- 1Department of Oral Immunology and Infectious Diseases, School of Dentistry, University of Louisville, Louisville, KY, United States
- 2Department of Preventive Dentistry, Osaka University Graduate School of Dentistry, Suita, Osaka, Japan
Accumulating microbiome data and mechanistic studies in vitro and in vivo have refined our understanding of the oral microbiota as a functionally integrated polymicrobial community. Emergent properties of these communities are driven to a large extent by interspecies communication which can be based on physical association, secreted small molecules or nutritional exchange. Porphyromonas gingivalis is a consensus periodontal pathogen; however, virulence is only expressed in the context of a polymicrobial community. Multivalent fimbriae mediate attachment to other oral species which can initiate a distinct transcriptional program in both constituents of the binding pair. P. gingivalis also responds to small molecules and nutritional cues produced by partner organisms. Physiological interdependence forms the basis of complex networks of cooperating organisms which begin to resemble an organismal entity exhibiting a spectrum of pathogenic potential.
Introduction
Appreciation of the oral microbiome as polymicrobial harkens back to the foundation of microbiology and the meticulous observations of van Leeuwenhoek. As the new discipline of microbiology developed, however, emphasis became focused on the classical microbial diseases of the time, most of which had a single species etiology. The spectacular success of Koch's postulates which related the presence of a single exogenous organism to a single disease further entrenched the notion of individual species as etiological entities. These prevailing concepts influenced the early studies into the etiology of periodontal diseases. While it was recognized that disease was associated with an increased diversity of endogenous subgingival species, the overall goal of research was often to fulfill Koch's postulates, either in the classical form or as modified to accommodate endogenous pathogens, for periodontal microorganisms (1). This approach had some limited success with the induction of disease in non-human primates infected with P. gingivalis (2), albeit at a high inoculation dose unlikely to be representative of human disease. Adoption of models with extreme parameters has recently been termed, with only a hint of hyperbole, “Koch's curse” (3). Nonetheless, there was general agreement in the field that P. gingivalis along with a limited number of other species, functioned as periodontal pathogens (4). The introduction of the concept that complexes of organisms (color-coded depending on association with health or disease) tended to co-occur in the periodontal microbiome (5) was a significant step toward the notion of a polymicrobial etiology, particularly when integrated in a framework whereby environmental factors drive the selection and enrichment of endogenous pathogens (6). In germ-free animal models of disease, however, P. gingivalis remained obstinately non-pathogenic as a single species infection (7), and the idea of a polymicrobial community as the fundamental etiological unit thus took hold (8). Within this community organisms can have specialized roles, and P. gingivalis has emerged as a keystone pathogen which elevates overall community pathogenicity, or nososymbiocity (9), while receiving support from other community constituents which can be categorized as accessory pathogens (10). The interactions among P. gingivalis and other organisms that control nososymbiocity can thus define health or disease in the oral cavity to a large extent (11), and the nature of these interactions is the focus of this perspective. It is important to note, however, that periodontal tissue destruction is effectuated mainly by a dysregulated and uncontrolled inflammatory response to bacterial colonizers, a topic not addressed here but comprehensively reviewed elsewhere (12–15).
Physical interactions
A requisite early step in the development of polymicrobial communities is inter-species adherence. Oral bacteria express a number of adhesins with specificity for genetically distinct organisms, indicating that adaptation to a polymicrobial environment has been evolutionarily selected. Study of inter-species adherence was based, for several decades, on the ability of partner organisms to clump or coaggregate in suspension, and this approach revealed complex networks of co-adherence (16, 17). In these assays, however, P. gingivalis displayed a very limited range of binding partners, predominantly fusobacteria. Such data contributed to the idea that Fusobacterium nucleatum was a bridging organism, linking so-called “late” colonizers such as P. gingivalis with “early” colonizers including streptococci and actinomyces (18). The development of more sensitive assays, involving adherence of P. gingivalis to substrates of partner species on a solid support, demonstrated that P. gingivalis can in fact adhere to a number of other organisms including streptococci, actinomyces, veillonellae, and treponemes (19–23). Importantly, image analyses of ex-vivo biofilm samples did not substantiate a major role for F. nucleatum as a physical bridging organism (24), and its role may relate more to metabolic integration (discussed below). Further, more recent evidence indicates that colonization of the tooth surface may not follow a rigid temporal progression of early, middle and late arrivals, but rather polymicrobial aggregates are preformed in saliva prior to surface attachment (25).
P. gingivalis deploys two, at least, types of fimbriae, comprising the structural subunit proteins FimA (major fimbriae) or Mfa1 (minor fimbriae), which account for most of the interspecies adherence activity (26). While both fimbriae also possess an accessory protein complex, the structural proteins themselves possess adhesin domains (26). FimA binds to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) on streptococcal cell surfaces, as well as to the surface of Actinomyces oris (19, 27). In the case of GAPDH, a C-terminal domain of FimA binds to amino acid residues 166–183 of GAPDH (28, 29). Mfa1 also mediates attachment to streptococci, and a cleft spanning the central to C-terminal region of Mfa1 interacts with the SspA/B major surface proteins, with specificity being defined by the amino acid sequence of a protruding BAR domain at the C-terminus (30–32).
