
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 19 May 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.846322
 Xiabing Chen1*
Xiabing Chen1* Zhiyong Shao1
Zhiyong Shao1 Lijun Wu1
Lijun Wu1 Bin He1
Bin He1 Wenhai Yang1
Wenhai Yang1 Jie Chen1
Jie Chen1 Erguang Jin1
Erguang Jin1 Qi Huang2,3
Qi Huang2,3 Liancheng Lei4
Liancheng Lei4 Jiajia Xu2,3
Jiajia Xu2,3 Haotian Li2,3
Haotian Li2,3 Hui Zhang2,3
Hui Zhang2,3 Yun Wan5
Yun Wan5 Wu Liu1*
Wu Liu1* Rui Zhou2,3*
Rui Zhou2,3*Actinobacillus pleuropneumoniae causes porcine pleuropneumonia. The function of the outer membrane protein W gene (ompW) of A. pleuropneumoniae has not been evaluated. Thus a deletion mutant of ompW, ΔompW, was constructed to explore the effect of ompW gene deletion on bacterial growth, biofilm formation, bacterial morphology, oxidative tolerance, susceptibility to antibiotics, and the expression of ribosome synthesis and ABC transporter related genes. Results showed that the ompW gene deletion did not affect biofilm formation and the growth of A. pleuropneumoniae but did affect bacterial morphology during steady growth, oxidative tolerance, and bacterial susceptibility to polymyxin B, kanamycin, and penicillin. The ompW gene deletion also affected the expression of ribosome synthesis and ABC transporter related genes. These results suggested that ompW may regulate the biological phenotype of A. pleuropneumoniae.
Actinobacillus pleuropneumoniae causes porcine pleuropneumonia and is one of the most important bacterial respiratory pathogens of pigs (1). In large-scale pig breeding farms, economic loss due to A. pleuropneumoniae is a serious concern. Based on the growth requirement for nicotinamide adenine dinucleotide (NAD), A. pleuropneumoniae are identified as biotype I or biotype II (2). Biotype I strictly requires NAD for growth, while biotype II does not require NAD as a growth supplement (3). Based on the nature of the bacterial capsule and lipopolysaccharide, A. pleuropneumoniae is classified into 19 serotypes, with serotypes 1, 5, 9, 10, and 11 highly virulent (4, 5). In China, serotypes 1, 3, and 7 are the dominant serovars (6). Immunological protection does not exist across serotypes (7), hence A. pleuropneumoniae infections are difficult clinical problems.
Recently, important virulence related factors have been identified that are involved in the pathogenesis of A. pleuropneumoniae. Repeats-in-toxin (RTX) and ApxIIIA toxin have been shown to target host cell β2 integrins killing many types of leukocytes and phagocytic cells (8). Deletion of PEP-carboxylase (PEPC) and PEP-carboxykinase (PEPCK) attenuate A. pleuropneumoniae virulence in a pig infection model (9). Disruption of TolC1 significantly reduces A. pleuropneumoniae virulence in a murine intraperitoneal injection model (10). Inactivation of flp1 and tadD genes affected A. pleuropneumoniae biofilm formation, cell adhesion, and resistance to phagocytosis (11). The hfq gene of A. pleuropneumoniae is helpful to regulate the adhesion to biological and abiotic surfaces, resistance to various stress conditions, and virulence (12). Deletion of outer membrane lipoprotein, Lip40, significantly attenuates adherence to St. Jude porcine lung cells and colonization of mouse lung tissue, indicating that Lip40 participates in the virulence of A. pleuropneumoniae (13). CpxA/CpxR affect A. pleuropneumoniae biofilm formation with mutation of cpxA/cpxR reducing mortality and bacterial load in a murine experimental lung infection model (14). Although these virulence factors have been intensively investigated, the mechanistic basis for A. pleuropneumoniae pathogenicity remains unclear.
Outer membrane proteins (OMPs) of bacteria play core roles in bacterial pathogenesis (15). The ompW gene is found in many bacteria, such as Escherichia coli (16), Aeromonas hydrophila (17), and Vibrio harveyi (18). E. coli OmpW knockout contributes to oxidative stress resistance (19). Response to the OmpW protein provides immunity to Aeromonas veronii challenge, with an immunization strategy a potential means by which to control Aeromonas veronii infection (20). OmpW regulates biofilm formation of Cronobacter sakazakii during NaCl stress (21). Deletion of ompW attenuates Vibrio cholerae growth in hypersaline culture conditions (22). Caulobacter crescentus ompW is an outer membrane cation channel (23). It serves as a multidrug resistance transporter involved in the efflux of ethidium multidrug resistance protein E (EmrE) substrates across the outer membrane (24). The function of A. pleuropneumoniae ompW has not been identified.
The purpose of this study was to investigate the role of A. pleuropneumoniae ompW in bacterial pathogenicity. To do so we constructed a deletion mutant of ompW, strain ΔompW, and a complemented strain, CΔompW. Deletion of the ompW gene did not affect the growth and biofilm formation of A. pleuropneumoniae, but did influence steady state growth bacterial morphology, as well as bacterial susceptibility to polymyxin B, kanamycin, and penicillin. In addition, ompW gene inactivation modified expression of ribosome synthesis related genes and ABC transporter genes. These results provide a better understanding of A. pleuropneumoniae ompW gene function during infection.
