- 1Department of Medical Microbiology, Faculty of Health Sciences, University of Pretoria, Pretoria, South Africa
- 2Department of Medical Laboratory Science, College of Health Sciences, Woldia University, Woldia, Ethiopia
- 3Department of Microbiology, School of Life Sciences, College of Agriculture, Engineering and Science, University of KwaZulu-Natal, Durban, South Africa
Purpose: This study examined the patterns and frequency of genetic changes responsible for resistance to first-line (rifampicin and isoniazid), fluoroquinolones, and second-line injectable drugs in drug-resistant Mycobacterium tuberculosis (MTB) isolated from culture-positive pulmonary tuberculosis (PTB) symptomatic attendees of spiritual holy water sites (HWSs) in the Amhara region.
Patients and methods: From June 2019 to March 2020, a cross-sectional study was carried out. A total of 122 culture-positive MTB isolates from PTB-suspected attendees of HWSs in the Amhara region were evaluated for their drug resistance profiles, and characterized gene mutations conferring resistance to rifampicin (RIF), isoniazid (INH), fluoroquinolones (FLQs), and second-line injectable drugs (SLIDs) using GenoType®MTBDRplus VER2.0 and GenoType®MTBDRsl VER2.0. Drug-resistant MTB isolates were Spoligotyped following the manufacturer’s protocol.
Results: Genetic changes (mutations) responsible for resistance to RIF, INH, and FLQs were identified in 15/122 (12.3%), 20/122 (16.4%), and 5/20 (25%) of MTB isolates, respectively. In RIF-resistant, rpoB/Ser531Lue (n = 12, 80%) was most frequent followed by His526Tyr (6.7%). Amongst INH-resistant isolates, katG/Ser315Thr1 (n = 19, 95%) was the most frequent. Of 15 MDR-TB, the majority (n = 12, 80%) isolates had mutations at both rpoB/Ser531Leu and katG/Ser315Thr1. All 20 INH and/or RIF-resistant isolates were tested with the MTBDRsl VER 2.0, yielding 5 FLQs-resistant isolates with gene mutations at rpoB/Ser531Lue, katG/Ser315Thr1, and gyrA/Asp94Ala genes. Of 20 Spoligotyped drug-resistant MTB isolates, the majority (n = 11, 55%) and 6 (30%) were SIT149/T3-ETH and SIT21/CAS1-Kili sublineages, respectively; and they were any INH-resistant (mono-hetero/multi-). Of 15 RIF-resistant (RR/MDR-TB) isolates, 7 were SIT149/T3-ETH, while 6 were SIT21/CAS1-Kili sublineages. FLQ resistance was detected in four SIT21/CAS1-Kili lineages.
Conclusion: In the current study, the most common gene mutations responsible for resistance to INH, RIF, and FLQs were identified. SIT149/T3-ETH and SIT21/CAS1-Kili constitute the majority of drug-resistant TB (DR-TB) isolates. To further understand the complete spectrum of genetic changes/mutations and related genotypes, a sequencing technology is warranted.
Background
Drug-resistant Mycobacterium tuberculosis (MTB) is an escalating health problem that presents substantial challenges for public health systems across the world (1–3). The advent and transmission of drug-resistant MTB strains, especially multidrug-resistant/RIF-resistant TB (MDR/RR-TB)-which is resistant to the two key anti-mycobacterial drugs, INH and RIF- and “extensively drug-resistant TB (XDR-TB)”-which is an MDR/RR isolate that is also resistant to any FLQs and bedaquiline and/or linezolid-has made it more challenging to contain and eradicate the disease globally (1–4). It makes TB control programs less effective since resistant TB strains require specialized laboratory facilities, diagnostic tools with higher accuracy, and treatment regimens that aren’t usually available in resource-constrained settings (1, 2). Globally, 10.6 million people contracted TB, and about “410,000 developed MDR or RIF-resistant TB” (5). The prevalence of DR-TB varies by geography, with India, China, and the Russian Federation having the largest burden (1). Ethiopia bears a substantial burden of TB, and the rising incidence of DR-TB has detrimental effects on the country’s health systems and efforts to contain the disease (1, 6–8). Moreover, although Ethiopia is on track to transition out of the thirty nations with high rates of MDR/RR-TB (1), the burden of resistant TB is still a problem, where 0.71% of new TB cases and 12.0% of retreated TB patients were reported to have MDR-TB (9).
Drug-resistant TB strain has a severe impact on both individual and societal levels, with resistant TB being associated with higher patient mortality rates than susceptible TB strains (1, 3, 10). The treatment outcomes for DR-TB are often unfavorable due to the limited accessibility and effectiveness of second-line anti-TB medicines, lengthy treatment durations, and higher toxicity of the drugs (2, 10). Thus, patients experience prolonged suffering and there is an increasing risk of transmitting the disease to others (3, 11). Accordingly, DR-TB necessitates an accurate and timely diagnosis for the effective management of TB patients (3).
Molecular diagnostic methods have transformed the detection and identification of drug-resistance-associated genetic changes in MTB in a rapid and precise manner (2, 12), providing accurate evidence for patient-centered treatment decisions (13). The application of “PCR-based tools, like the GeneXpert®MTB/RIF (Cepheid Inc., Sunnyvale, CA, USA),” has significantly transformed the process of detecting TB and drug-resistant TB and enhanced the treatment of TB patients (14). “Line Probe Assays (LPAs) (Hain Lifescience, Germany)” are another widely used tool for the simultaneous diagnosis of TB and the anti-mycobacterial resistance profile of TB strains (14, 15). These assays target specific genetic regions linked with resistance to INH and RIF, and FLQs and SLIDs, respectively. Although LPAs have certain limitations that only detect the gene mutations within the target of interest (15), “MTBDRplus VER 2.0” (16), and “MTBDRsl VER 2.0” (17), have higher sensitivity and specificity than the first version to diagnose anti-TB drug resistance, and associated gene mutations (16, 17). Ethiopia is one of the nations that has benefited from the use of LPA diagnostic technologies to improve the management of TB patients. LPAs (“MTBDRplus and MTBDRsl”) tests were integrated into the Ethiopian national TB diagnostic algorithm (18, 19). Since the rollout of LPAs in Ethiopia, TB diagnostic referral centers at the national and regional levels have been authorized to undertake those assays (19).
Genetic mutations play a crucial role in conferring anti-TB drug resistance in MTB (2). The presence of those genetic changes conferring resistance to key anti-mycobacterial drugs differs geographically and is influenced by factors like the prevalence of DR-TB strains, treatment regimens, and patient adherence to therapy (20). Thus, understanding the types and frequency of those genetic changes is critical for effective drug-resistant strain detection, guiding treatment strategies, and monitoring the emergence of new resistance patterns (20, 21).
Furthermore, it is paramount to identify populations at high risk for TB transmission and conduct TB case finding in hotspot settings (22, 23). The Ethiopian National TB Control Program identified prisoners, HIV-positive people, diabetic patients, the older adult, University residents, TB patients’ contacts, the homeless, healthcare workers, and refugees as key high-risk groups for TB (22); thereby earlier studies reported the burden of TB in prison settings (24, 25), refugees (26), homeless (27, 28), and University residents (29), in Ethiopia. Although Ethiopia has identified key populations for TB and implemented WHO’s policy on TB infection control in high-risk settings, targeting spiritual holy water sites as high-priority settings for TB control activities is not given much attention (30). The first national TB prevalence survey (6), and an earlier community-based TB study among key populations in hotspot settings in Ethiopia have been conducted (31). However, these studies excluded high-risk places such as holy water sites and other congregate settings, which can be potential hotspots for TB transmission. The current study was guided or aimed to answer the question, “What is the prevalence of TB, drug-resistant rate, the genotype distribution of resistant MTB isolates, and the frequency of gene mutations responsible for resistance for first and second-line anti-TB drugs among PTB suspects attending holy water site in the Amhara region?” As a baseline information, an earlier study on the prevalence of smear-positive TB among holy water site attendees was conducted in the Amhara region and found that the prevalence of TB was 7.4-fold higher in those study cohorts than in the general population in Ethiopia (32). However, this study was geographically limited and used acid-fast bacilli (AFB) smear microscopy to detect TB suspects, which has low sensitivity, thus the findings may not reflect the true TB burden among holy water site attendees in the region (32).
Spiritual holy water sites in Ethiopia are congregate settings where many people, particularly Orthodox Christian followers, come to receive holy water treatment blessed by priests for several kinds of illnesses, including TB and other respiratory diseases (32). Although studies showed that TB patients seek treatment from holy water sites, the prevalence of TB among individuals attending those settings in Ethiopia has not been thoroughly investigated. There is also scarce information on the genetic diversity of drug-resistant MTB genotypes and the types and frequency of genetic changes conferring resistance to commonly prescribed anti-TB drugs among TB isolates from PTB-symptomatic attendees of holy water sites in Ethiopia. Therefore, this study aimed to evaluate drug susceptibility profiles of TB isolates and characterize the types and frequency of mutations responsible for resistance to INH, RIF, FLQs, and SLIDs in MTB isolates, and to Spoligotype those isolates from culture-positive PTB-symptomatic attendees of holy water sites in the Amhara region, Ethiopia.
Methods
Setting and study period
A cross-sectional study was performed from June 2019 to March 2020 at nine purposively selected holy water sites found across nine administrative zones in the Amhara region. The Amhara region is found in Northwestern Ethiopia, which comprises thirteen zones and three administrative towns. Bahir Dar is the regional capital, which is 567 km far from the national capital, Addis Ababa. One holy water site was chosen from each administrative zone based on its consistent popularity for spiritual holy water treatment, its ability to accommodate a large number of attendees, and where many people visit and reside for an extended time (32). According to the criteria mentioned above, the selected holy water site from each study zone is deemed to be representative (Supplementary Table S1).
