- 1School of Biology and Food Engineering, Changshu Institute of Technology, Changshu, Suzhou, Jiangsu, China
- 2Pharma Technology A/S, Køge, Denmark
- 3State Key Laboratory of Dampness Syndrome of Chinese Medicine, The Second Affiliated Hospital of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong, China
- 4Second Clinical Medical College, Guangzhou University of Chinese Medicine, Guangzhou, China
- 5Guangdong-Hong Kong-Macau Joint Lab on Chinese Medicine and Immune Disease Research, Guangzhou, Guangdong, China
An increasing population, climate change, and diminishing natural resources present severe threats to global food security, with traditional breeding and genetic engineering methods often falling short in addressing these rapidly evolving challenges. CRISPR/Cas systems have emerged as revolutionary tools for precise genetic modifications in crops, offering significant advancements in resilience, yield, and nutritional value, particularly in staple crops like rice and maize. This review highlights the transformative potential of CRISPR/Cas technology, emphasizing recent innovations such as prime and base editing, and the development of novel CRISPR-associated proteins, which have significantly improved the specificity, efficiency, and scope of genome editing in agriculture. These advancements enable targeted genetic modifications that enhance tolerance to abiotic stresses as well as biotic stresses. Additionally, CRISPR/Cas plays a crucial role in improving crop yield and quality by enhancing photosynthetic efficiency, nutrient uptake, and resistance to lodging, while also improving taste, texture, shelf life, and nutritional content through biofortification. Despite challenges such as off-target effects, the need for more efficient delivery methods, and ethical and regulatory concerns, the review underscores the importance of CRISPR/Cas in addressing global food security and sustainability challenges. It calls for continued research and integration of CRISPR with other emerging technologies like nanotechnology, synthetic biology, and machine learning to fully realize its potential in developing resilient, productive, and sustainable agricultural systems.
Introduction
The ever-increasing global population and the consequent demand for food have placed immense pressure on agricultural systems worldwide. This challenge is compounded by the escalating impacts of climate change, which include extreme weather events, shifting pest and disease patterns, and declining arable land (Bibi and Rahman, 2023). These changes threaten crop yields and disrupt agricultural stability, making the task of ensuring global food security increasingly daunting. Traditional breeding methods, while having significantly contributed to past agricultural advancements, are often too slow to respond to these rapid environmental changes, while genetic engineering has faced issues of precision and public acceptance (Afzal et al., 2023; Ambika et al., 2024).
Staple crops such as rice, wheat, maize, and soybeans are the backbone of global food security, providing the primary source of calories for a large portion of the world’s population (Morrow et al., 2023). These crops are crucial not only for direct human consumption but also for animal feed and industrial uses. However, the productivity and resilience of these staple crops are increasingly threatened by climate change, pests, and diseases. Improving the yield, nutritional content, and stress tolerance of staple crops is therefore essential for ensuring food security, particularly in the face of a growing global population and diminishing arable land.
The evolution of agricultural technology from selective breeding to sophisticated genetic tools underscores our ongoing efforts to address these challenges. CRISPR/Cas technology as a revolutionary genome-editing tool has emerged as a game-changer in agricultural biotechnology (Muha-Ud-Din et al., 2024). CRISPR/Cas systems, a groundbreaking tool for targeted genome editing, have revolutionized both basic and applied research in agriculture. Originally derived from the adaptive immune systems of bacteria and archaea, the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) mechanism uses a guide RNA (gRNA) to direct the Cas (CRISPR-associated) nuclease to a specific DNA sequence, where it creates a precise double-strand break. This break is subsequently repaired by the cell’s natural DNA repair mechanisms, allowing for targeted modifications to the genome (Ghosh and Chatterjee, 2024). Unlike earlier genome editing tools like Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs), CRISPR/Cas systems are easier to design, more efficient, and less expensive, making them highly accessible for a wide range of applications in crop improvement (Ghoshal, 2024). The discovery of the CRISPR/Cas system as a genome editing tool was not just about identifying it in bacteria, but also about understanding how it could be harnessed and refined for use in more complex organisms. Early research clarified the critical roles of crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA) in guiding the Cas9 protein for precise DNA cleavage, which was pivotal in developing CRISPR/Cas into a versatile genome editing tool (Jung et al., 2024). The mechanism of action begins with the formation of an RNA-DNA hybrid, where the guide RNA binds to the target DNA sequence, directing the Cas9 protein to the specific genomic site. Once there, Cas9 introduces a double-strand break, which is then repaired by the cell’s natural DNA repair pathways—either non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Yuan et al., 2024). This detailed understanding of the CRISPR/Cas mechanism underscores its effectiveness in enabling precise and efficient genome modifications, making it a cornerstone technology for advancing crop traits and addressing global challenges such as food insecurity and climate change (Raza et al., 2024).
CRISPR technology has emerged as a transformative tool, allowing for the rapid development of crop varieties with enhanced traits such as improved resistance to biotic and abiotic stresses, increased nutritional value, and greater yield potential (Verma et al., 2023). Moreover, unlike traditional genetic modification techniques, CRISPR/Cas systems enhance agricultural productivity and sustainability through their simplicity, adaptability, cost-effectiveness, and publicly acceptable approach due to its ability to make precise alterations without introducing foreign DNA (Ali et al., 2023). Recent advancements, such as prime editing and base editing, have further refined the precision and scope of genome editing, enabling more complex genetic enhancements with fewer off-target effects (Naeem and Alkhnbashi, 2023; Saber Sichani et al., 2023). These innovations are paving the way for next-generation crops that can thrive in changing environmental conditions and meet the nutritional needs of a growing population.
This review aims to provide a comprehensive overview of CRISPR/Cas technology in enhancing crop resilience and productivity of staple grains amidst climate challenges. By exploring the latest research and technological advancements, this article highlights the transformative potential of CRISPR/Cas systems in modern agriculture. It would provide comprehensive insights for understanding current innovations and identifying strategic directions for future research and development, ultimately contributing to global food security and sustainable agricultural practices.
CRISPR/Cas technological innovations and advancements
Recent advancements in CRISPR technology have significantly enhanced the specificity and efficiency of genome editing, crucial for agricultural applications (Figure 1). Innovations like prime editing and base editing represent groundbreaking developments in precise genetic alterations. Prime editing combines CRISPR-Cas9 with a reverse transcriptase which has the potential to correct up to 89% of known genetic variants, enabling direct editing of target DNA sequences (Chen and Liu, 2023). Studies have demonstrated its effectiveness in enhancing disease resistance in rice by correcting specific point mutations without causing double-strand breaks (Gupta et al., 2023). Conversely, base editing facilitates the direct and irreversible conversion of one DNA base into another, increasing the precision of point mutations (Pfeiffer and Stafforst, 2023). Applications include altering flavor profiles in pea and tomatoes and improving cold tolerance in soybeans by modifying genes responsible for fatty acid desaturation and cold response pathways (Nizampatnam et al., 2024). Novel CRISPR-associated proteins, such as Cas12 and Cas13, expand the toolkit available for agricultural biotechnology. Cas12 offers advantages for multiplex editing, allowing simultaneous manipulation of multiple traits, for example, facilitate several disease resistance genes in soybeans (Sun et al., 2024). Cas13d offers a particularly robust solution for multiplex RNA virus interference in potato crops, making it a valuable asset in the ongoing efforts to enhance agricultural productivity and sustainability (Zhan et al., 2023).
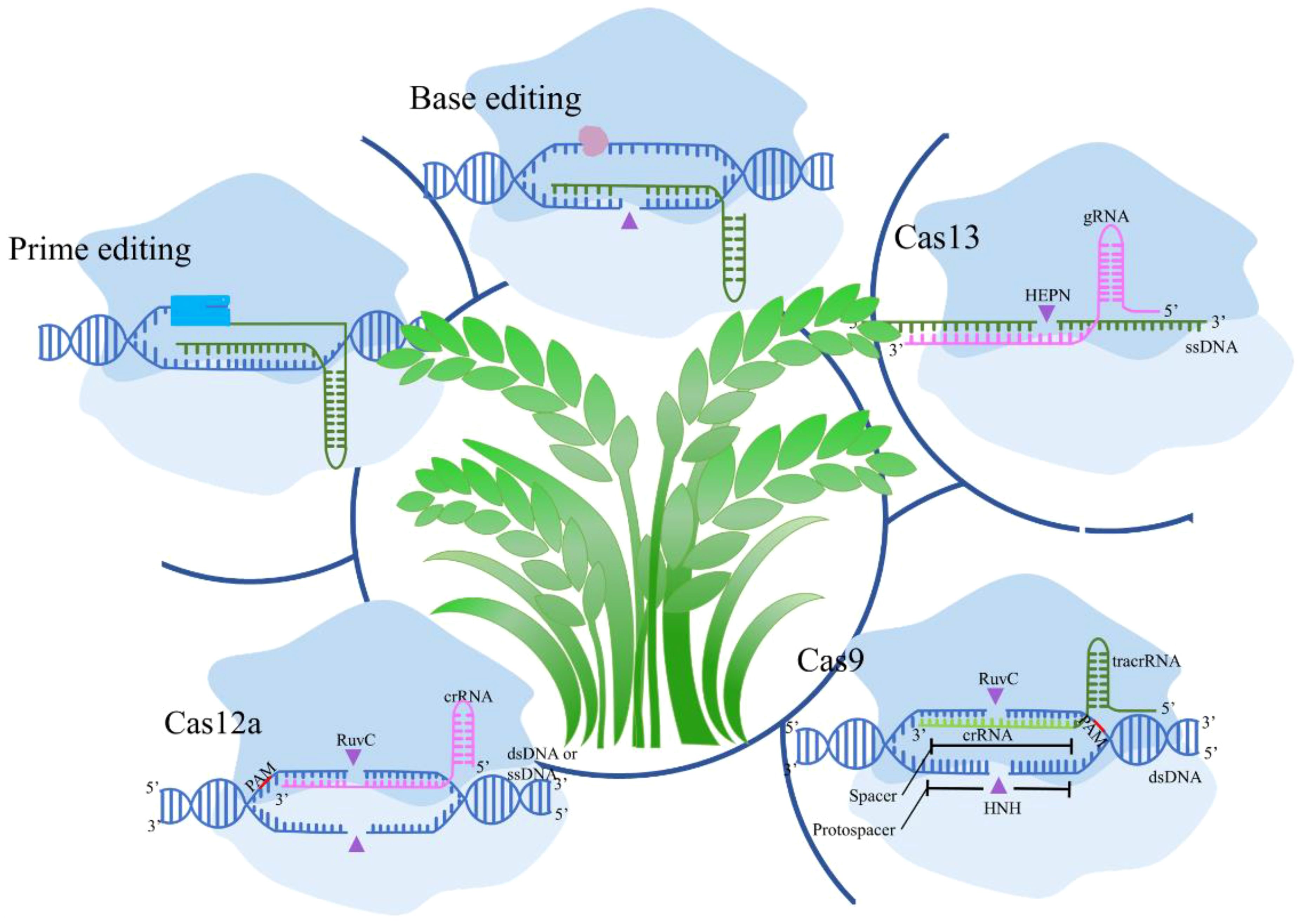
Figure 1. Diverse CRISPR/Cas systems mediated biotechnologies in crops. Cas9 in crops introduces double-strand breaks (DSBs) for targeted gene knockouts or insertions; Cas12a creates staggered DNA cuts and allows for multiplexed editing; Cas13 targets RNA for post-transcriptional regulation without altering the genome; Base Editing enables precise single-nucleotide changes without DSBs; and Prime Editing allows for small insertions, deletions, and base conversions with high precision.
Efficient delivery of CRISPR components is essential for successful genome editing in plants (Figure 2). Recent methodologies include nanoparticle-mediated delivery, which protects CRISPR components from degradation and enhances cellular uptake, significantly improving trait enhancement in maize (Chakraborty et al., 2023; Yau et al., 2024). Viral vectors, leveraging natural viral infection mechanisms, have shown improved efficiency and safety in transient expression projects, such as inducing virus resistance traits in tobacco and tomato (Jogam et al., 2023; Wang et al., 2024e). The ribonucleoprotein (RNP) complex delivery method delivers CRISPR components directly as proteins and RNA, reducing off-target effects. This method has been effective in crops like wheat for disease resistance and yield enhancement (Poddar et al., 2023). Enhanced computational tools for precise guide RNA design and the development of high-fidelity Cas variants exhibit reduced off-target activity (Zhang et al., 2023b). High-fidelity Cas9 variants have been used in wheat to reduce unintended mutations while enhancing drought tolerance (Poddar et al., 2023).These advancements not only improve the safety of genetic edits but also broaden CRISPR’s applicability in developing climate-resilient crops. The continuous refinement of CRISPR technologies, including the development of novel delivery methods and editing techniques is paving the way for transformative advances in agriculture in Table 1. By increasing the precision and efficiency of these tools, researchers are expanding the potential applications of CRISPR, facilitating the creation of more resilient, productive, and sustainable agricultural systems.
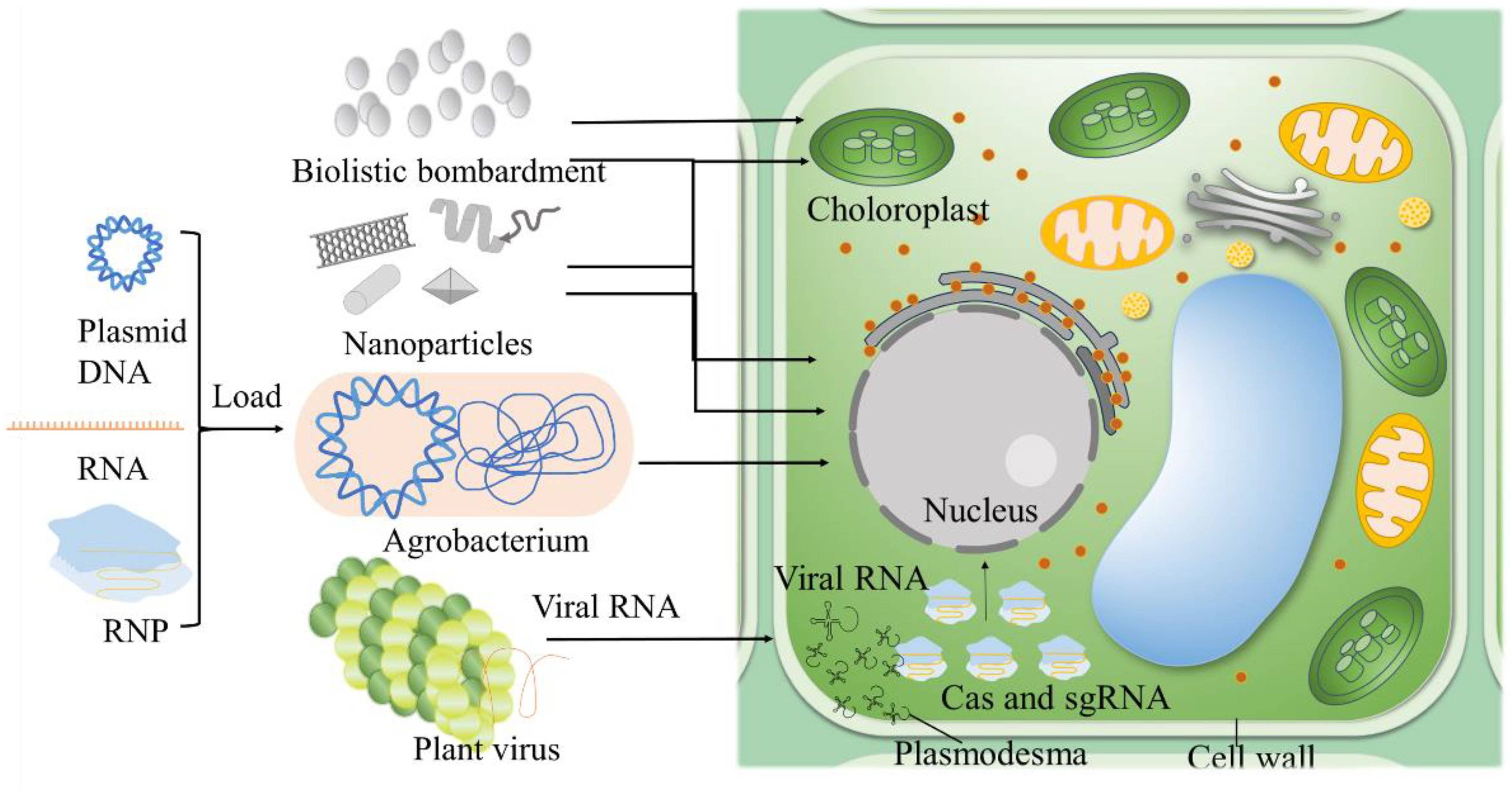
Figure 2. Strategies for CRIPSR/Cas delivery. Agrobacterium-mediated transformation exploits the bacterium's ability to transfer genetic material into plant genomes. Nanoparticles serve as carriers for CRISPR/Cas components, easily passing through plant cell walls. Biolistic bombardment which physically shoots DNA, RNA, or RNP-coated particles into plant tissues, useful for targeting chloroplasts and difficult-to-transform species. Virus-mediated delivery incorporates CRISPR/Cas into plant viruses, enabling systemic delivery across the plant. Partial figures are modified from previous publications (Zhu et al., 2020).