The A. pleuropneumoniae 4074 strain, serovar 1, is highly virulent and cultured on tryptic soy agar (TSA; Difco Laboratories, USA) or in tryptic soy broth (TSB, Difco Laboratories, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 10 μg/mL NAD (Sigma-Aldrich, USA). The ΔompW and CΔompW strains were constructed in this study, and the culture conditions were the same as the A. pleuropneumoniae 4074 strain. The pEMOC2 plasmid was from the laboratory of Prof. gerald-f. Gerlach and the pJFF224-XN plasmid was stored in our laboratory. The pEMΔompW and pJFF-ompW plasmid were constructed in this study. The E. coli β2155 strain was cultured on Luria-Bertani (LB, Oxoid Ltd. UK) agar or in broth. The A. pleuropneumoniae and E. coli strains were cultured at 37°C. If necessary, 2 μg/mL or 50 μg/mL of chloramphenicol (Sigma-Aldrich, USA) was added to the culture medium.
The method for construction of ΔompW has been reported previously (25). Briefly, a 648 bp internal deletion of the ompW gene was amplified with primers ompWUF/ompWUR and ompWDF/ompWDR utilizing single-overlap extension polymerase chain reaction (SOE PCR). Then a 2073 bp PCR product (pEompWUF/pEompWUR) was cloned into the suicide plasmid vector pEMOC2 to obtain the plasmid pEMΔompW (26), which included the 648 bp deletion fragment in frame. The pEMΔompW plasmid in E. coli β2155 strain was transformed into the wild-type strain of A. pleuropneumoniae with a single-step transconjugation system (27, 28). The ΔompW mutant was obtained by two homologous recombination steps and was identified with ompWMF/ompWMR primers.
The ompW gene was amplified with ompWF/ompWR primers for construction of CΔompW. The ompW gene was then connected to the shuttle vector pJFF224-XN and was electrically transferred into the ΔompW mutant to construct the corresponding complemented strain. Voltage was 2.5 KV, capacitance 25 μFD, and pulse resistance 800 Ω using an electroporation apparatus (Bio-Rad, USA). The complemented strain was screened with 2 μg/mL chloramphenicol and was identified with PJompWR/ PJompWF primers. The bacterial strains, plasmids used in this study are listed in Table 1 and the primers used in this study are listed in Table 2.
The effect of the ompW gene deletion on A. pleuropneumoniae growth was assessed as described previously (29). Briefly, the A. pleuropneumoniae, the ΔompW, and CΔompW strains were cultured in TSB supplemented with 10% FBS and 10 μg/mL NAD at 37°C overnight. The bacterial were then transferred to fresh TSB supplemented with 10% FBS and 10 μg/mL NAD at a 1:100 (V/V) dilution for logarithmic growth. The initial OD600 value was adjusted to 0.2 and the OD600 value of the bacterial suspension determined 1 h to 8 h after culture initiation for kinetic analysis.
The A. pleuropneumoniae and the ΔompW biofilm formation were as described previously with minor modifications (30). Briefly, an overnight bacterial culture was transferred to fresh TSB supplemented with 10% FBS and 10 μg/mL NAD at a 1:100 (V/V) dilution. Then a 100 μL bacterial suspension was added to the wells of a 96-well microplate and incubated for 12 h, 24 h, 36 h, and 48 h at 37°C. The bacterial culture liquid was removed, each well washed with PBS, and 100 μL of Crystal Violet dye solution (0.1%) added for 30 min. The wells were washed with PBS to remove unbound crystal violet dye and dried for 5 h at 37°C. Then 100 μL of a glacial acetic acid solution (33%, v/v) was added to each well and the microplate was shaken for 1 h. The absorbance at 590 nm was measured.
The bacterial morphologies of the A. pleuropneumoniae and the ΔompW strains were examined by SEM and TEM as described previously with minor modifications (31, 32). Briefly, the strains were cultured to logarithmic and steady state phases of growth. The bacteria were collected, washed three times with PBS, and fixed with 4% glutaraldehyde. Specimens were dehydrated, dried, and sprayed with gold. Bacterial morphologies were observed with a HITACHI SU8010 scanning electron microscope (Hitachi, Japan). For TEM, the strains were fixed with 0.1 M cacodylate buffer containing 5% glutaraldehyde and 0.15% ruthenium red at 37°C for 4 h and then treated with 1 mg/mL polycationic ferritin. Thin sections were prepared with a Tecnai G2 20 TWIN transmission electron microscope (FEI, Portland, OR, USA).
Stress resistance tests were conducted using the A. pleuropneumoniae, the ΔompW, and CΔompW strains. These strains were grown in TSA supplemented with 10% FBS and 10 μg/mL NAD at 37°C overnight. The bacterial were then transferred to fresh TSB supplemented with 10% FBS and 10 μg/mL NAD at a 1:100 (V/V) dilution for logarithmic growth. At an OD600 value of approximately 0.6, cells from 1 ml of broth cultures were centrifuged at 5000 g for 5 min. For the oxidative tolerance test, the cells were resuspended in 1 ml of TSB supplemented with 10% FBS, 10 μg/mL NAD and 1 mM hydrogen peroxide for 10 min or 20 min, respectively. The control samples of each strain were resuspended in 1 ml of TSB supplemented with 10% FBS and 10 μg/mL NAD without any treatment. Then, the cultures from oxidative stress resistance test were serially diluted in TSB, and spread on TSB plates supplemented with 10% FBS and 10 μg/mL NAD for CFU counting. Oxidative stress resistance was calculated as ((oxidative stressed sample CFU mL−1)/(control sample CFU mL−1)) × 100. The experiments were carried out in triplicate (20).
