaden, JT , Fowler, VG , Sexton, DJ , and Anderson, DJ . Increasing incidence of extended-Spectrum beta-lactamase-producing Escherichia coli in community hospitals throughout the southeastern United States. Infect Control Hosp Epidemiol. (2016) 37:49–54. doi: 10.1017/ice.2015.239
37. Mollenkopf, DF , Weeman, MF , Daniels, JB , Abley, MJ , Mathews, JL , Gebreyes, WA, et al. Variable within- and between-herd diversity of CTX-M Cephalosporinase-bearing Escherichia coli isolates from dairy cattle. Appl Environ Microbiol. (2012) 78:4552–60. doi: 10.1128/AEM.00373-12
38. Winokur, PL , Brueggemann, A , Desalvo, DL , Hoffmann, L , Apley, MD , Uhlenhopp, EK, et al. Animal and human multidrug-resistant, cephalosporin-resistant Salmonella isolates expressing a plasmid-mediated CMY-2 AmpC β-lactamase. Antimicrob Agents Chemother. (2000) 44:2777–83. doi: 10.1128/AAC.44.10.2777-2783.2000
39. Heider, LC , Funk, JA , Hoet, AE , Meiring, RW , Gebreyes, WA , and Wittum, TE . Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am J Vet Res. (2009) 70:389–93. doi: 10.2460/ajvr.70.3.389
40. HMNF, JDP . Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production – a threat around the world. Heliyon. (2020) 6:e03206. doi: 10.1016/j.heliyon.2020.e03206
41. Thrusfield, M , Christley, R , Brown, H , Diggle, PJ , French, N , Howe, K, et al. Veterinary Epidemiology. 4th ed Wiley-Blackwell (2018) Toronto, Ontario, Canada.
42. Doern, CD , and Butler-Wu, SM . Emerging and future applications of matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry in the clinical microbiology laboratory: a report of the Association for Molecular Pathology. J Mol Diagn. (2016) 18:789–802. doi: 10.1016/j.jmoldx.2016.07.007
43. Benti, D , Gelalcha, RIM , Gelgie, A , and Dego, OK . Molecular epidemiology and pathogenomics of extended-spectrum beta-lactamases producing E. coli and K. pneumoniae isolates from bulk tank milk in Tennessee, USA. Food Microbiol. (2023) 14:1283165. doi: 10.3389/fmicb.2023.1283165
44. Institute CLs . M100.Performance Standards for Antimicrobial Susceptibility Testing. 33rd Edition, (2023) CLSI, Malvern, PA, USA.
45. Magiorakos, AP , Srinivasan, A , Carey, RB , Carmeli, Y , Falagas, ME , Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
46. Sawant, AA , Sordillo, LM , and Jayarao, BM . A survey on antibiotic usage in dairy herds in Pennsylvania. J Dairy Sci. (2005) 88:2991–9. doi: 10.3168/jds.S0022-0302(05)72979-9
47. Guri, A , Flaks-Manov, N , Ghilai, A , Hoshen, M , Flidel Rimon, O , Ciobotaro, P, et al. Risk factors for third-generation cephalosporin resistant Enterobacteriaceae in gestational urine cultures: a retrospective cohort study based on centralized electronic health records. PLoS One. (2020) 15:e0226515. doi: 10.1371/journal.pone.0226515
48. Pol, M , and Ruegg, PL . Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J Dairy Sci. (2007) 90:249–61. doi: 10.3168/jds.S0022-0302(07)72626-7
49. Kumar, N , Manimaran, A , Kumaresan, A , Sreela, L , Patbandha, TK , Tiwari, S, et al. Episodes of clinical mastitis and its relationship with duration of treatment and seasonality in crossbred cows maintained in organized dairy farm. Vet World. (2016) 9:75–9. doi: 10.14202/vetworld.2016.75-79
50. USDA. (United States Department of Agriculture) . Dairy 2014, Milk Quality, Milking Procedures, and Mastitis in the United States, 2014 USDA–APHIS–VS–CEAH– NAHMS (2016) Fort Collins, CO, USA.