Strategic applications of CRISPR/Cas in enhancing staple crop resilience
The application of CRISPR/Cas technology in agriculture holds immense potential for improving the resilience of grain crops against various abiotic and biotic stresses (Yadav et al., 2023). This section explores how recent advancements in CRISPR/Cas technology have enhanced grain crop tolerance to these stresses, thereby supporting sustainable agricultural productivity in the face of climate change and other environmental challenges. CRISPR/Cas genome editing has become a mature tool for improving crop growth, development, and stress responses, as illustrated in Figure 3. In this context, we reviewed recent advances in CRISPR-mediated crop enhancement under abiotic and biotic stresses and improvements in various growth-related traits.
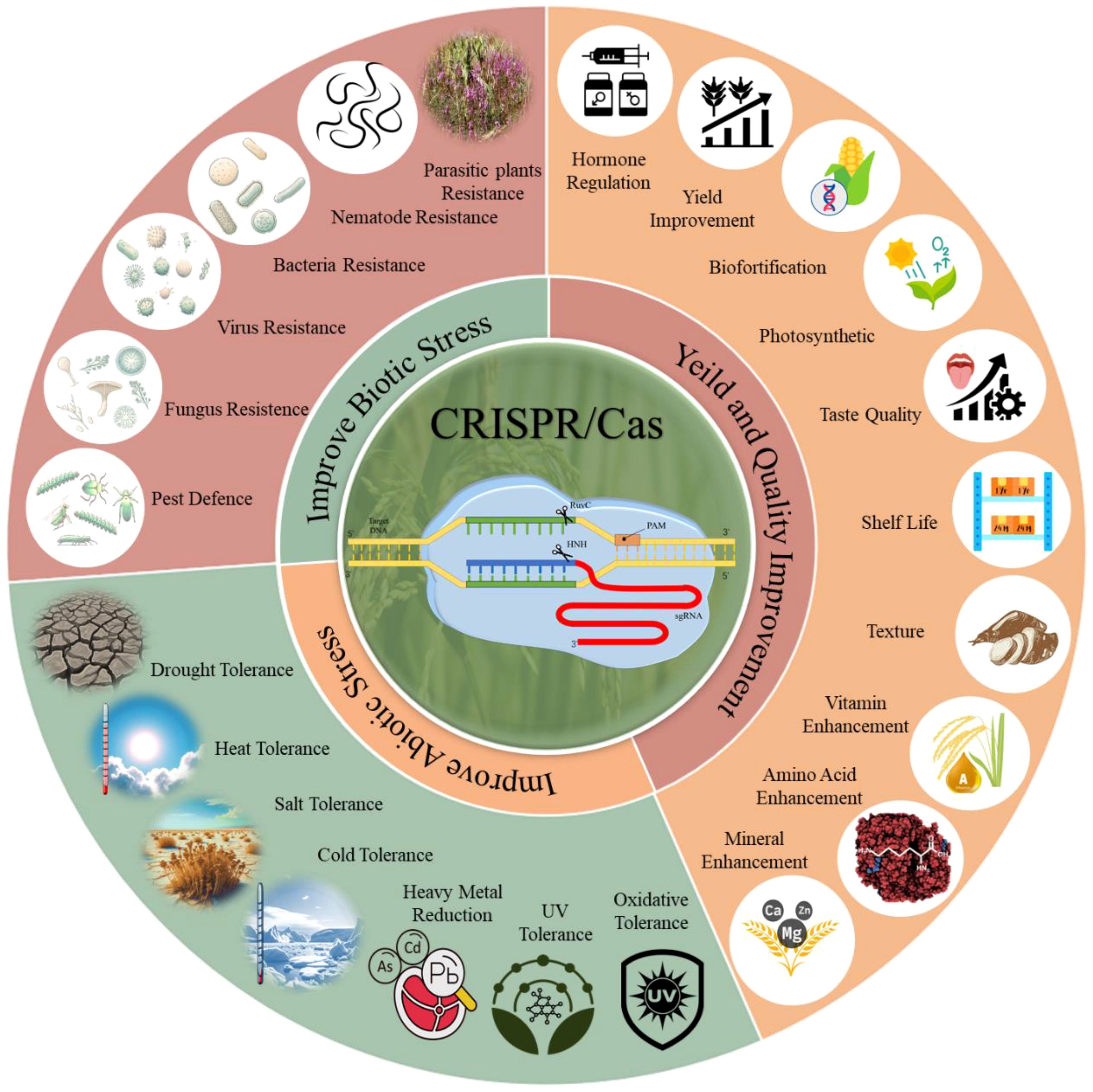
Figure 3. CRISPR/Cas genome editing is a powerful tool with immense potential for crop improvement. It enables precise modifications of individual genes to enhance plant tolerance to biotic and abiotic stresses, as well as yield and quality improvement. CRISPR/Cas has been used to develop resistance against various biotic stresses, including bacteria, viruses, fungi, pests, nematodes, and parasitic plants, and to improve resilience to abiotic stresses like drought, heat, salt, cold, heavy metals, UV exposure, and oxidative stress. Additionally, CRISPR/Cas enhances crop yield and quality by regulating hormone production, increasing photosynthetic efficiency, supporting biofortification, and improving shelf life, texture, and taste.
Enhancing abiotic stress tolerance
Abiotic stresses significantly reduce agricultural productivity and crop yields, but CRISPR/Cas technology provides precise tools to enhance grain crop tolerance by targeting specific stress response genes (Table 2).
Drought stress
Staple grain crops such as rice, wheat, and maize are particularly vulnerable to water scarcity, which poses a major challenge to food security. CRISPR/Cas technology allows precise genetic modifications to improve drought tolerance by targeting genes that regulate water use efficiency and osmotic balance (Shelake et al., 2022). A notable breakthrough in this area is the modification of the ZmHDT103 gene, a key component of the abscisic acid (ABA) signaling pathway, which has been shown to improve drought tolerance in maize by enhancing the plant’s ability to withstand water scarcity without compromising growth and yield under non-stress conditions (Wang et al., 2024f). Another promising application of CRISPR technology in combating drought stress involves engineering the TaRPK1 gene in wheat to enhance water absorption (Abdul Rahim et al., 2024). By targeting genes that influence root growth patterns and depth, CRISPR/Cas has been used to develop plants with deeper root systems capable of accessing water from further soil layers. Another innovative approach involves using CRISPR/Cas to manipulate Sal1 genes to improve the production of osmoprotectants, such as proline, which has been shown to enhance drought resistance in wheat by allowing plants to withstand dry periods more effectively through precise modulation of gene expression (Mohr et al., 2022).
Heat stress
Elevated temperatures can impair plant growth, reduce photosynthetic efficiency, and ultimately decrease crop yields, leading to a focus on using CRISPR/Cas technology to modify heat shock factors (HSFs) and heat shock proteins (HSPs) to enhance heat tolerance (Younas et al., 2024). These proteins protect cells from heat damage by refolding denatured proteins and preventing the aggregation of unfolded proteins. For instance, enhancing HsfA1b expression in wheat improves heat tolerance, maintaining growth and yields under high temperatures (Tian et al., 2020). Researchers are also editing genes involved in the synthesis of protective osmolytes and antioxidants to mitigate oxidative stress caused by high temperatures (Yadav et al., 2023). Furthermore, integrating CRISPR/Cas with other biotechnological tools enhances the robustness of crops against heat stress. For instance, gene drives engineered through CRISPR technology can spread heat tolerance traits rapidly through plant populations, potentially transforming the resilience of an entire crop in a few generations (Mohan et al., 2022).
Salt stress
Salt stress, an increasing agricultural concern particularly in regions affected by soil salinization due to improper irrigation practices and rising sea levels, is being addressed by CRISPR/Cas technology through the enhancement of salt tolerance by targeting genes that regulate ion homeostasis and osmotic balance (Hualpa-Ramirez et al., 2024). One strategy involves modifying transporters involved in sodium uptake and compartmentalization. For example, editing genes like knocking out AKT1, WRKYs reduces sodium accumulation, thereby improving salt tolerance and maintain better growth and yield in soybean and barley (Price, 2022; Feng et al., 2023). Additionally, by upregulating the DREB2A transcription factor has successfully enhanced salt tolerance in various crops, including soybeans and rice (Farhat et al., 2019; Feng et al., 2023). By fine-tuning the expression of these transcription factors, crops can activate comprehensive stress response pathways that confer enhanced tolerance to saline conditions.
Cold stress
CRISPR/Cas technology facilitates targeted genomic edits to confer cold tolerance by targeting the CBF (C-repeat Binding Factor) pathway, a well-documented regulatory mechanism in plants that enhances cold tolerance. CBF transcription factors activate a suite of genes that confer cold tolerance by enhancing the plant’s cellular machinery to cope with freezing stress (Perez-Garcia et al., 2023). CRISPR/Cas has been used to increase the expression of fatty acid desaturase genes, leading to changes in membrane lipid composition that better support cellular processes during cold exposure. For instance, studies have demonstrated that OsKASI-2 is required for increasing unsaturated fatty acids in rice membranes via CRISPR/Cas to improve cold tolerance (Zhang et al., 2024b). Another innovative approach involves using CRISPR/Cas for epigenetic modifications that influence gene expression related to cold stress (Jogam et al., 2022). This method offers a flexible approach to crop improvement that can be adjusted as environmental conditions change.
Heavy metal stress
Heavy metals such as cadmium (Cd), arsenic (As), and lead (Pb) are toxic to plants, causing stunted growth and reduced yields, but CRISPR/Cas technology can be used to enhance plant tolerance to heavy metals by modifying genes involved in metal transport and detoxification. For instance, editing the OsLCD gene in rice reduces cadmium uptake, thereby increasing cadmium tolerance and reducing its accumulation in the edible parts of the plant (Chen et al., 2023). Similarly, targeting genes like Lsi1 and Lsi2 in rice can decrease arsenic accumulation in plant tissues, thereby improving tolerance to arsenic-contaminated soils (Xu et al., 2024). Furthermore, targeting the ZmHMA3 gene in maize has shown to increase zinc tolerance by enhancing the plant’s ability to compartmentalize and detoxify these metals (Lv et al., 2023). These advancements contribute to the development of crops capable of growing in contaminated soils and producing safer food products.
UV radiation stress
CRISPR/Cas technology offers a promising solution to the significant threat of UV radiation to crop health by enabling precise genetic modifications that enhance tolerance to DNA damage, oxidative stress, and impaired photosynthesis. Targeting the OsCOP1 gene has demonstrated potential in improving UV tolerance in rice, enhancing their resistance to UV-B radiation (Hu et al., 2024). By boosting the plant’s protective mechanisms against UV damage, CRISPR/Cas technology can help develop crops that maintain productivity and growth under high UV exposure.
Oxidative stress
Oxidative stress results from the accumulation of reactive oxygen species (ROS) under various stress conditions. CRISPR/Cas technology provides a means to enhance oxidative stress tolerance in crops by targeting genes involved in ROS detoxification and antioxidant defense mechanisms. For example, Editing the CAT (catalase) gene family, particularly OsCAT3, which is crucial for detoxifying superoxide radicals and hydrogen peroxide, can enhance rice’s ability to mitigate oxidative damage (Jiang et al., 2023). Additionally, targeting regulatory genes such as zinc finger proteins, which modulate the expression of multiple antioxidant genes, can offer a comprehensive approach to improving oxidative stress resilience in crops (Qu et al., 2024).
Improving biotic stress resistance
Biotic stresses threaten crop health and productivity, and CRISPR/Cas technology enables precise genetic modifications to enhance crop resistance.
Viruses stress
CRISPR/Cas systems, particularly Cas13 have shown targeting and degrading the RNA genomes of RNA viruses, preventing their replication within the host plant (Sarkar et al., 2024). This approach has been effectively demonstrated in crops such as potato, where Cas13 was engineered to target and cleave the RNA of sweet potato virus disease (Zhan et al., 2023). Researchers have expanded the capabilities of CRISPR/Cas systems in viral defense by using them not only to target pathogens directly but also to modify the host plant’s genome to enhance its natural virus defense mechanisms (Uranga and Daròs, 2023). For instance, in crops like wheat and rice, CRISPR/Cas9 has been employed to knock out susceptibility genes such as TaPDIL5 or OsDjA2 and OsERF that facilitate viral infection, thus providing broad-spectrum virus resistance (Kan et al., 2022; Távora et al., 2022). Additionally, CRISPR/Cas technology was also employed to knock out ZmPDRP1 in maize, revealing that the loss of this gene significantly reduced the ability of sugarcane mosaic virus (SCMV) to replicate and spread within the plant (Xie et al., 2024). This highlights the utility of CRISPR/Cas not only for plant trait improvement but also as a powerful tool for dissecting gene functions in plant-pathogen interactions.
Bacteria stress
Enhancing bacterial resistance in crops using CRISPR/Cas technology involves targeting bacterial virulence genes and enhancing the plant’s immune response by disrupting key genes in bacterial pathogens to significantly reduce their virulence. For instance, knocking out the StNRL1 gene in potatoes using CRISPR/Cas9 increases resistance to late blight caused by Phytophthora infestans while simultaneously increasing susceptibility to early blight caused by Alternaria alternate (Norouzi et al., 2024). CRISPR/Cas technology has also been employed to modify plant immune receptors to recognize bacterial pathogens more effectively. Editing the FERONI and SlWak1 depend on FLS gene in rice and wheat, which encodes a receptor involved in pathogen recognition, has improved the plants’ ability to detect and respond to bacterial infections, thereby enhancing resistance (Huang et al., 2020; Zhang et al., 2020). This genetic modification has resulted in rice varieties with enhanced resistance to bacterial blight, leading to healthier plants and higher yields.
