
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 15 July 2022
Sec. Parasitology
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.933691
This article is part of the Research Topic Women in Parasitology: 2021 View all 10 articles
 Zhen-Huan Zhang1
Zhen-Huan Zhang1 Rui-Lin Qin1
Rui-Lin Qin1 Ya-Ya Liu1
Ya-Ya Liu1 Yang Zou2
Yang Zou2 Jin-Jin Mei1
Jin-Jin Mei1 Qing Liu1
Qing Liu1 Wen-Wei Gao1
Wen-Wei Gao1 Xing-Quan Zhu1,3,4
Xing-Quan Zhu1,3,4 Yu-Hong Ren1*
Yu-Hong Ren1* Shi-Chen Xie1,3*
Shi-Chen Xie1,3*Enterocytozoon bieneusi is a common opportunistic intestinal pathogen that can cause acute diarrhea in immunosuppressed humans and animals. Though E. bieneusi has been widely detected in pigs around the world, little is known of its prevalence and genotype distribution in pigs in Shanxi province, north China. In this study, a total of 362 fecal samples were collected from pigs in three representative counties in north, south, and central Shanxi province, China. The prevalence and genotypes of E. bieneusi were investigated by nested PCR amplification of the ribosomal internal transcribed spacer (ITS) region of the ribosomal RNA (rRNA) gene. Overall, the prevalence of E. bieneusi in pigs in Shanxi province was 54.70% (198/362). Statistical analysis showed the difference in prevalence was statistically significant between regions (χ2 = 41.94, df = 2, P < 0.001) and ages (χ2 = 80.37, df = 1, P < 0.001). In addition, 16 genotypes of E. bieneusi were identified in this study by sequence analysis of the ITS region, including 15 known genotypes (EbpC, EbpA, EbpB, pigEb4, PigEBITS5, I, Henan-I, G, WildBoar 7, SH10, EbpD, CHC5, PigSpEb1, PigSpEb2, and CHG19) and one novel genotype (designated as PigSX-1). Phylogenetic analysis revealed that 14 known genotypes and the novel genotype were clustered into Group 1, whereas genotype I belonged to Group 2. To the best of our knowledge, this is the first report on the prevalence and genotypes of E. bieneusi in pigs in Shanxi province. These findings enrich the genetic diversity of E. bieneusi and provide the baseline data for the prevention and control of E. bieneusi in pigs in the study regions.
The phylum Microsporidia contains a large group of single-celled, obligate intracellular spore-forming parasites (more than 220 genera and 1,700 species). Of which, Enterocytozoon bieneusi is the most frequently detected species in humans (1). Although E. bieneusi infection in immunocompetent individuals is usually asymptomatic (2), acute diarrhea can occur in immunocompromised individuals, such as patients with AIDS (3). In addition, E. bieneusi has also been detected in a variety of mammals and birds (4). Humans and animals can be infected by E. bieneusi through contact with infected hosts or by ingesting spore-contaminated water or food (5).
Genotyping of E. bieneusi is based on amplification and sequencing of the ribosomal internal transcribed spacer (ITS) region of the rRNA gene, which has high single nucleotide polymorphisms (SNPs) (6). At present, over 500 genotypes of E. bieneusi have been identified, which are divided into 11 phylogenetic groups (5). Group 1 is the largest human-pathogenic group containing more than 300 genotypes (5). The prevalence of E. bieneusi in pigs varied, ranging from 10 to 93.70% worldwide (5). A number of genotypes within Group 1 identified in humans have also been found in pigs, suggesting that pigs could serve as a potential reservoir for E. bieneusi transmission to humans (5, 7, 8).
According to data from the National Bureau of Statistics of China, approximately 8 million pigs were produced and consumed in Shanxi province annually (http://www.stats.gov.cn/tjsj/ndsj/2019/indexeh.htm). However, little is known about the epidemiology of E. bieneusi in pigs in Shanxi province. In this study, the prevalence and genotypes of E. bieneusi in pigs in Shanxi province were investigated by using nested PCR amplification of the ribosomal ITS region. Meanwhile, phylogenetic analysis was conducted to evaluate the zoonotic potential of the E. bieneusi isolates.