While close physical association between organisms facilitates small molecule or nutrition interactions, there is evidence that bacterial cells can sense and respond to the binding event itself. Outer membranes can sense stressors including mechanical changes resulting from adhesion (33). In E. coli for example, a surface lipoprotein can sense surface adhesion and activates two-component systems, which initiate an adhesion-dependent pattern of gene expression (34–36). Although the mechanisms have not been defined, a number of studies have shown that washed non-growing cells of P. gingivalis exhibit dramatic differential expression of mRNA and proteins when physically associated with other species. Contact with T. denticola upregulates the expression of P. gingivalis adhesins and proteases (37), and the organisms are synergistically pathogenic in murine models of periodontal disease (38). Time-coursed RNA-Seq showed that genes encoding a number of potential virulence determinants in P. gingivalis had higher expression in the presence of S. gordonii, including adhesins, the type IX secretion (T9SS) apparatus, and tetratricopeptide repeat (TPR) motif proteins. In contrast, genes encoding conjugation systems and many of the stress responses showed reduced expression (39). Expression of fimA is also increased, and stress-associated genes decreased, by Streptococcus oralis (40), and genes encoding T9SS components are upregulated in communities with Candida albicans (41). Proteomic analyses also show that partner species such as S. gordonii and F. nucleatum provide metabolic support to P. gingivalis in heterotypic communities (42–44), as is discussed further below. Collectively, this body of work is consistent with the notion of P. gingivalis virulence emerging as part of an evolutionary adaptation to a polymicrobial environment.
Small molecule and chemical effectors
Many bacterial species utilize Autoinducer (AI)-2, an indirect product of the LuxS enzyme, to coordinate behavior, particularly in the context of homo- or hetero- typic biofilm communities (45, 46). P. gingivalis possesses LuxS and produces AI-2 without which heterotypic community formation with S. gordonii does not occur (47). Expression of luxS is transcriptionally upregulated in communities with S. oralis (40), and AI-2 may participate in a positive feedback loop controlling expression of the Mfa1 fimbrial adhesin (48). The precise role of AI-2/LuxS is difficult to discern, however, as P. gingivalis lacks a recognized receptor for detection of extracellular AI-2. S-adenosyl methionine, a precursor of AI-2, is a product of the one carbon metabolism (OCM) pathway, and this may provide a link with pathogenicity, as discussed further below. Additionally, growth and virulence of P. gingivalis require a non-AI endogenous diffusible small molecule. However, P. gingivalis can overcome this requirement by utilizing a soluble molecule provided by V. parvula (49).
Many species of oral streptococci produce the reactive oxygen species (ROS) hydrogen peroxide which can damage both DNA and proteins, and as such is toxic to other organisms (50). P. gingivalis is a fairly aerotolerant anaerobe and can mount a vigorous protective oxidative stress response (51, 52), which is particularly effective in the context of a community with other aerotolerant organisms such as Filifactor alocis, F. nucleatum and Aggregatibacter actinomycetemcomitans (53–55). However, as a reducing environment is required for gingipain activity, in order to maintain the cysteine in the catalytic domain, oxidation by hydrogen peroxide can impair gingipain function in the absence of an effect on bacterial viability (56). Perhaps unsurprisingly, the toxic action of hydrogen peroxide is scale dependent, with toxicity being lost at longer distances between organisms (57, 58). Indeed, at longer distances peroxide may enhance P. gingivalis pathogenicity through the oxidation of oxyhemoglobin (oxyHb) to methemoglobin (metHb), an early step in heme acquisition (59).
Nutritional interactions
The importance of interspecies exchange of nutrients in the metabolism of P. gingivalis has been recognized for some time. For example, P. gingivalis produces isobutyric acid which stimulates growth of T. denticola, and reciprocally T. denticola produces succinic acid which enhances growth of P. gingivalis (60). More recent investigations have uncovered a web of nutritional connections which drive the interdependence of community constituents and are intimately involved in both enhancing and suppressing virulence.
