- 1Norwegian Structural Biology Center and Department of Chemistry, Faculty of Science and Technology, UiT The Arctic University of Norway, Tromsø, Norway
- 2Centre for Bioinformatics, Department of Chemistry, Faculty of Science and Technology, UiT The Arctic University of Norway, Tromsø, Norway
For bacteria to thrive in an environment with competitors, phages and environmental cues, they use different strategies, including Type VI Secretion Systems (T6SSs) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) to compete for space. Bacteria often use quorum sensing (QS), to coordinate their behavior as the cell density increases. Like other aliivibrios, Aliivibrio wodanis 06/09/139 harbors two QS systems, the main LuxS/LuxPQ system and an N-acyl homoserine lactone (AHL)-mediated AinS/AinR system and a master QS regulator, LitR. To explore the QS and survival strategies, we performed genome analysis and gene expression profiling on A. wodanis and two QS mutants (ΔainS and ΔlitR) at two cell densities (OD600 2.0 and 6.0) and temperatures (6 and 12°C). Genome analysis of A. wodanis revealed two CRISPR systems, one without a cas loci (CRISPR system 1) and a type I-F CRISPR system (CRISPR system 2). Our analysis also identified three main T6SS clusters (T6SS1, T6SS2, and T6SS3) and four auxiliary clusters, as well about 80 potential Type VI secretion effectors (T6SEs). When comparing the wildtype transcriptome data at different cell densities and temperatures, 13–18% of the genes were differentially expressed. The CRISPR system 2 was cell density and temperature-independent, whereas the CRISPR system 1 was temperature-dependent and cell density-independent. The primary and auxiliary clusters of T6SSs were both cell density and temperature-dependent. In the ΔlitR and ΔainS mutants, several CRISPR and T6SS related genes were differentially expressed. Deletion of litR resulted in decreased expression of CRISPR system 1 and increased expression of CRISPR system 2. The T6SS1 and T6SS2 gene clusters were less expressed while the T6SS3 cluster was highly expressed in ΔlitR. Moreover, in ΔlitR, the hcp1 gene was strongly activated at 6°C compared to 12°C. AinS positively affected the csy genes in the CRISPR system 2 but did not affect the CRISPR arrays. Although AinS did not significantly affect the expression of T6SSs, the hallmark genes of T6SS (hcp and vgrG) were AinS-dependent. The work demonstrates that T6SSs and CRISPR systems in A. wodanis are QS dependent and may play an essential role in survival in its natural environment.
Introduction
Quorum sensing (QS) is a cell density-dependent cell-to-cell communication system in which bacteria produce and respond to signaling molecules called autoinducers (AIs), which subsequently activates the QS transcriptional regulator to control specific functions such as bioluminescence, motility, biofilm, secretion and virulence (1–5). Multiple QS systems have been described in several Vibrio species (6). Two QS systems, AinS/AinR and LuxS/LuxPQ that are believed to work through phosphorelay mechanism have been identified in the genome of Aliivibrio wodanis (7). The AinS/AinR QS system produces AI-1 known as N-acyl homoserine lactones (AHLs) and is present in many Gram-negative bacteria (2, 8). These AHL-mediated QS systems are used for intra-species communication (8, 9). The LuxS/LuxPQ QS system is present in a wide variety of Gram-negative and Gram-positive bacteria, and produces the AI-2 called furanosyl borate diester and is involved in inter-species communication (10, 11). These two QS systems are known to work in parallel in Vibrio harveyi, Aliivibrio fischeri and Vibrio cholerae (12–14). At low cell density, when the AI concentrations are low, the AI receptors act as kinases and relay phosphate to the RpoN (σ54)-dependent activator LuxO via phosphotransferase LuxU. This, in turn, activates the expression of qrr sRNAs, which together with RNA chaperone Hfq represses translation of the mRNA encoding the master regulator LitR (9, 14, 15). At high cell density, the signaling molecules reach a threshold concentration and bind to the receptors to dephosphorylate LuxO, which terminates the qrr sRNA transcription. In the absence of Qrr sRNA, litR is activated to regulate hundreds of genes (1, 14, 16).
In the environment, bacteria co-exist in communities with multiple competitors, including other bacterial species and phages, and have to respond to various cues such as changes in temperature, nutrient and iron availability, pH, osmolarity and salinity (17–22). Hence, bacteria have developed various strategies such as protein secretion, contact-dependent growth inhibition, bacteriocin production and antibiotic production to survive and thrive (23–27). Some strategies are not necessarily harmful to competitors, such as adhesion, exopolysaccharide production, siderophore production, motility, biofilm formation, heat shock response and quorum quenching (28–34). Other strategies developed, such as defense mechanisms against phages or mobile genetic elements (MGEs), including restriction-modification, receptor modification and clustered regularly interspaced short palindromic repeats-CRISPR associated (CRISPR-Cas) are for protection (35–37).
The CRISPR-Cas system is an adaptive immune system against invading nucleic acids from phages and other MGEs and is composed of cas genes, a leader sequence and a CRISPR array with repeats separated by several spacer sequences (35, 38). Two CRISPR classes have been identified with six main types and several subtypes, which are categorized based on the types of cas genes, direct repeats and gene arrangement where class 1 includes the type I, III and IV whereas type II, V and VI belong to class 2 (39–41). Several Vibrio species harbor the type I CRISPR system classified into subtypes such as type I-A, I-B, I-C, I-D, I-E and I-F (40, 42, 43). QS regulation of CRISPR system has been described in bacteria like Pseudomonas aeruginosa, Serratia sp. and Chromobacterium violaceum (44–47).
QS regulates type VI secretion systems (T6SSs) in several vibrios such as V. cholerae, Vibrio parahaemolyticus, Vibrio anguillarum, Vibrio fluvialis and Vibrio alginolyticus (48–52). T6SS is one of the largest contact-dependent secretion system bacteria use to transport T6SS effectors (T6SEs) into eukaryotic hosts, bacterial competitors or the environment (53–56). In some bacteria, T6SSs are also known to be involved in the uptake of metal ion (57, 58). The T6SS was first identified in V. cholerae as a virulence-associated secretion (vas) gene cluster and later in many other bacteria (59–61). The T6SEs are toxin molecules with anti-bacterial or anti-eukaryotic activity (62–64). Several anti-bacterial effector molecules such as amidases, glycoside hydrolases, lipases, phospholipases and nucleases and anti-eukaryotic effectors such as VasX, the Multifunctional-autoprocessing repeats-in-toxin and EvpP have been identified (65–69). T6SS gene clusters often encode immunity proteins close to the effector genes in order to neutralize their effector molecules to prevent self or sibling-killing (70). For instance, immunity proteins such as antitoxin TsaB in V. cholerae have been reported to protect self-killing against effectors VgrG-3 and Tse2, respectively (71). In addition, immunity protein-independent mechanisms like envelope stress response and two-component systems can facilitate self-protection (72).
A. wodanis is a Gram-negative, rod-shaped, non-luminescent and motile bacterium with multiple polar flagella (73). A. wodanis strains grow in the range of temperatures and salt concentrations between 4–25°C and 1–4% respectively (73). The genome of A. wodanis 06/09/139 contains two chromosomes and 4 plasmids (7). A. wodanis has been repeatedly isolated together with Moritella viscosa (M. viscosa) from Atlantic salmon (Salmo salar) during outbreaks of winter ulcer that mainly occurs at a temperature below 8°C (74, 75). Infected fish can survive when the temperature rises above 8°C (74, 76). Winter ulcer causes mortality and significant losses in farming industry and is characterized by large ulcers, hemorrhages and internal tissue necrosis in the infected fish (74, 77). Although M. viscosa is the primary agent for the disease, the role of A. wodanis and the mechanism behind the co-existence with A. wodanis in the winter ulcers are still unclear (74, 77). Experimental study reproducing field observation reveal that A. wodanis affects the progression of M. viscosa infection and is responsible for the chronic pathogenesis in fish (78). A. wodanis adheres to Atlantic salmon head kidney cells and in a bath challenge, A. wodanis separately produces clinical symptoms such as fin rot and other internal pathological symptoms in Atlantic salmon and it can co-infect Atlantic salmon together with M. viscosa (78). In a co-cultivation experiment, A. wodanis impedes the growth of M. viscosa and when both the bacteria were implanted together in fish abdomen, A. wodanis alters the gene expression of M. viscosa (7). It has been further hypothesized that A. wodanis perhaps uses bacteriocin to impede the growth and virulence of M. viscosa (7). In our previous studies we have reported that A. wodanis produces one AHL and encodes two QS systems (7, 79). In the cell culture studies, A. wodanis is known to be cytotoxic to different salmon cell lines when treated with supernatants harvested at cell densities higher than OD600 (optical density measured at 600 nm) of 6.0 (78, 80). Moreover, in a HPLC-MS/MS analysis, the AHL production in A. wodanis begins at the early log phase and increases with increase in cell density along the growth curve (80). Hence in this transcriptomics study, we chose two cell densities one at the early log phase (OD600 2.0) and the other at the cell density close to the end log phase (OD600 6.0) to study the role of cell densities in gene expression. Furthermore, in our recent study, we found that the temperature 6°C which is lower than the winter ulcer disease threshold temperature 8°C, has more impact on AHL production and cytotoxicity in CHSE cell line than 12°C (80). Therefore, in this study, we wanted to analyze the effects of 6°C and 12°C in gene expression. We have also shown that A. wodanis uses the QS to regulate growth, motility, siderophore- and protease production, hemolysis as well as cytotoxicity in the Chinook salmon embryo (CHSE) cell line (80). Considering the importance of understanding the QS and survival strategies in A. wodanis, we performed genome analysis and RNA sequencing (RNA-Seq) to reveal the global gene expression in the wild type and its QS mutant strains ΔainS and ΔlitR.
Materials and Methods
Bacterial Strains and Growth Conditions
The A. wodanis 06/09/139 used was originally isolated from the head kidney of an Atlantic salmon on the west coast of Norway in 2006 (78). The construction of the ΔainS and ΔlitR in-frame mutants by allelic exchange has been described in a recent study (80). A. wodanis 06/09/139 and the mutants from glycerol stocks were grown at 12°C for 3 days on Luria-Bertani Agar (Difco BD Diagnostics Sparks, MD, USA) with a total concentration of 1.0% (wt/vol) peptone (Sigma-Aldrich, St. Louis, MO, USA), 0.5% (wt/vol) yeast extract (Merck, Darmstadt, Germany), 2.5% NaCl (wt/vol) (Sigma-Aldrich, St. Louis, MO, USA) and 1.5% agar (Sigma-Aldrich, St. Louis, MO, USA). The pH of the media was adjusted to 7.5. Three biological replicates of pre-cultures of A. wodanis 06/09/139 and the mutants ΔainS and ΔlitR were grown from a single colony in 2 ml of Luria-Bertani Broth (LB2.5) overnight at 12°C, 220 rpm.
RNA Extraction and rRNA Depletion
Pre-culture biological triplicates of A. wodanis 06/09/139, ΔainS and ΔlitR were diluted to a start OD600 of 0.01 in LB2.5 in a final volume of 60 ml using a 250 ml baffled flasks. The cultures were grown further in parallel using two Infors HT Multitron incubators set at 6 and 12°C at 220 rpm. The cultures were diluted 1:10 for OD600 measurement and harvested at low cell density (LCD) OD600 of 2.0 and high cell density (HCD) OD600 of 6.0. A small culture volume (1 ml) was harvested and mixed with two volumes of RNAprotect Bacteria reagent (Qiagen, Hilden, Germany). The treated cultures were then incubated for 5 min at room temperature and vortexed for a few seconds before centrifuging at 13,000 rpm for 2 min in a cold Heraeus fresco 21 centrifuge (Thermo Scientific, Waltham, MA, USA). The pellets were flash-frozen with liquid nitrogen and stored at −80°C until RNA isolation. The total RNA from the cell pellets was isolated using the MasterpureTM complete DNA/RNA purification kit (Epicenter, Madison, WI, USA) according to the manufacturer's instructions. The RNA concentration was measured in NanoDropTM 2000c spectrophotometer (Thermo Scientific, Waltham, MA, USA). Ribosomal RNA (rRNA) was depleted from the total RNA using a Ribo-zero rRNA removal kit for bacteria (Illumina, San Diego, CA, USA). The RNA quality before and after rRNA depletion was determined using Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA).
