- 1Genetics and Physiology of Microalgae, InBios/Phytosystems Research Unit, University of Liège, Liège, Belgium
- 2Redox Signaling Lab, VIB-VUB Center for Structural Biology, Vlaams Instituut voor Biotechnologie, Brussels, Belgium
- 3Messens Lab, Brussels Center for Redox Biology, Brussels, Belgium
- 4Structural Biology Brussels, Vrije Universiteit Brussel, Brussels, Belgium
- 5Departamento de Genética, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil
We review the newly classified ascorbate peroxidase–related (APX-R) proteins, which do not use ascorbate as electron donor to scavenge H2O2. We summarize recent discoveries on the function and the characterization of the APX-R protein of the green unicellular alga Chlamydomonas reinhardtii and the land plant Arabidopsis thaliana. Additionally, we conduct in silico analyses on the conserved MxxM motif, present in most of the APX-R protein in different organisms, which is proposed to bind copper. Based on these analyses, we discuss the similarities between the APX-R and the class III peroxidases.
Introduction
Ascorbate peroxidases (APXs) are heme-containing peroxidases classified as class I peroxidases. They detoxify H2O2 by oxidizing ascorbate into monodehydroascorbate and are exclusive to photosynthetic organisms. Although their roles have been known for years (Asada, 1992; de Montellano, 2010; Foyer and Noctor, 2011; Mittler and Poulos, 2005; Shigeoka et al., 2002), recent studies have reclassified APX enzymes on the basis of their amino acid sequences (Granlund et al., 2009; Lazzarotto et al., 2021a). APX enzymes are categorized into three groups: classic APX (Figure 1A); APX-related (APX-R) (Figure 1B), which lack only the amino acids necessary for ascorbate binding; and APX-like (APX-L), which lack all the amino acids required for ascorbate and heme binding, as well as those for peroxidase activity (Figure 1C) (Lazzarotto et al., 2021a). Crystal structures of the classic APX [examples: (Patterson and Poulos, 1995; Wada et al., 2003; Zhang et al., 2023a)] and APX-L (Lundberg et al., 2011) are available, whereas only structural predictions exist for the APX-R (Caccamo et al., 2023). The locations of critical amino acids are indicated in the protein structures of the three APX classes in Figure 1.
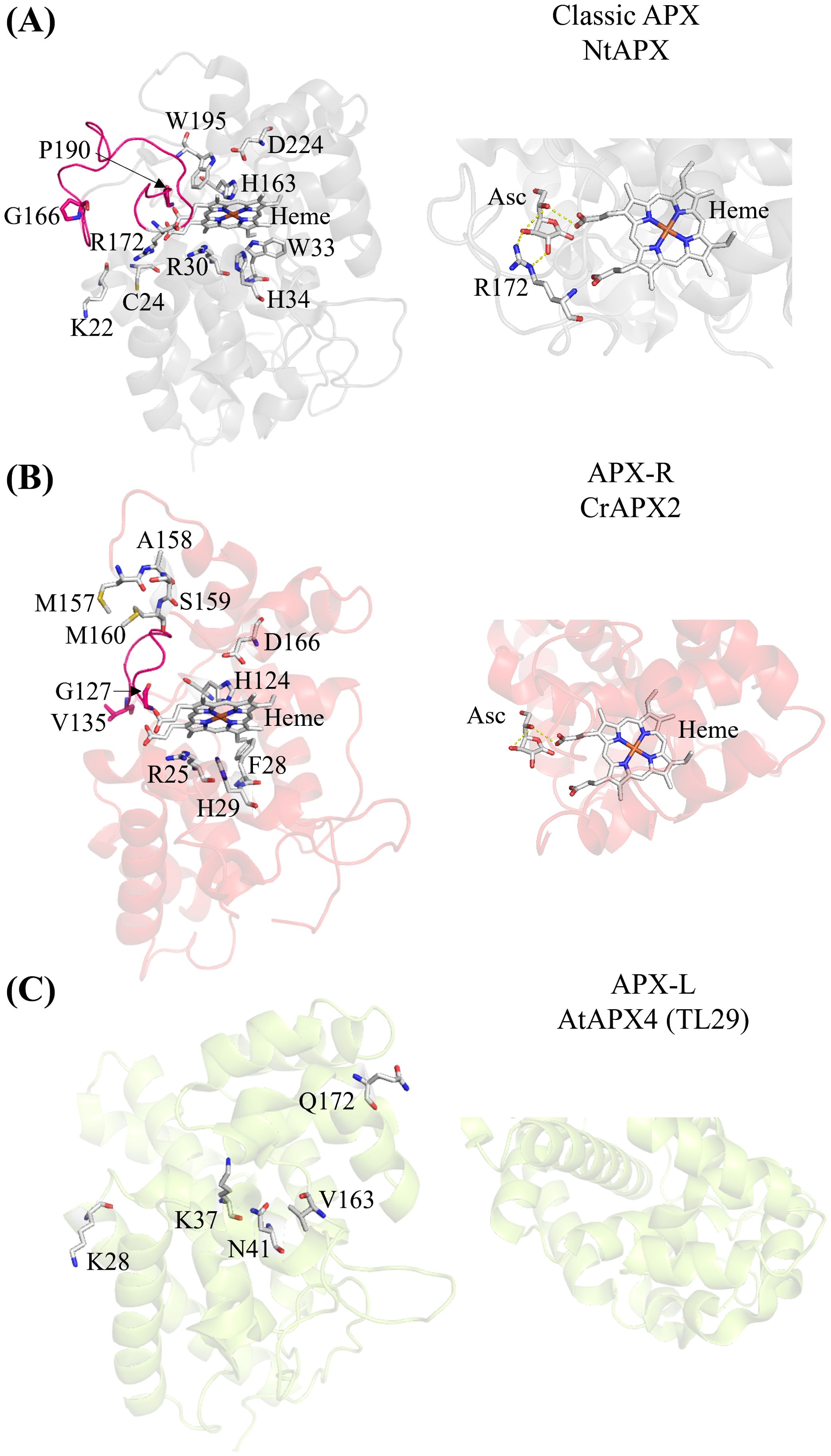
Figure 1. The three APX classes show structural differences. (A) Crystal structure of the chloroplastic APX from Nicotiana tabacum (Protein Data Bank (PDB) ID: 1IYN). The loop connecting two alpha helices present in the classic APX, spanning residues G166 to P190, is highlighted in pink. Key conserved amino acid residues for heme binding (H163), ascorbate binding (R172 with the contribution from K22 and C24), and catalytic activity (R30, W33, H34, H163, W195, and D224) are indicated (Lazzarotto et al., 2021a; Raven, 2003). The zoomed-in view of the predicted interaction between ascorbate (Asc), heme, and arginine (R) in classic NtAPX. (B) Predicted structure of the APX-R (APX2) from Chlamydomonas using AlphaFold2. The loop between G127 and