- Department of Biopsychology, Institute of Cognitive Neuroscience, Faculty of Psychology, Ruhr-University Bochum, Bochum, Germany
Handedness and language lateralization are partially determined by genetic influences. It has been estimated that at least 40 (and potentially more) possibly interacting genes may influence the ontogenesis of hemispheric asymmetries. Recently, it has been suggested that analyzing the genetics of hemispheric asymmetries on the level of gene ontology sets, rather than at the level of individual genes, might be more informative for understanding the underlying functional cascades. Here, we performed gene ontology, pathway and disease association analyses on genes that have previously been associated with handedness and language lateralization. Significant gene ontology sets for handedness were anatomical structure development, pattern specification (especially asymmetry formation) and biological regulation. Pathway analysis highlighted the importance of the TGF-beta signaling pathway for handedness ontogenesis. Significant gene ontology sets for language lateralization were responses to different stimuli, nervous system development, transport, signaling, and biological regulation. Despite the fact that some authors assume that handedness and language lateralization share a common ontogenetic basis, gene ontology sets barely overlap between phenotypes. Compared to genes involved in handedness, which mostly contribute to structural development, genes involved in language lateralization rather contribute to activity-dependent cognitive processes. Disease association analysis revealed associations of genes involved in handedness with diseases affecting the whole body, while genes involved in language lateralization were specifically engaged in mental and neurological diseases. These findings further support the idea that handedness and language lateralization are ontogenetically independent, complex phenotypes.
Introduction
Handedness and language lateralization are complex phenotypes and represent different aspects of functional brain asymmetries. Hemispheric asymmetries are a major principle of brain organization in many vertebrate (Ocklenburg et al., 2013d; Ströckens et al., 2013; Güntürkün and Ocklenburg, 2017) and invertebrate species (Frasnelli, 2013). In humans, handedness and language lateralization are related to some extent. Both are mostly controlled for by the left hemisphere in right-handed individuals. Moreover, left-handedness is associated with a higher probability for right-hemispheric language lateralization (Knecht et al., 2000; Somers et al., 2015). The predominance of the left hemisphere in processing fast temporal changes makes it ideally suited to process both complex motor function (Barber et al., 2012) and language (Slevc et al., 2011; Scott and McGettigan, 2013). This association prompted some authors to assume that one single gene determines both handedness and language lateralization: For example, the ‘Right-Shift Theory’ (Annett, 1975) proposes a single dominant allele (RS+), which increases the chance of being right-handed with a left-hemispheric dominance for language. The alternative recessive allele (RS-) does not influence lateralization, which reduces the ‘right-shift’ in RS+- individuals. In homozygous RS-- individuals, the direction of handedness and language lateralization is determined by chance. A similar single gene model has been conceived by McManus (1984, 1985), who proposed a dextral allele (D), which results in 100% right-handedness and left-hemispheric language dominance in homozygotes (DD). The chance allele (C) does not affect lateralization, so that right- and left-handedness occur with a probability of 50% each in the homozygote variant (CC). The heterozygote phenotype (DC) was proposed to result in a 75% probability of right-handedness. However, these early genetic theories are solely phenotype-driven and are not supported by molecular genetic evidence. In contrast, a number of twin studies estimated that around 25% of variance in handedness data is due to additive genetic effects. The remainder is suggested to be influenced by non-genetic factors (Medland et al., 2006, 2009; Vuoksimaa et al., 2009). In fact, no single gene has been identified as a potential exclusive determinant of handedness and language lateralization. Despite sample sizes allowing for adequate statistical power, evidence from genome-wide association studies (GWASs) strongly argues against the existence of such a gene (Eriksson et al., 2010; Ocklenburg et al., 2013c; Armour et al., 2014). However, these studies do not disprove the existence of a genetic component in handedness development per se. As suggested by McManus et al. (2013), a key biological model for the genetics of handedness is primary ciliary dyskinesia (PCD), which results in situs inversus, a mirror reversal of visceral organs, in 50% of all cases. Not surprisingly for a complex phenotype, at least 16 loci involved in PCD have been found so far. Similarly, molecular genetic studies suggest that multi-locus models might be a more suitable explanation for the ontogenesis of hemispheric asymmetries. Armour et al. (2014) suggest that at least 40 and potentially up to 100 genes are involved in the determination of functional lateralization.
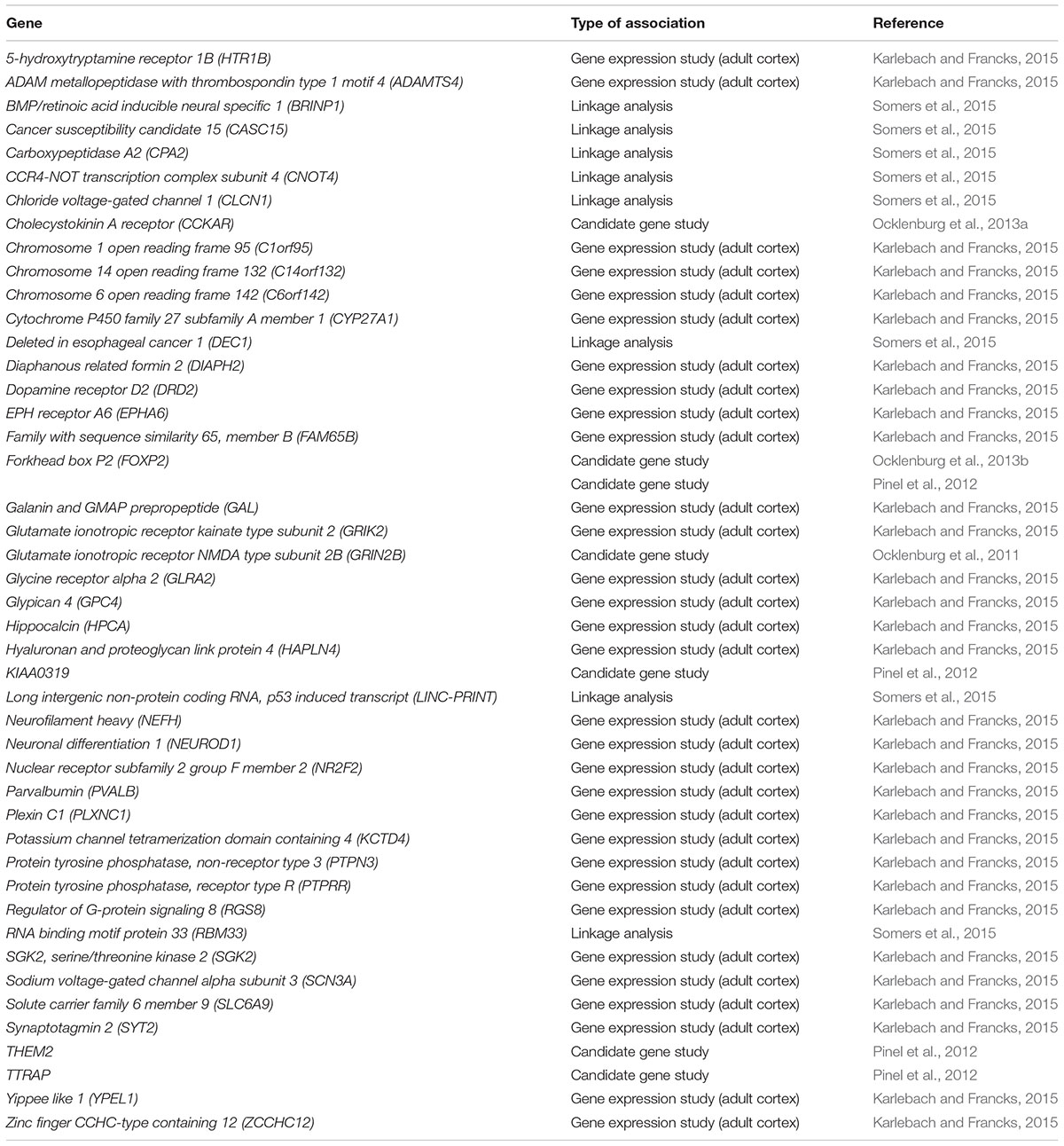
Genes associated with handedness include LRRTM1 (Francks et al., 2007), PCSK6 (Scerri et al., 2011; Arning et al., 2013; Brandler et al., 2013; Robinson et al., 2016), AR (Medland et al., 2005; Hampson and Sankar, 2012; Arning et al., 2015), COMT (Savitz et al., 2007), APOE (Bloss et al., 2010; but see Hubacek et al., 2013; Piper et al., 2013), and SETDB2 (Ocklenburg et al., 2015a). Genes associated with language lateralization include FOXP2 (Pinel et al., 2012; Ocklenburg et al., 2013b), CCKAR (Ocklenburg et al., 2013a), GRIN2B (Ocklenburg et al., 2011), and others (see below). However, these genes explain only a fraction of the variance in the respective phenotype. To this date, no study could reveal an association of one gene with both language lateralization and handedness that would point towards a shared genetic basis. Therefore, Ocklenburg et al. (2014) proposed that handedness and language lateralization differ in both their neurophysiological basis and genetic correlates. The authors suggest a relationship of partial pleiotropy between both phenotypes, i.e., handedness and language lateralization have shared as well as independent ontogenetic influencing factors contributing to their development.