In November 2020, with the permission of the farm owners, a total of 362 fresh fecal samples were randomly collected from pigs in three farms each in Shanyin county (39°52′ N, 112°81′ E) located in northern Shanxi province, Qi county (37°35′ N, 112°33′ E) located in central Shanxi province, and Jishan county (35°59′ N, 110°97′ E) located in southern Shanxi province. Approximately, 5–15% of samples were collected from each farm. All fecal samples were transported to the laboratory in a styrofoam box with ice packs immediately and stored at −20°C until genomic DNA extraction.
The genomic DNA was extracted from each fecal sample (approximately 200 mg) using the E.Z.N.A.® Stool DNA Kit (Omega Bio-tek Inc., Norcross, GA, USA) and stored at −20°C until used for subsequent PCR amplification. A nested PCR was performed to amplify the ITS region by using E. bieneusi-specific primers described in a previous study (9). Briefly, the reaction mixture (25 μl) contained 2.5 μl of 10×PCR Buffer (Mg2+ free), 1.5 mM of MgCl2, 2 μl of dNTP mixture (2.5 mM each), 1.25 U of Ex-Taq polymerase (Takara, Dalian, China), 1 μM of each primer, 14.75 μl of ddH2O, and 2 μl of DNA template. The conditions and cycling parameters were as follows: initial denaturation at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, annealing at 55°C for 30 s, 72°C for 40 s, and a final extension at 72°C for 10 min. To ensure the reliability of the results, each PCR amplification included a negative control (reagent-grade water) and a positive control (DNA of the E. bieneusi BEB6 genotype from sheep). Then, secondary products were checked by using 2.5% agarose gel and visualized under UV light after staining in ethidium bromide.
All PCR products were sent to Sangon Biotech Co. Ltd (Shanghai, China) for two-directional sequencing on an ABI PRISM DNA Analyzer (Applied Biosystems, Foster City, CA, USA) using relevant internal primers for PCR amplification. The obtained sequences were aligned with the relevant sequences available in the GenBank database using Basic Local Alignment Search Tool (BLAST) and Clustal X to determine the genotypes of E. bieneusi. All samples with novel genotypes were sequenced two times to ensure the reliability of the data. The novel genotype was denominated according to the nomenclature established by Santin and Fayer (6). The phylogenetic tree was constructed by MEGA 7 using the Neighbor-Joining (NJ) method and Kimura 2-parameter model with 1,000 bootstraps (9).
In this study, the software SPSS V26.0 (SPSS Inc., Chicago, IL, USA) was used to analyze the correlation between prevalence and risk factors of E. bieneusi in pigs by Chi-square (χ2) test. Odds ratios (ORs) and their 95% confidence intervals (95%CIs) were calculated to identify risk factors. There was a significant difference in prevalence when the p-value was <0.05.
In this study, 198 of 362 fecal samples were detected to be positive for E. bieneusi, and the prevalence of E. bieneusi in pigs in Shanxi province was 54.70% (Table 1). Statistical analysis showed that the prevalence of E. bieneusi in pigs aged <6 months was 71.73% (170/237), which was significantly higher than that in pigs aged more than 6 months (22.40%, 28/125) (χ2 = 80.37, df = 1, P < 0.001). The prevalence of E. bieneusi in Qi county (22.06%, 15/68) was significantly lower than that of Shanyin county (53.15%, 59/111) and Jishan county (67.76%, 124/183) (χ2 = 41.94, df = 2, P < 0.001), respectively.
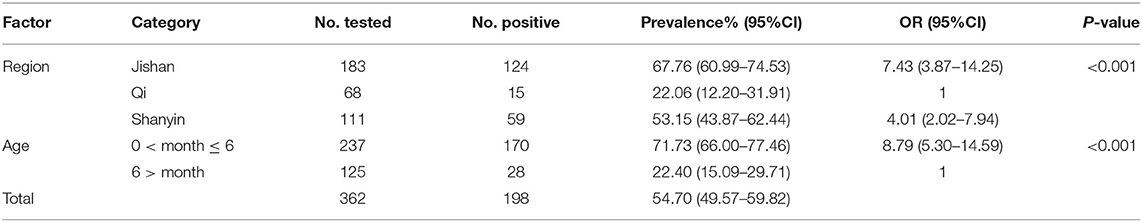
Table 1. Factors associated with prevalence of Enterocytozoon bieneusi in pigs in Shanxi province, China.
