- Department of Animal and Poultry Science, College of Aburaihan, University of Tehran, Tehran, Iran
Introduction: Fat-tail in sheep is considered as an important energy reservoir to provide energy as a survival buffer during harsh challenges. However, fat-tail is losing its importance in modern sheep industry systems and thin-tailed breeds are more desirable. Using comparative transcriptome analysis to compare fat-tail tissue between fat- and thin-tailed sheep breeds provides a valuable approach to study the complex genetic factors associated with fat-tail development. However, transcriptomic studies often suffer from issues with reproducibility, which can be improved by integrating multiple studies based on a meta-analysis.
Methods: Hence, for the first time, an RNA-Seq meta-analysis on sheep fat-tail transcriptomes was performed using six publicly available datasets.
Results and discussion: A total of 500 genes (221 up-regulated, 279 down-regulated) were identified as differentially expressed genes (DEGs). A jackknife sensitivity analysis confirmed the robustness of the DEGs. Moreover, QTL and functional enrichment analysis reinforced the importance of the DEGs in the underlying molecular mechanisms of fat deposition. Protein-protein interactions (PPIs) network analysis revealed the functional interactions among the DEGs and the subsequent sub-network analysis led to identify six functional sub-networks. According to the results of the network analysis, down-regulated DEGs in green and pink sub-networks (like collagen subunits IV, V, and VI, integrins 1 and 2, SCD, SCD5, ELOVL6, ACLY, SLC27A2, and LPIN1) may impair lipolysis or fatty acid oxidation and cause fat accumulation in tail. On the other hand, up-regulated DEGs, especially those are presented in green and pink sub-networks (like IL6, RBP4, LEPR, PAI-1, EPHX1, HSD11B1, and FMO2), might contribute to a network controlling fat accumulation in the tail of sheep breed through mediating adipogenesis and fatty acid biosynthesis. Our results highlighted a set of known and novel genes/pathways associated with fat-tail development, which could improve the understanding of molecular mechanisms behind fat deposition in sheep fat-tail.
Introduction
In recent years, with the growth of world population, there is a considerable demand for the agricultural improvement, especially for meat and milk of animals. In modern agriculture, sheep play an important role in the production of meat, wool and milk. Currently, there are a lot of sheep breeds over a wide geographical range worldwide with a large variation in many phenotypic traits. One of the most important phenotypic traits in sheep is the ability to store of fat in the tail. In this regard, generally sheep breeds can be divided into two main groups (thin- and fat-tailed breeds) and five subgroups including fat-long, fat-short, fat-rumped, thin-long, and thin-short tailed sheep breeds (1). It is well-known that the fat-tailed sheep breeds are evolved from its thin-tailed ancestor (~5,000 years ago), by the principle of selective breeding (natural or artificial selection) and following initial domestication, in response to hazardous environments (2, 3). Fat-tail is considered as an important energy reservoir for the animal to provide energy as a survival buffer during harsh challenges such as drought and food deprivation periods like winter, when pasture is dormant (4). Nowadays, fat-tail is losing its importance and lean-tailed breeds are more desirable, which can be explained by providing three reasons: (1) In modern sheep farming systems, intensive or semi-intensive feeding systems are preferred, which cause fat-tail be no longer important as an energy source; (2) fat deposition led to a higher energetic cost than accretion of an equivalent amount of lean tissue and decreases the feed efficiency (5); and (3) in modern society, consumers prefer low-fat foods to be healthy. Therefore, both customers and producers prefer thin-tailed sheep (6). It is estimated that the fat-tailed sheep breeds constitute about 26% of the world sheep population (7). Hence, study of the fat deposition in sheep tail is of great importance to understand the background molecular mechanisms of the fat-tail development as well as to develop the new breeding strategies to produce improved breeding sheep herds.
Up to now, several transcriptome-based studies have been conducted to elucidate the factors and molecular mechanisms responsible for the differences in fat deposition between different sheep breeds (6, 8–17). Earlier studies were focused on single gene to identify the candidate genes involved in regulating the fat-tail formation. In this regard, higher expression of the leptin (LEP) gene in fat lines of Coopworth sheep was reported for different fat tissues when comparing fat lines and lean lines in Coopworth sheep breed (18). In our previous study, fatty acid binding protein 4 (FABP4) gene was suggested as a candidate gene relevant to fat deposition in sheep breeds (8). Moreover, uncoupling protein 1 (UCP1) (15) and cell death-inducing DFFA-like effector c (CIDEC) (19) genes were reported as other candidate genes involved in regulation of fat deposition and mobilization in tail of sheep breeds.
In recent years, rapid development of next generation sequencing (NGS) technologies has lowered the barriers to perform high-throughput gene expression profiling through RNA sequencing (RNA-Seq) approach. This method made it possible to measure the expression of thousands of genes simultaneously, which enables us to better understand the genetic factors associated with phenotypic differences in sheep fat-tail. To date, several studies have been employed this approach to identify the genes linked to fat-tail deposition by conducting a comparative transcriptome study on the fat- and thin-tailed sheep breeds (6, 8–15, 17, 19).
A review of these studies indicates the variability in their results including different set of DEGs or inconsistent gene expression patterns, which can be attributed to different bioinformatics pipeline used, number of biological replicates and other limitations coming from the different nature of the samples. In this context, a meta-analysis of several independent studies focused on a specific biological question, offers a useful approach to increase the statistical power. Additionally, combining evidence from several studies increases the reliability and robustness of the results (6, 11–14, 17, 20). In this study, for the first time a meta-analysis of six independent RNA-Seq studies was performed to provide a basis for the identification of mechanisms underlying fat deposition in sheep tail, which can be exploited in future breeding strategies.
Materials and methods
RNA-Seq datasets
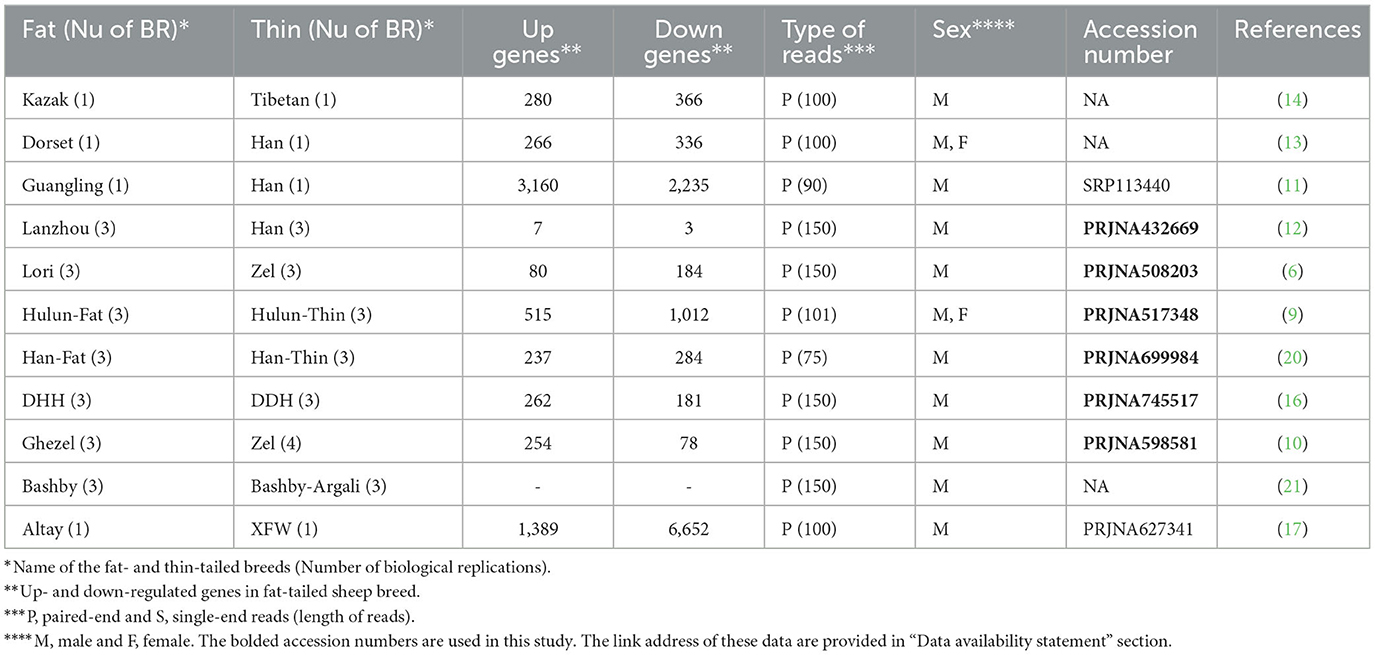
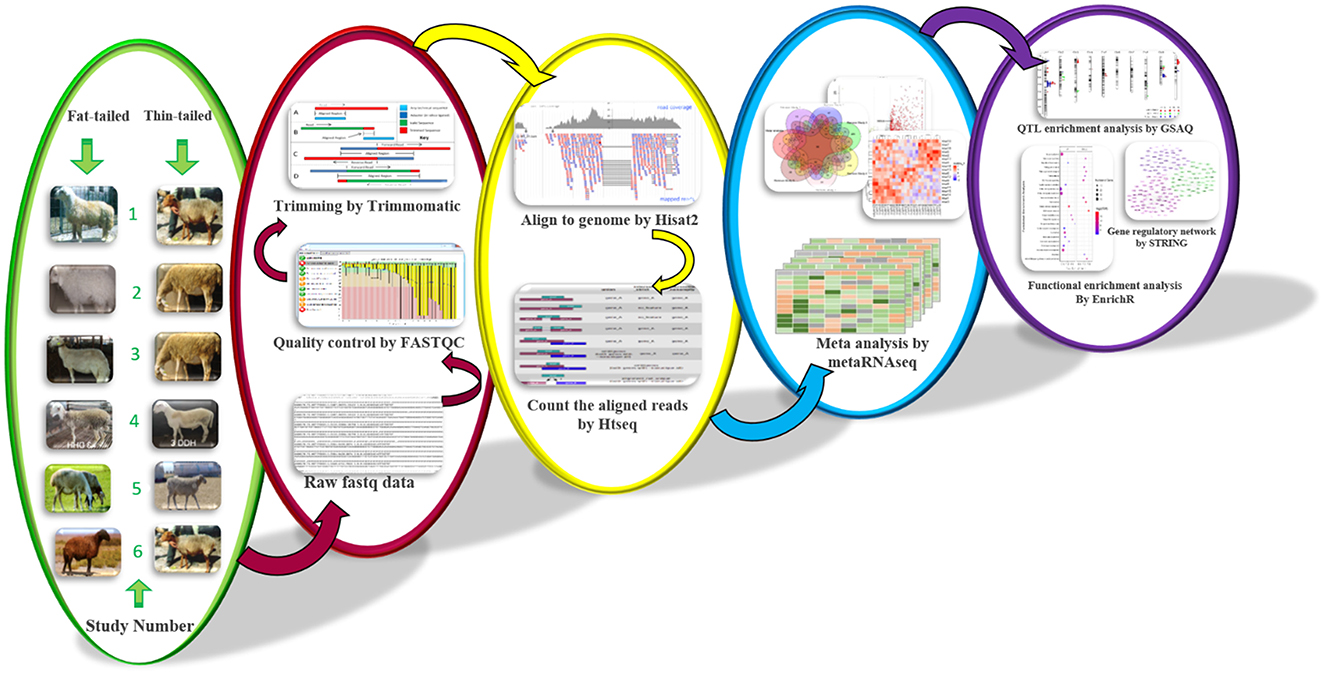
A comprehensive review of authoritative papers was conducted to find the studies that performed gene expression profiling by RNA-Seq method to identify molecular mechanisms affecting sheep fat-tails (Table 1, Supplementary material 1). Out of 11 identified studies, four studies (11, 13, 14, 17) had one biological replication per breed and data of one study was not available (17) in the public databases. Therefore, six datasets were remained to be used in this study. The samples of these datasets were related to fat-tail tissue of adult male sheep with at least three biological replications per breed (accession numbers of these studies are bolded in Table 1). Only male samples of Fan et al. (9) study were used. RNA-Seq reads for the selected studies (Table 1) were retrieved from the NCBI's sequence read archive database (SRA: https://trace.ncbi.nlm.nih.gov/Traces/sra/). All samples of each study were processed with the same bioinformatic pipeline, explained as follow (Figure 1).