One carbon metabolism (OCM)
OCM is an integral part of cellular intermediary metabolism, producing a number of one-carbon unit intermediates (formyl, methylene, methenyl, methyl) which are required for the synthesis of various amino acids and other biomolecules such as purines, thymidylate, folate and redox regulators (61, 62). Flux through the OCM cycle requires amino acid substrates as well as para-amino benzoic acid (pABA) which can be acquired from the extracellular milieu or produced de novo from chorismate by aminodeoxychorismate synthase (PabB) and 4-amino-4-deoxychorismate lyase (PabC) as part of the shikimate pathway (63). Exogenous pABA, which diffuses freely in and out of bacterial cells, can be salvaged by P. gingivalis, and oral streptococci such as S. gordonii produce exogenous pABA, which in the absence of cell-cell contact suppresses P. gingivalis pathogenicity. Indeed, dramatically, dual species communities of P. gingivalis and a pabC mutant of S. gordonii are significantly more pathogenic than a combination with the S. gordonii parental strain or P. gingivalis alone (64). The mechanisms of action of pABA remains to be fully determined; however, as an analogue of amino-benzoic acid, pABA is a competitive inhibitor of low molecular weight tyrosine phosphatases (LMW-PTPs) and consequently impacts tyrosine phosphorylation/dephosphorylation dependent signaling (65). In P. gingivalis, Ltp1 is a LMW-PTP and its cognate kinase, Ptk1, controls processing and secretion of gingipain proteases (66, 67), which in addition to their role as cardinal virulence factors also generate substrate amino acids for OCM flux (Figure 1). Tyrosine phosphorylation may thus serve as a metabolic couple linking OCM flux with substrate availability which in turn impacts virulence potential. Moreover, tyrosine phosphorylation based signaling in P. gingivalis may integrate and coordinate input from a number of organisms as Ptk1 is required for in vivo fitness with F. nucleatum (68). Further, synergistic interactions between P. gingivalis and other species may also involve OCM-based responses in P. gingivalis, as several genes in the OCM pathway are transcriptionally regulated when P. gingivalis is grown in T. denticola conditioned medium (69).
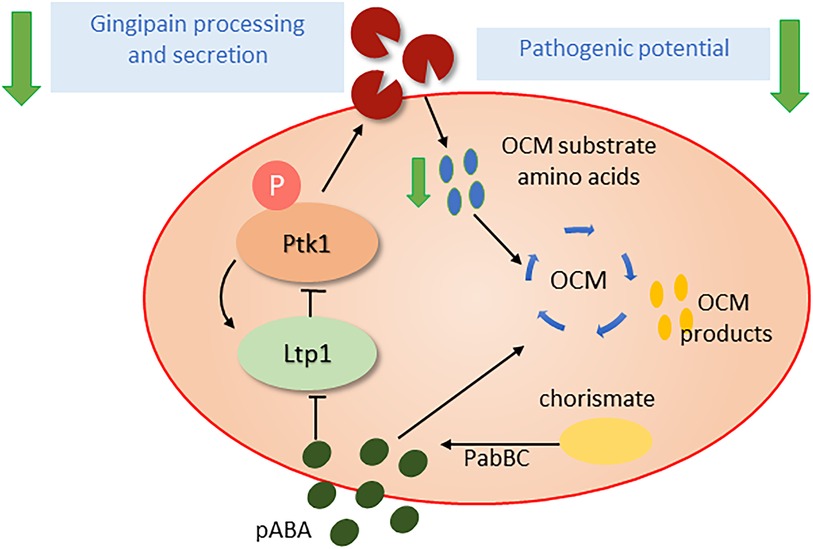
Figure 1. Influence of pABA (para-amino benzoic acid) on the pathogenic potential of P. gingivalis. pABA, an essential precursor for the one carbon metabolism (OCM) pathway, can be scavenged from partner organisms or synthesized endogenously from chorismate by the PabBC enzymes. pABA can inhibit the tyrosine phosphatase Ltp1 and thus disrupt activation state of the tyrosine kinase Ptk1 and tyrosine phosphorylation dependent signaling. Ptk1 phosphorylation of gingipains is required for optimal processing and secretion and when this is impeded the availability of amino acid substrates for OCM is diminished. Thus, the Ltp1-Ptk1 axis may function as a feedback mechanism to ensure balanced OCM flux. As gingipains are required for the provision of amino acid substrates for OCM and are major virulence factors, pABA levels regulate pathogenic potential.
Regulation of biofilm life-cycle
Metabolic integration of oral organisms also impacts community lifestyle. S. gordonii possesses an arginine deiminase system (ADS), which catalyzes the conversion of arginine to ammonia and CO2, along with the production of ATP. The ADS comprises three core enzymes: ArcA, ArcB (catabolic ornithine carbamoyltransferase), and ArcC (carbamate kinase) (70). S. gordonii also harbors a gene encoding the arginine-ornithine antiporter (ArcD), which is commonly located in the same locus with the arcABC genes. ArcD, a transmembrane protein, mediates the uptake of arginine and the concomitant export of ornithine in an ATP-independent manner, thus providing a substrate for the ADS pathway. Exported ornithine induces physiological and morphological alterations of planktonic F. nucleatum cells and the development of heterotypic biofilms with S. gordonii. Thus, S. gordonii ArcD modulates not only alkali and energy production, but also interspecies interactions with F. nucleatum, initiating a middle-stage of periodontopathic biofilm formation by metabolic cross-feeding (71).
