
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry, 23 September 2020
Sec. Behavioral and Psychiatric Genetics
Volume 11 - 2020 | https://doi.org/10.3389/fpsyt.2020.559210
This article is part of the Research TopicGenetic Mechanisms of Biomarkers in Schizophrenia, Bipolar Disorder and DepressionView all 18 articles
 Wenqiang Li1,2†
Wenqiang Li1,2† Xi Su1,2†
Xi Su1,2† Tengfei Chen1,2
Tengfei Chen1,2 Zhen Li1,2
Zhen Li1,2 Yongfeng Yang1,2
Yongfeng Yang1,2 Luwen Zhang1,2
Luwen Zhang1,2 Qing Liu1,2
Qing Liu1,2 Minglong Shao1,2
Minglong Shao1,2 Yan Zhang1,2
Yan Zhang1,2 Minli Ding1
Minli Ding1 Yanli Lu1
Yanli Lu1 Hongyan Yu1
Hongyan Yu1 Xiaoduo Fan3
Xiaoduo Fan3 Meng Song1,2*
Meng Song1,2* Luxian Lv1,2*
Luxian Lv1,2*Objective: Schizophrenia (SZ) is a common and complex psychiatric disorder that has a significant genetic component. The glutamate hypothesis describes one possible pathogenesis of SZ. The solute carrier family 1 gene (SLC1A1) is one of several genes thought to play a critical role in regulating the glutamatergic system and is strongly implicated in the pathophysiology of SZ. In this study, we identify polymorphisms of the SLC1A1 gene that may confer susceptibility to SZ in the Han Chinese population.
Methods: We genotyped 36 single-nucleotide polymorphisms (SNPs) using Illumina GoldenGate assays on a BeadStation 500G Genotyping System in 528 paranoid SZ patients and 528 healthy controls. Psychopathology was rated by the Positive and Negative Symptom Scale.
Results: Significant associations were found in genotype and allele frequencies for SNPs rs10815017 (p = 0.002, 0.030, respectively) and rs2026828 (p = 0.020, 0.005, respectively) between SZ and healthy controls. There were significant associations in genotype frequency at rs6476875 (p = 0.020) and rs7024664 (p = 0.021) and allele frequency at rs3780412 (p = 0.026) and rs10974573 (p = 0.047) between SZ and healthy controls. Meanwhile, significant differences were found in genotype frequency at rs10815017 (p = 0.015), rs2026828 (p = 0.011), and rs3780411 (p = 0.040) in males, and rs7021569 in females (p = 0.020) between cases and controls when subdivided by gender. Also, significant differences were found in allele frequency at rs2026828 (p = 0.003), and rs7021569 (p = 0.045) in males, and rs10974619 in females (p = 0.044). However, those associations disappeared after Bonferroni’s correction (p’s > 0.05). Significant associations were found in the frequencies of four haplotypes (AA, CA, AGA, and GG) between SZ and healthy controls (χ2 = 3.974, 7.433, 4.699, 4.526, p = 0.046, 0.006, 0.030, 0.033, respectively). There were significant associations between rs7032326 genotypes and PANSS total, positive symptoms, negative symptoms, and general psychopathology in SZ (p = 0.002, 0.011, 0.028, 0.008, respectively).
Conclusion: The present study provides further evidence that SLC1A1 may be not a susceptibility gene for SZ. However, the genetic variations of SLC1A1 may affect psychopathology symptoms.
Schizophrenia (SZ) is a complex disease with multiple susceptibility genes (1). The pathogenesis of SZ is unknown, and the glutamate hypothesis is one possible suggestion (2). Previously, our studies have revealed susceptibility genes (e.g., SCL1A6) in the glutamate pathway (3). This suggests that research on the glutamate pathway can provide important evidence for the pathogenesis of SZ. At present, a large number of genome-wide association study (GWAS) have revealed that SZ is a complex disease involving multiple genes (4–8). However, there are no consistent results for the important genetic susceptibility genes of SZ. Therefore, identifying SZ susceptibility genes from numerous candidates is an ongoing challenge.
Glutamate is a key primary excitatory neurotransmitter that plays a critical role in synaptic plasticity, neuronal toxicity, neuronal development, and signal transduction in the brain (9), and glutamatergic dysfunction could be involved in the pathogenesis of SZ (2, 10). The glutamatergic dysfunction hypothesis, supported by previous and our recent studies that involved glutamate receptor genes, such as GRIN2A (11), GRIN2B (12), and NRG1 (13, 14), and genes related to glutamatergic transmission, e.g., SCL1A6 (3) and SLC1A3 (15). Thus, further exploration of the genes of the glutamate pathway is important for the research of susceptibility genes for SZ.