RNA Sequencing and Data Analysis
The mRNA libraries were prepared using the TrueSeq stranded mRNA library kit (Illumina, San Diego, CA, USA) and sequenced on Nextseq 500 (Illumina, USA) using 150 cycles mid-output kit and run as a 75 bp paired-end reads at Norwegian Sequencing Center. The image analysis and base calling were performed using Illumina's RTA software version 2.4.6. Reads with low base call quality were removed using Illumina's default chastity criteria. The quality of the raw sequencing data was controlled using FastQC version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The gene expression levels were determined using EDGE-pro v1.0.1 (81) and DESeq2 (82) with default parameters. EDGE-pro was used to align the reads to the reference genome (GCA_000953695.1) and convert the raw coverage into reads per kilobase of gene per million reads mapped (RPKM). DEseq2 was used to estimate the comparative differential gene expression values and provide the output as a log2fold change value with p-value. The log2fold change values were converted into fold change (FC) values, and the cutoff values of ≥ 2.0 or ≤ −2.0 with padj values of < 0.05 were counted as significantly differentially expressed genes (DEGs). The transcriptome profiles of each strain was compared as high cell density against low cell density (tpHCD/LCD), high growth temperature 12°C against low temperature 6°C (tp12°C/6°C) and mutants against wild type (tpΔlitR/WT and tpΔainS/WT). These abbreviations of comparisons are used throughout this paper. The RNA sequence data of wild type A. wodanis, ΔainS and ΔlitR have been deposited in the European Nucleotide Archive (www.ebi.ac.uk/ena) under study accession number PRJEB34433.
Functional Gene Family and Pathway Analysis
KEGG BRITE and KEGG pathway mapper were used to map the DEGs (FC values ≥ 2.0 or ≤−2.0) found when wild type was compared against cell densities (tpHCD/LCD) and temperatures (tp12°C/6°C) and also when mutants were compared against wild type (tpΔlitR/WT and tpΔainS/WT) at different cell densities and temperatures against the reference organism A. wodanis 06/09/139 (83).
Identification of CRISPR Systems, Protospacers and Prophage
The CRISPR-Cas operon in A. wodanis 06/09/139 genome was identified using CRISPRCasFinder (84), and a homology search was performed using KEGG Sequence Similarity Database (SSDB) (83). An intergenic distance of < 100 bp between genes was considered as a single operon in this study. The CRISPR arrays with direct repeats and spacers were identified from the genome of A. wodanis using the CRISPR finder tool (85), while the protospacers were identified using CRISPR target (86). The prophages in A. wodanis were identified using the phage search tool (PHASTER) (87, 88).
Identification of Type VI Secretion Systems and Type VI Secretion Effectors
The T6SS gene clusters in A. wodanis 06/09/139 were identified using SecReT6 (89) and a homology search against other T6SS gene clusters using KEGG SSDB database (83). The naming of T6SS genes of A. wodanis 06/09/139 in this study follows the naming of V. cholerae T6SS (90). Potential T6SEs in A. wodanis were predicted using the Bastion 6 tool (91).
Results
We have previously shown that A. wodanis 06/09/139 produces one AHL (3OHC10-HSL) and encodes genes for QS (7, 79). Moreover, in a recent study, the QS system in A. wodanis 06/09/139 was found to affect various phenotypes that are possibly linked to the winter ulcer disease development (80). Genome analysis and transcriptome profiling was therefore performed to further study the QS and survival strategies in A. wodanis by comparing wild type and its ΔlitR and ΔainS mutants at low (OD600 of 2.0) and high cell density (OD600 of 6.0) and two different temperatures, 6 and 12°C.
Differential Expressed Genes
The transcriptome profiles of A. wodanis and mutant strains ΔlitR and ΔainS at different cell densities and temperatures are listed in Supplementary Table 1. The average of mapped reads to the reference genome of A. wodanis was above 90% in all samples suggesting that the transcriptome data were sufficient for further analysis.
The total number of DEGs at all tested conditions are shown in Figure 1, and the complete lists of all DEGs are given in Supplementary Table 2. When comparing the transcriptome profiles at HCD relative to LCD (tpHCD/LCD) of the wild type after growth at 6°C, 18% of the total genes (n = 4,282) of A. wodanis were differentially expressed. Increasing the growth temperature to 12°C lowered this number of DEGs to 15%. Among the tested tpHCD/LCD conditions of wildtype, ΔlitR and ΔainS (Figure 1A), the highest numbers of DEGs were observed for the ΔlitR mutant at 6°C, where the number of DEGs account for about 32% of the total genes in of A. wodanis. On the other side, the lowest number of DEGs (8% of the total 4282 genes) was observed in the ΔlitR mutant tpHCD/LCD at 12°C. The ainS mutant tpHCD/LCD at 6°C and 12°C showed 15 and 18% DEGs, respectively. Further, the DEGs in tpHCD/LCD (WT_12°C) and tpHCD/LCD (WT_6°C) were sorted using KEGG BRITE and KEGG pathway mapper using A. wodanis 06/09/139 as a reference organism. The functional families with at least 15 DEGs were enzymes, transporters, secretion system, bacterial motility proteins and ribosome as shown in Supplementary Figure 1A. In addition, in the wild type tpHCD/LCD comparison, at 6°C, a few DEGs (n = 5) with FC values ≥ 2.0 were also mapped into the functional families prokaryotic defense system, which includes the CRISPR related genes. The KEGG pathway mapping revealed that the DEGs (n ≥ 30) in the wild type tpHCD/LCD were involved in metabolic pathways, microbial metabolism in diverse environments, biosynthesis of secondary metabolites, ABC transporters, biosynthesis of antibiotics, two-component system, carbon metabolism and the ribosome. The numbers of DEGs involved in these pathways were similar in wild type tpHCD/LCD at both temperatures except in the ribosome pathway. At tpHCD/LCD (6°C), 42 DEGs mapped into the ribosome pathway, whereas only one DEG mapped at tpHCD/LCD (12°C) (Supplementary Figure 2A).
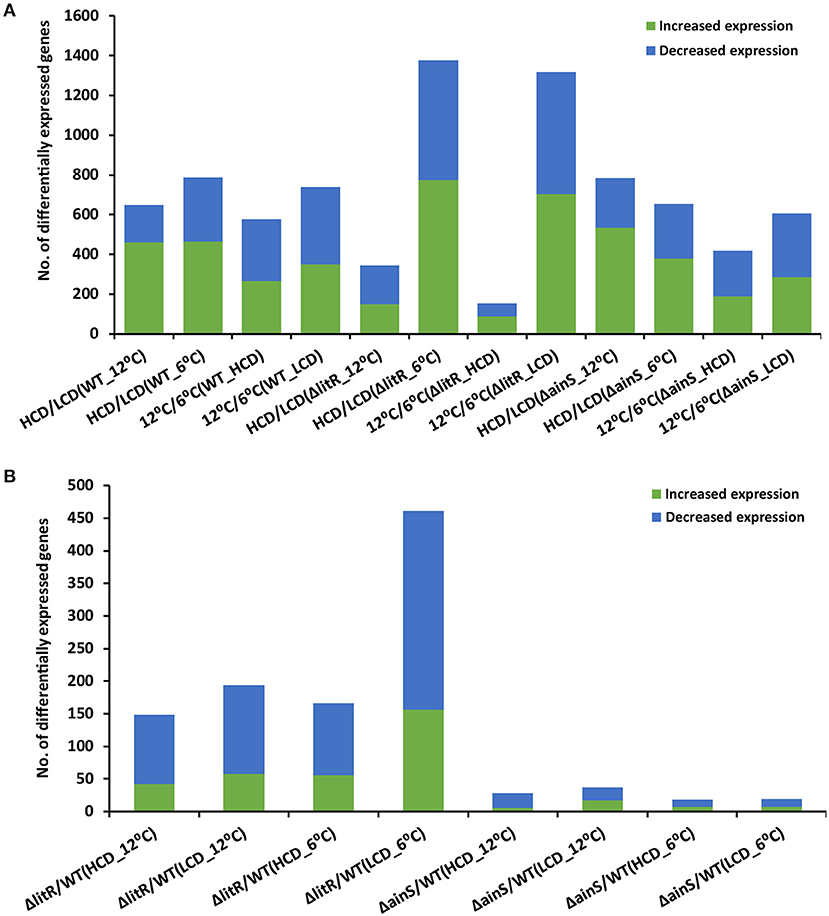
Figure 1. The total number of differentially expressed genes in A. wodanis 06/09/139, ΔainS and ΔlitR. (A) The bar chart shows the number of increased (green) and decreased (blue) expression of genes when comparing high cell density (HCD) to low cell density (LCD) and growth temperature at 12 to 6°C. (B) The bar chart shows the number of increased and downregulated genes in comparisons ΔlitR/WT and ΔainS/WT.
Comparison of the transcriptome profiles of the wild type grown at 12°C and 6°C (tp12°C/6°C) revealed DEGs, accounting for 13 and 17% of the total 4282 A. wodanis genes at HCD and LCD, respectively (Figure 1A). In the ΔainS mutant (tp12°C/6°C), 10 and 14% of total genes were differentially expressed at HCD and LCD, respectively. Compared to the wild type and the ΔainS mutant, 31% of the total genes in the ΔlitR mutant were differentially expressed (tp12°C/6°C) at LCD, whereas at HCD, 4% of the total genes were differentially expressed. Further, the DEGs in wild type (tp12°C/6°C) with FC values ≥ 2.0 and ≤−2.0 were sorted into KEGG BRITE and KEGG pathway mapper. The functional families with DEGs of ≥ 15 were enzymes, transporters, ribosome, ribosome biogenesis and non-coding RNAs (Supplementary Figure 1B). Few DEGs (n < 10) in wild type (tp12°C/6°C) mapped into families such as secretion system and prokaryotic defense system. The KEGG pathway mapping revealed that the DEGs (n ≥ 30) in the WT(tp12°C/6°C) were involved in pathways such as metabolic pathways, microbial metabolism in diverse environments, biosynthesis of secondary metabolites, ABC transporters, biosynthesis of antibiotics, two-component system, carbon metabolism, purine metabolism, Aminoacyl-tRNA biosynthesis and ribosome. The numbers of DEGs involved in these pathways were higher at tp12°C/6°C (LCD) than at tp12°C/6°C (HCD). For example, 43 DEGs at LCD mapped to the ribosome pathway while only 6 DEGs mapped at HCD (Supplementary Figure 2B).
When comparing the profile of the ΔlitR mutant to the wild type tpΔlitR/WT at HCD, 3 and 4% of the total genes were differentially expressed at 12 and 6°C, respectively. In the tpΔlitR/WT (HCD_6°C), 4% of the total genes were differentially expressed. In tpΔlitR/WT (LCD_12°C), 5% of total genes whereas in tpΔlitR/WT (LCD_ 6°C), 11% of the total genes were found to be differentially expressed. The number of DEGs was two times higher at 6°C and LCD compared to 12°C and HCD (Figure 1A). When comparing the expression profile of the ΔainS mutant with the wild type (tpΔainS/WT) at HCD, 0.7 and 0.4% of the total genes were differentially expressed at 12 and 6°C, respectively. In tpΔainS/WT, during LCD and 12°C, 0.9% of the total genes and at LCD and 6°C, 0.4% of the total genes were found to be differentially expressed. The DEGs in ΔlitR and ΔainS mutants were sorted using KEGG BRITE (Supplementary Figure 3A) and mapped into KEGG reference pathway for A. wodanis 06/09/139 (Supplementary Figure 4A). The DEGs (n ≥ 5) in the ΔlitR and ΔainS mutants compared to wild type with FC values of ≥ 2.0 and ≤−2.0 were associated with functional families such as enzymes, secretion system, bacterial motility proteins, transporters and prokaryotic defense system. The KEGG pathway mapping revealed that the DEGs in the tpΔlitR/WT and tpΔainS/WT mutants affected several pathways, including metabolic pathways, two-component system, biosynthesis of secondary metabolites, Quorum sensing and bacterial chemotaxis (Supplementary Figures 3B, 4B). Together our results suggest that, LitR is a crucial global regulator of genes at 6°C and at low cell density. As well, LitR has more impact on gene expression than AinS.
