- 1Department of Electronic Engineering, Core Research Institute of Intelligent Robots, Jeonbuk National University, Jeonju, Republic of Korea
- 2College of Artificial Intelligence, Tianjin University of Science and Technology, Tianjin, China
- 3Department of Computer Engineering, Mokpo National University, Mokpo, Republic of Korea
Although plant disease recognition has witnessed a significant improvement with deep learning in recent years, a common observation is that current deep learning methods with decent performance tend to suffer in real-world applications. We argue that this illusion essentially comes from the fact that current plant disease recognition datasets cater to deep learning methods and are far from real scenarios. Mitigating this illusion fundamentally requires an interdisciplinary perspective from both plant disease and deep learning, and a core question arises. What are the characteristics of a desired dataset? This paper aims to provide a perspective on this question. First, we present a taxonomy to describe potential plant disease datasets, which provides a bridge between the two research fields. We then give several directions for making future datasets, such as creating challenge-oriented datasets. We believe that our paper will contribute to creating datasets that can help achieve the ultimate objective of deploying deep learning in real-world plant disease recognition applications. To facilitate the community, our project is publicly available at https://github.com/xml94/PPDRD with the information of relevant public datasets.
1 Introduction
Having enough food is a basic requirement for human beings. However, more than 600 million people worldwide are estimated to be exposed to hunger in 2030 according to the United Nations. However, many things threaten the availability of food, and plant disease is one of the most essential. It is estimated that $220 billion is lost due to plant disease according to the Food and Agriculture Organization of the United Nations. It is therefore eager to mitigate them, and recognizing plant diseases is a fundamental mission. However, a traditional way is that human experts have to go to the farm to see the plants and then make decisions. This paradigm is expensive and noisy because training experts takes time and many factors play a role in human’s making decisions such as mood and the time taken to complete work (Kahneman et al., 2021).
Deep learning has shown the potential to recognize plant diseases automatically in recent years (Singh et al., 2018; Liu and Wang, 2021; Thakur et al., 2022; Xu et al., 2022b; Salman et al., 2023; Xu, 2023). To access deep learning models, the dataset is one of the most essential considerations (Krishna et al., 2017; Cui and Athey, 2022; Wright and Ma, 2022; Xu et al., 2023b). High-quality training datasets are expected to achieve decent test performance and superior generalization capability. However, plant disease recognition datasets have received relatively less attention in recent years. We argue that it is worth focusing on these datasets for the following reasons.
First, the current datasets exaggerate the performance of existing deep-learning models. From a general computer vision perspective, a common observation is that deep learning models tend to degrade non-trivially when the training and test datasets are not in the same distribution (Arjovsky et al., 2019; Bengio et al., 2021; Cui and Athey, 2022; Corso et al., 2023), known as poor generalization. In the context of plant disease recognition, the ultimate goal is to secure superior performance in the test process. When the test datasets are heterogeneous from the training datasets, the trained models do not have reliable performance when deploying. For example, very recent papers suggested that a model trained with controlled background images degrades in performance when tested with uncontrolled background images (Ahmad et al., 2023; Guth et al., 2023; Wu et al., 2023). Therefore, making reliable TEST datasets to test the performance of the model is a fundamental issue for real-world applications.
Second, high-quality TRAINING datasets for plant disease recognition are relatively difficult to collect as this requires an essential understanding in both the deep learning and agricultural fields. Compared to generic benchmarks in computer vision such as ImageNet (Deng et al., 2009) and COCO (Lin et al., 2014) that are related to daily-available objects, strong domain knowledge about agriculture and plants is required to create a plant disease recognition dataset. Simultaneously, knowledge about deep learning should be involved, such as the challenges related to current deep learning methods and data annotation strategies. For example, data annotation should be compatible with the application’s objective and deep learning methods, as detailed in Section 2.6. Hence, considering dataset characteristics from both deep learning and plant disease perspectives is another essential issue.
To address such issues, this paper aims to enhance the understanding of the creation of datasets, evaluate the reliability of deep learning models in close to real-world scenarios, and further facilitate the deployment of deep learning for plant disease recognition. Our ambitious objective is to deploy deep learning models in real-world applications effectively, efficiently, reliably, and robustly. Our study is inspired by a current paradigm in deep learning, data-centric AI1 (Whang et al., 2023; Zha et al., 2023). To our knowledge, this study is the first to address these issues. In this way, our paper is in the PERSPECTIVE style heterogeneous from the REVIEW papers (Liu et al., 2021; Ouhami et al., 2021; Singh et al., 2021; Thakur et al., 2022; Shoaib et al., 2023) with the aim of investigating the deep learning methods used and presenting current datasets. In summary, this study has the two main contributions:
● It proposes an informative taxonomy for plant disease recognition datasets.
● It presents future directions for creating plant disease datasets using deep learning.
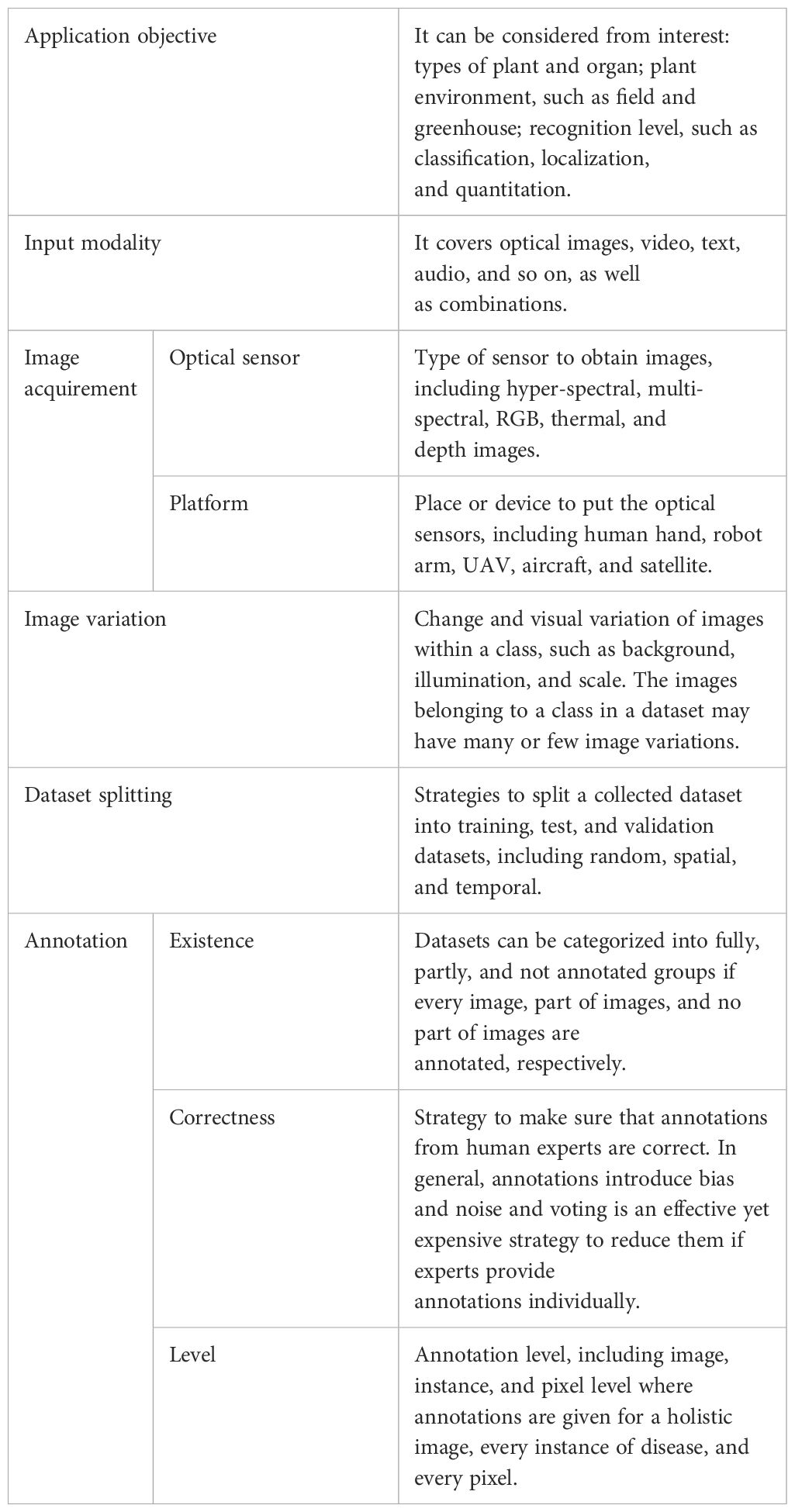
2 Taxonomy
Using deep learning to recognize plant disease is an interdisciplinary challenge. Such a holistic application should be considered from both perspectives, which is the motivation of this section. As shown in Table 1, a taxonomy is proposed. We hope that it will enhance the understanding of the community with the objectives of real-world applications, the collection of suitable training datasets and reliable test datasets, and the deployment of compatible deep learning methods.