The solute carrier family 1 gene (SLC1A1), a member of the neuronal high-affinity glutamate transporter family, is located at 9p24.2 and codes for the excitatory amino acid transporter (EAAT) 3 (16), and is expressed throughout the central nervous system, especially in the forebrain (17). Previous studies have reported that SLC1A1 is associated with risk of SZ (16, 18–20). Expression changes of SLC1A1 transcripts in SZ have strongly implicated it in SZ pathophysiology (16). Moreover, a 5-generation Palauan family study revealed that an SLC1A1 mutation and co-segregation correlated with the pathophysiology of SZ (20). Recent GWAS also suggested this gene as an SZ susceptibility gene (21). Some studies have reported that SLC1A1 SNP rs2228622 (15) and rs7022369 (19) were susceptibility markers involved in the pathogenesis of SZ. Meanwhile, rs16921385 of SLC1A1 was found to be associated with treatment response to risperidone in SZ (22). Those studies provide more evidence that variation of SLC1A1 may play a critical role in SZ pathogenesis. However, the findings were inconsistent and scarce (15, 19, 23). Therefore, we further explored the association between SLC1A1 and SZ in the Chinese Han population.
SZ patients were recruited as inpatients of the Second Affiliated Hospital of Xinxiang Medical University (China) from March of 2005 to December of 2008. The diagnostic criteria of SZ were according to the Diagnostic and Statistical Manual of Mental Disorders-Fourth Edition (DSM-IV). Psychopathology symptoms were measured by Positive and Negative Symptom Scale (PANSS) (24). As in our previous studies (3, 12), family history (FH) was used to explore genetic susceptibility. Inclusion and exclusion criteria of SZ patients and healthy controls were in line with our previous studies (12, 25). Inclusion of SZ: 1) patients with paranoid SZ according to DSM-IV; 2) PANSS ≥ 60; 3) male vs. female = 1:1; 4) Age range from 18 to 55 years old; 5) Han Chinese population. Inclusion of healthy controls: 1) Han Chinese population; 2) male vs. female = 1:1; 3) Age range from 18 to 55 years old. Both SZ and healthy controls were born and lived in the north of Henan Province (China), and unrelated individuals of Chinese Han population. Individuals with other psychiatric disorders, substance dependence, organic brain disease, and severe medical complications were excluded. In the sample collection, the clinical raters had rich experience in administering psychopathological tests and ensure the inter-rater consistency of diagnoses and test results through the training of every 6 months. This study protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Xinxiang Medical University (China). All subjects were informed and signed a written informed consent form.
In this study, we used the FASTSNP online service (26) to perform functional analysis, and all 36 SNPs covering the genomic region chr9: 4477575 - 4576808. Meanwhile, those SNPs were a minor allele frequency ≥0.05 and highly ranked risk in the Chinese Beijing population at the HapMap database.
Further, explore significant SNPs affect the expression level of SLC1A1 gene in brain tissues according to public eQTL databases (BrainSeq Phase 1: http://eqtl.brainseq.org/phase1/eqtl/; BrainSeq Phase 2: http://eqtl.brainseq.org/phase2/eqtl/; Brain xQTL: http://mostafavilab.stat.ubc.ca/xQTLServe/snp_query/).
Peripheral blood samples were collected from SZ and healthy controls by using evacuated tubes containing EDTA anticoagulant. RelaxGene Blood DNA System (Tiangen Biotech, Beijing, China) was used to extract genomic DNA from white blood cells. The method of genotyping was described in our previous studies that use the Illumina GoldenGate assays on a BeadStation 500G Genotyping System (Illumina, San Diego, CA, USA) (3, 12, 25).
Statistical analyses were in line with and described in our previous studies (3, 12, 25). G*Power software was used to calculate power (http://www.gpower.hhu.de/). The Haploview V4.1 program was used to assess alleles, genotypes, and haplotype frequency (27). One-way analysis of variance (ANOVA) tests were used for association analyses between PANSS scores and different genotype (SPSS version 25.0, SPSS, Inc. Chicago, IL, USA). Statistical significance was set as p < 0.05. Bonferroni correction was used to adjust for multiple testing.
A total of 1,056 subjects, 528 SZ and 528 healthy controls, were included in this study. There were no significant differences in age or gender between cases and healthy controls (p = 0.095, 1.000, respectively) (Table 1).
