- 1College of Animal Science and Technology, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, Northwest A&F University, Yangling, Shaanxi, China
- 2Shaanxi Provincial Engineering and Technology Research Center of Cashmere Goats, Yulin University, Yulin, Shaanxi, China
- 3Life Science Research Center, Yulin University, Yulin, Shaanxi, China
- 4Institute of Animal Husbandry and Veterinary, Fujian Academy of Agricultural Sciences, Fuzhou, Fujian, China
- 5Animal Husbandry and Veterinary Science Institute of Bijie City, Bijie, Guizhou, China
- 6College of Animal Science and Technology, Hunan Agricultural University, Changsha, Hunan, China
As a member of the SNX family, the goat sorting nexin 29 (SNX29) is initially identified as a myogenesis gene. Therefore, this study aimed to examine the polymorphism in the SNX29 gene and its association with growth traits. In this study, we used an online platform to predict the structures of the SNX29 protein and used quantitative real-time PCR to detect potential copy number variation (CNV) in Shaanbei white cashmere (SBWC) goats (n = 541), Guizhou black (GB) goats (n = 48), and Nubian (NB) goats (n = 39). The results showed that goat SNX29 protein belonged to non-secretory protein. Then, five CNVs were detected, and their association with growth traits was analyzed. In SBWC goats, CNV1, CNV3, CNV4, and CNV5 were associated with chest width and body length (P < 0.05). Among them, the CNV1 individuals with gain and loss genotypes were superior to those individuals with a median genotype, but CNV4 and CNV5 of individuals with the median genotype were superior to those with the loss and gain genotypes. In addition, individuals with the gain genotype had superior growth traits in CNV3. In brief, this study suggests that the CNV of SNX29 can be used as a molecular marker in goat breeding.
Introduction
Members of the SNX family are located in membrane-binding cytoplasm and can bind to phosphatidylinositol via the PX domain and interact with membrane-associated protein complexes, which play an important role in regulating endocytosis and protein transport through cell membrane compartments (1, 2). To date, 32 members have been identified, and they are divided into five subgroups based on protein domain. Among them, the SNX29 gene belongs to the SNX-PX subfamily (3), which has been reported to be involved in disease, nervous system development, and animal growth. Studies have linked the SNX29 gene to schizophrenia (SCZ), autism, and other psychiatric disorders (4, 5). The deletion of SNX29 intron 14 may lead to primary testicular lymphoma (6). Zhu et al. found that downregulation of the SNX29 gene was associated with epithelial ovarian carcinoma cells (7). Furthermore, Sparks et al. showed a strong association between IgA levels and the region between 6.89 and 14.95 Mb on sheep chromosome 24, which corresponds to the SNX29 gene (8). A circRNA of the SNX29 gene regulated the proliferation and differentiation of muscle cells (9). Studies have shown that the SNX29 gene plays a key role in subcutaneous fat deposition in Xiangdong black (XDB) goats, and the SNX29 CNV is significantly associated with the chest and abdominal girth of XDB goats (P < 0.01) (10). Based on the above, the SNX29 gene was selected to be studied in this study.
Copy number variation (CNV) exists widely in the genomes of organisms, and it is considered to be an important source of genetic differences between individuals (11, 12). In recent years, some studies reported that CNV was significantly correlated with the economic traits of livestock, such as litter size (13), meat quality (14), milk production (15), weight gain rate (16), and feed conversion rate (17). The advantages of CNV-promoting population diversity, simplicity, and efficiency were discovered by more people (18). As a applicable molecular marker, CNV can make marker-assisted selection (MAS) better play the advantages of convenience, simplicity, and so on. In short, it provides new ideas and methods for breeding work.
Shaanbei white cashmere (SBWC) goats were bred from Liaoning white cashmere goat and Ziwuling black goat (19), which has high cashmere value and meat value (20). Guizhou black (GB) goats are an excellent local breed with good meat quality and coarse feeding tolerance (21). Nubian (NB) goats have good value for meat and milk and have higher meat content than other dual-purpose goats (22). However, their growth performance fails to achieve the expected results, so it is helpful to increase the economic value of goats by improving their growth traits through MAS.
Currently, the CNVs of the SNX29 gene and its association with growth traits in SBWC goats have not been reported. Therefore, this study is characterized based on the aspects of protein structure, physicochemical properties, and DNA variation. Next, we explored five potential CNVs, which were detected in SBWC goats, GB goats, and NB goats by quantitative real-time PCR (qRT-PCR). An association analysis was carried out between the SNX29 gene and the growth traits of goats. These results will have a deeper understanding of gene variation and livestock growth traits, in order to lay a theoretical foundation for MAS breeding of goats.
Materials and methods
Animal welfare explanation
The samples used in this experiment comply with the Regulations on the Administration of Experimental Animals at Northwest A&F University (NWAFU-314020038).
Prediction of SNX29 protein physicochemical properties and structure
Using NCBI-searched SNX29 protein sequences, the goats' SNX29 protein amino acid number, molecular weight, and isoelectric point were calculated using the Expasy online platform, and the ProtScale application and ProtParam were used to predict the protein hydrophobicity. The SNX29 protein of transmembrane signal peptide was predicted using the TMHMM database and SignalP 4.1. The AlphaFold and SOPMA online platforms were used to predict the advanced structure of the SNX29 protein (23) (Supplementary Table S1).