Uncovering the ontogenesis of hemispheric asymmetries requires deeper knowledge of genes involved in their development. However, specifically investigating individual genes gives rise to different methodological difficulties: First, genes can never be interpreted on their own, but have to be regarded in the context of other genes (Zhang et al., 2015) and environmental factors (Asor and Ben-Shachar, 2016; Gattere et al., 2016). Second, another promising way to shed light on the development of hemispheric asymmetries is comparing gene expression between the left and right hemisphere. Grouping of genes into functional sets could manifest hemispheric asymmetries that are too subtle to uncover on the level of individual genes (Karlebach and Francks, 2015). Accordingly, gene ontology (GO) sets classify genes into functional groups depending on their biological effects. Applying GO analysis on a certain list of genes reveals information on shared molecular functions of these genes, their contributions to biological processes and their corresponding cellular locations (Gene Ontology Consortium, 2015). Here, we applied GO analyses on genes previously associated with handedness on the one hand and genes previously associated with language lateralization on the other hand to identify functional gene groups associated with the respective phenotype. We hypothesized that functional gene groups between phenotypes are mainly independent from each other. This study will provide additional evidence opposing models that assume 100% pleiotropy (the same ontogenetic factors determine both handedness and language lateralization), but instead is in line with a model of partial pleiotropy (shared and individual ontogenetic factors determine handedness and language lateralization) as suggested by Ocklenburg et al. (2014).
Materials and Methods
Identification of Relevant Genes
In order to identify genes associated with handedness or language lateralization, we performed literature search using the database PubMed1. Molecular genetic studies were included if performed on human subjects.
We included individual genes previously identified in candidate gene studies on handedness or language lateralization into analysis (Medland et al., 2005; Francks et al., 2007; Bloss et al., 2010; Ocklenburg et al., 2011, 2013a,b; Hampson and Sankar, 2012; Pinel et al., 2012; Arning et al., 2013, 2015; Robinson et al., 2016). Furthermore, we included all genes reaching p < 10-5 in a GWAS by Scerri et al. (2011) and a GWAS meta-analysis by Brandler et al. (2013). We further included differentially expressed genes from gene expression studies (p < 0.01; Sun et al., 2005; Karlebach and Francks, 2015) and top hits identified by family-based genetic association analysis (Savitz et al., 2007) and manual segregation analysis (van Agtmael et al., 2002). Lastly, we included all genes with LOD > 1.5 from a linkage analysis published by Somers et al. (2015). Table 1 shows the list of 63 genes previously associated with handedness ontogenesis. The list of 45 genes previously associated with the formation of language lateralization is listed in Table 2. Importantly, most of these genes do not reach conventional levels of significance or do not replicate. However, it is still likely that GO analysis reveals certain clusters of genes contributing to each of the phenotypes.
Gene Ontology Analysis
We used WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) (Zhang et al., 2005; Wang et al., 2013) to identify shared functional groups of all genes associated with handedness (see Table 1). The list containing 63 genes was inserted to WebGestalt to identify GO sets associated with handedness. A GO set is a pre-defined list of genes that share either molecular functions (biochemical activity of a gene product), cellular components (place in the cell where a gene product is active), or biological processes (biological objective of a gene or gene product). For example, the GO set ‘determination of left/right symmetry’ contains 82 genes and gene products whose biological objective is involved in body formation in a symmetric or asymmetric pattern (Ashburner et al., 2000).
For each GO set, WebGestalt calculated a ratio of enrichment (RE) by comparing the observed number of genes in the inserted gene list and also in the GO set (O) to the expected number of genes in the inserted gene list and also in the GO set (E). This expected value (E) was based on the number of genes in the inserted gene list (L) multiplied with the number of genes in the GO set (GO) and divided by the number of genes in the reference gene set (RG). If the observed value (O) exceeded the expected value (E), the GO set was enriched with a ratio of enrichment RE = O/E (Wang et al., 2013). WebGestalt then used the hypergeometric test to evaluate the significance of enrichment for GO sets in the list of genes. The significance level was set to 0.05 after Benjamini–Hochberg correction for multiple comparisons (Benjamini and Hochberg, 1995). WebGestalt only reported GO sets with corrected p-values smaller than 0.05.
In addition to statistical results, WebGestalt’s output included a visualization of relationships between GO sets. This hierarchical structure of GO sets included high level GO sets representing broad molecular functions/cellular components/biological processes, e.g., ‘signal transduction (GO:0007165).’ These broader GO sets were subdivided into more specific lower level GO sets, e.g., ‘regulation of postsynaptic neurotransmitter receptor activity (GO:0098962)’ (Ashburner et al., 2000). In order to improve the results’ transparency, significant lower level GO sets were clustered in superordinate groups of high level GO sets by visual inspection of this hierarchical structure.
The same procedure was applied on the gene list containing 45 genes associated with ontogenesis of language lateralization (see Table 2).
KEGG Pathway Analysis
Using WebGestalt, we performed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses (Kanehisa et al., 2008) to identify biological pathways including genes associated with the gene list of either handedness or language lateralization. Each list of genes (see Tables 1, 2) was entered to WebGestalt separately. KEGG pathways are pre-defined lists of genes that are involved in biological pathways. A RE was calculated for each KEGG pathway analogous to GO analysis. The significance of enrichment for each KEGG pathway was calculated with the hypergeometric test. The significance level was set to 0.05 after Benjamini–Hochberg correction for multiple comparisons (Benjamini and Hochberg, 1995).
Disease Association Analysis
In order to identify diseases associated with gene sets involved in either handedness or language lateralization, we conducted disease association analyses using WebGestalt (Wang et al., 2013). Gene-disease associations were inferred using GLAD4U (Gene List Automatically Derived For You) (Jourquin et al., 2012). Both gene lists (see Tables 1, 2) were entered to WebGestalt separately. A RE was calculated for each disease. The significance of enrichment was calculated using hypergeometric test with a significance level of 0.05 after Benjamini–Hochberg correction (Benjamini and Hochberg, 1995). Using ICD-10 (World Health Organization, 1992), we identified diseases categorized under “V: Mental and behavioral disorders” or “VI: Diseases of the nervous system” as disorders related to the central nervous system (CNS).