The genotypes of 36 SNPs were detected in 1056 samples, with a genotyping success rate of 99.79%. There were significant associations in genotype frequency and allele frequency at rs10815017 (p = 0.002, 0.030; respectively) and rs2026828 (p = 0.020, 0.005, respectively) between cases and healthy controls. Additionally, there were significant associations in genotype frequency at rs6476875 and rs7024664 between cases and healthy controls (p = 0.020, 0.021, respectively), and significant associations in allele frequency at rs3780412 and rs10974573 between cases and healthy controls (p = 0.026, 0.047, respectively). Meanwhile, there was an association trend in genotype frequency at rs7021569 and rs4742007 between SZ and health controls (p = 0.05 for both). Those associations disappeared after Bonferroni’s correction (p’s > 0.05). There were no significant associations in genotype frequency or allele frequency at the other 28 SNPs between the two groups (Table 2).
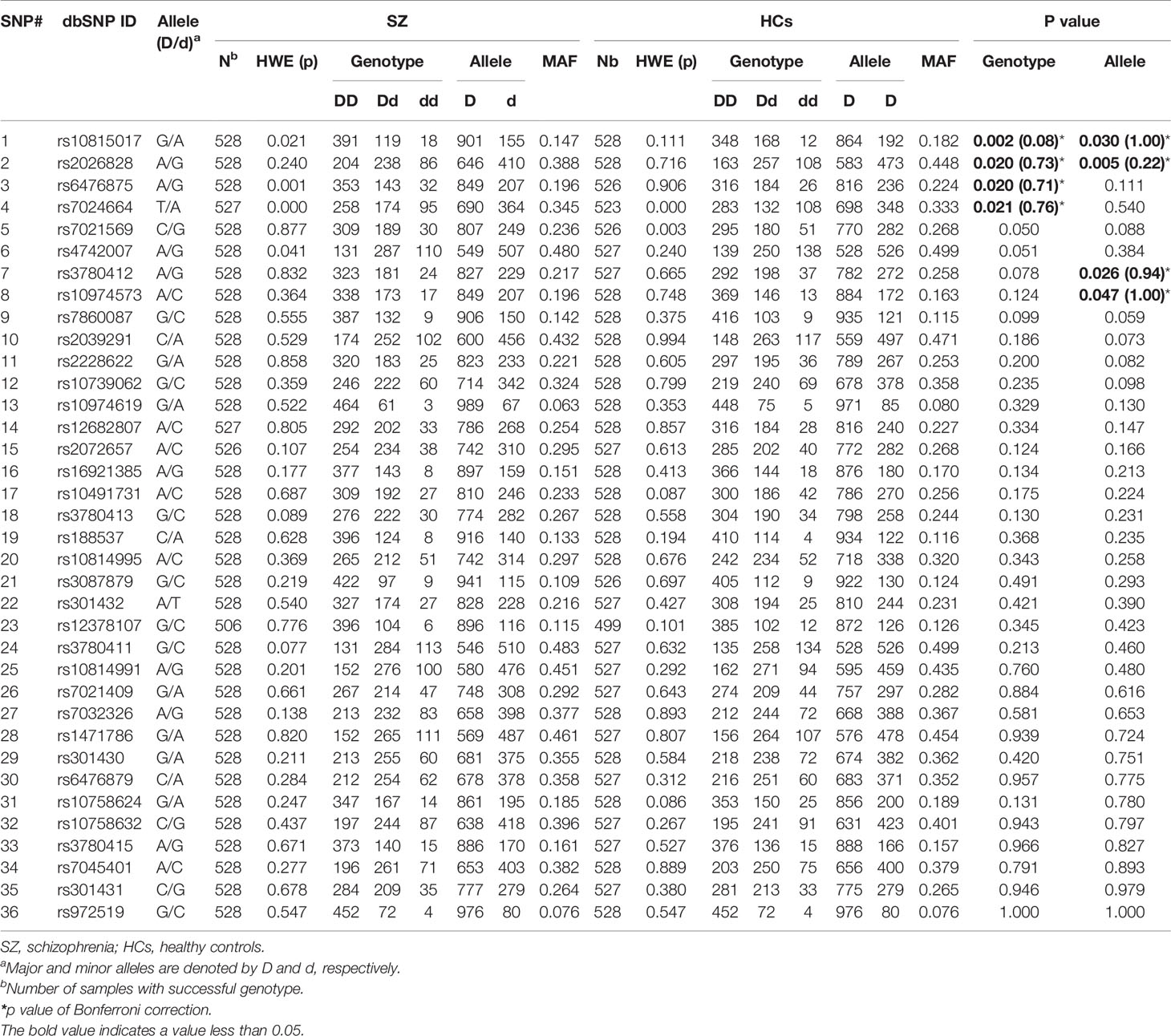
Table 2 Genotype and allele frequencies of 36 SNPs in the SLC1A1 gene in SZ patients and healthy controls.