A total of 16 genotypes were identified by ITS sequence analysis, including 15 known genotypes (EbpC, EbpA, EbpB, pigEb4, PigEBITS5, I, Henan-I, G, WildBoar7, SH10, EbpD, CHC5, PigSpEb1, CHG19, and PigSpEb2) and one novel genotype (named as PigSX-1) (Table 2). Of which, EbpA (5.05%, 10/198), EbpC (34.34%, 68/198), and PigSpEb2 (22.22%, 44/198) were the predominant genotype in Qi county, Jishan county, and Shanyin county, respectively. Notably, genotype PigSpEb2 was detected in Shanyin county (55.70%, 44/79) and Jishan county (44.30%, 35/79), but not in Qi county. A comparison between the two age groups showed that PigSpEb2 was mainly distributed in young pigs (<6 months) (94.94%, 75/79). Almost all the genotypes identified in Jishan county were EbpC (98.55%, 68/69), which was mainly detected in young pigs (91.30%, 63/69). In addition, the novel genotype pigSX-1 (2.53%, 5/198) was only detected in pigs in Jishan county. Sequence analysis revealed that the novel genotype pigSX-1 showed a 98.71% similarity to the genotype EbpB (AF076041), with five SNPs.
A phylogenetic tree was used to evaluate the genetic relationship of the 16 genotypes of E. bieneusi obtained in this study. The results showed that all 15 genotypes were clustered into Group 1, except for genotype I, which belonged to Group 2 (Figure 1).
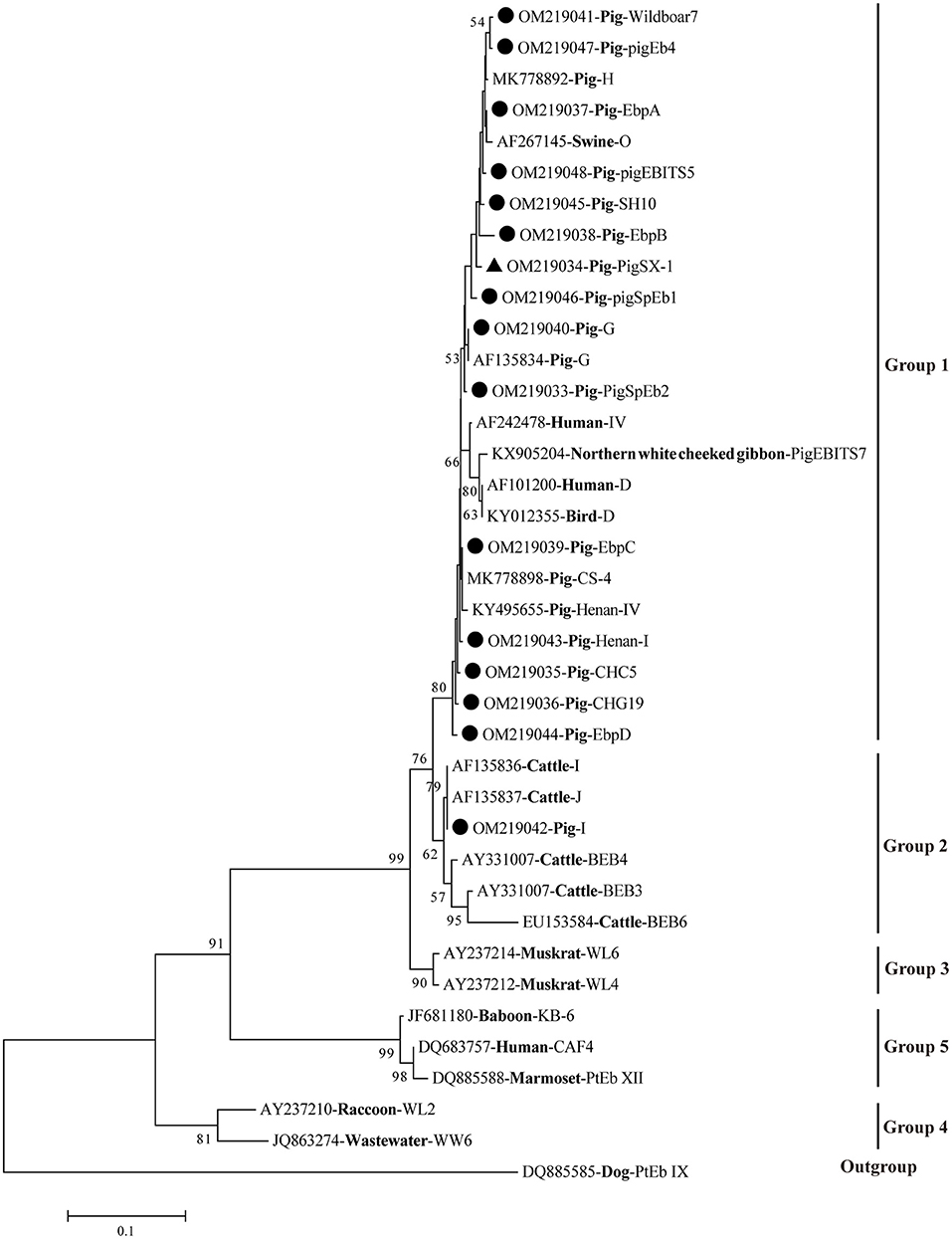
Figure 1. Phylogenetic relationships of E. bieneusi genotypes based on ITS sequences. Obtained ITS sequences in the present study were marked with black circles (• known genotypes) and black triangle (▴ novel genotype), respectively. The bootstrap value <50% was hide.