2.1 Application objective
In terms of plant disease recognition, different applications may have specific interests, such as the type of plants and organs. For example, some applications focus on one specific crop such as tomatoes (Fuentes et al., 2017b; Xu et al., 2022a) and apples (Thapa et al., 2020) whereas others may consider multiple crops (Hughes et al., 2015; Liu et al., 2021). Similarly, diseases exist in different organs, such as leaves, fruits, and stems.
Moreover, applications require different recognition levels. When diseases appear, one may wonder what it is, referring to classification. Sometimes, multiple abnormal patterns may occur simultaneously, and localizing them individually is beneficial. Specifically, a plant may have more than one unhealthy symptom where the locations give more precise information. Furthermore, some decisions and remedies can be adopted based on their magnitudes, termed quantitation, such as the number of infected leaves and the severity of an unhealthy leaf. To some extent, the complexities of the aforementioned analysis gradually improve. Fortunately, these analyses can be implemented by choosing the appropriate deep learning methods, such as image classification, object detection, and segmentation (Xu et al., 2023a).
Furthermore, plants may grow in either in a controlled environment, such as a greenhouse, or a field. In general, diverse environmental settings suggest differences that should be considered when developing datasets and deep learning methods.
2.2 Input modality
To recognize plant diseases, human experts use multiple senses such as vision and smell. In addition, knowledge from other experts and their own experience also provide benefits. In terms of machines equipped with deep learning, similar scenarios exist. Optical images, a type of vision, are one of the most fundamental modalities of information to recognize plant diseases. They can be obtained with different devices and with multiple sub-categories, as described in the next subsection. Videos and time-series images provide additional information compared to images alone. To be more specific, videos can capture visual patterns of plant diseases from different perspectives and distances that can be taken as accumulated observations. In a similar way, time series images resemble the actions of human experts who investigate the transformation of plant diseases over time to make decisions. In addition, texts are also beneficial for semantic information in nature because they are created by human beings. For example, text can depict the characteristics of plant diseases such as color and their temporal changes. Text can describe images such as the location of diseases and their magnitudes of severity (Fuentes et al., 2019; Wang et al., 2022; Cao et al., 2023). Furthermore, text can be replaced with audio to provide human knowledge. More modalities are possible and encouraged, such as smell and other new ones, because new types of sensors can also be employed in the future (Zhang et al., 2023).
2.3 Image acquirement
Although there are heterogeneous input modalities to recognize plant diseases as mentioned above, optical images are the most widely used. This subsection aims to probe the ways to obtain them since there are multiple types for optical images that are beneficial for diverse cases (Oerke et al., 2014; Mahlein, 2016). As shown in Figure 1, optical image acquirement is grouped into two factors, sensors that produce the images and the platform to hold the sensors. This paper focuses on passive sensing, and active remote sensing techniques such as radar are not considered.
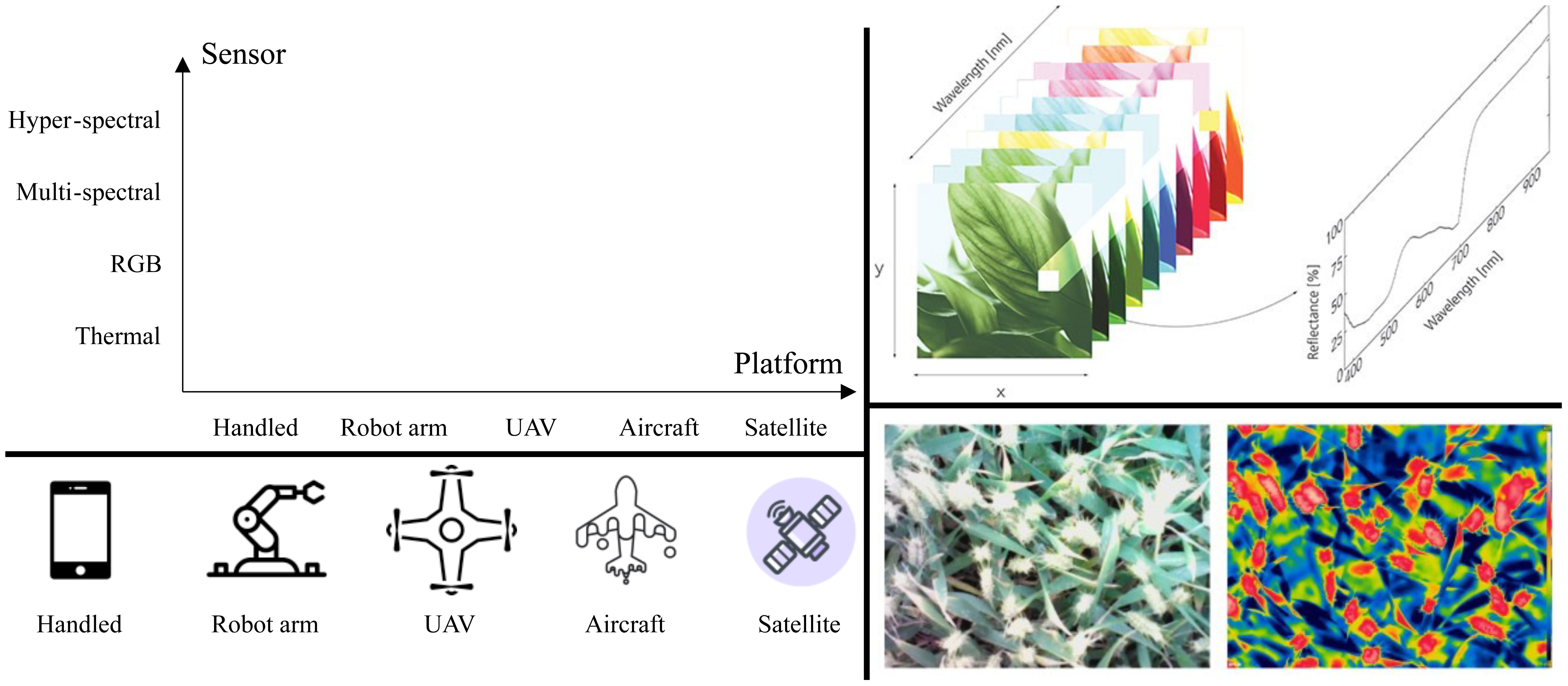
Figure 1. Top-Left: platforms and sensors can be used to obtain plant disease images in different combinations for different purposes such as for plants in fields and greenhouses, adapted from (Xu, 2023) and inspired by (Oerke et al., 2014; Mahlein, 2016). The distances between the plant and platform and the number of channels increase along the horizontal and vertical axes. Further types of sensors and platforms are possible and encouraged. For the advantages and disadvantages of the sensors and platform, please refer to (Oerke et al., 2014; Mahlein, 2016). Bottom-left: visual examples of platforms. Top-right2: an illustration of a hyper-spectral image and a multi-spectral image. Bottom-right: an RGB image and the corresponding thermal image using an infrared camera of a wheat canopy (Feng et al., 2021).
The most widely used type of optical sensor is the RGB camera (red, green, and blue) which captures a range of visible wavelengths. Humans understand RGB images very well. One of the reasons for the popularity of RGB images is the great availability resulting from relatively cheap mobile phones. Such phones handled by human beings can produce an enormous number of RGB images. Imagine a scenario where anyone with a mobile phone can take pictures if they are interested in abnormal plants. Furthermore, the resolution is significantly large with clear details. Unlike humans, RGB cameras can be fixed to monitor the growth of plants. RGB cameras placed in robot arms that can move automatically would be an efficient way to free human beings. We argue that this type of image capture is superior to plants in greenhouses. In spite of the super-comfort of RGB images, extra information is also required. For example, thermal sensors are light-free and thus can be employed at night when RGB cameras fail to work. Fluorescence is another possible type although, to the best of our knowledge, there is no related dataset.