Results
Lower Level GO Sets Involved in Handedness and Language Lateralization
After correction for multiple comparisons, GO analysis revealed 64 significant lower level GO sets for the 63 genes associated with handedness, among them 40 biological processes (see Table 3), 20 molecular functions, and 4 cellular components (see Supplementary Figure S1 for full hierarchical GO set overview). Top hits were ‘epithelial tube morphogenesis (GO:0060562)’ (p < 0.001), ‘tube development (GO:0035295)’ (p < 0.001), ‘tube morphogenesis (GO: 0035239)’ (p < 0.001) as well as ‘determination of left/right symmetry (GO:0007368)’/‘determination of bilateral symmetry (GO:0009855)’/‘specification of symmetry (GO:0009799)’ (all p < 0.001). GO sets with the most genes involved were ‘protein binding (GO:0005515)’ (p < 0.05) with 20 handedness genes involved and ‘anatomical structure development (GO:0048856)’ (p < 0.01) and ‘multicellular organismal development (GO:0007275)’ (p < 0.01) with 18 handedness genes involved.
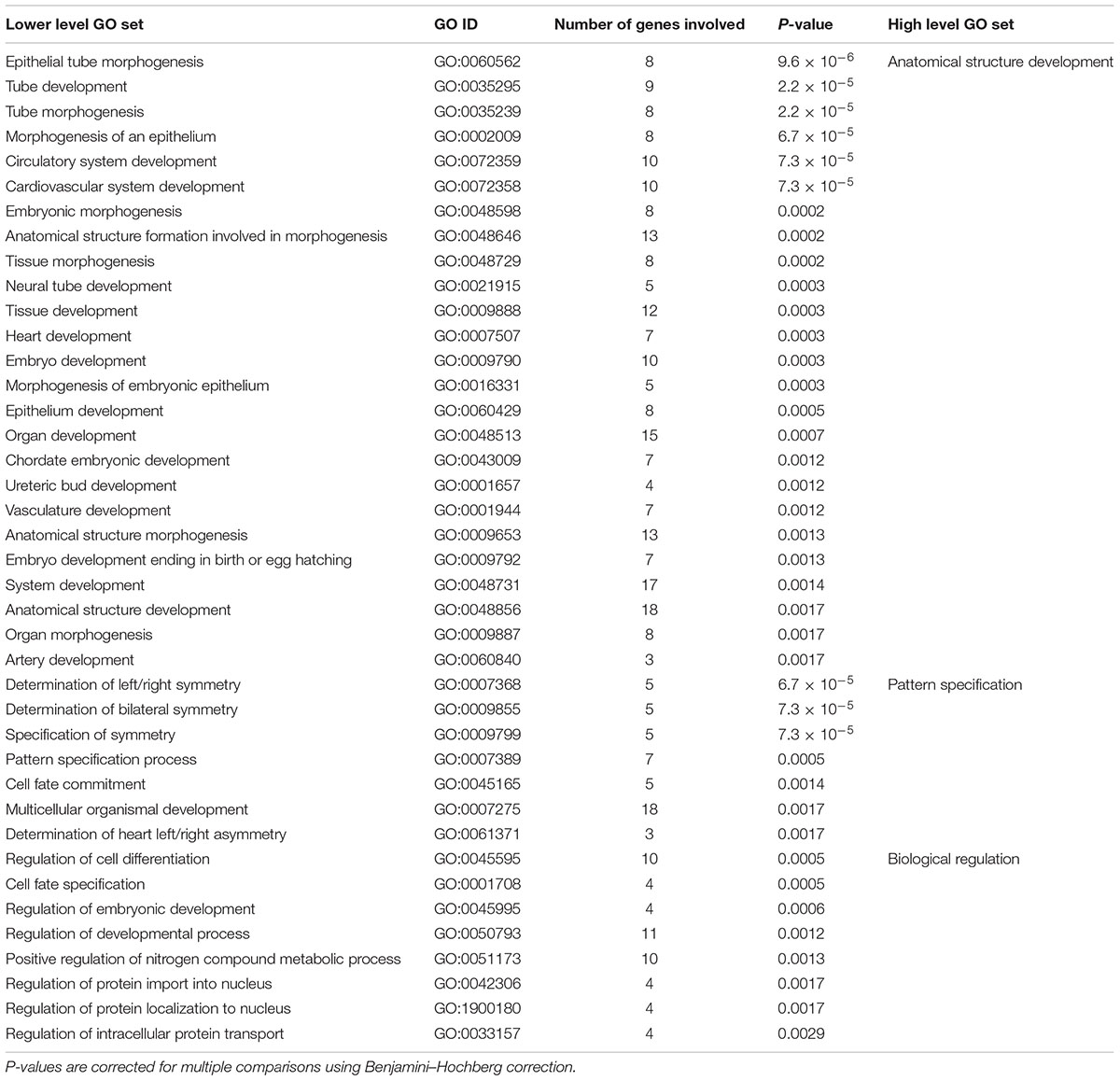
TABLE 3. Lower level and high level gene ontology (GO) sets enriched in genes associated with handedness ontogenesis.
For the 45 genes associated with language lateralization, GO analysis revealed 97 significant lower level GO sets. Among these GO sets were 40 biological processes (see Table 4), 29 molecular functions, and 28 cellular components (see Supplementary Figure S2 for full hierarchical GO set overview). Top hits of GO sets were ‘negative regulation of synaptic transmission, glutamatergic (GO:0051967)’ (p < 0.001), ‘feeding behavior (GO:0007631)’ (p < 0.001), and ‘signal release (GO:0023061)’ (p < 0.01). Most genes were involved in the cellular components ‘plasma membrane (GO:0005886)’ (p < 0.05), ‘cell periphery (GO:0071944)’ (p < 0.05) with 17 genes each and in the biological process ‘nervous system development (GO:0007399)’ (p < 0.01) with 13 genes involved.