The minimal inhibitory concentration (MIC) of the A. pleuropneumoniae, ΔompW, and CΔompW strains for 19 antibiotics (rifampicin, Ofloxacin, polymyxin B, vancomycin, acriflavine, cefalexin, lincomycin, gentamicin, kanamycin, neomycin, penicillin, amoxicillin, tetracycline, streptomycin, vibramycin, enrofloxacin, florfenicol, sulfamethoxazole, and ceftiofur sodium; Yuanye biology Co., Ltd, China) was determined as described previously with minor modifications (33). Briefly, 50 μL of a TSB culture supplemented with 10% FBS and 10 μg/mL NAD of each strain were added to 96-well plates. The appropriate concentration of antibiotics was diluted 12 times continuously, and then 50 μL of diluted antibiotics were added to the 96 well plate to make the final concentration of 0.125 to 256 μg/mL or 0.015625 to 32 μg/mL. Bacterial concentration was adjusted to a 0.5 Macbeth turbidity using 5 mL of sterile normal saline. Then 100 μL the bacterial suspension was added to 10 mL TSB supplemented with 10% FBS and 10 μg/mL NAD for measurement. The positive control (with bacteria and without drugs) and the negative control (without bacteria and drugs) were assessed by adding 50 μL of the tested bacterial solution to each antibiotic well. The tested bacterial solution was cultured at 37°C for 24 h. The highest concentration without bacterial growth was observed as the MIC of the antibiotic and recorded.
To explore the effect of the ompW gene deletion on the expression levels of chose genes, total RNA of bacteria was extracted by TRIzol reagent (Invitrogen, USA) (34). The strains cultured in TSB supplemented with 10% fetal bovine serumand 10 μg/mL NAD. The RNA was reverse-transcribed to cDNA with a PrimeScriptTM II 1st Strand cDNA kit (Takara, Dalian, China). Amplification of cDNA was carried out using the SYBR Premix Ex Taq kit (Takara, Dalian, China). The rpmA, rpmB, xylG, modA, rplT, rpsA, malK_1, phnS_2, rpsC, rplU, fhuB, prokka_00896, rplC genes were analyzed and the kdsB gene encoding 3-deoxy-manno-octulosonate cytidylyltransferase was used as the internal control (35). The primers used for RT-PCR are found in Table 2.
Experimental data are presented as means ± SD. The difference between two groups was analyzed by the two-tailed Student's t-test. P values of < 0.05 were considered significant, *p < 0.05 and **p < 0.01.
The ompW gene deletion mutant, ΔompW, was constructed by homologous recombination and verified by PCR (Figure 1A). The CΔompW strain was confirmed with PJompWF/PJompWR (Figure 1A).
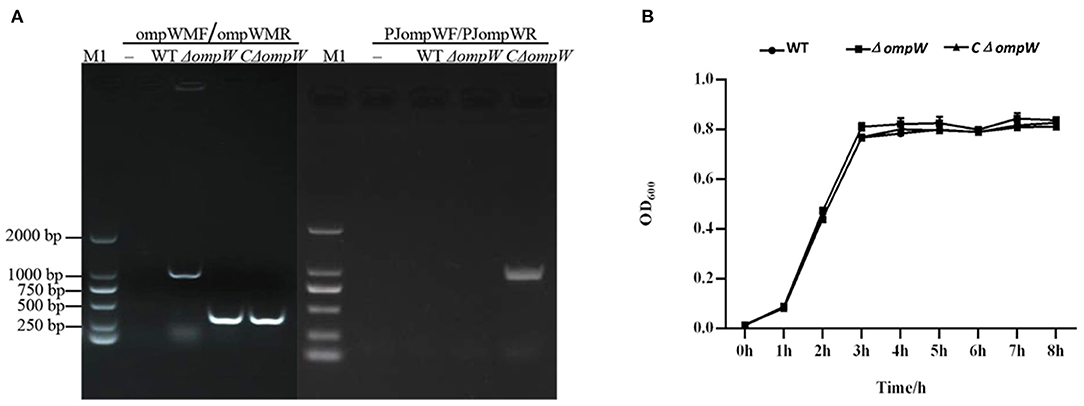
Figure 1. (A) Confirmation of the ΔompW mutant and CΔompW by PCR using primer pairs. M1: 2000 bp marker; WT: wild type strain of A. pleuropneumoniae. (B) Growth curves for the A. pleuropneumoniae strain, the ΔompW mutant strain, and the CΔompW strain, cultured to logarithmic growth phase. The OD600 values of bacterial solutions are presented.
The in vitro effect of the ompW gene deletion on 8 h growth was assessed for the A. pleuropneumoniae strain, the ΔompW mutant strain, and the CΔompW strain. Results showed the growth kinetics of the A. pleuropneumoniae, the ΔompW mutant strain, and the CΔompW strain to not differ significantly for this incubation period (Figure 1B).
We explored the effect of ompW gene deletion on biofilm formation of A. pleuropneumoniae and ΔompW using a quantitative assay. The results showed that deletion of ompW gene reduced the biofilm formation ability of A. pleuropneumoniae at 12 h to 48 h, but the difference is not significant (Figure 2).