Differential expression analysis
The quality of RNA-Seq reads was checked using FastQC (v0.11.5) software (22). Trimmomatic (v0.38) software (23) was applied to remove low-quality reads/bases and adaptor sequences based on its adaptive trimming algorithm, maximum information (MAXINFO), to balance the benefits of retaining longer reads against the drawback of having low-quality bases. The other trimming criteria were TRAILING: 20, MAXINFO: 80: 0.8, MINLEN: 80. FastQC was used again to re-evaluate the quality of the clean reads. The clean reads were then aligned to the ENSEMBL ovine reference genome (rambouillet_v1.0.104) using HISAT2 (24) software (version 2.1.0) based on the default parameters. A list of exon-exon junctions extracted from the Ensembl ovine GTF file (rambouillet_v1.0.104) was applied to guide the read mapping. To quantify the aligned reads to annotated genes, python script HTSeq-count (version 2.7.3) tool (25) was used (union intersection mode) according to Ensembl ovine GTF file (rambouillet_v1.0.104) and a gene expression matrix was created for each study. Genes with low read counts (< 10 reads across all samples) were filtered out. DESeq2 (v 1.32.0) was used to normalize and perform differential expression analysis for individual studies using the median-of-ratios method as a normalization factor (26). DESeq2 used Cox-Reid adjusted profile likelihood approach to estimate dispersions (27).
RNA-Seq meta-analysis
Raw p-values obtained from previous step were applied to perform meta-analysis using metaRNAseq R package (v1.0.7) (28). There are two p-value combination methods to combine p-values from different studies in this package including Fisher's and inverse normal methods. It is reported that both methods provide similar results. Using these methods enable us to combine results from heterogeneous datasets directly to identify commonly regulated genes among all studies. Here, to minimize the false positive results, both methods were used and differentially expressed genes (DEGs) with a false discovery rate (FDR) < 0.05 found in common between two methods were considered statistically significant. One of the limitations of p-value-based methods is that the combined p-values are estimated regardless of the expression patterns in the different studies. In other words, some genes are significantly up-regulated in some studies, while in the other studies they are significantly down-regulated, which may not reflect the biological reality (28). This limitation can be overcome by selecting the DEGs that showed the same expression pattern in most of studies. In the present study, out of the DEGs that were common between the two methods, only those genes that showed the same expression trend (up or down regulated) in more than half of the datasets, were finally considered as DEGs.
Jackknife analysis
One of the concerns in meta-analysis is that the identified DEGs can be affected by the properties of a single dataset (29). Hence, to evaluate the robustness of the meta-analysis results, a Jackknife sensitivity analysis was applied (30). To do this, meta-analysis was repeated (as described above) six times, each time including all the datasets except one and six sets of DEGs were generated. Finally, number of presence/absence of DEGs in the six sub meta-analysis was compared with the main meta-analysis.
Functional enrichment analysis
To gain further insight into the biological processes and pathways significantly enriched in DEGs, functional enrichment analysis was performed. To do this, EnrichR web-based tool was applied, using default parameters, focusing on gene ontology (GO; biological process) and the kyoto gene and genome encyclopedia (KEGG) (31). Also, up- and down-regulated DEGs were submitted separately and FDR < 0.05 was considered for the selection of the significantly enriched GO terms and pathways.
QTL enrichment analysis
To determine if the DEGs are located in the QTLs associated with fat metabolism, a co-localization analysis was applied. To do this, all QTLs related to fatness were obtained from sheep AnimalQTLdb database (32). Then, enrichment of the DEGs within the QTLs were analyzed using GSAQ R package based on Gene Set Validation with QTL (GSVQ) method (33).
Network analysis
To reveal the protein-protein interactions (PPIs) among the DEGs, STRING (34) database (v11.5) was applied using Ovis aries as the reference organism. The PPIs networks were constructed for up- and down-regulated DEGs, separately. Confidence score < 0.4 (6) was considered to discard unreliable PPIs. Moreover, sub-networks within the networks were identified based on the k-means clustering approach (three clusters) in the database. Cytoscape software (version 3.9) was used to visualize the networks (35).
Results
RNA-Seq data analysis
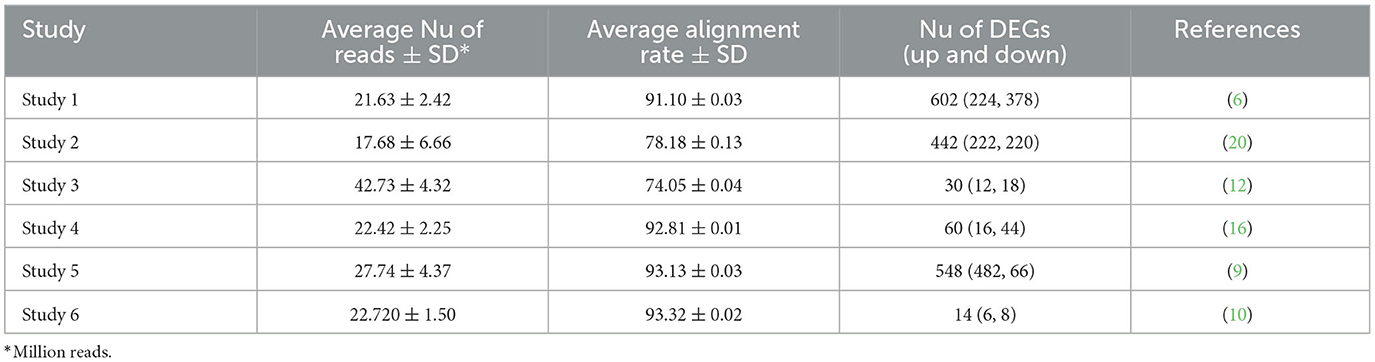
Six RNA-Seq datasets were analyzed including 18 fat-tailed and 19 thin-tailed sheep breeds (Table 1). In total, more than 952 million paired-end raw reads were obtained for the six datasets, ranging from 7.5 to 50.5 million per sample. Among these, 468 and 484 million reads were belonged to fat- and thin-tailed breeds, respectively. Only 1.6 million reads (~0.002%) were removed after quality control and filtering, which shows the quality of the datasets. The clean reads were then aligned to the sheep genome with an average of 87.26% read mapping rate (Table 2 and Supplementary material 1).
Meta-analysis
Differential gene expression analysis was performed for each study and the raw p-values of all studies were integrated based on our meta-analysis approach. After multiple testing correction, 761 DEGs were identified. Inspecting the DEGs to have the same log-fold change direction across most of the studies led to identification of 500 DEGs including 221 up- and 279 down-regulated genes in fat- against thin-tailed sheep breeds. Results of the differential expression analysis per study and meta-analysis can be found in Table 2 and Supplementary material 2. To illustrate the common DEGs shared among the individual studies and meta-analysis, a venn diagram was constructed (Figure 2). Out of 500 DEGs, 330 DEGs were shared with the other studies and 170 DEGs were identified only through the meta-analysis. Moreover, most of the identified DEGs from each individual study were not shared with the other studies. No DEG was found to be common among all the studies and meta-analysis.
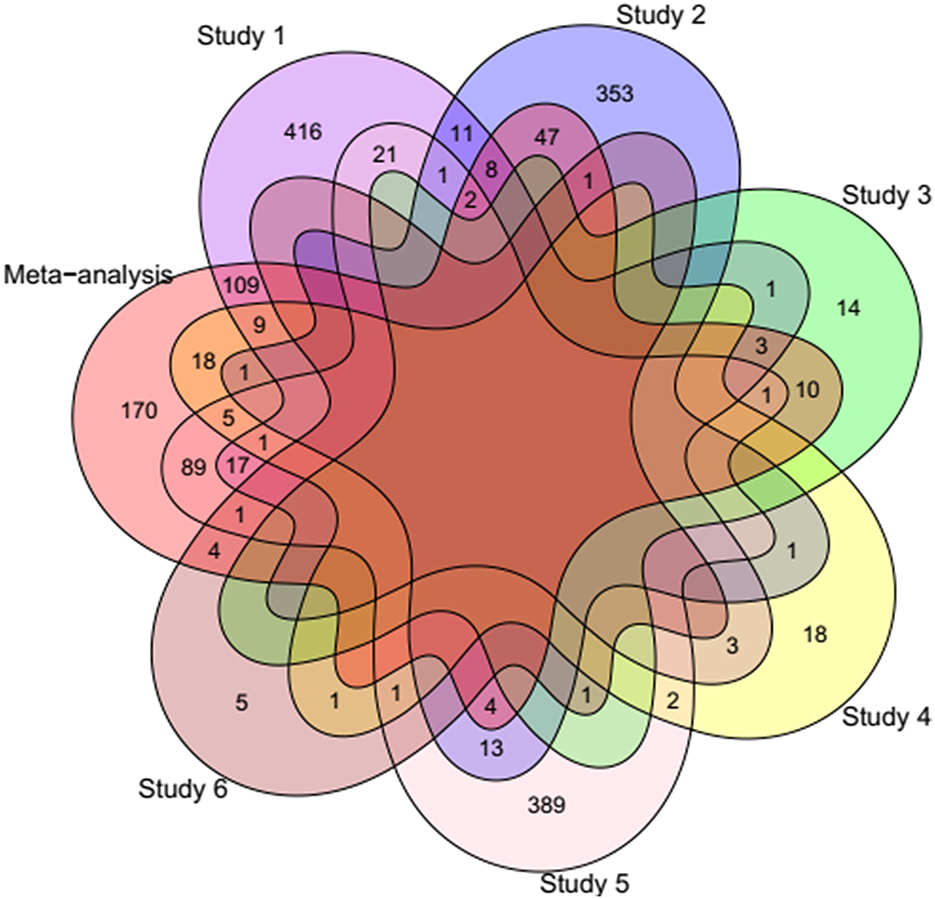
Figure 2. Venn diagram showing the number of common DEGs shared among the individual studies and meta-analysis. Study 1 (6), Study 2 (20), Study 3 (12), Study 4 (16), Study 5 (9), and Study 6 (10).