Conversely, when cocultured with S. gordonii, F. nucleatum increases amino acid availability to enhance the production of butyrate and putrescine, a polyamine produced by ornithine decarboxylation. Coculture with Veillonella parvula also increases lysine availability, promoting cadaverine production by F. nucleatum. Interestingly, both putrescine and cadaverine can enhance P. gingivalis biofilm formation, and further, putrescine also stimulates biofilm dispersal (Figure 2) (71). A corollary to this is that the species composition and ratio of early colonizers within multi-species biofilms affect the polyamine profile produced by F. nucleatum, and the behavior of the subsequently colonizing periodontal pathogens. Analyses of clinical specimens has confirmed not only the co-occurrence of P. gingivalis with genetic modules responsible for putrescine production by S. gordonii and F. nucleatum in plaque samples, but also distinct salivary metabolome profiles characterized by elevated levels of cadaverine in severe periodontitis patients and putrescine in the middle stages of periodontitis (72, 73). Furthermore, ornithine emerged as the salivary metabolite most notably associated with the periodontal inflamed surface area, as indicated by its high variable importance in projection (VIP) value, which underscores its significance in constructing predictive models for periodontal disease (74). Several other metabolomic studies employing saliva or gingival crevicular fluid as clinical specimens support these findings, with polyamines and their derivatives being identified as biomarkers for gingivitis, periodontitis or dysbiotic alteration of the periodontal microbiome (75–80).
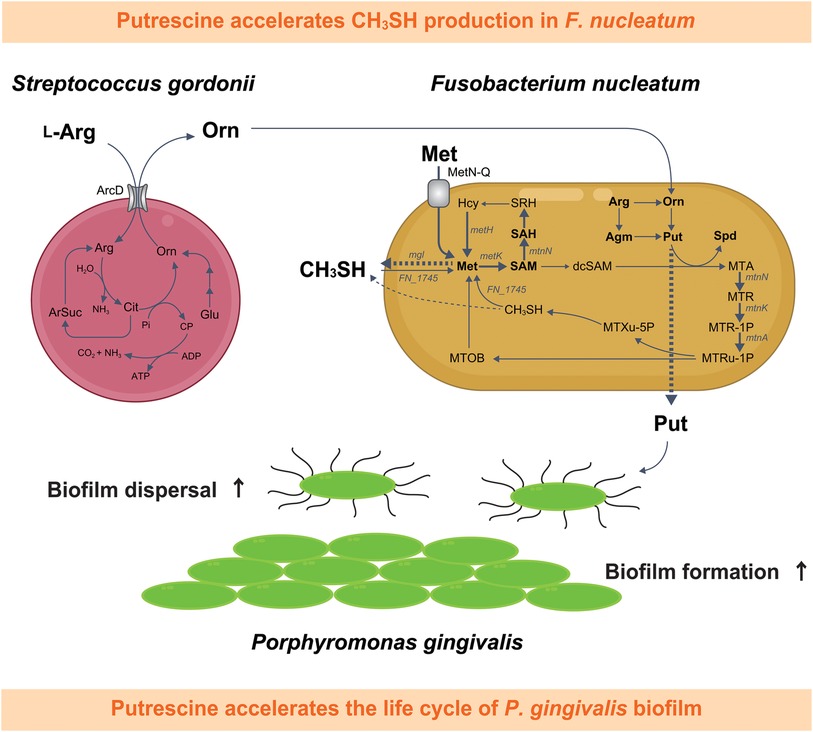
Figure 2. Schematic depiction of the metabolic interaction between Fusobacterium nucleatum and Streptococcus gordonii, highlighting the impact of putrescine, a metabolic byproduct synthesized during this interaction, on the life cycle of Porphyromonas gingivalis biofilms and CH3SH methyl mercaptan production in F. nucleatum. This diagram illustrates the pathway beginning with the uptake of L-arginine by S. gordonii, followed by the extracellular release of ornithine,which is subsequently metabolized by F. nucleatum into polyamines such as putrescine and spermidine. Putrescine released by F. nucleatum influences the biofilm dynamics of P. gingivalis. Furthermore, spermidine biosynthesis from putrescine in F. nucleatum is accompanied by an enhanced uptake of extracellular methionine, leading to a notable increase in CH3SH production. Detected metabolites are shown in bold, with dashed arrows for excretion and bold arrows for confirmed upregulation of bacterial metabolism. Met, l-methionine; SAM, S-adenosyl-l-methionine; SAH, S-adenosyl-l-homocysteine; SRH, S-ribosyl-l-homocysteine; Hcy, l-homocysteine; dcSAM, S-adenosylmethioninamine; MTA, 5'-methylthioadenosine; Arg, l-arginine; Agm, agmatine; Orn; l-ornithine; Cit, citrulline; Glu, glutamate; ArSuc, arginosuccinate; Pi, inorganic phosphate; CP, carbamoyl phosphate; Put, putrescine; Spd, spermidine; MTR, 5-methylthio-d-ribose; MTR-1P, S-methyl-5-thio-d-ribose 1-phosphate; MTRu-1P, S-methyl-5-thio-d-ribulose 1-phosphate; MTXu-5P, 1-(methylthio)xylulose 5-phosphate; CH3SH, methyl mercaptan; MTOB, 4-methylthio-2-oxobutanoic acid.