51. Becker, C , and Stone, A . Dry Cow Therapy: Choosing the Best Protocol for your Dairy, Mississippi State University Extension publication 3290 (POD-11-18), Department of Animal and Dairy Science, Mississippi State University, (2018) Available at:https://extension.msstate.edu/sites/default/files/publications/publications/p3290.pdf, (Accessed July 10, 2023) (Online).
52. Singer, RS , Patterson, SK , and Wallace, RL . Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl Environ Microbiol. (2008) 74:6956–62. doi: 10.1128/AEM.01241-08
53. USDA . Dairy14 Part III. Health and Management Practices on U.S. Dairy Operations, 2014; USDA-APHIS-VS-CEAHNAHMS, Fort Collins, CO. Washington, DC, USA: USDA (2018).
54. Schrag, NFD , Apley, MD , Godden, SM , Lubbers, BV , and Singe, SR . Antimicrobial use quantification in adult dairy cows – part 1 – standardized regimens as a method for describing antimicrobial use. Zoonoses Public Health. (2020) 67:51–68. doi: 10.1111/zph.12766
55. Aarestrup, FM , Seyfarth, AM , Emborg, HD , Pedersen, K , Hendriksen, RS , and Bager, F . Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrobial Agents Chemomother. (2001) 45:2054–9. doi: 10.1128/AAC.45.7.2054-2059.2001
56. Berge, ACB , Atwill, ER , and Sischo, WM . Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young calves. Prev Vet Med. (2005) 69:25–38. doi: 10.1016/j.prevetmed.2005.01.013
57. Poppe, C , Martin, LC , Gyles, CL , Reid-Smith, R , Boerlin, P , McEwen, SA, et al. Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the Turkey poult intestinal tract. Appl Environ Microbiol. (2005) 71:1184–92. doi: 10.1128/AEM.71.3.1184-1192.2005
58. Fan, S , Foster, D , Miller, WG , Osborne, J , and Kathariou, S . Impact of ceftiofur administration in steers on the prevalence and antimicrobial resistance of Campylobacter spp. Microorganisms. (2021) 9:318. doi: 10.3390/microorganisms9020318
59. Foster, DM , Jacob, ME , Farmer, KA , Callahan, BJ , Theriot, CM , Kathariou, S, et al. Ceftiofur formulation differentially affects the intestinal drug concentration, resistance of fecal Escherichia coli, and the microbiome of steers. PLoS One. (2019) 14:e0223378. doi: 10.1371/journal.pone.0223378
60. Lalak, A , Wasyl, D , Zajac, M , Skarzynska, M , Hoszowski, A , Samcik, I, et al. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet Microbiol. (2016) 194:69–73. doi: 10.1016/j.vetmic.2016.01.023
61. Cao, H , Pradhan, AK , Karns, JS , Hovingh, E , Wolfgang, DR , Vinyard, BT, et al. Age-associated distribution of antimicrobial-resistant Salmonella enterica and Escherichia coli isolated from dairy herds in Pennsylvania, 2013-2015. Foodborne Pathog Dis. (2019) 16:60–7. doi: 10.1089/fpd.2018.2519
62. Ohnishi, MOA , Esaki, H , Harada, K , Sawada, T , Murakami, M , Marumo, K, et al. Herd prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2 β-lactamases among Japanese dairy farms. J Appl Microbiol. (2013) 115:282–9. doi: 10.1111/jam.12211
63. Schmid, AHS , Messelhäusser, U , Käsbohrer, A , Sauter-Louis, C , and Mansfeld, R . Prevalence of extended-spectrum β-lactamase-producing Escherichia coli on Bavarian dairy and beef cattle farms. Appl Environ Microbiol. (2013) 79:3027–32. doi: 10.1128/AEM.00204-13
64. Agga, GE , Galloway, HO , Netthisinghe, AMP , Schmidt, JW , and Arthur, TM . Tetracycline-resistant, third-generation cephalosporin-resistant, and extended-Spectrum beta-lactamase-producing Escherichia coli in a beef cow-calf production system. J Food Prot. (2022) 85:1522–30. doi: 10.4315/JFP-22-178