Jackknife sensitivity analysis
To confirm that our meta-analysis results were not affected by any single study, a jackknife sensitivity analysis was conducted by iteratively removing one dataset at a time. Figure 3 shows the common DEGs among the six Jackknife tests and the full meta-analysis results. All the identified DEGs by meta-analysis (except two genes ASIC3 and MYBPC1) were identified in at least one Jackknife test. On the other hand, more than 80% of the DEGs (403 genes including 232 down- and 171 up-regulated) were present in at least 50% of the Jackknife test results (oval circles in Figures 4, 5 represent these genes). Sixty-eight DEGs pass all six jackknife analyses and showed complete consistency over the tests, indicative of their importance as they can be considered as a more robust set of potential candidates.
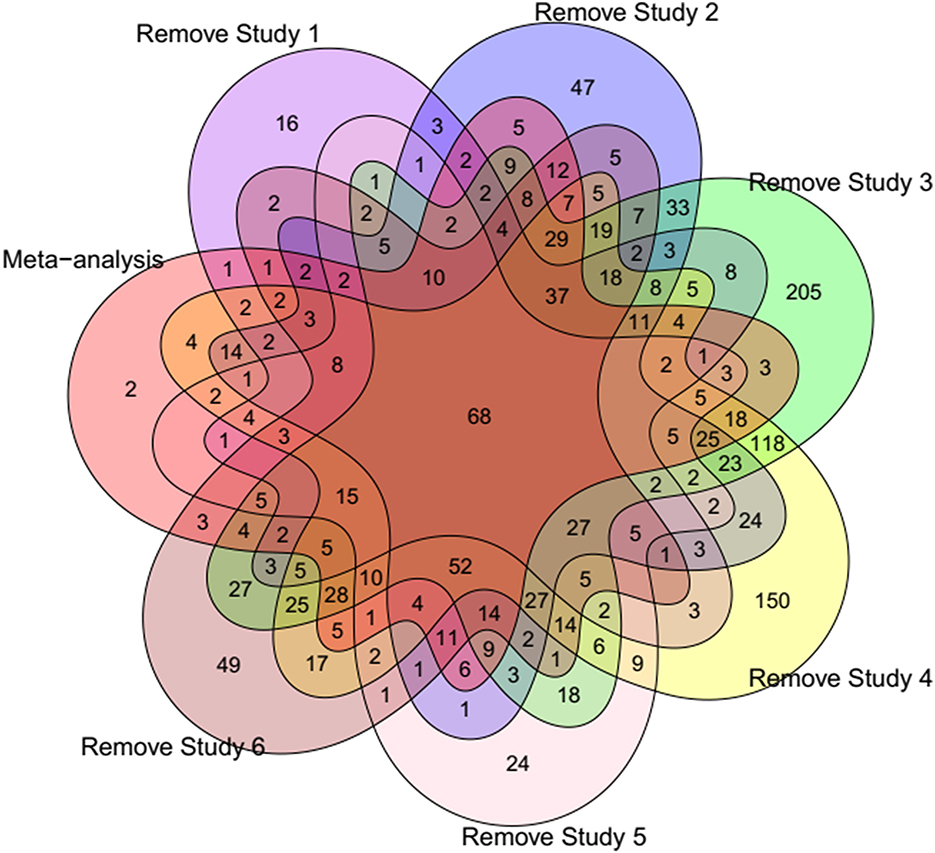
Figure 3. Venn diagram showing the number of common DEGs among different Jackknife tests and meta-analysis results. Study 1 (6), Study 2 (20), Study 3 (12), Study 4 (16), Study 5 (9), and Study 6 (10).
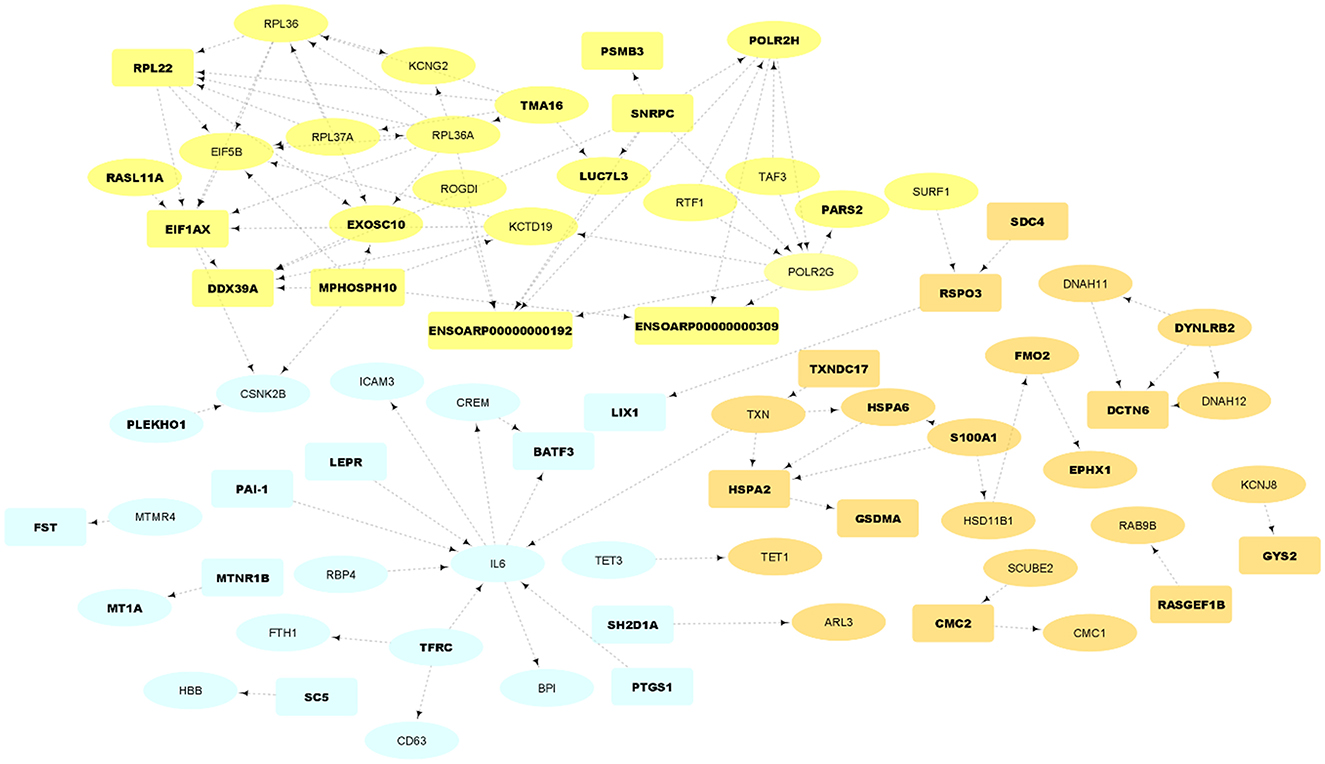
Figure 4. PPI network and functional clusters of the up-regulated DEGs. Orange, yellow and light blue nodes represent orange, yellow and light blue sub-networks, respectively. Oval circles indicate the DEGs that were presented in at least 50% of the Jackknife test results. The DEGs that were located in QTL regions related to fatness are represented as bold texts.