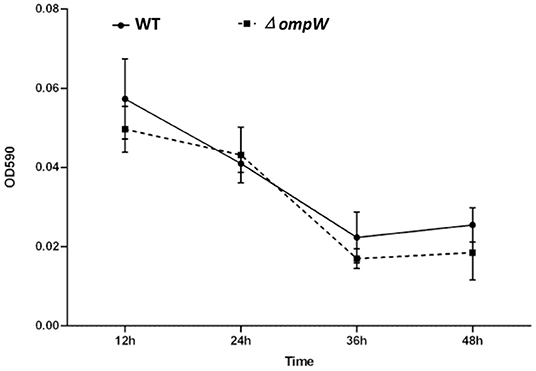
Figure 2. Biofilms formation by the A. pleuropneumoniae strain and the ΔompW mutant strain. Liquid was removed after static incubation at 37°C followed by the addition of a crystal violet dye solution. The biofilms were dissolved by adding a glacial acetic acid solution and the OD590 absorbance of each well determined. The experiment has at least three biological repetitions.
SEM and TEM morphologies were assessed for the A. pleuropneumoniae strain, the ΔompW mutant strain, and the CΔompW strain during logarithmic and steady state growth. There were no significant morphological differences for any of the strains during the logarithmic growth phase as judged by SEM (Figure 3A). However, for steady growth, the cell surface of the ΔompW mutant became rough, while the surfaces of the A. pleuropneumoniae strain and the CΔompW strain were smooth by SEM (Figure 3A), which indicated absence of the ompW gene resulted in a rough surface for A. pleuropneumoniae during steady growth. These surface morphologies were confirmed by TEM (Figure 3B).
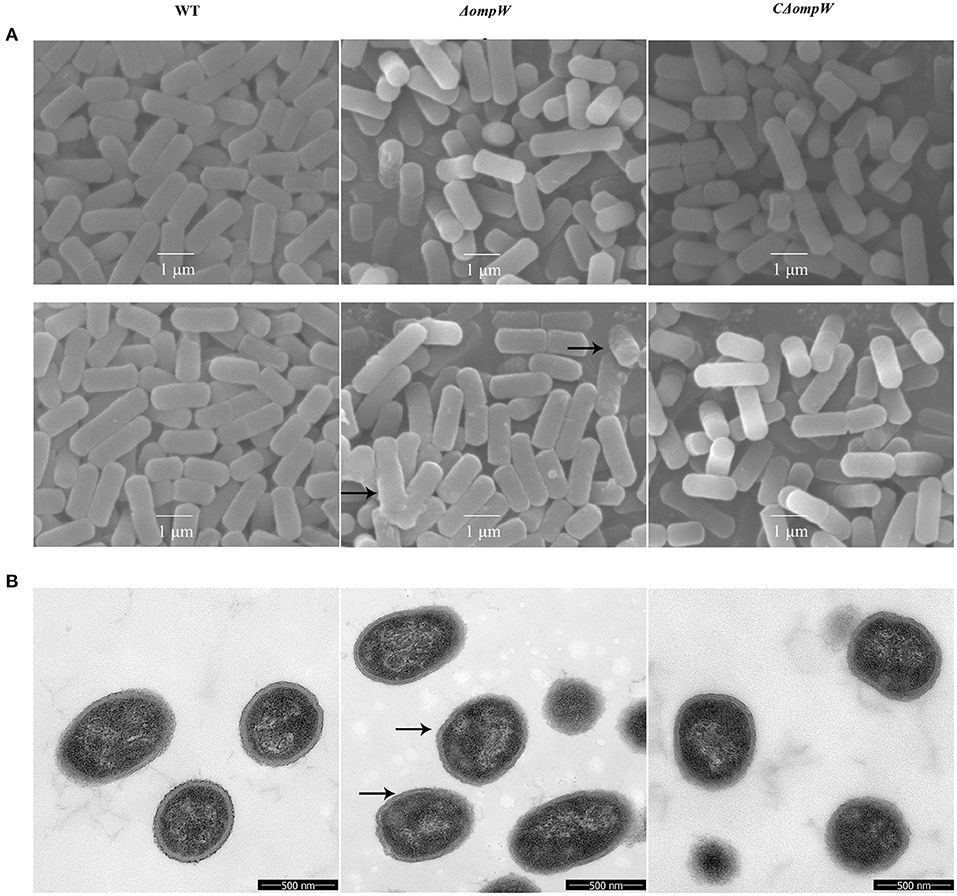
Figure 3. (A) SEM and (B) TEM morphology of the A. pleuropneumoniae, ΔompW mutant strain, and CΔompW strain during logarithmic and steady state growth.
The A. pleuropneumoniae, ΔompW, and CΔompW strains were exposed to oxidative stress conditions. When the strains were treated with 1 mM hydrogen peroxide for 10 min, the ΔompW strain survival rate was 0.21%, which was much lower than that of the A. pleuropneumoniae strain (0.64%) and the CΔompW strain (0.58%; Figure 4). When the strains were treated with 1 mM hydrogen peroxide for 20 min, the ΔompW strain survival rate was 0.18%, which was lower than that of the A. pleuropneumoniae strain (0.46%) and the CΔompW strain (0.44%). These results suggest that OmpW has a role in the tolerance of A. pleuropneumoniae to oxidative stress.