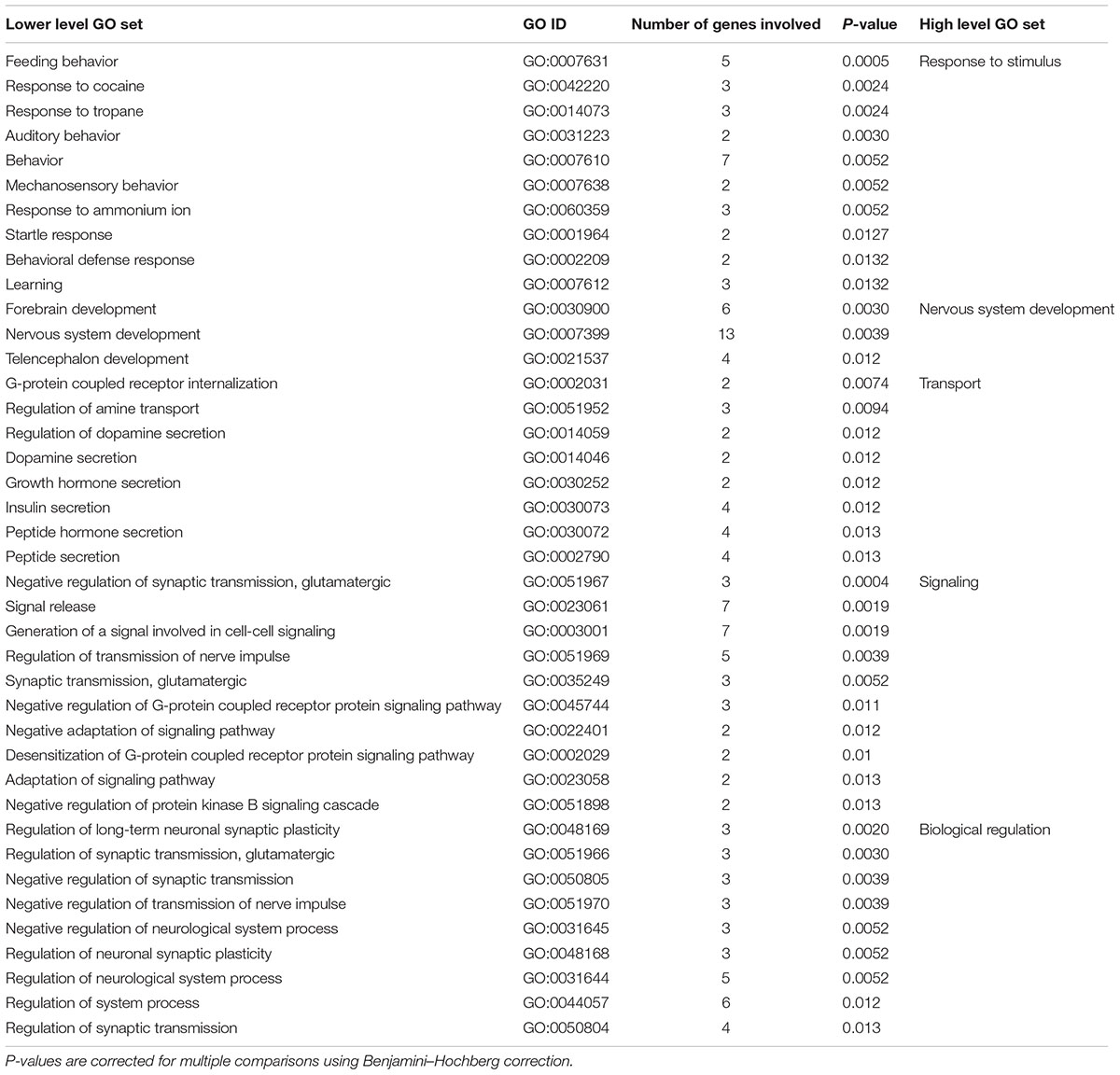
TABLE 4. Lower level and high level GO sets enriched in genes associated with the ontogenesis of language lateralization.
Two lower level GO sets concerning cellular components overlap between the gene lists for handedness and language lateralization: ‘cell projection (GO:0042995)’ (p < 0.05) and ‘neuron projection (GO:0043005)’ (p < 0.05). There was no overlap in biological processes.
The distribution of raw p-values for all significantly enriched GO sets for handedness and language lateralization is displayed in Supplementary Figure S3.
High Level GO Sets Involved in Handedness and Language Lateralization
Visual inspection of the hierarchical relationship between GO sets involved in handedness revealed that significant lower level GO sets regarding biological processes are clustered into three high level GO sets. First, 25 enriched lower level GO sets are involved in anatomical structure development. ‘Epithelial tube morphogenesis (GO:0060562)’ was the most significantly enriched GO set overall. Lower level GO sets contain not only ‘neural tube development (GO:0021915),’ but also ‘cardiovascular system development (GO:0072358),’ ‘artery development (GO:0060840),’ and ‘ureteric bud development (GO:0001657).’ Moreover, 6 lower level GO sets involve pattern specification, for example in terms of ‘specification of symmetry (GO:0009799),’ ‘determination of left/right symmetry (GO:0007368),’ and ‘determination of bilateral symmetry (GO:0009855).’ Lastly, 9 lower level GO sets involve biological regulation. These GO sets include ‘regulation of developmental process (GO:0050793)’ and ‘regulation of cell differentiation (GO:0045595).’ High level GO sets for genes associated with handedness are visualized in Supplementary Figure S4.
In contrast, significant lower level GO sets regarding biological processes in language lateralization are clustered into five high level GO sets. First, 10 enriched lower level GO sets can be described by the high level GO set ‘response to stimuli.’ These GO sets range from ‘feeding behavior (GO:0007631)’ to external stimuli like ‘behavioral defense response (GO:0002209)’ or ‘learning (GO:0007612)’ and organic substances like ‘response to cocaine (GO:0042220).’ Second, 3 lower level GO sets are involved in the high level GO set ‘nervous system development (GO:0007399),’ more specifically ‘forebrain development (GO:0030900),’ ‘telencephalon development (GO:0021537),’ and ‘nervous system development (GO:0007399).’ The third high level GO set with 8 lower level GO sets describes different forms of transport like ‘dopamine secretion (GO:0014046),’ ‘insulin secretion (GO:0030073)’ or ‘regulation of amine transport (GO:0051952).’ The fourth high level GO set includes 10 lower level GO sets involved in signaling, for example ‘regulation of transmission of nerve impulse (GO:0051969)’ or ‘synaptic transmission, glutamatergic (GO:0035249).’ Lastly, 9 lower level GO sets describe biological regulation, for example ‘regulation of long-term neuronal synaptic plasticity (GO:0048169)’ and ‘regulation of neurological system process (GO:0031644).’ High level GO sets for genes involved in language lateralization are visualized in Supplementary Figure S4.
Among the high level GO sets, biological regulation is involved in both handedness and language lateralization (see Supplementary Figure S4).
KEGG Pathway Analysis
For genes involved in handedness, KEGG analysis yielded six KEGG pathways significantly enriched after correction for multiple comparisons: ‘Pathways in cancer’ (p < 0.001), ‘Basal cell carcinoma’ (p < 0.01), ‘ECM-receptor interaction’ (p < 0.01), ‘TGF-beta signaling pathway’ (p < 0.01), ‘Cell adhesion molecules (CAMs)’ (p < 0.01), and ‘Focal adhesion’ (p < 0.05).
For genes involved in language lateralization, KEGG analysis yielded four KEGG pathways significantly enriched after correction for multiple comparisons: ‘Neuroactive ligand-receptor interaction’ (p < 0.001), ‘Amyotrophic lateral sclerosis (ALS)’ (p < 0.01), ‘Pancreatic secretion’ (p < 0.001), and ‘Axon guidance’ (p < 0.01). The distribution of corresponding raw p-values is displayed in Supplementary Figure S3.
Disease Association Analysis
Genes associated to handedness ontogenesis were involved in 156 diseases, among them 61 CNS-related diseases (39.10%). The most significantly enriched diseases were ‘Craniofacial Abnormalities’ (p < 0.001), ‘Amnesia’ (p < 0.001), and ‘Bone Diseases, Developmental’ (p < 0.01). Most genes were involved in ‘Craniofacial Abnormalities’ (p < 0.001) and ‘Congenital Abnormalities’ (p < 0.01) (six genes involved) and ‘Gilbert Disease’ (p < 0.01), ‘Epithelial cancers’ (p < 0.01), ‘Musculoskeletal Abnormalities’ (p < 0.01), and ‘Cancer or viral infections’ (p < 0.05) with five genes involved.
Genes involved in language lateralization were mostly associated to CNS-related diseases. 81 of 94 (86.17%) significantly enriched diseases were involved in mental or psychiatric states. The disease categories ‘Mental Disorders’ (p < 0.001), ‘Substance-Related Disorders’ (p < 0.001), and ‘Alcoholism’ (p < 0.001) were most significantly enriched. ‘Mental Disorders’ (p < 0.001) was enriched with 10 genes involved in language lateralization, followed by ‘Substance-Related Disorders’ (p < 0.001) and ‘Nervous System Diseases’ (p < 0.001) with seven genes involved. Associations between diseases and gene lists were much stronger in terms of p-values for genes involved in language lateralization than for genes involved in handedness (see Supplementary Figure S3).