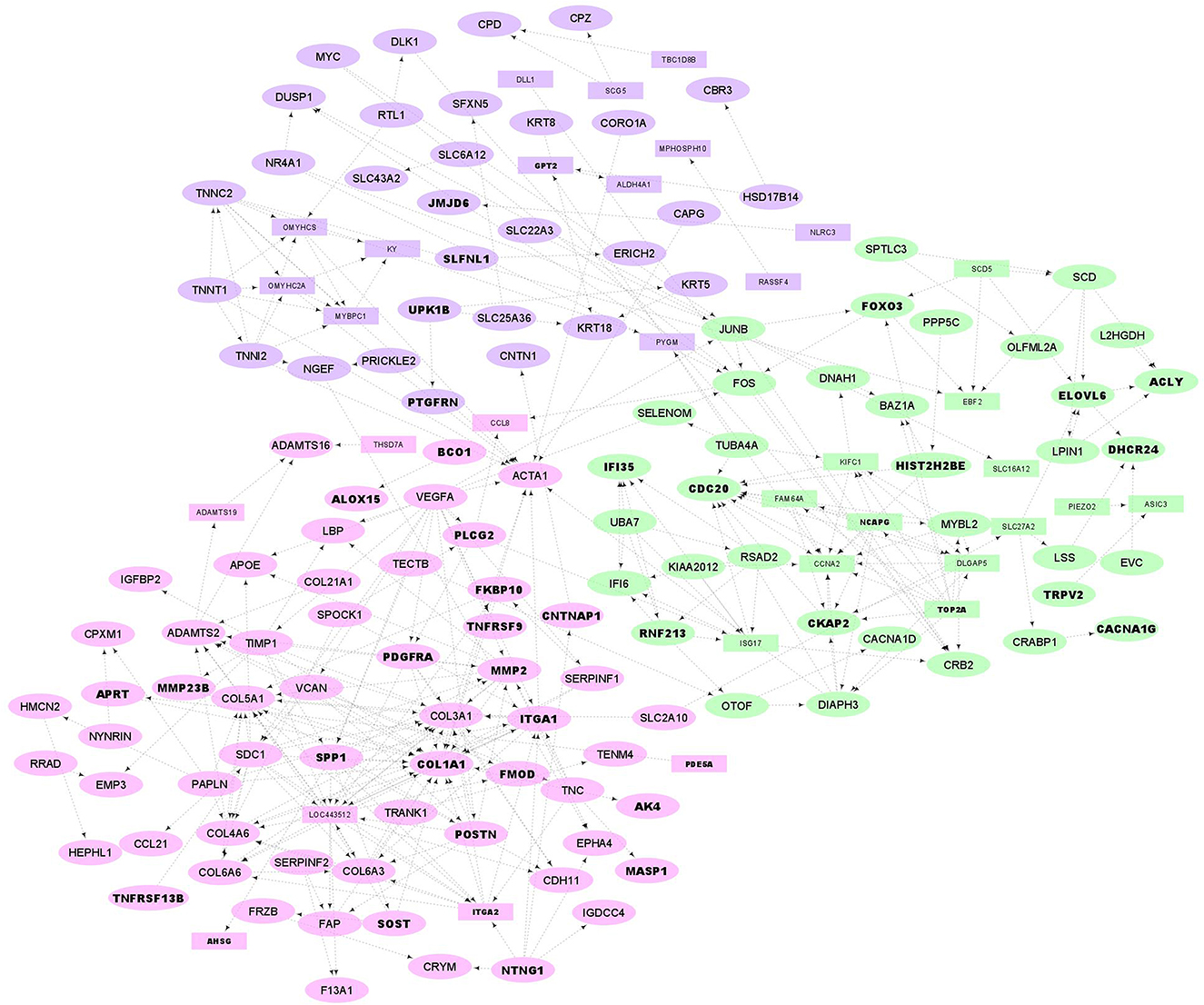
Figure 5. PPI network and functional clusters of the down-regulated DEGs. Green, pink and purple nodes represent green, pink and purple clusters, respectively. Oval circles indicate the DEGs that were presented in at least 50% of the Jackknife test results. The DEGs that were located in QTL regions related to fatness are represented as bold texts.
Functional enrichment analysis
To identify the main biological processes or pathways that the 500 DEGs (including 221 up- and 279 down-regulated genes in fat- against thin-tailed sheep breeds) may be involved in, functional enrichment analysis was performed. In up-regulated genes (higher expression in fat-tailed breeds), none of them were significant at FDR < 0.05 (Supplementary material 3). In total, 26 biological processes and seven KEGG were significantly enriched in down-regulated DEGs (lower expression in fat-tailed breeds) at FDR < 0.05 (Supplementary material 3). It is worthy to highlight the significant GO terms related to fat metabolism including “unsaturated fatty acid biosynthetic process” and “fatty-acyl-CoA biosynthetic process” in the down-regulated DEGs. Thus, it is reasonable to infer that these genes might be related to fat-tail development.
QTL enrichment analysis
A total of 98 annotated QTLs related to fatness were obtained from the sheep AnimalQTLdb database (32). Totally, 113 genes (including 50 up- and 63 down-regulated as represented in bold text in Figures 4, 5) were significantly (p-value = 0.018) located within coordinates of 16 QTLs (Supplementary material 4), which were located in chromosomes 1, 6, 8, 10, 11, 12, 18, 14, 16, 19, and 23. These QTLs pertained to nine different traits including internal fat amount, fat weight in carcass, carcass fat percentage, muscle depth at third lumbar, dressing percentage, subcutaneous fat area, subcutaneous fat thickness, carcass length and lean meat yield percentage. Out of 113 genes, 35 up- and 55 down-regulated DEGs were present in at least 50% of the Jackknife test results (~80%). To check whether the identified DEGs per individual study are significantly enriched in the QTLs, this analysis was performed on the DEGs of each study, separately. Out of six study, only DEGs of the study one was significantly enriched in the QTLs.
PPI network analysis
To construct an PPI network of DEGs (500 DEGs including 221 up- and 279 down-regulated genes in fat- against thin-tailed sheep breeds), where the nodes are proteins and the edges represent the predicted functional associations, STRING database (34) was used. ENSEMBL accession numbers of DEGs were first annotated and 361 gene names (135 up and 226 down) were found. Of these, 125 and 218 genes were matched with the database and applied to construct the PPI network in up- and down-regulated DEGs, respectively. A total of 72 nodes with 98 edges (known or predicted interactions) composed the final PPIs network of up-regulated DEGs (PPI enrichment p-value = 0.002, Figure 4). Further, the gene network interaction of down-regulated DEGs included 157 nodes with 337 edges (PPI enrichment p-value = < 1.0e−16, Figure 4).
The resulting PPIs networks were subjected to module analysis with k means clustering approach and three significant sub-networks were identified in each network. In this regard, three sub-networks including 22 nodes and 20 edges in sub-network orange (p-value = 6.51E−09), 24 nodes and 55 edges in yellow sub-network (p-value = 5.49E−11) and 20 nodes and 16 edges in light-blue sub-network (p-value = 2.89E−06) were found in up-regulated DEGs (Figure 4). Functional enrichment analysis of the clusters showed only six significant biological processes in orange sub-network including “gene expression,” “RNA metabolic process,” and “translation.” On the other hand, 34 nodes and 41 edges in green sub-network (p-value < 1.0e−16), 64 nodes and 90 edges in pink sub-network (p-value < 1.0e−16) and 51 nodes and 105 edges in purple sub-network (p-value < 1.0e−16) were found in the down-regulated DEGs (Figure 5). Genes of green sub-network were significantly enriched in “Lipid biosynthetic process” biological process (FDR < 0.04) and “Biosynthesis of unsaturated fatty acids” KEGG pathway (FDR < 0.01). Moreover, 17 biological processes (such as “Extracellular matrix organization” and “Anatomical structure morphogenesis”) and 11 KEEG (such as “ECM-receptor interaction,” “PI3K-Akt signaling pathway,” and “Focal adhesion”) were identified in pink sub-network (Figure 6). No significant term was found in purple sub-network.
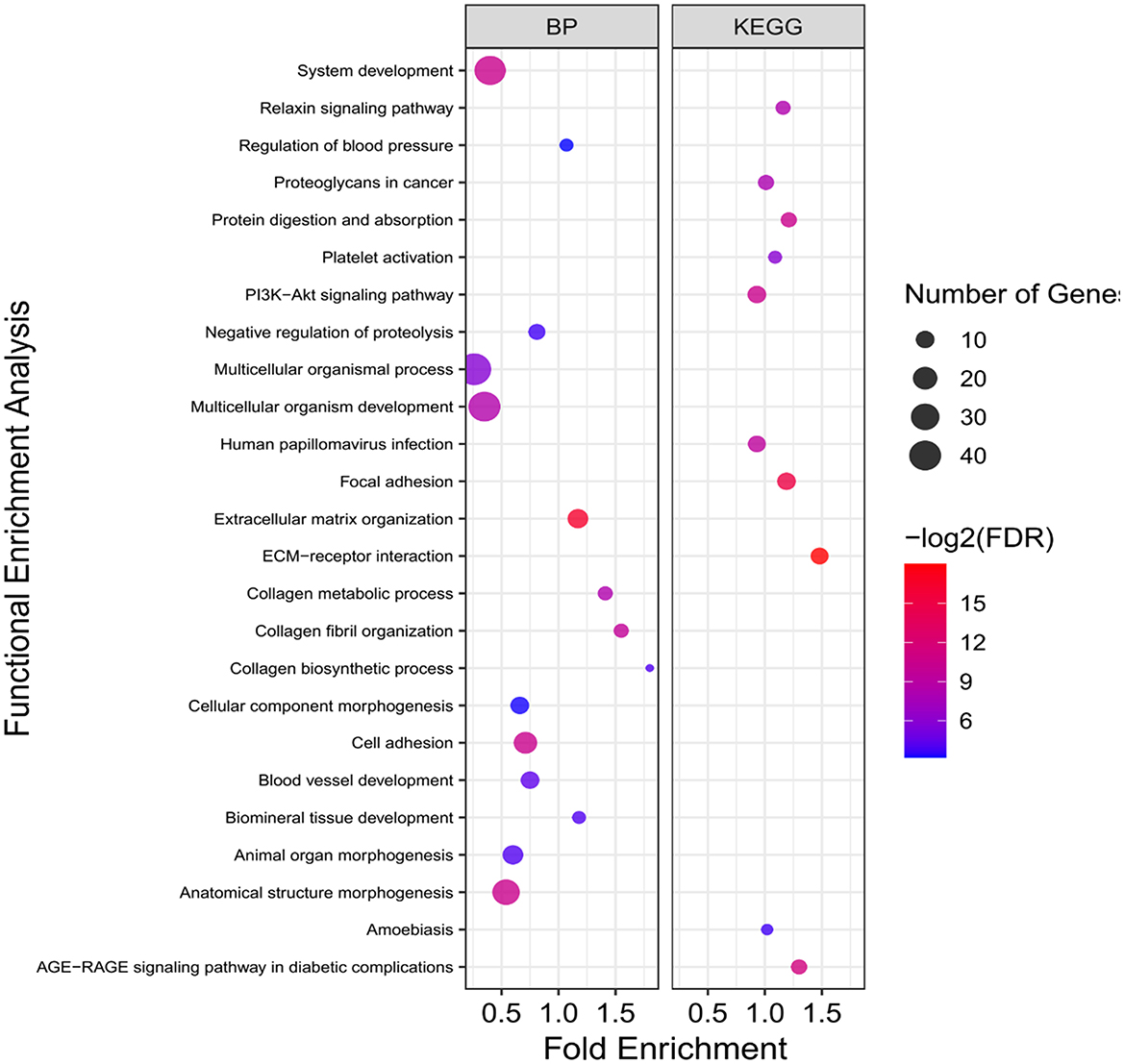
Figure 6. Functional enrichment analysis results of pink sub-network. Size and color of points represent -Log2 of FDR and number of genes associated with each term, respectively. BP, Biological processes.
Discussion
To date, several transcriptome-based studies have been conducted to identify DEGs related to fat-tail formation through comparison of fat- and thin-tailed sheep breeds (Table 1). However, one of the main problems of these studies is a small number of biological replicates due to the cost of sequencing, which can lead to decrease the statistical power to detect DEGs. In this regard, the variation among the identified DEGs by different datasets (Figure 2) emphasized the importance of meta-analysis to obtain core genes that are consistently DEGs across multiple studies. Some of the lack of agreement from one study to another can be attributed to differences in the experimental conditions such as environmental variables, bioinformatics pipeline, etc.