When the subjects were divided by sex, we found significant differences in genotype frequency at rs10815017 (p = 0.015), rs2026828 (p =0.011), and rs3780411 (p = 0.040) in males, and rs7021569 in females (p = 0.020) between cases and controls. Also, significant differences were found in allele frequency at rs2026828 and rs7021569 in males (p = 0.003, 0.045, respectively), and rs10974619 in females (p = 0.044) (Supplementary Table 1).
We also observed significant differences in genotype frequency at rs10814991 (p = 0.015) and rs1471786 (p = 0.046) between patients with and without family histories (FH) of SZ. A difference trend was found in genotype frequency at rs3780411 between FH (+) and FH (−) in SZ but was not significant (Supplementary Table 2).
As shown in Figure 1, 9 LD blocks and 32 haplotypes were formed from 36 SNPs. There were significant associations in the frequencies of four haplotypes (AA, CA, AGA, and GG) between SZ and healthy control (χ2 = 3.974, 7.433, 4.699, 4.526, p = 0.046, 0.006, 0.030, 0.033, respectively) (Table 3).
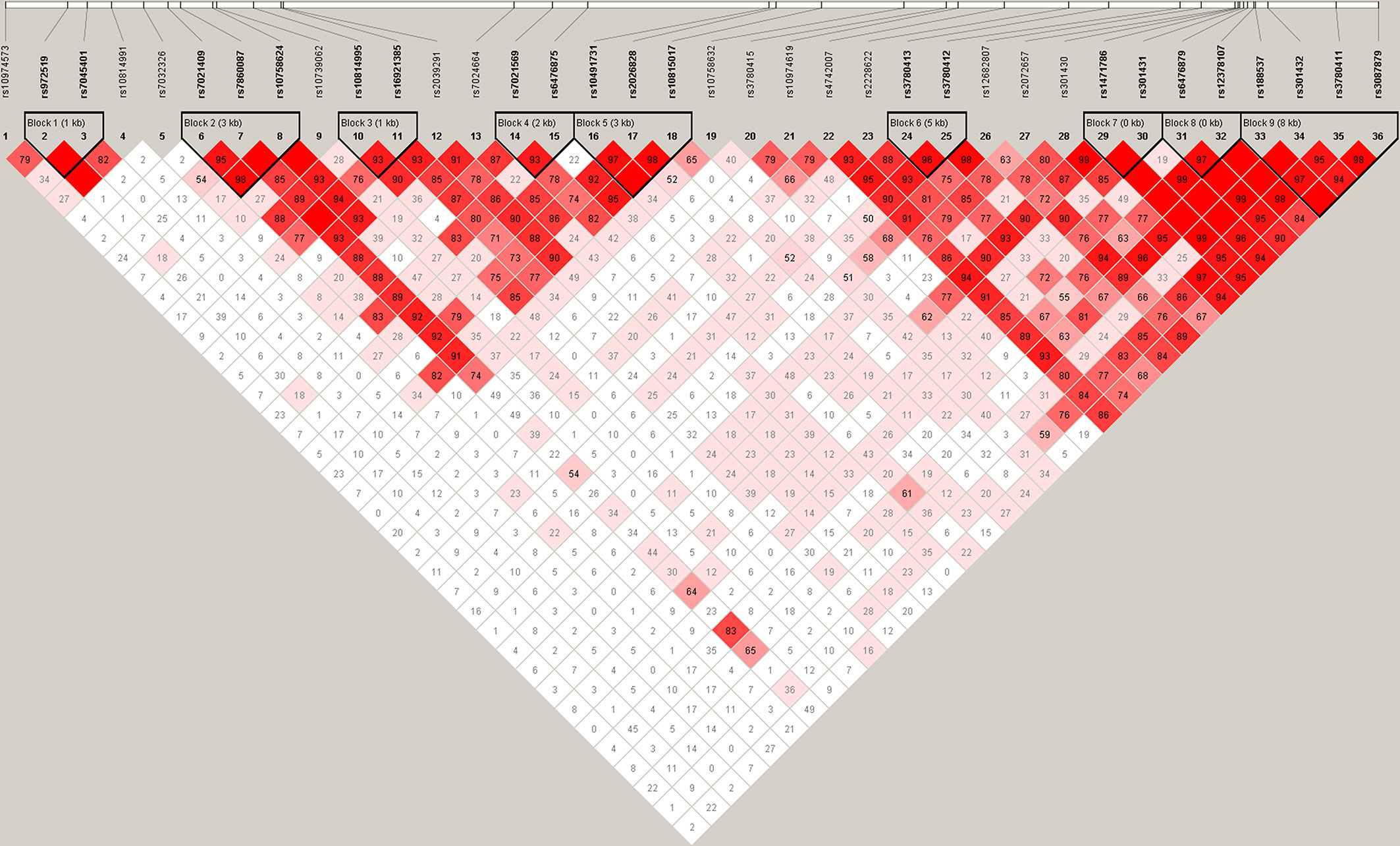
Figure 1 Haplotype block structure of the SLC1A1 gene in both SZ patients and HCs. The index association SNP is represented by a diamond. The colors of the remaining SNPs (circles) indicate LD with the index SNP based on pairwise r2 values from our data.