Key QS Genes of A. wodanis are Cell Density and Temperature Dependent
Only a few genes known to be involved in the A. wodanis QS system were differentially expressed under the experimental conditions tested (Supplementary Table 3). When comparing the wild type transcriptome profile at HCD to LCD (tpHCD/LCD), the AI synthase gene ainS (AWOD_I_1040) and master QS regulator gene litR (AWOD_I_0419) showed FC values of 2.01 and 2.24, at 6°C, respectively, while no significant differences were observed at 12°C.
When the wild type profile at high temperature was compared at low temperature (tp12°C/6°C), the qrr sRNA gene (AWOD_I_sRNA_054) was significantly higher with FC value of 2.92 at LCD, whereas no difference in its expression was observed at HCD. Temperature alone did not significantly impact other genes of the QS system at either HCD or LCD (Supplementary Table 3).
Comparing the profile of the ΔlitR mutant relative to the wild type (tpΔlitR/WT), only the FC value of ainS at LCD and 12°C was significant (FC = −1.93, padj value < 0.05), although slightly lower than the cutoff FC value used in this study. The deletion of litR did not affect other QS related genes in A. wodanis under the conditions tested. On the other hand, when comparing the ΔainS mutant profile to the wildtype (tpΔainS/WT), the litR expression was significantly lower at all conditions examined. The FC values of litR were notably lower at HCD compared to LCD. At HCD, the FC values for litR at 6°C and 12°C were −2.53 and −2.87, respectively. At LCD, the FC values at 6°C and 12°C were −1.93 and −2.04, respectively, indicating that the expression of litR in the ΔainS mutant increases with increased density and independent of the temperature. Furthermore, the phosphorelay protein-encoding gene luxU (AWOD_I_0921) expression was significantly higher in the ΔainS mutant at HCD (FC = 2.81) and LCD (FC = 2.17) at 6°C, whereas at 12°C no differences were observed. Hence, the luxU expression in the ΔainS mutant was slightly higher at HCD compared to LCD and its expression was affected only at low temperature.
Differential Expression of CRISPR-Cas System in A. wodanis
We identified two CRISPR systems in the genome of A. wodanis using CRISPRCas finder (84), where CRISPR system 1, located on chromosome 1, did not contain any cas loci, whereas CRISPR system 2, located in chromosome 2, contains the cas loci (Figure 2). The CRISPR system 2 was classified as a Type I-F CRISPR system. A total number of 25 and 40 spacers with a length of 32-33 nucleotides were identified in CRISPR system 1 and 2, respectively (Supplementary File S4). When the spacers were searched against the phage databases using CRISPR Target, spacers matched to the protospacers with at least 5 single nucleotide polymorphisms. For example, spacer 11 of CRISPR system 1 matched to the Vibrio phage CTX plasmid pCTX-2, whereas spacer 19 of CRISPR system 2 matched to the Staphylococcus phage phiSP38-1 with a score of 20.
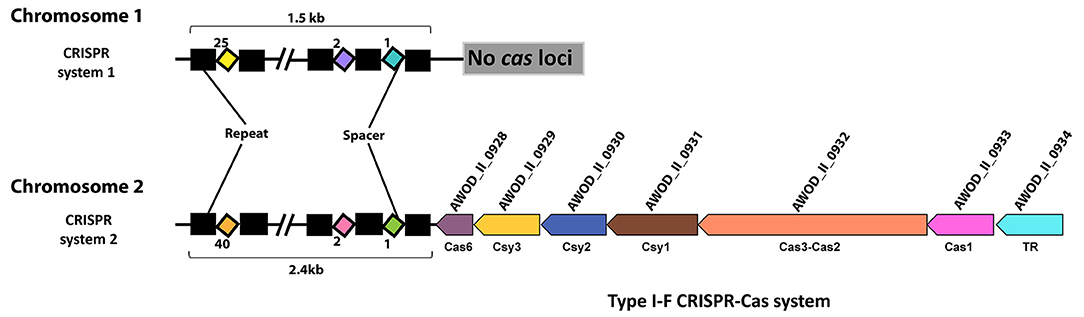
Figure 2. CRISPR systems in A. wodanis 06/09/139. The figure shows the CRISPR arrays with repeats (black rectangle) and spacers (colored diamonds). In CRISPR system 2, the CRISPR array is located close to the cas operon (arrows) of the Type I-F CRISPR-Cas system. TR denotes transcriptional regulator.
The data shows that in the wild type, the csy3, cas1, and cas3 genes of CRISPR system 2 have significantly higher FC values (FC > 2.0) at HCD than at LCD after growth at 6°C. No significant differences in FC values of the cas genes were observed in wild type (tpHCD/LCD) at 12°C (Supplementary Table 5). Cell density had no impact on the expression of CRISPR arrays of CRISPR systems 1 and 2 at either temperature.
When the expression profile at 12°C was compared to 6°C, at LCD and HCD, the CRISPR arrays of system 1 displayed significantly lower FC values (FC value = ~−2.0), suggesting temperature difference influences CRISPR system 1. However, temperature had no effect on the CRISPR system 2.
Further, the expression data showed that the expression of cas genes of CRISPR system 2 were strongly reduced in the ΔlitR mutant at all conditions examined. The FC values of the cas genes in the ΔlitR mutant increased with increasing cell density at both temperatures. Notably, in the ΔlitR mutant at 6°C, the FC values of csy3 and csy1 were twice as high at HCD (FC = − approx. 10.0) compared to LCD (FC = − approx. 5.0). At LCD, the genes csy1, csy2, csy3, and csy4 had lower FC values at 6°C compared to 12°C, whereas the cas1 and cas3 showed higher FC values (Supplementary Table 5). However, at HCD, the highest FC values for cas genes were observed at 6°C than at 12°C. The CRISPR array of system 2 in the ΔlitR mutant showed significantly lower expression at LCD and 12°C (FC value = −1.96) and LCD and 6°C (FC value = −2.69). However, at HCD, the expression of the CRISPR array of system 2 was not significantly different. The data also showed that the CRISPR array of system 1 was highly expressed at HCD and 12°C, whereas the FC values were not significantly different at HCD and 6°C or at LCD at 6°C and 12°C. In the ΔainS mutant compared to wild type, genes such as csy2, csy3, and csy4 showed significantly lower expression at two experimental conditions, LCD, 12°C and HCD, 6°C. No significant differences were observed in expressions of the CRISPR array of system 1 and 2 in the ΔainS mutant.
A. wodanis Harbors Three Main T6SSs and Four Auxiliary Clusters
A. wodanis was found to harbor three T6SSs, each encoding the conserved core and accessory T6SS genes. T6SS1 was located on chromosome 1, whereas the T6SS2 and T6SS3 were on chromosome 2. The T6SS1 gene clusters (AWOD_I_0981-0995) were composed of 3 operons with 15 consecutive genes, whereas the T6SS2 gene clusters consisted of 1 operon with 15 consecutive genes (AWOD_II_0111-0125) and T6SS3 gene clusters with 2 operons with 20 consecutive genes (AWOD_II_1008-1027) as shown in Figure 3. The main T6SS clusters comprised the core components vasABDEFGHJKLQRS, valine-glycine repeat protein G (vgrG) and Hemolysin Coregulated protein (hcp) and the accessory genes vasCIUVX (60, 92, 93). The genes vasABEJL and vasDFK may form the base-plate and membrane complex, respectively (94–96). The genes vasRQ and vasC may form the outer sheath and the FHA domain, respectively, while vasG is believed to act as a chaperone (97, 98). The gene vasH may serve as a sigma-54 dependent transcriptional regulator and vasIS as lysozyme-related proteins (99). The T6SS3 in A. wodanis does not encode the transcriptional regulator vasH (99). Four vgrG paralogous genes were identified in A. wodanis, and using them as markers, four auxiliary clusters were predicted, such as Aux-1 (AWOD_I_1435-1440, AWOD_I_2030), Aux-2 (AWOD_II_0126-0141), Aux-3 (AWOD_II_1028-1032) and Aux-4 (AWOD_II_1054-1060). The auxiliary clusters were located together with the core T6SS genes encoding Proline-alanine-alanine-arginine repeats (PAAR), VgrG and Hcp. The Aux-1 cluster does not encode Hcp, and Aux-4 does not encode a PAAR protein. Four genes encoding adaptor proteins of the DUF4123 family (unknown function) were identified adjacent to vgrG1, vgrG2 and vgrG4, whereas no adaptor protein-encoding gene was present adjacent to vgrG3.
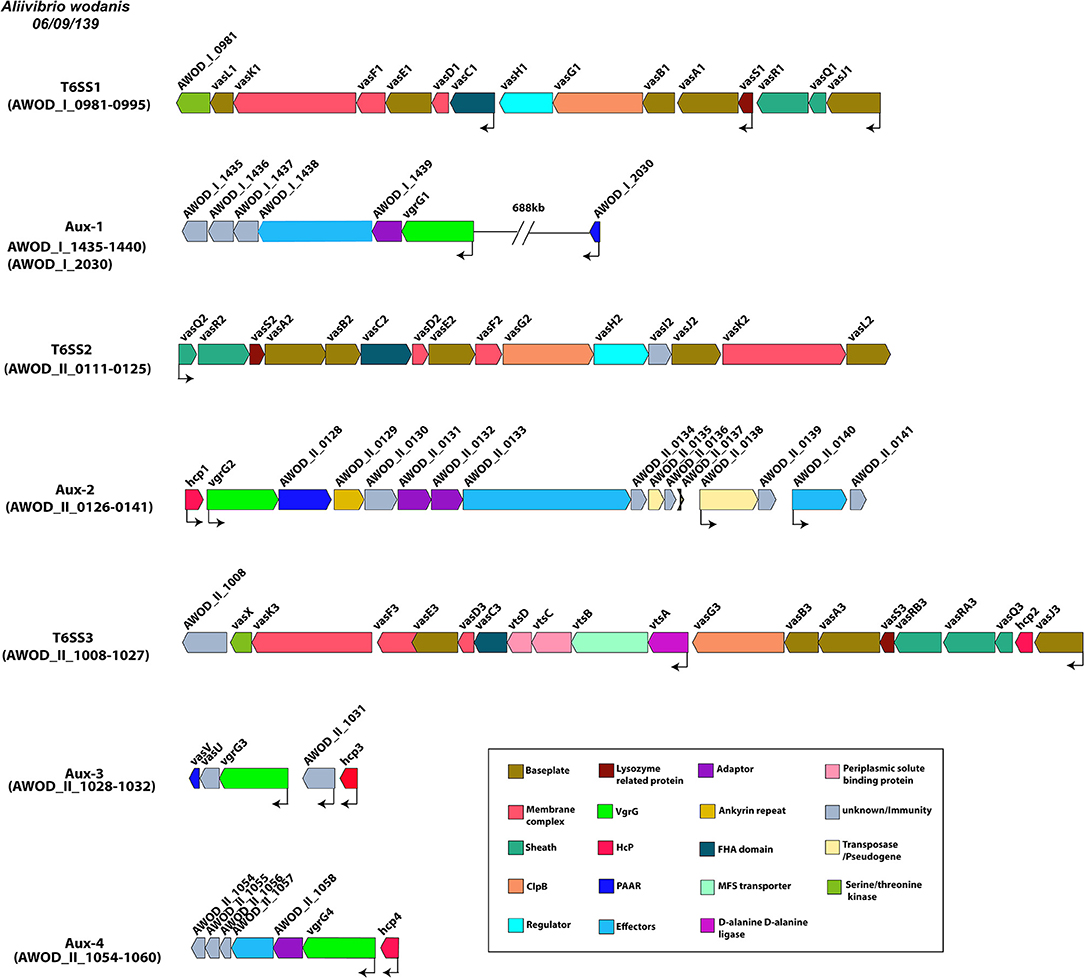
Figure 3. Type VI secretion systems in A. wodanis 06/09/139. Graphical representation of the three main T6SSs of A. wodanis and the four auxiliary T6SS clusters. The thin black arrows indicate the direction and start of the transcription sites.