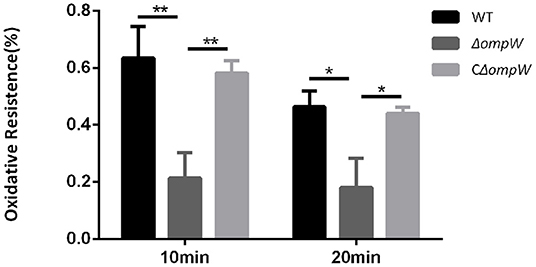
Figure 4. Impaired oxidative stress tolerance of the A. pleuropneumoniae strain, the ΔompW mutant strain, and the CΔompW strain. Overnight cultures were inoculated into fresh TSB supplemented with 10% FBS and 10 μg/mL NAD and grown to an OD600 value of approximately 0.6. Cells were then treated with 1 mM hydrogen peroxide for 10 min or 20 min, respectively. The experiment has at least three biological repetitions. Results are expressed as means ± SD of three independent experiments. ‘*' indicates significance at p < 0.05 and ‘**' indicates significance at p < 0.01.
The A. pleuropneumoniae, ΔompW, and CΔompW strains were assessed for bacterial susceptibility to; rifampicin, ofloxacin, polymyxin B, vancomycin, acriflavine, cefalexin, lincomycin, gentamicin, kanamycin, neomycin, penicillin, amoxicillin, tetracycline, streptomycin, vibramycin, enrofloxacin, florfenicol, sulfamethoxazole, and ceftiofur sodium. As judged by MIC, deletion of the ΔompW gene did not affect bacterial susceptibility to most assessed antibiotics (Table 3). However the deletion resulted in two times sensitive to polymyxin B, kanamycin, and penicillin (Table 3).
The effect of the A. pleuropneumoniae and ΔompW strains on ribosome synthesis related genes and ABC transporter genes at the mRNA level was determined by RT-PCR. Three genes, rpmA, rpmB, and rplT were significantly upregulated and three genes, rpsC, rplU, and rplC were significantly downregulated within the ribosome synthesis pathway (Figure 5). For the ABC transporter pathway, deletion of the ompW gene upregulated xylG, and modA, while phnS_2, PROKKA_00896, and fhuB were downregulated (Figure 5).
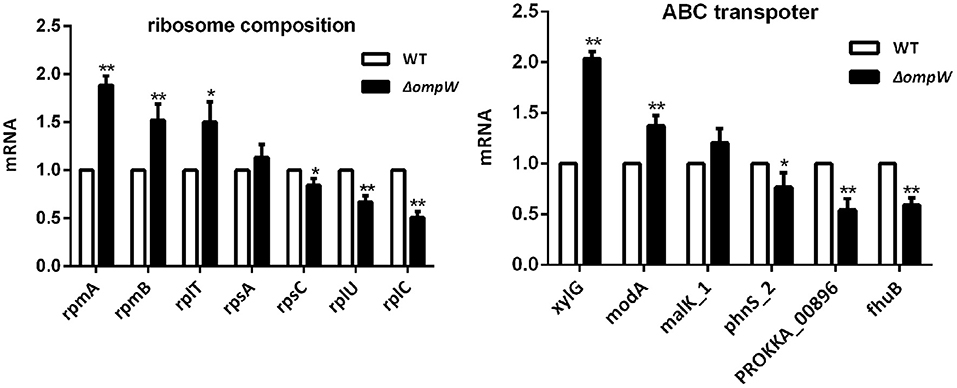
Figure 5. RT-PCR determination of the expression levels of ribosome synthesis related genes and ABC transporter genes following ompW gene deletion. Total RNA was extracted from bacterial strains and reverse-transcribed to cDNA. Amplification of cDNA was carried out using SYBR Premix Ex Taq kit. The experiment has at least three biological repetitions. Results are expressed as means ± SD of three independent experiments. ‘*' indicates significance at p < 0.05 and ‘**' indicates significance at p < 0.01.
A. pleuropneumoniae is considered one of the most important bacterial respiratory pathogens of pigs with epidemic disease resulting in serious economic loss (36). There are many studies on the infection mechanism and the resistance to environmental pressure of A. pleuropneumoniae, but there are still many unknowns to be researched. In this study, we explored the role of the A. pleuropneumoniae ompW gene in biofilm formation, resistance to environmental pressure and gene regulation, with the concept that this exploration may provide a novel new strategy for the control of A. pleuropneumoniae infections.
Biofilms are a complex community of microorganisms thought to be a sessile mode of life that permits attachment and growth of microorganisms on surfaces (37). Microorganisms comprising biofilms are highly resistant to antibiotics and are capable of prolonged persistence within a host (38), and are involved in virulence and pathogenicity (39). Device-related and chronic infections are often associated with biofilm formation (40). A. pleuropneumoniae can produce biofilm that confers bacterial resistance to antibiotics and increased pathogenicity (41–43). Interestingly, the transcription of ompW gene and the biofilm formation of A. pleuropneumoniae were inhibited by zinc (44). The transcription of ompW gene of A. pleuropneumoniae is down regulated under iron restriction (45). Studies have shown that ompW gene is involved in the protection of bacteria against various forms of environmental pressure. The OmpW was related to salt stress and that ompW gene transcription and expression were up-regulated in cultures containing high NaCl concentrations. Deletion of ompW gene did not affect the cell morphology of V. cholerae (22). Furthermore, iron regulates OmpW by binding to Fur and SoxS, a transcriptional factor involved in oxidative stress, was found to negatively regulate OmpW of E. coli (19). In this study, the ompW gene was demonstrated to protect A. pleuropneumoniae from oxidative stress and affect the cell morphology during steady growth. Whether the ompW gene is involved in A. pleuropneumoniae virulence requires further study.