The aforementioned sensors take images with a certain range of wavelengths. In contrast, multi-and hyper-spectral sensors can record images with multiple and many ranges of wavelength, resulting in images with many channels. Many vegetation indexes can be obtained with the two sensors (Adão et al., 2017; Lu et al., 2019; Lu et al., 2020; Wan et al., 2022). One of the main advantages is that they can capture images of a large area, beyond a single leaf and plant (Oerke et al., 2014; Mahlein, 2016; Xu, 2023). These two sensors are generally placed in UAVs (unmanned aerial vehicles) and aircraft to surveil many plants. However, their disadvantages are the non-trivial computations resulting from the many channels and being inconvenient to use.
2.4 Image variation
In the age of traditional machine learning, engineers and researchers carefully consider data collection, and thus the collected data are relatively small, but informative (Wright and Ma, 2022). This situation has changed in the era of deep learning, where the datasets have become much larger yet non-informative. Sometimes, datasets are collected without any specific objective in advance (Wright and Ma, 2022). Analyzing these datasets is therefore essential, and variation is arguably one of the most important variables for image-based datasets (Fuentes et al., 2017a; Singh et al., 2020; Wu et al., 2023; Xu, 2023; Xu et al., 2023b; Xu et al., 2023a). To achieve decent generalization performance and a basic assumption of machine learning and deep learning, the training and test datasets must be in an identical and independent distribution (i.i.d) (Vapnik, 1991). However, this assumption does not hold in many real-world applications. Hence, we contend that understanding variation within a collected dataset is beneficial for robust applications, and this study focuses on RGB image variation because of its prevalence in recent years.
Officially, image variations consist of inter-class, the diversity between two classes, and intra-class, the diversity within one class (Xu et al., 2023a). One of the basic assumptions in distinguishing plant diseases is that different diseases have different visual patterns even if they are similar (Xu et al., 2023a); otherwise, pattern recognition and classification methods fail. However, recognizing diseases that share some visual patterns, i.e., smaller inter-class image variation, is difficult. Images from one class but with disparate visual patterns, i.e., larger intra-class image variation, such as the flower colors in different growth stages, are also challenging to classify. From the perspective of agriculture, it is inevitable that we have smaller inter-class and larger intra-class image variations. Therefore, deep learning methods are expected to mitigate this challenge. In general, testing models with test images that have similar image variations as the training images tends to lead to high performances. In contrast, deep learning methods are expected to have a poor generalization ability, such that models training only with images from controlled imaging environments will have a low performance when tested with images from uncontrolled ones (Guth et al., 2023; Wu et al., 2023).
The main image variations are summarized in Table 2 and Figure 2 illustrates some image variations. Some variations are closely related. For example, images of the plants in the field may have a much larger diversity in illumination than images from the greenhouse and laboratory. Similarly, canopies tend to have smaller scales than leaves and fruits. Furthermore, additional factors may be the source of multiple variations, for example, a person’s habits when they take pictures could result in diversity in scales and viewpoints. We emphasize that we group backgrounds as either uncontrolled or controlled. For example, leaves are put on homogeneous materials such as paper in the laboratories or field. In the field, plant organs of interest can also be moved to have a simple background. In contrast, with an uncontrolled background, the images are taken without considering the background. Therefore, backgrounds vary significantly and can be controlled when taking pictures of the plants in fields.
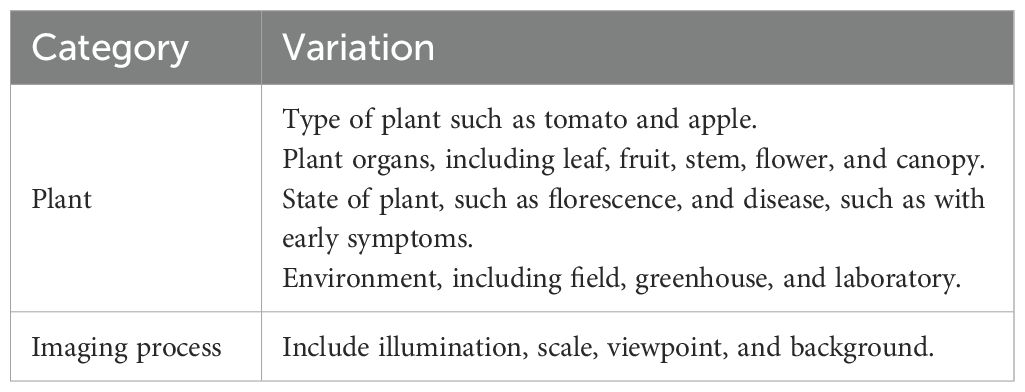
Table 2. Factors of image variation, partially summarized from (Xu et al., 2023a).

Figure 2. Examples of some image variations from the first to last row: disease stage, illumination, scale, and background. The images are taken from the corresponding datasets. In this paper, the image background is grouped into three groups: simple (the first two images), medium (the third and fourth images), and complex (the last image).
2.5 Dataset splitting
In general, three types of datasets are adopted to develop deep learning models. Training datasets are used to train models and validation datasets are used to choose the optimal set of hyper-parameters such as the architecture of the models. After training and validating, test datasets are finally used to check the performance of the trained models. In practical applications, the training and validation datasets are available in the training process, whereas the test dataset is not available until users use the trained model. In such a case, the test performance is not known and thus the model developers cannot assess the trained models. An alternative scheme is to split a holdout dataset as a test before training the models, resembling the real test data. Empirical results suggest that splitting results in a different performance, and thus the splitting strategies should be considered. To be clear, the collected dataset is referred to as the original one that will be split into three parts, training, validation, and test. The real test data from model users are distinguished from the split test dataset.
The most widely used strategy is random splitting. This is when an image in the original data set is randomly placed in one of the three datasets. One of the main issues with this strategy is that multiple images taken for the same observation (e.g., a symptom of plant disease in nature) with only slight differences can be assigned to the training and test datasets, by which the test performance could be overestimated. Moreover, this strategy ignores the generalization challenge (Arjovsky et al., 2019; Bengio et al., 2021; Corso et al., 2023; Xu et al., 2023a), which commonly exists in real-world applications, such that the real test data do not fall into the same distribution as the training dataset.
To mitigate the issues, splitting the data spatially and temporally is appealing. For example, images taken in a place are put into either the training or test datasets (Beery et al., 2022). Similarly, images on the same day or in the same year can be used for only one of the three datasets. In spite of being invariant to the type of plant diseases, the spatial and temporal factors explicitly allow the training and test datasets to be in different distributions. Although the strategy introduces new challenges, such as domain shift (Xu et al., 2023a) as suggested in (Beery et al., 2022), it is worthwhile. Specifically, our objective is to achieve the best performance in the real test process, rather than in the training or the split test datasets. For example, a model trained on the collected data from several farms this year is probably desired to be deployed on different farms for the next couple of years. Beyond spatial and temporal splitting, more things can be considered and encouraged, such as the images being taken by the same person who may have certain habits when taking pictures.
2.6 Annotation strategy
To begin with, we explicitly propose the rules for annotating datasets as follows:
● Annotations are difficult, time-consuming, and expensive to obtain.
● Annotations from humans have bias and noise.
● Images and their annotations should simultaneously satisfy the requirements of deep learning methods and agricultural tasks.
The first rule triggers the first question of whether an image is annotated or not, termed the existence of annotation. Usually, all images are fully labeled in related public datasets. On the contrary, all images in a collected dataset are not completely annotated. A more reasonable case is partial annotations where some images are labeled whereas other images are not, mainly because the images are more easily available in a relative manner. In such a scenario, two more factors should be considered: image level, whether an image should be annotated, and class level, how many images should be annotated for a class. We argue that partially annotated datasets should be promoted considering the characteristics of practical applications. In addition, theory also supports it as a marginal distribution of images can be useful with the learned conditional distribution or joint distribution, between labels and images (Bengio et al., 2013).
Furthermore, bias and noise appear when humans make decisions and the magnitude may be underestimated (Kahneman et al., 2021). For example, the validation dataset of ImageNet (Deng et al., 2009), used to perform image classification of generic objects such as dogs and cats, has approximately 6% incorrect labels (Northcutt et al., 2021). Compared to this case, plant disease annotation requires more domain knowledge and it may be more difficult to be precise. For example, three experts had only 85.9% accuracy on average when labeling 999 wheat images (Long et al., 2023). Noisy annotations in the training datasets could result in an unstable training process and inferior test performance, whereas the noise in the validation datasets may lead to the incorrect selection of hyper-parameters (Patrini et al., 2017). Deep learning generally assumes that the annotations are correct and tends to obtain better performance if the annotation noise is smaller (Patrini et al., 2017). Based on this observation, making precise annotations is worthwhile, yet it requires more resources. For example, independent voting by multiple experts tends to be beneficial (Kahneman et al., 2021). In addition, polymerase chain reaction (PCR) may also contribute (Pereira et al., 2023). We emphasize that we are not trying to say that bias and noise should be avoided completely but that they should be noticed when annotating and decreased considering the trade-offs both in the model training and validation stages.