Differential Expression of T6SSs and Auxiliary Clusters of A. wodanis
In the wild type, at HCD, the main clusters T6SS1 (vasR1Q1J1) and T6SS2 (vasH1I1J1K1L1) were highly expressed at both 6 and 12°C, compared to LCD. No differential expression was observed for the T6SS3 (Supplementary Table 6). Furthermore, the complete auxiliary cluster Aux-1 showed higher expression at HCD at both temperatures. The Aux-2, Aux-3 and Aux-4 showed only significant differences in some genes such as AWOD_II_0126 - 0139, AWOD_II_1031 - 1032 and AWOD_II_1054 - 1059, respectively.
The temperature did not affect the expression of the T6SS1 and T6SS3 in the wild type. However, a few genes (vasQ2, vasR2, vasS2, and vasD2) of T6SS2 were differentially expressed at HCD, while at LCD, only vasQ2 was differentially expressed. As for the auxiliary clusters, only the genes encoding membrane protein and DUF4123 (AWOD_I_1435 and AWOD_I_1439) of Aux-1, hcp1 of Aux-2 and a putative uncharacterized protein (AWOD_II_1031) of Aux-3 were differentially expressed.
When the ΔlitR mutant was compared to wild type (tpΔlitR/WT), the data revealed that the vasR1Q1J1 operon of T6SS1 was differentially expressed at all tested conditions. The FC values of genes vasR1Q1J1 were marginally higher at HCD compared to LCD. Particularly the FC values of vasQ1 gene in ΔlitR mutant were twice as high at HCD compared to LCD at both temperatures. The FC values of vasR1Q1J1 operon in the ΔlitR mutant were significantly higher at 6°C compared to 12°C at both cell densities (Supplementary Table 6). The genes vasF1D1C1 showed higher FC values in ΔlitR mutant at LCD than HCD. At 6°C, the complete T6SS2 cluster was differentially expressed in the ΔlitR mutant at both HCD and LCD. At 12°C, differential expression was only observed in some of the genes in the T6SS2 cluster. Moreover, the FC values of T6SS2 genes were higher at low (6°C) compared to high (12°C) temperature. The T6SS3 gene cluster was only differentially expressed at LCD and 6°C with an average FC value of 2.0. In ΔlitR mutant, some genes of Aux-1, Aux-2 and Aux-3 were differentially expressed, whereas the Aux-4 showed no differential expression (Supplementary Table 6). In Aux-1, the expression of vgrG1 was lower in ΔlitR mutant with an FC value of−4.90 at LCD and 6°C. The genes encoding Hcp1, VgrG2, PAAR motif-containing protein, a protein with ankyrin repeats and the rearrangement hotspot (RHS) protein in the Aux-2 were differentially expressed, where the FC values (FC = −22.75) of hcp1 at HCD and 6°C was approximately thrice compared to LCD (FC = −8.32). Similarly, three times higher expression of hcp1 was observed at 6°C compared to 12°C at both HCD and LCD. About 6 of the 16 total genes in the Aux-2 cluster showed significantly lower expression at LCD and 6°C. The Aux-3 was highly expressed in the ΔlitR mutant, relative to wild type, at HCD and 12°C and at LCD and 6°C. When comparing the ΔainS mutant with the wild type (tpΔainS/WT), only the expression of vasR1 and vasQ1 genes of the T6SS1 cluster were significantly reduced, whereas no differences in T6SS2 and T6SS3 gene clusters were observed. The vgrG1 of Aux-1 showed lower expression (FC value = −2.07) at LCD and 6°C while the hcp1 of Aux-2 showed significant lower expression (FC < −2.0) at all conditions except at LCD and 12°C. Additionally, the genes AWOD_II_1031 and hcp3 of Aux-3 showed significant higher expression at HCD and 12°C. Other genes of auxiliary clusters in ΔainS mutant did not show significant differential expression.
Potential T6SE Molecules Identified in A. wodanis 06/09/139
We predicted 80 potential effectors proteins using the T6SE prediction tool Bastion 6 (Supplementary Table 7) (91). Of the 80 potential effectors, 29 could be annotated, while 51 were identified as putative proteins. Of the potential effectors, 44 were located on chromosome 1, 33 effectors on chromosome 2 and 2 effectors were predicted to be located on plasmid p20. Among the annotated T6SEs, we found potential cell wall degrading effectors such as N-acetylmuramoyl-L-alanine amidase and hydrolases, membrane degrading effectors including phospholipases, and several nucleotide degrading nucleases. Only a few effectors such as a porin-like protein H (AWOD_I_1000), a RHS protein (AWOD_II_0133) and type VI secretion system secreted protein Hcp (AWOD_II_1032) were located close to the main T6SS clusters, whereas the rest were located in different locations on both chromosomes.
Differential Expression of T6SEs in A. wodanis
In wild type, 24 effector genes were differentially expressed at HCD (tpHCD/LCD) at 6°C, while 19 effectors were differentially expressed at 12°C (Supplementary Table 7). These effector genes encoded several proteins including outer membrane proteins, membrane proteins, proteins with PAAR motif, lipoproteins with LysM domain, a sulfite reductase [NADPH] flavoprotein alpha-component, a secreted endonuclease I and a choline dehydrogenase. Interestingly, in the wild type at HCD and 6°C, an FC value of 173.29 was found for choline dehydrogenase (AWOD_II_1235), which was 20 times higher than at 12°C (FC = 8.99). When the wild type profiles at high and low temperatures were compared (tp12°C/6°C), 18 effectors and 22 effectors were differentially expressed at HCD and LCD, respectively. These effectors comprised putative exported protein, putative lipoprotein, putative beta-lactamase, amine oxidase and choline dehydrogenase.
In the ΔlitR mutant, genes encoding effector molecules with significantly lower expression (FC < −2.0) were putative exported proteins (AWOD_I_0175, AWOD_I_1184, AWOD_II_0501, and AWOD_II_0656), outer membrane protein (AWOD_I_1120), putative lipoprotein (AWOD_I_1186, AWOD_II_0440, AWOD_II_0804, and AWOD_II_1206), putative polysaccharide deacetylase (AWOD_I_1338), membrane protein (AWOD_I_1567), amine oxidase (AWOD_II_0852) and phospholipase (AWOD_II_1212). The effector molecules genes differentially expressed in ΔainS mutants included a putative histidine decarboxylase (AWOD_I_1509), and a secreted endonuclease (AWOD_II_0252). Additionally, genes encoding effectors such as a porin-like protein H (AWOD_I_1000), an endonuclease I precursor (AWOD_I_2248) and a 6-phospho-beta-glucosidase (AWOD_I_0029) were differentially expressed in both ΔlitR and ΔainS mutants. The differential expression of effector molecules in ΔlitR mutant was seen more often during LCD and 6°C than at other experimental conditions like HCD and 12°C (Supplementary Table 7).
Discussion
A. wodanis is frequently isolated together with M. viscosa during the winter ulcer outbreaks and believed to be involved in the progression of winter ulcer disease (7, 73, 74, 78). Although M. viscosa is the main agent causing the disease, the reason for its co-existence with A. wodanis is not yet clear. Bacteria use several strategies to compete for niche adaptions, which are known to be regulated by various mechanisms, including QS (49, 100–102). In our previous study we have shown that QS in A. wodanis regulates various phenotypic traits and cytotoxicity on CHSE cell line (80). In this study, we performed genome analysis and gene expression profiling of A. wodanis to explore the QS system and its role in regulating the survival strategies.
Total DEGs
When comparing the transcriptome profile of the A. wodanis wild type between cell densities (tpHCD/LCD) and temperatures (tp12°C/6°C), the strongest effect in terms of number of DEGs were at LCD (164 more genes than at HCD) and at 6°C (140 more genes than at 12°C). Furthermore, the functional mapping of DEGs revealed that families such as secretion system, prokaryotic defense system and transporters were highly expressed at HCD compared to LCD. As expected, the gene families related to protein synthesis such as ribosomes, ribosome biogenesis and transfer RNA biogenesis were less expressed at HCD compared to LCD (tpHCD/LCD). Similar cell density-dependent gene expression has been reported in Aliivibrio salmonicida, where about 1,000 genes were differentially expressed in response to increase in cell density (103). In the ΔlitR mutant, 1.8 times more DEGs were observed at tpHCD/LCD at 6°C than for the wild type grown at the same condition, suggesting LitR is an important regulator in A. wodanis at low temperature. Furthermore, in A. wodanis, the lowly expressed genes in comparison tp12°C/6°C were related to protein synthesis such as non-coding RNAs, ribosomes, transfer RNA biogenesis and ribosome biogenesis. This suggests that genes related to protein synthesis are less expressed at 12°C compared to 6°C. Temperature is one of the major environmental stress factors that bacteria encounter in nature and many genes respond to temperature changes. In addition, the functional families such as the secretion system and prokaryotic defense system were also regulated by temperature in A. wodanis. For example, in V. parahaemolyticus 13% of DEGs were observed when the bacteria were grown at 15°C and 42°C compared to its optimal temperature 37°C, where genes related to energy metabolism were highly affected due to temperature change (104). In A. wodanis, more DEGs were identified in tpΔlitR/WT (LCD) and tpΔlitR/WT (6°C) compared to at HCD and 12°C, suggesting LitR is a global regulator when the temperature is low and the cells are in the early log phase. This is in contrast to A. salmonicida, where the numbers of DEGs in ΔlitR mutant were higher at HCD compared to LCD at 12°C (103). The comparison between the transcriptome profiles of mutants and wildtype (tpΔlitR/WT and tpΔainS/WT) showed about 5 to 24 times more DEGs in tpΔlitR/WT compared to tpΔainS/WT at both cell densities and temperatures. This indicates that only a few genes seem to be regulated through the AinS-dependent QS system. This is similar to A. salmonicida, where only about 20 genes were differentially expressed in ΔainS mutant when compared to wild type, whereas in litR mutant compared to wild type, 3 to 10 times more DEGs were found (103, 105). Moreover, in this study, the transcriptomics was performed at OD600 of 6.0, which is a late log phase in A. wodanis while it can reach an OD600 of ~8.0. Therefore, the AHL-mediated gene regulation in A. wodanis may differ at OD600 higher than 6.0. The functional mapping of DEGs from comparisons tpΔlitR/WT and tpΔainS/WT showed that the genes affected by LitR and AinS in A. wodanis mainly belongs to the families such as secretion system and prokaryotic defense mechanism, which contained the T6SS and CRISPR genes, respectively. Nevertheless, other families such as enzymes, bacterial motility proteins and transporters were also found to be affected by LitR and AinS in this study. LitR and AinS in A. wodanis also influenced various pathways such as metabolic pathways, Quorum sensing, and two-component systems, suggesting their role in signaling mechanisms and metabolic activities. QS regulation of metabolic functions such as glucose uptake, phosphoenolpyruvate-dependent sugar and nucleotide biosynthesis are known to enhance the co-operative behavior of bacteria to survive under limited nutrient conditions (101). Similarly, the LitR and AinS regulation of metabolic pathways in A. wodanis may play a role in co-operative behavior under limited nutrient conditions.
QS System in A. wodanis
In wild type, we found that an increase in cell density resulted in a higher expression of the AHL autoinducer synthase ainS and the master QS regulator litR when grown at 6°C and not at 12°C. This demonstrates that cell density only affects the expression of litR and ainS genes at low temperature. Similar increased expression of ainS was also observed in the ΔlitR mutant strain when comparing HCD to LCD (tpHCD/LCD), indicating that the ainS expression is not entirely LitR-dependent. These results confirm our previous work, where we showed that the 3OHC10-HSL production is cell density- and temperature-dependent (80). Similar to AinS in A. salmonicida (105), the AinS in A. wodanis negatively affected the luxU expression. This suggests that the decreased expression of luxU could repress qrr and activate litR in A. wodanis. Similarly, in this study, luxU was affected by AinS only at 6°C, which could suggest that low temperature affects the phosphorelay function of luxU. On the other hand, the qrr gene was more expressed at 12°C compared to 6°C. Qrr sRNAs in vibrios are known to regulate several target mRNAs, including the master QS regulator LuxR (106). In A. fischeri, Qrr sRNAs negatively regulates the production of LitR (16). Therefore, the increased expression of qrr in A. wodanis at 12°C may negatively affect the production of LitR and, subsequently, the QS dependent gene expression. Our results illustrate that litR, ainS, luxU, qrr sRNA in the A. wodanis QS system were differentially expressed in a cell density and temperature-dependent manner. The proposed QS model in A. wodanis and its regulation of target genes are presented in Figure 4.