Sample collection and genomic DNA extraction
Under the same feeding conditions, ear tissues of 541 SBWC goats, 48 GB goats, and 39 NB goats were selected from the Yulin goat farm in Shaanxi province, the Bijie goat farm in Guizhou province, and the Zhangzhou Nubian goat breeding cooperative in Fujian province. All the individuals were female goats (2–3 years) and were not related to each other. Genomic DNA was extracted from goat ear tissue using the high salt extraction method (24) and stored at 70% alcohol at −80°C (25). A NanoDrop™2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) was used to measure the OD260/280 ratio, and a ratio between 1.8 and 2.0 means that the nucleic acid concentration is qualified (26). Then, the extracted DNA was placed at −40°C.
Primer designing
We searched the Animal Omics database (27) (Supplementary Table S1) and found five CNV loci of the SNX29 gene in goats. Five pairs of amplified primers were referenced in a previous article (28).
CNV genotyping detection of the SNX29 gene
To ensure that the primers can amplify the target fragment, the primers are detected through the mixed pool (CNV1 = 137 bp, CNV2 = 138 bp, CNV3 = 104 bp, CNV4 = 151 bp, and CNV5 = 109 bp). Next, 541 SBWC goat samples, 48 GB goat samples, and 39 NB goat samples were used to detect the CNV loci. qRT-PCR amplification systems and procedures refer to previous laboratory articles (29, 30). The result was processed using method 2 *2(−δCt) (31).
Statistical analyses
The association between the variants and growth traits was explored using the analysis of variance (ANOVA) and independent sample t-test in SPSS 26.0 (IBM, USA), and the chi-square (χ2) test was used to analyze the significance between the three breeds (32). And the line model was used as a reference by Liu et al. (33). Where Yijk = αi + βj + eijk + u acts as an analysis model, Yijk is the evaluation of growth traits at the i level of fixed factor age (αi) and j level of fixed factor genotype (βj), u is the overall mean, and eijk is the random error.
Results
Prediction of SNX29 protein physicochemical properties and structure
To characterize the functions of the SNX29 gene, the protein structure and physicochemical properties were predicted. The results showed that the protein contained 817 amino acids, the molecular weight was 9,143.14, and the isoelectric point was 5.90 by the Expasy online platform. ProtScale online software predicted the hydrophobicity of the protein, and the results showed that there were more hydrophilic residues in the goat SNX29 protein, which indicated that this protein was hydrophilic (Figure 1A). The results were consistent with ProtParam online software predictions. TMHMM prediction results showed that the protein encoded by the SNX29 gene did not have transmembrane helix (Figure 1B). SignalP 4.1 prediction results showed that the