Study participants
For this study, the source population was all individuals who attended holy water sites during the data collection time. However, attendees who had PTB suggestive symptoms (33), particularly cough ≥2 weeks and having productive cough, and other symptoms such as night sweating, fatigue, fever, loss of appetite, shortness of breath, chest pain, unexplained weight loss, had contact history with active TB patients, and previous TB disease were recruited and included as the sample population. During the course of the study, at nine selected holy water sites, a total of 10,313 attendees (≥18 years of age) were screened, and 560 individuals with symptoms of PTB participated. Supplementary Table S1 illustrates the study settings, total number of attendees screened for PTB-suggestive symptoms, number of PTB suspected cases, and bacteriologically confirmed TB cases.
Eligibility criteria
Attendees (≥18 years of age) who had coughs ≥2 weeks plus the above-mentioned PTB suggestive symptoms were included, whereas individuals who were under 18 years and those who were seriously ill to provided sputum sample and necessary information were excluded. Furthermore, attendees who were on TB medications during the sample collection time were excluded.
Sociodemographic data collection
After obtaining participants’ informed consent to be included in the study, their sociodemographic data including sex, age, residence, marital status, educational level, family size per household, occupation, and study area were recorded using an interviewer-administered questionnaire. We followed the national TB screening guideline during the screening of PTB suggestive symptoms (33).
Sputum specimen collection and Mycobacterium tuberculosis complex isolates
Using a leak-proof, sterile screw-capped falcon tube (50 mL capacity), the sputum samples were received from PTB-symptomatic attendees. We used an ice-pack carrier to transport the sputum specimens to the regional research laboratory center, “Amhara Public Health Institute, Bahir-Dar, Ethiopia.” We followed the standard procedures during the mycobacterial culturing steps. In brief, the “N-acetyl-L-cysteine (NALC-NaOH)” solution was utilized to decontaminate the sputum, following the neutralization process using a phosphate buffer solution (pH 6.8). The mixture was centrifuged to prepare the inoculums for culture. Then after, the inoculum/sediment from the bottom of the tube was taken and inoculated into Löwenstein-Jensen (LJ) slant culture tubes, and incubated at a temperature of 37°C for at least 8 weeks. The growth of MTB colonies on LJ culture media was inspected weekly for up to 8 weeks, and the growth was confirmed by Ziehl-Neelsen (ZN) smear staining” (34). “Capilia TB-Neo (Tauns laboratories, Japan)” was utilized to differentiate MTB complex species (35). In each step of the test, a known H37Rv strain and sterile molecular-grade water were utilized as positive and negative controls, respectively.
Specimen preparation from MTB colonies grown on LJ culture media
From MTB colonies grown from LJ culture medium, suspensions were prepared and transferred into a transport media, 1.5 mL of “PrimeStore Molecular Transport Medium (PS-MTM; Longhorn Vaccine & Diagnostics, San Antonio, TX, USA).” The MTB suspension preparations from a positive LJ culture medium depend on the culture state (36). In summary, for intact slopes, using a sterile inoculation loop, MTB colonies were carefully scraped off and washed down (suspended) in 1 mL sterile molecular grading water in the original slant culture bottle. After pipetting off the prepared suspension, it was transferred into a 1.5 mL Eppendorf tube. The Eppendorf tubes holding suspensions were centrifuged at 13,000 g for 5 min, the supernatant was discarded (36), and the deposit was then transferred into the PS-MTM. The PrimeStore tubes holding the MTB suspension were shipped non-refrigerated to South Africa by air, where DNA extraction, genotyping drug susceptibility testing (gDST), and Spoligotyping procedures were performed.
Mycobacterium tuberculosis genomic DNA extraction
From all LJ culture-positive isolates, the “MTB genomic DNA was extracted using the PrimeXtract™ kit (Longhorn Vaccines and Diagnostics, San Antonio, TX, USA)” following the manufacturer’s protocol. In summary, MTB inoculum (200 μL), 100% ethanol (200 μL), and lysis buffer (200 μL) were transferred into a 1.5 mL microcentrifuge tube, then thorough vortexing and subsequent centrifugation was performed. Using a micro-extraction column, the entire supernatant was transferred, following centrifugation at 13,000 rpm for 1 min, and then the follow-through material was discarded. After adding wash buffer 1 (500 μL) to the extraction column, it was then centrifuged at 13,000 rpm for 1 min, followed by further addition of wash buffer 2 (500 μL) to the extraction columns, and subsequently centrifuged as described above, and the follow-through material was discarded. The extracted total MTB genomic DNA was eluted by centrifugation at 13,000 rpm for 1 min using preheated (60–70°C) 50 μL elution solution. Then, the extracted total DNA was stored at −20°C fridge for future use. Genomic MTB DNA concentration and quality were measured using a spectrophotometer at optimal densities of 269 and 280 nm.
Drug susceptibility testing
The genotype DST (gDST), MTBDRplus VER2.0 (“Hain Lifescience, Germany”) was performed on 122 culture-positive MTB isolates (16). MTB isolates that were found RIF and/or INH-resistant were subjected to MTBDRsl VER2.0 to detect FLQs and second-line injectable anti-mycobacterial drug resistance (17). The entire techniques that included the preparation of the master mix, polymerase chain reaction (PCR) amplification, and hybridization were done following the manufacturer’s protocol (“Hain Lifescience, Germany”) (16, 17).
Interpretation of results
The MTBDRplus assay can detect the presence and absence of wild-type (WT) and mutant (MUT) DNA sequences (bands) within particular regions of the three resistance-conferring genes: the rpoB gene, which encodes the “β-subunit of the RNA polymerase,” was used to determine RIF-resistance; whereas the katG gene, which encodes the “catalase-peroxidase,” was used to detect high-level INH resistance; and the inhA promoter region (encodes enoyl ACP NADH reductase), was used to identify low-level INH resistance. Rifampicin resistance was demonstrated by the absence of bands in the rpoB probes, whereas resistance to INH was demonstrated by the absence of katG and inhA bands. On the other hand, when the mutant probes’ bands are as strong as or stronger than the existing amplification control (AC) bands, they are considered resistant. In most cases, the absence of the WT band is associated with the presence of a MUT band, indicating resistance. Rarely, the absence of WT band (s) without the presence of a corresponding MUT band could be observed, which was attributed to “unknown” genetic changes in the probe region or mutations that exist outside of the drug-resistance-determining regions, which the assay cannot detect. The coexistence of WT and MUT bands within a single stripe may indicate the existence of hetero-resistance or mixed TB strain infection. In general, resistance was recorded where one or more WT bands were absent or where one or more WTs were missing with a corresponding mutation.
The MTBDRsl VER2.0 test determined the presence and absence of WT and MUT DNA sequences within a particular region of four genes: gyrA, which encodes the “A-subunit protein of DNA gyrase; gyrB (it encodes β -subunit protein of DNA gyrase)” were utilized to determine resistance to FLQs; and the rrs (encodes 16S rRNA) was utilized for the detection of cross-resistance to Kanamycin (KAM) and Amikacin (AMK), and Capreomycin (CPM) and Viomycin (VIO), whereas the eis, which encodes (aminoglycoside acetyltransferase), was used to identify low-level resistance to KAM. In short, resistance was recorded where one or more WT probes were absent or where one or more WTs were missing with a corresponding mutation type. However, the probability of strains developing mutations outside of the test regions cannot be ruled out, since the assay cannot detect genetic changes that exist outside the regions of interest.
Quality control
In each of the LPAs tests (MTBDRplus VER2.0 and MTBDRsl VER2.0), sterile water was utilized as the negative control, while the universal reference H37Rv strain, which is sensitive to all anti-TB medicines, was utilized as a positive control.
Spoligotyping
All 20 drug-resistant MTB isolates were subjected to Spoligotyping “following the manufacturer’s protocol” (37), and the Spoligotyping kit supplier’s instructions (“Ocimum Biosolutions Company, IJsselstein, The Netherlands”). In brief, the MTB isolate’s “direct repeat (DR) region was amplified by a thermal cycler PCR machine” (“VWR International, UK”) utilizing the oligonucleotide primers derived from the MTB direct repeat region. A total volume of 25 μL constituting the PCR amplification reaction mixture of 12.5 μL of “HotStarTaq Master Mix (Qiagen, UK),” 2 μL of forward primer (DRa), and 2 μL of reverse primer (DRb), 5 μL of extracted DNA, and 3.5 μL sterile molecular grading water. Then, the mixture was subjected to heat at 96°C for 3 min, and then subjected to 30 cycles for 1 min at 96°C, 1 min at 55°C, 30 s at 72°C, and 5 min at 72°C for 1 cycle.
The amplified PCR product underwent hybridization with a series of 43 immobilized oligonucleotides that are covalently bound to a membrane (“Animal and Plant Health Agency, Great Britain”). Each of these oligonucleotides corresponds to a different spacer sequence of DNA located within the direct repeat (DR) locus. The membrane was washed twice for 10 min in 2XSSPE (pH 7.7)-0.5% sodium dodecyl sulfate at 60°C after hybridization, then incubated in 1:4000 diluted streptavidin-peroxidase (“HotStar, UK”) for 45–60 min at 42°. The membrane was then washed twice for 10 min in 2X SSPE-0.5% sodium dodecyl sulfate at 42°C and rinsed for 5 min in 2XSSPE at room temperature. The detection of hybridized DNA was done using the enhanced chemiluminescence method (“Amersham Biosciences, UK”) and by exposing it to X-ray film (“Hyperfilm ECL, Amersham Biosciences, UK”), following the manufacturer’s instruction. The presence or absence of spacers was visualized on X-ray film through the depiction of black and white squares, respectively. Reference strains, M. bovis BCG and MTB H37Rv used as positive controls, while sterile Qiagen water (“Qiagen Company, Germany”) was utilized as negative control.
