
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Psychol. , 11 September 2018
Sec. Perception Science
Volume 9 - 2018 | https://doi.org/10.3389/fpsyg.2018.01695
This article is part of the Research Topic Contemporary Neural Network Modeling in Cognitive Science View all 12 articles
This article is a correction to:
Recurrent Convolutional Neural Networks: A Better Model of Biological Object Recognition
A Corrigendum on
Recurrent Convolutional Neural Networks: A Better Model of Biological Object Recognition
by Spoerer, C. J., McClure, P., and Kriegeskorte, N. (2017). Front. Psychol. 8:1551. doi: 10.3389/fpsyg.2017.01551
In the original article, there was a mistake in Table 1 as published. A small error was made in the calculation of the number of parameters in the networks. This caused the original figures to be inflated in all models by fewer than 100 parameters. The corrected Table 1 appears below.
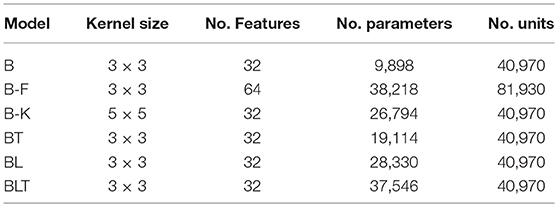
Table 1. Brief descriptions of the models used in these experiments including the number of learnable parameters and the number of units in each model.
Additionally, heading 3.1 was incorrectly titled as “Recognition of Sights under Debris”. The correct heading is “Recognition of Digits under Debris”.
Also, there was an error in the text. When McNemar's test was used to test for a significant difference in multiple digit recognition tasks, we treated each digit as an independent sample. However, the probabilities for correctly identifying multiple digits in the same image are not independent. Therefore, we fail to meet the assumption of independence between samples required for McNemar's test. Instead, a test that corrects for dependence between samples should have been used, such as the variation on McNemar's test proposed by Durkalski et al. (2003). Using this corrected test produces a marginal difference in the results and leads to no change in the significance of the tests.
A correction has been made to Results, Recognition of Multiple Digits, Paragraphs 1 and 2:
To examine the ability of the networks to handle occlusion when the occluder is not a distractor, the networks were trained and tested on their ability to recognize multiple overlapping digits. In this case, when testing for significance, we used a variant of McNemar's test that corrects for dependence between predictions (Durkalski et al., 2003), which can arise when identifying multiple digits in the same image.
When recognizing three digits simultaneously, recurrent networks generally outperformed feedforward networks (Figure 7), with the exception of BT and B-K where no significant difference was found [χ2(1, N = 30, 000) = 3.82, p = 0.05]. All other differences were found to be significant (FDR = 0.05). The error rates for all models are shown in Table 4. A similar pattern is found when recognizing both four and five digits simultaneously. However, in both four and five digit tasks, all pairwise differences were found to be significant, with B-K outperforming BT (Figure 7). This suggests that, whilst recurrent networks generally perform better at this task, they do not exclusively outperform feedforward models.
The authors apologize for these errors and state that this does not change the scientific conclusions of the article in any way.
The original article has been updated.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Keywords: object recognition, occlusion, top-down processing, convolutional neural network, recurrent neural network
Citation: Spoerer CJ, McClure P and Kriegeskorte N (2018) Corrigendum: Recurrent Convolutional Neural Networks: A Better Model of Biological Object Recognition. Front. Psychol. 9:1695. doi: 10.3389/fpsyg.2018.01695
Received: 09 August 2018; Accepted: 22 August 2018;
Published: 11 September 2018.
Edited and reviewed by: James L. McClelland, Stanford University, United States
Copyright © 2018 Spoerer, McClure and Kriegeskorte. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Courtney J. Spoerer, Y291cnRuZXkuc3BvZXJlckBtcmMtY2J1LmNhbS5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.