Ribosomes are essential to cellular protein production (46). Impairment or abnormalities in the ribosome biogenesis pathways have been related to disease (47). ATP-binding cassette (ABC) transporters use energy derived from ATP hydrolysis for molecular transport across membranes (48) and are involved in many physiological and pathological processes (49). In this study, the ribosome synthesis and ABC transporter pathways were found to have significant involvement. The genes rpmA, rpmB, and rplT were significantly upregulated and rpsC, rplU, and rplC were significantly downregulated for the ribosome synthesis pathway. The genes xylG, and modA were upregulated and phnS_2, PROKKA_00896, fhuB were downregulated for the ABC transporter pathway. These genes were chose by our previous RNA-seq results (not mentioned in this study). Research have been shown that these genes have important roles involved in the pathogenicity of A. pleuropneumoniae. Thus, we speculate that deletion of the ompW gene affected protein synthesis and ion transport, which may affect the toxicity and drug sensitivity of A. pleuropneumoniae to a certain extent. ompW gene deletion did affect bacterial susceptibility to polymyxin B, kanamycin, and penicillin. The effect on bacterial susceptibility to polymyxin B, kanamycin, and penicillin may also be due to the change of cell membrane permeability. However, the mechanistic basis for these observations requires further investigation.
In summary, data suggested that ompW gene deletion did not affect the growth of A. pleuropneumoniae, but did affect; morphology during steady growth, oxidative tolerance, and bacterial susceptibility to polymyxin B, kanamycin, and penicillin. The ompW gene deletion also altered the expression of ribosome synthesis and ABC transporter related genes. Thus, the ompW gene may regulate the biological phenotype of A. pleuropneumoniae during A. pleuropneumoniae infections.
The authors acknowledge that the data presented in this study must be deposited and made publicly available in an acceptable repository, prior to publication. Frontiers cannot accept a manuscript that does not adhere to our open data policies.
XC, WL, and RZ: designed the experiments. XC, ZS, and LW: performed the experiments. XC, BH, WY, JC, EJ, QH, LL, JX, HL, HZ, YW, WL, and RZ: analyzed the data. XC: wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was supported by the Natural Science Foundation of Hubei Province (No. 2017CFB236), the Hubei Provincial Central Guidance Local Science and Technology Development Project of China (No. 2020ZYYD029), the National Natural Science Foundation of China (No. 31520103917), and Science and Technology List System Science and Technology Project of HubeiProvince (2019AEE006).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.846322/full#supplementary-material
1. Prado IGO, da Silva GC, Crispim JS, Vidigal PMP, Nascimento M, Santana MF, et al. Comparative genomics of Actinobacillus pleuropneumoniae serotype 8 reveals the importance of prophages in the genetic variability of the species. Int J Genomics. (2020) 2020:9354204. doi: 10.1155/2020/9354204
2. Nielsen R, Andresen LO, Plambeck T. Serological characterization of Actinobacillus pleuropneumoniae biotype 1 strains antigenically related to both serotypes 2 and 7. Acta Vet Scand. (1996) 37:327–36. doi: 10.1186/BF03548098
3. Jacobsen MJ, Nielsen JP, Nielsen R. Comparison of virulence of different Actinobacillus pleuropneumoniae serotypes and biotypes using an aerosol infection model. Vet Microbiol. (1996) 49:159–68. doi: 10.1016/0378-1135(95)00184-0
4. Bao CT, Xiao JM, Liu BJ, Liu JF, Zhu RN, Jiang P, et al. Establishment and comparison of Actinobacillus pleuropneumoniae experimental infection model in mice and piglets. Microb Pathog. (2019) 128:381–9. doi: 10.1016/j.micpath.2019.01.028
5. Stringer OW, Bosse JT, Lacouture S, Gottschalk M, Fodor L, Angen O, et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet Microbiol. (2021) 255:109021. doi: 10.1016/j.vetmic.2021.109021
6. Zhang B, Ku X, Yu X, Sun Q, Wu H, Chen F, et al. Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci Rep. (2019) 9:9908. doi: 10.1038/s41598-019-45482-8
7. Haesebrouk F, Van de Kerkhof A, Dom P, Chiers K, Ducatelle R. Cross-protection between Actinobacillus pleuropneumoniae biotypes-serotypes in pigs. Vet Microbiol. (1996) 52:277–84. doi: 10.1016/S0378-1135(96)00075-2
8. Ristow LC, Welch RA. RTX Toxins ambush immunity's first cellular responders. Toxins. (2019) 11. doi: 10.3390/toxins11120720