Fungi stress
Fungal diseases are a major concern for crop health, and CRISPR/Cas technology offers new ways for enhancing fungal resistance in crops through precise genetic modifications. One strategy involves using CRISPR/Cas to knock out susceptibility genes that fungi exploit, such as MLO (Mildew Locus O) genes in soybean and wheat, which has been shown to confer resistance to powdery mildew by making the plants less susceptible to fungal infections (Li et al., 2022a; Bui et al., 2023). Enhancing plant defense genes involved in recognizing and responding to fungal attacks also improves resistance. For instance, editing the CNL (Coiled-Coil, Nucleotide-Binding, Leucine-Rich Repeat) gene family and MeRPPL1 in cassava has led to enhanced fungal resistance (Ramulifho and Rey, 2024). Another innovative application of CRISPR/Cas involves editing the genomes of fungal pathogens themselves. This approach has been explored in various fungal species, including Fusarium and Botrytis, which are responsible for significant agricultural losses.
Pests stress
CRISPR/Cas technology is also being utilized to enhance pest resistance in crops by knocking out susceptibility genes and enhancing plant defense mechanisms. For example, editing the ABC transporter gene in soybean has been shown to confer resistance to bollworms by disrupting the insect’s ability to digest plant tissues (Amezian et al., 2024).Another strategy is to enhance the expression of plant defense genes involved in producing secondary metabolites that deter insect feeding. For instance, increasing the expression of genes involved in the biosynthesis of phenolic compounds has been shown to reduce insect herbivory in crops like maize and soybean (Razzaq et al., 2023; Kumari et al., 2024). CRISPR/Cas technology has also been used to modify genes encoding insecticidal proteins, such as Cry proteins, VIP proteins improving pest resistance in crops (Dubovskiy et al., 2024). These genetic modifications result in plants that produce higher levels of natural insecticidal compounds, providing an effective defense against pests.
Nematode resistance
Nematodes, such as root-knot nematodes (Meloidogyne spp.), cause root damage and reduce nutrient and water uptake. CRISPR/Cas technology can enhance nematode resistance by targeting genes that facilitate nematode infection and reproduction. For instance, editing a susceptibility gene OsHPP04 in rice has conferred resistance to root-knot nematodes (Huang et al., 2023). Similarly, modifying the GmSNAP02 and an α-SNAP gene in soybean has enhanced resistance to cyst nematodes (Usovsky et al., 2023, Usovsky et al., 2023). Through disrupting the molecular pathways that nematodes exploit, CRISPR/Cas can develop crops with robust nematode resistance, reducing yield losses and the need for chemical nematicides.
Parasitic plants
Parasitic plants, such as Striga and Orobanche, attach to the roots of host plants and extract water and nutrients, significantly reducing crop yields. CRISPR/Cas technology can enhance resistance to parasitic plants by targeting genes involved in host-parasite interactions. For instance, editing the LGS1 gene in sorghum has conferred resistance to Striga by disrupting the production of strigolactones, which are essential for Striga seed germination and attachment (Makaza et al., 2023). By modifying specific signaling pathways and defense mechanisms, CRISPR/Cas can develop crops less susceptible to parasitic plants, thereby improving yield and sustainability, while highlighting the technology’s versatility in managing various biotic stresses to ensure better crop health and productivity.
Yield and quality improvement
Improving crop yield and quality is essential to meet growing global food demand, and CRISPR/Cas technology provides precise tools for enhancing these traits by targeting specific genes and pathways, as explored in this section (Table 3).
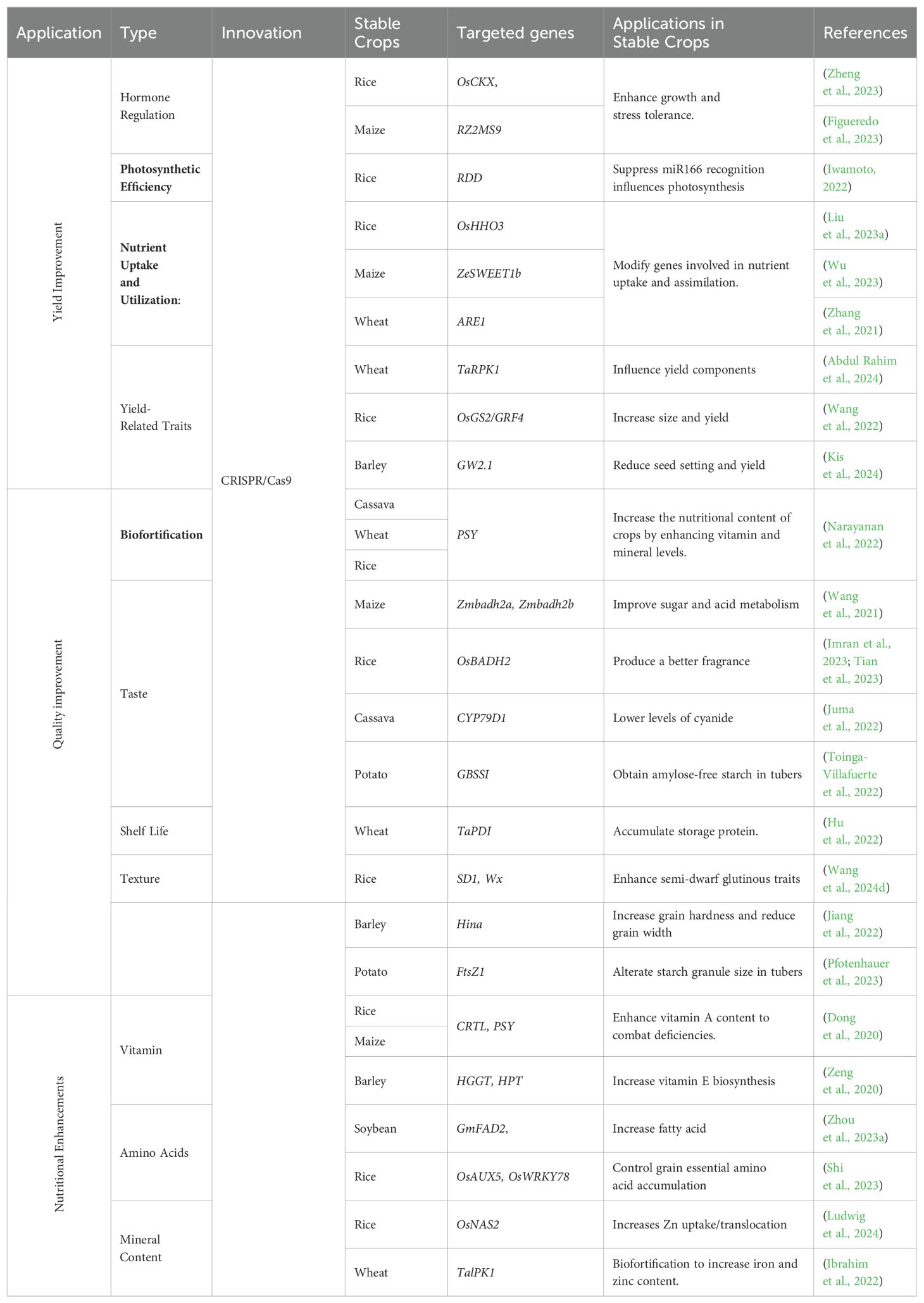
Table 3. Comprehensive overview of CRISPR/Cas applications in enhancing yield, quality, and nutritional value of stable crops.
Increasing crop yield
CRISPR/Cas technology offers new opportunities to enhance crop yield by directly targeting genes that regulate plant growth and development. For example, editing the OsAPL involved in nutrient transport has been shown to increase yield in rice (Zhang et al., 2024a). Enhancing photosynthetic efficiency by targeting genes involved in chlorophyll synthesis and light capture, such as the OsSXK1 gene in rice, has improved photosynthetic rates and increased grain yield (Zheng et al., 2021). Additionally, editing genes involved in nutrient uptake and utilization, such as the ARE genes in barley or wheat, enhances nitrogen use efficiency and leads to higher yields under low nitrogen conditions (Karunarathne et al., 2022). Recent studies have demonstrated the potential of CRISPR/Cas technology in enhancing yield-related traits in various crops. Editing the DEP1 gene in rice has led to the development of semi-dwarf varieties with improved lodging resistance and higher grain yield (Zhang et al., 2023a).
Improving crop quality
CRISPR/Cas9 technology has dramatically advanced agricultural biotechnology by enabling precise genome editing to improve various crop quality attributes, including safety, taste, texture, shelf life, and industrial applicability. In cassava, CRISPR/Cas9 has been used to edit the CYP79D1 gene, significantly reducing cyanogenic glycosides, which lowers the risk of cyanide toxicity, enhancing the safety of this staple crop without affecting its agronomic performance (Juma et al., 2022). In rice, the technology has been employed to enhance aromatic qualities by editing the OsBADH2 gene, leading to increased production of 2-acetyl-1-pyrroline (2-AP), a compound that imparts a desirable fragrance, thereby catering to consumer preferences (Tian et al., 2023). In potatoes, CRISPR/Cas9 has modified the gbss gene responsible for granule-bound starch synthase, resulting in amylose-free starch that provides a smoother texture, which is highly valued in both culinary applications and industrial processes (Toinga-Villafuerte et al., 2022). The technology has also been pivotal in extending the shelf life of various crops by targeting genes involved in the ripening process, such as those regulating ethylene production, allowing for slower ripening, reduced post-harvest losses, and improved economic viability. Furthermore, in barley, CRISPR/Cas9 has been used to enhance grain hardness by editing the Hina gene, producing grains with a higher hardness index that are better suited for industrial applications, though this has also resulted in reduced grain width and thousand-grain weight (Jiang et al., 2022). Additionally, in potatoes, targeting the FtsZ1 gene has led to the development of lines with larger starch granules, significantly increasing the final viscosity of starch paste, making these potatoes more suitable for specific industrial processes, all achieved without compromising the plant’s overall phenotype or nutritional quality (Pfotenhauer et al., 2023). These diverse applications of CRISPR/Cas9 underscore its transformative potential in crop improvement, enabling tailored modifications that meet consumer demands, enhance safety, and address specific industrial needs while ensuring the sustainability and economic viability of agricultural practices.
Nutritional enhancements
Addressing nutritional deficiencies through crop biofortification is a key goal in agricultural biotechnology, and CRISPR/Cas technology plays a pivotal role in achieving this. For instance, CRISPR/Cas has been employed to increase pro-vitamin A content in rice, a vital intervention in combating vitamin A deficiency in populations that rely heavily on rice as a staple food (Maiti and Banik, 2023). Biofortification aims to increase the content of essential nutrients in crops, thereby improving their nutritional value. The development of “Golden Rice,” which contains higher levels of beta-carotene, was accomplished by modifying genes involved in pro-vitamin A biosynthesis (Dong et al., 2020; Datta et al., 2021). RISPR/Cas technology has also been applied to enhance the mineral content of crops. In rice and wheat, genes such as OsNAS have been edited to increase iron and zinc levels, addressing micronutrient deficiencies that often lead to anemia and impaired immune function (Dueñas et al., 2021). Similarly, in maize, targeting the PSY1, Crtl, and LCYB genes has boosted the biosynthesis of pro-vitamin A, resulting in the creation of “Golden Maize” (Sobrino-Mengual et al., 2024).The technology is also instrumental in improving the amino acid content of crops. In cassava, CRISPR/Cas has been used to enhance the levels of essential amino acids and vitamins, significantly improving the nutritional quality of this staple crop (Otun et al., 2023). In maize, editing genes involved in lysine biosynthesis has increased lysine content, addressing a common deficiency in cereal grains (Hasan, 2024). Improving the protein quality of crops is another significant area of focus for CRISPR/Cas technology. For example, targeted mutagenesis of the OsAAP6 and OsAAP10 genes in rice can reduce grain protein content, thereby improving the eating and cooking quality of the crop (Wang et al., 2020). CRISPR/Cas technology has been used to reduce antinutritional factors such as phytic acid in soybean by targeting the GmIPK1 gene, enhancing the bioavailability of iron and zinc and thereby improving the overall nutritional quality of the soybean (Song et al., 2022).
Cases study of applications in staple crops
As the application of CRISPR/Cas technology is broad and impactful across various staple crops, focusing on specific case studies such as rice and maize allows us to delve deeper into its transformative role in enhancing resilience (Figure 4).
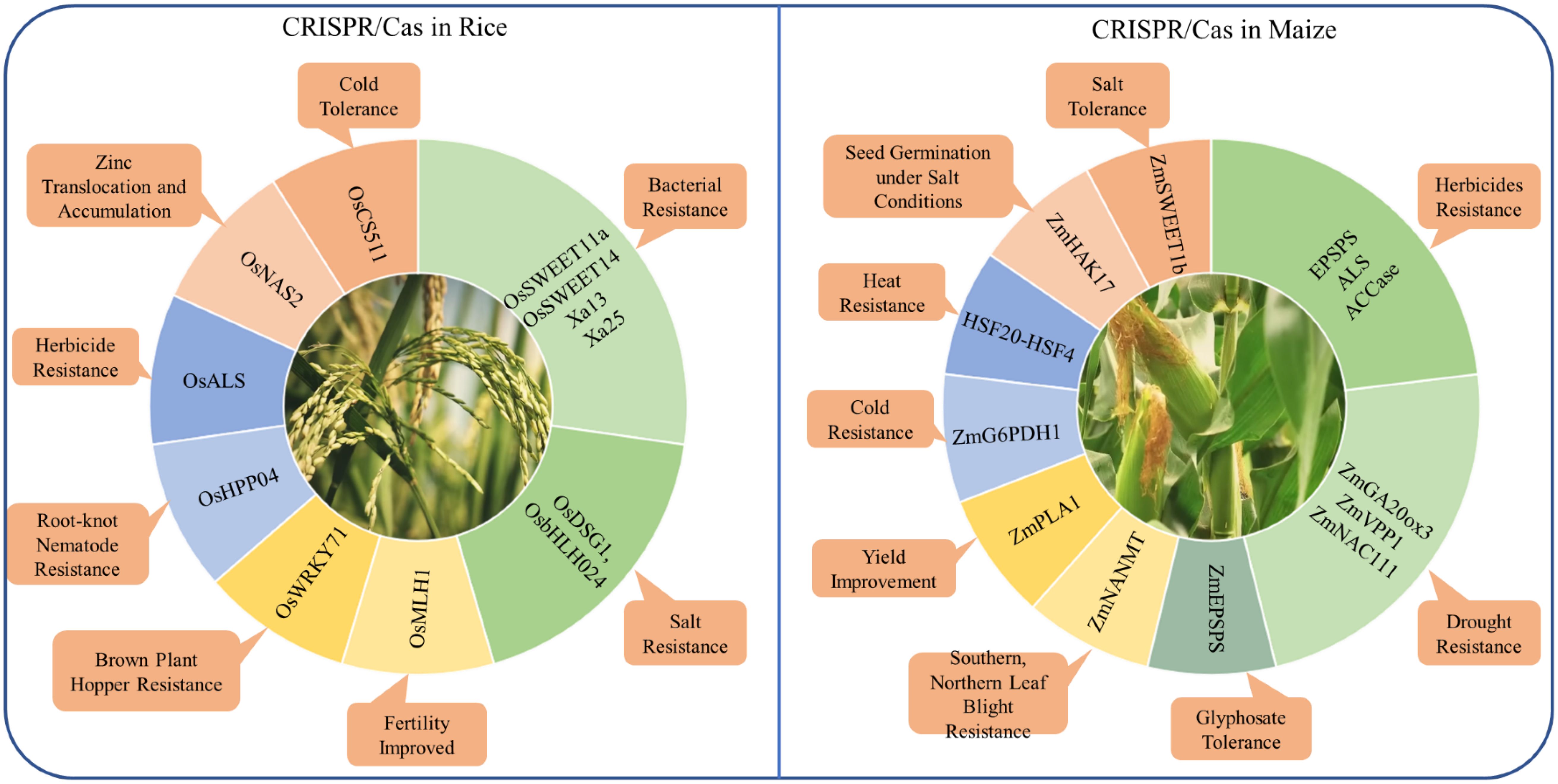
Figure 4. Application of CRISPR/Cas genome editing in rice and maize for the improvement of different traits.