65. Masse, J , Larde, H , Fairbrother, JM , Roy, JP , Francoz, D , Dufour, S, et al. Prevalence of antimicrobial resistance and characteristics of Escherichia coli isolates from fecal and manure pit samples on dairy farms in the province of Quebec, Canada. Front Vet Sci. (2021) 8:654125. doi: 10.3389/fvets.2021.654125
66. Springer, HR , Denagamage, TN , Fenton, GD , Haley, BJ , Van Kessel, JAS , and Hovingh, EP . Antimicrobial resistance in fecal Escherichia coli and Salmonella enterica from dairy calves: a systematic review. Foodborne Pathog Dis. (2019) 16:23–34. doi: 10.1089/fpd.2018.2529
67. Markland, S , Weppelmann, TA , Ma, Z , Lee, S , Mir, RA , Teng, L, et al. High prevalence of cefotaxime resistant bacteria in grazing beef cattle: a cross sectional study. Front Microbiol. (2019) 10:176. doi: 10.3389/fmicb.2019.00176
68. Carey, AM , Capik, SF , Giebel, S , Nickodem, C , Pineiro, JM , Scott, HM, et al. Prevalence and profiles of antibiotic resistance genes mph(a) and qnrB in extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from dairy calf feces. Microorganisms. (2022) 10:411. doi: 10.3390/microorganisms10020411
69. Duse, A , Waller, KP , Emanuelson, U , Unnerstad, HE , Persson, Y , and Bengtsson, B . Risk factors for antimicrobial resistance in fecal Escherichia coli from preweaned dairy calves. J Dairy Sci. (2015) 98:500–16. doi: 10.3168/jds.2014-8432
70. Watson, E , Jeckel, S , Snow, L , Stubbs, R , Teale, C , Wearing, H, et al. Epidemiology of extended spectrum beta-lactamase E. coli (CTX-M-15) on a commercial dairy farm. Vet Microbiol. (2012) 154:339–46. doi: 10.1016/j.vetmic.2011.07.020
71. Heuvelink, AE , Gonggrijp, MA , Buter, RGJ , Ter Bogt-Kappert, CC , van Schaik, G , Velthuis, AGJ, et al. Prevalence of extended-spectrum and AmpC beta-lactamase-producing Escherichia coli in Dutch dairy herds. Vet Microbiol. (2019) 232:58–64. doi: 10.1016/j.vetmic.2019.04.005
72. Awosile, B , McClure, J , Sanchez, J , Rodriguez-Lecompte, JC , Keefe, G , and Heider, LC . Salmonella enterica and extended-spectrum cephalosporin-resistant Escherichia coli recovered from Holstein dairy calves from 8 farms in New Brunswick, Canada. J Dairy Sci. (2018) 101:3271–84. doi: 10.3168/jds.2017-13277
73. Khachatryan, AR , Hancock, DD , Besser, TE , and Call, DR . Role of calf-adapted Escherichia coli in maintenance of antimicrobial drug resistance in dairy calves. Appl Environ Microbiol. (2004) 70:752–7. doi: 10.1128/AEM.70.2.752-757.2004
74. Khachatryan, AR , Besser, TE , Hancock, DD , and Call, DR . Use of a nonmedicated dietary supplement correlates with increased prevalence of streptomycin-sulfa-tetracycline-resistant Escherichia coli on a dairy farm. Appl Environ Microbiol. (2006) 72:4583–8. doi: 10.1128/AEM.02584-05
75. Davis, GS , and Price, LB . Recent research examining links among Klebsiella pneumoniae from food, food animals, and human extraintestinal infections. Curr Environ Health Rep. (2016) 3:128–35. doi: 10.1007/s40572-016-0089-9
76. Snow, LC , Warner, RG , Cheney, T , Wearing, H , Stokes, M , Harris, K, et al. Risk factors associated with extended spectrum beta-lactamase Escherichia coli (CTX-M) on dairy farms in north West England and North Wales. Prev Vet Med. (2012) 106:225–34. doi: 10.1016/j.prevetmed.2012.03.009
77. Sato, T , Okubo, T , Usui, M , Yokota, S , Izumiyama, S , and Tamura, Y . Association of veterinary third-generation cephalosporin use with the risk of emergence of extended-Spectrum-cephalosporin resistance in Escherichia coli from dairy cattle in Japan. PLoS One. (2014) 9:e96101. doi: 10.1371/journal.pone.0096101
78. Chambers, L , Yang, Y , Littier, H , Ray, P , Zhang, T , Pruden, A, et al. Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin. PLoS One. (2015) 10:e0133764. doi: 10.1371/journal.pone.0133764