There was considerable overlap in the enriched diseases for genes involved in handedness and language lateralization. Forty-two diseases were involved in both phenotypes, among them 39 (92.86%) CNS-related diseases.
Discussion
Handedness and language lateralization have been proposed to share a common ontogenetic basis (Annett, 1975), but single genes involved in the formation of both phenotypes have not been identified (Ocklenburg et al., 2014). Here we show that the GO sets enriched in language lateralization barely overlap with those found for handedness. Thus, in addition to the fact that individual genes involved in handedness and language lateralization development are independent from each other, functional gene products also differ fundamentally with no shared biological processes. This indicates different functional cascades underlying handedness and language lateralization.
For genes involved in ontogenesis of handedness, significant lower level GO sets of biological processes are clustered into three high level GO sets (see Supplementary Figure S4). First, most lower level GO sets describe anatomical structure development in different body parts. This implies that genes involved in handedness development exert their effect at an early embryonic stage and their functional gene products do not only contribute to the CNS, but also to the whole body. This is in line with the suggestion by Brandler et al. (2013), who claim that handedness is partially controlled by the molecular mechanisms that establish body asymmetry during early development. This finding has been supported by neuroimaging studies of patients with situs inversus, who displayed atypical patterns of frontal and occipital cerebral asymmetries (Kennedy et al., 1999; Ihara et al., 2010). However, situs inversus patients display the standard pattern of handedness, which rather supports a dissociation between visceral and brain asymmetries (Matsumoto et al., 1997; McManus et al., 2004; Afzelius and Stenram, 2006). It might be that genes associated with handedness are not necessarily involved in body asymmetry formation, but rather in anatomical structure development per se. Interestingly, most of the significant lower level GO sets involved in anatomical structure development include the androgen receptor (AR) gene. Prenatal testosterone has been shown to affect handedness and language lateralization in opposite directions (Lust et al., 2011). Our findings suggest that the capacity of binding testosterone in the developing fetal brain might induce differences in anatomical structure development that affect handedness, but not language lateralization. This finding is highly interesting in the context of sex differences in hemispheric asymmetries. While it is more or less undisputed that there is a 1.23 higher rate of male compared to female left-handers (Papadatou-Pastou et al., 2008), there are not necessarily sex differences in language lateralization (McManus, 2010). If that is the case, the findings from GO analysis may contribute to the explanation of this effect. Another high level GO set involved in handedness development is ‘pattern specification process (GO:0007389).’ As expected, the significant GO sets indicate the involvement of handedness genes on symmetry and asymmetry development. This result comes to no surprise, as there may likely be an ascertainment bias, since several of the original studies were candidate gene studies. Interestingly, KEGG pathway analysis revealed that genes involved in handedness ontogenesis are associated to the TGF-beta signaling pathway involved in bodily left-right asymmetry (Mittwoch, 2008; Shiratori and Hamada, 2014). While ACVR2B is involved in gonadal growth, embryo differentiation, and placenta formation, NODAL is involved in left-right axis determination and mesoderm and endoderm induction (see Supplementary Figure S5). This finding indicates an involvement of the TGF-beta signaling pathway on handedness ontogenesis at an early stage of development. In a recent study, asymmetrical gene expression was found between left and right human spinal cord at 8 weeks post conception. Besides DNA methylation patterns, gene expression asymmetries were epigenetically regulated by miRNAs involved in the TGF-beta signaling pathway. Since preliminary forms of handedness are already visible at this time point before the spinal cord and the motor cortex are functionally connected, the TGF-beta signaling pathway might have an impact on early behavioral asymmetries in arm movements (Ocklenburg et al., 2017). This in line with our finding that the TGF-beta signaling pathway is involved in handedness, but not in language lateralization. The last high level GO set of biological processes enriched in handedness genes is comprised of biological regulation, for example on developmental processes as well as cell differentiation. This indicates a regulatory function of genes associated with handedness on all levels of developmental control and cell fate determination.
For genes involved in ontogenesis of language lateralization, four high level GO sets were identified. Many lower level GO sets describe responses to different stimuli. Especially the role of the GO sets ‘startle response (GO:0001964)’ and ‘behavioral defense response (GO:0002209)’ are in line with a relation between stress and the ontogenesis of hemispheric asymmetries that has been reported in many vertebrate species (see Ocklenburg et al., 2016). It has been shown that both acute and chronic stress can affect different forms of lateralization in the human brain. Our findings here suggest that genetic predispositions for certain response patterns may also play a role in the ontogenesis of language lateralization, implying a role for gene-environment interactions during asymmetry development. Another highly interesting GO set involved in the formation of language lateralization is ‘learning (GO:0007612).’ Compared to handedness, language is more closely related to cognition, which is in line with the role of genes associated with language lateralization on neuronal signaling, e.g., neurotransmitters like glutamate and dopamine (Ocklenburg et al., 2011, 2013a). Also, the involvement of learning processes in the ontogenesis of language lateralization (Thomas et al., 1997) indicates a greater role of neuronal plasticity processes for this phenotype than for handedness. Secondly, lower level GO sets are involved in nervous system development. Compared to GO sets enriched in genes involved in handedness, which comprise cerebral, but also body development, this result suggests that genes involved in language lateralization are specifically engaged within the CNS. This is also supported by our finding that genes involved in language lateralization are significantly enriched in the axon guidance pathway including EPHA6 and PLXNC1, two receptors involved in axonal outgrowth, repulsion and attraction (see Supplementary Figure S6). In addition to their effect on basic cell metabolic processes, genes associated with language lateralization seem to be involved in neuronal signaling. ‘Negative regulation of G-protein coupled receptor protein signaling pathway (GO:0045744)’ or ‘desensitization of G-protein coupled receptor protein signaling pathway (GO:0002029)’ are important lower level GO sets within this category. The G-protein coupled receptor protein signaling pathway has been identified as asymmetrically expressed in adult human language related areas: Superior Temporal Gyrus (STS) and Heschl’s Gyrus (HG). Moreover, in our study many GO sets are involved in transmission of nerve impulse, a GO set asymmetrically expressed in STS, but not in HG (Karlebach and Francks, 2015). Lastly, lower level GO sets significantly enriched in genes associated with language lateralization are involved in the high level GO set of biological regulation. Although individual GO sets of language lateralization and handedness do not overlap in terms of biological processes, biological regulation represents a high level GO set within genes involved in both phenotypes. This can be considered as a minimal overlap between biological processes of gene products involved in handedness and those involved in language lateralization.
Overall, gene lists for handedness and language lateralization resulted in similar numbers of enriched GO sets. However, the distribution of genes differed between phenotypes. For genes associated with handedness, there were many GO sets with 10 or more genes enriched in. Thus, products of genes involved in handedness formation seems to be less complex compared to products of genes involved in language lateralization. The latter are more heterogenous with maximally seven genes enriched in the same GO set (with the exception of ‘nervous system development (GO:0007399)’ with 13 genes enriched) and less strong associations in terms of p-values.
In contrast, associations between diseases and gene lists were much stronger for genes involved in language lateralization than for genes involved in handedness. For language lateralization, many disease categories were enriched with high numbers of genes involved, mostly categorized in mental and neurological diseases. Among the diseases significantly associated with genes involved in language lateralization are schizophrenia (Ocklenburg et al., 2013e, 2015b) and autism spectrum disorders (Knaus et al., 2010; Tager-Flusberg, 2016). Language lateralization seems more strongly connected to disorders of neurological system development, which is completely in line with our finding that associated genes are enriched in nervous system development rather than anatomical structure development. In contrast, genes associated with handedness ontogenesis are involved in diseases affecting the whole body, which supports our findings from GO analyses and the argumentation pointed out by Brandler et al. (2013). Among the significantly enriched diseases were many that had been associated with handedness before, specifically depression (Denny, 2009), bipolar disorder (Nowakowska et al., 2008), language and learning disorders (Geschwind and Behan, 1982), anxiety disorders (Logue et al., 2015), attention deficit hyperactivity disorder (Brandler and Paracchini, 2014), and schizophrenia (Hirnstein and Hugdahl, 2014).