It should be noted that various sheep breeds have been used in the six studies, however these breeds can be used to identify core genes involved in fat deposition in the fat-tail of sheep. While vertebrate species differ in many phenotypic traits, however, organ physiology in mammals are conserved. Furthermore, it is well-known that many biological processes and pathways are deeply conserved among the species (36). Based on this idea that many developmental gene expression patterns are conserved across mammals, rodents can be applied as models of human tissues physiology (37). Previous studies in this area have shown that inter-study distances between similar tissues of various species were generally less than intra-study distances among various tissues, implying that meta-analysis of using RNA-Seq data from different breeds can provide insights into the molecular mechanisms behind the mammalian tissues (38). Here, a meta-analysis was performed to find the common core DEGs across six studies and remove the inconsistencies in these datasets to obtain a deeper biological insight associated with fat deposition in tail of sheep, compared to that achieved through single dataset. These core genes can be considered as putative candidates involved in fat-tail development regardless of the different characteristics of the studies. Therefore, our aim was to shed light into the molecular mechanisms conserved across the sheep breeds instead of identifying specific mechanisms in each breed.
To better integrate the datasets, a standard and similar bioinformatics workflow was applied to analyze the raw RNA-Seq data from the studies, in which different pipelines were used. To further evaluate the consistency of the DEGs across the datasets, Jackknife sensitivity analysis was applied and revealed that most of the DEGs (>80%) passed at least 50% of the Jackknife tests, which indicates the robustness of the results and independency of these genes to a single study. It is worth to note that in this study only DEGs with the same pattern of expression over half of the datasets were considered (500 DEGs including 221 up- and 279 down-regulated genes in fat- against thin-tailed sheep breeds), which make them more robust and constitutes a suitable way for better understanding the regulatory mechanisms of fat-tail development. To assess potential functions of DEGs in fat-tail development, we investigate their relationships with the QTLs related to sheep fatness and found that they were significantly enriched in these QTLs. These findings reinforced that the identified DEGs may play important roles in fat-tail development in sheep.
GO terms as well as KEGG pathways associated with fat metabolism were the most enriched terms in the DEGs, which explains the fact that these genes can be related to fat-tail formation (Supplementary material 3). Some of the DEGs grouped under terms that were directly or indirectly related to fat metabolism, including LEPRRBP4, CSNK2B, EPHX1, SCD, ALOX15, SCD5, HSPA6, ACLY, ELOVL6, FOXO3, PPP5C, DAGLA, ACSF2, DHCR24, JUNB, FNDC5, FMO2, FOS, FRZB, ZBTB16, LSS, COL5A1, SLC27A2, SPTLC3, PLPP3, ZBTB20, SMPDL3B, LBP, LPIN1, IL6, PDGFRA, CCL21, SPP1, PAI-1, HSD11B1, MTMR4, S100A1, COL6A3, TNNC2, ITGA1, SCG5, NR4A1, DUSP1, and APOE (6, 8–14, 17, 39–41). Most of these genes passed at least 50% of the Jackknife tests or reside in the QTLs previously found to be linked to sheep fatness (Supplementary material 2, 4). Some of these genes require particular attention considering that they have been previously linked to fat deposition.
PPIs network construction by STRING database confirmed that up- and down-regulated DEGs were members of functional interaction networks. It is well-known that genes from the same sub-network in a PPI network more probably play similar roles and are implicated in the same biological functions. To investigate this hypothesis, sub-network analysis on the PPI network results was performed and two of the three identified sub-networks in the down-regulated DEGs were significantly enriched in different GO terms and KEGG pathways. Hence, it might be possible to hypothesize that genes of green sub-network may be involved in fat deposition in tail of sheep breeds, as “Biosynthesis of unsaturated fatty acids” and “Lipid biosynthetic process” were significantly enriched (Figure 5). Some genes of this sub-network that may be of particular interests are SCD, SCD5, ELOVL6, ACLY, DHCR24, SPTLC3, LSS, LPIN1, SLC27A2, CKAP2, FOS, JUNB, FOXO3, EBF2, and MYBL2, which are located in the fatness-related QTLs regions or passed at least 50% of the Jackknife tests (Supplementary material 2, 4). In our previous study we hypothesized that down-regulation of the related genes to lipid metabolism in fa-tailed breed may be associated with other pathways than fat deposition such as fat composition (6) or fatty acid oxidation. It has been demonstrated that the breed has significant effect on fatty acid composition of tail fat (42). Here, the similar results were obtained as some members of green sub-network were involved in the pathways associated with fat composition or fatty acid oxidation such as SCD, SCD5, ELOVL6, ACLY, SLC27A2, and LPIN1.
Isoforms of stearoyl CoA desaturase (SCD and SCD5) play important roles in desaturation of saturated fatty acids. Lower expression of these genes has been previously reported in fat-tailed than thin-tailed sheep breeds (11, 13), which can cause lower content of saturated fatty acids in fat-tailed breeds. In chicken, SCD5 expression was significantly correlated with levels of stearidonic acid (43), which might lead to suppress adipocyte differentiation (44). Very long-chain fatty acids protein 6 (ELOVL6) is well-known as a major regulator of fatty acid composition in mammals (45). The first step of de novo lipogenesis, the conversion of citrate into acetyl-CoA, is catalyzed by ACLY. This product, acetyl-CoA, can then be used as building block for long chain fatty acids, cholesterol synthesis and/or histone acetylation (46). It has been shown that ACLY-deficient adipocytes accumulate lipid in vivo and display some differences in fatty acid content and synthesis (47). Mammals express three lipin genes (LPIN1-3) that their functions are evolutionarily conserved. One of the important roles of these genes is transcriptional co-regulators of gene expression in the nucleus to promote fatty acid oxidation (48). In accordance with our results, it was reported that the role of LPIN1 might be closely associated with fatty acid oxidation in the bovine liver (49). Solute carrier family 27 member 2 (SLC27A2) is a transmembrane transporter coenzyme that participates in the long-chain fatty acid beta-oxidation (50) as well as plays a key role in fatty acid degradation (51). SLC27A2 was identified to be closely associated with tail phenotype in Zhang et al. study that investigated the transcriptome profiles of fat deposition in tail of sheep (17). A recent study investigated the correlation of slow-growing meat type chicken liver gene expression with abdominal fat deposition using a time-course transcriptomic study (from the embryonic to the egg-producing period) and reported CKAP2 as one of the important candidates involved in fat metabolism (52). Furthermore, in green sub-network, five genes including JUNB, FOXO3, EBF2, FOS, and MYBL2 were transcription factors with known roles in lipid metabolism, which can be considered as regulators of this sub-network. A genome-wide association study showed that MYBL2 may be involved in abdominal fat deposition in chickens (53). JUNB is a transcription factor whose role in lipid metabolism and fat cell differentiation has been documented (54). Important roles of FOXO protein family in energy homeostasis and lipid metabolism have been highlighted in previous studies (55). Based on our findings, lower expression of members of the green sub-network in fat-tailed sheep breeds are reasonable and might be potential candidate contributing to shaping fat-tail phenotype through regulating fat composition or fatty acid oxidation.
On the other hand, some pathways related to the fat metabolism including “Extracellular matrix (ECM)-receptor interaction,” “Focal adhesion,” and “PI3K-Akt signaling pathway” were significantly enriched in the pink sub-network, which can further contribute to understanding the fat-tail development in sheep (Figure 6). All the genes associated with these pathways (SPP1, COL1A1, ITGA2, ITGA1, VEGFA, COL6A6, COL4A6, PDGFRA, COL6A3, and SDC1) were located in the QTLs related to fatness or passed at least 50% of the Jackknife tests (Supplementary material 2, 4), which indicated their potential involvement in fat-tail morphogenesis. Focal adhesion is related to ECM and act as mechanical linkages to the extracellular matrix (56). Hence, the DEGs belonging to these two pathways were very similar (Supplementary material 3). ECM is essential for tissue homeostasis and consists of a complex mixture of functional macromolecules in adipose tissue, such as glycosaminoglycans, collagen, elastin, fibronectin, and lammin (57). The ECM communicates with cells through cell surface-related elements such as integrin and regulates cell activities such as differentiation, proliferation, migration, adhesion and apoptosis (58). ECM receptor interaction signaling pathway has been reported as a significant enriched pathway in the DEGs between fat- and thin-tailed sheep breeds in several previous studies (6, 11, 16) as well as in the DEGs between lean and obese human (59). Moreover, interaction between ECM components and transmembrane receptors of fat cells have been demonstrated to be associated with depot-specific adipogenesis in bovine (60). However, we believed that research studies in animal filed have paid no sufficient attention to investigate the role of the ECM receptor interaction pathway in fat metabolism as well as fat-tail formation in sheep. ECM was detected as an inhibited KEGG pathway during differentiation of human mesenchymal stromal-cells into adipocytes. Down-regulated genes belonging to this pathway were in agreement with our results like collagen subunits IV, V and VI (COL1A1, COL6A6, COL4A6, and COL6A3), different integrins 1 and 2 (ITGA1 and ITGA2) and proteoglycans like syndecan 1 and 4 (SDC1). Down-regulation of these genes are attributed to cytoskeleton reorganization during adipogenesis (61). Collagen type IV denaturation is reported to be associated with adipogenic differentiation (62). In a comprehensive assessment to study the role of COL6A3 in human obesity and diabetes, it was revealed that COL6A3 expression increased after weight loss and showed a negative correlation with obesity, which is in good agreement with our findings in the current study (63). Moreover, it has been shown that integrin α5 is down-regulated during adipogenesis in 3T3-L1 cells. Therefore, up regulation of this gene inhibits cellular differentiation (64). Accordingly, our findings let hypothesize that ECM might mediates a mechanism involved in the differentiation of fat-tail adipocytes and lipid metabolism, thereby changing the fat-tail morphology of various sheep breeds.
Phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway, as the other enriched pathway in pink sub-network, is a key pathway that specifically phosphorylates phosphatidylinositol, generate an intracellular second messengers and mediates glucose and lipid metabolism (65). It is reported that PI3K/AKT signaling pathway has different functions in various adipocytes, as promote and inhibit the proliferation/differentiation in human adipocytes and 3T3-L1, respectively (66, 67). It has been shown that inhibition of this pathway in children with simple obesity participates in the occurrence and progression of obesity (68). In a recent study, PI3K/AKT signaling pathways was enriched in the target genes of differentially expressed miRNAs between fat- and thin-tailed sheep breeds (69) and suggested to be involved in biological processes related to fat deposition in fat-tail tissue. In the current study, enrichment of this pathway in the down-regulated genes in fat-tailed breeds suggesting that it may inhibits the proliferation and differentiation of lipid metabolism in fat-tail tissue and leads to differences in fat deposition between the different sheep breeds. Some of the DEGs belonging to PI3K/AKT signaling pathway in this study were included SPP1, PDGFRA, VEGFA, and TNC. Positive and negative effects of SPP1 in fat deposition had been demonstrated in previous reports. Studies have established that this gene is synthesized by adipocytes and its higher expression is related to fat deposition (70). In contrast, it is reported that interaction of SPP1 with integrin αv/β1 inhibits the adipogenesis of mesenchymal stem cells (71). In agreement with our results, a recent study found that SPP1 negatively regulated adipogenic differentiation of peripheral blood-derived mesenchymal stem cells and interaction between novel-miR-659 and SPP1 coregulate fat deposition in castrated male pigs (40). All these findings support the potential function of PI3K/AKT signaling pathway and its related genes in mediating lipolysis and energy expenditure, which can be led to lower fat deposition.
Some of the up-regulated DEGs in this study have been previously reported as candidate genes involved in fat-tail/fatness development or as DEG associated with fat metabolism in animal. Moreover, they were found in the fatness-related QTLs or passed at least 50% of the Jackknife tests (Supplementary material 2, 4). Some of the most important genes in light blue sub-network (Figure 4) were IL6, RBP4, LEPR, PAI-1, CSNK2B, and MTMR4. Recent studies highlighted the role of interleukins in lipid metabolism (49). Interleukin-6 (IL-6) is known as a key regulator of adipose homeostasis in obesity. It is worth to note that in Farhadi et al., study this gene was down-regulated in fat-tailed breed and they suggested it as a potential candidate gene in fat-tail formation. However, our analysis revealed that this gene was up-regulated in fat-tailed breed in all studies, except Farhadi et al., study. In this regard, results of a study showed that expression of IL-6 in lymphedema [a morbid disease characterized by adipose deposition (72)] is associated with adipose deposition. Since, IL-6 was identified as a highly connected gene in light blue sub-network (Figure 4), the other genes in this cluster are expected to be potential candidate genes in fat-tail formation. Interestingly, most of the connected genes with IL-6 were reported to be involved in fat deposition processes, which reinforce the importance of light blue sub-network in fat deposition. For example, retinol binding protein 4 (RBP4), which is a novel adipokine, is mainly secreted by adipocytes and is related to obesity. This gene contributes to systemic insulin resistance that can lead to fat deposition (73). Leptin receptor (LEPR), as other gene in light blue sub-network that is also located in fatness-related QTLs regions, was reported as candidate gene affecting fat deposition in pig (74). Higher levels of plasminogen activator inhibitor-1 (PAI-1) in plasma was reported to be a biochemical marker of obesity (75). The association between PAI-1 and fat deposition had been well-established in animal studies and the location of this gene in the QTLs linked to sheep fatness make it interesting for further functional investigations (76).
EPHX1, HSD11B1, FMO2, S100A1, and HSPA6 were some of the important up-regulated genes in orange sub-network (Figure 4). Association between EPHX1 and adipogenesis in mesenchymal stem cells through the activation of cryoprotective lipid mediators had been explained previously (77). Rosu-Myles et al., reported that EPHX1 expressing cells in human stromal cultures can be led to increased numbers of cells that have committed to the adipocyte lineage (78). FMO2 is a member of the FMO gene family and catalyze the NADPH-dependent oxidative metabolism of a wide array of foreign chemicals as well as is involved in fat deposition, adipogenesis and fatty acid biosynthesis (12). FMO3, as other member of this family, was reported as important candidate in fat metabolism of sheep through inhibiting fatty acid oxidation (14). Consistent with these observations, FMOs 1, 2, and 4 knockout mice exhibited a lean phenotype and stored less triglycerides in their white adipose tissue compared to wild-type mice, despite similar food intake (79). HSD11B1 is well-known to be closely related to the accumulation of abdominal fat (80). Recently, an RNA-Seq study was performed to explore the effects of castration on fat deposition in different parts of pigs and HSD11B1 was reported as a factor affects glucose uptake by adipocytes and leads to obesity (81). HSPA6 was reported as an important regulator of fatty acid metabolism in the skeletal muscles of sheep (39). Adipogenesis regulatory factor (ADIRF), as other important up-regulated gene, promotes adipocyte differentiation by enhancing the expression of peroxisome proliferator-activated receptor gamma (PPARG) and CCAAT-enhancer-binding protein alpha (CEBPA) in 3T3 L1 cells and play an important role in fat cell development. Higher expression of this gene in obese individuals, suggesting a role for ADIRF in the development of obesity (82). Angiopoietin-like 8 (ANGPTL8) exhibits its effects by inducing insulin receptor to inhibits lipolysis and controls post-prandial fat storage in white adipose tissue. This gene directs fatty acids to adipose tissue for storage during the fed state. Serpin family E member 1 (SERPINE1) was highly expressed in obese individuals and demonstrated as a key gene associated with the network pathway analysis of obesity (83). Altogether, these results support the potential role of up-regulated genes, especially light blue and orange sub-networks in fat deposition in tail of sheep.
Totally, 170 unique DEGs (77 up- and 93 down-regulated) were found in meta-analysis that were not identified as DEGs in individual studies (Figure 2), which can be attributed to the more statistical power of meta-analysis than individual studies for identifying new candidate genes associated with fat-tail formation. Of which, 38 DEGs (17 up and 21 down-regulated) were located in QTLs regions related to fatness of sheep, which further support that their functions might be relevant. Some of these genes have been previously reported to be related to lipid metabolism including NR4A1, ACSF2, MYC, SPP1, PLPP3, PHOSPHO1, and ACP6. For example, NR4A1 encodes a nuclear receptor (transcription factor) that is involved in regulation of lipid metabolism and modulating lipolysis in muscle (41). It is reported that female NR4A1 deficient mice exhibited higher fat mass compared to wild-type mice, under the same high-fat diet (84). This gene plays a vital role in the regulation of liver fat content (41). Expression of NR4A1 is reported to be negatively correlated with body-fat content and insulin sensitivity, as its expression was significantly lower in the muscle of obese men in comparison to lean men (85). MYC, which is a transcription factor, plays vital roles in lipid metabolism (86). Furthermore, ACSF2 is involved in the acyl-CoA metabolic process and malonyl-CoA metabolic process in mammals as well as reported to be associated with avian lipid metabolism (87, 88). These findings suggested a link between differentially expressed of these genes and fat-tail development in sheep.
Conclusion
In this study, a meta-analysis of six RNA-Seq datasets that compared transcriptome profiles of fat- and thin-tailed sheep breeds, were performed. By considering the expression pattern of DEGs across the different studies and performing Jackknife analysis, a list of robust DEGs were obtained, which were enriched in the QTLs related to sheep fatness. Moreover, functional interactions among the DEGs were confirmed using PPIs network analysis. Functional enrichment analysis showed that the DEGs were enriched in the GO terms/KEGG pathways related to fat metabolism such as “fatty-acyl-CoA biosynthetic process,” “ECM-receptor interaction,” and “PI3K-Akt signaling pathway.” Cluster analysis of the PPIs network led to identification several sub-networks that were directly or indirectly involved in fat deposition. Some down-regulated DEGs in green and pink sub-networks were SCD, SCD5, ELOVL6, ACLY, SLC27A2, LPIN1, COL1A1, COL6A6, COL4A6, COL6A3, ITGA1, ITGA2, SDC1, and SPP1, which probably promote the development of fat-tail through regulating lipolysis or fatty acid oxidation. In contrast, up-regulated DEGs such as IL6, RBP4, LEPR, PAI-1, CSNK2B, MTMR4, EPHX1, HSD11B1, FMO2, S100A1, and HSPA6 were mainly enriched in green and pink sub-networks and may contribute to a network controlling fat accumulation in tail of sheep breed through mediating adipogenesis and fatty acid biosynthesis. Overall, our meta-analysis successfully identified a core set of DEGs associated with lipid metabolism including well-known genes related to fat deposition as well as newly identified genes such as NR4A1 and ACSF2. Therefore, it is reasonable to infer that the suggested sub-networks and their gene members might be potential candidate contributing to shaping fat-tail phenotype. Although our findings were in good agreement with previous studies, however they are in transcriptome level and follow-up functional studies are required to investigate the mechanisms by which these genes contribute to the fat deposition in tail of sheep breeds.
Data availability statement
Publicly available datasets were analyzed in this study. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
MB conceived the ideas. MB and SH designed study and analyzed the data. MB, SH, and AS interpreted the data and wrote the main manuscript text. All authors read and approved the final manuscript.
Acknowledgments
We would like to thank the University of Tehran for the support of this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1159921/full#supplementary-material
References
1. Kalds P, Luo Q, Sun K, Zhou S, Chen Y, Wang X. Trends towards revealing the genetic architecture of sheep tail patterning: Promising genes and investigatory pathways. Anim Genet. (2021) 52:799–812. doi: 10.1111/age.13133
2. Beynon SE, Slavov GT, Farre M, Sunduimijid B, Waddams K, Davies B, et al. Population structure and history of the Welsh sheep breeds determined by whole genome genotyping. BMC Genet. (2015) 16:1–15. doi: 10.1186/s12863-015-0216-x
3. Muigai AW, Hanotte O. The origin of African sheep: archaeological and genetic perspectives. Afri Archaeol Rev. (2013) 30:39–50. doi: 10.1007/s10437-013-9129-0
4. Atti N, Hamouda MB. Relationships among carcass composition and tail measurements in fat-tailed Barbarine sheep. Small Ruminant Res. (2004) 53:151–5. doi: 10.1016/j.smallrumres.2003.08.016
5. Vatankhah M, Moradi-Shahrbabak M, Nejati-Javaremi A, Miraei-Ahtiani SR, Vaez-Torshizi R. A study of external fat-tail dimensions and their relationships with fat-tail weight in lori-bakhtiari breed of sheep. JWSS Isfahan Univ Technol. (2006) 10:445–56.