79. Barbosa, TM , and Levy, SB . The impact of antibiotic use on resistance development and persistence. Drug Resist Updat. (2000) 3:303–11. doi: 10.1054/drup.2000.0167
80. Kanwar, N , Scott, HM , Norby, B , Loneragan, GH , Vinasco, J , Cottell, JL, et al. Impact of treatment strategies on cephalosporin and tetracycline resistance gene quantities in the bovine fecal metagenome. Sci Rep. (2014) 4:5100. doi: 10.1038/srep05100
81. Jamali, H , Barkema, HW , Jacques, M , Lavallee-Bourget, EM , Malouin, F , Saini, V, et al. Invited review: incidence, risk factors, and effects of clinical mastitis recurrence in dairy cows. J Dairy Sci. (2018) 101:4729–46. doi: 10.3168/jds.2017-13730
82. Elghafghuf, A , Dufour, S , Reyher, K , Dohoo, I , and Stryhn, H . Survival analysis of clinical mastitis data using a nested frailty cox model fit as a mixed-effects Poisson model. Prev Vet Med. (2014) 117:456–68. doi: 10.1016/j.prevetmed.2014.09.013
83. Silva, AC , Laven, R , and Benites, NR . Risk factors associated with mastitis in smallholder dairy farms in Southeast Brazil. Animals (Basel). (2021) 11:2089. doi: 10.3390/ani11072089
84. Bradford, PA . Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. (2001) 14:933–51. doi: 10.1128/CMR.14.4.933-951.2001
85. Grohs, P , Tillecovidin, B , Caumont-Prim, A , Carbonnelle, E , Day, N , Podglajen, I, et al. Comparison of five media for detection of extended-spectrum Beta-lactamase by use of the wasp instrument for automated specimen processing. J Clin Microbiol. (2013) 51:2713–6. doi: 10.1128/JCM.00077-13
86. Rawat, D , and Nair, D . Extended-spectrum beta-lactamases in gram negative bacteria. J Glob Infect Dis. (2010) 2:263–74. doi: 10.4103/0974-777X.68531
87. Castanheira, M , Simner, PJ , and Bradford, PA . Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob Resist. (2021) 3:dlab092. doi: 10.1093/jacamr/dlab092
88. Okello, E , Williams, DR , ElAshmawy, WR , Adams, J , Pereira, RV , Lehenbauer, TW, et al. Survey on antimicrobial drug use practices in California preweaned dairy calves. Front Vet Sci. (2021) 8:636670. doi: 10.3389/fvets.2021.636670
89. Wener, KM , Schechner, V , Gold, HS , Wright, SB , and Carmeli, YT . Treatment with fluoroquinolones or with beta-lactam-beta-lactamase inhibitor combinations is a risk factor for isolation of extended-spectrum-beta-lactamase-producing Klebsiella species in hospitalized patients. Antimicrob Agents Chemother. (2010) 54:2010–6. doi: 10.1128/AAC.01131-09
90. Paterson, DL , Mulazimoglu, L , Casellas, JM , Ko, WC , Goossens, H , Von Gottberg, A, et al. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum beta-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin Infect Dis. (2000) 30:473–8. doi: 10.1086/313719
Keywords: ESBL Escherichia coli, prevalence, dairy farm, multidrug resistance, antimicrobial susceptibility, antimicrobial resistance, dairy cattle, calves
Citation: Gelalcha BD, Gelgie AE and Kerro Dego O (2023) Prevalence and antimicrobial resistance profiles of extended-spectrum beta-lactamase-producing Escherichia coli in East Tennessee dairy farms. Front. Vet. Sci. 10:1260433. doi: 10.3389/fvets.2023.1260433
Received: 17 July 2023; Accepted: 16 November 2023;
Published: 18 December 2023.
Edited by:
Toni L. Poole, United States Department of Agriculture, United StatesReviewed by:
Gizem Levent, Texas Tech University, United StatesCopyright © 2023 Gelalcha, Gelgie and Kerro Dego. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oudessa Kerro Dego, b2tlcnJvZGVAdXRrLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.