The last law highlights the format of annotation, called levels of the annotation. In general, image classification can be performed at image-level annotation, i.e., an image with a label. Multi-label image classification is also possible if an image contains multiple plant diseases. However, bounding boxes can point out the location of every instance of plant disease in an image and thus is called instance-level annotation. Moreover, pixel-level annotations are desired for the task of segmentation which assigns a label to every pixel. For every level of annotation, extra strategies exist, such as the EEP (Xu et al., 2023a): Exclusion, every annotation includes only one specific visual pattern of plant disease; Extensiveness, every plant disease in the images should have been annotated; Precision, annotation is expected to be precise for different tasks such as the correct labels and precise location of bounding boxes. Again, we highlight that incompatible formats of annotation become feasible with the concept of weak supervision (Zhou, 2018; Xu et al., 2023a) and have a negative impact on test performance. In addition, new types of annotations have emerged and new models may embrace different types of annotation.
Considering the advantages of a localization task using object detection with a weaker image assumption than image classification and a lower annotation workload than segmentation (Xu et al., 2023a), we give more details about it beyond the EEP (Xu et al., 2023a) strategy. To be more specific, how can we give the boundary box for different diseases in a consistent manner, as illustrated in Figure 3? In general, three independent strategies can be used for a bounding box. First, every instance of fruit or leaf with diseased symptoms is labeled, termed the global level. The problem is that an instance may have multiple diseases and thus the corresponding bounding boxes will have diverse labels and include the healthy parts, which may confuse deep learning models or cause challenges for model optimization. Second, every single symptom gets a bounding box and the symptom is assumed to be dense without a non-trivial gap, termed the local level. In this case, many bounding boxes may exist in an instance, such as the third and fourth images in the first row of Figure 3, which makes annotation harder and more time-consuming. Furthermore, different annotators may have diverse definitions of what constitutes “dense”. Third, the semi-level is a trade-off of the previous two, allowing a gap between symptoms, especially for those that are tiny but many. Based on our understanding and experimental results, an adaptive strategy (Dong et al., 2022) is recommended so that different diseases have different levels of annotation. Another issue is the inconsistency mentioned by Andrew Ng in a video. The underlying issue is that different annotators or even the same annotator at a different time would use different levels of bounding boxes, as shown when comparing annotations 1 and 2 for same image in Figure 3. This inconsistency in training datasets gives different information to models, resulting in unstable learning. In addition, inconsistency in the test process may give us an inaccurate evaluation.

Figure 3. Bounding box annotation strategies in object detection, useful for the localization task. Top row: three strategies for bounding boxes: global (light yellow), which covers one instance such as an instance of fruit or leaf; local (light blue), which covers local areas with dense and intensive symptoms; and semi-global (dotted red), which is a trade-off of previous two, covering local areas yet allowing sparse symptoms such as the case in the first image. Middle row: recommended disease-adaptive strategy in which different diseases may use either local or semi-global strategies. The global strategy is not recommended because an instance may include more than one type of diseases and may include healthy part. Bottom row: inconsistent annotation when the bounding boxes for the same disease are given using different strategies in a dataset, which may confuse the deep learning model and result in optimizing issues. The picture is adapted from (Dong et al., 2022).
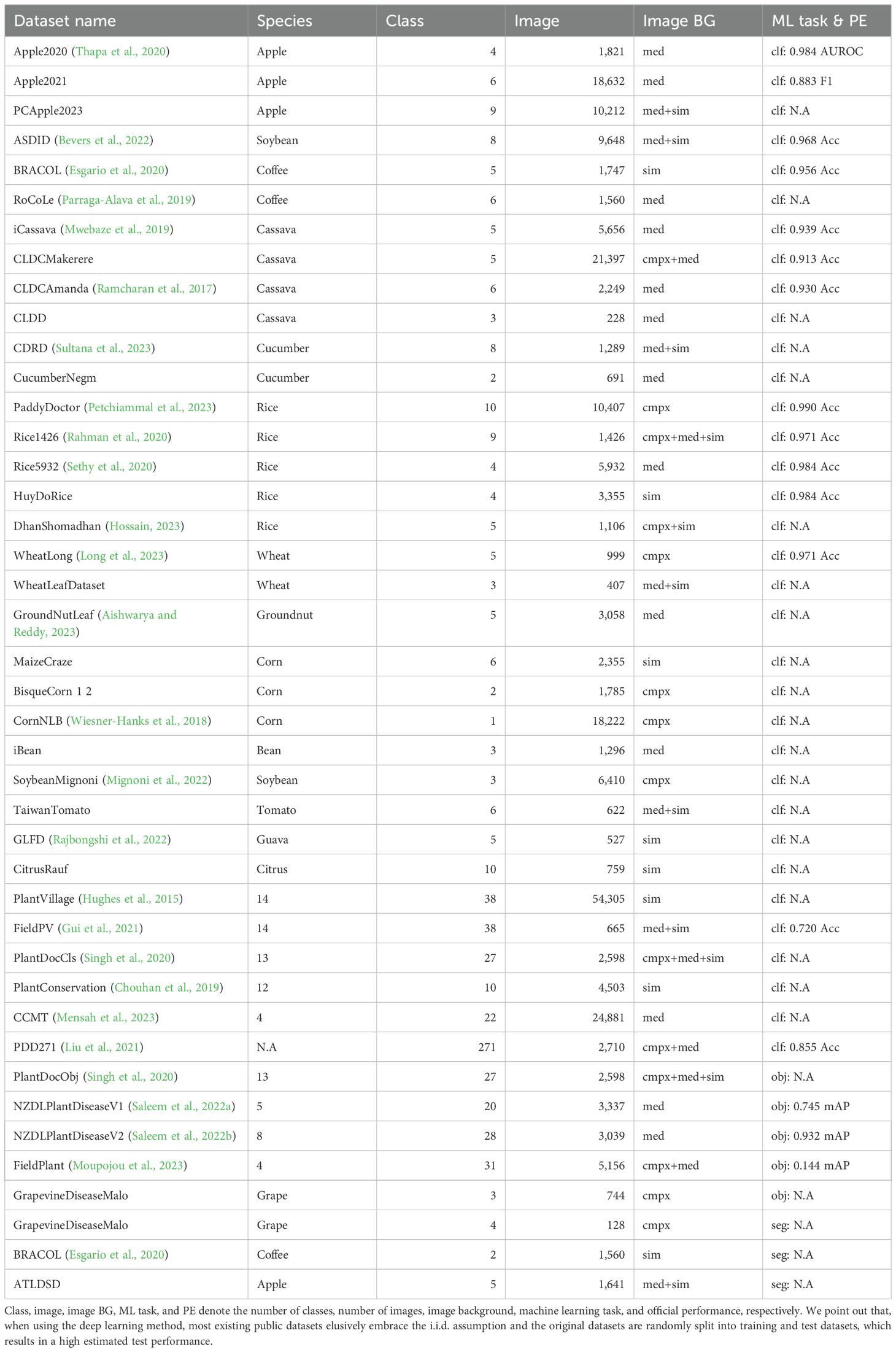
3 Public plant disease recognition datasets
Based on our preliminary survey, RGB images taken by hand-held cameras dominate in public plant disease recognition datasets. Other types of datasets are rarely utilized or are public. Therefore, we aim to provide a survey on the use of RGB images to recognize plant diseases in this section. We did not do a complete survey, as that is impossible to some extent, and rather focused on the datasets with a relatively higher frequency of utilization or which were released recently, which suggests the tendency in this field. For a dataset, the following tags were considered:
● Dataset name. We will assign a name for the dataset if it was not given one in the original material. The datasets are by default publicly available. Some partial public datasets are also included.
● Plant species. If only one crop is included, the name is given. Otherwise, the number of plant species is given.
● Number of classes. Disease classes and healthy ones are included.
● Number of images. Only the images with publicly available annotations are counted.
● Image background (BG). We split the image background into three categories as shown in the last row in Figure 2. The simple one was taken in the environment of a laboratory where the region of interest (RoI) is put on the controlled material. The complex one is taken in the field with a complex background. The medium one is also taken in the field but the RoI may be moved to have a simpler background. Their corresponding abbreviations are sim, med, and cmpx.