The 16 representative ITS sequences of E. bieneusi obtained in this study were deposited in the GenBank database under accession numbers OM219033-OM219048.
E. bieneusi has caused economic losses to the pig industry worldwide since it was detected in Sweden in 1996 for the first time (10). The results obtained in this study showed that the overall prevalence of E. bieneusi in pigs in Shanxi province was 54.70% (198/362), which was higher than that in pigs in most provinces in China (11–17) (Table 3), Thailand (14.75%, 36/244) (18), Japan (33.33%, 10/30) (19), Spain (22.58%, 42/186) (20), Massachusetts, USA (31.68%, 64/202) (21), and Malaysia (40.67%, 183/450) (22). However, the prevalence of E. bieneusi in pigs in Shanxi province was lower than that in two provinces in China (23, 24) (Table 3) and Brazil (59.34%, 54/91) (25). Regional differences in the prevalence of E. bieneusi may be related to geographical locations, sample volumes, breeding management, and ecological factors.
There were significant differences in E. bieneusi prevalence between the two age groups, which was consistent with the results of a previous study (14). Some researchers argue that the probable reason for the higher prevalence of E. bieneusi in young pigs (<6 months) might be due to their imperfect immune system (23). However, a high prevalence of E. bieneusi was also found in older pigs in different areas of China (12, 14). The difference in E. bieneusi prevalence among these age groups indicated that geoecology, rearing conditions, and stocking density may be partially responsible for the variations in prevalence.
In this study, 15 known genotypes (PigSpEb2, EbpC, EbpA, EbpB, pigEb4, PigEBITS5, I, Henan-I, G, WildBoar7, SH10, EbpD, CHC5, PigSpEb1, and CHG19) and a novel genotype (PigSX-1) were identified in pigs in Shanxi province. Of which, genotype PigSpEb2 (39.90%, 79/198) was the predominant genotype, followed by EbpC (34.85%, 69/198) (synonyms: E, WL13, WL17, and Peru4) and EbpA (9.60%, 19/198) (synonym: F). This finding was not consistent with the results of previous studies, in which EbpC was detected as the predominate genotype in pigs in Zhejiang province, Guangdong province, Jilin province, and Tibet Autonomous Region in China (13, 14, 26). So far, the reasons for the difference in predominate genotypes of E. bieneusi in pigs from different study regions are still unknown. We reasoned that the geographical locations, pig breeds, and hygiene conditions might be responsible for the variations in predominate genotypes. Hence, more samples from diverse hosts in the study areas should be examined in the future to further clarify the possible patterns of prevalent genotypes of E. bieneusi.