9. Konze SA, Abraham WR, Goethe E, Surges E, Kuypers MMM, Hoeltig D, et al. Link between heterotrophic carbon fixation and virulence in the porcine lung pathogen Actinobacillus pleuropneumoniae. Infect. Immun. (2019) 87. doi: 10.1128/IAI.00768-18
10. Li Y, Cao S, Zhang L, Yuan J, Zhao Q, Wen Y, et al. A requirement of TolC1 for effective survival, colonization and pathogenicity of Actinobacillus pleuropneumoniae. Microb Pathog. (2019) 134:103596. doi: 10.1016/j.micpath.2019.103596
11. Li T, Zhang Q, Wang R, Zhang S, Pei J, Li Y, et al. The roles of flp1 and tadD in Actinobacillus pleuropneumoniae pilus biosynthesis and pathogenicity. Microb Pathog. (2019) 126:310–7. doi: 10.1016/j.micpath.2018.11.010
12. Crispim JS, da Silva TF, Sanches NM, da Silva GC, Pereira MF, Rossi CC, et al. Serovar-dependent differences in Hfq-regulated phenotypes in Actinobacillus pleuropneumoniae. Pathog Dis. (2020) 78. doi: 10.1093/femspd/ftaa066
13. Liu J, Cao Y, Gao L, Zhang L, Gong S, Yang J, et al. Outer membrane lipoprotein Lip40 modulates adherence, colonization, and virulence of Actinobacillus pleuropneumoniae. Front Microbiol. (2018) 9:1472. doi: 10.3389/fmicb.2018.01472
14. Li H, Liu F, Peng W, Yan K, Zhao H, Liu T, et al. The CpxA/CpxR Two-component system affects biofilm formation and virulence in Actinobacillus pleuropneumoniae. Front Cell Infect Microbiol. (2018) 8:72. doi: 10.3389/fcimb.2018.00072
15. Wang Z, Zhu X, Su Y, Xu W, Liu H, Liu Z, et al. Dimethyl phthalate damaged the cell membrane of Escherichia coli K12. Ecotoxicol Environ Saf. (2019) 180:208–14. doi: 10.1016/j.ecoenv.2019.05.009
16. Ojima Y, Sawabe T, Konami K, Azuma M. Construction of hypervesiculation Escherichia coli strains and application for secretory protein production. Biotechnol Bioeng. (2020) 117:701–9. doi: 10.1002/bit.27239
17. Zeng H, Xie M, Ding C, Ma J, Xu D, Wang X, et al. Attenuated Listeria monocytogenes as a vaccine vector for the delivery of OMPW, the outer membrane protein of Aeromonas hydrophila. Front Microbiol. (2020) 11:70. doi: 10.3389/fmicb.2020.00070
18. Huang P, Cai J, Yu D, Tang J, Lu Y, Wu Z, et al. An IL-6 gene in humphead snapper (Lutjanus sanguineus): identification, expression analysis and its adjuvant effects on Vibrio harveyi OmpW DNA vaccine. Fish Shellfish Immunol. (2019) 95:546–55. doi: 10.1016/j.fsi.2019.11.013
19. Zhang P, Ye Z, Ye C, Zou H, Gao Z, Pan J. OmpW is positively regulated by iron via Fur, and negatively regulated by SoxS contribution to oxidative stress resistance in Escherichia coli. Microb Pathog. (2020) 138:103808. doi: 10.1016/j.micpath.2019.103808
20. Zhang L, Li Z, Li Y, Tian J, Jia K, Zhang D, et al. OmpW expressed by recombinant Lactobacillus casei elicits protective immunity against Aeromonas veronii in common carp. Microb Pathog. (2019) 133:103552. doi: 10.1016/j.micpath.2019.103552
21. Ye Y, Ling N, Gao J, Zhang X, Zhang M, Tong L, et al. Roles of outer membrane protein W (OmpW) on survival, morphology, and biofilm formation under NaCl stresses in Cronobacter sakazakii. J Dairy Sci. (2018) 101:3844–50. doi: 10.3168/jds.2017-13791
22. Fu X, Zhang J, Li T, Zhang M, Li J, Kan B. The outer membrane protein OmpW enhanced V. cholerae growth in hypersaline conditions by transporting carnitine. Front Microbiol. (2017) 8:2703. doi: 10.3389/fmicb.2017.02703
23. Benz R, Jones MD, Younas F, Maier E, Modi N, Mentele R, et al. OmpW of caulobacter crescentus functions as an outer membrane channel for cations. PLoS ONE. (2015) 10:e0143557. doi: 10.1371/journal.pone.0143557
24. Beketskaia MS, Bay DC, Turner RJ. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux. J Bacteriol. (2014) 196:1908–14. doi: 10.1128/JB.01483-14
25. Senanayake SD, Brian DA. Precise large deletions by the PCR-based overlap extension method. Mol Biotechnol. (1995) 4:13–5. doi: 10.1007/BF02907467
26. Baltes N, Tonpitak W, Hennig-Pauka I, Gruber AD, Gerlach GF. Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol Lett. (2003) 220:41–8. doi: 10.1016/S0378-1097(03)00064-8
27. Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. (1997) 179:538–40. doi: 10.1128/jb.179.2.538-540.1997
28. Oswald W, Tonpitak W, Ohrt G, Gerlach G. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol Lett. (1999) 179:153–60. doi: 10.1111/j.1574-6968.1999.tb08721.x
29. Jarma E, Corradino G, Regassa LB. Anaerobiosis, growth phase and Actinobacillus pleuropneumoniae RTX toxin production. Microb Pathog. (2004) 37:29–33. doi: 10.1016/j.micpath.2004.04.004
30. Guo L, Guo J, Liu H, Zhang J, Chen X, Qiu Y, et al. Tea polyphenols suppress growth and virulence-related factors of Haemophilus parasuis. J Vet Med Sci. (2018) 80:1047–53. doi: 10.1292/jvms.18-0085