Our results support the idea of a model of partial pleiotropy for handedness and language lateralization as suggested by Ocklenburg et al. (2014). However, biological and statistical issues remain to be solved: First, two or more lists of genes could result in different GO sets that might still be highly intercorrelated and therefore related to one another. However, this may rather concern low level GO sets. In our data, high level superordinate GO sets between phenotypes are distinct from each other, but this limitation should nonetheless be kept in mind. Second, since most of the included genes of both lists do not reach conventional levels of significance or do not replicate in association studies or GWASs we cannot rule out that statistical noise could have had an impact on the results. Low pleiotropy between genes associated with handedness and language lateralization could therefore partly represent measurement error.
Taken together, our findings further suggest that handedness and language lateralization are ontogenetically independent, complex phenotypes (Ocklenburg et al., 2014). Relative independence of these phenotypes has also recently been concluded in terms of genetic background (Corballis, 2017) as well as in terms of neuroanatomy (Króliczak et al., 2016). Compared to genes involved in handedness ontogenesis, which mostly contribute to structural development, genes involved in language lateralization rather contribute to activity-dependent cognitive processes partly associated to mental and neurological disorders. When searching for overlapping genetic contributions to the ontogenesis of these two traits, our results indicate that particularly genes within the high level GO set of ‘biological regulation’ may represent promising candidate genes. Revealing further candidate genes for handedness and language lateralization will not only contribute to important insights into the development of hemispheric asymmetries, but also to a better understanding of disorders related to atypical lateralization, e.g., schizophrenia (Levchenko et al., 2014).
Author Contributions
JS performed data collection, analyzed data and wrote the manuscript, SL analyzed data, RK analyzed data, OG designed the study, and SO designed the study. All authors discussed the results and edited the manuscript.
Funding
This research was partly supported by the Mercator Research Center Ruhr (Project number GZ: An-2015-0061).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We acknowledge support by the DFG Open Access Publication Funds of the Ruhr-Universität Bochum.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpsyg.2017.01144/full#supplementary-material
FIGURE S1 | Full hierarchical GO set overview for genes involved in handedness ontogenesis.
FIGURE S2 | Full hierarchical GO set overview for genes involved in the ontogenesis of language lateralization.
FIGURE S3 | Distribution of raw p-values for all significant lower level GO sets involved in handedness and language lateralization.
FIGURE S4 | High level GO sets involved in handedness and language lateralization.
FIGURE S5 | Output of KEGG analysis for the TGF-beta signaling pathway. Genes involved in handedness ontogenesis are highlighted in red.
FIGURE S6 | Output of KEGG analysis for the axon guiding pathway. Genes involved in language lateralization are highlighted in red.
Footnotes
References
Afzelius, B. A., and Stenram, U. (2006). Prevalence and genetics of immotile-cilia syndrome and left-handedness. Int. J. Dev. Biol. 50, 571–573. doi: 10.1387/ijdb.052132ba
Annett, M. (1975). Hand preference and the laterality of cerebral speech. Cortex 11, 305–328. doi: 10.1016/S0010-9452(75)80024-4
Armour, J. A. L., Davison, A., and McManus, I. C. (2014). Genome-wide association study of handedness excludes simple genetic models. Heredity 112, 221–225. doi: 10.1038/hdy.2013.93
Arning, L., Ocklenburg, S., Schulz, S., Ness, V., Gerding, W. M., Hengstler, J. G., et al. (2013). PCSK6 VNTR polymorphism is associated with degree of handedness but not direction of handedness. PLoS ONE 8:e67251. doi: 10.1371/journal.pone.0067251
Arning, L., Ocklenburg, S., Schulz, S., Ness, V., Gerding, W. M., Hengstler, J. G., et al. (2015). Handedness and the X chromosome: the role of androgen receptor CAG-repeat length. Sci. Rep. 5:8325. doi: 10.1038/srep08325
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Asor, E., and Ben-Shachar, D. (2016). Gene environment interaction in periphery and brain converge to modulate behavioral outcomes: insights from the SP1 transient early in life interference rat model. World J. Psychiatry 6, 294–302. doi: 10.5498/wjp.v6.i3.294
Barber, A. D., Srinivasan, P., Joel, S. E., Caffo, B. S., Pekar, J. J., and Mostofsky, S. H. (2012). Motor “dexterity”? Evidence that left hemisphere lateralization of motor circuit connectivity is associated with better motor performance in children. Cereb. Cortex 22, 51–59. doi: 10.1093/cercor/bhr062
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300.
Bloss, C. S., Delis, D. C., Salmon, D. P., and Bondi, M. W. (2010). APOE genotype is associated with left-handedness and visuospatial skills in children. Neurobiol. Aging 31, 787–795. doi: 10.1016/j.neurobiolaging.2008.05.021
Brandler, W. M., Morris, A. P., Evans, D. M., Scerri, T. S., Kemp, J. P., Timpson, N. J., et al. (2013). Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 9:e1003751. doi: 10.1371/journal.pgen.1003751
Brandler, W. M., and Paracchini, S. (2014). The genetic relationship between handedness and neurodevelopmental disorders. Trends Mol. Med. 20, 83–90. doi: 10.1016/j.molmed.2013.10.008
Corballis, M. C. (2017). The evolution of lateralized brain circuits. Front. Psychol. 8:386. doi: 10.3389/fpsyg.2017.01021
Denny, K. (2009). Handedness and depression: evidence from a large population survey. Laterality 14, 246–255. doi: 10.1080/13576500802362869
Eriksson, N., Macpherson, J. M., Tung, J. Y., Hon, L. S., Naughton, B., Saxonov, S., et al. (2010). Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 6:e1000993. doi: 10.1371/journal.pgen.1000993
Francks, C., Maegawa, S., Lauren, J., Abrahams, B. S., Velayos-Baeza, A., Medland, S. E., et al. (2007). LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry 12, 1129–1139. doi: 10.1038/sj.mp.4002053
Frasnelli, E. (2013). Brain and behavioral lateralization in invertebrates. Front. Psychol. 4:939. doi: 10.3389/fpsyg.2013.00939
Gattere, G., Stojanovic-Perez, A., Monseny, R., Martorell, L., Ortega, L., Montalvo, I., et al. (2016). Gene-environment interaction between the brain-derived neurotrophic factor Val66Met polymorphism, psychosocial stress and dietary intake in early psychosis. Early Interv. Psychiatry doi: 10.1111/eip.12371 [Epub ahead of print].