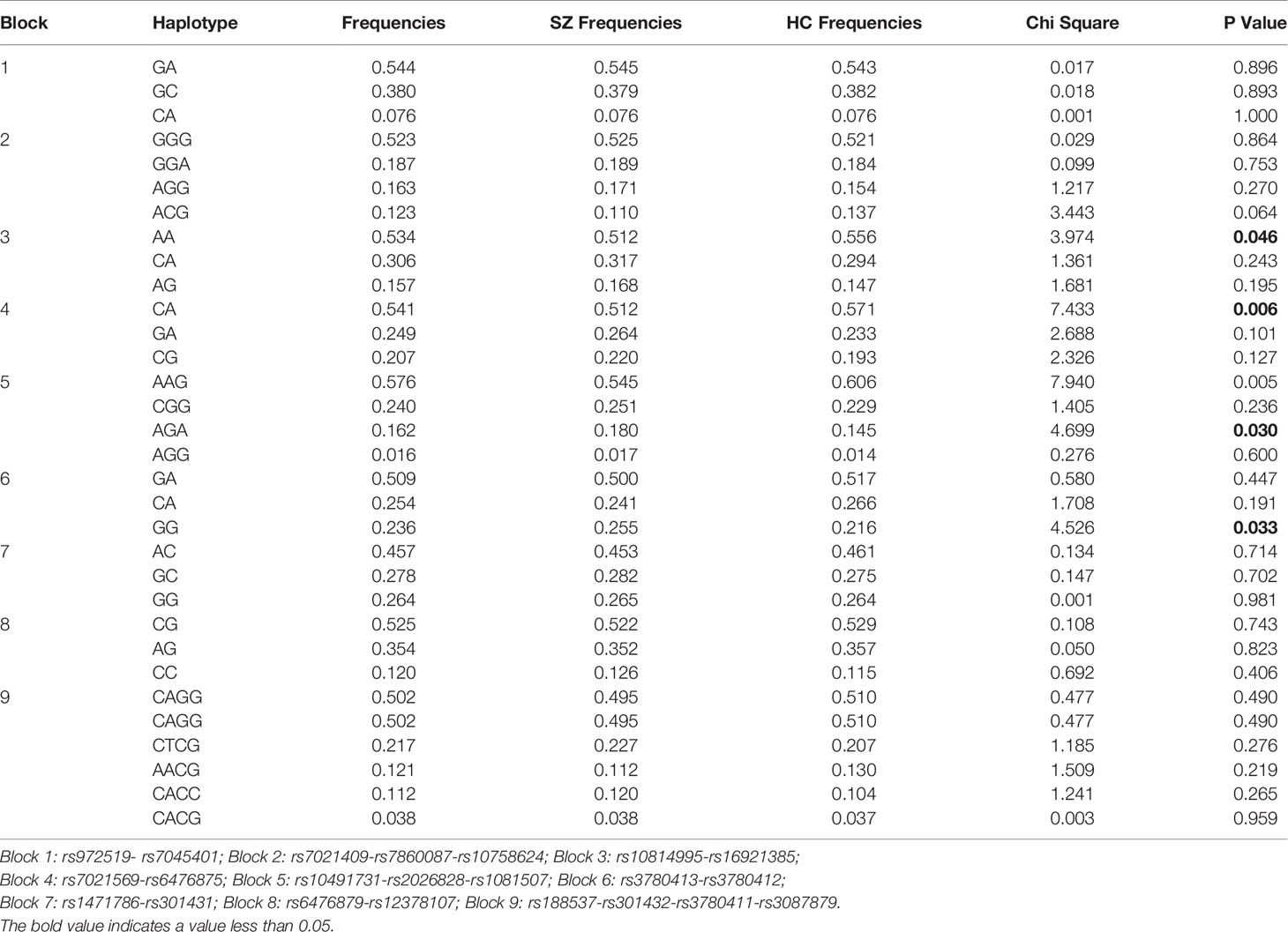
Table 3 Associated haplotype frequencies of 36 SNPs in the SLC1A1 gene between SZ and healthy controls.
Further, we make association analysis in six significant SNPs of the SLC1A1 gene in the PGC samples, including European and East Asian population (Supplementary Table 3). We found significant associations at rs10974573 in European population (p = 0.008, OR=1.031). Meanwhile, there no significant associations at six SNPs affect the expression level of SLC1A1 gene in main brain tissues, including frontal cortex and hippocampus (p’s > 0.05, Supplementary Table 4).