V135 connecting two alpha helices highlighted in pink is shorter than the one of the classic APX (A). The MxxM (MASM in Chlamydomonas) is indicated. Conserved amino acid residues for heme binding (H124) and catalytic activity (R25, F28, H29, H124, and D166) are indicated (Lazzarotto et al., 2021a; Raven, 2003). The zoomed-in view shows the predicted interaction between ascorbate (Asc) and heme. Note the absence of arginine in the APX-R of Chlamydomonas. (C) Crystal structure of the APX4 (TL29; APX-L) of Arabidopsis (PDB ID: 3RRW). The conserved amino acids differ from the classic APX are indicated. (A) heme binding (V163), the catalytic activity (K28, K37, N41) and the ascorbate binding (Q172) (Granlund et al., 2009). The zoomed-in view of APX-L (AtAPX4) reveals that there are no amino acid residues involved in binding ascorbate or heme. The prediction of ascorbate binding with arginine was aligned with the crystal structure of APX and ascorbate of Glycine max (PDB ID: 1OAF) using PyMol 2.5.2 (Janson et al., 2017).
Classical APX, found in chloroplasts, mitochondria, peroxisomes and the cytosol, function as antioxidant enzymes and redox regulators, and the cytosol function as antioxidant enzyme and redox regulator. These enzymes will not be discussed here [see recent reviews by Foyer and Hanke (2022) and Yoshimura and Ishikawa (2024)].
For APX-L, data are available for both Arabidopsis thaliana (hereafter Arabidopsis) and the green alga Chlamydomonas reinhardtii (hereafter Chlamydomonas). In Arabidopsis, APX-L (AtAPX4) is associated with photosystem II and likely involved in photosystem II photoprotection (Granlund et al., 2009), whereas, in Chlamydomonas, APX-L (CrAPX4) is crucial for cell protection under very high light conditions (Kuo et al., 2020). Granlund and colleagues (Granlund et al., 2009) found that recombinant AtAPX4 lacks ascorbate activity and does not bind heme. Additionally, peroxidase activity was unchanged between the wild-type and the AtAPX4 knockout line, suggesting that the protein does not play a stress response role. However, other studies on apx4 mutants of both Arabidopsis (Wang et al., 2014) and Chlamydomonas (Kuo et al., 2020) reported higher H2O2 levels, suggesting a H2O2-scavenging role (Li, 2023).
For APX-R, the function has been recently described for both Chlamydomonas and Arabidopsis.
APX-R in Chlamydomonas
In Chlamydomonas, APX2 is the APX-R protein that has been studied both in vitro and in vivo. Caccamo and co-workers (Caccamo et al., 2023) found that recombinant APX2 can reduce H2O2 without ascorbate, although the in vivo electron donor remains unknown. By analyzing the predicted structure of APX2 and comparing it with APX-R proteins from other organisms, they identified a conserved MxxM or MxxH motif (Figure 1B) typically associated with metal binding, such as copper or silver (Rubino et al., 2010). APX2 expressed in E. coli was shown to bind copper. The authors also noted that the heme-covering loop in classic APX differs in size (Figure 1A), suggesting potential protein–protein interactions.