31. Liu F, Sun Z, Wang F, Liu Y, Zhu Y, Du L, et al. Inhibition of biofilm formation and exopolysaccharide synthesis of Enterococcus faecalis by phenyllactic acid. Food Microbiol. (2020) 86:103344. doi: 10.1016/j.fm.2019.103344
32. Patel J, Yin HB, Bauchan G, Mowery J. Inhibition of Escherichia coli O157:H7 and Salmonella enterica virulence factors by benzyl isothiocyanate. Food Microbiol. (2020) 86:103303. doi: 10.1016/j.fm.2019.103303
33. Jia J, Cheng M, Xue X, Guan Y, Wang Z. Characterization of tetracycline effects on microbial community, antibiotic resistance genes and antibiotic resistance of Aeromonas spp. in gut of goldfish Carassius auratus Linnaeus. Ecotoxicol Environ Saf. (2020) 191:110182. doi: 10.1016/j.ecoenv.2020.110182
34. Fu S, Xu L, Li S, Qiu Y, Liu Y, Wu Z, et al. Baicalin suppresses NLRP3 inflammasome and nuclear factor-kappa B (NF-kappaB) signaling during Haemophilus parasuis infection. Vet Res. (2016) 47:80. doi: 10.1186/s13567-016-0359-4
35. Li L, Chen Z, Bei W, Su Z, Huang Q, Zhang L, et al. Catecholamines promote Actinobacillus pleuropneumoniae growth by regulating iron metabolism. PLoS ONE. (2015) 10:e0121887. doi: 10.1371/journal.pone.0121887
36. Cao Y, Gao L, Zhang L, Zhou L, Yang J, Deng L, et al. Genome-wide screening of lipoproteins in Actinobacillus pleuropneumoniae identifies three antigens that confer protection against virulent challenge. Sci Rep. (2020) 10:2343. doi: 10.1038/s41598-020-58968-7
37. Kim SK, Lee JH. Biofilm dispersion in Pseudomonas aeruginosa. J Microbiol. (2016) 54:71–85. doi: 10.1007/s12275-016-5528-7
38. Algharib SA, Dawood A, Xie S. Nanoparticles for treatment of bovine Staphylococcus aureus mastitis. Drug Deliv. (2020) 27:292–308. doi: 10.1080/10717544.2020.1724209
39. Hill A, Beitelshees M, Pfeifer BA, Jones CH. Standardization of pneumococcal biofilm release to PncO expression, a predictive measurement of virulence. Infect. Immun. (2018) 86. doi: 10.1128/IAI.00494-18
40. Del Pozo JL. Biofilm-related disease. Expert Rev Anti Infect Ther. (2018) 16:51–65. doi: 10.1080/14787210.2018.1417036
41. Hathroubi S, Loera-Muro A, Guerrero-Barrera AL, Tremblay YDN, Jacques M. Actinobacillus pleuropneumoniae biofilms: role in pathogenicity and potential impact for vaccination development. Anim Health Res Rev. (2018) 19:17–30. doi: 10.1017/S146625231700010X
42. Hathroubi S, Beaudry F, Provost C, Martelet L, Segura M, Gagnon CA, et al. Impact of Actinobacillus pleuropneumoniae biofilm mode of growth on the lipid A structures and stimulation of immune cells. Innate Immun. (2016) 22:353–62. doi: 10.1177/1753425916649676
43. Pereira MF, Rossi CC, Seide LE, Martins Filho S, Dolinski CM, Bazzolli DMS. Antimicrobial resistance, biofilm formation and virulence reveal Actinobacillus pleuropneumoniae strains' pathogenicity complexity. Res Vet Sci. (2018) 118:498–501. doi: 10.1016/j.rvsc.2018.05.003
44. Deslandes V, Nash JH, Harel J, Coulton JW, Jacques M. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics. (2007) 8:72. doi: 10.1186/1471-2164-8-72
45. Labrie J, Pelletier-Jacques G, Deslandes V, Ramjeet M, Auger E, Nash JH, et al. Effects of growth conditions on biofilm formation by Actinobacillus pleuropneumoniae. Vet Res. (2010) 41:3. doi: 10.1051/vetres/2009051
46. Catez F, Dalla Venezia N, Marcel V, Zorbas C, Lafontaine DLJ, Diaz JJ. Ribosome biogenesis: an emerging druggable pathway for cancer therapeutics. Biochem Pharmacol. (2019) 159:74–81. doi: 10.1016/j.bcp.2018.11.014
47. Vadivel Gnanasundram S, Fahraeus R. Translation stress regulates ribosome synthesis and cell proliferation. Int. J. Mol. Sci. (2018) 19:3757. doi: 10.3390/ijms19123757
48. Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Curr Opin Struct Biol. (2008) 18:726–33. doi: 10.1016/j.sbi.2008.09.007
Keywords: A. pleuropneumoniae, ompW, susceptibility, phenotype, infections
Citation: Chen X, Shao Z, Wu L, He B, Yang W, Chen J, Jin E, Huang Q, Lei L, Xu J, Li H, Zhang H, Wan Y, Liu W and Zhou R (2022) Involvement of the Actinobacillus pleuropneumoniae ompW Gene in Confrontation of Environmental Pressure. Front. Vet. Sci. 9:846322. doi: 10.3389/fvets.2022.846322
Received: 24 January 2022; Accepted: 28 April 2022;
Published: 19 May 2022.
Edited by:
Vassilis Papatsiros, University of Thessaly, GreeceReviewed by:
Abraham Loera Muro, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoCopyright © 2022 Chen, Shao, Wu, He, Yang, Chen, Jin, Huang, Lei, Xu, Li, Zhang, Wan, Liu and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiabing Chen, YmluZ3gxMjA1QHNpbmEuY29t; Wu Liu, bGl1d3U4MTc3QDE2My5jb20=; Rui Zhou, cnpob3VAbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.