There were significant associations between rs7032326 genotypes and PANSS total, positive symptoms, negative symptoms, or general psychopathology in SZ (p = 0.002, 0.011, 0.028, 0.008, respectively). There were significant associations between rs7860087 genotypes and PANSS total, negative symptoms, or general psychopathology in SZ (p = 0.011, 0.015, 0.041, respectively). Genotypes of rs2039291 and rs4742007 were associated with positive symptoms (p = 0.029, 0.039, respectively). Genotypes of rs301430 were associated with negative symptoms (p = 0.031) (Table 4).
We investigated SLC1A1 mutations associated with the pathogenesis and psychopathology symptoms of SZ in a Han Chinese population. None significant differences were found in genotype and allele frequencies of rs10815017 and rs2026828 between SZ patients and healthy controls after Bonferroni’s correction. Therefore, our study suggests that SLC1A1 may be not a susceptibility gene for SZ. However, we found that rs7032326 genotypes were associated with psychopathology symptoms of SZ.
Previous studies reported the decreased EAAT3 (encoded product of SLC1A1) transcript expression (28), and increased expression of transcripts encoding EAAT1 and EAAT2 (29) in SZ. These findings suggested that the glutamate receptors and related molecules were abnormally expressed in glutamatergic synapses in SZ. In addition, expression of SLC1A1 was decreased following chronic antipsychotic treatment in animal models. Some studies have also reported that SNP rs2228622 and rs7022369 in the SLC1A1 gene were susceptibility gene sites for SZ (15, 19). Moreover, the SNP rs16921385, located in an intron of SLC1A1, was found to be associated with risperidone treatment response (22, 30). Therefore, SLC1A1 variation was associated with the pathogeny of SZ. However, there were little studies to explore this association (15, 19, 23) and the mechanism of variation in SLC1A1 still unknown.
In the present study, there were no significant association with SZ at SNPs rs10815017, rs6476875, rs7024664, and rs10974573. These SNPs are novel gene site findings that have not been reported previously. Further, there had a consistent finding in East Asian population of PGC samples. However, significant association was found at rs10974573 in European population indicated that this SNP may be a susceptibility gene site for SZ. Our study also found that rs3087879, rs301430, rs972519, rs10814991, rs7032326, rs7860087, rs3780415, rs3780413, and rs2072657 were not the susceptibility gene sites for SZ, a result that is consistent with previous reports (15, 19, 23). Meanwhile, we found SNPs rs2026828 and rs3780412 were not susceptibility markers for SZ, which is consistent with previous studies (15, 23). Previous studies reported rs2228622 (15) and rs10814995 (19) of SLC1A1 were susceptibility markers for SZ in a Japanese population, which is inconsistent with our present finding. However, our finding for rs2228622 is in line with research in the Chinese population (23).
In current, there are only three studied (15, 19, 23) reported SNPs of SLC1A1 in SZ. Thus, our study provides further evidences for the susceptibility of SLC1A1 in SZ. As is well known, SZ is highly heterogeneous at genetic and symptomatic levels. Haplotypes of AA, CA, AGA, and GG were also associated with SZ and provided further evidence for the susceptibility of SLC1A1 in SZ. Compared with previous studies (15, 19, 23), our study has the following innovation and advantages: 1) SZ patient selection: only paranoid SZ patients were included and examined; 2) all subjects were living in the north Henan provinces of China and belonged to the same population group, this ensure the consistency of genetic background; 3) more SNP sites (36 SNPs) were tested than previous studies were including 8 SNPs (15), 19 SNPs (19), and 4 SNPs (23). Therefore, our studies were not only reduced the influence of phenotypic heterogeneity, improved the numbers of SNPs for examination.
SZ is characterized by positive symptoms, negative symptoms, disorganization of thoughts, behaviors, and cognitive deficits. Our previous studies observed the genetic basis for SZ psychopathology symptoms, such as GRIN2B was related to cognition deficit symptoms (12), and CDNF2 was related to negative symptoms (25). However, there are few studies regarding the association between SZ psychopathology symptoms and SLC1A1 (15, 19, 23). In this study, we found SNP rs7032326 was related to positive symptoms, negative symptoms, and general psychopathology. In addition, another four SNPs (rs7860087, rs2039291, rs4742007, and rs301430) were also related to clinical subtype symptoms. Although these SNPs were not the susceptibility markers for SZ, our finding also provided some evidence for the genetic basis for SZ psychopathology symptoms.
The present study also had some limitations. First, independent samples are needed to verify the finding. Second, the sample size that included PANSS scores was small and insufficient, and should be expanded to further explore the association between genotypes and psychopathology symptoms.