1H-Nuclear Magnetic Resonance (NMR) analyses were performed to explore possible interactions between recombinant APX2 and plastocyanin, a copper-containing protein involved in electron transfer during photosynthesis. Plastocyanin, located at the luminal side of the chloroplast, is likely to interact with APX2, which has a twin-arginine-translocator motif specific for the lumen (Caccamo et al., 2023). Although no direct interaction was observed in vitro, the findings suggest that APX2 might affect copper insertion into plastocyanin. Indeed, Chlamydomonas mutants lacking the APX2 protein display lower plastocyanin levels compared to the wild type, impacting electron transport and the redox state of photosystem I during photosynthesis (Caccamo et al., 2024). This difference is absent under copper-deficient conditions, indicating that Chlamydomonas apx2 mutants can switch to using the iron-containing protein cytochrome c6 as an alternative electron carrier. This result supports the link between the APX2 protein and plastocyanin.
These studies lay the foundation for understanding APX-R protein structure and function, but further research is needed to confirm the predicted structure and elucidate APX-R’s role, especially concerning the MxxM motif. Therefore, we conducted in silico analyses based on the APX-R sequences of Chlamydomonas and Arabidopsis to examine the presence of the MxxM motif among the green photosynthetic eukaryotes (Figure 2). Interestingly, the MxxM motif is widely present among the APX-R proteins in algae and plants, except for some sequences (Figure 2A). Some algal sequences, such as those of Chlorella sorokiniana (CsorAPxR), Bathycoccus prasinos (BprAPxR01), Ostreococcus tauri (OtAPxR), and Ostreococcus lucimarinus (OlAPxR), have a histidine instead of a second methionine (MxxH), which can also bind metals as described by Caccamo et al. (2023). Histidine is known to bind copper in various proteins, such as the pocket binding site in plastocyanin (Hill et al., 1996; Zhang et al., 2023b) or the P-type Adenosine Tri Phosphatase (ATPase) copper transporter in the thylakoid (PAA2 in Arabidopsis) (Abdel-Ghany et al., 2005).
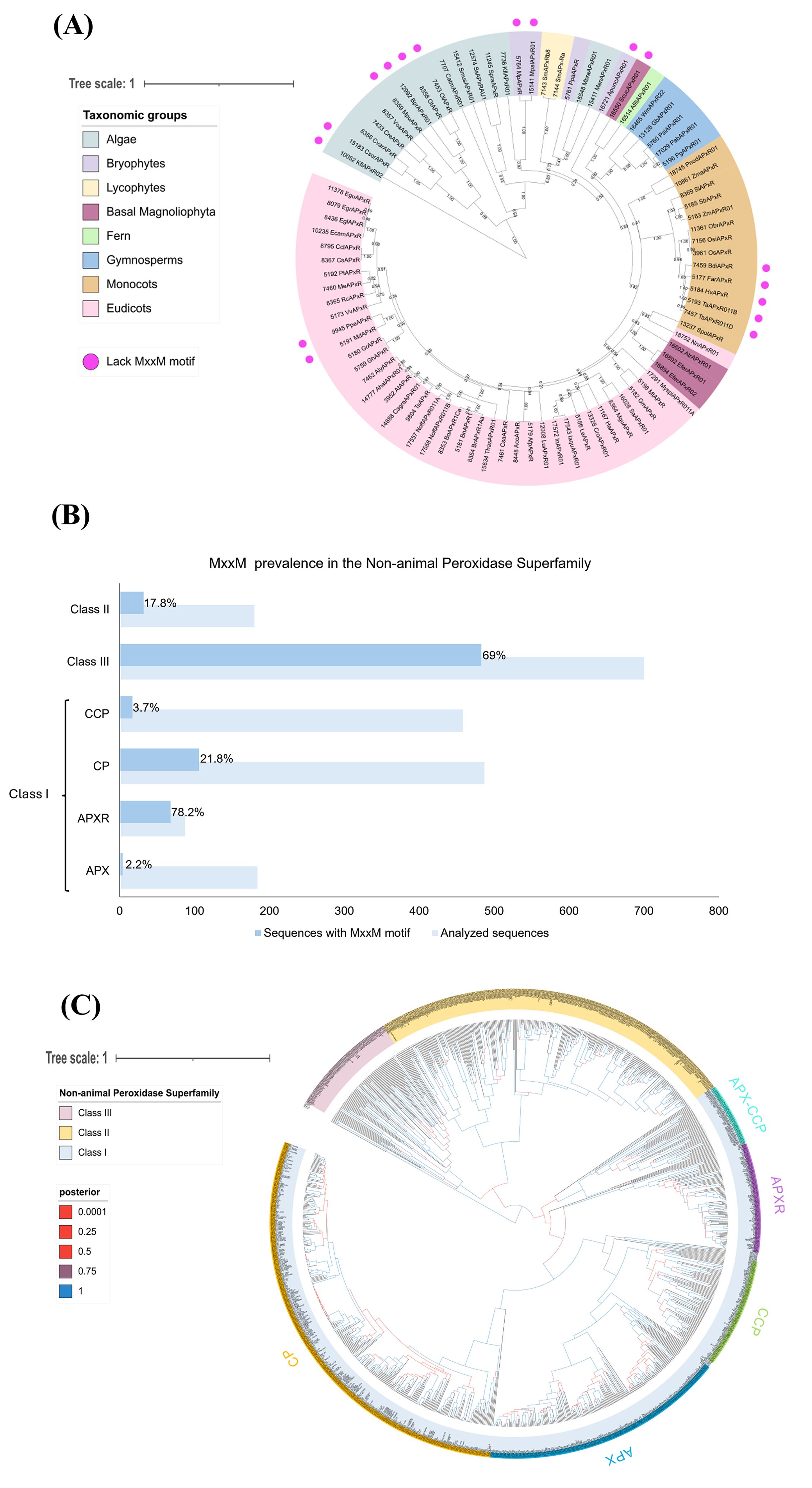
Figure 2. The MxxM motif is predominantly found in APX-R and class III peroxidases. (A) The analysis shows the phylogenetic relationship of 86 APX-R sequences, reconstructed through Bayesian inference. Out of these, 17 lack the MxxM/H motif, as indicated by pink dots. The absence of this motif occurs across sequences belonging to Algae, bryophytes, basal Magnoliophyta, monocots, and eudicots, indicating that this feature is not related to a specific taxonomic group. Posterior probabilities are indicated. (B) The graph shows that the MxxM motif is predominantly present in the class I APX-R and class III peroxidases. A total of 2,096 protein sequences belonging to the superfamily were examined for the presence of MxxM. In the figure, the light blue bars represent the total number of sequences analyzed for each family, whereas the darker blue bars indicate the sequences containing the MxxM motif. The percentage of sequences with the motif is displayed within the bars for each family. Protein sequences in FASTA format were analyzed for the degenerate motif MxxM using R with the Biostrings package. Sequence headers were parsed to extract labels via regular expressions, enabling grouping by predefined categories. The gregexpr function identified and quantified motif occurrences across