● Machine learning (ML) task and official performance (PE). This paper focuses on three types of machine learning tasks as discussed before: image classification (clf), object detection (obj), and segmentation (seg). A dataset may support more than one task. We only report official performance, either in the original publications or from the leaderboard of official challenges. Otherwise, NA denotes not available.
Table 3 summarizes the related public datasets and our project is publicly available on GitHub with more detailed information. Although we tried to do our best, some beneficial datasets may be not included and thus any new contribution is welcome. One of the main observations of enough descriptions about objectives and usages are lacking. As mentioned above, localization and quantitation analysis are also beneficial, but there are a few relevant datasets with relatively high quality that are available to the public. Another observation is that a decent performance is achieved in most of the reported performance datasets. An exception is that a model trained in the PlantVillage dataset with simple image backgrounds suffers in the FieldPV dataset with different levels of background. A similar situation appears in the FieldPlant dataset. In addition, we found that the majority of the research is unable to use or compare datasets, except the Plant Village dataset. Finally, we point out that, when using the deep learning method, most existing public datasets embrace the i.i.d. assumption 3 where the training and test datasets are identical and independent. In this assumption, the original datasets are randomly split into training and test datasets, and thus the reported test performance is more highly estimated than is the case when deploying the trained model.
4 Future direction of plant disease recognition datasets
● Stage one: verification, where deep learning methods are verified to be useful in recognizing plant diseases.
● Stage two: implementation, where deep learning methods are deployed in real-world applications of plant disease recognition with decent performance.
● Stage three: connection, where plant disease recognition using deep learning methods is connected to downstream applications.
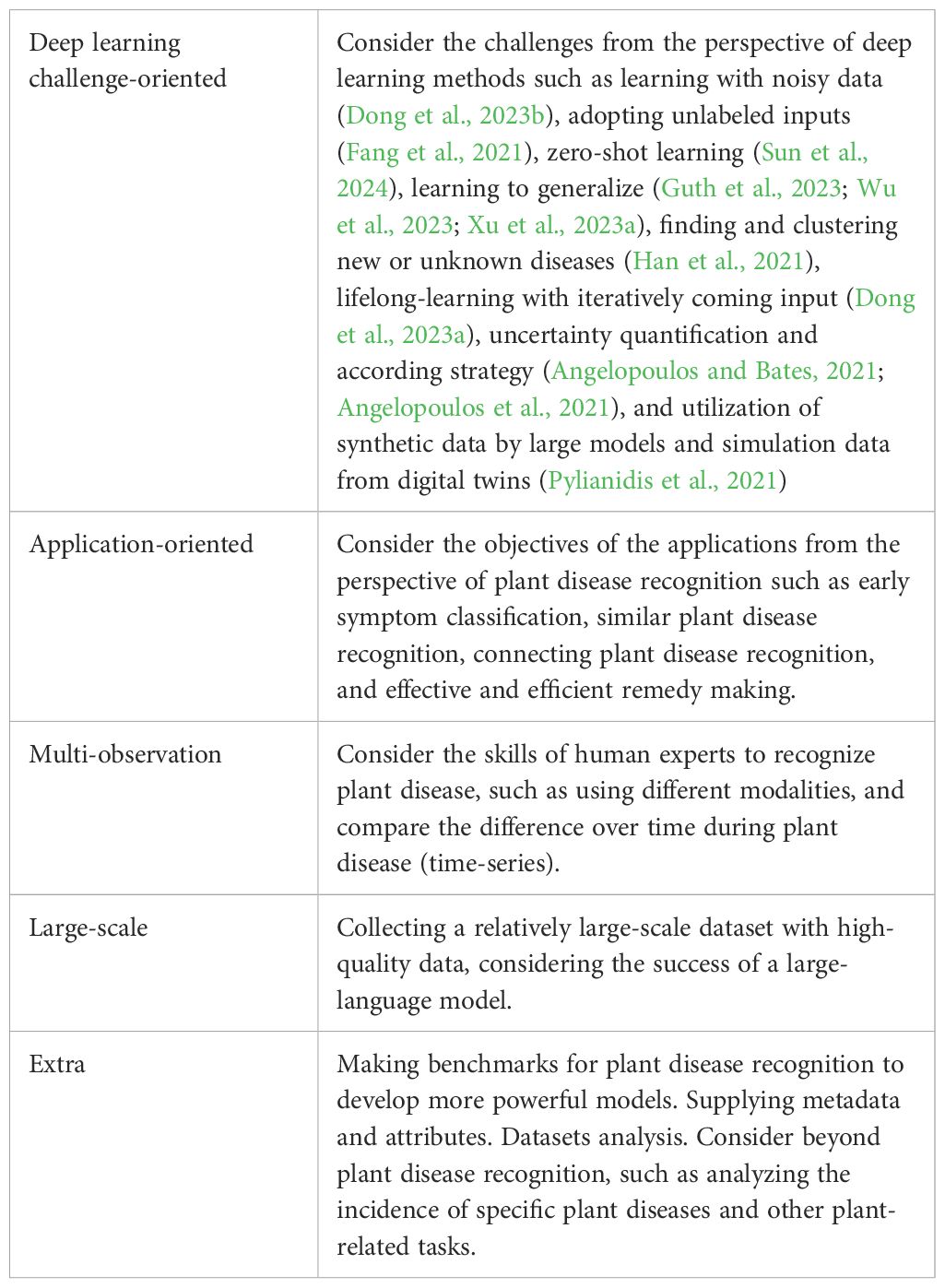
To probe future directions, we first declare three stages of plant disease recognition using deep learning. The first stage is straightforward and almost finished in recent years. However, the second and third stages are still in their infancy. Currently, few publications have mentioned the successful implementation in real-world applications. One of the main reasons for this comes from the assumptions embraced by deep learning methods that generally do not hold in real-world applications. From this perspective, existing datasets accommodated the assumptions. Therefore, one of the future directions of plant disease recognition is to make datasets that violate the assumptions, termed deep learning challenge-oriented datasets. Furthermore, we argue that recognition of plant disease is not the final objective and should be connected with the downstream work, arriving at the third stage. From such a perspective, we argue that another future direction is to make the datasets oriented to downstream applications. Besides achieving better performance in general, two inspirations are discussed, multi-observation and large-scale datasets. These are outlined in the following section with an additional discussion. Table 4 summarizes our thinking.
4.1 Deep learning challenge-oriented dataset
Although decent performance is achieved in most datasets, the corresponding trained models may suffer when deploying them in real-world applications. One of the reasons is that the assumptions to achieve good performance are not always valid (Xu et al., 2023a). Violating those assumptions results in challenges when deploying deep learning models. For example, a model trained in the datasets of several farms is desired to give better results when deployed in other farms, termed spatial generalization. In a similar spirit, it is desirable for a model trained in the datasets collected in a particular time duration to be decent when deployed in another time duration, termed temporal generalization. Additional invariant disease characteristics are also expected to have no impacts. However, current datasets do not support this kind of verification. Formally, deep learning challenge-oriented datasets are highlighted to test and develop models for plant disease recognition. Simultaneously, we argue that datasets should have meta-data, such as position and time stamp. In other words, current datasets assume that something in the training and test datasets is shared (Meng et al., 2023). For example, a new plant disease may exist in the testing stage and is desired to be classified from the known classes that exist in the training dataset (Meng et al., 2023), a challenge termed open set recognition. Furthermore, we highlight that realizing the assumptions of deep learning models and incorporating them into the dataset collection stage needs the cooperation of researchers from the agriculture and deep learning fields. Please refer to the detailed challenges regarding the datasets in (Xu et al., 2023a).
4.2 Application-oriented dataset
From the perspective of agriculture, recognizing plant diseases may not be the final objective, and downstream work may follow. For example, early visual pattern recognition is beneficial in making some remedies to reduce loss. Therefore, collecting such datasets is appealing. Although some papers aimed to focus on this issue, there is no agreement on the definition of early disease recognition. We contend that such data have two primary characteristics: recognizable patterns and effective remedies. One of the core assumptions embraced by deep learning models is that different plant diseases have their own patterns; otherwise, they cannot be distinguished. Considering that data modalities have heterogeneous advantages and disadvantages, selecting a suitable input modality is essential. However, disease states cause varying degrees of losses and there are difficulties in providing remedies. In an extreme scenario, when a plant disease explodes on a farm and the plants all die, recognizing the corresponding disease is not useful. More applications in the field of agriculture are possible. Although the objectives are from agriculture, we highlight that trade-offs exist such as in the case of early disease recognition which requires the cooperation from engineers in the deep learning field.