Statistical analysis
All the laboratory test data were first documented in a prepared “Microsoft Excel spreadsheet,” and checked if errors existed during recording (cleared), properly coded, then entered into STATA 15 (“Stata Corp, College Station, TX, USA”) for analysis. Descriptive statistical analysis, frequency, and percentage were used to summarize the key findings, and the Chi-square test’s p-value was reported. Findings were presented in tables and figures. The “shared international spoligotypes (SIT) number and the corresponding lineages/sublineages were obtained using the open-source spoligotype database (SpolDB4)1” (38). MTB Strains that exhibited similarity to a pre-existing spoligotype pattern in the database were assigned an SIT number, while TB strains without SIT numbers were categorized as “new.” MTB trains with the same spoligotype patterns was called a “cluster,” while a single pattern was defined as “unique” to this study.
Research ethics
The study received ethical clearance from the Human Research Ethics Committee of the University of Pretoria, Faculty of Health Sciences, South Africa (Ref: No. 600/2018) and the National Research Ethics Review Committee, Ethiopia (Ref: No. SHE/SM/14.4/708/2019). Furthermore, a written official permission letter was obtained from the Ethiopian Orthodox Tewaheido Church (Ref: No.2478/6275/2011). A verbal and/or written consent declaration with full details about the study was given to participants and signed. Participants were assured that their data would remain confidential by using the non-personal identifier. Attendees who tested TB positive were linked with nearby healthcare facilities to commence the appropriate treatment and further management.
Results
Characteristics of study subjects
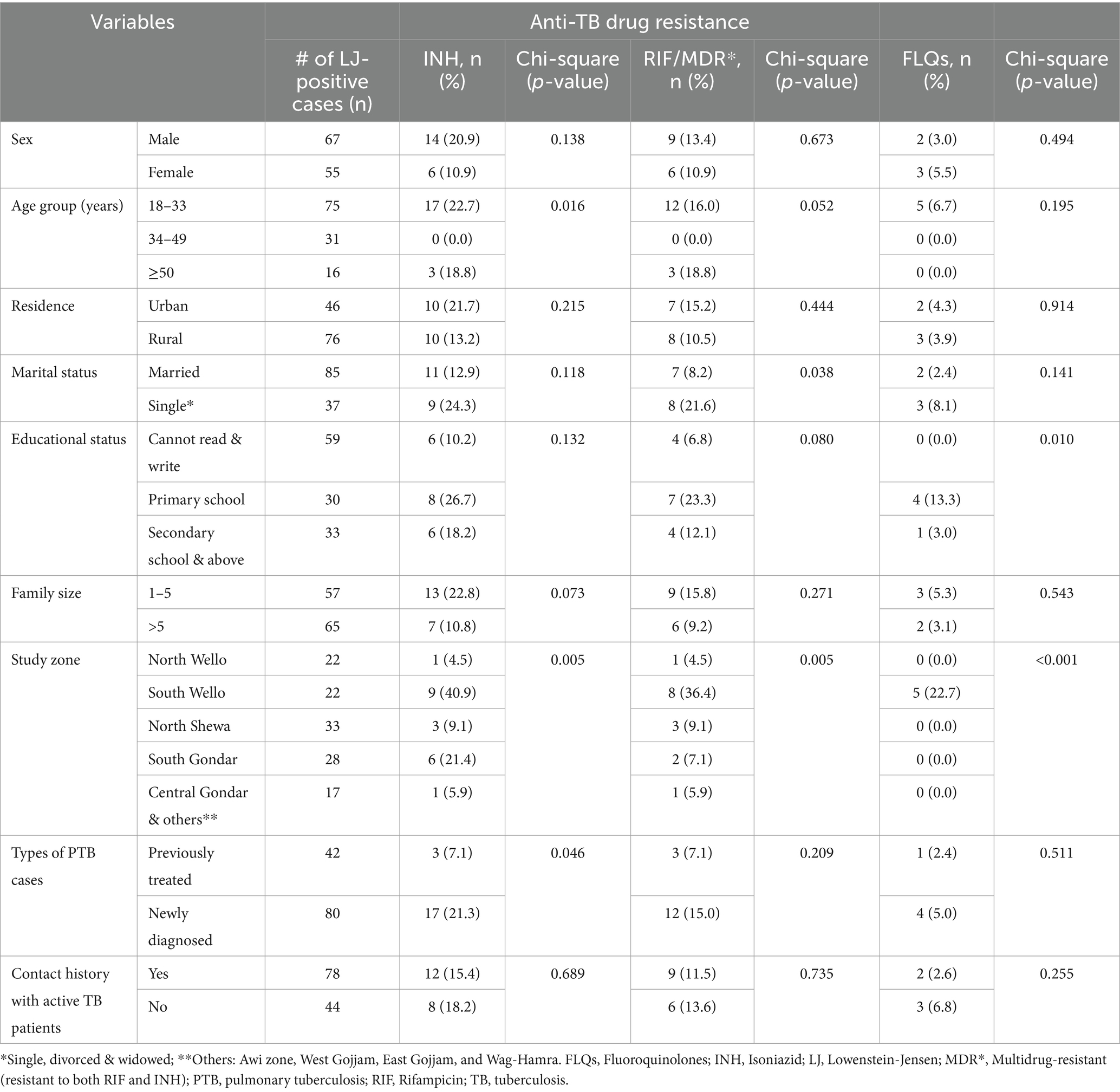
Five hundred and sixty PTB suspected cases participated in the study. Of these, 122 participants were bacteriologically confirmed (LJ culture-positive) (Supplementary Tables S1, S2). The median age of PTB suspected participants was 35 years, and male individuals constituted the majority (n = 308, 55.0%). One hundred twelve (20.0%), and 191 (31.1%) were previously treated and had a contact history with active TB patients, respectively (Supplementary Table S2). Of 122 bacteriologically confirmed TB cases, the majority (n = 33, 34.0%) and (n = 28, 26.7%) of the attendees were from the North Shewa and South Gondar zones, respectively. Over one-third (37.5 and 40.8%) of participants were previously treated and had contact history with active TB patients, respectively (Supplementary Tables S2, S3).
Drug susceptibility testing
In this study, we have done gDST using “MTBDRplus VER2.0” on 122TB isolates. Of which 20 (16.4%%), and 15 (12.3%) were resistant to INH and RIF, respectively. Of those resistant to INH, 15 (75%) were also resistant to RIF and 5 (25%) were INH-monoresistant, while 1 (5%) isolate was INH-hetero-resistant. Three-fourths of drug-resistant isolates (n = 15, 75%) were MDR-TB (both RIF and INH resistant). The majority (n = 12, 16.0%) of them were identified from participants aged between 18 and 33 years, and male accounts (n = 9, 13.4%). Furthermore, 8 (36.4%) of those isolates identified as MDR-TB were detected from the South Wollo zone study site (Table 1).
The MTBDRsl VER2.0 was done on 20 any INH-resistant (mono-hetero-multi/−) isolates and 15 RIF-resistant (mono-hetero-multi/−) MTB isolates. All of the isolates yielded interpretable results and were included in the study. Further resistance to FLQs was detected only in MDR-TB isolates. Accordingly, 25% (5/20) were identified as FLQs-resistant. Interestingly, all five (6.7%) FLQ-resistant isolates were detected from attendees aged between 18 and 33 years and from the South Wollo zone study area (Table 1).
Frequency of gene mutations conferring resistance to INH, RIF, and FLQs
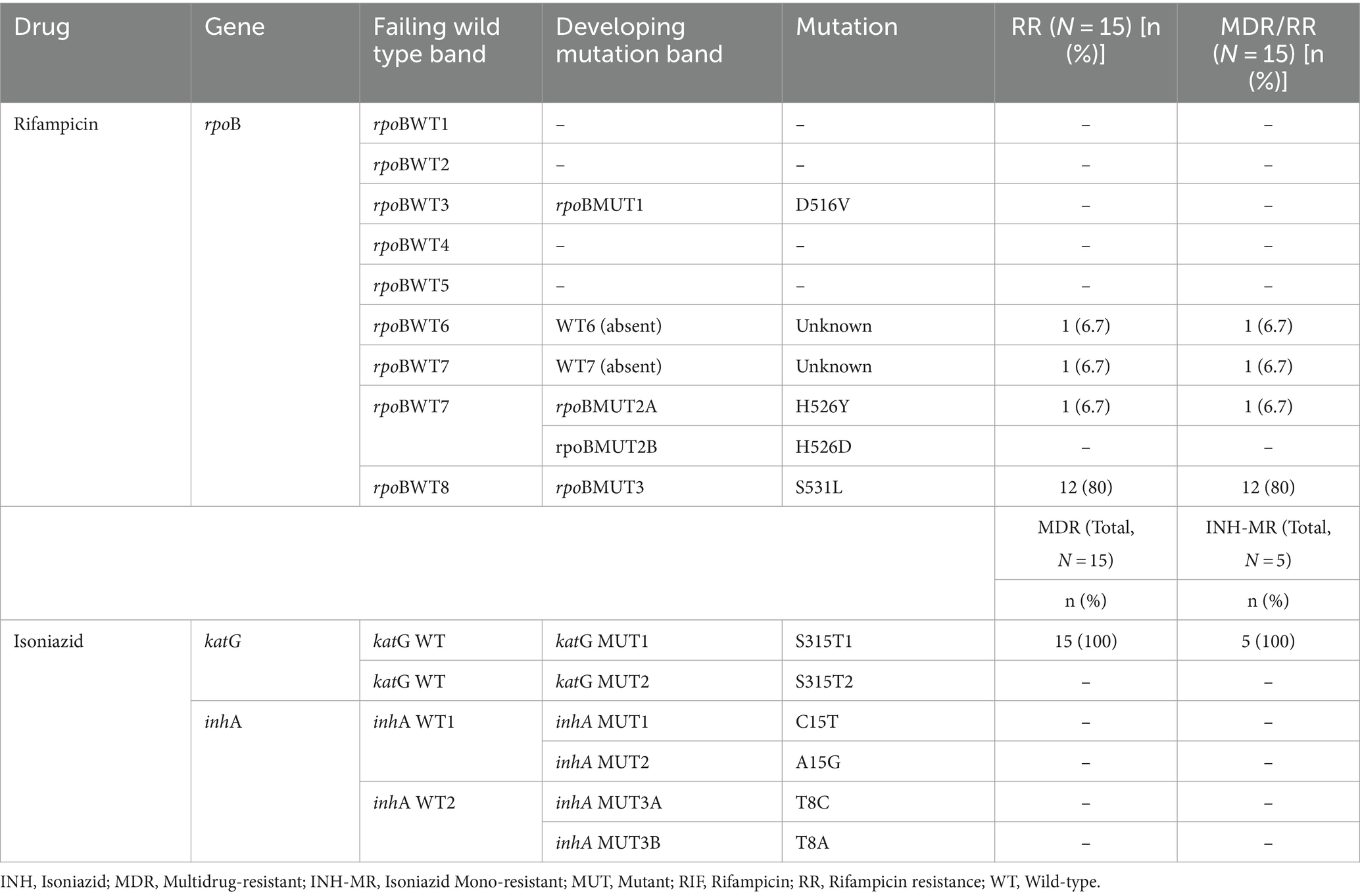
Mutations in the RIF-resistance determining region (rpoB)
In RIF-resistant isolates, the gene mutations in the rpoB were most frequent at codon Ser531Lue (80%); the next mutation was seen in His526Tyr (6.7%). The remaining two RIF-resistant isolates (13.3%) had shown a mutation or missing wild-type band (probe absent), but no corresponding MUT band, and were classified as “unknown” mutations. Those were rpoBWT6 and rpoBWT7 (6.7% each). This could be due to the gene mutations occurring outside the analyzed codon regions which LPAs cannot detect (Table 2).