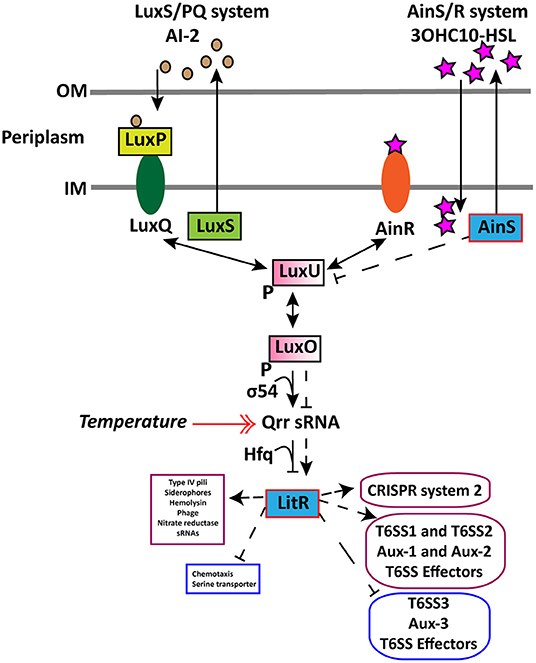
Figure 4. Proposed model for regulation of CRISPR and T6SS in A. wodanis 06/09/139 at 6°C. The model includes the two QS systems LuxS/PQ and AinS/R in A. wodanis. At low cell density (LCD), when the AIs concentration is low, the receptors (AinR and LuxPQ) may act as kinases and relay phosphate to LuxO via LuxU, which activates the expression of qrr sRNA. The Qrr sRNA inhibits the expression of litR. When the AI concentration is high at high cell density (HCD), they bind to the receptors to dephosphorylate LuxO and inactivate qrr sRNA. Inactivation of qrr sRNA, in turn, activates litR, which regulates many genes. Arrowheads at both ends indicate phosphorylation relay, and the symbol “P” indicates the phosphorylated state. AinS seems to affect luxU negatively and the litR expression positively. LitR seems to positively affect the genes involved in T6SS1, T6SS2, Aux-1, Aux-2, T6SS effectors and CRISPR system 2 and, conversely, affect T6SS3, Aux-3 and T6SS effectors negatively. Thin line and dashed line with arrowhead indicate regulation at LCD and HCD, respectively, where arrowheads indicate increased expression (+) and bars indicate decreased expression (-). A double arrowhead red line indicates the increased expression of qrr sRNA at 12°C compared to 6°C. Genes with significantly higher and lower expression are presented in purple and blue boxes, respectively.
In this study, LitR in A. wodanis did not affect the ainS expression, except at LCD and 12°C (FC = −1.93) in the ΔlitR mutant. This confirms our previous work where we showed that the 3OHC10-HSL production is not completely controlled through LitR (80). However, LitR in A. fischeri and A. salmonicida has been shown to significantly upregulate AinS (11, 107). In A. fischeri, several LitR-independent regulatory mechanisms have been reported to regulate ainS, such as ainS auto-regulation or cyclic AMP receptor protein- and glucose-mediated mechanisms (11, 108). Similarly, in A. wodanis, other regulatory mechanisms may regulate the expression of ainS. The deletion of ainS in A. wodanis negatively affected the litR expression. In the ΔainS mutants, the FC values of litR were slightly higher at HCD compared to LCD at both temperatures, which suggests that 3OHC10-HSL partly activates litR independently of the temperature. Similarly, the autoinducer synthase C8-HSL in A. fischeri positively affected litR in a cell density-dependent manner (14).
Regulation of CRISPRs and Spacers
A. wodanis harbors a type I-F CRISPR system and a CRISPR system without cas loci, the CRISPR system 1 and CRISPR system 2, respectively. CRISPR systems without cas loci are known as orphan arrays, where most of these are known to be non-functional (39). However, some of the orphan CRISPR arrays may function together with invading cas genes or the cas genes present in a different location within the same genome (109). In A. wodanis, the CRISPR system 1 consists of 25 spacers, whereas CRISPR system 2 consists of 40 spacers. The difference in the number of spacers in A. wodanis suggests that the CRISPR system 1 is less active than the CRISPR system 2 or is a remnant of a previously active CRISPR system. In addition, the spacers were not identical between CRISPR systems 1 and 2, indicating both are working independently of each other or that CRISPR system 2 has been introduced later than CRISPR system 1. The cas genes in A. wodanis shows ~90% amino acid similarity to the cas genes in M. viscosa, suggesting that the CRISPR system 2 has been horizontally transferred from M. viscosa. We speculate here that both bacteria can defend themselves against the same agents, which may favor their co-existence during the development of the winter ulcer disease. Similarity search of the spacers in A. wodanis against the phage databases revealed Vibrio phage CTX plasmid pCTX-2 and Staphylococcus phage phiSP38-1.
The expressions of cas genes (cas1, cas3, and csy3) in wild type seems to be cell density-dependent at 6°C, where the cas genes showed increased expression at HCD compared to LCD. However, cell density had no significant effect on the expression of CRISPR arrays of CRISPR systems 1 and 2 regardless of temperatures. Therefore, the CRISPR systems are partially dependent on cell density. Previous studies suggest that bacteria are at higher phage risk at higher cell density densities, and thus protection against phage is important at HCD (45). This suggests that A. wodanis positively influences cas genes as the cell density increases, and this might provide protection upon phage infection at HCD.
The CRISPR arrays of CRISPR system 1 in A. wodanis, was more highly expressed at 6°C when compared to 12°C, suggesting that the temperature 6°C plays a significant role in CRISPR array 1 expression. Temperature is known to regulate CRISPR-Cas genes in P. aeruginosa (45, 102). However, temperature difference did not regulate the expression of neither the cas operon nor the CRISPR array of the CRISPR system 2.
The complete cas operon of CRISPR system 2 in A. wodanis showed decreased expression in the ΔlitR mutant irrespective of cell densities and temperatures, which demonstrates that the CRISPR system 2 is regulated through LitR. In the ΔlitR mutant, the expression of the cas genes was higher at 6°C compared to 12°C at HCD. At LCD, only the expression values of the cas1 and cas3 were higher at 6°C compared to 12°C, while other genes such as csy1234 were higher at 12°C. Although LitR in A. wodanis activates the CRISPR system 2, the QS regulation is not completely cell density and temperature-dependent. Furthermore, the data shows that the CRISPR system 1 was more highly expressed in ΔlitR mutant compared to wild type, implying that LitR negatively affects CRISPR system 1.
In the ΔainS mutant, although a lower expression was observed on genes csy2, csy3, and csy4, the expression of CRISPR arrays of CRISPR system 2 and CRISPR system 1 were not changed by inactivation of ainS. Hence, the activation of the CRISPR system 2 in A. wodanis seems to be partly dependent on the AHL-mediated QS system. The QS activation of the CRISPR system has been reported as an efficient mechanism to enhance the benefit-to-cost ratio in P. aeruginosa (44, 45). Although several vibrios and aliivibrios comprise both QS and CRISPR systems, the QS regulation of the CRISPR system has not been described yet. Expression of the CRISPR system can be costly to the bacteria, and in some bacteria like Streptococcus thermophilus, the CRISPR system is constitutively expressed and confers a fitness cost (110). Therefore, the CRISPR system may only be expressed during phage infection or due to environmental cues (111–113). Similarly, the cell density, temperature and QS dependent regulation of CRISPR may enforce a benefit for A. wodanis during the development of winter ulcer.
Despite exhibiting CRISPR systems, one intact prophage was identified in A. wodanis chromosome 1 and several incomplete prophages in both chromosomes and the plasmids. Phages that infect P. aeruginosa are known to escape the type I-F and I-E CRISPR systems using anti-CRISPR proteins (114, 115). Therefore, we searched for anti-CRISPR proteins in A. wodanis using ArcFinder (116). No anti-CRISPR proteins were predicted, suggesting no self-targeted protospacers that prevent CRISPR-Cas response in A. wodanis. Although defective in their function, these incomplete phages are known to have adaptive functions in their hosts (117). Several of these incomplete phages are a putative source for phage-derived molecular products such as gene transfer agents, bacteriocins, phage killer particles, and they can also interfere with assembly of other phages (117, 118). The hypothetical proteins of intact and incomplete prophages from PHASTER output were searched against non-redundant protein database using BLASTP with default parameters. The hypothetical proteins present in the intact prophage were annotated as structural phage-related proteins. However, the incomplete prophages encoded several conserved proteins (with ≥ 90% amino acid identity and query coverage) such as type I restriction endonuclease subunit M, ClbS/DfsB family four-helix bundle protein, DUF559 domain-containing protein, type II toxin-antitoxin system RelE/ParE family toxin and other conserved hypothetical proteins (Supplementary Table 8). These proteins encoded by the incomplete prophage may play a role in adaptive functions of A. wodanis. However, further research is required to verify this.
Regulation of T6SSs, Aux, and T6SE Molecules
Bacteria secrete many virulence factors during the host-pathogen interface, not only to overcome the host's immune system but also for inter-bacterial competition (119). T6SS is an important virulence and survival factor present in about a quarter of known Gram-negative bacteria (120). A. wodanis genome revealed three T6SSs and four auxiliary clusters. Many bacteria possess more than one T6SS, which likely have different functions. For example, Burkholderia thailandensis possesses five T6SSs copies with different functions, where T6SS1 enhances the growth in the presence of other competing bacteria and T6SS4 is involved in the manganese transport to survive under oxidative stress, and the T6SS5 is involved in virulence in the murine model of pneumonic melioidosis (121, 122). Similarly, the multiple T6SSs in A. wodanis may have different cellular functions.
The T6SS1 in A. wodanis is highly similar (71–88%) to the T6SS system 1 in A. fischeri MJ11, while the T6SS2 shows high similarity (54–92%) to V. cholerae O1E1 T6SS and V. fluvialis T6SS2 (90, 123, 124). The T6SS3 in A. wodanis shows similarity (33–84%) to M. viscosa, V. anguillarum, A. salmonicida and Vibrio tapetis (48, 125, 126). Hcp and VgrG are essential components for the proper functioning of T6SS (127). A. wodanis encodes vgrG1 in Aux-1, however, it contains hcp in neither T6SS1 nor Aux-1 clusters. In A. fischeri MJ11, the T6SS1, which is similar to the T6SS1 of A. wodanis, is believed to interact with eukaryotic cells and is not involved in inter-strain killing (124). Moreover, in A. wodanis Aux-1 is located close to the heme uptake and utilization related genes. Metals such as iron are important for many cellular processes, and the genes located close to T6SS genes are known to be involved in iron uptake, for example, in P. aeuroginosa (58). Thus, the T6SS1 along with the Aux-1 may possibly be involved in iron uptake from the host or environment. Except for Hcp, the other structural T6SS proteins share low-level homology between each T6SSs. This indicates that the multiple T6SSs in A. wodanis is not a result of a recent duplication. Four copies of hcp were identified in the main and auxiliary T6SS clusters of chromosome 2. The proteins Hcp1 and Hcp4 showed 60% homology to each other, whereas Hcp3 and Hcp4 showed 100% homology to each other. Besides the structural role of Hcp, it is involved in the inter-bacterial competition, bacterial invasion, adherence, and cytotoxicity against host cells, also known to have other functions in different bacteria (127, 128). Therefore, the multiple copies of hcp in A. wodanis may have different functions.
V. cholerae utilizes T6SS to compete against diverse eukaryotic and prokaryotic organisms (66, 129). In V. fluvialis, the T6SS2 is anti-bacterial and provides a better competitive fitness in the marine environment (123). The T6SS2 in A. wodanis, which shows higher homology to V. fluvialis and V. cholerae, may enhance A. wodanis through inter-bacterial competition and virulence.