Gene Ontology Consortium (2015). Gene ontology consortium: going forward. Nucleic Acids Res. 43, D1049–D1056. doi: 10.1093/nar/gku1179
Geschwind, N., and Behan, P. (1982). Left-handedness: association with immune disease, migraine, and developmental learning disorder. Proc. Natl. Acad. Sci. U.S.A. 79, 5097–5100. doi: 10.1073/pnas.79.16.5097
Güntürkün, O., and Ocklenburg, S. (2017). Ontogenesis of lateralization. Neuron 94, 249–263. doi: 10.1016/j.neuron.2017.02.045
Hampson, E., and Sankar, J. S. (2012). Hand preference in humans is associated with testosterone levels and androgen receptor gene polymorphism. Neuropsychologia 50, 2018–2025. doi: 10.1016/j.neuropsychologia.2012.04.027
Hirnstein, M., and Hugdahl, K. (2014). Excess of non-right-handedness in schizophrenia: meta-analysis of gender effects and potential biases in handedness assessment. Br. J. Psychiatry 205, 260–267. doi: 10.1192/bjp.bp.113.137349
Hubacek, J. A., Piper, B. J., Pikhart, H., Peasey, A., Kubinova, R., and Bobak, M. (2013). Lack of an association between left-handedness and APOE polymorphism in a large sample of adults: results of the Czech HAPIEE study. Laterality 18, 513–519. doi: 10.1080/1357650X.2012.715164
Ihara, A., Hirata, M., Fujimaki, N., Goto, T., Umekawa, Y., Fujita, N., et al. (2010). Neuroimaging study on brain asymmetries in situs inversus totalis. J. Neurol. Sci. 288, 72–78. doi: 10.1016/j.jns.2009.10.002
Jourquin, J., Duncan, D., Shi, Z., and Zhang, B. (2012). GLAD4U: deriving and prioritizing gene lists from PubMed literature. BMC Genomics 13(Suppl. 8):S20. doi: 10.1186/1471-2164-13-S8-S20
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Karlebach, G., and Francks, C. (2015). Lateralization of gene expression in human language cortex. Cortex 67, 30–36. doi: 10.1016/j.cortex.2015.03.003
Kennedy, D. N., O’Craven, K. M., Ticho, B. S., Goldstein, A. M., Makris, N., and Henson, J. W. (1999). Structural and functional brain asymmetries in human situs inversus totalis. Neurology 53, 1260–1265. doi: 10.1212/WNL.53.6.1260
Knaus, T. A., Silver, A. M., Kennedy, M., Lindgren, K. A., Dominick, K. C., Siegel, J., et al. (2010). Language laterality in autism spectrum disorder and typical controls: a functional, volumetric, and diffusion tensor MRI study. Brain Lang. 112, 113–120. doi: 10.1016/j.bandl.2009.11.005
Knecht, S., Drager, B., Deppe, M., Bobe, L., Lohmann, H., Floel, A., et al. (2000). Handedness and hemispheric language dominance in healthy humans. Brain 123(Pt 12), 2512–2518. doi: 10.1093/brain/123.12.2512
Króliczak, G., Piper, B. J., and Frey, S. H. (2016). Specialization of the left supramarginal gyrus for hand-independent praxis representation is not related to hand dominance. Neuropsychologia 93, 501–512. doi: 10.1016/j.neuropsychologia.2016.03.023
Levchenko, A., Davtian, S., Petrova, N., and Malashichev, Y. (2014). Sequencing of five left-right cerebral asymmetry genes in a cohort of schizophrenia and schizotypal disorder patients from Russia. Psychiatr. Genet. 24, 75–80. doi: 10.1097/YPG.0000000000000021
Logue, D. D., Logue, R. T., Kaufmann, W. E., and Belcher, H. M. E. (2015). Psychiatric disorders and left-handedness in children living in an urban environment. Laterality 20, 249–256. doi: 10.1080/1357650X.2014.961927
Lust, J. M., Geuze, R. H., van de Beek, C., Cohen-Kettenis, P. T., Bouma, A., and Groothuis, T. G. G. (2011). Differential effects of prenatal testosterone on lateralization of handedness and language. Neuropsychology 25, 581–589. doi: 10.1037/a0023293
Matsumoto, T., Kuriya, N., Akagi, T., Ohbu, K., Toyoda, O., Morita, J., et al. (1997). Handedness and laterality of the viscera. Neurology 49:1751. doi: 10.1212/WNL.49.6.1751
McManus, I. C. (1984). Genetics of handedness in relation to language disorder. Adv. Neurol. 42, 125–138.
McManus, I. C. (1985). Handedness, language dominance and aphasia: a genetic model. Psychol. Med. Monogr. Suppl. 8, 1–40. doi: 10.1017/S0264180100001879
McManus, I. C. (2010). Precisely wrong? The problems with the Jones and Martin genetic model of sex differences in handedness and language lateralisation. Cortex 46, 700–702. doi: 10.1016/j.cortex.2009.08.008
McManus, I. C., Davison, A., and Armour, J. A. L. (2013). Multilocus genetic models of handedness closely resemble single-locus models in explaining family data and are compatible with genome-wide association studies. Ann. N. Y. Acad. Sci. 1288, 48–58. doi: 10.1111/nyas.12102
McManus, I. C., Martin, N., Stubbings, G. F., Chung, E. M. K., and Mitchison, H. M. (2004). Handedness and situs inversus in primary ciliary dyskinesia. Proc. Biol. Sci. 271, 2579–2582. doi: 10.1098/rspb.2004.2881
Medland, S. E., Duffy, D. L., Spurdle, A. B., Wright, M. J., Geffen, G. M., Montgomery, G. W., et al. (2005). Opposite effects of androgen receptor CAG repeat length on increased risk of left-handedness in males and females. Behav. Genet. 35, 735–744. doi: 10.1007/s10519-005-6187-3
Medland, S. E., Duffy, D. L., Wright, M. J., Geffen, G. M., Hay, D. A., Levy, F., et al. (2009). Genetic influences on handedness: data from 25,732 Australian and Dutch twin families. Neuropsychologia 47, 330–337. doi: 10.1016/j.neuropsychologia.2008.09.005
Medland, S. E., Duffy, D. L., Wright, M. J., Geffen, G. M., and Martin, N. G. (2006). Handedness in twins: joint analysis of data from 35 samples. Twin Res. Hum. Genet. 9, 46–53. doi: 10.1375/183242706776402885
Mittwoch, U. (2008). Different gene expressions on the left and the right: a genotype/phenotype mismatch in need of attention. Ann. Hum. Genet. 72, 2–9. doi: 10.1111/j.1469-1809.2007.00402.x
Nowakowska, C., Sachs, G. S., Zarate, C. A., Marangell, L. B., Calabrese, J. R., Goldberg, J. F., et al. (2008). Increased rate of non-right-handedness in patients with bipolar disorder. J. Clin. Psychiatry 69, 866–867. doi: 10.4088/JCP.v69n0522g
Ocklenburg, S., Arning, L., Gerding, W. M., Epplen, J. T., Güntürkün, O., and Beste, C. (2013a). Cholecystokinin A receptor (CCKAR) gene variation is associated with language lateralization. PLoS ONE 8:e53643. doi: 10.1371/journal.pone.0053643
Ocklenburg, S., Arning, L., Gerding, W. M., Epplen, J. T., Güntürkün, O., and Beste, C. (2013b). FOXP2 variation modulates functional hemispheric asymmetries for speech perception. Brain Lang. 126, 279–284. doi: 10.1016/j.bandl.2013.07.001
Ocklenburg, S., Arning, L., Gerding, W. M., Hengstler, J. G., Epplen, J. T., Güntürkün, O., et al. (2015a). Left-right axis differentiation and functional lateralization: a haplotype in the methyltransferase encoding gene SETDB2 might mediate handedness in healthy adults. Mol. Neurobiol. 53, 6355–6361. doi: 10.1007/s12035-015-9534-2