Gene mutations in the katG and inhA
Amongst INH-resistant strains (n = 20), mutations in the katG (indicating high-level resistance) were most frequent at codon Ser315Thr1 (95%), while no inhA gene mutation was observed. From 20 isolates with INH resistance, 5 (25.0%) were detected as INH-hetero-resistance and had mutations at codon Ser315Thr1 (100%) (Table 2).
Of 20 any drug-resistant isolates, 15 (75%) were MDR-TB strains (both RIF and INH- resistant). Of the total 15 MDR-TB strains, mutation in the rpoB and katG genes were most frequent at Ser531Leu (n = 12, 80%) and Ser315Thr1 (n = 15, 100%) (Table 2). The remaining 3 MDR-TB isolates had shown a gene mutation at rpoBWT6 (probe absent) and Ser315Thr1, rpoBWT7 (probe absent) and Ser315Thr1, and His526Tyr and Ser315Thr1 (6.7% each) (Table 2).
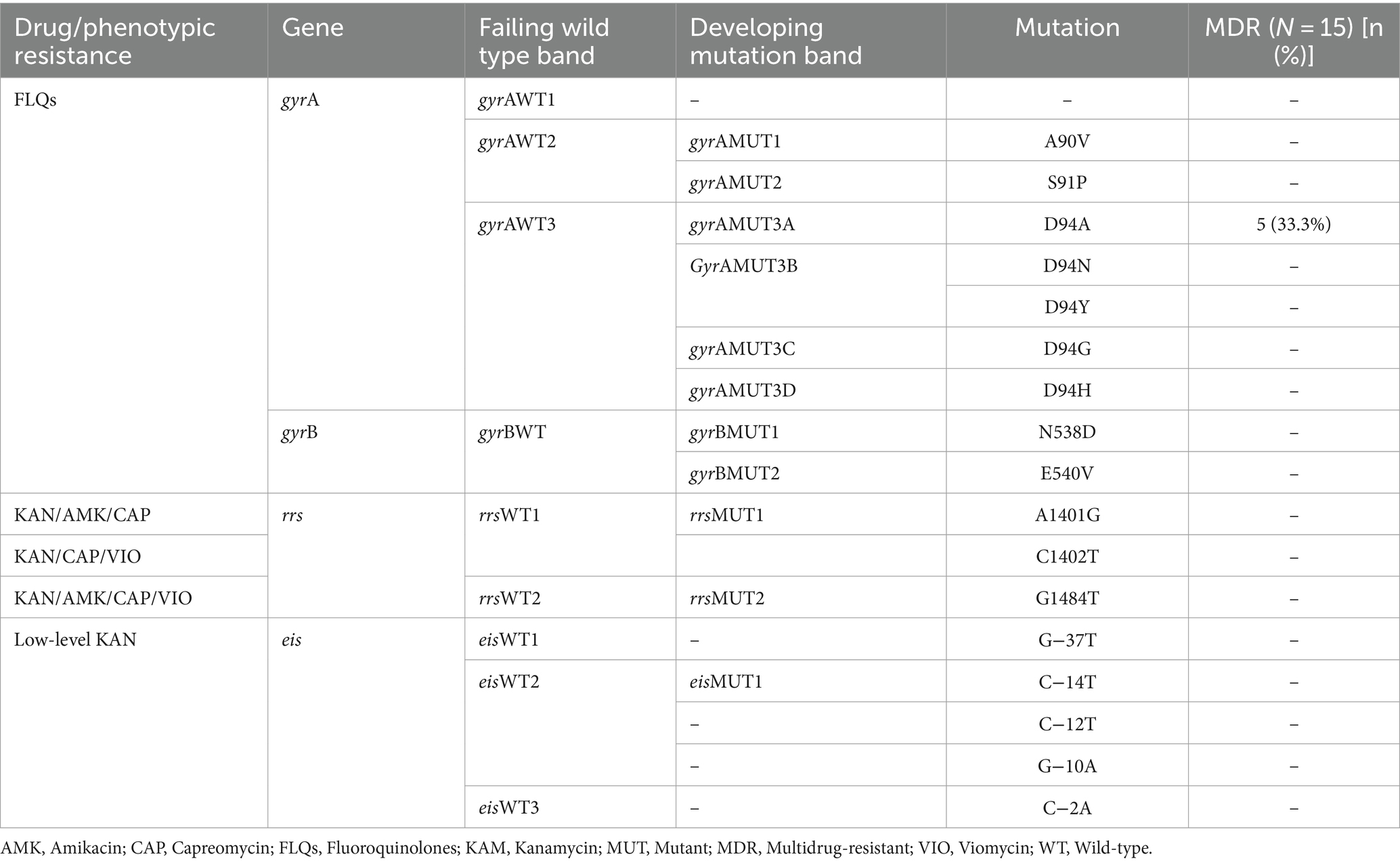
Gene mutations in the FLQ-resistance-determining region
The MTBDRsl VER2.0 was performed on 20 any drug-resistant TB strains to further assess the second-line injectable drugs (SLIDs) resistance. Accordingly, five MDR-TB strains had shown FLQs resistance, which is defined as pre-XDR-TB. All 5 FLQs-resistant isolates had mutations only in the gyrA genes (at codon Asp94Ala). No mutations were detected at gyrB, rrs, and eis high-level and low-level drug-resistance conferring gene mutations to SLIDs (Table 3).
Mycobacterium tuberculosis genotypes and profiles of genetic changes conferring resistance to anti-TB drugs
To further analyze the genotype and clustering of 20 drug-resistant MTB strains, we performed a genotyping technique using the established Spoligotyping method, following the manufacturer’s protocol. All 20 drug-resistant MTB strains were successfully genotyped and have interpretable spoligo-patterns. The majority (n = 11, 55%) drug-resistant strains were found to be SIT149/T3-ETH sublineages followed by SIT21/CAS1-Kili (n = 6, 30%). The remaining isolates were CAS1-Delhi, CAS1-family, and SIT54/MANU2. Of the total 11 SIT149//T3-ETH sublineages, 4 strains were detected from the South Gondar zone study site, while 2 and 3 isolates were detected from the South Wello and North Shewa zones, respectively. Also, from six SIT21/CAS1-Kili sublineages, four isolates were identified from the South Wello zone study site, while the remaining two isolates were from the North Shewa and North Wello zone study area (Table 4).