6. Bakhtiarizadeh MR, Salehi A, Alamouti AA, Abdollahi-Arpanahi R, Salami SA. Deep transcriptome analysis using RNA-Seq suggests novel insights into molecular aspects of fat-tail metabolism in sheep. Sci Rep. (2019) 9:1–14. doi: 10.1038/s41598-019-45665-3
7. Eryavuz A, Avci G, Kuculkkurt I, Fidan A. Comparison of plasma leptin, insulin and thyroid hormone concentrations and some biochemical parameters between fat-tailed and thin-tailed sheep breeds. Revue de médecine vétérinaire. (2007) 158:244–9.
8. Bakhtiarizadeh MR, Moradi-Shahrbabak M, Ebrahimie E. Underlying functional genomics of fat deposition in adipose tissue. Gene. (2013) 521:122–8. doi: 10.1016/j.gene.2013.03.045
9. Fan H, Hou Y, Sahana G, Gao H, Zhu C, Du L, et al. A transcriptomic study of the tail fat deposition in two types of Hulun Buir sheep according to tail size and sex. Animals. (2019) 9:655. doi: 10.3390/ani9090655
10. Farhadi S, Shodja Ghias J, Hasanpur K, Mohammadi SA, Ebrahimie E. Molecular mechanisms of fat deposition: IL-6 is a hub gene in fat lipolysis, comparing thin-tailed with fat-tailed sheep breeds. Archiv Anim Breed. (2021) 64:53–68. doi: 10.5194/aab-64-53-2021
11. Li B, Qiao L, An L, Wang W, Liu J, Ren Y, et al. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics. (2018) 19:1–13. doi: 10.1186/s12864-018-4747-1
12. Ma L, Zhang M, Jin Y, Erdenee S, Hu L, Chen H, et al. Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep. Front Genet. (2018) 9:365. doi: 10.3389/fgene.2018.00365
13. Miao X, Luo Q, Qin X, Guo Y, Zhao H. Genome-wide mRNA-seq profiling reveals predominant down-regulation of lipid metabolic processes in adipose tissues of Small Tail Han than Dorset sheep. Biochem Biophys Res Commun. (2015) 467:413–20. doi: 10.1016/j.bbrc.2015.09.129
14. Wang X, Zhou G, Xu X, Geng R, Zhou J, Yang Y, et al. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene. (2014) 549:252–7. doi: 10.1016/j.gene.2014.07.072
15. Yuan Y-N, Liu W-Z, Liu J-H, Qiao L-Y, Wu J-L. Cloning and ontogenetic expression of the uncoupling protein 1 gene UCP1 in sheep. J Appl Genet. (2012) 53:203–12. doi: 10.1007/s13353-012-0086-0
16. Yuan Z, Ge L, Sun J, Zhang W, Wang S, Cao X, et al. Integrative analysis of Iso-Seq and RNA-seq data reveals transcriptome complexity and differentially expressed transcripts in sheep tail fat. PeerJ. (2021) 9:e12454. doi: 10.7717/peerj.12454
17. Zhang W, Xu M, Wang J, Wang S, Wang X, Yang J, et al. Comparative transcriptome analysis of key genes and pathways activated in response to fat deposition in two sheep breeds with distinct tail phenotype. Front Genet. (2021) 12:639030. doi: 10.3389/fgene.2021.639030
18. Kumar B, Francis SM, Suttie JM, Thompson MP. Expression of obese mRNA in genetically lean and fat selection lines of sheep. Comparat Biochem Physiol B. (1998) 120:543–8. doi: 10.1016/S0305-0491(98)10041-X
19. Gao L, Xu R, Zhao W, Yang J, Liang Y, Liu S, et al. Cloning of sheep cell death-inducing DFFA-like effector c (CIDEC) cDNA and its differential expression in tail fat tissue of Altay sheep (Ovis aries) in persistent starvation. J Agricul Biotechol. (2015) 23:227–35.
20. Guangli Y, Huan Z, Shuhong Z, Zhiqiang L, Fengyi G, Guan W, et al. Identification of the genetic basis for the large-tailed phenotypic trait in Han sheep through integrated mRNA and miRNA analysis of tail fat tissue samples. Research. (2020) 2020:107294. doi: 10.21203/rs.3.rs-107294/v1
21. Su X, He H, Fang C, Liu L, Liu W. Screening and identification of the key LncRNAs associated with fat deposition in ovine tail. Research. (2021) 2021:1008169. doi: 10.21203/rs.3.rs-1008169/v1
22. Brown J, Pirrung M, Mccue LA. FQC dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics. (2017) 33:3137–9. doi: 10.1093/bioinformatics/btx373
23. Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
24. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
25. Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. bioinformatics. (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
26. Love ML, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:1–21. doi: 10.1186/s13059-014-0550-8
27. McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. (2012) 40:4288–97. doi: 10.1093/nar/gks042
28. Rau A, Marot G, Jaffrezic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinformatics. (2014) 15:1–10. doi: 10.1186/1471-2105-15-91
29. Rogic S, Wong A, Pavlidis P. Meta-analysis of gene expression patterns in animal models of prenatal alcohol exposure suggests role for protein synthesis inhibition and chromatin remodeling. Alcoholism. (2016) 40:717–27. doi: 10.1111/acer.13007
31. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. (2016) 44:90–7. doi: 10.1093/nar/gkw377
32. Hu Z-L, Park CA, Wu X-L, Reecy JM. Animal QTLdb: An improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. (2013) 41:D871–9. doi: 10.1093/nar/gks1150
33. Das S, Rai A, Mishra DC, Rai SN. Statistical approach for gene set analysis with trait specific quantitative trait loci. Sci Rep. (2018) 8:1–12. doi: 10.1038/s41598-018-19736-w
35. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
36. Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. (2012) 338:1593–9. doi: 10.1126/science.1228186
37. Chan ET, Quon GT, Chua G, Babak T, Trochesset M, Zirngibl RA, et al. Conservation of core gene expression in vertebrate tissues. J Biol. (2009) 8:1–17. doi: 10.1186/jbiol130
38. Skinnider MA, Foster LJ. Meta-analysis defines principles for the design and analysis of co-fractionation mass spectrometry experiments. Nat Methods. (2021) 18:806–15. doi: 10.1038/s41592-021-01194-4
39. Arora R, Fairoze MN, Kaur M, Sharma A, Girdhar Y, Devatkal SK, et al. Transcriptome profiling of longissimus thoracis muscles identifies highly connected differentially expressed genes in meat type sheep of India. PLoS ONE. (2019) 14:e0217461. doi: 10.1371/journal.pone.0217461
40. Xiao L, Xu Q, Liu X, Chan S, Luo Y, He S, et al. The novel-miR-659/SPP1 interaction regulates fat deposition in castrated male pigs. Animals. (2022) 12:944. doi: 10.3390/ani12080944
41. Zhang L, Wang Q, Liu W, Liu F, Ji A, Li Y. The orphan nuclear receptor 4A1: a potential new therapeutic target for metabolic diseases. J Diabet Res. (2018) 2018:9363461. doi: 10.1155/2018/9363461
42. Moharrery A. Effect of docking and energy of diet on carcass fat characteristics in fat-tailed Badghisian sheep. Small Ruminant Res. (2007) 69:208–16. doi: 10.1016/j.smallrumres.2005.12.021
43. Mihelic R, Winter H, Powers J, Das S, Lamour K, Campagna S, et al. Genes controlling polyunsaturated fatty acid synthesis are developmentally regulated in broiler chicks. Br Poult Sci. (2020) 61:508–17. doi: 10.1080/00071668.2020.1759788
44. Li Y, Rong Y, Bao L, Nie B, Ren G, Zheng C, et al. Suppression of adipocyte differentiation and lipid accumulation by stearidonic acid (SDA) in 3T3-L1 cells. Lipids Health Dis. (2017) 16:1–10. doi: 10.1186/s12944-017-0574-7
45. Shi H, Wu M, Zhu J, Zhang C, Yao D, Luo J, et al. Fatty acid elongase 6 plays a role in the synthesis of long-chain fatty acids in goat mammary epithelial cells. J Dairy Sci. (2017) 100:4987–95. doi: 10.3168/jds.2016-12159
46. Chypre M, Zaidi N, Smans K. ATP-citrate lyase: A mini-review. Biochem Biophys Res Commun. (2012) 422:1–4. doi: 10.1016/j.bbrc.2012.04.144
47. Zhao S, Torres A, Henry RA, Trefely S, Wallace M, Lee JV, et al. ATP-citrate lyase controls a glucose-to-acetate metabolic switch. Cell Rep. (2016) 17:1037–52. doi: 10.1016/j.celrep.2016.09.069
48. Peterfy M, Harris TE, Fujita N, Reue K. Insulin-stimulated interaction with 14-3-3 promotes cytoplasmic localization of lipin-1 in adipocytes. J Biol Chem. (2010) 285:3857–64. doi: 10.1074/jbc.M109.072488
49. Baumgard L, Hausman G, Fernandez MS. Insulin: Pancreatic secretion and adipocyte regulation. Domest Anim Endocrinol. (2016) 54:76–84. doi: 10.1016/j.domaniend.2015.07.001
50. Feng Y, Li S, Zhang R, Liu F, Xu Q, Ding H, et al. FOXM1 as a prognostic biomarker promotes endometrial cancer progression via transactivation of SLC27A2 expression. Int J Clin Exp Pathol. (2018) 11:3846.