D critical value of signal peptide and non-signal peptide of this protein was 0.450, and the D critical value of the SNX29 protein was 0.155 (Figure 1C). According to the signal peptide hypothesis, the SNX29 protein had no signal peptide and belonged to non-secretory protein. The SOPMA online platform predicted the detailed information on the secondary structure of SNX29 protein, and the results showed that alpha helix accounted for 47.98%, extended strand accounted for 12.24%, β-turn accounted for 4.04%, and random coil accounted for 35.74% (Figure 1E). AlphaFold online software predicted the three-dimensional structure of the SNX29 protein (Figure 1D).
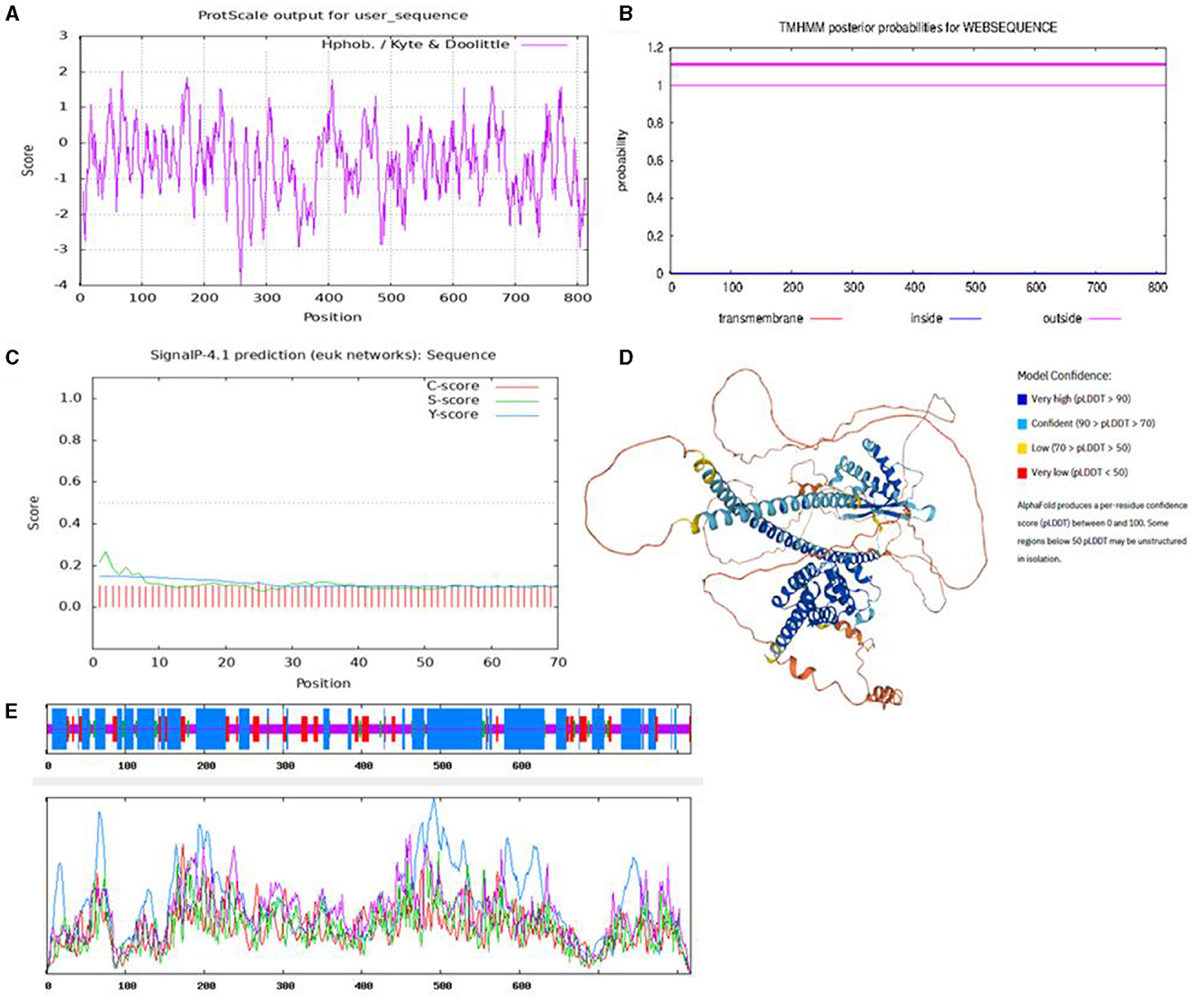
Figure 1. Prediction of physicochemical properties and structure of the SNX29 protein. (A) Hydrophobicity of goat SNX29 protein. (B) Goat SNX29 protein transmembrane signal peptide. (C) Goat SNX29 protein transmembrane signal peptide prediction. The abscissa axis represents the sequence number of amino acid residues corresponding to the submitted protein sequence; the value of the ordinate axis is the probability value of each amino acid located on the inside, outside, and TMhelix on the abscissa axis. (D) Three-dimensional model of goat SNX29 protein tertiary structure. (E) Secondary structural parameters of goat SNX29 protein. Blue means a-helix, red means extended backbone, green means β-folding, and yellow means random crimping.
Frequency of CNV genotypes in goats
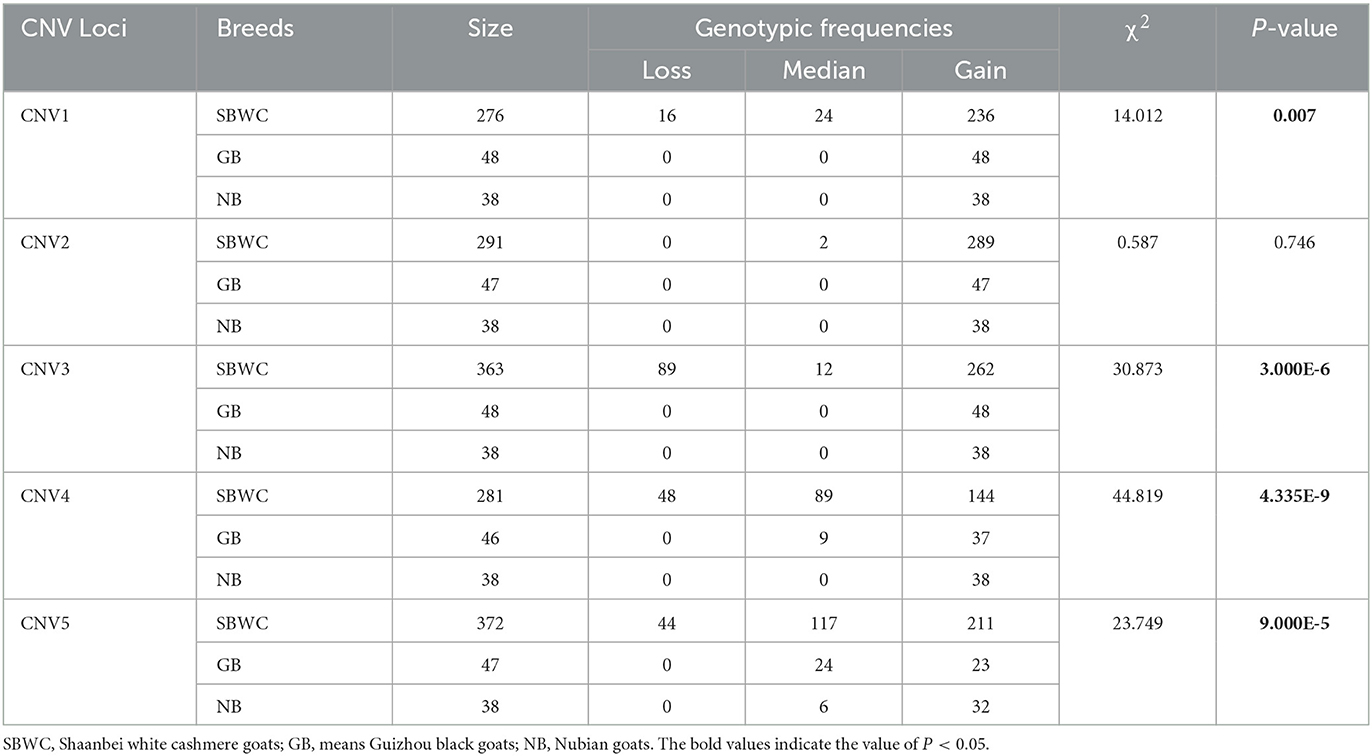
After mixed pool detection, it was found that the five CNVs were consistent with the target band (Figure 2). Then, by expanding the sample size for testing, the following results were obtained: In CNV1, the proportion of gain genotype was greater than that of median and loss genotypes in goats. There were 85.61% individuals with gain genotypes in the SBWC goats; however, and all individuals in the GB goats and NB goats were gain genotypes; in CNV2, all three goat breeds were gain genotype; in CNV3, there were 72.18% individuals of gain genotype, 3.31% individuals of median genotype, and 24.52% individuals of loss genotype in SBWC goats, and all GB goats and NB goats were gain genotype; in CNV4, there were 51.25% individuals of gain genotype, 31.67% individuals of median genotype, and 17.08% individuals of loss genotype in SBWC goats, there were 80.43% individuals of gain genotype, 19.57% individuals of median genotype in GB goats, and NB goats were all gain genotype; and in CNV5, there were 56.72% individuals of gain genotype, 31.45% individuals of median genotype, and 11.83% individuals of loss genotype in SBWC goats, there were 48.94% individuals of gain genotype, 51.06% individuals of median genotype in GB goats, there were 84.21% individuals of gain genotype, and 15.79% individuals of median genotype in NB goats (Figure 3).