sequences, which were then aggregated by label. The results, including total counts and percentage of sequences containing the motif, were exported as CSV files. CCP, cytochrome c peroxidase; CP, catalase peroxidase; APX-R, ascorbate peroxidase–related; and APX, ascorbate peroxidase. (C) The phylogenetic relationship between class I, class II, and class III peroxidases of the non-animal superfamily was reconstructed using the Bayesian method. A total of 1,102 protein sequences were included in the analysis, and ambiguous positions were removed from the alignment, with exception to amino acids implicated in the loop and in the MxxM domains. Classes I, II, and III are highlighted in blue, yellow, and pink, respectively. Class I families (CP, CCP, APX, and APX-CCP) are indicated. The posterior probabilities are shown according to the following color scheme: blue branches indicate posterior probabilities of 0.9 to 1.0; purple branches indicate posterior probabilities of 0.75 to 0.89; red branches indicate posterior probabilities inferior to 0.6. The phylogenetic analyses presented in (A, C) were reconstructed using conserved domains of protein sequences retrieved from RedOxiBase (Savelli et al., 2019) by Bayesian inference using BEAST (Vaughan et al., 2014). After manual inspection of the alignments, 86 sequences and 228 sites were used in the analysis (A) and 1,102 sequences and 336 sites in (C). The best fit model of amino acid replacement was Le-Gascuel (LG) with invariable sites and gamma-distributed rates, which was selected after analyses performed on ProtTest (Abascal et al., 2005). The birth and death model was selected as tree prior, and 50,000,000 generations were performed with Markov chain Monte Carlo algorithm (MCMC) (Gilks, 2005) to evaluate posterior distributions. Convergence was verified with Tracer (Rambaut et al., 2018), and the consensus trees were generated using TreeAnnotator, part of the BEAST package. The resulting trees were analyzed and edited using FigTree v.1.4.3 (http://tree.bio.ed.ac.uk/software/figtree) and iTOL (https://itol.embl.de/).
Conversely, some sequences of plants highlighted in bryophytes, basal Magnoliophyta, monocots and eudicots, lack both methionine and histidine, suggesting that these proteins might not be involved in copper sensing. Figure 2B shows the prevalence of the MxxM motif in non-animal peroxidase superfamily, which includes three classes. The class I peroxidase family contains APX-Rs together with the classic APXs (found in green photosynthetic eukaryotes), catalase peroxidases (CP) (mainly found in bacteria), and the cytochrome c peroxidases (CCP) (found in all organisms containing mitochondria except plants). Class II includes lignin peroxidases (LiPs), manganese peroxidases (MnPs), and versatile peroxidases (VPs) found only in fungi. Class III peroxidases, found in plants, are multifunctional proteins involved in pathways such as auxin metabolism, cell wall elongation and stiffening, and pathogen protection (Mbadinga Mbadinga et al., 2020). Their phylogenetic relationship is presented in Figure 2C.
Notably, the MxxM motif is highly prevalent in the APX-R, present in 78.2% in 90 sequences analyzed, compared to only 2.2% in 180 classic APX sequences (Figure 2B). Additionally, the MxxM motif is found in class III peroxidases with 69% in 700 sequences. This analysis could provide insights into the function of APX-R, particularly in relation to the role of the MxxM motif, by comparing it to class III peroxidases.