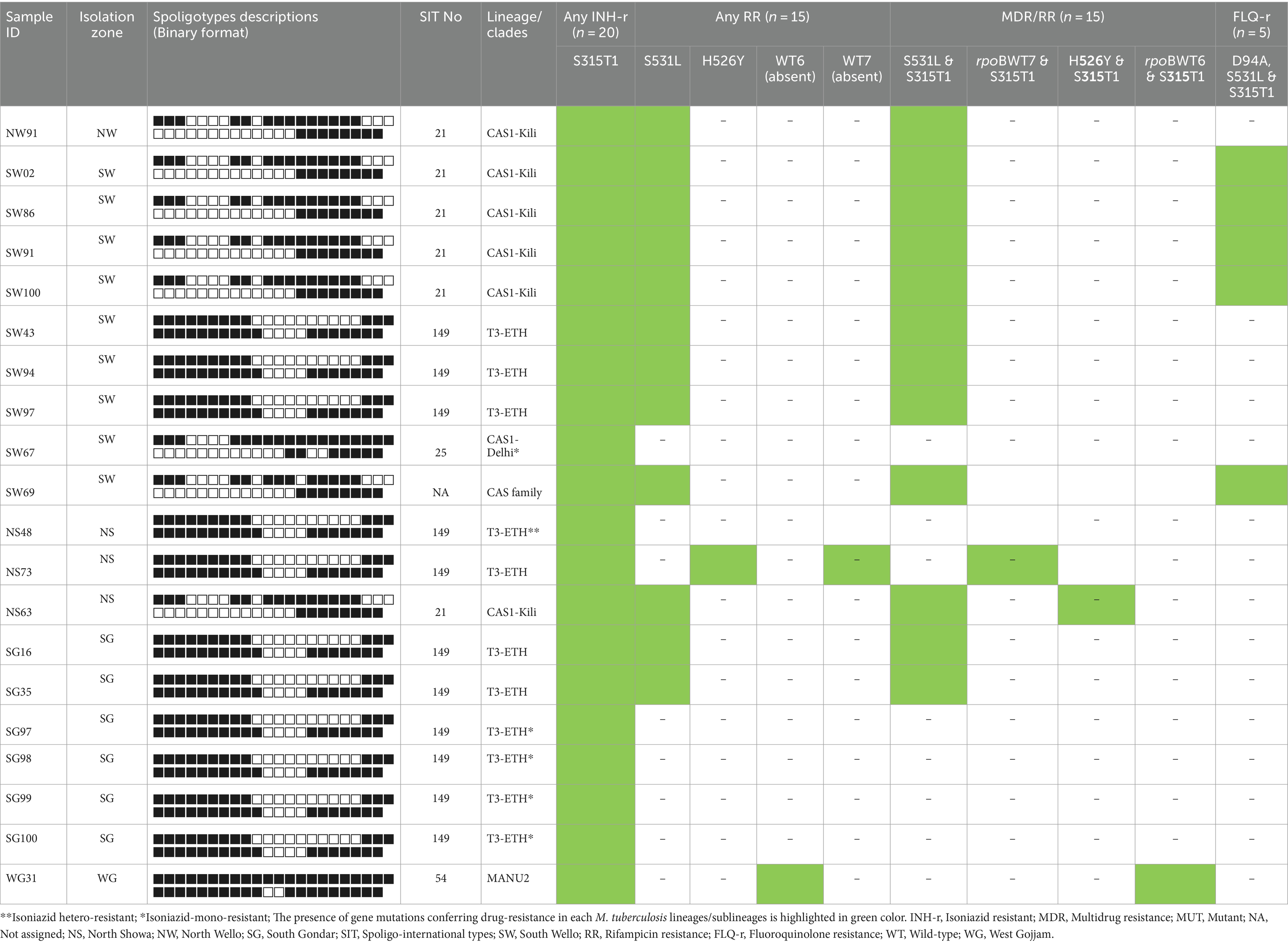
Table 4. Drug-resistant M. tuberculosis lineages and profiles of the gene mutations conferring resistance to INH, RIF, MDR/RR, and FLQs.
Furthermore, of 11 SIT149/T3-ETH sublineages, 7 strains were MDR-TB, while four were INH-mono-resistant. Amongst 6 CAS1-Kili/SIT21 and one CAS1-family sublineages, all were MDR-TB, while four of these were FLQs-resistant strains. The remaining two sublineages, SIT54/MANU2 and SIT25/CAS1-Delhi were MDR and INH-mono-resistant, respectively (Table 4).
Amongst 7 MDR SIT149 genotypes, five had genetic changes in rpoB and katG genes (at codon Ser531Leu and Ser315Thr1), while one isolate had a gene mutation at codon His526Tyr and Ser315Thr, and four isolates were INH-mono-resistant with a gene mutation in katG (at codon Ser315Thr1). All the six MDR SIT21/CAS1-Kili and CAS1-family spoligotypes had drug resistance-conferring gene mutations in rpoB and katG genes (at codon Ser531Leu and Ser315Thr1), of these, four isolates were FLQs-resistant with a drug-resistance conferring gene mutations in rpoB, katG, and gyrA genes (at codon Ser531Leu, Ser315Thr1, D94A, respectively) (Table 4).
Discussion
Early diagnosis and effective anti-mycobacterial drugs targeting the infecting MTB isolate are critical for improving patient management, increasing cure rates, and limiting further transmission of the disease (1, 3). Moreover, it is crucial to identify the key genetic changes responsible for anti-TB drug resistance and the associated drug-resistant MTB genotypes to handle patients with DR-TB effectively (2, 3, 39). Nowadays, the innovation and improvements of gDST methods capable of evaluating genes harboring antimicrobial drug resistance mutations have substantially improved the detection and management of drug-resistant TB (14, 15). In addition to XpertMTB/RIF assay, LPAs are commonly used in developing countries including Ethiopia, and come at the forefront in performing gDST and evaluating the occurrence of specific gene mutations resulting in drug resistance to some key anti-mycobacterial medicines used in the treatment of drug-susceptible and resistant TB strains (14–16, 40). Thus, the widespread gDST service is very important for tailoring TB patient treatment and hindering the transmission of the disease.
The most common gene mutations linked with RIF resistance occur in the 81 bp RIF-resistance determining region (RRDR) (codons 507–533) of the rpoB gene (95%), which encodes the “RNA polymerase beta-subunit” (2, 41). Those gene mutations alter the binding site of RIF, preventing its inhibitory action on RNA synthesis (13, 41, 42). Earlier studies have shown that specific mutations, at codons S531L, H526Y, and D516V are frequently observed in RIF-resistant MTB (13, 21, 43–45). In this study, among RIF-resistant TB isolates, the most frequent mutation (80%) associated with RIF resistance was at codon S531L, followed by His526Tyr (6.7%). The gene mutation at codon S531L was noted as the dominant mutation of the rpoB gene responsible for RIF resistance in several previous studies done in different parts of Ethiopia; which includes the Amhara region (73%) (46), Tigray region (70%) (44), Somali region (80%) (47), Addis Ababa (81.3%) (48), Southwest Ethiopia (82.4%) (49), a meta-analysis report, Ethiopia (74.2%) (21), and a recent multicenter study report, Ethiopia (59.1%) (45). In line with our result, a higher frequency of mutation at codon S531L was reported in other high TB burden countries like China (58%) (50), India (62%) (51), Pakistan (64%) (52), Sudan (64%) (53), and Iran (66%) (54). The other mutation in our study, His526Tyr was previously reported in Ethiopia, which ranged from 6.6–17.2% (21, 44, 45, 55), Sudan (12.8%) (53), India (11.1%) (51), and China (8.9%) (50). However, a similar or higher proportion to our results was noted in Sudan (12.8%) (53), India (11.1%) (51), China (8.9%) (50), South Africa, at amino acid position 526 (27.9%) (36). In this study, two RIF-resistant isolates (rpoBWT6 and rpoBWT7) had shown a mutation or missing WT band, but no corresponding MUT band, and were classified as unknown genetic changes. This proportion of isolates with unknown mutations is similar to other studies from a recent multicenter study, Ethiopia (13.6%) (45), Addis Ababa, Ethiopia (15.8%) (55), Southern Ethiopia (14.7%) (56), and elsewhere, a large, multisite diagnostic study (13%) (57). This could be due to the mutations occurring outside the analyzed codon regions (drug-resistance determining regions) which LPAs unable to evaluate/detect (58).
Isoniazid resistance is mostly due to the genetic change in the katG gene (50–95%) and the inhA promoter region (20–35%) (2, 13, 41, 51), depending on geographical distributions (59). The gene mutations in the katG gene inhibit INH activation, while the gene mutations in the inhA promoter region result in overexpression of the inhA gene, which encodes the INH target enzyme (13, 41). Prominent genetic changes associated with INH resistance include katG Ser315Thr1/2, and inhA -15C/T (44, 60). Similarly, in the current study, amongst INH-resistant isolates, mutations in the katG was most frequent at codon Ser315Thr1 (95%), while no inhA gene mutation was observed. In agreement with our findings, a recent multicenter study in Ethiopia reported a prevalence of 91.8% for katG/Ser315Thr1 mutation (45), and a meta-analysis study that examined INH conferring gene mutations noted a prevalence of 95.8% for katG315 mutation (21). The katG/Ser315Thr1 gene mutation, causing high-level INH resistance was predominant (95%) in the current study and other earlier studies in different parts of Ethiopia (21, 45–48, 55, 56, 61). “Mutations in the inhA promoter region which are associated with low-level INH resistance are usually less frequent when compared with katG mutations” (41). The inhA gene mutation is not observed in our study; as well as other studies in Ethiopia also reported no or low proportion of mutation in the inhA promotor region (21, 46, 48, 55, 61).
The indiscriminate use of FLQs in many countries including Ethiopia for various common infectious disease treatments, which might result in the development of antimicrobial resistance against these key antibiotics (62–64). In FLQ-resistant MTB isolates, mutations in the gyrA and gyrB genes, which encode the DNA gyrase enzyme, are commonly observed (20, 42, 65). These mutations alter the binding site of FLQs, reducing their inhibitory effect on DNA replication. Specific mutations, at codons, such as D94G, A90V, and S91P of gyrA (quinoline resistance-determining region, QRDR) have been frequently reported in FLQ-resistant TB (11, 45, 65). In the current study, amongst 15 MDR-TB isolates, 5 (33.3%) were FLQs-resistant, and all isolates had gyrA gene mutations at codon Asp94Ala(D/A). Supporting our finding, a recent multisite study conducted in Ethiopia reported that gyrA/Asp94Ala gene mutation (14.3%) was detected in FLQs-resistant TB isolates in Ethiopia (45). However, in contrast to our observation, studies in Ethiopia reported that gyrA/D94G gene mutations were predominant in FLQs-resistant TB isolates (44, 66, 67), while few other studies indicated that gyrA/A90V gene mutations are prevalent and responsible for FLQs-resistance in Ethiopia (45, 62). Several previous studies have indicated that gyrA/D94G gene mutation is predominant across the corners of the globe (68–72). However, further molecular study data is necessary to fully understand the spectrum of gene mutations that confer resistance to FLQs (gyrA/B gene mutations) on MTB in Ethiopia. In our study, gyrB gene mutation was not observed, which is in concordance with previous studies in Ethiopia (44, 45, 62).