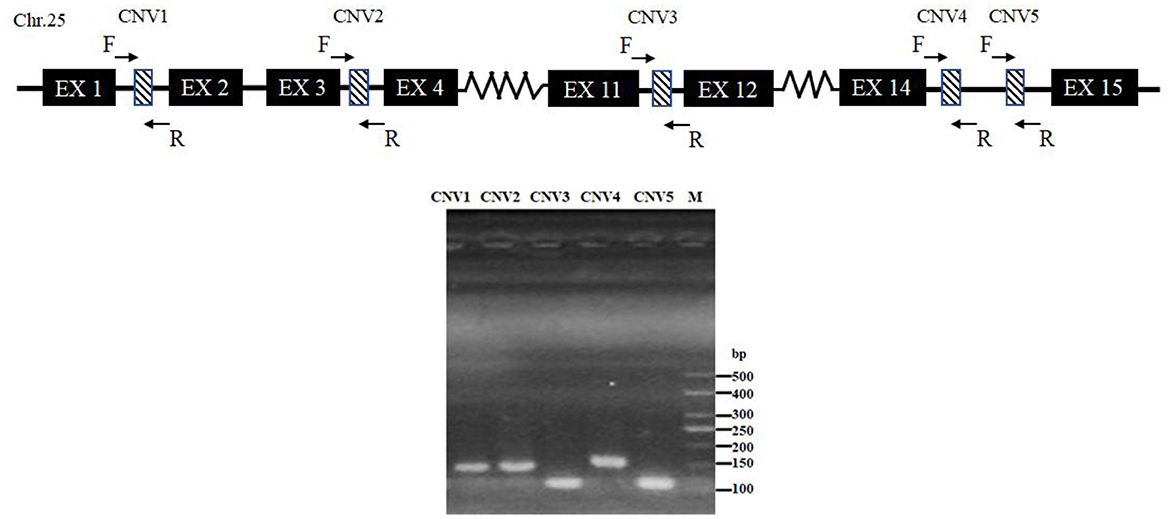
Figure 2. Schematic diagram of the PCR assay for CNVs of the goat SNX29 gene. Chr, chromosome; EX, exon; F, forward primer; R, reverse primer; M, means DNA marker.
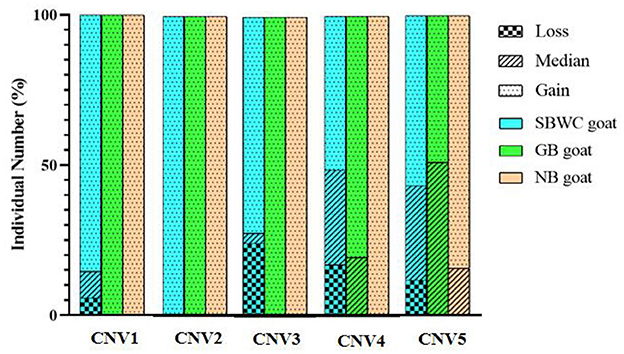
Figure 3. Genotyping proportion of CNVs in SBWC goat, GB goat, and NB goat. In CNV1: the total individual number of SBWC goats was 278, GB goats was 48, and NB goats was 38; in CNV2: the total individual number of SBWC goats was 290, GB goats was 48, and NB goats was 39; in CNV3: the total individual number of SBWC goats was 363, GB goats was 48, and NB goats was were 281, GB goats was 46, and NB goats was 38; in CNV5: the total individual number of SBWC goats was 372, GB goats was 48, and NB goats was 39.