APX-R in land plants: focus on Arabidopsis
Because the initial identification of APX-R as a distinct peroxidase family, significant efforts have been made to clarify its role in plant antioxidant metabolism. Preliminary in silico analyses indicated the presence of chloroplast-targeted peptides in all examined plant APX-R proteins (Lazzarotto et al., 2011). This was later confirmed through transient expression studies in rice (Oryza sativa) and Arabidopsis protoplasts, as well as in transgenic Arabidopsis overexpression lines. Proteomic analyses further identified APX-R in the stroma, plastoglobuli, and thylakoid membrane, validating its subcellular localization (Lazzarotto et al., 2021b).
In Arabidopsis, the gene encoding APX-R was initially annotated as ASCORBATE PEROXIDASE6 (APX6), suggesting that it was a putative cytosolic APX gene. However, phylogenetic and biochemical evidence later demonstrated that APX-R is not part of the APX family. Although functions as a peroxidase, it does not use ascorbate as an electron donor (Lazzarotto et al., 2021a, 2021b).
Functional characterization of two knockout mutants (apx6-1 and apx6-3) highlights the critical role of APX-R in oxidative protection during seed development and germination. Seeds lacking APX-R (apx6-1) show elevated levels of reactive oxygen species (ROS), increased oxidative damage, and reduced germination rates, particularly under stress conditions such as osmotic, salt, or heat stress. Metabolic profiling of apx6-1 seeds revealed alterations in tricarboxylic acid cycle activity, changes in amino acid levels, and increased metabolism of abscisic acid (ABA) and auxin (Chen et al., 2014).
Studies on Arabidopsis overexpressing lines have shown that APX-R-YFP accumulates in seeds and during early development stages, underscoring its crucial role in seed metabolism and germination. APX-R is believed to protect seeds from oxidative damage during desiccation and early germination by modulating ROS and hormone signaling (Chen et al., 2014). As the plant transitions from skotomorphogenic to photomorphogenic development, APX-R-YFP undergoes degradation in most plant tissues. However, APX-R-YFP has also been observed in plant roots and stomata, suggesting that its stability may be influenced by plastid type, tissue specificity, and developmental stage (Lazzarotto et al., 2021b).
The presence of APX-R in plastoglobuli-derived samples suggests its involvement in processes related to these cellular structures, which are linked to plastid transitions during de-etiolation, senescence, and plant responses to abiotic stresses (van Wijk and Kessler, 2017). Recent studies have associated APX-R with plant senescence, showing that the Arabidopsis APX-R gene is induced in aging leaves and in response to senescence-promoting stimuli such as ABA, extended darkness, and osmotic stress. Knockout mutants exhibited early developmental senescence and increased sensitivity to dark stress (Chen et al., 2021).
Additionally, Arabidopsis APX-R mRNA has been identified as a potential target of miR398, a key regulator of plant copper distribution. The expression of miR398 is induced during copper deficiency and is controlled by the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE7 (SPL7) transcription factor, which binds to the GTAC motif found in the miR398 promoter (Yamasaki et al., 2009). In AtSPL7 mutants, APX-R levels are higher compared to those of the wild-type plants, and this difference is further increased under copper deficiency conditions (Chen et al., 2021). These data strengthen the link between APX-R and copper, which could be relevant for understanding the role of copper in APX-R catalytic activity.
Discussion
In this mini-review, we summarize the latest findings on the newly classified APX-R protein in algae and plants. Our comparison highlights a key common feature: the presence of the MxxM motif and its association with copper. In Chlamydomonas, this relation is linked to the electron carrier plastocyanin (Caccamo et al., 2024), a major copper storage protein in the green alga. In Arabidopsis, the APX-R-copper relation has been proposed to be dependent on copper concentrations (Chen et al., 2021), with both APX-R and copper playing roles in the senescence process (Chen et al., 2021; Hao et al., 2022). In Arabidopsis, copper is important during senescence, activating the plantacyanin-senescence associated gene (PCY-SAG14) module, with copper being redistributed from plastocyanin in the chloroplast to the cell membrane (Hao et al., 2022).
Additionally, the predicted structure of APX-R (Figure 1) shows another difference from classic APX: a distinctive smaller loop facing the heme group. This could also imply a possible link to metal sensitivity. Interestingly, the MxxM motif is also present in the class III peroxidases (Figure 2), suggesting that these proteins might bind metals as well. Class III peroxidases, found in Streptophyta, are involved in H2O2 scavenging and various processes such as germination, senescence, lignification, cell elongation (Shigeto and Tsutsumi, 2016). Predominantly, class III peroxidases can use lignin precursors, and auxin as electron donors (Hiraga et al., 2001). Our studies on the APX-R in Chlamydomonas (Caccamo et al., 2023) and Arabidopsis (Lazzarotto et al., 2021b) demonstrated that in vitro this protein prefers phenolic compounds like guaiacol and pyrogallol over ascorbate. Moreover, the APX-R of Arabidopsis has been showed to participate in seed germination and senescence, suggesting similarities between these two classes of peroxidases (Chen et al., 2021, 2014; Lazzarotto et al., 2021b).