Case study: CRISPR/Cas in rice
CRISPR/Cas technology has significantly advanced rice improvement by enabling precise genome modifications, enhancing traits such as yield, stress resistance, and nutritional value. This technology has been instrumental in improving resistance to both biotic and abiotic stresses in rice. For example, the clade III subfamily of OsSWEETs, including OsSWEET11a and OsSWEET14, plays a crucial role in susceptibility to bacterial blight by mediating sucrose efflux for bacterial proliferation. These genes are targeted by specific transcription activator-like effectors (TALEs) from Xoo (Wang et al., 2024g). This approach demonstrates the potential of CRISPR/Cas9 to address other pathogen-related challenges in rice. Similarly, the OsCS511 gene has been edited to improve cold tolerance in rice, which is critical for expanding rice cultivation to cooler climates (Park et al., 2024). It highlights the adaptability of CRISPR/Cas technology in enabling rice to grow in suboptimal conditions, thereby expanding its cultivation area. Furthermore, base editing has been used to increase zinc uptake and plant yield by editing the promoter region of the OsNAS2 gene, which plays a crucial role in zinc translocation and accumulation (Ludwig et al., 2024). This precise editing has led to a significant increase in zinc concentration in rice grains, addressing both yield improvement and nutritional enhancement in a single genetic modification. Additionally, the knockout of the OsDSG1 and OsbHLH024 transcription factor has been shown to significantly enhance salt stress resistance, allowing rice to thrive in saline soils (Alam et al., 2022; Ly et al., 2024). This advancement is particularly important in regions where soil salinity limits agricultural productivity. Furthermore, CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 has conferred enhanced resistance to rice root-knot nematode, a significant pest that causes major yield losses in rice crops, demonstrating CRISPR/Cas9’s potential in sustainable rice production (Huang et al., 2023).
Despite these successes, challenges such as off-target effects, potentially lead to undesirable mutations. For instance, when editing the OsWRKY71 gene to enhance resistance against brown plant hopper, careful analysis was necessary to ensure that no off-target effects compromised the plant’s health (Li et al., 2023b). This case underscores the importance of precision in gene editing to avoid unintended consequences that could negate the benefits of the modifications. Another challenge is the relatively low efficiency of HDR in rice, which is required for precise gene insertions or complex modifications. This limitation has been addressed in part by developing more precise methods like prime editing, which does not rely on HDR. Prime editing has been successfully applied to the OsMLH1 gene to enhance the efficiency of the editing process without disturbing rice fertility, offering a promising approach for future genetic improvements (Liu et al., 2024a).
Recent advancements in CRISPR technology, particularly base editing and prime editing, have opened new avenues for improving specific traits in rice. Base editing has been successfully applied to the OsALS gene to confer herbicide resistance, allowing rice to resistant to herbicides such as imazamox, which is crucial for effective weed management (Zafar et al., 2023). This modification allows for more targeted and sustainable weed control strategies, reducing the need for broad-spectrum herbicides that can harm the environment. Additionally, prime editing has been applied to the Xa13 and Xa25 genes, creating rice varieties with enhanced resistance to bacterial blight by targeting and disrupting these susceptibility genes (Zhu et al., 2024). These innovations, including the application of base editing and prime editing to improve traits like herbicide resistance and bacterial blight resistance, are crucial for developing resilient rice varieties suited to climate change and global food security challenges.
Case study: CRISPR/Cas in maize
The application of CRISPR/Cas systems, including advanced techniques like prime editing and base editing, has revolutionized the ability to enhance maize’s tolerance to various abiotic and biotic stresses. Prime editing, particularly the optimized ePE5max system, has been instrumental in generating heritable mutations in maize, enabling resistance to herbicides by targeting key enzymes such as EPSPS, ALS, and ACCase. This has significantly improved the resilience of maize to herbicide stress. Base editing has also shown potential in this area, with the introduction of specific nucleotide substitutions, such as the triple amino acid substitution in the ZmEPSPS gene, leading to heightened glyphosate tolerance (Kaul et al., 2024). Additionally, the editing of ZmSWEET1b, a sugar transporter crucial for assimilate allocation, has demonstrated improved salt stress response, showcasing the effectiveness of these gene-editing techniques in enhancing maize’s abiotic stress tolerance (Wu et al., 2023).
Further advancements in maize stress tolerance have been achieved through the targeted editing of specific genes associated with drought, salinity, and disease resistance. For example, the gene editing of ZmGA20ox3 has not only improved drought tolerance but also optimized plant architecture, while the co-expression of ZmVPP1 with ZmNAC111 has conferred robust drought resistance by enhancing the plant’s stress response mechanisms (Liu et al., 2023d; Liu et al., 2024b). Similarly, genes like ZmHAK17, which encodes a Na+-selective transporter, and the HKT1 family sodium transporter have been pivotal in improving maize’s tolerance to salinity by regulating sodium influx and promoting seed germination under salt conditions (Zhang et al., 2023d; Wang et al., 2024a). Moreover, the genome editing of the ZmNANMT gene has conferred multiple disease resistance without agronomic penalties, addressing major diseases like southern leaf blight, northern leaf blight, and Fusarium stalk rot (Li et al., 2023c). These targeted genetic modifications underline the versatility of CRISPR/Cas systems in addressing both biotic and abiotic stresses in maize.
However, despite these promising developments, the application of CRISPR/Cas systems in maize faces several challenges, including off-target effects, regulatory hurdles, and efficiency in gene editing techniques. For instance, while the DNA-free genome editing of the ZmPLA1 gene via ribonucleoprotein complexes has been successful in increasing haploid induction rates in tropical maize, optimizing these methods further remains crucial to enhancing efficiency and reducing unintended mutations (Rangari et al., 2023). Similarly, the generation of maize lines with enhanced cold stress tolerance through the overexpression of ZmG6PDH1 in glucose-6-phosphate dehydrogenase family, and the regulation of heat stress tolerance through the HSF20-HSF4-cellulose synthase A2 module, illustrate the complexity of stress responses in maize that require precise and efficient gene editing tools (Li et al., 2023a, Li et al., 2024b). Addressing these challenges through ongoing research and technological advancements will be key to fully harnessing the potential of CRISPR/Cas systems in improving maize resilience and productivity under various environmental stresses.
Challenges and future prospect
Off-target effects
Off-target effects in CRISPR/Cas systems present a significant challenge in genome editing, where unintended cuts by the Cas enzyme at non-target sites can lead to adverse consequences. In crops, such unintended mutations can affect traits, reducing yield or introducing unwanted characteristics. Therefore, minimizing off-target effects is critical to ensuring the safety and efficacy of CRISPR-based technologies, making precision in genome editing a top priority. To address these challenges, considerable progress has been made in developing high-fidelity Cas variants and optimizing gRNA design. High-fidelity Cas9 variants, such as eSpCas9, SpCas9-HF1 and FrCas9, have been engineered to enhance precision by reducing nonspecific DNA interactions (Guo et al., 2023; He et al., 2024). These variants demonstrate lower off-target activity while maintaining robust on-target efficacy, making them suitable for applications. Additionally, studies have shown that Cas12b, with its strict PAM requirements and low tolerance for mismatches, has minimal sgRNA-dependent off-target effects, showing great promise in rice genome editing (Gurel et al., 2023). This system’s ability to precisely target specific genomic sites with minimal off-target activity underscores the importance of optimizing both the Cas enzyme and gRNA design to enhance the specificity of CRISPR/Cas systems. Future research should focus on enhancing accuracy, minimizing off-target effects, and developing novel variants or engineered Cas proteins that can better handle the complexities of multiplex editing, especially in crops with polyploid genomes. This will not only improve precision in genome editing but also reduce unintended consequences, making CRISPR technology safer and more reliable for agricultural applications.
Delivery methods
Delivering CRISPR components into plant cells, particularly in recalcitrant staple crops like wheat, maize, and certain rice varieties, presents significant challenges due to their complex genetic makeup and poor tissue culture responses. Traditional methods, such as Agrobacterium-mediated transformation and biolistic particle delivery, are often ineffective in these crops. Agrobacterium-mediated transformation is limited by its host range and often results in random transgene integration, leading to gene silencing and unpredictable expression patterns. Similarly, while biolistic delivery bypasses some limitations of Agrobacterium, it frequently causes multiple, unstable transgene insertions and physical damage to plant tissues, reducing transformation efficiency. Nanoparticle-mediated delivery, a more recent approach, shows promise but faces challenges related to the precise release and stability of CRISPR components within plant cells, as well as potential cytotoxicity concerns (Antony Ceasar and Ignacimuthu, 2023).
To overcome these challenges, research is increasingly focused on developing novel delivery methods that enhance the efficiency and specificity of CRISPR delivery in recalcitrant crops (Kocsisova and Coneva, 2023). Viral vectors offer a promising avenue by utilizing natural plant infection mechanisms to deliver CRISPR components, though their limited cargo capacity remains a significant hurdle (Liu et al., 2023c). Enhancements in nanoparticle systems, such as surface functionalization with specific ligands and the development of biodegradable particles, could improve the specificity and safety of CRISPR delivery. Protoplast transfection, combined with optimized regeneration protocols, offers a direct method for introducing CRISPR components, though its application is currently limited to species with efficient protoplast regeneration systems (Yang et al., 2024). Grafting techniques, which use transgenic rootstocks to deliver CRISPR components to wild-type scions, represent another innovative approach for achieving transgene-free genome editing in recalcitrant crops (Yang et al., 2023b). To achieve sustainable improvements, future research should not only focus on developing novel delivery methods but also on ensuring the long-term stability and heritability of CRISPR-induced modifications across multiple generations. Understanding mechanisms such as chromosomal rearrangements and DNA repair fidelity will be crucial to maintaining the integrity of these genetic edits. Longitudinal studies that track the persistence and effects of these modifications over time are essential to confirm their stability and effectiveness, ensuring that the benefits of CRISPR technology are retained through successive crop generations.
Ethical, and regulatory issue and public acceptance
CRISPR technology offers significant potential in crop improvement, but it also presents challenges in ethics, regulation, and socioeconomic impact. Ethically, the precision and speed of CRISPR modifications, such as those used in rice and wheat to enhance yield and nutritional content, raise concerns about the moral acceptability of altering plant genomes. These concerns are especially pronounced in regions like Europe, where public sentiment is cautious about genetic modifications. The ethical debate includes potential unintended consequences, the creation of “unnatural” organisms, and the long-term ecological impacts (Marone et al., 2023). Additionally, the regulatory landscape for CRISPR-edited crops varies significantly across countries. In European Union classifies CRISPR-edited crops as genetically modified organisms (GMOs), subjecting them to rigorous regulations that have hindered the commercialization of crops like CRISPR-edited wheat. Countries like Argentina and Brazil have adopted more flexible regulatory frameworks, focusing on the final product rather than the process, allowing for the development and commercialization of crops like CRISPR-edited sugarcane with less regulatory burden. Meanwhile, China’s significant investment in CRISPR research, particularly in crops like rice, positions it as a major player, although commercialization remains tightly regulated (Ghouri et al., 2023; Kumawat et al., 2024). Socioeconomic challenges further complicate the integration of CRISPR technology into global agriculture. The contentious patent landscape, dominated by the U.S. and European nations, restricts access to CRISPR technology, particularly for smaller entities or developing countries. This is evident in the development of CRISPR-edited crops like tomatoes with enhanced GABA levels, where patent issues could limit access in less affluent regions (Akhtar et al., 2023). Additionally, the concentration of CRISPR technology within a few large corporations could exacerbate inequalities in the agricultural sector, particularly for staple crops like maize and wheat, crucial for food security in developing regions (Molinari et al., 2024). Countries like India and several African nations face the challenge of ensuring that the benefits of CRISPR technology, such as drought-resistant maize or disease-resistant cassava, are accessible to all farmers, not just large agribusinesses (Munawar et al., 2024). As CRISPR technology advances, ensuring that regulatory frameworks keep pace with these developments is essential. Future research must focus on addressing safety concerns, including off-target effects and the long-term ecological impacts of CRISPR-modified crops. This requires the development of comprehensive risk assessment models and strong collaboration between scientists, regulators, and policymakers. By aligning technological advancements with robust regulatory measures, we can ensure the safe and responsible integration of CRISPR into global agricultural practices, ultimately fostering public trust and acceptance.
Synergy of CRISPR with advancement technologies
The intersection of CRISPR technology with emerging fields such as nanotechnology, synthetic biology, and machine learning (ML) offers a transformative potential to advance genome editing in staple crops like rice, maize, wheat, and potato. Nanotechnology, in particular, addresses one of the critical challenges in CRISPR-based genome editing: the efficient and precise delivery of CRISPR components into plant cells. Nanomaterials like carbon nanotubes and mesoporous silica nanoparticles can bypass the plant cell wall, enabling targeted and controlled delivery of CRISPR components, which increases transformation efficiency and reduces off-target effects (Khanna et al., 2023). Additionally, nanoparticle-mediated delivery systems are species-independent, democratizing CRISPR technology across diverse crops (Naik et al., 2022). Future research should focus on optimizing these nanotechnologies for larger CRISPR complexes, organelle-specific editing, and direct transformation of germline cells, potentially bypassing tissue culture. This synergy holds great potential for advancing sustainable agriculture, improving crop resilience, nutritional content, and reducing chemical input dependency.
The intersection of CRISPR and Synthetic Biology offers a promising path to enhance genome editing efficiency and precision in staple crops. CRISPR/Cas systems enable targeted modifications, but challenges such as off-target effects, variable efficiency, and polyploid genome complexity persist. Synthetic Biology addresses these issues by providing tools to design and control genetic circuits, thereby reducing off-target effects and improving adaptability across different species (Yang and Reyna-Llorens, 2023). Key advancements include regulatory circuits that fine-tune CRISPR activity and feedback loops that adjust editing in real-time, enhancing precision (Wang and Demirer, 2023). Integrating CRISPR with synthetic metabolic pathways could yield crops that are higher-yielding and more resilient to environmental stressors, crucial for addressing global food security challenges posed by climate change. Future research should focus on developing synthetic promoters for efficient editing in polyploid crops and combining CRISPR with metabolic engineering to produce bioactive compounds, advancing sustainable agriculture in both traditional and controlled environments like vertical farming.
Machine learning (ML) adds another layer of innovation by exponentially enhancing CRISPR’s potential for precise genome modifications. However, when combined with the predictive capabilities of ML, the potential of CRISPR can be vastly expanded. One of the key challenges in CRISPR genome editing is ensuring specificity and efficiency in targeting the correct genomic sites while minimizing off-target effects. ML models can address this challenge by analyzing large datasets from CRISPR experiments to predict the most effective guide RNA sequences, thereby enhancing the precision of the Cas9 enzyme and reducing unintended consequences (Chen et al., 2024). Additionally, ML can predict the phenotypic outcomes of specific gene edits, which is particularly complex due to the multifactorial nature of traits like yield and stress tolerance (Das et al., 2023). In crops like rice and maize, where multiple genes interact to influence traits such as drought resistance, ML can identify the most impactful gene edits by considering various genetic and environmental factors. Future research should focus on developing sophisticated ML algorithms capable of handling the complexity of polygenic traits and creating extensive datasets to train these models. Integrating CRISPR-ML into precision agriculture systems could provide tailored recommendations, optimizing crop performance in specific field conditions and ultimately contributing to more sustainable and efficient agricultural practices.