In conclusion, our study provides further evidence that SLC1A1 may be not a susceptibility gene for SZ in Chinese Han population. These suggesting the variation of SLC1A1 may be not a genetic mechanism of SZ. However, the genetic variations of SLC1A1 may affect psychopathology symptoms. Therefore, further studies need to explore other susceptibility genes of SZ.
The original contributions presented in the study are publicly available. This data can be found here: https://www.oebiotech.com/Article/slc1a1jyyj.html.
The studies involving human participants were reviewed and approved by Ethics Committee of the Second Affiliated Hospital of Xinxiang Medical University (China). The patients/participants provided their written informed consent to participate in this study.
Author LL designed the study protocol. Authors TC, YY, YZ, MD, YL, and HY conducted sample selection and data management. Authors XS, ZL, LZ, QL, MLS, and XF undertook the genotyping identify and statistical analysis, and authors WL and MS wrote the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported in part by the National Natural Science Foundation of China (81671330 and 81971252 to LL), Medical Science and Technology Research Project of Henan Province (2018020373 to HY), National Key Research and Development Program of China (2016YFC1307001), High Scientific and Technological Research Fund of Xinxiang Medical University (2017ZDCG-04 to LL), and the support project for the Disciplinary group of Psychiatry and Neuroscience, Xinxiang Medical University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the patients, their families, and the healthy volunteers for their participation, and the physicians who collect clinical data and blood samples in the Second Affiliated Hospital of Xinxiang Medical University.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2020.559210/full#supplementary-material
1. Flint J, Munafo M. Schizophrenia: genesis of a complex disease. Nature (2014) 511:412–3. doi: 10.1038/nature13645
2. Hu W, MacDonald ML, Elswick DE, Sweet RA. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Ann N Y Acad Sci (2015) 1338:38–57. doi: 10.1111/nyas.12547
3. Li W, Yang Y, Lin J, Wang S, Zhao J, Yang G, et al. Association of serotonin transporter gene (SLC6A4) polymorphisms with schizophrenia susceptibility and symptoms in a Chinese-Han population. Prog Neuropsychopharmacol Biol Psychiatry (2013) 44:290–5. doi: 10.1016/j.pnpbp.2013.04.003
4. Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet (2011) 43:1228–31. doi: 10.1038/ng.979
5. Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet (2011) 43:1224–7. doi: 10.1038/ng.980
6. Schizophrenia Psychiatric Genome-Wide Association Study C. Genome-wide association study identifies five new schizophrenia loci. Nat Genet (2011) 43:969–76. doi: 10.1038/ng.940
7. Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature (2014) 511:421–7. doi: 10.1038/nature13595
8. Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, et al. Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet (2017) 49:1576–83. doi: 10.1038/ng.3973
9. Goff DC, Coyle JT. The emerging role of glutamate in the pathophysiology and treatment of schizophrenia. Am J Psychiatry (2001) 158:1367–77. doi: 10.1176/appi.ajp.158.9.1367
10. Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell (1999) 98:427–36. doi: 10.1016/s0092-8674(00)81972-8
11. Iwayama Y, Hashimoto K, Nakajima M, Toyota T, Yamada K, Shimizu E, et al. Analysis of correlation between serum D-serine levels and functional promoter polymorphisms of GRIN2A and GRIN2B genes. Neurosci Lett (2006) 394:101–4. doi: 10.1016/j.neulet.2005.10.025
12. Yang Y, Li W, Zhang H, Yang G, Wang X, Ding M, et al. Association Study of N-Methyl-D-Aspartate Receptor Subunit 2B (GRIN2B) Polymorphisms and Schizophrenia Symptoms in the Han Chinese Population. PloS One (2015) 10:e0125925. doi: 10.1371/journal.pone.0125925
13. Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, et al. Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet (2002) 71:877–92. doi: 10.1086/342734
14. Loh HC, Tang PY, Tee SF, Chow TJ, Choong CY, Lim SY, et al. Neuregulin-1 (NRG-1) and its susceptibility to schizophrenia: a case-control study and meta-analysis. Psychiatry Res (2013) 208:186–8. doi: 10.1016/j.psychres.2013.01.022