The observed association between MTB genotypes and drug resistance in Ethiopia highlights the importance of understanding the genetic diversity of the pathogen in combating drug-resistant TB (63, 73, 74). The diversity of MTB genotypes circulating in the country, coupled with specific mutations conferring drug resistance, underscores the need for a comprehensive approach to TB control (73, 75). A lack of comprehensive data on the molecular epidemiology of DR-TB strains in Ethiopia further complicates the implementation of molecular diagnostics. Yet, in Ethiopia, the link between resistance to anti-TB drugs and MTB genotypes is a complex and multifactorial phenomenon (75). Thus, characterizing DR-TB genotypes and understanding the prevalence and distribution of resistance-associated gene mutations for key anti-TB drugs is essential for guiding treatment strategies and monitoring the emergence of new resistance patterns in the country (21).
In the current study, 20 drug-resistant MTB isolates were successfully Spoligotyped, in which the majority (55%) were SIT149/T3-ETH Sublineages followed by SIT21/CAS1-Kili (30%). Furthermore, from 11 SIT149/T3-ETH sublineages, 7 were MDR-TB, while four were INH-hetero-resistant. Supporting our findings, the high clustering rate and predominance of drug-resistant SIT149/T3-ETH sublineages were reported in Ethiopia (56, 63, 76, 77). In agreement with our findings, earlier studies in Ethiopia reported that SIT149/T3-ETH sublineages were predominantly linked with drug resistance, particularly MDR/XDR-TB (63, 73–75, 78). However, in contrast to our findings, a recent study in Ethiopia noted that “TUR-genotype (54%) was predominant in MDR-TB strains” (79). This could be the fact that the T3-ETH/SIT149 Sublineages is an Ethiopian-specific genotype that predominantly circulates in the highlands of the country and plays an important role in TB disease transmission (73). Thus, a comprehensive study using improved genotyping techniques such as sequencing methods with high discriminatory power is warranted to tailor the clustering of these sublineages and their association with drug resistance. However, the predominance and its association with drug-resistance, SIT149/T3-ETH sublineages could indicate that these strains are becoming more important in TB disease transmission and developing drug resistance in Ethiopia. On the other hand, high proportions of holy water site attendees infected with clustered TB strains, suggest probable recent transmission of MDR-TB in the study area. Furthermore, SIT21/CAS1-Kili sublineages were the second most prevalent in the current study, in which among 6 CAS1-Kili and one CAS-family sublineages, all strains were MDR-TB, and four of these were FLQs-resistant. In line with our observation, a few studies conducted in Ethiopia (63, 80), and elsewhere, in Zambia (81), reported that SIT21 sublineages were frequently detected in MDR/XDR-TB isolates. The emergence of drug resistance in the Ethiopian strains, CAS1-Kili/SIT21 and T3-ETH/SIT149, could potentially be attributed to local factors including delayed diagnosis, inadequate compliance, insufficient contact investigations, or other unidentified factors TB prevention and care system.
Limitations
However, this study has limitations. The number of TB isolates included in our study is relatively very small, and study population selection bias might impede generalizing the findings. Since LPAs can only detect the presence/absence of specific gene mutations responsible for drug resistance for key anti-TB drugs, it will not be enough to describe the spectrum of gene mutations in the country or study region. We used only the Spoligotyping technique to characterize the genetic diversity of drug-resistant MTB isolates, which may have resulted in low discriminatory capacity and hampered identification of transmission chains.
Conclusion
The finding of our study revealed that canonical drug resistance-conferring gene mutations at rpoB/S531L, katG/S315T1, and gyrA/D94G were the most frequent in RIF, INH, and FLQs-resistant isolates, respectively. In this study, higher clustering rates of drug-resistant MTB lineages were observed. Furthermore, two sublineages, SIT149/T3-ETH and CAS1-Kili showed a higher proportion of drug resistance, particularly MDR/pre-XDR-TB. However, to comprehend better the association of SIT149/T3-ETH and SIT21/CAS1-Kili sublineages with drug resistance in Ethiopia, improved genotyping techniques with high discriminatory power such as sequencing methods and a large number of molecular data is warranted to further elucidate such genotypes and mutations and predict drug-resistance. TB screening and surveillance for drug resistance among key populations are essential to effectively control the disease in the country.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Human Research Ethics Committee of the University of Pretoria, Faculty of Health Sciences, South Africa (Ref: No.600/2018). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MAR: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. NEM: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. PBF: Writing – review & editing, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Woldia University, Ethiopia, and University of Pretoria, South Africa (only for scholarship and laboratory fee). The funding institution had no role in the study design, data analysis and interpretation, or decision to prepare the manuscript and submit for publication.
Acknowledgments
Our heartfelt gratitude goes to University of Pretoria, South Africa, and Woldia University, Ethiopia, for their financial and material support for this study. The authors gratefully acknowledge all the study participants who voluntarily participated in this study. We express our appreciation to the Ethiopian Orthodox Tewaheido Church, Patriarchate Head Office, and zonal ecclesiastical office for their permission to conduct this study at the spiritual holy water sites and their unreserved overall support during sample collection. We highly appreciate the invaluable support of the Amhara Public Health Institute and TB laboratory staff. Last but not least, we thank the National Institute for Communicable Diseases (NICD), Centre for Tuberculosis, Johannesburg, South Africa for invaluable support during conducting Spoligotyping at their molecular laboratory center.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1356826/full#supplementary-material
Footnotes
References
1. WHO . Global tuberculosis report 2022. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO. 2022 (2022).
2. Walker, TM, Miotto, P, Köser, CU, Fowler, PW, Knaggs, J, Iqbal, Z, et al. The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe. (2022) 3:e265–73. doi: 10.1016/S2666-5247(21)00301-3
3. WHO . WHO consolidated guidelines on tuberculosis. Module 4: treatment-drug-resistant tuberculosis treatment, 2022 update. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO. 2022 (2022).
4. WHO . Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis, 27–29 October 2020. Geneva: World Health Organization (2021) CC BY-NC-SA 3.0 IGO. 2021.
5. WHO . Global tuberculosis report. World Health Organization 2023. (2023). Available at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023.
6. Kebede, AH, Alebachew, Z, Tsegaye, F, Lemma, E, Abebe, A, Agonafir, M, et al. The first population-based national tuberculosis prevalence survey in Ethiopia, 2010-2011. Int J Tuberc Lung Dis. (2014) 18:639. doi: 10.5588/ijtld.13.0417
7. Reta, MATBA, Abate, BB, Mensah, E, Maningi, NE, and Fourie, PB. Mycobacterium tuberculosis drug resistance in Ethiopia: an updated systematic review and meta-analysis. Trop Med Infect Dis. (2022) 7:300. doi: 10.3390/tropicalmed7100300
8. Sekyere, JO, Reta, MA, Maningi, NE, and Fourie, PB. Antibiotic resistance of Mycobacterium tuberculosis complex in Africa: a systematic review of current reports of molecular epidemiology, mechanisms and diagnostics. J Infect. (2019) 79:550–71. doi: 10.1016/j.jinf.2019.10.006
9. WHO . Global tuberculosis report 2020. Geneva: World Health Organization. License: CC BY-NC-SA 3.0 IGO (2020).
10. Koch, A, Cox, H, and Mizrahi, V. Drug-resistant tuberculosis: challenges and opportunities for diagnosis and treatment. Curr Opin Pharmacol. (2018) 42:7–15. doi: 10.1016/j.coph.2018.05.013
11. Dookie, N, Rambaran, S, Padayatchi, N, Mahomed, S, and Naidoo, K. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother. (2018) 73:1138–51. doi: 10.1093/jac/dkx506
12. Saravanan, M, Niguse, S, Abdulkader, M, Tsegay, E, Hailekiros, H, Gebrekidan, A, et al. Review on the emergence of drug-resistant tuberculosis (MDR & XDR-TB) and its molecular diagnosis in Ethiopia. Microb Pathog. (2018) 117:237–42. doi: 10.1016/j.micpath.2018.02.047
13. Xu, G, Liu, H, Jia, X, Wang, X, and Xu, P. Mechanisms and detection methods of Mycobacterium tuberculosis rifampicin resistance: the phenomenon of drug resistance is complex. Tuberculosis. (2021) 128:102083. doi: 10.1016/j.tube.2021.102083
14. Unitaid,. Tuberculosis: diagnosis technology landscape. 5th ed. Geneva, Switzerland: World Health Organization (2017).
15. WHO . The use of molecular line probe assays for the detection of resistance to second-line anti-tuberculosis drugs: policy guidance. Geneva, Switzerland: World Health Organization. Report No.: 9241516135 (2016).
16. HAIN-Lifescience . GenoType MTBDRplus v2, Molecular genetic assay for identification of the M. tuberculosis complex and its resistance to rifampicin and isoniazid from clinical specimens and cultivated samples. Nehren, Germany: Hain Lifescience GmbH Hardwiesenstraße. (2015).
17. HAIN Lifescience . GenoType MTBDRsl VER 2.0; Molecular genetic assay for identification of the M. tuberculosis complex and its resistance to fluoroquinolones and aminoglycosides/cyclic peptides from sputum specimens or cultivated samples. Nehren, Germany: Hain Lifescience GmbH Hardwiesenstraße. (2017).
18. Ethiopia-Ministry of Health . Guidelines for clinical and programmatic management of TB, leprosy and TB/HIV in Ethiopia, Addis Ababa, Ethiopia: Federal Democratic Republic of Ethiopia, Ministry of Healt. 5th edition. vol. 1 (2012). 149 p.
19. Ethiopia-Ministry of Health . Guidelines on programmatic management of drug-resistant tuberculosis in Ethiopia. 2nd ed. Addis Ababa, Ethiopia: Ethiopia-Ministry of Health (2014).