Future applications with CRISPR/Cas knock-in system
While the CRISPR/Cas9 system has predominantly been used for gene knockouts, the development of CRISPR/Cas-based knock-in strategies has significantly expanded its potential, enabling precise gene integration and enhancing crop traits with high accuracy. This system allows for the insertion of desired genes into specific genomic locations, facilitating complex genetic modifications such as the introduction of large DNA sequences or multiple genes, which are crucial for stacking beneficial traits like disease resistance, stress tolerance, and improved nutritional content. However, the knock-in approach relies primarily on HDR, which is less efficient in plants compared to NHEJ. To address this limitation, researchers are developing strategies to enhance HDR efficiency, including the use of HDR enhancers, dual-gRNA systems, and advanced delivery methods like nanoparticle-mediated delivery and viral vectors, making the knock-in process more feasible for large-scale agricultural applications.
The application of the CRISPR/Cas knock-in system in specific crops has demonstrated its versatility and promise in sustainable agriculture. For instance, in rice, this system has been used to confer glyphosate resistance by precisely editing the acetolactate synthase (ALS) gene and to upregulate genes involved in key metabolic pathways, significantly enhancing the crop’s nutritional content and stress tolerance (Sony et al., 2023). In wheat, researchers are focusing on improving HDR efficiency using fusion proteins like Cas9-VirD2 to enhance traits such as disease resistance and grain quality (Schreiber et al., 2024). Similarly, in maize, the knock-in of regulatory elements to more precisely control gene expression has been explored to improve yield, nutrient use efficiency, and stress tolerance (Kaul et al., 2024). These examples underscore the knock-in system’s crucial role in developing next-generation crops that are more resilient, productive, and adaptable to changing environmental conditions.
Despite its potential, the CRISPR/Cas knock-in system faces challenges, including low HDR efficiency and off-target effects. To overcome these, researchers are exploring various strategies, such as enhancing HDR pathways, developing modified CRISPR/Cas variants, and utilizing alternative genome editing tools like CRISPR nickase and prime editing, which reduce the risk of off-target mutations. Improving delivery systems, such as nanoparticle-based methods, is also critical for increasing the efficiency and precision of gene insertion. Future directions for this technology include further engineering of Cas proteins for enhanced specificity, optimizing guide RNA design to minimize off-target effects, and integrating the knock-in system with emerging technologies like base editing and prime editing. These advancements are essential for refining the CRISPR/Cas knock-in system and maximizing its impact on agricultural innovation.
Conclusion
In conclusion, this review underscores the transformative potential of CRISPR/Cas technology in improving the resilience, yield, and nutritional value of staple crops like rice and maize. Through precise genome modifications, CRISPR/Cas systems have revolutionized crop breeding by enhancing stress tolerance, disease resistance, and overall productivity. Recent advancements, including base editing, prime editing, and high-fidelity Cas variants, have significantly increased the specificity and efficiency of genome editing, reducing off-target effects and expanding its agricultural applications. Furthermore, CRISPR/Cas technology has played a crucial role in biofortification efforts, such as boosting pro-vitamin A content in rice and increasing iron and zinc levels in wheat, addressing critical global challenges like food security and malnutrition, particularly in developing regions. To fully harness the potential of CRISPR/Cas technology, future research should focus on improving HDR efficiency, expanding the CRISPR toolkit, addressing ethical and regulatory challenges, and integrating CRISPR into traditional breeding programs to accelerate the development of high-yielding, climate-resilient crops, thereby contributing to sustainable and resilient global food systems.
Author contributions
CL: Funding acquisition, Writing – original draft, Writing – review & editing. LC: Methodology, Writing – original draft. ZY: Formal analysis, Funding acquisition, Methodology, Writing – original draft. JX: Funding acquisition, Methodology, Supervision, Writing – original draft. LF: Methodology, Writing – original draft. NH: Writing – original draft. MG: Methodology, Writing – original draft. JZ: Methodology, Writing – original draft. ZC: Data curation, Methodology, Writing – original draft. HC: Methodology, Supervision, Writing – original draft. GY: Funding acquisition, Methodology, Supervision, Writing – original draft. FC: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Science Foundation for Young Scientists of China (82003885, 22308032) and Science and Technology Projects in Guangzhou (No.2023A03J0230).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul Rahim, A., Uzair, M., Rehman, N., Fiaz, S., Attia, K. A., Abushady, A. M., et al. (2024). CRISPR/Cas9 mediated TaRPK1 root architecture gene mutagenesis confers enhanced wheat yield. J. King Saud Univ. - Sci. 36, 103063. doi: 10.1016/j.jksus.2023.103063
Abeuova, L., Kali, B., Tussipkan, D., Akhmetollayeva, A., Ramankulov, Y., Manabayeva, S. (2023). CRISPR/Cas9-mediated multiple guide RNA-targeted mutagenesis in the potato. Transgenic Res. 32, 383–397. doi: 10.1007/s11248-023-00356-8
Afzal, S., Mubeen, M., Hussain, S., Ali, M., Javeed, H. M. R., Al-Ashkar, I., et al. (2023). “Modern Breeding Approaches for Climate Change,” in Climate Change Impacts on Agriculture: Concepts, Issues and Policies for Developing Countries. Eds. Jatoi, W. N., Mubeen, M., Hashmi, M. Z., Ali, S., Fahad, S., Mahmood, K. (Springer International Publishing, Cham), 299–313.
Akhtar, S., Ahmed, R., Begum, K., Das, A., Banu, S. (2023). “CRISPR/Cas in Improvement of Food Crops for Feeding the World into the Future,” in Advanced Crop Improvement, Volume 2: Case Studies of Economically Important Crops. Eds. Raina, A., Wani, M. R., Laskar, R. A., Tomlekova, N., Khan, S. (Springer International Publishing, Cham), 529–566.
Alam, M. S., Kong, J., Tao, R., Ahmed, T., Alamin, M., Alotaibi, S. S., et al. (2022). CRISPR/Cas9 Mediated Knockout of the OsbHLH024 Transcription Factor Improves Salt Stress Resistance in Rice (Oryza sativa L.). Plants 11, 1184. doi: 10.3390/plants11091184
Ali, A., Zafar, M. M., Farooq, Z., Ahmed, S. R., Ijaz, A., Anwar, Z., et al. (2023). Breakthrough in CRISPR/Cas system: Current and future directions and challenges. Biotechnol. J. 18, 2200642. doi: 10.1002/biot.202200642
Ambika, Bhati, S., Kumar, R. (2024). “Plant Breeding Using the CRISPR-Cas9 System for Food Security and Facing Climate Change,” in Plant Genome Editing Technologies: Speed Breeding, Crop Improvement and Sustainable Agriculture. Eds. Chen, J.-T., Ahmar, S. (Springer Nature Singapore, Singapore), 149–181.
Amezian, D., Nauen, R., Van Leeuwen, T. (2024). The role of ATP-binding cassette transporters in arthropod pesticide toxicity and resistance. Curr. Opin. Insect Sci. 63, 101200. doi: 10.1016/j.cois.2024.101200
Antony Ceasar, S., Ignacimuthu, S. (2023). CRISPR/Cas genome editing in plants: Dawn of Agrobacterium transformation for recalcitrant and transgene-free plants for future crop breeding. Plant Physiol. Biochem. 196, 724–730. doi: 10.1016/j.plaphy.2023.02.030
Arra, Y., Auguy, F., Stiebner, M., Chéron, S., Wudick, M. M., Miras, M., et al. (2024). Rice Yellow Mottle Virus resistance by genome editing of the Oryza sativa L. ssp. japonica nucleoporin gene OsCPR5.1 but not OsCPR5.2. Plant Biotechnol. J. 22, 1299–1311. doi: 10.1111/pbi.14266
Bibi, F., Rahman, A. (2023). An Overview of Climate Change Impacts on Agriculture and Their Mitigation Strategies. Agriculture 13, 1508. doi: 10.3390/agriculture13081508
Biswas, S., Bridgeland, A., Irum, S., Thomson, M. J., Septiningsih, E. M. (2022). Optimization of Prime Editing in Rice, Peanut, Chickpea, and Cowpea Protoplasts by Restoration of GFP Activity. Int. J. Mol. Sci. 23, 9809. doi: 10.3390/ijms23179809
Bui, T. P., Le, H., Ta, D. T., Nguyen, C. X., Le, N. T., Tran, T. T., et al. (2023). Enhancing powdery mildew resistance in soybean by targeted mutation of MLO genes using the CRISPR/Cas9 system. BMC Plant Biol. 23, 533. doi: 10.1186/s12870-023-04549-5
Chakraborty, M., Munshi, S. K., Haque, A., Azad, M. A. K., Islam, T., Alam, M., et al. (2023). “Current Biotechnological Approaches in Maize Improvement,” in Maize Improvement: Current Advances in Yield, Quality, and Stress Tolerance under Changing Climatic Scenarios. Eds. Wani, S. H., Dar, Z. A., Singh, G. P. (Springer International Publishing, Cham), 137–180.
Chen, H., Ye, R., Liang, Y., Zhang, S., Liu, X., Sun, C., et al. (2023). Generation of low-cadmium rice germplasms via knockout of OsLCD using CRISPR/Cas9. J. Environ. Sci. 126, 138–152. doi: 10.1016/j.jes.2022.05.047
Chen, L., Liu, G., Zhang, T. (2024). Integrating machine learning and genome editing for crop improvement. aBIOTECH 5, 262–277. doi: 10.1007/s42994-023-00133-5
Chen, P. J., Liu, D. R. (2023). Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 24, 161–177. doi: 10.1038/s41576-022-00541-1
Cheng, J., Hill, C., Han, Y., He, T., Ye, X., Shabala, S., et al. (2023). New semi-dwarfing alleles with increased coleoptile length by gene editing of gibberellin 3-oxidase 1 using CRISPR-Cas9 in barley (Hordeum vulgare L.). Plant Biotechnol. J. 21, 806–818. doi: 10.1111/pbi.13998
Dai, Z., Yang, Q., Chen, D., Li, B., Que, J., Hu, L., et al. (2023). ZmAGO18b negatively regulates maize resistance against southern leaf blight. Theor. Appl. Genet. 136, 158. doi: 10.1007/s00122-023-04405-y
Das, J., Kumar, S., Mishra, D. C., Chaturvedi, K. K., Paul, R. K., Kairi, A. (2023). Machine learning in the estimation of CRISPR-Cas9 cleavage sites for plant system. Front. Genet. 13, 1085332. doi: 10.3389/fgene.2022.1085332
Datta, S. K., Majumder, S., Datta, K. (2021). “Molecular Breeding for Improved β-carotene Synthesis in Golden Rice: Recent Progress and Future Perspectives,” in Molecular Breeding for Rice Abiotic Stress Tolerance and Nutritional Quality, 287–303. doi: 10.1002/9781119633174.ch15
Ding, J., Ji, C., Yu, L., Wang, C., Ding, G., Wang, S., et al. (2024). OsMYB84, a transcriptional regulator of OsCOPT2 and OsHMA5, modulates copper uptake and transport and yield production in rice. Crop J. 12, 456–469. doi: 10.1016/j.cj.2024.01.012
Dong, O. X., Yu, S., Jain, R., Zhang, N., Duong, P. Q., Butler, C., et al. (2020). Marker-free carotenoid-enriched rice generated through targeted gene insertion using CRISPR-Cas9. Nat. Commun. 11, 1178. doi: 10.1038/s41467-020-14981-y
Dubovskiy, I. M., Grizanova, E. V., Gerasimova, S. V. (2024). “Plant Recombinant Gene Technology for Pest Control in the Twenty-First Century: From Simple Transgenesis to CRISPR/Cas,” in Gene Editing in Plants: CRISPR-Cas and Its Applications. Eds. Kumar, A., Arora, S., Ogita, S., Yau, Y.-Y., Mukherjee, K. (Springer Nature Singapore, Singapore), 17–72.
Dueñas, C., Jr., Mota, G. M. B., Oliva, N., Slamet-Loedin, I. H. (2021). “Genome Engineering for Enriching Fe and Zn in Rice Grain and Increasing Micronutrient Bioavailability,” in Genome Engineering for Crop Improvement., 69–82. doi: 10.1002/9781119672425.ch4
Farhat, S., Jain, N., Singh, N., Sreevathsa, R., Dash, P. K., Rai, R., et al. (2019). CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin. Cell Dev. Biol. 96, 91–99. doi: 10.1016/j.semcdb.2019.05.003
Feng, C., Gao, H., Zhou, Y., Jing, Y., Li, S., Yan, Z., et al. (2023). Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production. Front. Plant Sci. 14, 1162014. doi: 10.3389/fpls.2023.1162014
Figueredo, E. F., Cruz, T. A. D., Almeida, J. R. D., Batista, B. D., Marcon, J., Andrade, P. A. M. D., et al. (2023). The key role of indole-3-acetic acid biosynthesis by Bacillus thuringiensis RZ2MS9 in promoting maize growth revealed by the ipdC gene knockout mediated by the CRISPR-Cas9 system. Microbiological Res. 266, 127218. doi: 10.1016/j.micres.2022.127218
Gaillochet, C., Peña Fernández, A., Goossens, V., D’halluin, K., Drozdzecki, A., Shafie, M., et al. (2023). Systematic optimization of Cas12a base editors in wheat and maize using the ITER platform. Genome Biol. 24, 6. doi: 10.1186/s13059-022-02836-2
Ghosh, S. (2024). Current approaches and future potential for delivering CRISPR/Cas components in oilseeds and millets. Nucleus 67, 141–156. doi: 10.1007/s13237-024-00486-2
Ghosh, S. K., Chatterjee, T. (2024). “Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Associated Proteins (Cas) [CRISPR–Cas]: An Emerging Technique in Plant Disease Detection and Management,” in Gene Editing in Plants: CRISPR-Cas and Its Applications. Eds. Kumar, A., Arora, S., Ogita, S., Yau, Y.-Y., Mukherjee, K. (Springer Nature Singapore, Singapore), 589–645.