Of those 16 identified genotypes, seven known genotypes (EbpC, EbpA, EbpB, PigEBITS5, I, EbpD, and CHG19) were commonly observed in humans (27), livestock (7, 28–30), non-human primates (NHPs) (31), wild animals (32), and water (33), posing a great threat to the public health. Particularly, genotypes EbpC and I were also found in squirrels and pet rabbits in China, respectively, which have close contact with humans (34, 35). Genotypes PigSpEb1 and PigSpEb2 were first identified in pigs in Spain in 2020 and 2021, respectively, but there was no data regarding the age patterns of the two genotypes in pigs (20, 36). Although our results revealed that younger pigs (<6 months) were more susceptible to PigSpEb1 and PigSpEb2, more investigations are still needed to confirm this in the future. A few studies have reported the presence of PigEb4, Henan-I, CHC5, Wildboar7, and SH10 in pigs, and further studies are warranted to clarify the host specificity and public health implications of these genotypes (24, 30, 37, 38). Phylogenetic analysis showed that 15 known genotypes were clustered into Group 1 and Group 2 (Figure 1). The novel genotype (PigSX-1) was clustered into Group 1, and was genetically closely related to zoonotic genotype EbpB, suggesting its importance and zoonotic potential.
This study reported, for the first time, the prevalence of E. bieneusi (54.70%) in pigs in Shanxi province, north China, and a higher prevalence was observed in young pigs. Fifteen known E. bieneusi genotypes and one novel genotype (PigSX-1) were identified. Fifteen genotypes were clustered into Group 1, suggesting that these infections may not only be a veterinary issue but also a public health concern. These findings enriched the global genetic diversity of E. bieneusi and provided baseline data for the prevention and control of E. bieneusi infection in pigs in the study regions.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethical review and approval was not required for the animal study because this is not applicable. Written informed consent was obtained from the owners for the participation of their animals in this study.
S-CX, X-QZ, and Y-HR conceived and designed the experiments. Z-HZ performed the experiments, analyzed the data, and wrote the paper. J-JM, R-LQ, and Y-YL participated in the collection of fecal samples. W-WG and Y-HR participated in the implementation of the study. S-CX, QL, YZ, and X-QZ critically revised the manuscript. All authors have read and approved the final version of the manuscript.
Project support was provided by Fund for Shanxi 1331 Project (Grant No. 20211331-13), the Special Research Fund of Shanxi Agricultural University for High-level Talents (Grant No. 2021XG001), the Yunnan Expert Workstation (Grant No. 202005AF150041), and the Veterinary Public Health Innovation Team of Yunnan Province (Grant No. 202105AE160014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Han B, Pan G, Weiss LM. Microsporidiosis in humans. Clin Microbiol Rev. (2021) 34:e0001020. doi: 10.1128/CMR.00010-20
2. Sak B, Brady D, Pelikánová M, Květonová D, Rost M, Kostka M, et al. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J Clin Microbiol. (2011) 49:1064–70. doi: 10.1128/JCM.01147-10
3. Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. (2012) 2012:981424. doi: 10.1155/2012/981424
4. Santin M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. (2011) 90:363–71. doi: 10.1016/j.rvsc.2010.07.014
5. Li W, Feng Y, Santin M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. (2019) 35:436–51. doi: 10.1016/j.pt.2019.04.004
6. Santin M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. (2009) 56:34–8. doi: 10.1111/j.1550-7408.2008.00380.x
7. Hu Y, Feng Y, Huang C, Xiao L. Occurrence, source, and human infection potential of Cryptosporidium and Enterocytozoon bieneusi in drinking source water in Shanghai, China, during a pig carcass disposal incident. Environ Sci Technol. (2014) 48:14219–27. doi: 10.1021/es504464t