Ornithine and mercaptan production
The major oral odor compound methyl mercaptan (CH3SH) is strongly associated with both halitosis and periodontitis. CH3SH production stems from metabolism of polymicrobial communities in periodontal pockets and on the tongue dorsum. Using a large-volume anaerobic non-contact coculture system, F. nucleatum was found to be a potent producer of CH3SH, with synthesis stimulated by metabolic interactions with S. gordonii (80). Analysis of extracellular amino acids using an S. gordonii ArcD mutant demonstrated that ornithine excreted from S. gordonii is a key contributor to increased CH3SH production by F. nucleatum (80). Furthermore, a metabolic tracing analysis with 13C, 15N-methionine, as well as gene expression analysis, revealed that ornithine secreted by S. gordonii increased the demand for methionine through accelerated polyamine synthesis by F. nucleatum, leading to elevated methionine pathway activity and CH3SH production. Acetylated polyamines were also detected in F. nucleatum cells, although their presence was dependent on the levels of putrescine and spermidine, suggesting that, in excess, putrescine and spermidine may cause polyamine acetylation which will help maintain constant intracellular levels (80). Acetylation of polyamines by diamine N-acetyltransferase in F. nucleatum requires acetyl-CoA. In silico analysis of the acetyl-CoA (m + 1) biosynthetic pathway from 13C/15N-labeled L-methionine (m + 6) revealed only one pathway for incorporating a 13C into acetyl-CoA and no pathway for a 15N. Notably, F. nucleatum cannot complete this pathway alone; it requires the enzymatic reaction of glycine hydroxymethyltransferase in the one carbon pool, which is encoded by S. gordonii (80). It is also likely that elevated polyamine-synthesis in the context of coexistence with S. gordonii increases the demand for methionine, following enhancement of methionine metabolism and CH3SH generation.
Discussion
While P. gingivalis is commonly and correctly considered a pathogen in periodontal disease, it has become increasingly apparent that, unlike classic infections of single microbial etiology, periodontitis is not caused by a single organism but rather by polymicrobial communities of endogenous microbes acting in concert. Within these communities, organisms can have specialized roles, and although P. gingivalis can be seen as a driver of pathogenicity it is only in the context of a synergistic polymicrobial environment. Mapping of interspecies interactions spatially, temporally, and in molecular detail has revealed the basis of the emergent overall functions that promote or destabilize periodontal tissue homeostasis. This information provides the basis for new opportunities for intervention, not to eliminate P. gingivalis, which is likely a futile endeavor, but rather to restrain pathogenic potential through community engineering.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
RL: Writing – original draft, Writing – review & editing. MK: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
Primary research by the authors is supported by NICR grants DE012505, DE011111, DE023193, DE031756, DE033452 (RL); and KAKENHI Grants from the Japan Society for the Promotion of Science (JSPS) 22H03300 and 22K19622 (MK).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Socransky SS. Criteria for the infectious agents in dental caries and periodontal disease. J Clin Periodontol. (1979) 6:16–21. doi: 10.1111/j.1600-051X.1979.tb02114.x
2. Holt SC, Ebersole J, Felton J, Brunsvold M, Kornman KS. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science. (1988) 239:55–7. doi: 10.1126/science.3336774
3. Dewan KK, Harvill ET. Koch’s curse: how models of extreme pathology bias studies of host-pathogen interactions. PLoS Pathog. (2024) 20:e1011997. doi: 10.1371/journal.ppat.1011997
4. Socransky SS, and Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. (1992) 63 Suppl 4S, 322–31. doi: 10.1902/jop.1992.63.4s.322
5. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. (1998) 25, 134–44. doi: 10.1111/j.1600-051X.1998.tb02419.x
6. Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. (2003) 149:279–94. doi: 10.1099/mic.0.26082-0
7. Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. (2011) 10:497–506. doi: 10.1016/j.chom.2011.10.006
8. Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. (2012) 27:409–19. doi: 10.1111/j.2041-1014.2012.00663.x
9. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. (2012) 10:717–25. doi: 10.1038/nrmicro2873
10. Hajishengallis G, Lamont RJ. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. (2016) 24:477–89. doi: 10.1016/j.tim.2016.02.010
11. Hajishengallis G, Lamont RJ. Polymicrobial communities in periodontal disease: their quasi-organismal nature and dialogue with the host. Periodontol 2000. (2021) 86, 210–30. doi: 10.1111/prd.12371
12. Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol. (2015) 15:30–44. doi: 10.1038/nri3785
13. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. (2018) 16:745–59. doi: 10.1038/s41579-018-0089-x
14. Uriarte SM, Hajishengallis G. Neutrophils in the periodontium: interactions with pathogens and roles in tissue homeostasis and inflammation. Immunol Rev. (2022) 314:93–110. doi: 10.1111/imr.13152
15. Eltay EG, Van Dyke T. Resolution of inflammation in oral diseases. Pharmacol Ther. (2023) 247:108453. doi: 10.1016/j.pharmthera.2023.108453
16. Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. Communication among oral bacteria. Microbiol Mol Biol Rev. (2002) 66, 486–505. doi: 10.1128/MMBR.66.3.486-505.2002
17. Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. (2010) 8, 471–80. doi: 10.1038/nrmicro2381
18. Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. (1993) 175:3247–52. doi: 10.1128/jb.175.11.3247-3252.1993
19. Goulbourne PA, Ellen RP. Evidence that Porphyromonas (Bacteroides) gingivalis fimbriae function in adhesion to Actinomyces viscosus. J Bacteriol. (1991) 173:5266–74. doi: 10.1128/jb.173.17.5266-5274.1991
20. Lamont RJ, Hersey SG, Rosan B. Characterization of the adherence of Porphyromonas gingivalis to oral streptococci. Oral Microbiol Immunol. (1992) 7:193–7. doi: 10.1111/j.1399-302X.1992.tb00024.x
21. Yao ES, Lamont RJ, Leu SP, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. (1996) 11:35–41. doi: 10.1111/j.1399-302X.1996.tb00334.x
22. Zhou P, Liu J, Merritt J, Qi F. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis, and human oral buccal cells. Mol Oral Microbiol. (2015) 30:269–79. doi: 10.1111/omi.12091
23. Lamont RJ, Hajishengallis G, Koo H. Social networking at the microbiome-host interface. Infect Immun. (2023) 91:e0012423. doi: 10.1128/iai.00124-23
24. Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. (2016) 113:E791–800. doi: 10.1073/pnas.1522149113
25. Simon-Soro A, Ren Z, Krom BP, Hoogenkamp MA, Cabello-Yeves PJ, Daniel SG, et al. Polymicrobial aggregates in human saliva build the oral biofilm. mBio. (2022) 13:e0013122. doi: 10.1128/mbio.00131-22
26. Hasegawa Y, Nagano K. Porphyromonas gingivalis FimA and Mfa1 fimbriae: current insights on localization, function, biogenesis, and genotype. Jpn Dent Sci Rev. (2021) 57:190–200. doi: 10.1016/j.jdsr.2021.09.003
27. Maeda K, Nagata H, Yamamoto Y, Tanaka M, Tanaka J, Minamino N, et al. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect Immun. (2004) 72:1341–8. doi: 10.1128/IAI.72.3.1341-1348.2004
28. Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar HT, et al. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. (1997) 76:852–7. doi: 10.1177/00220345970760040601
29. Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, et al. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun. (2009) 77:5130–8. doi: 10.1128/IAI.00439-09
30. Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun. (2001) 69:5736–41. doi: 10.1128/IAI.69.9.5736-5741.2001
31. Forsgren N, Lamont RJ, Persson K. Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci. (2009) 18:1896–905. doi: 10.1002/pro.200
32. Roky M, Trent JO, Demuth DR. Identification of functional domains of the minor fimbrial antigen involved in the interaction of Porphyromonas gingivalis with oral streptococci. Mol Oral Microbiol. (2020) 35:66–77. doi: 10.1111/omi.12280
33. Saha S, Lach SR, Konovalova A. Homeostasis of the gram-negative cell envelope. Curr Opin Microbiol. (2021) 61:99–106. doi: 10.1016/j.mib.2021.03.008
34. Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc Natl Acad Sci U S A. (2002) 99:2287–92. doi: 10.1073/pnas.042521699
35. Shimizu T, Ichimura K, Noda M. The surface sensor NlpE of enterohemorrhagic Escherichia coli contributes to regulation of the type III secretion system and flagella by the Cpx response to adhesion. Infect Immun. (2016) 84:537–49. doi: 10.1128/IAI.00881-15
36. Feng L, Yang B, Xu Y, Xiong Y, Wang F, Liu B, et al. Elucidation of a complete mechanical signaling and virulence activation pathway in enterohemorrhagic Escherichia coli. Cell Rep. (2022) 39:110614. doi: 10.1016/j.celrep.2022.110614