There is evidence that class III peroxidases in plants respond to elevated copper levels by promoting lignin biosynthesis, which help to protect from metal stress (Ali et al., 2006; Chen et al., 2002; Lequeux et al., 2010; Lin et al., 2005; Tugbaeva et al., 2022). However, the presence and role of the MxxM motif in class III peroxidases remains unexplored, making it an exciting topic for further exploration. This raises questions whether APX-R protects cells through direct H2O2 scavenging, copper-binding, or a combination of both. Additionally, understanding why some sequences lack the MxxM (or MxxH) motif could help elucidate additional roles of APX-R (Figure 2A).
Additionally, information on APX-R gene expression in plants under various stress conditions provides valuable insights into its role (Chen et al., 2021, 2014; Lazzarotto et al., 2021a, 2011; Tyagi et al., 2020; Verma et al., 2022). In rice, APX-R expression is upregulated in response to aluminum stress, water deficiency, and 24 h-cold stress. In Brassica rapa and B. juncea, APX-R responds to heat and drought stresses. In Triticum aestivum, transcriptomic profiles of six APX-R genes show involvement in developmental stages and various biotic and abiotic stresses (e.g., fungal infection, salt, heat, and drought stresses). Moreover, Tyagi and co-authors observed that APX-R mRNA could be miRNA target (Tyagi et al., 2020). Similar regulation has been already suggested and discussed for Arabidopsis. In crops, targeting APX-R activation could be of interest. However, APX-R is a single-copy gene (Dunand et al., 2011), and overexpression in Arabidopsis leads to APX6 degradation (Lazzarotto et al., 2021b), whereas overexpressing in rice showed no alterations (Lazzarotto et al., 2011).
Further investigations are needed to explore the diversity of APX-R functions in plants and algae, the significance of the MxxM motif, and its role in copper binding. The analyses presented here offer an intriguing starting point for future research.
Author contributions
AC: Writing – original draft, Writing – review & editing. FL: Writing – review & editing. MM-P: Funding acquisition, Writing – review & editing. JM: Funding acquisition, Supervision, Writing – review & editing. CR: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. MM-P is supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). CR and JM acknowledge Fonds de la Recherche Scientifique—the Research Foundation Flanders—Excellence of Science project number 30829584. CR acknowledges F.R.S.-FNRS (CDR J.0149.23). JM acknowledges VIB.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abascal, F., Zardoya, R., Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. doi: 10.1093/bioinformatics/bti263
Abdel-Ghany, S. E., Muller-Moule, P., Niyogi, K. K., Pilon, M., Shikanai, T. (2005). Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 17, 1233–1251. doi: 10.1105/tpc.104.030452
Ali, M. B., Singh, N., Shohael, A. M., Hahn, E. J., Paek, K.-Y. (2006). Phenolics metabolism and lignin synthesis in root suspension cultures of Panax ginseng in response to copper stress. Plant Sci. 171, 147–154. doi: 10.1016/j.plantsci.2006.03.005
Asada, K. (1992). Ascorbate peroxidase – a hydrogen peroxide-scavenging enzyme in plants. Physiologia Plantarum 85, 235–241. doi: 10.1111/j.1399-3054.1992.tb04728.x
Caccamo, A., Vega de Luna, F., Misztak, A. E., Pyr dit Ruys, S., Vertommen, D., Cardol, P., et al. (2024). APX2 is an ascorbate peroxidase–related protein that regulates the levels of plastocyanin in chlamydomonas. Plant Cell Physiol 65, 644–656. doi: 10.1093/pcp/pcae019
Caccamo, A., Vega de Luna, F., Wahni, K., Volkov, A. N., Przybyla-Toscano, J., Amelii, A., et al. (2023). Ascorbate peroxidase 2 (APX2) of chlamydomonas binds copper and modulates the copper insertion into plastocyanin. Antioxidants 12 (11), 1946. doi: 10.3390/antiox12111946
Chen, C., Galon, Y., Rahmati Ishka, M., Malihi, S., Shimanovsky, V., Twito, S., et al. (2021). ASCORBATE PEROXIDASE6 delays the onset of age-dependent leaf senescence. Plant Physiol. 185, 441–456. doi: 10.1093/plphys/kiaa031
Chen, C., Letnik, I., Hacham, Y., Dobrev, P., Ben-Daniel, B. H., Vankova, R., et al. (2014). ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol. 166, 370–383. doi: 10.1104/pp.114.245324
Chen, E.-L., Chen, Y.-A., Chen, L.-M., Liu, Z.-H. (2002). Effect of copper on peroxidase activity and lignin content in Raphanus sativus. Plant Physiol. Biochem. 40, 439–444. doi: 10.1016/S0981-9428(02)01392-X
de Montellano, P. R. O. (2010). “Catalytic mechanisms of heme peroxidases,” in Biocatalysis based on heme peroxidases: peroxidases as potential industrial biocatalysts. Eds. Torres, E., Ayala, M. (Springer Berlin Heidelberg, Berlin, Heidelberg), 79–107.