20. Zignol, M, Cabibbe, AM, Dean, AS, Glaziou, P, Alikhanova, N, Ama, C, et al. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis. (2018) 18:675–83. doi: 10.1016/S1473-3099(18)30073-2
21. Reta, MA, Alemnew, B, Abate, BB, and Fourie, PB. Prevalence of drug resistance-conferring mutations associated with isoniazid- and rifampicin-resistant Mycobacterium tuberculosis in Ethiopia: a systematic review and meta-analysis. J Glob Antimicrob Resist. (2021) 26:207–18. doi: 10.1016/j.jgar.2021.06.009
22. National Tuberculosis Control Program . National operation guide on TB key affected populations in Ethiopia and implementation plan 2018–2020. Addis Ababa: FMOH (2017).
23. Stop TB Partnership . Leave no one behind. Stop TB Partnership launches seven key population briefs. Geneva: WHO (2016).
24. Genet, A, and Girma, A. Magnitude, associated risk factors, and trend comparisons of identified tuberculosis types among prisons in Ethiopia: a systematic review and meta-analysis. Health Sci. Rep. (2024) 7:e1789. doi: 10.1002/hsr2.1789
25. Melese, A, and Demelash, H. The prevalence of tuberculosis among prisoners in Ethiopia: a systematic review and meta-analysis of published studies. Arch Public Health. (2017) 75:37. doi: 10.1186/s13690-017-0204-x
26. Meaza, A, Yenew, B, Amare, M, Alemu, A, Hailu, M, Gamtesa, DF, et al. Prevalence of tuberculosis and associated factors among presumptive TB refugees residing in refugee camps in Ethiopia. BMC Infect Dis. (2023) 23:498. doi: 10.1186/s12879-023-08469-5
27. Shamebo, T, Mekesha, S, Getahun, M, Gumi, B, Petros, B, and Ameni, G. Prevalence of pulmonary tuberculosis in homeless individuals in Addis Ababa City, Ethiopia. Front Public Health. (2023) 11:1128525. doi: 10.3389/fpubh.2023.1128525
28. Semunigus, T, Tessema, B, Eshetie, S, and Moges, F. Smear positive pulmonary tuberculosis and associated factors among homeless individuals in Dessie and Debre Birhan towns, Northeast Ethiopia. Ann Clin Microbiol Antimicrob. (2016) 15:50. doi: 10.1186/s12941-016-0165-x
29. Mekonnen, A, Collins, J, Aseffa, A, Ameni, G, and Petros, B. Prevalence of pulmonary tuberculosis among students in three eastern Ethiopian universities. Int J Tuberc Lung Dis. (2018) 22:1210–5. doi: 10.5588/ijtld.18.0029
30. WHO . WHO policy on TB infection control in health-care facilities, congregate settings, and households. Geneva, Switzerland: World Health Organization (2009).
31. Dememew, ZG, Jerene, D, Datiko, DG, Hiruy, N, Tadesse, A, Moile, T, et al. The yield of community-based tuberculosis and HIV among key populations in hotspot settings of Ethiopia: a cross-sectional implementation study. PLoS One. (2020) 15:e0233730. doi: 10.1371/journal.pone.0233730
32. Derseh, D, Moges, F, and Tessema, B. Smear positive pulmonary tuberculosis and associated risk factors among tuberculosis suspects attending spiritual holy water sites in Northwest Ethiopia. BMC Infect Dis. (2017) 17:100. doi: 10.1186/s12879-017-2211-5
33. Ethiopia-Ministry of Health . Guidelines for clinical and programmatic management of TB, leprosy and TB/HIV in Ethiopia. Addis Ababa: Federal Ministry of Health (2012).
34. Weyer, K, Kantor, I, Kim, S, Frieden, T, Laszlo, A, Luelmo, F, et al. Laboratory services in tuberculosis control. Part II: Microscopy. Geneva: World Health Organization (1998).
35. Shen, GH, Chen, CH, and Hung, CH. Combining the Capilia TB assay with smear morphology for the identification of Mycobacterium tuberculosis complex. Int J Tuberc Lung Dis. (2009) 13:371–6.
36. Maningi, NE, Daum, LT, Rodriguez, JD, Said, HM, Peters, RP, Sekyere, JO, et al. Multi-and extensively drug-resistant Mycobacterium tuberculosis in South Africa: a molecular analysis of historical isolates. J Clin Microbiol. (2018) 56:e01214–7. doi: 10.1128/JCM.01214-17
37. Kamerbeek, J, Schouls, L, Kolk, A, Van Agterveld, M, Van Soolingen, D, Kuijper, S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. (1997) 35:907–14. doi: 10.1128/jcm.35.4.907-914.1997
38. Brudey, K, Driscoll, R, Rigouts, L, Prodinger, M, Gori, A, and Al-Hajoj, A. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. (2006) 6:23. doi: 10.1186/1471-2180-6-23
39. WHO . The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide. (WHO/CDS/TB/2018.19). Licence: CC BY-NC-SA 3.0 IGO. Geneva: World Health Organization (2018).
40. Hain-Lifescience . GenoType MTBDRplus VER 2.0: Molecular genetic assay for identification of the M. tuberculosis complex and its resistance to rifampicin and isoniazid from clinical specimens and cultivated samples. Nehren: Hain Lifescience GmbH (2012).
41. Zhang, Y, and Yew, WW. Mechanisms of drug resistance in Mycobacterium tuberculosis: update 2015. Int J Tuberc Lung Dis. (2015) 19:1276–89. doi: 10.5588/ijtld.15.0389
42. Zhang, Y, Yew, WW, and Barer, MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother. (2012) 56:2223–30. doi: 10.1128/AAC.06288-11
43. Abebe, G, Paasch, F, Apers, L, Rigouts, L, and Colebunders, R. Tuberculosis drug resistance testing by molecular methods: opportunities and challenges in resource-limited settings. J Microbiol Methods. (2011) 84:155–60. doi: 10.1016/j.mimet.2010.11.014
44. Welekidan, LN, Skjerve, E, Dejene, TA, Gebremichael, MW, Brynildsrud, O, Tønjum, T, et al. Frequency and patterns of first-and second-line drug resistance-conferring mutations in Mycobacterium tuberculosis isolated from pulmonary tuberculosis patients in a cross-sectional study in Tigray Region, Ethiopia. J Glob Antimicrob Resistance. (2021) 24:6–13. doi: 10.1016/j.jgar.2020.11.017
45. Agonafir, M, Belay, G, Feleke, A, Maningi, N, Girmachew, F, Reta, M, et al. Profile and Frequency of Mutations Conferring Drug-Resistant Tuberculosis in the Central, Southeastern and Eastern Ethiopia. Infect Drug Resistance. (2023) 16:2953–61. doi: 10.2147/IDR.S408567
46. Tessema, B, Beer, J, Emmrich, F, Sack, U, and Rodloff, AC. Analysis of gene mutations associated with isoniazid, rifampicin, and ethambutol resistance among Mycobacterium tuberculosis isolates from Ethiopia. BMC Infect Dis. (2012) 12:37. doi: 10.1186/471-2334-12-37
47. Brhane, M, Kebede, A, and Petros, Y. Molecular detection of multidrug-resistant tuberculosis among smear-positive pulmonary tuberculosis patients in Jigjiga town, Ethiopia. Infect Drug Resist. (2017) 10:75. doi: 10.2147/IDR.S127903
48. Damena, D, Tolosa, S, Hailemariam, M, Zewude, A, Worku, A, Mekonnen, B, et al. Genetic diversity and drug susceptibility profiles of Mycobacterium tuberculosis obtained from Saint Peter’s TB Specialized Hospital, Ethiopia. PLoS One. (2019) 14:e0218545. doi: 10.1371/journal.pone.0218545
49. Tadesse, M, Abebe, G, Bekele, A, Bezabih, M, de Rijk, P, Meehan, CJ, et al. The predominance of Ethiopian-specific Mycobacterium tuberculosis families and minimal contribution of Mycobacterium bovis in tuberculous lymphadenitis patients in Southwest Ethiopia. Infect Genet Evol. (2017) 55:251–9. doi: 10.1016/j.meegid.2017.09.016
50. Jian, J, Yang, X, Yang, J, and Chen, L. Evaluation of the GenoType MTBDRplus and MTBDRsl for the detection of drug-resistant Mycobacterium tuberculosis on isolates from Beijing, China. Infect Drug Resistance. (2018) 11:1627–34. doi: 10.2147/IDR.S176609
51. Maurya, A, Singh, A, Kant, S, Umrao, J, Kumar, M, Kushwaha, R, et al. Use of GenoType® MTBDRplus assay to assess drug resistance and mutation patterns of multidrug-resistant tuberculosis isolates in northern India. Indian J Med Microbiol. (2013) 31:230–6. doi: 10.4103/0255-0857.115625
52. Farooqi, JQ, Khan, E, Alam, SMZ, Ali, A, Hasan, Z, and Hasan, R. Line probe assay for detection of rifampicin and isoniazid-resistant tuberculosis in Pakistan. J Pakistan Med Assoc. (2012) 62:767.