Association analysis between CNVs and the goat SNX29 gene
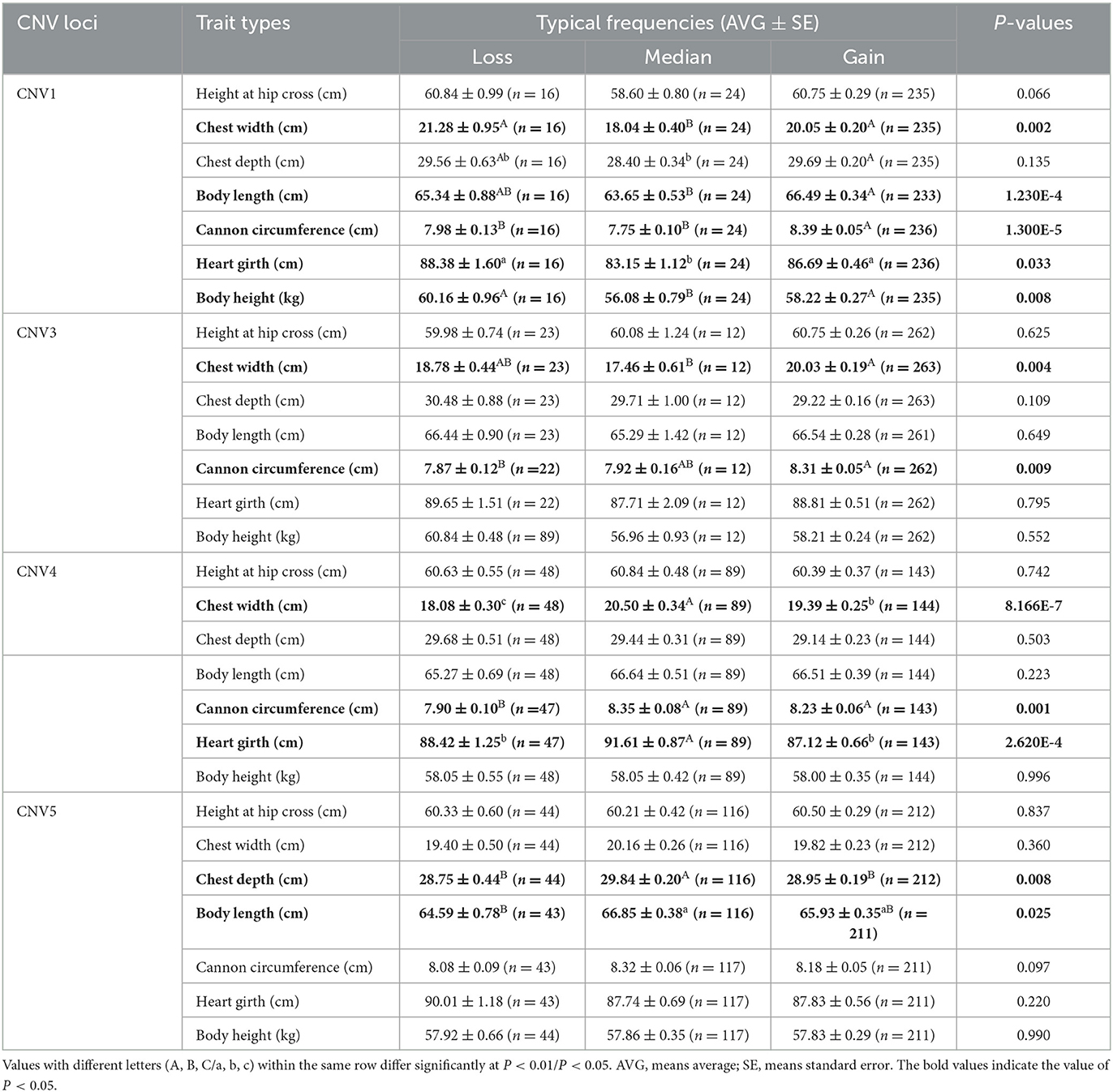
The association analysis results showed that four CNVs were related to growth traits in SBWC goats. CNV1 was significantly associated with chest width (P = 0.002), body length (P = 1.230E-4), body height (P = 0.008), cannon circumference (P = 1.300E-5), and heart girth (P = 0.033). CNV3 was significantly associated with chest width (P = 0.004) and cannon circumference (P = 0.009). CNV4 was significantly associated with chest width (P = 8.166E-7), heart girth (P = 2.620E-4), and cannon circumference (P = 0.001). CNV5 was significantly associated with chest depth (P = 0.008) and body length (P = 0.025). Additionally, in the association analysis between growth traits of SBWC goats and CNVs, we found that in CNV1 individuals, gain and loss genotypes were superior to those with median genotype on the aspect of growth traits, but in CNV4 and CNV5 individuals, median genotypes were superior to loss and gain. In addition, in the CNV3, the gain genotype performed better growth traits (Table 1). The χ2 test results showed that except for CNV2, the remaining CNV loci were significantly associated among the SBWC goats, GB goats, and NB goats (P < 0.01) (Table 2).
Discussion
Relevant studies have shown that SNX7 (34, 35), SNX8 (36), SNX9 (37), SNX10 (38), and SNX19 genes (39) were associated with animal growth traits. As a member of the same family, we speculated that the SNX29 CNVs may have a remarkable influence on growth traits. To preliminarily explore the function of the SNX29 gene, the goat SNX29 protein structure was predicted using an online platform. The results showed that the SNX29 protein was hydrophilic and had no transmembrane helix and signal peptide, and it is a non-secretory protein and performed a relevant function in the cytoplasm, which was consistent with the previous description (40).
To further explore the relationship between this gene and growth traits, we conducted population validation. Five CNVs were retrieved from the database. After population distribution detection, it was found that the genotypes of goats of the three breeds were different at different loci. This is because genetic variations vary from breed to breed (41). In the three goat breeds, more individuals performed gain genotype. This may be because the gain genotype showed better economic efficiency and was retained in artificial selection. Notably, the association analysis showed that four CNVs were observably associated with chest width, body length, body height, cannon circumference, and chest circumference (P < 0.05) in SBWC goats, which supports our conjecture. Moreover, we found that in CNV1 individuals, the gain and loss genotypes were superior to those with the median genotypes in terms of growth traits, but in CNV4 and CNV5 individuals, the median genotypes were better than the loss and gain genotypes. In addition, in the CNV3, the gain genotype performed better growth traits, which could be due to mutation, selection, gene recombination, and genetic drift migration (42). These outcomes suggest that the gain/loss genotype of CNV1, the gain genotype of CNV3, and the median genotype of CNV4 and CNV5 have a positive effect on growth traits (43).