Ocklenburg, S., Arning, L., Hahn, C., Gerding, W. M., Epplen, J. T., Güntürkün, O., et al. (2011). Variation in the NMDA receptor 2B subunit gene GRIN2B is associated with differential language lateralization. Behav. Brain Res. 225, 284–289. doi: 10.1016/j.bbr.2011.07.042
Ocklenburg, S., Beste, C., Arning, L., Peterburs, J., and Güntürkün, O. (2014). The ontogenesis of language lateralization and its relation to handedness. Neurosci. Biobehav. Rev. 43, 191–198. doi: 10.1016/j.neubiorev.2014.04.008
Ocklenburg, S., Beste, C., and Güntürkün, O. (2013c). Handedness: a neurogenetic shift of perspective. Neurosci. Biobehav. Rev. 37, 2788–2793. doi: 10.1016/j.neubiorev.2013.09.014
Ocklenburg, S., Güntürkün, O., Hugdahl, K., and Hirnstein, M. (2015b). Laterality and mental disorders in the postgenomic age–A closer look at schizophrenia and language lateralization. Neurosci. Biobehav. Rev. 59, 100–110. doi: 10.1016/j.neubiorev.2015.08.019
Ocklenburg, S., Korte, S. M., Peterburs, J., Wolf, O. T., and Güntürkün, O. (2016). Stress and laterality - the comparative perspective. Physiol. Behav. 164, 321–329. doi: 10.1016/j.physbeh.2016.06.020
Ocklenburg, S., Schmitz, J., Moinfar, Z., Moser, D., Klose, R., Lor, S., et al. (2017). Epigenetic regulation of lateralized fetal spinal gene expression underlies hemispheric asymmetries. Elife 6:e22784. doi: 10.7554/eLife.22784
Ocklenburg, S., Ströckens, F., and Güntürkün, O. (2013d). Lateralisation of conspecific vocalisation in non-human vertebrates. Laterality 18, 1–31. doi: 10.1080/1357650X.2011.626561
Ocklenburg, S., Westerhausen, R., Hirnstein, M., and Hugdahl, K. (2013e). Auditory hallucinations and reduced language lateralization in schizophrenia: a meta-analysis of dichotic listening studies. J. Int. Neuropsychol. Soc. 19, 410–418. doi: 10.1017/S1355617712001476
Papadatou-Pastou, M., Martin, M., Munafo, M. R., and Jones, G. V. (2008). Sex differences in left-handedness: a meta-analysis of 144 studies. Psychol. Bull. 134, 677–699. doi: 10.1037/a0012814
Pinel, P., Fauchereau, F., Moreno, A., Barbot, A., Lathrop, M., Zelenika, D., et al. (2012). Genetic variants of FOXP2 and KIAA0319/TTRAP/THEM2 locus are associated with altered brain activation in distinct language-related regions. J. Neurosci. 32, 817–825. doi: 10.1523/JNEUROSCI.5996-10.2012
Piper, B. J., Yasen, A. L., Taylor, A. E., Ruiz, J. R., Gaynor, J. W., Dayger, C. A., et al. (2013). Non-replication of an association of Apolipoprotein E2 with sinistrality. Laterality 18, 251–261. doi: 10.1080/1357650X.2012.660164
Robinson, K. J., Hurd, P. L., Read, S., and Crespi, B. J. (2016). The PCSK6 gene is associated with handedness, the autism spectrum, and magical ideation in a non-clinical population. Neuropsychologia 84, 205–212. doi: 10.1016/j.neuropsychologia.2016.02.020
Savitz, J., van der Merwe, L., Solms, M., and Ramesar, R. (2007). Lateralization of hand skill in bipolar affective disorder. Genes Brain Behav. 6, 698–705. doi: 10.1111/j.1601-183X.2006.00299.x
Scerri, T. S., Brandler, W. M., Paracchini, S., Morris, A. P., Ring, S. M., Richardson, A. J., et al. (2011). PCSK6 is associated with handedness in individuals with dyslexia. Hum. Mol. Genet. 20, 608–614. doi: 10.1093/hmg/ddq475
Scott, S. K., and McGettigan, C. (2013). Do temporal processes underlie left hemisphere dominance in speech perception? Brain Lang. 127, 36–45. doi: 10.1016/j.bandl.2013.07.006
Shiratori, H., and Hamada, H. (2014). TGFbeta signaling in establishing left-right asymmetry. Semin. Cell Dev. Biol. 32, 80–84. doi: 10.1016/j.semcdb.2014.03.029
Slevc, L. R., Martin, R. C., Hamilton, A. C., and Joanisse, M. F. (2011). Speech perception, rapid temporal processing, and the left hemisphere: a case study of unilateral pure word deafness. Neuropsychologia 49, 216–230. doi: 10.1016/j.neuropsychologia.2010.11.009
Somers, M., Aukes, M. F., Ophoff, R. A., Boks, M. P., Fleer, W., de Visser, K. C. L., et al. (2015). On the relationship between degree of hand-preference and degree of language lateralization. Brain Lang. 144, 10–15. doi: 10.1016/j.bandl.2015.03.006
Ströckens, F., Güntürkün, O., and Ocklenburg, S. (2013). Limb preferences in non-human vertebrates. Laterality 18, 536–575. doi: 10.1080/1357650X.2012.723008
Sun, T., Patoine, C., Abu-Khalil, A., Visvader, J., Sum, E., Cherry, T. J., et al. (2005). Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science 308, 1794–1798. doi: 10.1126/science.1110324
Tager-Flusberg, H. (2016). Risk factors associated with language in autism spectrum disorder: clues to underlying mechanisms. J. Speech Lang. Hear Res. 59, 143–154. doi: 10.1044/2015_JSLHR-L-15-0146
Thomas, C., Altenmüller, E., Marckmann, G., Kahrs, J., and Dichgans, J. (1997). Language processing in aphasia: changes in lateralization patterns during recovery reflect cerebral plasticity in adults. Electroencephalogr. Clin. Neurophysiol. 102, 86–97.
van Agtmael, T., Forrest, S. M., and Williamson, R. (2002). Parametric and non-parametric linkage analysis of several candidate regions for genes for human handedness. Eur. J. Hum. Genet. 10, 623–630. doi: 10.1038/sj.ejhg.5200851
Vuoksimaa, E., Koskenvuo, M., Rose, R. J., and Kaprio, J. (2009). Origins of handedness: a nationwide study of 30,161 adults. Neuropsychologia 47, 1294–1301. doi: 10.1016/j.neuropsychologia.2009.01.007
Wang, J., Duncan, D., Shi, Z., and Zhang, B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. doi: 10.1093/nar/gkt439
World Health Organization (1992). The ICD-10 Classification of Mental and Behavioural Disorders: Clinical Descriptions and Diagnostic Guidelines. Geneva: World Health Organization.
Zhang, B., Kirov, S., and Snoddy, J. (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748. doi: 10.1093/nar/gki475
Keywords: handedness, language lateralization, ontogenesis, gene ontology, asymmetry, genetics
Citation: Schmitz J, Lor S, Klose R, Güntürkün O and Ocklenburg S (2017) The Functional Genetics of Handedness and Language Lateralization: Insights from Gene Ontology, Pathway and Disease Association Analyses. Front. Psychol. 8:1144. doi: 10.3389/fpsyg.2017.01144
Received: 02 December 2016; Accepted: 22 June 2017;
Published: 06 July 2017.
Edited by:
David Peter Carey, Bangor University, United KingdomReviewed by:
James Danckert, University of Waterloo, CanadaChris McManus, University College London, United Kingdom
Diego Forero, Universidad Antonio Nariño, Colombia
Copyright © 2017 Schmitz, Lor, Klose, Güntürkün and Ocklenburg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Ocklenburg, c2ViYXN0aWFuLm9ja2xlbmJ1cmdAcnViLmRl
 Judith Schmitz
Judith Schmitz Stephanie Lor
Stephanie Lor Rena Klose
Rena Klose Onur Güntürkün
Onur Güntürkün Sebastian Ocklenburg
Sebastian Ocklenburg