Ghoshal, B. (2024). A birds-eye-view on CRISPR-Cas system in agriculture. Nucleus 67, 89–96. doi: 10.1007/s13237-023-00462-2
Ghouri, M. Z., Munawar, N., Aftab, S. O., Ahmad, A. (2023). “Chapter 15 - Regulation of CRISPR edited food and feed: legislation and future,” in GMOs and Political Stance. Eds. Nawaz, M. A., Chung, G., Golokhvast, K. S., Tsatsakis, A. M. (Academic Press), 261–287. doi: 10.1016/B978-0-12-823903-2.00004-4
Guo, C., Ma, X., Gao, F., Guo, Y. (2023). Off-target effects in CRISPR/Cas9 gene editing. Front. Bioengineering Biotechnol. 11, 1143157. doi: 10.3389/fbioe.2023.1143157
Gupta, A., Liu, B., Chen, Q.-J., Yang, B. (2023). High-efficiency prime editing enables new strategies for broad-spectrum resistance to bacterial blight of rice. Plant Biotechnol. J. 21, 1454–1464. doi: 10.1111/pbi.14049
Gurel, F., Wu, Y., Pan, C., Cheng, Y., Li, G., Zhang, T., et al. (2023). On- and Off-Target Analyses of CRISPR-Cas12b Genome Editing Systems in Rice. CRISPR J. 6, 62–74. doi: 10.1089/crispr.2022.0072
Hao, J., Yang, Y., Futrell, S., Kelly, E. A., Lorts, C. M., Nebie, B., et al. (2023). CRISPR/Cas9-Mediated Mutagenesis of Carotenoid Cleavage Dioxygenase (CCD) Genes in Sorghum Alters Strigolactone Biosynthesis and Plant Biotic Interactions. Phytobiomes J. 7, 339–351. doi: 10.1094/PBIOMES-08-22-0053-R
Hasan, M. M. (2024). Towards programming-based synthetic genetic circuit enabled high-lysine maize. Curr. Plant Biol. 39, 100355. doi: 10.1016/j.cpb.2024.100355
He, F., Wang, C., Sun, H., Tian, S., Zhao, G., Liu, C., et al. (2023). Simultaneous editing of three homoeologues of confers broad-spectrum resistance to stripe rust in wheat. Plant Biotechnol. J. 21, 354–368. doi: 10.1111/pbi.13956
He, Y., Han, Y., Ma, Y., Liu, S., Fan, T., Liang, Y., et al. (2024). Expanding plant genome editing scope and profiles with CRISPR-FrCas9 systems targeting palindromic TA sites. Plant Biotechnol. J. 22, 2488–2503. doi: 10.1111/pbi.14363
Hu, S., Chen, Y., Qian, C., Ren, H., Liang, X., Tao, W., et al. (2024). Nuclear accumulation of rice UV-B photoreceptors is UV-B- and OsCOP1-independent for UV-B responses. Nat. Commun. 15, 6396. doi: 10.1038/s41467-024-50755-6
Hu, J., Yu, M., Chang, Y., Tang, H., Wang, W., Du, L., et al. (2022). Functional analysis of TaPDI genes on storage protein accumulation by CRISPR/Cas9 edited wheat mutants. Int. J. Biol. Macromolecules 196, 131–143. doi: 10.1016/j.ijbiomac.2021.12.048
Hualpa-Ramirez, E., Carrasco-Lozano, E. C., Madrid-Espinoza, J., Tejos, R., Ruiz-Lara, S., Stange, C., et al. (2024). Stress salinity in plants: New strategies to cope with in the foreseeable scenario. Plant Physiol. Biochem. 208, 108507. doi: 10.1016/j.plaphy.2024.108507
Huang, Q., Lin, B., Cao, Y., Zhang, Y., Song, H., Huang, C., et al. (2023). CRISPR/Cas9-mediated mutagenesis of the susceptibility gene OsHPP04 in rice confers enhanced resistance to rice root-knot nematode. Front. Plant Sci. 14, 1134653. doi: 10.3389/fpls.2023.1134653
Huang, Y.-Y., Liu, X.-X., Xie, Y., Lin, X.-Y., Hu, Z.-J., Wang, H., et al. (2020). Identification of FERONIA-like receptor genes involved in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 2, 14. doi: 10.1186/s42483-020-00052-z
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., Khan, M. R. (2022). CRISPR/Cas9 mediated disruption of Inositol Pentakisphosphate 2-Kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Advanced Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006
Imran, M., Shafiq, S., Tang, X. (2023). CRISPR-Cas9-mediated editing of BADH2 gene triggered fragrance revolution in rice. Physiologia Plantarum 175, e13871. doi: 10.1111/ppl.v175.1
Iwamoto, M. (2022). In-frame editing of transcription factor gene RDD1 to suppress miR166 recognition influences nutrient uptake, photosynthesis, and grain quality in rice. Sci. Rep. 12, 10795. doi: 10.1038/s41598-022-14768-9
Jiang, Y., Li, J., Liu, B., Cao, D., Zong, Y., Chang, Y., et al. (2022). Novel Hina alleles created by genome editing increase grain hardness and reduce grain width in barley. Transgenic Res. 31, 637–645. doi: 10.1007/s11248-022-00324-8
Jiang, W., Ye, Q., Wu, Z., Zhang, Q., Wang, L., Liu, J., et al. (2023). Analysis of CAT Gene Family and Functional Identification of OsCAT3 in Rice. Genes 14, 138. doi: 10.3390/genes14010138
Jianing, G., Yuhong, G., Yijun, G., Rasheed, A., Qian, Z., Zhiming, X., et al. (2022). Improvement of heat stress tolerance in soybean (Glycine max L), by using conventional and molecular tools. Front. Plant Sci. 13, 993189. doi: 10.3389/fpls.2022.993189
Jogam, P., Sandhya, D., Alok, A., Peddaboina, V., Allini, V. R., Zhang, B. (2022). A review on CRISPR/Cas-based epigenetic regulation in plants. Int. J. Biol. Macromolecules 219, 1261–1271. doi: 10.1016/j.ijbiomac.2022.08.182
Jogam, P., Sandhya, D., Alok, A., Peddaboina, V., Singh, S. P., Abbagani, S., et al. (2023). Editing of TOM1 gene in tobacco using CRISPR/Cas9 confers resistance to Tobacco mosaic virus. Mol. Biol. Rep. 50, 5165–5176. doi: 10.1007/s11033-023-08440-2
Juma, B. S., Mukami, A., Mweu, C., Ngugi, M. P., Mbinda, W. (2022). Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front. Plant Sci. 13, 1009860. doi: 10.3389/fpls.2022.1009860
Jung, W. J., Park, S.-J., Cha, S., Kim, K. (2024). Factors affecting the cleavage efficiency of the CRISPR-Cas9 system. Anim. Cells Syst. 28, 75–83. doi: 10.1080/19768354.2024.2322054
Kan, J., Cai, Y., Cheng, C., Jiang, C., Jin, Y., Yang, P. (2022). Simultaneous editing of host factor gene TaPDIL5-1 homoeoalleles confers wheat yellow mosaic virus resistance in hexaploid wheat. New Phytol. 234, 340–344. doi: 10.1111/nph.v234.2
Karmacharya, A., Li, D., Leng, Y., Shi, G., Liu, Z., Yang, S., et al. (2023). Targeting Disease Susceptibility Genes in Wheat Through wide Hybridization with Maize Expressing Cas9 and Guide RNA. Mol. Plant-Microbe Interactions® 36, 554–557. doi: 10.1094/MPMI-01-23-0004-SC
Karunarathne, S. D., Han, Y., Zhang, X.-Q., Li, C. (2022). CRISPR/Cas9 gene editing and natural variation analysis demonstrate the potential for HvARE1 in improvement of nitrogen use efficiency in barley. J. Integr. Plant Biol. 64, 756–770. doi: 10.1111/jipb.13214
Kaul, T., Thangaraj, A., Jain, R., Bharti, J., Kaul, R., Verma, R., et al. (2024). CRISPR/Cas9-mediated homology donor repair base editing system to confer herbicide resistance in maize (Zea mays L.). Plant Physiol. Biochem. 207, 108374. doi: 10.1016/j.plaphy.2024.108374
Khanna, K., Ohri, P., Bhardwaj, R. (2023). Nanotechnology and CRISPR/Cas9 system for sustainable agriculture. Environ. Sci. pollut. Res. 30, 118049–118064. doi: 10.1007/s11356-023-26482-8
Kim, M.-S., Ko, S.-R., Jung, Y. J., Kang, K.-K., Lee, Y.-J., Cho, Y.-G. (2023). Knockout Mutants of OsPUB7 Generated Using CRISPR/Cas9 Revealed Abiotic Stress Tolerance in Rice. Int. J. Mol. Sci. 24, 5338. doi: 10.3390/ijms24065338
Kim, M.-S., Le, V. T., Jung, Y. J., Kang, K.-K., Cho, Y.-G. (2024). OsPUB9 Gene Edited by CRISPR/Cas9 Enhanced Resistance to Bacterial Leaf Blight in Rice (Oryza sativa L.). Int. J. Mol. Sci. 25, 7145. doi: 10.3390/ijms25137145
Kis, A., Polgári, D., Dalmadi, Á., Ahmad, I., Rakszegi, M., Sági, L., et al. (2024). Targeted mutations in the GW2.1 gene modulate grain traits and induce yield loss in barley. Plant Sci. 340, 111968. doi: 10.1016/j.plantsci.2023.111968
Kloc, Y., Dmochowska-Boguta, M., Żebrowska-Różańska, P., Łaczmański, Ł., Nadolska-Orczyk, A., Orczyk, W. (2024). HvGSK1.1 Controls Salt Tolerance and Yield through the Brassinosteroid Signaling Pathway in Barley. Int. J. Mol. Sci. 25, 998. doi: 10.3390/ijms25020998
Kocsisova, Z., Coneva, V. (2023). Strategies for delivery of CRISPR/Cas-mediated genome editing to obtain edited plants directly without transgene integration. Front. Genome Editing 5, 1209586. doi: 10.3389/fgeed.2023.1209586
Krishna, T. P. A., Maharajan, T., Ceasar, S. A. (2023). The Role of Membrane Transporters in the Biofortification of Zinc and Iron in Plants. Biol. Trace Element Res. 201, 464–478. doi: 10.1007/s12011-022-03159-w
Kumari, R., Saha, T., Kumar, P., Singh, A. K. (2024). CRISPR/Cas9-mediated genome editing technique to control fall armyworm (Spodoptera frugiperda) in crop plants with special reference to maize. Physiol. Mol. Biol. Plants 30, 1161–1173. doi: 10.1007/s12298-024-01486-x
Kumawat, T., Agarwal, A., Saxena, S., Arora, S. (2024). “An Analysis of Global Policies and Regulation on Genome Editing in Plants,” in ene Editing in Plants: CRISPR-Cas and Its Applications. Eds. Kumar, A., Arora, S., Ogita, S., Yau, Y.-Y., Mukherjee, K. (Springer Nature Singapore, Singapore), 775–793.
Lawrenson, T., Clarke, M., Kirby, R., Forner, M., Steuernagel, B., Brown, J. K. M., et al. (2024). An optimised CRISPR Cas9 and Cas12a mutagenesis toolkit for Barley and Wheat. Plant Methods 20, 123. doi: 10.1186/s13007-024-01234-y
Lee, J. S., Bae, S.-J., Kim, J.-S., Kim, C., Kang, B.-C. (2023). A streamlined guide RNA screening system for genome editing in Sorghum bicolor. Plant Methods 19, 90. doi: 10.1186/s13007-023-01058-2
Lei, R., Kuang, R., Peng, X., Jiao, Z., Zhao, Z., Cong, H., et al. (2023). Portable rapid detection of maize chlorotic mottle virus using RT-RAA/CRISPR-Cas12a based lateral flow assay. Front. Plant Sci. 14, 1088544. doi: 10.3389/fpls.2023.1088544
Li, X., Cai, Q., Yu, T., Li, S., Li, S., Li, Y., et al. (2023a). ZmG6PDH1 in glucose-6-phosphate dehydrogenase family enhances cold stress tolerance in maize. Front. Plant Sci. 14, 1116237. doi: 10.3389/fpls.2023.1116237
Li, Y. J., Gu, J. M., Ma, S., Xu, Y., Liu, M., Zhang, C., et al. (2023c). Genome editing of the susceptibility gene ZmNANMT confers multiple disease resistance without agronomic penalty in maize. Plant Biotechnol. J. 21, 1525–1527. doi: 10.1111/pbi.14078
Li, Z., Huang, Q., Lin, B., Guo, B., Wang, J., Huang, C., et al. (2022b). CRISPR/Cas9-targeted mutagenesis of a representative member of a novel PR10/Bet v1-like protein subfamily significantly reduces rice plant height and defense against Meloidogyne graminicola. Phytopathol. Res. 4, 38. doi: 10.1186/s42483-022-00143-z
Li, Z., Li, Z., Ji, Y., Wang, C., Wang, S., Shi, Y., et al. (2024b). The heat shock factor 20-HSF4-cellulose synthase A2 module regulates heat stress tolerance in maize. Plant Cell 36, 2652–2667. doi: 10.1093/plcell/koae106
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022a). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi: 10.1038/s41586-022-04395-9
Li, S., Zhang, Y., Liu, Y., Zhang, P., Wang, X., Chen, B., et al. (2024a). The E3 ligase TaGW2 mediates transcription factor TaARR12 degradation to promote drought resistance in wheat. Plant Cell 36, 605–625. doi: 10.1093/plcell/koad307
Li, X., Zhang, J., Shangguan, X., Yin, J., Zhu, L., Hu, J., et al. (2023b). Knockout of OsWRKY71 impairs Bph15-mediated resistance against brown planthopper in rice. Front. Plant Sci. 14, 1260526. doi: 10.3389/fpls.2023.1260526
Liu, Y., Chen, Z., Zhang, C., Guo, J., Liu, Q., Yin, Y., et al. (2024b). Gene editing of ZmGA20ox3 improves plant architecture and drought tolerance in maize. Plant Cell Rep. 43, 18. doi: 10.1007/s00299-023-03090-x
Liu, X., Gu, D., Zhang, Y., Jiang, Y., Xiao, Z., Xu, R., et al. (2024a). Conditional knockdown of OsMLH1 to improve plant prime editing systems without disturbing fertility in rice. Genome Biol. 25, 131. doi: 10.1186/s13059-024-03282-y
Liu, S., Liu, X., Zhang, X., Chang, S., Ma, C., Qin, F. (2023d). Co-Expression of ZmVPP1 with ZmNAC111 Confers Robust Drought Resistance in Maize. Genes 14, 8. doi: 10.3390/genes14010008
Liu, K., Sakuraba, Y., Ohtsuki, N., Yang, M., Ueda, Y., Yanagisawa, S. (2023a). CRISPR/Cas9-mediated elimination of OsHHO3, a transcriptional repressor of three AMMONIUM TRANSPORTER1 genes, improves nitrogen use efficiency in rice. Plant Biotechnol. J. 21, 2169–2172. doi: 10.1111/pbi.v21.11
Liu, L., Wang, J., Zhang, Q., Sun, T., Wang, P. (2023b). Cloning of the Soybean GmNHL1 Gene and Functional Analysis under Salt Stress. Plants 12, 3869. doi: 10.3390/plants12223869
Liu, Q., Zhao, C., Sun, K., Deng, Y., Li, Z. (2023c). Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant 16, 616–631. doi: 10.1016/j.molp.2023.02.003
Lu, M., Mingfeng, T., Yuxing, Z., Longtao, T. (2023). Knocking-Out OsPDR7 Triggers Up-Regulation of OsZIP9 Expression and Enhances Zinc Accumulation in Rice. Rice Sci. 30, 36–49. doi: 10.1016/j.rsci.2022.05.004
Ludwig, Y., Dueñas, C., Arcillas, E., Macalalad-Cabral, R. J., Kohli, A., Reinke, R., et al. (2024). CRISPR-mediated promoter editing of a cis-regulatory element of OsNAS2 increases Zn uptake/translocation and plant yield in rice. Front. Genome Editing 5, 1308228. doi: 10.3389/fgeed.2023.1308228
Lv, G.-H., Li, Y.-Q., Chen, J.-J., Wu, Z.-X., Wu, W.-M., Wu, X.-H., et al. (2023). ZmHMA3 enhances heavy mental Zn stress tolerance and mediates Zn transport in Maize. Res. Square. doi: 10.21203/rs.3.rs-2970282/v1
Ly, L. K., Ho, T. M., Bui, T. P., Nguyen, L. T., Phan, Q., Le, N. T., et al. (2024). CRISPR/Cas9 targeted mutations of OsDSG1 gene enhanced salt tolerance in rice. Funct. Integr. Genomics 24, 70. doi: 10.1007/s10142-024-01347-6
Maiti, S., Banik, A. (2023). Strategies to fortify the nutritional values of polished rice by implanting selective traits from brown rice: A nutrigenomics-based approach. Food Res. Int. 173, 113271. doi: 10.1016/j.foodres.2023.113271
Makaza, W., En-Nahli, Y., Amri, M. (2023). Harnessing plant resistance against Striga spp. parasitism in major cereal crops for enhanced crop production and food security in Sub-Saharan Africa: a review. Food Secur. 15, 1127–1149. doi: 10.1007/s12571-023-01345-9
Mali, S., Zinta, G. (2024). “Genome Editing for Trait Improvement in Potato (Solanum tuberosum L.),” in Genetic Engineering of Crop Plants for Food and Health Security: Volume 2. Eds. Tiwari, S., Koul, B. (Springer Nature Singapore, Singapore), 405–423.