The T6SS3, unlike T6SS1 and T6SS2 in addition to structural components, contains additional genes vtsABCD encoding transporter proteins and does not contain vasH. VtsA-D plays a role in stress responses, transport function and hcp expression in V. anguillarum, and VasH is essential to drive the expression of the T6SS operon by inducing hcp and vgrG expression (48, 130). The mutation in vasH repressed hcp expression and impaired its anti-bacterial activity in other vibrios (123, 131). In V. anguillarum, VtsA-D proteins are involved in stress responses, however, plays no role in virulence (48). Similarly, T6SS3 in A. wodanis is probably involved in stress responses. After V. anguillarum, the T6SS similar to T6SS3 in A. wodanis is in M. viscosa (mts1). The genes encoding Hcp (AWOD_II_1028 and AWOD_II_1032), which are located close to each other, shows 71% similarity to M. viscosa hcp, MVIS_3030 (7). This implies that the T6SS3 in A. wodanis might have a similar function as mts1 of M. viscosa during the winter ulcer disease development.
In this study, expression of the T6SS1 and T6SS2 genes and their auxiliary clusters in A. wodanis are dependent on cell density. Such cell density-dependent T6SS expression has been reported in V. parahaemolyticus (50). Some of the genes in the T6SS2 and auxiliary clusters (Aux-2 and Aux-3) were found to be altered by temperature. The expression was found to be higher at 6°C than at 12°C. In V. fluvialis, the T6SS2 is regulated by temperature (123). Temperature-dependent regulation of virulent factor genes may be an essential feature for many bacteria to survive in harsh environments (132).
LitR in A. wodanis seemed to positively affect two T6SSs (T6SS1 and T6SS2) and negatively affect the expression of T6SS3 gene cluster. This demonstrates that QS regulation of the T6SSs in A. wodanis is very complex. The regulation of T6SS3 by LitR in A. wodanis indicates T6SS3 may play different roles than T6SS1 and T6SS2. Such reciprocal regulation has also been shown in V. parahaemolyticus, where the QS regulator OpaR downregulates T6SS1 and upregulates T6SS2 where T6SS1 functions as anti-bacterial and T6SS2 as anti-eukaryotic (133).
LitR in A. wodanis seemed to positively affect only the genes encoding the outer sheath and base-plate proteins of T6SS1, suggesting these genes are QS dependent. Interestingly, LitR was involved in activating the expression of the whole apparatus of the T6SS2 system. Moreover, LitR also repressed the entire T6SS3 gene clusters but only at LCD at 6°C. Therefore, T6SS2 is ultimately QS dependent, whereas T6SS1 and T6SS3 are not completely QS dependent. Regulation of T6SS by LuxR homologs have been described in V. cholerae (HapR), V. alginolyticus (LuxR) and V. anguillarum (VanT) (48, 49, 51, 134). Furthermore, we observed that LitR in A. wodanis is a strong activator of hcp1 expression at 6°C than at 12°C. Hence, this possibly implies that hcp1 of T6SS2 is may be involved in the cytotoxicity in CHSE cell line (80). Temperature has been shown to influence hcp expression in other bacteria such as Yersinia pestis and V. parahaemolyticus (133, 135). Similarly, in A. wodanis, the high expression of hcp at 6°C indicates that the T6SS2 could be more active at low temperature (6°C).
We identified that the genes in Aux-2 encoding ankyrin repeat-containing proteins and RHS proteins were differentially expressed in tpΔlitR/WT at 6°C. Ankyrin repeat proteins are known to be involved in pathogenesis by imitating and impeding host function (136). RHS proteins are toxins that are exported to the cell surface through T6SS, and it mediates anti-bacterial activity (137–139). AinS had no significant effect on the main and auxiliary clusters of T6SSs like LitR. However, AinS positively influenced the expression of the hcp1 and vgrG1 genes and may indicate that only these T6SS genes are dependent on AHL-mediated QS. We predicted several T6SS effectors in A. wodanis, including lipoprotein, nucleases, membrane proteins, amidases and succinylglutamate desuccinylase. However, most of them are putative and hypothetical proteins, which require further research to confirm. From the transcriptomics data, we found that cell density regulated several predicted T6SEs in wild type. Choline dehydrogenase, an osmoprotectant enzyme that protects bacteria from adverse temperatures and other stresses, was more highly expressed at 6°C (140, 141). The temperature has affected expression of several T6SEs indicating a temperature-dependent production of effector proteins. Similar to the LitR regulation of T6SS main and auxiliary clusters, it also controlled several T6SEs. Few T6SEs were also found to be affected by AinS. This may indicate that several effectors are dependent on QS, where LitR influenced the expression higher at LCD and 6°C. Some of the known effectors included porin-like protein H (AWOD_I_1000), a molecular filter for hydrophilic compounds and bacteriocin (AWOD_p920_0063), which is a virulent factor in A. wodanis that modulates the growth and virulence of M. viscosa (7, 17). Genes encoding T6SS immunity proteins are usually located close to the genes encoding effector proteins (63). The potential immunity protein of Aux-1 (AWOD_I_1437) in A. wodanis shows 30% amino acid similarity to the immunity protein in V. cholerae strain O1E1 (90), while in Aux-2, the immunity protein (AWOD_II_0134) shows 29% amino acid similarity to Mucilaginibacter gotjawali (142). The immunity protein (AWOD_II_1056) in Aux-4 shows 77% amino acid similarity to a hypothetical protein in A. fischeri MJ11 (124).
Conclusion
In this present study we show that cell densities and temperatures influenced the expression of many genes in A. wodanis. Moreover, the QS related genes in A. wodanis are cell density- and temperature-dependent, where 6°C plays an essential role in activating the AHL-mediated QS system. A. wodanis harbors two CRISPR systems, three T6SSs and four auxiliary T6SSs in its genome. We show that the CRISPR system 2 and T6SS3 of A. wodanis are similar to those in M. viscosa, the bacteria with which A. wodanis co-exists during winter ulcer disease. The low-temperature 6°C at which winter ulcer occurs exerts a significant effect on the expression pattern of A. wodanis than at high-temperature 12°C. We demonstrate here that LitR regulates CRISPR-Cas and T6SSs in a cell density- and temperature manner. Moreover, QS is found to regulate several potential T6SEs in A. wodanis. Thus, the QS regulation of T6SSs and CRISPR-Cas system of A. wodanis could be essential to understand the possible mechanisms used by A. wodanis during its co-existence with other bacteria like M. viscosa or the host.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
AM, EH, HH, and NW conceived and designed the experiments. AM performed the experiments. AM, EH, and NW analyzed the transcriptomics data. AM and NW wrote the manuscript. All authors contributed to reviewing and proof reading the final manuscript.
Funding
This work was funded by the Research council of Norway (#270068: ELIXIR2) and UiT The Arctic University of Tromsø. The publication charges for this article have been funded by UiT The Arctic University of Tromsø.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The sequencing service was provided by the Norwegian Sequencing Centre (http://www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the Functional Genomics and Infrastructure programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.799414/full#supplementary-material
Abbreviations
AHL, N-acyl homoserine lactone; Aux, Auxiliary; Bp, Base pair; CHSE, Chinook salmon embryo; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; DEGs, Differentially expressed genes; FC, Fold change; HCD, High cell density; LCD, Low cell density; MGE, Mobile genetic element; Min, minutes; OD600, Optical density measured at 600 nm; QS, Quorum sensing; RHS, Rearrangement hotspot protein; RPKM, reads per kilobase of gene per million reads mapped; T6SE, Type VI Secretion Effector; T6SS, Type VI Secretion System; tp, Transcriptome profile.
References
1. Dunlap PV, Kuo A. Cell density-dependent modulation of the Vibrio fischeri luminescence system in the absence of autoinducer and LuxR protein. J Bacteriol. (1992) 174:2440–8. doi: 10.1128/jb.174.8.2440-2448.1992
2. Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. (1994) 176:269–75. doi: 10.1128/jb.176.2.269-275.1994
3. Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. (1998) 280:295–8. doi: 10.1126/science.280.5361.295
4. Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. (2002) 99:3129–34. doi: 10.1073/pnas.052694299
5. Pena RT, Blasco L, Ambroa A, Gonzalez-Pedrajo B, Fernandez-Garcia L, Lopez M, et al. Relationship Between Quorum Sensing and Secretion Systems. Front Microbiol. (2019) 10:1100. doi: 10.3389/fmicb.2019.01100
6. Girard L. Quorum sensing in Vibrio spp.: the complexity of multiple signalling molecules in marine and aquatic environments. Crit Rev Microbiol. (2019) 45:451–471. doi: 10.1080/1040841X.2019.1624499
7. Hjerde E, Karlsen C, Sorum H, Parkhill J, Willassen NP, Thomson NR. Co-cultivation and transcriptome sequencing of two co-existing fish pathogens Moritella viscosa and Aliivibrio wodanis. BMC Genomics. (2015) 16:447. doi: 10.1186/s12864-015-1669-z
8. Greenberg EP. Quorum sensing in Gram-negative bacteria: acylhomoserine lactone signalling and cell-cell communication. Symp Soc Gen Microbi. (1999) 57:71–84.