Dunand, C., Mathe, C., Lazzarotto, F., Margis, R., Margis-Pinheiro, M. (2011). Ascorbate peroxidase-related (APx-R) is not a duplicable gene. Plant Signal Behav. 6, 1908–1913. doi: 10.4161/psb.6.12.18098
Foyer, C. H., Hanke, G. (2022). ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J. 111, 642–661. doi: 10.1111/tpj.15856
Foyer, C. H., Noctor, G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18. doi: 10.1104/pp.110.167569
Gilks, W. R. (2005). Markov chain monte carlo. Encyclopedia Biostatistics. doi: 10.1002/0470011815.b2a14021
Granlund, I., Storm, P., Schubert, M., Garcia-Cerdan, J. G., Funk, C., Schroder, W. P. (2009). The TL29 protein is lumen located, associated with PSII and not an ascorbate peroxidase. Plant Cell Physiol. 50, 1898–1910. doi: 10.1093/pcp/pcp134
Hao, C., Yang, Y., Du, J., Deng, X. W., Li, L. (2022). The PCY-SAG14 phytocyanin module regulated by PIFs and miR408 promotes dark-induced leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 119 (3), e2116623119. doi: 10.1073/pnas.2116623119
Hill, K. L., Hassett, R., Kosman, D., Merchant, S. (1996). Regulated copper uptake in Chlamydomonas reinhardtii in response to copper availability. Plant Physiol. 112, 697–704. doi: 10.1104/pp.112.2.697
Hiraga, S., Sasaki, K., Ito, H., Ohashi, Y., Matsui, H. (2001). A large family of class III plant peroxidases. Plant Cell Physiol. 42, 462–468. doi: 10.1093/pcp/pce061
Janson, G., Zhang, C., Prado, M. G., Paiardini, A. (2017). PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33, 444–446. doi: 10.1093/bioinformatics/btw638
Kuo, E. Y., Cai, M. S., Lee, T. M. (2020). Ascorbate peroxidase 4 plays a role in the tolerance of Chlamydomonas reinhardtii to photo-oxidative stress. Sci. Rep. 10, 13287. doi: 10.1038/s41598-020-70247-z
Lazzarotto, F., Menguer, P. K., Del-Bem, L. E., Zamocky, M., Margis-Pinheiro, M. (2021a). Ascorbate peroxidase neofunctionalization at the origin of APX-R and APX-L: evidence from basal archaeplastida. Antioxidants (Basel) 10 (4), 597. doi: 10.3390/antiox10040597
Lazzarotto, F., Teixeira, F. K., Rosa, S. B., Dunand, C., Fernandes, C. L., de Vasconcelos Fontenele, A., et al. (2011). Ascorbate peroxidase-related (APx-R) is a new heme-containing protein functionally associated with ascorbate peroxidase but evolutionarily divergent. New Phytol. 191, 234–250. doi: 10.1111/j.1469-8137.2011.03659.x
Lazzarotto, F., Wahni, K., Piovesana, M., Maraschin, F., Messens, J., Margis-Pinheiro, M. (2021b). Arabidopsis APx-R is a plastidial ascorbate-independent peroxidase regulated by photomorphogenesis. Antioxidants (Basel) 10 (1), 65. doi: 10.3390/antiox10010065
Lequeux, H., Hermans, C., Lutts, S., Verbruggen, N. (2010). Response to copper excess in Arabidopsis thaliana: Impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 48, 673–682. doi: 10.1016/j.plaphy.2010.05.005
Li, S. (2023). Novel insight into functions of ascorbate peroxidase in higher plants: More than a simple antioxidant enzyme. Redox Biol. 64, 102789. doi: 10.1016/j.redox.2023.102789
Lin, C.-C., Chen, L.-M., Liu, Z.-H. (2005). Rapid effect of copper on lignin biosynthesis in soybean roots. Plant Sci. 168, 855–861. doi: 10.1016/j.plantsci.2004.10.023
Lundberg, E., Storm, P., Schroder, W. P., Funk, C. (2011). Crystal structure of the TL29 protein from Arabidopsis thaliana: an APX homolog without peroxidase activity. J. Struct. Biol. 176, 24–31. doi: 10.1016/j.jsb.2011.07.004
Mbadinga Mbadinga, D. L., Li, Q., Ranocha, P., Martinez, Y., Dunand, C. (2020). Global analysis of non-animal peroxidases provides insights into the evolution of this gene family in the green lineage. J. Exp. Bot. 71, 3350–3360. doi: 10.1093/jxb/eraa141
Mittler, R., Poulos, T. L. (2005). Ascorbate peroxidase. Antioxidants reactive oxygen species Plants, 87–100. doi: 10.1002/9780470988565
Patterson, W. R., Poulos, T. L. (1995). Crystal structure of recombinant pea cytosolic ascorbate peroxidase. Biochemistry 34, 4331–4341. doi: 10.1021/bi00013a023