15. Deng X, Shibata H, Takeuchi N, Rachi S, Sakai M, Ninomiya H, et al. Association study of polymorphisms in the glutamate transporter genes SLC1A1, SLC1A3, and SLC1A6 with schizophrenia. Am J Med Genet B Neuropsychiatr Genet (2007) 144B:271–8. doi: 10.1002/ajmg.b.30351
16. Afshari P, Myles-Worsley M, Cohen OS, Tiobech J, Faraone SV, Byerley W, et al. Characterization of a Novel Mutation in SLC1A1 Associated with Schizophrenia. Mol Neuropsychiatry (2015) 1:125–44. doi: 10.1159/000433599
17. Shilling PD, Saint Marie RL, Shoemaker JM, Swerdlow NR. Strain differences in the gating-disruptive effects of apomorphine: relationship to gene expression in nucleus accumbens signaling pathways. Biol Psychiatry (2008) 63:748–58. doi: 10.1016/j.biopsych.2007.10.015
18. Nudmamud-Thanoi S, Piyabhan P, Harte MK, Cahir M and Reynolds GP. Deficits of neuronal glutamatergic markers in the caudate nucleus in schizophrenia. J Neural Transm Suppl (2007) 72:281–5. doi: 10.1007/978-3-211-73574-9_34
19. Horiuchi Y, Iida S, Koga M, Ishiguro H, Iijima Y, Inada T, et al. Association of SNPs linked to increased expression of SLC1A1 with schizophrenia. Am J Med Genet B Neuropsychiatr Genet (2012) 159B:30–7. doi: 10.1002/ajmg.b.31249
20. Myles-Worsley M, Tiobech J, Browning SR, Korn J, Goodman S, Gentile K, et al. Deletion at the SLC1A1 glutamate transporter gene co-segregates with schizophrenia and bipolar schizoaffective disorder in a 5-generation family. Am J Med Genet B Neuropsychiatr Genet (2013) 162B:87–95. doi: 10.1002/ajmg.b.32125
21. Rees E, Walters JT, Chambert KD, O’Dushlaine C, Szatkiewicz J, Richards AL, et al. CNV analysis in a large schizophrenia sample implicates deletions at 16p12.1 and SLC1A1 and duplications at 1p36.33 and CGNL1. Hum Mol Genet (2014) 23:1669–76. doi: 10.1093/hmg/ddt540
22. Yu H, Yan H, Wang L, Li J, Tan L, Deng W, et al. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. Lancet Psychiatry (2018) 5:327–38. doi: 10.1016/S2215-0366(18)30049-X
23. Lu W, Wu H, Cai J, Wang Z, Yi Z, Yu S, et al. Lack of association of SLC1A1 variants with schizophrenia in Chinese Han population. Psychiatry Res (2013) 210:669–71. doi: 10.1016/j.psychres.2013.07.009
24. Kay SR, Fiszbein A and Opler LA. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr Bull (1987) 13:261–76. doi: 10.1093/schbul/13.2.261
25. Yang Y, Yu H, Li W, Liu B, Zhang H, Ding S, et al. Association between cerebral dopamine neurotrophic factor (CDNF) 2 polymorphisms and schizophrenia susceptibility and symptoms in the Han Chinese population. Behav Brain Funct (2018) 14:1. doi: 10.1186/s12993-017-0133-4
26. Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res (2006) 34:W635–41. doi: 10.1093/nar/gkl236
27. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (2005) 21:263–5. doi: 10.1093/bioinformatics/bth457
28. McCullumsmith RE, Meador-Woodruff JH. Striatal excitatory amino acid transporter transcript expression in schizophrenia, bipolar disorder, and major depressive disorder. Neuropsychopharmacology (2002) 26:368–75. doi: 10.1016/S0893-133X(01)00370-0
29. Smith RE, Haroutunian V, Davis KL, Meador-Woodruff JH. Expression of excitatory amino acid transporter transcripts in the thalamus of subjects with schizophrenia. Am J Psychiatry (2001) 158:1393–9. doi: 10.1176/appi.ajp.158.9.1393
Keywords: schizophrenia, SLC1A1, single-nucleotide polymorphisms, psychopathology symptoms, association
Citation: Li W, Su X, Chen T, Li Z, Yang Y, Zhang L, Liu Q, Shao M, Zhang Y, Ding M, Lu Y, Yu H, Fan X, Song M and Lv L (2020) Solute Carrier Family 1 (SLC1A1) Contributes to Susceptibility and Psychopathology Symptoms of Schizophrenia in the Han Chinese Population. Front. Psychiatry 11:559210. doi: 10.3389/fpsyt.2020.559210
Received: 12 May 2020; Accepted: 31 August 2020;
Published: 23 September 2020.
Edited by:
Xiancang Ma, First Affiliated Hospital of Xi’an Jiaotong University, ChinaReviewed by:
Wenbin Guo, Central South University, ChinaCopyright © 2020 Li, Su, Chen, Li, Yang, Zhang, Liu, Shao, Zhang, Ding, Lu, Yu, Fan, Song and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Song, c29uZy5tZW5nLjIwMUAxNjMuY29t; Luxian Lv, bHZ4OTI4QDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.