37. Meuric V, Martin B, Guyodo H, Rouillon A, Tamanai-Shacoori Z, Barloy-Hubler F, et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol Oral Microbiol. (2013) 28:40–53. doi: 10.1111/omi.12004
38. Orth RK, O'brien-Simpson NM, Dashper SG, Reynolds EC. Synergistic virulence of Porphyromonas gingivalis and Treponema denticola in a murine periodontitis model. Mol Oral Microbiol. (2011) 26:229–40. doi: 10.1111/j.2041-1014.2011.00612.x
39. Hendrickson EL, Beck DA, Miller DP, Wang Q, Whiteley M, Lamont RJ, et al. Insights into dynamic polymicrobial synergy revealed by time-coursed RNA-Seq. Front Microbiol. (2017) 8:261. doi: 10.3389/fmicb.2017.00261
40. Maeda K, Nagata H, Ojima M, Amano A. Proteomic and transcriptional analysis of interaction between oral microbiota Porphyromonas gingivalis and Streptococcus oralis. J Proteome Res. (2015) 14:82–94. doi: 10.1021/pr500848e
41. Sztukowska MN, Dutton LC, Delaney C, Ramsdale M, Ramage G, Jenkinson HF, et al. Community development between Porphyromonas gingivalis and Candida albicans mediated by InlJ and Als3. MBio. (2018) 9:e00202–18. doi: 10.1128/mBio.00202-18
42. Kuboniwa M, Hendrickson EL, Xia Q, Wang T, Xie H, Hackett M, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. (2009) 9:98. doi: 10.1186/1471-2180-9-98
43. Bostanci N, Grant M, Bao K, Silbereisen A, Hetrodt F, Manoil D, et al. Metaproteome and metabolome of oral microbial communities. Periodontol 2000. (2021) 85:46–81. doi: 10.1111/prd.12351
44. Shokeen B, Dinis MDB, Haghighi F, Tran NC, Lux R. Omics and interspecies interaction. Periodontol 2000. (2021) 85:101–11. doi: 10.1111/prd.12354
45. Mcnab R, Lamont RJ. Microbial dinner-party conversations: the role of LuxS in interspecies communication. J Med Microbiol. (2003) 52:541–5. doi: 10.1099/jmm.0.05128-0
46. Pereira CS, Thompson JA, and Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. (2013) 37, 156–81. doi: 10.1111/j.1574-6976.2012.00345.x
47. Mcnab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol. (2003) 185:274–84. doi: 10.1128/JB.185.1.274-284.2003
48. Chawla A, Hirano T, Bainbridge BW, Demuth DR, Xie H, Lamont RJ. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. (2010) 78:1510–22. doi: 10.1111/j.1365-2958.2010.07420.x
49. Hoare A, Wang H, Meethil A, Abusleme L, Hong BY, Moutsopoulos NM, et al. A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J. (2021) 15:1490–504. doi: 10.1038/s41396-020-00865-y
50. Baty JJ, Stoner SN, Scoffield JA. Oral commensal streptococci: gatekeepers of the oral cavity. J Bacteriol. (2022) 204:e0025722. doi: 10.1128/jb.00257-22
51. Henry LG, Mckenzie RM, Robles A, Fletcher HM. Oxidative stress resistance in Porphyromonas gingivalis. Future Microbiol. (2012) 7:497–512. doi: 10.2217/fmb.12.17
52. Leclerc J, Rosenfeld E, Trainini M, Martin B, Meuric V, Bonnaure-Mallet M, et al. The cytochrome bd oxidase of Porphyromonas gingivalis contributes to oxidative stress resistance and dioxygen tolerance. PLoS One. (2015) 10, e0143808. doi: 10.1371/journal.pone.0143808
53. Bradshaw DJ, Marsh PD, Watson GK, and Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. (1998) 66, 4729–32. doi: 10.1128/IAI.66.10.4729-4732.1998
54. Diaz PI, Zilm PS, and Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. (2002) 148, 467–72. doi: 10.1099/00221287-148-2-467
55. Zhu B, Macleod LC, Newsome E, Liu J, Xu P. Aggregatibacter actinomycetemcomitans mediates protection of Porphyromonas gingivalis from Streptococcus sanguinis hydrogen peroxide production in multi-species biofilms. Sci Rep. (2019) 9:4944. doi: 10.1038/s41598-019-41467-9
56. Fitzsimonds ZR, Liu C, Stocke KS, Yakoumatos L, Shumway B, Miller DP, et al. Regulation of olfactomedin 4 by Porphyromonas gingivalis in a community context. ISME J. (2021) 15:2627–42. doi: 10.1038/s41396-021-00956-4
57. Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A. (2014) 111:7819–24. doi: 10.1073/pnas.1400586111
58. Duan D, Scoffield JA, Zhou X, Wu H. Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ Microbiol. (2016) 18:40023–4036. doi: 10.1111/1462-2920.13425
59. Brown JL, Yates EA, Bielecki M, Olczak T, Smalley JW. Potential role for Streptococcus gordonii-derived hydrogen peroxide in heme acquisition by Porphyromonas gingivalis. Mol Oral Microbiol. (2018) 33:322–35. doi: 10.1111/omi.12229
60. Grenier D. Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infect Immun. (1992) 60:5298–301. doi: 10.1128/iai.60.12.5298-5301.1992
61. Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. (2017) 25:27–42. doi: 10.1016/j.cmet.2016.08.009
62. Shetty S, Varshney U. Regulation of translation by one-carbon metabolism in bacteria and eukaryotic organelles. J Biol Chem. (2021) 296:100088. doi: 10.1074/jbc.REV120.011985
63. Stocke KS, Lamont RJ. One-carbon metabolism and microbial pathogenicity. Mol Oral Microbiol. (2023). doi: 10.1111/omi.12417
64. Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. (2017) 2:1493–9. doi: 10.1038/s41564-017-0021-6
65. Jung YJ, Miller DP, Perpich JD, Fitzsimonds ZR, Shen D, Ohshima J, et al. Porphyromonas gingivalis tyrosine phosphatase Php1 promotes community development and pathogenicity. mBio. (2019) 10:e02004–02019. doi: 10.1128/mBio.02004-19
66. Nowakowska Z, Madej M, Grad S, Wang T, Hackett M, Miller DP, et al. Phosphorylation of major Porphyromonas gingivalis virulence factors is crucial for their processing and secretion. Mol Oral Microbiol. (2021) 36:316–26. doi: 10.1111/omi.12354
67. Lamont RJ, Miller DP. Tyrosine kinases and phosphatases: enablers of the Porphyromonas gingivalis lifestyle. Front Oral Health. (2022) 3:835586. doi: 10.3389/froh.2022.835586
68. Perpich JD, Yakoumatos L, Stocke KS, Lewin GR, Ramos A, Yoder-Himes DR, et al. Porphyromonas gingivalis tyrosine kinase is a fitness determinant in polymicrobial infections. Infect Immun. (2022) 90:e0017022. doi: 10.1128/iai.00170-22
69. Kin LX, Butler CA, Slakeski N, Hoffmann B, Dashper SG, Reynolds EC. Metabolic cooperativity between Porphyromonas gingivalis and Treponema denticola. J Oral Microbiol. (2020) 12:1808750. doi: 10.1080/20002297.2020.1808750
70. Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. (1986) 50:314–52. doi: 10.1128/mr.50.3.314-352.1986
71. Sakanaka A, Kuboniwa M, Shimma S, Alghamdi SA, Mayumi S, Lamont RJ, et al. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems. (2022) 7:e0017022. doi: 10.1128/msystems.00170-22
72. Sakanaka A, Kuboniwa M, Hashino E, Bamba T, Fukusaki E, Amano A. Distinct signatures of dental plaque metabolic byproducts dictated by periodontal inflammatory status. Sci Rep. (2017) 7:42818. doi: 10.1038/srep42818
73. Sakanaka A, Kuboniwa M, Katakami N, Furuno M, Nishizawa H, Omori K, et al. Saliva and plasma reflect metabolism altered by diabetes and periodontitis. Front Mol Biosci. (2021) 8:742002. doi: 10.3389/fmolb.2021.742002
74. Kuboniwa M, Sakanaka A, Hashino E, Bamba T, Fukusaki E, Amano A. Prediction of periodontal inflammation via metabolic profiling of saliva. J Dent Res. (2016) 95:1381–6. doi: 10.1177/0022034516661142
75. Lamster IB, Mandella RD, Zove SM, Harper DS. The polyamines putrescine, spermidine and spermine in human gingival crevicular fluid. Arch Oral Biol. (1987) 32:329–33. doi: 10.1016/0003-9969(87)90087-2
76. Barnes VM, Teles R, Trivedi HM, Devizio W, Xu T, Mitchell MW, et al. Acceleration of purine degradation by periodontal diseases. J Dent Res. (2009) 88:851–5. doi: 10.1177/0022034509341967
77. Ozeki M, Nozaki T, Aoki J, Bamba T, Jensen KR, Murakami S, et al. Metabolomic analysis of gingival crevicular fluid using gas chromatography/mass spectrometry. Mass Spectrom (Tokyo. (2016) 5:A0047. doi: 10.5702/massspectrometry.A0047
78. Liebsch C, Pitchika V, Pink C, Samietz S, Kastenmuller G, Artati A, et al. The saliva metabolome in association to oral health status. J Dent Res. (2019) 98:642–51. doi: 10.1177/0022034519842853
79. Andorfer L, Holtfreter B, Weiss S, Matthes R, Pitchika V, Schmidt CO, et al. Salivary metabolites associated with a 5-year tooth loss identified in a population-based setting. BMC Med. (2021) 19:161. doi: 10.1186/s12916-021-02035-z
Keywords: periodontal, community, interspecies communication, biofilm, nutritional integration
Citation: Lamont RJ and Kuboniwa M (2024) The polymicrobial pathogenicity of Porphyromonas gingivalis. Front. Oral. Health 5:1404917. doi: 10.3389/froh.2024.1404917
Received: 21 March 2024; Accepted: 12 April 2024;
Published: 26 April 2024.
Edited by:
Georgios N. Belibasakis, Karolinska Institutet (KI), SwedenReviewed by:
Justin Merritt, Oregon Health and Science University, United States© 2024 Lamont and Kuboniwa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard J. Lamont cmljaC5sYW1vbnRAbG91aXN2aWxsZS5lZHU= Masae Kuboniwa a3Vib25pd2EubWFzYWUuZGVudEBvc2FrYS11LmFjLmpw
†These authors have contributed equally to this work
 Richard J. Lamont
Richard J. Lamont Masae Kuboniwa
Masae Kuboniwa