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Systematic Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Raven, E. L. (2003). Understanding functional diversity and substrate specificity in haem peroxidases: what can we learn from ascorbate peroxidase? Nat. Prod Rep. 20, 367–381. doi: 10.1039/b210426c
Rubino, J. T., Riggs-Gelasco, P., Franz, K. J. (2010). Methionine motifs of copper transport proteins provide general and flexible thioether-only binding sites for Cu(I) and Ag(I). J. Biol. Inorg Chem. 15, 1033–1049. doi: 10.1007/s00775-010-0663-9
Savelli, B., Li, Q., Webber, M., Jemmat, A. M., Robitaille, A., Zamocky, M., et al. (2019). RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 26, 101247. doi: 10.1016/j.redox.2019.101247
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. doi: 10.1093/jexbot/53.372.1305
Shigeto, J., Tsutsumi, Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytol. 209, 1395–1402. doi: 10.1111/nph.13738
Tugbaeva, A., Ermoshin, A., Wuriyanghan, H., Maleva, M., Borisova, G., Kiseleva, I. (2022). Copper stress enhances the lignification of axial organs in zinnia elegans. Horticulturae 8, 558. doi: 10.3390/horticulturae8060558
Tyagi, S., Shumayla, Verma, P. C., Singh, K., Upadhyay, S. K. (2020). Molecular characterization of ascorbate peroxidase (APX) and APX-related (APX-R) genes in Triticum aestivum L. Genomics 112, 4208–4223. doi: 10.1016/j.ygeno.2020.07.023
van Wijk, K. J., Kessler, F. (2017). Plastoglobuli: plastid microcompartments with integrated functions in metabolism, plastid developmental transitions, and environmental adaptation. Annu. Rev. Plant Biol. 68, 253–289. doi: 10.1146/annurev-arplant-043015-111737
Vaughan, T. G., Kuhnert, D., Popinga, A., Welch, D., Drummond, A. J. (2014). Efficient Bayesian inference under the structured coalescent. Bioinformatics 30, 2272–2279. doi: 10.1093/bioinformatics/btu201
Verma, D., Upadhyay, S. K., Singh, K. (2022). Characterization of APX and APX-R gene family in Brassica juncea and B. rapa for tolerance against abiotic stresses. Plant Cell Rep. 41, 571–592. doi: 10.1007/s00299-021-02726-0
Wada, K., Tada, T., Nakamura, Y., Ishikawa, T., Yabuta, Y., Yoshimura, K., et al. (2003). Crystal Structure of Chloroplastic Ascorbate Peroxidase from Tobacco Plants and Structural Insights into its Instability. J. Biochem. 134, 239–244. doi: 10.1093/jb/mvg136
Wang, Y. Y., Hecker, A. G., Hauser, B. A. (2014). The APX4 locus regulates seed vigor and seedling growth in Arabidopsis thaliana. Planta 239, 909–919. doi: 10.1007/s00425-014-2025-2
Yamasaki, H., Hayashi, M., Fukazawa, M., Kobayashi, Y., Shikanai, T. (2009). SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in arabidopsis. Plant Cell 21, 347–361. doi: 10.1105/tpc.108.060137
Yoshimura, K., Ishikawa, T. (2024). Physiological function and regulation of ascorbate peroxidase isoforms. J. Exp. Bot. 75, 2700–2715. doi: 10.1093/jxb/erae061
Zhang, B., Lewis, J. A., Vermerris, W., Sattler, S. E., Kang, C. (2023a). A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids. Plant Physiol. 192, 102–118. doi: 10.1093/plphys/kiac604
Keywords: Chlamydomonas, Arabidopsis, APX-R, H2O2, phylogeny, MxxM motif, copper
Citation: Caccamo A, Lazzarotto F, Margis-Pinheiro M, Messens J and Remacle C (2024) The ascorbate peroxidase–related protein: insights into its functioning in Chlamydomonas and Arabidopsis. Front. Plant Sci. 15:1487328. doi: 10.3389/fpls.2024.1487328
Received: 27 August 2024; Accepted: 17 September 2024;
Published: 09 October 2024.
Edited by:
Yanjie Xie, Nanjing Agricultural University, ChinaReviewed by:
Quan Gu, Hefei University, ChinaCopyright © 2024 Caccamo, Lazzarotto, Margis-Pinheiro, Messens and Remacle. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claire Remacle, c.remacle@uliege.be; Joris Messens, joris.messens@vub.be
†ORCID: Anna Caccamo, orcid.org/0000-0002-3147-313X
Fernanda Lazzarotto, orcid.org/0000-0003-3257-5952
Marcia Margis-Pinheiro, orcid.org/0000-0001-5356-3497
Joris Messens, orcid.org/0000-0002-2128-8264
Claire Remacle, orcid.org/0000-0002-5016-9547
 Anna Caccamo1,2,3,4†
Anna Caccamo1,2,3,4† Marcia Margis-Pinheiro
Marcia Margis-Pinheiro Joris Messens
Joris Messens Claire Remacle
Claire Remacle