51. Jiang Z, Michal JJ, Chen J, Daniels TF, Kunej T, Garcia MD, et al. Discovery of novel genetic networks associated with 19 economically important traits in beef cattle. Int J Biol Sci. (2009) 5:528. doi: 10.7150/ijbs.5.528
52. Xing S, Liu R, Zhao G, Groenen MA, Madsen O, Liu L, et al. Time course transcriptomic study reveals the gene regulation during liver development and the correlation with abdominal fat weight in chicken. Front Genet. (2021) 2021:1593. doi: 10.3389/fgene.2021.723519
53. Wu X, Zhang Q, Xu S, Jin P, Luan P, Li Y, et al. Differential expression of six chicken genes associated with fatness traits in a divergently selected broiler population. Mol Cell Probes. (2016) 30:1–5. doi: 10.1016/j.mcp.2015.12.003
54. Luther J, Ubieta K, Hannemann N, Jimenez M, Garcia M, Zech C, et al. Fra-2/AP-1 controls adipocyte differentiation and survival by regulating PPARγ and hypoxia. Cell Death Different. (2014) 21:655–64. doi: 10.1038/cdd.2013.198
55. Binh TD, Pham TL, Men TT, Dang TT, Kamei K. LSD-2 dysfunction induces dFoxO-dependent cell death in the wing of Drosophila melanogaster. Biochem Biophys Res Commun. (2019) 509:491–7. doi: 10.1016/j.bbrc.2018.12.132
56. Romer LH, Birukov KG, Garcia JG. Focal adhesions: Paradigm for a signaling nexus. Circ Res. (2006) 98:606–16. doi: 10.1161/01.RES.0000207408.31270.db
57. Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, et al. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol Cell Biol. (2009) 29:1575–91. doi: 10.1128/MCB.01300-08
58. Thomas K, Engler AJ, Meyer GA. Extracellular matrix regulation in the muscle satellite cell niche. Connect Tissue Res. (2015) 56:1–8. doi: 10.3109/03008207.2014.947369
59. Henegar C, Tordjman J, Achard V, Lacasa D, Cremer I, Guerre-Millo M, et al. Adipose tissue transcriptomic signature highlights the pathological relevance of extracellular matrix in human obesity. Genome Biol. (2008) 9:1–32. doi: 10.1186/gb-2008-9-1-r14
60. Lee H-J, Jang M, Kim H, Kwak W, Park W, Hwang JY, et al. Comparative transcriptome analysis of adipose tissues reveals that ECM-receptor interaction is involved in the depot-specific adipogenesis in cattle. PLoS ONE. (2013) 8:e66267. doi: 10.1371/journal.pone.0066267
61. Casado-Díaz A, Anter J, Mueller S, Winter P, Quesada-Gómez JM, Dorado G. Transcriptomic analyses of adipocyte differentiation from human mesenchymal stromal-cells (MSC). J Cell Physiol. (2017) 232:771–84. doi: 10.1002/jcp.25472
62. Mauney J, Volloch V. Human bone marrow-derived stromal cells show highly efficient stress-resistant adipogenesis on denatured collagen IV matrix but not on its native counterpart: implications for obesity. Matrix Biol. (2010) 29:9–14. doi: 10.1016/j.matbio.2009.09.002
63. Mcculloch LJ, Rawling TJ, Sjöholm K, Franck N, Dankel SN, Price EJ, et al. COL6A3 is regulated by leptin in human adipose tissue and reduced in obesity. Endocrinology. (2015) 156:134–46. doi: 10.1210/en.2014-1042
64. Liu J, Deyoung SM, Zhang M, Zhang M, Cheng A, Saltiel AR. Changes in integrin expression during adipocyte differentiation. Cell Metab. (2005) 2:165–77. doi: 10.1016/j.cmet.2005.08.006
65. Huang W, Guo Y, Du W, Zhang X, Li A, Miao X. Global transcriptome analysis identifies differentially expressed genes related to lipid metabolism in Wagyu and Holstein cattle. Sci Rep. (2017) 7:1–11. doi: 10.1038/s41598-017-05702-5
66. Choi EO, Park C, Shin SS, Cho EJ, Kim BW, Hwang JA, et al. Zanthoxylum schinifolium leaf ethanol extract inhibits adipocyte differentiation through inactivation of the extracellular signal regulated kinase and phosphoinositide 3-kinase/Akt signaling pathways in 3T3-L1 pre-adipocytes. Mol Med Rep. (2015) 12:1314–20. doi: 10.3892/mmr.2015.3463
67. Xu Y, Wang N, Tan H-Y, Li S, Zhang C, Zhang Z, et al. Panax notoginseng saponins modulate the gut microbiota to promote thermogenesis and beige adipocyte reconstruction via leptin-mediated AMPKα/STAT3 signaling in diet-induced obesity. Theranostics. (2020) 10:11302. doi: 10.7150/thno.47746
68. Su X, Gu D, Xu L, Liang Z, Luo X, Yang P, et al. PI3K/Akt pathway expression in children with different obesity degrees and its relationship with glucolipid metabolism and insulin resistance. Am J Transl Res. (2021) 13:6592.
69. Fei X, Jin M, Wang Y, Li T, Lu Z, Yuan Z, et al. Transcriptome reveals key microRNAs involved in fat deposition between different tail sheep breeds. PLoS ONE. (2022) 17:e0264804. doi: 10.1371/journal.pone.0264804
70. Chapman J, Miles PD, Ofrecio JM, Neels JG, Yu JG, Resnik JL, et al. Osteopontin is required for the early onset of high fat diet-induced insulin resistance in mice. PLoS ONE. (2010) 5:e13959. doi: 10.1371/journal.pone.0013959
71. Chen Q, Shou P, Zhang L, Xu C, Zheng C, Han Y, et al. An osteopontin-integrin interaction plays a critical role in directing adipogenesis and osteogenesis by mesenchymal stem cells. Stem Cells. (2014) 32:327–37. doi: 10.1002/stem.1567
72. Cuzzone DA, Weitman ES, Albano NJ, Ghanta S, Savetsky IL, Gardenier JC, et al. IL-6 regulates adipose deposition and homeostasis in lymphedema. Am J Physiol Heart Circulat Physiol. (2014) 306:H1426–34. doi: 10.1152/ajpheart.01019.2013
73. Cheng J, Li Y, Wu G, Zheng J, Lu H, Shi XE, et al. Ectopic expression of RBP4 impairs the insulin pathway and inguinal fat deposition in mice. J Physiol Biochem. (2014) 70:479–86. doi: 10.1007/s13105-014-0326-3
74. Xing K, Liu H, Zhang F, Liu Y, Shi Y, Ding X, et al. Identification of key genes affecting porcine fat deposition based on co-expression network analysis of weighted genes. J Anim Sci Biotechnol. (2021) 12:1–16. doi: 10.1186/s40104-021-00616-9
75. Barnard SA, Pieters M, De Lange Z. The contribution of different adipose tissue depots to plasma plasminogen activator inhibitor-1 (PAI-1) levels. Blood Rev. (2016) 30:421–9. doi: 10.1016/j.blre.2016.05.002
76. Ekström M, Liska J, Eriksson P, Sverremark-Ekström E, Tornvall P. Stimulated in vivo synthesis of plasminogen activator inhibitor-1 in human adipose tissue. Thromb Haemost. (2012) 108:485–92. doi: 10.1160/TH11-11-0822
77. Tomita Y, Matsumura K, Wakamatsu Y, Matsuzaki Y, Shibuya I, Kawaguchi H, et al. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J Cell Biol. (2005) 170:1135–46. doi: 10.1083/jcb.200504061
78. Rosu-Myles M, She Y-M, Fair J, Muradia G, Mehic J, Menendez P, et al. Identification of a candidate proteomic signature to discriminate multipotent and non-multipotent stromal cells. PLoS ONE. (2012) 7:e38954. doi: 10.1371/journal.pone.0038954
79. Veeravalli S, Omar BA, Houseman L, Hancock M, Malagon SGG, Scott F, et al. The phenotype of a flavin-containing monooyxgenase knockout mouse implicates the drug-metabolizing enzyme FMO1 as a novel regulator of energy balance. Biochem Pharmacol. (2014) 90:88–95. doi: 10.1016/j.bcp.2014.04.007
80. Baudrand R, Carvajal CA, Riquelme A, Morales M, Solis N, Pizarro M, et al. Overexpression of 11β-hydroxysteroid dehydrogenase type 1 in hepatic and visceral adipose tissue is associated with metabolic disorders in morbidly obese patients. Obes Surg. (2010) 20:77–83. doi: 10.1007/s11695-009-9937-0
81. Liu X, Bai Y, Cui R, He S, Ling Y, Wu C, et al. Integrated analysis of the ceRNA network and M-7474 function in testosterone-mediated fat deposition in pigs. Genes. (2022) 13:668. doi: 10.3390/genes13040668
82. Ni Y, Ji C, Wang B, Qiu J, Wang J, Guo X. A Novel pro-adipogenesis factor abundant in adipose tissues and over-expressed in obesity acts upstream of PPARγ and C/EBPα. J Bioenerg Biomembr. (2013) 45:219–28. doi: 10.1007/s10863-012-9492-6
83. Kaur P, Reis MD, Couchman GR, Forjuoh SN, Greene JF, Asea A. SERPINE 1 links obesity and diabetes: A pilot study. J Proteomics Bioinform. (2010) 3:191. doi: 10.4172/jpb.1000139
84. Perez-Sieira S, Martinez G, Porteiro B, Lopez M, Vidal A, Nogueiras R, et al. Female Nur77-deficient mice show increased susceptibility to diet-induced obesity. PLoS ONE. (2013) 8:e53836. doi: 10.1371/journal.pone.0053836
85. Kanzleiter T, Preston E, Wilks D, Ho B, Benrick A, Reznick J, et al. Overexpression of the orphan receptor Nur77 alters glucose metabolism in rat muscle cells and rat muscle in vivo. Diabetologia. (2010) 53:1174–83. doi: 10.1007/s00125-010-1703-2
86. Wahlström T, Henriksson MA. Impact of MYC in regulation of tumor cell metabolism. Biochim Biophys Acta. (2015) 1849:563–9. doi: 10.1016/j.bbagrm.2014.07.004
87. Tian W, Zheng H, Yang L, Li H, Tian Y, Wang Y, et al. Dynamic expression profile, regulatory mechanism and correlation with egg-laying performance of ACSF gene family in chicken (Gallus gallus). Sci Rep. (2018) 8:1–10. doi: 10.1038/s41598-018-26903-6
Keywords: adipose tissue, lipid metabolism, comparative transcriptome, gene expression—tools and techniques, RNA-Seq
Citation: Hosseini SF, Bakhtiarizadeh MR and Salehi A (2023) Meta-analysis of RNA-Seq datasets highlights novel genes/pathways involved in fat deposition in fat-tail of sheep. Front. Vet. Sci. 10:1159921. doi: 10.3389/fvets.2023.1159921
Received: 06 February 2023; Accepted: 11 April 2023;
Published: 12 May 2023.
Edited by:
Sara Pegolo, University of Padua, ItalyReviewed by:
Aroa Suarez Vega, Universidad de León, SpainGan Shangquan, Xinjiang Academy of Agricultural and Reclamation Sciences (XAARS), China
Copyright © 2023 Hosseini, Bakhtiarizadeh and Salehi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammad Reza Bakhtiarizadeh, bXJiYWtodGlhcmlAdXQuYWMuaXI=
†ORCID: Mohammad Reza Bakhtiarizadeh orcid.org/0000-0001-5336-6987
 Seyedeh Fatemeh Hosseini
Seyedeh Fatemeh Hosseini Mohammad Reza Bakhtiarizadeh
Mohammad Reza Bakhtiarizadeh Abdolreza Salehi
Abdolreza Salehi