4.3 Multi-observation dataset
In general, human experts make superior decisions to recognize plant diseases through multiple observations rather than single observations. Inspired by this situation, we contend that deep learning methods can also be improved with multi-observation. Essentially, multiple observations distribute different information. Multi-modal datasets refer to datasets with various modalities for the same plant diseases. For example, given a leaf with a plant disease, various optical images and texts can be made. In addition, datasets can be in a time series, such as taking images of plant diseases at different times. In particular, visual patterns become clearer and easier to recognize when diseases gradually involve. Time-series datasets may mitigate the challenge of the early recognition of plant diseases. For image data, higher test performance can also be due to multi-spatial datasets, such as taking images in different scales and perspectives. For example, some plant diseases have different patterns on the front and back of leaves.
4.4 Large-scale dataset
Large-scale datasets tend to be beneficial for model generalization in many general computer vision tasks and datasets (Kaplan et al., 2020; Zhai et al., 2022; Xu et al., 2023c). Therefore, collecting large-scale datasets for plant disease recognition is appealing and worthwhile although it is time-consuming, difficult, and expensive. One way in which this could be done is crowdsourcing (Coletta et al., 2020) by which related people in different locations take images and then upload them to a platform. These images would then be annotated by the community. In this way, the collected datasets have enormous variations and thus contribute to model generalization (Xu et al., 2023a).
4.5 Extra discussion
Benchmarks. In recent years, plant disease recognition has witnessed a significant improvement (Singh et al., 2018; Liu and Wang, 2021; Thakur et al., 2022; Salman et al., 2023; Xu et al., 2023a), as well as the number of related publications. However, a relative comparison is relatively lacking to evaluate different models in diverse applications. One of the main reasons is the shortage of benchmarks, i.e., public and widely used high-quality datasets. We argue that this kind of benchmark will facilitate the community and speed up the deployment of deep learning methods in the real-world applications of plant disease recognition.
Meta-data is the information used to describe datasets from different perspectives, usually with tags. Most of the current relevant public datasets only have the types of plant disease. Other types of information are expected to be beneficial, such as spatial and temporary tags. The datasets with meta-data can be used for different applications by making new datasets.
Analysis of datasets. In general, different applications have heterogeneous difficulties and challenges. Datasets show the faces of applications and therefore, analysis of datasets are essential to understand the applications and further to achieve a better performance. However, few datasets have corresponding analysis and one of the expected future research directions is automatic analysis, such as for intra- and inter-class image variations. Furthermore, dataset analysis can be used in an iterative way to make high-quality datasets.
Beyond plant disease recognition. Recognizing plant disease is just one of the fundamental requirements to have decent crop yields. This objective may be further facilitated by incorporating disease recognition and more things. For example, plants may be infected by specific diseases or viruses in some conditions where finding the correlated factors are beneficial to prevent the plants from succumbing to those diseases. In addition, plant disease recognition is plant-related and thus, from a wider perspective, its recognition can be connected to other tasks such as plant species recognition (Xu et al., 2022b; Meng et al., 2023).
5 Concluding Remarks
Using deep learning to recognize plant disease is an interdisciplinary challenge and thus requires a unified perspective. Compared to making deep learning models, we highlighted that datasets are also essential if our objective is to deploy models in real-world applications. Making a plant disease recognition dataset reliable and close to real-world applications a requires superior understanding of both the deep learning and agriculture fields. A systematic taxonomy for related datasets was first provided. We specially emphasize dataset splitting and the annotation strategies that are scarcely discussed in the literature and suggest possible challenges in real-world applications. Further, RGB images are observed as the dominant input modality and an extensive summarization was given. Finally, four types of dataset are described as future directions: deep learning challenge-oriented, application-oriented, multi-observation, and large-scale, with an additional discussion.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
MX: Conceptualization, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. JP: Investigation, Writing – original draft, Conceptualization. JL: Writing – review & editing, Investigation. JY: Conceptualization, Writing – review & editing. SY: Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) and Korea Smart Farm R&D Foundation (KosFarm) through Smart Farm Innovation Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) and Ministry of Science and ICT (MSIT), Rural Development Administration (RDA) (RS-2021-IP42027). This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2019R1A6A1A09031717). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (RS-2024-00360581).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ https://spectrum.ieee.org/andrew-ng-data-centric-ai.
- ^ https://www.greenhousecanada.com/expanding-sight-plant-health-beyond-the-naked-eye/.
- ^ https://www.statisticshowto.com/iid-statistics/.
References
Adão, T., Hruška, J., Pádua, L., Bessa, J., Peres, E., Morais, R., et al. (2017). Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 9, 1110. doi: 10.3390/rs9111110
Ahmad, A., El Gamal, A., Saraswat, D. (2023). Toward generalization of deep learning-based plant disease identification under controlled and field conditions. IEEE Access 11, 9042–9057. doi: 10.1109/ACCESS.2023.3240100
Aishwarya, M., Reddy, A. P. (2023). Dataset of groundnut plant leaf images for classification and detection. Data Brief 48, 109185.
Angelopoulos, A. N., Bates, S. (2021). A gentle introduction to conformal prediction and distribution-free uncertainty quantification. arXiv preprint arXiv:2107.07511.
Angelopoulos, A. N., Bates, S., Jordan, M., Malik, J. (2021). “Uncertainty Sets for Image Classifiers using Conformal Prediction,” in International Conference on Learning Representations. (Vienna, Austria). Available at: https://openreview.net/forum?id=eNdiU_DbM9.
Arjovsky, M., Bottou, L., Gulrajani, I., Lopez-Paz, D. (2019). Invariant risk minimization. arXiv preprint arXiv:1907.02893.
Beery, S., Wu, G., Edwards, T., Pavetic, F., Majewski, B., Mukherjee, S., et al. (2022). The auto arborist dataset: A large-scale benchmark for multi-view urban forest monitoring under domain shift. Proc. IEEE/CVF Conf. Comput. Vision Pattern Recognit., 21294–21307. doi: 10.1109/CVPR52688.2022.02061
Bengio, Y., Courville, A., Vincent, P. (2013). Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828. doi: 10.1109/TPAMI.2013.50
Bengio, Y., Lecun, Y., Hinton, G. (2021). Deep learning for AI. Commun. ACM 64, 58–65. doi: 10.1145/3448250
Bevers, N., Sikora, E. J., Hardy, N. B. (2022). Soybean disease identification using original field images and transfer learning with convolutional neural networks. Comput. Electron. Agric. 203, 107449. doi: 10.1016/j.compag.2022.107449
Cao, Y., Chen, L., Yuan, Y., Sun, G. (2023). Cucumber disease recognition with small samples using image-text-label-based multi-modal language model. Comput. Electron. Agric. 211, 107993. doi: 10.1016/j.compag.2023.107993
Chouhan, S. S., Singh, U. P., Kaul, A., Jain, S. (2019). “A data repository of leaf images: Practice towards plant conservation with plant pathology,” in 2019 4th International Conference on Information Systems and Computer Networks (ISCON). 700–707 (Mathura: IEEE).
Coletta, A., Bartolini, N., Maselli, G., Kehs, A., McCloskey, P., Hughes, D. P. (2020). Optimal deployment in crowdsensing for plant disease diagnosis in developing countries. IEEE Internet Things J. 9, 6359–6373. doi: 10.1109/JIOT.2020.3002332
Corso, A., Karamadian, D., Valentin, R., Cooper, M., Kochenderfer, M. J. (2023). A holistic assessment of the reliability of machine learning systems. arXiv preprint arXiv:2307.10586.
Cui, P., Athey, S. (2022). Stable learning establishes some common ground between causal inference and machine learning. Nat. Mach. Intell. 4, 110–115. doi: 10.1038/s42256-022-00445-z
Deng, J., Dong, W., Socher, R., Li, L. J., Li, K., Fei-Fei, L. (2009). “Imagenet: A large-scale hierarchical image database,” in 2009 IEEE conference on computer vision and pattern recognition. 248–255 (Miami, Florida, USA: Ieee).