8. Leelayoova S, Piyaraj P, Subrungruang I, Pagornrat W, Naaglor T, Phumklan S, et al. Genotypic characterization of Enterocytozoon bieneusi in specimens from pigs and humans in a pig farm community in central Thailand. J Clin Microbiol. (2009) 47:1572–4. doi: 10.1128/JCM.00187-09
9. Ma YT, Zou Y, Liu Q, Xie SC, Li RL, Zhu XQ, et al. Prevalence and multilocus genotypes of Enterocytozoon bieneusi in alpacas (Vicugna pacos) in Shanxi province, northern China. Parasitol Res. (2019) 118:3371–5. doi: 10.1007/s00436-019-06503-7
10. Deplazes P, Mathis A, Muller C, Weber R. Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J Eukaryot Microbiol. (1996) 43:93S. doi: 10.1111/j.1550-7408.1996.tb05018.x
11. Zhang N, Wu R, Ji T, Cui LL, Cao HX, Li D, et al. Molecular detection, multilocus genotyping, and population genetics of Enterocytozoon bieneusi in pigs in southeastern China. J Eukaryot Microbiol. (2020) 67:107–14. doi: 10.1111/jeu.12759
12. Wan Q, Lin Y, Mao Y, Yang Y, Li Q, Zhang S, et al. High prevalence and widespread distribution of zoonotic Enterocytozoon bieneusi genotypes in swine in northeast China: implications for public health. J Eukaryot Microbiol. (2016) 63:162–70. doi: 10.1111/jeu.12264
13. Zou Y, Zheng WB, Song HY, Xia CY, Shi B, Liu JZ, et al. Prevalence and genetic characterization of Enterocytozoon bieneusi and Giardia duodenalis in Tibetan pigs in Tibet, China. Infect Genet Evol. (2019) 75:104019. doi: 10.1016/j.meegid.2019.104019
14. Zou Y, Hou L, Li FC, Zou FC, Lin RQ, Ma JG, et al. Prevalence and genotypes of Enterocytozoon bieneusi in pigs in southern China. Infect Genet Evol. (2018) 66:52–6. doi: 10.1016/j.meegid.2018.09.006
15. Feng S, Jia T, Huang J, Fan Y, Chang H, Han S, et al. Identification of Enterocytozoon bieneusi and Cryptosporidium spp. in farmed wild boars (Sus scrofa) in Beijing, China. Infect Genet Evol. (2020) 80:104231. doi: 10.1016/j.meegid.2020.104231
16. Li DF, Zhang Y, Jiang YX, Xing JM, Tao DY, Zhao AY, et al. Genotyping and zoonotic potential of Enterocytozoon bieneusi in pigs in Xinjiang, China. Front Microbiol. (2019) 10:2401. doi: 10.3389/fmicb.2019.02401
17. Zhou HH, Zheng XL, Ma TM, Qi M, Zhou JG, Liu HJ, et al. Molecular detection of Enterocytozoon bieneusi in farm-raised pigs in Hainan province, China: infection rates, genotype distributions, and zoonotic potential. Parasite. (2020) 27:12. doi: 10.1051/parasite/2020009
18. Thathaisong U, Siripattanapipong S, Leelayoova S, Mungthin M. Prevalence and molecular characterization of Enterocytozoon bieneusi among pigs in Chonburi province, eastern Thailand. Am J Trop Med Hyg. (2019) 101:1392–6. doi: 10.4269/ajtmh.19-0569
19. Abe N, Kimata I. Molecular survey of Enterocytozoon bieneusi in a Japanese porcine population. Vector Borne Zoonotic Dis. (2010) 10:425–7. doi: 10.1089/vbz.2009.0039
20. Dashti A, Rivero-Juárez A, Santín M, López-López P, Caballero-Gómez J, Frías-Casas M, et al. Enterocytozoon bieneusi (Microsporidia): identification of novel genotypes and evidence of transmission between sympatric wild boars (Sus scrofa ferus) and Iberian pigs (Sus scrofa domesticus) in southern Spain. Transbound Emerg Dis. (2020) 67:2869–80. doi: 10.1111/tbed.13658
21. Buckholt MA, Lee JH, Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. (2002) 68:2595–9. doi: 10.1128/AEM.68.5.2595-2599.2002
22. Ruviniyia K, Abdullah DA, Sumita S, Lim YAL, Ooi PT, Sharma RSK. Molecular detection of porcine Enterocytozoon bieneusi infection in Peninsular Malaysia and epidemiological risk factors associated with potentially zoonotic genotypes. Parasitol Res. (2020) 119:1663–74. doi: 10.1007/s00436-020-06648-w
23. Wang SS, Li JQ, Li YH, Wang XW, Fan XC, Liu X, et al. Novel genotypes and multilocus genotypes of Enterocytozoon bieneusi in pigs in northwestern China: a public health concern. Infect Genet Evol. (2018) 63:89–94. doi: 10.1016/j.meegid.2018.05.015