53. Elbir, H, and Ibrahim, NY. Frequency of mutations in the rpo B gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from Sudan. J Infect Dev Ctries. (2014) 8:796–8. doi: 10.3855/jidc.4496
54. Hamed, Z, Mohajeri, P, Farahani, A, Shamseddin, J, Zandi, M, Izadi, B, et al. The frequency of point mutations associated with resistance to isoniazid and rifampin among clinical isolates of multidrug-resistant Mycobacterium tuberculosis in the west of Iran. Gene Rep. (2021) 22:100981. doi: 10.1016/j.genrep.2020.100981
55. Abate, D, Tedla, Y, Meressa, D, and Ameni, G. Isoniazid and rifampicin resistance mutations and their effect on second-line anti-tuberculosis treatment. Int J Tuberc Lung Dis. (2014) 18:946–51. doi: 10.5588/ijtld.13.0926
56. Tadesse, M, Aragaw, D, Dimah, B, Efa, F, Abdella, K, Kebede, W, et al. Drug resistance-conferring mutations in Mycobacterium tuberculosis from pulmonary tuberculosis patients in Southwest Ethiopia. Int J Mycobacteriol. (2016) 5:185–91. doi: 10.1016/j.ijmyco.2016.02.009
57. Seifert, M, Georghiou, SB, Catanzaro, D, Rodrigues, C, Crudu, V, Victor, TC, et al. MTBDR plus and MTBDR sl assays: absence of wild-type probe hybridization and implications for detection of drug-resistant tuberculosis. J Clin Microbiol. (2016) 54:912–8. doi: 10.1128/JCM.02505-15
58. Cuella-Martin, I, Ngabonziza, JCS, Torrea, G, Meehan, CJ, Mulders, W, Ushizimpumu, B, et al. Rifampicin resistance-conferring mutations among Mycobacterium tuberculosis strains in Rwanda. Int J Mycobacteriol. (2023) 12:274–81. doi: 10.4103/ijmy.ijmy_103_23
59. Bostanabad, S, Titov, L, Bahrmand, A, and Nojoumi, S. Detection of mutation in isoniazid-resistant Mycobacterium tuberculosis isolates from tuberculosis patients in Belarus. Indian J Med Microbiol. (2008) 26:143–7. doi: 10.4103/0255-0857.40528
60. Solo, ES, Nakajima, C, Kaile, T, Bwalya, P, Mbulo, G, Fukushima, Y, et al. Mutations in rpoB and katG genes and the inhA operon in multidrug-resistant Mycobacterium tuberculosis isolates from Zambia. J Glob Antimicrob Resistance. (2020) 22:302–7. doi: 10.1016/j.jgar.2020.02.026
61. Biadglegne, F, Tessema, B, Rodloff, AC, and Sack, U. Magnitude of gene mutations conferring drug resistance in Mycobacterium tuberculosis isolates from lymph node aspirates in Ethiopia. Int J Med Sci. (2013) 10:1589–94. doi: 10.7150/ijms.6806
62. Diriba, G, Alemu, A, Tola, HH, Yenew, B, Amare, M, Eshetu, K, et al. Pre-extensively drug-resistant tuberculosis among multidrug-resistant tuberculosis patients in Ethiopia: a laboratory-based surveillance study. IJID Regions. (2022) 5:39–43. doi: 10.1016/j.ijregi.2022.08.012
63. Agonafir, M, Lemma, E, Wolde-Meskel, D, Goshu, S, Santhanam, A, Girmachew, F, et al. Phenotypic and genotypic analysis of multidrug-resistant tuberculosis in Ethiopia. Int J Tuberc Lung Dis. (2010) 14:1259–65.
64. Shibabaw, A, Gelaw, B, Gebreyes, W, Robinson, R, Wang, SH, and Tessema, B. The burden of pre-extensively and extensively drug-resistant tuberculosis among MDR-TB patients in the Amhara region, Ethiopia. PLoS One. (2020) 15:e0229040. doi: 10.1371/journal.pone.0229040
65. Bakuła, Z, Napiórkowska, A, Kamiński, M, Augustynowicz-Kopeć, E, Zwolska, Z, Bielecki, J, et al. Second-line anti-tuberculosis drug resistance and its genetic determinants in multidrug-resistant Mycobacterium tuberculosis clinical isolates. J Microbiol Immunol Infect. (2016) 49:439–44. doi: 10.1016/j.jmii.2015.04.003
66. Welekidan, LN, Yimer, SA, Skjerve, E, Dejene, TA, Homberset, H, Tønjum, T, et al. Whole genome sequencing of drug-resistant and drug susceptible Mycobacterium tuberculosis isolates from tigray region, Ethiopia. Front Microbiol. (2021) 12:743198. doi: 10.3389/fmicb.2021.743198
67. Ejo, M, Torrea, G, Diro, E, Abebe, A, Kassa, M, Girma, Y, et al. Strain diversity and gene mutations associated with presumptive multidrug-resistant Mycobacterium tuberculosis complex isolates in Northwest Ethiopia. J Glob Antimicrob Resistance. (2023) 32:167–75. doi: 10.1016/j.jgar.2022.11.012
68. Ajbani, K, Nikam, C, Kazi, M, Gray, C, Boehme, C, Balan, K, et al. Evaluation of genotype MTBDRsl assay to detect drug resistance associated with fluoroquinolones, aminoglycosides, and ethambutol on clinical sediments. PLoS One. (2012) 7:e49433. doi: 10.1371/journal.pone.0049433
69. Jnawali, HN, Hwang, SC, Park, YK, Kim, H, Lee, YS, Chung, GT, et al. Characterization of mutations in multi-and extensive drug resistance among strains of Mycobacterium tuberculosis clinical isolates in the Republic of Korea. Diagn Microbiol Infect Dis. (2013) 76:187–96. doi: 10.1016/j.diagmicrobio.2013.02.035
70. Kabir, S, Tahir, Z, Mukhtar, N, Sohail, M, Saqalein, M, and Rehman, A. Fluoroquinolone resistance and mutational profile of gyrA in pulmonary MDR tuberculosis patients. BMC Pulm Med. (2020) 20:1–6. doi: 10.1186/s12890-020-1172-4
71. Singh, PK, Singh, U, and Jain, A. Emergence of specific gyrA mutations associated high-level fluoroquinolone-resistant Mycobacterium tuberculosis among multidrug-resistant tuberculosis cases in North India. Microb Drug Resist. (2021) 27:647–51. doi: 10.1089/mdr.2020.0240
72. Avalos, E, Catanzaro, D, Catanzaro, A, Ganiats, T, Brodine, S, Alcaraz, J, et al. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolate: a systematic review. PLoS One. (2015) 10:e0120470. doi: 10.1371/journal.pone.0120470
73. Mekonnen, D, Munshea, A, Nibret, E, Adnew, B, Getachew, H, Kebede, A, et al. Mycobacterium tuberculosis sub-lineage 4.2. 2/SIT149 as dominant drug-resistant clade in Northwest Ethiopia 2020–2022: in-silico whole-genome sequence analysis. Infect Drug Resistance. (2023) 16:6859–70. doi: 10.2147/IDR.S429001
74. Worku, G, Gumi Donde, B, Abdela, MG, Mohammedbirhan, B, Diriba, G, Seid, G, et al. Drug Sensitivity of Clinical Isolates of Mycobacterium tuberculosis and Its Association with Bacterial Genotype in the Somali Region, Eastern Ethiopia. Front Public Health. (2022) 10:942618. doi: 10.3389/fpubh.2022.942618
75. Diriba, G, Kebede, A, Tola, HH, Alemu, A, Yenew, B, Moga, S, et al. Mycobacterial lineages associated with drug resistance in patients with extrapulmonary tuberculosis in Addis Ababa, Ethiopia. Tuberculosis Res Treatment. (2021) 2021:1–7. doi: 10.1155/2021/5239529
76. Zewdie, O, Mihret, A, Abebe, T, Kebede, A, Desta, K, Worku, A, et al. Genotyping and molecular detection of multidrug-resistant Mycobacterium tuberculosis among tuberculosis lymphadenitis cases in Addis Ababa, Ethiopia. New Microbes New Infect. (2018) 21:36–41. doi: 10.1016/j.nmni.2017.10.009
77. Diriba, G, Kebede, A, Tola, HH, Yenew, B, Moga, S, Addise, D, et al. Molecular characterization and drug resistance patterns of Mycobacterium tuberculosis complex in extrapulmonary tuberculosis patients in Addis Ababa, Ethiopia. PLoS One. (2020) 15:e0243493. doi: 10.1371/journal.pone.0243493
78. Bekele, S, Derese, Y, Hailu, E, Mihret, A, Dagne, K, Yamuah, L, et al. Line-probe assay and molecular typing reveal a potential drug-resistant clone of Mycobacterium tuberculosis in Ethiopia. Trop Dis Travel Med Vacc. (2018) 4:15. doi: 10.1186/s40794-018-0075-3
79. Shibabaw, A, Gelaw, B, Ghanem, M, Legall, N, Schooley, AM, Soehnlen, MK, et al. Molecular epidemiology and transmission dynamics of multi-drug resistant tuberculosis strains using whole genome sequencing in the Amhara region, Ethiopia. BMC Genomics. (2023) 24:400. doi: 10.1186/s12864-023-09502-2
80. Diriba, B, Berkessa, T, Mamo, G, Tedla, Y, and Ameni, G. Spoligotyping of multidrug-resistant Mycobacterium tuberculosis isolates in Ethiopia. Int J Tuberc Lung Dis. (2013) 17:246–50. doi: 10.5588/ijtld.12.0195
Keywords: drug-resistant TB, rpoB, katG, gyrA, mutations, spoligotypes, pulmonary tuberculosis, spiritual holy water site attendees
Citation: Reta MA, Maningi NE and Fourie PB (2024) Patterns and profiles of drug resistance-conferring mutations in Mycobacterium tuberculosis genotypes isolated from tuberculosis-suspected attendees of spiritual holy water sites in Northwest Ethiopia. Front. Public Health. 12:1356826. doi: 10.3389/fpubh.2024.1356826
Edited by:
Pacifique Ndishimye, Dalhousie University, CanadaReviewed by:
Michael Jeroen Adjabeng, WHO Country Office, GhanaJames Millard, University of Liverpool, United Kingdom
Copyright © 2024 Reta, Maningi and Fourie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melese Abate Reta, melese1985@gmail.com
 Melese Abate Reta
Melese Abate Reta