Dong, J., Fuentes, A., Yoon, S., Kim, H., Jeong, Y., Park, D. S. (2023a). A new deep learning-based dynamic paradigm towards open-world plant disease detection. Front. Plant Sci. 14, 1243822. doi: 10.3389/fpls.2023.1243822
Dong, J., Fuentes, A., Yoon, S., Kim, H., Park, D. S. (2023b). An iterative noisy annotation correction model for robust plant disease detection. Front. Plant Sci. 14, 1238722. doi: 10.3389/fpls.2023.1238722
Dong, J., Lee, J., Fuentes, A., Xu, M., Yoon, S., Lee, M. H., et al. (2022). Data-centric annotation analysis for plant disease detection: Strategy, consistency, and performance. Front. Plant Sci. 13, 1037655. doi: 10.3389/fpls.2022.1037655
Esgario, J. G., Krohling, R. A., Ventura, J. A. (2020). Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput. Electron. Agric. 169, 105162. doi: 10.1016/j.compag.2019.105162
Fang, U., Li, J., Lu, X., Gao, L., Ali, M., Xiang, Y. (2021). Self-supervised cross-iterative clustering for unlabeled plant disease images. Neurocomputing 456, 36–48. doi: 10.1016/j.neucom.2021.05.066
Feng, Z., Song, L., Duan, J., He, L., Zhang, Y., Wei, Y., et al. (2021). Monitoring wheat powdery mildew based on hyperspectral, thermal infrared, and RGB image data fusion. Sensors 22, 31. doi: 10.3390/s22010031
Fuentes, A., Im, D. H., Yoon, S., Park, D. S. (2017a). “Spectral analysis of CNN for tomato disease identification,” in Artificial Intelligence and Soft Computing: 16th International Conference, ICAISC 2017, Zakopane, Poland, June 11-15, 2017. 40–51 (Springer), Proceedings, Part I 16.
Fuentes, A., Yoon, S., Kim, S. C., Park, D. S. (2017b). A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 17, 2022. doi: 10.3390/s17092022
Fuentes, A., Yoon, S., Park, D. S. (2019). Deep learning-based phenotyping system with glocal description of plant anomalies and symptoms. Front. Plant Sci. 10, 1321. doi: 10.3389/fpls.2019.01321
Gui, P., Dang, W., Zhu, F., Zhao, Q. (2021). Towards automatic field plant disease recognition. Comput. Electron. Agric. 191, 106523. doi: 10.1016/j.compag.2021.106523
Guth, F. A., Ward, S., McDonnell, K. (2023). From lab to field: an empirical study on the generalization of convolutional neural networks towards crop disease detection. Eur. J. Eng. Technol. Res. 8, 33–40. doi: 10.24018/ejeng.2023.8.2.2773
Han, K., Rebuffi, S. A., Ehrhardt, S., Vedaldi, A., Zisserman, A. (2021). Autonovel: Automatically discovering and learning novel visual categories. IEEE Trans. Pattern Anal. Mach. Intell. 44, 6767–6781. doi: 10.1109/TPAMI.2021.3091944
Hossain, M. F. (2023). Dhan-Shomadhan: A dataset of rice leaf disease classification for Bangladeshi local rice. arXiv preprint arXiv:2309.07515.
Hughes, D., Salathé, M. (2015). An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv preprint arXiv:1511.08060.
Kaplan, J., McCandlish, S., Henighan, T., Brown, T. B., Chess, B., Child, R., et al. (2020). Scaling laws for neural language models. arXiv preprint arXiv:2001.08361.
Krishna, R., Zhu, Y., Groth, O., Johnson, J., Hata, K., Kravitz, J., et al. (2017). Visual genome: Connecting language and vision using crowdsourced dense image annotations. Int. J. Comput. Vision 123, 32–73. doi: 10.1007/s11263-016-0981-7
Lin, T. Y., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., et al. (2014). “Microsoft coco: Common objects in context,” in Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014. 740–755 (Springer), Proceedings, Part V 13.
Liu, X., Min, W., Mei, S., Wang, L., Jiang, S. (2021). Plant disease recognition: A large-scale benchmark dataset and a visual region and loss reweighting approach. IEEE Trans. Image Process. 30, 2003–2015. doi: 10.1109/TIP.83
Liu, J., Wang, X. (2021). Plant diseases and pests detection based on deep learning: a review. Plant Methods 17, 1–18. doi: 10.1186/s13007-021-00722-9
Long, M., Hartley, M., Morris, R. J., Brown, J. K. (2023). Classification of wheat diseases using deep learning networks with field and glasshouse images. Plant Pathol. 72, 536–547. doi: 10.1111/ppa.13684
Lu, B., Dao, P. D., Liu, J., He, Y., Shang, J. (2020). Recent advances of hyperspectral imaging technology and applications in agriculture. Remote Sens. 12, 2659. doi: 10.3390/rs12162659
Lu, B., He, Y., Dao, P. D. (2019). Comparing the performance of multispectral and hyperspectral images for estimating vegetation properties. IEEE J. selected topics Appl. Earth observations Remote Sens. 12, 1784–1797. doi: 10.1109/JSTARS.4609443
Mahlein, A. K. (2016). Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 100, 241–251. doi: 10.1094/PDIS-03-15-0340-FE
Meng, Y., Xu, M., Kim, H., Yoon, S., Jeong, Y., Park, D. S. (2023). Known and unknown class recognition on plant species and diseases. Comput. Electron. Agric. 215, 108408. doi: 10.1016/j.compag.2023.108408
Mensah, P. K., Akoto-Adjepong, V., Adu, K., Ayidzoe, M. A., Bediako, E. A., Nyarko-Boateng, O., et al. (2023). CCMT: Dataset for crop pest and disease detection. Data Brief 49, 109306. doi: 10.1016/j.dib.2023.109306
Mignoni, M. E., Honorato, A., Kunst, R., Righi, R., Massuquetti, A. (2022). Soybean images dataset for caterpillar and Diabrotica speciosa pest detection and classification. Data Brief 40, 107756. doi: 10.1016/j.dib.2021.107756
Moupojou, E., Tagne, A., Retraint, F., Tadonkemwa, A., Wilfried, D., Tapamo, H., et al. (2023). FieldPlant: A dataset of field plant images for plant disease detection and classification with deep learning. IEEE Access 11, 35398–35410. doi: 10.1109/ACCESS.2023.3263042
Mwebaze, E., Gebru, T., Frome, A., Nsumba, S., Tusubira, J. (2019). iCassava 2019 fine-grained visual categorization challenge. arXiv preprint arXiv:1908.02900.
Northcutt, C. G., Athalye, A., Mueller, J. (2021). Pervasive label errors in test sets destabilize machine learning benchmarks. arXiv preprint arXiv:2103.14749.
Oerke, E. C., Mahlein, A. K., Steiner, U. (2014). Proximal sensing of plant diseases. Detection diagnostics Plant Pathog., 55–68.
Ouhami, M., Hafiane, A., Es-Saady, Y., El Hajji, M., Canals, R. (2021). Computer vision, IoT and data fusion for crop disease detection using machine learning: A survey and ongoing research. Remote Sens. 13, 2486. doi: 10.3390/rs13132486
Parraga-Alava, J., Cusme, K., Loor, A., Santander, E. (2019). RoCoLe: A robusta coffee leaf images dataset for evaluation of machine learning based methods in plant diseases recognition. Data Brief 25, 104414. doi: 10.1016/j.dib.2019.104414
Patrini, G., Rozza, A., Krishna Menon, A., Nock, R., Qu, L. (2017) Making deep neural networks robust to label noise: A loss correction approach. Proc. IEEE Conf. Comput. Vision Pattern Recognit., 1944–1952. doi: 10.1109/CVPR.2017.240
Pereira, M. R., Dos Santos, F. N., Tavares, F., Cunha, M. (2023). Enhancing host-pathogen phenotyping dynamics: early detection of tomato bacterial diseases using hyperspectral point measurement and predictive modeling. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1242201
Petchiammal, A., Kiruba, B., Murugan, D., Arjunan, P. (2023). “Paddy doctor: A visual image dataset for automated paddy disease classification and benchmarking,” in Proceedings of the 6th Joint International Conference on Data Science & Management of Data (10th ACM IKDD CODS and 28th COMAD). (Mumbai India) 203–207.