In this study, we found that the CNVs of SNX29 were associated with the growth traits of goats, which is consistent with the function of SNX29 in previous studies associated with growth. A genome-wide scan identified the growth-related SNP markers of SNX29 in Chinese Wagyu cattle (44). Genome-wide association analysis showed that CNV27 of the SNX29 gene was associated with growth traits of African goats (45), and also two InDels within this gene are significantly correlated with chest width, hip width, and other growth traits in goats (46). In addition, this gene has shown growth-related functions in different species. In York pigs, genome-wide association analysis of five meat quality traits found that 12 intron SNPs of the SNX29 gene were associated with intramuscular fat content (47). Therefore, the SNX29 has been identified as a candidate gene associated with growth traits, whose CNVs can also act as an influence on the growth traits of livestock. We will continue to explore the molecular mechanism between this gene and growth traits in further studies.
Conclusion
In this study, the growth effect of the SNX29 gene was elucidated from the aspects of protein structure, physicochemical properties, and DNA variation. The protein encoded by SNX29 was a non-secreted protein, whose five CNVs were identified in SBWC goats, GB goats, and NB goats. Moreover, CNVs were found to be associated with growth traits in SBWC goats. The CNV1, CNV3, CNV4, and CNV5 were significantly associated with the SBWC goats, GB goats, and NB goats (P < 0.01). Thus, the SNX29 gene may be an essential functional candidate gene for growth traits.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Northwest A&F University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
XS, HZ, XW, ZG, and CP: sample collection. QW and YB: experimental operation. QW, YB, HZ, XW, and ZG: data collation and analysis. QW: article writing. QW, YB, and CP: manuscript revision and editing. ML and CP: project management. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (No.32002166).
Acknowledgments
The authors sincerely thank the Shaanxi Yulin goat farm, Guizhou Bijie goat farm, and Fujian ZhangZhou goat cooperative for providing them with samples. The authors would also like to thank Lei Qu, Hailong Yan, and Jinwang Liu from Yulin University for their help in sample collection, and XW for her help at the Institute of Animal Husbandry and Veterinary, Fujian Academy of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1132833/full#supplementary-material
References
1. Hao X, Wang Y, Ren F, Zhu S, Ren Y, Jia B, et al. SNX25 regulates TGF-β signaling by enhancing the receptor degradation. Cell Signal. (2011) 23:935–46. doi: 10.1016/j.cellsig.2011.01.022
2. Lin YJ, Chang JS, Liu X, Lin TH, Huang SM, Liao CC, et al. Sorting nexin 24 genetic variation associates with coronary artery aneurysm severity in Kawasaki disease patients. Cell Biosci. (2013) 3:44. doi: 10.1186/2045-3701-3-44
3. Amatya B, Lee H, Asico LD, Konkalmatt P, Armando I, Felder RA, et al. Subfamily of SNXs in the regulation of receptor-mediated signaling and membrane trafficking. Int J Mol Sci. (2021) 22:2319. doi: 10.3390/ijms22052319
4. Xia L, Ou J, Li K, Guo H, Hu Z, Bai T, et al. Genome-wide association analysis of autism identified multiple loci that have been reported as strong signals for neuropsychiatric disorders. Autism Res. (2020) 13:382–96. doi: 10.1002/aur.2229
5. Chen JH, Zhao Y, Khan RAW, Li ZQ, Zhou J, Shen JW, et al. SNX29, a new susceptibility gene shared with major mental disorders in Han Chinese population. World J Biol Psychiatry. (2021) 22:526–34. doi: 10.1080/15622975.2020.1845793
6. Twa DD, Mottok A, Chan FC, Ben-Neriah S, Woolcock BW, Tan KL, et al. Recurrent genomic rearrangements in primary testicular lymphoma. J Pathol. (2015) 236:136–41. doi: 10.1002/path.4522
7. Zhu L, Hu Z, Liu J, Gao J, Lin B. Gene expression profile analysis identifies metastasis and chemoresistance-associated genes in epithelial ovarian carcinoma cells. Med Oncol. (2015) 32:426. doi: 10.1007/s12032-014-0426-5
8. Sparks AM, Watt K, Sinclair R, Pilkington JG, Pemberton JM, McNeilly TN, et al. The genetic architecture of helminth-specific immune responses in a wild population of Soay sheep (Ovis aries). PLoS Genet. (2019) 15:e1008461. doi: 10.1371/journal.pgen.1008461
9. Peng S, Song C, Li H, Cao X, Ma Y, Wang X, et al. Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of myoblasts by activating the Wnt5a/Ca2+ signaling pathway. Mol Therapy Nucl Acids. (2019) 16:481–93. doi: 10.1016/j.omtn.2019.03.009