9. Milton DL. Quorum sensing in vibrios: complexity for diversification. Int J Med Microbiol. (2006) 296:61–71. doi: 10.1016/j.ijmm.2006.01.044
10. Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, et al. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. (2002) 415:545–9. doi: 10.1038/415545a
11. Lupp C, Ruby EG. Vibrio fischeri LuxS and AinS: Comparative study of two signal synthases. J Bacteriol. (2004) 186:3873–81. doi: 10.1128/JB.186.12.3873-3881.2004
12. Freeman JA, Bassler BL. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J Bacteriol. (1999) 181:899–906. doi: 10.1128/JB.181.3.899-906.1999
13. Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. (2002) 110:303–14. doi: 10.1016/S0092-8674(02)00829-2
14. Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol Microbiol. (2003) 50:319–31. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x
15. Millikan DS, Ruby EG. FlrA, a sigma(54)-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol. (2003) 185:3547–57. doi: 10.1128/JB.185.12.3547-3557.2003
16. Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol. (2010) 77:1556–67. doi: 10.1111/j.1365-2958.2010.07309.x
17. Benz R, Bauer K. Permeation of Hydrophilic Molecules through the Outer-Membrane of Gram-Negative Bacteria - Review on Bacterial Porins. Eur J Biochem. (1988) 176:1–19. doi: 10.1111/j.1432-1033.1988.tb14245.x
18. Konkel ME, Tilly K. Temperature-regulated expression of bacterial virulence genes. Microbes Infect. (2000) 2:157–66. doi: 10.1016/S1286-4579(00)00272-0
19. Ramos JL, Gallegos MT, Marques S, Ramos-Gonzalez MI, Espinosa-Urgel M, Segura A. Responses of Gram-negative bacteria to certain environmental stressors. Curr Opin Microbiol. (2001) 4:166–71. doi: 10.1016/S1369-5274(00)00183-1
20. Sandy M, Butler A. Microbial Iron Acquisition: Marine and Terrestrial Siderophores. Chem Rev. (2009) 109:4580–95. doi: 10.1021/cr9002787
21. Tan HJ, Liu SH, Oliver JD, Wong HC. Role of RpoS in the susceptibility of low salinity-adapted Vibrio vulnificus to environmental stresses. Int J Food Microbiol. (2010) 137:137–42. doi: 10.1016/j.ijfoodmicro.2009.12.006
22. Quinn MJ, Resch CT, Sun J, Lind EJ, Dibrov P, Hase CC. NhaP1 is a K+(Na+)/H+ antiporter required for growth and internal pH homeostasis of Vibrio cholerae at low extracellular pH. Microbiol-Sgm. (2012) 158:1094–105. doi: 10.1099/mic.0.056119-0
23. Kohanski MA, Dwyer DJ, Collins JJ. How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol. (2010) 8:423–35. doi: 10.1038/nrmicro2333
24. Mercy C, Ize B, Salcedo SP, De Bentzmann S, Bigot S. Functional Characterization of Pseudomonas Contact Dependent Growth Inhibition (CDI) Systems. PLoS ONE. (2016) 11:e0150538. doi: 10.1371/journal.pone.0150538
25. Lasica AM, Ksiazek M, Madej M, Potempa J. The Type IX Secretion System (T9SS): Highlights and recent insights into its structure and function. Front Cell Infect Mi. (2017) 7:215. doi: 10.3389/fcimb.2017.00215
26. O'sullivan JN, Rea MC, O'connor PM, Hill C, Ross RP. Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. Fems Microbiol Ecol. (2019) 95:241. doi: 10.1093/femsec/fiy241
27. Yu KW, Xue P, Fu Y, Yang L. T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int J Mol Sci. (2021) 22:478. doi: 10.3390/ijms22020478
28. West SA, Buckling A. Cooperation, virulence and siderophore production in bacterial parasites. P Roy Soc B-Biol Sci. (2003) 270:37–44. doi: 10.1098/rspb.2002.2209
29. Reichenbach T, Mobilia M, Frey E. Mobility promotes and jeopardizes biodiversity in rock-paper-scissors games. Nature. (2007) 448:1046–9. doi: 10.1038/nature06095
30. Natrah FMI, Defoirdt T, Sorgeloos P, Bossier P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar Biotechnol. (2011) 13:109–26. doi: 10.1007/s10126-010-9346-3
31. Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm Formation As a Response to Ecological Competition. PLoS Biol. (2015) 13:e1002191. doi: 10.1371/journal.pbio.1002191
32. Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. Isme J. (2015) 9:139–49. doi: 10.1038/ismej.2014.174
33. Roncarati D, Scarlato V. Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. Fems Microbiol Rev. (2017) 41:549–74. doi: 10.1093/femsre/fux015
34. Toska J, Ho BT, Mekalanos JJ. Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc Natl Acad Sci USA. (2018) 115:7997–8002. doi: 10.1073/pnas.1808469115
35. Mojica FJM, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. (2005) 60:174–82. doi: 10.1007/s00239-004-0046-3
36. Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. (2014) 42:70–86. doi: 10.1093/nar/gkt711
37. Rostol JT, Marraffini L. (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe. (2019) 25:184–94. doi: 10.1016/j.chom.2019.01.009
38. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. (2011) 9:467–77. doi: 10.1038/nrmicro2577
39. Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. (2015) 13:722–36. doi: 10.1038/nrmicro3569
40. Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. (2017) 37:67–78. doi: 10.1016/j.mib.2017.05.008
41. Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. (2020) 18:67–83. doi: 10.1038/s41579-019-0299-x
42. Mcdonald ND, Regmi A, Morreale DP, Borowski JD, Boyd EF. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genomics. (2019) 20:5439. doi: 10.1186/s12864-019-5439-1
43. Liu TY, Doudna JA. Chemistry of Class 1 CRISPR-Cas effectors: Binding, editing, and regulation. J Biol Chem. (2020) 295:14473–87. doi: 10.1074/jbc.REV120.007034
44. Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GPC, Przybilski R, et al. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell. (2016) 64:1102–8. doi: 10.1016/j.molcel.2016.11.012
45. Hoyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, et al. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci USA. (2017) 114:131–5. doi: 10.1073/pnas.1617415113
46. Mion S, Plener L, Remy B, Daude D, Chabriere E. Lactonase SsoPox modulates CRISPR-Cas expression in Gram-negative proteobacteria using AHL-based quorum sensing systems. Res Microbiol. (2019) 170:296–9. doi: 10.1016/j.resmic.2019.06.004
47. Broniewski JM, Chisnall M, Hoyland-Kroghsbo NM, Buckling A, Westra ER. The effect of Quorum sensing inhibitors on the evolution of CRISPR-based phage immunity in Pseudomonas aeruginosa. Isme J. (2021) 15:2465–73. doi: 10.1038/s41396-021-00946-6
48. Weber B, Hasic M, Chen C, Wai SN, Milton DL. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ Microbiol. (2009) 11:3018–28. doi: 10.1111/j.1462-2920.2009.02005.x
49. Sheng LL, Gu D, Wang QY, Liu Q, Zhang YX. Quorum sensing and alternative sigma factor RpoN regulate type VI secretion system I (T6SSVA1) in fish pathogen Vibrio alginolyticus. Arch Microbiol. (2012) 194:379–90. doi: 10.1007/s00203-011-0780-z
50. Wang L, Zhou DS, Mao PY, Zhang YQ, Hou J, Hu Y, et al. Cell density- and quorum sensing-dependent expression of type VI secretion system 2 in Vibrio parahaemolyticus. PLoS ONE. (2013) 8:e073363. doi: 10.1371/journal.pone.0073363
51. Shao Y, Bassler BL. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol Microbiol. (2014) 92:921–30. doi: 10.1111/mmi.12599
52. Liu X, Pan J, Gao H, Han Y, Zhang A, Huang Y, et al. CqsA/LuxS-HapR Quorum sensing circuit modulates type VI secretion system V fl T6SS2 in Vibrio fluvialis. Emerg Microbes Infect. (2021) 10:589–601. doi: 10.1080/22221751.2021.1902244
53. Leung KY, Siame BA, Snowball H, Mok YK. Type VI secretion regulation: crosstalk and intracellular communication. Curr Opin Microbiol. (2011) 14:9–15. doi: 10.1016/j.mib.2010.09.017
54. Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. (2014) 15:9–21. doi: 10.1016/j.chom.2013.11.008
55. Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. (2016) 24:51–62. doi: 10.1016/j.tim.2015.10.005
56. Wang J, Brodmann M, Basler M. Assembly and subcellular localization of bacterial type VI secretion systems. Annu Rev Microbiol. (2019) 73:621–38. doi: 10.1146/annurev-micro-020518-115420
57. Wang TT, Si MR, Song YH, Zhu WH, Gao F, Wang Y, et al. Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. (2015) 11:1005020. doi: 10.1371/journal.ppat.1005020
58. Lin JS, Zhang WP, Cheng JL, Yang X, Zhu KX, Wang Y, et al. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun. (2017) 8:14888. doi: 10.1038/ncomms14888
59. Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. (2006) 312:1526–30. doi: 10.1126/science.1128393
60. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, et al. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. (2006) 103:1528–33. doi: 10.1073/pnas.0510322103
61. Schell MA, Ulrich RL, Ribot WJ, Brueggemann EE, Hines HB, Chen D, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. (2007) 64:1466–85. doi: 10.1111/j.1365-2958.2007.05734.x
62. Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. (2014) 12:137–48. doi: 10.1038/nrmicro3185
63. Lien YW, Lai EM. Type VI secretion effectors: methodologies and biology. Front Cell Infect Microbiol. (2017) 7:254. doi: 10.3389/fcimb.2017.00254
64. Ma J, Pan Z, Huang J, Sun M, Lu C, Yao H. The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence. (2017) 8:1189–202. doi: 10.1080/21505594.2017.1279374
65. Prochazkova K, Shuvalova LA, Minasov G, Voburka Z, Anderson WF, Satchell KJF. Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites. J Biol Chem. (2009) 284:26557–68. doi: 10.1074/jbc.M109.025510
66. Miyata ST, Kitaoka M, Brooks TM, Mcauley SB, Pukatzki S. Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun. (2011) 79:2941–9. doi: 10.1128/IAI.01266-10
67. Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, et al. Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem. (2013) 288:26616–24. doi: 10.1074/jbc.M113.488320
68. Jiang F, Waterfield NR, Yang J, Yang GW, Jin Q. A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe. (2014) 15:600–10. doi: 10.1016/j.chom.2014.04.010
69. Chen H, Yang DH, Han FJ, Tan JC, Zhang LZ, Xiao JF, et al. The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-Jnk pathway. Cell Host Microbe. (2017) 21:47–58. doi: 10.1016/j.chom.2016.12.004
70. Hood RD, Singh P, Hsu FS, Guvener T, Carl MA, Trinidad RRS, et al. A type VI secretion system of Pseudomonas aeruginosa targets, a toxin to bacteria. Cell Host Microbe. (2010) 7:25–37. doi: 10.1016/j.chom.2009.12.007
71. Brooks TM, Unterweger D, Bachmann V, Kostiuk B, Pukatzki S. Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem. (2013) 288:7618–25. doi: 10.1074/jbc.M112.436725
72. Hersch SJ, Watanabe N, Stietz MS, Manera K, Kamal F, Burkinshaw B, et al. Envelope stress responses defend against type six secretion system attacks independently of immunity proteins. Nat Microbiol. (2020) 5:706–14. doi: 10.1038/s41564-020-0672-6
73. Lunder T, Sorum H, Holstad G, Steigerwalt AG, Mowinckel P, Brenner DJ. Phenotypic and genotypic characterization of Vibrio viscosus sp nov and Vibrio wodanis sp nov isolated from Atlantic salmon (Salmo salar) with 'winter ulcer'. Int J Syst Evol Micr. (2000) 50:427–50. doi: 10.1099/00207713-50-2-427
74. Lunder T, Evensen Ø, Holstad G, Hastein T. Winter ulcer in the atlantic salmon salmo salar- pathological and bacteriological investigations and transmission experiments. Dis Aquat Organ. (1995) 23:39–49. doi: 10.3354/dao023039
75. Benediktsdottir E, Helgason S, Sigurjonsdottir H. Vibrio spp. isolated from salmonids with shallow skin lesions and reared at low temperature. J Fish Dis. (1998) 21:19–28. doi: 10.1046/j.1365-2761.1998.00065.x
76. Lovoll M, Wiik-Nielsen CR, Tunsjo HS, Colquhoun D, Lunder T, Sorum H, et al. Atlantic salmon bath challenged with Moritella viscosa–pathogen invasion and host response. Fish Shellfish Immunol. (2009) 26:877–84. doi: 10.1016/j.fsi.2009.03.019
77. Bruno DW, Griffiths J, Petrie J, Hastings TS. Vibrio viscosus in farmed Atlantic salmon Salmo salar in Scotland: field and experimental observations. Dis Aquat Organ. (1998) 34:161–6. doi: 10.3354/dao034161
78. Karlsen C, Vanberg C, Mikkelsen H, Sorum H. Co-infection of Atlantic salmon (Salmo salar), by Moritella viscosa and Aliivibrio wodanis, development of disease and host colonization. Vet Microbiol. (2014) 171:112–21. doi: 10.1016/j.vetmic.2014.03.011
79. Purohit AA, Johansen JA, Hansen H, Leiros HKS, Kashulin A, Karlsen C, et al. Presence of acyl-homoserine lactones in 57 members of the Vibrionaceae family. J Appl Microbiol. (2013) 115:835–47. doi: 10.1111/jam.12264
80. Maharajan A, Hansen H, Khider M, Willassen N. Quorum sensing in Aliivibrio wodanis 06/09/139 and its role in controlling various phenotypic traits. Peerj. (2021) 9:11980. doi: 10.7717/peerj.11980