Pylianidis, C., Osinga, S., Athanasiadis, I. N. (2021). Introducing digital twins to agriculture. Comput. Electron. Agric. 184, 105942. doi: 10.1016/j.compag.2020.105942
Rahman, C. R., Arko, P. S., Ali, M. E., Iqbal Khan, M. A., Apon, S. H., Nowrin, F., et al. (2020). Identification and recognition of rice diseases and pests using convolutional neural networks. Biosyst. Eng. 194, 112–120. doi: 10.1016/j.biosystemseng.2020.03.020
Rajbongshi, A., Sazzad, S., Shakil, R., Akter, B., Sara, U. (2022). A comprehensive guava leaves and fruits dataset for guava disease recognition. Data Brief 42, 108174. doi: 10.1016/j.dib.2022.108174
Ramcharan, A., Baranowski, K., McCloskey, P., Ahmed, B., Legg, J., Hughes, D. P. (2017). Deep learning for image-based cassava disease detection. Front. Plant Sci. 8, 1852. doi: 10.3389/fpls.2017.01852
Saleem, M. H., Potgieter, J., Arif, K. M. (2022a). A performance-optimized deep learning-based plant disease detection approach for horticultural crops of New Zealand. IEEE Access 10, 89798–8 822. doi: 10.1109/ACCESS.2022.3201104
Saleem, M. H., Potgieter, J., Arif, K. M. (2022b). A weight optimization-based transfer learning approach for plant disease detection of New Zealand vegetables. Front. Plant Sci. 13, 1008079. doi: 10.3389/fpls.2022.1008079
Salman, Z., Muhammad, A., Piran, M. J., Han, D. (2023). Crop-saving with AI: latest trends in deep learning techniques for plant pathology. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1224709
Sethy, P. K., Barpanda, N. K., Rath, A. K., Behera, S. K. (2020). Deep feature based rice leaf disease identification using support vector machine. Comput. Electron. Agric. 175, 105527. doi: 10.1016/j.compag.2020.105527
Shoaib, M., Shah, B., Ei-Sappagh, S., Ali, A., Ullah, A., Alenezi, F., et al. (2023). An advanced deep learning models-based plant disease detection: A review of recent research. Front. Plant Sci. 14, 1158933. doi: 10.3389/fpls.2023.1158933
Singh, A. K., Ganapathysubramanian, B., Sarkar, S., Singh, A. (2018). Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 23, 883–898. doi: 10.1016/j.tplants.2018.07.004
Singh, D., Jain, N., Jain, P., Kayal, P., Kumawat, S., Batra, N. (2020) PlantDoc: A dataset for visual plant disease detection. In. Proc. 7th ACM IKDD CoDS 25th COMAD., 249–253. doi: 10.1145/3371158
Singh, D., Jain, N., Jain, P., Kayal, P., Kumawat, S., Batra, N. (2020). “PlantDoc: A Dataset for Visual Plant Disease Detection,” in Proceedings of the 7th ACM IKDD CoDS and 25th COMAD. CoDS COMAD 2020 (Association for Computing Machinery, Hyderabad, India), 249–253. doi: 10.1145/3371158.3371196
Singh, A., Jones, S., Ganapathysubramanian, B., Sarkar, S., Mueller, D., Sandhu, K., et al. (2021). Challenges and opportunities in machine-augmented plant stress phenotyping. Trends Plant Sci. 26, 53–69. doi: 10.1016/j.tplants.2020.07.010
Sultana, N., Shorif, S. B., Akter, M., Uddin, M. S. (2023). A dataset for successful recognition of cucumber diseases. Data Brief 109320. doi: 10.1016/j.dib.2023.109320
Sun, J., Cao, W., Fu, X., Ochi, S., Yamanaka, T. (2024). Few-shot learning for plant disease recognition: A review. Agron. J. 116, 1204–1216. doi: 10.1002/agj2.21285
Thakur, P. S., Khanna, P., Sheorey, T., Ojha, A. (2022). Trends in vision-based machine learning techniques for plant disease identification: A systematic review. Expert Syst. Appl. 118117. doi: 10.1016/j.eswa.2022.118117
Thapa, R., Zhang, K., Snavely, N., Belongie, S., Khan, A. (2020). The Plant Pathology Challenge 2020 data set to classify foliar disease of apples. Appl. Plant Sci. 8, e11390. doi: 10.1002/aps3.11390
Vapnik, V. (1991). Principles of risk minimization for learning theory. Adv. Neural Inf. Process. Syst. 4.
Wan, L., Li, H., Li, C., Wang, A., Yang, Y., Wang, P. (2022). Hyperspectral sensing of plant diseases: principle and methods. Agronomy 12, 1451. doi: 10.3390/agronomy12061451
Wang, C., Zhou, J., Zhang, Y., Wu, H, Zhao, C, Teng, G, et al. (2022). A plant disease recognition method based on fusion of images and graph structure text. Front. Plant Sci. 12, 731688. doi: 10.3389/fpls.2021.731688
Whang, S. E., Roh, Y., Song, H., Lee, J. G. (2023). Data collection and quality challenges in deep learning: A data-centric ai perspective. VLDB J. 32, 791–813. doi: 10.1007/s00778-022-00775-9
Wiesner-Hanks, T., Stewart, E. L., Kaczmar, N., DeChant, C., Wu, H., Nelson, R. J., et al. (2018). Image set for deep learning: field images of maize annotated with disease symptoms. BMC Res. Notes 11, 1–3. doi: 10.1186/s13104-018-3548-6
Wright, J., Ma, Y. (2022). High-dimensional data analysis with low-dimensional models: Principles, computation, and applications (Cambridge, University Printing House Shaftesbury Road, United Kingdom: Cambridge University Press).
Wu, X., Fan, X., Luo, P., Choudhury, S. D., Tjahjadi, T., Hu, C. (2023). From laboratory to field: unsupervised domain adaptation for plant disease recognition in the wild. Plant Phenom. 5, 0038. doi: 10.34133/plantphenomics.0038
Xu, M. (2023). Enhanced Plant Disease Recognition with Limited Training Dataset Using Image Translation and Two-Step Transfer Learning. Jeonju-si: Jeonbuk National University.
Xu, M., Kim, H., Yang, J., Fuentes, A., Meng, Y., Yoon, S., et al. (2023a). Embracing limited and imperfect training datasets: opportunities and challenges in plant disease recognition using deep learning. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1225409
Xu, M., Yoon, S., Fuentes, A., Park, D. S. (2023b). A comprehensive survey of image augmentation techniques for deep learning. Pattern Recogn. 109347. doi: 10.1016/j.patcog.2023.109347
Xu, M., Yoon, S., Fuentes, A., Yang, J., Park, D. S. (2022a). Style-consistent image translation: A novel data augmentation paradigm to improve plant disease recognition. Front. Plant Sci. 12, 3361. doi: 10.3389/fpls.2021.773142
Xu, M., Yoon, S., Jeong, Y., Park, D. S. (2022b). Transfer learning for versatile plant disease recognition with limited data. Front. Plant Sci. 13, 1010981. doi: 10.3389/fpls.2022.1010981
Xu, M., Yoon, S., Wu, C., Baek, J., Park, D. S. (2023c). Plantclef2023: A bigger training dataset contributes more than advanced pretraining methods for plant identification. Work. Notes CLEF.
Zha, D., Bhat, Z. P., Lai, K. H., Yang, F., Jiang, Z., Zhong, S., et al. (2023). Data-centric artificial intelligence: A survey. arXiv preprint arXiv:2303.10158.
Zhai, X., Kolesnikov, A., Houlsby, N., Beyer, L. (2022). “Scaling vision transformers,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. (New Orleans Louisiana) 12104–12113.
Zhang, C., Kong, J., Wu, D., Guan, Z., Ding, B., Chen, F. (2023). Wearable sensor: an emerging data collection tool for plant phenotyping. Plant Phenom. 5, 0051. doi: 10.34133/plantphenomics.0051
Keywords: plant disease recognition, deep learning, dataset making, smart agriculture, precision agriculture
Citation: Xu M, Park J-E, Lee J, Yang J and Yoon S (2024) Plant disease recognition datasets in the age of deep learning: challenges and opportunities. Front. Plant Sci. 15:1452551. doi: 10.3389/fpls.2024.1452551
Received: 21 June 2024; Accepted: 04 September 2024;
Published: 27 September 2024.
Edited by:
Ning Yang, Jiangsu University, ChinaReviewed by:
Xing Yang, Anhui Science and Technology University, ChinaChao Qi, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2024 Xu, Park, Lee, Yang and Yoon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jucheng Yang, amN5YW5nQHR1c3QuZWR1LmNu; Sook Yoon, c3lvb25AbW9rcG8uYWMua3I=
 Mingle Xu
Mingle Xu Ji-Eun Park1
Ji-Eun Park1 Jaehwan Lee
Jaehwan Lee Jucheng Yang
Jucheng Yang Sook Yoon
Sook Yoon