10. Chen Y, Yang L, Lin X, Peng P, Shen W, Tang S, et al. Effects of genetic variation of the sorting nexin 29 (SNX29) gene on growth traits of xiangdong black goat. Animals. (2022) 12:3461. doi: 10.3390/ani12243461
11. Wright D, Boije H, Meadows JR, Bed'hom B, Gourichon D, Vieaud A, et al. (2009). Copy number variation in intron 1 of SOX5 causes the Pea-comb phenotype in chickens. iPLoS Geneti. 5, e1000512. doi: 10.1371/journal.pgen.1000512
12. Henkel J, Saif R, Jagannathan V, Schmocker C, Zeindler F, Bangerter E, et al. Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS Genet. (2019) 15:e1008536. doi: 10.1371/journal.pgen.1008536
13. Zhang RQ, Wang JJ, Zhang T, Zhai HL, Shen W. Copy-number variation in goat genome sequence: a comparative analysis of the different litter size trait groups. Gene. (2019) 696:40–6. doi: 10.1016/j.gene.2019.02.027
14. Wang, L, Xu L., Liu X., Zhang T., Li N., Hay el H., et al. Copy number variation-based genome wide association study reveals additional variants contributing to meat quality in Swine. Sci Rep. (2015) 5:12535. doi: 10.1038/srep12535
15. Kang X, Li M, Liu M, Liu S, Pan MG, Wiggans GR, et al. Copy number variation analysis reveals variants associated with milk production traits in dairy goats. Genomics. (2020) 112:4934–7. doi: 10.1016/j.ygeno.2020.09.007
16. Fernandes AC, da Silva VH, Goes CP, Moreira GC, Godoy TF, Ibelli AM, et al. Genome-wide detection of CNVs and their association with performance traits in broilers. BMC Genom. (2021) 22:354. doi: 10.1186/s12864-021-07676-1
17. Strillacci MG, Gorla E, Ríos-Utrera A, Vega-Murillo VE, Montaño-Bermudez M, Garcia-Ruiz A, et al. Copy number variation mapping and genomic variation of autochthonous and commercial turkey populations. Front Genet. (2019) 10:982. doi: 10.3389/fgene.2019.00982
18. Pös O, Radvanszky J, Buglyó G, Pös Z, Rusnakova D, Nagy B, et al. Copy number variation: main characteristics, evolutionary significance, and pathological aspects. Biomed J. (2021) 44:548–59. doi: 10.1016/j.bj.2021.02.003
19. Bi Y, Feng B, Wang Z, Zhu H, Qu L, Lan X, et al. Myostatin (MSTN) gene indel variation and its associations with body traits in Shaanbei White Cashmere Goat. Animals. (2020) 10:168. doi: 10.3390/ani10010168
20. Wei Z, Wang K, Wu H, Wang Z, Pan C, Chen H, et al. Detection of 15-bp deletion mutation within PLAG1 gene and its effects on growth traits in goats. Animals. (2021) 11:2064. doi: 10.3390/ani11072064
21. Cai HF, Chen Z, Luo WX. Associations between polymorphisms of the GFI1B gene and growth traits of indigenous Chinese goats. Genet Mol Res. (2014) 13:872–80. doi: 10.4238/2014.February.13.5
22. Kholif AE, Gouda GA, Hamdon HA. Performance and milk composition of Nubian goats as affected by increasing level of nannochloropsis oculata microalgae. Animals. (2020) 10:2453. doi: 10.3390/ani10122453
23. Li X, Ding X, Liu L, Yang P, Yao Z, Lei C, et al. Copy number variation of bovine DYNC1I2 gene is associated with body conformation traits in Chinese beef cattle. Gene. (2022) 810:146060. doi: 10.1016/j.gene.2021.146060
24. Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. (1997) 25:4692–3. doi: 10.1093/nar/25.22.4692
25. Cui Y, Chen R, Lv X, Pan C. Detection of coding sequence, mRNA expression and three insertions/deletions (indels) of KDM6A gene in male pig. Theriogenology. (2019) 133:10–21. doi: 10.1016/j.theriogenology.2019.04.023
26. Zhang X, Yu S, Yang Q, Wang K, Zhang S, Pan C, et al. Goat boule: isoforms identification, mRNA expression in testis and functional study and promoter methylation profiles. Theriogenology. (2018) 116:53–63. doi: 10.1016/j.theriogenology.2018.05.002
27. Fu W, Wang R, Yu J, Hu D, Cai Y, Shao J, et al. A goat genome variation database for tracking the dynamic evolutionary process of selective signatures and ancient introgressions. J Genet Genom. (2021) 48:248–56. doi: 10.1016/j.jgg.2021.03.003
28. Wang Q, Bi Y, Wang Z, Zhu H, Liu M, Wu X, et al. Goat SNX29: mRNA expression, indel and CNV detection, and their associations with litter size. Front Vet Sci. (2022) 9:981315. doi: 10.3389/fvets.2022.981315
29. Li J, Zhang S, Erdenee S, Sun X, Dang R, Huang Y, et al. Nucleotide variants in prion-related protein (testis-specific) gene (PRNT) and effects on Chinese and Mongolian sheep phenotypes. Prion. (2018) 12:185–96. doi: 10.1080/19336896.2018.1467193
30. Yang Q, Zhang S, Li J, Wang X, Peng K, Lan X, et al. Development of a touch-down multiplex PCR method for simultaneously rapidly detecting three novel insertion/deletions (indels) within one gene: an example for goat GHR gene. Anim Biotechnol. (2019) 30:366–71. doi: 10.1080/10495398.2018.1517770