Malzahn, A. A., Tang, X., Lee, K., Ren, Q., Sretenovic, S., Zhang, Y., et al. (2019). Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 17, 9. doi: 10.1186/s12915-019-0629-5
Marone, D., Mastrangelo, A. M., Borrelli, G. M. (2023). From Transgenesis to Genome Editing in Crop Improvement: Applications, Marketing, and Legal Issues. Int. J. Mol. Sci. 24, 7122. doi: 10.3390/ijms24087122
Mohan, C., Satish, L., Muthubharathi, B. C., Selvarajan, D., Easterling, M., Yau, Y.-Y. (2022). “CRISPR-Cas Technology: A Genome-Editing Powerhouse for Molecular Plant Breeding,” in Biotechnological Innovations for Environmental Bioremediation. Eds. Arora, S., Kumar, A., Ogita, S., Yau, Y. Y. (Springer Nature Singapore, Singapore), 803–879.
Mohr, T., Horstman, J., Gu, Y. Q., Elarabi, N. I., Abdallah, N. A., Thilmony, R. (2022). CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat. Plants 11, 2259. doi: 10.3390/plants11172259
Molinari, M. D. C., Pagliarini, R. F., Florentino, L. H., Lima, R. N., Arraes, F. B. M., Abbad, S. V., et al. (2024). “Navigating the Path from Lab to Market: Regulatory Challenges and Opportunities for Genome Editing Technologies for Agriculture,” in Plant Genome Editing Technologies: Speed Breeding, Crop Improvement and Sustainable Agriculture. Eds. Chen, J.-T., Ahmar, S. (Springer Nature Singapore, Singapore), 25–63.
Morrow, N., Borrell, J. S., Mock, N. B., Büchi, L., Gatto, A., Lulekal, E. (2023). Measure of indigenous perennial staple crop, Ensete ventricosum, associated with positive food security outcomes in southern Ethiopian highlands. Food Policy 117, 102451. doi: 10.1016/j.foodpol.2023.102451
Muha-Ud-Din, G., Ali, F., Hameed, A., Naqvi, S. A. H., Nizamani, M. M., Jabran, M., et al. (2024). CRISPR/Cas9-based genome editing: A revolutionary approach for crop improvement and global food security. Physiol. Mol. Plant Pathol. 129, 102191. doi: 10.1016/j.pmpp.2023.102191
Munawar, N., Ahsan, K., Ahmad, A. (2024). “Chapter 18 - CRISPR-edited plants’ social, ethical, policy, and governance issues,” in Global Regulatory Outlook for CRISPRized Plants. Eds. Abd-Elsalam, K. A., Ahmad, A. (Academic Press), 367–396.
Naeem, M., Alkhnbashi, O. S. (2023). Current Bioinformatics Tools to Optimize CRISPR/Cas9 Experiments to Reduce Off-Target Effects. Int. J. Mol. Sci. 24, 6261. doi: 10.3390/ijms24076261
Nagy, B., Öktem, A., Ferenc, G., Ungor, D., Kalac, A., Kelemen-Valkony, I., et al. (2023). CRISPR/Cas9 Mutagenesis through Introducing a Nanoparticle Complex Made of a Cationic Polymer and Nucleic Acids into Maize Protoplasts. Int. J. Mol. Sci. 24, 16137. doi: 10.3390/ijms242216137
Naik, B. J., Shimoga, G., Kim, S.-C., Manjulatha, M., Subramanyam Reddy, C., Palem, R. R., et al. (2022). CRISPR/Cas9 and Nanotechnology Pertinence in Agricultural Crop Refinement. Front. Plant Sci. 13, 843575. doi: 10.3389/fpls.2022.843575
Narayanan, N., Cueto-Reaño, M. F., Eroğlu, S., Ludwig, Y., Okwuonu, I., Taylor, N. J., et al. (2022). Iron biofortification through genetic modification in rice, wheat, and cassava and its potential contribution to nutritional security. CABI Rev. 11, 1–17. doi: 10.1079/cabireviews202217011
Nguyen, T. T., Pham, D. T., Nguyen, N. H., Do, P. T., To, H. T. M. (2023). The Germin-like protein gene OsGER4 is involved in heat stress response in rice root development. Funct. Integr. Genomics 23, 271. doi: 10.1007/s10142-023-01201-1
Nizampatnam, N. R., Sharma, K., Gupta, P., Pamei, I., Sarma, S., Sreelakshmi, Y., et al. (2024). Introgression of a dominant phototropin1 mutant enhances carotenoids and boosts flavour-related volatiles in genome-edited tomato RIN mutants. New Phytol. 241, 2227–2242. doi: 10.1111/nph.v241.5
Norouzi, M., Nazarain-Firouzabadi, F., Ismaili, A., Ahmadvand, R., Poormazaheri, H. (2024). CRISPR/Cas StNRL1 gene knockout increases resistance to late blight and susceptibility to early blight in potato. Front. Plant Sci. 14, 1278127. doi: 10.3389/fpls.2023.1278127
Otun, S., Escrich, A., Achilonu, I., Rauwane, M., Lerma-Escalera, J. A., Morones-Ramírez, J. R., et al. (2023). The future of cassava in the era of biotechnology in Southern Africa. Crit. Rev. Biotechnol. 43, 594–612. doi: 10.1080/07388551.2022.2048791
Park, J.-R., Kim, E.-G., Jang, Y.-H., Jan, R., Farooq, M., Asif, S., et al. (2024). CRISPR/Cas9-mediated genome editing of OsCS511 enhances cold tolerance in Oryza sativa L. Environ. Exp. Bot. 226, 105932. doi: 10.1016/j.envexpbot.2024.105932
Perez-Garcia, P., Pucciariello, O., Sanchez-Corrionero, A., Cabrera, J., Del Barrio, C., Del Pozo, J. C., et al. (2023). The cold-induced factor CBF3 mediates root stem cell activity, regeneration, and developmental responses to cold. Plant Commun. 4, 100737. doi: 10.1016/j.xplc.2023.100737
Pfeiffer, L. S., Stafforst, T. (2023). Precision RNA base editing with engineered and endogenous effectors. Nat. Biotechnol. 41, 1526–1542. doi: 10.1038/s41587-023-01927-0
Pfotenhauer, A. C., Occhialini, A., Harbison, S. A., Li, L., Piatek, A. A., Luckett, C. R., et al. (2023). Genome-Editing of FtsZ1 for Alteration of Starch Granule Size in Potato Tubers. Plants 12, 1878. doi: 10.3390/plants12091878
Poddar, S., Tanaka, J., Running, K. L. D., Kariyawasam, G. K., Faris, J. D., Friesen, T. L., et al. (2023). Optimization of highly efficient exogenous-DNA-free Cas9-ribonucleoprotein mediated gene editing in disease susceptibility loci in wheat (Triticum aestivum L.). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1084700
Price, L. J. (2022). Investigating barley WRKYs for salinity tolerance by scrutinising CRISPR edited mutants. (College of Science, Health, Engineering and Education, Murdoch University, Perth, WA 6150, Australia).
Qu, Z., Zhang, B., Kong, L., Zhang, Y., Zhao, Y., Gong, Y., et al. (2024). Myeloid zinc finger 1 knockdown promotes osteoclastogenesis and bone loss in part by regulating RANKL-induced ferroptosis of osteoclasts through Nrf2/GPX4 signaling pathway. J. Leukocyte Biol. 115, 946–957. doi: 10.1093/jleuko/qiae011
Ramulifho, E., Rey, C. (2024). A Coiled-Coil Nucleotide-Binding Domain Leucine-Rich Repeat Receptor Gene MeRPPL1 Plays a Role in the Replication of a Geminivirus in Cassava. Viruses 16, 941. doi: 10.3390/v16060941
Rangari, S. K., Kaur Sudha, M., Kaur, H., Uppal, N., Singh, G., Vikal, Y., et al. (2023). DNA-free genome editing for ZmPLA1 gene via targeting immature embryos in tropical maize. GM Crops Food 14, 1–7. doi: 10.1080/21645698.2023.2197303
Raza, H., Abdullah, M. A., Khaliq, S., Sajjad, M. H., Elahi, M., Khan, M. R., et al. (2024). Role of CRISPR in Crop Improvement: A Review. Asian J. Res. Agric. Forestry 10, 12–19. doi: 10.9734/ajraf/2024/v10i2280
Razzaq, M. K., Hina, A., Abbasi, A., Karikari, B., Ashraf, H. J., Mohiuddin, M., et al. (2023). Molecular and genetic insights into secondary metabolic regulation underlying insect-pest resistance in legumes. Funct. Integr. Genomics 23, 217. doi: 10.1007/s10142-023-01141-w
Saber Sichani, A., Ranjbar, M., Baneshi, M., Torabi Zadeh, F., Fallahi, J. (2023). A Review on Advanced CRISPR-Based Genome-Editing Tools: Base Editing and Prime Editing. Mol. Biotechnol. 65, 849–860. doi: 10.1007/s12033-022-00639-1
Sarkar, J., Jyoti, T. P., Sahana, S., Bhattacharya, A., Chandel, S., Singh, R. (2024). CRISPR–Cas13d in plant biology: an insight. Plant Biotechnol. Rep. 18, 301–311. doi: 10.1007/s11816-024-00893-6
Schreiber, T., Prange, A., Schäfer, P., Iwen, T., Grützner, R., Marillonnet, S., et al. (2024). Efficient scar-free knock-ins of several kilobases in plants by engineered CRISPR-Cas endonucleases. Mol. Plant 17, 824–837. doi: 10.1016/j.molp.2024.03.013
Shaibu, A. S., Zhang, S., Ma, J., Feng, Y., Huai, Y., Qi, J., et al. (2022). The GmSNAP11 Contributes to Resistance to Soybean Cyst Nematode Race 4 in Glycine max. Front. Plant Sci. 13, 939763. doi: 10.3389/fpls.2022.939763
Shelake, R. M., Kadam, U. S., Kumar, R., Pramanik, D., Singh, A. K., Kim, J.-Y. (2022). Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 3, 100417. doi: 10.1016/j.xplc.2022.100417
Shen, Y., Ye, Q., Wu, Z., Jiang, W., Wang, L., Zhang, Q., et al. (2024). Functional characterization of OsCAT2 gene in rice that regulates ROS scavenging and plant growth and development. Plant Growth Regul. 103, 165–175. doi: 10.1007/s10725-023-01097-8
Shi, Y., Zhang, Y., Sun, Y., Xie, Z., Luo, Y., Long, Q., et al. (2023). Natural variations of OsAUX5, a target gene of OsWRKY78, control the neutral essential amino acid content in rice grains. Mol. Plant 16, 322–336. doi: 10.1016/j.molp.2022.12.013
Sobrino-Mengual, G., Alvarez, D., Twyman, R. M., Gerrish, C., Fraser, P. D., Capell, T., et al. (2024). Activation of the native PHYTOENE SYNTHASE 1 promoter by modifying near-miss cis-acting elements induces carotenoid biosynthesis in embryogenic rice callus. Plant Cell Rep. 43, 118. doi: 10.1007/s00299-024-03199-7
Song, J. H., Shin, G., Kim, H. J., Lee, S. B., Moon, J. Y., Jeong, J. C., et al. (2022). Mutation of GmIPK1 Gene Using CRISPR/Cas9 Reduced Phytic Acid Content in Soybean Seeds. Int. J. Mol. Sci. 23, 10583. doi: 10.3390/ijms231810583
Sony, S. K., Kaul, T., Motelb, K. F. A., Thangaraj, A., Bharti, J., Kaul, R., et al. (2023). CRISPR/Cas9-mediated homology donor repair base editing confers glyphosate resistance to rice (Oryza sativa L.). Front. Plant Sci. 14, 1122926. doi: 10.3389/fpls.2023.1122926
Sun, X., Lei, R., Zhang, H., Chen, W., Jia, Q., Guo, X., et al. (2024). Rapid and sensitive detection of two fungal pathogens in soybeans using the recombinase polymerase amplification/CRISPR-Cas12a method for potential on-site disease diagnosis. Pest Manage. Sci. 80, 1168–1181. doi: 10.1002/ps.v80.3
Tang, Y., Zhang, Z., Yang, Z., Wu, J. (2023). CRISPR/Cas9 and Agrobacterium tumefaciens virulence proteins synergistically increase efficiency of precise genome editing via homology directed repair in plants. J. Exp. Bot. 74, 3518–3530. doi: 10.1093/jxb/erad096
Távora, F. T. P. K., Meunier, A. C., Vernet, A., Portefaix, M., Milazzo, J., Adreit, H., et al. (2022). CRISPR/Cas9-Targeted Knockout of Rice Susceptibility Genes OsDjA2 and OsERF104 Reveals Alternative Sources of Resistance to Pyricularia oryzae. Rice Sci. 29, 535–544. doi: 10.1016/j.rsci.2022.04.001
Tian, X., Wang, F., Zhao, Y., Lan, T., Yu, K., Zhang, L., et al. (2020). Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol. J. 18, 1109–1111. doi: 10.1111/pbi.13268
Tian, Y., Zhou, Y., Gao, G., Zhang, Q., Li, Y., Lou, G., et al. (2023). Creation of Two-Line Fragrant Glutinous Hybrid Rice by Editing the Wx and OsBADH2 Genes via the CRISPR/Cas9 System. Int. J. Mol. Sci. 24, 849. doi: 10.3390/ijms24010849
Tiwari, J. K., A, J., Tuteja, N., Khurana, S. M. P. (2022). Genome editing (CRISPR-Cas)-mediated virus resistance in potato (Solanum tuberosum L.). Mol. Biol. Rep. 49, 12109–12119. doi: 10.1007/s11033-022-07704-7
Toinga-Villafuerte, S., Vales, M. I., Awika, J. M., Rathore, K. S. (2022). CRISPR/Cas9-Mediated Mutagenesis of the Granule-Bound Starch Synthase Gene in the Potato Variety Yukon Gold to Obtain Amylose-Free Starch in Tubers. Int. J. Mol. Sci. 23, 4640. doi: 10.3390/ijms23094640
Tuo, D., Yao, Y., Yan, P., Chen, X., Qu, F., Xue, W., et al. (2023). Development of cassava common mosaic virus-based vector for protein expression and gene editing in cassava. Plant Methods 19, 78. doi: 10.1186/s13007-023-01055-5
Uranga, M., Daròs, J.-A. (2023). Tools and targets: The dual role of plant viruses in CRISPR–Cas genome editing. Plant Genome 16, e20220. doi: 10.1002/tpg2.20220
Usovsky, M., Gamage, V. A., Meinhardt, C. G., Dietz, N., Triller, M., Basnet, P., et al. (2023). Loss-of-function of an α-SNAP gene confers resistance to soybean cyst nematode. Nat. Commun. 14, 7629. doi: 10.1038/s41467-023-43295-y