81. Magoc T, Wood D, Salzberg SL. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol Bioinform Online. (2013) 9:127–36. doi: 10.4137/EBO.S11250
82. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
83. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. (2012) 40:D109–14. doi: 10.1093/nar/gkr988
84. Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Neron B, et al. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. (2018) 46:W246–51. doi: 10.1093/nar/gky425
85. Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. (2007) 35:W52–7. doi: 10.1093/nar/gkm360
86. Biswas A, Gagnon JN, Brouns SJJ, Fineran PC, Brown CM. CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. Rna Biol. (2013) 10:817–27. doi: 10.4161/rna.24046
87. Zhou Y, Liang YJ, Lynch KH, Dennis JJ, Wishart DS. PHAST: A fast phage search tool. Nucleic Acids Res. (2011) 39:W347–52. doi: 10.1093/nar/gkr485
88. Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. (2016) 44:W16–21. doi: 10.1093/nar/gkw387
89. Li J, Yao YF, Xu HH, Hao LM, Deng ZX, Rajakumar K, et al. SecReT6: a web-based resource for type VI secretion systems found in bacteria. Environ Microbiol. (2015) 17:2196–202. doi: 10.1111/1462-2920.12794
90. Joshi A, Kostiuk B, Rogers A, Teschler J, Pukatzki S, Yildiz FH. Rules of engagement: The type VI secretion system in Vibrio cholerae. Trends Microbiol. (2017) 25:267–79. doi: 10.1016/j.tim.2016.12.003
91. Wang JW, Yang BJ, Leier A, Marquez-Lago TT, Hayashida M, Rocker A, et al. Bastion6: a bioinformatics approach for accurate prediction of type VI secreted effectors. Bioinformatics. (2018) 34:2546–55. doi: 10.1093/bioinformatics/bty155
92. Unterweger D, Kostiuk B, Otjengerdes R, Wilton A, Diaz-Satizabal L, Pukatzki S. Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. (2015) 34:2198–210. doi: 10.15252/embj.201591163
93. Spinola-Amilibia M, Davo-Siguero I, Ruiz FM, Santillana E, Medrano FJ, Romero A. The structure of VgrG1 from Pseudomonas aeruginosa, the needle tip of the bacterial type VI secretion system. Acta Crystallogr D Struct Biol. (2016) 72:22–33. doi: 10.1107/S2059798315021142
94. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. (2007) 104:15508–13. doi: 10.1073/pnas.0706532104
95. Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, et al. TssK is a trimeric cytoplasmic protein interacting with components of both phage-like and membrane anchoring complexes of the type VI secretion system. J Biol Chem. (2013) 288:27031–41. doi: 10.1074/jbc.M113.499772
96. Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. (2015) 11:1005545. doi: 10.1371/journal.pgen.1005545
97. Bonemann G, Pietrosiuk A, Diemand A, Zentgraf H, Mogk A. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. (2009) 28:315–25. doi: 10.1038/emboj.2008.269
98. Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, et al. Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem. (2011) 286:30010–21. doi: 10.1074/jbc.M111.253377
99. Kitaoka M, Miyata ST, Brooks TM, Unterweger D, Pukatzki S. VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic vibrio cholerae. J Bacteriol. (2011) 193:6471–82. doi: 10.1128/JB.05414-11
100. Sana TG, Hachani A, Bucior I, Soscia C, Garvis S, Termine E, et al. The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and fur and modulates internalization in epithelial cells. J Biol Chem. (2012) 287:27095–105. doi: 10.1074/jbc.M112.376368
101. An JH, Goo E, Kim H, Seo YS, Hwang I. Bacterial quorum sensing and metabolic slowing in a cooperative population. P Natl Acad Sci USA. (2014) 111:14912–7. doi: 10.1073/pnas.1412431111
102. Hoyland-Kroghsbo NM, Munoz KA, Bassler BL. Temperature, by controlling growth rate, regulates CRISPR-Cas activity in Pseudomonas aeruginosa. Mbio. (2018) 9:2184. doi: 10.1128/mBio.02184-18
103. Khider M, Hjerde E, Hansen H, Willassen NP. Differential expression profiling of litR and rpoQ mutants reveals insight into QS regulation of motility, adhesion and biofilm formation in Aliivibrio salmonicida. BMC Genomics. (2019) 20:5594. doi: 10.1186/s12864-019-5594-4
104. Urmersbach S, Aho T, Alter T, Hassan SS, Autio R, Huehn S. Changes in global gene expression of Vibrio parahaemolyticus induced by cold- and heat-stress. Bmc Microbiol. (2015) 15:565. doi: 10.1186/s12866-015-0565-7
105. Khider M, Hansen H, Hjerde E, Johansen JA, Willassen NP. Exploring the transcriptome of luxI(-) and Delta ainS mutants and the impact of N-3-oxo-hexanoyl-L- and N-3-hydroxy-decanoyl-L-homoserine lactones on biofilm formation in Aliivibrio salmonicida. Peerj. (2019) 7:6845. doi: 10.7717/peerj.6845
106. Feng L, Rutherford ST, Papenfort K, Bagert JD, Van Kessel JC, Tirrell DA, et al. A Qrr noncoding RNA deploys four different regulatory mechanisms to optimize quorum-sensing dynamics. Cell. (2015) 160:228–40. doi: 10.1016/j.cell.2014.11.051
107. Hansen H, Purohit AA, Leiros HKS, Johansen JA, Kellermann SJ, Bjelland AM, et al. The autoinducer synthases LuxI and AinS are responsible for temperature-dependent AHL production in the fish pathogen Aliivibrio salmonicida. Bmc Microbiol. (2015) 15:402. doi: 10.1186/s12866-015-0402-z
108. Lyell NL, Colton DM, Bose JL, Tumen-Velasquez MP, Kimbrough JH, Stabb EV. Cyclic AMP receptor protein regulates pheromone-mediated bioluminescence at multiple levels in Vibrio fischeri ES114. J Bacteriol. (2013) 195:5051–63. doi: 10.1128/JB.00751-13
109. Almendros C, Guzman NM, Garcia-Martinez J, Mojica FJM. Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR-Cas I-F systems. Nat Microbiol. (2016) 1:81. doi: 10.1038/nmicrobiol.2016.81
110. Vale PF, Lafforgue G, Gatchitch F, Gardan R, Moineau S, Gandon S. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. P Roy Soc B-Biol Sci. (2015) 282:41–49. doi: 10.1098/rspb.2015.1270
111. Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J Mol Biol. (2010) 395:270–81. doi: 10.1016/j.jmb.2009.10.057
112. Perez-Rodriguez R, Haitjema C, Huang QQ, Nam KH, Bernardis S, Ke AL, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. (2011) 79:584–99. doi: 10.1111/j.1365-2958.2010.07482.x
113. Meaden S, Capria L, Alseth E, Gandon S, Biswas A, Lenzi L, et al. Phage gene expression and host responses lead to infection-dependent costs of CRISPR immunity. Isme J. (2021) 15:534–44. doi: 10.1038/s41396-020-00794-w
114. Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. (2013) 493:429–181. doi: 10.1038/nature11723
115. Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR. A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. Mbio. (2014) 5:869. doi: 10.1128/mBio.00896-14
116. Yi H, Huang L, Yang B, Gomez J, Zhang H, Yin Y. AcrFinder: genome mining anti-CRISPR operons in prokaryotes and their viruses. Nucleic Acids Res. (2020) 48:W358–65. doi: 10.1093/nar/gkaa351
117. Bobay LM, Touchon M, Rocha EPC. Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci USA. (2014) 111:12127–32. doi: 10.1073/pnas.1405336111
118. Czajkowski R. The phage be with you? Prophage-like elements in the genomes of soft rot Pectobacteriaceae: Pectobacterium spp. and Dickeya spp. Front Microbiol. (2019) 10:138. doi: 10.3389/fmicb.2019.00138
119. Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. (2012) 40:D641–5. doi: 10.1093/nar/gkr989
120. Bingle LEH, Bailey CM, Pallen MJ. Type VI secretion: a beginner's guide. Curr Opin Microbiol. (2008) 11:3–8. doi: 10.1016/j.mib.2008.01.006
121. Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, et al. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. (2010) 6:e1001068. doi: 10.1371/journal.ppat.1001068
122. Si MR, Zhao C, Burkinshaw B, Zhang B, Wei DW, Wang Y, et al. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci USA. (2017) 114:E2233–42. doi: 10.1073/pnas.1614902114
123. Huang Y, Du P, Zhao M, Liu W, Du Y, Diao B, et al. Functional characterization and conditional regulation of the type VI secretion system in Vibrio fluvialis. Front Microbiol. (2017) 8:528. doi: 10.3389/fmicb.2017.00528
124. Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci USA. (2018) 115:E8528–37. doi: 10.1073/pnas.1808302115
125. Bjornsdottir B, Hjerde E, Bragason BT, Gudmundsdottir T, Willassen NP, Gudmundsdottir BK. Identification of type VI secretion systems in Moritella viscosa. Vet Microbiol. (2012) 158:436–42. doi: 10.1016/j.vetmic.2012.02.030
126. Rodrigues S, Paillard C, Van Dillen S, Tahrioui A, Berjeaud JM, et al. Relation between biofilm and virulence in Vibrio tapetis: a transcriptomic study. Pathogens. (2018) 7:92. doi: 10.3390/pathogens7040092
127. Peng Y, Wang XR, Shou J, Zong BB, Zhang YY, Tan J, et al. Roles of Hcp family proteins in the pathogenesis of the porcine extraintestinal pathogenic Escherichia coli type VI secretion system. Sci Rep-Uk. (2016) 6:26816. doi: 10.1038/srep26816
128. Zhou Y, Tao J, Yu H, Ni J, Zeng L, Teng Q, et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect Immun. (2012) 80:1243–51. doi: 10.1128/IAI.05994-11
129. Ma AT, Mekalanos JJ. In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci USA. (2010) 107:4365–70. doi: 10.1073/pnas.0915156107
130. Seibt H, Aung KM, Ishikawa T, Sjöström A, Gullberg M, Atkinson GC, et al. Elevated levels of VCA0117 (VasH) in response to external signals activate the type VI secretion system of Vibrio cholerae O1 El Tor A1552. Environ Microbiol. (2020) 22:4409–23. doi: 10.1111/1462-2920.15141
131. Metzger LC, Stutzmann S, Scrignari T, Van Der Henst C, Matthey N, Blokesch M. Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep. (2016) 15:951–8. doi: 10.1016/j.celrep.2016.03.092
132. Ishikawa T, Sabharwal D, Broms J, Milton DL, Sjostedt A, Uhlin BE, et al. Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun. (2012) 80:575–84. doi: 10.1128/IAI.05510-11
133. Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE. (2013) 8:e061086. doi: 10.1371/journal.pone.0061086
134. Yang Z, Zhou X, Ma Y, Zhou M, Waldor MK, Zhang Y, et al. Serine/threonine kinase PpkA coordinates the interplay between T6SS2 activation and quorum sensing in the marine pathogen Vibrio alginolyticus. Environ Microbiol. (2018) 20:903–19. doi: 10.1111/1462-2920.14039
135. Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, et al. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiol-Sgm. (2009) 155:498–512. doi: 10.1099/mic.0.022160-0
136. Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. (2010) 18:132–9. doi: 10.1016/j.tim.2009.11.004
137. Koskiniemi S, Lamoureux JG, Nikolakakis KC, De Roodenbeke CT, Kaplan MD, Low DA, et al. Rhs proteins from diverse bacteria mediate intercellular competition. P Natl Acad Sci USA. (2013) 110:7032–7. doi: 10.1073/pnas.1300627110
138. Diniz JA, Coulthurst SJ. Intraspecies competition in serratia marcescens is mediated by type VI-Secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol. (2015) 197:2350–60. doi: 10.1128/JB.00199-15
139. Pei TT, Li H, Liang X, Wang ZH, Liu G, Wu LL, et al. Intramolecular chaperone-mediated secretion of an Rhs effector toxin by a type VI secretion system. Nat Commun. (2020) 11:1865. doi: 10.1038/s41467-020-15774-z
140. Gadda G, Mcallister-Wilkins EE. Cloning, expression, and purification of choline dehydrogenase from the moderate halophile Halomonas elongata. Appl Environ Microbiol. (2003) 69:2126–32. doi: 10.1128/AEM.69.4.2126-2132.2003
141. Tang D, Wang X, Wang J, Wang M, Wang Y, Wang W. Choline-betaine pathway contributes to hyperosmotic stress and subsequent lethal stress resistance in Pseudomonas protegens SN15-2. J Biosci. (2020) 45:60. doi: 10.1007/s12038-020-00060-3
Keywords: CRISPR, T6SS, QS, Aliivibrio wodanis 06/09/139, LitR and AinS
Citation: Maharajan AD, Hjerde E, Hansen H and Willassen NP (2022) Quorum Sensing Controls the CRISPR and Type VI Secretion Systems in Aliivibrio wodanis 06/09/139. Front. Vet. Sci. 9:799414. doi: 10.3389/fvets.2022.799414
Received: 21 October 2021; Accepted: 12 January 2022;
Published: 08 February 2022.
Edited by:
Najiah Musa, University of Malaysia Terengganu, MalaysiaReviewed by:
Natrah Fatin Mohd Ikhsan, Putra Malaysia University, MalaysiaQingpi Yan, Jimei University, China
Copyright © 2022 Maharajan, Hjerde, Hansen and Willassen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amudha Deepalakshmi Maharajan, YW11ZGhhLmQubWFoYXJhamFuQHVpdC5ubw==; Nils Peder Willassen, bmlscy1wZWRlci53aWxsYXNzZW5AdWl0Lm5v
 Amudha Deepalakshmi Maharajan
Amudha Deepalakshmi Maharajan Erik Hjerde
Erik Hjerde Hilde Hansen1
Hilde Hansen1 Nils Peder Willassen
Nils Peder Willassen