31. Bi Y, Feng W, Kang Y, Wang K, Yang Y, Qu L, et al. Detection of mRNA expression and copy number variations within the goat FecB gene associated with litter size. Front Vet Sci. (2021) 8:758705. doi: 10.3389/fvets.2021.758705
32. Yang Y, Hu H, Mao C, Jiang F, Lu X, Han X, et al. Detection of the 23-bp nucleotide sequence mutation in retinoid acid receptor related orphan receptor alpha (RORA) gene and its effect on sheep litter size. Anim Biotechnol. (2020) 33:70–8. doi: 10.1080/10495398.2020.1770273
33. Liu H, Xu H, Lan X, Cao X, Pan C. The InDel variants of sheep IGF2BP1 gene are associated with growth traits. Anim Biotechnol. (2021) 13:1–9. doi: 10.1080/10495398.2021.1942029
34. Edea Z, Hong JK, Jung JH, Kim DW, Kim YM, Kim ES, et al. Detecting selection signatures between Duroc and Duroc synthetic pig populations using high-density SNP chip. Anim Genet. (2017) 48:473–7. doi: 10.1111/age.12559
35. Lin S, Zhang H, Hou Y, Liu L, Li W, Jiang J, et al. discovery and functional candidate gene identification for milk composition based on whole genome resequencing of Holstein bulls with extremely high and low breeding values. PLoS ONE. (2019) 14:e0220629. doi: 10.1371/journal.pone.0220629
36. Muirhead G, Dev KK. The expression of neuronal sorting nexin 8 (SNX8) exacerbates abnormal cholesterol levels. J Mol Neurosci. (2014) 53:125–34. doi: 10.1007/s12031-013-0209-z
37. An B, Xu L, Xia J, Wang X, Miao J, Chang T, et al. Multiple association analysis of loci and candidate genes that regulate body size at three growth stages in Simmental beef cattle. BMC Genet. (2020) 21:32. doi: 10.1186/s12863-020-0837-6
38. Castillejo-Lopez C, Pjanic M, Pirona AC, Hetty S, Wabitsch M, Wadelius C, et al. Detailed functional characterization of a waist-hip ratio locus in 7p15.2 defines an enhancer controlling adipocyte differentiation. iScience. (2019) 20:42–59. doi: 10.1016/j.isci.2019.09.006
39. Guo L, Sun H, Zhao Q, Xu Z, Zhang Z, Liu D, et al. Positive selection signatures in Anqing six-end-white pig population based on reduced-representation genome sequencing data. Anim Genet. (2021) 52:143–54. doi: 10.1111/age.13034
40. Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. (2000) 28:231–4. doi: 10.1093/nar/28.1.231
41. Zhang Y, Wang K, Liu J, Zhu H, Qu L, Chen H, et al. An 11-bp Indel polymorphism within the CSN1S1 gene is associated with milk performance and body measurement traits in Chinese Goats. Animals. (2019) 9:1114. doi: 10.3390/ani9121114
42. Huang Y, Su P, Akhatayeva Z, Pan C, Zhang Q, Lan X, et al. Novel InDel variations of the Cry2 gene are associated with litter size in Australian White sheep. Theriogenology. (2022) 179:155–61. doi: 10.1016/j.theriogenology.2021.11.023
43. Zhou T, Wei H, Li D, Yang W, Cui Y, Gao J, et al. Novel missense mutation within the domain of lysine demethylase 4D (KDM4D) gene is strongly associated with testis morphology traits in pigs. Anim Biotechnol. (2020) 31:52–8. doi: 10.1080/10495398.2018.1531880
44. Wang Z, Ma H, Xu L, Zhu B, Liu Y, Bordbar F, et al. Genome-wide scan identifies selection signatures in Chinese Wagyu cattle using a high-density SNP array. Animals. (2019) 9:296. doi: 10.3390/ani9060296
45. Liu M, Woodward-Greene J, Kang X, Pan MG, Rosen B, Van Tassell CP, et al. Genome-wide CNV analysis revealed variants associated with growth traits in African indigenous goats. Genomics. (2020) 112:1477–80. doi: 10.1016/j.ygeno.2019.08.018
46. Bi Y, Chen Y, Xin D, Liu T, He L, Kang Y, et al. Effect of indel variants within the sorting nexin 29 (SNX29) gene on growth traits of goats. Anim Biotechnol. (2020) 19:1–6. doi: 10.1080/10495398.2020.1846547
Keywords: sorting nexin 29 (SNX29) gene, copy number variation (CNV), growth traits, goats, marker-assisted selection (MAS)
Citation: Wang Q, Song X, Bi Y, Zhu H, Wu X, Guo Z, Liu M and Pan C (2023) Detection distribution of CNVs of SNX29 in three goat breeds and their associations with growth traits. Front. Vet. Sci. 10:1132833. doi: 10.3389/fvets.2023.1132833
Received: 28 December 2022; Accepted: 17 July 2023;
Published: 29 August 2023.
Edited by:
Anupama Mukherjee, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Zhuanjian Li, Henan Agricultural University, ChinaTatiana Deniskova, L.K. Ernst Federal Science Center for Animal Husbandry (RAS), Russia
Copyright © 2023 Wang, Song, Bi, Zhu, Wu, Guo, Liu and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuanying Pan, Y2h1YW55aW5ncGFuQDEyNi5jb20=; Mei Liu, TWVpLkxpdUBodW5hdS5lZHUuY24=
†These authors have contributed equally to this work
 Qian Wang1†
Qian Wang1† Yi Bi
Yi Bi Haijing Zhu
Haijing Zhu Xianfeng Wu
Xianfeng Wu Mei Liu
Mei Liu Chuanying Pan
Chuanying Pan