Verma, V., Kumar, A., Partap, M., Thakur, M., Bhargava, B. (2023). CRISPR-Cas: A robust technology for enhancing consumer-preferred commercial traits in crops. Front. Plant Sci. 14, 1122940. doi: 10.3389/fpls.2023.1122940
Wang, M., Cheng, J., Wu, J., Chen, J., Liu, D., Wang, C., et al. (2024b). Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1;5-D while preserving yield-related traits. Nat. Genet. 56, 1257–1269. doi: 10.1038/s41588-024-01762-2
Wang, Y., Demirer, G. S. (2023). Synthetic biology for plant genetic engineering and molecular farming. Trends Biotechnol. 41, 1182–1198. doi: 10.1016/j.tibtech.2023.03.007
Wang, H., Feng, M., Jiang, Y., Du, D., Dong, C., Zhang, Z., et al. (2023). Thermosensitive SUMOylation of TaHsfA1 defines a dynamic ON/OFF molecular switch for the heat stress response in wheat. Plant Cell 35, 3889–3910. doi: 10.1093/plcell/koad192
Wang, Q., Gao, H., Liu, K., Wang, H., Zhang, F., Wei, L., et al. (2024d). CRISPR/Cas9-mediated enhancement of semi-dwarf glutinous traits in elite Xiangdaowan rice (Oryza sativa L.): targeting SD1 and Wx genes for yield and quality improvement. Front. Plant Sci. 15, 1333191. doi: 10.3389/fpls.2024.1333191
Wang, X., Guo, Y., Wang, Y., Peng, Y., Zhang, H., Zheng, J. (2024f). ZmHDT103 Negatively Regulates Drought Stress Tolerance in Maize Seedlings. Agronomy 14, 134. doi: 10.3390/agronomy14010134
Wang, X., Ju, Y., Wu, T., Kong, L., Yuan, M., Liu, H., et al. (2024g). The clade III subfamily of OsSWEETs directly suppresses rice immunity by interacting with OsHMGB1 and OsHsp20L. Plant Biotechnol. J. 22, 2186–2200. doi: 10.1111/pbi.14338
Wang, Y., Liu, X., Zheng, X., Wang, W., Yin, X., Liu, H., et al. (2021). Creation of aromatic maize by CRISPR/Cas. J. Integr. Plant Biol. 63, 1664–1670. doi: 10.1111/jipb.13105
Wang, W., Wang, W., Pan, Y., Tan, C., Li, H., Chen, Y., et al. (2022). A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield. Crop J. 10, 1207–1212. doi: 10.1016/j.cj.2022.01.004
Wang, L., Wang, Y., Yin, P., Jiang, C., Zhang, M. (2024a). ZmHAK17 encodes a Na+-selective transporter that promotes maize seed germination under salt conditions. New Crops 1, 100024. doi: 10.1016/j.ncrops.2024.100024
Wang, P., Yamaji, N., Mitani-Ueno, N., Ge, J., Ma, J. F. (2024c). Knockout of a rice K5.2 gene increases Ca accumulation in the grain. J. Integr. Plant Biol. 66, 252–264. doi: 10.1111/jipb.13587
Wang, S., Yang, Y., Guo, M., Zhong, C., Yan, C., Sun, S. (2020). Targeted mutagenesis of amino acid transporter genes for rice quality improvement using the CRISPR/Cas9 system. Crop J. 8, 457–464. doi: 10.1016/j.cj.2020.02.005
Wang, Q., Zhang, D., Dai, Y.-R., Liu, C.-C. (2024e). Efficient tobacco rattle virus-induced gene editing in tomato mediated by the CRISPR/Cas9 system. Biotechnol. J. 19, 2400204. doi: 10.1002/biot.202400204
Wu, Y., Wang, S., Du, W., Ding, Y., Li, W., Chen, Y., et al. (2023). Sugar transporter ZmSWEET1b is responsible for assimilate allocation and salt stress response in maize. Funct. Integr. Genomics 23, 137. doi: 10.1007/s10142-023-01062-8
Wulff-Vester, A., Andersson, M., Brurberg, M. B., Hofvander, P., Alsheikh, M., Harwood, W., et al. (2024). Colour change in potato (Solanum tuberosum L.) tubers by disruption of the anthocyanin pathway via ribonucleoprotein complex delivery of the CRISPR/Cas9 system. Plant Cell Tissue Organ Culture (PCTOC) 157, 25. doi: 10.1007/s11240-024-02743-3
Xie, J., Fei, X., Yan, Q., Jiang, T., Li, Z., Chen, H., et al. (2024). The C4 photosynthesis bifunctional enzymes, PDRPs, of maize are co-opted to cytoplasmic viral replication complexes to promote infection of a prevalent potyvirus sugarcane mosaic virus. Plant Biotechnol. J. 22, 1812–1832. doi: 10.1111/pbi.14304
Xu, X., Sun, S.-K., Zhang, W., Tang, Z., Zhao, F.-J. (2024). Editing Silicon Transporter Genes to Reduce Arsenic Accumulation in Rice. Environ. Sci. Technol. 58, 1976–1985. doi: 10.1021/acs.est.3c10763
Yadav, R. K., Tripathi, M. K., Tiwari, S., Tripathi, N., Asati, R., Chauhan, S., et al. (2023). Genome Editing and Improvement of Abiotic Stress Tolerance in Crop Plants. Life 13, 1456. doi: 10.3390/life13071456
Yang, J., Fang, Y., Wu, H., Zhao, N., Guo, X., Mackon, E., et al. (2023a). Improvement of resistance to rice blast and bacterial leaf streak by CRISPR/Cas9-mediated mutagenesis of Pi21 and OsSULTR3;6 in rice (Oryza sativa L.). Front. Plant Sci. 14, 1209384. doi: 10.3389/fpls.2023.1209384
Yang, S. H., Kim, S. W., Lee, S., Koo, Y. (2024). Optimized protocols for protoplast isolation, transfection, and regeneration in the Solanum genus for the CRISPR/Cas-mediated transgene-free genome editing. Appl. Biol. Chem. 67, 21. doi: 10.1186/s13765-024-00870-1
Yang, L., Machin, F., Wang, S., Saplaoura, E., Kragler, F. (2023b). Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat. Biotechnol. 41, 958–967. doi: 10.1038/s41587-022-01585-8
Yang, J.-S., Reyna-Llorens, I. (2023). Plant synthetic biology: exploring the frontiers of sustainable agriculture and fundamental plant biology. J. Exp. Bot. 74, 3787–3790. doi: 10.1093/jxb/erad220
Yasmeen, A., Shakoor, S., Azam, S., Bakhsh, A., Shahid, N., Latif, A., et al. (2022). CRISPR/Cas-mediated knockdown of vacuolar invertase gene expression lowers the cold-induced sweetening in potatoes. Planta 256, 107. doi: 10.1007/s00425-022-04022-x
Yau, Y.-Y., Easterling, M., Kumar, A. (2024). Advances in Delivery of CRISPR–Cas Reagents for Precise Genome Editing in Plants. Gene Editing Plants 543-569.
Ye, W., Hossain, R., Pröbsting, M., Ali, A. A. M., Han, L., Miao, Y., et al. (2024). Knock-out of BnHva22c reduces the susceptibility of Brassica napus to infection with the fungal pathogen Verticillium longisporum. Crop J. 12, 503–514. doi: 10.1016/j.cj.2024.02.012
Ye, N., Wang, Y., Yu, H., Qin, Z., Zhang, J., Duan, M., et al. (2023). Abscisic Acid Enhances Trehalose Content via OsTPP3 to Improve Salt Tolerance in Rice Seedlings. Plants 12, 2665. doi: 10.3390/plants12142665
Younas, A., Riaz, N., Rashid, M., Tufail, A., Hyder, S., Noreen, Z. (2024). “CRISPR /Cas System for Achieving Abiotic Stress Tolerance,” in OMICs-based Techniques for Global Food Security, 213–231. doi: 10.1002/9781394209156.ch11
Yuan, B., Bi, C., Tian, Y., Wang, J., Jin, Y., Alsayegh, K., et al. (2024). Modulation of the microhomology-mediated end joining pathway suppresses large deletions and enhances homology-directed repair following CRISPR-Cas9-induced DNA breaks. BMC Biol. 22, 101. doi: 10.1186/s12915-024-01896-z
Zafar, K., Khan, M. Z., Amin, I., Mukhtar, Z., Zafar, M., Mansoor, S. (2023). Employing template-directed CRISPR-based editing of the OsALS gene to create herbicide tolerance in Basmati rice. AoB Plants 15, plac059. doi: 10.1093/aobpla/plac059
Zeng, Z., Han, N., Liu, C., Buerte, B., Zhou, C., Chen, J., et al. (2020). Functional dissection of HGGT and HPT in barley vitamin E biosynthesis via CRISPR/Cas9-enabled genome editing. Ann. Bot. 126, 929–942. doi: 10.1093/aob/mcaa115
Zhan, X., Liu, W., Nie, B., Zhang, F., Zhang, J. (2023). Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Commun. Biol. 6, 855. doi: 10.1038/s42003-023-05205-2
Zhang, Y., Blahut-Beatty, L., Zheng, S., Clough, S. J., Simmonds, D. H. (2022a). The Role of a Soybean 14-3-3 Gene (Glyma05g29080) on White Mold Resistance and Nodulation Investigations Using CRISPR-Cas9 Editing and RNA Silencing. Mol. Plant-Microbe Interact. 36, 159–164. doi: 10.1094/MPMI-07-22-0157-R
Zhang, Y., Guo, W., Chen, L., Shen, X., Yang, H., Fang, Y., et al. (2022b). CRISPR/Cas9-Mediated Targeted Mutagenesis of GmUGT Enhanced Soybean Resistance Against Leaf-Chewing Insects Through Flavonoids Biosynthesis. Front. Plant Sci. 13, 802716. doi: 10.3389/fpls.2022.802716
Zhang, L., He, W., Fu, R., Wang, S., Chen, Y., Xu, H. (2023b). Guide-specific loss of efficiency and off-target reduction with Cas9 variants. Nucleic Acids Res. 51, 9880–9893. doi: 10.1093/nar/gkad702
Zhang, M., Li, Y., Liang, X., Lu, M., Lai, J., Song, W., et al. (2023d). A teosinte-derived allele of an HKT1 family sodium transporter improves salt tolerance in maize. Plant Biotechnol. J. 21, 97–108. doi: 10.1111/pbi.13927
Zhang, H., Liu, M., Yin, K., Liu, H., Liu, J., Yan, Z. (2024a). A novel OsHB5-OsAPL-OsMADS27/OsWRKY102 regulatory module regulates grain size in rice. J. Plant Physiol. 295, 154210. doi: 10.1016/j.jplph.2024.154210
Zhang, N., Pombo, M. A., Rosli, H. G., Martin, G. B. (2020). Tomato Wall-Associated Kinase SlWak1 Depends on Fls2/Fls3 to Promote Apoplastic Immune Responses to Pseudomonas syringae. Plant Physiol. 183, 1869–1882. doi: 10.1104/pp.20.00144
Zhang, L., Wang, S., Bai, B., Chen, Y., Xiang, Z., Chen, C., et al. (2024b). OsKASI-2 is required for the regulation of unsaturation levels of membrane lipids and chilling tolerance in rice. Plant Biotechnol. J. 22, 2157–2172. doi: 10.1111/pbi.14336
Zhang, A., Wang, F., Kong, D., Hou, D., Huang, L., Bi, J., et al. (2023a). Mutation of DEP1 facilitates the improvement of plant architecture in Xian rice (Oryza sativa). Plant Breed. 142, 338–344. doi: 10.1111/pbr.v142.3
Zhang, N., Wang, S., Zhao, S., Chen, D., Tian, H., Li, J., et al. (2023e). Global crotonylatome and GWAS revealed a TaSRT1-TaPGK model regulating wheat cold tolerance through mediating pyruvate. Sci. Adv. 9, eadg1012. doi: 10.1126/sciadv.adg1012
Zhang, L., Xiang, Z., Li, J., Wang, S., Chen, Y., Liu, Y., et al. (2023c). bHLH57 confers chilling tolerance and grain yield improvement in rice. Plant Cell Environ. 46, 1402–1418. doi: 10.1111/pce.14513
Zhang, J., Zhang, H., Li, S., Li, J., Yan, L., Xia, L. (2021). Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9. J. Integr. Plant Biol. 63, 1649–1663. doi: 10.1111/jipb.13151
Zhao, J., Yao, B., Peng, Z., Yang, X., Li, K., Zhang, X., et al. (2024). Splicing defect of StDRO2 intron 1 promotes potato root growth by disturbing auxin transport to adapt to drought stress. Hortic. Plant J. In press. doi: 10.1016/j.hpj.2023.11.003
Zheng, S., Ye, C., Lu, J., Liufu, J., Lin, L., Dong, Z., et al. (2021). Improving the Rice Photosynthetic Efficiency and Yield by Editing OsHXK1 via CRISPR/Cas9 System. Int. J. Mol. Sci. 22, 9594. doi: 10.3390/ijms22179554
Zheng, X., Zhang, S., Liang, Y., Zhang, R., Liu, L., Qin, P., et al. (2023). Loss-function mutants of OsCKX gene family based on CRISPR-Cas systems revealed their diversified roles in rice. Plant Genome 16, e20283. doi: 10.1002/tpg2.20283
Zhou, J., Li, Z., Li, Y., Zhao, Q., Luan, X., Wang, L., et al. (2023a). Effects of Different Gene Editing Modes of CRISPR/Cas9 on Soybean Fatty Acid Anabolic Metabolism Based on GmFAD2 Family. Int. J. Mol. Sci. 24, 4769. doi: 10.3390/ijms24054769
Zhou, J., Liu, G., Zhao, Y., Zhang, R., Tang, X., Li, L., et al. (2023b). An efficient CRISPR–Cas12a promoter editing system for crop improvement. Nat. Plants 9, 588–604. doi: 10.1038/s41477-023-01384-2
Zhu, H., Li, C., Gao, C. (2020). Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9
Keywords: climate change, CRISPR/Cas system, genetic modifications, abiotic stress, global food security, sustainable agricultural
Citation: Chen F, Chen L, Yan Z, Xu J, Feng L, He N, Guo M, Zhao J, Chen Z, Chen H, Yao G and Liu C (2024) Recent advances of CRISPR-based genome editing for enhancing staple crops. Front. Plant Sci. 15:1478398. doi: 10.3389/fpls.2024.1478398
Received: 09 August 2024; Accepted: 03 September 2024;
Published: 23 September 2024.
Edited by:
Avinash Mishra, Central Salt & Marine Chemicals Research Institute (CSIR), IndiaReviewed by:
Engin Tilkat, Batman University, TürkiyeMst. Muslima Khatun, National Institute of Biotechnology (NIB), Bangladesh
Copyright © 2024 Chen, Chen, Yan, Xu, Feng, He, Guo, Zhao, Chen, Chen, Yao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Chen, Y2hlbmZlbmdiZWVAZ21haWwuY29t
 Feng Chen
Feng Chen Lu Chen2
Lu Chen2 Huiqi Chen
Huiqi Chen Chunping Liu
Chunping Liu