24. Li D, Zheng S, Zhou C, Karim MR, Wang L, Wang H, et al. Multilocus typing of Enterocytozoon bieneusi in pig reveals the high prevalence, zoonotic potential, host adaptation and geographical segregation in China. J Eukaryot Microbiol. (2019) 66:707–18. doi: 10.1111/jeu.12715
25. Fiuza VR, Oliveira FC, Fayer R, Santín M. First report of Enterocytozoon bieneusi in pigs in Brazil. Parasitol Int. (2015) 64:18–23. doi: 10.1016/j.parint.2015.01.002
26. Li W, Diao R, Yang J, Xiao L, Lu Y, Li Y, et al. High diversity of human-pathogenic Enterocytozoon bieneusi genotypes in swine in northeast China. Parasitol Res. (2014) 113:1147–53. doi: 10.1007/s00436-014-3752-9
27. Wang SS, Wang RJ, Fan XC, Liu TL, Zhang LX, Zhao GH. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. (2018) 183:142–52. doi: 10.1016/j.actatropica.2018.04.017
28. Li J, Luo N, Wang C, Qi M, Cao J, Cui Z, et al. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit Vectors. (2016) 9:142. doi: 10.1186/s13071-016-1425-5
29. Shi K, Li M, Wang X, Li J, Karim MR, Wang R, et al. Molecular survey of Enterocytozoon bieneusi in sheep and goats in China. Parasit Vectors. (2016) 9:23. doi: 10.1186/s13071-016-1304-0
30. Li W, Li Y, Li W, Yang J, Song M, Diao R, et al. Genotypes of Enterocytozoon bieneusi in livestock in China: high prevalence and zoonotic potential. PLoS ONE. (2014) 9:e97623. doi: 10.1371/journal.pone.0097623
31. Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS ONE. (2015) 10:e0117991. doi: 10.1371/journal.pone.0117991
32. Lin X, Xin L, Cao Y, Hou M, Qiao F, Li J, et al. Common occurrence of Enterocytozoon bieneusi genotypes SHR1 and PL2 in farmed masked palm civet (Paguma larvata) in China. Int J Parasitol Parasites Wildl. (2021) 16:99–102. doi: 10.1016/j.ijppaw.2021.08.009
33. Ye J, Ji Y, Xu J, Ma K, Yang X. Zoonotic Enterocytozoon bieneusi in raw wastewater in Zhengzhou, China. Folia Parasitol. (2017) 64:2017.002. doi: 10.14411/fp.2017.002
34. Deng L, Chai Y, Luo R, Yang L, Yao J, Zhong Z, et al. Occurrence and genetic characteristics of Cryptosporidium spp. and enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Sci Rep. (2020) 10:1026. doi: 10.1038/s41598-020-57896-w
35. Deng L, Chai Y, Xiang L, Wang W, Zhou Z, Liu H, et al. First identification and genotyping of Enterocytozoon bieneusi and Encephalitozoon spp. in pet rabbits in China. BMC Vet Res. (2020) 16:212. doi: 10.1186/s12917-020-02434-z
36. Dashti A, Rivero-Juárez A, Santín M, George NS, Köster PC, López-López P, et al. Diarrhoea-causing enteric protist species in intensively and extensively raised pigs (Sus scrofa domesticus) in southern Spain. part I: prevalence and genetic diversity. Transbound Emerg Dis. (2021). doi: 10.1111/tbed.14388. [Epub ahead of print].
37. Rinder H, Thomschke A, Dengjel B, Gothe R, Löscher T, Zahler M. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J Parasitol. (2000) 86:185–8. doi: 10.1645/0022-3395(2000)086[0185:CGRBEB]2.0.CO;2
Keywords: Enterocytozoon bieneusi, pigs, prevalence, genotypes, Shanxi province
Citation: Zhang Z-H, Qin R-L, Liu Y-Y, Zou Y, Mei J-J, Liu Q, Gao W-W, Zhu X-Q, Ren Y-H and Xie S-C (2022) Molecular Detection and Genotyping of Enterocytozoon bieneusi in Pigs in Shanxi Province, North China. Front. Vet. Sci. 9:933691. doi: 10.3389/fvets.2022.933691
Received: 01 May 2022; Accepted: 16 June 2022;
Published: 15 July 2022.
Edited by:
Simona Gabrielli, Sapienza University of Rome, ItalyReviewed by:
Iraj Mohammadpour, Shiraz University of Medical Sciences, IranCopyright © 2022 Zhang, Qin, Liu, Zou, Mei, Liu, Gao, Zhu, Ren and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Hong Ren, renyuhong1963@163.com; Shi-Chen Xie, xieshichen221@163.com
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.